 Open Access
Open Access
ARTICLE
Efficient Wound Classification Using YOLO11n: A Lightweight Deep Learning Approach
1 Department of Computer Science, College of Engineering and Computer Science, Jazan University, Jazan, 45142, Saudi Arabia
2 Engineering and Technology Research Center, Jazan University, P.O. Box 114, Jazan, 82817, Saudi Arabia
3 Department of Computer Science, University of Wah, Wah Cantt, 47040, Pakistan
4 Department of Computer Science, Air University, Islamabad, 44000, Pakistan
* Corresponding Author: Ayesha Siddiqa. Email:
(This article belongs to the Special Issue: Emerging Trends and Applications of Deep Learning for Biomedical Signal and Image Processing)
Computers, Materials & Continua 2025, 85(1), 955-982. https://doi.org/10.32604/cmc.2025.065853
Received 23 March 2025; Accepted 16 June 2025; Issue published 29 August 2025
Abstract
Wound classification is a critical task in healthcare, requiring accurate and efficient diagnostic tools to support clinicians. In this paper, we investigated the effectiveness of the YOLO11n model in classifying different types of wound images. This study presents the training and evaluation of a lightweight YOLO11n model for automated wound classification using the AZH dataset, which includes six wound classes: Background (BG), Normal Skin (N), Diabetic (D), Pressure (P), Surgical (S), and Venous (V). The model’s architecture, optimized through experiments with varying batch sizes and epochs, ensures efficient deployment in resource-constrained environments. The model’s architecture is discussed in detail. The visual representation of different blocks of the model is also presented. The visual results of training and validation are shown. Our experiments emphasize the model’s ability to classify wounds with high precision and recall, leveraging its lightweight architecture for efficient computation. The findings demonstrate that fine-tuning hyperparameters has a significant impact on the model’s detection performance, making it suitable for real-world medical applications. This research contributes to advancing automated wound classification through deep learning, while addressing challenges such as dataset imbalance and classification intricacies. We conducted a comprehensive evaluation of YOLO11n for wound classification across multiple configurations, including 6, 5, 4, and 3-way classification, using the AZH dataset. YOLO11n acquires the highest F1 score and mean Average Precision of 0.836 and 0.893 for classifying wounds into six classes, respectively. It outperforms the existing methods in classifying wounds using the AZH dataset. Moreover, Gradient-weighted Class Activation Mapping (Grad-CAM) is applied to the YOLO11n model to visualize class-relevant regions in wound images.Keywords
According to a survey conducted in 2018, more than 8 million people are dealing with wounds, with Medicare expenses for wound care estimated between 28.1 billion and 96.8 billion. This staggering figure highlights the scale of wound care and management. Chronic wounds are not only painful but also require extensive care, including regular cleaning, dressing changes, and the use of antibiotics to ensure proper healing.
Chronic wounds include Diabetic Foot Ulcers (DFU), Venous Leg Ulcers (VLU), Pressure Ulcers (PU), and Surgical Wounds (SW). Approximately 34% of individuals with diabetes face a lifetime risk of developing a DFU, and more than half of these ulcers become infected. Globally, around 0.15% to 0.3% of people suffer from active VLU. Pressure ulcers affect 2.5 million individuals annually, while roughly 4.5% of people who undergo surgery develop a surgical wound each year [1]. DFUs are a caused due to serious complications in diabetes. Two common challenges in treating DFUs are infection and ischemia, both of which can lead to limb amputation and hospital admission. Following amputation, a patient’s quality of life often declines significantly, with life expectancy typically dropping to under three years. Infection affects 40%–80% of DFUs, while ischemia occurs in nearly 50%. Infection arises when bacteria in the wound cause cell death, particularly in lower limb areas like foot soles. A chronic complication of diabetes causes ischemia due to poor blood circulation [2,3].
Classifying wound severity is a crucial aspect of the diagnosis process, as it aids physicians in making swift and accurate treatment decisions. The simplest method of monitoring wounds is a visual inspection, which has several limitations, including inter-observer variability, visual impairments, and obesity, due to which the user may struggle to detect subtle changes, making it difficult to assess wound progression accurately [4]. With the rising prevalence of chronic wounds, there is a growing need for automated solutions that can support healthcare providers, enhance treatment efficiency, and help reduce the overall cost of care. Furthermore, these systems can detect wounds on time and reduce the risk of amputation.
Diagnosing and treating chronic wounds presents a significant challenge for healthcare professionals. Hence, the physicians must identify the type of wound and then prescribe the correct medication and treatment plan. This careful monitoring is crucial in managing the healing process over time. The traditional method includes medical practitioners visually inspecting the wound to classify it, but with advancements in automation, several Machine Learning (ML)-based systems have been developed to streamline this process. These systems typically rely on handcrafted feature extraction followed by classification. However, they have limitations, as manual feature extraction can be time-consuming, prone to human error, and may not capture the full complexity of wounds. Furthermore, these systems also fail to perform due to poor illumination and contrast in images. Hence, the performance of these systems is limited, thus showcasing the need for more advanced approaches like deep learning, which can automatically learn and extract features from data without human intervention [5]. With the advancement of AI, this process has become more efficient. AI not only saves time and reduces costs but can also outperform human predictions in certain cases. Unlike earlier rule-based AI systems that heavily relied on expert knowledge, modern AI algorithms have evolved into data-driven systems that operate independently, without the need for human or expert input [6,7].
Among deep learning approaches, the You Only Look Once (YOLO) family of object detection models has gained popularity for its balance of speed and accuracy. YOLO performs object detection in a single forward pass, making it suitable for real-time applications. Since its introduction, YOLO has evolved through several versions (YOLOv1 to YOLOv8), each improving in accuracy, architectural efficiency, and adaptability to diverse tasks. While widely applied in general object detection and autonomous systems, its application in specialized domains like medical imaging remains underexplored.
In this paper, we leverage the latest lightweight variant, YOLO11n, to perform automated wound classification using the AZH wound dataset. The dataset includes six classes: Background (BG), Normal Skin (N), Diabetic (D), Pressure (P), Surgical (S), and Venous (V). To evaluate the adaptability and robustness of YOLO11n, we conducted a series of classification experiments under multiple configurations. These include 6-way classification for identifying all wound and background classes, and 5-way classification excluding the background class. Additionally, 4-way classification that focuses on core wound categories, and 3-way classification to distinguish between three types of wound images. This multi-scenario approach provides a comprehensive understanding of YOLO11n’s performance across diverse levels of classification complexity.
The contributions of this paper are as follows:
• Exploring the potential of the lightweight YOLO11n model for wound classification under various class configurations, including six-class, five-class, four-class, and three-class wound classification.
• Analyzing the adaptability and robustness of YOLO11n in addressing different levels of wound classification complexity.
• Providing a detailed evaluation of YOLO11n’s performance metrics, such as accuracy, precision, recall, and F1 Score, across all classification settings to offer insights into how class reduction affects model efficiency and effectiveness.
• Visual explainability for wound classification using Gradient-weighted Class Activation Mapping (Grad-CAM) at different stages of YOLO11n architecture.
The rest of the paper is organized as follows: Section 2 provides background on wound classification, Section 3 critically reviews existing systems, Section 4 elaborates the proposed methodology, Section 5 presents experimental results, Section 6 presents discussion on the results, and Section 7 concludes the study.
Wound care is a critical aspect of healthcare, as poorly managed wounds can lead to severe complications such as infections or amputations. Hence, timely identification and classification of these wounds can save patients from developing serious complications. The advanced wound care market is projected to exceed $22 billion by 2024, with a high ratio of patients suffering from wound infections and chronic wounds [8]. Thus, managing postoperative wounds remains a challenging and resource-intensive task for healthcare professionals and patients alike.
Traditional wound identification methods rely heavily on the expertise of medical professionals who manually analyze the wound size, depth, and tissue type. This approach, however, is subject to variability in accurate identification and delayed treatment, especially in resource-constrained settings or among non-specialist healthcare providers. Another approach to wound detection involves using traditional machine-learning techniques to identify wounds from images. However, these methods rely on manually crafted features [9], which limit their ability to effectively handle a wide variety of wound types, especially those with similar features. Due to the growing global burden of chronic wounds, including diabetic ulcers, pressure ulcers, and post-surgical wounds, there is an urgent need for more reliable and scalable wound assessment methods that require almost no human intervention. Hence, DL based automated wound image classification systems hold immense promise here, as they aim to detect and categorize different types of wounds using advanced computational techniques [1].
Deep learning, a type of AI, has emerged as a powerful tool for image classification and analysis due to its ability to learn complex patterns and features directly from data without extensive manual feature engineering. These systems are deployed to solve various computer vision tasks, such as image classification or localization, often performing comparable to or better than a trained pathologist. Hence, these automated systems can perform automated learning of complex features, including tissue composition, edge sharpness, inflammation, color, texture, size, etc., which are essential for determining wound type, severity, and healing progress [10]. Due to their efficacy, several AI-based medical diagnostic systems have already attained FDA approval, including software for identifying prostate cancer [11], skin cancer [12], and breast cancer [13].
However, despite the performance, the development of robust deep-learning-based wound classification systems faces challenges mainly due to the scarcity of high-quality wound datasets, slight variability in the wound appearances, as well as poor illumination and contrast in images. As of now, several studies have explored the application of deep learning for wound image classification, however, gaps remain in terms of real-world implementation and deployment. Many existing approaches are limited to specific wound types or rely on small, homogeneous, or imbalanced datasets, making it difficult to deploy these systems in diverse clinical settings. Some of these studies are critically analyzed in Section 3. Hence, robust and scalable solutions should be presented to overcome these challenges so that these systems can be integrated into clinical workflows and automate the process of wound classification.
With the advancement of deep learning techniques, several models have been proposed for wound detection and classification from medical images. This section presents a critical review of existing literature and highlights the research gap that our study addresses.
Some existing works are primarily focused on classifying wound types using transfer learning. For instance, Ahsan et al. [4] proposed a wound classification system using several transfer-learned frameworks such as AlexNet, VGG16, VGG19, GoogLeNet, ResNet50, ResNet101, MobileNet, SqueezeNet, and DenseNet, to classify infection and ischemia from the DFU2020 dataset. To address the data limitation, the applied data augmentation techniques were used. ResNet50 achieved the highest accuracy, with 99.49% for ischemia classification and 84.76% for infection classification. In another study, Almufadi et al. [14] also used various transfer learning architectures such as EfficientNetB0, DenseNet121, ResNet101, VGG16, InceptionV3, MobileNetV2, and InceptionResNetV2 as head models along with various Machine Learning classifiers. They also classified infection and ischemia using the DFU2020 dataset. Their system attained accuracy of 92.7% on Infection class samples and 96.7% on Ischemia class samples.
Other researchers have proposed custom CNN architectures to address domain-specific challenges. Alzubaidi et al. [15] proposed a novel Deep Convolutional Neural Network called DFU QUTNet for the automatic classification of DFU. Rather than increasing the network depth, which could result in gradient-related issues, their model focused on increasing network width while maintaining depth. The features extracted from DFU QUTNet were then supplied to various ML classifiers, Support Vector Machine (SVM) and K-Nearest Neighbors (KNN) classifiers. The system attained highest F1 Score of 94.5%. They used a custom database containing normal and abnormal (DFU) images to conduct the study.
Anisuzzaman et al. [1] developed a multi-modal wound classification network using both wound images and their corresponding locations (body map) to classify wounds into different types, such as DFU, pressure ulcers, surgical wounds, venous ulcers, and normal skin. A body map was also designed to assist clinicians in documenting wound locations consistently. The study was conducted using different databases, i.e., Metdec, custom dataset (AZH), and AZHMT (a combination of Metdec and AZH). They performed classification using VGG16, VGG19, Long Short Term Memory (LSTM), Multi-Layer Perceptron (MLP), ResNet50, InceptionV3, and AlexNet. Their system using VGG19 + MLP attained 82.48% accuracy on the AZH database with 6 classes, namely Background, Normal skin, DFU, Pressure ulcer, Surgical wound, and venous ulcer. In another study, Anisuzzaman et al. [6] developed a custom CNN for wound severity classification based on their color, i.e., green, yellow, and red, with green representing wounds in the early stages of healing, yellow indicating wounds that require more attention, and red denoting the most severe cases needing urgent care. They conducted this study on a custom database gathered from AZH Wound and Vascular Center and created with the assistance of wound specialists. They used various transfer learned architectures, i.e., VGG16-19, Inception-V3, NasNetLarge, ResNet50, DenseNet101, XceptionNet, MobileNetV2, InceptionResNetV2. They stacked nine models each containing four different models, for better feature extraction and classification than the individual models. They attained an accuracy of 68.49% for multi-class classification and 77.57% to 81.40% accuracy for binary class classification. The system, however, performed poorly due to the very few images.
Eldem et al. [16] proposed a classification system for pressure and diabetic wounds from images by developing six variations of the AlexNet architecture. These models utilized different combinations of Convolution, Pooling, and ReLU layers, with classification performance evaluated using both Softmax and SVM classifiers. They also used a custom database to evaluate the model’s performance, on which their system attained 98.85% accuracy. Moreover, on the Medetec database, they obtained a 95.3% accuracy. Goyal et al. [17] collected a comprehensive dataset containing normal and abnormal images of DFU from various patients. They proposed DFUNet, a CNN-based framework containing convolutional, parallel convolutional, max-pooling, and fully connected layers for the classification of images into normal and DFU. Their system obtained 92.5% accuracy on the custom database. In another study, Goyal et al. [18] created a new dataset named DFU dataset to identify infection and ischemia in DFUs. They introduced a novel feature descriptor called the Superpixel Colour Descriptor for a handcrafted machine-learning approach. They evaluated the performance of various state-of-the-art ML and DL classifiers and also proposed an Ensemble CNN for classification. They obtained the highest accuracy of 90% for ischemia classification and 73% for infection class classification using Ensemble-CNN. Giridhar et al. [19] presented a DL-based approach for DFU detection using transfer learned CNN and image processing techniques. Their system attained F1 Scores of 98%, 98%, and 97% for ischemia, none, and infection stages, respectively. However the system is computationally complex. Huong et al. [20] proposed a Particle Swarm Optimization (PSO)-incorporated DL framework for classifying infection and ischemia from the DFUC2021 database, using three deep learning models: AlexNet, GoogleNet, and EfficientNet-B0. They obtained the highest accuracy of 91% for the infection class and 99% for Ischemia class. Liu et al. [2] utilized geometric and color image augmentation techniques to enhance the DFU dataset and performed binary classification of infection and ischemia using EfficientNetB1, ResNet50, Inception v3, VGG16, and CNN. They obtained F1 Score of 99.3% on Ischemia samples and 97.14% on Infection using EfficientNet-B5 and EfficientNet-B1 on the DFUC2021 database, respectively.
Narang et al. [21] used ResNet-50 with a Channel Attention (CA) Network for the classification of DFU and healthy images. The CA module extracts channel-wise features, improving the accuracy of the ResNet-50 model, which uses a 3-layer bottleneck architecture. They utilized a DFU dataset from the Kaggle repository consisting of 1048 images. Data augmentation was applied to avoid overfitting, and the model achieved 90% validation accuracy. Patel et al. [22] introduced a multi-modal deep CNN for classifying wounds into four categories, i.e., diabetic, pressure, surgical, and venous ulcers. Their approach combined wound images with corresponding body location data for precise wound location tagging. They used state-of-the-art DL models like VGG16, ResNet152, and EfficientNetB2 by using Squeeze-and-Excitation modules, Axial Attention, and an Adaptive Gated Multi-Layer Perceptron. They conducted their study on the images obtained from on AZH and Metedec databases. Their model obtained 87.50% accuracy on the classification of 6 classes from the AZH database. An ensemble Deep Convolutional Neural Network (CNN)-based classifier was developed by Rostami et al. [23] to categorize wound images into surgical, diabetic, and venous ulcers. They combined patch-wise and image-wise classification scores obtained via Multilayer Perceptron. The system achieved 84.9% accuracy on the AZH database.
Object detection frameworks like YOLO have also been applied in wound classification. Aldughayfiq et al. [24] presented a YOLOv5-based wound classification model that was trained and validated on the Medetec database along with some random samples collected from the internet. They trained their system for 500 epochs with a patience of 100. The model achieved an overall mAP50 of 76.9% and mAP50-95 of 39.8% on the validation set. Sarmun et al. [25] proposed a robust deep learning-based system for detecting DFU images acquired from the DFUC2020 database. The system employed advanced ensemble techniques like Non-Maximum Suppression (NMS), Soft-NMS, and Weighted Bounding box Fusion (WBF) to combine predictions from state-of-the-art object detection models. By integrating YOLOv8 and FRCNN-ResNet101, their method achieved a mAP50 score of 86.4%. These studies demonstrate that while YOLO-based architectures hold promise, their application to wound classification, especially in lightweight configurations, remains underexplored.
Despite these advancements, gaps remain: Most existing studies focus on binary classification (e.g., infection vs. ischemia) and do not address the full diversity of wound types. YOLO-based methods for wound classification are limited and often computationally heavy, lacking deployment readiness for resource-constrained environments. Few works investigate performance across different class granularities (e.g., 6-class to 3-class classification) for practical adaptation. To address these gaps, our work proposes a lightweight wound classification system using YOLO11n. Unlike previous studies, we evaluated the performance of the YOLO11n at different stages using the Grad-CAM technique. The performance is compared with other architectures as well, using the AZH dataset. An overview of prior techniques, datasets, and their limitations is summarized in Table 1, which provides context for the development and evaluation of our proposed framework.
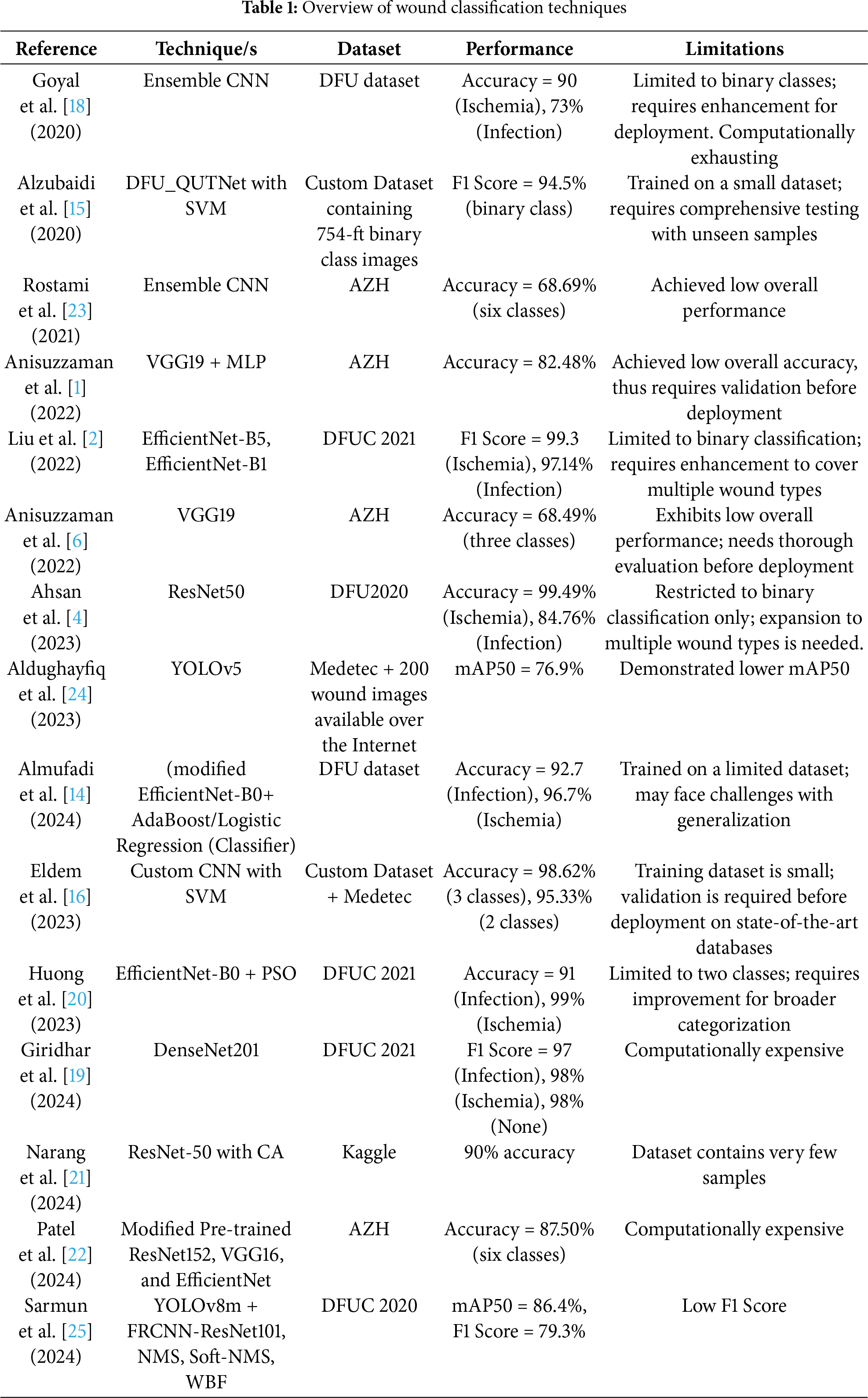
In this section, the detail of the proposed methodology for the detection of different types of wounds is presented. The nano model of YOLO version 11 is fine-tuned on a dataset that has five categories of wound images and one category containing background images. YOLO11n is utilized in this work because it is smaller and lightweight as it is designed with fewer layers and parameters compared to the YOLO11 standard model. It is suitable for real-time applications on edge devices or low-powered hardware (e.g., mobile phones, drones). The dataset contains images of various sizes. During the prepossessing of the dataset, images are resized to

Figure 1: Overview of the proposed methodology for wound classification
Fig. 2 illustrates the detailed architecture of the YOLO11n used in this work. It consists of convolution (Conv), C3k2, Spatial Pyramid Pooling Fast (SPPF), C2PSA, concatenation, up-sampling, and detection head blocks. The C3k2 block is more efficient in terms of computation, and it is a custom implementation of the Cross Stage Partial (CSP) Bottleneck. It uses two convolutions instead of one large convolution, which speeds up feature extraction. CSP networks work by splitting the input feature map into two parts. One part is passed through a bottleneck layer, which reduces the dimensionality and complexity of the data. The other part bypasses the bottleneck and is merged with the output of the bottleneck layer. This design reduces computational complexity while improving the network’s ability to learn and represent features effectively.

Figure 2: YOLO11n layer architecture
In YOLO11n, there are a total of eight C3k2 blocks. Each C3k2 block starts with a convolution layer with a kernel size of 1 × 1 and ends with it. Blocks 1, 2, 5, 6, and 7 consist of one bottleneck module containing two consecutive convolution layers with a kernel size of 3 × 3. Whereas, Block 3, Block 4, and Block 8 consist of one C3k module (containing three convolution layers of kernel size 1 × 1) and two bottleneck modules in sequence. Each of these bottleneck modules contains two convolution layers of kernel size 3 × 3. Every convolution layer in this architecture is followed by a Batch Normalization layer and a Sigmoid Linear Unit (SiLU) activation layer. Hence, it is sometimes referred to as the CBS block. Fig. 3 shows the architecture of C3k2 Block 1 and Block 3.

Figure 3: C3k2 blocks without and with C3k module
The characteristic of the SPPF block is that it concatenates after multi-scale pooling, which is crucial for capturing diverse information and expanding the number of channels temporarily for richer feature aggregation. SPPF is used to combine information from different scales. The detection head outputs predictions based on 6 classes with a bounding box showing the model’s confidence in the predicted class label.
C2PSA is a hybrid module designed to enhance feature extraction by combining convolutional operations and Parallel Spatial Attention (PSA). Its work integrates key principles of feature transformation, attention mechanisms, and spatial encoding. The core functionality of the C2PSA block comes from the PSA mechanism within the Block. PSA helps the model focus on critical spatial regions of the feature map. The feature map is divided into two parallel branches (usually split channel-wise). One branch processes spatial attention, while the other may bypass the attention for a residual-like structure. A convolution projects the split feature map to derive query, key, and value embeddings. These embeddings are processed to calculate attention weights. Positional encoding (via depthwise convolution) introduces information about spatial arrangements, essential for tasks like object detection or localization. The use of 1 × 1 convolutions, channel splitting, and attention mechanisms ensures that the module remains computationally efficient and suitable for the lightweight model YOLO11n. The details of 24 blocks utilized in the YOLO11n model for the classification of different types of wounds are shown in Table 2. The index column shows the number of the block in sequential order. The input column shows the input to the block, and
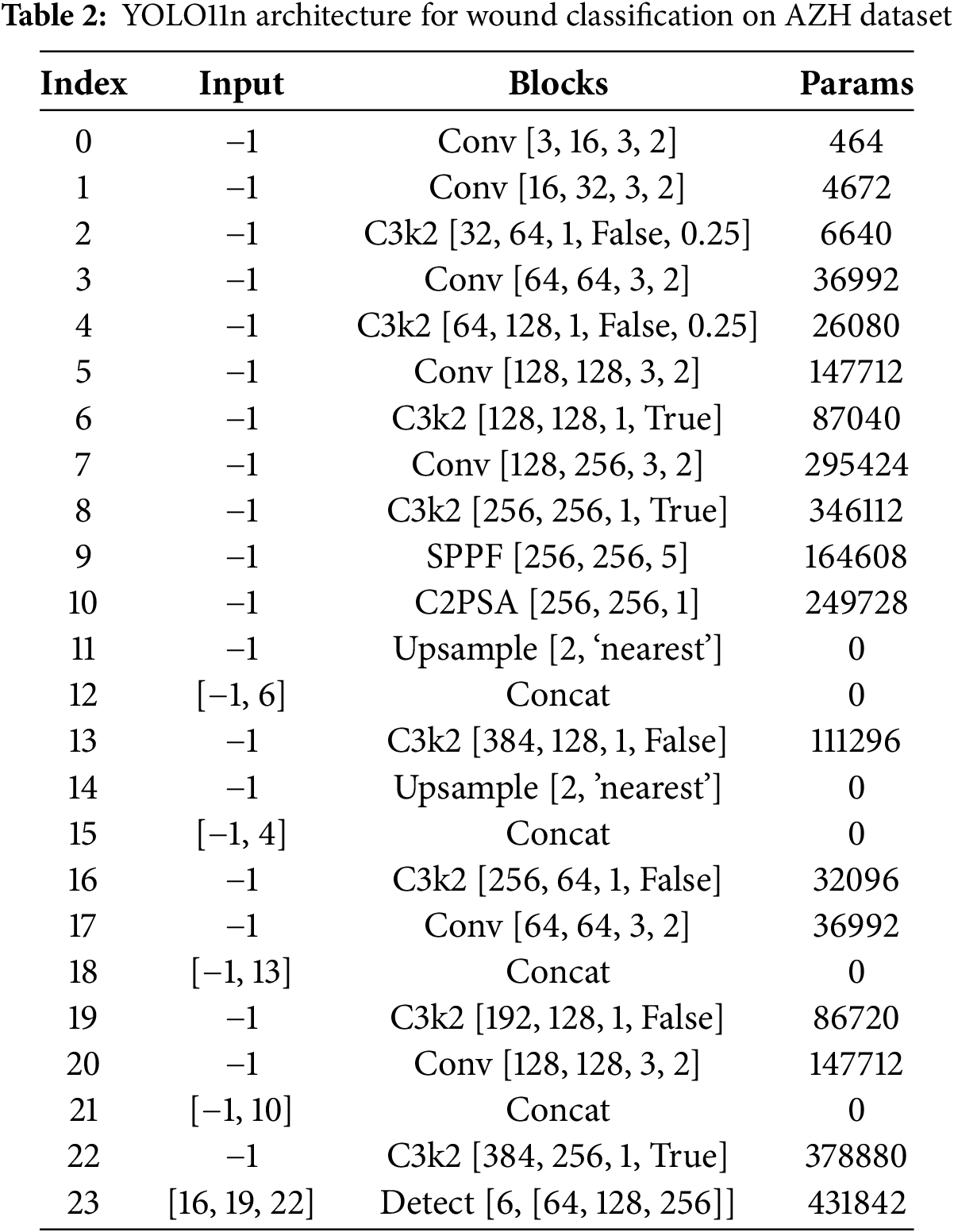
This study utilizes the AZH Wound dataset [6,22], a clinically curated collection of 930 wound images collected over two years at the AZH Wound and Vascular Center in Milwaukee, Wisconsin, USA. The images are in.jpg format and vary in resolution, with widths ranging from 320 to 700 pixels and heights from 240 to 525 pixels. The training folder contains 696 images and test folder contains 234 images belonging to six classes. Each image corresponds to one of four primary wound types: diabetic, venous, pressure, and surgical wounds. The dataset was acquired using two imaging devices: an iPad Pro (software version 13.4.1) and a Canon SX620 HS digital camera. Most images in the dataset were taken from unique patients; however, in some cases, multiple images were captured from the same patient at different body locations or stages of healing. In such cases, since the wound shapes differ, these are treated as independent samples. Labeling was conducted by certified wound specialists. Although the original dataset includes only four wound classes, additional categories such as Normal Skin (N) and Background (BG) were incorporated during preprocessing to facilitate object detection and multi-class classification in this study. The resulting six-class dataset includes: BG: Background, Non-wound, non-skin region, N: Normal healthy skin with no visible wounds, D: Diabetic foot ulcer wounds, P: Pressure wounds, S: Surgical, post-operative wounds, and V: Venous wounds.
The images were divided into training and testing sets, with a balanced distribution maintained across all classes. Due to the absence of ground-truth bounding box annotations, dummy bounding boxes covering the entire image were used to adapt the dataset for YOLO-based object detection. All images were resized to

Figure 4: Sample images from each class of AZH dataset
5.2 Experimental Setup and Evaluation
The YOLO11n model was trained on the AZH Wound dataset, which consists of 930 wound images divided into six classes: Background (BG), Diabetic (D), Pressure (P), Venous (V), Surgical (S), and Normal Skin (N). The training was performed on the Kaggle platform, utilizing a computational environment with access to GPUs for faster training.
During the training, the model’s architecture was modified to handle six wound types, overriding its original configuration designed for 80 classes. The model’s training spanned from 75 to 125 epochs, using batch sizes of 4, 8, and 16. Input images were resized to
Fig. 5 displays the learning curves of the YOLO11n model on the AZH wound dataset over 75 epochs with a batch size of 8. Whereas, Figs. 6 and 7 show the training curves of the model over 100 and 125 epochs, respectively. These learning curves show training and validation loss as well as precision, recall, and

Figure 5: Learning curves of YOLO11n over 75 epochs with 8 batch size

Figure 6: Learning curves of YOLO11n over 100 epochs with 8 batch size

Figure 7: Learning curves of YOLO11n over 125 epochs with 8 batch size

Figure 8: Different training batches of size 8 during training of YOLO11n model

Figure 9: Validation performance of YOLO11n model over 75, 100, and 125 epochs during training
During the first set of experiments, we set the number of epochs to 100 and changed the batch size value to 4, 8, and 16. Table 3 shows the performance of the YOLO11n on the test images at a batch size of 4, and the model was trained for 100 epochs. Similarly, Table 4 shows the results when the model was trained using a batch size of 8. The results using batch size 16 are shown in Table 5. It can be seen that the YOLO11n model demonstrated strong performance across the six wound classes of the AZH Wound dataset, achieving an overall
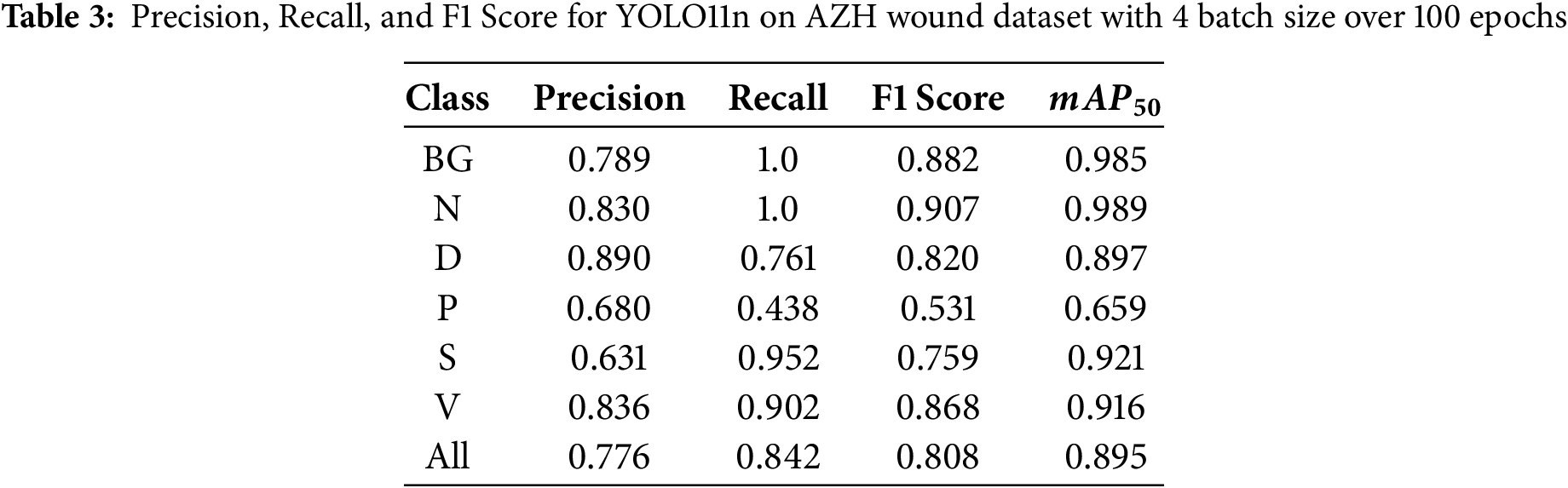

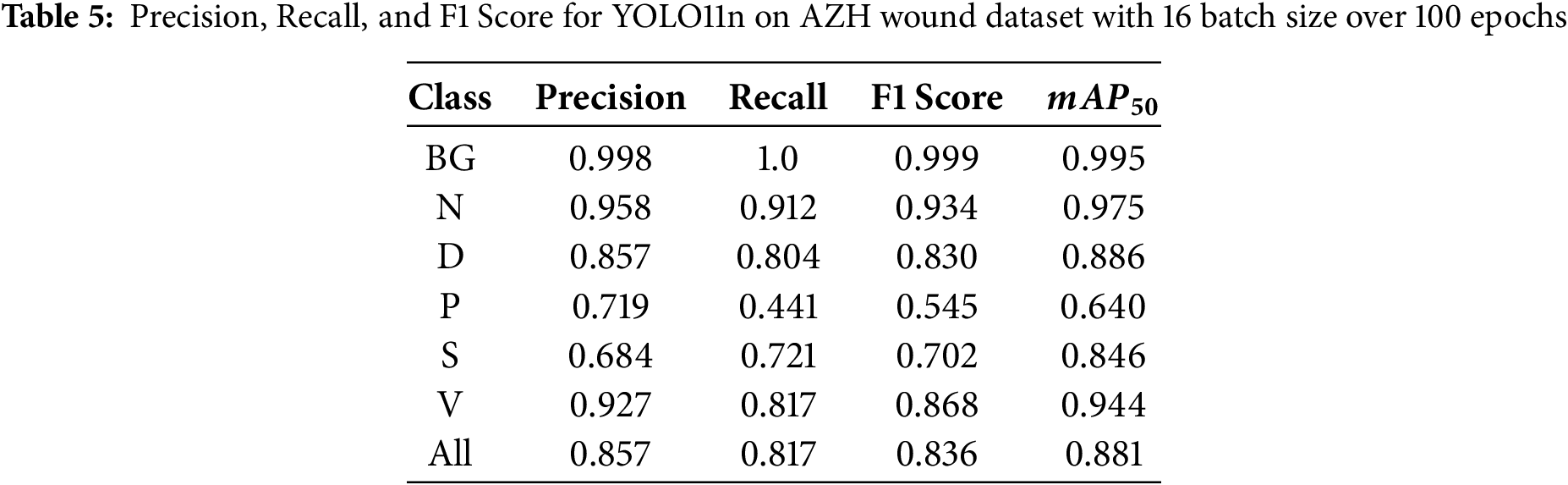
During the second set of experiments, we kept the batch size to 8 and changed the number of epochs to 75, 100, and 125. With a batch size of 8, training images were fed to the YOLO11n model during learning. Tables 6 and 7 show the performance of the YOLO11n model on the classification of wound images after training the model for 75 and 125 epochs, respectively. Overall, the highest recall of 0.863 is obtained at 100 epochs, the highest precision of 0.826 is achieved at 125 epochs, and the highest F1 Score of 0.828 is also obtained at 125 epochs.


Fig. 10 displays results obtained from a YOLO11n model applied to test images from the six classes in the AZH Wound dataset. It shows the performance of the YOLO11 model after training it on the AZH dataset over 100 epochs with 8 batch sizes. Each row corresponds to a specific class, with four examples per class. The model’s predictions are shown along with confidence scores (ranging from 0 to 1), which indicate how confident the model is in classifying each wound type. The first row contains images from the Normal Skin class of the dataset. The model shows near-perfect confidence with scores of 0.99 to 1.00. This strong performance suggests that the model can differentiate between wound and healthy skin very effectively, likely due to clear visual differences. The second row displays images from the Diabetic class of the dataset. The model has high confidence for diabetic wounds, with scores like 0.93 and 0.97. This demonstrates that the model reliably identifies the characteristic features of diabetic ulcers. The third row has images from the Pressure class. The model’s confidence for pressure wounds is also relatively high, with scores of 0.85 and 0.88. The third image, with a confidence of 0.82, suggests consistent recognition across various pressure wound cases, even though it sometimes exhibits slightly lower certainty. Surgical class images are shown in the fourth row. The model generally performs well with confidence scores above 0.70. The second image has a slightly lower confidence (0.71), while others range from 0.79 to 0.97. This indicates good recognition of surgical wounds, though it occasionally shows moderate confidence. Venous class images are displayed in the fifth row. The model performs quite well, with scores ranging from 0.53 to 1.00. However, the first image of the venous wound shows a notably lower confidence score (0.53), indicating that the model may struggle with certain variations or unclear visual features in this class. The last rows has images from the Background class of the dataset. The background class has high confidence scores, with most images close to 1.00. The last image in the row, however, has a slightly lower score (0.93), but overall, the model performs well at distinguishing wound sites from background scenes.

Figure 10: Performance of YOLO11n across different classes of wounds with training on 100 epochs with 8-batch size
It is observed through the results that the YOLO11n model shows high confidence in distinguishing clear cases of healthy skin and background, as well as certain wound types like diabetic and venous wounds, especially when the wound features are prominent. However, the model shows slightly reduced confidence in some cases, such as the first venous wound (V 0.53) and the second surgical wound (S 0.71), where wound features may overlap with characteristics from other wound types or the background. Overall, the model appears to be consistent in predicting the correct classes, with most predictions yielding high confidence levels. The lower confidence cases are exceptions and may require further investigation, such as adjusting model thresholds or improving the quality of training data for those wound types. The confidence curves of YOLO11n on testing images from the AZH dataset are shown in Fig. 11 with six classes of wounds. Similarly, the precision-recall plot is shown in Fig. 12.
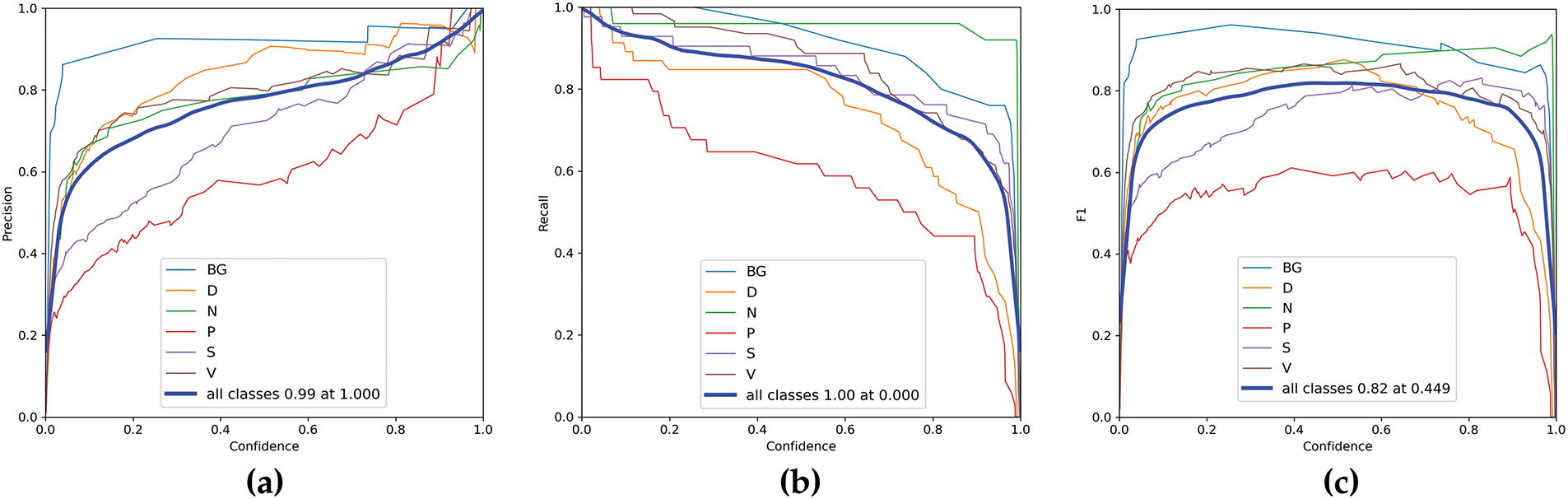
Figure 11: Confidence curves of YOLO11n on testing images from AZH dataset (a) Precision, (b) Recall, (c) F1 Score

Figure 12: Precision-Recall curves of YOLO11n on AZH dataset
The results demonstrate that the YOLO11n model performs well across most classes, with a few challenging cases where confidence levels drop slightly. Improving the dataset’s diversity and refining model hyperparameters could further enhance its ability to distinguish between visually similar wound types or background noise. The model demonstrates a promising capacity to detect and classify different wound types. Further analysis of the confusion matrix and detection results provides valuable insight into potential areas for refinement, such as the handling of background or unannotated regions.
The model completed training and testing, with all six classes represented in both the training and validation sets. However, during the generation of the confusion matrix, a 7
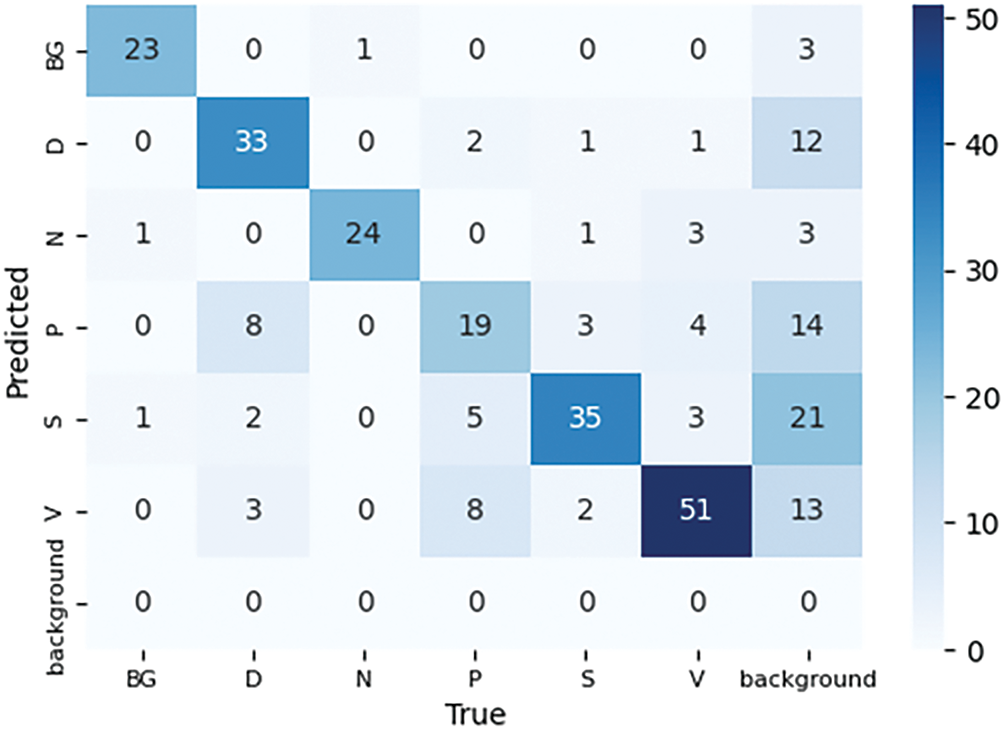
Figure 13: Confusion matrix of trained YOLO11n over 100 epochs with 8 batch size on AZH dataset
To further assess the flexibility and adaptability of the YOLO11n model, we performed wound classification using the AZH dataset under three additional scenarios, i.e., 5-way Classification, 4-way Classification, and 3-way Classification. For each scenario, the dataset was appropriately reorganized, and YOLO11n was retrained to ensure optimal model performance. The performance of the model is shown in the following subsections.
In this experiment, the YOLO11n model is trained without the BG class. The model achieved promising results across different wound types as shown in Table 8. The model attained a Precision of 0.776, a Recall of 0.796, and an


Figure 14: Confusion matrix of five-way wound classification on AZH dataset
5.4 Four-Way Wound Classification
In this experiment, the BG and N classes were excluded from the training and testing datasets. The wound classification is performed using only four classes of wounds. The model achieved a precision of 0.757 and a recall of 0.737 across all classes, with a
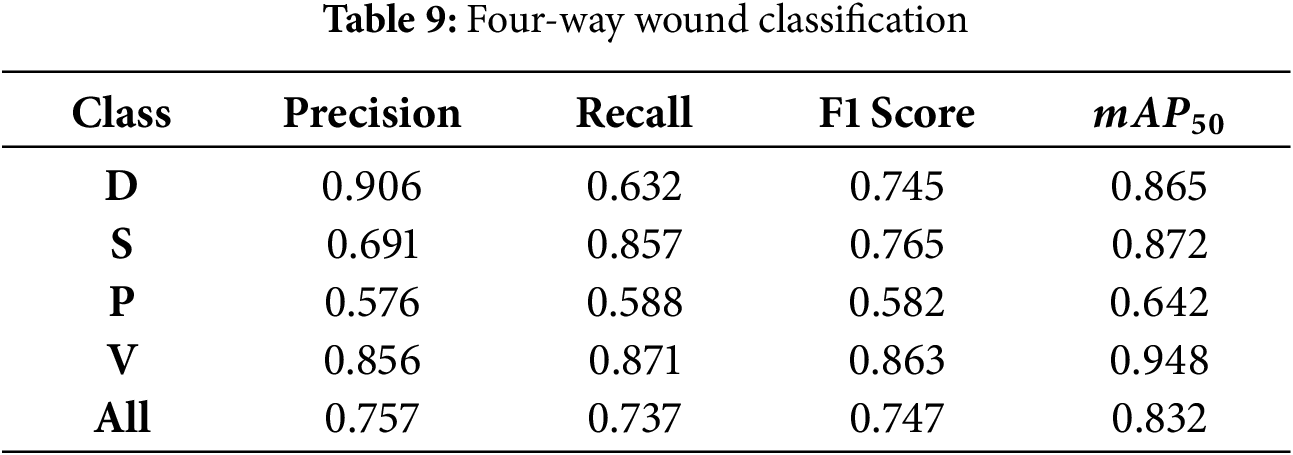

Figure 15: Confusion matrix of four-way wound classification on AZH dataset
In three-way classification, we conducted two experiments. In experiment I, we trained the model on D, S, and P classes, whereas in experiment II, D, S, and V classes were used. In experiment I, the model achieved a precision of 0.711 and recall of 0.729, with a


Figure 16: Confusion matrix of 3-way wound classification on AZH dataset (a) D vs S vs P, (b) D vs S vs V
5.6 Comparison with Existing Models
Table 11 presents a comparative evaluation of existing deep learning models on the AZH dataset using six wound classes. The model proposed by Anisuzzaman et al. [1] employed various VGG-based architectures, with the VGG+MLP configuration achieving the highest accuracy at 82.48%. The EfficientNet-based model with Swish-ELU activations, SEEN-B4, introduced by Aldoulah et al. [26], reported an improved accuracy of 83.19%. In contrast, the YOLO11n approach achieved a significantly higher performance with a mAP@0.5 of 89.3%. This demonstrates the advantage of using an object detection framework for fine-grained wound localization and classification, especially in scenarios where multiple wound types or background elements may coexist.

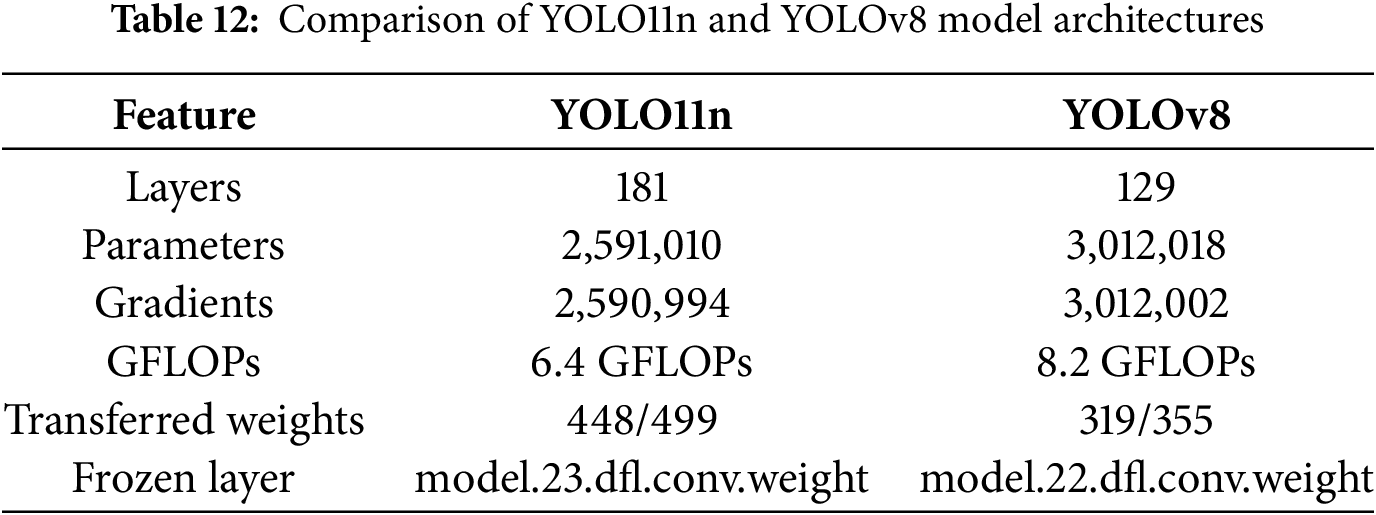
The architecture-based comparison is also carried out with the YOLOv8 model using the AZH dataset. The YOLO11n architecture comprises 181 layers with approximately 2.6 million parameters and 6.4 GFLOPs, indicating a lightweight yet efficient model design. In contrast, YOLOv8 consists of 129 layers but includes a larger number of parameters (over 3 million) and a higher computational complexity of 8.2 GFLOPs. Despite YOLOv8’s architectural complexity and increased resource demands, YOLO11n demonstrates competitive detection performance on the AZH wound dataset. Its smaller footprint in terms of parameters and floating-point operations makes YOLO11n particularly attractive for deployment in resource-constrained environments, such as portable edge devices or embedded clinical systems. Table 12 shows the summary of the comparison in terms of number of layers, parameters, etc. It was observed that YOLOv8 offers marginally higher performance but incurs a higher computational cost. While YOLO11n is a faster and lighter alternative that maintains competitive accuracy and is ideal for real-time applications in low-power clinical settings.
5.7 Architectural Component Evaluation and Robustness
To investigate the contribution of intermediate architectural blocks in YOLO11n, we conducted additional classification experiments using feature outputs from Block 9 (SPPF) and Block 10 (C2PSA). These features were passed into a custom classification head comprising batch normalization, dropout, and a two-layer MLP. Results indicated that Block 10 yielded superior classification performance (79.0% accuracy) compared to Block 9 (70.0% accuracy). We also evaluated model robustness under common clinical degradations such as blur, low-light, and high-light conditions. Block 10 features exhibited improved resilience across all perturbations (blur: 64.9%, low-light: 78.6%, high-light: 49.6%) relative to Block 9 (blur: 51.7%, low-light: 70.5%, high-light: 47.0%). These findings suggest that deeper attention-based modules like C2PSA contribute meaningfully to both classification accuracy and robustness in visually degraded scenarios. A comprehensive ablation with statistical testing will be pursued in future work to further isolate and quantify the impact of these architectural components. Table 13 shows the classification performance using the SPPF block and using the SPPF and C2PSA blocks of the YOLO11n architecture with and without degrading the test images.

To evaluate the spatial attention of the YOLO11n model on wound classification, we applied Grad-CAM at multiple depths of the backbone. Specifically, we selected three convolutional layers—Block 5 (shallow), Block 9 (intermediate, SPPF), and Block 22 (deep, near detection head). For each test image, we extracted the feature maps and gradients at these layers and computed class-agnostic Grad-CAM heatmaps by averaging the gradients over spatial dimensions. These maps were up-sampled and overlaid onto the original RGB image to visualize the regions influencing model decisions. Fig. 17 shows the Visualization of Grad-CAM activation maps at three different stages of the YOLO11n model for wound classification on the AZH dataset. Column (a) shows the original input image. Column (b) presents the YOLO11n detection results with predicted class labels and confidence scores. Columns (c), (d), and (e) display Grad-CAM overlays at layers Block 5, Block 9, and Block 22, respectively.

Figure 17: Visualization of Grad-CAM activation maps at three different stages of the YOLO11n model for wound classification on the AZH dataset
It is evident from Fig. 17 how the attention of the YOLO11n model evolves across different network depths. At Block 5 (column c), the model attends to low-level textures and edges, often highlighting background structures or skin folds. These early features exhibit broader and less localized responses. In contrast, Block 9 (column d), the SPPF module, exhibits mid-level semantic focus, with attention beginning to concentrate on wound shapes, exudate zones, and lesion boundaries. This layer captures richer contextual information compared to Block 5. By Block 22 (column e), the attention becomes highly discriminative and localized, tightly conforming to the wound core, borders, and inflammatory regions. These final-stage activations are most aligned with the YOLO detection heads and show strong agreement with the predicted bounding boxes in column (b). Across all wound types (diabetic, pressure, venous, and surgical), the Grad-CAM overlays at Block 22 consistently emphasize the model’s focus on pathologically relevant regions. This multi-layer visualization demonstrates the progressive refinement of spatial attention, affirming the hierarchical learning structure of the YOLO11n backbone and enhancing the interpretability of model predictions in a clinical setting.
The trend of results showed that as the number of classes decreased, both precision and recall increased. This is expected as the model faces less complexity in distinguishing between fewer categories. Classes like V and D consistently performed well across all experiments, indicating robust feature extraction for these wound types. In contrast, the P class remained challenging, showing lower precision and recall, suggesting the need for more diverse or higher-quality training data. Excluding the BG and N classes led to better performance, as these classes might introduce noise or ambiguity in the training process. Removing them allowed the model to focus on wound-specific features.
While the proposed YOLO11n-based framework achieved strong performance across several wound classes, certain limitations must be acknowledged. The dataset contains an uneven number of samples per class, especially fewer Pressure wound images, which affected the model’s ability to generalize across all categories equally. The original dataset lacked localized wound annotations. As a workaround, synthetic bounding boxes covering the entire image were used, which may have reduced the effectiveness of localization and Explainable AI techniques. The dataset consists of only 930 images. Although these were collected in a clinical setting, a larger dataset with richer variations would help improve the robustness of deep learning models.
First, the dataset contains an uneven number of samples per class, particularly fewer pressure wound images, which affected the model’s ability to generalize equally across all categories. As a result, the model achieved strong overall performance but exhibited comparatively lower precision and recall for underrepresented classes such as pressure wounds. This suggests that class imbalance in the training data influenced the model’s performance. Although standard augmentation techniques (e.g., mosaic, color jittering, horizontal flipping) were automatically applied during YOLO11n training, no explicit class-rebalancing strategies-such as class-aware oversampling or targeted augmentations used to mitigate this imbalance. In future work, we plan to incorporate targeted data augmentation and sampling methods to improve recognition of minority wound classes. Second, the original dataset lacked region-level ground-truth wound annotations, which are critical for training and evaluating object detectors. As a workaround, synthetic bounding boxes were generated to cover the entire image, allowing the YOLO11n model to function in a classification-like manner. However, this approach limits the model’s ability to learn precise spatial localization features and may reduce the validity of explainable AI techniques such as Grad-CAM, which rely on spatial gradients to highlight class-discriminative regions. Consequently, the generated attention maps may not accurately reflect clinically relevant wound features. This limitation underscores the importance of properly annotated datasets for both accurate localization and interpretable decision-making in medical imaging applications. Additionally, although Grad-CAM visualizations were included to provide interpretability, the analysis remains qualitative due to the absence of expert-annotated wound region masks in the AZH dataset. As a result, we were unable to compute region-level agreement metrics such as Intersection Over Union (IoU) or Dice scores to quantitatively validate the alignment of attention maps with wound regions. This is acknowledged as a key limitation, and future work will involve validating explainability outputs against expert annotations to ensure clinical reliability. Future work may include expert evaluation of explainability outputs to ensure clinical trust and usability. Furthermore, this study includes an ablation analysis to isolate the contributions of architectural components such as C3k2, SPPF, and C2PSA. However, incorporating more experiments in future work would help clarify the impact of each component on overall model performance.
Third, the dataset size remains relatively small (930 images), even though images were captured in a clinical environment. A larger dataset with richer intra-class and inter-class variation, including differences in wound shape, size, lighting, and anatomical location, would likely improve the robustness and generalizability of deep learning models in real-world clinical scenarios. Addressing these limitations through more balanced and diverse datasets, inclusion of precise wound region annotations, and dedicated class-balancing augmentation strategies will be important for future work. These improvements would enhance the model’s localization capability, improve interpretability via explainable AI, and ensure reliable performance across all wound types.
In this work, we highlighted the effectiveness of the latest light version of YOLO, known as the YOLO11n model, in classifying wounds across six categories using the AZH dataset. Extensive experiments are carried out to evaluate the effectiveness of YOLO11n on wound classification. The experimental results underscore the importance of tuning hyperparameters, particularly batch size and epochs, in achieving optimal performance. The lightweight architecture of the model and optimization through hyperparameter tuning resulted in competitive performance, with an overall F1 score of 0.83 and
This study encountered several challenges, including class imbalance, inter-class visual similarity, and the absence of bounding box annotations, all of which affected classification accuracy for certain wound types. In future work, we intend to apply class-rebalancing techniques such as data augmentation and oversampling, and to expand the dataset with localized wound annotations. Incorporating multimodal inputs (e.g., body location) and testing on larger, real-world datasets will further validate and extend the applicability of our proposed model.
Beyond technical performance, real-world deployment of AI models requires attention to regulatory compliance, ethical considerations such as patient privacy, and seamless integration into clinical workflows. These factors are essential for safe and effective clinical translation. Future work can focus on validating the model in real-world settings and addressing these practical challenges.
Acknowledgement: The authors gratefully acknowledge the funding of the Deanship of Graduate Studies and Scientific Research, Jazan University, Saudi Arabia, through project number: (RG24-S0150).
Funding Statement: This research was funded by the Deanship of Graduate Studies and Scientific Research, Jazan University, Saudi Arabia, through project number: (RG24-S0150).
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Fathe Jeribi and Ali Tahir; methodology, Ayesha Siddiqa and Hareem Kibriya; software, Ayesha Siddiqa and Hareem Kibriya; validation, Fathe Jeribi, Ali Tahir and Nadim Rana; formal analysis, Fathe Jeribi; investigation, Ayesha Siddiqa and Hareem Kibriya; resources, Ayesha Siddiqa and Hareem Kibriya; data curation, Hareem Kibriya and Ayesha Siddiqa; writing—original draft preparation, Hareem Kibriya and Ayesha Siddiqa; writing—review and editing, Hareem Kibriya and Ayesha Siddiqa; visualization, Hareem Kibriya and Ayesha Siddiqa; supervision, Fathe Jeribi and Ali Tahir; project administration, Fathe Jeribi; funding acquisition, Fathe Jeribi, Ali Tahir and Nadim Rana. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Dataset used in this work are openly available in a public repository.
Ethics Approval: This study utilized the publicly available AZH Wound Dataset, collected and curated by the AZH Wound and Vascular Center in Milwaukee, Wisconsin, and made accessible through the University of Wisconsin–Milwaukee Big Data Lab’s GitHub repository https://github.com/uwm-bigdata/wound-classification-using-images-and-locations (accessed on 15 June 2025). The dataset was released for academic use and contains de-identified wound images annotated by clinical experts. No personally identifiable information (PII) is included, and no interaction with patients was conducted by the authors. Therefore, ethical approval and informed consent were not required for this study.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Anisuzzaman D, Patel Y, Rostami B, Niezgoda J, Gopalakrishnan S, Yu Z. Multi-modal wound classification using wound image and location by deep neural network. Sci Rep. 2022;12(1):20057. doi:10.1038/s41598-022-21813-0. [Google Scholar] [PubMed] [CrossRef]
2. Liu Z, John J, Agu E. Diabetic foot ulcer ischemia and infection classification using EfficientNet deep learning models. IEEE Open J Eng Med Biol. 2022;3(2):189–201. doi:10.1109/ojemb.2022.3219725. [Google Scholar] [PubMed] [CrossRef]
3. Sadaf D, Amin J, Sharif M, Mussarat Y. Detection of diabetic foot ulcer using machine/deep learning. In: Mire A, editor. Advances in deep learning for medical image analysis. Abingdon (UKTaylor & Francis; 2022. p. 101–23. doi:10.1201/9781003230540-7. [Google Scholar] [CrossRef]
4. Ahsan M, Naz S, Ahmad R, Ehsan H, Sikandar A. A deep learning approach for diabetic foot ulcer classification and recognition. Information. 2023;14(1):36. doi:10.3390/info14010036. [Google Scholar] [CrossRef]
5. Saeed T, Khan MA, Hamza A, Shabaz M, Khan WZ, Alhayan F, et al. Neuro-XAI: explainable deep learning framework based on deeplabV3+ and Bayesian optimization for segmentation and classification of brain tumor in MRI scans. J Neurosci Methods. 2024;410(6):110247. doi:10.1016/j.jneumeth.2024.110247. [Google Scholar] [PubMed] [CrossRef]
6. Anisuzzaman D, Patel Y, Niezgoda J, Gopalakrishnan S, Yu Z. Wound severity classification using deep neural network. arXiv:2204.07942. 2022. [Google Scholar]
7. Kibriya H, Siddiqa A, Khan WZ. Melanoma lesion localization using UNet and explainable AI. Neural Comput Appl. 2025;37(16):10175–96. doi:10.1007/s00521-025-11080-1. [Google Scholar] [CrossRef]
8. Sen CK. Human wounds and its burden: an updated compendium of estimates. Adv Wound Care. 2019;8(2):39–48. doi:10.1089/wound.2019.0946. [Google Scholar] [PubMed] [CrossRef]
9. Ezugwu AE, Ho YS, Egwuche OS, Ekundayo OS, Van Der Merwe A, Saha AK, et al. Classical machine learning: seventy years of algorithmic learning evolution. arXiv:2408.01747. 2024. [Google Scholar]
10. Cheng J, Schmidt C, Wilson A, Wang Z, Hao W, Pantanowitz J, et al. Artificial intelligence for human gunshot wound classification. J Pathol Inf. 2024;15:100361. doi:10.1016/j.jpi.2023.100361. [Google Scholar] [PubMed] [CrossRef]
11. Stephens K. FDA authorizes prostate AI software [Internet]. Overland Park (KSAXIS Imaging News; 2021 [cited 2024 Nov 25]. Available from: https://axisimagingnews.com/radiology-products/radiology-software/ai-machine-learning/fda-authorizes-prostate-ai-software. [Google Scholar]
12. FDA Approves First AI-Powered Skin Cancer Diagnostic Tool—targetedonc.com. [cited 2024 Nov 25]. Available from: https://www.targetedonc.com/view/fda-approves-first-ai-powered-skin-cancer-diagnostic-tool. [Google Scholar]
13. FDA clears AI software that ups cancer detection in dense breasts by 50. [cited 2024 Nov 25]. Available from: https://healthimaging.com/topics/artificial-intelligence/fda-clears-ai-software-ups-cancer-detection-dense-breasts-50. [Google Scholar]
14. Almufadi N, Alhasson HF. Classification of diabetic foot ulcers from images using machine learning approach. Diagn. 2024;14(16):1807. doi:10.3390/diagnostics14161807. [Google Scholar] [PubMed] [CrossRef]
15. Alzubaidi L, Fadhel MA, Oleiwi SR, Al-Shamma O, Zhang J. DFU_QUTNet: diabetic foot ulcer classification using novel deep convolutional neural network. Multimed Tools Appl. 2020;79(21):15655–77. doi:10.1007/s11042-019-07820-w. [Google Scholar] [CrossRef]
16. Eldem H, Ülker E, Işıklı OY. Alexnet architecture variations with transfer learning for classification of wound images. Eng Sci Technol Int J. 2023;45(13):101490. doi:10.1016/j.jestch.2023.101490. [Google Scholar] [CrossRef]
17. Goyal M, Reeves ND, Davison AK, Rajbhandari S, Spragg J, Yap MH. DFUNet: convolutional neural networks for diabetic foot ulcer classification. IIEEE Trans Emerg Top Comput Intell. 2018;4(5):728–39. doi:10.1109/tetci.2018.2866254. [Google Scholar] [CrossRef]
18. Goyal M, Reeves ND, Rajbhandari S, Ahmad N, Wang C, Yap MH. Recognition of ischaemia and infection in diabetic foot ulcers: dataset and techniques. Comput Biol Med. 2020;117(7):103616. doi:10.1016/j.compbiomed.2020.103616. [Google Scholar] [PubMed] [CrossRef]
19. Giridhar C, Akhila B, Kumar SP, Sumalata G. Detection of multi stage diabetes foot ulcer using deep learning techniques. In: 2024 3rd International Conference on Applied Artificial Intelligence and Computing (ICAAIC). Salem, India: IEEE; 2024. p. 553–60. [Google Scholar]
20. Huong A, Tay KG, Jumadi NA, Mahmud WMHW, Ngu X. DFU infection and ischemia classification: PSO-optimized deep learning networks. AMS. 2023;7:111–21. [Google Scholar]
21. Narang K, Gupta M, Kumar R, Obaid AJ. Channel attention based on ResNet-50 model for image classification of DFUs using CNN. In: 2024 5th International Conference for Emerging Technology (INCET); 2024 May 24-26; Belagavi, India. Piscataway (NJIEEE; 2024. p. 1–6. doi:10.1109/INCET61516.2024.10593169. [Google Scholar] [CrossRef]
22. Patel Y, Shah T, Dhar MK, Zhang T, Niezgoda J, Gopalakrishnan S, et al. Integrated image and location analysis for wound classification: a deep learning approach. Sci Rep. 2024;14(1):7043. doi:10.1038/s41598-024-56626-w. [Google Scholar] [PubMed] [CrossRef]
23. Rostami B, Anisuzzaman D, Wang C, Gopalakrishnan S, Niezgoda J, Yu Z. Multiclass wound image classification using an ensemble deep CNN-based classifier. Comput Biol Med. 2021;134(5):104536. doi:10.1016/j.compbiomed.2021.104536. [Google Scholar] [PubMed] [CrossRef]
24. Aldughayfiq B, Ashfaq F, Jhanjhi N, Humayun M. Yolo-based deep learning model for pressure ulcer detection and classification. Healthcare. 2023;11(9):1222. doi:10.3390/healthcare11091222. [Google Scholar] [PubMed] [CrossRef]
25. Sarmun R, Chowdhury ME, Murugappan M, Aqel A, Ezzuddin M, Rahman SM, et al. Diabetic foot ulcer detection: combining deep learning models for improved localization. Cogn Comput. 2024;16(3):1413–31. doi:10.1007/s12559-024-10267-3. [Google Scholar] [CrossRef]
26. Aldoulah ZA, Malik H, Molyet R. A novel fused multi-class deep learning approach for chronic wounds classification. Appl Sci. 2023;13(21):11630. doi:10.3390/app132111630. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools