 Open Access
Open Access
ARTICLE
SSANet-Based Lightweight and Efficient Crop Disease Detection
1 Shandong Province University Laboratory for Protected Horticulture, Weifang University of Science and Technology, Weifang, 262700, China
2 Department of Computer Engineering, Dongseo University, Busan, 47011, Republic of Korea
* Corresponding Author: Dae-Ki Kang. Email:
Computers, Materials & Continua 2025, 85(1), 1675-1692. https://doi.org/10.32604/cmc.2025.067675
Received 09 May 2025; Accepted 11 July 2025; Issue published 29 August 2025
Abstract
Accurately identifying crop pests and diseases ensures agricultural productivity and safety. Although current YOLO-based detection models offer real-time capabilities, their conventional convolutional layers involve high computational redundancy and a fixed receptive field, making it challenging to capture local details and global semantics in complex scenarios simultaneously. This leads to significant issues like missed detections of small targets and heightened sensitivity to background interference. To address these challenges, this paper proposes a lightweight adaptive detection network—StarSpark-AdaptiveNet (SSANet), which optimizes features through a dual-module collaborative mechanism. Specifically, the StarNet module utilizes Depthwise separable convolutions (DW-Conv) and dynamic star operations to establish multi-stage feature extraction pathways, enhancing local detail perception within a lightweight framework. Moreover, the Multi-scale Adaptive Spatial Attention Gate (MASAG) module integrates cross-layer feature fusion and dynamic weight allocation to capture multi-scale global contextual information, effectively suppressing background noise. These modules jointly form a “local enhancement-global calibration” bidirectional optimization mechanism, significantly improving the model’s adaptability to complex disease patterns. Furthermore, the proposed Scale-based Dynamic Loss (SD Loss) dynamically adjusts the weight of scale and localization losses, improving regression stability and localization accuracy, especially for small targets. Experiments on the eggplant fruit disease dataset demonstrate that SSANet achieves an mAP50 of 83.9% and a detection speed of 273.5 FPS with only 2.11 M parameters and 5.1 GFLOPs computational cost, outperforming the baseline YOLO11 model by reducing parameters by 18.1%, increasing mAP50 by 1.3%, and improving inference speed by 9.1%. Ablation studies further confirm the effectiveness and complementarity of the modules. SSANet offers a high-accuracy, low-cost solution suitable for real-time pest and disease detection in crops, facilitating edge device deployment and promoting precision agriculture.Keywords
Eggplant is important in global agricultural production, especially in Asia, where approximately 74% of the world’s eggplants are grown. Pest and disease infestations significantly affect eggplant yield and quality. Diseases such as fruit borer (Leucinodes orbonalis), yellow spot disease (Alternaria solani), and fruit rot disease (Phytophthora capsici) can lead to yield losses of 30%–50% in a single growing season. Early detection and precise control of these pests and diseases can significantly reduce agricultural economic losses. However, traditional disease detection methods rely on manual field surveys, which are inefficient and poorly adapted to dynamic changes across large-scale planting environments and varying growth conditions. Therefore, developing lightweight and efficient disease detection technologies has become essential for improving pest and disease monitoring effectiveness and safeguarding agricultural production.
In recent years, deep learning technology has demonstrated substantial potential in agricultural disease detection, especially in disease identification and control. For instance, Beikmohammadi et al. [1] developed SWP-LeafNET, employing multi-stage deep convolutional neural networks (CNN) to effectively identify leaf diseases. Liu et al. [2] proposed PSOC-DRCNet, optimizing rice disease detection in complex backgrounds. Deep learning-based disease detection methods have gradually evolved from general object detection frameworks (e.g., Faster R-CNN, SSD) to more lightweight and specialized architectures. Models from the YOLO series, due to their real-time single-stage detection architecture, have become preferred for field deployment. For example, YOLOv5 optimizes feature extraction efficiency through Cross Stage Partial Networks (CSPNet), while YOLOv8 introduces dynamic label assignment strategies to further improve small-target detection performance. The recently released YOLO11 integrates dynamic attention modules and multi-scale fusion techniques, maintaining high accuracy while reducing computational complexity, making it suitable for edge deployment.
Adaptive attention mechanisms have also shown superior performance in other deep learning applications. For example, Vo et al. [3] first introduced adaptive attention mechanisms in AOE-Net for temporal action proposal generation, significantly enhancing model responsiveness. Luo et al. [4] developed AMAnet, flexibly balancing visual and textual information in image and text generation tasks, demonstrating strong adaptability. Additionally, Wang et al. [5] utilized graph self-attention in Graphformer to capture complex spatial-temporal dependencies in long sequences, broadening the applications of adaptive attention mechanisms. Nevertheless, studies by Deng et al. [6], Xu et al. [7], and Prudviraj et al. [8], among others, improved performance via multi-scale feature fusion and attention mechanisms but still face limitations in cross-task scalability, global modeling, and feature alignment precision, indicating the need for further research and optimization [9].
Traditional deep learning models are typically trained on limited environments or disease samples and lack intrinsic adaptability to the complex variations encountered in real-world conditions. This deficiency results in significant performance degradation in new scenarios. This cross-scenario “domain shift” issue is particularly pronounced in outdoor agriculture, where uncertainties in lighting and background conditions frequently interfere with the extraction of critical disease characteristics. Methods relying solely on color, texture, or single-type features struggle to maintain robustness, thus limiting their potential applications in diverse disease identification tasks. Therefore, consistently maintaining accuracy under highly variable field conditions remains a critical challenge to resolve.
Currently, eggplant disease detection accuracy research has noticeable shortcomings. Kaniyassery et al. [10] proposed a triangular model analyzing environmental, pathological, and plant characteristic factors influencing eggplant diseases. Ma et al. [11] introduced a multimodal data fusion and embedded attention mechanism-based method for eggplant disease detection, but its real-time application in complex environments still requires improvement. Liu et al. [12] proposed an eggplant disease detection model based on YOLOv8, enhancing accuracy and speed via FasterNet and TAM modules, yet still experienced errors detecting small objects. These studies provide significant technical support for eggplant disease detection and management, but still require further optimization. Many existing methods improve accuracy by widening network structures and introducing complex attention mechanisms, leading to high computational costs, parameter redundancy, and limited dynamic adaptability [13]. Conversely, traditional fixed receptive fields and static feature fusion methods struggle to capture both local details and global semantics effectively and cannot dynamically adjust multi-scale information. These issues not only increase model complexity and resource consumption but also restrict their practical application in complex scenarios. Thus, a highly efficient and lightweight approach is urgently needed to achieve dynamic multi-scale feature fusion and high-dimensional nonlinear mappings, meeting the dual requirements of accuracy and efficiency.
Fixed receptive fields and simplistic feature fusion methods in traditional skip connections limit the effective utilization of local details and global semantics. Recent research [14] introduced a new method named Multi-Scale Adaptive Spatial Attention Gate (MASAG), which utilizes multi-scale feature fusion and dynamic receptive field adjustments to highlight critical regions while suppressing background noise, effectively balancing local detail with global semantics. MASAG adaptively weights features from both encoder and decoder sides, and employs spatial interaction and cross-modulation strategies to mutually complement local and global contexts. Compared with traditional methods relying on fixed receptive fields or simple feature concatenation, MASAG significantly improves predictive accuracy while avoiding excessive parameters and computational overhead. Studies show that the MASAG module can be flexibly embedded in CNN-Transformer hybrid networks, leveraging dynamic attention to capture data variations and structural integrity.
Furthermore, this study finds that YOLO11 can balance high detection accuracy with real-time inference efficiency, particularly in resource-constrained and complex target distribution scenarios. Specifically, YOLO11’s lightweight backbone network efficiently captures disease features across multiple scales, effectively reducing redundant calculations and parameter size without sacrificing high-dimensional expressive capabilities. To enable lightweight adaptability, we introduced a Star module based on the “Rewrite the Stars” theory into the YOLO11 network. The Star Operation performs element-wise multiplication on low-dimensional input features, mapping them into high-dimensional nonlinear feature spaces.
In numerous highly complex tasks, Star Operation coordinates multi-node or multi-scale features globally through a central node. On one hand, it aggregates and coordinates local information from child nodes, fully exploring multi-scale patterns. On the other hand, the central node quickly feeds back key features to each branch based on adaptive strategies, enhancing identification and processing in critical areas. Consequently, Star Operation has demonstrated enhanced fusion efficiency and generalization capabilities in fields like video deraining [15], pedestrian trajectory prediction [16], multi-processor network diagnosis [17], brain-computer interfaces [18], automatic modulation recognition [19], and local network design [20]. Star Operation enables high-dimensional nonlinear mapping through compact computation, significantly enhancing feature discrimination while maintaining lightweight characteristics. Differing from traditional methods that widen networks to increase feature dimensions, Star Operation maintains network compactness and lightweight features, bringing stronger adaptability and efficiency to YOLO11, thus improving detection performance in agricultural contexts [21].
Leveraging the YOLO framework, this research seamlessly integrates MASAG and Star Operation modules, constructing an efficient and lightweight object detection model tailored for eggplant disease detection. Our primary contributions include:
• Star operation completes high-dimensional nonlinear mapping under compact computing, which not only significantly enhances feature discrimination but also maintains the network’s lightweight. Compared with relying on network widening to improve expression dimensions, star operation can retain rich discriminant information in a limited model scale, greatly improving the detection efficiency and robustness of YOLO11.
• The MASAG module dynamically adjusts spatial receptive fields, balancing local detail with global semantics, reducing redundant computation, and improving adaptability to complex disease patterns.
• The Scale-based Dynamic Loss (SD Loss) adaptively adjusts weights, assigning higher loss weights to small targets, enhancing sensitivity to challenging pests and diseases in complex agricultural scenarios.
• Utilizing Roboflow for dataset augmentation, we expanded the training set to 7521 samples while maintaining validation and test sets at 744 and 365 images, respectively, enhancing the model’s generalization capability and reliability.
• Dataset: This study utilized a public dataset containing different categories of eggplant fruit diseases, specifically: healthy, fruit borer, yellow spot, and fruit rot disease [22]. The dataset comprises a total of 3616 images, divided into 2507 training images, 744 validation images, and 365 test images. To improve the model’s generalization and detection performance, we applied data augmentation using the Roboflow platform, expanding the training set to 7521 images, while the validation and test sets remained at 744 and 365 images, respectively. The Fig. 1 shows examples of the four categories of eggplant fruit diseases: (a) healthy, (b) fruit borer, (c) yellow spot, (d) fruit rot disease.
Since the specific number of images and their distribution ratios among various categories (health diseases, fruit decay diseases, macular diseases and fruit rot diseases) were not clearly marked in the original data release, we are still unable to accurately grasp their category distribution at the current stage. When the categories in the dataset are unevenly distributed, the model tends to predict the majority of categories, thereby resulting in poor prediction effects for the minority of categories. In the future, in our research, we will give priority to selecting more comprehensive public datasets with clear category distribution annotations and conduct systematic analysis in combination with category balance. Meanwhile, we will also initiate the construction of our own high-quality dataset to ensure the balance of sample category distribution and provide reliable data resources for relevant researchers.
• Experimental environment: In this study, PyTorch was used as the deep learning framework. In Table 1, the experiments were conducted on a system equipped with an Intel Core i9-13900K processor and an NVIDIA GeForce RTX 4090 24 GB GPU, with CUDA version 12.1. The model was trained for 100 epochs with a batch size of 16 per GPU, and the initial learning rate was set to 0.02.
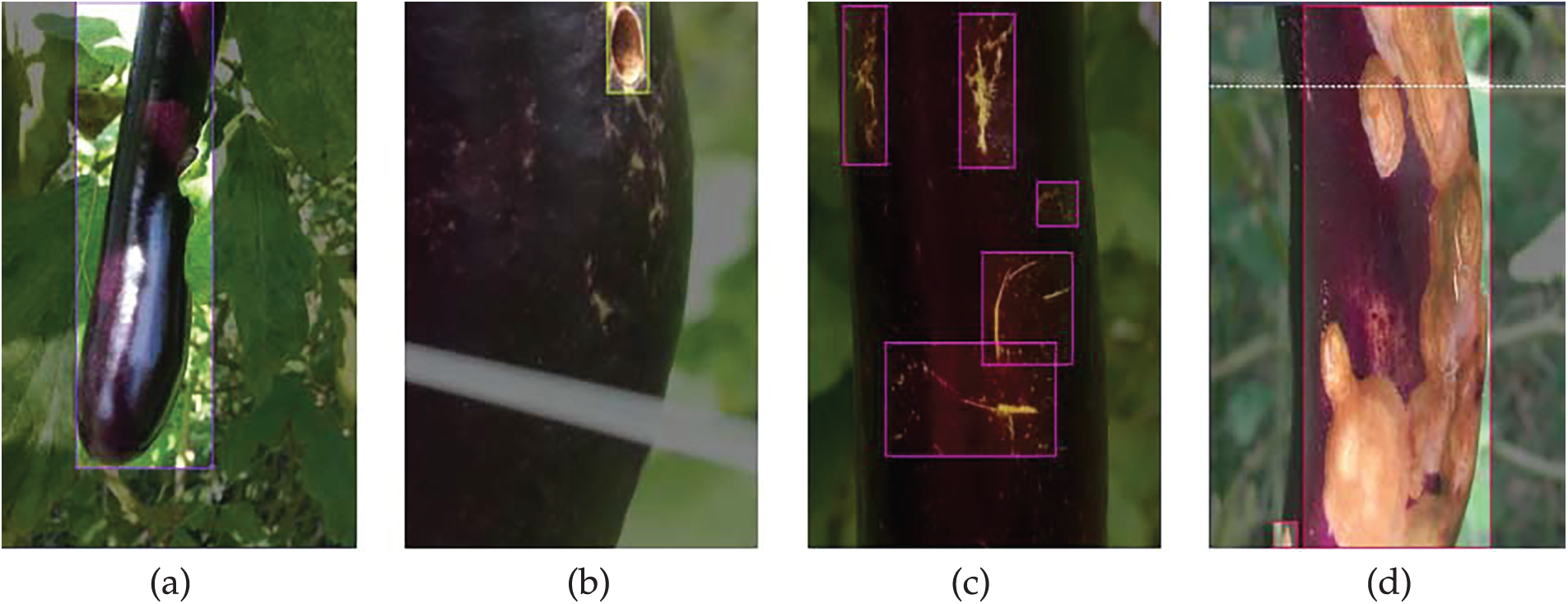
Figure 1: Display of four eggplant fruit diseases. Example images showing eggplant conditions: (a) healthy fruit, (b) fruit borer, (c) yellow spot, and (d) fruit rot disease
To achieve lightweight and efficient eggplant disease detection, this study proposes an improved adaptive model based on the YOLO11 framework, named StarSpark-AdaptiveNet (SSANet). This section first provides an overview of the SSANet architecture and then describes its two core components: the StarNet and the Multi-Scale Adaptive Spatial Attention Gate (MASAG).
• SSANet:
As shown in Fig. 2, SSANet builds upon the basic architecture of YOLO11, replacing its feature extraction component with StarNet, which dynamically transforms input features and enables mapping into a high-dimensional nonlinear feature space. The neck of the network primarily consists of C3k2 modules, which utilize upsampling and downsampling operations to fuse low-level detailed features with high-level semantic features, resulting in richer and more comprehensive feature representations. This unique design significantly enhances the model’s ability to detect multi-scale targets. In addition, we integrate the MASAG module, which dynamically fuses the features from StarNet and the C3k2 fusion module via cross-layer skip connections and weighted combination. This allows for dynamic receptive field adjustment (capturing both local and global contextual information), ensuring the model selectively highlights spatially relevant features while minimizing background interference. As a result, SSANet effectively addresses the issue of weak feature representation in complex scenes and promotes the development of lightweight detection frameworks.
• StarNet:
As the improved feature extraction layer, StarNet is designed with a multi-stage cascading architecture. It employs a staged hierarchical structure, where each stage performs spatial downsampling, channel expansion, and dynamic feature optimization. This setup progressively extracts feature maps at multiple scales, transitioning from low-level visual features to high-level semantic information and forming a complete feature extraction pipeline from local details to global semantics. This enables multi-scale, multi-level understanding of image content.
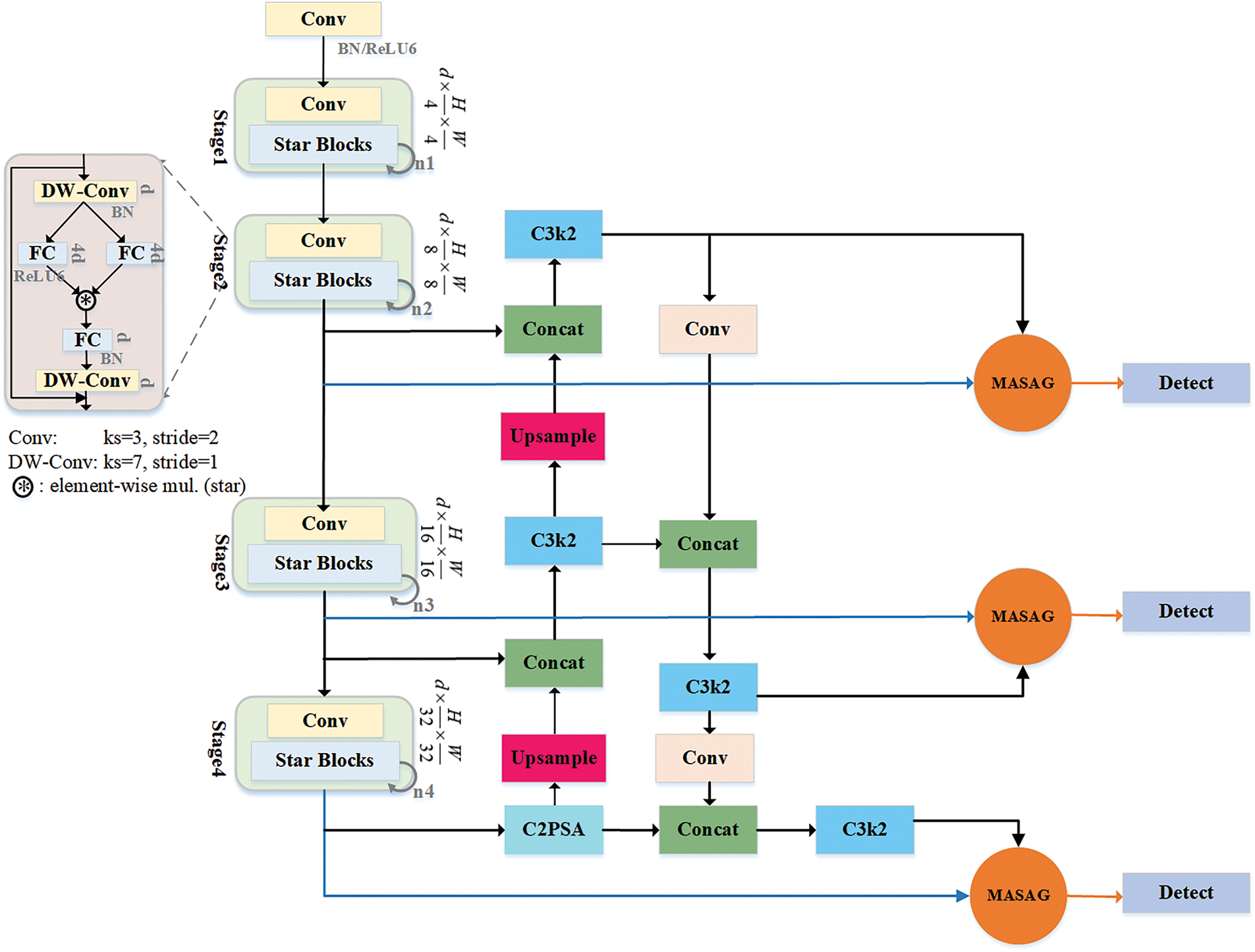
Figure 2: StarSpark-AdaptiveNet
The Star Blocks in StarNet progressively refine features across four stages by increasing iteration depth (e.g., from 1 to 8). Each block combines DW-Conv, channel attention, and residual connections to balance semantic abstraction with efficiency. This enables StarNet to maintain high accuracy and lightweight performance in complex multi-scale detection scenarios.
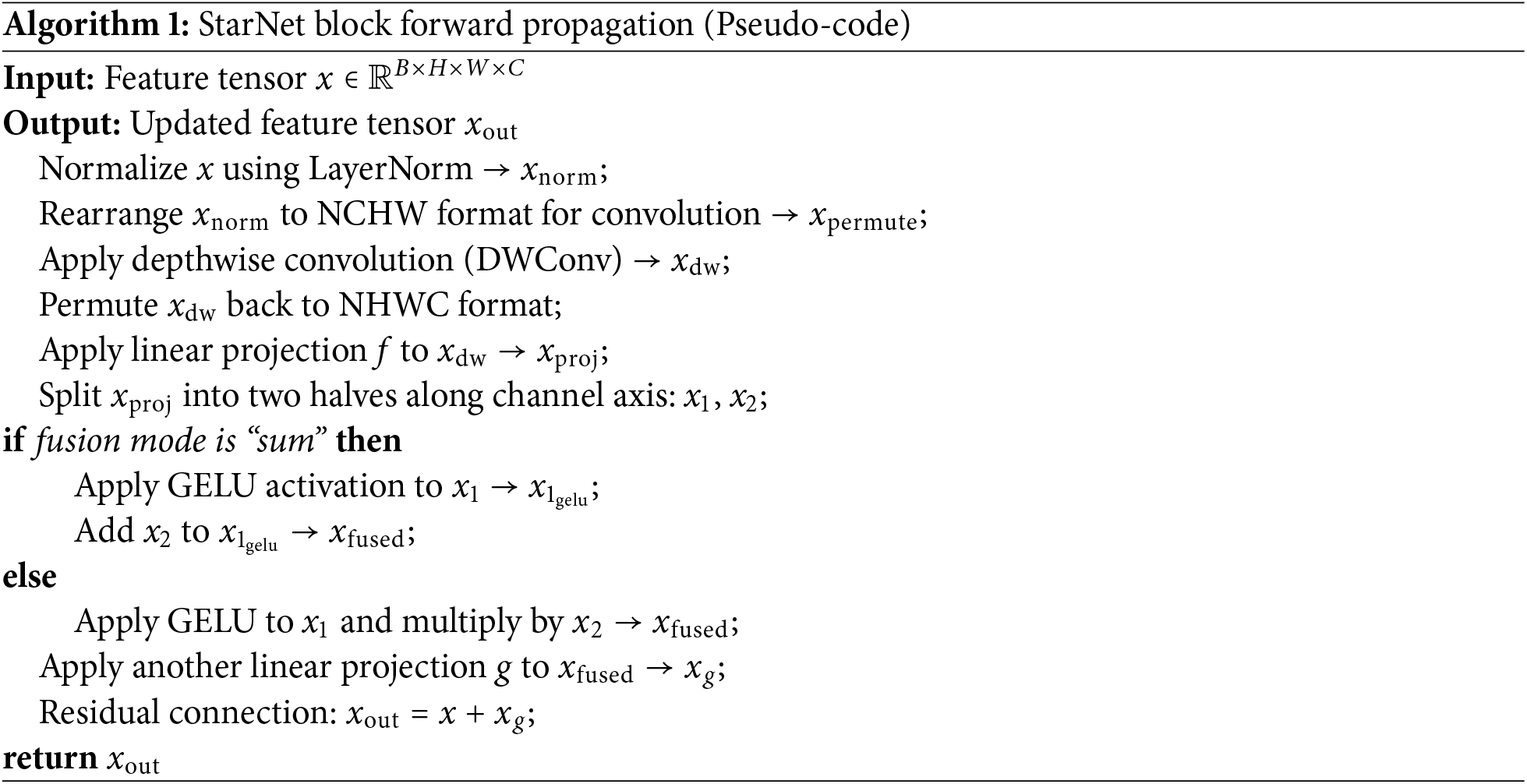
The core unit of the Star Blocks module is the star operation, which can map inputs to extremely high-dimensional nonlinear feature spaces. Its method of achieving nonlinear high-dimensionality is different from traditional neural networks, which are achieved by increasing the network width (i.e., the number of channels). Star operations are similar to kernel functions, which multiply features from different channels in pairs. In particular, the polynomial kernel function combines different subspace features to enhance feature expression capabilities, while improving the ability to capture details and semantics, and can accurately calibrate features under a lightweight computing framework. The star operation is a closed-loop design of “dynamic calibration-weighted fusion-residual retention”, which enables StarNet to achieve detection accuracy close to complex models while maintaining lightweight, providing efficient solutions for difficult target detection tasks in real-time scenarios.
Specifically, assume the input image has dimensions H, W and C, forming the input tensor
Step 1: Downsampling is performed through the initial convolution layer, and batch normalization (BN) and ReLU6 activation functions are used to extract low-level features such as edge texture of the feature map, reducing the amount of computation to meet efficiency requirements. The output is
Step 2-1: In Stage 1, another downsampling is applied to focus on high-level semantic information, enhancing the model’s capacity to handle complex features. The output is
Step 2-2: Enter the Star Blocks module again, the number of cycles N = 1, and the output is
where C represents the input value of the star operation,
Step 3: Complete the sampling of subsequent stages in turn. Finally, the output of the satellite network is
• Multi-scale Adaptive Spatial Attention Mechanism (MASAG):
The Multi-Scale Adaptive Spatial Attention Mechanism (MASAG) exists as an independent cross-level feature fusion interface, introducing a spatial interaction stage into the model, enabling bidirectional processes between local and global features, thereby providing detailed context information for the feature map. The module strategically highlights spatially related features across multiple scales, allowing it to effectively outline and locate complex structures for effective modeling. It promotes spatial interaction between local and global features and enriches the feature map with nuanced contextual information.
MASAG fuses outputs from StarNet and the feature fusion module by dynamically adjusting their weights. It integrates high-resolution low-level semantic features
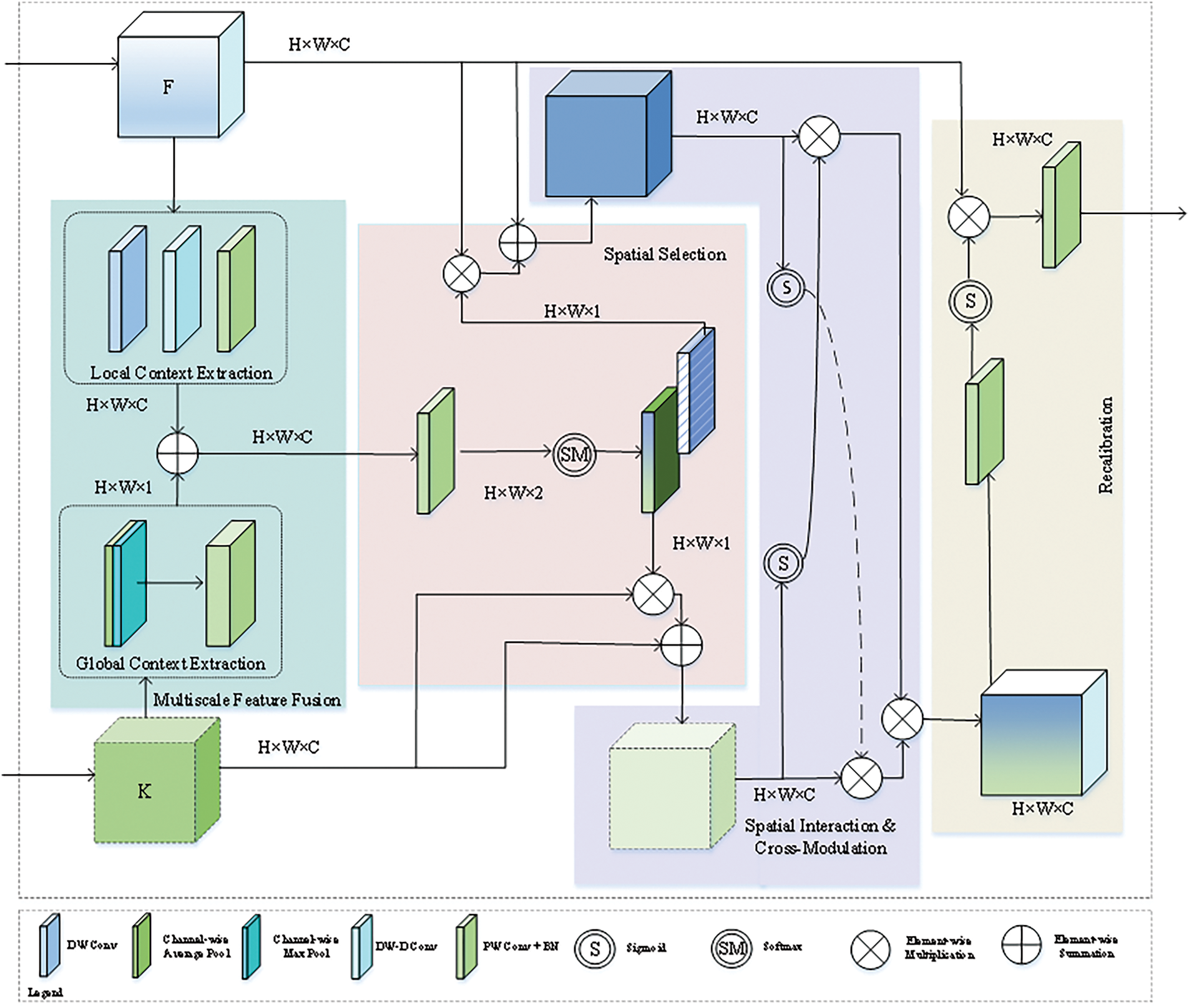
Figure 3: Multi-Scale adaptive spatial attention gate (MASAG)
Multi-scale feature fusion: In the multi-scale feature fusion stage of MASAG, local context extraction is performed on low-level features F through deep convolutions and dilated convolutions, and global context extraction is performed on high-level features K using maximum pooling and average pooling. The two are combined to form a fused feature map U, and its formula is as follows.
DW and DW−D represent depth and unfolding convolutions,
Spatial selection: In the spatial selection stage, the fused feature map U is convoluted to calculate spatial selective weights. These weights are calculated using the softmax function to yield spatially selective versions
S represents the softmax operation performed on each channel and is used to calculate the weights.
Spatial interaction and cross-modulation: This stage achieves mutual enhancement between features
where
The MASAG module processes features from different modules through four key stages (multi-scale feature fusion, spatial selection, spatial interaction and cross-modulation, and recalibration) and dynamically adjusts their weights to generate enhanced fused features. This mechanism can retain detailed information during the feature fusion process and integrate global context information, thereby improving the recognition performance of the model.
• Scale-based Dynamic Loss (SD Loss):
We introduce SD Loss to address the challenge of multi-scale targets and class imbalance in agricultural disease detection. Unlike traditional losses, SD Loss dynamically adjusts weights based on target scale, enabling better focus on both small spots and large lesions. This improves robustness in field environments with variable lighting and background noise.
SD Loss consists of multiple components, each of which is suitable for target detection of different scales. We define total SD Loss as
where N is the number of scales,
where
This dynamic scaling ensures that the loss function adjusts itself based on the characteristics of the data being processed. Smaller objects or fine-grained features gain higher weights in the loss function, prompting models to pay more attention to these key details during training. Larger objects are given relatively low weights to ensure that the model does not overemphasize objects that are easier to detect, thereby improving the stability and accuracy of the model regression process and improving detection results.
This paper uses several indicators to evaluate the performance of the model, including Parameters, computational complexity (GFLOPs), mean precision (mAP50-90) with IoU thresholds from 0.5 to 0.95, mean precision (mAP50) with IoU thresholds of 0.5, and frame rate (FPS). The method for calculating the average accuracy is detailed in the following formula.
K represents the total number of unique object classes in the dataset. The accuracy of each class is evaluated using its corresponding average accuracy (AP) score. Key indicators in performance evaluation include: true positives (TP), which are instances of correctly identifying target conditions; false positives (FP), which occur when the algorithm mistakenly identifies conditions that do not exist; and false negatives (FN), which refer to actual conditions that the system misses.
To highlight the breakthrough of the SSANet model in terms of lightweight and efficiency, this paper conducts comparative experimental analysis based on the public data set of eggplant fruit diseases from three dimensions: parameter compression, precision-speed balance, and calculation efficiency optimization. Combined with the experimental data in Table 2, the performance comparison results between the SSANet model and the current mainstream model are shown. With 2,114,081 parameters and 5.1 GFLOPs, the model achieves a detection speed of 49.7% mAP50-95, 83.9% mAP50, and 273.5 FPS, achieving an optimal balance between accuracy and efficiency.
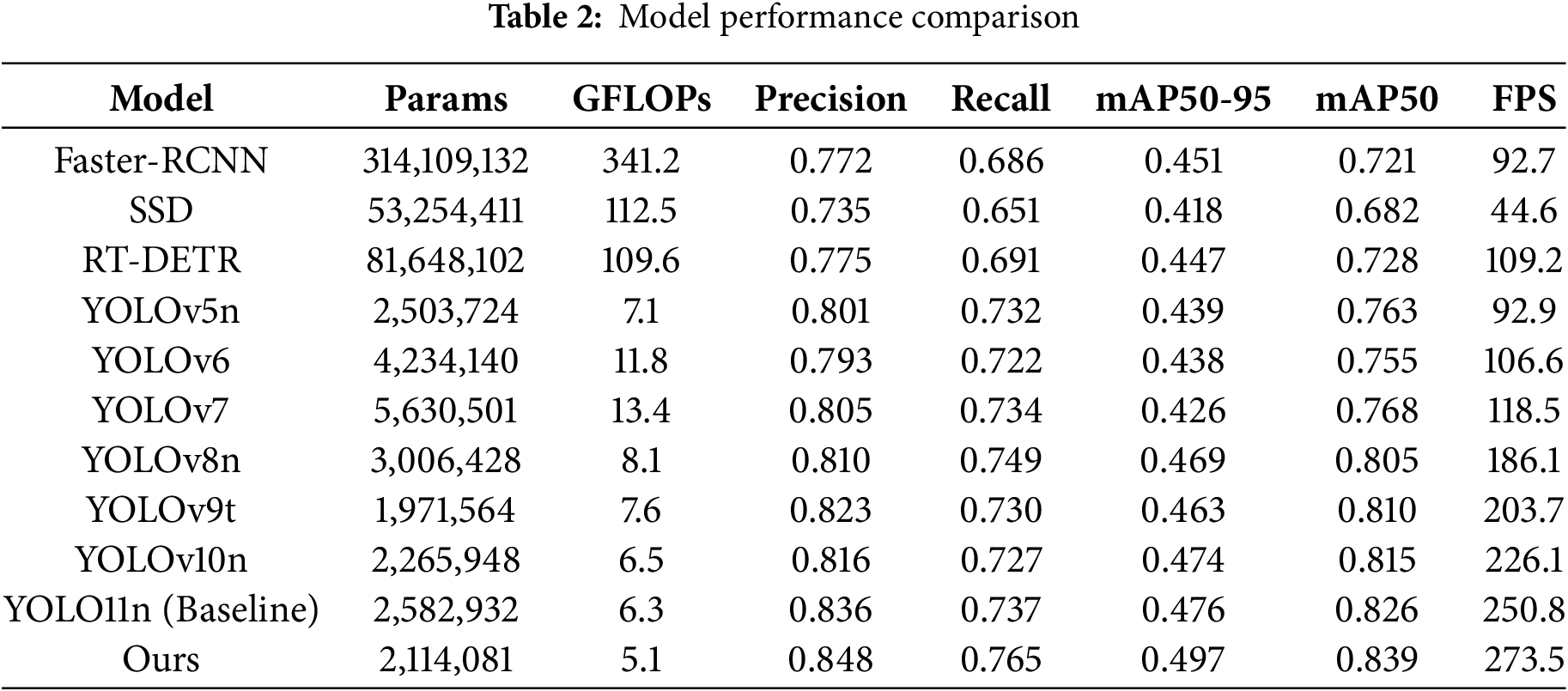
In terms of lightweight, SSANet has the lowest parameter amount and calculation amount among the comparative models, reducing 18.1% parameters and 19.0% calculation amount compared to YOLO11n, and reducing 6.8% parameters and 32.9% calculation amount compared to YOLOv9t; Its efficiency is that at the same parameter level, the inference speed of 273.5 FPS far exceeds that of the comparison model, which is better than Faster-RCNN (45.1%) is 4.6%, which is 5.0% higher than RT-DETR (44.7%), 2.1% higher than YOLO11n, and 9.1% higher than YOLO11n; its computing efficiency optimization is reflected in that compared with the traditional model SSD (44.6 FPS/41.8% mAP50-95), SSANet achieves 6.1 times acceleration and 7.9% accuracy improvement, verifying its excellent adaptability in edge computing scenarios.
Furthermore, in the comparative experiments, we have included the RT-DETR model as a representative Transformer detector for performance evaluation. RT-DETR was proposed by Baidu in 2023, integrating the Transformer structure with a fast decoding strategy, representing the exploration of the Transformer model in the direction of lightweighting. However, the DINO network structure is complex, the FLOPs increase significantly, and the training and inference costs are much higher than those of the YOLO series, which is not conducive to achieving lightweight deployment. Therefore, it was not included in the comparative experiments.
In addition to the numerical comparison, we conducted a statistical significance analysis on three key performance indicators—Params, GFLOPs, and mAP50—to rigorously evaluate the advantages of the proposed SSANet model. In Table 3, using paired
Fig. 4 visually shows the dynamic comparison of mAP_50 and mAP_50-95 indicators between the SSANet model and YOLO11 during the training process on the eggplant disease dataset. Experimental results show that as the training cycles (epochs) gradually increase from 20 to 100, the SSANet model is significantly better than YOLO11 in both indicators. Among them, SSANet’s mAP_50 is always higher than YOLO11, which verifies the model’s ability to continuously optimize target positioning accuracy; the mAP_50-95 curve is located above YOLO11 throughout the entire process, especially after 60 rounds, the improvement trend is more significant, indicating that it is more robust to multi-scale disease targets. This performance advantage stems from SSANet’s innovative architectural design: the StarNet module is used to efficiently extract multi-scale global features, and the MASAG mechanism is introduced to avoid information redundancy and improve feature expression consistency, and output weights are dynamically adjusted to generate enhanced fusion features. This collaborative design effectively combines global feature extraction capabilities and local feature enhancement capabilities, reduces the loss of feature information, and improves the overall performance of the model in accurately capturing the target task of eggplant diseases in complex contexts.
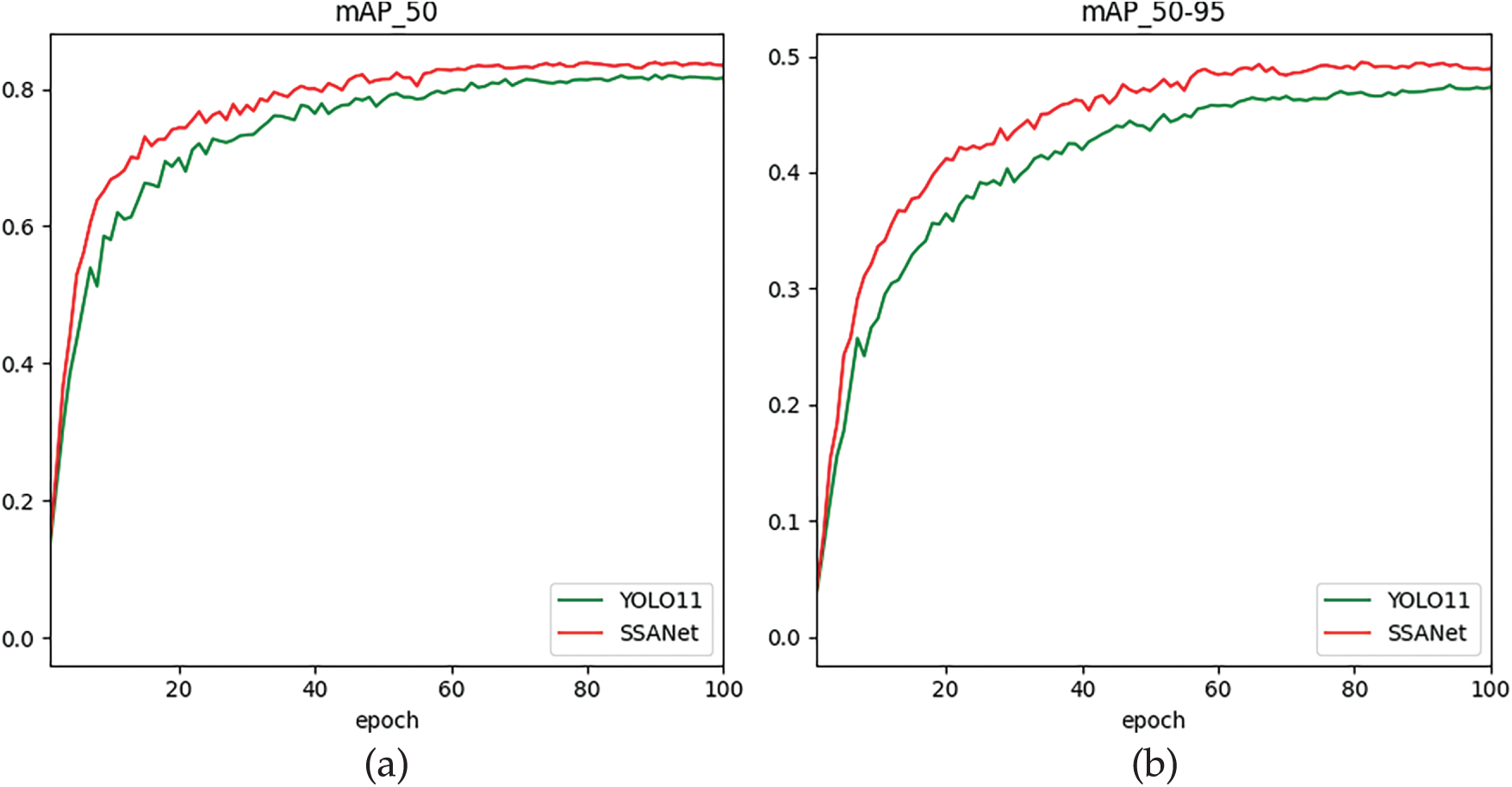
Figure 4: The Model Performance Comparison of mAP. (a) mAP50, (b) mAP50-95
The eggplant disease categories (healthy, fruit borer, yellow spot, and fruit rot) were evaluated using the confusion matrix and its normalized matrix to assess the detection performance of the SSANet model. The model achieved a detection accuracy of 83.9%, demonstrating good performance in detecting individual eggplant diseases. Fig. 5 shows the confusion matrix of SSANet.
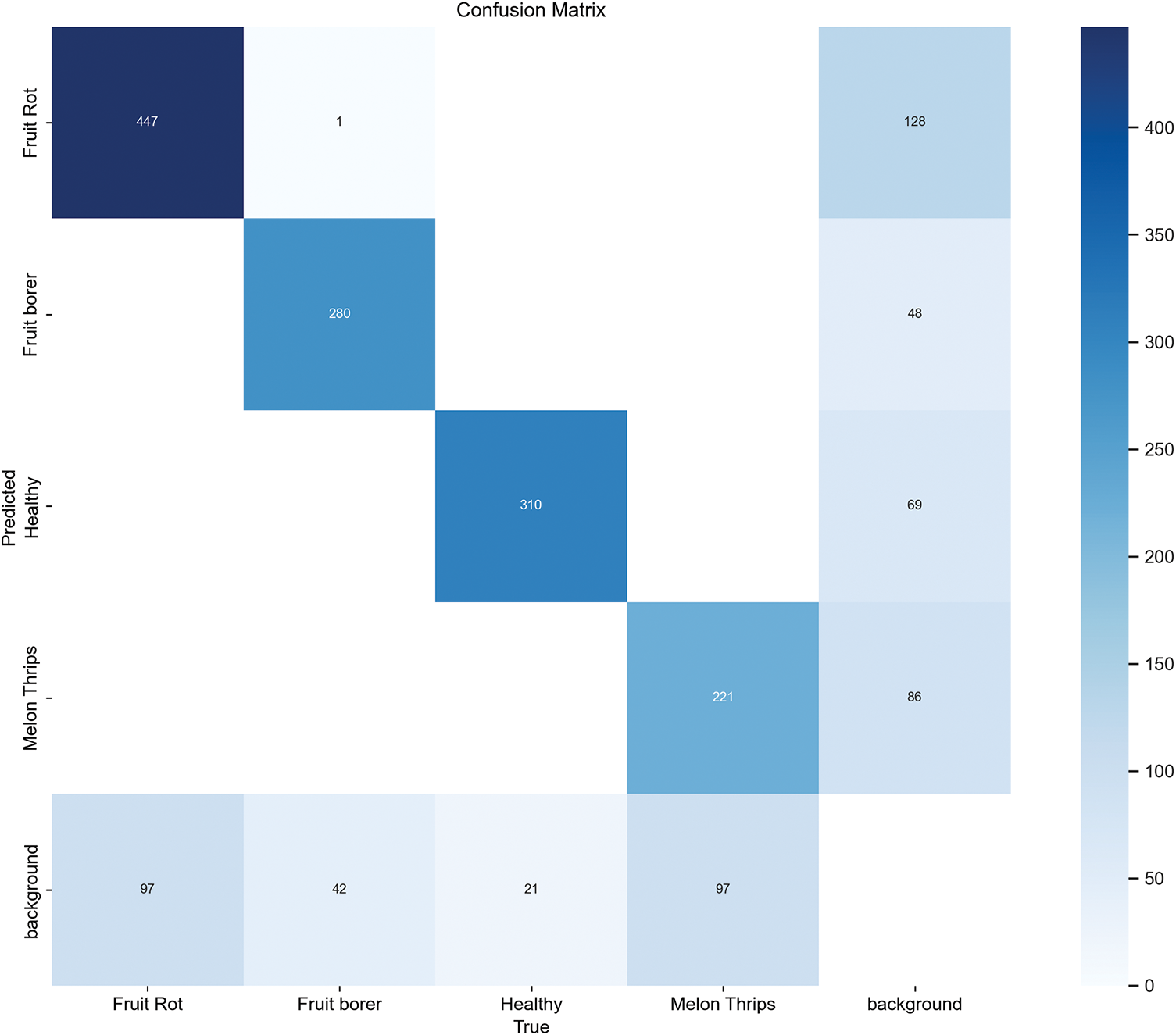
Figure 5: The confusion matrix of SSANet
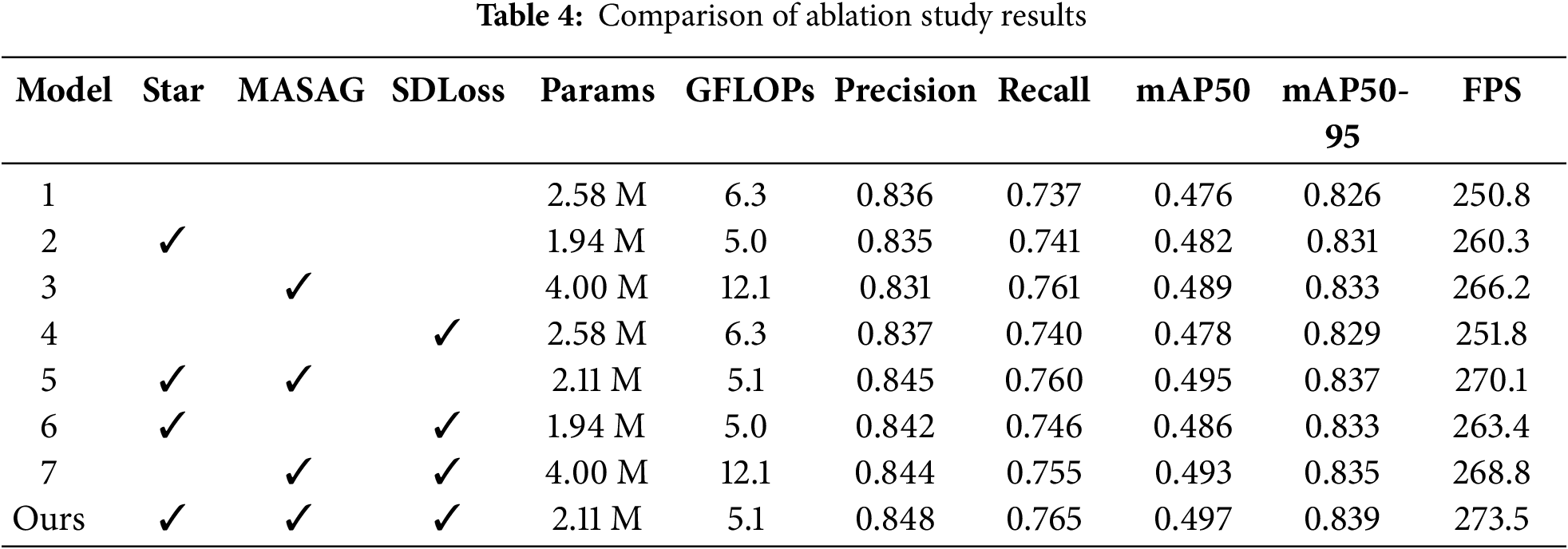
To verify the effectiveness of each innovative module in the SSANet model, this paper carried out a systematic ablation experiment on the eggplant fruit disease dataset. The experimental results are shown in Table 4. As shown in Model 1, when no new modules were introduced, the model achieved performance of 0.476 for mAP50, 0.826 for mAP50-95, and 250.8 for FPS. When introducing individual modules separately (Model2 Model4), the three indicators of mAP50, mAP50-95, and FPS are all improved compared with the corresponding indicators of Model1, further explaining the effectiveness of each new module to the model; when two new modules are introduced each time. (Model5 Model7), The performance indicators of the entire model have been further improved; when three new modules were introduced at the same time (Model Ours), the model achieved optimal values in all three indicators, reaching mAP50 of 0.497, mAP50-95 of 0.839, and FPS of 273.5, respectively. Moreover, the parameter amount and calculation amount are lower than most comparative configurations, proving the complementarity between the modules. These results show that each module has a unique focus, and combining these focuses can significantly enhance the overall disease detection ability of eggplant.
To more intuitively demonstrate the detection performance of our proposed SSANet model, we performed a visual detection comparison between the SSANet model and the baseline model. As shown in Fig. 6, for the detection of the same healthy eggplant, the confidence of the SSANet model increased by approximately 7.4% compared with the baseline model; for the detection of moth-eaten eggplants, the confidence increased by approximately 3.8%–12.5%; and for the detection of eggplant fruit rot disease, the confidence of the SSANet model increased by approximately 4.5%–6.7% compared with the baseline model. It can also be seen from the figure that our proposed SSANet model is more accurate and sensitive in detecting eggplant fruit diseases than the baseline model. Therefore, our proposed SSANet model is significantly better than the baseline model in detecting eggplant diseases and provides a feasible solution for efficiently detecting other vegetable diseases.

Figure 6: Comparison of the visualization. (a) Baseline, (b) SSANet
This paper verifies the efficiency and robustness of SSANet in dynamic agricultural scenarios through experiments. The StarNet module implements high-dimensional nonlinear mapping in low-dimensional space through star operations, avoiding parameter redundancy caused by traditional network widening. At the same time, it takes into account local details extraction and computational efficiency through the co-design of (DW-Convv) and residual connection. The core advantage of the MASAG module lies in its multi-scale dynamic weighting mechanism, which achieves adaptive calibration of cross-layer features by fusing the local context features of deep convolution and expanded convolution, as well as the global semantic information of pooling operations. Experiments show that MASAG’s spatial interaction and cross-modulation strategy can effectively suppress background noise and improve the feature saliency of small targets.
It is worth noting that SSANet’s lightweight performance benefits from the complementary design of StarNet and MASAG. StarNet optimizes links through progressive feature optimization to gradually enhance semantic abstraction capabilities, while MASAG dynamically adjusts receptive fields through a recalibration mechanism, making up for the shortcomings of shallow features in global modeling. Ablation experiments further showed that although introducing any module alone can improve performance, the synergy of the two can increase mAP50-95 by 1.3%, verifying the necessity of the “local-global” bidirectional optimization mechanism.
However, SSANet still faces challenges in actual deployment. Future research can be carried out by exploring knowledge distillation and model quantification technologies, using SSANet as a teacher model to guide ultra-lightweight student networks, further compressing the amount of parameters and computing costs, and adapting resource-limited equipment such as drones and field sensors to further optimize the model’s Lightweight deployment optimization. In addition, we will consider model pruning and hardware acceleration as potential optimization directions to further compress the model size on edge devices while maintaining its reasoning speed. In addition, the current experiment only targets eggplant diseases, and the model’s migration ability on tomatoes, rice, and other crops needs to be verified in the future. Consider introducing a common feature extraction module and cross-task distillation technology, which may become a key solution that can further improve the cross-crop scalability of the model.
In addition, cross-domain robustness and Test-Time Adaptation (TTA) capability are the research directions that our research team is actively planning, especially for the migration and adaptation of crop pest and disease identification tasks across different regions and imaging conditions. Currently, we are working on constructing a dataset with heterogeneous real-field characteristics and plan to systematically evaluate the performance of SSANet and its improved variants under cross-dataset domain shift scenarios. The ultimate goal is to stably deploy the SSANet model in edge computing devices at our local vegetable research base, enabling real-time pest and disease detection and intelligent decision-making in open-field agricultural settings.
In this paper, a lightweight adaptive detection network, StarSpark-AdaptiveNet (SSANet), is proposed to address the challenges of large model parameters, missed detection of small targets, and complex background interference in crop disease detection in dynamic agricultural environments. By integrating the StarNet module with the MASAG, a two-way optimization mechanism of “local detail enhancement-global semantic calibration” is constructed. StarNet is based on DW-Convv and Star Operation to achieve efficient extraction and nonlinear mapping of multi-scale features under a lightweight framework; the MASAG module uses cross-layer dynamic weighting and multi-scale context fusion to effectively suppress background noise and enhance the semantic representation capabilities of small targets. The collaborative design of the two significantly improves the adaptability of the model to complex disease patterns.
Experimental results show that SSANet achieved a detection speed of 83.9% of mAP50 and 273.5 FPS on the eggplant fruit disease dataset with a parameter quantity of 2.11M and a calculation cost of 5.1 GFLOPs, which reduced the parameter quantity of the benchmark model YOLO11 by 18.1%, increased mAP50 by 1.3%, and increased the inference speed by 9.1%. This model provides a high-precision and low-cost solution for real-time disease detection in field scenarios, adapts to edge equipment deployment, and helps the intelligent development of precision agriculture.
Acknowledgement: Thanks to all the authors cited in this article and the referee for their helpful comments and suggestions.
Funding Statement: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2022R1A2C2012243).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Hao Sun; data collection: Di Cai; analysis and interpretation of results: Hao Sun, Di Cai and Dae-Ki Kang; draft manuscript preparation: Hao Sun, Di Cai and Dae-Ki Kang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data available within the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Beikmohammadi A, Faez K, Motallebi A. SWP-LeafNET: a novel multistage approach for plant leaf identification based on deep CNN. Expert Syst Appl. 2022;202(8):117470. doi:10.1016/j.eswa.2022.117470. [Google Scholar] [CrossRef]
2. Liu Z, Zhou G, Zhu W, Chai Y, Li L, Wang Y, et al. Identification of rice disease under complex background based on PSOC-DRCNet. Expert Syst Appl. 2024;249(11):123643. doi:10.1016/j.eswa.2024.123643. [Google Scholar] [CrossRef]
3. Vo K, Truong S, Yamazaki K, Raj B, Tran M-T, Le N. AOE-Net: entities interactions modeling with adaptive attention mechanism for temporal action proposals generation. Int J Comput Vis. 2023;131(1):302–23. doi:10.1007/s11263-022-01702-9. [Google Scholar] [CrossRef]
4. Luo C, Feng S, Quan Y, Ye Y, Xu Y, Li X, et al. AMANet: an adaptive memory attention network for video cloud detection. Pattern Recognit. 2024;155(2):110616. doi:10.1016/j.patcog.2024.110616. [Google Scholar] [CrossRef]
5. Wang Y, Long H, Zheng L, Shang J. Graphformer: adaptive graph correlation transformer for multivariate long sequence time series forecasting. Knowl Based Syst. 2024;285(6):111321. doi:10.1016/j.knosys.2023.111321. [Google Scholar] [CrossRef]
6. Deng L, Wang S, Zhang Y. ELMGAN: a GAN-based efficient lightweight multi-scale-feature-fusion multi-task model. Knowl Based Syst. 2022;252(4):109434. doi:10.1016/j.knosys.2022.109434. [Google Scholar] [CrossRef]
7. Xu Z, Tian B, Liu S, Wang X, Yuan D, Gu J, et al. Collaborative attention guided multi-scale feature fusion network for medical image segmentation. IEEE Trans Netw Sci Eng. 2024;11(2):1857–71. doi:10.1109/TNSE.2023.3332810. [Google Scholar] [CrossRef]
8. Prudviraj J, Vishnu C, Mohan CK. M-FFN: multi-scale feature fusion network for image captioning. Appl Intell. 2022;52(13):14711–23. doi:10.1007/s10489-022-03463-x. [Google Scholar] [CrossRef]
9. Chi Y, Li J, Fan H. Pyramid-attention based multi-scale feature fusion network for multispectral pan-sharpening. Appl Intell. 2022;52(5):5353–65. doi:10.1007/s10489-021-02732-5. [Google Scholar] [CrossRef]
10. Kaniyassery A, Goyal A, Thorat SA, Rao MR, Chandrashekar HK, Murali TS, et al. Association of meteorological variables with leaf spot and fruit rot disease incidence in eggplant and YOLOv8-based disease classification. Ecol Inform. 2024;83(3):102809. doi:10.1016/j.ecoinf.2024.102809. [Google Scholar] [CrossRef]
11. Ma X, Dai X, Bai Y, Wang Y, Fu Y. Rewrite the stars. In: 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2024; Seattle, WA, USA. p. 5694–703. doi:10.1109/CVPR52733.2024.00544. [Google Scholar] [CrossRef]
12. Liu J, Wang X. EggplantDet: an efficient lightweight model for eggplant disease detection. Alex Eng J. 2025;115(1):308–23. doi:10.1016/j.aej.2024.12.037. [Google Scholar] [CrossRef]
13. Wang CY, Liao HYM, Wu YH, Chen PY, Hsieh JW, Yeh IH. CSPNet: a new backbone that can enhance learning capability of CNN. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2020; Seattle, WA, USA. p. 1571–80. doi:10.1109/CVPRW50498.2020.00203. [Google Scholar] [CrossRef]
14. Kolahi SG, Chaharsooghi SK, Khatibi T, Bozorgpour A, Azad R, Heidari M, et al. MSA2Net: multi-scale adaptive attention-guided network for medical image segmentation. arXiv:2407.21640. 2024. [Google Scholar]
15. Zhong W, Zhang X, Ma L, Liu R, Fan X, Luo Z. Star-Net: spatial-temporal attention residual network for video deraining. In: 2021 IEEE International Conference on Multimedia and Expo (ICME). Shenzhen, China; 2021. p. 1–6. [Google Scholar]
16. Zhu Y, Qian D, Ren D, Xia H. Starnet: pedestrian trajectory prediction using deep neural network in star topology. In: 2019 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS). Macau, China; 2019. p. 8075–80. [Google Scholar]
17. Song J, Lin L, Huang Y, Hsieh SY. Intermittent fault diagnosis of split-star networks and its applications. IEEE Trans Parallel Distrib Syst. 2023;34(4):1253–64. doi:10.1109/tpds.2023.3242089. [Google Scholar] [CrossRef]
18. Wang X, Yang W, Qi W, Wang Y, Ma X, Wang W. STaRNet: a spatio-temporal and Riemannian network for high-performance motor imagery decoding. Neural Netw. 2024;178(16):106471. doi:10.1016/j.neunet.2024.106471. [Google Scholar] [PubMed] [CrossRef]
19. Zhang X, Wang Z, Wang X, Luo T, Xiao Y, Fang B, et al. Starnet: an efficient spatiotemporal feature sharing reconstructing network for automatic modulation classification. IEEE Trans Wirel Commun. 2024;23(10):13300–12. doi:10.1109/TWC.2024.3400754. [Google Scholar] [CrossRef]
20. Wu CL, Feng TY, Lin MC. Star: a local network system for real-time management of imagery data. IEEE Trans Comput. 1982;100(10):923–33. doi:10.1109/TC.1982.1675901. [Google Scholar] [CrossRef]
21. Wang X, Yan F, Li B, Yu B, Zhou X, Tang X, et al. A multimodal data fusion and embedding attention mechanism-based method for eggplant disease detection. Plants. 2024;14(5):786. doi:10.3390/plants14050786. [Google Scholar] [PubMed] [CrossRef]
22. BSCS. Eggplant disease detection computer vision project [Internet]. [cited 2025 May 9]. Available from: https://universe.roboflow.com/bohol-island-state-university-vgjlb/eggplant-disease-detection. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools