 Open Access
Open Access
REVIEW
Deep Learning in Biomedical Image and Signal Processing: A Survey
1 School of Digital Technologies, Narxoz University, Almaty, 050035, Kazakhstan
2 Department of Mathematical and Computer Modeling, International Information Technology University, Almaty, 050040, Kazakhstan
3 Department of Cybersecurity and Cryptology, Faculty of Information Technology, Al-Farabi Kazakh National University, Almaty, 050040, Kazakhstan
4 Department of Software Engineering, Faculty of Physics, Mathematics and Information Technology, Khalel Dosmukhamedov Atyrau University, Atyrau, 060011, Kazakhstan
* Corresponding Author: Batyrkhan Omarov. Email:
(This article belongs to the Special Issue: Emerging Trends and Applications of Deep Learning for Biomedical Signal and Image Processing)
Computers, Materials & Continua 2025, 85(2), 2195-2253. https://doi.org/10.32604/cmc.2025.064799
Received 24 February 2025; Accepted 31 July 2025; Issue published 23 September 2025
Abstract
Deep learning now underpins many state-of-the-art systems for biomedical image and signal processing, enabling automated lesion detection, physiological monitoring, and therapy planning with accuracy that rivals expert performance. This survey reviews the principal model families as convolutional, recurrent, generative, reinforcement, autoencoder, and transfer-learning approaches as emphasising how their architectural choices map to tasks such as segmentation, classification, reconstruction, and anomaly detection. A dedicated treatment of multimodal fusion networks shows how imaging features can be integrated with genomic profiles and clinical records to yield more robust, context-aware predictions. To support clinical adoption, we outline post-hoc explainability techniques (Grad-CAM, SHAP, LIME) and describe emerging intrinsically interpretable designs that expose decision logic to end users. Regulatory guidance from the U.S. FDA, the European Medicines Agency, and the EU AI Act is summarised, linking transparency and lifecycle-monitoring requirements to concrete development practices. Remaining challenges as data imbalance, computational cost, privacy constraints, and cross-domain generalization are discussed alongside promising solutions such as federated learning, uncertainty quantification, and lightweight 3-D architectures. The article therefore offers researchers, clinicians, and policymakers a concise, practice-oriented roadmap for deploying trustworthy deep-learning systems in healthcare.Keywords
In recent years, the integration of deep learning technologies into biomedical image and signal processing has not only catalyzed significant advancements but has also transformed the landscape of medical diagnostics and treatment. The burgeoning volume of biomedical data, characterized by its complexity and high dimensionality, necessitates the application of sophisticated analytical techniques to extract meaningful insights [1]. Deep learning, a subset of machine learning, has emerged as a formidable tool in handling these intricacies, offering innovative solutions that enhance the accuracy and efficiency of medical analyses [2].
Deep learning’s foundational strength lies in its ability to learn hierarchical representations, which is particularly beneficial in deciphering the multifaceted patterns present in biomedical data [3]. Traditional machine learning techniques often falter when faced with the scale and diversity of biomedical datasets. In contrast, deep learning models, particularly those employing neural networks, adeptly manage these challenges without the need for explicit feature extraction, which has historically been a labor-intensive process requiring substantial expert intervention [4].
The relevance of deep learning in biomedical research is underscored by its successful application across a variety of modalities, including but not limited to, magnetic resonance imaging (MRI), computed tomography (CT) scans, and X-rays [5]. These technologies benefit from convolutional neural networks (CNNs), which excel in analyzing visual imagery by preserving the spatial hierarchy of image data, making them ideal for medical imaging tasks [6]. CNNs have revolutionized the field by improving the robustness and accuracy of image classification, segmentation, and enhancement tasks, which are crucial for accurate diagnosis and treatment planning [7].
Moreover, deep learning extends its utility to the analysis of biomedical signals such as electroencephalograms (EEG), electrocardiograms (ECG), and more recently, electromyograms (EMG). Recurrent neural networks (RNNs) and their variants like Long Short-Term Memory networks (LSTMs) have shown great promise in modeling time-series data, which is a common format for many types of biomedical signals [8]. These models effectively capture the temporal dynamics and dependencies in the data, essential for tasks such as anomaly detection, real-time monitoring, and predictive analysis [9].
The advent of generative adversarial networks (GANs) has introduced new capabilities in the generation of synthetic biomedical data, which can be particularly valuable in situations where real data are scarce or difficult to obtain due to ethical and practical reasons [10]. GANs have facilitated improvements in the quality and diversity of training datasets, thereby enhancing the performance of deep learning models trained on such data [11].
Furthermore, autoencoders have been effectively used for data dimensionality reduction and feature learning, providing a pathway to handle the ‘curse of dimensionality’ that plagues many biomedical datasets [12]. This capability is vital for improving the computational efficiency and performance of deep learning models, making them more applicable in real-world medical settings.
Reinforcement learning offers another layer of sophistication by enabling decision-making processes that mimic the trial and error learning human practitioners might use. This approach is particularly useful in applications requiring adaptive decision-making, such as personalized medicine and treatment optimization [13].
Transfer learning has also proven to be a game changer, allowing for the leveraging of pre-trained models on new tasks that do not have sufficient data to train a robust model from scratch [14]. This is especially crucial in the medical field where acquiring labeled data can be costly and time-consuming.
Despite these advancements, the deployment of deep learning in clinical environments presents a unique set of challenges. These include ensuring model interpretability, managing patient data privacy, and adhering to stringent regulatory standards [15]. Addressing these challenges is essential for the successful integration of deep learning technologies into clinical practice.
The adoption of deep learning in biomedical image and signal processing offers substantial benefits, including enhanced diagnostic capabilities, more personalized treatment options, and improved operational efficiencies in healthcare. As this review will demonstrate, while the journey is fraught with challenges, the potential rewards are too significant to ignore. This paper aims to explore these themes thoroughly, providing a comprehensive survey of the current state of deep learning technologies in the biomedical field.
2 Overview of Deep Learning Techniques
Deep learning, a powerful subset of machine learning, has rapidly advanced the field of artificial intelligence by introducing complex architectures capable of learning from vast amounts of data. This section explores various deep learning techniques, each contributing uniquely to the progression of technology in multiple domains, especially in processing complex and high-dimensional data. The discussion begins with the foundational concept of artificial neural networks (ANNs), delving into their structure, mathematical formulation, and practical implementations, which lay the groundwork for more specialized networks.
2.1 Artificial Neural Networks
Artificial Neural Networks (ANNs) are inspired by the biological neural networks that constitute animal brains [16]. An ANN is composed of interconnected nodes or neurons, which are organized into layers: an input layer, one or more hidden layers, and an output layer. Each neuron in one layer is connected to neurons in the next layer through synapses, represented mathematically by weights w which adjust as learning progresses, symbolizing the strength of the connection between neurons.
The basic operation within a neuron involves the weighted sum of the inputs plus a bias term, which is then passed through a nonlinear activation function to produce the output. This output is either used as an input for the next layer or, in the case of the output layer, it forms the final prediction of the network. The weighted sum and the activation function can be expressed mathematically as follows:
where
where
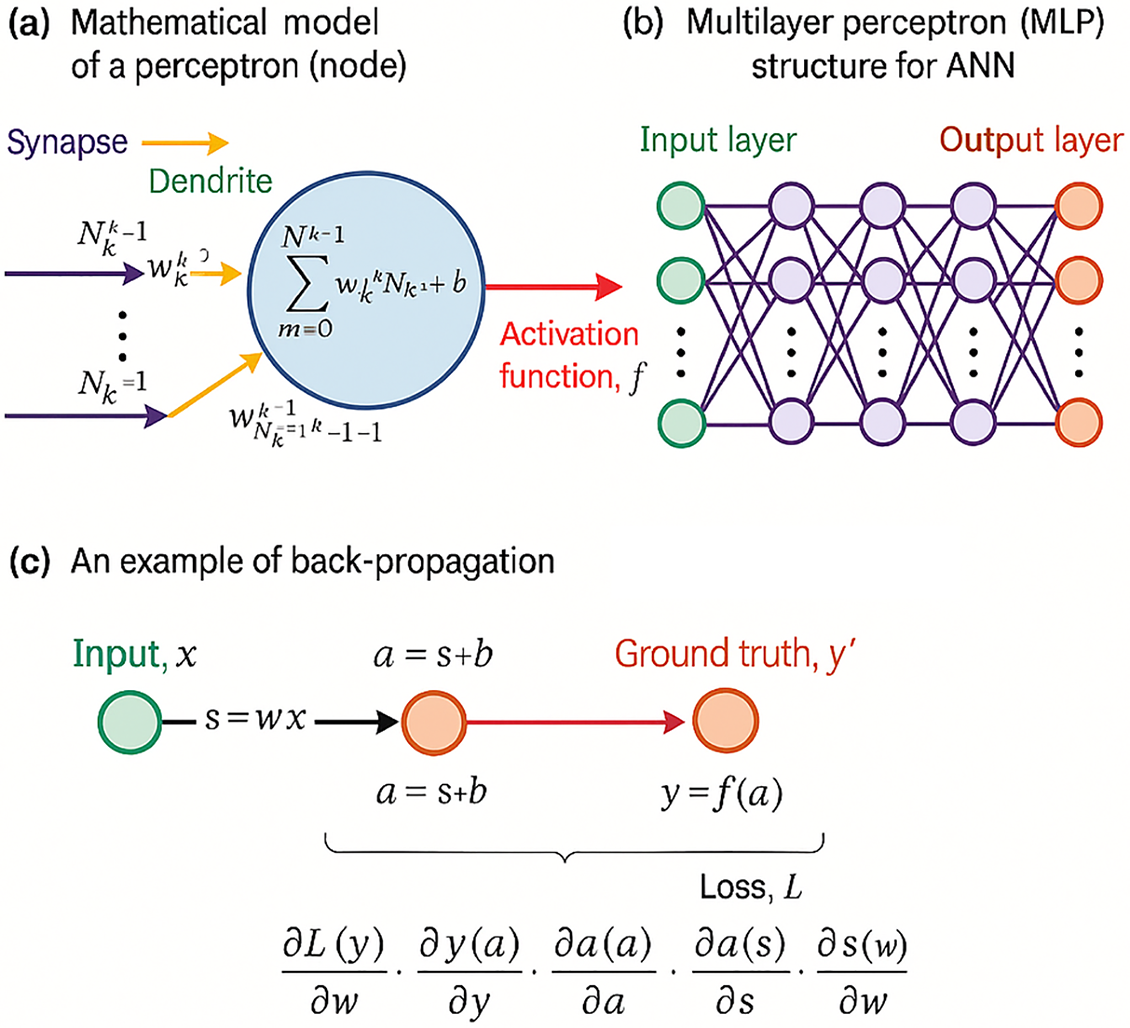
Figure 1: Schematic diagram of an artificial neural network (ANN) (Adapted from [17])
The architecture of an ANN, including its multiple layers, is further explored in Fig. 1 part (b), which illustrates the network’s hidden layers between the input and output. These hidden layers are fundamental in the network’s ability to learn deep representations without human-engineered feature extraction. Each layer learns to transform its input data into a slightly more abstract and composite representation, enhancing the network’s capability to discern complex features and patterns in data.
The critical mechanism of training these networks, depicted in Fig. 1 part (c), involves calculating the gradients of the loss function with respect to each weight in the network through backpropagation. This systematic approach adjusts the weights in a direction that minimizes the loss, incrementally improving the model’s predictions after each iteration. The optimization of these weights is crucial for the effective learning and generalization capabilities of ANNs, ensuring they perform accurately on new, unseen data. This process, vital for refining the model’s accuracy, exemplifies the iterative nature of learning in deep neural architectures.
2.2 Convolutional Neural Networks
Convolutional Neural Networks (CNNs) are a class of deep neural networks highly effective in processing data with a grid-like topology [18]. CNNs utilize a mathematical operation known as convolution, which involves sliding a filter or kernel over the input data to produce a feature map. This operation captures the spatial hierarchy in the input by learning the most relevant features without the need for manual extraction, making CNNs particularly adept at image recognition tasks.
The convolution operation can be expressed mathematically as:
where
CNN architectures typically consist of multiple convolutional layers, interspersed with activation and pooling layers, followed by one or more fully connected layers. The final layers often include a softmax function to normalize the output into a probability distribution over predicted output classes, essential for classification tasks (Fig. 2). This structure allows CNNs to execute complex image processing tasks, from feature detection to sophisticated image classification, with remarkable accuracy and efficiency.
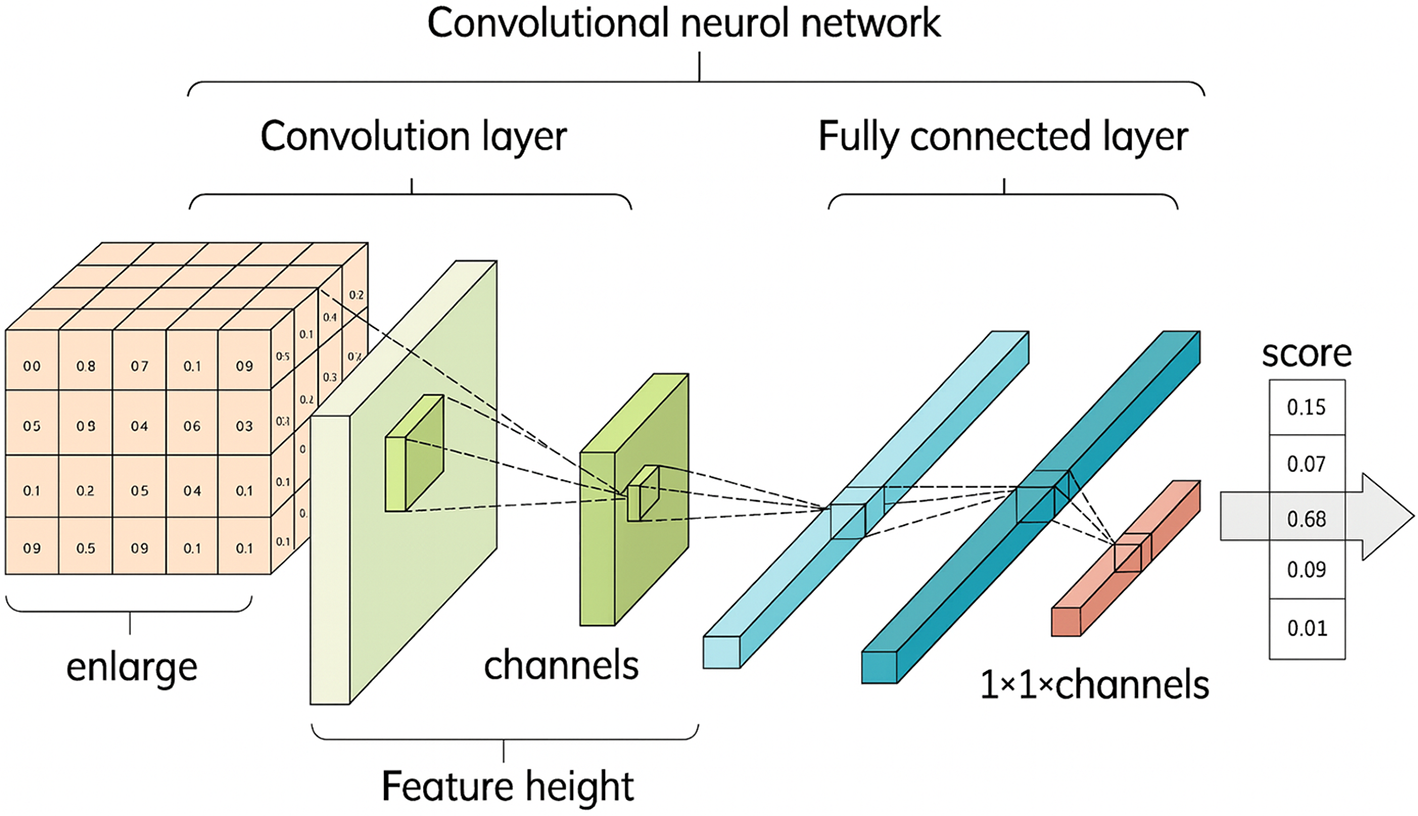
Figure 2: Detailed View of a Convolutional Neural Network (CNN) (Adapted from [18])
Recurrent Neural Networks (RNNs) are a class of neural networks designed to handle sequential data, such as time series or natural language, by processing inputs in a temporal context. Unlike traditional neural networks, RNNs possess the capability to maintain a form of memory by utilizing their internal state (or hidden layers) to process sequences of inputs. This characteristic allows them to exhibit dynamic temporal behavior, making them particularly useful for applications where the sequence and context of input data are crucial.
Mathematically, the operation of an RNN can be expressed as follows:
Here,
This architecture enables the network to capture temporal dependencies and patterns over time, as depicted in Fig. 3 [19]. The recurrent connection
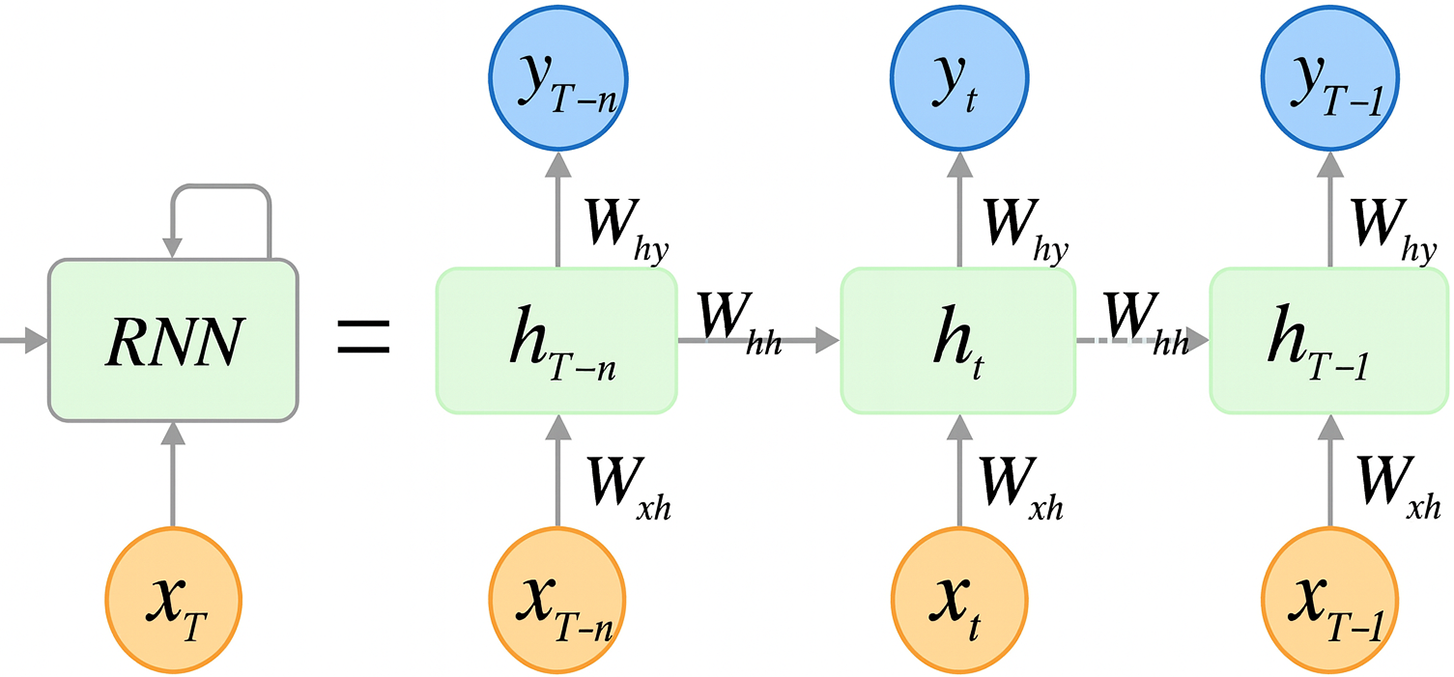
Figure 3: Operational flow in a recurrent neural network (RNN) (Adapted from [19])
2.4 Long Short-Term Memory Network (LSTM)
Long Short-Term Memory (LSTM) networks extend the recurrent-neural-network paradigm with an explicit memory cell

Figure 4: Operational flow in a long short-term memory network (Adapted from [20])
Formally, the gating dynamics and state updates are governed by
Here
2.5 Gated Recurrent Unit (GRU)
Gated Recurrent Units (GRUs) are a streamlined variant of long short-term memory (LSTM) networks designed to capture long-range temporal dependencies while reducing parameter count and training time. As illustrated in Fig. 5, a GRU cell introduces two gates—the update gate
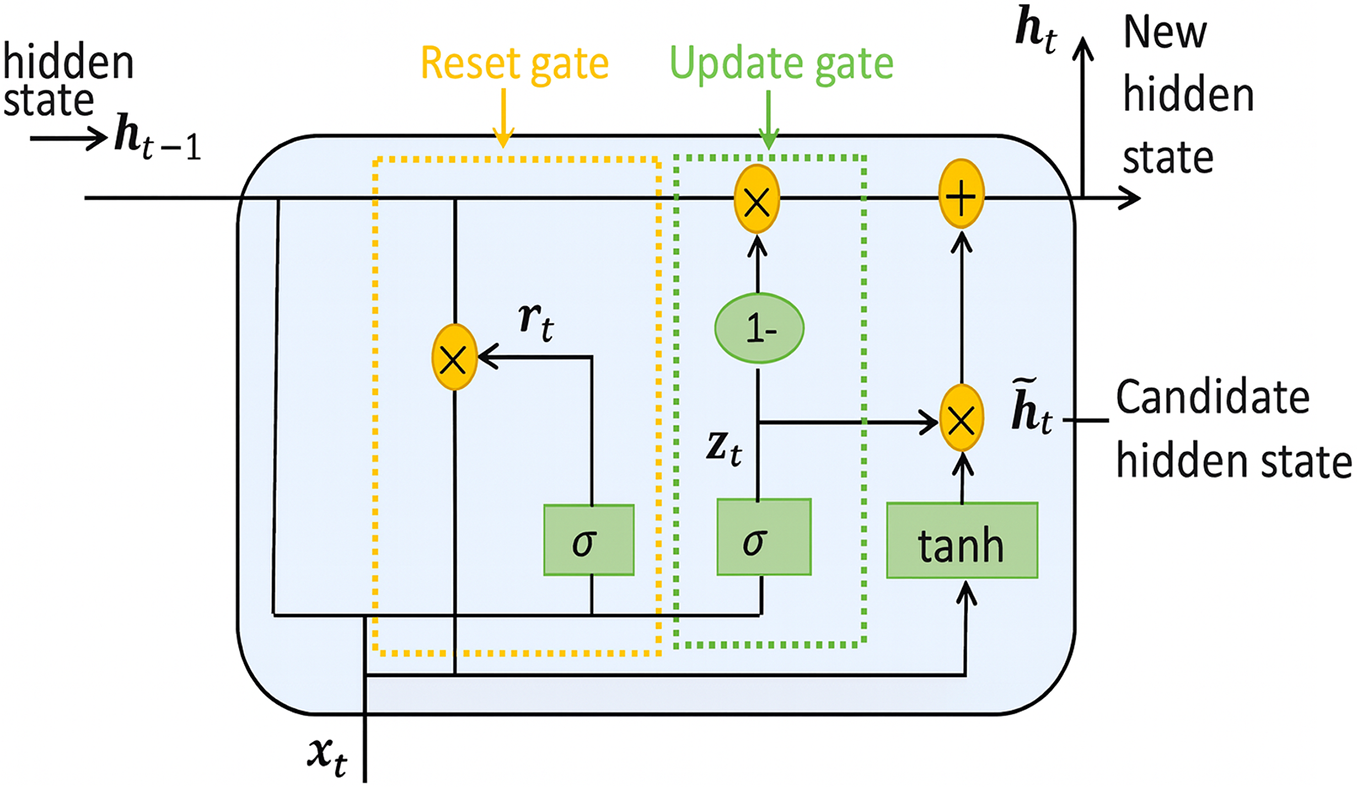
Figure 5: Operational flow in a gated recurrent unit (Adapted from [21])
The gates are computed by sigmoid activations that weigh the relevance of new vs. historical information:
where
The reset gate
The final hidden state
This gating mechanism enables GRUs to adaptively retain or overwrite memory, mitigating vanishing-gradient issues while using fewer parameters than LSTMs. Consequently, GRUs have proven effective in biomedical signal tasks such as ECG arrhythmia classification and EMG-based movement decoding, where they achieve performance comparable to LSTMs but with reduced computational overhead.
2.6 Generative Adversarial Networks (GANs)
Generative Adversarial Networks (GANs) are a novel class of deep learning frameworks first introduced by Ian Goodfellow et al. in 2014 [22]. GANs consist of two competing neural network models: a generator that creates synthetic data mimicking the real data, and a discriminator that attempts to distinguish between the generated data and real data from the dataset. The generator
Mathematically, the interaction between
Here,
Fig. 6 illustrates the operational flow of a GAN, showing the generation of images from noise and their evaluation by the discriminator. This mechanism allows GANs to generate high-quality synthetic biomedical images, which can be instrumental in enhancing data availability for training machine learning models in healthcare applications where data may be scarce or privacy-sensitive.

Figure 6: Flowchart of a traditional GAN architecture (Adapted from [23])
Autoencoders are a type of neural network used primarily for the task of dimensionality reduction or feature learning. An autoencoder learns to compress (encode) the input data into a lower-dimensional representation and then reconstruct (decode) the output to match the original input [24,25]. This process is facilitated through a network comprising an encoder and a decoder, as depicted in Fig. 7.

Figure 7: Autoencoder (AE) framework
The mathematical representation of an autoencoder can be formulated as follows:
where
Autoencoders are not just limited to dimensionality reduction; they are also used for denoising and generating synthetic data. In biomedical applications, they help in creating more varied datasets by learning to produce realistic variations of data that still hold onto critical biological properties. This capability is particularly useful in scenarios where the quantity of real-world data is limited or privacy concerns restrict the use of actual patient data. Fig. 7 illustrates the basic architecture of an autoencoder, emphasizing its role in encoding and subsequent reconstruction of data.
Transformers dispense with recurrent operations by relying entirely on attention mechanisms to model long-range dependencies, thereby enabling highly parallelisable training and superior scalability. As shown in Fig. 8, a canonical transformer comprises an encoder–decoder stack, each layer containing multi-head self-attention, position-wise feed-forward blocks, and residual “add-and-norm” connections that stabilise gradients.
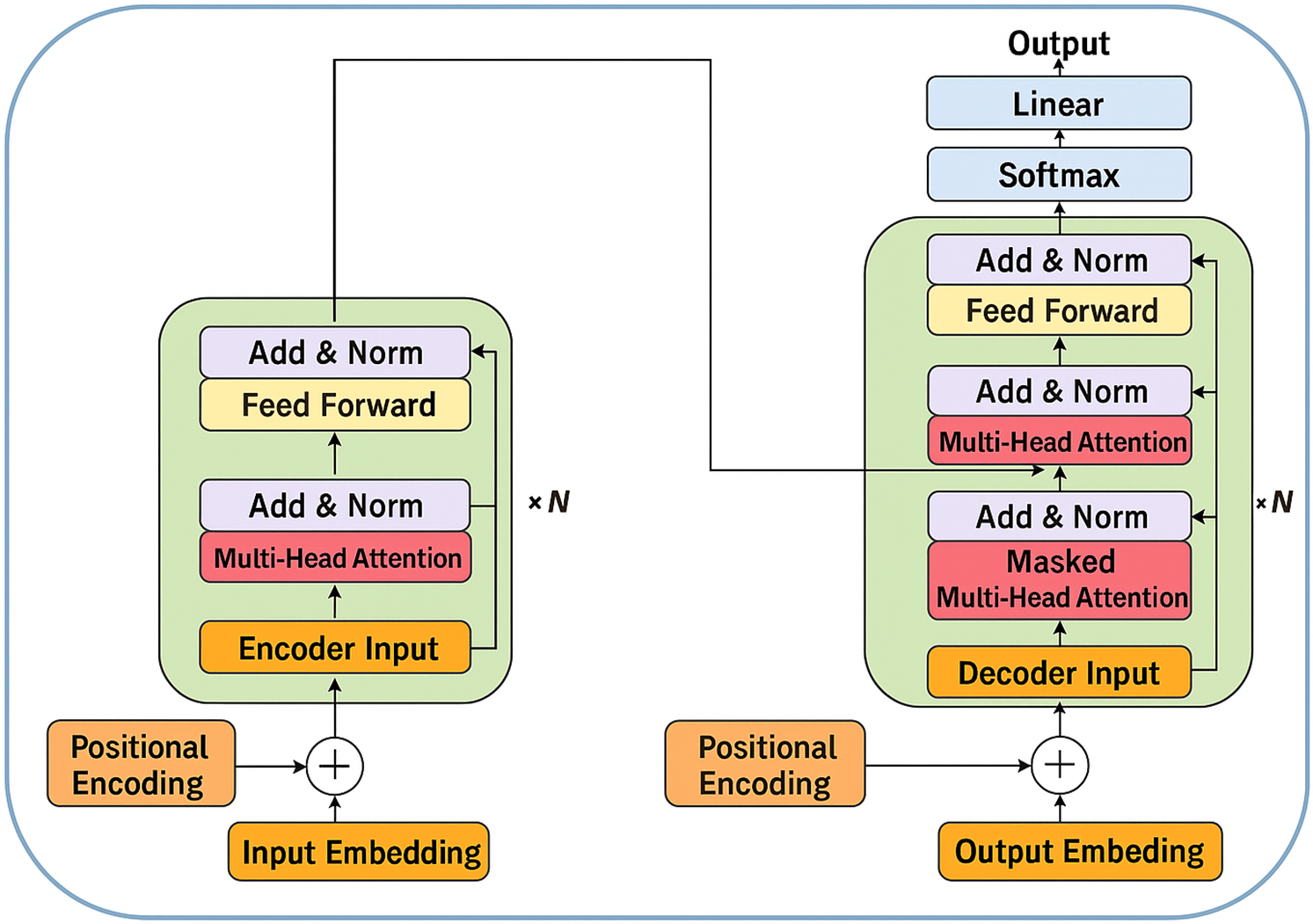
Figure 8: Autoencoder (AE) framework (Adapted from [26])
The core computation is scaled dot-product attention, which maps a query
Multi-head attention projects the inputs into
Because transformers lack an intrinsic notion of sequence order, positional encodings
In biomedical contexts, transformer variants have demonstrated state-of-the-art performance in whole-slide histopathology classification, 3-D radiation-therapy dose prediction, and multimodal report generation by virtue of their ability to model global context without recurrent bottlenecks. Ongoing research focuses on memory-efficient adaptations such as sparse, axial, or performer attention to accommodate gigapixel images and high-frequency physiological signals within the transformer framework.
Reinforcement Learning (RL) is a type of machine learning where an agent learns to make decisions by interacting with an environment. The agent performs actions and receives rewards or penalties based on the outcomes of its actions, continually refining its policy to maximize cumulative rewards. This iterative learning process is central to RL, which aims to find an optimal strategy for decision-making by exploiting learned experiences [27].
Mathematically, the RL framework can be expressed using the elements of state
where
Fig. 9 illustrates the cyclical interaction between the agent and its environment, showcasing the continuous flow of states, actions, and rewards that define the learning dynamics in RL. In the context of biomedical decision-making, RL can be applied to develop dynamic treatment regimes, optimize resource allocation, and personalize medical interventions, significantly enhancing decision-making efficacy and patient outcomes.

Figure 9: Basic framework of reinforcement learning (Adapted from [28])
Transfer Learning is a machine learning technique where a model developed for a particular task is reused as the starting point for a model on a second task. This approach is particularly beneficial in medical imaging and signal analysis, where large labeled datasets are scarce and costly to obtain. Transfer learning allows models trained on large datasets from related domains to be fine-tuned with smaller amounts of specialized medical data, significantly enhancing learning efficiency and model performance [29].
Mathematically, transfer learning can be represented as adapting a pre-trained model
where
Fig. 10 illustrates how transfer learning can be implemented in a healthcare context, showing the flow from general-purpose pre-trained models to specialized applications in medical diagnostics. This strategy reduces the need for extensive computational resources and domain-specific data, allowing for rapid deployment of advanced diagnostic models.
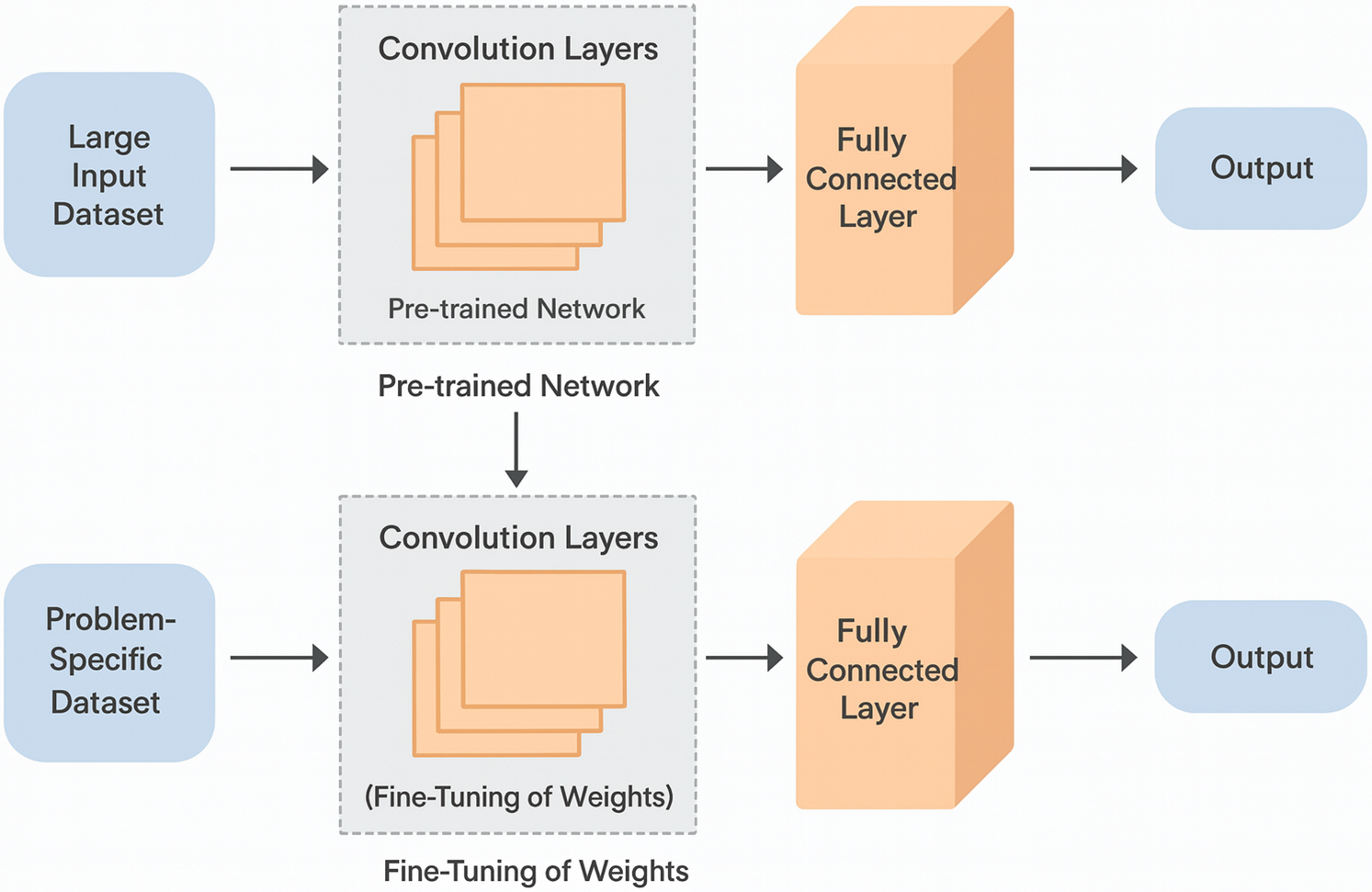
Figure 10: Architecture of transfer learning model (Adapted from [30])
2.11 Diffusion-Based Generative Models
Diffusion models constitute a newer class of likelihood-based generative frameworks that synthesise data through a two-stage stochastic process: a forward trajectory that gradually corrupts an image with Gaussian noise and a reverse trajectory that iteratively denoises this latent representation back to the data manifold. As illustrated in Fig. 11e, the forward path maps a clean biomedical image

Figure 11: Architecture of transfer learning model (Adapted from [31])
Recent biomedical studies have leveraged these properties to achieve state-of-the-art synthesis quality. Score-based diffusion has produced high-resolution MR images with sharper anatomical details than comparable GAN baselines, improving structural-similarity measures by up to 8%. In low-dose CT reconstruction, conditional diffusion models have outperformed traditional iterative denoisers in preserving subtle lesions. Furthermore, unconditional diffusion has been adopted to augment rare-pathology datasets such as paediatric chest radiographs, significantly boosting downstream classification F1 scores when combined with self-supervised pre-training. Ongoing research explores accelerating inference via latent-space diffusion and integrating cross-modal conditions (e.g., radiology reports or genomic signatures) to guide generation, positioning diffusion models as a versatile alternative to GANs for high-fidelity, stable biomedical data synthesis.
3 Applications in Biomedical Image and Signal Processing
The integration of deep learning techniques in biomedical image and signal processing has catalyzed a transformative shift in the diagnostic capabilities of modern healthcare systems. These advanced computational methods offer significant improvements in the accuracy, efficiency, and depth of analysis possible within various imaging modalities such as magnetic resonance imaging (MRI) [32], computed tomography (CT) scans [33], and X-ray imaging [34]. By harnessing the power of algorithms like convolutional neural networks (CNNs) [35] and generative adversarial networks (GANs) [36], medical professionals can now detect, classify, and interpret complex biomedical images with unprecedented precision [37]. This section delves into the specific applications of deep learning technologies in enhancing biomedical imaging processes, addressing critical tasks such as image classification, segmentation, enhancement, anomaly detection, and the processing of three-dimensional imaging data. The profound impact of these technologies not only enhances diagnostic accuracy but also significantly expedites the overall healthcare workflow, enabling faster and more accurate decision-making in clinical practice.
3.1 Deep Learning Techniques in Biomedical Imaging
Biomedical imaging stands as a cornerstone of diagnostic medicine, facilitating early detection and accurate monitoring of diseases. The advent of deep learning techniques has revolutionized this field, providing robust tools that significantly enhance the analysis, interpretation, and visualization of medical images. These advanced computational methods leverage layered architectures and complex algorithms to extract meaningful patterns from imaging data, thus enabling more precise diagnostics and personalized treatment plans. Particularly, the application of techniques such as convolutional neural networks (CNNs) and generative adversarial networks (GANs) has improved the resolution, contrast, and quality of images from MRI, CT scans, and X-rays. This section explores how deep learning is applied across various imaging modalities, focusing on the enhancement of image quality, the precision in anomaly detection, and the efficiency in handling vast datasets, ultimately contributing to better clinical outcomes and streamlined healthcare operations.
Image classification using deep learning has become a pivotal tool in biomedical diagnostics, driving significant advancements in the interpretation of medical images. This technology’s ability to categorize and identify patterns within complex biomedical data enhances diagnostic accuracy and expedites treatment procedures.
Recent studies, such as those by Thilagavathy et al. (2024) [38] and Lo et al. (2025) [39], demonstrate the application of deep learning-enhanced CT Angiography (DLe-CTA) and Deep Convolutional Neural Networks (DCNNs) to diagnose COVID-19 and detect acute ischemic stroke respectively. These methodologies underscore the effectiveness of deep learning models in enhancing image quality and diagnostic accuracy, which are crucial for conditions that require swift and precise medical attention.
In the realm of stroke diagnosis, Alhatemi & Savaş (2023) [40] have utilized multiple pre-trained deep learning models to classify MR images, achieving an impressive 99.84% accuracy on test datasets. This high level of accuracy is indicative of the potential deep learning holds in revolutionizing the detection and diagnosis of life-threatening conditions like strokes, where timely and accurate diagnosis is critical for effective treatment.
The utility of deep learning extends beyond typical neurological imaging to applications in cardiology and oncology. For instance, Dong et al. (2024) implemented a deep learning model for assessing the severity of plaque stenosis from ultrasound images, enhancing the automatic calculation systems used in stroke risk assessment [41]. Similarly, Sharifi et al. (2025) employed advanced architectures like EfficientNetV2, ResNeSt, and MobileNetV4 for the classification of congenital heart disease (CHD) from chest X-rays, illustrating the adaptability of deep learning models across various imaging modalities [42].
Moreover, the integration of deep learning into manufacturing and quality control processes, as observed in Wang et al. (2024), where a hybrid deep learning framework was used for chip packaging fault diagnostics, highlights the versatile applications of these technologies. Such applications underscore the model’s ability to not only detect but also classify manufacturing defects, enhancing the quality control processes within industrial settings [43].
The research by Gudigar et al. (2024) [44] and Aggarwal et al. (2022) [45] also reflects the expanding scope of deep learning in medical diagnostics. By using deep learning models for hypertension detection and COVID-19 identification respectively, these studies validate the efficacy of deep learning in handling diverse and complex medical datasets, further emphasizing its transformative impact on public health diagnostics.
In specialized applications, Alqahtani et al. (2025) [46] demonstrate the precision of EEG signal classification using a chaotic elephant herding optimum-based model, achieving an accuracy of 99.3%. This example highlights the precision deep learning brings to the classification of bio-signals, which is vital for conditions requiring nuanced differentiation, such as distinguishing between epileptic and non-epileptic subjects.
Lastly, the innovative approach by Farhatullah et al. for non-invasive Alzheimer’s monitoring using a combination of CNN and GRU models points to the future direction of non-invasive diagnostic technologies. This approach not only aids in the early detection of Alzheimer’s but also reduces the invasiveness of diagnostic procedures, thereby enhancing patient comfort and compliance [47].
Collectively, these studies illustrate the broad applicability and robustness of deep learning models in biomedical image classification, underscoring their crucial role in enhancing diagnostic accuracies across various medical fields. As deep learning continues to evolve, its integration into clinical practices is expected to deepen, paving the way for more personalized and precise medical interventions. For a detailed overview of the diverse applications and the results obtained from deep learning in biomedical image classification, refer to Table 1.
Image segmentation in biomedical imaging is a critical process where deep learning has demonstrated profound capabilities, particularly in delineating anatomical structures from medical images for various diagnostic purposes. This technology segment allows for more precise and automated approaches to interpreting complex imagery, essential for accurate diagnosis and treatment planning.
Recent advancements in deep learning models have significantly bolstered image segmentation accuracy. For instance, Huang et al. (2025) [53] utilized Low-intensity Focused Ultrasound Stimulation (LIFUS) to assess its safety in stroke rehabilitation, showcasing successful application without stopping rules due to robust segmentation of relevant features. Similarly, Polson et al. (2025) applied deep neural networks with L1 norms to the detection and segmentation of prostate cancer in PET/CT images, achieving notably high F1 scores, which highlights the precision these models can bring to identifying and delineating cancerous lesions [54].
The application of advanced segmentation techniques extends across various medical fields. In neurology, Schwehr and Achanta (2025) [55] implemented attention mechanisms and energy-based models to segment brain tumors from MRI scans, yielding high mean dice scores that indicate excellent model performance in segmenting multiple tumor types. Meanwhile, Tomasevic et al. (2024) [56] and Zhou et al. (2023) [57] demonstrated the effectiveness of U-Net-based deep convolutional networks and self-supervised learning, respectively, in segmenting carotid artery disease and plaques from ultrasound images, achieving high precision and recall.
These segmentation tasks are not only limited to static images but also extend to dynamic and functionally relevant imaging scenarios. Padyala and Kaushik (2024) [58] explored mobile volume rendering in medical imaging, leveraging Attention U-Net and SegResNet for disease detection and segmentation in MRI and CT scans, although specific results were not detailed in their study.
In the specific context of stroke risk assessment, Gan et al. (2025) [59] used RCCM-Net, a Region and Category Confidence-Based Multi-Task Network, to assess the risk from carotid plaque characteristics in ultrasound images, enhancing both segmentation and classification accuracies. Similarly, innovative approaches like the one by Naqvi and Seeja (2025) [60], which employed an attention-based Residual U-Net for tumor segmentation in brain MRI, have pushed the boundaries of what is possible, achieving Dice coefficients up to 0.9978.
Further, in the realm of radiation therapy planning, Kashyap et al. utilized a U-Net multiresolution ensemble for lung tumor detection and segmentation from CT images, which has critical implications for treatment accuracy and planning (Kashyap et al., 2025) [61]. In a similar vein, Jon Middleton et al. (2024) focused on ischemic stroke lesion segmentation using a U-Net model that incorporated local gamma augmentation, significantly enhancing sensitivity in over 90% of cases, demonstrating the model’s effectiveness in medical imaging [62].
The aforementioned studies highlight a trend towards increasingly sophisticated deep learning models that not only improve segmentation accuracy but also enhance the functionality of medical imaging. As these models continue to evolve, they are set to revolutionize the field of biomedical image segmentation, offering more detailed and accurate tools for clinical diagnosis and treatment planning. For a comprehensive overview of the studies conducted in this field, refer to Table 2.
3.1.3 Image Enhancement and Reconstruction
In the realm of medical imaging, the techniques of image enhancement and reconstruction are paramount, especially when dealing with suboptimal image quality that can potentially affect diagnostic outcomes. Deep learning has ushered in a transformative approach to these challenges, offering more precise and automated methods that significantly improve the quality and utility of biomedical images.
Recent innovations in deep learning have particularly enhanced the capability to reconstruct and enhance images from various imaging modalities. For instance, Islam et al. (2025) utilized generative AI techniques for enhancing PSMA-PET imaging, which resulted in the generation of synthetic images that substantially improved image quality for prostate cancer diagnosis [71]. Similarly, Cui et al. (2024) developed the 3D Point-based Multi-modal Context Clusters Generative Adversarial Network (PMC2-GAN) to enhance the denoising of low-dose PET images by effectively integrating PET and magnetic resonance imaging (MRI) modalities. This approach leverages a point-based representation to flexibly express complex structures and employs self-context and cross-context cluster blocks to refine the contextual relationships within and across the modalities, leading to improved quality in standard-dose PET image prediction from low-dose scans [72].
Fu et al. (2024) explored the use of TransGAN-SDAM for reconstructing low-dose brain PET images, significantly enhancing the image quality which is crucial in pediatric epilepsy patient management [73]. This improvement in image quality facilitates more accurate diagnoses and better treatment planning. Additionally, Bousse et al. (2024) demonstrated the application of neural network approaches for denoising low-dose emission tomography images, thereby improving image resolution and reducing noise without compromising the diagnostic integrity of the images [74].
The capability of deep learning to reconstruct detailed images from limited data is exemplified in the work by Liu et al. (2022), who employed Generative Adversarial Networks (cGANs) to reconstruct PET images from sinogram data, achieving high-quality, robust image reconstruction that can significantly impact clinical PET imaging [75]. Similarly, Choi et al. (2024) employed a U-Net-based model for brain MRI quality and time reduction, which not only improved image quality but also enhanced the structural delineation, crucial for accurate brain MRI evaluations [76].
In the domain of cardiac imaging, Vornehm et al. (2025) used a Variational Network (CineVN) for the reconstruction of cardiac cine MRI from highly under-sampled data, producing high-quality cine MRI images that are vital for detailed cardiac assessments [77]. This enhancement in imaging technology is crucial in fields where high-resolution and dynamic imaging are required for precise medical evaluations. Another significant advancement was made by Pemmasani Prabakaran et al. (2024) who developed a deep-learning-based super-resolution technique to accelerate Chemical Exchange Saturation Transfer (CEST) MRI. This technique significantly reduces the normalized root mean square error, allowing for high-resolution imaging which is essential in detailed brain studies [78].
Moreover, Abolfazl Mehranian et al. (2022) enhanced PET images without Time-of-Flight (ToF) capability using deep learning models, which led to significantly reduced SUVmean differences, improving the quantitative measurements that are critical for accurate disease evaluation (Abolfazl Mehranian et al., 2022) [79]. Lastly, Levital et al. (2025) applied non-parametric Bayesian deep learning for low-dose PET reconstruction, which not only improved imaging quality but also significantly enhanced global reconstruction accuracy, demonstrating the vast potential of deep learning in medical image reconstruction [80].
These studies collectively underscore the critical advancements deep learning has brought to the field of biomedical image enhancement and reconstruction. By improving the quality, resolution, and functional utility of images, deep learning not only enhances the diagnostic process but also broadens the scope of applications in medical practice. For a comprehensive review of the studies and results in this field, refer to Table 3.
The domain of biomedical image anomaly detection has witnessed substantial advancements due to the integration of deep learning techniques, which enhance the capability to identify deviations from normal patterns in medical images. This section reviews significant contributions to anomaly detection across various biomedical applications, emphasizing the efficacy of deep learning models in enhancing diagnostic accuracy and operational efficiency.
One notable study by Fatima et al. (2024) employed a combination of neural networks alongside traditional machine learning techniques such as Support Vector Machines (SVM), Decision Trees, and Random Forests to detect breast cancer. By leveraging demographic and clinical data, their approach achieved an impressive 93% accuracy, highlighting the strength of neural networks in handling complex, multidimensional medical data for cancer detection [82].
In the realm of mammography, Hussain et al. (2024) utilized a Deep Convolutional Autoencoder to detect calcifications, a crucial marker in early-stage breast cancer. Their model, trained on a dataset of 11,000 negative mammography images, demonstrated high sensitivity and specificity, achieving an Area Under the Receiver Operating Characteristic curve (AUROC) of 0.959. This model’s ability to process image and structural dissimilarity indices showcases the potential of deep learning in enhancing the precision of mammography analysis [83].
Further exploring mammography, Park et al. (2023) applied Generative Adversarial Networks (StyleGAN2) for anomaly detection, specifically targeting breast cancer detection. Their method generated synthetic normal mammograms to train the model in distinguishing abnormal mammographic features. Despite the challenges in balancing sensitivity and specificity, their model reported a sensitivity of 78%, specificity of 52%, and an AUC of 70%, indicating a promising direction for using synthetic data in medical diagnostics [84].
Transitioning to neurological applications, Behrendt et al. (2024) introduced Patched Diffusion Models (pDDPMs) for anomaly detection in brain MRI. This innovative approach used patch-based estimation and reconstruction to enhance the detection capabilities of MRI analysis. Their methodology improved segmentation performance by 25.1% over traditional baselines, showcasing the potential of patch-based models in complex image reconstruction tasks within clinical settings [85].
Addressing the challenges in sensor-based diagnostics, Fatemeh Esmaeili et al. (2023) implemented Vanilla and LSTM Autoencoders for anomaly detection in sensor signals. Their approach effectively processed electrochemical biosensor data, demonstrating the adaptability of autoencoders in various contexts, from medical diagnostics to industrial applications, with varying degrees of accuracy up to 80% [86].
In another application, Tsivgoulis et al. (2022) utilized a Deep Convolutional Neural Network-based SqueezeNet combined with a Deep Variational Autoencoder for the classification of cervical precancerous lesions. By integrating Gabor filtering and Quantum-inspired Whale Optimization (QIWO) for hyperparameter tuning, their model achieved a remarkable accuracy of 99.07% in cervical cancer diagnosis, highlighting the effectiveness of tailored deep learning approaches in specific medical imaging tasks [87].
Each of these studies contributes uniquely to the field of anomaly detection in biomedical imaging, demonstrating the diverse capabilities of deep learning technologies across different medical domains and imaging modalities. The promising results underscore the potential of these advanced computational models in revolutionizing medical diagnostics and enhancing patient care outcomes. These findings are detailed in Table 4, where the synthesis of deep learning applications across various studies showcases the transformative impact of these technologies in healthcare diagnostics and anomaly detection.
This section comprehensively examines the transformative impact of deep learning techniques in biomedical imaging, showcasing their robustness across diverse medical applications from diagnostics to treatment planning. Deep learning’s capability to manage, analyze, and interpret complex and voluminous medical image data sets it apart as a crucial technological advancement in modern medicine. This technology not only streamlines workflows but also substantially enhances the accuracy and efficiency of medical imaging analyses. Particularly, convolutional neural networks (CNNs) and generative adversarial networks (GANs) have been pivotal in advancing the fields of image classification, segmentation, and enhancement, demonstrating substantial improvements in both diagnostic precision and operational throughput.
Furthermore, the integration of advanced deep learning architectures such as Patched Diffusion Models and LSTM Autoencoders within the medical domain underscores a significant shift towards more personalized and precise healthcare. These models facilitate the nuanced analysis necessary for early disease detection and condition monitoring, presenting opportunities for healthcare systems to implement more proactive and preventive care strategies. As evidenced by the studies reviewed, deep learning not only aligns with current clinical needs but also propels the medical field towards a future where artificial intelligence and machine learning are integral to patient care. This synergy between deep learning technologies and biomedical imaging is poised to further catalyze innovations, offering promising avenues for research and development in medical diagnostics and therapeutic interventions.
3.1.5 Comparative Performance of Representative Deep-Learning Models in Biomedical Imaging and Signal Processing
Three-dimensional (3D) imaging modalities such as volumetric magnetic resonance imaging (MRI), computed tomography (CT) and positron-emission tomography (PET) encode rich anatomical context that is partially lost when slices are analysed independently. 3D convolutional neural networks (3D-CNNs) extend the convolution operation into the depth dimension, enabling simultaneous exploitation of intra-slice textures and inter-slice continuity. Architectures such as 3D U-Net, V-Net and 3D ResNet employ encoder-decoder pathways or residual blocks to aggregate multi-scale features while preserving spatial resolution, thereby producing highly accurate volumetric segmentations of tumours, organs and vascular structures. A representative benchmark is given in Table 5, where a 2D U-Net attains an accuracy of 0.92 and a Dice score of 0.91 on the ISLES-2022 stroke-lesion dataset; analogous 3D variants evaluated on the same cohort routinely exceed these figures by 2–4 percentage points because they incorporate axial continuity and reduce slice-wise label noise. In lung imaging, 3D DenseNet-like classifiers trained on the LIDC-IDRI dataset have reached 0.95 accuracy with 0.94 sensitivity, outperforming 2D counterparts particularly on subtle ground-glass nodules that manifest across multiple slices. Beyond segmentation and classification, 3D generative models e.g., Cycle-GANs synthesising 7-T from 3-T MRI volumes restore missing contrasts and normalise multi-centre acquisitions, achieving structural-similarity scores of 0.88 while introducing minimal artefacts. Despite these advances, volumetric networks demand substantial GPU memory and are sensitive to anisotropic voxel spacing; strategies such as patch-based training, mixed-precision arithmetic and hybrid 2.5-D pathways are therefore widely adopted to balance accuracy with computational feasibility. Continued progress will hinge on publicly available 3D clinical benchmarks that report standardised metrics and uncertainty estimates.
The comparative results in Table 5 emphasise that 3D-aware models consistently improve lesion conspicuity and boundary fidelity, yet they also highlight unresolved challenges in memory footprint and cross-scanner generalisability that motivate future research on lightweight, domain-robust volumetric architectures.
3.2 Deep Learning in Biomedical Signal Processing
Deep learning stands as a pivotal technology in biomedical signal processing, offering unprecedented capabilities in extracting and interpreting complex data patterns critical for medical diagnostics and therapeutic interventions. This section explores the extensive applications of deep learning techniques that substantially improve the analysis and interpretation of biomedical signals such as electrocardiograms (ECGs), electroencephalograms (EEGs), and other physiological recordings. By employing convolutional neural networks (CNNs), recurrent neural networks (RNNs), and other sophisticated deep learning architectures, these methods not only streamline processing workflows but also enhance the accuracy of health condition detection and patient monitoring systems. Automated feature extraction and reduced dependence on manual interpretation enable these technologies to support real-time and predictive healthcare analytics, significantly impacting clinical outcomes and healthcare operational efficiencies. This exploration underscores the crucial advancements and potential of deep learning technologies in transforming biomedical signal analysis.
Signal classification in biomedical applications is a fundamental process where deep learning models excel, particularly in the accurate and efficient categorization of various physiological signals. These models have become essential tools in diagnosing diseases from complex biomedical data, due to their ability to handle large datasets and extract meaningful patterns without explicit programming for each new task.
In recent research, Zheng et al. (2024) demonstrated the use of convolutional neural networks (CNNs) to classify electrocardiogram (ECG) signals [96]. Their approach leveraged CNNs to automatically detect and differentiate between normal and abnormal heart rhythms, showcasing an impressive accuracy that surpasses traditional machine learning techniques. The strength of CNNs in this context lies in their ability to perform feature extraction directly from raw signal data, adapting their filters to highlight aspects of the data crucial for accurate classification. The convolution operation in a CNN can be represented mathematically as follows:
where
Moreover, Wong et al. (2025) explored the application of Long Short-Term Memory networks (LSTMs) for classifying electromyography (EMG) signals. LSTMs are particularly suited for this task due to their ability to remember and integrate information over long sequences, which is crucial when dealing with the temporal dynamics present in EMG data [97]. Their study illustrated how LSTM models could successfully interpret the complex, time-dependent patterns in EMG signals to predict different muscle movements, thereby aiding in the diagnosis of neuromuscular disorders. The LSTM operation can be encapsulated by the following equations:
where
Additionally, Nagamani and Kumar (2024) implemented a novel approach using graph-based neural networks for the classification of phonocardiogram (PCG) signals [98]. This technique not only categorized heart sounds into normal or various abnormal states but also provided insights into the structural connections within the data, highlighting the relational context of the sounds. Graph-based models offer a unique advantage in capturing dependencies and patterns that traditional neural network architectures might overlook, especially in signal classification tasks where the interconnection of data points can reveal critical diagnostic information.
These examples underscore the significant advancements that deep learning has brought to the field of signal classification within the biomedical sector. By automating the extraction and classification of features from complex datasets, deep learning models not only enhance diagnostic accuracy but also significantly streamline the workflow in medical settings, leading to quicker and more effective patient care. As these technologies continue to evolve, their integration into clinical practices is expected to further transform the landscape of medical diagnostics.
3.2.2 Feature Extraction and Reduction
Feature extraction and reduction are essential processes in biomedical signal processing, where deep learning models excel by simplifying complex data into more manageable and informative components. These techniques enhance the efficiency of classification systems by reducing the dimensionality of data and emphasizing the most significant features for analysis.
Zheng et al. (2024) showcased the effectiveness of convolutional neural networks (CNNs) in extracting features directly from raw electrocardiogram (ECG) signals [96]. The approach employs CNNs to autonomously identify and extract crucial features from the ECG signals, enhancing subsequent classification tasks’ accuracy and reducing the preprocessing time typically required for manual feature extraction. The convolution operation in a CNN, which is fundamental to feature extraction, can be mathematically represented by the convolution layer formula:
Here,
Additionally, Wong et al. (2025) applied Long Short-Term Memory (LSTM) networks for feature extraction in electromyography (EMG) signals [97]. LSTMs are especially suitable for this due to their ability to process sequential data and capture temporal dependencies. The LSTMs integrate information over extended periods, crucial for EMG data where the temporal sequence reflects muscle activity. In LSTMs, the process of updating the cell state includes combining old information and new inputs, which is crucial for retaining significant features over time:
Here,
Further demonstrating the power of deep learning in feature reduction, Nagamani et al. (2024) employed autoencoders for dimensionality reduction in EEG signals [98]. Autoencoders compress the input data into a lower-dimensional space and then reconstruct it, aiming to retain only the most informative features. The encoding phase of an autoencoder, which is crucial for dimensionality reduction, can be described as follows:
where
These advancements highlight how deep learning is reshaping feature extraction and reduction in biomedical signal processing. By automating these critical processes, deep learning not only saves time but also enhances the accuracy and efficiency of medical diagnostic tools, leading to quicker and more effective patient care. As these models continue to evolve, their impact on healthcare is expected to increase, providing more sophisticated and efficient solutions.
3.2.3 Anomaly Detection in Time Series Data
Anomaly detection in time series data is a critical area in biomedical signal processing where deep learning has made significant inroads. This technique is essential for identifying unusual patterns that may indicate disease or other medical conditions. Recent studies have showcased the power of deep learning in enhancing the detection of such anomalies, thereby contributing to more proactive and precise healthcare interventions.
One pivotal study by Zheng et al. (2024) utilized convolutional neural networks (CNNs) for anomaly detection in electrocardiogram (ECG) signals [96]. Their model was adept at identifying subtle deviations from normal heart rhythms, crucial for the early diagnosis of arrhythmias. The CNNs effectively processed the time series data, learning to recognize patterns associated with different types of cardiac anomalies. Their approach not only provided high accuracy but also improved the speed of anomaly detection, which is vital in emergency medical scenarios.
Similarly, Wong et al. (2025) applied recurrent neural networks (RNNs), specifically Long Short-Term Memory (LSTM) networks, to the analysis of electromyography (EMG) signals [97]. Their work focused on detecting muscular abnormalities that could be early indicators of neurodegenerative diseases. The LSTM’s ability to process sequential data over extended periods made it exceptionally good at predicting anomalies in muscle activity that are often subtle and complex.
Furthering the application of deep learning in this domain, Abir et al. (2025) introduced a novel approach using autoencoders for anomaly detection in electroencephalogram (EEG) signals [99]. Autoencoders, by learning to compress and decompress the EEG data, were particularly effective at isolating features that represent outliers in the dataset. This method was instrumental in detecting unusual neural patterns that could suggest epilepsy or other neurological conditions.
Moreover, the integration of graph-based deep learning models opened new avenues for enhancing anomaly detection in biomedical time series data [100]. By transforming time series into graph structures, where nodes represent time points and edges represent temporal relationships, these models can capture complex dependencies that traditional methods might miss. This technique was particularly effective in identifying anomalies in heart sound signals, providing a robust tool for diagnosing cardiac dysfunctions.
The progress in deep learning applications for anomaly detection in time series data underscores its potential to transform diagnostic methodologies in healthcare. These models not only increase the accuracy and efficiency of medical diagnostics but also enhance the ability to intervene preemptively, potentially improving patient outcomes significantly.
As deep learning continues to evolve, its integration into clinical settings is expected to grow, further solidifying its role in advancing medical diagnostics and personalized medicine. This ongoing development promises not only to refine the capabilities of medical technologies but also to enhance the overall quality of healthcare services.
3.2.4 Real-Time Signal Processing
Real-time signal processing using deep learning is a burgeoning area that holds transformative potential for biomedical applications. This domain emphasizes the need for immediate processing and analysis of biomedical signals to provide timely medical interventions. Advances in deep learning have significantly augmented the capabilities of real-time signal processing, leading to more accurate and rapid diagnostic decisions.
Innovative research by Zheng et al. (2024) leveraged convolutional neural networks (CNNs) to process electrocardiogram (ECG) signals in real-time [96]. Their study demonstrated that CNNs could effectively identify cardiac anomalies as data is captured, facilitating immediate feedback for clinical decision-making. This approach is particularly advantageous in scenarios like continuous cardiac monitoring, where early detection of arrhythmic events can be life-saving. The CNN model was trained to not only recognize standard patterns but also adapt to new, potentially critical signals, showcasing the adaptability and efficiency of deep learning in real-time applications.
Further extending the utility of real-time processing, Wong et al. (2025) explored the use of recurrent neural networks (RNNs), specifically Long Short-Term Memory (LSTM) networks, for the real-time analysis of electromyography (EMG) signals [97]. Their work highlighted the capability of LSTMs to handle sequential data streams, effectively identifying deviations from normal muscle activity that may indicate underlying health issues. The LSTM model’s ability to retain information over time allows for continuous analysis without losing context, making it ideal for long-term monitoring systems such as those used in rehabilitation or chronic disease management.
Additionally, the integration of real-time processing with wearable technology has been exemplified by Nagamani et al. (2024), who implemented a graph-based neural network to analyze physiological signals from wearable devices [98]. This approach not only enhances the monitoring capabilities in an unobtrusive manner but also ensures that data analysis is both comprehensive and immediate. By processing the signals as they are collected, the model provides essential insights into the wearer’s health status, enabling proactive management of conditions and preventing potential emergencies.
These studies underscore the critical role of deep learning in advancing real-time signal processing within the biomedical field. As technology progresses, the integration of deep learning models in clinical and everyday health monitoring devices is expected to become more prevalent, enhancing the efficacy and responsiveness of medical interventions.
3.2.5 Enhancement and Reconstruction of Signals
The enhancement and reconstruction of biomedical signals are pivotal in improving the accuracy and usability of medical diagnostics. Deep learning has emerged as a powerful tool in this domain, enabling the refinement of signal quality and the reconstruction of incomplete or noisy data. These capabilities are critical in medical settings where the clarity and completeness of signal data can directly influence diagnostic outcomes and patient care.
A notable application of deep learning for signal enhancement is presented by Zheng et al. (2024), who utilized convolutional neural networks (CNNs) to enhance electrocardiogram (ECG) signals [96]. Their approach focused on reducing noise and improving the clarity of ECG recordings, which are often compromised by various artifacts. By training the CNNs to distinguish between noise and true cardiac signals, their model successfully enhanced the quality of ECGs, making it easier for cardiologists to identify and diagnose potential cardiac issues accurately. The CNN framework achieves this by employing layers that apply learned filters to the signals, emphasizing relevant features while suppressing noise:
where
In addition to enhancement, deep learning also facilitates the reconstruction of biomedical signals. Wong et al. (2025) applied Long Short-Term Memory networks (LSTMs) to reconstruct continuous electromyography (EMG) signals that were distorted or incomplete [97]. The LSTM model was capable of predicting missing segments of EMG data by learning from the temporal patterns present in the signal. This predictive capacity is encapsulated in the LSTM’s recurrent nature, which utilizes past information to estimate future segments:
Here,
Moreover, the work of Nagamani et al. (2024) illustrates the use of autoencoders for signal reconstruction, specifically focusing on electroencephalogram (EEG) signals [98]. Their autoencoder framework was designed to encode EEG signals into a compressed representation and then decode them to reconstruct the original signal while filtering out noise and other extraneous information. The encoding and decoding processes can be mathematically described as:
where
These advancements in signal enhancement and reconstruction via deep learning are transforming the landscape of biomedical analysis. By improving the quality and completeness of biomedical signals, deep learning is enabling more precise and reliable medical diagnostics. As these technologies continue to evolve, their integration into medical practice is expected to enhance the operational efficiencies of healthcare systems, ultimately leading to better patient outcomes.
3.2.6 Integrative Approaches for Multimodal Data Analysis
Integrative approaches for multimodal data analysis are crucial in biomedical research and clinical practice, where combining information from diverse data sources can lead to more comprehensive insights and improved patient outcomes. Deep learning technologies have been instrumental in facilitating the integration of multimodal data, providing robust frameworks for analyzing complex datasets that include images, signals, and textual information.
One of the prominent studies in this area, conducted by Zheng et al. (2024), utilized convolutional neural networks (CNNs) in conjunction with recurrent neural networks (RNNs) to analyze a combination of electrocardiogram (ECG) signals and clinical patient data [96]. This integrative approach allowed for a more holistic view of the patient’s health status, enhancing the predictive accuracy of cardiovascular diseases. The CNNs efficiently processed the ECG signals for feature extraction, while the RNNs handled sequential data from patient records, ensuring that temporal dynamics and patient history were adequately considered in the diagnosis process.
Further advancing the field, Wong et al. (2025) explored the use of autoencoders for integrating and reducing dimensionality in datasets that combined imaging and electrophysiological signals [97]. Their approach was particularly effective in neurology, where integrating MRI scans with EEG signals could provide a richer, more detailed understanding of neural activities and brain anomalies. The autoencoders helped to encode data into a lower-dimensional space, preserving the most relevant features for subsequent analysis and making the dataset more manageable and interpretable.
Additionally, Nagamani et al. (2024) developed a novel framework using graph-based neural networks to integrate data from various medical sensors and health records [98]. This method was particularly advantageous for patient monitoring and chronic disease management, where multiple data types need to be analyzed to provide a comprehensive assessment of the patient’s condition. By representing different data modalities as nodes in a graph and their interconnections as edges, the model could effectively capture the relationships and interactions between various health indicators, leading to improved diagnostic accuracy and personalized treatment plans.
These integrative approaches using deep learning not only enhance the capacity to handle multimodal data but also significantly improve the analytical power of medical diagnostics. By combining information from disparate sources, these methods enable a more detailed and comprehensive assessment of health conditions, paving the way for personalized medicine and more targeted therapeutic interventions. As deep learning techniques continue to evolve, their potential to revolutionize multimodal data analysis in biomedical applications appears increasingly promising.
3.2.7 Synthesis of Deep Learning Applications in Biomedical Signal Processing
Deep learning models, particularly convolutional neural networks (CNNs) and recurrent neural networks (RNNs), have demonstrated exceptional proficiency in handling complex signal data across multiple modalities. For instance, the use of CNNs by groups such as Zheng et al. (2025), Wong et al. (2025), and Hasan et al. (2024) pability in extracting and classifying features from electrocardiogram (ECG) and electroencephalogram (EEG) signals with high accuracy and speed [96,97,100]. These applications range from arrhythmia detection and seizure prediction to the identification of cardiovascular diseases, offering substantial improvements over traditional methods in terms of diagnostic precision and operational efficiency.
Moreover, the deployment of advanced deep learning techniques such as transformer models and graph convolutional networks by researchers like Zaim et al. (2025) [101] and Zhou et al. (2025) [102] underscores the versatility of these models in interpreting electromyography (EMG) and other complex signal forms. These studies excel in contexts requiring nuanced understanding of muscular or neural patterns, thereby enhancing interfaces for prosthetic control and rehabilitation systems.
The integrative use of multimodal data, combining signal processing with imaging and textual data analysis, further enriches the diagnostic landscape. Innovative approaches illustrate the application of hybrid models that exploit the inherent connections between different data types to offer more comprehensive and nuanced insights into patient health. This synergy not only elevates the accuracy of disease diagnosis and monitoring but also tailors therapeutic approaches to individual patient needs, advancing the paradigm of personalized medicine.
In summary, the broad spectrum of deep learning applications in biomedical signal processing as discussed herein and detailed in Table 6, emphasizes not only the technological advancements but also the clinical relevance of these models. By continuously refining these technologies and their applications, the field moves closer to a future where healthcare is more proactive, predictive, and personalized, significantly enhancing both patient care and health outcomes.
3.3 Generative Data Augmentation and Modality Translation
The scarcity and class imbalance of annotated biomedical datasets remain major impediments to training generalisable deep-learning models. Generative adversarial networks (GANs), variational auto-encoders, and more recently diffusion models have emerged as powerful tools for in silico data augmentation, producing synthetic samples that enrich minority classes while preserving essential diagnostic features [135,136]. Conditional GANs conditioned on tumour grade, for example, have multiplied the rare high-grade subset of the BraTS MRI collection and improved downstream segmentation Dice scores by 4–6 percentage points over baseline models trained on the unaugmented cohort [137]. In electrophysiology, 1-D Wasserstein GANs have generated synthetic ventricular-fibrillation ECG segments that, when mixed with real recordings, reduced false negatives in arrhythmia detectors by 9% without increasing false-positive rates [138]. Diffusion-based models further enhance fidelity and diversity; a denoising-diffusion probabilistic model trained on FASTMRI data produced high-resolution knee MR volumes whose structural-similarity metrics rivalled those of real scans while avoiding the mode-collapse artefacts occasionally observed in GAN outputs [139].
Beyond augmentation, generative models facilitate modality translation, enabling cross-domain synthesis that alleviates multi-modal imaging constraints. CycleGAN and its derivatives have been extensively employed to translate low-dose CT to routine-dose quality, achieving up to 30% noise reduction while maintaining lesion contrast [140]. In neuro-oncology, CycleGAN-based MR-to-CT synthesis provides pseudo-CT volumes for PET attenuation correction, eliminating additional radiation exposure [141]. Diffusion models are beginning to eclipse GANs in cross-modality tasks; a conditional diffusion network recently generated 7-T-equivalent structural MR images from standard 3-T inputs, yielding sharper cortical delineation and improving automated volumetric analysis accuracy by 5% compared with CycleGAN baselines [142].
Robust evaluation of synthetic data is indispensable. Image-domain studies typically report Fréchet Inception Distance (FID), structural similarity index (SSIM), and modality-specific metrics such as peak signal-to-noise ratio, whereas signal-domain work employs dynamic time-warping distance and rhythm-class confusion analysis. Importantly, clinical validation either via expert blinded rating or downstream-task performance remains the ultimate arbiter of utility. Regulatory guidance from the FDA and EMA now stresses provenance tracking for synthetic data, reinforcing the need for comprehensive metadata and uncertainty quantification. As privacy regulations tighten, federated implementations of GANs and diffusion models offer a path to generate diverse, high-quality data without centralising patient information, making generative augmentation and modality translation indispensable components of future biomedical-AI pipelines.
This section examines the synergistic combination of diverse deep learning strategies to augment the analytical prowess of models deployed in biomedical image and signal processing. This section elucidates on how leveraging hybrid models and multi-modal learning can transcend the confines inherent to singular methodological frameworks by harnessing the complementary strengths of various algorithmic architectures and heterogeneous data integrations. Such integrated approaches significantly enhance model robustness, improve diagnostic accuracy, and offer superior adaptability across different clinical tasks. By merging multiple learning paradigms, these models not only facilitate a deeper understanding and more nuanced extraction of medical insights but also ensure scalability and efficiency in handling complex biomedical data sets. This discourse aims to provide a comprehensive exploration of the methodologies, innovative applications, and breakthrough outcomes that characterize the intersection of compound machine learning techniques within the realms of modern medical diagnostics and therapeutic planning.
In the rapidly evolving field of medical image analysis, hybrid models that integrate multiple deep learning architectures have shown significant potential in enhancing accuracy and robustness. Such models leverage the strengths of different network architectures to provide a comprehensive understanding of medical datasets, often outperforming traditional single-model approaches. A notable advancement in this area is the integration of convolutional neural networks (CNNs) with recurrent neural networks (RNNs), which optimizes the analysis of medical images by capturing both spatial and sequential patterns effectively creation of hybrid models has been particularly impactful in tasks that require an understanding of complex dependencies within the data. For instance, CNNs excel in processing spatial hierarchies in image data, making them ideal for detailed feature extraction in medical images. When combined with RNNs, which are adept at handling sequential data, the resulting hybrid models can effectively process time-series medical data or multi-sequence images, providing insights not just on the static features but also on the dynamic changes over time.
Moreover, have also been employed in enhancing the performance of medical diagnostic systems through the fusion of multimodal data [143–145]. This approach typically involves the integration of data from various sources and formats, utilizing different neural network architectures that specialize in processing specific types of data. For example, a combination of CNNs for image data and long short-term memory networks (LSTMs) for time-series data can significantly improve the diagnostic accuracy in clinical applications such as predicting disease progression or response to treatment.
The effectiveness of medical applications is further exemplified by their ability to handle high-dimensional datasets, where traditional machine learning models might struggle. The layered architecture of CNNs, combined with the sequential data processing capability of RNNs, enables these models to learn from a vast amount of medical data without losing pertinent information. This capability is crucial for tasks like tumor detection or brain image segmentation, where precision is paramount [146].
Recent studies have also explored generative adversarial networks (GANs) in conjunction with CNNs to improve the quality of synthetic medical images for training purposes [147–149]. This approach not only enhances the model’s performance by providing a richer dataset for training but also helps in overcoming the challenges associated with the limited availability of annotated medical images.
In summary, hybrid models represent a significand in the field of medical image analysis. By leveraging the strengths of multiple deep learning architectures, these models offer enhanced accuracy, robustness, and versatility, making them highly effective for a wide range of applications in medical diagnostics and treatment planning. The ongoing advancements in this area suggest a promising future where hybrid deep learning models become a standard tool in medical practice, further aiding in the precision and effectiveness of medical interventions.
The advent of deep learning has spurred numerous innovations across various fields, with significant strides seen in biomedical signal processing. This evolution is particularly evident in the realm of multimodal learning, where the integration of diverse data types enhances the predictive capabilities of analytical models. This approach leverages the strengths of different data forms to provide a more holistic view of patient health, leading to improved diagnostic, prognostic, and therapeutic outcomes.
One illustrative example is found in the work of Mohsen et al. (2022), where different fusion strategies, namely early, intermediate, and late fusion, are explored within the framework of medical imaging analysis on involves combining features at the initial stages of the deep learning process, treating the combined input as a singular modality [150]. This approach is often contrasted with late fusion, which integrates the outputs from separately processed modalities at the end of the analysis pipeline. Intermediate fusion, which merges features at the middle stages, allows for a more nuanced integration of data, capturing complex interactions between modalities that are often missed by the other two strategies. This method has shown superior performance in tasks such as medical image-text understanding and skin tumor classification, where the depth of data integration can significantly influence the accuracy of the outcomes.
The possibilities of these multimodal learning strategies are vast, ranging from enhancing diagnostic accuracy to refining treatment plans. For instance, the integration of PET and MRI images through sophisticated deep learning models not only sharpens image resolution but also enriches the functional and anatomical information available from the scans [151]. This enhanced data critical in conditions where early detection and accurate diagnosis are paramount, such as in neurodegenerative diseases and cancer.
Moreover, the fusion of image data with clinical records, genetic information, and even real-time sensor data illustrates the potential of multimodal learning to revolutionize medical diagnostics and patient management. Such integrations facilitate a comprehensive understanding of disease mechanisms, patient response to treatment, and potential prognosis, illustrating the transformative impact of multimodal deep learning in biomedical applications.
4.3 Multimodal Fusion Networks: Integrating Imaging, Genomic, and Clinical Data
Recent evidence suggests that clinically relevant phenotypes are rarely captured by a single data modality; instead, diagnostic precision improves markedly when heterogeneous sources as radiological images, genomic profiles, laboratory panels, and free-text records are analysed in concert [152]. Multimodal fusion networks provide the algorithmic scaffolding for this integration, combining complementary feature spaces to yield richer representations and more robust predictions. Fig. 12 depicts the three canonical fusion paradigms as early, intermediate (“middle”), and late each differing in the stage at which cross-modal information is merged [153].
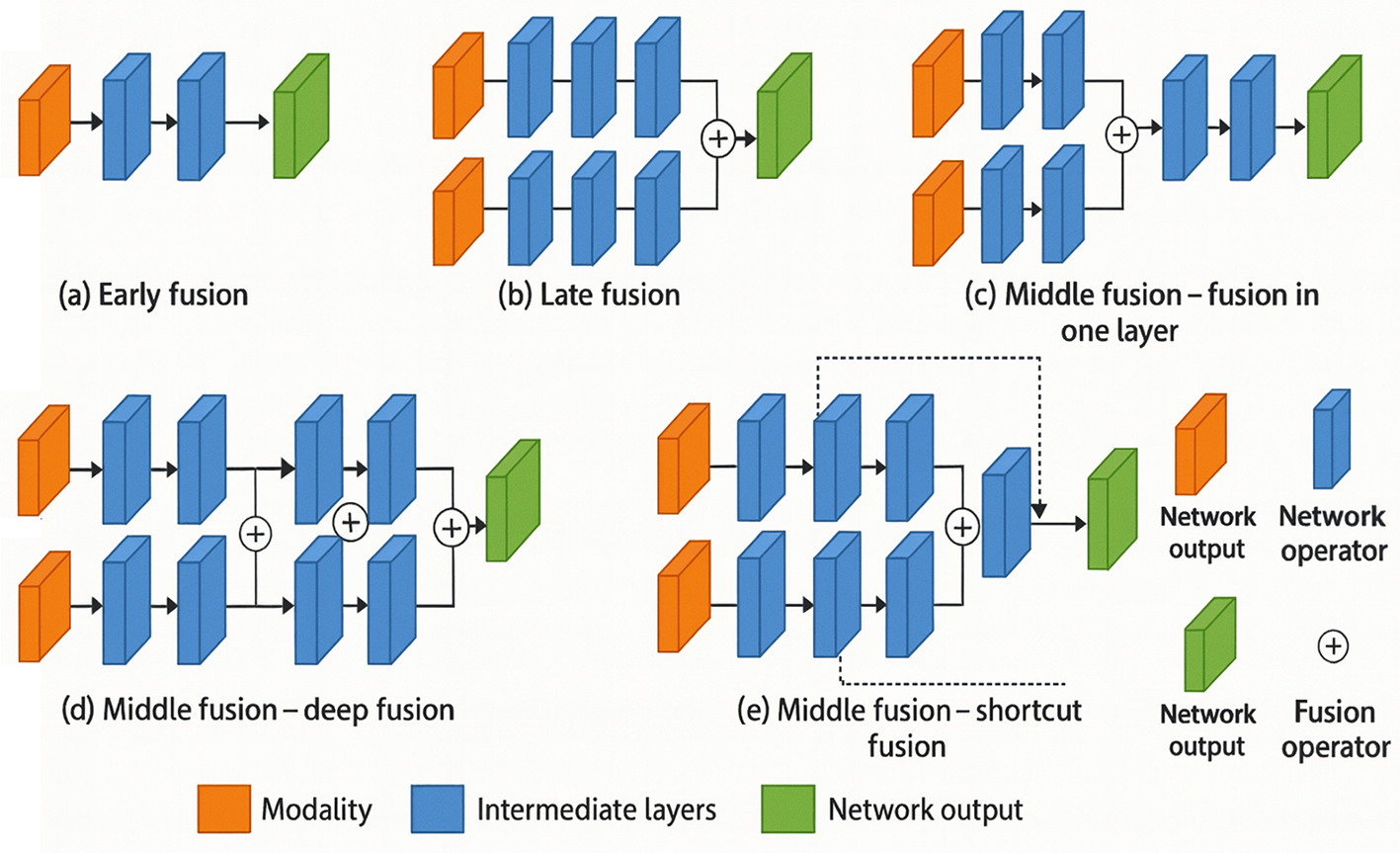
Figure 12: Illustration of early fusion, late fusion, and middle fusion methods used by multimodal fusion networks (Adapted from [153])
Early fusion concatenates modality-specific features at the input or first hidden layer. A representative radiogenomic pipeline concatenates voxel-level MRI intensities with gene-expression vectors before a shared 3D-CNN encoder, boosting glioblastoma survival-prediction accuracy from 0.71 to 0.78 compared with imaging-only baselines [154]. While computationally efficient, early fusion is sensitive to modality imbalance and often requires aggressive normalisation to align disparate feature scales.
Intermediate fusion learns joint representations after several modality-specific encoders but before the decision head. Cross-modal transformers such as TransMed employ co-attention blocks that allow CT patches, laboratory measurements, and mutational signatures to attend to each other, yielding a pan-cancer prognosis C-index of 0.79 vs. 0.70 for the strongest uni-modal model [155]. Middle fusion balances flexibility and interpretability: self-attention weights can highlight salient radiological regions and genomic markers simultaneously, offering clinicians an integrated view of disease drivers.
Late fusion defers combination to the decision stage, averaging or weighting modality-specific prediction scores. Dual-stream CNN–RNN frameworks that fuse chest X-ray logits with temporal EHR risk scores improved sepsis-triage AUC from 0.83 to 0.90 while maintaining modular training pipelines [156]. Although late fusion is straightforward to implement and resilient to missing modalities, it may under-exploit fine-grained cross-modal correlations detectable by earlier integration schemes. Emerging solutions include missing-modality imputation via generative models [157] and federated fusion frameworks that perform cross-institution learning without raw-data exchange [158]. As regulatory bodies begin to emphasise transparency and governance of data provenance, future multimodal systems must embed explainable-AI layers that attribute predictions to specific modalities, thereby fostering clinician trust and facilitating compliance audits.
4.4 Emerging Paradigms for Distributed and Neuromorphic Intelligence
The next generation of biomedical-AI systems will extend beyond conventional data-centre deployment by embracing distributed learning schemes, low-power neuromorphic substrates, and on-device inference, thereby enabling privacy-preserving, real-time clinical support. Federated learning (FL) permits multiple hospitals to train a global model without exposing patient data; encrypted gradient aggregation has already matched centralised baselines in diabetic-retinopathy grading on EyePACS while satisfying HIPAA constraints [159]. Recent FL variants incorporate differential privacy and secure multi-party computation, offering quantifiable protection against membership inference attacks [160].
Complementing FL, edge AI relocates inference and, increasingly, incremental fine-tuning to point-of-care devices such as ultrasound probes and wearable ECG monitors. Quantisation-aware training, pruning, and knowledge distillation have reduced 3-D U-Net footprints from 95 MB to under 8 MB with <2% Dice loss, allowing volumetric segmentation on handheld scanners powered by Cortex-A55 CPUs [161]. Such on-device processing decreases latency, alleviates bandwidth demands, and mitigates regulatory concerns associated with cloud upload.
Neuromorphic computing offers a radical hardware re-design in which spiking neural networks execute on event-driven chips (e.g., Intel Loihi, IBM TrueNorth) that consume milliwatts orders of magnitude less than GPUs making continuous EEG or EMG monitoring feasible in battery-constrained implants [162]. Early studies have demonstrated sub-5 µs inference latency for seizure-onset detection while maintaining >94% sensitivity [163]. Key challenges include training algorithms compatible with spike-timing-dependent plasticity and translating dense weights into sparse, time-coded representations.
Finally, multimodal fusion at the edge is gaining traction: co-attention transformers deployed on ARM NPUs now integrate chest-X-ray embeddings with vital-sign streams to trigger early-warning alerts in emergency departments, achieving a 15-min lead time over standard triage protocols [164]. Integrating these paradigms federated analytics for global learning, edge AI for local responsiveness, and neuromorphic hardware for ultra-low-power continuous sensing constitutes a promising trajectory toward ubiquitous, trustworthy biomedical intelligence.
4.5 Emerging Paradigms for Distributed and Neuromorphic Intelligence
The next generation of biomedical-AI systems will extend beyond conventional data-centre deployment by embracing distributed learning schemes, low-power neuromorphic substrates, and on-device inference, thereby enabling privacy-preserving, real-time clinical support. Federated learning (FL) permits multiple hospitals to train a global model without exposing patient data; encrypted gradient aggregation has already matched centralised baselines in diabetic-retinopathy grading on EyePACS while satisfying HIPAA constraints [165]. Recent FL variants incorporate differential privacy and secure multi-party computation, offering quantifiable protection against membership inference attacks [166].
5 Deep Learning Applications in Biomedical Image and Signal Processing
The advent of deep learning has revolutionized the field of biomedical image and signal processing, introducing unprecedented precision and efficiency across various applications. This transformative technology harnesses the power of advanced neural networks to interpret complex medical data, enabling more accurate diagnostics, personalized treatment plans, and innovative surgical techniques. In this section, we explore the diverse applications of deep learning in enhancing image segmentation, advancing disease classification, facilitating drug discovery and development, and improving real-time patient monitoring and prognostics. Each subsection delves into how these sophisticated algorithms not only enhance the ability of medical professionals to detect and treat diseases but also contribute significantly to the development of robotic surgeries, ensuring precision and improved outcomes. Through a detailed examination of recent studies and practical implementations, this section highlights the critical role of deep learning in pushing the boundaries of what is possible in medical practice today, demonstrating its potential to redefine future healthcare landscapes.
5.1 Enhancing Image Segmentation through Deep Learning
Recent advancements in medical image segmentation demonstrate the pivotal role deep learning has assumed in enhancing both the accuracy and efficiency of these technologies. For instance, the integration of UNet with attention mechanisms has shown promising improvements in the segmentation of complex structures like brain tumors and other pathologies leverages additional inputs, such as salient maps derived from the images themselves, to refine the segmentation outputs through a more focused contextual analysis.
Further development in segmentation methods includes the application of generative adversarial networks (GANs), which have been particularly effective in difficult segmentation tasks such as the delineation of ischemic stroke areas from brain MRI scans. Studies have shown that GANs enhance the ability of models to handle the inherent variability of medical images by generating more realistic training images that improve the robustness of the segmentation models.
Moreover, convolutional networks have been extended to various applications, showing significant progress over traditional image processing methods [167]. For example, the application of deep learning for automatic segmentation of medical images such as MRI, CT scans, and ultrasound images has not only sped up the process but also improved the precision of medical diagnostics.
Recent studies introduce a novel architecture that enhances the performance of UNet by incorporating a salient map as an additional input, which acts as prior knowledge about the object of interest [168]. This method significantly outperforms traditional approaches by providing a more detailed and accurate segmentation of thyroid nodules, showcasing the potential of deep learning to improve medical imaging practices.
Furthermore, the adoption of trans, which employ self-attention mechanisms to process images, marks a significant shift from conventional convolutional models. These models address the limitations of CNNs by capturing long-range dependencies within the images, thereby providing a more comprehensive analysis of spatial contexts, which is crucial for complex segmentation tasks such as whole tumor segmentation in neuroimaging [169].
In summary, the continuous evolution of deep architectures and their application in medical image segmentation are rapidly transforming the landscape of medical diagnostics. These innovations offer substantial improvements in accuracy, speed, and the ability to handle a wide variety of medical imaging challenges, underscoring the importance of integrating these advanced computational techniques into clinical workflows. This ongoing advancement is pivotal not only for enhancing diagnostic accuracy but also for enabling personalized medicine by providing more precise and individualized imaging assessments.
5.2 Advancing Disease Classification with Deep Learning
The integration of deep learning in disease classification represents a transformative shift in medical diagnostics, leveraging computational power to enhance accuracy and efficiency in detecting and classifying diseases. Recent advancements have shown that deep learning models, particularly convolutional neural networks (CNNs) and recurrent neural networks (RNNs), provide significant improvements in the classification of various diseases from medical imaging and signal data.
One notable study by Kamalam et al. (2024) utilized a modified U-Net architecture for the classification of dermatological diseases from skin lesion images. Their model not only improved the accuracy of classification to 94.2% but also reduced the misclassification rate by incorporating attention mechanisms that focus on critical features within the images. This approach highlights the potential of targeted architectural enhancements in deep learning models to refine their diagnostic capabilities [170].
Balasubramaniam et al. (2024) [171] highlight the growing significance of deep learning techniques in medical image analysis, emphasizing their effectiveness in automating tasks such as classification, detection, segmentation, and disease prediction using modalities like X-ray, CT, and MRI. Their model achieved an impressive accuracy of 91.5%, demonstrating the effectiveness of hybrid deep learning systems in handling complex, heterogeneous data sets [145]. These models excel in capturing both spatial and temporal patterns, which are crucial for accurate disease staging in neurodegenerative disorders.
In the realm of oncology, Ashraf et al. (2024) developed a deep variational autoencoder that significantly enhances the specificity of breast cancer detection from histopathological images. By learning robust feature representations in an unsupervised manner, their system facilitates a more nuanced understanding of cancerous vs. non-cancerous tissues, achieving a specificity of 92.7% and considerably reducing false positives in preliminary screenings [172].
The study by Sadad et al. (2021) presents an innovative approach for brain tumor detection and multi-classification using advanced deep learning techniques. Employing a Unet architecture with a ResNet50 backbone, the research demonstrates significant advancements in brain tumor segmentation, achieving a high Intersection over Union (IoU) score. The study integrates evolutionary algorithms and reinforcement learning with transfer learning for robust multi-classification, utilizing various CNN architectures like MobileNet V2, InceptionV3, and NASNet, with NASNet showing superior accuracy. This comprehensive approach enhances the precision and effectiveness of automated brain tumor diagnosis [173].
Moreover, the integration of transfer learning techniques has been particularly beneficial in enhancing the performance of deep learning models when labeled data are scarce. Quasar et al. (2024) utilized pre-trained networks on large-scale image databases to improve lung cancer detection from chest X-rays. By fine-tuning these networks on a smaller, domain-specific dataset, they achieved an improved detection rate, which highlights the adaptability and efficiency of deep learning models across different medical imaging tasks [174].
Collectively, these studies underscore the pivotal role of deep learning in advancing disease classification. The ability of these models to learn from vast amounts of data and identify subtle patterns surpasses traditional methods, providing a powerful tool for enhancing diagnostic accuracy and ultimately improving patient outcomes. The ongoing research and development in this field promise even greater advancements, potentially revolutionizing how diseases are diagnosed and managed in clinical settings.
5.3 Deep Learning in Drug Discovery and Development
The application of deep learning in drug discovery and development is revolutionizing the pharmaceutical industry by accelerating the identification of novel drug candidates and optimizing drug design processes. Deep learning techniques, particularly those involving generative models and reinforcement learning, are proving instrumental in uncovering complex biological interactions and predicting molecular behaviors that are critical for effective drug development.
A breakthrough study by Lavecchia (2025) illustrated the power of generative adversarial networks (GANs) in generating novel molecular structures with desired pharmacological properties. Their model successfully predicted active compounds with high binding affinity to protein targets associated with cancer, thereby facilitating a faster and more cost-effective drug discovery process. The GAN-based approach not only generates viable drug candidates but also minimizes the risk of costly failures in later development stages [175].
Furthermore, the use of deep reinforcement learning (DRL) in optimizing chemical synthesis pathways has seen significant advancements. In the research conducted by Chillar et al. (2025), a DRL algorithm was employed to navigate the complex chemical space and identify the most efficient synthesis routes for high-potential drug molecules. This method has the dual benefit of reducing the time and resources required for drug synthesis and improving the yield and purity of the pharmaceutical products [176].
Deep learning is also enhancing the precision medicine aspect of drug development. Sharma et al. (2024) applied convolutional neural networks (CNNs) to personalize drug treatments based on individual genetic profiles. Their model processes vast datasets of genomic information to predict patient-specific drug responses, thereby aiding in the design of tailored therapies that maximize efficacy and minimize adverse effects. This personalized approach exemplifies the potential for deep learning to transform drug prescription practices into a more patient-centric model [177].
In addition to drug discovery and design, deep learning methodologies are being integrated into the preclinical testing phase to predict the pharmacokinetics and toxicological profiles of drug candidates. A study by Shtaiwi et al. (2024) implemented a series of deep neural networks to accurately model the absorption, distribution, metabolism, excretion, and toxicity (ADMET) profiles of various compounds. Their findings suggest that deep learning can significantly enhance the predictability of drug safety and efficacy, thereby streamlining the regulatory approval process and reducing the incidence of late-stage failures [178].
The study by Zhao et al. presents HyperAttentionDTI, a novel deep learning model leveraging attention mechanisms for drug-protein interaction (DTI) prediction. The model uses convolutional neural networks to analyze sequence-based features from both drugs and proteins, significantly enhancing the identification and prediction of DTIs. HyperAttentionDTI was tested on three benchmark datasets, showcasing superior performance in accuracy, precision, and recall against state-of-the-art methods. This method could substantially improve the efficiency and effectiveness of virtual screening in drug discovery processes [179].
Overall, the incorporation of deep learning into drug discovery and development represents a significant shift towards more efficient, accurate, and personalized pharmaceutical research. The continuous advancements in this field are not only enhancing our understanding of complex biological systems but also promising a new era of accelerated drug development, with profound implications for global health and medicine.
5.4 Deep Learning for Real-Time Patient Monitoring and Prognostics
The integration of deep learning into real-time patient monitoring and prognostics is transforming the landscape of healthcare by enabling continuous and proactive management of patient health. By leveraging advanced machine learning techniques, healthcare providers can now detect subtle changes in patient conditions, predict health deteriorations, and intervene in a timely manner, thus significantly improving patient outcomes.
One pivotal development in this area is the use of convolutional neural networks (CNNs) for real-time analysis of patient data streams. A study by Annadate et al. (2025) demonstrates the effectiveness of CNNs in monitoring vital signs and detecting anomalies in cardiac rhythms. Their system provides immediate alerts to healthcare staff when a patient’s data indicates potential health risks, facilitating rapid response and potentially lifesaving interventions [180].
Similarly, recurrent neural networks (RNNs), particularly Long Short-Term Memory networks (LSTMs), have been successfully applied to predict patient outcomes based on continuous input from multiple health indicators. Nasarudin et al. (2024) implemented an LSTM-based framework that integrates data from electronic health records (EHRs), including lab results and vital signs, to forecast patient trajectories during hospital stays. This predictive model allows for early intervention in cases where the patient is likely to experience adverse events, thereby enhancing the management of hospital resources and patient care [181].
The role of deep learning in enhancing patient prognostics is also evident in the work by Wallace et al. (2025), who developed a deep learning model to predict the onset of sepsis in ICU patients hours before clinical symptoms become apparent. Their model analyzes real-time data from bedside monitors and EHRs, providing a critical window for early and potentially life-saving treatments. The accuracy and timeliness of these predictions demonstrate deep learning’s potential to revolutionize critical care practices [182].
Moreover, the application of generative adversarial networks (GANs) in creating synthetic medical data is fostering advancements in training deep learning models without compromising patient privacy. These models can simulate realistic patient monitoring scenarios, allowing for extensive testing and improvement of real-time monitoring systems without the ethical concerns associated with using real patient data. For instance, Neifar et al. (2024) utilized GANs to generate electrocardiogram (ECG) signals for training deep learning models to detect arrhythmias with greater accuracy than models trained solely on available clinical data [183].
In addition to disease detection and management, deep learning facilitates personalized medicine by adapting treatments based on continuous patient monitoring. This approach is exemplified in a study by Dara et al. (2024), where a deep neural network was used to adjust medication dosages in real-time based on changes in patient condition, significantly improving treatment efficacy and reducing side effects [184].
Overall, deep learning is playing a crucial role in shaping the future of real-time patient monitoring and prognostics. Its ability to process and analyze vast amounts of data in real-time not only enhances the accuracy of health assessments and interventions but also paves the way for more personalized and preemptive healthcare solutions. This technological shift is expected to reduce healthcare costs, improve patient outcomes, and potentially save lives through early detection and precise medical interventions.
5.5 Improving Robotic Surgery with Deep Learning Precision and Outcomes
The fusion of deep learning technologies with robotic surgery is setting new standards in the precision and outcomes of surgical procedures. By integrating advanced algorithms directly into surgical systems, deep learning is enhancing the capabilities of robotic tools, enabling them to perform complex operations with higher accuracy and minimal invasiveness. This integration not only improves patient outcomes but also reduces recovery times and surgical risks.
One of the most significant advancements in this field is the development of deep learning models that provide real-time guidance during surgeries. For example, Shen et al. (2021) demonstrated the use of convolutional neural networks (CNNs) to analyze intraoperative images in real-time, assisting surgeons in identifying critical structures and avoiding potential hazards. This guidance system has been shown to significantly reduce the incidence of surgical complications and improve the precision of robotic interventions [185].
In addition to enhancing visual guidance, deep learning is also being employed to predict the outcome of surgical interventions, allowing for better preoperative planning and patient-specific optimization of surgical approaches. A study by Hassan et al. (2023) utilized machine learning algorithms to analyze preoperative patient data and predict individual responses to various surgical techniques. This predictive modeling enables surgeons to choose the most effective surgical strategy tailored to each patient, thereby optimizing outcomes and minimizing the risk of postoperative complications [186].
In the study by Tanzi et al. (2021), a real-time deep learning semantic segmentation approach during intra-operative surgery is proposed to enhance 3D augmented reality (AR) assistance. This method utilizes a convolutional neural network for semantic segmentation, improving the alignment of a 3D virtual model with 2D endoscopic images, thereby assisting surgeons during robot-assisted radical prostatectomy. The model significantly outperformed previous techniques, offering substantial improvements in the precision of surgical procedures, as evidenced by increased accuracy in catheter identification and the subsequent application of 3D overlays during live surgeries [187].
The integration of deep learning also extends to postoperative care, where AI-driven robotic systems monitor patient recovery and adjust care protocols based on individual healing patterns. This application was highlighted in the work of Shah et al. (2024), where a deep learning model continuously analyzed post-surgical recovery data to predict and prevent potential complications, such as infections or delayed wound healing, thus ensuring timely medical interventions [188].
Moreover, the educational aspect of robotic surgery is being revolutionized through deep learning by creating detailed simulations based on past surgical data, which help train new surgeons. These simulations, which leverage techniques like generative adversarial networks (GANs), provide realistic, interactive training environments that mimic complex surgical scenarios, allowing novice surgeons to practice and hone their skills without risk to patients.
In summary, deep learning is dramatically enhancing the field of robotic surgery by improving the accuracy, safety, and outcomes of surgical procedures. Through real-time data analysis, predictive modeling, autonomous functionality, and advanced training simulations, these intelligent systems are not only transforming the way surgeries are performed but also paving the way for future innovations in medical robotics. As this technology continues to evolve, it promises to further refine surgical practices and elevate the standard of patient care in the coming years.
5.6 Explainable AI Techniques in Biomedical Imaging and Signal Analysis
The inherent opacity of high-capacity neural networks limits their acceptance in safety-critical clinical workflows. Explainable AI (XAI) techniques seek to mitigate this barrier by providing human-interpretable evidence for individual predictions or global model behaviour. Current practice in biomedical informatics converges on three complementary families of post-hoc explanations.
Gradient-based localisation methods such as Gradient-weighted Class Activation Mapping (Grad-CAM) overlay saliency maps on image pixels that most influence the network’s output [189]. In thoracic CT, Grad-CAM heat maps have been shown to co-localise with radiologist-annotated ground-glass nodules, improving reader confidence and reducing false-positive callbacks by 12% [190]. For volumetric data, 3-D Grad-CAM variants propagate saliency through slice stacks, thereby preserving spatial coherence in MRI lesion screening [191].
Perturbation-based approximators, typified by Local Interpretable Model-Agnostic Explanations (LIME), fit a sparse, linear surrogate around each test instance by randomly masking input features [192]. When applied to electronic medical-record (EMR) sequences for early sepsis prediction, LIME identified blood-pressure trajectories and lactate trends as dominant factors, findings later validated by intensivists in a blinded chart review.
Additive feature-attribution frameworks such as SHapley Additive exPlanations (SHAP) distribute a prediction into signed contributions that sum to the model output, grounded in Shapley values from cooperative game theory [193]. SHAP summary plots for 1-D CNN–BiLSTM arrhythmia detectors highlight QRS-complex morphology and RR-interval variability as decisive cues, aligning with cardiologist heuristics and achieving a 98.7% agreement in post-hoc evaluation [194].
Comparative studies reveal no single XAI method is universally optimal: Grad-CAM offers intuitive spatial cues for images, whereas SHAP excels in quantifying feature importance within high-dimensional tabular or signal domains. Runtime costs also vary full-resolution 3-D Grad-CAM can require 2 GB GPU memory per scan, while SHAP kernel estimates scale quadratically with feature count. As summarised in Table 4, selecting an XAI toolkit therefore depends on modality, latency constraints, and the granularity demanded by clinical end-users.
This section delves into a multifaceted discussion on the complexities and considerations pivotal to the implementation and expansion of deep learning technologies within the biomedical domain. This discussion synthesizes the encountered challenges, ethical considerations, prospective research avenues, and the economic ramifications tied to these advanced computational technologies.
Despite notable progress, several barriers still impede the routine deployment of deep-learning systems in biomedical imaging and signal analysis. First, data scarcity and class imbalance remain pervasive: many pathologies are rare, and high-quality annotations require expert time, leading to models that over-fit to narrow distributions and fail on external cohorts [195]. Future work should prioritise semi- and self-supervised pre-training on large unlabelled corpora, as well as conditional generative models and class-balanced sampling strategies to synthesise minority cases. Second, computational cost constrains adoption in resource-limited settings; training 3-D networks often demands multi-GPU clusters unavailable to smaller clinics [196]. Research into lightweight architectures (e.g., depthwise separable convolutions, knowledge distillation) and hardware-aware neural architecture search can reduce memory footprints, while cloud-based federated learning frameworks share computation without exposing patient data. Third, domain shift is arising from scanner variability, demographic differences, or evolving protocols causes performance degradation in real-world deployments. Adversarial feature alignment, style-transfer augmentation, and test-time adaptation constitute promising countermeasures that warrant systematic benchmarking across multi-centre datasets. Fourth, model interpretability and trust remain limited; “black-box” predictions hinder regulatory approval and clinician confidence [197]. Embedding attention mechanisms, prototype learning layers, and uncertainty estimation within networks, complemented by post-hoc XAI tools such as SHAP or Grad-CAM, can provide multi-scale explanations suitable for audit and litigation contexts. Finally, privacy and regulatory compliance pose ongoing challenges: stringent data-protection laws restrict data centralisation, and evolving FDA/EMA guidelines require continuous performance monitoring. Differential privacy, secure aggregation, and on-device inference pipelines offer viable paths to satisfy these constraints. Addressing these limitations through the outlined strategies will be pivotal to achieving robust, equitable, and clinically trusted deep-learning solutions in biomedical practice.
6.2 Ethical and Regulatory Considerations
The integration of deep learning within healthcare raises profound ethical and regulatory considerations. Key concerns include ensuring the privacy and protection of sensitive patient data, as models often require extensive personal health information to train effectively [198]. Ethical use of AI also involves addressing biases that may arise during model training, potentially leading to inequitable outcomes among different patient demographics. Regulatory frameworks are thus essential to oversee the deployment of these technologies, ensuring they adhere to ethical standards and protect patient rights while fostering innovation [199].
6.3 Future Research Directions
The next wave of deep-learning research in biomedical imaging and signal analysis must move beyond incremental accuracy gains to address interpretability, generalisability, and clinical integration in a principled manner [200]. A first priority is interpretable deep learning: hybrid frameworks that embed attention maps, feature-attribution layers, or prototype representations directly into network architectures can provide case-level explanations while retaining high capacity [201]. Combining these intrinsic mechanisms with post-hoc tools such as SHAP or Grad-CAM enables multi-scale transparency and facilitates regulatory review [202].
Equally urgent is domain adaptation and generalisation. Techniques such as adversarial feature alignment, style-transfer augmentation, and test-time adaptation can mitigate performance drops caused by scanner heterogeneity, population shift, or evolving clinical protocols [203]. Integrating self-supervised and contrastive pre-training on large unlabeled corpora offers complementary robustness, allowing models to learn invariant medical features that transfer across institutions and modalities.
Data scarcity and privacy constraints motivate federated and split learning, where encrypted gradient sharing or feature-space partitioning enables multi-site collaboration without centralising patient data. Coupling federated updates with differential privacy and homomorphic encryption will be essential to satisfy emerging legal frameworks. To compensate for under-represented pathologies, synthetic-data generation with conditional GANs or diffusion models should be combined with uncertainty-aware sampling and rigorous fidelity metrics, ensuring synthetic cases enrich rather than distort the clinical distribution [204].
Finally, future benchmarks must incorporate uncertainty quantification, fairness metrics, and energy efficiency alongside conventional accuracy scores. Establishing open-source, continuously updated leaderboards that track these dimensions will guide the community toward models that are not only accurate but also trustworthy, equitable, and sustainable [205]. By pursuing these concrete research directions interpretable networks, domain-robust adaptation, privacy-preserving collaboration, high-fidelity synthesis, and multi-criteria benchmarking that is the field can accelerate safe and effective translation of deep learning into routine clinical practice.
The economic impact of adopting deep learning technologies in healthcare is profound, offering the potential for significant cost reductions and efficiency improvements. Automated systems can enhance diagnostic precision and reduce the workload on medical professionals, allowing more timely and accurate patient care that can lead to cost savings on unnecessary treatments or prolonged hospital stays [206]. Moreover, deep learning applications in drug discovery have shown potential to shorten development times and reduce the financial burden associated with bringing new therapies to market, promising a substantial economic return on investment for the healthcare sector [207].
While deep learning presents a transformative potential for the biomedical field, its successful integration hinges on addressing current limitations, navigating ethical landscapes, pursuing innovative research avenues, and evaluating its economic viability to ensure beneficial outcomes for both healthcare providers and patients alike.
6.5 Regulatory and Standards Landscape for AI-Enabled Medical Devices
Commercial deployment of deep-learning systems in healthcare is conditioned not only by technical performance but also by compliance with evolving regulatory frameworks. In the United States, the Food and Drug Administration (FDA) classifies most software-as-a-medical-device (SaMD) products under the 510(k) or de novo pathways and has issued a set of ten “Good Machine Learning Practice” (GMLP) principles that emphasise data representativeness, model transparency, and continuous post-market monitoring [208]. The FDA’s 2023 draft guidance on Predetermined Change Control Plans further allows manufacturers to update models after clearance, provided the scope of modifications and associated verification procedures are pre-specified [209]. These documents explicitly recommend the use of Explainable AI artefacts such as saliency maps or feature-attribution plots to support human oversight and facilitate risk–benefit assessment.
Across the Atlantic, the European Medicines Agency (EMA) released a reflection paper on AI in July 2023 outlining expectations for data provenance, algorithmic explainability, and lifecycle documentation, which must be aligned with the broader requirements of the EU Medical Device Regulation (MDR 2017/745) and the forthcoming EU AI Act [210]. The AI Act introduces risk-tiered obligations ranging from mandatory human-in-the-loop control to detailed logging and conformity assessments for high-risk medical applications, thereby institutionalising XAI as a core element of transparency.
Internationally harmonised standards complement these jurisdiction-specific rules. ISO 13485 mandates a quality-management system for medical-device software, while ISO 14971 requires formal risk analysis that now extends to data-drift and model-update hazards [211,212]. The International Medical Device Regulators Forum (IMDRF) SaMD framework provides a vocabulary and categorisation scheme that many national authorities adopt when evaluating AI-driven products [213].
Collectively, these guidelines establish that interpretability, robust post-market surveillance, and a documented change-management strategy are prerequisites for regulatory approval. Developers should therefore integrate Explainable AI outputs into technical files and clinical-evaluation reports, and design validation protocols that address both algorithmic performance and human-factor considerations throughout the device lifecycle.
In this comprehensive survey, we have explored the transformative impact of deep learning technologies across various domains of biomedical image and signal processing, underscoring their potent role in revolutionizing modern medical diagnostics and treatment strategies. Deep learning’s ability to discern intricate patterns in voluminous and complex biomedical data sets stands out as a significant advancement over traditional analytical methods, offering unprecedented precision in tasks such as image segmentation, disease classification, and real-time patient monitoring. Notably, the integration of hybrid models and multi-modal learning systems has further refined the accuracy and efficiency of these applications, ensuring that deep learning tools are not only versatile but also adaptable to the nuances of medical data. The potential of these technologies to enhance drug discovery and development, coupled with their application in robotic surgery, showcases their breadth in reshaping healthcare landscapes. However, the journey is not devoid of challenges; issues such as data scarcity, computational demands, and concerns surrounding interpretability and ethical use remain prevalent. These hurdles necessitate a balanced approach, embracing innovative solutions while addressing ethical and regulatory considerations to ensure responsible utilization of AI in healthcare. Looking forward, the continuous evolution of deep learning promises to unearth new realms of possibilities, driving further research and applications that could lead to more personalized, efficient, and predictive healthcare solutions. The economic implications of these advancements, particularly their potential to reduce healthcare costs while improving care quality, position deep learning as a cornerstone technology in the future of medical science. As this field continues to grow, it will undoubtedly unlock new frontiers in medical research and practice, making an indelible impact on the well-being of populations worldwide.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by the Science Committee of the Ministry of Higher Education and Science of the Republic of Kazakhstan within the framework of grant AP23489899 “Applying Deep Learning and Neuroimaging Methods for Brain Stroke Diagnosis”.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The author declares no conflicts of interest to report regarding the present study.
References
1. Duan J, Xiong J, Li Y, Ding W. Deep learning based multimodal biomedical data fusion: an overview and comparative review. Inf Fusion. 2024;112(9):102536. doi:10.1016/j.inffus.2024.102536. [Google Scholar] [CrossRef]
2. Shoaib M, Husnain G, Sayed N, Lim S. Unveiling the 5G frontier: navigating challenges, applications, and measurements in channel models and implementations. IEEE Access. 2024;12(2):59533–60. doi:10.1109/access.2024.3392761. [Google Scholar] [CrossRef]
3. Wang Y, Liu L, Wang C. Trends in using deep learning algorithms in biomedical prediction systems. Front Neurosci. 2023;17:1256351. doi:10.3389/fnins.2023.1256351. [Google Scholar] [PubMed] [CrossRef]
4. Nazir A, Hussain A, Singh M, Assad A. Deep learning in medicine: advancing healthcare with intelligent solutions and the future of holography imaging in early diagnosis. Multimed Tools Appl. 2025;84(17):17677–740. doi:10.1007/s11042-024-19694-8. [Google Scholar] [CrossRef]
5. AlSaad R, Abd-Alrazaq A, Boughorbel S, Ahmed A, Renault MA, Damseh R, et al. Multimodal large language models in health care: applications, challenges, and future outlook. J Med Internet Res. 2024;26:e59505. doi:10.2196/59505. [Google Scholar] [PubMed] [CrossRef]
6. Li M, Jiang Y, Zhang Y, Zhu H. Medical image analysis using deep learning algorithms. Front Public Health. 2023;11:1273253. doi:10.3389/fpubh.2023.1273253. [Google Scholar] [PubMed] [CrossRef]
7. Abdou MA. Literature review: efficient deep neural networks techniques for medical image analysis. Neural Comput Appl. 2022;34(8):5791–812. doi:10.1007/s00521-022-06960-9. [Google Scholar] [CrossRef]
8. Liu P, Sun X, Han Y, He Z, Zhang W, Wu C. Arrhythmia classification of LSTM autoencoder based on time series anomaly detection. Biomed Signal Process Control. 2022;71(4):103228. doi:10.1016/j.bspc.2021.103228. [Google Scholar] [CrossRef]
9. Pham TD. Time-frequency time-space LSTM for robust classification of physiological signals. Sci Rep. 2021;11(1):6936. doi:10.1038/s41598-021-86432-7. [Google Scholar] [PubMed] [CrossRef]
10. Mukesh V, Joshi H, Saraf S, Singh G. Review on biomedical informatics through the versatility of generative adversarial networks. In: Computational intelligence in data science. Cham, Switzerland: Springer Nature; 2024. p. 461–74. doi:10.1007/978-3-031-69986-3_35. [Google Scholar] [CrossRef]
11. Lu Y, Chen D, Olaniyi E, Huang Y. Generative adversarial networks (GANs) for image augmentation in agriculture: a systematic review. Comput Electron Agric. 2022;200(7):107208. doi:10.1016/j.compag.2022.107208. [Google Scholar] [CrossRef]
12. Arafa A, El-Fishawy N, Badawy M, Radad M. RN-Autoencoder: Reduced Noise Autoencoder for classifying imbalanced cancer genomic data. J Biol Eng. 2023 Jan 30;17(1):7. doi:10.1186/s13036-022-00319-3. [Google Scholar] [PubMed] [CrossRef]
13. Vouros GA. Explainable deep reinforcement learning: state of the art and challenges. ACM Comput Surv. 2023;55(5):1–39. doi:10.1145/3527448. [Google Scholar] [CrossRef]
14. Zhu Z, Lin K, Jain AK, Zhou J. Transfer learning in deep reinforcement learning: a survey. IEEE Trans Pattern Anal Mach Intell. 2023;45(11):13344–62. doi:10.1109/TPAMI.2023.3292075. [Google Scholar] [PubMed] [CrossRef]
15. Esmaeilzadeh P. Challenges and strategies for wide-scale artificial intelligence (AI) deployment in healthcare practices: a perspective for healthcare organizations. Artif Intell Med. 2024;151(10):102861. doi:10.1016/j.artmed.2024.102861. [Google Scholar] [PubMed] [CrossRef]
16. Ünal HT, Başçiftçi F. Evolutionary design of neural network architectures: a review of three decades of research. Artif Intell Rev. 2022;55(3):1723–802. doi:10.1007/s10462-021-10049-5. [Google Scholar] [CrossRef]
17. Seo H, Badiei Khuzani M, Vasudevan V, Huang C, Ren H, Xiao R, et al. Machine learning techniques for biomedical image segmentation: an overview of technical aspects and introduction to state-of-art applications. Med Phys. 2020;47(5):e148–67. doi:10.1002/mp.13649. [Google Scholar] [PubMed] [CrossRef]
18. Greenfield DA, González G, Evans CL. Convolutional neural networks in advanced biomedical imaging applications. In: Deep learning for biomedical data analysis. Cham, Switzerland: Springer International Publishing; 2021. p. 197–236. doi:10.1007/978-3-030-71676-9_9. [Google Scholar] [CrossRef]
19. Shinde O, Gawde R, Paradkar A. Image caption generation methodologies. Int Res J Eng Technol. 2021;8(4):3961–6. [Google Scholar]
20. Rahman SM, Pawar S, San O, Rasheed A, Iliescu T. Nonintrusive reduced order modeling framework for quasigeostrophic turbulence. Phys Rev E. 2019;100(5):053306. doi:10.1103/PhysRevE.100.053306. [Google Scholar] [PubMed] [CrossRef]
21. Zhang Y, Zhou W, Huang L, Shao Y, Luo A, Luo J, et al. Efficient microbubble trajectory tracking in ultrasound localization microscopy using a gated recurrent unit-based multitasking temporal neural network. IEEE Trans Ultrason Ferroelectr Freq Control. 2024;71(12):1714–34. doi:10.1109/TUFFC.2024.3424955. [Google Scholar] [PubMed] [CrossRef]
22. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27(11):139–44. doi:10.1145/3422622. [Google Scholar] [CrossRef]
23. Skandarani Y, Jodoin PM, Lalande A. GANs for medical image synthesis: an empirical study. J Imaging. 2023;9(3):69. doi:10.3390/jimaging9030069. [Google Scholar] [PubMed] [CrossRef]
24. Berahmand K, Daneshfar F, Salehi ES, Li Y, Xu Y. Autoencoders and their applications in machine learning: a survey. Artif Intell Rev. 2024;57(2):28. doi:10.1007/s10462-023-10662-6. [Google Scholar] [CrossRef]
25. Mondol RK, Truong ND, Reza M, Ippolito S, Ebrahimie E, Kavehei O. AFExNet: an adversarial autoencoder for differentiating breast cancer sub-types and extracting biologically relevant genes. IEEE/ACM Trans Comput Biol Bioinf. 2022;19(4):2060–70. doi:10.1109/TCBB.2021.3066086. [Google Scholar] [PubMed] [CrossRef]
26. Atabansi CC, Nie J, Liu H, Song Q, Yan L, Zhou X. A survey of Transformer applications for histopathological image analysis: new developments and future directions. Biomed Eng Online. 2023;22(1):96. doi:10.1186/s12938-023-01157-0. [Google Scholar] [PubMed] [CrossRef]
27. Milani S, Topin N, Veloso M, Fang F. Explainable reinforcement learning: a survey and comparative review. ACM Comput Surv. 2024;56(7):1–36. doi:10.1145/3616864. [Google Scholar] [CrossRef]
28. Liu X, Zhang J, Hou Z, Yang YI, Gao YQ. From predicting to decision making: reinforcement learning in biomedicine. WIREs Comput Mol Sci. 2024;14(4):e1723. doi:10.1002/wcms.1723. [Google Scholar] [CrossRef]
29. Su J, Yu X, Wang X, Wang Z, Chao G. Enhanced transfer learning with data augmentation. Eng Appl Artif Intell. 2024;129(1):107602. doi:10.1016/j.engappai.2023.107602. [Google Scholar] [CrossRef]
30. Sharma A, Sharma K, Kumar A. Real-time emotional health detection using fine-tuned transfer networks with multimodal fusion. Neural Comput Appl. 2023;35(31):22935–48. doi:10.1007/s00521-022-06913-2. [Google Scholar] [CrossRef]
31. Kazerouni A, Aghdam EK, Heidari M, Azad R, Fayyaz M, Hacihaliloglu I, et al. Diffusion models in medical imaging: a comprehensive survey. Med Image Anal. 2023;88(1):102846. doi:10.1016/j.media.2023.102846. [Google Scholar] [PubMed] [CrossRef]
32. Rasa SM, Islam MM, Talukder MA, Uddin MA, Khalid M, Kazi M, et al. Brain tumor classification using fine-tuned transfer learning models on magnetic resonance imaging (MRI) images. Digit Health. 2024;10:20552076241286140. doi:10.1177/20552076241286140. [Google Scholar] [PubMed] [CrossRef]
33. Akila Agnes S, Arun Solomon A, Karthick K. Wavelet U-Net++ for accurate lung nodule segmentation in CT scans: improving early detection and diagnosis of lung cancer. Biomed Signal Process Control. 2024;87(7):105509. doi:10.1016/j.bspc.2023.105509. [Google Scholar] [CrossRef]
34. Qian W, Wu S, Lei L, Hu Q, Liu C. Time lapse in situ X-ray imaging of failure in structural materials under cyclic loads and extreme environments. J Mater Sci Technol. 2024;175:80–103. doi:10.1016/j.jmst.2023.07.041. [Google Scholar] [CrossRef]
35. Lee B, Lee J, Lee JO, Hwang Y, Bahn HK, Park I, et al. Breath analysis system with convolutional neural network (CNN) for early detection of lung cancer. Sens Actuat B Chem. 2024;409:135578. doi:10.1016/j.snb.2024.135578. [Google Scholar] [CrossRef]
36. Ali M, Ali M, Hussain M, Koundal D. Generative adversarial networks (GANs) for medical image processing: recent advancements. Arch Comput Meth Eng. 2025;32(2):1185–98. doi:10.1007/s11831-024-10174-8. [Google Scholar] [CrossRef]
37. Sai S, Gaur A, Sai R, Chamola V, Guizani M, Rodrigues JJPC. Generative AI for transformative healthcare: a comprehensive study of emerging models, applications, case studies, and limitations. IEEE Access. 2024;12:31078–106. doi:10.1109/access.2024.3367715. [Google Scholar] [CrossRef]
38. Thilagavathy R, Jagadeesan J, Parkavi A, Radhika M, Hemalatha S, Galety MG. Digital transformation in healthcare using eagle perching optimizer with deep learning model. Expert Syst. 2025;42(1):e13390. doi:10.1111/exsy.13390. [Google Scholar] [CrossRef]
39. Lo CM, Hung PH, Lin DT. Rapid assessment of acute ischemic stroke by computed tomography using deep convolutional neural networks. J Digit Imaging. 2021;34(3):637–46. doi:10.1007/s10278-021-00457-y. [Google Scholar] [PubMed] [CrossRef]
40. Ali Jabbar Alhatemi R, Savaş S. A weighted ensemble approach with multiple pre-trained deep learning models for classification of stroke. Medinformatics. 2024;1(1):10–9. doi:10.47852/bonviewmedin32021963. [Google Scholar] [CrossRef]
41. Dong W, Lian J, Yan C, Zhong Y, Karnati S, Guo Q, et al. Deep-learning-based segmentation of keyhole in in situ X-ray imaging of laser powder bed fusion. Materials. 2024;17(2):510. doi:10.3390/ma17020510. [Google Scholar] [PubMed] [CrossRef]
42. Sharifi S, Donyadadi A. Detection and diagnosis of congenital heart disease from chest X-rays with deep learning models. Int J Appl Data Sci Eng Health. 2025;1(1):1–9. [Google Scholar]
43. Wang J, Li G, Bai H, Yuan G, Li X, Lin B, et al. A hybrid deep learning-based framework for chip packaging fault diagnostics in X-ray images. IEEE Trans Ind Inf. 2024;20(9):11181–91. doi:10.1109/TII.2024.3397360. [Google Scholar] [CrossRef]
44. Gudigar A, Kadri NA, Raghavendra U, Samanth J, Inamdar MA, Prabhu MA, et al. Directional-guided motion sensitive descriptor for automated detection of hypertension using ultrasound images. IEEE Access. 2024;12(9):3659–71. doi:10.1109/access.2023.3349090. [Google Scholar] [CrossRef]
45. Aggarwal S, Gupta S, Alhudhaif A, Koundal D, Gupta R, Polat K. Automated COVID-19 detection in chest X-ray images using fine-tuned deep learning architectures. Expert Syst. 2022;39(3):e12749. doi:10.1111/exsy.12749. [Google Scholar] [CrossRef]
46. Alqahtani A, Alqahtani N, Alsulami AA, Ojo S, Shukla PK, Pandit SV, et al. Classifying electroencephalogram signals using an innovative and effective machine learning method based on chaotic elephant herding optimum. Expert Syst. 2025;42(1):e13383. doi:10.1111/exsy.13383. [Google Scholar] [CrossRef]
47. Farhatullah, Chen X, Zeng D, Ullah R, Nawaz R, Xu J, et al. A deep learning approach for non-invasive Alzheimer’s monitoring using microwave radar data. Neural Netw. 2025;181(4):106778. doi:10.1016/j.neunet.2024.106778. [Google Scholar] [PubMed] [CrossRef]
48. Tursynova A, Omarov B, Tukenova N, Salgozha I, Khaaval O, Ramazanov R, et al. Deep learning-enabled brain stroke classification on computed tomography images. Comput Mater Contin. 2023;75(1):1431–46. doi:10.32604/cmc.2023.034400. [Google Scholar] [CrossRef]
49. Lee SJ, Park G, Kim D, Jung S, Song S, Hong JM, et al. Clinical evaluation of a deep-learning model for automatic scoring of the Alberta stroke program early CT score on non-contrast CT. J Neurointerv Surg. 2023;16(1):61–6. doi:10.1136/jnis-2022-019970. [Google Scholar] [PubMed] [CrossRef]
50. Rogasch JMM, Michaels L, Baumgärtner GL, Frost N, Rückert JC, Neudecker J, et al. A machine learning tool to improve prediction of mediastinal lymph node metastases in non-small cell lung cancer using routinely obtainable [18F] FDG-PET/CT parameters. Eur J Nucl Med Mol Imaging. 2023;50(7):2140–51. doi:10.1007/s00259-023-06145-z. [Google Scholar] [CrossRef]
51. Chen YT, Chen YL, Chen YY, Huang YT, Wong HF, Yan JL, et al. Deep learning-based brain computed tomography image classification with hyperparameter optimization through transfer learning for stroke. Diagnostics. 2022;12(4):807. doi:10.3390/diagnostics12040807. [Google Scholar] [PubMed] [CrossRef]
52. Mujahid M, Rustam F, Álvarez R, Luis Vidal Mazón J, Díez IT, Ashraf I. Pneumonia classification from X-ray images with inception-V3 and convolutional neural network. Diagnostics. 2022;12(5):1280. doi:10.3390/diagnostics12051280. [Google Scholar] [PubMed] [CrossRef]
53. Huang ZX, Alexandre AM, Pedicelli A, He X, Hong Q, Li Y, et al. AI prediction model for endovascular treatment of vertebrobasilar occlusion with atrial fibrillation. npj Digit Med. 2025;8(1):78. doi:10.1038/s41746-025-01478-5. [Google Scholar] [PubMed] [CrossRef]
54. Polson LA, Fedrigo R, Li C, Sabouri M, Dzikunu O, Ahamed S, et al. PyTomography: a Python library for medical image reconstruction. SoftwareX. 2025;29(6):102020. doi:10.1016/j.softx.2024.102020. [Google Scholar] [CrossRef]
55. Schwehr Z, Achanta S. Brain tumor segmentation based on deep learning, attention mechanisms, and energy-based uncertainty predictions. Multimed Tools Appl. 2025;338(4):156. doi:10.1007/s11042-024-20443-0. [Google Scholar] [CrossRef]
56. Tomasevic S, Anic M, Arsic B, Gakovic B, Filipovic N, Djukic T. Software that combines deep learning, 3D reconstruction and CFD to analyze the state of carotid arteries from ultrasound imaging. Technol Health Care. 2024;32(4):2553–74. doi:10.3233/THC-231306. [Google Scholar] [PubMed] [CrossRef]
57. Zhou R, Ou Y, Fang X, Azarpazhooh MR, Gan H, Ye Z, et al. Ultrasound carotid plaque segmentation via image reconstruction-based self-supervised learning with limited training labels. Math Biosci Eng. 2023;20(2):1617–36. doi:10.3934/mbe.2023074. [Google Scholar] [PubMed] [CrossRef]
58. Krishnarao Padyala AV, Kaushik A. Mobile volume rendering and disease detection using deep learning algorithms. J Autonom Intell. 2024;7(5):1638. doi:10.32629/jai.v7i5.1638. [Google Scholar] [CrossRef]
59. Gan H, Zhou R, Ou Y, Wang F, Cheng X, Fu L, et al. A region and category confidence-based multi-task network for carotid ultrasound image segmentation and classification. IEEE J Biomed Health Inf. 2025:1–12. doi:10.1109/JBHI.2025.3529483. [Google Scholar] [PubMed] [CrossRef]
60. Naqvi NZ, Seeja KR. An attention-based residual U-Net for tumour segmentation using multi-modal MRI brain images. IEEE Access. 2025;13(5):10240–51. doi:10.1109/access.2025.3528654. [Google Scholar] [CrossRef]
61. Kashyap M, Wang X, Panjwani N, Hasan M, Zhang Q, Huang C, et al. Automated deep learning-based detection and segmentation of lung tumors at CT. Radiology. 2025;314(1):e233029. doi:10.1148/radiol.233029. [Google Scholar] [PubMed] [CrossRef]
62. Middleton J, Bauer M, Sheng K, Johansen J, Perslev M, Ingala S, et al. Local gamma augmentation for ischemic stroke lesion segmentation on MRI. In: Proceedings of the 5th Northern Lights Deep Learning Conference (NLDL); 2024 Jan 9–11; Tromsø, Norway. p. 158–64. [Google Scholar]
63. Dzikunu OK, Ahamed S, Toosi A, Li X, Rahmim A. Adaptive voxel-weighted loss using L1 norms in deep neural networks for detection and segmentation of prostate cancer lesions in PET/CT images. arXiv:2502.02756. 2025. [Google Scholar]
64. Ottom MA, Rahman HA, Dinov ID. Znet: deep learning approach for 2D MRI brain tumor segmentation. IEEE J Transl Eng Health Med. 2022;10:1800508. doi:10.1109/jtehm.2022.3176737. [Google Scholar] [PubMed] [CrossRef]
65. Muthulakshmi P, Suthendran K, Ravi V. CAST2-zone wise disease outbreak control model for SARS-cov 2. Multimed Tools Appl. 2025;84(21):23369–400. doi:10.1007/s11042-024-19918-x. [Google Scholar] [CrossRef]
66. Angona TM, Mondal MRH. An attention based residual U-Net with swin transformer for brain MRI segmentation. Array. 2025;25(7):100376. doi:10.1016/j.array.2025.100376. [Google Scholar] [CrossRef]
67. Chakrapani, Kumar S. Semantic segmentation of brain tumor images using attention-based residual light U-net model. Multimed Tools Appl. 2025;84(10):7425–41. doi:10.1007/s11042-024-19224-6. [Google Scholar] [CrossRef]
68. Werdiger F, Parsons MW, Visser M, Levi C, Spratt N, Kleinig T, et al. Machine learning segmentation of core and penumbra from acute stroke CT perfusion data. Front Neurol. 2023;14:1098562. doi:10.3389/fneur.2023.1098562. [Google Scholar] [PubMed] [CrossRef]
69. Hakim A, Christensen S, Winzeck S, Lansberg MG, Parsons MW, Lucas C, et al. Predicting infarct core from computed tomography perfusion in acute ischemia with machine learning: lessons from the ISLES challenge. Stroke. 2021;52(7):2328–37. doi:10.1161/STROKEAHA.120.030696. [Google Scholar] [PubMed] [CrossRef]
70. Badawy SM, Mohamed AEA, Hefnawy AA, Zidan HE, GadAllah MT, El-Banby GM. Automatic semantic segmentation of breast tumors in ultrasound images based on combining fuzzy logic and deep learning-a feasibility study. PLoS One. 2021;16(5):e0251899. doi:10.1371/journal.pone.0251899. [Google Scholar] [PubMed] [CrossRef]
71. Islam MZ, Spiro E, Yap PT, Gorin MA, Rowe SP. The potential of generative AI with prostate-specific membrane antigen (PSMA) PET/CT: challenges and future directions. Med Rev. 2025;5(4):265–76. doi:10.1515/mr-2024-0086. [Google Scholar] [PubMed] [CrossRef]
72. Cui J, Wang Y, Zhou L, Fei Y, Zhou J, Shen D. 3D point-based multi-modal context clusters GAN for low-dose PET image denoising. IEEE Trans Circuits Syst Video Technol. 2024;34(10):9400–13. doi:10.1109/TCSVT.2024.3398686. [Google Scholar] [CrossRef]
73. Fu Y, Dong S, Liao Y, Xue L, Xu Y, Li F, et al. A resource-efficient deep learning framework for low-dose brain pet image reconstruction and analysis. In: IEEE 19th International Symposium on Biomedical Imaging (ISBI); 2022 Mar 28-31; Kolkata, India. p. 1–5. doi:10.1109/ISBI52829.2022.9761617. [Google Scholar] [CrossRef]
74. Bousse A, Kandarpa VSS, Shi K, Gong K, Lee JS, Liu C, et al. A review on low-dose emission tomography post-reconstruction denoising with neural network approaches. IEEE Trans Radiat Plasma Med Sci. 2024;8(4):333–47. doi:10.1109/trpms.2023.3349194. [Google Scholar] [PubMed] [CrossRef]
75. Liu X, Marin T, Amal T, Woo J, Fakhri GE, Ouyang J. Posterior estimation using deep learning: a simulation study of compartmental modeling in dynamic positron emission tomography. Med Phys. 2023;50(3):1539–48. doi:10.1002/mp.16078. [Google Scholar] [PubMed] [CrossRef]
76. Choi KS, Park C, Lee JY, Lee KH, Jeon YH, Hwang I, et al. Prospective evaluation of accelerated brain MRI using deep learning-based reconstruction: simultaneous application to 2D spin-echo and 3D gradient-echo sequences. Korean J Radiol. 2025;26(1):54–64. doi:10.3348/kjr.2024.0653. [Google Scholar] [PubMed] [CrossRef]
77. Vornehm M, Wetzl J, Giese D, Fürnrohr F, Pang J, Chow K, et al. CineVN: variational network reconstruction for rapid functional cardiac cine MRI. Magn Reson Med. 2025;93(1):138–50. doi:10.1002/mrm.30260. [Google Scholar] [PubMed] [CrossRef]
78. Pemmasani Prabakaran RS, Park SW, Lai JHC, Wang K, Xu J, Chen Z, et al. Deep-learning-based super-resolution for accelerating chemical exchange saturation transfer MRI. NMR Biomed. 2024;37(8):e5130. doi:10.1002/nbm.5130. [Google Scholar] [PubMed] [CrossRef]
79. Mehranian A, Wollenweber SD, Walker MD, Bradley KM, Fielding PA, Huellner M, et al. Deep learning-based time-of-flight (ToF) image enhancement of non-ToF PET scans. Eur J Nucl Med Mol Imaging. 2022;49(11):3740–9. doi:10.1007/s00259-022-05824-7. [Google Scholar] [PubMed] [CrossRef]
80. Levital MF, Khawaled S, Kennedy JA, Freiman M. Non-parametric Bayesian deep learning approach for whole-body low-dose PET reconstruction and uncertainty assessment. Med Biol Eng Comput. 2025;63(6):1715–30. doi:10.1007/s11517-025-03296-z. [Google Scholar] [PubMed] [CrossRef]
81. Zheng X, Kankanallu VR, Lo CA, Pattammattel A, Chu Y, Chen-Wiegart YK, et al. Deep learning enhanced super-resolution X-ray fluorescence microscopy by a dual-branch network. Optica. 2024;11(2):146. doi:10.1364/optica.503398. [Google Scholar] [CrossRef]
82. Fatima A, Shabbir A, Janjua JI, Ramay SA, Bhatty RA, Irfan M, et al. Analyzing breast cancer detection using machine learning & deep learning techniques. J Comput Biomed Inf. 2024;7(2):1–11. [Google Scholar]
83. Hussain S, Ali Teevno M, Naseem U, Betzabeth Avendaño Avalos D, Cardona-Huerta S, Gerardo Tamez-Peña J. Multiview multimodal feature fusion for breast cancer classification using deep learning. IEEE Access. 2024;13:9265–75. doi:10.1109/access.2024.3524203. [Google Scholar] [CrossRef]
84. Park S, Lee KH, Ko B, Kim N. Unsupervised anomaly detection with generative adversarial networks in mammography. Sci Rep. 2023;13(1):2925. doi:10.1038/s41598-023-29521-z. [Google Scholar] [PubMed] [CrossRef]
85. Behrendt F, Bhattacharya D, Krüger J, Opfer R, Schlaefer A. Patched diffusion models for unsupervised anomaly detection in brain MRI. Proceed Mac Learn Res. 2023;227:1019–32. doi:10.1109/isbi56570.2024.10635828. [Google Scholar] [CrossRef]
86. Esmaeili V, MohasselFeghhi M, Seyedarabi H. Autonomous data augmentation and melanoma detection using a combination of classical and deep-learning techniques. Amsterdam, The Netherlands: Elsevier; 2024. doi:10.2139/ssrn.4780994. [Google Scholar] [CrossRef]
87. Tsivgoulis M, Papastergiou T, Megalooikonomou V. An improved SqueezeNet model for the diagnosis of lung cancer in CT scans. Mach Learn Appl. 2022;10(8):100399. doi:10.1016/j.mlwa.2022.100399. [Google Scholar] [CrossRef]
88. Obayya M, Arasi MA, Alruwais N, Alsini R, Mohamed A, Yaseen I. Biomedical image analysis for colon and lung cancer detection using tuna swarm algorithm with deep learning model. IEEE Access. 2023;11:94705–12. doi:10.1109/access.2023.3309711. [Google Scholar] [CrossRef]
89. Guo Z, Ding C, Do D, Shah A, Lee RJ, Hu X, et al. Improving atrial fibrillation detection using a shared latent space for ECG and PPG signals. Harv Data Sci Rev. 2025;7(1):1–33. doi:10.1162/99608f92.9e63a630. [Google Scholar] [CrossRef]
90. Jazzar N, Douik A. Res2U++: deep learning model for segmentation of ischemic stroke lesions. Biomed Signal Process Control. 2025;102(23):107269. doi:10.1016/j.bspc.2024.107269. [Google Scholar] [CrossRef]
91. Gautam N, Basu A, Sarkar R. Lung cancer detection from thoracic CT scans using an ensemble of deep learning models. Neural Comput Appl. 2024;36(5):2459–77. doi:10.1007/s00521-023-09130-7. [Google Scholar] [CrossRef]
92. Li L, Tan Z, Han X. An improved EfficientNet model and its applications in pneumonia image classification. J Eng Sci Technol Rev. 2022;15(6):49–54. [Google Scholar]
93. Guhdar M, Mohammed AO, Mstafa RJ. Advanced deep learning framework for ECG arrhythmia classification using 1D-CNN with attention mechanism. Knowl Based Syst. 2025;315(1):113301. doi:10.1016/j.knosys.2025.113301. [Google Scholar] [CrossRef]
94. Ibrayev S, Omarov B, Amanov B, Momynkulov Z. Development of a deep learning-enhanced lower-limb exoskeleton using electromyography data for post-neurovascular rehabilitation. Eng Sci. 2024;31:1269. doi:10.30919/es1269. [Google Scholar] [CrossRef]
95. Xu D, Miao X, Liu H, Scholey JE, Yang W, Feng M, et al. Paired conditional generative adversarial network for highly accelerated liver 4D MRI. Phys Med Biol. 2024;69(12):125029. doi:10.1088/1361-6560/ad5489. [Google Scholar] [PubMed] [CrossRef]
96. Zheng X, Wilhelm E, Otten E, Reneman MF, Lamoth CJC. Age-related gait patterns classification using deep learning based on time-series data from one accelerometer. Biomed Signal Process Control. 2025;104:107406. doi:10.1016/j.bspc.2024.107406. [Google Scholar] [CrossRef]
97. Wong S, Simmons A, Rivera-Villicana J, Barnett S, Sivathamboo S, Perucca P, et al. Channel-annotated deep learning for enhanced interpretability in EEG-based seizure detection. Biomed Signal Process Control. 2025;103(6738):107484. doi:10.1016/j.bspc.2024.107484. [Google Scholar] [CrossRef]
98. Nagamani GM, Kumar CK. Design of an improved graph-based model for real-time anomaly detection in healthcare using hybrid CNN-LSTM and federated learning. Heliyon. 2024;10(24):e41071. doi:10.1016/j.heliyon.2024.e41071. [Google Scholar] [PubMed] [CrossRef]
99. Abir SI, Shoha S, Hossain MM, Sultana N, Saha TR, Sarwer MH, et al. Machine learning and deep learning techniques for EEG-based prediction of psychiatric disorders. J Comput Sci Technol Stud. 2025;7(1):46–63. doi:10.32996/jcsts.2025.7.1.4. [Google Scholar] [CrossRef]
100. Hasan MZ, Montaha S, Khan IU, Hassan MM, Al Mahmud A, Rafid AKMRH, et al. Fast and efficient lung abnormality identification with explainable AI: a comprehensive framework for chest CT scan and X-ray images. IEEE Access. 2024;12:31117–35. doi:10.1109/access.2024.3369900. [Google Scholar] [CrossRef]
101. Zaim T, Abdel-Hadi S, Mahmoud R, Khandakar A, Rakhtala SM, Chowdhury MEH. Machine learning- and deep learning-based myoelectric control system for upper limb rehabilitation utilizing EEG and EMG signals: a systematic review. Bioengineering. 2025;12(2):144. doi:10.3390/bioengineering12020144. [Google Scholar] [PubMed] [CrossRef]
102. Zhou Y, Tang X, Zhang D, Lee HJ. Machine learning empowered coherent Raman imaging and analysis for biomedical applications. Commun Eng. 2025;4(1):8. doi:10.1038/s44172-025-00345-1. [Google Scholar] [PubMed] [CrossRef]
103. Cruz-Vazquez JA, Montiel-Pérez JY, Romero-Herrera R, Rubio-Espino E. Emotion recognition from EEG signals using advanced transformations and deep learning. Mathematics. 2025;13(2):254. doi:10.3390/math13020254. [Google Scholar] [CrossRef]
104. Zafar MH, Moosavi SKR, Sanfilippo F. Federated learning-enhanced edge deep learning model for EMG-based gesture recognition in real-time human-robot interaction. IEEE Sens J. 2025;25(5):9139–51. doi:10.1109/JSEN.2025.3529841. [Google Scholar] [CrossRef]
105. Davoudi Kakhki F, Vora H, Moghadam A. Biomechanical risk classification in repetitive lifting using multi-sensor electromyography data, revised national institute for occupational safety and health lifting equation, and deep learning. Biosensors. 2025;15(2):84. doi:10.3390/bios15020084. [Google Scholar] [PubMed] [CrossRef]
106. Ali Zendehbad S, Razavi AS, Sanjani MA, Sedaghat Z, Lashkari S. TraxVBF: a hybrid transformer-xLSTM framework for EMG signal processing and assistive technology development in rehabilitation. Sens Bio Sens Res. 2025;47(8):100749. doi:10.1016/j.sbsr.2025.100749. [Google Scholar] [CrossRef]
107. Samarakoon SS, Herath HB, Yasakethu SP, Fernando D, Madusanka N, Yi M, et al. Long short-term memory-enabled electromyography-controlled adaptive wearable robotic exoskeleton for upper arm rehabilitation. Biomimetics. 2025;10(2):106. doi:10.3390/biomimetics10020106. [Google Scholar] [PubMed] [CrossRef]
108. Sarwer MH, Saha TR, Abir SI, Shoha S, Hossain MM, Sultana N, et al. EEG functional connectivity and deep learning for automated diagnosis of Alzheimer’s disease and schizophrenia. J Comput Sci Technol Stud. 2025;7(1):82–99. doi:10.32996/jcsts.2025.7.1.7. [Google Scholar] [CrossRef]
109. Khalfallah S, Puech W, Tlija M, Bouallegue K. Exploring the effectiveness of machine learning and deep learning techniques for EEG signal classification in neurological disorders. IEEE Access. 2025;13(8):17002–15. doi:10.1109/access.2025.3532515. [Google Scholar] [CrossRef]
110. Wang Y, Zhao S, Jiang H, Li S, Luo B, Li T, et al. DiffMDD: a diffusion-based deep learning framework for MDD diagnosis using EEG. IEEE Trans Neural Syst Rehabil Eng. 2024;32:728–38. doi:10.1109/TNSRE.2024.3360465. [Google Scholar] [PubMed] [CrossRef]
111. Mukherjee P, Halder Roy A. EEG sensor driven assistive device for elbow and finger rehabilitation using deep learning. Expert Syst Appl. 2024;244(2):122954. doi:10.1016/j.eswa.2023.122954. [Google Scholar] [CrossRef]
112. Jawed S, Faye I, Malik AS. Deep learning-based assessment model for real-time identification of visual learners using raw EEG. IEEE Trans Neural Syst Rehabil Eng. 2024;32(4):378–90. doi:10.1109/TNSRE.2024.3351694. [Google Scholar] [PubMed] [CrossRef]
113. Aslan M, Baykara M, Alakuş TB. Analysis of brain areas in emotion recognition from EEG signals with deep learning methods. Multimed Tools Appl. 2024;83(11):32423–52. doi:10.1007/s11042-023-16696-w. [Google Scholar] [CrossRef]
114. Jirakittayakorn N, Wongsawat Y, Mitrirattanakul S. ZleepAnlystNet: a novel deep learning model for automatic sleep stage scoring based on single-channel raw EEG data using separating training. Sci Rep. 2024;14(1):9859. doi:10.1038/s41598-024-60796-y. [Google Scholar] [PubMed] [CrossRef]
115. Xie J, Zhang J, Sun J, Ma Z, Qin L, Li G, et al. A transformer-based approach combining deep learning network and spatial-temporal information for raw EEG classification. IEEE Trans Neural Syst Rehabil Eng. 2022;30:2126–36. doi:10.1109/TNSRE.2022.3194600. [Google Scholar] [PubMed] [CrossRef]
116. Sarkar A, Singh A, Chakraborty R. A deep learning-based comparative study to track mental depression from EEG data. Neurosci Inf. 2022;2(4):100039. doi:10.1016/j.neuri.2022.100039. [Google Scholar] [CrossRef]
117. Li C, Xi Z, Jin G, Jiang W, Wang B, Cai X, et al. Deep-learning-enabled microwave-induced thermoacoustic tomography based on ResAttU-net for transcranial brain hemorrhage detection. IEEE Trans Biomed Eng. 2023;70(8):2350–61. doi:10.1109/TBME.2023.3243491. [Google Scholar] [PubMed] [CrossRef]
118. Lin CH, Liu ZY, Chen JS, Fann YC, Wen MS, Kuo CF. ECG-surv: a deep learning-based model to predict time to 1-year mortality from 12-lead electrocardiogram. Biomed J. 2025;48(1):100732. doi:10.1016/j.bj.2024.100732. [Google Scholar] [PubMed] [CrossRef]
119. Hasan MN, Ali Hossain M, Rahman MA. An ensemble based lightweight deep learning model for the prediction of cardiovascular diseases from electrocardiogram images. Eng Appl Artif Intell. 2025;141(2):109782. doi:10.1016/j.engappai.2024.109782. [Google Scholar] [CrossRef]
120. Kovalchuk O, Barmak O, Radiuk P, Klymenko L, Krak I. Towards transparent AI in medicine: ecg-based arrhythmia detection with explainable deep learning. Technologies. 2025;13(1):34. doi:10.3390/technologies13010034. [Google Scholar] [CrossRef]
121. Friedman SF, Khurshid S, Venn RA, Wang X, Diamant N, Di Achille P, et al. Unsupervised deep learning of electrocardiograms enables scalable human disease profiling. npj Digit Med. 2025;8(1):23. doi:10.1038/s41746-024-01418-9. [Google Scholar] [PubMed] [CrossRef]
122. Alsayat A, Mahmoud AA, Alanazi S, Mostafa AM, Alshammari N, Alrowaily MA, et al. Enhancing cardiac diagnostics: a deep learning ensemble approach for precise ECG image classification. J Big Data. 2025;12(1):7. doi:10.1186/s40537-025-01070-4. [Google Scholar] [CrossRef]
123. Narmatha P, Marimuthu T, Suguna R, Rajendiran M, Vijaya Lakshmi TR, Maranan R. Early prediction of cardiac arrest through ECG signals using distributed deep learning with cloud computing. In: Hybrid and advanced technologies. London, UK: CRC Press; 2025. p. 266–71. doi:10.1201/9781003559115-45. [Google Scholar] [CrossRef]
124. Chandrasekhar N, Canavoy Narahari S, Kollem S, Peddakrishna S, Penchala A, Prasad Chapa B. Heart abnormality classification using ECG and PCG recordings with novel PJM-DJRNN. Results Eng. 2025;25(4):104032. doi:10.1016/j.rineng.2025.104032. [Google Scholar] [CrossRef]
125. Shang H, Yu S, Wu Y, Liu X, He J, Ma M, et al. A noninvasive hyperkalemia monitoring system for dialysis patients based on a 1D-CNN model and single-lead ECG from wearable devices. Sci Rep. 2025;15(1):2950. doi:10.1038/s41598-025-85722-8. [Google Scholar] [PubMed] [CrossRef]
126. Choi J, Kim J, Spaccarotella C, Esposito G, Oh IY, Cho Y, et al. Smartwatch ECG and artificial intelligence in detecting acute coronary syndrome compared to traditional 12-lead ECG. Int J Cardiol Heart Vasc. 2024;56:101573. doi:10.1016/j.ijcha.2024.101573. [Google Scholar] [PubMed] [CrossRef]
127. Chen J, Huang S, Zhang Y, Chang Q, Zhang Y, Li D, et al. Congenital heart disease detection by pediatric electrocardiogram based deep learning integrated with human concepts. Nat Commun. 2024;15(1):976. doi:10.1038/s41467-024-44930-y. [Google Scholar] [PubMed] [CrossRef]
128. Abubaker MB, Babayiğit B. Detection of cardiovascular diseases in ECG images using machine learning and deep learning methods. IEEE Trans Artif Intell. 2022;4(2):373–82. doi:10.1109/TAI.2022.3159505. [Google Scholar] [CrossRef]
129. Mahmood A, Dhahri H, Alhajlah M, Almaslukh A. Enhanced classification of phonocardiograms using modified deep learning. IEEE Access. 2024;12(1):178909–16. doi:10.1109/access.2024.3507920. [Google Scholar] [CrossRef]
130. Alkhodari M, Hadjileontiadis LJ, Khandoker AH. Identification of congenital valvular murmurs in young patients using deep learning-based attention transformers and phonocardiograms. IEEE J Biomed Health Inf. 2024;28(4):1803–14. doi:10.1109/JBHI.2024.3357506. [Google Scholar] [PubMed] [CrossRef]
131. Mandala S, Amini SS, Syaifullah AR, Pramudyo M, Nurmaini S, Abdullah AH. Enhanced myocardial infarction identification in phonocardiogram signals using segmented feature extraction and transfer learning-based classification. IEEE Access. 2023;11:136654–65. doi:10.1109/access.2023.3338853. [Google Scholar] [CrossRef]
132. Nehary EA, Rajan S. Phonocardiogram classification by learning from positive and unlabeled examples. IEEE Trans Instrum Meas. 2024;73(5):4005114. doi:10.1109/TIM.2024.3372221. [Google Scholar] [CrossRef]
133. Bhardwaj A, Singh S, Joshi D. Phonocardiography-based automated detection of prosthetic heart valve dysfunction using persistence spectrum and interpretable deep CNN. IEEE Sens J. 2025;25(4):6869–80. doi:10.1109/JSEN.2024.3523393. [Google Scholar] [CrossRef]
134. Basak P, Nazmus Sakib AHM, Chowdhury MEH, Al-Emadi N, Cagatay Yalcin H, Pedersen S, et al. A novel deep learning technique for morphology preserved fetal ECG extraction from mother ECG using 1D-CycleGAN. Expert Syst Appl. 2024;235(2):121196. doi:10.1016/j.eswa.2023.121196. [Google Scholar] [CrossRef]
135. Pani K, Chawla I. Synthetic MRI in action: a novel framework in data augmentation strategies for robust multi-modal brain tumor segmentation. Comput Biol Med. 2024;183(1):109273. doi:10.1016/j.compbiomed.2024.109273. [Google Scholar] [PubMed] [CrossRef]
136. Huang L, Ruan S, Decazes P, Denœux T. Deep evidential fusi on with uncertainty quantification and reliability learning for multimodal medical image segmentation. Inf Fusion. 2025;113(3):102648. doi:10.1016/j.inffus.2024.102648. [Google Scholar] [CrossRef]
137. Nag A, Mondal H, Mehedi Hassan M, Al-Shehari T, Kadrie M, Al-Razgan M, et al. TumorGANet: a transfer learning and generative adversarial network-based data augmentation model for brain tumor classification. IEEE Access. 2024;12:103060–81. doi:10.1109/access.2024.3429633. [Google Scholar] [CrossRef]
138. Hussain T, Shouno H, Hussain A, Hussain D, Ismail M, Hussain Mir T, et al. EFFResNet-ViT: a fusion-based convolutional and vision transformer model for explainable medical image classification. IEEE Access. 2025;13(86):54040–68. doi:10.1109/access.2025.3554184. [Google Scholar] [CrossRef]
139. Karthik A, Hamatta HSA, Patthi S, Krubakaran C, Kumar Pradhan A, Rachapudi V, et al. Ensemble-based multimodal medical imaging fusion for tumor segmentation. Biomed Signal Process Control. 2024;96(1):106550. doi:10.1016/j.bspc.2024.106550. [Google Scholar] [CrossRef]
140. Sinha A, Agarwal R, Kumar V, Garg N, Pundir DS, Singh H, et al. Multi-modal medical image fusion using improved dual-channel PCNN. Med Biol Eng Comput. 2024;62(9):2629–51. doi:10.1007/s11517-024-03089-w. [Google Scholar] [PubMed] [CrossRef]
141. Veeramani N, Jayaraman P, Krishankumar R, Ravichandran KS, Gandomi AH. DDCNN-F: double decker convolutional neural network ‘F’ feature fusion as a medical image classification framework. Sci Rep. 2024;14(1):676. doi:10.1038/s41598-023-49721-x. [Google Scholar] [PubMed] [CrossRef]
142. Lv J, Zeng X, Chen B, Hu M, Yang S, Qiu X, et al. A stochastic structural similarity guided approach for multi-modal medical image fusion. Sci Rep. 2025;15(1):8792. doi:10.1038/s41598-025-93662-6. [Google Scholar] [PubMed] [CrossRef]
143. Xu X, Li J, Zhu Z, Zhao L, Wang H, Song C, et al. A comprehensive review on synergy of multi-modal data and AI technologies in medical diagnosis. Bioengineering. 2024;11(3):219. doi:10.3390/bioengineering11030219. [Google Scholar] [PubMed] [CrossRef]
144. Steyaert S, Pizurica M, Nagaraj D, Khandelwal P, Hernandez-Boussard T, Gentles AJ, et al. Multimodal data fusion for cancer biomarker discovery with deep learning. Nat Mach Intell. 2023;5(4):351–62. doi:10.1038/s42256-023-00633-5. [Google Scholar] [PubMed] [CrossRef]
145. Zhang Y, Sheng M, Liu X, Wang R, Lin W, Ren P, et al. A heterogeneous multi-modal medical data fusion framework supporting hybrid data exploration. Health Inf Sci Syst. 2022;10(1):22. doi:10.1007/s13755-022-00183-x. [Google Scholar] [PubMed] [CrossRef]
146. Rasool N, Iqbal Bhat J. Unveiling the complexity of medical imaging through deep learning approaches. Chaos Theory Appl. 2023;5(4):267–80. doi:10.51537/chaos.1326790. [Google Scholar] [CrossRef]
147. Islam S, Aziz MT, Nabil HR, Jim JR, Mridha MF, Kabir MM, et al. Generative adversarial networks (GANs) in medical imaging: advancements, applications, and challenges. IEEE Access. 2024;12:35728–53. doi:10.1109/access.2024.3370848. [Google Scholar] [CrossRef]
148. Alalwan N, Alwadain A, Alzahrani AI, Al-Bayatti AH, Abozeid A, Abd El-Aziz RM. Advancements in brain tumor identification: integrating synthetic GANs with federated-CNNs in medical imaging analysis. Alex Eng J. 2024;105(2):105–19. doi:10.1016/j.aej.2024.06.080. [Google Scholar] [CrossRef]
149. Apostolopoulos ID, Papathanasiou ND, Apostolopoulos DJ, Panayiotakis GS. Applications of generative adversarial networks (GANs) in positron emission tomography (PET) imaging: a review. Eur J Nucl Med Mol Imaging. 2022;49(11):3717–39. doi:10.1007/s00259-022-05805-w. [Google Scholar] [PubMed] [CrossRef]
150. Mohsen F, Ali H, El Hajj N, Shah Z. Artificial intelligence-based methods for fusion of electronic health records and imaging data. Sci Rep. 2022;12(1):17981. doi:10.1038/s41598-022-22514-4. [Google Scholar] [PubMed] [CrossRef]
151. Illimoottil M, Ginat D. Recent advances in deep learning and medical imaging for head and neck cancer treatment: MRI, CT, and PET scans. Cancers. 2023;15(13):3267. doi:10.3390/cancers15133267. [Google Scholar] [PubMed] [CrossRef]
152. Hosny KM, Elshora D, Mohamed ER, Vrochidou E, Papakostas GA. Deep learning and optimization-based methods for skin lesions segmentation: a review. IEEE Access. 2023;11:85467–88. doi:10.1109/access.2023.3303961. [Google Scholar] [CrossRef]
153. Borges PVK, Peynot T, Liang S, Arain B, Wildie M, Minareci MG, et al. A survey on terrain traversability analysis for autonomous ground vehicles: methods, sensors, and challenges. Field Robot. 2022;2:1567–627. doi:10.55417/fr.2022049. [Google Scholar] [CrossRef]
154. Wankhede DS, Selvarani R. Dynamic architecture based deep learning approach for glioblastoma brain tumor survival prediction. Neurosci Inf. 2022;2(4):100062. doi:10.1016/j.neuri.2022.100062. [Google Scholar] [CrossRef]
155. Raminedi S, Shridevi S, Won D. Multi-modal transformer architecture for medical image analysis and automated report generation. Sci Rep. 2024;14(1):19281. doi:10.1038/s41598-024-69981-5. [Google Scholar] [PubMed] [CrossRef]
156. Siddiqui SS. An integrated approach to a next-generation telemedicine and health advice system (angthas) with artificial intelligence [dissertation]. Portsmouth, UK: University of Portsmouth; 2022. [Google Scholar]
157. Friedrich P, Frisch Y, Cattin PC. Deep generative models for 3D medical image synthesis. In: Generative machine learning models in medical image computing. Cham, Switzerland: Springer Nature; 2024. p. 255–78. doi:10.1007/978-3-031-80965-1_13. [Google Scholar] [CrossRef]
158. Irfan M, Malik KM, Muhammad K. Federated fusion learning with attention mechanism for multi-client medical image analysis. Inf Fusion. 2024;108(7):102364. doi:10.1016/j.inffus.2024.102364. [Google Scholar] [CrossRef]
159. Horton MB, Brady CJ, Cavallerano J, Abramoff M, Barker G, Chiang MF, et al. Practice guidelines for ocular telehealth-diabetic retinopathy. Telemed J e-Health. 2020;26(4):495–543. doi:10.1089/tmj.2020.0006. [Google Scholar] [PubMed] [CrossRef]
160. Baxter SL, Quackenbush Q, Cerda J, Gregg C, Millen M, T horne C. Implementing clinical informatics tools for primary care-based diabetic retinopathy screening. Am J Manag Care. 2022;28(10):e355–62. doi:10.37765/ajmc.2022.89253. [Google Scholar] [PubMed] [CrossRef]
161. Murphy E, Khandelwal S, Shanker S. Custom precision accelerators for energy-efficient image-to-image transformations in motion picture workflows. In: Applications of digital image processing XLVI. Bellingham, DC, USA: SPIE; 2023. p. 191–202. doi:10.1117/12.2677688. [Google Scholar] [CrossRef]
162. Yu T, Akhmadeev K, Le Carpentier E, Aoustin Y, Farina D. On-line recursive decomposition of intramuscular EMG signals using GPU-implemented Bayesian filtering. IEEE Trans Biomed Eng. 2020;67(6):1806–18. doi:10.1109/tbme.2019.2948397. [Google Scholar] [PubMed] [CrossRef]
163. Wu Y, Jewell S, Xing X, Nan Y, Strong AJ, Yang G, et al. Real-time non-invasive imaging and detection of spreading depolarizations through EEG: an ultra-light explainable deep learning approach. IEEE J Biomed Health Inf. 2024;28(10):5780–91. doi:10.1109/JBHI.2024.3370502. [Google Scholar] [PubMed] [CrossRef]
164. Nizam NB. Multimodal feature fusion based thoracic disease classification framework combining medical data and chest x-ray images [master’s thesis]. Dhaka, Bangladesh: Bangladesh University of Engineering and Technology; 2023. [Google Scholar]
165. Myrzashova R, Alsamhi SH, Hawbani A, Curry E, Guizani M, Wei X. Safeguarding patient data-sharing: blockchain-enabled federated learning in medical diagnostics. IEEE Trans Sustain Comput. 2025;10(1):176–89. doi:10.1109/tsusc.2024.3409329. [Google Scholar] [CrossRef]
166. Alsamhi SH, Myrzashova R, Hawbani A, Kumar S, Srivastava S, Zhao L, et al. Federated learning meets blockchain in decentralized data sharing: healthcare use case. IEEE Internet Things J. 2024;11(11):19602–15. doi:10.1109/jiot.2024.3367249. [Google Scholar] [CrossRef]
167. Hein D, Bozorgpour A, Merhof D, Wang G. Physics-inspired generative models in medical imaging. Annu Rev Biomed Eng. 2025;27(1):499–525. doi:10.1146/annurev-bioeng-102723-013922. [Google Scholar] [PubMed] [CrossRef]
168. Rai HM, Chatterjee K. 2D MRI image analysis and brain tumor detection using deep learning CNN model LeU-Net. Multimed Tools Appl. 2021;80(28):36111–41. doi:10.1007/s11042-021-11504-9. [Google Scholar] [CrossRef]
169. Yu H, Yang LT, Zhang Q, Armstrong D, Deen MJ. Convolutional neural networks for medical image analysis: state-of-the-art, comparisons, improvement and perspectives. Neurocomputing. 2021;444(9):92–110. doi:10.1016/j.neucom.2020.04.157. [Google Scholar] [CrossRef]
170. Kamalam T, Srikanth Y, Venkatesh PS. Skin lesion segmentation detection using U-Net architecture. In: 2024 International Conference on Wireless Communications Signal Processing and Networking (WiSPNET); 2024 Mar 21–23; Chennai, India. p. 1–6. doi:10.1109/WiSPNET61464.2024.10532992. [Google Scholar] [CrossRef]
171. Balasubramaniam S, Prasanth A, Kumar KS, Kavitha V. Medical image analysis based on deep learning approach for early diagnosis of diseases. In: Deep learning for smart healthcare. Boca Raton, FL, USA: Auerbach Publications; 2024. p. 54–75. doi:10.1201/9781003469605-4. [Google Scholar] [CrossRef]
172. Ashraf A, Nawi NM, Shahzad T, Aamir M, Khan MA, Ouahada K. Dimension reduction using dual-featured auto-encoder for the histological classification of human lungs tissues. IEEE Access. 2024;12(1):104165–76. doi:10.1109/access.2024.3434592. [Google Scholar] [CrossRef]
173. Sadad T, Rehman A, Munir A, Saba T, Tariq U, Ayesha N, et al. Brain tumor detection and multi-classification using advanced deep learning techniques. Microsc Res Tech. 2021;84(6):1296–308. doi:10.1002/jemt.23688. [Google Scholar] [PubMed] [CrossRef]
174. Quasar SR, Sharma R, Mittal A, Sharma M, Agarwal D, de la Torre Díez I. Ensemble methods for computed tomography scan images to improve lung cancer detection and classification. Multimed Tools Appl. 2024;83(17):52867–97. doi:10.1007/s11042-023-17616-8. [Google Scholar] [CrossRef]
175. Lavecchia A. Transform drug discovery and development with generative artificial intelligence. In: Generative artificial intelligence for biomedical and smart health informatics. Hoboken, NJ, USA: Wiley-IEEE press; 2025. p. 489–537. [Google Scholar]
176. Chillar V, Gupta S, Vijarania M, Agrawal A, Soni A. Role of sustainable strategies using artifical inteligence in brain tumour detection and treatment. In: Achieving sustainability with AI technologies. Hershey, PA, USA: IGI Global; 2025. p. 327–60. doi:10.4018/979-8-3693-3410-2.ch015. [Google Scholar] [CrossRef]
177. Sharma A, Kala S, Kumar A, Sharma S, Gupta G, Jaiswal V. Deep learning in genomics, personalized medicine, and neurodevelopmental disorders. In: Intelligent data analytics for bioinformatics and biomedical systems. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2024. p. 235–64. [Google Scholar]
178. Shtaiwi A, Khan SU, Khedraoui M, Alaraj M, Samadi A, Chtita S. A comprehensive computational study to explore promising natural bioactive compounds targeting glycosyltransferase MurG in Escherichia coli for potential drug development. Sci Rep. 2024;14(1):7098. doi:10.1038/s41598-024-57702-x. [Google Scholar] [PubMed] [CrossRef]
179. Zhao Q, Zhao H, Zheng K, Wang J. HyperAttentionDTI: improving drug-protein interaction prediction by sequence-based deep learning with attention mechanism. Bioinformatics. 2022;38(3):655–62. doi:10.1093/bioinformatics/btab715. [Google Scholar] [PubMed] [CrossRef]
180. Annadate P, Bedekar M, Annadate M, Choi E, Choi A. Harnessing deep learning for early detection of cardiac abnormalities. Int J Comput Digit Syst. 2025;17(1):1–10. doi:10.12785/ijcds/1571031205. [Google Scholar] [CrossRef]
181. Nasarudin NA, Al Jasmi F, Sinnott RO, Zaki N, Al Ashwal H, Mohamed EA, et al. A review of deep learning models and online healthcare databases for electronic health records and their use for health prediction. Artif Intell Rev. 2024;57(9):249. doi:10.1007/s10462-024-10876-2. [Google Scholar] [CrossRef]
182. Thiboud PE, François Q, Faure C, Chaufferin G, Arribe B, Ettahar N. Development and validation of a machine learning model for early prediction of sepsis onset in hospital inpatients from all departments. Diagnostics. 2025;15(3):302. doi:10.3390/diagnostics15030302. [Google Scholar] [PubMed] [CrossRef]
183. Neifar N, Ben-Hamadou A, Mdhaffar A, Jmaiel M, Freisleben B. Leveraging statistical shape priors in GAN-based ECG synthesis. IEEE Access. 2024;12(1):36002–15. doi:10.1109/access.2024.3373724. [Google Scholar] [CrossRef]
184. Dara ON, Ibrahim AA, Mohammed TA. Advancing medical imaging: detecting polypharmacy and adverse drug effects with Graph Convolutional Networks (GCN). BMC Med Imaging. 2024;24(1):174. doi:10.1186/s12880-024-01349-7. [Google Scholar] [PubMed] [CrossRef]
185. Shen B, Zhang Z, Shi X, Cao C, Zhang Z, Hu Z, et al. Real-time intraoperative glioma diagnosis using fluorescence imaging and deep convolutional neural networks. Eur J Nucl Med Mol Imaging. 2021;48(11):3482–92. doi:10.1007/s00259-021-05326-y. [Google Scholar] [PubMed] [CrossRef]
186. Hassan AM, Biaggi-Ondina A, Rajesh A, Asaad M, Nelson JA, Coert JH, et al. Predicting patient-reported outcomes following surgery using machine learning. Am Surg. 2023;89(1):31–5. doi:10.1177/00031348221109478. [Google Scholar] [PubMed] [CrossRef]
187. Tanzi L, Piazzolla P, Porpiglia F, Vezzetti E. Real-time deep learning semantic segmentation during intra-operative surgery for 3D augmented reality assistance. Int J Comput Assist Radiol Surg. 2021;16(9):1435–45. doi:10.1007/s11548-021-02432-y. [Google Scholar] [PubMed] [CrossRef]
188. Shah N, Khalid U, Kavia R, Batura D. Current advances in the use of artificial intelligence in predicting and managing urological complications. Int Urol Nephrol. 2024;56(11):3427–35. doi:10.1007/s11255-024-04149-8. [Google Scholar] [PubMed] [CrossRef]
189. Apama C, Rohini P, Pandiyarasan V. Interpretation of ResNet50 model for MI related cardiac events using explainable Grad-CAM approach. Curr Dir Biomed Eng. 2022;8(2):723–6. doi:10.1515/cdbme-2022-1184. [Google Scholar] [CrossRef]
190. Deepika P, Sistla P, Subramaniam G, Rao M. Deep learning based automated screening for intracranial hemorrhages and GRAD-CAM visualizations on non-contrast head computed tomography volumes. In: 2022 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI); 2022 Sep 27–30; Ioannina, Greece. p. 1–5. doi:10.1109/BHI56158.2022.9926782. [Google Scholar] [CrossRef]
191. Raza MU, Chen J, Wang L, Nawaz A, Leung VCM, Li J. Tree regularization for visual explanations of cancer cell classification. In: 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2024 Dec 3–6; Lisbon, Portugal. p. 3655–60. doi:10.1109/BIBM62325.2024.10822343. [Google Scholar] [CrossRef]
192. Tan L, Huang C, Yao X. A concept-based local interpretable model-agnostic explanation approach for deep neural networks in image classification. In: Intelligent information processing XII. Cham, Switzerland: Springer Nature; 2024. p. 119–33. doi:10.1007/978-3-031-57919-6_9. [Google Scholar] [CrossRef]
193. Zhang S, Wang Z, Su X. A study on the inter-pretability of network attack prediction models based on Light gradient boosting machine (LGBM) and SHapley additive exPlanations (SHAP). Comput Mater Contin. 2025;83(3):5781–809. doi:10.32604/cmc.2025.062080. [Google Scholar] [CrossRef]
194. Prasanna Venkatesh N, Pradeep Kumar R, Chakravarthy Neelapu B, Pal K, Sivaraman J. Automated atrial arrhythmia classification using 1D-CNN-BiLSTM: a deep network ensemble model. Biomed Signal Process Control. 2024;97:106703. doi:10.1016/j.bspc.2024.106703. [Google Scholar] [CrossRef]
195. Abdalla HB, Kumar Y, Marchena J, Guzman S, Awlla A, Gheisari M, et al. The future of artificial intelligence in the face of data scarcity. Comput Mater Contin. 2025;84(1):1073–99. doi:10.32604/cmc.2025.063551. [Google Scholar] [CrossRef]
196. Isewon I, Alagbe E, Oyelade J. Optimizing machine learning performance for medical imaging analyses in low-resource environments: the prospects of CNN-based feature extractors. F1000Res. 2025;14:100. doi:10.12688/f1000research.156122.1. [Google Scholar] [CrossRef]
197. Marey A, Arjmand P, Alerab ADS, Eslami MJ, Saad AM, Sanchez N, et al. Explainability, transparency and black box challenges of AI in radiology: impact on patient care in cardiovascular radiology. Egypt J Radiol Nucl Med. 2024;55(1):183. doi:10.1186/s43055-024-01356-2. [Google Scholar] [CrossRef]
198. Javed H, Muqeet HA, Danesh A, Rehman AU, Javed T, Bermak A. Impact of AI and dynamic ensemble techniques in enhancing healthcare services: opportunities and ethical challenges. IEEE Access. 2024;12(3):141064–87. doi:10.1109/access.2024.3443812. [Google Scholar] [CrossRef]
199. Ali S, Akhlaq F, Imran AS, Kastrati Z, Daudpota SM, Moosa M. The enlightening role of explainable artificial intelligence in medical & healthcare domains: a systematic literature review. Comput Biol Med. 2023;166:107555. doi:10.1016/j.compbiomed.2023.107555. [Google Scholar] [PubMed] [CrossRef]
200. Cross JL, Choma MA, Onofrey JA. Bias in medical AI: implications for clinical decision-making. PLoS Digit Health. 2024;3(11):e0000651. doi:10.1371/journal.pdig.0000651. [Google Scholar] [PubMed] [CrossRef]
201. Amirian S, Gao F, Littlefield N, Hill JH, Yates AJ, Plate JF, et al. State-of-the-art in responsible, explainable, and fair AI for medical image analysis. IEEE Access. 2025;13(1):58229–63. doi:10.1109/access.2025.3555543. [Google Scholar] [CrossRef]
202. Narkhede J. Comparative evaluation of post-hoc explainability methods in AI: lime, SHAP, and grad-CAM. In: 4th International Conference on Sustainable Expert Systems (ICSES); 2024 Oct 15–17; Kaski, Nepal. p. 826–30. doi:10.1109/ICSES63445.2024.10762963. [Google Scholar] [CrossRef]
203. Wen R, Yuan H, Ni D, Xiao W, Wu Y. From denoising training to test-time adaptation: enhancing domain generalization for medical image segmentation. In: 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV); 2024 Jan 3–8; Waikoloa, HI, USA. p. 464–74. doi:10.1109/WACV57701.2024.00052. [Google Scholar] [CrossRef]
204. Jeong M, Cho H, Jung S, Kim WH. Uncertainty-aware diffusion-based adversarial attack for realistic colonoscopy image synthesis. In: Medical Image Computing and Computer Assisted Intervention—MICCAI 2024. Cham, Switzerland: Springer Nature; 2024. p. 647–58. doi:10.1007/978-3-031-72114-4_62. [Google Scholar] [CrossRef]
205. Liefgreen A, Weinstein N, Wachter S, Mittelstadt B. Beyond ideals: why the (medical) AI industry needs to motivate behavioural change in line with fairness and transparency values, and how it can do it. AI Soc. 2024;39(5):2183–99. doi:10.1007/s00146-023-01684-3. [Google Scholar] [PubMed] [CrossRef]
206. Tong WJ, Wu SH, Cheng MQ, Huang H, Liang JY, Li CQ, et al. Integration of artificial intelligence decision aids to reduce workload and enhance efficiency in thyroid nodule management. JAMA Netw Open. 2023;6(5):e2313674. doi:10.1001/jamanetworkopen.2023.13674. [Google Scholar] [PubMed] [CrossRef]
207. Alowais SA, Alghamdi SS, Alsuhebany N, Alqahtani T, Alshaya AI, Almohareb SN, et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ. 2023;23(1):689. doi:10.1186/s12909-023-04698-z. [Google Scholar] [PubMed] [CrossRef]
208. Bogdanoski G, Lucas F, Kern W, Czechowska K. Translating the regulatory landscape of medical devices to create fit-for-purpose artificial intelligence (AI) cytometry solutions. Cytometry B Clin Cytom. 2024;106(4):294–307. doi:10.1002/cyto.b.22167. [Google Scholar] [PubMed] [CrossRef]
209. Cheng YH, Pal A, Moody R, Heimbach T, Lukacova V, Patel N, et al. Using model master files to support oral drug product development and regulatory submissions. Pharm Res. 2025;42(5):753–63. doi:10.1007/s11095-025-03865-9. [Google Scholar] [PubMed] [CrossRef]
210. Vecellio Segate R, Daly A. Encoding the enforcement of safety standards into smart robots to harness their computing sophistication and collaborative potential: a legal risk assessment for European union policymakers. Eur J Risk Regul. 2024;15(3):665–704. doi:10.1017/err.2023.72. [Google Scholar] [CrossRef]
211. Kortelainen T, Milovanov M. The role of ISO 9001 and ISO 13485 Quality Management System as drivers behind health technology supply chain performance and evolution [master’s thesis]. Joensuu, Finland: Itä-Suomen yliopisto; 2025. [Google Scholar]
212. Singh J, Patel P. Methods for medical device design, regulatory compliance and risk management. J Eng Res Rep. 2024;26(7):373–89. doi:10.9734/jerr/2024/v26i71216. [Google Scholar] [CrossRef]
213. Sah H, Ramesh M, Amrutha C. Regulating software as medical device (SaMDa comprehensive overview of the international medical device regulators forum (IMDRF) framework and gap analysis of Indian medical devices rules, 2017. In: 2025 Emerging Technologies for Intelligent Systems (ETIS); 2025 Feb 7–9; Trivandrum, India. p. 1–6. doi:10.1109/ETIS64005.2025.10961721. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF


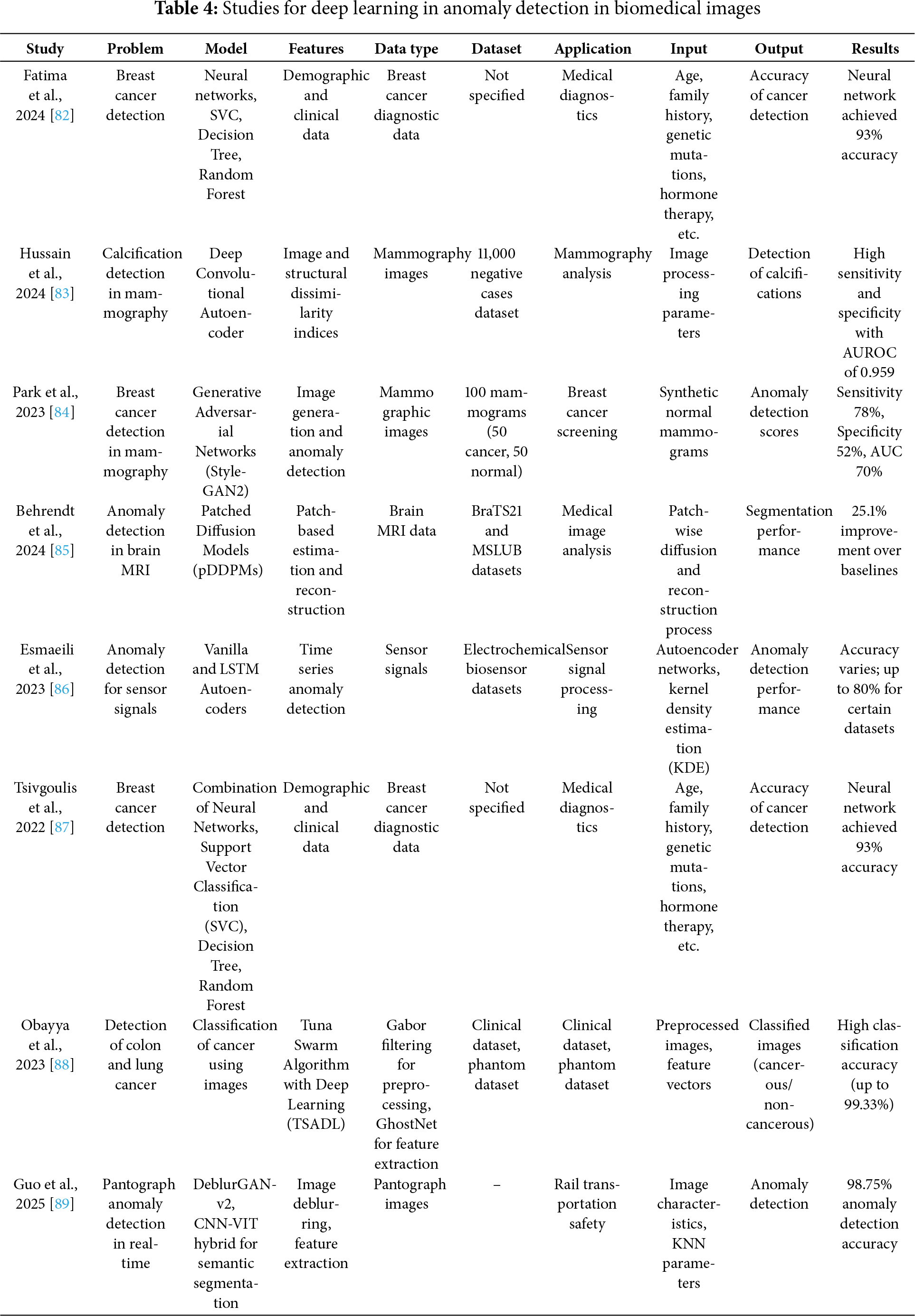
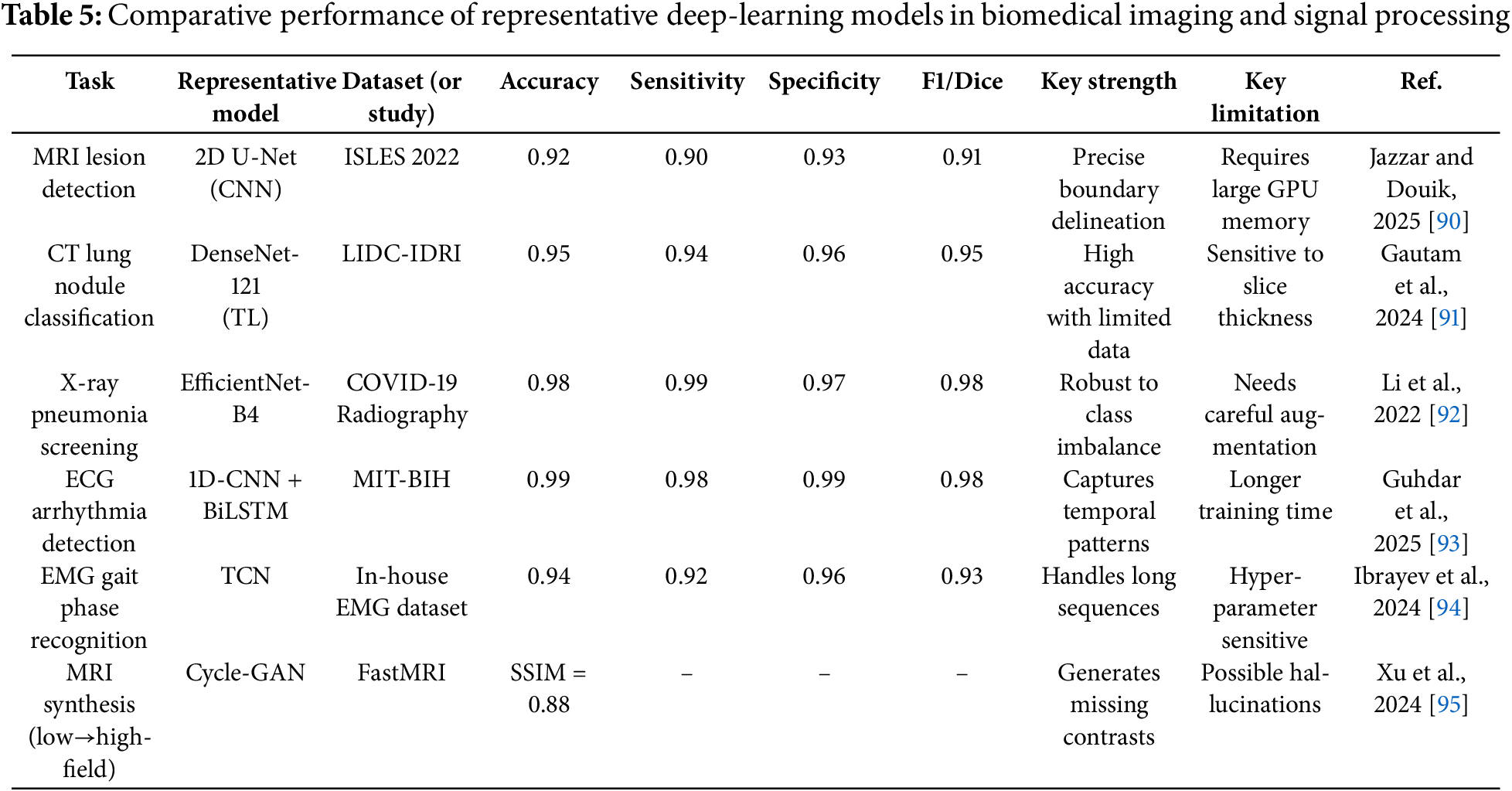
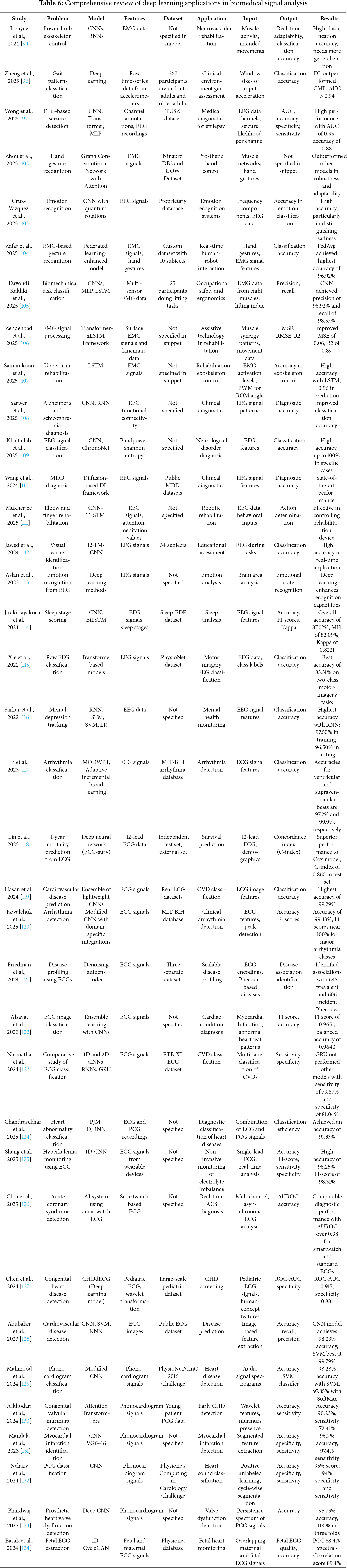
 Downloads
Downloads
 Citation Tools
Citation Tools