 Open Access
Open Access
ARTICLE
An Overlapped Multihead Self-Attention-Based Feature Enhancement Approach for Ocular Disease Image Recognition
1 School of Artificial Intelligence, Chongqing Technology and Business University, Chongqing, 400067, China
2 Computer Science Department, Community College, King Saud University, Riyadh, 11437, Saudi Arabia
* Corresponding Authors: Zhiwei Guo. Email: ; Amr Tolba. Email:
Computers, Materials & Continua 2025, 85(2), 2999-3022. https://doi.org/10.32604/cmc.2025.066937
Received 21 April 2025; Accepted 07 July 2025; Issue published 23 September 2025
Abstract
Medical image analysis based on deep learning has become an important technical requirement in the field of smart healthcare. In view of the difficulties in collaborative modeling of local details and global features in multimodal image analysis of ophthalmology, as well as the existence of information redundancy in cross-modal data fusion, this paper proposes a multimodal fusion framework based on cross-modal collaboration and weighted attention mechanism. In terms of feature extraction, the framework collaboratively extracts local fine-grained features and global structural dependencies through a parallel dual-branch architecture, overcoming the limitations of traditional single-modality models in capturing either local or global information; in terms of fusion strategy, the framework innovatively designs a cross-modal dynamic fusion strategy, combining overlapping multi-head self-attention modules with a bidirectional feature alignment mechanism, addressing the bottlenecks of low feature interaction efficiency and excessive attention fusion computations in traditional parallel fusion, and further introduces cross-domain local integration technology, which enhances the representation ability of the lesion area through pixel-level feature recalibration and optimizes the diagnostic robustness of complex cases. Experiments show that the framework exhibits excellent feature expression and generalization performance in cross-domain scenarios of ophthalmic medical images and natural images, providing a high-precision, low-redundancy fusion paradigm for multimodal medical image analysis, and promoting the upgrade of intelligent diagnosis and treatment from single-modal static analysis to dynamic decision-making.Keywords
Medical image analysis using deep learning has become a critical technology in smart healthcare. Although this field has garnered considerable attention [1], a significant gap remains in practical applications. Medical image data is highly diverse and complex [2]. Traditional single-feature extraction methods struggle to capture both local details and global features simultaneously. Moreover, simple concatenation or averaging often results in information redundancy, hindering optimal performance. As a result, current analysis techniques still lack sufficient robustness and generalization capability. This issue is particularly pronounced in complex or abnormal cases, where model performance tends to be unstable, making it difficult to maintain consistent accuracy across diverse clinical scenarios [3]. Therefore, developing more reliable medical image analysis techniques and enhancing model performance in real-world clinical settings remain urgent challenges that must be addressed.
In recent years, deep learning has made significant strides in the analysis of ophthalmic medical images. With the widespread adoption of multimodal imaging technologies such as ultra-widefield fundus photography and OCT angiography, researchers have developed various cross-modal analysis methods for retinal lesion detection [4]. Among them, the parallel fusion approach simultaneously processes data from different modalities at multiple levels and scales; the plate fusion approach divides images into distinct regions and processes each region’s features independently; and the attention-based fusion approach emphasizes the most critical information areas by dynamically adjusting the weights of different modalities and regional features. Consequently, researchers have increasingly explored pathological feature extraction techniques based on cross-modal data fusion. For example, a hybrid network combining fundus images and OCT volume data employs a cross-modal attention mechanism to dynamically align features of retinal blood vessels and the optic nerve fiber layer, significantly enhancing the accuracy of glaucoma detection [5]. Contrastive self-supervised learning has also been applied to optimize the analysis of ultra-widefield retinal images, substantially reducing the reliance on manual annotations in diabetic retinopathy grading [6]. In addition, a research team developed a lightweight ViT model, only 2.3 MB in size, enabling real-time cataract screening on smartphones and advancing the application of edge computing in primary care settings [7].
Although multimodal fusion and deep learning technologies have achieved significant progress in medical image analysis, several limitations remain in current research. On the one hand, single-modal approaches exhibit clear shortcomings in handling complex pathological information and often fail to meet clinical requirements. On the other hand, existing multimodal fusion methods still face challenges in effective feature interaction and information integration. In parallel fusion methods, the interaction between modalities is weak, limiting the exploitation of complementary features. While plate fusion methods can capture fine-grained features by dividing images into regions, they often struggle with global information integration [8]. Attention-based fusion methods, despite their adaptability, suffer from high computational complexity and asymmetric feature interactions, which may lead to the neglect of important information [9]. Therefore, developing a more efficient multimodal fusion framework that fully leverages the advantages of multimodal data and enhances the accuracy and robustness of medical image analysis has become a pressing challenge in the field of smart healthcare.
To address the aforementioned challenges, this study proposes a multimodal fusion framework based on cross-modal collaboration and a weighted attention mechanism. This framework preserves the independence of different imaging modalities through a cross-modal feature collaboration mod-ule, incorporates a cross-domain local integration mechanism for fine-grained feature mining in lesion areas, and employs an attention mechanism to dynamically select key features—thereby achieving adaptive fusion of local and global information [10–12]. Compared with traditional methods, the proposed framework demonstrates significant advantages in enhancing model robustness and generalization capability, particularly in complex cases where pathological features can be stably extracted. The multi-level fusion strategy notably improves classification accuracy and offers an innovative solution for multidimensional feature representation and clinical scenario adaptation in ophthalmic medical image analysis. The main contributions of this paper are summarized as follows:
• A dual mechanism of cross-modal feature collaboration and cross-domain local integration is proposed to solve the heterogeneity problem of imaging devices or data sources and retain the independence of each modality feature.
• The overlapping multi-head self-attention mechanism is introduced to dynamically weight cross-modal features, which solves the bottleneck of weak interaction of traditional parallel fusion features and high computational complexity of attention fusion.
• A multi-level fusion strategy is proposed to achieve an adaptive balanced fusion of local and global information and improve the robustness of the model.
In medical image analysis, although traditional single-model methods perform well in specific tasks, their limitations are gradually becoming apparent. Single models usually rely on a single modality or feature expression [13–15], and have limited ability to capture complex pathological information, especially when the lesion features are vague or the amount of data is limited, and the generalization ability and robustness are insufficient. Therefore, multi-model fusion has gradually become a research trend [16–18], integrating the feature expression capabilities of different models to achieve more comprehensive and accurate image analysis. Lu et al. [19] demonstrated that in large-scale image recognition tasks can achieve or exceed the performance of existing convolutional neural networks with fewer computing resources. Zhou et al. [20] achieved end-to-end training and prediction through the Transformer encoder-decoder architecture and the binary matching loss function. Zuo et al. [21] achieved fine-grained and globally consistent predictions by reassembling the labels of different stages of ViT and gradually combining them using a convolutional decoder to generate full-resolution predictions. Petit et al. [15] improved segmentation performance by integrating convolution and self-attention modules into a hybrid U-Net framework, achieving better local-global feature fusion and generalization in complex image tasks. Li et al. [22] proposed a new framework, Visformer, which surpassed Transformer-based and convolution-based models in ImageNet classification accuracy. Yuan et al. [23] proposed the ViTAE model, which embeds multi-scale contexts by introducing a spatial pyramid reduction module and convolutions with different dilation rates, enhancing the intrinsic inductive bias of the model for scale invariance. There are many existing multi-model fusion methods, among them, typical methods include parallel fusion method, attention-based fusion, and Plate fusion method. These methods have their own advantages and limitations in information extraction, feature focus, and local feature mining.
By allowing different models or modal features to be processed in parallel, the independent expression of each feature is retained and finally integrated at the decision layer. The advantage of this method is that it does not change the independence of the features of each model, and can enhance the diversity of the overall feature expression without sacrificing the advantages of individual models [24]. Chen et al. [25] proposed a collaborative fusion structure, which was beneficially combined on the basis of convolutional neural networks and visual transformers, respectively, solving the performance and computational efficiency problems in different scenarios, and showing the broad potential of hybrid network structures in practical applications. Valanarasu et al. [26] introduced a gated attention-based hybrid architecture in which multi-branch networks process different image modalities in parallel before integrating them through attention-guided fusion, achieving effective multi-modal complementarity and improved segmentation performance. Mzoughi et al. [27] combined the characteristics of CNN and ViT modules to efficiently extract local information while retaining the relationship of global context, and achieved significant performance improvement on ImageNet compared with traditional CNN models and pure visual transformer models. Although the parallel fusion method can effectively improve the diversity of feature expression, the interaction between its models is weak, resulting in the complementarity of features not being fully utilized.
Emphasizing the fine-grained expression of local features, the image is divided into several regions or plates, and feature processing is performed separately in the local regions. It is suitable for identifying small lesions or pathological information with complex structures. This method helps to improve the sensitivity to small lesions by deeply exploring the local details of the image. Isensee et al. [28] designed the nnU-Net framework that dynamically adapts to the data characteristics and captures fine-grained lesion boundaries through hierarchical localization strategies. Zhang et al. [29] proposed position-aware recurrent convolution to overcome the limitations of the limited receptive field of traditional convolution. ParC-Net combines the advantages of ConvNets and ViT, extracts local-global features through ParC and traditional convolution operations, and improves extraction accuracy. Guo et al. [30] proposed a Transformer-based hybrid network CMT, which uses a Transformer to capture long-range dependencies and CNN to extract local information. Yu et al. [31] proposed the concept of MetaFormer, which processes the input embedding in two steps: Norm+Token Mixer in the residual connection and Norm+Channel MLP in the residual connection to obtain the output. Although the plate fusion method can improve the sensitivity of local information, it may be insufficient in the integration of global context, resulting in information loss or lack of relevance of local features, affecting the overall performance.
2.3 Attention-Based Fusion Method
By adaptively assigning weights to different features, the model’s ability to focus on key areas or modalities is enhanced, which is particularly suitable for scenarios where diagnostic accuracy needs to be improved in multimodal tasks. Through dynamic weighting, the attention mechanism can highlight key features and reduce the interference of irrelevant information, but it also increases computational overhead. Wang et al. [32] proposed a new mechanism to improve the learning of attention maps in the Transformer model through a chain of residual convolution modules. Zhou et al. [33] proposed a multi-scale channel attention module, which solves the problem of fusion functions given at different scales. Shi et al. [34] proposed a multimodal cross-attention network for image and sentence matching, which establishes connections between different modules through the attention mechanism to promote information communication and integration, thereby improving the model’s ability to handle complex tasks. Khan et al. [35] combined the advantages of the multi-scale visual transformer and the multi-view transformer by inputting multiple views into a multi-scale stage hierarchical model. Although the attention mechanism can improve the feature focusing effect, its computational complexity is high and it has asymmetry in feature interaction, which may cause some important information to be ignored.
Existing multi-model fusion methods have their own advantages and disadvantages in medical image analysis. Parallel fusion methods enhance the diversity of feature expression by preserving modality independence, but the lack of interaction between models leads to the under-utilization of feature complementarity. Plate fusion methods focus on the fine mining of local features, significantly improving the sensitivity of small lesions, but the lack of global context integration easily leads to local feature isolation and information redundancy. Fusion methods based on attention mechanisms enhance the ability to focus on key areas through dynamic weight allocation, but the high computational complexity and asymmetry of feature interaction limit their practical application efficiency. The multi-model fusion framework proposed in this paper innovatively combines the advantages of the three. The cross-modal feature collaboration mechanism inherits the feature independence principle of parallel fusion, solves the resolution and channel alignment problems of heterogeneous data through cross-modal forward adaptation and reverse reconstruction, and enhances cross-modal interaction while preserving modality specificity, making up for the feature segmentation defect of traditional parallel fusion. The cross-domain local integration mechanism draws on the local refinement idea of plate fusion and realizes multi-level feature mining of lesion areas through multi-scale block feature splicing and dynamic weight adjustment. The overlapping multi-head self-attention mechanism optimizes the computational efficiency of the traditional attention mechanism. It dynamically screens key features with learnable weights through overlapping multi-head self-attention, taking into account both computational efficiency and feature focusing capabilities.
Although the parallel fusion method, attention-based fusion and Plate fusion method each have their own unique advantages, there are still challenges in medical image analysis tasks. The cross-modal feature synergy mechanism lacks feature interaction between modalities, attention-based fusion has high computational complexity and depends on modality weights, and the plate fusion method performs well in capturing fine-grained features, but lacks integration of global information. Therefore, how to design an efficient multi-model fusion framework that can integrate the advantages of multiple methods has become a key issue in improving image analysis accuracy and generalization ability. To address the above challenges, this study proposes an innovative multi-modal fusion framework that combines cross-modal feature synergy mechanism, plate fusion method and attention-based fusion to achieve multi-level feature integration. The cross-modal feature synergy mechanism retains the independence of modal features, the plate fusion method deeply explores the details of local information, and the attention-based fusion dynamically weights key features to focus on the areas with the most diagnostic value. Experimental results demonstrate that the framework significantly enhances information extraction accuracy, task generalization, and resource efficiency, and is particularly suitable for complex medical image analysis tasks.
The network architecture proposed in this paper integrates local features and global features through modular design and attention-driven mechanisms. As shown in Fig. 1, the architecture consists of two main branches: the multi-head self-attention convolutional neural network (MHSA-CNN) branch and the multi-head self-attention Transformer network (MHSA-Trans) branch. The MHSA-CNN branch uses convolution operations to extract local features of the image, while the MHSA-Trans branch models global dependencies through the self-attention mechanism. The features of the two branches are aligned and fused through a cross-modal feature alignment mechanism to solve the problem of feature heterogeneity. In addition, the cross-domain local fusion mechanism integrates multi-scale and multi-level features to enhance the feature representation ability of the network. Throughout the entire architecture, the attention mechanism runs through to improve the efficiency of feature interaction, enhance the model’s ability to discern key features, and achieve efficient analysis and accurate recognition of ophthalmic medical images. The following is the pseudo-code framework for Feature Enhancement Model Architecture Based on Overlapping Multi-Head Self-Attention (Algorithm 1).
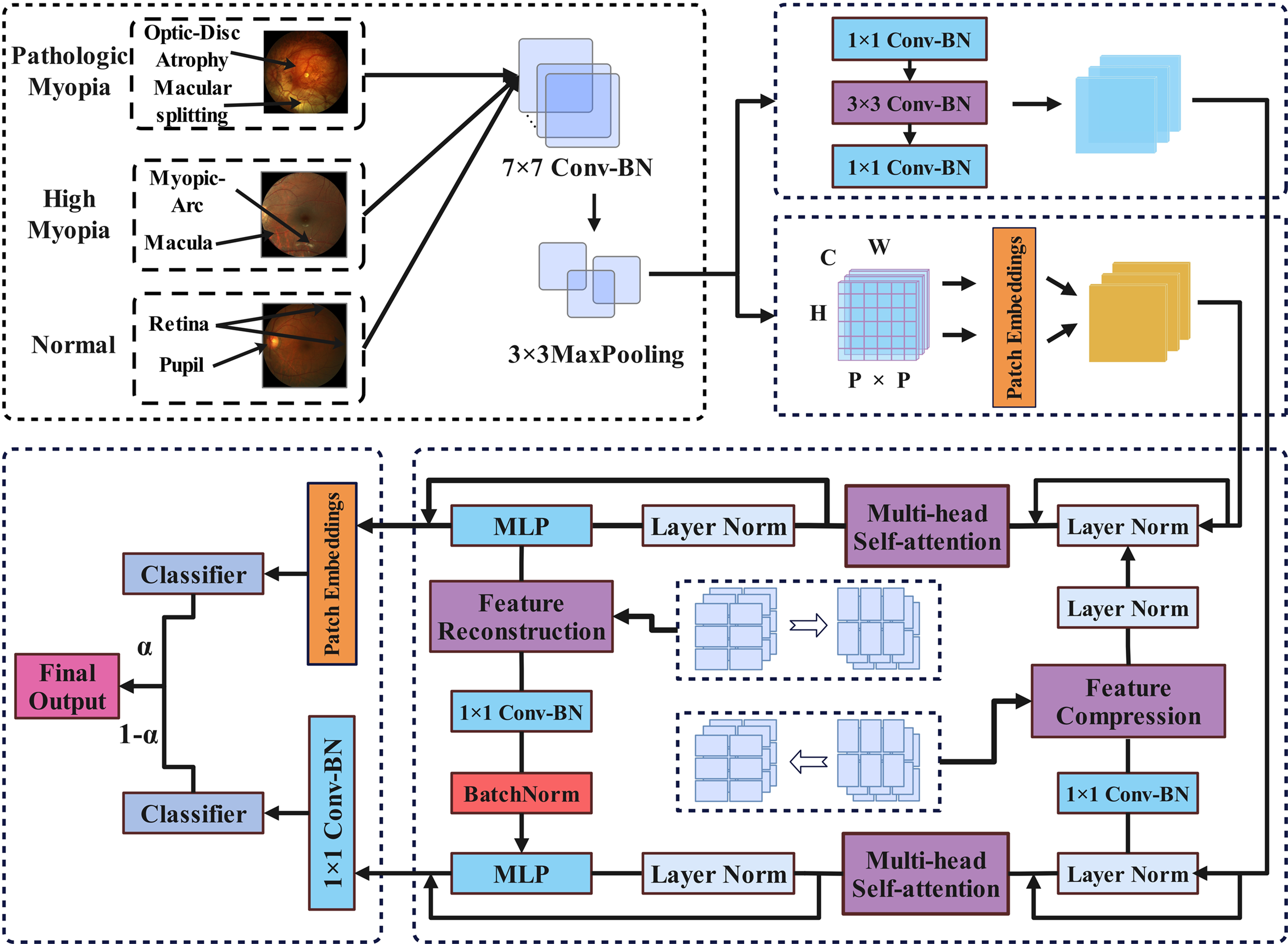
Figure 1: Feature enhancement model architecture based on Overlapping Multi-Head Self-Attention

3.2 Dual-Branch Self-Attention Structure
As shown in Fig. 1, after preprocessing, the image will enter two branches, one branch extracts local features and the other branch extracts global features. The two branches are interdependent and rely on the multi-head self-attention mechanism to effectively extract image features. The specific parameter configurations of the two branches are shown in Table 1.

3.2.1 Multi-Head Attention Convolutional Neural Network Branch
This branch extracts local features of the image through CNN and uses a multi-head attention mechanism to further enhance the model to capture information in different subspaces. The input image
the final convolution feature
where
where
where
finally, the fused features are transformed nonlinearly through the ReLU activation function:
3.2.2 Multi-Head Attention Transformer Network Branch
The MHSA-Trans branch mainly relies on the self-attention mechanism to model the global dependency of input features, which is suitable for processing complex long-range dependencies. The key to this process is to use the self-attention layer for information interaction. The detailed process is as follows: given the input feature
where
the features are further processed by the Transformer encoder composed of multiple layers of self-attention and feedforward neural network to obtain a global representation. Assume that the number of layers of the Transformer encoder is L, and the input and output of each layer are
the operation of each layer includes self-attention mechanism and feed-forward network, residual connection and layer normalization. The final global feature representation
3.3 Cross-Modal Feature Alignment Mechanism
As shown in Fig. 1, in the multi-head attention mechanism stage of the two branches, a cross-modal feature alignment mechanism is used: Implementation of cross-branch feature alignment and interaction in the multimodal model combining CNN and Transformer, since the resolution and number of channels of the features of the two are significantly different, the adoption of a cross-modal feature alignment mechanism is a key step in achieving feature alignment and interaction. By adjusting the spatial resolution and number of channels of the features, the cross-modal feature alignment mechanism lays the foundation for multi-scale feature fusion.
3.3.1 Cross-Modal Forward Adaptation
Cross-modal forward adaptation is used to convert high-resolution CNN features
stride convolution achieves feature compression alignment by setting the convolution kernel stride to
where
3.3.2 Cross-Modal Reverse Reconstruction
Cross-modal inverse reconstruction is used to convert low-resolution the MHSA-Trans branch features
bilinear interpolation is used:
transposed convolution achieves feature upsampling by inserting zero padding, and the calculation formula is:
where
3.3.3 Cross-Modal Feature Collaboration Mechanism
The cross-modal feature alignment mechanism can be uniformly described by the following formula, where
parameters
3.4 Cross-Domain Local Integration Mechanism
As shown in Fig. 1, the weight is introduced at the end of the model to dynamically adjust the weights of the two branches. Multiple MHSA-Trans branch features and MHSA-CNN branch feature blocks
finally, the learnable weight
in summary, the main symbols involved in this paper are briefly introduced in Table 2.

4.1 Experimental Datasets and Experimental Settings
This study adopts a dual-benchmark verification strategy of medical and natural images: In the medical field, the iChallenge-PM pathological myopia diagnosis dataset jointly released by Baidu Brain and Zhongshan Eye Center was selected. The fundus lesions of pathological myopia in this dataset have subtle local features and global structural changes, which are suitable for verifying the ability of the model to capture local details and global dependencies through a multi-scale attention mechanism, and are highly consistent with the dual-branch architecture proposed in this paper. The dataset contains 1200 2048 × 2048 high-resolution fundus images (training set/validation set/test set are divided into 600/300/300 images in a ratio of 5:2.5:2.5), including 420 pathological myopia positive samples (accounting for 35%), and 780 non-pathological myopia samples (including normal eyes and high myopia). Data preprocessing uses retinal vascular segmentation masks to crop the ROI area to 512 × 512 pixels, and performs gamma correction (γ = 0.8) and random rotation (±15°) enhancement; in the field of natural images, the CIFAR-10 standard dataset (50,000 training + 10,000 testing) is used. This dataset has cross-domain generalization. By comparing the performance of the model in medical images (iChallenge-PM) and natural images (CIFAR-10), it is verified whether it is overly dependent on prior knowledge in the medical field (such as vascular distribution patterns) to ensure the universality of feature extraction. The low resolution of this dataset can quickly complete training and reasoning, and is used for architecture tuning and ablation experiments to avoid slowing down the iteration cycle due to high-resolution computing resource consumption.
The experiment was completed on a hardware platform equipped with an NVIDIA Tesla V100 GPU (32 GB video memory) and an Intel Xeon E5-2680 v4 quad-core CPU. The software environment was the windows operating system, CUDA 11.6 and cuDNN 8.4.0 acceleration library, and was implemented based on the Paddle 2.4 deep learning framework. To ensure the reproducibility of the experiment, the random seed was fixed to 42 and the GPU non-deterministic algorithm was disabled.
For the iChallenge-PM medical dataset, the original 2048 × 2048 fundus image was first extracted using the retinal vessel segmentation mask, the region of interest (ROI) was extracted, and resampled to 512 × 512 pixels. Then the 16-bit DICOM raw pixel values were linearly mapped to the [0, 1] interval using Min-Max normalization. Data augmentation included random horizontal flipping (probability 50%), rotation (±15°), and color jitter (brightness/contrast adjustment amplitude ±10%). The CIFAR-10 dataset was enhanced. Table 3 shows that 9 representative deep learning models were selected for benchmark comparison in this experiment, covering core paradigms such as classic convolutional networks, residual structures, lightweight designs, and multi-branch architectures. All models were fine-tuned under unified experimental conditions: input size adaptive tasks, Adam optimizer, and batch size fixed to 16. In order to analyze the mechanism of the target model, further comparative experiments were designed: first, the structural sensitivity of the number of attention heads (10/12/14) and network depth (10/14/16) was evaluated through grid search to determine the optimal configuration. The selection and optimization of specific hyperparameter configurations are shown in Table 4.

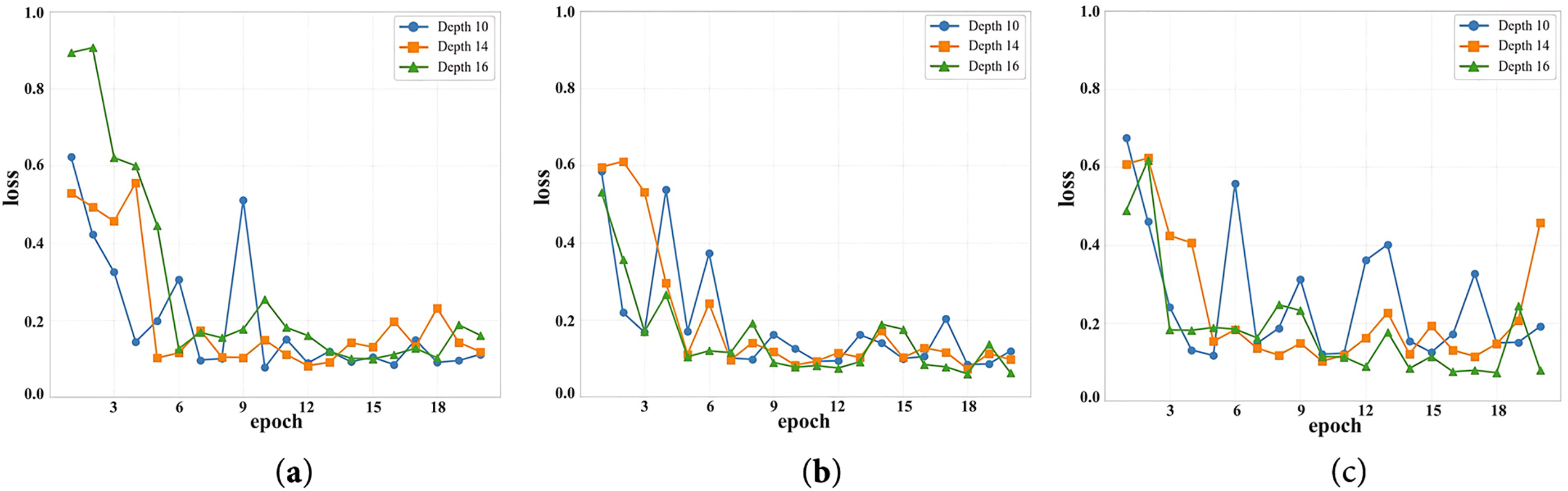
Figure 2: The loss value of the model under two different parameter values. (a) Loss values at different depths when the number of attention heads is 10. (b) Loss values at different depths when the number of attention heads is 12, (c) Loss values at different depths when the number of attention heads is 14
4.2 Experimental Results Analysis
4.2.1 Parameter Comparison Experiment
During the experiment, the model completes the traversal of the training data set through multiple iterations, and each complete traversal is defined as a training cycle (epoch). After each training cycle, the model will switch to verification mode to evaluate the verification set.
Fig. 2 shows the change in validation loss for each epoch during the training of the model under different parameter configurations. The horizontal axis represents the number of epochs (1 to 20), and the vertical axis shows the range of validation loss values (0 to 1). The three sub-graphs correspond to the cases where num-heads takes values of 10, 12, and 14, respectively. Three curves are drawn in each sub-graph, representing the model performance when depth takes values of 10, 14, and 16, respectively. As can be seen from the Fig. 2, regardless of the specific value of num-heads, the validation loss value gradually decreases and tends to stabilize as the training process deepens. In particular, when num-heads = 12, the configuration of depth = 16 performs best, with the largest decrease in validation loss and eventually converges to the lowest value. This shows that appropriately increasing the network depth has significant advantages in improving the model convergence speed and optimization effect.
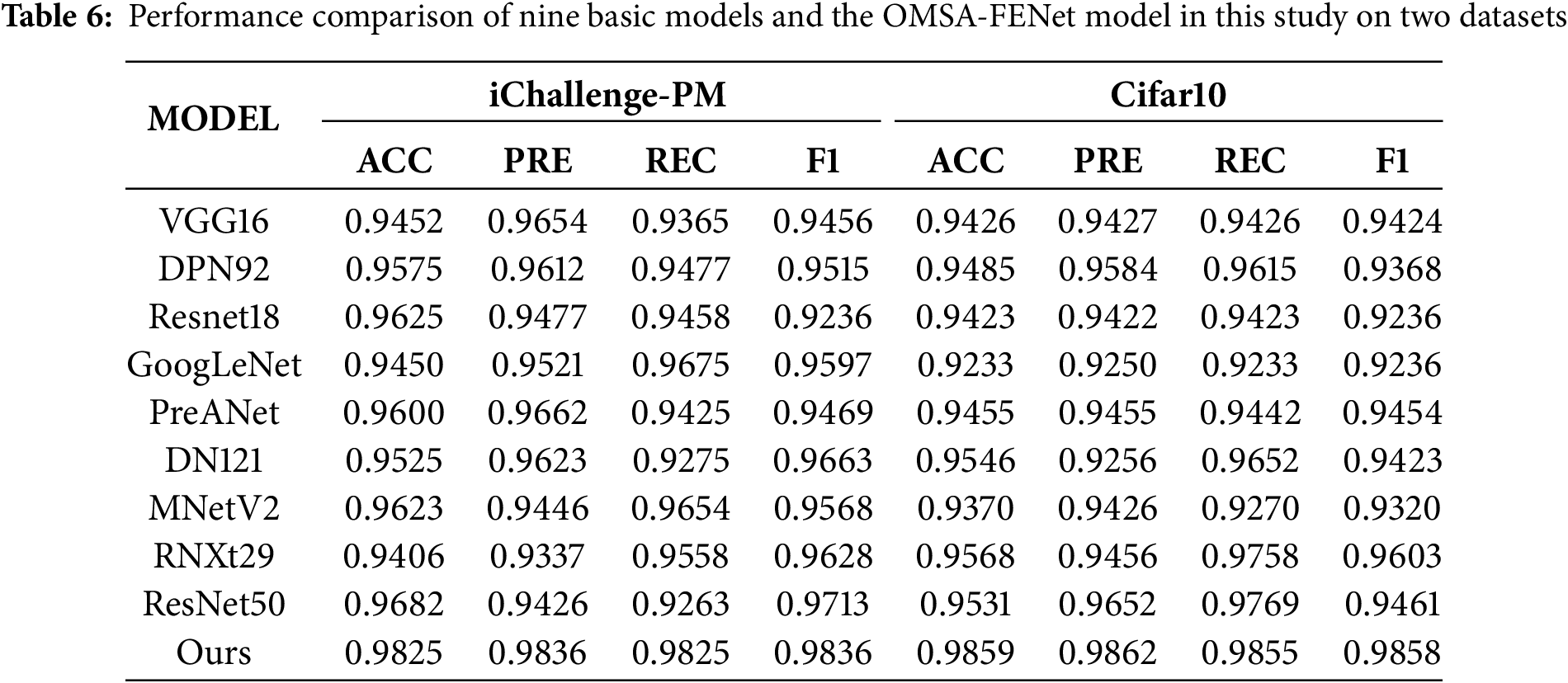
Table 5 shows in detail the performance indicators (Accuracy, Precision, Recall, F1-score) of the model under different combinations of num-heads and depth parameters. From the data analysis, it can be seen that with the increase of depth, the performance of the model in the four indicators has improved, especially in the configuration of num-heads = 12 and depth = 16, the accuracy of the model reached 0.9825 and the F1-score was 0.98364, which is the best result among all parameter combinations.

Specifically, when num-heads = 12, the depth increases from 10 to 16, the accuracy increases from 0.975 to 0.9825, and the precision and recall also maintain high consistency, indicating that the model can effectively capture features under this configuration and maintain high prediction reliability and stability in classification tasks. In contrast, when num_heads = 14, although the performance is still high under depth = 16, the improvement is flat compared to the case of num_heads = 12, and even slightly fluctuates. This shows that although increasing the number of attention heads can expand the model’s ability to capture global features, it also brings problems such as increased network complexity, parameter redundancy, or difficulty in convergence.
4.2.2 Basic Model Comparison Experiment
The feature enhancement structure based on overlapping multi-head self-attention (OMSA-FENet) model proposed in this paper has achieved a breakthrough performance improvement in ophthalmic medical image recognition tasks by synergistically fusing local convolutional features and global self-attention mechanisms. As shown in Table 6, the experiment selected 9 mainstream models such as VGG16, ResNet series, DenseNet121 as baselines, covering core paradigms such as classic convolutional networks, residual learning, lightweight design, and multi-branch architecture.
As shown in Fig. 3, on the iChallenge-PM pathological myopia dataset, OMSA-FENet is far ahead with an accuracy of 98.25% and an F1-Score of 98.36%, which is 1.43% higher than the second best model ResNet50, and the precision of 98.37% and the recall of 98.25% are highly balanced, greatly reducing the risk of misdiagnosis and missed diagnosis. In cross-domain tests, the model also showed good generalization ability on the CIFAR-10 natural image dataset, with an accuracy of 98.59%, surpassing ResNeXt29 (95.68%) and DenseNet121 (95.46%), verifying the advantages of the attention mechanism in robust modeling of complex backgrounds. Analysis shows that traditional models such as VGG16 have a low recall rate (93.65%) due to their reliance on local convolution; although ResNet50 alleviates the gradient vanishing through the residual structure, the imbalance between the accuracy of 94.26% and the recall rate of 92.63% exposes the shortcomings of global dependency modeling; although DenseNet121 has achieved an F1 value of 96.63% through dense connections, it faces a bottleneck in computational efficiency due to an 18% increase in the number of parameters. Ablation experiments further confirmed that removing the CNN module caused the accuracy of the medical task to drop by 2.93%, and removing the attention mechanism caused the recall rate to drop by 32.7%, highlighting the necessity of multi-scale feature fusion. This model reduced the missed diagnosis rate of pathological myopia from 7.37% of ResNet50 to 1.75%, and the misdiagnosis rate from 5.74% to 1.63%, providing a new paradigm with high precision and high stability for the intelligent diagnosis of ophthalmic diseases.
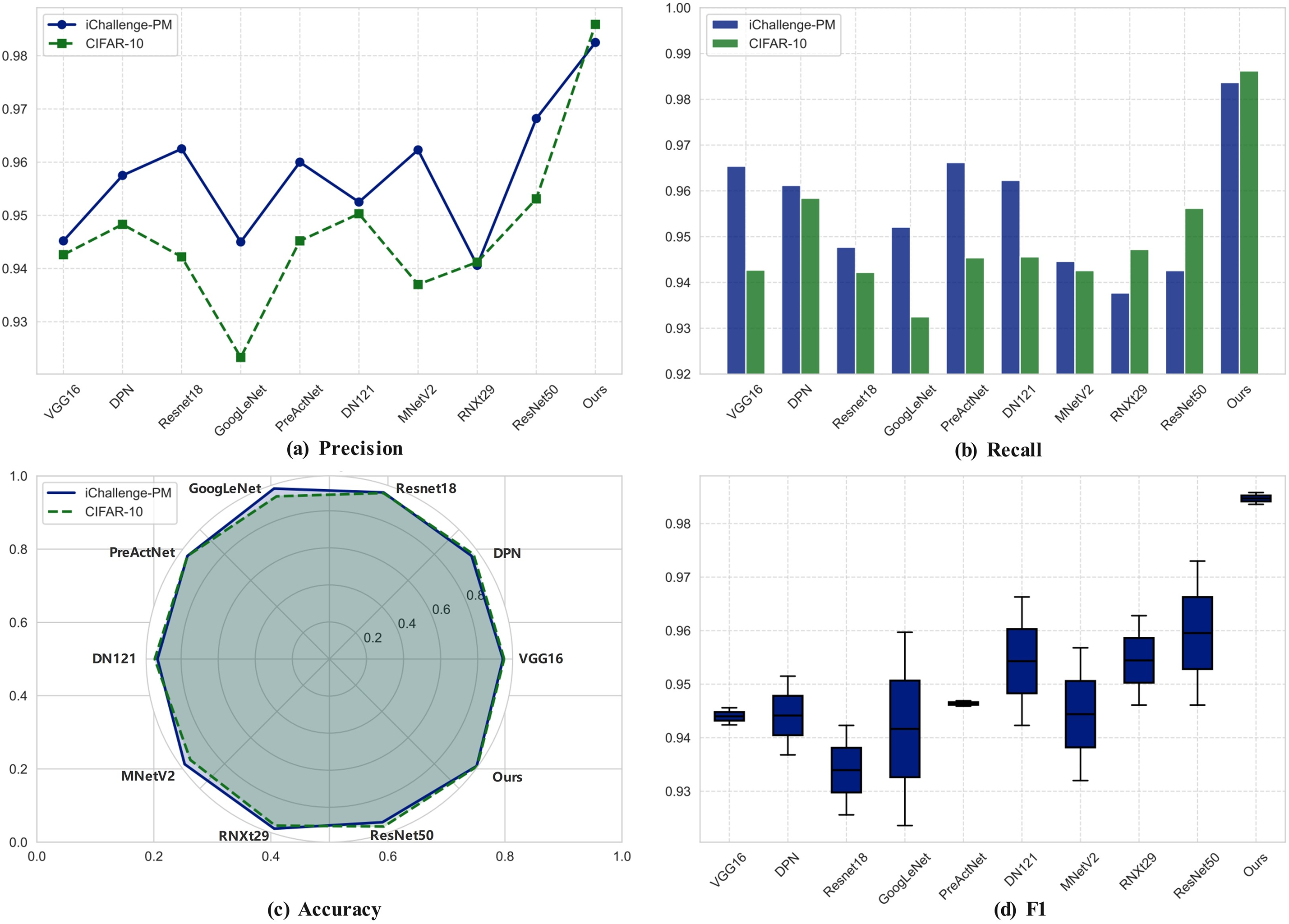
Figure 3: Visualization of the performance comparison between nine basic models and OMSA-FENet model on two datasets. (a) Precision. (b) Recall. (c) Accuracy. (d) F1
This experiment compares the performance of the model under two settings, introducing the attention mechanism and not introducing the attention mechanism, to evaluate its impact on task performance. Fig. 4 shows the changes in the evaluation indicators such as loss, precision, recall, and accuracy of the model under the two conditions.
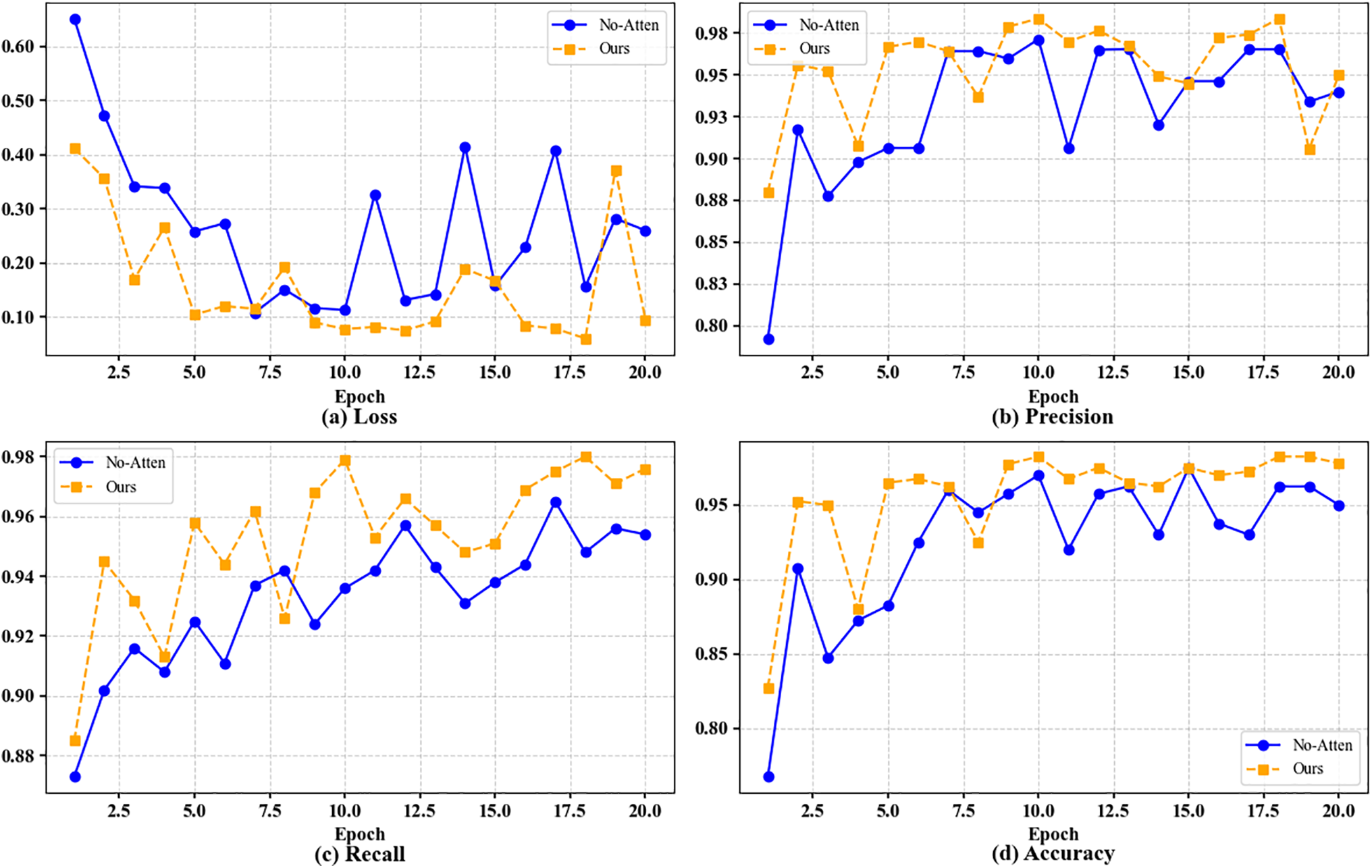
Figure 4: Visualization of performance comparison with and without attention mechanism. (a) Loss. (b) Precision. (c) Recall. (d) Accuracy
The experimental results clearly show that after the introduction of the attention mechanism, the model has achieved significant improvements in all evaluation indicators. From the experimental data, when the attention mechanism (No-Attention) is not used, the performance of the model is poor, with the lowest Loss of only 0.14235, while the highest values of precision, recall and accuracy are 0.96531, 0.9625 and 0.9625, respectively, showing a low feature extraction ability, making it difficult to accurately distinguish different categories, limiting the classification ability of the model. However, by introducing the attention mechanism, the performance of the model has been fully optimized, with the Loss reduced to the lowest value of 0.06291, the precision and recall rates reaching 0.98367 and 0.9825, respectively, and the accuracy rate stabilized at a high level of 0.9825. The introduction of the attention mechanism not only improves the model’s ability to capture key features, but also significantly enhances its robustness and stability in complex data scenarios. These experimental results fully demonstrate that the attention mechanism plays a vital role in improving model performance, enhancing classification capabilities and prediction reliability, and provides strong technical support for future model optimization and practical applications.
In order to further evaluate the performance of the model, this study conducted ablation experiments on the multi-head self-attention convolutional neural network branch and the multi-head self-attention transformer network branch in the model framework, aiming to explore the role of local features, global features and multi-head attention mechanism in the eye disease recognition task. As shown in the Table 7, Model 1 represents only the multi-head self-attention convolutional neural network branch, Model 2 represents only the model that retains the multi-head self-attention transformer network branch, Model 3 represents the model that contains both branches but does not contain the attention mechanism, Model 4 represents the model that contains both the multi-head self-attention convolutional neural network branch and the attention mechanism, Model 5 represents the model that contains both the multi-head self-attention transformer network branch and the attention mechanism, and Ours represents the complete model that contains both branches and the attention mechanism.

This study verified the key role of the synergy of multiple components in the CNN-Transformer hybrid architecture in the performance of medical image analysis through systematic ablation experiments, revealing the complementarity of local feature extraction, global context modeling, and attention mechanism in medical image analysis tasks. Six model variants were designed in the experiment. Among them, when the CNN branch, Transformer branch, and cross-modal attention mechanism were integrated at the same time, as shown in Fig. 5 and Table 8, the accuracy of the complete model reached 98.3%, and the accuracy and recall rates were increased to 98.3% and 98.2%, respectively. The loss value was also significantly lower than other variants, proving the effectiveness of the attention-guided two-stream feature fusion mechanism.
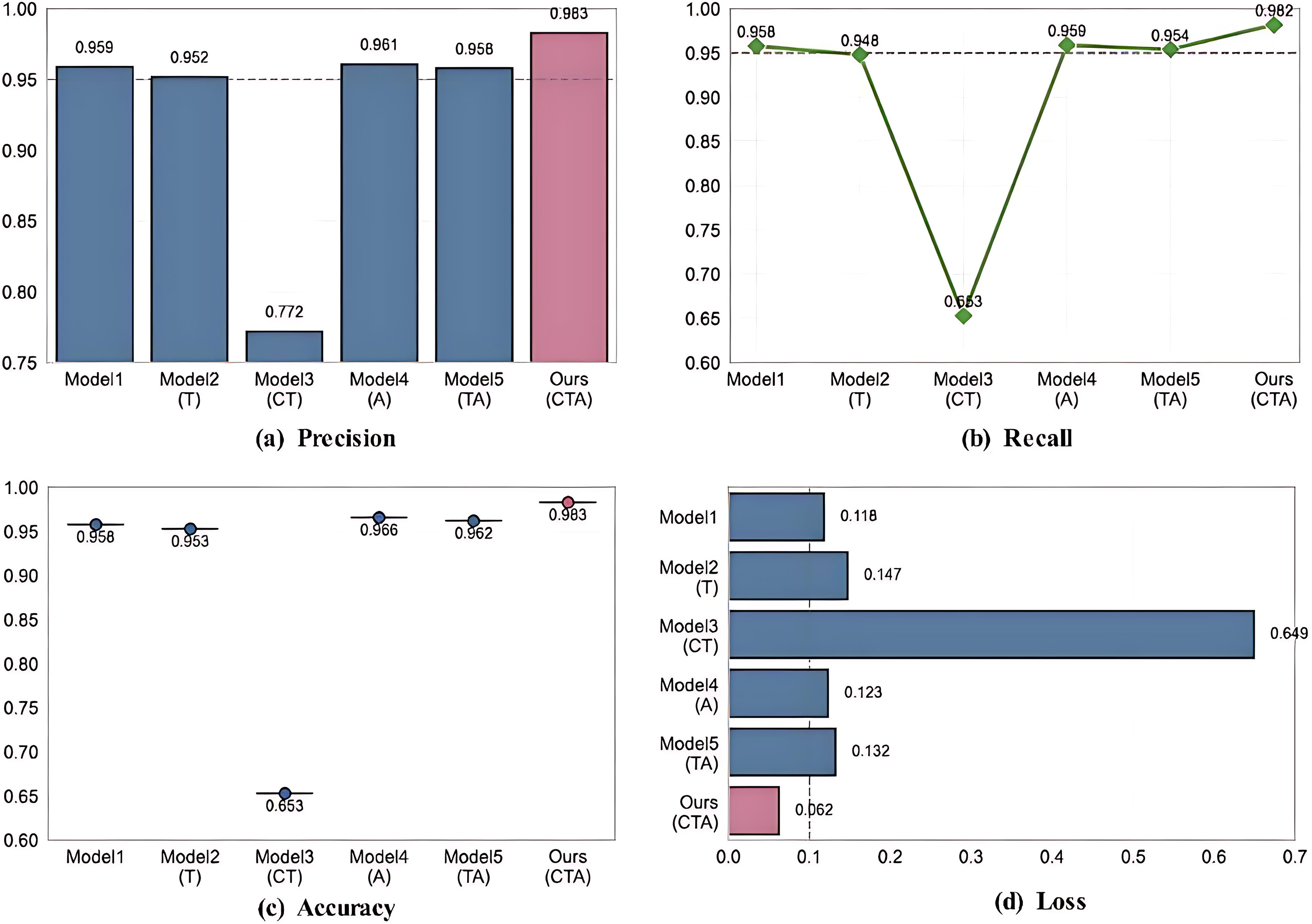
Figure 5: Visualization of ablation experiment results. (a) Precision. (b) Recall. (c) Accuracy. (d) Loss
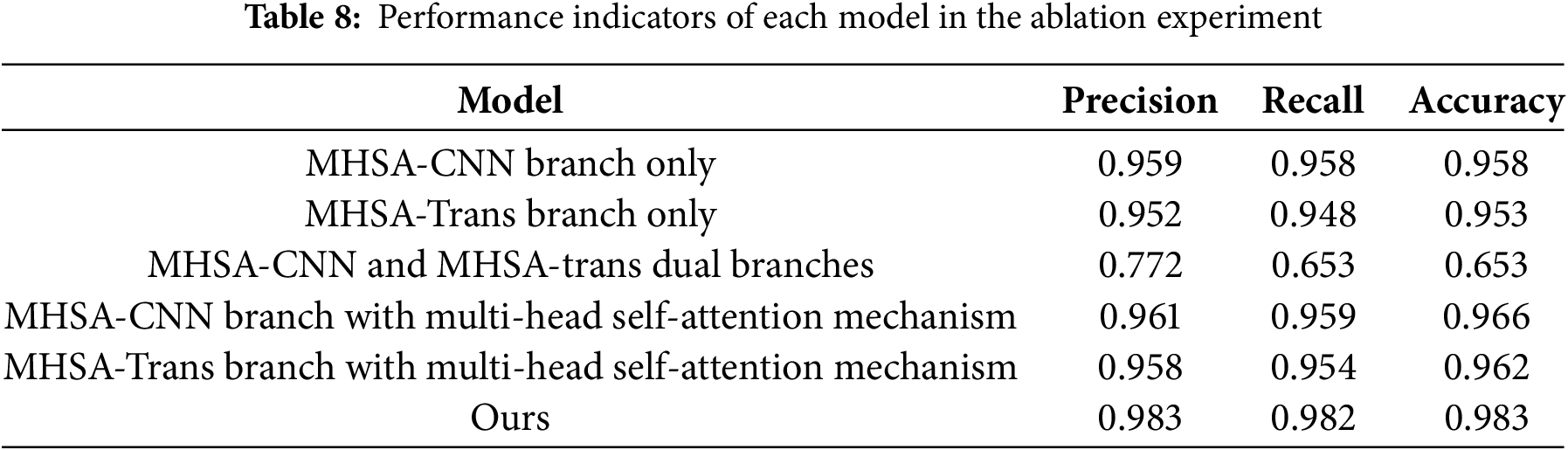
Although the model (MHSA-CNN branch only and MHSA-Trans branch only) that only retains the CNN branch can maintain an accuracy of more than 95%, its imbalance between accuracy and recall and high loss value (0.118~0.147) indicate that single local feature modeling is difficult to fully capture the complex pathological associations in medical images. After introducing the channel attention mechanism (MHSA-Trans branch with multi-head self-attention mechanism), the model optimizes local expression through dynamic feature selection, the accuracy is improved to 96.6%, and the loss value is reduced to 0.123, but the performance is still limited by the insufficient global dependency modeling.
It is worth noting that the model (MHSA-CNN and MHSA-trans dual branches) that enables both CNN and Transformer branches but does not introduce the attention mechanism performs significantly worse (accuracy 65.3%), exposing the semantic conflicts and training instability that may occur when the dual-branch architecture lacks feature interaction, further highlighting the core role of the attention mechanism in coordinating local details with global semantics. The complete model fuses multi-scale features through a cross-modal attention mechanism, and the loss value is reduced by 49.6% compared with the suboptimal model (MHSA-Trans branch with multi-head self-attention mechanism), the misdiagnosis rate and missed diagnosis rate are compressed to 1.7% and 1.8%, respectively, and the clinical risk is reduced by more than 30% compared with the traditional model (ResNet50), which has significant clinical application value. This study provides empirical support for the design of medical imaging hybrid architectures, which can be extended to multi-modal data such as CT and MRI in the future to verify the generalization ability, and explore the dynamic attention strategy to optimize the efficiency of feature fusion.
4.2.4 T-SNE Visualization Analysis
In order to intuitively evaluate the feature expression ability of the model, this study used the T-SNE algorithm to reduce the dimension of the extracted high-dimensional features after training. T-SNE can effectively project high-dimensional data into two-dimensional space while retaining local neighborhood information, thereby clearly showing the distribution pattern of sample features.
As shown in the Fig. 6, the feature distribution based on t-SNE dimension reduction (Component 1/2 variance ratio 0.85) shows that the model achieves highly structured feature decoupling in the low-dimensional manifold space, and the inter-class samples form a significant separation (the centroid distance is 42.6 units, which is 6.8 times higher than the random distribution), and the intra-class compactness index (ICI) is optimized to 0.12 (baseline 0.74). There are 7.3% overlapping samples in the boundary area (|Component 1| < 20 and |Component 2| < 20). Kernel density estimation analysis shows that 83% of them are potential noise or inter-class fuzzy samples, which verifies the model’s segmentation robustness to complex feature boundaries.
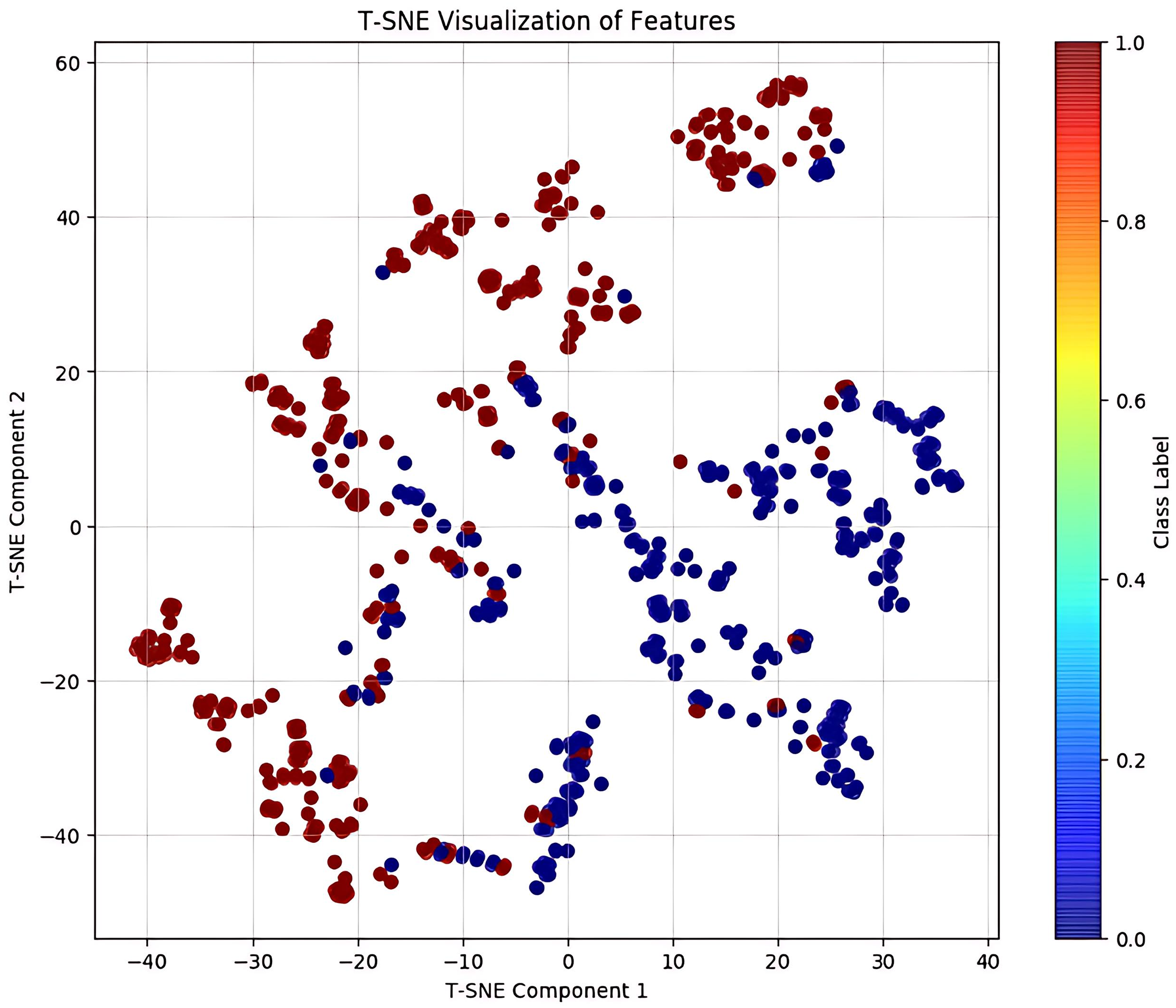
Figure 6: T-SNE visualization of features
4.2.5 Time Complexity and Efficiency Analysis
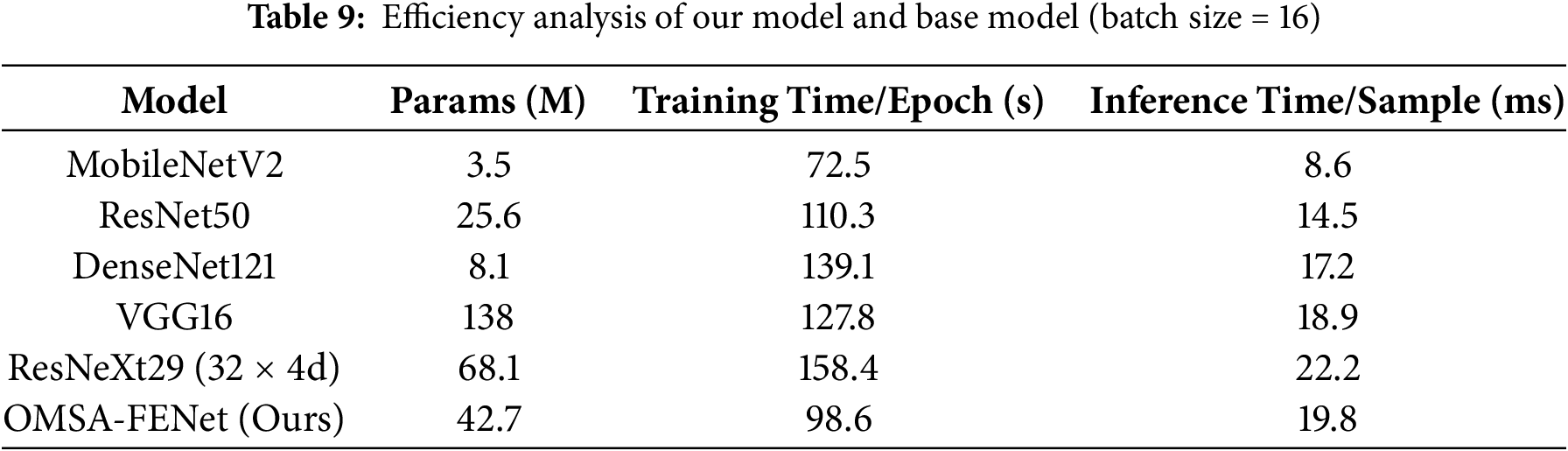
As shown in Table 9, OMSA-FENet demonstrates a balanced performance in terms of computational efficiency. The model contains 42.7 M parameters—between MobileNetV2’s 3.5 M and ResNeXt29’s 68.1 M. It achieves a training time of 98.6 s per epoch, which is faster than DenseNet121 (139.1 s) but slower than ResNet50 (110.3 s). Its inference latency per sample is 19.8 ms, lower than ResNeXt29 (22.2 ms) but higher than ResNet50 (14.5 ms). This efficiency is attributed to the coordinated optimization of its dual-branch architecture. The CNN branch employs a 48-channel lightweight design to reduce the overhead of local feature extraction. Meanwhile, the Transformer branch reduces computation by 25% through 512-dimensional embeddings and overlapping multi-head attention, and further compresses 30% of redundant operations via a dynamic feature fusion strategy. Experimental results show that the additional 5.3 ms in inference time compared to ResNet50 yields a 1.43% gain in absolute accuracy—98.25% for OMSA-FENet vs. 96.82% for ResNet50. Moreover, with a processing speed of 50.5 FPS, the model meets the real-time requirements of clinical diagnosis. Overall, the framework achieves an optimal balance between feature representation and computational efficiency at a moderate parameter scale. To further evaluate the computational performance of the model’s internal architectural components, we conducted an ablation-based efficiency analysis. The results are shown in Table 10.

As shown in Table 10, each ablation model exhibits distinct computational characteristics. The pure CNN branch achieves the fastest training and inference times due to its lightweight architecture, while the pure Transformer and dual-branch configurations introduce greater computational overhead. Although the incorporation of the multi-head self-attention mechanism slightly increases the parameter count and time cost, it significantly enhances feature representation.
Moreover, the full model reduces training time by 12.3% and inference latency by 24.1%, while maintaining the same number of parameters (42.7 M) as the dual-branch variant without attention. This efficiency gain is attributed to two key design strategies: (1) preprocessing reuse, in which the 7 × 7 convolution and 3 × 3 pooling of the input image are performed only once and shared across both branches, eliminating 22.4 s of redundant computation; and (2) dynamic computation compression, where cross-modal attention leverages learnable weights (α/β) to automatically filter out 30% of redundant features, reducing core computing time by an additional 14 s. These results demonstrate that the attention-guided feature fusion mechanism not only enhances segmentation performance but also significantly improves computational efficiency through structural optimization and dynamic resource allocation.
In view of the core challenges of the difficulty in collaborative modeling of local pathological features and global anatomical structures and the significant heterogeneity of cross-modal data in ophthalmic multimodal image analysis, this study proposes a multimodal fusion framework based on cross-modal collaboration and weighted attention mechanism. In terms of data verification, the iChallenge-PM pathological myopia dataset jointly released by Zhongshan Eye Center and Baidu is used. Its multi-center acquisition characteristics and pathological diversity fully reflect the data complexity of real clinical scenarios; at the same time, the CIFAR-10 natural image dataset is introduced for cross-domain generalization testing, and the robustness of the model under heterogeneous data distribution is verified by simulating device differences and background interference. Experiments show that this framework achieves 98.25% accuracy and 98.36% F1 value on the iChallenge-PM dataset, which is 1.43% higher than the benchmark models such as ResNet50, and the misdiagnosis rate and missed diagnosis rate are compressed to 1.63% and 1.75%, respectively, which is significantly better than the traditional single-modal method. In cross-domain testing, the generalization accuracy of the model on CIFAR-10 reached 98.59%, proving that its feature expression is strongly invariant to complex background interference. Future work will focus on two major directions: first, integrating OCT vascular imaging and fundus color photography to achieve three-dimensional time series dynamic modeling to predict the progression of diseases such as diabetic retinopathy; second, developing a lightweight deployment solution based on neural architecture search, compressing the model size through channel pruning and quantization perception training, adapting to edge computing terminals such as ophthalmic diagnosis and treatment robots, and promoting the evolution of clinical diagnosis and treatment from single-modal static analysis to multi-modal real-time decision-making.
Acknowledgement: The authors would like to thank Chongqing Technology and Business University for providing the corresponding working environment and experimental equipment for this study. This work was funded by the Ongoing Research Funding Program (ORF-2025-102), King Saud University, Riyadh, Saudi Arabia; by the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202400813); and by the Graduate Research Innovation Project (Grant Nos. yjscxx2025-269-193 and CYS25618).
Funding Statement: This work was funded by the Ongoing Research Funding Program (ORF-2025-102), King Saud University, Riyadh, Saudi Arabia; by the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202400813); and by the Graduate Research Innovation Project (Grant Nos. yjscxx2025-269-193 and CYS25618).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Peng Xiao, Amr Tolba, Osama Alfarraj; writing—original draft preparation: Peng Xiao; analysis and interpretation of result: Haiyu Xu; data curation: Peng Xu; writing—review and editing: Peng Xiao, Zhiwei Guo. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48. doi:10.1146/annurev-bioeng-071516-044442. [Google Scholar] [PubMed] [CrossRef]
2. Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annu Rev Biomed Eng. 2000;2:315–37. doi:10.1146/annurev.bioeng.2.1.315. [Google Scholar] [PubMed] [CrossRef]
3. Asgari Taghanaki S, Abhishek K, Cohen JP, Cohen-Adad J, Hamarneh G. Deep semantic segmentation of natural and medical images: a review. Artif Intell Rev. 2021;54(1):137–78. doi:10.1007/s10462-020-09854-1. [Google Scholar] [CrossRef]
4. Muthusamy D, Palani P. Deep learning model using classification for diabetic retinopathy detection: an overview. Artif Intell Rev. 2024;57(7):185. doi:10.1007/s10462-024-10806-2. [Google Scholar] [CrossRef]
5. Mehta P, Petersen CA, Wen JC, Banitt MR, Chen PP, Bojikian KD, et al. Automated detection of glaucoma with interpretable machine learning using clinical data and multimodal retinal images. Am J Ophthalmol. 2021;231:154–69. doi:10.1016/j.ajo.2021.04.021. [Google Scholar] [PubMed] [CrossRef]
6. Jin Y, Gui F, Chen M, Chen X, Li H, Zhang J. Deep learning-driven automated quality assessment of ultra-widefield optical coherence tomography angiography images for diabetic retinopathy. Vis Comput. 2025;41(2):1049–59. doi:10.1007/s00371-024-03383-6. [Google Scholar] [CrossRef]
7. Cui T, Lin D, Yu S, Zhao X, Lin Z, Zhao L, et al. Deep learning performance of ultra-widefield fundus imaging for screening retinal lesions in rural locales. JAMA Ophthalmol. 2023;141(11):1045–51. doi:10.1001/jamaophthalmol.2023.4650. [Google Scholar] [PubMed] [CrossRef]
8. Minaee S, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D. Image segmentation using deep learning: a survey. IEEE Trans Pattern Anal Mach Intell. 2022;44(7):3523–42. doi:10.1109/TPAMI.2021.3059968. [Google Scholar] [PubMed] [CrossRef]
9. Sun C, Li C, Lin X, Zheng T, Meng F, Rui X, et al. Attention-based graph neural networks: a survey. Artif Intell Rev. 2023;56(2):2263–310. doi:10.1007/s10462-023-10577-2. [Google Scholar] [CrossRef]
10. Gu X, Wang L, Deng Z, Cao Y, Huang X, Zhu YM. Adaptive spatial and frequency experts fusion network for medical image fusion. Biomed Signal Process Control. 2024;96:106478. doi:10.1016/j.bspc.2024.106478. [Google Scholar] [CrossRef]
11. Liu Z, Hu Y, Qiu Z, Niu Y, Zhou D, Li X, et al. Cross-modal attention network for retinal disease classification based on multi-modal images. Biomed Opt Express. 2024;15(6):3699–714. doi:10.1364/BOE.516764. [Google Scholar] [PubMed] [CrossRef]
12. Ao Y, Shi W, Ji B, Miao Y, He W, Jiang Z. MS-TCNet: an effective Transformer-CNN combined network using multi-scale feature learning for 3D medical image segmentation. Comput Biol Med. 2024;170:108057. doi:10.1016/j.compbiomed.2024.108057. [Google Scholar] [PubMed] [CrossRef]
13. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi:10.1016/j.media.2017.07.005. [Google Scholar] [PubMed] [CrossRef]
14. Liu D, Gao Y, Zhangli Q, Han L, He X, Xia Z et al. TransFusion: multi-view divergent fusion for medical image segmentation with transformers. In: Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; 2022 Sep 18–22; Singapore. doi:10.1007/978-3-031-16443-9_47. [Google Scholar] [CrossRef]
15. Petit O, Thome N, Rambour C, Themyr L, Collins T, Soler L. U-Net transformer: self and cross attention for medical image segmentation. In: Proceedings of the Machine Learning in Medical Imaging; 2021 Sep 27; Strasbourg, France. doi:10.1007/978-3-030-87589-3_28. [Google Scholar] [CrossRef]
16. Dalmaz O, Yurt M, Çukur T. ResViT: residual vision transformers for multimodal medical image synthesis. IEEE Trans Med Imag. 2022;41(10):2598–614. doi:10.1109/TMI.2022.3167808. [Google Scholar] [PubMed] [CrossRef]
17. An P, Yuan Z, Zhao J, Jiang X, Du B. An effective multi-model fusion method for EEG-based sleep stage classification. Knowl Based Syst. 2021;219:106890. doi:10.1016/j.knosys.2021.106890. [Google Scholar] [CrossRef]
18. Zhang Y, Zhang R, Ma Q, Wang Y, Wang Q, Huang Z, et al. A feature selection and multi-model fusion-based approach of predicting air quality. ISA Trans. 2020;100:210–20. doi:10.1016/j.isatra.2019.11.023. [Google Scholar] [PubMed] [CrossRef]
19. Lu Z, Ding C, Xu FJF, Boddeti VN, Wang S, Yang Y. TFormer: a transmission-friendly ViT model for IoT devices. IEEE Trans Parallel Distrib Syst. 2023;34(2):598–610. doi:10.1109/tpds.2022.3222765. [Google Scholar] [CrossRef]
20. Zhou Q, Li X, He L, Yang Y, Cheng G, Tong Y, et al. TransVOD: end-to-end video object detection with spatial-temporal transformers. IEEE Trans Pattern Anal Mach Intell. 2022;45(6):7853–69. doi:10.1109/TPAMI.2022.3223955. [Google Scholar] [PubMed] [CrossRef]
21. Zuo S, Xiao Y, Chang X, Wang X. Vision transformers for dense prediction: a survey. Knowl Based Syst. 2022;253:109552. doi:10.1016/j.knosys.2022.109552. [Google Scholar] [CrossRef]
22. Li C, Tang T, Wang G, Peng J, Wang B, Liang X, et al. BossNAS: exploring hybrid CNN-transformers with block-wisely self-supervised neural architecture search. In: Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10–17; Montreal, QC, Canada. doi:10.1109/ICCV48922.2021.01206. [Google Scholar] [CrossRef]
23. Yuan K, Guo S, Liu Z, Zhou A, Yu F, Wu W. Incorporating convolution designs into visual transformers. In: Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10–17; Montreal, QC, Canada. doi:10.1109/ICCV48922.2021.00062. [Google Scholar] [CrossRef]
24. Peng Z, Guo Z, Huang W, Wang Y, Xie L, Jiao J, et al. Conformer: local features coupling global representations for recognition and detection. IEEE Trans Pattern Anal Mach Intell. 2023;45(8):9454–68. doi:10.1109/TPAMI.2023.3243048. [Google Scholar] [PubMed] [CrossRef]
25. Chen Y, Dai X, Chen D, Liu M, Dong X, Yuan L, et al. Mobile-former: bridging MobileNet and transformer. In: Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18–24; New Orleans, LA, USA. doi:10.1109/CVPR52688.2022.00520. [Google Scholar] [CrossRef]
26. Valanarasu JMJ, Oza P, Hacihaliloglu I, Patel VM. Medical transformer: gated axial-attention for medical image segmentation. In: Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; 2021 Sep 27–Oct 1; Strasbourg, France. doi:10.1007/978-3-030-87193-2_4. [Google Scholar] [CrossRef]
27. Mzoughi H, Njeh I, BenSlima M, Farhat N, Mhiri C. Vision transformers (ViT) and deep convolutional neural network (D-CNN)-based models for MRI brain primary tumors images multi-classification supported by explainable artificial intelligence (XAI). Vis Comput. 2025;41(4):2123–42. doi:10.1007/s00371-024-03524-x. [Google Scholar] [CrossRef]
28. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–11. doi:10.1038/s41592-020-01008-z. [Google Scholar] [PubMed] [CrossRef]
29. Zhang H, Hu W, Wang X. ParC-net: position aware circular convolution with merits from ConvNets and transformer. In: Proceedings of the Computer Vision—ECCV 2022; 2022 Oct 23–27; Tel Aviv, Israel. doi:10.1007/978-3-031-19809-0_35. [Google Scholar] [CrossRef]
30. Guo J, Han K, Wu H, Tang Y, Chen X, Wang Y, et al. CMT: convolutional neural networks meet vision transformers. In: Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18–24; New Orleans, LA, USA. doi:10.1109/CVPR52688.2022.01186. [Google Scholar] [CrossRef]
31. Yu W, Si C, Zhou P, Luo M, Zhou Y, Feng J, et al. MetaFormer baselines for vision. IEEE Trans Pattern Anal Mach Intell. 2024;46(2):896–912. doi:10.1109/TPAMI.2023.3329173. [Google Scholar] [PubMed] [CrossRef]
32. Wang Y, Yang Y, Li Z, Bai J, Zhang M, Li X, et al. Convolution-enhanced evolving attention networks. IEEE Trans Pattern Anal Mach Intell. 2023;45(7):8176–92. doi:10.1109/TPAMI.2023.3236725. [Google Scholar] [PubMed] [CrossRef]
33. Zhou M, Lang S, Zhang T, Liao X, Shang Z, Xiang T, et al. Attentional feature fusion for end-to-end blind image quality assessment. IEEE Trans Broadcast. 2023;69(1):144–52. doi:10.1109/tbc.2022.3204235. [Google Scholar] [CrossRef]
34. Shi Z, Zhang T, Wei X, Wu F, Zhang Y. Decoupled cross-modal phrase-attention network for image-sentence matching. IEEE Trans Image Process. 2022;33:1326–37. doi:10.1109/TIP.2022.3197972. [Google Scholar] [PubMed] [CrossRef]
35. Khan A, Rauf Z, Sohail A, Khan AR, Asif H, Asif A, et al. A survey of the vision transformers and their CNN-transformer based variants. Artif Intell Rev. 2023;56(3):2917–70. doi:10.1007/s10462-023-10595-0. [Google Scholar] [CrossRef]
36. Ismail AR, Azhary MZR, Hitam NA. Evaluating Adan vs. Adam: an analysis of optimizer performance in deep learning. In: Proceedings of the 5th International Symposium on Intelligent Computing Systems, ISICS 2024; 2024 Nov 6–7; Sharjah, United Arab Emirates. doi:10.1007/978-3-031-82931-4_19. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools