 Open Access
Open Access
ARTICLE
Attention U-Net for Precision Skeletal Segmentation in Chest X-Ray Imaging: Advancing Person Identification Techniques in Forensic Science
1 Laboratory of Mathematics, Informatics and Systems (LAMIS), Echahid Cheikh Larbi Tebessi University, Tebessa, 12002, Algeria
2 Department of Electrical Engineering, Umm Al-Qura University, Makkah, 21955, Saudi Arabia
3 College of Computer Science and Engineering, Taibah University, Medina, 41477, Saudi Arabia
4 Department of Information Technology, Aylol University College, Yarim, 547, Yemen
* Corresponding Authors: Akram Bennour. Email: ; Mohammed Al-Sarem. Email:
Computers, Materials & Continua 2025, 85(2), 3335-3348. https://doi.org/10.32604/cmc.2025.067226
Received 27 April 2025; Accepted 10 July 2025; Issue published 23 September 2025
Abstract
This study presents an advanced method for post-mortem person identification using the segmentation of skeletal structures from chest X-ray images. The proposed approach employs the Attention U-Net architecture, enhanced with gated attention mechanisms, to refine segmentation by emphasizing spatially relevant anatomical features while suppressing irrelevant details. By isolating skeletal structures which remain stable over time compared to soft tissues, this method leverages bones as reliable biometric markers for identity verification. The model integrates custom-designed encoder and decoder blocks with attention gates, achieving high segmentation precision. To evaluate the impact of architectural choices, we conducted an ablation study comparing Attention U-Net with and without attention mechanisms, alongside an analysis of data augmentation effects. Training and evaluation were performed on a curated chest X-ray dataset, with segmentation performance measured using Dice score, precision, and loss functions, achieving over 98% precision and 94% Dice score. The extracted bone structures were further processed to derive unique biometric patterns, enabling robust and privacy-preserving person identification. Our findings highlight the effectiveness of attention mechanisms in improving segmentation accuracy and underscore the potential of chest bone-based biometrics in forensic and medical imaging. This work paves the way for integrating artificial intelligence into real-world forensic workflows, offering a non-invasive and reliable solution for post-mortem identification.Keywords
Biometric identification has become a cornerstone in forensic science, enabling the recognition of individuals in complex and challenging conditions [1]. Traditional modalities, including fingerprints, facial recognition, and DNA profiling, have demonstrated high accuracy. However, their reliability diminishes in cases involving advanced decomposition, trauma, or environmental degradation [2]. When the human body is subjected to severe damage, prolonged decomposition, or exposure to extreme environmental conditions, many of these modalities fail to provide accurate results [3,4]. For instance, fingerprint recognition becomes unreliable when the skin is burned or decayed, and facial recognition is ineffective when the face has been disfigured due to trauma [5,6]. Similarly, DNA analysis is often hindered by contamination or the degradation of biological samples [7]. Moreover, methods such as iris or vein pattern recognition are impractical when the eyes or limbs have sustained irreversible damage. These limitations not only complicate the identification process but also introduce delays in forensic investigations, particularly when time-sensitive results are required. In many cases, forensic experts are compelled to rely on multiple biometric modalities to ensure a more reliable identification, but even this approach may not be sufficient under extreme conditions. Compounding these challenges is the surge in large-scale fatalities caused by ongoing conflicts, particularly in war-torn regions such as Gaza, Ukraine, and other countries. The sheer number of casualties, combined with the catastrophic injuries sustained in these environments, underscores the pressing need for alternative and more robust identification methods. Given the inadequacies of conventional biometric techniques in such contexts, researchers have turned their attention to the use of chest X-ray images as a viable solution for post-mortem identification [8–12]. Chest X-ray imaging has gained increasing attention due to the structural durability of skeletal features, which remain intact even when soft tissues deteriorate. Capitalizing on this resilience, researchers have begun exploring computational methods to extract and analyse skeletal structures for biometric identification. Chest X-ray imaging has gained increasing attention due to the structural durability of skeletal features, which remain intact even when soft tissues deteriorate. This makes the use of skeletal features particularly effective in cases of body decomposition, as the soft tissue is often decomposed or significantly altered, while the skeletal structure remains relatively stable and provides reliable diagnostic or biometrics information [13–16]. This study presents a novel framework utilizing the Attention U-Net architecture to achieve precise skeletal segmentation from chest X-ray images, with the aim of advancing person identification techniques. The Attention U-Net model employs gated attention mechanisms to selectively highlight anatomically relevant regions while suppressing irrelevant details, enhancing segmentation accuracy and ensuring the extracted skeletal structures are well-defined and consistent. By focusing on the stability of bones as biometric markers, especially in post-mortem scenarios, this approach addresses key challenges faced by traditional methods, offering a reliable and scalable alternative for identity verification. The proposed framework integrates specialized encoder-decoder structures with attention gates to refine feature extraction, enabling adaptation to varying image qualities and anatomical differences. To further evaluate the effectiveness of the proposed approach, we conducted an ablation study assessing the impact of different architectural choices. Specifically, we compared the performance of the Attention U-Net with and without attention mechanisms, as well as the influence of data augmentation on segmentation accuracy. This analysis provides deeper insights into the contributions of attention-based feature selection and data variability in enhancing model robustness. Extensive evaluations on curated datasets of chest X-rays demonstrated over 90% precision in segmentation tasks, underscoring the model’s ability to isolate skeletal structures effectively.
This paper makes the following contributions.
• Introduces a novel framework using the Attention U-Net architecture for precise skeletal segmentation from chest X-ray images, enhancing the accuracy of isolating skeletal structures.
• Utilizes gated attention mechanisms to selectively highlight anatomically relevant regions while suppressing irrelevant details, improving segmentation precision and consistency.
• Addresses the limitations of traditional biometric modalities (e.g., fingerprints, facial recognition, DNA) in cases of decomposition, trauma, or environmental degradation by leveraging the durability of skeletal features.
• Conducts an ablation study to assess the impact of architectural choices, including the role of attention mechanisms and data augmentation, providing insights into the model’s robustness and performance.
While this study primarily focuses on skeletal segmentation, the extracted bone structures will serve as a foundation for future research aimed at developing advanced algorithms for person identification specially in body and soft tissue decomposition. By combining state-of-the-art deep learning techniques with medical imaging, this work lays the groundwork for a scalable and privacy-preserving identification framework, bridging the gap between modern AI applications and practical forensic challenges. The remainder of this paper is structured as follows: Section 2 presents a review of related works, highlighting recent advancements in deep learning-based medical image segmentation and biometric identification using skeletal structures. Section 3 details the materials and methodology, describing the dataset, preprocessing techniques, model architecture, and evaluation metrics used for bone segmentation. Section 4 discusses the results and analysis, comparing our proposed attention-enhanced U-Net with conventional segmentation methods, alongside a detailed performance evaluation. Finally, Section 5 concludes the study by summarizing key findings, discussing potential applications in forensic identification, and outlining directions for future research.
Accurate bone segmentation from chest X-ray images is crucial for various medical and forensic applications. This section reviews existing studies on segmentation techniques, categorizing them into three primary areas: (1) Classical and Alternative Segmentation Techniques, (2) U-Net-Based Approaches, and (3) Hybrid and Multitask Models. The objective is to highlight the evolution of segmentation methods, from traditional approaches to deep learning-based innovations, emphasizing their strengths and limitations.
2.1 Classical and Alternative Segmentation Techniques
Several studies have attempted rib and clavicle segmentation using classical computer vision techniques. Template-matching methods were employed in [17–19] for rib detection, while a Hybrid Dynamic Programming, Active Shape Model, and Pixel Classification (HDAP) algorithm was proposed in [20] for clavicle segmentation. Despite their contributions, these approaches struggled with low accuracy and robustness in complex anatomical structures. To improve posterior rib segmentation, Reference [21] proposed a fully automated algorithm integrating the Generalized Hough Transform (GHT) and a Bilateral Dynamic Programming Algorithm. This method improved rib localization and lesion visibility, enhancing computer-aided detection (CAD) systems for lung nodules. The study reported over 90% of segmented ribs rated as high quality, demonstrating clinical utility in radiological assessments.
U-Net and its variants have been extensively used for bone segmentation due to their effectiveness in capturing fine anatomical details. In [22], a U-Net-based deep learning model was employed to automate the extraction of bone features from chest radiographs. The model demonstrated strong performance, achieving an IoU of 0.834, a Dice coefficient of 0.909, a sensitivity of 0.944, and a specificity of 0.895. By improving segmentation accuracy in forensic applications, this work highlighted the feasibility of using deep learning for analyzing skeletal structures in post-mortem scenarios. An enhanced approach was introduced in [23], where a modified U-Net with attention mechanisms was proposed for precise bone segmentation in both X-ray and DEXA images. The method achieved Dice scores of 0.94 and 0.92 on two different datasets and successfully classified bone mineral density (BMD) levels into osteopenia, osteoporosis, and normal categories. These results reinforce the potential of integrating attention-based enhancements in deep learning architectures to refine bone segmentation accuracy. Else in [24] proposes a novel multiclass teeth segmentation architecture combining an M-Net-like structure, Swin Transformers, and a Teeth Attention Block (TAB). The Swin Transformer captures long-range dependencies through self-attention, while the TAB refines segmentation by focusing on tooth boundaries. The U-Net-like framework, with encoder-decoder skip connections and multiscale supervision, preserves spatial information and enhances feature representation. A squared Dice loss mitigates class imbalance, ensuring robust segmentation. Evaluated on a panoramic teeth X-ray dataset, the method achieves a Dice Coefficient of 0.9102, outperforming state-of-the-art approaches and demonstrating significant potential for improving dental image analysis and applications.
2.3 Hybrid and Multitask Models
To address segmentation challenges such as artifacts and overlapping soft tissues, hybrid deep learning architectures have been explored. The MDU-Net model introduced in [25] combines multiscale feature fusion, DenseNet’s dense connections, and a multitasking mechanism to enhance segmentation performance. Evaluated on 88 chest X-ray images using fourfold cross-validation, the model achieved Dice similarity coefficients (DSC) of 93.78% for clavicle segmentation, 80.95% for anterior ribs, 89.06% for posterior ribs, and 88.38% for all bones. However, the study emphasized that the relatively small dataset size posed a challenge for generalizability. Another hybrid approach was presented in [26], where U-Net was combined with DeepLab v3+ for segmenting bones in severely unhealthy cases. The model utilized ResNet50 with an Atrous Spatial Pyramid Pooling (ASPP) block, improving feature extraction and boundary preservation. Trained on an in-house dataset including healthy and pulmonary disease cases, the model achieved a Dice coefficient of 92.68% for clavicle segmentation, setting a new benchmark for abnormal chest X-ray analysis. Across the reviewed studies, U-Net-based models and hybrid architectures with attention mechanisms consistently outperformed classical approaches in bone segmentation tasks. However, challenges such as dataset limitations, overfitting, and segmentation artifacts remain key issues requiring further investigation. Our study builds on these findings by introducing an Attention U-Net model optimized for forensic applications, leveraging gated attention mechanisms to improve segmentation precision and biometric feature extraction from skeletal structures. Additionally, Table 1 provides a comparative overview of the reviewed segmentation techniques, summarizing their key contributions and performance metrics.
Our study introduces a deep learning-driven approach for bone segmentation in chest radiographs, leveraging a U-Net architecture augmented with attention mechanisms. The subsequent sections elaborate on the data preparation strategy, architectural design, training methodology, and dataset specifications. Our approach is designed to enhance segmentation accuracy and robustness, offering valuable support in forensic and clinical applications, particularly in postmortem identification scenarios.
Following the methodology outlined in [23], the dataset used for this study was derived from the LIDC-IDRI dataset. Specifically, an approach based on Hounsfield Unit (HU)-based segmentation was employed to extract bone structures from CT scans. Bone voxels were identified as those within the HU range of [300, 700], while non-bone voxels (e.g., soft tissues and air) were assigned a value of −1024 HU, representing air in CT imaging. From these segmented bone voxels, synthetic 2D “bone X-ray” images were generated, serving as the target masks for training our model. This technique enabled the creation of a robust set of synthetic training data consisting of paired synthetic X-ray images (Digitally Reconstructed Radiographs, or DRRs) and corresponding bone masks, facilitating the effective training of our deep learning model for bone extraction and enhancement tasks. The dataset comprises 386 images with their corresponding masks. Fig. 1 illustrates a sample chest X-ray image alongside its corresponding mask. The Fig. 1 illustrates a sample of DRR dataset.

Figure 1: Sample of chest X-ray with mask
In the preprocessing step of our bone segmentation process from chest X-ray images, we first analyze the Digitally Reconstructed Radiograph (DRR) dataset to ensure consistency and quality in the input data. We then utilize the masks generated by the method in [23] to accurately delineate the bone structures. To further enhance the quality of these masks, we apply histogram equalization, which improves contrast and highlights important structural details as shown in Fig. 2. This preprocessing step is crucial as it optimizes the input data before feeding it into our segmentation architecture, ultimately contributing to a more effective training process and improved segmentation performance.

Figure 2: Preprocessing results
In this work, we introduce a deep learning framework for bone segmentation that leverages an attention-enhanced U-Net architecture. This approach is specifically designed to address the challenges inherent in segmenting bone structures from X-ray images. X-ray imaging is particularly challenging due to low contrast between bone and surrounding soft tissue, imaging noise, and complex anatomical variability. Moreover, precise delineation of cortical and trabecular structures is critical for accurate diagnosis and treatment planning [28,29]. Our framework overcomes these limitations through a symmetric encoder–decoder design with skip connections, augmented by attention mechanisms that selectively emphasize relevant bone features. The encoder systematically extracts hierarchical features while progressively reducing spatial resolution, thereby capturing both global context and fine structural details. Although skip connections ensure the preservation of essential spatial information, not all transmitted features are equally informative for bone delineation. To resolve this, attention gates are integrated into the skip connections. These gates compute spatial attention masks by fusing contextual cues from deeper layers with the original encoder features. The resulting masks selectively amplify salient bone structures and attenuate irrelevant background noise, enhancing the network’s ability to delineate intricate bone boundaries accurately. This attention-gate mechanism is particularly advantageous in clinical scenarios, where the accurate segmentation of bone is crucial for orthopaedic surgery, trauma assessment, and diagnostic applications [30]. Furthermore, the robust extraction of skeletal features enables potential use in biometric applications, where bone structure can serve as a unique identifier for identity verification. In summary, our attention-enhanced U-Net framework offers a powerful and resilient solution for bone segmentation in X-ray imaging, facilitating reliable automated analysis and improved clinical outcomes.
The proposed architecture comprises three primary components Encoder Blocks, Decoder Blocks, and Attention Gates as illustrated in Fig. 3.
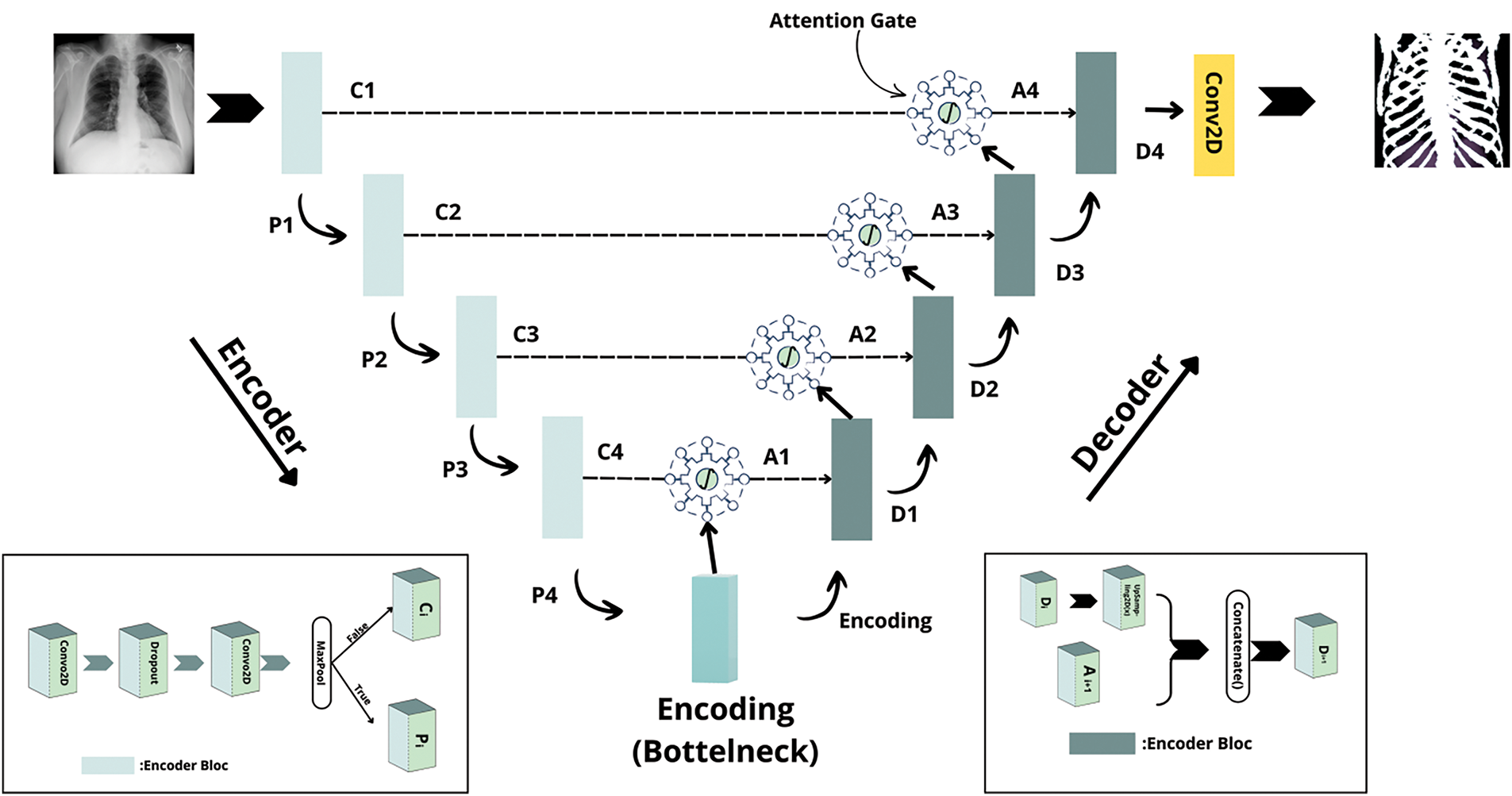
Figure 3: The proposed architecture
A. Encoder Blocks
Each encoder block is designed to progressively extract hierarchical features while reducing the spatial dimensions of the input image. In practice, an encoder block consists of two successive convolutional layers with a 3 × 3 kernel and ReLU activations, with a dropout layer inserted between them to help mitigate overfitting. Optionally, a max pooling operation is applied after these convolutions to further reduce the spatial resolution. The block outputs both the downsampled feature map, used for subsequent encoding, and the pre-pooled feature map, which serves as a skip connection.
B. Decoder Blocks
The decoder path reconstructs the segmentation map from the compressed representation generated by the encoder. Each decoder block starts with an upsampling operation that restores the spatial resolution. The upsampled feature map is then concatenated with the corresponding refined skip connection from the encoder. This combined feature set is processed through a convolutional block similar to an encoder block but without the pooling operation to yield a refined output. The upsampling factor is chosen to match the resolution of the corresponding encoder feature map.
C. Attention Gates
To further improve the quality of the skip connections, attention gates are integrated into the network. These gates selectively emphasize regions of interest namely, the bone structures while suppressing extraneous background information. Each attention gate receives two inputs: a gating signal from deeper layers of the network and a skip connection from the encoder. The gating signal is processed using a convolutional layer with ReLU activation, while the skip connection is downsampled using a similar convolution with a stride of 2. The outputs of these operations are fused and passed through an additional convolutional layer with a sigmoid activation function to generate an attention mask. This mask is then up sampled to the spatial dimensions of the original skip connection and used to filter the skip connection via element-wise multiplication. Optionally, batch normalization can be applied to stabilize the refined features. Attention Gate illustrated in Fig. 4.
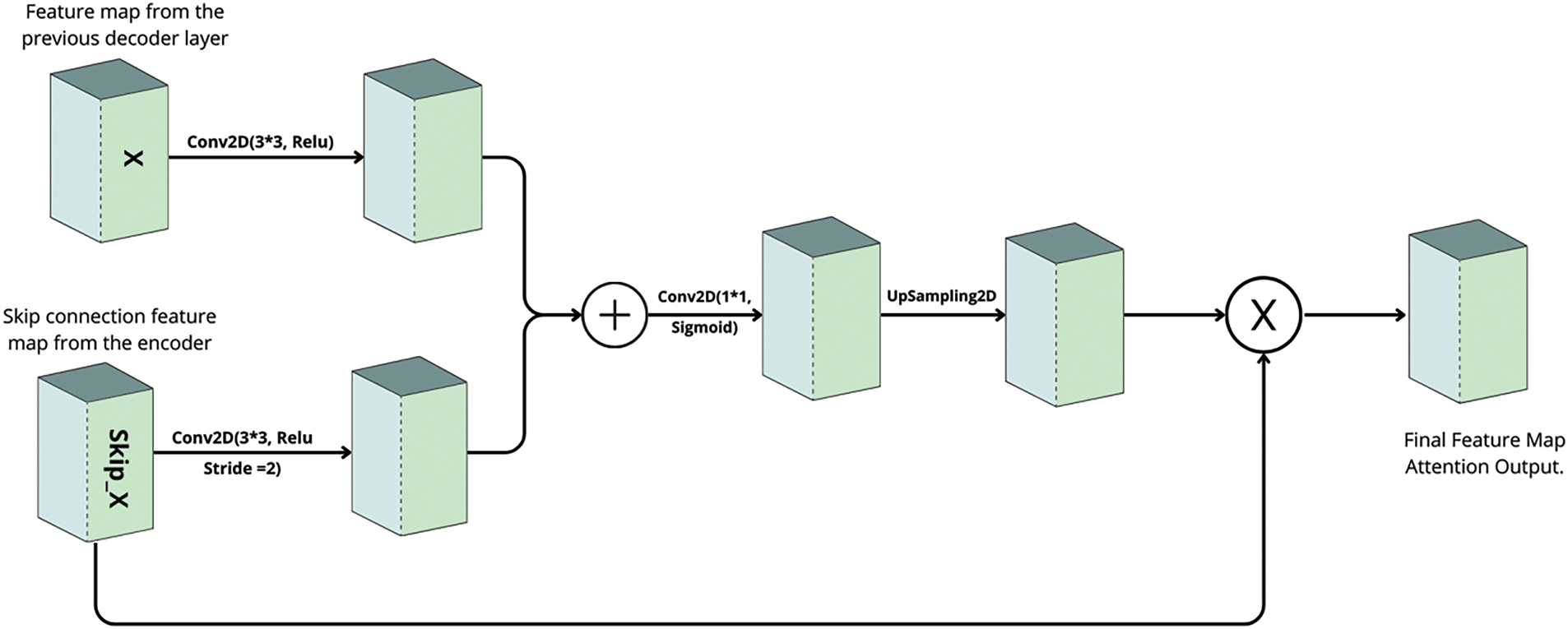
Figure 4: Attention gate architecture
The complete network begins by encoding the input X-ray image through a series of encoder blocks, which progressively reduce spatial dimensions and extract multi-scale features. The deepest encoder block, which does not apply pooling, forms a bottleneck that serves as a comprehensive representation of the input. In the decoding phase, upsampling operations restore the original image resolution, and attention gates refine the skip connections by filtering out irrelevant features. The refined features are concatenated with the upsampled decoder features and further processed by decoder blocks, culminating in a final 1 × 1 convolution with a sigmoid activation that produces the bone segmentation mask. This attention-guided U-Net architecture is ideally suited for bone segmentation in X-ray imaging, where the precise delineation of fine bone structures is critical for clinical diagnosis and treatment planning. Furthermore, its robust extraction of skeletal features extends its utility to biometric applications, such as identity verification. Future work will focus on benchmarking this method against conventional U-Net models and exploring the integration of additional imaging modalities to further enhance segmentation performance.
4.1 Experimental Setup and Evaluation
To thoroughly evaluate our proposed U-Net model with an attention mechanism for bone segmentation, we designed a structured experimental framework. The model was implemented using the Keras library in Python and executed on Google Colab. The dataset, derived from LIDC-IDRI, consists of 386 X-ray images paired with corresponding bone masks. The dataset was split into 80% training and 20% validation to ensure a balanced and reproducible evaluation.
To assess the contributions of the attention mechanism and data augmentation, we conducted a comprehensive ablation study with four experimental configurations. The first experiment involved a standard U-Net trained on the original dataset without the attention mechanism or data augmentation, serving as a benchmark to evaluate the model’s baseline performance. The second experiment integrated the attention mechanism into the U-Net while maintaining the original dataset, allowing us to isolate the effect of attention on segmentation accuracy. The third experiment applied data augmentation techniques, including rotation, scaling, contrast adjustment, and horizontal flipping, to expand the dataset from 386 to 3088 images while excluding the attention mechanism. This setup provided insight into the impact of data augmentation on the model’s ability to generalize. The final experiment combined both the attention mechanism and data augmentation, forming our proposed model, to evaluate the potential synergy between these enhancements. In addition to quantitative evaluation, qualitative comparisons were conducted by overlaying predicted bone masks on the original X-ray images. This visual assessment highlighted segmentation accuracy and anatomical consistency, further demonstrating the effects of each experimental variation. Through this structured ablation study, we systematically analyzed the contributions of the attention mechanism and data augmentation, ensuring a robust evaluation of our proposed approach.
4.2 Performance Evaluation Metrics
To rigorously assess the performance of our proposed bone segmentation system, we employ a comprehensive set of evaluation metrics specifically designed for segmentation tasks. These include loss, Dice coefficient (Dice), and precision, each providing unique insights into model effectiveness. The loss function, typically binary cross-entropy or Dice loss, quantifies the discrepancy between the predicted segmentation and the ground truth, serving as a crucial optimization guide during training. While loss tracks model convergence, it does not fully capture segmentation quality, necessitating additional evaluation metrics. The Dice coefficient, defined in Eq. (1), measures the overlap between predicted and ground truth regions. This metric is particularly valuable in medical imaging due to its robustness against class imbalance. The Dice score ranges from 0 to 1, where higher values indicate greater alignment between predictions and actual annotations.
where X represents the set of predicted pixels, Y represents the ground truth pixels, and |X ∩ Y| denotes their intersection. Additionally, precision, defined in Eq. (2), evaluates the model’s ability to minimize false positives by quantifying the proportion of correctly predicted positive pixels relative to all predicted positives. This metric is particularly crucial in medical applications, where false positives can lead to misdiagnoses or unnecessary interventions.
To provide a holistic evaluation of the model’s performance, we analyze loss, Dice coefficient, and precision across multiple iterations. The graphical representations of evaluation loss and precision coefficient over training iterations are presented in Figs. 3, and 4, respectively. By leveraging these metrics, we ensure a rigorous assessment of our approach, validating its capability to achieve accurate and reliable bone segmentation in chest radiographs.
The effectiveness of our approach was assessed through multiple experimental configurations, evaluating the impact of both attention gates (AG) and data augmentation (DA) on bone segmentation performance. Table 2 presents the segmentation results, demonstrating the contribution of each component to model accuracy and robustness. The baseline U-Net, without any modifications, achieved a Dice similarity coefficient of 83% and a precision of 87%. While this performance aligns with traditional segmentation approaches, the introduction of data augmentation significantly improved generalization, leading to an increase in Dice score to 89% and a reduction in segmentation loss from 0.21 to 0.19. The integration of attention gates, without augmentation, resulted in a Dice score of 92%, underscoring the effectiveness of AG in refining feature selection by suppressing irrelevant regions. The best performance was achieved when combining both AG and DA, reaching a Dice score of 94%, the highest among all configurations. Additionally, this setup achieved the highest precision (98%) and the lowest segmentation loss (0.06), highlighting its capability to generate well-defined and accurate segmentation maps. These results demonstrate the complementary effect of AG and DA. Attention gates dynamically refine feature extraction, enhancing the model’s ability to focus on relevant structures, while data augmentation further improves generalization and robustness against variations in image acquisition conditions.
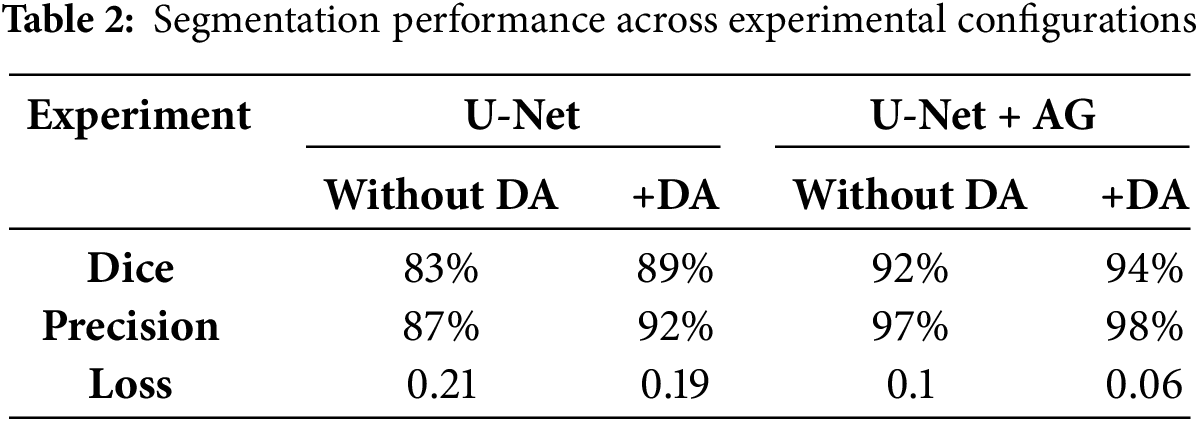
A deeper analysis of the segmentation maps reveals that models incorporating AG exhibit superior boundary delineation, particularly in complex anatomical regions where bones overlap with soft tissues. The baseline U-Net struggles with such regions, often leading to over-segmentation or fragmented contours. In contrast, AG enhances spatial awareness by prioritizing bone structures and suppressing background noise, contributing to more reliable segmentation. Furthermore, the significant drop in Dice score (from 94% to 83%) when neither AG nor DA is used underscores the necessity of these enhancements for robust segmentation performance. The high precision achieved with AG also indicates a substantial reduction in false-positive detections, which is crucial for medical imaging applications where misclassification can impact clinical decision-making.
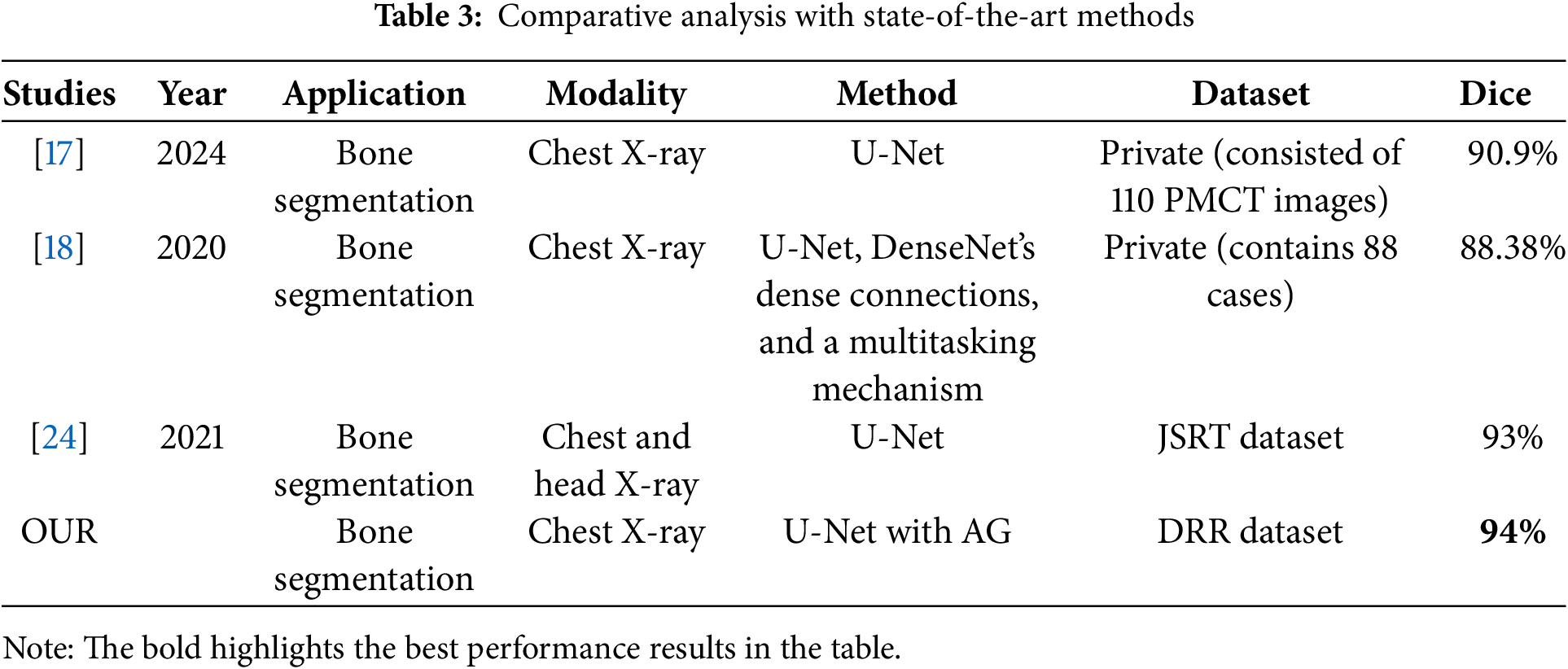
Figs. 3 and 4 illustrate the loss and precision curves, respectively. The loss curve demonstrates a steady decrease during training, converging at lower values for the attention-based U-Net. The precision curve further highlights the stability and improved performance of the proposed approach, where the validation precision remains consistently high across epochs. A qualitative assessment is presented in Fig. 5, showing the predicted segmentation masks compared to the ground truth. The results confirm that AG enhances the segmentation quality by reducing noise and refining structural boundaries, particularly in challenging regions of the ribcage. To further contextualize our results, we compare the performance of our proposed method against state-of-the-art approaches for bone segmentation in chest X-rays. Table 3 provides a summary of related works, highlighting differences in methodologies, datasets, and performance. Our approach achieves the highest Dice score among the compared methods, outperforming previous U-Net-based implementations. The DenseNet-based multitasking approach proposed in [18] achieved 88.38%, demonstrating the effectiveness of hybrid architectures, but still falling short of our AG-based refinement. Similarly, the standard U-Net in [17] attained 90.9% Dice score, but without explicit attention mechanisms, it lacked the ability to dynamically suppress background artifacts. The best-performing prior method [24] achieved 93% Dice score, using the JSRT dataset, which is smaller than the DRR dataset used in our study.
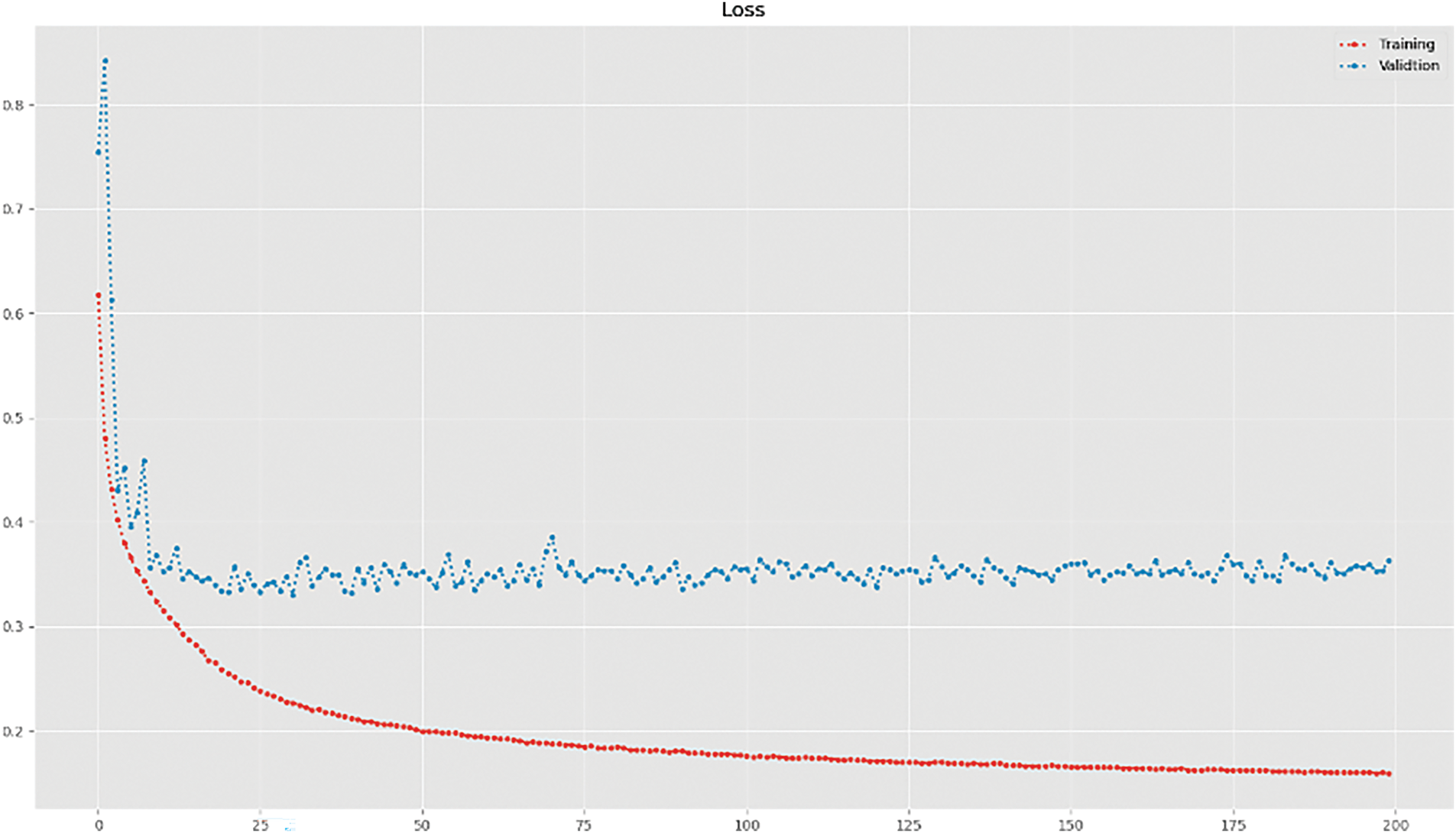
Figure 5: Training and validation loss curve
Our results indicate that incorporating attention gates effectively enhances segmentation precision and structural awareness beyond conventional U-Net implementations. The DRR dataset, characterized by synthetic yet anatomically accurate representations of bone structure, provides a robust training environment, allowing the model to learn fine-grained spatial relationships within chest X-ray images. This dataset advantage, coupled with the attention-driven feature selection mechanism, enables our approach to achieve state-of-the-art performance. While these results are promising, certain challenges remain. The computational complexity of AG-based architectures is higher than standard U-Net models, requiring additional optimization for real-time clinical deployment. Future research should focus on reducing inference time while preserving segmentation accuracy, possibly by integrating lightweight attention modules or transformer-based mechanisms. Furthermore, while the DRR dataset has been instrumental in training the model, evaluating the approach on real-world clinical datasets will be essential for assessing its generalizability across different imaging conditions. In summary, our method advances the field of bone segmentation in chest X-rays by integrating attention gates, significantly improving segmentation precision and generalization. These findings pave the way for the development of more robust and interpretable deep learning models for medical image analysis, with potential applications in automated diagnosis and forensic identification. The findings of this study have several important implications for both medical image analysis and future biometric systems. From a clinical perspective, our attention-enhanced U-Net architecture offers a robust and accurate tool for automated bone segmentation in chest X-ray images, which can assist radiologists in diagnostics, preoperative planning, or post-mortem analysis. The high precision and reliability of our model reduce the need for manual annotation and support scalable processing of large medical datasets. Additionally, the successful extraction of detailed skeletal structures lays the foundation for future research in skeletal-based person identification, offering a potential path toward non-invasive, privacy-aware biometric systems. This is particularly relevant in scenarios such as patient re-identification in medical databases, forensic investigations, or identity identification in cases where traditional biometric modalities are not applicable. By providing a strong segmentation backbone, our work contributes to advancing both medical imaging tools and secure, anatomy-based identification technologies.
In this study, we proposed an attention-enhanced U-Net for bone segmentation from chest X-ray images, achieving a Dice score of 94% and a precision of 98%, surpassing conventional U-Net architectures. By integrating attention gate mechanisms, our approach enhances feature extraction, enabling more precise localization of bone structures while effectively suppressing irrelevant regions. Compared to standard deep learning models, our method demonstrates superior segmentation accuracy and interpretability, addressing key challenges such as anatomical overlap and false-positive detections. Beyond medical imaging, our findings open new avenues for biometric identification, particularly in postmortem human recognition. The skeletal structures of the ribs and clavicles, which remain intact even after significant soft tissue degradation, could serve as unique anatomical markers for identifying individuals in forensic investigations, disaster victim identification, and missing person cases. This research represents the first stage of a broader effort to explore the feasibility of using automated bone-based identification in postmortem forensic applications. Future work will focus on extending this approach by integrating advanced deep learning architectures, including hybrid models that combine transformers or self-supervised learning techniques to enhance segmentation performance. Cross-validation on diverse datasets will be critical to ensuring model robustness across varied clinical and forensic settings. Further investigations will also explore 3D reconstruction techniques from 2D X-rays to enhance the accuracy of postmortem identification. Moreover, implementing federated learning frameworks will be explored to facilitate collaborative model training across multiple institutions while preserving patient privacy and data security. This approach can leverage distributed datasets without centralizing sensitive medical data, thereby improving model generalization and applicability in real-world clinical and forensic environments. Overall, this study contributes to the advancement of attention-based deep learning for medical image segmentation, offering a high-performance, interpretable, and scalable solution with promising applications in both healthcare and forensic biometrics.
Acknowledgement: The authors extend their appreciation to Umm Al-Qura University, Saudi Arabia for funding this research work through grant number: 25UQU4300346GSSR08.
Funding Statement: This research work was funded by Umm Al-Qura University, Saudi Arabia under grant number: 25UQU4300346GSSR08.
Author Contributions: The authors confirm contributions to the paper as follows: Study conception and design: Hazem Farah, and Hama Soltani; Methodology, Data curation draft manuscript preparation: Akram Bennour, and Mohammed Al-Sarem; Supervision, Methodology, Draft preparation, Final paper revision: Mouaaz Nahas, and Rashiq Rafiq Marie; Funding acquisition and project administration, Data curation, Validation: Mouaaz Nahas, and Rashiq Rafiq Marie. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The original CT scans used in this study were obtained from the publicly available LIDC-IDRI dataset (https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI, accessed on 09 July 2025). The synthetic bone X-ray images (Digitally Reconstructed Radiographs, DRRs) and corresponding bone masks generated from this dataset are available from the corresponding authors upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Warren CP. Personal identification of human remains: an overview. J Forensic Sci. 1978;23(3):388–95. doi:10.1520/jfs10774j. [Google Scholar] [CrossRef]
2. Bogicevic D, Petrovic LJ, Kostic D, Zivanovic M, Tomic S, Radojicic S. Biometric standards and methods. Vojnoteh Glas. 2021;69(4):963–77. [Google Scholar]
3. Jain AK, Ross A. Bridging the gap: from biometrics to forensics. Philos Trans R Soc Lond B Biol Sci. 2015;370(1674):20140254. doi:10.1098/rstb.2014.0254. [Google Scholar] [PubMed] [CrossRef]
4. Saini M, Kapoor AK. Biometrics in forensic identification: applications and challenges. J Forensic Med. 2016;1(108):2. [Google Scholar]
5. Drahansky M, Brezinova I, Lodrova Z, Dolezel M, Urbanek J, Mezl M, et al. Influence of skin diseases on fingerprint recognition. Biomed Res Int. 2012;2012(4):626148. doi:10.1155/2012/626148. [Google Scholar] [PubMed] [CrossRef]
6. Sarfraz N. Adermatoglyphia: barriers to biometric identification and the need for a standardized alternative. Cureus. 2019;11(2):e4040. doi:10.7759/cureus.4040. [Google Scholar] [PubMed] [CrossRef]
7. Wells JD, Linville JG. Biology/DNA/entomology: overview. In: James SH, Nordby JJ, editors. Encyclopedia of forensic sciences. 2nd ed. Amsterdam, The Netherlands: Elsevier; 2013. p. 387–93. [Google Scholar]
8. Emergency Care Research Institute. Patient identification errors [Internet]. [cited 2021 Sep 28]. Available from: https://www.ecri.org/Resources/HIT/Patient%20ID/Patient_Identification_Evidence_Based_Literature_final.pdf. [Google Scholar]
9. Morishita J, Katsuragawa S, Kondo K, Doi K. An automated patient recognition method based on an image-matching technique using previous chest radiographs. Med Phys. 2001;28(6):1093–7. [Google Scholar] [PubMed]
10. Danaher LA, Herts BR, Einstein DM, Obuchowski NA, McCoy JM, Azevedo RM, et al. Is it possible to eliminate patient identification errors in medical imaging? J Am Coll Radiol. 2011;8(8):568–74. [Google Scholar] [PubMed]
11. Hazem F, Bennour A, Mekhaznia T, Neesrin AK, Khalaf OI, Belkacem S. Beyond traditional biometrics: harnessing chest X-ray features for robust person identification. Acta Inf Pragensia. 2024;13(2):234–50. doi:10.18267/j.aip.238. [Google Scholar] [CrossRef]
12. Farah H, Akram B, Kurdi NA, Khalaf OI, Mekhaznia T, Boulaknadel S. Channel and spatial attention in chest X-ray radiographs: advancing person identification and verification with self-residual attention network. Diagnostics. 2024;14(23):2655. doi:10.3390/diagnostics14232655. [Google Scholar] [PubMed] [CrossRef]
13. Jain R, Naranje P, Gupta N, Patel V, Mehta R, Sharma M. Chest X-ray as the first pointer in various skeletal dysplasia and related disorders. Indographics. 2022;1(2):196–207. doi:10.1055/s-0042-1759845. [Google Scholar] [CrossRef]
14. Hazem F, Akram B, Mekhaznia T, Neesrin AK, Belkacem S, Boulaknadel S. X-ray insights: a Siamese with CNN and spatial attention network for innovative person identification. In: Proceedings of the Intelligent Systems and Pattern Recognition 2024; 2024 Jun 26–28; Istanbul, Türkiye. [Google Scholar]
15. Hazem F, Akram B, Khalaf OI, Mekhaznia T, Kurdi NA, Belkacem S. X-ray insights: innovative person identification through Siamese and Triplet networks. IET Conf Proc. 2023;2023(39):40–9. doi:10.1049/icp.2024.0463. [Google Scholar] [CrossRef]
16. Soltani H, Bendib I, Haouam MY, Amroune M. Advancements in breast cancer diagnosis: a comprehensive review of mammography datasets, preprocessing and classification techniques. Acta Inf Pragensia. 2024;13(2):308–26. doi:10.18267/j.aip.244. [Google Scholar] [CrossRef]
17. Ogul H, Ogul BB, Agildere AM, Bayrak T, Sumer E. Eliminating rib shadows in chest radiographic images providing diagnostic assistance. Comput Methods Programs Biomed. 2015;122(2):208–14. doi:10.1016/j.cmpb.2015.12.006. [Google Scholar] [PubMed] [CrossRef]
18. Lee JS, Wang JW, Wu HH, Yuan MZ. A nonparametric-based rib suppression method for chest radiographs. Comput Math Methods Med. 2012;64(8):1390–9. doi:10.1016/j.camwa.2012.03.084. [Google Scholar] [CrossRef]
19. Li XC, Luo SH, Hu QM, Li JM, Wang DD. Rib suppression in chest radiographs for lung nodule enhancement. In: Proceedings of the 2015 IEEE International Conference on Information and Automation; 2015 Aug 8–10; Lijiang, China. [Google Scholar]
20. Ding L, Zhao K, Zhang X, Wang X, Zhang J. A lightweight U-Net architecture multi-scale convolutional network for pediatric hand bone segmentation in X-ray image. IEEE Access. 2019;7:68436–45. doi:10.1109/access.2019.2918205. [Google Scholar] [CrossRef]
21. Cong L, Guo W, Li Q. Segmentation of ribs in digital chest radiographs. Proc SPIE. 2016;9788:463–8. [Google Scholar]
22. Kim Y, Yoon Y, Matsunobu Y, Seo M, Chung J, Kim D, et al. Gray-scale extraction of bone features from chest radiographs based on deep learning technique for personal identification and classification in forensic medicine. Diagnostics. 2024;14(16):1778. doi:10.3390/diagnostics14161778. [Google Scholar] [PubMed] [CrossRef]
23. Singh A, Lall B, Panigrahi BK, Mittal P, Sinha S, Mehta V. Semantic segmentation of bone structures in chest X-rays including unhealthy radiographs: a robust and accurate approach. Int J Med Inform. 2022;165(6):104831. doi:10.1016/j.ijmedinf.2022.104831. [Google Scholar] [PubMed] [CrossRef]
24. Ghafoor A, Moon SY, Lee B. Multiclass segmentation using teeth attention modules for dental X-ray images. IEEE Access. 2023;11(1):123891–903. doi:10.1109/access.2023.3329364. [Google Scholar] [CrossRef]
25. Wang W, Feng H, Bu Q, Zhang J, Li L, Xu H. MDU-Net: a convolutional network for clavicle and rib segmentation from a chest radiograph. J Healthc Eng. 2020;2020(1):2785464. doi:10.1155/2020/2785464. [Google Scholar] [PubMed] [CrossRef]
26. Gómez Ó, Mesejo P, Ibáñez Ó. Automatic segmentation of skeletal structures in X-ray images using deep learning for comparative radiography. Forensic Imaging. 2021;26(5):200458. doi:10.1016/j.fri.2021.200458. [Google Scholar] [CrossRef]
27. Fathima NS, Tamilselvi R, Beham MP, Shanmugasundaram R, Deepa P, Premalatha B. Diagnosis of osteoporosis using modified U-Net architecture with attention unit in DEXA and X-ray images. J Xray Sci Technol. 2020;28(5):953–73. doi:10.3233/xst-200692. [Google Scholar] [PubMed] [CrossRef]
28. Irrera P, Bloch I, Delplanque M. A flexible patch based approach for combined denoising and contrast enhancement of digital X-ray images. Med Image Anal. 2016;28(4):33–45. doi:10.1016/j.media.2015.11.002. [Google Scholar] [PubMed] [CrossRef]
29. Dodamani PS, Danti A. Grey wolf optimization guided non-local means denoising for localizing and extracting bone regions from X-ray images. Biomed Pharmacol J. 2023;16(2):935–46. doi:10.13005/bpj/2676. [Google Scholar] [CrossRef]
30. Xie Y, Chen L, Zhou L, Liu Y, Han Z, Li Y, et al. Attention mechanisms in medical image segmentation: a survey. arXiv:2305.17937. 2023. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools