 Open Access
Open Access
ARTICLE
An Efficient CSP-PDW Approach for ECG Signal Compression and Reconstruction for IoT-Based Healthcare
1 School of Computing, Gachon University, Seongnam-si, 13120, Republic of Korea
2 Department of Electronics and Communication Engineering, Indian Institute of Technology (IIT), Dhanbad, 826004, India
3 Department of Information Technology, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
4 School of Computer Science and Engineering, Yeungnam University, Gyeongsan, 38541, Republic of Korea
* Corresponding Authors: Saurabh Agarwal. Email: ; Wooguil Pak. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Recent Advancements in Machine Learning and Data Analysis for Disease Detection)
Computers, Materials & Continua 2025, 85(3), 5723-5745. https://doi.org/10.32604/cmc.2025.070391
Received 15 July 2025; Accepted 10 September 2025; Issue published 23 October 2025
Abstract
A hybrid Compressed Sensing and Primal-Dual Wavelet (CSP-PDW) technique is proposed for the compression and reconstruction of ECG signals. The compression and reconstruction algorithms are implemented using four key concepts: Sparsifying Basis, Restricted Isometry Principle, Gaussian Random Matrix, and Convex Minimization. In addition to the conventional compression sensing reconstruction approach, wavelet-based processing is employed to enhance reconstruction efficiency. A mathematical model of the proposed algorithm is derived analytically to obtain the essential parameters of compression sensing, including the sparsifying basis, measurement matrix size, and number of iterations required for reconstructing the original signal and determining the type and level of wavelet processing. The low time complexity of the proposed algorithm makes it an ideal candidate for ECG monitoring systems in IoT-based e-healthcare applications. A feature extraction algorithm is also developed to show that the important ECG peaks remain unaltered after reconstruction. The clinical relevance of the reconstructed signal and the efficiency of the developed algorithm are evaluated using four validation parameters at three different compression ratios.Keywords
Smart healthcare services are one of the most important aspects of biomedical research, serving humanity. For the last few decades, information and communication technology has played a vital role in the healthcare sector. External or implantable tiny sensors for remote health monitoring have evolved in tandem with the rapid growth of the embedded system and semiconductor industries. Remote healthcare is a technological platform that enables the recording and analysis of vital body parameters, regardless of the patient’s location. This may change the way rural diagnostics and treatment are done. The conventional approach to sampling a signal reliably follows Shannon’s theorem, which states that ‘the sampling rate must be at least twice the maximum frequency present in the signal.’ Breakthrough research [1,2] has been conducted in the field of Compression sensing (CS), and it posed a challenge to the conventional sampling method, stating that ‘sampling frequency can be equal to the maximum frequency component in the signal’. This methodology compresses data while capturing it, ensuring the originality of the data remains intact upon reception. The CS concept is based on two fundamental principles: sparsity and incoherence. Compression algorithms [3] can be classified as lossy and lossless. CS uses the point that most physical signals can be represented as sparse when their sparse basis (
A typical ECG signal is semi-periodic, and the variance throughout the signal is non-uniform [4]. The standard deviation of ECG suggests that 70%–80% of the region has low information, 15%–20% has high information, and the rest has medium information. This semi-sparse nature of the ECG signal makes it an ideal choice for CS algorithms. Incoherence means that the signal should be incoherent among its measurements and have a sparsifying basis. It should spread across the measurement basis. CS undergoes two steps—compression and recovery. CS represents the signals on a linear functional or orthonormal basis. Initially, a framework for CS using the basis pursuit theorem and non-adaptive measurements was discussed, along with the methods to find out the essential parameters for Compressive Sensing [5,6]. The Least Mutual coherence between the measurement and the compressed matrix is one of the vital requirements for a successful reconstruction [7]. The absolute diagonal elements of the identical matrix were considered to reduce the coherence. The main limitation of this framework was its mathematical complexity, as implementing complex mathematical equations overloads the system’s main processor, thereby decreasing processing speed, due to the increased time required for memory read/write cycles. Biomedical signals are very dynamic, as each change has a significant contribution to feature extraction. Hence, processing speed and accurate reconstruction are the key factors in biomedical signal processing [8]. General dictionaries, as well as specific dictionaries, can be built using normal and pathological cardiac patterns and can be used for CS [9]. The method ensures the processing of fewer bits of information, reducing memory and power requirements [10]. An ECG signal can be accurately represented as a sparse combination of a few significant bands. Measurement matrix and sensing matrix construction are important parts of CS. The size of the measurement matrix and its lesser coherence with the sensing matrix are key requirements of CS [3]. Authors in [11,12] explained the size of the measurement matrix based on the Robust Uncertainty principle and suggested using an optimized measurement matrix to improve reconstruction efficiency. ECG compression on an embedded Shimmer wireless mote discusses the importance of the Restricted Isometry Property [13] and the Basic Pursuit algorithm in constructing the sensing matrix for successful compression and reconstruction. Another hurdle to processing the ECG signal is its localized behavior. Block Sparse Bayesian Learning frameworks were utilized to enhance patient mobility and mitigate the location dependency of ECG signals [14]. Furthermore, the wavelet transform was suggested to enhance the reconstruction efficiency. It enhances the application of CS on the ECG signal by decomposing the signal into sub-bands, thereby preserving spatial resolution, frequency, and directional characteristics [13,15–17].
Huang et al. [18] utilized down-sampling techniques to reduce and compress the ECG data signals. For this purpose, they have employed the China Physiological Signal Challenge 2018 dataset and root mean square error metrics to evaluate the obtained compression result. Singhai et al. [19] presented nature-inspired algorithms with the discrete wavelet transform (DWT) for the compression of ECG signal data. They utilized DWT to decompose the signals and optimization techniques to select specific and suitable threshold values. They have also calculated various parameters, including PRD, QS, SNR, and CR, to validate the performance of their algorithm. Yildirim et al. [20] presented a 27-layer deep learning structure comprising encoder and decoder sections for compressing large ECG datasets. They utilized 4800 ECG segments from 48 patients to validate the compression result. They obtained a 32.25 compression rate and a 2.73% average percent root mean square difference (PRD) value using the model. The hybrid lossy/lossless compression system Chang and Sobelman [21] suggest has high signal quality and CR properties. After decompression, deep neural networks are used to assess performance. Fathi et al. [22] presented an algorithm for compressing ECG signals from the MIT-BIH arrhythmias database and the ECG-ID database. In their algorithm, they used discrete Krawtchouk moments and Ant Lion Optimizer as feature extractors and feature selectors. They obtained the following average values of quality score (QS), Peak signal-to-noise ratio (PSNR), CR, signal-to-noise ratio (SNR), and PRD: 23.92, 49.04, 15.56, 44.52, and 0.69, respectively. Shi et al. [23] proposed an ECG signal compression method utilizing residual error compensation based on a binary convolutional autoencoder. In this work, they utilized the MIT-BIH dataset to validate their methodology, obtaining a compression ratio of 117.33 and a PRD of 7.76%. Kuldeep and Zhang [24] introduced a lightweight ECG signal compression technique for transmitting signals from the MIT-BIH dataset. In this work, the authors have utilized two pruning methods, sign and binary pruning, which require less computation. The Gaussian elimination method was employed for the reconstruction of ECG signals, and its performance was validated using the metrics QS, PRD, and CR. Pal et al. [25] presented a method for compressing ECG signals using the hybrid coronavirus herd immunity optimizer and tunable-Q wavelet transform (TQWT). To validate their algorithm, they utilized the MIT-BIH dataset and multiple evaluation metrics, CR, SNR, PRD, QS, and correlation coefficient. A Very-Large-Scale Integration architecture for an ECG compression engine is introduced [26]. The processor’s efficiency was confirmed on MIT BIH databases. The authors [27] introduced a compression algorithm that combines an adaptive TQWT with a modified dead-zone quantizer. A new Sparse Grey Wolf Optimization algorithm is presented to optimize TQWT parameters. In [28], a 2-D ECG compression technique whereby DCT and TQWT were combined with both row-wise and column-wise operations employed to grow sparsity in the transform domain. The TQWT and quantizer parameters were tuned using the COOT bird optimization algorithm.
Prior studies on ECG compression have focused on either compressed sensing (CS) or wavelet-based methods, which have limited clinical applicability due to limitations in achieving high compression rates and efficient, high-fidelity reconstruction. The CS-PDW framework combines compressive sensing (CS) with a Primal-Dual wavelet-based reconstruction method that utilizes wavelet-domain sparsity to achieve high compression and high-fidelity reconstruction in a wide range of challenging compression ratios. In contrast to the works above, where parameters are set empirically, the proposed work enables the analytically determination of correct CS and wavelet settings, promising better fidelity and lower computational cost. Aimed at IoT healthcare, CSP-PDW has a low time complexity and achieves excellent performance in preserving diagnostic features. The combination of these features—compression efficiency, robustness, and real-time readiness—makes the proposed scheme a feasible solution for IoT-enabled e-healthcare applications.
CS brought a revolution in the way signals are handled and analyzed worldwide. It enhances the ability of a device to utilize allocated bandwidth effectively, while also improving the battery life of implantable devices and making them more portable. This paper presents a mathematically hardware-realizable and straightforward CS algorithm for ECG signals with moderate compression efficiency. The offered CSP-PDW techniques can also be applied to other biomedical signals. This paper has two segments. The first one is compression, and the second one is the reconstruction and wavelet processing procedure. In the compression procedure, the Fast Fourier Transform (FFT) is applied to generate the sparse representation of the ECG signal. A random Gaussian matrix is used to generate the measurement matrix. Iterative algorithms and optimization algorithms are used to reconstruct the original ECG signal. Additionally, wavelet-based processing is performed to improve the overall reconstruction efficiency.
The paper is organized as follows: Section 2 describes some preliminary state-of-the-art concepts used in this work. Section 3 illustrates the proposed scheme. Section 4 defines the simulation result and performance analysis. Finally, the conclusion is discussed in the last section.
This section explains the methods used to design the algorithm for efficiently compressing ECG signals. The algorithm consists of two segments: the first compresses the signal, and the second reconstructs without losing information. For this reason, compression sensing (CS), restricted isometry property, and DWT were utilized, along with selected optimization techniques. These methods are briefed in the preceding subsections.
CS relies on two major principles: sparsity and incoherence [5,29]. A sparse signal is a special type of signal that comprises a small amount of information compared to its total length. Consider a vector
where X is the coefficient sequence of the original ECG signal c. Generally, CS utilizes the measurement matrix
where
Considering sparsity
where
Consider a signal
If c is compressible and sparse, the approximation will be accurate and the error
here the columns of
The limit of the coherence is given by:
Random numbers adhere to the least mutual coherence. Independent Identically Distributed (i.i.d.) entries of a random Gaussian matrix are used in this paper to construct the measurement matrix. As the signal is represented as sparse, it contains many zero entries, but the actual locations of the non-zero points are unknown. Fixed basis Gaussian random numbers were used in this paper to randomly choose the rows and columns with non-zero entries to generate a sensing matrix. A PDF of i.i.d. entries of a random Gaussian matrix can be written mathematically as
where,
where, ‘
2.2 Restricted Isometry Property
The Restricted Isometry Property (RIP) provides sufficient conditions to approximate large matrices for the implementation of the CS concept. The existence of inimitability of the solution can be guaranteed if the sensing matrix (A) follows RIP in order of s as:
where,
Given the RIP condition, the relationship between
For the matrix
where
In compressed sensing, the
In this paper, a convex problem is relaxed by using
Compressed sensing often deals with underdetermined systems, where the number of unknowns, n, exceeds the number of measurements, m. In this case, infinite (maximum) solutions exist due to the problem’s underdetermined nature. To obtain the sparsest solution, the problem can be reformulated as a dual problem and solved using the primal-dual approach.
The primal problem seeks to maximize the objective function
here,
The Dual problem involves minimizing the objective function
here,
The Backline search approach is used to find the step size
where,
The prime dual approach, combined with a backtrack line search algorithm, is utilized for compressed sensing to solve underdetermined systems and ensure convergence to sparse solutions, as outlined in Algorithm 1.

2.4 Discrete Wavelet Transformation
The Wavelet transform provides a multiresolution analysis of the signal, making it useful for enhancing signal reconstruction. The Continuous Wavelet Transform (CWT) provides a redundant representation of the signal, which may not be necessary for full signal reconstruction. The CWT of a function
where,
where, Dilation factor:
The
where
This section describes the details of the methods used for ECG signal compression and reconstruction using hybrid compressed sensing and primal-dual wavelet (CSP-PDW) techniques. Fig. 1 shows the block diagram of the proposed methodology. In the first step, data were collected from two sources: the PTB-DB and MIT-BIH databases, both of which are benchmarked and open-access datasets containing various types of ECG signals, available on the PhysioNet website. In the second step, the collected datasets were denoised using the DWT. In the third step, baseline wander was removed using the polynomial fitting technique. ECG signal compression is performed after signal preprocessing, utilizing modified compressed sensing algorithms for the compression process. In the next step, greedy iterative optimization algorithms were applied to reconstruct the compressed ECG signal. Wavelet transform techniques were employed to enhance further the quality and originality of the reconstructed ECG signal. Finally, postprocessing was conducted using various metrics to validate the suggested methodology. Each step is described in detail in the preceding subsections.
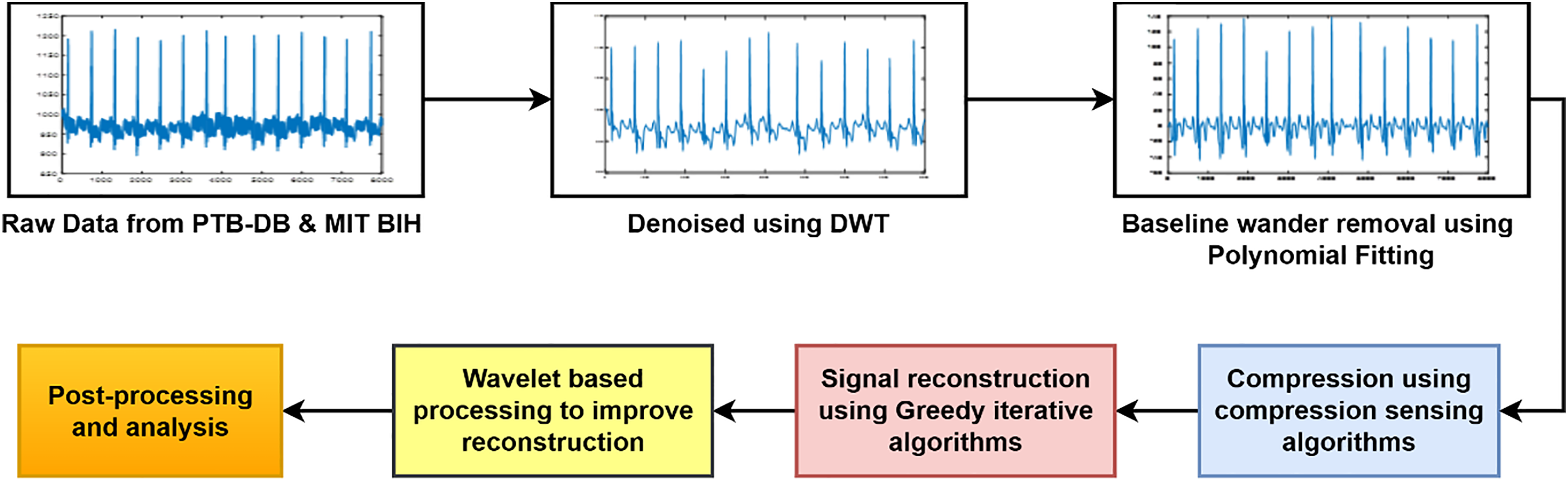
Figure 1: Block diagram of the process
3.1 ECG Data Collection and Preprocessing
ECG signal data are acquired from the PhysioNet database, which serves as a benchmark and open-access directory for ECG datasets. The two ECG datasets, the PTB Diagnostic ECG Database (PTB-DB) and the MIT-BIH arrhythmias database (MIT-BIH), are considered to maintain the generalizability of the methodology. The PTB-BD dataset contains records of 549 patients, consisting of 290 subjects, and includes 15-lead ECG signals. In comparison, the MIT-BIH database contains 48 records, each consisting of 30 min with 2-channel recordings. The dataset is digitized at 360 Hz per channel with an amplitude range of up to 10 mV. As the two datasets differ entirely in both content and length, they were segmented into 2-s recordings. The acquired data is contaminated with several sources of noise. Since the wavelet is a very powerful and efficient tool for noise removal, a wavelet-based denoising algorithm is utilized to denoise the acquired ECG data. The best part of the WT is that it preserves important features while denoising, resulting in no loss of integrity to the original signal. The DWT of 10 levels of decomposition is used to obtain the approximation and detail coefficients of the pre-processed ECG signals, as given by Eq. (22).
where A represents the approximation coefficients and D represents the detail coefficients. The noise level was estimated from the detail coefficients at the highest level using the median absolute deviation (MAD) estimator, as given in Eq. (23).
After that, soft thresholding was applied at each level of the detail coefficients, as given in Eq. (24).
Ultimately, the signals are reconstructed by applying the inverse DWT to the threshold coefficients, thereby obtaining the denoised ECG signal, as shown in Eq. (25).
The denoised ECG signals exhibited baseline wander, which was corrected using the polynomial fitting technique. A polynomial of degree 5 was fitted to the denoised ECG signals obtained in Eq. (25) to capture the baseline wander, as shown in Eq. (26).
where
The obtained ECG signal (
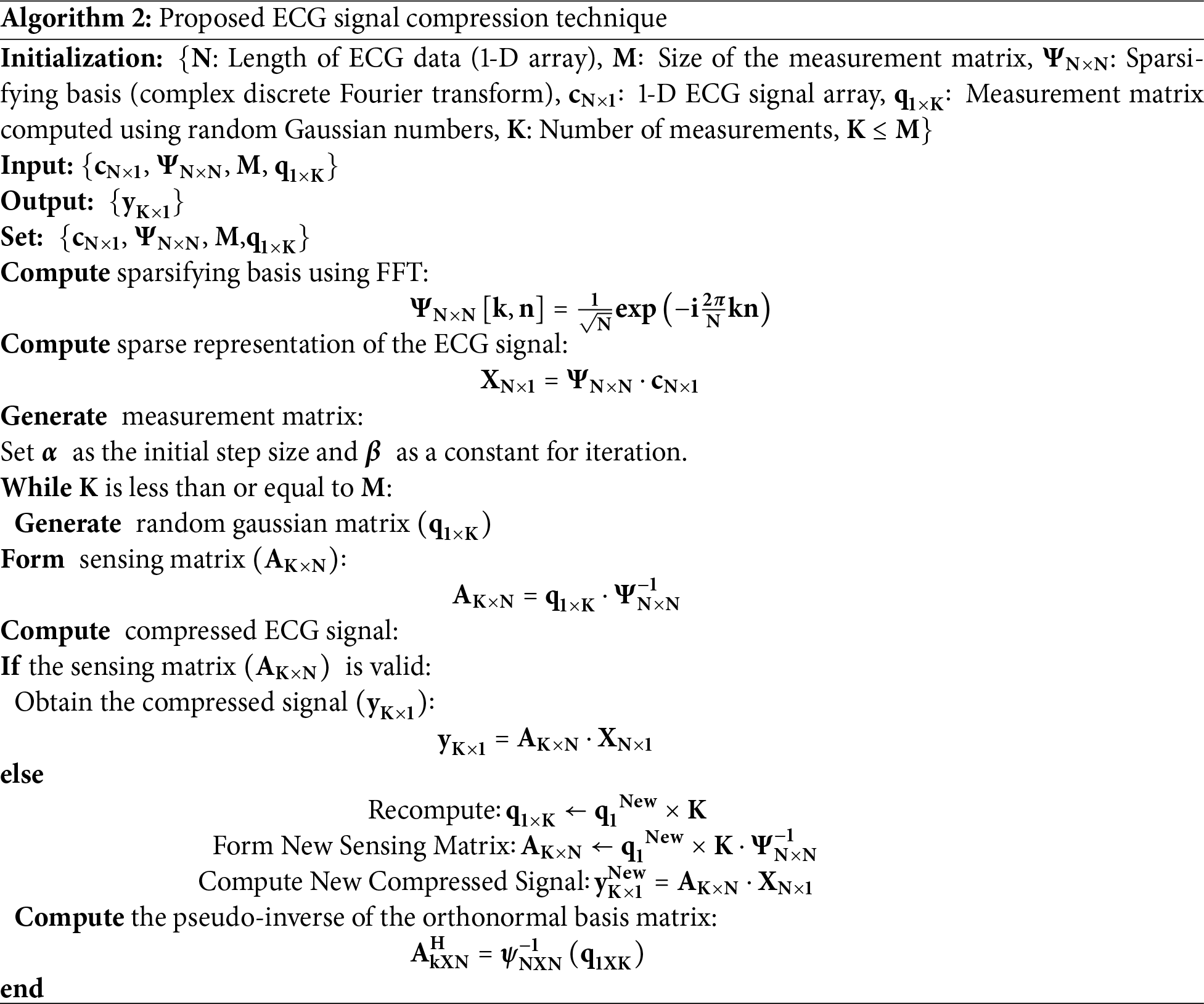
3.2 Proposed ECG Signal Compression Technique
The proposed compression algorithm leverages the features of CS and sparse representation principles to obtain high compression rates while maintaining signal integrity with accurate reconstruction. The stepwise visual representation of the enhanced ECG compression algorithm is presented in Fig. 2. After choosing the ECG signal length and creating a 1D array, the FFT is used to calculate orthonormal bases. The RIP technique uses optimal size selection to build a random Gaussian matrix. The compressed matrix is created by multiplying the sensing matrix by the orthonormal bases. The compressed signal is calculated once the correctness of the sensing matrix is verified. The measurement matrix is recalculated otherwise. Lastly, the signal is reconstructed by pseudo-inverting the orthonormal basis.
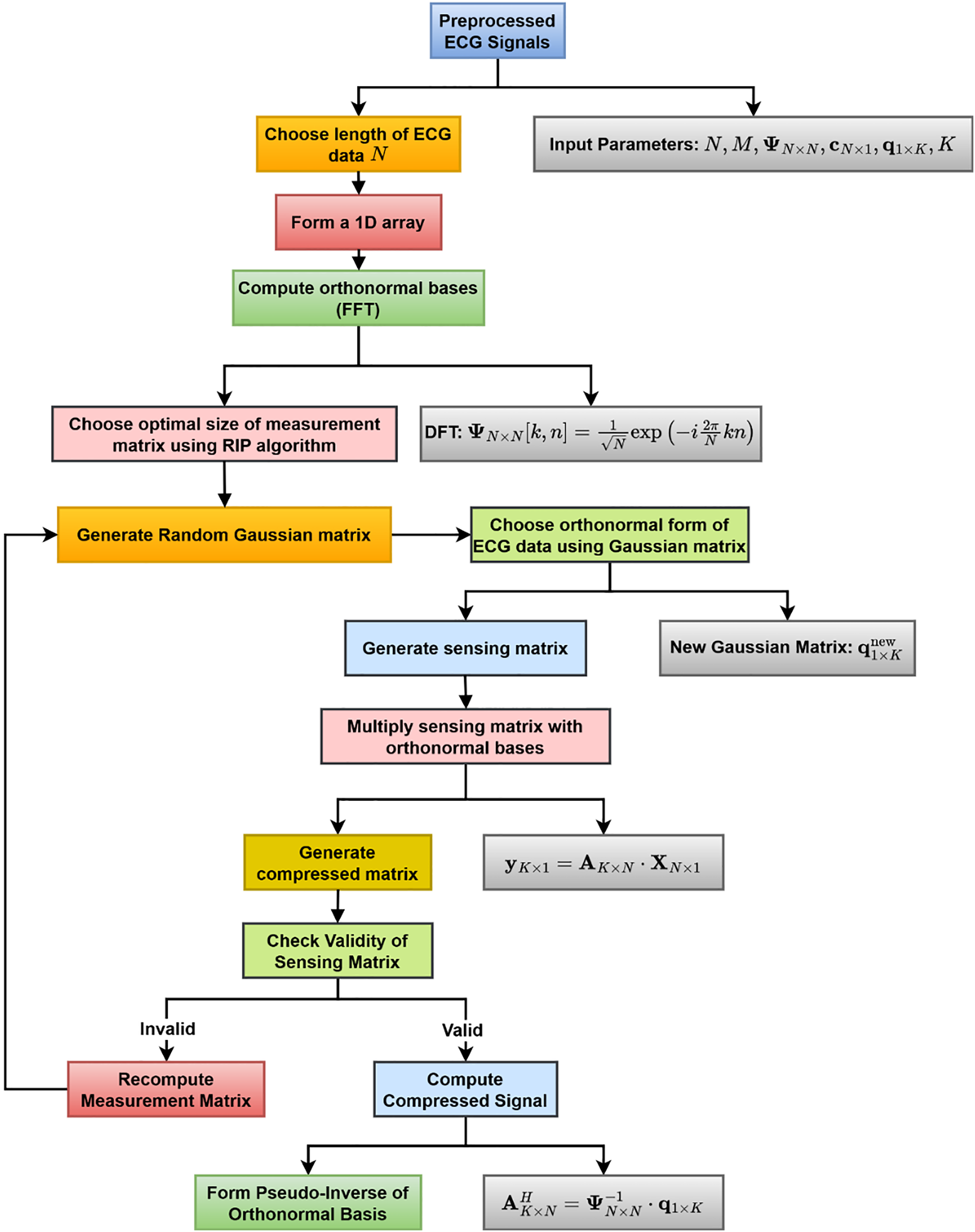
Figure 2: Block diagram of the proposed ECG signal compression technique
In this work, a length (N) of 2000 samples of the ECG signal was chosen for compression. ECG signal is semi-sparse in nature, so FFT is computed to convert the data into its sparse form by computing sparsifying basis matrix
This computation provides a minimal number of non-zero coefficients in the Fourier domain of the ECG signal with a high degree of accuracy. In the next step, the sparse representation of the pre-processed ECG signal
In the following step, the measurement matrix is generated, where the
The sensing matrix was subsequently validated, and upon validation, the compressed ECG signal
If the validation of the sensing matrix is unsuccessful, the measurement matrix is recomputed, a new sensing matrix is formed, and a new compressed ECG signal is calculated.
Finally, the pseudo-inverse of the orthonormal basis matrix is computed to ensure the accurate reconstruction of the original ECG signal, as represented in Eq. (33).
This approach effectively compresses the ECG signal while preserving the essential features necessary for accurate and reliable reconstruction. The algorithm’s robustness in handling the sparse representation of the ECG signal in the Fourier domain, along with its efficient measurement matrix generation and computation, makes it a highly effective solution for compressing ECG signals. The complete steps of the ECG compression method are presented in Algorithm 2.
3.3 Proposed ECG Signal Reconstruction Technique
Fig. 3 presents the flowchart of the proposed sparse representation and primal-dual wavelet-based ECG reconstruction technique. Optimization techniques, namely Primal-Dual and basis pursuit, were used to recover the coefficients of the original signal for reconstruction. One of the major limitations of the algorithms is that they are only applicable to convex problems. A standardization algorithm was applied to reduce signal variance. Once the master solution set is obtained, greedy iterative algorithms (Backtracking line search) were used to get the optimum solution set. To improve reconstruction efficiency, wavelet-based processing was applied to the reconstructed signal.
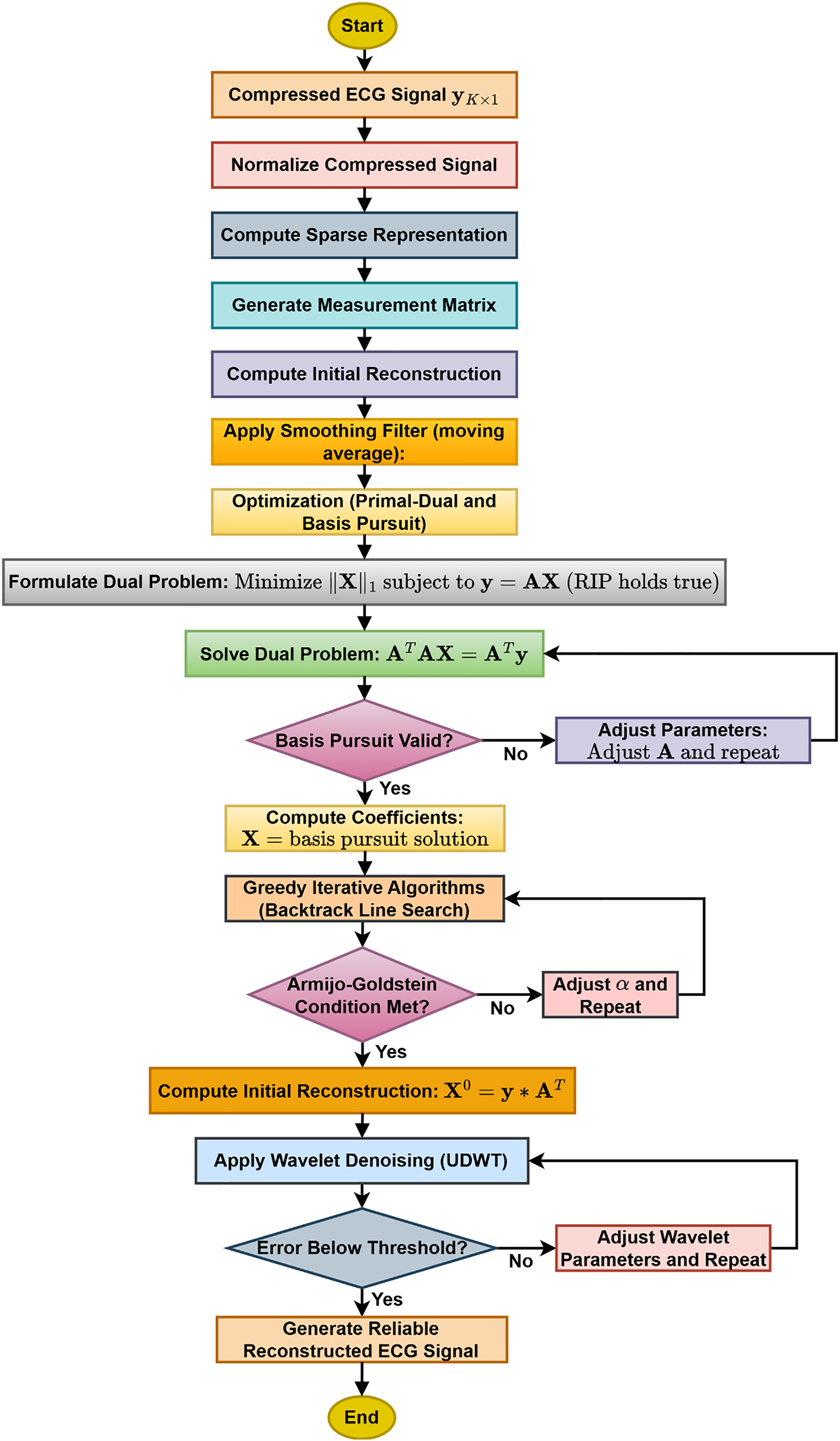
Figure 3: Flowchart of the proposed sparse representation and primal-dual wavelet-based ECG reconstruction technique
The signal
The signal
A large initial estimate of
Eq. (16) simplifies to Eq. (17), providing the estimated ECG input signal

4 Simulation Result and Performance Analysis
In this section, the experimental results and performance of the proposed compression and reconstruction algorithm are discussed. The algorithm that was developed was tested on the PhysioNet database (PTB-DB and MIT-BIH arrhythmia). For the PTB-DB database, the sampling frequency is 1000 Hz with a 16-bit resolution over a ±16 mV range. For the MIT-BIH arrhythmia database, the sampling frequency is 360 Hz with 11-bit resolution over a 10 mV range. Five standard performance metrics were used to quantify the efficiency of the compression and decompression algorithms. Compression ratio (CR) (Eq. (18)) to estimate the amount of compression and the remaining four parameters, namely Mean square Error (MSE) (Eq. (19)), PRD (Eq. (20)), Correlation Coefficient (CoC) (Eq. (21)), and Signal to noise ratio (SNR) (Eq. (22)), reflect on the efficiency of the decompression algorithm to preserve the clinical relevance of ECG data.
where, N = Length of the signal, (i) = Input Reference signal, x′(i) = reconstructed signal output. The correlation between the PRD and the clinical relevance of the reconstructed signal was suggested by Zigei et al. [30]. Table 1 presents the percentage of PRD corresponding to different classes of signal reconstruction quality, categorized by diagnostic relevance.
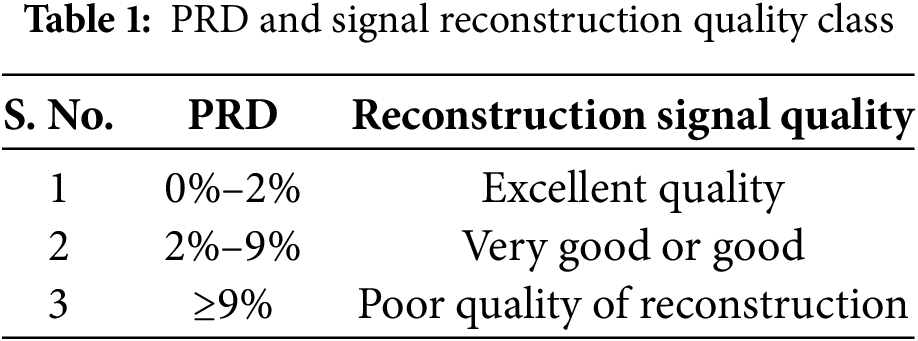
4.1 Simulation Result for Compression and Reconstruction Algorithm
Fig. 4 illustrates the test results of the developed ECG signal compression and reconstruction algorithm using a healthy record (s-0464) from the PTB-DB database. Fig. 5 illustrates the performance analysis of the proposed reconstruction algorithm at various compression ratios using the same record. Fig. 6 shows some deviations in the isoelectric part of the ECG signal, as Fourier-based denoising techniques suffer from this error, although peaks remain unaltered.
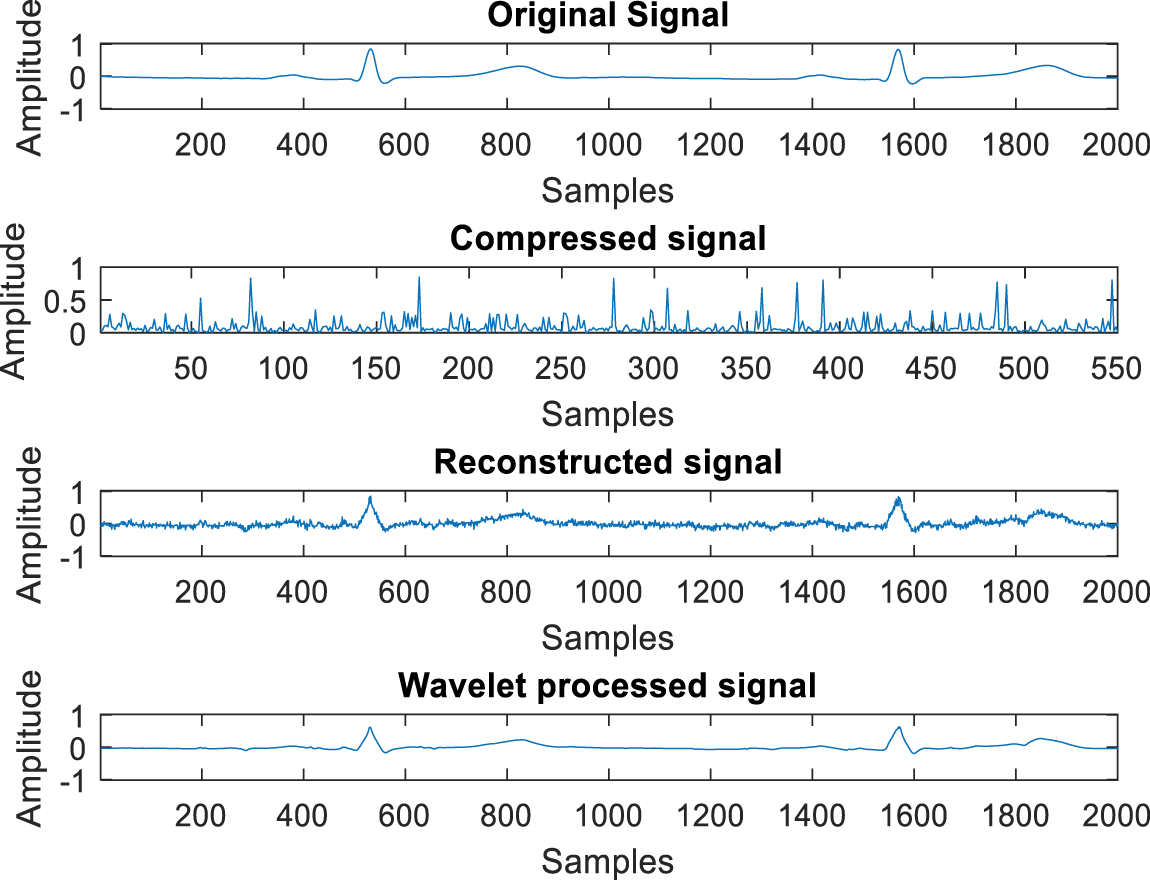
Figure 4: The test result of compression and reconstruction with rec. s_0464
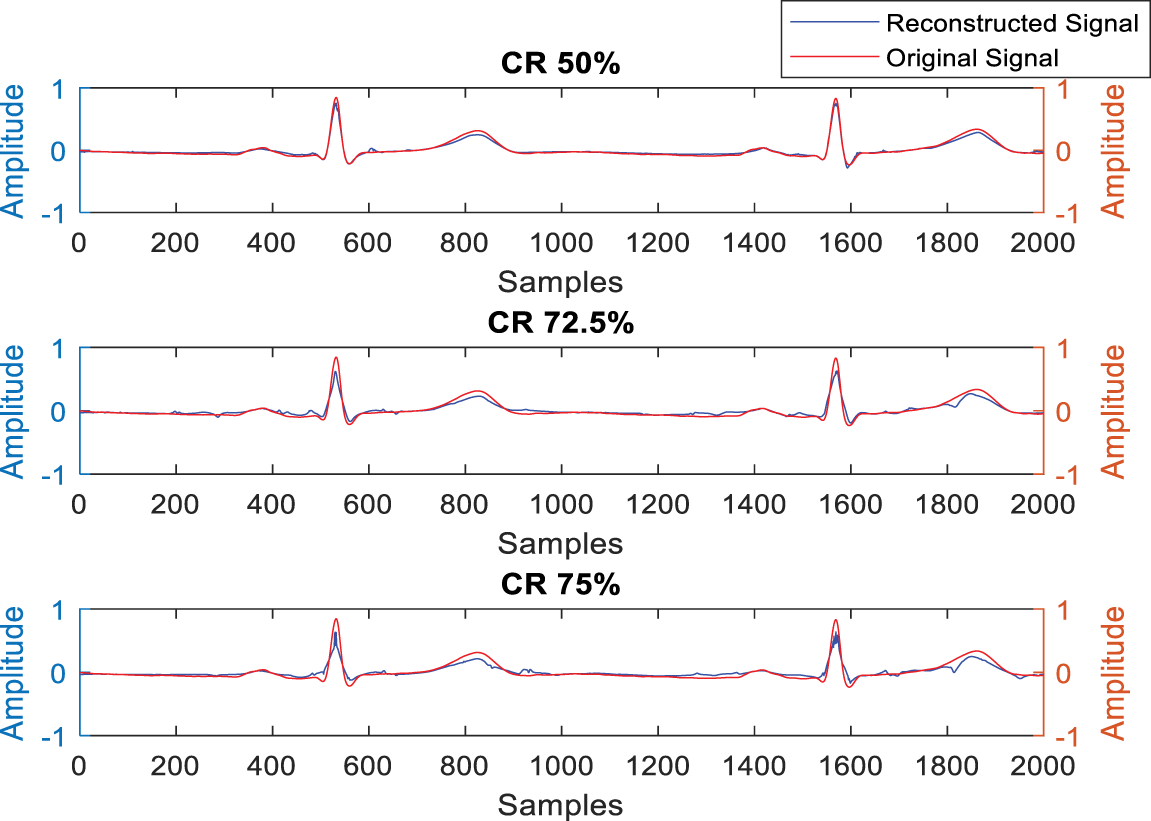
Figure 5: Test result for different compression ratios with rec. s_0464
Figure 6: Test result for QRS complex detection after reconstruction with rec. s_0464
4.2 Performance Metrics Calculation
Table 2 indicates the performance analysis of different wavelets. Column I lists the different wavelets used in this comparative study, while columns II, III, IV, V, and VI present five assessment parameters. According to the results obtained in Table 2, the Level-10 Coif-1-based wavelet filtering method showed the most satisfactory reconstruction of the ECG data.
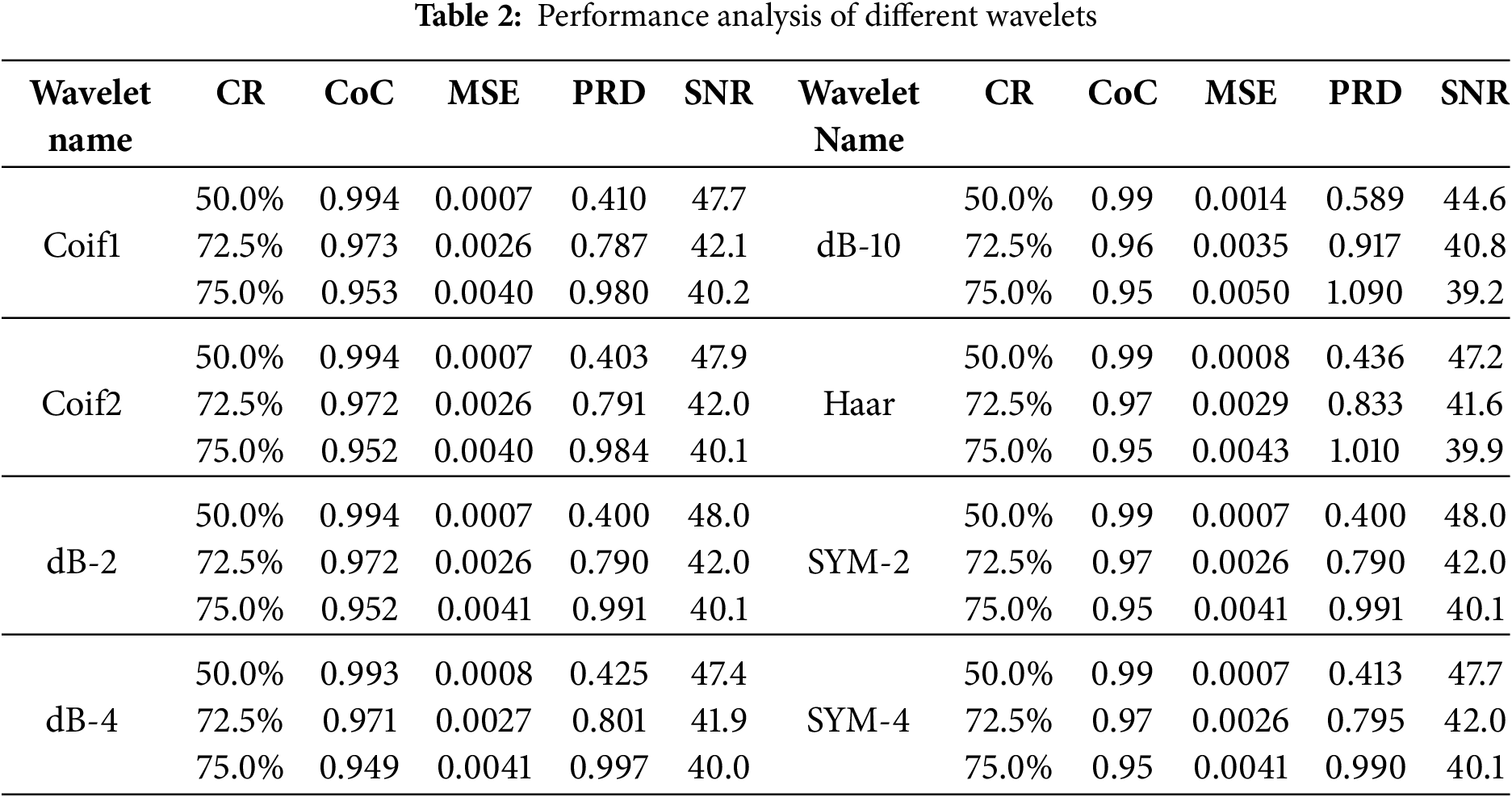
Table 3 presents the estimated assessment parameters for six different ECG signals. Different validation parameters were calculated to justify reliable reconstruction with different compression ratios. Column II indicates compression ratios, while the remaining columns display standard performance metrics used to evaluate the quality of reconstruction. Table 3 indicates a satisfactory reconstruction as per the criteria listed in Table 1.
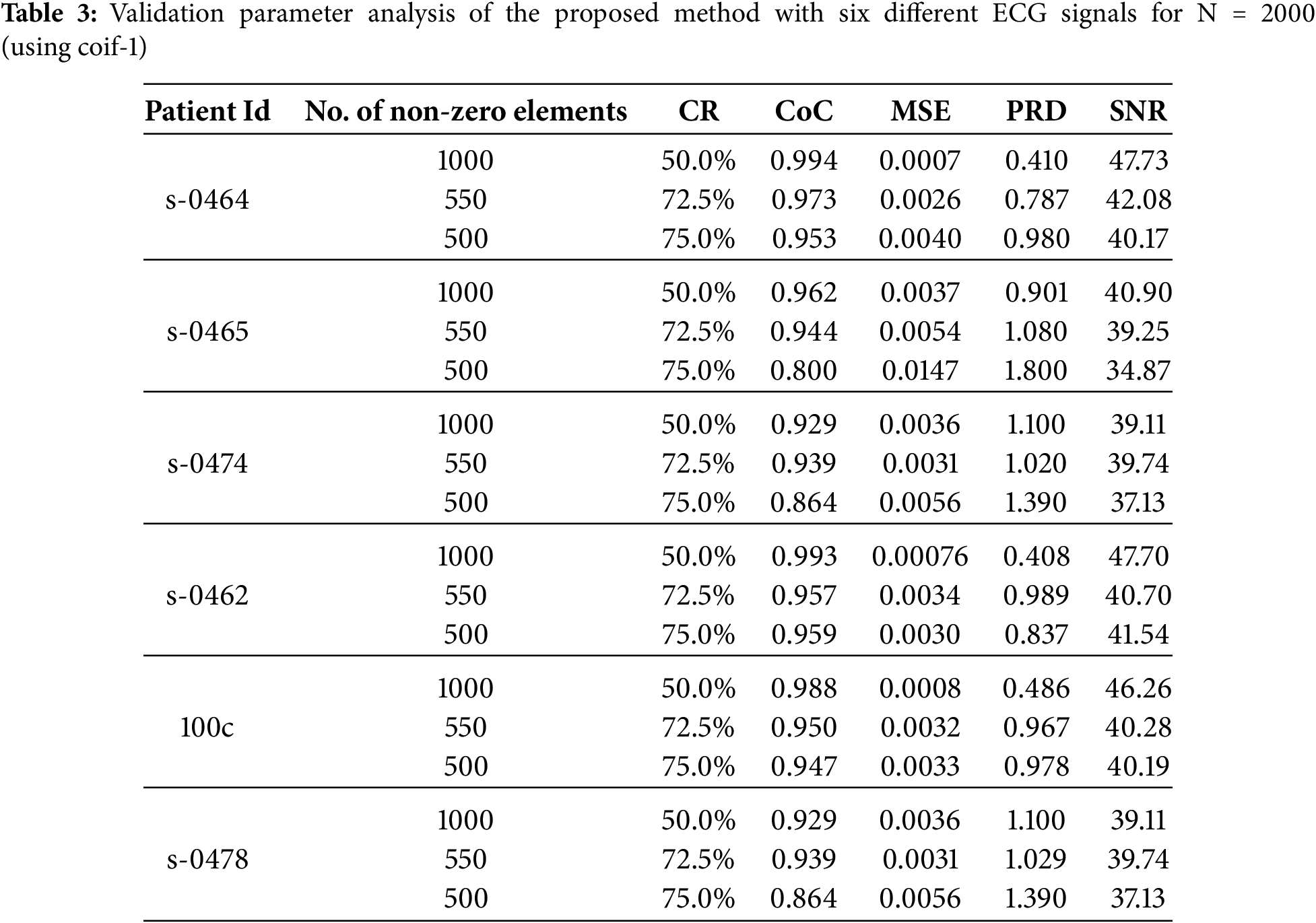
It is observed from Fig. 7 that with an increase in compression ratio, MSE and PRD increase, so to retain the clinical usability of the reconstructed data, the compression ratio should be adjusted accordingly. Fig. 8 shows that increasing the number of non-zero elements in the sensing matrix improves CoC. PRD values in Fig. 9 indicate the comparative analysis of all validation parameters before and after applying wavelet-based filtering. All validation parameters suggested satisfactory results for all tested samples.
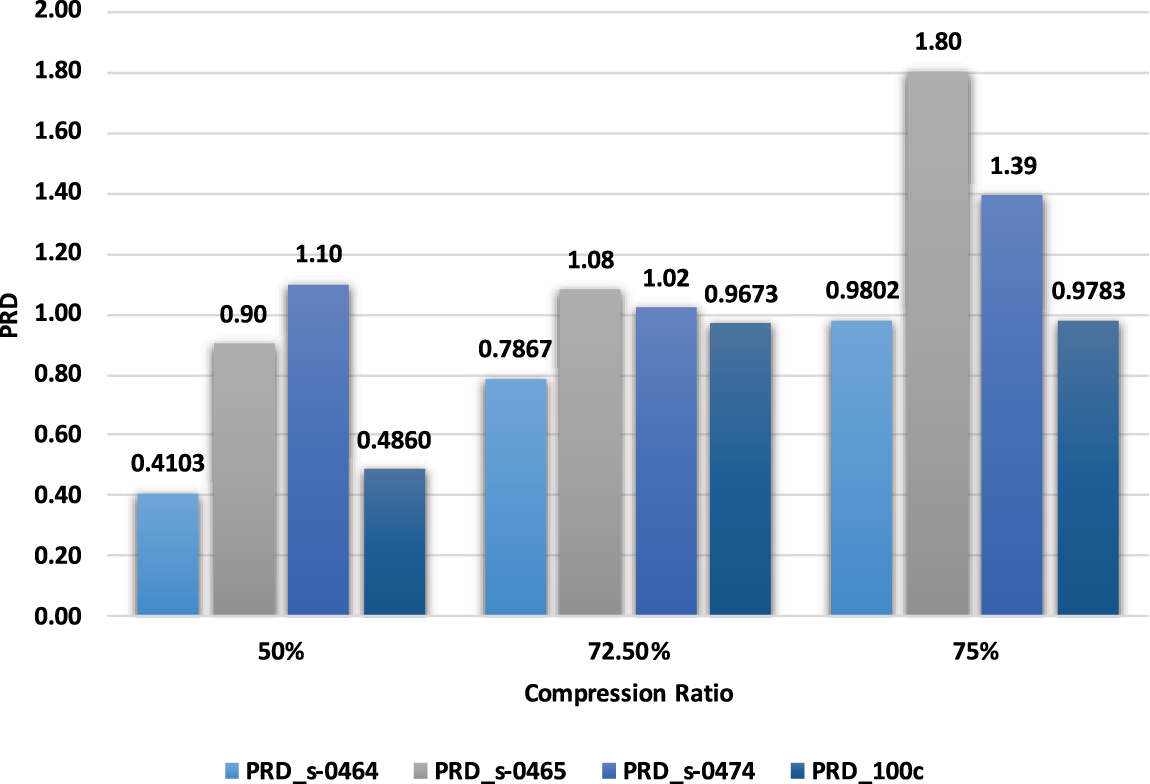
Figure 7: Comparison of PRD with Compression ratio
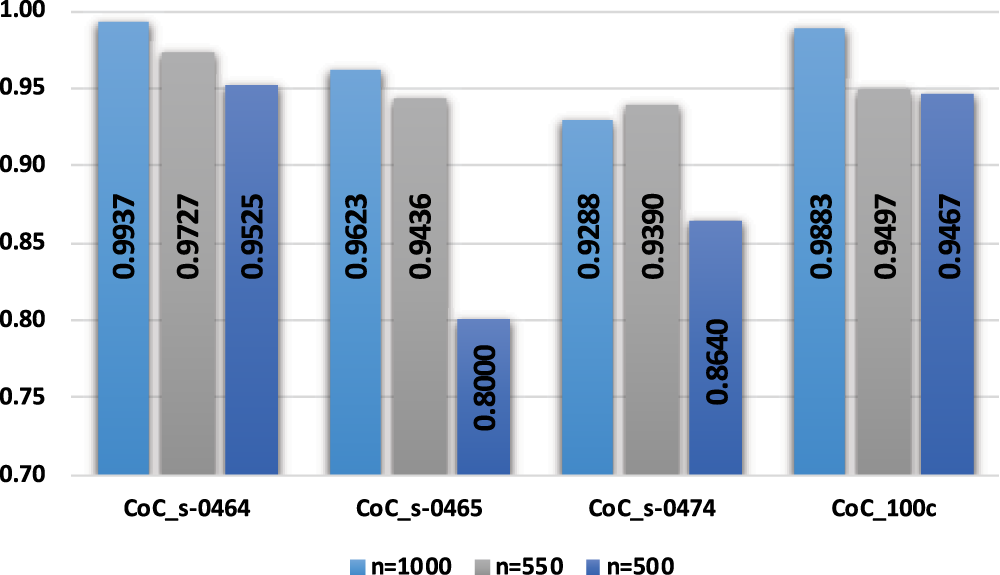
Figure 8: Comparison of CoC with non-zero elements
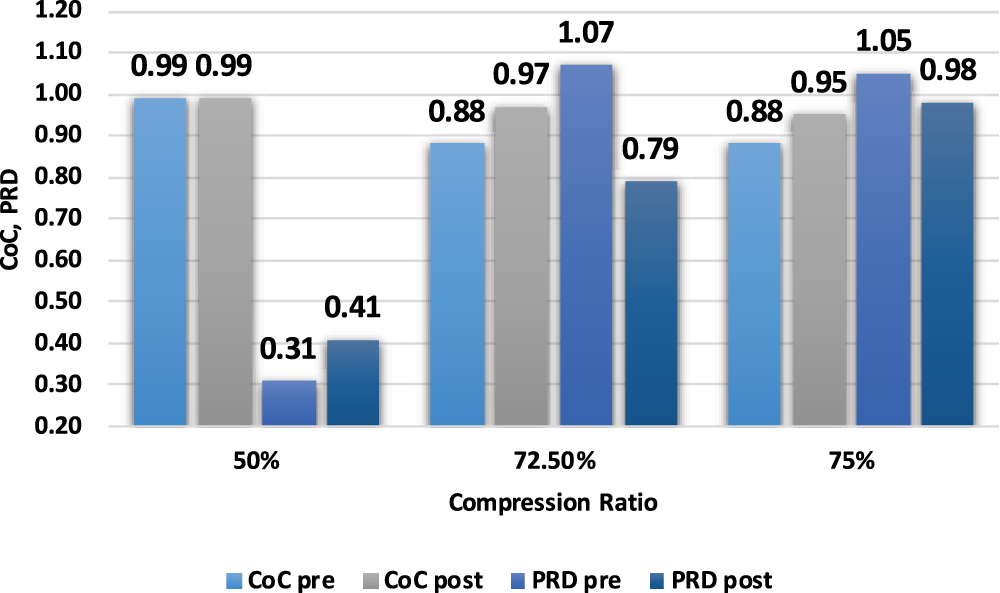
Figure 9: Pre-analysis and post-analysis of validation parameters
The CSP-PDW framework has been designed to meet the needs of real-time ECG analysis in IoT-based e-healthcare, where low latency and energy efficiency are of paramount importance. Through the merger of compressed sensing with wavelet-domain processing, it reduces the number of samples as well as the reconstruction dimension, allowing for diagnostic insights with millisecond precision and without a high burden of postprocessing. We have O(MN) (Gaussian projection) complexity for compression (M ≪ N), O(N) complexity for wavelet transforms, and O(kMN) complexity for Primal-Dual reconstruction, where k is a small number of iterations due to rapid convergence. The approach runs in below 50 ms per 1 s ECG segment (on a mid-range CPU). It achieves a good trade-off between compression efficiency, reconstruction accuracy, and speed, making it practical for embedded medical devices and wireless ECG monitors with scarce computing resources.
4.3 Limitations and Future Directions
Apart from several significant findings, our proposed CSP-PDW method also has some limitations. At extremely high compression ratios, fine morphological details, especially the ECG signals with complex morphological variations or arrhythmias, may be partially lost. The reconstruction quality of the ECG signal is sensitive to the selection of parameters, such as the sparsifying basis, measurement matrix size, and number of iterations. However, the use of a fixed mother wavelet may not be optimal for all patient records or pathological conditions. Although the proposed method is computationally efficient for standard IoT hardware, ultra-low power devices with minimal resources may require further optimization. The evaluation and validation of the proposed method were limited to standard ECG databases, and broader validation on diverse real-world datasets under varying noise conditions remains necessary.
In future work, the CSP-PDW algorithm will also be enhanced for robust processing of multi-class ECG signals across various morphologies and pathological conditions, with automatic parameter tuning and adaptive selection of mother wavelets. We will also extend our approach to utilize hardware-level optimizations for ultra-low-power IoT devices and validate it using larger, realistic datasets with real-world noise and motion artifacts, ensuring future reproducibility for continuous, real-time clinical monitoring.
This paper has presented a hybrid CSP-PDW technique for ECG signal compression and reconstruction of original ECG signals. The simplicity of the suggested algorithm makes it suitable for hardware-level implementation. The number of bits during measurement and transmission has been reduced by the suggested algorithm, which in turn has reduced the overall energy consumption. The algorithm presented has been less complex and has thus provided an opportunity to design standalone embedded systems using low-cost embedded boards for various applications. The average value of the correlation coefficient and the validation parameters suggests that the retention of clinical relevance is maintained after reconstruction. Obtained values for PRD, SNR, MSE, and CoC at a maximum compression ratio of 75% were 1.23, 38.05, 0.006, and 0.89, respectively. Fast data acquisition has addressed bandwidth and power issues in rural India, making it suitable for smart healthcare applications.
Acknowledgement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R749), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R749), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: Methodology, Hari Mohan Rai, Chandra Mukherjee, Joon Yoo, Hanaa A. Abdallah, Saurabh Agarwal, Wooguil Pak; Conceptualization, Hari Mohan Rai, Chandra Mukherjee, Joon Yoo, Hanaa A. Abdallah, Saurabh Agarwal, Wooguil Pak; Visualization, Hari Mohan Rai, Chandra Mukherjee, Joon Yoo, Hanaa A. Abdallah, Saurabh Agarwal, Wooguil Pak; Writing—original draft preparation, Hari Mohan Rai, Chandra Mukherjee, Joon Yoo, Hanaa A. Abdallah, Saurabh Agarwal, Wooguil Pak; Writing—review and editing, Hari Mohan Rai, Chandra Mukherjee, Joon Yoo, Hanaa A. Abdallah, Saurabh Agarwal, Wooguil Pak. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset used in this work is publicly available and may be freely downloaded from the given link: MIT-BIH dataset: https://www.physionet.org/content/mitdb/1.0.0/; PTB-DB dataset: https://www.physionet.org/content/ptbdb/1.0.0/ (accessed on 09 September 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Donoho DL. Compressed sensing. IEEE Trans Inf Theory. 2006;52(4):1289–306. doi:10.1109/TIT.2006.871582. [Google Scholar] [CrossRef]
2. Candes EJ, Wakin MB. An introduction to compressive sampling. IEEE Signal Process Mag. 2008;25(2):21–30. doi:10.1109/MSP.2007.914731. [Google Scholar] [CrossRef]
3. Lal B, Gravina R, Spagnolo F, Corsonello P. Compressed sensing approach for physiological signals: a review. IEEE Sens J. 2023;23(6):5513–34. doi:10.1109/jsen.2023.3243390. [Google Scholar] [CrossRef]
4. Gupta V, Chaturvedi Y, Saxena NK, Sharma AK, Salim, Gupta V. ECG signal analysis using emerging tools in current scenario of health informatics. In: 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence); 2021 Jan 28–29; Noida, India. doi:10.1109/confluence51648.2021.9377119. [Google Scholar] [CrossRef]
5. Hassan AMA, Mohsen S, Abo-Zahhad MM. ECG signals compression using dynamic compressive sensing technique toward IoT applications. Multimed Tools Appl. 2023;83(12):35709–26. doi:10.1007/s11042-023-17099-7. [Google Scholar] [CrossRef]
6. Hui C, Zhang S, Cui W, Liu S, Jiang F, Zhao D. Rate-adaptive neural network for image compressive sensing. IEEE Trans Multimed. 2023;26:2515–30. doi:10.1109/TMM.2023.3301213. [Google Scholar] [CrossRef]
7. Mavaddati S. Classification of ECG arrhythmia using wavelet packet transform analysis and sparse learning method. Iran J Sci Technol Trans Electr Eng. 2023;47(4):1583–93. doi:10.1007/s40998-023-00620-x. [Google Scholar] [CrossRef]
8. Alqudah AM, Moussavi Z. A review of deep learning for biomedical signals: current applications, advancements, future prospects, interpretation, and challenges. Comput Mater Contin. 2025;83(3):3753–841. doi:10.32604/cmc.2025.063643. [Google Scholar] [CrossRef]
9. Fira M, Costin HN, Goraş L. A study on dictionary selection in compressive sensing for ECG signals compression and classification. Biosensors. 2022;12(3):146. doi:10.3390/bios12030146. [Google Scholar] [PubMed] [CrossRef]
10. Ansari-Ram F, Hosseini-Khayat S. ECG signal compression using compressed sensing with nonuniform binary matrices. In: The 16th CSI International Symposium on Artificial Intelligence and Signal Processing (AISP 2012); 2012 May 2–3; Shiraz, Iran. doi:10.1109/AISP.2012.6313763. [Google Scholar] [CrossRef]
11. Kot A, Nawrocka A. Measurement uncertainty for biological signals. In: Proceedings of the 2025 26th International Carpathian Control Conference (ICCC); 2025 May 19–21; Starý Smokovec, High Tatras, Slovakia. doi:10.1109/ICCC65605.2025.11022854. [Google Scholar] [CrossRef]
12. Patel S, Vaish A. An efficient optimization of measurement matrix for compressive sensing. J Vis Commun Image Represent. 2023;95(4):103904. doi:10.1016/j.jvcir.2023.103904. [Google Scholar] [CrossRef]
13. Singhai P, Ateek A, Kumar A, Ahmad Ansari I, Bhalerao S. ECG signal compression based on wavelet parameterization and thresholding using PSO. In: 2020 International Conference on Communication and Signal Processing (ICCSP); 2020 Jul 28–30; Chennai, India. doi:10.1109/iccsp48568.2020.9182407. [Google Scholar] [CrossRef]
14. Zhang Z, Jung TP, Makeig S, Rao BD. Compressed sensing for energy-efficient wireless telemonitoring of noninvasive fetal ECG via block sparse Bayesian learning. IEEE Trans Biomed Eng. 2013;60(2):300–9. doi:10.1109/TBME.2012.2226175. [Google Scholar] [PubMed] [CrossRef]
15. Lu Z, Kim DY, Pearlman WA. Wavelet compression of ECG signals by the set partitioning in hierarchical trees algorithm. IEEE Trans Biomed Eng. 2000;47(7):849–56. doi:10.1109/10.846678. [Google Scholar] [PubMed] [CrossRef]
16. Rajoub BA. An efficient coding algorithm for the compression of ECG signals using the wavelet transform. IEEE Trans Biomed Eng. 2002;49(4):355–62. doi:10.1109/10.991163. [Google Scholar] [PubMed] [CrossRef]
17. Ali Hassan AM, Mohsen S. Compression of electrocardiogram signals using compressive sensing technique based on curvelet transform toward medical applications. Multimed Tools Appl. 2025;84(12):11203–19. doi:10.1007/s11042-024-19328-z. [Google Scholar] [CrossRef]
18. Huang R, Xue X, Xiao R, Bu F. A novel method for ECG signal compression and reconstruction: down-sampling operation and signal-referenced network. Electronics. 2023;12(8):1760. doi:10.3390/electronics12081760. [Google Scholar] [CrossRef]
19. Singhai P, Kumar A, Ateek A, Ahmad Ansari I, Singh GK, Lee HN. ECG signal compression based on optimization of wavelet parameters and threshold levels using evolutionary techniques. Circuits Syst Signal Process. 2023;42(6):3509–37. doi:10.1007/s00034-022-02280-4. [Google Scholar] [CrossRef]
20. Yildirim O, Tan RS, Acharya UR. An efficient compression of ECG signals using deep convolutional autoencoders. Cogn Syst Res. 2018;52:198–211. doi:10.1016/j.cogsys.2018.07.004. [Google Scholar] [CrossRef]
21. Chang Y, Sobelman GE. Lightweight lossy/lossless ECG compression for medical IoT systems. IEEE Internet Things J. 2024;11(7):12450–8. doi:10.1109/JIOT.2023.3336995. [Google Scholar] [CrossRef]
22. Fathi IS, Makhlouf MAA, Osman E, Ali Ahmed M. An energy-efficient compression algorithm of ECG signals in remote healthcare monitoring systems. IEEE Access. 2022;10(1):39129–44. doi:10.1109/ACCESS.2022.3166476. [Google Scholar] [CrossRef]
23. Shi J, Wang F, Qin M, Chen A, Liu W, He J, et al. New ECG compression method for portable ECG monitoring system merged with binary convolutional auto-encoder and residual error compensation. Biosensors. 2022;12(7):524. doi:10.3390/bios12070524. [Google Scholar] [PubMed] [CrossRef]
24. Kuldeep G, Zhang Q. Lightweight electrocardiogram signal compression. Biomed Signal Process Control. 2023;85(9):105012. doi:10.1016/j.bspc.2023.105012. [Google Scholar] [CrossRef]
25. Pal HS, Kumar A, Vishwakarma A, Singh GK, Lee HN. An effective ECG signal compression algorithm with self controlled reconstruction quality. Comput Methods Biomech Biomed Engin. 2024;27(7):849–59. doi:10.1080/10255842.2023.2206933. [Google Scholar] [PubMed] [CrossRef]
26. Ez-ziymy S, Hatim A, Hammia S. Real-time hardware architecture of an ECG compression algorithm for IoT health care systems and its VLSI implementation. Multimed Tools Appl. 2024;83(10):30937–61. doi:10.1007/s11042-023-16631-z. [Google Scholar] [CrossRef]
27. Pal HS, Kumar A, Vishwakarma A, Lee HN. Electrocardiogram signal compression using adaptive tunable-Q wavelet transform and modified dead-zone quantizer. ISA Trans. 2023;142:335–46. doi:10.1016/j.isatra.2023.07.033. [Google Scholar] [PubMed] [CrossRef]
28. Pal HS, Kumar A, Vishwakarma A, Singh GK. Optimized tunable-Q wavelet transform-based 2-D ECG compression technique using DCT. IEEE Trans Instrum Meas. 2023;72:1–13. doi:10.1109/TIM.2023.3279885. [Google Scholar] [CrossRef]
29. Polania LF, Carrillo RE, Blanco-Velasco M, Barner KE. Compressed sensing based method for ECG compression. In: 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2011 May 22–27; Prague, Czech Republic. doi:10.1109/ICASSP.2011.5946515. [Google Scholar] [CrossRef]
30. Zigel Y, Cohen A, Katz A. The weighted diagnostic distortion (WDD) measure for ECG signal compression. IEEE Trans Biomed Eng. 2000;47(11):1422–30. doi:10.1109/TBME.2000.880093. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools