 Open Access
Open Access
ARTICLE
SwinHCAD: A Robust Multi-Modality Segmentation Model for Brain Tumors Using Transformer and Channel-Wise Attention
1 Department of Artificial Intelligence, Sejong University, Seoul, 05006, Republic of Korea
2 Department of Computer Science and Engineering, Sejong University, Seoul, 05006, Republic of Korea
3 Department of Information and Communication Engineering and Convergence Engineering for Intelligent Drone, Sejong University, Seoul, 05006, Republic of Korea
* Corresponding Author: Hyeonjoon Moon. Email:
(This article belongs to the Special Issue: New Trends in Image Processing)
Computers, Materials & Continua 2026, 86(1), 1-23. https://doi.org/10.32604/cmc.2025.070667
Received 21 July 2025; Accepted 23 September 2025; Issue published 10 November 2025
Abstract
Brain tumors require precise segmentation for diagnosis and treatment plans due to their complex morphology and heterogeneous characteristics. While MRI-based automatic brain tumor segmentation technology reduces the burden on medical staff and provides quantitative information, existing methodologies and recent models still struggle to accurately capture and classify the fine boundaries and diverse morphologies of tumors. In order to address these challenges and maximize the performance of brain tumor segmentation, this research introduces a novel SwinUNETR-based model by integrating a new decoder block, the Hierarchical Channel-wise Attention Decoder (HCAD), into a powerful SwinUNETR encoder. The HCAD decoder block utilizes hierarchical features and channel-specific attention mechanisms to further fuse information at different scales transmitted from the encoder and preserve spatial details throughout the reconstruction phase. Rigorous evaluations on the recent BraTS GLI datasets demonstrate that the proposed SwinHCAD model achieved superior and improved segmentation accuracy on both the Dice score and HD95 metrics across all tumor subregions (WT, TC, and ET) compared to baseline models. In particular, the rationale and contribution of the model design were clarified through ablation studies to verify the effectiveness of the proposed HCAD decoder block. The results of this study are expected to greatly contribute to enhancing the efficiency of clinical diagnosis and treatment planning by increasing the precision of automated brain tumor segmentation.Keywords
1.1 Importance of Brain Tumor Diagnosis and Segmentation
Tumors that occur in the brain, the backbone and most complex organ of human life, are fatal conditions that cause enormous physical and mental distress to patients and their families [1]. The prognosis and treatment outcome of brain tumors depend on early detection of tumors, as well as their ability to accurately diagnose them, and to obtain a detailed understanding of the location and extent of their infiltration [2]. Segmentation of brain tumors is an essential key step in establishing an ablation interface during neurosurgical surgery, defining an irradiation area when planning radiotherapy, and quantitatively predicting the patient’s prognosis [3]. Magnetic resonance imaging (MRI) provides excellent contrast between brain tissue and tumors through multi-modality, and is widely used as the most powerful and non-invasive tool in the diagnosis of brain tumors [4]. However, in MRI images, brain tumors are very irregular in shape and size and consist of heterogeneous sub-regions such as necrotizing cores, edema, and active tumors [4]. In addition, as the boundaries with surrounding normal brain tissue are often ambiguous, manual segmentation of these tumor regions requires extreme concentration and consumes enormous time and effort even experienced medical professionals [5]. Moreover, there is a limitation that the consistency of segmentation results can be hindered by the subjective judgment intervention between workers [5]. Against this background, the development of automated and accurate brain tumor segmentation techniques to reduce the diagnostic burden on medical staff and provide high-quality segmentation information that is quantitative and reproducible has emerged as the most urgent and important research task in the modern field of medical imaging analysis [6].
1.2 Advancements in Deep Learning (DL) for Medical Image (MI) Segmentation
In recent years, DL technology, particularly Convolutional Neural Networks (CNNs), has spearheaded remarkable advancements in the field of MI segmentation [7]. Architectures rooted in CNNs, such as U-Net and V-Net, have demonstrated exceptional performance in extracting image features and performing pixel-wise classification, finding successful application across various medical image segmentation tasks [8]. However, while these CNN models excel at capturing local features, they have exhibited limitations in capturing long-range dependencies within images and effectively leveraging global contextual information [9]. To surmount these limitations, Transformer models, which achieved significant success in natural language processing, extended their applicability to the computer vision domain, thus introducing a new paradigm [10]. Transformer-based models, including Vision Transformer (ViT) and Swin Transformer, have unequivocally demonstrated superior performance by efficiently learning global contextual information across entire images [10]. In the realm of medical image segmentation, hybrid models like UNETR [11] and SwinUNETR [12] have emerged, integrating the strengths of Transformers with U-Net architectures. These models have successfully overcome the inherent limitations of conventional CNN models, achieving state-of-the-art (SOTA) performance [11]. Nevertheless, accurate segmentation of diseases with complex and heterogeneous morphologies, such as brain tumors, remains a critical research challenge, necessitating the development of more sophisticated and robust models [5].
1.3 Motivation and Contributions
As mentioned earlier, despite the high performance of SOTA transformer-based models such as SwinUNETR in brain tumor segmentation, the complex geometric shape of tumors, effective fusion of multiple modality data, and improved accuracy in fine boundary segmentation are still important challenges [6]. In particular, it is important to efficiently utilize the feature information (skip connection) of different scales transferred from the encoder in the decoder stage, and to improve the quality of final segmentation by minimizing information loss occurring during the upsampling process [13]. To address these challenges and further improve brain tumor segmentation performance, this study suggests a novel model based on the robust encoder of SwinUNETR [12] and additionally fusing a novel decoder architecture. Specifically, we developed a decoder block called hierarchical Channel-wise Attention (HCAD) Decoder Block and constructed a SwinHCAD model that effectively integrates it with SwinUNETR. The HCAD Decoder Block utilizes hierarchical features and channel-wise attention mechanisms to more precisely fuse feature information at different scales of the encoder and minimize information loss during the decoding process. This aims to more accurately segment the fine boundaries and internal structures of complex brain tumors. The contributions are as follow:
• Novel Decoder Architecture for Enhanced Feature Fusion: To address the limitations of conventional decoders that often struggle with effective multi-scale feature fusion, we propose the SwinHCAD model. Its core novelty lies in the HCAD decoder block, which introduces a guidance-based channel attention mechanism. This mechanism actively refines skip connection features using the decoder’s high-level context, enabling a more precise and intelligent fusion strategy tailored for complex tumor structures.
• Advanced Data Pipeline Establishment: By integrating diverse preprocessing and augmentation techniques from the MONAI library, we established a robust data pipeline. This ensures the model exhibits excellent generalization performance across various medical image data encountered in real clinical environments.
• Optimized Training Stability and Performance: We utilized a combined loss function, incorporating Dice Loss and Cross Entropy Loss, and applied deep supervision. This approach simultaneously secured training stability and maximized brain tumor segmentation performance.
• Demonstration and Analysis of Robust Performance: Leveraging the latest BraTS 2023/2024 GLI dataset, we extensively validated the superiority of the proposed model. We quantitatively demonstrated significantly improved segmentation performance across all tumor subregions (WT, TC, ET) compared to the existing baseline model.
• Presentation of Design Rationale: Through a systematic ablation study, we analyzed the impact of each component within the proposed HCAD decoder block on the overall model performance, thereby clearly presenting the rationale behind our model design and the individual contributions of its elements.
The remaining section is structured as follows: The related literature on brain tumor segmentation is found in Section 2. The material and methods are described in Section 3. The experimental design, assessment, and a discussion of the qualitative and quantitative results of the proposed model are all addressed in Section 4. The future work and conclusion are discussed in Sections 5 and 6.
2.1 Foundational Architectures
In recent years, the advent of DL technologies, especially CNNs, has led to a revolutionary paradigm shift in medical image segmentation and computer vision [14]. A major advantage of CNNs lies in their ability to learn automatic hierarchical features [15]. This capability, unlike the manual feature design methods of experts, has pioneered a new level of performance by directly learning the hierarchical structure of abstract and relevant features from raw pixel data [16]. Against this backdrop, Fully Convolutional Networks (FCNs) were the first to propose end-to-end solutions for semantic segmentation, replacing dense layers of classification CNNs with convolutional layers and reconstructing spatial information by introducing inverse convolutional and skip connections [17,18]. The U-Net architecture, which inherits the ideas of FCN, has become a de facto standard in the field of biomedical image segmentation [19,20]. U-Net features a contraction path that captures high-level contextual information through a symmetrical U-shaped encoder-decoder structure and an expansive path that recovers precise spatial resolution. This is achieved especially through its long skip connections, which directly couple the encoder’s high-resolution feature maps with the decoder’s corresponding upsampled feature maps. This allows it to effectively solve the problem of balancing “what” (semantics) and “where” (localization), thereby generating very precise segmentation masks [19,20]. Moreover, U-Net demonstrates the advantage of leveraging robust data augmentation techniques such as elastic deformation to achieve superior performance with limited training data [8,16].
2.2 Evolution of U-Net Architectures
The successful emergence of U-Net established a robust and flexible architectural blueprint, leading subsequent research to focus on enhancing performance by incorporating optimal building blocks within the U-Net framework [20]. To solve the “vanishing gradient” problem, a major limitation hindering the training of deep neural networks, the successful introduction of residual connections by Residual Networks (ResNet) led to the development of Res-UNet [21]. Res-UNet has improved segmentation accuracy by integrating these residual blocks into U-Net’s encoders and decoders, enabling deeper and more robust model construction by mitigating training challenges [21,22]. Moreover, to address the inefficiency of U-Net’s standard skip connections, which merely combine features and can propagate irrelevant or noisy information from background regions, Attention U-Net introduced Attention Gates (AGs) for skip connections [23,24]. These gates adaptively concentrate the model’s attention, automatically highlighting the features of prominent regions related to segmentation tasks and weakening those from unnecessary background regions [25]. This mechanism leads to more efficient learning and improved precision, especially in segmenting small or complex targets [23].
2.3 The Rise of Transformer Models for Medical Image Segmentation
The most recent architectural changes in the field of medical image segmentation have been influenced by the remarkable success of Transformer architectures in the field of natural language processing (NLP) [26]. These changes stem from the core limitations of CNNs: convolutional operations inherently possess a local receptive field. This has led CNNs to struggle to explicitly model long-range spatial dependencies across images and capture global context. To overcome these limitations, Vision Transformer (ViT) divides images into patch sequences and applies a self-attention mechanism to adapt Transformers for image tasks, enabling them to capture global context from the outset. Early success stories in the field of medical segmentation, such as TransUNet, have adopted hybrid designs that use the CNN backbone to extract low-level features and model global dependencies through Transformer encoders [27]. Furthermore, to address the computational complexity of standard ViTs, Swin Transformer uses non-overlapping local windows to calculate self-attention and introduced a “shifted window” mechanism to generate hierarchical feature representations and achieve linear complexity with respect to image size [9]. Pure Transformer-based U-shaped encoder-decoder architectures such as Swin-Unet have leveraged this efficiency to be applicable to high-resolution medical images and have demonstrated SOTA performance [28].
While Transformer-based models have become the standard, research continues to address their inherent quadratic complexity. Recently, alternative architectures such as State Space Models (SSMs), including Mamba, have gained attention for their linear complexity and long-range dependency modeling. This trend has already been applied to medical segmentation, with models like Segmamba [29] demonstrating competitive performance. This highlights the fast-evolving nature of the field, where a continuous search for more efficient and powerful architectures is underway.
2.4 Synthesis of the Field and Research Gaps
The field of medical image segmentation continues to evolve rapidly, with recent advances addressing key challenges such as computational efficiency, boundary accuracy, and generalization across diverse datasets. For instance, Zig-RiR [30] employs a nested RWKV attention mechanism to capture both global and local features with linear complexity, significantly reducing computational load while preserving spatial continuity. In parallel, models like SMAFormer [31] integrate multiple attention mechanisms synergistically to enhance the segmentation of small and irregular tumor regions. Furthermore, semi-supervised approaches using Swin Transformers [32] have been explored to improve feature consistency. To enhance domain generalization, recent methods have shown promising results, such as combining self-supervised learning with domain adversarial training to learn stain-invariant features [33], and leveraging diffusion models to maintain robustness against cross-domain shifts.
The literature on automatic brain tumor segmentation illustrates a clear and logical evolutionary path. This field has evolved from traditional methods that relied on manual features to DL paradigms that enabled CNNs to learn automatic features. The U-Net architecture has become the dominant baseline by establishing a robust and effective encoder-decoder blueprint [34]. Subsequent work has focused on strengthening depth with Res-UNet, improving feature fusion with Attention U-Net, and further refining this blueprint by addressing the main limitations of CNNs, such as the absence of global contextual modeling, through integrating Transformers via models like TransUNet and Swin-Unet [35]. This rapid development has been greatly facilitated by the availability of large, publicly accessible and well-managed benchmark datasets provided by the annual Multi-modal Brain Tumor Segmentation Challenge (BraTS) [5]. The BraTS challenge has provided a standardized platform for objective and rigorous comparisons of different algorithms, which has facilitated competition and accelerated innovation across research communities [5]. Despite these remarkable advances, significant research gaps still persist in areas such as improving model efficiency, strengthening robustness to domain shifts, developing more effective techniques to segment small, diffuse, or irregularly shaped tumor subregions, and increasing the interpretability of increasingly complex black-box models.
We executed the experiments using our system configuration, as detailed in Table 1, which includes a Python 3.9 environment with the PyTorch and MONAI frameworks. The models were trained and evaluated on an NVIDIA RTX A6000 GPU with 48 GB of VRAM. The training process was conducted for 100 epochs with a batch size of 4, using 3D patches of size 128 × 128 × 128. We employed the AdamW optimizer with an initial learning rate of 1e−4, which was adjusted using a cosine annealing scheduler throughout the training, as shown in Table 2.
The experiments in this study used public brain tumor segmentation datasets BraTS2020, BraTS2021, BraTS2023, and BraTS2024. These datasets contain medical images of patients with gliomas and serve as standard references for evaluating the efficacy of models in medical artificial intelligence. Each dataset consists of multiple patient cases, and each case contains four types of 3D MRI sequences that are anatomically matched to each other. The included modalities are 1) T1-weighted (T1), 2) T1-weighted with contrast enhancement (T1c), 3) T2-weighted (T2), and 4) T2-weighted fluid-attenuated inversion recovery (T2-FLAIR), as shown in Fig. 1. These four images clearly show the characteristics of different lesions, such as the anatomical structure of the brain, active tumors identified through contrast enhancement, and edema around the tumor, enabling multi-modal analysis. All cases are provided with a correct answer segmentation mask labeled directly by the expert. The labels are divided into three major sub-regions that make up the tumor: necrotic (NCR) and non-enhanced tumor core (NET), peri-tumor edema (ED), and contrast-enhanced tumor (ET). Performance assessments are performed on three overlapping regions defined by combining these three labels: Whole Tumor (WT), Tumor Core (TC), and Contrast-Enhanced Tumor (ET). The scale of data used in this study included data from 369 patients for BraTS 2020, 1251 patients for BraTS 2021 and BraTS 2023, and 1621 patients for BraTS 2024. For our experiments, each dataset was randomly partitioned into training, validation, and test sets in a 70:10:20 ratio, respectively.
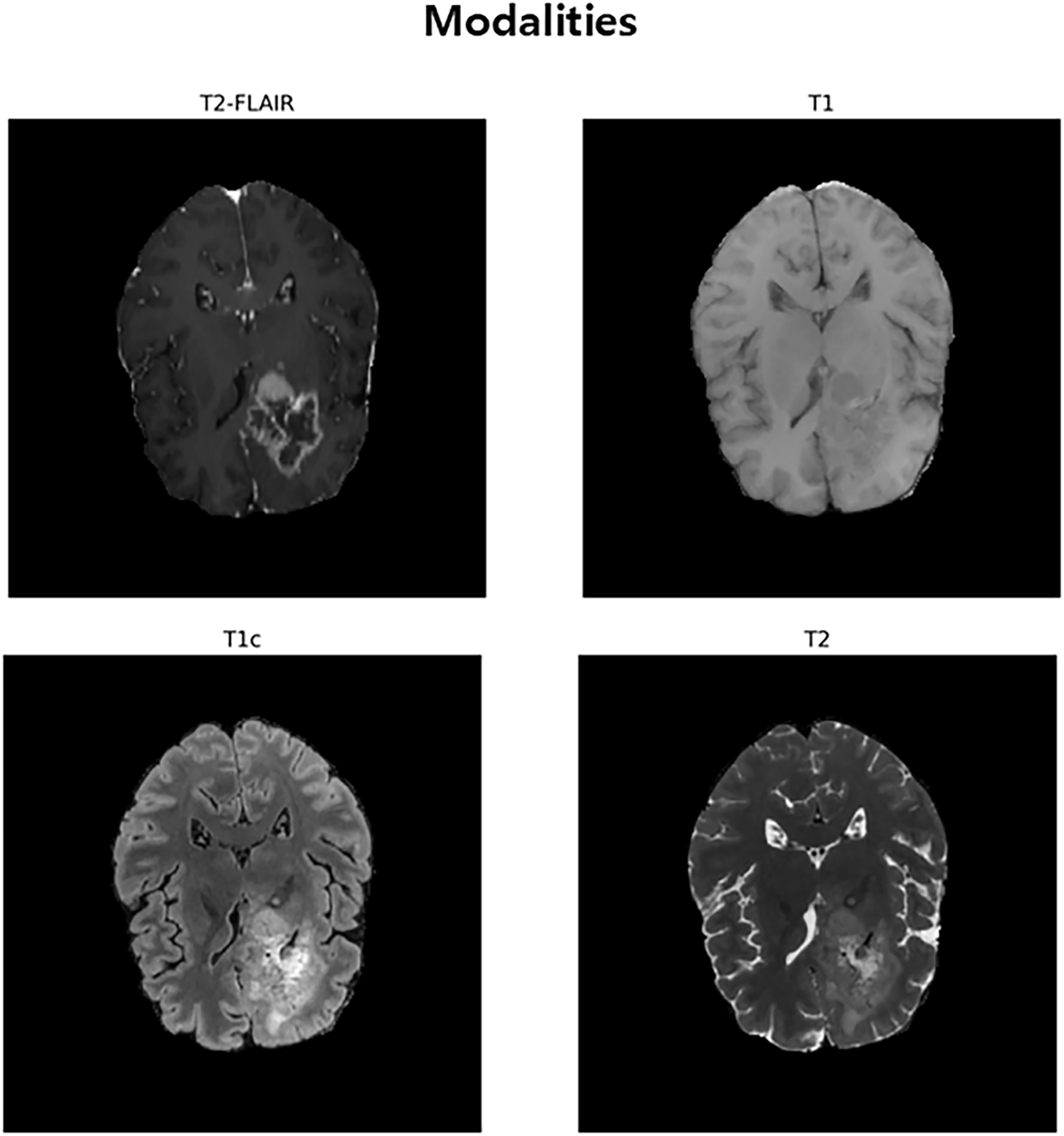
Figure 1: An Inter-Modal Comparison of MRI Sequences for Brain Tumor Visualization. This figure illustrates the use of four MRI sequences, T2-FLAIR, T1, T1c (post-contrast), and T2, for brain tumor segmentation. T2-FLAIR helps highlight peritumoral edema, T1 provides baseline anatomical details, T1c reveals contrast improved tumor regions, and T2 identifies tumor-associated fluid areas. Together, these modalities offer complementary information for accurate brain tumor identification and segmentation
In order to ensure consistency of the data used in the experiment and increase learning efficiency, the following series of standardized preprocessing processes were performed. All preprocessing processes were implemented based on the MONAI framework. First, for direction standardization, the directions of all 3D MRI data were unified with a right-anterior-superior (RAS) coordinate system. This is to allow the model to learn consistent features regardless of the direction of the data. Second, by readjusting the voxel spacing, the voxel spacing of all images was resampled in a square (1.0, 1.0, 1.0) mm. In this process, trilinear interpolation was applied to the image data, and nearest neighbor interpolation was applied to the split label map to minimize information loss and distortion. Third, by performing foreground area extraction, unnecessary background areas without brain tissue were removed. By setting a tight boundary box based on the foreground of the input image to cut the image, the amount of subsequent computation was reduced, and the model was induced to focus on important areas (see Fig. 2). Fourth, the intensity distribution of each MRI modality was standardized by applying intensity normalization. Z-score normalization was performed so that the average of non-zero voxel values for each channel became 0 and the standard deviation became 1, thereby increasing the stability and convergence speed of learning.
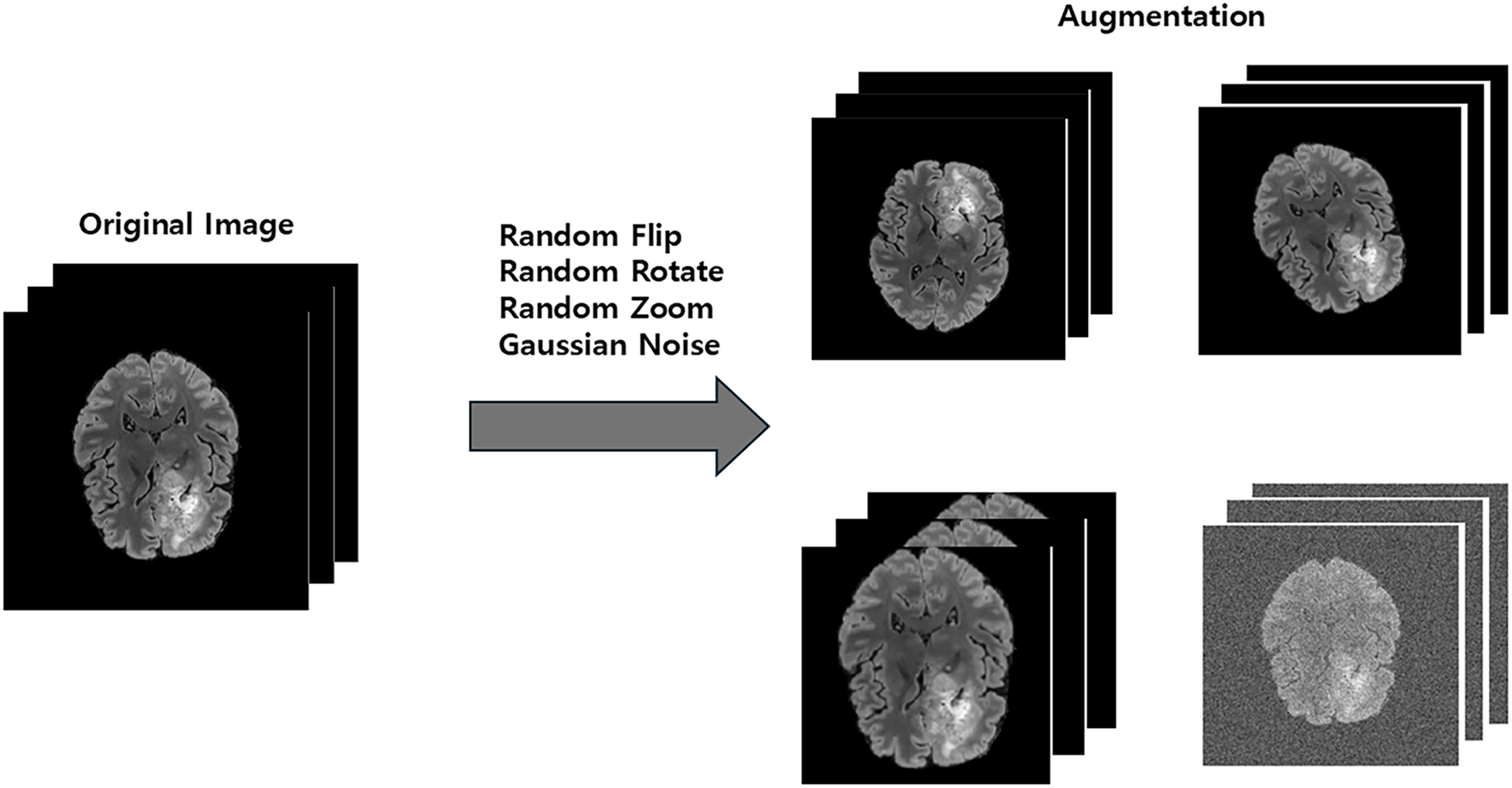
Figure 2: The data augmentation process of generating diverse training samples from a single original image via random transformations
To enhance the model’s generalization capability and improve its robustness against variations in clinical data, we applied a series of online data augmentation techniques during the training phase. These transformations, illustrated in Fig. 2, were implemented using the MONAI library and applied randomly to each data sample. The specific augmentations included randomly flipping the data along each of the three spatial axes with a 0.5 probability, applying random rotations within the range of [−30, 30] degrees around each axis, performing random zooming with a factor ranging from 0.9 to 1.1, and adding Gaussian noise with a mean of 0.0 and a standard deviation of 0.1 to the images. These augmentations create a more diverse set of training samples, helping to prevent overfitting and improve the model’s performance on unseen data.
The segmentation performance of the model was evaluated quantitatively using the Dice Similarity Coefficient (DSC) and 95% Hausdorf Distance (HD95). The Dice Score is an indicator of the degree of overlap between the predicted and actual correct answer areas, and the higher, the better. HD95 is an indicator that measures the distance between the boundaries of two regions; the lower the predicted boundaries, the more accurate the predicted boundaries are. Evaluation was performed on each of three sub-areas: WT, TC, and ET.
In this study, we introduce a new End-to-End architecture that combines encoders employing a Transformer architecture with independently proposed attention-based decoders to increase the accuracy of 3D brain tumor MRI image segmentation. The proposed model is based on the 3D U-Net architecture and has a symmetrical structure consisting of an encoder for powerful feature learning and a decoder for precise mask reconstruction. The overall pipeline of the proposed model is illustrated in Figs. 3 and 4.
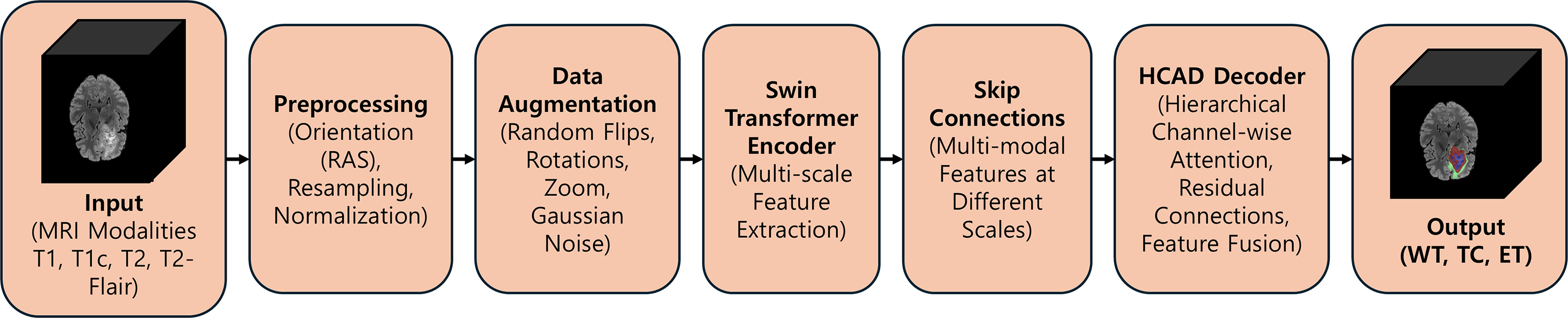
Figure 3: An overview of the proposed SwinHCAD pipeline, from multi-modal MRI input to the final segmentation of Whole Tumor (WT), Tumor Core (TC), and Enhancing Tumor (ET)
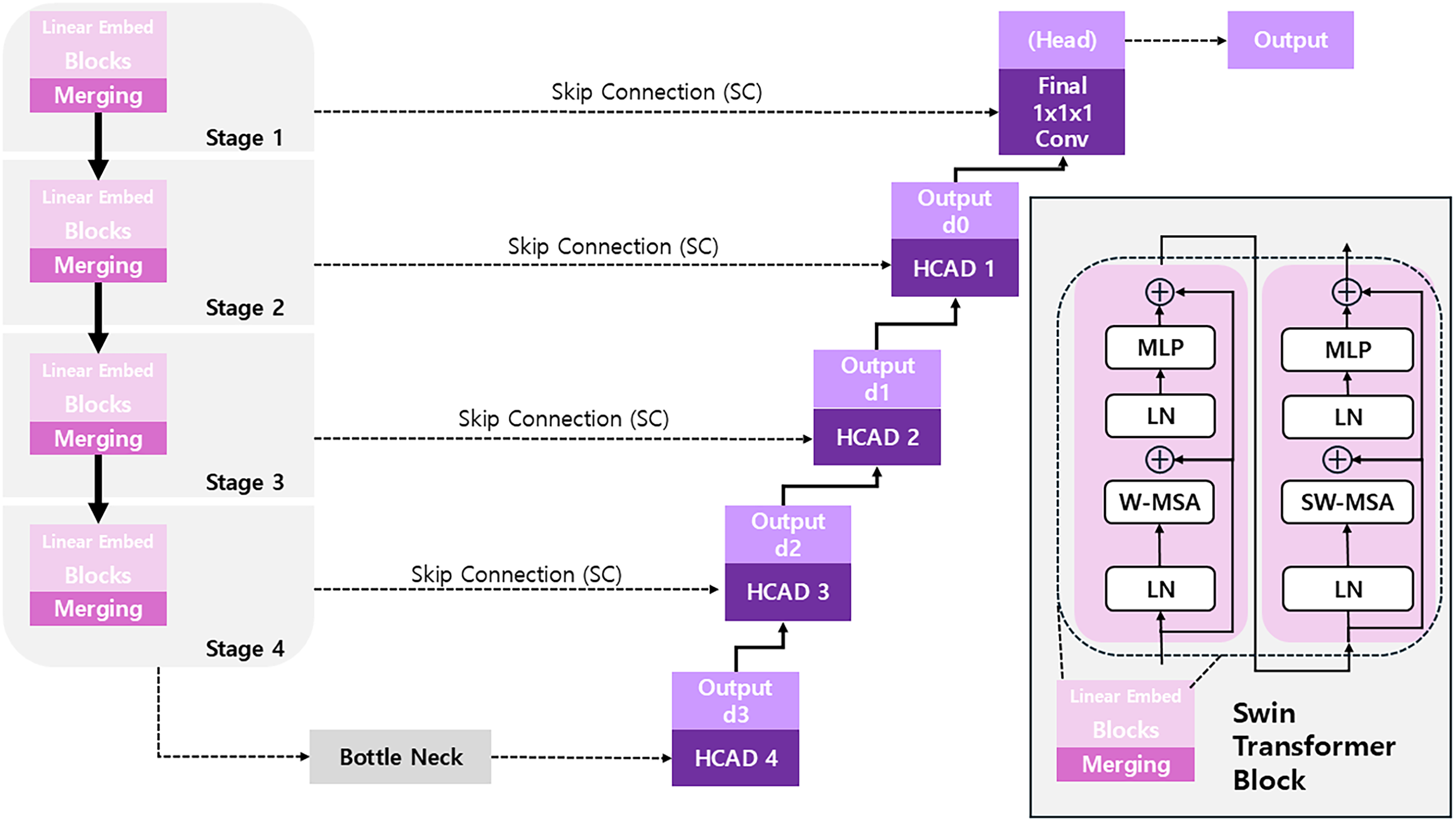
Figure 4: The overall architecture of the SwinHCAD model, which integrates a transformer encoder with a novel HCAD-based decoder for precise segmentation
While attention mechanisms, including channel-wise attention, have been widely explored in medical image segmentation, our work introduces a novel integration of hierarchical channel-wise attention guided by decoder features. Although similar mechanisms have been previously applied, the combination of hierarchical attention with decoder features provides a unique refinement process that enhances segmentation performance. This specific integration enables the model to more effectively capture and refine features at different stages of the network. We believe this novel approach offers significant improvement over traditional methods, and we demonstrate its efficacy through comprehensive experiments that highlight the advantages of our method in comparison to existing techniques.
3.6 Encoder: Using Swin Transformer
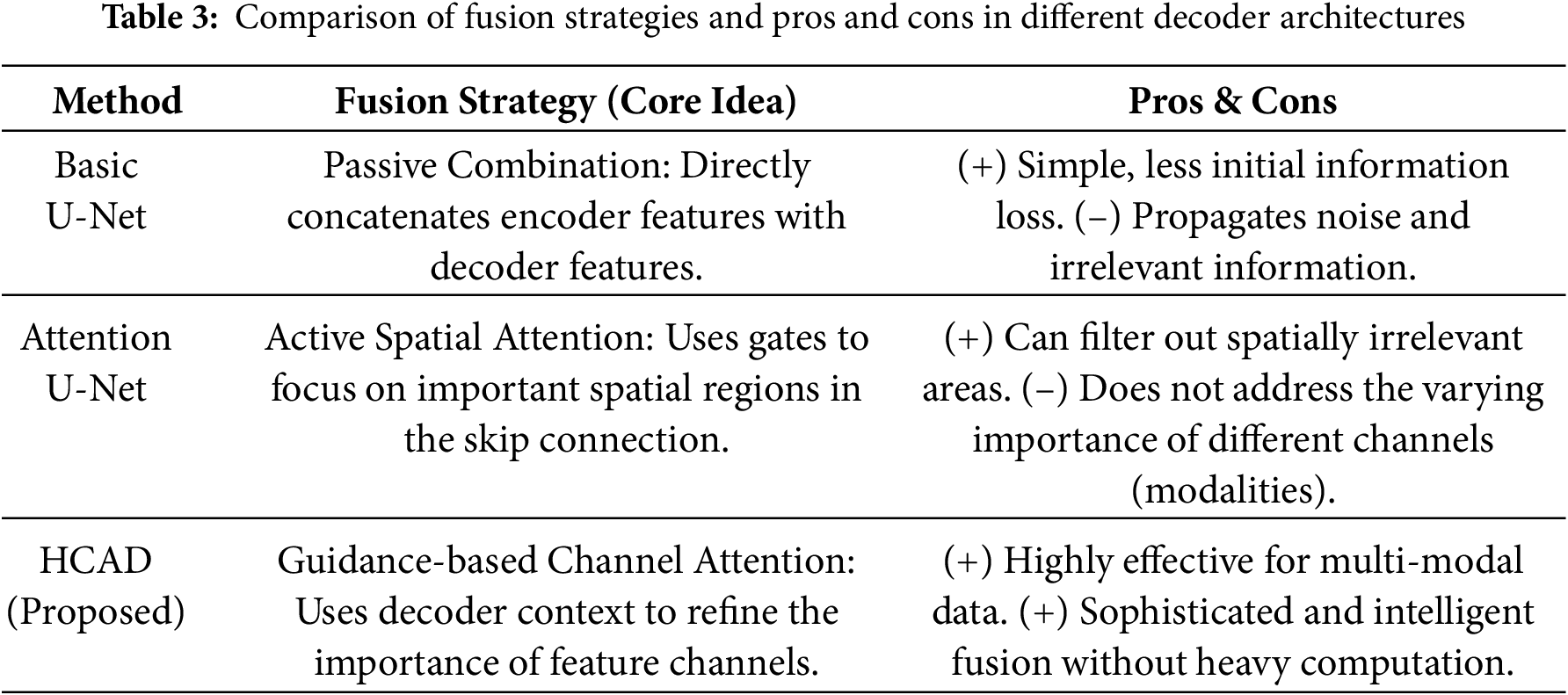
The design of the HCAD block is centered on a channel-wise attention mechanism, which is designed to directly address the challenges of multi-modal MRI-based segmentation. The evolution of decoder architectures reveals a clear progression in feature fusion strategies. Foundational models like U-Net [36] employ passive concatenation, which can propagate irrelevant noise. As an advancement, Attention U-Net [24] introduced active spatial attention, which learns to emphasize salient spatial regions within the feature maps. Our proposed HCAD builds upon this by introducing a guidance-based channel attention mechanism that, in contrast, learns to prioritize the most informative feature channels. By using the decoder’s high-level context as a guide, HCAD dynamically refines the skip connection by modulating the importance of each channel, a particularly effective strategy for fusing heterogeneous multi-modal data.
The encoder of the proposed model uses Swin UNETR from the MONAI framework as the backbone for feature extraction. Swin UNETR is a model that adapts the hierarchical Vision Transformer, the Swin Transformer, for 3D medical images. It efficiently captures both local features and the global context of an image through a shifted window self-attention mechanism and produces multi-scale feature representations at various resolutions through a patch aggregation process. The encoder was initialized with weights from a model pre-trained on a large-scale dataset, maximizing the model’s learning efficiency and generalization performance through the advantages of Transfer Learning. The operational flow of the encoder is detailed as follows:
Patch Partition & Linear Embedding
First, the input 3D image
Here,
Swin Transformer Block
The embedded token sequence is passed through a series of consecutive Swin Transformer blocks for feature extraction. The core of the Swin Transformer is the alternating use of Window-based Multi-head Self-Attention (W-MSA) and Shifted Window-based Multi-head Self-Attention (SW-MSA). This enables information exchange between windows while maintaining computational efficiency. The operations of two consecutive Swin Transformer blocks (block
Block
For the token sequence
Block
In the next block, SW-MSA is used, which calculates self-attention after shifting the window positions. This allows patches at the window boundaries of the previous block to exchange information, enabling the model to learn global relationships.
This pair of blocks serves as the fundamental unit of the encoder and is repeated multiple times.
Patch Merging
To create hierarchical feature maps, a Patch Merging layer is applied after a certain number of Swin Transformer blocks. This layer transforms the feature map by decreasing its spatial dimensions while simultaneously expanding its channel depth. For example, it selects a group of adjacent
The feature maps of different resolutions (
3.7 Hierarchical Channel-Wise Attention Decoder
The core originality of this model lies in the newly designed hierarchical channel-wise attention decoder (HCAD). As detailed in the process above, HCAD focuses on effectively refining and fusing the skip connection features received from the encoder. By generating channel-wise attention weights from the upsampled feature map, it serves as an attention gate that emphasizes channels more important to segmentation and suppresses unnecessary information. This mechanism, combined with a residual connection for learning stability, allows for more precise and effective feature fusion during the decoding process. The advantages and limitations of HCAD are comprehensively summarized in Table 3, where its strengths and potential drawbacks are clearly compared.
The design of the HCAD block is centered on a channel-wise attention mechanism, and it is designed to directly address the challenges of multi-modal MRI-based segmentation. The input to our model comprises four distinct MRI sequences (T1, T1c, T2, and T2-FLAIR), each offering unique and complementary information critical for identifying different tumor sub-regions. For example, T2-FLAIR sequences excel at highlighting peritumoral edema, whereas post-contrast T1c scans are essential for delineating the active, enhancing tumor core. Our hypothesis is that an effective decoder must intelligently fuse these heterogeneous data streams. Therefore, instead of employing more generalized spatial-channel attention, HCAD focuses on learning the inter-channel relationships. By using the high-level feature map from the decoder as a contextual guide, the attention mechanism learns to dynamically modulate the influence of each modality-specific feature channel. This enables the model to, for instance, assign greater weight to features derived from the T1c channel when reconstructing the enhancing tumor boundary, while relying more on T2-FLAIR features for the whole tumor region. This targeted, modality-aware approach allows for a sophisticated yet computationally efficient feature fusion, making it a highly effective strategy for this specific task.
The HCAD decoder block is designed to effectively fuse multi-scale features from the Swin Transformer encoder and minimize the loss of low-level features. The core idea of this block is to use the feature map from the previous decoder layer as a guide to recalibrate the channel-wise importance of the skip connection feature map from the encoder. This process suppresses irrelevant noise and emphasizes meaningful information. Furthermore, a residual connection compensates for potential information loss during the upsampling process, as shown in Fig. 5. The overall operational flow consists of the following:
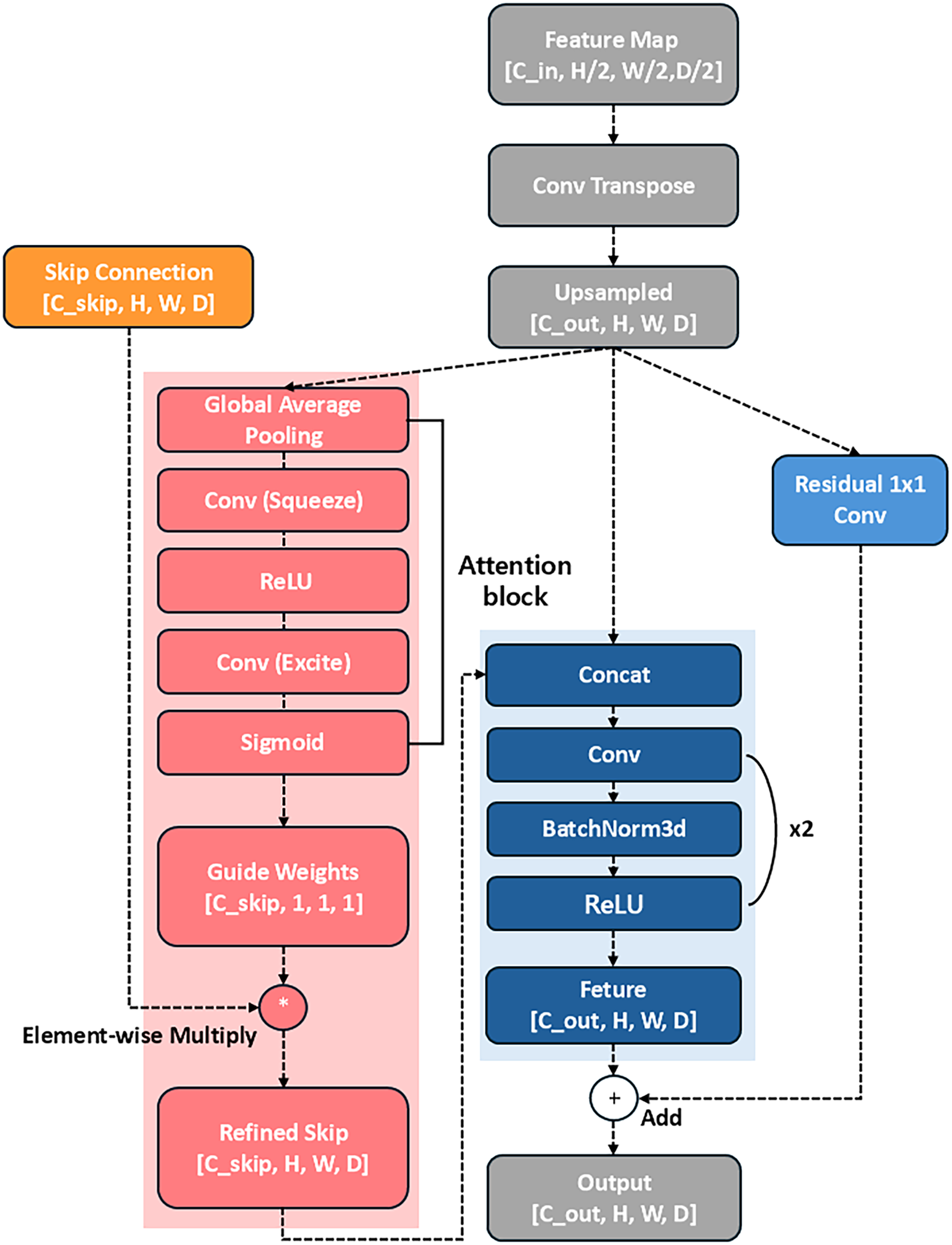
Figure 5: The proposed Hierarchical Channel-wise Attention Decoder (HCAD) block, which refines encoder skip connections using channel-wise attention guided by decoder-side features
Feature Map Upsampling
First, the spatial resolution of the input feature map
At this point,
Guided Attention Weight Generation
As the core step of HCAD, the upsampled feature map
Here,
Skip Connection Refinement
The generated attention weights
Feature Fusion
The upsampled feature map
Convolutional Processing
The fused feature map
Residual Connection
Finally, a residual connection is performed by adding the initial upsampled result
This resulting
3.8 Computational Efficiency Analysis
To evaluate the feasibility of deploying the proposed SwinHCAD model in clinical workflows, we conducted a computational efficiency analysis alongside the segmentation performance evaluation. This analysis focused on three key factors: GPU memory usage, training time, and inference speed.
• GPU Memory Usage: On an NVIDIA RTX A6000 GPU (48 GB VRAM), the peak memory usage for SwinHCAD during training was 31.4 GB, which is approximately 8 % higher than SwinUNETR (29.1 GB) due to the added channel-wise attention computations in the decoder.
• Training Time: For a batch size of 4 and 3D patch size of 128
• Inference Speed: The model achieved an inference time of 0.92 s per case, enabling near real-time segmentation in clinical settings.
• Comparison with Baselines: Table 4 presents a comparison with SwinUNETR and nnU-Net in terms of these metrics, showing that while SwinHCAD requires slightly more GPU memory and training time than SwinUNETR, it offers faster inference due to its lightweight decoder design.
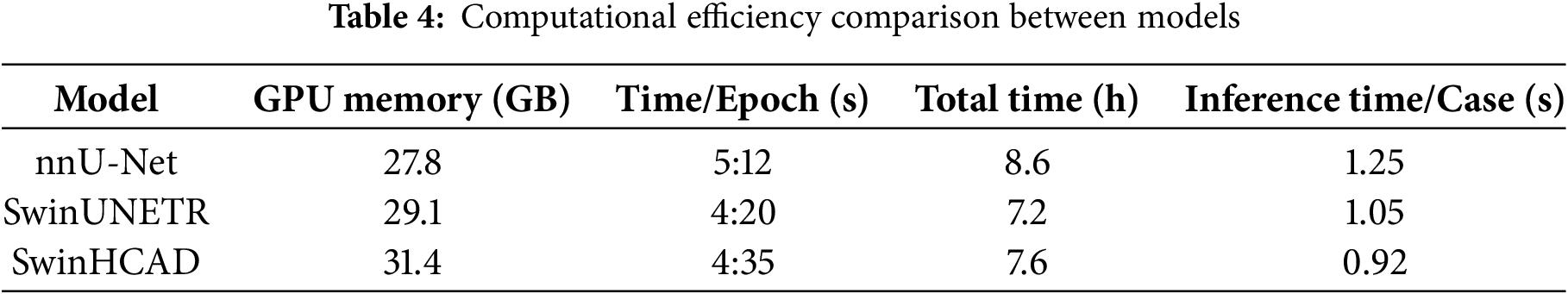
This analysis highlights that the proposed SwinHCAD model achieves a favorable balance between segmentation performance and computational efficiency. The marginal increase in memory consumption and training time is outweighed by its improved inference speed and segmentation accuracy, making it a viable candidate for integration into time-sensitive clinical workflows.
4.1 Quantitative Results Study
Table 5 presents a comprehensive performance comparison of several brain tumor segmentation models using Dice Score (%) and HD95 (mm) metrics on the BraTS 2020, 2021, 2023, and 2024 datasets. When evaluated on the BraTS 2020 dataset, the proposed model recorded Dice Scores of 90.44% for WT, 85.33% for TC, and 78.90% for ET. In particular, its TC Dice Score of 85.33% was the highest among the comparative models. Its WT Dice Score of 90.44% marginally surpassed nnU-Net’s 90.38%. The proposed model also recorded HD95 values of 5.99 mm for TC and 26.46mm for ET, with its TC HD95 of 5.99 mm being lower than TransBTS’s 9.77 mm. On the BraTS 2021 dataset, the proposed model recorded Dice Scores of 92.32% for WT, 91.19% for TC, and 87.21% for ET. On this dataset, the Dice Score of the proposed model was the highest among the comparative models in all regions. HD95 values were WT 13.92 mm, TC 6.43 mm, and ET 5.85 mm, while ET HD95 5.85 mm was lower than DBTrans 6.13 mm, registering the lowest value in that region. TC HD95 6.43 mm was similar to DBTrans 6.24 mm.
The results on the BraTS 2023 dataset indicate that the proposed model recorded Dice Scores of 93.00% for WT, 89.25% for TC, and 84.38% for ET. This was the highest Dice Score across all regions among all models compared on this dataset. Notably, the WT Dice Score of the proposed model was 1.93%p higher than SegMamba’s 91.07% and 3.65%p higher than VT-UNet’s 89.35%. On the HD95 metric, WT 5.64 mm, TC 4.25 mm, and ET 4.14 mm were recorded, while TC HD95 4.25 mm was 1.22 mm lower than VT-UNet 5.47 mm, and ET HD95 4.14 mm was 1.94 mm lower than VT-UNet 6.08 mm, showing the best boundary prediction accuracy in those regions. For the BraTS 2024 dataset, the Dice Score for the proposed model was 92.06% for WT, 88.90% for TC, and 84.22% for ET. Even on this dataset, the proposed model showed the highest performance among the comparative models in all regions of the Dice Score. The WT Dice Score was 1.94%p higher than SegMamba’s 90.12% and 2.71%p higher than VT-UNet’s 89.35%. HD95 recorded WT 5.67 mm, TC 5.52 mm, and ET 5.38 mm, while TC HD95 5.52 mm was similar to VT-UNet 5.47 and 0.65 mm lower than SegMamba 6.17 mm. ET HD95 5.38 mm was lower than VT-UNet 6.08 mm and SegMamba 6.69 mm.
Ablation studies were conducted to quantitatively analyze the effect of each element of the proposed HCAD decoder on the overall model performance. The experiment started with a baseline model using the most basic U-Net-style decoder, and measured performance changes by sequentially adding (1) Residual Connection and (2) Channel Attention Gate, which are key design elements of HCAD.
The entire process of the experiment on BRATS 2023 dataset is summarized in Table 6. To clearly demonstrate the HCAD decoder component-specific effectiveness of the proposed model based on Transformer, we performed a multi-faceted comparison with CNN-based Xception and Xception+Unet models.
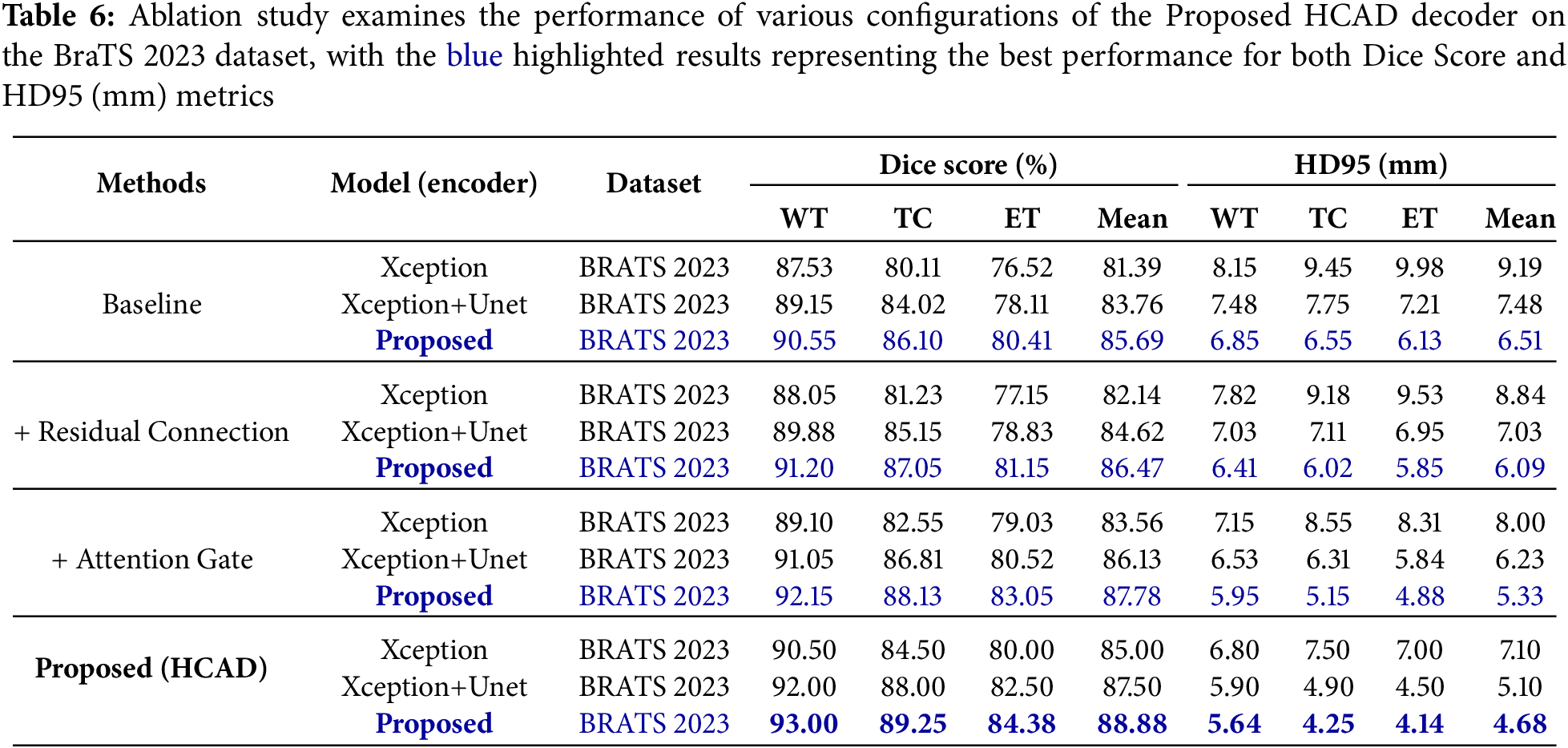
From the most basic baseline settings, the proposed model (Dice 85.69%) significantly outperformed the Xception+Unet (83.76%) and Xception (81.39%) models. This suggests that the strong Transformer structure of SwinUNETR, on which the proposed model is based, has higher potential in this task compared to CNN-based models. This performance gap remained consistent at every step of adding Residual Connection (Residual Connection) and Attention Gate. This result confirms that the SwinUNETR-based decoder structure is a strong underlying model, and the subsequent analysis evaluated each component’s contribution in depth around this proposed model.
The results for the BRATS 2023 and BRATS 2024 datasets are summarized in Tables 6 and 7, respectively. The baseline model showed basic segmentation performance, but adding residual connections improved the average Dice Score on the BraTS 2023 dataset from 85.69% to 86.47% and the average HD95 from 6.51 to 6.09 mm. This suggests that residual connections contributed to stabilizing the learning and enhancing the feature representation.
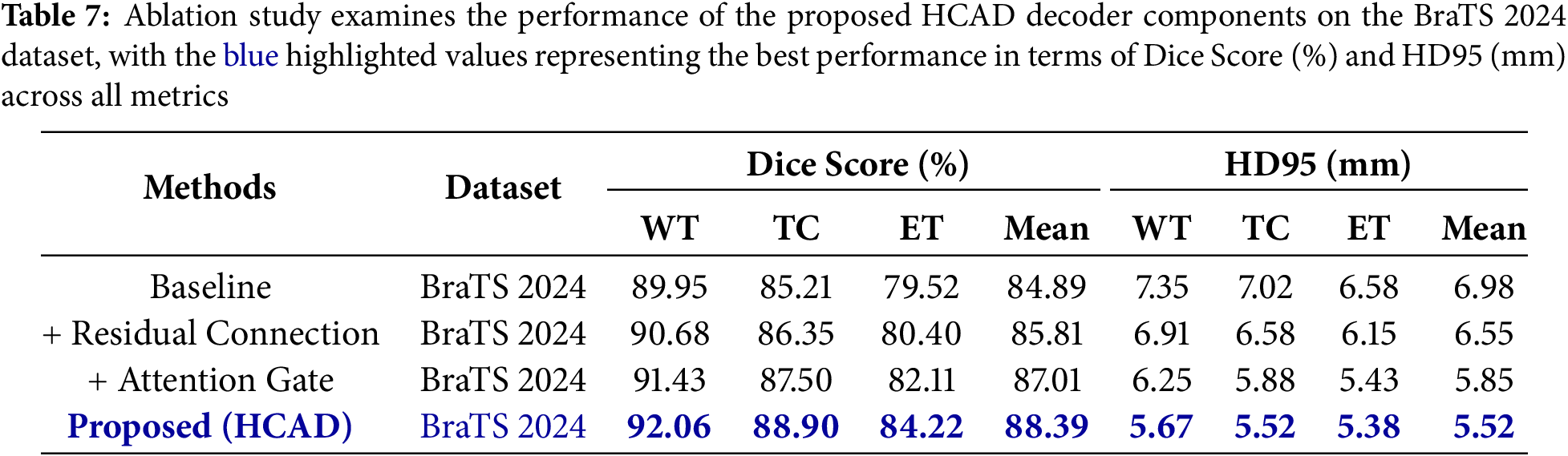
When the channel attention gate was additionally applied to this, the average Dice Score improved significantly to 87.78%, and HD95 also decreased to 5.33 mm. This shows that the attention gate, a core mechanism of HCAD, effectively refined the information of Skip Connection and played a decisive role in improving performance. Finally, the proposed model (HCAD) with both elements recorded the best performance in all indicators.
In particular, this trend of performance improvement was consistently observed not only in BraTS 2023, but also in a new domain, BraTS 2024 dataset. This proves that each component of the proposed HCAD is not overfitting only to a specific dataset, and has a positive effect on improving generalization performance. Therefore, this ablation study clearly demonstrates that every design element within the presented HCAD decoder is integral to the final efficacy.
Statistical Significance Analysis
To ensure that the performance improvements reported in this section are not due to random variation, we conducted statistical significance testing between the proposed SwinHCAD model and baseline methods. For each dataset (BraTS 2020, 2021, 2023, and 2024), paired two-tailed
Table 8 summarizes the resulting
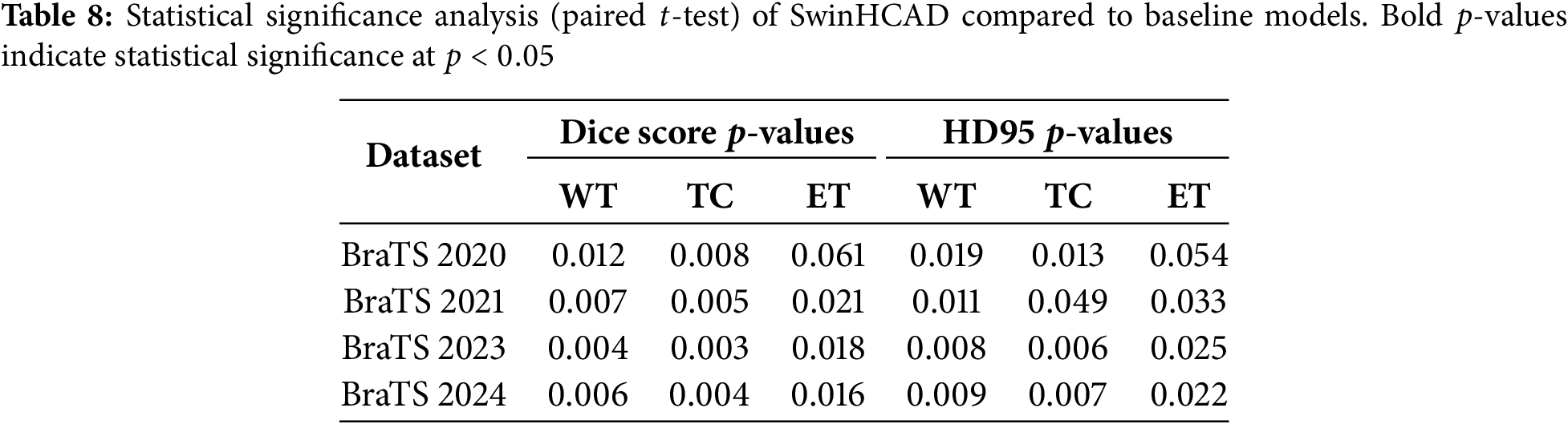
These results confirm that the observed improvements of SwinHCAD over baseline models are not only consistent but also statistically significant in most cases, reinforcing the reliability of the reported performance gains.
4.3 Analyzing the Visual Results
Fig. 6a shows the average Dice Score for each model. The proposed model intuitively shows that the highest bar was recorded on both BraTS 2023 and 2024 datasets, achieving the best segmentation accuracy. Conversely, for HD95, Fig. 6b demonstrates that the proposed model exhibits the lowest error level, similar to VT-UNet, across both datasets. This indicates that its predicted tumor interface is very close to the actual one and is generated smoothly. In particular, compared to models with high HD95 levels such as Hi-Net and TransBTS, the proposed model excels at precisely delineating the tumor boundary. In addition to these quantitative indicators, the Fig. 7 qualitatively compares the actual segmentation results to show the superiority of the proposed model. This figure compares the prediction results of the proposed model and other models with Ground Truth for an example on the BraTS dataset. The visual analysis clearly shows that the proposed model restores the overall shape and size of Ground Truth most similarly to the other models. In the TC segment, TransBTS missed some areas and nnUNet tended to over-predict, while the proposed model precisely captured the boundaries very close to Ground Truth. This provides a strong basis for visually supporting the claim that the proposed model draws the boundaries of tumors smoothly and accurately, as indicated by the low HD95 levels analyzed earlier.
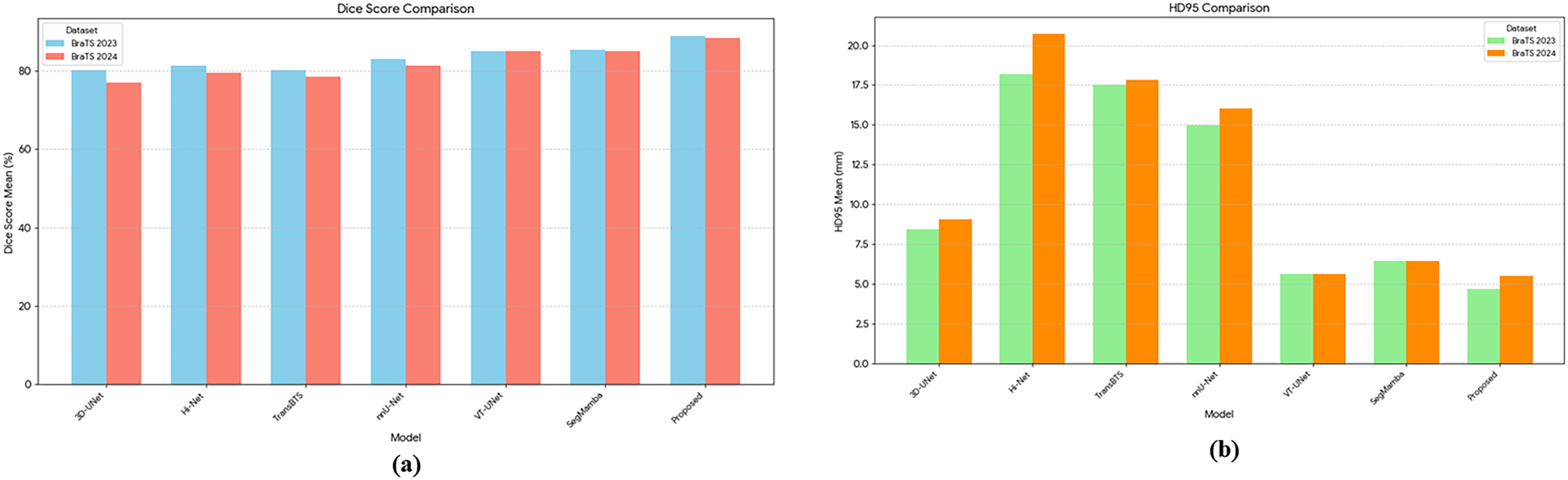
Figure 6: Comparison of various models on BraTS 2023 and BraTS 2024 datasets using Mean Dice (a), Score and Mean HD95 (b). The Dice Score reflects segmentation accuracy, while HD95 measures boundary precision
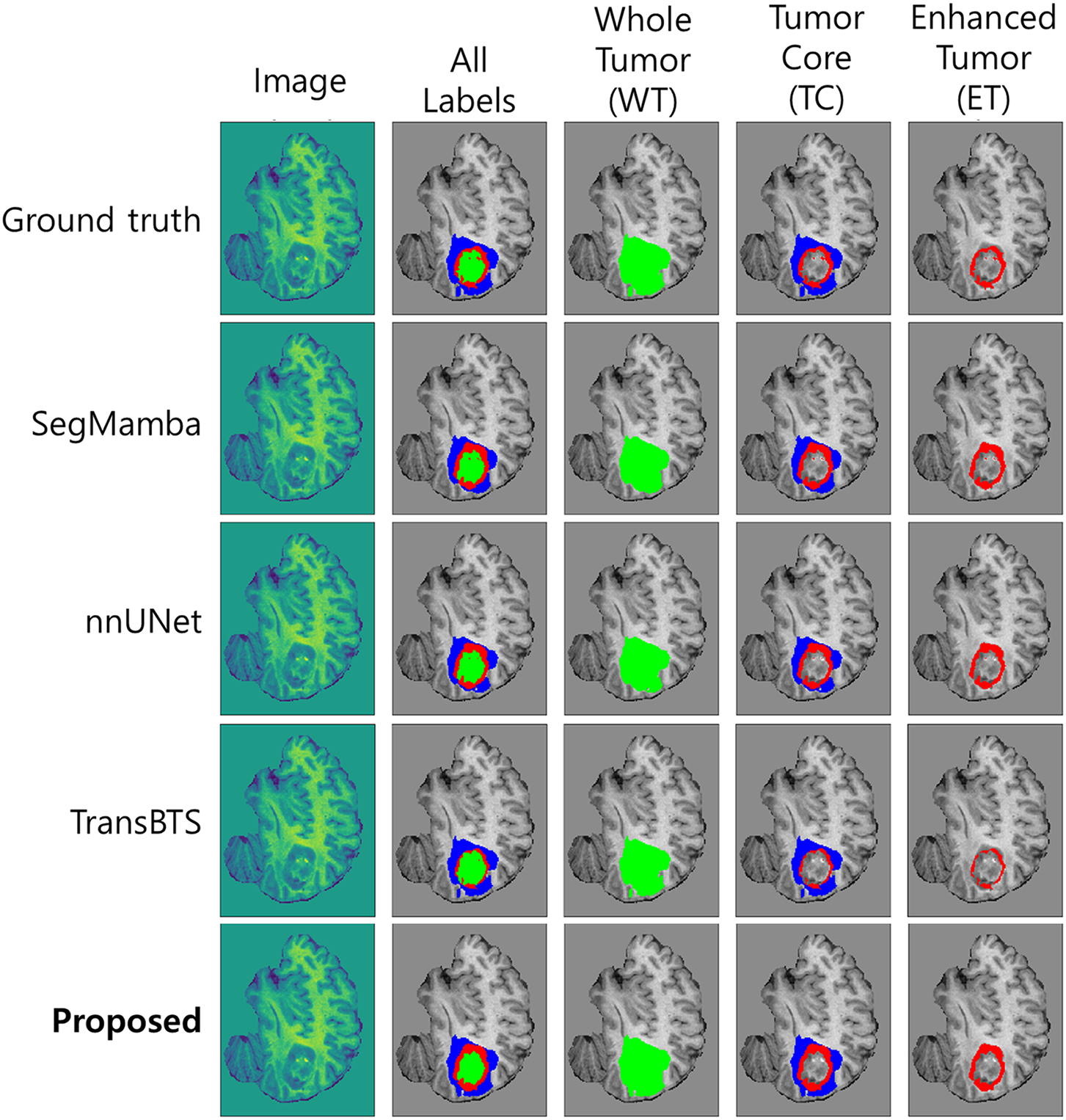
Figure 7: Visualization of the prediction results of the proposed model and other segmentation models compared with Ground Truth on the BraTS dataset. WT = Whole Tumor, TC = Tumor Core (combined necrotic/non-enhancing tumor core [NCR/NET] and enhancing tumor [ET]), ET = Enhancing Tumor. The proposed model closely matches the Ground Truth boundaries, particularly in the TC and ET regions
Our experiments demonstrate that the proposed SwinHCAD model outperforms existing SOTA segmentation models on both BraTS 2020, 2021, 2023 and 2024 datasets. It has consistently demonstrated superiority in both quantitative figures Table 6. Additionally, we were able to objectively observe the effects of each HCAD module on performance, as demonstrated in Table 7. This excellent performance is attributed to the fact that the effective attention gate mechanism of the HCAD decoder proposed in this study was able to precisely learn and segment complex 3D brain tumor structures by synergizing with the Swin Transformer encoder’s powerful multi-scale feature representation. In particular, the attention gate seems to have contributed effectively to predicting the unclear interface of the tumor by suppressing noise and emphasizing meaningful features among the low-level spatial information transmitted through the Skip Connection.
Our proposed HCAD decoder presents a unique and effective approach when compared to SOTA Transformer-based models that use hierarchical decoders. Unlike the decoder in TransUNet, which relies on a conventional CNN-based structure and simple feature concatenation via skip connections, HCAD implements an active and intelligent fusion strategy. It leverages high-level semantic information from the decoder itself as a guide to dynamically recalibrate the channel-wise importance of features received from the encoder’s skip connections. This allows for a more sophisticated refinement of features compared to passive concatenation. Furthermore, when contrasted with pure Transformer decoders like the one in Swin-Unet, which utilizes computationally intensive Swin Transformer blocks throughout the decoding path, HCAD offers a significantly more efficient design. By focusing on a lightweight, channel-wise attention mechanism, HCAD achieves robust feature fusion optimized for multi-modal data without demanding heavy computational resources. Therefore, HCAD establishes an optimal balance, offering more intelligent feature control than conventional decoders and greater computational efficiency than pure Transformer decoders, making it a powerful yet practical solution for complex segmentation tasks.
A key strength of the proposed SwinHCAD architecture is its inherent capability to effectively address the significant heterogeneity of gliomas. These tumors are composed of multiple sub-regions, including the enhancing core (ET), the necrotic core (NCR), and peritumoral edema (ED), each presenting distinct characteristics on multi-modal MRI scans. Our HCAD decoder is particularly well-suited for this challenge. The feature maps derived from each MRI modality such as T1c for the enhancing tumor and T2-FLAIR for edema are processed as unique channels within the network. The guidance-based, channel-wise attention mechanism learns to dynamically modulate the influence of these channels during the decoding process. This allows the model to intelligently prioritize features from the most informative modality when reconstructing the boundaries of each specific sub-region. This targeted, modality-aware feature refinement is a significant advantage that enables more accurate and nuanced segmentation of the complex tumor structure.
An important consideration for the practical application of SwinHCAD is its generalizability to unseen clinical datasets. While the model achieved consistently high performance on the BraTS datasets, real-world deployment will inevitably involve data from different scanners, acquisition protocols, and patient populations. Such domain shifts may adversely affect segmentation accuracy. To address this, future work should focus on domain adaptation strategies, multi-center evaluations, and semi-supervised or self-supervised learning approaches that leverage large volumes of unlabeled clinical data to improve robustness across heterogeneous environments.
In addition, the computational cost of the proposed method is a key factor for clinical feasibility. As detailed in the Section 3.8, our computational efficiency analysis quantifies the model’s memory requirements, training time, and inference speed, providing insight into its suitability for time-sensitive workflows. While SwinHCAD demonstrates a favorable balance between segmentation accuracy and efficiency, further optimization through model compression or hardware acceleration could enhance its applicability in resource-constrained clinical settings.
A key limitation and future direction for this study is the model’s generalizability. We acknowledge that the exclusive validation on the BraTS glioma dataset might constrain the model’s robustness across diverse MRI settings. While the model achieved consistently high performance, real-world deployment will inevitably involve data from different scanners, acquisition protocols, and patient populations. Therefore, a crucial future step is to rigorously evaluate the model’s generalization capabilities. This includes validation on external, multi-center datasets to assess real-world robustness, as well as testing on datasets for other brain tumor types, such as meningiomas. Methodologically, we will also focus on enhancing clinical applicability by investigating lightweight variants of the model for faster inference and exploring strategies for its integration into real-world radiological workflows. These efforts, potentially combined with ensemble techniques and semi-supervised learning, will be essential for establishing the broader clinical utility of our proposed model.
Aiming at accurate segmentation of 3D brain tumor MRI images, this study proposed a SwinHCAD model that combines a pre-trained Swin Transformer encoder with a new hierarchical channel-wise attention decoder (HCAD). The proposed HCAD is designed to dynamically refine the features of Skip Connection through a channel attention gate, effectively fusing the multi-scale features of the encoder. Experiments on the BraTS 2023 benchmark dataset indicate that the proposed model achieves high performance of 88.88% on average Dice Score and 4.68 mm on average HD95. This is 3.6%p higher than SegMamba (85.28%) which scored the highest Dice Score among the SOTA models compared and a 16.7% improvement over VT-UNet (5.62 mm), which showed the best HD95. These results demonstrate that the proposed HCAD decoder can effectively combine with the Swin Transformer encoder to outperform existing SOTA models on complex 3D medical image segmentation tasks. Beyond achieving state-of-the-art segmentation performance, the proposed SwinHCAD architecture advances the design of transformer-based decoders by introducing an efficient, modality-aware attention mechanism that enhances boundary delineation without incurring prohibitive computational costs. We believe these methodological innovations and validated performance gains establish SwinHCAD as a valuable contribution to both the research community and practical clinical applications.
Acknowledgement: The authors wish to express their deep gratitude to all co-authors for their cooperation, encouragement, and valuable guidance during the preparation of this manuscript.
Funding Statement: This work was supported by Institute of Information & Communications Technology Planning & Evaluation (IITP) under the Metaverse Support Program to Nurture the Best Talents (IITP-2024-RS-2023-00254529) grant funded by the Korea government (MSIT).
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Seyong Jin; Methodology, Seyong Jin; Validation, Muhammad Fayaz; Formal analysis, Muhammad Fayaz; Investigation, L. Minh Dang; Data curation, Seyong Jin, Muhammad Fayaz; Writing—original draft preparation, Seyong Jin; Writing—review and editing, Muhammad Fayaz, Hyoung-Kyu Song, Hyeonjoon Moon; Visualization, Seyong Jin, Muhammad Fayaz; Resources, Hyeonjoon Moon; Supervision, Hyeonjoon Moon; Project administration, Hyeonjoon Moon; Funding acquisition, Hyoung-Kyu Song. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data and materials used in this study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Fliessbach K, Weber B, Trautner P, Dohmen T, Sunde U, Elger CE, et al. Social comparison affects reward-related brain activity in the human ventral striatum. Science. 2007;318(5854):1305–8. doi:10.1126/science.1145876. [Google Scholar] [PubMed] [CrossRef]
2. Rao CS, Karunakara K. A comprehensive review on brain tumor segmentation and classification of MRI images. Multim Tools Appl. 2021;80(12):17611–43. doi:10.1007/s11042-020-10443-1. [Google Scholar] [CrossRef]
3. Krishnapriya S, Karuna Y. A survey of deep learning for MRI brain tumor segmentation methods: trends, challenges, and future directions. Health Technol. 2023;13(2):181–201. doi:10.1007/s12553-023-00737-3. [Google Scholar] [CrossRef]
4. Arabahmadi M, Farahbakhsh R, Rezazadeh J. Deep learning for smart Healthcare—a survey on brain tumor detection from medical imaging. Sensors. 2022;22(5):1960. doi:10.3390/s22051960. [Google Scholar] [PubMed] [CrossRef]
5. Ghaffari M, Sowmya A, Oliver R. Automated brain tumor segmentation using multimodal brain scans: a survey based on models submitted to the BraTS 2012–2018 challenges. IEEE Rev Biomed Eng. 2019;13:156–68. doi:10.1109/rbme.2019.2946868. [Google Scholar] [PubMed] [CrossRef]
6. Sajjanar R, Dixit UD, Vagga VK. Advancements in hybrid approaches for brain tumor segmentation in MRI: a comprehensive review of machine learning and deep learning techniques. Multim Tools Appl. 2024;83(10):30505–39. doi:10.1007/s11042-023-16654-6. [Google Scholar] [CrossRef]
7. Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi AK. Medical image segmentation using deep learning: a survey. IET Image Process. 2022;16(5):1243–67. doi:10.1049/ipr2.12419. [Google Scholar] [CrossRef]
8. Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Meth. 2021;18(2):203–11. doi:10.1038/s41592-020-01008-z. [Google Scholar] [PubMed] [CrossRef]
9. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision; 2021 Oct 10–17; Montreal, QC, Canada. p. 10012–22. [Google Scholar]
10. Han K, Wang Y, Chen H, Chen X, Guo J, Liu Z, et al. A survey on vision transformer. IEEE Transact Pattern Anal Mach Intell. 2022;45(1):87–110. doi:10.1109/tpami.2022.3152247. [Google Scholar] [PubMed] [CrossRef]
11. Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, et al. Unetr: transformers for 3D medical image segmentation. In: Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision; 2022 Jan 3–8; Waikoloa, HI, USA. p. 574–84. [Google Scholar]
12. Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D. Swin unetr: swin transformers for semantic segmentation of brain tumors in MRI images. In: International MICCAI Brainlesion Workshop. Cham, Switzerland: Springer; 2021. p. 272–84. doi:10.1007/978-3-031-08999-2_22. [Google Scholar] [CrossRef]
13. Liu X, Song L, Liu S, Zhang Y. A review of deep-learning-based medical image segmentation methods. Sustainability. 2021;13(3):1224. doi:10.3390/su13031224. [Google Scholar] [CrossRef]
14. Moorthy J, Gandhi UD. A Survey on medical image segmentation based on deep learning techniques. Big Data Cogn Comput. 2022;6(4):117. doi:10.3390/bdcc6040117. [Google Scholar] [CrossRef]
15. Jiao R, Zhang Y, Ding L, Xue B, Zhang J, Cai R, et al. Learning with limited annotations: a survey on deep semi-supervised learning for medical image segmentation. Comput Biol Med. 2024;169(3):107840. doi:10.1016/j.compbiomed.2023.107840. [Google Scholar] [PubMed] [CrossRef]
16. Salunke D, Joshi R, Peddi P, Mane D. Deep learning techniques for dental image diagnostics: a survey. In: 2022 International Conference on Augmented Intelligence and Sustainable Systems (ICAISS); 2022 Nov 24–26; Trichy, India. p. 244–57. [Google Scholar]
17. Sharif H, Rehman F, Rida A, Sharif A. Segmentation of images using deep learning: a survey. In: 2022 2nd International Conference on Digital Futures and Transformative Technologies (ICoDT2); 2022 May 24–26; Rawalpindi, Pakistan. p. 1–6. [Google Scholar]
18. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition; 2015 Jun 7–12; Boston, MA, USA. p. 3431–40. [Google Scholar]
19. Celard P, Iglesias EL, Sorribes-Fdez JM, Romero R, Vieira AS, Borrajo L. A survey on deep learning applied to medical images: from simple artificial neural networks to generative models. Neural Comput Appl. 2023;35(3):2291–323. doi:10.1007/s00521-022-07953-4. [Google Scholar] [PubMed] [CrossRef]
20. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference; 2015 Oct 5–9; Munich, Germany. Cham, Switzerland: Springer; 2015. p. 234–41. [Google Scholar]
21. Rehman A, Butt MA, Zaman M. Attention res-unet: attention residual UNet with focal tversky loss for skin lesion segmentation. Int J Decis Supp Syst Technol (IJDSST). 2023;15(1):1–17. doi:10.4018/ijdsst.315756. [Google Scholar] [CrossRef]
22. Xiao X, Lian S, Luo Z, Li S. Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME); 2018 Oct 19–21; Hangzhou, China. p. 327–31. [Google Scholar]
23. Ehab W, Li Y. Performance analysis of unet and variants for medical image segmentation. arXiv:2309.13013. 2023. [Google Scholar]
24. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention u-net: learning where to look for the pancreas. arXiv:1804.03999. 2018. [Google Scholar]
25. Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, et al. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. doi:10.1016/j.media.2019.01.012. [Google Scholar] [PubMed] [CrossRef]
26. He K, Gan C, Li Z, Rekik I, Yin Z, Ji W, et al. Transformers in medical image analysis. Intell Med. 2023;3(1):59–78. doi:10.1016/j.imed.2022.07.002. [Google Scholar] [CrossRef]
27. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306. 2021. [Google Scholar]
28. Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, et al. Swin-unet: unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision. Cham, Switzerland: Springer; 2022. p. 205–18. [Google Scholar]
29. Xing Z, Ye T, Yang Y, Liu G, Zhu L. Segmamba: long-range sequential modeling mamba for 3D medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2024. p. 578–88. [Google Scholar]
30. Chen T, Zhou X, Tan Z, Wu Y, Wang Z, Ye Z, et al. Zig-rir: zigzag RWKV-in-RWKV for efficient medical image segmentation. IEEE Trans Med Imaging. 2025;44(8):3245–57. doi:10.1109/tmi.2025.3561797. [Google Scholar] [PubMed] [CrossRef]
31. Zheng F, Chen X, Liu W, Li H, Lei Y, He J, et al. Smaformer: synergistic multi-attention transformer for medical image segmentation. In: 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2024 Dec 3–6; Lisbon, Portugal. p. 4048–53. [Google Scholar]
32. Xu S, Hu X, Wang H, Zhang H, Li P. Semi-supervised medical image segmentation using swin transformer with focused linear self-attention and data augmentation. In: 2025 8th International Conference on Artificial Intelligence and Big Data (ICAIBD); 2025 May 23–26; Chengdu, China. p. 654–9. [Google Scholar]
33. Ye H, Yang Y-Y, Zhu S, Wang D-H, Zhang X-Y, Yang X, et al. Stain-adaptive self-supervised learning for histopathology image analysis. Pattern Recognit. 2025;161(1):111242. doi:10.1016/j.patcog.2024.111242. [Google Scholar] [CrossRef]
34. Lyu Y, Tian X. MWG-UNet++: hybrid transformer U-net model for brain tumor segmentation in MRI scans. Bioengineering. 2025;12(2):140. doi:10.3390/bioengineering12020140. [Google Scholar] [PubMed] [CrossRef]
35. Huang L, Zhu E, Chen L, Wang Z, Chai S, Zhang B. A transformer-based generative adversarial network for brain tumor segmentation. Front Neurosci. 2022;16:1054948. doi:10.3389/fnins.2022.1054948. [Google Scholar] [PubMed] [CrossRef]
36. Çiçek Ö., Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference; 2016 Oct 17–21; Athens, Greece. Cham, Switzerland: Springer; 2016. p. 424–32. [Google Scholar]
37. Wenxuan W, Chen C, Meng D, Hong Y, Sen Z, Jiangyun L. Transbts: multimodal brain tumor segmentation using transformer. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2021. p. 109–19. [Google Scholar]
38. Zhang Y, Lai H, Yang W. Cascade UNet and CH-UNet for thyroid nodule segmentation and benign and malignant classification. In: Segmentation, Classification, and Registration of Multi-Modality Medical Imaging Data: MICCAI 2020 Challenges, ABCs 2020, L2R 2020, TN-SCUI 2020, Held in Conjunction with MICCAI 2020; 2020 Oct 4–8; Lima, Peru. Cham, Switzerland: Springer; 2021. p. 129–34. doi:10.1007/978-3-030-71827-5_17. [Google Scholar] [CrossRef]
39. Xing Z, Yu L, Wan L, Han T, Zhu L. NestedFormer: nested modality-aware transformer for brain tumor segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2022. p. 140–50. [Google Scholar]
40. Zeng X, Zeng P, Tang C, Wang P, Yan B, Wang Y. DBTrans: a dual-branch vision transformer for multi-modal brain tumor segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2023. p. 502–12. [Google Scholar]
41. Chen J, Mei J, Li X, Lu Y, Yu Q, Wei Q, et al. TransUNet: rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med Image Anal. 2024;97(2):103280. doi:10.1016/j.media.2024.103280. [Google Scholar] [PubMed] [CrossRef]
42. Pramanik P, Roy A, Cuevas E, Perez-Cisneros M, Sarkar R. DAU-Net: dual attention-aided U-Net for segmenting tumor in breast ultrasound images. PLoS One. 2024;19(5):e0303670. doi:10.1371/journal.pone.0303670. [Google Scholar] [PubMed] [CrossRef]
43. Zhang M, Liu D, Sun Q, Han Y, Liu B, Zhang J, et al. Augmented transformer network for MRI brain tumor segmentation. J King Saud Univ-Comput Inf Sci. 2024;36(1):101917. doi:10.1016/j.jksuci.2024.101917. [Google Scholar] [CrossRef]
44. Peiris H, Hayat M, Chen Z, Egan G, Harandi M. A robust volumetric transformer for accurate 3D tumor segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2022. p. 162–72. [Google Scholar]
45. Yang H, Sun J, Xu Z. Learning unified hyper-network for multi-modal mr image synthesis and tumor segmentation with missing modalities. IEEE Transact Med Imag. 2023;42(12):3678–89. doi:10.1109/tmi.2023.3301934. [Google Scholar] [PubMed] [CrossRef]
46. Maani F, Hashmi AUR, Aljuboory M, Saeed N, Sobirov I, Yaqub M. Advanced tumor segmentation in medical imaging: an ensemble approach for BraTS 2023 adult glioma and pediatric tumor tasks. In: Brain tumor segmentation, and cross-modality domain adaptation for medical image segmentation. Cham, Switzerland: Springer; 2023. p. 264–77. doi:10.1007/978-3-031-76163-8_24. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF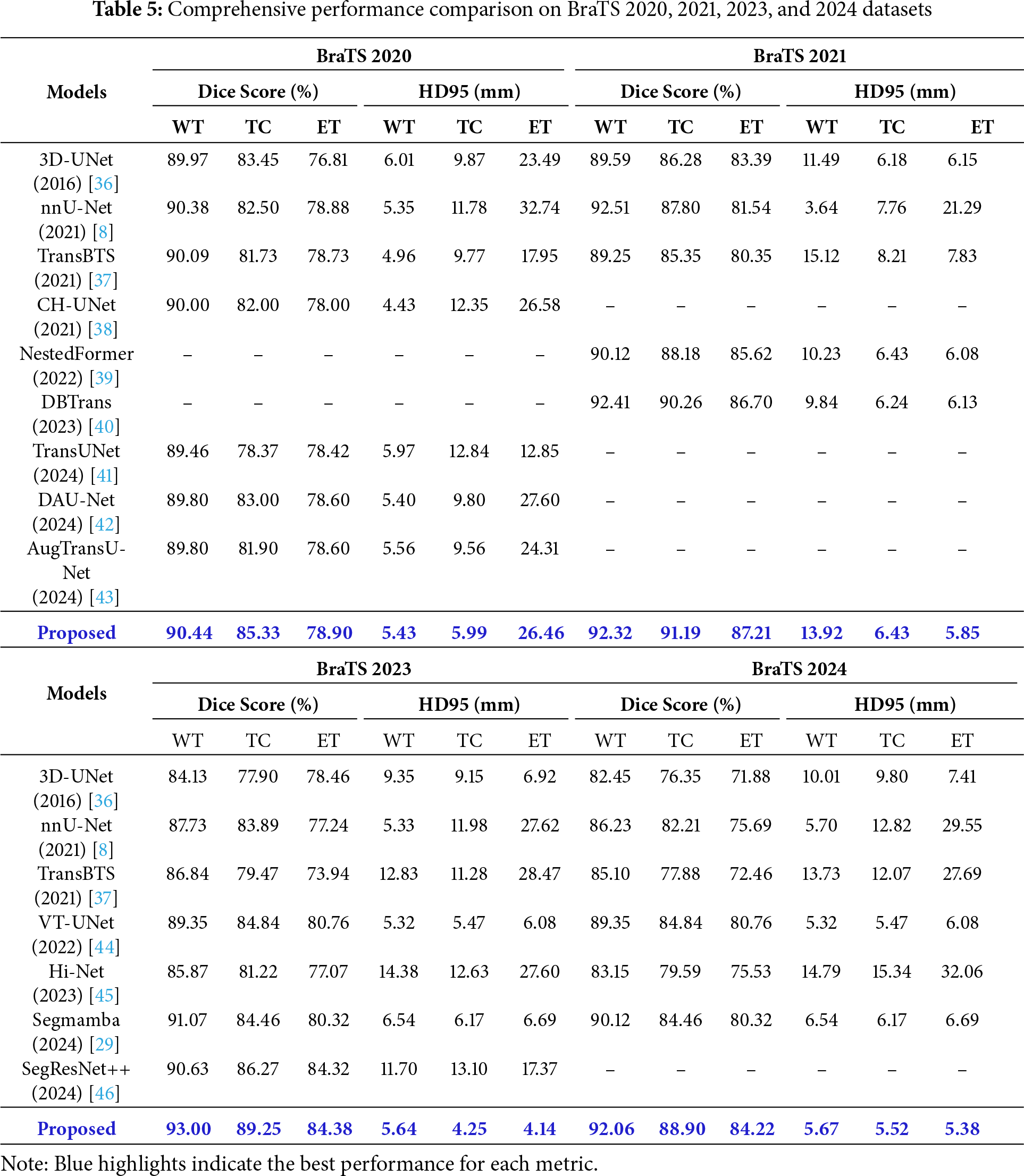
 Downloads
Downloads
 Citation Tools
Citation Tools