 Open Access
Open Access
REVIEW
Transforming Healthcare with State-of-the-Art Medical-LLMs: A Comprehensive Evaluation of Current Advances Using Benchmarking Framework
1 Department of Computer Science, SNEC, University of Calcutta, Kolkata, 700073, India
2 Department of Computer Science, PRTGC, West Bengal State University, Barasat, 700126, India
3 Department of Computer Science & Engineering, The Neotia University, Kolkata, 743368, India
4 Department of Computer Science & Engineering, University College of Science and Technology, University of Calcutta, Kolkata, 700009, India
5 Department of Food Science, University of Copenhagen, Copenhagen, 1165, Denmark
6 Department of Computer Science, Illinois Institute of Technology, 10 West 35th Street, Chicago, IL 60616, USA
7 Department of Computer Science, San Francisco State University, 1600 Holloway Avenue, San Francisco, CA 94132, USA
* Corresponding Author: Dipanwita Chakraborty Bhattacharya. Email:
Computers, Materials & Continua 2026, 86(2), 1-56. https://doi.org/10.32604/cmc.2025.070507
Received 17 July 2025; Accepted 16 September 2025; Issue published 09 December 2025
Abstract
The emergence of Medical Large Language Models has significantly transformed healthcare. Medical Large Language Models (Med-LLMs) serve as transformative tools that enhance clinical practice through applications in decision support, documentation, and diagnostics. This evaluation examines the performance of leading Med-LLMs, including GPT-4Med, Med-PaLM, MEDITRON, PubMedGPT, and MedAlpaca, across diverse medical datasets. It provides graphical comparisons of their effectiveness in distinct healthcare domains. The study introduces a domain-specific categorization system that aligns these models with optimal applications in clinical decision-making, documentation, drug discovery, research, patient interaction, and public health. The paper addresses deployment challenges of Medical-LLMs, emphasizing trustworthiness and explainability as essential requirements for healthcare AI. It presents current evaluation techniques that improve model transparency in high-stakes medical contexts and analyzes regulatory frameworks using benchmarking datasets such as MedQA, MedMCQA, PubMedQA, and MIMIC. By identifying ongoing challenges in bias mitigation, reliability, and ethical compliance, this work serves as a resource for selecting appropriate Med-LLMs and outlines future directions in the field. This analysis offers a roadmap for developing Med-LLMs that balance technological innovation with the trust and transparency required for clinical integration, a perspective often overlooked in existing literature.Keywords
Large Language Models (LLMs) have garnered the majority of attention in healthcare AI research due to their unparalleled ability to process, generate, and contextualize natural language. Natural language is the primary medium of medical knowledge exchange [1]. While other paradigms such as federated learning have shown promise for preserving privacy and mitigating risks such as model poisoning in distributed healthcare settings [2–6], and Explainable AI (XAI) [7–9], Agentic AI [10,11], and traditional machine learning approaches continue to play important roles in specialized applications, LLMs offer a unifying capability. They can seamlessly integrate clinical text, patient records, and biomedical literature, enabling a wide range of applications, from decision support to patient communication. Their pretraining on massive, diverse corpora enables them to generalize across domains with minimal task-specific tuning, providing a practical edge over more narrowly focused methods. Consequently, while federated learning and explainable AI address specific challenges such as privacy and interpretability, LLMs have become the central focus due to their versatility, scalability, and translational potential across nearly every aspect of healthcare delivery. To foster the integration of LLMs into real-world healthcare and to overcome regulatory hurdles, this work also emphasizes the complementary role of paradigms such as Explainable AI in enabling safe, interpretable, and trustworthy clinical decision-making. LLMs may thus be envisioned as the “brain” for medical knowledge and reasoning, with Federated Learning, Explainable AI, and Agentic AI serving as supportive technologies that enhance privacy, trust, and practical deployment in healthcare.
In 2024, according to the study in MedRxiv [12] out of 28,180 AI-related healthcare publications indexed in PubMed (https://pubmed.ncbi.nlm.nih.gov/ (accessed on 15 September 2025)), 1693 were identified as mature, with 1551 selected for detailed analysis. Notably, 479 publications (30.9%) employed Large Language Models (LLMs), surpassing traditional deep learning models (372 publications). The use of text data also rose significantly, accounting for 33.1% (525 publications) of the total, reflecting a growing shift toward LLM-driven research in healthcare—particularly for educational and administrative applications. Leveraging domain-specific data, medical LLMs can assist in diverse tasks such as medical note generation, patient interaction, and medical question answering (QA) [13]. One significant opportunity offered by medical LLMs lies in automating medical documentation. For instance, HEAL, a 13B-parameter model [14], excels in generating physician-quality SOAP notes directly from patient-doctor interactions, significantly reducing the administrative burden on healthcare professionals. Similarly, retrieval-augmented models like JMLR improve factual accuracy in medical QA by integrating domain-specific retrievers, ensuring that responses are both contextually relevant and grounded in credible data [15]. Such innovations promise to enhance efficiency and precision in clinical workflows. Multimodal Medical LLMs integrate diverse inputs such as text, images, and EHR data for holistic reasoning such as Med-PaLM Multimodal [16]. Another key advancement is the introduction of multilingual medical LLMs, such as BiMediX [17], which addresses the linguistic diversity in healthcare. By supporting seamless interaction in both English and Arabic, BiMediX caters to underrepresented populations, bridging gaps in access to medical knowledge and services [18]. Agentic Medical LLMs like ArgMed-Agents [19] act as autonomous clinical assistants that perform multi-step reasoning and decision support.
The adoption of LLMs in healthcare is not without challenges, such as ensuring reliability, minimizing biases, and addressing ethical concerns. Despite these advancements, medical LLMs face significant challenges. One of the critical issues is the potential for generating hallucinated or inaccurate information, which can lead to adverse clinical outcomes [20]. Some state-of-the-art models such as JMLR [15] and BiMediX require careful evaluation to ensure their recommendations align with established medical guidelines [21,22]. Another concern is the bias inherent in training data, which may disproportionately affect certain demographics, exacerbating health disparities [23]. Ethical considerations such as patient data privacy and model transparency remain pivotal in clinical adoption [24]. A crucial aspect of deploying LLMs in medicine is interpretability, which ensures that healthcare professionals can understand and trust model-generated outputs. Deep learning models typically operate as black boxes, in contrast to traditional rule-based systems, which makes understanding the rationale behind their specific predictions challenging. This lack of transparency raises concerns about patient safety, accountability, and clinical decision-making [25]. Techniques such as attention visualization, saliency mapping, and explainable AI (XAI) frameworks can enhance interpretability by providing insights into how models process medical information [26]. Ensuring that LLMs offer clear and justifiable outputs is essential for regulatory compliance, ethical considerations, and broader acceptance in clinical practice. Addressing these challenges requires a multi-faceted approach, combining advancements in model architectures, robust evaluation frameworks, and interdisciplinary collaboration. As medical LLMs continue to evolve, they offer an exciting frontier for improving healthcare delivery while underscoring the need for careful implementation and oversight.
While numerous surveys published in 2024–2025 [24,27–31] explore the development, architecture, and clinical applications of Medical Large Language Models (Med-LLMs), a critical gap remains in evaluating these models from the perspective of real-world deployment readiness. Existing reviews largely focus on performance metrics from static benchmarks and NLP tasks—such as clinical summarization and question answering—but do not examine challenges related to regulatory compliance, Electronic Health Record (EHR) integration, explainability, or safety in clinical workflows. Based on the of recent literature, no existing paper systematically addresses Med-LLM evaluation through the lens of clinical risk, operational deployment barriers, and alignment with global regulatory standards (e.g., FDA, HIPAA, EU AI Act, WHO ethics).
To address this gap, this work provides the following key contributions:
• In this work, the transformation of LLM to Medical LLMs, evolution of Med-LLMs are traced and framing of LLMs as a paradigm shift in the digital healthcare ecosystem has been exhibited.
• This paper analyzes the state-of-the-art Med-LLMs by their underlying architecture, clinical tasks, and benchmark performance.
• Different evaluation techniques has been discussed, how current models are assessed using real-world criteria, going beyond accuracy to include robustness, hallucination risk, factual consistency, and clinician alignment.
• This paper synthesizes international regulations and ethical frameworks to examine the deployment readiness of Med-LLMs and the benchmarking practices of different countries around the world.
• The major deployment barriers—including technical, regulatory, and infrastructural challenges are identified in this work with the future direction and scope in this research domain.
This paper leads to a valuable insight of the architectural evolution of Med-LLMs, the current state of real-world readiness, and the practical scope and limitations that must be addressed for safe and effective deployment in clinical settings. The organization of this paper is captured in Fig. 1.
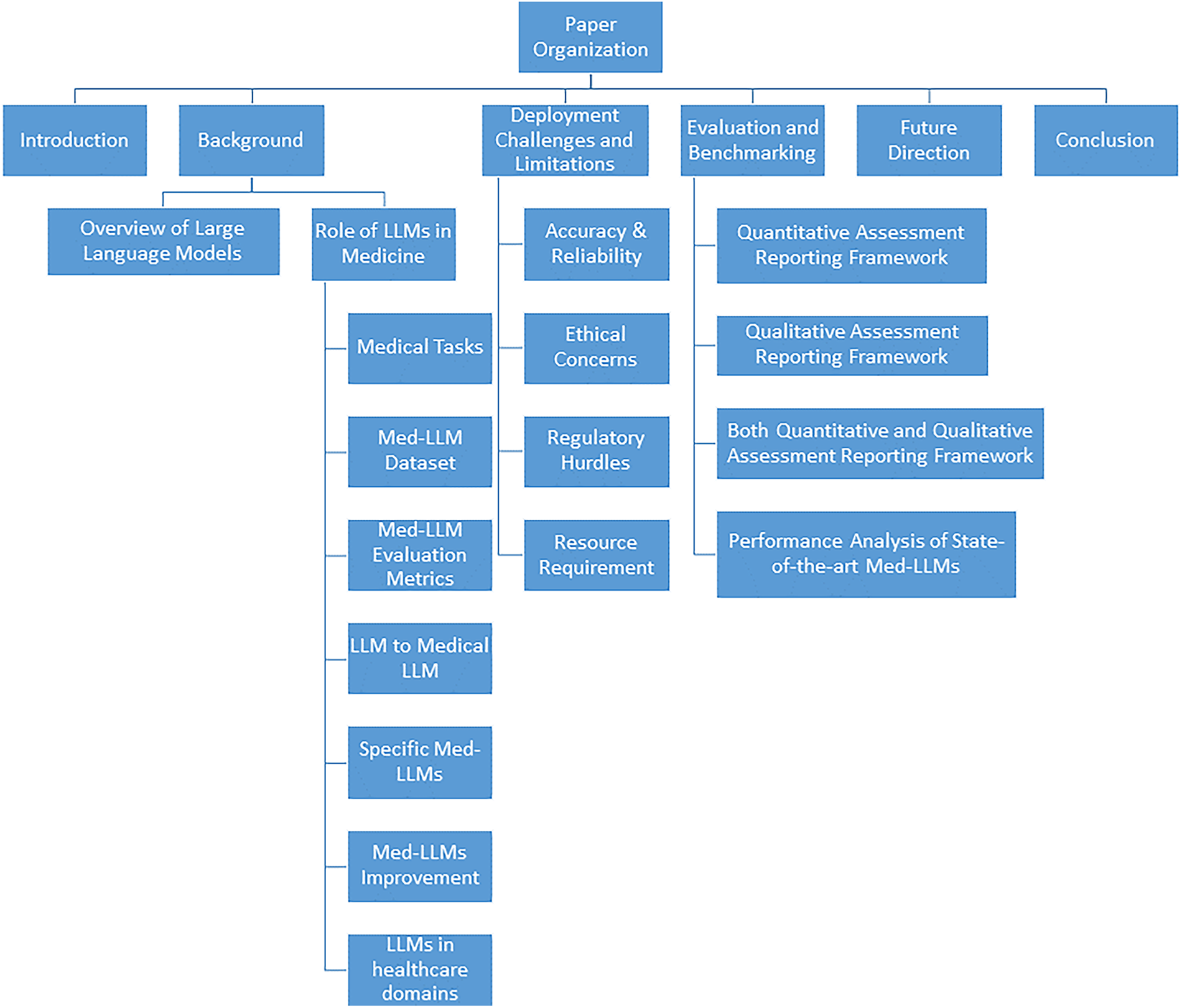
Figure 1: A comprehensive study of Medical-LLMs, its deployment challenges and benchmarking techniques towards providing transparency
2.1 Overview of Large Language Models
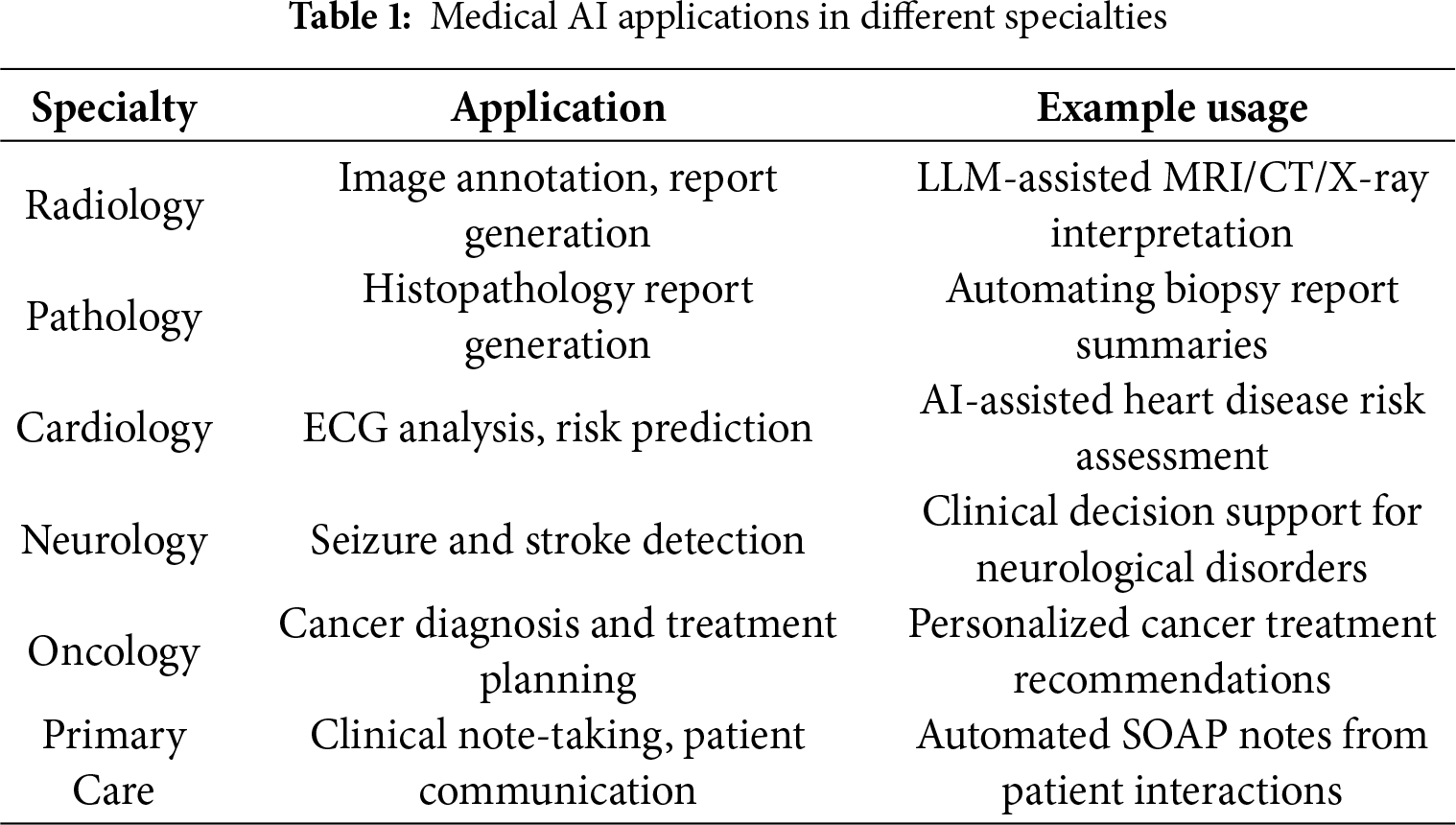
Large Language Models (LLMs) are advanced AI systems built on deep learning techniques and trained on vast collections of text data to comprehend and produce natural language. These models, which utilize transformer-based architectures like GPT [14], BERT [18,32], and LLaMA [33–35], have shown remarkable capabilities in various language tasks including answering questions, creating summaries, translating text, and engaging in conversations. Through self-attention mechanisms and extensive pre-training combined with specialized fine-tuning, LLMs can grasp intricate patterns in meaning, grammar, and context within language. Recent developments have introduced Multimodal LLMs [36–39], which extend beyond text processing to integrate and understand multiple data types simultaneously, including images, audio, and video alongside textual information. These models can analyze medical images, interpret diagnostic scans, and correlate visual findings with clinical text, significantly expanding their utility in healthcare applications. Similarly, Multilingual [40,41] LLMs have been developed to process and generate content across multiple languages, breaking down language barriers in global healthcare delivery and enabling cross-cultural medical communication and research. Within healthcare, LLMs are being adapted through training on medical literature, patient records, clinical documentation, and specialized healthcare datasets to tackle medical applications such as supporting clinical decisions, automating medical documentation, facilitating patient communications, and enhancing medical training. The integration of multimodal capabilities allows these systems to process medical imaging data, laboratory results, and clinical photographs alongside textual information, while multilingual capabilities enable them to serve diverse patient populations and facilitate international medical collaboration. Advanced variants like Med-PaLM, BioMistral, Med-GPT, and BiMediX (as illustrated in Table 1)—which incorporate instruction-following capabilities, multiple languages, and various data types—have broadened the potential uses of LLMs in medical settings. These specialized systems, known as Medical LLMs (Med-LLMs), prioritize key aspects like task-specific optimization, safety protocols, accuracy, and interpretability in their development and assessment. While LLMs continue to advance rapidly and expand the possibilities for medical AI applications, they also present ongoing challenges related to dependability, addressing bias, and practical implementation in healthcare environments.
Model Architecture
The Transformer technology brings new possibilities to handling sequence-to-sequence tasks such as machine translation. Transformer models have developed at an extraordinary rate in recent times through growth to large parameter spaces. The models contain parameter counts ranging from billions to hundreds of billions [14,15]. The fundamental parts of the transformer design have the following features:
1. Encoder-Decoder Structure: The Transformer functions as two separate systems, processing input sequences through an encoder while producing output sequences through a decoder. This design enables flexible input-output mappings, which benefits translations and other related tasks.
2. Self-Attention Mechanism: Self-attention allows each input token to compute attention scores with all other tokens in the sequence. The model performs self-attention operations across multiple projection layers simultaneously, capturing intricate relationships within the input [18].
3. Positional Encoding: Since standard transformer operations lack an inherent understanding of sequence order, positional encoding plays a critical role. Common methods include sinusoidal encodings, learned embeddings, and more advanced techniques like rotary and hybrid encodings [20].
4. Residual Connections and Layer Normalization: Residual connections mitigate the vanishing gradient problem, ensuring stable model updates during training. After each residual connection, layer normalization improves model convergence, stability, and training efficiency [22].
a) Discussion on Pre-Training Stage: During LLM pre-training a model learns from basic text training before getting specific application tasks. Pre-training allows models to extract fundamental language patterns before fine-tuning for domain-specific tasks [42,43]. The following are commonly used pre-training tasks:
i Next Word Prediction (NWP): NWP is a core language modeling task where the model predicts the next word based on preceding words [13,44]. This is a crucial step in training LLMs as it helps models understand contextual dependencies in text [45].
ii Masked Language Modeling (MLM): MLM, as used in BERT, trains a model to predict masked words in a given sentence, enhancing its understanding of syntactic and semantic relationships [32,46,47].
iii Replaced Token Detection (RTD): RTD enables models to distinguish between naturally occurring tokens and deliberately replaced ones, improving their ability to detect subtle shifts in meaning [48–50].
iv Next Sentence Prediction (NSP): NSP helps models capture sentence coherence by predicting whether a given sentence logically follows the previous one. This aids in discourse-level understanding.
v Sentence Order Prediction (SOP): SOP refines a model’s ability to maintain narrative structure by ensuring correct sentence sequencing, which is beneficial for processing long-form medical texts [33,51].
b) Discussion on Fine-Tuning Stage: Fine-tuning adapts pre-trained models for specific tasks using domain-specific data [52–55]. The major fine-tuning techniques are:
i Supervised Fine-Tuning (SFT): SFT improves LLMs by training them on labeled datasets, ensuring they perform well on specific downstream tasks [56,57]. This is widely used in healthcare applications.
ii Instruction Fine-Tuning (IFT): IFT enhances models by training them on instruction-based datasets, allowing them to generalize across multiple tasks [36,58,59]. This technique has been effective in aligning models with human preferences.
iii Parameter-Efficient Fine-Tuning (PEFT): Fine-tuning becomes computationally efficient through PEFT, which modifies only a small portion of model parameters rather than the entire model [18,60,61]. This technique is critical for deploying LLMs in resource-constrained environments.
c) Reinforcement Learning from Human Feedback (RLHF): LLMs undergo additional refinement through RLHF, where human evaluators assess and guide model responses to enhance accuracy and alignment with expert knowledge. This ensures that the models remain reliable in high-stakes applications such as healthcare.
d) In-Context Learning (ICL): By offering a few examples within the input prompt, ICL enables LLMs to adapt across different tasks without requiring explicit fine-tuning. This is particularly useful in medical question-answering applications.
Large Language Models play a pivotal role across multiple domains of healthcare, including clinical decision support, medical documentation, patient communication, biomedical research, and education, by enabling advanced reasoning over vast medical knowledge sources. LLMs, when integrated with EHR data—including clinical notes, lab reports, medical images, and patient interactions—can support radiology, pathology, and clinical planning according to Table 1, enabling AI-driven applications such as diagnosis, prognosis, surgical assistance, telehealth, and hospital guidance as demonstrated in Fig. 2.
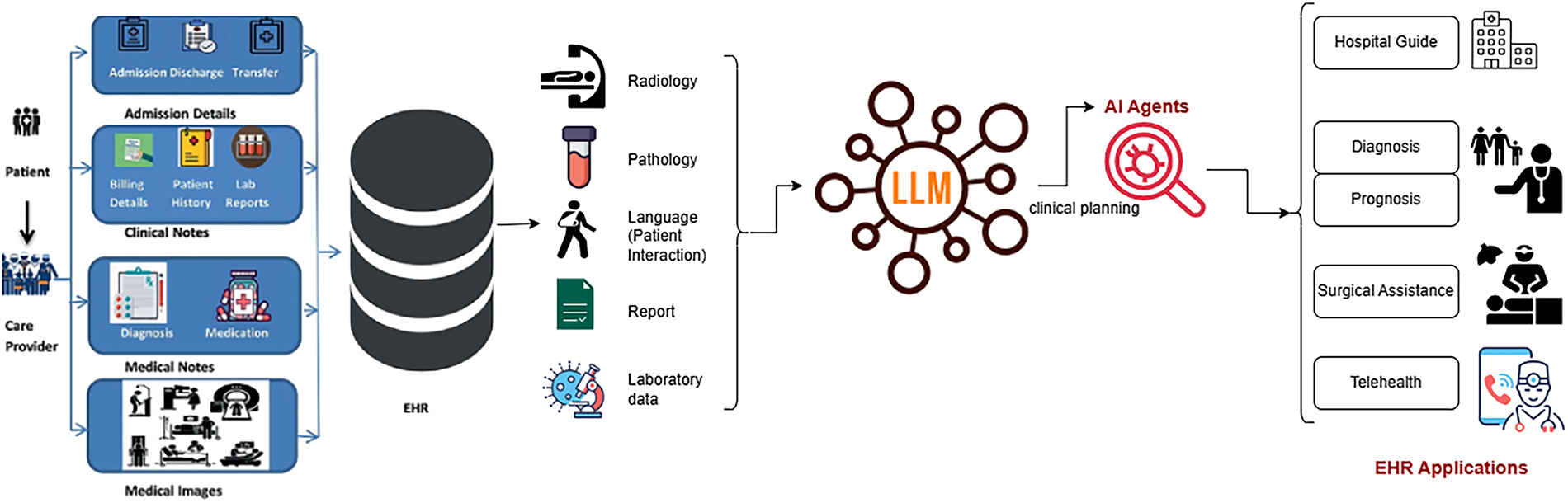
Figure 2: LLM in healthcare
2.2.1 NLP Tasks under Medical Domain
NLP is used to extract medical data (Med-IE), answer medical questions (Med-QA) [45], recognize medical relationships (Med-NLI), and generate medical texts (Med-Gen) [62]. Some medical NLP datasets have been mentioned in Table 2.
1. Medical Information Extraction: Medical Information Extraction (Med-IE) helps to find important medical details from all types of healthcare records without structure [63]. The effort aims to convert text data from medical information into organized forms ready for analysis through decision support tools and research programs [42].
2. Medical Question Answering: Through the Medical Question Answering (Med-QA) task, users build artificial intelligence systems (especially LLMs) to process medical questions from different stakeholders [64]. The system needs structured and unstructured medical data like healthcare records to create precise and dependable responses [65].
3. Medical Natural Language Inference: Medical Natural Language Inference (Med-NLI) is designed to evaluate medical text relationships [59]. The system needs to stay balanced when assessing one segment against another [66]. This task forms a sub-domain within Natural Language Processing where it is referred to as Recognizing Textual Entailment (RTE) [67].
This section seeks to gather and present important medical datasets for language models. The research includes basic dataset information such as publication year, data size, medical task, and available language versions. Medical datasets serve a dual purpose for Med-LLMs by making available expert-level knowledge for training and testing these systems against specific clinical tasks [68]. The rapid development of LLMs in recent years’ achievements drives a strong need for large and well-organized datasets, especially in intelligent medical research [69,70].
While most benchmarks such as MedQA [71] and NEET are limited to multiple-choice or exam-style QA, newer benchmarks are emerging to capture real-world tasks like summarization and patient communication. Datasets such as MIMIC-III/IV [75] for clinical note summarization, MedDialog [74] for doctor–patient dialogues, and HealthSearchQA for consumer-friendly explanations are increasingly being used to evaluate models on tasks beyond QA, aligning more closely with clinical practice.
2.2.3 Med-LLM Evaluation Metrics
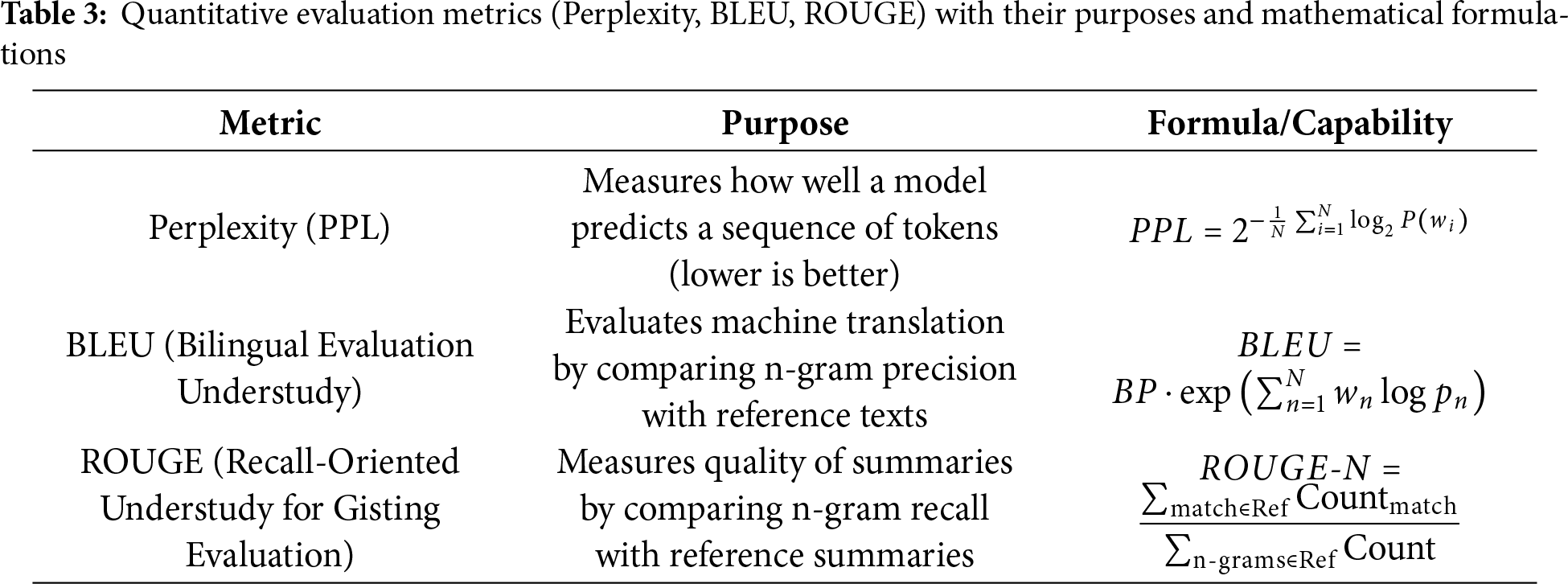
1) Quantitative Evaluation Metrics: The detailed key metrics used for evaluation are listed below:
• Perplexity: Perplexity serves as a basic standard to check how well a probability model predicts input samples. Higher prediction certainty in turn reduces a model’s perplexity score [22].
• ROUGE: A test of text similarity measures how many overlapping units (such as n-grams or chunks) exist between generated output and reference content. ROUGE offers several variants for evaluating text overlaps from its primary four assessments: ROUGE-N, ROUGE-L, ROUGE-W, and ROUGE-SU [18].
• BLEU: According to BLEU analysis, a translation’s similarity to multiple reference translations establishes its effectiveness. A translation quality should repeat small word groupings in the target language just like what appears in available reference texts [80].
However, BLEU [81] and ROUGE [82] summarized in Table 3 continue to be widely applied for evaluating Med-LLM outputs,their reliance on exact n-gram matches has been shown to inadequately capture semantic equivalence, particularly in the presence of medical synonyms, abbreviations, and clinically critical paraphrases. For example, terms such as “myocardial infarction” and “heart attack” or “acetylsalicylic acid” and “aspirin” are semantically equivalent but are penalized under n-gram-based scoring schemes. These challenges necessitate the adoption of more sophisticated evaluation methods that better capture semantic and clinical correctness. Recent works have explored the following approaches:
• Embedding-based semantic metrics: BERTScore leverages contextual embeddings to evaluate semantic similarity beyond surface form matching, thus addressing terminological variability more effectively [83].
• Ontology-aware evaluation: Resources such as the Unified Medical Language System (UMLS) have been employed to normalize entity variation and align synonymous medical expressions across outputs, improving evaluation in terminology-rich clinical contexts [84].
• Task-specific factuality metrics: PlainQAFact introduces a factuality evaluation benchmark for medical summarization, where factual questions are automatically generated from model outputs and verified against source documents, providing stronger alignment with expert judgments [85].
Together, these approaches demonstrate that while BLEU and ROUGE remain useful for comparability, future evaluation of Med-LLMs requires the integration of semantic, ontology-aware, and task-specific metrics to more accurately assess clinical utility.
2) Qualitative Evaluation Metrics: In NLP settings, qualitative evaluations help analyze how humans interpret language output instead of using numerical scores [32]. Human experts use their evaluation skills to examine LLM results because quantitative metrics depend on numerical value measurements. Here are some common qualitative evaluation approaches:
• Human Evaluation: Expert evaluators examine generated text against criteria that measure readability, organized flow, writing quality, topic match, and accurate information [63].
• Error Analysis: Researchers study all system mistakes to identify its effective aspects and problem areas. System errors also need to discover strengths and weaknesses, which guides us in updating the model [64].
• Case Studies: A deep study of how systems perform in real-world scenarios shows what problems arise when dealing with unique situations that statistics cannot detect [86].
• User Feedback: Researchers need to talk with users who use LLM applications to learn if they get helpful responses that help them perform tasks [87].
• Thematic Analysis: The analysis studies how LLMs process language context to create accurate meaning [59].
• Aesthetic Judgments: This is hard to assign numbers to how well generated text looks when read [65].
• Ethical and Societal Impact Assessment: Studies both ethical issues and social outcomes of LLM system functions [25].
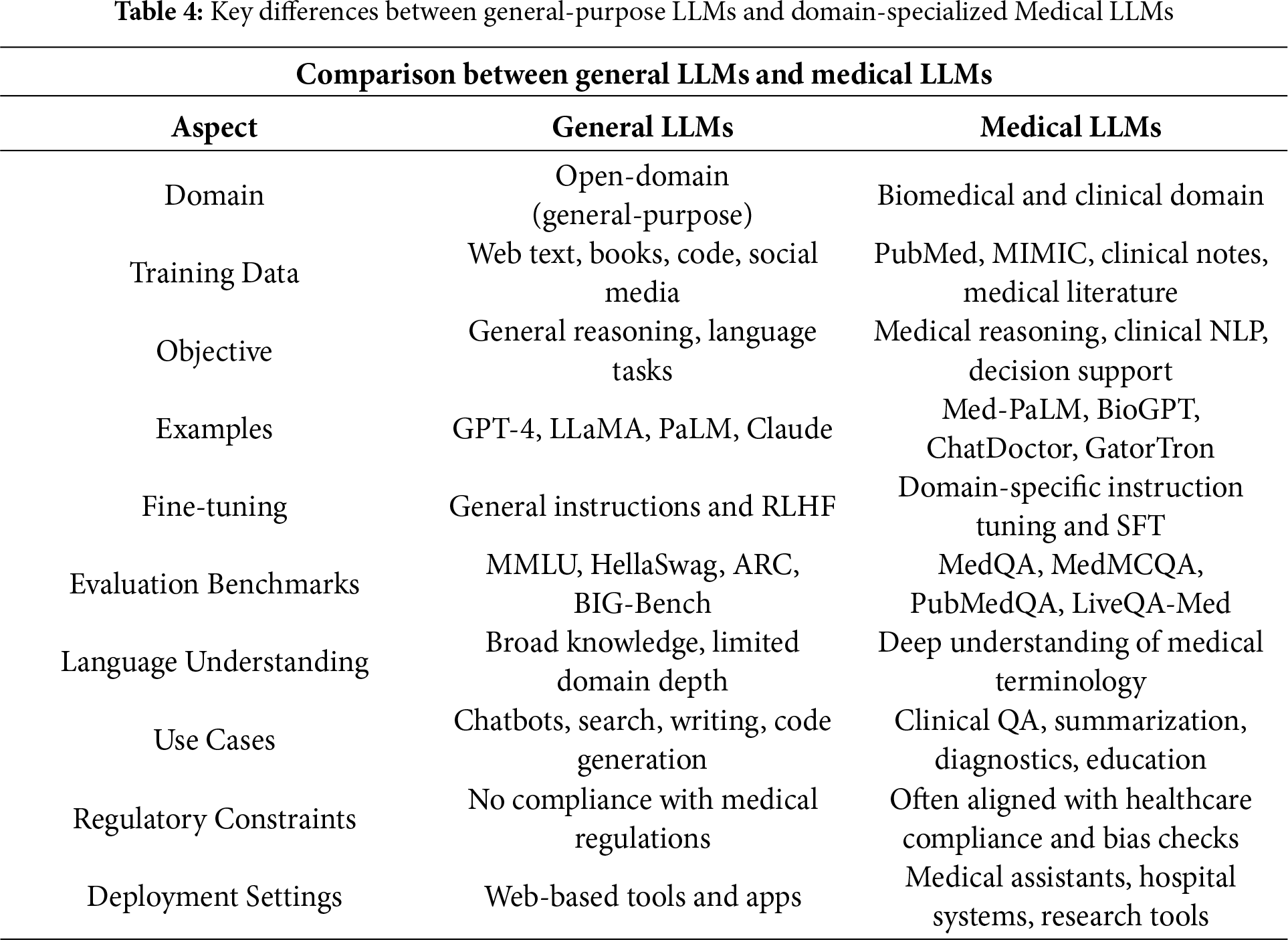
The procedure to enhance a basic large language model into a medical LLM, key differences stated in Table 4, entails complex techniques that surpass basic parameter tuning. The overall process contains a multi-step approach: methods use prompt engineering, medical training, and Retrieval-Augmented Generation (RAG) [54,59].
• Prompt Engineering: Building prompts to ensure the model generates medical responses that align with healthcare standards [32]. The initial stage designs medical-specific prompts that feature structured instructions, real medical context, and predefined response patterns. Prompts guide the model to diagnose based on symptoms, recommend treatments, and simplify complex medical processes for human understanding [64].
• Medical-Specific Fine-Tuning: Choosing high-quality medical datasets begins with clinical notes, textbooks, anonymized patient records, and peer-reviewed research. The dataset must cover essential topics such as pathology, pharmacology, anatomy, diagnostics, therapeutic procedures, and patient treatment [63]. Proper fine-tuning enhances the model’s ability to provide accurate and reliable medical recommendations [66].
• Medical-Specific RAG: Developing a medical RAG system requires integrating retrieval mechanisms to access precise medical information efficiently. The system enables the model to fetch details from up-to-date medical sources, improving accuracy and reliability [65]. By combining generative capabilities with real-time retrieval from trusted databases, LLMs can produce more evidence-based responses [42].
Training Techniques Medical LLMs including Med-PALM,Codex-Med, and MedAlpaca have delivered healthcare progress while focusing on distinct design features with their specialized structures and operational abilities which are mentioned in the Table 5. Medical LLMs were systematically grouped into BERT-based, GPT-based [88], and LLaMA-based [89] families, enabling a structured comparison of their design and performance characteristics highlighted in Figs. 3, 4.
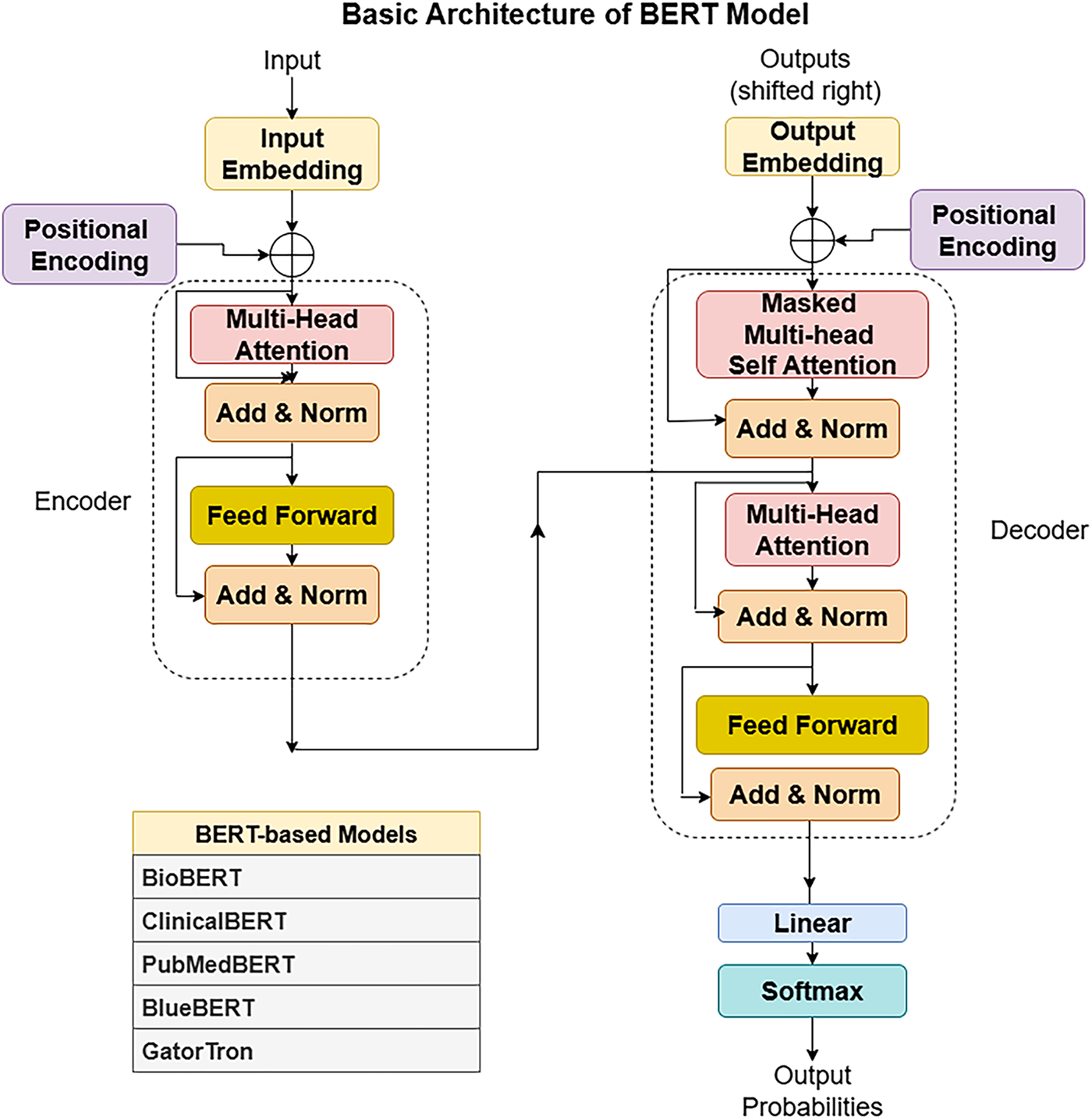
Figure 3: BERT architecture
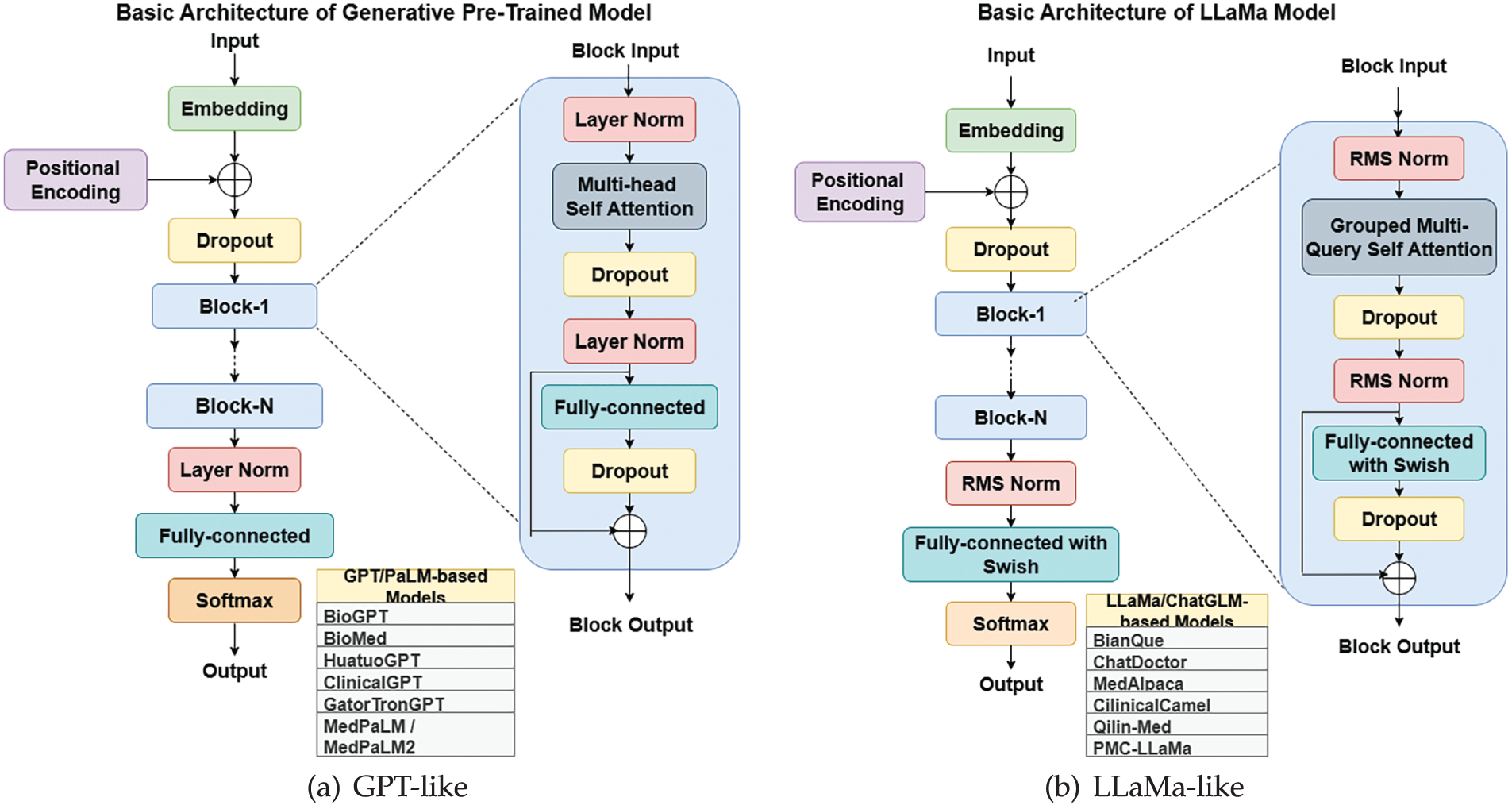
Figure 4: Rise of Medical-LLM from basic architecture of LLMs
A. BERT-Based Models
• BioBERT: BioBERT functions as a BERT variant adapted to process medical and biological text. The system focuses on enhancing medical and biological content comprehension beyond standard text processing designs [32].
• PubMedBERT: PubMedBERT trains a BERT model using abstracts found in PubMed and the architecture is. This approach shows strong results when processing biomedical publications, especially during literature analysis, retrieval, and abstraction tasks [18].
• GatorTron: GatorTron is a large language model developed for various tasks in the medical domain. Research shows this model successfully processes medical documents while producing excellent results [90].
• ClinicalT5: ClinicalT5 uses T5 technology to process clinical healthcare data. This system supports both clinical text generation and documentation summarization. It processes medical records, generates professional reports, creates patient summaries, and supports doctors in medical decision-making [91].
B. GPT/MedPaLM Based Models
• Med-PaLM: Med-PaLM is a language model developed for medical use cases. It undergoes extensive medical data training to assist healthcare professionals with text generation and data retrieval tasks [92].
• GPT-4-Med: GPT-4-Med is an adaptation of GPT-4 for specialized medical applications. It processes complex medical records and generates high-quality medical reports [66].
• BianQue: BianQue is designed to analyze medical text and retrieve medical information for various healthcare applications [62].
• Med-PaLM 2: Med-PaLM 2 is an upgraded version of Med-PaLM, offering improved performance in medical documentation and information extraction [54].
• GatorTronGPT: GatorTronGPT is a medical-oriented extension of GatorTron. It is optimized for medical text generation and information retrieval, producing comprehensive reports and assisting doctors in clinical decision-making [90].
• HuatuoGPT: HuatuoGPT is a medical language model used by doctors for generating detailed medical reports and aiding in clinical decisions [93].
• MedicalGPT: MedicalGPT is a multi-purpose medical LLM that assists in generating comprehensive reports, supporting healthcare providers in decision-making, and facilitating patient interactions [94].
• QiZhenGPT: QiZhenGPT is specialized in Traditional Chinese Medicine diagnostics and treatment planning. It helps physicians detect diseases, recommend therapies, and educate patients about preventive healthcare [95].
• HuatuoGPT-II: HuatuoGPT-II is an improved version of HuatuoGPT, optimized for more reliable and precise medical applications [93].
• Codex-Med: The Codex-Med medical variant delivers extensive medical reports and selects relevant clinical data for processing [96].
• Galactica: Galactica demonstrates strong performance in handling complex medical and scientific content across multiple fields [97].
C. LLaMa/ChatGLM Based Models
• Meditron: Meditron [98] is an open-source suite of multimodal foundation models built on Meta LLaMA, tailored for medical applications such as clinical decision support and diagnosis. Developed through collaboration between EPFL, Yale School of Medicine, and humanitarian organizations like the ICRC, it is trained on curated medical datasets with clinician input, ensuring domain alignment. Notably, the release of LLaMA-3[8B]-MeditronV1.0 demonstrated state-of-the-art performance within its parameter class on benchmarks such as MedQA and MedMCQA, highlighting its impact, particularly for low-resource healthcare settings.
• ChatDoctor: ChatDoctor allows AI-powered medical consultations, providing symptom-based recommendations as initial medical guidance [99].
• BenTsao: BenTsao enhances healthcare system operations by efficiently processing medical text and extracting relevant information [64].
• PMC-LLaMA: PMC-LLaMA is trained on research findings from PubMed Central. Its specialized functions facilitate medical literature searches and biomedical text generation [33].
• Med-ChatGLM: Med-ChatGLM is designed for patient interaction, offering tailored health recommendations and tracking ongoing treatments through user feedback [100].
• Med-Flamingo: Med-Flamingo is designed to support medical systems by generating detailed reports and assisting healthcare professionals in treatment decisions [13].
• ShenNong-TCM-LLM: ShenNong-TCM-LLM focuses on Traditional Chinese Medicine (TCM). It assists practitioners in diagnosing and recommending herbal treatments based on TCM principles [101].
D. Bilingual Model
• Taiyi-LLM: Taiyi-LLM specializes in Traditional Chinese Medicine, identifying medical conditions and recommending herbal remedies while educating patients on TCM practices [34].
The development of medical large language models (LLMs) has advanced through distinct stages as demonstrated in Fig. 5. Initial applications from 2020 to 2022 addressed clinical reasoning by answering medical examination questions and supporting diagnostic reasoning. Subsequently, from 2021 to 2023, LLMs integrated knowledge graphs and structured biomedical data networks to improve factual accuracy. The emergence of retrieval-augmented generation (RAG) between 2022 and 2023 enabled models to access external sources such as PubMed and electronic health records (EHRs), enhancing output reliability. Joint Medical LLM and Retrieval Training (JMLR, 2023–2024) further refined this process by optimizing both the retriever, which locates relevant data, and the generator, which produces responses, to ensure evidence aligned with clinical reasoning. Most recently, domain-specific RAG (2023–2024) restricted retrieval to trusted biomedical datasets, and multimodal RAG (2024–2025) has begun integrating text, imaging, and structured EHR data to provide more comprehensive clinical decision support. Emerging directions include the development of medical agents (2024–2025), which equip LLMs with planning, reasoning, and task-execution capabilities within clinical workflows. There is also growing emphasis on aligning LLMs with human expertise (2024–2025) through physician-in-the-loop evaluation, reinforcement learning with human feedback (RLHF), and consensus-driven fine-tuning to ensure outputs are safe, trustworthy, and clinically actionable.
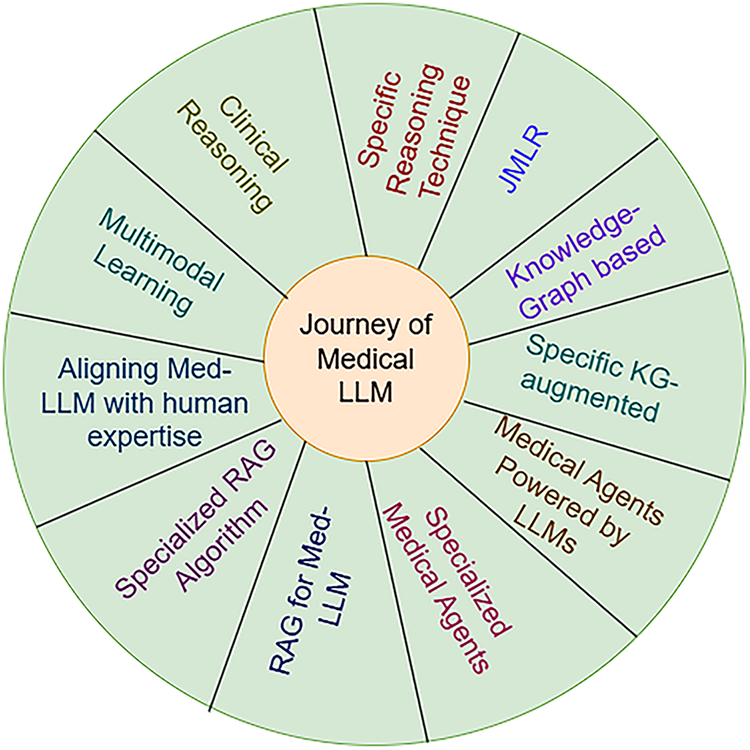
Figure 5: Structural Improvements over Medical LLMs
1. Medical Large Language Models (Med-LLMs) and Clinical Reasoning
Through clinical reasoning, LLMs demonstrate their ability to replicate and assist medical professionals in making decisions [110], similar to how a human doctor applies logical reasoning [54,66].
(a) General Algorithm: When someone works through a series of mental connections, they follow a Chain-of-Thought process, which helps them systematically analyze problems and responsibilities [25]. A healthcare model follows structured steps to break down complex medical questions, assess possible solutions, eliminate incorrect options based on prior knowledge, and explain the final reasoning behind the decision [65].
(b) Specific Reasoning Techniques: The improvement of clinical reasoning of Med-LLMs depends on certain specialized approaches such as:
In-Context Padding (ICP): ICP recommends four actions to improve LLM reasoning in clinical settings [86]:
• The first step is to identify key medical terminology in clinical texts and determine the primary purpose of the document [59].
• The system retrieves important medical entities from knowledge graph (KG) databases based on relevant medical source information [42].
• The model generates both an answer and a detailed medical explanation to support clinical decision-making [87].
2. Joint Medical LLM and Retrieval Training (JMLR)
The JMLR approach stands apart from other Retrieval-Augmented Generation (RAG) systems, which train their retrieval and LLM models separately, because it trains both elements together at the fine-tuning stage [15]. By training LLMs in synchronization with retrieval systems for medical guidelines, JMLR enhances decision-making in patient care while reducing computational requirements [42,65].
3. Knowledge Graph for Med-LLMs
Although LLMs excel at various tasks, they struggle with knowledge base limitations, such as fabricating information and lacking specialized domain expertise [65]. Knowledge graphs (KGs) store extensive structured data that LLMs can leverage to enhance their performance by retrieving precise and relevant medical knowledge [42].
(a) Standard Algorithms: Academic investigations have identified three fundamental approaches for combining knowledge graphs with large language models: establishing LLM-KG Connectivity and developing LLM-KG Compatibility. The implementation of KG-enhanced LLMs alongside LLM-augmented KGs creates complementary structural arrangements that mutually strengthen the capabilities and effectiveness of both knowledge graphs and large language models [66]. The synergized LLM+KG model strengthens the connection between LLMs and knowledge graphs to optimize both systems.
• Inference-Time KG Augmentation: With a retrieval system, the LLM incorporates relevant knowledge from the KG based on the surrounding data to improve performance. By leveraging external knowledge from structured databases, the model reduces the likelihood of generating erroneous outputs and enhances domain-specific responses [63].
• Training-Time KG Augmentation: The LLM undergoes specialized training that integrates natural language processing tasks with structured knowledge graph data. This training process enables the model to utilize KGs more effectively for improved decision-making [64].
4. Specific KG-Augmented Med-LLMs
Healthcare LLM systems achieve superior performance in medical tasks when they receive access to structured medical information. The following are medical applications of LLM systems after integrating medical knowledge graphs [65,111].
• DR.KNOWS: The clinical diagnostic standards form the foundation for introducing DR.KNOWS, which enhances LLMs’s diagnostic capacity. Medical data retrieval reaches new standards by merging medical knowledge graphs with the National Library of Medicine’s UMLS system. DR.KNOWS utilizes medical knowledge graphs to assist doctors in analyzing complex medical cases [42,112].
• KG-Rank: KG-Rank enhances LLM performance in handling medical question inconsistencies and mitigating information bias. This approach improves the accuracy of free-text medical question-answering through ranking and re-ranking techniques applied within a medical knowledge graph [45]. The knowledge graph provides foundational information by extracting relevant triplet data when processing specific medical queries.
• MedKgConv: Conventional dialogue generation models tend to produce repetitive and unengaging interactions. MedKgConv improves natural medical dialogue by integrating multiple pretrained language models with UMLS and utilizing the MedDialog-EN database [63].
• ChiMed: The ChiMed system prepares Chinese medicine datasets tailored for the Qilin-med framework. It integrates patient inquiries, textual data, knowledge graphs, and medical interaction transcripts to enhance multilingual medical comprehension [40].
• DISC-MedLLM: DISC-MedLLM developed a sample creation method guided by medical knowledge graphs to build supervised fine-tuning (SFT) datasets. This approach ensures that generated medical responses remain accurate and trustworthy. The system selects relevant triples from a medical knowledge base based on patient queries through a department-oriented approach [62].
5. Medical Agents Powered by LLMs
LLMs extend beyond text generation by serving as control systems for advanced autonomous AI agents at the forefront of artificial intelligence development [113]. This new AI model moves beyond passive question-answering systems to create active agents capable of handling complex medical tasks.
• Planning Component: AI agents require robust planning systems for strategic reasoning. This mechanism allows the agent to systematically decompose tasks into subtasks and make dynamic adjustments during execution, mimicking human-like problem-solving techniques [55].
• Memory Component: Autonomous medical AI systems need efficient memory architectures to ensure long-term data retention and retrieval. These systems go beyond temporary chatbot memory, incorporating stable data storage and knowledge retrieval mechanisms for improved performance [114].
• Tool Utilization: The integration of planning and memory allows AI systems to iteratively refine inputs and outputs within a given environment. By leveraging external tools, these systems develop adaptive strategies for specialized medical tasks while utilizing memory to execute complex plans [42].
• Evaluation: AgentBench provides a standardized framework for assessing how well large language models function in agent-based tasks. This platform evaluates AI performance across multiple domains, including medical knowledge management, interactive diagnostic reasoning, and clinical decision support [115].
6. Specialized Medical Agents
Several medical applications have been built using large language models as depicted in Fig. 6. These AI-driven medical agents are designed to work alongside healthcare providers, ensuring that critical human skills such as empathy, intuition, and patient-centered care are not lost [42].
• CT-Agent: CT-Agent represents an integration between advanced LLM capabilities and multi-agent systems to improve availability and efficiency in clinical settings. CT-Agent enhances clinical workflows by leveraging GPT-4, multi-agent architectures, and advanced reasoning technologies such as LEAST-TO-MOST and ReAct [113].
• AutoGen: The open-source framework AutoGen enables users to build highly efficient AI programs capable of handling mathematical problems, coding tasks, and online decision-making. AutoGen allows for the creation of multi-agent setups, where different agents collaborate dynamically to complete complex tasks [55].
• ArgMed-Agents: ArgMed-Agents supports medical professionals in making explainable clinical decisions by utilizing multiple AI agents. The system captures the steps of clinical reasoning and generates human-readable explanations. Through self-argumentation cycles guided by a structured reasoning mechanism, ArgMed-Agents models human-like clinical thought processes. The framework employs a directed graph to represent conflicting interactions between different medical concepts, enabling logical rule evaluation [19].
• MAD: The MAD framework is designed to enhance the factual accuracy of LLM-generated responses while optimizing cost, time, and accuracy trade-offs. This system operates through a debate-based platform, where competing AI models assess and refine responses. However, achieving optimal performance with MAD remains challenging due to its dependency on fine-tuned model adjustments and case-specific configurations [116].
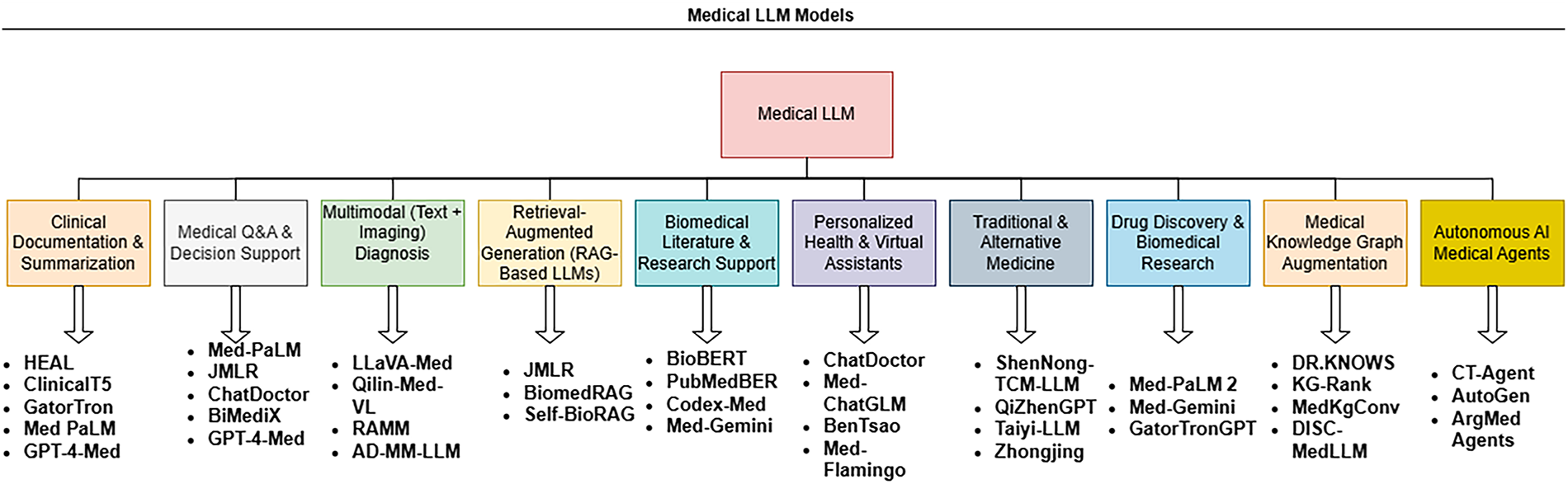
Figure 6: Task-specific medical LLM models
7. RAG for Med-LLMs
Retrieval-Augmented Generation (RAG) represents an innovative machine learning framework that merges retrieval mechanisms with generative capabilities to improve both the precision and variety of produced responses. The methodology has attracted considerable interest within natural language processing circles, finding particular utility in conversational AI systems, platforms designed for answering questions, and tools that condense document content [42].
• Retrieval Component: The information retrieval process starts by accessing relevant data from extensive storage repositories. The system efficiently locates prior information by performing query-based searches against stored conversations, medical records, and scientific literature [65].
• Generation Component: After retrieving relevant data, a generative model synthesizes this context to formulate responses. The generation model significantly improves its accuracy when trained with retrieved medical knowledge, as it can leverage structured input to produce clinically relevant outputs [63].
• Component Integration: The model first integrates retrieved data with input queries and forwards this enhanced context to the generator. The retrieval and generation components then collaborate iteratively, refining both input context and output accuracy across multiple processing stages [64].
8. Specific RAG Algorithms
Combining external knowledge systems with generative models enables RAG to retrieve more precise information that matches user context better than standard systems [42,117] whose architecture has been shown in Fig. 7.
• Clinfo.ai: Clinfo.ai is an open-source WebApp that utilizes scientific literature to provide medical answers. It evaluates both information retrieval methods and summary generation techniques to assess retrieval-augmented LLM systems [55].
• Almanac: LLMs often produce incorrect or harmful outputs. Almanac addresses these limitations by connecting to medical databases, offering users medically verified guidelines and protocols [66].
• BiomedRAG: Unlike traditional retrieval augmentation approaches, BiomedRAG directly integrates retrieved documents into LLMs without applying cross-attention encoding. This design reduces noise in search results while maintaining the integrity of retrieved medical information [63].
• Self-BioRAG: Self-BioRAG is a framework for biomedical text processing that includes specialized explanation generation, domain-specific document retrieval, and self-reflection on model-generated outputs. It is trained on 84,000 biomedical instruction sets [64].
• ECG-RAG: ECG-RAG employs zero-shot retrieval-augmented diagnosis, accessing LLM knowledge while incorporating expert knowledge through specialized prompts [46].
• ChatENT: Standard LLMs suffer from unpredictable outputs, dependence on conditional guidance, and a tendency to generate misleading information. ChatENT refines parameter tuning for medical applications, improving accuracy and reliability [113].
• MIRAGE: MIRAGE evaluates medical RAG systems using a dataset of 7663 medical QA questions collected from five specialized medical datasets. It demonstrates superior performance compared to Chain-of-Thought prompting, GPT-3.5, Mixtral, and GPT-4, improving LLM reasoning capabilities [20].
• MedicineQA: Companies face challenges when applying LLMs in medical domains due to a lack of domain-specific medical knowledge. MedicineQA was developed to test LLMs in real-world medical scenarios, requiring them to retrieve and use evidence from medical datasets to answer questions accurately. It features 300 multi-turn question-answer dialogues [45].
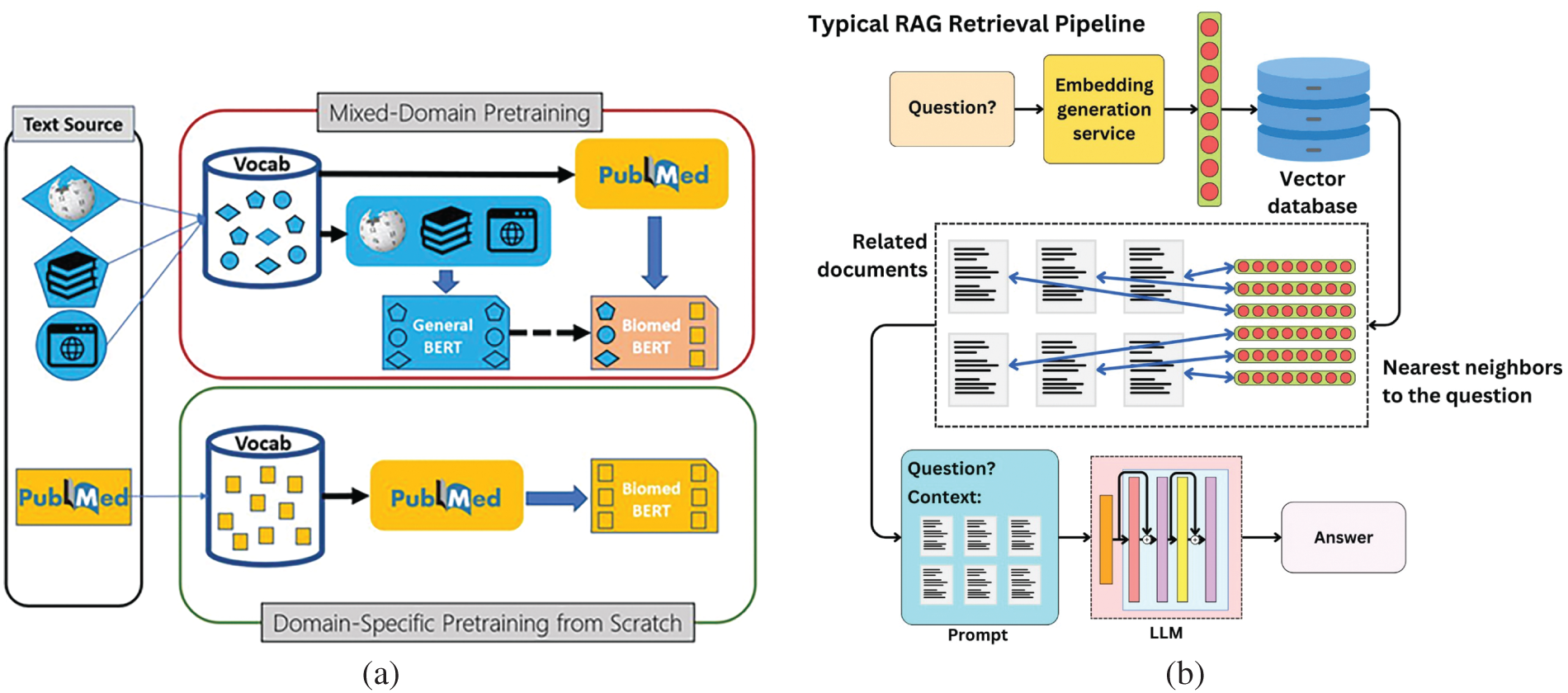
Figure 7: Advancement in architecture of Med-LLMs. (a) RAG Architecture adapted from Microsoft Research Blog [118]; (b) Multi-Modal RAG Architecture adapted from the AI Edge [119]
9. Aligning Med-LLMs with Human Expertise
(a) General Algorithm: LLMs trained on vast text corpora have become the standard for numerous natural language processing applications. However, these state-of-the-art models often struggle with accurately interpreting human instructions, generating biased outputs, and producing incorrect responses due to limitations in their reasoning capabilities. Research groups worldwide are focusing on improving LLMs to better align with human expectations [42,64].
• Data: Developing high-quality training datasets at scale requires access to widely recognized language benchmarks and professional annotators. Advanced language models assist in crafting task-specific instructions to improve the dataset’s reliability and diversity [63].
• Training: Machine learning experts have devised optimization techniques that reduce data requirements while training LLMs to respond more effectively to human feedback. These methods enhance model adaptability and minimize computational overhead [65].
• Evaluation: Researchers have introduced multiple evaluation frameworks and automated testing methods to assess various aspects of LLM alignment, such as factual accuracy, response coherence, and bias mitigation [46].
(b) Specific Alignment Algorithm: Achieving human alignment in medical LLM development is essential, as it directly impacts patient safety and public trust in AI-driven healthcare systems. The following alignment approaches help ensure the safe and responsible deployment of medical LLMs [25,42].
• Safety Alignment: Conducting initial safety assessments is a critical step in developing Medical LLMs. These evaluations establish foundational safety protocols and alignment standards for medical AI applications, ensuring models provide reliable and non-harmful recommendations [65].
• SELF-ALIGN: Addressing the challenges of expensive human supervision and variable quality, the SELF-ALIGN method integrates principle-driven reasoning with LLM-generated refinements. This approach seeks to minimize human intervention while maximizing model reliability [63]. SELF-ALIGN consists of four key steps:
1. The system employs synthetic prompt creation techniques to increase prompt diversity.
2. It utilizes human-written instructions to generate structured responses.
3. The model incorporates self-alignment mechanisms to produce high-quality fine-tuning outcomes.
• EGR: Enhancing Generalization via Retrieval (EGR) optimizes LLMs by incorporating multiple prompt-generation strategies. This method is particularly effective when models are provided with limited input samples, ensuring robust performance despite constrained data availability [115].
10. Multimodal Learning
Multi-Modal Large Language Models (MM-LLMs) are sophisticated systems that can process diverse data types—text, images, audio, video, and sensor information [42]. By incorporating various sensory inputs, these models expand beyond traditional text-based language processing to improve contextual understanding [63]. In this context, Multi-modal fusion denotes the process of integration of multiple data types-such as clinical text, medical images, and structured records-into a single, cohesive model for comprehensive reasoning. In Medical LLMs, this fusion is typically realized through:
• Feature Concatenation (Early Fusion): Simply merging embeddings from different modalities into one vector prior to processing.
• Attention-Based Fusion: Leveraging cross-attention layers, where tokens from one modality selectively attend to elements in another, enabling context-driven intermodal grounding.
• Hierarchical Fusion: Combining modality-specific encoders followed by joint attention layers, facilitating alignment at multiple abstraction levels.
General Algorithms: MM-LLMs are designed to function as unified platforms that process multimedia data across various communication modalities, enabling deeper and more context-aware interactions. Several general-purpose MM-LLMs exemplify these capabilities [115].
• Med-PaLM Multimodal (Med-PaLM M): Med-PaLM M [16] is a generalist, multimodal extension of Google’s medical LLM, designed to process and interpret diverse biomedical data—such as clinical notes, images, and genomic inputs—within a single model framework. Evaluated on the MultiMedBench benchmark (14 tasks), it achieves performance competitive with or exceeding that of task-specific specialist models, often outperforming expert systems.
• PaLM-E: Developed under Google’s Pathways initiative, PaLM-E is designed to process multiple modalities, including visual and textual data. It generates content responses based on images and text-based instructions, enabling automatic image descriptions, visual question answering, and advanced reasoning without requiring extensive specialized training datasets [39].
• LLaVA: LLaVA integrates a vision encoder with a language decoder (Vicuna) to create a foundational vision-to-language MM-LLM. The model undergoes fine-tuning on 158,000 image-text pairs, sourced from the MS-COCO dataset, allowing it to perform visual reasoning and multimodal understanding tasks.
• mPLUG-OWL: In response to parameter flexibility challenges, researchers from Alibaba DAMO Academy introduced mPLUG-OWL. This model combines the ViT-L/14 visual encoder from CLIP with the LLaMA-7B language decoder from mPLUG, enhancing multimodal comprehension and response generation.
11. Specialized Multimodal Med-LLMs
Recent research extensively explores medical vision-language models [117,120] by analyzing multimodal medical datasets, architectural frameworks, and training methodologies [63].
• BiomedCLIP: BiomedCLIP [121] is a biomedical vision-language foundation model pretrained on PMC-15M, a dataset containing 15 million image–caption pairs from biomedical literature. It was trained using contrastive learning to align image and text modalities and demonstrates state-of-the-art performance across diverse biomedical vision tasks (e.g., retrieval, classification, VQA). Notably, it surpasses even specialized radiology models like BioViL on tasks such as pneumonia detection.
• MMed-RAG (Multimodal Retrieval-Augmented Generation): MMed-RAG [122] is a retrieval-augmented generation system tailored for medical vision-language models. It introduces several key enhancements—domain-aware retrieval selection, adaptive context calibration, and RAG-based preference finetuning—to improve factual alignment in responses. Across datasets spanning radiology, pathology, and ophthalmology, MMed-RAG achieves up to 43.8% improvement in factual accuracy, particularly in VQA and report generation tasks.
• V-RAG (Visual Retrieval-Augmented Generation): V-RAG [120] is a framework developed to mitigate hallucinations in medical multimodal LLMs by incorporating both textual and visual evidence in retrieval steps. Evaluations on chest X-ray captioning and VQA tasks show that V-RAG significantly reduces hallucination errors and improves entity-level accuracy compared to baseline RAG methods.
• AD-MM-LLM: This model leverages LLMs for embedding textual data while utilizing ConvNeXt to process medical images, specifically for Alzheimer’s disease detection [25].
• RAMM: RAMM introduces a hybrid training framework that retrieves and integrates medical text and image data during fine-tuning, addressing challenges posed by small biomedical datasets. The model leverages PubMed-based PMCPM data to create image-text pairs, using contrastive learning for pretraining [45].
• LLaVA-Med: LLaVA-Med exhibits strong performance across diverse medical communication scenarios. During training, it associates biomedical terminology with figures, captions and employs instruction-following data to facilitate open-ended semantic comprehension [42].
• Qilin-Med-VL: As the first Chinese vision-language medical model, Qilin-Med-VL integrates a vision transformer core with an LLM foundation. Its training dataset comprises 1 million image-text pairs, sourced from the ChiMed-VL corpus [40].
2.2.7 LLMs in Healthcare Domains
Large Language Models (LLMs) are transforming the entire medical field as exhibited in Fig. 8 by enhancing clinical decision-making, improving patient interaction, and accelerating biomedical research. These applications range from increasing diagnostic accuracy and automating documentation to expediting drug discovery. By leveraging structured reasoning and explainable AI techniques, medical LLMs provide healthcare professionals with more reliable and interpretable insights [42,63].
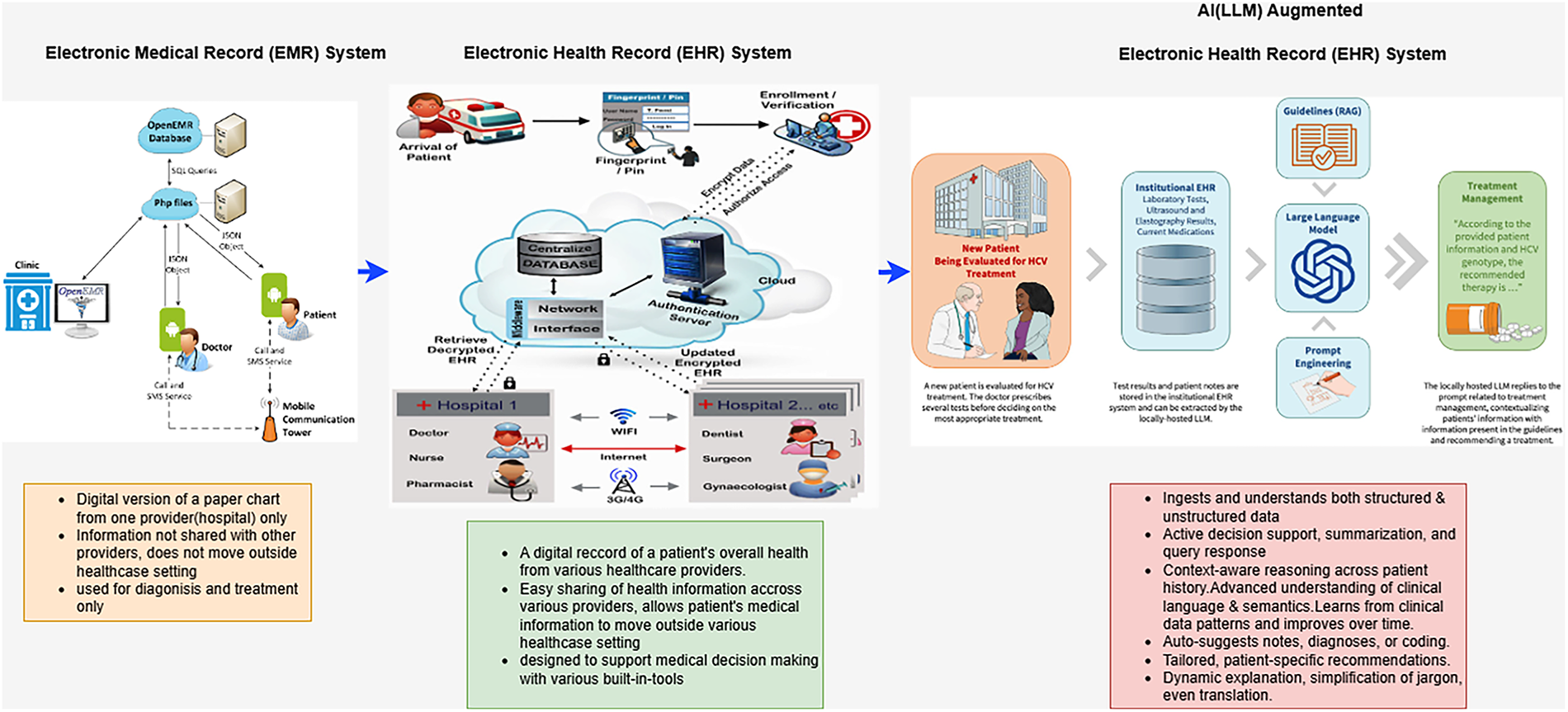
Figure 8: Transformation of Healthcare system
A. Clinical Decision Support
Clinical reasoning serves as the foundation of sound medical practice, equipping clinicians with the ability to diagnose accurately and implement effective management interventions [113,123]. It integrates theoretical knowledge, in-depth analysis, and contextual understanding to reach decisive conclusions and formulate optimal treatment plans. Med-LLMs enhance clinical decision support systems by providing data-driven insights that assist medical teams in making more informed treatment decisions. Specific applications include:
• Symptom Analysis: By analyzing symptom data from healthcare professionals or patients, Med-LLMs access extensive medical knowledge to propose potential diagnoses, assisting in the detection of rare or complex conditions [54].
• Risk Assessment: Med-LLMs evaluate patient demographics and health data to forecast potential medical risks, allowing healthcare teams to take preventive actions and develop personalized treatment plans [45,124].
• Treatment Recommendations: Based on a patient’s diagnosis, Med-LLMs suggest treatment plans that align with existing health guidelines and medication interactions, helping doctors optimize patient care [65].
With the increasing complexity of patient cases, particularly those involving multiple comorbidities and atypical presentations, advancements in AI and LLMs play a crucial role in refining clinical reasoning. The integration of LLMs into medical workflows enables physicians to navigate complex medical scenarios more effectively, ensuring high-quality patient care [65,115]. The following discussion explores the fundamental components, enablers, and emerging technologies that are reshaping clinical reasoning, with a particular focus on how AI and LLMs are driving these transformative changes.
Some Core Elements of Clinical Reasoning
Clinical reasoning can be categorized into two primary approaches: intuitive reasoning and analytical reasoning [65,115].
• Intuitive Reasoning: Commonly referred to as “System 1 thinking,” this approach heavily relies on pattern recognition and experiential knowledge. Clinicians use prior experiences and mental shortcuts to rapidly suggest possible diagnoses. While this method is fast and efficient, it is also prone to cognitive biases such as anchoring bias, confirmation bias, and the availability heuristic [113].
• Analytical Reasoning: Also known as “System 2 thinking,” this approach involves a more deliberate, systematic analysis of clinical data. It is particularly useful for complex or unfamiliar cases that require differential diagnosis, analysis of both favorable and unfavorable factors, and identification of gaps in available information to support accurate conclusions [64].
Both approaches are inherently interconnected. Experienced clinicians frequently alternate between intuitive and analytical reasoning—using the former to generate diagnostic hypotheses and the latter to validate and refine them. The synthesis of these modes results in a well-rounded diagnostic process that enhances reliability and minimizes diagnostic errors [42].
B. Medical Documentation, Summarization and Report Generation
The advancement of artificial intelligence (AI) and large language models (LLMs) has revolutionized medical documentation and summarization [125,126]. In the healthcare sector, Electronic Health Records (EHRs) serve as vital repositories of patient information, yet their vast and unstructured nature often hinders efficient retrieval and analysis. AI-driven models, particularly those leveraging advanced reasoning and summarization techniques, offer promising solutions for enhancing the accessibility and interpretability of EHRs [42,65].
Medical documentation demands precision and contextual understanding, making it a complex task for AI systems. Large language models, such as GPT-4, have demonstrated the capability to generate clinical notes, summarize patient histories, and assist in diagnostic reasoning. Research on diagnostic reasoning prompts for LLMs highlights how prompt engineering refines AI performance in clinical tasks. By structuring inputs with diagnostic reasoning frameworks, LLMs can produce more interpretable and clinically relevant outputs, improving trust and usability among healthcare professionals [54].
Efficient summarization of EHRs is essential for reducing clinicians’ cognitive load and ensuring timely decision-making. A knowledge-seed-based framework introduces a novel approach to guiding AI models in clinical reasoning. By integrating knowledge graphs and domain-specific insights, this method enhances the summarization process, ensuring that critical medical information is retained while minimizing redundancy. This structured approach aligns AI outputs with the cognitive pathways of clinicians, improving accuracy and reliability in medical decision-making [64].
One of the key challenges in AI-driven medical summarization is the interpretability of generated outputs. The ArgMed-Agents framework addresses this issue by employing argumentation-based reasoning. By engaging in self-argumentation and verifying clinical decisions through multi-agent interactions, this framework enhances the explainability of AI-generated summaries. Such systems not only improve accuracy but also foster greater trust among medical practitioners, ensuring that AI recommendations align with established medical knowledge [19].
Med-LLMs streamline clinical documentation by automating report generation, improving both speed and accuracy in medical record keeping.
• Automated Report Writing: Med-LLMs compile data from various medical sources to generate comprehensive reports, reducing manual workload. These systems can generate discharge summaries, radiology interpretations, pathology reports, and surgical notes [66].
• Customizable Templates: By integrating institutional reporting standards, Med-LLMs produce clinical reports that adhere to regulatory and professional guidelines, ensuring consistency and relevance [46].
• Consistency and Accuracy: Med-LLMs rely on extensive medical databases to maintain standardized and reliable documentation, minimizing errors and ensuring high-quality reporting [64].
Beyond documentation, AI is revolutionizing drug discovery and biomedical research by accelerating hypothesis generation, literature mining, and molecular simulations. Processing large-scale biomedical data, AI models can identify promising drug targets, analyze biomolecular interactions, and predict therapeutic efficacy. Advanced natural language processing (NLP) techniques enable researchers to extract critical insights from vast biomedical literature, leading to new discoveries and greater efficiency [45]. Moreover, AI-driven optimization in drug design and repurposing significantly reduces development time and costs, streamlining the drug discovery pipeline [46].
By making EHRs more actionable and facilitating innovation in drug discovery, AI is poised to transform healthcare delivery. Enhanced models can substantially reduce the time and effort required for documentation by leveraging structured reasoning, knowledge-based summarization, and explainability frameworks. As these capabilities continue to evolve, AI will play an increasingly critical role in augmenting medical decision-making, expediting biomedical research, and ultimately improving patient outcomes while optimizing healthcare workflows [115].
C. Drug Discovery and Biomedical Research
The introduction of artificial intelligence into drug discovery and biomedical research has significantly accelerated scientific discovery and hypothesis generation. The advent of large language models (LLMs) and advanced reasoning frameworks has fueled AI-driven approaches for mining vast biomedical literature, generating original hypotheses, and refining research methodologies. Several studies highlight the role of AI in biomedical research and clinical decision-making, with a focus on explainability, reasoning, and the organization of information retrieval [42,63].
In biomedical research, AI facilitates hypothesis generation and testing, expediting the research process. Knowledge-seed-based frameworks introduce structured methodologies to guide LLMs, integrating critical knowledge from clinical datasets to enhance reasoning. These frameworks are particularly relevant in drug discovery [127,128], aiding in the identification of molecular interactions, potential therapeutic targets, and drug-repurposing opportunities. By embedding both structured and unstructured biomedical knowledge, AI can bridge gaps in existing research and uncover new possibilities for pharmaceutical innovation [46].
Mining literature across vast biomedical databases would be infeasible without AI-driven automation. The ArgMed-Agents framework provides a structured clinical reasoning model using argumentation schemes, improving interpretability in decision-making. When applied to drug discovery, this approach enables AI to evaluate conflicting research, weigh evidence, and identify the most promising research avenues. Automated text-mining techniques further facilitate the extraction of key insights, allowing researchers to stay up-to-date with the latest scientific advancements [19].
One of the primary challenges in AI-driven research is ensuring the interpretability of generated insights. Studies on diagnostic reasoning prompts for LLMs demonstrate that structured prompts enhance AI reasoning capacities, improving alignment with expert decision-making processes. When applied to biomedical research, these structured reasoning methods enable AI to formulate hypotheses based on logical implications and supporting evidence, ensuring reliability and validity in generated outputs [54].
Additionally, AI has demonstrated significant success in drug repurposing by identifying new therapeutic applications for existing medications. By integrating structured reasoning techniques with knowledge-seed frameworks, AI models can make informed predictions regarding drug interactions, side effects, and mechanisms of action. Literature mining tools further enhance this process by cross-referencing studies on drug efficacy, thereby optimizing clinical trial design and drug development pipelines [45].
AI continues to expand its role in drug discovery and biomedical research, providing powerful tools for hypothesis generation, literature mining, and structured reasoning. Research indicates that AI-driven approaches enhance accuracy, explainability, and efficiency in biomedical research, ultimately leading to faster and more reliable scientific breakthroughs. As AI technology evolves, its capacity to address complex biomedical challenges will be crucial in shaping the future of healthcare and pharmaceutical innovation [115].
D. Patient Interaction and Health Education
Breakthroughs in artificial intelligence (AI) and large language models (LLMs) have dramatically reshaped the landscape of patient engagement and health-related educational approaches. The integration of chatbots and virtual health assistants into healthcare systems is increasingly undertaken to enhance patient engagement, provide medical guidance, and improve health literacy [129]. Recent studies illustrate how AI-driven clinical reasoning and argumentation frameworks contribute to developing more effective and explainable virtual health assistants [42,65].
LLMs have been employed to create conversational AI agents that assist patients with medical inquiries, appointment scheduling, and basic triage. The ArgMed-Agents framework provides an approach for enhancing AI-driven decision-making in virtual health assistants by leveraging argumentation schemes, allowing for more structured and transparent responses to patient concerns. Structured reasoning also helps mitigate misinformation and fosters greater trust in digital health interactions [19].
Health dietetics is a crucial aspect of preventive medicine, and AI-driven chatbots serve as personalized tutors, guiding patients toward healthier lifestyles. The knowledge-seed-based framework demonstrates how structured medical knowledge can enhance AI-powered reasoning, particularly in virtual health assistants. By tailoring health education materials to individual patient histories and risk factors, this approach improves adherence to medical advice and empowers patients to make informed health decisions [46].
Med-LLMs revolutionize medical education by providing interactive and efficient learning resources.
• Question-Answering Systems: Med-LLMs respond to medical queries with precise and evidence-based answers, improving access to expert-level knowledge for students and professionals [115].
• Simulation of Clinical Cases: Med-LLMs facilitate medical training by simulating patient cases, allowing students to practice diagnostic reasoning and treatment planning [20].
• Summarization of Research Papers: By continuously analyzing recent medical publications, Med-LLMs generate concise summaries that help students and researchers stay updated on the latest advancements [40].
One of the primary challenges in AI-based medical assistants is ensuring the interpretability of their recommendations. Studies on diagnostic reasoning prompts for LLMs emphasize the importance of structured prompts in improving AI-generated responses. Applying similar structured reasoning techniques in virtual health assistants ensures that patient information is communicated in a clear, simple, and easily understandable manner. Transparent information exchange fosters trust and acceptance of AI-driven healthcare assistants [54].
Despite these advancements, AI health assistants face several challenges, including biases in training data, risks of misinformation, and policy regulation concerns. Many of these issues can be mitigated through an argumentation-based reasoning framework that requires chatbots to ground their responses in evidence and comply with established medical guidelines. Additionally, real-time feedback mechanisms can refine chatbot interactions based on patient inputs, further enhancing their accuracy and reliability [45].
LLM-enabled chatbots and virtual health assistants are rapidly expanding their role in patient interaction and health education. The use of structured reasoning frameworks, knowledge-seed methodologies, and explainable AI techniques enables these tools to provide accurate, transparent, and personalized health guidance. As AI technologies continue to evolve, they will further empower patients by improving access to trustworthy health information, ultimately advancing healthcare outcomes and patient satisfaction [115].
Med-LLMs are opening new possibilities for integrating AI-driven robotics in medical training and clinical care.
• Personalized Surgical Planning: Med-LLMs develop customized surgical strategies based on a patient’s medical history and physiological characteristics [19]. Medical LLMs can contribute to personalized surgical planning by synthesizing multimodal patient data, including clinical notes, imaging reports, and laboratory findings, into coherent decision support. By reasoning over patient-specific factors and comparing them with prior clinical evidence, LLMs can generate tailored surgical strategies that align with individual anatomical and risk profiles. When integrated with multimodal models [16,121,122], they enable more precise preoperative guidance and support dynamic intraoperative decision-making.
• Robotic Training for Surgeons: In robotic surgical training, Medical LLMs serve as interactive tutors capable of delivering real-time, natural language guidance during simulation-based practice. They can summarize performance metrics such as errors, timing, and motion smoothness into actionable feedback, while also adapting training difficulty to a learner’s progress. This positions LLMs as valuable complements to robotic platforms, enhancing skill acquisition and ensuring evidence-based learning support. Med-LLMs support interactive procedural simulations, providing performance feedback to improve surgical training [65].
E. Public Health and Epidemiology
Public health and epidemiology are essential fields for monitoring, preventing, and controlling diseases at the population level. The integration of artificial intelligence (AI) and large language models (LLMs) has opened new avenues for real-time disease surveillance, predictive modeling, and informed decision-making. Recent studies highlight the role of AI-based clinical reasoning, literature mining, and structured argumentation frameworks in shaping public health strategies and epidemiological research [42,65].
The next frontier in disease surveillance involves AI-driven real-time data mining, which has the potential to revolutionize automated health monitoring. The knowledge-seed-based framework illustrates how structured knowledge generation enables AI to recognize sequential data patterns, trends, and anomalies within large datasets. Applied to epidemiological data, AI can help uncover emerging health threats, provide early outbreak warnings, and optimize responses to public health crises [46].
Timely access to relevant research on epidemic-prone diseases is crucial for epidemiologists in designing effective interventions. The ArgMed-Agents framework introduces an argumentation-based methodology that enhances AI-assisted literature mining for public health applications. AI systems can sift through vast biomedical research databases, extract critical findings, and synthesize evidence-based recommendations. This ensures that policymakers and researchers have the most up-to-date scientific knowledge to guide public health initiatives [19].
One of the primary challenges AI-powered public health initiatives face is ensuring transparency and explainability in decision-making. Studies on diagnostic reasoning prompts emphasize the importance of structured prompt engineering in guiding AI-generated recommendations. Implementing structured reasoning methods in public health models can enhance clarity and justification for policy decisions, including vaccination strategies, preventive programs, and epidemiological intervention plans [54].
By integrating AI-driven surveillance, literature mining, and structured reasoning, public health and epidemiology can leverage LLMs to improve decision-making, enhance early disease detection, and optimize intervention strategies. As AI technologies evolve, their ability to process and interpret large-scale epidemiological data will be instrumental in advancing global health security and public health preparedness [115].
3 Deployment Challenges and Limitations
Despite extensive research into LLM applications as shown in Fig. 9 in healthcare demonstrating their potential for improving diagnostics, patient management, and clinical decision-making, global adoption remains heavily concentrated in high-income countries like the United States and China [130]. This uneven distribution stems from significant deployment challenges including inadequate technological infrastructure, insufficient data governance frameworks, regulatory complexities, and concerns over bias and privacy [131]. While LLMs could potentially address healthcare inequalities in underserved regions of the Global South by automating patient screening and supporting frontline health workers (Fig. 2), barriers such as limited internet connectivity, computational resources, and quality training data prevent widespread implementation [131].
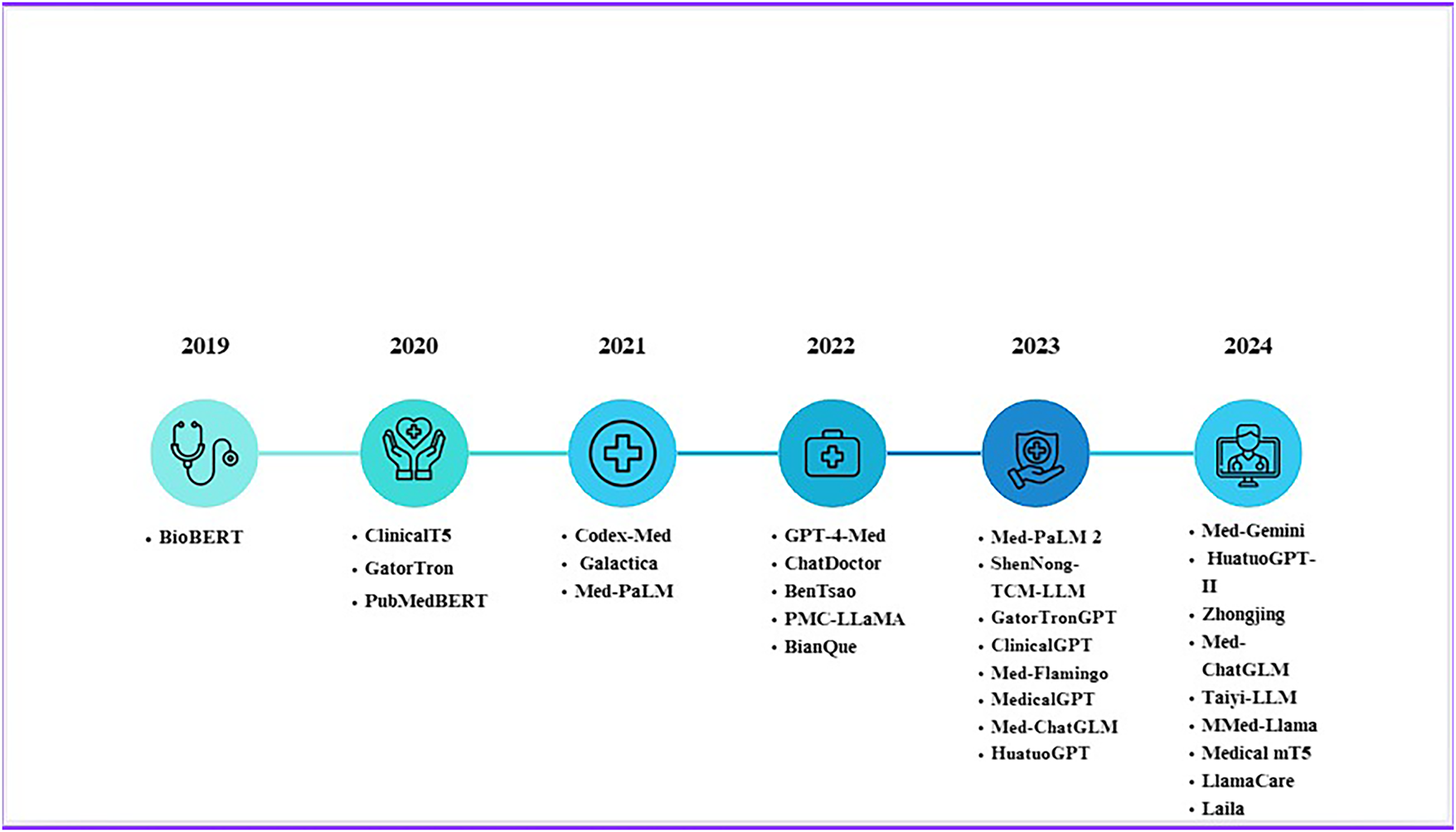
Figure 9: Evolution of Med LLMs from 2019 to 2024
While clinical reasoning is at the core of medical practice, various challenges contribute to diagnostic errors, delays, clinical incompetence, and potential patient harm:
• Cognitive Load: With the rapidly expanding body of medical knowledge and the vast amounts of patient data that must be assessed, clinicians often experience cognitive overload, increasing the risk of diagnostic errors [65].
• Diagnostic Uncertainty: Incomplete medical histories, ambiguous test results, and atypical presentations often introduce uncertainty into the diagnostic reasoning process, complicating decision-making [42].
• Cognitive Biases: Even experienced clinicians are prone to biases that distort their reasoning processes. These include anchoring bias, where clinicians become fixated on an initial diagnosis, and availability bias, where salient but irrelevant details disproportionately influence decisions [19].
• Time Constraints: In high-pressure environments such as emergency departments, clinicians may have limited time to reflect on complex cases thoroughly, increasing the likelihood of diagnostic errors [46].
Recent advancements in artificial intelligence (AI) have introduced transformative tools to support clinical reasoning. Large language models (LLMs) such as GPT-4 have demonstrated potential in improving diagnostic accuracy, facilitating information retrieval, and supporting clinical decision-making [54].
• Augmenting Differential Diagnosis: LLMs assist clinicians by generating comprehensive differential diagnoses based on vast medical knowledge and contextual awareness. For instance, AI models trained on medical datasets can recognize rare and atypical presentations, thereby reducing diagnostic uncertainty [45].
• Facilitating Information-Seeking: Tools such as MediQ leverage LLMs to generate follow-up questions and extract critical missing information during clinical interactions. This approach aligns with the sequential reasoning process clinicians follow in practice [113].
• Mitigating Cognitive Biases: AI-driven systems enhance diagnostic objectivity by synthesizing medical evidence and highlighting inconsistencies in reasoning. By surfacing overlooked differential diagnoses, these models help counteract cognitive biases such as premature closure and confirmation bias [115].
• Education and Training: LLMs provide valuable educational tools for medical trainees by simulating diagnostic reasoning processes. Interactive platforms guide learners through cases with structured explanations and real-time feedback, reinforcing clinical reasoning skills [64].
While AI presents significant opportunities for improving clinical reasoning, challenges remain. LLMs occasionally generate plausible but incorrect responses, and their reasoning processes are not always inherently transparent. Ensuring interpretability, trustworthiness, and ethical compliance—especially regarding patient privacy, data security, and equitable access—remains critical for their integration into clinical practice [115].
Clinical reasoning is an active and evolving skill set that plays a crucial role in patient care. Advances in AI and LLMs provide unique opportunities to enhance diagnostic accuracy and alleviate clinicians’ cognitive burden. However, careful consideration must be given to ensuring that these tools support, rather than replace, human judgment, empathy, and ethical responsibility. By addressing limitations and fostering collaboration between clinicians and AI systems, healthcare can advance in a manner that prioritizes patient-centered innovation [65].
Providing accurate and interpretable medical documentation is a challenging task for AI systems. Large language models (LLMs) such as GPT-4 have demonstrated their ability to comprehend clinical notes, summarize patient histories, and assist in diagnostic reasoning. Research on diagnostic reasoning prompts for LLMs illustrates how prompt engineering can enhance AI performance in clinical settings. By structuring inputs based on diagnostic reasoning frameworks, LLMs generate more interpretable and clinically relevant outputs, improving trust and usability among healthcare professionals [54].
Effectively summarizing electronic health records (EHRs) is critical for reducing clinicians’ cognitive load and enabling timely decision-making. The knowledge-seed-based framework presents a novel approach for enhancing AI models in clinical reasoning. By integrating knowledge graphs and domain-specific insights, this method refines the summarization process, ensuring that essential medical information is preserved while minimizing redundancy. Such an organized approach aligns AI outputs with clinicians’ cognitive pathways, thereby improving accuracy and reliability in medical decision-making [13,19,46].
The interpretability of AI-generated outputs remains a significant challenge in medical summarization. The ArgMed-Agents framework addresses this issue by employing argumentation-based reasoning. Self-argumentation enhances decision-making by enabling AI models to test clinical recommendations against established medical knowledge, ensuring that generated summaries are both accurate and aligned with professional standards. Multi-agent argumentation systems further improve accuracy and foster trust among medical practitioners, reinforcing that AI-driven recommendations align with best practices in clinical decision-making [65,113,115].
Integrating AI into medical summarization and documentation has the potential to transform healthcare delivery by making EHRs more user-friendly and actionable. Advanced models that incorporate structured reasoning, knowledge-guided summarization, and explainability frameworks can significantly enhance the efficiency and reliability of clinical documentation. As AI technology continues to evolve, its role in optimizing medical documentation processes will become increasingly vital for improving patient outcomes and healthcare workflows [42,45,63].
The ethical challenges surrounding the trustworthiness of large language models (LLMs) in the medical field and diagnostic applications remain a pressing concern. Some of the major challenges have been discussed in Table 6. This issue is extensively explored in two key studies: CARES: A Comprehensive Benchmark of Trustworthiness in Medical Vision Language Models and CoD: Towards an Interpretable Medical Agent Using Chain of Diagnosis. The primary concerns regarding the integration of LLMs in healthcare involve trustworthiness, fairness, safety, privacy, and interpretability, all of which are crucial for the responsible deployment of AI in medicine [132–134].
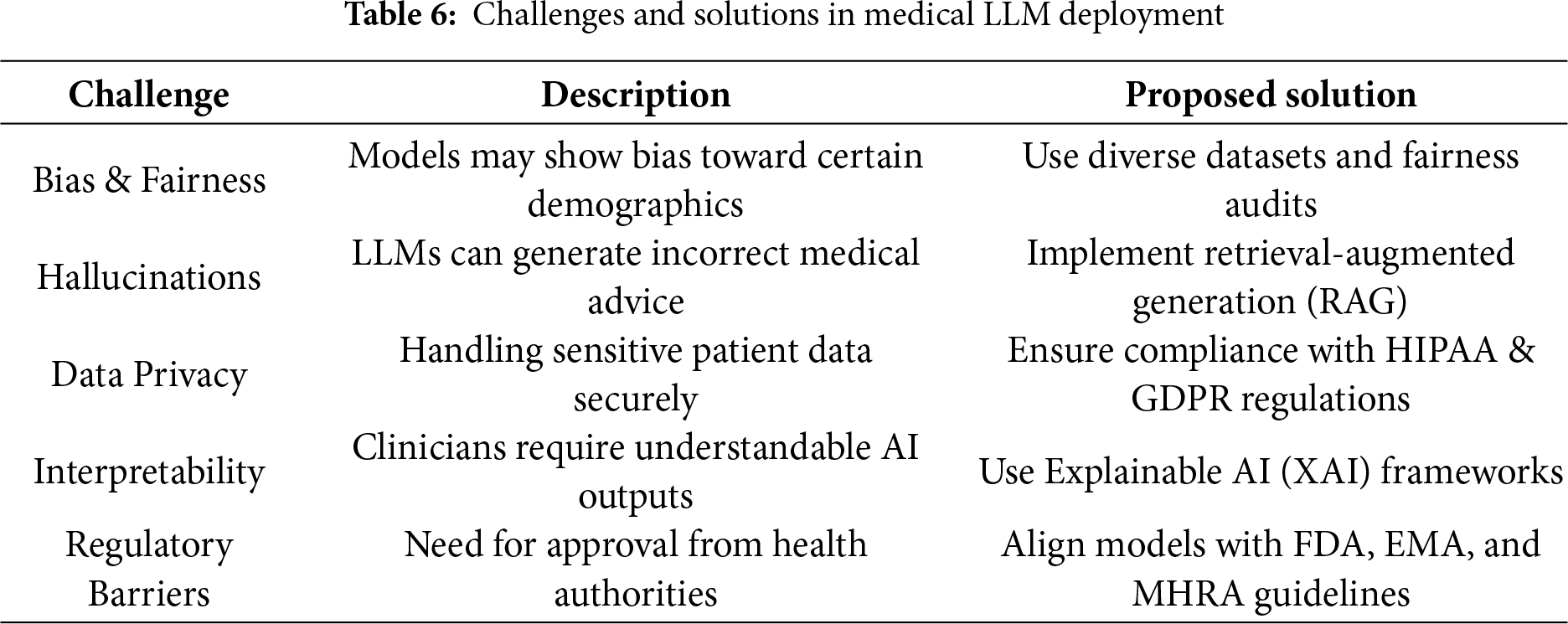
A significant point of concern regarding medical LLMs is their trustworthiness, which, as highlighted in the CARES paper, is primarily defined by factual accuracy and uncertainty estimation [54,135]. Evidence suggests that factual hallucinations occur in nearly all Med-LVLMs (Medical Large Vision Language Models), leading to incorrect medical diagnoses with excessive confidence. This poses a serious ethical issue, as AI-generated diagnoses that are delivered with misplaced certainty can result in unnecessary interventions or even patient harm [136,137]. Robustness, hallucination risk, and factuality were quantitatively assessed through normalized expert scoring. Robustness was measured by evaluating performance on perturbed inputs (e.g., synonym substitution, noisy inputs). Hallucination risk was quantified by calculating the proportion of responses flagged as unsupported by authoritative sources. Factuality was evaluated through expert verification against clinical references as shown in Table 7.
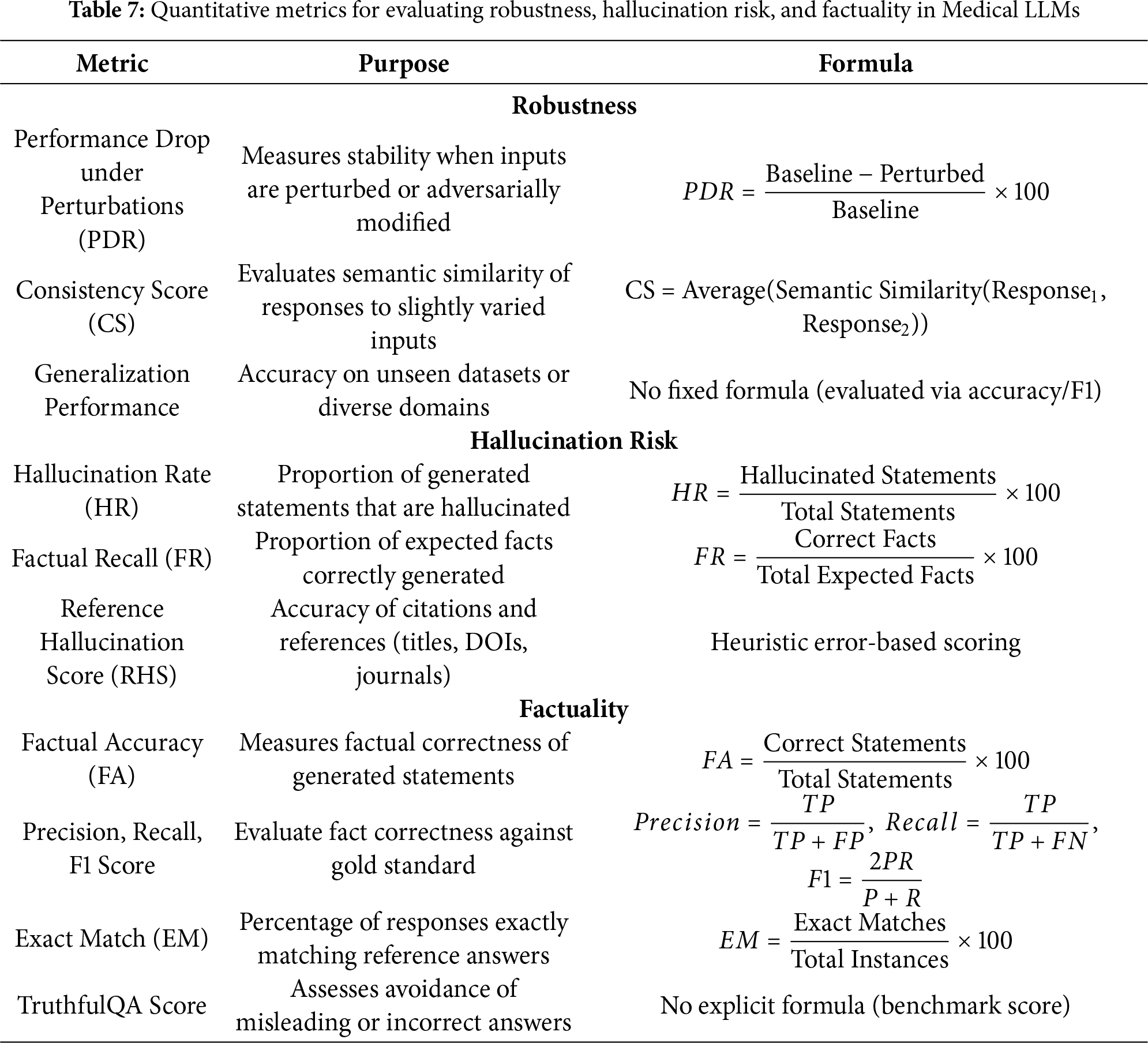
Similarly, CoD highlights the risks posed by fixed LLMs producing arbitrary diagnoses without sufficient query refinement. This limitation raises concerns regarding medical validity, as rationality, caution, and justification should be fundamental principles in clinical decision-making [138,139].
Disparities in AI models can exacerbate health inequities and contribute to unfair treatment across demographic groups. CARES identifies systematic inaccuracies in Med-LVLMs based on patient demographics, including age, gender, and race [38,140]. For instance:
• Accuracy tends to be lower for elderly patients while performing better for middle-aged groups (40–60 years).
• Some models exhibit notable biases against Hispanic and Caucasian populations, resulting in less accurate predictions compared to other racial groups.
• While gender-based biases appear to be less pronounced, disparities exist in dermatology and CT scan models.
These biases reinforce structural inequalities within the healthcare system, making it ethically imperative to enhance fairness in AI models before their clinical deployment [141].
Another major ethical dilemma relates to the safety of AI in medical applications. CARES evaluates safety based on the following criteria:
• Jailbreaking Attacks: Specially crafted prompts can be used to manipulate Med-LVLMs into providing false, misleading, or unsafe medical advice.
• Toxicity Risks: Some models produce harmful or inappropriate content when prompted with biased or misleading inputs. The CoD study further indicates that LLMs often lack proper search procedures for symptoms, leading to reckless or incomplete decisions that could delay or misguide clinical treatment [142,143].
Privacy plays a crucial role in healthcare AI, as the improper handling of sensitive patient data can lead to confidentiality breaches. CARES highlights concerns that Med-LVLMs often fail to safeguard personal health information. These models have been observed leaking private patient data when queried or even generating fabricated personal details, raising serious ethical and legal challenges.
To address these concerns, CoD proposes the use of synthetic patient cases rather than real patient data, thus enhancing privacy at the cost of some model efficacy. However, synthetic data itself presents ethical risks, particularly when poorly designed datasets fail to provide an accurate approximation of real-world medical scenarios. Ensuring that AI models uphold privacy, fairness, and safety remains critical to their ethical integration into clinical practice [132].
ICE-T represents a novel interpretive examination framework designed to improve Large Language Model (LLM) classification outcomes. The system advances both interpretability and performance, making it highly relevant to medical and legal applications. ICE-T employs a controlled multi-prompt strategy that transforms LLM responses into numerical feature vectors, which can then be processed by standard classifiers [138,144]. This method enables compact models to perform at levels comparable to or even surpassing larger models in zero-shot settings. Performance evaluations indicate that ICE-T outperforms zero-shot baselines across medical, legal, and climate-focused datasets based on F1-score assessments. By facilitating structured decision-making and allowing experts to track model reasoning, ICE-T represents a significant advancement in interpretable AI applications, particularly in high-stakes environments [19,42].
Another diagnostic framework, Chain-of-Diagnosis (CoD), enhances both interpretability and operational efficiency in automated medical diagnostics [145]. CoD replicates a physician’s diagnostic workflow through a process involving symptom acquisition, candidate disease retrieval, diagnostic processing, confidence measurement, and final decision implementation [132,133]. This system employs an entropy-reduction mechanism to assess uncertainty, ensuring that diagnoses are accompanied by confidence estimates. DiagnosisGPT, a model trained using synthetic patient scenarios derived from a database of 9604 diseases, achieves over 90% accuracy in high-confidence settings. CoD bridges LLM reasoning capabilities with real-world clinical applications while addressing privacy concerns associated with synthetic patient data [139].
Despite these advancements, autonomous healthcare solutions based on Medical Large Vision Language Models (Med-LVLMs) face significant trust issues due to imperfect factual accuracy, demographic biases, security vulnerabilities, and inadequate privacy protections. The CARES system systematically evaluates Med-LVLMs using 18,000 images and 41,000 question-answer pairs covering 16 imaging modalities and 27 body regions. Findings indicate that these models frequently produce erroneous outputs with excessive confidence, exhibit demographic biases, and are susceptible to jailbreak attacks and toxic prompt manipulations [54,135]. This highlights the necessity for robust evaluation frameworks such as CARES, which help detect reliability and fairness issues in AI-driven medical diagnostics. Publicly releasing evaluation benchmarks and code will enable researchers to enhance medical AI systems that prioritize accuracy, safety, and equitable treatment across diverse patient populations.
One study assesses the application of LLMs such as ChatGPT in healthcare diagnostics, particularly for zero-shot and few-shot learning scenarios. This research explores heart disease risk prediction using a dataset of 920 patient records and compares LLMs to traditional machine learning (ML) systems. Researchers analyzed diagnostic interactions between Numerical Conversational (NC) and Natural Language Single-Turn (NL-ST) models, incorporating domain knowledge enrichment through ML-derived prompts [38,140]. Results indicate that while ML models maintain superior performance in zero-shot settings, LLMs achieve comparable diagnostic accuracy in few-shot learning conditions, particularly when paired with explainable AI (XAI) methods. The study demonstrates that LLMs can serve as effective tools for minimizing false negatives while improving operational efficiency. Further refinements in prompt engineering and explanatory reasoning functions could significantly enhance performance, supporting the development of collaborative healthcare frameworks that integrate ML and LLM technologies [141].
LLMs’ ability to emulate clinical reasoning in medical diagnostics has also been evaluated. A study compared GPT-3.5 and GPT-4 in diagnostic reasoning tasks, assessing their ability to replicate clinician reasoning processes such as differential diagnosis, analytical reasoning, intuitive reasoning, and Bayesian inference. Two benchmark datasets were used: a modified MedQA USMLE dataset and the NEJM case series. Results indicate that GPT-4 outperforms GPT-3.5, demonstrating improved diagnostic reasoning capabilities. However, the study also notes that diagnostic reasoning prompts did not necessarily enhance diagnostic accuracy. Moreover, challenges such as data hallucinations and market-specific limitations (e.g., a focus on U.S.-based medical cases) were identified. These findings underscore the urgent need for model refinement, particularly through engineering improvements that facilitate safe clinical workflow integration [142,143].
Integrating large language models (LLMs) into healthcare diagnostics and medical applications creates substantial regulatory hurdles related to establishing trust, ensuring equitable outcomes, maintaining safety protocols, protecting sensitive information, and providing clear operational visibility, discussed in [146]. Research initiatives like CARES: A Comprehensive Benchmark of Trustworthiness in Medical Vision-Language Models and CoD: Towards an Interpretable Medical Agent Using Chain of Diagnosis illuminate critical obstacles that require resolution prior to the safe implementation of LLMs within medical environments [132,133]. Table 8 is presented to show different regulatory frameworks for AI in Healthcare for different regions.

3.3.1 Lack of Standardized Trustworthiness Benchmarks
The absence of uniformity in assessing trustworthiness across large language models is one of the main regulatory challenges. While CARES is a benchmark to evaluate trustfulness, fairness, safety, privacy, and robustness, it points out that such models don’t exhibit consistent performance, creating problems for regulatory oversight [63,147].
A regulator, therefore, is left with the task of deciding the minimum performance indicators that must be met before the deployment of the LLMs. Should a model have to achieve a threshold rate of factual accuracy? How can uncertainty estimation be assessed by regulators? Due to a lack of commonly accepted guidelines, certification and compliance enforcement become difficult [137].
3.3.2 Bias & Fairness Regulations
Bias in medical datasets remains a critical barrier for reliable Medical LLMs [148]. Demographic bias is the most pervasive, arising when specific populations such as women, racial or ethnic minorities, or rural communities are underrepresented, leading to systematic performance gaps. Gender bias often results from male-dominant datasets, while racial and ethnic bias emerges from Western-centric corpora that fail to generalize to regions such as South Asia, Africa, and the Middle East. Additional categories include data bias, resulting from skewed sampling across diseases or care settings; measurement bias, caused by inconsistent coding and diagnostic criteria; algorithmic bias, introduced when models are optimized for overall accuracy but fail on minority subgroups; and deployment bias, which occurs when models are applied in contexts different from those where they were trained.
Mitigation strategies include diversifying datasets with inclusive regional data, applying fairness-aware learning methods such as reweighting, resampling, or adversarial debiasing, performing subgroup-specific evaluation to audit performance across demographics, using prompt engineering to reduce biased outputs in generative tasks, and adopting transparent reporting standards such as CONSORT-AI [149] and MINIMAR. Collectively, these approaches are essential to improve the fairness, equity, and trustworthiness of Medical LLMs in global healthcare practice.
The CARES study reveals considerable demographic biases in medical LLMs based specifically on age, sex, and ethnicity [38,46]. Regulatory bodies such as the FDA in the United States, EMA in Europe, and MHRA in the United Kingdom currently lack guidelines related to formalizing fairness in AI-mediated medical applications and then adjusting for those inequalities [147].
For example, should these models show similar accuracy across every defined demographic category before obtaining approval? What would be the fair thresholds under which the regulators would operate? In the absence of express legal frameworks, developers may ignore fairness considerations, contributing to health disparities once the model is applied in real time.
3.3.3 Data Privacy & Compliance Issues
Privacy regulations, such as HIPAA (U.S.), GDPR (Europe), and India’s DPDP Act, impose strict requirements on managing patient data. CARES emphasizes that Med-LVLMs fail to maintain confidentiality of personal information and either hallucinate or expose sensitive patient details [115].
CoD attempts to put this into action by using synthetic patient cases rather than real patient cases. However, this raises more regulatory questions.
Could it be alleged Hypothetically:
• Does privacy law allow for the substitution of synthetic data in place of real data?
• Do clinically valid diagnoses arise from models trained on synthetic data [137]?
• What could provide regulators with assurance that biases are not being introduced by synthetic data?
In the absence of definitive and clear laws, developers are caught in a swamp of near-insurmountable regulations that ultimately slow down approvals and increase compliance costs [38].
3.3.4 Safety and Liability Challenges
One vital area regulating AI in health care is by defining liability. CARES demonstrates Med-LVLMs are susceptible to jailbreaking attacks which can fool models to produce unsafe medical advice. In this case, who will be liable if an LLM advises someone not to seek medical care?
• Is it then the AI developer? [141]
• Or the healthcare provider using it? [144]
• Or the regulators that gave it the thumbs up?
Without clear laws on liability, the deployment of LLMs may be stalled since hospitals and doctors fear being dragged to court. The present regulations do not unequivocally distinguish AI-caused medical errors, and this creates ambiguity regarding insurances, malpractice claims, and patient safety protocols
The development of trustworthy LLMs for medical applications demands substantial computational, data, and human resources. The two papers—CARES: A Comprehensive Benchmark of Trustworthiness in Medical Vision Language Models and CoD: Towards an Interpretable Medical Agent Using Chain of Diagnosis—highlight the resource-guzzling endeavor in building LLMs usable, fair, and safe for real-world medical applications [150].
3.4.1 Computational Infrastructure
It can also be computationally intensive to adopt the methods to improve and fine-tune the Medicine Foundation Medicine LLMs, considering the mix of text, image, and report medical data.
• Training Models: CARES assesses Med-LVLMs on a scale of various medical imaging modalities across 27 anatomical regions for training on baseline medical datasets. Models of this nature were able to be trained only with additional resources such as high-memory-capacity GPUs/TPUs, for example, NVIDIA A100 or H100 GPUs.
• Fine-Tuning: CoD trains DiagnosisGPT on 48,020 patient cases and fine-tunes it on Yi-34B-Base, a big model based on transformers. The fine-tuning on synthetic patient cases reduces privacy risks while still requiring substantial computational resources.
• Inference Costs: Deploying real-time medical diagnosis systems on edge devices (i.e., on hospital servers) continues to be challenging because of the high memory and processing demands of LLMs for use in clinical settings [13,38].
Without high-performance computing resources, it is difficult to train or deploy trustworthy medical AI models at scale.
3.4.2 Data Collection & Annotation Efforts
Building trustworthy LLMs requires high-quality medical data, which is expensive and time-consuming to collect.
• Diverse Medical Datasets: CARES integrates many datasets provided from different sources such as MIMIC-CXR, IU-Xray, Harvard-FairVLMed, PMC-OA, and HAM10000 to enable complete evaluation [115].
• Synthetic Data Generation: CoD builds a training set by generating synthetic patient cases using disease encyclopedias. The anonymity of the dataset is improved, but careful validation is required to ensure the synthetic data reflect reality.
• Manual Annotation: Medical data need to be annotated by medical professionals, thus rendering its creation expensive. When manually verifying model outputs, CARES relies on expert consultation to check for correctness, equity, and robustness [38].
The key ingredient to training medical LLMs worth an interest is, however, large, diverse, and well-annotated patient datasets.
3.4.3 Impact of Data Pre-Processing and Splitting on Reproducibility
Dataset preprocessing and splitting strategies have often lacked standardization in the literature, undermining the reproducibility of Med-LLM evaluations. In biomedical contexts, data complexity—marked by heterogeneity, missingness, class imbalance, and multimodality—complicates preprocessing pipelines and can introduce variability between runs when workflows are not rigorously documented (e.g., unit harmonization, imputation, normalization) [151]. Moreover, inadvertent data leakage—such as applying feature scaling or imputation across the entire dataset before train-test splitting—can lead to over-optimistic performance estimates and diminish generalizability [152,153]. In digital pathology, unintentional leakage caused by including tiles from the same patient in both training and validation sets has been demonstrated to inflate performance by up to 41% [154]. To mitigate such issues, standardized pipelines—like MIMIC-Extract, which applies pre-processing after splitting and preserves time-series structures in EHR data—have been proposed to bolster transparency and reproducibility [155]. In practice, best practices include locking validation sets until model configuration is completed, using stratified or grouped splits to prevent patient overlap, and thoroughly documenting all pre-processing steps to enable fair and reproducible comparisons across studies.
3.4.4 Human Expertise & Multidisciplinary Collaboration
Requisite to ensure trustworthiness of medical LLMs are collaboration between AI researchers, medical professionals, and regulatory experts [70].
• Medical Experts: Doctors, radiologists, and pathologists are required to verify the AI-given diagnosis, provide fairness, and flag potential biases present in marked training data [150].
• Ethics & Compliance Teams: There are privacy concerns as raised by CARES; hence, union between hospitals and AI developers with legal and regulatory professionals is required for compliance as per laws like HIPAA, GDPR, and other healthcare data protection laws [60].
• AI Engineers & Model Developers: Developing advanced LLMs with uncertainty estimation (CoD) and bias mitigation techniques (CARES) requires the involvement of domain-specific AI researchers [43].
Failure to have any of the required domain expertise could jeopardize the safety and reliability of LLM-based medical tools [38].
3.4.5 Deployment and Maintenance Costs
Requisite to ensure trustworthiness of medical LLMs are collaboration between AI researchers, medical professionals, and regulatory experts [43,139].
• Medical Experts: Doctors, radiologists, and pathologists are required to verify the AI-given diagnosis, provide fairness, and flag potential biases present in marked training data [38,147].
• Ethics and Compliance Teams: There are privacy concerns as raised by CARES; hence, union between hospitals and AI developers with legal and regulatory professionals is required for compliance as per laws like HIPAA, GDPR, and other healthcare data protection laws [54,63].
• AI Engineers and Model Developers: Developing advanced LLMs with uncertainty estimation (CoD) and bias mitigation techniques (CARES) requires the involvement of domain-specific AI researchers [60,150].
Failure to have any of the required domain expertise could jeopardize the safety and reliability of LLM-based medical tools [66].
The important evaluation of the Medical Large Language Models (Med-LLMs) will help understand their efficacy in Named Entity Recognition (NER), Relation Extraction (RE), Question Answering (QA), Sentiment Analysis, Text Classification, and Text Generation. Different countries have different benchmarking practices as stated in Table 9. These experiments often employ different metrics to assess performance, including but not limited to the F1 Score, Accuracy, Exact Match (EM), Mean Reciprocal Rank (MRR), ROC-AUC, and BLEU scores [147]. These evaluation systems demand high LLM interaction rates to produce diagnostic dialogues while experts have to rate their quality at a high price.
While quantitative metrics like accuracy and F1-score provide essential benchmarks for Medical LLM performance, they represent only the foundational layer of evaluation, failing to address the complex clinical realities these models must navigate. Qualitative reporting frameworks—including TRIPOD-LLM, MEDIC, and MedHELM—serve as complementary evaluation approaches that illuminate the clinical reasoning pathways, contextual appropriateness, and ethical implications that quantitative measures cannot capture. These structured frameworks enable systematic assessment of model behavior in nuanced clinical scenarios, revealing how LLMs handle ambiguous cases, manage uncertainty, and maintain consistency across diverse patient populations. Rather than replacing quantitative metrics, qualitative reporting creates a comprehensive evaluation ecosystem that examines not just what the model outputs, but how it processes clinical information, when it recognizes its limitations, and why its reasoning aligns with clinical best practices. This dual approach—combining numerical performance indicators with structured qualitative analysis—provides the rigorous, multi-dimensional assessment necessary for establishing clinical trust and ensuring safe deployment of Medical LLMs in healthcare environments where both precision and interpretability are paramount.
4.1 Quantitative Assessment Reporting Frameworks
• MedBench: MedBench provides a complete medical evaluation platform that contains 40,000+ test questions in multiple medical specialties [70]. The main components of MedBench involve: Medical students take four key evaluations to prepare them for clinical practice through official licensing tests and real patient case studies. Extensive experiments provided the following findings: The results show that HuaTuoGPT fails on MedBench tasks because clinical knowledge and accurate medical judgment matter. Meanwhile, GPT-4 shows unexpected clinical knowledge while performing these tasks [63].
• AutoEval: AutoEval helps test LLMs at diagnosing medical cases automatically based on actual doctor-patient conversations. AutoEval transforms USMLE exams into multiple-choice medical questions as a first step in testing LLM doctor functions [42]. Four metrics are used to evaluate Doctor LLMs in multi-turn consultation scenarios: Medical Information Coverage Rate, Accuracy of the Final Task, Average Turn, and Average Length measurements [65].
• LLM-Mini-CEX: MiniCEX leads us to build LLM-specific Mini-CEX as a strong evaluation system that judges LLMs’s medical skills [58].
• MedGPTEval: MedGPTEval offers medical datasets in Chinese and public comparison standards [63]. The evaluation criteria derive from medical professional and social research skills combined with context awareness and computing strength. They contain detailed measurements across 16 expertise aspects [66].
• LLM-Human Evaluation: The system proves to handle new issues surpassing its training data, although it trains only on certain task instructions. The study tests whether using this feature offers a better solution than using people to review medical content [64].
• RJUA-SPs: An RJUA-SPs evaluation plan has three core parts:
– Metric: Using professional healthcare practice standards develops practice-specific abilities a doctor needs, called LLM-specific clinical capabilities, within a clinical pathway (LCP) [22].
– Data: The use of Standardized Patients (SPs) as workspace teachers for improved data collection practices [25].
– Algorithm: The model creates different software agents to simulate doctor-patient interactions. The retrieval-augmented algorithm tests if an LLM doctor follows established LCP standards in their work [87].
4.1.1 Open Medical-LLM Leaderboard
The Open Medical LLM Leaderboard [159] is a public platform dedicated to tracking, evaluating, and ranking large language models (LLMs) on a wide range of medical question-answering tasks. Its goal is to provide a transparent and consistent benchmark to assess how well these models perform in medical contexts—an area that demands high accuracy, domain expertise, and reliable reasoning. The leaderboard is powered by the Eleuther AI Language Model Evaluation Harness, ensuring rigorous, automated, and standardized testing across all submitted models.
• What It Evaluates The leaderboard assesses model performance across several critical medical datasets (shown in Table 2), including:
– MedQA (USMLE)—medical licensing exam questions
– PubMedQA—biomedical research Q&A
– MedMCQA—clinical medicine MCQs
– MMLU-Med Subsets—including topics like anatomy, genetics, and clinical knowledge
Together, these datasets test a model’s breadth of medical knowledge and its ability to reason through complex, domain-specific problems.
• Evaluation Metric The primary evaluation metric is Accuracy (ACC), and most results are reported in the zero-shot setting (no example given before answering), with the exception of Med-PaLM-2, which uses 5-shot accuracy from its original paper.
Models listed on the leaderboard [159] are for research and comparison only. They are not approved for clinical use, deployment, or decision-making in medical settings. For full advisory guidance, users should consult the Advisory Notice. While most results are evaluated in-house, benchmark scores for models like GPT-4, Med-PaLM-2, and Gemini are taken directly from peer-reviewed papers and conferences (e.g., NAACL 2024 for Gemini). Scores of the Models evaluated on benchmark datasets are presented as a comparative chart in Fig. 10.
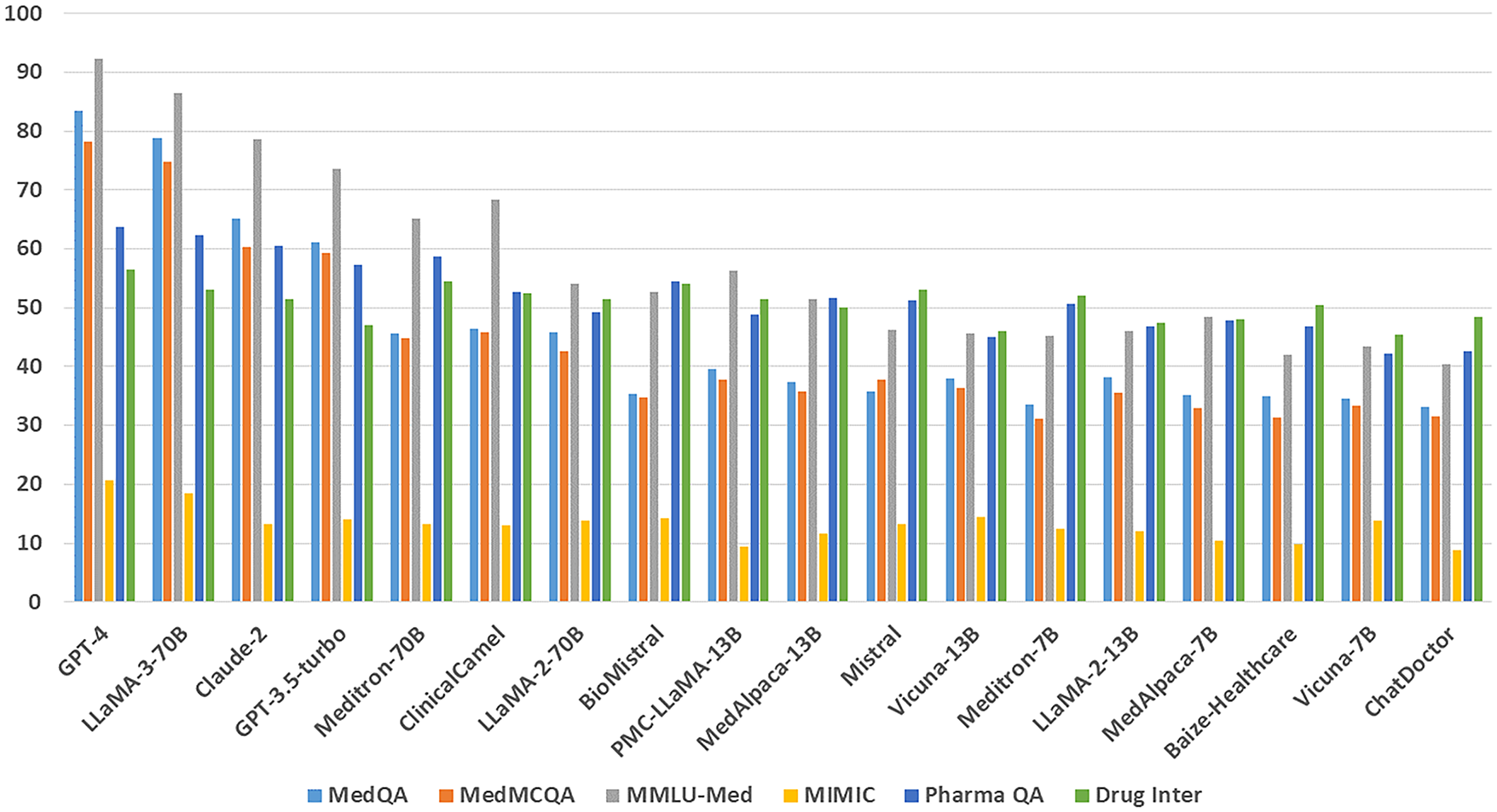
Figure 10: Performance comparison of Med-LLMs over benchmark datasets according to Open Med-LLM Leaderboard by Huggingface
PubMedQA (from the original paper [73]) is a benchmark dataset designed to evaluate the reasoning capabilities of medical LLMs on biomedical research questions. It consists of yes/no/maybe questions derived from PubMed abstracts, requiring models to understand and reason over complex scientific content—especially quantitative findings.
Each instance includes a question (often the article title), context (abstract excluding the conclusion), a long answer (the conclusion), and a summarized yes/no/maybe response. The benchmark challenges LLMs to go beyond superficial text matching and engage in nuanced biomedical reasoning.
The chart (Fig. 11) reveals a significant performance gap between general-purpose LLMs with medical prompting (like GPT-4 MediPrompt and Med-PaLM 2) and traditional biomedical models (like PubMedBERT and BioBERT) on the PubMedQA dataset. Despite being trained on biomedical corpora, older domain-specific models struggle with complex research question answering, indicating limitations in reasoning and contextual understanding. In contrast, state-of-the-art LLMs, especially those fine-tuned or prompted for medical tasks, not only match but surpass human performance. This underscores a shift where model architecture, scale, and prompt strategy outweigh mere domain pretraining, signaling the need to evolve biomedical LLMs beyond vocabulary familiarity toward deeper comprehension.
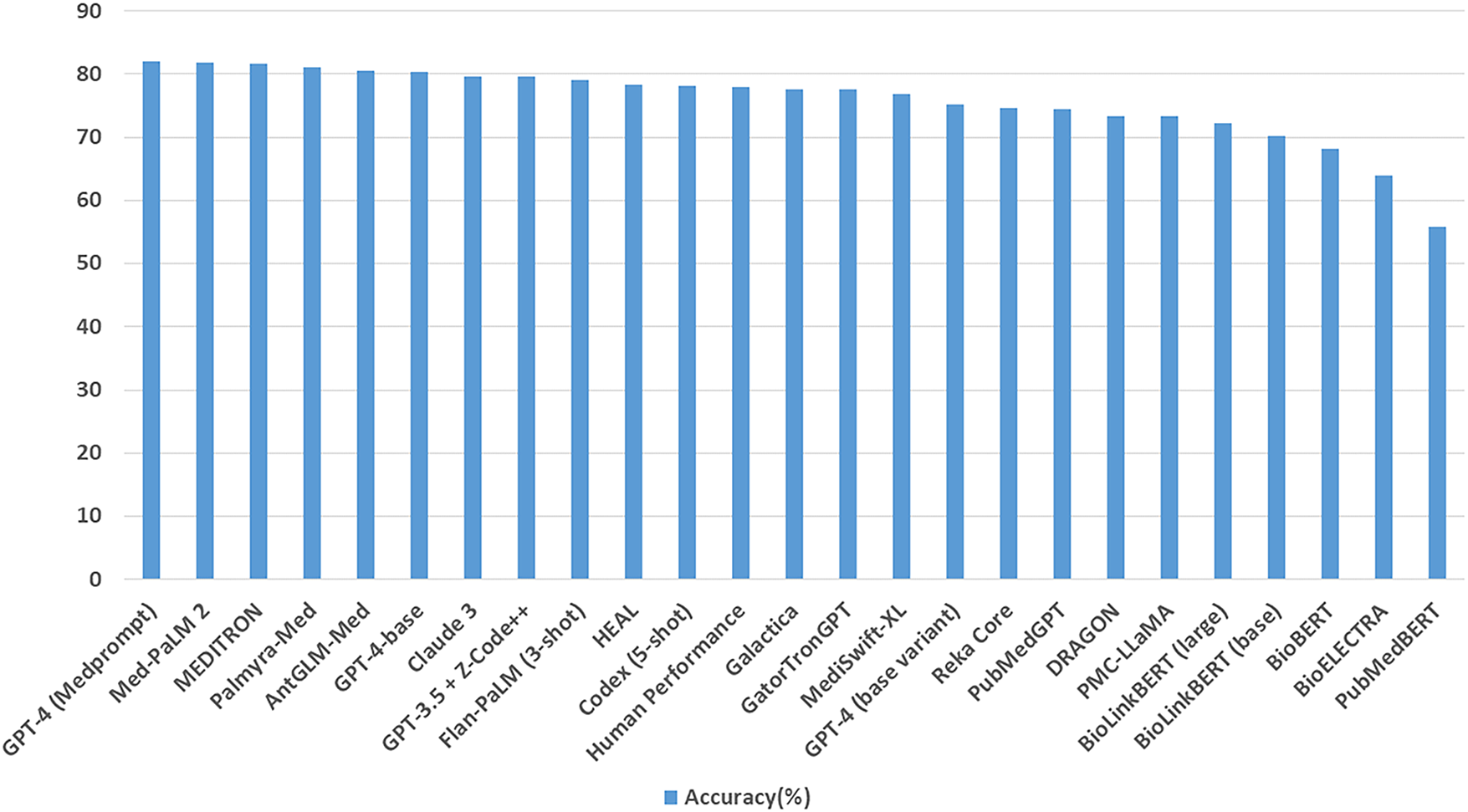
Figure 11: Performance comparison of Med-LLMs on PubMedQA dataset
These benchmarks indicate progress in Med-LLMs and their various applications in clinical, research, and patient-interaction areas. Research in medical AI will focus on further progress concerning explainability, reliability, and ethical characteristics [32,54,90].
4.2 Qualitative Assessment Reporting Framework
• TRIPOD-LLM: Tripod LLM represents a novel benchmarking framework that emerged in 2025 [160], developed by Gallifant et al. to evaluate large language models within medical applications. The system utilizes a comprehensive three-dimensional assessment approach—Knowledge, Reasoning, and Communication—designed to capture essential capabilities needed for reliable medical AI deployment (workflow shown in Fig. 12). The framework draws from an extensive collection of medical datasets spanning various specialties including medical question-answering (such as MedQA, PubMedQA), clinical report generation (including MIMIC-CXR), patient communication, and diagnostic imaging analysis. Performance measurement relies on standardized, domain-appropriate metrics (including accuracy for assessment tasks, ROUGE/BLEU for text generation), facilitating robust cross-model evaluation. Although Tripod LLM has gained significant traction within academic research communities and institutions (including Stanford CRFM, Tsinghua MedAI), it has not received official endorsement from major healthcare authorities or regulatory agencies like the FDA or MHRA. Nevertheless, its systematic, transparent methodology is generating considerable momentum among AI practitioners and emerging companies, positioning it as a strong contender for eventual standardization in medical LLM assessment protocols.
• QUEST [161]: QUality Evaluation Scoring Tool operates by evaluating LLM outputs across six domains: authorship, attribution, conflict of interest, currency, complementarity, and tone. Reviewers manually or semi-automatically score outputs based on predefined criteria, determining whether responses are transparent, balanced, up-to-date, and ethically presented. This helps identify hallucinations, misinformation, and non-evidence-based recommendations, ensuring content aligns with patient-centered and trustworthy communication standards.
• S.C.O.R.E: Scoring Clinical Output with Reasoning Evaluation works by assessing the reasoning structure behind an LLM’s clinical response [162]. Evaluators score outputs along five axes: Soundness (clinical accuracy), Completeness (coverage of relevant details), Originality (non-template thinking), Rationale (clarity of reasoning), and Explainability (ease of understanding for human users). This rubric is particularly effective in tasks like diagnosis justification, case triage, and treatment planning, where the path to the answer is as vital as the answer itself.
• MEDIC [163]: MEDIC is a multi-dimensional evaluation framework that assesses medical large language models across five core aspects: medical reasoning, ethical alignment and bias, language and data comprehension, in-context learning, and clinical safety. Instead of relying on reference answers, MEDIC uses a set of high-quality prompts designed to test a model’s ability to reason, maintain factual consistency, and adhere to medical ethics. It leverages cross-examination techniques and prompt variation to simulate clinical complexity and measure hallucination frequency. The system helps identify weaknesses in real-world generalization without requiring ground-truth datasets or human labeling, making it scalable and robust for longitudinal tracking of model improvements.
• MedHELM [164]: MedHELM adapts Stanford’s HELM (Holistic Evaluation of Language Models) for healthcare by incorporating 98 distinct clinical tasks across categories such as decision support, medical report generation, and patient interaction. It involves task definitions validated by 29 licensed clinicians and provides cost-effectiveness analysis for each model’s output. MedHELM enables researchers to evaluate models’ behavior not only based on accuracy but also on their ability to balance clinical reasoning with safety, fairness, and efficiency. The framework emphasizes transparency and replicability by publishing open-access task definitions, inputs, and expected outcomes.
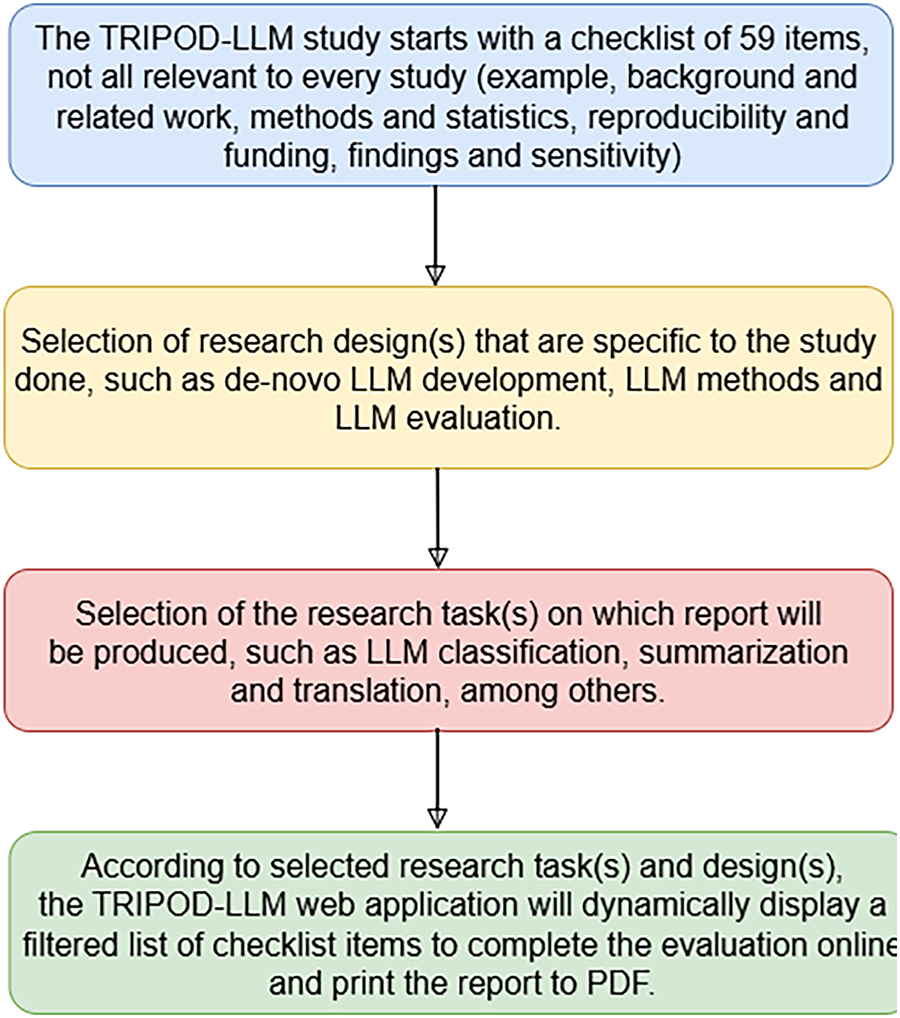
Figure 12: TRIPOD-LLM: A benchmarking framework for LLMs
4.3 Both Quantitative and Qualitative Reporting Framework
• LLMEval-Med: LLMEval-Med introduces a physician-supervised benchmark that evaluates LLMs using 2996 real-world clinical cases [165]. These include tasks from EHR notes, consultation histories, and diagnostic reports. Its evaluation system combines automatic LLM-based scoring with a human-machine agreement loop, ensuring reliable performance validation. Unlike static MCQ formats, LLMEval-Med captures model abilities in nuanced clinical reasoning and contextual understanding, aiming to reflect the complexity of healthcare decision-making. The benchmark encourages development of medical AI systems that generalize well across real patient data and diverse medical environments.
• MedQA-CS: MedQA-CS is inspired by the Objective Structured Clinical Examination (OSCE) format and tests LLMs in dual roles—as both medical student and examiner [166]. This framework emphasizes the development of clinical soft skills, reasoning, and real-time patient interaction capabilities. Scenarios include verbal diagnosis, treatment suggestion, and peer explanation tasks. MedQA-CS evaluates model output using rubrics similar to those used in real medical board exams. It provides a more interactive and structured assessment compared to traditional multiple-choice question formats, aligning with the evolving expectations of AI-assisted clinical practice.
• MedAgentBench: MedAgentBench [167] is a comprehensive benchmarking suite developed to evaluate the agentic capabilities of large language models (LLMs) within clinical environments. Unlike traditional chatbot-style assessments, MedAgentBench focuses on complex, real-world medical tasks by simulating an interactive FHIR-compliant EMR environment. It includes 300 physician-authored tasks across 10 clinical categories and 100 patient profiles with over 700,000 data points, reflecting realistic healthcare scenarios. The benchmark reveals that even state-of-the-art models like Claude 3.5 Sonnet v2 achieve only 69.67% success, highlighting both existing strengths and the considerable room for improvement. By providing standardized tasks, APIs, and reproducible code, MedAgentBench fills a crucial gap in medical AI evaluation and paves the way for more robust, clinically useful LLM agents.
Unlike traditional MCQ-style QA datasets, emerging benchmarks such as LLMEval-Med [165] and MedAgentBench [167] are beginning to evaluate Medical LLMs on realistic clinical data from EHR systems, reflecting ongoing research efforts toward real-world applicability. Since there is no universally accepted composite evaluation formulation for Med-LLMs currently exists, a weighted scheme inspired by multi-criteria decision analysis [168] and NLP evaluation practices [81,82] has been adopted in existing platforms. The overall evaluation score has therefore been formalized as:
where
represents normalized scores from automated metrics such as BLEU, ROUGE, and accuracy, and
represents averaged expert ratings of qualitative dimensions such as robustness, hallucination risk, and factuality.
The parameters
4.4 Performance Evaluation of Key Med-LLMs
Several state-of-the-art Med-LLMs have been benchmarked across multiple datasets, yielding significant insights into their capabilities:
• BioBERT (2019) stands out in biomedical Named Entity Recognition (NER), attaining an F1 score of 85.6 on the BC5CDR dataset. It also exhibits a notable F1 score of 71.3 in Relation Extraction (RE) within the ChemProt dataset [32].
• ClinicalBERT (2020), which is trained using Electronic Health Records (EHR), achieves a ROC-AUC of 0.76 when predicting hospital readmissions and reaches 0.78 for mortality prediction [113].
• PubMedBERT (2021) surpasses earlier biomedical models by acquiring an impressive F1 score of 89.7 in NER, along with a score of 75.1 in RE, showcasing its effectiveness for extracting information from scientific literature [18].
• Med-PaLM (2022) demonstrates a solid performance with an accuracy rate of 81.2% in medical question answering and registers a pass rate of 67% on the United States Medical Licensing Examination (USMLE), highlighting its proficiency in medical reasoning [92].
• GPT-4-Med (2023) takes precedence over earlier models, reflecting an exemplary pass rate on the USMLE at 84.3% and achieving an accuracy level of 87.1% in medical QA, underscoring its advantages in long-form medical reasoning tasks.
• GatorTron (2022) attains an F1 score of 91.2 in Clinical NER, establishing itself as one of the top performers for processing structured clinical texts effectively [90,115].
• PMC-LLaMA (2023) displays commendable abilities in understanding biomedical literature, obtaining a score of 83.6% on PubMedQA assessments.
• Med-PaLM 2 (2023) surpasses its predecessor significantly with a remarkable USMLE pass rate reaching 92.1%. Additionally, it shows an accuracy rating of 88.7% in HealthSearchQA evaluations [54].
• ChatDoctor (2023) has been tailored specifically for patient interactions; it achieves an accuracy rate of 79% in medical dialogue alongside maintaining coherence rated at 88.3% during human evaluations [40].
• GatorTronGPT reflects progress made across several parameters, particularly exhibiting enhancements in medical QA with an accuracy of 88.4% and drug interaction identification with an F1 score of 76.3 [90,147,170].
• HuatuoGPT-II does well in Chinese medical QA, scoring 85.4% in MedQA and 83.9% in CMMLU (Chinese Medical Multi-task Learning Understanding) [106].
5 Discussion and Future Directions
The integration of Medical Large Language Models (Med-LLMs) into healthcare has already shown great potential, but many key aspects require more research and development to make them more efficient, trustworthy, and widely deployable [171].
5.1 Enhancing Explainability and Trustworthiness
One of the foremost challenges posed by Med-LLMs is their lack of interpretability, a problem that hinders clinical decision-making and suspends acceptance from patients. Future developments seem to arise along lines of enhancing interpretability in interpretive reasoning packed into approaches such as argumentation-based models enabling self-reflection upon AI-generated clinical suggestions. Other frameworks, such as ICE-T, the “Interpretable Cross-Examination Technique,” operate in ways that offer prospects of multi-prompt strategies to enhance model classification and decision-making.
5.2 Mitigating Bias and Ensuring Fairness
Medical AI systems must be built with the concept of fairness to avoid any demographic biases that might create healthcare disparities. Recent studies show that Med-LLMs have been exhibiting biases concerning age, gender, and ethnicity that affect the degree of diagnostic accuracy [150]. Future research should focus on fairness-aware training techniques, domain adaptation methods, and continuous auditing utilizing datasets that are well-represented and diverse [32,54,115].
5.3 Secure and Ethical AI Deployment
Ensuring the safety and accountability of Med-LLMs remains a significant challenge. Future work should involve the development of robust monitoring and validation frameworks (example: Table 8), such as the CARES benchmark, which evaluates AI models for safety, bias, and privacy risks. Additionally, ethical considerations surrounding AI-driven medical recommendations necessitate clearer legal frameworks that outline liability, patient consent protocols, and compliance with healthcare data protection laws like HIPAA and GDPR.
5.4 Regulatory Fragmentation Across Regions
Deployment of Medical LLMs faces a patchwork of regulatory frameworks that vary in scope, enforceability, and focus. For example, as discussed in [146]:
• The EU enforces the GDPR and the EU AI Act, classifying AI in healthcare as high-risk and mandating rigorous governance.
• In Japan, AI governance remains largely voluntary, with no binding legislation on medical AI yet in place.
• In China, regulatory specificity is increasing through new guidelines and proposed laws targeting AI-enabled medical software, but consistency remains limited.
5.5 Integration with Clinical Workflows
In order for medical language models (Med-LLMs) to be successfully introduced and adopted within real-world healthcare environments, they must be easily integrated into the existing workflow of clinical practice [49,147,172]. It requires collaboration among AI developers, medical practitioners, and hospital administrators to align AI capabilities with the operational needs of these stakeholders [58,137,139]. Some issues that must be addressed to enhance efficiency by lowering the cognitive load on healthcare professionals include the automation of clinical note generation, real-time decision support, and interoperability with electronic health records.
5.6 Benchmark Misalignment with Real-World Settings
Existing evaluation datasets tend to focus on structured, exam-style tasks rather than chaotic, real-world clinical environments—such as unstructured EHRs or patient–doctor interactions. A systematic review of clinical NLP benchmarks found that most benchmarks inadequately cover the workflow tasks clinicians prioritize, particularly those related to clinical documentation and routine care tasks [173].
5.7 Addressing Computational and Resource Constraints
Developing Med-LLMs in resource-limited terrains continues to pose challenges, especially owing to the enormous computational complexity involved in training and inference [43]. Future research must devote an effort toward exploring better model architectures, low-resource fine-tuning techniques, and edge AI for these models in order to make them more accessible in marginalized regions [174]. Furthermore, methods for generating synthetic data could be further developed to generate diverse, privacy-preserving datasets for training the models without compromising data quality. Tackling such challenges will allow Med-LLMs to transition from research experimentation to being trustworthy and scalable solutions for enhanced medical practice, with improved patient outcomes and equitable access to AI-infused healthcare [175,176].
A pronounced lack of publicly available clinical datasets from regions such as the Middle East and India hinders the fair testing of Medical LLMs in locally relevant scenarios. This imbalance limits both model validation and tailoring for regional healthcare contexts, potentially exacerbating disparities in AI effectiveness. For instance, AI-driven eye disease diagnostics have been shown to underperformed in underrepresented populations from South Asia and Africa due to dataset biases toward North America, Europe, and China [published in WIRED, American Magazine]. Some strategies are adopted to address the gaps—(i) developing localized datasets through regional collaborations with hospitals and research institutes, (ii) applying federated learning frameworks that enable model training on sensitive patient data without centralizing it, thereby respecting privacy while incorporating underrepresented populations [6], (iii) leveraging cross-lingual transfer learning and domain adaptation to fine-tune existing Med-LLMs for local languages and practices, and (iv) incentivizing open data initiatives under appropriate governance models to improve transparency and inclusiveness.
5.9 Language and Cultural Nuances
While some regions like the EU and Korea have developed localized language datasets, many global AI models still rely primarily on English-adapted corpora as stated in Table 9. This reliance fails to capture region-specific medical practices (e.g., Ayurveda in India, Traditional Chinese Medicine), leading to culturally contextually misaligned outputs and reduced clinical accuracy.
Medical Large Language Models (LLMs) have the potential to transform healthcare by combining advanced natural language processing with domain-specific medical knowledge [137]. They can improve diagnostics, automate clinical documentation, support decision-making, and facilitate patient communication [96]. The contribution of this work lies in examining how multimodal architectures—integrating text and imaging data—can enhance accuracy and usability in real-world applications [49,172]. Despite these opportunities, medical LLMs face challenges including hallucinations, bias, interpretability, and data privacy [150,177]. Future research should prioritize integrating knowledge graphs, retrieval-augmented generation (RAG), and explainable AI (XAI) [116,178]. Furthermore, developing unified pipelines that combine domain-specific fine-tuning, multimodal processing, and human-in-the-loop validation can improve both reliability and trustworthiness [58,141].
Potential areas for future exploration include:
1. Personalized Medicine—Using LLMs to tailor treatment recommendations based on patient history and genetics.
2. Telemedicine & Virtual Assistants—Improving AI-driven chatbots for patient consultation and remote healthcare.
3. Clinical Trial Optimization—Enhancing drug discovery through AI-assisted literature mining and predictive modelling.
Furthermore, models such as GPT-4Med, Med-PaLM, MEDITRON, PubMedGPT, and MedAlpaca have demonstrated strong performance; however, challenges remain in ensuring reliable decision-making. Future research should prioritize reducing bias, enhancing adaptability across diverse populations, and ensuring regulatory compliance. Integrating the strengths of multiple AI methodologies has the potential to yield a more robust and efficient healthcare system that supports both practitioners and patients.
Acknowledgement: The authors would like to thank all contributors and institutions that supported this research. This article was generously supported by Saptarshi Banerjee and Jon Turdiev.
Funding Statement: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Himadri Nath Saha and Satyasaran Changdar; Literature review, Survey, Evaluation mechanisms, Dipanwita Chakraborty Bhattacharya, Sancharita Dutta, Arnab Bera, Srutorshi Basuray; Validation, Himadri Nath Saha, Satyasaran Changdar, Saptarshi Banerjee, Jon Turdiev; Formal analysis, Dipanwita Chakraborty Bhattacharya; Data curation, Dipanwita Chakraborty Bhattacharya and Arnab Bera; writing—original draft preparation, Dipanwita Chakraborty Bhattacharya and Sancharita Dutta; writing—review and editing, Himadri Nath Saha; Visualization, Dipanwita Chakraborty Bhattacharya and Arnab Bera; supervision, Himadri Nath Saha and Satyasaran Changdar. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: This paper is predominantly a review that synthesizes existing methods and literature findings. This investigation utilized only data obtained from publicly accessible sources. These datasets are accessible via the sources listed in the References section of this paper. As the data originates from publicly accessible repositories, its accessibility is unrestricted.
Ethics Approval: This study did not involve human or animal participants and therefore did not require ethical approval.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Zhang H, Shao H. Exploring the latest applications of OpenAI and ChatGPT: an in-depth survey. Comput Model Eng Sci. 2024;138(3):2061–102. doi:10.32604/cmes.2023.030649. [Google Scholar] [CrossRef]
2. Nabavirazavi S, Taheri R, Shojafar M, Iyengar SS. Impact of aggregation function randomization against model poisoning in federated learning. In: 2023 IEEE 22nd International Conference on Trust, Security and Privacy in Computing and Communications (TrustCom); 2023 Nov 1–3; Exeter, UK; 2024. p. 165–72. doi:10.1109/TrustCom60117.2023.00043. [Google Scholar] [CrossRef]
3. Upreti D, Yang E, Kim H, Seo C. A comprehensive survey on federated learning in the healthcare area: concept and applications. Comput Model Eng Sci. 2024;140(3):2239–74. doi:10.32604/cmes.2024.048932. [Google Scholar] [CrossRef]
4. Li M, Xu P, Hu J, Tang Z, Yang G. From challenges and pitfalls to recommendations and opportunities: implementing federated learning in healthcare. arXiv:2409.09727. 2024. [Google Scholar]
5. Nabavirazavi S, Taheri R, Iyengar SS. Enhancing federated learning robustness through randomization and mixture. Future Gener Comput Syst. 2024;158(4):28–43. doi:10.1016/j.future.2024.04.009. [Google Scholar] [CrossRef]
6. Polap D, Srivastava G, Yu K. Agent architecture of an intelligent medical system based on federated learning and blockchain technology. J Inf Secur Appl. 2021;58(11):102748. doi:10.1016/j.jisa.2021.102748. [Google Scholar] [CrossRef]
7. Bharati S, Mondal MRH, Podder P. A review on explainable artificial intelligence for healthcare: why, how, and when? arXiv:2304.04780. 2023. [Google Scholar]
8. Nasarian E, Alizadehsani R, Acharya UR, Tsui KL. Designing interpretable ML system to enhance trust in healthcare: a systematic review to proposed responsible clinician-AI-collaboration framework. arXiv:2311.11055. 2023. [Google Scholar]
9. Houssein EH, Gamal AM, Younis EMG, Mohamed E. Explainable artificial intelligence for medical imaging systems using deep learning: a comprehensive review. Clust Comput. 2025;28(7):469. doi:10.1007/s10586-025-05281-5. [Google Scholar] [CrossRef]
10. Neupane S, Mittal S, Rahimi S. Towards a HIPAA compliant agentic AI system in healthcare. arXiv:2504.17669. 2025. [Google Scholar]
11. Hinostroza Fuentes VG, Karim HA, Tan MJT, AlDahoul N. AI with agency: a vision for adaptive, efficient, and ethical healthcare. Front Digit Health. 2025;7:1600216. doi:10.3389/fdgth.2025.1600216. [Google Scholar] [PubMed] [CrossRef]
12. Awasthi R, Ramachandran SP, Mishra S, Mahapatra D, Arshad H, Atreja A, et al. Artificial intelligence in healthcare: 2024 year in review. In: MedRxiv. Woodbury, NY, USA: Cold Spring Harbor Laboratory Press; 2025. doi:10.1101/2025.02.26.25322978. [Google Scholar] [CrossRef]
13. Moor M, Huang Q, Wu S, Yasunaga M, Dalmia Y, Leskovec J, et al. Med-flamingo: a multimodal medical few-shot learner. In: Machine learning for health (ML4H). Westminster, UK: PMLR; 2023. p. 353–67. [Google Scholar]
14. Yuan D, Rastogi E, Naik G, Rajagopal SP, Goyal S, Zhao F, et al. A continued pretrained LLM approach for automatic medical note generation. arXiv:2403.09057. 2024. [Google Scholar]
15. Wang J, Yang Z, Yao Z, Yu H. JMLR: joint medical LLM and retrieval training for enhancing reasoning and professional question answering capability. arXiv:2402.17887. 2024. [Google Scholar]
16. Tu T, Azizi S, Driess D, Schaekermann M, Amin M, Chang P, et al. Towards generalist biomedical AI. arXiv:2307.14334. 2023. [Google Scholar]
17. Pieri S, Mullappilly SS, Khan FS, Anwer RM, Khan S, Baldwin T, et al. BiMediX: bilingual medical mixture of experts LLM. In: Al-Onaizan Y, Bansal M, Chen Y-N, editors. Findings of the association for computational linguistics: EMNLP 2024. Stroudsburg, PA, USA: Association for Computational Linguistics; 2024. p. 16984–7002. [Google Scholar]
18. Wang S, Bian J, Huang X, Zhou H, Zhu S. PubLabeler: enhancing automatic classification of publications in UniProtKB using protein textual description and PubMedBERT. IEEE J Biomed Health Inform. 2025;29(5):3782–91. doi:10.1109/JBHI.2024.3520579. [Google Scholar] [PubMed] [CrossRef]
19. Hong S, Xiao L, Zhang X, Chen J. ArgMed-agents: explainable clinical decision reasoning with LLM disscusion via argumentation schemes. arXiv:2403.06294. 2024. [Google Scholar]
20. Ye H, Liu T, Zhang A, Hua W, Jia W. Cognitive mirage: a review of hallucinations in large language models. arXiv:2309.06794. 2023. [Google Scholar]
21. Li H, Wei W, Xu H. Drug discovery is an eternal challenge for the biomedical sciences. Acta Mater Med. 2022;1(1):1–3. doi:10.15212/amm-2022-1001. [Google Scholar] [CrossRef]
22. Deng J, Zubair A, Park YJ. Limitations of large language models in medical applications. Postgrad Med J. 2023;99(1178):1298–9. doi:10.1093/postmj/qgad069. [Google Scholar] [PubMed] [CrossRef]
23. Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Bender A, et al. A review of biomedical datasets relating to drug discovery: a knowledge graph perspective. Brief Bioinform. 2022;23(6):bbac404. doi:10.1093/bib/bbac404.doi:. [Google Scholar] [CrossRef]
24. Ullah E, Parwani A, Baig MM, Singh R. Challenges and barriers of using large language models (LLM) such as ChatGPT for diagnostic medicine with a focus on digital pathology—a recent scoping review. Diagn Pathol. 2024;19(1):43. doi:10.1186/s13000-024-01464-7. [Google Scholar] [PubMed] [CrossRef]
25. Tian S, Jin Q, Yeganova L, Lai PT, Zhu Q, Chen X, et al. Opportunities and challenges for ChatGPT and large language models in biomedicine and health. Brief Bioinform. 2023;25(1):bbad493. doi:10.1093/bib/bbad493. [Google Scholar] [PubMed] [CrossRef]
26. Sohn J, Park Y, Yoon C, Park S, Hwang H, Sung M, et al. Rationale-guided retrieval augmented generation for medical question answering. arXiv:2411.00300. 2024. [Google Scholar]
27. Shool S, Adimi S, Saboori Amleshi R, Bitaraf E, Golpira R, Tara M. A systematic review of large language model (LLM) evaluations in clinical medicine. BMC Med Inform Decis Mak. 2025;25(1):117. doi:10.1186/s12911-025-02954-4. [Google Scholar] [PubMed] [CrossRef]
28. Lin C, Kuo CF. Roles and potential of large language models in healthcare: a comprehensive review. Biomed J. 2025;48(5):100868. doi:10.1016/j.bj.2025.100868. [Google Scholar] [PubMed] [CrossRef]
29. Bedi S, Liu Y, Orr-Ewing L, Dash D, Koyejo S, Callahan A, et al. Testing and evaluation of health care applications of large language models: a systematic review. JAMA. 2025;333(4):319. doi:10.1001/jama.2024.21700. [Google Scholar] [PubMed] [CrossRef]
30. Kamath U, Keenan K, Somers G, Sorenson S. LLMs: evolution and new frontiers. In: Large language models: a deep dive. Cham, Switzerland: Springer Nature Switzerland; 2024. p. 423–38. doi:10.1007/978-3-031-65647-7_10. [Google Scholar] [CrossRef]
31. Zheng Y, Gan W, Chen Z, Qi Z, Liang Q, Yu PS. Large language models for medicine: a survey. Int J Mach Learn Cybern. 2025;16(2):1015–40. doi:10.1007/s13042-024-02318-w. [Google Scholar] [CrossRef]
32. McInnes BT, Tang J, Mahendran D, Nguyen MH. BioBERT-based deep learning and merged chemprot-drugprot for enhanced biomedical relation extraction. arXiv:2405.18605. 2024. [Google Scholar]
33. Wu C, Lin W, Zhang X, Zhang Y, Xie W, Wang Y. PMC-LLaMA: toward building open-source language models for medicine. J Am Med Inform Assoc. 2024;31(9):1833–43. doi:10.1093/jamia/ocae045. [Google Scholar] [PubMed] [CrossRef]
34. Luo L, Ning J, Zhao Y, Wang Z, Ding Z, Chen P, et al. Taiyi: a bilingual fine-tuned large language model for diverse biomedical tasks. J Am Med Inform Assoc. 2024;31(9):1865–74. doi:10.1093/jamia/ocae037. [Google Scholar] [PubMed] [CrossRef]
35. Qiu P, Wu C, Zhang X, Lin W, Wang H, Zhang Y, et al. Towards building multilingual language model for medicine. arXiv:2402.13963. 2024. [Google Scholar]
36. Siragusa I, Contino S, Pirrone R. DR-Minerva: a multimodal language model based on Minerva for diagnostic information retrieval. In: AIxIA 2024—advances in artificial intelligence. Cham, Switzerland: Springer Nature Switzerland; 2025. p. 288–300. doi:10.1007/978-3-031-80607-0_22. [Google Scholar] [CrossRef]
37. Zhang S, Fang Q, Yang Z, Feng Y. LLaVA-Mini: efficient image and video large multimodal models with one vision token. arXiv:2501.03895. 2025. [Google Scholar]
38. Panagoulias DP, Virvou M, Tsihrintzis GA. Evaluating LLM-generated multimodal diagnosis from medical images and symptom analysis. arXiv:2402.01730. 2024. [Google Scholar]
39. Driess D, Xia F, Sajjadi MS, Lynch C, Chowdhery A, Ichter B, et al. Palm-e: an embodied multimodal language model. arXiv:2303.03378. 2023. [Google Scholar]
40. Liu J, Wang Z, Ye Q, Chong D, Zhou P, Hua Y. Qilin-med-VL: towards Chinese large vision-language model for general healthcare. arXiv:2310.17956. 2023. [Google Scholar]
41. García-Ferrero I, Agerri R, Atutxa Salazar A, Cabrio E, de la Iglesia I, Lavelli A, et al. Medical mT5: an open-source multilingual text-to-text LLM for the medical domain. arXiv:2404.076. 2024. [Google Scholar]
42. Wang C, Long Q, Xiao M, Cai X, Wu C, Meng Z, et al. Biorag: a RAG-LLM framework for biological question reasoning. arXiv:2408.01107. 2024. [Google Scholar]
43. Su C, Wen J, Kang J, Wang Y, Su Y, Pan H, et al. Hybrid RAG-empowered multimodal LLM for secure data management in Internet of medical things: a diffusion-based contract approach. IEEE Internet Things J. 2025;12(10):13428–40. doi:10.1109/JIOT.2024.3521425. [Google Scholar] [CrossRef]
44. Kudo A. DeepMedcast: a deep learning method for generating intermediate weather forecasts among multiple NWP models. J Meteorolog Soc Japan. 2025;103(5):613–28. doi:10.2151/jmsj.2025-031. [Google Scholar] [CrossRef]
45. Miao H, Jia J, Cao Y, Zhou Y, Jiang Y, Liu Z, et al. Ultrasound-QBench: can LLMs aid in quality assessment of ultrasound imaging? arXiv:2501.02751. 2025. [Google Scholar]
46. Tan Y, Zhang Z, Li M, Pan F, Duan H, Huang Z, et al. MedChatZH: a tuning LLM for traditional Chinese medicine consultations. Comput Biol Med. 2024;172:108290. doi:10.1016/j.compbiomed.2024.108290. [Google Scholar] [PubMed] [CrossRef]
47. Abdullah MHA, Aziz N, Abdulkadir SJ, Akhir EAP, Talpur N. Event detection and information extraction strategies from text: a preliminary study using GENIA corpus. In: Proceedings of the 2nd International Conference on Emerging Technologies and Intelligent Systems. Cham, Switzerland: Springer International Publishing; 2022. p. 118–27. doi:10.1007/978-3-031-20429-6_12. [Google Scholar] [CrossRef]
48. Caballero-Oteyza A, Crisponi L, Peng XP, Yauy K, Volpi S, Giardino S, et al. GenIA, the Genetic Immunology Advisor database for inborn errors of immunity. J Allergy Clin Immunol. 2024;153(3):831–43. doi:10.1016/j.jaci.2023.11.022. [Google Scholar] [PubMed] [CrossRef]
49. Bai F, Du Y, Huang T, Meng MQH, Zhao B. M3d: advancing 3D medical image analysis with multi-modal large language models. arXiv:2404.00578. 2024. [Google Scholar]
50. Chen Z, Peng W, Zhang D, Liu X, Wang Z. Application, challenges, and prospects of large language model in the field of traditional Chinese Medicine. Med J Peking Union Med Coll Hosp. 2024;16(1):83–9. [Google Scholar]
51. Abd-Alrazaq A, AlSaad R, Alhuwail D, Ahmed A, Healy PM, Latifi S, et al. Large language models in medical education: opportunities, challenges, and future directions. JMIR Med Educ. 2023;9:e48291. doi:10.2196/48291. [Google Scholar] [PubMed] [CrossRef]
52. Nazi ZA, Peng W. Large language models in healthcare and medical domain: a review. Informatics. 2024;11(3):57. doi:10.3390/informatics11030057. [Google Scholar] [CrossRef]
53. Maharjan J, Garikipati A, Singh NP, Cyrus L, Sharma M, Ciobanu M, et al. OpenMedLM: prompt engineering can out-perform fine-tuning in medical question-answering with open-source large language models. Sci Rep. 2024;14(1):14156. doi:10.1038/s41598-024-64827-6. [Google Scholar] [PubMed] [CrossRef]
54. Singhal K, Tu T, Gottweis J, Sayres R, Wulczyn E, Amin M, et al. Toward expert-level medical question answering with large language models. Nat Med. 2025;31(3):943–50. doi:10.1038/s41591-024-03423-7. [Google Scholar] [PubMed] [CrossRef]
55. Lozano A, Fleming SL, Chiang CC, Shah N. Clinfo.ai: an open-source retrieval-augmented large language model system for answering medical questions using scientific literature. Pac Symp Biocomput. 2024;29:8–23. doi:10.1142/9789811286421_0002. [Google Scholar] [CrossRef]
56. Fan Z, Wei L, Tang J, Chen W, Wang S, Wei Z, et al. AI hospital: benchmarking large language models in a multi-agent medical interaction simulator. In: Proceedings of the 31st International Conference on Computational Linguistics; 2025 Jan 19–24; Abu Dhabi, United Arab Emirates. p. 10183–213. [Google Scholar]
57. Jung D, Butler A, Park J, Saperstein Y. Evaluating the impact of a specialized LLM on physician experience in clinical decision support: a comparison of Ask Avo and ChatGPT-4. arXiv:2409.15326. 2024. [Google Scholar]
58. Chang Y, Yin JM, Li JM, Liu C, Cao LY, Lin SY. Applications and future prospects of medical LLMs: a survey based on the M-KAT conceptual framework. J Med Syst. 2024;48(1):112. doi:10.1007/s10916-024-02132-5. [Google Scholar] [PubMed] [CrossRef]
59. Wilhelm TI, Roos J, Kaczmarczyk R. Large language models for therapy recommendations across 3 clinical specialties: comparative study. J Med Internet Res. 2023;25:e49324. doi:10.2196/49324. [Google Scholar] [PubMed] [CrossRef]
60. Veeramachaneni V. Large language models: a comprehensive survey on architectures, applications, and challenges. Adv Innovat Comput Programm Lang. 2025;7(1):20–39. [Google Scholar]
61. Dong X, Zhang X, Bu W, Zhang D, Cao F. A survey of LLM-based agents: theories, technologies, applications and suggestions. In: 2024 3rd International Conference on Artificial Intelligence, Internet of Things and Cloud Computing Technology (AIoTC); 2024 Sep 13–15; Wuhan, China. p. 407–13. doi:10.1109/AIoTC63215.2024.10748304. [Google Scholar] [CrossRef]
62. Chen Y, Wang Z, Xing X, Xu Z, Fang K, Wang J, et al. Bianque: balancing the questioning and suggestion ability of health LLMs with multi-turn health conversations polished by ChatGPT. arXiv:2310.15896. 2023. [Google Scholar]
63. Xu J, Lu L, Peng X, Pang J, Ding J, Yang L, et al. Data set and benchmark (MedGPTEval) to evaluate responses from large language models in medicine: evaluation development and validation. JMIR Med Inform. 2024;12(2):e57674. doi:10.2196/57674. [Google Scholar] [PubMed] [CrossRef]
64. Ng KKY, Matsuba I, Zhang PC. RAG in health care: a novel framework for improving communication and decision-making by addressing LLM limitations. NEJM AI. 2025;2(1):AIra2400380. doi:10.1056/aira2400380. [Google Scholar] [CrossRef]
65. Jiang Z, Zhong L, Sun M, Xu J, Sun R, Cai H, et al. Efficient knowledge infusion via KG-LLM alignment. arXiv:2406.03746. 2024. [Google Scholar]
66. Wang Z, Sun Y, Li Z, Yang X, Chen F, Liao H. LLM-RG4: flexible and factual radiology report generation across diverse input contexts. arXiv:2412.12001. 2024. [Google Scholar]
67. Chu Z, Wang Y, Cui Q, Li L, Chen W, Qin Z, et al. LLM-guided multi-view hypergraph learning for human-centric explainable recommendation. arXiv:2401.08217. 2024. [Google Scholar]
68. Chawla M, Panda SN, Khullar V, Garg KD, Angurala M. Deep learning based next word prediction aided assistive gaming technology for people with limited vocabulary. Entertain Comput. 2024;50(1):100661. doi:10.1016/j.entcom.2024.100661. [Google Scholar] [CrossRef]
69. Cai Y, Wang L, Wang Y, De Melo G, Zhang Y, Wang Y, et al. MedBench: a large-scale Chinese benchmark for evaluating medical large language models. Proc AAAI Conf Artif Intell. 2024;38(16):17709–17. doi:10.1609/aaai.v38i16.29723. [Google Scholar] [CrossRef]
70. Liu M, Hu W, Ding J, Xu J, Li X, Zhu L, et al. MedBench: a comprehensive, standardized, and reliable benchmarking system for evaluating Chinese medical large language models. Big Data Min Anal. 2024;7(4):1116–28. doi:10.26599/BDMA.2024.9020044. [Google Scholar] [CrossRef]
71. Jin Z, Dhingra Y, Cohen W, White RW, Liu X. What disease does this patient have? A large-scale open domain question answering dataset from medical exams. Adv Neural Inform Process Syst. 2021. doi:10.3390/app11146421. [Google Scholar] [CrossRef]
72. Pal S, Pandey R, Raj P, Balasubramanian VN. MedMCQA: a large-scale Indian medical multiple choice question dataset. In: Findings of the ACL 2022. Stroudsburg, PA, USA: ACL; 2022. p. 3427–36. [Google Scholar]
73. Jin Q, Dhingra B, Liu Z, Cohen W, Liu X. PubMedQA: a dataset for biomedical research question answering. In: Proceedings of the BioNLP Workshop 2019; 2019 Aug 1; Florence, Italy. p. 1–10. [Google Scholar]
74. Zeng G, Yang W, Ju Z, Yang Y, Wang S, Zhang R, et al. MedDialog: large-scale medical dialogue datasets. In: Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP). Stroudsburg, PA, USA: ACL; 2020. p. 9241–50. doi:10.18653/v1/2020.emnlp-main.743. [Google Scholar] [CrossRef]
75. Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035. doi:10.1038/sdata.2016.35. [Google Scholar] [PubMed] [CrossRef]
76. Huang MS, Lai PT, Lin PY, You YT, Tsai RT, Hsu WL. Biomedical named entity recognition and linking datasets: survey and our recent development. Brief Bioinform. 2020;21(6):2219–38. doi:10.1093/bib/bbaa054. [Google Scholar] [PubMed] [CrossRef]
77. Segura-Bedmar H, Martínez P, Pazos. MT. The DDI corpus: drug–drug interaction extraction. J Biomed Inform. 2013;46(5):914–20. [Google Scholar]
78. Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36(5):431–2. doi:10.1038/ng0504-431. [Google Scholar] [PubMed] [CrossRef]
79. Baker S, Silins I, Guo Y, Ali I, Högberg J, Stenius U, et al. Automatic semantic classification of scientific literature according to the hallmarks of cancer. Bioinformatics. 2015;32(3):432–40. doi:10.1093/bioinformatics/btv584. [Google Scholar] [CrossRef]
80. Ghassemiazghandi M. An evaluation of ChatGPT’s translation accuracy using BLEU score. TPLS. 2024;14(4):985–94. doi:10.17507/tpls.1404.07. [Google Scholar] [CrossRef]
81. Papineni K, Roukos S, Ward T, Zhu WJ. BLEU: a method for automatic evaluation of machine translation. In: Proceedings of the 40th Annual Meeting on Association for Computational Linguistics-ACL '02; 2002 Jul 7–12; Philadelphia, PA, USA. Morristown, NJ, USA: ACL; 2002. p. 311–8. doi:10.3115/1073083.1073135. [Google Scholar] [CrossRef]
82. Lin CY. ROUGE: a package for automatic evaluation of summaries. In: Text Summarization Branches Out: Proceedings of the ACL-04 Workshop. Stroudsburg, PA, USA: ACL; 2004. p. 74–81. [Google Scholar]
83. Zhang T, Kishore V, Wu F, Weinberger KQ, Artzi Y. BERTScore: evaluating text generation with BERT. arXiv:1904.09675. 2019. [Google Scholar]
84. Bodenreider O. The Unified Medical Language System (UMLSintegrating biomedical terminology. Nucleic Acids Res. 2004;32(90001):D267–70. doi:10.1093/nar/gkh061. [Google Scholar] [PubMed] [CrossRef]
85. You H, Guo Y. PlainQAFact: a factuality evaluation benchmark for medical plain language summarization. arXiv:2503.08890. 2025. [Google Scholar]
86. Hartman V, Zhang X, Poddar R, McCarty M, Fortenko A, Sholle E, et al. Developing and evaluating large language model–generated emergency medicine handoff notes. JAMA Netw Open. 2024;7(12):e2448723. doi:10.1001/jamanetworkopen.2024.48723. [Google Scholar] [PubMed] [CrossRef]
87. Sanaei MJ, Ravari MS, Abolghasemi H. ChatGPT in medicine: opportunity and challenges. Iranian J Blood Cancer. 2023;15(3):60–7. doi:10.61186/ijbc.15.3.60. [Google Scholar] [CrossRef]
88. OpenAI. GPT-4 technical report. arXiv:2303. 08774. 2023. [Google Scholar]
89. Meta AI. LLaMA 3: open foundation and instruction models. Meta AI Blog; 2024. [cited 2025 Sep 1]. Available from: https://ai.meta.com/blog/meta-llama-3/. [Google Scholar]
90. Peng C, Yang X, Lyu M, Smith KE, Costa A, Flores MG, et al. GatorTron and GatorTronGPT: large language models for clinical narratives. In: AAAI 2024 Spring Symposium on Clinical Foundation Models; 2024 Mar 25–27; Stanford, CA, USA. [Google Scholar]
91. Lu Q, Dou D, Nguyen T. ClinicalT5: a generative language model for clinical text. In: Findings of the association for computational linguistics: EMNLP 2022. Stroudsburg, PA, USA: ACL; 2022. p. 5436–43. doi:10.18653/v1/2022.findings-emnlp.398. [Google Scholar] [CrossRef]
92. Qian J, Jin Z, Zhang Q, Cai G, Liu B. A liver cancer question-answering system based on next-generation intelligence and the large model med-PaLM 2. Int J Comput Sci Inf Technol. 2024;2(1):28–35. doi:10.62051/ijcsit.v2n1.04. [Google Scholar] [CrossRef]
93. Chen J, Wang X, Ji K, Gao A, Jiang F, Chen S, et al. HuatuoGPT-II, one-stage training for medical adaption of LLMs. arXiv:2311.09774. 2023. [Google Scholar]
94. shibing624. MedicalGPT: medical large language model via ChatGPT pipeline (Pretraining, SFT, RLHF, DPO). GitHub repository; 2024 [Internet]. [cited 2025 Sep 1]. Available from: https://github.com/shibing624/MedicalGPT. [Google Scholar]
95. Liu Z, Zhong T, Li Y, Zhang Y, Pan Y, Zhao Z, et al. Evaluating large language models for radiology natural language processing. arXiv:2307.13693. 2023. [Google Scholar]
96. Liu L, Yang X, Lei J, Shen Y, Wang J, Wei P, et al. A survey on medical large language models: technology, application, trustworthiness, and future directions. arXiv:2406.03712. 2024. [Google Scholar]
97. Taylor R, Kardas M, Cucurull G, Scialom T, Hartshorn A, Saravia E, et al. Galactica: a large language model for science. arXiv:2211.09085. 2022. [Google Scholar]
98. Zakka M, Wang Y, Boag J. Meditron: a transformer-based language model trained on biomedical data. arXiv:2310.00017, 2023. [Google Scholar]
99. Li Y, Li Z, Zhang K, Dan R, Jiang S, Zhang Y. ChatDoctor: a medical chat model fine-tuned on a large language model meta-AI (LLaMA) using medical domain knowledge. Cureus. 2023;15(6):e40895. doi:10.7759/cureus.40895. [Google Scholar] [PubMed] [CrossRef]
100. Tan Y, Li M, Huang Z, Yu H, Fan G. MedChatZH: a better medical adviser learns from better instructions. arXiv:2309.01114. 2023. [Google Scholar]
101. Tian H, Yang K, Dong X, Zhao C, Ye M, Wang H, et al. TCMLLM-PR: evaluation of large language models for prescription recommendation in traditional Chinese medicine. Digit Chin Med. 2024;7(4):343–55. doi:10.1016/j.dcmed.2025.01.007. [Google Scholar] [CrossRef]
102. Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large language models encode clinical knowledge. Nature. 2023;620(7972):172–80. doi:10.1038/s41586-023-06291-2. [Google Scholar] [PubMed] [CrossRef]
103. Taori R, Gulrajani I, Zhang T, Dubois Y, Li X, Guestrin C, et al. Hashimoto, Stanford Alpaca: an instruction-following LLaMA model. GitHub repository; 2023 [Internet]. [cited 2025 Sep 1]. Available from: https://github.com/tatsu-lab/stanford_alpaca. [Google Scholar]
104. Zhang K, Zhou R, Adhikarla E, Yan Z, Liu Y, Yu J, et al. A generalist vision–language foundation model for diverse biomedical tasks. Nat Med. 2024;30(11):3129–41. doi:10.1038/s41591-024-03185-2. [Google Scholar] [PubMed] [CrossRef]
105. Saab K, Tu T, Weng W-H, Tanno R, Stutz D, Wulczyn E, et al. Capabilities of gemini models in medicine. arXiv:2404.18048. 2024. [Google Scholar]
106. Tian Y, Gan R, Song Y, Zhang J, Zhang Y. ChiMed-GPT: a Chinese medical large language model with full training regime and better alignment to human preferences. arXiv:2311.06025. 2023. [Google Scholar]
107. Liu J, Wang Y, Du J, Zhou JT, Liu Z. MedCoT: medical chain of thought via hierarchical expert. arXiv.2412.13736. 2024. [Google Scholar]
108. Labrak Y, Bazoge A, Morin E, Gourraud P-A, Rouvier M, Dufour R. BioMistral: a collection of open-source pretrained large language models for medical domains. arXiv:2402.10373. 2024. [Google Scholar]
109. Zekaoui NE, Mikram M, Rhanoui M, Yousfi S. BioMed-LLaMa-3: instruction-efficient fine-tuning of large language models for improved biomedical language understanding. In: Multi-disciplinary trends in artificial intelligence. Singapore: Springer Nature Singapore; 2025. p. 399–410. doi:10.1007/978-981-96-0695-5_32. [Google Scholar] [CrossRef]
110. Goh E, Gallo R, Hom J, Strong E, Weng Y, Kerman H, et al. Large language model influence on diagnostic reasoning: a randomized clinical trial. JAMA Netw Open. 2024;7(10):e2440969. doi:10.1001/jamanetworkopen.2024.40969. [Google Scholar] [PubMed] [CrossRef]
111. Li D, Yang S, Tan Z, Baik JY, Yun S, Lee J, et al. DALK: dynamic co-augmentation of LLMs and KG to answer Alzheimer’s Disease questions with scientific literature. arXiv:2405.04819. 2024. [Google Scholar]
112. Jiang B, Wang Y, Luo Y, He D, Cheng P, Gao L. Reasoning on efficient knowledge paths: knowledge graph guides large language model for domain question answering. In: 2024 IEEE International Conference on Knowledge Graph (ICKG); 2024 Dec 11–12; Abu Dhabi, United Arab Emirates. p. 142–9. doi:10.1109/ickg63256.2024.00026. [Google Scholar] [CrossRef]
113. Yue L, Xing S, Chen J, Fu T. ClinicalAgent: clinical trial multi-agent system with large language model-based reasoning. In: Proceedings of the 15th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. New York, NY, USA: The Association for Computing Machinery (ACM); 2024. p. 1–10. doi:10.1145/3698587.3701359. [Google Scholar] [CrossRef]
114. Li SS, Balachandran V, Feng S, Ilgen JS, Pierson E, Koh PW, et al. MediQ: question-asking LLMs and a benchmark for reliable interactive clinical reasoning. In: The Thirty-Eighth Annual Conference on Neural Information Processing Systems; 2024 Dec 9–15; Vancouver, BC, Canada. [Google Scholar]
115. Fleming SL, Lozano A, Haberkorn WJ, Jindal JA, Reis E, Thapa R, et al. MedAlign: a clinician-generated dataset for instruction following with electronic medical records. Proc AAAI Conf Artif Intell. 2024;38(20):22021–30. doi:10.1609/aaai.v38i20.30205. [Google Scholar] [CrossRef]
116. Feng Y, Zhou L, Ma C, Zheng Y, He R, Li Y. Knowledge graph–based thought: a knowledge graph–enhanced LLM framework for pan-cancer question answering. GigaScience. 2025;14:giae082. doi:10.1093/gigascience/giae082. [Google Scholar] [PubMed] [CrossRef]
117. Nguyen DM, Diep NT, Nguyen TQ, Le HB, Nguyen T, Nguyen T, et al. LoGra-Med: long context multi-graph alignment for medical vision-language model. arXiv:2410.02615. 2024. [Google Scholar]
118. Microsoft Research Blog. Title of the Blog Post [Internet]. [cited 2025 Aug 25]. Available from: https://www.microsoft.com/en-us/research/blog/domain-specific-language-model-pretraining-for-biomedical-natural-language-processing/. [Google Scholar]
119. The AI Edge. Typical RAG Retrieval Pipeline [Internet]. [cited 2025 Aug 26]. Available from: https://learn.theaiedge.io/. [Google Scholar]
120. Chu YW, Zhang K, Malon C, Min MR. Reducing hallucinations of medical multimodal large language models with visual retrieval-augmented generation. arXiv:2502.15040. 2025. [Google Scholar]
121. Zhang S, Xu Y, Usuyama N, Xu H, Bagga J, Tinn R, et al. BiomedCLIP: a multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. arXiv:2303.00915. 2023. [Google Scholar]
122. Xia P, Zhu K, Li H, Wang T, Shi W, Wang S, et al. MMed-RAG: versatile multimodal RAG system for medical vision-language models. arXiv:2410.13085. 2024. [Google Scholar]
123. Rajashekar NC, Shin YE, Pu Y, Chung S, You K, Giuffre M, et al. Human-algorithmic interaction using a large language model-augmented artificial intelligence clinical decision support system. In: Proceedings of the CHI Conference on Human Factors in Computing Systems. New York, NY, USA: The Association for Computing Machinery (ACM); 2024. p. 1–20. doi:10.1145/3613904.3642024. [Google Scholar] [CrossRef]
124. Ong JCL, Jin L, Elangovan K, Lim GYS, Lim DYZ, Sng GGR, et al. Development and testing of a novel large language model-based clinical decision support systems for medication safety in 12 clinical specialties. arXiv:2402.01741. 2024. [Google Scholar]
125. Van Veen D, Van Uden C, Blankemeier L, Delbrouck JB, Aali A, Bluethgen C, et al. Clinical text summarization: adapting large language models can outperform human experts. Research Square. 2023. doi:10.21203/rs.3.rs-3483777/v1. [Google Scholar] [PubMed] [CrossRef]
126. Tang L, Sun Z, Idnay B, Nestor JG, Soroush A, Elias PA, et al. Evaluating large language models on medical evidence summarization. npj Digit Med. 2023;6(1):158. doi:10.1038/s41746-023-00896-7. [Google Scholar] [PubMed] [CrossRef]
127. Karara A, Nan A, Dang Y, Shukla R. A drug discovery and biomedical research training program for underserved minority youth. J STEM Outreach. 2023;6(2):1–17. doi:10.15695/jstem/v6i2.05. [Google Scholar] [CrossRef]
128. Ramakrishnan S, Weerakkody JS. Suspended lipid bilayer: a versatile platform for nextgen drug discovery and biomedical applications. Acc Mater Res. 2022;3(10):996–8. doi:10.1021/accountsmr.2c00157. [Google Scholar] [CrossRef]
129. Sallam M. The utility of ChatGPT as an example of large language models in healthcare education, research and practice: systematic review on the future perspectives and potential limitations. MedRxiv. 2023. doi:10.1101/2023.02.19.23286155. [Google Scholar] [CrossRef]
130. JosephNg PS, Fu Z, Zhang R, Phan KY. The impact and acceptance of large language models in healthcare: a perspective from China. J Adv Res Appl Sci Eng Tech. 2024;59(2):110–58. doi:10.37934/araset.59.2.110158. [Google Scholar] [CrossRef]
131. Gangavarapu A. LLMs: a promising new tool for improving healthcare in low-resource nations. In: 2023 IEEE Global Humanitarian Technology Conference (GHTC); 2023 Oct 12–15; Radnor, PA, USA. p. 252–5. doi:10.1109/GHTC56179.2023.10354650. [Google Scholar] [CrossRef]
132. Chen J, Gui C, Gao A, Ji K, Wang X, Wan X, et al. Towards an interpretable medical agent using chain of diagnosis. arXiv:2407.13301. 2024. [Google Scholar]
133. Xia P, Chen Z, Tian J, Gong Y, Hou R, Xu Y, et al. CARES: a comprehensive benchmark of trustworthiness in medical vision language models. arXiv:2406.06007. 2024. [Google Scholar]
134. Huang Y, Sun L, Wang H, Wu S, Zhang Q, Li Y, et al. Trustllm: trustworthiness in large language models. arXiv:2401.05561. 2024. [Google Scholar]
135. Wu J, Wu X, Yang J. Guiding clinical reasoning with large language models via knowledge seeds. arXiv:2403.06609. 2024. [Google Scholar]
136. Movva R, Koh PW, Pierson E. Annotation alignment: comparing LLM and human annotations of conversational safety. arXiv:2406.06369. 2024. [Google Scholar]
137. Zhang Q, Dong J, Chen H, Zha D, Yu Z, Huang X. Knowgpt: knowledge graph based prompting for large language models. In: The Thirty-eighth Annual Conference on Neural Information Processing Systems; 2024 Dec 10–15; Vancouver, BC, Canada. [Google Scholar]
138. Muric G, Delay B, Minton S. Interpretable Cross-Examination Technique (ICE-Tusing highly informative features to boost LLM performance. arXiv:2405.06703. 2024. [Google Scholar]
139. Liu J, Wang W, Ma Z, Huang G, Y. SU, Chang KJ, et al. Medchain: bridging the gap between LLM agents and clinical practice through interactive sequential benchmarking. arXiv:2412.01605. 2024. [Google Scholar]
140. Tian D, Jiang S, Zhang L, Lu X, Xu Y. The role of large language models in medical image processing: a narrative review. Quant Imaging Med Surg. 2024;14(1):1108–21. doi:10.21037/qims-23-892. [Google Scholar] [PubMed] [CrossRef]
141. Gubanov M, Pyayt A, Karolak A. CancerKG.ORG-a web-scale, interactive, verifiable knowledge graph-LLM hybrid for assisting with optimal cancer treatment and care. In: Proceedings of the 33rd ACM International Conference on Information and Knowledge Management. New York, NY, USA: The Association for Computing Machinery (ACM); 2024. p. 4497–4505. doi:10.1145/3627673.3680094. [Google Scholar] [CrossRef]
142. Jia M, Duan J, Song Y, Wang J. medIKAL: integrating knowledge graphs as assistants of LLMs for enhanced clinical diagnosis on EMRs. arXiv:2406.14326. 2024. [Google Scholar]
143. Bao Z, Chen W, Xiao S, Ren K, Wu J, Zhong C, et al. Disc-MedLLM: bridging general large language models and real-world medical consultation. arXiv:2308.14346. 2023. [Google Scholar]
144. Tang X, Zou A, Zhang Z, Li Z, Zhao Y, Zhang X, et al. Medagents: large language models as collaborators for zero-shot medical reasoning. arXiv:2311.10537. 2023. [Google Scholar]
145. Savage T, Nayak A, Gallo R, Rangan E, Chen JH. Diagnostic reasoning prompts reveal the potential for large language model interpretability in medicine. npj Digit Med. 2024;7(1):20. doi:10.1038/s41746-024-01010-1. [Google Scholar] [PubMed] [CrossRef]
146. Busch F, Geis R, Wang Y-C, Kather JN, Al Khori N, Makowski MR, et al. AI regulation in healthcare around the world: what is the status quo?. 2025. doi:10.1101/2025.01.25.25321061. [Google Scholar] [CrossRef]
147. Wang Z, Zhu Y, Zhao H, Zheng X, Wang T, Tang W, et al. ColaCare: enhancing electronic health record modeling through large language model-driven multi-agent collaboration. arXiv:2410.02551. 2024. [Google Scholar]
148. Omar M, Sorin V, Agbareia R, Apakama DU, Soroush A, Sakhuja A, et al. Evaluating and addressing demographic disparities in medical large language models: a systematic review. Int J Equity Health. 2025;24(1):57. doi:10.1186/s12939-025-02419-0. [Google Scholar] [PubMed] [CrossRef]
149. Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK, Darzi A, et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26(9):1364–74. doi:10.1038/s41591-020-1034-x. [Google Scholar] [PubMed] [CrossRef]
150. Fayyaz H, Poulain R, Beheshti R. Enabling scalable evaluation of bias patterns in medical LLMs. arXiv:2410.14763. 2024. [Google Scholar]
151. Han H. Challenges of reproducible AI in biomedical data science. BMC Med Genom. 2025;18(1):8. doi:10.1186/s12920-024-02072-6. [Google Scholar] [PubMed] [CrossRef]
152. Pfob A, Lu SC, Sidey-Gibbons C. Machine learning in medicine: a practical introduction to techniques for data pre-processing, hyperparameter tuning, and model comparison. BMC Med Res Methodol. 2022;22(1):282. doi:10.1186/s12874-022-01758-8. [Google Scholar] [PubMed] [CrossRef]
153. Kapoor S, Cantrell EM, Peng K, Pham TH, Bail CA, Gundersen OE, et al. REFORMS: consensus-based recommendations for machine-learning-based science. Sci Adv. 2024;10(18):eadk3452. doi:10.1126/sciadv.adk3452. [Google Scholar] [PubMed] [CrossRef]
154. Bussola N, Marcolini A, Maggio V, Jurman G, Furlanello C. AI slipping on tiles: data leakage in digital pathology. arXiv:1909.06539. 2019. [Google Scholar]
155. Wang S, McDermott MBA, Chauhan G, Hughes MC, Naumann T, Ghassemi M. MIMIC-extract: a data extraction, preprocessing, and representation pipeline for MIMIC-III. arXiv:1907.08322. 2019. [Google Scholar]
156. Jin D, Pan E, Oufattole N, Weng WH, Fang H, Szolovits P. What disease does this patient have? A large-scale open domain question answering dataset from medical exams. arXiv:2009.13081. 2020. [Google Scholar]
157. So B, Byun K, Kang K, Cho S. JaQuAD: Japanese question answering dataset for machine reading comprehension. arXiv:2202.01764. 2022. [Google Scholar]
158. Kweon S, Choi B, Chu G, Song J, Hyeon D, Gan S, et al. KorMedMCQA: multi-choice question answering benchmark for Korean healthcare professional licensing examinations. arXiv:2403.01469. 2024. [Google Scholar]
159. Pal A, Minervini P, Motzfeldt AG, Alex B. Open medical LLM leaderboard. Hugging face; 2024 [Internet]. [cited 2025 Jun 30]. Available from: https://huggingface.co/spaces/openlifescienceai/open_medical_llm_leaderboard. [Google Scholar]
160. Gallifant J, Afshar M, Ameen S, Aphinyanaphongs Y, Chen S, Cacciamani G, et al. The TRIPOD-LLM reporting guideline for studies using large language models. Nat Med. 2025;31(1):60–9. doi:10.1038/s41591-024-03425-5. [Google Scholar] [PubMed] [CrossRef]
161. Tam TYC, Sivarajkumar S, Kapoor S, Stolyar AV, Polanska K, McCarthy KR, et al. A framework for human evaluation of large language models in healthcare derived from literature review. npj Digit Med. 2024;7(1):258. doi:10.1038/s41746-024-01258-7. [Google Scholar] [PubMed] [CrossRef]
162. Cianciolo AT, LaVoie N, Parker J. Machine scoring of medical students’ written clinical reasoning: initial validity evidence. Acad Med. 2021;96(7):1026–35. doi:10.1097/acm.0000000000004010. [Google Scholar] [PubMed] [CrossRef]
163. Kanithi PK, Christophe C, PimentelMAF, Raha T, Saadi N, Javed H, et al. MEDIC: towards a comprehensive framework for evaluating LLMs in clinical applications. arXiv:2409.12345. 2024. [Google Scholar]
164. Bedi S, Cui H, Fuentes M, Unell A, Wornow M, Banda JM, et al. MedHELM: holistic evaluation of large language models for medical tasks. arXiv:2505.98765. 2025. [Google Scholar]
165. Zhang M, Shen Y, Li Z, Sha H, Hu B, Wang Y, et al. LLMEval-Med: a real-world clinical benchmark for medical LLMs with physician validation. arXiv:2506.02594. 2025. [Google Scholar]
166. Yao Z, Zhang Z, Tang C, Bian X, Zhao Y, Yang Z, et al. MedQA-CS: benchmarking large language models clinical skills using an AI-SCE framework. arXiv:2410.01234. 2024. [Google Scholar]
167. Jiang Y, Black KC, Geng G, Park D, Zou J, Ng AY, et al. MedAgentBench: a realistic virtual EHR environment to benchmark medical LLM agents. arXiv:2501.12345. 2025. [Google Scholar]
168. Keeney RL, Raiffa H. Decisions with multiple objectives: preferences and value tradeoffs. Cambridge, UK: Cambridge University Press; 1993. [Google Scholar]
169. Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28(1):31–8. doi:10.1038/s41591-021-01614-0. [Google Scholar] [PubMed] [CrossRef]
170. Lyu M, Peng C, Paredes D, Chen Z, Chen A, Bian J, et al. UF-HOBI at “discharge me!”: a hybrid solution for discharge summary generation through prompt-based tuning of GatorTronGPT models. In: Proceedings of the 23rd Workshop on Biomedical Natural Language Processing. Stroudsburg, PA, USA: ACL; 2024. p. 685–95. doi:10.18653/v1/2024.bionlp-1.60. [Google Scholar] [CrossRef]
171. Clusmann J, Kolbinger FR, Muti HS, Carrero ZI, Eckardt JN, Laleh NG, et al. The future landscape of large language models in medicine. Commun Med. 2023;3(1):141. doi:10.1038/s43856-023-00370-1. [Google Scholar] [PubMed] [CrossRef]
172. Xu L, Sun H, Ni Z, Li H, Zhang S. MedViLaM: a multimodal large language model with advanced generalizability and explainability for medical data understanding and generation. arXiv:2409.19684. 2024. [Google Scholar]
173. Blagec K, Kraiger J, Fruehwirt W, Samwald M. Benchmark datasets driving artificial intelligence development fail to capture the needs of medical professionals. arXiv:2201.07040. 2022. [Google Scholar]
174. Hu X, Gu L, Kobayashi K, Liu L, Zhang M, Harada T, et al. Interpretable medical image Visual Question Answering via multi-modal relationship graph learning. Med Image Anal. 2024;97(11):103279. doi:10.1016/j.media.2024.103279. [Google Scholar] [PubMed] [CrossRef]
175. Nagoor S, Hederman L, Koidl K, Martin-Loeches I. Distinguishing clinical sentiment in intensive care unit clinical notes. In: 2024 IEEE 37th International Symposium on Computer-Based Medical Systems (CBMS); 2024 Jun 26–28; Guadalajara, Mexico; 2024. p. 249–59. doi:10.1109/CBMS61543.2024.00049. [Google Scholar] [CrossRef]
176. Monaco S, Monaco L, Apiletti D. Uncertainty-aware segmentation for rainfall prediction post processing. arXiv:2408.16792. 2024. [Google Scholar]
177. Strika Z, Petkovic K, Likic R, Batenburg R. Bridging healthcare gaps: a scoping review on the role of artificial intelligence, deep learning, and large language models in alleviating problems in medical deserts. Postgrad Med J. 2024;101(1191):4–16. doi:10.1093/postmj/qgae122. [Google Scholar] [PubMed] [CrossRef]
178. Nazary F, Deldjoo Y, Di Noia T, di Sciascio E. XAI4LLM: let machine learning models and LLMs collaborate for enhanced in-context learning in healthcare. arXiv:2405.06270. 2024. [Google Scholar]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF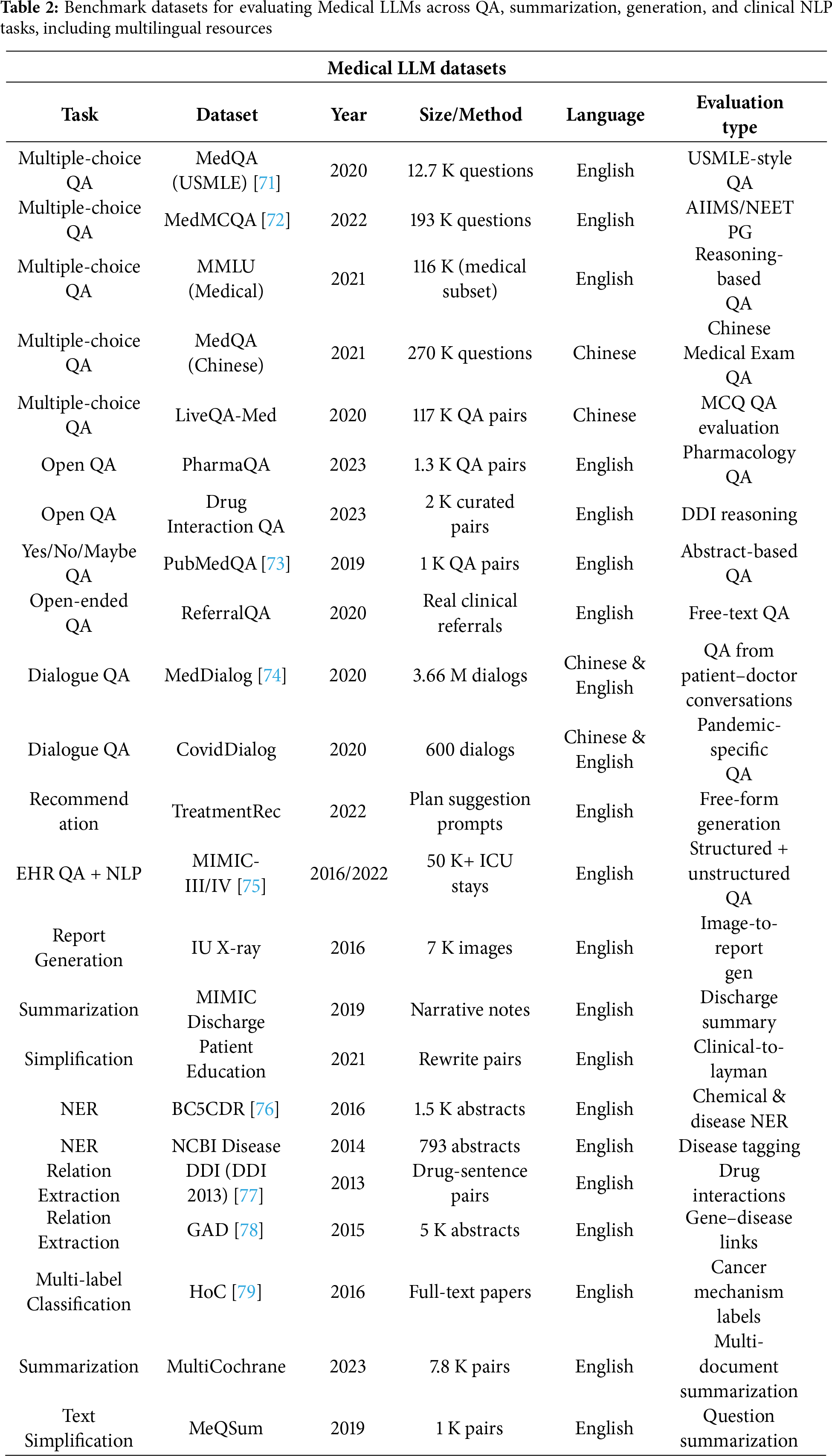
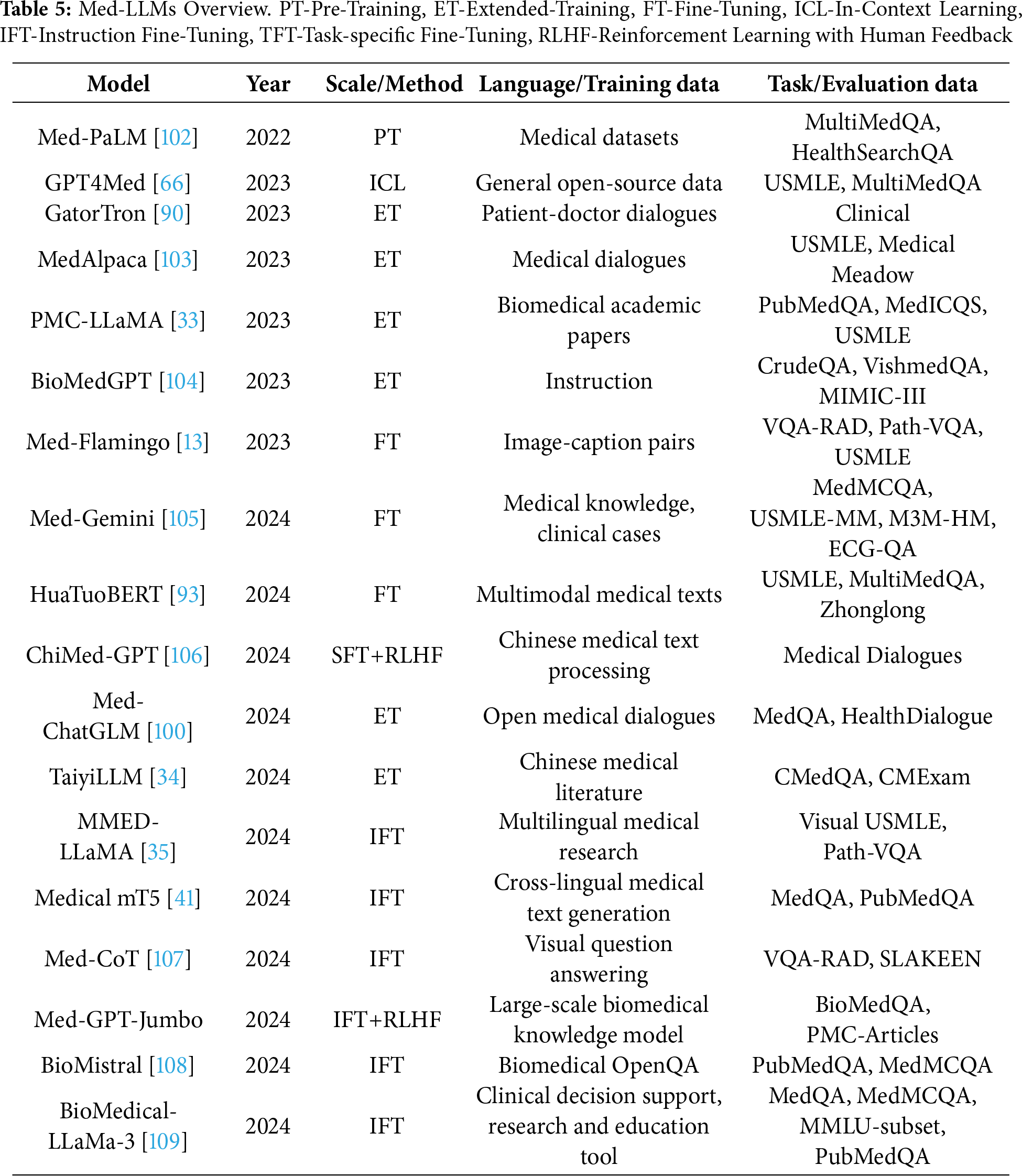
 Downloads
Downloads
 Citation Tools
Citation Tools