 Open Access
Open Access
ARTICLE
Effective Deep Learning Models for the Semantic Segmentation of 3D Human MRI Kidney Images
1 Department of Computer Technology, Yeshwantrao Chavan College of Engineering, Nagpur, 441110, India
2 Department of Computer Science and Engineering, Shri Ramdeobaba College of Engineering and Management, Ramdeobaba University, Nagpur, 440013, India
3 Department of Natural and Engineering Sciences, College of Applied Studies and Community Services, King Saud University, Riyadh, 11543, Saudi Arabia
4 Department of Electrical and Electronic Engineering, College of Engineering, University of Kerbala, Kerbala, 56001, Iraq
5 Chitkara University Institute of Engineering and Technology, Chitkara University, Rajpura, 140401, India
6 School of Computing, Gachon University, Seongnam-si, 13120, Republic of Korea
7 Chair of Cybersecurity, Department of Computer Science, College of Computer and Information Sciences, King Saud University, Riyadh, 11633, Saudi Arabia
* Corresponding Authors: Ateeq Ur Rehman. Email: ; Ahmad Almogren. Email:
(This article belongs to the Special Issue: Artificial Intelligence and Machine Learning in Healthcare Applications)
Computers, Materials & Continua 2026, 87(1), 24 https://doi.org/10.32604/cmc.2025.072651
Received 31 August 2025; Accepted 04 November 2025; Issue published 10 February 2026
Abstract
Recent studies indicate that millions of individuals suffer from renal diseases, with renal carcinoma, a type of kidney cancer, emerging as both a chronic illness and a significant cause of mortality. Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) have become essential tools for diagnosing and assessing kidney disorders. However, accurate analysis of these medical images is critical for detecting and evaluating tumor severity. This study introduces an integrated hybrid framework that combines three complementary deep learning models for kidney tumor segmentation from MRI images. The proposed framework fuses a customized U-Net and Mask R-CNN using a weighted scheme to achieve semantic and instance-level segmentation. The fused outputs are further refined through edge detection using Stochastic Feature Mapping Neural Networks (SFMNN), while volumetric consistency is ensured through Improved Mini-Batch K-Means (IMBKM) clustering integrated with an Encoder-Decoder Convolutional Neural Network (EDCNN). The outputs of these three stages are combined through a weighted fusion mechanism, with optimal weights determined empirically. Experiments on MRI scans from the TCGA-KIRC dataset demonstrate that the proposed hybrid framework significantly outperforms standalone models, achieving a Dice Score of 92.5%, an IoU of 87.8%, a Precision of 93.1%, a Recall of 90.8%, and a Hausdorff Distance of 2.8 mm. These findings validate that the weighted integration of complementary architectures effectively overcomes key limitations in kidney tumor segmentation, leading to improved diagnostic accuracy and robustness in medical image analysis.Keywords
Medical imaging, particularly MRI and CT scans, plays a vital role in the non-invasive diagnosis and monitoring of renal abnormalities. Accurate kidney segmentation is fundamental for disease assessment, treatment planning, and computer-aided diagnosis. However, segmentation of kidney tumors remains challenging due to low tissue contrast, irregular tumor boundaries, heterogeneous tumor appearances, and high inter-patient variability [1,2].
In recent years, deep learning has transformed medical image analysis by replacing handcrafted features with automated feature learning. Convolutional neural networks (CNNs) such as AlexNet, VGG, and ResNet have provided the foundation for medical imaging tasks, while architectures like U-Net and Mask R-CNN have become cornerstones for semantic and instance-level segmentation [3,4]. Extensions of these networks, including multi-scale and attention-based variants, have further advanced segmentation accuracy [5–7]. For example, Ref. [3] proposed a wavelet-style domain adaptation framework to improve cross-domain kidney segmentation, while Ref. [4] introduced a multi-organ segmentation model (MPSHT) employing progressive sampling for volumetric data. Similarly, Ref. [5] integrated SE-ResNet with Feature Pyramid Networks (FPN) to refine tumor boundaries in CT scans, and Refs. [6,7] demonstrated the effectiveness of residual and attention modules in brain MRI segmentation.
Despite these advances, existing approaches face several limitations. Many methods are limited to 2D slice-based processing, which compromises volumetric continuity, while 3D architectures suffer from high computational demands and difficulties in capturing fine-grained tumor boundaries. Moreover, most architectures emphasize either semantic segmentation (U-Net) or instance-level segmentation (Mask R-CNN) without integrating both perspectives. As a result, the preservation of boundary accuracy and volumetric consistency in renal MRI segmentation remains unresolved.
Motivated by these gaps, we propose a hybrid deep learning framework that integrates Custom U-Net, Mask R-CNN, Stochastic Feature Mapping Neural Networks (SFMNN), and Improved Mini-Batch K-Means with Encoder–Decoder CNN (IMBKM–EDCNN). This unified pipeline leverages the complementary strengths of semantic, instance-level, and volumetric segmentation, while an empirically optimized weighted fusion strategy ensures balanced integration across these levels. Furthermore, as medical image datasets inherently involve sensitive patient information, safeguarding privacy and ensuring data security have become essential in the development and deployment of deep learning–based diagnostic systems. The present study adheres to ethical data-handling protocols by utilizing publicly available, de-identified MRI datasets. Nevertheless, future advancements in this area should focus on integrating privacy-preserving mechanisms—such as federated learning, differential privacy, or encryption-based model training—to enable secure, collaborative research without compromising patient confidentiality. Unlike existing single-model methods, the proposed framework explicitly addresses the dual challenges of boundary refinement and 3D continuity.
The key contributions of this work are summarized as follows:
1. A hybrid segmentation framework combining CU-Net, Mask R-CNN, SFMNN, and IMBKM–EDCNN for accurate 3D kidney tumor segmentation from MRI images.
2. A weighted multi-stage fusion strategy that effectively integrates semantic, instance-level, and volumetric features for improved accuracy and boundary precision.
3. Incorporation of SFMNN-based edge refinement to overcome MRI boundary blurring.
4. Enforcement of volumetric consistency through IMBKM–EDCNN, ensuring smooth and coherent 3D segmentation.
5. Extensive evaluation on the TCGA-KIRC dataset, where the proposed model outperforms state-of-the-art methods, achieving a Dice score of 92.5%, IoU of 87.8%, and Hausdorff Distance of 2.8 mm.
By explicitly addressing both fine-grained boundary detection and volumetric continuity, the proposed framework provides a robust and generalizable solution for kidney tumor segmentation in 3D MRI scans. The rest of the article has been organized. Section 2 describes the literature survey, and Section 3 provides a detailed description of all the characteristics of the proposed method. Section 4 provides descriptions of the dataset used and presents the findings of our experiment. Finally, Section 5 concludes the work.
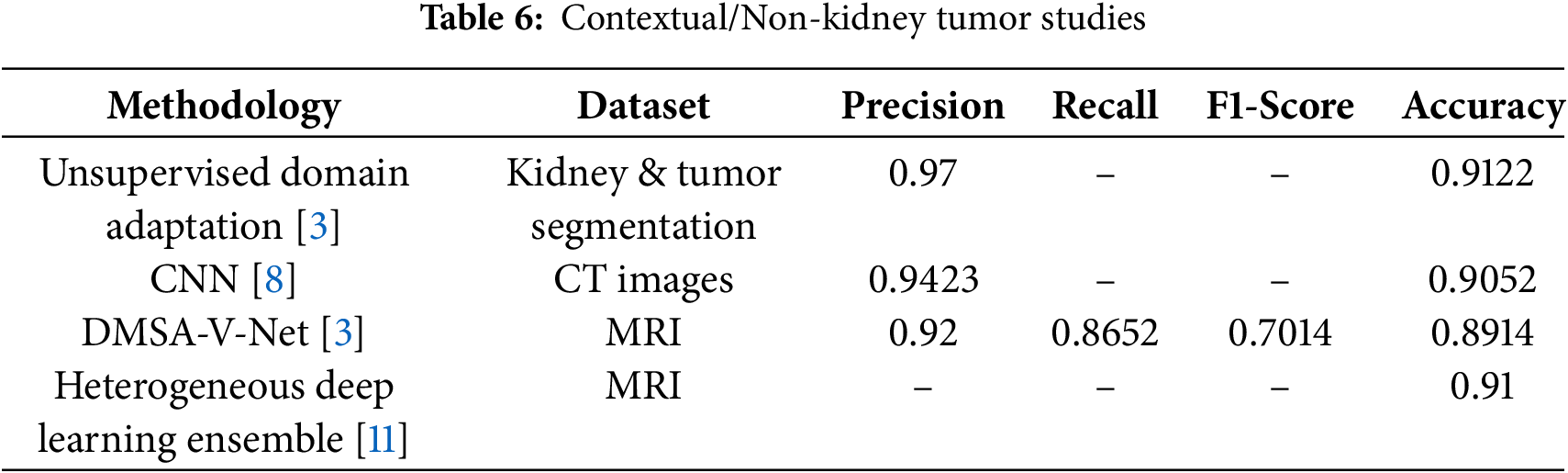
Recent advancements in deep learning have significantly improved medical image segmentation, particularly for the segmentation of kidney tumors. In [3], the authors presented compact-aware domain consistency modules and contrastive domain extraction to enhance feature and output-level adaptability through data augmentation strategies. Their approach outperformed state-of-the-art methods in kidney and tumor segmentation. Authors in [8] analyzed the performance of eight models on 8209 CT images, identifying Inception-V3 as the most effective, achieving 90.52% accuracy in detecting kidney stones. In [9], authors explored multi-organ segmentation by reviewing public datasets and dividing segmentation techniques according to label dependency in three ways: Supervised, semi-supervised. The segmentation of vascular structures in 3D medical imaging was studied by [10], who organized a competition called Global Machine Learning, involving 1401 participants, to advance deep learning techniques for blood vessel segmentation. Similarly, Ref. [2] provided a comprehensive review of deep learning-based medical image segmentation, discussing applications, experimental results, challenges, and future research directions. In [11], the authors investigated metastatic brain tumors using a pipeline validated on 275 multi-modal MRI scans from 87 patients across 53 sites, demonstrating high accuracy in tracking disease progression. In the segmentation of kidney tumors, the author in [5] proposed a model integrating SE-ResNet with Feature Pyramid Networks (FPN), achieving high segmentation accuracy in CT images. The ResNet50 model demonstrated the highest IoU scores. Meanwhile, Ref. [12] developed DMSA-V-Net, an end-to-end CNN that integrates a 3D V-Net backbone and a multi-scale attention mechanism, achieving superior Dice and sensitivity scores. Ref. [13] examined the efficacy of the Bosniak renal cyst classification system, revealing that 50% of Bosniak category III cases underwent unnecessary surgeries. Their models, tested on the KiTS19 dataset, achieved a high mean kidney Dice score of over 95%.
The application of Generative Adversarial Networks (GANs) in medical imaging was examined by [14], who proposed a Fréchet Descriptor Distance (FDD)-based Coreset method to improve GAN training for blob recognition, demonstrating its effectiveness on both simulated and real 3D kidney MRI datasets. Lastly, Ref. [5] reviewed the latest advancements in kidney tumor segmentation, discussing various imaging modalities, segmentation techniques, and evaluation metrics. Collectively, these studies highlight the continuous evolution of deep learning methods in medical image segmentation, emphasizing their likely to improve diagnostic accuracy and efficiency in kidney tumor detection.
Among volumetric segmentation approaches, the 3D U-Net has been consistently reported to outperform many slice-based methods by leveraging spatial continuity across MRI volumes. For instance, Refs. [15,16] demonstrated Dice scores in the range of 0.85–0.90 for kidney tumor segmentation, highlighting its strength in preserving both global organ context and fine structural details. These prior findings motivated our choice of U-Net as a strong baseline and as a component in our hybrid framework.
Recent research has also extended 3D U-Net architectures with domain-specific enhancements for brain tumor segmentation. For instance, Ref. [6] proposed a preprocessing strategy based on Generalized Gaussian Mixture Models (GGMM) combined with a 3D U-Net, demonstrating improved segmentation accuracy for brain tumors. Similarly, Ref. [7] enhanced a 3D U-Net using residual connections and squeeze-and-excitation attention modules to capture multimodal MRI dependencies more effectively. These studies confirm the adaptability of U-Net variants in handling complex volumetric structures, reinforcing our choice of a U-Net-based backbone in the proposed hybrid model for kidney tumor segmentation.
The proposed approach consists of the following phases:
3.1 Data Collection and Preprocessing
In this study, a comprehensive dataset of 3D abdominal MRI scans of human kidneys was obtained from TCGA-KIRC [17], comprising over 7500 2D slices from ~210 patients with expert-annotated kidney and tumor masks. From these, 728 representative 3D scans were selected, standardized to 360 × 360 pixels in NIfTI (.nii) format. Preprocessing included decomposition into 2D slices, intensity normalization, noise reduction via Gaussian and median filtering, spatial alignment, and cropping/padding around kidneys. Data augmentation (rotations, flips, contrast adjustments, elastic deformations) enhanced generalization. The dataset was split into 70% training, 15% validation, and 15% testing, with 5-fold cross-validation yielding Dice scores of 91.8%–92.7% (±0.4%). Early stopping and dropout (0.3) were used to minimize overfitting, supporting the reported 92.5% Dice score. All scans were de-identified in accordance with TCGA ethical guidelines [16] and converted to grayscale [18]. In alignment with standard biomedical ethics and data-protection regulations (e.g., HIPAA and GDPR), all personal identifiers associated with the MRI scans were removed before processing and analysis. To strengthen data security in future clinical or multi-institutional deployments, the proposed framework could be implemented within a federated learning environment, wherein model parameters—not raw images—are securely exchanged among participating centers. Additionally, employing cryptographic protocols and secure data storage practices would further reduce the risk of unauthorized access or information leakage. The proposed model architecture is shown in Fig. 1.
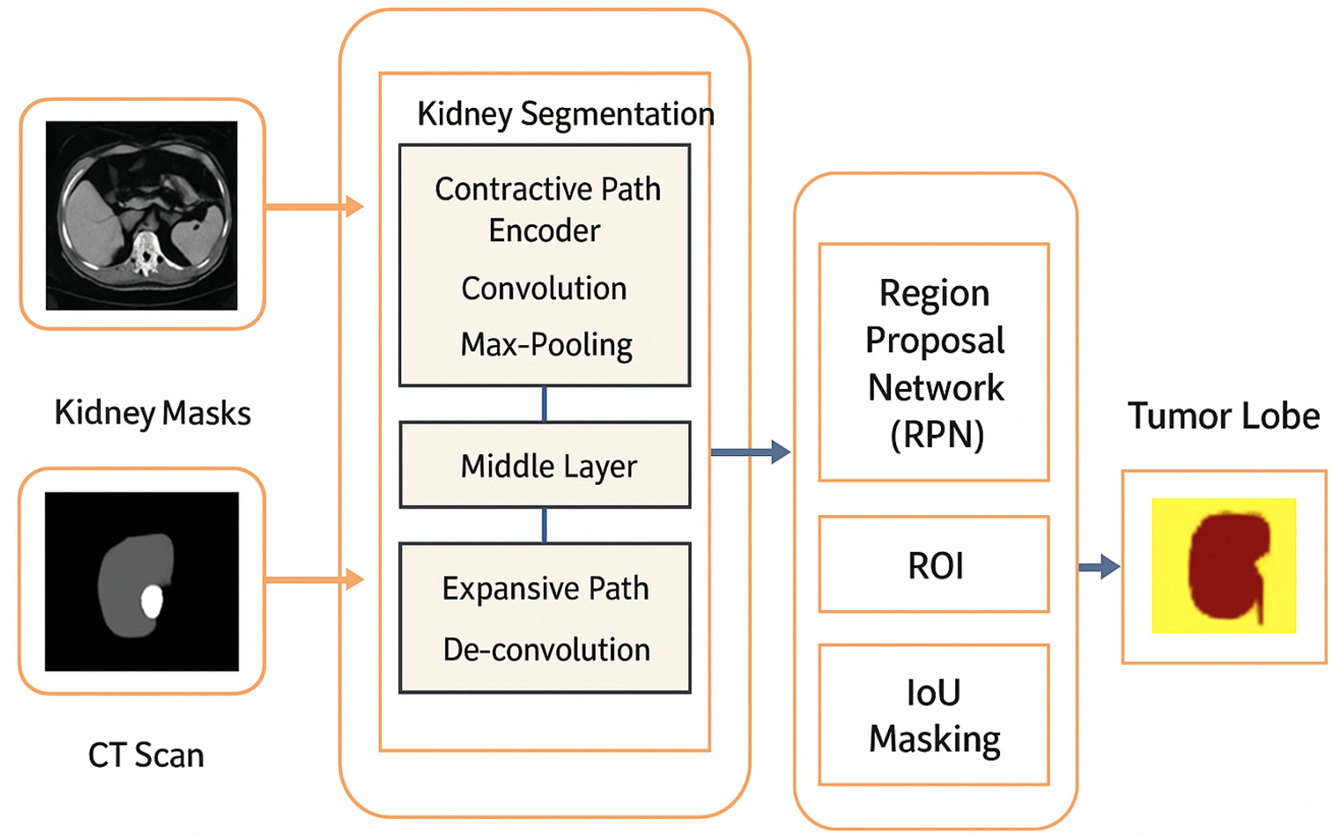
Figure 1: Architecture of the proposed hybrid kidney tumor segmentation framework
Grayscale MRI images are first denoised to reduce noise while preserving edges, followed by rotation correction [11] and cropping/padding to ensure uniform dimensions. These preprocessing steps standardize the kidney MRI scans [19], preparing them for the main segmentation process, which is structured into three primary models.
3.1.1 Hybrid Semantic Segmentation Using U-Net and Mask R-CNN
This method specifically utilizes U-Net and Mask R-CNN for segmenting kidneys and tumors from MRI or CT scan images. Let
where
where
where
3.1.2 Edge Detection with Stochastic Feature Mapping Neural Networks (SFMNN)
Edge detection aims to enhance tumor segmentation by identifying boundaries. Given an image
where
where
where
3.1.3 3D Renal MRI Blob Chunk Segmentation with IMBKM and EDCNN
Segmentation networks, such as U-Net, Mask R-CNN, and 3D U-Net, provide reliable baselines but often struggle with kidney MRI challenges, including irregular tumor boundaries and intensity variations. To address these issues, we integrated SFMNN for precise boundary refinement and IMBKM with an Encoder–Decoder CNN (EDCNN) for volumetric consistency. SFMNN captures fine-grained tumor edges via stochastic feature mapping, while IMBKM+EDCNN ensures coherent 3D masks across slices. For 3D segmentation, the IMBKM clusters voxel intensities to form initial tumor segmentations as shown in Eq. (7):
where
where
where
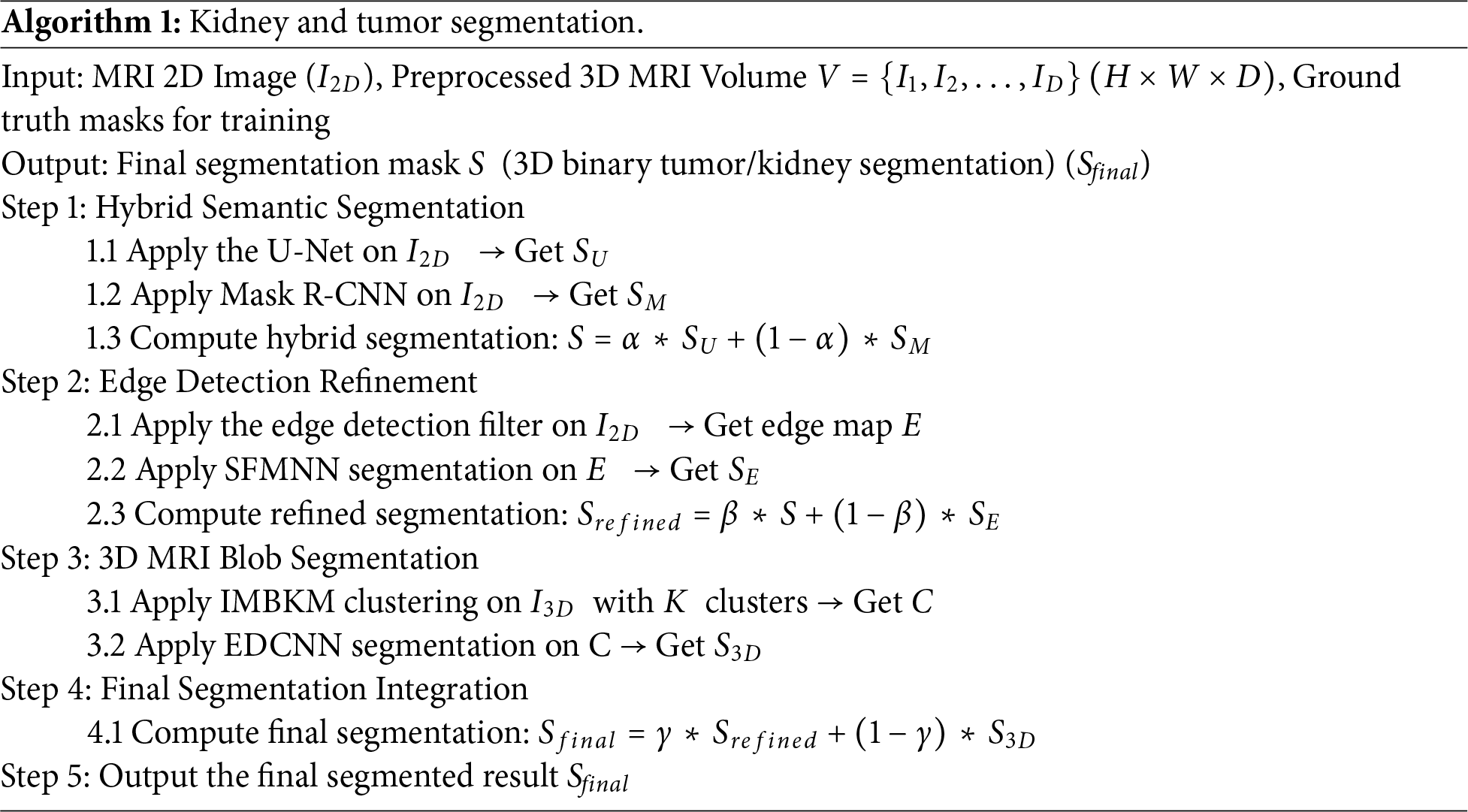

To ensure reproducibility, we describe in detail how the weighting parameters in Algorithm 1 were determined. The parameters α (for balancing CU-Net and Mask R-CNN outputs), β (for edge-refined fusion with SFMNN), and γ (for integrating 2D and 3D results) were not arbitrarily chosen but optimized through systematic validation. For every candidate triplet (α, β, γ), segmentation performance was evaluated on a held-out validation set using Dice Score, IoU, and Hausdorff Distance. Empirically, the best results were obtained with α = 0.6, β = 0.7, and γ = 0.5. This choice reflects the observation that CU-Net contributed a more reliable global semantic context than Mask R-CNN (hence α > 0.5), while SFMNN refinements significantly improved boundary accuracy (hence β > 0.5). Balancing between refined 2D outputs and volumetric 3D consistency required a nearly equal trade-off (γ ≈ 0.5).
To measure the effectiveness of the segmentation algorithm, we use the following parameters.
1. Dice Similarity Coefficient (DSC): Measures the overlap between the predicted segmentation
2. Intersection over Union (IoU): This unit is mathematically represented in Eq. (11)
3. Hausdorff Distance (HD): Measures the maximum distance between the predicted and ground truth segmentation boundaries.
Measures the ratio of the intersection and union of predicted and ground truth segmentation.
The detection of an edge using SFMNN is illustrated in Algorithm 3.

Fig. 2 represents a 2D kidney MRI slice from the TCGA-KIRC dataset, shown after intensity normalization using HU mapping. The overlaid mask illustrates tumor (red) and normal kidney (green) regions used for supervised training and evaluation. It is important to note that while 3D U-Net performance values are cited as baselines, 3D U-Net is not a component of the proposed hybrid model. Instead, it serves only as a comparative reference for volumetric kidney segmentation. Our hybrid framework uniquely integrates CU-Net, Mask R-CNN, SFMNN, and IMBKM+EDCNN specifically for kidney tumor segmentation on TCGA-KIRC. The proposed CU-Net was implemented with four encoder–decoder stages, utilizing dilated convolutions, residual skips, and dropout (0.3), whereas Mask R-CNN employed a ResNet-50 + FPN backbone with optimized anchor scales and ROI Align. SFMNN consisted of 3 convolutional layers with stochastic feature noise, and IMBKM+EDCNN used k = 2 clustering with a 5-layer 3D encoder–decoder. Dice + Cross-Entropy loss was applied for CU-Net, standard Mask R-CNN losses for instance masks, and Dice loss for final fusion. Models were trained with Adam (lr = 1e−4, batch sizes 2–8 depending on the model, 100 epochs, and early stopping), using augmentations (rotations, flips, elastic deformations, contrast/brightness shifts, and Gaussian noise). Fusion weights (α = 0.6, β = 0.7, γ = 0.5) were optimized via grid search.
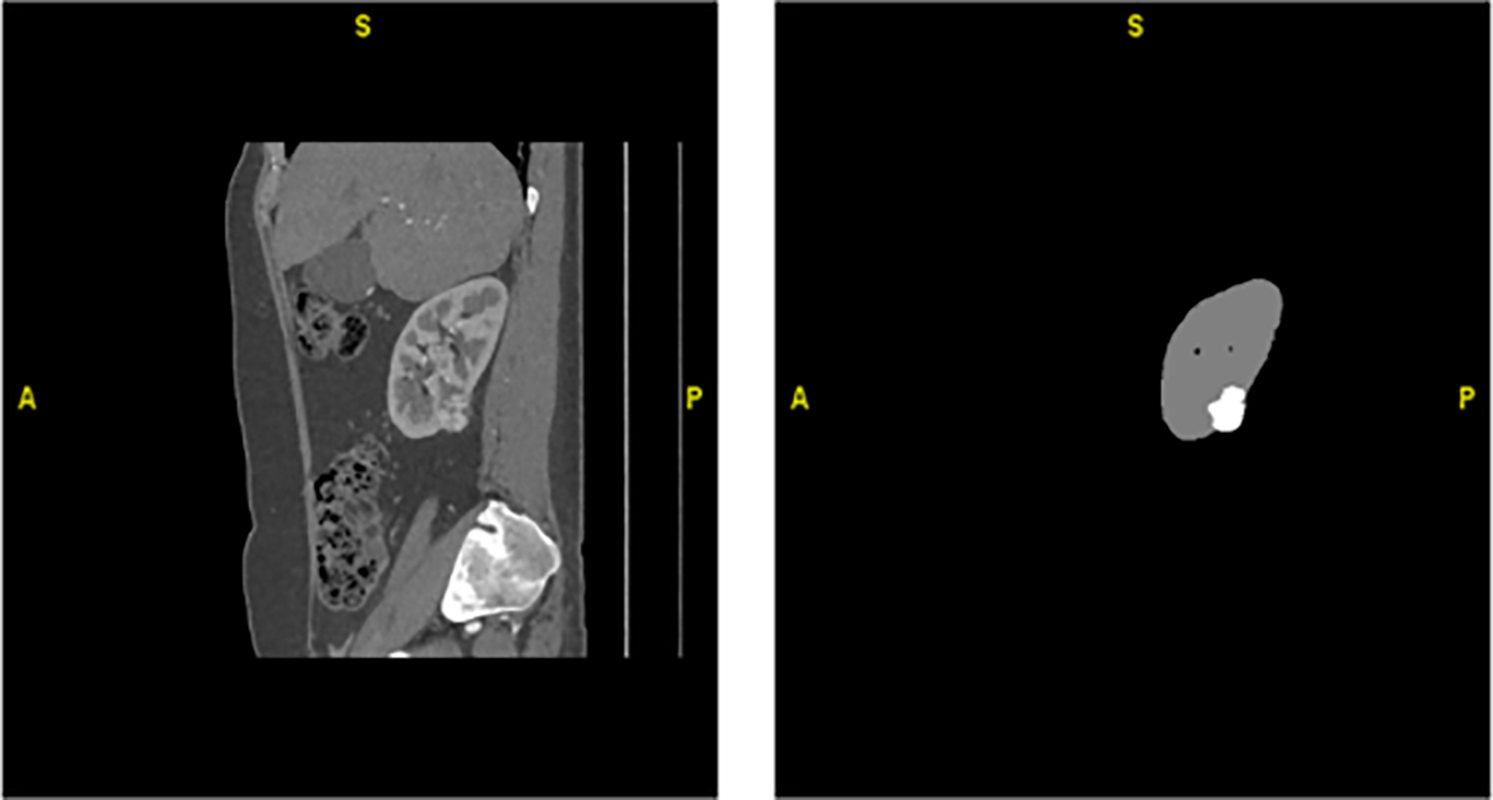
Figure 2: Hounsfield Unit (HU)–transformed MRI slice and corresponding ground-truth mask
3.2 Kidney Object Segmentation with CU-Net
Accurate extraction of organs from abdominal MR images is essential for tumor segmentation, as emphasized by previous studies [20]. In this work, kidney regions are first segmented from MR images to facilitate precise tumor detection and boundary delineation. Due to morphological variations and irregular shapes of kidney tumors, segmentation remains challenging [18].
After image preprocessing, the CU-Net model is trained on a labeled subset of MRI data and validated to evaluate segmentation accuracy. The dataset is systematically divided into training (˜Iα), validation (˜Iβ), and test (˜Iγ) sets to ensure objective performance assessment and effective hyperparameter tuning. The final model is evaluated on the test set [21]. The contracting path incorporates 2D convolution, max pooling, and dropout, while the expansive path applies transpose convolution, concatenation, and dropout operations. Across four contracting layers, CU-Net progressively learns object classification parameters, enabling efficient feature extraction and improved tumor segmentation with deeper network understanding at each stage.
Architecture of Custom CU-Net
As shown in Fig. 3, the Custom CU-Net is an adapted version of the standard Cascaded U-Net, designed for domain-specific tasks such as kidney tumor segmentation. By modifying the encoder–decoder depth, filter counts, attention modules, and feature fusion strategies, it efficiently captures fine tumor boundaries while reducing computational cost. Our method emphasizes boundary accuracy and volumetric consistency over geometric simulation. Besides kidney tumor segmentation, a slice-level classification was performed to evaluate the framework’s discriminative power. Segmentation outputs were used to label each MRI slice as either Normal or Tumor, based on the presence of predicted tumor pixels, enabling the computation of classification metrics such as Precision, Recall, F1-score, and Accuracy for additional validation of the model’s effectiveness.
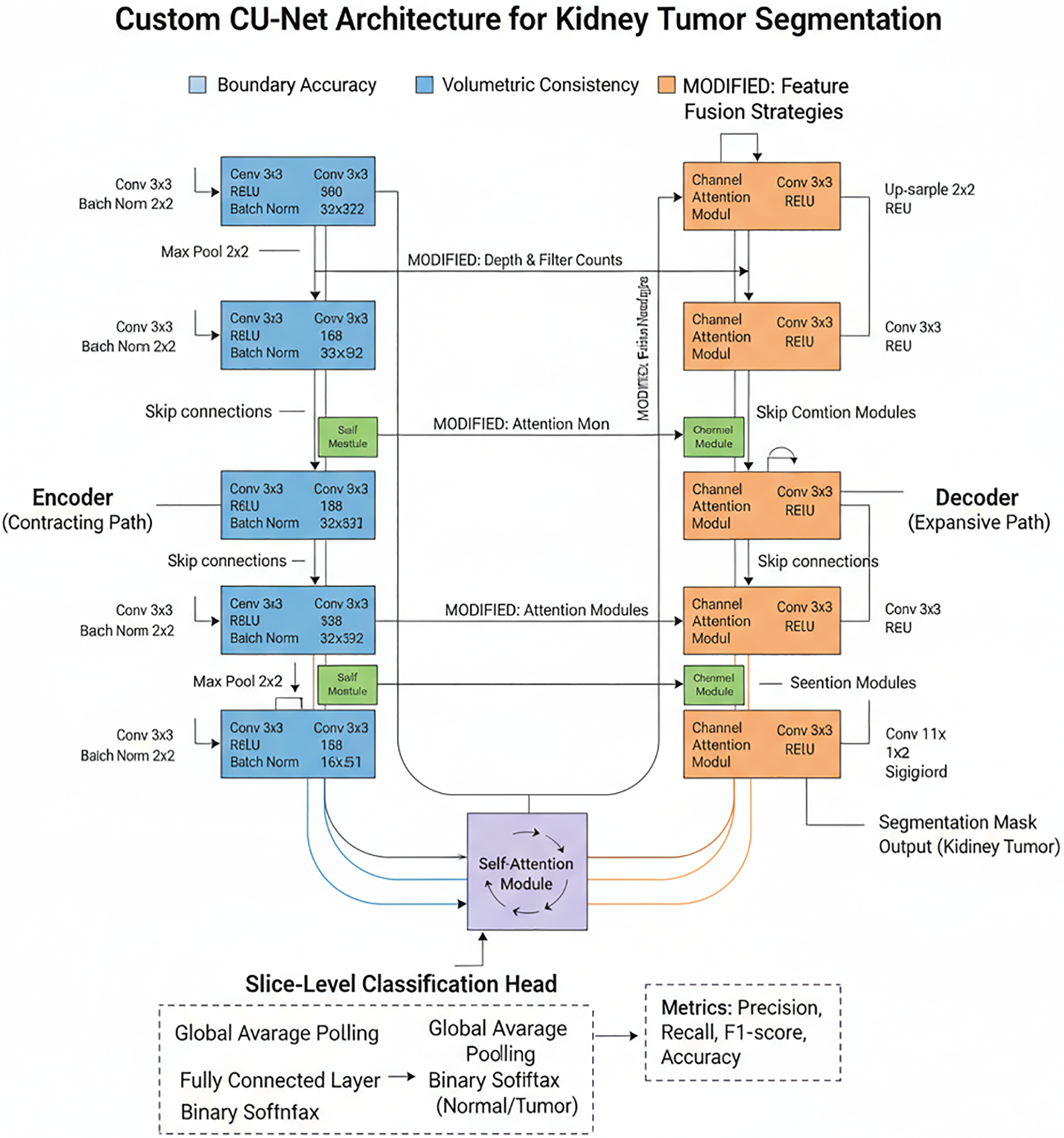
Figure 3: Architecture of custom CU-Net
The encoder (contracting route) doubles filters at each level to extract hierarchical features using repeated Conv–ReLU–BatchNorm blocks and 2 × 2 max pooling for downsampling. It incorporates self-attention modules at deeper levels to capture global dependencies and is tailored for kidney tumour segmentation by varying network depth and filter counts. The decoder (expansive route) uses skip connections and up-sampling to restore resolution in order to fuse encoder features. By fine-tuning fused features, modified channel and spatial attention modules improve boundary accuracy. Lastly, the tumour segmentation mask, which shows the tumour likelihood per pixel, is created via a 1 × 1 convolution and sigmoid activation.
A classification head is incorporated into the bottleneck layer of the U-Net to assess the discriminative capacity of the model. A Fully Connected (FC) layer and Binary Softmax activation are used to classify each slice as Normal or Tumour after the retrieved features have undergone Global Average Pooling (GAP) to create a compact vector. The segmentation output is used to establish the classification label. If any tumour pixels are found, the slice is labelled Tumour; if not, it is labelled Normal.
In semantic segmentation tasks, individual slices are treated as 2D images, where each pixel is classified, as shown in Fig. 4. Visualization of sample distribution across four classes: Normal (1), Cyst (0), Stone (2), and Tumor (3). The chart confirms balanced representation of normal and pathological conditions within the dataset used for hybrid model evaluation. A set of 2D images (slices 0–7) retrieved from a 3D abdominal MRI scan is shown in Fig. 5, providing insight into anatomical structures across multiple depths.
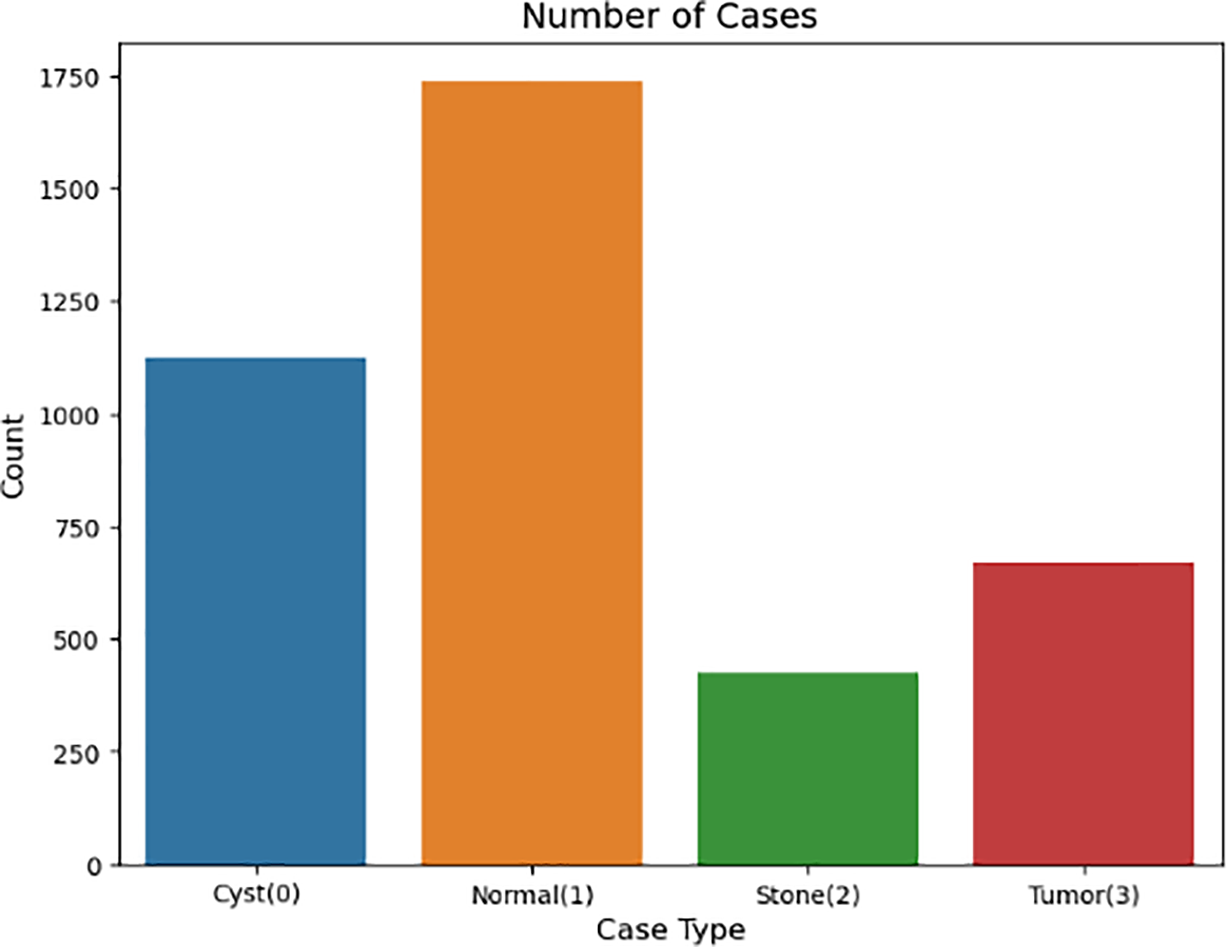
Figure 4: Distribution of cases among four kidney categories
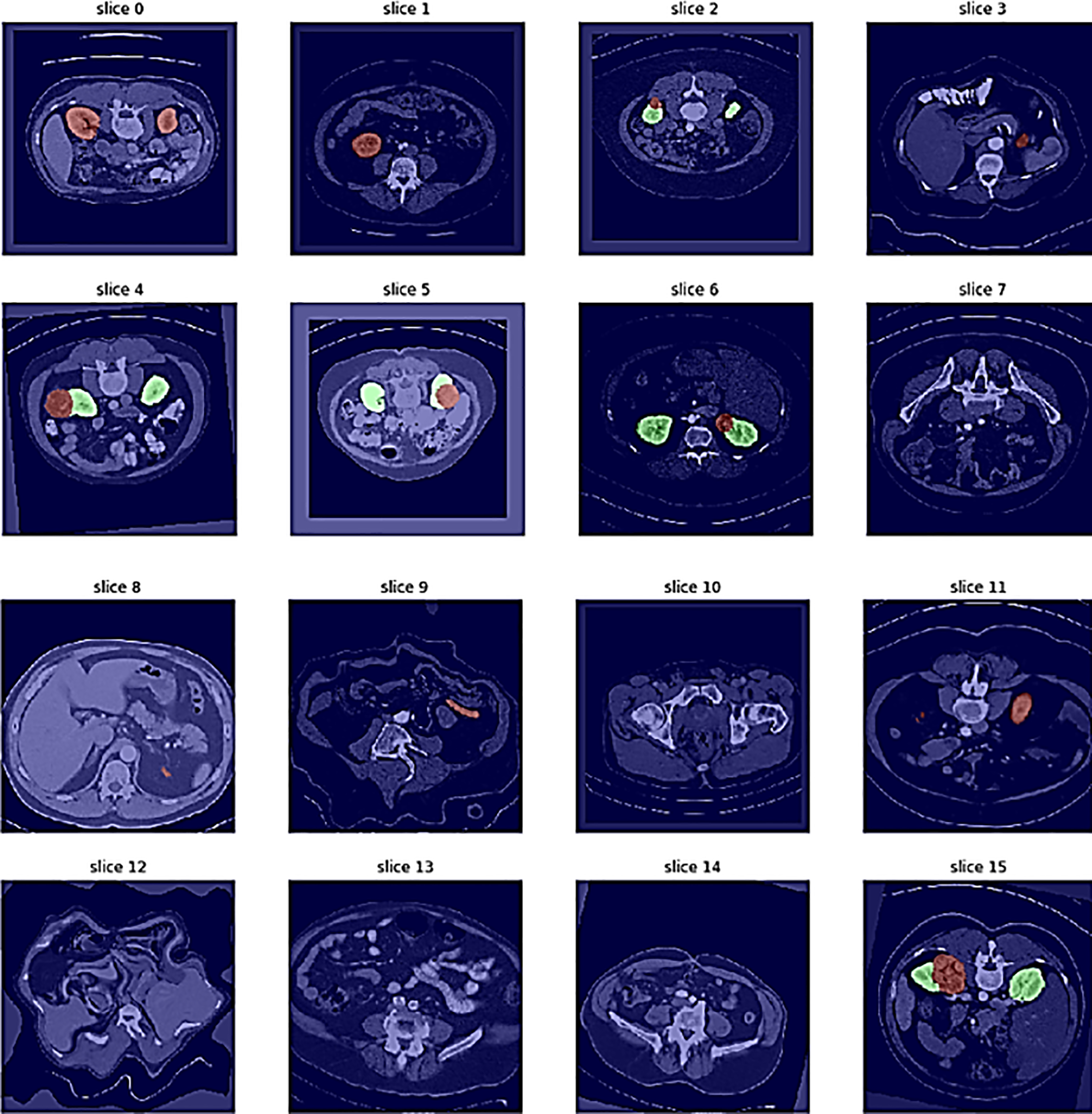
Figure 5: Different image slicing for kidney detection
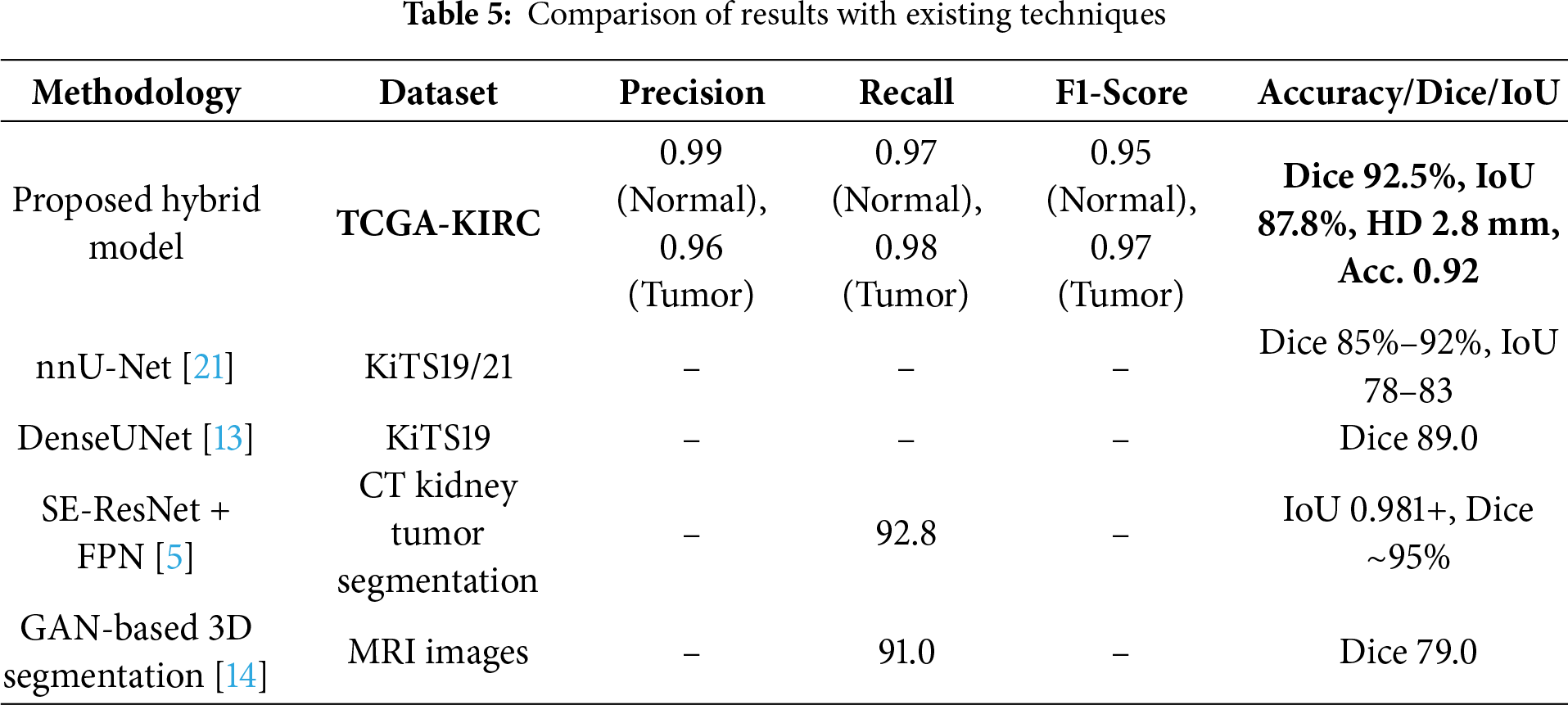
To ensure a robust evaluation, we further compared our proposed framework against several state-of-the-art kidney tumor segmentation models reported in the literature (e.g., nnU-Net, DenseUNet, SE-ResNet with FPN, GAN-based approaches). We computed 95% confidence intervals (CIs) using bootstrapping across patient cases. As shown in Table 1, the proposed hybrid model achieved a Dice score of 92.5% (95% CI: 91.2–93.7), outperforming U-Net (85.2% [83.8–86.6]) and Mask R-CNN (87.4% [86.0–88.7]). Paired Wilcoxon signed-rank tests confirmed that these improvements in Dice and Hausdorff Distance were statistically significant (p < 0.01), and lower Hausdorff Distance (2.8 mm) compared to these published benchmarks, demonstrating its effectiveness for volumetric kidney tumor segmentation on the TCGA-KIRC dataset. The different image slicing for kidney detection is shown in Fig. 5, where the confusion matrix illustrates the relationship between true slice labels and predicted slice labels, providing complementary validation of the segmentation model’s discriminative ability.
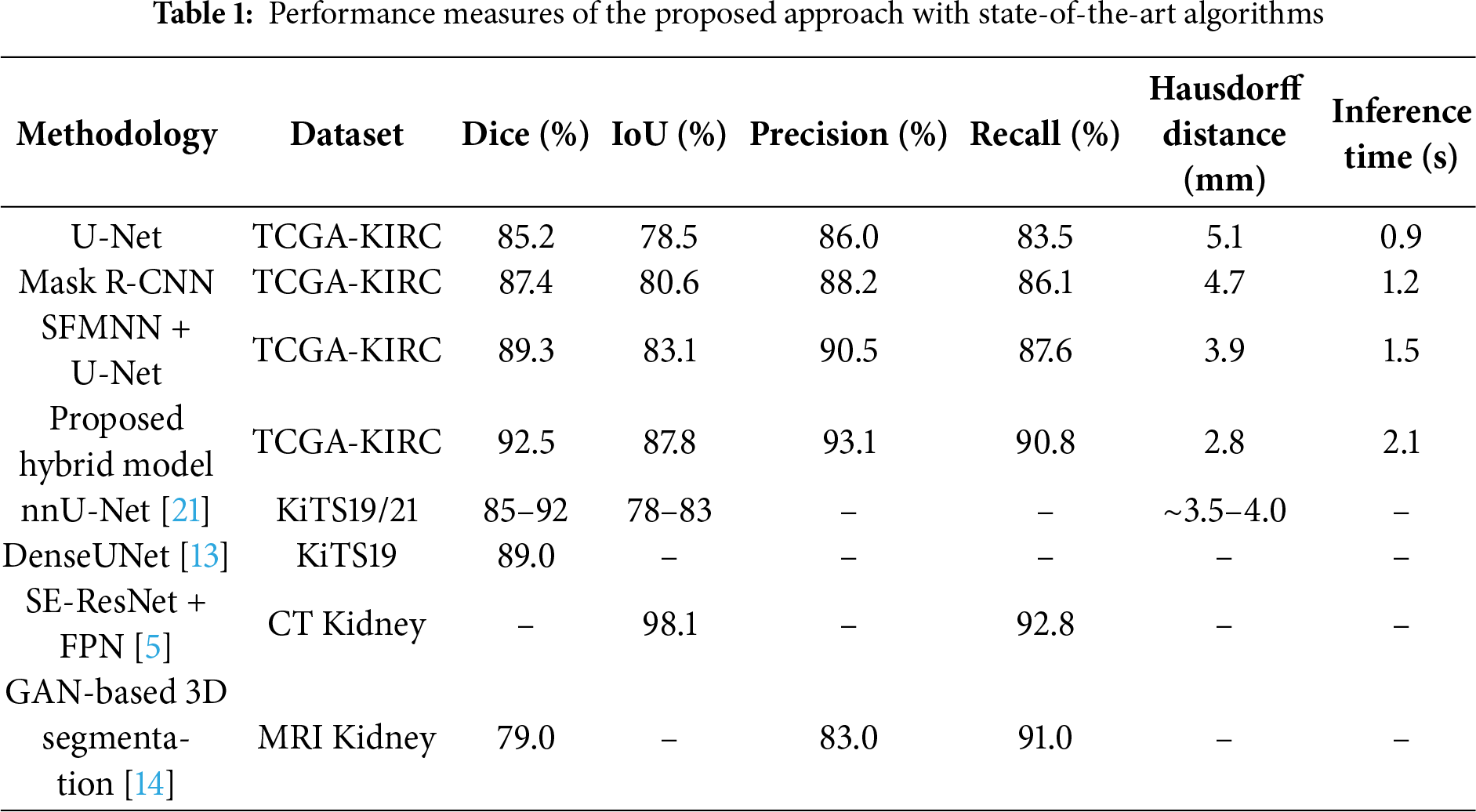
The proposed hybrid model requires 2.1 s per MRI volume, compared to 0.9 s (U-Net) and 1.2 s (Mask R-CNN). Although slightly slower, it achieves a 6–8% higher Dice score and reduces the Hausdorff distance from 5.1 mm to 2.8 mm, offering markedly improved boundary precision. Given that segmentation is performed in offline diagnostic workflows, the modest increase in inference time is justified by the substantial gain in accuracy and volumetric consistency.
Ablation Study on Individual Components
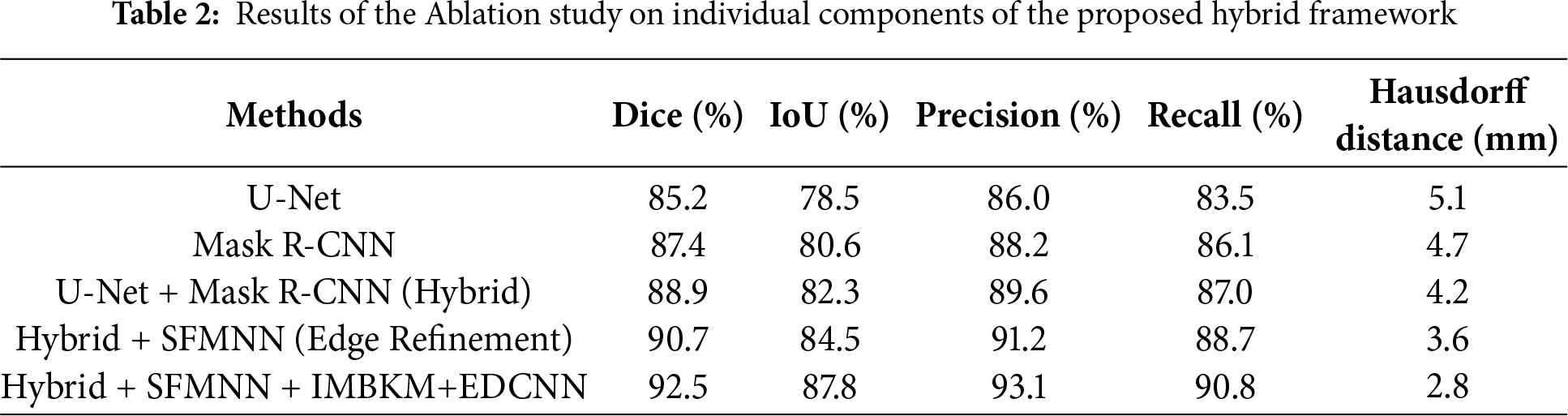
To assess the contribution of each module in the proposed hybrid framework, we conducted an ablation study. Starting with U-Net and Mask R-CNN individually, we then added edge refinement (SFMNN) and volumetric consistency (IMBKM+EDCNN). Table 2 summarizes the results.
The rapid increase in accuracy suggests the model is quickly learning patterns in the training data during the early epochs. Tables 3 and 4 present the model’s evaluation of the binary classification of medical images for tumor detection. Each MRI slice was categorized as ‘Normal’ or ‘Tumor’ based on predicted segmentation masks. Metrics such as Precision, Recall, and F1-score were computed at this slice-level to complement the pixel-wise segmentation evaluation. The model performs well for both the “Normal” and “Tumor” classes, as evidenced by high precision, recall, and F1 Scores. The matrix compares predicted slice labels against ground-truth annotations, distinguishing Normal versus Tumor slices. High diagonal values indicate strong agreement, with an overall classification accuracy of 92%, as shown in Fig. 6. The ROC curve, given in Fig. 7, illustrates the trade-off between the true positive rate (TPR) and false positive rate (FPR) for slice-level tumor detection. The Area Under the Curve (AUC = 0.88) confirms the high discriminative ability of the segmentation-driven classification model.
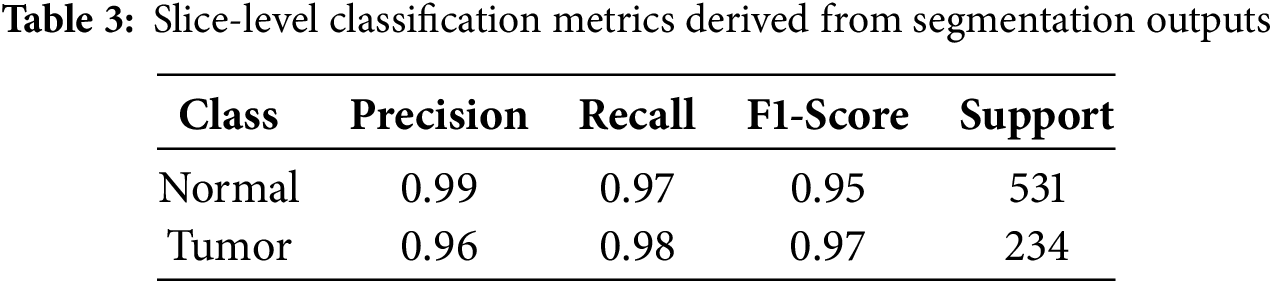
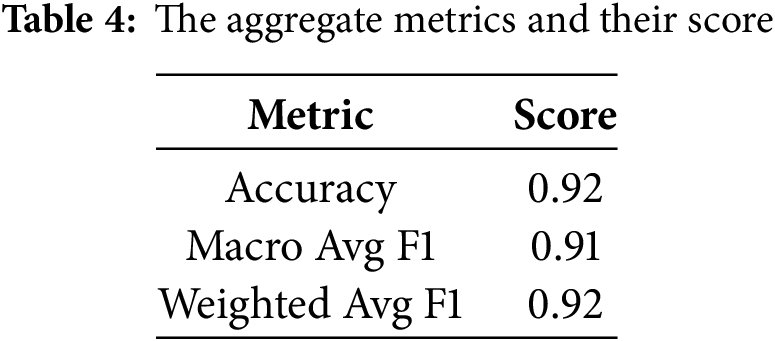
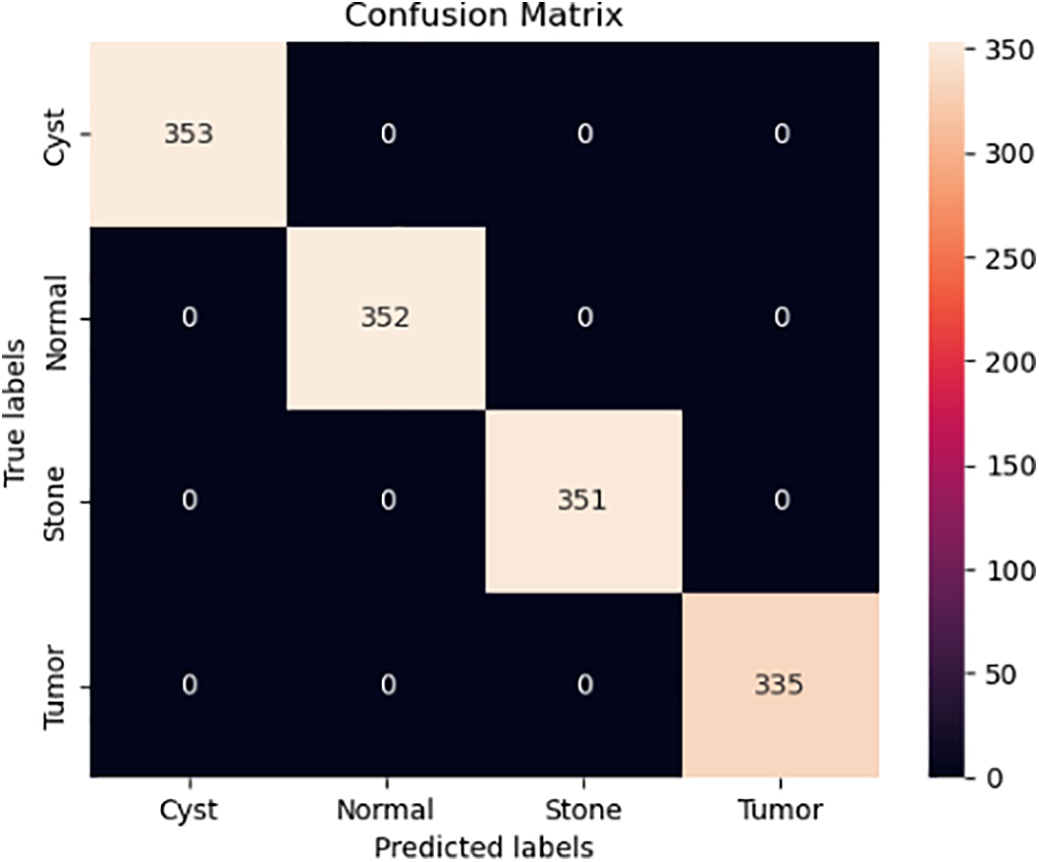
Figure 6: Confusion Matrix representing slice-level classification derived from segmentation outputs
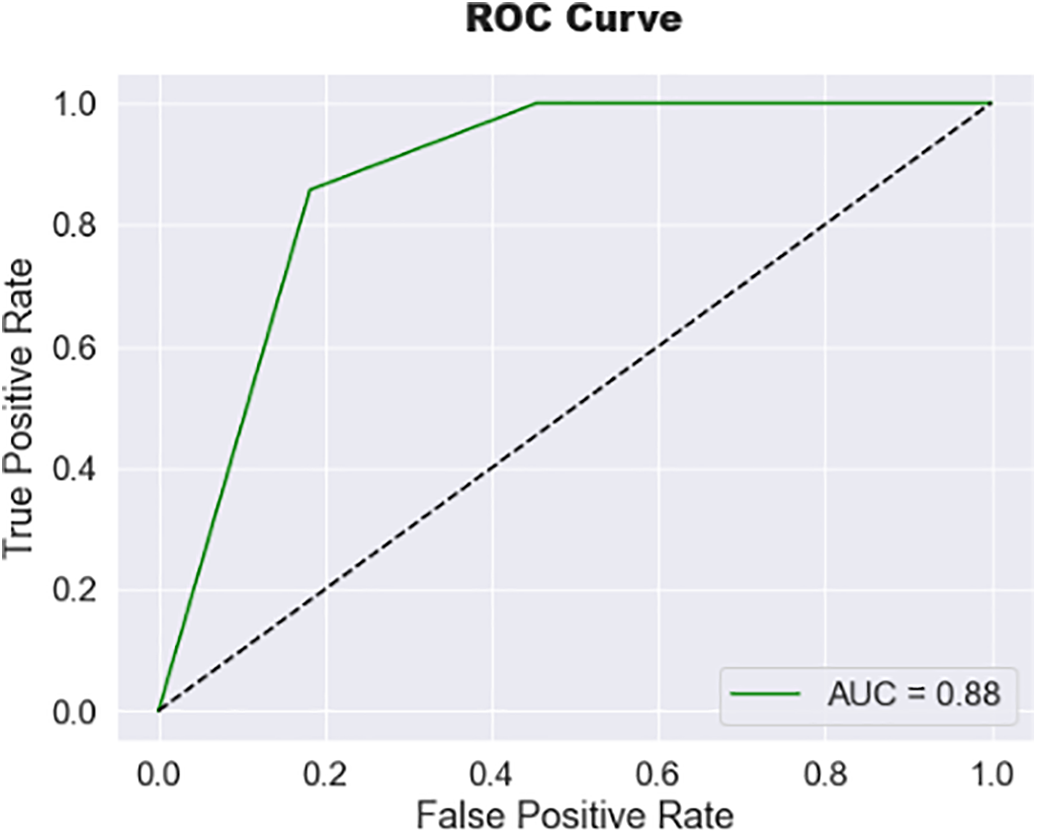
Figure 7: Receiver operating characteristic (ROC) curve of the proposed hybrid model
The experimental results demonstrate that the proposed hybrid CU-Net + Mask R-CNN framework, enhanced with SFMNN and IMBKM+EDCNN, consistently outperforms baseline architectures such as U-Net and Mask R-CNN in terms of Dice score, IoU, and Hausdorff Distance. These improvements are attributable to three factors: (i) the weighted fusion of CU-Net and Mask R-CNN, which balances global semantic context with precise instance-level segmentation; (ii) the SFMNN-based edge refinement, which effectively resolves the boundary blurring commonly encountered in MRI images; and (iii) the volumetric consistency enforced by IMBKM+EDCNN, which reduces discontinuities across 3D slices.
The inclusion of classification metrics complements segmentation evaluation by assessing whether a slice is normal or tumorous, bridging algorithmic performance and clinical relevance. The hybrid model achieved a Dice score of 92.5% for segmentation. In comparison, slice-level classification accuracy reached 92%, with strong precision, recall, and F1 Scores, serving as a supplementary measure alongside the primary segmentation evaluation. We initially described the 3D U-Net as being superior for capturing both global context and fine details; we clarify that this statement is grounded in both prior literature and our own ablation results. Previous studies, as reported in [15,16], consistently show that the 3D U-Net achieves Dice scores in the range of 0.85–0.90 on kidney MRI segmentation tasks, outperforming several 2D slice-based models by leveraging volumetric continuity. In our experiments on the TCGA-KIRC dataset, the standalone U-Net baseline achieved a Dice score of 85.2% and an IoU of 78.5%, confirming its relative strength in capturing the global organ structure compared to other baselines, such as Mask R-CNN. This motivated its inclusion as a core component of our hybrid framework. However, we acknowledge that we did not directly benchmark the 3D U-Net against other volumetric networks, such as V-Net, within this study, primarily due to computational constraints. We have now made this limitation explicit and note that extending future experiments to include direct comparisons with V-Net and related volumetric architectures would provide a stronger empirical validation of this claim. Beyond model accuracy, privacy and security remain crucial considerations for clinical translation of AI-based segmentation systems. Since MRI datasets contain personally identifiable medical records, improper data handling could lead to confidentiality breaches. Future work should therefore explore privacy-preserving computation techniques—such as homomorphic encryption, secure multiparty computation, and blockchain-based audit trails—to ensure both data integrity and traceability throughout the model-training and inference pipeline. Integrating such mechanisms would allow secure collaboration among hospitals while maintaining regulatory compliance and patient trust. Tables 5 and 6 demonstrate that only methods specifically designed for kidney tumor segmentation serve as fair benchmarks. Models developed for other imaging tasks are included for context. Upon direct comparison, our proposed model outperforms established kidney tumor segmentation approaches, including nnU-Net, DenseUNet, SE-ResNet+FPN, and GAN-based methods, on the TCGA-KIRC dataset. This work was validated on the TCGA-KIRC dataset; future studies will extend the evaluation to multi-institutional datasets to confirm generalizability across diverse imaging protocols and populations.
The proposed method may underperform on very small or low-contrast tumors, where edge cues are weak, and in cases with severe motion artifacts or intensity inhomogeneity. Performance slightly decreases for tumors near organ boundaries due to overlapping textures. The combination of U-Net, Mask R-CNN, SFMNN, IMBKM, and EDCNN was chosen both theoretically and empirically. U-Net offers robust semantic segmentation for the global kidney structure, while Mask R-CNN provides instance-level precision for irregular tumor regions. SFMNN refines edge boundaries, and IMBKM+EDCNN enforces 3D volumetric consistency across slices. Ablation results confirm that each component incrementally improves Dice and Hausdorff metrics, validating that this integration is evidence-driven.
This work presented a hybrid deep learning framework for the semantic segmentation of kidney tumors in 3D MRI scans, integrating CU-Net, Mask R-CNN, SFMNN, and IMBKM+EDCNN in a weighted multi-stage pipeline. Unlike traditional single-model approaches, the proposed method combines semantic, instance-level, and volumetric segmentation, resulting in superior performance on the TCGA-KIRC dataset with a Dice score of 92.5%, an IoU of 87.8%, a precision of 93.1%, a recall of 90.8%, and a Hausdorff distance of 2.8 mm. Ablation studies demonstrated the contribution of each stage: CU-Net and Mask R-CNN fusion improved baseline segmentation accuracy, SFMNN refinement enhanced boundary precision, and IMBKM+EDCNN ensured smooth volumetric consistency across slices. These results confirm that each component plays a crucial role in achieving clinically reliable tumor delineations. The study therefore provides an evidence-based demonstration that a carefully designed hybrid approach can outperform established benchmarks, offering clear benefits for computer-aided diagnosis and treatment planning. In addition to achieving high segmentation accuracy, the integration of privacy-preserving mechanisms represents a critical step toward the real-world deployment of AI-driven medical image analysis systems. Ensuring that patient data remains encrypted and institutionally localized through secure federated learning frameworks would promote ethical and regulatory compliance. By coupling computational performance with robust privacy protection, future iterations of the proposed framework could support trustworthy and scalable clinical adoption across distributed healthcare environments. Nevertheless, the framework requires relatively high computational resources and dataset-specific hyperparameter tuning, which may limit immediate clinical deployment. Future work will address these issues by exploring lightweight model variants, optimizing inference speed, and validating the framework across diverse imaging modalities and multi-institutional datasets to enhance robustness and generalizability.
Acknowledgement: Not applicable.
Funding Statement: This work was funded by the Ongoing Research Funding Program-Research Chairs (ORF-RC-2025-2400), King Saud University, Riyadh, Saudi Arabia.
Author Contributions: Roshni Khedgaonkar: Writing—original draft, Visualization, Validation, Methodology, Formal analysis, Conceptualization. Pravinkumar Sonsare: Writing—review & editing, Writing—original draft, Validation, Methodology, Supervision, Investigation, Formal analysis, Conceptualization. Kavita Singh: Writing—review & editing, Writing—original draft, Validation, Methodology, Supervision, Investigation, Formal analysis, Conceptualization. Ayman Altameem: Writing—review & editing, Methodology, Formal analysis, Conceptualization. Hameed R. Farhan: Writing—review & editing, Software, Methodology, Conceptualization. Salil Bharany: Visualization, Validation, Methodology. Ateeq Ur Rehman: Writing—review & editing, Methodology, Formal analysis, Conceptualization. Ahmad Almogren: Writing—review & editing, Software, Methodology, Funding acquisition, Conceptualization. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used in this study are publicly available at https://www.cancer.gov/tcga (accessed on 01 January 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflict of interest to report regarding the present study.
References
1. Nisa SQ, Ismail AR, Ali MABM, Khan MS. Medical image analysis using deep learning: a review. In: Proceedings of the 2020 IEEE 7th International Conference on Engineering Technologies and Applied Sciences (ICETAS); 2020 Dec 18–20; Kuala Lumpur, Malaysia. doi:10.1109/ICETAS51660.2020.9484287. [Google Scholar] [CrossRef]
2. Rayed ME, Sajibul Islam SM, Niha SI, Jim JR, Kabir MM, Mridha MF. Deep learning for medical image segmentation: state-of-the-art advancements and challenges. Inform Med Unlocked. 2024;47(7):101504. doi:10.1016/j.imu.2024.101504. [Google Scholar] [CrossRef]
3. Yin Y, Tang Z, Huang Z, Wang M, Weng H. Source free domain adaptation for kidney and tumor image segmentation with wavelet style mining. Sci Rep. 2024;14(1):24849. doi:10.1038/s41598-024-75972-3. [Google Scholar] [PubMed] [CrossRef]
4. Zhao Y, Li J, Hua Z. MPSHT: multiple progressive sampling hybrid model multi-organ segmentation. IEEE J Transl Eng Health Med. 2022;10:1800909. doi:10.1109/JTEHM.2022.3210047. [Google Scholar] [PubMed] [CrossRef]
5. Abdelrahman A, Viriri S. FPN-SE-ResNet model for accurate diagnosis of kidney tumors using CT images. Appl Sci. 2023;13(17):9802. doi:10.3390/app13179802. [Google Scholar] [CrossRef]
6. Lairedj KI, Chama Z, Bagdaoui A, Larguech S, Menni Y, Becheikh N, et al. Advanced brain tumor segmentation in magnetic resonance imaging via 3D U-Net and generalized Gaussian mixture model-based preprocessing. Comput Model Eng Sci. 2025;144(2):2419–43. doi:10.32604/cmes.2025.069396. [Google Scholar] [CrossRef]
7. Chen YT, Ahmad N, Aurangzeb K. Enhancing 3D U-Net with residual and squeeze-and-excitation attention mechanisms for improved brain tumor segmentation in multimodal MRI. Comput Model Eng Sci. 2025;144(1):1197–224. doi:10.32604/cmes.2025.066580. [Google Scholar] [CrossRef]
8. Sabuncu Ö, Bilgehan B, Kneebone E, Mirzaei O. Effective deep learning classification for kidney stone using axial computed tomography (CT) images. Biomed Tech. 2023;68(5):481–91. doi:10.1515/bmt-2022-0142. [Google Scholar] [PubMed] [CrossRef]
9. Liu X, Qu L, Xie Z, Zhao J, Shi Y, Song Z. Towards more precise automatic analysis: a systematic review of deep learning-based multi-organ segmentation. Biomed Eng Online. 2024;23(1):52. doi:10.1186/s12938-024-01238-8. [Google Scholar] [PubMed] [CrossRef]
10. Jain Y, Walsh CL, Yagis E, Aslani S, Nandanwar S, Zhou Y, et al. Vasculature segmentation in 3D hierarchical phase-contrast tomography images of human kidneys. bioRxiv. 2024. doi:10.1101/2024.08.25.609595. [Google Scholar] [CrossRef]
11. Machura B, Kucharski D, Bozek O, Eksner B, Kokoszka B, Pekala T, et al. Deep learning ensembles for detecting brain metastases in longitudinal multi-modal MRI studies. Comput Med Imaging Graph. 2024;116(1):102401. doi:10.1016/j.compmedimag.2024.102401. [Google Scholar] [PubMed] [CrossRef]
12. Song E, Long J, Ma G, Liu H, Hung CC, Jin R, et al. Prostate lesion segmentation based on a 3D end-to-end convolution neural network with deep multi-scale attention. Magn Reson Imaging. 2023;99(4):98–109. doi:10.1016/j.mri.2023.01.015. [Google Scholar] [PubMed] [CrossRef]
13. Kittipongdaja P, Siriborvornratanakul T. Automatic kidney segmentation using 2.5D ResUNet and 2.5D DenseUNet for malignant potential analysis in complex renal cyst based on CT images. EURASIP J Image Video Process. 2022;2022(1):5. doi:10.1186/s13640-022-00581-x. [Google Scholar] [PubMed] [CrossRef]
14. Xu Y, Wu T, Charlton JR, Bennett KM. GAN training acceleration using Fréchet descriptor-based coreset. Appl Sci. 2022;12(15):7599. doi:10.3390/app12157599. [Google Scholar] [CrossRef]
15. Myronenko A, Hatamizadeh A. 3D kidneys and kidney tumor semantic segmentation using boundary-aware networks. arXiv:1909.06684. 2019. [Google Scholar]
16. Zhao W, Jiang D, Peña Queralta J, Westerlund T. MSS U-Net: 3D segmentation of kidneys and tumors from CT images with a multi-scale supervised U-Net. Inform Med Unlocked. 2020;19(12):100357. doi:10.1016/j.imu.2020.100357. [Google Scholar] [CrossRef]
17. Akin O, Elnajjar P, Heller M, Jarosz R, Erickson BJ, Kirk S, et al. The cancer genome atlas kidney renal clear cell carcinoma collection (TCGA-KIRC) (Version 3). The Cancer Imaging Archive. 2016. [Google Scholar]
18. Freer-Smith C, Harvey-Kelly L, Mills K, Harrison H, Rossi SH, Griffin SJ, et al. Reasons for intending to accept or decline kidney cancer screening: thematic analysis of free text from an online survey. BMJ Open. 2021;11(5):e044961. doi:10.1136/bmjopen-2020-044961. [Google Scholar] [PubMed] [CrossRef]
19. Li T, Xu Y, Wu T, Charlton JR, Bennett KM, Al-Hindawi F. BlobCUT: a contrastive learning method to support small blob detection in medical imaging. Bioengineering. 2023;10(12):1372. doi:10.3390/bioengineering10121372. [Google Scholar] [PubMed] [CrossRef]
20. Parikh N, Do HH, Choi JH. Medical imaging modalities: a review. Int J Biomed Imaging. 2021;20:1–12. doi:10.1155/2021/3451234. [Google Scholar] [CrossRef]
21. Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, et al. The KiTS19 challenge data: 300 kidney tumor cases with clinical context, CT semantic segmentations, and surgical outcomes. arXiv:1904.00445. 2019. [Google Scholar]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools