 Open Access
Open Access
ARTICLE
Structure-Based Virtual Sample Generation Using Average-Linkage Clustering for Small Dataset Problems
School of Management, National Taiwan University of Science and Technology, No. 43, Sec. 4, Keelung Rd., Taipei, 106335, Taiwan
* Corresponding Author: Chih-Chieh Chang. Email:
Computers, Materials & Continua 2026, 87(1), 34 https://doi.org/10.32604/cmc.2025.073177
Received 12 September 2025; Accepted 25 December 2025; Issue published 10 February 2026
Abstract
Small datasets are often challenging due to their limited sample size. This research introduces a novel solution to these problems: average linkage virtual sample generation (ALVSG). ALVSG leverages the underlying data structure to create virtual samples, which can be used to augment the original dataset. The ALVSG process consists of two steps. First, an average-linkage clustering technique is applied to the dataset to create a dendrogram. The dendrogram represents the hierarchical structure of the dataset, with each merging operation regarded as a linkage. Next, the linkages are combined into an average-based dataset, which serves as a new representation of the dataset. The second step in the ALVSG process involves generating virtual samples using the average-based dataset. The research project generates a set of 100 virtual samples by uniformly distributing them within the provided boundary. These virtual samples are then added to the original dataset, creating a more extensive dataset with improved generalization performance. The efficacy of the ALVSG approach is validated through resampling experiments and t-tests conducted on two small real-world datasets. The experiments are conducted on three forecasting models: the support vector machine for regression (SVR), the deep learning model (DL), and XGBoost. The results show that the ALVSG approach outperforms the baseline methods in terms of mean square error (MSE), root mean square error (RMSE), and mean absolute error (MAE).Keywords
Recent advances in machine learning and artificial intelligence have enabled powerful data-driven models in many domains. However, these models typically rely on large, representative datasets. In many practical scenarios, only a small number of samples are available due to high data collection costs, privacy regulations, or the rarity of the underlying phenomenon. Such small datasets often lead to overfitting, poor generalization, and unstable model behavior.
Virtual sample generation (VSG) has emerged as a promising strategy to mitigate small-data limitations by synthetically augmenting the training set. Early work by Cho and Cha [1] introduced the idea of generating virtual samples in population networks, and Niyogi et al. [2] showed that prior knowledge–driven virtual examples can improve object recognition accuracy. Chen et al. [3] proposed a particle swarm optimization–based VSG (PSOVSG) to enhance forecasting models trained on small datasets. More recently, He et al. [4] developed t-SNE-VSG for data-driven soft sensors, demonstrating substantial accuracy gains in data-scarce industrial settings. Several other variants and applications of VSG have also been reported in the literature [5,6]. Together, these studies confirm that well-designed virtual samples can effectively reinforce learning in small-data regimes. In related small-data applications, clustering-based approaches have also been shown to play an important role in learning from limited samples.
In this work, we focus on two common small-data conditions. The first is the genuinely low-sample setting, where the available dataset is too small to capture sufficient variability, as in rare-disease studies with only a handful of cases. The second is the high-dimensional, small-sample scenario, where the number of attributes is large relative to the number of instances. In both cases, the core difficulty lies in the limited information content of the original data set. Recent studies on decision support systems have emphasized that, in the presence of data scarcity, transparent and interpretable modeling is essential for reliable decision-making. To address these challenges, we propose an average-linkage virtual sample generation method (ALVSG) that explicitly exploits the underlying data structure before generating virtual samples.
Cluster analysis provides a natural tool for uncovering latent structure in data by grouping similar instances [7]. Li et al. [8], for example, used DBSCAN to reveal structure and improve prediction on small datasets. Inspired by such structure-aware approaches, ALVSG employs hierarchical clustering with average linkage (UPGMA) to construct a dendrogram of the original data. From the merging process, we derive an average-based representation that reflects how frequently each data point participates in cluster formation. This representation is then used as a sampling prior to generate virtual samples within data-driven attribute bounds. The virtual samples are finally combined with the original data to form an enriched training set that can be used with arbitrary predictive models.
We evaluate the proposed ALVSG method on two real small datasets. The first concerns medical records for predicting the success of radiotherapy treatment for bladder cancer cells, while the second involves multi-layer ceramic capacitors (MLCCs) commonly used in electronic devices. We benchmark ALVSG against baseline models using support vector regression (SVR), a deep learning model (DL), and XGBoost, and assess performance using mean absolute error (MAE), mean square error (MSE), root mean square error (RMSE), and paired statistical
The rest of this paper is organized as follows. Section 2 reviews related work on small datasets, virtual sample generation, and forecasting models. Section 3 presents the proposed ALVSG methodology. Section 4 reports the experimental setup and results on the two case-study datasets. Section 5 concludes the paper and outlines directions for future work.
Contributions
This paper makes the following contributions: (i) we propose ALVSG, a structure-informed virtual sample generator that derives average-based weights from agglomerative average-linkage (UPGMA) clustering and uses them as an interpretable sampling prior; (ii) we bound the generator using data-driven
This section reviews prior work related to small-dataset learning, virtual sample generation (VSG), and the forecasting models used to evaluate the proposed ALVSG method.
Small datasets pose fundamental challenges for machine learning models, including high variance, overfitting, and unreliable generalization [9]. Recent surveys have examined this “small-data dilemma” from different disciplinary perspectives. For example, Xu et al. [10] summarized how data scarcity limits model performance in materials science and grouped existing remedies into three levels: data-source level (e.g., database construction, high-throughput computation), algorithm level (e.g., imbalanced learning, specialized small-data models), and learning-strategy level (e.g., active learning, transfer learning). These reviews highlight that small-data issues are widespread and require both data-centric and model-centric solutions. In addition to methodological studies, practical machine learning workflows have also been developed to accommodate small and heterogeneous datasets; for instance, Zhang and Deng introduced a data-driven machine learning interface for materials science that explicitly targets limited-sample settings and supports model development under data scarcity [11].
In practice, small datasets arise in many engineering and scientific problems, such as rare-disease analysis, optimization of real-world engineering systems [12]. When the number of instances is limited relative to the dimensionality, it becomes difficult to extract stable patterns and to build robust predictive models. Various approaches have been explored to mitigate these issues, including dimensionality reduction (e.g., linear discriminant analysis, LDA [13]) and structure-aware data partitioning for classification under class imbalance [14]. Complementary to these methods, virtual sample generation has emerged as an effective strategy to augment small datasets, and it is the main focus of this work.
Virtual sample generation (VSG) aims to enrich small datasets with synthetic samples that are consistent with the underlying data distribution. Early work, Niyogi et al. [2] introduced the basic idea of generating virtual examples to improve network training and object recognition. These pioneering studies established VSG as a viable tool for strengthening learning under data scarcity.
Subsequent research has proposed more sophisticated VSG mechanisms tailored to specific domains. Li et al. [8] used a mega-trend diffusion membership function that applies DBSCAN clustering and fuzzy membership functions to construct new attributes for small datasets. Chen et al. [3] developed PSOVSG, which employs particle swarm optimization to generate virtual samples that improve forecasting performance on small data. Zhu et al. He et al. [4] introduced t-SNE-VSG, which interpolates manifold features obtained from t-SNE and estimates virtual outputs via random forests to enhance soft-sensor performance in process industries. More recent work has further advanced VSG toward adaptive and statistically constrained frameworks. Zhu et al. [15] presented a co-training-based VSG (CTVSG) that employs two
Compared with these methods, the proposed ALVSG adopts a simpler, purely structure-informed strategy. Instead of relying on complex surrogate models or deep generative networks, ALVSG uses agglomerative average-linkage (UPGMA) hierarchical clustering to construct a dendrogram of the original data. From the merging process, we derive an average-based representation that quantifies how frequently each instance contributes to cluster formation. This representation serves as an interpretable sampling prior for generating virtual samples within data-driven attribute bounds. As a result, ALVSG provides a lightweight, model-agnostic, and interpretable VSG mechanism that is particularly suitable for small tabular datasets.
To evaluate the effectiveness of ALVSG, we consider representative forecasting models that are commonly adopted in practical prediction tasks under limited data conditions. Motivated by such practical settings, we consider three representative forecasting models for tabular regression: support vector regression (SVR), a feed-forward deep learning (DL) regressor, and XGBoost. These models span margin-based learning, neural networks, and tree-based ensembles, and they are widely used in practice due to their strong performance on small to medium-sized structured datasets. Their behavior on original vs. ALVSG-augmented data provides a comprehensive benchmark for our method.
2.3.1 Support Vector Regression (SVR)
Support vector machines (SVMs) were originally proposed for classification based on the principle of structural risk minimization, aiming to find a maximum-margin separating hyperplane in a transformed feature space. Support vector regression (SVR) extends this idea to regression by introducing an
that is as flat as possible while keeping prediction errors within an
Here
Deep learning models based on feed-forward neural networks can approximate complex nonlinear relationships by stacking multiple layers of linear transformations and nonlinear activation functions. In this study, we employ a fully connected multilayer perceptron (MLP) as a generic deep learning regressor. The network consists of an input layer, several hidden layers with rectified linear unit (ReLU) activations, and a single output neuron for regression. Model parameters are learned by minimizing a mean-squared-error loss using gradient-based optimization with backpropagation. Although deep networks are often associated with large datasets, carefully regularized shallow architectures can still serve as competitive baselines in small-data scenarios.
XGBoost is a gradient boosting framework that builds an ensemble of regression trees in a stage-wise manner, optimizing a regularized objective that balances data fit and model complexity. At each iteration, a new tree is added to correct the residual errors of the current ensemble, and explicit regularization terms on leaf weights and tree structure help prevent overfitting. XGBoost has demonstrated strong performance on a wide range of structured-data tasks and is particularly effective when the number of samples is limited but informative features are available. In our experiments, we use XGBoost as a representative tree-based ensemble to assess how ALVSG-augmented data affect boosted decision-tree models.
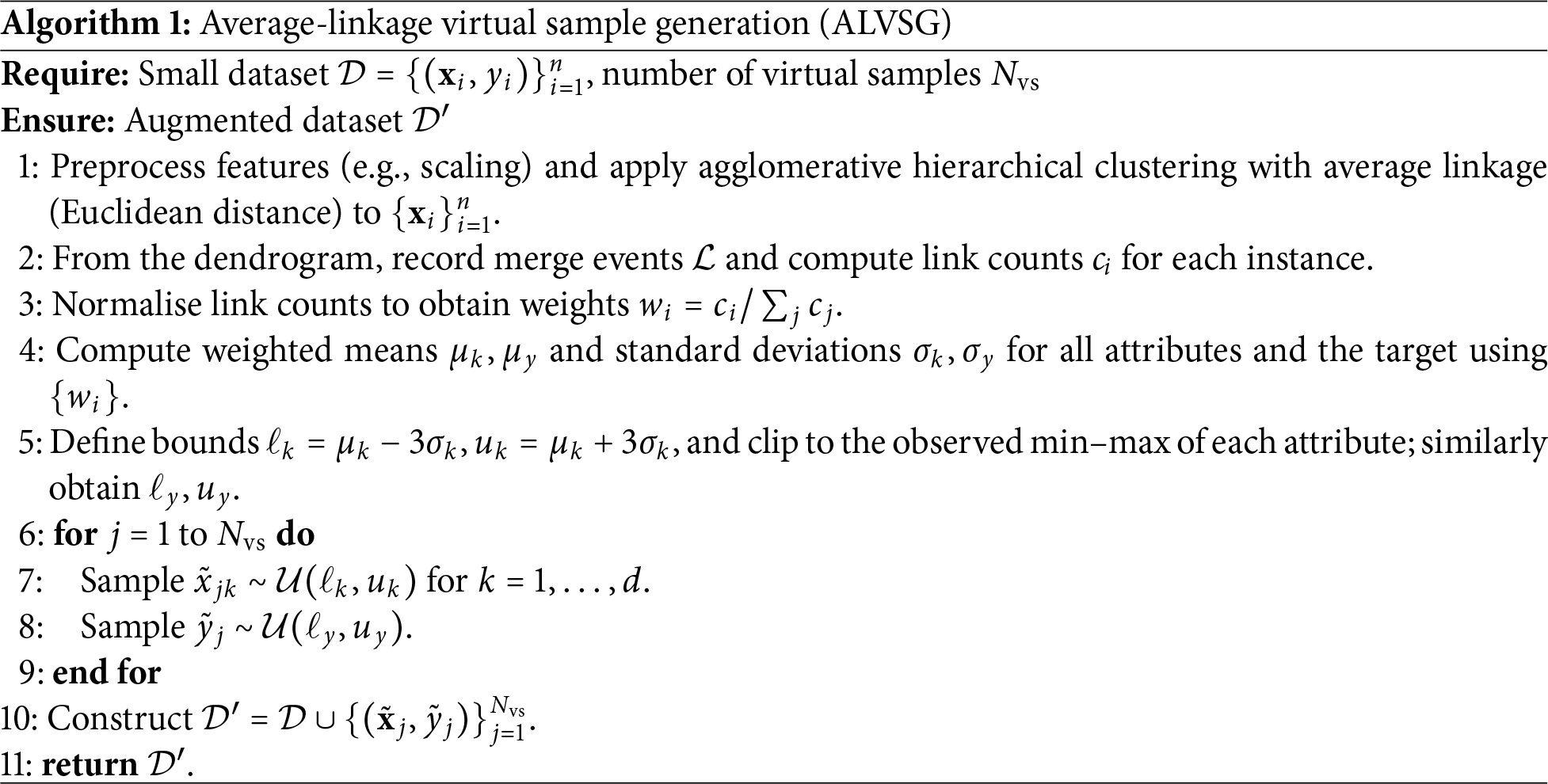
3 Methodology: Average-Linkage Virtual Sample Generation (ALVSG)
This section describes the proposed average-linkage virtual sample generation (ALVSG) method. Given a small regression dataset
3.1 Average-Linkage Clustering and Notation
To exploit the latent structure of the small dataset, we first apply agglomerative hierarchical clustering with average linkage (UPGMA) using the Euclidean distance. Each instance
Let
that is, the number of linkages in which instance
which serve as a structure-informed importance measure for each data point.
3.2 Average-Based Representation
Instead of working directly with the original dataset, ALVSG constructs an average-based representation that emphasizes structurally important instances. Conceptually, this can be viewed as forming a “virtual” dataset in which each instance
For each attribute
and the corresponding weighted standard deviations
These statistics summarise the average-based dataset implied by the hierarchical clustering structure without explicitly materialising replicated samples. They will be used to define the sampling region for virtual samples in the next step.
The goal of virtual sample generation is to sample new points that (1) respect the empirical scale of each attribute and (2) preserve the structure-informed variability captured by the weights
For each attribute
Under mild distributional assumptions, approximately 99.7% of the mass lies within
Given the hyper-rectangle
we generate
In this study, we set
For the output variable, two options are commonly used in VSG frameworks: (1) assigning virtual outputs via a surrogate model, or (2) sampling directly from a bounded distribution. In this work, we follow the latter and independently sample
The resulting virtual samples
Algorithm 1 summarises the overall ALVSG procedure.
In the experiments (Section 4), we compare forecasting models trained on the original dataset
We empirically evaluate the proposed ALVSG method on two real small datasets. The first case concerns radiotherapy response in bladder cancer cell lines, where the goal is to predict resistance to Cobalt-60 treatment from protein expression profiles. The second case involves multilayer ceramic capacitors (MLCC), where the task is to predict the K-value of ceramic powder based on process and material descriptors. In both cases, we compare baseline models (SVR, deep learning, XGBoost) with their ALVSG-augmented counterparts across a range of small training sizes.
4.1 Experimental Setup and Parameter Settings
Table 1 summarizes the hyperparameters used in this study. For SVR we set
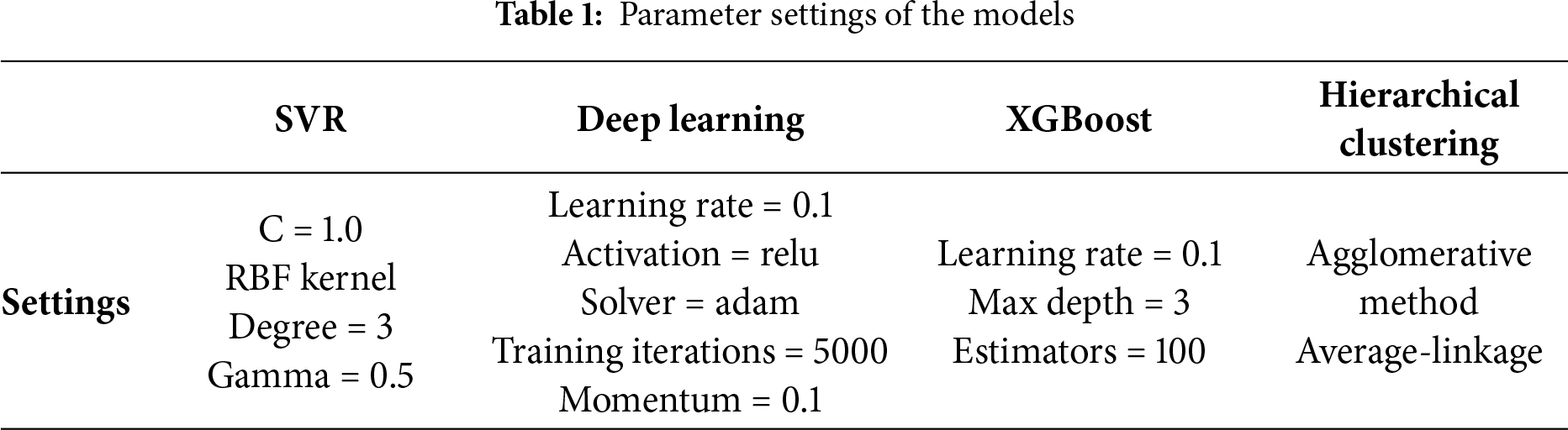
All features are standardized (zero mean, unit variance) using statistics computed on the training split only. To emulate small-dataset regimes, we consider training sizes
Unless otherwise specified, all statistical comparisons between baseline and ALVSG-augmented models (SVR, DL, XGBoost) use a two-tailed paired
4.2 Case 1: Radiotherapy Treatment of Bladder Cancer
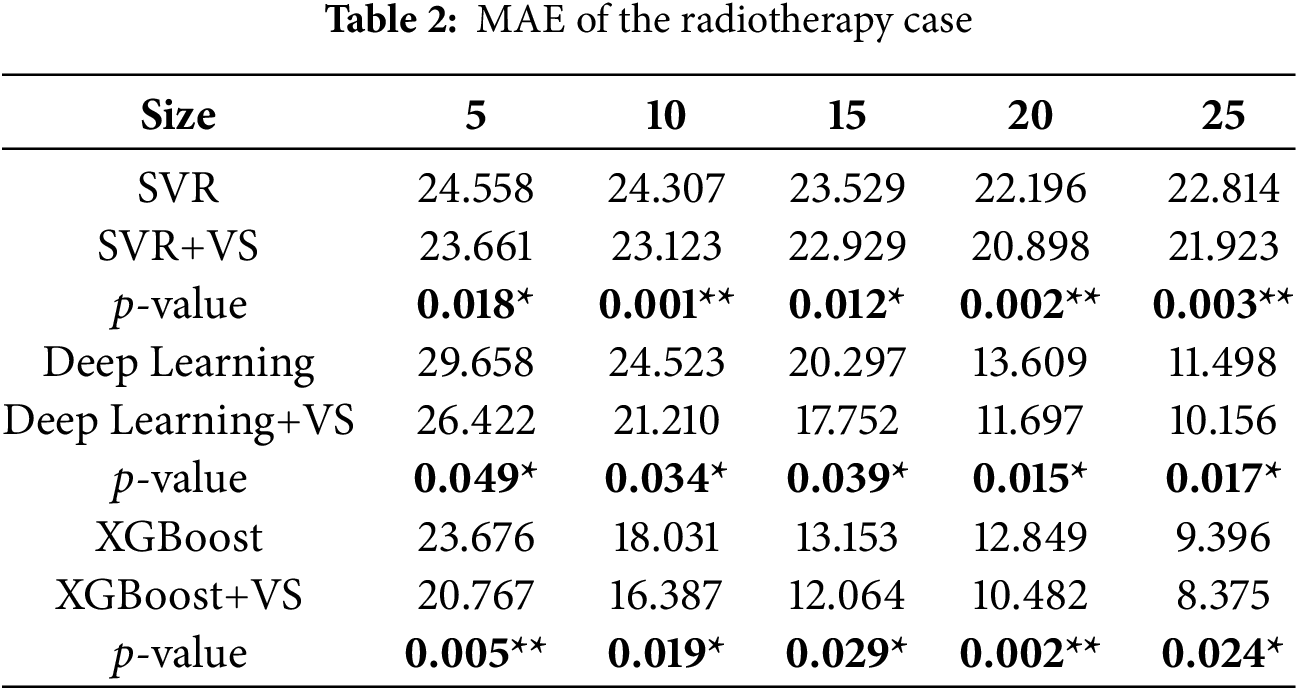
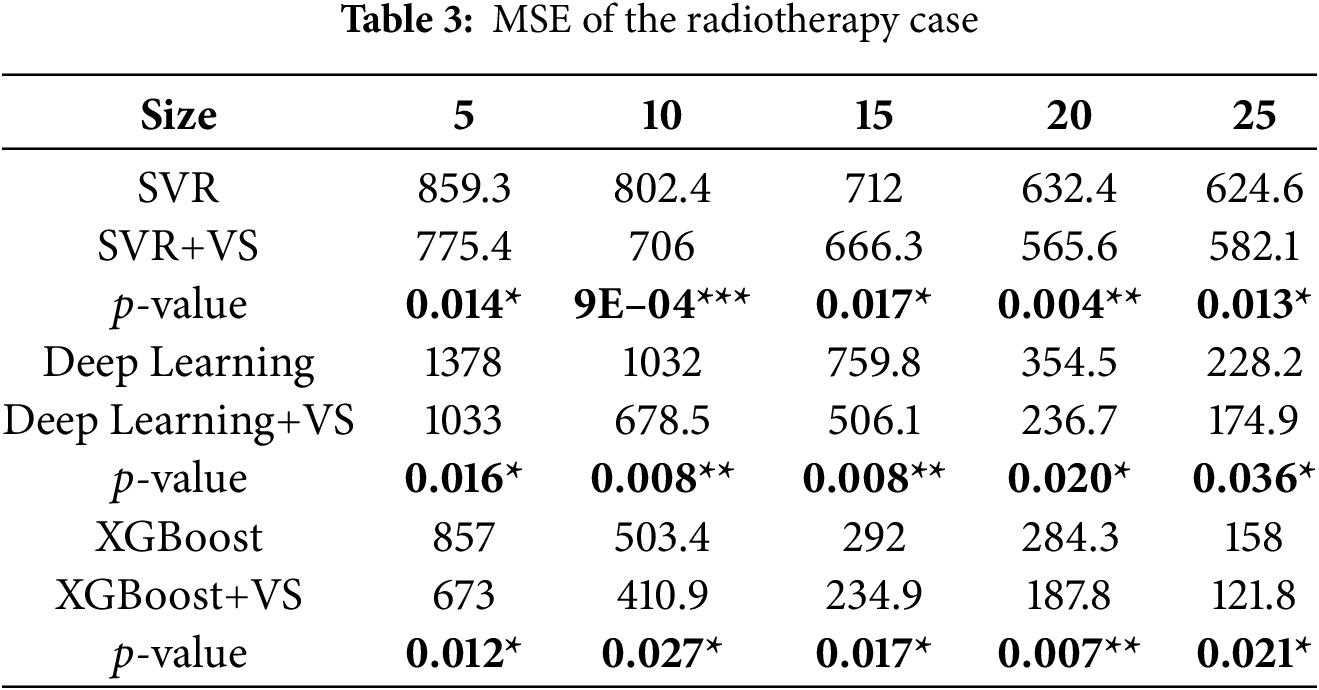
Radiotherapy is a common non-surgical treatment for bladder cancer that uses high-energy radiation to destroy tumor cells. In the dataset considered here, nine immortal bladder cancer cell lines were subjected to Cobalt-60 treatment at doses of 5, 10, 20, and 30 Gy. Each observation consists of the expression levels of thirteen proteins related to radiotherapy resistance (MDR, Topo II, EGFR, Neu, c-ErbB-3, c-ErbB-4, cyclin A, cyclin D1, Cdc2, Bcl2, Rb, P16, Bax) plus two additional inputs, for a total of fifteen inputs and one continuous output representing resistance to radiotherapy. The dataset contains 36 valid instances; further details can be found in Chao et al. [22]. Complete per-resample results are omitted here for brevity but are available from the corresponding author upon request.
Results
Tables 2–4 report MAE, MSE, and RMSE for SVR, DL, and XGBoost with and without ALVSG across training sizes. For all three metrics, error decreases as the training size
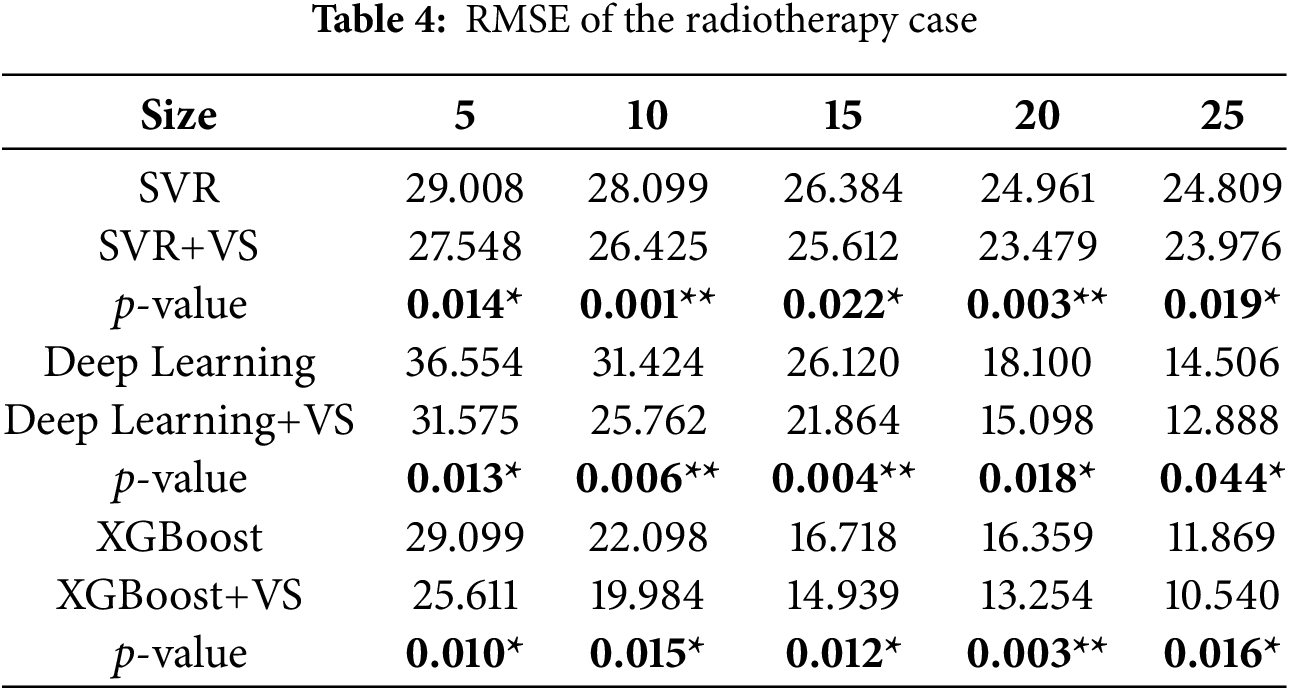
4.3 Case 2: Multilayer Ceramic Capacitors (MLCC)
Multilayer ceramic capacitors (MLCC) are widely used in electronic devices due to their high efficiency and compact form factor. Ceramic powder, a key material in MLCCs, accounts for about 40% of the overall production cost, and its batch-to-batch variability can substantially affect the dielectric constant (K-value) and downstream yield. Manufacturers typically perform pilot runs for each new powder batch to measure the K-value, which increases lead time and cost. In this case study, the goal is to predict the K-value of AD143 ceramic powder from twelve input variables, including surface area (SA), particle size distribution (PSD-90, PSD-50, PSD-10), moisture (Mois), sintering temperature (Sinter Temp), potassium content (K), dissipation factor (DF), and several temperature-coefficient–related descriptors (TC-min, TC-max, TC-peak, D-50). The dataset contains 44 pilot runs. Recent work has shown that ensemble models such as XGBoost are effective for MLCC reliability and degradation modeling [23], providing additional motivation for including XGBoost as a benchmark in this study. Per-resample results are omitted here for brevity; detailed metrics for all resamples can be obtained from the authors upon request.
Results
Tables 5–7 summarize MAE, MSE, and RMSE for the MLCC case. As in the radiotherapy case, ALVSG consistently improves all three models across all training sizes. For SVR, DL, and XGBoost, the +VS variants yield lower errors than their baselines, and the corresponding
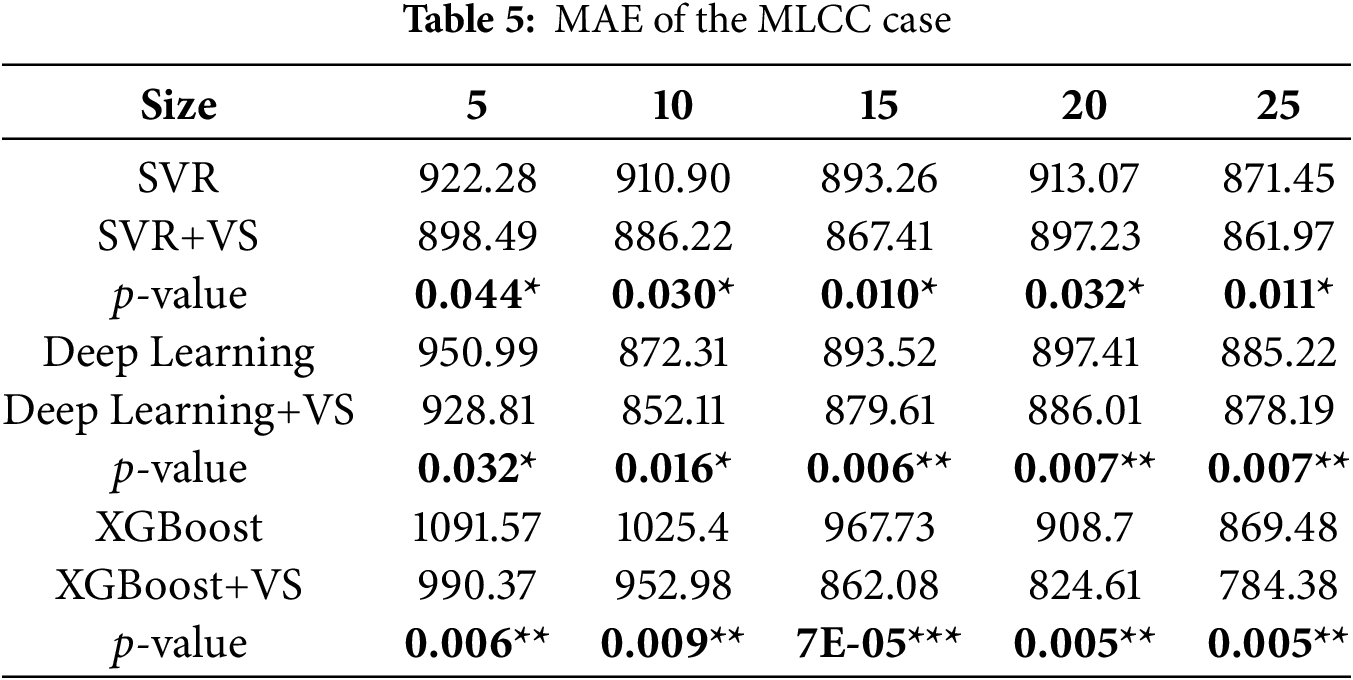
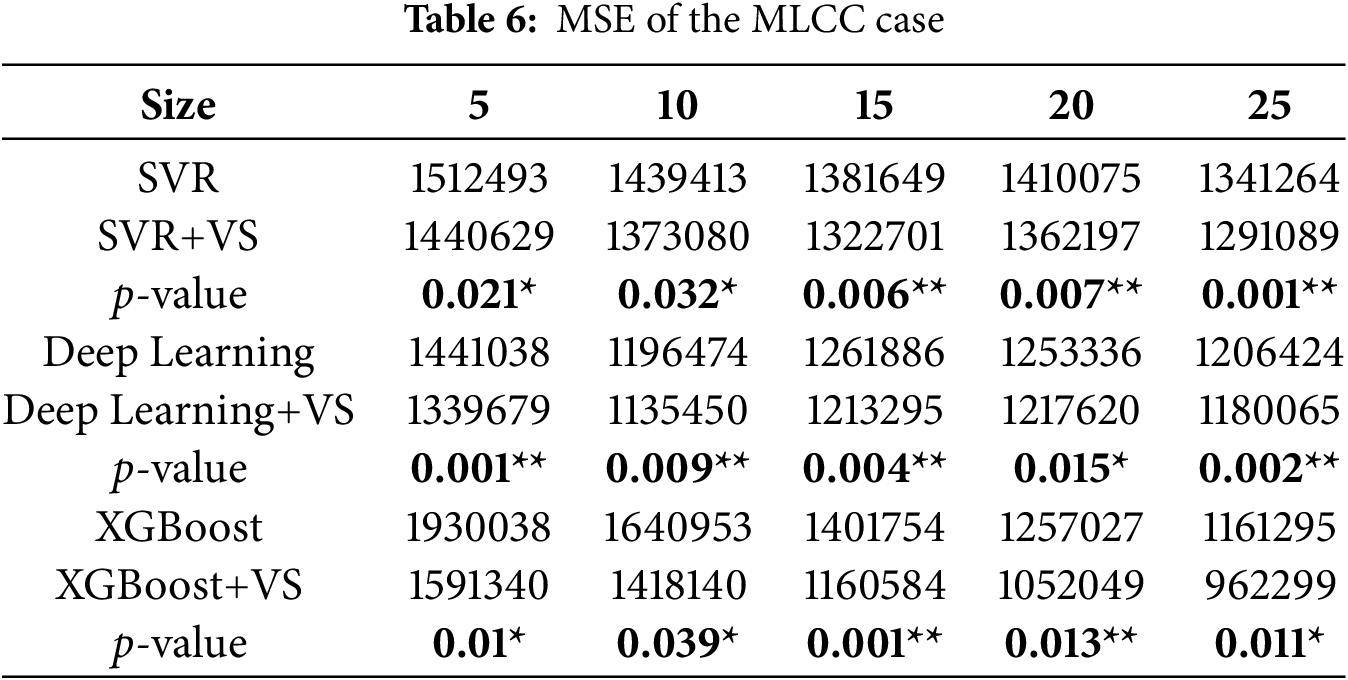
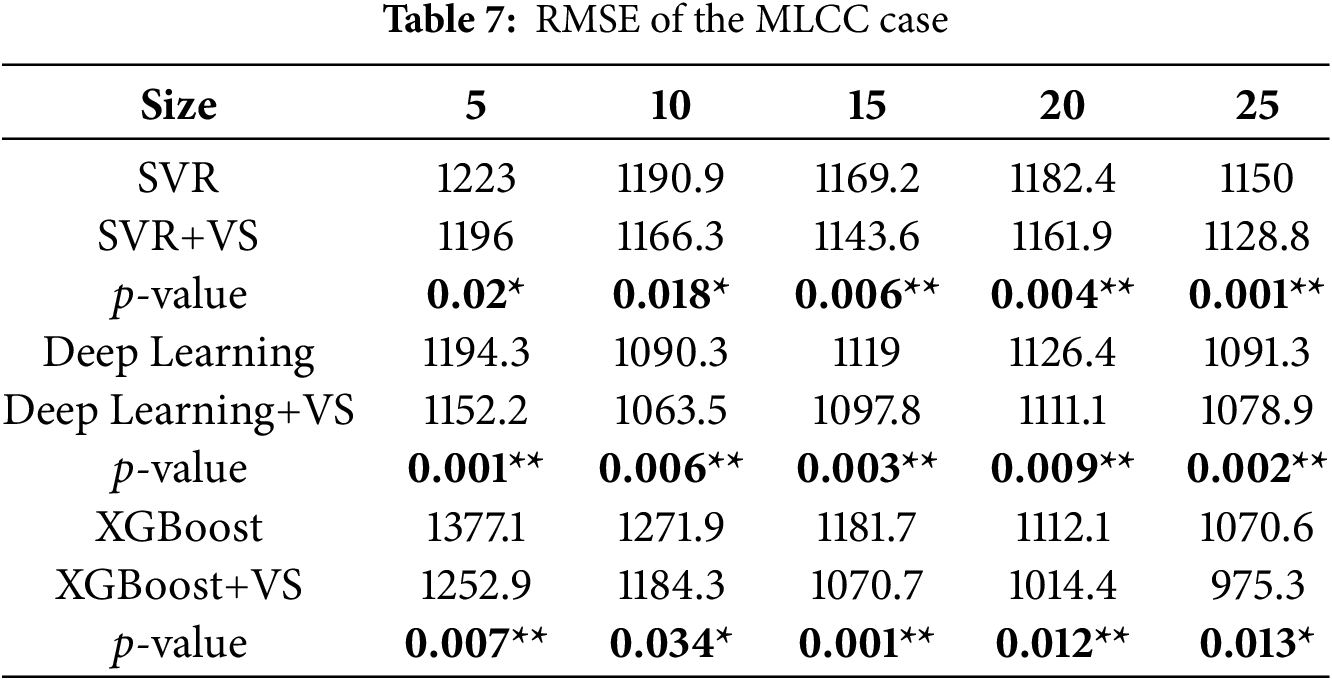
4.4 Baselines and Alternatives (Discussion)
Virtual sample generation can be realized via multiple paradigms. Manifold/interpolation-based t-SNE-VSG [4] leverages low-dimensional embeddings; GAN/active-learning hybrids [16] and statistically constrained APS-VSG [17] emphasize generative fidelity and validity; and co-training VSG [15] iteratively accepts samples passing consistency checks. Compared with these, ALVSG is (i) structure-aware yet lightweight, relying only on average-linkage dendrograms; (ii) hyperparameter-lean (no adversarial training or heavy surrogates); and (iii) transparent, since link counts translate directly into sampling weights. On the two real small datasets, ALVSG consistently improves three diverse predictors under paired resampling. A full ablation against the above generative families would be valuable but data-hungry; we therefore leave a calibrated, multi-dataset head-to-head as future work and position ALVSG as a strong, interpretable baseline for very small tabular
This paper proposed average-linkage virtual sample generation (ALVSG), a structure-aware yet lightweight approach for small tabular regression datasets. ALVSG first applies agglomerative hierarchical clustering with average linkage and turns dendrogram link counts into instance weights, yielding an average-based representation that emphasizes structurally central points. It then defines conservative, data-driven
The method was evaluated on two real small-data cases: radiotherapy response in bladder cancer and K-value prediction for multilayer ceramic capacitors (MLCC). Across both datasets, three forecasting models (SVR, deep learning, XGBoost), and five training sizes (
Overall, ALVSG shows that exploiting hierarchical clustering structure to construct a weighted representation, combined with bounded uniform sampling, is an effective and practical way to improve predictive performance when data are scarce. Future work will extend the evaluation to additional domains and compare ALVSG head-to-head with more complex virtual-sample generators under carefully controlled small-data benchmarks.
Practical Implications and Limitations
In practice, ALVSG is most useful when (i) the number of samples is very small (
Limitations include: (a) the method does not explicitly model the full data density; (b) uniform sampling within
Acknowledgement: This research was supported by the National Science and Technology Council (NSTC), Taiwan.
Funding Statement: This work received funding support from the National Science and Technology Council (NSTC), Taiwan, under Grant No. 114-2410-H-011-026-MY3.
Author Contributions: Chih-Chieh Chang, Khairul Izyan Bin Anuar and Yu-Hwa Liu contributed to the writing of the main sections, including the introduction, literature review, methodology, experiments, and conclusion. Chih-Chieh Chang and Khairul Izyan Bin Anuar organized the related work and implemented the experimental setup. Chih-Chieh Chang and Yu-Hwa Liu revised the manuscript and discussed the experimental results. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: No datasets were generated or analyzed during the current study.
Ethics Approval: This paper does not contain any studies with human participants or animals performed by any of the authors.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Cho S, Cha K. Evolution of neural network training set through addition of virtual samples. In: Proceedings of IEEE International Conference on Evolutionary Computation; 1996 May 20–22; Nagoya, Japan. p. 685–8. [Google Scholar]
2. Niyogi P, Girosi F, Poggio T. Incorporating prior information in machine learning by creating virtual examples. Proc IEEE. 1998;86(11):2196–209. doi:10.1109/5.726787. [Google Scholar] [CrossRef]
3. Chen ZS, Zhu B, He YL, Yu LA. A PSO based virtual sample generation method for small sample sets: applications to regression datasets. Eng Appl Artif Intell. 2017;59:236–43. doi:10.1016/j.engappai.2016.12.024. [Google Scholar] [CrossRef]
4. He YL, Hua Q, Zhu QX, Lu S. Enhanced virtual sample generation based on manifold features: applications to developing soft sensor using small data. ISA Trans. 2022;126(4):398–406. doi:10.1016/j.isatra.2021.07.033. [Google Scholar] [PubMed] [CrossRef]
5. Wedyan M, Crippa A, Al-Jumaily A. A novel virtual sample generation method to overcome the small sample size problem in computer aided medical diagnosing. Algorithms. 2019;12(8):160. doi:10.3390/a12080160. [Google Scholar] [CrossRef]
6. Zhu QX, Hou KR, Chen ZS, Gao ZS, Xu Y, He YL. Novel virtual sample generation using conditional GAN for developing soft sensor with small data. Eng Appl Artif Intell. 2021;106(2):104497. doi:10.1016/j.engappai.2021.104497. [Google Scholar] [CrossRef]
7. Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit Lett. 2010;31(8):651–66. doi:10.1016/j.patrec.2009.09.011. [Google Scholar] [CrossRef]
8. Li DC, Chang CC, Liu CW. Using structure-based data transformation method to improve prediction accuracies for small data sets. Decis Support Syst. 2012;52(3):748–56. doi:10.1016/j.dss.2011.11.021. [Google Scholar] [CrossRef]
9. Lu J, Gong P, Ye J, Zhang J, Zhang C. A Survey on Machine Learning from Few Samples. Pattern Recognit. 2023;139(2):109480. doi:10.1016/j.patcog.2023.109480. [Google Scholar] [CrossRef]
10. Xu P, Ji X, Li M, Lu W. Small data machine learning in materials science. npj Comp Mater. 2023;9(42):42. doi:10.1038/s41524-023-01000-z. [Google Scholar] [CrossRef]
11. Zhang L, Deng H. NJmat 2.0: user instructions of data-driven machine learning interface for materials science. Comput Mater Contin. 2025;83(1):1–11. doi:10.32604/cmc.2025.062666. [Google Scholar] [CrossRef]
12. Fan C, Hou B, Zheng J, Xiao L, Yi L. A surrogate-assisted particle swarm optimization using ensemble learning for expensive problems with small sample datasets. Appl Soft Comput. 2020;91:106242. doi:10.1016/j.asoc.2020.106242. [Google Scholar] [CrossRef]
13. Sharma A, Paliwal KK. Linear discriminant analysis for the small sample size problem: an overview. Int J Mach Learn Cybern. 2015;6(3):443–54. doi:10.1007/s13042-013-0226-9. [Google Scholar] [CrossRef]
14. Doan QH, Mai SH, Do QT, Thai DK. A cluster-based data splitting method for small sample and class imbalance problems in impact damage classification. Appl Soft Comput. 2022;120:108628. doi:10.1016/j.asoc.2022.108628. [Google Scholar] [CrossRef]
15. Zhu QX, Zhang HT, Tian Y, Zhang N, Xu Y, He YL. Co-training based virtual sample generation for solving the small sample size problem in process industry. ISA Trans. 2023;134:290–301. doi:10.1016/j.isatra.2022.08.021. [Google Scholar] [PubMed] [CrossRef]
16. Cui C, Tang J, Xia H, Wang D, Yu G. Virtual Sample Generation Method Based on GAN for Process Data with Its Application. In: Proceedings of the 2022 34th Chinese Control and Decision Conference (CCDC); 2022 Aug 12–17; Hefei, China. p. 242–7. [Google Scholar]
17. Chen Z, Lv Z, Di R, Wang P, Li X, Sun X, et al. A novel virtual sample generation method to improve the quality of data and the accuracy of data-driven models. Neurocomputing. 2023;548(01):126380. doi:10.1016/j.neucom.2023.126380. [Google Scholar] [CrossRef]
18. Wang Z, Wang P, Liu K, Wang P, Fu Y, Lu CT, et al. A comprehensive survey on data augmentation. IEEE Trans Knowl Data Eng. 2026;38(1):47–66. doi:10.1109/tkde.2025.3622600. [Google Scholar] [CrossRef]
19. Dong Y, Luo M, Li J, Liu Z, Zheng Q. Semi-supervised graph contrastive learning with virtual adversarial augmentation. IEEE Trans Knowl Data Eng. 2024;36(8):4232–44. doi:10.1109/tkde.2024.3366396. [Google Scholar] [CrossRef]
20. Drucker H, Wu D, Vapnik VN. Support vector machines for spam categorization. IEEE Trans Neural Netw. 1999;10(5):1048–54. doi:10.1109/72.788645. [Google Scholar] [PubMed] [CrossRef]
21. Yang J, Yu X, Xie ZQ, Zhang JP. A novel virtual sample generation method based on Gaussian distribution. Knowl-Based Syst. 2011;24(6):740–8. doi:10.1016/j.knosys.2010.12.010. [Google Scholar] [CrossRef]
22. Chao GY, Tsai TI, Lu TJ, Hsu HC, Bao BY, Wu WY, et al. A new approach to prediction of radiotherapy of bladder cancer cells in small dataset analysis. Expert Syst Appl. 2011;38(7):7963–9. doi:10.1016/j.eswa.2010.12.035. [Google Scholar] [CrossRef]
23. Yousefian P, Sepehrinezhad A, van Duin ACT, Randall CA. Improved prediction for failure time of multilayer ceramic capacitors (MLCCsa physics-based machine learning approach. APL Mach Learn. 2023;1(3):036107. doi:10.1063/5.0221988. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools