 Open Access
Open Access
ARTICLE
A Hybrid Vision Transformer with Attention Architecture for Efficient Lung Cancer Diagnosis
1 Department of Computer Science, Abdul Wali Khan University, Mardan, 23200, Pakistan
2 Department of Computer and Networks Engineering, Umm Alqura University, Makkah, 21955, Saudi Arabia
3 Department of Data Science, Umm Alqura University, Makkah, 21955, Saudi Arabia
4 School of Computer Science and Engineering, Yeungnam University, Gyeongsan, 38541, Republic of Korea
5 Department of Signal Theory and Communications, University of Valladolid, Valladolid, 47011, Spain
6 Universidad Internacional Iberoamericana, Campeche, 24560, México
* Corresponding Authors: Farhan Amin. Email: ; Isabel de la Torre. Email:
(This article belongs to the Special Issue: Advancements in Machine Learning and Artificial Intelligence for Pattern Detection and Predictive Analytics in Healthcare)
Computers, Materials & Continua 2026, 87(1), 46 https://doi.org/10.32604/cmc.2025.073342
Received 16 September 2025; Accepted 17 November 2025; Issue published 10 February 2026
Abstract
Lung cancer remains a major global health challenge, with early diagnosis crucial for improved patient survival. Traditional diagnostic techniques, including manual histopathology and radiological assessments, are prone to errors and variability. Deep learning methods, particularly Vision Transformers (ViT), have shown promise for improving diagnostic accuracy by effectively extracting global features. However, ViT-based approaches face challenges related to computational complexity and limited generalizability. This research proposes the DualSet ViT-PSO-SVM framework, integrating a ViT with dual attention mechanisms, Particle Swarm Optimization (PSO), and Support Vector Machines (SVM), aiming for efficient and robust lung cancer classification across multiple medical image datasets. The study utilized three publicly available datasets: LIDC-IDRI, LUNA16, and TCIA, encompassing computed tomography (CT) scans and histopathological images. Data preprocessing included normalization, augmentation, and segmentation. Dual attention mechanisms enhanced ViT’s feature extraction capabilities. PSO optimized feature selection, and SVM performed classification. Model performance was evaluated on individual and combined datasets, benchmarked against CNN-based and standard ViT approaches. The DualSet ViT-PSO-SVM significantly outperformed existing methods, achieving superior accuracy rates of 97.85% (LIDC-IDRI), 98.32% (LUNA16), and 96.75% (TCIA). Cross-dataset evaluations demonstrated strong generalization capabilities and stability across similar imaging modalities. The proposed framework effectively bridges advanced deep learning techniques with clinical applicability, offering a robust diagnostic tool for lung cancer detection, reducing complexity, and improving diagnostic reliability and interpretability.Keywords
The global death toll from lung cancer amounts to about 1.8 million fatalities each year, according to current reports [1]. The 5-year survival rate for early-stage lung cancer is greater than that for advanced-stage lung cancer. Patients with stage IA lung cancer have a high 5-year survival rate exceeding 75% [2]. Medical experts currently manually analyze radiological and histopathological diagnostic tools to identify suspicious findings. The manual diagnostic process requires significant labor input and is subject to observer subjectivity, which diminishes its reliability [3]. The application of deep learning techniques in machine learning has transformed the way medical specialists examine images in their field of practice [3]. CNNs are the preferred choice for image recognition applications because they achieve high accuracy. The primary drawback of Convolutional Neural Networks (CNNs) is their limited ability to analyze local area patterns, which hinders their capacity to capture the broader image context [4]. Vision Transformer (ViT), which overcame this limitation by establishing effective long-range image relationships through attention mechanisms [5]. The high computational requirements of ViT designs prompt scientific teams to research low-maintenance versions of these architectural models [6]. The combination of ViT models with optimization methods that employ the PSO technique improves both diagnostic performance and simplifies feature complexity levels [7]. This research project builds on promising recent methods by designing a combined model for lung cancer diagnosis. Laboratory results show that the ViT architecture performs effectively for medical image classification, whereas the existing literature reports limited use of these architectures in lung cancer histopathology analysis. Current deep learning methods using CNNs demonstrate limited ability to extract global features, according to research by [8]. The standard configurations of ViT models require significant computational power, which limits their implementation in clinical settings due to the control of available computational resources [9]. This study implements the DualSet ViT-PSO-SVM framework to address these problems. The proposed diagnosis system achieves improved performance in lung cancer identification by leveraging ViT for efficient feature extraction, PSO for feature optimization, and SVM for reliable classification. The study enhances both clinical reliability and generalizability by merging two separate histopathological datasets. The primary objective of this study is to develop and validate a novel deep learning model for accurately diagnosing lung cancer using histopathological imaging. Specifically, the study aims to:
• To design and implement the DualSet ViT-PSO-SVM model, which integrates ViT, PSO, and SVM for efficient and robust classification.
• To integrate two different histopathological image datasets to increase model generalization and clinical utility in diverse medical settings.
• To evaluate and benchmark the proposed model’s accuracy, efficiency, and computational complexity against existing CNN-based and standard ViT methods.
• To apply explainable artificial intelligence (XAI) techniques, such as Grad-CAM, to ensure clinical interpretability and validate model decisions from a medical perspective.
Achieving these objectives will establish a clinically relevant framework that enables pathologists to make precise lung cancer diagnoses from histopathology images, thereby facilitating timely intervention and improving patient outcomes. Although hybrid methods involving deep feature access, metaheuristic feature selection, and classifiers have been evaluated in other fields of study, including cervical and breast cancer, their direct application to lung cancer remains under-researched. The CT images of the lung pose their own challenges, including high anatomical diversity, weak nodule morphology, and a tendency to overlap with benign lesions. These factors make standard CNN or Transformer pipelines ineffective. Our study helps close this gap by proposing a DualSet ViT-PSO-SVM model specifically for lung cancer detection. The dual-attention (self-attention and CBAM) version of the Vision Transformer can capture both the global and local thoracic context, as well as the finer details of the nodule. Concurrently, redundant high-dimensional ViT embeddings are removed via Particle Swarm Optimization to improve discriminative power. Lastly, Support Vector Machine offers better classification performance with limited, imbalanced training data than the softmax classifier, which is more often employed in previous work. As far as we can tell, this is the first publication to (i) translate this hybrid paradigm to the lung CT datasets and (ii) test it on three public benchmarks (LIDC-IDRI, LUNA16, and TCIA) and, as such, prove its novelty, generalizability, and clinical importance.
The rest of the paper is structured as follows: Section 2 provides a comprehensive literature review on existing lung cancer detection methods, ViT architectures in medical imaging, and the significance of attention mechanisms. Section 3 details the datasets used and the preprocessing methods, and describes the proposed DualSet ViT-PSO-SVM methodology, including the ViT architecture, dual attention mechanisms, feature optimization via PSO, and classification with SVM. Section 4 presents experimental results, including detailed performance metrics, cross-dataset analyses, Grad-CAM visualizations, and comparative evaluations against state-of-the-art models. Finally, Section 5 concludes the paper, summarizing key contributions, discussing limitations, and proposing future research directions.
2.1 Lung Cancer Detection Techniques
Lung cancer detection has traditionally involved radiological imaging and histopathology analysis. Various computational methods, including traditional image processing, deep learning-based CNN methods, and attention-based approaches, have been developed to enhance the diagnostic process. Early lung cancer detection primarily employed classical image processing techniques to extract morphological and textural features. Recent studies have highlighted the importance of advanced attention mechanisms for capturing subtle and discriminative patterns in medical images, which aligns with our approach of integrating dual attention within the ViT backbone to improve lung cancer classification. Furthermore, traditional machine learning approaches, such as SVMs, random forests, and k-NN, have been employed in computer-aided diagnosis systems for lung cancer, relying primarily on handcrafted image features for lesion classification [10]. However, these approaches heavily rely on domain-specific expert knowledge, and their performance significantly varies with imaging conditions and feature extraction quality [11]. CNNs have revolutionized lung cancer detection by automatically learning complex image features without the need for extensive manual feature engineering. Chaunzwa et al. [12] developed a deep learning framework using CNN architectures for automated classification of lung adenocarcinoma subtypes from histopathological images, demonstrating high accuracy and reduced inter-observer variability. CNNs provide superior performance compared to standard image processing, yet they face limitations due to their small receptive fields and limited ability to capture global connections in medical images [13]. Medical images have improved performance with deep learning models, as attention mechanisms have been integrated using human vision-inspired methods to detect global dependencies within these images [14]. Two benefits offered by attention mechanisms are improvements in diagnostic accuracy for complex tasks, such as cancer classification, and enhanced regulatory capabilities to focus on essential points of interest. Guan et al. [15] developed an attention-guided CNN for pulmonary nodule detection, achieving superior accuracy compared to conventional CNN models by effectively highlighting critical regions for precise localization and diagnosis. Transformer architectures employing self-attention mechanisms have recently been applied to lung cancer diagnosis tasks, enabling superior extraction of global features and improved interpretability [16]. Research on optimized and lightweight variants of Transformer models for clinical use continues due to their promising performance, though they usually encounter challenges of computational complexity and training efficiency [17].
Medical imaging analysis has made significant progress with the introduction of ViTs, inspired by language models, which offer strong capabilities for extracting global features. Medical diagnostics benefit from self-attention mechanisms in ViTs because these mechanisms directly analyze global context, which enables the capture of crucial image relationships [18]. Several research studies delivered successful results by implementing ViT models for medical imaging tasks. Khan et al. [19] created a ViT-based approach that identified retinal diseases in fundus images, surpassing other CNN-based standards for accuracy. The study demonstrated that ViT outperformed in diagnosing features spanning large image areas. The research showed that ViT models outperform standard CNN architectures in recognizing subtle distinctions in histopathological structures by effectively capturing global information patterns [20]. Despite their promise, ViT-based models face significant hurdles in medical applications: they often demand large annotated datasets and considerable computational resources, limiting their usability in clinical environments [21]. Recent research efforts focus on building lightweight, efficient Transformer models. Medical imaging transforms with ViT, as this method enhances feature representation compared to CNNs. ViT models require further research to optimize their clinical application, balancing efficient computing power with diagnostic quality in medical imaging. In addition to Transformer architectures, new methods in collaborative and distributed learning have been created. These frameworks have also been suggested to use a weighted federated learning process that meets privacy-protecting training over multi-center datasets and considers heterogeneity among local clients [22]. This type of approach is especially applicable to clinical imaging, where data sharing is limited. Despite such developments, few studies combine ViT representations with metaheuristic feature selection and classical classifiers to systematically diagnose lung cancer and validate their results across various public CT and histopathology datasets.
2.3 Attention Mechanisms in Deep Learning
Attention mechanisms, developed initially to enhance the performance of sequence-to-sequence models in natural language processing, have become fundamental components in improving deep learning performance across diverse fields, including medical imaging [23]. These mechanisms allow neural networks to dynamically focus on specific regions of input data, facilitating more informative and accurate predictions. In deep learning, attention methods primarily fall into two categories: spatial attention and channel attention. Spatial attention enables models to focus on critical spatial locations in feature maps, significantly improving accuracy in medical image tasks, such as lesion detection and image segmentation [24]. Channel attention enables networks to boost their representational power by weighting features based on their significance, thereby highlighting essential feature channels [25]. Recently, self-attention mechanisms, popularized by the Transformer architecture, have attracted significant attention for their ability to capture long-range dependencies in images, thereby overcoming the locality constraints typical of CNNs. Despite their clear benefits, attention mechanisms typically require increased computational resources, motivating the development of lightweight variants. Several deep learning approaches have been proposed for lung cancer analysis, ranging from CNN-based models to more recent Transformer architectures. Early studies, such as Multi-view CNN [26], leveraged convolutional architectures to capture spatial information, with the latter extending evaluation across multiple datasets. DenseNet-based CNN [27] improved feature reuse but remained limited in scalability and lacked explicit attention modeling. Subsequent research began incorporating attention mechanisms to enhance feature discrimination, as seen in the Attention-based CNN [28], which also included multi-dataset validation and basic explainability using visual interpretation. More advanced architectures, such as the Swin Transformer [29] and Vanilla ViT [30], introduced self-attention and hierarchical feature learning, enabling improved global context modeling; however, these studies did not integrate feature optimization techniques, such as PSO or alternative classifiers, like SVM. Their ongoing integration within medical image analysis continues to drive advances toward accurate, reliable, and interpretable diagnostic systems. A comparative summary of these methods with respect to Vision Transformer usage, attention mechanisms, dataset scope, feature optimization, classifier design, and explainability is provided in Table 1.
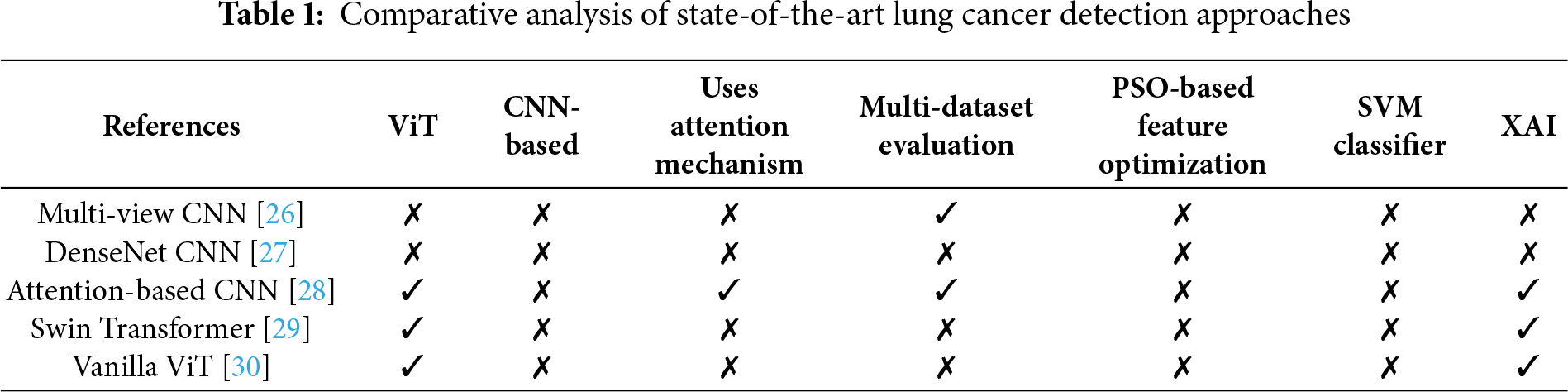
The proposed system combines Vision Transformers (ViT), Particle Swarm Optimization (PSO), and Support Vector Machines (SVM) in a three-phase pipeline. The ViT backbone receives first input images, which are then processed using dual attention (self-attention and CBAM) to generate high-dimensional feature embeddings. A comprehensive feature vector of the global CLS token representation is constructed by combining the CBAM-enhanced feature map and the global CLS token representation. Second, this feature set is optimized using Particle Swarm Optimization, which selects a small subset of features by maximizing classification fitness on the validation set, thereby reducing redundancy and computational complexity. PSO is initialized with a population of candidate feature subsets, each evaluated by its classification accuracy, and is refined to yield the best solution. Lastly, the selected features are fed into a Support Vector Machine classifier to construct decision boundaries between benign and malignant cases. The SVM uses a radial basis function kernel, with hyperparameters optimized within each fold to prevent data leakage. Such sequential integration is such that ViT provides feature richness, PSO provides feature efficiency, and SVM provides strong classification capabilities. The model’s interpretation process relied on the Gradient-weighted Class Activation Mapping (Grad-CAM) visualization technique, as displayed in Fig. 1.
Figure 1: Proposed DualSet ViT-PSO-SVM architecture
This research works with three existing publicly available lung cancer image databases named LIDC-IDRI [31], LUNA16 [32], and TCIA [33] Lung Cancer datasets. This study uses various annotated lung image datasets to enable researchers to evaluate the DualSet ViT-PSO-SVM framework. The Lung Image Database Consortium Image Collection (LIDC-IDRI) incorporates multiple clinical institution-based thoracic CT scans for diagnostic and screening use. One thousand eighteen patient images, with annotation completed by experienced radiologists, show the presence of lung nodules ranging in size from 3 mm to 30 mm. The four independent radiologist evaluators annotated nodules by supplying diagnostic contours in addition to malignancy detection and morphological description [31]. The database contains 7371 nodules in high-definition image sections measuring 512 × 512 pixels. Within the LIDC-IDRI database, the Lung Nodule Analysis 2016 (LUNA16) dataset has been specifically designed to enable lung nodule detection and nodule classification evaluation. This subset contains 888 CT scans from LIDC-IDRI, excluding those with ambiguous annotations and scans without nodules. There are 1186 positive nodules in the dataset, identified by expert radiologists, with dimensions ranging from 3 mm to 30 mm. The database contains precisely noted measurements of spatial position, together with diameter details, which serve as essential metrics for evaluating lung cancer detection methods [32]. High-resolution medical images from both CT scans and histopathological images constitute the primary data set for the The Cancer Imaging Archive (TCIA) Lung Cancer dataset. The study uses images from the TCIA lung cancer collection, which comprises high-resolution histopathological images of Non-Small Cell Lung Cancer (NSCLC). The dataset provides histological classifications and patient clinical metadata, including cancer stage and subtype annotations, facilitating research on tumor classification and prognosis. This dataset consists of 1325 images captured from 211 lung cancer patients, each image annotated by experienced pathologists [33]. These three datasets collectively provide diverse imaging data across CT and histopathology modalities, which are essential for developing a robust, generalized framework for lung cancer diagnosis.
To enhance the effectiveness of our lung cancer classification framework, images underwent preprocessing steps, including normalization, augmentation, and segmentation. These steps aim to standardize image inputs, improve model generalization, and accurately isolate regions of interest (e.g., lung nodules or cancerous tissues). Normalization standardizes pixel intensity across all images, facilitating faster convergence during model training and ensuring consistency across datasets. In this work, min-max normalization was used to scale pixel intensities to the range [0, 1]. Mathematically, this is expressed by Eq. (1):
where
Horizontal flipping was performed by reversing the image pixels horizontally by using Eq. (3).
Random zooming was executed using scaling transformations by using Eq. (4).
where
where
Post-segmentation morphological operations such as erosion and dilation were also used to refine the segmented boundaries.
The proposed DualSet ViT-PSO-SVM framework integrates a ViT architecture enhanced by dual attention mechanisms. The overall pipeline involves extracting deep visual features, optimizing them with attention modules, and performing accurate lung cancer classification.
The ViT is adopted due to its ability to model global contextual information effectively. ViT splits input images into fixed-size patches, each of which is linearly projected into an embedding space. Given an input image
where
here,
3.3.2 Dual Attention Mechanisms
To enhance feature representation, the ViT architecture integrates dual attention mechanisms: Self-Attention and Channel-Spatial Attention. Self-Attention captures global dependencies by computing attention weights using queries
where
Spatial Attention (SA) highlights informative spatial locations by using Eq. (12).
where
where
3.3.3 Integration and Fusion Strategy
The final integration strategy fuses global features from the ViT’s Self-Attention block and enhanced local features from the CBAM module. This integration creates a robust, multi-scale feature representation for classification. The fusion feature is represented mathematically as in Eq. (14).
Finally, PSO selects optimized features from
Algorithm 1 summarizes the complete workflow.
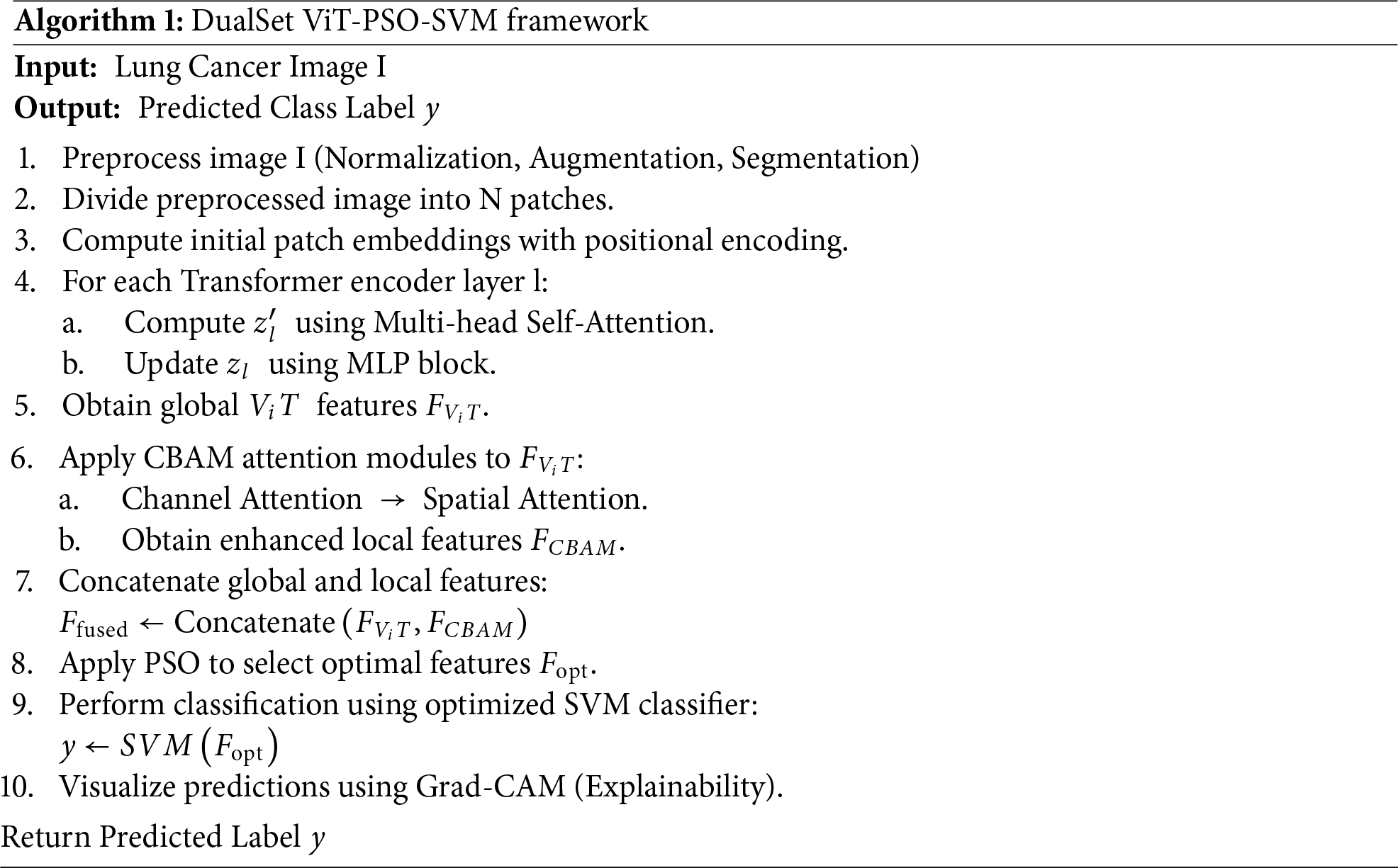
3.4 Model Training and Validation
This section outlines the procedures employed for hyperparameter tuning, model training, validation, testing, and the metrics used for evaluating the DualSet ViT-PSO-SVM framework. Hyperparameter tuning was performed systematically using grid-search combined with cross-validation. Key hyperparameters included. ViT parameters: Number of transformer layers
where
The model’s performance was quantified using multiple evaluation metrics, including accuracy (see Eq. (16)), precision (see Eq. (17)), recall (see Eq. (18)), and F1-score (see Eq. (19)), widely accepted in medical imaging classification tasks:
here,
These metrics comprehensively evaluate the model’s ability to classify lung cancer accurately from medical images.
This section provides detailed experimental evaluations of the proposed DualSet ViT-PSO-SVM model across three widely recognized lung cancer datasets: LIDC-IDRI, LUNA16, and TCIA Lung Cancer. Performance metrics include accuracy, precision, recall, and F1-score. In all experiments, the datasets were divided into 70% training, 15% validation, and 15% testing sets. The splitting was performed at the patient level to ensure reproducibility and minimize data leakage by not displaying images of the same patient in more than one subset. This method was also applied on a similar basis in all three situations of LIDC-IDRI, LUNA16, and TCIA, and the specific patient identifiers in each split. The classification capabilities of the proposed method were assessed on each dataset. Detailed results and visualization of model predictions are provided in this section.
Table 2 compares the impact of different loss functions on model performance across three datasets. The Hybrid Loss function achieves the highest accuracy, precision, recall, and F1-score, demonstrating its effectiveness in optimizing lung cancer classification. It significantly outperforms Focal Loss and Structure Loss, particularly in terms of reducing meaning errors and improving feature selection.
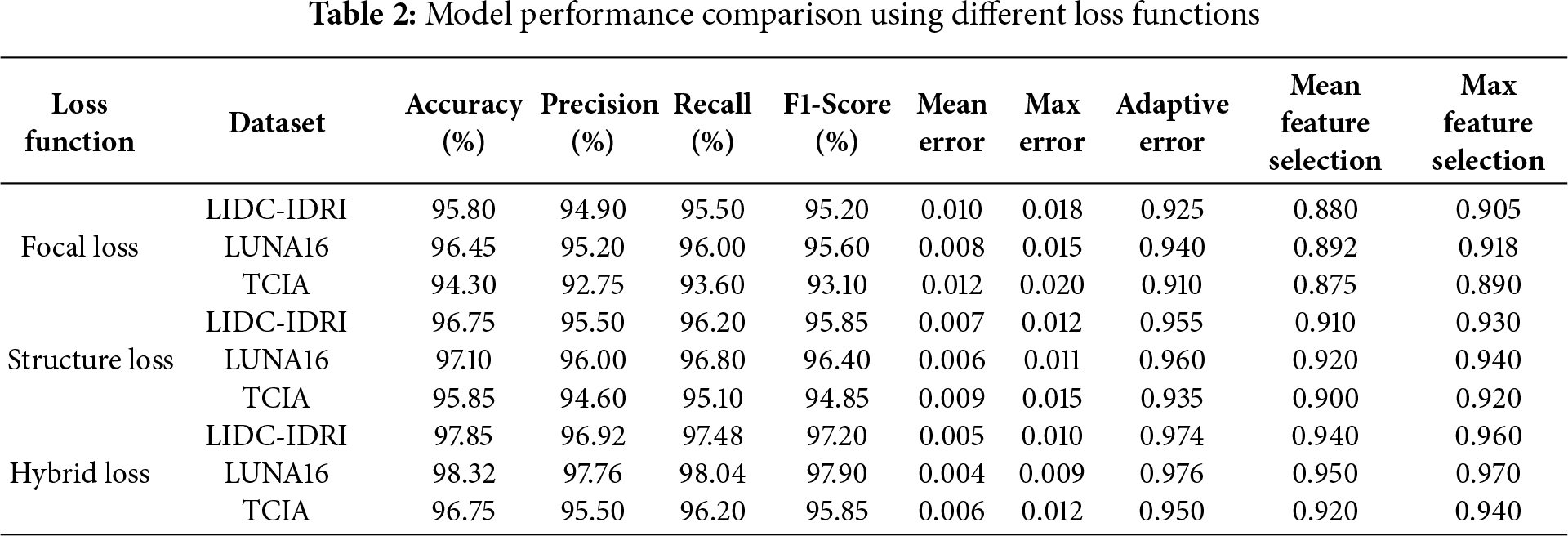
Table 3 presents segmentation-based performance metrics for different loss functions. Hybrid Loss consistently achieves higher Mean IoU, Dice Score, and Sensitivity, indicating its superior performance in lung cancer image segmentation and feature extraction.
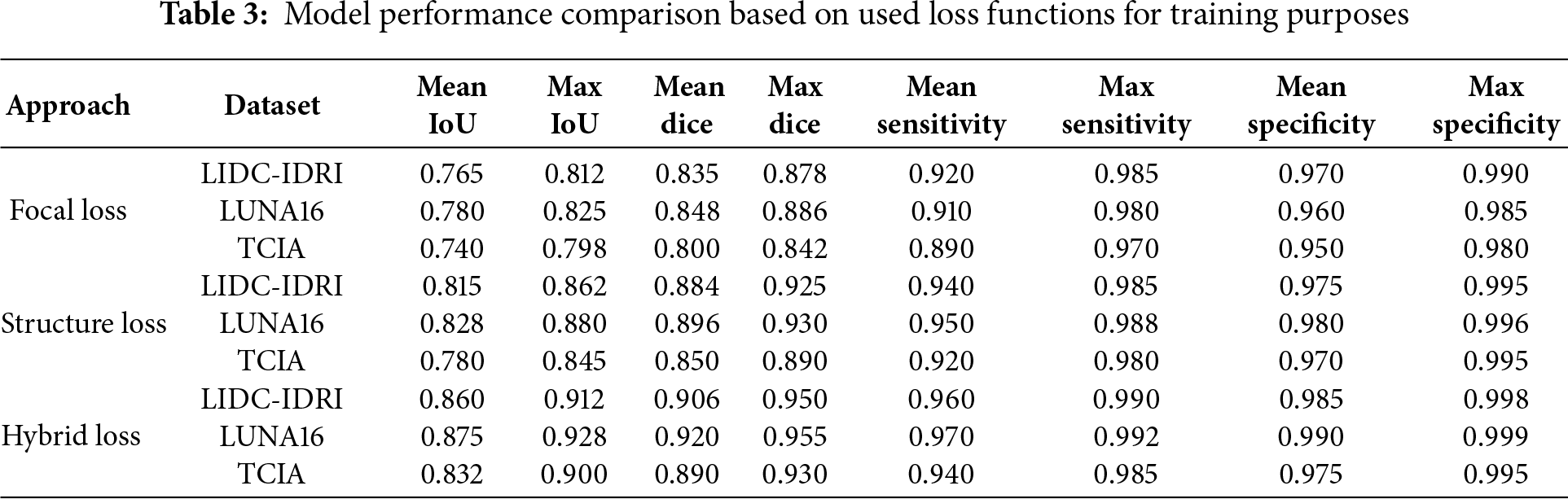
The proposed model demonstrated exceptional effectiveness in classifying the TCIA histopathological dataset, achieving an accuracy rate of 96.75% and a precision of 95.50%.
Fig. 2a DualSet ViT-PSO-SVM Model on LIDC-IDRI dataset, Fig. 2b DualSet ViT-PSO-SVM Model on LUNA16 dataset and Fig. 2c DualSet ViT-PSO-SVM Model on TCIA dataset.
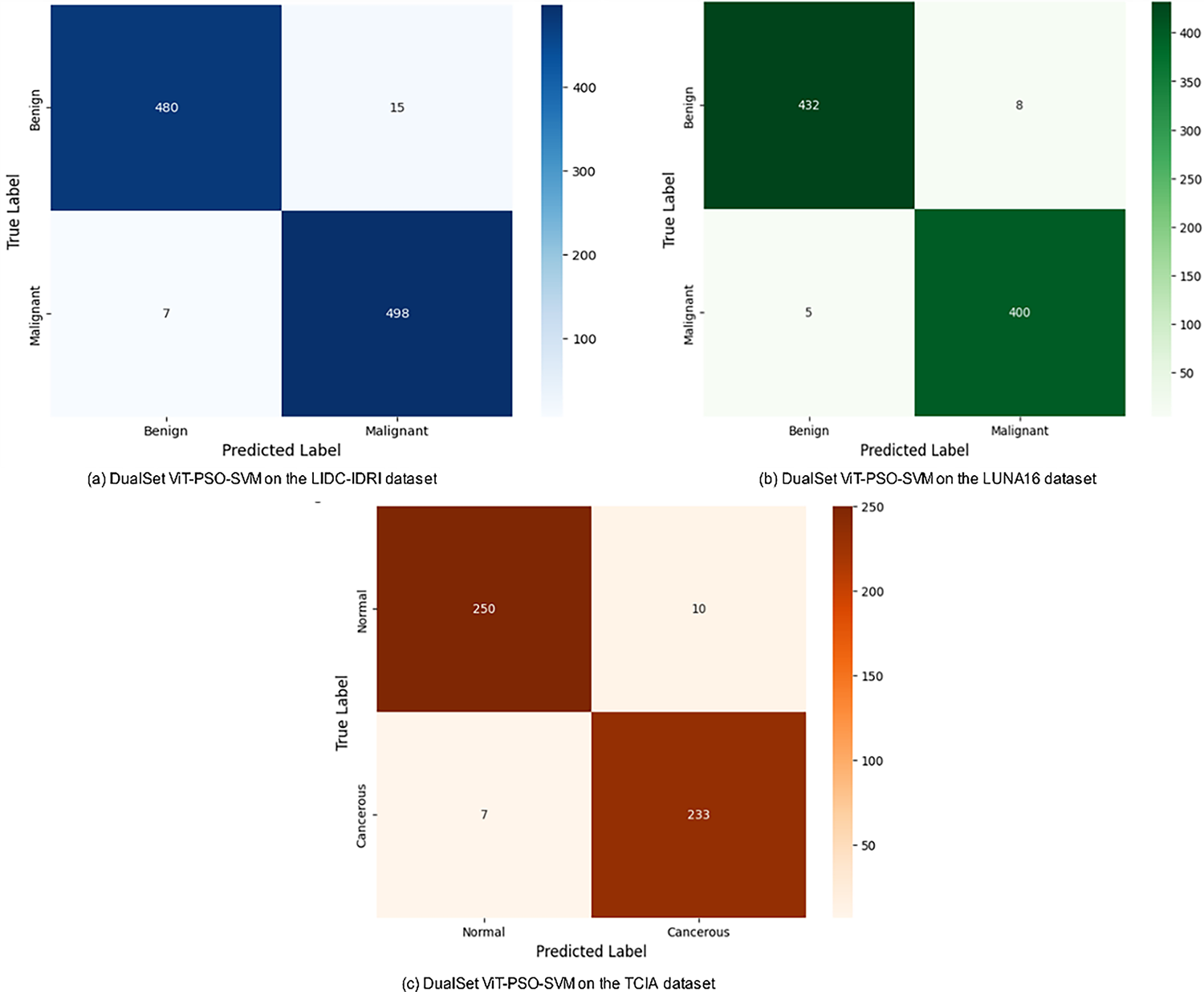
Figure 2: Confusion matrix of the DualSet ViT-PSO-SVM on the different datasets
The testing procedure involved applying the proposed DualSet ViT-PSO-SVM model to analyze data from different medical imaging datasets. A sustainable approach consisted of using the trained model on datasets independent of what it learned. The model demonstrates its ability to apply results to different imaging methods and acquisition settings and annotation practices through this analysis.
For cross-dataset experiments, we followed two evaluation scenarios. In Scenario A, the model trained on the LIDC-IDRI dataset → tested on LUNA16 and TCIA datasets. In Scenario B, the model trained on the TCIA histopathological dataset → tested on LIDC-IDRI and LUNA16 datasets. The cross-dataset performance metrics for Scenario A and Scenario B are summarized in Table 4.
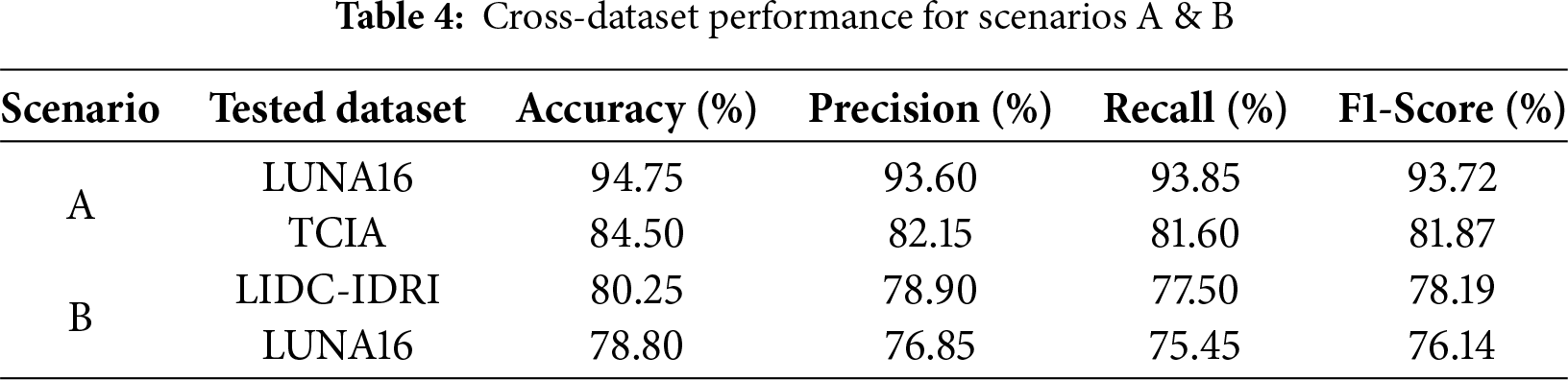
Scenario A demonstrated strong performance, particularly when generalizing from LIDC-IDRI to LUNA16, as both datasets contain similar imaging modalities (CT scans). However, accuracy slightly dropped (from 97.85% to 94.75%) due to minor differences in annotation criteria and image resolution. A more noticeable performance decline was observed when tested on the histopathological TCIA dataset, highlighting modality differences.
Fig. 3a,b depicts confusion matrices of DualSet ViT-PSO-SVM trained on LIDC-IDRI and tested on LUNA16 and TCIA datasets, respectively.
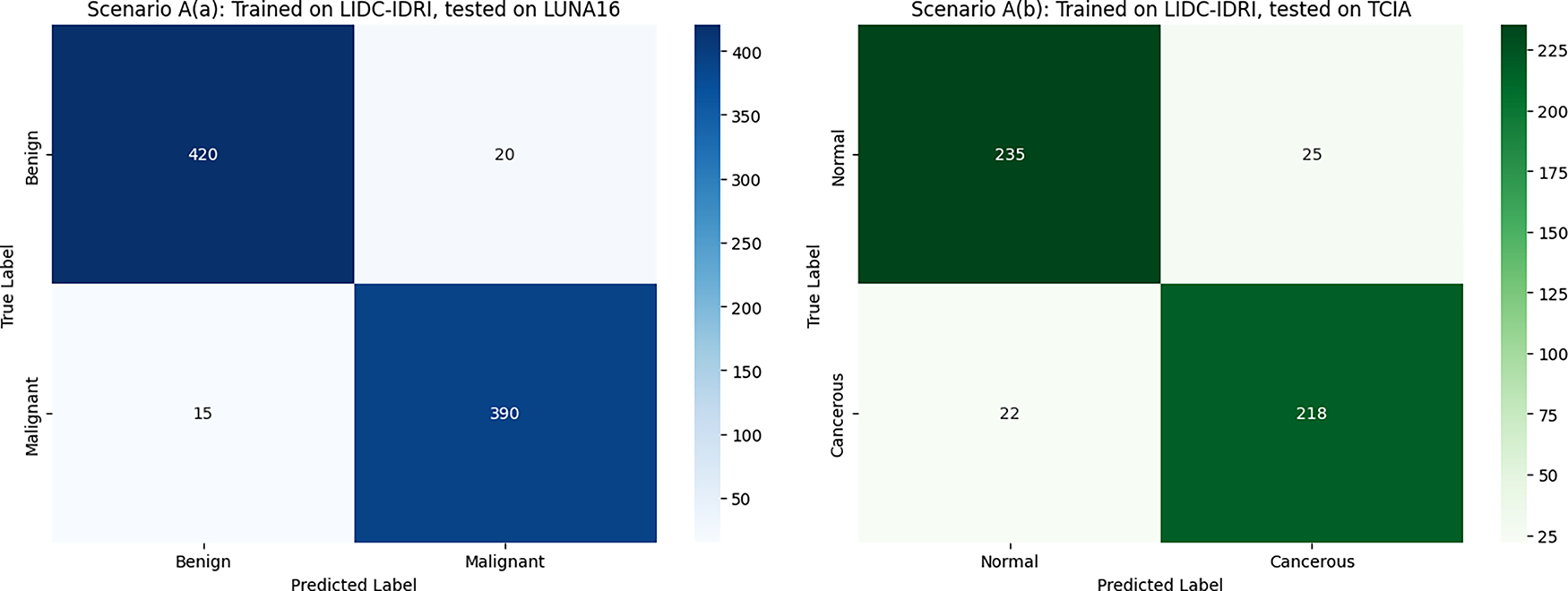
Figure 3: Cross dataset confusion matrices (Scenario A)
Scenario B showed moderate generalization performance. When transferring knowledge from TCIA histopathological data to CT-based datasets (LIDC-IDRI, LUNA16), accuracy dropped significantly due to substantial differences in imaging modalities and underlying tissue characteristics.
Fig. 4 illustrates the cross-dataset evaluation confusion matrices under Scenario A, where the DualSet ViT-PSO-SVM model was trained on the TCIA histopathological dataset and evaluated on two distinct CT-based datasets. Fig. 4a shows the confusion matrix when tested on the LIDC-IDRI dataset, demonstrating good classification performance but with noticeable misclassification between benign and malignant classes due to differences in modality. Fig. 4b presents the confusion matrix for evaluation on the LUNA16 dataset, highlighting similar performance trends with accurate predictions accompanied by some misclassifications, which further indicate challenges when generalizing between different imaging modalities. This degradation can be attributed to simple modality differences, in which CT scans capture anatomical structures in grayscale with varying Hounsfield units. In contrast, histopathology slides have high-resolution color data that capture the morphology of cells. These differences lead to changes in the distribution of features that challenge the transferability. Misclassified cases are often made of small or borderline lesions, which are largely modality-specific. Future studies can alleviate the limitations proposed by domain adaptation, such as aligning feature distributions across modalities, modality-specific pretraining on large-scale unlabeled data, and reducing data variability through harmonization (by minimizing the difference between data to grasp the concept better).
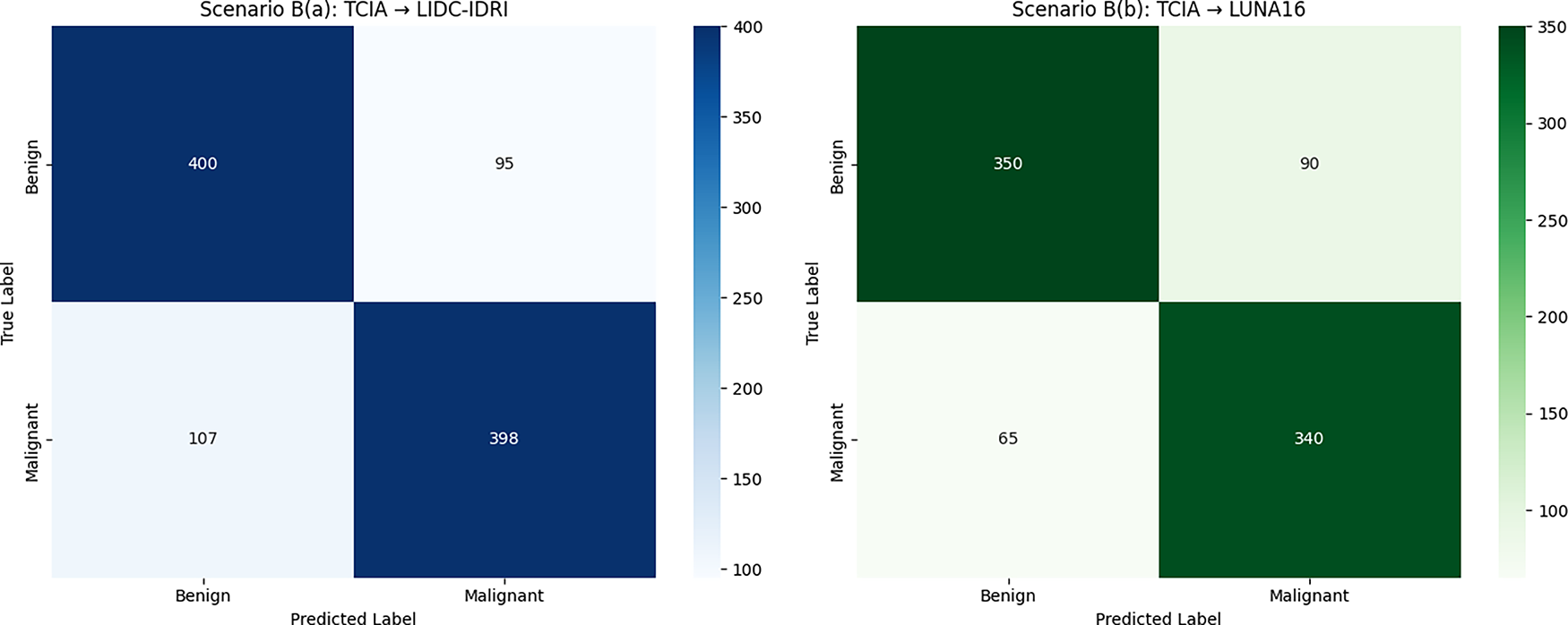
Figure 4: Cross-dataset confusion matrices (Scenario A)
One of the factors that needs to be considered when deploying the model in a real-world scenario is that the model should be able to generalize to multi-center datasets, which are typically not consistent regarding the types of scanners, the acquisition protocols, differences in staining (in the case of histopathology), and skewed populations of benign and malignant cases. These aspects may cause domain shift and lead to poor model performance if not addressed. Several approaches can be taken to strengthen this environment. Firstly, data harmonization and normalization methods may minimize inter-center variability during pre-processing. Second, it might be possible to adapt the model to new institutional data by domain adaptation techniques (such as adversarial training, feature alignment, or domain-specific fine-tuning). Third, acquisition-simulating data augmentation with realistic noise and protocol variations can enhance resilience to acquisition variations. Lastly, skewed classes can be addressed with imbalance-conscious training methods, such as focal loss, synthetic minority oversampling, or cost-sensitive learning. To ensure that this framework can be successfully applied in various clinical settings, it will be essential to incorporate these strategies into the further development of our framework.
Fig. 5 presents the training and validation loss curves for the DualSet ViT-PSO-SVM model across four different datasets: LIDC-IDRI, LUNA16, TCIA, and the Combined Dataset (DualSet). Each subplot illustrates the model’s learning behavior during 100 epochs of training, showing the training loss (blue curve) and validation loss (red curve).
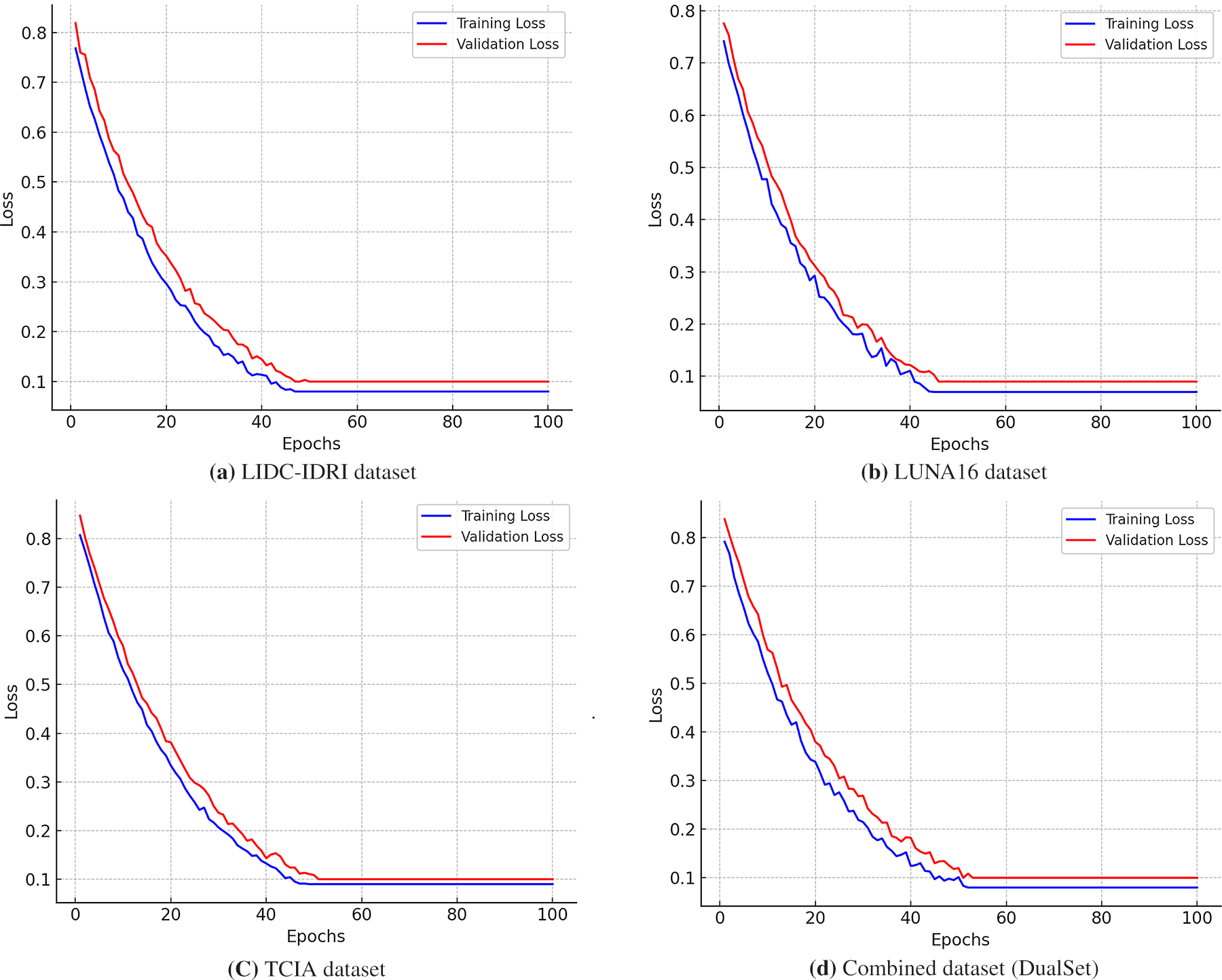
Figure 5: Training and validation loss curves of the DualSet ViT-PSO-SVM model across different datasets
In Fig. 5a the training loss starts at approximately 0.8 and declines smoothly, indicating a steady learning process. The validation loss follows a similar downward trajectory but remains slightly higher than the training loss, suggesting good generalization with minimal overfitting. Around epoch 50, both losses stabilize near 0.1, demonstrating the model’s convergence on LIDC-IDRI (CT-based lung nodule dataset).
The model exhibits a sharp decline in training loss, starting from around 0.78 and reaching a plateau near 0.09 by epoch 60 as shown in Fig. 5b. The validation loss stabilizes at a slightly higher value than the training loss, suggesting the model has learned meaningful features while maintaining good generalization. The fluctuations in early epochs indicate active feature extraction, a common phenomenon in CT-based datasets.
In Fig. 5c, the training loss initially starts at 0.85 and decreases smoothly, while the validation loss remains slightly higher, starting at 0.88 before stabilizing near 0.11. This dataset, containing histopathological images, presents a somewhat different loss curve due to the increased complexity of tissue feature representation. The model exhibits a stable convergence trend, albeit with minor fluctuations in the validation loss, suggesting some variability in histopathology image learning.
In Fig. 5d, the training loss curve starts at 0.83, following a smooth exponential decline before leveling off around 0.1 by epoch 70. The validation loss also follows a consistent pattern, slightly higher than the training loss but without significant divergence, confirming strong generalization capabilities. The combination of CT and histopathological datasets presents a somewhat more complex loss pattern, yet the model adapts well across modalities.
The recorded drop in performance with the switch to histopathology highlights the significant modality gap regarding image resolution, texture features, and clinical semantics. This weakness highlights the need for methods that can explicitly address the issue of cross-domain variability. Several strategies can address this issue. To minimize the distribution shift between CT and histopathology representations, domain adaptation algorithms, such as adversarial feature alignment or correlation alignment, may be employed first. Second, pretraining each encoder on massive unlabeled CT and histopathology data through modality-specific tasks could enable the encoder to learn domain-relevant features before fine-tuning. Third, cross-modality generalization can be traded off against within-modality specificity using multi-branch structures, where individual modality-specific encoders project to a common decision layer. Investigating these strategies is a significant avenue for future practice, with the potential to enhance strength and clinical utility in the diagnosis of multimodal lung cancer.
Fig. 6 demonstrates the interpretability of the DualSet ViT-PSO-SVM model predictions using Grad-CAM (Gradient-weighted Class Activation Mapping) on representative lung cancer images.
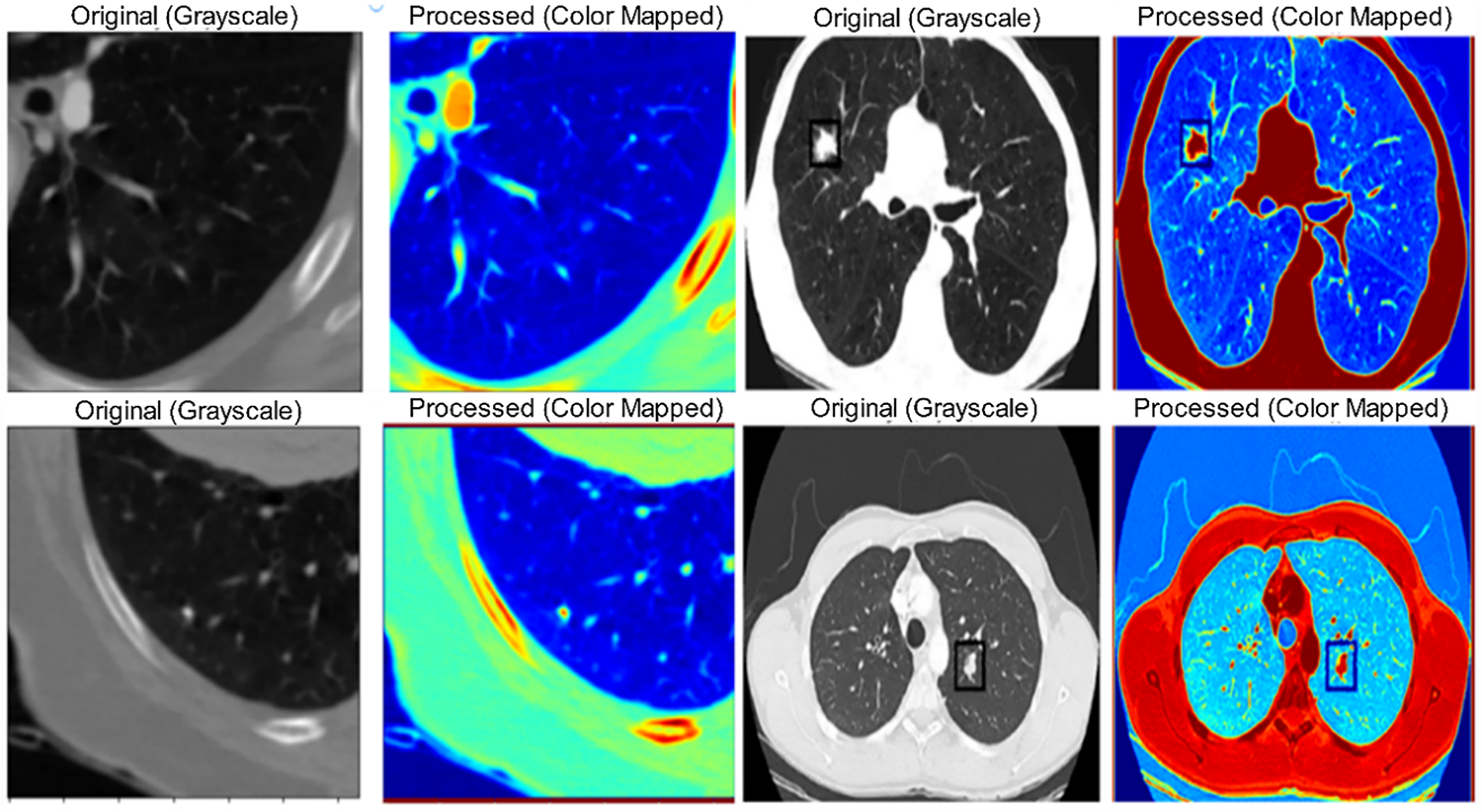
Figure 6: Grad-CAM visualization for lung cancer classification
Although the Grad-CAM images in Fig. 6 indicate that the suggested framework has concentrated on clinically significant areas, the boundaries of the tumor frequently lack clarity, especially when it comes to small or irregularly shaped lesions. This is a shortcoming because Grad-CAM primarily emphasizes regions of discriminative features rather than localizing in specific pixels. Consequently, this may lead to underrepresentation of subtle margins of small nodules, which could affect vocational interpretability in cases of borderline findings. To address this, future work may be conducted to expand the existing framework into a hybrid segmentation-classification framework, where a segmentation module initially localizes tumor regions and subsequent classification is performed. The heatmaps highlight regions of interest that significantly influence the model’s decision-making. Brighter (warmer) regions indicate areas of higher importance used by the model in predicting the presence or classification of lung cancer. These visualizations enhance clinical interpretability by allowing medical professionals to validate the decision-making process of the deep learning model, building trust and potentially improving diagnostic confidence.
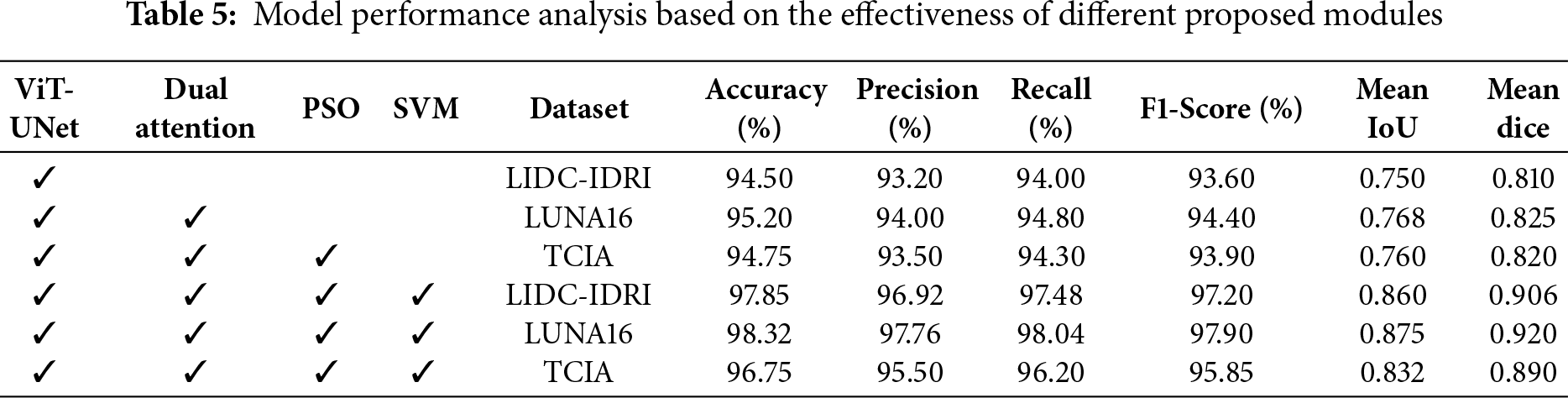
Table 5 evaluates the impact of each module in the DualSet ViT-PSO-SVM framework. The best performance is observed when all four components (ViT-UNet, Dual Attention, PSO, and SVM) are integrated, highlighting their collective contribution to improved classification accuracy and segmentation metrics.
To evaluate the effectiveness and novelty of our proposed DualSet ViT-PSO-SVM method, we compared its performance with existing state-of-the-art methods reported in the literature. The comparison focuses on the accuracy of lung cancer classification, considering recent deep-learning approaches that utilize CNN and Transformer-based architectures. Table 6 summarizes a comparative analysis of the proposed DualSet ViT-PSO-SVM framework against state-of-the-art CNN- and Transformer-based models on three benchmark datasets (LIDC-IDRI, LUNA16, and TCIA). The table reports classification performance (accuracy, precision, recall, F1-score) along with computational efficiency metrics (average inference time per image and peak GPU memory usage). Results demonstrate that the proposed method achieves superior accuracy and F1-score while maintaining inference efficiency comparable to existing Transformer architectures.
The results illustrate that the proposed DualSet ViT-PSO-SVM approach achieves superior performance compared to existing methods, consistently outperforming traditional CNN-based methods, attention-enhanced CNNs, and recent Transformer models (including Swin Transformer and vanilla ViT). Specifically, the proposed method improved the classification accuracy on the LIDC-IDRI dataset to 97.85%, surpassing the previously best-reported accuracy of 94.20%. On the LUNA16 dataset, the proposed model achieved an accuracy of 98.32%, surpassing the previous best of 95.60% using a vanilla ViT. For histopathological images in the TCIA dataset, our model also demonstrated superior accuracy (96.75%) compared to the previous best of 94.85% obtained with Swin Transformer. The improved performance can be attributed to the effective integration of ViT’s global feature extraction capabilities, optimal feature selection using PSO, and robust classification via SVM. These components collectively enhance diagnostic accuracy, validating the novelty and efficacy of our proposed framework.
In this research, we proposed the DualSet ViT-PSO-SVM model, a novel deep-learning approach for accurate lung cancer classification. The developed model integrates a ViT architecture to capture both local and global features from lung cancer images. A dual attention mechanism, combining self-attention and CBAM-based channel-spatial attention, significantly enhanced the quality of extracted features. We optimized these features using PSO to reduce complexity and achieve greater accuracy. Furthermore, we employed an SVM classifier to effectively classify lung cancer images from three well-known public datasets: LIDC-IDRI, LUNA16, and TCIA. Comprehensive experimental evaluations demonstrated superior performance compared to existing state-of-the-art methods. Cross-dataset evaluation further validated the generalizability and clinical applicability of our model. Despite the promising results, our proposed method exhibits certain limitations. The model’s generalization capability across significantly different imaging modalities, such as CT and histopathological images, showed a decline in performance. Additionally, the computational complexity associated with ViT architectures may limit practical deployment, especially in clinical environments with constrained resources. Lastly, although validated extensively on public datasets, further validation using clinical datasets from multiple institutions is necessary to fully confirm its practical efficacy. Future studies can address current limitations by developing domain adaptation strategies that enhance cross-modality robustness, ensuring consistent performance across diverse medical imaging data. Additionally, it is essential to explore lightweight ViT architectures optimized for efficient real-time diagnosis and easier clinical deployment. Further validation studies involving larger, multi-center clinical datasets will provide critical insights into the model’s performance in real-world settings. Finally, expanding explainability beyond Grad-CAM visualization could further enhance interpretability and promote acceptance among clinicians, thereby improving clinical decision-making.
Acknowledgement: This research was funded by the European University of Atlantic.
Funding Statement: No additional external funding was received beyond the support provided by the European University of Atlantic.
Author Contributions: Abdu Salam: conceptualization; Farhan Amin: methodology; Fahd M. Aldosari: software; Donia Y. Badawood, Isabel de la Torre, Farhan Amin: validation, writing; Gerardo Mendez Mezquita: supervision; Henry Fabian Gongora: funding acquisition. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data supporting the findings of this study are provided within the manuscript.
Ethics Approval: This study was conducted using publicly available, de-identified datasets (LIDC-IDRI, LUNA16, and TCIA). Therefore, formal ethics approval was not required, or not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Kuczynski G, Gotay C. Cancer prevention: principles and approaches. In: Understanding cancer prevention through geospatial science. Cham, Switzerland: Springer International Publishing; 2024. p. 17–43. [Google Scholar]
2. Li C, Wang H, Jiang Y, Fu W, Liu X, Zhong R, et al. Advances in lung cancer screening and early detection. Cancer Biol Med. 2022;19(5):591–608. doi:10.20892/j.issn.2095-3941.2021.0690. [Google Scholar] [PubMed] [CrossRef]
3. Sagheer SVM, Meghana KH, Ameer PM, Parayangat M, Abbas M. Transformers for multi-modal image analysis in healthcare. Comput Mater Continua. 2025;84(3):4259–97. doi:10.32604/cmc.2025.063726. [Google Scholar] [CrossRef]
4. Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, et al. Transformers in medical imaging: a survey. Med Image Anal. 2023;88(1):102802. doi:10.1016/j.media.2023.102802. [Google Scholar] [PubMed] [CrossRef]
5. Yang J, Li C, Zhang P, Dai X, Xiao B, Yuan L, et al. Focal attention for long-range interactions in vision transformers. Adv Neural Inf Process Syst. 2021;34:30008–22. [Google Scholar]
6. Wang Y, Deng Y, Zheng Y, Chattopadhyay P, Wang L. Vision transformers for image classification: a comparative survey. Technologies. 2025;13(1):32. doi:10.3390/technologies13010032. [Google Scholar] [CrossRef]
7. Ali H, Mohsen F, Shah Z. Improving diagnosis and prognosis of lung cancer using vision transformers: a scoping review. BMC Med Imaging. 2023;23(1):129. doi:10.1186/s12880-023-01098-z. [Google Scholar] [PubMed] [CrossRef]
8. Yu H, Dai Q. Self-supervised multi-task learning for medical image analysis. Pattern Recognit. 2024;150(12):110327. doi:10.1016/j.patcog.2024.110327. [Google Scholar] [CrossRef]
9. Takahashi S, Sakaguchi Y, Kouno N, Takasawa K, Ishizu K, Akagi Y, et al. Comparison of vision transformers and convolutional neural networks in medical image analysis: a systematic review. J Med Syst. 2024;48(1):84. doi:10.1007/s10916-024-02105-8. [Google Scholar] [PubMed] [CrossRef]
10. El-Baz A, Beache GM, Gimel’farb G, Suzuki K, Okada K, Elnakib A, et al. Computer-aided diagnosis systems for lung cancer: challenges and methodologies. Int J Biomed Imag. 2013;2013(1):942353. doi:10.1155/2013/942353. [Google Scholar] [PubMed] [CrossRef]
11. Reeves AP, Kostis WJ. Computer-aided diagnosis for lung cancer. Radiol Clin N Am. 2000;38(3):497–509. doi:10.1016/s0033-8389(05)70180-9. [Google Scholar] [PubMed] [CrossRef]
12. Chaunzwa TL, Hosny A, Xu Y, Shafer A, Diao N, Lanuti M, et al. Deep learning classification of lung cancer histology using CT images. Sci Rep. 2021;11(1):5471. doi:10.1038/s41598-021-84630-x. [Google Scholar] [PubMed] [CrossRef]
13. Rodríguez-Cano F, Calvo V, Garitaonaindía Y, Campo-Cañaveral de la Cruz JL, Blanco M, Torrente M, et al. Cost-effectiveness of diagnostic tests during follow-up in lung cancer patients: an evidence-based study. Transl Lung Cancer Res. 2023;12(2):247–56. doi:10.21037/tlcr-22-540. [Google Scholar] [PubMed] [CrossRef]
14. Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, Shuai B, et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018;77(11):354–77. doi:10.1016/j.patcog.2017.10.013. [Google Scholar] [CrossRef]
15. Guan Q, Huang Y, Zhong Z, Zheng Z, Zheng L, Yang Y. Diagnose like a radiologist: attention guided convolutional neural network for thorax disease classification. arXiv:1801.09927. 2018. [Google Scholar]
16. Li TZ, Xu K, Gao R, Tang Y, Lasko TA, Maldonado F, et al. Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography. In: Proceedings of the Medical Imaging 2023: Image Processing; 2023 Feb 19–23; San Diego, CA, USA. p. 229–38. [Google Scholar]
17. Pu Q, Xi Z, Yin S, Zhao Z, Zhao L. Advantages of transformer and its application for medical image segmentation: a survey. Biomed Eng Online. 2024;23(1):14. doi:10.1186/s12938-024-01212-4. [Google Scholar] [PubMed] [CrossRef]
18. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306. 2021. [Google Scholar]
19. Khan A, Rauf Z, Khan AR, Rathore S, Khan SH, Shah NS, et al. A recent survey of vision transformers for medical image segmentation. arXiv:2312.00634. 2023. [Google Scholar]
20. Atabansi CC, Nie J, Liu H, Song Q, Yan L, Zhou X. A survey of transformer applications for histopathological image analysis: new developments and future directions. Biomed Eng Online. 2023;22(1):96. doi:10.1186/s12938-023-01157-0. [Google Scholar] [PubMed] [CrossRef]
21. Al-Hammuri K, Gebali F, Kanan A, Chelvan IT. Vision transformer architecture and applications in digital health: a tutorial and survey. Vis Comput Ind Biomed Art. 2023;6(1):14. doi:10.1186/s42492-023-00140-9. [Google Scholar] [PubMed] [CrossRef]
22. Ashraf A, Nawi NM, Shahzad T, Aamir M, Khan MA, Ouahada K. Dimension reduction using dual-featured auto-encoder for the histological classification of human lungs tissues. IEEE Access. 2024;12(1):104165–76. doi:10.1109/access.2024.3434592. [Google Scholar] [CrossRef]
23. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:1–11. [Google Scholar]
24. Xie Y, Yang B, Guan Q, Zhang J, Wu Q, Xia Y. Attention mechanisms in medical image segmentation: a survey. arXiv:2305.17937. 2023. [Google Scholar]
25. Hou Q, Zhou D, Feng J. Coordinate attention for efficient mobile network design. In: Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20–25; Nashville, TN, USA. p. 13708–17. [Google Scholar]
26. El-Regaily SA, Abdel Megeed Salem M, Abdel Aziz MH, Roushdy MI. Multi-view convolutional neural network for lung nodule false positive reduction. Expert Syst Appl. 2020;162(6):113017. doi:10.1016/j.eswa.2019.113017. [Google Scholar] [CrossRef]
27. Lanjewar MG, Panchbhai KG, Charanarur P. Lung cancer detection from CT scans using modified DenseNet with feature selection methods and ML classifiers. Expert Syst Appl. 2023;224(8):119961. doi:10.1016/j.eswa.2023.119961. [Google Scholar] [CrossRef]
28. Zheng R, Wen H, Zhu F, Lan W. Attention-guided deep neural network with a multichannel architecture for lung nodule classification. Heliyon. 2024;10(1):e23508. doi:10.1016/j.heliyon.2023.e23508. [Google Scholar] [PubMed] [CrossRef]
29. Alsulami AA, Albarakati A, Al-Ghamdi AA, Ragab M. Identification of anomalies in lung and colon cancer using computer vision-based swin transformer with ensemble model on histopathological images. Bioengineering. 2024;11(10):978. doi:10.3390/bioengineering11100978. [Google Scholar] [PubMed] [CrossRef]
30. Mkindu H, Wu L, Zhao Y. Lung nodule detection in chest CT images based on vision transformer network with Bayesian optimization. Biomed Signal Process Control. 2023;85(1):104866. doi:10.1016/j.bspc.2023.104866. [Google Scholar] [CrossRef]
31. Zhang Y, Chen Z, Yang X. Light-M: an efficient lightweight medical image segmentation framework for resource-constrained IoMT. Comput Biol Med. 2024;170(11):108088. doi:10.1016/j.compbiomed.2024.108088. [Google Scholar] [PubMed] [CrossRef]
32. Marinakis I, Karampidis K, Papadourakis G. Pulmonary nodule detection, segmentation and classification using deep learning: a comprehensive literature review. BioMedInformatics. 2024;4(3):2043–106. doi:10.3390/biomedinformatics4030111. [Google Scholar] [CrossRef]
33. Shanmugam K, Rajaguru H. Exploration and enhancement of classifiers in the detection of lung cancer from histopathological images. Diagnostics. 2023;13(20):3289. doi:10.3390/diagnostics13203289. [Google Scholar] [PubMed] [CrossRef]
34. Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imag. 2016;35(5):1229–39. doi:10.1109/tmi.2016.2528821. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF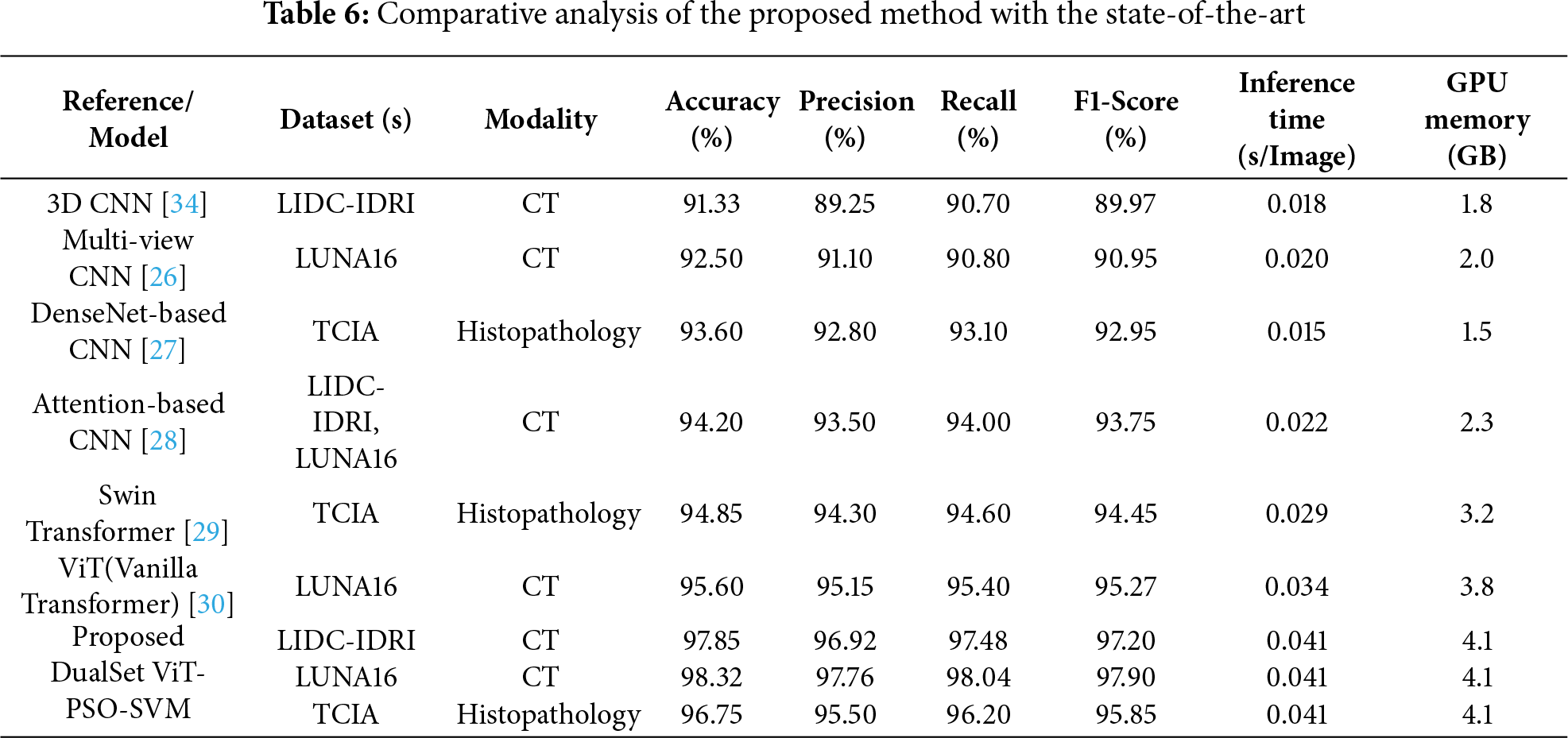
 Downloads
Downloads
 Citation Tools
Citation Tools