 Open Access
Open Access
ARTICLE
A Robust Image Encryption Method Based on the Randomness Properties of DNA Nucleotides
1 Cardiff School of Technologies, Cardiff Metropolitan University, Cardiff, CF23 6XD, UK
2 Networks and Cybersecurity Department, Amman Al Ahliyya University, Amman, 19328, Jordan
3 Faculty of Engineering, Al-Balqa Applied University, Al-Salt, 19117, Jordan
* Corresponding Author: Bassam Al-Shargabi. Email:
Computers, Materials & Continua 2026, 87(1), 11 https://doi.org/10.32604/cmc.2025.074550
Received 13 October 2025; Accepted 17 December 2025; Issue published 10 February 2026
Abstract
The advent of 5G technology has significantly enhanced the transmission of images over networks, expanding data accessibility and exposure across various applications in digital technology and social media. Consequently, the protection of sensitive data has become increasingly critical. Regardless of the complexity of the encryption algorithm used, a robust and highly secure encryption key is essential, with randomness and key space being crucial factors. This paper proposes a new Robust Deoxyribonucleic Acid (RDNA) nucleotide-based encryption method. The RDNA encryption method leverages the unique properties of DNA nucleotides, including their inherent randomness and extensive key space, to generate a highly secure encryption key. By employing transposition and substitution operations, the RDNA method ensures significant diffusion and confusion in the encrypted images. Additionally, it utilises a pseudorandom generation technique based on the random sequence of nucleotides in the DNA secret key. The performance of the RDNA encryption method is evaluated through various statistical and visual tests, and compared against established encryption methods such as 3DES, AES, and a DNA-based method. Experimental results demonstrate that the RDNA encryption method outperforms its rivals in the literature, and achieves superior performance in terms of information entropy, avalanche effect, encryption execution time, and correlation reduction, while maintaining competitive values for NMAE, PSNR, NPCR, and UACI. The high degree of randomness and sensitivity to key changes inherent in the RDNA method offers enhanced security, making it highly resistant to brute force and differential attacks.Keywords
The advent of cloud computing technologies has facilitated the computation and storage of a substantial volume of image data through cloud platforms. The advent of the 5G era is poised to significantly augment the utilization of images, thereby necessitating an elevated emphasis on the security of image storage and transmission. Given the pervasive practice of transmitting images across diverse networks in numerous applications, the imperative to safeguard the contents of digital images against unauthorized access has become a prominent concern. However, the escalating reliance on digital images across various domains, including healthcare and surveillance, has underscored the critical need for a dedicated focus on preserving the confidentiality and integrity of visual data. In specific applications, such as within healthcare, images of patients are considered highly sensitive information. The improper usage of such images carries the potential for incorrect diagnoses and compromised medical decisions [1–5], further accentuating the imperative for robust protective measures in this context.
However, there has been a concerning rise in data security breaches perpetrated by hackers and criminals, in such IBM reported in 2019 that the health industry had the highest number of data breaches, and that data can be misappropriated in a variety of ways [6]. Furthermore, over 46 million individuals were affected by health data hacking attempts in the United States between the years 2009 and 2021 [7]. As a result, there is an ongoing need for persistent endeavours in developing novel security methods. Hence, significant endeavours are dedicated by both industry and academic research to safeguard business and governmental data, which is either conveyed over diverse digital communication channels or saved in computer systems [8,9]. To ensure the confidentiality of images transferred through various networks, numerous different security measures are utilised. Data concealing, watermarking images, and encryption are some examples of these methods.
Conventional cryptographic approaches have historically constituted the cornerstone for ensuring the security of data. Prominent among these are efficient cryptographic methodologies, including the Advanced Encryption Standard (AES), the Data Encryption Standard (DES), and triple DES, among others [6,10]. These methods have achieved widespread adoption and application for encrypting various forms of multimedia, including texts, images, and video files. However, they can be computationally costly and less suitable for certain application scenarios, particularly those involving resource-constrained devices and the Internet of Things (IoT) [11].
DNA cryptography is an emerging technological advancement that has promise to enhance the security and efficiency of image encryption [2,12,13]. DNA cryptography refers to a cryptographic technique that leverages the distinctive characteristics of DNA for encoding and decoding data [14]. Deoxyribonucleic acid (DNA) is a macromolecule that encompasses the hereditary information utilised in the growth and operation of all recognized living entities. The DNA is made up of a long, random chain of nucleotides, which act as the basic components of DNA. The four nucleotides present in DNA are adenine (A), thymine (T), cytosine (C), and guanine (G). Each two of the four nucleotides are associated with a bond in DNA, where adenine (A) is linked with thymine (T) and cytosine (C) is linked with guanine (G). Based on these three characteristics of DNA (extremely long length and the randomness in its contents and the special relationship between nucleotides), it can be exploited for key generation. Leveraging DNA in key generation offers several distinct advantages over traditional cryptographic methods. Originally, DNA’s vast and complex structure enables the creation of highly intricate encryption keys, bolstering security against sophisticated decryption attempts [15,16]. Additionally, the inherent randomness of DNA sequences enhances the unpredictability and entropy of generated keys, further fortifying the encryption process [17]. Moreover, the specific nucleotide pairings within DNA provide a built-in mechanism for error detection and correction, contributing to the robustness and reliability of the encryption scheme [18,19]. Furthermore, DNA-based encryption methods exhibit potential for scalability and adaptability across diverse application domains, from healthcare to finance, owing to DNA’s universal presence and applicability [18].
A wide range of DNA-based encryption methods have been utilised in the field of data security. However, numerous DNA cryptography predominantly revolve around the utilization of logistic maps and fundamental biological manipulations on DNA sequences, such as logical XOR, subtraction, and addition [2,10,20]. One potential method is the integration of DNA sequence operations alongside chaos-based approaches to encrypt data, particularly images. In addition, the integration of the SHA 256 hash function has been implemented as a fundamental element of this encryption process. Different approaches utilize a system that incorporates a collection of rules based on concepts drawn from DNA sequences to carry out transposition operations to encrypt images, along with generating the encryption key from the DNA sequence [21,22]. However, some issues related to using DNA nucleotides as secret keys include the complexity of accurately mapping biological sequences to cryptographic keys and potential vulnerabilities in key distribution and management. In this paper, we propose the RDNA encryption method, which exploits the random nature of DNA nucleotides, with the following main contributions as presented in this research, including:
• The use of DNA nucleotides as a secret code for encrypting images. The digital encoding of DNA nucleotides, represented by the characters A, T, C, and G, is extended into a long sequence with random duplication, guaranteeing the generation of a highly sensitive and durable encryption key.
• The RDNA encryption method incorporates a unique XOR operation during the preparation step, where the source image is combined with the DNA secret key. This innovative advancement enhances the ability to detect small changes in the input image, therefore strengthening the security of the encryption process.
• The RDNA approach involves carrying out transposition and substitution operations on the source image. These activities, driven by a pseudorandom generation method, provide diffusion and confusion effects that are essential for hindering decryption efforts by potential attackers.
• Dependence on random sequences of DNA nucleotides, where the seed value for the pseudorandom generation algorithm depends on the values in the DNA key, adding another layer of security. This, combined with the confidentiality of the DNA key and the algorithm used, poses a significant challenge for attackers attempting to crack the encrypted image.
This paper presents a robust encryption method that is based on leveraging DNA nucleotides and structure for generating encryption keys with a high level of security. A novel method for image encryption has been established that efficiently incorporates encryption keys, achieving a security level comparable to existing encryption methods. The rest of the paper is structured as follows: Section 2 examines background and related work in the area of DNA image encryption. Section 3 presents the details of the proposed RDNA encryption method. Section 4 presents the experimental evaluation results of the proposed RDNA and compares them against its best rivals in the literature. Section 5 concludes the paper.
Cryptography is a distinct field of scientific study that pertains to the process of encoding data to conceal the content of messages. Recent advancements in the field of cryptography have demonstrated the potential for leveraging DNA computation. This presents an opportunity for the utilization of DNA computing as a novel approach to enhance the development of a robust and efficient cryptographic system. This section presents a comprehensive overview of the most recent research findings in the field of cryptography systems specifically designed for encrypting image data.
An image encryption method that utilises a hyperchaotic system in conjunction with a Fibonacci Q-matrix is introduced in [1]. The initial phase of the method involves permuting the original image through the utilization of 6D hyperchaotic. This phase is effective in enhancing encryption efficiency and increasing the level of security due to its complicated high-dynamic behaviours and possession of two positive Lyapunov exponents. Afterward, the permuted image is diffused by employing the Fibonacci Q-matrix. The same applied to similar method but with the integration of the Fibonacci Q-matrix along with fractional-order four-dimensional hyperchaotic Chen system [23].
DNA computing was integrated with double-chaos systems for encrypting colour images at the bit level [24,25], where the Arnold algorithm was employed to disarrange the three constituents of the colour image. The total number of rounds was established based on the mean of the three components. This method enhanced the efficacy of the Arnold algorithm in achieving a more pronounced scrambling effect. Additionally, they proposed the use of a dual-chaos system that consists of the Lorenz chaotic mapping with parameters that are adjustable and the fourth-order Rossler hyperchaotic mapping. This method is employed to produce three distinct sets of chaotic sequences for the purpose of diffusion operation [20,26]. The double-chaos system is used to mitigate the inherent pseudo-randomness of two distinct types of chaotic mappings, thereby enhancing the unpredictability of chaotic sequences. Next, the disordered component images and disordered sequences are converted into DNA sequences using eight DNA coding principles. These coding principles are established based on either plaintext information or generated chaotic sequences. In addition to performing addition, subtraction, and XOR operations on them. Another alternative method is to encrypt coloured images by utilizing DNA sequences and chaotic sequences as the underlying mechanism [27]. The method being presented utilises a DNA substitution box to encrypt the pixels of a coloured image. Additionally, a chaotic sequence is employed to effectively jumble the encrypted pixels. Another chaotic image encryption techniques with DNA sequence functions introduced in [28]. The encryption process is independently implemented on the three channels (R, G, and B) of the colour image using an appropriate chaotic map. The technique for map selection is used to choose a suitable chaotic map by considering various image features and additional parameters. After the process of chaotic encryption followed by DNA encoding.
Furthermore, numerous image encryption methods that rely on the use of DNA encoding and chaos systems have been devised. Most of these methods make use of chaotic systems that rely on the use of periodic windows or patterns in the bifurcation diagrams but are still prone to various chaos-specific attacks. As in [29], they presented a method for image encryption through the utilisation of a conservative chaos standard map generated from the DNA coding scheme. This method relies on employing distinct rules for encoding, addition/subtraction, and decoding for each pixel. These rules are randomly chosen by utilizing pseudorandom sequences generated through the conservative chaotic standard map. Another method relies on the same concept by using the fractional-order Hyperchaotic Lorenz System and DNA Computing [30]. The method comprises four components: firstly, the generations of a pseudorandom sequence and the use of a fractional order hyperchaotic Lorenz system. subsequently, the implementation of a straightforward diffusion scheme to propagate minimal changes in one pixel to all other pixels; thirdly, the images encoded using rules extracted from the DNA sequence along and executing the appropriate DNA operations.
Another image encryption method that combines chaos theory and DNA computing [31], where image permutation is established by applying a series of Pseudo-Random Numbers (PRN), which are subsequently encrypted via DNA sequence. They exploited two PRN sequences produced using a pseudo-number generator (PRNG) that relies on a cross-coupled chaotic logistic map and utilises two distinct sets of keys. The initial PRN sequence is employed to perform permutation on the plaintext image, whereas the second PRN sequence is utilized for the generation of a random DNA sequence. Such a method was applied to grayscale-labeled images, with the potential for extension to encompass colour images and textual data.
A method that involves both cryptography and DNA-based steganography is proposed in [32]. The method demonstrates a way for encrypting images before they are embedded into DNA sequences in order to protect the images’ confidentiality while they are being sent. The method involves the conversion of an image into a DNA sequence, which is subsequently utilized to embed encrypted data within an input image. The data is encrypted using a randomly generated 256-bit secret key. Subsequently, the encrypted data is concealed within a Steganographic Image (SI). Subsequently, the System Information (SI) is kept on the cloud server. In addition, a study aims to explore the domain of DNA-based image steganography [33]. The technique encompasses the covert embedding of concealed images into host images. As they demonstrated the utilisation of DNA sequences as a clandestine communication route for the secure concealment of visually sensitive data. While in [34], an approach involves the utilization of a hybrid DNA encoding scheme with Fuzzy Integral sequences to implement encryption and steganography methods. The approach involves taking advantage of a map of a chaos system to encode a distorted representation of an image. Subsequently, four distinct coded images are generated by employing different four DNA sequences. Additionally, four pseudo-random sequences are generated through the application of Choquet’s fuzzy integral sequence. The encoded images are then diffused using the complementary DNA XOR rules. Finally, the resulting fuzzy-DNA encoded images are combined using the wavelet-based fusion system, leading to the generation of the encrypted image.
Moreover, in another method [35], the Genetic Algorithm (GA) is employed to modify the pixel coordinates to minimise the correlation between neighbouring pixels during the transposition operation. Followed by an XOR operation between the binary representations of the pixel values and DNA substrings is obtained from a randomly generated DNA string. DNA sequences are employed to generate the encryption key.
A selective medical image encryption method utilizing DNA cryptography is presented to reduce the amount of computational time required in the medical industry [2]. To encrypt digital medical images, the encryption method relies on applying permutation and diffusion techniques to specific pixel values. The DNA setup for the image is created by applying all the rules for DNA encoding based on the position of the pixel, while the cipher image is acquired by applying all the rules for DNA decoding based on the value of the pixel in a similar method, the image was transformed into a DNA sequence, which was subsequently encrypted using a DNA computing method [3].
A framework for the encryption of images, employing DNA and Recursive Cellular Automata (RCA) in distinct phases was introduced in [36]. Their method implied applying a permutation step that encompasses the manipulation of image rows and columns, followed by the application of DNA and RCA techniques to alter the shade of gray of pixels, resulting in new values. The method is also founded upon the utilisation of the SHA-256 algorithm, which produces a hash key of 256 bits in length that establishes a straight association with the distinct key. Moreover, another method is designated for colour image encryption through the utilisation of DNA tape and a replacement box based on chaos. They used an S-box generated directly from the DNA sequence, Arnold transformation, and a chaotic dynamic system [27].
A hash encoding with DNA computing principles is presented [37], where they utilise the DNA hash encoding module to facilitate the mapping of image pixels onto DNA computing bases. Content-aware encryption is employed to restructure the image data organization to enhance its resistance against potential attacks.
It is clear from related works that all encryption methods developed by the researchers that use DNA are based on finding an innovative approach to use the distinctive features of the DNA sequence. The first group of researchers represents the DNA nucleotides as digital codes and uses them to perform substitution operations to produce a confusion effect in the encrypted data. The second group of researchers exploits the unique arrangement of nucleotides in the DNA sequence to perform the transposition operations necessary to produce the diffusion effect in the encrypted data. The third group focuses on the large size of DNA used as a secret key in the encryption method to raise the level of security and increase the difficulties against attackers. The RDNA encryption method proposed in this paper integrates the three methods used in related works to produce an innovative approach to using DNA for image encryption. This is a distinctive point in the proposed method.
In summary, the field of DNA-based image encryption has evolved from simple encoding operations to sophisticated hybrids combining chaos theory, cellular automata, and hash functions. A common trend is the reliance on external chaotic systems to generate randomness. While effective, this can introduce complexity and potential vulnerabilities specific to the chaotic maps used. This observation motivates the development of RDNA, which seeks to simplify the architecture by deriving all necessary cryptographic randomness directly from the DNA key material itself.
3 The Proposed RDNA Encryption Method
While existing DNA-based methods often rely on external chaotic systems for randomness, the core innovation of RDNA is its direct and exclusive reliance on the intrinsic randomness of a user-provided DNA nucleotide sequence itself for all cryptographic operations (seed generation, transposition, and substitution). This eliminates the need for complex chaotic maps and derives all security directly from the biological key.
Unlike hybrid DNA-chaos systems, RDNA uses the DNA key not just for encoding but as the sole source of entropy for a pseudo-random generator that drives a unique transposition operation based on nucleotide bonding rules (A-T, C-G).
The RDNA encryption method proposed in this paper adopted the DNA nucleotides as a secret key for image encryption. In the proposed RDNA encryption method, the digital DNA nucleotides (key) are used and manipulated as a very long string of four characters (A, T, C, and G) that are repeated randomly in the string, as depicted in Fig. 1.

Figure 1: A hypothetical example of the nucleotide sequence in a digital DNA file used in the proposed RDNA encryption method
As a preparatory stage, RDNA initially applies an XOR operation to both the user’s source image and the DNA secret key to make them highly sensitive to every small change. In the encryption stage, the RDNA performs transposition and substitution operations on the source image to generate the necessary diffusion and confusion effects in the encrypted image. All of this is done using a pseudorandom generation algorithm and relying on the property of the random sequence of nucleotides in the DNA secret key.
Below are a set of definitions and data structures prescribed to facilitate understanding of the operations performed in the encryption stage:
S: refers to the source bitmap image that is used as input to the proposed RDNA encryption method. In the preparation stage, the 2D source image, entered by the user, is represented as a 1D array of bytes where every three consecutive bytes represent the red, green, and blue colour values for each pixel in the source image. The size of the 1D array that represents S (in bytes) is calculated using Eq. (1), where Palette = 3 (three colour values: red, green, and blue in each pixel).
In this preparation stage, an XOR operation is performed between all bytes in S. This operation helps to produce a different encrypted image when only one byte is changed in the source image S. Eq. (2) shows how this process is performed.
KN: refers to the digital DNA nucleotides file that represents a random string of the four nucleotides (A, T, C, and G) that is used as the secret key in the proposed RDNA method. The total number of characters in KN is KNSize, which is usually ≥(SSize × 4).
KB: refers to the digital DNA file after being represented as a string of bytes by merging every four successive nucleotides in KN to form a single byte. This is carried out using Table 1. For example, for the following segment of KN: A C A T G C A C..., the byte representation KB of this segment is: 41 226, where: (A C A T) = (0010 0001) = 41 and (G C A C) = (1110 0010) = 226. The total number of bytes in KB is KBSize and is calculated using Eq. (3).
An XOR operation is performed between all bytes in KB. The goal of performing this process is to make the RDNA encryption method use a different key whenever any change is made to the entered digital DNA file and this will produce a different encrypted image. After performing this XOR operation, a new KN containing a new string of nucleotides is generated, and this process is performed using Table 1.
E: refers to the encrypted coloured bitmap image that is produced by the proposed RDNA encryption method after completing the execution of the encryption operations. Both Source (S) and encrypted (E) images are of the same size.
In this stage, the bytes in the source image S are processed by performing two operations (Transposition and Substitution) to create the encrypted image E. The proposed RDNA encryption method adopts a pseudo-random generation algorithm to generate a random sequence of numbers that is used to perform these two operations.
To conduct preliminary tests to evaluate the performance of the proposed RDNA encryption method, the pseudo-random generation library function available in the C# programming language was used to write the programming code required to test the proposed RDNA encryption method. The seed value of the pseudo-random generation algorithm mainly depends on the DNA key used by the user and is calculated using Eq. (4).
These two operations are explained below:
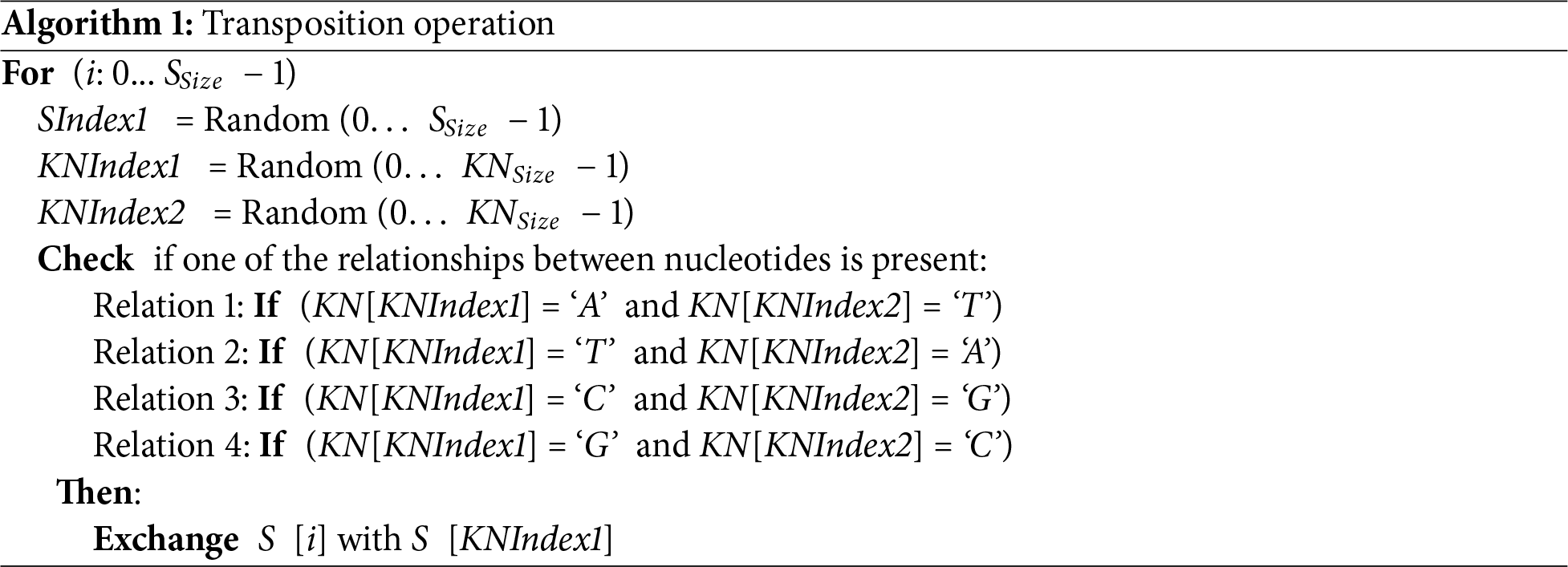
1. Transposition Operation
The implementation of this operation aims to change the position of each byte at index i (where i: 0… SSize − 1) in the source image S, and this will achieve the necessary diffusion effect in the source image S as shown in Algorithm 1. This is done by random selection of three indices SIndex1, KNIndex1, and KNIndex2. Where the first index (SIndex1) represents an index for a byte in the source image S and the other two indices (KNIndex1 and KNIndex2) represent indices for two nucleotides in the KN.
After the three indices are obtained, the exchange operation is performed between two bytes at i and SIndex1 in the source image S based on the presence or absence of a relationship between the two nucleotides at the indices KNIndex1 and KNIndex2 in KN. As mentioned earlier (i.e., A-T and C-G). The bellow Algorithm 1 demonstrates the implementation of the transposition operation.
To clarify the transposition operation for readers, the following example is provided below with these assumptions:
• KN which is used as a key to implement the transposition operation:

• S before implementing the transposition operation.

• i = 0, SIndex1 = 7, KNIndex1 = 2, KNIndex2 = 8.
• Now, when the transposition operation is performed based on these assumptions, the new
S becomes:

It is necessary to mention here that:
• Dependence of the seed value on the values in the DNA key.
• Not knowing of the pseudo-random generation algorithm used in the RDNA encryption method.
• In addition to the confidentiality of the DNA key used.
All of these points will pose a huge challenge for attackers to crack the encrypted image.
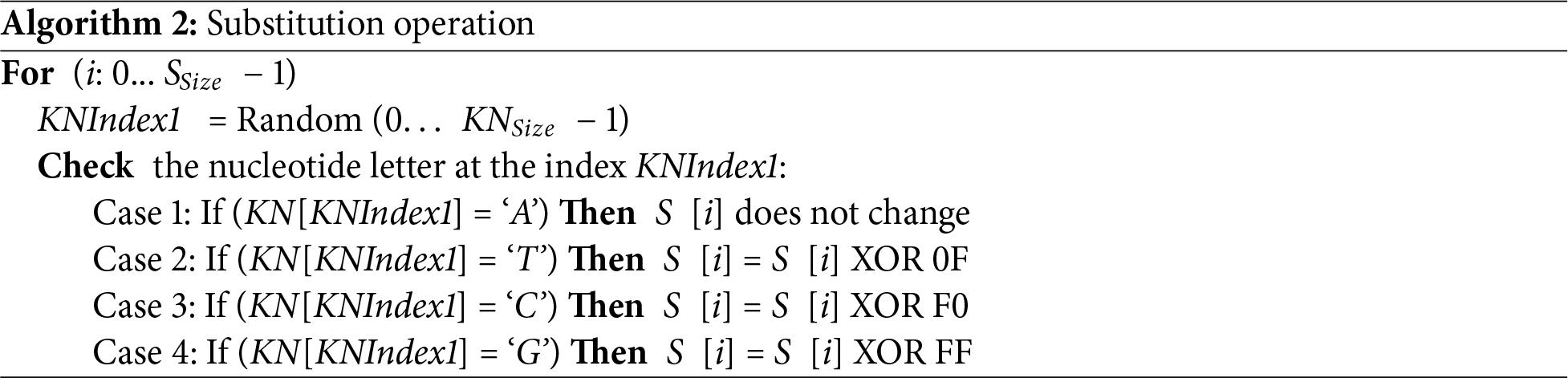
2. Substitution Operation
The goal of performing the substitution operation on the byte values of the source image S is to change the byte values and create a confusion effect in the source image S. This is done by random selection of an index KNIndex1 of a nucleotide in the KN as shown in Algorithm 2. Then perform an XOR operation between each byte at index i in the source image S and one of the four bytes in Table 2 based on the nucleotide letter at the index KNIndex1 in the KN.

To help understand the substitution operation more clearly, an example is provided below with the following assumptions:
• KN which is used as a key to implement the substitution operation:

• S before implementing the transposition operation:

• i = 3, KNIndex1 = 6.
• Now, when the substitution operation is performed based on these assumptions, the new
S becomes:

It should be noted here that using DNA as a key makes the XOR operation performed between each byte in the source image S and one of four truly random bytes in Table 2 based on the nucleotide letter in the KB. Due to the random and sufficiently vast nature of the DNA, the task of attackers to deduce the DNA key and decrypt the encoded image becomes considerably more challenging. The encryption stage will result in an encrypted image that is generated by combining every three consecutive bytes in the source image S to form a single pixel in the encrypted image E.
To recover the source image S from the encrypted image E in the decryption stage, the same DNA key must be entered. Decryption requires the exact same DNA key. The preparation stage is identical. The core encryption operations are then reversed in inverse order: first, the inverse substitution (which is identical to the substitution operation due to the properties of XOR) is applied, followed by the inverse transposition. The inverse transposition uses the same pseudo-random sequence (generated from the same seed) to identify and reverse the swap operations performed during encryption.
To provide a formal foundation for the RDNA algorithm, this section outlines the mathematical models for its core operations and analyzes its computational complexity.
The transposition and substitution operations can be formally defined as follows:
Let S be the source image represented as a vector of bytes, where S[i] denotes the byte at index i, and i ∈ [0, SSize − 1]. Let KN be the key nucleotide sequence, and PRNG(Seed) be the pseudorandom number generator initialized with the Seed from Eq. (4).
Transposition Operation: For each byte S[i] in the source image, the transposition is defined by:
1. Generate three random indices using the PRNG:
• SIndex1 ∈ [0, SSize − 1]
• KNIndex1, KNIndex2 ∈ [0, KNSize − 1]
2. Let isBonded (X, Y) be a function that returns True if nucleotides X and Y are a complementary pair (i.e., (X, Y) ∈ {(A, T), (T, A), (C, G), (G, C)}), and False otherwise.
3. The swap condition is then:
If isBonded (KN[KNIndex1], KN[KNndex2]) is True, then swap (S[i], S[SIndex1])
Substitution Operation: For each byte S[i] in the source image (post-transposition), the substitution is defined by:
1. Generate a random index KNIndex1 ∈ [0, KNSize − 1] using the PRNG.
2. Let f: {A,T, C, G} → {0x00, 0x0F, 0xF0, 0xFF} be the mapping function defined in Table 2.
3. The substitution is performed using the XOR operation:
S[i] = S[i] ⊕ f(KN[KNIndex1])
This operation is its own inverse, which is crucial for the decryption process.
Complexity Analysis: The time complexity of the RDNA algorithm is linear, O(n), with respect to the number of bytes n in the source image S. This is because both the transposition and substitution operations perform a single pass over the image, and the operations within the loop (PRNG calls, table lookups, and conditional swaps) are executed in constant time, O(1). The preparation stage also involves linear-time passes for the XOR chaining of S and KB. Therefore, the overall complexity remains O(n), making the algorithm efficient for images of various sizes.
To evaluate the proposed RDNA encryption method, different sets of images of different sizes were used in the experiments. The performance of the RDNA method was evaluated by employing a comprehensive set of statistical and visual tests. Additionally, a comparative analysis was conducted, wherein the findings obtained from the RDNA method were contrasted with those obtained from various other methods, including 3DES, AES, and another approach that relies on the utilization of DNA [38]. The performance evaluation process relied on many criteria, such as information entropy, avalanche effect, key space, encryption execution time, correlation analysis, Normalised Mean Absolute Error (NMAE), Peak Signal-to-Noise Ratio (PSNR), Number of Changing Pixel Rates (NPCR), and Unified Averaged Changed Intensity (UACI). Fig. 2 shows some selected images used in the experiments. While our current set of standard test images demonstrates the algorithm’s efficacy. Future work will include more extensive testing on specialized image types [e.g., medical MRI scans] to further validate performance across domains. The measurements values are listed in the results tables as an average of values calculated for the RGB channels of the images.

Figure 2: Selected images used in the experiments
All programming codes were written using C# language and executed on a computer system with specifications: Processor (11th Gen Intel(R) Core (TM) i7-1165G7-2.80 GHz), RAM (16.0 GB), Operating System (Windows 10, 64-bit operating system). Various standard and non-standard images were used in the experiments, and some of these images and their results are included in the manuscript.
4.1 Statistical and Visual Tests
An image histogram is a statistical metric that attackers can use to obtain information about the colour/byte intensity in the image. The high degree of flatness of the colour/byte histogram of the encrypted image indicates the efficiency of the RDNA encryption method and the achievement of a high level of protection for the encrypted image, this will add more difficulties against attackers who use statistical analysis. Fig. 3 shows the histograms of the source and encrypted image of two images chosen as examples, Irbid University and the Peppers images in Fig. 2.
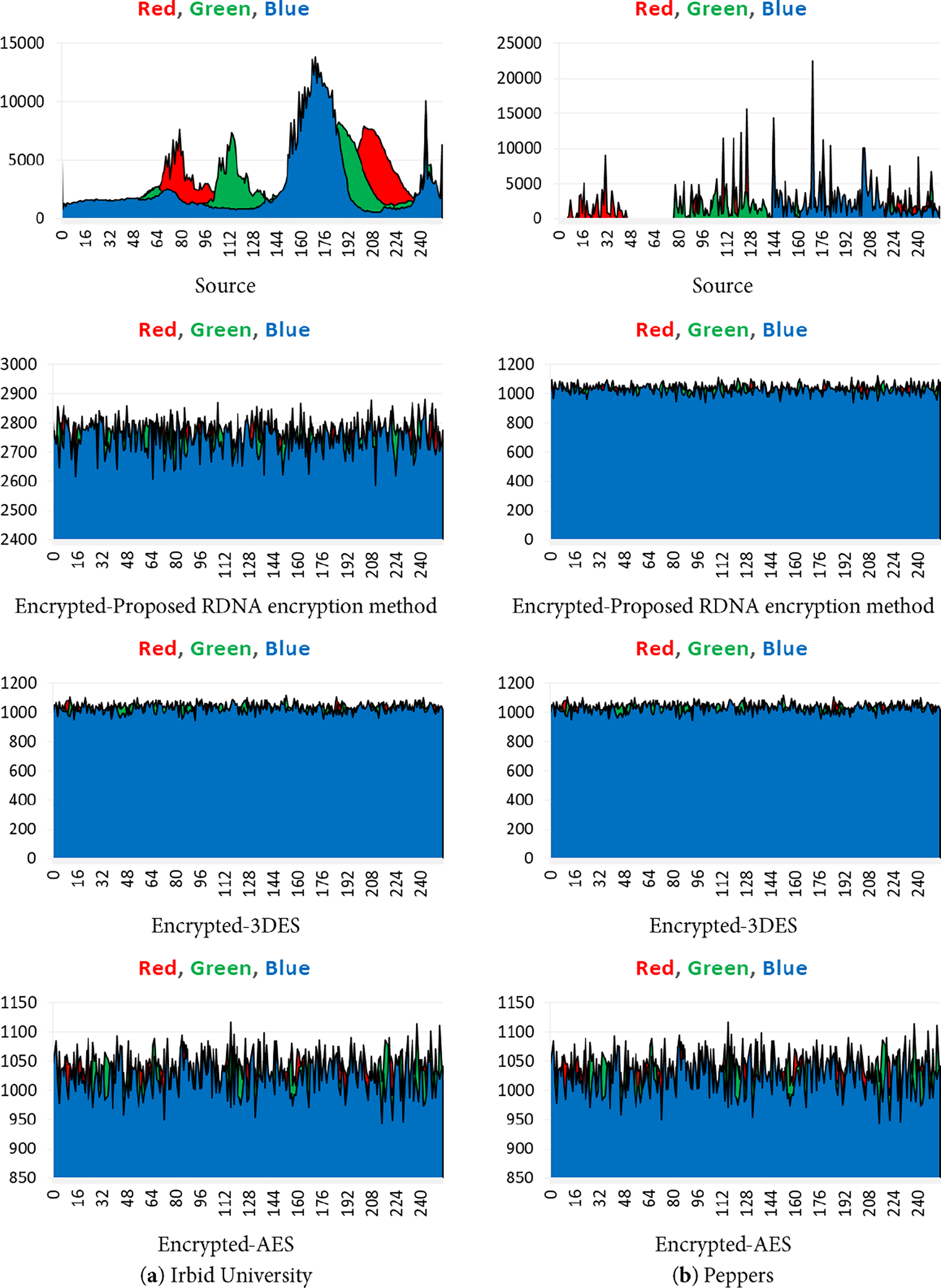
Figure 3: Histograms of the source and encrypted images, (a) Irbid University (b) Peppers
The encrypted images that resulted from the experiments proved that the proposed RDNA encryption method produces a high level of diffusion and confusion effects (i.e., distortion) in the encrypted images. Fig. 4 shows the resulting encrypted images after implementing the proposed RDNA encryption method for the images in Fig. 2.
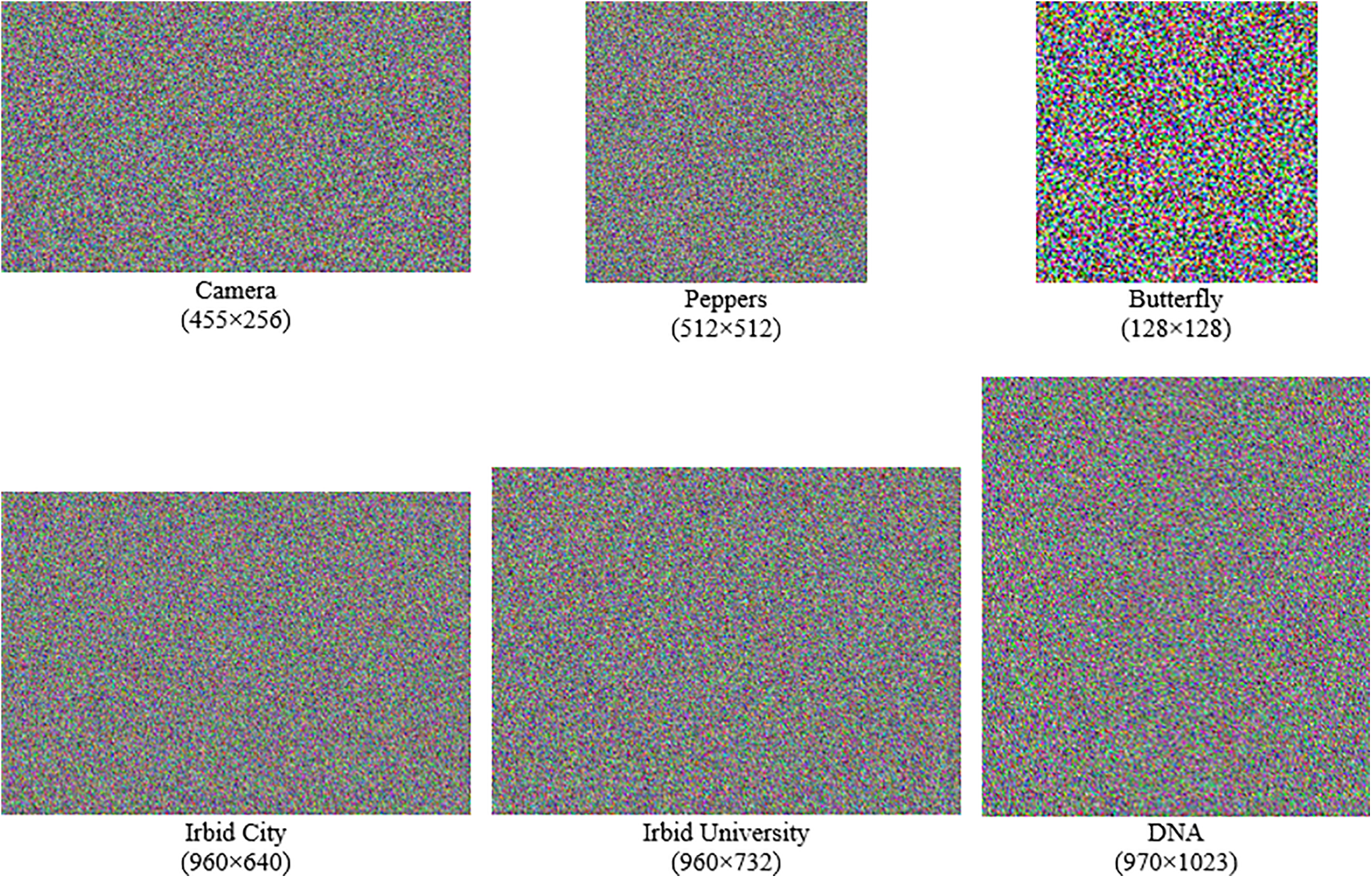
Figure 4: The resulting encrypted images from the source images in Fig. 2
Information entropy is the measure used to find the degree of randomness in an image. Entropy is the average amount of information in an image. It is difficult for attackers to predict the content of the image when the information entropy of the image is high. The information entropy value is calculated using Eq. (5) [39,40].
where Pi is the probability of occurrence of a data value will occur and n is the number of different data values. The calculated Information entropy values for the 3DES, AES, and DNA-based [38] encryption methods are listed in Table 3 to facilitate a comparison of different encryption methods.
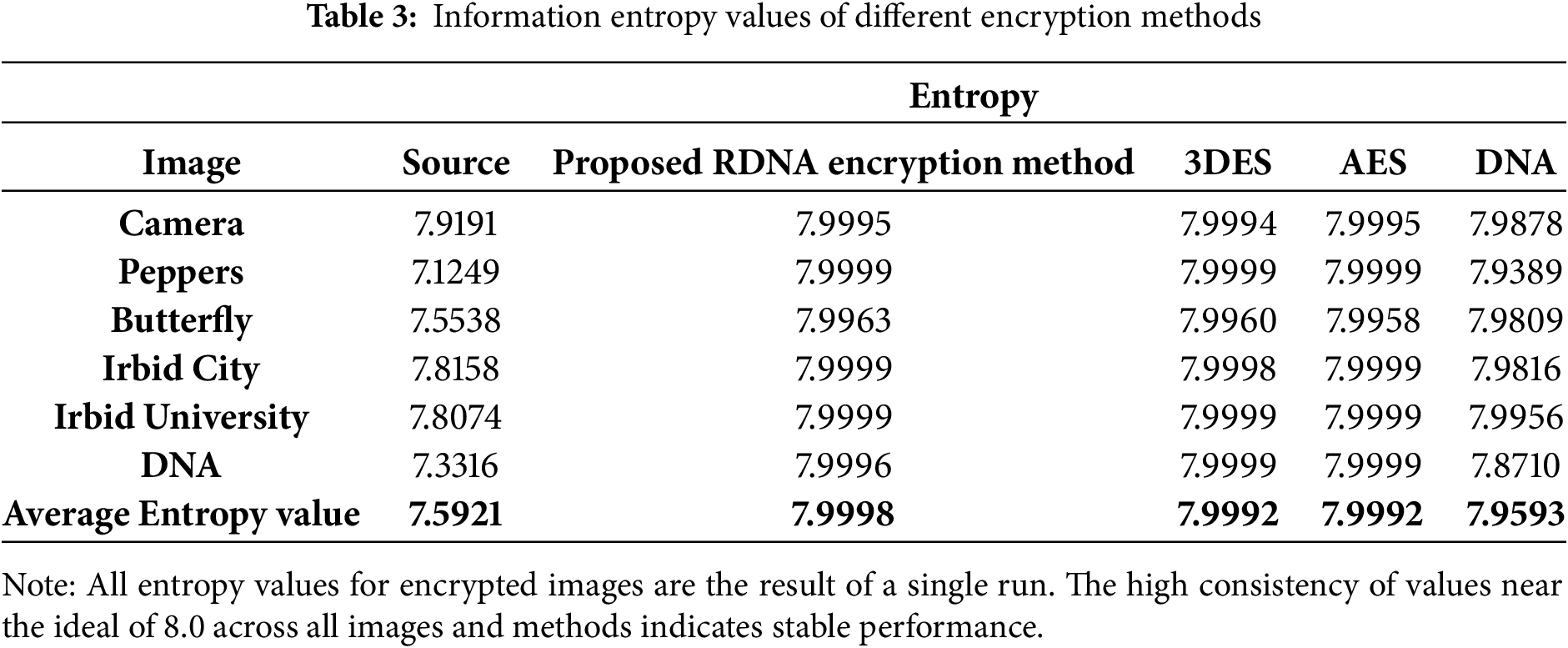
The average entropy value in Table 3 proves that the proposed RDNA encryption method achieves a competitive level of randomness in the encrypted image compared to other methods. Therefore, encrypted images generated by the proposed RDNA method have a high level of immunity against attacks.
One of the main goals of developing any encryption method is to make the method highly sensitive to any small changes in its parameters, for example, any slight change in the source data or the key will cause a significant change in the encrypted data.
The seed value of the pseudo-random generation algorithm calculated at the beginning of the encryption stage depends entirely on the values of the nucleotides in the DNA key used in the proposed method. Therefore, any small changes in the DNA key will produce a completely different encrypted image. The avalanche effect is used to test the sensitivity of the proposed RDNA encryption method. The sensitivity property can be tested by changing different numbers of bits in the key and recording the numerical and visual effects of these changes on the generated encrypted image and the recovered source image.
4.3.1 Avalanche Effect on the Encrypted Image
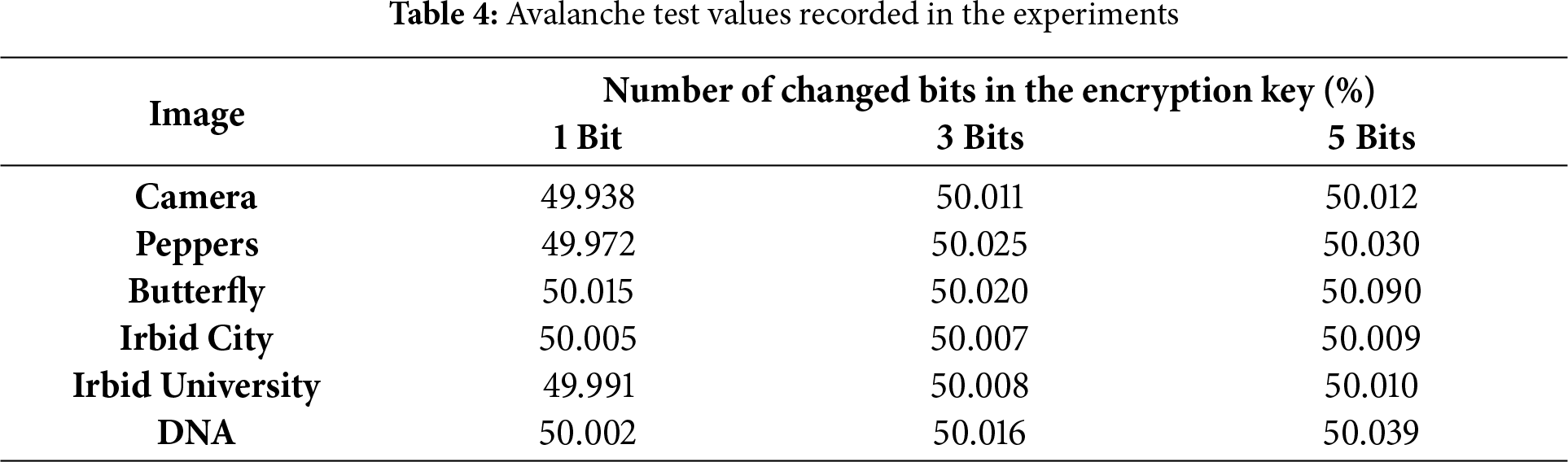
The avalanche effect test is calculated using Eq. (6) [41], where the avalanche effect value is the percentage of bits in the encrypted image change when a few bits are changed in the key (1 bit to 7 bits changed during experiments for example).
The avalanche test values recorded in the experiments for the images in Fig. 2, are listed in Table 4.
4.3.2 Avalanche Effect on the Recovered Source Image
The avalanche effect can also be seen in the recovery of the source image where any slight changes in the key will result in an image that is completely different from the source image. Fig. 5 shows the images that are recovered from the images in Fig. 4, after changing some bits in the key.
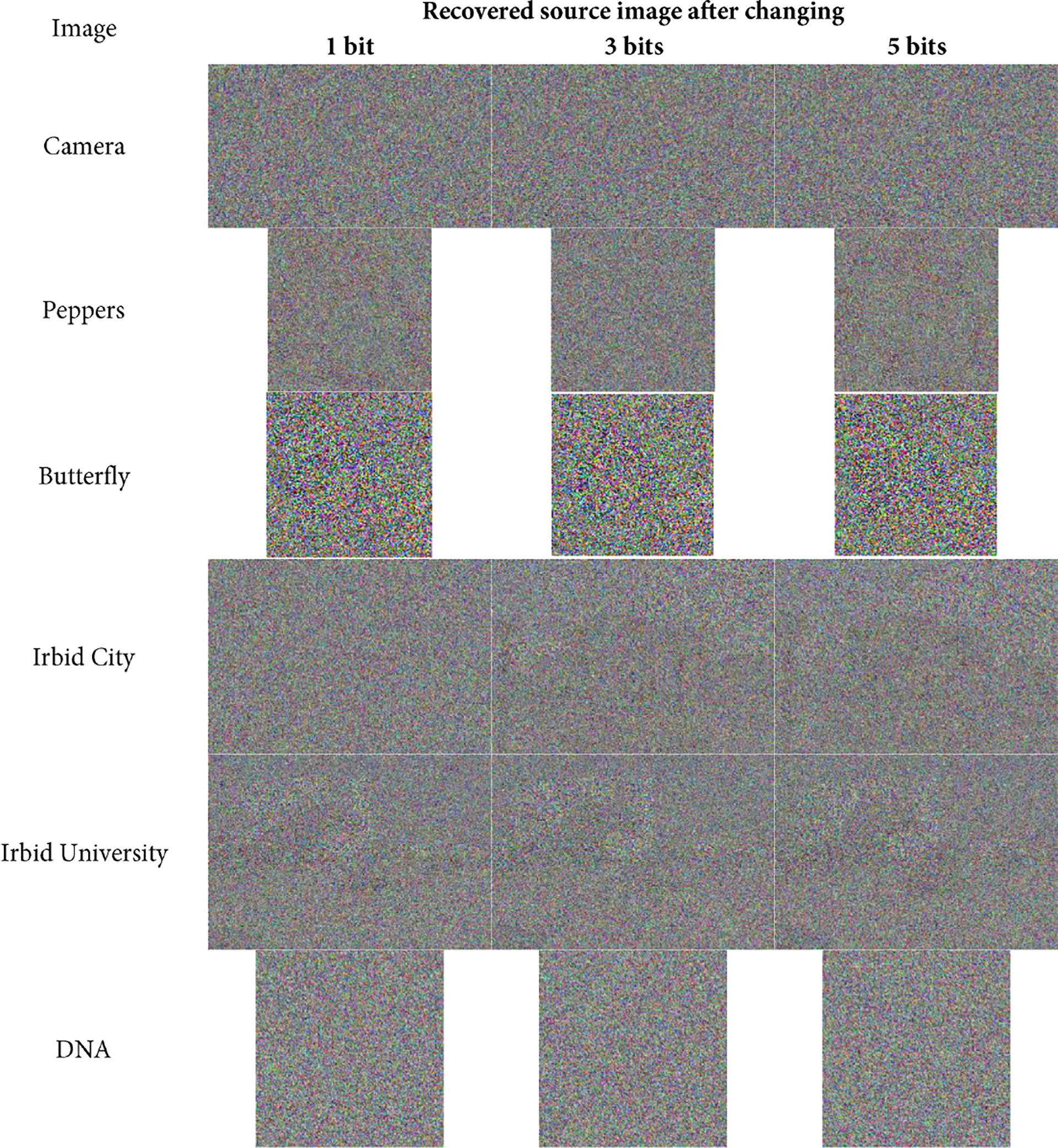
Figure 5: The recovered source images from the images in Fig. 4
It is evident from the recovered images in Fig. 5 that they are completely different from the source images in Fig. 2. This indicates the high level of sensitivity of the proposed RDNA encryption method to any slight changes in the key.
4.4 Key Size and Its Complexity
The large length and high degree of randomness of the encryption key make it very difficult to guess the key. Where attackers using brute force attack need a long time to crack the key. The use of DNA as a key in the proposed RDNA encryption method helps to increase the difficulty against attackers due to the long length and randomness properties of the DNA. The key length used in any encryption method is usually measured in bits.
In the proposed RDNA encryption method, the user can select any digital DNA file of any size to use as an encryption key. The key length (in bits) used in the proposed RDNA encryption method is equal to the total number of bits in the source image and is calculated using Eq. (7).
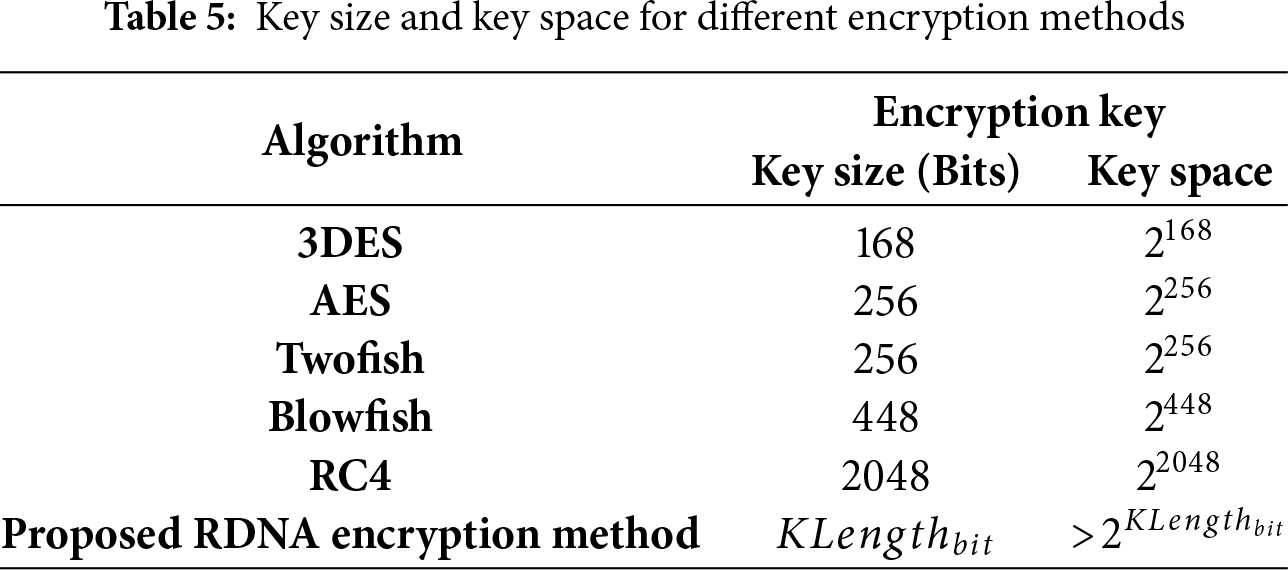
This encryption key in the proposed RDNA encryption method results in a huge keyspace compared to other encryption methods, such as 3DES, AES, Twofish, Blowfish, and RC4 [42], as shown in Table 5, which indicates a high level of security.
Encryption execution time is one of the important factors in evaluating the performance of an encryption method. Table 6 lists the encryption execution time recorded when encrypting images in Fig. 2, using the proposed RDNA encryption method and other known encryption methods (3DES, AES, and DNA-based method).
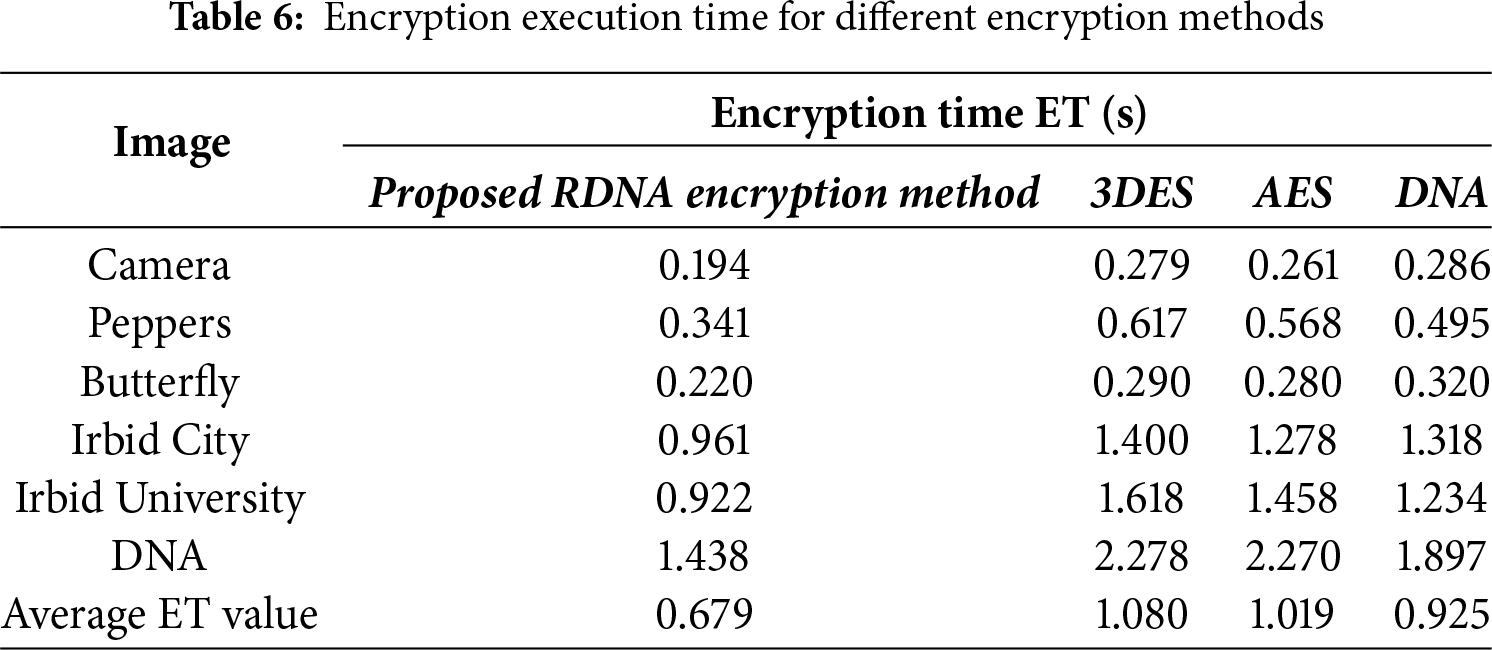
The average encryption execution time in Table 6 for the proposed method is less than other encryption methods. This indicates that the proposed RDNA encryption method has a distinct strength point compared to other encryption methods.
The amount of distortion produced in the encrypted image can be quantified numerically using two metrics: the normalized mean absolute error NMAE and the PSNR. Eqs. (8) and (9) are used to calculate NMAE and PSNR, respectively [41].
where S and E are the source image and the encrypted image, respectively, and Max is the maximum byte value in S. The NMAE and PSNR for the proposed RDNA encryption method and other known methods (3DES, AES, and DNA-based) were calculated for the images in Fig. 2, and the obtained results are listed in Tables 7 and 8. It is clear from the average values of NMAE and PSNR in Tables 7 and 8 that the proposed RDNA method succeeded in achieving competitive values compared to 3DES, AES, and DNA-based encryption methods. Figs. 6 and 7 show graphically the values recorded in Tables 7 and 8, respectively.
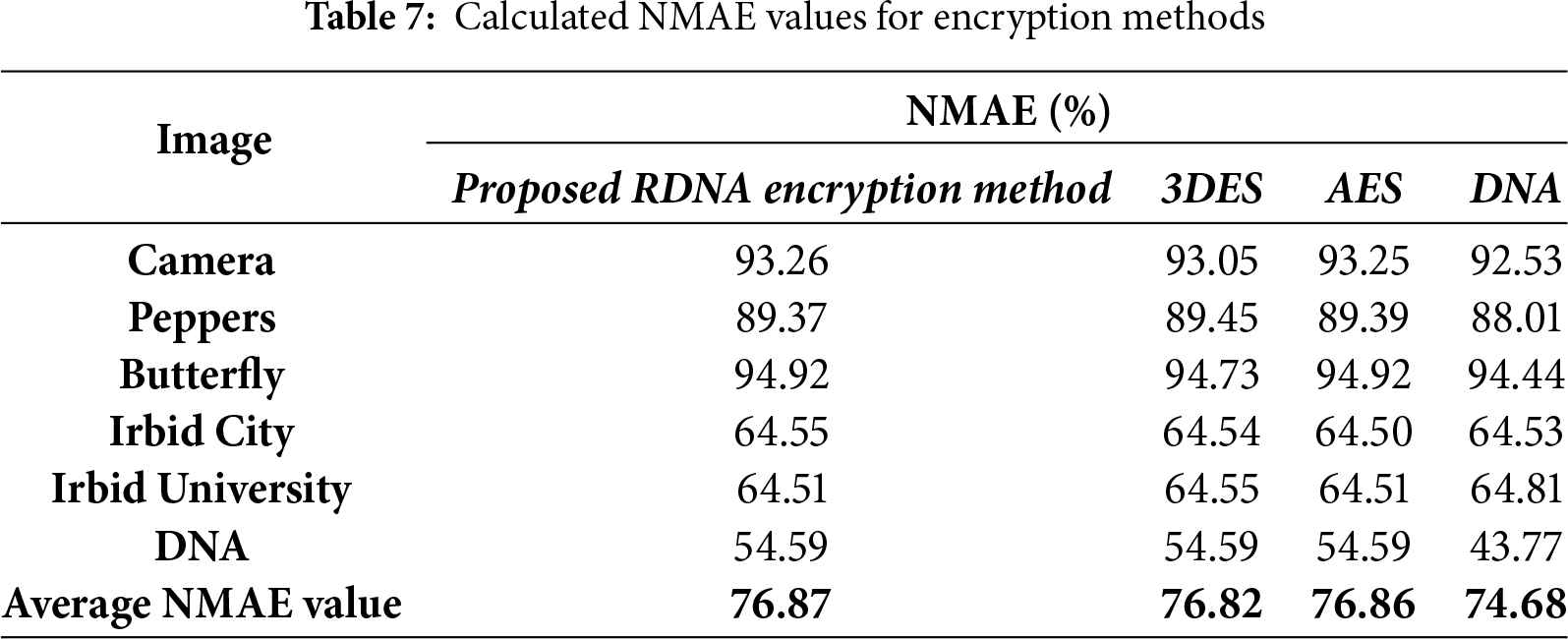
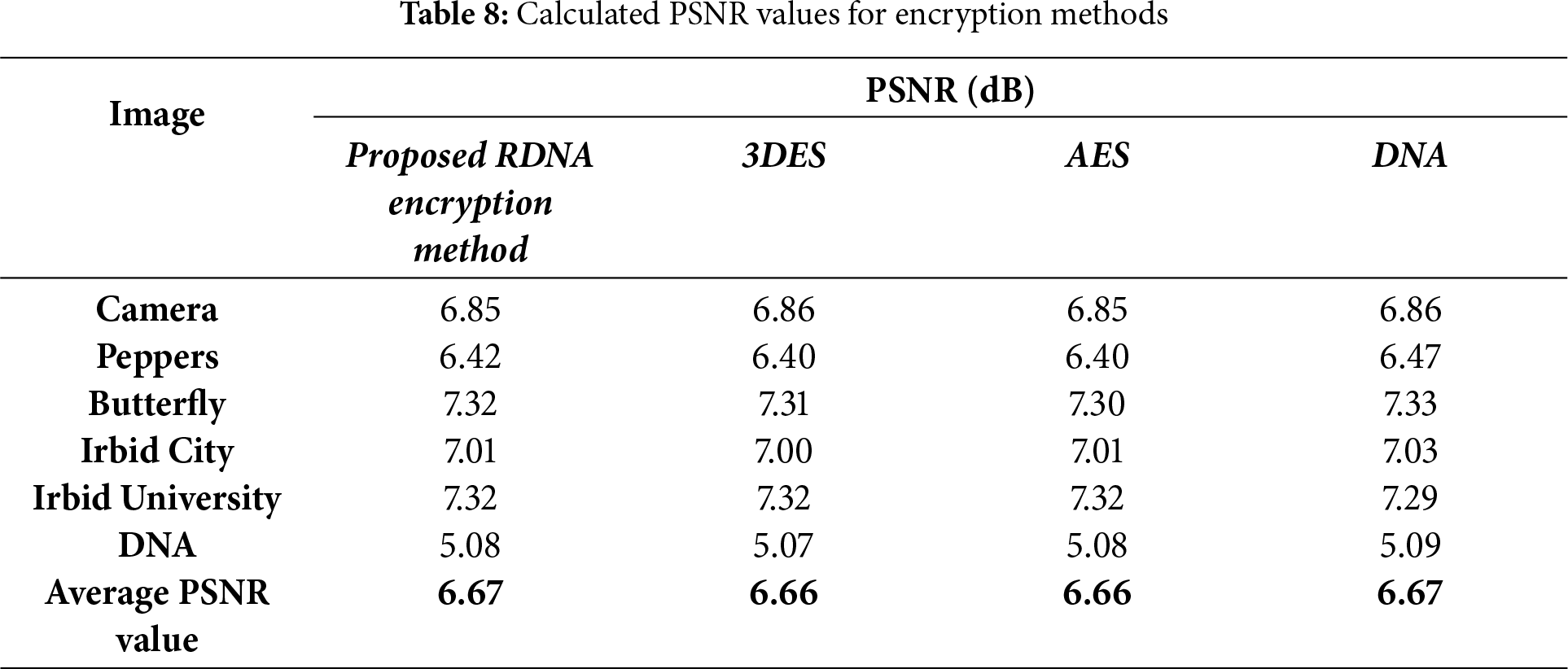
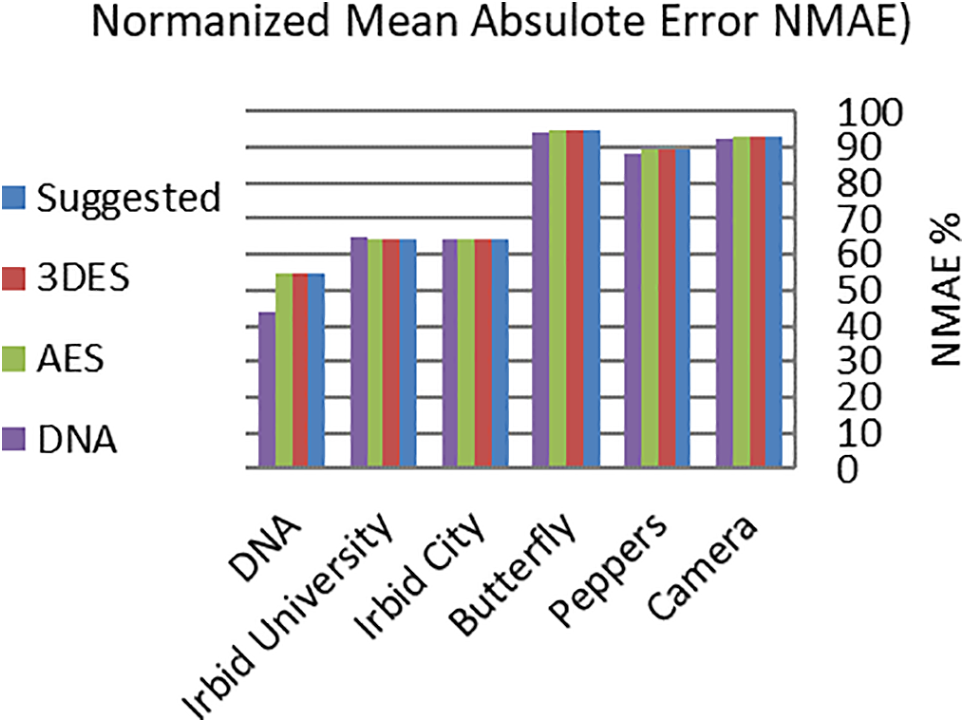
Figure 6: Graphical representation of NAME values in Table 7
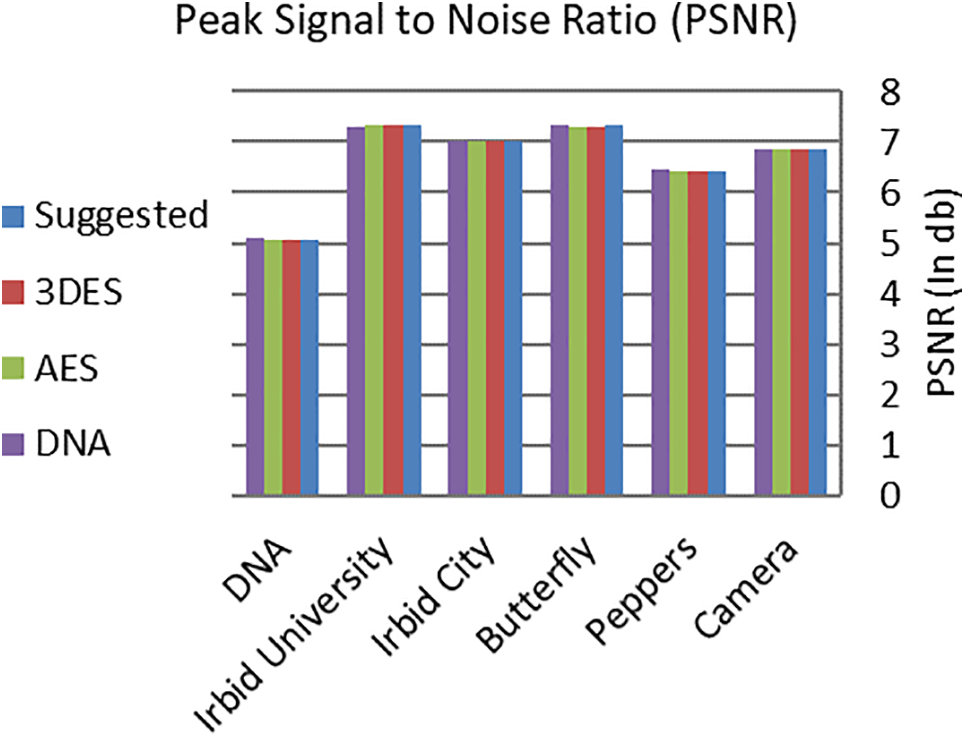
Figure 7: Graphical representation of PSNR values in Table 8
An efficient encryption method is a method that can reduce the relationship between adjacent pixel values in the encrypted image. This is the result of the distortion effect that occurs in the encryption image. Typically, correlation is calculated between every two adjacent pixels in four directions: horizontal, vertical, diagonal, and anti-diagonal using Eq. (10).
where X and Y are the colour values of two adjacent pixels in the encrypted image and N is the number of adjacent pixels selected from the image for to correlation calculation.
The correlation values were calculated by randomly selecting 5000 pairs of two adjacent pixels. Correlation values that were calculated in the experiments in four directions: vertical, horizontal, diagonal, and anti-diagonal are listed in Tables 9–12.
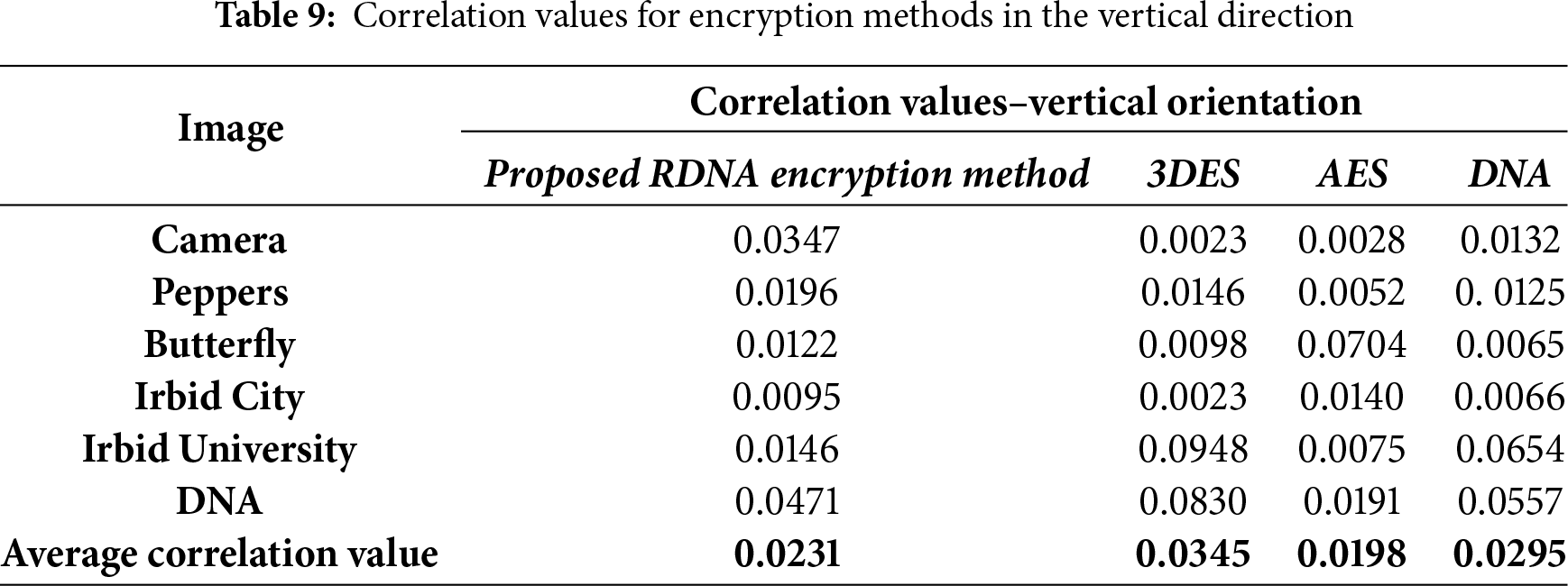
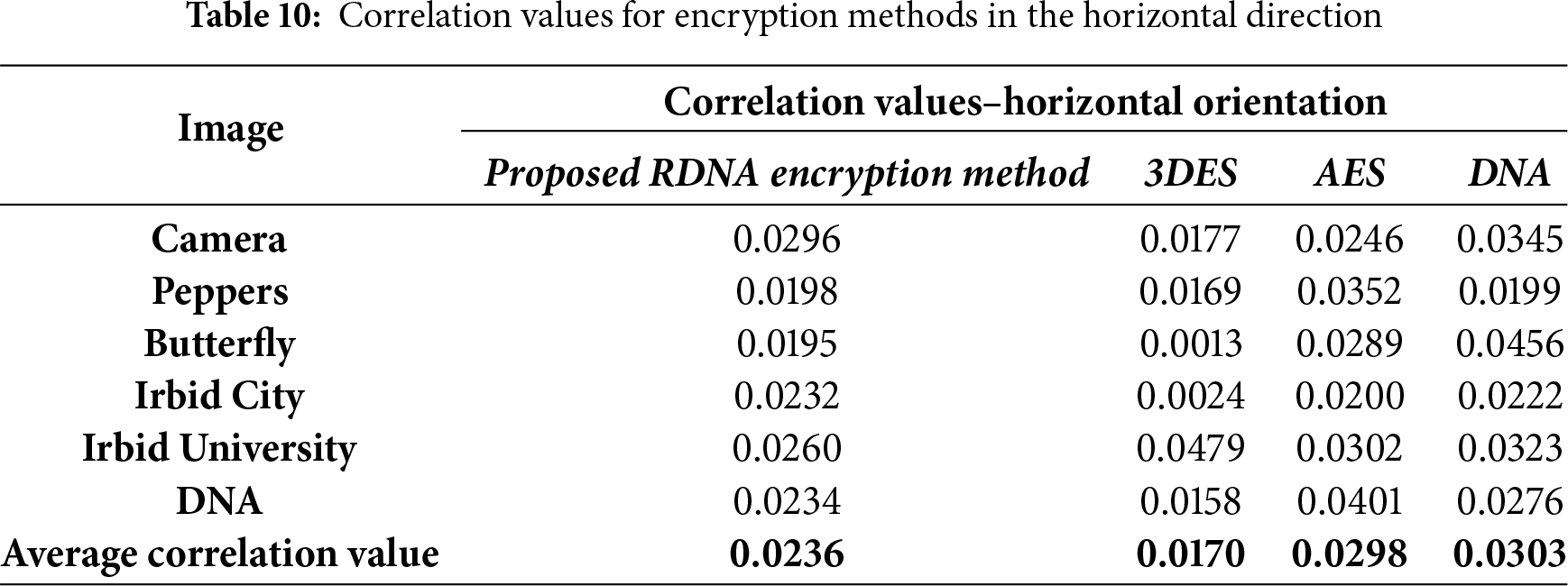
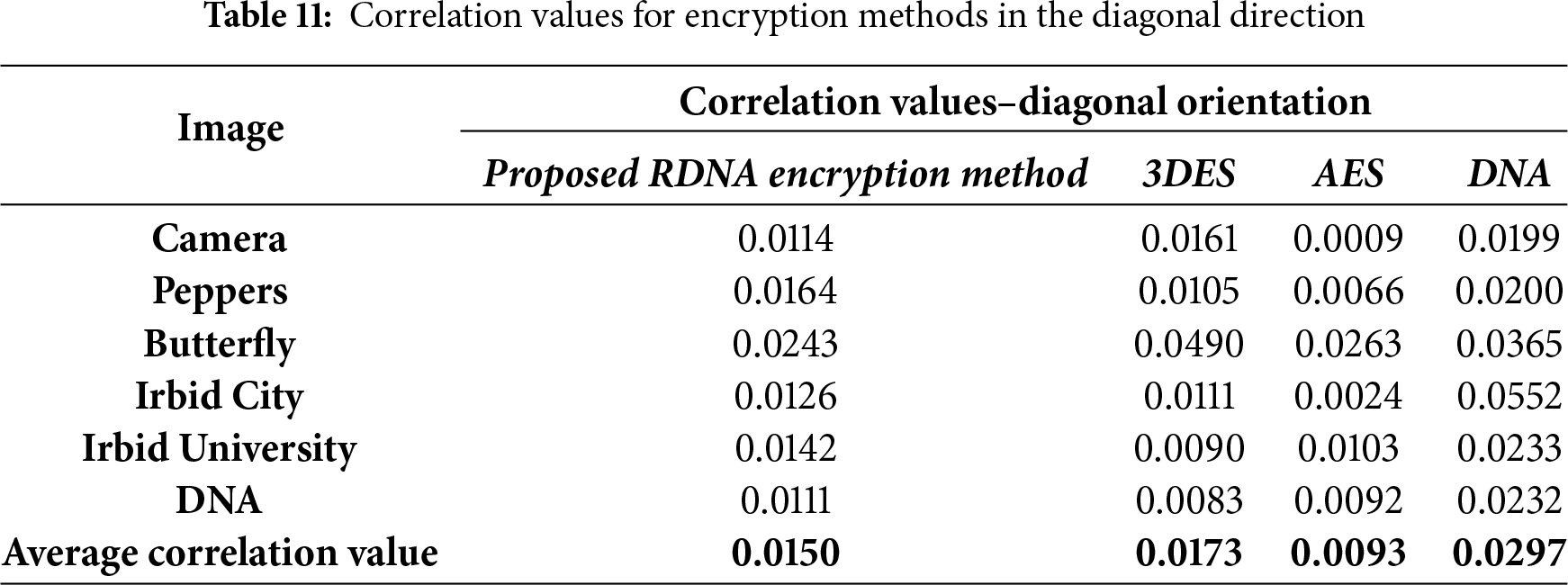
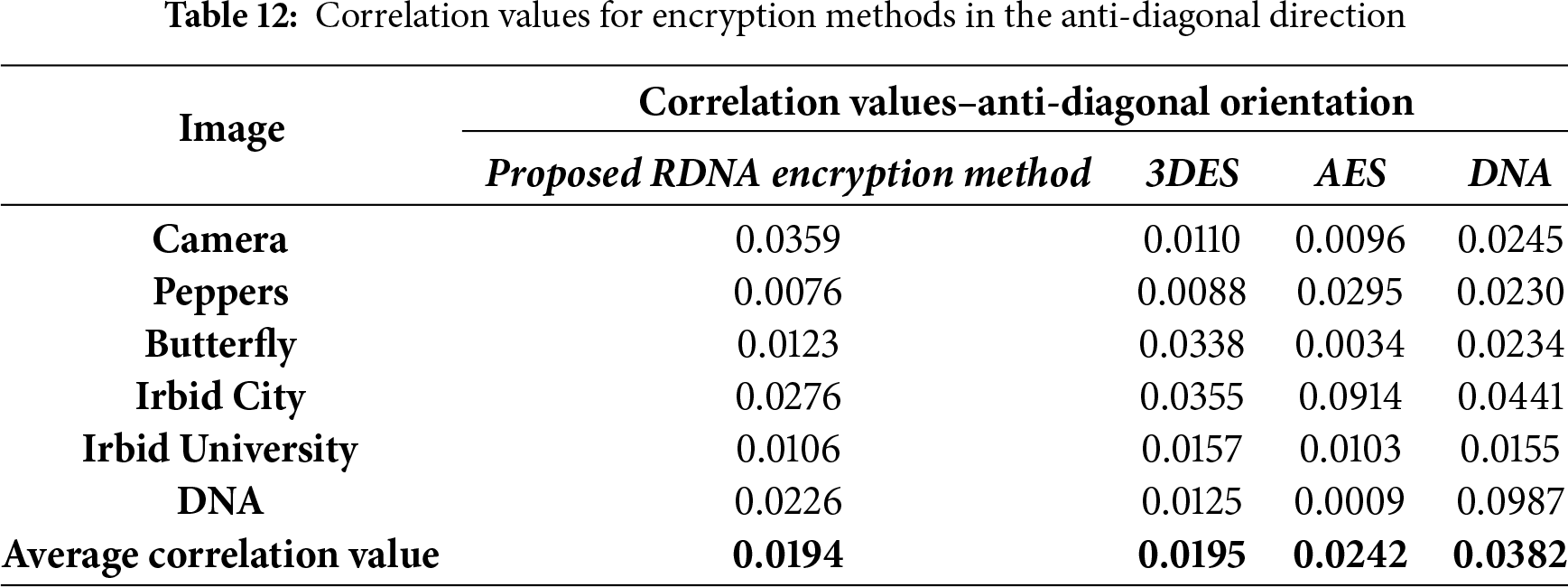
It should be noted here that the average correlation values of the proposed RDNA method in Tables 9–12 are close to other methods.
4.8 NPCR and UACI Randomness Tests
The immunity of image encryption methods to differential attacks can be tested using NPCR and UACI metrics. Eqs. (12) and (13) are used to calculate the NPCR and UACI values, respectively.
where
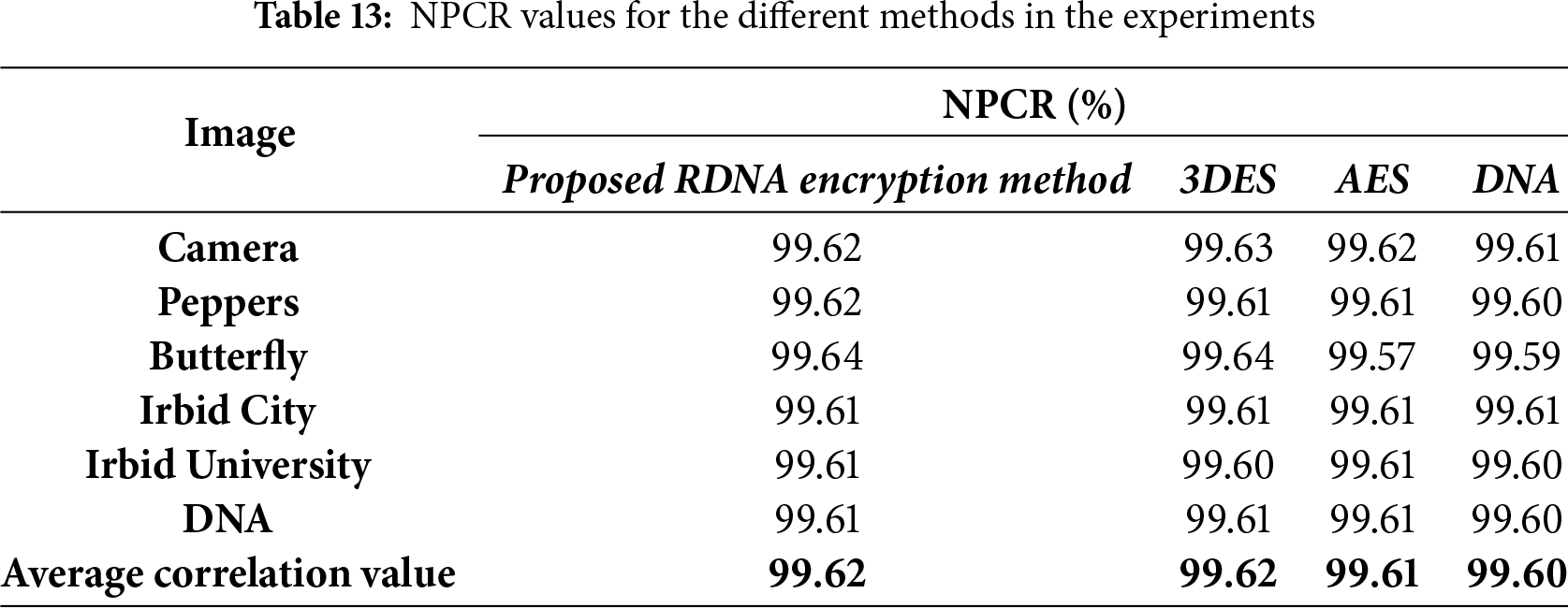
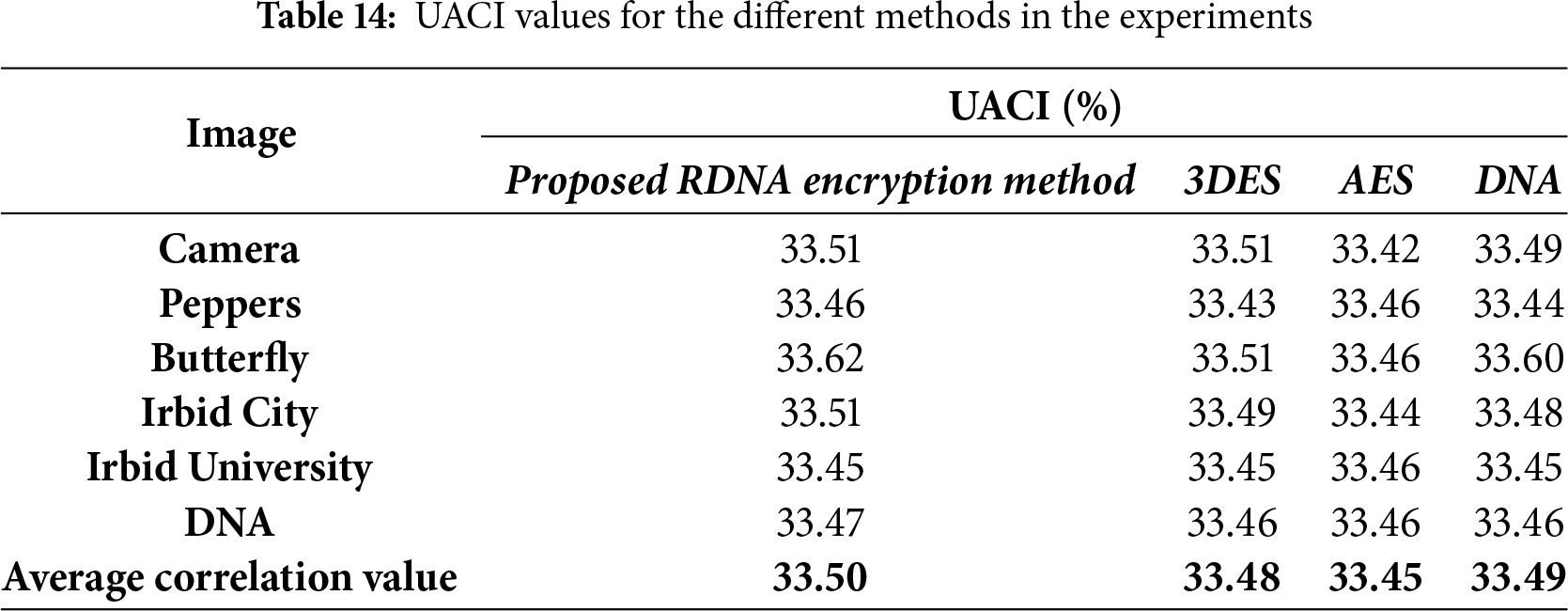
When comparing the values of NPCR and UACI for the proposed RDNA method with the other encryption methods in Tables 13 and 14, we conclude that when only one pixel in the source image is changed, the proposed method produces two different encrypted images. This means that the proposed RDNA method is very sensitive to any small changes in the source image.
4.9 Security Analysis and Resistance to Common Attacks
A robust encryption algorithm must demonstrate resilience against a wide range of cryptanalytic attacks. The proposed RDNA method is evaluated below against several common threats, with its defense substantiated by the experimental results presented in this paper.
• Resistance to Statistical Attacks: An encryption scheme is vulnerable to statistical attacks if the ciphertext reveals any patterns or statistical biases from the plaintext. The RDNA method effectively mitigates this threat. The uniformly flat histograms of the encrypted images (Fig. 3) demonstrate that the distribution of pixel intensities is nearly random. Furthermore, the average correlation coefficients between adjacent pixels in the encrypted images are exceptionally close to zero (Tables 9–12), indicating the successful removal of spatial redundancy. These two factors confirm that RDNA is highly resistant to statistical analysis.
• Resistance to Differential Attacks: In a differential attack, an adversary analyzes the difference between two ciphertexts resulting from a slight modification in the plaintext to deduce the key. The high sensitivity of RDNA to minor changes in the input is validated by the NPCR and UACI tests. The average NPCR value of 99.62% and UACI value of 33.50% (Tables 13 and 14) are very close to the ideal values, indicating that a change in a single pixel in the source image propagates throughout the entire ciphertext, producing a completely different encrypted image. This strong avalanche effect makes differential attacks infeasible.
• Resistance to Brute-Force Attacks: The security of RDNA against exhaustive key search is grounded in the immense size of its key space. As shown in Table 5, the effective key space is > 2KLengthbit, where KLengthbit is directly proportional to the image size and is typically several orders of magnitude larger than the key spaces of AES or 3DES. This vast key space, stemming from the use of a long, random DNA sequence, renders brute-force attacks computationally impractical.
• Resistance to Known/Chosen-Plaintext Attacks: The RDNA algorithm is designed to be highly sensitive to the secret key. The preparation stage (Eq. (2)) ensures that the encryption process is dependent on the entire source image, while the initial XOR operation on the DNA key (Section 3.1) ensures that any minor change in the key produces a completely different pseudo-random sequence for transposition and substitution. As evidenced by the avalanche effect on the recovered image (Fig. 5), even a 1-bit change in the key results in a total decryption failure. This strong key sensitivity, combined with the large key space, ensures that even if an attacker obtains pairs of plaintexts and ciphertexts, they cannot feasibly derive the key or decrypt other messages.
In summary, the comprehensive statistical, differential, and key space analyses confirm that the proposed RDNA method provides a high level of security against the most prevalent cryptanalytic threats.
This paper has proposed a novel RDNA encryption method based on the exploitation of structural attributes inherent in DNA nucleotides. The RDNA method utilises the inherent randomness and structural properties of DNA nucleotides to generate a highly robust encryption key with a vast number of possible combinations. A thorough performance evaluation of the proposed RDNA encryption method has been conducted through comprehensive security assessments, encompassing statistical and visual testing, avalanche effects analysis, and entropy analysis. The outcomes of the evaluation elucidate that the proposed RDNA encryption method outperforms its rivals in the literature, including other established cryptosystems such as DES, 3DES, AES, and another DNA-based method. The empirical findings articulated in this paper substantiate the high security level and resilience of the RDNA encryption method, making it a promising advancement within the specialized domain of image encryption methodologies. As a promising avenue for future research, we intend to study the effects of multi-round encryption, where different segments of the same long DNA key are used for each round, potentially further increasing security. This entails generating distinct keys for each encryption round, offering valuable insights into potential enhancements and variations in security. These investigations will not only contribute to further refining the RDNA encryption method but also extend its applicability within the realm of high-performance parallel distributed computing systems. Furthermore, as part of our future work, We will also explore integrating asymmetric key techniques, such as Elliptic Curve Cryptography, for secure DNA key exchange, with the goal of achieving a >10% increase in resistance to differential attacks without a significant increase in computational overhead, further enhancing its robustness and security. Additionally, we plan to include more NIST tests to further evaluate and enhance our encryption method’s robustness, specifically incorporating side-channel attack analysis to assess and mitigate vulnerabilities related to implementation characteristics, and the chi-square test to rigorously evaluate the statistical properties of our encryption method.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Conceptualization: Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi and Abdelrahman Abuarqoub; methodology: Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi, Omar Albahbouh Aldabbas and Abdelrahman Abuarqoub; software: Mohammed Abbas Fadhil Al-Husainy and Bassam Al-Shargabi; formal analysis: Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi and Abdelrahman Abuarqoub; writing: Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi, Omar Albahbouh Aldabbas and Abdelrahman Abuarqoub; review and editing: Bassam Al-Shargabi, Mohammed Abbas Fadhil Al-Husainy and Abdelrahman Abuarqoub; visualization: Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi and Abdelrahman Abuarqoub. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Hosny KM, Kamal ST, Darwish MM, Papakostas GA. New image encryption algorithm using hyperchaotic system and fibonacci Q-matrix. Electronics. 2021;10(9):1066. doi:10.3390/electronics10091066. [Google Scholar] [CrossRef]
2. Akkasaligar PT, Biradar S. Selective medical image encryption using DNA cryptography. Inf Secur J A Glob Perspect. 2020;29(2):91–101. doi:10.1080/19393555.2020.1718248. [Google Scholar] [CrossRef]
3. Wu Y, Zhang L, Berretti S, Member S, Wan S. Medical image encryption by content-aware DNA computing for secure healthcare. IEEE Trans Industr Inform. 2023;19(2):2089–98. doi:10.1109/TII.2022.3194590. [Google Scholar] [CrossRef]
4. Radhakrishnan I, Jadon S, Honnavalli PB. Efficiency and security evaluation of lightweight cryptographic algorithms for resource-constrained IoT devices. Sensors. 2024;24(12):4008. doi:10.3390/s24124008. [Google Scholar] [PubMed] [CrossRef]
5. Kumar S, Pillai CS. An analysis of light weight symmetric encryption algorithms for secure data transmission in IoT. In: Proceedings of the 2024 International Conference on Intelligent Algorithms for Computational Intelligence Systems (IACIS); 2024 Aug 23–24; Hassan, India. doi:10.1109/IACIS61494.2024.10721701. [Google Scholar] [CrossRef]
6. Priyanka, Singh AK. A survey of image encryption for healthcare applications. Evol Intell. 2023;16(3):801–18. doi:10.1007/s12065-021-00683-x. [Google Scholar] [PubMed] [CrossRef]
7. Ugwu AO, Gao X, Ugwu JO, Chang V. Ethical implications of AI in healthcare data: a case study using healthcare data breaches from the US department of health and human services breach portal between 2009–2021. In: Proceedings of the 2022 International Conference on Industrial IoT, Big Data and Supply Chain (IIoTBDSC); 2022 Sep 23–25; Beijing, China. doi:10.1109/IIoTBDSC57192.2022.00070. [Google Scholar] [CrossRef]
8. Fadhil Al-Husainy MA. A novel image encryption algorithm based on the extracted mapof overlapping paths from the secret key. RAIRO-Theor Inf Appl. 2016;50(3):241–9. doi:10.1051/ita/2016023. [Google Scholar] [CrossRef]
9. Kaur M, Kumar V. A comprehensive review on image encryption techniques. Arch Comput Meth Eng. 2020;27(1):15–43. doi:10.1007/s11831-018-9298-8. [Google Scholar] [CrossRef]
10. Fang P, Liu H, Wu C, Liu M. A survey of image encryption algorithms based on chaotic system. Vis Comput. 2023;39(5):1975–2003. doi:10.1007/s00371-022-02459-5. [Google Scholar] [CrossRef]
11. Sabri O, Al-Shargabi B, Abuarqoub A, Ali Hakami T. A lightweight encryption method for IoT-based healthcare applications: a review and future prospects. IoT. 2025;6(2):23. doi:10.3390/iot6020023. [Google Scholar] [CrossRef]
12. Xiao G, Lu M, Qin L, Lai X. New field of cryptography: DNA cryptography. Chin Sci Bull. 2006;51(12):1413–20. doi:10.1007/s11434-006-2012-5. [Google Scholar] [CrossRef]
13. Al-Shargabi B, Fadhil Al-Husainy MA. Multi-round encryption for COVID-19 data using the DNA key. Int J Electr Comput Eng. 2022;12(1):478. doi:10.11591/ijece.v12i1.pp478-488. [Google Scholar] [CrossRef]
14. Chu L, Su Y, Yao X, Xu P, Liu W. A review of DNA cryptography. Intell Comput. 2025;4:0106. doi:10.34133/icomputing.0106. [Google Scholar] [CrossRef]
15. Namasudra S, Devi D, Kadry S, Sundarasekar R, Shanthini A. Towards DNA based data security in the cloud computing environment. Comput Commun. 2020;151(1):539–47. doi:10.1016/j.comcom.2019.12.041. [Google Scholar] [CrossRef]
16. Al-Shargabi B, Sabri O, Albahbouh Aldabbas O, Abuarqoub A. A survey on lightweight encryption methods for IoT-enabled healthcare applications. In: Proceedings of the 7th International Conference on Future Networks and Distributed Systems; 2023 Dec 21–22; Dubai, United Arab Emirates. p. 753–7. doi:10.1145/3644713.3644839. [Google Scholar] [CrossRef]
17. Huang X, Ye G. An image encryption algorithm based on hyper-chaos and DNA sequence. Multimed Tools Appl. 2014;72(1):57–70. doi:10.1007/s11042-012-1331-6. [Google Scholar] [CrossRef]
18. Hazra A, Ghosh S, Jash S. A review on DNA based cryptographic techniques. Int J Netw Secur. 2018;20:1093–104. [Google Scholar]
19. De Silva PY, Ganegoda GU. New trends of digital data storage in DNA. Biomed Res Int. 2016;2016:8072463. doi:10.1155/2016/8072463. [Google Scholar] [PubMed] [CrossRef]
20. Alawida M. A novel DNA tree-based chaotic image encryption algorithm. J Inf Secur Appl. 2024;83:103791. doi:10.1016/j.jisa.2024.103791. [Google Scholar] [CrossRef]
21. AL-Shargabi B, Dar Assi A. A modified lightweight DNA-based cryptography method for Internet of Things devices. Expert Syst. 2023;40(6):e13270. doi:10.1111/exsy.13270. [Google Scholar] [CrossRef]
22. Al-Shargabi B, Assi AD. An improved DNA based encryption algorithm for Internet of Things devices. In: Proceedings of the 2022 International Conference on Engineering & MIS (ICEMIS); 2022 Jul 4–6; Istanbul, Turkey. doi:10.1109/ICEMIS56295.2022.9914290. [Google Scholar] [CrossRef]
23. Hosny KM, Kamal ST, Darwish MM. Novel encryption for color images using fractional-order hyperchaotic system. J Ambient Intell Humaniz Comput. 2022;13(2):973–88. doi:10.1007/s12652-021-03675-y. [Google Scholar] [PubMed] [CrossRef]
24. Liu Q, Liu L. Color image encryption algorithm based on DNA coding and double chaos system. IEEE Access. 2020;8:83596–610. doi:10.1109/ACCESS.2020.2991420. [Google Scholar] [CrossRef]
25. Mirzajani S, Moafimadani SS, Roohi M. A new encryption algorithm utilizing DNA subsequence operations for color images. AppliedMath. 2024;4(4):1382–403. doi:10.3390/appliedmath4040073. [Google Scholar] [CrossRef]
26. Huang L, Ding C, Bao Z, Chen H, Wan C. A DNA encoding image encryption algorithm based on chaos. Mathematics. 2025;13(8):1330. doi:10.3390/math13081330. [Google Scholar] [CrossRef]
27. Masood F, Masood J, Zhang L, Jamal SS, Boulila W, Rehman SU, et al. A new color image encryption technique using DNA computing and chaos-based substitution box. Soft Comput. 2022;26(16):7461–77. doi:10.1007/s00500-021-06459-w. [Google Scholar] [CrossRef]
28. Jithin KC, Sankar S. Colour image encryption algorithm combining Arnold map, DNA sequence operation, and a Mandelbrot set. J Inf Secur Appl. 2020;50(2):102428. doi:10.1016/j.jisa.2019.102428. [Google Scholar] [CrossRef]
29. Akgul A, He S, Patidar V. A novel conservative chaos driven dynamic DNA coding for image encryption. Front Appl Math Stat. 2023;8:1100839. doi:10.3389/fams.2022.1100839. [Google Scholar] [CrossRef]
30. Li T, Yang M, Wu J, Jing X. A novel image encryption algorithm based on a fractional-order hyperchaotic system and DNA computing. Complexity. 2017;2017:9010251. doi:10.1155/2017/9010251. [Google Scholar] [CrossRef]
31. Mondal B, Mandal T. A light weight secure image encryption scheme based on chaos & DNA computing. J King Saud Univ Comput Inf Sci. 2017;29(4):499–504. doi:10.1016/j.jksuci.2016.02.003. [Google Scholar] [CrossRef]
32. Namasudra S. A secure cryptosystem using DNA cryptography and DNA steganography for the cloud-based IoT infrastructure. Comput Electr Eng. 2022;104:108426. doi:10.1016/j.compeleceng.2022.108426. [Google Scholar] [CrossRef]
33. Aljahdali AO, Al-Harbi OA. Double layer steganography technique using DNA sequences and images. PeerJ Comput Sci. 2023;9(1):e1379. doi:10.7717/peerj-cs.1379. [Google Scholar] [PubMed] [CrossRef]
34. El-Khamy SE, Korany NO, Mohamed AG. A new fuzzy-DNA image encryption and steganography technique. IEEE Access. 2020;8:148935–51. doi:10.1109/ACCESS.2020.3015687. [Google Scholar] [CrossRef]
35. Pujari SK, Bhattacharjee G, Bhoi S. A hybridized model for image encryption through genetic algorithm and DNA sequence. Procedia Comput Sci. 2018;125:165–71. doi:10.1016/j.procs.2017.12.023. [Google Scholar] [CrossRef]
36. Babaei A, Motameni H, Enayatifar R. A new permutation-diffusion-based image encryption technique using cellular automata and DNA sequence. Optik. 2020;203:164000. doi:10.1016/j.ijleo.2019.164000. [Google Scholar] [CrossRef]
37. Li H, Zhang L, Cao H, Wu Y. Hash based DNA computing algorithm for image encryption. Appl Sci. 2023;13(14):8509. doi:10.3390/app13148509. [Google Scholar] [CrossRef]
38. Al Husainy MAF, Al Sewadi HA, Masadeh SR. Using a DNA tape as a key for encrypt images. Int J Electron Secur Digit Forensics. 2022;14(4):373. doi:10.1504/ijesdf.2022.123868. [Google Scholar] [CrossRef]
39. Zhang H, Fritts JE, Goldman SA. An entropy-based objective evaluation method for image segmentation. In: Storage and retrieval methods and applications for multimedia 2004. San Jose, CA, USA: SPIE; 2003. p. 38–49. doi:10.1117/12.527167. [Google Scholar] [CrossRef]
40. Zhang W, Wang S, Han W, Yu H, Zhu Z. An image encryption algorithm based on random Hamiltonian path. Entropy. 2020;22(1):73. doi:10.3390/e22010073. [Google Scholar] [PubMed] [CrossRef]
41. Alabdullah B, Beloff N, White M. E-ART: a new encryption algorithm based on the reflection of binary search tree. Cryptography. 2021;5(1):4. doi:10.3390/cryptography5010004. [Google Scholar] [CrossRef]
42. Kumar Kushwaha P, Singh MP, Kumar P. A survey on lightweight block ciphers. Int J Comput Appl. 2014;96(17):1–7. doi:10.5120/16883-6923. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools