 Open Access
Open Access
ARTICLE
Classification of Gastric Lesions Using Gabor Block Local Binary Patterns
1 College of Computing and Informatics, Saudi Electronic University, Riyadh, 11673, Saudi Arabia
2 College of Electrical and Mechanical Engineering, National University of Sciences and Technology, Islamabad, Pakistan
3 Department of Computer Science, National University of Sciences and Technology, Balochistan Campus, Quetta, Pakistan
* Corresponding Author: Muhammad Tahir. Email:
Computer Systems Science and Engineering 2023, 46(3), 4007-4022. https://doi.org/10.32604/csse.2023.032359
Received 15 May 2022; Accepted 04 August 2022; Issue published 03 April 2023
Abstract
The identification of cancer tissues in Gastroenterology imaging poses novel challenges to the computer vision community in designing generic decision support systems. This generic nature demands the image descriptors to be invariant to illumination gradients, scaling, homogeneous illumination, and rotation. In this article, we devise a novel feature extraction methodology, which explores the effectiveness of Gabor filters coupled with Block Local Binary Patterns in designing such descriptors. We effectively exploit the illumination invariance properties of Block Local Binary Patterns and the inherent capability of convolutional neural networks to construct novel rotation, scale and illumination invariant features. The invariance characteristics of the proposed Gabor Block Local Binary Patterns (GBLBP) are demonstrated using a publicly available texture dataset. We use the proposed feature extraction methodology to extract texture features from Chromoendoscopy (CH) images for the classification of cancer lesions. The proposed feature set is later used in conjuncture with convolutional neural networks to classify the CH images. The proposed convolutional neural network is a shallow network comprising of fewer parameters in contrast to other state-of-the-art networks exhibiting millions of parameters required for effective training. The obtained results reveal that the proposed GBLBP performs favorably to several other state-of-the-art methods including both hand crafted and convolutional neural networks-based features.Keywords
Texture is an important visual characteristic of image classification and has drawn the attention of researchers during the past three decades. Texture analysis has found a number of potential applications including biomedical imaging [1], image retrieval [2], remote sensing [3,4], aerial imaging [5], computer vision [6,7] etc. Anticipating the significance of visual texture description, numerous techniques have been developed by different researchers in the field. The applicability of the designed methodologies is restricted to the requirements of intended application domain. Textures from Real-world are usually dynamic in nature owing to the disparities in illumination, orientation, scale, and other visual appearances of the image. One such application where these requirements are very crucial is gastroenterology (GE) imaging.
With the advancement of machines and modern technology, sedentary lifestyle in human societies is a common trend. Moreover, consumption of high caloric and unhealthy food has led to rapid increase in the diseases related to digestive system. Some of the major diseases related to GE include gastric cancer, colon cancer, ulcers of various kinds, inflammation, polyps, abnormal lesions, and bleeding. The stomach and its associated gastrointestinal (GI) tract are not visible through normal non-invasive procedures. Usually, endoscopy of the GI tract is used to diagnose potential problems. In this procedure, an endoscope mounted with a camera and light source at its tip is inserted through the GI tract. It thus, can help the physicians in visual examination of the GI tract and finding abnormalities. If need arises, a biopsy of the suspected region is required which is an invasive procedure. Invariance property of image texture is desired in this particular scenario due to the low-level control of the camera where it is incapable of maintaining a constant distance from the visualized tissue, giving rise to a visual analysis of the tissue at different scales under various homogeneous illuminations. For some organs such as stomach where the tissue is soft, it is not possible to view the tissue while keeping it orthogonal to the camera giving rise to illumination gradients appearing in the visualized tissue. Additionally, the lack of sense of a specific direction creates the possibility of viewing the same tissue from various perspectives. It is expected that a reliable assisted diagnosis system should be able to analyze the visual patterns in the images uniformly without getting effected by the above-mentioned imaging dynamics. Additionally, with the latest technological advancements, many new imaging modalities are being introduced having distinct complementary visual characteristics. In the specific scenario of GE imaging, these new imaging technologies include (but are not limited to) chromoendoscopy, narrow-band imaging, capsule endoscopy etc. These modalities require extensive validation before they can be deployed for effective clinical decision making. With the design of more generic image descriptors, the assisted decision-making systems have the potential to cater for the imaging dynamics which are specific to this technological growth, thus bridging the widening gap between computer vision and technology.
In this article, we put forward novel features based on Gabor filters. The key contributions of this work include:
1. We introduce novel GBLBP texture descriptors, which are multiresolution features owing to the ability of Gabor filters to perform multiresolution analysis, and have the ability to perform an analysis of microtexture features; a trademark of the techniques based on local binary patterns. The features are useful for texture analysis in methods that require more robust texture-based features.
2. We investigate the capabilities of a shallow Convolutional Neural Network (CNN) architecture in exploiting the GBLBP feature maps for the classification of images from an underlying imaging scenario.
3. We validate the GBLBP method on a gastrointestinal imaging modality, named as Chromoendoscopy. Empirical results demonstrated the superiority of the GBLBP technique.
Rest of the article is structured as follows. First, Section 2 reviews the existing literature. Next, Section 3 discusses the gastroenterology datasets used for our experiments. Then, Section 4 describes the methods adopted for feature extraction. After that, Section 5 presents the experimental results and their analysis. Finally, Section 6 draws the conclusive remarks.
Extraction of features from GE imagery is a challenging task that has been addressed by various researchers using different techniques as summarized in Table 1. Hwang et al. [8] developed a technique to detect bleeding in images of capsule endoscopy (CE) by manually selecting blood and non-blood pixels for training their respective probability models. Their results are highly dependent on the training data. Mackewicz et al. [9] employed adaptive color histograms to track color distributions of bleeding in CE videos over time. They utilized Hue-Saturation-Intensity (HSI) and Local Binary Patterns (LBP) to extract features from input images. The features extracted this way are sensitive to illumination due to the HSI components and therefore, the methodology does not discriminate between dark and normal bleeding regions. Li et al. [10] used LBP and chrominance moments for feature extraction. For chrominance, only hue and saturation components were used for illumination invariance of features. Their experimental validation is limited given that for the enhancement of dataset (for statistically relevant results). They divided the images into blocks and formed training and testing sets from these blocks.
Hwang et al. [11] devised a novel method based on initial marker selection and watershed segmentation techniques to detect polyps. Their proposed method demonstrated good efficacy in the identification of polyps with high sensitivity values but lower specificity values because non-polyp regions contain rich texture and led the classifier to wrongly classify them as polyps. Karargyris et al. [12] proposed a framework for the detection of polyps in the bowel. The authors used a synergistic approach based on filtering, edge detection and active contours to do segmentation of polyps. This method exhibits rule-based decision making (method is data driven) which strongly effects the specificity of the algorithm. Therefore, this method is not considered generic enough for a broader range of datasets. Li et al. [13] proposed a texture descriptor based on acquiring LBPs of the wavelet subbands giving good results for tumor detection. Hafner et al. [14] applied pit-pattern classification for detection of colonic polyps. They have extracted texture features in the wavelet domain using Dual Tree Complex Wavelet Transform (DTCWT). They have performed classification using the nearest neighborhood classifier. The authors in their conclusion have argued that due to the lighting conditions, shift and rotation invariant features are more effective during endoscopy examination, a conclusion that we have used as a starting point for our research. Coimbra et al. [15] carried out statistical analysis of MPEG-7 visual description to classify CE images. Their experiments showed that the multiresolution analysis of images exhibits definitive advantages over normal image descriptors. Kumar et al. [16] performed analysis of Crohn’s disease lesions in CE images. The authors used Homogeneous Texture (HT), Dominant Color Descriptor (DCD) and Edge Histogram Descriptor (EHD) in all their possible permutations for lesion classification. The collective usage of all the descriptors has demonstrated superior performance in the classification task. Hafner et al. [17] utilized ring filters to extract features, which are exploited by Support Vector Machine (SVM) for classification. They showed that their results are slightly inferior to the combination of DTCWT and LBP features. Riaz et al. [18] extracted features using Gabor filters from GE imagery that are invariant to scale, rotation as well as homogeneous illumination.
Recently, the emergence of wireless capsule endoscopy (WCE) has given rise to more research challenges and potential solutions. Caroppo et al. [19] employed three pre-trained Convolutional Neural Networks (CNNs) namely, VGG19, InceptionV3 and ResNet50 as part of a feature extraction method for bleeding detection. The minimum Redundancy Maximum Relevance method and different fusion rules are then employed to select and fuse these features, which are finally classified into bleeding and nonbleeding categories using supervise machine learning classifiers. Tsuboi et al. [20] developed a CNN based technique formulated on Single Shot Multibox Detector for the detection of small-bowel angioectasia in CE images. In another work by Laiz et al. [21], deep learning in combination with metric learning is used to detect polyps in WCE images. Their work is especially beneficial for extraction of features from small datasets using Triplet Loss function. Yamada et al. [22] proposed a system for the detection of colorectal neoplasms in colon capsule endoscopy (CCE) images. They used CNNs with Single Shot Multibox Detector to automatically detect lesions such as polyps and cancers of various types. Deep learning is also used to detect mucosal ulcers, or small-bowel ulcers, in Crohn’s disease (CD) on capsule endoscopy (CE) images by Klang et al. [23]. They used a large dataset of images and have reported reasonably good accuracy on the test images. However, as is the case with all deep learning-based algorithms, the algorithm is time consuming when it comes to the training phase. Several generic conclusions based on this brief review are as follows:
1. Multiresolution analysis is the key to achieving good performance for extracting features from GE images.
2. The feature design should be generic enough to cater for the imaging dynamics arising from different imaging modalities.
3. Invariance of features to scale, rotation, homogeneous illumination as well as illumination gradients are the most important requirements for the design of image descriptors due to the lack of strong control over the endoscopic probe.
A GI tract is composed of different organs including mouth, esophagus, stomach, small intestine, and large intestine. The anatomy and structure of these organs pose different complications in examining GI. For instance, small intestine and esophagus restricts the field of view during the examination of internal probe due to their tube-like structures that allows fewer possibilities of positioning the camera inside these organs to observe the tissues. In contrast, stomach is a sac-like structure that allows numerous perspectives while scanning the same tissue however, leading the camera to rotate unpredictably. Hence, imaging conditions in stomach are unfavorable and difficult to control. Different imaging conditions may give rise to the need for rotation invariance, scale, and average amount of illumination due to the appearance of inappropriate rotation, scale, and brightness or darkness. Illumination and scale variances are caused by varying distances between the light source attached to the tip of endoscope and the GI tract wall. Note that the tract structure may cause shadows that lead to non-homogeneous components. This issue can be addressed with image segmentation techniques. In this article, such complexities are avoided by annotating the data with the help of expert physicians. Images obtained through vital stained magnification endoscopy (Chromoendoscopy-CH) have been used to analyze the proposed GBLBP method under dynamic imaging conditions for diagnosing stomach cancer. The images from this modality are categorized as normal, pre-cancerous, and cancerous following the Dinis-Ribeiro classification scheme for CH images [24], which employs the medical taxonomies for defining the clinical interpretations of tissues. CH images are grouped into their respective categories according to regularity of pit patterns, color, and shape attributes as shown in Fig. 1. Images are placed under Group I category if the images exhibit regular patterns of gastric mucosal and do not change color after staining with methylene blue. Similarly, images are grouped into Group II category if the mucosa are regular after the tissues are stained in blue. Likewise, images are arranged in Group III category if there is no clear noticeable pattern as well as change in color.
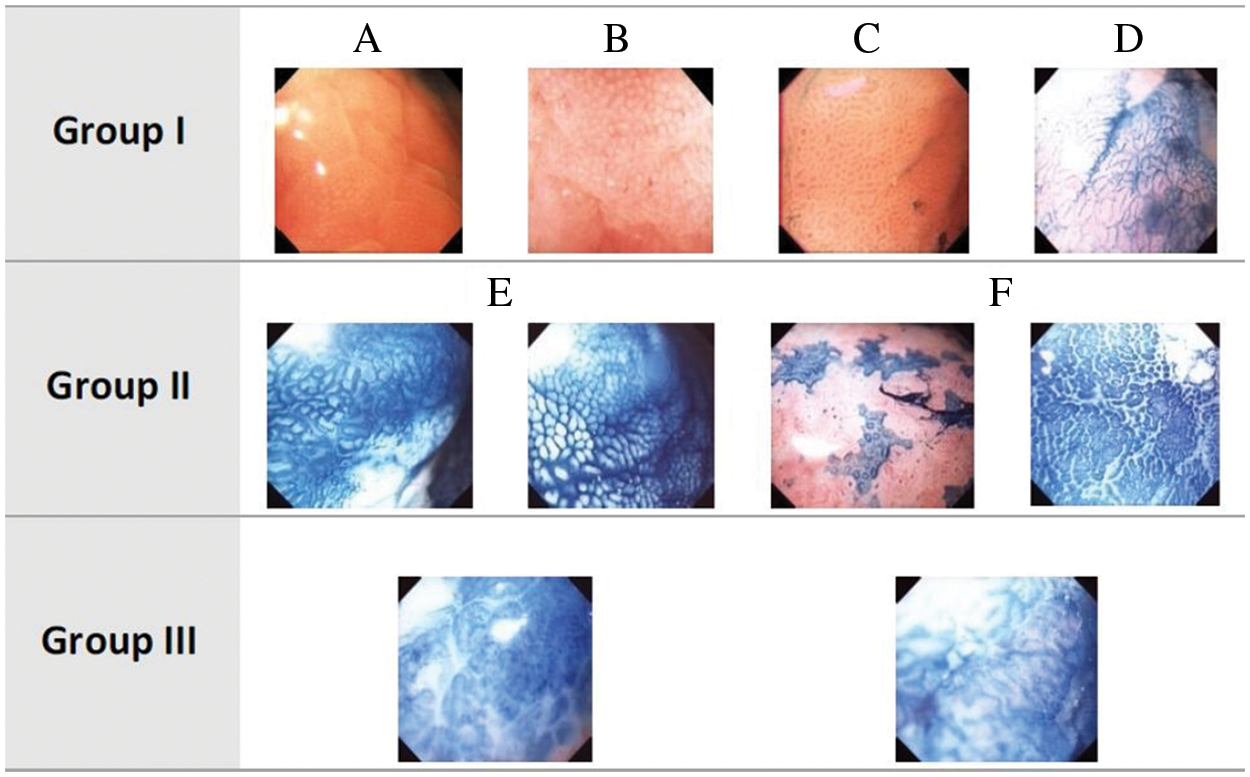
Figure 1: Dinis-Ribeiro classification of CH images. CH images: Normal (Group I), Pre-cancerous (Group II) and Cancerous (Group III)
Olympus GIF-H180 was used to obtain CH images at the Portuguese Institution of Oncology (IPO) Porto, Portugal during routine clinical procedures. Interested readers may refer to [18] for more details. Initially, 176 images, from a four-hour long video, have been excerpted and provided to two physicians for an independent analysis. The two physicians were equipped with an annotation software for Region of Interest (ROI) identification in an image, its appropriate categorization into group/subgroup based on the Dinis-Ribeiro proposal and confidence level measurement of the physicians for allocation to a group. Consequently, a gold standard of 135 images was acquired demonstrating high confidence of both the physicians and their agreement. Regarding the disagreements between the physicians, it was observed that they annotated different areas for their respective conclusions. The ultimate classification of the disagreement sets was determined in consultation with clinical practitioners where worst-case analysis for fail-safe screening of the patients was taken into account. As a result, a dataset of 176 images with high quality annotations was obtained. Further refinements were made after careful analysis by removing images with similar patterns from the dataset that resulted into 126 images with the following distribution. Group I contains 37 images (29.36%), Group II contains 74 images (58.73%), and Group III has 15 images (11.90%). It should be noted that the number of Group III images is quite small. For a computer assisted diagnosis (CAD) system, training reliable models for this class could be very challenging even with the most well-known methods. Moreover, the main aim of designing a CAD system is to be able to raise an alarm on any anomaly that is detected by the system. Given this, we aim to design a CAD system in the subsequent sections of the paper, which has the ability to ‘detect’ anomalies in the images giving rise to a 2-class problem. Group A (only Group I images) and Group B (Group II and III images).
In this section, we have proposed the GBLBP framework for the extraction of features and classification of the images as shown in Fig. 2. The images are subject to Gabor filtering, followed by the usage of a variant of Local Binary Patterns for feature extraction, and subsequently, classification.
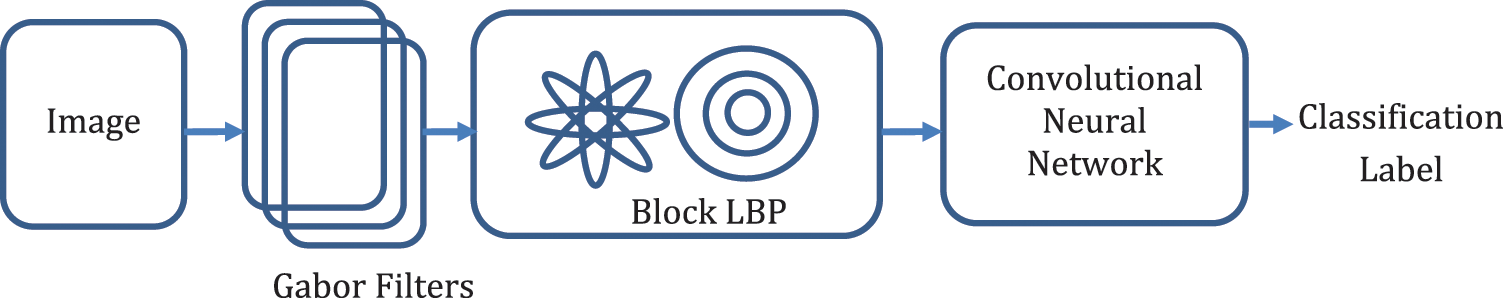
Figure 2: The proposed framework
In the proposed GBLBP framework, feature extraction is the first step that involves application of Gabor filters to the input images. Gabor filters have been widely used by researchers in related fields of texture classification during the past two decades. The key motivation of using the two-dimensional Gabor filters is that they are closely similar to the visual cortex of mammals [25]. Additionally, Gabor filters are capable of achieving optimal localization in spatial as well as frequency domains [26]. A sinusoidal plane wave, having a particular frequency and orientation, modulated by a Gaussian envelope, constitutes a Gabor filter. Gabor transformation of an image can be obtained by convolving the image with a bank of Gabor filters as shown in Eq. (1).
where
where
This filter is regarded as a bandpass filter in the frequency domain. The feature vector thus obtained is of size
4.2 Block Local Binary Patterns
The basic LBP operator was proposed by Ojala et al. [27] in 2002. The LBP patterns are obtained by calculating the difference of a central pixel and each of its neighboring pixels in a certain neighborhood [28]. Mathematically, this concept can be expressed as given in Eq. (6).
where
A visual illustration of the shapes of patterns found in endoscopy images (Fig. 3) shows that primarily, the normal and abnormal images are differentiated based on the shapes of the patterns found in the images. Typically, these can be either round or villous (elliptical) in shape, or the image might present the pathology with a pattern of very random nature, not characterized by a specific shape. Appropriately, a texture descriptor having the capability to encode the shape inherently in the features can be more useful in finding the underlying descriptive content in the images. The block LBPs proposed by Wan et al. [29] can be effectively employed in such a scenario.
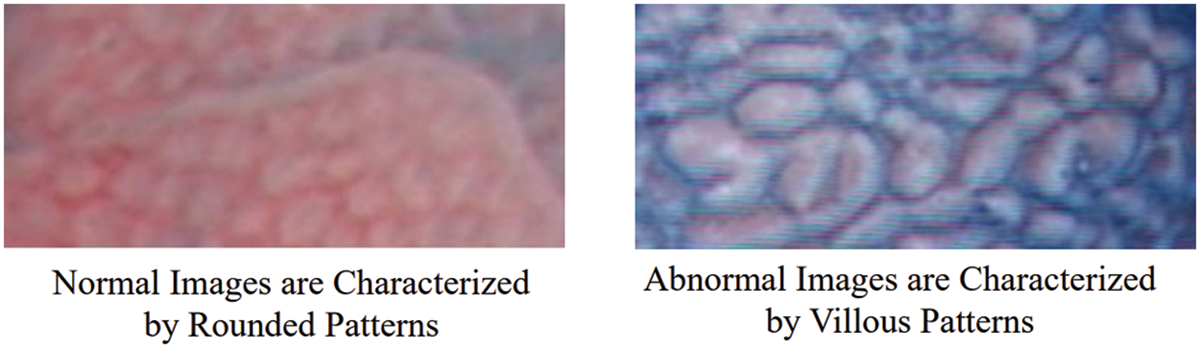
Figure 3: Shape patterns of normal and abnormal CH images
In block LBPs, a comparison of the average intensity values of pixels lying in a block of a specific shape around a certain neighborhood is performed (Fig. 4).
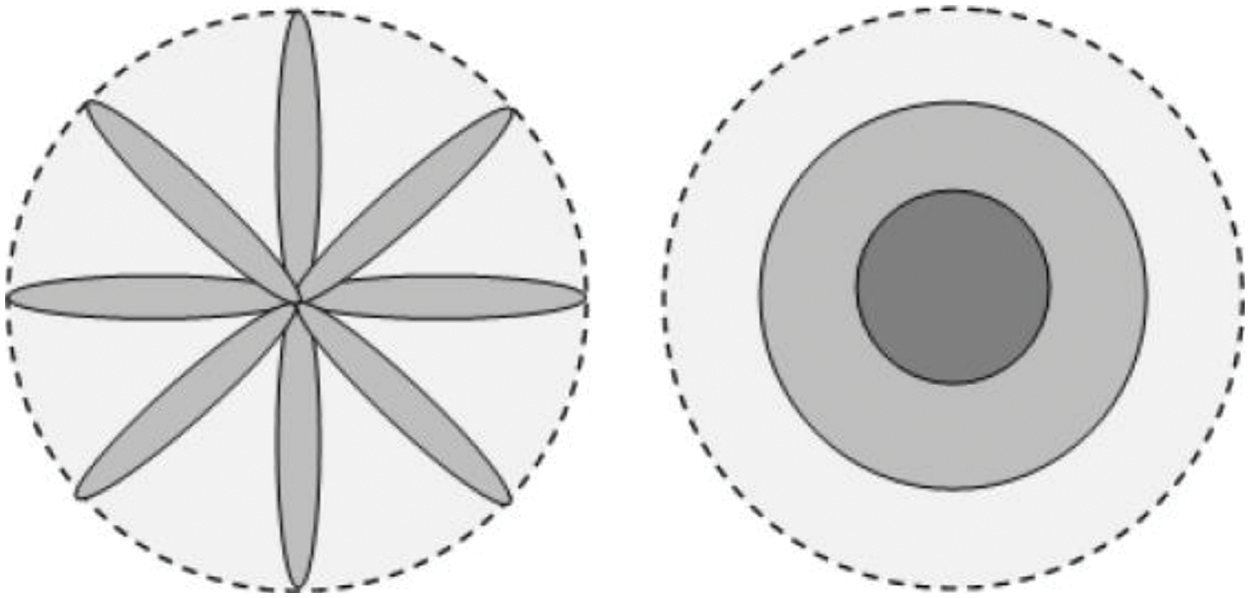
Figure 4: The spoke (left) and ring (right) patterns, indicating the capability to extract the shape-based features from the images (figure adapted from [29])
In this context, two specific patterns to perform the comparisons are done in this implementation: comparison of intensities of images lying in a ring, and around an ellipsoid also known as spoke LBP. The ring LBP is calculated by comparison of the intensity values of the center pixel with the average intensity of neighbors in a ring around the center pixel as shown in Eq. (7).
where
where
where
where
4.3 Convolutional Neural Networks
After the extraction of LBP feature maps, we feed them for the purpose of classification to a convolutional neural network. With the advent of high processing capabilities and trainable algorithms, the convolutional neural networks have gained extreme popularity in solving various image/video processing and computer vision tasks [30–32]. These networks specifically provide good results on image and video data as the inherent architecture of these networks is well aligned for extracting information from such data. The conventional convolutional neural network architecture comprises of convolution layers, pooling layers and a classification layer as illustrated in Fig. 5. The convolution layers extract useful information/features from the input in a hierarchical manner. The abstraction and complexity of the features increase with the number of convolution layers. The pooling layers decrease the spatial resolution of the features for faster processing. These layers also add some degree of invariance to the network. At the end of these layers a fully connected layer is presented followed by the classification of the respective sample at the final layer.
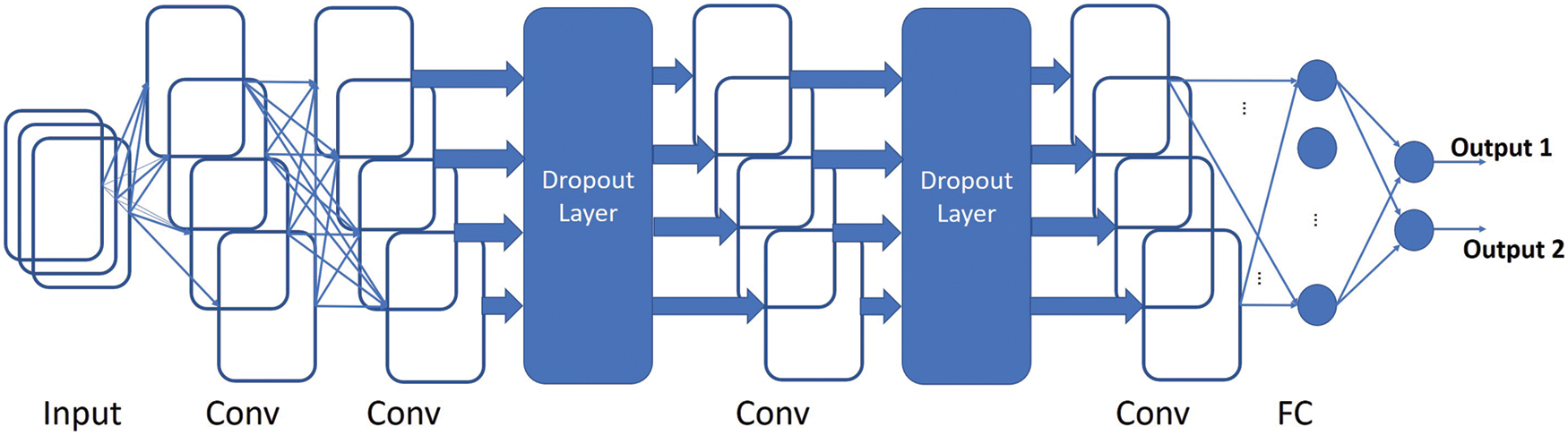
Figure 5: A visual demonstration of the architecture of the Fully Convolutional Network that has been used in this paper
We have designed a network to accept variable sized 2D images. We slightly modified the CNN architecture for the classification of this dataset. Our first convolution layer employs filters of size 4 × 4 and produces 16 feature maps. We skip the pooling layer and directly feed these 16 feature maps to the 2nd of the convolution layers. The 2nd convolution layer generates 32 feature maps and employs filters of size 4 × 4. After that we employ a drop out of 50% to improve the generalization capability of the network. The output is fed to the 3rd convolution layer that also employs filters of size 4 × 4 and generates 64 feature maps. A further drop out of 50% is used and the last convolution layer gives us 128 feature maps by employing filters of size 4 × 4. We employed the exponential linear unit (elu) activation function in all of our convolution layers. This function is known to have faster convergence rate to zero and provide better classification accuracy. After the last convolutional layer, we use the global max pooling layer to flatten our 2D features and then the densely connected softmax layer is employed for the classification purpose.
The optimization algorithm which is chosen for designing a particular CNN is critical since it helps in initialization of kernels, the weights and biases of neurons in a NN. Subsequently, the weight and bias values are adjusted on the basis of a loss function performance. In this work, we have used RMSprop (Root Mean Square Propagation) as the optimizer. It is a powerful optimizer and has proved to be very useful in recent works. It exhibits advantages over the other optimizers such as SGD (Stochastic Gradient Descent) related to the diminishing learning rates. This is because, it uses the moving average over root mean squared gradients to normalize the present gradients. This effectively means that RMSprop can automatically adjust for a small or diminishing gradients and decrease for exploding gradients.
We have used binary cross entropy (BCE) as the loss function for the proposed CNN. The loss function determines the performance of our model with a known set of labels. It is used along with the optimizer to determine the weights and biases of the neurons. The binary cross entropy can be defined as given in Eq. (11).
where
In this section, we analyze the proposed GBLBP descriptor for its performance in classifying gastroenterology (GE) images. We aim at empirical demonstration of the invariance property of the proposed GBLBP and its comparison with state-of-the-art existing approaches. Later, we will use the novel descriptor for identifying cancer in GE images. We have employed Gabor filters with six different orientations and eight scales that have been determined using the grid search technique. GBLBP demonstrated consistent performance irrespective of the selected parameters.
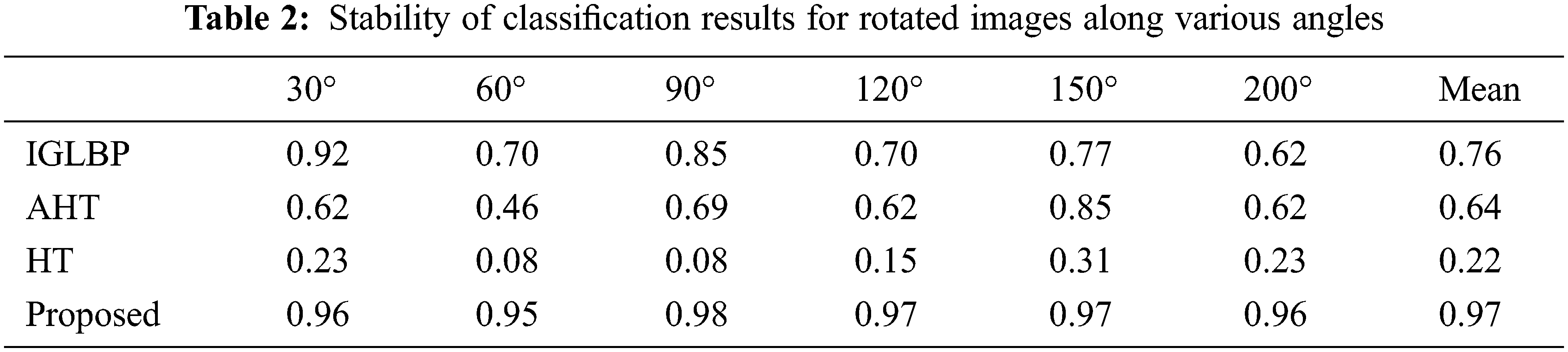
We employed USC-SIPI image database [33] to analyze the rotation invariant behavior of our proposed GBLBP technique. Interested readers may refer to [34–36] that utilized this database for the same purpose. The USC-SIPI database is composed of 13 texture images from Brodatz texture dataset where each of them is digitized along seven distinct angles including 0°, 30°, 60°, 90°, 120°, 150°, and 200° that produce a database of 91 images of size 512 × 512 each in terms of pixels resolution with 8-bit depth. The rotation invariance of GBLBP is demonstrated by training the classifier with the original images without rotation and then testing the same classifier with the rotated versions of the same images. We adopted the overall classification accuracy as a performance indicator to GBLBP. The experimental evaluations revealed that the proposed GBLBP method attributes better invariance compared to other feature-based techniques (Table 2).
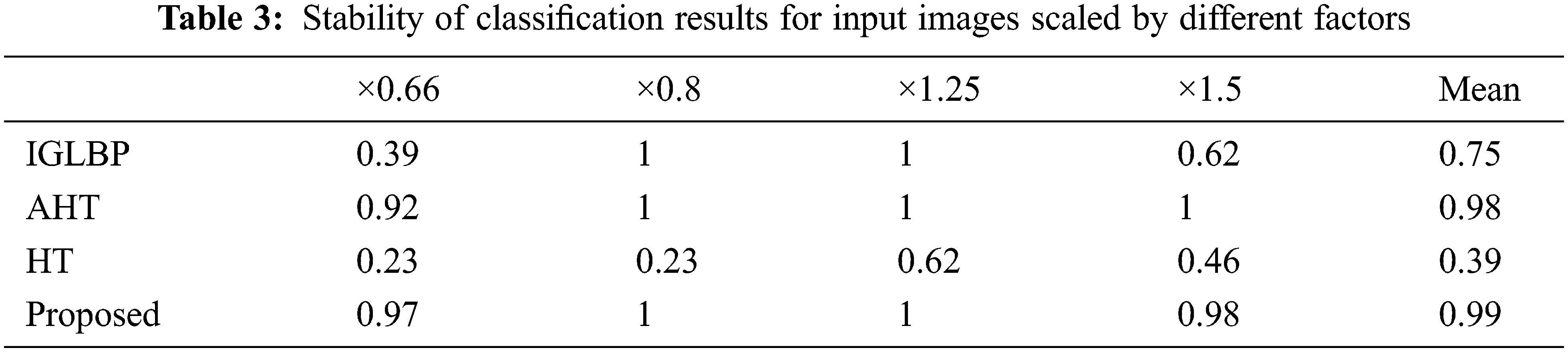
We performed another set of experiments to demonstrate the scale invariance property of GBLBP. We first trained the classification model using 13 unrotated texture images followed by testing the same classifier using the features extracted from the resampled versions of the texture images with scaling values of ×0.66, ×0.8, ×1.25 and ×1.5. We employed bicubic interpolation for producing the scaled versions of input images. The experimental results reveal that GBLBP demonstrates excellent invariance characteristics to scale changes in the images (Table 3).
In order to further strengthen our assertions about GBLBP, we present another set of experiments where we analyze efficacy of both the rotation and scaling on the performance of GBLBP (Table 4). In this connection, we use 13 unrotated texture images to train the classification model whereas rotated versions (along 30°, 60°, 90°, 120°, 150°, and 200°) and scaled versions (for values ×0.66, ×0.8, ×1.25, and ×1.5) are used to test the classification model.
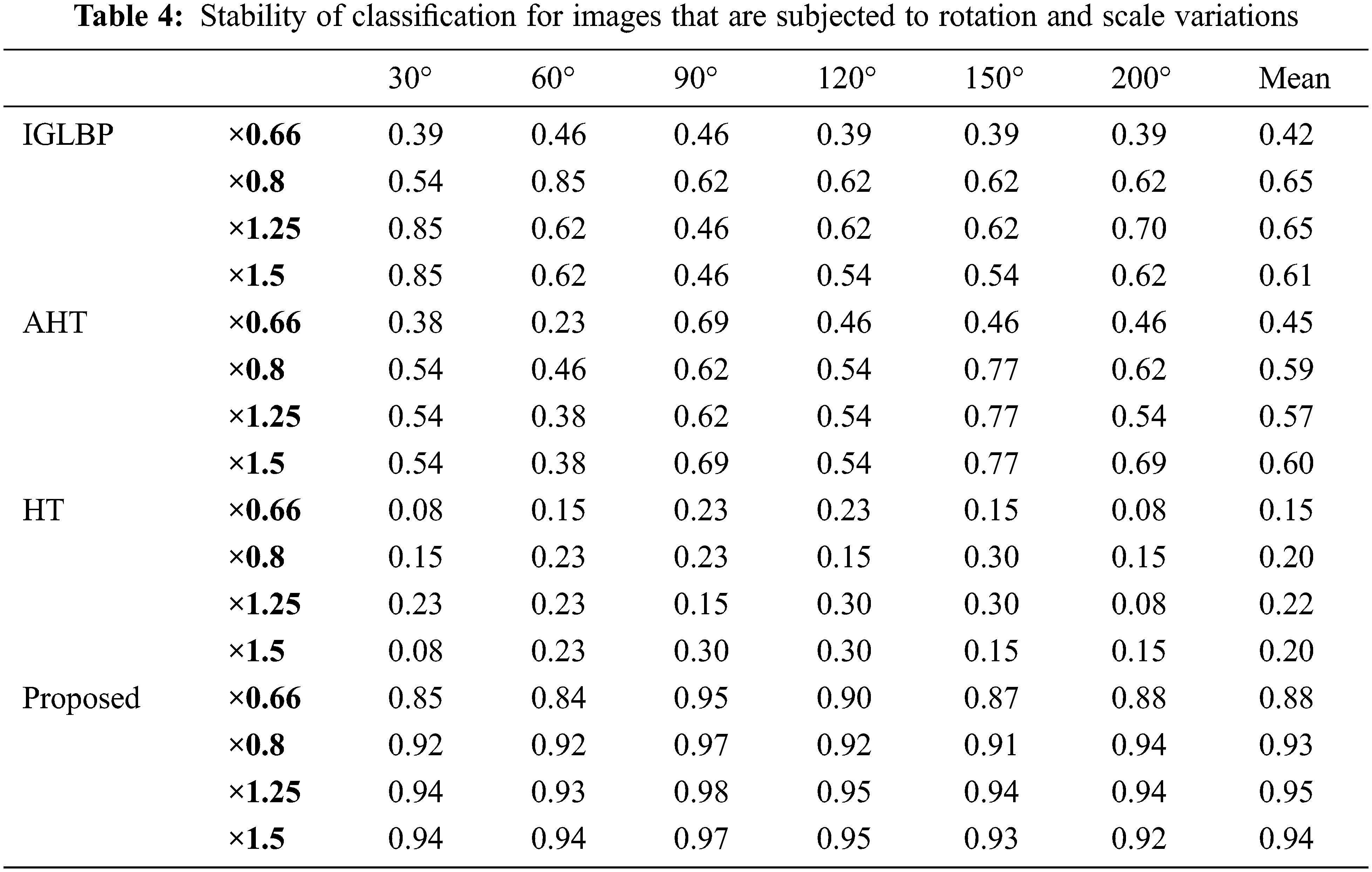
As evident from the obtained results, the proposed GBLBP technique demonstrated superior performance in the presence of rotation and scale variations of input images. The scaling factor 0.6 is the only exception where the performance is relatively low (still better than other methods) due to the subsampling of input imagery by losing important details. In case of immense subsampling, the image details are lost affecting the constructed block LBP patterns.
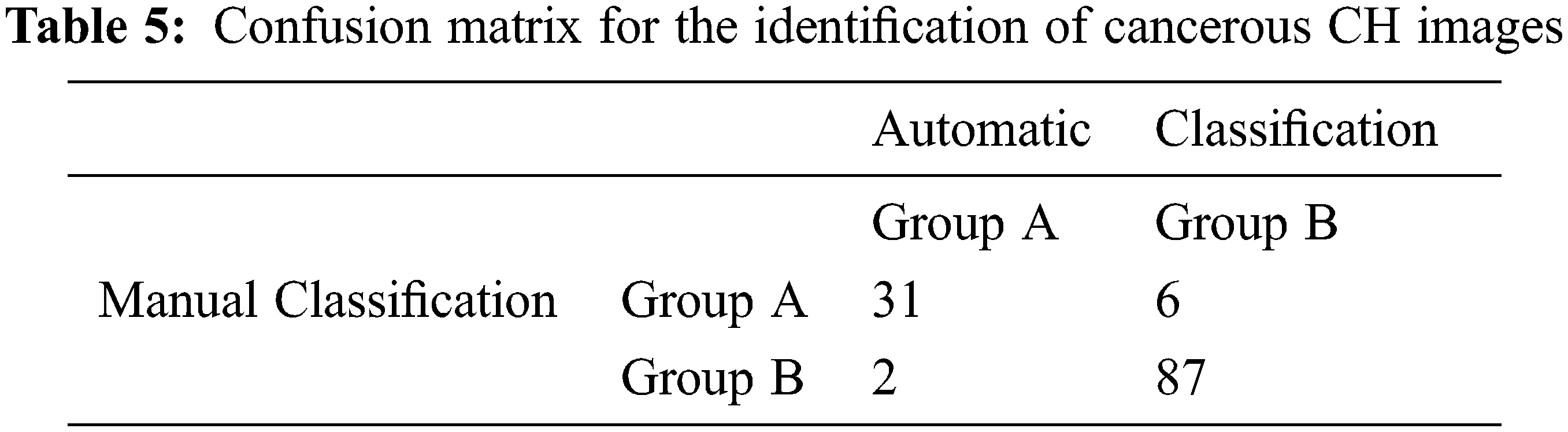
We utilized only manually annotated image patches for the image classification task assuming perfect segmentation of the input images. For feature-based methods, Support Vector Machines (SVM) with Radial Basis Function (RBF) kernel is used as classifier where the parameters for SVM are optimized through the 10-fold cross-validation technique. We use the confusion matrix as shown in Table 5 to analyze the performance of our proposed GBLBP methodology. We have obtained an overall classification rate of around 93.6% as shown in Table 6 for classification of CH images. The outputs of the classification model in the confusion matrix reveals that 2 among the 8 misclassified images are abnormal but identified as normal. From these experiments, we can deduce that the GBLBP is capable of effectively raising alarms during the endoscopy procedure with high accuracy.
5.3 Comparison with Handcrafted Features
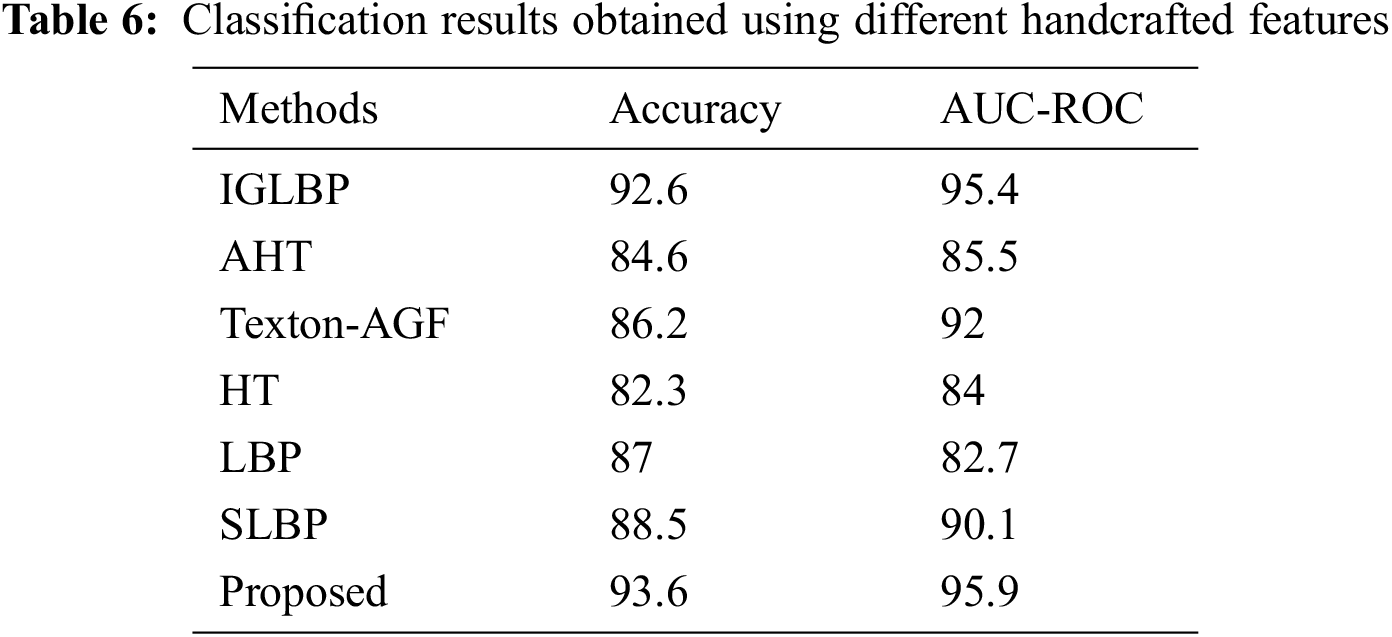
In this article, we performed comparison of our classification results with several other methods based on Gabor filters and LBP. In specific, we have performed comparison of GBLBP (histogram of feature maps are classified using SVMs) with Invariance Gabor Local Binary Patterns (IGLBP) [37], Autocorrelation Homogeneous Texture (AHT) [38], Texton Autocorrelation Gabor Features (Texton-AGF) [38], Homogeneous Texture (HT) [39] and Local Binary Patterns (LBP) [27]. Our experiments show that we have obtained the best classification results using GBLBP (Table 6). This shows the superior discriminative power of the propose descriptor in comparison to its state-of-the-art counterparts.
This is because GBLBP is not only rotation and scale invariant, it is also illumination invariant due to the usage of local binary patterns in the process of feature extraction. Moreover, the GBLBP extract shape features from the images, unlike the other descriptors which are considered in this paper. LBP compensates for homogeneous and non-homogeneous illumination changes in the images. The proposed GBLBP method is also robust to noise given that the difference of gray level values for calculating LBPs is done on an average intensity of a neighborhood. The AHT and Texton-AGF are based on autocorrelation of Gabor filter responses for normalizing rotation and scale changes in the images but researchers have found that autocorrelation is not considered a good discriminator of isotropy. In GE, the pit patterns are round (isotropic) thus, these features do not show as good results on classification of GE images as the GBLBP do. The HT features are not invariant and thus exhibit low classification rates as compared to the other rotation and scale invariant feature sets.
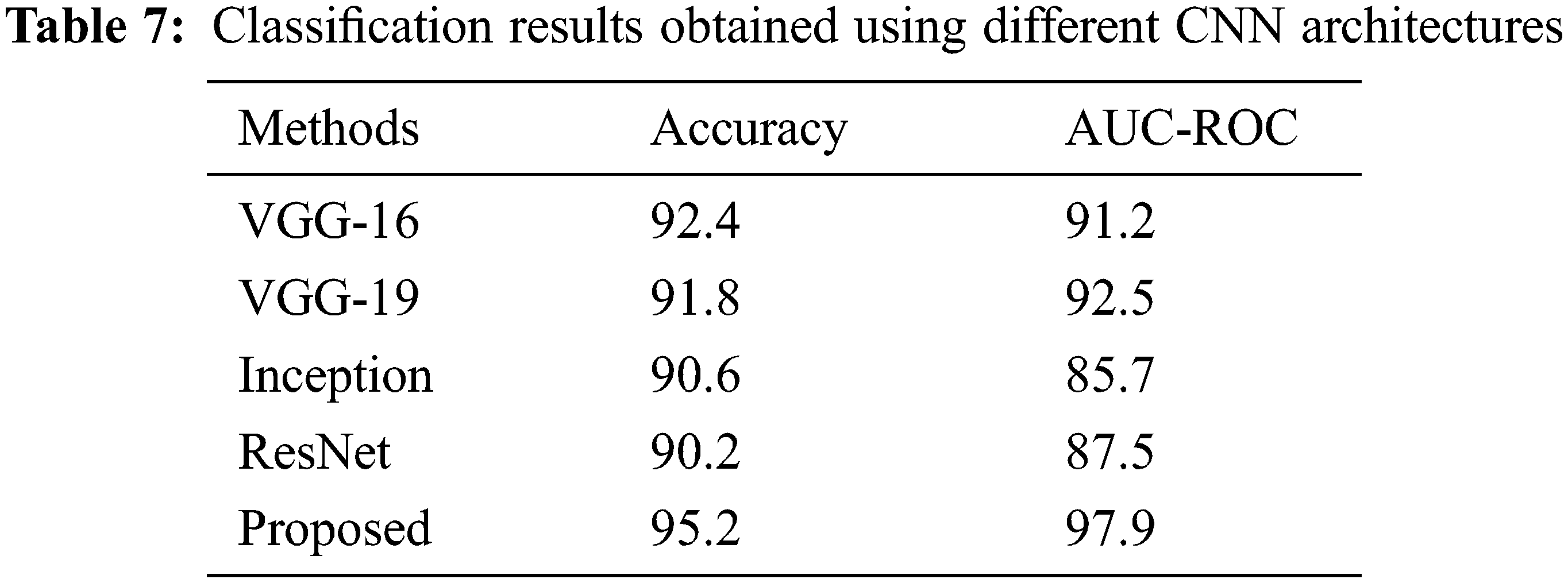
When used in a CNN setting (GBLBP feature maps are input to the proposed CNN), the proposed GBLBP framework also performs better as compared to the other methods based on convolutional neural networks (Table 7). We attribute this better performance to two important factors; the multiresolution feature set obtained as a result of Gabor filtering and later the block LBPs extract the clinically relevant information from the images. Therefore, the relevant feature maps are extracted by this section. After the extraction of these feature maps, a relatively shallow network (like the one designed in this paper) can yield very good classification results. Moreover, the deeper CNNs require parameter training of millions of parameters whereas the proposed network is shallow and adapts itself more aggressively to the application scenario leading to better results.
It is indeed true that the CNNs are used for feature extraction but typically, once a CNN is designed, the depth of the network is typically kept large. Increasing the depth of the network allows the parameters to be tweaked effectively and a reasonable depth will give rise to better features. Among the different layers of the network, the initial layers are used to extract the salient micro features from the images, thereby learning the low-level information from the images. Subsequently, the layers which are closer to the classification layer incorporate the high-level information (application-level information) from the images. A visual illustration of the respective filters is shown in Fig. 6. Given this, it is feasible to replace the initial layers (and even the mid layers) with some handcrafted features and later use a shallow CNN, training only the application layer information in the network. This is the main rationale behind using handcrafted in conjuncture with CNNs.
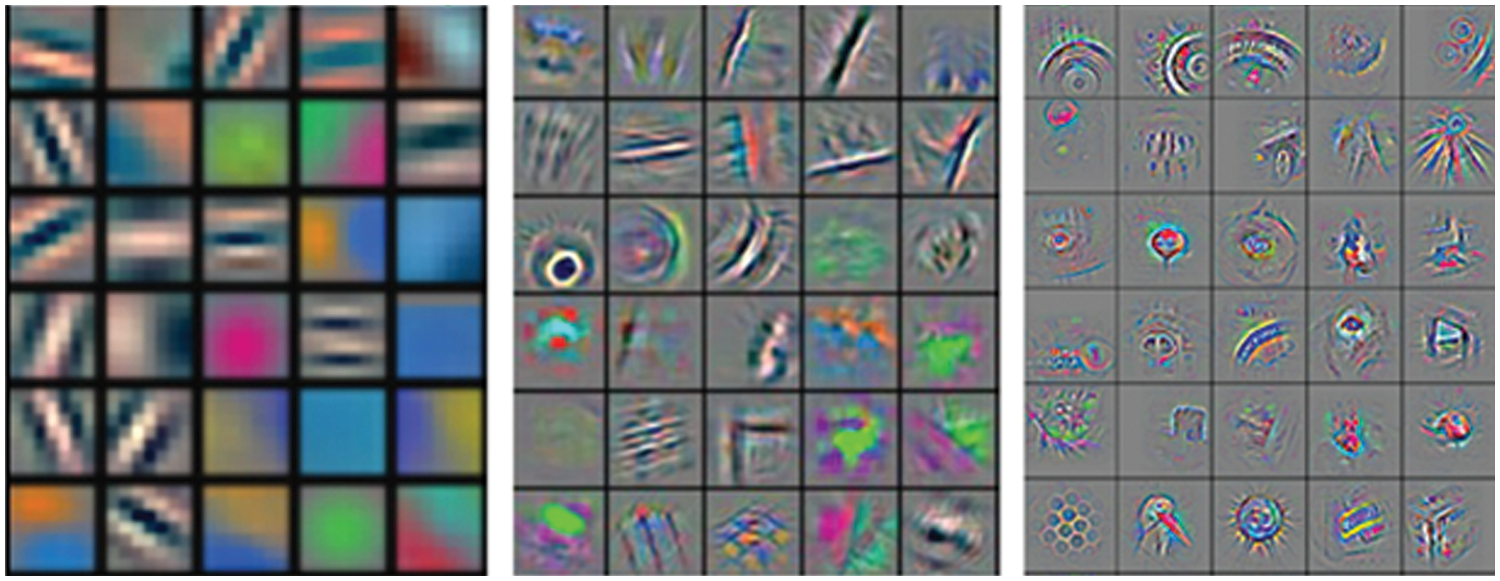
Figure 6: The filter bank representing initial layers (left), middle layers (middle) and high level (right) information int the images
It is well known that the CNNs are used for feature extraction and subsequently classification. For applications in which contextual information is not given, they can be a good source of features as they have the ability to auto-learn the features. However, if some contextual information is known, it is possible to propose better features which are more typical of the presented scenarios. Nevertheless, in this paper, we have proposed a feature map that has been used in both, a traditional machine learning setting and CNNs for classification. Results show the superior performance given by the feature maps.
In this article, we developed a novel texture classification technique that is proven to be invariant to homogeneous illumination, rotation, scale, and illumination gradients in the images. The proposed GBLBP methodology decomposes the input image with Gabor filters into multiple filter responses that are sensitive to image scale and orientation. Next, block LBP of the obtained responses are computed where both homogeneous illuminations and illumination gradients have been removed. Then the classification network is designed to classify the images. GBLBP is used for the classification of Chromoendoscopy (CH) images from the stomach. This is a challenging dataset given that the lack of control over the endoscopic probe presents a practical challenge potentially creating multiple visualizations of the same image. The invariance of GBLBP is empirically demonstrated on the USC-SIPI dataset composed of Brodatz texture images digitized along various rotations. The images are synthetically scaled to reveal the invariance characteristics of the devised technique. Experimental results validated the robustness of GBLBP to image transformations. The proposed GBLBP technique is later adopted for the classification of GE images that outperformed state-of-the-art methods under consideration. It is worth mentioning that the network was kept shallow due to the availability of a small number of images. Experiments show that there is promise in adopting the CNNs for classification. However, due to a large number of parameters, an extensive validation of the use of CNNs for the subject problem is an interesting area for future research.
Although the proposed GBLBP descriptor shows very good classification results, we intend to reduce the number of false negatives (FN) even further in our future work. This could possibly be achieved by utilizing the color characteristics of the images. In future, we intend to continue our research on the classification of GE images by researching on more adequate color spaces which could potentially help us in improving our results even further.
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number 7906.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Tahir, S. Anwar, A. Mian and A. W. Muzaffar, “Deep localization of subcellular protein structures from fluorescence microscopy images,” Neural Computing and Applications, vol. 34, no. 7, pp. 5701–5714, 2022. [Google Scholar]
2. M. K. I. Rahmani, M. Ansari and A. K. Goel, “An efficient indexing algorithm for cbir,” in Int. Conf. on Computational Intelligence & Communication Technology, Ghaziabad, India, IEEE, pp. 73–77, 2015. [Google Scholar]
3. Y. Dong, T. Liang, Y. Zhang and B. Du, “Spectral-spatial weighted kernel manifold embedded distribution alignment for remote sensing image classification,” IEEE Transactions on Cybernetics, vol. 51, no. 6, pp. 3185–3197, 2020. [Google Scholar]
4. L. Ding, H. Tang and L. Bruzzone, “Lanet: Local attention embedding to improve the semantic segmentation of remote sensing images,” IEEE Transactions on Geoscience and Remote Sensing, vol. 59, no. 1, pp. 426–435, 2020. [Google Scholar]
5. X. Tao, D. Zhang, Z. Wang, X. Liu, H. Zhang et al., “Detection of power line insulator defects using aerial images analyzed with convolutional neural networks,” IEEE Transactions on Systems, Man, and Cybernetics: Systems, vol. 50, no. 4, pp. 1486–1498, 2018. [Google Scholar]
6. W. Sun, X. Chen, X. R. Zhang, G. Z. Dai, P. S. Chang et al., “A multi-feature learning model with enhanced local attention for vehicle re-identification,” Computers, Materials & Continua, vol. 69, no. 3, pp. 3549–3560, 2021. [Google Scholar]
7. W. Sun, G. C. Zhang, X. R. Zhang, X. Zhang and N. N. Ge, “Fine-grained vehicle type classification using lightweight convolutional neural network with feature optimization and joint learning strategy,” Multimedia Tools and Applications, vol. 80, no. 20, pp. 30803–30816, 2021. [Google Scholar]
8. S. Hwang, J. Oh, J. Cox, S. J. Tang and H. F. Tibbals, “Blood detection in wireless capsule endoscopy using expectation maximization clustering,” in Medical Imaging 2006; Image Processing, vol. 6144. San Diego, California, USA: SPIE, pp. 577–587, 2006. [Google Scholar]
9. M. Mackiewicz, M. Fisher and C. Jamieson, “Bleeding detection in wireless capsule endoscopy using adaptive color histogram model and support vector classification,” in Medical Imaging 2008; Image Processing, vol. 6914. San Diego, California, USA: SPIE, pp. 69140R.1–69140R.12, 2008. [Google Scholar]
10. B. Li and M. H. Meng, “Computer-based detection of bleeding and ulcer in wireless capsule endoscopy images by chromaticity moments,” Journal of Computers in Biology and Medicine, vol. 39, no. 2, pp. 141–147, 2009. [Google Scholar] [PubMed]
11. S. Hwang and M. Celebi, “Polyp detection in wireless capsule endoscopy videos based on image segmentation and geometric feature,” in Int. Conf. on Acoustics, Speech and Signal Processing, Dallas, TX, USA, IEEE, pp. 678–681, 2010. [Google Scholar]
12. A. Karargyris and N. Bourbakis, “Detection of small bowel polyps and ulcers in wireless capsule endoscopy videos,” IEEE transactions on Biomedical Engineering, vol. 58, no. 10, pp. 2777–2786, 2011. [Google Scholar] [PubMed]
13. B. Li, M. Meng and J. Lau, “Computer-aided small bowel tumor detection for capsule endoscopy,” Artificial Intelligence in Medicine, vol. 52, no. 1, pp. 11–16, 2011. [Google Scholar] [PubMed]
14. M. Hafner, R. Kwitt, A. Ulh, F. Wrba, A. Gangl et al., “Computer-assisted pit-pattern classification in different wavelet domains for supporting dignity assessment of colonic polyps,” Journal on Pattern Recognition, vol. 42, no. 6, pp. 1180–1191, 2009. [Google Scholar]
15. M. T. Coimbra and J. S. Cunha, “Mpeg-7 visual descriptors-contributions for automated feature extraction in capsule endoscopy,” IEEE Transactions on Circuits and Systems for Video Technology, vol. 16, no. 5, pp. 628–637, 2006. [Google Scholar]
16. R. Kumar, Z. Qian, S. Seshamani, G. Mullin, G. Hager et al., “Assessment of chron’s disease lesions in wireless capsule endoscopy images,” IEEE Transactions on Biomedical Engineering, vol. 59, no. 2, pp. 355–362, 2011. [Google Scholar] [PubMed]
17. M. Hafner, L. Brunauer, H. Payer, R. Resch, A. Gangl et al., “Computer-aided classification of zoom endoscopical images using fourier filters,” IEEE Transactions on Information Technology in Biomedicine, vol. 14, no. 4, pp. 958–970, 2010. [Google Scholar] [PubMed]
18. F. Riaz, F. Silva, M. Ribeiro and M. Coimbra, “Invariant gabor texture descriptors for classification of gastroenterology images,” IEEE Transactions on Biomedical Engineering, vol. 59, no. 10, pp. 2893–2904, 2012. [Google Scholar] [PubMed]
19. A. Caroppo, A. Leone and P. Siciliano, “Deep transfer learning approaches for bleeding detection in endoscopy images,” Computerized Medical Imaging and Graphics, vol. 88, no. 11, pp. 101852, 2021. [Google Scholar] [PubMed]
20. A. Tsuboi, S. Oka, K. Aoyama, H. Saito, T. Aoki et al., “Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images,” Digestive Endoscopy, vol. 32, no. 3, pp. 382–390, 2020. [Google Scholar] [PubMed]
21. P. Laiz Treceño, J. Vitrià i Marca, H. Wenzek, C. Malagelada Prats, F. Azpiroz Vidaur et al., “Wce polyp detection with triplet based embeddings,” Computerized Medical Imaging and Graphics, vol. 86, pp. 101794, 2020. [Google Scholar]
22. A. Yamada, R. Niikura, K. Otani, T. Aoki and K. Koike, “Automatic detection of colorectal neoplasia in wireless colon capsule endoscopic images using a deep convolutional neural network,” Endoscopy, vol. 53, no. 8, pp. 832–836, 2021. [Google Scholar] [PubMed]
23. E. Klang, Y. Barash, R. Y. Margalit, S. Soffer, O. Shimon et al., “Deep learning algorithms for automated detection of crohn’s disease ulcers by video capsule endoscopy,” Gastrointestinal Endoscopy, vol. 91, no. 3, pp. 606–613, 2020. [Google Scholar] [PubMed]
24. P. Pimentel-Nunes, M. Dinis-Rebeiro, J. B. Soares, R. Marcos-Pinto, C. Santos et al., “A multicenter validation of an endoscopic classification with narrow band imaging for gastric precancerous and cancerous lesions,” Endoscopy, vol. 44, no. 3, pp. 236–246, 2012. [Google Scholar] [PubMed]
25. T. Yoshida and K. Ohki, “Natural images are reliably represented by sparse and variable populations of neurons in visual cortex,” Nature Communications, vol. 11, no. 1, pp. 1–19, 2020. [Google Scholar]
26. H. S. Munawar, R. Aggarwal, Z. Qadir, S. I. Khan, A. Z. Kouzani et al., “A gabor filter-based protocol for automated image-based building detection,” Buildings, vol. 11, no. 7, pp. 302, 2021. [Google Scholar]
27. T. Ojala, M. Pietikainen and T. Maenpaa, “Multiresolution gray-scale and rotation invariant texture classification with local binary patterns,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 24, no. 7, pp. 971–987, 2002. [Google Scholar]
28. M. Tahir and A. Idris, “Md-lbp: An efficient computational model for protein subcellular localization from hela cell lines using SVM,” Current Bioinformatics, vol. 15, no. 3, pp. 204–211, 2020. [Google Scholar]
29. S. Wan, X. Huang, H. -C. Lee, J. G. Fujimoto and C. Zhou, “Spoke-lbp and ring-lbp: New texture features for tissue classification,” in Int. Symp. on Biomedical Imaging (ISBI), Brooklyn, NY, USA, IEEE, pp. 195–199, 2015. [Google Scholar]
30. M. Mahrishi, S. Morwal, A. W. Muzaffar, S. Bhatia, P. Dadheech et al., “Video index point detection and extraction framework using custom yolov4 darknet object detection model,” IEEE Access, vol. 9, pp. 143378–143391, 2021. [Google Scholar]
31. R. Sharma, B. Kaushik, N. K. Gondhi, M. Tahir and M. K. I. Rahmani, “Quantum particle swarm optimization based convolutional neural network for handwritten script recognition,” Computers, Materials & Continua, vol. 71, no. 3, pp. 5855–5873, 2022. [Google Scholar]
32. K. Bhalla, D. Koundal, S. Bhatia, M. K. I. Rahmani and M. Tahir, “Fusion of infrared and visible images using fuzzy based siamese convolutional network,” Computers, Materials & Continua, vol. 70, no. 3, pp. 5503–5518, 2022. [Google Scholar]
33. S. Hossain and S. Serikawa, “Texture databases–A comprehensive survey,” Pattern Recognition Letters, vol. 34, no. 15, pp. 2007–2022, 2013. [Google Scholar]
34. H. Zhou, R. Wang and C. Wang, “A novel extended local-binary-pattern operator for texture analysis,” Information Sciences, vol. 178, no. 22, pp. 4314–4325, 2008. [Google Scholar]
35. Z. Li, R. Hayward, R. Walker and Y. Liu, “A biologically inspired object spectral-texture descriptor and its application to vegetation classification in power-line corridors,” IEEE Geoscience and Remote Sensing Letters, vol. 8, no. 4, pp. 631–635, 2011. [Google Scholar]
36. T. Ojala, M. Pietikäinen and T. Mäenpää, “Gray scale and rotation invariant texture classification with local binary patterns,” in Computer Vision ECCV, Lecture Notes in Computer Science, vol. 182. Berlin, Heidelberg: Springer, pp. 404–420, 2000. [Google Scholar]
37. F. Riaz, A. Hassan and S. Rehman, “Texture classification framework using gabor filters and local binary patterns,” in Intelligent Computing, Advances in Intelligent Systems and Computing, vol. 858, Switzerland: Springer Cham, pp. 569–580, 2019. [Google Scholar]
38. F. Riaz, F. B. Silva, M. D. Ribeiro and M. T. Coimbra, “Invariant gabor texture descriptors for classification of gastroenterology images,” IEEE Transactions on Biomedical Engineering, vol. 59, no. 10, pp. 2893–2904, 2012. [Google Scholar] [PubMed]
39. B. S. Manjunath, J. -R. Ohm, V. V. Vasudevan and A. Yamada, “Color and texture descriptors,” IEEE Transactions on Circuits and Systems for Video Technology, vol. 11, no. 6, pp. 703–715, 2001. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF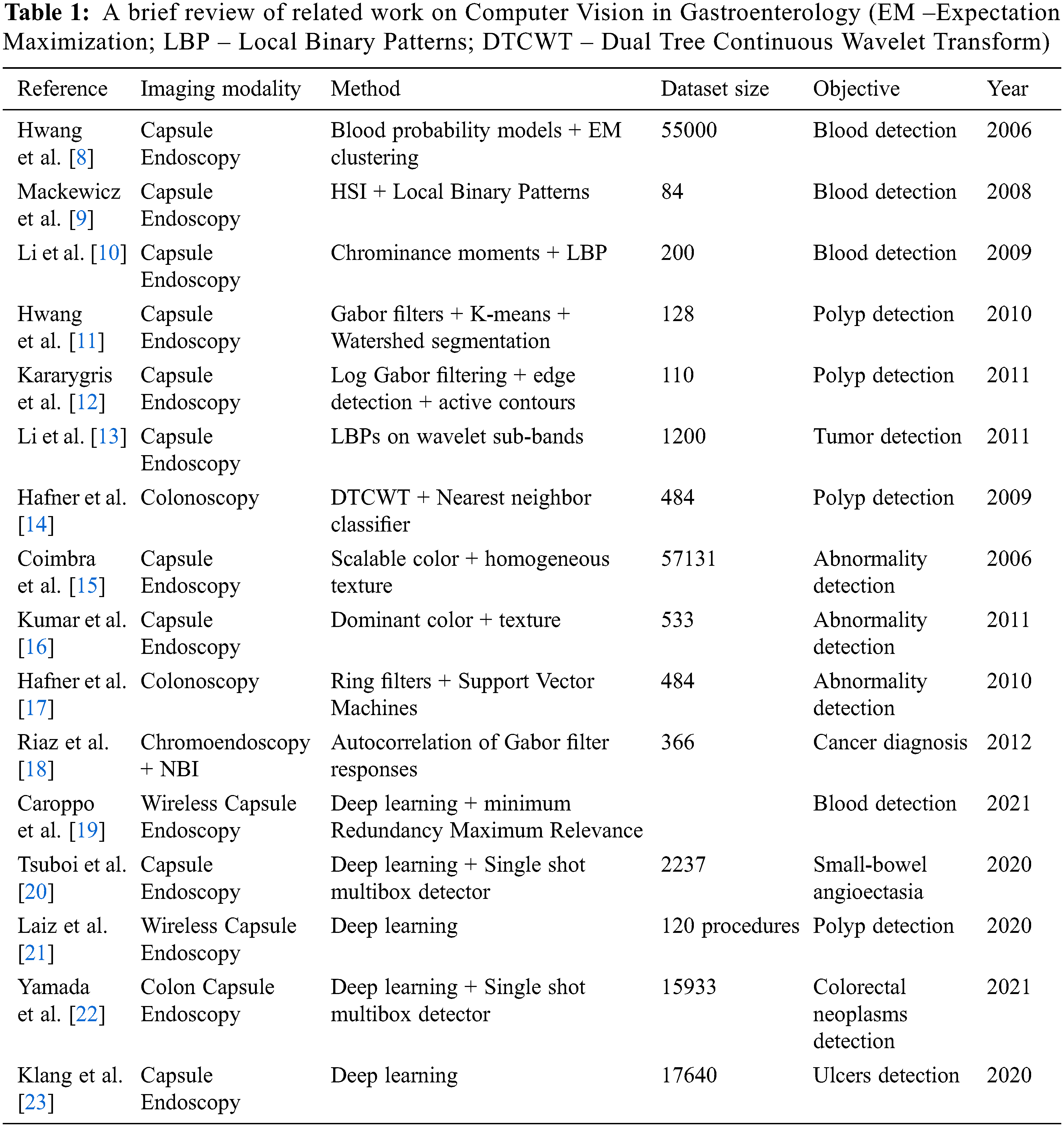
 Downloads
Downloads
 Citation Tools
Citation Tools