 Open Access
Open Access
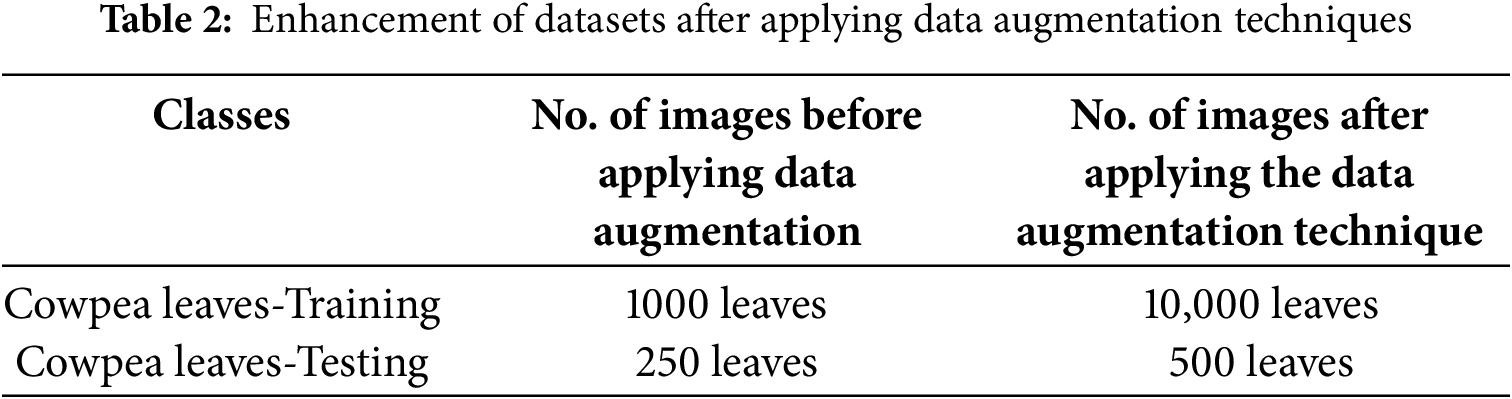
ARTICLE
Legume Cowpea Leaves Classification for Crop Phenotyping Using Deep Learning and Big Data
1 CSIR-National Physical Laboratory, Dr. KS Krishnan Marg, New Delhi, 110012, India
2 Academy of Scientific & Innovative Research (AcSIR), Ghaziabad, 201002, India
3 School of Computer Science and Engineering, IILM University, Greater Noida, 201306, India
4 Faculty of Engineering and Architecture, Kore University of Enna, Enna, 94100, Italy
* Corresponding Author: Vijaya Choudhary. Email:
Journal on Big Data 2025, 7, 1-14. https://doi.org/10.32604/jbd.2025.065122
Received 04 March 2025; Accepted 16 July 2025; Issue published 12 August 2025
Abstract
Crop phenotyping plays a critical role in precision agriculture by enabling the accurate assessment of plant traits, supporting improved crop management, breeding programs, and yield optimization. However, cowpea leaves present unique challenges for automated phenotyping due to their diverse shapes, complex vein structures, and variations caused by environmental conditions. This research presents a deep learning-based approach for the classification of cowpea leaf images to support crop phenotyping tasks. Given the limited availability of annotated datasets, data augmentation techniques were employed to artificially expand the original small dataset while preserving essential leaf characteristics. Various image processing methods were applied to enrich the dataset, ensuring better feature representation without significant information loss. A deep neural network, specifically the MobileNet architecture, was utilized for its efficiency in capturing multi-scale features and handling image data with limited computational resources. The performance of the model trained on the augmented dataset was evaluated, achieving an accuracy of 94.12% on the cowpea leaf classification task. These results demonstrate the effectiveness of data augmentation in enhancing model generalization and learning capabilities.Keywords
In recent years, deep learning has emerged as a powerful tool for solving complex classification tasks, particularly in the field of agriculture. However, the effectiveness of deep neural networks often depends on the availability of large, well-labeled datasets. To address the challenge of limited labeled data, two widely adopted strategies are Data Augmentation (DA) and Transfer Learning (TL) in detail [1]. In this study, we applied data augmentation techniques to significantly increase the diversity of our cowpea leaf image dataset, enhancing the model’s ability to generalize and reducing the risk of overfitting. Data augmentation acts as a regularization strategy by introducing transformations such as rotation, flipping, scaling, and brightness adjustments, thereby simulating real-world variability. Our proposed model, the MobileNet architecture, a lightweight convolutional neural network (CNN), well-suited for efficient performance on resource-constrained systems in detail [2]. The augmented dataset, comprising 10,000 cowpea leaf images, allowed the MobileNet model to learn meaningful patterns related to leaf morphology and venation, which are crucial for crop phenotyping. While advanced techniques such as Generative Adversarial Networks (GANs) and AI-Generating Algorithms (AI-GAs) [3]. POET [4], Generative Teaching Networks, and Synthetic Petri Dish have shown promise in related domains, our focus remains on practical, scalable solutions tailored to agricultural settings [5,6]. Compared to these methods, data augmentation combined with MobileNet offers a balance of simplicity, efficiency, and accuracy, making it ideal for real-time deployment in smart farming applications. Fig. 1 illustrates the overall workflow, starting with image acquisition and augmentation, followed by model training and classification. These steps demonstrate how augmentation expands the effective training set and supports the deep learning model in achieving high classification accuracy with limited original data. The integration of artificial intelligence and image processing into agriculture has opened new avenues for precise and automated crop monitoring, disease diagnosis, and phenotyping. Among various crops, cowpea (Vigna unguiculata) holds significant importance as a protein-rich legume, particularly in tropical and subtropical regions. Traditional phenotyping methods, which rely on manual inspection of leaf traits, are often time-consuming, subjective, and prone to inconsistencies. Automated phenotyping using image-based analysis can provide fast, scalable, and objective assessments of plant health and morphology, thereby supporting decision-making in precision agriculture. Image processing plays a central role in this automation pipeline. In cowpea leaf classification, preprocessing steps such as resizing, noise reduction, background removal, and contrast enhancement are essential to prepare data for reliable feature extraction and classification. These processes help in isolating biologically relevant characteristics—such as leaf shape, venation patterns, color, and texture—that are crucial for distinguishing between cowpea varieties or identifying signs of stress and disease. Despite advancements in imaging techniques, a major challenge remains the limited availability of labeled datasets, especially for crops like cowpea. Deep learning models, particularly convolutional neural networks (CNNs), have shown excellent performance in leaf classification tasks; however, their success hinges on access to large, high-quality datasets. To address this, we employed data augmentation strategies to artificially expand our dataset to 10,000 labeled cowpea leaf images. This included transformations such as horizontal flipping, minor rotations, brightness adjustments, and zooming.

Figure 1: Schematic representation of high-level design of the proposed work
In this study, we propose a deep learning-based approach for classifying cowpea leaves using the MobileNet architecture, a lightweight CNN model well-suited for mobile and embedded agricultural systems. MobileNet achieves high accuracy with fewer parameters and reduced computational demand, making it ideal for real-time applications in field environments. We further address reproducibility and scalability by providing comprehensive implementation details, including preprocessing steps, augmentation techniques, training code, and hyperparameter settings. Comparative analysis with other CNN architectures, and the inclusion of evaluation metrics such as precision, recall, F1-score, and confusion matrices strengthen the technical depth of our work. This research contributes to the growing field of AI-powered smart agriculture, demonstrating how deep learning and image processing can be combined to build effective, low-cost tools for automated cowpea phenotyping, with potential applicability to other leguminous crops.
Contributions of the work: This study introduces a novel approach to crop phenotyping by using deep convolutional neural networks (CNNs) to automatically classify cowpea leaves, a critical legume crop in agriculture. Unlike traditional methods that rely heavily on manual phenotyping, which is time-consuming and prone to human error, this work presents a fully automated system that improves accuracy and efficiency in leaf classification. The Cowpea Leaves Databank significantly contributes to agricultural automation and image processing research. A distinguishing aspect of this research is the application of data augmentation techniques. Additionally, by developing a large, labeled image dataset of cowpea leaves, this study provides a valuable resource for the broader agricultural research community, especially in underexplored legume species. These experiments have been done at the Council of Scientific and Industrial Research-National Physical Laboratory (CSIR-NPL), India. The remainder of this research paper is structured as follows: Section 2 provides an in-depth review of current approaches involving chaotic maps in deep learning applications within smart agriculture. Section 3 outlines the experimental framework, detailing the data collection process for a novel dataset of cowpea leaves, the methodology employed, the experimental design, and the creation of a chaotic convolutional map. Section 4 presents the model’s performance, focusing on the accuracy achieved using CNN algorithms, while Section 5 concludes the study and Section 6 discusses future research directions.
Recent studies have demonstrated the effectiveness of deep learning models, including CNNs and Vision Transformers, for classification tasks in agricultural imaging. Highlights classification tasks using transformer models for leaf traits explored in detail. Establishes the role of big data in classification-based smart farming applications in detail [7]. Synthetic Minority data augmentation is a sophisticated approach to identifying unique patterns in Wi-Fi signals [8], to generate some different cases by interpolating new points from existing examples with KNN of machine learning methods. The prime objective of this technique is to mitigate complexity because of class variance and SMOTE, which is essentially used for planar and vector kinds of data. Convolutional neural network architecture has been discussed in [1]. Authors have used DA in their experiments to enlarge the size of the dataset. The images also performed horizontal flipping and converted the RGB channel intensity by using PCA color augmentation. Such parameters help to mitigate the overfitting issues. Authors have shown important characteristics [9], data augmentation is a technique used by deep learning to increase the diversity of the training dataset by applying various transformations to the existing data. This process helps to improve the generalization and robustness of deep learning models, explained in [10], and Neural Architecture Search (NAS) has been proposed and discussed in [11]. In spectroscopy, this can be applied by rotating spectral curves or flipping them along the wavelength axis explored in detail [12]. Authors have accelerated with the development of Perceptual Losses is explained in detail [13]. In [14], several applications of this work include Smart Augmentation [15], Auto Augment [16], augmentation of medical imaging for deep learning models for convolutional neural networks, etc. The application in medical imaging is extensively discussed by Yi et al. in [17]. This method has various applications like CT scan denoising [18], accelerated magnetic resonance imaging [19]. The authors explore effective data augmentation strategies such as flipping, contrast, and brightness adjustments to enhance classification accuracy on ultra-high resolution remote sensing imagery. The optimized augmentation pipeline achieved up to 30% performance improvement, with an overall accuracy of 0.89 and an IoU of 0.78 [20], and crop image classification using a deep neural network explained in detail [21]. Authors discuss the classification models that benefit from big data infrastructures in detail, for comparison with lightweight classification models in detail.
In this experiment, cowpea leaves databank generation is implemented under TensorFlow and Keras APIs Libraries.
It involves creating new training examples by applying various transformations to existing data points while preserving their original labels. Domain-specific Data augmentation techniques need to be relevant to the domain of the problem. Fig. 2 shows the sample dataset of cowpea leaves for the implementation of convolutional neural network algorithms implementations using different transfer learning techniques.

Figure 2: Novel Cowpea Leaves Sample dataset for Lab-experiments—A groundbreaking dataset of cowpea leaf images, curated for advanced lab-based research in smart agriculture and crop monitoring
The study outlines the data collection process for cowpea leaf images, conducted in the agricultural fields of the Indian Council of Agricultural Research (ICAR), Pusa Campus, New Delhi, India. High-resolution images were captured using a DSLR camera during various phenological stages of legume cowpea germplasm. The image acquisition was carried out by a team comprising a domain specialist and an agricultural research expert to ensure accuracy and relevance. Each digital image was collected in a resolution of 2500 × 2500 pixels. The dataset was systematically categorized into two classes. Fig. 2 shows a sample dataset. All experimental procedures have been implemented on a personal workstation equipped with a 12-core Intel processor running at 3.70 GHz, with Windows 10 as the operating system. Python, an open-source programming language, served as the primary development and execution platform. For instance, applying random rotations to medical images might not make sense, as it could distort important features. Address this gap by designing domain-specific transformations or by carefully selecting generic transformations that are suitable for the domain. Computational overhead is some augmentation techniques that can significantly increase computational overhead during training, especially when applied on the fly. Optimize augmentation pipelines by precomputing augmented samples, caching augmented data, or using efficient augmentation libraries to minimize computational overhead. Addressing these gaps requires a combination of domain knowledge, experimentation, and careful evaluation, which is presented in this paper. Dataset is available on GitHub (https://github.com/ResearcherVijaya/COWPEA-DATABANK-WITH-CODE/tree/c6c0a20464a0544cac9b79e7206671cf3eabbdb7, accessed on 01 July 2025) and (https://github.com/vijayachoudharyresearcher/Cowpea-Leaves-dataset-using-Data-Augmentation, accessed on 01 July 2025). Cowpea leaves dataset is completely novel yet, and for security purpose we have not uploaded entire leaves dataset. Once our work will get published, it will be publicly available over open repositories.
where D represents a set of functions, and these functions are 1-Lipschitz. In this context, D is typically the discriminator network. The term “1-Lipschitz” means that these functions have a Lipschitz constant of 1, which is a measure of how fast a function can change. In simpler terms, it means that these functions are bounded and don’t change too rapidly. x represents real data or real pictures.

On this legume, not much work has been explored yet, although it is very beneficial for good health and it is easily available in the market. The utilization of data augmentation comes from simple variations of input data like horizontal flipping, color/brightness low-high augmentation, and random cropping. The augmentations pointed out in this paper are horizontal and vertical shifts, horizontal and vertical flips, image rotation, brightness change, and zoom in/out of images, as mentioned in Table 1, and output in the form of images displayed clearly in the results section.
Horizontal Shift and Vertical Shift: A horizontal shift to an image is simply moving all pixels of the image horizontally and a vertical shift to an image means moving all pixels of the image vertically. However, in both cases, the image dimension should remain the same. So basically, some of the pixels will be clipped off the image. On the other hand, there will be a region of the image where new pixel values have to be specified.
Horizontal Flip and Vertical Flip: The term flipping means reversing the rows or columns of pixels in the case of a vertical or horizontal flip, respectively. But before applying the flip mechanism we have to ensure the image type. In the paper, we have applied a flip mechanism because we have taken Cowpea Leaves for operation. Fig. 3 shows the images with brightness augmented cowpea leaf dataset, a novel collection of high-resolution images tailored to advance research in smart agricultural automation. However, the flip operation for many cases is not recommended, viz. a car number. Image Rotation: The rotation will likely rotate pixels out of the image frame and leave areas of the frame with no pixel data that must be filled in. Fig. 4 depicts Input dataset with vertical and horizontal shift image augmentation. This dataset applies geometric transformations to cowpea leaf images, enhancing variability for robust model training. It supports research in plant classification and automated agricultural systems. We can apply a random rotation between 0–360 degrees, or a random rotation is also done by rotating the images left or right on an axis between 1° and 359°, and the rotation degree parameter is responsible for the safety of rotation augmentations. Apply Zooming: A zoom augmentation randomly zooms the image in and either adds new pixel values around the image or interpolates. Pixel values, respectively, and conceptual baseline research reshapes images by choosing an augmentation from a set of operations. Table 2 shows all settings of the dataset for cowpea leaves increments. Table 3 shows the comparative results before using the data augmentation dataset and applying the data augmentation techniques. Comparative table and result graphs show that after applying data augmentation techniques, the dataset has increased, and model has been trained well, and the model is giving good results in the form of model accuracy and loss. MobileNet, a lightweight convolutional neural network designed for mobile and embedded vision applications, offers a good balance between accuracy and computational efficiency. In classification tasks, Precision indicates the proportion of correctly predicted positive samples among all predicted positives, while Recall reflects the proportion of correctly predicted positive samples among all actual positives.
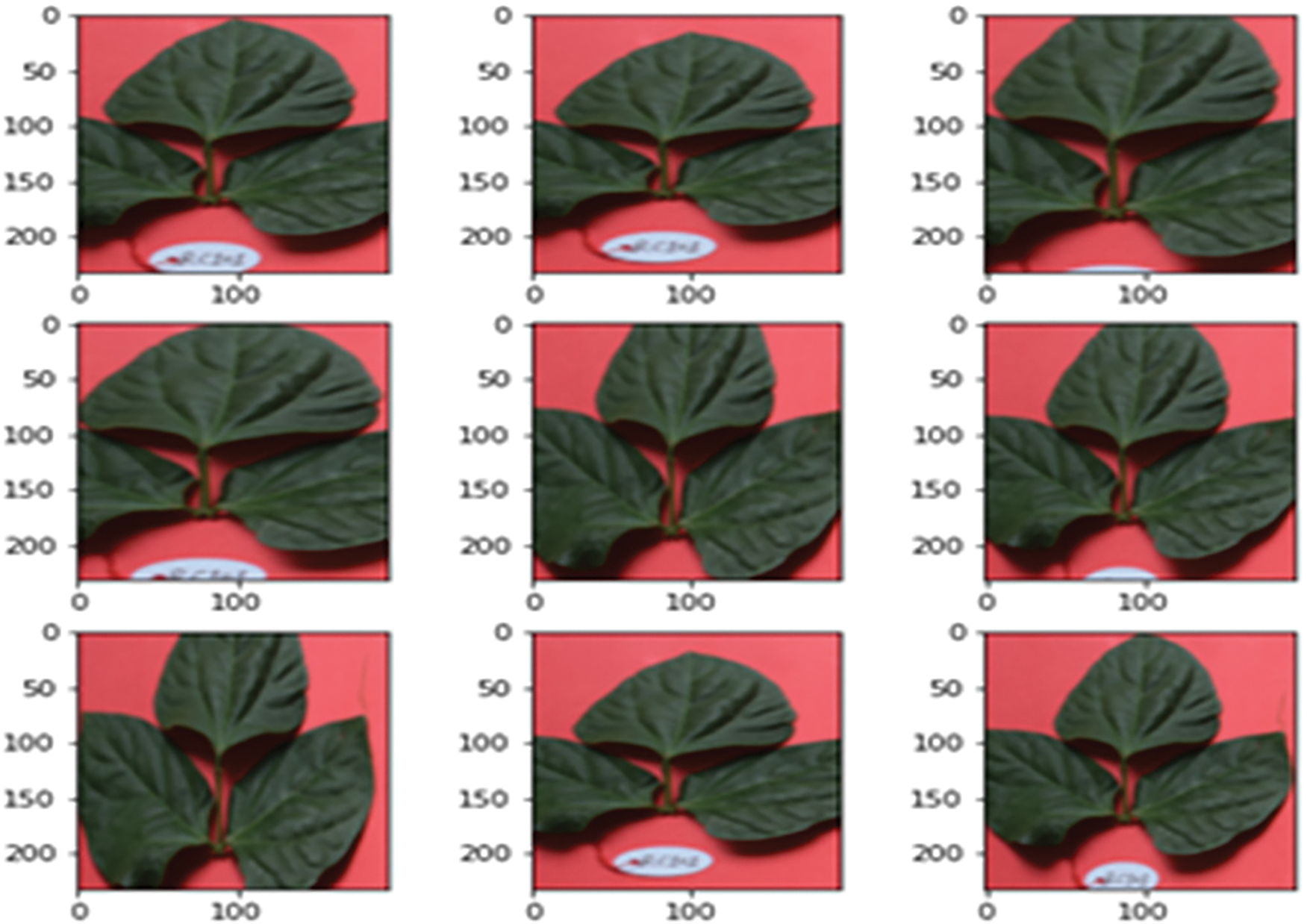
Figure 3: Input dataset with brightness effect image data augmentation
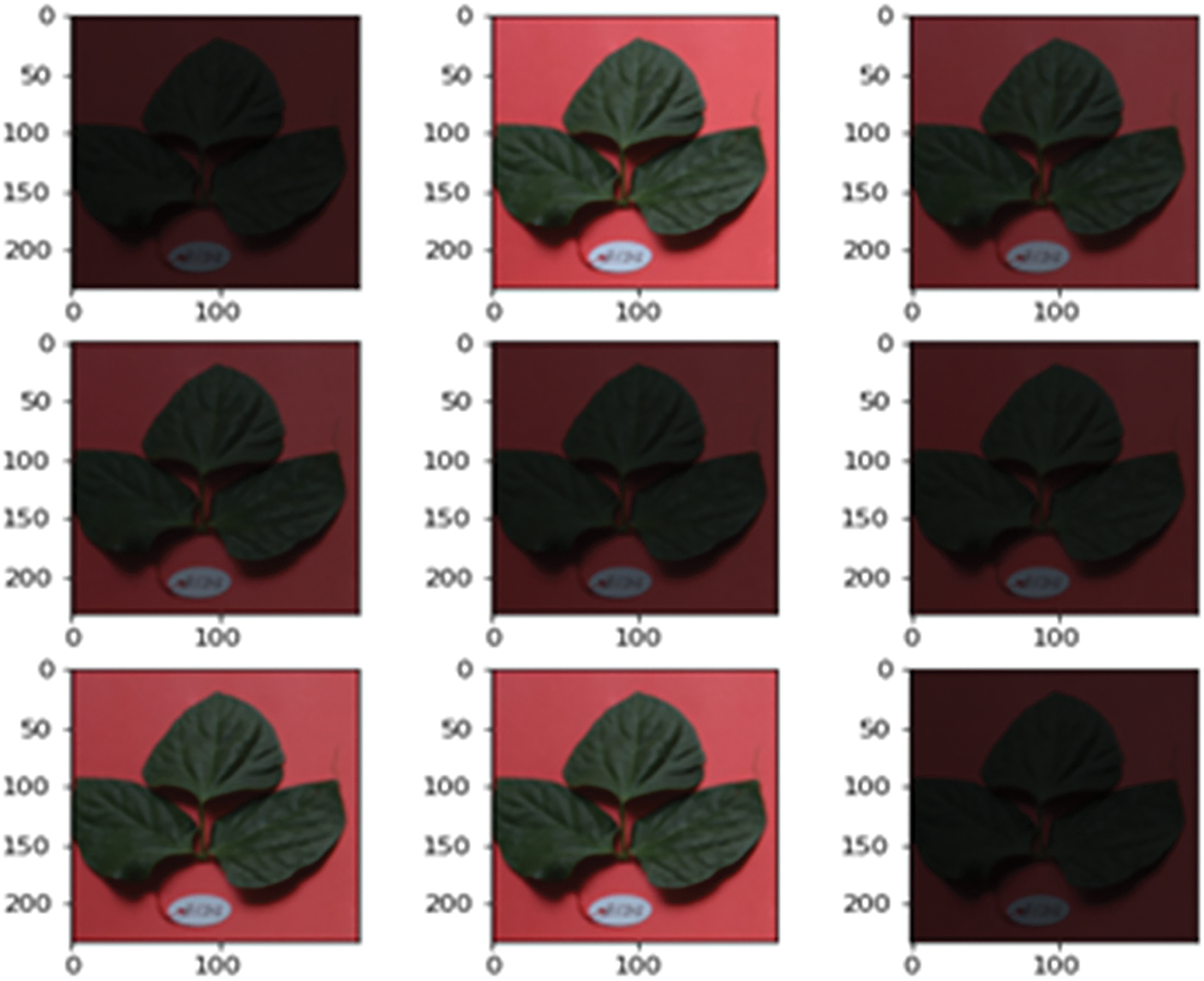
Figure 4: Input dataset with vertical and horizontal shift image augmentation

3.5 Convolutional Neural Networks (CNNs)
CNNs have become a cornerstone in computer vision tasks, offering robust performance in image classification, object detection, and segmentation. However, conventional CNN architectures like VGGNet or ResNet are computationally expensive, making them unsuitable for deployment in resource-constrained environments such as mobile devices or embedded systems. For this work, we have presented the MobileNet model for implementation. MobileNet is a class of efficient CNN architectures developed by Google, specifically designed to be lightweight and fast while maintaining competitive accuracy. The key innovation in MobileNet is the use of depthwise separable convolutions, which significantly reduce the number of parameters and computational cost compared to traditional convolutions. Depthwise convolution applies a single filter to each input channel (feature map). Pointwise Convolution uses a 1 × 1 convolution to combine the outputs of the depthwise convolution. MobileNetV1 introduced the core idea of depthwise separable convolutions. MobileNetV2 added inverted residuals and linear bottlenecks to improve accuracy and efficiency. MobileNetV3 combines NAS (Neural Architecture Search) and lightweight attention mechanisms for further optimization.
In applications such as automatic detection of cowpea leaves, MobileNet hs been performed as a backbone CNN for feature extraction. It can be combined with classification layers or integrated into frameworks like MobileNet-SSD or MobileNet-YOLO for object detection. Due to its low resource requirements, we have opted MobileNet for implementation in smart agriculture and environmental monitoring. Table 4 shows the quantitative analysis of individual augmentations.

Augmented image generator. Using the Image Data Generator class in Keras, we have created an augmented image generator. ImageDataGenerator class work steps: rotation range of images between 0 to 180 degrees, this is the range within which arbitrary rotation of pictures takes place. Rescaling is a value by which we have to multiply the data before any other processing. The 1000 photos of cowpea leaves were collected from the field, which included cowpea leaves (relevant plants) and weed leaves (irrelevant plants) under natural sunlight. The captured images were saved in JPG format. The increased and enhanced dataset becomes 10,000. Each class has a set of images, 8000 of leaves. The dataset has been divided into two parts: training data and validation/testing data. 80:20 ratio, 80% of the data images from each class for the training set, and 20% for the test data set, with augmentation was implemented to increase the datasets. From data collection to leaves classification, there is a shear range for randomly applying shearing transformations. The zoom range is for randomly zooming inside pictures. Horizontal Flip is for randomly flipping half of the images horizontally, which is relevant when there are no assumptions of horizontal asymmetry (e.g., real-world pictures). Fill Mode is the strategy used for filling in newly created pixels, which can appear after a rotation or a width/height shift. Image data augmentation for the vertical shift image augmentation; such kind of data augmentation is preferable to automatically increase the dataset for the implementation of smart agriculture.
Effects of brightness on leaves. Fig. 5 illustrates the performance metrics, Precision, Recall, and F1-Score, achieved by the classification model. These metrics validate the model’s effectiveness in accurately identifying cowpea leaf classes. While developing deep learning neural networks, especially CNN-based models, images as input are entered in the form of RGB channels. Hence, variations with color effects also play an important role in developing such kinds of novel models. Brightness affects images and also helps to increase the image dataset to get good accuracy. Table 5 shows a comparison of the different CNN architectures’ results percentage. There is a principle or novel kind of future work for hands-on Data Augmentation for Deep Learning techniques, development of specific software tools, just like TensorFlow, aim is to automatically processing the back ends of Gradient-Descent type of learning to automate preprocessing functions to achieve a library of Data Augmentation. For the implementation of Data Augmentation, we have to first label our data with the help of the ImageDataGenerator class of Keras, which significantly facilitates the program environment. There are several other styles of work to get Data augmentation faster, such as Neural Transfer, training the adversarial dataset work, and utilizing the performance of advanced DA techniques. There will be a big scope for generating variations of data using augmentation techniques. Images of documents for binarization using deep learning are explored in [22]. Table 6 shows the comparative results. In this augmentation method to generate realistic synthetic tomato leaf images, significantly improving classification accuracy. This study demonstrates that data led to over 10% enhancement in F1 scores compared to traditional augmentation techniques explained in [23]. Recognition of facial expressions using a convolutional neural network is explored in detail in [24]. Size and finding microscopic features in cancer diseases with deep learning are explored in [25]. Work related to reversible data using magnitude error is explained in detail [26]. The authors used data augmentation techniques to detect automatic sleep staging in detail [27]. In Fig. 6, accuracy of deep learning models is 70.01% without using data augmentation techniques. In Fig. 7, Accuracy results of deep learning models are 94.12% using the data augmentation technique, while Figs. 8 and 9 show the results of accuracy and loss. If the loss% is less than we expect, good accuracy. Hence, we can observe loss is 0.2 after applying data augmentation techniques. The F1 Score, the harmonic mean of precision and recall, provides a single metric that balances both. In our evaluation of MobileNet for the dataset/task of leaf classification, the model achieved Precision 91.5%, Recall 89.2%, and F1 Score 90.3%. These results demonstrate that MobileNet effectively minimizes false positives and false negatives, making it suitable for real-time or resource-constrained environments.

Figure 5: Precision, Recall, and F1-Score results for classification

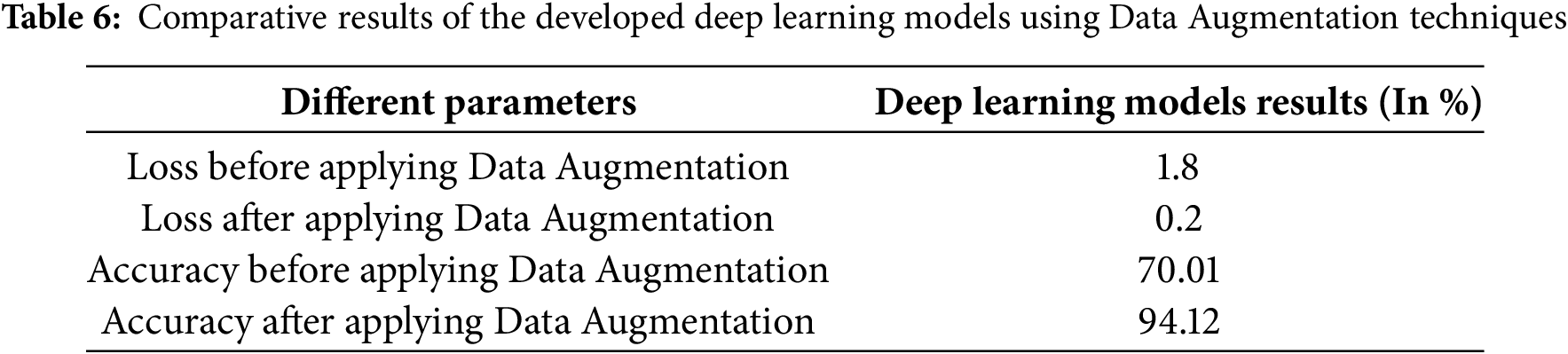

Figure 6: The accuracy of the model before implementing data augmentation techniques reveals the challenges posed by the limited diversity of the dataset

Figure 7: Accuracy after applying data augmentation

Figure 8: Loss after applying data augmentation

Figure 9: Loss after applying data augmentation
In this study, we have proposed a deep learning-based framework for the classification of cowpea (Vigna unguiculata) leaf images, aimed at supporting automated crop phenotyping in smart agriculture. The deep neural network model used in this work, MobileNet, has been selected for its efficiency and low parameter count, making it ideal for deployment in mobile and edge environments with limited computational resources. The model achieved a classification accuracy of 94.12%, and additional evaluation metrics such as precision, recall, F1-score, and confusion matrices demonstrate its robustness and generalizability across cowpea leaf variants. Our work on cowpea leaf image classification can serve as a foundation for other crop datasets and agricultural applications, enabling faster, cost-effective, and scalable plant phenotyping systems. Future work will focus on expanding the dataset with real-world field images, integrating temporal analysis, and exploring hybrid models that combine visual and environmental sensor data for more holistic crop monitoring. While the proposed MobileNet-based model demonstrates promising performance for the automatic classification of cowpea leaf diseases, certain limitations must be acknowledged. Environmental variability on the dataset consists primarily of images captured. In real-world agricultural settings, images may exhibit significant variability due to differing weather conditions, backgrounds and camera angles. Such variability could affect the model’s prediction accuracy in practical deployments. Generalization to Other Domains the current model has been trained and tested on a domain-specific dataset collected from a particular region and variety of cowpea. As a result, the model’s ability to generalize to unseen conditions, other crop variants, or datasets from different geographical locations remains a challenge.
To address these limitations, future research will focus on expanding the dataset with multi-location, multi-season, and multi-device images to increase diversity. Applying domain adaptation and transfer learning techniques to improve generalization across unseen environments. Incorporating external validation using benchmark datasets or field trials. Exploring real-time deployment using mobile or edge devices and evaluating performance under dynamic conditions. Future research will focus on capturing and analyzing time-series images of cowpea leaves to understand the temporal dynamics of disease progression. Incorporating temporal data can improve early detection and distinguish between visually similar symptoms that evolve differently over time. For this purpose, models such as Recurrent Neural Networks (RNNs), Long Short-Term Memory (LSTM) networks, or Temporal Convolutional Networks (TCNs) will be considered. A major challenge in this direction will be the collection of longitudinal field data under consistent imaging conditions and the annotation of temporal stages of each disease. While such models have demonstrated improved accuracy in other domains, their higher computational complexity may impact real-time deployment in field conditions, particularly on mobile or edge devices. Strategies for model compression and optimization will also be investigated.
Acknowledgement: The authors would like to thank CSIR-NPL, Delhi, India, for the collection of leaves of the legume Cowpea for experimental infrastructure.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Conceptualization, Data Collection, Methodology, Validation, Resources and Formal Analysis: Vijaya Choudhary. Data Curation and Writing: Vijaya Choudhary. Investigation: Paramita Guha. Review, Editing and Supervision: Paramita Guha and Giovanni Pau. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data collected from CSIR, Delhi, India. Codes may be available on request. Few of them could for available with this link. https://github.com/ResearcherVijaya/COWPEA-DATABANK-WITH-CODE/tree/c6c0a20464a0544cac9b79e7206671cf3eabbdb7 (accessed on 01 July 2025). On-demand data could be available.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Bird JJ, Barnes CM, Manso LJ, Ekárt A, Faria DR. Fruit quality and defect image classification with conditional GAN data augmentation. Sci Hortic. 2022;293(9):110684. doi:10.1016/j.scienta.2021.110684. [Google Scholar] [CrossRef]
2. Kukačka J, Volkov V, Cremers D. Regularization for deep learning: a taxonomy. arXiv:1710.10686. 2017. [Google Scholar]
3. Clune J. AI-GAs: AI-generating algorithms, an alternate paradigm for producing general artificial intelligence. arXiv:1905.10985. 2019. [Google Scholar]
4. Wang R, Lehman J, Clune J, Stanley KO. POET: open-ended coevolution of environments and their optimized solutions. In: Proceedings of the Genetic and Evolutionary Computation Conference; 2019 Jul 13–17; Prague, Czech Republic. p. 142–51. doi:10.1145/3321707.3321799. [Google Scholar] [CrossRef]
5. Bahmei B, Birmingham E, Arzanpour S. CNN-RNN and data augmentation using deep convolutional generative adversarial network for environmental sound classification. IEEE Signal Process Lett. 2022;29:682–6. doi:10.1109/LSP.2022.3150258. [Google Scholar] [CrossRef]
6. Park SS, Kwon HJ, Baek JW, Chung K. Dimensional expansion and time-series data augmentation policy for skeleton-based pose estimation. IEEE Access. 2022;10:112261–72. doi:10.1109/access.2022.3214659. [Google Scholar] [CrossRef]
7. Wolfert S, Ge L, Verdouw C, Bogaardt MJ. Big data in smart farming—a review. Agric Syst. 2017;153:69–80. doi:10.1016/j.agsy.2017.01.023. [Google Scholar] [CrossRef]
8. Raviv T, Shlezinger N. Data augmentation for deep receivers. IEEE Trans Wireless Commun. 2023;22(11):8259–74. doi:10.1109/twc.2023.3261782. [Google Scholar] [CrossRef]
9. Gracia Moisés A, Vitoria Pascual I, Imas González JJ, Ruiz Zamarreño C. Data augmentation techniques for machine learning applied to optical spectroscopy datasets in agrifood applications: a comprehensive review. Sensors. 2023;23(20):8562. doi:10.3390/s23208562. [Google Scholar] [PubMed] [CrossRef]
10. Luis P, Jason W. The effectiveness of data augmentation in image classification using deep learning. In: Stanford University research report. Stanford, CA, USA: Stanford University; 2017. [Google Scholar]
11. Lemley J, Bazrafkan S, Corcoran P. Smart augmentation learning an optimal data augmentation strategy. IEEE Access. 2017;5:5858–69. doi:10.1109/access.2017.2696121. [Google Scholar] [CrossRef]
12. Ekin DC, Barret Z, Dandelion M, Vijay V, Quoc VL. AutoAugment: learning augmentation policies from data. arXiv:1805.09501. 2018. [Google Scholar]
13. Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. arXiv:1809.07294. 2018. [Google Scholar]
14. Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36(12):2536–45. doi:10.1109/tmi.2017.2708987. [Google Scholar] [PubMed] [CrossRef]
15. Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174(7):550–62. doi:10.1016/j.neuroimage.2018.03.045. [Google Scholar] [PubMed] [CrossRef]
16. Calimeri F, Marzullo A, Stamile C, Terracina G. Biomedical data augmentation using generative adversarial neural networks. In: Proceedings of 26th International Conference on Artificial Neural Networks; 2017 Sep 11–14; Alghero, Italy. p. 626–34. doi:10.1007/978-3-319-68612-7_71. [Google Scholar] [CrossRef]
17. Baur C, Albarqouni S, Navab N. MelanoGANs: high-resolution skin lesion synthesis with GANs. arXiv:1804.04338. 2018. [Google Scholar]
18. Moradi M, Madani A, Karargyris A, Syeda-Mahmood TF. Chest X-ray generation and data augmentation for cardiovascular abnormality classification. In: Medical Imaging 2018: Image Processing; 2018 Feb 10–15; Houston, TX, USA. doi:10.1117/12.2293971. [Google Scholar] [CrossRef]
19. Frid‐Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. arXiv:1801.02385. 2018. [Google Scholar]
20. Sierra S, Ramo R, Padilla M, Cobo A. Optimizing deep neural networks for high-resolution land cover classification through data augmentation. Environ Monit Assess. 2025;197(4):423. doi:10.1007/s10661-025-13870-5. [Google Scholar] [PubMed] [CrossRef]
21. Gupta YP, Mukul, Gupta N. Deep learning model based multimedia retrieval and its optimization in augmented reality applications. Multimed Tools Appl. 2023;82(6):8447–66. doi:10.1007/s11042-022-13555-y. [Google Scholar] [PubMed] [CrossRef]
22. Brahma SR, Singh S, Gupta DK, Malik A. A reversible data hiding technique using lower magnitude error channel pair selection. Multimed Tools Appl. 2023;82(6):8467–88. doi:10.1007/s11042-022-13554-z. [Google Scholar] [CrossRef]
23. Cho SB, Cheng Y, Sul S. Enhanced classification performance through GauGAN-based data augmentation for tomato leaf images. IET Image Process. 2024;18(14):4887–97. doi:10.1049/ipr2.13069. [Google Scholar] [CrossRef]
24. Islam N, Shin S. Robust deep learning models for OFDM-based image communication systems in intelligent transportation systems (ITS) for smart cities. Electronics. 2023;12(11):2425. doi:10.3390/electronics12112425. [Google Scholar] [CrossRef]
25. Gong Y, Wang F, Lv Y, Liu C, Li T. Automatic sleep staging using BiRNN with data augmentation and label redirection. Electronics. 2023;12(11):2394. doi:10.3390/electronics12112394. [Google Scholar] [CrossRef]
26. Xing L, Wang K, Wu H, Ma H, Zhang X. FL-MAAE: an intrusion detection method for the Internet of Vehicles based on federated learning and memory-augmented autoencoder. Electronics. 2023;12(10):2284. doi:10.3390/electronics12102284. [Google Scholar] [CrossRef]
27. Zeng MZ. Research on crop disease image data augmentation method based on generative adversarial network [master’s thesis]. Hefei, China: University of Science and Technology of China; 2021. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools