 Open Access
Open Access
ARTICLE
Rectal Cancer Stages T2 and T3 Identification Based on Asymptotic Hybrid Feature Maps
1 R&D Center of Artificial Intelligence Systems and Applications, School of Artificial Intelligence and Computer Science, Jiangnan University, Wuxi, 214122, China
2 Shanghai Enterprise Information Operation Center, China Telecom Group Co., Ltd., Shanghai, 201315, China
3 Department of Radiology, Dushu Lake Hospital Affiliated to Soochow University, Suzhou, 215123, China
4 Suzhou Yunmai Software Technology Co., Ltd., Suzhou, 215100, China
* Corresponding Author: Pengjiang Qian. Email:
(This article belongs to the Special Issue: Intelligent Biomedical Image Processing and Computer Vision)
Computer Modeling in Engineering & Sciences 2023, 137(1), 923-938. https://doi.org/10.32604/cmes.2023.027356
Received 26 October 2022; Accepted 19 December 2022; Issue published 23 April 2023
Abstract
Many existing intelligent recognition technologies require huge datasets for model learning. However, it is not easy to collect rectal cancer images, so the performance is usually low with limited training samples. In addition, traditional rectal cancer staging is time-consuming, error-prone, and susceptible to physicians’ subjective awareness as well as professional expertise. To settle these deficiencies, we propose a novel deep-learning model to classify the rectal cancer stages of T2 and T3. First, a novel deep learning model (RectalNet) is constructed based on residual learning, which combines the squeeze-excitation with the asymptotic output layer and new cross-convolution layer links in the residual block group. Furthermore, a two-stage data augmentation is designed to increase the number of images and reduce deep learning’s dependence on the volume of data. The experiment results demonstrate that the proposed method is superior to many existing ones, with an overall accuracy of 0.8583. Oppositely, other traditional techniques, such as VGG16, DenseNet121, EL, and DERNet, have an average accuracy of 0.6981, 0.7032, 0.7500, and 0.7685, respectively.Keywords
Rectal cancer is a type of colorectal cancer that is a common malignant tumor of the digestive tract, and its fatality rate is second only to lung cancer [1]. Among malignant tumors of the digestive system, it is followed by gastric cancer [2,3], and esophageal cancer [4], all of which pose a serious threat to a patient’s life and health. To a large extent, the description of the disease statute, the determination of the tumor treatment plan, the prediction of the disease progression, and the likelihood of disease recovery are all dependent on accurate rectal cancer staging. Thus, it plays a crucial role in the entire diagnosis and treatment process, such as determining which treatments and prognosis are appropriate for which stages. From a medical perspective, tumors are classified as T staging, N staging, or another staging, and T staging is employed in this paper. Rectal cancer tumors in the T2 stage penetrate the submucosa, whereas those in the T3 stage penetrate not only the submucosa but also the intrinsic muscle layer, as seen in Fig. 1. Hence, T3 is a more severe stage than T2, and there is a significant difference in their therapeutic interventions. When tumors are discovered at the T2 stage, they can be treated surgically right away. However, if metastatic lymph nodes are present above or around the T3 stages, neoadjuvant chemoradiotherapy should be used. As a result, the T2 and T3 stages mark the beginning of the transition from benign to malignant rectal cancer. The incorrect staging can result in unnecessary over-treatment and may lead to unanticipated consequences and expenses while providing no therapeutic benefit [5]. Correctly measuring the difference in T2 and T3 stages is a topic of cardinal importance and has great clinical significance.

Figure 1: Different stages of rectal cancer
Deep learning is one of the advantageous tools for medical data analysis, which involves image classification [6,7], medical image segmentation [8–10]. and so on. At the same time, the evolution of deep learning in the medical field is dependent on the accumulation of larger amounts of medical datasets, which have multi-modal properties and provide a large amount of rich data. Besides, Deep learning has made brilliant achievements in the processing of medical images, such as tumor detection, prediction, classification, segmentation, and registration of medical images, especially in gastric [11], brain [12], lung [13], histopathology [14], and image synthesis [15–17] or the other aspects of diagnosis and treatment [18–21]. Furthermore, differential feature maps [22], rough set theory [23], discrete gravity search algorithm (DGSA) [24], and other techniques are also employed effectively. Among them, ResNet [25] also achieves excellent performance in image classification [26,27] and detection [28,29]; Moreover, we also gain some understanding of few-shot learning tasks and learning their minds. Although the development of deep learning methods has achieved remarkable results with adequate data sets, there are some problems with small sample tasks [30]. Kim et al. [31] performed classification by computing the similarity between the multi-scale representation of the image and the label features of each class. Dong et al. [32] used low-level information to improve the classification accuracy for the few-shot task. For the problem of insufficient training samples [33–38], various methods are employed to handle specific challenges.
At present, deep learning for rectal cancer involves the following aspects: segmentation of the rectum and rectal cancer [39], analysis of rectal cancer lymphatic metastasis based on computerized tomography images [40], automatic detection of the invasion of vessels outside the rectal wall [41], assisting patients’ treatment programs. and detection or classification of intestinal polyps [42]. These deep learning methods have specific requirements on the dataset and lack studies when the sample size is scarce. Traditional machine learning methods, such as radiomics [43–48], can exploit potential differences in radiomics features of rectal cancer patients to predict patient survival, but feature extraction in radiomics requires manual involvement. Deep learning continues to evolve in treatment problems and medical diagnosis. The medical perspective includes a new classification based on magnetic resonance imaging (MRI), which determines the relationship between tumors and their fixed parameters on MRm [49] or using support vector machines as well as RNA test data for classification [50]. There are enough cases for doctors to analyze for this condition. In response to the lack of training datasets, Wang, Cui et al. proposed a multi-branch hybrid model [51], which makes full use of the information contained in the data. Our method can deal with the same situation but in a different way.
In this paper, we propose an accurate and reliable deep-learning model, which effectively addresses the challenge of traditional rectal cancer staging and data collection. First, we make full use of the advantages of deep learning, where feature extraction is extracted automatically via convolution kernels rather than manual selection. Meanwhile, it reduces the time and labor cost of collecting and manually annotation to a certain extent. To address the issue of the limited and unbalanced dataset, we present a new classification model with appropriate modifications based on ResNet34. Apart from squeezing and extracting between residual blocks, we add this structure to residual groups and re-create the residual architecture by applying the idea of feature fusion to establish cross-convolution layer links. T2WI images, both annotated and unannotated, were selected and preprocessed. Then, the training set consists of the original images and the preprocessed images. Our image preprocessing mainly includes noise, contrast, shift tumor location, and so on; besides, the images obtained after preprocessing must ensure the integrity of the tumor and avoid the loss of tumor features. There is a correlation between the preprocessed samples. To alleviate the influence of the relevance of the data in the training model, we use an asymptotic output layer (AOL) at the end of the model. Additionally, due to personnel shortages and unequal distribution, our approach may be used in the future to help doctors prevent delays and bias in imaging results [52]. The contributions of our work are as follows:
1) We develop a new network framework for T2WI image classification of rectal cancer, which incorporates the recalibrating feature and AOL organically; Meanwhile, we widen the network width further by using cross-convolution layers, reducing the loss of feature information. Compared with the existing classification models, our model achieves the best.
2) We use effective data augmentation methods to avoid problems, which could be generated by small and unbalanced data in the process of model training, and it is conducive to training a model.
3) The proposed classification model is based on significant data expansion and model innovation. Its accuracy can reach 0.8583, which helps doctors make correct judgments, avoid subjectivity, save labor, and achieve better treatment benefits.
The rest of our manuscript is organized as follows: Section 2 summarizes and reviews the relevant works, such as SENet, SpianlNet, and others. The proposed RectalNet method and the processing of the rectal cancer dataset are stated detailedly in Section 3. The details of the experimental setup, the analysis, and the discussion of experimental results are presented in Section 4; the conclusion is given in Section 5.
In this paper, we fully draw the advantages of feature recalibration [53] to improve the performance of our model. The basic parameter propagation mechanism is depicted in Fig. 2, and the fundamental idea behind this network’s work is that it can learn attention weights for its channels in an adaptable manner given an input. The following are the primary three processes utilized to recalibrate the previously acquired feature maps:

Figure 2: The main mechanism is squeeze and excitation
(1) Squeeze Operation.
(2) Excitation Operation. This operation’s role is primarily reflected in the use of the fully connected layer, whose purpose is to perform a nonlinear transformation of the previous stage’s result. A channel scaling parameter is set, representing a correction between channels, and the weights are continuously updated in the backpropagation.
(3) Scale Operation.
Deep neural networks bring high performance to engineering and science fields and have attracted attention for their success in various cognitive tasks of machine learning. Asymptotic output is a neural network module that imitates the properties of the human somatosensory system, which mainly refers to how the spine receives external input signals and outputs bodily responses; This module can be divided into an input layer, a hidden layer, and an output layer. The input layer mainly receives external signals, the hidden layer can be regarded as the joint processing of input signals by neurons, and the output layer is used to present the processing results of the signals. The working principle is shown in Fig. 3: (1) Each time a portion of the input data is presented as the first neuron’s input; (2) The other part remaining data and the output of the first neuron are considered as the input of the second neuron; (3) Repeat the second step until all the input data is used up; (4) The output of each neuron is concatenated together as the final output. This process is expressed in Eq. (4). The goal of a deep learning network is to minimize the loss of input feature mapping and thus obtain better classification results. To achieve this goal, this paper utilizes multiple fully connected layers to process input data with inter-processing between fully connected layers.

Figure 3: The workflow of asymptotic output
At present, many scholars apply ResNet as the benchmark for a variety of tasks in the field of computer vision because ResNet can build residual blocks by adding a shortcut connection between input and output. It can be understood that this network is a combination of several parallel sub-networks by combining multiple paths rather than stacking. In this paper, an expanded residual learning network, named RectalNet, to classify T2WI images of rectal cancer. It can increase its depth and width at the same time and learn image information correctly and repeatedly.
3.1 Augmentation of Rectal Cancer Dataset (RECD)
In this paper, the RECD was collected by the Imaging Department of Changshu First People’s Hospital and has been approved by the ethics committee. Patients with colorectal cancer who underwent curative resection, patients with early-stage rectal cancer (Early stage: T1 or T2 and N0) who underwent total mesorectal excision (TME), and patients with progressive-stage rectal cancer (Advanced stage: T3, T4, N+ or CRM+) who underwent neoadjuvant chemoradiotherapy followed by TME were enrolled in this paper. T and N staging were determined based on postoperative histopathological findings in the first type of patients. For the second type of patients, T and N staging was evaluated based on a pre-neoadjuvant MRI report by a radiologist (Zhihua Lu) experienced in MRI diagnosis of gastrointestinal tumors. It should be noted that histopathological specimens did not correspond to MR images. The images in this dataset were collected from 67 patients. These patients were diagnosed with rectal T1, T2, T3, and T4 stages, as well as malignant lymphoma and mucinous adenocarcinoma, and all the labeled data were annotated by experienced doctors. According to these diagnostic results and the number of patients with corresponding diseases, there are only five patients with rectal cancer T1, only three patients with rectal cancer T4, and a total of four patients with other tumors. Thus, we abandoned those seriously unbalanced diseases and finally chose to classify T2 (19 patients) and T3 (36 patients) rectal cancer. Because this has a certain clinical significance, subsequent treatment and recovery are different depending on the stages. Computerized tomography (CT) and magnetic resonance (MR) imaging are performed on each patient. MR imaging, as opposed to CT imaging, images at the molecular level and offers superior soft-tissue resolution with no radiation. MR scans can provide physiological, pathological, and metabolic information in addition to the anatomical structure of the human body. Accordingly, MR(T2WI) has the most reference value.
When a deep learning model is used to categorize the T2 and T3 of rectal cancer, the training dataset’s volume significantly impacts the model’s performance. Due to our limited data amount, data augmentation technology is indispensable. In this paper, our workflow is depicted in Fig. 4, and n represents the number of cycles (n is 20 in our experiments). In the first phase, we conduct numerous fundamental data alterations, such as horizontal and vertical mirror inversion, pepper and salt noise, contrast enhancement, and so on. After this phase, the model’s generalization capability is improved, and the data amount is enhanced. In addition, we exploit random clipping, random offset, and zca-whitening processing in the second and most significant step. Meanwhile, we ensure the tumor’s integrity in the image regions. As shown in Fig. 5, the purpose of this procedure is to expand the diversity of training images while also assisting the model in extracting the features of the tumor fully. Finally, the training set contains 2000 images, where T2 and T3 are 800 and 1200 images, respectively.
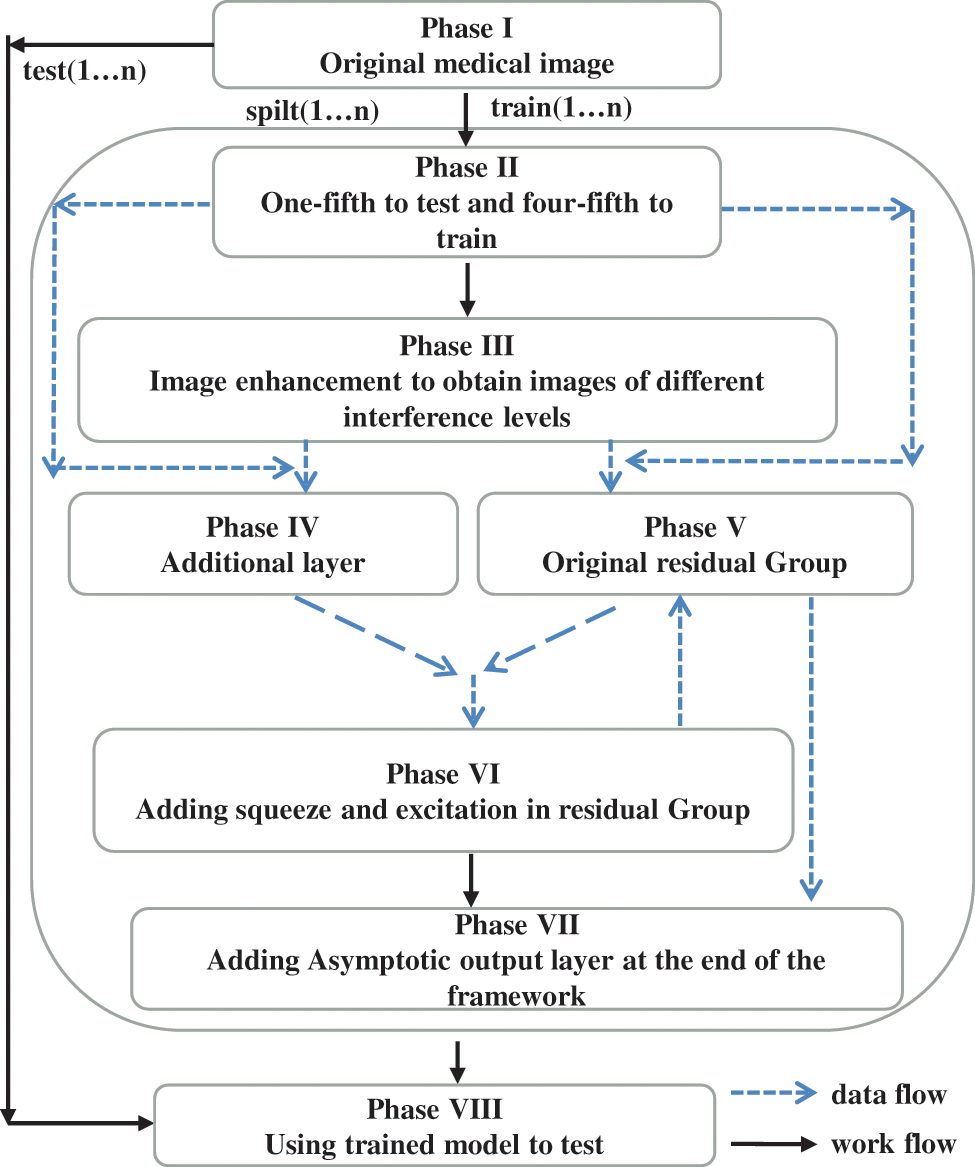
Figure 4: The workflow

Figure 5: T2WI images of one patient after processing
3.2 Proposed Network Framework
3.2.1 Reduce the Loss of Feature Information in Residual Group(SE)
We introduce squeeze-excitation into the residual group, which primarily compresses and extracts the feature maps. It is also proved in [37] that this operation, combined with some basic network models, can improve network performance even further. What distinguishes our network in this paper is that it employs this operation in residual blocks as well as in residual groups. We constructed this substructure to alleviate the following issues: Firstly, the semantic information of feature maps will be lost after repeated convolution operations between each residual block. Secondly, there is no discernible difference between our data categories, and it is difficult for our model to distinguish information. Thirdly, compared with natural images, medical images are more likely to lose some vital information; After preprocessing, the images input our model, which not only compresses and extracts each residual block, recalibrates the features, but also performs global average pooling and dimensionality reduction between each residual block. The importance of each feature channel is automatically acquired through repeated learning, and the corresponding parameters of each channel number are constantly updated to achieve multiple captures of shallow information, reduce feature loss, and provide meaningful information supplements. Fig. 6 depicts the sub-model, whereas Part II of Fig. 7 depicts the inter-structure. This mechanism is added between the third and fourth residual blocks, and the performance is improving greatly. The following is the flow of our algorithm:

Figure 6: The structure of SE between the residual group

Figure 7: The overall structure of the RectalNet
3.2.2 Asymptotic Output Avoid Information Loss (AOL)
We use the asymptotic output block as our classifier at the end of the model, which is composed of full connection layers. We use it as our classifier because it can gradually integrate useful information, avoiding the feature of information redundancy. As shown in Fig. 8, this block inputs information step by step to avoid accepting too much information at once in the final fully connected layer and to efficiently output differentiated information. Furthermore, the asymptotic output layer consists of three elements: the input layer, the hidden layer, and the output layer; the process is as follows: when the tensor propagates forward to the hidden layer, each hidden layer selects the pixel values in different dimensions of the input tensor for processing, and there is information interaction between each hidden layer; The use of hidden layers will increase the depth of the network; The overall structure of the asymptotic output layer is depicted in Fig. 8 and is Part III of Fig. 7. The application of adaptive average pooling can compress spatial dimensions and remove the mean values of corresponding dimensions, which can suppress some useless features to a certain extent. In addition, the final experimental results are diverse depending on the number of hidden layers in the asymptotic input layer. The selection of the number of relevant layers will be given in the experimental part. As illustrated in formula (1), we use five hidden layers as a classifier.
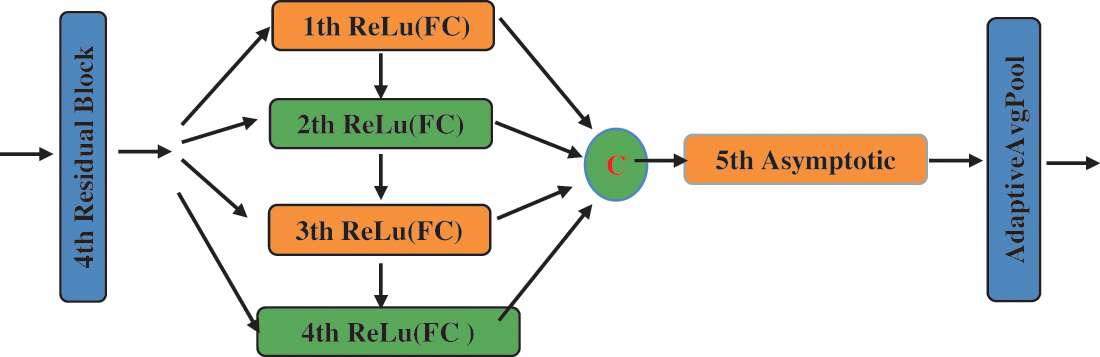
Figure 8: The framework of the asymptotic layer
3.2.3 Get Different Receptive Fields between Residual Groups(ADD)
To reduce the loss of low-lever feature information in medical images, we construct an additional block, including Batch Normalization, ReLu activation function, and convolution with a 2 * 2 kernel. It can be inserted between the residual blocks to reuse the feature map of the previous group of residual blocks. The structure can be seen in Fig. 6. The additional layer has a different receptive field and can acquire more information from limited data, thereby increasing the richness of the feature map. The convolution kernels of 1 * 1, 3 * 3, and 7 * 7 are used in most network models, whereas our framework employs an even-numbered convolution kernel of 2 * 2. Diverse convolution kernels have different receptive fields so that they can obtain more feature information from distinct convolution processes. Their fundamental function is to use the receptive field of the even convolution kernel to extract the shallow feature information that is ignored by the odd receptive field. This process corresponds to Part I of Fig. 7.
3.3 The Overall Structure of the Network Framework
RectalNet is a feature representation learning model, which repeatedly uses shallow semantic information in the process of feature extraction. As for complex rectal cancer T2WI images, the model has a strong multi-level abstraction ability to learn distinguishing features of T2 and T3 stages, and spatial distance constraint capability. Fig. 7 depicts the proposed network’s outline. Inspired by [43,44], the model solves the problem of too large correlations between sample categories during the data expansion phase and continuously utilizes the advantages of [43] networks to solve our dilemmas. Moreover, the main features of the RectalNet model are as follows:
(1) We draw on the characteristics of feature fusion and the feature output of the residual group structure to reduce the loss of the residual block from the previous layer, as shown in Part I of Fig. 7;
(2) We use the SE channel attention mechanism in the last two sets of residual structures to recalibrate the features multiple times in the channel dimension, as shown in Part II of Fig. 7;
(3) We add an asymptotic output layer at the end of the network to avoid overfitting, as shown in Part III of Fig. 7.
Our experimental settings are as follows: the development system uses Ubuntu 16.04 LTS (64 bits), the processor is the CPU E5-2640 V4, the memory is 64 G, the GPU is the GeForce RTX 2080 Ti, the editor uses PyCharm, and the development language used is Python, and the deep learning framework uses PyTorch version 1.8.0. During model training, our learning rate is set to 0.005, the maximum number of iterations is 100, and the batch size is 20. In the experimental part, we conduct comparison experiments and ablation experiments and also investigate the number of hidden layers in the asymptotic input layer. As illustrated in Fig. 9, the final accuracy of our proposed model can converge to around 0.85 and its loss to around 0.3. To better train our model, the optimizer of our choice during the training process is Adam. To control the update range of model parameters during the experiment, L2 regularization is added, and the weight decay parameter is set to 0.01. To verify the generalization of our model, each method is run 20 times for random data, and we take the mean of all the criteria plus or minus the standard deviation.
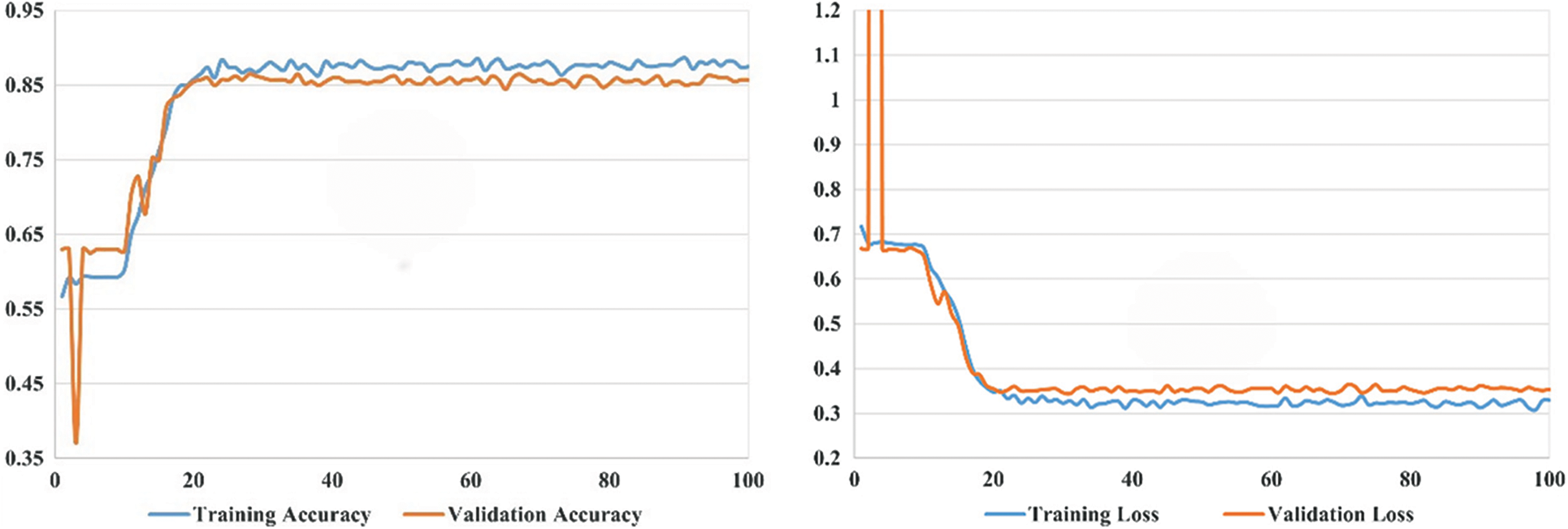
Figure 9: The curve of the proposed model’s accuracy and loss
4.2 Experiment Result and Discussion
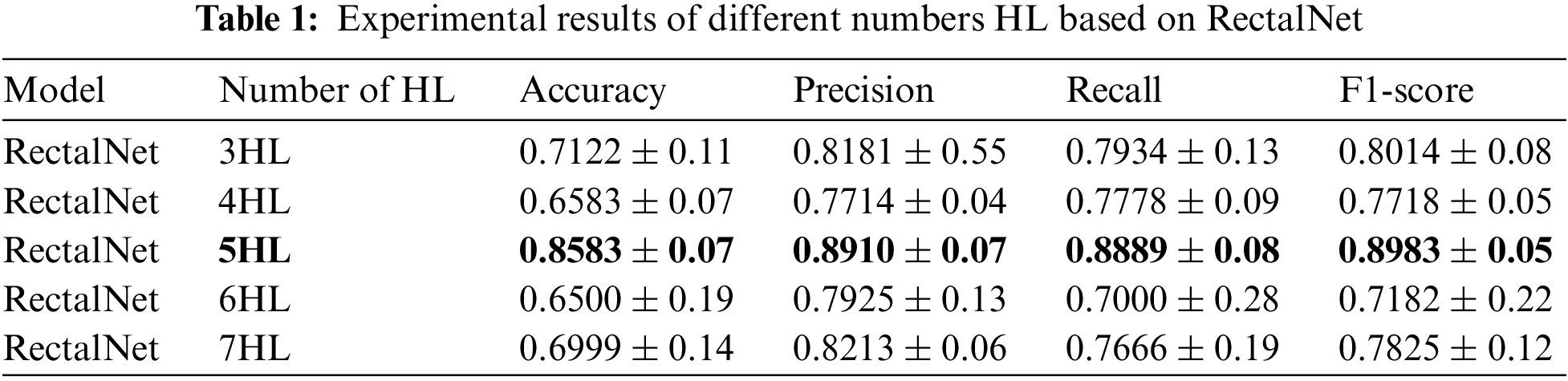
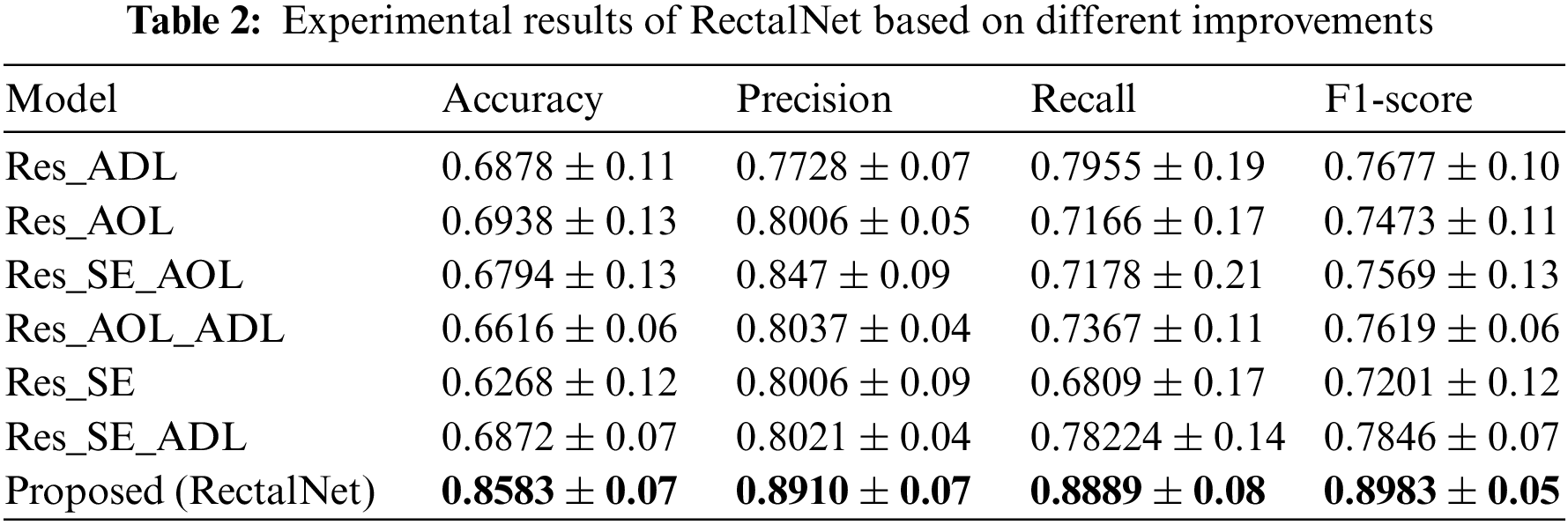
In this section, we assess the effectiveness of the proposed RectalNet method for rectal cancer classification. We use accuracy, precision, recall, and F1-score to evaluate our model, where F1-score can balance recall and precision, avoiding either being too high or too low. Firstly, we affirm the number of hidden layers in the asymptotic output block. Different numbers of hidden layers have diverse influences on the model. In addition to more layers bringing more variables, it also harms the final performance of the model. As available from Table 1, when the number of hidden layers (HL) is 6 or 7, the final test accuracy is less than HL = 5. Similarly, if HL is 3 or 4, the performance is unsatisfactory too. While the number of HL is 5, the ultimate test accuracy of the model achieves the best at 0.8583. Therefore, comprehensively considering the accuracy and parameters, the number of hidden layers of our model is 5 in the subsequent experiments. Besides, we can find that fewer parameters will not improve performance, and we should choose the appropriate number of hidden layers. Fig. 9 illustrates the training process of our model and the training accuracy converges to 0.85, while the training loss is closer to 0.2. Secondly, we conduct the ablation experiment to confirm our final model is valid in Table 2. Our proposed model has three main novel highlights: squeeze-excitation (SE) block, additional layer (ADL), and asymptotic output layer (AOL). They can recalibrate features, and obtain information from different acceptive fields across multi-layer convolutional connections. In our model, we apply a squeeze-excitation (SE) block to each set of residuals with an accuracy of 0.6268, we utilize an additional layer (ADL) with an accuracy of 0.7616, and we employ an asymptotic output layer (AOL) with an accuracy of 0.8583. Oppositely, the performance combination of any two of the three alterations or one change drop dramatically. Furthermore, as known from Table 2, each highlight is meaningful. Finally, we conduct the comparison experiment to ensure our model exceeds existing deep learning models. We performed all the models 20 times to verify their stability. It is clear from Table 3 that the proposed network has the highest accuracy. Although the recall values of VGG16, AlexNet, and EL1 models are higher than ours, their precision values are lower. In addition to this, the accuracy, precision, recall, and F1-score of our model are higher than other models. Therefore, our model is superior to existing deep learning models. Besides, our model is sensitive to the RECD, while other models are unfavorable to RECD.
In this paper, we propose a method for rectal cancer classification. Despite being a part of the T stages, few reporters researched it, and our method exhibits superior performance. As we can see from the experimental tables, the accuracy has increased, as have the other three indicators, and the recall has decreased acceptably. To better determine the treatment plan from doctors, it is crucial for our model to accurately recognize the status of patients’ tumors from medical images. Currently, there are three major challenges in rectal cancer classification. The first problem is the lack of labeled data because only a few radiologists and physicians can accurately identify the location and shape of rectal cancer from medical images; Collecting sufficient data and labeling data is time-consuming. Another challenge is that the results of disease diagnoses tend to be skewed towards a certain category, resulting in imbalanced data; Collecting medical images with a comparable number of categories is tougher. The last one is that the shape of the tumors and rectum is varied, depending on the object and location being depicted, so it is challenging to establish a universal model for classifying rectal tumors.
When developing a deep learning model for medical image classification, we must take into account the characteristics of medical images. Medical images have fewer distinguishing features and are more difficult to identify than conventional images. In this study, we focused on classifying the T2 and T3 stages of rectal cancer and compensated for the deficiencies of our dataset by constructing a model. Based on the results of the experiments, we can conclude that the proposed model has some validity. Our model outperforms typical deep learning models in terms of results. We have successfully treated restricted datasets in this paper to decrease the reliance of deep learning models on the amount of data; Furthermore, the method of data augmentation may result in significant correlations between images, causing the model to overfit during the training process. We may need to limit the scope of data preprocessing, and cannot blindly expand the quantity. Our method can be applied to other small sample datasets as a model. Besides, the dominant sources of our errors are as follows: The patient’s age, gender, tumor distribution site, and other factors influence tumor characteristics, but our data had a small sample size, resulting in a lack of variation in tumor features; Another source is our model; in the meantime, our model may lack generalizability. One of the main reasons is that tumors have a variety of strange shapes. From a medical perspective, the more a tumor seems irregular, the more likely it is a malignant tumor.
We presented the asymptotic-feature-map-hybrid approach as constructive and practical based on feature information from combination preprocessing, identifying rectal cancer stages between T2 and T3. Asymptotic-hybrid-feature-map has incorporated multiple techniques and strategies, including uncomplicated data preprocessing, feature compression and extraction, the asymptotic output of feature information, and adding cross-multi-layer convolution information interaction. Consequently, our experimental results demonstrate the superiority of our approach over the other networks. This raises the likelihood of being used in clinical settings. In future work, we will try to apply the proposed method to other medical image classification problems to test whether it is suitable for a more limited sample of medical datasets and whether the superior performance of our method generalizes to larger groups, and we will further combine deep learning with the medical field to compare the performance of the tool with the performance of radiologists and postoperative histology. Likewise, we will take into account tumor segmentation because lump size affects treatment decisions for rectal cancer and then use our model to classify the carcinoma according to its morphology.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China under Grants 62172192, U20A20228, and 62171203, in part by the 2018 Six Talent Peaks Project of Jiangsu Province under Grant XYDXX-127, and in part by the Science and Technology Demonstration Project of Social Development of Jiangsu Province under Grant BE2019631.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ibrahim, D. M., Elshennawy, N. M., Sarhan, A. M. (2021). Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Computers in Biology and Medicine, 132(2), 104348. https://doi.org/10.1016/j.compbiomed.2021.104348 [Google Scholar] [PubMed] [CrossRef]
2. Hamada, K., Kawahara, Y., Tanimoto, T., Ohto, A., Toda, A. et al. (2022). Application of convolutional neural networks for evaluating the depth of invasion of early gastric cancer based on endoscopic images. Journal of Gastroenterology and Hepatology, 37(2), 352–357. https://doi.org/10.1111/jgh.15725 [Google Scholar] [PubMed] [CrossRef]
3. Ling, T., Wu, L., Fu, Y., Xu, Q., An, P. et al. (2021). A deep learning-based system for identifying differentiation status and delineating the margins of early gastric cancer in magnifying narrow-band imaging endoscopy. Endoscopy, 53(5), 469–477. https://doi.org/10.1055/a-1229-0920 [Google Scholar] [PubMed] [CrossRef]
4. Lee, S. L., Yadav, P., Starekova, J., Christensen, L., Chandereng, T. et al. (2021). Diagnostic performance of MRI for esophageal carcinoma: A systematic review and meta-analysis. Radiology, 299(3), 583–594. https://doi.org/10.1148/radiol.2021202857 [Google Scholar] [PubMed] [CrossRef]
5. Dietz, D. W. (2013). Multidisciplinary management of rectal cancer: The OSTRICH. Journal of Gastrointestinal Surgery, 17(10), 1863–1868. https://doi.org/10.1007/s11605-013-2276-4 [Google Scholar] [PubMed] [CrossRef]
6. Qian, P., Xi, C., Xu, M., Jiang, Y., Su, K. H. et al. (2018). SSC-EKE: Semi-supervised classification with extensive knowledge exploitation. Information Sciences, 422, 51–76. https://doi.org/10.1016/j.ins.2017.08.093 [Google Scholar] [PubMed] [CrossRef]
7. Cai, L., Gao, J., Zhao, D. (2020). A review of the application of deep learning in medical image classification and segmentation. Annals of Translational Medicine, 8(11). https://doi.org/10.21037/atm.2020.02.44 [Google Scholar] [PubMed] [CrossRef]
8. Tran, S. T., Cheng, C. H., Nguyen, T. T., Le, M. H., Liu, D. G. (2021). TMD-Unet: Triple-Unet with multi-scale input features and dense skip connection for medical image segmentation. In: Healthcare, vol. 954. Basel: MDPI. [Google Scholar]
9. Latif, U., Shahid, A. R., Raza, B., Ziauddin, S., Khan, M. A. (2021). An end-to-end brain tumor segmentation system using multi-inception-UNET. International Journal of Imaging Systems and Technology, 31(4), 1803–1816. https://doi.org/10.1002/ima.22585 [Google Scholar] [CrossRef]
10. Yao, Y., Gou, S., Tian, R., Zhang, X., He, S. (2021). Automated classification and segmentation in colorectal images based on self-paced transfer network. BioMed Research International, 2021(2), 1–7. https://doi.org/10.1155/2021/6683931 [Google Scholar] [PubMed] [CrossRef]
11. Meier, A., Nekolla, K., Earle, S., Hewitt, L., Aoyama, T. et al. (2018). End-to-end learning to predict survival in patients with gastric cancer using convolutional neural networks. Annals of Oncology, 29, VIII23. https://doi.org/10.1093/annonc/mdy269.075 [Google Scholar] [CrossRef]
12. Ertosun, M. G., Rubin, D. L. (2015). Automated grading of gliomas using deep learning in digital pathology images: A modular approach with ensemble of convolutional neural networks. AMIA Annual Symposium Proceedings, vol. 2015, pp. 1899–1908. Bethesda, MD, USA, American Medical Informatics Association. [Google Scholar]
13. Teramoto, A., Tsukamoto, T., Kiriyama, Y., Fujita, H. (2017). Automated classification of lung cancer types from cytological images using deep convolutional neural networks. BioMed Research International, 2017(1), 1–6. https://doi.org/10.1155/2017/4067832 [Google Scholar] [PubMed] [CrossRef]
14. Chang, H. Y., Jung, C. K., Woo, J. I., Lee, S., Cho, J. et al. (2019). Artificial intelligence in pathology. Journal of Pathology and Translational Medicine, 53(1), 1–12. https://doi.org/10.4132/jptm.2018.12.16 [Google Scholar] [PubMed] [CrossRef]
15. Qian, P., Chen, Y., Kuo, J. W., Zhang, Y. D., Jiang, Y. et al. (2019). mDixon-based synthetic CT generation for PET attenuation correction on abdomen and pelvis jointly using transfer fuzzy clustering and active learning-based classification. IEEE Transactions on Medical Imaging, 39(4), 819–832. https://doi.org/10.1109/TMI.2019.2935916 [Google Scholar] [PubMed] [CrossRef]
16. Yang, H., Xia, K., Anqi, B., Qian, P., Khosravi, M. R. (2019). Abdomen MRI synthesis based on conditional GAN. 2019 International Conference on Computational Science and Computational Intelligence (CSCI), pp. 1021–1025. Las Vegas, NV, IEEE. [Google Scholar]
17. Yang, H., Qian, P., Fan, C. (2020). An indirect multimodal image registration and completion method guided by image synthesis. Computational and Mathematical Methods in Medicine, 2020(1), 1–10. https://doi.org/10.1155/2020/2684851 [Google Scholar] [PubMed] [CrossRef]
18. Mushtaq, M. F., Shahroz, M., Aseere, A. M., Shah, H., Majeed, R. et al. (2021). BHCNet: Neural network-based brain hemorrhage classification using head CT Scan. IEEE Access, 9, 113901–113916. https://doi.org/10.1109/ACCESS.2021.3102740 [Google Scholar] [CrossRef]
19. Rajesh, T., Malar, R., Geetha, M. R. (2019). Brain tumor detection using optimisation classification based on rough set theory. Cluster Computing, 22(6), 13853–13859. https://doi.org/10.1007/s10586-018-2111-5 [Google Scholar] [CrossRef]
20. Abd El Kader, I., Xu, G., Shuai, Z., Saminu, S., Javaid, I. et al. (2021). Differential deep convolutional neural network model for brain tumor classification. Brain Sciences, 11(3). https://doi.org/10.3390/brainsci11030352 [Google Scholar] [PubMed] [CrossRef]
21. Islam, K. T., Wijewickrema, S., O’Leary, S. (2019). A rotation and translation invariant method for 3D organ image classification using deep convolutional neural networks. PeerJ Computer Science, 5(2), e181. https://doi.org/10.7717/peerj-cs.181 [Google Scholar] [PubMed] [CrossRef]
22. Kowsari, K., Sali, R., Ehsan, L., Adorno, W., Ali, A. et al. (2020). HMIC: Hierarchical medical image classification, a deep learning approach. Information, 11(6), 318. https://doi.org/10.3390/info11060318 [Google Scholar] [PubMed] [CrossRef]
23. Helaly, H. A., Badawy, M., Haikal, A. Y. (2022). Deep learning approach for early detection of Alzheimer’s disease. Cognitive Computation, 14(5), 1711–1727. https://doi.org/10.1007/s12559-021-09946-2 [Google Scholar] [PubMed] [CrossRef]
24. Dai, Y., Gao, Y., Liu, F. (2021). TransMed: Transformers advance multi-modal medical image classification. Diagnostics, 11(8), 1384. https://doi.org/10.3390/diagnostics11081384 [Google Scholar] [PubMed] [CrossRef]
25. He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778. Las Vegas, NV, USA. [Google Scholar]
26. Budhiman, A., Suyanto, S., Arifianto, A. (2019). Melanoma cancer classification using resnet with data augmentation. 2019 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), pp. 17–20. Yogyakarta, Indonesia, IEEE. [Google Scholar]
27. Li, X., Rai, L. (2020). Apple leaf disease identification and classification using resnet models. 2020 IEEE 3rd International Conference on Electronic Information and Communication Technology (ICEICT), pp. 738–742. Shenzhen, China, IEEE. [Google Scholar]
28. Zhu, Z., Zhai, W., Liu, H., Geng, J., Zhou, M. et al. (2021). Juggler-ResNet: A flexible and high-speed ResNet optimization method for intrusion detection system in software-defined industrial networks. IEEE Transactions on Industrial Informatics, 18(6), 4224–4233. https://doi.org/10.1109/TII.2021.3121783 [Google Scholar] [CrossRef]
29. Zheng, Z., Zhang, H., Li, X., Liu, S., Teng, Y. (2021). Resnet-based model for cancer detection. 2021 IEEE International Conference on Consumer Electronics and Computer Engineering (ICCECE), pp. 325–328. Guangzhou, China, IEEE. [Google Scholar]
30. Qi, G. J., Luo, J. (2020). Small data challenges in big data era: A survey of recent progress on unsupervised and semi-supervised methods. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(4), 2168–2187. https://doi.org/10.1109/TPAMI.2020.3031898 [Google Scholar] [PubMed] [CrossRef]
31. Kim, J., Chi, M. (2021). SAFFNet: Self-attention-based feature fusion network for remote sensing few-shot scene classification. Remote Sensing, 13(13), 2532. https://doi.org/10.3390/rs13132532 [Google Scholar] [CrossRef]
32. Dong, B., Wang, R., Yang, J., Xue, L. (2021). Multi-scale feature self-enhancement network for few-shot learning. Multimedia Tools and Applications, 80(25), 33865–33883. https://doi.org/10.1007/s11042-021-11205-3 [Google Scholar] [CrossRef]
33. Tong, C., Liang, B., Su, Q., Yu, M., Hu, J. et al. (2020). Pulmonary nodule classification based on heterogeneous features learning. IEEE Journal on Selected Areas in Communications, 39(2), 574–581. https://doi.org/10.1109/JSAC.2020.3020657 [Google Scholar] [CrossRef]
34. Han, C., Rundo, L., Murao, K., Noguchi, T., Shimahara, Y. et al. (2021). MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinformatics, 22(2), 1–20. https://doi.org/10.1186/s12859-020-03936-1 [Google Scholar] [PubMed] [CrossRef]
35. Li, X., Wu, J., Sun, Z., Ma, Z., Cao, J. et al. (2020). BSNet: Bi-similarity network for few-shot fine-grained image classification. IEEE Transactions on Image Processing, 30, 1318–1331. https://doi.org/10.1109/TIP.2020.3043128 [Google Scholar] [PubMed] [CrossRef]
36. Chen, Z., Zhang, X., Huang, W., Gao, J., Zhang, S. (2021). Cross modal few-shot contextual transfer for heterogenous image classification. Frontiers in Neurorobotics, 15, 56. https://doi.org/10.3389/fnbot.2021.654519 [Google Scholar] [PubMed] [CrossRef]
37. Fabian, H. S., Kang, D. K. (2019). Learning deep representation by increasing ConvNets depth for few shot learning. International Journal of Advanced Smart Convergence, 8(4), 75–81. https://doi.org/10.7236/IJASC.2019.8.4.75 [Google Scholar] [CrossRef]
38. Yu, Z., Raschka, S. (2020). Looking back to lower-level information in few-shot learning. Information, 11(7), 345. https://doi.org/10.3390/info11070345 [Google Scholar] [CrossRef]
39. Lee, J., Oh, J. E., Kim, M. J., Hur, B. Y., Sohn, D. K. (2019). Reducing the model variance of a rectal cancer segmentation network. IEEE Access, 7, 182725–182733. https://doi.org/10.1109/ACCESS.2019.2960371 [Google Scholar] [CrossRef]
40. Wang, H., Wang, H., Song, L., Guo, Q. (2019). Automatic diagnosis of rectal cancer based on CT images by deep learning method. 2019 12th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), pp. 1–5. Suzhou, China, IEEE. [Google Scholar]
41. Li, S., Zhang, Z., Lu, Y. (2020). Efficient detection of EMVI in rectal cancer via richer context information and feature fusion. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 1464–1468. Iowa City, IA, USA, IEEE. [Google Scholar]
42. Sabol, P., Sinčák, P., Hartono, P., Kočan, P., Benetinová, Z. et al. (2020). Explainable classifier for improving the accountability in decision-making for colorectal cancer diagnosis from histopathological images. Journal of Biomedical Informatics, 109(4), 103523. https://doi.org/10.1016/j.jbi.2020.103523 [Google Scholar] [PubMed] [CrossRef]
43. Rosati, S., Gianfreda, C. M., Balestra, G., Giannini, V., Mazzetti, S. et al. (2018). Radiomics to predict response to neoadjuvant chemotherapy in rectal cancer: Influence of simultaneous feature selection and classifier optimization. 2018 IEEE Life Sciences Conference (LSC), pp. 65–68. Montreal, QC, Canada, IEEE. [Google Scholar]
44. Li, H., Boimel, P., Janopaul-Naylor, J., Zhong, H., Xiao, Y. et al. (2019). Deep convolutional neural networks for imaging data based survival analysis of rectal cancer. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 846–849. Venice, Italy, IEEE. [Google Scholar]
45. Liang, M., Cai, Z., Zhang, H., Huang, C., Meng, Y. et al. (2019). Machine learning-based analysis of rectal cancer MRI radiomics for prediction of metachronous liver metastasis. Academic Radiology, 26(11), 1495–1504. https://doi.org/10.1016/j.acra.2018.12.019 [Google Scholar] [PubMed] [CrossRef]
46. Zhang, X., Zhang, B., Wang, B., Zhang, F. (2022). Automatic prediction of T2/T3 staging of rectal cancer based on radiomics and machine learning. Big Data Research, 30(2), 100346. https://doi.org/10.1016/j.bdr.2022.100346 [Google Scholar] [CrossRef]
47. Detering, R., Oostendorp, S. E., Meyer, V. M., Dieren, S., Bos, A. C. R. K. et al. (2020). MRI cT1-2 rectal cancer staging accuracy: A population-based study. Journal of British Surgery, 107(10), 1372–1382. https://doi.org/10.1002/bjs.11590 [Google Scholar] [PubMed] [CrossRef]
48. Shin, J., Seo, N., Baek, S. E., Son, N. H., Lim, J. S. et al. (2022). MRI radiomics model predicts pathologic complete response of rectal cancer following chemoradiotherapy. Radiology, 303(2), 351–358. https://doi.org/10.1148/radiol.211986 [Google Scholar] [PubMed] [CrossRef]
49. Alasari, S., Lim, D., Kim, N. K. (2015). Magnetic resonance imaging based rectal cancer classification: Landmarks and technical standardization. World Journal of Gastroenterology, 21(2), 423–431. https://doi.org/10.3748/wjg.v21.i2.423 [Google Scholar] [PubMed] [CrossRef]
50. Zhang, Y., Wu, Y., Gong, Z. Y., Ye, H. D., Zhao, X. K. et al. (2021). Distinguishing rectal cancer from colon cancer based on the support vector machine method and RNA-sequencing data. Current Medical Science, 41(2), 368–374. https://doi.org/10.1007/s11596-021-2356-8 [Google Scholar] [PubMed] [CrossRef]
51. Wang, J., Cui, Y., Shi, G., Zhao, J., Yang, X. et al. (2020). Multi-branch cross attention model for prediction of KRAS mutation in rectal cancer with t2-weighted MRI. Applied Intelligence, 50(8), 2352–2369. https://doi.org/10.1007/s10489-020-01658-8 [Google Scholar] [CrossRef]
52. Gao, Y., Lu, Y., Li, S., Dai, Y., Feng, B. et al. (2021). Chinese guideline for the application of rectal cancer staging recognition systems based on artificial intelligence platforms (2021 edition). Chinese Medical Journal, 134, 1261–1263. [Google Scholar] [PubMed]
53. Hu, J., Shen, L., Sun, G. (2018). Squeeze-and-excitation networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7132–7141. Salt Lake City, UT, USA. [Google Scholar]
54. Krizhevsky, A., Sutskever, I., Hinton, G. E. (2017). Imagenet classification with deep convolutional neural networks. Communications of the ACM, 60(6), 84–90. https://doi.org/10.1145/3065386 [Google Scholar] [CrossRef]
55. Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K. Q. (2017). Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 4700–4708. Honolulu, HI, USA. [Google Scholar]
56. Simonyan, K., Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
57. Zheng, Z., Dong, X., Yao, J., Zhou, L., Ding, Y. et al. (2021). Identification of epileptic EEG signals through TSK transfer learning fuzzy system. Frontiers in Neuroscience, 15, 738268. https://doi.org/10.3389/fnins.2021.738268 [Google Scholar] [PubMed] [CrossRef]
58. Sultan, H., Owais, M., Park, C., Mahmood, T., Haider, A. et al. (2021). Artificial intelligence-based recognition of different types of shoulder implants in X-ray scans based on dense residual ensemble-network for personalized medicine. Journal of Personalized Medicine, 11(6), 482. https://doi.org/10.3390/jpm11060482 [Google Scholar] [PubMed] [CrossRef]
59. Uysal, F., Hardalaç, F., Peker, O., Tolunay, T., Tokgöz, N. (2021). Classification of shoulder X-ray images with deep learning ensemble models. Applied Sciences, 11(6), 2723. https://doi.org/10.3390/app11062723 [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF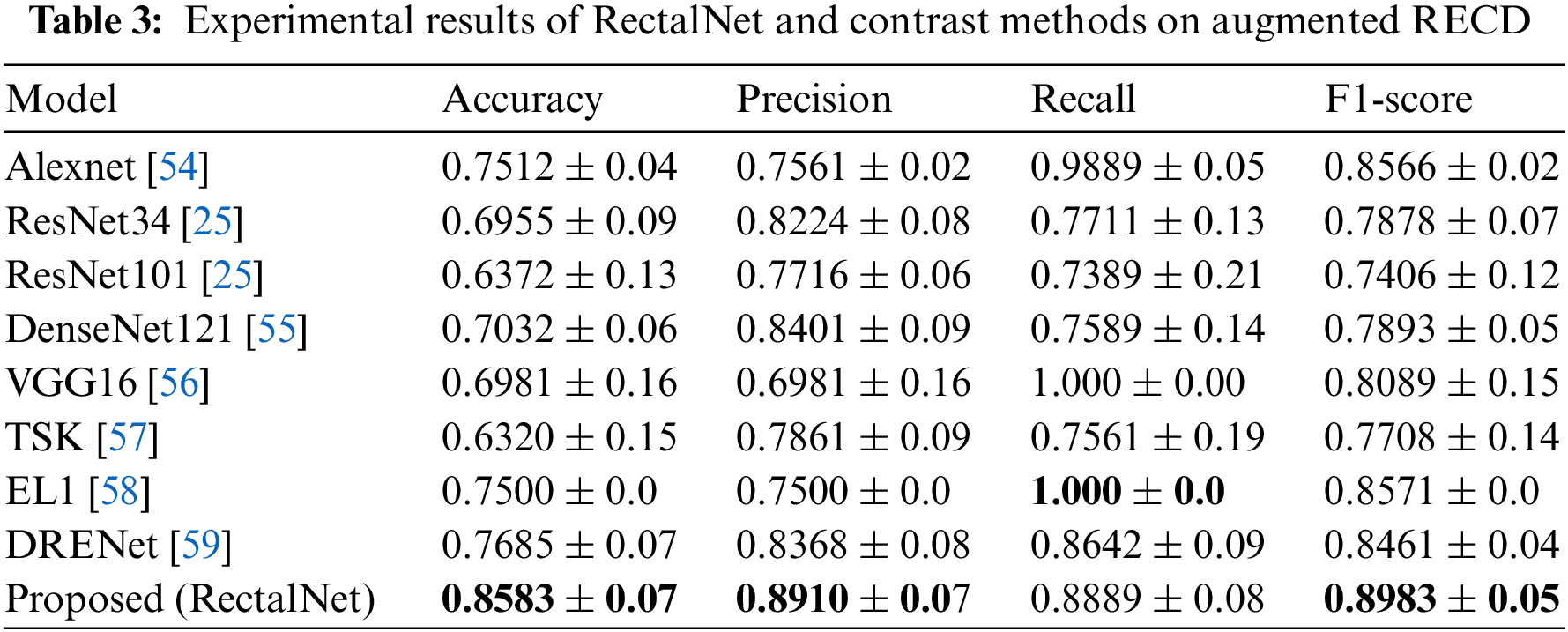
 Downloads
Downloads
 Citation Tools
Citation Tools