 Open Access
Open Access
ARTICLE
Enhanced Multimodal Physiological Signal Analysis for Pain Assessment Using Optimized Ensemble Deep Learning
1 Department of Computer Science, College of Computer and Information Sciences, Jouf University, Sakaka, 72388, Saudi Arabia
2 Faculty of Computing and Information, Al-Baha University, Al-Baha, 65528, Saudi Arabia
3 REGIM-Lab, National School of Engineers of Sfax, University of Sfax, Sfax, 3038, Tunisia
4 College of Computing and Information Technology, University of Bisha, Bisha, 61922, Saudi Arabia
5 Cybersecurity Department, College of Engineering and Information Technology, Buraydah Private Colleges, Buraydah, 51418, Saudi Arabia
6 Department of Computer Science, Prince Sattam bin Abdulaziz University, Al-Kharj, 11942, Saudi Arabia
* Corresponding Author: Olfa Hrizi. Email:
(This article belongs to the Special Issue: Artificial Intelligence Models in Healthcare: Challenges, Methods, and Applications)
Computer Modeling in Engineering & Sciences 2025, 143(2), 2459-2489. https://doi.org/10.32604/cmes.2025.065817
Received 22 March 2025; Accepted 30 April 2025; Issue published 30 May 2025
Abstract
The potential applications of multimodal physiological signals in healthcare, pain monitoring, and clinical decision support systems have garnered significant attention in biomedical research. Subjective self-reporting is the foundation of conventional pain assessment methods, which may be unreliable. Deep learning is a promising alternative to resolve this limitation through automated pain classification. This paper proposes an ensemble deep-learning framework for pain assessment. The framework makes use of features collected from electromyography (EMG), skin conductance level (SCL), and electrocardiography (ECG) signals. We integrate Convolutional Neural Networks (CNN), Long Short-Term Memory Networks (LSTM), Bidirectional Gated Recurrent Units (BiGRU), and Deep Neural Networks (DNN) models. We then aggregate their predictions using a weighted averaging ensemble technique to increase the classification’s robustness. To improve computing efficiency and remove redundant features, we use Particle Swarm Optimization (PSO) for feature selection. This enables us to reduce the features’ dimensionality without sacrificing the classification’s accuracy. With improved accuracy, precision, recall, and F1-score across all pain levels, the experimental results show that the suggested ensemble model performs better than individual deep learning classifiers. In our experiments, the suggested model achieved over 98% accuracy, suggesting promising automated pain assessment performance. However, due to differences in validation protocols, comparisons with previous studies are still limited. Combining deep learning and feature selection techniques significantly improves model generalization, reducing overfitting and enhancing classification performance. The evaluation was conducted using the BioVid Heat Pain Dataset, confirming the model’s effectiveness in distinguishing between different pain intensity levels.Keywords
Experiencing pain is a complicated and multidimensional experience that includes sensory, emotional, cognitive, and social elements. From a biological point of view, it is an essential defensive system that notifies the body of any possible or actual tissue injury. A person’s psychological condition, cultural background, and previous experiences all shape their impression of it, which is subjective and can vary significantly from person to person. By the definition provided by the International Association for the Study of Pain (IASP), pain is described as “an unpleasant sensory and emotional experience associated with, or resembling, that associated with actual or potential tissue damage.” This formulation brings to the forefront the physiological and emotional aspects of pain. Acute pain and chronic pain are the two primary types of pain that people experience. The onset of acute pain is often rapid and lasts relatively short. Acute pain is a warning signal that indicates an injury or illness. It often disappears when the underlying cause begins to heal. On the other hand, chronic pain is different from acute pain in that it lasts beyond the typical recovery period, typically more than twelve weeks, and can develop into a chronic condition. Chronic pain is challenging, which can lead to a decrease in quality of life, higher medical costs, and notable social and economic consequences. Among other things, it is related to a wide range of comorbidities, including anxiety, sadness, insomnia, and reduced mobility. According to the findings of epidemiological studies, chronic pain is one of the leading causes of disability in the world and affects a large fraction of the world’s population.
Clinical practice requires a practical evaluation of pain for diagnosis, monitoring, and determining the effectiveness of treatment. However, the assessment of pain involves several obstacles, particularly in those who cannot communicate adequately, such as babies, patients with cognitive impairment, or those who are in critical care settings. Currently regarded as the gold standard for evaluating pain, self-report measures include the Visual Analog Scale (VAS), the Numeric Rating Scale (NRS), and other questionnaire-based tools. Though helpful, these techniques are naturally subjective and might not sufficiently represent the intensity or character of the pain felt. In circumstances like these, experts usually decide whether or not a patient is in pain based on physiological indicators and observational cues. An extensive study has been done on the use of behavioral indicators as substitute signals for pain. These behavioral indicators include facial expressions, vocalizations (like moaning and sobbing), body posture, and motor responses. Like the previous example, physiological signals—such as heart rate, skin conductance, muscle tension (measured by electromyography), and breathing rate—provide objective markers showing the body’s autonomic and somatic reactions to painful stimuli. Still, interpreting these signals remains challenging because of the inter-individual heterogeneity and the impact of external factors like stress, weariness, or medication. The complexity and evaluation of pain have led to a substantial amount of research on automated pain detection systems that make use of current advances in artificial intelligence (AI) and machine learning (ML) [1,2]. These systems examine behavioral and physiological data using computer models to increase the objectivity, repeatability, and scalability of pain assessment. Deep learning, a subfield of artificial intelligence, has been essential to this subject. DL methods, including convolutional neural networks (CNNs) and recurrent neural networks (RNNs), which feature long-short-term memory (LSTM) and bidirectional gated recurrent units (BiGRUs), have demonstrated the possibility of learning temporal and spatial patterns connected to pain. These methods can extract hierarchical characteristics from raw data and simulate complicated, non-linear interactions. These methods can extract hierarchical characteristics from raw data and simulate complex, non-linear interactions. When used in the framework of physiological signal analysis, electromyography (EMG), electrocardiogram (ECG), and skin conductance level (SCL) are beneficial. Electromyography (EMG) shows the muscle activation and tension, which may change in response to pain. SCL shows variations in sweat gland activity, usually linked to mental and physical stress. On the other hand, electrocardiograms measure heart rate variation, a fundamental sign of the autonomic nervous system activity. It is feasible to gather these signals non-invasively and then examine them to find minor physiological changes linked to the feeling of pain. Though artificial intelligence is improving at evaluating pain, many challenges still exist. A significant issue has to be resolved given the high dimensionality and redundancy of the extracted characteristics, which can lead to overfitting and higher computational expenses. Feature selection is crucial to improve the model’s performance and render it more understandable. There have been times when metaheuristic optimization techniques, such as Particle Swarm Optimization (PSO), have been effectively used to select the most relevant features from large datasets, reducing the noise, and enhancing the classification accuracy. Using ensemble learning, which integrates the strengths of different learning models, classification task robustness and generalization have also been enhanced. Weighted ensemble methods, in particular, let individual model outputs distinguish their contributions to the final decision depending on their performance, so improving the general accuracy of the system.
This study presents a multimodal framework for automatic pain assessment that integrates deep learning models with optimization-based feature selection. Specifically, we propose a pipeline that processes EMG, ECG, and SCL signals through an ensemble of CNN, LSTM, BiGRU, and Deep Neural Network (DNN) architectures. We apply particle swarm optimization (PSO) to feature selection to improve efficiency and reduce dimensionality. A weighted averaging strategy combines the outputs of the individual models, leading to a robust and accurate pain classification system. The primary contributions of this work are: (1) the design of a multimodal framework based on deep learning for pain classification using physiological signals; (2) the application of PSO for optimal feature selection to improve model efficiency; (3) the integration of a weighted ensemble learning strategy to improve classification robustness; and (4) the experimental validation of the proposed method on the BioVid Heat Pain dataset, which demonstrates superior performance compared to baseline models.
Our paper consists of four main sections. Section 1: Related work reviews previous studies on pain assessment using machine learning and deep learning, highlighting different classification models and feature selection techniques. Section 2: The proposed approach to pain assessment describes our methodology. In Section 3: Results and Discussions, we discuss the experimental evaluation that used the BioVid Heat Pain Dataset to compare how well individual models worked and how well the proposed ensemble approach worked. Finally, Section 4: Conclusion highlights the benefits of the suggested approach and potential future developments in automated pain assessment by summarizing our results.
2 Related Work for Pain Assessment
Many studies have been conducted on classification using deep learning and machine learning for pain assessment. To increase accuracy, several researchers have looked at deep learning architectures, ensemble approaches, and traditional machine learning techniques [3]. This section examines current pain assessment techniques, emphasizing their benefits, limitations, and advancements.
2.1 Traditional Machine Learning Approaches for Pain Assessment
Conventional machine learning models are widely used in pain assessment because of their capacity to identify crucial patterns from behavioral and physiological data. In [4], the researchers looked at how electrocardiographic features (ECG) might reveal pain following surgery. They discovered that features of manually extracted ECG signals might serve as reliable indicators of pain. The authors of [5] investigated gradient-boosting models and random forests for pain recognition. They found that boosting methods like XGBoost produce better classification results when they used continuous error reduction.
Blood volume pulse (BVP) signals can be used with machine learning to accurately measure pain levels, as shown by Khan et al. [6] and Pouromran et al. [7]. Khan et al. [6] recorded 22 healthy people’s electrically stimulated BVP signals. This study classified (no pain vs. high pain) with 96.6% accuracy using artificial neural networks (ANN). The classification of (no pain vs. low pain) achieved an accuracy of 83.3% using the AdaBoost classifier. The multiclass experiment classified without low or high pain using artificial neural networks (ANN) and had a 69% overall accuracy. Furthermore, researchers in [8] validated their findings by demonstrating that BVP signals can precisely detect pain levels. The researchers carefully examined different physiological signals to measure pain intensity in a study [9] and found that EDA was the most helpful signal to measure pain intensity over time. A different study [10] examined the EDA signals in detail to determine how to measure pain. It suggested that EDA’s sympathetic responses are more related to stimulus pain than perceived pain.
Chu et al. [11] concentrated on feature processing due to the novelty of the dataset. After the features were extracted from the raw signals, a genetic algorithm (GA) eliminated the features that did not matter. Next, they applied Principal Component Analysis (PCA) to transform the features into a space devoid of linear relationships. They tested three different ways to classify the data: linear discriminant analysis (LDA), the K-nearest neighbor (KNN) algorithm, and the support vector machine (SVM). These were used on single-signal, multi-signal, multi-subject, and multi-day datasets.
Kächele et al. [12] presented the personalization of pain intensity recognition systems through several strategies utilizing various sources of information. The underlying concept of penalization is to assess pain levels by identifying the most analogous individuals in the training set to the new test subject. Methods for identifying commonalities among topics are categorized into three primary types. Meta-information refers to general data regarding attributes such as age and gender. Secondly, distance-based measurements include the proposed K-Nearest-Neighbor and Hausdorff distance techniques. Ultimately, the machine learning-based method categorizes this group into metrics based on supervised learning, metrics based on unsupervised machine learning, and proxy categorization.
In a recent study, Naseri et al. [13] developed a Natural Language Processing (NLP) pipeline in clinical notes to extract and find pain that physicians automatically reported, even when not recorded by structured data input. The NLP pipeline achieved an 85% F1 score in detecting pain from clinical notes on radiation oncology. Numerous research studies utilize comprehensive electronic medical records to improve clinical evaluations by applying natural language processing. A survey by Dave et al. [14] developed an NLP pipeline to extract nine pain-related factors from electronic medical records. Parameters encompass location, onset, quality, amount, severity, radiation, intervention, prior treatment, and frequency. The model created by the author in [14] achieved a 90% F1 score in assessing the severity of pain in medical records.
A study by Bartlett et al. [15] looked at support vector machines (SVM) with linear and RBF kernels and Adaboost classifiers (with a maximum of 200 characteristics selected per AU) to classify facial actions across 20 AUs. The RU-FACS (spontaneous) and CK (posed) datasets provided the Gabor wavelet features. Chew et al. [16] use a linear support vector machine (SVM) to compare pixel-based features and more complex appearance features (HOG, Gabor Magnitudes) from several datasets (CK+, M3, UNBC-McMaster, and GEMEP-FERA), each with a different level of face alignment accuracy. According to the experiment results, more elaborate visual descriptors are resistant to alignment mistakes in the AU identification; nevertheless, the benefits of these descriptors are limited when the alignment is virtually perfect.
Ding et al. [17] proposed a multi-stage framework for facial Action Unit (AU) detection using a Cascade of Tasks (COT) architecture. They performed AU detection at the frame, segment, and transition levels, integrating the outputs sequentially. Frame-level detection, trained via Support Vector Machines (SVM) trained on Scale-Invariant Feature Transform (SIFT) features, enhanced the quality of segment-level data. A weighted-margin SVM emphasized segments with more positive frame-level responses for segment detection, while a bag of words with geometric features represented segments. The transition detectors were then trained to refine the onset and offset boundaries of the AU segments. Finally, scores from segment and transition detectors were pooled linearly to identify AU events, with dynamic programming and branch-and-bound techniques used for temporal optimization within video sequences.
2.2 Deep Learning Models for Pain Assessment
Deep learning techniques have greatly improved pain assessment [18]. These techniques involve automatically developing hierarchical representations from raw physiological signals. Gkikas et al. [1] propose a systematic review of deep learning techniques for automatic pain assessment, highlighting the role of multimodal and temporal data. The authors evaluated 110 studies and showed that combining facial, physiological, and behavioral modalities improves pain detection. They emphasize that deep learning models, particularly CNNs and LSTMs, outperform traditional methods in clinical settings. Their review also discusses dataset limitations, validation protocols, and the importance of explainability. This paper lays the foundation for creating robust real-time pain identification systems for use in healthcare. In the same context of pain assessment, Gkikas et al. [3] propose a multimodal transformer-based framework for automatic evaluation of acute pain using facial videos and heart rate signals. Their architecture combines four components: a Spatial Module for video embeddings, a Heart Rate Encoder, an AugmNet for latent space augmentation, and a Temporal Module for final assessment. Feature extraction is improved by pretraining on tasks such as face and emotion recognition. Experiments on the BioVid dataset show state-of-the-art performance, with 82.74% accuracy for binary and 39.77% for multi-level pain classification. The research emphasizes the efficiency of quick pain detection by combining behavioral and physiological data. Walter et al. [19] trained a deep convolutional neural network (CNN) model on multimodal datasets to improve the accuracy of pain categorization. The paper’s authors, who examined multilayer perceptrons (MLP), sought to classify pain using biosignals. The writers showed notable gains.
Furthermore, the study authors in [20,21] applied long-short-term memory (LSTM) networks to characterize temporal relationships in pain expression, outperforming static classification tools.
The author constructed a hybrid deep learning model in [22]. This model combines the benefits of LSTM, an algorithm for capturing temporal dynamics, with those of CNN, an algorithm for extracting spatial features. The signals were then classified into “pain” or “no pain” categories by processing the recovered traits with a Fully Connected Layer (FCL).
Using graph neural networks (GNNs) in [23], the author showed better accuracy than spontaneous cases while setting new standards by using them creatively to assess pain scenarios. The author developed the Cross-Stream Attention (CSA) mechanism in 2023 in the publication [24]. This mechanism was used in conjunction with the DFEPN dataset, which resulted in a considerable improvement. The mechanism attained an accuracy of 66.20 percent when applied to challenging tasks such as recognizing pain in newborns.
The authors of [25] built a complete pain monitoring network using a custom data set and special acquisition equipment. This network reached at least the same level of accuracy as clinical specialists, while simultaneously simplifying the workload of medical personnel. It surpasses current models in accuracy by using adaptive learning to handle seven different pain intensity levels. This suggests that CNNs are promising for clinical pain assessment. In their study published in [26], Talaat et al. developed a technology that utilizes facial expressions and deep learning to provide a real-time emotion recognition system for children with autism. The method has an accuracy rate of 95.23 percent, potentially improving your quality of life.
Ghosh et al. showed in their paper [27] a good way to determine how people feel by using the 2D Face-Set database of the UNBC-McMaster shoulder pain dataset with Pain expression. Their method combined advanced techniques, including the Histogram of Oriented Gradients (HoG), Local Binary Patterns (LBP), and Convolutional Neural Networks (CNN), to accurately detect pain-related expressions. This system makes use of cutting-edge methodologies to detect discomfort. CNN and bidirectional long- and short-term memory (BiLSTM) were utilized by Wu et al. in their study with video data from critically ill patients, which was published in [28]. Real-time pain recognition was performed by Pikulkaew et al. in their work published in [29]. The researchers used Deep CNN to analyze the UNBC-McMaster Shoulder Pain Expression Archive and JAFFE databases.
Ismail and Waseem proposed implementing patient-centric pain treatment [30]. This implementation includes two approaches: CNN and LSTM. They accomplish this using the UNBC-McMaster Shoulder Pain Expression Database. Their goal was to implement patient-centered pain treatment. The goal of Kumar et al. [31] was to make it possible for computers to automatically detect pain based on facial expressions. They did this using machine learning on the UNBC-McMaster Shoulder Pain Expression Database, the BUCC Facial Pain Image Database, and the FFP-Pain Database. However, Alghamdi et al. [32] obtained amazingly high accuracy rates in their study, using two CNNs to automatically evaluate pains that were recorded in the UNBC-McMaster Shoulder Pain Expression Databases.
Using the UNBC-McMaster Shoulder Pain Expression Archive Dataset and the BioVid Heat Pain Dataset, Morabit and Rivenq researched transformers and distillation-based facial expression analysis for pain identification. Their findings were published in [33]. A deep graph neural network, referred to as D-GNN, was proposed by Patania et al. in their paper titled “patania2022deep.” This network is designed to perform video-based assessment of facial pain expression and uses the FACS pain expression data set. In their study, Rathee et al. [34] used a model-agnostic meta-learning technique in conjunction with deep learning pre-training on the UNBC-McMaster Shoulder Pain Expression Dataset.
Another study aimed to identify the prevalence of pain upon arrival in the Emergency Department by applying a clinical text deep learning model to unstructured nursing assessments stored in electronic health records [35]. The model was proven to have an accuracy of 93.2% on average.
A recent study on the evaluation of low back pain (LBP) proposed a unique hybrid deep learning framework called GLEAM (GAN-convolution-self-attention-Enhanced Temporal LSTM) to categorize the severity of LBP using surface electromyography (sEMG) and electroencephalography (EEG) signals [36]. The model uses a CNN to extract localized temporal features, a denoising GAN to improve signal quality, and a self-attention mechanism to catch global dependencies. Using an ETLSTM architecture, the last classification is done with a detection accuracy of 98.95% over four LBP intensity levels. This study emphasizes the possibility of using sophisticated deep learning architectures and multimodal physiological signals to assess pain intensity consistently. It provides a valuable guide for creating intense, non-invasive pain-measuring tools.
The insufficient size of pain-oriented datasets poses a significant challenge to effectively train a deep neural network for the Automatic Pain Detection (APD) problem. Egede et al. [37] solved this problem using the UNBC-McMaster data set and hand-crafted and deep-learned features together to measure pain intensity as a multimodal fusion problem that was designed for small sample settings. They took deep learning features from two CNN models already trained for the AU detection task on the BP4D dataset [38]. These features came from the eye and mouth regions of the face, respectively.
Despite promising advances in pain assessment using machine learning and deep learning, several limitations remain in existing approaches. Many studies rely on single-modal data, which can reduce robustness in real-world applications due to the limited representation of physiological responses. In addition, hand-made feature extraction methods and static models often struggle with generalization across various topics and pain intensities. Some techniques also lack efficient feature selection techniques, which leads to high computational expenses and superfluous input data. Moreover, model ensembles are underexplored; they usually lack the best integrating strategies that balance prediction strengths among models.
Moreover, model ensembles are underexplored; when applied, they frequently lack optimal integration strategies balancing prediction strengths among models. The proposed approach addresses these deficiencies by using multimodal physiological signals—EMG, ECG, and SCL—to capture complementary components of the pain reaction. A weighted averaging ensemble improves performance and includes automatic feature learning via several deep architectures (CNN, LSTM, BiGRU, and DNN). Furthermore, it uses particle swarm optimization (PSO) to select features, significantly reducing redundancy and increasing computational efficiency. These elements create a strong, scalable, precise system for detecting objective pain levels.
This study presents a multimodal pain assessment system that combines deep learning models and feature selection to enhance classification accuracy. Feature extraction processes EMG, SCL, and ECG; an ensemble of CNN, LSTM, BiGRU, and DNN models classifies them. By choosing the most relevant features, Particle Swarm Optimization (PSO) reduces feature redundancy, improving efficiency. For resilient categorization, a weighted average ensemble method combines model predictions as shown in Algorithm 1. The proposed method allows for a precise automatic pain assessment. Fig. 1 presents all the steps of our approach.

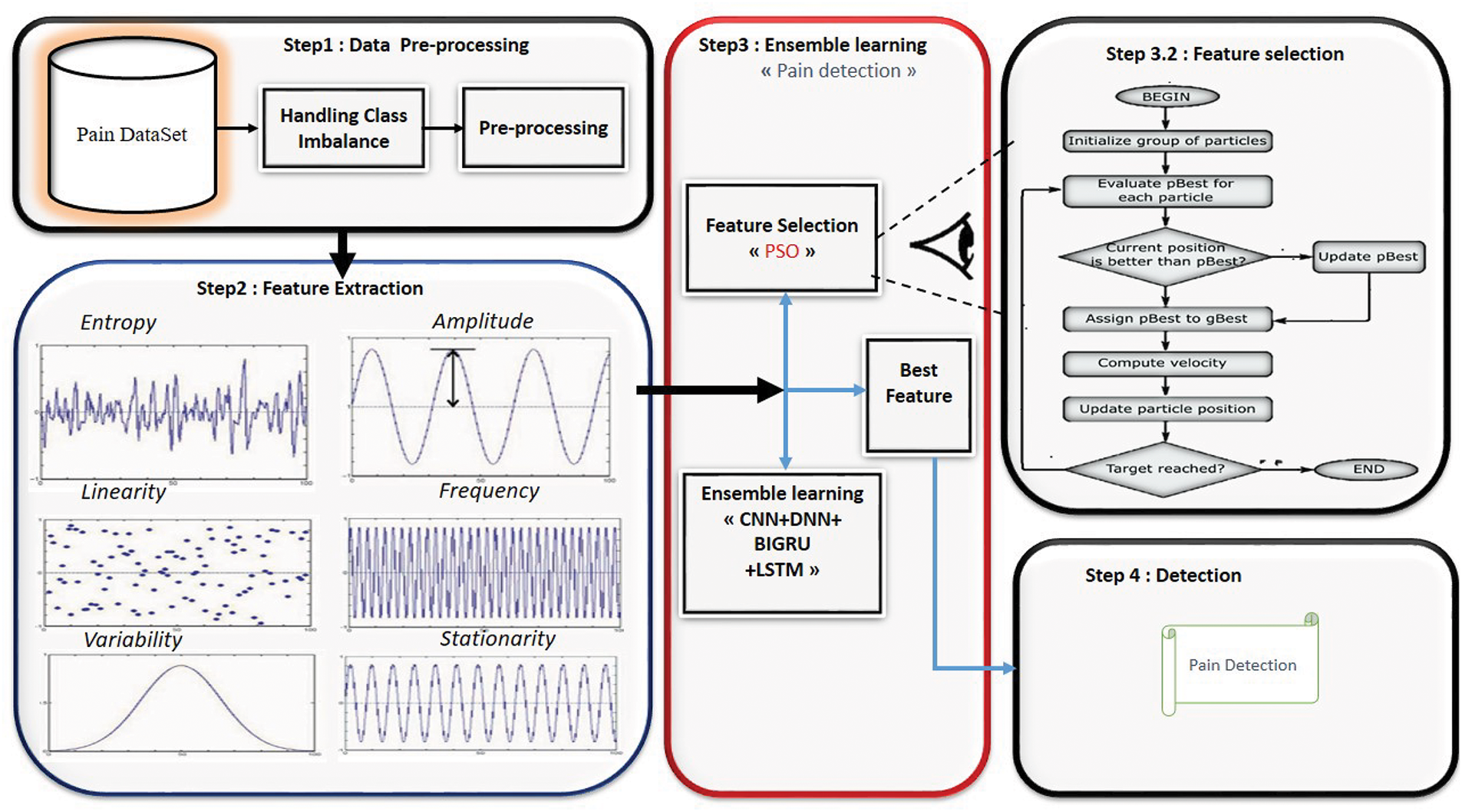
Figure 1: Proposed model for pain assessment
This study uses a data set derived from the BioVid Heat Pain Database [39]1. This database was developed to capture different levels of pain through the use of controlled stimulation. This study focuses primarily on biopotential signals (EMG, ECG, and SCL), although the dataset also includes behavioral data such as facial videos, which are not utilized in the proposed approach. The BioVid Heat Pain dataset used in this study offers several essential features: it involves highly controlled pain stimulation procedures and provides synchronized multimodal physiological signals from 87 participants, with each subject contributing 20 samples per pain level across five pain classes, resulting in a balanced dataset of 8600. While the dataset provides the raw biosignals, our study performs systematic feature extraction based on six statistical domains—amplitude, frequency, stationarity, entropy, linearity, and variability—generating 135 distinct features. These extracted features are detailed in Table 1, and form the input to the proposed classification framework.

Electromyography (EMG), skin conductance level (SCL), and electrocardiography (ECG) are the physiological signals that are examined, among other physiological signals. EMG is a method that monitors muscle activity and is associated with the activation of the sympathetic nervous system. Sensors were implanted in the muscles of the zygomaticus, corrugator, and trapezius, which are the muscles that react when they are in pain. During frowning, the corrugator supercilii muscle contracts, the zygomaticus major muscle elevates the corners of the mouth, and the activity of the trapezius muscle signals that the individual is experiencing significant amounts of stress. The SCL, which is recorded by electrodes placed on the index and ring fingers, reflects electrodermal activity, which is exclusively regulated by the sympathetic nervous system. Alterations in the SCL that occur suddenly and last one to three seconds indicate responses to stress, deep breathing, or emotional arousal. Heart rate (HR), interbeat interval (IBI), and heart rate variability (HRV) are some of the essential parameters that are provided by electrocardiograms (ECGs), which are recorded by placing electrodes on the upper right and lower left sides of the body. As a result of its widespread application in the evaluation of mental exertion and stress, HRV is an essential component for the detection of pain.
Although Table 1 presents a selection of representative characteristics, the complete set includes 135 distinct characteristics extracted in three physiological modalities: EMG, ECG, and SCL. These features are derived from six statistical domains—amplitude, frequency, stationarity, entropy, linearity, and variability. Specifically, each signal modality contributes 45 features, bringing the total to 135. The features listed in Table 1 highlight commonly used descriptors such as peak, RMS, and entropy measures, which are applied consistently across the three modalities. A complete list of all features extracted per modality is available upon request.
The classification step in our pain assessment system uses deep learning models to go through a file that already has features from electromyography (EMG) Zygomaticus, EMG Corrugator, EMG Trapezius, ECG, and EDA/SCL/GSR that have already been calculated. They use advanced architectures such as CNN, LSTM, DNN, and BiGRU structures. The following sections will discuss the role of each classifier, mathematical formulation, and pain intensity classification.
3.3.1 Convolutional Neural Network (CNN) for Feature Correlation Learning
Although convolutional neural networks (CNNs) are most commonly associated with image processing, they can be effectively adapted to handle structured tabular or sequential physiological data using a 1D CNN architecture. In our approach, we employ a 1D CNN to learn temporal or spatial correlations between extracted physiological features (e.g., EMG, ECG, SCL) organized as a one-dimensional sequence.
The 1D convolution operation is defined as:
where:
•
•
•
•
Following the convolutional layer, we apply the ReLU activation function:
To reduce the dimensionality and enhance the model’s generalization capability, we apply a 1D pooling operation (e.g., max pooling). This helps to capture the most salient features while reducing computational complexity.
3.3.2 Long Short-Term Memory (LSTM) for Sequential Feature Learning
LSTM networks specialize in time-dependent data analysis, making them excellent for assessing pain from physiological signals. Some extracted features preserve sequential aspects, such as the characteristics of the ECG and EMG time domains. LSTM gates are defined as:
where the model’s generalization capability is:
•
•
•
•
•
•
•
•
•
•
•
•
3.3.3 Deep Neural Network (DNN) for High-Dimensional Classification
Deep neural networks (DNNs), composed of multiple fully connected (dense) layers, are used to model high-dimensional relationships between extracted pain-related features. Each layer in the network transforms the input space through a learned linear mapping followed by a nonlinear activation function. This process allows the model to capture complex patterns and interactions among features.
The transformation at each layer is defined as:
where:
•
•
•
•
For binary pain classification, the final Softmax function assigns class probabilities:
3.3.4 Bidirectional Gated Recurrent Unit (BiGRU) for Contextual Pain Learning
BiGRU is an optimized RNN variant that captures dependencies in both forward and backward directions, improving contextual pain classification.
The GRU (Gated Recurrent Unit) architecture simplifies the LSTM by using only the update and reset gates. The GRU equations are defined as follows:
where:
•
•
•
•
•
•
•
•
•
•
3.3.5 Ensemble Learning for Pain Assessment Using Weighted Averaging
A good approach is ensemble learning, which combines several classifiers to increase the general predictive accuracy. This work improves pain assessment capacity by combining CNN, LSTM, BiGRU, and DNN models with a weighted averaging approach. Ensemble learning does not depend on only one model to improve and produce more consistent outcomes when classifying items. Instead, it combines the advantages of many models.
Convolutional neural networks (CNN) identify patterns in the classification process; long-short-term memory (LSTM) provides sequential learning; deep neural networks (DNN) support high-dimensional decision-making; and bidirectional feature dependency learning (BiGRU) guarantees a strong and accurate pain classification. Based on physiological signals, this hybrid deep learning technique best serves automated pain assessment.
The weighted averaging technique gives each classifier a different importance depending on its performance. Given the prediction probabilities of the classifiers, the final ensemble prediction is computed as follows:
where
The weights are adjusted to reflect each model’s particular performance criteria, including accuracy, precision, and recall, ensuring that models with better performance have more influence on the final forecast.
The ensemble learning approach increases the dependability of pain assessment by combining deep learning models, overcoming the constraints of individual classifiers, and enhancing the robustness of the whole classification system. Each model in the ensemble—CNN, LSTM, BiGRU, and DNN—contributes uniquely depending on the signal characteristics it processes. Every model in the ensemble—CNN, LSTM, BiGRU, and DNN—contributes significantly depending on the signal characteristics it processes. While LSTM and BiGRU models are better for temporal dependencies, CNNs capture local spatial features better. The DNN adds generic non-linear decision boundaries. The weighted averaging strategy is designed to leverage their strengths dynamically. Empirically, BiGRU and CNN showed the highest individual accuracy, thus receiving higher ensemble weights.
3.4 Feature Selection Using Particle Swarm Optimization (PSO)
We can greatly improve classification performance and simplify the computation process by using a feature selection step based on the Particle Swarm Optimization (PSO) method. Because the dataset comprises 135 extracted characteristics, it is essential to select the subset that is the most pertinent to minimize redundancy and maximize classification accuracy.
The particle swarm optimization (PSO) algorithm is based on nature. It uses possible solutions (particles) moving through a multidimensional search space. The position of each particle is updated based on the best solution it has discovered for itself and the best solution that its neighbors have discovered. In the context of this investigation, the purpose of PSO is to identify the optimal subset of characteristics that maximizes classification accuracy while simultaneously limiting the number of features available for selection.
The fitness function used in PSO is formulated as follows:
where:
•
• N is the total number of iterations,
• |S| is the number of selected features,
• |F| is the total number of features (135),
•
At each iteration, the velocity and position of each particle are updated using:
where:
•
•
•
•
•
•
•
This step in feature selection guarantees that only the most pertinent physiological signal features are used in the last classification phase. Applying PSO helps us to find the most informative elements that help to classify pain. This increases the model’s performance and lowers the load of computational duty at the same time.
This part describes the results of our suggested pain assessment approach using different classification models and assessment situations. In the test, various machine learning and deep learning classifiers are used, and an ensemble learning approach is included to improve performance. Moreover, we use Particle Swarm Optimization (PSO) to hone feature selection and assess its effect on classification performance. Several measures, including accuracy, precision, recall, F1 score, Cohen’s Kappa, and Receiver Operating Characteristic Area Under the Curve (ROC-AUC), allow one to evaluate the effectiveness of each model. These measures provide a complete picture of how well the model can distinguish between pain levels of different intensity (L1, L2, L3, L4) and painless conditions (NP).
The following metrics are used to assess classification performance:
- Accuracy:
- Precision:
- Recall (Sensitivity):
- F1 Score:
- Cohen’s Kappa:
where
The findings are organized into four subsections.
• Pain assessment using machine learning: This subsection shows the performance of conventional machine learning classifiers-including decision trees, logistic regression, K-nearest neighbors (KNN), random forests, AdaBoost, and support vector machines (SVM)-and assesses their efficacy in pain evaluation.
• Deep Learning for Pain Assessment: This subsection examines deep neural networks (DNN), bidirectional gated recurrent units (BiGRU), long-short-term memory (LSTM), convolutional neural networks (CNN), and other deep learning models. It evaluates their capacity to acquire intricate patterns from physiological signals for pain categorization.
• Deep Learning Ensemble Learning for Pain Assessment: We use an ensemble learning technique that combines the CNN, LSTM, BiGRU, and DNN predictions to improve classification performance. Each model’s contribution is optimized using a weighted averaging technique, guaranteeing a more potent and generalized pain assessment tool.
• Feature Selection Using PSO for Pain Assessment: Feature Selection This section looks at how Particle Swarm Optimization (PSO) feature selection affects pain classification. We ran four experimental scenarios to assess how well PSO selects the most pertinent features while maintaining strong classification performance.
The comparative study of these situations reveals the need to choose the best characteristics to enhance classification accuracy and computational efficiency in pain assessment.
4.2 Validation Protocol and Study Limitations
In this work, we applied a hold-out validation technique in which the data set was randomly divided into 80% for training and 20% for testing. This division was done without subject separation; thus, data from the same subjects could have appeared in both the training and testing sets. Although this method simplifies the assessment, it increases the possibility of information leakage since the model could unknowingly learn subject-specific patterns repeated in both sets. The stated performance might therefore exaggerate the generalization capacity of the model. Consequently, the performance stated could exaggerate the generalizability of the model. We recognize this as a constraint of our present research. Initially intended for Leave-One-Subject-Out (LOSO) validation, the BioVid Heat Pain dataset guarantees subject independence and a more rigorous evaluation of generalizability. Future projects will seek to more accurately assess model performance in real-world environments where unseen subjects are anticipated using LOSO or other subject-independent validation techniques. Notwithstanding this constraint, the findings shown offer valuable analysis of the performance of our suggested multimodal ensemble framework and underline its promise when trained on appropriate data distributions. Notwithstanding this constraint, the findings shown offer valuable analysis of the performance of our suggested multimodal ensemble framework and underline its promise when trained on appropriate data distributions.
4.3 Performance Analysis of Machine Learning Models for Pain Classification
The dataset classifies pain by separating nonpain (NP) from various degrees of pain: L1, L2, L3, and L4, each level indicating an increase in pain intensity related to the temperature used during data collection. While NP vs. L2, NP vs. L3, and NP vs. L4 indicate higher pain levels, the classification NP vs. L1 defines the difference between nonpain and pain at level 1. In addition, the NP vs. P-all classification combines all pain levels, so there is a binary difference between non-pain and any degree of pain.
Taking into account different degrees of pain (L1, L2, L3, L4) and an aggregated scenario (P-all L), the findings in Table 2 offer a thorough assessment of several machine learning models for binary pain evaluation. Several key insights emerge from the results.
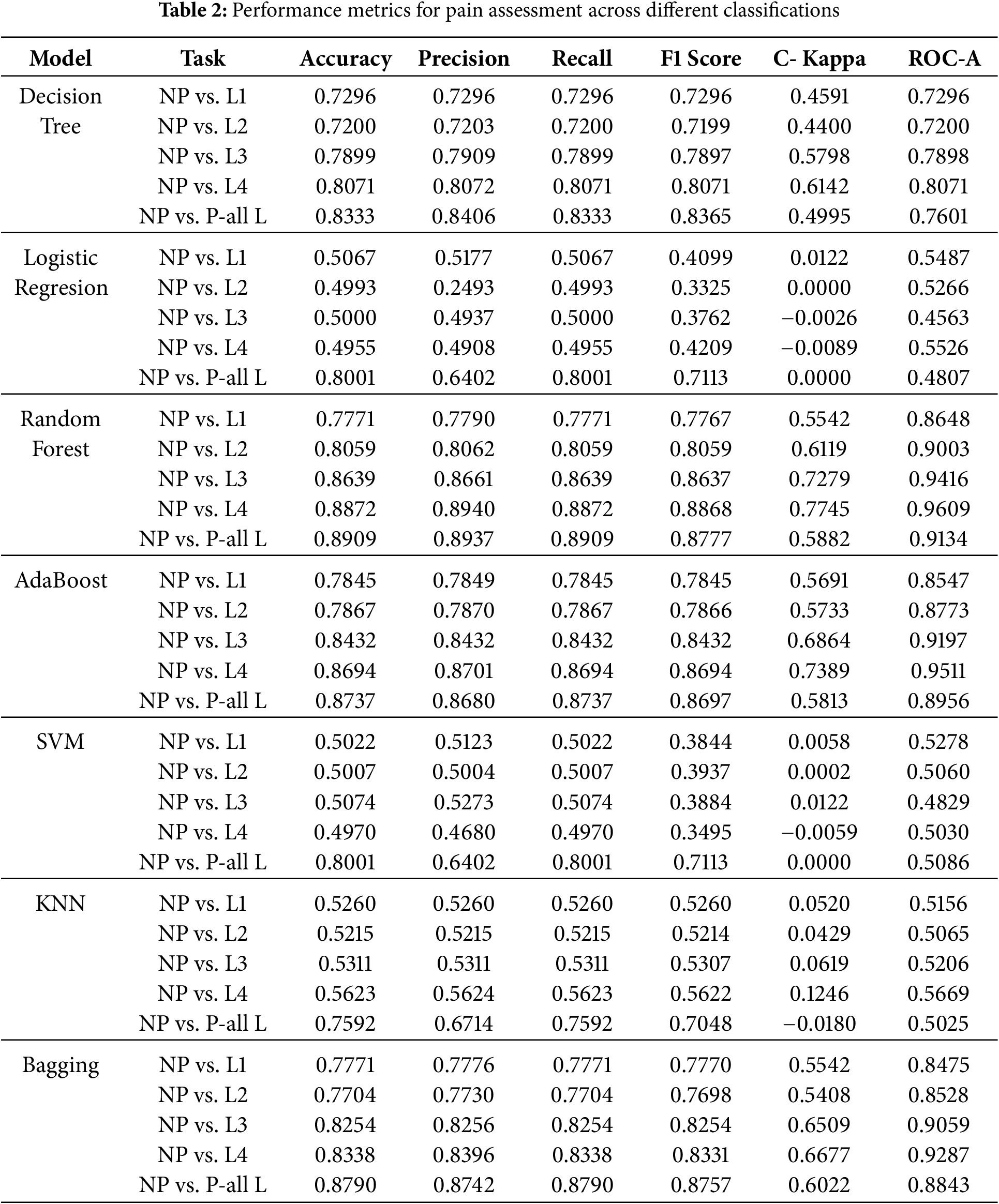
Performance Trends across Pain Levels:
Generally speaking, most models show improved accuracy, recall, and F1 scores as the intensity of pain increases from L1 to L4. This evidence indicates that higher pain levels are more straightforward to classify due to stronger physiological reactions caused by pain. This pattern is especially noticeable in Random Forest (0.8872 for NP vs. L4) and AdaBoost (0.8694 for NP vs. L4), the categorization methods that obtain the highest performance.
Best Performing Models:
Random Forest, AdaBoost, and Bagging are examples of ensemble learning models that outperform individual classifiers. These models demonstrate higher accuracy and ROC AUC scores across the board for all different levels of pain. Random Forest always does the best job, with an ROC AUC of 0.9609 for NP vs. L4. This shows that it can reliably find nonlinear relationships in physiological data. Random Forest is the best way to analyze physiological data.
Weak Performance of Traditional Models:
Neither logistic regression nor support vector machines (SVMs) work well. This is especially true when trying to tell the difference between lower pain levels (L1, L2), where they are only about fifty percent accurate. The result indicates that these models have difficulty dealing with complicated and overlapping variables when classifying pain.
KNN and Decision Tree Limitations:
KNN demonstrates moderate performance, with accuracy between 0.5215 (NP vs. L2) and 0.5623 (NP vs. L4), reflecting a constrained generalization for pain classification. Although the decision tree outperforms KNN, it presents lower Cohen Kappa values, indicating a lower reliability in its predictions.
Bagging and Overall Performance:
Bagging provides competitive results, but slightly underperforms compared to Random Forest and AdaBoost, demonstrating that simple ensemble techniques alone are insufficient for optimal classification.
Generalized Pain Classification (NP vs. P-all L):
When consolidating all pain levels into a singular category (NP vs. P-all L), the models exhibit a marginal decline in accuracy and ROC AUC, presumably attributable to increased class overlap and variability. However, ensemble models continue to represent the most effective methodologies. As shown in Fig. 2, the results underscore the efficacy of ensemble learning models (Random Forest, AdaBoost, and Bagging) in pain assessment, while conventional models, such as Logistic Regression and SVM, encounter difficulties in complex feature spaces. Improvement in performance at elevated pain levels indicates that more intense physiological responses are more easily classified. Future advancements may investigate hybrid deep learning methodologies and sophisticated feature selection techniques to enhance the precision of pain assessment further.
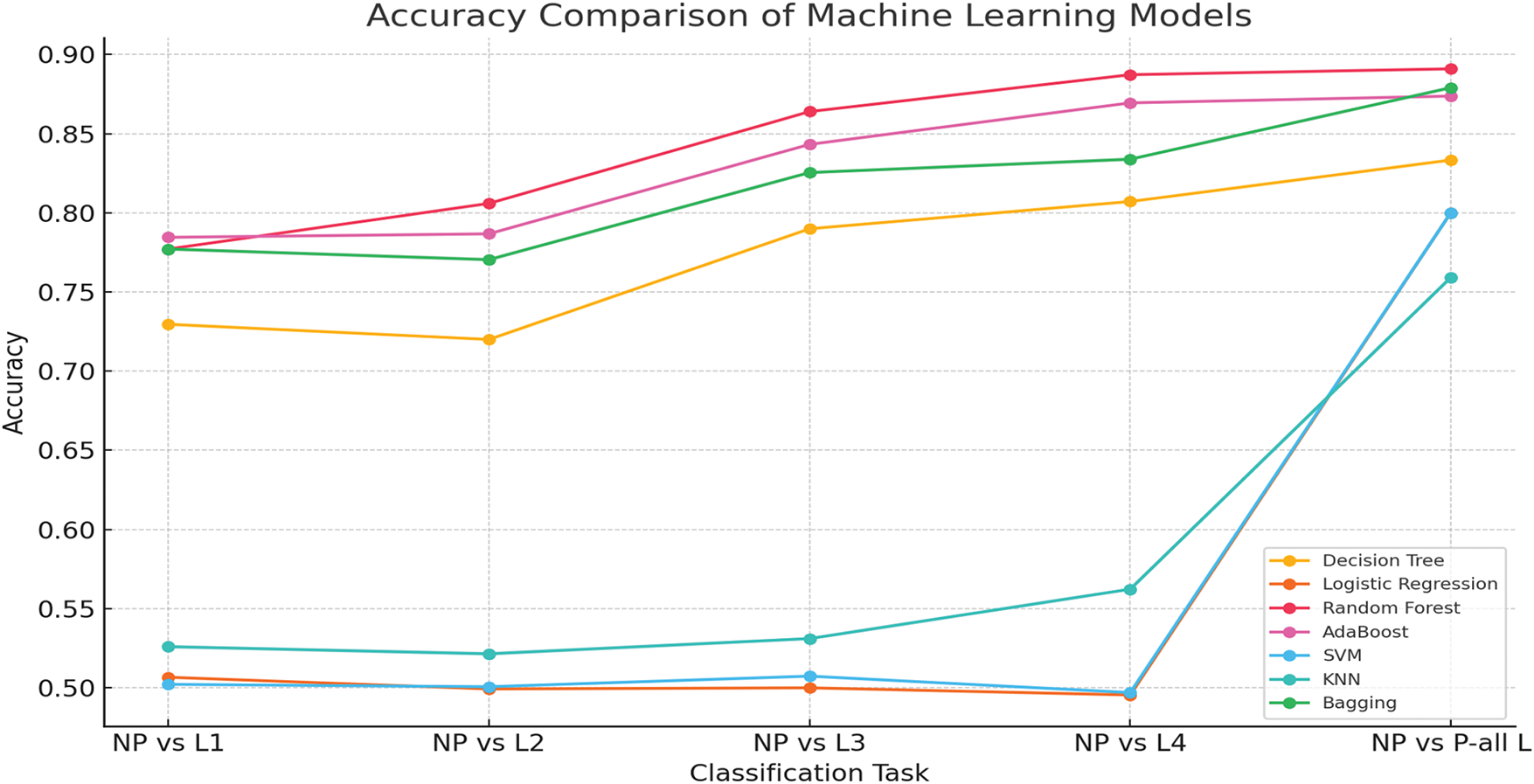
Figure 2: Accuracy comparison of machine learning models for pain assessment
4.4 Performance Analysis of Multi-Modal Feature Extraction for Pain Classification
We conducted a comparative analysis in five configurations to evaluate the effectiveness of each physiological modality and its combined impact. EMG Zygomaticus, EMG Corrugator, EMG Trapecius, ECG, and multimodal fusion of all signals. Each modality was tested using various deep learning models to ensure consistent benchmarking. The results are presented in Table 3 and reveal clear trends in the model’s performance.
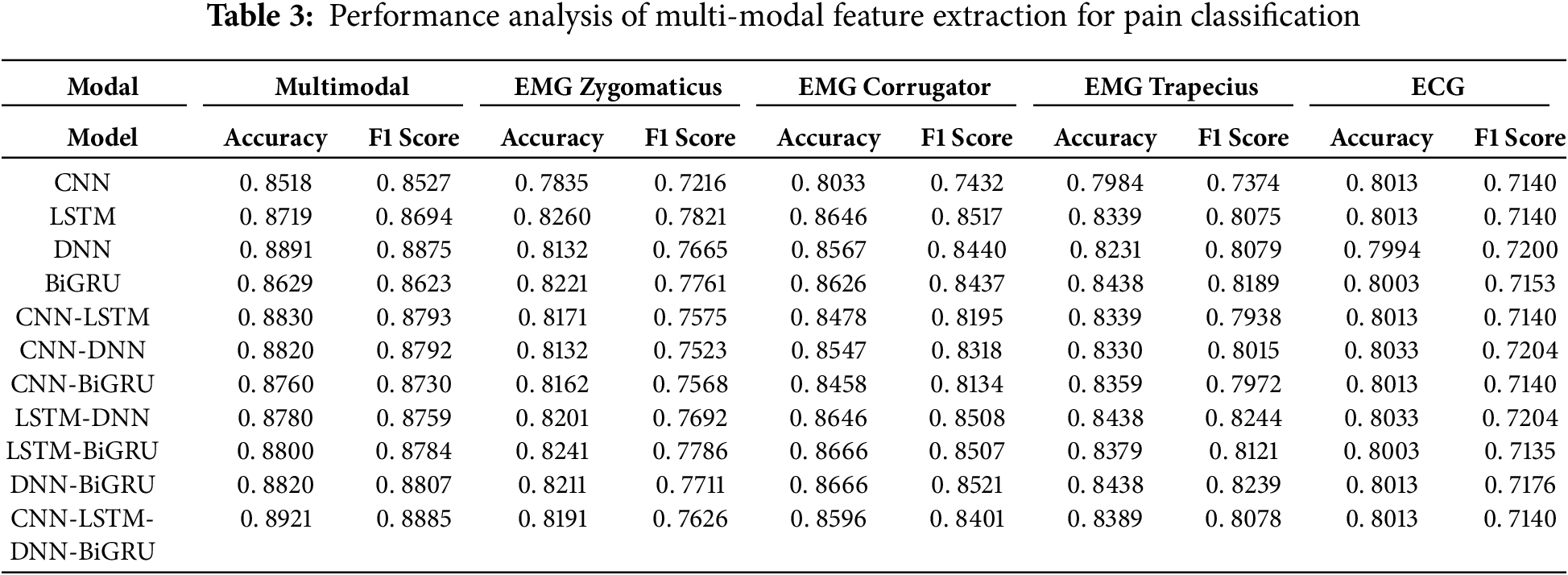
The comparative results show the superiority of the multimodal approach over individual modalities for pain assessment. Across all deep learning models—CNN, LSTM, DNN, BiGRU, and their combinations—the multimodal configuration consistently achieves the highest accuracy and F1-scores.
For instance, the ensemble model CNN-LSTM-DNN-BiGRU achieves an accuracy of 0.8921 and an F1-score of 0.8885 in the multimodal setup, significantly higher than the best scores obtained using any single channel alone. Specifically, when applied to the EMG Zygomaticus, EMG Corrugator, EMG Trapecius, or ECG channels individually, none of the models surpasses this multimodal performance, with the highest F1 score among single modalities reaching only 0.8522 for the corrugator channel using the DNN-BiGRU model.
This performance gain is attributed to the complementary nature of the physiological signals. Each modality captures a distinct aspect of the body’s response to pain: muscle tension, heart rate variability, skin conductance, and their integration provides a richer and more holistic representation of the pain experience. Ensemble deep learning models further amplify this effect by leveraging the strengths of different architectures, improving robustness and generalization.
In contrast, relying solely on a single channel limits the model’s ability to capture the complexity of physiological responses and increases susceptibility to noise or signal artifacts. Thus, the findings strongly support using multimodal fusion as a more reliable and accurate strategy for automatic pain recognition systems.
4.5 Performance Analysis of Deep Learning Models for Pain Classification
Deep learning models—CNN, LSTM, DNN, and BiGRU—were evaluated for pain assessment at different levels. Several key findings can be derived from Table 4:
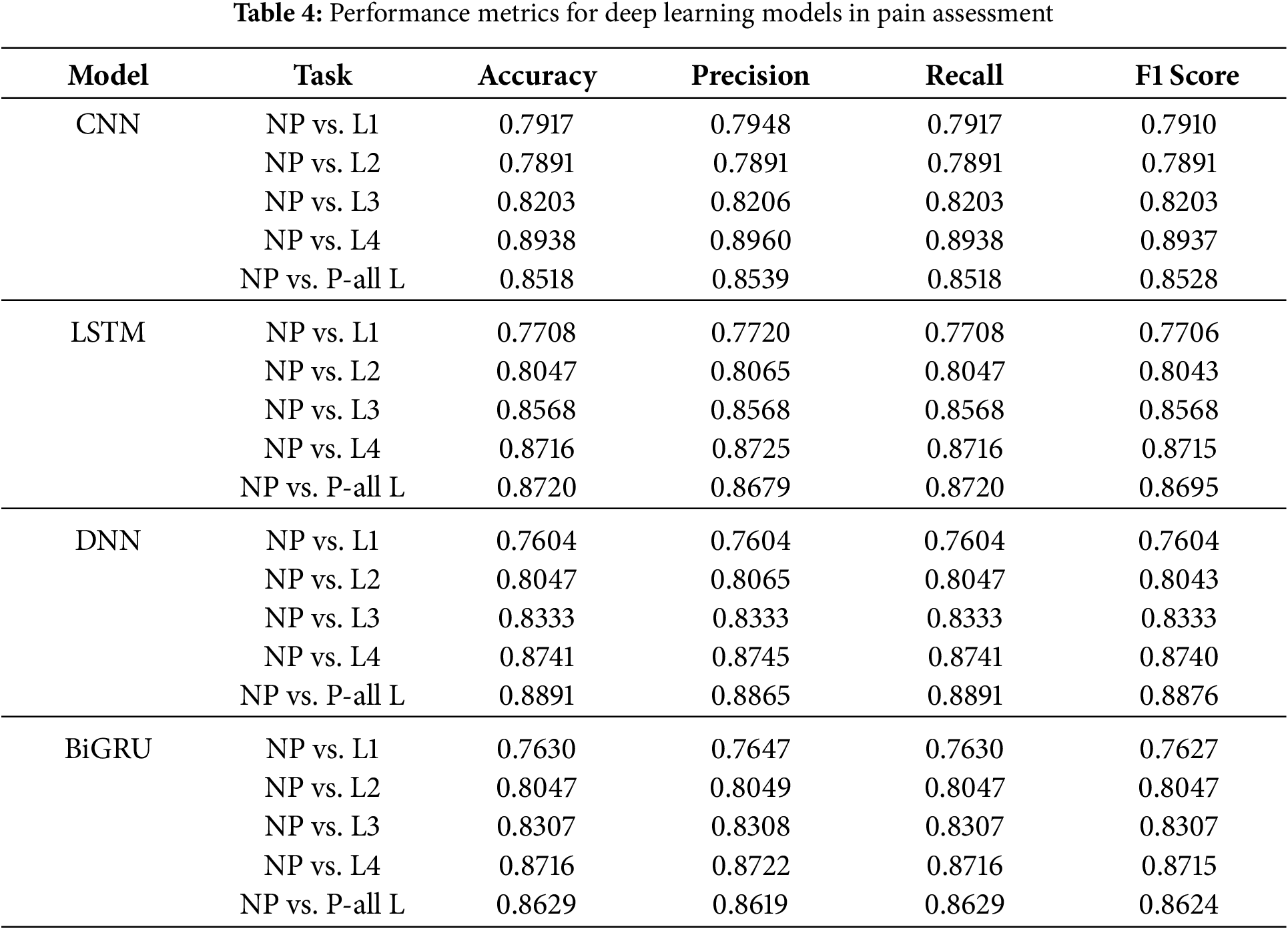
DNN Achieved the Highest Performance:
The deep Neural Network (DNN) model consistently outperformed the other models across all pain levels. It achieved the highest accuracy (0.8891 for NP vs. P-all L) and was the most accurate overall. It seems more likely that fully connected architectures can correctly detect complex feature correlations in physiological pain signals.
CNN vs. LSTM Performance:
CNN performed well compared to LSTM and BiGRU at intermediate pain levels (L2 and L3), but it performed somewhat worse than LSTM and BiGRU when compared to NP vs. L4 (0.8938 accuracy).
LSTM demonstrated a higher recall (0.8716 for NP compared to L4), highlighting the model’s effectiveness in identifying temporal connections in sequential physiological data.
BiGRU’s Competitive Performance:
The BiGRU model produced competitive results, particularly in the areas of NP vs. L3 (0.8307) and NP vs. L4 (0.8716).
Because it is bidirectional, it can account for both past and future dependencies in sequences. This makes it an excellent choice for classifying physiological data.
General Pain Assessment (NP vs. P-all L):
The DNN model did the best (0.8891 accuracy) at telling the difference between people who were not in pain and people who were in pain at all levels. This shows that it can extract many features.
BiGRU and LSTM also did a good job, showing that models that use sequential dependencies work best to find pain.
According to the findings, DNN appears to be the best model for pain categorization. LSTM and BiGRU are two more models that offer excellent alternatives for sequential analysis of physiological data. Regarding high-intensity pain (L4), CNN performs admirably, but it provides marginally better results in other situations. Future research could investigate hybrid deep learning architectures, ensemble learning, and attention mechanisms to increase classification performance further.
Deep Neural Networks (DNN) perform better in pain categorization due to several critical variables. These characteristics include their architecture, capacity to capture complicated feature correlations, and ability to generalize. In the next section, we will present an in-depth analysis of why DNN performed better than CNN, LSTM, and BiGRU in our experimental findings.
- DNN’s fully connected layers allow for better utilization of all extracted features, whereas CNN struggles with spatial data constraints.
- LSTM and BiGRU, while adequate for time-dependent tasks, are not well-suited for this dataset, where pre-extracted statistical features do not require sequential modeling.
- DNN generalizes well, distinguishing pain and non-pain across all intensity levels.
- Its superior recall scores indicate that it minimizes false negatives, making it an ideal choice for pain assessment applications where accurate early detection is crucial for patient care.
- This analysis confirms that DNN is our dataset’s best deep learning model for pain classification and should be prioritized for future enhancements.
4.6 General Discussion for Deep Learning Performance
A. Fully Connected Architecture and Feature Representation:
To learn detailed patterns from the high-dimensional feature set retrieved from physiological signals (EMG, ECG, and SCL), a deep neural network (DNN) is composed of numerous fully coupled layers.
Different neural networks (DNN) process the full feature space, catching subtle correlations across various physiological parameters. This is in contrast to convolutional neural networks (CNN), which primarily concentrate on the extraction of spatial features.
The deep neural network (DNN) could tell the difference between non-pain and pain by using all the collected features without having to depend on spatial or sequential dependencies. This was shown by the high accuracy of 0.8891 for NP against P-all L.
B. Comparison with CNN: Feature Extraction vs. Full Feature Utilization:
The pain dataset is organized as tabular data and contains features already retrieved from EMG, ECG, and SCL. CNNs are very good at extracting features from spatially structured data, such as pictures.
CNN may not be the best pain categorization method because it relies on convolutional filters to extract local features.
Digital neural networks, on the other hand, make full use of all features, which results in improved classification performance at all pain levels, particularly in the case of NP vs. P-all L.
C. Comparison with LSTM and BiGRU: Sequential Learning Limitations:
The LSTM and BiGRU models are designed to work with sequential data, which makes them particularly useful for tasks such as speech recognition and time series analysis. However, our data set is made up of statistical features that have already been extracted, which means that there is no explicit sequential dependency between the features.
However, LSTM achieved satisfactory performance (0.8720 for NP vs. P-all L), but was outperformed by DNN. The difference is most likely because its memory-based architecture introduces unnecessary complexity when it comes to classifying independent feature sets.
Although BiGRU performs better than CNN, it is still less effective than DNN. This difference is because bidirectionality is advantageous for time-dependent tasks, but it does not provide any advantages regarding tabular data classification.
D. Generalization and Overfitting Considerations:
One major issue that could arise with deep learning models is overfitting, which is especially problematic in datasets with small sample sizes.
DNN probably did better at generalization than CNN, LSTM, and BiGRU because it used regularization methods such as dropout and batch normalization and updated its weights more efficiently.
Higher recall scores in DNN show that it is less likely to produce false negatives, which means it is more effective at catching pain signals at various frequencies and intensities.
E. Performance across Pain Levels:
CNN, LSTM, and BiGRU have trouble because there are fewer clear physiological responses at lower pain levels (NP vs. L1, L2), but DNN keeps up its performance by picking up small changes in features. This is because DNN can distinguish between physiological responses.
CNN is superior to L4 in terms of performance at higher pain levels (NP vs. L4) because higher intensity pain causes stronger physiological signals, which in turn makes identification of local features more effective. Despite this, dynamic neural networks continue to beat CNN, suggesting that a fully connected architecture is more adaptive.
When comparing NP to P-all L, DNN performs better than any other model, demonstrating that it is the most effective method for managing different levels of pain.
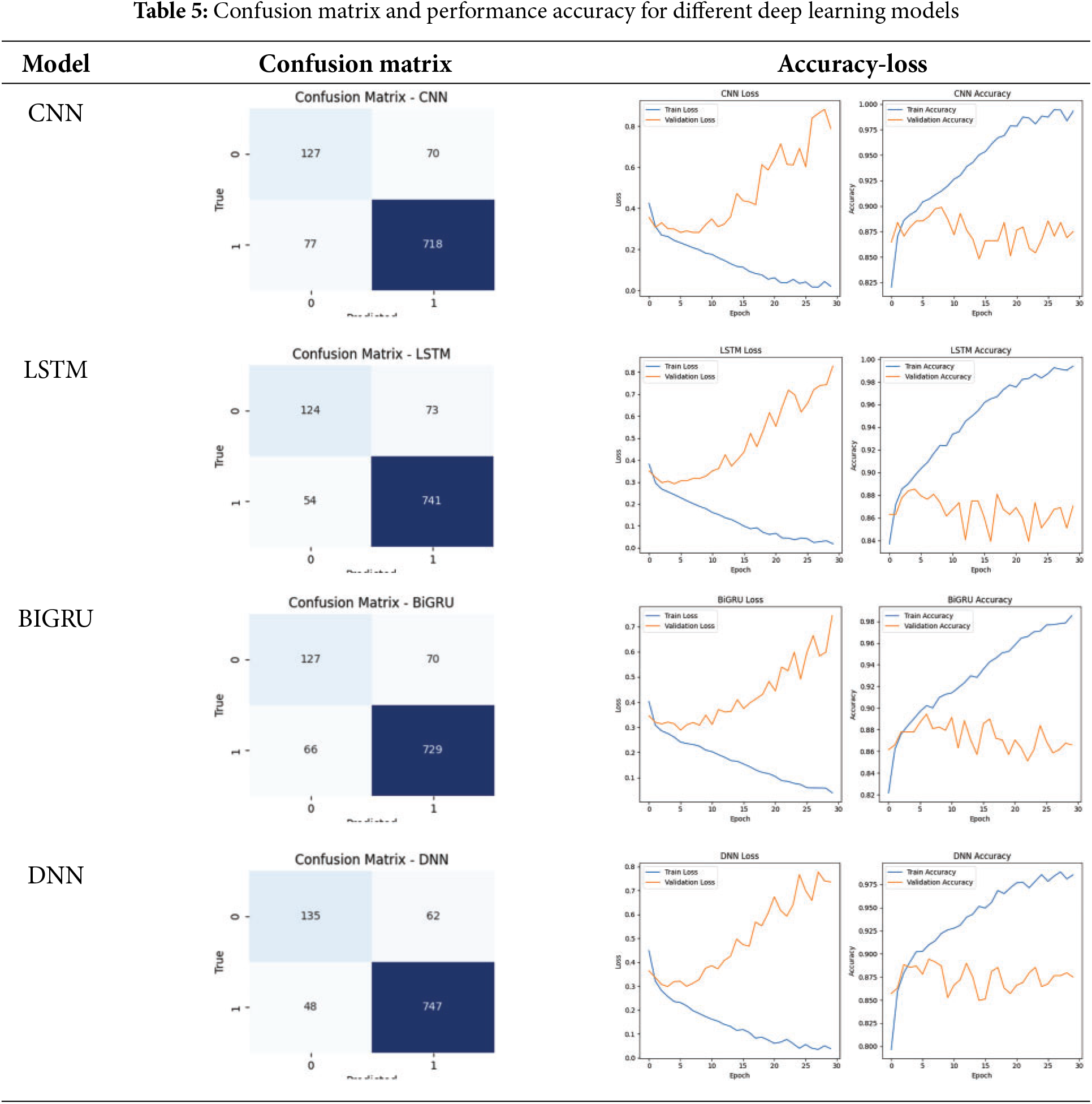
There are several patterns that emerge when one examines the outcomes of the deep learning models for pain assessment (NP vs. P-all L), about the degree to which the models were successful. In light of the fact that the confusion matrices, which are displayed in the Table 5, demonstrate that DNN outperforms other models by displaying a smaller number of false positives and false negatives, the evidence also substantiates the idea that it offers great capabilities in terms of feature extraction. Even though CNN has somewhat higher misclassification rates than DNN, it still performs very well and makes use of spatial feature learning. Although LSTM and BiGRU effectively capture temporal dependencies, they show a greater degree of fluctuation in validation accuracy and a more significant number of incorrect predictions. Based on the results presented here, it appears that temporal dependencies are not sufficient to identify pain in themselves. Overfitting is a problem in LSTM, BiGRU, and DNN, as seen by the loss and accuracy curves, which support the assertion above. It is possible to deduce this from these three models experiencing an increasing verification loss. CNN remains steadier in maintaining its validation accuracy, which shows that it has a higher generalization. CNN keeps its validation accuracy. By combining the two models through the ensemble learning process, it is feasible to enhance the pain assessment system’s resilience and accuracy. It would be possible to achieve this by correctly balancing the spatial and sequential feature representations, which would ultimately result in a more reliable application.
4.7 Performance Analysis of Ensemble Learning Models for Pain Classification
When compared to standalone models, ensemble learning models, which combine many deep learning architectures, are much better at classifying things. Several critical insights arise from Tables 6 and 7.
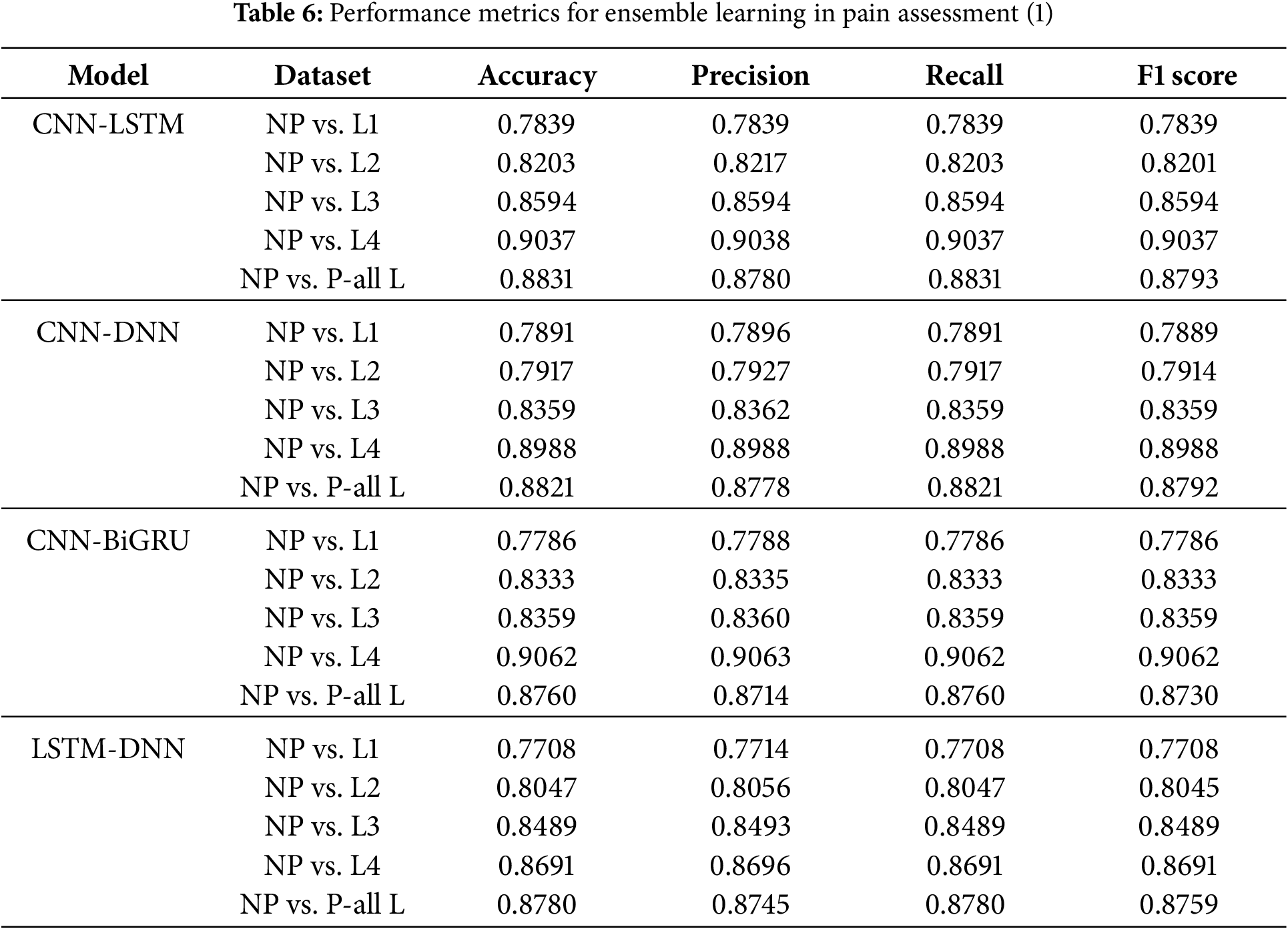
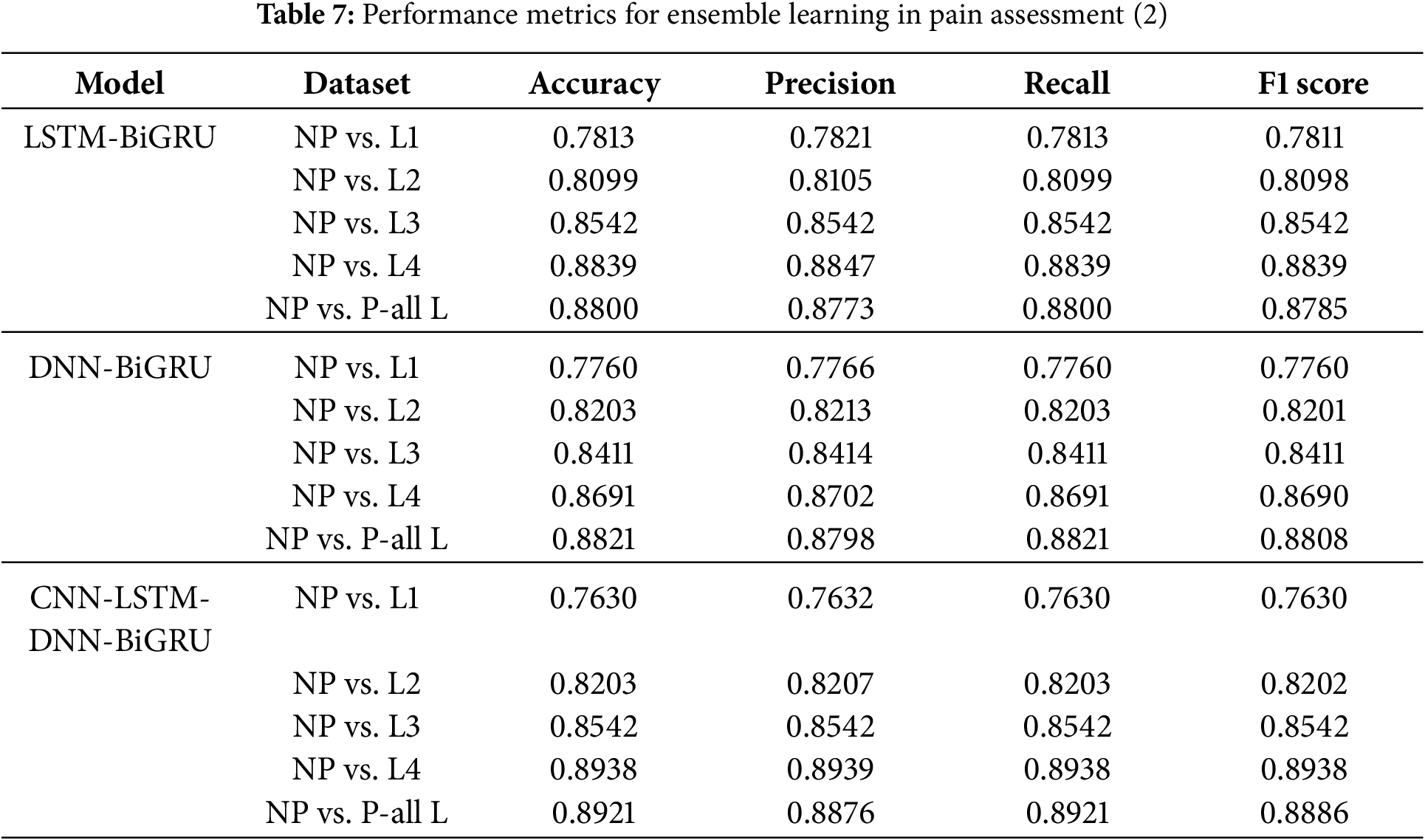
CNN-LSTM-DNN-BiGRU had the highest precision across all levels of pain, especially in the NP vs. all L scenario, with 0.8921. This suggests that several designs capture unique physiological information, improving the understanding of the pain pattern.
CNN-LSTM-DNN-BiGRU works effectively because it uses the best qualities of each part. CNN excels at extracting spatial features and localizing pain-related physiological data. LSTM and BiGRU record temporal relationships, which are needed to understand physiological changes over time. In contrast, DNN uses all the extracted features to improve representation learning throughout the set of features. These structures allow the ensemble model to simultaneously incorporate geographical, temporal, and statistical dependencies. This improves pain classification.
It is interesting that the CNN-LSTM and CNN-DNN models performed well at higher pain levels (NP against L4, 0.9037 and 0.8988, respectively). These results show that convolutional designs can capture the powerful physiological effects of extreme pain. Body changes increase with pain. CNNs can detect these discrepancies better, as they can extract hierarchical feature representations. CNN, however, cannot maintain sequential dependencies. This is why the LSTM and BiGRU ensembles improve recall. Both the LSTM-BiGRU and DNN-BiGRU models exhibited better recall scores, reducing false negatives and the likelihood that pain was mislabeled. In therapeutic contexts, an incorrect evaluation of pain can lead to insufficient therapy and patient distress.
Ensemble models outperform individual deep learning models. Multimodel fusion is crucial for pain diagnosis. Ensemble learning minimizes model shortcomings by creating a balanced and robust classification approach. It provides a balanced and robust alternative to CNN, LSTM, and DNN. There appears to be no optimal deep learning model for pain assessment due to improved classification accuracy across all pain levels. This indicates that superior performance requires a mix of architectures.
Ensemble learning models also performed better across a wider range of pain intensities than individual models, which struggled with lower pain intensities (NP against L1, NP vs. L2). Due to the ensemble’s ability to integrate feature representations and decision patterns, learning becomes more stable and adaptive. Compared with NP to L4, the CNN-BiGRU model classified high-intensity pain best (0.9062). Thus, bidirectional recurrent networks extract deeper features better. It suggests that adaptive weight-based ensemble learning processes may be future enhancements. These solutions dynamically adjust model contributions based on pain control.
As shown in Fig. 3, we found that CNN-LSTM-DNN-BiGRU is the best pain categorization model. This model gives the best accuracy, recall, and F1 score in all intensity of pain. Spatial, sequential, and fully connected architectures improve pain assessment categorization. This approach accurately captures the complex physiological changes of pain. Future research may include attention mechanisms, transformer-based topologies, and adaptive ensemble weighting to improve the accuracy of pain categorization and clinical utility.
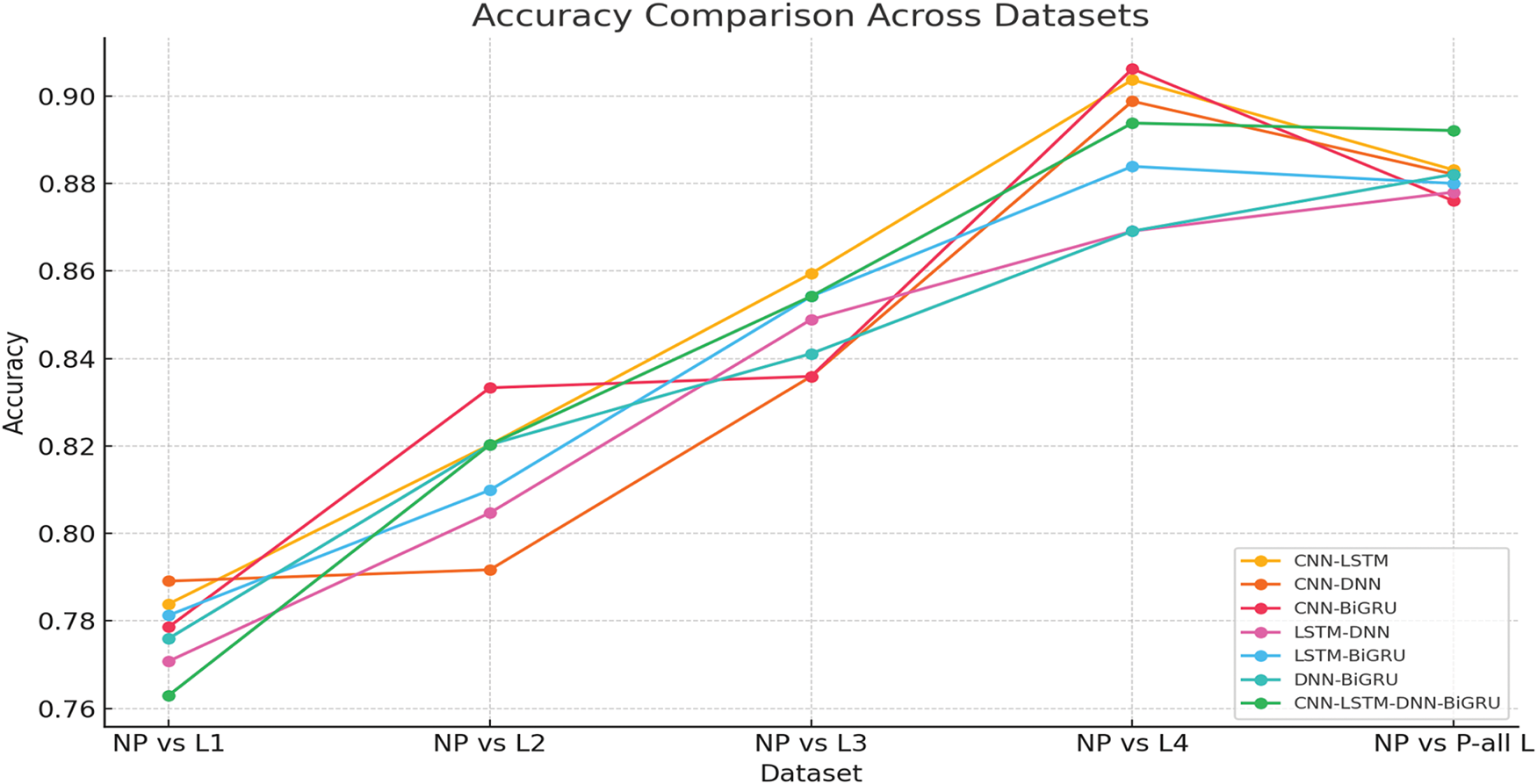
Figure 3: Performance metrics for ensemble learning in pain assessment
4.8 Performance Analysis of Feature Selection Based on PSO Algorithm and Ensemble Learning Models for Pain Classification
By reducing the dimensionality of the feature space while preserving the relevant information, the selection of features is a crucial component in improving classification performance. An initial collection of 135 extracted features was collected from a variety of physiological signals, such as electromyography (EMG) (Zygomaticus, Corrugator, and Trapezius), electrocardiogram (ECG), and electrodermal sclerosis (EDA/SCL/GSR). This work applied Particle Swarm Optimization (PSO) to select the features that were the most discriminative using these features. This model’s ability to properly discriminate between pain and non-pain states is significantly impacted by the features that were selected.
Several noteworthy observations are presented in Table 8. Certain qualities appear to continuously improve classification performance, as evidenced by the fact that the number of picked features varies throughout several PSO cycles. The best subsets consist of between 71 and 93 features. The performance of the models was maintained or improved, despite the fact that the number of features was reduced from 135 to 71. This suggests that a significant number of the original features were unnecessary or lacked any informative contribution.
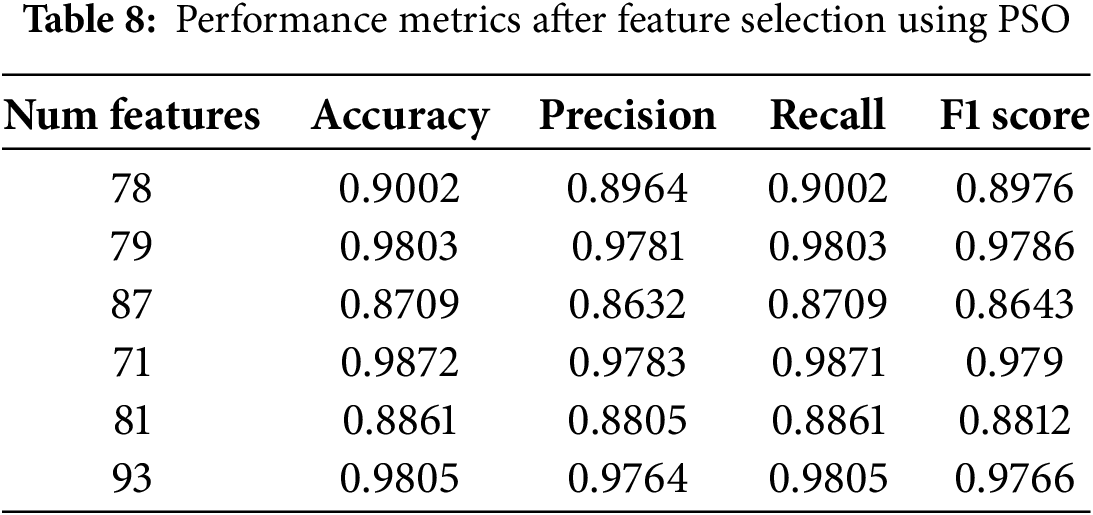
Particularly within statistical categories such as amplitude (A), frequency (F), stationarity (S), and entropy (E), it should be noted that the characteristics of EMG Zygomaticus (CH22) and EMG Corrugator (CH23) are consistently chosen throughout a variety of iterations. This is a notable observation. By the existing body of research on pain assessment, which states that facial muscle contractions are closely associated with the level of pain, this shows that face EMG signals bear a striking resemblance to pain responses. When determining how the autonomic nervous system reacts to pain stimuli, the fact that the characteristics of ECG (CH25) and EDA (CH26) are included in various optimal feature sets demonstrates the value of these characteristics.
The association between the selected feature subset and efficient data classification is an important observation. The classifier was able to achieve a perfect separation between pain and non-pain states, as evidenced by the fact that particular feature sets, such as 79 and 71 characteristics, produced an optimal accuracy of 98%. With larger feature sets (for example, 93 features), comparable performance levels were achieved, which may indicate the possibility of overfitting. Intricate non-linear correlations in physiological signals are shown to significantly improve classification accuracy when specific statistical properties are incorporated into the analysis. These attributes include approximate entropy, sample entropy, and mutual information-based features.
The significance of inter-channel interactions in pain identification is highlighted by the fact that coherence and correlation-based properties (Sim-cohe and Sim-corr) are present across many channels. Pain responses can simultaneously originate across several sensors, which is particularly relevant in multimodal physiological data. As shown using frequency-domain features (F-cf, F-bw, and F-fmed) in several iterations, spectral characteristics are crucial for differentiating pain from non-pain states. Changes in muscle activity and autonomic nervous system responses to pain stimulation may explain this fact.
Ultimately, PSO showed its capacity to increase the efficiency of the computation without sacrificing accuracy in the classification process by successfully lowering the number of features while preserving a high degree of classification performance. Together with the ECG and EDA data, the Zygomaticus and Corrugator EMG derived the most commonly chosen traits. This implies that a combination of facial muscle activity and autonomic responses influences the recognition of pain. Future research, which might look at adaptive feature selection techniques or hybrid optimization strategies, could concentrate on improving the feature selection process while maintaining its resilience across several datasets.
4.9 Comparison with Existing Methods
This part aims to compare the outcomes of our method with those of other approaches already present in the literature.
In Table 9, we compare our proposed approaches with several existing state-of-the-art methods for pain detection. Earlier works, such as those by Walter et al. [40] and Kächele et al. [41], relied on traditional machine learning techniques like SVMs, achieving accuracies of 77.05% and 76.60% respectively. Thiam et al. [42] explored deep learning with CNNs, leading to a modest performance gain (79.48%). Meanwhile, Gouverneur et al. [43] obtained a higher accuracy of 84.22% (
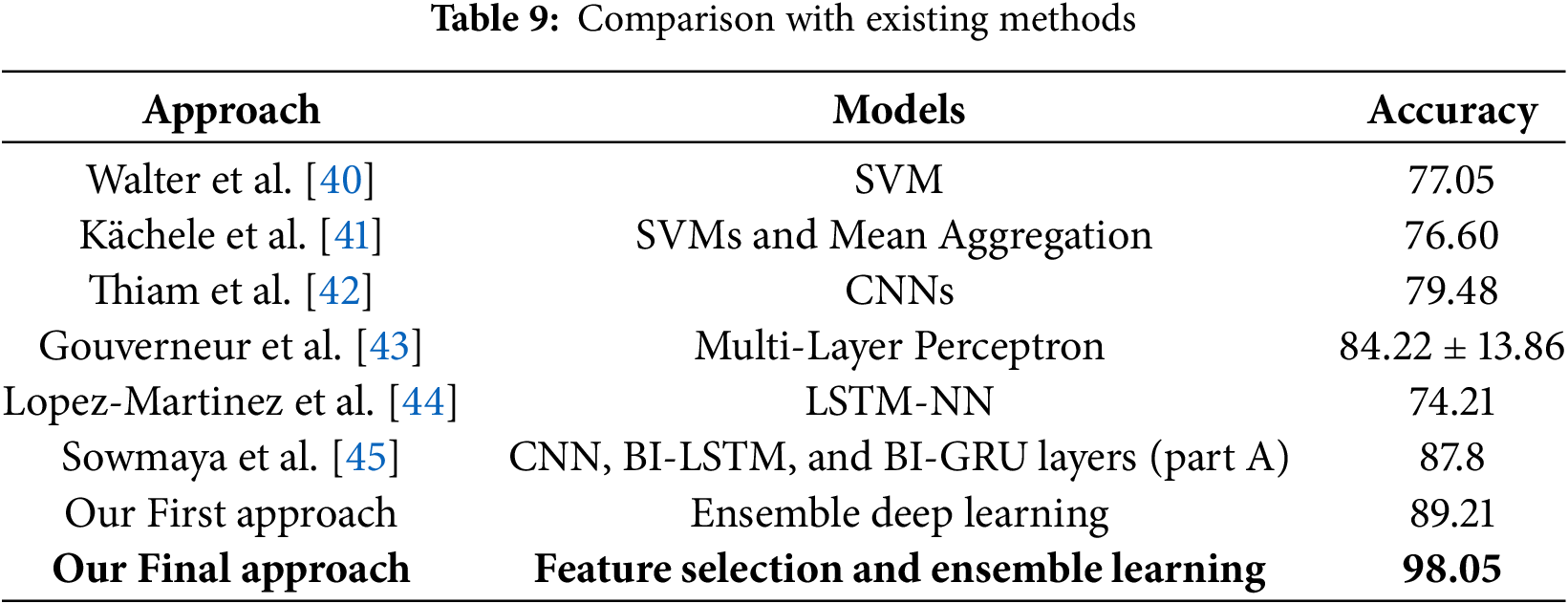
Our first proposed method, which leverages ensemble deep learning, outperformed previous works with an accuracy of 89.21%. The final proposed approach, integrating feature selection with optimized ensemble learning, achieved a significantly higher accuracy of 98.05%. This result highlights the effectiveness of combining metaheuristic-driven feature selection with ensemble strategies, offering superior generalization and predictive performance. Our method substantially advances traditional and deep learning-based baselines, emphasizing the importance of integrating feature engineering and ensemble modeling for robust pain assessment systems.
4.10 Computational Efficiency and Optimization Trade-Offs
Though particle swarm optimization (PSO) improves feature selection performance, it also increases total computational load during training. The lower dimensionality, which increases the inference speed, helps offset this expense. Alternative optimization techniques such as Genetic Algorithms or Bayesian Optimization could provide similar performance with varied computational profiles. Investigating these techniques could be beneficial for certain embedded systems or real-time applications.
4.11 Clinical Applications and Ethical Considerations
Automated pain assessment systems, such as the one suggested in this article, show great promise in clinical settings, especially for non-verbal patients, ICU monitoring or telemedicine. However, ethical issues, including patient consent, physiological data privacy, and risk of algorithmic bias, must be discussed first before use. Future efforts should guarantee responsible integration into healthcare systems by working with ethicists and doctors.
4.12 Limitations and Generalizability
Although the suggested approach shows promising results on the BioVid Heat Pain dataset, its applicability to various populations and clinical situations remains unknown. Its relevance to patients with multiple pain profiles or comorbidities needs further research, since the model was trained and evaluated only on this data set comprising healthy individuals in controlled experimental settings. Future studies will focus on clinical validation and cross-dataset evaluation to determine population robustness.
In this research, a multimodal pain assessment approach is presented. The system uses feature extraction, deep learning, ensemble learning, and PSO-based feature selection. We collected 135 statistical features from EMG signals (Zygomaticus, Corrugator, and Trapezius), ECG, and GSR signals using our methods. These features encompassed six primary mathematical domains. We used four deep learning models: CNN, LSTM, BiGRU, and DNN to improve classification accuracy. Subsequently, we used an ensemble learning strategy to combine the forecasting skills of these models.
We used particle swarm optimization (PSO) for feature selection to improve the model’s performance and the computations’ efficiency. This resulted in a significant reduction in the dimensionality of the features while maintaining the most discriminative characteristics. Ensemble learning was found to be superior to individual models. In contrast, PSO-based feature selection improved classification accuracy while reducing the amount of feature redundancy. The integration of fully connected, temporal and spatial architectures, in conjunction with an optimized subset of features, led to improvements in accuracy, precision, recall, and F1 score, proving the robustness of the methodology described here.
For actual clinical applications, there are still challenges in terms of scalability and real-time execution, even though performance is lacking. Future research will study hybrid optimization methods, attention-driven deep learning architectures, and real-time multimodal signal processing to improve the precision and relevance of pain identification in healthcare settings. This study highlights the potential of automated pain assessment through multimodal physiological data, which can promote improved patient monitoring, faster intervention, and individualized pain management. By combining physiological signals with features extracted from medical images (such as thermal or facial expression images), we want to incorporate image-based data into our multi-modal pain assessment system. This will involve a combination of the two types of data. Because of this, the work that we have been doing will naturally continue in this manner. As a result of this combination of image-based and physiologically-based pain assessment, it will be feasible to gain a more thorough understanding of individuals’ responses to pain. The potential to result in even improved classification accuracy and real-time applicability in clinical settings is currently being explored. Furthermore, we intend to examine hybrid optimization strategies and advanced deep learning architectures, such as attention mechanisms and transformers, to further improve the interpretability and durability of the model in the context of real-world healthcare applications.
Acknowledgement: This work was funded by the Deanship of Graduate Studies and Scientific Research at Jouf University under grant No. (DGSSR-2023-02-02341).
Funding Statement: This work was funded by the Deanship of Graduate Studies and Scientific Research at Jouf University under grant No. (DGSSR-2023-02-02341).
Author Contributions: The authors confirm their contributions to the paper as follows: Conceptualization: Karim Gasmi, Olfa Hrizi and Najib Ben Aoun; methodology: Karim Gasmi, Olfa Hrizi, Ali Alqazzaz and Ibrahim Alrashdi; software: Omer Hamid, Lassaad Ben Ammar, Manel Mrabet and Omrane Necibi; validation: Lassaad Ben Ammar, Mohamed O. Altaieb and Alameen E. M. Abdalrahman; formal analysis: Karim Gasmi, Najib Ben Aoun and Mohamed O. Altaieb; resources: Omer Hamid, Ali Alqazzaz and Ibrahim Alrashdi; data curation: Karim Gasmi and Olfa Hrizi; writing—original draft preparation: Karim Gasmi, Olfa Hrizi, Najib Ben Aoun and Ali Alqazzaz; writing—review and editing: Alameen E. M. Abdalrahman, Manel Mrabet, Omrane Necibi and Mohamed O. Altaieb; visualization: Olfa Hrizi, Ibrahim Alrashdi, Mohamed O. Altaieb and Ali Alqazzaz; funding acquisition: Karim Gasmi and Olfa Hrizi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Dataset available on request from the authors: https://www.nit.ovgu.de/BioVid.html, accessed on 3 February 2025.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
1https://www.nit.ovgu.de/BioVid.html (accessed on 3 February 2025)
References
1. Gkikas S, Tsiknakis M. Automatic assessment of pain based on deep learning methods: a systematic review. Comput Methods Programs Biomed. 2023;231(3):107365. doi:10.1016/j.cmpb.2023.107365. [Google Scholar] [PubMed] [CrossRef]
2. Ben Aoun N. A review of automatic pain assessment from facial information using machine learning. Technologies. 2024;12(6):92. doi:10.3390/technologies12060092. [Google Scholar] [CrossRef]
3. Gkikas S, Tachos NS, Andreadis S, Pezoulas VC, Zaridis D, Gkois G, et al. Multimodal automatic assessment of acute pain through facial videos and heart rate signals utilizing transformer-based architectures. Front Pain Res. 2024;5:1372814. doi:10.3389/fpain.2024.1372814. [Google Scholar] [CrossRef]
4. Kasaeyan Naeini E, Subramanian A, Calderon MD, Zheng K, Dutt N, Liljeberg P, et al. Pain recognition with electrocardiographic features in postoperative patients: method validation study. J Med Internet Res. 2021;23(5):e25079. doi:10.1101/2023.06.07.23291094. [Google Scholar] [PubMed] [CrossRef]
5. Shi H, Chikhaoui B, Wang S. Tree-based models for pain detection from biomedical signals. In: International Conference on Smart Homes and Health Telematics. Paris, France: Springer; 2022. p. 183–95. doi:10.1007/978-3-031-09593-1_14. [Google Scholar] [CrossRef]
6. Khan MU, Aziz S, Hirachan N, Joseph C, Li J, Fernandez-Rojas R. Experimental exploration of multilevel human pain assessment using blood volume pulse (BVP) signals. Sensors. 2023;23(8):3980. doi:10.3390/s23083980. [Google Scholar] [PubMed] [CrossRef]
7. Pouromran F, Lin Y, Kamarthi S. Automatic pain recognition from Blood Volume Pulse (BVP) signal using machine learning techniques. arXiv:230310607. 2023. [Google Scholar]
8. Lin Y, Xiao Y, Wang L, Guo Y, Zhu W, Dalip B, et al. Experimental exploration of objective human pain assessment using multimodal sensing signals. Front Neurosci. 2022;16:831627. doi:10.3389/fnins.2022.831627. [Google Scholar] [PubMed] [CrossRef]
9. Pouromran F, Radhakrishnan S, Kamarthi S. Exploration of physiological sensors, features, and machine learning models for pain intensity estimation. PLoS One. 2021;16(7):e0254108. doi:10.1371/journal.pone.0254108. [Google Scholar] [PubMed] [CrossRef]
10. Posada-Quintero HF, Kong Y, Chon KH. Objective pain stimulation intensity and pain sensation assessment using machine learning classification and regression based on electrodermal activity. Am J Physiol-Regulat, Integrat Comparat Physiol. 2021;321(2):R186–96. doi:10.1152/ajpregu.00094.2021. [Google Scholar] [CrossRef]
11. Chu Y, Zhao X, Han J, Su Y. Physiological signal-based method for measurement of pain intensity. Front Neurosci. 2017;11:279. doi:10.3389/fnins.2017.00279. [Google Scholar] [PubMed] [CrossRef]
12. Kächele M, Thiam P, Amirian M, Schwenker F, Palm G. Methods for person-centered continuous pain intensity assessment from bio-physiological channels. IEEE J Select Top Signal Process. 2016;10(5):854–64. doi:10.1109/jstsp.2016.2535962. [Google Scholar] [CrossRef]
13. Naseri H, Kafi K, Skamene S, Tolba M, Faye MD, Ramia P, et al. Development of a generalizable natural language processing pipeline to extract physician-reported pain from clinical reports: generated using publicly-available datasets and tested on institutional clinical reports for cancer patients with bone metastases. J Biomed Inform. 2021;120(4):103864. doi:10.1016/j.jbi.2021.103864. [Google Scholar] [PubMed] [CrossRef]
14. Dave AD, Ruano G, Kost J, Wang X. Automated extraction of pain symptoms: a natural language approach using electronic health records. Pain Physician. 2022;25(2):E245. doi:10.1101/2024.03.08.24304011. [Google Scholar] [PubMed] [CrossRef]
15. Bartlett MS, Littlewort G, Frank MG, Lainscsek C, Fasel IR, Movellan JR, et al. Automatic recognition of facial actions in spontaneous expressions. J Multim. 2006;1(6):22–35. doi:10.1109/fgr.2006.55. [Google Scholar] [CrossRef]
16. Chew SW, Lucey P, Lucey S, Saragih J, Cohn JF, Matthews I, et al. In the pursuit of effective affective computing: the relationship between features and registration. IEEE Transact Syst Man Cybern Part B (Cybern). 2012;42(4):1006–16. doi:10.1109/tsmcb.2012.2194485. [Google Scholar] [CrossRef]
17. Ding X, Chu WS, De la Torre F, Cohn JF, Wang Q. Cascade of tasks for facial expression analysis. Image Vis Comput. 2016;51(1):36–48. doi:10.1016/j.imavis.2016.03.008. [Google Scholar] [CrossRef]
18. Ben Aoun N. Deep learning-based pain intensity estimation from facial expressions. In: 23th Proceedings of the 23th International Conference on Intelligent Systems Design and Applications (ISDA 2023). Porto, Portugal: Springer; 2023. p. 484–93. doi:10.1007/978-3-031-64836-6_47. [Google Scholar] [CrossRef]
19. Walter S, Al-Hamadi A, Gruss S, Frisch S, Traue H, Werner P. Multimodal recognition of pain intensity and pain modality with machine learning. Der Schmerz. 2020;34(5):400–9. doi:10.1007/s00482-020-00468-8. [Google Scholar] [PubMed] [CrossRef]
20. Wang R, Xu K, Feng H, Chen W. Hybrid RNN-ANN based deep physiological network for pain recognition. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). Montréal, BC, Canada: IEEE; 2020. p. 5584–7. [Google Scholar]
21. De S, Mukherjee P, Roy AH. A hybrid pain assessment approach with stacked autoencoders and attention-based CP-LSTM. In: 2023 International Conference on Ambient Intelligence, Knowledge Informatics and Industrial Electronics (AIKIIE). Ballari, India: IEEE; 2023. p. 1–6. doi:10.1109/aikiie60097.2023.10390223. [Google Scholar] [CrossRef]
22. Subramaniam SD, Dass B. Automated nociceptive pain assessment using physiological signals and a hybrid deep learning network. IEEE Sensors J. 2020;21(3):3335–43. doi:10.1109/jsen.2020.3023656. [Google Scholar] [CrossRef]
23. Patania S, Boccignone G, Buršić S, D’Amelio A, Lanzarotti R. Deep graph neural network for video-based facial pain expression assessment. In: Proceedings of the 37th ACM/SIGAPP Symposium on Applied Computing. Brno, Czech Republic: ACM; 2022. p. 585–91. doi:10.1145/3477314.3507094. [Google Scholar] [CrossRef]
24. Lu G, Chen H, Wei J, Li X, Zheng X, Leng H, et al. Video-based neonatal pain expression recognition with cross-stream attention. Multim Tools Applicat. 2024;83(2):4667–90. doi:10.1007/s11042-023-15403-z. [Google Scholar] [CrossRef]
25. Wu W, Bi L, Ren W, Nie W, Lin R, Liu Z, et al. Detecting temporal pain status of postoperative children from facial expression. In: International Conference on Intelligent Robotics and Applications. Kyoto, Japan: Springer; 2022. p. 700–11. doi:10.1007/978-3-031-13841-6_63. [Google Scholar] [CrossRef]
26. Talaat FM, Ali ZH, Mostafa RR, El-Rashidy N. Real-time facial emotion recognition model based on kernel autoencoder and convolutional neural network for autism children. Soft Comput. 2024;28(9–10):6695–708. doi:10.1007/s00500-023-09477-y. [Google Scholar] [CrossRef]
27. Ghosh A, Dhara BC, Pero C, Umer S. A multimodal sentiment analysis system for recognizing person aggressiveness in pain based on textual and visual information. J Ambient Intelli Human Comput. 2023;14(4):4489–501. doi:10.1007/s12652-023-04567-z. [Google Scholar] [CrossRef]
28. Wu CL, Liu SF, Yu TL, Shih SJ, Chang CH, Yang Mao SF, et al. Deep learning-based pain classifier based on the facial expression in critically ill patients. Front Med. 2022;9:851690. doi:10.3389/fmed.2022.851690. [Google Scholar] [CrossRef]
29. Pikulkaew K, Boonchieng W, Boonchieng E. Real-time pain detection using deep convolutional neural network for facial expression and motion. In: Proceedings of Seventh International Congress on Information and Communication Technology: ICICT 2022. London, UK: Springer; 2022. p. 341–9. doi:10.1007/978-981-19-1610-6_29. [Google Scholar] [CrossRef]
30. Ismail L, Waseem MD. Towards a deep learning pain-level detection deployment at UAE for patient-centric-pain management and diagnosis support: framework and performance evaluation. Procedia Comput Sci. 2023;220(5):339–47. doi:10.1016/j.procs.2023.03.044. [Google Scholar] [PubMed] [CrossRef]
31. Kumar V, Dhapola P, Kushwaha AN. Automatic pain detection through facial expression. In: Investigations in pattern recognition and computer vision for Industry 4.0. Hershey, PA, USA: IGI Global; 2023. p. 81–9. [Google Scholar]
32. Alghamdi T, Alaghband G. Facial expressions based automatic pain assessment system. Appl Sci. 2022;12(13):6423. doi:10.3390/app12136423. [Google Scholar] [CrossRef]
33. El Morabit S, Rivenq A. Pain detection from facial expressions based on transformers and distillation. In: 2022 11th International Symposium on Signal, Image, Video and Communications (ISIVC). Eljadida, Morocco: IEEE; 2022. p. 1–5. doi:10.1109/isivc54825.2022.9800746. [Google Scholar] [CrossRef]
34. Rathee N, Pahal S, Sheoran P. Pain detection from facial expressions using domain adaptation technique. Pattern Analy Applicat. 2022;25(3):567–74. doi:10.1007/s10044-021-01025-4. [Google Scholar] [CrossRef]
35. Hughes JA, Wu Y, Jones L, Douglas C, Brown N, Hazelwood S, et al. Analyzing pain patterns in the emergency department: leveraging clinical text deep learning models for real-world insights. Int J Med Inform. 2024;190(10):105544. doi:10.1016/j.ijmedinf.2024.105544. [Google Scholar] [PubMed] [CrossRef]
36. De S, Mukherjee P, Roy AH. GLEAM: a multimodal deep learning framework for chronic lower back pain detection using EEG and sEMG signals. Comput Biol Med. 2025;189(12):109928. doi:10.1016/j.compbiomed.2025.109928. [Google Scholar] [PubMed] [CrossRef]
37. Egede J, Valstar M, Martinez B. Fusing deep learned and hand-crafted features of appearance, shape, and dynamics for automatic pain estimation. In: 2017 12th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2017). Washington, DC, USA: IEEE; 2017. p. 689–96. [Google Scholar]
38. Jaiswal S, Valstar M. Deep learning the dynamic appearance and shape of facial action units. In: 2016 IEEE Winter Conference on Applications of Computer Vision (WACV). Lake Placid, NY, USA: IEEE; 2016. p. 1–8. doi:10.1109/wacv.2016.7477625. [Google Scholar] [CrossRef]
39. Walter S, Gruss S, Ehleiter H, Tan J, Traue HC, Werner P, et al. The biovid heat pain database data for the advancement and systematic validation of an automated pain recognition system. In: 2013 IEEE International Conference on Cybernetics (CYBCO). Lausanne, Switzerland: IEEE; 2013. p. 128–31. doi:10.1109/cybconf.2013.6617456. [Google Scholar] [CrossRef]
40. Walter S, Gruss S, Limbrecht-Ecklundt K, Traue HC, Werner P, Al-Hamadi A, et al. Automatic pain quantification using autonomic parameters. IEEE Transact Affect Comput. 2014;8(3):286–99. doi:10.1109/taffc.2016.2537327. [Google Scholar] [CrossRef]
41. Kächele M, Werner P, Al-Hamadi A, Palm G, Walter S, Schwenker F. Bio-visual fusion for person-independent recognition of pain intensity. In: Multiple Classifier Systems: 12th International Workshop, MCS 2015; 2015 Jun 29–Jul 1; Günzburg, Germany. p. 220–30. [Google Scholar]
42. Thiam P, Bellmann P, Kestler HA, Schwenker F. Exploring deep physiological models for nociceptive pain recognition. Sensors. 2019;19(20):4503. doi:10.1101/622431. [Google Scholar] [PubMed] [CrossRef]
43. Gouverneur P, Li F, Adamczyk WM, Szikszay TM, Luedtke K, Grzegorzek M. Comparison of feature extraction methods for physiological signals for heat-based pain recognition. Sensors. 2021;21(14):4838. doi:10.3390/s21144838. [Google Scholar] [PubMed] [CrossRef]
44. Lopez-Martinez D, Picard R. Continuous pain intensity estimation from autonomic signals with recurrent neural networks. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Honolulu, HI, USA: IEEE; 2018. p. 5624–7. doi:10.1109/embc.2018.8513575. [Google Scholar] [CrossRef]
45. Sowmya M, Dinakar N, Heiner A. Pain recognition via multi-context integration of physiological signals. In: 2025 International Conference on Multi-Agent Systems for Collaborative Intelligence (ICMSCI). Tamil Nadu, India: IEEE; 2025. p. 1559–64. doi:10.1109/icmsci62561.2025.10894617. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools