 Open Access
Open Access
REVIEW
A Survey of Large-Scale Deep Learning Models in Medicine and Healthcare
School of Computer Science, Nanjing University of Information Science and Technology, Nanjing, 210044, China
* Corresponding Author: Yangyang Guo. Email:
# These authors contribute equally to this work and share first authorship
Computer Modeling in Engineering & Sciences 2025, 144(1), 37-81. https://doi.org/10.32604/cmes.2025.067809
Received 13 May 2025; Accepted 15 July 2025; Issue published 31 July 2025
Abstract
The rapid advancement of artificial intelligence technology is driving transformative changes in medical diagnosis, treatment, and management systems through large-scale deep learning models—a process that brings both groundbreaking opportunities and multifaceted challenges. This study focuses on the medical and healthcare applications of large-scale deep learning architectures, conducting a comprehensive survey to categorize and analyze their diverse uses. The survey results reveal that current applications of large models in healthcare encompass medical data management, healthcare services, medical devices, and preventive medicine, among others. Concurrently, large models demonstrate significant advantages in the medical domain, especially in high-precision diagnosis and prediction, data analysis and knowledge discovery, and enhancing operational efficiency. Nevertheless, we identify several challenges that need urgent attention, including improving the interpretability of large models, strengthening privacy protection, and addressing issues related to handling incomplete data. This research is dedicated to systematically elucidating the deep collaborative mechanisms between artificial intelligence and the healthcare field, providing theoretical references and practical guidance for both academia and industry.Keywords
The convergence of AI and medicine, particularly the incorporation of large-scale deep learning models, introduces unparalleled opportunities and challenges in the domains of medical diagnosis, treatment, and management [1,2]. Recent reviews, such as Nazi and Peng, further consolidate the growing body of research on the potential and constraints of large language models in medical applications, underscoring their transformative potential [3]. In this article, ‘large models’ refer to a class of deep learning neural networks that are characterized by large parameter sizes (usually ranging from billions to trillions) and have demonstrated excellent performance in various tasks such as natural language processing (NLP) and computer vision (CV). The training of these models usually includes two main stages: pretraining and fine-tuning. In the pretraining stage, the model learns general data representations and patterns on large unlabeled datasets through self-supervised learning methods; the fine-tuning stage adapts the model to specific tasks.
Large models obtain rich and general data representations through pretraining, which enables them to effectively adapt to different tasks in the fine-tuning stage, achieving multitask learning and reducing the need for task-specific models. These models demonstrate versatility across domains, executing natural language processing (text generation, machine translation, sentiment analysis), visual computing (image categorization, object recognition, pixel-wise segmentation), and multimodal applications spanning healthcare and finance. Although LLMs have shown promising prospects in healthcare, they also face challenges, including data privacy, model interpretability, and ethical issues. As a cutting-edge representative of AI, large models are swiftly penetrating the healthcare landscape, furnishing more precise, expeditious, and dependable support for clinical decision-making. On November 30, 2022, OpenAI, the American AI company, launched a notable Large Language Model (LLM) named ChatGPT, distinguished by its outstanding natural language processing (NLP) capabilities. The advent of ChatGPT marks a major breakthrough in the application of artificial intelligence in the field of healthcare, providing new possibilities for patient consultation, case analysis, and health education [4]. Currently, research efforts have explored the utilization of authentic clinical texts to develop a generative LLM termed Ga-torTronGPT, assessing its efficacy in biomedical NLP and healthcare text generation [5]. Models trained with synthetic medical text generated by GatorTronGPT outperformed models trained with real clinical text on some tasks. Yet, concurrent with this technological progression, a gamut of contemplative issues has surfaced, scrutinizing not only the prowess of technology but also the quality and enduring nature of healthcare services [6].
The healthcare sector processes a large amount of data, including patient records, medical images, and laboratory tests. However, traditional manual analysis methods can no longer meet the needs of fast processing and accurate acquisition of key information [7]. Although large-scale deep learning models have achieved remarkable success in natural language processing and image recognition, their complexity and specificity still bring challenges to healthcare applications [8]. The intricacy of medical data is not only evident in its massive volume but also in its heterogeneity and dynamism, presenting unprecedented complexity in the design and application of models [9]. In this context, there is an urgent need to explore how to better leverage large models to serve patients, support medical decision-making, and propel the development of medical research. In practical applications, the complexity of large models and their computational resource requirements pose a series of challenges [10]. For many healthcare institutions and research teams, achieving efficient applications of large models entails overcoming not only technical challenges but also addressing limitations in hardware and resources. Consequently, the pressing issue at hand is how to better apply large models in resource-constrained environments within medicine and healthcare.
Large-scale deep learning models have become a pivotal catalyst for transformative advancements in medical and healthcare domains [11,12]. From the intelligent processing of medical images to the analysis of patient records, these models play an increasingly vital role in clinical decision-making, disease diagnosis, and the formulation of treatment plans [2]. Our work focuses on various applications of large-scale deep learning models in medicine and healthcare, systematically exploring and summarizing the advantages of large models in healthcare. Through a comprehensive analysis of existing research and practical cases, our aim is to provide a thorough understanding of the potential value of large models for healthcare practitioners, researchers, and decision-makers in medicine and healthcare. To ensure comprehensive coverage of relevant literature, we systematically searched multiple databases, notably PubMed and Google Scholar. In the case of Google Scholar, only the first 50 most pertinent studies were included, as further results exhibited diminishing relevance and deviated from the study’s core objectives. Additionally, the technical search was based on abstracts and titles, with our search scope limited to English articles published between 2019 and 2023.
This review will systematically discuss the applications of large models in medicine and healthcare and their unique potential in the realm of medicine. As shown in Fig. 1 below, in Section 2, we will present a comprehensive examination of large-scale models, including their fundamental principles and developmental history. In Section 3, we will primarily analyze the overall application of large models in medicine and healthcare, with Table 1 summarizing existing studies for comparison with our paper. Subsequently, in Section 4, we will concentrate on the advantages of LLMs in healthcare, exploring their specific performances in high-precision diagnosis, data analysis, and knowledge discovery, and enhancing work efficiency. Finally, in Section 5, we will review the current challenges faced by the healthcare sector to gain a more comprehensive understanding of the application prospects of large models in healthcare. This paper will conclude with a conclusion section serving as an overall summary of the research, emphasizing the potential value of large models in the healthcare domain, and proposing directions for further investigation.
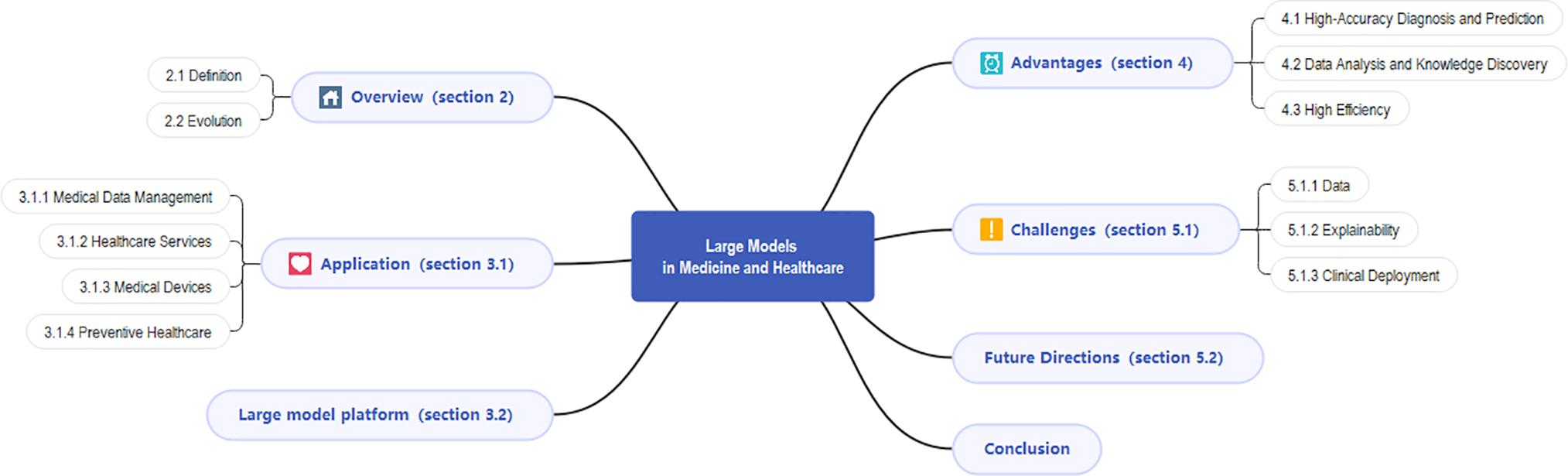
Figure 1: Diagram of the content structure
2.1 Definition of the Large Model
Large models, as characterized by their extensive parameter scale ranging from billions to trillions, have become a defining feature of modern deep learning neural networks [13]. Such models require domain-specific adaptations in high-stakes fields like emergency medicine, where real-time interpretation of complex data (e.g., ECG waveforms or trauma imaging) is critical for clinical decision-making [14,15]. To align with the current consensus within the AI community, we recognize models such as SAM for computer vision and Contrastive Language-Image Pre-training (CLIP) for vision-language tasks as more appropriate examples of large models. SAM, a recent advancement in CV, demonstrates the capabilities of large models in handling complex image data, while CLIP showcases the potential of large models in understanding and correlating visual and textual information. Large models have demonstrated versatility beyond NLP and CV, such as Spectrum-BERT (Wang et al.) which adapts bidirectional transformers for spectral data classification, suggesting potential extensions to medical spectral analysis (e.g., spectroscopy-based diagnostics) [16].
Large models have revolutionized the way we approach multimodal tasks, such as medical diagnosis and financial analysis, by providing a more comprehensive and nuanced understanding of data. Their ability to learn rich and universal representations through pretraining on massive datasets enables them to adapt to a wide array of tasks during the fine-tuning stage, reducing the need for task-specific models and enhancing multitask learning. As shown in Fig. 2, the development of medical big models covers a complete technical chain from multisource data collection to iterative optimization. Among them, the pretraining stage extracts general representations through self-supervised learning, while the fine-tuning stage adapts to specific clinical tasks (such as image classification), and ultimately drives model optimization through performance evaluation.
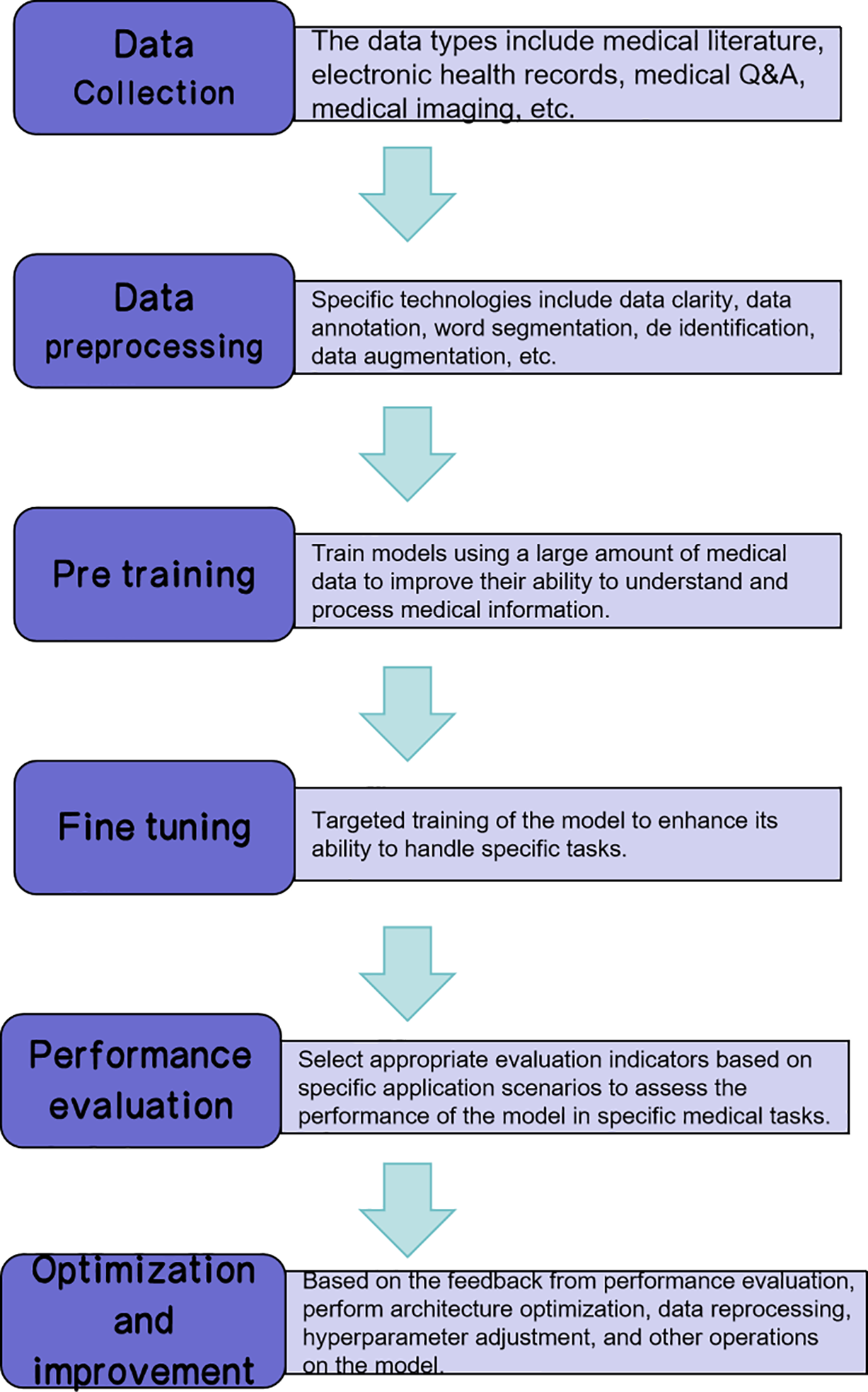
Figure 2: Technical pipeline of medical large model development
2.2 Evolution of the Large Model
Fig. 3 shows the key developments of large-scale AI models from 2005 to 2024. Each marked event represents a technological breakthrough or the release of an important model. The advancements in these models and technologies have not only accelerated the rapid development of AI but also laid the foundation for its application in the healthcare field. The following timeline outlines the influential models and architectures, particularly significant breakthroughs in the fields of natural language processing (NLP) and computer vision (CV).
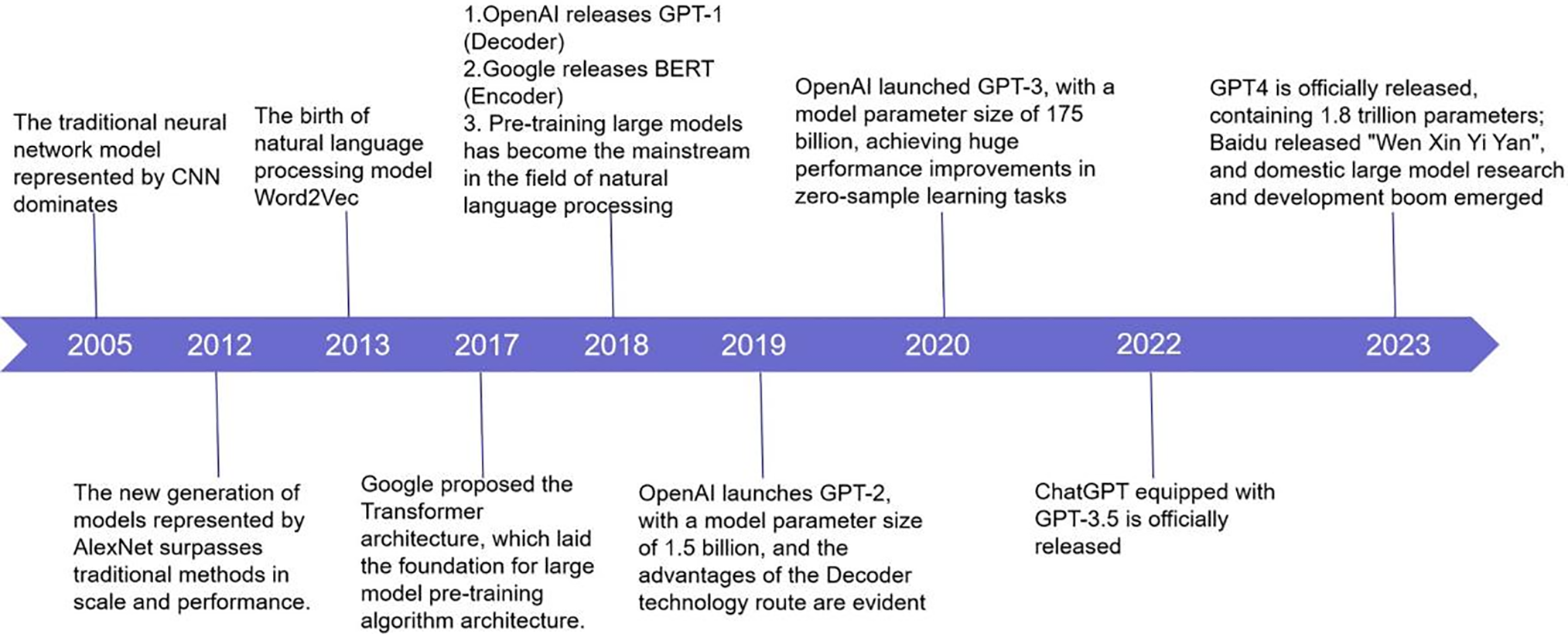
Figure 3: The evolution of large models
In Section 2.2.1, we will discuss in detail how the Transformer architecture lays the foundation for a large model pre-training algorithm architecture. Section 2.2.2 will explore the development of the GPT series models, especially the release of GPT-1, GPT-2, and GPT-3, and their performance in natural language processing tasks. Finally, in Section 2.2.3, we will examine current challenges and future directions, while referring to the timeline in Fig. 2 to highlight where these discussion points fit into the evolution of large-scale models.
2.2.1 From Transformer to Large Model
The Transformer architecture represents a paradigm shift in large model development. Departing from traditional Bidirectional Recurrent Neural Networks(RNNs)/Convolutional Neural Networks(CNNs), its purely attention-based design enables parallel sequence processing and superior training efficiency. The architecture’s core innovation-the self-attention mechanism-establishes direct dependencies between arbitrary sequence positions (see Fig. 4). Transformer achieves global modeling of long texts through the attention mechanism and is widely used in medical NLP tasks such as case understanding and symptom analysis [17]. Compared with traditional models such as RNN, its parallel processing capability and modeling efficiency are more suitable for building high-performance medical large models (such as GPT series, BERT series, etc.) [18].
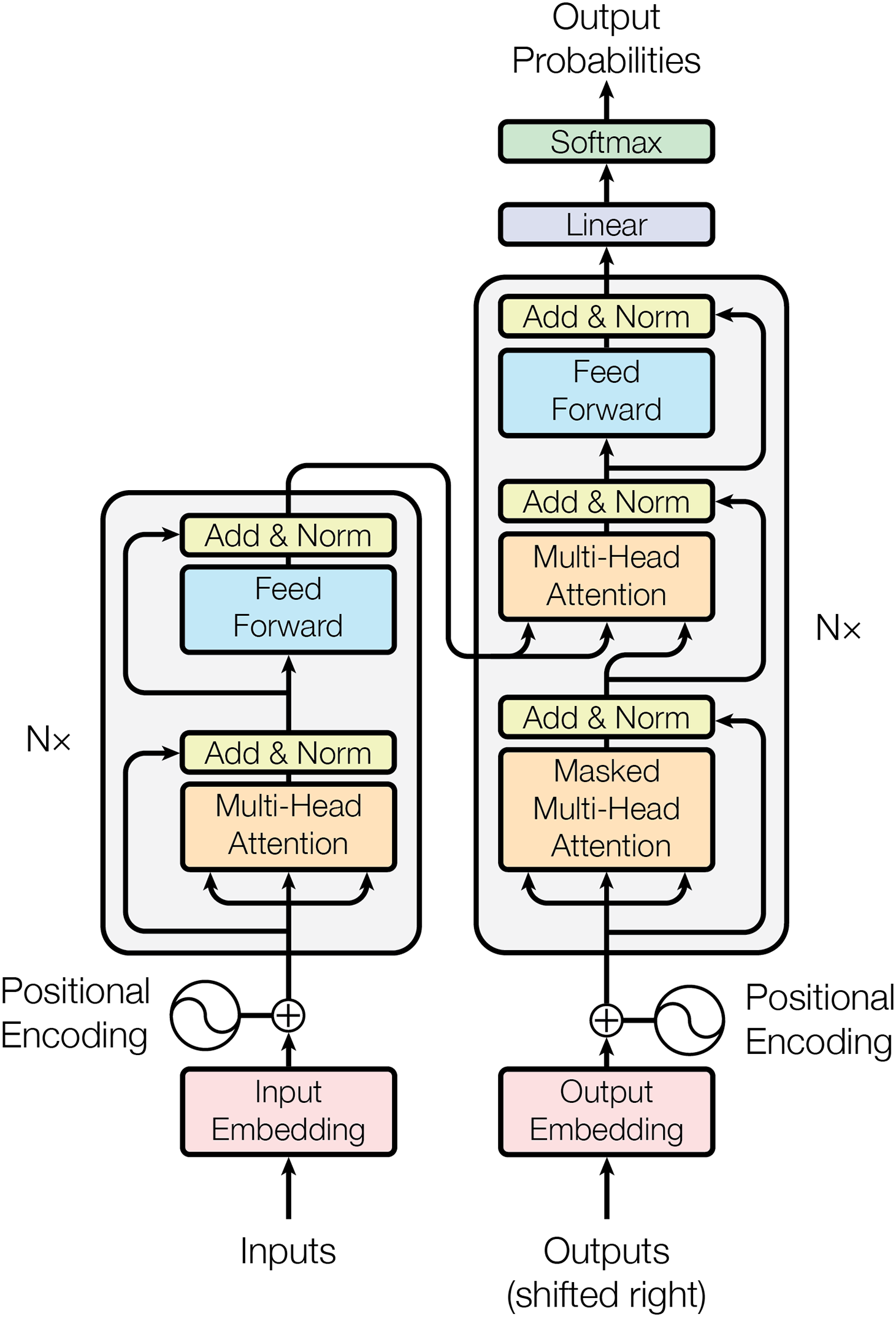
Figure 4: The Transformer’s model architecture
The introduction of the Transformer model provides a theoretical basis and architectural support for building large models [19]. Since Transformers can effectively handle a large number of parameters without being affected by the gradient vanishing or exploding problems in traditional models, researchers began to explore the possibility of improving performance by increasing the size of the model [20].
The Transformer architecture has achieved remarkable success through implementations like GPT (Generative Pre-trained Transformer) [21]. The initial GPT-1 model established the effectiveness of Transformer-based pre-training across diverse NLP applications. This breakthrough was followed by progressively scaled versions—GPT-2 and GPT-3—with the latter’s unprecedented 175 billion parameters demonstrating exceptional few-shot learning and natural language comprehension capabilities [22,23].
In 2018, the research community introduced BERT (Bidirectional Encoder Representations from Transformers), which revolutionized language model pre-training through its innovative Masked Language Modeling (MLM) and Next Sentence Prediction (NSP) objectives. These training mechanisms enable the model to learn deep bidirectional language representations. BERT’s success not only advanced large model development but also established foundational techniques that influenced nearly all subsequent language model architectures in NLP research [24].
With the success of BERT and GPT series models, large models have begun to expand their applications in various fields, including healthcare, financial analysis, etc. These models can adapt to and solve problems in specific fields by fine-tuning on data in specific fields [25]. At the same time, this has also brought about discussions on model generalization ability, data privacy, and ethical issues.
The introduction of the Transformer architecture and the progress of BERT and GPT have greatly promoted the development of the field of artificial intelligence, especially in the construction and application of large-scale models. However, with the increase in model size, new challenges have also been brought about in terms of computing resources, model interpretability, and ethical issues. Future progress in large models needs to not only boost performance but also resolve these accompanying problems. This includes developing more efficient training algorithms, reducing the carbon footprint of the model, improving the interpretability of the model, and ensuring that the decision-making process of the model meets ethical standards. Through these efforts, large models are expected to contribute to the sustainable development of society while promoting the advancement of artificial intelligence technology.
2.2.2 Development of GPT Series Models
The GPT series of models is a large-scale pre-trained language model based on the Transformer architecture launched by OpenAI. Since 2018, the GPT series of models has undergone several important iterations, and each iteration has achieved significant improvements in model scale and performance [26,27].
GPT-1: The first model in the GPT series, which introduced a Transformer-based pre-trained language model and demonstrated its potential in various NLP tasks. GPT-1 is trained by predicting the next word in a text sequence, using a large amount of book and web page data as training material [28].
GPT-2: Based on GPT-1, GPT-2 significantly expanded the model size, with the number of parameters reaching 1.5 billion. GPT-2 uses a more diverse data source during training, including Wikipedia, books, and web pages, which further improves its ability to understand and generate text [29–31].
GPT-3: As a significant advancement in the GPT lineage, GPT-3 marked a breakthrough with its unprecedented scale, boasting 175 billion parameters, ranking it among the world’s largest language models upon release [32]. Beyond sheer size, GPT-3 demonstrated substantial performance gains, particularly in few-shot learning and natural language comprehension. Its versatility enabled zero-shot applications like translation, text summarization, and Question Answering(QA) systems, achieving competitive results without task-specific fine-tuning.
GPT-4o: GPT-4o is OpenAI’s latest model in the GPT series, released on May 13, 2024, where “o” stands for “omni,” highlighting its omnipotent multimodal capabilities. GPT-4o not only processes text, but also can understand and generate voice and visual content, providing a more natural interactive experience. It has a fast response speed, averaging 320 milliseconds, close to the speed of human conversation, and reduces costs by 50% and increases speed by 2 times, significantly improving efficiency [33,34]. In terms of security, GPT-4o addresses the risks of new modalities by filtering training data and adjusting model behavior. The launch of this model heralds an important step forward in artificial intelligence’s simulation of human interaction and demonstrates a new trend in the future development of AI technology. Hurst et al. provided a detailed system card for GPT-4o, highlighting its advanced capabilities and potential applications in various fields, including healthcare.
The progression of the GPT model series has not only accelerated breakthroughs in natural language processing but also unlocked novel potentials for diverse AI applications [35]. As the model size increases, the GPT series models have demonstrated outstanding capabilities in handling complex language understanding and generation tasks [36]. However, as the size of models grows, new challenges also arise, such as the demand for computing resources, model interpretability, and ethical issues. Future research needs to address these accompanying problems while improving model performance to ensure the sustainable development of the technology.
2.2.3 Existing Limitations and Prospective Developments
The application of large-scale models in healthcare presents significant opportunities but is also accompanied by some challenges that need to be addressed through future research and practice [37]. Although large models have great potential in the medical field, they still face challenges such as computing resource consumption, data privacy, and model interpretability. Professor Shen pointed out that efficient training strategies and algorithm optimization are the key to improving training efficiency [38]. Since medical data is highly sensitive, ensuring its privacy and security has become a prerequisite for the actual deployment of the model. For instance, Demelius et al. provide a systematic review of differential privacy in centralized deep learning, outlining improvements in noise calibration and training efficiency [39]. A critical challenge lies in ensuring algorithmic interpretability, particularly for clinical applications where decision transparency is paramount. The medical community requires comprehensible rationales behind model outputs to establish trust and facilitate appropriate adoption in healthcare settings. At the same time, data quality and diversity are also important factors affecting model performance. Data bias may lead to errors in model output, affecting fairness and accuracy. Training data regional bias is a significant limitation of current large-scale models. Most models are trained on large public datasets from North America or Europe, which may result in poor performance in other regions (such as Africa, Asia, etc.). For example, language differences, varying disease spectra, and diverse medical practices can reduce the model’s generalization ability, thus limiting its fair application worldwide. Additionally, training and deploying large-scale models require substantial computational resources, leading to significant carbon emissions. Studies have shown that the energy consumed to train a model similar in scale to GPT-3 is equivalent to the lifetime carbon emissions of a car. To achieve sustainable development, future research should explore green AI strategies, including model compression, knowledge distillation, and more efficient hardware. Finally, regulatory compliance and technical integration issues are also challenges that need to be overcome in large-scale model applications [40]. The strict regulatory requirements in the medical field and the technical integration issues of existing systems need to be fully considered during the development and deployment of the model.
Faced with these challenges, future research needs to explore in multiple directions [41,42]. First of all, enhancing model interpretability is the key to improving clinical deployment of AI models. Researchers need to develop new techniques to improve the interpretability of LLMs and make their decision-making process more transparent. Secondly, interdisciplinary collaboration is also an important way to promote the role of advanced deep learning systems in healthcare and medical applications. Through the collaboration of experts in computer science, data science, medicine, and other fields, we can jointly solve the problems of applying large models in the field of healthcare. In addition, technological innovations, including algorithm optimization, computing efficiency improvement, and the development of new hardware, will promote the application of large models in the healthcare field. At the same time, updating and improving relevant ethical standards and laws, and regulations to ensure that the application of large-scale models complies with ethical standards and legal requirements is also an important direction for future research.
As large-scale models become more widely used in healthcare, ethical and regulatory compliance becomes increasingly important. This requires that patient privacy, data security and compliance must be taken into consideration during the development and application of the model. Global cooperation is also an important direction for the future, especially when facing global public health challenges such as epidemics, which require global cooperation and sharing of data and models to improve response capabilities. In addition, considering the environmental impact of large models and developing more energy-efficient training and operation methods to achieve sustainable development is also a key issue in the future. Overcoming these limitations could enable large-scale models to enhance healthcare delivery significantly, ultimately improving patient outcomes.
To address the challenges of medical data privacy and model interpretability, researchers have proposed a number of specific technical solutions and achieved initial results in the medical field [43].
In terms of privacy protection, Federated Learning is widely used in medical artificial intelligence systems. This method allows models to be trained on local devices without the need to centrally store sensitive data, thereby effectively reducing the risk of data leakage. For example, in a diabetic retinopathy screening project carried out by Google in cooperation with medical institutions, federated learning was successfully used to improve model performance without collecting user data [44]. In addition, Differential Privacy, as a mechanism for adding random noise, is also used in medical text modeling to prevent the reverse inference of individual data [45].
In terms of model interpretability, explainable artificial intelligence (XAI) methods such as LIME (Local Interpretable Model-Agnostic Explanations) and SHAP (SHapley Additive Explanations) are integrated into medical decision support systems to help doctors understand model outputs. For example, SHAP is used in breast cancer prediction and critical care scoring models to demonstrate how the model makes risk judgments based on specific physiological indicators [46]. In addition, models that combine attention mechanisms with medical ontology structures (such as Clinical Attention Networks) are emerging, which can highlight key features and cross-validate with medical knowledge graphs, thereby enhancing the medical rationality of explanations.
As technology evolves, future research should continue to promote the deep integration of privacy-enhancing technologies (PETs) and interpretable frameworks to enhance the security and trust of large models in actual medical scenarios.
2.3 Literature Selection Methodology
To ensure the rigor of the research method, this paper adopts the systematic literature screening process recommended by the PRISMA guidelines (see Table 1). The literature published between 2005 and 2023 was searched in five major databases (PubMed, IEEE Xplore, Web of Science, China National Knowledge Infrastructure, and arXiv) using a combined search formula ((“big model” OR “LLM”) AND (“medical” OR “clinical”)), which is consistent with the big model development timeline shown in Fig. 3.
3 Integration of Medicine and Large Models
3.1 Application of Large Models in Medicine and Healthcare
The applications of large models in medicine and healthcare span across various domains, including medical data management, healthcare services, preventive medicine, and medical device innovation, providing novel possibilities for medical research, clinical practices, and patient care [47]. These application areas can intersect, forming comprehensive medical solutions, enhancing healthcare services for patients, expediting medical research, and improving the efficiency of healthcare, as illustrated in Fig. 5. Based on this, Table 2 sums up the existing papers, with four areas of application highlighted below.
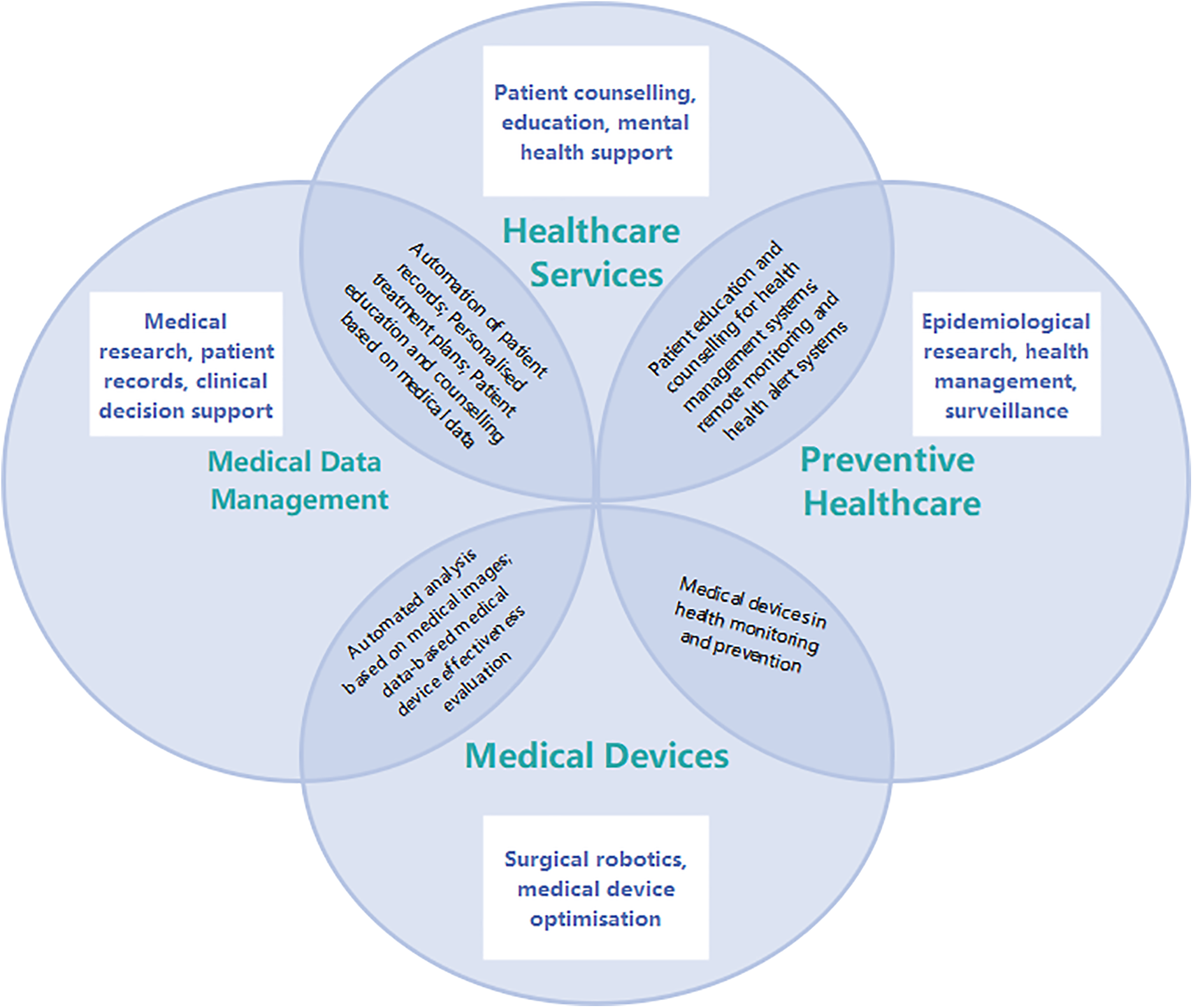
Figure 5: Framework of the application of the large model in medicine and healthcare
In order to improve the systematicness and comparability of the literature review, we classified the existing research according to its core tasks. The classification criteria are as follows: (1) If the research focuses on clinical text, image or multimodal data processing, it is classified as “medical data management”; (2) If the research focuses on improving patient service experience or supporting clinical processes, it is classified as “health care services”; (3) If the purpose of the research is disease prediction, epidemic monitoring or health risk intervention, it is classified as “preventive medicine”; (4) Research involving physical tools or systems such as medical equipment, surgical assistance, and remote monitoring is classified as “medical devices”. Some literature involves multiple application directions and may be checked in multiple categories [48]. Table 2 shows the distribution of representative studies under the above classification framework.
The progress of artificial intelligence heavily depends on high-quality data. However, in clinical AI research, a major challenge is the scarcity of large-scale, accurately annotated training datasets, which hinders further development [64]. The application of large models in handling healthcare data is increasingly gaining attention [49–51]. Large models can govern, analyze, and apply healthcare data using cutting-edge methods and technologies, forming multimodal databases that include various data types such as scales, text, images, waveforms, and omics. Recent advances in clinical concept annotation, such as the active transfer learning approach proposed by Abbas et al., demonstrate how contextual word embeddings can improve the efficiency of extracting structured information from unstructured medical records [65]. Here are some ways in which large models are constructed and utilized for multimodal healthcare databases [2]:
• Association of Text and Image Data: Large models can analyze textual medical records, such as case histories and reports, along with medical images like CT scans or X-ray images [66]. This allows doctors and researchers to understand the patient’s condition more easily, enabling accurate diagnosis and treatment.
• Annotation of Images and Text: Large models can be used to add textual descriptions to medical images, making them more understandable and manageable. This is useful for building image databases and research, enhancing the accessibility of medical images [67]. As shown in Fig. 6, the integration of multimodal data through large models enables comprehensive annotation and analysis, which is critical for improving medical image interpretability.
• Multimodal Analysis: Large models can correlate and analyze information from different data types, assisting doctors in identifying potential health issues in patients [68]. For example, combining genetic information, clinical data, and medical images can provide more comprehensive diagnostic and treatment recommendations.
• Data Integration and Interaction: Large models can integrate multiple data sources to construct comprehensive patient profiles. This helps healthcare professionals better understand a patient’s medical history and condition, enabling wiser decision-making.
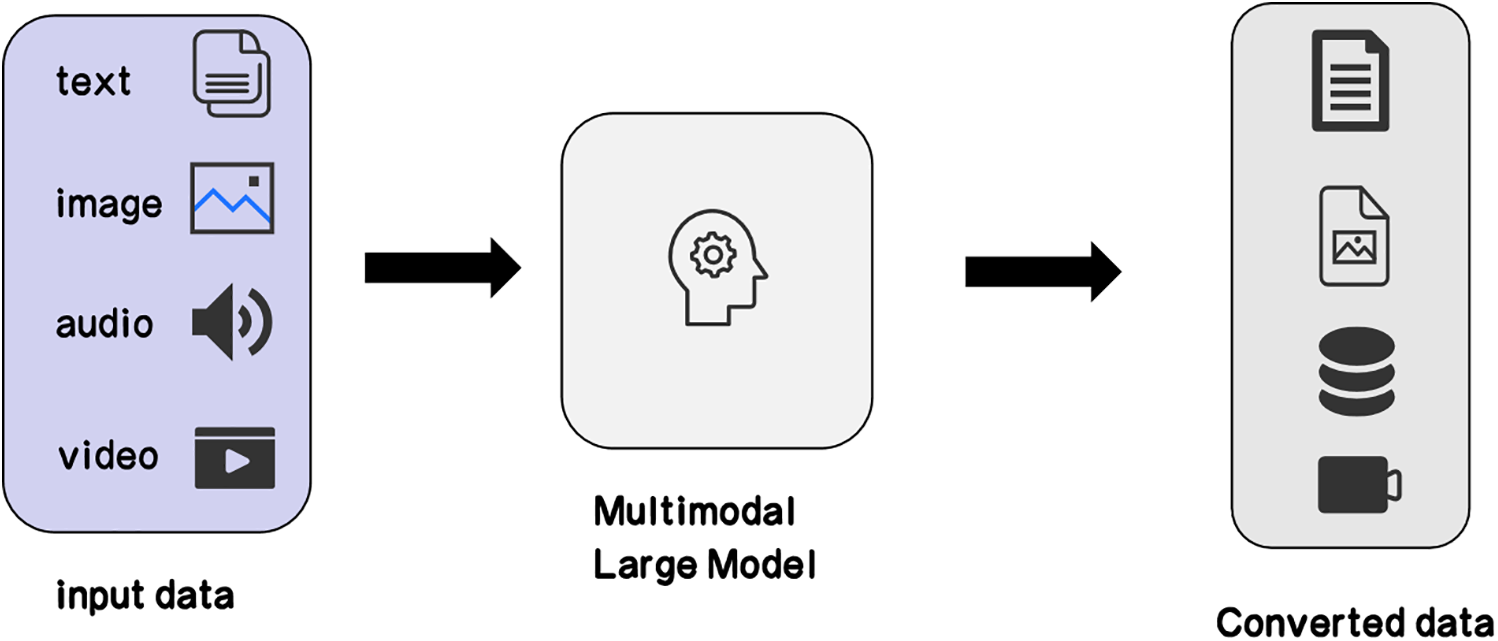
Figure 6: Multimodal large language model
In addition, the exponential growth of medical publications poses significant challenges for healthcare professionals to stay current with cutting-edge research and evolving clinical standards. Large models can automatically read, understand, and summarize medical literature, providing timely information to assist doctors in making wiser diagnostic and treatment decisions [69]. Furthermore, by analyzing patients’ clinical data, genetic information, and lifestyles, large models can help doctors formulate personalized treatment plans. This can improve the effectiveness of treatment and reduce unnecessary medications or therapies. They can also analyze large-scale health data to identify potential epidemic risks and provide early warnings. This is crucial for healthcare institutions and government decision-makers to better prepare for and respond to epidemic outbreaks.
The use of large models in the healthcare sector provides new opportunities to enhance efficiency, accuracy, and accessibility of healthcare [52]. Large models can be utilized to build intelligent healthcare assistants, answering patient queries, providing diagnostic reports, improving recommendations, and even explaining medical reports [53]. Recent clinical trials have demonstrated that AI-powered diagnostic tools, such as mobile phone-based skin cancer analysis, can achieve comparable or even superior accuracy to human specialists in certain scenarios [70–72]. This enhances the accessibility and efficiency of healthcare. The potential for utilizing large language models in assisting medical education and employing large language models (such as ChatGPT) in general practice knowledge testing falls within the healthcare domain [54,55]. These studies explore how advanced language model technology can elevate medical education and its practical applications, introducing new possibilities to the healthcare [73]. Huang highlighted the transformative potential of multi-modal LLMs (e.g., ChatGPT) in dental practice, demonstrating their capacity to improve diagnostic accuracy and therapeutic efficacy [74]. Then Alqahtani et al. highlighted the emergent role of AI and LLMs in higher education and research, demonstrating their potential to transform medical education and research practices [75]. These applications aim to improve medical professional training and enhance the efficiency of healthcare.
In everyday healthcare, large models can be employed to monitor individuals’ health, providing early warnings to prevent health risks or complications that may arise during the recovery process [76]. Large models can contribute to the development of intelligent health assistants or virtual doctors to answer personal health-related queries and provide medical information and advice [77,78]. Recent studies, such as Hua and Eastwood, highlight the growing role of LLMs in mental health care, demonstrating their ability to assist in psychological counseling, symptom screening, and personalized therapy recommendations [79,80]. This aids in enhancing public health knowledge, helping individuals better understand diseases, symptoms, and preventive measures. Through smartphone apps or wearable devices, large models can assist individuals in recording and monitoring their health data, such as step count, heart rate, and sleep quality. This helps people better understand their health status and identify potential problems immediately. By analyzing personal health data, large models can assist individuals in tracking health trends, such as weight management, blood sugar control, and blood pressure management. This facilitates the creation of suitable health plans. Moreover, large models can provide personalized dietary advice based on individuals’ dietary preferences, health goals, and nutritional needs, promoting healthy lifestyles [81]. For patients in the recovery phase, large models can be used to monitor their recovery progress. By analyzing lifestyle data, exercise data, and biological indicators, the model can offer customized feedback and suggestions to help individuals achieve their recovery goals and predict potential outcomes of the recovery process. This aids healthcare professionals and patients in understanding long-term recovery trends and adjusting treatment plans based on these predictions. During the recovery process, large models can assist patients in managing medications, providing medication reminders, monitoring medication adherence, and addressing questions related to medication therapy. Additionally, large models can offer psychological support to patients in recovery. They can answer questions about emotional health, coping with stress and anxiety, or provide resources and advice to help patients address emotional and mental health issues.
Large models analyze CT, Magnetic Resonance Imaging (MRI) and other medical images to identify anatomical structures and provide navigation and path planning support for intelligent surgical robots. They can assist in developing personalized surgical plans, optimizing incision locations and instrument trajectories, and thus improve surgical accuracy and safety [56]. They can identify and segment a patient’s anatomical structures, including vessels, nerves, and tissues, providing accurate navigation and target localization for surgical robots. Based on personalized anatomical structures, large models can assist in planning surgical paths and operational steps. This involves determining optimal incision locations, organ positioning, and the best trajectories for tool movement. This helps predict potential difficulties and risks in advance, optimizing surgical plans. Surgical robots equipped with sensors and cameras provide real-time feedback of the surgical scene. Large models can analyze this data, assisting surgical teams in understanding the surgery’s progress in real-time and making adjustments based on patient physiological features [82,83]. This real-time intelligent feedback helps avoid potential risks and enhances surgical safety [84]. Utilizing the computational capabilities of large models, surgical robots can more accurately locate a patient’s anatomical structures. This is crucial for minimally invasive surgeries, precise cutting, and suturing. Through real-time navigation, surgical teams can execute surgical operations more accurately. Large models can optimize the control systems of surgical robots. Using deep learning algorithms, the model can learn and adapt to the physiological changes of different patients, enabling more precise and intelligent control of surgical tools, ensuring the best treatment outcomes [85]. Virtual reality scenarios created using large models allow doctors and surgical teams to undergo high-quality, realistic training [57]. This helps improve surgeons’ skills, reduce operation risks, and provides a platform for the application of new technologies and instruments. Large models can analyze extensive surgical data, extract lessons learned, and help improve surgical techniques and processes. This data-driven approach contributes to establishing safer and more efficient surgical standards.
Large models play a key role in optimizing medical devices, covering aspects such as design, performance optimization, and user experience enhancement. Large models can be used to simulate and optimize the design process of medical devices [86]. Recent advancements in AI-driven training tools have shown significant potential in the education of emergency medicine doctors, where realistic simulations and virtual environments can improve diagnostic accuracy and clinical skills [87]. For example, in the design of imaging diagnostic devices, the model can consider factors such as patient anatomy and organ structure, optimizing the geometric construction of X-ray or magnetic resonance imaging devices to improve image quality and diagnostic accuracy. Large models can simulate the performance of medical devices, including signal processing, image reconstruction, sensor response, and more. By simulating different working conditions and parameter settings, the performance of the device can be optimized to ensure outstanding performance in various practical scenarios. Large models can analyze the user interface of medical devices, providing improvement suggestions to optimize the user experience. This includes simplifying operation procedures, optimizing the layout of control buttons, improving alarm systems, and reducing the workload and risks for healthcare personnel. For imaging diagnostic devices, large models can be used to optimize image processing algorithms, enhancing image clarity, contrast, and even assisting in automatic anomaly detection. This helps improve the diagnostic accuracy and speed of doctors. Large models can evaluate the energy consumption of medical devices, propose energy-saving solutions, and design more environmentally friendly devices. This is particularly important for devices that operate for extended periods, such as those in operating rooms, as it reduces energy expenses and complies with environmental standards. Large models can simulate the performance changes of medical devices after prolonged use and provide optimization suggestions to extend the device’s lifespan. Additionally, the model can analyze the maintainability of the device, offering designs that are easier to repair and maintain.
Overall, large models provide powerful computational and learning capabilities for the development of intelligent surgical robots, improving surgical precision and safety. This not only helps enhance patient treatment outcomes but also drives technological innovation of surgical medicine. Furthermore, the application of large models in optimizing medical devices not only improves device performance and reliability but also drives innovation in medical technology, providing a better medical experience for patients and healthcare professionals. As technology advances, it is essential to ensure the interpretability of models and their compliance with medical regulations.
The application of large models in epidemiology and genetics covers a wide range of scenarios, with the primary advantage lying in the handling and analysis of large-scale medical data to better understand disease spread, predict disease trends, and formulate effective public health strategies [58]. Here are detailed applications of large models in areas like epidemiological research:
• Disease Spread Prediction: Large models can analyze extensive medical data, including patient records, symptoms, and treatment plans, to predict the spread trends of diseases. This is crucial for early detection and response to emerging viruses or disease pandemics [88].
• Infectious Disease Modeling: Large models can be used to construct models simulating the spread of viruses or bacteria within populations. This helps understand the transmission pathways, speed, and scale of diseases, providing a scientific basis for public health decisions.
• Real-time Epidemic Monitoring: By monitoring medical data, social media information, and other relevant data in real-time, large models provide timely epidemic monitoring. This continuous surveillance enables prompt intervention and timely execution of containment strategies to mitigate disease transmission.
• Optimizing Epidemiological Surveys: Large models assist in designing and optimizing epidemiological surveys, improving data collection efficiency. Through deep learning, models can identify potential risk factors and correlations from multiple data sources, offering a more comprehensive perspective for disease research.
• Assessing Treatment Efficacy: During outbreaks, large models can assess the effectiveness of different treatment options, including drug treatments and vaccinations. By analyzing patient responses and disease progression, models can provide better treatment recommendations for clinical doctors.
• Risk Assessment Modeling: Large models can integrate multidimensional data to build risk assessment models, providing early warnings of potential public health risks [89]. This helps decision-makers take necessary preventive and control measures before an outbreak.
• Genomic Data Analysis: Large models can combine genomics data to study the correlation between individual genes and infectious diseases. This enhances the understanding of why certain individuals may be more susceptible or recover more easily.
The use of large models in epidemiological research provides the medical community with deeper and more comprehensive insights, aiding in the better understanding of diseases, optimizing medical decisions, and providing a scientific basis for public health work [59,60]. Recent advances in AI-driven drug repurposing (e.g., Singh) demonstrate the potential of large models to accelerate therapeutic discovery for infectious diseases by screening existing drug libraries against pathogen targets [90].
With the in-depth application of AI technology in the healthcare field, large model platforms have become key drivers of progress. Through comprehensive medical data synthesis and analytical processing, these platforms enable enhanced clinical decision support, facilitating accurate diagnostics and optimized treatment strategies. In the literature screening of platform applications, studies on publicly available large-scale model-based healthcare platforms were included, focusing on precision diagnosis and treatment optimization. Exclusion criteria included: 1) studies where the platform did not report clinical effects; 2) studies that did not involve specific medical tasks or data applications. This section aims to explore the specific applications of large model platforms in medicine and healthcare, and how they improve the quality of medical services by offering precise diagnostic support, optimizing treatment plans, and enhancing patient care.
Although large general models such as GPT-3 possess powerful language understanding and generation capabilities through pre-training on massive datasets [91], their performance and credibility in the medical domain are still subject to limitations due to the highly specialized and sensitive nature of medical data. To better address the needs of medicine, models specifically designed for healthcare have emerged. These models leverage extensive medical data, including clinical records, medical literature, and medical imaging, during the pretraining phase to gain a better understanding of medical terminologies, complex disease relationships, and medical practices [92]. In the fine-tuning phase, models designed for medicine collaborate with healthcare professionals to learn more accurate clinical judgments and decision-making processes.
One such model is GatorTronGPT, developed for clinical applications using the GPT-3 architecture with 200 billion parameters [5]. Evaluating its performance in medical research and healthcare, with a focus on key functionalities in text generation, revealed that GatorTronGPT achieved state-of-the-art performance on four biomedical NLP benchmark datasets (out of six evaluated). This demonstrates the benefits of GatorTronGPT in biomedical research, generating synthetic clinical text for the development of synthetic clinical NLP models (GatorTronS). Evaluations by medical professionals indicate that GatorTronGPT can generate clinically relevant content with language readability comparable to real-world clinical records. Recent studies propose hybrid evaluation frameworks combining human expertise and automated metrics (e.g., Sblendorio et al.) to dynamically assess LLMs’ clinical feasibility, which aligns with the validation approaches of domain-specific models like GatorTronGPT [93].
This development brings about more precise and reliable applications of artificial intelligence in medicine and healthcare, supporting doctors in making more accurate choices in diagnosis, treatment, and decision-making [61]. Models designed specifically for medicine can not only handle the complexity of medical knowledge but also better adapt to the uniqueness of medical practices [94,95]. This customized approach is expected to enhance the efficiency of healthcare, reduce error rates, and provide patients with more personalized and precise medical services. In the future, as concerns about medical data privacy and security continue to rise, the development of models specifically designed for medicine will likely yield more significant results driven by the advancement of medical artificial intelligence. Fig. 7 provides a brief summary illustration, showcasing the diverse applications and potential impact of large models in medicine and healthcare.
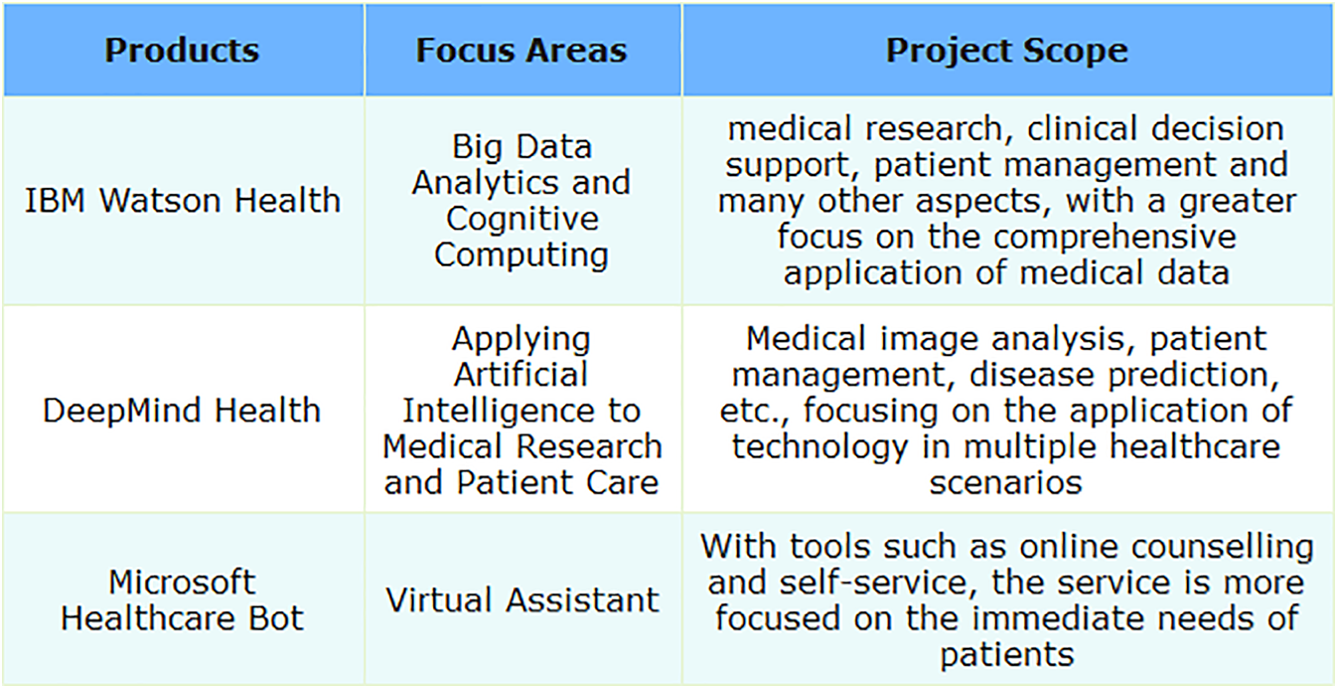
Figure 7: Large model platform designed for medicine
IBM’s healthcare-focused division, Watson Health, utilizes cutting-edge AI and cognitive computing to foster advancements in medical services and enhance industry standards. The platform integrates technologies such as big data analytics, machine learning, and deep learning to process and analyze extensive medical datasets, including clinical data, genetic information, and medical images. IBM Watson Health aims to provide comprehensive solutions to empower healthcare professionals in making more accurate diagnoses, devising personalized treatment plans, and supporting innovation in medical research and health management. By introducing advanced computing capabilities into medicine, IBM Watson Health strives to enhance patients’ medical experiences, reduce healthcare costs, and foster the collaborative development of the entire healthcare ecosystem. It includes multiple tools and solutions such as Watson for Oncology and Watson for Drug Discovery.
DeepMind Health is a department specifically dedicated to applying artificial intelligence to healthcare, covering various aspects including medical image analysis and patient data management. DeepMind Health is a branch of DeepMind, an AI company that is now a subsidiary of Alphabet, focusing on exploring and applying AI technologies to improve medicine. Established in 2016, the mission of DeepMind Health is to use advanced technologies, including deep learning and machine learning, to address complex issues in medicine and healthcare. The department’s research and projects cover areas such as medical image analysis, optimization of patient management systems, disease prediction, and treatment. Notably, DeepMind Health has achieved significant milestones in medical image recognition and processing and has collaborated closely with healthcare professionals in various medical projects, aiming to drive innovative applications of AI in healthcare. What’s more, DeepMind Health’s projects have sparked some controversy regarding medical data privacy, raising concerns about technology and medical ethics.
Microsoft Healthcare Bot is an intelligent robot platform designed specifically for healthcare, launched by Microsoft. The platform combines AI and NLP technologies to provide powerful virtual assistants for healthcare institutions and service providers, aiming to improve the interaction experience between patients and the healthcare system. Microsoft Healthcare Bot can be integrated into healthcare institutions websites, applications, and other digital channels. Through automated responses to common questions, providing health information, and facilitating functionalities like appointment scheduling, it effectively supports patients’ self-service and management. Its flexibility and customizability enable healthcare institutions to tailor the robot’s features according to their needs, better meeting patients’ personalized requirements, and enhancing the efficiency and accessibility of healthcare services. Microsoft Healthcare Bot is a comprehensive virtual assistant platform, offering a wide range of healthcare services with high customizability, allowing institutions to tailor features based on their specific needs and achieve integration across multiple digital channels.
This section introduces several large-scale platforms that have had a significant impact in the medical field. They mainly rely on advanced deep learning and artificial intelligence technologies and are applied to medical data analysis, disease prediction, patient management, and other fields. The following Table 3 summarizes the core technologies and practical applications of these platforms.
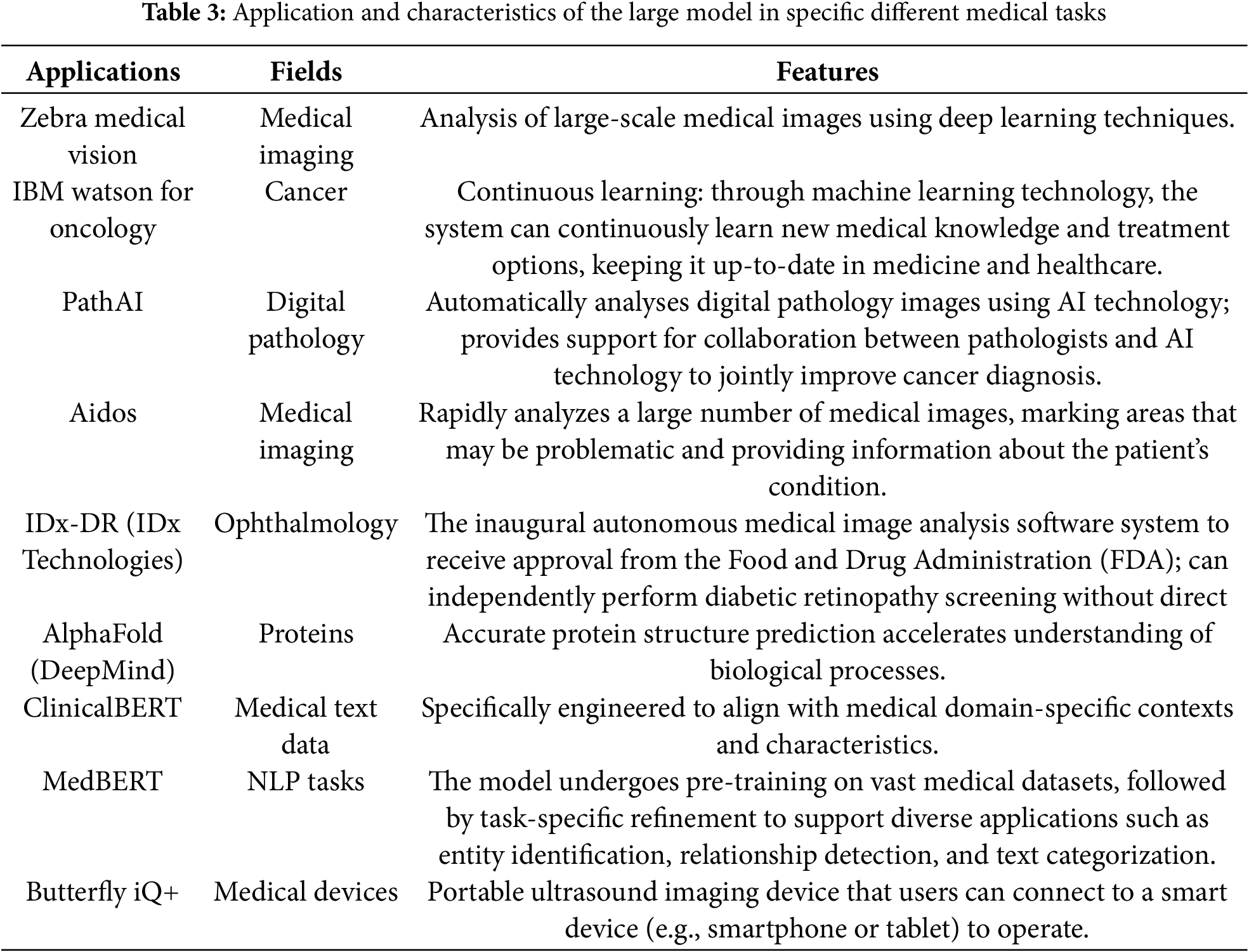
ClinicalBERT and MedBERT are pre-trained language models focused on the medical domain to enhance the efficiency of processing medical texts. Both ClinicalBERT and MedBERT are built on the BERT architecture, incorporating pre-training capabilities. ClinicalBERT is an extension of BioBERT, specialized in the biomedical domain, with a primary focus on clinical medical texts. In contrast, MedBERT aims to adapt more broadly to medicine, encompassing biomedical and clinical medicine. ClinicalBERT is primarily used for processing clinical medical texts such as electronic health records and clinical reports, while MedBERT aims for a broader adaptation, covering biomedical literature and clinical medical texts. Due to differences in their design objectives, their performance may vary across specific tasks, and performance comparisons typically require evaluations based on specific tasks and datasets. In summary, ClinicalBERT and MedBERT exhibit differences in design objectives, foundational datasets, and application scope, and the choice between them may depend on researchers’ needs and application scenarios.
Zebra Medical Vision employs deep learning technology to analyze medical images, providing automated diagnostic and screening tools for tasks such as breast cancer screening and lung nodule detection. Caption Health utilizes deep learning and artificial intelligence to offer intelligent ultrasound imaging, enabling physicians to conduct ultrasound examinations without the direct intervention of ultrasound specialists. The emergence of these platforms signifies advancements in large model technology within the healthcare domain, providing more advanced tools and methods for medical diagnosis and research.
Watson for Oncology is a specific application under the IBM Watson Health department. IBM Watson Health is dedicated to leveraging artificial intelligence and cognitive computing technologies to improve healthcare and life sciences. Watson for Oncology, focusing on cancer treatment, aims to provide personalized treatment recommendations to doctors based on patient medical records and extensive medical literature knowledge.
AlphaFold is a model designed for predicting protein structures and is a project by DeepMind, specializing in the biological domain. Its goal is to address significant biological challenges, such as the protein folding problem, using machine learning methods. AlphaFold aims to advance the scientific understanding of protein structures and provide more accurate predictions, which is crucial for drug discovery and disease understanding. AlphaFold has achieved notable success in the Critical Assessment of Structure Prediction (CASP) competition, gaining global attention and being considered a milestone on protein structure prediction.
4 Advantages of Large Models in Healthcare
4.1 High-Accuracy Diagnosis and Prediction
In the healthcare domain, achieving high-accuracy diagnosis and prediction is a crucial advantage of applying large models. This advantage has profound implications for elevating the standards of medical diagnosis and improving patient treatment outcomes. ChatGPT, particularly GPT-4, demonstrated significant comprehension of complex surgical clinical information, achieving an accuracy rate of 76.4% on the Korean Board of General Surgery exam [44]. As shown in Fig. 8 below, the overall accuracy of the GPT-3.5 was 46.8%, while the GPT-4 showed significant improvement with an overall accuracy of 76.4%. Through deep learning and extensive data analysis, large models exhibit a high degree of sensitivity and accuracy in handling medical images, physiological data, and clinical information. Large models, when analyzing patient data, can identify potential risk factors, assisting healthcare professionals in achieving early disease diagnosis and prediction. Consequently, this provides doctors with more reliable auxiliary information, enhancing the precision of medical diagnoses and the accuracy of predictions. Although large language models perform well in text generation, their “hallucination” phenomenon raises serious concerns in the medical context. Models may generate information that is inconsistent with actual medical knowledge, and if doctors are unaware of it, it may lead to misdiagnosis or treatment errors. This risk is particularly critical in clinical decision support systems. Therefore, rigorous validation should be carried out before deployment, supplemented by a manual review mechanism to ensure the accuracy and reliability of the information.
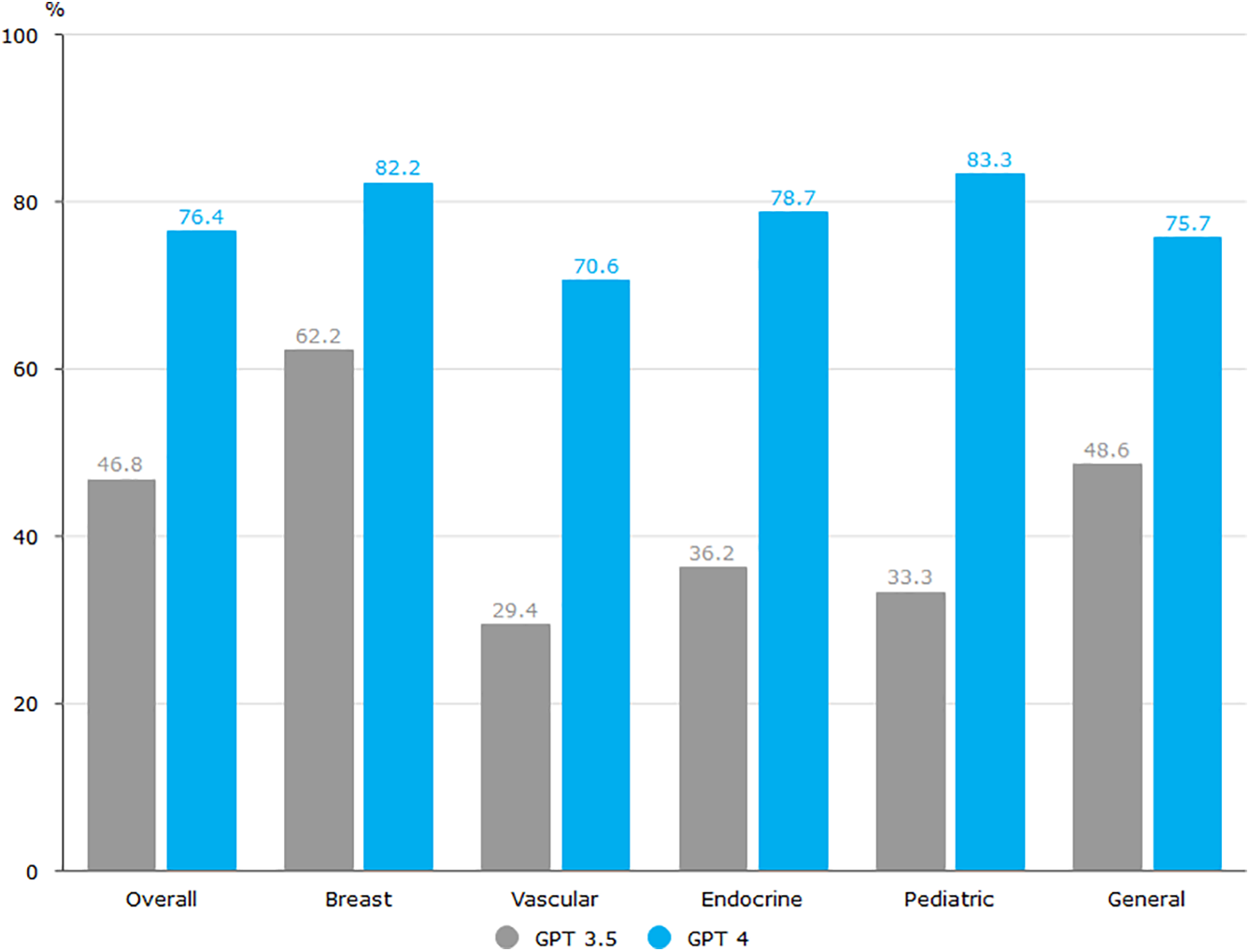
Figure 8: Comparative performance evaluation of CPT-3.5 and CPT-4 across surgical subspecialties
4.1.1 High Precision in Medical Data Analysis
Medical imaging, such as CT scans, MRI, and X-ray images, constitutes a crucial component of clinical diagnosis. Large models, leveraging deep learning techniques, can learn complex image features and patterns, thereby achieving high-precision diagnosis in medical image analysis [96]. Esteva demonstrated that deep learning-enabled computer vision systems can achieve expert-level performance in tasks such as radiology and pathology image interpretation, highlighting the transformative potential of AI in medical imaging [97]. For instance, in tumor detection, large models can accurately label and classify abnormal cells, assisting doctors in early detection of patient lesions and improving the early diagnosis rates of diseases like cancer [98]. Large, accurately annotated datasets are crucial for successful deep learning applications in medical imaging [99]. However, collecting such high-quality labels is particularly difficult for histopathology images due to their unique characteristics, including gigapixel resolutions, diverse cancer subtypes, and significant staining variations. Self-supervised learning (SSL) offers a viable alternative by learning meaningful representations directly from unlabeled data, which can then be adapted to various downstream tasks. Self-supervised learning has shown promise in medical image analysis, particularly in scenarios with limited labeled data. For instance, Ouyang et al. demonstrated that SSL can significantly improve few-shot medical image segmentation by leveraging unlabeled data to learn robust representations [100]. Additionally, data augmentation techniques play a crucial role in enhancing the performance of deep learning models by artificially increasing the size and diversity of training datasets. Visual representations based on Self-Representational Contrastive Learning (SRCL) not only achieved state-of-the-art performance on each dataset but also demonstrated robustness and transferability compared to other SSL methods and ImageNet pretraining (both supervised and self-supervised methods) [101].
While self-supervised pre-training with supervised fine-tuning has demonstrated success in image recognition with limited labeled data, its adoption in medical imaging remains underexplored. Azizi et al. [62] investigated this approach for medical image classification, evaluating performance on two distinct tasks: dermatological condition identification from photographs and multi-label chest X-ray interpretation [102]. Their findings revealed that domain-specific self-supervised pre-training following ImageNet initialization substantially enhanced classifier performance [103]. The researchers further proposed Multi-Instance Contrastive Learning (MICLe), which leverages multiple patient images to create enriched positive pairs for contrastive learning. Notably, the study also demonstrated the resilience of large self-supervised models to distributional variations.
Beyond medical images, large models can also process and analyze clinical text data, such as health records and doctors’ diagnostic reports. Through deep learning, large models can understand and extract key information from textual data, providing comprehensive patient information to healthcare professionals. This textual data analysis aids in accurately identifying a patient’s medical history, symptoms, and treatment feedback, thereby enhancing the overall assessment accuracy of patients.
Pathology reports serve as critical sources of clinical and research data, yet their unstructured narrative format poses significant challenges for qualitative data extraction. Automated keyword extraction offers an efficient solution for summarizing these complex documents and improving processing efficiency. Kim developed a supervised deep learning model incorporating natural language processing to identify three key categories of pathological information: specimen details, procedures, and pathology types [104]. Their study evaluated the model’s performance against conventional extraction methods using 3115 expert-annotated reports, then applied it to analyze 36,014 unlabeled reports with validation through biomedical terminology standards. The findings confirmed the model’s effectiveness in practical clinical data extraction scenarios.
4.1.2 Enhanced Predictive Capability through Multimodal Data Fusion
Large models possess the capability to handle multimodal data, combining medical images with patients’ genetic information and clinical records, among others. The fusion of such data provides a more comprehensive reflection of a patient’s condition, offering more accurate information for diagnosis and prediction [105]. Recent studies further demonstrate the effectiveness of deep learning-based multi-modal fusion in neurodegenerative disease classification, achieving high accuracy in Alzheimer’s disease diagnosis by integrating longitudinal MRI, PET, and clinical data [106]. Leveraging the capabilities of large models, they demonstrate unique advantages in early diagnosis and disease prediction [107]. By learning from extensive disease data, large models can identify potential disease signs and risk factors. For instance, combining imaging and genetic data, large models can predict the risk of certain genetic diseases, providing personalized health management recommendations for patients. Fu provided a comprehensive survey on the evaluation of multimodal large language models (MLLMs), highlighting the importance of multimodal data fusion in enhancing model performance and predictive capabilities [70]. In predicting cardiovascular diseases, large models can analyze various data aspects, including physiological indicators, lifestyle, and genetic information, helping physicians predict the probability that a patient develops a disease and take early intervention measures to reduce the risks of the disease.
Modern precision medicine increasingly relies on multimodal data integration to enhance clinical decision-making across diagnosis, treatment planning, and outcome prediction [108]. Common quantitative evaluation methods for multimodal large language models (MLLMs) have their limitations, making it difficult to comprehensively evaluate performance. To address these challenges, Fig. 9 illustrates a comprehensive multimodal data processing workflow, which systematically integrates medical data from heterogeneous sources (e.g., imaging, genomics, and clinical records) through critical stages including secure data collection, standardized preprocessing, domain-specific feature extraction, and adaptive fusion strategies. This framework not only highlights the synergy between data diversity and model robustness but also provides a scalable approach to enhance predictive accuracy in real-world clinical settings.
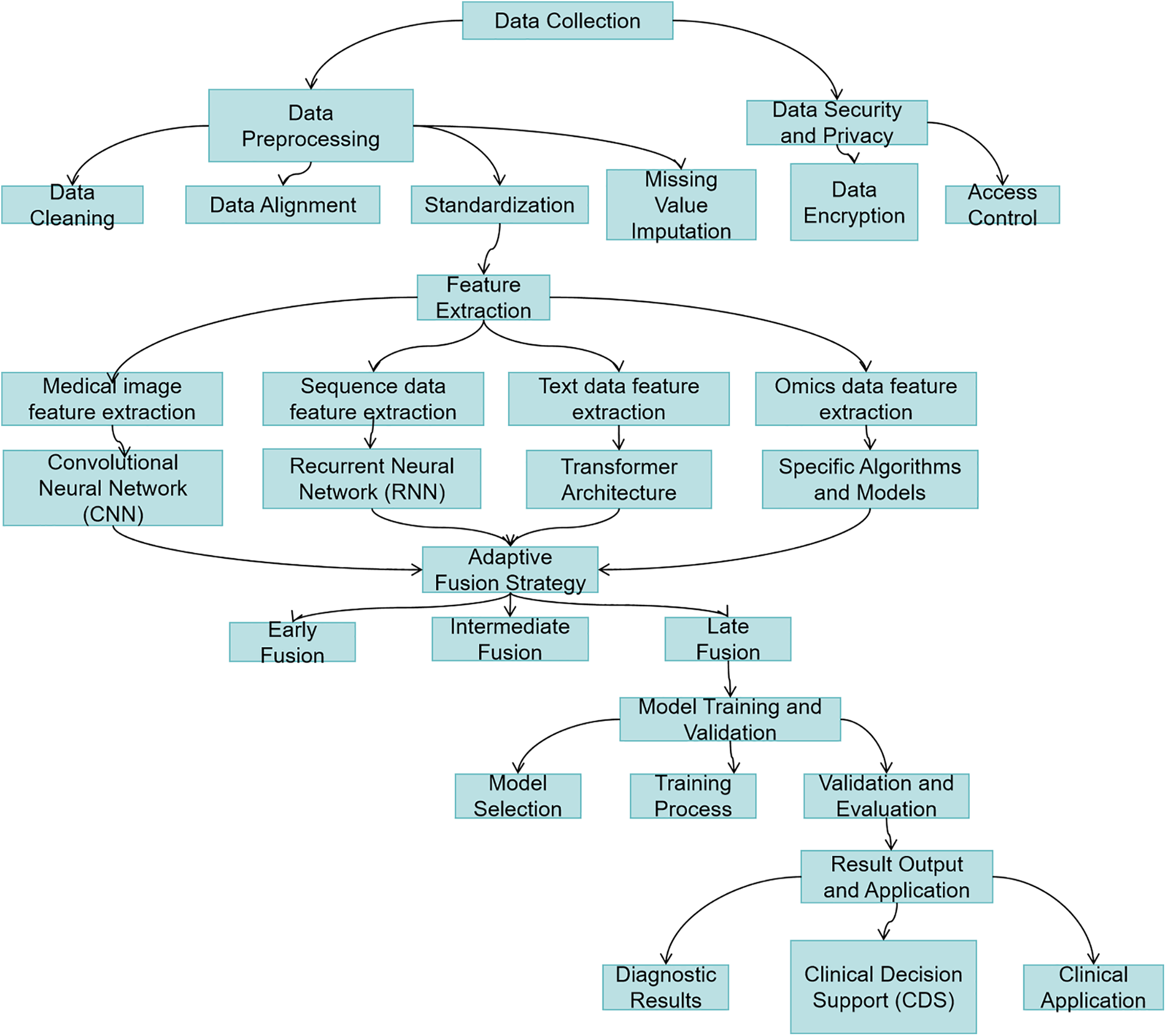
Figure 9: Multimodal healthcare data processing workflow
Multimodal Evaluation Benchmark (MME) is the first comprehensive Multimodal Large Language Model(MLLM) assessment benchmark with rich experiments on 14 subtasks to comprehensively assess MLLM, and Fig. 10 shows the results of three MLLM assessments [20]. Mohsen conducted a systematic review of AI-based multimodal data fusion in clinical settings, emphasizing the integration of electronic health records (EHR) with medical imaging [109]. Their work evaluated diverse fusion strategies, disease-specific applications, and machine learning algorithms, while also cataloging available multimodal datasets. The findings revealed that combined EHR and imaging models consistently surpassed unimodal approaches in diagnostic and predictive performance [110]. In a related study, the author developed an end-to-end deep learning model to predict neurodevelopmental impairments (cognitive, language, and motor) in 2-year-olds using multimodal MRI (T2-weighted, DTI, resting-state fMRI) and clinical data [111]. The model achieved prediction accuracies of 88.4%, 87.2%, and 86.7% for each domain, respectively, significantly outperforming single-modality baselines.
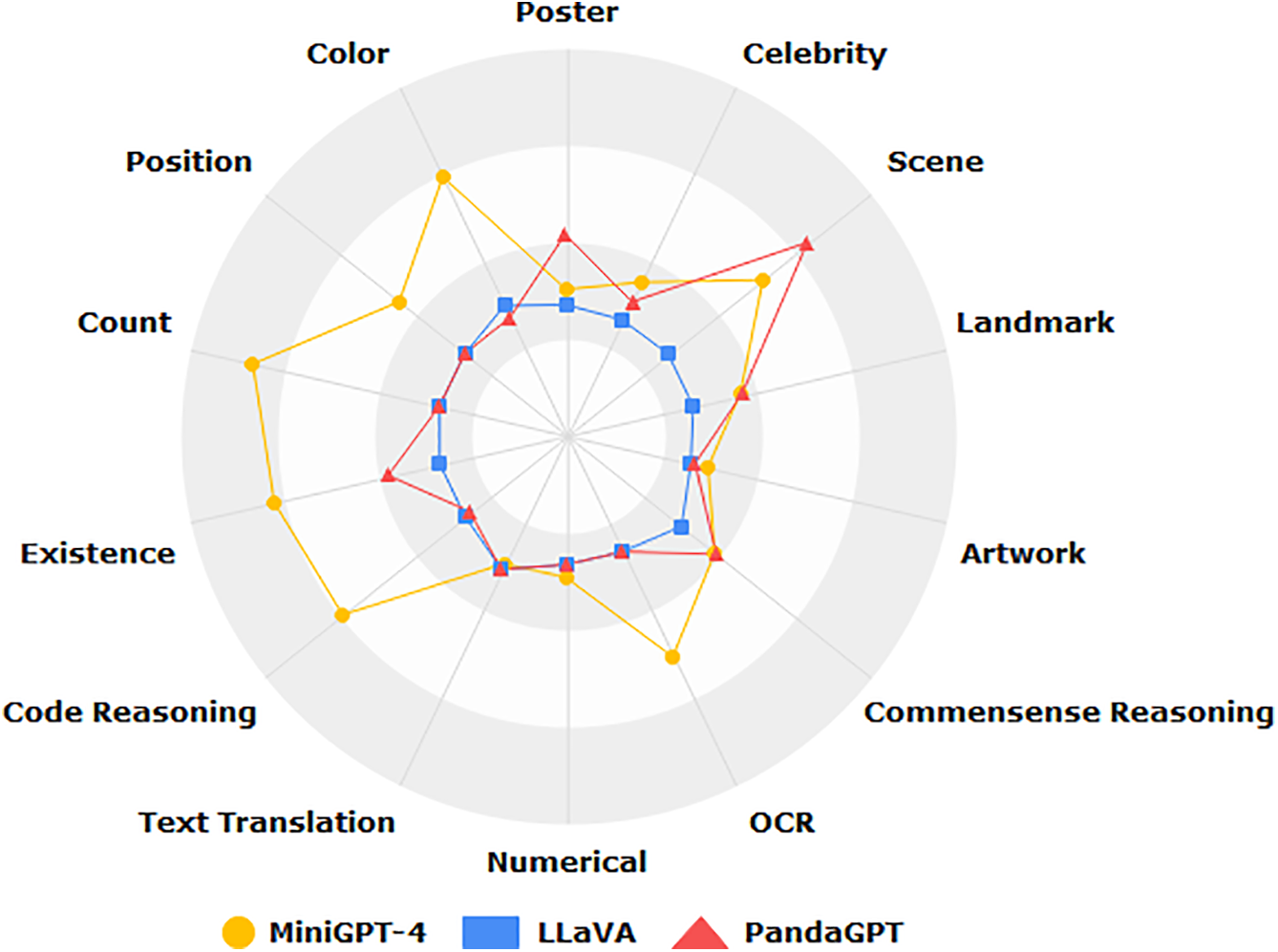
Figure 10: Benchmarking multimodal LLMs: a comprehensive evaluation across 14 vision-language tasks
Although multimodal data fusion technology has shown great potential in improving diagnostic accuracy and predictive ability, it still faces many challenges in actual clinical deployment and urgently needs in-depth discussion.
First, the interoperability bottleneck is a major obstacle to achieving multimodal data integration. Data generated by different hospitals or devices often have inconsistent formats and different collection frequencies, and lack common data standards and interfaces, which seriously limit the deployability of the model in a cross-institutional environment. In addition, the time alignment and semantic consistency issues between different modalities often make information fusion difficult, affecting model performance and stability.
Secondly, the annotation cost of high-quality multimodal data is extremely high. Clinical images, texts, genetic data, etc. require a large number of experts to manually annotate, but the resources of medical professionals are extremely limited. This not only increases the cost of research but also limits the scalability of the model in real clinical environments.
Thirdly, deployment in clinical environments is difficult. Large multimodal models usually require high-performance computing resources and complex integrated deployment solutions, and many primary care institutions are limited by technical infrastructure and find it difficult to support the operation of such models. At the same time, clinical staff lack understanding of the internal mechanisms of the model, which also exacerbates its distrust and usage barriers in actual applications.
In addition, real-time and data privacy issues also limit the implementation of multimodal fusion technology. Clinical decisions are usually highly time-sensitive, and the processing of multimodal models is relatively complex and may not meet the requirements of real-time feedback. At the same time, integrating multi-source data may increase the risk of privacy leakage, especially in a medical environment that lacks a sound data governance mechanism.
In summary, the future development of multimodal data fusion systems needs to focus on solving practical obstacles such as standardization, annotation automation, resource optimization, and compliance while improving algorithm performance, so as to achieve large-scale promotion and implementation in clinical practice.
4.1.3 Individualized Treatment Feedback
AI and machine learning are revolutionizing healthcare by sifting through massive amounts of patient data, from medical histories to genetic profiles. These smart systems spot hidden trends and connections, helping doctors predict how different patients might respond to particular therapies. Large models play a crucial role in analyzing treatment feedback and formulating personalized plans. By monitoring patients’ physiological data, treatment history, and genetic information, large models can more accurately predict patient responses to specific medications, providing support for doctors to formulate individualized treatment plans and thereby improving treatment efficacy [84]. Andrews suggested that ML and AI are being used to analyze large datasets containing patient information, medical records, genetic data, and treatment outcomes [112]. And Landi proposed a deep learning-based unsupervised framework for processing heterogeneous EHR and deriving patient representations, efficiently and effectively achieving large-scale patient stratification [113]. This study analyzed electronic health records (EHRs) from 1,608,741 patients across multiple hospital cohorts, encompassing 57,464 clinical concepts. We developed ConvAE, an innovative representation learning model that integrates word embeddings, convolutional neural networks (CNNs), and autoencoders to encode patient trajectories into low-dimensional latent vectors. Hierarchical clustering analysis validated that the model demonstrates effective patient stratification capabilities for both general and disease-specific populations. Notably, when applied to conditions like type 2 diabetes, Parkinson’s disease, and Alzheimer’s disease, ConvAE identified clinically meaningful subtypes that correlated strongly with disease progression patterns, symptom severity, and comorbidity profiles. The findings show that ConvAE effectively creates patient profiles that reveal clinically useful information. This adaptable approach enables deeper analysis of diverse patient subgroups with varying disease causes, while uncovering new opportunities for personalized treatment research using electronic health records.
While large models hold great potential in medicine, they come with important ethical risks. One major concern is algorithmic bias, especially when the training data underrepresents the global population. Data bias can lead to differences in diagnostic accuracy and treatment recommendations, especially for marginalized groups. For example, if the training data comes primarily from North America and Europe, the model may not be able to effectively address different disease prevalence, medical practices, or language differences in other regions when applied to those regions.
For example, a model trained primarily on male and European data may overlook or misunderstand health problems that are more common in women or other racial groups, leading to misdiagnosis or inappropriate treatment. This bias can exacerbate health inequalities and prevent marginalized groups from receiving the same accurate medical services as mainstream groups in terms of diagnosis and treatment. Therefore, ensuring the diversity and inclusiveness of training data is critical to the responsible application of AI in healthcare.
4.2 Data Analysis and Knowledge Discovery
4.2.1 Diversity and Complexity of Data
In healthcare, we are confronted with an ever-expanding and diverse ocean of data, encompassing various types of information across multiple layers, from clinical records to medical images and genomic data [109]. This diversity provides doctors with more comprehensive and in-depth patient information, offering rich material for the development of personalized medical care and treatment plans. However, accompanying this diversity is the complexity of the data, rendering traditional manual analysis methods inadequate when dealing with such vast and complex medical datasets.
Clinical records stand as one of the fundamental and extensively collected data types in the medical domain, incorporating patients’ medical histories, symptom descriptions, diagnostic information, and more [114]. These records are typically unstructured text, covering a wide range of medical knowledge. Yet, their unstructured nature makes traditional manual organization and analysis exceptionally cumbersome. Simultaneously, medical imaging data provides rich visual information, such as X-rays, MRIs, CT scans, etc., playing a crucial role in disease diagnosis and treatment processes. Nevertheless, large-scale medical imaging data requires not only highly specialized interpretation but also effective tools to extract key information. Genomic data has rapidly emerged in recent years, presenting unprecedented opportunities for medical research and treatment. Individual genetic information can reveal the genetic basis of many diseases and provide a theoretical foundation for devising personalized treatment plans [115–117]. AI-driven approaches, as demonstrated by Ozaybi et al., enable efficient analysis of complex biochemical interactions, facilitating novel drug target identification [118]. However, the complexity and high-dimensional features of genomic data make traditional analysis methods insufficient, requiring the assistance of advanced computer technology and algorithms for precise interpretation and analysis.
The emergence of large-scale models opens up brand new possibilities for processing and analyzing medical data, as shown in Fig. 11. Traditional manual analysis methods prove inadequate when faced with these multi-source, high-dimensional, and unstructured data. Firstly, manually processing medical data consumes a significant amount of time and effort, resources that should ideally be allocated more towards patient diagnosis and care. Secondly, manual processing is susceptible to subjective factors, leading to inconsistency and inaccuracy in results. In this era of information, we need more efficient, accurate, and intelligent methods to address the challenges posed by medical data. Deep learning provides a wide array of tools, techniques, and frameworks to tackle these challenges [119]. Deep learning technologies, especially models like CNNs (Convolutional Neural Networks) and NLP (Natural Language Processing), excel in the processing of medical images and text data. These models can learn complex patterns and associations within the data, providing doctors with more comprehensive diagnostic information.
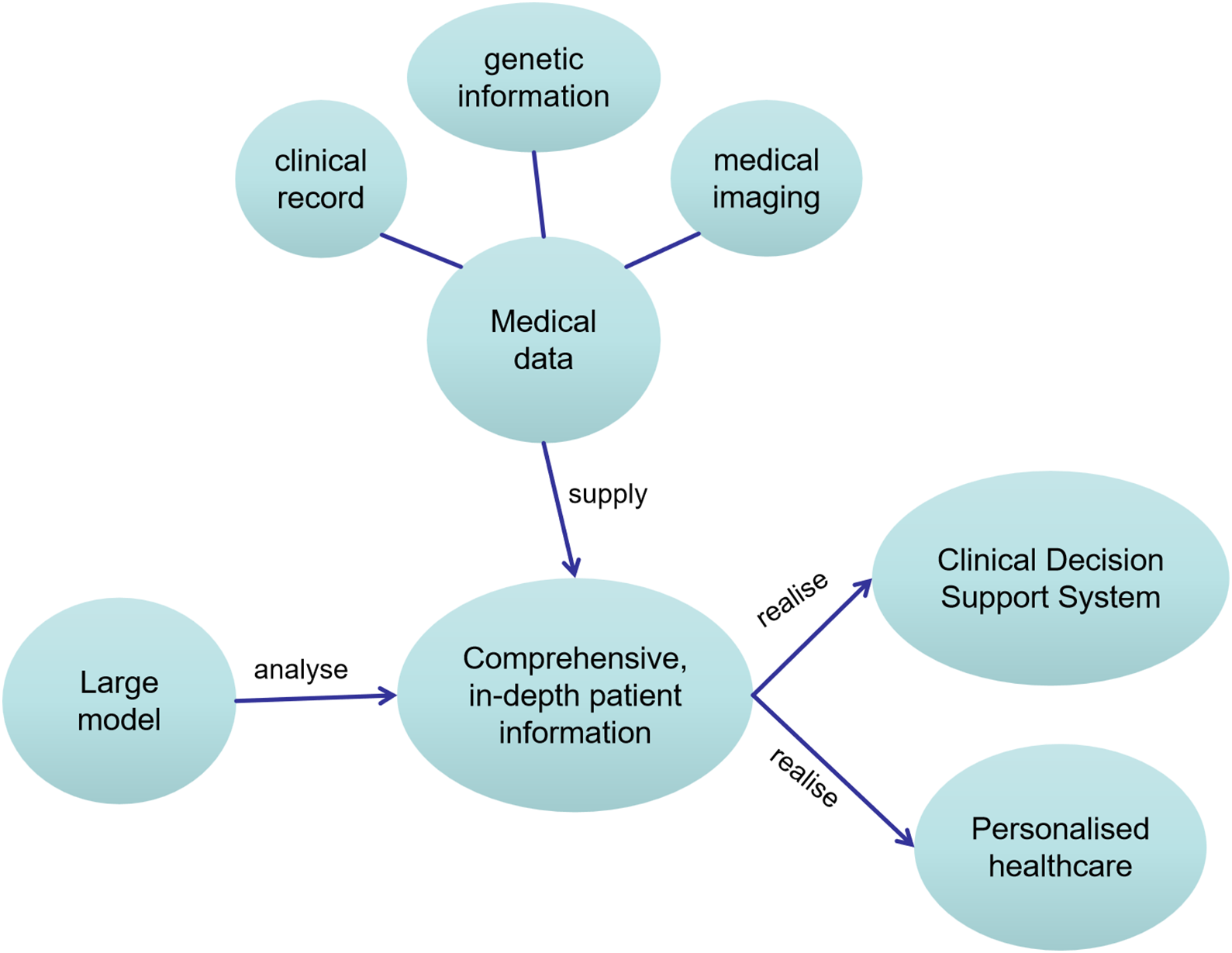
Figure 11: Schematic of knowledge graph for healthcare data analysis and knowledge discovery
4.2.2 Analytical Capabilities of Large Models
Medical data is often multimodal and complex, including medical images, genetic data, and biomarkers. Large models can capture complex relationships and patterns within such data [120]. Through deep learning algorithms, large models can extract valuable information from large-scale, multimodal medical data. For medical images, large models can perform image segmentation, feature extraction, and even automatically annotate abnormal areas. In clinical text, large models can understand context, extract entity information, aiding doctors in better comprehending patient medical histories and symptoms [121]. One core idea of deep learning is to learn abstract representations of data through hierarchical non-linear transformations. Each layer performs a series of complex non-linear operations on the input, gradually extracting high-level features from the data [122]. This layered non-linear transformation allows large models to adapt to complex data distributions and relationships, thereby better modeling latent patterns in the data. Large models, by learning representations of data during the training process, automatically discover key features within input data. This parameter learning approach enables large models to extract useful information from massive datasets, including complex relationships and patterns hidden in the data. Adjusting model weights, large models can optimize the loss function, improving their fit to the training data [123].
4.2.3 Advantages of Large Models in Knowledge Discovery
Large AI models have become powerful tools for medical knowledge discovery, leveraging advanced deep learning and NLP methods to uncover valuable insights from complex healthcare datasets. These models excel at identifying hidden patterns, novel correlations, and emerging trends across diverse medical data sources, significantly enhancing both research capabilities and clinical decision-making. Their ability to process massive datasets and detect intricate relationships makes them particularly valuable for:
• Text Mining and Knowledge Extraction: Large models can deeply understand and analyze text by learning from vast amounts of medical literature and clinical records. Through natural language processing techniques, they can extract keywords, entities, relationships, and events, establishing connections between concepts and forming medical knowledge graphs. These graphs not only assist doctors in better understanding literature but also uncover medical relationships that might exist in literature but have not been explicitly expressed.
• Data Associations and Pattern Recognition: Large models can process various types of medical data, including images, biomarkers, clinical records, etc. Through deep learning techniques, they can identify complex relationships and patterns within this data. This aids in discovering common features in patient populations, potential risk factors for diseases, and the effectiveness of drugs on different patients (e.g., uncovering hidden drug-disease relationships through AI, as in Islam et al.) [124].
• Automated Experiment Design and Research Planning: When analyzing large-scale biomedical data, large models can automatically identify potential research directions and experiment designs [125]. They can guide researchers in choosing appropriate biomarkers, sample sizes, and research methods, thus enhancing research efficiency. This automated research planning helps accelerate progress in medical research, enabling scientists to explore new treatment methods and disease mechanisms more rapidly.
• Large Models and Knowledge Graphs: Knowledge graphs are structured representations of medical knowledge, depicting different entities and their relationships in a graph format. When large models understand and analyze medical text, they can generate meaningful embedding vectors, contributing to the construction of richer knowledge graphs [123,126]. Recent reviews (e.g., Perdomo-Quinteiro & Belmonte-Hernández) systematically analyze knowledge graph-based approaches for drug repurposing, demonstrating their potential in accelerating biomedical discovery by integrating heterogeneous data sources [127].
• Large-Scale Literature Mining: Traditional literature mining methods often face challenges related to the complexity of text and the vast volume of data. Large models, through deep learning, can better capture semantic information in text, discovering correlations and trends in medical literature. This provides researchers with a more extensive and in-depth understanding of literature, expediting the progress of medical research.
Improving efficiency is a key advantage of applying large models in medicine and healthcare. Through various technological advantages such as automation, data processing, and decision support, large models significantly enhance medical research, clinical diagnosis, treatment decision-making, and more. In this process, large models play crucial roles in automating medical research, analyzing image data, processing and integrating data, optimizing clinical workflows, managing and monitoring patients, and decision support systems. Table 4 presents a performance comparison between GatorTronGPT and four other biomedical transformer models for end-to-end relation extraction tasks, including drug-drug interactions and chemical-disease relations. The results indicate that GatorTronGPT achieved superior performance across all three benchmark datasets when compared to existing models [5].
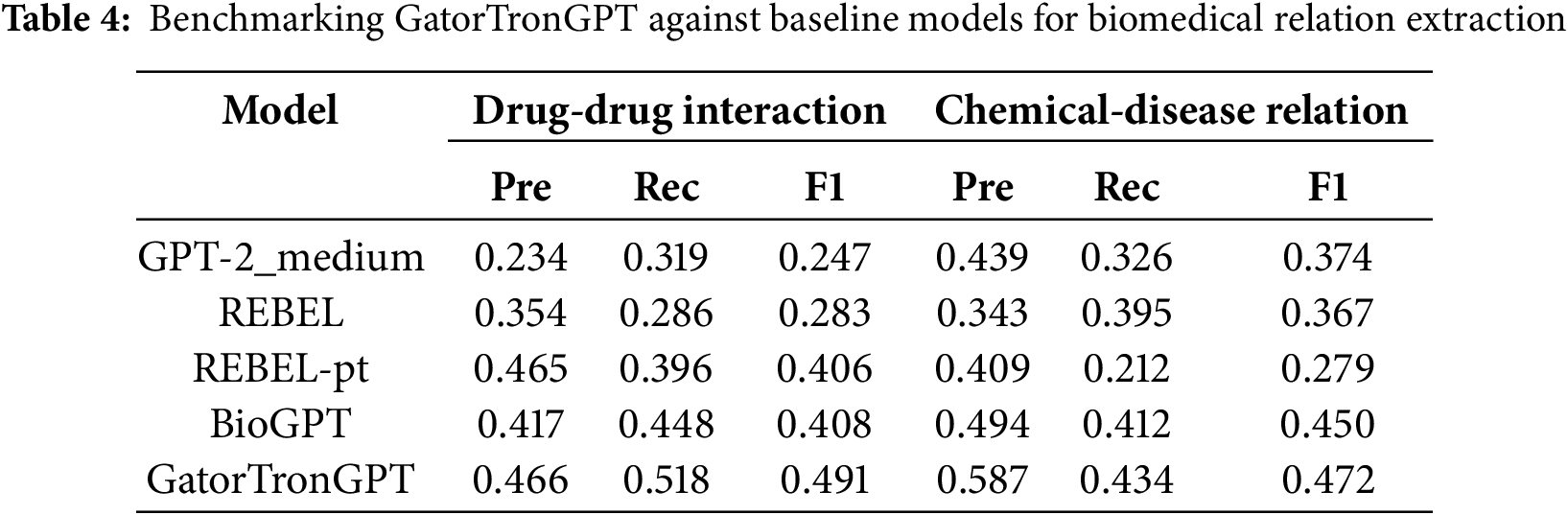
To enhance the understanding of the performance advantages of GatorTronGPT, this article supplements the task details and indicator settings involved in the evaluation in Table 3. The experiment covers multiple biomedical natural language processing (NLP) tasks, including drug-drug interaction identification (DDI), chemical-disease relations, etc. The evaluation datasets used include BioCreative V Chemical-Disease Relations(CDR), DDIExtraction 2013 and NCBI Disease datasets. The main evaluation indicators include accuracy, precision, recall and F1 score.
GatorTronGPT outperforms existing models such as BioBERT, ClinicalBERT, and PubMedBERT on all datasets. Its performance advantage can be attributed to the following key factors:
• Model architecture advantage: GatorTronGPT is based on the GPT-3 architecture and has 200 billion parameters, which gives it a significant advantage in processing long texts and modeling complex language structures.
• Pre-training data quality: The model is pre-trained using high-quality medical corpus from real clinical records, medical literature, and standardized terminology libraries, thereby enhancing the ability to understand medical semantics.
• Task fine-tuning strategy: During the fine-tuning stage, GatorTronGPT was fine-tuned for different NLP tasks, so that it can effectively adapt to various information extraction and relationship recognition tasks.
• Synthetic data generation capability: GatorTronGPT also has the ability to synthesize medical texts. Studies have shown that models trained with synthetic texts generated by it (GatorTronS) are even better than models trained with real data in specific tasks. This capability improves the robustness and generalization ability of the model in data-scarce scenarios.
In summary, GatorTronGPT’s optimizations in parameter scale, training corpus quality, and training strategy have jointly contributed to its outstanding performance in medical NLP tasks.
4.3.1 Automation in Medical Research
Medical research has always faced enormous challenges in information processing, ranging from vast literature and case reports to complex clinical trial data [63]. This process involves an in-depth analysis of diverse and heterogeneous data. Traditional research methods are often time-consuming and heavily dependent on human resources, limiting the speed and efficiency of research progress. However, automation in medical research has made significant strides, offering new possibilities for faster and more accurate exploration of medical knowledge.
Large models can mine key information, extract knowledge, and build knowledge graphs from extensive medical literature, playing a vital role in medical research. By learning from various medical data, these models can not only understand the semantics and contextual relationships of medical text but also gradually develop a deep understanding of medical knowledge. This understanding goes beyond a single information source, encompassing literature, case reports, clinical trials, and more, providing a more comprehensive perspective on medical research.
Through the automated analysis of extensive medical literature, large models aid in discovering potential patterns and associations in the text, accelerating new research findings. This ability is crucial for rapidly advancing the knowledge boundaries in medicine and healthcare, especially in rapidly evolving scientific research, where researchers need to quickly access and understand the latest medical knowledge. Large models can automatically identify key terms in literature, discover potential research directions, and provide timely summaries of the latest research, enabling researchers to track and engage in cutting-edge work more efficiently.
The automated research capabilities of large models become a significant driving force in medicine and healthcare. This automation extends beyond the processing of textual data to other forms of data, such as biomarkers, enabling medical researchers to understand existing medical knowledge more comprehensively and rapidly, thus accelerating scientific progress. In medical automation, the widespread application of large models provides researchers with exceptional tools, helping them gain a deeper understanding of patients’ physiological conditions, disease development processes, and treatment effects. Through the comprehensive analysis of diverse data sources, including clinical text, medical images, and biomarkers, large models can automatically extract and correlate potential key information, offering a new perspective for medical research. This automated data analysis capability not only improves research efficiency but also provides scientists and doctors with more comprehensive information to better understand and interpret medical data.
4.3.2 Data Automation, Processing, and Integration
Medical imaging plays an indispensable role in clinical diagnosis and treatment, but traditional image analysis processes often require manual interpretation by doctors [13]. Large models make medical image analysis more automated and efficient. These models can learn and identify complex features in images, such as tumors, anomalies, and organ structures, enabling automated disease detection and diagnosis [37]. Taking medical imaging as an example, the application of large models in automated image analysis allows doctors to quickly and accurately obtain key information. For instance, in X-rays, CT scans, or MRI images, large models can rapidly and accurately identify abnormalities, providing detailed explanations for doctors, thus shortening the diagnosis time and improving patient treatment outcomes. Recent advancements in segmentation models, such as the Segment Anything Model (SAM) proposed by Huang et al., have shown significant potential in improving the accuracy and efficiency of medical image segmentation.
The medicine involves various data types, including images, text, laboratory data, and more. Integrating and processing this heterogeneous data has always been a challenge, and large models, with their powerful computing capabilities and multimodal learning, provide solutions for this [68]. Large models can efficiently handle large-scale, complex medical datasets, accelerating the process of data integration and providing doctors with more comprehensive patient information. By integrating information from multiple data sources, large models offer doctors a more comprehensive view of patients. Patient records, images, laboratory results, and other data can be seamlessly combined through large models, allowing doctors to better understand the overall health status of patients. Large models can automate many medical tasks, such as record management, report generation, and data entry, enhancing the efficiency of healthcare systems. This comprehensive patient information helps formulate more personalized treatment plans, thereby improving treatment outcomes.
4.3.3 Clinical Workflow Optimization and Decision Support Systems
The provision of medical services typically involves complex clinical workflows, including patient visits, medical examinations, diagnoses, and treatments. Large models, through the automation and optimization of these processes, significantly enhance the efficiency of medical services. In electronic health record management, the application of large models makes tasks such as record-keeping and report generation more efficient [112,113]. By automating these processes, doctors can focus more on communicating with patients and providing diagnoses, improving clinical work efficiency. Moreover, large models can be applied to automate diagnostic assistance, speeding up patient visits and providing more timely medical services.
As decision support systems, large models provide doctors with more comprehensive and accurate information, assisting them in making diagnoses and treatment decisions. Recent studies (e.g., Hager et al.) have systematically evaluated limitations of LLMs in clinical decision-making, highlighting the need for mitigation strategies to ensure safe deployment [128]. These models can integrate patient medical records, medical knowledge databases, and the latest research findings, offering doctors a more holistic view of patients. This comprehensive information helps doctors better understand the patient’s condition and formulate more effective treatment plans. Decision support systems can also provide real-time clinical guidance for doctors. For example, in disease diagnosis, large models can analyze a large number of similar cases to provide doctors with diagnostic suggestions [91]. This real-time decision support helps improve the accuracy of diagnoses and provides patients with more precise treatment plans.
4.3.4 Patient Management and Monitoring
The application of large models in patient management and monitoring enables healthcare teams to more effectively track the health status of patients [5]. By monitoring patients’ physiological parameters, medical images, and real-time medical records, large models can automatically identify potential health risks and provide timely alerts. This real-time monitoring not only helps improve patient health management but also allows healthcare teams to intervene more quickly, improving patient treatment outcomes. In the management of chronic diseases, the application of large models provides doctors with more tools for remotely monitoring patients’ health conditions [52]. By integrating various information such as patients’ lifestyles, medication history, physiological parameters, and more, large models can generate more comprehensive health assessments, supporting the formulation of personalized treatment plans. Telemedicine and remote monitoring, as innovative technologies in medicine and healthcare, significantly improve the efficiency of healthcare services, bringing many conveniences to both patients and healthcare professionals, making healthcare services more accessible, especially in remote areas or crisis situations. Key factors contributing to the efficiency improvement in telemedicine include:
• Real-time Medical Consultation and Remote Diagnosis: Telemedicine platforms allow patients to have real-time medical consultations with doctors anytime, anywhere. Through video calls or online chats, patients can promptly receive advice from doctors, address daily health issues, and reduce unnecessary outpatient visits. For common ailments, doctors can quickly assess the condition through remote diagnosis, providing effective treatment plans for patients and avoiding cumbersome waiting times and commuting.
• Long-term Management of Patients with Chronic Diseases: For patients with chronic diseases, telemedicine offers a more convenient way for long-term management. Doctors can use remote monitoring devices to obtain patients’ physiological data, such as blood pressure, blood sugar, heart rate, etc., to keep track of their health status in real-time. By setting thresholds, doctors can promptly intervene in case of anomalies, adjust treatment plans, achieve early intervention, and reduce the risk of patients being readmitted due to the recurrence of chronic diseases.
• Remote Surgical Guidance and Professional Collaboration: Telemedicine technology also plays a crucial role in surgery. Expert doctors can provide remote guidance for surgeries in distant areas, facilitating remote surgical collaboration using high-definition cameras and real-time communication tools. This provides professional technical support to medical institutions in remote areas and allows doctors to participate in international medical teams, improving the precision and success rate of surgeries.
• Optimization of Medical Resources: Telemedicine effectively optimizes the allocation of medical resources. Through online consultations, a doctor can simultaneously serve multiple patients without the need for face-to-face appointments. This allows doctors to use their time more efficiently, alleviating the burden on the healthcare system. Furthermore, through telemedicine, it is possible to share medical resources across different regions, extending high-quality healthcare services to a broader population.
Remote monitoring, through advanced technological means, achieves real-time collection and analysis of patients’ physiological data, bringing significant advantages to medical care. This monitoring method not only allows doctors to remotely grasp patients’ health conditions, make timely interventions, and adjust treatment plans but also stimulates patients’ self-management awareness, increasing their focus on their health. By reducing medical costs, promoting the formulation of personalized treatment plans, and effectively responding to emergencies, remote monitoring improves medical efficiency and provides patients with a more convenient and personalized healthcare experience. The above analysis, as illustrated in Fig. 12, demonstrates that large models are reshaping the efficiency standards of healthcare services through three key advantages: high-precision diagnosis, multimodal data analysis, and process automation. However, it is important to note that these technological breakthroughs must be implemented alongside solutions to challenges such as privacy protection and interpretability (see Section 5 for details).
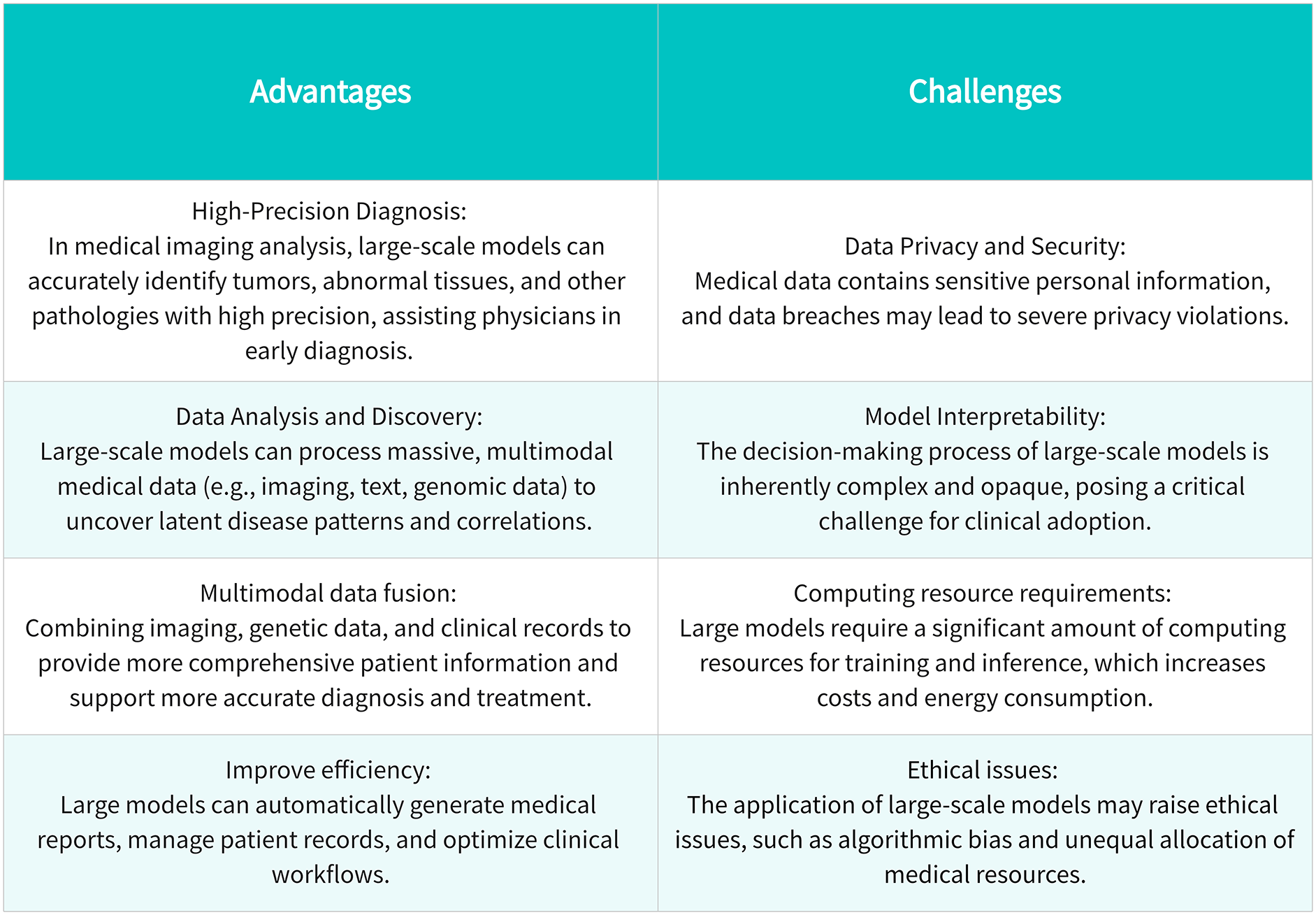
Figure 12: The dual facets of healthcare AI: capabilities and constraints of large-scale models
5 Challenges of Large Models and Future Directions
5.1 Challenges in the Healthcare Sector
The heterogeneity of healthcare data complicates the integration and management of data; complex deep learning models often lack intuitive interpretability, adding to the difficulty of explaining model decisions; and the application of large-scale models to different healthcare scenarios needs to take into account this diversity, including differences in the ways of interacting with healthcare professionals, patients, and medical equipment [129,130]. From the challenges of managing large and complex healthcare data to the pursuit of interpretability in healthcare decision-making and ultimately the application of advanced algorithms to real-world clinical environments, the healthcare industry inevitably faces many important and complex issues as it embraces this innovation. However, deploying and running large models in low-resource settings, such as rural hospitals or primary care clinics in developing countries, faces huge challenges. These include insufficient hardware, unstable network connections, lack of technical support, and other issues. In addition, introducing advanced AI systems into such environments may exacerbate inequality in medical resources. Therefore, developing lightweight models and local adaptation strategies is key to achieving technological equity. The complexity of the big model challenges in healthcare can be visualised through Fig. 13.
Figure 13: Schematic of the challenges faced by big models in medicine and healthcare
5.1.1 Data Challenges in Healthcare
For large models specifically, the data challenges in healthcare are magnified due to the immense volume and complexity of data these models require [131]. The sector’s struggle with data silos and privacy concerns is exacerbated by the need for large, diverse, and high-quality datasets to train and refine these models [132]. When selecting data for training the model, we explicitly included public datasets that contain complete patient medical records and medical images, such as Medical Information Mart for Intensive Care(MIMIC)-III. Exclusion criteria included: 1) records with serious data missing; 2) studies where the dataset was not preprocessed and consistency could not be guaranteed. Additionally, the heterogeneity of medical data, including its structured and unstructured forms, poses a significant challenge for large models that rely on comprehensive data preprocessing and feature engineering.
In this context, the issue of data sharing becomes more pronounced. The unique requirements for confidentiality and privacy of medical data exacerbate the challenges in data integration. As highlighted by Iserson, obtaining informed consent for AI applications—especially in high-stakes fields like emergency medicine—is critical to ensure ethical compliance and patient trust, even when dealing with large-scale data silos [133]. Moreover, existing business systems often use different data formats, standards, and coding systems, making data fusion and interaction complex and difficult. The diversity and complexity of medical data necessitate more intelligent and efficient methods to address this. Dealing with the processing, statistics, and analysis of massive and discrete medical data, particularly in the medical profession, poses not only technical challenges but also requires the interdisciplinary application of data science and artificial intelligence [134]. The discreteness of massive data implies the need to handle a large amount of heterogeneous and unstructured information, where traditional data processing methods may prove inadequate. Solving the challenge of data integration and model application in the healthcare domain requires the comprehensive application of knowledge from computer science, data science, medicine, and artificial intelligence. On one hand, there is a need to develop more intelligent algorithms capable of finding patterns and mining information in data silos for efficient data integration. On the other hand, advanced computing technologies, including distributed computing, cloud computing, and edge computing, should be applied to enhance the processing speed and efficiency of algorithms on large-scale datasets. Recent studies further highlight the need for interdisciplinary collaboration to establish trust in AI-driven healthcare. For instance, Starke et al. (2025) proposed a consensus framework from international experts, emphasizing transparent model design and ethical guidelines to bridge the gap between technical capabilities and clinical acceptance [135].
5.1.2 Explainability of Medical Decisions
The opaque decision-making process of complex AI systems poses significant challenges in medical applications, given the potentially grave outcomes of erroneous judgments. However, the lack of transparency in decision-making is not only a trust issue but also a matter of patient safety. Therefore, developing explainable AI for large models is crucial to ensure that medical professionals can understand and trust the model’s recommendations, which is especially important in life-critical applications [136].
Firstly, the inherent opacity of deep learning architectures hinders traceability of clinical decision-making processes. In traditional medical practice, doctors use their years of experience and professional knowledge to make diagnoses and treatment recommendations, providing explanations for their decisions [137]. However, with deep learning models, these decisions may be confusing, as the models form patterns by learning vast amounts of data, patterns that may be difficult for humans to interpret. This situation can raise doubts among doctors and patients, as they need to trust the model’s decisions while also understanding why the decision is reasonable. Secondly, transparency is a crucial element in medical practice, particularly for diagnostics and treatment. Patients and doctors want to know why a specific diagnosis is made or why a particular treatment plan is recommended. This interpretability is not only essential for patient trust but also helps doctors better understand the limitations and scope of the model. For example, in tumor diagnosis, a deep learning model may identify a pattern, but doctors need to know if this pattern is relevant to the specific clinical background of a patient to make data-driven decisions.
Addressing these challenges requires the development of interpretable deep learning architectures with mathematically grounded explanations. This involves designing new model structures, developing interpretable algorithms, and proposing visualization methods aimed at enabling healthcare professionals and patients to understand the internal workings of the model. In this process, a challenge is to maintain model performance while increasing its interpretability. Typically, enhancing model interpretability may come with some performance loss. Therefore, researchers need to find a balance between the two to ensure that the model provides both high accuracy in clinical practice and convincing, understandable explanations [138]. Moreover, improving the interpretability of large deep learning models also contributes to strengthening medical research and knowledge dissemination. Transparent models help medical researchers better understand disease mechanisms, driving advancements in medical science. Additionally, interpretability facilitates the sharing and dissemination of knowledge, making it easier for the medical community to accept and adopt new research findings.
5.1.3 Clinical Deployment of Healthcare Practices
The clinical deployment of large models is hindered by specific challenges related to their scale and complexity. These include ensuring the models’ robustness in real-world scenarios, integrating them with existing healthcare systems, and addressing the resistance from healthcare professionals due to the models’ complexity [139]. Moreover, the continuous evolution of these models requires ongoing validation and regulatory approval, which can be a lengthy process. AlShannaq et al. discussed the mechanisms and evolution of GPT models, emphasizing the need for robust validation and regulatory compliance to ensure their successful integration into healthcare practices.
Firstly, technological challenges primarily include aspects such as data integration, model interpretability, security, and performance (as discussed in Section 5.1.1 and Section 5.1.2). Secondly, regulatory obstacles are a major challenge in introducing new technologies into the healthcare sector. The healthcare industry is subject to numerous regulations and ethical standards to ensure the full protection of patients’ rights and privacy. Introducing large models into clinical practice requires compliance with regulations governing the use and sharing of medical data. Additionally, adapting to regulatory changes and ensuring compliance are prerequisites for healthcare institutions to successfully use large models. Cultural barriers are equally important. Doctors and other healthcare practitioners are often trained in traditional medical education and practice, and accepting new technologies may require some time [140,141]. Therefore, promoting the application of large models in healthcare practices requires creating an environment that fosters a digital healthcare culture. Training doctors and other healthcare practitioners is also a key factor in the successful deployment of large models. This involves providing training on how large models work, their scope of application, and limitations. Successful clinical integration requires physicians to comprehend model outputs and seamlessly incorporate them into clinical workflows. Tailored training curricula must align with physicians’ cognitive processes and practice environments to maximize the utility of large-scale AI systems.
5.2 Emerging Directions for Foundation Models in Medical Practice
The rapid development of artificial intelligence technology in the medical field has made large-scale models a key catalyst for transformative medical breakthroughs. In the future, the development of these models will focus more on improving their capabilities in biomedical data analysis, especially in the fields of genomics and personalized medicine [142]. Researchers will work to develop models that can predict treatment responses based on individual genetic profiles, thereby significantly improving the accuracy and effectiveness of treatment [143,144]. In addition, the combination of large models with wearable technology will enable these advanced models to act as health monitors in non-clinical settings, providing real-time health data and early warnings, and providing new possibilities for remote patient care and early disease detection [145].
In terms of data privacy and security, future developments will include strengthening privacy protection measures, such as using blockchain technology to enhance digital identity systems and password authentication mechanisms, and implementing strict access control and authorization systems to protect privacy at a higher level [146]. It is particularly important to establish and adhere to strict data protection standards, which involve not only technical protection measures but also legal and ethical considerations. Promoting the establishment of internationally recognized medical data protection standards will provide a clear and consistent legal environment for the cross-border flow of medical data, thereby improving the overall quality and efficiency of global medical services. The integration of multimodal data (such as imaging, genomics, and electronic health records (EHR)) is the cornerstone of future large-scale models, as shown in Fig. 14. Combined with privacy-preserving technologies such as federated learning, these models can achieve real-time remote monitoring while complying with global data protection standards such as the General Data Protection Regulation (GDPR) and Health Insurance Portability and Accountability Act (HIPAA).
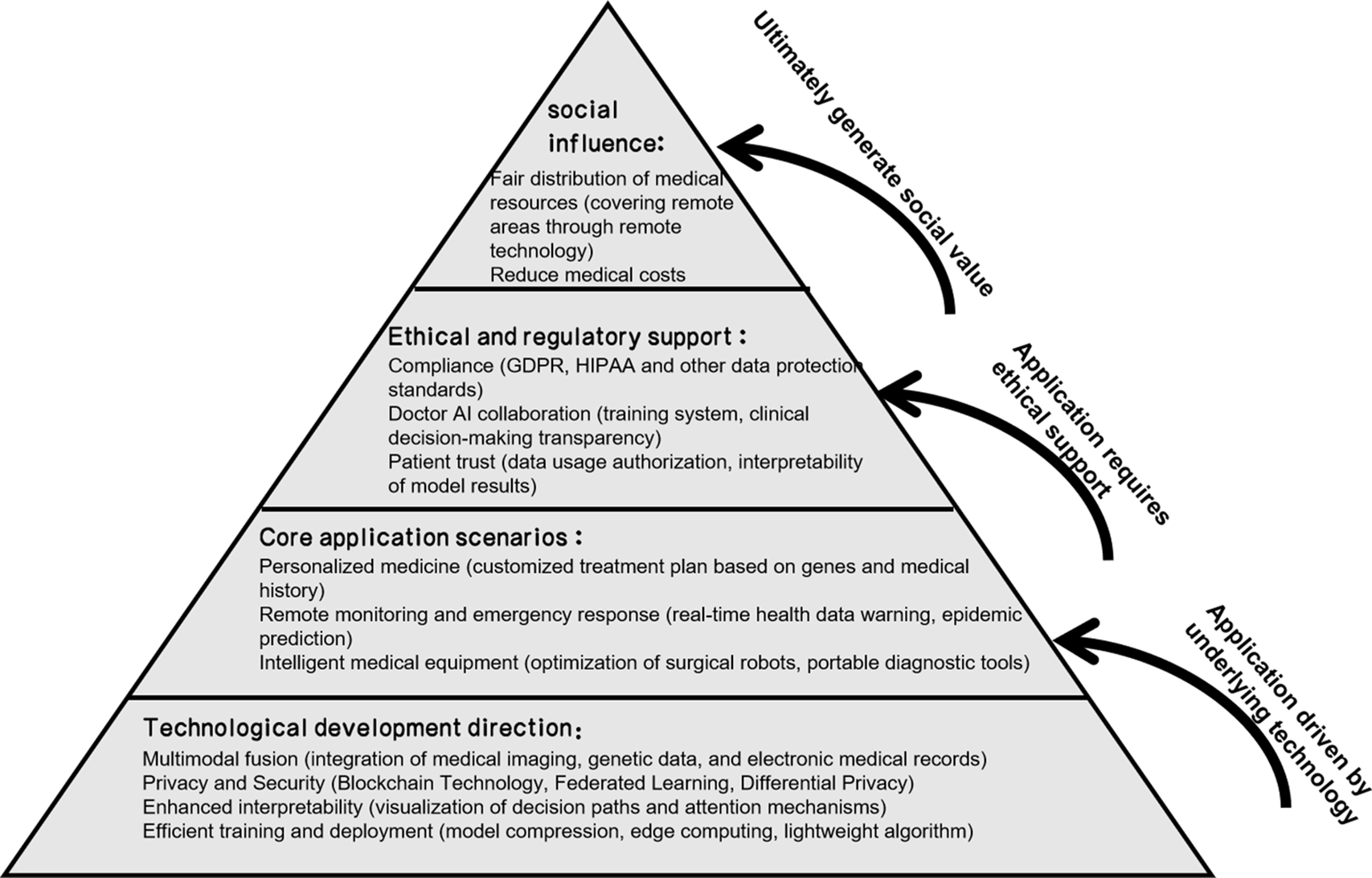
Figure 14: Future trends and key frameworks of LLMs in healthcare
In order to further solve core bottlenecks such as model deployment, privacy, and explainability, future research should also focus on the following key technical paths and prioritize them according to their correspondence with current challenges:
• Lightweight models and model compression: Large models often have huge parameter sizes, high deployment costs, and poor real-time performance, which limits their widespread application in low-resource environments. Future research needs to focus on lightweight architectures (such as MobileBERT, TinyML) and model compression technologies (such as pruning and knowledge distillation) to improve reasoning efficiency and reduce dependence on hardware. This direction will help promote the popularization of large models in scenarios such as wearable devices and mobile terminals.
• Federated Learning and Differential Privacy: Medical data is extremely sensitive. Federated learning allows the model to be trained on local devices and only shares gradient information. Combined with differential privacy mechanisms, it can further prevent reverse reasoning and reduce the risk of data leakage. This approach is of great value in joint modeling between hospitals, cross-regional disease prediction, and other tasks.
• Explainable Artificial Intelligence (XAI): The “black box” nature of large models hinders their trust in clinical decision-making. In the future, it is necessary to introduce XAI tools such as LIME and SHAP, and combine medical knowledge graphs and attention mechanisms to improve interpretability, so that doctors can understand the reasoning path and decision-making basis of the model, and promote its application in scenarios such as auxiliary diagnosis and treatment recommendation.
• Multimodal fusion and semantic integration: Faced with heterogeneous data sources such as images, texts, genes, and time series, multimodal fusion has become a key direction to improve diagnostic accuracy and personalized intervention effects. Further research should be conducted on methods such as semantic consistency modeling, cross-modal alignment, and attention mechanism fusion to achieve intelligent medical systems driven by data collaboration.
• Technology priority table for trends and challenges, see Table 5.
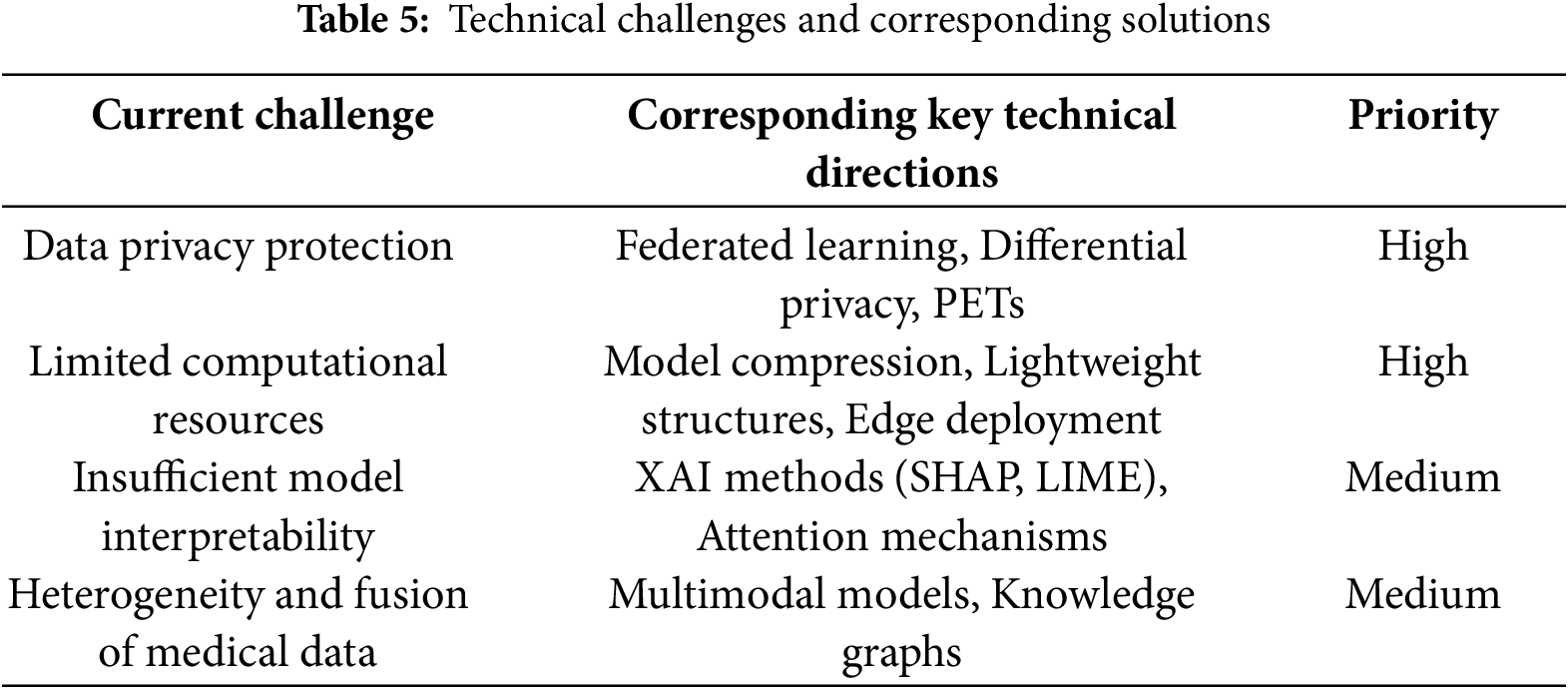
5.3 Future Development Roadmap of Medical Big Models
As large-scale models penetrate the medical and health fields, their clinical application potential coexists with risks and challenges. To promote the steady development of this technology, this paper proposes a future development roadmap covering research gaps, policy needs, and technical priorities (see Fig. 15) to guide the focus of future research and practice.
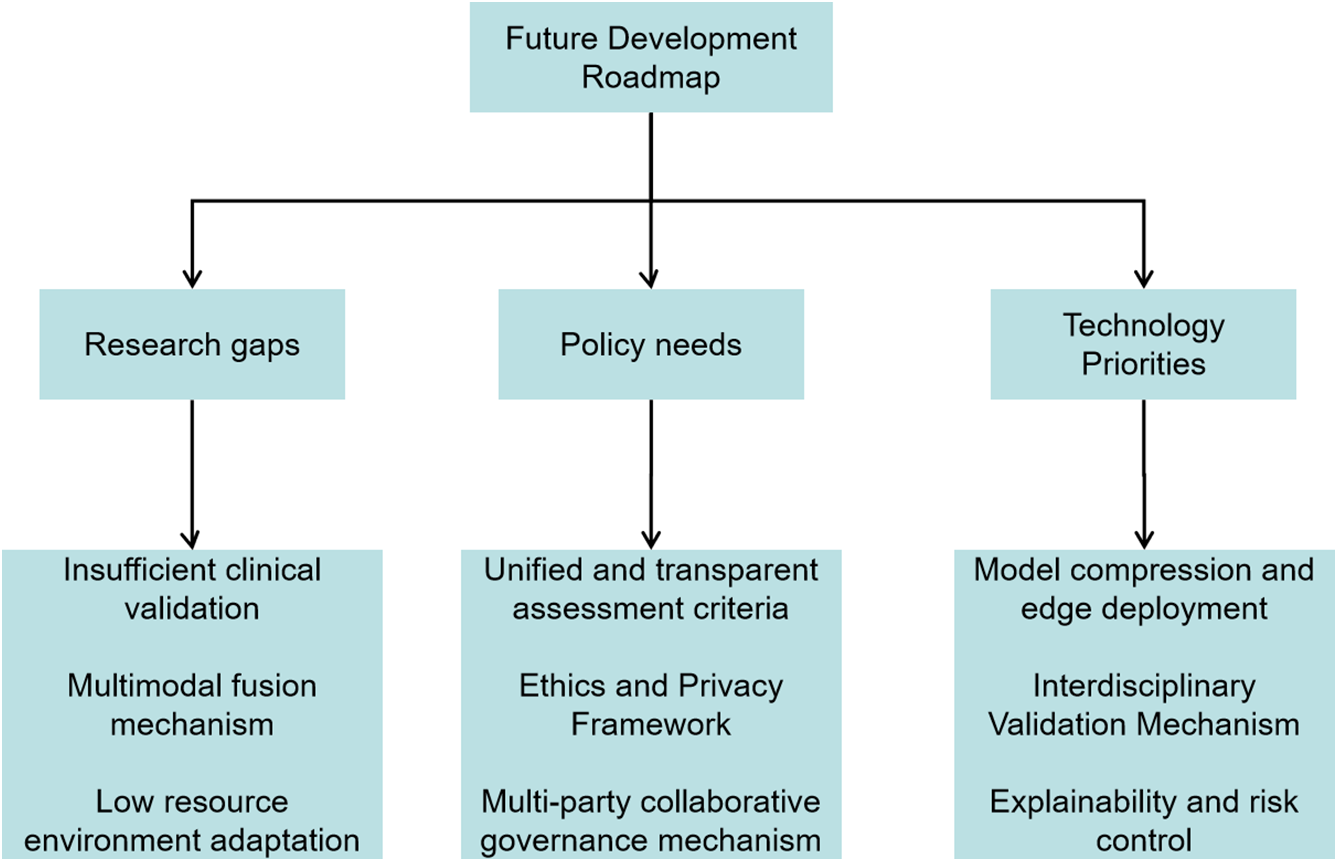
Figure 15: Future trends and key frameworks of LLMs in healthcare
First, at the research level, there are still many key gaps in large-scale medical models that need to be filled. Most current models lack continuous verification in real clinical environments, resulting in their stability and reliability in complex clinical scenarios not being fully evaluated. In addition, although multimodal data fusion has been widely used in medical tasks, its fusion mechanism is still in the exploratory stage, and the information complementarity between modalities has not been effectively mined. It is particularly noteworthy that most models rely on high-performance computing resources and have not yet formed a lightweight deployment solution suitable for low-resource environments such as mobile devices and edge nodes, which limits their application expansion in primary care.
Secondly, at the policy and governance level, the development of large-scale models urgently needs to be supported by a supporting institutional framework and ethical norms. The current lack of a unified and transparent performance evaluation benchmark makes it difficult to compare and regulate models, which in turn affects their clinical trust. In addition, the privacy sensitivity of medical data requires the establishment of a strict data protection and review mechanism throughout the entire process of model training, deployment, and application to ensure that patient rights are not violated. At the same time, we should promote the construction of a collaborative governance mechanism involving multiple parties, guide doctors, patients, AI researchers, and policymakers to jointly participate in the evaluation, feedback, and optimization of the model, and build a medical AI ecosystem with trust and security at its core.
Finally, at the level of technological evolution, several priority development directions need to be identified to improve the practicality and sustainability of the model. The primary task is to advance research on technologies such as model compression, knowledge distillation, and energy efficiency optimization, so that the model can perform well in a low-computing environment. Secondly, an interdisciplinary collaborative verification system should be established, integrating the strengths of medical, computer science, and ethics experts to promote scientific evaluation of the entire process from model design to deployment. In addition, enhancing the interpretability, transparency, and error prevention and control capabilities of the model is also key to future technological evolution. For example, the introduction of causal reasoning mechanisms and adversarial sample detection methods can help improve the model’s ability to recognize “hallucination phenomena” and the overall robustness of the system.
In addition, future research can further conduct systematic performance comparisons around large language models in healthcare tasks (such as ChatGPT, Med-PaLM, BioGPT, etc.), covering core tasks such as question answering, text generation, and auxiliary diagnosis. The performance differences of different models on specific tasks deserve in-depth discussion. At the same time, conducting a meta-analysis of existing research results will not only help clarify the applicable boundaries of various models but also promote the standardization of evaluation indicators, thereby enhancing the practicality of the model and the overall influence of this research direction.
In order to more clearly define the target audience of this article and enhance the application and transformation value of the research results, Table 6 summarizes the roles and interests of different stakeholders in the development of large-scale medical models. This matrix aims to show how the research framework, development roadmap and challenge analysis proposed in this article can serve various actual decision-makers, thereby enhancing the practicality and strategic guidance of the research.
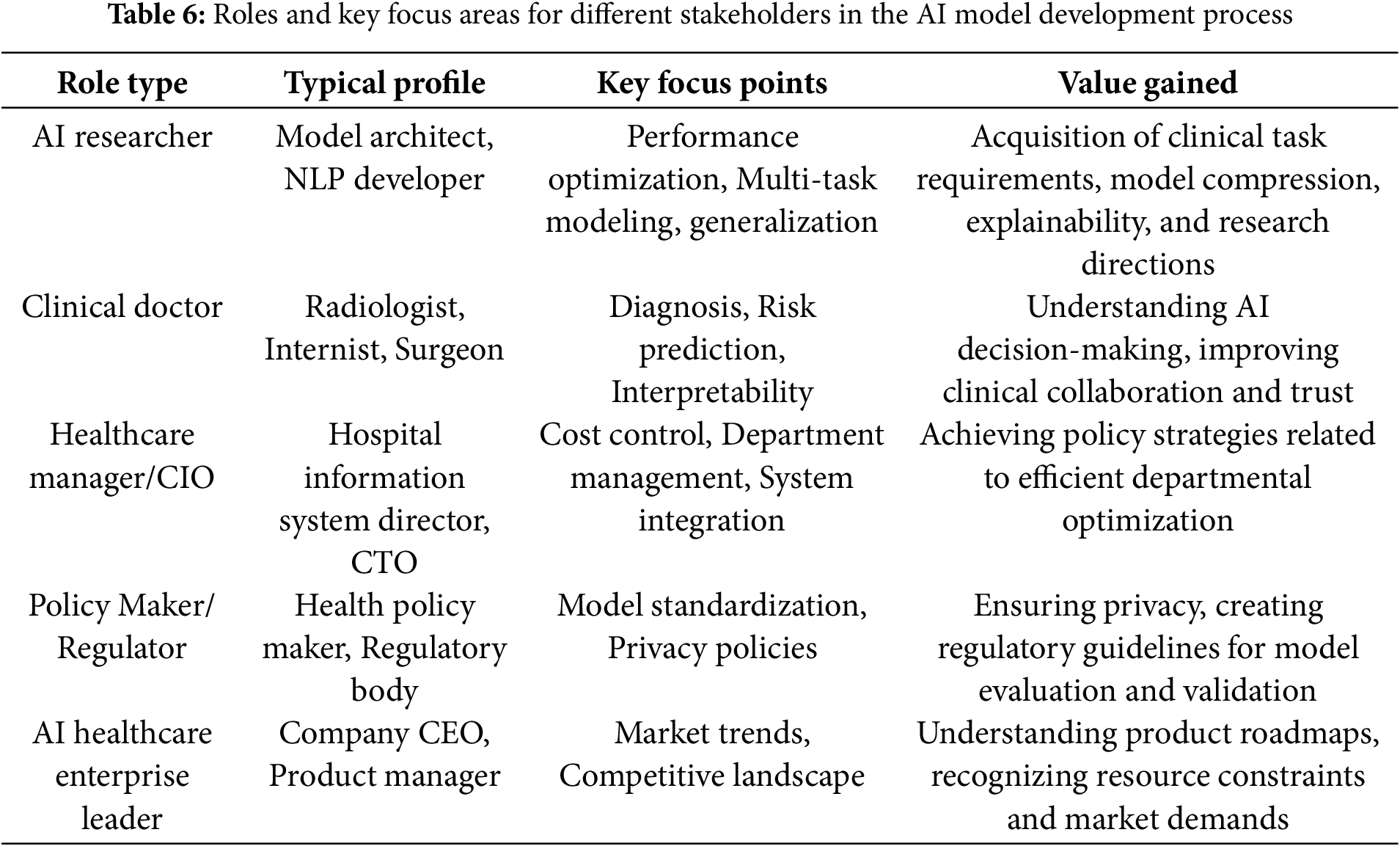
Although this article tries its best to cover the latest progress in the field of medicine and healthcare, there are still some limitations. First, some of the references are preprints, technical reports, or conference papers, which have not yet undergone formal peer review. Although these documents can reflect the most cutting-edge research trends in the field, their research conclusions have not yet been fully verified by the scientific community.
This may affect the rigor and reproducibility of the conclusions of the review to a certain extent. In particular, readers should interpret some key claims (such as the performance of the model in real clinical settings, generalization ability and safety, etc.) with caution. In addition, some studies lack large-scale clinical trial data or systematic evaluation, which may cause bias or limitations in the conclusions.
Future work can focus more on integrating published peer-reviewed literature when updating reviews, and combine more real clinical data or multicenter validation results to enhance the authority and practicality of review research.
This study explores the potential and applications of large-scale deep learning models in healthcare, especially their significant advantages in improving the accuracy of medical diagnosis, accelerating disease prediction, and optimizing treatment options. However, despite the great application prospects of these technologies, their responsible deployment still faces a series of challenges, especially in terms of data privacy protection, model interpretability, ethical norms, and cross-cultural adaptability. Therefore, to promote the healthy development of large-scale models in the medical field, researchers and practitioners should take concrete steps to ensure the transparency, fairness, and sustainability of the technology.
First, data privacy protection is the cornerstone of ensuring patient trust. To this end, future research should explore privacy-enhancing technologies such as differential privacy, homomorphic encryption, and federated learning to ensure the security and privacy of patient data. Second, improving the interpretability of models is crucial to promoting doctors and patients’ understanding of AI decisions. Therefore, it is recommended to widely apply interpretability tools (such as LIME and SHAP) to help medical professionals understand how AI models work and ensure their transparency in clinical applications. In addition, interdisciplinary collaboration and global data sharing are also crucial. Only through multi-party collaboration can a unified ethical framework and technical standards be developed to promote the widespread application of medical AI technology. Finally, given the differences in global medical practices, future research should focus on the adaptability of large models in different cultures and regions to ensure their fairness and effectiveness on a global scale. These measures will help promote the responsible application of AI technology in healthcare and ultimately improve patient health and global medical services.
Acknowledgement: The authors thank the National Natural Science Foundation of China and the Natural Science Foundation of Jiangsu Province for supporting this study.
Funding Statement: This study is funded by the National Natural Science Foundation of China (Grant No. 62272236) and the Natural Science Foundation of Jiangsu Province (Grant No. BK20201136).
Author Contributions: Conceptualization, Runze Liu; methodology, Zhiwei Chen; validation, Runze Liu and Zhiwei Chen; investigation, Yangyang Guo; resources, Shitao Huang; writing—original draft preparation, Zhiwei Chen; writing—review and editing, Zhiwei Chen and Runze Liu; visualization, Zhiwei Chen; supervision, Yongjun Ren; project administration, Shitao Huang; funding acquisition, Yongjun Ren; Zhiwei Chen and Runze Liu share first authorship. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data used in this study were obtained from Google Scholar and Web of Science. Restrictions apply to the availability of these data, which were used under license for the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28(1):31–8. doi:10.1038/s41746-023-00958-w. [Google Scholar] [CrossRef]
2. Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large language models encode clinical knowledge. arXiv:2212.13138. 2022. doi:10.48550/arXiv.2212.13138. [Google Scholar] [CrossRef]
3. Nazi ZA, Peng W. Large language models in healthcare and medical domain: a review. Informatics. 2024;11(3):57. doi:10.3390/informatics11030057. [Google Scholar] [CrossRef]
4. Pahune S, Rewatkar N. Healthcare: a growing role for large language models and generative AI. Int J Res Appl Sci Eng Technol. 2023;11(8):2288–301. [Google Scholar]
5. Peng C, Yang X, Chen A, Smith KE, PourNejatian N, Costa AB, et al. A study of generative large language model for medical research and healthcare. npj Digi Med. 2023;6(1):210. doi:10.1038/s41746-023-00958-w. [Google Scholar] [PubMed] [CrossRef]
6. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–9. doi:10.1038/s41591-018-0316-z. [Google Scholar] [PubMed] [CrossRef]
7. Ren Y, Leng Y, Qi J, Sharma PK, Wang J, Almakhadmeh Z, et al. Multiple cloud storage mechanism based on blockchain in smart homes. Future Gen Comput Syst. 2021;115(3):304–13. doi:10.1016/j.future.2020.09.019. [Google Scholar] [CrossRef]
8. Zhang H, Wu S, Wang S, Xia N. Application of blockchain technology in pet medical industry. IOP Conf Series Earth Environ Sci. 2020;440(4):042049. doi:10.1088/1755-1315/440/4/042049. [Google Scholar] [CrossRef]
9. Karabacak M, Ozkara BB, Margetis K, Wintermark M, Bisdas S. The advent of generative language models in medical education. JMIR Med Educ. 2023;9:e48163. doi:10.2196/48163. [Google Scholar] [PubMed] [CrossRef]
10. Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, et al. The future of digital health with federated learning. npj Digi Med. 2020;3(1):119. doi:10.1038/s41746-020-00323-1. [Google Scholar] [PubMed] [CrossRef]
11. Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236–46. doi:10.1093/bib/bbx044. [Google Scholar] [PubMed] [CrossRef]
12. Qiu J, Li L, Sun J, Peng J, Shi P, Zhang R, et al. Large AI models in health informatics: applications, challenges, and the future. IEEE J Biomed Health Inform. 2023;27(12):6074–87. doi:10.1109/JBHI.2023.3316750. [Google Scholar] [PubMed] [CrossRef]
13. Chlap P, Min H, Vandenberg N, Dowling J, Holloway L, Haworth A. A review of medical image data augmentation techniques for deep learning applications. J Med Imag Radiat Oncol. 2021;65(5):545–63. doi:10.1111/1754-9485.13261. [Google Scholar] [PubMed] [CrossRef]
14. Shen L, Sun Y, Yu Z, Ding L, Tian X, Tao D. On efficient training of large-scale deep learning models: a literature review. arXiv:2304.03589. 2023. doi:10.48550/arXiv.2304.03589. [Google Scholar] [CrossRef]
15. Zhang H, Shao H. Exploring the latest applications of OpenAI and ChatGPT: an in-depth survey. Comput Model Eng Sci. 2024;138(3):2061–102. doi:10.32604/cmes.2023.030649. [Google Scholar] [CrossRef]
16. Wang Y, Sun Y, Fu Y, Zhu D, Tian Z. Spectrum-BERT: pre-training of deep bidirectional transformers for spectral classification of Chinese liquors. IEEE Trans Instrum Meas. 2024;73(6):2516713. doi:10.1109/TIM.2024.3374300. [Google Scholar] [CrossRef]
17. Sun L, Wang Y, Ren Y, Xia F. Path signature-based xai-enabled network time series classification. Sci China Inf Sci. 2024;67(7):170305. doi:10.1007/s11432-023-3978-y. [Google Scholar] [CrossRef]
18. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Advances in neural information processing systems (NeurIPS 2017). 2017. p. 6000–10. [Google Scholar]
19. Luo R, Sun L, Xia Y, Qin T, Zhang S, Poon H, et al. BioGPT: generative pre-trained transformer for biomedical text generation and mining. Brief Bioinform. 2022;23(6):bbac409. doi:10.1093/bib/bbac409. [Google Scholar] [PubMed] [CrossRef]
20. Fu C, Zhang YF, Yin S, Li B, Fang X, Zhao S, et al. Mme-survey: a comprehensive survey on evaluation of multimodal llms. arXiv:2411.15296. 2024. doi:10.48550/arXiv.2411.15296. [Google Scholar] [CrossRef]
21. Zhou S, Xu Z, Zhang M, Xu C, Guo Y, Zhan Z, et al. Large language models for disease diagnosis: a scoping review. npj Artif Intell. 2025;1(1):1–17. doi:10.1038/s44387-025-00011-z. [Google Scholar] [PubMed] [CrossRef]
22. Lwakatare LE, Raj A, Crnkovic I, Bosch J, Olsson HH. Large-scale machine learning systems in real-world industrial settings: a review of challenges and solutions. Inf Softw Tech. 2020;127(2):106368. doi:10.1016/j.infsof.2020.106368. [Google Scholar] [CrossRef]
23. Mir MM, Mir GM, Raina NT, Mir SM, Mir SM, Miskeen E, et al. Application of artificial intelligence in medical education: current scenario and future perspectives. J Adv Med Educ Prof. 2023;11(3):133. doi:10.30476/JAMP.2023.98655.1803. [Google Scholar] [PubMed] [CrossRef]
24. Alomari EA. Unlocking the potential: a comprehensive systematic review of ChatGPT in natural language processing tasks. Comput Model Eng Sci. 2024;141(1):43–85. doi:10.32604/cmes.2024.052256. [Google Scholar] [CrossRef]
25. Kowsher M, Sami AA, Prottasha NJ, Arefin MS, Dhar PK, Koshiba T. Bangla-bert: transformer-based efficient model for transfer learning and language understanding. IEEE Access. 2022;10(8):91855–70. doi:10.1109/ACCESS.2022.3197662. [Google Scholar] [CrossRef]
26. Chen C, Miao J, Wu D, Zhong A, Yan Z, Kim S, et al. MA-SAM: modality-agnostic sam adaptation for 3d medical image segmentation. Med Image Anal. 2024;98:103310. doi:10.1016/j.media.2024.103310. [Google Scholar] [PubMed] [CrossRef]
27. Yang J, Jin H, Tang R, Han X, Feng Q, Jiang H, et al. Harnessing the power of llms in practice: a survey on chatgpt and beyond. ACM Trans Knowl Discov Data. 2024;18(6):1–32. doi:10.1145/3649506. [Google Scholar] [CrossRef]
28. Yenduri G, Ramalingam M, Selvi GC, Supriya Y, Srivastava G, Maddikunta PKR, et al. Gpt (generative pre-trained transformer)—a comprehensive review on enabling technologies, potential applications, emerging challenges, and future directions. IEEE Access. 2024;12:54608–49. doi:10.48550/arXiv.2305.10435. [Google Scholar] [CrossRef]
29. Raiaan MAK, Mukta MSH, Fatema K, Fahad NM, Sakib S, Mim MMJ, et al. A review on large language models: architectures, applications, taxonomies, open issues and challenges. IEEE Access. 2024;12(8):26839–74. doi:10.1109/ACCESS.2024.3365742. [Google Scholar] [CrossRef]
30. AlShannaq FB, Shehab MM, Al-Assaf AH, Alhenawi E, Awawdeh S. An exploration into the mechanisms and evolution of GPT models. In: Impacts of generative AI on the future of research and education. Hershey, PA, USA: IGI Global; 2025. p. 477–98. doi:10.4018/979-8-3693-0884-4.ch018. [Google Scholar] [CrossRef]
31. Briouya A, Briouya H, Choukri A. Overview of the progression of state-of-the-art language models. Telecommun Comput Electron Control. 2024;22(4):897–909. doi:10.12928/telkomnika.v22i4.25936. [Google Scholar] [CrossRef]
32. Petrosanu DM, Pîrjan A, Tăbuscă A. Tracing the influence of large language models across the most impactful scientific works. Electronics. 2023;12(24):4957. doi:10.3390/electronics12244957. [Google Scholar] [CrossRef]
33. Wu Y, Hu X, Fu Z, Zhou S, LiJ. GPT-4o: visual perception performance of multimodal large language models in piglet activity understanding. arXiv:2406.09781. 2024. doi:10.48550/arXiv.2406.09781. [Google Scholar] [CrossRef]
34. Hurst A, Lerer A, Goucher AP, Perelman A, Ramesh A, Clark A, et al. Gpt-4o system card. arXiv:2410.21276. 2024. doi:10.48550/arXiv.2410.21276. [Google Scholar] [CrossRef]
35. Alqahtani T, Badreldin HA, Alrashed M, Alshaya AI, Alghamdi SS, Bin Saleh K, et al. The emergent role of artificial intelligence, natural learning processing, and large language models in higher education and research. Res Soc Adminis Pharm. 2023;19(8):1236–42. doi:10.1016/j.sapharm.2023.05.016. [Google Scholar] [PubMed] [CrossRef]
36. Ye J, Chen X, Xu N, Zu C, Shao Z, Liu S, et al. A comprehensive capability analysis of gpt-3 and gpt-3.5 series models. arXiv:2303.10420. 2023. doi:10.48550/arXiv.2303.10420. [Google Scholar] [CrossRef]
37. Huang Y, Yang X, Liu L, Zhou H, Chang A, Zhou X, et al. Segment anything model for medical images? Med Image Anal. 2024;92(5):103061. doi:10.1016/j.media.2023.103061. [Google Scholar] [PubMed] [CrossRef]
38. Shen L, Sun Y, Yu Z, Ding L, Tian X, Tao D. On efficient training of large-scale deep learning models. ACM Comput Surv. 2024;57(3):1–36. doi:10.1145/3700439. [Google Scholar] [CrossRef]
39. Demelius L, Kern R, Trügler A. Recent advances of differential privacy in centralized deep learning: a systematic survey. ACM Comput Surv. 2025;57(6):1–28. doi:10.1145/3712000. [Google Scholar] [CrossRef]
40. Alon I, Haidar H, Haidar A, Guimón J. The future of artificial intelligence: insights from recent Delphi studies. Futures. 2024;165(1):103514. doi:10.1016/j.futures.2024.103514. [Google Scholar] [CrossRef]
41. Marcus G. The next decade in AI: four steps towards robust artificial intelligence. arXiv:2002.06177. 2020. doi:10.48550/arXiv.2002.06177. [Google Scholar] [CrossRef]
42. Singla R, Pupic N, Ghaffarizadeh SA, Kim C, Hu R, Forster BB, et al. Developing a Canadian artificial intelligence medical curriculum using a Delphi study. npj Digital Medicine. 2024;7(1):323. doi:10.1038/s41746-024-01307-1. [Google Scholar] [PubMed] [CrossRef]
43. Yang L, Tian M, Xin D, Cheng Q, Zheng J. AI-driven anonymization: protecting personal data privacy while leveraging machine learning. arXiv:2402.17191. 2024. doi:10.48550/arXiv.2402.17191. [Google Scholar] [CrossRef]
44. Wang T, Du Y, Gong Y, Choo KKR, Guo Y. Applications of federated learning in mobile health: scoping review. J Med Internet Res. 2023;25:e43006. doi:10.2196/43006. [Google Scholar] [PubMed] [CrossRef]
45. Abadi M, Chu A, Goodfellow I, McMahan HB, Mironov I, Talwar K, et al. Deep learning with differential privacy. In: Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security; 2016 Oct 24–28; Vienna, Austria. p. 308–18. doi:10.1145/2976749.2978318. [Google Scholar] [CrossRef]
46. Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, et al. Explainable AI for trees: from local explanations to global understanding. arXiv:1905.04610. 2019. doi:10.48550/arXiv.1905.04610. [Google Scholar] [CrossRef]
47. Chang Y, Wang X, Wang J, Wu Y, Yang L, Zhu K, et al. A survey on evaluation of large language models. ACM Trans Intell Syst Technol. 2024;15(3):1–45. doi:10.1145/3641289. [Google Scholar] [CrossRef]
48. Wang X, Chen G, Qian G, Gao P, Wei XY, Wang Y, et al. Large-scale multi-modal pre-trained models: a comprehensive survey. Mach Intell Res. 2023;20(4):447–82. doi:10.1007/s11633-022-1410-8. [Google Scholar] [CrossRef]
49. Hamidi A, Roberts K. Evaluation of AI chatbots for patient-specific ehr questions. arXiv:2306.02549. 2023. doi:10.48550/arXiv.2306.02549. [Google Scholar] [CrossRef]
50. Jahan I, Laskar MTR, Peng C, Huang J. Evaluation of ChatGPT on biomedical tasks: a zero-shot comparison with fine-tuned generative transformers. arXiv:2306.04504. 2023. doi:10.48550/arXiv.2306.04504. [Google Scholar] [CrossRef]
51. Johnson D, Goodman R, Patrinely J, Stone C, Zimmerman E, Donald R, et al. Assessing the accuracy and reliability of AI-generated medical responses: an evaluation of the Chat-GPT model. Res Square. Forthcoming 2023. doi:10.21203/rs.3.rs-2566942/v1. [Google Scholar] [PubMed] [CrossRef]
52. Cascella M, Montomoli J, Bellini V, Bignami E. Evaluating the feasibility of ChatGPT in healthcare: an analysis of multiple clinical and research scenarios. J Med Syst. 2023;47(1):33. doi:10.1007/s10916-023-01925-4. [Google Scholar] [PubMed] [CrossRef]
53. Lyu Q, Tan J, Zapadka ME, Ponnatapura J, Niu C, Myers KJ, et al. Translating radiology reports into plain language using ChatGPT and GPT-4 with prompt learning: results, limitations, and potential. Vis Comput Indus Biomed Art. 2023;6(1):9. doi:10.1186/s42492-023-00136-5. [Google Scholar] [PubMed] [CrossRef]
54. Kung TH, Cheatham M, Medenilla A, Sillos C, De Leon L, Elepaño C, et al. Performance of ChatGPT on USMLE: potential for AI-assisted medical education using large language models. PLoS Dig Health. 2023;2(2):e0000198. doi:10.1371/journal.pdig.0000198. [Google Scholar] [PubMed] [CrossRef]
55. Thirunavukarasu AJ, Hassan R, Mahmood S, Sanghera R, Barzangi K, El Mukashfi M, et al. Trialling a large language model (ChatGPT) in general practice with the applied knowledge test: observational study demonstrating opportunities and limitations in primary care. JMIR Med Educ. 2023;9(1):e46599. doi:10.2196/46599. [Google Scholar] [PubMed] [CrossRef]
56. Chervenak J, Lieman H, Blanco-Breindel M, Jindal S. The promise and peril of using a large language model to obtain clinical information: ChatGPT performs strongly as a fertility counseling tool with limitations. Fertil Steril. 2023;120(3):575–83. doi:10.1016/j.fertnstert.2023.05.151. [Google Scholar] [PubMed] [CrossRef]
57. Oh N, Choi GS, Lee WY. ChatGPT goes to the operating room: evaluating GPT-4 performance and its potential in surgical education and training in the era of large language models. Ann Surg Treat Res. 2023;104(5):269–73. doi:10.4174/astr.2023.104.5.269. [Google Scholar] [PubMed] [CrossRef]
58. Duong D, Solomon BD. Analysis of large-language model versus human performance for genetics questions. Europ J Human Genet. 2024;32(4):466–8. doi:10.1038/s41431-023-01396-8. [Google Scholar] [PubMed] [CrossRef]
59. Holmes J, Liu Z, Zhang L, Ding Y, Sio TT, McGee LA, et al. Evaluating large language models on a highly-specialized topic, radiation oncology physics. Front Oncol. 2023;13:1219326. doi:10.3389/fonc.2023.1219326. [Google Scholar] [PubMed] [CrossRef]
60. Lahat A, Shachar E, Avidan B, Shatz Z, Glicksberg BS, Klang E. Evaluating the use of large language model in identifying top research questions in gastroenterology. Sci Rep. 2023;13(1):4164. doi:10.1038/s41598-023-31412-2. [Google Scholar] [PubMed] [CrossRef]
61. Ruan J, Chen Y, Zhang B, Xu Z, Bao T, Mao H, et al. TPTU: task planning and tool usage of large language model-based AI agents. In: NeurIPS 2023 Foundation Models for Decision Making Workshop; 2023 Dec 15; New Orleans, LA, USA. [Google Scholar]
62. Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, et al. Big self-supervised models advance medical image classification. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021 Oct 11–17; Montreal, BC, Canada. p. 3478–88. doi:10.48550/arXiv.2101.05224. [Google Scholar] [CrossRef]
63. Khan YA, Hokia C, Xu J, Ehlert B. covLLM: large language models for COVID-19 biomedical literature. arXiv:2306.04926. 2023. doi:10.48550/arXiv.2306.04926. [Google Scholar] [CrossRef]
64. Van Veen D, Van Uden C, Blankemeier L, Delbrouck JB, Aali A, Bluethgen C, et al. Adapted large language models can outperform medical experts in clinical text summarization. Nat Med. 2024;30(4):1134–42. doi:10.1038/s41591-024-02855-5. [Google Scholar] [PubMed] [CrossRef]
65. Abbas A, Lee M, Shanavas N, Kovatchev V. Clinical concept annotation with contextual word embedding in active transfer learning environment. Digit Health. 2024;10:20552076241308987. doi:10.1177/20552076241308987. [Google Scholar] [PubMed] [CrossRef]
66. Huang Z, Bianchi F, Yuksekgonul M, Montine TJ, Zou J. A visual-language foundation model for pathology image analysis using medical twitter. Nat Med. 2023;29(9):2307–16. doi:10.1038/s41591-023-02504-3. [Google Scholar] [PubMed] [CrossRef]
67. Subha S, Shanmugathai M, Prasanth A, Varagi SS, Dhanashree V. Digital transformation in the pharmaceutical and biotech industry: challenges and research directions. In: Dhanaraj RK, Balusamy B, Samuel P, Bashir AK, Kadry S, editors. Digital twins in industrial production and smart manufacturing: an understanding of principles, enhancers, and obstacles. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2024. p. 297–324. doi:10.1002/9781394195336.ch13. [Google Scholar] [CrossRef]
68. Huang H, Zheng O, Wang D, Yin J, Wang Z, Ding S, et al. ChatGPT for shaping the future of dentistry: the potential of multi-modal large language model. Int J Oral Sci. 2023;15(1):29. doi:10.1038/s41368-023-00239-y. [Google Scholar] [PubMed] [CrossRef]
69. Müller A, Christmann LS, Kohler S, Eils R, Prasser F. Machine learning for medical data integration. Caring is sharing—exploiting the value in data for health and innovation. In: Hägglund M, Blusi M, Bonacina S, editors. Amsterdam, The Netherlands: IOS Press; 2023. p. 691–5. doi:10.3233/SHTI230241. [Google Scholar] [CrossRef]
70. Menzies SW, Sinz C, Menzies M, Lo SN, Yolland W, Lingohr J, et al. Comparison of humans versus mobile phone-powered artificial intelligence for the diagnosis and management of pigmented skin cancer in secondary care: a multicentre, prospective, diagnostic, clinical trial. Lancet Digit Health. 2023;5(10):e679–91. doi:10.1016/S2589-7500(23)00130-9. [Google Scholar] [PubMed] [CrossRef]
71. Jovy-Klein F, Stead S, Salge TO, Sander J, Diehl A, Antons D. Forecasting the future of smart hospitals: findings from a real-time delphi study. BMC Health Serv Res. 2024;24(1):1421. doi:10.1186/s12913-024-11895-z. [Google Scholar] [PubMed] [CrossRef]
72. Paslı S, Şahin AS, Beşer MF, Topçuoğlu H, Yadigaroğlu M, İmamoğlu M. Assessing the precision of artificial intelligence in emergency department triage decisions: insights from a study with ChatGPT. Am J Emerg Med. 2024;78(4):170–5. doi:10.1016/j.ajem.2024.01.037. [Google Scholar] [PubMed] [CrossRef]
73. Borg A, Jobs B, Huss V, Gentline C, Espinosa F, Ruiz M, et al. Enhancing clinical reasoning skills for medical students: a qualitative comparison of LLM-powered social robotic versus computer-based virtual patients within rheumatology. Rheumatol Int. 2024;44(12):3041–51. doi:10.1007/s00296-024-05731-0. [Google Scholar] [PubMed] [CrossRef]
74. Harris E. Study tests large language models’ ability to answer clinical questions. JAMA. 2023;330(6):496. doi:10.1001/jama.2023.12553. [Google Scholar] [PubMed] [CrossRef]
75. Chenais G, Lagarde E, Gil-Jardiné C. Artificial intelligence in emergency medicine: viewpoint of current applications and foreseeable opportunities and challenges. J Med Internet Res. 2023;25(8):e40031. doi:10.2196/40031. [Google Scholar] [PubMed] [CrossRef]
76. Huang J, Jin W, Duan X, Liu X, Shu T, Fu L, et al. Twenty-eight-day in-hospital mortality prediction for elderly patients with ischemic stroke in the intensive care unit: interpretable machine learning models. Front Public Health. 2023;10:1086339. doi:10.3389/fpubh.2022.1086339. [Google Scholar] [PubMed] [CrossRef]
77. Alsabri M, Aderinto N, Mourid MR, Laique F, Zhang S, Shaban NS, et al. Artificial intelligence for pediatric emergency medicine. J Med Surg Public Health. 2024;3(2):100137. doi:10.1016/j.glmedi.2024.100137. [Google Scholar] [CrossRef]
78. Kim JK, Chua M, Rickard M, Lorenzo A. ChatGPT and large language model (LLM) chatbots: the current state of acceptability and a proposal for guidelines on utilization in academic medicine. J Pediatric Urol. 2023;19(5):598–604. doi:10.1016/j.jpurol.2023.05.018. [Google Scholar] [PubMed] [CrossRef]
79. Hua Y, Liu F, Yang K, Li Z, Na H, Sheu Y, et al. Large language models in mental health care: a scoping review. arXiv:2401.02984. 2024. doi:10.48550/arXiv.2401.02984. [Google Scholar] [CrossRef]
80. Eastwood KW, May R, Andreou P, Abidi S, Abidi SSR, Loubani OM. Needs and expectations for artificial intelligence in emergency medicine according to Canadian physicians. BMC Health Serv Res. 2023;23(1):798. doi:10.1186/s12913-023-09740-w. [Google Scholar] [PubMed] [CrossRef]
81. Han X, Hu Z, Wang S, Zhang Y. A survey on deep learning in COVID-19 diagnosis. J Imaging. 2022;9(1):1. doi:10.3390/jimaging9010001. [Google Scholar] [PubMed] [CrossRef]
82. Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv:1711.05225. 2017. doi:10.48550/arXiv.1711.05225. [Google Scholar] [CrossRef]
83. Guo Y, Liu H, Sun Y, Ren Y. Virtual human pose estimation in a fire education system for children with autism spectrum disorders. Multimed Syst. 2024;30(2):84. doi:10.1007/s00530-024-01274-3. [Google Scholar] [CrossRef]
84. Piliuk K, Tomforde S. Artificial intelligence in emergency medicine. A systematic literature review. Int J Med Inform. 2023;180(6):105274. doi:10.1016/j.ijmedinf.2023.105274. [Google Scholar] [PubMed] [CrossRef]
85. Liu N, Zhang Z, Ho AFW, Ong MEH. Artificial intelligence in emergency medicine. J Emerg Critic Care Med. 2018;2:82. doi:10.21037/jeccm.2018.10.08. [Google Scholar] [CrossRef]
86. Nuka ST. Leveraging AI and generative AI for medical device innovation: enhancing custom product development and patient specific solutions. J Neonatal Surg. 2025;14(4s):511–22. doi:10.52783/jns.v14.1825. [Google Scholar] [CrossRef]
87. Basnawi A, Koshak A. Application of artificial intelligence in advanced training and education of emergency medicine doctors: a narrative review. Emerg Care and Med. 2024;1(3):247–59. doi:10.3390/ecm1030026. [Google Scholar] [CrossRef]
88. Alam MA, Nabil AR, Uddin MM, Sarker MTH, Mahmud S. The role of predictive analytics in early disease detection: a data-driven approach to preventive healthcare. Front Appl Eng Technol. 2024;1(1):105–23. doi:10.70937/faet.v1i01.22. [Google Scholar] [CrossRef]
89. Ren Y, Liu R, Sang H, Yu X. Avatar-based picture exchange communication system enhancing joint attention training for children with autism. IEEE J Biomed Health Inform. 2024;1–12. doi:10.1109/JBHI.2024.3487589. [Google Scholar] [PubMed] [CrossRef]
90. Singh A. Artificial intelligence for drug repurposing against infectious diseases. Artif Intell Chem. 2024;2(2):100071. doi:10.1016/j.aichem.2024.100071. [Google Scholar] [CrossRef]
91. Wang Z, Li R, Dong B, Wang J, Li X, Liu N, et al. Can LLMs like GPT-4 outperform traditional AI tools in dementia diagnosis? arXiv:2306.01499. 2023. doi:10.48550/arXiv.2306.01499. [Google Scholar] [CrossRef]
92. Tan Y, Zhang Z, Li M, Pan F, Duan H, Huang Z, et al. MedChatZH: a tuning LLM for traditional Chinese medicine consultations. Comput Biol Med. 2024;172:108290. doi:10.1016/j.compbiomed.2024.108290. [Google Scholar] [PubMed] [CrossRef]
93. Sblendorio E, Dentamaro V, Cascio AL, Germini F, Piredda M, Cicolini G. Integrating human expertise & automated methods for a dynamic and multi-parametric evaluation of large language models’ feasibility in clinical decision-making. Int J Med Inform. 2024;188(9):105501. doi:10.1016/j.ijmedinf.2024.105501. [Google Scholar] [PubMed] [CrossRef]
94. Xie Q, Schenck EJ, Yang HS, Chen Y, Peng Y, Wang F. Faithful AI in medicine: a systematic review with large language models and beyond. MedRxiv. 2023;2:719. doi:10.1101/2023.04.18.23288752. [Google Scholar] [PubMed] [CrossRef]
95. Thirunavukarasu AJ, Ting DSJ, Elangovan K, Gutierrez L, Tan TF, Ting DSW. Large language models in medicine. Nature Med. 2023;29(8):1930–40. doi:10.1038/s41591-023-02448-8. [Google Scholar] [PubMed] [CrossRef]
96. Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal. 2020;63(4):101693. doi:10.1016/j.media.2020.101693. [Google Scholar] [PubMed] [CrossRef]
97. Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. npj Digi Med. 2021;4(1):5. doi:10.1038/s41746-020-00376-2. [Google Scholar] [PubMed] [CrossRef]
98. Yuan D, Rastogi E, Naik G, Rajagopal SP, Goyal S, Zhao F, et al. A continued pretrained llm approach for automatic medical note generation. arXiv:2403.09057. 2024. doi:10.48550/arXiv.2403.09057. [Google Scholar] [CrossRef]
99. Ren Y, Sang H, Huang S, Qin X. Multistream adaptive attention-enhanced graph convolutional networks for youth fencing footwork training. Pedia Exerc Sci. 2024;1(aop):1–15. doi:10.1123/pes.2024-0025. [Google Scholar] [PubMed] [CrossRef]
100. Ouyang C, Biffi C, Chen C, Kart T, Qiu H, Rueckert D. Self-supervised learning for few-shot medical image segmentation. IEEE Trans Med Imag. 2022;41(7):1837–48. doi:10.1109/TMI.2022.3150682. [Google Scholar] [PubMed] [CrossRef]
101. Wang X, Yang S, Zhang J, Wang M, Zhang J, Yang W, et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med Image Anal. 2022;81(1):102559. doi:10.1016/j.media.2022.102559. [Google Scholar] [PubMed] [CrossRef]
102. Makkar A, Santosh K. SecureFed: federated learning empowered medical imaging technique to analyze lung abnormalities in chest X-rays. Int J Mach Learn Cybern. 2023;14(8):2659–70. doi:10.1007/s13042-023-01789-7. [Google Scholar] [PubMed] [CrossRef]
103. Xu H, Usuyama N, Bagga J, Zhang S, Rao R, Naumann T, et al. A whole-slide foundation model for digital pathology from real-world data. Nature. 2024;630(8015):181–8. doi:10.1038/s41586-024-07441-w. [Google Scholar] [PubMed] [CrossRef]
104. Kim Y, Lee JH, Choi S, Lee JM, Kim JH, Seok J, et al. Validation of deep learning natural language processing algorithm for keyword extraction from pathology reports in electronic health records. Sci Rep. 2020;10(1):20265. doi:10.1038/s41598-020-77258-w. [Google Scholar] [PubMed] [CrossRef]
105. Ren Y, Leng Y, Cheng Y, Wang J. Secure data storage based on blockchain and coding in edge computing. Math Biosci Eng. 2019;16(4):1874–92. doi:10.3934/mbe.2019091. [Google Scholar] [PubMed] [CrossRef]
106. Muksimova S, Umirzakova S, Baltayev J, Cho YI. Multi-modal fusion and longitudinal analysis for Alzheimer’s disease classification using deep learning. Diagnostics. 2025;15(6):717. doi:10.3390/diagnostics15060717. [Google Scholar] [PubMed] [CrossRef]
107. Braconi D, Nadwa H, Bernardini G, Santucci A. Omics and rare diseases: challenges, applications, and future perspectives. Expert Rev Proteom. 2025;22(3):107–22. doi:10.1080/14789450.2025.2468300. [Google Scholar] [PubMed] [CrossRef]
108. Chen S. Crossing disease boundaries: how AI drives rare disease drug discovery. Biol Evid. 2024;14(1):21–8. doi:10.5376/be.2024.14.0003. [Google Scholar] [CrossRef]
109. Mohsen F, Ali H, El Hajj N, Shah Z. Artificial intelligence-based methods for fusion of electronic health records and imaging data. Sci Rep. 2022;12(1):17981. doi:10.1038/s41598-022-22514-4. [Google Scholar] [PubMed] [CrossRef]
110. Lu Q, Wen A, Nguyen T, Liu H. Enhancing clinical relevance of pretrained language models through integration of external knowledge: case study on cardiovascular diagnosis from electronic health records. JMIR AI. 2024;3(1):e56932. doi:10.2196/56932. [Google Scholar] [PubMed] [CrossRef]
111. He L, Li H, Chen M, Wang J, Altaye M, Dillman JR, et al. Deep multimodal learning from MRI and clinical data for early prediction of neurodevelopmental deficits in very preterm infants. Front Neurosci. 2021;15:753033. doi:10.3389/fnins.2021.753033. [Google Scholar] [PubMed] [CrossRef]
112. Andrews SM. Emerging role of artificial intelligence and machine learning in precision medicine. Int J Eng Technol Manage Sci. 2023;7(4):622–6. [Google Scholar]
113. Landi I, Glicksberg BS, Lee HC, Cherng S, Landi G, Danieletto M, et al. Deep representation learning of electronic health records to unlock patient stratification at scale. npj Digi Med. 2020;3(1):96. doi:10.1038/s41746-020-0301-z. [Google Scholar] [PubMed] [CrossRef]
114. Chaabene S, Boudaya A, Bouaziz B, Chaari L. An overview of methods and techniques in multimodal data fusion with application to healthcare. Int J Data Sci Anal. 2025;2025(1):1–25. doi:10.1007/s41060-025-00715-0. [Google Scholar] [CrossRef]
115. Alberts IL, Mercolli L, Pyka T, Prenosil G, Shi K, Rominger A, et al. Large language models (LLM) and ChatGPT: what will the impact on nuclear medicine be? Euro J Nucl Med Mol Imag. 2023;50(6):1549–52. doi:10.1007/s00259-023-06172-w. [Google Scholar] [PubMed] [CrossRef]
116. Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al. Language models are few-shot learners. Adv Neural Inform Process Syst. 2020;33:1877–901. [Google Scholar]
117. Cortial L, Montero V, Tourlet S, Del Bano J, Blin O. Artificial intelligence in drug repurposing for rare diseases: a mini-review. Front Med. 2024;11:1404338. doi:10.3389/fmed.2024.1404338. [Google Scholar] [PubMed] [CrossRef]
118. Liebman M. The role of artificial intelligence in drug discovery and development. Chem Int. 2022;44(1):16–9. doi:10.1515/ci-2022-0105. [Google Scholar] [CrossRef]
119. Muniasamy A, Tabassam S, Hussain MA, Sultana H, Muniasamy V, Bhatnagar R. Deep learning for predictive analytics in healthcare. In: The International Conference on Advanced Machine Learning Technologies and Applications. Cham, Switzerland: Springer International Publishing; 2020. p. 32–42. doi:10.1007/978-3-030-14118-9. [Google Scholar] [CrossRef]
120. Gu Y, Zalkikar A, Liu M, Kelly L, Hall A, Daly K, et al. Predicting medication adherence using ensemble learning and deep learning models with large scale healthcare data. Sci Rep. 2021;11(1):18961. doi:10.1038/s41598-021-98387-w. [Google Scholar] [PubMed] [CrossRef]
121. Ekundayo F. Reinforcement learning in treatment pathway optimization: a case study in oncology. Int J Sci Res Arch. 2024;13(2):2187–205. doi:10.30574/ijsra.2024.13.2.2450. [Google Scholar] [CrossRef]
122. Devlin J, Chang MW, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; Minneapolis, MN, USA; 2019. p. 4171–86. doi:10.18653/v1/N19-1423. [Google Scholar] [CrossRef]
123. Tian Y, Gan R, Song Y, Zhang J, Zhang Y. Chimed-gpt: a chinese medical large language model with full training regime and better alignment to human preferences. arXiv:2311.06025. 2023. doi:10.48550/arXiv.2311.06025. [Google Scholar] [CrossRef]
124. Islam MT, Newaz AAH, Paul R, Hassan Melon MM, Hussen M. AI-driven drug repurposing: uncovering hidden potentials of established medications for rare disease treatment. Libr Prog-Libr Sci Inform Technol Comput. 2024;44(3):21949–65. [Google Scholar]
125. Huang SC, Pareek A, Jensen M, Lungren MP, Yeung S, Chaudhari AS. Self-supervised learning for medical image classification: a systematic review and implementation guidelines. npj Digital Med. 2023;6(1):74. doi:10.1038/s41746-023-00811-0. [Google Scholar] [PubMed] [CrossRef]
126. Ma X, Yang X, Xiong W, Chen B, Yu L, Zhang H, et al. Megalodon: efficient llm pretraining and inference with unlimited context length. Adv Neu Infom Process Syst. 2024;37:71831–54. [Google Scholar]
127. Perdomo-Quinteiro P, Belmonte-Hernández A. Knowledge graphs for drug repurposing: a review of databases and methods. Brief Bioinform. 2024;25(6):bbae461. doi:10.1093/bib/bbae461. [Google Scholar] [PubMed] [CrossRef]
128. Hager P, Jungmann F, Holland R, Bhagat K, Hubrecht I, Knauer M, et al. Evaluation and mitigation of the limitations of large language models in clinical decision-making. Nature Med. 2024;30(9):2613–22. doi:10.1038/s41591-024-03097-1. [Google Scholar] [PubMed] [CrossRef]
129. Zhou K, Zhu Y, Chen Z, Chen W, Zhao WX, Chen X, et al. Don’t make your LLM an evaluation benchmark cheater. arXiv:2311.01964. 2023. doi:10.48550/arXiv.2311.01964. [Google Scholar] [CrossRef]
130. Zhou H, Liu F, Gu B, Zou X, Huang J, Wu J, et al. A survey of large language models in medicine: progress, application, and challenge. arXiv:2311.05112. 2023. doi:10.48550/arXiv.2311.05112. [Google Scholar] [CrossRef]
131. Ramkumar PN, Masotto AF, Woo JJ. Off-the-shelf large language models (LLM) are of insufficient quality to provide medical treatment recommendations, while customization of LLMs result in quality recommendations. Arthroscopy. 2025 Feb;41(2):276–8. doi:10.1016/j.arthro.2024.09.047. [Google Scholar] [PubMed] [CrossRef]
132. Bhatt C, Kumar I, Vijayakumar V, Singh KU, Kumar A. The state of the art of deep learning models in medical science and their challenges. Multimed Syst. 2021;27(4):599–613. doi:10.1007/s00530-020-00694-1. [Google Scholar] [CrossRef]
133. Iserson KV. Informed consent for artificial intelligence in emergency medicine: a practical guide. Am J Emerg Med. 2024;76(6):225–30. doi:10.1016/j.ajem.2023.11.022. [Google Scholar] [PubMed] [CrossRef]
134. Ullah E, Parwani A, Baig MM, Singh R. Challenges and barriers of using large language models (LLM) such as ChatGPT for diagnostic medicine with a focus on digital pathology-a recent scoping review. Diagn Pathol. 2024;19(1):43. doi:10.1186/s13000-024-01464-7. [Google Scholar] [PubMed] [CrossRef]
135. Starke G, Gille F, Termine A, Aquino YSJ, Chavarriaga R, Ferrario A, et al. Finding consensus on trust in AI in health care: recommendations from a panel of international experts. J Med Internet Res. 2025;27(7):e56306. doi:10.2196/56306. [Google Scholar] [PubMed] [CrossRef]
136. Takale DG, Mahalle PN, Sule B. Generative large language models for healthcare applications: opportunities, challenges, and future directions. J Compute Based Paral Program. 2024;9(2):25–30. [Google Scholar]
137. Yao Y, Duan J, Xu K, Cai Y, Sun Z, Zhang Y. A survey on large language model (LLM) security and privacy: the good, the bad, and the ugly. High-Confiden Comput. 2024;4(2):100211. doi:10.1016/j.hcc.2024.100211. [Google Scholar] [CrossRef]
138. Shamshirband S, Fathi M, Dehzangi A, Chronopoulos AT, Alinejad-Rokny H. A review on deep learning approaches in healthcare systems: taxonomies, challenges, and open issues. J Biomed Inform. 2021;113:103627. doi:10.1016/j.jbi.2020.103627. [Google Scholar] [PubMed] [CrossRef]
139. Nori H, King N, McKinney SM, Carignan D, Horvitz E. Capabilities of gpt-4 on medical challenge problems. arXiv: 2303.13375. 2023. doi:10.48550/arXiv.2303.13375. [Google Scholar] [CrossRef]
140. Perez-Downes JC, Tseng AS, McConn KA, Elattar SM, Sokumbi O, Sebro RA, et al. Mitigating bias in clinical machine learning models. Current Treat Options Cardiovas Med. 2024;26(3):29–45. doi:10.1007/s11936-023-01032-0. [Google Scholar] [CrossRef]
141. Du X, Novoa-Laurentiev J, Plasek JM, Chuang YW, Wang L, Marshall GA, et al. Enhancing early detection of cognitive decline in the elderly: a comparative study utilizing large language models in clinical notes. EBioMedicine. 2024;109(1):105401. doi:10.1016/j.ebiom.2024.105401. [Google Scholar] [PubMed] [CrossRef]
142. Hamadi R. Large language models meet computer vision: a brief survey. arXiv:2311.16673. 2023. doi:10.48550/arXiv.2311.16673. [Google Scholar] [CrossRef]
143. Okada Y, Ning Y, Ong MEH. Explainable artificial intelligence in emergency medicine: an overview. Clin Exp Emerg Med. 2023;10(4):354–62. doi:10.15441/ceem.23.145. [Google Scholar] [PubMed] [CrossRef]
144. Zheng Y, Gan W, Chen Z, Qi Z, Liang Q, Yu PS. Large language models for medicine: a survey. Int J Mach Learn Cybern. 2025;16(2):1015–40. doi:10.1007/s13042-024-02318-w. [Google Scholar] [CrossRef]
145. Gupta NS, Kumar P. Perspective of artificial intelligence in healthcare data management: a journey towards precision medicine. Comput Biol Med. 2023;162(1988):107051. doi:10.1016/j.compbiomed.2023.107051. [Google Scholar] [PubMed] [CrossRef]
146. Suresh T, Kaliappan S, Ali HM, Kumar BV. AI-driven drug discovery and therapeutic target identification for rare genetic diseases. In: 2024 International Conference on Advancements in Smart, Secure and Intelligent Computing (ASSIC). Bhubaneswar, India: IEEE; 2024. p. 1–6. doi:10.1109/ASSIC60049.2024.10507989. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF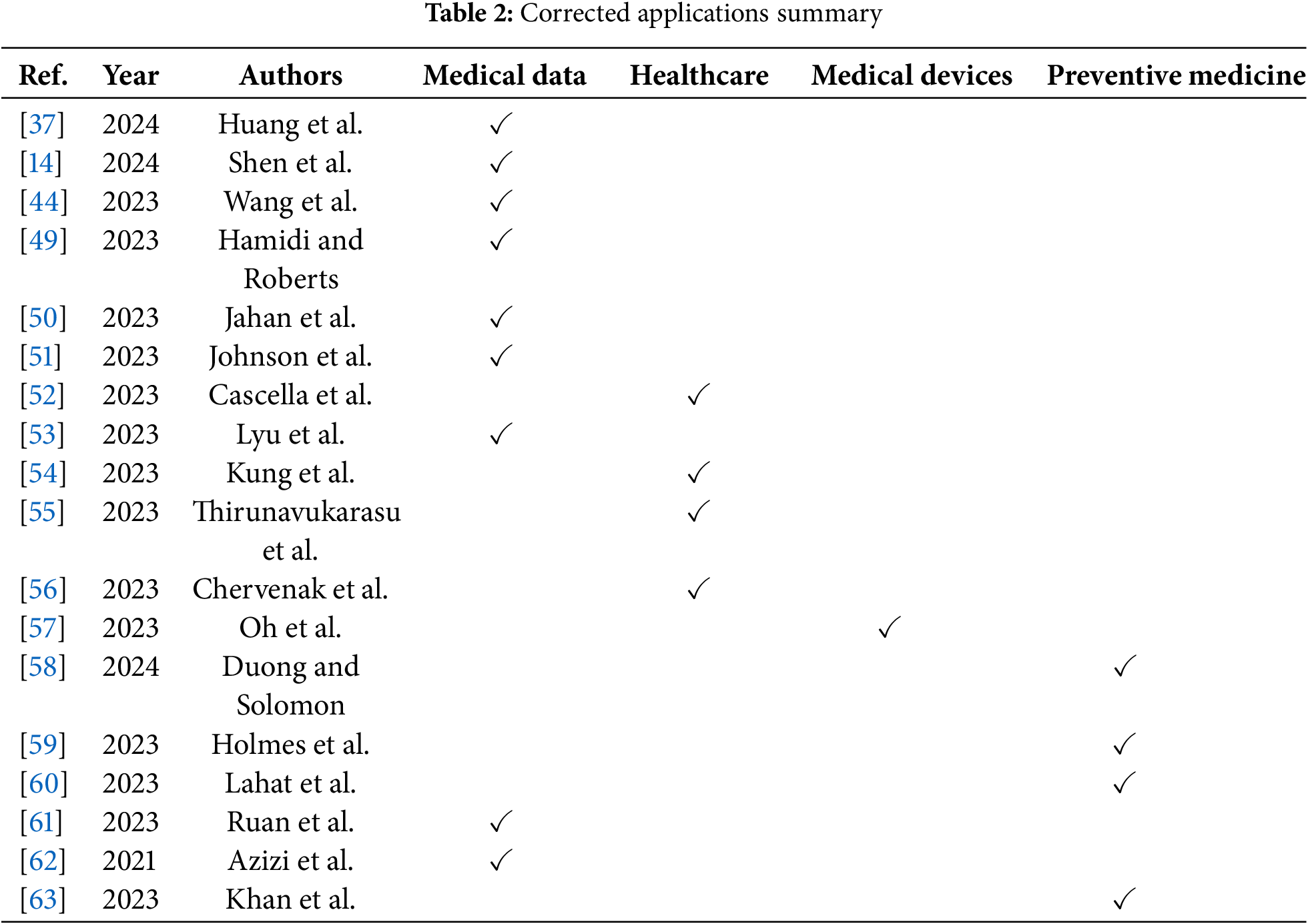
 Downloads
Downloads
 Citation Tools
Citation Tools