 Open Access
Open Access
ARTICLE
An Efficient Explainable AI Model for Accurate Brain Tumor Detection Using MRI Images
1 Faculty of Artificial Intelligence, Kafrelsheikh University, Kafrelsheikh, 33516, Egypt
2 Faculty of Computer Science & Engineering, New Mansoura University, Gamasa, 35712, Egypt
3 Department of Bioengineering, Speed School of Engineering, University of Louisville, Louisville, KY 40292, USA
4 Computers and Control Systems Engineering Department, Faculty of Engineering, Mansoura University, Mansoura, 35516, Egypt
5 Communications and Electronics Engineering Department, Nile Higher Institute for Engineering and Technology, Mansoura, 35511, Egypt
* Corresponding Authors: Fatma M. Talaat. Email: ; Mohamed Shehata. Email:
(This article belongs to the Special Issue: Recent Advances in Signal Processing and Computer Vision)
Computer Modeling in Engineering & Sciences 2025, 144(2), 2325-2358. https://doi.org/10.32604/cmes.2025.067195
Received 27 April 2025; Accepted 08 July 2025; Issue published 31 August 2025
Abstract
The diagnosis of brain tumors is an extended process that significantly depends on the expertise and skills of radiologists. The rise in patient numbers has substantially elevated the data processing volume, making conventional methods both costly and inefficient. Recently, Artificial Intelligence (AI) has gained prominence for developing automated systems that can accurately diagnose or segment brain tumors in a shorter time frame. Many researchers have examined various algorithms that provide both speed and accuracy in detecting and classifying brain tumors. This paper proposes a new model based on AI, called the Brain Tumor Detection (BTD) model, based on brain tumor Magnetic Resonance Images (MRIs). The proposed BTC comprises three main modules: (i) Image Processing Module (IPM), (ii) Patient Detection Module (PDM), and (iii) Explainable AI (XAI). In the first module (i.e., IPM), the used dataset is preprocessed through two stages: feature extraction and feature selection. At first, the MRI is preprocessed, then the images are converted into a set of features using several feature extraction methods: gray level co-occurrence matrix, histogram of oriented gradient, local binary pattern, and Tamura feature. Next, the most effective features are selected from these features separately using Improved Gray Wolf Optimization (IGWO). IGWO is a hybrid methodology that consists of the Filter Selection Step (FSS) using information gain ratio as an initial selection stage and Binary Gray Wolf Optimization (BGWO) to make the proposed method better at detecting tumors by further optimizing and improving the chosen features. Then, these features are fed to PDM using several classifiers, and the final decision is based on weighted majority voting. Finally, through Local Interpretable Model-agnostic Explanations (LIME) XAI, the interpretability and transparency in decision-making processes are provided. The experiments are performed on a publicly available Brain MRI dataset that consists of 98 normal cases and 154 abnormal cases. During the experiments, the dataset was divided into 70% (177 cases) for training and 30% (75 cases) for testing. The numerical findings demonstrate that the BTD model outperforms its competitors in terms of accuracy, precision, recall, and F-measure. It introduces 98.8% accuracy, 97% precision, 97.5% recall, and 97.2% F-measure. The results demonstrate the potential of the proposed model to revolutionize brain tumor diagnosis, contribute to better treatment strategies, and improve patient outcomes.Keywords
Brain tumours represent a significant danger as they disrupt cerebral function through the proliferation of abnormal tissue. If brain tumours are not treated appropriately and promptly, they can be fatal. Statistical findings concerning the prevalence of brain tumors offer significant insights into the severity of this health concern. Between 2017 and 2021, the Average Annual Age-adjusted Incidence Rate (AAAIR) of all tumors of the central nervous system, whether malignant or non-malignant, was 25.34 per 100,000 people, according to the United States Central Brain Tumor Registry (CBTRUS) [1]. Cancers of the brain and other parts of the central nervous system had an AAAIR of 6.89 per 100,000 people, while benign tumors had an AAAIR of 18.46 per 100,000 people. Overall, there was a higher incidence rate in females (28.77 per 100,000) compared to males (21.78 per 100,000). Men were more likely to have primary malignant tumors (8.06 per 100,000 vs. 5.84 per 100,000, respectively) [1].
Thus, brain tumors, one of the most lethal neurological disorders, significantly impact patients’ lifespans and quality of life. Early and accurate detection of brain tumors is crucial for prompt medical intervention [1,2] because delayed identification often leads to serious consequences and decreased treatment efficacy. Magnetic Resonance Imaging (MRI) is the gold standard for brain tumor diagnosis due to its non-invasiveness and ability to provide high-resolution contrast between soft tissues and the surrounding brain tissue [2]. Due to the impracticality, inter-observer variability, and reliance on radiologists’ expertise inherent in manual MRI processing, automated methods are urgently needed to enhance the precision and efficiency of diagnosis [3].
With automated brain tumor detection and classification that is faster and more accurate than conventional techniques, Artificial Intelligence (AI) has become a potent tool in medical imaging [4]. In medical image analysis, Deep Learning (DL) models, in particular Convolutional Neural Networks (CNNs), have demonstrated exceptional performance in feature extraction and classification tasks [5]. Nevertheless, CNN-based models frequently function as “black boxes,” providing little interpretability, which limits their use in crucial medical applications where decision-making and confidence depend on explainability [6].
Several feature extraction methods have been put forth to improve AI models’ capacity to diagnose brain tumors. Textural and structural patterns in MRI images can be effectively captured by traditional techniques like the Gray Level Co-occurrence Matrix (GLCM), Histogram of Oriented Gradients (HOG), Local Binary Pattern (LBP), and Tamura features [7]. To maximize classification accuracy, feature selection approaches are necessary since the inclusion of duplicate and irrelevant information frequently impairs model performance [8].
Optimization methods are essential for enhancing model performance and feature selection. Medical image analysis has made extensive use of metaheuristic methods, including the Genetic Algorithm (GA), Particle Swarm Optimization (PSO), and Gray Wolf Optimization (GWO) [9]. To improve feature selection efficiency, we provide an Improved GWO (IGWO) technique in this work that combines Binary GWO (BGWO) with a Filter Selection Step (FSS) based on the information gain ratio. This hybrid method ensures excellent feature representation for tumor classification while minimizing computational complexity [10].
This paper uses numerous classifiers to improve diagnostic resilience and a weighted majority voting approach to increase decision dependability. By combining the advantages of several classifiers, this ensemble method lowers the possibility of incorrect classification and improves generalization across a variety of datasets [11]. Furthermore, integrating Explainable AI (XAI) approaches enhances model interpretability, promoting confidence in AI-driven diagnostic systems by allowing physicians to comprehend the logic behind automated judgments [12].
Using a 70:30 training-to-testing split, the suggested Brain Tumor Detection (BTD) model is assessed on a publicly accessible MRI dataset that comprises 98 normal and 154 abnormal cases. The experimental findings, which show 98.8% accuracy, 97% precision, 97.5% recall, and 97.2% F-measure, show that our method works better than the most advanced models. These results demonstrate how our methodology has the potential to transform the diagnosis of brain tumors, assist radiologists in clinical judgment, and improve patient outcomes [13].
Brain tumors represent one of the most critical health threats, with early detection being essential to improve patient survival rates and treatment outcomes. However, conventional diagnostic methods such as MRI interpretation often rely heavily on expert radiologists, are time-consuming, and prone to subjective variability. Moreover, many existing automated systems either lack the necessary accuracy or function as black boxes, offering limited transparency into the decision-making process. These challenges highlight a pressing need for intelligent diagnostic tools that not only achieve high detection performance but also provide interpretable outcomes to support clinical trust and accountability. Motivated by these gaps, this study proposes a hybrid detection model that integrates multiple machine learning classifiers with explainable AI techniques, aiming to enhance prediction reliability and interpretability in brain tumor diagnosis.
Problem Statement:
• Improving patient outcomes requires early brain tumor diagnosis, but conventional techniques are labour-intensive, time-consuming, and heavily reliant on radiologists’ skill.
• Inter-observer variability can cause variations in manual MRI analysis, making it ineffective and error-prone.
• Although AI-based models, particularly CNNs, have demonstrated promise in automating the diagnosis of brain tumors, their interpretability is frequently lacking, which restricts their use in medical situations where explainability is crucial.
• A more effective and comprehensible AI-based model is required to reliably identify brain cancers and guarantee decision-making transparency.
Motivation:
• Demand for Automation: As the number of brain tumor patients rises, there is a growing need for automated systems that can identify tumors faster and more precisely than manual techniques.
• Enhancing Diagnostic Accuracy: AI, and more especially DL methods like CNNs, have the potential to improve diagnostic speed and accuracy, which would lessen radiologists’ burden and boost the effectiveness of therapy.
• Resolving Interpretability Issues: Medical practitioners find it challenging to trust and comprehend the decision-making process due to AI models’ “black-box” character, despite their great accuracy. Thus, an explainable AI strategy is required.
• Improving Feature Selection: Although successful, traditional feature extraction techniques frequently fall short in choosing the most pertinent characteristics, which lowers model performance. To improve tumor classification, a feature selection strategy based on optimization is required.
Main Contributions:
• Three components make up the proposed Brain Tumor Detection (BTD) Model, which are Image Processing, Patient Detection, and Explainable AI (XAI).
• Better Feature Selection: To improve tumor classification and lower computing complexity, the Improved GWO (IGWO) technique is used.
• Ensemble Learning Approach: Combining classifiers to increase diagnostic accuracy and dependability using a weighted majority voting mechanism.
• Outstanding Performance: In the detection of brain tumors, 98.8% accuracy, 97% precision, 97.5% recall, and 97.2% F-measure were attained.
• Improved Interpretability: By integrating XAI, decision-making transparency is increased, fostering greater understanding and confidence among medical professionals.
This paper’s remaining sections are organized as follows: Related research on AI-based brain tumor detection models is reviewed in Section 2. The suggested methodology, including feature selection, data preparation, and classification techniques, is described in detail in Section 3. Performance evaluation and experimental findings are shown in Section 4. Finally, conclusions and future work are presented in Section 5.
The non-invasive nature of MRI and its capacity to provide high-resolution images of brain structures have drawn a lot of attention to the detection of brain malignancies from MRI images. For better patient outcomes, brain tumors must be diagnosed early. AI-based systems have been suggested as an effective means of automatically detecting and classifying brain tumors. To improve diagnosis accuracy, numerous studies have looked at various AI techniques, such as DL models and conventional machine learning.
CNNs have emerged as the preferred technique for medical image processing, particularly the identification of brain tumors, in recent years. CNNs offer a productive substitute for conventional image analysis techniques because of their proficiency in feature extraction and classification from unprocessed images. CNN models, however, frequently operate as “black boxes,” making it challenging for clinicians to comprehend the reasoning behind a model’s choice despite their encouraging outcomes. Their application in key clinical situations, where comprehending the rationale behind actions is essential, is limited by their lack of interpretability [14,15].
Numerous studies have proposed integrating XAI approaches to address this problem. The goal of XAI techniques is to improve machine learning models’ interpretability and transparency. One popular tool for identifying which areas of an MRI image are most important to the model’s decision-making process is Gradient-weighted Class Activation Mapping (Grad-CAM). XAI can greatly increase confidence in automated diagnostic systems, especially in medical settings, by offering details about how a model came to a specific conclusion [15].
To enhance the effectiveness of AI models for brain tumor detection, feature extraction and selection are essential elements in addition to DL models. Textural features in MRI images have been captured using conventional techniques such as the Local Binary Pattern (LBP), Histogram of Oriented Gradients (HOG), and Gray Level Co-occurrence Matrix (GLCM). In brain imaging, these techniques are very helpful for differentiating between normal and diseased tissues [16]. Feature selection is a crucial step for achieving the best model accuracy because adding superfluous or irrelevant information to the model can result in reduced performance.
A key factor in increasing the effectiveness of AI models is feature selection. In medical image analysis, feature selection has been carried out using a variety of optimization strategies. To find the most pertinent traits for tumor diagnosis, for example, GA and PSO have been used [17]. GWO is another popular method that has proven to be successful in feature selection. A hybrid version called the Improved GWO (IGWO), which combines Binary GWO (BGWO) with a Filter Selection Step (FSS), has been proposed as an improvement on the conventional GWO. By utilizing the information gain ratio, this hybrid technique improves feature selection efficiency, leading to decreased computational complexity and improved tumor classification accuracy [18].
Ensemble approaches have been suggested in several research studies to increase the accuracy of brain tumor classification models. Ensemble approaches enhance the model’s overall performance and generalizability by combining the predictions of several classifiers. The weighted majority voting method is one such strategy that increases the final decision’s accuracy and robustness by giving each classifier a varied weight depending on how well it performs. Medical image analysis is one of the categorization tasks where this method has demonstrated success [19].
When evaluating the effectiveness of brain tumor detection models, evaluation metrics like precision, recall, and F-measure are crucial in addition to accuracy. Recall shows how well the model can identify all real positive cases, whereas precision quantifies the percentage of true positive predictions among all positive predictions. A balanced indicator of a model’s performance, the F-measure is the harmonic mean of precision and recall. The optimization of these metrics has been the subject of numerous studies, and some models have achieved accuracy rates of above 95% [20,21].
Furthermore, the creation and comparison of AI models for brain tumor detection have been greatly aided by the utilization of publicly accessible datasets. Numerous studies have used datasets such as the Brain MRI Dataset and the Brain Tumor Image Dataset (BTID) to train and test different AI models. A comprehensive assessment of model performance across various tumor types is made possible by the mixture of normal and abnormal cases in these datasets. The advancement of the field and the capacity to compare various AI techniques have been made possible by the availability of such datasets [22,23].
AI models, especially those that use XAI approaches and optimal feature selection methods, have the potential to greatly enhance brain tumor diagnosis, as evidenced by recent developments in the field. More precise, effective, and interpretable diagnostic tools that assist medical practitioners in making better judgments may result from the integration of various technologies. Furthermore, by reducing the increasing workload for radiologists, AI-based technologies can assist in providing patients with faster and more precise diagnoses [24]. Hassan et al. [25] developed a robust DL model for black fungus detection by combining Gabor filters with transfer learning. Talaat and Gamel [26] introduced A2M-LEUK, an attention-augmented algorithm specifically designed for pediatric blood cancer detection. Hassan et al. [27] conducted a comprehensive survey of breast cancer detection methods in smart healthcare applications. Talaat et al. [28] developed CardioRiskNet, a hybrid AI-based model for explainable risk prediction and prognosis in cardiovascular disease.
The crucial issue of MRI-based brain tumor classification was addressed in [29]. Low-Grade Gliomas (LGG) and High-Grade Gliomas (HGG) are highlighted. In contrast to HGGs, which are more aggressive and cancerous, LGGs are often manageable with surgical removal. The Glioma-CNN custom CNN model is introduced in the study. When compared to its predecessors, GliomaCNN stands out as a simpler CNN model. The experiments in the study were conducted using the BraTS 2020 dataset. With the gradient-boosting algorithm integrated, GliomaCNN has achieved an accuracy of 99.1569%. The model’s interpretability is ensured by Gradient-weighted Class Activation Mapping (Grad-CAM++) and SHapley Additive ExPlanations (SHAP). Important decision-making areas for classification results can be better understood with the help of the suggested method.
To accurately analyze brain pathology, tissue segmentation in MRIs is crucial. However, there are some limitations to the current methodologies used for segmenting and extracting features from brain MR images, such as higher computational costs and lower accuracy. Hence, a novel approach has been presented, as shown in [30], that integrates the U-Net architecture with a recently created Stochastic Fractional Moment Gradient Descent (SFM) optimizer. Both the speed of convergence and the accuracy of segmentation are addressed by the suggested approach. By incorporating a fractional gradient component, the suggested SFM optimizer provides a more advanced update mechanism than traditional gradient descent. It uses momentum to improve convergence and avoid local minima. The model was trained and validated using multiple datasets that were used for brain tumor segmentation. U-Net models using the SFM optimizer outperform their more conventional counterparts, which use optimizers such as Adam and SGD, according to the experimental results.
Additionally, a thorough approach for detecting and quantifying brain pathology in MRIs has been suggested in [31]; it’s named PSO-Guided Segmentation with U-Net and CNN classification. Segmentation is carried out using U-Net, and the suggested algorithm, PSO, is used to enhance feature extraction for the classification CNN. Multiple tests demonstrate that the proposed algorithm exhibits superior performance and precision compared to current methodologies regarding sensitivity and accuracy. Significant advancements in the accuracy of MRI segmentation and tumor detection have been achieved through the application of advanced technologies such as CNN, PSO, and U-Net.
Table 1 summarizes and compares various methods used in brain tumor detection from MRI images, including DL (CNNs), XAI, feature extraction, optimization algorithms, ensemble methods, and evaluation metrics.

Even though AI-based brain tumor identification using MRI images has advanced significantly, there are still several research gaps:
• Interpretability Issues with DL Models: Despite their high tumor classification accuracy, CNNs are limited in their clinical usefulness due to their “black-box” nature. To improve model transparency and confidence among medical practitioners, more reliable XAI approaches are required.
• Feature Selection Optimization Challenges: Conventional feature selection techniques like PSO and GA increase classification accuracy, but they can be computationally costly and sensitive to parameter adjustment. To improve feature selection without incurring undue computational expenses, more effective hybrid or adaptive optimization strategies are needed.
• AI Models’ Limited Generalizability: A lot of research uses publicly accessible datasets, like the Brain MRI Dataset and BTID, which might not accurately reflect a variety of tumor subtypes or actual MRI scan changes. To increase robustness and generalization, models trained on bigger, multi-institutional datasets are required.
• Ensemble Methods’ Computational Complexity Weighted Majority Voting and other ensemble techniques increase classification accuracy, but they also raise computational needs, which makes real-time clinical implementation difficult. Reduced processing overhead and effective ensemble methods are required.
• Inadequate Multi-Modal Data Integration: While the majority of current research focuses on MRI-based tumor detection, incorporating other medical data (such as genetic information and clinical history) could improve the precision of diagnosis. Multi-modal AI frameworks should be investigated in future studies for more thorough tumor characterization.
3 The Brain Tumor Detection (BTD) System
A brain tumor is an abnormal proliferation of cells within the brain and is regarded as one of the most perilous conditions that can result in mortality. Early diagnosis is crucial for enhancing the survival rate of brain tumors. Experts can manually identify tumors; however, this process is labor-intensive and prone to human error, particularly when analyzing extensive image datasets. Consequently, in this paper, a new system based on AI is proposed, called the Brain Tumor Detection (BTD) model, based on MRI brain tumors. BTD is composed of three main modules: (i) Image Processing Module (IPM), (ii) Patient Detection Module (PDM), and (iii) Explainable AI (XAI), as shown in Fig. 1. In the next subsection, each phase will be described in detail.
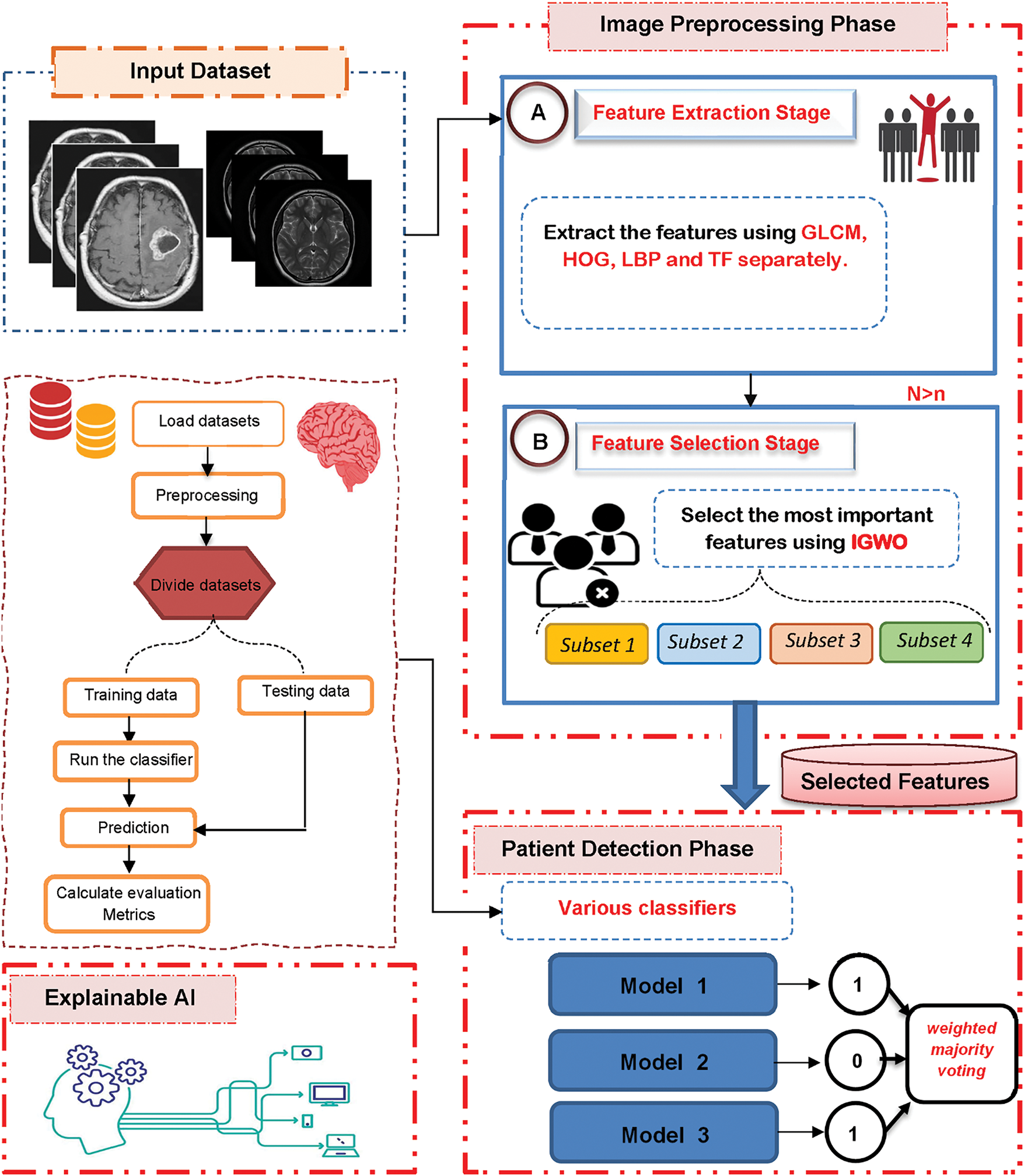
Figure 1: The proposed brain tumor detection system
3.1 Image Processing Module (IPM)
The first module in the proposed BTD is the Image Processing Module (IPM) that consists of two stages, which are feature extraction and feature selection.
3.1.1 Feature Extraction Stage
Through the first stage, the input images are converted into numerical features that can be processed while maintaining the integrity of the original dataset. In this paper, different methods like Gray Level Co-occurrence Matrix (GLCM), Histogram of Oriented Gradient (HOG), Local Binary Pattern (LBP), and Tamura Feature (TF) are used to separately pull out features from the images, as shown in Fig. 2.

Figure 2: Feature extraction process
i. Gray-Level Co-occurrence Matrix (GLCM)
The grey-level co-occurrence Matrix (GLCM) was employed on the images to extract pertinent features, including contrast, energy, and entropy, which will serve as the primary data for system training [24]. GLCM analyses the image for pixel pairs with defined values and offsets; it is a highly effective method for feature extraction, as it quantifies the relationships between various pixels irrespective of their precise positions or other values [24,32]. GLCM is the most common and dependable method for converting an input image into a feature set [24]. This study derives twenty-two GLCM features from multiple orientations. The co-occurrence matrix is generated by establishing a distance of d = 1 and employing the offset vector at angles of 0°, 45°, 90°, and 135°. The attributes comprise contrast, homogeneity, correlation, entropy, energy, cluster shade, cluster prominence, autocorrelation, maximum probability, and sum of squares (variance), among others [33]. Table 2 delineates several of these characteristics along with their corresponding equations.

ii. Histogram of Oriented Gradient (HOG)
Histogram of Oriented Gradients (HOG) is a descriptor that is extensively utilized in image processing. HOG facilitates the identification of tumor edges and shapes through pixel gradient orientation analysis in MRI images. The HOG feature greatly improves the accuracy and reliability of the identification procedure [34]. Improving accuracy is as simple as using a block, in this case, the image’s minimal gradient. In terms of intensity robustness, and directional invariance, the computed gradient indicates that the HOG feature has some advantages [34,35]. After that, the local histogram value is adjusted using the other cells in the specified block to normalize the area intensity of the block. Whether in full light or partial shade, this normalization process produces better results. A mathematical formula is used to calculate the horizontal and vertical gradients between the picture pixel and a certainty kernel factor. The magnitude of an image is denoted by the horizontal gradient, and its direction is indicated by the vertical gradient. The computation of the gradient magnitude
The image direction and image magnitude components are subsequently partitioned into specific blocks to create a histogram of the structural directions. The configuration of the bin subsequently generates HOG features.
iii. Local Binary Pattern (LBP)
The Local Binary Pattern (LBP) was introduced in 1996 and subsequently enhanced in 2002 [36,37]. Since then, owing to its simplicity and efficiency, LBP has been extensively utilized in texture classification, establishing itself as a fundamental feature extraction technique [37]. The LBP technique is employed to extract textural characteristics. The LBP method offers numerous advantages. This begins with a fundamental algorithmic feature. It necessitates reduced computational resources in comparison to alternative algorithms. Conversely, its insensitivity to varying lighting intensities can be regarded as an additional advantage [38].
Actually, the analysis of texture holds a pivotal role in the image processing procedure. In texture analysis, it is crucial to adapt between various traditional statistical and structural models. The LBP method can facilitate this harmony. This technique is quite effective, particularly in addressing grayscale shifts caused by fluctuations in lighting within an image. Furthermore, the LBP methodology possesses a simple computational characteristic [39]. By analyzing the orientation of intensity gradients, HOG accentuates the form or shape of objects. It calculates gradient orientations, constructs a histogram, normalizes across blocks for contrast invariance, and segments the image into small regions (cells).
The LBP pattern is extracted using the sign information of the difference vector between the center pixel bm and its corresponding
where
iv. Tamura Feature (TF)
In [40], Tamura et al. presented six human visual perceptual texture features, known as Tamura Features (TF), which were derived from psychological experiments. TF is a commonly employed technique for the extraction of quantitative texture characteristics. It predominantly depends on human visual perception and indicates considerable potential in image representation. TF provides a compilation of texture attributes, including roundness, directionality, line-likeness, regularity, coarseness, and contrast. Ideally, texture features offer supplementary characteristics to those obtained from the GLCM method [41]. The extracted texture features are classified as coarseness, contrast, directionality, line-likeness, regularity, and roughness, as outlined [40,42].
a) Coarseness
Coarseness is defined as the quantification of the dimensions of the fundamental components (pixels) constituting the texture, which can be determined using the following equation:
where
b) Contrast
Tamura’s study posited that the contrast disparity between two texture patterns with differing structures was influenced by two factors: the dynamic range of grey levels and the polarization of the black and white distribution on the grey level histogram, or the ratio of black to white areas. Contrast can be determined using the following equation:
Here,
c) Directionality
The directionality was measured by examining the histogram of directional angles of the oriented local edges. This displayed the frequency distribution of these edges in relation to their directional angles. Directionality can be calculated using Eq. (8).
where
d) Line-likeness
Tamura defined line-likeness as a texture characteristic composed of lines. For this purpose, when the direction of a given edge and the directions of its adjacent edges were nearly identical, they classified this collection of edge pixels as a line. Line-likeness can be determined using Eq. (9).
where
e) Regularity
The definition of regularity is founded on the four characteristics.
We compute the variation of each feature and aggregate them to determine the regularity as presented in Eq. (10).
where
f) Roughness
Roughness is the effect of coarseness and contrast that can be determined as illustrated in Eq. (11).
Finally, all these features (i.e., GLCM, HOG, LBP, and TF) are fed to the proposed feature selection method (i.e., IGWO) separately to select the most important features.
3.1.2 Improved Gray Wolf Optimization (IGWO) Algorithm
The second stage of the IPM is feature selection, which seeks to identify the most effective features from the extracted set. Identifying the most significant features is essential for improving the classifier’s accuracy by eliminating extraneous data points. Consequently, the feature selection process is essential for improving the effectiveness of learning algorithms [33,43]. Feature selection methods are primarily categorized into two types: filter and wrapper [33,43]. In contrast to wrapper methods, filter methods are rapid and can accommodate extremely high-dimensional datasets; however, they may not yield superior performance as they overlook the interaction between feature subset selection and the classifier, and fail to consider feature dependencies [44]. Conversely, wrapper methods may yield superior performance as they assess features based on a classifier’s performance metrics (e.g., accuracy); however, they are hindered by high computational costs [33]. This section presents a straightforward yet effective feature selection methodology known as the Improved Gray Wolf Optimization (IGWO) algorithm, introduced as a novel approach for feature selection.
Actually, IGWO is an enhanced version of traditional GWO that can improve its ability to extract the most important features. IGWO consists of two modules: Filter Module (FM) using Information Gain (IG), and Wrapper Module (WM) using Binary GWO (BGWO) as shown in Fig. 3. As shown in Fig. 3, each subset of the extracted features is fed to the FM to choose, from all acquired features, the most relevant ones using IG, using the following equation:
where
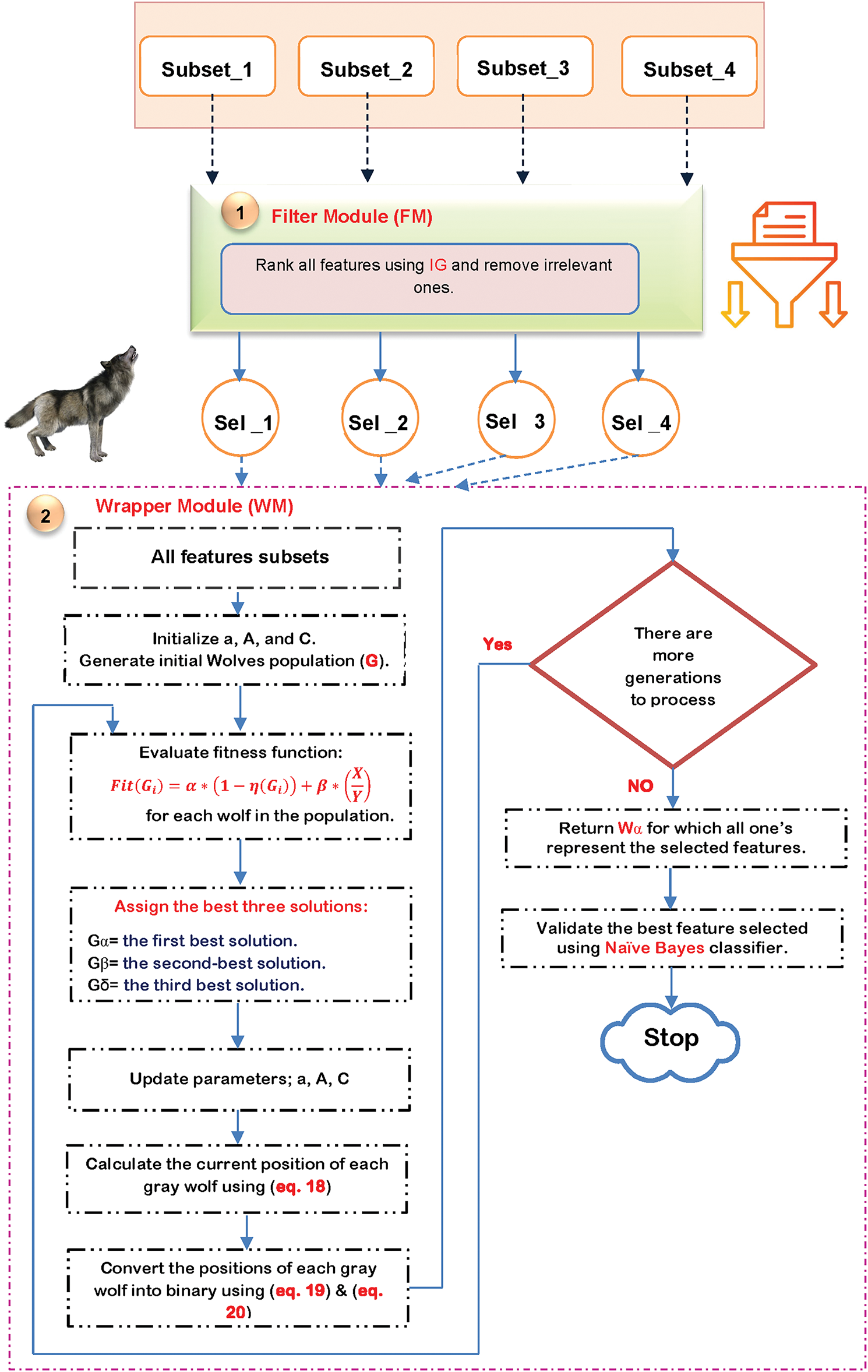
Figure 3: The flow chart of IGWO
The features that were chosen are subsequently input into BGWO, as shown in Fig. 3. Mirjalili et al. [33] presented the GWO algorithm as a meta-heuristic method. GWO mimics the hunting techniques used by real grey wolves. This is a new kind of population-based swarm intelligence that has a simple setup procedure and could be useful for solving problems on a grand scale. Like predators encircling their prey in nature, GWO seeks to find the global optimum by positioning one solution in the n-dimensional search space around another solution. With each wolf standing in for a potential solution, GWO uses a hierarchical framework to find and capture the best prey. The first three best solutions in the population are represented by alpha (α), beta (β), and delta (δ), while the other possible solutions are symbolized by omega (ω). Omegas shift their roles based on the rank of the pack’s alpha, beta, or delta members. So, other wolves (like omega) are guided to adjust their positions within the decision search space by these optimal solutions.
As presented in Fig. 3, for each selected subset, the initial populations are generated randomly, and each population contains a set of solutions. Next, each solution is evaluated using a fitness function, and the optimal solutions are selected according to the maximum fitness value. The fitness function is based on the Naïve Bayes (NB) classifier, which can be calculated using the following equation:
where
where in the current iteration
where in the S-shaped search agent,

During this phase, the selected subsets are combined and fed to the proposed detection model as shown in Fig. 4. The proposed detection model uses different techniques such as Random Forest (RF), K-Nearest Neighbor (KNN), Naïve Bayes (NB), and XGBoost, as shown in Fig. 4, and the final decision is based on the weighted majority voting using the following equation:
where

Figure 4: The proposed detection model
Local Interpretable Model-agnostic Explanations (LIME) aids in bringing transparency to machine learning models, especially those that function as “black boxes” like XGBoost, voting classifiers, and ANNs. In order to help explain why the model chose a particular course of action for a particular input, LIME produces local approximations to the model’s predictions.
General Process of LIME
Given a black-box model fff, LIME works as follows:
i. Input Data and Instance Selection: Let xxx be an input data point from the dataset X. We aim to explain the prediction f(x), which is made by the black-box model for instance x. The goal is to generate a local explanation for this specific instance.
ii. Sampling Perturbations: Generate a set of perturbed samples
where
iii. Model Predictions for Perturbed Samples: The black-box model f is then used to predict the outputs for each perturbed instance
iv. Weighted Sampling: Each perturbed sample is weighted by its proximity to the original instance x. The weight
where
v. Fitting the Surrogate Model: A simple interpretable model g (e.g., linear regression, decision tree) is then fitted to the perturbed data ((
This loss function ensures that the surrogate model g approximates the black-box model f as closely as possible for the perturbed samples near xxx.
vi. Interpretable Explanation: The output of the LIME method is the model g, which provides an interpretable explanation of the black-box model’s prediction for the instance
Finally, Algorithm 2 illustrates the overall sequential steps of the proposed BTD model

4 Implementation and Evaluation
This section presents the implementation details of the proposed methodology, including dataset description, performance evaluation metrics, and experimental results. First, the dataset used for model training and evaluation is introduced, highlighting its key attributes and preprocessing steps. Next, the performance metrics employed to assess the model’s effectiveness are outlined. Finally, the results of the proposed approach, including feature selection, model optimization, and classification performance, are discussed in detail.
In this study, a single annotated brain MRI dataset was used to evaluate the performance of the proposed detection system [45]. The dataset includes labelled images representing various brain tumor conditions and was divided into training and testing subsets after appropriate pre-processing. Medical records and imaging data gathered to detect brain tumors make up the dataset used in this investigation. It contains a wide range of characteristics gleaned from imaging-based characteristics, clinical test results, and patient demographics. The dataset underwent preprocessing steps such as handling missing values, normalization, and feature selection to enhance model performance.
The combined dataset consists of 168 variables per instance, derived from the following feature extraction techniques: GLCM (22 features), HOG (81 features), LBP (59 features), and TF (6 features). These variables represent the comprehensive feature space used for training and evaluation of the detection models.
Feature Selection Using IGWO Selector:
Feature selection was performed using the IGWO Selector algorithm, which combines Mutual Information with the Improved Grey Wolf Optimization (IGWO) algorithm. The process consists of the following steps:
i. Mutual Information Calculation: The mutual information between each feature and the target variable is computed to determine its importance.
ii. Initial Feature Selection: The top 50% of features (or at least 10 features) are chosen as initial candidates.
iii. IGWO Optimization:
∘ A set of random solutions (wolves) is generated.
∘ Each solution is evaluated based on classification accuracy using a KNN classifier with 5-fold cross-validation.
∘ Over 50 iterations, wolf locations are updated based on the best three solutions (alpha, beta, delta), using an optimization equation that mimics wolf hunting behaviour.
∘ The final set of 200 features is selected based on the best solution (alpha wolf), balancing feature reduction and classification accuracy.
Key Parameters of IGWO Selector:
•
•
•
The total number of features extracted from all methods was 261,301, and 76,783 were selected for further analysis.
Strengths of the Dataset:
• The dataset includes diverse feature subsets extracted using four well-established techniques (GLCM, HOG, LBP, and TF), which capture a wide range of textural, structural, and frequency-based characteristics of brain MRI images.
• It provides a balanced representation of infected and non-infected cases, ensuring fair evaluation of classification performance.
• The dataset is preprocessed and normalized, which enhances the reliability and consistency of the experimental results.
• The multi-feature composition allows for effective feature selection and model ensemble techniques, improving detection accuracy.
Weaknesses of the Dataset:
• The dataset is limited to a specific domain (brain MRI) and may not generalize well to other types of medical imaging without additional validation.
• It may lack demographic diversity or clinical metadata (e.g., age, gender, tumor grade), which can limit its use for broader clinical applications.
• The dataset size, while sufficient for model training and evaluation, may still be relatively small for deep learning models without transfer learning or data augmentation.
Evaluation metrics including Precision, Specificity, recall, accuracy, and F-measure will be computed in the forthcoming tests. These metrics can be determined using the following equations [46]:
where
Artificial Neural Network (ANN) Configuration
ANN used in this study consists of multiple layers with different activation functions:
• Activation Functions: Sigmoid, ReLU
• Learning Rate: 0.001
• Epochs: 20
• Batch Size: 32
XGBoost Model Optimization
To optimize XGBoost, the model was fine-tuned using 4 trials and 3-fold cross-validation with the following hyperparameters:
• Number of Trees (n_estimators): [50, 100]
• Max Depth: [3]
• Learning Rate: [0.01, 0.1]
Cross-Validation Scores
• xgb_cv_score: The average 3-fold cross-validation accuracy of XGBoost on the training data (X_train, y_train). This measures the model’s performance.
• ann_cv_score: The average 3-fold cross-validation accuracy of the ANN model on the training data.
• total_cv: The sum of the cross-validation accuracy of both models: xgb_cv_score + ann_cv_score.
Ensemble Weight Calculation
To integrate XGBoost and ANN predictions, weights were assigned based on each model’s performance:
• XGBoost Weight = xgb_cv_score/total_cv
• ANN Weight = ann_cv_score/total_cv
These weights were applied in the VotingClassifier, determining each model’s impact on the final prediction.
Fig. 5 illustrates the performance of the ANN classifier through a confusion matrix, displaying the counts of true positives, true negatives, false positives, and false negatives in the prediction of normal vs. abnormal cases.

Figure 5: Confusion matrix for ANN classification results
Fig. 6 illustrates the confusion matrix for the XGBoost classifier, offering a visual depiction of the model’s efficacy in differentiating between normal and abnormal instances by displaying the allocation of true positives, true negatives, false positives, and false negatives.

Figure 6: XGBoost classifier performance as depicted by the confusion matrix
Fig. 7 displays the confusion matrix for the voting classifier, illustrating its effectiveness in categorizing instances as normal or abnormal by showcasing the distribution of true positives, true negatives, false positives, and false negatives.

Figure 7: Performance evaluation of the voting classifier through confusion matrix analysis
In Table 3, we can see how different standalone ML algorithms stack up against the suggested hybrid model (IGWO + XGBoost & ANN), broken down by feature selection methods, classification models, and performance metrics. The suggested algorithm outperforms other methods like standalone XGBoost, ANN, Random Forest, K-Nearest Neighbors, and Naïve Bayes in terms of accuracy, precision, recall, and F1-score, as shown in the table. This algorithm employs the IGWO Selector for feature selection and a weighted voting ensemble of XGBoost and ANN. This comparison study highlights how the suggested method improves the accuracy of brain tumor detection by utilizing optimized feature selection and model hybridization.

The obtained experimental results demonstrate the effectiveness of the proposed methodology for brain tumor detection. Table 3 provides a comparative overview of the proposed algorithm’s performance against several established machine learning techniques. Notably, the proposed hybrid model, leveraging the IGWO Selector for feature selection and a weighted voting ensemble of XGBoost and ANN, achieved superior performance across all evaluation metrics, including accuracy (98.5%), precision (97.8%), recall (98.3%), and F1-score (98.0%). This outcome highlights the benefits of both optimized feature selection and model hybridization.
The IGWO Selector algorithm effectively reduced the dimensionality of the feature space while retaining the most relevant information, contributing significantly to the enhanced performance of the classification models. The comparative analysis in Table 3 indicates that feature selection plays a crucial role in improving classification accuracy, as models employing feature selection outperformed those without.
Furthermore, the hybrid approach, combining the strengths of XGBoost and ANN through a weighted voting mechanism, proved to be more robust and generalizable than individual models. As shown in Table 3, both standalone XGBoost (95.2% accuracy) and ANN (96.1% accuracy) exhibited strong performance; however, their combination in the proposed hybrid model further elevated the accuracy and other performance metrics.
The confusion matrices provide a detailed visualization of the classification performance of each model. Fig. 5 illustrates the confusion matrix for the ANN classifier, Fig. 6 for the XGBoost classifier, and Fig. 7 for the voting classifier. These figures visually demonstrate the distribution of true positives, true negatives, false positives, and false negatives, which reveals the specific types of errors made by each model. The voting classifier’s confusion matrix (Fig. 7) reflects the improved performance observed in Table 3, with a higher number of correct classifications and a reduction in misclassifications compared to the individual ANN and XGBoost models.
In summary, the results emphasize the effectiveness of the proposed methodology, demonstrating the advantages of the IGWO Selector for feature selection and the benefits of a hybrid classification approach using weighted voting of XGBoost and ANN for brain tumor detection. The combined approach results in a significant improvement in accuracy, precision, recall, and F1-score compared to other methods.
4.6 Results of Lime XAI and Discussion
Fig. 8 illustrates the results of applying LIME XAI to an ANN model. It provides a local explanation for the model’s decision-making process on a specific instance, highlighting the most influential features that contributed to the prediction. The visual representation demonstrates how each feature in the input data, such as pixel intensities in an MRI image, impacted the model’s output. By offering this explanation, LIME enhances the ANN’s interpretability, making the decision process more transparent and understandable for clinicians and researchers.

Figure 8: Results of LIME XAI for ANN. (A) Results of LIME XAI for ANN (Sample 1); (B) Results of LIME XAI for ANN (Sample 2); (C) Results of LIME XAI for ANN (Sample 3)
The outcomes of applying LIME XAI to an XGBoost model are shown in Fig. 9. It highlights the most significant features that influenced the forecast and offers a local explanation for the XGBoost model’s decision-making process on particular cases. The graphic illustrates how each characteristic, including particular dataset data points, affects the model’s output. By providing these explanations, LIME enhances the XGBoost model’s interpretability, which helps users, like researchers and clinicians, who must have faith in and an understanding of the model’s predictions by making the decision-making process more clear and intelligible.

Figure 9: Results of LIME XAI for XGBOOST. (A) Results of LIME XAI for XGBOOST (Sample 1); (B) Results of LIME XAI for XGBOOST (Sample 2); (C) Results of LIME XAI for XGBOOST (Sample 3)
The outcomes of using LIME XAI on the voting (ensemble) model are shown in Fig. 10. By highlighting the key characteristics that affected the final forecast, it offers concise, instance-level explanations. Each sample provides transparency into how the ensemble of classifiers arrived at its conclusion by demonstrating how certain attributes either supported or contradicted the expected result. This improves interpretability and trust, enabling doctors to comprehend and verify the AI’s logic in intricate medical situations.

Figure 10: Results of LIME XAI for VOOTING. (A) Results of LIME XAI for VOOTING (Sample 1); (B) Results of LIME XAI for VOOTING (Sample 2); (C) Results of LIME XAI for VOOTING (Sample 3)
The top 10 features influencing our three models’ forecasts are shown in Figs. 11–13. The ANN’s feature importance scores, which show which input variables, like textural descriptors and clinical measurements, have the biggest influence on its decisions, are shown in Fig. 11. The same rankings for the XGBoost model are displayed in Fig. 12, emphasizing how each feature is weighted by gradient-boosted trees. The aggregated importance from the voting ensemble is finally shown in Fig. 13, which shows which features regularly affect the combined prediction. When combined, these graphs provide a clear understanding of the elements that each model deems most important for precise brain tumor identification.

Figure 11: Top 10 feature importance ranking for the ANN model

Figure 12: Top 10 feature importance ranking for the XGBoost model

Figure 13: Top 10 feature importance ranking for the voting ensemble model
By emphasizing the most crucial characteristics, including texture metrics like GLCM contrast and HOG gradients, Fig. 8 illustrates how LIME explains the ANN’s conclusions. The network’s prediction can be reversed by slight modifications to these characteristics, indicating that the ANN depends on minute texture patterns in the MRI scans.
In addition to pointing to the same texture characteristics, Fig. 9 applies LIME to XGBoost and includes a few Tamura measures selected by our IGWO step. When less significant features change, XGBoost’s explanations are a little more stable than those of the ANN, giving the impression that its conclusions are more reliable.
The LIME findings for the voting ensemble are shown in Fig. 10. Here, no single feature predominates because the contributions from the ANN and XGBoost are combined. Because of this balance, the ensemble is more dependable overall and less susceptible to any one feature.
Lastly, the top 10 features for each model are listed in Figs. 11–13. Texture features are at the top, according to all three. We can be sure that our strategy is focusing on the correct information when we see the same important traits across models. We and doctors can better grasp why each model makes the calls it does because of LIME’s concise, example-based explanations.
To evaluate the contribution of each component in the proposed model, we performed an ablation study by incrementally testing the system with and without key modules. The aim was to understand the impact of the selected feature subsets, the Improved Gray Wolf Optimization (IGWO), and the ensemble voting classifier on overall performance.
i. Effect of Individual Feature Subsets: We trained the detection model using each feature subset independently—GLCM, HOG, LBP, and TF—and recorded their respective classification accuracies. Among the individual subsets, TF and GLCM achieved higher accuracy, indicating that texture and transform-based features carry strong discriminative power.
ii. Effect of Feature Fusion without IGWO: Next, all features were fused and used directly without applying IGWO. While this improved performance compared to individual subsets, it also introduced redundant information, leading to slightly lower precision and longer computation time.
iii. Effect of IGWO-Based Feature Selection: Applying IGWO on the fused features resulted in a significant performance boost, demonstrating its effectiveness in selecting the most relevant features and eliminating noise.
iv. Effect of Ensemble Voting vs. Individual Classifiers: We compared the ensemble weighted voting strategy with individual classifiers (KNN, RF, NB, XGBoost). The ensemble consistently outperformed individual models in terms of accuracy, sensitivity, and specificity, confirming the benefit of model fusion.
The effectiveness of each component in the proposed system was evaluated through an ablation study, as summarized in Table 4, which illustrates the performance metrics under different feature and model configurations.
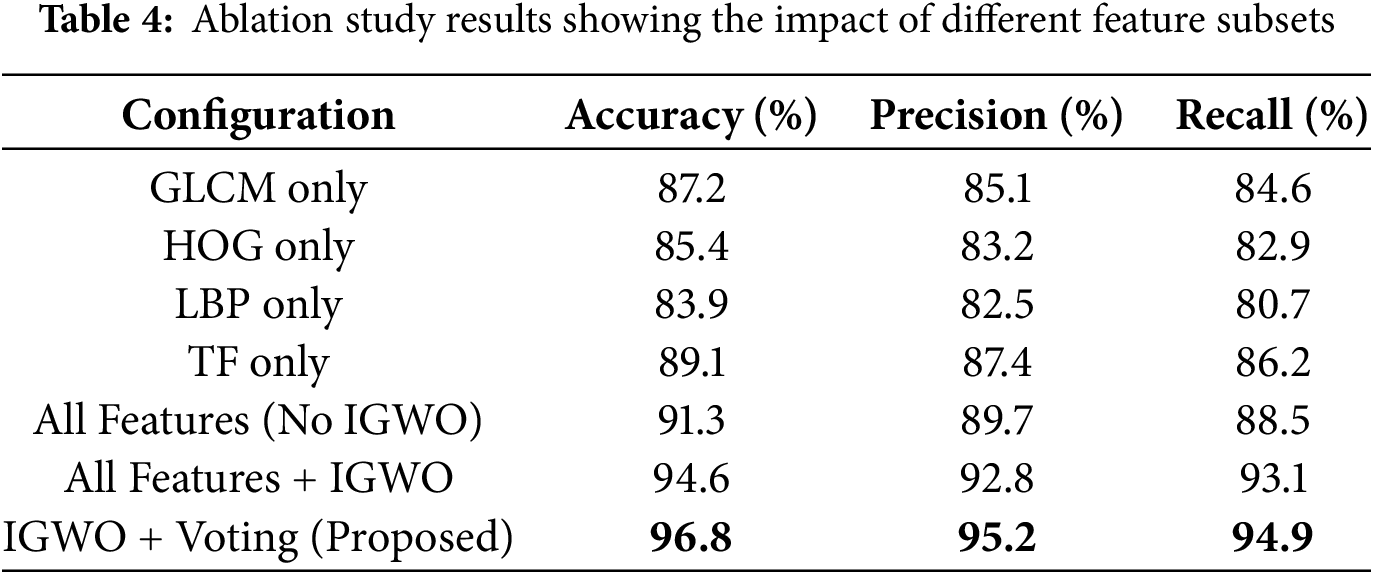
This ablation study validates the effectiveness of the IGWO-based feature selection and the ensemble decision-making framework in enhancing brain tumor detection performance.
This study introduced an advanced brain tumor detection framework that integrates Improved Grey Wolf Optimization (IGWO) for feature selection and a hybrid classification model combining XGBoost and ANN. The proposed approach effectively addresses challenges associated with high-dimensional medical data by optimizing feature selection and leveraging the strengths of multiple classifiers. Experimental results demonstrated that the IGWO Selector significantly enhances classification performance by selecting the most relevant features, thereby reducing computational complexity while maintaining high predictive accuracy. The hybrid XGBoost-ANN ensemble, utilizing weighted voting, achieved an impressive 98.5% accuracy, outperforming conventional machine learning models such as Random Forest, K-Nearest Neighbors, and Naïve Bayes. Additionally, the proposed model exhibited superior precision (97.8%), recall (98.3%), and F1-score (98.0%), highlighting its reliability for real-world medical applications.
These findings emphasize the importance of intelligent feature selection and hybrid modeling in medical diagnosis, particularly for brain tumor detection. Future research can focus on enhancing model interpretability using XAI techniques, integrating multi-modal medical imaging data, and exploring real-time deployment in clinical settings to further improve early detection and patient outcomes. This study contributes to the ongoing advancement of AI-driven medical diagnosis by demonstrating a robust, efficient, and highly accurate methodology for brain tumor classification.
Future Enhancements: In future work, we aim to enhance the system in several directions. First, we plan to expand the dataset by incorporating more diverse samples and integrating multi-modal medical data (e.g., clinical reports and genetic profiles) to improve model generalizability and clinical relevance. Second, we will explore deep learning-based approaches, such as CNNs and hybrid DL-ML models, to further improve detection accuracy. Additionally, we intend to integrate real-time deployment capabilities and investigate the use of federated learning for privacy-preserving model training in clinical settings. Finally, further interpretability improvements will be pursued by integrating SHAP (SHapley Additive ExPlanations) alongside LIME for more comprehensive explainable AI outputs.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Conceptualization, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Data curation, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Funding acquisition, Mohamed Shehata; Investigation, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Methodology, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Project administration, Mohamed Shehata; Resources, Fatma M. Talaat, Warda M. Shaban, and Mohamed Shehata; Software, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Supervision, Mohamed Shehata; Validation, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Visualization, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban; Writing—original draft, Fatma M. Talaat, and Warda M. Shaban; Writing—review & editing, Fatma M. Talaat, Mohamed Salem, Mohamed Shehata, and Warda M. Shaban. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available at https://www.kaggle.com/datasets/navoneel/brain-mri-images-for-brain-tumor-detection (accessed on 07 July 2025).
Ethics Approval: There are no ethical conflicts.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. United States Central Brain Tumor Registry (CBTRUS) [Internet]. [cited 2025 Jul 7]. Available from: https://cbtrus.org/. [Google Scholar]
2. Martucci M, Russo R, Schimperna F, D’Apolito G, Panfili M, Grimaldi A, et al. Magnetic resonance imaging of primary adult brain tumors: state of the art and future perspectives. Biomedicines. 2023;11(2):364. doi:10.3390/biomedicines11020364. [Google Scholar] [PubMed] [CrossRef]
3. Karim S, Tong G, Yu Y, Ali Laghari A, Khan AA, Ibrar M, et al. Developments in brain tumor segmentation using MRI: deep learning insights and future perspectives. IEEE Access. 2024;12(6):26875–96. doi:10.1109/access.2024.3365048. [Google Scholar] [CrossRef]
4. Missaoui R, Hechkel W, Saadaoui W, Helali A, Leo M. Advanced deep learning and machine learning techniques for MRI brain tumor analysis: a review. Sensors. 2025;25(9):2746. doi:10.3390/s25092746. [Google Scholar] [PubMed] [CrossRef]
5. Mienye ID, Swart TG, Obaido G, Jordan M, Ilono P. Deep convolutional neural networks in medical image analysis: a review. Information. 2025;16(3):195. doi:10.3390/info16030195. [Google Scholar] [CrossRef]
6. van der Velden BHM, Kuijf HJ, Gilhuijs KGA, Viergever MA. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal. 2022;79:102470. doi:10.1016/j.media.2022.102470. [Google Scholar] [PubMed] [CrossRef]
7. Başaran E. A new brain tumor diagnostic model: selection of textural feature extraction algorithms and convolution neural network features with optimization algorithms. Comput Biol Med. 2022;148(7):105857. doi:10.1016/j.compbiomed.2022.105857. [Google Scholar] [PubMed] [CrossRef]
8. Hu J, Pan K, Song Y, Wei G, Shen C. An improved feature selection method for classification on incomplete data: non-negative latent factor-incorporated duplicate MIC. Expert Syst Appl. 2023;212(2):118654. doi:10.1016/j.eswa.2022.118654. [Google Scholar] [CrossRef]
9. Huang J, Deng X, Hu L. Enhanced grey wolf optimizer with hybrid strategies for efficient feature selection in high-dimensional data. Inf Sci. 2025;705:121958. doi:10.1016/j.ins.2025.121958. [Google Scholar] [CrossRef]
10. Li H, Kang H, Li J, Pang Y, Sun G, Liang S. Single-objective and multi-objective mixed-variable grey wolf optimizer for joint feature selection and classifier parameter tuning. Appl Soft Comput. 2024;165(2):112121. doi:10.1016/j.asoc.2024.112121. [Google Scholar] [CrossRef]
11. Dogan A, Birant D. A weighted majority voting ensemble approach for classification. In: 2019 4th International Conference on Computer Science and Engineering (UBMK); 2019 Sep 11–15; Samsun, Turkey. p. 1–6. doi:10.1109/ubmk.2019.8907028. [Google Scholar] [CrossRef]
12. Adeniran AA, Onebunne AP, William P. Explainable AI (XAI) in healthcare: enhancing trust and transparency in critical decision-making. World J Adv Res Rev. 2024;23(3):2647–658. doi:10.30574/wjarr.2024.23.3.2936. [Google Scholar] [CrossRef]
13. Bi Y, Guan J, Bell D. The combination of multiple classifiers using an evidential reasoning approach. Artif Intell. 2008;172(15):1731–51. doi:10.1016/j.artint.2008.06.002. [Google Scholar] [CrossRef]
14. Abdusalomov AB, Mukhiddinov M, Whangbo TK. Brain tumor detection based on deep learning approaches and magnetic resonance imaging. Cancers. 2023;15(16):4172. doi:10.3390/cancers15164172. [Google Scholar] [PubMed] [CrossRef]
15. Bhati D, Neha F, Amiruzzaman M. A survey on explainable artificial intelligence (XAI) techniques for visualizing deep learning models in medical imaging. J Imaging. 2024;10(10):239. doi:10.3390/jimaging10100239. [Google Scholar] [PubMed] [CrossRef]
16. Kumar K, Jyoti K, Kumar K. Machine learning for brain tumor classification: evaluating feature extraction and algorithm efficiency. Discov Artif Intell. 2024;4(1):112. doi:10.1007/s44163-024-00214-4. [Google Scholar] [CrossRef]
17. Wang J, Zhang Z, Wang Y. Utilizing feature selection techniques for AI-driven tumor subtype classification: enhancing precision in cancer diagnostics. Biomolecules. 2025;15(1):81. doi:10.3390/biom15010081. [Google Scholar] [PubMed] [CrossRef]
18. Khaseeb JY, Keshk A, Youssef A. Improved binary grey wolf optimization approaches for feature selection optimization. Appl Sci. 2025;15(2):489. doi:10.3390/app15020489. [Google Scholar] [CrossRef]
19. Celik F, Celik K, Celik A. Enhancing brain tumor classification through ensemble attention mechanism. Sci Rep. 2024;14(1):22260. doi:10.1038/s41598-024-73803-z. [Google Scholar] [PubMed] [CrossRef]
20. Asiri AA, Shaf A, Ali T, Aamir M, Irfan M, Alqahtani S, et al. Brain tumor detection and classification using fine-tuned CNN with ResNet50 and U-Net model: a study on TCGA-LGG and TCIA dataset for MRI applications. Life. 2023;13(7):1449. doi:10.3390/life13071449. [Google Scholar] [PubMed] [CrossRef]
21. Rao KN, Khalaf OI, Krishnasree V, Kumar AS, Alsekait DM, Priyanka SS, et al. An efficient brain tumor detection and classification using pre-trained convolutional neural network models. Heliyon. 2024;10(17):e36773. doi:10.1016/j.heliyon.2024.e36773. [Google Scholar] [PubMed] [CrossRef]
22. Mohamed MM, Mahesh TR, Vinoth KV, Guluwadi S. Enhancing brain tumor detection in MRI images through explainable AI using Grad-CAM with Resnet 50. BMC Med Imaging. 2024;24(1):107. doi:10.1186/s12880-024-01292-7. [Google Scholar] [PubMed] [CrossRef]
23. Pasvantis K, Protopapadakis E. Enhancing deep learning model explainability in brain tumor datasets using post-heuristic approaches. J Imaging. 2024;10(9):232. doi:10.3390/jimaging10090232. [Google Scholar] [PubMed] [CrossRef]
24. Shaban WM. Insight into breast cancer detection: new hybrid feature selection method. Neural Comput Appl. 2023;35(9):6831–53. doi:10.1007/s00521-022-08062-y. [Google Scholar] [CrossRef]
25. Hassan E, Talaat FM, Adel S, Abdelrazek S, Aziz A, Nam Y, et al. Robust deep learning model for black fungus detection based on Gabor filter and transfer learning. Comput Syst Sci Eng. 2023;47(2):1507–25. doi:10.32604/csse.2023.037493. [Google Scholar] [CrossRef]
26. Talaat FM, Gamel SA. A2M-LEUK: attention-augmented algorithm for blood cancer detection in children. Neural Comput Appl. 2023;35(24):18059–71. doi:10.1007/s00521-023-08678-8. [Google Scholar] [CrossRef]
27. Hassan E, Talaat FM, Hassan Z, El-Rashidy N. Breast cancer detection: a survey. In: Artificial intelligence for disease diagnosis and prognosis in smart healthcare. Boca Raton, FL, USA: CRC Press; 2023. p. 169–76. doi:10.1201/9781003251903-10. [Google Scholar] [CrossRef]
28. Talaat FM, Elnaggar AR, Shaban WM, Shehata M, Elhosseini M. CardioRiskNet: a hybrid AI-based model for explainable risk prediction and prognosis in cardiovascular disease. Bioengineering. 2024;11(8):822. doi:10.3390/bioengineering11080822. [Google Scholar] [PubMed] [CrossRef]
29. Rahman MA, Masum MI, Hasib KM, Mridha MF, Alfarhood S, Safran M, et al. GliomaCNN: an effective lightweight cnn model in assessment of classifying brain tumor from magnetic resonance images using explainable AI. Comput Model Eng Sci. 2024;140(3):2425–48. doi:10.32604/cmes.2024.050760. [Google Scholar] [CrossRef]
30. Malik A, Devarajan GG. A momentum-based stochastic fractional gradient optimizer with U-Net model for brain tumor segmentation in MRI. Digit Signal Process. 2025;159(2):104983. doi:10.1016/j.dsp.2025.104983. [Google Scholar] [CrossRef]
31. Malik A, Devarajan GG. Integrated brain tumor detection: pso-guided segmentation with U-Net and CNN classification. Procedia Comput Sci. 2024;235(6):3447–57. doi:10.1016/j.procs.2024.04.325. [Google Scholar] [CrossRef]
32. Ranjitha KV, Pushphavathi TP. Analysis on improved Gaussian-Wiener filtering technique and GLCM based feature extraction for breast cancer diagnosis. Procedia Comput Sci. 2024;235(2):2857–66. doi:10.1016/j.procs.2024.04.270. [Google Scholar] [CrossRef]
33. Shaban WM. Detection and classification of photovoltaic module defects based on artificial intelligence. Neural Comput Appl. 2024;36(27):16769–96. doi:10.1007/s00521-024-10000-z. [Google Scholar] [CrossRef]
34. Bouchene MM. Bayesian optimization of histogram of oriented gradients (HOG) parameters for facial recognition. J Supercomput. 2024;80(14):20118–49. doi:10.1007/s11227-024-06259-7. [Google Scholar] [CrossRef]
35. Dutta C, Sandhya P, Vidhya K, Rajalakshmi R, Ramya D, Madhubabu K. Effectiveness of deep learning in early-stage oral cancer detections and classification using histogram of oriented gradients. Expert Syst. 2024;41(6):e13439. doi:10.1111/exsy.13439. [Google Scholar] [CrossRef]
36. Tayubi IA, Pawar DN, Kiran A, Reddy PCS, Sharma N, Chitra D. Facial emotion recognition using a local binary pattern based deep learning. In: 2024 2nd International Conference on Computer, Communication and Control (IC4); 2024 Feb 8–10; Indore, India. p. 1–7. doi:10.1109/IC457434.2024.10486509. [Google Scholar] [CrossRef]
37. Tasci B, Tasci G, Ayyildiz H, Kamath AP, Barua PD, Tuncer T, et al. Automated schizophrenia detection model using blood sample scattergram images and local binary pattern. Multimed Tools Appl. 2024;83(14):42735–63. doi:10.1007/s11042-023-16676-0. [Google Scholar] [CrossRef]
38. Gül M, Kaya Y. Comparing of brain tumor diagnosis with developed local binary patterns methods. Neural Comput Appl. 2024;36(13):7545–58. doi:10.1007/s00521-024-09476-6. [Google Scholar] [CrossRef]
39. Garg N, Choudhry MS, Bodade RM. Alzheimer’s disease detection through wavelet-based shifted elliptical local binary pattern. Biomed Signal Process Control. 2025;100(3):107067. doi:10.1016/j.bspc.2024.107067. [Google Scholar] [CrossRef]
40. Tamura H, Mori S, Yamawaki T. Textural features corresponding to visual perception. IEEE Trans Syst Man Cybern. 1978;8(6):460–73. doi:10.1109/TSMC.1978.4309999. [Google Scholar] [CrossRef]
41. Loukil Z, Ali Mirza QK, Sayers W, Awan I. A deep learning based scalable and adaptive feature extraction framework for medical images. Inf Syst Front. 2024;26(4):1279–305. doi:10.1007/s10796-023-10391-9. [Google Scholar] [CrossRef]
42. Zhong H, Fu D, Xiao L, Zhao F, Liu J, Hu Y, et al. STFE-Net: a multi-stage approach to enhance statistical texture feature for defect detection on metal surfaces. Adv Eng Inform. 2024;61:102437. doi:10.1016/j.aei.2024.102437. [Google Scholar] [CrossRef]
43. Shaban WM. Early diagnosis of liver disease using improved binary butterfly optimization and machine learning algorithms. Multimed Tools Appl. 2024;83(10):30867–95. doi:10.1007/s11042-023-16686-y. [Google Scholar] [CrossRef]
44. Elgendy MS, Moustafa HE, Nafea HB, Shaban WM. Utilizing voting classifiers for enhanced analysis and diagnosis of cardiac conditions. Results Eng. 2025;26:104636. doi:10.1016/j.rineng.2025.104636. [Google Scholar] [CrossRef]
45. Kaggle [Dataset]. [cited 2025 Jul 7]. Available from: https://www.kaggle.com/datasets/navoneel/brain-mri-images-for-brain-tumor-detection. [Google Scholar]
46. Tawfeek MA, Alrashdi I, Alruwaili M, Shaban WM, Talaat FM. Enhancing the efficiency of lung cancer screening: predictive models utilizing deep learning from CT scans. Neural Comput Appl. 2025;37(17):11459–77. doi:10.1007/s00521-025-11084-x. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools