 Open Access
Open Access
REVIEW
Deep Learning in Medical Image Analysis: A Comprehensive Review of Algorithms, Trends, Applications, and Challenges
1 Symbiosis Artificial Intelligence Institute, Symbiosis International (Deemed University), Pune, 412115, Maharashtra, India
2 Department of Electronics and Communication Engineering, Faculty of Engineering & Technology, Marwadi University Research Center, Marwadi University, Rajkot, 360003, Gujarat, India
3 Faculty of Engineering, Sohar University, Sohar, 311, Oman
4 Centre for Research Impact & Outcome, Chitkara University Institute of Engineering and Technology, Chitkara University, Rajpura, 140401, Punjab, India
5 College of Technical Engineering, The Islamic University, Najaf, 54001, Iraq
6 Department of Computer Science and Information Technology, Siksha ‘O’ Anusandhan (Deemed to be University), Bhubaneswar, 751030, Odisha, India
7 Department of Biotechnology, University Centre for Research and Development, Chandigarh University, Mohali, 140413, Punjab, India
* Corresponding Author: Dawa Chyophel Lepcha. Email:
(This article belongs to the Special Issue: Artificial Intelligence Models in Healthcare: Challenges, Methods, and Applications)
Computer Modeling in Engineering & Sciences 2025, 145(2), 1487-1573. https://doi.org/10.32604/cmes.2025.070964
Received 28 July 2025; Accepted 24 October 2025; Issue published 26 November 2025
Abstract
Medical image analysis has become a cornerstone of modern healthcare, driven by the exponential growth of data from imaging modalities such as MRI, CT, PET, ultrasound, and X-ray. Traditional machine learning methods have made early contributions; however, recent advancements in deep learning (DL) have revolutionized the field, offering state-of-the-art performance in image classification, segmentation, detection, fusion, registration, and enhancement. This comprehensive review presents an in-depth analysis of deep learning methodologies applied across medical image analysis tasks, highlighting both foundational models and recent innovations. The article begins by introducing conventional techniques and their limitations, setting the stage for DL-based solutions. Core DL architectures, including Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), Generative Adversarial Networks (GANs), Vision Transformers (ViTs), and hybrid models, are discussed in detail, including their advantages and domain-specific adaptations. Advanced learning paradigms such as semi-supervised learning, self-supervised learning, and few-shot learning are explored for their potential to mitigate data annotation challenges in clinical datasets. This review further categorizes major tasks in medical image analysis, elaborating on how DL techniques have enabled precise tumor segmentation, lesion detection, modality fusion, super-resolution, and robust classification across diverse clinical settings. Emphasis is placed on applications in oncology, cardiology, neurology, and infectious diseases, including COVID-19. Challenges such as data scarcity, label imbalance, model generalizability, interpretability, and integration into clinical workflows are critically examined. Ethical considerations, explainable AI (XAI), federated learning, and regulatory compliance are discussed as essential components of real-world deployment. Benchmark datasets, evaluation metrics, and comparative performance analyses are presented to support future research. The article concludes with a forward-looking perspective on the role of foundation models, multimodal learning, edge AI, and bio-inspired computing in the future of medical imaging. Overall, this review serves as a valuable resource for researchers, clinicians, and developers aiming to harness deep learning for intelligent, efficient, and clinically viable medical image analysis.Keywords
In modern clinical practice, the accuracy of cancer and other disease detection and diagnosis relies on the expertise of particular clinicians (e.g., radiologists, pathologists), leading to significant inter-reader variability in the interpretation of medical images [1]. To handle and resolve this clinical dilemma, many computer-aided diagnosis (CAD) systems [2] have been developed and evaluated with the intention of assisting physicians in understanding medical images more efficiently and making diagnostic decisions with greater accuracy and objectivity [3]. This practice is scientifically justified as it enables computer-aided, objective analysis of image features which can address several challenges in clinical practice, including the shortage of skilled clinicians, the risk of fatigue among human experts and the limited availability of medical resources [4]. While initial computer-aided diagnosis systems were established in the 1970s [5], advancements in CAD systems have accelerated since the mid-1990s due to the incorporation of more sophisticated machine learning practices into these systems [6–8]. Traditional CAD practices typically involve a three-step development process: image segmentation, feature computation, and disease classification. For example, Sahiner et al. [9] devised a CAD system for facilitating mass categorization on digital mammograms [10]. The regions of interest comprising the object masses were initially classified from the background utilizing a modified active contour system.
A substantial array of imaging elements was utilized for computing the lesions characteristics in terms of dimension, morphology, margin geometry, texture, and other factors. Thus, the unprocessed pixel data are transformed into a vector of representative characteristics. A classification model based on linear discriminant analyses (LDA) was ultimately employed on the feature vector to ascertain bulk malignancy. In contrast, deep learning models progressively identify and learn hidden patterns within regions of interest using the hierarchical structure of deep neural networks [11]. Throughout the process, significant characteristics of the input image will be progressively recognized and enhanced for specific tasks (e.g., detection, classification) while unnecessary features will be reduced and eliminated. An MRI image illustrating questionable liver lesions consists of a pixel array [12] with each entry serving as an individual source feature for the deep learning model. The initial layers of the design may acquire fundamental lesion information, including tumour morphology, position, and orientation [13]. The subsequent layer batch may identify and maintain traits consistently associated with lesion malignancy, while ignoring unimportant fluctuations. Relevant features will undergo additional processing and integration by following high levels in a more intangible manner. Increasing the numbers of layers enhances the feature representations level [14–18]. Throughout the complete process, significant attributes hidden within the raw input are identified by the general neural network structure in an unsupervised manner, eliminating the necessity for manual feature extraction [19–21]. Alom et al. proposed an improved U-Net architecture that maintains the same number of network parameters while achieving superior performance in medical image segmentation. The model was evaluated on multiple benchmark datasets, including blood vessel segmentation in retinal images, skin cancer segmentation, and lung lesion segmentation. Experimental results demonstrated its enhanced segmentation accuracy compared to existing models such as U-Net and residual U-Net (ResU-Net). The study presents an optimized architecture that achieves optimal performance without increasing network complexity. Due to its significant advantages, deep learning methodologies have emerged as the predominant technology in the CAD domain and have been extensively utilized across various tasks, such as disease classification, region of interest (ROI) segmentation, medical object detection, and image registration [22–25]. Supervised learning was the initial deep learning technique utilized in medical image analysis [26–28]. Despite its successful application in numerous contexts [29,30], the broader implementation of supervised algorithms in various scenes is significantly hindered by the limited dimension of several medical datasets. In contrast to traditional datasets in computer visions, the dataset typically comprises a limited number of images, with only a negligible portion of these images annotated by specialists. Fig. 1 depicts the taxonomy of deep learning methods, applications, and challenges in medical image analysis.
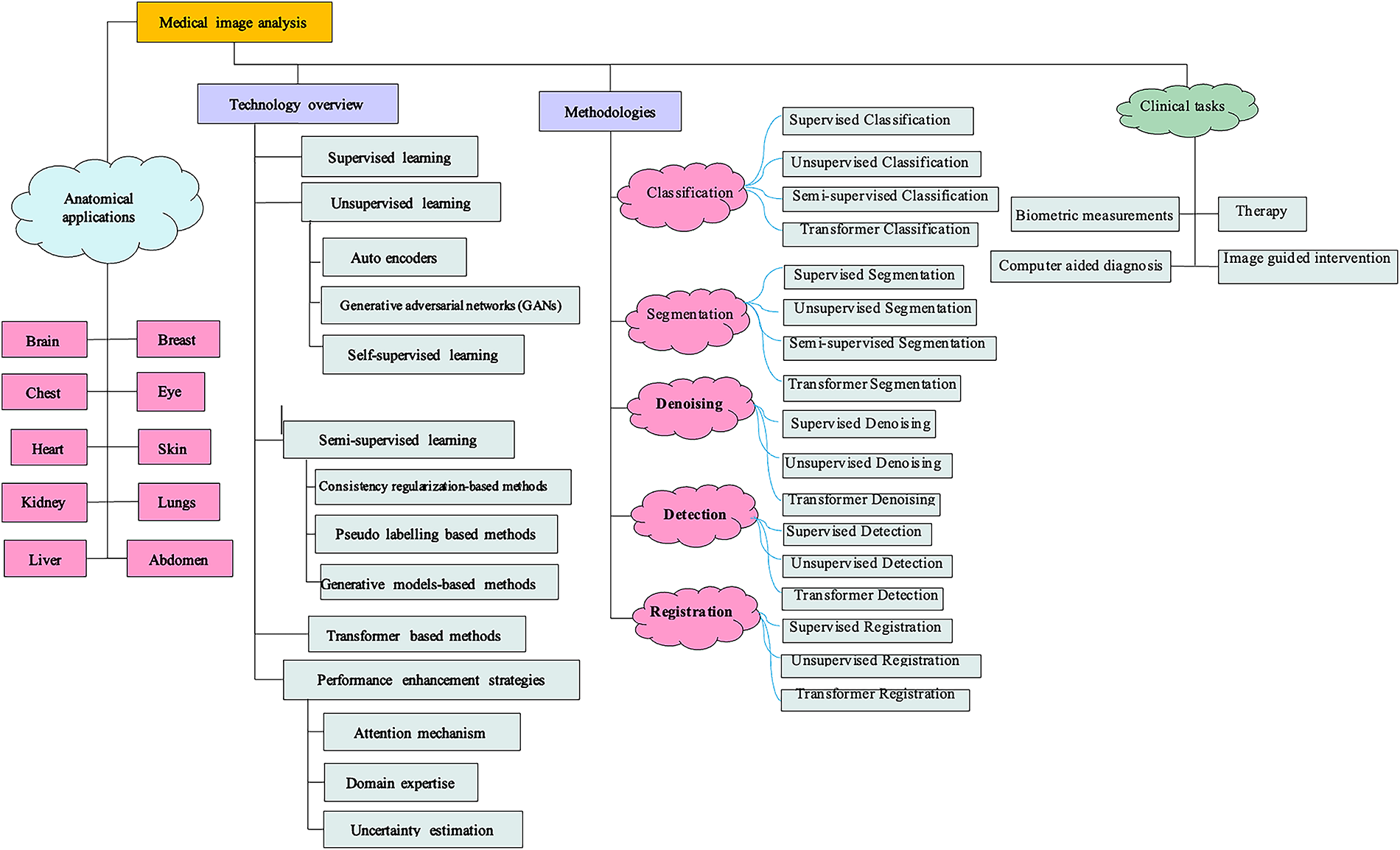
Figure 1: Overall taxonomy of this study outlining deep learning methods, applications, and challenges in medical image analysis
To address these restrictions, unsupervised as well as semi-supervised learning models have garnered significant attention in the past few years, enabling (1) the generation of additional labelled images from model optimisation, (2) the extraction of significant hidden information from unlabelled image data, and (3) the creation of pseudo labels for the unlabelled data. The number of outstanding survey publications summarizing deep learning usages in medical image analysis presently exist. References [31,32] examined early deep learning methodologies, primarily grounded in supervised procedures. Recently, References [33,34] examined the use of GANs in several medical imaging tasks. Reference [35] examined the application of semi-supervised learning and multiple instance deep learning in segmentation tasks. Reference [36] examined various methods to address dataset constraints (e.g., limited or inadequate annotations) specifically in image segmentation. These points echo earlier discussions, as they remain fundamental barriers for applying supervised learning at scale.
This study aims to elucidate how the medical image analysis domain, sometimes limited by scarce annotated data, may benefit from recent advancements in deep learning. Our study is notable from recent publications by its comprehensiveness and technical alignment. Firstly, we emphasize the applications of many promising methodologies under the machine learning paradigms, encompassing self-supervised, unsupervised, and semi-supervised learning simultaneously. Secondly, instead of focusing just on a particular task, we present the applicability of the above-mentioned learning methodologies across several important medical image analysis techniques. We precisely studied deep learning in detail, which is a topic that is hardly addressed in recent survey articles. We concentrated on the utilization of chest X-rays, mammograms, CT scans, MRI, PET images, and so on. All these image types share numerous common properties, which are analysed by radiologists within the same department (Radiology). We also referenced some general methodologies utilized in other image domains (e.g., histopathology, skin lesion, ultrasound, Dermoscopy, pneumonia, etc.) that may be applicable to radiographic or MRI images. Third, existing models for these techniques are explained thoroughly. We precisely examine the recent advancements in machine learning paradigm methodologies. This survey may benefit a broad audience, including researchers specializing in deep learning, big data, artificial intelligence, machine learning, and physicians or medical researchers.
The remainder of the article is structured as follows: Section 2 outlines the research process in medical image analysis. Section 3 presents deep learning paradigms relevant to medical imaging, while Section 4 offers a critical evaluation of prominent DL models. Section 5 explores clinical suitability and adoption barriers of emerging techniques. Section 6 emphasizes scalability and generalizability beyond benchmark datasets. Section 7 discusses evaluation metrics, regulatory frameworks, and interpretability. Section 8 introduces model maturity mapping for clinical readiness. Sections 9 and 10 address the research–clinic gap and deployment best practices. Sections 11 to 14 analyze model validation, clinical trends, dataset challenges, and security concerns. Finally, Sections 15 to 18 present open challenges, discussion, future directions, and the conclusion.
This study meticulously examined relevant documents that generally investigated the application of deep learning models in medical image analysis. This section thoroughly covers the domain of medical image analysis by using the systematic literature review exercise. This study involves a thorough evaluation of all research conducted on a pivotal topic. The study concludes with a comprehensive analysis of machine learning models in the domain of medical image analysis. The reliability of the research selection procedures is examined. The subsequent subsections contain further information regarding research methodologies, including selection criteria. The primary objectives of the research are to discover, evaluate, and distinguish all significant publications in the domain of deep learning systems for medical image analysis. The practice of systematic literature review can be employed to analyse the components and attributes of methodologies for achieving the specified objectives. Besides, a systematic literature review enables the attainment of a deep understanding of the critical issues and obstacles in this field.
2.1 The Process of Paper Analysis
The search and selection processes used in this inquiry are divided into five stages, as shown in Fig. 2. The total of 584 papers is obtained by using an electronic database to retrieve relevant materials such as journals, chapters, technical studies, notes, conference papers, and special issues. Following a thorough analysis of these papers in accordance with a set of predefined criteria, only those that satisfied the requirements shown in Fig. 3 are selected for additional assessment. Fig. 2 displays the distribution of publications in this first phase. There were 494 articles remaining at the end of the first phase. The titles and abstracts of the selected papers were carefully studied in the following step with an emphasis on the methodology, analysis, discussion, and conclusion of the publications to make sure they were pertinent to the research. Following this stage, only 429 papers were kept, and 315 papers were further processed and selected for a more thorough assessment with the final goal of selecting publications that satisfied the predefined parameters of the study.
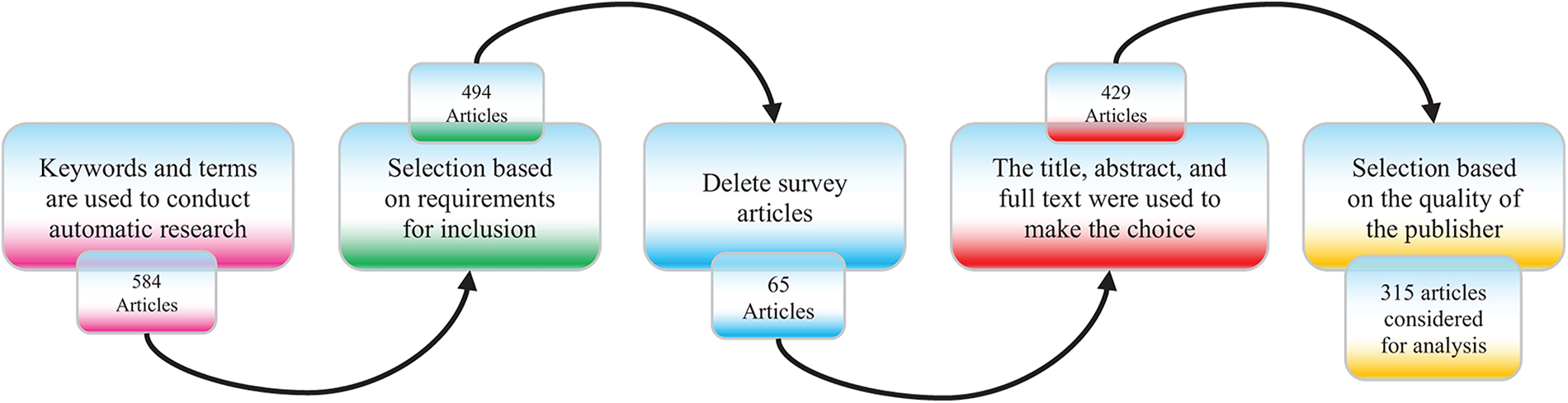
Figure 2: Phases of the article searching and selection process used in the systematic literature review
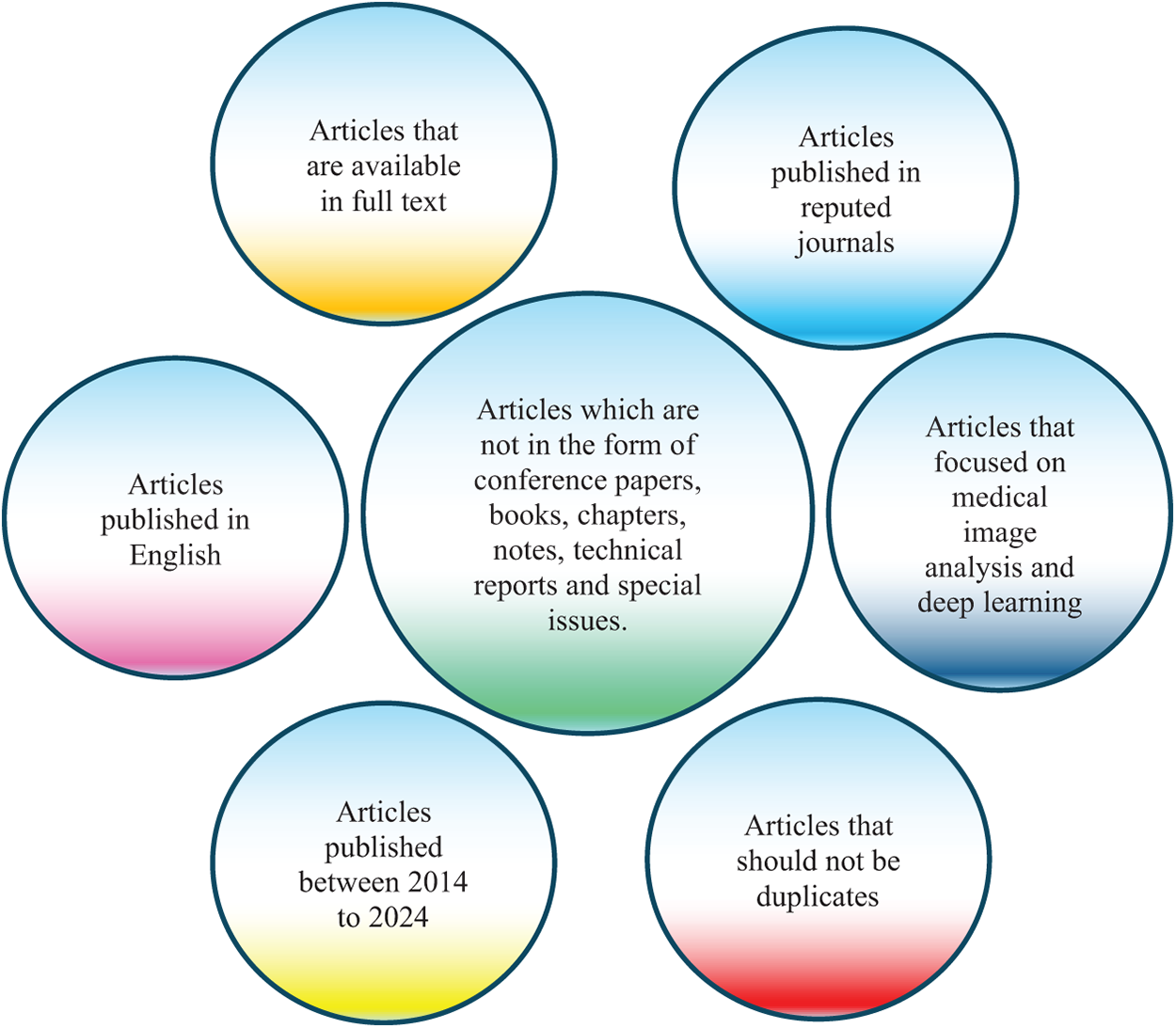
Figure 3: Criteria for inclusion in the article selection process used for identifying relevant studies
Fig. 4 provides a top-down roadmap of the study. The central theme of Deep Learning in Medical Image Analysis is first expanded into four major learning paradigms. These paradigms support a range of medical imaging applications, which are further contextualized in clinical domains such as oncology, cardiology, neurology, and infectious diseases. At the foundation, key challenges and future directions highlight emerging issues such as data scarcity, interpretability, federated and edge AI, ethical concerns, and the role of foundation models. Together, this overview helps readers navigate the structure and flow of the review.
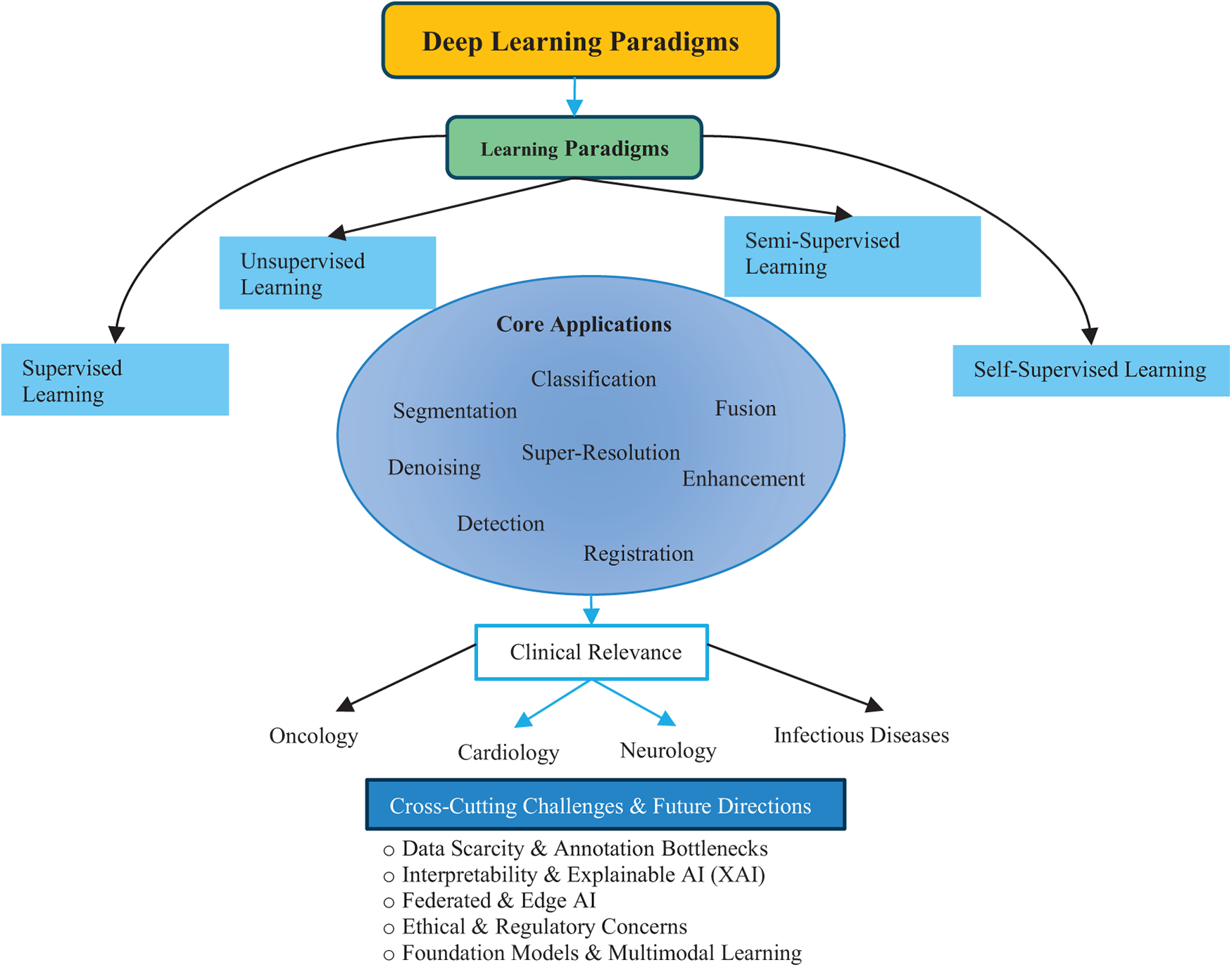
Figure 4: Overview of the review structure illustrating the relationship between deep learning paradigms, applications, and clinical domains
3 Deep Learning Paradigms in Medical Imaging
Deep learning (DL) has profoundly transformed medical image analysis, offering powerful tools for disease diagnosis, prognosis, and therapeutic planning; however, its methodological landscape encompassing supervised, unsupervised, semi-supervised, and self-supervised learning necessitates a critical examination not only of their theoretical formulations but also of their limitations, real-world relevance, and potential for generalization. Supervised learning remains the most established approach in the field, particularly through the use of Convolutional Neural Networks (CNNs) [37], which excel at hierarchically extracting features from images and yielding highly accurate classification and segmentation outputs when large, annotated datasets are available. Nonetheless, the dependence on extensive, high-quality labelled data severely restricts the scalability of this paradigm, especially in medical domains where data annotation demands expert radiologists, incurs high costs, and raises privacy concerns. In addition, CNNs are prone to performance degradation under class imbalance, a common issue in rare disease datasets, and often suffer from poor generalization to out-of-distribution data, thus undermining their utility in diverse clinical settings. Furthermore, their fixed receptive field size and pooling operations can result in the loss of subtle anatomical details that are critical for accurate medical interpretation, exacerbated by their black-box nature, which complicates clinical trust and transparency. In contrast, unsupervised learning offers an alluring promise of label-free discovery by modeling underlying data distributions using techniques such as autoencoders (AEs) [38], stacked autoencoders (SAEs) [39], and various regularized variants including sparse [40], denoising [41], and contractive autoencoders [42], which aim to extract compact and generalizable latent features. Yet, these models often fail to yield clinically useful representations without extensive tuning, and while deep generative frameworks like Variational Autoencoders (VAEs) [43] and their extensions using Gaussian mixture priors [44] or conditional architectures [45] attempt to address expressiveness, they struggle with reconstructing sharp and realistic medical images, undermining diagnostic reliability. Generative Adversarial Networks (GANs) [46] represent a more expressive alternative and are widely applied for image synthesis, domain translation, and augmentation, yet they are notoriously difficult to train, with frequent issues such as mode collapse and sensitivity to hyperparameters, although architectural innovations like Wasserstein GANs (WGANs) [47], conditional GANs (cGANs) [48], and auxiliary classifier GANs (ACGANs) [49] have significantly improved training stability and controllability. Nevertheless, the metrics typically used to evaluate these models’ reconstruction error or adversarial success do not always align with clinically meaningful performance, and as a result, their adoption in medical pipelines remains limited due to concerns around interpretability and reliability. Meanwhile, self-supervised learning (SSL) emerges as an innovative middle ground by automatically generating supervisory signals (pseudo-labels) from unlabelled data using pretext tasks such as image inpainting [50], colorization [51], relative patch prediction [52], jigsaw puzzle solving [53], and rotation recognition [54] inspired by its success in natural language processing with models like Bidirectional Encoder Representations from Transformers (BERT) [55]. Among SSL techniques, contrastive learning frameworks such as Momentum Contrast for Unsupervised Visual Representation Learning (MoCo) [56] and Simple Framework for Contrastive Learning of Visual Representations (SimCLR) [57] have gained significant traction by training models to maximize agreement between different augmented views of the same image while minimizing agreement with views from other images, using Information Noise-Contrastive Estimation (InfoNCE) loss [58] and projection heads to create well-separated latent spaces. However, while contrastive SSL has shown promise in natural image tasks, its application in medical imaging is still in its infancy, constrained by domain-specific challenges such as limited variability in anatomical structures, class imbalance, and the need for clinically appropriate augmentations. Recent studies highlight the potential of hybrid approaches that combine SSL with classical machine learning classifiers. Such frameworks can improve robustness and interpretability compared to fully end-to-end deep learning pipelines. For instance, a study demonstrated that SSL features coupled with conventional classifiers achieved superior performance in biomedical image analysis tasks [59,60]. These results suggest that hybrid SSL–Machine Learning (ML) paradigms may serve as strong alternatives in scenarios with limited labeled data and the need for greater interpretability.
Furthermore, the potential of SSL remains largely untapped in medical applications due to a lack of large-scale domain-specific studies and benchmark datasets, although it holds significant promise in overcoming annotation bottlenecks that plague other learning paradigms. Semi-supervised learning (also abbreviated as SSL) offers a practical compromise between supervised and unsupervised approaches by combining small labeled datasets with abundant unlabelled data, thus reducing the dependency on expert annotation. Early criticisms, such as those highlighted in [61], which demonstrated that unlabelled data could degrade performance, are now being revisited in light of more sophisticated deep semi-supervised architectures that consistently outperform their supervised counterparts in medical imaging tasks [62]. Consistency regularization methods, including Π-model and temporal ensembling [63], as well as ladder networks [64], enforce stability under perturbations by minimizing the difference in model outputs when noise or augmentation is applied to the same input, while mean-teacher models [65] improve robustness through a student-teacher framework optimized with dual loss components. Pseudo-labelling strategies [66], which generate soft labels for unlabelled data based on model confidence, are further enhanced through Mixup augmentation [67], unsupervised data augmentation (UDA) [68], and co-training techniques [69], each helping to refine model predictions iteratively with minimal supervision. Meanwhile, semi-supervised extensions of generative models, particularly GANs adapted for classification tasks [70,71], and the more modular Triple-GAN [72], show considerable promise in integrating both generative fidelity and discriminative accuracy, although training these multi-objective architectures remains challenging and computationally intensive. Across all paradigms, enhancing deep learning models for medical use necessitates strategic architectural interventions, such as attention mechanisms that enable models to focus on salient regions of interest. These mechanisms, inspired by visual cognition [73] and widely applied in NLP [74,75] have found success in vision tasks like image captioning [76–78], object recognition, and medical image segmentation [79,80] with specific implementations like spatial attention [81], channel attention [82], self-attention [83], and the hybrid convolutional block attention module (CBAM) [84,85] each offering tailored benefits in highlighting diagnostic features. The Transformer model built entirely on self-attention layers, epitomizes the trend towards attention-dominant architectures, although its high computational demands and dependency on large training corpora remain barriers to adoption in medical imaging. Furthermore, attention mechanisms, while improving localization and interpretability, add architectural complexity and are often dependent on large datasets to learn meaningful attention maps, which can limit their application in resource-constrained healthcare environments. Another crucial area is the integration of domain expertise, as many pre-trained models derived from natural images perform poorly on medical data due to limited texture diversity, subtle inter-class differences, and smaller dataset sizes. Effective DL applications in medicine often involve embedding anatomical priors [86,87], 3D spatial dependencies, and multimodal metadata [88] into network architectures, either via auxiliary channels, feature fusion, or custom loss functions; for instance, radiomic features and pathology reports can provide complementary information that boosts model performance and interpretability. While these integrations improve clinical relevance and model accuracy, they often require highly specialized architectural designs and limit cross-domain generalizability. Finally, the estimation of predictive uncertainty is essential for clinical trustworthiness, especially in high-stakes domains like oncology, where decisions based on erroneous predictions can have life-threatening consequences. Bayesian deep learning methods, such as Monte Carlo dropout (MC-dropout) [89], approximate posterior distributions over network weights to quantify uncertainty, while model ensembles [90] capture variance across independently trained models to provide robust confidence estimates. These uncertainty estimation techniques, although computationally expensive, enable critical functionalities such as error detection, selective prediction, and triage decision support [91], helping bridge the trust gap between AI systems and clinical practitioners. Despite their theoretical appeal, the practical implementation of uncertainty-aware models is still rare in deployed systems due to added computational burden, interpretability challenges, and a lack of standardized evaluation protocols. In conclusion, the landscape of deep learning for medical image analysis is marked by both remarkable progress and persistent challenges, with supervised learning offering strong performance under ideal data conditions, unsupervised learning pushing the boundaries of label-free modeling, semi-supervised learning providing data-efficient compromises, and self-supervised learning presenting a new frontier for leveraging unlabelled data through creative supervisory signals. Enhancing these paradigms through the strategic use of attention mechanisms, domain-specific features, and uncertainty estimation not only improves accuracy and generalization but also fosters clinical acceptance by addressing the foundational concerns of trust, transparency, and reproducibility. As the field evolves, a hybrid methodology that dynamically integrates these learning strategies in a modular, data-aware, and context-specific manner is likely to drive the next wave of breakthroughs in medical AI, moving beyond algorithmic performance metrics to prioritize real-world utility, ethical compliance, and patient safety. Bio-inspired models, including swarm intelligence and evolutionary algorithms, are increasingly applied in medical imaging. These approaches can optimize feature extraction, segmentation, and classification processes while offering improved adaptability to complex datasets. Recent works demonstrate their effectiveness in medical domains, reinforcing their role as complementary paradigms alongside conventional deep learning [92].
4 Critical Evaluation of Deep Learning Models
Deep learning-based classification in computer-aided diagnosis (CADx) has made significant strides in disease identification and lesion characterization from medical images [93]. However, rather than presenting a purely descriptive overview, it is critical to assess these methods in light of their performance bottlenecks, data dependencies, and clinical reliability, especially when deployed in diverse, real-world healthcare settings. Supervised learning models, such as AlexNet [94], Visual Geometry Group (VGG) [95], GoogleLeNet [96], ResNet [97], and DenseNet [98] have become foundational in CADx. These architectures provide deep hierarchical representations that excel in capturing complex spatial features in medical images. However, their reliance on extensive, high-quality annotated datasets poses a significant barrier, particularly in domains like 3D MRI and CT imaging, where expert-labeled data is scarce and difficult to acquire due to clinical workload, patient privacy, and cost [99,100]. Transfer learning has emerged as a practical solution to this challenge. By leveraging pretrained weights from large-scale datasets like ImageNet [101] or domain-specific medical image datasets, models can be effectively fine-tuned for target clinical tasks with significantly fewer training examples, consistently outperforming models trained from scratch [102]. These strategies have proven particularly effective across modalities such as CT [103], MRI [104], mammography [105], and X-ray [106], and are further enhanced by attention mechanisms [107–109] which enable models to focus on the most diagnostically relevant regions. For instance, Huo et al.’s Hierarchical Fusion Network (HiFuse) [110] employs a hierarchical multi-scale fusion of local and global features using an adaptive fusion block to improve representational power and classification accuracy across diverse imaging conditions. Fig. 5 demonstrated schematic flow diagram of a deep learning–based image classification network.
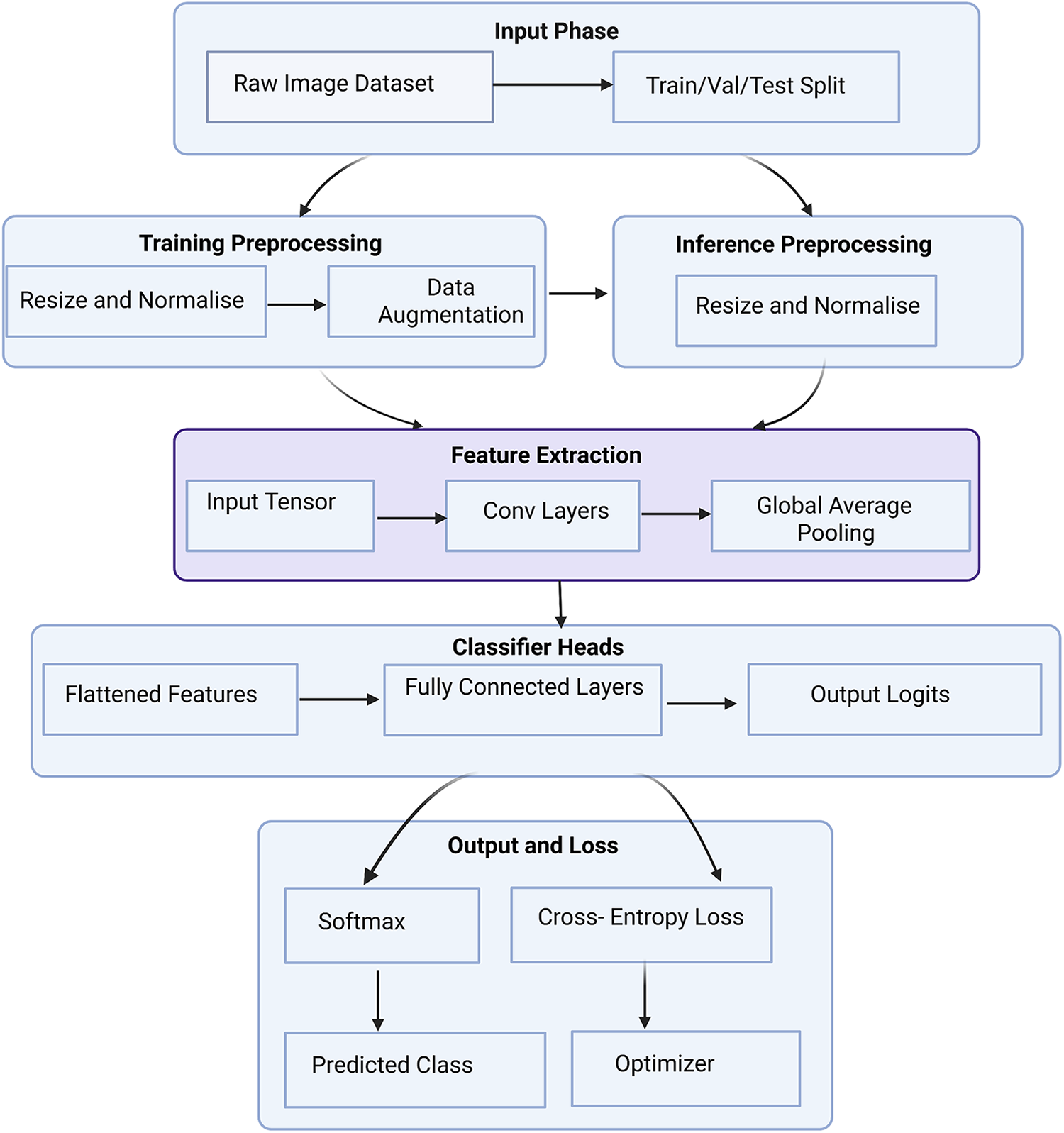
Figure 5: Schematic flow diagram of a deep learning–based image classification network
The supervised models dominate clinical research, while unsupervised methods are gaining grip for their potential to reduce dependency on labeled data. Generative Adversarial Networks (GANs) have been extensively explored for data augmentation, offering the ability to synthesize realistic pathological variations. Frid-Adar et al. [111] demonstrated improved liver lesion classification performance using Deep Convolutional Generative Adversarial Network (DCGAN)-generated samples, later extending to ACGAN [17] for class-conditional generation, though with mixed results. Similarly, conditional GANs have been used in mammogram classification to generate lesion-specific samples with marginal accuracy improvements [112]. Complementing GANs, few-shot learning approaches like PFEMed [113] use dual-encoder strategies and Variational Autoencoders to extract both general and task-specific features, enhancing classification with limited samples. In addition to GANs and VAEs, diffusion models have emerged as powerful generative tools for medical imaging. These models are capable of synthesizing high-fidelity medical images, enabling data augmentation, anomaly detection, and simulation of rare conditions. Their integration into generative frameworks enhances the ability to train robust models under limited data conditions. Table 1 summarizes the quantitative performance of different classification models.
More recently, self-pretraining using Masked Autoencoders (MAE) [114] has shown promise in vision transformer (ViT)-based architectures, where models learn to reconstruct missing image regions, leveraging contextual relationships inherent in medical images. Self-supervised learning (SSL) has also emerged as a transformative approach. Frameworks like MoCo [15,56], SimCLR [122] and others [123–127] rely on contrastive learning or pretext tasks to extract robust representations from unlabelled data, significantly boosting classification performance in tasks like diabetic retinopathy, chest X-ray analysis [128], and COVID-19 detection [129]. These models benefit from clever pretraining strategies such as Rubik’s cube recovery [130,131], rotation prediction [132], and context restoration [133], which enhance the feature extraction process and improve transferability to downstream tasks. Efficient hybrid models like Eff-CTNet [134] and CNN-transformer frameworks [135] combine global attention with local texture learning while mitigating computational burden and vulnerability to adversarial attacks. In parallel, semi-supervised learning methods such as consistency regularization via Mean Teacher [65], semi-supervised GANs [115,121], and knowledge-aware SSL frameworks like Unsupervised Knowledge-guided Self-Supervised Learning (UKSSL) [136] integrate unlabeled and labeled data for performance gains. UKSSL incorporates Medical Contrastive Learning of Representations (MedCLR) and Unsupervised Knowledge-guided Multi-Layer Perceptron (UKMLP) modules for feature extraction and classification using only 50% labeled data while achieving near state-of-the-art accuracy on diverse benchmarks. Taken together, while supervised models provide a strong baseline, unsupervised, self-supervised, and semi-supervised methods critically address data limitations and generalizability challenges, establishing themselves as vital components in the future of CADx-driven medical image classification. Yang et al. [137] present MedKAN, a medical image classification framework built upon KAN and its convolutional extensions. MedKAN features two core modules: The Local Information KAN (LIK) module for fine-grained feature extraction and the Global Information KAN (GIK) module for global context integration. Lai et al. [138] proposed a new Multi-instance Learning (MIL) framework integrating CNN Convolutional Neural Networks (CNN) and Broad Learning Systems (BLS). Hussain et al. [139] present Efficient Residual Network-Vision Transformer (EFFResNet-ViT), a novel hybrid deep learning (DL) model designed to address these challenges by combining EfficientNet-B0 and ResNet-50 CNN backbones with a vision transformer (ViT) module. The proposed architecture employs a feature fusion strategy to integrate the local feature extraction strengths of CNNs with the global dependency modeling capabilities of transformers. Regmi et al. [140] uses different CNNs and transformer-based methods with a wide range of data augmentation techniques. We evaluated their performance on three medical image datasets from different modalities. We evaluated and compared the performance of the vision transformer model with other state-of-the-art pre-trained CNN networks.
Medical image segmentation [1], a fundamental step in quantitative medical analysis, involves delineating organs, lesions, and tissues from complex imaging backgrounds [31,141,142]. While early studies often emphasized descriptive overviews of architectures such as U-Net [143], a critical evaluation is warranted to assess limitations, innovations, and the broader clinical applicability of recent advances. Segmentation tasks pose unique challenges, demanding pixel- or voxel-level precision, and thus requiring substantial annotated datasets for supervised models [36]. The original U-Net, designed for biomedical image segmentation, relies on an encoder-decoder architecture with skip connections [143], which enhance localization by fusing low-level and high-level features. However, traditional U-Net struggles with deeper semantic understanding and long-range dependency modeling, especially in high-resolution and 3D applications [144,145]. To address these, U-Net++ [146] introduced nested skip connections, improving feature propagation and semantic fusion, while 3D U-Net [147] extended the architecture to volumetric data. V-Net [148] advanced this further by incorporating residual units and proposing a Dice-based loss to tackle class imbalance. The Dense V-Net [149] improved multi-organ segmentation by integrating dense blocks, yielding superior Dice scores in abdominal CT scans. Hybrid networks, like the RU-Net [19] fused residual mappings from ResNet [97] and Recurrent Convolutional Layers (RCLs) from Region-based Convolutional Neural Network (RCNN) [150], improving training stability and segmentation accuracy. Attention U-Net [151] utilized attention gates to suppress irrelevant features, enhancing pancreas segmentation in CT. GAN-based adversarial training [152,153] and uncertainty quantification using variational autoencoders [154–156] further enhanced segmentation reliability and robustness. Recent works have leveraged Transformer-based models to overcome CNN limitations in capturing global context. Transformer-based U-Net (TransUNet) [157] pioneered the integration of CNNs with Transformer encoders, using self-attention to model long-range dependencies while maintaining spatial precision through skip connections. This hybrid approach achieved competitive results on multi-organ CT segmentation. Transformer-Fusion Network (TransFuse) [158] employed a parallel CNN-Transformer design, enhancing performance by fusing local and global features at multiple scales. Convolutional Transformer Network (CoTr) [159] adopted deformable self-attention to reduce computational complexity in 3D segmentation. Swin UNet [160], a pure Transformer model, replaced convolutions with hierarchical Swin Transformer blocks [161], offering high-resolution feature representation with low computational overhead. These Transformer-based models demonstrate improved generalization, though many rely on pretraining on large external datasets [162,163] which raises concerns regarding data leakage and generalization to unseen modalities. Mask R-CNN [164], originally developed for object detection, has been adapted for instance-level medical segmentation. It incorporates Region of Interest Alignment (RoIAlign), Feature Pyramid Network (FPN) [165], and a segmentation mask branch, providing multiscale representations. Volumetric adaptations with attention modules [166] improved contextual awareness and reduced false positives.
The Mask R-CNN++ [146] hybrid, combining UNet++’s nested skip connections yielded state-of-the-art results in complex segmentation tasks. In unsupervised settings, GANs and generative models were initially used to augment datasets [86,167], but self-supervised and semi-supervised approaches have emerged as more scalable alternatives. TransUNet [157] was extended with modality-agnostic 3D adapters (MA-SAM) [168], preserving pretrained weights while adapting to volumetric inputs. Vision Mamba U-Net (VM-Unet) [169] introduced a Vision Selective Scan (VSS) block and asymmetric architecture to enhance contextual understanding in ISIC and Synapse datasets. Self-supervised methods using pretext tasks, such as semantic inpainting [50], anatomical position prediction [125], and metadata integration [170] have demonstrated robust representation learning. Fig. 6 demonstrated schematic flow diagram of a deep learning–based segmentation network.
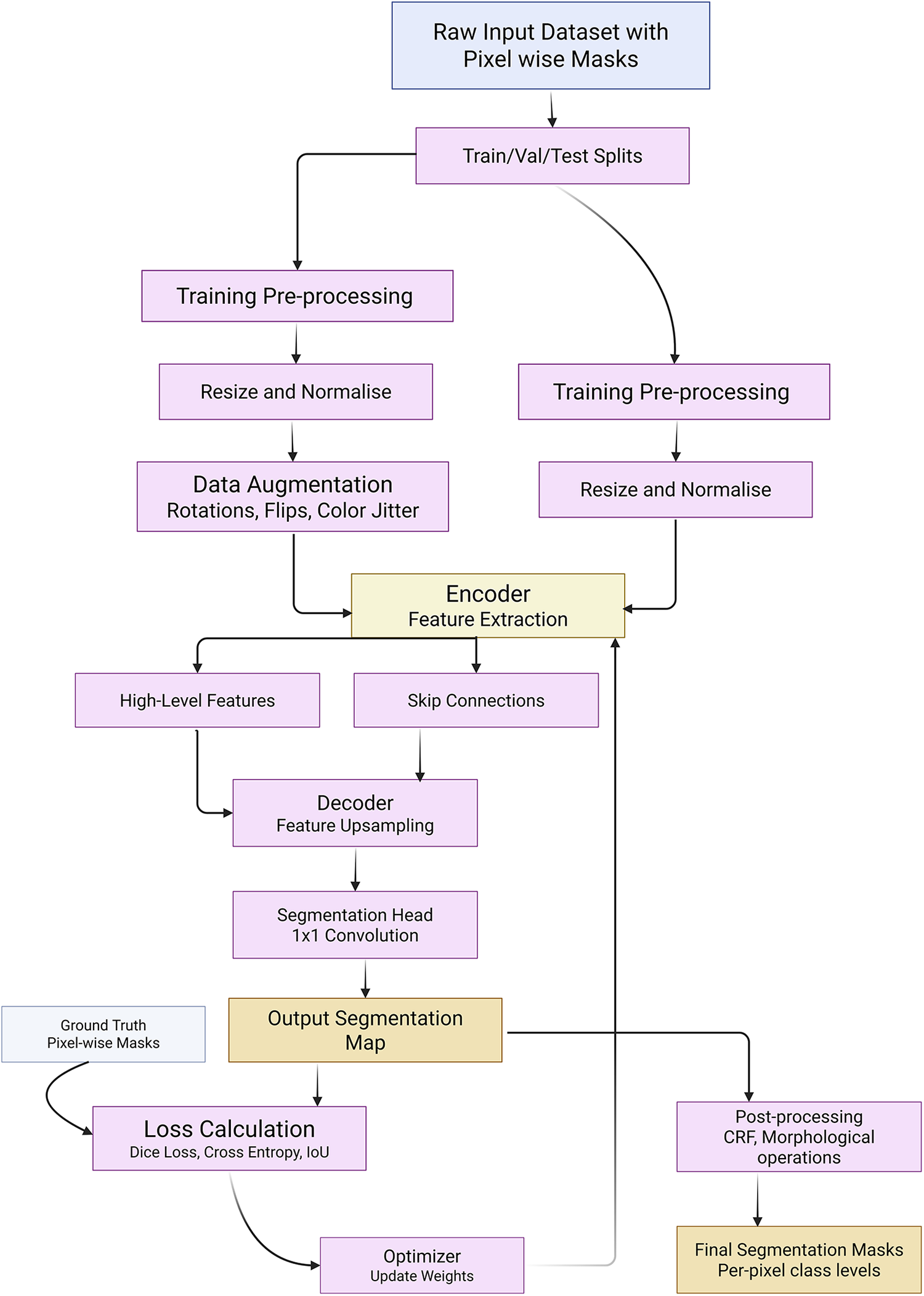
Figure 6: Schematic flow diagram of a deep learning–based segmentation network
Adaptations of 2D tasks (e.g., jigsaw, rotation) to 3D segmentation [171] have improved generalization, even surpassing supervised pretraining. Contrastive learning variants [172] have also emerged, with methods like local contrastive loss [173] capturing pixel-wise distinctions vital for segmentation. These techniques benefit from data augmentations like Mixup [67] to further enhance robustness. Semi-supervised segmentation strategies like Mean Teacher [20,174] leverage teacher-student frameworks with uncertainty modeling to guide learning from unlabeled data. COVID-19 segmentation tasks [175], utilized pseudo-labeling and iterative self-training in architectures like Semi-supervised Inf-Net (Semi-InfNet) incorporating reverse attention (RA), edge attention (EA), and parallel partial decoder (PPD) [176–178]. Federated semi-supervised learning (FSSL) [179] with pseudo-labeling addressed data heterogeneity across institutions. Dual-VAE frameworks [180] combined latent representation learning and mask prediction, while shared-encoder models reconstructed both foreground and background separately to enhance attention-based segmentation. Domain priors, such as anatomical [181], atlas [182], and topological [183], further refined segmentation consistency. Advanced architectures like nnFormer [184] used volume-aware attention with skip-attention mechanisms for 3D segmentation. Table 2 summarizes the quantitative performance of different segmentation models.
While H2Former [187] combined CNNs, multiscale attention, and Transformers to outperform existing models. Wu et al. [188] extend the adaptation of SS2D by proposing a High-order Vision Mamba UNet (H-vmunet) model for medical image segmentation. Among them, the H-vmunet model includes the proposed novel High-order 2D-selective-scan (H-SS2D) and Local-SS2D module. Zheng et al. [189] proposes an asymmetric adaptive heterogeneous network for multi-modality image feature extraction with modality discrimination and adaptive fusion. For feature extraction, it uses a heterogeneous two-stream asymmetric feature-bridging network to extract complementary features from auxiliary multi-modality and leading single-modality images, respectively. Iqbal et al. [190] propose a novel deep learning architecture for medical image segmentation, which takes advantage of CNNs and vision transformers. Our proposed model, named Transformer-Based Convolutional Long Short-Term Memory Network “TBConvL-Net”, involves a hybrid network that combines the local features of a CNN encoder–decoder architecture with long-range and temporal dependencies using biconvolutional long-short-term memory (LSTM) networks and vision transformers (ViT).
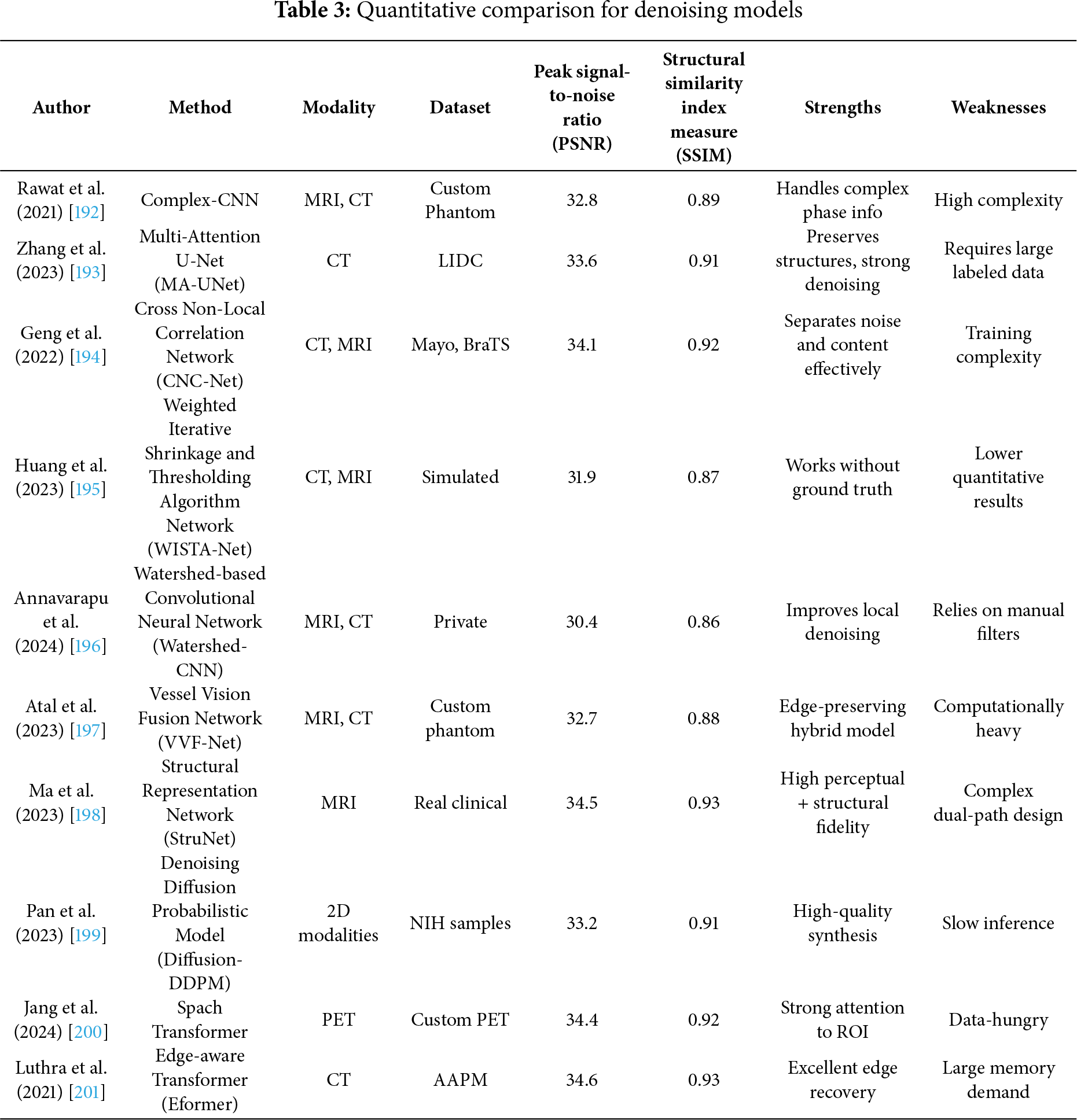
Medical image denoising remains a critical preprocessing step in diagnostic pipelines for modalities such as CT, MRI, X-rays, and ultrasound, which are frequently corrupted by noise, leading to potential misdiagnoses [5]. Traditional reliance on radiologist interpretation has evolved with the integration of intelligent deep learning-based systems [191]. However, a critical evaluation of modern denoising techniques reveals both advancements and ongoing challenges. Rawat et al. [192] proposed CVMIDNet, a complex-valued CNN leveraging residual learning and CReLU activations to predict residual noise from chest X-rays rather than estimating the clean image directly, thereby reducing signal distortion. Similarly, attention-based U-Net architectures, like that of [193] integrated local, channel, and task-adaptive attention mechanisms to better localize features and suppress irrelevant noise in CT images. Geng et al. [194] demonstrated that adversarial frameworks like Content-Noise Complementary Learning (CNCL), when combined with base models such as Universal Network (U-Net), Denoising Convolutional Neural Network (DnCNN), and Super-Resolution Dense Network (SRDenseNet), improved denoising across Computed Tomography (CT), Magnetic Resonance (MR), and Positron Emission Tomography (PET) data. While generative approaches provide flexibility, they often require extensive computational resources. In contrast, self-supervised sparse coding methods such as Weighted Iterative Shrinkage and Thresholding Algorithm (WISTA) and its deep-learning counterpart WISTA-Net [195] achieved competitive denoising without ground-truth images by exploiting lp-norm constraints and Deep Neural Network (DNN)-based parameter updates. Annavarapu et al. [196] introduced a denoising pipeline incorporating CNNs with adaptive watershed segmentation and contrast enhancement, validated on MRI and CT images, though their performance gains were limited by dependence on manually designed filters. Atal et al. [197] addressed CT noise using a hybrid approach that combines a deep CNN with an optimization-based vectorial variation filter, where pixel-wise noise maps are identified and cleaned using the Feedback Artificial Lion (FAL) algorithm. Fig. 7 demonstrated schematic flow diagram of a deep learning–based image denoising network. The hybrid learning-optimization method improved accuracy but introduced higher computational complexity. Ma et al. [198] advanced denoising via a dual-path encoder-decoder utilizing Swin Transformer blocks and residual units in parallel. This design effectively captured local and global features, while low-rank regularization and perceptual loss preserved feature consistency and structural fidelity. Transformer-based innovations have further proliferated. Pan et al. [199] employed a Swin-transformer-driven diffusion model for denoising and image synthesis across multiple imaging modalities. This model generated realistic synthetic images, verified using Inception Score (IS), Fréchet Inception Distance (FID), and visual Turing tests, and showed that synthesized data could complement real data in downstream classification tasks. Jang et al. [200] presented the Spach Transformer, validated across multiple PET tracers, and reported improved quantitative performance over leading networks. Meanwhile, Eformer by Luthra et al. [201] introduced a Transformer-based denoising network integrating learnable Sobel-Feldman edge enhancement operators to retain critical edge features while employing non-overlapping windowed self-attention for computational efficiency. These methods, although promising, highlight ongoing tensions between denoising performance, computational burden, and data requirements. Many approaches lack generalizability across modalities or underperform in low-signal to noise (SNR) settings without modality-specific tuning. Additionally, few studies assess clinical applicability through radiologist-in-the-loop evaluations or real-time performance metrics. A critical path forward will involve designing lightweight, explainable, and generalizable denoising models that balance accuracy, speed, and interpretability across diverse clinical environments.
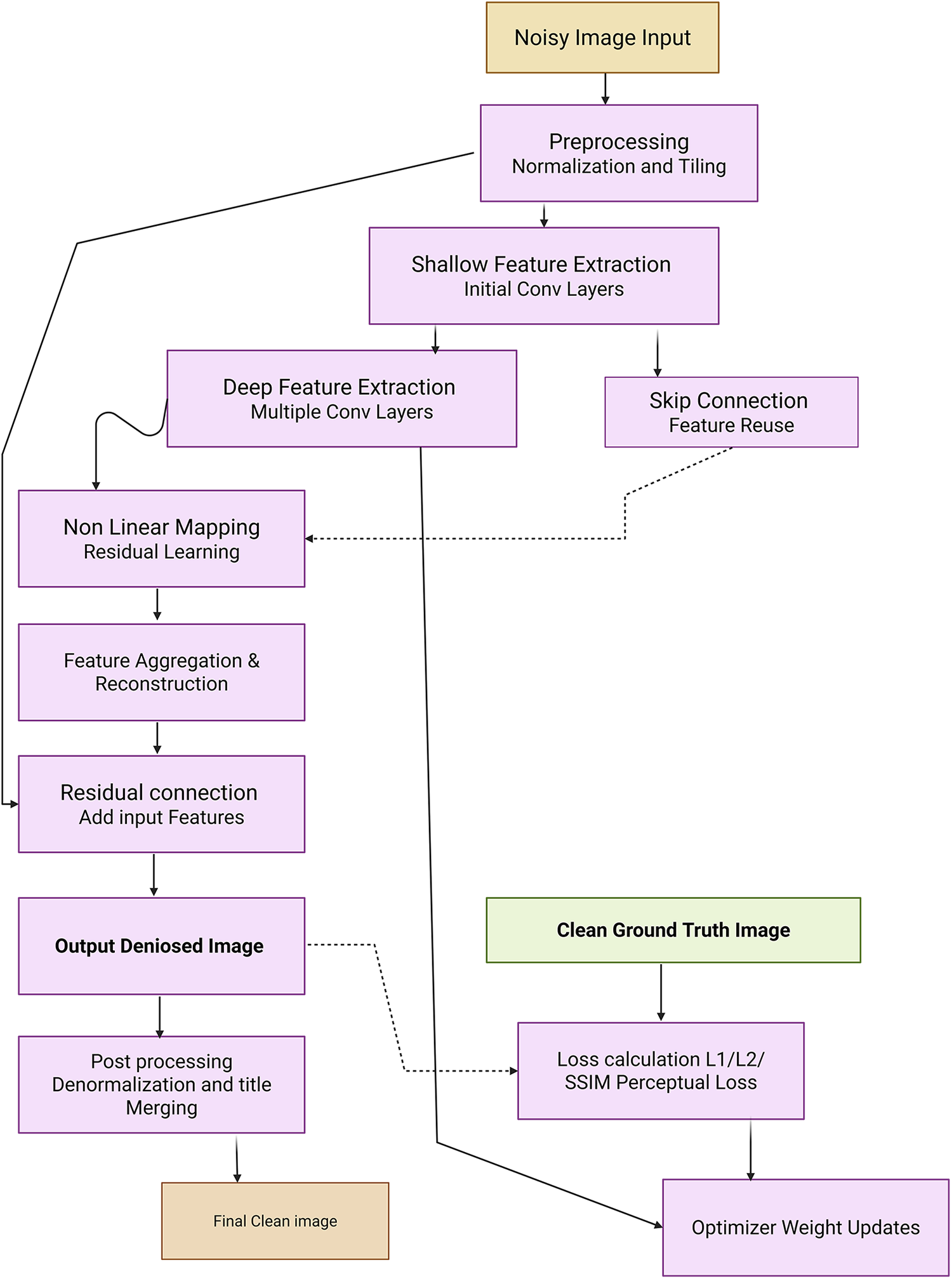
Figure 7: Schematic flow diagram of a deep learning–based image denoising network
Demir et al. [202] propose Diffusion-based Denoising Network (DiffDenoise), a powerful self-supervised denoising approach tailored for medical images, designed to preserve high-frequency details. Chen et al. [203] propose a task-based regularization strategy for use with the Plug-and-Play Learning Strategy (PLS) in medical image denoising. The proposed task-based regularization is associated with the likelihood of linear test statistics of noisy images for Gaussian noise models. Kathiravan et al. [204] proposed a hybrid approach to image denoising that makes use of elements of EfficientNetB3 and Pix2Pix models. The EfficientNetB3’s efficient scaling and feature extraction capabilities, combined with the Pix2Pix’s image-to-image translation capabilities, enable the model to effectively remove noise while preserving essential image features. And Table 3 summarizes the quantitative performance of different denoising models.
The evolution of object detection in medical imaging from classic region-based methods like RCNN [205] and OverFeat [206,207] to advanced CNN-based and Transformer-based models demands a critical re-evaluation of detection frameworks tailored for the complex structure of medical data. While general object detectors, such as Faster Region-Based -CNN [205], You Only Look Once (YOLO) [208], and RetinaNet [209] brought considerable improvements in efficiency and accuracy over their predecessors, their clinical deployment in computer-aided detection (CADe) remains constrained by domain-specific limitations like lesion size, variability, and class imbalance. Two-stage detectors, particularly Faster R-CNN [205,210], revolutionized medical image detection by introducing a Region Proposal Network (RPN) for efficient anchor-based region generation. Despite notable improvements, its dependency on anchor design and computational bottlenecks in ROI pooling persisted. To address privacy concerns, Liu et al. [211] extended Faster R-CNN into a secure detection framework (SecRCNN) that leverages secret sharing in edge environments, ensuring patient data confidentiality. Mask R-CNN [164], a derivative of Faster R-CNN, enhanced performance through instance segmentation using FPN [165], further improving object delineation in cluttered medical backgrounds. However, computational expense and limited real-time capability restrict its scalability. In contrast, YOLO [208], as a one-stage detector, gained popularity for its simplicity and speed, although with suboptimal accuracy in small lesion detection. Later YOLO iterations, including YOLOv2 and YOLO9000 integrated fine-grained features, anchor optimization, and multiscale training to alleviate initial weaknesses [208]. Fig. 8 demonstrated schematic flow diagram of a deep learning–based detection network. RetinaNet [209] addressed class imbalance in one-stage detectors via focal loss, boosting sensitivity to hard examples and improving detection rates. Nevertheless, the extensive use of anchor boxes across these models introduced challenges such as hyperparameter tuning, redundancy, and positive-negative imbalance. Anchor-free models like Corner-based Object Detection Network (CornerNet) [212,213] and CenterNet [214] emerged to address these issues, leveraging keypoint detection strategies, although CornerNet’s performance suffered due to weak regional context modeling. CenterNet improved upon this with triplet key points and contextual reasoning. For lesion-specific detection, especially of sclerosis lesions [24] and pulmonary nodules [215,216] and breast tumors [217,218], standard detectors struggled due to the subtle nature and size of lesions. Xu et al. [219] proposed Pyramid Attention and Context Network (PAC-Net), a multi-pathway FPN with position-attention-guided connections and vertex distance Intersection over Union (IoU) loss to increase sensitivity in universal lesion detection. Incorporating domain-specific adaptations, including 3D convolutional layers [220,221], deconvolution layers for resolution recovery [222] and spatial context aggregation, substantially enhanced nodule recognition accuracy, exemplified by 3D Faster R-CNN ranking first in LUNA16 [223,224]. In histopathology, streamlined YOLOv2 versions [208], modified for whole-slide lymphocyte detection that improved speed and F1 score but remained inferior to U-Net-based pixel-wise methods. Semi-supervised approaches including Mixed Sample Data Augmentation Technique (Mixup) [67], MixMatch Semi-Supervised Learning Framework (MixMatch) [225], and focal loss extensions [226] that improved generalizability with minimal labeled data. For instance, the PAC-Net’s modified focal loss and pseudo-labeling with Mixup led to substantial accuracy improvements in 3D lesion detection. Uncertainty estimation methods like MC dropout and predictive entropy [24,227,228] enhanced confidence calibration in detecting ambiguous small lesions, proving critical for sclerosis or tumor boundary remains in label completeness and cross-domain variability. Contextual and spatial attention modules [84,85,210] further enriched features by weighting informative slices and regions. Table 4 summarizes the quantitative performance of different detection models.
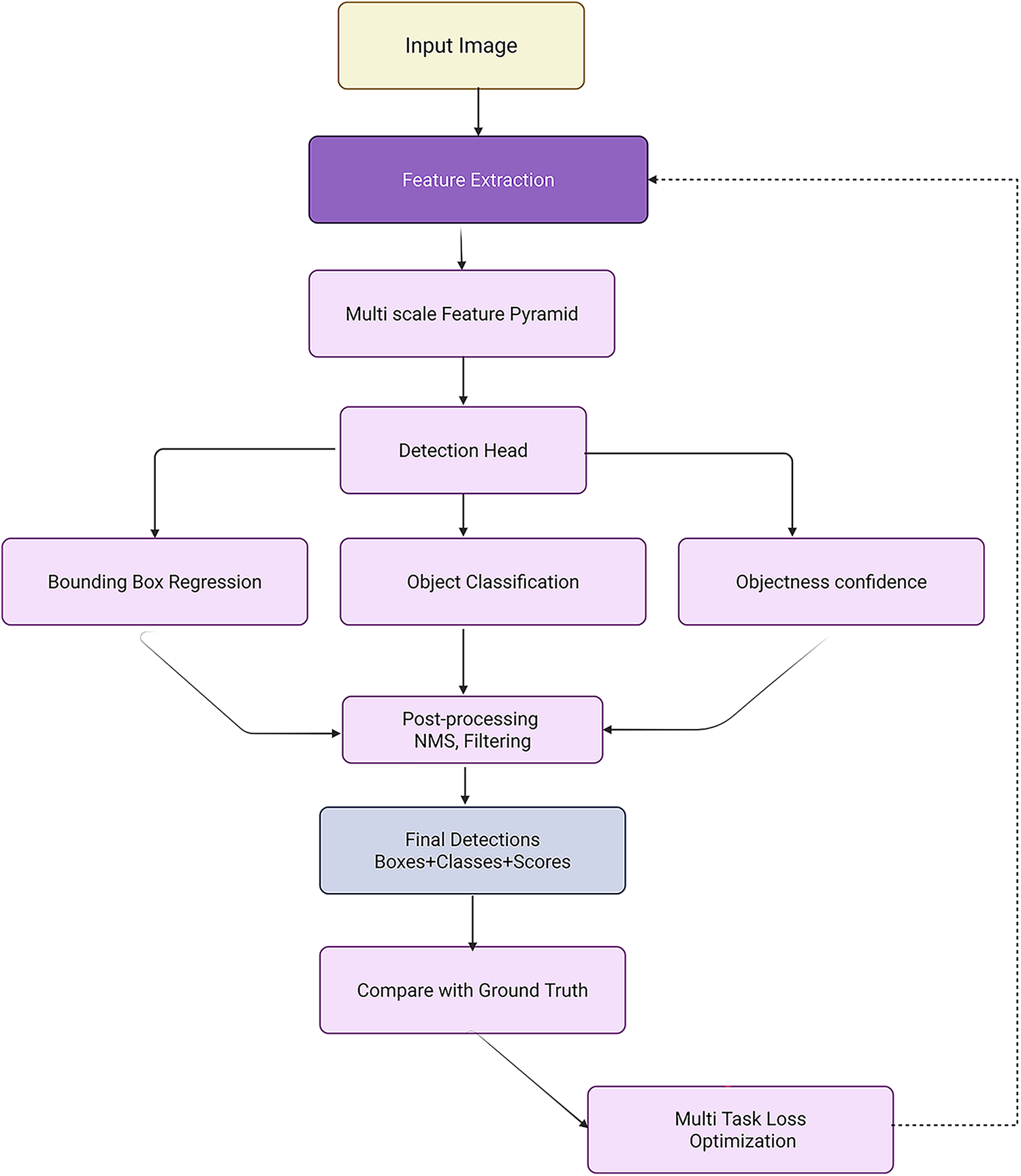
Figure 8: Schematic flow diagram of a deep learning–based detection network
Furthermore, methods incorporating radiologist-style window reweighting [239] improved interpretability and alignment with clinical practice. Meanwhile, unsupervised lesion detection methods primarily reconstruction- and restoration-based models’ normal anatomy to highlight anomalies. Anomaly Generative Adversarial Network (AnoGAN) [240], with iterative latent optimization using residual and discrimination losses, was effective but slow. Fast Anomaly Generative Adversarial Network (f-AnoGAN) [241] and AnoVAE-GAN [242] introduced encoders for rapid inverse mapping. Evidence Lower Bound (ELBO) approximation inaccuracies [243] were addressed by local gradient corrections, while 3D modeling improved spatial coherence. Additionally, Conditional Variational Autoencoder (CVAE) [244] incorporated spatial priors for delineation. Predecessor such as 3DCE [220] has been outperformed by Universal lesion detectors like ULDor [245] and MULAN [246] built upon Mask R-CNN with multitask learning integrated classification, segmentation, and detection. However, the challenge is the more precise segmentation of anomalous regions. Self-supervised detection models, such as One-Class Self-Supervised Learning (OC-SSL) with Dual-Domain Anomaly Detection (DDAD) [237] utilized reconstruction discrepancies across normal and unlabelled data to define anomaly scores, pushing unsupervised frameworks closer to supervised benchmarks. Altogether, while supervised detectors dominate clinical pipelines, their dependence on annotated data and sensitivity to domain shifts remain bottlenecks. Semi- and self-supervised frameworks, augmented with uncertainty quantification, task fusion, and Transformer attention, offer scalable, robust alternatives. As detection models evolve toward unified, anchor-free, and 3D-aware architectures, critical integration of clinical priors, interpretability tools, and multi-institutional datasets will be essential to drive real-world CADe adoption and effectiveness. Bi et al. [247] propose a novel unsupervised anomaly detection framework based on a diffusion model that incorporates a synthetic anomaly (Synomaly) noise function and a multi-stage diffusion process. Synomaly noise introduces synthetic anomalies into healthy images during training, allowing the model to effectively learn anomaly removal. Hoover et al. [248] investigate the robustness of pre-trained deep learning models for classifying bone fractures in X-ray images and seeks to address global healthcare disparity through the lens of technology. Lab-on-a-chip technology offers high-throughput, automated data generation that addresses the major bottleneck of AI [249]. The study explores the emerging synergy of ‘AI on a chip,’ highlighting key advances, challenges, and future opportunities.
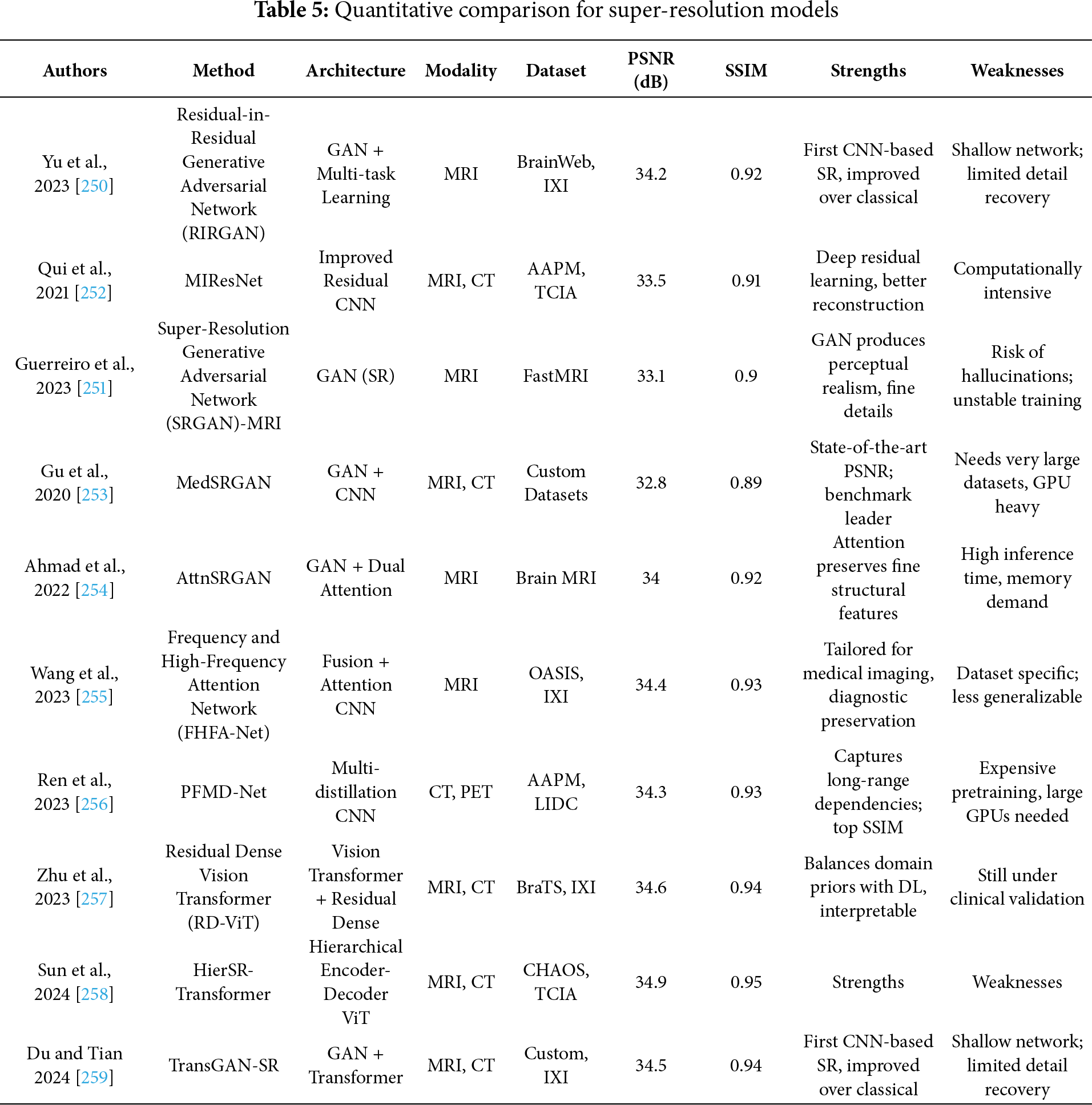
In medical imaging, the challenge of improving the resolution of diagnostically critical images without compromising accuracy or introducing artifacts has led to the rapid development of advanced super-resolution (SR) models. However, critical analysis reveals that while multiple architectures have demonstrated promising results, their applicability in real-world clinical settings remains bounded by computational complexity, generalization capacity, and integration with diagnostic pipelines. The strengths of Transformers are reiterated here to underline their significance across various imaging modalities and tasks. Yu et al. [250] proposed RIRGAN, a multi-task GAN using Residual-in-Residual (RIR) blocks for simultaneous denoising and super-resolution. While the model’s long skip connections facilitate deeper architectures to learn high-frequency information, the complexity of tuning adversarial losses and ensuring convergence remains a limitation. The reliance on relativistic average discriminators enhances realism but also increases training instability. Guerreiro et al. [251] introduced a perception consistency method using cycle-GANs to enforce reconstruction from LR-SR-LR (Low-Resolution → Super-Resolution → Low-Resolution) paths, eliminating the need for HR labels. This self-supervised method offers robustness but can be limited in datasets with significant anatomical variation. Similarly, the Multiple Improved Residual Network (MIRN) proposed by Qiu et al. [252] addresses residual feature correlation using deep skip connections; however, its dependence on adaptive learning schedules and complex residual aggregation raises questions about scalability. Gu et al. [253] developed MedSRGAN using a Residual Whole Map Attention Network (RWMAN) for channel-wise feature emphasis. Despite high performance, training with multiple adversarial and feature loss functions add complexity and computational overhead. Ahmad et al. [254] proposed a three-stage GAN model employing ResNet34 and multi-path extraction. While incremental upscaling addresses gradient propagation and authenticity, its deeper architecture may be unsuitable for low-resource clinical environments. Wang et al. [255] introduced a fuzzy hierarchical attention model with fuzzy logic integration. This hybrid model innovatively addresses pixel uncertainty but may overfit in diverse image scenarios due to the handcrafted fuzzy membership logic. Ren et al. [256] designed a pyramidal multi-distillation network incorporating entropy-based attention and gradient map supervision. Though perceptually effective, integrating multiple supervision paths increases inference time. Zhu et al. [257] introduced perceptual loss from a pretrained segmentation U-Net into super-resolution networks, including CNNs and Transformers. While the segmentation-aware supervision improves semantic preservation, it can bias models toward overfitting on the pretraining domain, especially in heterogeneous datasets. Transformer-based models like Transformer-based Hierarchical Encoder–Decoder Network (THEDNet) by Sun et al. [258] offer multi-scale attention via Exponential Moving Average (EMA) modules to extract global dependencies. However, Transformers demand high memory and are often over-parameterized for limited medical datasets. Du et al. [259] combined Transformers and T-GANs for texture-aware reconstruction using weighted multi-task loss. While effective in preserving texture details, balancing the trade-off between content and adversarial losses remains a challenge. Collectively, these models highlight the evolving landscape of SR in medical imaging; yet, their critical deployment demands addressing generalizability, interpretability, and efficiency. While adversarial and Transformer-based approaches enhance visual fidelity, their integration into practical CAD systems must consider training complexity, reproducibility, and dataset variance. Furthermore, benchmark datasets and standardized evaluation metrics are essential to avoid overclaiming model robustness. Ultimately, although numerous SR frameworks have improved medical image resolution across CT, MRI, and X-rays, critical bottlenecks persist in balancing visual enhancement with diagnostic reliability, especially in edge-device and real-time settings where resource constraints and model transparency are paramount. Fig. 9 demonstrated schematic flow diagram of a deep learning–based super-resolution network.
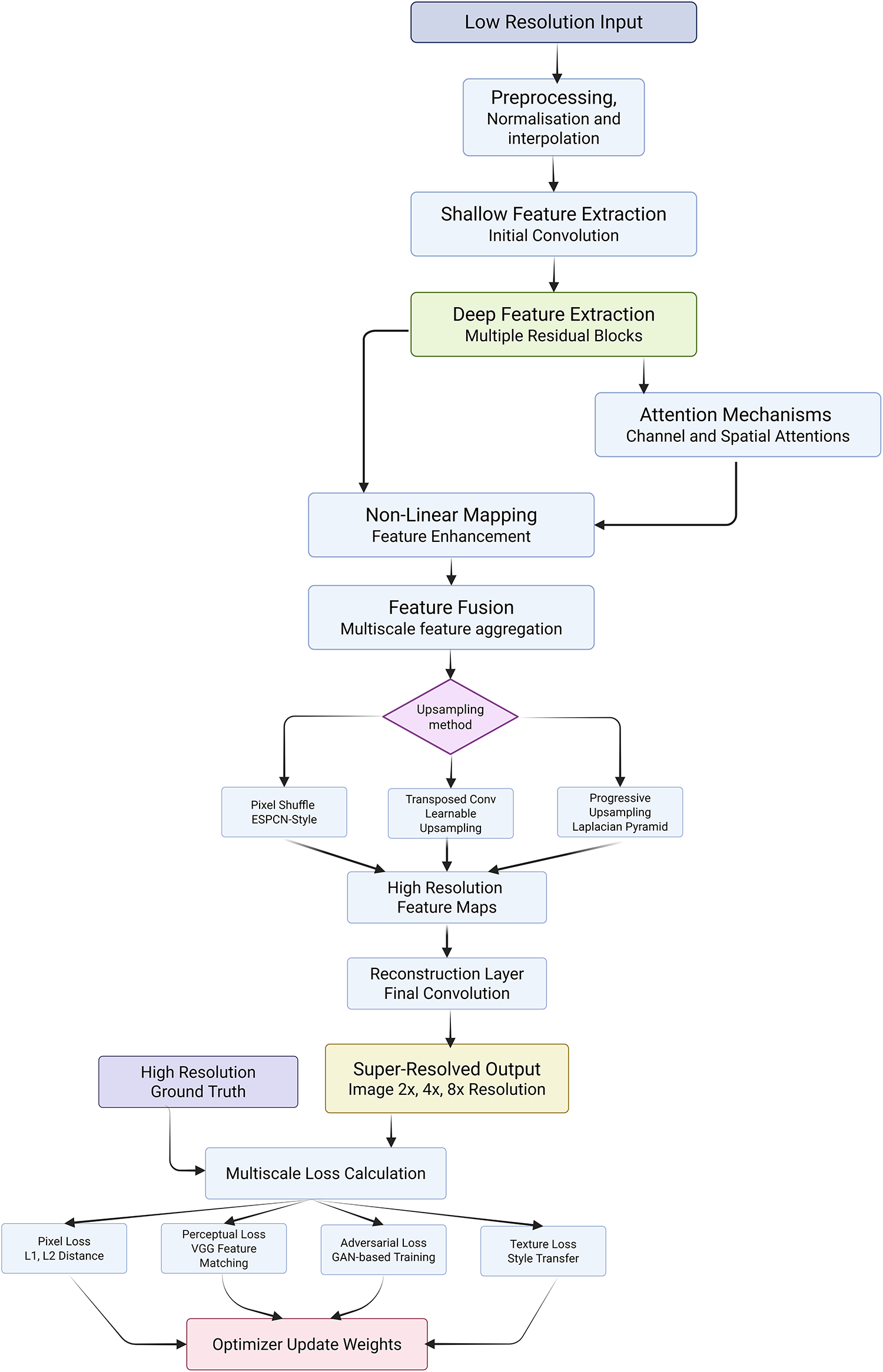
Figure 9: Schematic flow diagram of a deep learning–based super-resolution network
Goyal et al. [260] proposed an SR method that integrates multiscale CNNs with weighted least squares optimization (WLSO), leveraging wavelet decompositions. Although effective in capturing multiscale contextual information, wavelet-based models can struggle with learning fine semantic details across modalities. Lu et al. [261] propose a novel sparsity-guided medical image SR network, namely SG-SRNet, by exploiting the spatial sparsity characteristics of the medical images. SG-SRNet mainly consists of two components: a sparsity mask (SM) generator for image sparsity estimation, and a sparsity-guided Transformer (SGTrans) for high-resolution image reconstruction. Pang et al. [262] present Neural Explicit Representation (NExpR) for fast arbitrary-scale medical image SR. Our algorithm represents an image with an explicit analytical function, whose input is the low-resolution image and output is the parameterization of the analytical function. Ji et al. [263] propose a self-prior guided Mamba network with edge-aware constraint (SEMambaSR) for medical image super-resolution. Recently, State Space Models (SSMs), notably Mamba, have gained prominence for the ability to efficiently model long-range dependencies with low complexity. Li et al. [264] develop a Self-rectified Texture Supplementation network for Reference-based Super-Resolution (STS-SR) to enhance fine details in MRI images and support the expanding role of autonomous AI in healthcare. Our network comprises a texture-specified self-rectified feature transfer module and a cross-scale texture complementary network. Table 5 summarizes the quantitative performance of different super-resolution models.
Recent advances in medical image fusion have led to numerous innovations, yet a critical evaluation reveals considerable variance in effectiveness, generalizability, and interpretability across supervised, unsupervised, and transformer-based approaches. Traditionally, fusion strategies aimed to combine information from different modalities (e.g., MRI, CT, PET) or from different focal lengths to reduce the fragmentary nature of clinical data and enhance visualization [265–267]. While supervised fusion frameworks such as the one proposed by Li et al. [268] utilize Deep Boltzmann Machines (DBM) to learn fusion mappings, the requirement of extensive labeled datasets and perfect image registration limits scalability in clinical applications. Likewise, the Multi-Scale Enhanced Network (MSENet) [269] and Saliency-Driven Network (SDNet) [270] introduce modular enhancements like dilated convolutions, unique fusion modules, and gradient-intensity decomposition but their dependency on hand-crafted scoring metrics and fixed architectural assumptions may limit adaptability to new modalities or diagnostic contexts. In contrast, unsupervised fusion techniques attempt to bypass the annotation bottleneck but often sacrifice interpretability or robustness. Xu et al. [271] combine surface-level and deep constraints in an unsupervised setting but may be sensitive to feature extraction biases from pre-trained encoders. The Foveation-based Differentiable Architecture Search Framework (F-DARTS) framework by Ye et al. [272] innovatively leverages human visual saliency via a foveation operator and multi-component loss functions, but the computational overhead and lack of end-user transparency present hurdles for clinical integration. Similarly, LatLRR-GAN [273] incorporates Latent Low-Rank Representations with a dual discriminator GAN for detail preservation in low-rank regions, yet suffers from dependency on optimal thresholding and network tuning. MambaDFuse [274] attempts to balance shallow and deep fusion using Mamba blocks and CNNs, but the integration of long-range dependencies through channel exchange raises questions about feature redundancy and optimization stability. The MATR [275] refines semantic extraction through adaptive convolutions and regional mutual information loss, showcasing state-of-the-art performance in unsupervised multimodal fusion. However, its reliance on complex objective functions and multiscale learning complicates training convergence and practical deployment. Overall, while the field has progressed from early rule-based strategies to sophisticated deep and Transformer-based architectures, limitations persist in terms of scalability, interpretability, and the balance between global semantic context and local detail preservation. A critical takeaway is the need for future frameworks that are not only robust across diverse modalities but also interpretable, computationally efficient, and capable of adapting to varying levels of supervision without compromising diagnostic accuracy or clinical utility. Fig. 10 demonstrated schematic flow diagram of a deep learning–based fusion network.
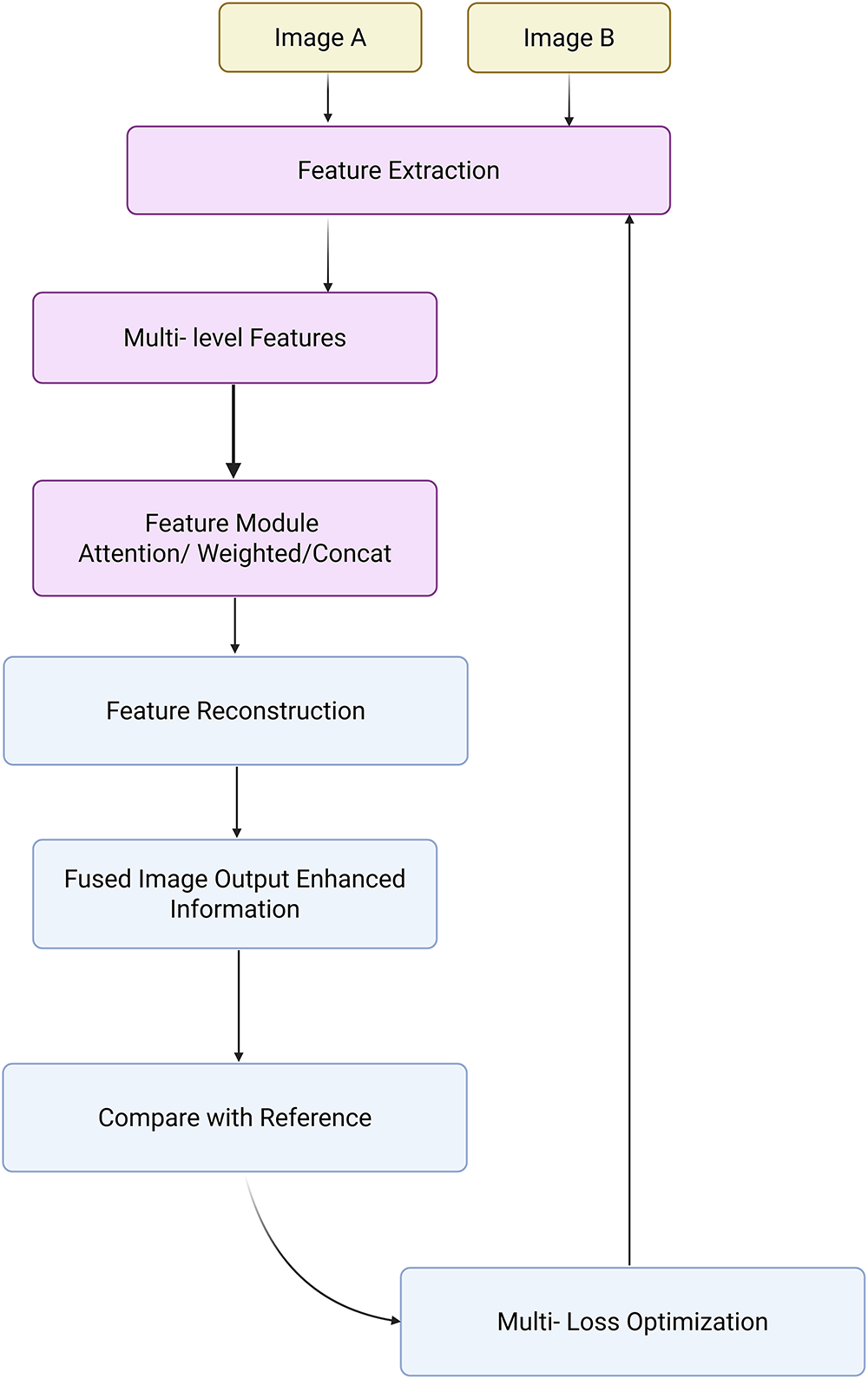
Figure 10: Schematic flow diagram of a deep learning–based fusion network
Table 6 summarizes the quantitative performance of different fusion models. In the context of Transformer-based fusion, recent models like the enhanced DenseNet-Transformer hybrid by Song et al. [276] present promising improvements in minimizing feature loss and edge blurring, but Dense concatenation architectures can be memory-intensive and may face difficulties in generalizing to smaller datasets. He et al. [277] propose a novel invertible fusion network (MMIF-INet) that accepts three-channel color images as inputs to the model and generates multichannel data distributions in a process-reversible manner. Specifically, the discrete wavelet transform (DWT) is utilized for downsampling, aiming to decompose the source image pair into high- and low-frequency components. Li et al. [278] propose an unaligned medical image fusion method called Bidirectional Stepwise Feature Alignment and Fusion (BSFA-F) strategy. PH Dinh [279] propose a novel medical image fusion method that combines the strengths of Bilateral Texture Filtering (BTF) and transfer learning with a modified ResNet-101 network (M_ResNet-101). The method begins with the application of BTF to decompose the input images into texture and detail layers, preserving structural integrity while effectively separating relevant features. Liu et al. [280] propose a novel salient semantic enhancement fusion (SSEFusion) framework, whose key components include a dual-branch encoder that combines Mamba and spiking neural network (SNN) models (Mamba-SNN encoder), feature interaction attention (FIA) blocks, and a decoder equipped with detail enhancement (DE) blocks.
Medical image enhancement has become an indispensable pre-processing step to augment the visibility of anatomical and pathological details across imaging modalities such as CT, MRI, and X-ray, thereby assisting radiologists and automated systems in accurate interpretation and diagnosis. However, a critical evaluation of the enhancement methods reveals several methodological strengths, algorithmic limitations, and practical challenges that persist in clinical application. While traditional enhancement algorithms focus on improving global contrast or noise suppression, recent approaches based on supervised deep learning exhibit improved robustness through learning complex intensity transformations. Rundo et al. [281] proposed MedGA, which leverages genetic algorithms to enhance images with bimodal gray-level histograms, demonstrating clinical applicability in contrast-enhanced MRI. However, its reliance on predefined histogram properties restricts adaptability across diverse image contexts. He et al. [282] tackled cross-domain enhancement through the Unsupervised Multi-domain Image Enhancement (UMIE) strategy, integrating high-quality guidance into low-quality image transformation via variational modelling. This framework addresses the domain gap between low- and high-quality image distributions but depends heavily on the availability and representativeness of high-quality prompts. Alenezi et al. [283] introduced a modified Hopfield Neural Network (MHNN) under cohomological constraints to regulate gradient vector flow, thus enhancing local-global feature correlations. Although theoretically promising, MHNN’s computational demands and convergence guarantees warrant further empirical validation. Ghandour et al. [284] evaluated feature-level fusion in Deep Learning–based Medical Image Fusion (DLMIF) using pretrained CNNs for enhancement without ground-truth supervision, highlighting CNN feature reliability but also underlining limitations in control over fused feature distribution due to static pretrained layers. Unsupervised strategies, while more flexible, present their own trade-offs between realism and interpretability. Structure and Illumination–aware Generative Adversarial Network (StillGAN) [285] exemplify GAN-based enhancement techniques treating image quality as a domain transfer task. StillGAN’s bi-directional architecture integrates structural and illumination priors, addressing both global coherence and local fidelity. Nevertheless, the training instability of GANs and the reliance on handcrafted domain constraints remain pertinent drawbacks. Similarly, Multimodal Adversarial Generative Adversarial Network (MAGAN) [286] exploits multi-scale attention mechanisms in an adversarial setting to capture feature hierarchies but inherits the GAN framework’s susceptibility to mode collapse. FS-GAN [287] innovatively employ a fuzzy domain discriminator and structure-retention modules to refine nerve fiber imagery and light distribution. While promising in preserving structural integrity, the fuzzy domain formulation may vague interpretability and reproducibility in clinical settings. Moreover, Transformer-based enhancement techniques are gaining traction due to their capability to model global dependencies. Fig. 11 demonstrated schematic flow diagram of a deep learning–based enhancement network.
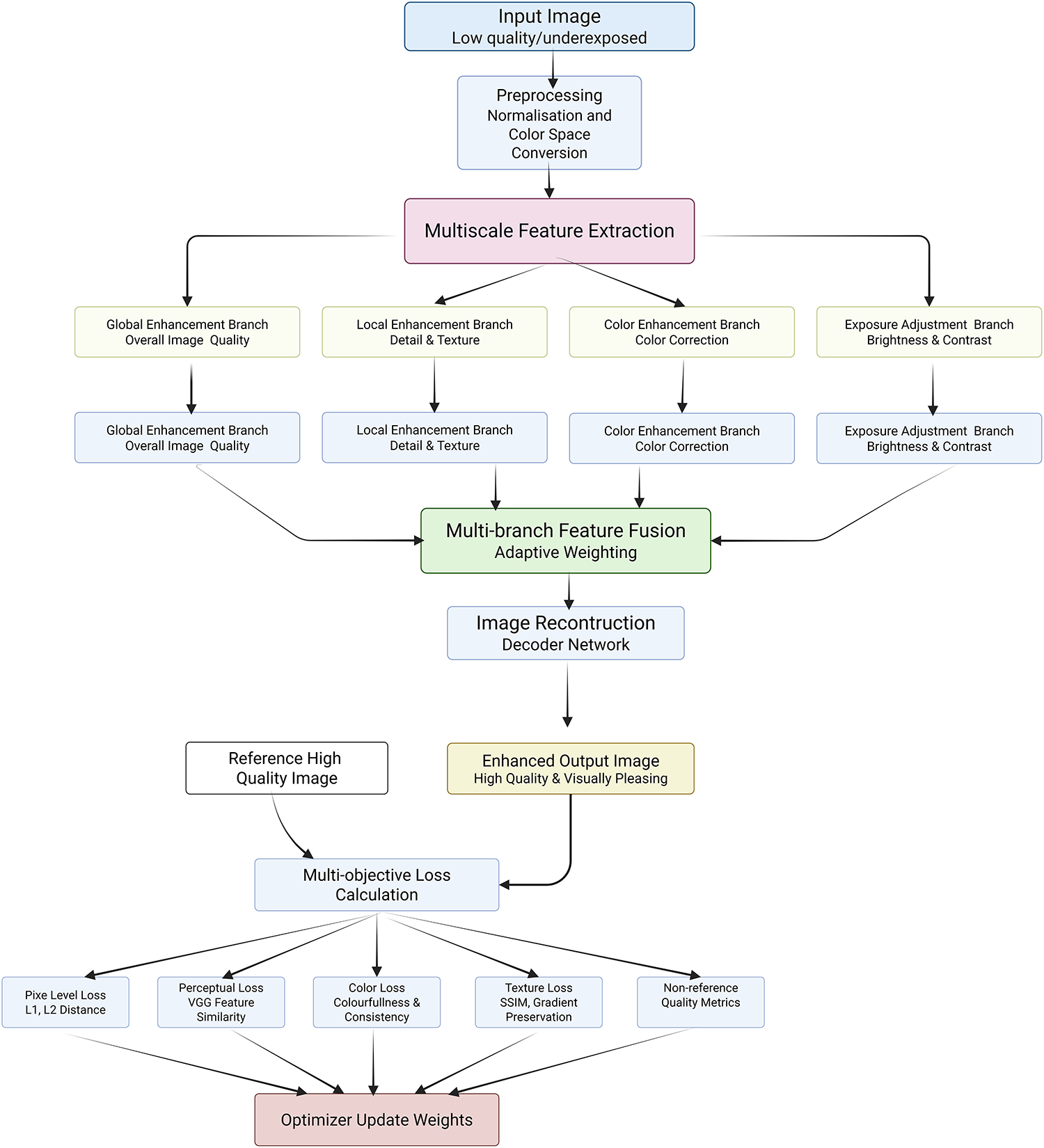
Figure 11: Schematic flow diagram of a deep learning–based enhancement network
Xia et al. [288] observed that the previous models necessitate large training data and complex tuning, making their deployment in data-constrained scenarios challenging. Thus, despite notable progress, critical gaps remain in generalizability, real-time processing, interpretability, and integration with downstream diagnostic tasks. Enhancement models must be evaluated not only based on visual quality but also on their impact on clinical decision-making and compatibility with classification, segmentation, and detection pipelines. Continued development of hybrid models combining CNNs, Transformers, and domain adaptation strategies, along with the adoption of evaluation metrics that reflect diagnostic relevance, is essential to transition from visually plausible to diagnostically reliable enhancement solutions in medical image analysis. Lei et al. [289] propose a general framework called Contrast-Driven Medical Image Segmentation (ConDSeg). They develop a contrastive training strategy called Consistency Reinforcement. It is designed to improve the encoder’s robustness in various illumination and contrast scenarios, enabling the model to extract high-quality features even in adverse environments. Table 7 summarizes the quantitative performance of different enhancement models.
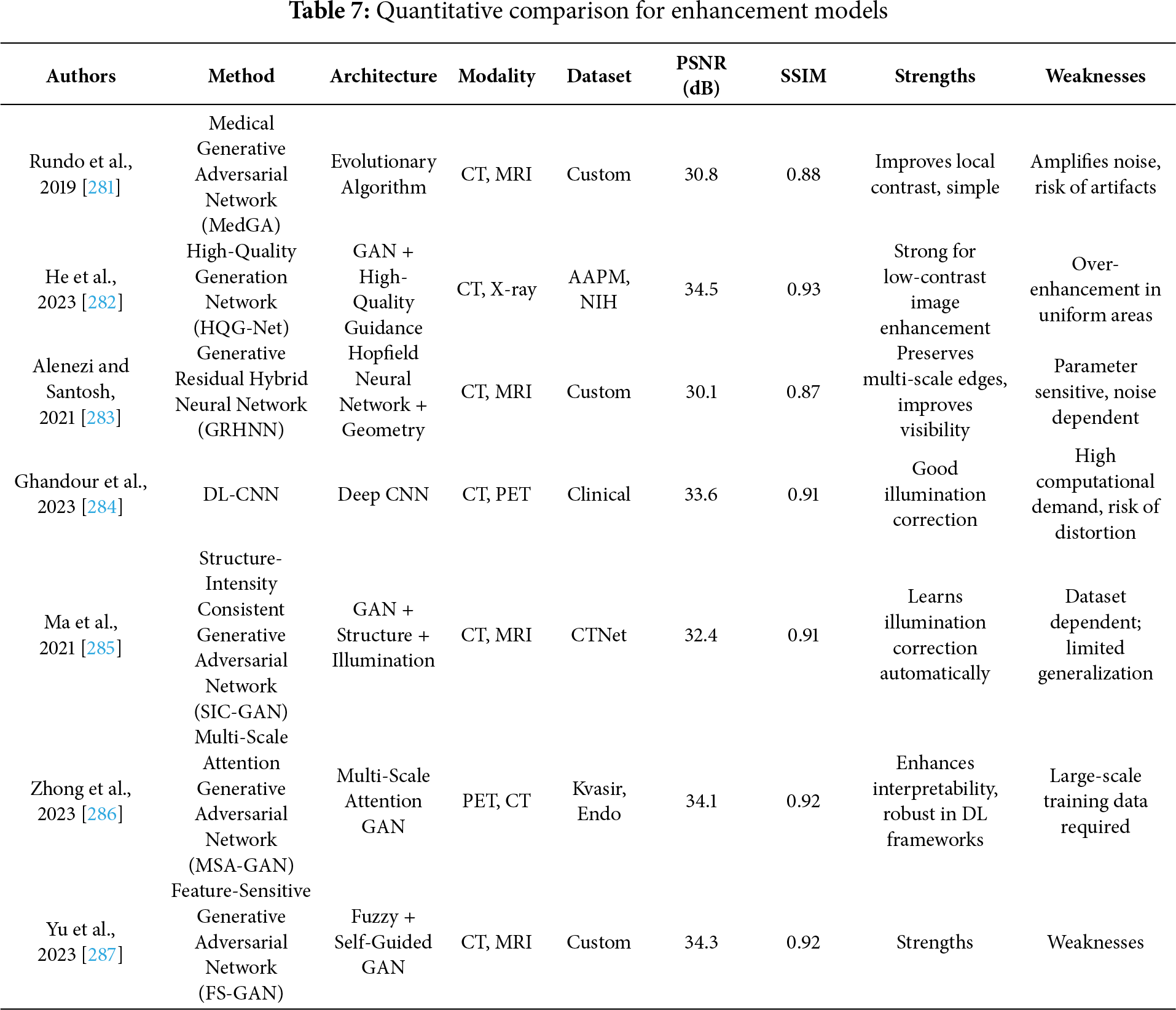
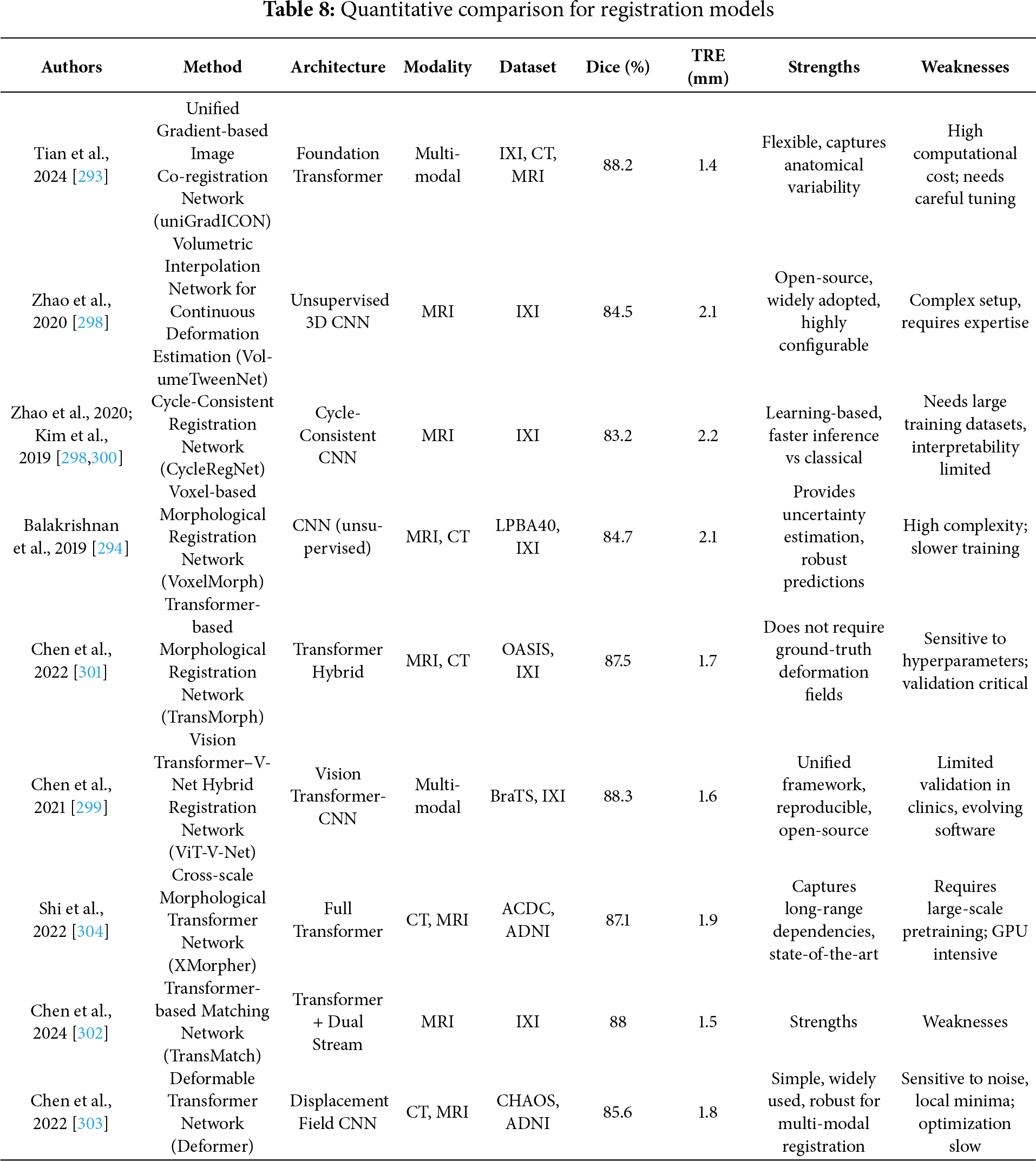
Image registration plays a pivotal role in medical image analysis by aligning images within a common coordinate space for accurate diagnosis and treatment planning [2]. However, the effectiveness of current deep learning-based registration methods must be critically evaluated across supervised, unsupervised, and Transformer-based paradigms. Traditional rigid registration techniques that apply consistent transformations lack adaptability in modeling patient-specific deformations, while deformable (non-rigid) registration allows localized spatial alignment but increases computational complexity and model instability. The deep iterative registration paradigm attempts to overcome limitations of handcrafted similarity metrics by learning task-specific similarity measures through CNNs. Yet, the robustness of such learned metrics across image modalities is inconsistent. For instance, CNN-based metrics [26] that outperformed mutual information in aligning T1-T2 MRI scans showed reduced performance when applied to anatomically dissimilar modalities like Magnetic Resonance–Transrectal Ultrasound Fusion Imaging (MR-TRUS) [290]. Additionally, these learned metrics are computationally expensive as they still require integration with classical iterative optimization algorithms. In supervised deep learning approaches, models like the multiscale CNN proposed in [27] effectively predict dense vector fields (DVFs) in one pass, reducing time complexity. However, this gain in efficiency comes at the cost of requiring large labeled datasets of ground truth deformation fields, which are often generated via traditional registration algorithms, thereby limiting their generalization potential [291,292]. While dual-supervision models, such as the one described in [292], provide performance boosts via complementary loss constraints, they remain data-hungry and constrained to specific modalities and deformation scenarios. The uniGradICON [293] is a foundation model for medical image registration that unites the speed of deep learning with the versatility of traditional methods, enabling cross-dataset performance and zero-shot generalization. While extensions of VoxelMorph that integrate anatomical [294] and weak supervision [295,296] improve performance and these additions often reintroduce domain-specific bias and complicate training. The foundation models like uniGradICON [295] demonstrate promise in zero-shot settings, but such methods still face interpretability issues and lack consensus in standardizing evaluation benchmarks across diverse medical datasets. On the other hand, unsupervised registration frameworks such as VoxelMorph [28], which utilize spatial transformer networks and CNNs to jointly predict and apply deformation fields without labeled data, represent a scalable solution for clinical use. However, their reliance on fixed similarity metrics (e.g., MSE or cross-correlation) in loss functions may degrade accuracy in multimodal settings. Moreover, adversarial frameworks [294] incorporating GANs for similarity estimation offer better cross-modality generalization, but they introduce training instability and require careful calibration of generator-discriminator dynamics. The Deep Learning Image Registration (DLIR) framework [297], with its progressive affine-to-deformable stages, exemplifies modularity and hierarchical refinement, yet its performance is sensitive to hyperparameter tuning across each stage, and it lacks robustness in handling large-scale variability across patients. A similar limitation exists in Volume Twinning Networks (VTN) [298], where cascaded architecture and invertibility constraints enhance alignment quality but may result in diminishing gains with each added stage. In terms of Transformer-based registration, architectures like ViT-V-Net [299] leverage global attention to capture long-range spatial dependencies. Fig. 12 demonstrated schematic flow diagram of a deep learning–based registration network.
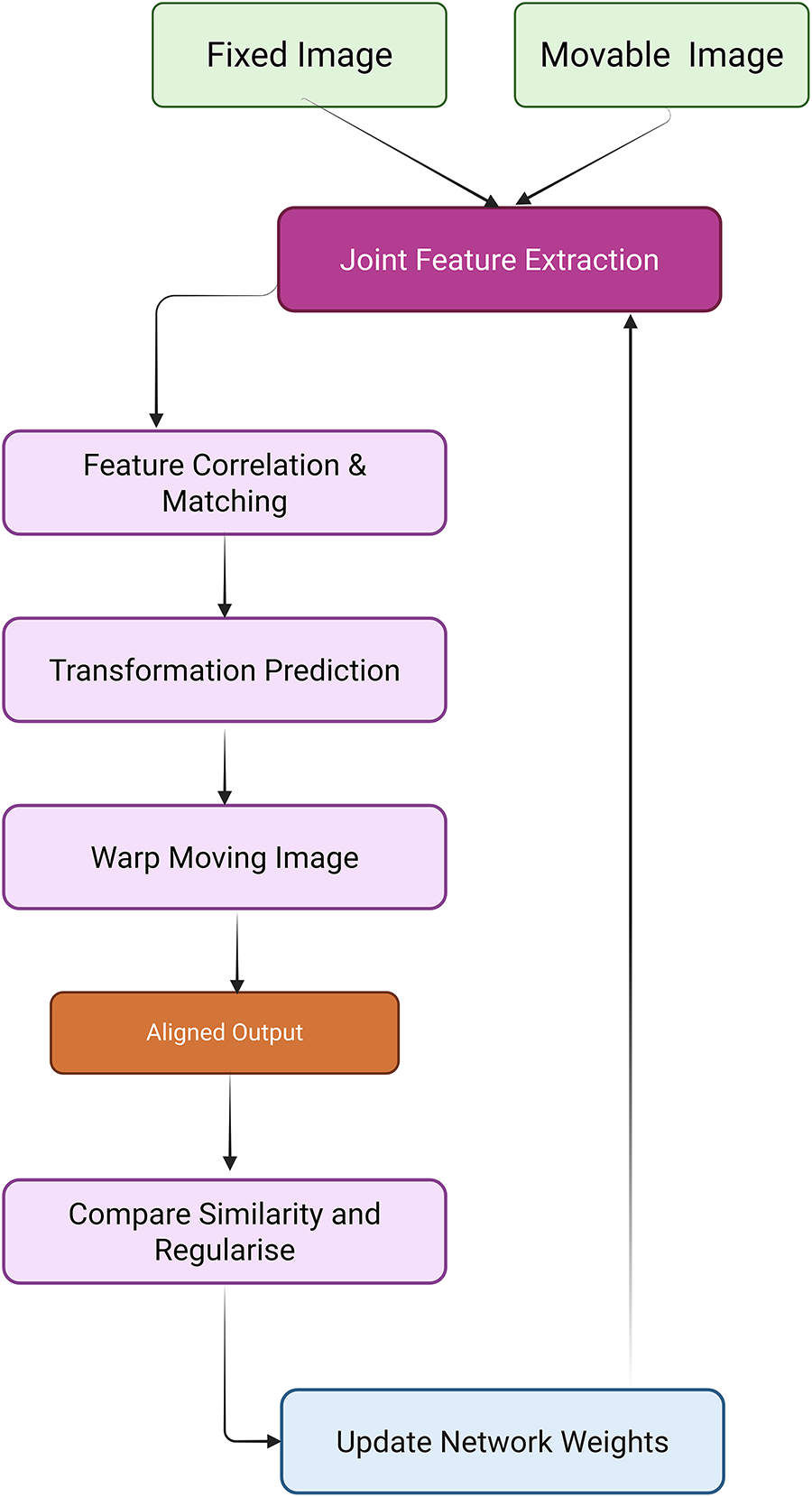
Figure 12: Schematic flow diagram of a deep learning–based registration network
Kim et al. [300] present an unsupervised cycle-consistent CNN for fast and accurate 3D deformable medical image registration, achieving precise alignment on multiphase liver CT images for improved cancer size estimation. TransMorph [301] further enhance this approach by effectively modeling large deformations, addressing the limitations of CNNs. Despite their superior performance across multiple benchmarks, these models often require substantial computational resources and pretrained weights to avoid overfitting. TransMatch [302] and Deformer-based multi-scale frameworks [303] refine displacement prediction using hierarchical matching, yet they often necessitate multiscale ground truth data for effective supervision and have yet to demonstrate consistent improvements across multimodal domains.
Models like XMorpher [304] and C2FViT [305] that use cross-attention or hierarchical feature alignment strategies exhibit better registration for high-resolution 3D images but lack robustness in low-resolution or sparse anatomical datasets. Overall, the field is rapidly evolving with hybrid approaches combining CNNs and Transformers, but most existing models struggle to balance precision, speed, generalizability, and interpretability. Furthermore, benchmarking remains inconsistent across datasets, making fair comparison difficult. While supervised methods provide high accuracy in controlled settings, they are data- intensive and prone to overfitting. Unsupervised methods offer scalability but struggle with multimodal alignment without additional anatomical priors or adversarial constraints. Transformer-based solutions provide a compelling direction but require significant computational overhead and careful architectural design. Critical gaps remain in generalization across imaging modalities, robustness in real-world clinical scenarios, and standardized evaluation protocols. Future research must prioritize model interpretability, cross-domain generalizability, and computational efficiency to transition these methods from proof-of-concept to practical clinical deployment. Meng et al. [306] propose an Automatic Fusion network (AutoFuse) that provides flexibility to fuse information at many potential locations within the network. A Fusion Gate (FG) module is also proposed to control how to fuse information at each potential network location based on training data. Chen et al. [307] propose a new method called Multi-scale Large Kernel Attention UNet (MLKA-Net), which combines a large kernel convolution with the attention mechanism using a multi-scale strategy, and uses a correction module to fine-tune the deformation field to achieve high-accuracy registration. Meyer et al. [308] proposed a hyperparameter perturbation approach to estimate an ensemble of deformation vector fields for a given computed tomography (CT) to cone beam CT (CBCT) Deformable Image Registration (DIR). For each voxel, a principal component analysis was performed on the distribution of homologous points to construct voxel-specific DIR uncertainty confidence ellipsoids. Jiang et al. [309] introduce Fast-DDPM, a simple yet effective approach capable of simultaneously improving training speed, sampling speed, and generation quality. Unlike DDPM, which trains the image denoiser across 1000-time steps, Fast-DDPM trains and samples using only 10-time steps. Table 8 summarizes the quantitative performance of different registration models.
5 Clinical Suitability and Adoption Barriers of Emerging Techniques
The emergence of Transformers, Generative Adversarial Networks (GANs), and contrastive learning has revolutionized the technical frontier of medical image analysis, offering state-of-the-art performance in tasks such as segmentation, detection, denoising, and classification. However, despite their academic appeal, a critical assessment reveals significant shortcomings in their clinical suitability, largely due to unmet requirements for robustness, explainability, scalability, and regulatory compliance. For instance, the widespread adoption of Transformer-based architectures such as TransUNet [157], CoTr [159] and Swin-Unet [160] has been praised for their capacity to capture long-range dependencies and global semantic context through self-attention mechanisms. These designs, particularly in tasks like organ segmentation and multi-modal image fusion, often outperform traditional convolutional architectures like U-Net 128 or V-Net [148] in benchmark datasets such as BraTS, Synapse, and CHAOS. However, the translational gap persists. As emphasized in [128], these models often rely on high-volume, well-curated data, perform poorly under domain shift, and lack transparency, making their integration into clinical pipelines questionable. Furthermore, their substantial computational requirements and sensitivity to adversarial perturbations limit practical deployment, particularly in resource-constrained settings. The promise of Transformers is tempered by their inability to ensure traceability, reproducibility, and interpretability factors critical to clinician trust and regulatory approval. While efforts like ViT-VNet [299] and XMorpher [304] attempt to incorporate multi-resolution strategies and cross-attention modules for improved volumetric registration, their evaluation remains restricted to retrospective studies on static datasets, with limited evidence of real-time inference stability, robustness to imaging artifacts, or adaptability to varying acquisition protocols. As clinical environments often deal with imperfect, noisy, or corrupted data, especially in modalities such as ultrasound and low-dose CT, the lack of robustness under such conditions is a major bottleneck. Additionally, studies rarely assess how well these models generalize across institutions or patient demographics, an issue that is particularly important in federated clinical systems. GANs represent another transformative trend, particularly valued for their utility in data augmentation, unsupervised representation learning, and super-resolution. Architectures like MedSRGAN [253], LatLRR-GAN [273] and FS-GAN [287] demonstrate powerful image synthesis capabilities, useful in generating realistic anatomical variations for training or simulating rare conditions. However, these techniques suffer from critical vulnerabilities. GAN training is notoriously unstable due to issues like mode collapse and sensitivity to hyperparameters. More importantly, clinical deployment of GANs is hindered by their black-box nature. For example, although FS-GAN [287] improves visual quality in nerve imaging using self-guided structure retention, it provides no guarantees against hallucinations that might mislead diagnosis. Moreover, studies such as [195] and [199] have noted that GAN-generated outputs can introduce artifacts that are difficult to distinguish from genuine pathology, leading to potential misinterpretation by clinicians. There is also a lack of systematic evaluation of how GAN-augmented datasets affect downstream tasks such as lesion classification or segmentation. While methods like f-AnoGAN [240] and AnoVAE-GAN [241] improve unsupervised anomaly detection through inverse mapping and latent space modeling, they remain largely experimental, with no Food and Drug Administration (FDA)-cleared applications for clinical decision support. The need for uncertainty quantification, model interpretability, and harmonization across diverse imaging systems remains unresolved. Contrastive learning, especially in self-supervised paradigms, offers an elegant solution to the scarcity of labeled medical data.
Methods such as SimCLR, MoCo, and their medical adaptations [170,171] have shown promise in learning semantically rich representations by maximizing agreement between positive pairs under varying augmentations. When applied to tasks like organ segmentation, tumor detection, and modality translation, contrastive learning improves generalization and reduces overfitting. For example, the local contrastive loss introduced in [173] incorporates structural similarity priors and shows superior performance on MRI segmentation under low-annotation settings. However, challenges abound. First, constructing meaningful positive and negative pairs in the medical domain is non-trivial due to subtle pathological variations, overlapping appearances of disease and normal tissue, and high inter-observer variability. Furthermore, models trained with instance-level contrastive objectives often overfit to pretext tasks, failing to transfer effectively to downstream clinical use cases. The inability of such models to quantify uncertainty or explain predictions poses additional risks in real-time diagnostic contexts. Moreover, the reliance on augmentations like MixUp [67] or rotation prediction [171], which are inherited from natural image tasks, often lacks biological relevance and may inadvertently distort medically significant features. Another major limitation across these paradigms is the lack of rigorous clinical validation. While performance metrics like Dice scores, PSNR, or SSIM are useful proxies, they fall short in assessing clinical outcomes such as diagnostic accuracy, time to diagnosis, or impact on therapeutic decision-making. For instance, segmentation models that excel on datasets like LiTS or CHAOS often show reduced accuracy when applied to external clinical scans due to variation in scanner type, acquisition protocol, and patient anatomy. Even advanced architectures like H2Former [187] and THEDNet [258] despite offering hierarchical multi-scale attention for improved segmentation, remain untested in prospective clinical trials. Moreover, models rarely account for the economic and ethical considerations of deployment, such as cost-effectiveness, bias amplification, or the risk of automation bias among clinicians. As noted in [128], many evaluations still occur on academic benchmarks, not real-world clinical systems where interpretability, safety, and reproducibility are paramount. Additionally, there is insufficient integration between these deep learning frameworks and existing hospital infrastructure, such as picture archiving and communication systems (PACSs) or electronic health records (EHRs). Models such as MedGA [281] or UMIE [282] may enhance image quality, but their outputs are not easily interpretable or traceable within clinical audit trails. Furthermore, techniques like attention U-Net [151] or MAGAN [286] provide heatmaps or attention maps as interpretability tools, yet their utility in actual clinical interpretation remains questionable. Studies such as [24] and [128] suggest that uncertainty estimation using methods like MC dropout or test-time augmentation could enhance trust in AI predictions, particularly for small or ambiguous lesions. However, such strategies are rarely implemented in production-grade systems, and their effect on clinician decision-making is understudied. It is also worth noting that most deep learning models fail to meet regulatory standards for medical devices. Models like uniGradICON [293], XMorpher [304], and TransMatch [302] offer strong performance in cross-modality registration or zero-shot generalization, but lack explainability mechanisms required by regulatory agencies like the FDA or EMA. These agencies mandate not only performance benchmarks but also traceability, robustness, and evidence of reproducibility across patient subgroups and imaging devices. Very few Transformer-based or GAN-powered systems have undergone such scrutiny. Furthermore, models trained in a single-center academic setting rarely generalize across populations, leading to algorithmic bias. A lesion detector trained on predominantly Caucasian populations may underperform in Asian or African cohorts, reinforcing health disparities rather than mitigating them. In summary, while Transformers, GANs, and contrastive learning represent transformative advances in medical image analysis, their clinical readiness is undermined by a convergence of challenges: lack of real-world validation, limited robustness to data variability, poor explainability, high computational demands, regulatory ineligibility, and misalignment with clinician needs. Future research must pivot toward clinically grounded AI, incorporating (1) model interpretability and explainability tools (e.g., saliency maps, uncertainty maps); (2) domain adaptation and continual learning for real-world variability; (3) integrated evaluation using clinical outcome metrics (e.g., diagnosis accuracy, radiologist trust scores); and (4) adherence to ethical, legal, and regulatory frameworks. Techniques like federated learning for multi-center generalization, self-supervised pretraining on real clinical data, and user-in-the-loop model design may help bridge this translational chasm. Without these enhancements, even the most technically advanced architectures will remain detached from the clinical reality they seek to serve.
6 Beyond Benchmark Datasets: Toward Scalable and Generalizable Deep Learning in Medical Imaging
The field of medical image analysis has traditionally leaned heavily on benchmark datasets such as BraTS, LUNA16, ISIC, and DeepLesion, which have indeed been instrumental in catalyzing progress across classification, segmentation, and detection tasks. However, this dependence, while foundational, has begun to reveal substantial limitations when assessing the clinical translatability and robustness of AI models. These datasets are typically derived from a single or limited number of institutions, exhibit uniform imaging protocols, and represent constrained demographic diversity, all of which contribute to data homogeneity and model overfitting. As a result, algorithms trained and validated exclusively on such curated datasets often fail to generalize to external clinical environments with differing acquisition devices, population characteristics, or pathological spectra [36,310]. This systemic limitation stems from the “closed-world” assumption embedded within benchmark-based evaluation, while the flawed notion that all clinically relevant variations are sufficiently captured within a finite, static dataset. In practice, the medical imaging landscape is riddled with distributional shifts due to inter-institutional discrepancies in scanner vendors, protocols, and labeling standards. For instance, skin lesion segmentation models developed using ISIC often degrade in performance when tested on images from underrepresented skin tones or acquired under variable lighting conditions, revealing stark performance disparities [311,312]. Moreover, rare diseases and atypical presentations are underrepresented in most benchmarks, thereby reducing sensitivity to long-tail cases that hold substantial clinical significance [313]. In response to these challenges, two key research directions, cross-institutional data harmonization and federated learning (FL), have emerged as promising avenues for achieving real-world robustness. Data harmonization involves aligning imaging statistics across institutions by applying standardization techniques such as histogram matching, intensity normalization, and domain adaptation. Notably, the ComBat method has been adapted for MRI harmonization by modeling batch effects while preserving biological variability [314]. Similarly, GAN-based style transfer approaches have been leveraged to synthetically bridge domain gaps while retaining pathological content, thereby enabling cross-site generalization [315]. Domain-adversarial frameworks like medical domain-adversarial neural network (MedDANN) and adversarial loss regularization strategies are also increasingly used to encourage domain-invariant feature learning during model training, significantly reducing cross-institutional variance in tasks like brain MRI segmentation and lung nodule classification [316]. However, the harmonization process is not without drawbacks. Excessive normalization may obscure subtle but clinically informative cues, particularly in disease subtyping or radiomic analysis. Moreover, harmonization assumes access to data from multiple sites, a constraint that is often infeasible due to privacy regulations or data sovereignty concerns. This motivates the integration of federated learning as a complementary strategy.
Federated learning offers a privacy-preserving approach by enabling decentralized training across institutions without requiring raw data sharing. The canonical FL paradigm trains local models on-site and periodically aggregate parameters into a global model, maintaining data residency at each institution and thereby satisfying privacy laws like General Data Protection Regulation (GDPR) and Health Insurance Portability and Accountability Act (HIPAA). In landmark work, Sheller et al. [317] demonstrated that FL could match the performance of centralized training for brain tumor segmentation across multiple centers, highlighting its potential in collaborative medical imaging applications. Recent advancements have also introduced personalized federated algorithms such as Federated Batch Normalization and Federated Proximal Algorithm (FedProx), which accommodate data heterogeneity by allowing partial model personalization or relaxed aggregation schemes, further enhancing accuracy across dissimilar client distributions [318,319]. More sophisticated FL variants, such as Federated Domain Generalization (FedDG), combine domain generalization principles with federated optimization to improve model robustness to unseen domains. In a multi-institutional study on COVID-19 chest X-ray classification, FedDG showed superior out-of-distribution performance compared to both FL and centralized models [320]. Despite these advantages, FL frameworks encounter practical bottlenecks, including communication latency, client synchronization failures, and vulnerability to gradient leakage attacks. Differential privacy techniques and secure multiparty computation protocols are being actively explored to mitigate these risks and bolster trustworthiness in federated pipelines [321].
Beyond harmonization and FL, synthetic data generation has gained traction as a third pillar of generalizable AI. Generative models like diffusion models and 3D GANs are being employed to simulate rare disease presentations or augment underrepresented imaging modalities. These synthetic examples enhance data diversity and help overcome annotation bottlenecks while avoiding privacy pitfalls. Nonetheless, synthetic realism and label fidelity remain ongoing challenges, especially in high-stakes diagnostic domains. Another pivotal concern relates to transparency and reproducibility. Many published models omit critical information about dataset origins, preprocessing steps, or demographic composition, hindering reproducibility and complicating cross-study comparison. Reporting frameworks such as CLAIM (Checklist for Artificial Intelligence in Medical Imaging) [322] and Consolidated Standards of Reporting Trials–Artificial Intelligence Extension (CONSORT-AI) [323] advocate for detailed documentation of data provenance, annotation protocols, and model evaluation pipelines to ensure clinical relevance and external validity. Without such disclosures, the scientific community risks proliferating models that are fundamentally opaque and clinically unreliable. Emerging research is also investigating continual and meta-learning paradigms that enable adaptive retraining as new institutional data become available. These paradigms allow pre-trained models to incrementally learn from new domains without catastrophic forgetting, a feature crucial in rapidly evolving healthcare settings. For example, hybrid frameworks that combine FL with meta-learning or continual learning enable adaptation to unseen domains with minimal fine-tuning [324]. To further enhance model interpretability and safety, Bayesian deep learning methods have been integrated into FL pipelines to quantify uncertainty and flag predictions requiring expert review. Epistemic uncertainty estimates, in particular, are useful for identifying low-confidence predictions stemming from data drift or underrepresented pathologies [89]. Recent studies also explore fairness-aware training by stratifying performance metrics across sensitive subgroups (e.g., age, ethnicity, scanner type), uncovering algorithmic bias that may otherwise remain undetected under benchmark-centric evaluation [325]. In summary, while benchmark datasets continue to play a valuable role in reproducible evaluation and method comparison, their overutilization risks producing brittle, overfitted models with poor external validity. Transitioning toward clinically viable AI requires a paradigm shift away from closed-set evaluation toward open-world, diverse, and distributed learning scenarios. Cross-institutional data harmonization, federated optimization, and synthetic augmentation offer synergistic solutions for addressing these challenges. Future research must prioritize robustness testing on external datasets, integration of uncertainty quantification, and fairness audits to move from theoretical accuracy to clinical impact.
7 Evaluation Metrics, Regulatory Frameworks, and Interpretability Considerations
Deep learning (DL) applications in medical image analysis have shown remarkable performance improvements in tasks such as classification, segmentation, and anomaly detection. However, their clinical deployment remains hindered by insufficient attention to evaluation rigor, interpretability, and alignment with regulatory frameworks. In this section, we critically examine the superficial treatment of evaluation metrics, regulatory concerns, and model interpretability, and propose comprehensive strategies based on recent literature to foster robust, trustworthy, and clinically acceptable DL systems.
7.1 Evaluation Metrics: Beyond Accuracy
Many studies evaluating DL models in medical imaging predominantly report metrics such as accuracy, precision, and recall, which may be insufficient for assessing clinical relevance. For instance, segmentation tasks demand more nuanced evaluation metrics such as the Dice Similarity Coefficient (DSC) and Hausdorff Distance. DSC measures the overlap between the predicted and ground-truth regions, particularly valuable in tumor or lesion segmentation tasks, while Hausdorff Distance quantifies boundary precision, which is crucial for surgical planning or radiotherapy [326,327]. Classification tasks require not only accuracy but also balanced metrics like F1-score, especially in cases of class imbalance. Furthermore, the Area Under the Receiver Operating Characteristic Curve (AUC-ROC) is used to assess model discrimination, especially in binary and multi-class disease detection scenarios. However, recent work has emphasized the importance of calibration metrics (e.g., Expected Calibration Error) to measure the confidence of predictions—a critical component for clinical decision-making [328]. Uncertainty quantification through Bayesian networks or ensemble techniques is gaining traction, enabling models to estimate when they are likely to be incorrect thus flagging vague cases for expert review [329].
7.2 Regulatory Frameworks: Navigating Software as a Medical Device (SaMD) and MDR Compliance
From a regulatory standpoint, AI models used in healthcare must conform to global guidelines such as the FDA’s SaMD framework, the European Union’s Medical Device Regulation (EU MDR 2021/2226), and the International Medical Device Regulators Forum (IMDRF) principles. These frameworks mandate transparent validation pipelines, robust performance metrics, risk management protocols, and reproducibility to ensure patient safety and model reliability [330,331]. The FDA’s Good Machine Learning Practice (GMLP) guidance stresses continuous learning, dataset diversity, and traceability of model updates. However, a significant bottleneck is the “black box” nature of DL models, which impedes approval due to interpretability and reproducibility concerns. Clinical trials, real-world performance studies, and external validation on multi-institutional datasets are increasingly demanded by regulatory bodies to demonstrate generalizability and safety [332]. Furthermore, standards such as International Organization for Standardization / International Electrotechnical Commission Technical Report 24028:2020 (ISO/IEC TR 24028:2020) and International Organization for Standardization 14971 (ISO 14971) provide frameworks for risk assessment and management in AI-driven medical devices. These require models to clearly define performance thresholds, failure modes, and response protocols, which most academic prototypes currently fail to deliver. The lack of standardized datasets and external validation also complicates direct model comparisons, leading to inconsistent performance claims [333].
7.3 Interpretability and Explainability: Key to Clinical Trust
Interpretability is central to clinical adoption. Without comprehensible explanations, clinicians are unlikely to trust AI-generated decisions, particularly in high-stakes environments. Explainable AI (XAI) tools such as Grad-CAM, SHapley Additive exPlanations (SHAP), and Local Interpretable Model-Agnostic Explanations (LIME) have been developed to visualize and quantify the contribution of input features to model predictions [334,335]. These tools help delineate the regions of interest in images and support clinicians in verifying the rationale behind model outputs. Recent studies have demonstrated how attention-based models and saliency maps enhance decision transparency, particularly in mammography and retinal imaging. For instance, the integration of anatomical priors into network architectures improves interpretability while preserving accuracy. Moreover, hybrid systems combining symbolic reasoning with DL allow for more transparent reasoning paths, especially in tasks requiring sequential decision-making like disease progression modeling [336]. However, current XAI tools often offer post-hoc approximations rather than faithful representations of the model’s decision-making process, raising concerns about reliability. Therefore, ongoing research is shifting toward inherently interpretable models and counterfactual explanations that simulate “what-if” scenarios to evaluate model stability and causality [337]. These developments not only assist clinicians but also fulfill regulatory requirements demanding explainability for risk classification and accountability.
7.4 Transparency, Documentation, and Reproducibility
Transparency in model development and documentation is critical for clinical integration. The CLAIM (Checklist for Artificial Intelligence in Medical Imaging) and CONSORT-AI guidelines emphasize the need for detailed reporting of dataset composition, annotation procedures, preprocessing steps, hyperparameter tuning, and evaluation metrics [322]. Lack of such documentation can lead to irreproducibility, inflated performance claims, and difficulty in translating research models into clinical practice. Moreover, the Minimum Information for Clinical Artificial Intelligence Modeling (MI-CLAIM) framework advocates for the documentation of model interpretability mechanisms, uncertainty quantification, and clinician-in-the-loop evaluations. These checklists serve as a bridge between academic innovation and real-world implementation, ensuring that models meet regulatory and ethical standards.
7.5 Clinical Validation and Human-in-the-Loop Evaluation
A critical component of deployment is clinical validation through prospective studies and randomized controlled trials (RCTs). Recent work by Wang et al. highlighted that models demonstrating excellent retrospective performance often underperform in live clinical settings due to differences in workflow integration and user interaction [338]. Hence, human-in-the-loop systems, where AI assists rather than replaces human decision-making, have shown improved outcomes in diagnostic accuracy and workflow efficiency. Such systems also help in bias detection and correction by enabling clinician oversight. They can identify model drift where performance degrades over time due to changes in population or data acquisition and initiate recalibration protocols. The incorporation of uncertainty flags allows clinicians to triage cases needing manual review, ensuring safety while maintaining efficiency.
7.6 Ethical and Societal Implications
The convergence of evaluation metrics, regulatory approval, and interpretability also intersects with broader ethical concerns. Inadequate evaluation and opaque decision-making can perpetuate biases, particularly when models are trained on non-representative datasets. Recent studies have shown that dermatological models trained on lighter skin tones perform poorly on darker skin types, leading to inequitable outcomes [339]. Hence, fairness-aware evaluation metrics such as demographic parity and equalized odds are being explored to assess and mitigate algorithmic bias. Privacy-preserving techniques like federated learning and differential privacy are also gaining regulatory support to allow secure model training across institutions without compromising patient confidentiality. These techniques are critical for complying with GDPR and HIPAA regulations while enabling large-scale, diverse model training [340].
8 From Research to Clinic: Model Readiness Assessment (Maturity Mapping)
The integration of deep learning into medical image analysis has led to unprecedented advancements in detection, segmentation, synthesis, and classification. However, the line between experimental models and those ready for clinical deployment is often blurred in research narratives. This lack of demarcation poses challenges in assessing the true readiness of models for real-world clinical integration, especially under the regulatory, ethical, and safety constraints imposed by healthcare systems. To address this, we propose a maturity mapping framework that categorizes models based on their research maturity, clinical validation, and regulatory readiness. This framework is essential to prevent overstatement of capabilities in academic literature and ensure translational integrity. Table 9 summarizes the maturity mapping table.
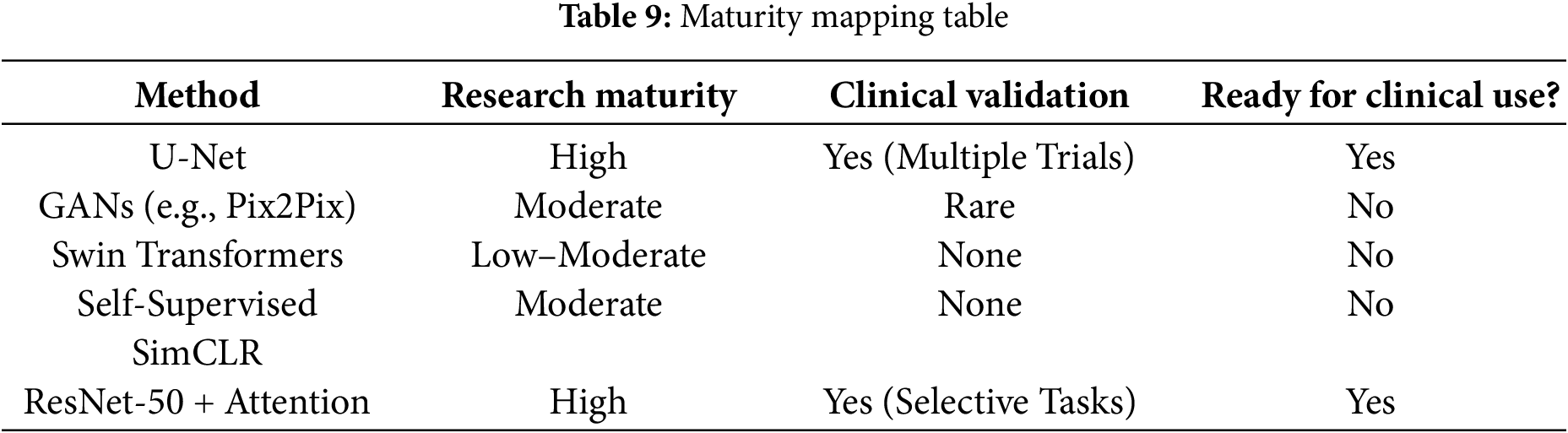
8.1 U-Net: A Clinically Vetted Backbone
U-Net, introduced in 2015 has become the gold standard for medical image segmentation tasks. Its symmetric encoder-decoder architecture with skip connections ensures the retention of spatial context, crucial for precise segmentation of organs and lesions. Its widespread adoption across modalities such as MRI, CT, and ultrasound, along with its extensive use in clinical trials, has validated its performance across various datasets and clinical conditions [143]. Numerous FDA-approved tools like ProFound AI for breast cancer detection are built upon U-Net or its derivatives, reinforcing its clinical maturity and reliability. The maturity mapping framework in this review serves as a structured tool to evaluate the progression of deep learning methods in medical imaging, ranging from early proof-of-concept demonstrations to large-scale validation, regulatory clearance, and eventual clinical adoption. While many approaches show strong experimental results, they remain at lower maturity levels due to several barriers. Key limitations include restricted dataset diversity, which reduces generalizability across populations; lack of external, multi-institutional validation; and high computational demands that hinder real-time clinical integration. In addition, the limited interpretability of deep learning models reduces clinician trust, while challenges in complying with regulatory and ethical standards delay approval and deployment. By mapping these stages and identifying the reasons for immaturity, the framework highlights both current gaps and the pathways necessary for advancing AI tools toward clinical readiness.
8.2 GANs: Transformative but Unstable
Generative Adversarial Networks (GANs), especially conditional variants like Pix2Pix, have gained traction in synthesizing realistic medical images, data augmentation, and domain adaptation. However, GANs are still considered experimental due to their sensitivity to training instabilities, mode collapse, and the lack of quantitative uncertainty modelling. While their utility in tasks such as cross-modality image synthesis (e.g., CT-to-MRI) is evident, GAN-generated content raises ethical concerns, especially in clinical decision support. Furthermore, clinical validation of GAN-based pipelines is sparse, and their black-box nature complicates interpretability and trust in critical diagnoses [341].
8.3 Swin Transformers: Promising but Immature
Transformer-based architectures, particularly Swin Transformers represent a paradigm shift in vision-based learning. With their hierarchical structure and non-local attention and they excel in modeling long-range dependencies in 2D and 3D imaging contexts. Swin-Unet and its variants have achieved impressive results in segmentation benchmarks like Synapse or BraTS, often surpassing traditional CNNs [161]. However, these architectures remain confined to experimental validations. Their large parameter space, training inefficiencies, and the absence of external validations in clinical trials hinder their real-world applicability. Regulatory frameworks such as the FDA’s SaMD demand explainability and reproducibility, both of which are currently unmet by transformer models [160].
8.4 Self-Supervised Learning: SimCLR and Representation Bottlenecks
SimCLR (Simple Framework for Contrastive Learning of Visual Representations) and its derivatives signify a shift towards label-efficient learning, essential for medical imaging, where annotations are costly and expertise-dependent. SimCLR learns visual embeddings using contrastive loss without requiring labeled data, enabling the utilization of large unlabeled datasets [57]. Applications in chest X-ray triage, pathology slide representation, and retinal disease classification have been proposed [122]. Despite their scalability, such methods lack task specificity and direct interpretability, limiting their standalone use in clinical diagnosis. No self-supervised model has yet passed regulatory checkpoints or clinical trials, although hybrid approaches combining self-supervised features with supervised heads show promise.
8.5 ResNet-50 + Attention: Practical in Clinical Subtasks
ResNet-50 [108] combined with attention mechanisms such as CBAM or SENet has demonstrated robust performance in classification and detection tasks. These architectures are interpretable, lightweight, and have been embedded into tools for ophthalmology, dermatology, and radiology screening [57]. For example, EyeArt, an AI system for diabetic retinopathy screening, incorporates a ResNet-based architecture and is FDA-approved [342]. Attention maps also improve clinician trust by providing heatmaps that localize decision-critical regions. Given their explainability, availability of pretrained weights, and generalization capacity, ResNet-based models are transitioning successfully from research to regulated clinical tools.
8.6 The Importance of Maturity Mapping
The maturity mapping table not only informs clinical stakeholders but also guides researchers in setting realistic goals. Research models with low maturity (e.g., transformers, GANs) should be presented with disclaimers, while clinically validated models should include external validations and documentation aligned with CLAIM and CONSORT-AI guidelines [322,343]. The separation also ensures compliance with regulations like the EU MDR 2021/2226 and the FDA’s SaMD guidelines, which emphasize performance transparency, interpretability, and real-world evidence. A growing concern is the misuse of experimental models in retrospective studies with exaggerated claims. Publications often do not distinguish between proof-of-concept validation and readiness for clinical integration, leading to potential mistrust and misuse. Initiatives such as the AI-Med Maturity Framework and the Digital Imaging and Communications (DICOM) AI profile are increasingly being adopted to standardize AI readiness assessment [344].
9 Bridging the Research–Clinic Gap
To bridge the persistent gap between experimental AI prototypes and their actual deployment in clinical settings, three pivotal pathways must be emphasized. First, external multi-center validation plays a foundational role in establishing model robustness and generalizability across diverse healthcare ecosystems. Clinical trials such as the CONFIRM-AI study underscore the necessity for testing AI systems across multiple institutions and patient populations, thereby minimizing the risk of bias stemming from overfitting to specific imaging equipment or localized datasets [323]. Second, explainability frameworks are indispensable in enhancing transparency and clinician trust. The integration of tools like Gradient-weighted Class Activation Mapping (Grad-CAM), SHapley Additive exPlanations (SHAP), and Local Interpretable Model-agnostic Explanations (LIME) provides critical insight into model decision-making processes, enabling both medical professionals and regulatory agencies to understand and audit algorithmic outputs effectively [345]. This interpretability is particularly crucial for post-deployment legal accountability and ethical compliance. Third, regulatory sandboxes offer a structured yet flexible environment for evaluating emerging AI technologies. Initiatives such as the UK’s Medicines and Healthcare products Regulatory Agency (MHRA) AI sandbox provide collaborative spaces where developers and regulators can jointly assess model performance under realistic clinical conditions without compromising patient safety. Collectively, these strategies form a triad that facilitates the responsible translation of AI research into trustworthy and usable clinical tools.
10 Clinical Deployment Challenges and Best Practices
The translation of deep learning (DL) models from controlled research environments to real-world clinical workflows remains a major bottleneck in medical image analysis. Despite remarkable algorithmic performance in benchmark studies, the deployment of AI systems in hospital settings often exposes critical weaknesses primarily concerning robustness, fairness, and explainability. These gaps risk undermining trust in AI-assisted diagnostics and slowing regulatory adoption. A systematic understanding of these challenges is essential to pave the path for practical and responsible clinical deployment.
10.1 Robustness and Generalization
In clinical environments, the robustness and generalization capabilities of deep learning (DL) models are profoundly challenged by the inherent heterogeneity of medical imaging data stemming from variations in scanner types, acquisition protocols, patient demographics, and institutional practices. This domain shift can significantly degrade performance when models trained on clean, homogeneous benchmark datasets are deployed in real-world, diverse clinical settings [346]. To mitigate these challenges, several robustness enhancing strategies have emerged. Test-Time Augmentation (TTA) modifies inputs at inference using transformations like rotations, flips, and intensity shifts and aggregates predictions to stabilize outputs against minor perturbations [347]. Adversarial training, which injects synthetically generated perturbations into training images, conditions models to handle worst-case scenarios, though it often increases computational overhead. Domain adaptation methods such as Cycle-Consistent Generative Adversarial Network (CycleGAN)-based style transfer, feature alignment, and domain-invariant representation learning allow models trained in one domain to generalize to unseen domains without labeled target data [348]. These are particularly important for adapting models across institutions or imaging devices. Additionally, ensemble models and uncertainty estimation techniques such as Monte Carlo dropout and deep ensembles are increasingly used to identify out-of-distribution inputs and quantify prediction confidence, guiding clinicians when predictions may be unreliable [349]. Despite these promising directions, achieving robust generalization across diverse patient populations, imaging modalities, and clinical protocols remains an open research frontier. No single strategy guarantees seamless deployment, highlighting the need for adaptive, hybrid approaches, rigorous cross-institutional validation, and continual monitoring in clinical workflows. Despite these promising directions, achieving robust generalization across diverse patient populations, imaging modalities, and clinical protocols remains an open research frontier, as no single strategy guarantees seamless deployment, highlighting the need for adaptive, hybrid approaches, rigorous cross-institutional validation, and continual monitoring in clinical workflows [350]. Although previously discussed, Transformer merits are restated here to highlight their relevance in disease-specific applications.
10.2 Fairness and Bias Mitigation
Bias and fairness remain critical challenges in clinical deployment of deep learning (DL) models, particularly in medical imaging, where societal inequities can be inadvertently encoded and amplified during model training. Bias in medical image segmentation can arise from several sources, including instrumental bias from variations in scanners and acquisition protocols, normalization bias introduced during preprocessing, annotation bias from subjective labeling, and population bias due to unbalanced datasets. Such biases can degrade segmentation accuracy and limit generalizability across clinical settings [351–353]. Addressing these challenges requires harmonized imaging protocols, standardized annotation practices, and bias-aware training strategies to ensure reliable clinical deployment.
Disparities in race, gender, age, and socioeconomic status within training datasets can lead to disproportionately inaccurate predictions for underrepresented subgroups, posing significant risks in clinical decision-making [325]. For instance, models trained on datasets like CheXpert or MIMIC-CXR have shown reduced diagnostic accuracy for Black and female patients compared to White male patients, reflecting latent demographic biases in the data [325]. Mitigation strategies are evolving to address this issue: re-weighting and re-sampling techniques attempt to balance subgroup representation during training, but can introduce trade-offs with overall model accuracy if not finely calibrated [354]. Adversarial de-biasing, where models are trained to maximize task performance while minimizing the ability to infer protected attributes such as gender or race, has emerged as an effective method for fairness preservation without severely compromising utility. Similarly, fair representation learning aims to extract features that are invariant to sensitive demographic variables, thereby enforcing equity across predictions [355]. Importantly, stratified evaluation, where performance is disaggregated by demographic groups rather than reported as global metrics, is increasingly recognized as a mandatory practice for transparency and accountability. Recent efforts also advocate for integrating fairness audits into clinical AI pipelines, including subgroup-specific calibration curves, error analysis, and equalized odds frameworks, to ensure that medical AI systems do not inadvertently harm vulnerable populations [356]. These developments underscore the need for fairness-aware model design, continuous monitoring, and inclusive dataset curation to foster trust and effectiveness in AI-driven healthcare.
10.3 Explainability in Practice
A persistent concern among clinicians regarding the adoption of deep learning (DL) systems in medical imaging is the “black-box” nature of such models, which often provide highly accurate predictions without intelligible justifications, particularly in high-stakes applications like cancer diagnosis, disease progression modeling, or surgical planning [357]. This opacity undermines clinician trust and impedes regulatory acceptance, prompting a growing emphasis on explainability. Tools like Local Interpretable Model-Agnostic Explanations (LIME) attempt to address this by perturbing input data to determine which features most influence the model’s decision; however, LIME can be unstable and computationally intensive, especially in high-dimensional medical data such as volumetric scans. Grad-CAM (Gradient-weighted Class Activation Mapping) has gained popularity in radiology by offering visualizations of image regions that contributed most to a model’s decision, thus aiding in tumor localization and interpretation of diagnostic cues. Concept bottleneck models introduce intermediate, human-interpretable variables (e.g., “mass shape,” “spiculated border,” or “calcification pattern”) into the prediction pipeline, allowing physicians to validate model outputs against known diagnostic criteria and improving auditability. In parallel, attention mechanisms in models like Vision Transformers (ViT) inherently highlight relevant spatial features during inference, offering implicit forms of interpretability that can be mapped visually or statistically [358]. Similarly, saliency maps and heatmaps provide overlays that help localize discriminative regions for classification or segmentation tasks, but these methods can be misleading or overly sensitive to model initialization, architecture, and noise [359]. To address this, a combination of multiple explainability tools that are integrated into clinician-facing interfaces, such as interactive dashboards or AI-assisted PACS, has shown promise in bridging the trust gap. Nevertheless, explainability alone should not be considered a proxy for reliability or trustworthiness; these tools must undergo rigorous clinical validation, user studies, and iterative feedback loops to align model explanations with clinical reasoning and decision-making workflows [360].
10.4 Clinical Workflow Integration
The successful clinical deployment of AI in medical imaging is frequently hindered at the integration stage, where even high-performing models encounter practical barriers in aligning with real-world hospital workflows. Interoperability issues persist due to the proprietary nature of existing systems like Hospital Information Systems (HIS), Picture Archiving and Communication Systems (PACS), and Electronic Medical Records (EMRs), which complicates the incorporation of AI modules. Although emerging interoperability standards such as Health Level Seven—Fast Healthcare Interoperability Resources (HL7 FHIR) and Digital Imaging and Communications in Medicine where Working Group 23 (Application Hosting) (DICOM-WG23) show promise for supporting AI integration, their adoption remains limited across institutions [361]. Moreover, latency and usability present critical concerns in time-sensitive environments like emergency or surgical care, where cloud-based AI solutions may not offer a real-time response or comply with stringent data privacy requirements under regulations such as GDPR and HIPAA. Human-in-the-loop (HITL) frameworks are now emphasized to maintain clinician oversight and accountability; these systems enable feedback refinement, foster trust, and ensure that AI serves as a decision support tool rather than a replacement [362]. However, achieving meaningful clinical adoption also demands education and upskilling: clinicians must understand AI principles, and developers must be versed in clinical constraints and ethical standards, underscoring the importance of interdisciplinary collaboration. Post-deployment, ongoing performance monitoring is essential to identify model drift and maintain safety across varied cohorts and imaging modalities [357]. A model that performs reliably in development may fail silently in new settings without adaptive mechanisms in place. These challenges emphasize that technical performance alone is insufficient for clinical readiness; robust system integration, governance, education, and continuous oversight must form the foundation for sustainable, trustworthy AI in healthcare.
10.5 Regulatory and Ethical Best Practices
Deployment of AI models in clinical settings necessitates rigorous adherence to regulatory frameworks and ethical standards to ensure patient safety, accountability, and public trust. Regulatory approval processes from authoritative bodies such as the FDA (USA), CE (Europe), and CDSCO (India) mandate comprehensive evidence of a model’s safety, efficacy, and clinical utility, particularly under high-risk categories. The FDA’s “AI/ML-based Software as a Medical Device (SaMD) Action Plan” (2021) and the European Union’s Medical Device Regulation (EU MDR 2021/2226) are prominent frameworks guiding AI regulation, both of which stress real-world validation, continual monitoring, and transparency in algorithmic updates [363]. Similar to pharmaceuticals, high-stakes AI applications must undergo prospective, multi-center clinical trials to demonstrate generalizability across populations and institutions. Ethical oversight is equally critical, with Institutional Review Boards (IRBs) ensuring that all aspects of AI development, data use, patient consent, and downstream application adhere to regional data protection regulations such as GDPR (Europe) and HIPAA (USA) [364]. Inadequate ethical safeguards risk violations of patient privacy, algorithmic discrimination, or the misuse of clinical predictions in care decisions. Moreover, transparent reporting practices have been strongly advocated to improve trust and reproducibility in AI systems. Model cards and datasheets, structured documentation tools introduced by Mitchell et al. (2019) are now encouraged for publication and regulatory submissions. These resources capture critical metadata, including dataset provenance, performance disaggregated by demographic subgroups, known failure modes, and intended use contexts. Recent initiatives like CONSORT-AI and SPIRIT-AI further codify the reporting standards for AI clinical trials, enhancing regulatory readiness and ethical transparency [365]. Ultimately, building clinically trustworthy AI demands not only technical performance but a strong foundation in ethical design, transparent evaluation, and robust regulatory alignment.
11 Model Validation and Regulatory Benchmarks: Ensuring Clinical Readiness of AI Tools in Radiology
Despite the growing momentum of artificial intelligence (AI) in medical imaging, real-world clinical deployment remains fraught with critical concerns regarding safety, reproducibility, and trustworthiness. Two essential elements to address these concerns are: (1) a rigorous model validation pipeline with key checkpoints and (2) transparency through benchmarking against FDA-approved AI tools in radiology. Together, they represent the gold standard for transitioning AI models from research prototypes to clinical-grade technologies. Model Validation Pipeline with Key Checkpoints serves as a structured framework for evaluating and certifying AI tools across different stages of development and deployment as demonstrated in Fig. 13. This pipeline typically comprises four critical checkpoints: data quality validation, model performance validation, interpretability assessment, and clinical trial validation. Each checkpoint acts as a safeguard to ensure that the AI model is not just statistically robust but also clinically relevant and ethically compliant. The first checkpoint involves curating high-quality, representative, and diverse datasets. Poor data quality due to label noise, under-representation of patient demographics, or unstandardized imaging protocols can lead to spurious patterns and biased outputs. Thus, validation at this stage emphasizes data de-identification, class balance, and harmonization across institutions (e.g., through ComBat or GAN-based style transfer). The second checkpoint, model performance validation, evaluates diagnostic metrics such as Dice Similarity Coefficient (DSC) for segmentation, AUC-ROC and F1-score for classification, and calibration curves for uncertainty assessment. Crucially, this phase also requires external validation across independent cohorts to ensure generalizability, an element missing in many academic publications but necessary for FDA approval. The third checkpoint, interpretability assessment, determines whether the AI’s decision-making process can be understood and trusted by clinicians. Techniques like Grad-CAM, SHAP, or concept bottlenecks help visualize model attention and highlight features influencing decisions. These tools are instrumental in building clinician trust and identifying algorithmic failures. The final checkpoint involves clinical trials, including prospective and multi-center validation, which align with regulatory standards like the FDA’s Software as a Medical Device (SaMD) framework or the EU MDR. Only after demonstrating reproducibility, efficacy, and safety in these trials can a model be considered for real-world use. To illustrate how models successfully progress through this pipeline, Table 10 presents examples of FDA-approved AI tools in radiology (2020–2024), providing valuable insights into their approval and implementation processes. For instance, Viz LVO by Viz.ai, approved in 2018 and updated in 2022 was among the first tools to automate large vessel occlusion detection using CT angiography. It underwent extensive real-world validation in stroke triage workflows across multiple institutions, justifying its FDA clearance [366]. Similarly, Aidoc’s suite of AI tools for pulmonary embolism and brain hemorrhage detection has secured multiple FDA approvals over the years, reflecting its strong model generalization and real-time performance [367]. Other tools, such as ProFound AI® for breast cancer detection and HeartFlow FFRct for coronary disease prediction, underscore the importance of task-specific optimization and integration within existing diagnostic workflows [368]. Notably, newer tools like DermaSensor, approved in 2023 for real-time skin lesion analysis, illustrate a trend toward point-of-care AI that must meet both interpretability and latency requirements [369].

Figure 13: Model validation pipeline with key checkpoints
In contrast, tools like Lunit INSIGHT and Qure.ai, which focus on chest X-ray and CT analytics, showcase how cross-national collaborations and federated learning frameworks support regulatory approval even in data-restrictive environments [371,373]. Together, the model validation pipeline and the curated list of FDA-approved AI tools provide a roadmap for future developers. They underscore the necessity of robust, explainable, and regulation-compliant AI systems not just for academic excellence, but for tangible clinical impact. As regulators grow more stringent and clinicians demand transparency, aligning with these best practices is not optional, but it is imperative. Table 10 summarizes the FDA-Approved AI Tools in Radiology (2020–2024).
12 Deep Learning Application in Medical Image Analysis: Trends and Challenges
A notable outcome was achieved in the ImageNet Challenge, the well-known competition for image classification and segmentation, with the development of multiple CNN-based deep neural networks [374]. The main advantage of CNN compared to its predecessors is its ability to autonomously identify critical elements without human intervention [375]. Utilizing these performance metrics, we may assess the optimal CNN model [376]. The prevalent issues of imbalanced data, absence of confidence intervals, and insufficiently annotated data in recent deep learning literature pertaining to medical imaging can be identified as the primary challenges currently hindering the field’s comprehensive exploration of deep learning advancements. The quantity of samples and patients in the presently accessible public databases for medical imaging jobs is restricted, with the exception of a few datasets. Medical imaging datasets are significantly constrained in comparison to generic computer vision datasets, which typically comprise hundreds of thousands to millions of annotated images [377]. Conversely, there is an increasing trend within the medical imaging industry to adopt the end-to-end learning procedures of the broader pattern recognition community. Nevertheless, the broader community has generally met such operations due to the accessibility of extensive annotated datasets, which are essential for developing precise deep models [378]. Consequently, it remains uncertain how efficiently end-to-end certified models could execute medical image processing tasks without yielding overfitting on the training datasets. Several researchers have developed fundamental data enhancement techniques, including image flipping, padding, principal component analysis (PCA), image cropping, and adversarial training. Nonetheless, these algorithms are inferior to GANs in terms of dataset enhancement [379–383]. A significant impediment may be the utilization of black boxes; the legal implications of black-box functionality could dissuade healthcare providers from depending on it. Who could be deemed accountable if the result was adverse? Given the sensitivity of this region, the hospital may be reluctant to employ a black-box system that would enable tracking of individual results attributed to an optometrist [382]. Unlocking the black box is a significant study topic, and deep learning scientists are endeavouring to address it [383]. Moreover, the intricate nature of data structures renders the instruction of deep learning models a highly costly undertaking. They occasionally require high-performance GPUs and several computers, resulting in increased costs for users [384]. The heightened complexity of multiple layers requires substantial processing resources, thereby impairing training performance. Advanced activation functions, cost function structures, and dropout techniques have been employed to address vanishing gradient and overfitting challenges [385]. The challenge of substantial computing demand has been mitigated through the utilization of highly parallel hardware, including GPUs and batch normalization. Table 1 delineates deep learning architectures associated with their respective applications from the past to the present [386]. The establishment of an interdisciplinary data repository is facilitated by the abundance of electronic medical record data.
The shortages of high-quality labelled data are significant challenges for deep learning systems employed in medical image interpretation. DL models need more labelled data to obtain optimal performance and generalization. Acquiring high-quality annotations for medical images presents numerous challenges. The expense and time required to record and annotate medical images limit the volume of data from annotated images. The annotation procedure necessitates medical specialists with specialized training and expertise, who are not consistently accessible. Medical images are complex and very diverse due to alterations in patient anatomy, imaging modalities, and disease pathology. Annotating medical images necessitates a high precision level and uniformity, which could be difficult for complex and diverse medical problems. Privacy and ethical concerns: The annotating procedure may render medical images with sensitive patient information susceptible to misuse or illegal access. Medical image analysis faces considerable challenges in safeguarding patient privacy and confidentiality while maintaining the quality of annotated data. Annotating medical images necessitates subjective evaluations, potentially leading to bias and inconsistency in the annotations. These characteristics may influence the efficiency and generalizability of deep learning models, particularly when annotations are inconsistent among datasets or annotations.
To overcome the persistent challenge posed by the limited availability and access to high-quality annotated medical data, several strategic approaches have been proposed and are increasingly being adopted in deep learning workflows for medical image analysis [387]. One prominent solution is transfer learning, which leverages models pretrained on large, generic datasets and fine-tunes them on specific medical tasks. This approach not only enhances model performance but also reduces the volume of domain-specific annotated data required, thereby improving generalizability and accelerating development cycles. Another effective technique is data augmentation, where existing annotated images are synthetically altered through transformations such as rotation, scaling, or contrast enhancement, to create diverse and expanded datasets. This helps mitigate overfitting and improves the model’s resilience to variations in imaging modalities and acquisition settings. A third method, active learning, prioritizes the annotation of only the most uncertain or informative samples rather than the entire dataset. By iteratively selecting data points that the model is least confident about, this approach reduces the annotation burden while maximizing training efficiency and model performance. Lastly, collaborative annotation strategies wherein radiologists, clinicians, patients, and domain experts collectively contribute to the labeling process can improve the accuracy, clinical relevance, and consistency of the annotations. Such inclusive collaboration ensures that annotated datasets reflect real-world clinical needs and standards, thus enhancing the downstream utility of AI systems in healthcare settings. Collectively, these methods form a robust framework for mitigating data scarcity and strengthening the foundation for reliable AI development in medical imaging. It is essential to implement a combination of technical approaches to address the challenge of limited high-quality annotated data. This challenge is revisited in later sections to emphasize its recurring impact across multiple applications in medical imaging. Ethical and social strategies that enhance the quality, quantity, and diversity of annotated data while safeguarding patient privacy and upholding ethical standards. DL models for medical image analysis often face significant issues related to data quality, where annotation efforts are better directed toward the most uncertain or informative samples rather than the entire dataset. Such strategies provide a robust framework for mitigating data scarcity and strengthening the foundation for reliable AI development in medical imaging [388]. Acquiring medical images can be challenging, and their quality may fluctuate due to various factors, including the imaging equipment utilized, resolution, noise, artefacts, and the imaging methodology. Moreover, the annotations or labels employed for training might significantly influence the data quality. Annotations may lack accuracy and may be affected by inter- and intra-observer variability, thereby resulting in biased models with suboptimal generalisation performance. To address the issue of data quality, researchers must implement stringent quality control protocols for image collecting and annotation. Furthermore, they must create algorithms capable of processing noisy or low-quality data to enhance annotation performance. In the end, they must establish methodologies to assess the data quality utilized for training the DL models [389]. Interpretability presents a considerable barrier in medical image analysis with deep learning models, mainly because of the traditional black box characteristic of these models that complicates the understanding of the rationale behind their predictions. The absence of interpretability hinders clinical acceptability as healthcare practitioners require understanding and confidence in a model’s decision-making process to use it effectively. Besides, interpretability is vital for recognizing and addressing biases in the data and model, guaranteeing that judgments remain unaffected by extraneous or discriminatory attributes. Addressing class imbalance remains a persistent challenge in medical image analysis. One promising direction is the use of one-class classifiers, which focus on modeling a single target class and identifying deviations as anomalies. Such methods are particularly well suited for rare disease detection, where positive samples are scarce [390].
Several strategies have been explored to improve the interpretability of DL models for medical image analysis. These methods include saliency maps, model explanations, and visualization tools. However, because it requires a trade-off between interpretability and model performance, full interpretability is still a challenge in this discipline. Finding the ideal balance between these elements is still a work in progress. The capacity of a deep learning model that has been trained on one dataset to generalise and perform well on additional datasets with different properties is known as transferability. The diversity of medical imaging data, including differences in image quality, imaging methods, and imaging modalities, makes transferability a major difficulty in the context of medical image analysis. When DL models are trained on one dataset, they might not work properly on other datasets with diverse imaging properties and data quality. Since it is frequently impractical to train a model for each dataset. This may be an issue when creating DL models for medical image analysis. Researchers have looked into methods like domain adaptation and transfer learning to address this issue. Transfer learning can increase performance and decrease the amount of training needed by initializing the model weights for the novel dataset using a pretrained model on a different but similar dataset. Domain adaptation is the process of altering the model to take into consideration variations across the source and target domains, including variations in imaging modalities or protocols [391]. Research is still being done to create more reliable and transferable DL models for medical image analysis, as the transferability problem is still a major concern in this field.
In DL based medical image analysis, overfitting is a common problem when a model becomes excessively complex and closely aligns with the training data, resulting in inadequate generalization to novel and hidden datasets. Numerous problems, such as the presence of noise in the training data, an imbalanced class distribution, or insufficient training data, may result in overfitting [392]. The latter constitutes a common issue in medical imaging due to the limited dataset size resulting from the lack of annotated data. Overfitting may yield false positive or negative results, as it might achieve high performance on training data while exhibiting subpar performance on validation or testing data. To prevent overfitting in deep learning, many strategies can be employed, including registration, initial stopping, and data augmentation. In medical image analysis, maintaining data quality and augmenting the dataset size are crucial to mitigate overfitting. Clinical adoption denotes the integration of novel technology or methods into clinical practice. Clinical adoption of deep learning algorithms in medical image analysis poses a challenge, necessitating a substantial shift in the diagnostic and therapeutic approaches of physicians and healthcare professionals [393]. Clinical adoption encompasses technical aspects, including the integration of algorithms into current systems and processes with legal, ethical, and regulatory factors, as well as the training of healthcare personnel for utilizing novel technology efficiently and safely. A primary difficulty in clinical adoption is guaranteeing the accuracy and reliability of deep learning algorithms for clinical decision making. This necessitates thorough validation and testing of the models, along with resolving issues related to the interpretability and generalizability of the outcomes. Moreover, healthcare practitioners and patients may express apprehensions over the utilization of these models in medical decision-making, especially if the models are perceived as supplanting or lessening the position of the human clinician. A further hurdle to clinical adoption is the necessity for regulatory permission, especially when algorithms are used to assist in diagnosis or treatment decisions. The regulatory agencies, including the FDA, may mandate clinical trials to validate the safety and efficiency of the models prior to their implementation in clinical practice. The implementation of these methods may be impeded by this process due to its time-consuming and costly nature. The clinical adoption is a significant obstacle in the development and implementation of medical image analysis utilizing DL models, as it influences the overall effectiveness of the technologies on patient care.
13 Dataset for Medical Image Analysis Using Deep Learning Methods
Data constitutes the foundation of deep learning. In medical image analysis, a dataset comprises a compilation of medical images utilized for training machine learning models for the detection and classification of anomalies. The dataset may be acquired from diverse sources, including imaging studies, clinical trials, or public respiratory. The size and quality of the dataset significantly influence the performance of the machine learning model. Thus, a dataset must be diverse and representative of the target sample to guarantee the accuracy and generalizability of the models. Also, datasets may necessitate pre-processing, including augmentation or normalisation to rectify problems such as data imbalance, low contrast, or abnormalities. In the field of medical imaging, extensive medical image datasets, often comprising hundreds of images or more, are being developed for enhancing the training and evaluation of novel models. A significant example is the annual Medical Image Computing and Computer-Assisted Intervention (MICCAI) challenges, which provide benchmark datasets for various diseases, substantially advancing the field of medical imaging. Nevertheless, we must exercise caution regarding the potential biases inherent in relying solely on the single public dataset as the whole community tries to attain the best performance, which may result in community-wide overfitting on this dataset [394]. This issue has been acknowledged by numerous academics, making it customary to utilize various public and/or private datasets to evaluate a novel model’s performance more thoroughly. This approach reduces community-wide bias, yet it is not adequate for extensive therapeutic applications. A critical challenge in medical image analysis is the detection and utilization of extensive, well-curated medical image databases. Efforts are being made to enhance the quality and accessibility of medical imaging datasets for researchers to further the advancement of machine learning models for medical diagnosis and therapy. In medical image analysis employing machine learning models, a dataset comprises a compilation of images utilized for training and evaluating ML models. Acquiring and annotating medical images from diverse sources, such as clinics, hospitals, and research institutions are a conventional procedure in dataset formation [395]. The images must be annotated to define the regions of interests or attributes that the ML model is required to learn. These labels may offer information regarding the known disease, anatomy of the image region, or additional relevant facts. The training set and test set are formed when the dataset is established. The machine learning model is trained on the training set and evaluated on the test set. Thus, there is continuous study in medical image analysis focused on enhancing dataset quality and size, alongside the development of superior technologies for gathering and annotating medical images. The most commonly used programming languages for learning algorithms in medical image analysis are demonstrated in Fig. 14.
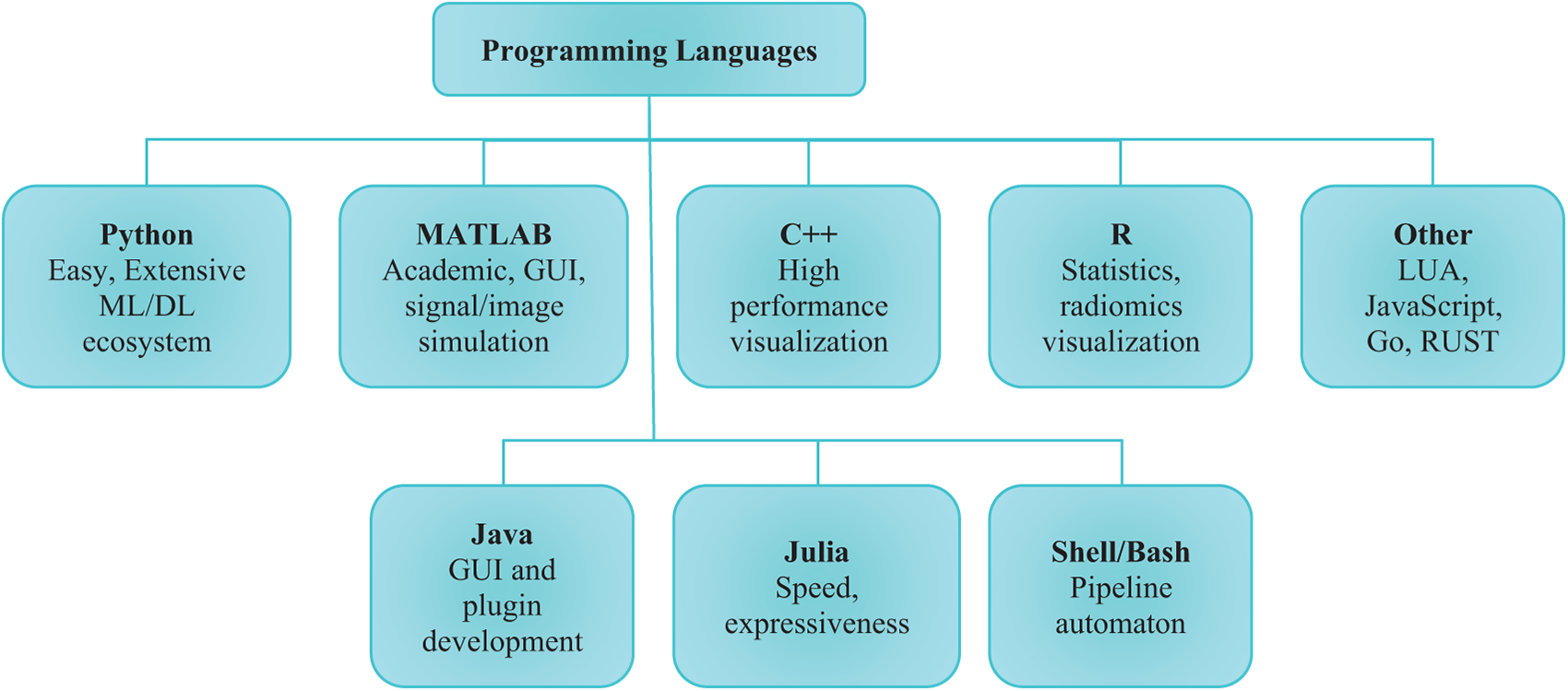
Figure 14: Programming languages used for medical image analysis
14 Security Concerns, Obstacles, and Hazards
Deep learning (DL) models for medical image analysis raise significant security, privacy, and ethical concerns that must be thoroughly addressed to ensure safe and trustworthy deployment in clinical environments. Medical images used to train DL models often contain sensitive patient data, including demographic details, medical histories, and personally identifiable information (PII), which necessitates strict privacy safeguards. A primary concern is the risk of dataset breaches during data collection, storage, or transfer, where malicious actors might exploit vulnerabilities in hardware or software infrastructure to access or intercept data [396]. Therefore, robust security mechanisms such as data encryption, stringent access control, and continuous monitoring systems must be deployed to safeguard data integrity and confidentiality. Another pressing issue is the susceptibility of DL models to adversarial attacks, where attackers deliberately input manipulated data to deceive models into making incorrect predictions, potentially compromising patient diagnosis and treatment outcomes. This underlines the critical need for designing DL models that are inherently secure and resistant to such threats. Moreover, several broader challenges accompany DL deployment in healthcare, including inaccuracy, where models may yield incorrect results leading to flawed clinical decisions; bias, which can result in unequal or erroneous outcomes for certain patient demographics; and lack of transparency, as many DL systems function as “black boxes,” limiting interpretability and reducing trust in their recommendations. Additionally, privacy concerns emerge from the risk of exposing sensitive patient information during the analytical process, and cybersecurity risks stem from potential attacks on network-connected medical systems. To mitigate these risks, it is vital to adopt robust cybersecurity strategies, enforce strict privacy regulations, and validate the models for fairness, accuracy, and transparency. Furthermore, the integration of the Internet of Things (IoT) in medical image analysis through connected imaging modalities such as MRI, CT, and ultrasound enables real-time data collection and cloud-based processing, significantly enhancing remote diagnosis and collaborative consultations. However, IoT connectivity introduces additional vulnerabilities, as sensitive patient data transmitted and stored over networks becomes susceptible to cyber threats. Consequently, securing IoT-based imaging workflows requires holistic strategies that ensure the confidentiality, availability, and integrity of both data and models throughout the digital healthcare ecosystem.
Several unresolved challenges associated with medical image analysis using DL systems. These involve:
Data privacy is a significant issue in medical image analysis employing DL systems. Medical images include sensitive patient info that must remain discreet and safe. To safeguard patient data from unauthorized access, utilization, or disclosure, any model or system employed for medical image analysis must adhere to this principle. The analysis of medical images can be particularly challenging due to the substantial volumes of data involved, increasing the risk of data breaches or unauthorized access. One of the principal challenges in data privacy within medical image analysis is achieving a balance between the requirements for data access and the safeguarding of patient privacy. Numerous medical image analysis methods depend on extensive datasets to attain elevated accuracy and performance, necessitating data sharing among various entities. This poses significant challenges when managing sensitive patient information due to the potential for data loss or misuse. This involves regulations and procedures for ensuring that data is accessed and utilized solely for lawful purposes, data anonymization, encryption, and access limitations. Besides, to ensure the protection and correct management of patient data, healthcare organizations must comply with relevant data privacy regulations.
Data bias is a significant unresolved issue when using deep learning systems to interpret medical images. It refers to the systematic inadequacies in the data utilized to train the DL models [397]. A number of factors, such as the training data selection, labelling, and data representativeness of the population of interest, might lead to these errors. Models that underperform on specific population segments, including members of underrepresented groups or individuals with uncommon medical conditions, can be produced as a result of data bias. If the models are utilized in clinical decision-making, this could have detrimental effects on patient safety as well as the precision and equity of medical image analysis systems. To guarantee that the models are representative and unbiased, addressing data bias necessitates giving careful thought to the data sources, data labelling, and model training methods.
15.3 Limited Availability of Annotated Data
DL methods in medical image analysis require substantial annotated data for efficient training. The annotated data denotes medical images that have been classified by specialists to specify the position and nature of anomalies, including tumours, lesions, or other diseases. Acquiring annotated medical image datasets is notably difficult due to various variables. Initially, annotating medical images necessitates considerable time and a profound comprehension. Only seasoned radiologists or doctors possess the expertise to effectively detect and annotate problems in medical images, hence limiting the availability of annotated data. Besides, there are privacy issues related to medical imaging data. Patient confidentiality is also an important issue in healthcare, and medical imaging data is seen as particularly sensitive. Subsequently, acquiring extensive annotated medical image datasets for deep learning is difficult due to privacy issues and the necessity to adhere to standards like HIPAA. The heterogeneity of medical imaging data can be an obstacle. Medical images exhibit significant variability in modality, acquisition techniques, and image quality, complicating the creation of extensive, diverse datasets for DL. Deep learning models for medical image analysis may face limitations in development and validation due to the difficulties in obtaining annotated medical image datasets. To reduce the quantity of labelled data required for training, researchers have addressed this challenge employing practices such as transfer learning, data augmentation, and semi-supervised learning [398]. Nevertheless, these practices may prove inadequate in certain instances, necessitating the availability of more annotated medical image datasets for academics to further the domain of medical image analysis.
15.4 Transparency and Interpretability
Transparency and interpretability are important considerations when using DL systems for medical image analysis. Because DL models can be hard to read and understand how they make decisions, they are sometimes referred to as black boxes. Interpretability is crucial in medical image analysis so that doctors can recognize possible biases or errors and comprehend and trust the algorithms. The ability to understand the logic underlying a model’s decision-making process is known as interpretability. Because of this intricacy, it may be challenging for physicians who are unfamiliar with deep learning to understand how the model came to specific conclusions. The capacity to look into the model and comprehend how its functions is referred to as transparency [399]. Put differently, transparency indicates that the model’s decision-making procedure is transparent, comprehensible, and amenable to auditing and validation. To make sure the model is operating accurately and not introducing biases or errors, transparency is crucial. Because doctors must comprehend how the algorithm arrived at its conclusions, interpretability and openness are essential in medical image analysis. The clinicians can use this knowledge to detect biases or mistakes and make sure the algorithm is generating choices that align with clinical practice. Several methods have been developed to improve the interpretability and transparency of DL models in medical image analysis. For example, visualization techniques can be used to create heatmaps that show which regions of an image the model is using to form conclusions. Attention processes can also be utilized to describe the model’s decision-making process and draw attention to key aspects of an image. Additional strategies include integrating domain knowledge into the models and applying explainable AI (XAI) approaches. These methods have demonstrated potential, but in order to increase their usefulness in clinical practice, more transparent and interpretable DL models in medical image processing are still required.
The major unresolved problem in DL based medical image analysis is generalizability. The ability of a model to perform efficiently on data that differs from its training data is termed generalizability. A trained model must generalize to other datasets while maintaining strong performance. Generalizability is vital since it guarantees that DL models may be used for novel patient groups or in diverse clinical environments. Nonetheless, DL algorithms may be susceptible to overfitting, a phenomenon where a model excels on its training data yet underperforms on novel data. This poses significant challenges in medical image processing, as an overfitting model may result in erroneous or inconsistent diagnoses. The generalizability of DL models in medical image processing may fluctuate based on several factors. The diversity of the training dataset can substantially influence a model’s ability to generalize [400]. The model may fail to recognize previously unseen anomalies if the training dataset lacks sufficient diversity. The model’s performance on various types of medical images is another element that can influence generalizability. A model trained on CT scans may exhibit suboptimal performance on MRI scans due to the differences in image modality and acquisition techniques. Researchers are investigating techniques such as transfer learning, domain adaptation, and data augmentation to enhance the generalizability of DL models in medical image analysis. Transfer learning involves refining a pretrained model by utilizing a new dataset as the initial reference. Data augmentation involves employing modifications like translations to artificially increase the size and diversity of the training datasets. Domain adaptation involves adjusting a model trained on one dataset to operate effectively on another dataset with diverse characteristics. The generalizability of DL models in medical image processing must be enhanced to ensure their safe and efficient deployment in clinical practice, notwithstanding their demonstrated potential.
15.6 Validation and Regulatory Authorization
Validation and regulatory approval are critical unresolved difficulties in the application of DL algorithms for medical image processing. Validation denotes the procedure of confirming that a model is precise and dependable. Regulatory approval denotes the procedure of securing authorization from regulatory agencies, such as the FDA in the United States, prior to the implementation of a model in clinical practice. Validation is important in medical image analysis as erroneous or unreliable models may result in wrong diagnosis and treatment planning. Validation entails assessing the model using an independent dataset that was excluded from the training process and evaluating its performance across various parameters. Validation may also entail juxtaposing the model’s performance with that of human experts. Regulatory approval is crucial in medical image analysis to guarantee that the models are safe and accurate for clinical use. Regulatory authorities necessitate proof of the model’s safety and performance prior to its approval for utilization. This evidence may include clinical trials. Real-world data analyses and further validation methods. Numerous issues exist regarding the validation and regulatory approval of DL models in medical image processing. A significant difficulty is the absence of defined validation techniques, complicating the comparison of various models’ performance. A further difficulty is the absence of transparency and interpretability in DL models, complicating the validation of their performance and the assurance of their safety and efficiency. Researchers and regulatory bodies are working together to establish uniform validation processes and criteria for the regulatory approval of DL models in medical image analysis to address these challenges. The FDA has issued recommendations for the development and approval of medical devices utilizing AI/ML. These guidelines offer advice for the design and validation of AI/ML-based medical devices, particularly those utilized for medical image analysis. Despite the promising nature of these initiatives, further study and coordination between academics and regulatory authorities is essential to guarantee the safe and successful application of DL models in medical image analysis.
15.7 Ethical and Legal Implications
DL models for medical image processing provide several critical unresolved issues regarding ethical and legal dilemmas [387]. These factors pertain to the utilization of patient data in research and the potential for models’ biases, and the obligation of researchers and healthcare practitioners to ensure the ethical and secure implementation of these technologies. The utilization of patient data in research constitutes an ethical concern. Extensive quantities of patient data are requisite for medical image processing, and the utilization of this data prompts inquiries around patient privacy and consent. Patient privacy must be preserved, and researchers and healthcare practitioners must ensure the responsible use of patient data [401]. The potential for bias in models represents another ethical concern. DL models can be trained on biased datasets, potentially resulting in distorted model outputs. Biases may lead to erroneous diagnosis and treatment planning in medical image processing, potentially resulting in dire consequences. Researchers must act to alleviate any potential biases in their datasets and models. DL models for medical image analogues analysis provide legal challenges with intellectual property, liability, and regulatory compliance. There are concerns over the potential for unauthorized access to patient data and the necessity to comply with data protection standards for maintaining patient privacy. To tackle these ethical and legal issues, researchers and healthcare practitioners should adhere to best practices for data protection and security, secure informed consent from patients and strive to minimize any biases in their models. Engaging with stakeholders such as patients, regulatory authorities, and legal professionals is vital to guarantee that the development and application of these technologies are safe, ethical, and consistent with applicable laws and regulations.
16.1 Moving toward More Effective Deep Learning and Medical Image Analysis Pairings
16.1.1 From a Task Specific Perspective
Advancements of DL based medical image analysis follow a remarkably robust but lagging trajectory for computer vision. Nevertheless, a direct application of computer vision techniques might not produce adequate results since medical images differ from natural images. It is necessary to solve the difficulties specific to medical imaging tasks to attain high performance. The secret to success in the classification process is to extract highly discriminative features pertaining to specific classes. This could be challenging for domains with high inter-class similarity, but it is rather simple for domains with considerable interclass variance. Since it is challenging to capture discriminative signals for breast tumours in the presence of overlapping, diverse fibroglandular tissues, the overall performance of mammography classification is not very excellent [402]. Given significant inter-class similarity, the idea of fine-grained visual categorization (FVGC) [403], which seeks to detect minute variations between visually similar items, may be appropriate for learning distinguishing characteristics. However, keep in mind that benchmark FVGC datasets are specifically gathered to ensure that every image sample consistently displays significant inter-class similarity. Because just a portion of the images in medical datasets show high inter-class similarity, methods created and tested on the datasets may not be easily transferable to medical datasets. However, we think FVGC techniques will be useful for learning features with strong discriminative power in medical image classification if they are properly adjusted. The usage of attention modules, local and global features, global knowledge, etc., is another potential strategy to improve the discrimination power of features.
The medical object detection is more complex than classification, as demonstrated by the process of bounding box prediction. Detection encounters the obstacles intrinsic to classification. In addition, there are further obstacles, particularly the detection of small-scale objects and class imbalance. One stage detector often exhibits comparable efficacy to two-stage detectors in identifying large things, although they encounter greater difficulties in detecting small objects. Recent research indicates that the utilization of multi-scale features significantly mitigates this problem in both one stage and two stage detectors. A simple yet efficient method is featured image pyramids [404], wherein features are independently retrieved from several scales of a similar image. The technique can augment diminutive items to enhance performance, although it is computationally intensive and sluggish. A more efficient method features pyramids that employ multiscale feature maps derived from various convolutional layers. While other methods for constructing feature pyramids are available, a general guideline is to integrate robust, high-level semantics with high-resolution feature maps. The class imbalance occurs when detectors must assess a vast number of candidate regions, whereas only a limited number contain relevant objects. The class distribution is significantly biased towards negative instances, mostly comprising easily identifiable negative examples such as backdrop regions. The abundance of easily obtainable negatives might inundate the training process, resulting in poor detection outcomes. Two-stage detectors effectively address the class imbalance problem more proficiently than one-stage detectors, as the majority of negative proposals are eliminated during the region proposal phase. Recent studies indicate that eliminating the predominant reliance on anchor boxes in one-stage detectors can significantly mitigate class imbalance [214]. Nonetheless, the majority of methodologies employed in medical image detection remain anchor-based. In the imminent future, we expect increases in investigations into anchor-free, one stage detectors for medical image detection.
Medical image segmentation integrates issues in classification and detection. Similar to detection, class imbalance is a prevalent challenge in both 2D and 3D medical image segmentation tasks. Another comparable problem is the segmentation of small lesions and organs. Moreover, these two difficulties are frequently evident as interconnected. These concerns have been predominantly alleviated by modifying metrics and losses to assess segmentation performance, such as the Dice coefficient [148], generalised Dice [405], and the incorporation of focused loss [406], among others. Nonetheless, these measures are region-specific. This may result in the loss of clinical info relating to structures, forms, and contours essential for diagnosis and prognosis in subsequent phases. Thus, we assert the necessity of developing non-region-based measures to furnish supplementary information alongside region-based metrics for enhanced segmentation performance. At present, only a limited number of studies are available in this region [407]. We anticipate observing an increase in the future. Besides, techniques that integrate local and global context, attention mechanisms, multiscale features, and anatomical signals typically enhance segmentation accuracy for both large and small objects. Despite the utility of long-range dependencies in attaining precise segmentation, most CNN-based approaches do not explicitly emphasize this factor. There are approximately two categories of dependencies: intra-slice reliance and inter-slice dependencies [408]. Recent research indicates that transformer-based methodologies are effective in both scenarios [157,163]. The use of vision transformers for medical image segmentation, particularly in three dimensions, remains in its nascent phase, with further developments in this area anticipated shortly. Medical image registration differs from prior tasks as its objective is to establish pixel-wise or voxel-wise correspondence between two images. A distinctive obstacle pertains to the difficulty of obtaining dependable ground truth registrations that are either synthetically derived from traditional registration techniques. Unsupervised techniques are considerable potential in addressing this problem. Nonetheless, numerous unsupervised registration frameworks (e.g., [297]) consist of multiple phases for registering images in a coarse-to-fine approach. Notwithstanding commendable performance, multistage frameworks can elevate computing complexity and complicate the training process. It is preferable to create registration frameworks that consist of minimal steps and may be trained in an end-to-end manner.
16.1.2 On the Viewpoint of Several Learning Paradigms
Despite the significant developments achieved by deep learning in radiological image processing, continual performance enhancement is mostly hindered by the necessity for extensive levelled datasets. Supervised transfer learning can significantly remove this problem by initializing the model’s weights for the target task with those of a model pretrained on pertinent or non-pertinent datasets (e.g., ImageNet). In addition to the prevalent application of transfer learning, two other avenues exist: (a) employing GAN models to augment the labelled dataset, (b) leveraging self-supervised and semi-supervised learning models to extract info from extensive unlabelled medical images. GANs have demonstrated significant potential in medical image synthesis and semi-supervised learning; nonetheless, an obstacle remains in establishing a robust linkage between the GANs generator and the target. The absence of this connection may result in a slight performance enhancement relative to traditional data augmentation technique [112]. The relationship between the generator and classifier can be enhanced by employing a semi-supervised GAN, wherein the discriminator is adapted to function as a classifier [70]. Numerous training procedures may be utilized to investigate a poor generator that can substantially enhance effective semi-supervised classification [409]; simultaneously optimising the three components of the generator, a discriminator, and a classifier. Exploring novel methods to establish links between the generator and a specific medical imaging task is essential for enhanced performance. Besides, GAN typically requires a minimum of thousands of training instances to achieve convergence, hence constraining its utility in small medical datasets. This problem could be partially alleviated with the application of traditional data augmentation techniques for adversarial learning [111]. Besides, if substantial quantities of medical images exhibit textural, structural, and semantic similarities with the target datasets, pretraining generators/ discriminators may enhance convergence speed and overall performance [120]. Recent novel augmentation mechanisms, including differentiable augmentation and adaptive discriminator augmentation [410] have empowered GANs to generate high-fidelity images in data-scarce environments; however, these techniques have yet to be utilized in medical image analysis tasks. We expect that these novel strategies will exhibit promising efficacy in forthcoming research within the medical image analysis domain.
Self-supervision can be established by pretext tasks or contrastive learning; however, the latter appears to be a more promising path for research. This is due to the fact that directly employing pretext tasks from computer vision is generally insufficient for guaranteeing the acquisition of robust feature representations for radiological images. Conversely, creating novel pretext activities can be challenging, requiring meticulous manipulation. Self-supervised contrastive learning, rather than employing diverse pretext tasks, instructs the network to acquire significant features by ensuring invariance to various augmented views, potentially surpassing supervised transfer in multiple downstream tasks, including classification and segmentation. Notwithstanding the promising efficiency of self-supervised contrastive learning, its implementation in radiological image processing remains in the exploratory phase, and determining the optimal application of this novel learning paradigm presents a significant challenge. To realize its potential, we present our recommendations from the following three perspectives. Using the advantages of contrastive learning and supervised learning. Analysis of existing studies reveals that most employ a two-step approach for medical image analysis: contrastive pretraining on unlabelled data followed by supervised fine-tuning with labelled data. During the pretraining phase, the majority of research depends on substantial, unlabelled datasets to facilitate the acquisition of high-quality, transferable features, which can result in enhanced performance when subsequently fine-tuned using a limited amount of labelled data. Nonetheless, dependence on extensive unlabelled data may provide challenges in projects that do not possess substantial quantities of such data.
To expand the application scope, it would be advantageous to get high-quality feature representations with less unlabelled data. A potential strategy involves consolidating the aforementioned two distinct phases into a single process to utilize the label information in contrastive learning. This resembles semi-supervised learning, which concurrently employs both unlabelled and labelled data for enhancing performance. Class labels can effectively facilitate the construction of positive and negative pairs by aligning images from the same class more closely in the lower-dimensional representation phase [411]. Features acquired with this method should necessitate less unlabelled input and exhibit reduced redundancy compared to features obtained only via self-supervised learning (i.e., in the absence of class labels). Consider specific attributes of contrastive learning to enhance performance. One study demonstrates that contrastive learning is highly advantageous when utilizing large clusters of comparable points rather than pairs [412]. This heuristic may be particularly effective for acquiring transferable features from 3D CT and MRI volumes that display consecutive anatomical similarities. Tailor data augmentation procedures for downstream tasks are sensitive to such modifications. The formulation of various data augmentation techniques is essential for acquiring representative features in the majority of current contrastive learning frameworks. SimCLR employs three transformation techniques on unlabelled images: random cropping, colour distortions, and Gaussian blur [57]. Nevertheless, several often-employed augmentation procedures may be unsuitable for medical images. In radiography, when the majority of images are displayed in grayscale, the colour distortion technique is probably inappropriate. Moreover, in instances when the fine-grained characteristics of unlabelled medical images contain significant info, the application of a Gaussian blur may compromise the detailed information and reduce the quality of features during the pretraining phase. Consequently, selecting suitable data augmentation techniques is crucial for achieving adequate downstream performance. Besides, self-supervised contrastive pretraining is hindered by the substantial computational complexity of big models, necessitating a significant range of multicore Tensor Processing Units (TPUs) [57]. Consequently, it is imperative to pursue the development of novel models or training procedures to improve computational efficiency. For instance, reference [413] introduced a hierarchical pretraining technique that accelerated the self-supervised pretraining process by up to 80 times while enhancing accuracy across various tasks.
Similar to self-supervised contrastive learning, recent semi-supervised techniques like FixMatch (A Semi-Supervised Learning Framework Combining Consistency Regularization and Pseudo-Labeling) [45] significantly depend on sophisticated data augmentation tactics to get optimal performance. To effectively apply semi-supervised learning in medical image analysis, it is crucial to develop suitable enhancement strategies tailored through either a dataset-driven or task-driven approach. A dataset-driven strategy focuses on identifying the optimal augmentation policy specific to the dataset in question. Traditionally, achieving this has been challenging due to the extensive parameter search space involved. Recently, automated data augmentation techniques [414] have been introduced to considerably decrease the search space. But the notion of automated enhancement is predominantly unexamined in the field of medical image analysis. Being task-driven entails detecting appropriate augmentation algorithms for a particular task that includes several datasets. This may be considered an extension of dataset-driven augmentation, rendering it more complexes however yet it can enhance the generalization of algorithms created on one dataset to other datasets pertaining to the same task. Another critical concern is the potential decline in model performance arising from the violation of a core assumption in semi-supervised learning, namely, that labelled and unlabeled data are collected from the same underlying distribution. Distribution mismatch is a common challenge when applying semi-supervised algorithms to medical image analysis. During the segmentation of COVID-19 lung infections from CT slices, the labelled dataset typically comprises CT volumes with a relatively balanced representation of infected and non-infected slices. However, the available unlabelled CT volumes may be less available. Moreover, these unlabelled images may include cases of other pulmonary diseases not represented in the annotated dataset. This discrepancy introduces a distribution mismatch between the labelled and unlabelled data, which can adversely affect the performance of semi-supervised learning models. Current research indicates that this will significantly impair the performance of semi-supervised approaches, occasionally resulting in outcomes inferior to those of a basic supervised baseline [415,416]. Thus, it is essential for modifying semi-supervised models to accommodate the distribution discrepancy between labelled and unlabelled medical data. The field of domain adaptation may offer valuable insights to achieve this objective.
16.1.3 Architecture Search and Pipeline Optimisation
Recent success of deep learning in medical image analysis can be attributed not only to the diversity of learning paradigms but, perhaps more significantly to the continuous evolution of architectures and models developed over time. Upon reflection, we observe that significant advancements are intricately related to the evolution of architectures with notable instances being AlexNet [94], residual connections [97], skip connections [143], and self-attention [417]. Considering this historical progression, it is indeed plausible that an improved neural architecture could independently address numerous existing limitations as noted by [418]. We examine two factors that may assist in identifying superior architectures. Biologically and cognitively inspired mechanisms will remain significant in architectural design. DL neural networks are initially inspired by the structure of the cerebral cortex. In recent years, the idea of attention, derived from monkeys’ visual attention mechanisms, has been effectively employed in natural language processing and computer vision to enable models to concentrate on significant aspects of input data, resulting in enhanced performance. Transformer-based designs excel in capturing global and long-term relationships between the input and the output sequence compared to mainstream CNN-based models. Also, the inductive biases inherent to CNNs, such as translation equivalence and localization are significantly reduced in [417]. In addition to attention mechanisms, several biological or cognitive processes, like dynamic hierarchies in human language and one-shot acquisition of novel objects and concepts without gradient descent [419] may inspire the development of more robust designs. Secondly, autonomous architecture engineering may illuminate the creation of superior architectures. The majority of the architectures in use today are created by human professionals, and the design process is error-prone and iterative. This is one of the reasons why medical image analysis models are mostly derived from computer vision models. Neural architecture search (NAS) is a similar field that researchers have proposed to automate architecture engineering in order to eliminate the requirement for manual designing [420]. Nevertheless, the majority of NAS research to now has been limited to image categorization [421], and no genuinely ground-breaking models capable of bringing about significant improvements have emerged from this procedure. However, NAS remains an area worth investigating. Pipelines with automatic configuration features would be ideal on a larger scale. Radiological image analysis will benefit from the development of automatic pipelines that can automatically configure its subcomponents to obtain better performance, even though architectural engineering still faces many challenges. These days, deep learning-based pipelines usually comprise a number of interdependent subcomponents, including data augmentation techniques, network architecture adaptation and training, image pre-processing and post-processing, and the selection of suitable losses. However, there are frequently too many design options for experimenters to manually determine the best pipeline. Furthermore, a pipeline that performs well when set up for one dataset of a particular task might not perform well when applied to another dataset of a similar task. Thus, to speed up empirical design, pipelines that can automatically set their subcomponents are required. Examples that fall under this category include U-Net [186], which is designed especially for medical image segmentation, and Nifty Network (NiftyNet) [422], which is a modular pipeline for various medical applications. We anticipate that this track will lead to additional study.
16.1.4 Integrating Domain Expertise
Domain expertise, a crucial yet often neglected element, can yield insights for the development of high-performing DL models in medical image processing. As earlier stated, most of the models employed in medical imaging are modifications of those created for natural images; yet medical images present greater complexities due to different challenges. When utilized effectively, domain expertise alleviates these problems with reduced time and computational expenses. Researchers with a robust background in deep learning can relatively easily leverage limited domain expertise, including patient metadata [88], anatomical information in CT and MRI images [89], data from similar patients [122], radiomic features and associated text reports. On the other hand, we note that it may be more challenging to successfully integrate the robust subject knowledge with which radiologists are acquainted. An instance is the detection of breast cancer by mammography. Each patient has four mammograms, comprising two CC (Cranio-Caudal) and two Mediolateral Oblique images for both the left and the right breast. In clinical practice, bilateral differences and unilateral correspondences are critical indicators for radiologists to identify troublesome areas and assess malignancy. Subsequently, additional research endeavours are required for optimizing the application of robust domain expertise.
16.2 Wide-Ranging Uses of Deep Learning in Clinical Environments
Despite its extensive application in the analysis of medical images within academic and industrial research settings, deep learning has not achieved the anticipated impact in clinical practice. This is plainly evident in the initial phases of combating COVID-19, the inaugural worldwide pandemic occurring within the era of DL. During the pandemic, owing to its extensive medical, social, and economic ramifications, it may mostly be viewed as a significant evaluation of the present state of DL models in clinical translation. Soon after the epidemic, researchers globally employed DL methodologies for analysing mostly chest X-rays and CT scans from patients with probable infections, with the objective of achieving precise and effective diagnoses and prognosis of the disease. Thus, many methodologies based on DL and machine learning have been developed. Wynants et al. [423] observed all these models exhibited a high or unclear risk of bias, rendering them unsuitable for clinical application. Although each model reported moderate to excellent performance, the optimistic results were significantly skewed due to issues such as model overfitting, inappropriate evaluation methods, and the use of inadequate data sources. Beyond the COVID-19 example, the high-risk bias inherent in deep learning methodologies is a persistent problem across many medical image analysis tasks and applications [424] which significantly limits the potential of deep learning in clinical radiography. While quantifying the inherent bias is challenging, it can be mitigated if managed correctly.
Future research in the rapidly evolving domain of medical image analysis using deep learning models holds significant potential to enhance the accuracy and efficiency of medical diagnosis and treatment. These fields involve:
17.1 Multimodal Image Analysis
Multimodal image analysis will be the main focus of future studies in DL models for medical image analysis. A more complete image of a patient’s anatomy and condition could be retrieved by using a range of imaging modalities, including a CT, MRI, PET, ultrasound, and optical imaging. This approach can help improve diagnostic accuracy and reduce the likelihood of inaccurate or missed diagnoses. Deep learning methods for a variety of tasks, such as segmentation, classification, registration, denoising, super resolution, a detection can be trained using multi-modal image data. For example, an algorithm developed using PET and MRI data may be utilised to locate the regions of the brain affected by Alzheimer’s disease. In a similar manner, liver cancers could be detected by training a deep learning system using CT and ultrasound data. Deep learning systems face a number of difficulties when analysing multimodal images. For instance, the algorithm’s performance may be impacted by the resolution, noise, and contrast characteristics of various imaging modalities. Furthermore, compared to single-modality data, multimodal data might be more complicated and challenging to analyse, requiring the use of more sophisticated algorithms and computer resources [425]. Researchers are developing novel deep learning models and algorithms that can combine and evaluate data from several modalities to overcome these difficulties. For instance, input from many imaging modalities can be combined using multi-modal fusion networks, and the algorithm’s attention can be directed onto pertinent characteristics in each modality using attention mechanisms. All things considered, multi-modal image analysis has the potential to increase the precision and effectiveness of deep learning algorithms used in medical diagnosis and therapy. It will be important to make sure that these technologies are being used responsibly, safely, and in compliance with all applicable rules and regulations as they develop further.
Explainable AI (XAI) will be the main focus of future studies on DL models for medical image analysis. The ability of an AI system to communicate its decision-making process in a form that a human could understand is known as XAI [426]. When used in the context of medical image analysis, XAI can help to boost trust in deep learning algorithms, ensure their safe and ethical use, and enable clinicians to make more well-versed decisions based on the algorithms’ output. In medical image analysis, XAI entails creating algorithms that are able to segment images or make correct predictions while also offering understandable justifications for their choices. This can be especially crucial when the AI system’s results conflict with or diverge from the clinician’s evaluation or past understanding. Making heatmaps or visual explanations that highlight the regions of an image that are most vital to the algorithm’s decision-making process is one method of using XAI in medical image analysis. These justifications can be useful in locating areas of interest, spotting small anomalies, and shedding light on the algorithm’s mental processes [427]. Using prior information or outside knowledge to inform the algorithm’s decision-making is another method of using XAI in medical image analysis. The algorithm that evaluates brain MRIs, for instance, might be developed to take into account anatomical landmarks or recognized patterns of illness progression. All things considered, XAI has the potential for enhancing the openness, interpretability, and reliability of DL algorithms in medical image analysis. It will be crucial to make sure that these technologies are being used responsibly, safely, and in compliance with all applicable rules and regulations as they develop further [428].
Transfer learning will be the main emphasis of upcoming research in deep learning-based medical image processing. Transfer learning refers to the utilization of pretrained deep learning models to enhance a model’s efficiency on a novel task or dataset. Transfer learning is particularly advantageous in the interpretation of medical images as it eliminates the necessity for extensive labelled data, which can be challenging and labour-intensive to gather. By leveraging the knowledge and representations acquired by pretrained models from extensive datasets, researchers can enhance the accuracy and effectiveness of their models. Transfer learning can significantly aid in the interpretation of medical images by reducing the necessity for extensive labelled data, which is sometimes difficult and time-consuming to obtain. Researchers can enhance the precision and efficiency of their models by utilizing the representations and data amassed by pretrained models that have been trained on extensive datasets [429]. The pretrained model facilitates learning from limited data and may alleviate the risk of overfitting, making it an advantageous starting point for the medical image analysis task. Besides, transfer learning may enhance the generalizability of deep learning models in the interpretation of medical images. Utilizing pretrained models that have acquired representations of real images, medical image analysis models can potentially yield more precise and generalizable representations of medical images relevant to a broader spectrum of tasks and datasets. Transfer learning may enhance the efficiency, accuracy, and applicability of deep learning models utilized for medical image interpretation. Ensuring the responsible, safe, and compliant utilization of these technologies will be essential as they continue to grow.
Federated learning will be the main focus of future studies on DL models for medical image analysis. Federated learning is the process of training machine learning algorithms on data that is spread across multiple devices or institutions without requiring the data to be moved to a central server. Since federated learning allows institutions to share knowledge and experience while protecting the privacy and security of sensitive patient data, it can be especially useful in the context of medical image analysis. This can be especially important in cases where patient data is governed by strict privacy laws such as HIPAA in the US. In order for federated learning to function, a central ML model is trained using a set of initial weights that are subsequently transmitted to all participating devices or institutions. Then, using the original weights as a starting point, each device or organization trains the model on its own local data. The central server then receives the new weights from every institution or device, which are combined for updating the central model. Iteratively, this approach is carried out until the model coverages. Medical institutions can increase the accuracy and generalizability of their models by utilizing the combined knowledge and experience of several institutions through the use of federated learning for model training. Additionally, because the data remains on local devices or organisations, patient privacy and confidentiality are maintained. In the domain of medical image analysis, federated learning generally exhibits promise for improving the generalizability, speed, and privacy of deep learning models. It will be crucial to make certain that these technologies are being utilised responsibly, safely, and in compliance with all applicable rules and regulations as they develop further.
17.5 EHR (Electronic Health Record) Integration
Integrations with EHRs will be the main focus of future developments in deep learning models for medical image analysis. Numerous clinical data, such as patient demographics, medical history, test results, and imaging studies, are included in EHRs. By combining DL models with EHRs, researchers and clinicians may be able to improve the accuracy and efficiency of medical image analysis. Using patient-specific data from EHRs to enhance the interpretation of medical images is one possible use for this integration. For instance, using a patient’s clinical history, test results, and imaging investigations, deep learning models can be trained to forecast the probability of specific diseases or ailments. This could improve the accuracy of medical image analysis and reduce the need for intrusive or expensive diagnostic procedures. Another potential use is to automatically extract data from medical images using deep learning models and integrate it into electronic health records [430]. Deep learning algorithms, for instance, might be trained to automatically detect and quantify cancers or lesions in medical imaging and enter this data into the patient’s electronic health record. This could improve the accuracy of medical image interpretation and reduce the need for intrusive or expensive diagnostic procedures. Another potential application is the automatic extraction of data from medical images using deep learning algorithms, which may then be integrated into electronic health records. This could enhance the efficiency and accuracy of clinical decision-making while also reducing the burden on doctors. Overall, there is a premise for enhancing the accuracy, effectiveness, and efficiency of medical image processing through the integration of deep learning models with EHRs. As these technologies develop, it will be critical to ensure their use is ethical, safe, and compliant with all relevant laws and relating to data security and patient privacy [431].
Future studies on deep learning models for medical image analysis should explore few-shot learning. This method has a lot of ability when labelled data is few or hard to come by, as is often the case in medical imaging. It will be crucial to look into methods that allow models to pick up knowledge from a limited number of annotated instances. The potential of meta learning techniques that train models to swiftly adapt to novel tasks with less data. Besides, techniques for data synthesis and augmentation that are especially suited for few-shot situations could be created. We can greatly expand the range of applications, making AI-driven healthcare solutions more accessible, and eventually raise the standard of patient care by developing few-shot learning in the context of medical imaging [432].
17.7 Federated Learning (FL) and Edge AI
Federated learning (FL) is emerging as a critical approach for collaborative medical image analysis, enabling institutions to train shared models without exchanging raw data and ensuring compliance with privacy regulations such as HIPAA and GDPR. By leveraging diverse, distributed datasets, FL improves model generalizability and reduces bias, supporting clinically robust AI development [433,434]. Complementing this, privacy-preserving AI methods such as differential privacy, secure multi-party computation, and homomorphic encryption strengthen data security while enabling safe cross-institutional collaboration [435]. In parallel, edge AI reduces reliance on cloud infrastructure by enabling real-time inference on local devices and imaging systems, which is vital for emergency diagnostics and resource-constrained settings [436]. Together, these advancements will be central to developing scalable, trustworthy, and clinically deployable AI systems, requiring close collaboration between researchers, clinicians, and policymakers to ensure technical robustness and ethical integration [437].
This study provides a comprehensive and forward-looking evaluation of deep learning applications in medical image analysis, highlighting both significant achievements and emerging opportunities. Advances in CNNs, GANs, Transformers, and hybrid architectures have demonstrated remarkable success in classification, segmentation, and enhancement tasks, establishing a strong foundation for future innovation. Yet, clinical translation requires addressing challenges such as robustness across diverse datasets, interpretability, and ethical deployment. The recurring themes of supervised learning challenges and Transformer advantages are restated here to reinforce their central role in shaping future research directions. Self-supervised and semi-supervised learning are gaining traction as scalable alternatives to conventional supervised approaches, while techniques like attention mechanisms and uncertainty estimation are improving transparency and clinical trust. Importantly, the field is shifting toward patient-centered evaluation frameworks, emphasizing real-world clinical impact over narrow performance metrics. Future progress will depend on greater dataset diversity, federated learning, domain adaptation, and closer collaboration among AI researchers, clinicians, and policymakers. With emerging architectures like Transformers and ongoing work on reducing complexity and enhancing interpretability, the field is poised to achieve equitable, generalizable, and ethically responsible AI solutions. By embracing this trajectory, deep learning is positioned to play a transformative role in the next generation of medical imaging.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Dawa Chyophel Lepcha: Conceptualization, Data curation, Writing—original draft, Formal analysis. Bhawna Goyal: Methodology, Supervision, Writing—original draft, Formal analysis, Validation. Ayush Dogra: Software, Investigation, Writing—review & editing, Resources. Prabhat Kumar Sahu: Validation & Formal analysis. Ahmed Alkhayyat: Investigation & Formal analysis. Aaliya Ali: Formal Analysis, Investigation & Project administration. Vinay Kukreja: Investigation & Validation. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All relevant data are included within the article.
Ethics Approval: This study did not involve human or animal participants; therefore, ethics approval was not required.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Rayed ME, Sajibul Islam SM, Niha SI, Jim JR, Kabir MM, Mridha MF. Deep learning for medical image segmentation: state-of-the-art advancements and challenges. Inform Med Unlocked. 2024;47:101504. doi:10.1016/j.imu.2024.101504. [Google Scholar] [CrossRef]
2. Chen J, Liu Y, Wei S, Bian Z, Subramanian S, Carass A, et al. A survey on deep learning in medical image registration: new technologies, uncertainty, evaluation metrics, and beyond. Med Image Anal. 2025;100:103385. doi:10.1016/j.media.2024.103385. [Google Scholar] [PubMed] [CrossRef]
3. Kaur N, Hans R. Transfer learning for cancer diagnosis in medical images: a compendious study. Int J Comput Intell Syst. 2025;18(1):62. doi:10.1007/s44196-025-00772-0. [Google Scholar] [CrossRef]
4. Lepcha DC, Goyal B, Dogra A, Goyal V. Image super-resolution: a comprehensive review, recent trends, challenges and applications. Inf Fusion. 2023;91(1):230–60. doi:10.1016/j.inffus.2022.10.007. [Google Scholar] [CrossRef]
5. Nazir N, Sarwar A, Saini BS. Recent developments in denoising medical images using deep learning: an overview of models, techniques, and challenges. Micron. 2024;180:103615. doi:10.1016/j.micron.2024.103615. [Google Scholar] [PubMed] [CrossRef]
6. Meyers PH, Nice CMJr, Becker HC, Nettleton WJJr, Sweeney JW, Meckstroth GR. Automated computer analysis of radiographic images. Radiology. 1964;83(6):1029–34. doi:10.1148/83.6.1029. [Google Scholar] [PubMed] [CrossRef]
7. Kruger RP, Townes JR, Hall DL, Dwyer SJ, Lodwick GS. Automated radiographic diagnosis via feature extraction and classification of cardiac size and shape descriptors. IEEE Trans Biomed Eng. 1972;BME-19(3):174–86. doi:10.1109/TBME.1972.324115. [Google Scholar] [PubMed] [CrossRef]
8. Sezaki N, Ukena K. Automatic computation of the cardiothoracic ratio with application to mass screening. IEEE Trans Biomed Eng. 1973;BME-20(4):248–53. doi:10.1109/TBME.1973.324188. [Google Scholar] [PubMed] [CrossRef]
9. Sahiner B, Petrick N, Chan HP, Hadjiiski LM, Paramagul C, Helvie MA, et al. Computer-aided characterization of mammographic masses: accuracy of mass segmentation and its effects on characterization. IEEE Trans Med Imaging. 2001;20(12):1275–84. doi:10.1109/42.974922. [Google Scholar] [PubMed] [CrossRef]
10. Doi K, MacMahon H, Katsuragawa S, Nishikawa RM, Jiang Y. Computer-aided diagnosis in radiology: potential and pitfalls. Eur J Radiol. 1999;31(2):97–109. doi:10.1016/S0720-048X(99)00016-9. [Google Scholar] [PubMed] [CrossRef]
11. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi:10.1038/nature14539. [Google Scholar] [PubMed] [CrossRef]
12. Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29(7):3338–47. doi:10.1007/s00330-019-06205-9. [Google Scholar] [PubMed] [CrossRef]
13. Umirzakova S, Ahmad S, Khan LU, Whangbo T. Medical image super-resolution for smart healthcare applications: a comprehensive survey. Inf Fusion. 2024;103:102075. doi:10.1016/j.inffus.2023.102075. [Google Scholar] [CrossRef]
14. Li X, Jia M, Islam MT, Yu L, Xing L. Self-supervised feature learning via exploiting multi-modal data for retinal disease diagnosis. IEEE Trans Med Imag. 2020;39(12):4023–33. doi:10.1109/TMI.2020.3008871. [Google Scholar] [PubMed] [CrossRef]
15. Shorfuzzaman M, Hossain MS. MetaCOVID: a Siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2021;113:107700. doi:10.1016/j.patcog.2020.107700. [Google Scholar] [PubMed] [CrossRef]
16. Zhang Y, Jiang H, Miura Y, Manning CD, Langlotz CP. Contrastive learning of medical visual representations from paired images and text. In: Proceedings of Machine Learning Research; 2022 Nov 28–Dec 2; New Orleans, LA, USA. [Google Scholar]
17. Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321(5):321–31. doi:10.1016/j.neucom.2018.09.013. [Google Scholar] [CrossRef]
18. Kumar A, Kim J, Lyndon D, Fulham M, Feng D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inform. 2017;21(1):31–40. doi:10.1109/JBHI.2016.2635663. [Google Scholar] [PubMed] [CrossRef]
19. Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK. Recurrent residual convolutional neural network based on U-Net (R2U-Net) for Medical image segmentation. arXiv:1802.06955. 2018. [Google Scholar]
20. Yu L, Wang S, Li X, Fu CW, Heng PA. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 605–13. doi:10.1007/978-3-030-32245-8_67. [Google Scholar] [CrossRef]
21. Fan DP, Zhou T, Ji GP, Zhou Y, Chen G, Fu H, et al. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging. 2020;39(8):2626–37. doi:10.1109/tmi.2020.2996645. [Google Scholar] [PubMed] [CrossRef]
22. Rijthoven Mv, Swiderska-Chadaj Z, Seeliger K, van der Laak J, Ciompi F. You only look on lymphocytes once. 2018 Apr 11. [cited 2025 Oct 23]. Available from: https://openreview.net/pdf?id=S10IfW2oz. [Google Scholar]
23. Mei J, Cheng MM, Xu G, Wan LR, Zhang H. SANet: a slice-aware network for pulmonary nodule detection. IEEE Trans Pattern Anal Mach Intell. 2021;2021:1. doi:10.1109/tpami.2021.3065086. [Google Scholar] [PubMed] [CrossRef]
24. Nair T, Precup D, Arnold DL, Arbel T. Exploring uncertainty measures in deep networks for Multiple sclerosis lesion detection and segmentation. Med Image Anal. 2020;59:101557. doi:10.1016/j.media.2019.101557. [Google Scholar] [PubMed] [CrossRef]
25. Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D. 3D deep learning for efficient and robust landmark detection in volumetric data. In: Medical image computing and computer-assisted intervention – MICCAI 2015. Cham, Switzerland: Springer International Publishing; 2015. p. 565–72. doi:10.1007/978-3-319-24553-9_69. [Google Scholar] [CrossRef]
26. Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N. A deep metric for multimodal registration. In: Medical image computing and computer-assisted intervention—MICCAI 2016. Cham, Switzerland: Springer International Publishing; 2016. p. 10–8. doi:10.1007/978-3-319-46726-9_2. [Google Scholar] [CrossRef]
27. Sokooti H, de Vos B, Berendsen F, Lelieveldt BPF, Išgum I, Staring M. Nonrigid image registration using multi-scale 3D convolutional neural networks. In: Medical image computing and computer assisted intervention—MICCAI 2017. Cham, Switzerland: Springer International Publishing; 2017. p. 232–9. doi:10.1007/978-3-319-66182-7_27. [Google Scholar] [CrossRef]
28. Balakrishnan G, Zhao A, Sabuncu MR, Dalca AV, Guttag J. An unsupervised learning model for deformable medical image registration. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. p. 9252–60. doi:10.1109/cvpr.2018.00964. [Google Scholar] [CrossRef]
29. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. doi:10.1038/nature21056. [Google Scholar] [PubMed] [CrossRef]
30. Long E, Lin H, Liu Z, Wu X, Wang L, Jiang J, et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat Biomed Eng. 2017;1(2):24. doi:10.1038/s41551-016-0024. [Google Scholar] [CrossRef]
31. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi:10.1016/j.media.2017.07.005. [Google Scholar] [PubMed] [CrossRef]
32. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19(1):221–48. doi:10.1146/annurev-bioeng-071516-044442. [Google Scholar] [PubMed] [CrossRef]
33. Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58(2):101552. doi:10.1016/j.media.2019.101552. [Google Scholar] [PubMed] [CrossRef]
34. Kazeminia S, Baur C, Kuijper A, van Ginneken B, Navab N, Albarqouni S, et al. GANs for medical image analysis. Artif Intell Med. 2020;109:101938. doi:10.1016/j.artmed.2020.101938. [Google Scholar] [PubMed] [CrossRef]
35. Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal. 2019;54:280–96. doi:10.1016/j.media.2019.03.009. [Google Scholar] [PubMed] [CrossRef]
36. Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal. 2020;63:101693. doi:10.1016/j.media.2020.101693. [Google Scholar] [PubMed] [CrossRef]
37. Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst. 2018;42(11):226. doi:10.1007/s10916-018-1088-1. [Google Scholar] [PubMed] [CrossRef]
38. Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7. doi:10.1126/science.1127647. [Google Scholar] [PubMed] [CrossRef]
39. Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. In: Advances in neural information processing systems 19. Cambridge, MA, USA: The MIT Press; 2007. p. 153–60. doi:10.7551/mitpress/7503.003.0024. [Google Scholar] [CrossRef]
40. Ranzato M, Poultney C, Chopra S, LeCun Y. Efficient learning of sparse representations with an energy-based model. In: Advances in neural information processing systems 19. Cambridge, MA, USA: The MIT Press; 2007. p. 1137–44. doi:10.7551/mitpress/7503.003.0147. [Google Scholar] [CrossRef]
41. Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol PA. Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion pascal vincent hugo larochelle yoshua bengio pierre-antoine manzagol. J Mach Learn Res. 2010;11:3371–408. [Google Scholar]
42. Rifai S, Vincent P, Muller X, Glorot X, Bengio Y. Contractive auto-encoders: explicit invariance during feature extraction. In: ICML'11: Proceedings of the 28th International Conference on International Conference on Machine Learning; 2018 Jun 28–Jul 2; Madison, WI, USA. p. 833–40. [Google Scholar]
43. Kingma DP, Welling M. Auto-encoding variational bayes. In: 2nd International Conference on Learning Representations, ICLR 2014—Conference Track Proceedings; 2014 Apr 14–16; Banff, AB, Canada. [Google Scholar]
44. Dilokthanakul N, Mediano PAM, Garnelo M, Lee MCH, Salimbeni H, Arulkumaran K, et al. Deep unsupervised clustering with gaussian mixture variational autoencoders. arXiv:1611.02648. 2016. [Google Scholar]
45. Sohn K, Lee H, Yan X. Learning structured output representation using deep conditional generative models. In: NIPS'15: Proceedings of the 29th International Conference on Neural Information Processing Systems; 2015 Dec 7–12; Montreal, QC, Canada. p. 3483–91. [Google Scholar]
46. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Commun ACM. 2020;63(11):139–44. doi:10.1145/3422622. [Google Scholar] [CrossRef]
47. Arjovsky M, Chintala S, Bottou L. Wasserstein generative adversarial networks. In: ICML'17: Proceedings of the 34th International Conference on Machine Learning; 2017 Aug 6–11; Sydney, NSW, Australia. p. 214–33. [Google Scholar]
48. Mirza M, Osindero S. Conditional generative adversarial nets. arXiv:1411.1784. 2014. [Google Scholar]
49. Odena A, Olah C, Shlens J. Conditional image synthesis with auxiliary classifier GANs. In: ICML'17: Proceedings of the 34th International Conference on Machine Learning; 2017 Aug 6–11; Sydney, NSW, Australia. p. 2642–51. [Google Scholar]
50. Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context encoders: feature learning by inpainting. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV, USA. doi:10.1109/cvpr.2016.278. [Google Scholar] [CrossRef]
51. Zhang R, Isola P, Efros AA. Colorful image colorization. In: Leibe B, Matas J, Sebe N, Welling M, editors. Computer Vision—ECCV 2016. Cham, Switzerland: Springer; 2016. p. 649–66. doi:10.1007/978-3-319-46487-9_40. [Google Scholar] [CrossRef]
52. Doersch C, Gupta A, Efros AA. Unsupervised visual representation learning by context prediction. In: 2015 IEEE International Conference on Computer Vision (ICCV); 2015 Dec 7–13; Santiago, Chile. doi:10.1109/iccv.2015.167. [Google Scholar] [CrossRef]
53. Noroozi M, Favaro P. Unsupervised learning of visual representations by solving jigsaw puzzles. In: Computer vision—ECCV 2016. Cham, Switzerland: Springer International Publishing; 2016. p. 69–84. doi:10.1007/978-3-319-46466-4_5. [Google Scholar] [CrossRef]
54. Gidaris S, Singh P, Komodakis N. Unsupervised representation learning by predicting image rotations. In: 6th International Conference on Learning Representations, ICLR 2018; 2018 Apr 30–May 3; Vancouver, BC, Canada. [Google Scholar]
55. Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: NAACL HLT 2019—2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2019 Jun 2–7; Minneapolis, MN, USA. p. 4171–86. [Google Scholar]
56. He K, Fan H, Wu Y, Xie S, Girshick R. Momentum contrast for unsupervised visual representation learning. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13–19; Seattle, WA, USA. p. 9726–35. doi:10.1109/cvpr42600.2020.00975. [Google Scholar] [CrossRef]
57. Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations. In: Proceedings of the 37th International Conference on Machine Learning; 2020 Jul 13–18. PMLR; 2020. p. 1597–67. [Google Scholar]
58. van den Oord A, Li Y, Vinyals O. Representation learning with contrastive predictive coding. arXiv:1807.03748. 2018. [Google Scholar]
59. Wenderoth L, Asemissen AM, Modemann F, Nielsen M, Werner R. Transferable automatic hematological cell classification: overcoming data limitations with self-supervised learning. Comput Methods Programs Biomed. 2025;260(7):108560. doi:10.1016/j.cmpb.2024.108560. [Google Scholar] [PubMed] [CrossRef]
60. Hussien A, Elkhateb A, Saeed M, Elsabawy NM, Elnakeeb AE, Elrashidy N. Explainable self-supervised learning for medical image diagnosis based on DINO V2 model and semantic search. Sci Rep. 2025;15(1):32174. doi:10.1038/s41598-025-15604-6. [Google Scholar] [PubMed] [CrossRef]
61. Chapelle O, Scholkopf B, Zien E. Semi-supervised learning (chapelle, O. et al., eds.; 2006). Book reviews. IEEE Trans Neural Netw. 2009;20(3):542. doi:10.1109/TNN.2009.2015974. [Google Scholar] [CrossRef]
62. Ouali Y, Hudelot C, Tami M. An overview of deep semi-supervised learning. arXiv:2006.05278. 2020. [Google Scholar]
63. Laine S, Aila T. Temporal ensembling for semi-supervised learning. In: 5th International Conference on Learning Representations, ICLR 2017; 2017 Apr 24–26; Toulon, France. [Google Scholar]
64. Rasmus A, Berglund M, Honkala M, Valpola H, Raiko T. Semi-supervised learning with ladder networks. In: NIPS'15: Proceedings of the 29th International Conference on Neural Information Processing Systems; 2015 Dec 7–12; Montreal, QC, Canada. [Google Scholar]
65. Tarvainen A, Valpola H. Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 1195–204. [Google Scholar]
66. Lee DH. Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on Challenges in Representation Learning; 2013 Jun 21; Atlanta, GA, USA. [Google Scholar]
67. Zhang H, Cisse M, Dauphin YN, Lopez-Paz D. mixup: beyond empirical risk minimization. In: 6th International Conference on Learning Representations, ICLR 2018—Conference Track Proceedings; 2018 Apr 30–May 3; Vancouver, BC, Canada. [Google Scholar]
68. Xie Q, Dai Z, Hovy E, Luong T, Le Q. Unsupervised data augmentation for consistency training. In: NIPS'20: Proceedings of the 34th International Conference on Neural Information Processing Systems; 2020 Dec 6–12; Vancouver, BC, Canada. p. 6256–68. [Google Scholar]
69. Qiao S, Shen W, Zhang Z, Wang B, Yuille A. Deep co-training for semi-supervised image recognition. In: Proceedings of the European Conference on Computer Vision (eccv); 2018 Sep 8–14; Munich, Germany. p. 135–52. [Google Scholar]
70. Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training gans. In: NIPS'16: Proceedings of the 30th International Conference on Neural Information Processing Systems; 2016 Dec 5–10; Barcelona, Spain. p. 2234–42. [Google Scholar]
71. Odena A. Semi-supervised learning with generative adversarial networks. arXiv:1606.01583. 2016. [Google Scholar]
72. Li CX, Xu T, Zhu J, Zhang B. Triple generative adversarial nets. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 4091–101. [Google Scholar]
73. Itti L, Koch C, Niebur E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans Pattern Anal Machine Intell. 1998;20(11):1254–9. doi:10.1109/34.730558. [Google Scholar] [CrossRef]
74. Bahdanau D, Cho KH, Bengio Y. Neural machine translation by jointly learning to align and translate. In: 3rd International Conference on Learning Representations, ICLR 2015; 2015 May 7–9; San Diego, CA, USA. [Google Scholar]
75. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 6000–10. [Google Scholar]
76. Xu K, Ba J, Kiros R, Cho K, Courville A, Salakhudinov R, et al. Show, attend and tell: neural image caption generation with visual attention. In: International Conference on Machine Learning; 2015 Jul 6–11; Lille, France. p. 2048–57. [Google Scholar]
77. You Q, Jin H, Wang Z, Fang C, Luo J. Image captioning with semantic attention. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV, USA. doi:10.1109/cvpr.2016.503. [Google Scholar] [CrossRef]
78. Anderson P, He X, Buehler C, Teney D, Johnson M, Gould S, et al. Bottom-up and top-down attention for image captioning and visual question answering. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. p. 6077–86. doi:10.1109/cvpr.2018.00636. [Google Scholar] [CrossRef]
79. Chen LC, Yang Y, Wang J, Xu W, Yuille AL. Attention to scale: scale-aware semantic image segmentation. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV, USA. p. 3640–9. doi:10.1109/cvpr.2016.396. [Google Scholar] [CrossRef]
80. Ren M, Zemel RS. End-to-end instance segmentation with recurrent attention. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI, USA. p. 293–301. doi:10.1109/cvpr.2017.39. [Google Scholar] [CrossRef]
81. Jaderberg M, Simonyan K, Zisserman A. Spatial transformer networks. In: NIPS'15: Proceedings of the 29th International Conference on Neural Information Processing Systems; 2015 Dec 7–12; Montreal, QC, Canada. p. 2017–25. [Google Scholar]
82. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. p. 7132–41. doi:10.1109/cvpr.2018.00745. [Google Scholar] [CrossRef]
83. Wang X, Girshick R, Gupta A, He K. Non-local neural networks. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. IEEE; 2018. p. 7794–803. doi:10.1109/cvpr.2018.00813. [Google Scholar] [CrossRef]
84. Wang F, Jiang M, Qian C, Yang S, Li C, Zhang H, et al. Residual attention network for image classification. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI, USA. p. 6450–8. doi:10.1109/cvpr.2017.683. [Google Scholar] [CrossRef]
85. Woo S, Park J, Lee JY, Kweon IS. CBAM: convolutional block attention module. In: Computer vision – ECCV 2018. Cham, Switzerland: Springer International Publishing; 2018. p. 3–19. doi:10.1007/978-3-030-01234-2_1. [Google Scholar] [CrossRef]
86. Zhao A, Balakrishnan G, Durand F, Guttag JV, Dalca AV. Data augmentation using learned transformations for one-shot medical image segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15–20; Long Beach, CA, USA. p. 8535–45. doi:10.1109/CVPR.2019.00874. [Google Scholar] [CrossRef]
87. Zhou Z, Sodha V, Pang J, Gotway MB, Liang J. Models genesis. Med Image Anal. 2021;67(4):101840. doi:10.1016/j.media.2020.101840. [Google Scholar] [PubMed] [CrossRef]
88. Vu YNT, Wang R, Balachandar N, Liu C, Ng AY, Rajpurkar P. MedAug: contrastive learning leveraging patient metadata improves representations for chest X-ray interpretation. In: Proceedings of the 6th Machine Learning for Healthcare Conference; 2021 Aug 6–7; Virtual. p. 755–69. [Google Scholar]
89. Gal Y, Ghahramani Z. Dropout as a bayesian approximation: appendix. In: 33rd International Conference on Machine Learning, ICML 2016; 2016 Jun 20–22; New York, NY, USA. p. 1661–80. [Google Scholar]
90. Lakshminarayanan B, Pritzel A, Deepmind CB. Simple and scalable predictive uncertainty estimation using deep ensembles. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 6405–16. [Google Scholar]
91. Abdar M, Pourpanah F, Hussain S, Rezazadegan D, Liu L, Ghavamzadeh M, et al. A review of uncertainty quantification in deep learning: techniques, applications and challenges. Inf Fusion. 2021;76(1):243–97. doi:10.1016/j.inffus.2021.05.008. [Google Scholar] [CrossRef]
92. Fiore P, Terlizzi A, Bardozzo F, Liò P, Tagliaferri R. Advancing label-free cell classification with connectome-inspired explainable models and a novel LIVECell-CLS dataset. Comput Biol Med. 2025;192(Pt B):110274. doi:10.1016/j.compbiomed.2025.110274. [Google Scholar] [PubMed] [CrossRef]
93. van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology. 2011;261(3):719–32. doi:10.1148/radiol.11091710. [Google Scholar] [PubMed] [CrossRef]
94. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. doi:10.1145/3065386. [Google Scholar] [CrossRef]
95. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations, ICLR 2015; 2015 May 7–9; San Diego, CA, USA. [Google Scholar]
96. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2015 Jun 7–12; Boston, MA, USA. p. 1–9. doi:10.1109/CVPR.2015.7298594. [Google Scholar] [CrossRef]
97. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV, USA. p. 770–8. doi:10.1109/cvpr.2016.90. [Google Scholar] [CrossRef]
98. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI, USA. p. 2261–9. doi:10.1109/cvpr.2017.243. [Google Scholar] [CrossRef]
99. Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–312. doi:10.1109/tmi.2016.2535302. [Google Scholar] [PubMed] [CrossRef]
100. Chen S, Ma K, Zheng Y. Med3D: transfer learning for 3D medical image analysis. arXiv:1904.00625. 2019. [Google Scholar]
101. Deng J, Dong W, Socher R, Li LJ, Kai L, Li FF. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009 Jun 20–25; Miami, FL, USA. p. 248–55. doi:10.1109/CVPR.2009.5206848. [Google Scholar] [CrossRef]
102. Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition; 2014 Jun 23–28; Columbus, OH, USA. doi:10.1109/cvpr.2014.81. [Google Scholar] [CrossRef]
103. Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–98. doi:10.1109/tmi.2016.2528162. [Google Scholar] [PubMed] [CrossRef]
104. Yuan Y, Qin W, Buyyounouski M, Ibragimov B, Hancock S, Han B, et al. Prostate cancer classification with multiparametric MRI transfer learning model. Med Phys. 2019;46(2):756–65. doi:10.1002/mp.13367. [Google Scholar] [PubMed] [CrossRef]
105. Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imag. 2016;3(3):034501. doi:10.1117/1.jmi.3.3.034501. [Google Scholar] [PubMed] [CrossRef]
106. Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi:10.1016/j.media.2020.101794. [Google Scholar] [PubMed] [CrossRef]
107. Zhou Y, He X, Huang L, Liu L, Zhu F, Cui S, et al. Collaborative learning of semi-supervised segmentation and classification for medical images. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15–20; Long Beach, CA, USA. p. 2074–83. doi:10.1109/CVPR.2019.00218. [Google Scholar] [CrossRef]
108. Guan Q, Huang Y, Zhong Z, Zheng Z, Zheng L, Yang Y. Diagnose like a radiologist: attention guided convolutional neural network for Thorax Disease classification. arXiv:1801.09927. 2018. [Google Scholar]
109. Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, et al. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. doi:10.1016/j.media.2019.01.012. [Google Scholar] [PubMed] [CrossRef]
110. Huo X, Sun G, Tian S, Wang Y, Yu L, Long J, et al. HiFuse: hierarchical multi-scale feature fusion network for medical image classification. Biomed Signal Process Control. 2024;87:105534. doi:10.1016/j.bspc.2023.105534. [Google Scholar] [CrossRef]
111. Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 Apr 4–7; Washington, DC, USA. p. 289–93. doi:10.1109/ISBI.2018.8363576. [Google Scholar] [CrossRef]
112. Wu E, Wu K, Cox D, Lotter W. Conditional infilling GANs for data augmentation in mammogram classification. In: Image analysis for moving organ, breast, and thoracic images. Cham, Switzerland: Springer International Publishing; 2018. p. 98–106. doi:10.1007/978-3-030-00946-5_11. [Google Scholar] [CrossRef]
113. Dai Z, Yi J, Yan L, Xu Q, Hu L, Zhang Q, et al. PFEMed: few-shot medical image classification using prior guided feature enhancement. Pattern Recognit. 2023;134:109108. doi:10.1016/j.patcog.2022.109108. [Google Scholar] [CrossRef]
114. Zhou L, Liu H, Bae J, He J, Samaras D, Prasanna P. Self pre-training with masked autoencoders for medical image classification and segmentation. In: 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI); 2023 Apr 18–21; Cartagena, Colombia. p. 1–6. doi:10.1109/ISBI53787.2023.10230477. [Google Scholar] [CrossRef]
115. Liu Q, Yu L, Luo L, Dou Q, Heng PA. Semi-supervised medical image classification with relation-driven self-ensembling model. IEEE Trans Med Imag. 2020;39(11):3429–40. doi:10.1109/TMI.2020.2995518. [Google Scholar] [PubMed] [CrossRef]
116. Shang H, Sun Z, Yang W, Fu X, Zheng H, Chang J, et al. Leveraging other datasets for medical imaging classification: evaluation of transfer, multi-task and semi-supervised learning. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 431–9. doi:10.1007/978-3-030-32254-0_48. [Google Scholar] [CrossRef]
117. Hassan M, Ali S, Alquhayz H, Safdar K. Developing intelligent medical image modality classification system using deep transfer learning and LDA. Sci Rep. 2020;10(1):12868. doi:10.1038/s41598-020-69813-2. [Google Scholar] [PubMed] [CrossRef]
118. Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021;51(2):854–64. doi:10.1007/s10489-020-01829-7. [Google Scholar] [PubMed] [CrossRef]
119. Rubin M, Stein O, Turko NA, Nygate Y, Roitshtain D, Karako L, et al. TOP-GAN: stain-free cancer cell classification using deep learning with a small training set. Med Image Anal. 2019;57:176–85. doi:10.1016/j.media.2019.06.014. [Google Scholar] [PubMed] [CrossRef]
120. Xie Y, Zhang J, Xia Y. Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal. 2019;57(2):237–48. doi:10.1016/j.media.2019.07.004. [Google Scholar] [PubMed] [CrossRef]
121. Kumar R, Kumbharkar P, Vanam S, Sharma S. Medical images classification using deep learning: a survey. Multimed Tools Appl. 2024;83(7):19683–728. doi:10.1007/s11042-023-15576-7. [Google Scholar] [CrossRef]
122. Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, et al. Big self-supervised models advance medical image classification. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10–17; Montreal, QC, Canada. p. 3458–68. doi:10.1109/iccv48922.2021.00346. [Google Scholar] [CrossRef]
123. Chen X, Fan H, Girshick R, He K. Improved baselines with momentum contrastive learning. arXiv:2003.04297. 2020. [Google Scholar]
124. Misra I, van der Maaten L. Self-supervised learning of pretext-invariant representations. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13–19; Seattle, WA, USA. p. 6706–16. doi:10.1109/cvpr42600.2020.00674. [Google Scholar] [CrossRef]
125. Bai W, Chen C, Tarroni G, Duan JM, Guitton F, Petersen SE, et al. Self-supervised learning for cardiac MR image segmentation by anatomical position prediction. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer; 2019. p. 541–9. doi:10.1007/978-3-030-32245-8_60. [Google Scholar] [CrossRef]
126. Tao X, Li Y, Zhou W, Ma K, Zheng Y. Revisiting rubik’s cube: self-supervised learning with volume-wise transformation for 3D medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2020. Cham, Switzerland: Springer International Publishing; 2020. p. 238–48. doi:10.1007/978-3-030-59719-1_24. [Google Scholar] [CrossRef]
127. Jing L, Tian Y. Self-supervised visual feature learning with deep neural networks: a survey. IEEE Trans Pattern Anal Mach Intell. 2021;43(11):4037–58. doi:10.1109/TPAMI.2020.2992393. [Google Scholar] [PubMed] [CrossRef]
128. Sowrirajan H, Yang JB, Ng AY, Rajpurkar P. Moco pretraining improves representation and transferability of chest x-ray models. In: Medical Imaging with Deep Learning; 2021 Jul 7; Lübeck, Germany. p. 728–44. [Google Scholar]
129. Chen X, Yao L, Zhou T, Dong J, Zhang Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021;113:107826. doi:10.1016/j.patcog.2021.107826. [Google Scholar] [PubMed] [CrossRef]
130. Zhuang X, Li Y, Hu Y, Ma K, Yang Y, Zheng Y. Self-supervised feature learning for 3D medical images by playing a rubik’s cube. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 420–8. doi:10.1007/978-3-030-32251-9_46. [Google Scholar] [CrossRef]
131. Zhu J, Li Y, Hu Y, Ma K, Zhou SK, Zheng Y. Rubik’s Cube+: a self-supervised feature learning framework for 3D medical image analysis. Med Image Anal. 2020;64:101746. doi:10.1016/j.media.2020.101746. [Google Scholar] [PubMed] [CrossRef]
132. Tajbakhsh N, Hu Y, Cao J, Yan X, Xiao Y, Lu Y, et al. Surrogate supervision for medical image analysis: effective deep learning from limited quantities of labeled data. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019 Apr 8–11; Venice, Italy. doi:10.1109/isbi.2019.8759553. [Google Scholar] [CrossRef]
133. Chen L, Bentley P, Mori K, Misawa K, Fujiwara M, Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med Image Anal. 2019;58:101539. doi:10.1016/j.media.2019.101539. [Google Scholar] [PubMed] [CrossRef]
134. Liu S, Yue W, Guo Z, Wang L. Multi-branch CNN and grouping cascade attention for medical image classification. Sci Rep. 2024;14(1):15013. doi:10.1038/s41598-024-64982-w. [Google Scholar] [PubMed] [CrossRef]
135. Manzari ON, Ahmadabadi H, Kashiani H, Shokouhi SB, Ayatollahi A. MedViT: a robust vision transformer for generalized medical image classification. Comput Biol Med. 2023;157:106791. doi:10.1016/j.compbiomed.2023.106791. [Google Scholar] [PubMed] [CrossRef]
136. Ren Z, Kong X, Zhang Y, Wang S. UKSSL: underlying knowledge based semi-supervised learning for medical image classification. IEEE Open J Eng Med Biol. 2024;5:459–66. doi:10.1109/OJEMB.2023.3305190. [Google Scholar] [PubMed] [CrossRef]
137. Yang Z, Zhang J, Luo X, Lu Z, Shen L. MedKAN: an advanced Kolmogorov-Arnold network for medical image classification. arXiv:2502.18416. 2025. [Google Scholar]
138. Lai Q, Vong CM, Yan T, Wong PK, Liang X. Hybrid multiple instance learning network for weakly supervised medical image classification and localization. Expert Syst Appl. 2025;260:125362. doi:10.1016/j.eswa.2024.125362. [Google Scholar] [CrossRef]
139. Hussain T, Shouno H, Hussain A, Hussain D, Ismail M, Hussain Mir T, et al. EFFResNet-ViT: a fusion-based convolutional and vision transformer model for explainable medical image classification. IEEE Access. 2025;13:54040–68. doi:10.1109/access.2025.3554184. [Google Scholar] [CrossRef]
140. Regmi S, Subedi A, Tomar NK, Bagci U, Jha D. Vision transformer for efficient chest X-ray and gastrointestinal image classification. In: Medical Imaging 2025: Computer-Aided Diagnosis; 2025 Feb 16–21; San Diego, CA, USA. doi:10.1117/12.3045810. [Google Scholar] [CrossRef]
141. Geng P, Lu J, Zhang Y, Ma S, Tang Z, Liu J. TC-fuse: a transformers fusing CNNs network for medical image segmentation. Comput Model Eng Sci. 2023;137(2):2001–23. doi:10.32604/cmes.2023.027127. [Google Scholar] [CrossRef]
142. Shao J, Chen S, Zhou J, Zhu H, Wang Z, Brown M. Application of U-Net and optimized clustering in medical image segmentation: a review. Comput Model Eng Sci. 2023;136(3):2173–219. doi:10.32604/cmes.2023.025499. [Google Scholar] [CrossRef]
143. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention – MICCAI 2015. Cham, Switzerland: Springer International Publishing; 2015. p. 234–41. doi:10.1007/978-3-319-24574-4_28. [Google Scholar] [CrossRef]
144. Hariharan B, Arbeláez P, Girshick R, Malik J. Hypercolumns for object segmentation and fine-grained localization. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 Jun 7–12; Boston, MA, USA. p. 447–56. doi:10.1109/CVPR.2015.7298642. [Google Scholar] [CrossRef]
145. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 Jun 7–12; Boston, MA, USA. p. 3431–40. doi:10.1109/CVPR.2015.7298965. [Google Scholar] [CrossRef]
146. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham, Switzerland: Springer International Publishing; 2018. p. 3–11. doi:10.1007/978-3-030-00889-5_1. [Google Scholar] [CrossRef]
147. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical image computing and computer-assisted intervention – MICCAI 2016. Cham, Switzerland: Springer International Publishing; 2016. p. 424–32. doi:10.1007/978-3-319-46723-8_49. [Google Scholar] [CrossRef]
148. Milletari F, Navab N, Ahmadi SA. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV); 2016 Oct 25–28; Stanford, CA, USA. p. 565–71. doi:10.1109/3DV.2016.79. [Google Scholar] [CrossRef]
149. Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans Med Imaging. 2018;37(8):1822–34. doi:10.1109/tmi.2018.2806309. [Google Scholar] [PubMed] [CrossRef]
150. Liang M, Hu X. Recurrent convolutional neural network for object recognition. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 Jun 7–12; Boston, MA, USA. p. 3367–75. doi:10.1109/CVPR.2015.7298958. [Google Scholar] [CrossRef]
151. Oktay O, Schlemper J, Le Folgoc L, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas. arXiv:1804.03999. 2018. [Google Scholar]
152. Xue Y, Xu T, Zhang H, Long LR, Huang X. SegAN: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics. 2018;16(3):383–92. doi:10.1007/s12021-018-9377-x. [Google Scholar] [PubMed] [CrossRef]
153. Zhang Y, Miao S, Mansi T, Liao R. Unsupervised X-ray image segmentation with task driven generative adversarial networks. Med Image Anal. 2020;62:101664. doi:10.1016/j.media.2020.101664. [Google Scholar] [PubMed] [CrossRef]
154. Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing. 2019;335(1):34–45. doi:10.1016/j.neucom.2019.01.103. [Google Scholar] [PubMed] [CrossRef]
155. Baumgartner CF, Tezcan KC, Chaitanya K, Hötker AM, Muehlematter UJ, Schawkat K, et al. PHiSeg: capturing uncertainty in medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer; 2019. p. 119–27. doi:10.1007/978-3-030-32245-8_14/TABLES/1. [Google Scholar] [CrossRef]
156. Mehrtash A, Wells WM, Tempany CM, Abolmaesumi P, Kapur T. Confidence calibration and predictive uncertainty estimation for deep medical image segmentation. IEEE Trans Med Imag. 2020;39(12):3868–78. doi:10.1109/TMI.2020.3006437. [Google Scholar] [PubMed] [CrossRef]
157. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306. 2021. [Google Scholar]
158. Zhang Y, Liu H, Hu Q. TransFuse: fusing transformers and CNNs for medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2021. Cham, Switzerland: Springer International Publishing; 2021. p. 14–24. doi:10.1007/978-3-030-87193-2_2. [Google Scholar] [CrossRef]
159. Xie Y, Zhang J, Shen C, Xia Y. CoTr: efficiently bridging CNN and transformer for 3D medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2021. Cham, Switzerland: Springer International Publishing; 2021. p. 171–80. doi:10.1007/978-3-030-87199-4_16. [Google Scholar] [CrossRef]
160. Cao H, Wang YY, Chen J, Jiang DS, Zhang XP, Tian Q, et al. Swin-Unet: unet-like pure transformer for medical image segmentation. In: Computer vision—ECCV 2022 workshops. Cham, Switzerland: Springer; 2023. p. 205–18. doi:10.1007/978-3-031-25066-8_9. [Google Scholar] [CrossRef]
161. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: hierarchical vision transformer using shifted windows. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10–17; Montreal, QC, Canada. p. 9992–10002. doi:10.1109/iccv48922.2021.00986. [Google Scholar] [CrossRef]
162. Wang H, Zhu Y, Green B, Adam H, Yuille A, Chen LC. Axial-DeepLab: stand-alone axial-attention for panoptic segmentation. In: Computer Vision—ECCV 2020. Cham, Switzerland: Springer International Publishing; 2020. p. 108–26. doi:10.1007/978-3-030-58548-8_7. [Google Scholar] [CrossRef]
163. Valanarasu JMJ, Oza P, Hacihaliloglu I, Patel VM. Medical transformer: gated axial-attention for medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2021. Cham, Switzerland: Springer International Publishing; 2021. p. 36–46. doi:10.1007/978-3-030-87193-2_4. [Google Scholar] [CrossRef]
164. He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. In: IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. IEEE; 2017. doi:10.1109/iccv.2017.322. [Google Scholar] [CrossRef]
165. Lin TY, Dollar P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI, USA. p. 936–44. doi:10.1109/cvpr.2017.106. [Google Scholar] [CrossRef]
166. Wang X, Han S, Chen Y, Gao D, Vasconcelos N. Volumetric attention for 3D medical image segmentation and detection. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 175–84. doi:10.1007/978-3-030-32226-7_20. [Google Scholar] [CrossRef]
167. Zhang Z, Yang L, Zheng Y. Translating and segmenting multimodal medical volumes with cycle- and shape-consistency generative adversarial network. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. p. 9242–51. doi:10.1109/CVPR.2018.00963. [Google Scholar] [CrossRef]
168. Chen C, Miao J, Wu D, Zhong A, Yan Z, Kim S, et al. MA-SAM: modality-agnostic SAM adaptation for 3D medical image segmentation. Med Image Anal. 2024;98:103310. doi:10.1016/j.media.2024.103310. [Google Scholar] [PubMed] [CrossRef]
169. Ruan J, Li J, Xiang S. VM-UNet: vision mamba UNet for medical image segmentation. arXiv:2402.02491. 2024. [Google Scholar]
170. Hu SY, Wang SH, Weng WH, Wang JC, Wang XH, Ozturk A, et al. Self-supervised pretraining with DICOM metadata in ultrasound imaging. In: Machine Learning for Healthcare Conference; 2020 Aug 7–8; Virtual. p. 732–49. [Google Scholar]
171. Taleb A, Loetzsch W, Noel D, Julius S, Thomas G, Benjamin B, et al. 3D self-supervised methods for medical imaging. In: NIPS’20: Proceedings of the 34th International Conference on Neural Information Processing Systems; 2020 Dec 6–12; Vancouver, BC, Canada. p. 18158–72. [Google Scholar]
172. Jamaludin A, Kadir T, Zisserman A. Self-supervised learning for spinal MRIs. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham, Switzerland: Springer International Publishing; 2017. p. 294–302. doi:10.1007/978-3-319-67558-9_34. [Google Scholar] [CrossRef]
173. Chaitanya K, Erdil E, Karani N, Konukoglu E. Contrastive learning of global and local features for medical image segmentation with limited annotations. In: NIPS'20: Proceedings of the 34th International Conference on Neural Information Processing Systems; 2020 Dec 6–12; Vancouver, BC, Canada. p. 12546–58. [Google Scholar]
174. Li X, Yu L, Chen H, Fu CW, Xing L, Heng PA. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Trans Neural Netw Learn Syst. 2021;32(2):523–34. doi:10.1109/TNNLS.2020.2995319. [Google Scholar] [PubMed] [CrossRef]
175. Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, et al. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans Med Imaging. 2020;39(8):2653–63. doi:10.1109/tmi.2020.3000314. [Google Scholar] [PubMed] [CrossRef]
176. Zhang Z, Fu H, Dai H, Shen J, Pang Y, Shao L. ET-net: a generic edge-aTtention guidance network for medical image segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 442–50. doi:10.1007/978-3-030-32239-7_49. [Google Scholar] [CrossRef]
177. Wu Z, Su L, Huang Q. Cascaded partial decoder for fast and accurate salient object detection. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15–20; Long Beach, CA, USA. p. 3902–11. doi:10.1109/CVPR.2019.00403. [Google Scholar] [CrossRef]
178. Chen SH, Tan XL, Wang B, Hu XL. Reverse attention for salient object detection. In: Proceedings of the European Conference on Computer Vision (ECCV); 2018 Sep 8–14; Munich, Germany. p. 234–50. [Google Scholar]
179. Qiu L, Cheng J, Gao H, Xiong W, Ren H. Federated semi-supervised learning for medical image segmentation via pseudo-label denoising. IEEE J Biomed Health Inform. 2023;27(10):4672–83. doi:10.1109/JBHI.2023.3274498. [Google Scholar] [PubMed] [CrossRef]
180. Sedai S, Mahapatra D, Hewavitharanage S, Maetschke S, Garnavi R. Semi-supervised segmentation of optic cup in retinal fundus images using variational autoencoder. In: Medical image computing and computer-assisted intervention—MICCAI 2017. Cham, Switzerland: Springer International Publishing; 2017. p. 75–82. doi:10.1007/978-3-319-66185-8_9. [Google Scholar] [CrossRef]
181. He Y, Yang G, Chen Y, Kong Y, Wu J, Tang L, et al. DPA-DenseBiasNet: semi-supervised 3D fine renal artery segmentation with dense biased network and deep priori anatomy. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 139–47. doi:10.1007/978-3-030-32226-7_16. [Google Scholar] [CrossRef]
182. Zheng H, Lin L, Hu H, Zhang Q, Chen Q, Iwamoto Y, et al. Semi-supervised segmentation of liver using adversarial learning with deep atlas prior. In: Medical image computing and computer assisted intervention – MICCAI 2019. Cham: Springer International Publishing; 2019. p. 148–56. doi:10.1007/978-3-030-32226-7_17. [Google Scholar] [CrossRef]
183. Clough JR, Byrne N, Oksuz I, Zimmer VA, Schnabel JA, King AP. A topological loss function for deep-learning based image segmentation using persistent homology. IEEE Trans Pattern Anal Mach Intell. 2022;44(12):8766–78. doi:10.1109/TPAMI.2020.3013679. [Google Scholar] [PubMed] [CrossRef]
184. Zhou HY, Guo J, Zhang Y, Han X, Yu L, Wang L, et al. nnFormer: volumetric medical image segmentation via a 3D transformer. IEEE Trans Image Process. 2023;32:4036–45. doi:10.1109/tip.2023.3293771. [Google Scholar] [PubMed] [CrossRef]
185. Wu J, Wang Z, Hong M, Ji W, Fu H, Xu Y, et al. Medical SAM adapter: adapting segment anything model for medical image segmentation. Med Image Anal. 2025;102:103547. doi:10.1016/j.media.2025.103547. [Google Scholar] [PubMed] [CrossRef]
186. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–11. doi:10.1038/s41592-020-01008-z. [Google Scholar] [PubMed] [CrossRef]
187. He A, Wang K, Li T, Du C, Xia S, Fu H. H2Former: an efficient hierarchical hybrid transformer for medical image segmentation. IEEE Trans Med Imag. 2023;42(9):2763–75. doi:10.1109/TMI.2023.3264513. [Google Scholar] [PubMed] [CrossRef]
188. Wu R, Liu Y, Liang P, Chang Q. H-vmunet: high-order Vision Mamba UNet for medical image segmentation. Neurocomputing. 2025;624:129447. doi:10.1016/j.neucom.2025.129447. [Google Scholar] [CrossRef]
189. Zheng S, Ye X, Yang C, Yu L, Li W, Gao X, et al. Asymmetric adaptive heterogeneous network for multi-modality medical image segmentation. IEEE Trans Med Imaging. 2025;44(4):1836–52. doi:10.1109/tmi.2025.3526604. [Google Scholar] [PubMed] [CrossRef]
190. Iqbal S, Khan TM, Naqvi SS, Naveed A, Meijering E. TBConvL-Net: a hybrid deep learning architecture for robust medical image segmentation. Pattern Recognit. 2025;158:111028. doi:10.1016/j.patcog.2024.111028. [Google Scholar] [CrossRef]
191. Chyophel Lepcha D, Goyal B, Dogra A. Low-dose CT image denoising using sparse 3D transformation with probabilistic non-local means for clinical applications. Imag Sci J. 2023;71(2):97–109. doi:10.1080/13682199.2023.2176809. [Google Scholar] [CrossRef]
192. Rawat S, Rana KPS, Kumar V. A novel complex-valued convolutional neural network for medical image denoising. Biomed Signal Process Control. 2021;69:102859. doi:10.1016/j.bspc.2021.102859. [Google Scholar] [CrossRef]
193. Zhang J, Niu Y, Shangguan Z, Gong W, Cheng Y. A novel denoising method for CT images based on U-Net and multi-attention. Comput Biol Med. 2023;152:106387. doi:10.1016/j.compbiomed.2022.106387. [Google Scholar] [PubMed] [CrossRef]
194. Geng M, Meng X, Yu J, Zhu L, Jin L, Jiang Z, et al. Content-noise complementary learning for medical image denoising. IEEE Trans Med Imaging. 2022;41(2):407–19. doi:10.1109/tmi.2021.3113365. [Google Scholar] [PubMed] [CrossRef]
195. Huang H, Zhang C, Zhao L, Ding S, Wang H, Wu H. Self-supervised medical image denoising based on WISTA-net for human healthcare in metaverse. IEEE J Biomed Health Inform. 2024;28(11):6329–37. doi:10.1109/JBHI.2023.3278538. [Google Scholar] [PubMed] [CrossRef]
196. Annavarapu A, Borra S. An adaptive watershed segmentation based medical image denoising using deep convolutional neural networks. Biomed Signal Process Control. 2024;93(1):106119. doi:10.1016/j.bspc.2024.106119. [Google Scholar] [CrossRef]
197. Atal DK. Optimal deep CNN-based vectorial variation filter for medical image denoising. J Digit Imag. 2023;36(3):1216–36. doi:10.1007/s10278-022-00768-8. [Google Scholar] [PubMed] [CrossRef]
198. Ma Y, Yan Q, Liu Y, Liu J, Zhang J, Zhao Y. StruNet: perceptual and low-rank regularized transformer for medical image denoising. Med Phys. 2023;50(12):7654–69. doi:10.1002/mp.16550. [Google Scholar] [PubMed] [CrossRef]
199. Pan S, Wang T, Qiu RLJ, Axente M, Chang CW, Peng J, et al. 2D medical image synthesis using transformer-based denoising diffusion probabilistic model. Phys Med Biol. 2023;68(10):105004. doi:10.1088/1361-6560/acca5c. [Google Scholar] [PubMed] [CrossRef]
200. Jang SI, Pan T, Li Y, Heidari P, Chen J, Li Q, et al. Spach transformer: spatial and channel-wise transformer based on local and global self-attentions for PET image denoising. IEEE Trans Med Imaging. 2024;43(6):2036–49. doi:10.1109/tmi.2023.3336237. [Google Scholar] [PubMed] [CrossRef]
201. Luthra A, Sulakhe H, Mittal T, Iyer A, Yadav S. Eformer: edge enhancement based transformer for medical image denoising. arXiv:2109.08044. 2021. [Google Scholar]
202. Demir B, Liu Y, Chen X, Chen EZ, Zhao L, Mailhe B, et al. DiffDenoise: self-supervised medical image denoising with conditional diffusion models. arXiv:2504.00264. 2025. [Google Scholar]
203. Chen W, Xu T, Zhou W. Task-based regularization in penalized least-squares for binary signal detection tasks in medical image denoising. In: Medical Imaging 2025: Image Perception, Observer Performance, and Technology Assessment; 2025 Feb 16–21; San Diego, CA, USA. doi:10.1117/12.3048886. [Google Scholar] [CrossRef]
204. Kathiravan M, Shyamala Devi R, Mary MMJ, Anandha Baskar A, Priyan RG, Gokulanathan J. Hybrid deep learning approach for medical image denoising with efficient NetB3 and Pix2Pi. In: 2025 IEEE International Students’ Conference on Electrical, Electronics and Computer Science (SCEECS); 2025 Jan 18–19; Bhopal, India. p. 1–6. doi:10.1109/SCEECS64059.2025.10940920. [Google Scholar] [CrossRef]
205. Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–49. doi:10.1109/tpami.2016.2577031. [Google Scholar] [PubMed] [CrossRef]
206. Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y. OverFeat: integrated recognition, localization and detection using convolutional networks. In: 2nd International Conference on Learning Representations, ICLR 2014; 2014 Apr 14–16; Banff, AB, Canada. [Google Scholar]
207. Ciompi F, de Hoop B, van Riel SJ, Chung K, Scholten ET, Oudkerk M, et al. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med Image Anal. 2015;26(1):195–202. doi:10.1016/j.media.2015.08.001. [Google Scholar] [PubMed] [CrossRef]
208. Redmon J, Farhadi A. YOLO9000: better, faster, stronger. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI, USA. p. 6517–25. doi:10.1109/cvpr.2017.690. [Google Scholar] [CrossRef]
209. Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. p. 2999–3007. doi:10.1109/ICCV.2017.324. [Google Scholar] [CrossRef]
210. Dai J, Li Y, He K, Sun J. R-FCN: object detection via region-based fully convolutional networks. In: 30th Conference on Neural Information Processing Systems (NIPS 2016); 2016 Dec 5–10; Barcelona, Spain. p. 379–87. doi:10.48550/arXiv.1605.06409. [Google Scholar] [CrossRef]
211. Liu Y, Ma Z, Liu X, Ma S, Ren K. Privacy-preserving object detection for medical images with faster R-CNN. IEEE Trans Inform Forensic Secur. 2022;17:69–84. doi:10.1109/tifs.2019.2946476. [Google Scholar] [CrossRef]
212. Newell A, Huang Z, Deng J. Associative embedding: end-to-end learning for joint detection and grouping. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 2274–84. [Google Scholar]
213. Tychsen-Smith L, Petersson L. DeNet: scalable real-time object detection with directed sparse sampling. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. IEEE; 2017. p. 428–36. doi:10.1109/ICCV.2017.54. [Google Scholar] [CrossRef]
214. Duan K, Bai S, Xie L, Qi H, Huang Q, Tian Q. CenterNet: keypoint triplets for object detection. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27–Nov 2; Seoul, Republic of Korea. p. 6568–77. doi:10.1109/iccv.2019.00667. [Google Scholar] [CrossRef]
215. Gu Y, Lu X, Yang L, Zhang B, Yu D, Zhao Y, et al. Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput Biol Med. 2018;103:220–31. doi:10.1016/j.compbiomed.2018.10.011. [Google Scholar] [PubMed] [CrossRef]
216. Xie H, Yang D, Sun N, Chen Z, Zhang Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019;85:109–19. doi:10.1016/j.patcog.2018.07.031. [Google Scholar] [CrossRef]
217. Akselrod-Ballin A, Karlinsky L, Hazan A, Bakalo R, Horesh AB, Shoshan Y, et al. Deep learning for automatic detection of abnormal findings in breast mammography. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham, Switzerland: Springer; 2017. p. 321–9. doi:10.1007/978-3-319-67558-9_37. [Google Scholar] [CrossRef]
218. Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with Deep Learning. Sci Rep. 2018;8(1):4165. doi:10.1038/s41598-018-22437-z. [Google Scholar] [PubMed] [CrossRef]
219. Xu Z, Li T, Liu Y, Zhan Y, Chen J, Lukasiewicz T. PAC-Net: multi-pathway FPN with position attention guided connections and vertex distance IoU for 3D medical image detection. Front Bioeng Biotechnol. 2023;11:1049555. doi:10.3389/fbioe.2023.1049555. [Google Scholar] [PubMed] [CrossRef]
220. Yan K, Bagheri M, Summers RM. 3D context enhanced region-based convolutional neural network for end-to-end lesion detection. In: Medical image computing and computer assisted intervention—MICCAI 2018. Cham, Switzerland: Springer International Publishing; 2018. p. 511–9. doi:10.1007/978-3-030-00928-1_58. [Google Scholar] [CrossRef]
221. Tao Q, Ge Z, Cai J, Yin J, See S. Improving deep lesion detection using 3D contextual and spatial attention. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 185–93. doi:10.1007/978-3-030-32226-7_21. [Google Scholar] [CrossRef]
222. Ding J, Li A, Hu Z, Wang L. Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In: Medical image computing and computer assisted intervention—MICCAI 2017. Cham, Switzerland: Springer International Publishing; 2017. p. 559–67. doi:10.1007/978-3-319-66179-7_64. [Google Scholar] [CrossRef]
223. Setio AAA, Traverso A, de Bel T, Berens MSN, van den Bogaard C, Cerello P, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal. 2017;42(4):1–13. doi:10.1016/j.media.2017.06.015. [Google Scholar] [PubMed] [CrossRef]
224. Zhu W, Liu C, Fan W, Xie X. DeepLung: deep 3D dual path nets for automated pulmonary nodule detection and classification. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV); 2018 Mar 12–15; Lake Tahoe, NV, USA. p. 673–81. doi:10.1109/WACV.2018.00079. [Google Scholar] [CrossRef]
225. Berthelot D, Carlini N, Goodfellow I, Oliver A, Papernot N, Nicolas P, et al. MixMatch: a holistic approach to semi-supervised learning. In: Proceedings of the 33rd International Conference on Neural Information Processing Systems; 2019 Dec 8–14; Vancouver, BC, Canada. p. 5049–59. [Google Scholar]
226. Wang D, Zhang Y, Zhang K, Wang L. FocalMix: semi-supervised learning for 3D medical image detection. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13–19; Seattle, WA, USA. p. 3950–9. doi:10.1109/cvpr42600.2020.00401. [Google Scholar] [CrossRef]
227. Ozdemir O, Woodward B, Berlin AA. Propagating uncertainty in multi-stage Bayesian convolutional neural networks with application to pulmonary nodule detection. arXiv:1712.00497. 2017. [Google Scholar]
228. Kendall A, Gal Y. What uncertainties do we need in bayesian deep learning for computer vision? In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 5580–90. [Google Scholar]
229. Baumgartner CF, Kamnitsas K, Matthew J, Fletcher TP, Smith S, Koch LM, et al. SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans Med Imaging. 2017;36(11):2204–15. doi:10.1109/tmi.2017.2712367. [Google Scholar] [PubMed] [CrossRef]
230. Zhu X, Su W, Lu L, Li B, Wang X, Dai J. Deformable DETR: deformable transformers for end-to-end object detection. In: ICLR 2021—9th International Conference on Learning Representations; 2021 May 5; Vienna, Austria. [Google Scholar]
231. Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, et al. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging. 2016;35(5):1182–95. doi:10.1109/tmi.2016.2528129. [Google Scholar] [PubMed] [CrossRef]
232. Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Išgum I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal. 2016;34(4):123–36. doi:10.1016/j.media.2016.04.004. [Google Scholar] [PubMed] [CrossRef]
233. Yan K, Cai J, Zheng Y, Harrison AP, Jin D, Tang Y, et al. Learning from multiple datasets with heterogeneous and partial labels for universal lesion detection in CT. IEEE Trans Med Imaging. 2021;40(10):2759–70. doi:10.1109/tmi.2020.3047598. [Google Scholar] [PubMed] [CrossRef]
234. Pisov M, Kondratenko V, Zakharov A, Petraikin A, Gombolevskiy V, Morozov S, et al. Keypoints localization for joint vertebra detection and fracture severity quantification. In: Medical image computing and computer assisted intervention—MICCAI 2020. Cham, Switzerland: Springer; 2020. p. 723–32. doi:10.1007/978-3-030-59725-2_70. [Google Scholar] [CrossRef]
235. Lung KY, Chang CR, Weng SE, Lin HS, Shuai HH, Cheng WH. ROSNet: robust one-stage network for CT lesion detection. Pattern Recognit Lett. 2021;144(7):82–8. doi:10.1016/j.patrec.2021.01.011. [Google Scholar] [CrossRef]
236. Zhu H, Yao Q, Xiao L, Zhou SK. You only learn once: universal anatomical landmark detection. In: Medical image computing and computer assisted intervention—MICCAI 2021. Cham, Switzerland: Springer International Publishing; 2021. p. 85–95. doi:10.1007/978-3-030-87240-3_9. [Google Scholar] [CrossRef]
237. Cai Y, Chen H, Yang X, Zhou Y, Cheng KT. Dual-distribution discrepancy with self-supervised refinement for anomaly detection in medical images. Med Image Anal. 2023;86:102794. doi:10.1016/j.media.2023.102794. [Google Scholar] [PubMed] [CrossRef]
238. Chen X, You S, Tezcan KC, Konukoglu E. Unsupervised lesion detection via image restoration with a normative prior. Med Image Anal. 2020;64:101713. doi:10.1016/j.media.2020.101713. [Google Scholar] [PubMed] [CrossRef]
239. Li Z, Zhang S, Zhang J, Huang K, Wang Y, Yu Y. MVP-net: multi-view FPN with position-aware attention for deep universal lesion detection. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 13–21. doi:10.1007/978-3-030-32226-7_2. [Google Scholar] [CrossRef]
240. Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In: Information processing in medical imaging. Cham, Switzerland: Springer International Publishing; 2017. p. 146–57. doi:10.1007/978-3-319-59050-9_12. [Google Scholar] [CrossRef]
241. Schlegl T, Seeböck P, Waldstein SM, Langs G, Schmidt-Erfurth U. F-AnoGAN: fast unsupervised anomaly detection with generative adversarial networks. Med Image Anal. 2019;54(3):30–44. doi:10.1016/j.media.2019.01.010. [Google Scholar] [PubMed] [CrossRef]
242. Baur C, Wiestler B, Albarqouni S, Navab N. Deep autoencoding models for unsupervised anomaly segmentation in brain MR images. In: Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Cham, Switzerland: Springer International Publishing; 2019. p. 161–9. doi:10.1007/978-3-030-11723-8_16. [Google Scholar] [CrossRef]
243. Chen X, Pawlowski N, Glocker B, Konukoglu E. Normative ascent with local Gaussians for unsupervised lesion detection. Med Image Anal. 2021;74:102208. doi:10.1016/j.media.2021.102208. [Google Scholar] [PubMed] [CrossRef]
244. Uzunova H, Schultz S, Handels H, Ehrhardt J. Unsupervised pathology detection in medical images using conditional variational autoencoders. Int J Comput Assist Radiol Surg. 2019;14(3):451–61. doi:10.1007/s11548-018-1898-0. [Google Scholar] [PubMed] [CrossRef]
245. Tang YB, Yan K, Tang YX, Liu J, Xiao J, Summers RM. Uldor: a universal lesion detector for ct scans with pseudo masks and hard negative example mining. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019 Apr 8–11; Venice, Italy. p. 833–6. doi:10.1109/isbi.2019.8759478. [Google Scholar] [CrossRef]
246. Yan K, Tang Y, Peng Y, Sandfort V, Bagheri M, Lu Z, et al. MULAN: multitask universal lesion analysis network for joint lesion detection, tagging, and segmentation. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 194–202. doi:10.1007/978-3-030-32226-7_22. [Google Scholar] [CrossRef]
247. Bi Y, Huang L, Clarenbach R, Ghotbi R, Karlas A, Navab N, et al. Synomaly noise and multi-stage diffusion: a novel approach for unsupervised anomaly detection in medical images. Med Image Anal. 2025;105(1):103737. doi:10.1016/j.media.2025.103737. [Google Scholar] [PubMed] [CrossRef]
248. Hoover R, Elsayed N, ElSayed Z, Li C. Pre-trained under noise: a framework for robust bone fracture detection in medical imaging. arXiv:2507.09731. 2025. [Google Scholar]
249. Isozaki A, Harmon J, Zhou Y, Li S, Nakagawa Y, Hayashi M, et al. AI on a chip. Lab Chip. 2020;20(17):3074–90. doi:10.1039/d0lc00521e. [Google Scholar] [PubMed] [CrossRef]
250. Yu M, Guo M, Zhang S, Zhan Y, Zhao M, Lukasiewicz T, et al. RIRGAN: an end-to-end lightweight multi-task learning method for brain MRI super-resolution and denoising. Comput Biol Med. 2023;167:107632. doi:10.1016/j.compbiomed.2023.107632. [Google Scholar] [PubMed] [CrossRef]
251. Guerreiro J, Tomás P, Garcia N, Aidos H. Super-resolution of magnetic resonance images using Generative Adversarial Networks. Comput Med Imag Graph. 2023;108(1):102280. doi:10.1016/j.compmedimag.2023.102280. [Google Scholar] [PubMed] [CrossRef]
252. Qiu D, Zheng L, Zhu J, Huang D. Multiple improved residual networks for medical image super-resolution. Future Gener Comput Syst. 2021;116(1):200–8. doi:10.1016/j.future.2020.11.001. [Google Scholar] [CrossRef]
253. Gu Y, Zeng Z, Chen H, Wei J, Zhang Y, Chen B, et al. MedSRGAN: medical images super-resolution using generative adversarial networks. Multimed Tools Appl. 2020;79(29–30):21815–40. doi:10.1007/s11042-020-08980-w. [Google Scholar] [CrossRef]
254. Ahmad W, Ali H, Shah Z, Azmat S. A new generative adversarial network for medical images super resolution. Sci Rep. 2022;12(1):9533. doi:10.1038/s41598-022-13658-4. [Google Scholar] [PubMed] [CrossRef]
255. Wang C, Lv X, Shao M, Qian Y, Zhang Y. A novel fuzzy hierarchical fusi on attention convolution neural network for medical image super-resolution reconstruction. Inf Sci. 2023;622:424–36. doi:10.1016/j.ins.2022.11.140. [Google Scholar] [CrossRef]
256. Ren S, Guo K, Ma J, Zhu F, Hu B, Zhou H. Realistic medical image super-resolution with pyramidal feature multi-distillation networks for intelligent healthcare systems. Neural Comput Appl. 2023;35(31):22781–96. doi:10.1007/s00521-021-06287-x. [Google Scholar] [PubMed] [CrossRef]
257. Zhu J, Yang G, Lio P. A residual dense vision transformer for medical image super-resolution with segmentation-based perceptual loss fine-tuning. arXiv:2302.11184. 2023. [Google Scholar]
258. Sun J, Zeng X, Lei X, Gao M, Li Q, Zhang H, et al. Medical image super-resolution via transformer-based hierarchical encoder-decoder network. Netw Model Anal Health Inform Bioinforma. 2024;13(1):34. doi:10.1007/s13721-024-00469-y. [Google Scholar] [CrossRef]
259. Du W, Tian S. Transformer and GAN-based super-resolution reconstruction network for medical images. Tsinghua Sci Technol. 2024;29(1):197–206. doi:10.26599/TST.2022.9010071. [Google Scholar] [CrossRef]
260. Goyal B, Lepcha DC, Dogra A, Wang SH. A weighted least squares optimisation strategy for medical image super resolution via multiscale convolutional neural networks for healthcare applications. Complex Intell Syst. 2022;8(4):3089–104. doi:10.1007/s40747-021-00465-z. [Google Scholar] [CrossRef]
261. Lu H, Mei J, Qiu Y, Li Y, Hao F, Xu J, et al. Information sparsity guided transformer for multi-modal medical image super-resolution. Expert Syst Appl. 2025;261:125428. doi:10.1016/j.eswa.2024.125428. [Google Scholar] [CrossRef]
262. Pang K, Zhao K, Hung ALY, Zheng H, Yan R, Sung K. NExpR: neural Explicit Representation for fast arbitrary-scale medical image super-resolution. Comput Biol Med. 2025;184:109354. doi:10.1016/j.compbiomed.2024.109354. [Google Scholar] [PubMed] [CrossRef]
263. Ji Z, Zou B, Kui X, Li H, Vera P, Ruan S. Generation of super-resolution for medical image via a self-prior guided Mamba network with edge-aware constraint. Pattern Recognit Lett. 2025;187:93–9. doi:10.1016/j.patrec.2024.11.020. [Google Scholar] [CrossRef]
264. Li Y, Hao W, Zeng H, Wang L, Xu J, Routray S, et al. Cross-scale texture supplementation for reference-based medical image super-resolution. IEEE J Biomed Health Inform. 2025:1–15. doi:10.1109/jbhi.2025.3572502. [Google Scholar] [PubMed] [CrossRef]
265. Basu S, Singhal S, Singh D. A systematic literature review on multimodal medical image fusion. Multimed Tools Appl. 2024;83(6):15845–913. doi:10.1007/s11042-023-15913-w. [Google Scholar] [CrossRef]
266. Goyal B, Dogra A, Lepcha DC, Koundal D, Alhudhaif A, Alenezi F, et al. Multi-modality image fusion for medical assistive technology management based on hybrid domain filtering. Expert Syst Appl. 2022;209:118283. doi:10.1016/j.eswa.2022.118283. [Google Scholar] [CrossRef]
267. Lepcha DC, Dogra A, Goyal B, Chohan JS, Koundal D, Zaguia A, et al. Multimodal medical image fusion based on pixel significance using anisotropic diffusion and cross bilateral filter. Hum Centric Comput Inf Sci. 2022;12:13. doi:10.22967/HCIS.2022.12.015. [Google Scholar] [CrossRef]
268. Li Y, Zhao J, Lv Z, Li J. Medical image fusion method by deep learning. Int J Cogn Comput Eng. 2021;2:21–9. doi:10.1016/j.ijcce.2020.12.004. [Google Scholar] [CrossRef]
269. Li W, Li R, Fu J, Peng X. MSENet: a multi-scale enhanced network based on unique features guidance for medical image fusion. Biomed Signal Process Control. 2022;74:103534. doi:10.1016/j.bspc.2022.103534. [Google Scholar] [CrossRef]
270. Zhang H, Ma J. SDNet: a versatile squeeze-and-decomposition network for real-time image fusion. Int J Comput Vis. 2021;129(10):2761–85. doi:10.1007/s11263-021-01501-8. [Google Scholar] [CrossRef]
271. Xu H, Ma J. EMFusion: an unsupervised enhanced medical image fusion network. Inf Fusion. 2021;76:177–86. doi:10.1016/j.inffus.2021.06.001. [Google Scholar] [CrossRef]
272. Ye S, Wang T, Ding M, Zhang X. F-DARTS: foveated differentiable architecture search based multimodal medical image fusion. IEEE Trans Med Imaging. 2023;42(11):3348–61. doi:10.1109/tmi.2023.3283517. [Google Scholar] [PubMed] [CrossRef]
273. Zhou T, Li Q, Lu H, Zhang X, Cheng Q. Hybrid multimodal medical image fusion method based on LatLRR and ED-D2GAN. Appl Sci. 2022;12(24):12758. doi:10.3390/app122412758. [Google Scholar] [CrossRef]
274. Li Z, Pan H, Zhang K, Wang Y, Yu F. MambaDFuse: a mamba-based dual-phase model for multi-modality image fusion. arXiv:2404.08406. 2024. [Google Scholar]
275. Tang W, He F, Liu Y, Duan Y. MATR: multimodal medical image fusion via multiscale adaptive transformer. IEEE Trans Image Process. 2022;31:5134–49. doi:10.1109/TIP.2022.3193288. [Google Scholar] [PubMed] [CrossRef]
276. Song Y, Dai Y, Liu W, Liu Y, Liu X, Yu Q, et al. DesTrans: a medical image fusion method based on Transformer and improved DenseNet. Comput Biol Med. 2024;174:108463. doi:10.1016/j.compbiomed.2024.108463. [Google Scholar] [PubMed] [CrossRef]
277. He D, Li W, Wang G, Huang Y, Liu S. MMIF-INet: multimodal medical image fusion by invertible network. Inf Fusion. 2025;114:102666. doi:10.1016/j.inffus.2024.102666. [Google Scholar] [CrossRef]
278. Li H, Su D, Cai Q, Zhang Y. BSAFusion: a bidirectional stepwise feature alignment network for unaligned medical image fusion. Proc AAAI Conf Artif Intell. 2025;39(5):4725–33. doi:10.1609/aaai.v39i5.32499. [Google Scholar] [CrossRef]
279. Dinh PH. MIF-BTF-MRN: medical image fusion based on the bilateral texture filter and transfer learning with the ResNet-101 network. Biomed Signal Process Control. 2025;100:106976. doi:10.1016/j.bspc.2024.106976. [Google Scholar] [CrossRef]
280. Liu S, Li W, He D, Wang G, Huang Y. SSEFusion: salient semantic enhancement for multimodal medical image fusion with Mamba and dynamic spiking neural networks. Inf Fusion. 2025;119:103031. doi:10.1016/j.inffus.2025.103031. [Google Scholar] [CrossRef]
281. Rundo L, Tangherloni A, Nobile MS, Militello C, Besozzi D, Mauri G, et al. MedGA: a novel evolutionary method for image enhancement in medical imaging systems. Expert Syst Appl. 2019;119:387–99. doi:10.1016/j.eswa.2018.11.013. [Google Scholar] [CrossRef]
282. He C, Li K, Xu G, Yan J, Tang L, Zhang Y, et al. HQG-net: unpaired medical image enhancement with high-quality guidance. IEEE Trans Neural Netw Learning Syst. 2024;35(12):18404–18. doi:10.1109/tnnls.2023.3315307. [Google Scholar] [PubMed] [CrossRef]
283. Alenezi F, Santosh KC. Geometric regularized Hopfield neural network for medical image enhancement. Int J Biomed Imaging. 2021;2021:6664569. doi:10.1155/2021/6664569. [Google Scholar] [PubMed] [CrossRef]
284. Ghandour C, El-Shafai W, El-Rabaie S. Medical image enhancement algorithms using deep learning-based convolutional neural network. J Opt. 2023;52(4):1931–41. doi:10.1007/s12596-022-01078-6. [Google Scholar] [CrossRef]
285. Ma Y, Liu J, Liu Y, Fu H, Hu Y, Cheng J, et al. Structure and illumination constrained GAN for medical image enhancement. IEEE Trans Med Imaging. 2021;40(12):3955–67. doi:10.1109/tmi.2021.3101937. [Google Scholar] [PubMed] [CrossRef]
286. Zhong G, Ding W, Chen L, Wang Y, Yu YF. Multi-scale attention generative adversarial network for medical image enhancement. IEEE Trans Emerg Top Comput Intell. 2023;7(4):1113–25. doi:10.1109/TETCI.2023.3243920. [Google Scholar] [CrossRef]
287. Yu YF, Zhong G, Zhou Y, Chen L. FS-GAN: fuzzy Self-guided structure retention generative adversarial network for medical image enhancement. Inf Sci. 2023;642:119114. doi:10.1016/j.ins.2023.119114. [Google Scholar] [CrossRef]
288. Xia K, Wang J. Recent advances of Transformers in medical image analysis: a comprehensive review. MedComm Future Medicine. 2023 Mar;2(1):e38. doi:10.1002/MEF2.38. [Google Scholar] [CrossRef]
289. Lei M, Wu H, Lv X, Wang X. ConDSeg: a general medical image segmentation framework via contrast-driven feature enhancement. Proc AAAI Conf Artif Intell. 2025;39(5):4571–9. doi:10.1609/aaai.v39i5.32482. [Google Scholar] [CrossRef]
290. Haskins G, Kruecker J, Kruger U, Xu S, Pinto PA, Wood BJ, et al. Learning deep similarity metric for 3D MR-TRUS image registration. Int J CARS. 2019;14(3):417–25. doi:10.1007/s11548-018-1875-7. [Google Scholar] [PubMed] [CrossRef]
291. Uzunova H, Wilms M, Handels H, Ehrhardt J. Training CNNs for image registration from few samples with model-based data augmentation. In: Medical image computing and computer assisted intervention—MICCAI 2017. Cham, Switzerland: Springer International Publishing; 2017. p. 223–31. doi:10.1007/978-3-319-66182-7_26. [Google Scholar] [CrossRef]
292. Fan J, Cao X, Yap PT, Shen D. BIRNet: brain image registration using dual-supervised fully convolutional networks. Med Image Anal. 2019;54:193–206. doi:10.1016/j.media.2019.03.006. [Google Scholar] [PubMed] [CrossRef]
293. Tian L, Greer H, Kwitt R, Vialard FX, San José Estépar R, Bouix S, et al. uniGradICON: a foundation model for medical image registration. In: Medical image computing and computer assisted intervention—MICCAI 2024. Cham, Switzerland: Springer Nature Switzerland; 2024. p. 749–60. doi:10.1007/978-3-031-72069-7_70. [Google Scholar] [CrossRef]
294. Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imag. 2019;38(8):1788–800. doi:10.1109/TMI.2019.2897538. [Google Scholar] [PubMed] [CrossRef]
295. Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, et al. Weakly-supervised convolutional neural networks for multimodal image registration. Med Image Anal. 2018;49:1–13. doi:10.1016/j.media.2018.07.002. [Google Scholar] [PubMed] [CrossRef]
296. Hu Y, Modat M, Gibson E, Ghavami N, Bonmati E, Moore CM, et al. Label-driven weakly-supervised learning for multimodal deformarle image registration. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 Apr 4–7; Washington, DC, USA. p. 1070–4. doi:10.1109/isbi.2018.8363756. [Google Scholar] [CrossRef]
297. de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal. 2019;52:128–43. doi:10.1016/j.media.2018.11.010. [Google Scholar] [PubMed] [CrossRef]
298. Zhao S, Lau T, Luo J, Chang EI, Xu Y. Unsupervised 3D end-to-end medical image registration with volume tweening network. IEEE J Biomed Health Inform. 2020;24(5):1394–404. doi:10.1109/JBHI.2019.2951024. [Google Scholar] [PubMed] [CrossRef]
299. Chen J, He Y, Frey EC, Li Y, Du Y. ViT-V-net: vision transformer for unsupervised volumetric medical image registration. arXiv:2104.06468. 2021. [Google Scholar]
300. Kim B, Kim J, Lee JG, Kim DH, Park SH, Ye JC. Unsupervised deformable image registration using cycle-consistent CNN. In: Medical image computing and computer assisted intervention—MICCAI 2019. Cham, Switzerland: Springer International Publishing; 2019. p. 166–74. doi:10.1007/978-3-030-32226-7_19. [Google Scholar] [CrossRef]
301. Chen J, Frey EC, He Y, Segars WP, Li Y, Du Y. TransMorph: transformer for unsupervised medical image registration. Med Image Anal. 2022;82:102615. doi:10.1016/j.media.2022.102615. [Google Scholar] [PubMed] [CrossRef]
302. Chen Z, Zheng Y, Gee JC. TransMatch: a transformer-based multilevel dual-stream feature matching network for unsupervised deformable image registration. IEEE Trans Med Imag. 2024;43(1):15–27. doi:10.1109/TMI.2023.3288136. [Google Scholar] [PubMed] [CrossRef]
303. Chen J, Lu D, Zhang Y, Wei D, Ning M, Shi X, et al. Deformer: towards displacement field learning for unsupervised medical image registration. In: Medical image computing and computer assisted intervention—MICCAI 2022. Cham, Switzerland: Springer Nature Switzerland; 2022. p. 141–51. doi:10.1007/978-3-031-16446-0_14. [Google Scholar] [CrossRef]
304. Shi J, He YT, Kong YY, Coatrieux JL, Shu HZ, Yang GY, et al. XMorpher: full transformer for deformable medical image registration via cross attention. In: Medical image computing and computer assisted intervention – MICCAI 2022. Cham, Switzerland: Springer; 2022. p. 217–26. doi:10.1007/978-3-031-16446-0_21. [Google Scholar] [CrossRef]
305. Mok TCW, Chung ACS. Affine medical image registration with coarse-to-fine vision transformer. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18–24; New Orleans, LA, USA. p. 20803–12. doi:10.1109/CVPR52688.2022.02017. [Google Scholar] [CrossRef]
306. Meng M, Fulham M, Feng D, Bi L, Kim J. AutoFuse: automatic fusion networks for deformable medical image registration. Pattern Recognit. 2025;161(1):111338. doi:10.1016/j.patcog.2024.111338. [Google Scholar] [CrossRef]
307. Chen Y, Hu X, Lu T, Zou L, Liao X. A multi-scale large kernel attention with U-Net for medical image registration. J Supercomput. 2024;81(1):70. doi:10.1007/s11227-024-06489-9. [Google Scholar] [CrossRef]
308. Meyer S, Hu YC, Rimner A, Mechalakos J, Cerviño L, Zhang P. Deformable image registration uncertainty-encompassing dose accumulation for adaptive radiation therapy. Int J Radiat Oncol. 2025;122(4):818–26. doi:10.1016/j.ijrobp.2025.04.004. [Google Scholar] [PubMed] [CrossRef]
309. Jiang H, Imran M, Zhang T, Zhou Y, Liang M, Gong K, et al. Fast-DDPM: fast denoising diffusion probabilistic models for medical image-to-image generation. IEEE J Biomed Health Inform. 2025;29(10):7326–35. doi:10.1109/jbhi.2025.3565183. [Google Scholar] [PubMed] [CrossRef]
310. Ying HR, Lia YJ, Fu ZD. Domain adaptation and generalization using foundation models in healthcare imaging; 2025 Mar. doi:10.2139/SSRN.5345726. [Google Scholar] [CrossRef]
311. Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5(1):180161. doi:10.1038/sdata.2018.161. [Google Scholar] [PubMed] [CrossRef]
312. Benčević M, Habijan M, Galić I, Babin D, Pižurica A. Understanding skin color bias in deep learning-based skin lesion segmentation. Comput Methods Programs Biomed. 2024;245:108044. doi:10.1016/j.cmpb.2024.108044. [Google Scholar] [PubMed] [CrossRef]
313. Ranschaert ER, Morozov S, Algra PR. Artificial intelligence in medical imaging: opportunities, applications and risks. Cham, Switzerland: Springer; 2019 Jan. p. 1–373. doi:10.1007/978-3-319-94878-2/COVER. [Google Scholar] [CrossRef]
314. Fortin JP, Cullen N, Sheline YI, Taylor WD, Aselcioglu I, Cook PA, et al. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage. 2018;167:104–20. doi:10.1016/j.neuroimage.2017.11.024. [Google Scholar] [PubMed] [CrossRef]
315. El-Gazzar A, Thomas RM, van Wingen G. Harmonization techniques for machine learning studies using multi-site functional MRI data. bioRxiv. 2023. doi:10.1101/2023.06.14.544758. [Google Scholar] [CrossRef]
316. Zhang L, Wang X, Yang D, Sanford T, Harmon S, Turkbey B, et al. Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans Med Imaging. 2020;39(7):2531–40. doi:10.1109/tmi.2020.2973595. [Google Scholar] [PubMed] [CrossRef]
317. Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep. 2020;10(1):12598. doi:10.1038/s41598-020-69250-1. [Google Scholar] [PubMed] [CrossRef]
318. Li X, Jiang M, Zhang X, Kamp M, Dou Q. FedBN: federated learning on Non-IID features via local batch normalization. In: ICLR 2021—9th International Conference on Learning Representations; 2021 May 3–7; Virtual Event, Austria. [Google Scholar]
319. Li T, Sahu AK, Zaheer M, Sanjabi M, Talwalkar A, Smith V. Federated optimization in heterogeneous networks. In: Proceedings of Machine Learning and Systems; 2020 Mar 2–4; Austin, TX, USA. p. 429–50. [Google Scholar]
320. Liu Q, Chen C, Qin J, Dou Q, Heng PA. FedDG: federated Doma in generalization on medical image segmentation via episodic learning in continuous frequency space. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20–25; Nashville, TN, USA. p. 1013–23. doi:10.1109/cvpr46437.2021.00107. [Google Scholar] [CrossRef]
321. Bonawitz K, Eichner H, Grieskamp W, Huba D, Ingerman A, Ivanov V, et al. Towards federated learning at scale: system design. In: Proceedings of Machine Learning and Systems; Palo Alto, CA, USA. p. 374–888. doi:10.48550/arXiv.1902.01046. [Google Scholar] [CrossRef]
322. Tejani AS, Klontzas ME, Gatti AA, Mongan JT, Moy L, Park SH, et al. Checklist for artificial intelligence in medical imaging (CLAIM2024 update. Radiol Artif Intell. 2024 Jul;6(4):240300. doi:10.1148/RYAI.240300/ASSET/IMAGES/LARGE/RYAI.240300.TBL1.JPEG. [Google Scholar] [CrossRef]
323. Group The CONSORT-AI and SPIRIT-AI Steering. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat Med. 2019;25(10):1467–8. doi:10.1038/s41591-019-0603-3. [Google Scholar] [PubMed] [CrossRef]
324. Finn C, Abbeel P, Levine S. Model-agnostic meta-learning for fast adaptation of deep networks. In: Proceedings of the 34th International Conference on Machine Learning; 2017 Aug 6–11; Sydney, NSW, Australia. p. 1126–35. [Google Scholar]
325. Seyyed-Kalantari L, Liu G, McDermott M, Chen IY, Ghassemi M. CheXclusion: fairness gaps in deep chest X-ray classifiers. In: Biocomputing 2021: Proceedings of the Pacific Symposium. Kohala Coast, HI, USA: World Scientific; 2020. p. 232–43. doi:10.1142/9789811232701_0022. [Google Scholar] [CrossRef]
326. Maier-Hein L, Reinke A, Godau P, Tizabi MD, Buettner F, Christodoulou E, et al. Metrics reloaded: recommendations for image analysis validation. Nat Methods. 2024;21(2):195–212. doi:10.1038/s41592-023-02151-z. [Google Scholar] [PubMed] [CrossRef]
327. Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imag. 2015;15(1):29. doi:10.1186/s12880-015-0068-x. [Google Scholar] [PubMed] [CrossRef]
328. Minderer M, Djolonga J, Romijnders R, Hubis F, Zhai X, Houlsby N, et al. Revisiting the calibration of modern neural networks. In: NIPS'21: Proceedings of the 35th International Conference on Neural Information Processing Systems; 2021 Dec 6–14, Red Hook, NY, USA. p. 15682–94. [Google Scholar]
329. Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv:2003.10769. 2020. [Google Scholar]
330. Food and Drug Administration. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD). 2019 [cited 2025 Oct 23]. Available from: https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm514737.pdf. [Google Scholar]
331. Neubauer R, Schröttner J, Baumgartner C. Safety requirements for medical devices in compliance with European standards. In: Medical devices and in vitro diagnostics. Cham, Switzerland: Springer International Publishing; 2022. p. 1–30. doi:10.1007/978-3-030-98743-5_4-1. [Google Scholar] [CrossRef]
332. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17(1):195. doi:10.1186/s12916-019-1426-2. [Google Scholar] [PubMed] [CrossRef]
333. Darwiesh A, El-Baz AH, Abualkishik AZ, Elhoseny M. Artificial intelligence model for risk management in healthcare institutions: towards sustainable development. Sustainability. 2023;15(1):420. doi:10.3390/su15010420. [Google Scholar] [CrossRef]
334. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. p. 618–26. doi:10.1109/iccv.2017.74. [Google Scholar] [CrossRef]
335. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 4768–77. [Google Scholar]
336. Holzinger A, Langs G, Denk H, Zatloukal K, Müller H. Causability and explainability of artificial intelligence in medicine. WIREs Data Min Knowl. 2019;9(4):e1312. doi:10.1002/widm.1312. [Google Scholar] [PubMed] [CrossRef]
337. Wachter S, Mittelstadt B, Russell C. Counterfactual explanations without opening the black box: automated decisions and the GDPR. Harvard J Law Technol (Harvard JOLT). 2017;31(2):841–87. [Google Scholar]
338. Savage CH, Abou Elkassem A, Hamki O, Sturdivant A, Benson D, Grumley S, et al. Prospective evaluation of artificial intelligence triage of incidental pulmonary emboli on contrast-enhanced CT examinations of the chest or abdomen. Am J Roentgenol. 2024;223(3):e2431067. doi:10.2214/ajr.24.31067. [Google Scholar] [PubMed] [CrossRef]
339. Buolamwini J, Gebru T. Gender shades: intersectional accuracy disparities in commercial gender classification. In: Conference on Fairness, Accountability and Transparency; 2018 Feb 2–3; New York, NY, USA. p. 77–91. [Google Scholar]
340. Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, et al. The future of digital health with federated learning. npj Digit Med. 2020;3(1):119. doi:10.1038/s41746-020-00323-1. [Google Scholar] [PubMed] [CrossRef]
341. Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: Simulation and synthesis in medical imaging. SASHIMI 2018. Cham, Switzerland: Springer; 2018. doi:10.1007/978-3-030-00536-8_1/TABLES/1. [Google Scholar] [CrossRef]
342. Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digit Med. 2018;1(1):39. doi:10.1038/s41746-018-0040-6. [Google Scholar] [PubMed] [CrossRef]
343. Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK, Ashrafian H, et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Lancet Digit Health. 2020;2(10):e537–48. doi:10.1016/S2589-7500(20)30218-1. [Google Scholar] [PubMed] [CrossRef]
344. Langlotz CP, Allen B, Erickson BJ, Kalpathy-Cramer J, Bigelow K, Cook TS, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/the academy workshop. Radiology. 2019;291(3):781–91. doi:10.1148/radiol.2019190613. [Google Scholar] [PubMed] [CrossRef]
345. Samek W, Wiegand T, Müller KR. Explainable artificial intelligence: understanding, visualizing and interpreting deep learning models. arXiv:1708.08296. 2017. [Google Scholar]
346. Bajwa J, Munir U, Nori A, Williams B. Artificial intelligence in healthcare: transforming the practice of medicine. Future Healthc J. 2021;8(2):e188–94. doi:10.7861/fhj.2021-0095. [Google Scholar] [PubMed] [CrossRef]
347. Kanca Gulsoy E, Ayas S, Baykal Kablan E, Ekinci M. Enhancing the adversarial robustness in medical image classification: exploring adversarial machine learning with vision transformers-based models. Neural Comput Appl. 2025;37(12):7971–89. doi:10.1007/s00521-024-10516-4. [Google Scholar] [CrossRef]
348. Armanious K, Jiang C, Fischer M, Küstner T, Hepp T, Nikolaou K, et al. MedGAN: medical image translation using GANs. Comput Med Imag Graph. 2020;79:101684. doi:10.1016/j.compmedimag.2019.101684. [Google Scholar] [PubMed] [CrossRef]
349. Ghesu FC, Georgescu B, Mansoor A, Yoo Y, Gibson E, Vishwanath RS, et al. Quantifying and leveraging predictive uncertainty for medical image assessment. Med Image Anal. 2021;68:101855. doi:10.1016/j.media.2020.101855. [Google Scholar] [PubMed] [CrossRef]
350. Hong Z, Yue Y, Chen Y, Cong L, Lin H, Luo Y, et al. Out-of-distribution detection in medical image analysis: a survey. arXiv:2404.18279. 2024. [Google Scholar]
351. Koçak B, Ponsiglione A, Stanzione A, Bluethgen C, Santinha J, Ugga L, et al. Bias in artificial intelligence for medical imaging: fundamentals, detection, avoidance, mitigation, challenges, ethics, and prospects. Bezmialem Sci. 2025; 31(2):75–88. doi:10.4274/dir.2024.242854. [Google Scholar] [PubMed] [CrossRef]
352. Galvez-Hernandez P, Kratz J. Annotator bias and its effect on deep learning segmentation of uncured composite micrographs. NDT E Int. 2024;144(3):103088. doi:10.1016/j.ndteint.2024.103088. [Google Scholar] [CrossRef]
353. Maleki F, Moy L, Forghani R, Ghosh T, Ovens K, Langer S, et al. RIDGE: reproducibility, integrity, dependability, generalizability, and efficiency assessment of medical image segmentation models. J Imaging Inform Med. 2025;38:2524–36. doi:10.1007/s10278-024-01282-9. [Google Scholar] [PubMed] [CrossRef]
354. Faghani S, Khosravi B, Zhang K, Moassefi M, Jagtap JM, Nugen F, et al. Mitigating bias in radiology machine learning: 3. performance metrics. Radiol Artif Intell. 2022;4(5):e220061. doi:10.1148/ryai.220061. [Google Scholar] [PubMed] [CrossRef]
355. Quadrianto N, Sharmanska V, Thomas O. Discovering fair representations in the data domain. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15–20; Long Beach, CA, USA. p. 8219–28. doi:10.1109/CVPR.2019.00842. [Google Scholar] [CrossRef]
356. Pierson E, Cutler DM, Leskovec J, Mullainathan S, Obermeyer Z. An algorithmic approach to reducing unexplained pain disparities in underserved populations. Nat Med. 2021;27(1):136–40. doi:10.1038/s41591-020-01192-7. [Google Scholar] [PubMed] [CrossRef]
357. Ghassemi M, Oakden-Rayner L, Beam AL. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health. 2021;3(11):e745–50. doi:10.1016/S2589-7500(21)00208-9. [Google Scholar] [PubMed] [CrossRef]
358. Chen RJ, Lu MY, Chen TY, Williamson DFK, Mahmood F. Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng. 2021;5(6):493–7. doi:10.1038/s41551-021-00751-8. [Google Scholar] [PubMed] [CrossRef]
359. Kindermans PJ, Hooker S, Adebayo J, Alber M, Schütt KT, Dähne S, et al. The (un)reliability of saliency methods. In: Explainable AI: interpreting, explaining and visualizing deep learning. Cham, Switzerland: Springer; 2019. p. 267–80. doi:10.1007/978-3-030-28954-6_14. [Google Scholar] [CrossRef]
360. Consortium TP, Amann J, Blasimme A, Vayena E, Frey D, Madai VI. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak. 2020;20(1):310. doi:10.1186/s12911-020-01332-6. [Google Scholar] [PubMed] [CrossRef]
361. Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28(1):31–8. doi:10.1038/s41591-021-01614-0. [Google Scholar] [PubMed] [CrossRef]
362. Holzinger A, Haibe-Kains B, Jurisica I. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur J Nucl Med Mol Imaging. 2019;46(13):2722–30. doi:10.1007/s00259-019-04382-9. [Google Scholar] [PubMed] [CrossRef]
363. Benjamens S, Dhunnoo P, Meskó B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. npj Digit Med. 2020;3(1):118. doi:10.1038/s41746-020-00324-0. [Google Scholar] [PubMed] [CrossRef]
364. Morley J, Floridi L, Kinsey L, Elhalal A. From what to how: an initial review of publicly available AI ethics tools, methods and research to translate principles into practices. Sci Eng Ethics. 2020;26(4):2141–68. doi:10.1007/s11948-019-00165-5. [Google Scholar] [PubMed] [CrossRef]
365. Ibrahim H, Liu X, Rivera SC, Moher D, Chan AW, Sydes MR, et al. Reporting guidelines for clinical trials of artificial intelligence interventions: the SPIRIT-AI and CONSORT-AI guidelines. Trials. 2021;22(1):11. doi:10.1186/s13063-020-04951-6. [Google Scholar] [PubMed] [CrossRef]
366. Viz.ai, the proven AI-powered care coordination platform. [cited 2025 Jul 27]. Available from: https://www.viz.ai/. [Google Scholar]
367. Aidoc Secures Landmark FDA Clearance for Foundation Model AI. [cited 2025 Jul 27]. Available from: https://www.aidoc.com/about/news/aidoc-secures-landmark-fda-clearance/. [Google Scholar]
368. Heartflow - Decisive Coronary Care. [cited 2025 Jul 27]. Available from: https://www.heartflow.com/. [Google Scholar]
369. Advanced Skin Cancer & Melanoma Detection Technology | DermaSensor. [cited 2025 Jul 27]. Available from: https://www.dermasensor.com/. [Google Scholar]
370. Oxipit | The world’s first autonomous radiology AI. [cited 2025 Jul 27]. Available from: https://oxipit.ai/. [Google Scholar]
371. Lunit - Conquer Cancer through AI. [cited 2025 Jul 27]. Available from: https://www.lunit.io/en. [Google Scholar]
372. Artificial Intelligence in Healthcare & Radiology | Quantib. [cited 2025 Jul 27]. Available from: https://www.quantib.com/. [Google Scholar]
373. Qure AI | AI assistance for Accelerated Healthcare. [cited 2025 Jul 27]. Available from: https://www.qure.ai/. [Google Scholar]
374. Suganyadevi S, Seethalakshmi V, Balasamy K. A review on deep learning in medical image analysis. Int J Multimed Inf Retr. 2022;11(1):19–38. doi:10.1007/s13735-021-00218-1. [Google Scholar] [PubMed] [CrossRef]
375. Jain G, Mittal D, Thakur D, Mittal MK. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybern Biomed Eng. 2020;40(4):1391–405. doi:10.1016/j.bbe.2020.08.008. [Google Scholar] [PubMed] [CrossRef]
376. Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi:10.1016/j.compbiomed.2020.103805. [Google Scholar] [PubMed] [CrossRef]
377. Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–40. doi:10.1007/s13246-020-00865-4. [Google Scholar] [PubMed] [CrossRef]
378. Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos Solitons Fractals. 2020;138:109944. doi:10.1016/j.chaos.2020.109944. [Google Scholar] [PubMed] [CrossRef]
379. Ouchicha C, Ammor O, Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest X-ray images. Chaos Solitons Fractals. 2020;140:110245. doi:10.1016/j.chaos.2020.110245. [Google Scholar] [PubMed] [CrossRef]
380. Sethy PK, Behera SK, Ratha PK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci. 2020;5(4):643–51. doi:10.33889/ijmems.2020.5.4.052. [Google Scholar] [CrossRef]
381. Jaiswal AK, Tiwari P, Kumar S, Gupta D, Khanna A, Rodrigues JJPC. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement. 2019;145:511–8. doi:10.1016/j.measurement.2019.05.076. [Google Scholar] [CrossRef]
382. Civit-Masot J, Luna-Perejón F, Domínguez Morales M, Civit A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci. 2020;10(13):4640. doi:10.3390/app10134640. [Google Scholar] [CrossRef]
383. Singh N, Sanjay P, Sonbhadra K, Agarwal S, Sonbhadra SK. COVID-19 epidemic analysis using machine learning and deep learning algorithms. medRxiv. 2020 Jun:2020.04.08.20057679. doi:10.1101/2020.04.08.20057679. [Google Scholar] [CrossRef]
384. Akselrod-Ballin A, Karlinsky L, Alpert S, Hasoul S, Ben-Ari R, Barkan E. A region based convolutional network for tumor detection and classification in breast mammography. In: Deep learning and data labeling for medical applications. Cham, Switzerland: Springer International Publishing; 2016. p. 197–205. doi:10.1007/978-3-319-46976-8_21. [Google Scholar] [CrossRef]
385. Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H. Visualizing and enhancing a deep learning framework using patients age and gender for chest X-ray image retrieval. In: Medical imaging 2016: computer-aided diagnosis. San Diego, CA, USA: SPIE; 2016. 978510 p. doi:10.1117/12.2217587. [Google Scholar] [CrossRef]
386. Andermatt S, Pezold S, Cattin P. Multi-dimensional gated recurrent units for the segmentation of biomedical 3D-data. In: Deep learning and data labeling for medical applications. Cham, Switzerland: Springer International Publishing; 2016. p. 142–51. doi:10.1007/978-3-319-46976-8_15. [Google Scholar] [CrossRef]
387. Li M, Jiang Y, Zhang Y, Zhu H. Medical image analysis using deep learning algorithms. Front Public Health. 2023;11:1273253. doi:10.3389/fpubh.2023.1273253. [Google Scholar] [PubMed] [CrossRef]
388. Istiake Sunny MA, Maswood MMS, Alharbi AG. Deep learning-based stock price prediction using LSTM and bi-directional LSTM model. In: 2020 2nd Novel Intelligent and Leading Emerging Sciences Conference (NILES); 2020 Oct 24–26; Giza, Egypt. p. 87–92. doi:10.1109/niles50944.2020.9257950. [Google Scholar] [CrossRef]
389. Dehghani F, Larijani A. A machine learning-jaya algorithm (Ml-Ijaya) approach for rapid optimization using high performance computing. 2023. doi: 10.2139/ssrn.4423338. [Google Scholar] [CrossRef]
390. Novotny A, Bebis G, Tavakkoli A, Nicolescu M. Ensembles of deep one-class classifiers for multi-class image classification. Mach Learn Appl. 2025;19:100621. doi:10.1016/j.mlwa.2025.100621. [Google Scholar] [CrossRef]
391. Li J, Yu Z, Du Z, Zhu L, Shen HT. A comprehensive survey on source-free domain adaptation. IEEE Trans Pattern Anal Mach Intell. 2024;46(8):5743–62. doi:10.1109/tpami.2024.3370978. [Google Scholar] [PubMed] [CrossRef]
392. Xiao M, Wu Y, Zuo G, Fan S, Yu H, Shaikh ZA, et al. Addressing overfitting problem in deep learning-based solutions for next generation data-driven networks. Wirel Commun Mob Comput. 2021;2021(1):8493795. doi:10.1155/2021/8493795. [Google Scholar] [CrossRef]
393. Ahmadi SS, Khotanlou H. Enhance support relation extraction accuracy using improvement of segmentation in RGB-D images. In: 2017 3rd International Conference on Pattern Recognition and Image Analysis (IPRIA); 2017 Apr 19–20; Shahrekord, Iran. p. 166–9. doi:10.1109/PRIA.2017.7983040. [Google Scholar] [CrossRef]
394. Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3(3):199–217. doi:10.1038/s42256-021-00307-0. [Google Scholar] [CrossRef]
395. Lambert B, Forbes F, Doyle S, Dehaene H, Dojat M. Trustworthy clinical AI solutions: a unified review of uncertainty quantification in Deep Learning models for medical image analysis. Artif Intell Med. 2024;150:102830. doi:10.1016/j.artmed.2024.102830. [Google Scholar] [PubMed] [CrossRef]
396. Chukwunweike JN, Yussuf M, Okusi O, Bakare TO, Abisola AJ. The role of deep learning in ensuring privacy integrity and security: applications in AI-driven cybersecurity solutions. World J Adv Res Rev. 2024;23(2):1778–90. doi:10.30574/wjarr.2024.23.2.2550. [Google Scholar] [CrossRef]
397. Ibrahim A, Thiruvady D, Schneider JG, Abdelrazek M. The challenges of leveraging threat intelligence to stop data breaches. Front Comput Sci. 2020;2:36. doi:10.3389/fcomp.2020.00036. [Google Scholar] [CrossRef]
398. Yang S, Li Q, Li W, Li X, Liu AA. Dual-level representation enhancement on characteristic and context for image-text retrieval. IEEE Trans Circuits Syst Video Technol. 2022;32(11):8037–50. doi:10.1109/tcsvt.2022.3182426. [Google Scholar] [CrossRef]
399. Liu AA, Zhai Y, Xu N, Nie W, Li W, Zhang Y. Region-aware image captioning via interaction learning. IEEE Trans Circuits Syst Video Technol. 2022;32(6):3685–96. doi:10.1109/tcsvt.2021.3107035. [Google Scholar] [CrossRef]
400. Wang Y, Xu N, Liu AA, Li W, Zhang Y. High-order interaction learning for image captioning. IEEE Trans Circuits Syst Video Technol. 2022;32(7):4417–30. doi:10.1109/tcsvt.2021.3121062. [Google Scholar] [CrossRef]
401. Lin Z, Wang H, Li S. Pavement anomaly detection based on transformer and self-supervised learning. Autom Constr. 2022;143(1):104544. doi:10.1016/j.autcon.2022.104544. [Google Scholar] [CrossRef]
402. Geras KJ, Mann RM, Moy L. Artificial intelligence for mammography and digital breast tomosynthesis: current concepts and future perspectives. Radiology. 2019;293(2):246–59. doi:10.1148/radiol.2019182627. [Google Scholar] [PubMed] [CrossRef]
403. Yang Z, Luo TG, Wang D, Hu ZQ, Gao J, Wang LW. Learning to navigate for fine-grained classification. In: Proceedings of the European Conference on Computer Vision (ECCV); 2018 Sep 8–14; Munich, Germany. p. 420–35. [Google Scholar]
404. Liu L, Ouyang W, Wang X, Fieguth P, Chen J, Liu X, et al. Deep learning for generic object detection: a survey. Int J Comput Vis. 2020;128(2):261–318. doi:10.1007/s11263-019-01247-4. [Google Scholar] [CrossRef]
405. Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham, Switzerland: Springer International Publishing; 2017. p. 240–8. doi:10.1007/978-3-319-67558-9_28. [Google Scholar] [CrossRef]
406. Abraham N, Khan NM. A novel focal tversky loss function with improved attention U-Net for lesion segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019 Apr 8–11; Venice, Italy. p. 683–7. doi:10.1109/isbi.2019.8759329. [Google Scholar] [CrossRef]
407. Kervadec H, Bouchtiba J, Desrosiers C, Granger E, Dolz J, Ayed IB. Boundary loss for highly unbalanced segmentation. In: Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning; 2019 Jul 8–10; London, UK. p. 285–96. [Google Scholar]
408. Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: self-attention convolutional neural network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans Med Imag. 2020;39(7):2289–301. doi:10.1109/TMI.2020.2968472. [Google Scholar] [PubMed] [CrossRef]
409. Dai Z, Yang Z, Yang F, Cohen WW, Salakhutdinov RR. Good semi-supervised learning that requires a bad GAN. In: NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017 Dec 4–9; Long Beach, CA, USA. p. 6513–23. [Google Scholar]
410. Karras T, Aittala M, Hellsten J, Laine S, Lehtinen J, Aila T. Training generative adversarial networks with limited data. In: NIPS'20: Proceedings of the 34th International Conference on Neural Information Processing Systems; 2020 Dec 6–12; Vancouver, BC, Canada. p. 12104–14. [Google Scholar]
411. Khosla P, Teterwak P, Wang C, Sarna A, Tian YL, Isola P, et al. Supervised contrastive learning. In: NIPS'20: Proceedings of the 34th International Conference on Neural Information Processing System; 2020 Dec 6–12; Vancouver, BC, Canada. p. 18661–73. [Google Scholar]
412. Saunshi N, Plevrakis O, Arora S, Khodak M, Khandeparkar H. A theoretical analysis of contrastive unsupervised representation learning. In: Proceedings of the 36th International Conference on Machine Learning; 2019 Jun 9–15; Long Beach, CA, USA. p. 5628–37. [Google Scholar]
413. Reed CJ, Yue X, Nrusimha A, Ebrahimi S, Vijaykumar V, Mao R, et al. Self-supervised pretraining improves self-supervised pretraining. In: 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV); 2022 Jan 3–8; Waikoloa, HI, USA. p. 1050–60. doi:10.1109/wacv51458.2022.00112. [Google Scholar] [CrossRef]
414. Cubuk ED, Zoph B, Shlens J, Le QV. Randaugment: practical automated data augmentation with a reduced search space. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2020 Jun 14–19; Seattle, WA, USA. p. 3008–17. doi:10.1109/cvprw50498.2020.00359. [Google Scholar] [CrossRef]
415. Oliver A, Odena A, Raffel C, Cubuk ED, Goodfellow Google Brain IJ. Realistic evaluation of deep semi-supervised learning algorithms. In: NIPS'18: Proceedings of the 32nd International Conference on Neural Information Processing Systems; 2018 Dec 3–8; Montréal, QC, Canada. p. 3239–50. [Google Scholar]
416. Guo LZ, Zhang ZY, Jiang Y, Li YF, Zhou ZH. Safe deep semi-supervised learning for unseen-class unlabeled data. PMLR. 2020;119:3897–906. doi:10.2139/ssrn.4423338. [Google Scholar] [CrossRef]
417. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16 x 16 Words: transformers for image recognition at scale. In: ICLR 2021—9th International Conference on Learning Representations; 2021 May 4; Vienna, Austria. [Google Scholar]
418. Yuille AL, Liu C. Deep nets: what have they ever done for vision? Int J Comput Vis. 2021;129(3):781–802. doi:10.1007/s11263-020-01405-z. [Google Scholar] [CrossRef]
419. Marblestone AH, Wayne G, Kording KP. Toward an integration of deep learning and neuroscience. Front Comput Neurosci. 2016;10:94. doi:10.3389/fncom.2016.00094. [Google Scholar] [PubMed] [CrossRef]
420. Zoph B, Le QV. Neural architecture search with reinforcement learning. In: 5th International Conference on Learning Representations, ICLR 2017; 2017 Apr 24–26; Toulon, France. [Google Scholar]
421. Elsken T, Metzen JH, Hutter F. Neural architecture search: a survey. J Mach Learn Res. 2018 Aug;20:1–21. [Google Scholar]
422. Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, et al. NiftyNet: a deep-learning platform for medical imaging. Comput Meth Programs Biomed. 2018;158:113–22. doi:10.1016/j.cmpb.2018.01.025. [Google Scholar] [PubMed] [CrossRef]
423. Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;6:m1328. doi:10.1136/bmj.m1328. [Google Scholar] [PubMed] [CrossRef]
424. Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020:m689. doi:10.1136/bmj.m689. [Google Scholar] [PubMed] [CrossRef]
425. Bayoudh K. A survey of multimodal hybrid deep learning for computer vision: architectures, applications, trends, and challenges. Inf Fusion. 2024;105:102217. doi:10.1016/j.inffus.2023.102217. [Google Scholar] [CrossRef]
426. Zhang Z, Wang L, Zheng W, Yin L, Hu R, Yang B. Endoscope image mosaic based on pyramid ORB. Biomed Signal Process Control. 2022;71:103261. doi:10.1016/j.bspc.2021.103261. [Google Scholar] [CrossRef]
427. Lu S, Yang B, Xiao Y, Liu S, Liu M, Yin L, et al. Iterative reconstruction of low-dose CT based on differential sparse. Biomed Signal Process Control. 2023;79:104204. doi:10.1016/j.bspc.2022.104204. [Google Scholar] [CrossRef]
428. Liu M, Zhang X, Yang B, Yin Z, Liu S, Yin L, et al. Three-dimensional modeling of heart soft tissue motion. Appl Sci. 2023;13(4):2493. doi:10.3390/app13042493. [Google Scholar] [CrossRef]
429. Dang W, Xiang L, Liu S, Yang B, Liu M, Yin Z, et al. A feature matching method based on the convolutional neural network. JIST. 2023;67(3):30402–1–11. doi:10.2352/j.imagingsci.technol.2023.67.3.030402. [Google Scholar] [CrossRef]
430. Gao Z, Pan X, Shao J, Jiang X, Su Z, Jin K, et al. Automatic interpretation and clinical evaluation for fundus fluorescein angiography images of diabetic retinopathy patients by deep learning. Br J Ophthalmol. 2023;107(12):1852–8. doi:10.1136/bjo-2022-321472. [Google Scholar] [PubMed] [CrossRef]
431. Jin K, Gao Z, Jiang X, Wang Y, Ma X, Li Y, et al. MSHF: a multi-source heterogeneous fundus (MSHF) dataset for image quality assessment. Sci Data. 2023;10(1):286. doi:10.1038/s41597-023-02188-x. [Google Scholar] [PubMed] [CrossRef]
432. Ye X, Wang J, Qiu W, Chen Y, Shen L. Excessive gliosis after vitrectomy for the highly myopic macular hole: a spectral domain optical coherence tomography study. Retina. 2023;43(2):200–8. doi:10.1097/IAE.0000000000003657. [Google Scholar] [PubMed] [CrossRef]
433. Guan H, Yap PT, Bozoki A, Liu M. Federated learning for medical image analysis: a survey. Pattern Recognit. 2024;151:110424. doi:10.1016/j.patcog.2024.110424. [Google Scholar] [PubMed] [CrossRef]
434. Haripriya R, Khare N, Pandey M. Privacy-preserving federated learning for collaborative medical data mining in multi-institutional settings. Sci Rep. 2025;15(1):12482. doi:10.1038/s41598-025-97565-4. [Google Scholar] [PubMed] [CrossRef]
435. Dong C, Li TZ, Xu K, Wang Z, Maldonado F, Sandler K, et al. Characterizing browser-based medical imaging AI with serverless edge computing: towards addressing clinical data security constraints. In: Medical Imaging 2023: Imaging Informatics for Healthcare, Research, and Applications; 2023 Feb 19–24. San Diego, CA, USA. 5 p. doi:10.1117/12.2653626. [Google Scholar] [PubMed] [CrossRef]
436. Eshwarappa NM, Baghban H, Hsu CH, Hsu PY, Hwang RH, Chen MY. Communication-efficient and privacy-preserving federated learning for medical image classification in multi-institutional edge computing. J Cloud Comput. 2025;14(1):44. doi:10.1186/s13677-025-00734-z. [Google Scholar] [CrossRef]
437. Zhan S, Huang L, Luo G, Zheng S, Gao Z, Chao HC. A review on federated learning architectures for privacy-preserving AI: lightweight and secure cloud-edge–end collaboration. Electronics. 2025;14(13):2512. doi:10.3390/electronics14132512. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF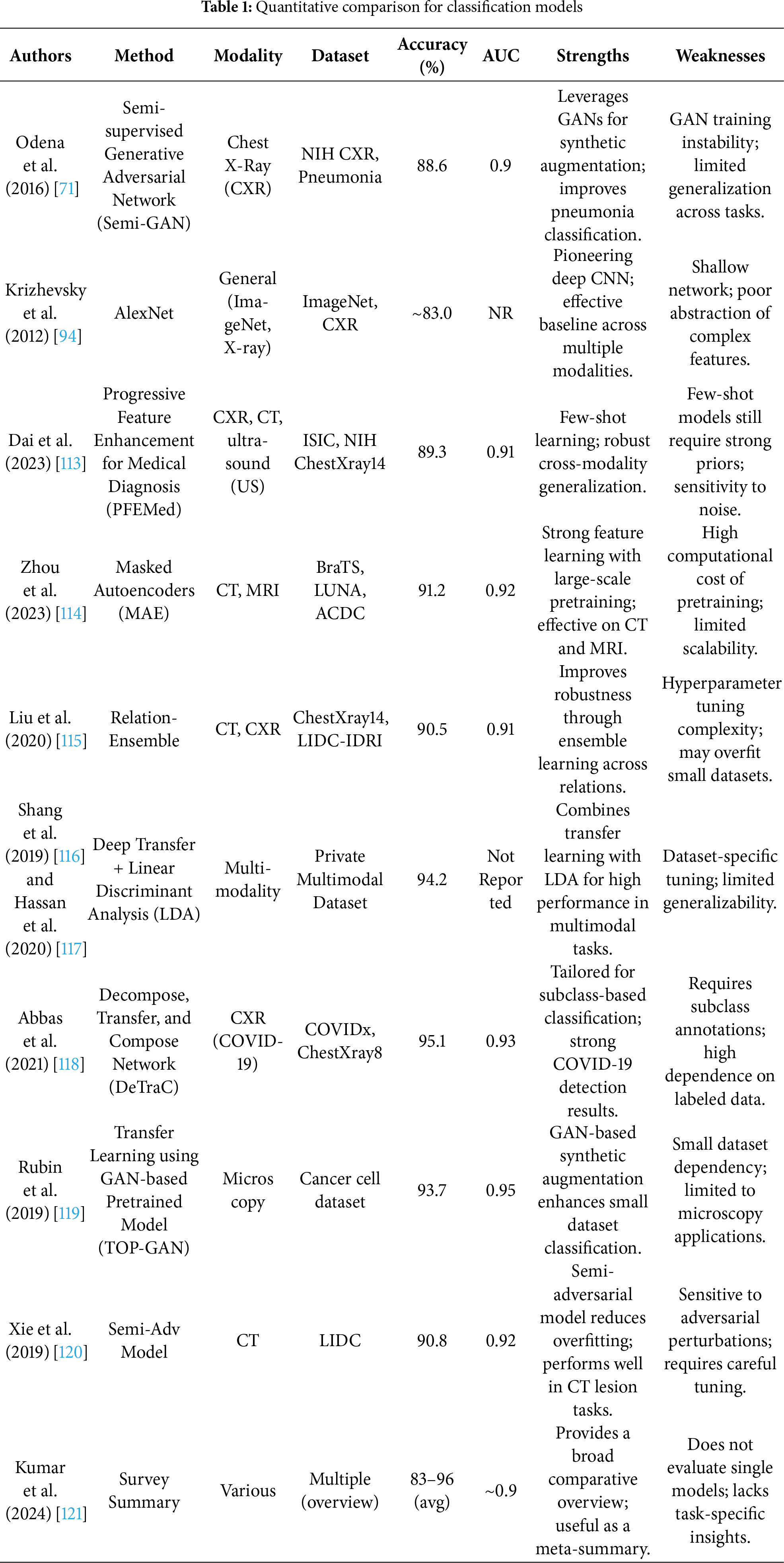
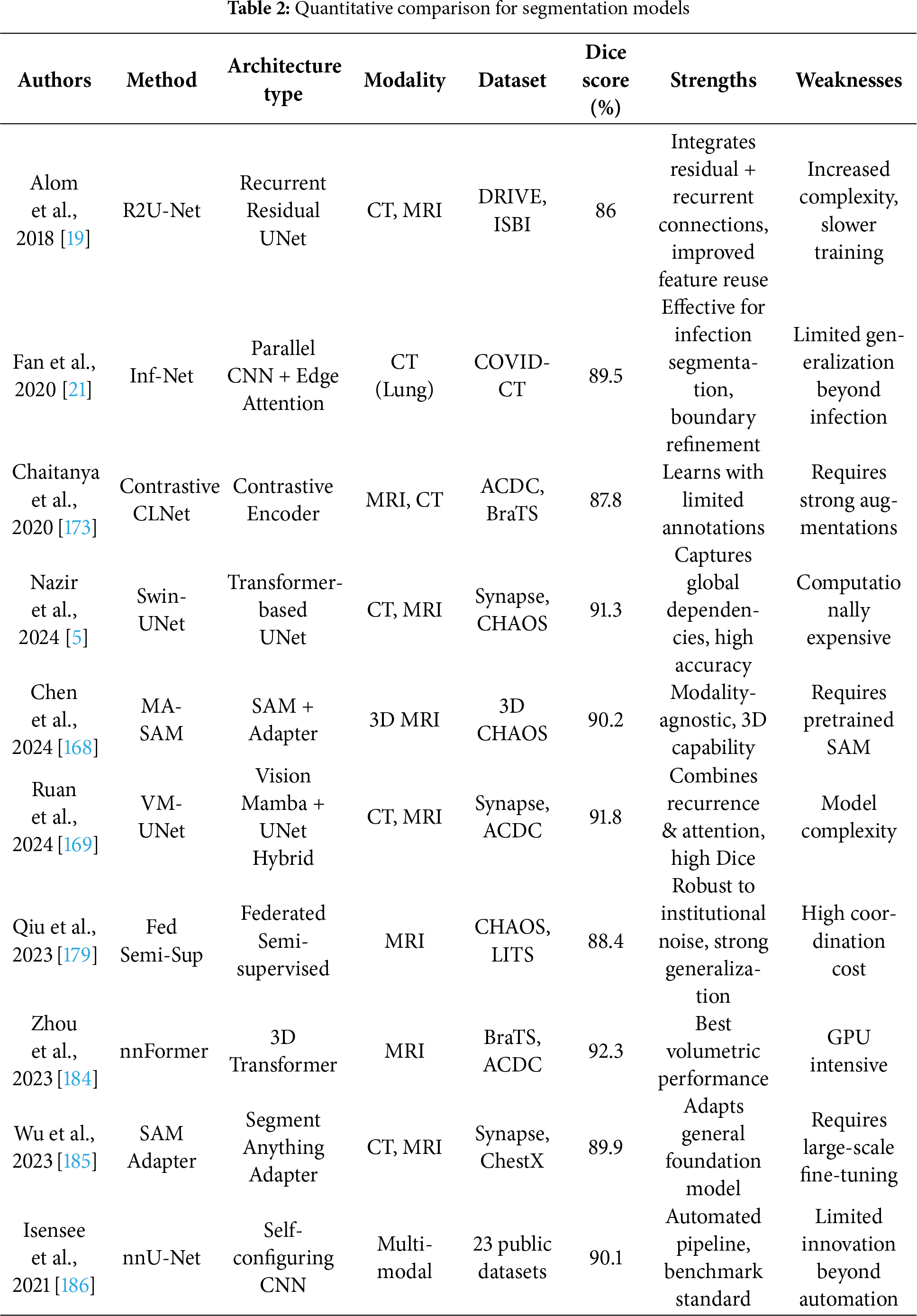
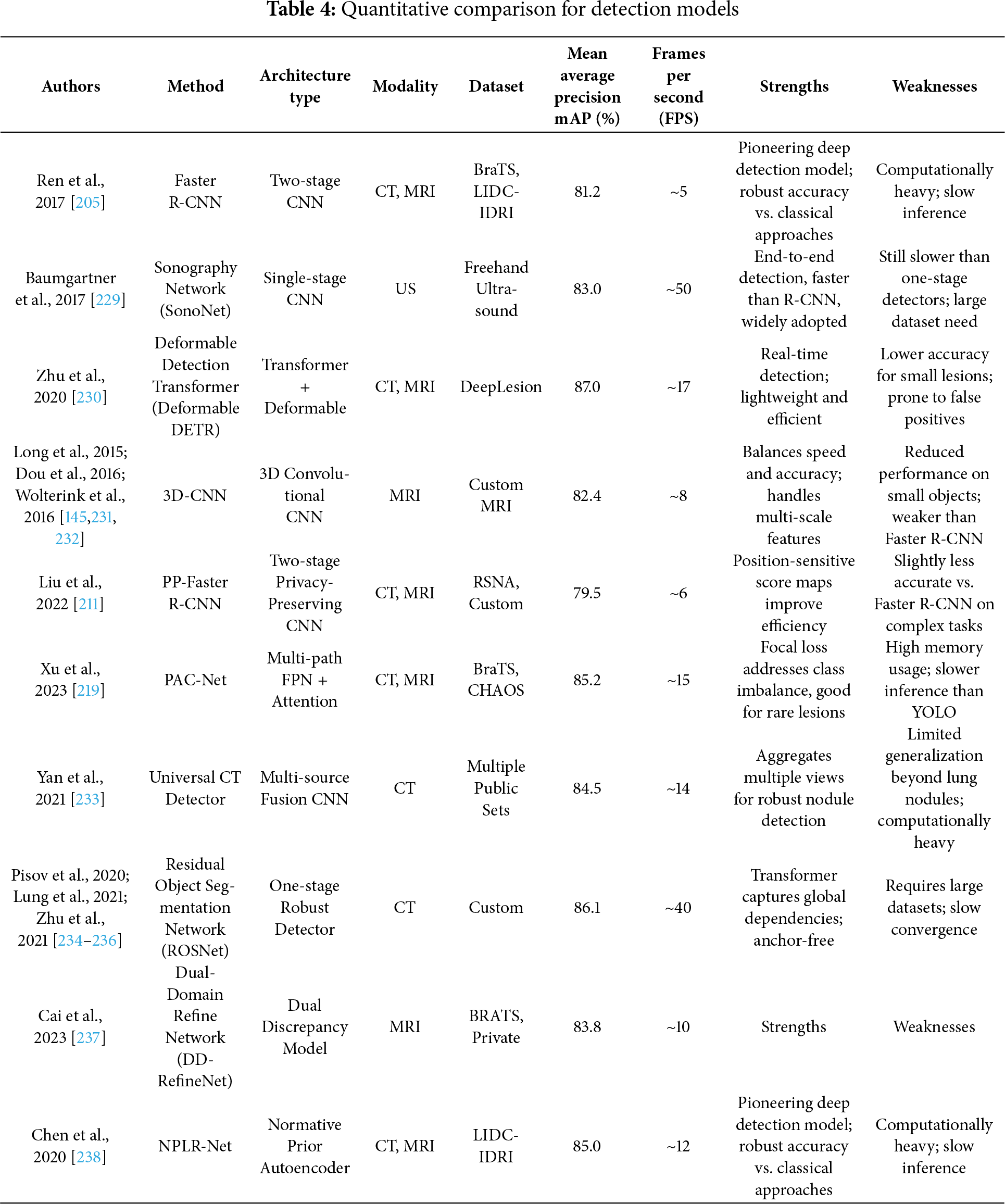
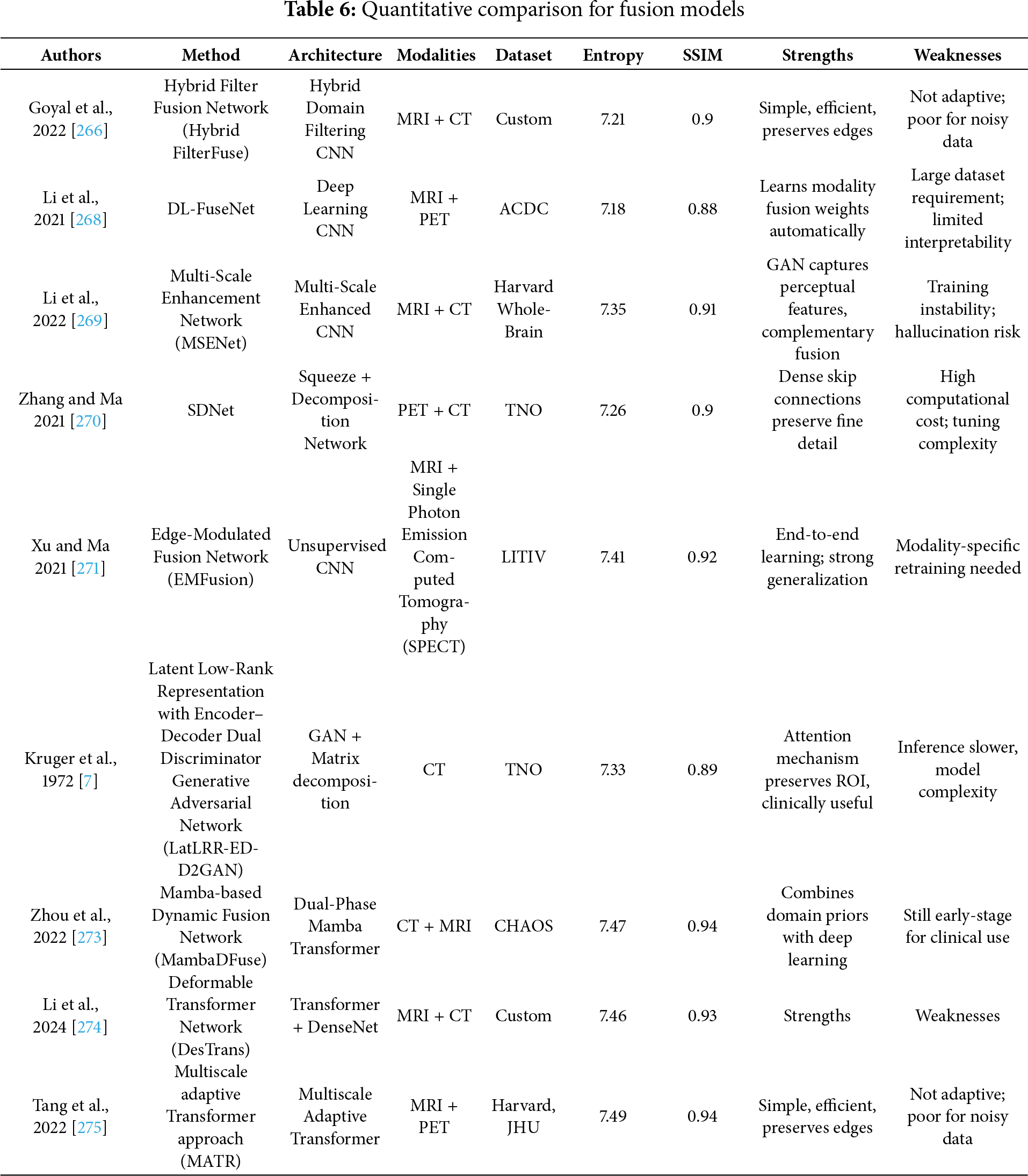
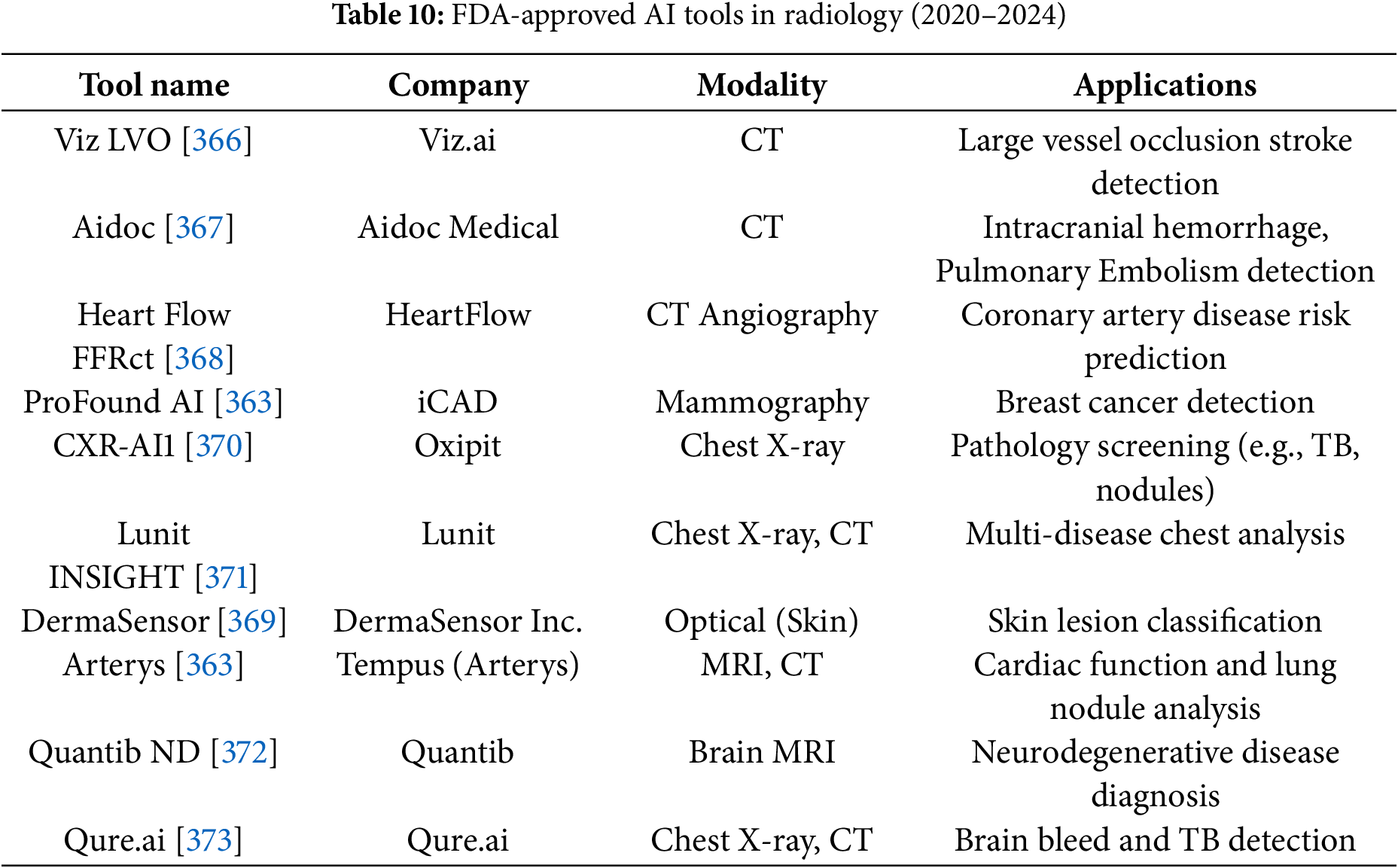
 Downloads
Downloads
 Citation Tools
Citation Tools