 Open Access
Open Access
ARTICLE
Inhibition of proliferation, migration, and invasiveness of bladder cancer cells through SAPCD2 knockdown
Department of Urology, Shaoxing People’s Hospital, Shaoxing Hospital, Zhejiang University School of Medicine, Shaoxing, China
* Corresponding Author: JIAJUN YAN. Email:
BIOCELL 2024, 48(1), 97-109. https://doi.org/10.32604/biocell.2023.045303
Received 23 August 2023; Accepted 23 November 2023; Issue published 30 January 2024
Abstract
Introduction: Bladder cancer (BC) has a high incidence and mortality rate worldwide. Suppressor anaphase-promoting complex domain containing 2 (SAPCDC2) is over-expressed in a variety of tumors. Objectives: This study investigated the effects of SAPCD2 knockdown on BC cells. Methods: T24 and UMUC3 cell models and the xenografted BC tumor model with SAPCD2 knockdown were established to observe the malignant phenotype of BC cells by cell counting kit-8 assay, colony formation test, wound healing, and Transwell assay, mRNA and proteins expressions were measured with quantitative real-time polymerase chain reaction, western blotting, and tissue immunohistochemistry. Lithium chloride agonist on the Wnt/β-catenin pathway was used to clarify the molecular mechanism of SAPCD2 knockdown. Results: SAPCD2 expression was significantly higher in BC cell lines than in SV-HUC-1 cells. SAPCD2 knockdown inhibited viability and cloning, hindered the G0/G1 phase of the cell cycle in UMUC3 and T24 cells, and decreased the migration and invasiveness of BC cells. SAPCD2 knockdown inhibited expression levels of cyclin D1, cyclin B1, N-cadherin, vimentin, Snail, β-catenin, c-Myc, and cyclin-dependent kinase 4, while the P21 and E-cadherin were raised by SAPCD2 knockdown. Furthermore, lithium chloride reversed the effects of SAPCD2 knockdown on the expression levels of the above proteins in UMUC3 and T24 cells. In vivo, SAPCD2 knockdown inhibited the volume, weight, and expression of Ki-67 and β-catenin in tumors and increased the E-cadherin expression. Conclusion: SAPCD2 knockdown inhibits the malignant phenotype of BC via a pathway involving β-catenin.Graphic Abstract
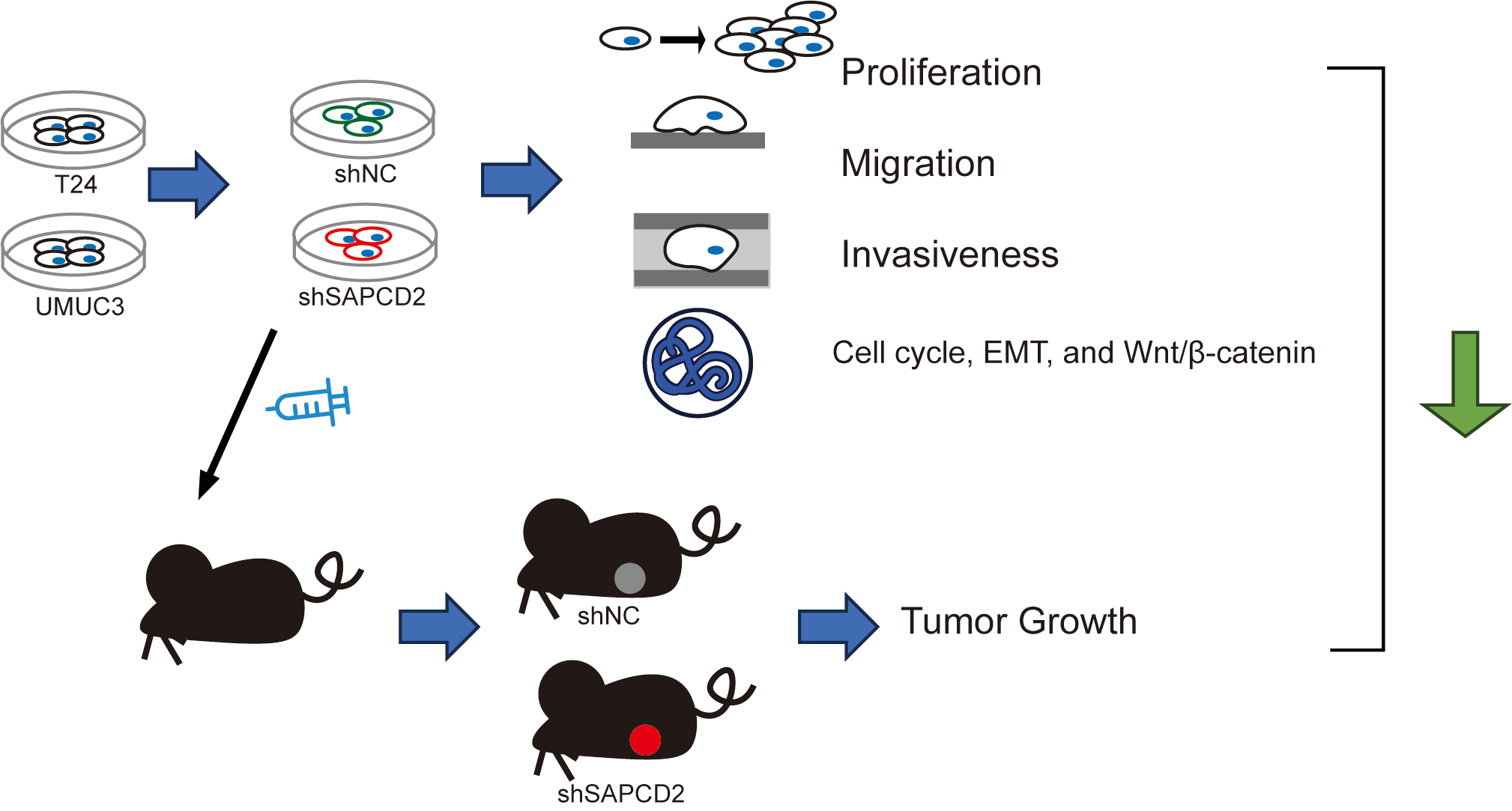
Keywords
Bladder cancer (BC) is one of the most common genitourinary malignancies. Global Cancer Statistics, 2020 reported more than 573000 new cases of BC and more than 213000 deaths worldwide [1]. Scientists found that sex, age, tobacco use, occupational carcinogens, environment, or diet, etc., are related to the incidence rate and mortality of BC [2,3]. From 2006 to 2016, the morbidity of BC decreased in Europe but increased in Asia [4,5]. Most patients are diagnosed with non-muscle invasive BC. In the United States, non-muscle-invasive BC accounts for the majority of new cases, approximately 75%, and muscle-invasive BC accounts for 25% [6]. Additionally, BC usually originates from urothelial cells, and the remaining are mainly small cell carcinoma, squamous cell carcinoma, and adenocarcinoma [7]. Currently, the main treatment is surgical resection in combination with radiotherapy and chemotherapy, and early diagnosis of biomarkers is insufficient [8]. However, the quality of life of patients is poor, and the 5-year survival rate of patients with advanced BC is far from that expected [9]. Therefore, it is essential to explore BC development mechanisms for the treatment and prognosis of patients with BC.
Suppressor anaphase-promoting complex domain containing 2 (SAPCDC2), a cell cycle-dependent gene, was initially discovered in gastric cancer [10]. The protein it encodes, SAPCDC2, is a novel axin1-interacting protein and is situated on human chromosome 9q42.3. Evidence shows that SAPCDC2 functions as a mediator in the repressive feedback mechanism of the Wnt/β-catenin signaling pathway [11]. SAPCDC2 is over-expressed in a variety of tumors, including carcinoma of the stomach, hepatocellular carcinoma, melanoma, glioblastoma, and renal cell carcinoma, and plays an essential role in proliferation, metastasis, and invasion of tumor cells [10,12]. Skeldon et al. reported that SAPCDC2 elevates the proliferation and metastasis of colorectal cancer cells [10] and the invasion and metastasis of breast cancer cells through the YAP/TAZ pathways [13]. In addition, SAPCD2 of knock-out inhibits the proliferation and lung metastasis of fibrosarcoma via the Hippo signaling pathway [14]. Previously, a study by Xu et al. combined the Gene Expression Omnibus (GEO) database (GSE7476 and GSE13507) and The Cancer Genome Atlas (TCGA) database analysis and reported that the level of SAPCD2 was up-regulated in BC tissue [15]. However, research on the effects of SAPCD2 knockdown on BC is still lacking. Consequently, we speculated that SAPCD2 is associated with the development of BC. To investigate this association, cell models with SAPCD2 knockdown and the BC xenografted tumor model were constructed in this study. The aim of this study was to explore the molecular mechanisms underlying the influence of SAPCD2 on the proliferation, migration, and invasiveness of BC cells.
The UALCAN network (http://ualcan.path.uab.edu) was used to compare BC tumor (Bladder Urothelial Carcinoma, BLCA) and ordinary tissues in terms of transcripts per million (TPM) expression levels of SAPCD2 (C90RF140) based on the TCGA database [16]. Overall survival was explored through GEPIA [17] (http://gepia.cancer-pku.cn/index.html).
SV-HUC-1 cells and BC cell lines (SWU780, T24, UMUC3, and BIU87) were supplied by the iCell Bioscience Inc. (Shanghai, China). All were developed in a complete medium consisting of Dulbecco’s modified Eagle medium (DMEM, SH30243.01, Hyclone, Logan, Utah, USA), as well as 10% fetal bovine serum (FBS, 11011-8615, Tianhang Biotechnology, Hangzhou, Zhejiang, China) at 37°C in 5% CO2.
Determination of cell viability
The manufacturer’s guidelines were followed to determine cell viability by employing the cell counting kit-8 (CCK-8) kit (CK04-11, DOJINDO, Kumamoto, Kyushu region, Japan). Cells in the logarithmic growth stage were evenly seeded on 96-well plates for 24, 48, 72, and 96 h. Cells were then subjected to incubation with 10 μL of CCK-8 in the incubator for 2 h. Following this, the optical density (OD) was determined at 450 nm with a CMaxPlus microtiter reader (Molecular Devices, Sunnyvale, California, USA).
Flow cytometry analysis was conducted using PI/RNase staining kits (C1052, Beyotime, Shanghai, China). UMUC3 and T24 cell suspensions, in their logarithmic growth phase, were seeded separately into 6-well plates and subjected to incubation for 24 h. Following manipulation of the cells as shNC or shSAPCD2 treatment groups, the resultant cells were fixed for 12 h and collected following digestion and centrifugation. Subsequently, cells were exposed to PI/RNase staining reagents (PI:RNase = 2.5:1) at 37°C for 30 min in darkness. Cell cycle profiling was fulfilled through the use of a dedicated flow cytometry (C6, BD Biosciences, Franklin Lake, New Jersey, USA).
Establishment of stably transfected cell lines
Oligos of SAPCD2-shRNAs were integrated into lentiviral vector GV248 (GeneChem, Shanghai, China) and empty vectors were employed as controls. Puromycin (0.5 mg/mL; ST551, Beyotime, Shanghai, China) was used to select cell lines expressing SAPCD2-shRNAs for 2 weeks.
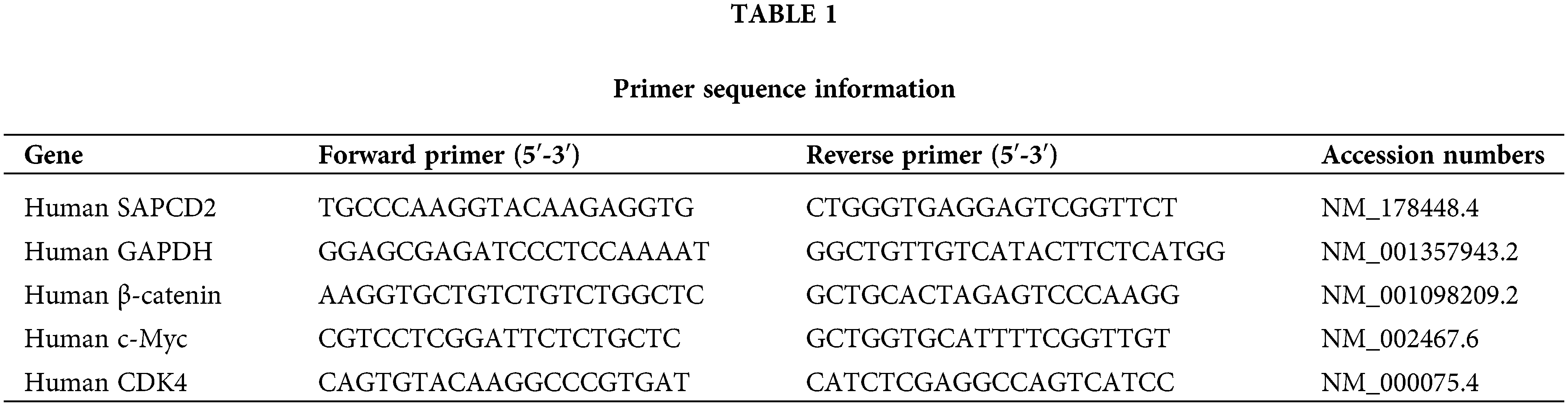
Gene expression was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Briefly, cell homogenate was added with Trizol (B511311, Sangon, Shanghai, China) and centrifuged to separate the total RNA. The supernatant was centrifuged in chloroform and isopropanol to get the precipitate. The precipitate was then rinsed with 75% ethanol and finally dissolved in 20 μL of water containing diethyl pyrocarbonate (R0021, Beyotime, Shanghai, China) for −80°C storage. It was reverse transcribed using a SYBR PCR kit (RR820A, Takara, Osaka, Japan). qRT-PCR was performed on a StepOne Plus real-time PCR system (Mastercycler, Eppendorf, Hamburg, Germany). The amplification protocol involved an initial incubation period at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s per cycle, and a final extension at 60°C for 60 s. Relative quantification of results using the 2−ΔΔCT method. The primer sequence information is detailed in Table 1.
UMUC3 and T24 cells were divided into shNC and shSAPCD2 groups to observe the role of SAPCD2 in BC. Additionally, cells were categorized into four groups: shNC, shSAPCD2, shSAPCD2+DMSO, and shSAPCD2+lithium chloride (LiCl). LiCl (20 mmol/L, treatment for 24 h) (ab120853, Sigma, Saint Louis, Missouri, USA) was used to examine the impact between SAPCD2 and β-catenin/c-Myc/cyclin-dependent kinase 4 (CDK4) pathway in UMUC3 and T24 cell lines according to the previous report [18]. Dimethyl sulfoxide (DMSO, D8371, Solarbio, Beijing, China) served as the control for LiCl.
WB was performed for the semi-quantitative analysis of protein expression levels. In brief, the total protein isolated from cells and tissues was extracted using a medium-strength radioimmunoprecipitation assay lysis buffer (P0013C, Beyotime, Shanghai, China). The ensuing concentration of proteins was then determined utilizing the BCA kit (pc0020, Solarbio, Beijing, China). Following this, electrophoresis and wet transfer were performed to transfer proteins to an immune-blot membrane (1620177, Bio-Rad, Hercules, California, USA) using a wet-blotting system. Subsequently, the membranes were blocked by 5% milk, and incubated overnight with primary antibodies, including rabbit anti-SAPCD2 (C-Term) polyclonal antibody (43 kD, 1:2000, abs105974, Absin, Shanghai, China), cyclin D1 antibody (33 kD, 1:2000, AF0931, Affinity, Cincinnati, Ohio, USA), cyclin B1 antibody (60 kD, 1:2000, AF6168, Affinity), β-catenin antibody (92 kD, 1:2000, AF6266, Affinity), c-Myc antibody (49 kD, 1:3000, AF0358, Affinity), CDK4 antibody (34 kD, 1:2000, DF6102, Affinity), p21 Cip1 antibody (21 kD, 1:2000, AF6290, Affinity), E-cadherin antibody (120 kD, 1:3000, AF0131, Affinity), N-cadherin antibody (140 kD, 1:1000, AF4039, Affinity), vimentin antibody (53 kD, 1:2000, AF7013, Affinity), Snail antibody (29 kD, 1:2000, AF6032, Affinity), and GAPDH antibody (29 kD, 1:2000, AF7021, Affinity). Following this, membranes were washed and incubated with the anti-rabbit IgG antibody (1:6000, #7074, cell signaling technology, Boston, Massachusetts, USA) at 25°C for 2 h. Finally, ECL kit (KF8003, Affinity) was used to visualize target bands. Bands were quantified using the grayscale value statistical function of ImageJ software (National Institutes of Health, Bethesda, Maryland, USA).
Colony formation test is used to measure BC cell proliferation by counting colonies. UMUC3 and T24 cells (500 cells/well) were seeded in 24-well plates with the complete medium of 30% FBS. The cell state was observed by optical microscope (AE2000, Motic, Xiamen, Fujian, China) each 3-day until clones were visible to the naked eye (about 2 weeks). Cell supernatants were discarded, and cells were washed once with phosphate-buffered saline (PBS, P1003, Solarbio, Beijing, China). Subsequently, 1 mL of 4% paraformaldehyde (P0099, Beyotime, Shanghai, China) was added to each well and maintained at 4°C for 60 min. Lastly, each well was added with 1 mL of crystal violet staining solution (548-62-9, Shunqiang, Shanghai, China) for 60 min, to acquire an image under a light microscope (AE2000, Motic, Xiamen, Fujian, China) and counting colonies using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA).
The purpose of the wound healing assay was to test the migration ability of BC cells. The cellular suspension was distributed into 6-well plates, each containing 5 × 105 cells per well, and was subsequently cultured for 24 h. Following this, using a pipette tip cut, a linear wound was created. Wounded cells were then maintained in a serum-free medium for one day. Wound gaps at 0 and 24 h time points were quantified by ImageJ (National Institutes of Health, Bethesda, Maryland, USA), and relative cell migration rates were calculated. The formula used to calculate these indices was: Relative cell migration distance (%) = (initial wound width-wound width at 24 h)/initial wound width × 100%.
A Transwell assay was performed to observe the invasiveness of UMUC3 and T24 cells. Cells (1 × 104 cells/well) were cultured in the upper layer chambers (3422, Corning, New York, USA) that were wrapped with Matrigel matrix (356234, BD, Columbus, Ohio, USA) and were cultured with 400 μL DMEM in 5% CO2, at 37°C for 24 h, and complete medium was added to lower layer chambers. After wiping cells from the upper chamber and matrix glue with a cotton swab, the cells were subsequently fixed utilizing a concentration of 4% paraformaldehyde (P0099, Beyotime, Shanghai, China) for a duration of 10 min, followed by washed cells with PBS. Cells were stained using a crystal violet staining solution (0.1%, 548-62-9, Shunqiang, Shanghai, China) for 30 min. The number of cells was counted under a light microscope (AE2000, Motic, Xiamen, Fujian, China).
Animals and xenografted tumor model
The experimental procedures of the animals were in line with the relevant animal ethics requirements. Shaoxing People’s Hospital, lacking the necessary facilities for animal experimentation, has established a collaborative public platform for medical science in partnership with Zhejiang Eyong Pharmaceutical R&D Co., Ltd. (China). All procedures involving animals in this study were approved and supervised by the Animal Experimentation Ethics Committee of Zhejiang Eyong Pharmaceutical Research and Development Center (ethics No. ZJEY-20211008-04). Five-week-old BALB/c nude mice were cultivated by Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animal experiments were performed to observe the effect of SAPCD2 silencing on BC tumor proliferation capacity in vivo. Mice were reared under standard conditions (light/dark cycle: 12/12 h, temp.: 25°C; humidity: 40%; free diet) and randomized into two groups (n = 6 each), namely shNC and shSAPCD2 groups. UMUC3 cells (5 × 106 cells) treated with shNC/shSAPCD2 were subcutaneously inoculated on one side of each mouse to establish BC xenografted tumor models. Tumor growth was tracked every 3 days from day 7 with a caliper estimating size via (Length × Width2)/2. By day 30, all subjects were euthanized by CO2, and tumors were removed for weighing and measurement. The tumor was isolated and fixed in 4% paraformaldehyde for subsequent research.
Immunohistochemical (IHC) staining
Serial sections of the tissues embedded in paraffin were prepared (4 µM thick). Sections were deparaffinized and rehydrated. Thin sections are placed in the box abounding with citric acid antigen repair buffer (pH 6.0) with microwave heating. After natural cooling, the slides were cleaned in PBS (pH 7.4) on a decolorization shaking table thrice, each time for 5 min. Endogenous peroxidase activity was stopped by adding 3% H2O2 for 25 min. Slides were incubated in BSA (3% concentration) for 30 min. Additionally, SAPCD2 antibody (1:200, PA5-60632, Invitrogen, Carlsbad, USA), Ki67 antibody (1:200, AF0198, Abcam, Cambridge, Cambridgeshire, UK), beta-catenin antibody (1:200, AF6266, Abcam) or E-cadherin antibody (1:200, AF0131, Abcam) were used to incubate slides at ordinary temperature all-night, followed by incubation with a secondary antibody, goat anti-rabbit IgG H & L (HRP) and pre-adsorption (1:1000, ab97080, Abcam), for 50 min at general temperature. After the slices were cleaned with PBS and dried, fresh DAB color development solution was added drop-wise on the sections, and the discoloration time under the microscope was controlled. The positive was brown-yellow. Slices were successively processed by hematoxylin, hematoxylin differentiation solution, and hematoxylin back blue. Water washing is required between every two steps. The slices were placed in 75% alcohol, 85% alcohol, absolute ethanol I–II, n-butanol, and xylene I each for 5 min. Post-drying, the slices were sealed with neutral resin and observed under a microscope (magnification, ×400). ImageJ software (National Institutes of Health, Bethesda, Maryland, USA) was used for IHC quantifications.
Every experimental procedure was replicated thrice or more. For data analysis and representation, SPSS statistical software (Version 16.0, SPSS Inc., Chicago, Illinois IL, USA) and GraphPad Prism 9 (GraphPad Software, San Diego, California, USA) were used, respectively. All data satisfied the normal distribution and homogeneity variance. One-way ANOVA was performed. Subsequently, the Tukey post hoc contrast was performed to analyze data across multiple groups. Additionally, data were analyzed using a student’s t-test to compare the two groups. All data are presented in mean values (±SD), and a statistical significance p-value of <0.05 is deemed significant (0.01 ≤ *p < 0.05, 0.001 ≤ **p or ##p < 0.01).
SAPCD2 knockdown suppressed proliferation of UMUC3 and T24 cells
Overexpression of SAPCD2 in BC tissues and cell lines
Bioinformatics analysis revealed that compared to normal samples, SAPCD2 was highly expressed in individual cancer stages II–IV, papillary, and non-papillary clinical samples. Furthermore, SAPCD2 was overexpressed in the N0–N3 nodal metastasis status of BC patients (p < 0.01) (Fig. 1A). Additionally, the use of GEPIA revealed that SAPCD2 was highly expressed in BLCA samples than in normal samples, and the overall survival of BLCA patients with low SAPCD2 versus high SAPCD2 differed significantly (p < 0.05, HR = 1.7) (Fig. 1B).
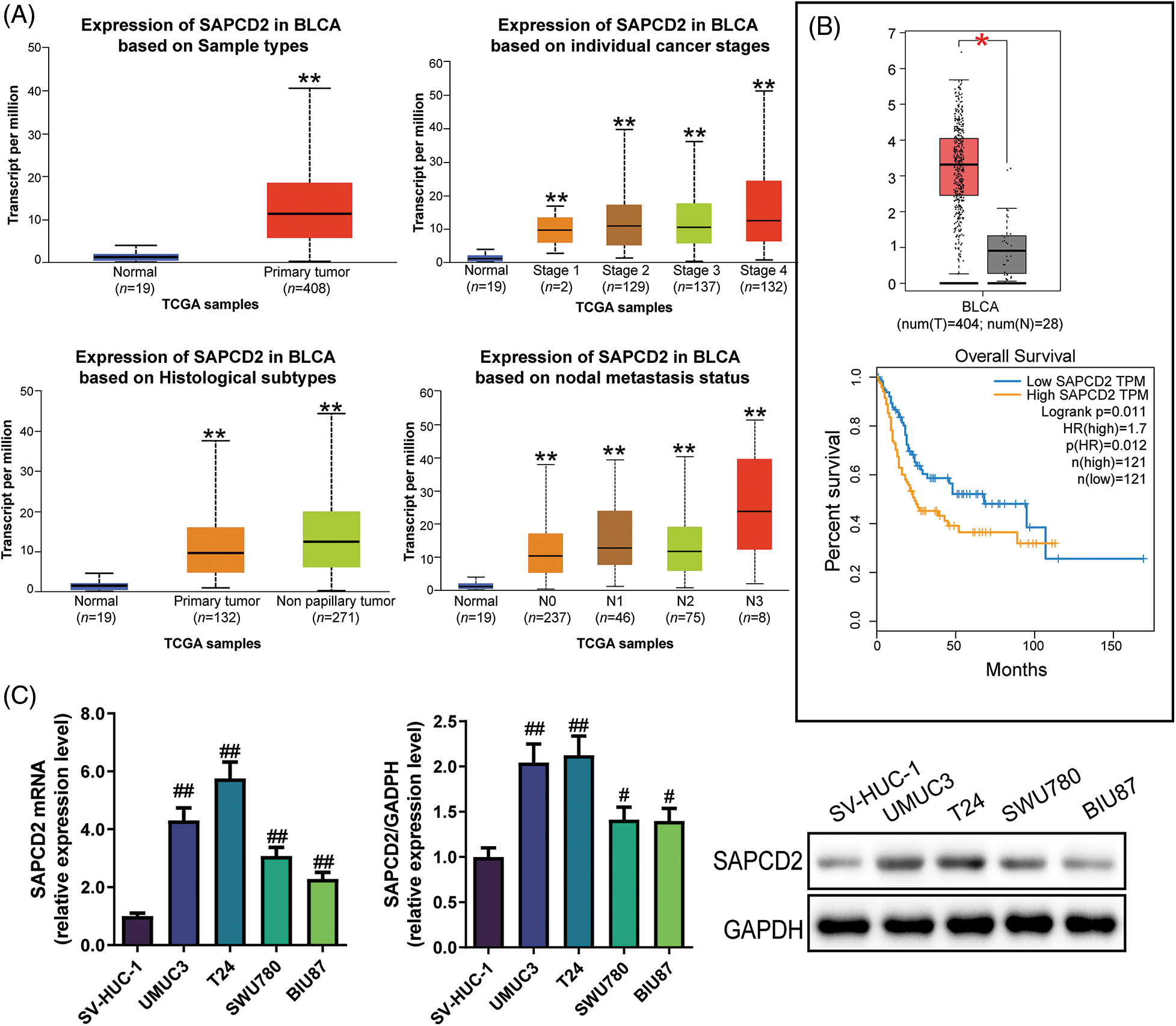
Figure 1: Expression of suppressor anaphase-promoting complex domain containing 2 (SAPCD2) in bladder cancer (BC) tissues and cells. (A) Transcripts per million (TPM) of SAPCD2 in bladder urothelial carcinoma (BLCA) on sample types, individual cancer stages, histological subtypes, and nodal metastasis status were derived from TCGA level 3 RNA-seq data and retrieved from UALCAN. **p < 0.01, vs. normal group; statistical analysis between the two groups was performed by t-test. (B) The log2 (TPM+1) of SAPCD2 was higher in the tumor (red) than in the normal group (grey). Additionally, data on the expression level and overall survival curve of SAPCD2 was obtained from the GEPIA website; the level of expression between the two groups was statistically analyzed by one-way ANOVA; the log-rank test was used for the survival analyses. The p-value of the overall survival curve was 0.011, and HR was 1.7. *p < 0.05, vs. normal group. (C) Relative expression levels of SAPCD2 mRNA and protein in SV-HUC-1, UMUC3, T24, SWU780, and BIU87 cell lines (n = 3); one-way ANOVA was performed, followed by Tukey post hoc contrast to analyze data across multiple groups. #p < 0.05, ##p < 0.01, vs. SV-HUC-1 group.
In addition, qRT-PCR and WB assay revealed that the SAPCD2 mRNA levels in SWU780, T24, UMUC3, and BIU87 cell lines were overexpressed compared to SV-HUC-1 cells (p < 0.05) (Fig. 1C). Results of WB were consistent with those of RT-qPCR (p < 0.05) (Fig. 1C). The top two SAPCD2 highly expressing cells, UMUC-3 and T24, were used for subsequent research.
Inhibition of SAPCD2 silencing on the proliferation and clone ability of bladder cells
First, the knockdown efficacy of shRNA#1 and shRNA#2 targeting SAPCD2 in UMUC3 and T24 cells was evaluated by both qRT-PCR and WB. The levels of SAPCD2 mRNA and protein were significantly reduced following treatment with shRNA#1 and shRNA#2 (p < 0.05) (Figs. 2A, 2B), and shRNA#1 had a better knockdown efficiency. Thus, shRNA#1 was selected for the subsequent experiments.
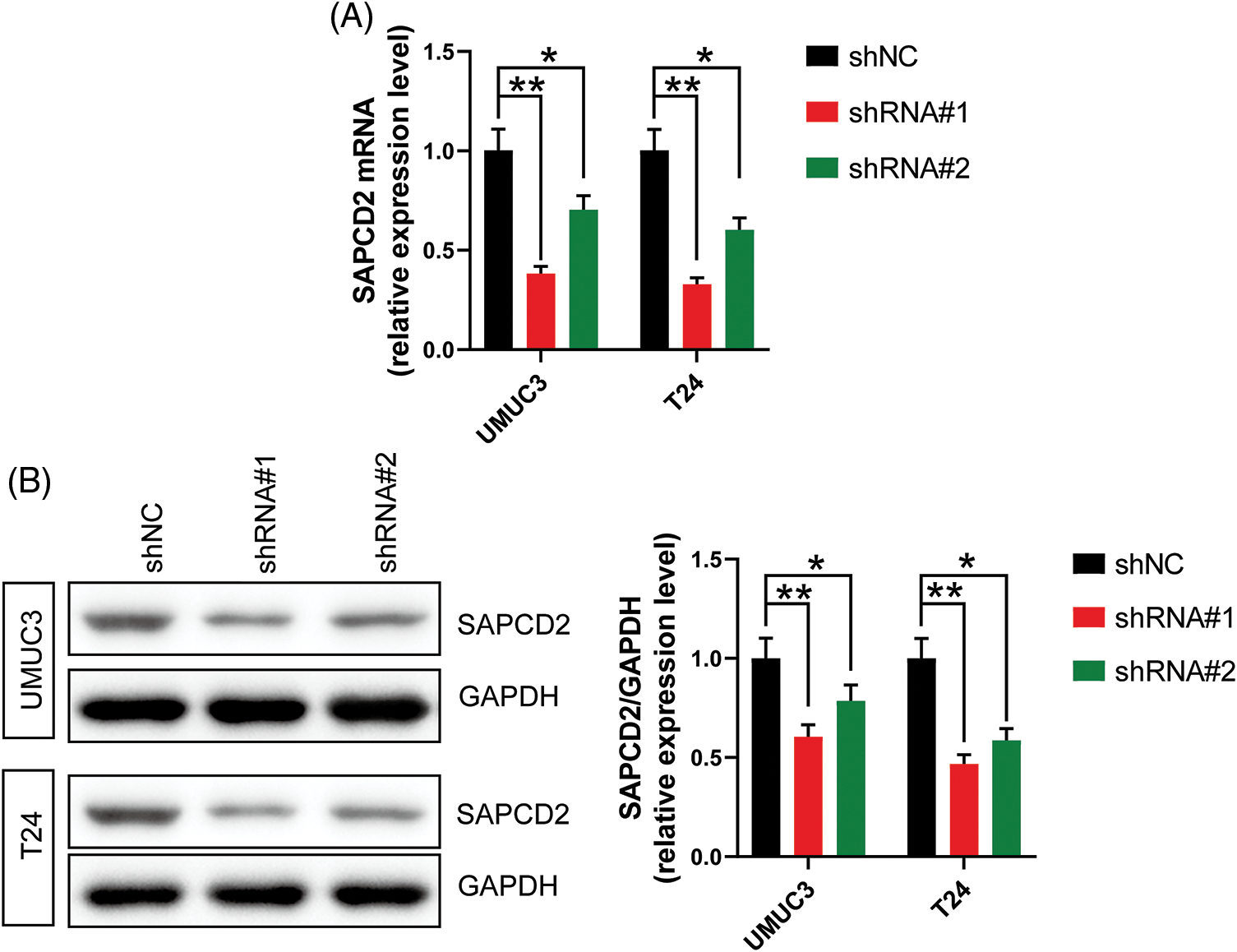
Figure 2: The transfection efficiency of shRNA#1 and shRNA#2 was analyzed by measuring suppressor anaphase-promoting complex domain containing 2 (SAPCD2) levels in UMUC3 and T24 cells using quantitative real-time polymerase chain reaction and western blotting. (A) and (B) relative expression levels of SAPCD2 mRNA and protein in UMUC3 and T24 cell lines (n = 3). Values are expressed as means ± SD, *p < 0.05, **p < 0.01, vs. SV-HUC-1 group.
The proliferation and cloning capacity of the cells were detected using a CCK-8 kit and colony formation test. Cell viabilities were expressed as OD values. In both UMUC3 and T24 cell lines, there was a significant decrease in OD values at 48, 72, and 96 h and the number of cell clones after SAPCD2 knockdown (p < 0.05) (Figs. 3A–3C).
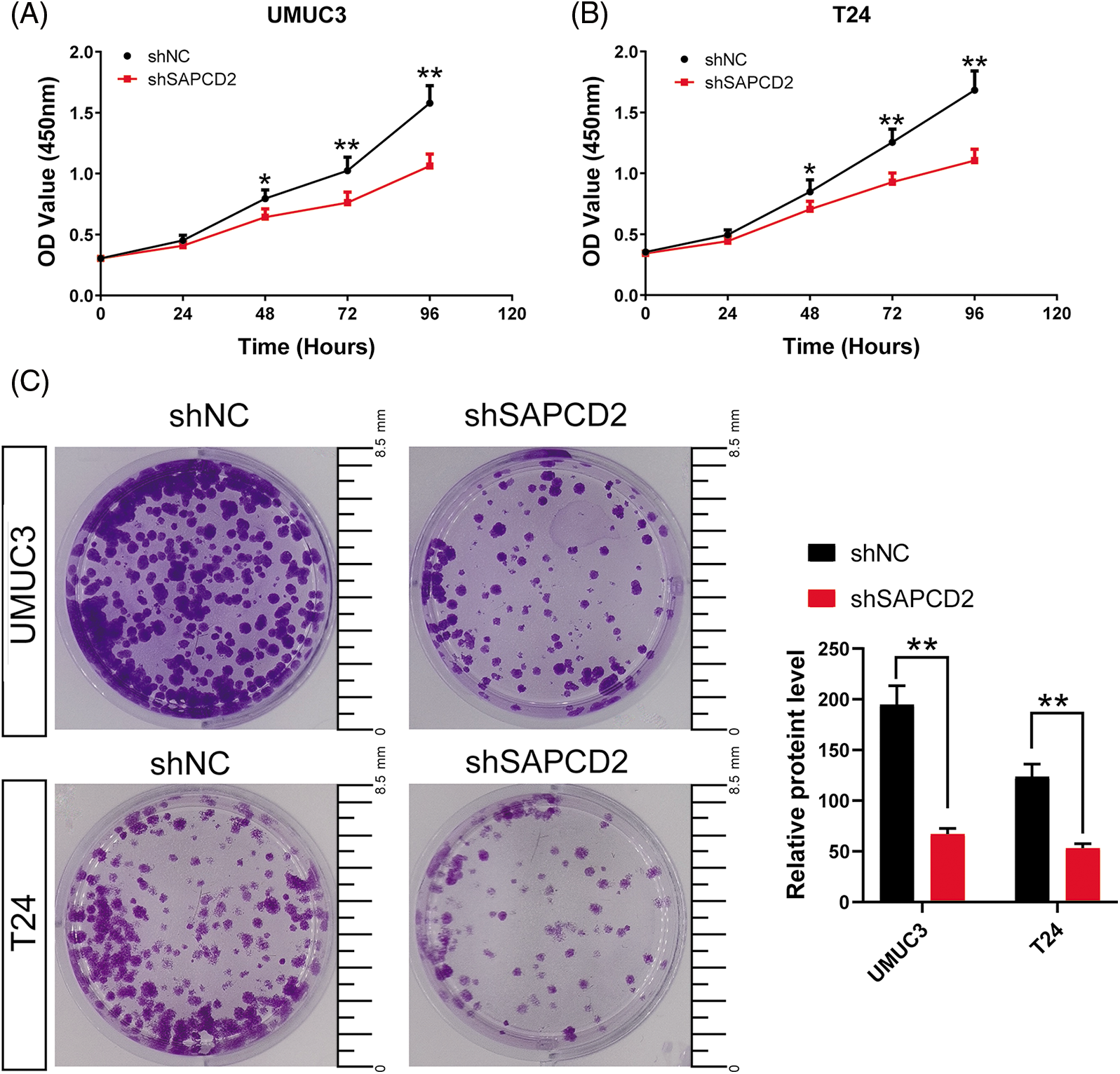
Figure 3: Knockdown of suppressor anaphase-promoting complex domain containing 2 (SAPCD2) inhibited the proliferation and cloning ability of bladder cancer (BC) cells. (A) and (B) The OD value of BC cells was detected by CCK-8 assay (n = 5). (C) The colony number of BC cells was detected by colony formation test (n = 3) (Scale bar = 8.5 mm). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
SAPCD2 knockdown blocked the cell cycle of bladder cancer cells
The cell cycle was examined by flow cytometry. G0/G1 cell number significantly increased in UMUC3 and T24 cells as a result of shSAPDCD2 treatment (p < 0.05), while the number of cells in S and G2/M decreased (p < 0.05) (Figs. 4A, 4B).
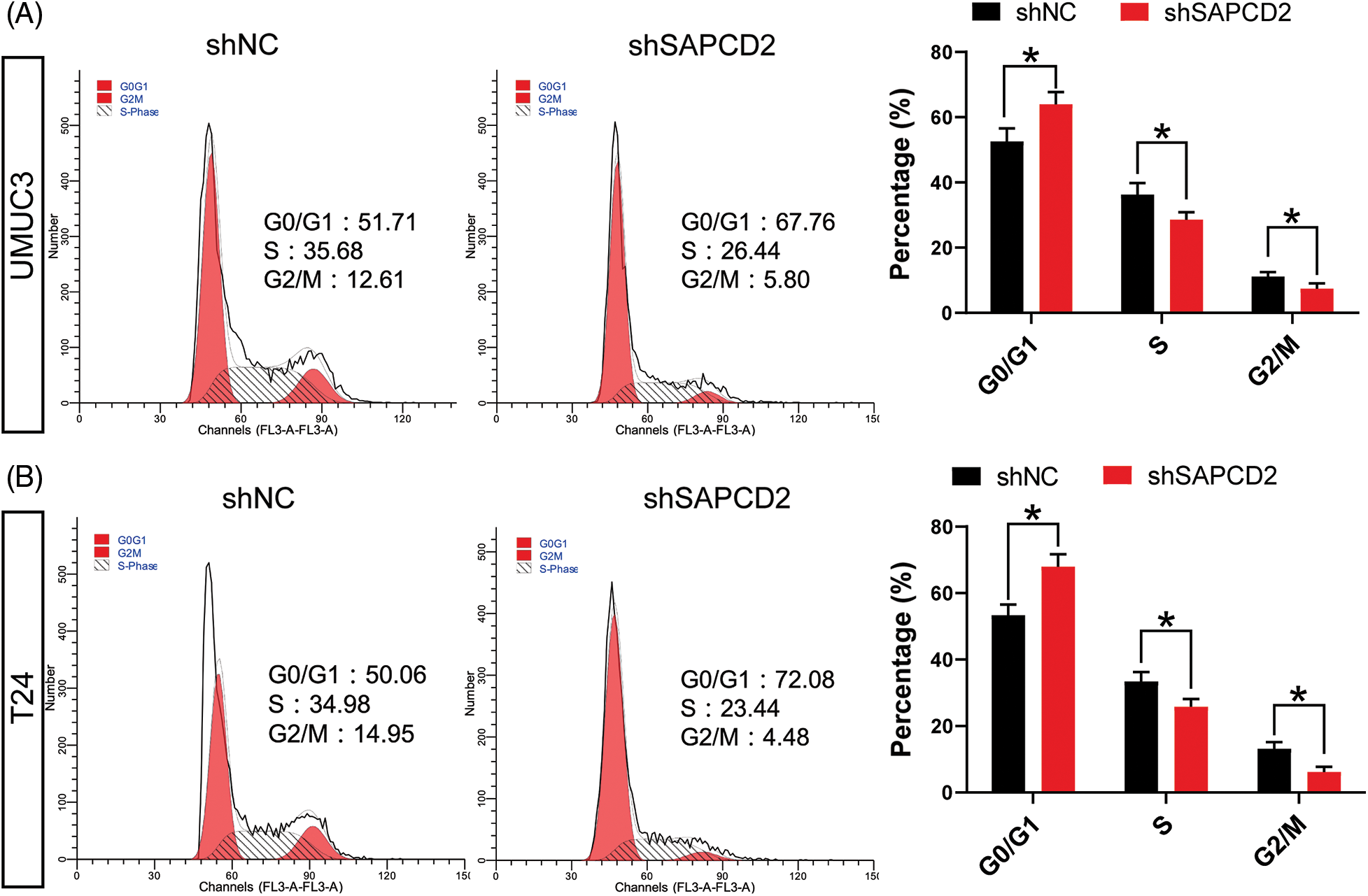
Figure 4: The knockdown of suppressor anaphase-promoting complex domain containing 2 (SAPCD2) blocked the cell cycle of bladder cancer (BC) cells. (A) and (B) The cell cycle of BC was detected by flow cytometry (n = 3). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
Analysis of cell cycle protein levels in BC cells by WB revealed that SAPCD2 knockdown decreased G1-related proteins in UMUC3 and T24 cells, including cyclin D1, cyclin B1, c-Myc, and CDK4 (p < 0.01) (Fig. 5). Besides, the level of P21 protein increased to 199% in UMUC3 cells and 68% in T24 cells after SAPCD2 knockdown (p < 0.01) (Fig. 5).
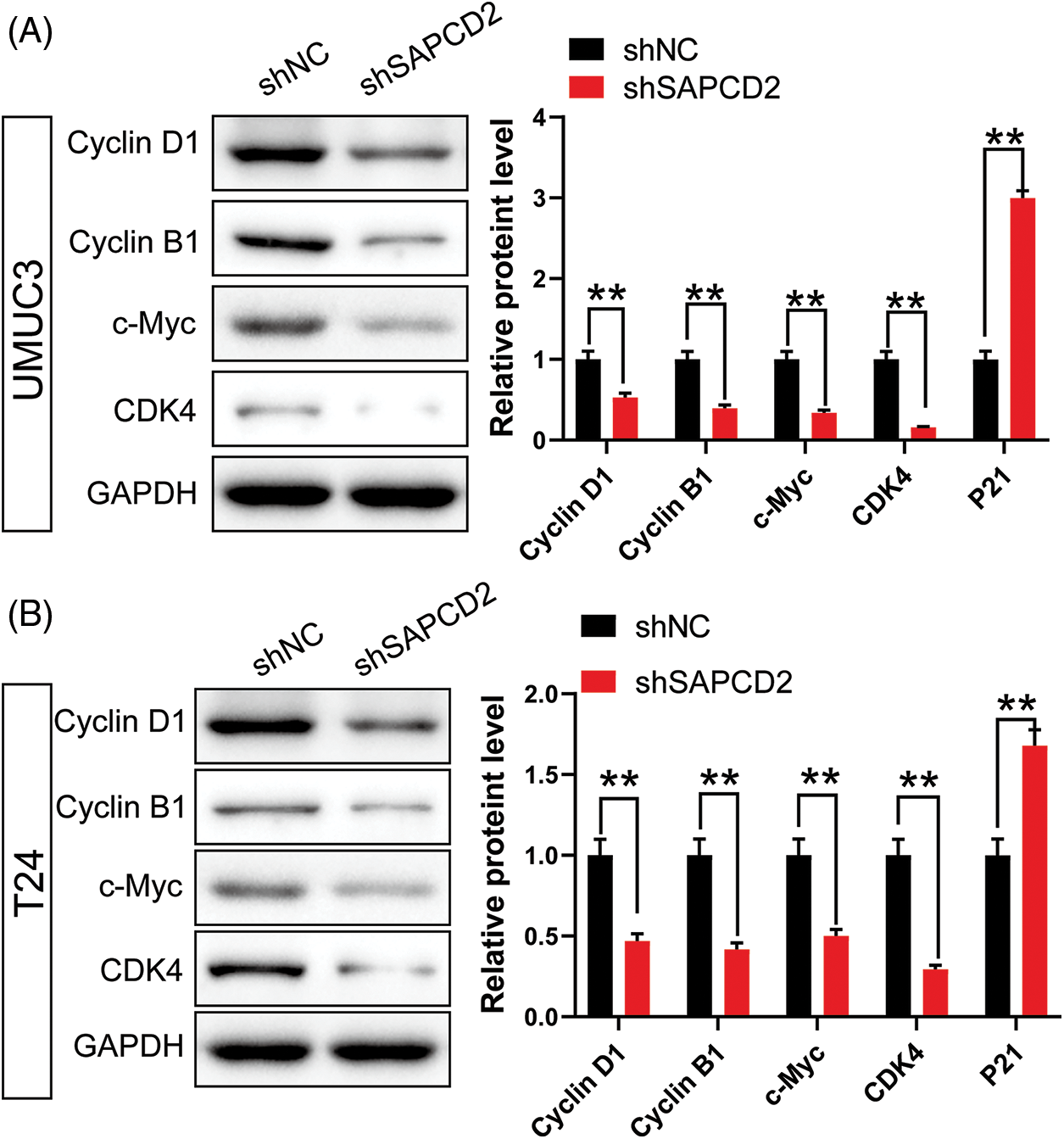
Figure 5: Knockdown of suppressor anaphase-promoting complex domain containing 2 (SAPCD2) inhibited the expression of G1 phase-related proteins in bladder cancer (BC) cells. (A, B) Relative protein levels of cyclin D1, cyclin B1, c-Myc, and cyclin-dependent kinase 4 (CDK4), which are related to G1 in the cell cycle in SAPCD2-silenced BC cells, were detected by western blot (n = 3). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
SAPCD2 knockdown limited the migration, invasiveness, and epithelial-mesenchymal transformation (EMT) in bladder cancer cells
Statistical analysis of wound healing and Transwell tests revealed significantly lower relative migration distance and number of invading cells in UMUC3 and T24 cells with SAPCD2 knockdown (p < 0.01) (Figs. 6A, 6B).
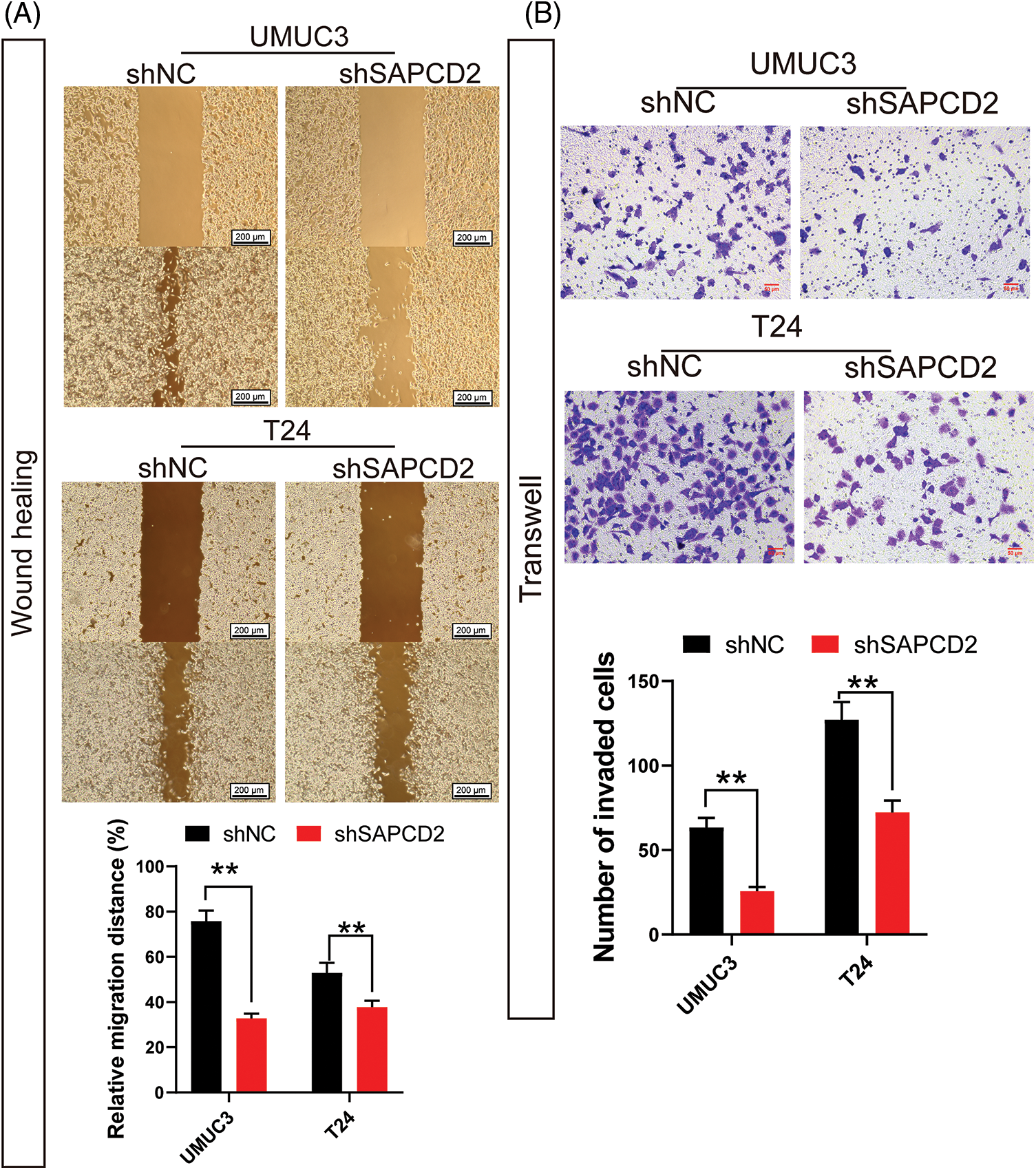
Figure 6: Knockdown of suppressor anaphase-promoting complex domain containing 2 (SAPCD2) inhibited the migration and invasiveness of bladder cancer cells. (A) Cell mobility was measured by the wound healing assay (n = 3) (scale bar = 200 μm). (B) Cell invasiveness was measured by the wound healing assay (n = 3) (×400, scaler bar = 50 μm). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
Considering the correlation between SAPCD2 and EMT, A comparison between the shNC and shSAPCD2 groups of UMUC3 and T24 cells revealed that following SAPCD2 knockdown, E-cadherin protein levels were up-regulated, whereas the levels of N-cadherin, vimentin, and Snail protein were notably downregulated (p < 0.01) (Figs. 7A, 7B).
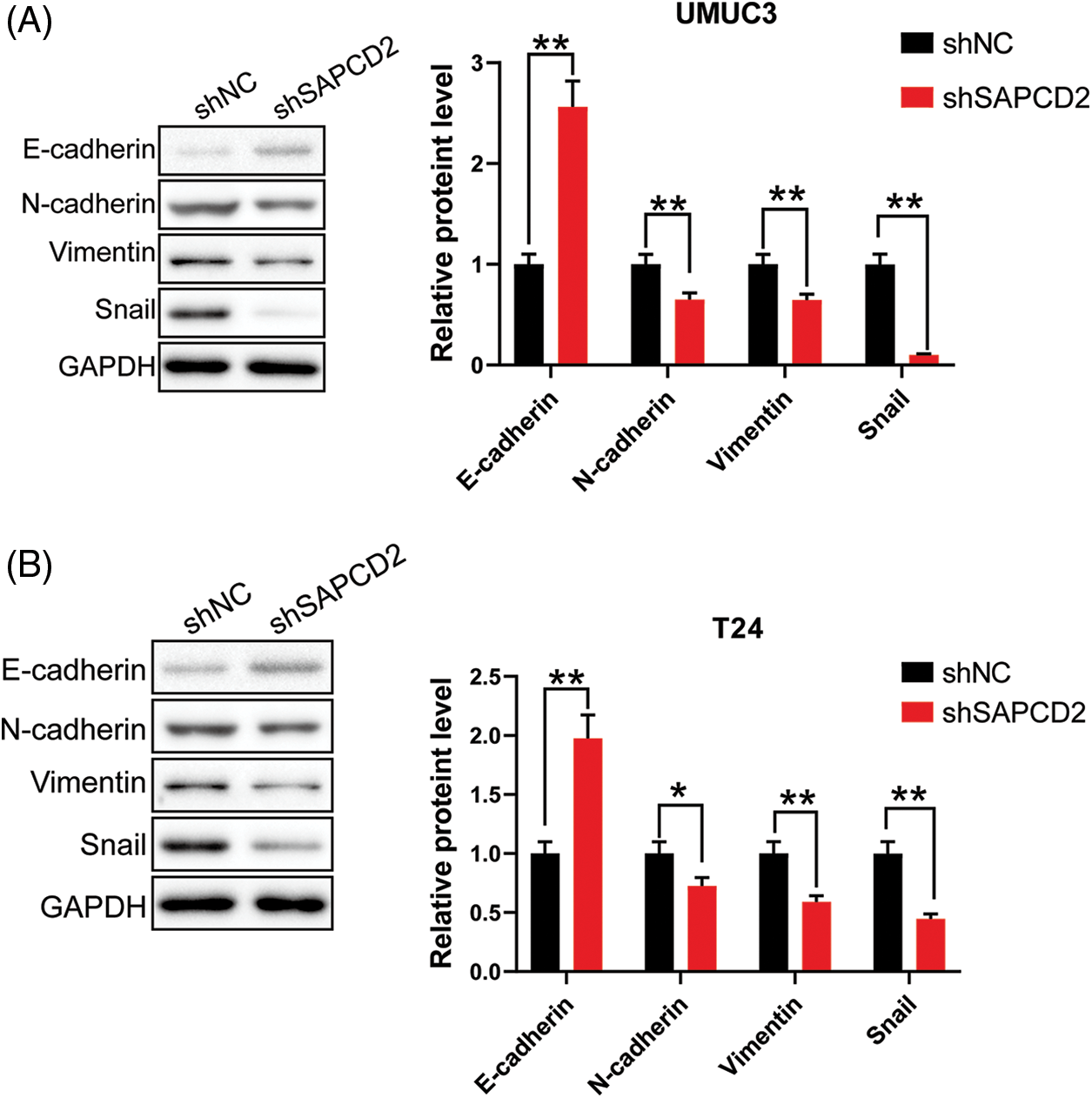
Figure 7: Knockdown of suppressor anaphase-promoting complex domain containing 2 (SAPCD2 inhibited epithelial-mesenchymal transformation of bladder cancer cells. (A) and (B) The relative levels of E-cadherin, N-cadherin, vimentin, and Snail proteins were detected by western blotting (n = 3). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
SAPCD2 knockdown modulated β-catenin/c-Myc/CDK4 signaling in bladder cancer cells
UMUC3 and T24 cells were treated with 20 mmol/L LiCl to examine the relationship between SAPCD2 and the β-catenin/c-Myc/CDK4 pathway in BC. As illustrated in Fig. 8A, following SAPCD2 knockdown in UMUC3 cells, the levels of SAPCD2, c-Myc, CDK4, cyclin D1, Snail, and N-cadherin protein were noticeably reduced while that of E-cadherin increased (p < 0.01). Furthermore, LiCl intervention essentially nullified the shSAPCD2-induced suppression of the β-catenin/c-Myc/CDK4 pathway by stimulating the expression of β-catenin, c-Myc, cyclin D1, CDK4, N-cadherin, Snail polypeptides and reducing the expression of E-cadherin protein (p < 0.01) (Fig. 8A).
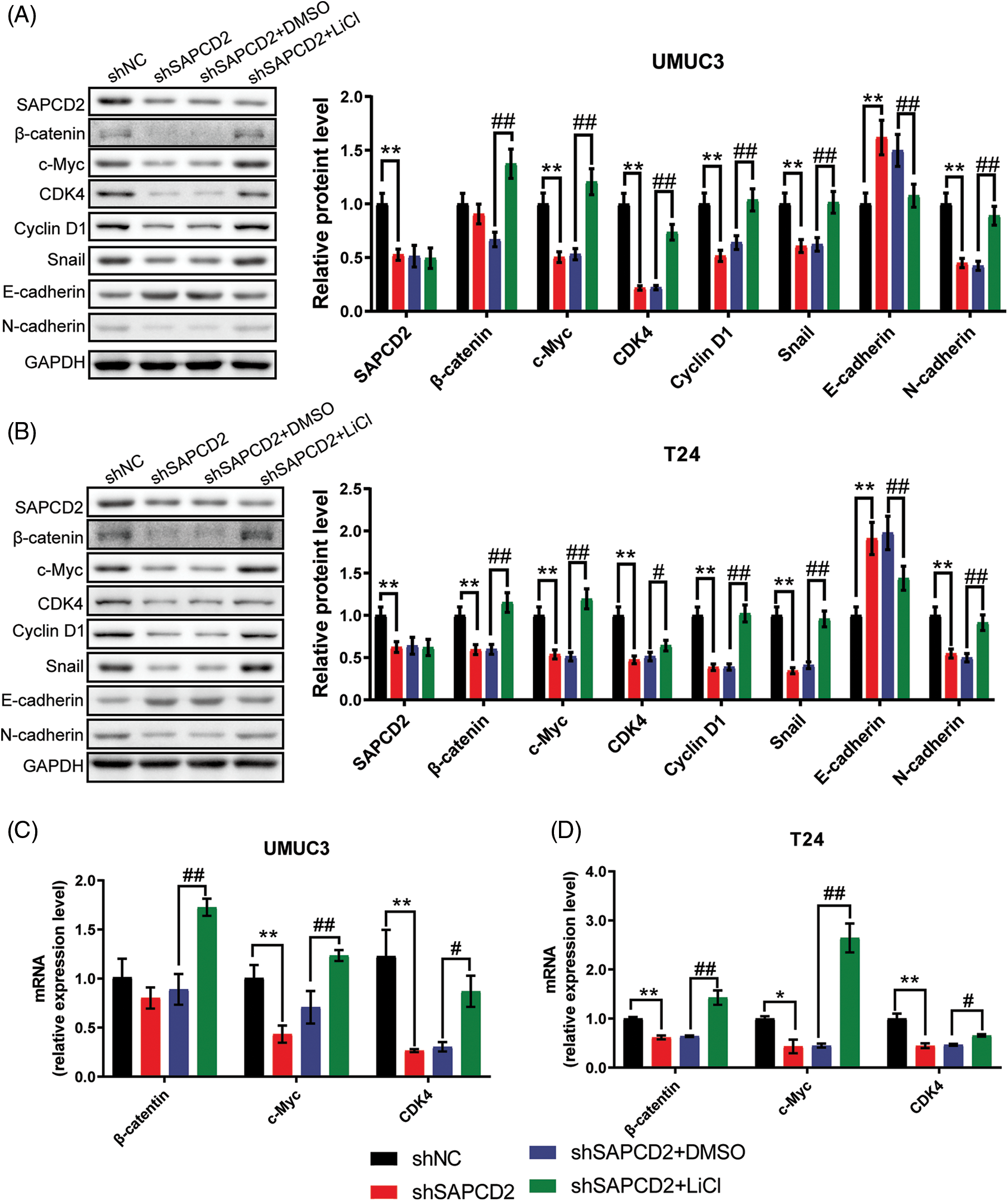
Figure 8: Suppressor anaphase-promoting complex domain containing 2 (SAPCD2) modulated the β-catenin/c-Myc/CDK4 signaling pathway in bladder cancer (BC) cells. (A, B) The relative protein levels of SAPCC2, β-catenin, c-Myc, CDK4, cyclin D1, Snail, E-cadherin, E-cadherin, and N-cadherin in UMUC3 and T24 cells were detected by western blotting (n = 3). (C, D) β-Catenin, c-Myc, and CDK4 mRNA were evaluated by real-time-quantitative polymerase chain reaction (n = 3). Values are means ± SD, *p < 0.05, **p < 0.01, vs. shNC group, #p < 0.05, ##p < 0.01, vs. shSAPCD2 group.
Likewise, knockdown of shSAPCD2 in T24 cells increased the expression of E-cadherin protein, but inhibited SAPCD2, β-catenin, c-Myc, CDK4, cyclin D1, and Snail expression (p < 0.01) (Fig. 8B). In the shSAPCD2+LiCl group of T24 cells, a visible disparity was noted between the shSAPCD2+DMSO group; the degrees of proteins in the Wnt signaling pathway such as β-catenin, c-Myc, cyclin D1, CDK4, N-cadherin, and Snail proteins increased significantly, conversely the level of E-cadherin was reduced markedly (p < 0.05) (Fig. 8B).
Additionally, β-catenin, c-Myc, and CDK4 mRNA levels were evaluated by RT-qPCR and significantly decreased by 20.7%, 56.9% (p < 0.01), and 74.5% (p < 0.01) in UMUC3 cells post-SAPCD2 knockdown, respectively. However, this reduction was reversed by LiCl intervention (Fig. 8C). Similarly, post-SAPCD2 knockdown in T24 cells, a decrease of 38.4%, 56.6%, and 55.6% was noted in mRNA levels of β-Catenin, c-Myc and CDK4, respectively; these effects were significantly reversed by LiCl treatment (p < 0.05) (Fig. 8D).
Knockdown of SAPCD2 reduced the tumorigenicity of UMUC3 cells in vivo
The correlation between SAPCD2 and the tumorigenicity of BC in vivo was observed by the xenografted tumor model and IHC. The growth of tumors with SAPCD2 knockdown was slower than those in the shNC group, and the tumor weight in the shSAPCD2 group was lighter (p < 0.05) (Figs. 9A, 9B).
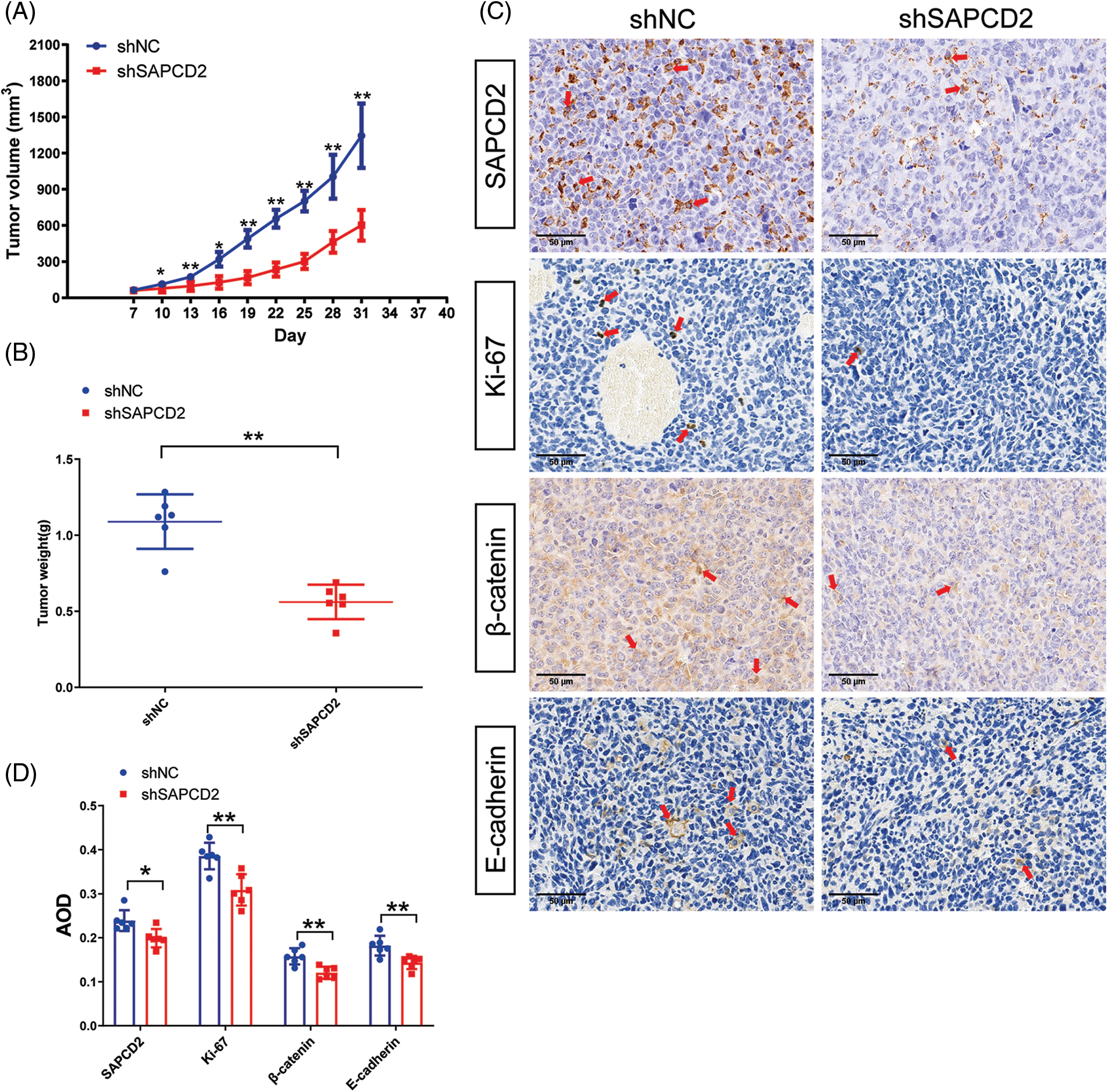
Figure 9: Suppressor anaphase-promoting complex domain containing 2 (SAPCD2) knockdown reduced tumorigenicity of bladder cancer (BC) in vivo. (A, B) Tumor volume and weight were noted by the xenografted tumor model. Tumor volumes were measured every 5 days (n = 6). (C) Representative images of SAPCD2, Ki67, β-catenin, and E-cadherin immunohistochemical staining in xenograft tumors from shNC and shSAPCD2 mice (×400, scaler bar = 50 μm) (n = 6). (D) Results of quantitative and statistical analysis of immunohistochemistry. Values are presented as means ± SD, *p < 0.05, **p < 0.01, vs. shNC group.
Furthermore, compared with the shNC, the degrees of SAPCD2, β-catenin, and Ki-67 protein were remarkably reduced, while the level of E-cadherin was immensely increased in the shSAPCD2 group (Figs. 9C, 9D).
SAPCD2 stands as a crucial chromosome-associated gene integral to cell cycle progression and is crucially involved in tumor proliferation, migration, and invasion [19]. The activation of SAPCD2 promotes proliferation and inhibits the death of triple negative breast cancer cells [20]; correspondingly, silencing SAPCD2 activates the Hippo signaling pathway and curtails fibrosarcoma proliferation [14]. Also, Luo et al. noticed that SAPCD2 encourages proliferation and invasion in colorectal cancer cells [10]. Moreover, TCGA and GEO Data revealed that SAPCD2 was up-regulated in BC tissue, implying its potential in the development of BC [15]. Therefore, in this study, statistical analysis data was collected from BC clinical samples within the TCGA database on UALCAN, and the gene and protein levels of SAPCD2 in BC cell lines (WU780, UMUC3, T24, and BIU87) were analyzed to clarify the high expression of SAPCD2 in BC investigating the biological role of SAPCD2 in BC malignant behavior.
The SAPCD2 knockdown BC cell model showed impaired cell viability and cloning ability of BC cells, as well as a stagnated cell cycle in the G0/G1 phase. Cyclin D1/B1, c-Myc, CDK4, and P21 are biomarkers of the cell cycle [21]. CDK4/6 inhibitors have been used to treat breast cancer [22]. This study showed that SAPCD2 knockdown inhibits the cell cycle signaling pathway. Furthermore, the up-regulation of P21 is facilitated by the pivotal tumor suppression factor p53, resulting in mitigated progression through the G1 phase of the cell cycle [23]. Consequently, in this study, the observed effects on cell cycle progression might be a result of the obstruction of p53 activity. Further exploration of the influence of p53 on BC proliferation post SAPCD2 knockdown could be an interesting topic for future research.
In this study, SAPCD2 knockdown inhibited the invasiveness and migration capability of BC cells and the EMT signaling pathway. One study reported that the genetic characteristics of the EMT signaling pathway can be used to predict BC patient prognosis [24]. SAPCD2 is notably correlated with poor prognosis [25], and the malignant progression of the tumor mainly involves sustained proliferation and high invasive capacity. We observed that SAPCD2 knockdown inhibited the malignant development of BC, suggesting the potential of SAPCD2 knockdown in improving the poor prognosis of BC.
In order to investigate the effects of SAPCD2 on BC, we assessed the expression of the related proteins in the β-catenin/c-Myc/CDK4 pathway. The β-catenin/c-Myc/CDK4 pathway participates in the regulation of oncogenic signaling communication, serving as a crucial factor in stimulating the rapid proliferation, differentiation, and transference of tumor cells [26]. The c-Myc/CDK4 axis is an important part of the Wnt pathway, and is one of the most abnormal pathways in the development of tumors [27]. In particular, scientists have proposed that this pathway is closely associated with BC [28]. In one recent research, inhibition of the β-catenin/c-Myc pathway suppressed the viability of cancer cells [29]. This study noticed that SAPCD2 silencing repressed the signal strength of the β-catenin/c-Myc/CDK4 pathway protein expression. SAPCD2 knockdown antagonized the development of BC by inhibiting the β-catenin/c-Myc/CDK4 pathway.
The correction of β-catenin/c-Myc/CDK4 elements is helpful to BC treatment. Li et al. reported a superior reduction of nuclear β-catenin in cells that obtained cisplatin treatment [30]. Previously, researchers had indicated dysregulation of the Wnt/β-catenin pathway within the neuronal subset of BC [31]. The present investigation disclosed that SAPCD2 affects β-catenin accumulation in neuroendocrine-like cell line UMUC3, while β-catenin expression was subdued in the basal squamous cell strain T24 upon SAPCD2 silencing. They all have mutations in p53 [32], suggesting the potential correlation between SAPCD2, p53, and the Wnt/β-catenin signaling pathway. Additionally, SAPCD2 may regulate β-catenin to activate the progress of EMT [11,33]. EMT leads to the transformation of cell morphology from polygonal to fusiform, making cells more prone to distant metastasis and leading to poor cancer prognosis. The β-Catenin/c-Myc/CDK4 pathway expression in the SAPCD2 knockdown BC cells is low, with reduced c-Myc and CDK4 proteins, up-regulated E-cadherin, and down-regulated mesenchymal markers (N-cadherin, vimentin, and Snail). These phenomena could be antagonized by LiCl, the agonist of the Wnt/β-catenin pathway [34]. LiCl does not change SAPCD2 expression and up-regulated the β-catenin/c-Myc/CDK4 signaling pathway, indicating that it is one of the key pathways for BC malignant behavior after SAPCD2 knockdown. An animal model further proved that SAPCD2 knockdown could inhibit the tumorigenicity of BC cells. Nonetheless, more evidence is still needed to prove the direct role of SAPCD2 in the β-catenin/c-Myc/CDK4 pathway of BC and the toxic side effects of knockdown SAPCD2 on normal cells; these will be considered in the future.
Furthermore, accumulating evidence indicates that the activation of the Wnt/β-catenin signaling pathway correlates with poor T cell infiltrates across human malignancies, suggesting this pathway may represent a potential mechanism for immune evasion [35]. Even though immune checkpoint blockade has become a common strategy for treating BC, it lacks significant clinical benefit with a poorly understood mechanism [36]. These studies indicated the involvement of SAPCD2 in β-catenin/c-Myc/CDK4 signaling, which may influence the immune escape of BC cells. Therefore, further investigation of the mechanisms underlying immune evasion in BC is necessary.
This study has several limitations. The current study preliminarily explores the role of SAPCD2 in BC, focusing on the suppression of BC malignancy after SAPCD2 knockdown. However, further research is needed to fully understand the implications of SAPCD2 in BC cells/tissues and normal cells. Further, although, the study primarily focused on the role of SAPCD2 suppression in BC malignancy, the impact of SAPCD2 overexpression on BC development remains unclear. Additionally, a xenografted tumor model was utilized, but clinical outcomes are better reflected by an orthotopic model of BC. The findings of this study, which are preliminary, are not yet suitable for clinical application and require additional animal experimentation and clinical usage evidence to support their clinical utility. Nonetheless, this research demonstrated the potential of SAPCD2 knockdown in suppressing BC proliferation, migration, and invasion, providing direction and a scientific basis for improving prognosis and clinical outcomes for BC patients.
The SAPCD2 knockdown suppressed the malignant phenotype of BC cells. Additionally, SAPCD2 knockdown blocked the cell cycle and EMT, including related protein expression. Besides, SAPCD2 knockdown inhibited the signal strength of the β-catenin/c-Myc/CDK4 pathway, which was reversed by LiCl. The β-catenin/c-Myc/CDK4 signaling pathway is one of the key pathways that inhibit the malignant behavior of BC cells after SAPCD2 knockdown. This study provides direction and scientific basis for improving prognosis and clinical outcomes for BC patients.
Acknowledgement: None.
Funding Statement: This study was supported by the Medical and Health Science and Technology Program of Zhejiang Province (No. 2021KY367).
Author Contributions: Jiajun Yan and Chong Shen conceived and designed this study. Chong Shen, Yu Ren, and Zhirong Zhu conducted experiments and collected data. Chong Shen, Zhirong Zhu, Xiaolong Zhang, Yu Ren, and Shuixiang Tao analyzed and interpreted data. Chong Shen drafted the manuscript. Jiajun Yan and Chong Shen reviewed the manuscript. Jiajun Yan obtained the funding.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: All animal tests were approved by the Animal Experimentation Ethics Committee of Zhejiang Eyong Pharmaceutical Research and Development Center (No. ZJEY-20211008-04).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [Google Scholar] [PubMed]
2. Lobo N, Afferi L, Moschini M, Mostafid H, Porten S, Psutka SP, et al. Epidemiology, screening, and prevention of bladder cancer. Eur Urol Oncol. 2022;5(6):628–39. [Google Scholar] [PubMed]
3. Aveta A, Cacciapuoti C, Barone B, di Zazzo E, Del Giudice F, Maggi M, et al. The impact of meat intake on bladder cancer incidence: is it really a relevant risk? Cancers. 2022;14(19):4775. [Google Scholar] [PubMed]
4. Seung SJ, Traore AN, Pourmirza B, Fathers KE, Coombes M, Jerzak KJ. A population-based analysis of breast cancer incidence and survival by subtype in Ontario women. Curr Oncol. 2020;27(2):e191–8. [Google Scholar] [PubMed]
5. Teoh JY, Huang J, Ko WY, Lok V, Choi P, Ng CF, et al. Global trends of bladder cancer incidence and mortality, and their associations with tobacco use and gross domestic product per capita. Eur Urol. 2020;78(6):893–906. [Google Scholar] [PubMed]
6. Ślusarczyk A, Zapała P, Zapała Ł., Borkowski T, Radziszewski P. Cancer-specific survival of patients with non-muscle-invasive bladder cancer: a population-based analysis. Ann Surg Oncol. 2023;30(12):7892–902. [Google Scholar] [PubMed]
7. Dyrskjøt L, Hansel DE, Efstathiou JA, Knowles MA, Galsky MD, Teoh J, et al. Bladder cancer. Nat Rev Dis Primers. 2023;9(1):58. [Google Scholar]
8. Aveta A, Cilio S, Contieri R, Spena G, Napolitano L, Manfredi C, et al. Urinary MicroRNAs as biomarkers of urological cancers: a systematic review. Int J Mol Sci. 2023;24(13):10846. [Google Scholar] [PubMed]
9. Wang Z, Zhou Y, Guan C, Ding Y, Tao S, Huang X, et al. The impact of previous cancer on overall survival of bladder cancer patients and the establishment of nomogram for overall survival prediction. Medicine. 2020;99(38):e22191. [Google Scholar] [PubMed]
10. Luo Y, Wang L, Ran W, Li G, Xiao Y, Wang X, et al. Overexpression of SAPCD2 correlates with proliferation and invasion of colorectal carcinoma cells. Cancer Cell Int. 2020;20:43. [Google Scholar] [PubMed]
11. Jiang J, Tang S, Xia J, Wen J, Chen S, Shu X, et al. C9orf140, a novel Axin1-interacting protein, mediates the negative feedback loop of Wnt/β-catenin signaling. Oncogene. 2018;37(22):2992–3005. [Google Scholar] [PubMed]
12. Zhang X, Nie X, Long J, Yu J, Zhang P, Liu Y, et al. Expression of p42.3 in non-small cell lung cancer. Ann Transl Med. 2020;8(13):819. [Google Scholar] [PubMed]
13. Zhang Y, Liu JL, Wang J. SAPCD2 promotes invasiveness and migration ability of breast cancer cells via YAP/TAZ. Eur Rev Med Pharmacol Sci. 2020;24(7):3786–94. [Google Scholar] [PubMed]
14. Zhu B, Wu Y, Niu L, Yao W, Xue M, Wang H, et al. Silencing SAPCD2 represses proliferation and lung metastasis of fibrosarcoma by activating hippo signaling pathway. Front Oncol. 2020;10:574383. [Google Scholar] [PubMed]
15. Xu Y, Wu G, Li J, Li J, Ruan N, Ma L, et al. Screening and identification of key biomarkers for bladder cancer: a study based on TCGA and GEO data. Biomed Res Int. 2020;2020:8283401. [Google Scholar] [PubMed]
16. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25(1):18–27. [Google Scholar] [PubMed]
17. Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–6. [Google Scholar] [PubMed]
18. Yokoo H, Nemoto T, Yanagita T, Satoh S, Yoshikawa N, Maruta T, et al. Glycogen synthase kinase-3β: homologous regulation of cell surface insulin receptor level via controlling insulin receptor mRNA stability in adrenal chromaffin cells. J Neurochem. 2007;103(5):1883–96. [Google Scholar] [PubMed]
19. Sun Z, Mao Y, Zhang X, Lu S, Wang H, Zhang C, et al. Identification of ARHGEF38, NETO2, GOLM1, and SAPCD2 associated with prostate cancer progression by bioinformatic analysis and experimental validation. Front Cell Dev Biol. 2021;9:718638. [Google Scholar] [PubMed]
20. Yang Z, Ding H, Pan Z, Li H, Ding J, Chen Q. YY1-inudced activation of lncRNA DUXAP8 promotes proliferation and suppresses apoptosis of triple negative breast cancer cells through upregulating SAPCD2. Cancer Biol Ther. 2021;22(3):216–24. [Google Scholar] [PubMed]
21. Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39(6):759–78. [Google Scholar] [PubMed]
22. Piezzo M, Cocco S, Caputo R, Cianniello D, Gioia GD, Lauro VD, et al. Targeting cell cycle in breast cancer: CDK4/6 inhibitors. Int J Mol Sci. 2020;21(18):6479. [Google Scholar] [PubMed]
23. Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29(5):946–60. [Google Scholar] [PubMed]
24. Cao R, Yuan L, Ma B, Wang G, Qiu W, Tian Y. An EMT-related gene signature for the prognosis of human bladder cancer. J Cell Mol Med. 2020;24(1):605–17. [Google Scholar] [PubMed]
25. Zhang ZM, Cao HB, Li ZH, Zhuo R, Tao YF, Li XL, et al. SAPCD2 promotes neuroblastoma progression by altering the subcellular distribution of E2F7. Cell Death Dis. 2022;13(2):174. [Google Scholar] [PubMed]
26. Liu T, Fan MQ, Xie XX, Shu QP, Du XH, Qi LZ, et al. Activation of CTNNB1 by deubiquitinase UCHL3-mediated stabilization facilitates bladder cancer progression. J Transl Med. 2023;21(1):656. [Google Scholar] [PubMed]
27. Li C, Li SZ, Huang XC, Chen J, Liu W, Zhang XD, et al. PTPN18 promotes colorectal cancer progression by regulating the c-MYC-CDK4 axis. Genes Dis. 2021;8(6):838–48. [Google Scholar] [PubMed]
28. Chehrazi-Raffle A, Dorff TB, Pal SK, Lyou Y. Wnt/β-catenin signaling and immunotherapy resistance: lessons for the treatment of urothelial carcinoma. Cancers. 2021;13(4):889. [Google Scholar] [PubMed]
29. Zhang W, Huang J, Tang Y, Yang Y, Hu H. Inhibition of fatty acid synthase (FASN) affects the proliferation and apoptosis of HepG2 hepatoma carcinoma cells via the β-catenin/C-myc signaling pathway. Ann Hepatol. 2020;19(4):411–6. [Google Scholar] [PubMed]
30. Li T, Tang Z, Li C, Liu X, Cheng L, Yang Z, et al. Magnesium-assisted cisplatin inhibits bladder cancer cell survival by modulating Wnt/β-catenin signaling pathway. Front Pharmacol. 2021;12:804615. [Google Scholar] [PubMed]
31. Eray A, Güneri PY, Yılmaz G, Karakülah G, Erkek-Ozhan S. Analysis of open chromatin regions in bladder cancer links β-catenin mutations and Wnt signaling with neuronal subtype of bladder cancer. Sci Rep. 2020;10(1):18667. [Google Scholar] [PubMed]
32. Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2020;77(4):420–33. [Google Scholar] [PubMed]
33. Zhang J, Cai H, Sun L, Zhan P, Chen M, Zhang F, et al. LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J Exp Clin Cancer Res. 2018;37(1):225. [Google Scholar] [PubMed]
34. Song P, Feng L, Li J, Dai D, Zhu L, Wang C, et al. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer. 2020;19(1):129. [Google Scholar] [PubMed]
35. Gao R, Zheng X, Jiang A, He W, Liu T. Modulating β-catenin/BCL9 interaction with cell-membrane-camouflaged carnosic acid to inhibit Wnt pathway and enhance tumor immune response. Front Immunol. 2023;14:1274223. [Google Scholar] [PubMed]
36. Chen X, Xu R, He D, Zhang Y, Chen H, Zhu Y, et al. CD8+ T effector and immune checkpoint signatures predict prognosis and responsiveness to immunotherapy in bladder cancer. Oncogene. 2021;40(43):6223–34. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools