 Open Access
Open Access
ARTICLE
Meiotic nuclear divisions 1 suppresses the proliferation and invasion of pancreatic cancer cells via regulating H2A.X variant histone
1 Anhui Province Key Laboratory of Clinical and Preclinical Research in Respiratory Disease, Molecular Diagnosis Center, The Department of Pulmonary Critical Care Medicine, First Affiliated Hospital of Bengbu Medical College, Bengbu, 233030, China
2 Department of Genetics, School of Life Sciences, Bengbu Medical College, Bengbu, 233000, China
3 Department of Clinical Medicine, Bengbu Medical College, Bengbu, 233030, China
4 Research Center of Clinical Laboratory Science, Bengbu Medical College, Bengbu, 233030, China
* Corresponding Authors: XIAOJING WANG. Email: ; CHAOQUN LIAN. Email:
BIOCELL 2024, 48(1), 111-122. https://doi.org/10.32604/biocell.2023.046903
Received 18 October 2023; Accepted 23 November 2023; Issue published 30 January 2024
Abstract
Introduction: Among all malignant tumors of the digestive system, pancreatic carcinoma exhibits the highest mortality rate. Currently, prevention and effective treatment are urgent issues that need to be addressed. Methods: The study focused on meiotic nuclear divisions 1 (MND1), integrating data from the Gene Expression Profiling Interactive Analysis (GEPIA) database with prognostic survival analysis. Simultaneously, experiments at cellular level were employed to demonstrate the effect of MND1 on the proliferation and migration of PC. The small-molecule inhibitor of MND1 was used to suppress the migration of PC cells by knocking down MND1 using small interfering RNA (siRNA) in Patu-8988 and Panc1 cell lines. Results: The results of Cell Counting Kit-8 indicated that the suppression of MND1 resulted in a decrease in cell proliferation. Wound healing and Transwell assays revealed that MND1 knockdown reduced cell migration and invasion. Flow cytometry revealed that inhibiting MND1 hindered the cell cycle. Furthermore, MND1 could stimulate the proliferation, migration, and invasion of Patu-8988 and Panc1 cells by increasing the expression of MND1. Notably, MND1 had a positive effect on H2AFX expression in PC cells. Elevated MND1 expression suggests the low overall survival rate of individuals diagnosed with PC. Conclusion: These findings suggest that MND1 has the potential to be a gene with the ability to accurately diagnose and treat PC.Keywords
Pancreatic carcinoma (PC) is one of the most dangerous cancers related to the digestive tract, ranking fourth and sixth in America and China, respectively, as the leading cause of cancer-related death [1], and it is projected to be the second leading cause of cancer-related death by 2030 [2]. PC is a malignant disease characterized by low survival and high recurrence rate, and patients are mostly at the stage of locally advanced or metastatic disease when first diagnosed [3]. Presently, imaging examination and CA19-9 (cancer antigen 19-9) were used to diagnose PC [4,5], however, the overall survival failed to improve due to resistance to chemotherapy drugs [6,7]. Therefore, it is crucial to define the fundamental molecular process and discover new therapies for PC.
Meiotic nuclear divisions 1 (MND1) plays an essential role in meiosis and associates with homologous pairing protein 2 (HOP2) to create a stable heterodimeric complex, facilitating the repair of homologous chromosome pairing DNA double-strand breaks (DSBs) repair during meiosis [8,9]. In squamous cell lung cancer, lung adenocarcinoma, kidney renal clear cell carcinoma, and hepatocellular carcinoma, MND1 has been highly shown to be demonstrated, serving as a potential biomarker for poor prognosis [9–13]. In previous studies, we confirmed the oncogenic function of MND1 in human PC cells. MND1 has the ability to stimulate cell growth and advance cell cycle progression. The current research seeked to confirm that H2AFX is a downstream regulatory molecule of MND1 through experimental methods, taking into account the earlier finding that MND1 has an oncogene role in PC.
H2A.X variant histone (H2AFX) is one part of the histone H2A gene family, essential for DNA repair, replication, transcription regulation, and chromosomal stability [14]. Previous studies have confirmed the presence of H2AFX in various types of cancers, such as hepatocellular carcinoma [14,15], Leukemia [16], head and neck squamous cell carcinoma [17], and non-Hodgkin lymphoma [18]; nevertheless, the effects of H2AFX on the progression of PC malignancy have not been documented. In order to achieve this objective, we evaluated how H2AFX was expressed in PC cells when MND1 was either inhibited or overexpressed.
Our comprehensive analysis revealed the function of MND1 in the cell growth, cell cycle, migration, and invasion in vitro. Additionally, H2AFX was pinpointed as a target for MND1 in PC cells. Furthermore, our research investigated if MND1 performs its biological role by controlling H2AFX in PC cells, demonstrating MND1’s tumor-enhancing effects through increased H2AFX levels in PC cells. The findings of our research offer a justification for focusing on MND1 as an innovative treatment target in PCs.
The TIMER2.0 database [19] was used to acquire the MND1 expression data of pancreatic. The survival prognosis data of MND1 were sourced from GEPIA2.0 [20] and Kaplan Meier Plotter databases [21].
Human PC cells, including Patu-8988 and PANC1 cells, were cultured in Dulbecco’s modified Eagle’s medium (Gibco, New York, USA) with 10% fetal bovine serum, kept in a water-jacketed incubator at 37°C with 5% CO2. The most common driver mutations in PC are in genes KRAS, TP53, CDKN2A, and SMAD4. There is an early molecular event about KRAS gene mutation in PC, and the most common mutation sites are G12D and G12V. In reference [22], G12D mutation was detected in PANC-1 cells, and G12V mutation was detected in Patu8988 cells. The growth of PANC-1 cells was inhibited by 1 U/mL levaspase. PANC-1 cells were able to grow on soft agar and to tumorize in nude mice, while Patu-8988 were not able to tumorize in nude mice.
Cell Counting Kit-8(CCK8) was purchased from Sigma (St. Louis, Mo, USA). Transwell inserts and Matrigel were bought from Corning (Corning, New York, USA). Proteintech Group provided us with anti-MND1 (Cat#: 11636-1-AP, Wuhan, China) and anti-Vinculin (66305-1-ig, Wuhan, China) antibodies. Bimake was the source of Anti-H2AFX (A5728, Houston, Texas, USA), whereas anti-Tublin (#2148S, Proteintech Group Wuhan, China) and secondary antibodies were procured from Cell Signaling Technology (Danvers, Massachusetts, USA).
Transfection of siRNA sequences and plasmids
The siRNA (GenePharma, Shanghai, China) sequences were transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The MND1 siRNA sequences were designed in the following manner: 5′-CGC AAG UUG UGG AAG AAA UTT-3 (sense) and 5′-AUU UCU UCC ACA ACU UGC GTT-3′ (antisense). Design of the H2AFX siRNA sequences was as such: H2AFX-Homo-286, 5′-GAC AAC AAG AAG ACG CGA ATT-3′(sense) and 5′-UUC GCG UCU UCU UGU UGU CTT-3′ (antisense); H2AFX-Homo-1181, 5′-CUC CCU CCA UCU UCA UUC ATT-3′(sense) and 5′-UGA AUG AAG AUG GAG GGA GTT-3′(antisense); MND1-Homo-1408, 5′-GCA AAC UCA ACU CGG CAA UTT-3′ (sense) and 5′-AUU GCC GAG UUG AGU UUG CTT-3′ (antisense). All oligodeoxynucleotides referenced previously were obtained from Shanghai GenePharma, China.
Transfection of H2AFX plasmid (Youbio Changsha, China) sequences was conducted using Lipofectamine 3000 and p3000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Subcloning of the H2AFX plasmid occurred into the mammalian expression structure pcDNA3.1. The cells were examined in accordance with the instructions provided in the results section.
Extraction of RNA and real-time polymerase chain reaction (RT-PCR)
Total cellular RNA was obtained using Magen Reagent (Biotechnology Co., Ltd., Guangzhou, China) following the guidelines provided by the manufacturer. The SYBR green RT-PCR assay (Vazyme, Nanjing, China) was employed to measure the levels of MND1 and H2AFX, which were then standardized against GAPDH. Primers used in PCR are as follows: MND1, forward primer, 5′-TGT GAG AGG ATC GGA ACT TCT-3′, Reverse Primer, 5′-CAC ATC GGC CAA TTT TAG CTT TC-3′, H2AFX, forward primer, 5′-TTC ACC GGT CTA CCT CGA TA-3′, reverse primer, 5′-CGG CTC AGC TCT TTC CAT GA-3′, GAPDH, forward primer, 5′-GGA GCG AGA TCC CTC CAA AAT-3′, reverse primer, 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′.
Total protein was isolated through a chilled radioimmunoprecipitation assay (RIPA, Beyotime Biotechnology, Beijing, China) solution containing proteinase inhibitors. Quantification of the protein was conducted using a BCA assay kit (Beyotime Biotechnology, Beijing, China), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene fluoride (PVDF, Massachusetts, USA) membranes and with a fast-blocking solution (Epizyme, Shanghai, China) for 15–30 min. Then, the PVDF membranes were incubated with the indicated antibodies as follows: MND1, H2AFX, vinculin, and tubulin, and were kept 4°C for at least 12 h, as described previously [22]. Subsequently, the membrane underwent three washes using Tris-buffered saline (Servicebio, Wuhan, China) containing Tween 20 (NeoFroxx GmbH, Einhausen, Germany), each for 10 min. And then it underwent incubation with the secondary antibody working solution for 2 h at ambient temperature.
PC cells underwent seeding in 6-well plates (Corning, New York, USA) overnight and transfected with MND1 siRNA or cDNA and H2AFX siRNA or cDNA for 24 h. Then, these cells were trypsinized and placed in 96-well plates (Corning, New York, USA) for 24, 48, and 72 h. Cell viability was detected by CCK8 assay as described previously [23].
Following the aforementioned transfection process, PC cells were planted in 6-well plates and left to incubate overnight until they reached 95% confluence. To create the scratch wound, the surface cells of the plates were scraped using a pipette tip, followed by cleansing the separated cells with PBS (Servicebio, Wuhan, China). Then, the residual connected cells in 6-well plates underwent a 24-h incubation. The wound healing in each well at 0 h and 24 h was photographed.
Transwell migration and invasion assay
Cell migration and invasion were detected by transwell assay as described before [22]. Cells were placed in transwell chambers with 8.0-mm pore membranes within 24-well plates (Corning, New York, USA). In the invasion assay, the chambers were set up by applying a slender coating of Matrigel (Corning, New York, USA) prior to cell seeding. Following a 24-h incubation period, the bottom-surfaced cells of the chamber were stained with Giemsa solution (Beyotime Biotechnology, Beijing, China), captured in photographs, and tallied using a microscope. Migration is the ability of a cell to move, while invasion is the ability of a cell to secrete proteins that digest the extracellular matrix (ECM) or basement membrane extract (BME) while moving, removing barriers to movement, an experiment that better mimics the in vivo environment of the human body.
PC cells were seeded in a 60 mm dish (Corning, New York, USA) at 5 × 105 cells/well and incubated overnight. After transfection with different cMND1 cDNA constructs, MND1 siRNA, H2AFX cDNA construct, and H2AFX siRNA as mentioned above. Over a period of 24 h, cells were gathered, washed with cold PBS, and stabilized using 70% ethanol (Macklin, Shanghai, China) for at least 4 h at 4°C. Following three cold PBS washes, the cell pellets were reconstituted in 500 μL of 1 × PI staining solution (Beyotime Biotechnology, Beijing, China) and left to incubate for 30 min at 37°C in darkness. Subsequently, the cell cycle was assessed using a BD Aria III Flow Cytometer (BD Biosciences, San Jose, California, USA), and each phase (sub-G1, G1, S, G2) was analyzed using FlowJo software (BD Biosciences, San Jose, California, USA).
Statistical examination of the data was conducted using the Student’s t-test for two-group comparisons and ANOVA for multiple group comparisons, employing GraphPad Prism 8.0 (GraphPad, La Jolla, CA, USA). A p-value less than 0.05 was deemed to hold statistical significance.
Pancreatic carcinoma has an increased expression of meiotic nuclear divisions 1
We employed the GEPIA database to investigate the presence of MND1 in both tumor and non-tumor tissues by examining its expression in PC. The GEPIA database indicates that MND1 is expressed at a high level, as depicted in (Fig. 1A). Concurrently, we used the Kaplan Meier analysis to investigate the prognosis of the high and low expression groups of MND1 in PC. The finding showed that the elevated MND1 expression predicted an unfavorable prognosis (Fig. 1B). In addition, we employed western blotting and qRT-PCR to explore MND1 expression in both normal pancreatic epithelial cells and PC. As illustrated in Figs. 1C and 1D, the MND1 expression was higher than the MND1 expression in the normal pancreatic epithelial cells Line HPDEC6-7.
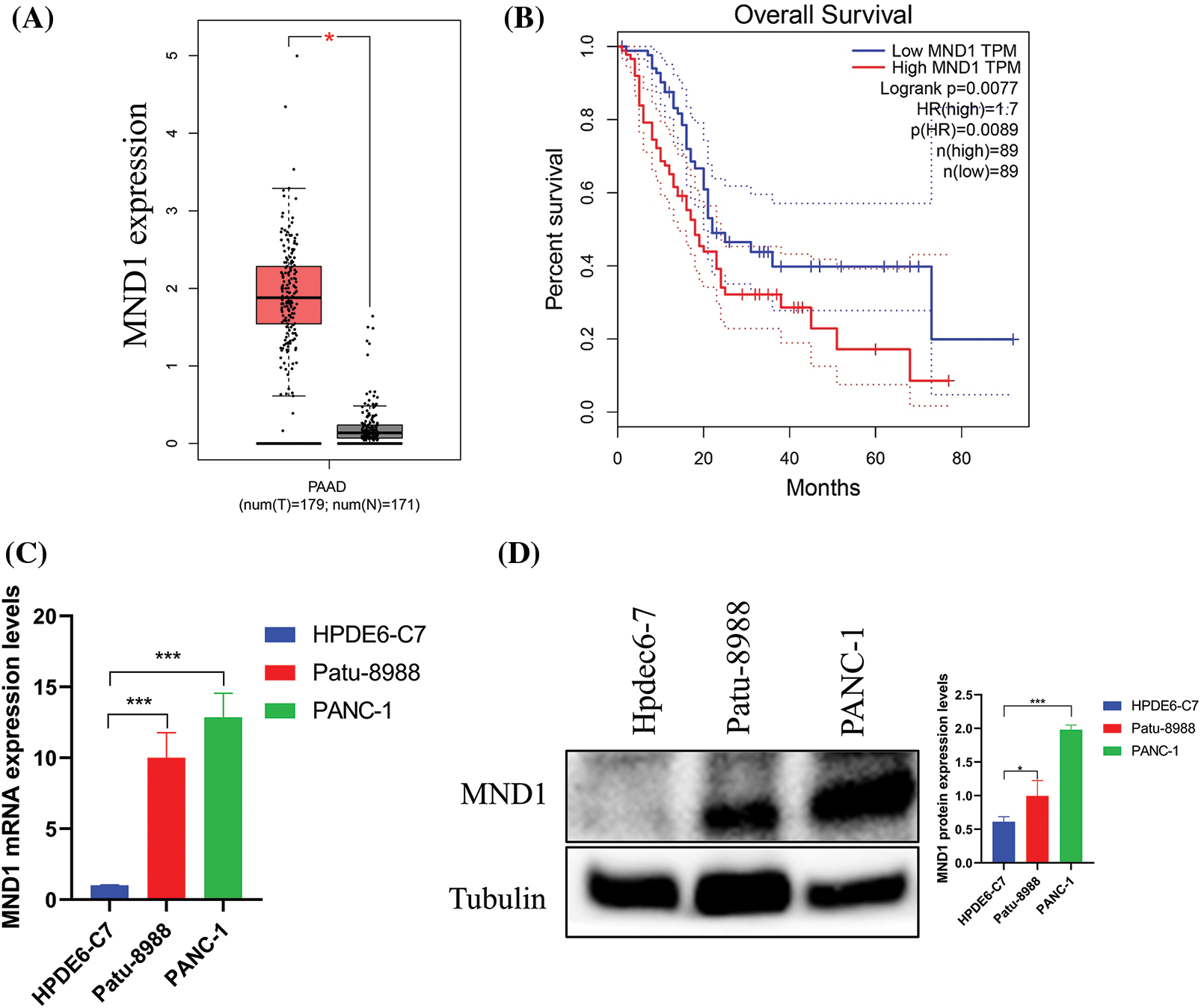
Figure 1: Meiotic nuclear divisions 1 (MND1) is overexpression in PC. (A) MND1 is highly expressed in the gene expression profiling interactive analysis (GEPIA) database. (B) KM survival curve for MND1 in GEPIA database. (C and D) Compared with normal pancreatic epithelial cells (HPDE6-C7), MND1 is highly expressed at the RNA and protein levels in Patu-8988 and PANC-1 cells. The data presentation was in the form of mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
Meiotic nuclear divisions 1 overexpression promoted cell proliferation, migration, and invasion in vitro
We introduced the MND1 cDNA construct into PC cells and noticed an increase in MND1 mRNA and protein levels when MND1 cDNA was transfected into PC cells (Figs. 2A and 2B). The CCK8 assay results (Fig. 2C) demonstrated that an increase in MND1 expression led to an increase in cell proliferation. We also performed a cell scratch test and a transwell test to ascertain the migration and invasion capabilities of PC cells. From the wound healing assay results, we saw that overexpressed MND1 promoted cell migration in PC cells (Fig. 2D). Meanwhile, we found that overexpression of MND1 enhanced the migration and invasion capacity in PC cells (Fig. 2E). Migration is the ability of a cell to move, while invasion is the ability of a cell to secrete proteins that digest the ECM or BME while moving, removing barriers to movement, an experiment that better mimics the in vivo environment of the human body. To put it succinctly, overexpression of MND1 in Patu-8988 and PANC-1 cells resulted in enhanced growth, migration, and invasion.
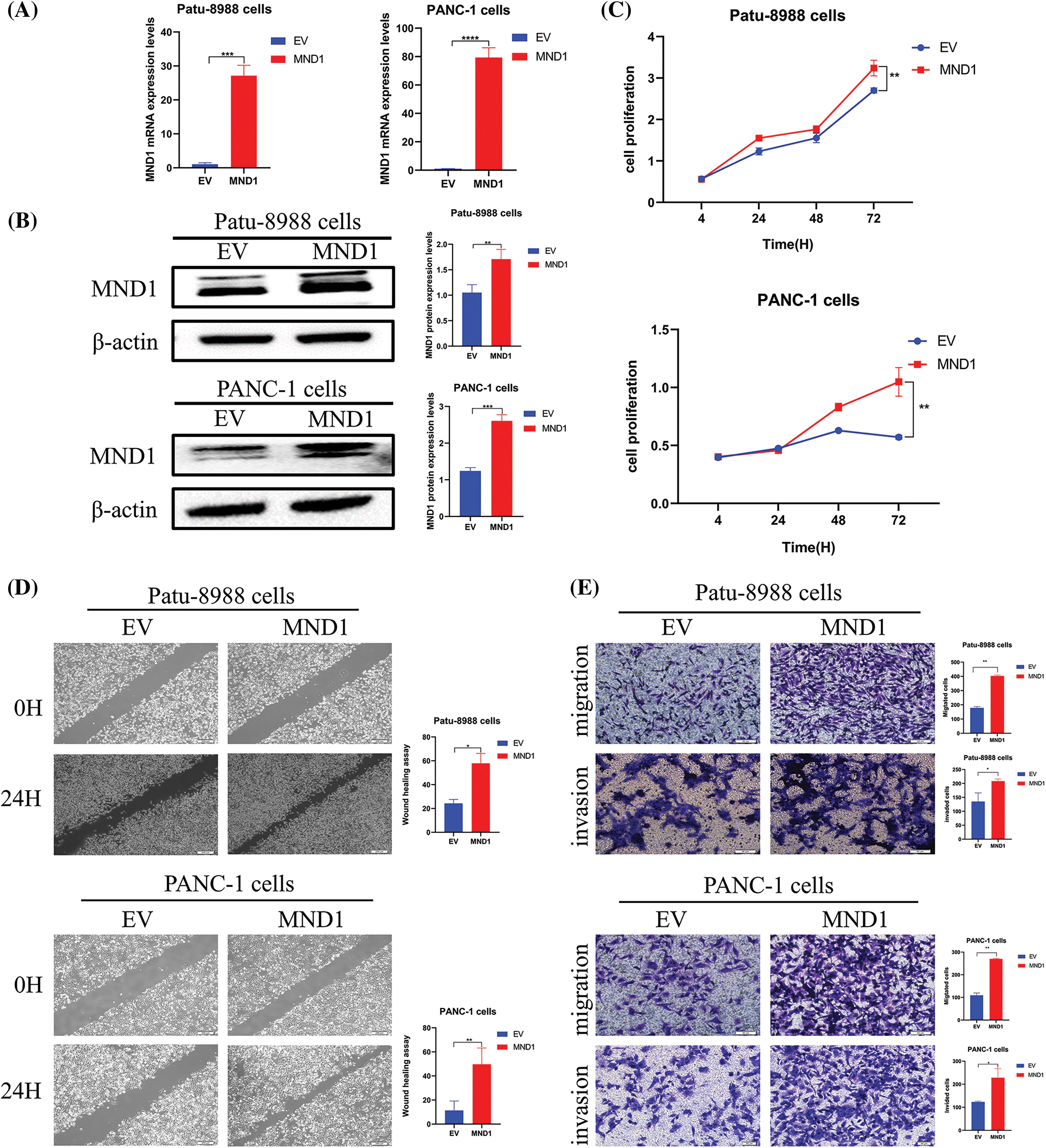
Figure 2: Meiotic nuclear divisions 1 (MND1) overexpression promoted cell proliferation, migration, and invasion in vitro. (A and B) Quantitative real-time polymerase chain reaction and western blotting validated the overexpression effect of MND1 plasmid. (C) CCK-8 assay was conducted in pancreatic cancer cells after MND1 overexpression. (D) Wound healing assay was conducted to assess the migratory capacity in PC cells following excessive MND1 overexpression. (E) Experiments on migration and invasion were conducted to assess the migratory and invasive capacity in Patu-8988 cells and PANC-1 cells in response to MND1 overexpression. Data were represented as mean ± SD. *p < 0.05; **p < 0.01, ***p < 0.001 ****p < 0.0001 vs. empty vector. EV: Empty vector. MND1: MND1 vector.
Meiotic nuclear divisions 1 controls the expression of H2A.X variant histone
We explored whether MND1 regulates other cellular signaling pathways in PC cells, impacting the cell cycle and migratory abilities. We observed that MND1 had a positive effect on H2AFX mRNA expression (Fig. 3A). Furthermore, western blotting showed that the downregulation of MND1 reduced protein levels of H2AFX in PC cells (Fig. 3B). In the meanwhile, a positive correlation was observed between MND1 and H2AFX, as evidenced by a correlation coefficient of 0.49 in the GEPIA database (Fig. 3C). Our findings provide evidence that MND1 plays a pivotal role in affecting the expression levels of H2AFX. In summary, our finding suggests that MND1 is responsible for governing H2AFX expression in PC cells.
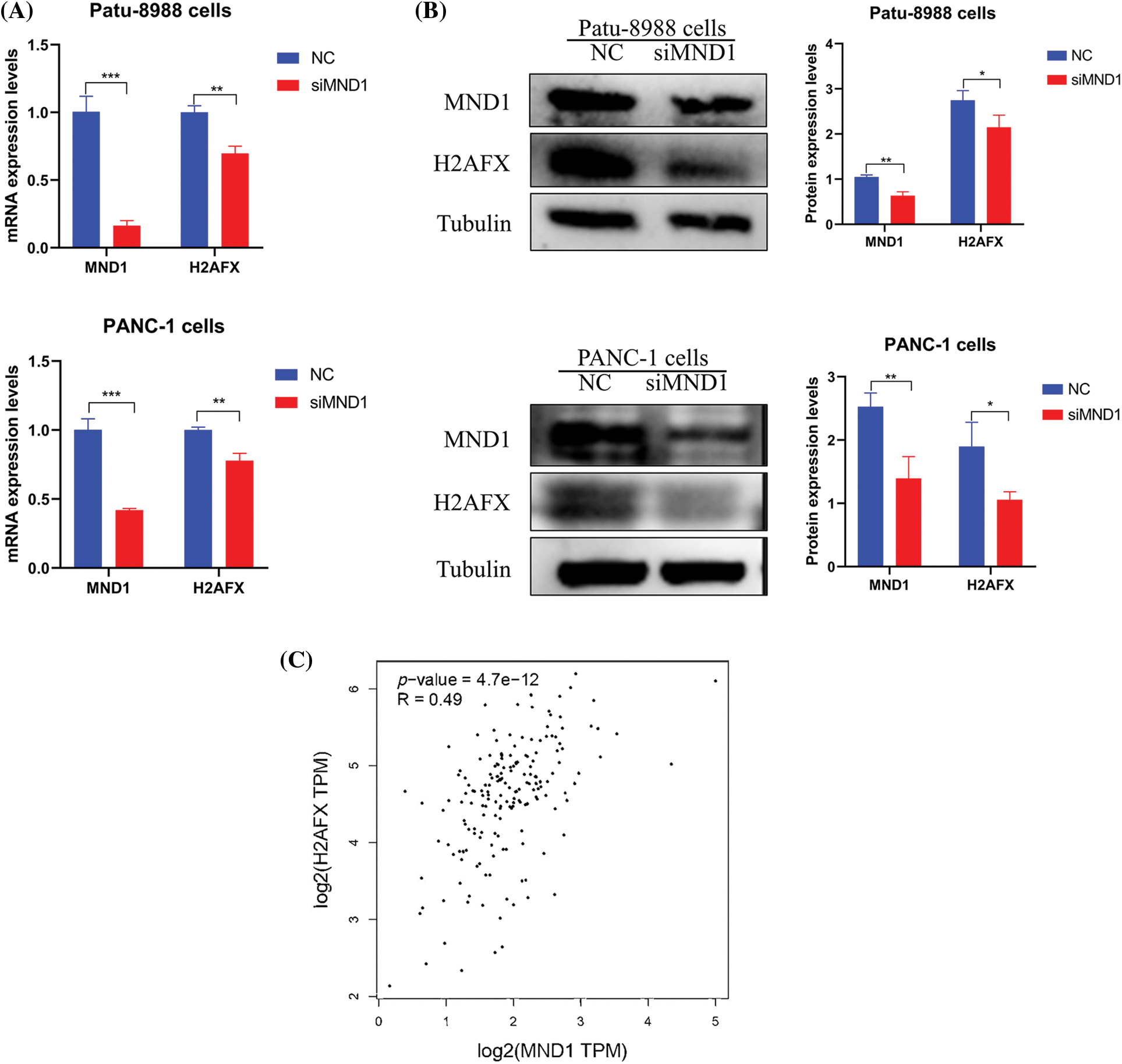
Figure 3: Meiotic nuclear divisions 1 (MND1) controls H2A.X variant histone (H2AFX) expression. (A) Real-time-polymerase chain reaction was used to measure the mRNA levels of MND1 and H2AFX in pancreatic cancer (PC) cells after MND1 siRNA transfection. (B) Western blotting analysis was employed to identify the expression of MND1 and H2AFX in PC cells after MND1 inhibition. (C) MND1 is positively correlated with H2AFX in the gene expression profiling interactive analysis database. Data were represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 vs. control. NC: Negative Control siRNA. si: siRNA-MND1.
H2A.X variant histone is overexpression overexpressed in Pancreatic Carcinoma
Investigating H2AFX expression in both tumor and non-tumor tissues, we used the GEPIA database and analyzed the expression of H2AFX in PC. The result is shown in (Fig. 4A). At the same time, we used western blot and qRT-PCR to explore H2AFX expression in normal pancreatic epithelial cells and PC. As shown in Figs. 4B and 4C, H2AFX expression was higher than the H2AFX expression in the normal pancreatic epithelial cells Line HPDEC6-7.
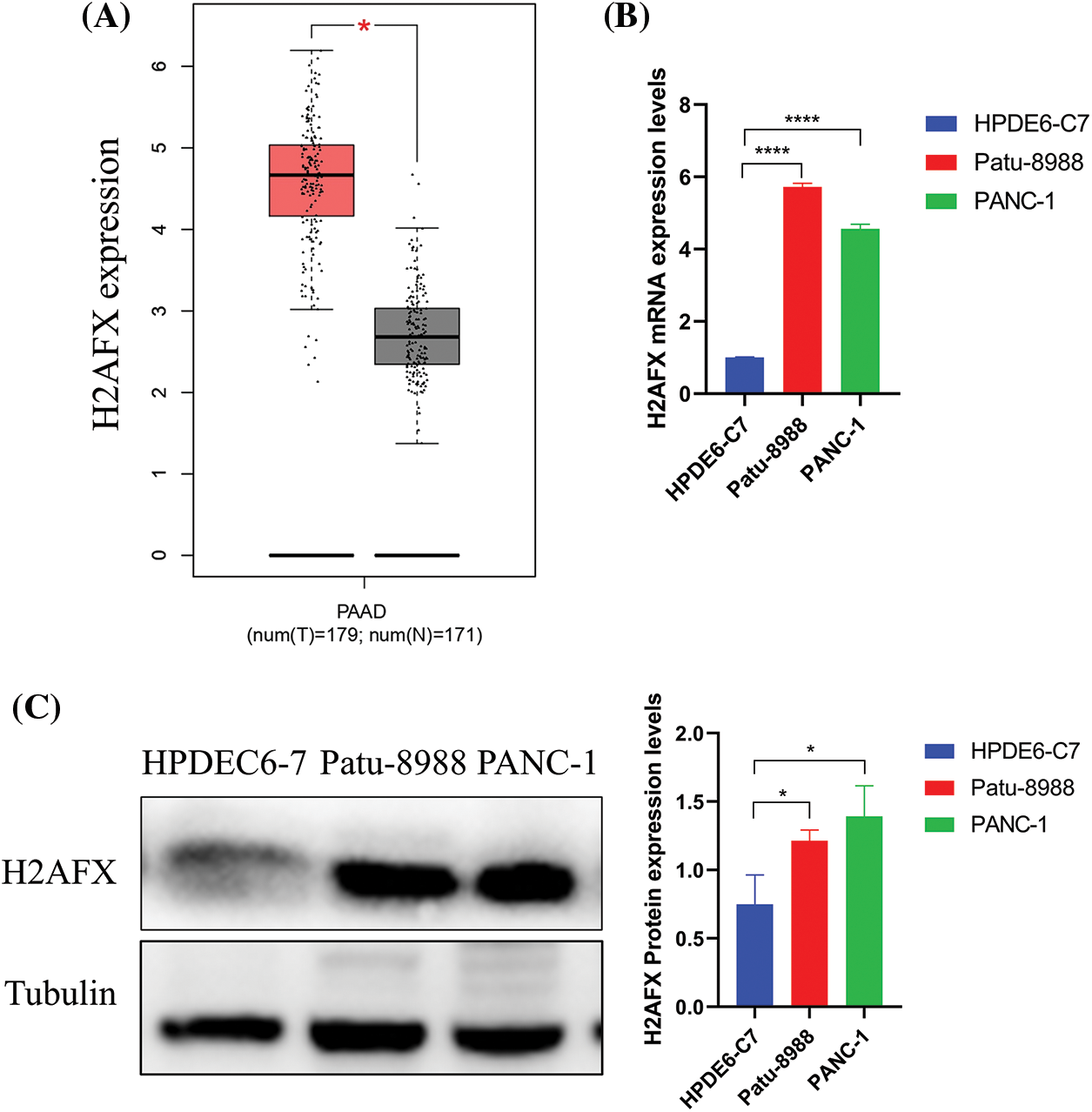
Figure 4: H2A.X variant histone (H2AFX) is overexpressed in pancreatic carcinoma. (A) H2AFX is highly expressed in the gene expression profiling interactive analysis database. (B and C) Compared with normal pancreatic epithelial cells (HPDE6-C7), H2AFX is highly expressed at the RNA and protein levels in Patu-8988 and PANC-1 cells. Data were represented as mean ± SD (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Downregulation of H2A.X variant histone (inhibited cell growth, migration, invasion, and cell cycle in PC cells
To investigate the part H2AFX plays in PC cell growth, we selected a single, most effective siRNA for H2AFX by analyzing the results of western blotting and qRT-PCR, as illustrated in Figs. 5A and 5B. SiH2AFX-3 knockdown was the most effective. PC cells transfected with H2AFX siRNA underwent the CCK8 assay. The downregulation of H2AFX inhibited the growth in PC cells (Fig. 5C). Findings from the wound healing assay revealed that H2AFX downregulation inhibited cell migration in both PANC-1 and Patu-8988 cells (Fig. 6A). Moreover, data of transwell chamber assay demonstrated that reducing H2AFX levels hindered the cell migration and invasion in PC cells (Fig. 6B). We further observed that H2AFX downregulation inhibited cell cycle progression in PC cells (Fig. 6C). Collectively, our data indicates that H2AFX plays a role in promoting tumor growth in PCs.
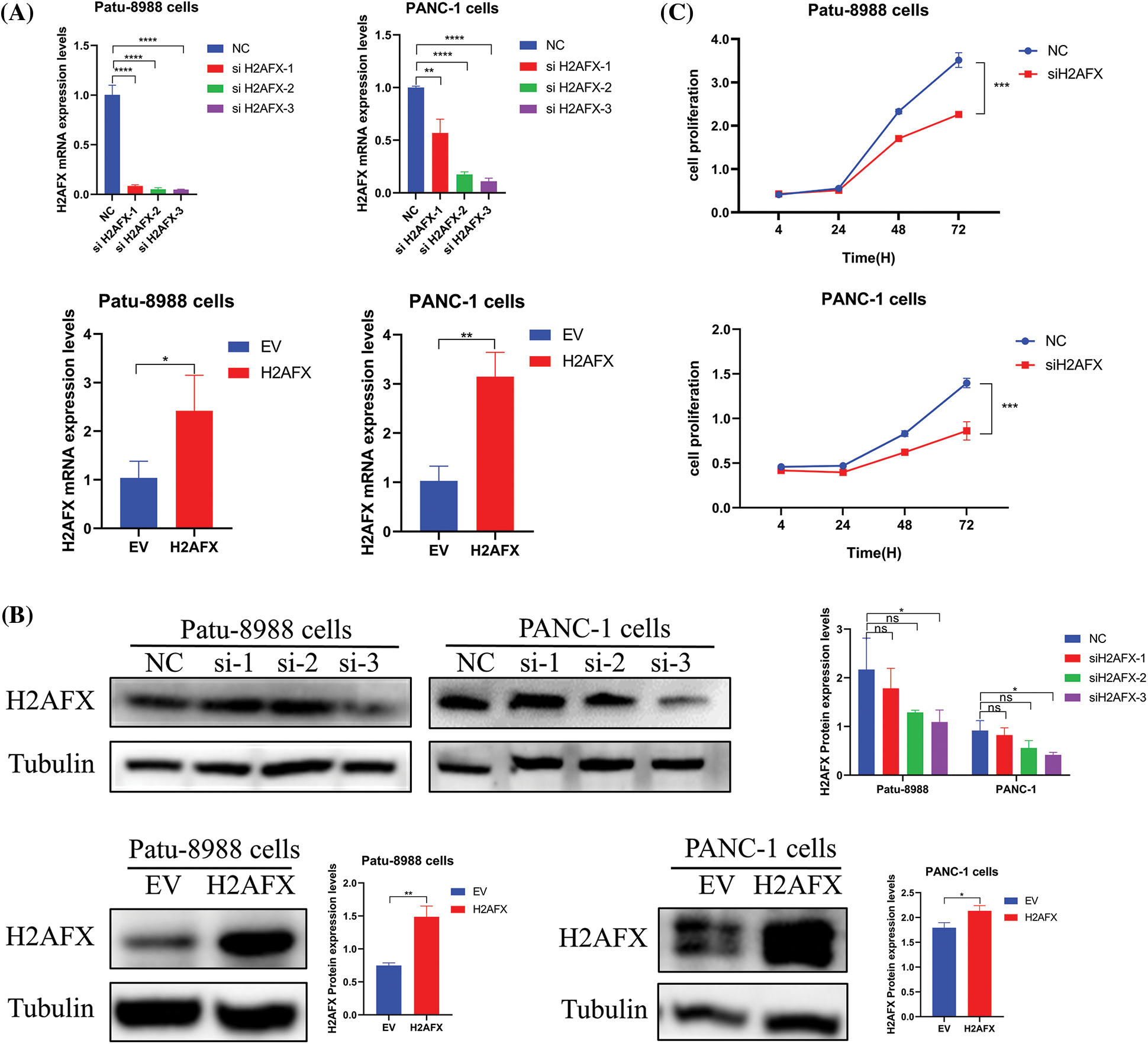
Figure 5: Downregulation of H2A.X variant histone (H2AFX) inhibited growth, migration, invasion, and cell cycle in pancreatic carcinoma (PC) cells. (A) Quantitative real-time-polymerase chain reaction confirmed that the expression of H2AFX was inhibited by H2AFX siRNA and overexpression. (B) The transfection efficiency of si-H2AFX and H2AFX overexpression in the Patu-8988 and Panc-1 cells was detected by western blotting. (C) Cell counting Kit-8 assay was conducted in PC cells after H2AFX was inhibited. Data were represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 vs. NC. NC: Negative Control siRNA. si: siRNA-H2AFX. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. empty vector. EV: Empty vector. H2AFX: H2AFX vector.
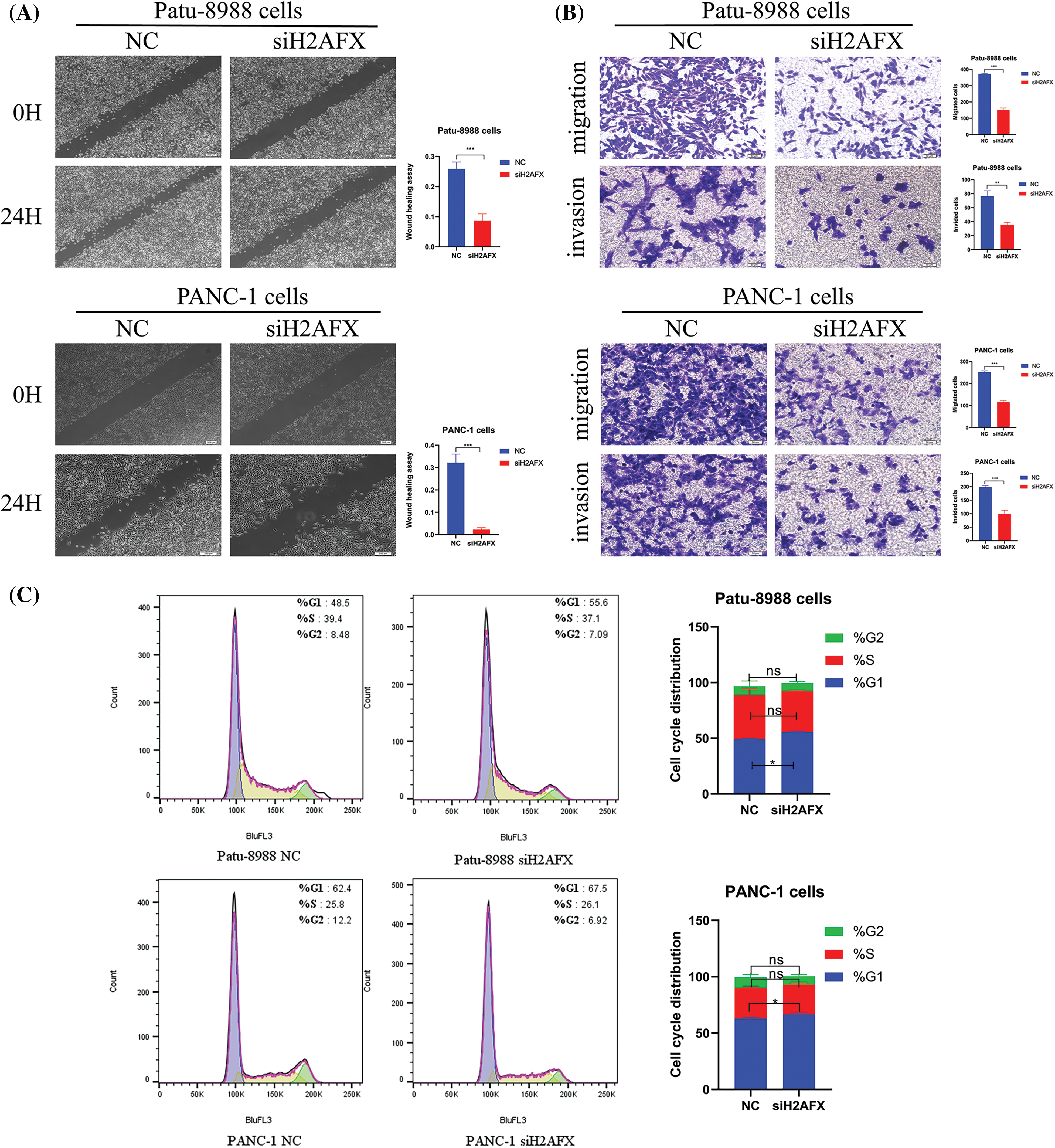
Figure 6: Downregulation of H2A.X variant histone (H2AFX) inhibited growth, migration, invasion, and cell cycle in pancreatic carcinoma (PC) cells. (A) Wound healing assay was conducted to assess the migratory capacity of PC cells when exposed to H2AFX inhibition. (B) Migration and invasion assays were conducted to assess the migratory capacity in Patu-8988 cells and PANC-1 cells in response to H2AFX inhibition. (C) Flow cytometry cell cycle experiment was measured in Patu-8988 cells and PANC-1 cells after H2AFX knockdown. Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Negative Control. NC: Negative Control. si: siRNA-H2AFX.
Upregulation of H2A.X variant histone rescues meiotic nuclear divisions 1 knockdown-induced tumor suppressive effects in PC cells
In an effort to ascertain if MND1 promotes tumor growth by increasing H2AFX levels, PC cells were transfected with siMND1 in combination with H2AFX cDNA. Our western blot data showed that the H2AFX protein expression level was the lowest in the MND1 interference group and the highest in the H2AFX overexpression group, whereas the H2AFX expression level in the combined action group was intermediate and about the same as that of the NC group, revealed that H2AFX upregulation was rescued by MND1 low expression in PC cells (Fig. 7A). The results of CCK8 assay indicated that the MND1 interference group had the weakest cell proliferation ability, and the H2AFX overexpression group had the strongest cell proliferation ability. The cell proliferation ability of the combined group was in the middle, and there was no statistically significant difference between the NC group and the combined group. We found that H2AFX upregulation abrogated MND1-induced cell growth inhibition in PC cells (Fig. 7B). Aligning with previous findings, the flow cell cycle results suggested that the MND1 interference group had the highest proportion of G1 phase, indicating that the cell cycle process was blocked, the H2AFX overexpression group had an increased proportion of S phase compared with the NC group, indicating that the cell cycle process was promoted. Meanwhile, the results of both groups were close to those of the NC group. We observed that H2AFX upregulation abolished MND1-triggered cell cycle (Fig. 7C). Findings from the wound healing assay and transwell assay demonstrated that the MND1 interference group had the weakest migratory and invasive ability, the H2AFX overexpression group had the strongest migratory and invasive ability, and both groups had results close to those of the NC group, as compared with the NC group. Strikingly, the upregulation of H2AFX rescued MND1-triggered inhibition of cell migration and invasion in PC cells (Figs. 8A and 8B). Altogether, these results coherently demonstrate that MND1’s tumor-promoting effects are partly due to the control of H2AFX in PC cells.
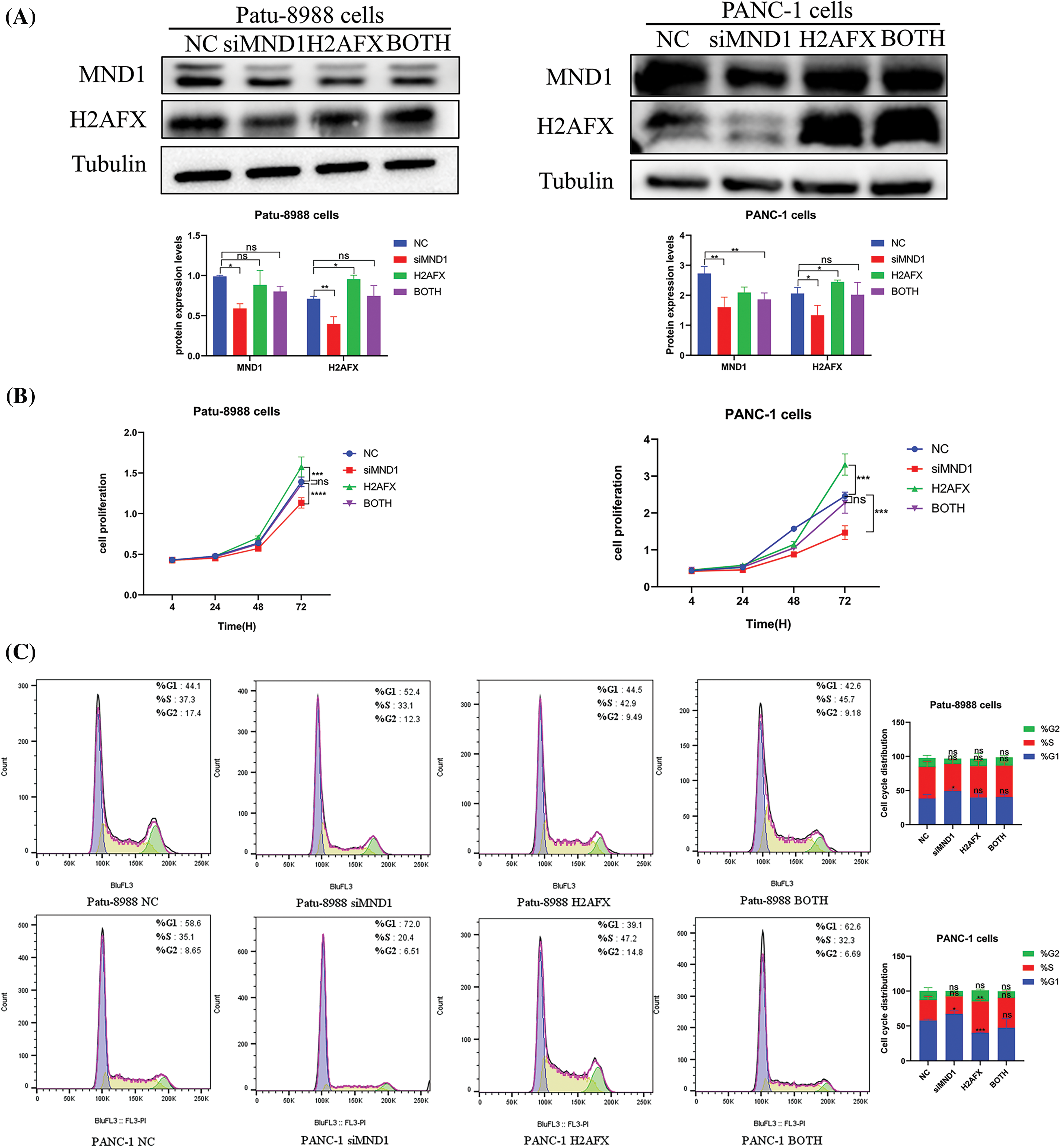
Figure 7: Upregulation of H2A.X variant histone (H2AFX) rescues meiotic nuclear divisions 1 (MND1) knockdown-induced tumor suppressive effects in pancreatic carcinoma (PC) cells. (A) To measure the levels of MND1 and H2AFX in PC cells following MND1 siRNA and H2AFX overexpression transfection, western blotting was performed (B) Cell counting Kit-8 test was performed on PC cells after MND1 siRNA transfection and H2AFX overexpression. (C) Flow cytometry was performed to examine the cell cycle in Patu-8988 cells and PANC-1 cells after MND1 siRNA transfection and H2AFX overexpression. Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. NC. NC: Negative Control siRNA. si: siRNA-MND1. H2AFX: H2AFX vector. BOTH: siRNA-MND1 and H2AFX vector.
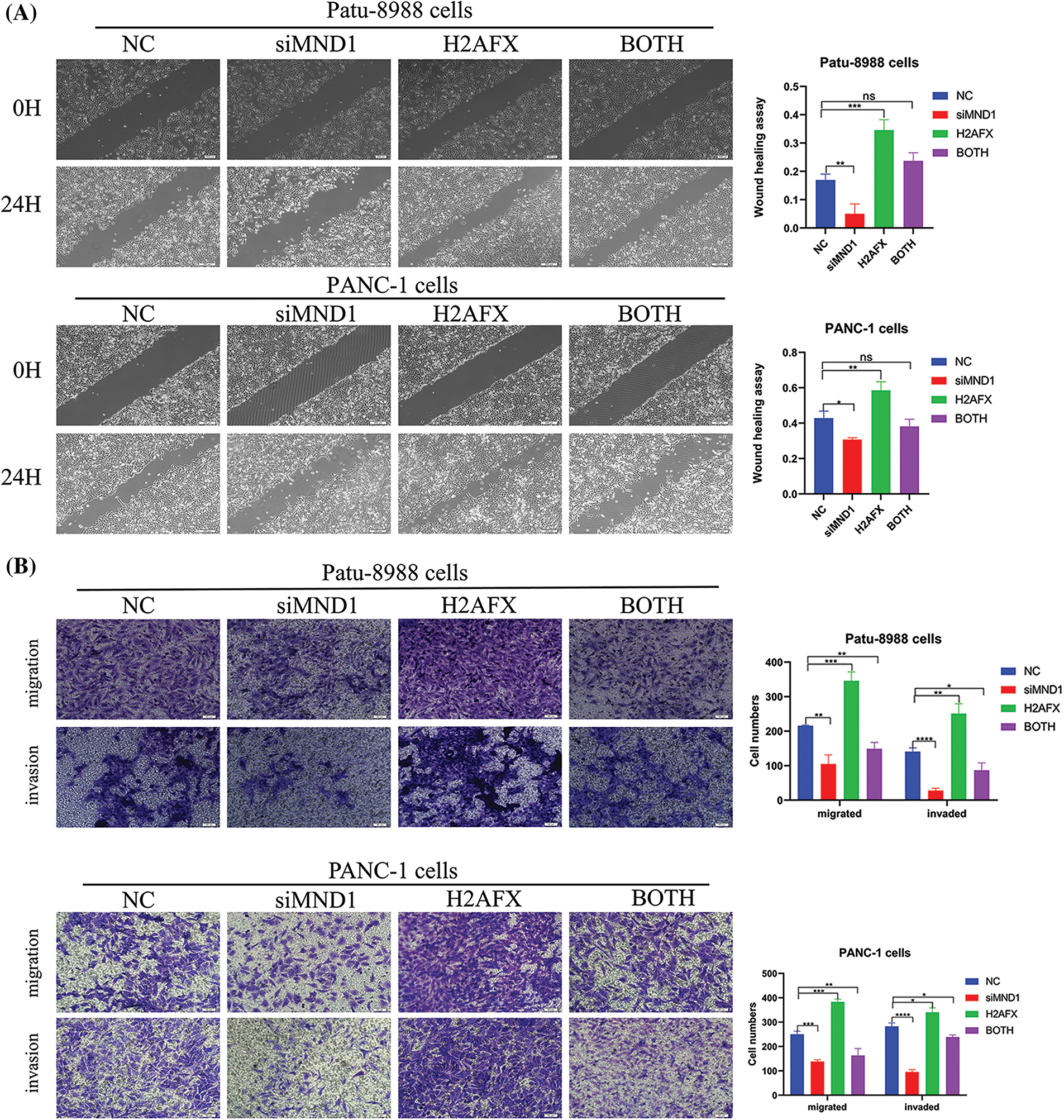
Figure 8: Upregulation of H2A.X variant histone (H2AFX) rescues meiotic nuclear divisions 1 (MND1) knockdown-induced tumor suppressive effects in pancreatic carcinoma (PC) cells. (A) Wound healing assay was conducted to assess the migratory capacity of PC cells following the introduction of MND1 siRNA and H2AFX overexpression. (B) Experiments on migration and invasion were conducted to assess the migratory abilities of Patu-8988 cells and PANC-1 cells in response to MND1 siRNA transfection and H2AFX overexpression. Data were represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001 vs. NC. NC: Negative Control siRNA. Si: siRNA-MND1. H2AFX: H2AFX vector. BOTH: siRNA-MND1 and H2AFX vector.
The role of MND1 has been a hot topic in recent years. MND1 is a meiosis-specific protein that specifically targets the pairing of homologous chromosomes [8,23]. The repair of DNA double-strand break (DSB) during meiosis by MND1 is significant and associates with HOP2 to form a stable heterodimeric complex and facilitates the repair of homologous chromosome pairing DNA double-strand break (DSB) repair during meiosis [24,25]. Koob [23] demonstrated that MND1 localized to DSBs, where it stimulated DNA repair through homologous recombination (HR). Importantly, MND1 is not involved in the response to replication-associated DSBs, which implies that it is dispensable for HR-mediated repair of one-ended DSBs. Hop2 and Mnd1 were discovered to work together as a complex that promotes homologous chromosome pairing and DSB repair during meiosis [8,24,26]. Conversely, numerous studies have demonstrated that MND1 facilitates tumor function in certain cancers. Corresponding to this view, Bao et al. [27] utilized bioinformatics tools and conducted experiments to validate the correlation between elevated MND1 expression and unfavorable Breast Cancer Survival. In lung adenocarcinoma, MND1 regulates the progression of the cell cycle by activating a KLF6/E2F1 positive feedback loop, while simultaneously functioning as a proto-oncogene [9]. MND1 has the potential to serve as a prognostic biomarker linked to the cell cycle and immune infiltration in kidney renal clear cell carcinoma [12]. Through genetic network and gene set enrichment analyses, MND1 has been identified as a promising gene for diagnosing and treating lung adenocarcinoma [11]. Recent studies have indicated that MND1 may be a viable gene for diagnosing and treating gastric cancer [28]. The above studies suggest that MND1 is involved in the process of malignant tumor progression. However, the role of MND1 in the development of PC and its underlying mechanism are yet to be fully understood. Therefore, we first analyzed the expression of MND1 in PC tumor tissues and normal tissues as well as the relationship between high and low MND1 expression groups in tumor tissues and survival prognosis using the GEPIA online database, and the results showed that MND1 was highly expressed in PC and that high expression of MND1 predicted a poorer prognosis (Figs. 1A and 1B). In addition, Western blot and qRT-PCR experiments were utilized to confirm that MND1 expression was increased in PC cells compared with normal epithelial cells of pancreatic ducts (Figs. 1C and 1D). Further in vitro experiments revealed that overexpression of MND1 promoted the proliferation, migration, and invasion of PC cells (Figs. 2A–2E). Our current research, which included CCK8 assay, wound healing assay, and transwell migration and invasion assay, demonstrated that MND1 had a considerable impact on PC cell growth and mobility in vitro. Our results revealed that MND1 may be a viable option for PC therapy due to its ability to stimulate tumor growth in PCs.
Latest research has underscored the importance of H2AFX in development of cancer. As an example, in non-small lung cancer, H2AFX serves as a novel DNA damage repair-related marker for forecasting prognostic and treatment response in non-small lung cancer [29]. H2AFX was considered as a necroptosis-related gene signature for the prediction of prognosis and tumor immunity in lung adenocarcinoma [30]. Al-Harazi et al. [15] speculated that H2AFX could potentially serve as a diagnostic and prognostic indicator for early-stage hepatocellular carcinoma. Studies have shown that H2AFX is the most critical gene linked to the primary Salivary gland carcinoma [31]. Additionally, miR-24-2 has been found to directly control H2AFX expression and induces apoptosis in MCF-7 and HeLa cells [32], while miR-328-3p has been shown to stimulate migration and invasion by targeting H2AFX in head and neck squamous cell carcinoma [17]. Nevertheless, the regulatory relationship between MND1 and H2AFX in PC had not been clarified, and our analysis using the GEPIA database revealed a positive correlation between MND1 and H2AFX, which was consistent with the results of Western blot and qRT-PCR experiments, which MND1 controls positively H2AFX expression (Figs. 3A–3C). Subsequently, the increased expression of H2AFX in PC tissues was analyzed using the GEPIA online database, and the Western blot and qRT-PCR results similarly demonstrated that H2AFX was more highly expressed in PC cells than in normal epithelial cells of the pancreas (Figs. 4A–4C). In vitro experiments further confirmed that silencing H2AFX inhibited the proliferation, migration, and invasive ability of PC cells (Figs. 5A–5C, 6A and 6B) and blocked the cell cycle at the G1 phase (Fig. 6C), suggesting that it may intervene in the malignant biological behaviors of PC cells by affecting the progression of the cell cycle. Our results revealed that H2AFX functions as a tumor promotion in PCs, suggesting that H2AFX might be a potential target for PC treatment.
In the present study, we report that MND1 positively controls the expression of H2AFX in PC cells. First, we used Western blotting to verify that the level of H2AFX protein expression in the combined effect group of interfering with MND1 and overexpressing H2AFX was intermediate between interfering and MND1 overexpressing H2AFX in PCs (Fig. 7A). Following that, the results of CCK8 assay, wound healing assay and transwell assay revealed that the proliferation, migration and invasion ability of the combined effector group of interfering with MND1 and overexpressing H2AFX were intermediate between that of interfering with MND1 and overexpressing H2AFX (Figs. 7B, 8A and 8B). Meanwhile, the flow cell cycle results suggested that H2AFX upregulation abolished the MND1-triggered cell cycle. It is noteworthy that the overexpression of MND1 abrogated H2AFX knockdown-induced tumor inhibition in PC cells, suggesting that H2AFX is involved in MND1-mediated promo-cancer activity in PC. Taken together, we conclude that the MND1/H2AFX axis plays a role in the advancement of PC. Focusing on this axis may represent a useful approach for treating PC. In summary, our findings unravel new molecular mechanisms of MND1-mediated tumor promotion in PC cells. This research further indicates that the reducing MND1 and H2AFX levels could be an innovative approach in treating PC.
Acknowledgement: None.
Funding Statement: This study was supported by grants from National Innovation Program for College Students (202210367076), Graduate Student Research Innovation Program of Bengbu Medical College (Byycxz22016), and the National Natural Science Foundation of China (82072585), and the Key Research Project of Bengbu Medical College (No. 2020byzd029).
Author Contributions: XW, CL, and JZ provided the idea and design of this article. Clinical data were collected and analyzed by YS, LW, ZW, SS, HH and MH. DW and XL drafted the first draft of the article and the drawing of charts. CL and JZ reviewed the revised paper. All authors read and approved the final manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no competing interests.
References
1. Fang X, Lan H, Jin K, Qian J. Pancreatic cancer and exosomes: role in progression, diagnosis, monitoring, and treatment. Front Oncol. 2023;13:1149551. doi:https://doi.org/10.3389/fonc.2023.1149551. [Google Scholar] [PubMed] [CrossRef]
2. Olajubutu O, Ogundipe OD, Adebayo A, Adesina SK. Drug delivery strategies for the treatment of pancreatic cancer. Pharmaceut. 2023;15(5):1318. doi:https://doi.org/10.3390/pharmaceutics15051318. [Google Scholar] [PubMed] [CrossRef]
3. Yang H, Li W, Ren L, Yang Y, Zhang Y, Ge B, et al. Progress on diagnostic and prognostic markers of pancreatic cancer. Oncol Res. 2023;31(2):83–99. doi:https://doi.org/10.32604/or.2023.028905. [Google Scholar] [PubMed] [CrossRef]
4. Baliyan V, Kordbacheh H, Parakh A, Kambadakone A. Response assessment in pancreatic ductal adenocarcinoma: role of imaging. Abdom Radiol. 2018;43(2):435–44. doi:https://doi.org/10.1007/s00261-017-1434-7. [Google Scholar] [PubMed] [CrossRef]
5. Goh SK, Gold G, Christophi C, Muralidharan V. Serum carbohydrate antigen 19-9 in pancreatic adenocarcinoma: a mini review for surgeons. ANZ J Surg. 2017;87(12):987–92. doi:https://doi.org/10.1111/ans.14131. [Google Scholar] [PubMed] [CrossRef]
6. Gaianigo N, Melisi D, Carbone C. EMT and treatment resistance in pancreatic cancer. Cancers. 2017;9(9):122. doi:https://doi.org/10.3390/cancers9090122. [Google Scholar] [PubMed] [CrossRef]
7. Bisht S, Brossart P, Feldmann G. Current therapeutic options for pancreatic ductal adenocarcinoma. Oncol Res Treat. 2018;41(10):590–4. doi:https://doi.org/10.1159/000493868. [Google Scholar] [PubMed] [CrossRef]
8. Kang HA, Shin HC, Kalantzi AS, Toseland CP, Kim HM, Gruber S, et al. Crystal structure of Hop2-Mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Res. 2015;43(7):3841–56. doi:https://doi.org/10.1093/nar/gkv172. [Google Scholar] [PubMed] [CrossRef]
9. Zhang Q, Shi R, Bai Y, Meng L, Hu J, Zhu H, et al. Meiotic nuclear divisions 1 (MND1) fuels cell cycle progression by activating a KLF6/E2F1 positive feedback loop in lung adenocarcinoma. Cancer Commun. 2021;41(6):492–510. doi:https://doi.org/10.1002/cac2.12155. [Google Scholar] [PubMed] [CrossRef]
10. Zhang N, Wang H, Xie Q, Cao H, Wu F, Di Wu DB, et al. Identification of potential diagnostic and therapeutic target genes for lung squamous cell carcinoma. Oncol Lett. 2019;18(1):169–80. doi:https://doi.org/10.3892/ol.2019.10300. [Google Scholar] [PubMed] [CrossRef]
11. Wei J, Meng G, Wu J, Zhang Q, Zhang J. Genetic network and gene set enrichment analyses identify MND1 as potential diagnostic and therapeutic target gene for lung adenocarcinoma. Sci Rep. 2021;11(1):9430. doi:https://doi.org/10.1038/s41598-021-88948-4. [Google Scholar] [PubMed] [CrossRef]
12. Fang J, Zhen J, Gong Y, Ke Y, Fu B, Jiang Y, et al. MND1 functions as a potential prognostic biomarker associated with cell cycle and immune infiltration in kidney renal clear cell carcinoma. Aging. 2022;14(18):7416–42. doi:https://doi.org/10.18632/aging.204280. [Google Scholar] [PubMed] [CrossRef]
13. Tan K, Wang K, Zhao A, Liu Z, Song W, Cheng Q, et al. Meiotic nuclear divisions 1 promotes proliferation and metastasis in hepatocellular carcinoma and is a potential diagnostic and therapeutic target gene. Med Oncol. 2022;40(1):14. doi:https://doi.org/10.1007/s12032-022-01875-w. [Google Scholar] [PubMed] [CrossRef]
14. Hu H, Zhong T, Jiang S. H2AFX might be a prognostic biomarker for hepatocellular carcinoma. Cancer Rep. 2023;6(1):e1684. doi:https://doi.org/10.1002/cnr2.1684. [Google Scholar] [PubMed] [CrossRef]
15. Al-Harazi O, Kaya IH, Al-Eid M, Alfantoukh L, Al Zahrani AS, Al Sebayel M, et al. Identification of gene signature as diagnostic and prognostic blood biomarker for early hepatocellular carcinoma using integrated cross-species transcriptomic and network analyses. Front Genet. 2021;12:710049. doi:https://doi.org/10.3389/fgene.2021.710049. [Google Scholar] [PubMed] [CrossRef]
16. de Sousa Portilho AJ, da Silva EL, Bezerra ECA, Moraes Rego Gomes CBS, Ferreira V, de Moraes MEA, et al. 1, 4-Naphthoquinone (CNN1) induces apoptosis through DNA damage and promotes upregulation of H2AFX in leukemia multidrug resistant cell line. Int J Mol Sci. 2022;23(15):8105. doi:https://doi.org/10.3390/ijms23158105. [Google Scholar] [PubMed] [CrossRef]
17. Ma H, Liu C, Zhang S, Yuan W, Hu J, Huang D, et al. miR-328-3p promotes migration and invasion by targeting H2AFX in head and neck squamous cell carcinoma. J Cancer. 2021;12(21):6519–30. doi:https://doi.org/10.7150/jca.60743. [Google Scholar] [PubMed] [CrossRef]
18. Bretherick KL, Schuetz JM, Morton LM, Purdue MP, Conde L, Gallagher RP, et al. Sex- and subtype-specific analysis of H2AFX polymorphisms in non-Hodgkin lymphoma. PLoS One. 2013;8(9):e74619. doi:https://doi.org/10.1371/journal.pone.0074619. [Google Scholar] [PubMed] [CrossRef]
19. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14. doi:https://doi.org/10.1093/nar/gkaa407. [Google Scholar] [PubMed] [CrossRef]
20. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. doi:https://doi.org/10.1093/nar/gkz430. [Google Scholar] [PubMed] [CrossRef]
21. Győrffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Gerosci. 2023;45:1889–1898. doi:https://doi.org/10.1007/s11357-023-00742-4. [Google Scholar] [PubMed] [CrossRef]
22. Sun K, Wang D, Zhang Z, Huang Y, Lian X, Hua J, et al. Columbianetin acetate inhibits the occurrence and development of pancreatic cancer cells by down-regulating the expression of Meiotic nuclear divisions 1. BIOCELL. 2023;47(2):297–307. doi:https://doi.org/10.32604/biocell.2023.023553. [Google Scholar] [CrossRef]
23. Koob L, Friskes A, Van Bergen L, Feringa FM, van den Broek B, Koeleman ES, et al. MND1 enables homologous recombination in somatic cells primarily outside the context of replication. Mol Oncol. 2023;17(7):1192–211. doi:https://doi.org/10.1002/1878-0261.13448. [Google Scholar] [PubMed] [CrossRef]
24. Moktan H, Guiraldelli MF, Eyster CA, Zhao W, Lee CY, Mather T, et al. Solution structure and DNA-binding properties of the winged helix domain of the meiotic recombination HOP2 protein. J Biol Chem. 2014;289(21):14682–91. doi:https://doi.org/10.1074/jbc.M114.548180. [Google Scholar] [PubMed] [CrossRef]
25. Ploquin M, Petukhova GV, Morneau D, Déry U, Bransi A, Stasiak A, et al. Stimulation of fission yeast and mouse Hop2-Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007;35(8):2719–33. doi:https://doi.org/10.1093/nar/gkm174. [Google Scholar] [PubMed] [CrossRef]
26. Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, et al. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42(2):906–17. doi:https://doi.org/10.1093/nar/gkt924. [Google Scholar] [PubMed] [CrossRef]
27. Bao Z, Cheng J, Zhu J, Ji S, Gu K, Zhao Y, et al. Using weighted gene co-expression network analysis to identify increased MND1 expression as a predictor of poor breast cancer survival. Int J Gen Med. 2022;15:4959–74. doi:https://doi.org/10.2147/IJGM.S354826. [Google Scholar] [PubMed] [CrossRef]
28. Hu X, Zhou S, Li H, Wu Z, Wang Y, Meng L, et al. FOXA1/MND1/TKT axis regulates gastric cancer progression and oxaliplatin sensitivity via PI3K/AKT signaling pathway. Cancer Cell Int. 2023;23(1):234. doi:https://doi.org/10.1186/s12935-023-03077-4. [Google Scholar] [PubMed] [CrossRef]
29. Li L, Zou BJ, Zhao JZ, Liang JB, She ZY, Zhou WY, et al. A novel DNA damage repair-related signature for predicting prognositc and treatment response in non-small lung cancer. Front Oncol. 2022;12:961274. doi:https://doi.org/10.3389/fonc.2022.961274. [Google Scholar] [PubMed] [CrossRef]
30. Lei K, Tan B, Liang R, Lyu Y, Wang K, Wang W, et al. Development and clinical validation of a necroptosis-related gene signature for prediction of prognosis and tumor immunity in lung adenocarcinoma. Am J Cancer Res. 2022;12(11):5160–82. [Google Scholar] [PubMed]
31. Bayat Z, Ahmadi-Motamayel F, Parsa MS, Taherkhani A. Potential biomarkers and signaling pathways associated with the pathogenesis of primary salivary gland carcinoma: a bioinformatics study. Genomics Inform. 2021;19(4):e42. doi:https://doi.org/10.5808/gi.21052. [Google Scholar] [PubMed] [CrossRef]
32. Srivastava N, Manvati S, Srivastava A, Pal R, Kalaiarasan P, Chattopadhyay S, et al. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 2011;13(2):R39. doi:https://doi.org/10.1186/bcr2861. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools