 Open Access
Open Access
ARTICLE
Novel Framework of Segmentation 3D MRI of Brain Tumors
1 Department of Computer Science, Faculty of Computers and Informatics, Zagazig University, Zagazig, Egypt
2 Department of Computer Science, Faculty of Computers and Informatics, Kafrelsheikh University, Kafrelsheikh, Egypt
3 Department of Embedded Network Sys. and Tec., Faculty of Artificial Intelligence, Kafrelsheikh University, Kafrelsheikh, Egypt
4 Computers Engineering and Control Systems Department, Faculty of Engineering, Mansoura University, Mansoura, Egypt
* Corresponding Author: Zainab H. Ali. Email:
Computers, Materials & Continua 2023, 74(2), 3489-3502. https://doi.org/10.32604/cmc.2023.033356
Received 15 June 2022; Accepted 19 August 2022; Issue published 31 October 2022
Abstract
Medical image segmentation is a crucial process for computer-aided diagnosis and surgery. Medical image segmentation refers to portioning the images into small, disjointed parts for simplifying the processes of analysis and examination. Rician and speckle noise are different types of noise in magnetic resonance imaging (MRI) that affect the accuracy of the segmentation process negatively. Therefore, image enhancement has a significant role in MRI segmentation. This paper proposes a novel framework that uses 3D MRI images from Kaggle and applies different diverse models to remove Rician and speckle noise using the best possible noise-free image. The proposed techniques consider the values of Peak Signal to Noise Ratio (PSNR) and the level of noise as inputs to the attention-U-Net model for segmentation of the tumor. The framework has been divided into three stages: removing speckle and Rician noise, the segmentation stage, and the feature extraction stage. The framework presents solutions for each problem at a different stage of the segmentation. In the first stage, the framework uses Vibrational Mode Decomposition (VMD) along with Block-matching and 3D filtering (Bm3D) algorithms to remove the Rician. Afterwards, the most significant Rician noise-free images are passed to the three different methods: Deep Residual Network (DeRNet), Dilated Convolution Auto-encoder Denoising Network (Di-Conv-AE-Net), and Denoising Generative Adversarial Network (DGAN-Net) for removing the speckle noise. VMD and Bm3D have achieved PSNR values for levels of noise (0, 0.25, 0.5, 0.75) for reducing the Rician noise by (35.243, 32.135, 28.214, 24.124) and (36.11, 31.212, 26.215, 24.123) respectively. The framework also achieved PSNR values for removing the speckle noise process for each level as follows: (34.146, 30.313, 28.125, 24.001), (33.112, 29.103, 27.110, 24.194), and (32.113, 28.017, 26.193, 23.121) for DeRNet, Di-Conv-AE-Net, and DGAN-Net, respectively. The experiments that have been conducted have proved the efficiency of the proposed framework against classical filters such as Bilateral, Frost, Kuan, and Lee according to different levels of noise. The attention gate U-Net achieved 94.66 and 95.03 in the segmentation of free noise images in dice and accuracy, respectively.Keywords
Image segmentation is a crucial challenge in computer vision and image processing alike. For images, segmentation generally means dividing them into mutually exclusive areas to make them easy for analysis and examination [1–3]. The trend of medical image segmentation is growing rapidly, and the area of stockholders has become very large. The stockholders, such as clinicians, engineers, developers, patients, etc., need to rapidly update their knowledge to stay updated with the new changes, especially in the new changes in the map of medicine in the world [4–6]. There are many images of this visual representation, such as magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), mammography, and molecular imaging, which can be used in the segmentation process [5,6]. MRI images have been widely used for the segmentation of brain tumors.
The brain is one of the vital organs in the human body, so brain diseases need quick examination and diagnosis. The early detection of brain tumors participates in the process of diagnosis. The main reason for this type of segmentation is to generate an accurate delineation of brain tumor areas with correctly located masks [7]. Noise such as speckle noise and Rician noise must be taken into consideration in the segmentation process [8]. The most common type of noise in MRI is called thermal noise, which is due to the thermal agitation of electrons from the machines or objects to be imaged. Minimizing different types of noise is very important for clinicians in the diagnosis and detection process, especially in the visualisation stage. Reducing or removing noise helps the models enhance the accuracy of the segmentation process. Many methods and filters have been introduced to remove the noise, such as Frost et al. [9], Bilateral [10], Kuan et al. [11], Lee [12], Mean, and Median Filters.
The main objective of this paper is to introduce a novel framework for brain tumor segmentation from MRI images, as well as to reduce the speckle and Rician noise. The novel’s framework consists of two principal stages. The first is image enhancement, which is used to reduce the speckle and Rician noise, and the second stage is MRI image segmentation using attention U-Net. The framework starts by reducing the Rician and speckle noise from MRI to remove the unnecessary signal from MRI images. Reducing noise stages helps to reduce the loss of vital information in MRI, especially in small images [13–16]. In this paper, many algorithms have been used and modified to remove speckle and Rician noise, and their efficiency has been compared to other available filters.
The second stage in the proposed framework is called the segmentation stage for the denoised MRI using the attention U-Net model. The recent research in applying learning approaches such as machine learning (ML) and deep learning (DL) to the segmentation process has become a significant direction in offering quick medical treatment to patients [17–19]. DL has presented many models for dealing with brain tumors and has proved their efficiency in the segmentation process, such as (Convolution Neural Network) CNNs, (Fully Convolution Neural Network) FCN, autoencoder, U-Net, etc. The proposed framework uses the attention U-Net due to a smaller number of parameters used in the segmentation process compared with the U-Net and the residual U-Net.
The contributions of this paper can be summarized as follows:
a) Demonstrating a novel framework for enhancing the quality of MRI images by reducing noises that are called speckle and Rician noise.
b) Introducing a hyper model of VMD-BM3D for Rician noise and (DeRNet, Di-Conv-AE-Net, DGAN-Net) methods for diminishing noise and increasing accuracy.
c) Minimizing noise using VMD and different modified methods has cooperated in the process of increasing the accuracy of attention U-Net compared to other available models.
d) Achieving better accuracy than other compared models such as U-Net and residual U-Net.
The rest of this paper is organized as follows: Section 2 introduces the methodology and describes the main stages in the proposed framework, which are: importing MRI images as a dataset, reducing both Rician and speckle noise; and the segmentation process using the U-Net model. Section 3 demonstrates the discussion and experimental results. Section 4 is the paper’s conclusion.
Steps of methodology:
First step: image acquisition: the novel framework used an MRI dataset from Kaggle.
Second step: image enhancement: this step contains two different stages.
• Stage A: reducing Rician noise using VMD or BM3D.
• Stage B: reducing speckle noise using different methods (Conv-AE-Net, D-U-Net, Br-U-Net, DGan-Net, and DeRNet)
Step3rd: segmentation: Using attention U-Net architecture as illustrated in Fig. 1.
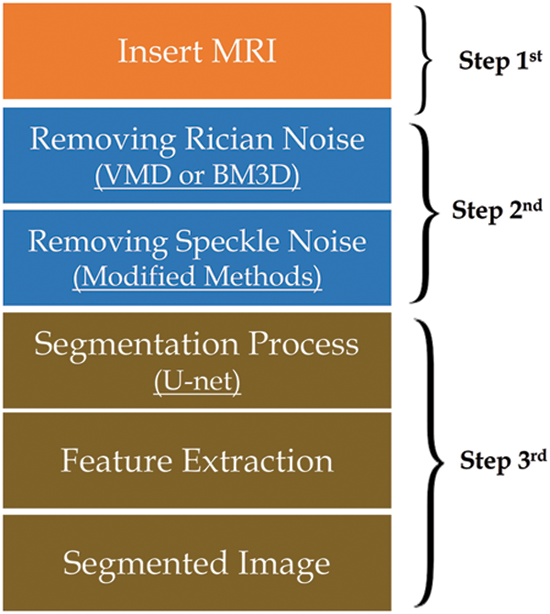
Figure 1: Methodology steps
The pseudocode of the segmentation process:
The novel methodology used a dataset from Kaggle called the Low Grad Glioma (LGG) Segmentation Dataset, which contains brain MRI images together with manual fluid-attenuated inversion recovery (FLAIR) abnormality masks. The medical images were collected from The Cancer Imaging Archive for a greater number of cancer cases contained in The Cancer Genome Atlas (TCGA) lower-grade glioma collection with at least FLAIR sequence and genomic cluster data available. Fig. 2 shows some images and masks from the LGG dataset.
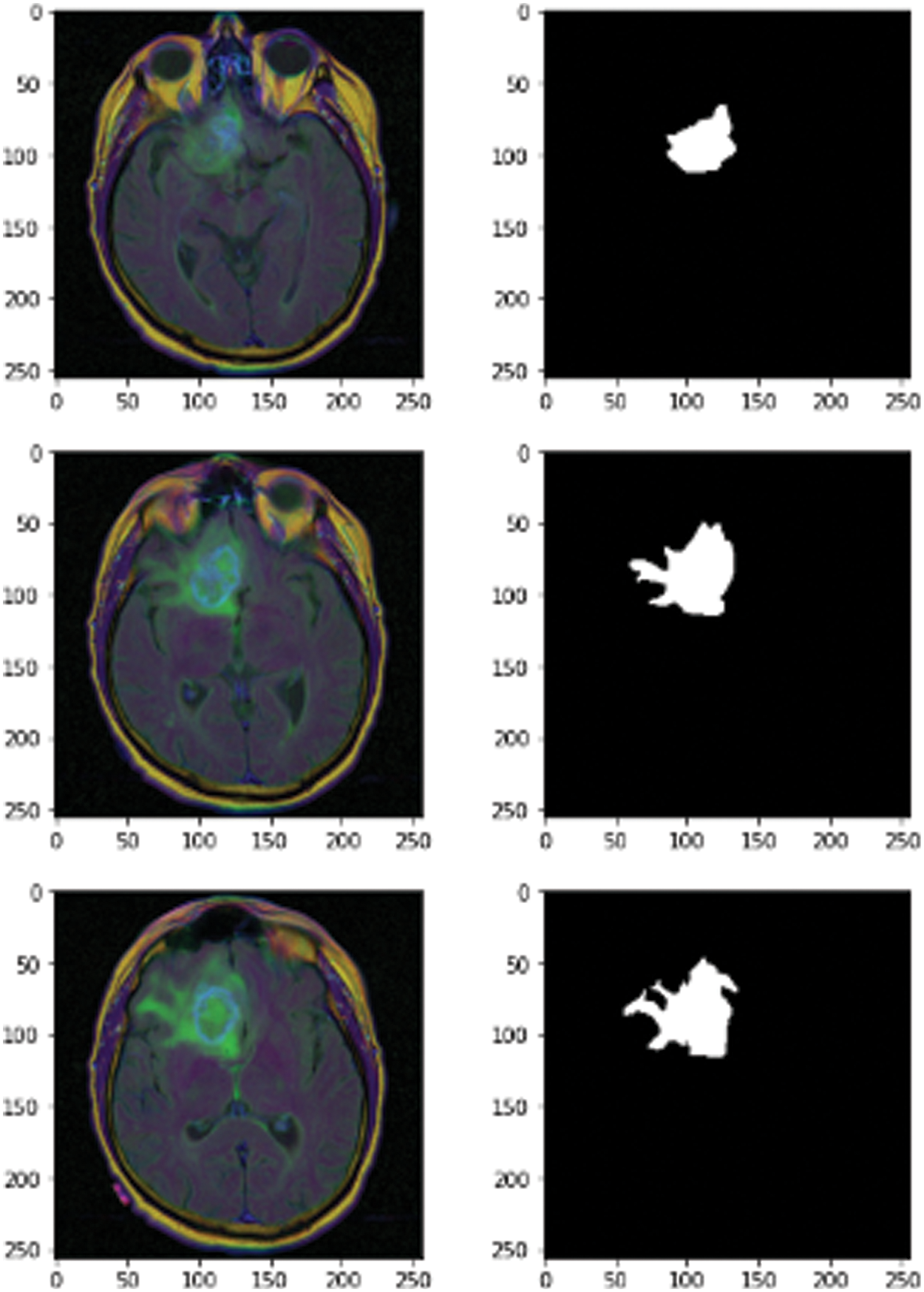
Figure 2: MRI images of 3D LGG
Reducing noise plays a vital role in the framework. This step is divided into two stages. The first stage is removing the Rician noise using VMD, and the second stage is removing the speckle noise using different modified models like BM3D, Di-Conv-AE-Net, D-U-Net, Br-U-Net, DGan-Net, and DeRNet.
In this subsection, we added speckle noise and Rician noise for MRI for testing and training. And the Rician noise is added using Eq. (1). Where is the image after adding Rician noise, is the image before adding the noise, and e1 and e2 are random numbers from a Gaussian distribution [20].
For adding the speckle noise, the original image expressed by G(x, y), f (x, y) is used to express noisy image and the multiplicative and additive noises are ηm(x, y) and ηa(x, y), respectively as shown in Eq. (2) [21].
Classification of MRI denoising methods 3-dimensional discrete wavelet transformation and wavelet shrinkage. The MRI denoising method is divided into two stages. The first stage is the filtering, and the second is called the Transform Domain. The filtering domain is also linear and nonlinear, the linear is for the spatial and transform domain, and the nonlinear is for the combination of domain and range filters, Anisotropic Diffusion Filtering (ADF), and Non-Local Means (NLM). The second part is called the transform domain, which is divided into wavelet, courvalet, and contourlet [22] as shown in Fig. 3.
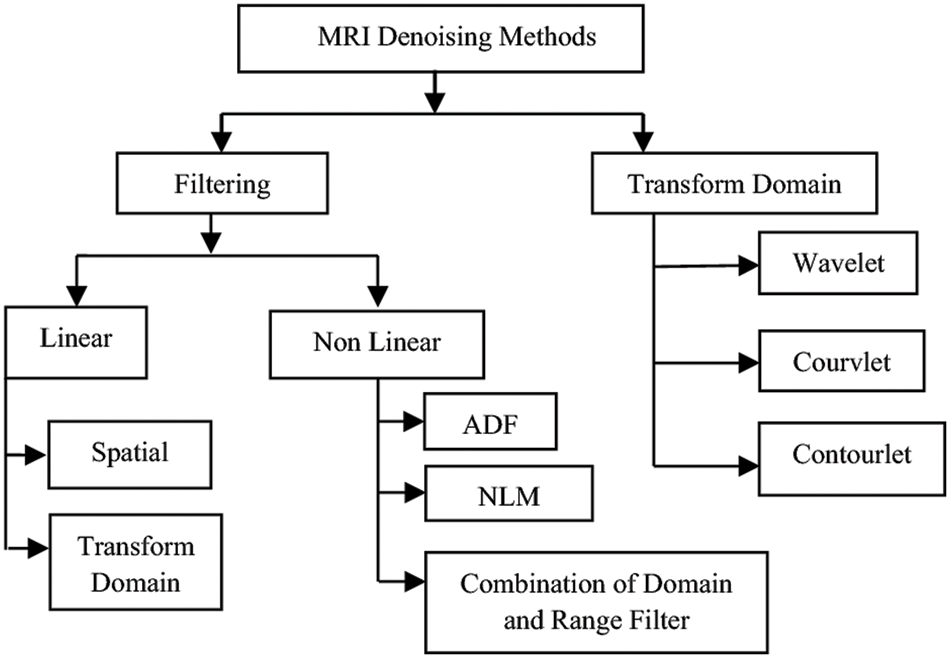
Figure 3: Classification of MRI denoising methods [22]
2.2.2 Vibrational Mode Decomposition
This method has been used in the novel methodology to reduce the Rician noise. The method divides the MRI images into different frequency components, such as low frequency and high frequency. The mechanism of the algorithm starts by removing the higher frequency and using the lower frequency to retrieve the image. The last step in the algorithm is to use a total variation (tv)-regularization to remove remnant noise details from the first stage [23], as shown in the next block diagram in Fig. 4.
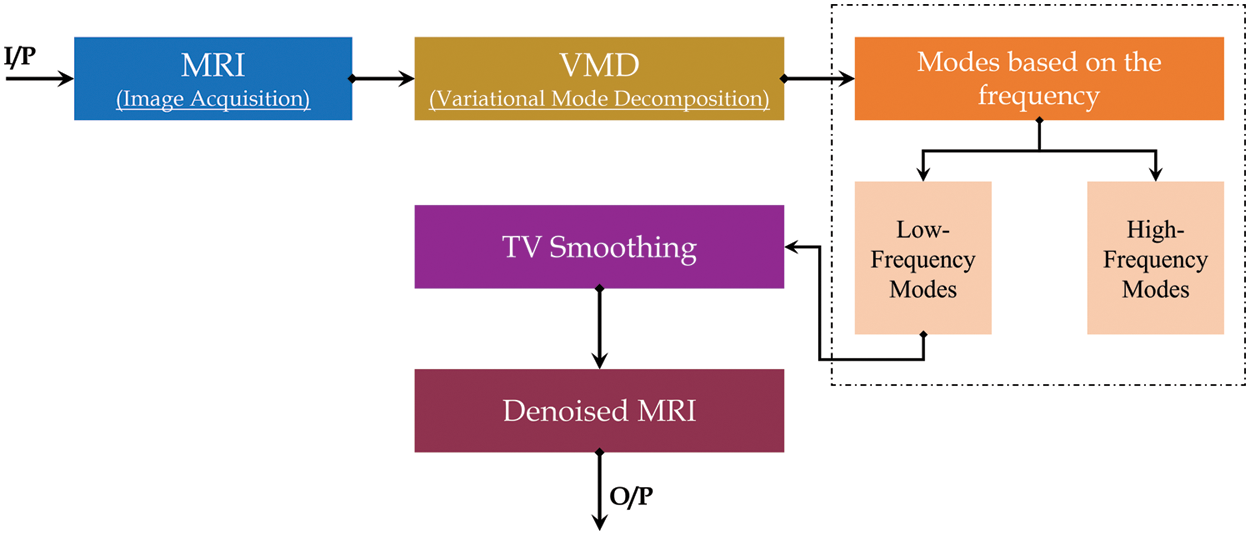
Figure 4: VMG block diagram
2.2.3 Block-Matching and 3-Dimensional Filtering
BM3D is a modified method for removing the Rician noise. It’s used to reduce the Rician from the MRI 3D images. The mechanism of BM3D is divided into two stages. The first stage is used to group wavelet transforms, noise invalidation denoising, and produce the inverse for the 3D wavelet transform. Stage 2 uses the signal and inverse signal with a wiener filter and winner coefficient to produce an inverse 3D wavelet. In the final step, the mechanism uses inverse variance stabilisation transform (VST) and clahe to produce the denoised image as shown in Fig. 5 [22].
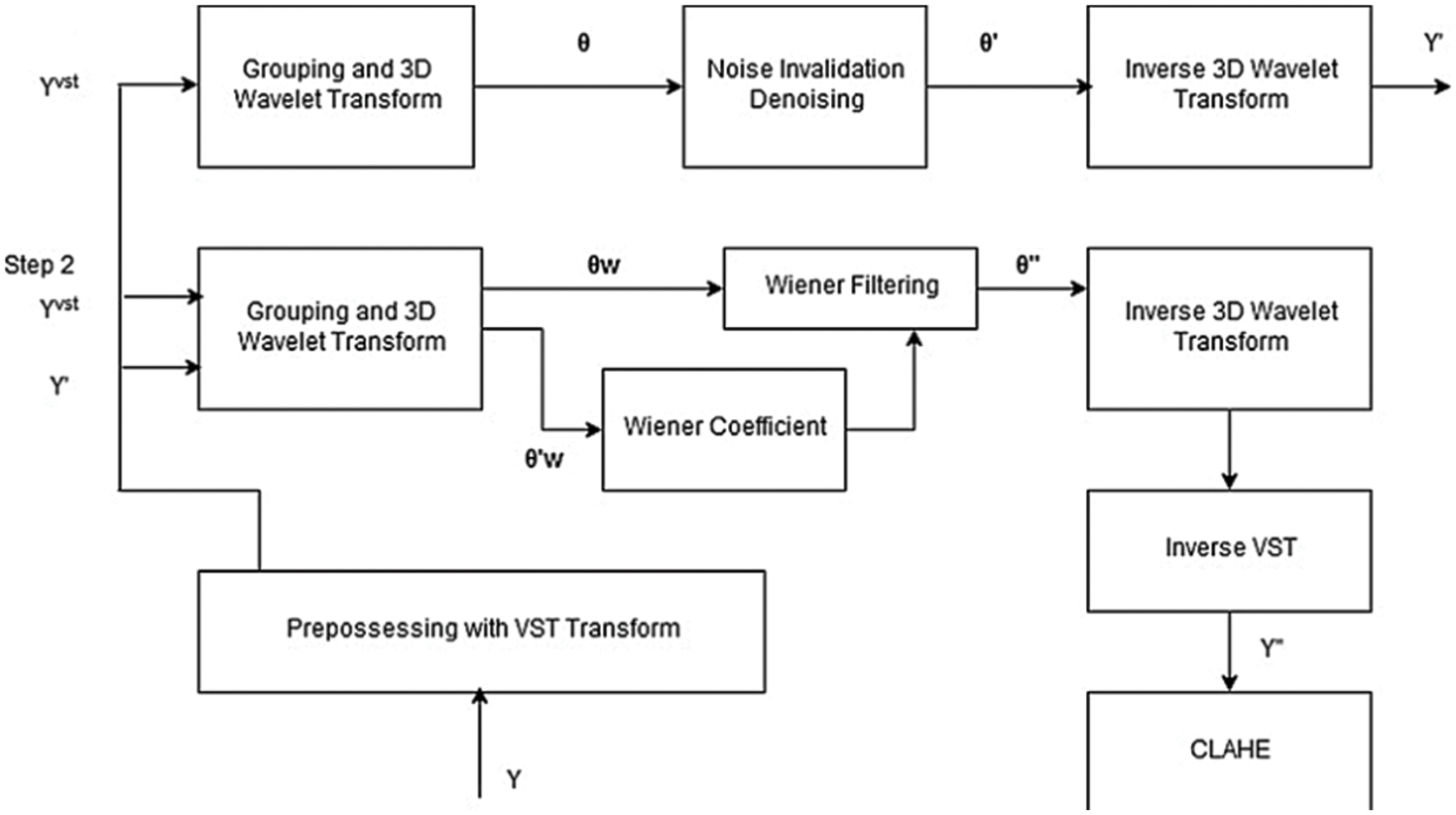
Figure 5: BM3D block digram
The DeRNet model has been proposed to handle the vanishing gradient issue. DeRNet minimizes gradients by creating shortcut connections between layers to resume the process of learning and training. This model uses 25 layers, and the overall block contains two stages: the res-block and the overall block [24] as shown in Fig. 6. This type of model can deal with the model at high depth and produce an MRI of the same size. The model contains a set of connected layers starting with a 3 × 3 convolution layer connected with batch normalization and rectified function as an activation function that can be activated by any input in the dataset. The dilated convolution in DeRNET is used for the exponential expansion of the receptive field without loss of resolution or coverage. DeRNET uses an extra-deep residual network for efficient computation rather than the classical number of residual networks.
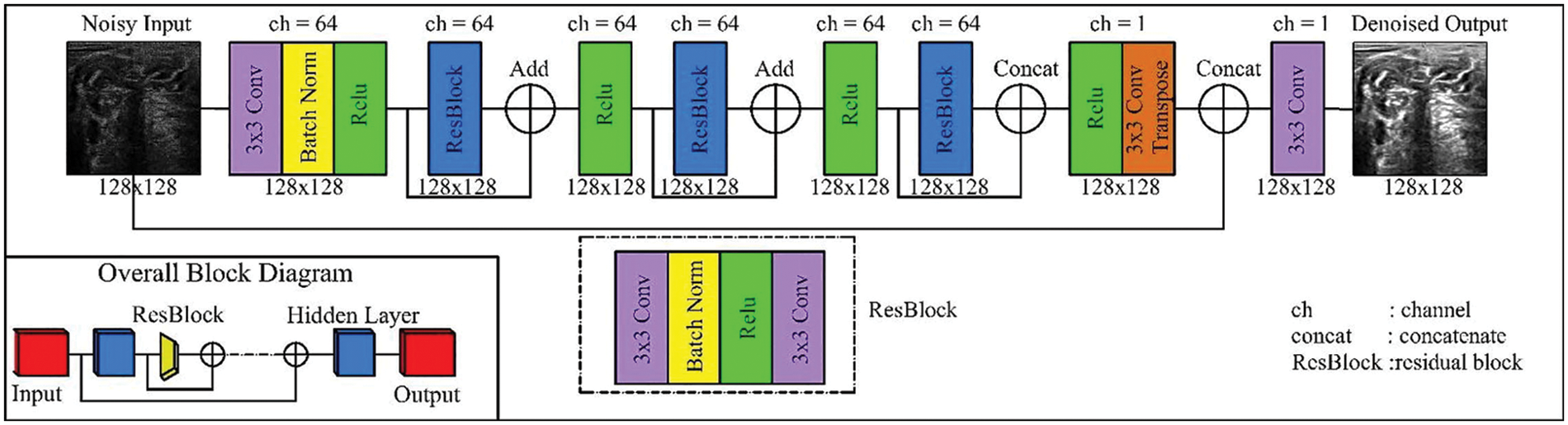
Figure 6: CNN-Residual model [25]
2.4 Generative Adversarial Denoising Network
The DGAN-Net model is considered a modified form of the traditional Generative Adversarial Network (GAN) model. DGAN-Net consists of two layers, which are the discriminator and the generator. The generator layer is used for generating fake MRI images to be used in the process of training, while the other part is used for differentiating between the fake and real MRI images. The Discriminator Deep Network uses zero padding, batch normalization, and rectifier (Relu) to differentiate between fake and real images [24,26,27], as shown in Fig. 7.
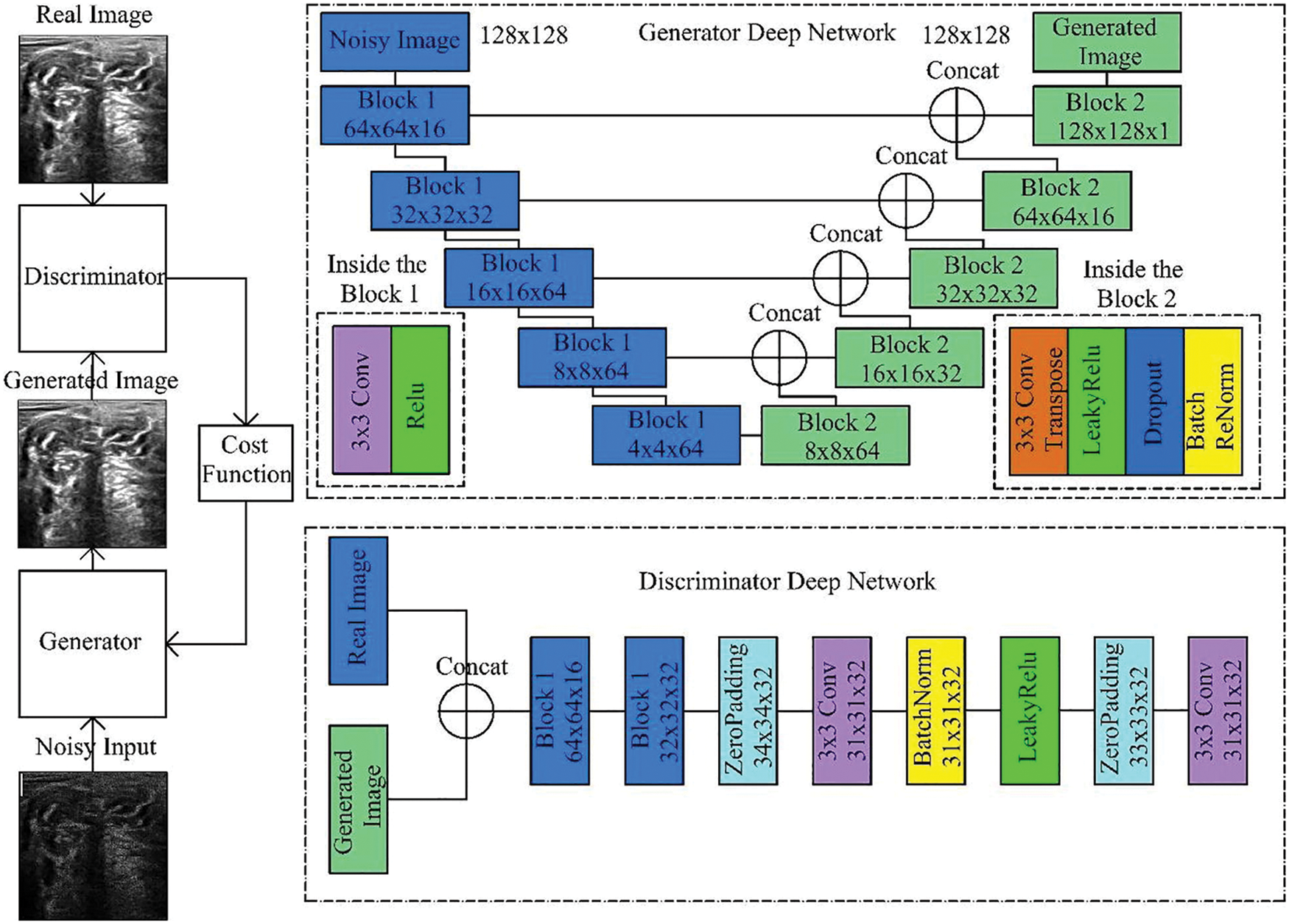
Figure 7: Generative adversarial denoising network [25]
2.5 Detailed Autoencoder Modified Method
This modified version of the autoencoder model is used to deal with the small image to maximize the image to avoid losing details of the MRI images. In the CNN model, operations are done in two stages; the first stage is called subsampling, and the second stage is called down-sampling. The subsampling layers represent the content of the image, while the down-sampling layers allow the receptive field to expand. This causes a big problem because the resolution of the MRI image is very important in removing the speckle noise process. The modified version of the auto-encoder avoids losing any details because it keeps the resolution of the MRI image by maximizing the number of parameters of the filter and the number of operations of location, which is still the same [28,29]. The model consists of two parts: an encoder and a decoder. The first part, which is called the encoder, consists of dilated convolution layers at each of the three levels, respectively, while the second part, which is called the decoder, has three dilated convolution layers at each of the three levels, respectively, as shown in Fig. 8 [30].
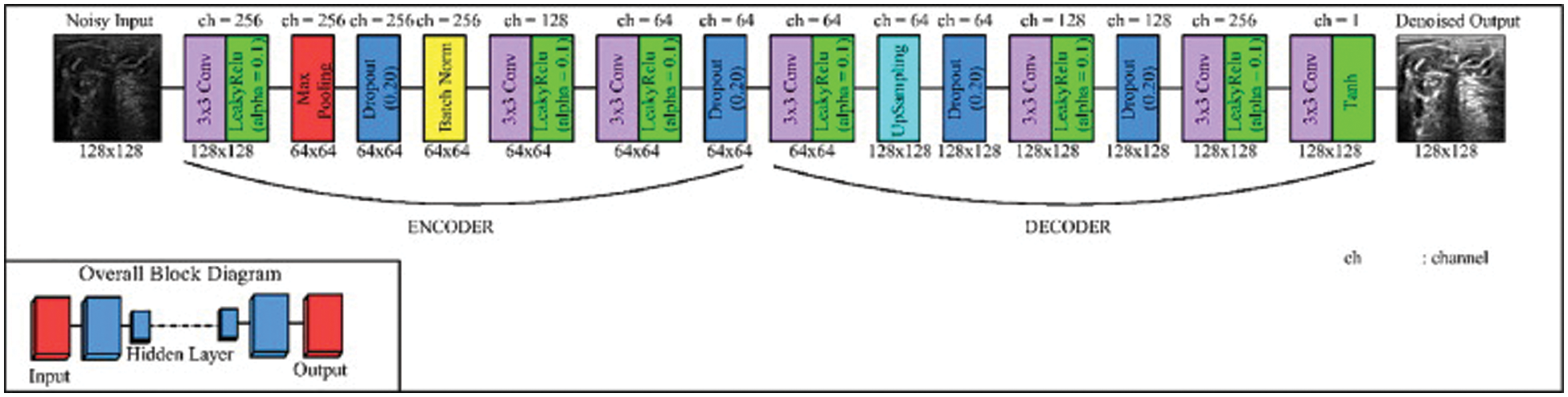
Figure 8: Modified version of autoencoder model [25]
In this step, the proposed framework uses the modified version from U-Net for segmenting the images after removing speckle and Rician noise in the previous steps. The modified version of U-Net, which is called the attention gate U-Net, can deal with different structures of medical images that may vary in size and shapes. Attention U-Net enables us to get over the need to use explicit external organ localization [31]. The Attention U-Net model is similar to the structure of the classical U-Net model but with some differences. The main advantages of the attention U-Net model over the classical U-Net model are the number of convolution filters and the number of parameters used by both models. Moreover, attention U-Net is better than the classical version of U-Net in simplicity. However, the consuming time of the attention U-Net is longer than that of the classical U-Net because of its dependency on a smaller number of parameters than the classical U-Net, as shown in Fig. 9 [32].
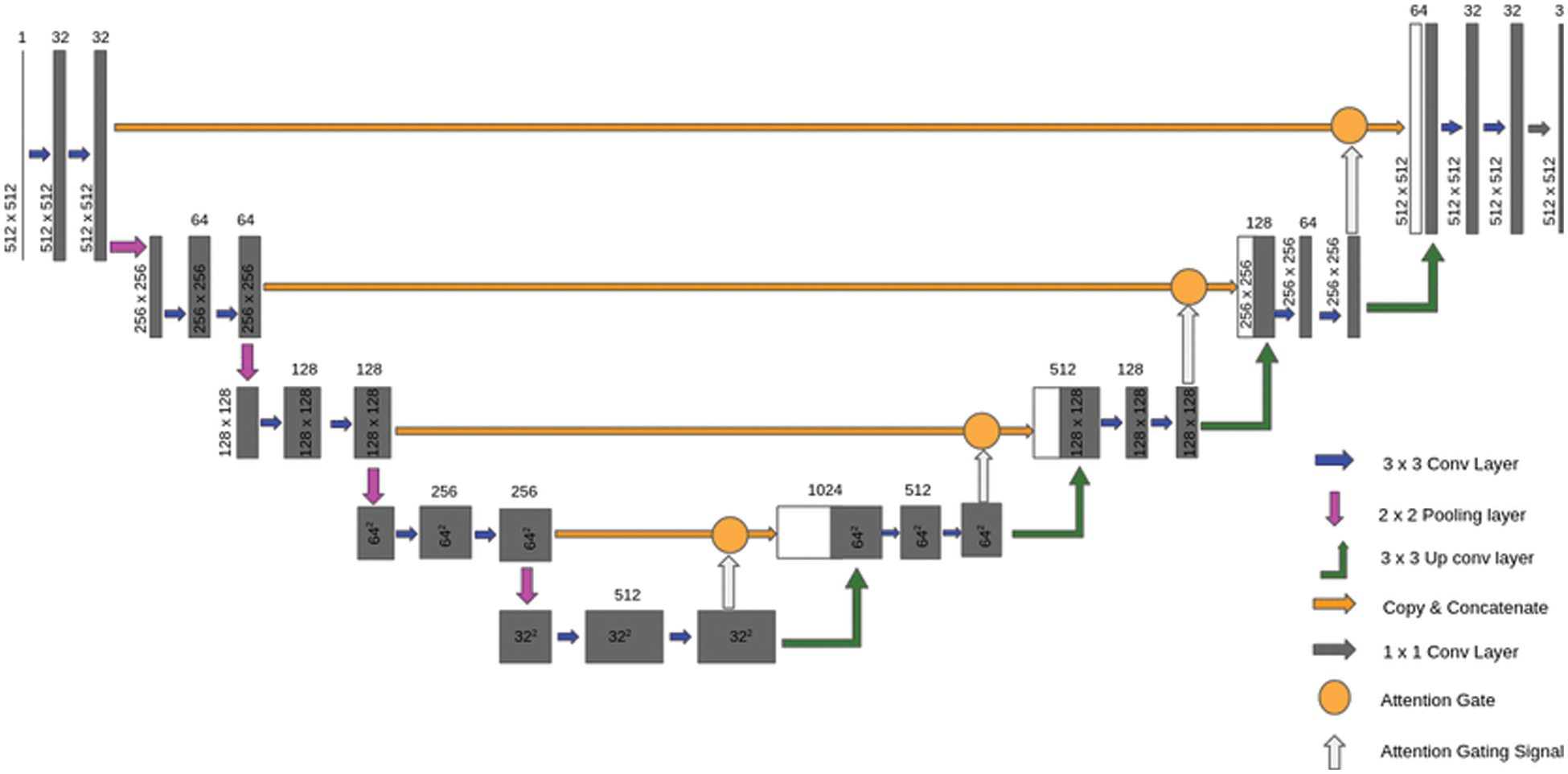
Figure 9: Attention U-Net [33]
The results and discussion section are divided into three sections: the first stage shows the results of reducing Rician noise; the second section shows the results of reducing the speckle noise, and the last section shows the results of segmentation of MRI images using the attention U-Net model.
3.1 Results of Removing Rician Noise
In this part of the paper, we show the results of removing the Rician noise using VMD and BM3D as mentioned in Tab. 1, and we also show the results of a comparison between different classical methods of removing noise and DeRNet, Di-Conv-AE-Net, and DGAN-Net as mentioned in Tab. 2. The comparison has been done for a different level of noise using PSNR using Mean Square Error (MSE) using Eqs. (3) and (4), since m × n monochrome image I and its noisy approximation K and Structural Similarity Index (SSIM) using Eq. (5), since
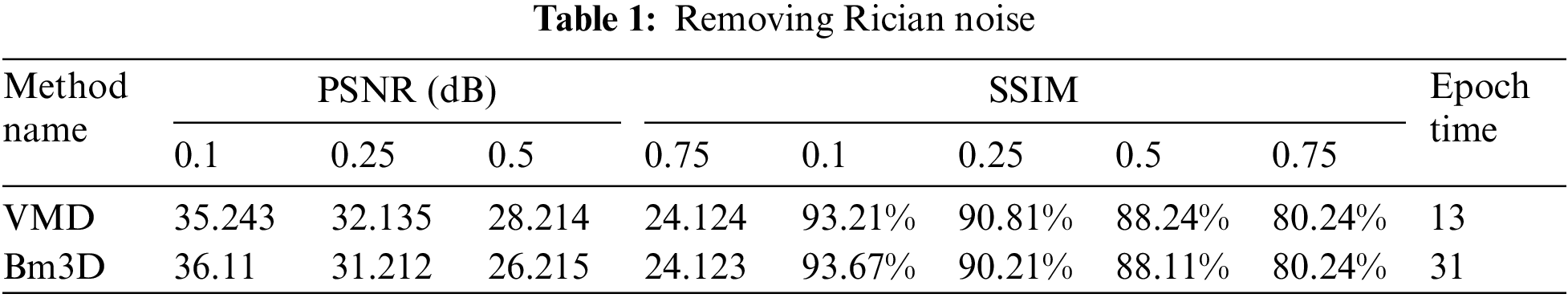
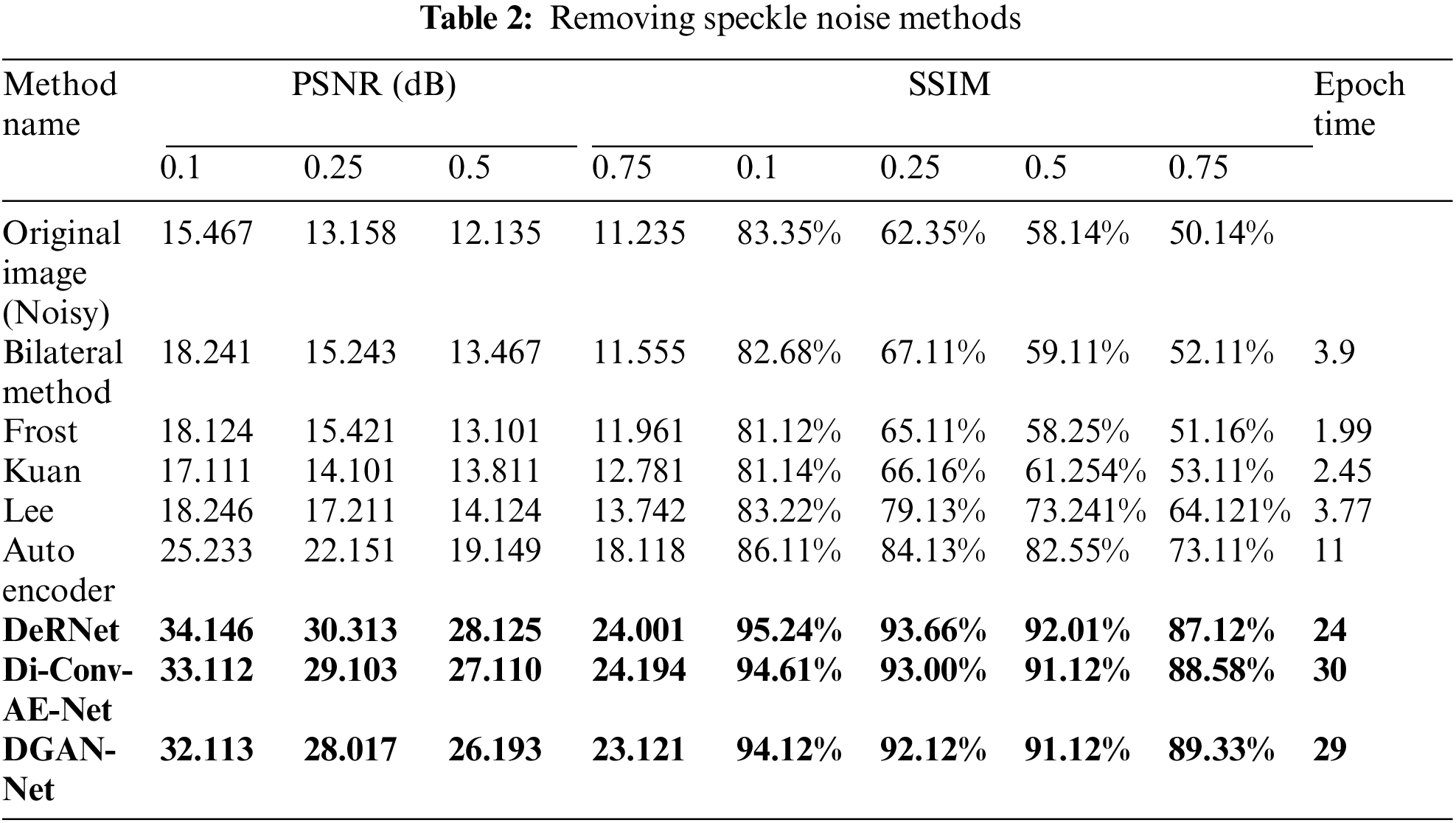
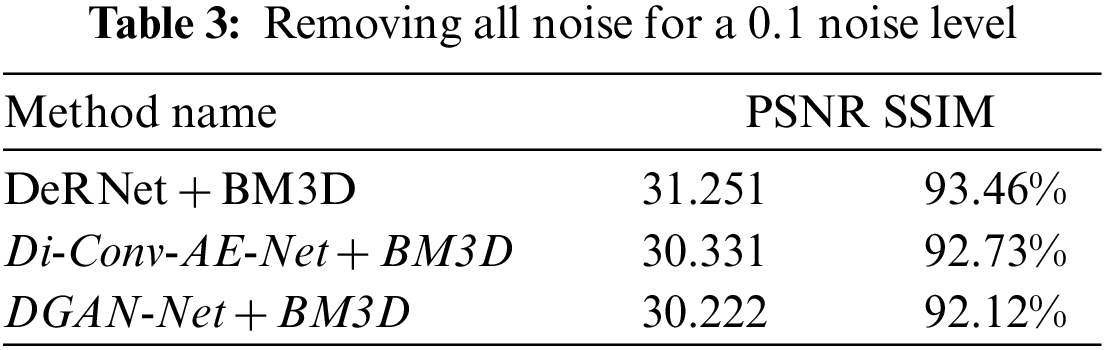

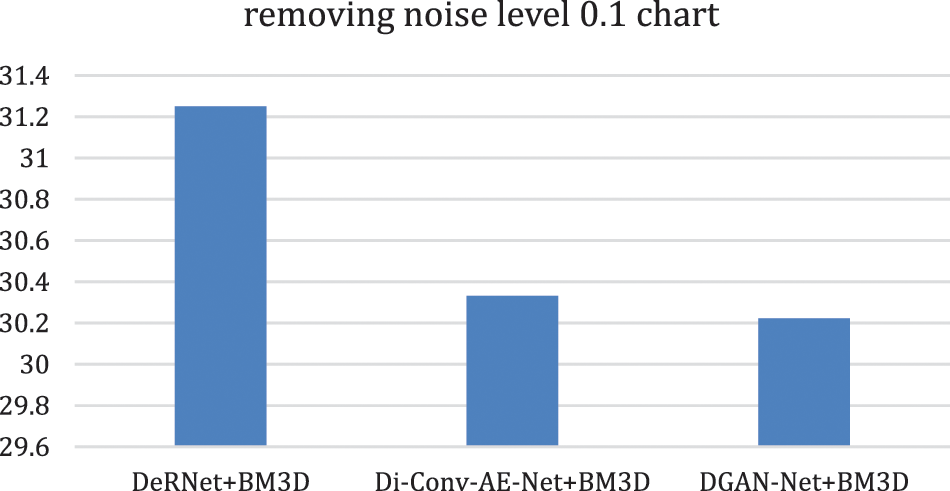
Figure 10: Removing overall noise at level 0.1 by the hybird algorithms
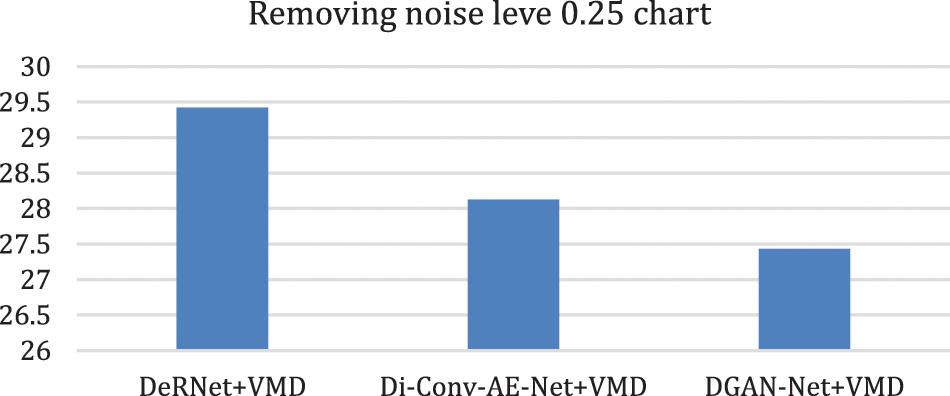
Figure 11: Chart of removing overall noise at level 0.25 by the hybird algorithms
3.2 Results of the Segmentation Step
In this subsection, the novel framework shows the final results after using the attention-U-Net deep learning model for segmentation of the MRI images after reducing the speckle and Rician noise in the previous stages. The attention U-Net with the previous method of reducing the noise has achieved an accuracy of nearly 94.66, which is better than the accuracy of U-Net and the accuracy without the previous model for removing the noise. Fig. 12 shows the predicted mast, mask, and the original image. The novel framework has achieved results in dice scores better than the previous versions of U-Net. The attention U-Net has achieved accuracy better than U-Net and residual U-Net. A U-Net and residual U-Net have been achieved 93.23 and 93.45 in accuracy, respectively.
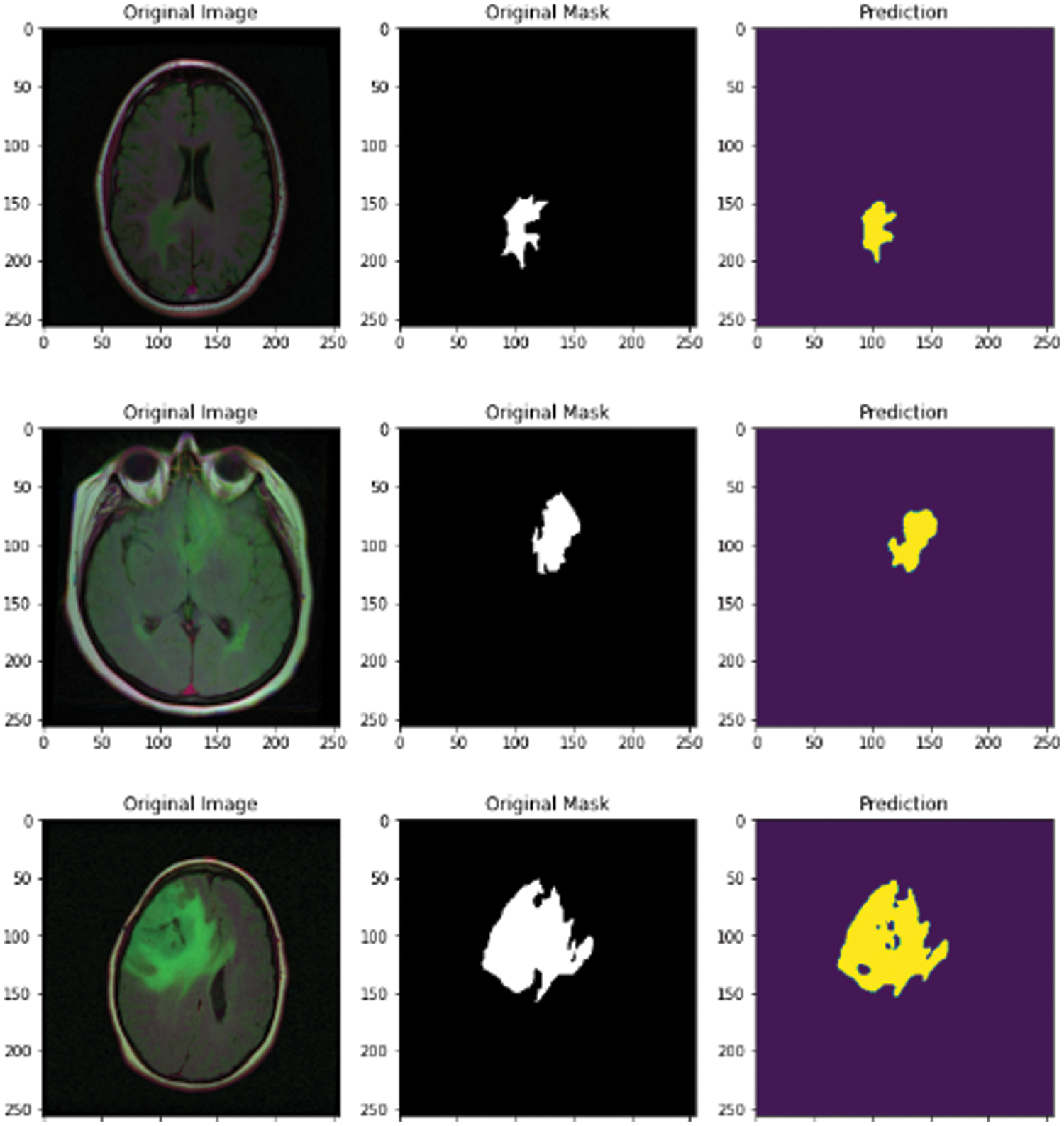
Figure 12: MRI original image, mask, and predicted mask
Segmentation of brain tumors is one of the most significant tasks for brain clinicians. Segmentation helps clinicians detect tumors in their early stages. The process of segmentation of brain tumors is divided into a set of stages such as image gathering, image enhancement, image segmentation, and feature extraction. To achieve final accuracy in the segmentation of MRI images, the methodology must take into consideration all details in each step because each stage’s accuracy can change the final result of segmentation. The process of MRI image enhancement and reducing all types of noise, such as speckle noise and Rician, plays a principal role in the accuracy of final segmentation. The novel framework has used more than one algorithm for reducing noise for each type of noise, such as VMD and BM3D for reducing Rician noise and more than one modified algorithm for reducing speckle noise. The framework chooses the best-qualified image in each stage. Finally, the framework used the attention U-Net model for segmenting the images from previous stages.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. E. Sasikala, P. Kanmani, R. Gopalakrishnan and R. Radha, “Identification of lesion using an efficient hybrid algorithm for MRI brain image segmentation,” Journal of Ambient Intelligence and Humanized Computing, vol. 12, no. 1, pp. 1–11, 2021. [Google Scholar]
2. A. S. Lundervold and A. Lundervold, “An overview of deep learning in medical imaging focusing on MRI,” Journal of Medical Physics, vol. 29, no. 2, pp. 102–127, 2019. [Google Scholar]
3. R. Ranjbarzadeh, A. B. Kasgari, S. J. Ghoushchi, S. Anari, M. Naseriet et al., “Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images,” International Journal of Scientific Reports, vol. 11, no. 1, pp. 10930–10945, 2021. [Google Scholar]
4. A. Bhargava and A. Bansal, “Novel coronavirus (COVID-19) diagnosis using computer vision and artificial intelligence techniques: A review,” Multimedia Tools and Applications, vol. 80, no. 13, pp. 19931–19946, 2021. [Google Scholar]
5. A. E. Hassanien, R. Bhatnagar, V. Snasel and M. Yasin Shams, “A deep learning based cockroach swarm optimization approach for segmenting brain MRI images medical informatics and bioimaging using artificial intelligence: Challenges, issues, innovations and recent developments,” Springer International Publishing, vol. 21, no. 1, pp. 3–13, 2022. [Google Scholar]
6. O. M. Elzeki, M. Abd Elfattah, H. Salem, A. E. Hassanien and M. Shams, “A novel perceptual two layer image fusion using deep learning for imbalanced COVID-19 dataset,” PeerJ Computer Science, vol. 7, no. 1, pp. 364–379, 2021. [Google Scholar]
7. H. Z. Zhihua, L. Chen, L. Tong and F. Zhou, “Deep learning based brain tumor segmentation: A survey,” arXiv preprint arXiv:2007.09479, vol. 20, no. 2, pp. 1–25, 2020. [Google Scholar]
8. J. Mohan, V. Krishnaveni and Y. Guo, “A survey on the magnetic resonance image denoising methods,” Biomedical Signal Process and Control, vol. 9, no. 2, pp. 56–69, 2014. [Google Scholar]
9. V. S. Frost, J. A. Stiles, K. S. Shanmugan and J. C. Holtzman, “A model for radar images and its application to adaptive digital filtering of multiplicative noise,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 8, no. 2, pp. 157–166, 1982. [Google Scholar]
10. M. Elad, “On the origin of the bilateral filter and ways to improve it,” IEEE Transiction on Image Processing, vol. 11, no. 10, pp. 1141–1151, 2002. [Google Scholar]
11. D. T. Kuan, A. A. Sawchuk and P. Chavel, “Adaptive noise smoothing filter for images with signal-dependent noise,” IEEE Transiction and Pattern Analusis Machine Intelligence, vol. 7, no. 2, pp. 165–177, 1985. [Google Scholar]
12. J. S. Lee, “Speckle analysis, and smoothing of synthetic aperture radar images,” Computer Graphics and Image Processing, vol. 17, no. 1, pp. 24–32, 1981. [Google Scholar]
13. H. Wang, C. Wu, J. Chi, X. Yu and Q. Hu, “Speckle noise removal in ultrasound images with stationary wavelet transform and canny operator,” in Proc. in the Chinese Control Conf. (CCC 2019), Guangzhou, China, pp. 7822–7827, 2019. [Google Scholar]
14. S. Wang, T. Z. Huang, X. L. Zhao, J. Mei and J. Huang, “Speckle noise removal in ultrasound images by first- and second-order total variation,” Numerical Algorithms, vol. 78, no. 2, pp. 513–533, 2018. [Google Scholar]
15. N. Rahimizadeh, R. Hasanzadeh and F. J. Sharifi, “An optimized non-local LMMSE approach for speckle noise reduction of medical ultrasound images,” Multimedia Tools and Applications, vol. 80, no. 6, pp. 9231–9253, 2021. [Google Scholar]
16. S. Karthikeyan, T. Manikandan, V. Nandalal, J. L. MazherIqbal and J. J. Babu, “A survey on despeckling filters for speckle noise removal in ultrasound umages,” in Proc. in the 3rd Int. Conf. on Electronics, Communication and Aerospace Technology (ICECA 2019), Coimbatore, India, pp. 605–609, 2019. [Google Scholar]
17. M. Havaei, A. Davi, D. W. Farely, A. Biard, A. Corville et al., “Brain tumor segmentation with deep neural networks,” Medical Image Analysis, vol. 35, no. 1, pp. 18–31, 2017. [Google Scholar]
18. X. Yao, X. Wang, S. -H. Wang and Y. D. Zhang, “A comprehensive survey on convolutional neural network in medical image analysis,” Multimedia Tools and Applications, vol. 11, no. 2, pp. 1–45, 2020. [Google Scholar]
19. L. Alzubaidi, J. Zhang, A. J. humidi, A. Aldujailli, Y. Duan et al., “Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions,” Journal of Big Data, vol. 8, no. 1, pp. 53–66, 2021. [Google Scholar]
20. D. W. Kim, C. Kim, D. H. Kim and D. H. Lim, “Rician nonlocal means denoising for MR images using nonparametric principal component analysis,” Journal on Image and Video Processing, SpringerOpen, vol. 2011, no. 1, pp. 15–29, 2011. [Google Scholar]
21. X. Y. Song, S. Zhang, K. O. Song, W. Yang and Y. Z. Chen, “Speckle supression for medical ultrasound images based on modelling speckle with Rayleigh distribution in contourlet domain,” in Proc. of the Int. Conf. on Wavelet Analysis and Pattern Recognition (ICWAPR2008), Hong Kong, China, vol. 1, pp. 194–199, 2008. [Google Scholar]
22. V. Hanchate and K. Joshi, “MRI denoising using BM3D equipped with noise invalidation denoising technique and VST for improved contrast,” SN Applied Sciences, vol. 2, no. 2, pp. 234–250, 2020. [Google Scholar]
23. D. Pankaj, D. Govind and K. A. Nayaranakautty, “A novel method for removing Rician noise from MRI based on variational mode decomposition,” Biomedical Signal Processing and Control, vol. 69, no. 1, pp. 102737–102750, 2021. [Google Scholar]
24. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in. Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition(CVPR 2016), Long Beach, CA, USA, pp. 770–778, 2016. [Google Scholar]
25. O. Karaoglu, H. S. Bilge and I. Uluer, “Removal of speckle noises from ultrasound images using five different deep learning networks,” Engineering Science and Technology an International Journal, vol. 29, no. 1, pp. 101030–101049, 2022. [Google Scholar]
26. Z. Chen, Z. Zeng, H. Shen, X. Zheng, P. Dai et al., “DN-GAN: Denoising generative adversarial networks for speckle noise reduction in optical coherence tomography images,” Biomedical Signal Processing and Control, vol. 55, no. 1, pp. 101632–101648, 2020. [Google Scholar]
27. S. Xun, D. Le, H. Zhu, M. chen, J. wang et al., “Generative adversarial networks in medical image segmentation: A review,” Computers in Biology and Medicine, vol. 140, no. 11, pp. 105063–105075, 2022. [Google Scholar]
28. R. Hamaguchi, A. Fujita, K. Nemoto, T. Imaizumi and S. Hikosaka, “Effective use of dilated convolutions for segmenting small object instances in remote sensing imagery,” in Proc. of the IEEE Winter Conf. on Applications of Computer Vision (WACV 2018), Lake Tahoe, NV, USA, pp. 1442–1450, 2018. [Google Scholar]
29. L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy and A. L. Yuille, “DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 40, no. 4, pp. 834–848, 2018. [Google Scholar]
30. Y. Li, X. Zhang and D. Chen, “CSRNet: Dilated convolutional neural networks for understanding the highly congested scenes,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition(CVPR 2018), Nashville, TN, USA, pp. 1091–1100, 2018. [Google Scholar]
31. O. Oktay, J. Shanbler, L. Fologoc, M. Lee, M. Hienrich et al., “Attention U-net: Learning where to look for the pancreas,” arXiv preprint arXiv:1804.03999, vol. 12, no. 3, pp. 16–32, 2018. [Google Scholar]
32. D. John and C. Zhang, “An attention-based U-Net for detecting deforestation within satellite sensor imagery,” International Journal of Applied Earth Observation and Geoinformation, vol. 107, no. 12, pp. 102685–102699, 2022. [Google Scholar]
33. S. Ai and J. Kwon, “Extreme low-light image enhancement for surveillance cameras using attention U-Net,” Sensors, Multidisciplinary Digital Publishing Institute, vol. 20, no. 2, pp. 495–511, 2020. [Google Scholar]
34. Z. Wang, A. C. Bovik, H. R. Sheikh and E. P. Simoncelli, “Image quality assessment: From error visibility to structural similarity,” IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600–612, 2004. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools