 Open Access
Open Access
ARTICLE
Attenuate Class Imbalance Problem for Pneumonia Diagnosis Using Ensemble Parallel Stacked Pre-Trained Models
School of Computer Science and Engineering, Vellore Institute of Technology, Chennai, 600127, India
* Corresponding Author: Harini Sriraman. Email:
Computers, Materials & Continua 2023, 75(1), 891-909. https://doi.org/10.32604/cmc.2023.035848
Received 06 September 2022; Accepted 19 November 2022; Issue published 06 February 2023
Abstract
Pneumonia is an acute lung infection that has caused many fatalities globally. Radiologists often employ chest X-rays to identify pneumonia since they are presently the most effective imaging method for this purpose. Computer-aided diagnosis of pneumonia using deep learning techniques is widely used due to its effectiveness and performance. In the proposed method, the Synthetic Minority Oversampling Technique (SMOTE) approach is used to eliminate the class imbalance in the X-ray dataset. To compensate for the paucity of accessible data, pre-trained transfer learning is used, and an ensemble Convolutional Neural Network (CNN) model is developed. The ensemble model consists of all possible combinations of the MobileNetv2, Visual Geometry Group (VGG16), and DenseNet169 models. MobileNetV2 and DenseNet169 performed well in the Single classifier model, with an accuracy of 94%, while the ensemble model (MobileNetV2+DenseNet169) achieved an accuracy of 96.9%. Using the data synchronous parallel model in Distributed Tensorflow, the training process accelerated performance by 98.6% and outperformed other conventional approaches.Keywords
The acute pulmonary infection known as pneumonia can be brought on by a virus that affects the lungs, causing inflammation and pleural effusion, which fills the lungs and makes breathing difficult. The majority of cases of pneumonia occur in impoverished and emerging nations, where there is a shortage of medical resources, excessive population, pollution, and unclean environmental conditions. Therefore, preventing the disease from turning fatal can be significantly aided by early diagnosis and care. The diagnosis of lung disorders typically involves radiological evaluation of the lungs using various imaging techniques like Computed Tomography (CT), X-ray, and Magnetic Resonance Imaging (MRI). Pneumonia is the leading cause of mortality attributed to respiratory illnesses. Medical professionals examine X-rays [1] to identify this illness in clinical practice, which is biased based on the radiologist’s experience and is highly time-consuming [2]. X-ray diagnostics are accurate when large-scale labeled datasets and deep learning algorithms are used. Deep learning enables the extraction of hierarchical features from adequate training datasets [3,4]. However, X-rays are misinterpreted due to the subjective variations in the investigation techniques used by health specialists. The impact of artificial intelligence in the medical field is widely studied [5].
In the medical sector, machine learning methods have been reported for pneumonia, cancer diagnosis, and real-time healthcare monitoring [6,7]. The robust feature extraction algorithms in Deep learning have shown considerable promise in medical image segmentation and prognosis [8–11]. In the medical and image domain, CNN is widely used and works effectively only with a large amount of data. The amount of labelled data available in the medical sector is less since creating the manually labelled dataset is highly time-consuming and requires much medical professionals’ experience. To get around this problem, use transfer learning. In this method, network weights established by a pre-trained model in a vast dataset are used to solve the problem in a small dataset. Many medical imaging problems are too complex to be solved using a single algorithm. Ensemble models use many models to tackle a particular problem. This technique aimed to overcome single models’ shortcomings and solidify their strengths.
The final result of the ensemble method is an aggregate of the different single-model outputs. Furthermore, the ensemble model reduces the variance for predictions and generalization error, which considerably improves computational learning and allows for the use of a minimal quantity of training examples. Typically, training these models on a standard processor takes weeks or months. Even though training neural networks has become hundreds of times faster thanks to contemporary Graphics Processing Units (GPU) and customized accelerators, training time still impacts both the accuracy of these methods’ predictions and their applicability. Many significant application areas can benefit from techniques that accelerate neural network training. By allowing professionals to train more on data and reducing the experimental iteration time, faster training can significantly increase model quality. This will enable researchers to test out new ideas and configurations more quickly. Accelerated training effectively uses neural networks when models and datasets are updated. Data parallelism is a simple and sound method for accelerated neural network training. Data parallelism refers to the distribution of training instances among several processors to compute gradient updates, followed by aggregating these locally computed updates. In this work, a data-parallel model accelerates the training process, and an extensive experimental study is conducted on the X-ray images. Pneumonia affects many people, especially children, and is most common in developing and impoverished nations, known for risk factors such as congestion, unsanitary living conditions, and poverty, in addition to the lack of adequate medical services. Most past studies focused on developing a separate network for detecting infected patients, and the application of ensemble methods with parallelism has not been explored.
Parallel and distributed implementation of medical models are the need of the hour due to the enormous size of the medical data. Medical data is growing daily, and these applications are inherently parallel since the data is stored locally at hospitals and cannot be shared due to security and ethical issues. So, the model needs to be parallelized to process the data locally and share only the results; this, in turn, makes the model faster than centralized processing, which needs to share data and is subjected to node failure. The memory footprint is almost the same when the data is processed parallelly among the nodes rather than sequentially. The model is replicated among nodes which is the only additional memory requirement, but it helps achieve faster and more accurate results. The computational complexity in the data parallel model is lesser than in the sequential model. Now healthcare applications are even developed using the 6G framework and the Internet of Things [12] with data security [13]. Existing methods are focused on a sequential approach and improving efficiency. The model improves efficiency using ensemble methods and is parallelized among multiple nodes for accelerated disease prediction.
The significant contributions in this work are listed below:
1. Three fined-tuned transfer learning models were implemented for the pneumonia prediction
2. An ensemble model using the three transfer learning models
3. Using regularization and augmentation techniques like SMOTE to reduce overfitting and remove the minority class imbalance problem that exists in the pneumonia dataset
4. Learning bias is reduced when anomalies and class imbalance are managed.
5. Data parallel model to reduce the training time and accelerate the training process
Detecting pneumonia using X-ray has been an unresolved issue for a long time, with a lack of publicly available data being the most significant obstacle. Chandra et al. [14] separated the pulmonary regions using the input lung images. They obtained eight characteristics from these areas to classify these. Using the Multi-Layer Perceptron (MLP), the technique was evaluated on 412 photographs and obtained a performance accuracy of 95.39%. Kuo et al. [15] used eleven variables for pneumonia detection in schizophrenia patients. Sharma et al. [16] created virtual CNN networks for categorizing pneumonic lung X-ray images. Stephen et al. [17] also developed a similar model. They used data enhancement to compensate for the absence of data. Janizek et al. [18] introduced an approach focused on adversarial evaluation to minimize models’ dependence on the origin of datasets and deliver accurate findings. In the source domain, they earned a Receiver Operating Curve (ROC) of 74.7%, while in the target domain, they achieved a score of 73.3%. Zhang et al. [19] designed a one-class, confidence-aware anomaly detection module for X-ray data. Their dataset achieved a ROC of 83.61 percent. Tuncer et al. [20] taught a computer an approach that performed the images’ fuzzy tree transformation and exemplar division. After retrieving features with a multichannel local binary pattern, the data were classified using standard classifiers. On a small dataset consisting of COVID-19 and pneumonia data, they established a 97.01 percent accuracy rate for the approach. The weights and model parameters learned from massive datasets are used for training the small dataset models in transfer learning. This method is mainly used to overcome the data shortage in the medical domain and make data processing faster and more efficient. The major existing works are shown in Table 1.
3 Proposed Work and Methodology
The proposed model has mainly four modules: Data Preprocessing and Augmentation, Transfer Learning models, Ensemble model, and Data Parallel Model, as shown in Fig. 1.

Figure 1: Overview of proposed model
The dataset is organized into three (training, testing, and Validation). There are 5,863 X-Ray images split into two classes (for standard and Pneumonia patients). The Guangzhou Women and Children’s Medical Center, Guangzhou, analyzed chest X-ray images from retrospective batches of children ages one to five.
Privacy regulations, the costly expense of acquiring annotations, and other issues limit the growth of medical imaging datasets. After dataset preprocessing and partition, data augmentation is used in the training process to supplement data in data-limited circumstances and minimize overfitting. In addition, our approaches used geometrical changes such as rescale, rotate, shifting, shearing, zooming, and flipping. Unfortunately, this location featured an uneven distribution of positive and negative observations, minimal data, and a significantly lower number of standard photographs than pneumonia photographs. This may result in poor post-training verification and generalization.
The prevalence of class imbalance issues in diagnosing diseases is high. A conventional classifier may favor the majority class and disregard the significance of the minority class. Therefore, this issue affects most supervised classification techniques, requiring researchers to exert much more effort to address it. The classification of outliers is a crucial aspect of deep learning. This issue arises when sample data seldom adhere to a distinct pattern. Techniques for managing outliers and unbalanced data have been presented, which may be categorized into two major categories: algorithm-based and data-level-based methods. The former seeks to adapt a learning algorithm toward the data and is considered to incur a high computational expense. The second is classifier-independent and straightforward to implement since it relies on approaches for data pre-treatment [36]. Several researchers address class disparity by under-sampling the dominant class or up-sampling the minority class [37]. In this work, Synthetic Minority Oversampling Technique is used to remove class imbalance.
Synthetic Minority Oversampling Technique [38]: Interpreting the minority group is one method for addressing uneven data sets. The most straightforward technique is duplicating instances from the minority class; however, these examples contribute no additional insight into the model. Alternatively, one may build new instances by synthesizing previous ones. The SMOTE is a method of supplementing data for this minority group. The augmented images generated using SMOTE analysis are shown in Fig. 2.

Figure 2: Augmented images
Deep Learning models are widely utilized for pneumonia diagnosis. However, owing to privacy regulations, the expensive expense of collecting annotations, and other factors, a considerable quantity of data is currently accessible in diagnostic imaging for deep learning models, even though they have shown tremendous performance in medical imaging. In light of the absence of medical datasets, transfer learning is used. Transfer learning is a deep learning approach in which a previously trained model from ImageNet is reused and transferred to a newly trained model. VGG16, Mobilenetv2, and Densenet169 are the transfer learning models used in this research. After analyzing the problem and dataset, the models are selected because the network structure significantly impacts the model’s performance [39].
With 3 × 3 convolutional kernels with 2 × 2 pooling layers, a VGGNet design may be considered an extended AlexNet. The network infrastructure can be further developed to improve deep features by utilizing a smaller convolution layer. VGGNet-16 and VGGNet-19 are now the two most popular VGGNet versions. The layers of the VGG model are given in Table 2.

MobileNet V2 is an upgraded form of MobileNet V1, CNN with 54 layers and a 224-by-224-pixel input image size. This approach does many convolutions using a single kernel instead of a two-dimensional convolution. Instead, it uses two 1-dimensional convolutions using two depth-separable kernels. Consequently, less memory and parameters are necessary for training, resulting in a small and effective model. The layers, along with the filter details of the Mobilenet model, are given in Table 2.
Huang et al. [40] suggestion for increasing CNN’s depth in the DenseNet. When CNNs’ model sizes increased in complexity, this strategy was initially used to address problems. The authors ensured sufficient information and gradient transfer by thoroughly connecting each layer to the one after it. One of the main advantages of using such a structure is that, through feature recycling, the DenseNet structure optimizes its capacity by using less of a deep or broad design. DenseNet does not learn duplicate features, unlike conventional CNNs. As a result, it needs fewer parameters. The structure only adds a few new feature maps because the layers are thin. It is important to note that there is no aggregation between the input feature maps and the output picture features of the layer because the DenseNet concatenates both. Dense Blocks are used to make sure that the size of extracted features remains constant within a block even though there are different numbers of filters in each one. In the Dense Blocks, layers of a specific type (referred to as transition layers) are placed. DenseNet is widely used in the medical diagnosis of critical illnesses like cancer [41]. The DenseNet layers and filter size are shown in Table 2.
The neural network model is distributed and trained in several High-Performance Computing (HPC) devices in the data parallelism. Training data are dispersed among the equipment to run synchronously or asynchronously. All-reduce is a method that reduces the target arrays throughout all machines to a tensor and returns the tensor to every device. Forward and backward propagation of the neural network are the two critical processes in the stochastic gradient learning of CNNs. An error about the desired results is computed after the forward pass calculates the outputs for a data set. In the so-called backward phase, this mistake or loss is then discriminated against about each parameter within CNN. The weights inside the network are then updated using the obtained gradients. These procedures are iteratively repeated until convergence, that is, until a local minimum inside the error function is reached. There are two modes of data parallelism. First, the model is replicated on each worker node and is used to process distinct data batches. Second, the parameter server nodes store and update the model parameters. Essentially, the worker will take the model’s parameters, run it on a batch of data, and transmit the gradients over to the Parameter Server (PS), where the model will be changed to improve it. However, multiple model update policies might be selected at the PS to carry out the training. The Parameter Server waits until all agent nodes have determined the gradients concerning their respective data sets in this scenario. The Parameter Server applies the gradients to the current weight after receiving them and then updates the model before sending it back to all worker nodes. This approach may result in variable connection speeds whenever other users share the cluster because updates are not made until all worker nodes have completed the calculation. It is only as fast as the slowest node. However, as more precise gradient estimates are produced, convergence occurs more quickly.
After using preprocessing, data segmentation, and data augmentation techniques, the quantity of our training data is increased, and it is ready to be given to the suggested method for extracting features to acquire suitable and relevant characteristics. The features acquired from each proposed system are merged to build the final, fully linked layer, which is then utilized to categorize each image into its corresponding class. In addition, each model within the ensemble is independently trained to address the specific issue. The ensemble model’s ultimate output is the mean or fusion of the various separate model outputs. In addition, ensemble models reduce the variance of prediction and generalization errors, considerably improve computational training, and may be implemented with a small amount of training data. This work developed three well-known CNN classification methods for pneumonia in pulmonary images using the literature and the suggested ensemble technique. Utilizing early termination prevents processes from getting overfitted. The model is built in Python using Tensorflow. Even though Tensorflow currently supports distributed repeated training, which has solved the problem of long training periods and may reduce training times, there is still room for development. The proposed model parameters are shown in Table 3.

The dataset has data and associated labels based on which the model accuracy and other evaluation parameters are calculated. Based on the label associated with data, the output can be grouped into four classes: True positives (correctly predicted output for positive labels), True Negative (Correctly predicted output for negative labels), False positive (False output for positive values, when the label is negative but got predicted as positive) and False Negative (False output for negative values, the label is positive but got predicted as unfavorable). These four factors provide the basis for most categorization task evaluation measures. Classification techniques are evaluated using the accuracy statistic. It is determined by dividing the number of correct predictions for both positive and negative labels by the total number of predictions. Precision is determined by dividing the number of actual positives by the total of real and false positives. When the impact of false negatives is considerable, it is reasonable to apply the recall statistic. The recall is determined by dividing the number of genuine positives by the total number of true positives and false negatives. The training time and speed up are also measured for model analysis.
5.1 Existing State-of-the-Art Transfer Learning CNN Model
Cohen’s Kappa score is a compelling performance statistic for datasets with imbalances. Various ranges of Kappa scores are used to examine the consistency with the acquired findings. If the score is lower than zero, it indicates a lack of data consistency. For example, if the score range is between 0.01 and 0.20, it indicates that there is just a modest degree of agreement, between 0.21 and 0.40, reasonable agreement, between 0.41 and 0.60, moderate agreement, and between 0.61 and 0.80, strong agreement. The range of Kappa scores between 0.81 and 1.00 represents an almost perfect degree of agreement. Precision and accuracy have also been included as a measure for the model’s evaluation. The dataset used for training has 5863 pictures. As most examples correspond to the negative samples, the models incorrectly forecasted most cases as belonging to the negative samples, resulting in much worse accuracy, recall, and F1 score values for positive class prediction. The model performance before SMOTE is given in Table 4.

The performance measures of the three base transfer learning models–Densenet 169, ResNet 50, and MobileNet after SMOTE are given in Table 5.

In Fig. 3, the performance of VGG16 is analyzed using accuracy, and loss is calculated using the training and validation phase. The confusion matrix is used to calculate the precision and recall. From Fig. 3, it is clear that accuracy gradually increases from 0.6056 to 0.8614 till epoch five, and after epoch 10, there is a stepwise improvement and reaches 0.89. The loss gradually decreases to five epochs and then reduces stepwise to 0.2511.

Figure 3: VGG16 performance
In Fig. 4, the performance of MobileNet v2 is analyzed using accuracy, and loss is calculated using the training and validation phase. The confusion matrix is used to calculate the precision and recall. From Fig. 4, it is clear that accuracy gradually increases 0.7753 to 0.9068 till epoch five, and after epoch 10, there is a stepwise improvement and reaches 0.9371. The loss gradually decreases till five epochs, and after that, it reduces stepwise to 0.1604. In Fig. 5, the performance of DenseNet169 is analyzed using accuracy, and loss is calculated using the training and validation phase. The confusion matrix is used to calculate the precision and recall. From Fig. 5, it is clear that accuracy gradually increases from 0.6578 to 0.9047 till epoch five, and after epoch 10, there is a stepwise improvement and reaches 0.9452. The loss gradually decreases till five epochs, and after that, it reduces stepwise to 0.1134.
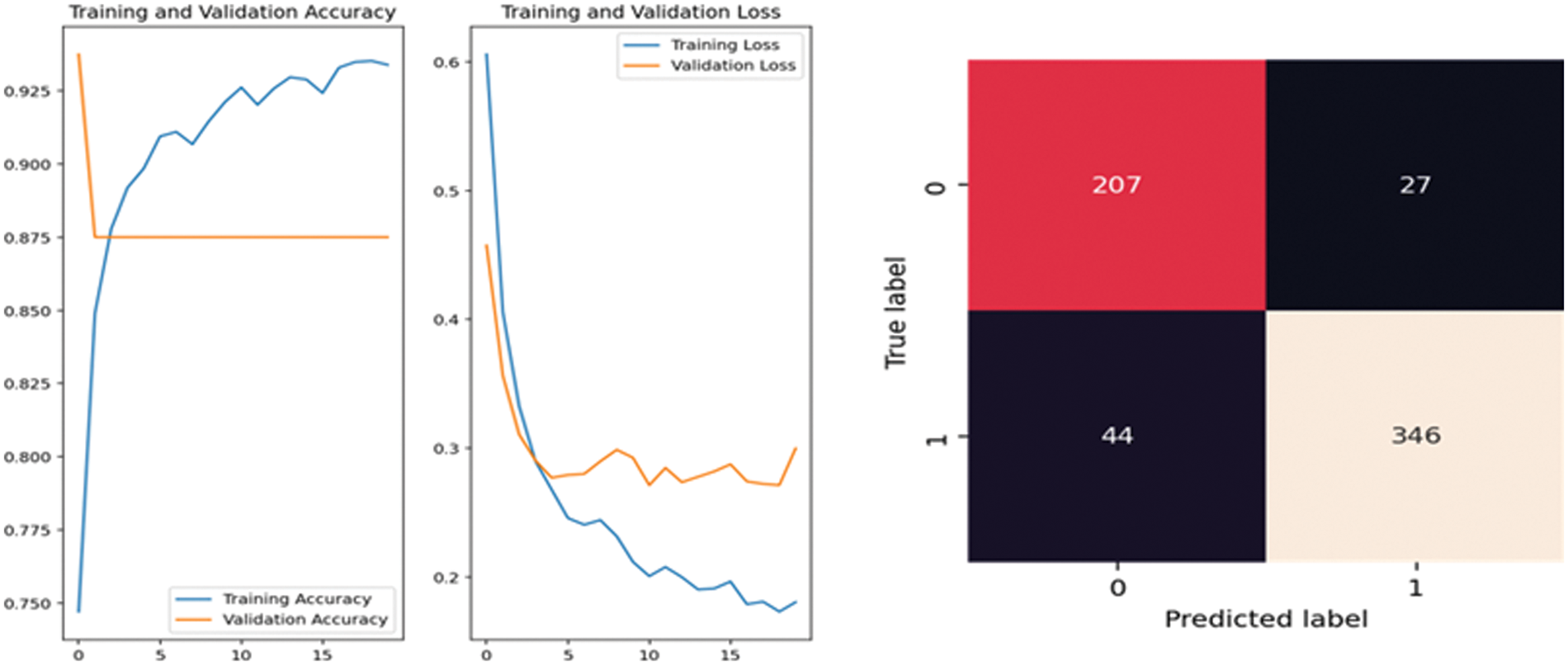
Figure 4: MobileNet V2 performance

Figure 5: DenseNet169 performance
The ensemble model’s output is preferable to the base learning models because it serves to include the discriminatory features of all of its component models. A practical approach for classifier merging is weighted average assembly. In this section, the ensemble learning models are studied to obtain improved performance, and the accuracy is measured. In Table 6, the ensemble models’ performance is shown. The Confusion matrix for the three-combination ensemble model created after stacking all three pre-trained model features is given in Fig. 6.


Figure 6: (a) MobileNetV2+DenseNet169+VGG16 (b) MobileNetV2+DenseNet169 (c) VGG16+ MobileNetV2 (d) VGG16+DenseNet169
5.3 Data Parallel Transfer Learning Models
There are two types of bandwidths in a data pipeline: data load bandwidth and model training bandwidth. The actual model training bandwidth is also constrained by the restricted on-device storage of the GPUs or other accelerators. From the flow viewpoint, it is false to say that the bigger input data size causes a longer time for training in single-node. From a system standpoint, the problem is the mismatch between data loading and model train bandwidth. The issue of the bandwidth mismatch between the data load bandwidth and the model building bandwidth is the fundamental cause of the lengthy single-node model training phase. We can boost the model-building bandwidth proportional to the number of processors used in the same training run by using data parallelism. The main steps of the data parallel model are shown in Fig. 7.

Figure 7: Data parallel model
The training pipeline in a data-parallel model mainly consists of six steps:
1. Data collection, Preprocessing of the image, and Augmentation
2. Data partition based on the number of devices/hardware accelerators available without bias
3. Loading data into the accelerators
4. Building identical models on the accelerators and training them
5. Model synchronization after the gradient calculation among all nodes/devices
6. Update the model with the updated parameter in each node/device
7. Repeat steps 4–6 till the end of an epoch
Table 7 shows the training time in the sequential and parallel models. It is found that the parallel mode is found to be faster compared to the serial model, which can be further accelerated using multiple nodes and multiple devices for parallel execution. Faster execution is very crucial in the case of medical applications which require timely detection of the disease and need an immediate cure. The performance and accuracy are also accelerated in parallel training, making the model more efficient. In the literature, the existing works mainly deal with sequential execution models for pneumonia prediction. All existing works mainly concentrate on the sequential ensemble models and increasing the accuracy of the models. The performance measures used in the current works are accuracy and, in this proposed model, both accuracy and training time. The proposed ensemble method gave an accuracy of 98.6% with accelerated training time in distributed data-parallel models in distributed TensorFlow.

The proposed model consists of a parallel ensemble model with accelerated training time and improved performance of 98.6%. The model is compared with the existing works, broadly classified as works on sequential ensemble techniques for pneumonia detection, parallel techniques for pneumonia detection, and parallel ensemble models. From Table 8, the proposed model is efficient compared to the existing parallel techniques for pneumonia detection. In Table 9, the existing sequential models are compared with the proposed model, and [26] was found to have higher accuracy. However, it is a sequential model, and accuracy will be reduced when executed in parallel. In Table 10, the existing parallel ensemble models are compared, and the proposed work was found to have better accuracy and performance.
Early detection of pneumonia has the potential to save more lives than later detection. Detecting pneumonia from X-Rays is difficult; thus, assistive tools and approaches may aid in diagnosing this illness. Medical data analysis demands a high degree of precision, and as a result, a great deal of research is being conducted to create novel diagnostic procedures. The proposed technique is based on a parallel ensemble model with 98.4%. The results demonstrate the efficacy of features derived from VGG16, MobileNetV2, and DenseNet169 in conjunction with a data-parallel model for successfully diagnosing pneumonia in a balanced enhanced dataset. Even though the strategy yielded promising outcomes, it must be validated on more datasets to ensure its robustness. After demonstrating its efficiency with larger real-world data sets, the approach may be used as a medical aid. The proposed model uses three transfer learning algorithms ensembled together, and it is a time-consuming task, but the distributed parallel framework effectively balances it. The main limitation of the proposed model is that the memory requirement is slightly higher since the model is replicated in each parallel node. High-performance computing systems will make the model faster, but it is costly. Even then, the GPU system’s performance outweighs its cost. In the future, the model will be scaled to HPC systems in Cloud, and a general framework needs to be created for deployment in real-time setup.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. C. P. West, V. M. Montori and P. Sampathkumar, “COVID-19 testing: The threat of false-negative results,” Mayo Clin. Proc., vol. 95, no. 6, pp. 1127–1129, 2020. [Google Scholar]
2. N. Liu, L. Wan, Y. Zhang, T. Zhou, H. Huo et al., “Exploiting convolutional neural networks with deeply local description for remote sensing image classification,” IEEE Access, vol. 6, pp. 11215–11228, 2018. [Google Scholar]
3. I. U. Khan and N. Aslam, “A deep-learning-based framework for automated diagnosis of COVID-19 using X-ray images,” Information, vol. 11, no. 9, Art. no. 9, pp. 1–13, 2020. [Google Scholar]
4. J. V. Pranav, R. Anand, T. Shanthi, K. Manju, S. Veni et al., “Detection and identification of COVID-19 based on chest medical image by using convolutional neural networks,” International Journal of Intelligent Networks, vol. 1, pp. 112–118, 2020. [Google Scholar]
5. Y. Kumar, A. Koul, R. Singla and M. F. Ijaz, “Artificial intelligence in disease diagnosis: A systematic literature review, synthesizing framework and future research agenda,” Journal of Ambient Intelligence and Humanized Computing, pp. 1–28, 2022. [Google Scholar]
6. A. Loddo, F. Pili and C. Di Ruberto, “Deep learning for COVID-19 diagnosis from CT images,” Applied Sciences, vol. 11, no. 17, Art. no. 17, pp. 1–16, 2021. [Google Scholar]
7. H. Alquran, M. Alsleti, R. Alsharif, I. A. Qasmieh, A. M. Alqudah et al., “Employing texture features of chest X-ray images and machine learning in COVID-19 detection and classification,” MENDEL, vol. 27, no. 1, Art. no. 1, pp. 9–17, 2021. [Google Scholar]
8. J. Redmon and A. Farhadi, “YOLO9000: Better, faster, stronger,” in Proc. of IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, pp. 6517–6525, 2017. [Google Scholar]
9. F. Chollet, “Xception: Deep learning with depthwise separable convolutions,” arXiv, 2017. [Online]. Available: http://arxiv.org/abs/1610.02357. [Google Scholar]
10. J. John, A. Ravikumar and B. Abraham, “Prostate cancer prediction from multiple pretrained computer vision model,” Health and Technology, vol. 11, no. 5, pp. 1003–1011, 2021. [Google Scholar]
11. M. Robin, J. John and A. Ravikumar, “Breast tumor segmentation using u-net,” in Proc. of 5th Int. Conf. on Computing Methodologies and Communication (ICCMC), Erode, India, pp. 1164–1167, 2021. [Google Scholar]
12. P. N. Srinivasu, M. F. Ijaz, J. Shafi, M. Woźniak and R. Sujatha, “6G diven fast computational networking framework for healthcare applications,” IEEE Access, vol. 10, pp. 94235–94248, 2022. [Google Scholar]
13. M. Kumar, S. Verma, A. Kumar, M. F. Ijaz, D. B. Rawat et al., “ANAF-IoMT: A novel architectural framework for iomt enabled smart healthcare system by enhancing security based on RECC-VC,” IEEE Transactions on Industrial Informatics, vol. 18, no. 12, pp. 1–8, 2022. [Google Scholar]
14. T. B. Chandra and K. Verma, “Pneumonia detection on chest X-ray using machine learning paradigm,” in Proc. of 3rd Int. Conf. on Computer Vision and Image Processing, Singapore, pp. 21–33, 2020. [Google Scholar]
15. K. M. Kuo, P. C. Talley, C. H. Huang and L. C. Cheng, “Predicting hospital-acquired pneumonia among schizophrenic patients: A machine learning approach,” BMC Medical Informatics and Decision Making, vol. 19, no. 1, Art. no. 42, pp. 1–8, 2019. [Google Scholar]
16. A. Sharma, A. Jain, P. Gupta and V. Chowdary, “Machine learning applications for precision agriculture: A comprehensive review,” IEEE Access, vol. 9, pp. 4843–4873, 2021. [Google Scholar]
17. O. Stephen, M. Sain, U. J. Maduh and D. -U. Jeong, “An efficient deep learning approach to pneumonia classification in healthcare,” Journal of Healthcare Engineering, vol. 2019, pp. e4180949, pp. 1–8, 2019. [Google Scholar]
18. J. D. Janizek, G. Erion, A. J. DeGrave and S. -I. Lee, “An adversarial approach for the robust classification of pneumonia from chest radiographs,” in Proc. of the ACM Conf. on Health, Inference, and Learning, New York, NY, USA, pp. 69–79, 2020. [Google Scholar]
19. Q. Zhang, X. Shen, L. Xu and J. Jia, “Rolling guidance filter,” in Proc. of Computer Vision–ECCV 2014, Zurich, Switzerland, pp. 815–830, 2014. [Google Scholar]
20. T. Tuncer, F. Ozyurt, S. Dogan and A. Subasi, “A novel covid-19 and pneumonia classification method based on f-transform,” Chemometr Intell. Lab Syst., vol. 210, pp. 104256, pp. 1–11, 2021. [Google Scholar]
21. S. Albahli, H. T. Rauf, A. Algosaibi and V. E. Balas, “AI-driven deep CNN approach for multi-label pathology classification using chest X-rays,” PeerJ Comput. Sci., vol. 7, pp. e495, 2021. [Google Scholar]
22. G. Liang and L. Zheng, “A transfer learning method with deep residual network for pediatric pneumonia diagnosis,” Computer Methods and Programs in Biomedicine, vol. 187, pp. 104964, 2020. [Google Scholar]
23. S. Albahli, H. Tayyab Rauf, M. Arif, M. Tabrez Nafis and A. Algosaibi, “Identification of thoracic diseases by exploiting deep neural networks,” Computers, Materials & Continua, vol. 66, no. 3, pp. 3139–3149, 2021. [Google Scholar]
24. O. N. Oyelade, A. E. -S. Ezugwu and H. Chiroma, “Covframenet: An enhanced deep learning framework for COVID-19 detection,” IEEE Access, vol. 9, pp. 77905–77919, 2021. [Google Scholar]
25. A. H. Alharbi and H. A. Hosni Mahmoud, “Pneumonia transfer learning deep learning model from segmented X-rays,” Healthcare (Basel), vol. 10, no. 6, pp. 987, 2022. [Google Scholar]
26. R. Kundu, R. Das, Z. W. Geem, G. -T. Han and R. Sarkar, “Pneumonia detection in chest X-ray images using an ensemble of deep learning models,” PLOS ONE, vol. 16, no. 9, pp. e0256630, 2021. [Google Scholar]
27. A. K. Jaiswal, P. Tiwari, S. Kumar, D. Gupta, A. Khanna et al., “Identifying pneumonia in chest X-rays: A deep learning approach,” Measurement, vol. 145, pp. 511–518, 2019. [Google Scholar]
28. I. Pan, A. Cadrin-Chênevert and P. M. Cheng, “Tackling the radiological society of North America pneumonia detection challenge,” American Journal of Roentgenology, vol. 213, no. 3, pp. 568–574, 2019. [Google Scholar]
29. S. Rajaraman, S. Candemir, I. Kim, G. Thoma and S. Antani, “Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs,” Applied Sciences, vol. 8, no. 10, Art. no. 10, pp. 1–17, 2018. [Google Scholar]
30. W. Ausawalaithong, S. Marukatat, A. Thirach and T. Wilaiprasitporn, “Automatic lung cancer prediction from chest X-ray images using deep learning approach,” in Proc. of 11th Biomedical Engineering Int. Conf. (BMEiCON), Chaing Mai, Thailand, pp. 1–5, 2018. [Google Scholar]
31. E. Ayan and H. M. Unver, “Diagnosis of pneumonia from chest X-ray images using deep learning,” in Proc. of 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), Istanbul, Turkey, pp. 1–5, 2019. [Google Scholar]
32. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Phys. Eng. Sci. Med., vol. 43, no. 2, pp. 635–640, 2020. [Google Scholar]
33. E. Ayan, B. Karabulut and H. M. Ünver, “Diagnosis of pediatric pneumonia with ensemble of deep convolutional neural networks in chest X-ray images,” Arab. J. Sci. Eng., vol. 47, no. 2, pp. 2123–2139, 2022. [Google Scholar]
34. S. Dey, R. Bhattacharya, S. Malakar, S. Mirjalili and R. Sarkar, “Choquet fuzzy integral-based classifier ensemble technique for COVID-19 detection,” Computers in Biology and Medicine, vol. 135, pp. 104585, 2021. [Google Scholar]
35. A. Mabrouk, R. P. Díaz Redondo, A. Dahou, M. Abd Elaziz and M. Kayed, “Pneumonia detection on chest X-ray images using ensemble of deep convolutional neural networks,” Applied Sciences, vol. 12, no. 13, Art. no. 13, pp. 1–15, 2022. [Google Scholar]
36. B. Krawczyk, “Learning from imbalanced data: Open challenges and future directions,” Prog. Artif. Intell., vol. 5, no. 4, pp. 221–232, 2016. [Google Scholar]
37. P. Skryjomski and B. Krawczyk, “Influence of minority class instance types on SMOTE imbalanced data oversampling,” in Proc. of the First Int. Workshop on Learning with Imbalanced Domains: Theory and Applications, Skopje, Macedonia, pp. 7–21, 2017. [Google Scholar]
38. N. V. Chawla, K. W. Bowyer, L. O. Hall and W. P. Kegelmeyer, “SMOTE: Synthetic minority over-sampling technique,” Jair, vol. 16, pp. 321–357, 2002. [Google Scholar]
39. A. Ravikumar, H. Sriraman, P. M. Sai Saketh, S. Lokesh and A. Karanam, “Effect of neural network structure in accelerating performance and accuracy of a convolutional neural network with GPU/TPU for image analytics,” PeerJ Computer Science, vol. 8, pp. e909, 2022. [Google Scholar]
40. G. Huang, Z. Liu, L. Van Der Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in Proc. of IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, pp. 2261–2269, 2017. [Google Scholar]
41. A. Vulli, P. N. Srinivasu, M. S. K. Sashank, J. Shafi, J. Choi et al., “Fine-tuned DenseNet-169 for breast cancer metastasis prediction using fastai and 1-cycle policy,” Sensors, vol. 22, no. 8, Art. no. 8, pp. 1, 2022. [Google Scholar]
42. G. De Melo, S. O. Macedo, S. L. Vieira and L. G. Leandro Oliveira, “Classification of images and enhancement of performance using parallel algorithm to detection of pneumonia,” in Proc. of IEEE Int. Conf. on Automation/XXIII Congress of the Chilean Association of Automatic Control (ICA-ACCA), Chile, pp. 1–5, 2018. [Google Scholar]
43. H. Moujahid, B. Cherradi, O. el Gannour, L. Bahatti, O. Terrada et al., “Convolutional neural network based classification of patients with pneumonia using X-ray lung images,” Advances in Science, Technology and Engineering Systems Journal, vol. 5, no. 5, pp. 167–175, 2020. [Google Scholar]
44. N. Dey, Y. -D. Zhang, V. Rajinikanth, R. Pugalenthi, and N. S. M. Raja, “Customized VGG19 architecture for pneumonia detection in chest X-rays,” Pattern Recognition Letters, vol. 143, pp. 67–74, 2021. [Google Scholar]
45. A. Manconi, G. Armano, M. Gnocchi and L. Milanesi, “A soft-voting ensemble classifier for detecting patients affected by COVID-19,” Applied Sciences, vol. 12, no. 15, Art. no. 15, pp. 1–23, 2022. [Google Scholar]
46. K. Zhang, X. Liu, J. Shen, Z. Li, Y. Sang et al., “Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, no. 6, pp. 1423–1433.e11, 2020. [Google Scholar]
47. N. K. Chowdhury, M. A. Kabir, M. M. Rahman and N. Rezoana, “ECOVNet: A highly effective ensemble based deep learning model for detecting COVID-19,” PeerJ Comput. Sci., vol. 7, pp. e551, 2021. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF



 Downloads
Downloads
 Citation Tools
Citation Tools