 Open Access
Open Access
ARTICLE
Managing Health Treatment by Optimizing Complex Lab-Developed Test Configurations: A Health Informatics Perspective
1 Department of Computer Science, Federal Urdu University of Arts Science and Technology, Karachi, Pakistan
2 Department of Computer Science, Institute of Business Administration, Karachi, Pakistan
3 Department of Computer Science, College of Computer, Qassim University, Buraydah, Saudi Arabia
4 Computer Engineering Department, King Fahd University of Petroleum and Minerals, Dhahran, Saudi Arabia
5 SDAIA-KFUPM Joint Research Center for Artificial Intelligence, Dhahran, 31261, Saudi Arabia
* Corresponding Author: Ali Mustafa Qamar. Email:
Computers, Materials & Continua 2023, 75(3), 6251-6267. https://doi.org/10.32604/cmc.2023.037653
Received 12 November 2022; Accepted 13 February 2023; Issue published 29 April 2023
Abstract
A complex Laboratory Developed Test (LDT) is a clinical test developed within a single laboratory. It is typically configured from many feature constraints from clinical repositories, which are part of the existing Laboratory Information Management System (LIMS). Although these clinical repositories are automated, support for managing patient information with test results of an LDT is also integrated within the existing LIMS. Still, the support to configure LDTs design needs to be made available even in standard LIMS packages. The manual configuration of LDTs is a complex process and can generate configuration inconsistencies because many constraints between features can remain unsatisfied. It is a risky process and can lead patients to undergo unnecessary treatments. We proposed an optimized solution (opt-LDT) based on Genetic Algorithms to automate the configuration and resolve the inconsistencies in LDTs. Opt-LDT encodes LDT configuration as an optimization problem and generates a consistent configuration that satisfies the constraints of the features. We tested and validated opt-LDT for a local secondary care hospital in a real healthcare environment. Our results, averaged over ten runs, show that opt-LDT resolves 90% of inconsistencies while taking between 6 and 6.5 s for each configuration. Moreover, positive feedback based on a subjective questionnaire from clinicians regarding the performance, acceptability, and efficiency of opt-LDT motivates us to present our results for regulatory approval.Keywords
A Laboratory Developed Test (LDT) is a relatively simple, non-commercial clinical test developed and validated within a single laboratory and regulated by the drug regulatory authorities (DRAs) [1–3]. In Pakistan, the Drug Regulatory Authority of Pakistan (DRAP) has several divisions to provide compliance with Pakistan Drugs Act., particularly the Quality Assurance and Laboratory Testing (QALT) division and Management Information Services (MAS) division. Although QALT and MAS are responsible for regulating LDTs in Pakistan, they have no standard operating procedure for such a regulation.
LDTs can be either simple or complex [4,5]. Typically, a simple LDT is configured by clinicians from a small repository of clinical testing features and measures single-analytes, such as a test to measure the sodium or calcium level in a patient. The configuration of simple LDTs is comparatively an easy process where clinicians can manually satisfy the limited number of constraints of clinical features. While a complex LDT is configured from hundreds of feature constraints. The manual configuration of complex LDTs is a complicated and error-prone process. It can generate configuration errors because many constraints between features can only be satisfied partially through the manual process. A limited number of hospitals configure (complex) LDTs in Pakistan. A team of clinicians configures an LDT, which requires selecting features from available medical repositories which are part of the Laboratory Information Management System (LIMS) (explained in Section 2). Each clinician configures the features related to their specialized field included in the final LDT configuration. During the LDT configuration, clinicians communicate extensively to resolve all feature constraints, which indicate clinical rules. This process is complicated, possibly taking many weeks, in which important features can be overlooked with many unresolved constraints in the final integrated LDT configuration. This could lead to an inconsistent LDT configuration. Two examples of this scenario could be the omission of the features concerning the timing and amount of drug administration, and the inability of the clinician to specify the method for collecting and handling specimens in the LIMS. The difficulties increase for those LDTs that require contributions from clinicians with various specializations. A physician with expertise in a particular field can focus on selecting the relevant features within their domain, without considering the features from other domains. However, for an LDT, the interpretation of these features must be combined to produce an accurate outcome. Further defining this complexity is that complex LDTs use large feature repositories, increasing the chances of an improper LDT configuration and potentially leading to faulty data analyses and unacceptable clinical performance. For instance, patients can undergo unnecessary treatment or run risks of extreme and irreversible interventions [6–9]. These issues have motivated DRAs to start a risk-based procedure to regulate high-risk complex LDTs [6–8]. In Pakistan, DRAP has yet to begin any regulatory process for LDTs, possibly because of a minimal number of LDT configurations due to a lack of required infrastructure. However, some rare diseases rarely became prevalent locally in Pakistan, such as Diastrophic Dysplasia [10], for which configuring a complex LDT became necessary. In this situation, local clinicians of LIMS-based hospitals resort to manual LDT configuration mechanisms. Regular medical lab test functions and automated support for storing patient information and sample results of an LDT are also integrated within the existing LIMS. However, the support to configure and resolve inconsistencies in complex LDT design currently needs to be made available. Even in standard healthcare software packages available to store, retrieve, and process data from LDTs, such as ClinQuan software [11], LDT configuration support with a focus on inconsistency resolution is not available. To the best of our knowledge, there is no standard automated support to date for designing and configuring an LDT within a LIMS.
Besides these issues, this paper’s work is mainly motivated by a recent rare disease in Gujrat’s suburban region (Pakistan). The clinicians of a local secondary care hospital in Gujrat were required to configure an LDT to detect the disease. This LDT required medical experts’ input from different domains, and the main issue was that a consensus could not be reached amongst the experts in resolving all feature constraints. Hence, the final LDT product remained inconsistent. This paper employs state-of-the-art health informatics concepts to manage and solve these LDT issues. Specifically, we propose opt-LDT (Optimized LDT), an automated LDT which implements evolutionary optimization to resolve feature constraints in an LDT configuration. In opt-LDT, Genetic Algorithms (GA) are used because it is widely applicable in many domains [4,7,8,12].
This paper’s purpose is to execute opt-LDT in a healthcare environment. The performance of opt-LDT is also compared with the manual LDT configuration process. For this, we are interested in answering the following two research questions:
Q1: Does GA have the potential to produce a consistent (conflicts-free) LDT configuration?
Q2: Is the efficiency of a GA higher than that of manual LDT configuration?
In this research, an opt-LDT prototype has been created comprising 100 test features in the repository to answer these questions. Our results, averaged over ten runs, show that GA resolves 90% of inconsistencies while taking between 6 and 6.5 s for each configuration. In other words, opt-LDT can autonomously resolve a large percentage of constraints (inconsistencies) in almost negligible time. The results show that opt-LDT can improve the LDT configuration process.
The main contributions of this paper are:
• An optimized solution (opt-LDT) based on GA to automate the configuration of complex LDT is presented.
• Testing and validation of opt-LDT prototype in a real-time hospital environment.
• Practical evaluation of opt-LDT based on clinicians’ feedback.
The rest of the paper is organized as follows: Section 2 discusses the background of the research with the motivation and challenges presented in Section 3. The theoretical framework and methodology are discussed in Section 4. The dataset is described in Section 5, and the details about the experiments and results. The questionnaire evaluation and limitations are provided in Section 6, and the paper is concluded in the next section.
This section describes a relevant background related to Laboratory Information Management Systems (LIMS) and Genetic Algorithms.
2.1 Laboratory Information Management Systems (LIMS)
LIMS is a collection of modules to store, process, and manage data from all stages of a medical lab test [13,14]. It helps clinicians supervise many patient tests, such as hematology, immunology, and microbiology. Typically, LIMS has the following three modules, which exchange patient, sample, and laboratory result data with each other.
• Master Data Module (MDM): It has features to store all the relevant data of patients or requesters. The patient’s name, phone number, address, and email can be included.
• Sample Data Module (SDM): It stores sample data features such as sample date, collection time, materials (e.g., blood, urine), and sample results.
• Report Generation Module (RGM): RGM stores report printing features such as report format, report header (name and address of the lab), contents of the report, comments on test results analysis, and interpretation. In a typical LIMS, RGM has proper interfacing with MDM and SDM to fetch timely data and produce an error-free and reliable report.
Like a regular medical lab test, automated support for storing patient information and sample results of an LDT is also integrated within the LIMS. Clinical Laboratory Improvement Amendments (CLIA) of 1988-compliant software is available to store, retrieve, and process data from LDTs, such as ClinQuan software [11].
GA is an optimization method that simulates the process of natural selection to generate effective solutions [15,16]. A data structure called chromosome represents the optimization task, with different task parameter values producing different chromosomes [17]. The fitness of each chromosome towards an optimized solution is determined using an objective function. GA initially generates an initial population of chromosomes. The two best chromosomes (identified through the objective function) are selected, and GA parameters called mutation and crossover are used to produce the next-generation chromosome [18].
The crossover combines the parent chromosomes to produce children’s chromosomes. The mutation changes the new children to produce a small diversity from their parents.
Moreover, the parameter crossover fraction specifies each population’s fraction, other than the best children, made up of crossover children. A crossover fraction of 1 means that all children other than the best are crossover children, while a crossover fraction of 0 means that all children are mutated. Both extremes are unacceptable, and a crossover fraction between 0 and 1 is selected. This process of generating children’s chromosomes is repeated until a new population is evolved.
After several iterations, one of the chromosomes is selected as the optimal solution for the problem designers. The optimal values for GA parameters, i.e., crossover function, crossover fraction, mutation rate, and population size, depend on the problem’s nature. Therefore, before running the actual GA experiments, it is necessary to tune these parameters to obtain their optimal values [19].
3 LDT Configuration: Challenges & Motivation
The management of laboratory testing is crucial in healthcare evaluation and patient treatment and is accomplished through the use of In Vitro Diagnostic (IVD) devices, which are commercially available medical products consisting of reagents, instruments, or systems used for patient diagnosis, prognosis, disease monitoring, and prevention [20]. Simple LDTs can be created by selecting testing features from a small repository and with the help of experts who can easily fulfill the limited number of clinical feature constraints, avoiding errors resulting from inconsistencies.
On the other hand, a complex LDT which involves configuring from larger repositories, often containing hundreds of feature constraints, can measure a vast range of analytes such as DNA variations or parameters related to ongoing epidemic diseases. The manual configuration of such complex LDTs is a complex process, leading to configuration errors due to partial satisfaction of constraints between features. This manual process can result in critical features being omitted in the final LDT product.
The configuration of complex LDT in Pakistan, using LIMS, is a challenging task. This is due to the fact that there are a limited number of laboratories performing the configuration and the process involves a team of clinicians handpicking features from medical databases, each within their area of expertise, such as patient behavior, microscopic analysis, microbiology cultures, tissue samples, laboratory reporting, etc. The manual configuration process can result in errors such as omitted critical features and partial satisfaction of constraints between features. The LIMS provides different modules for managing data across various stages of a medical test, including workflow management, scheduling, sample analysis, and reporting. Some examples of this scenario are listed next:
• The biological features in an LDT are selected incorrectly, e.g., diet and obesity features are included in a microscopic examination.
• A genetic test is applied to subjects of an incorrect age group.
• The value of a particular feature (e.g., hypercalcemia) is not noted during testing.
• There are no features associated with any standard template for reporting, possibly leading to the omission of important information or incorrect communication of LDT results.
The configuration of LDTs becomes complex when involving medical experts from various specialties. Each expert may select features within their area of expertise without considering the significance of features from other domains, leading to inconsistent LDT configurations that can result in improper data analysis and unacceptable clinical outcomes. However, to provide appropriate patient care, medical experts require the ability to configure LDTs. Despite the challenges posed by the configuration of complex LDTs, regulatory agencies such as DRAP have not yet established regulations for LDTs in Pakistan. In the absence of such regulations, the manual configuration of LDTs by medical experts using LIMS-based laboratories remains the only solution, particularly for rare diseases such as Diastrophic Dysplasia, which affects a large number of people globally, including children. Thus, resolving the issues associated with the configuration of LDTs becomes increasingly important.
In the context of this discussion, LDT designing is proposed as a feature selection problem along with automated configuration support (detailed in Section 4).
4 Theoretical Framework and Methodology
In this section, first, we discuss the LDT configuration as a feature selection problem and the framework of opt-LDT. The GA objective function’s formalization and the chromosome encoding scheme are presented next.
4.1 LDT Configuration as a Feature Selection Problem
In this subsection, the LDT design process is described in detail. An LDT is configured by selecting features from the available medical repository. A team of medical practitioners has participated in the configuration process. Each practitioner configures their particular field’s features and adds these to the LDT. Finally, these configured features produce a new LDT design. This integration can lead to several issues, notably inconsistencies across test features due to the non-satisfaction of constraints between features. For instance, consider that practitioner A adds feature P1, and practitioner B adds feature P2, and medical validation sources oppose the existence of P1 and P2 in a single LDT configuration. It is also possible that practitioner A adds P1 to the LDT and forgets to add feature P3, while medical regulations dictate that the interpretation of P1 is incomplete without the analysis of P3. Our research identified five types of constraints for an LDT configuration: mandatory, optional, exclude, include, and alternative. We provide examples of these constraints next (most of these examples are taken from [21]):
• Mandatory: Feature presence is compulsory in an LDT configuration. For instance, for molecular genetic testing for heritable diseases and conditions, the test method’s intended use, the test’s purpose, and the test method used are features with mandatory constraints. Some of CLIA’s mandatory attributes for a medical lab that performs non-waived (high and moderate complexity test) testing are: test requester identifier, patient unique identifier, and sex [22].
• Optional: Feature presence is optional in an LDT configuration; for instance, additional information is required for the specific test, the patient’s place of birth, date/time of the test, and patient racial details (for some tests, it is mandatory).
• OR: An LDT configuration can have a feature(s) from an OR-ed feature set, for instance, test requester name OR any suitable unique identifier, patient name OR patient identifier, and date of birth OR age.
• Alternative: An LDT configuration can contain only a single feature from a set of alternative features. For instance, the type of specimens retained (whole blood and DNA samples), test permission levels (lab director, supervisor, and technician), and type of testing (diagnostic, pre-symptomatic, predictive, carrier, susceptibility) are examples of alternative features.
• Include: F1 includes F2 means the existence of F1 in an LDT configuration implies the inclusion of F2 in the configuration, such as the patient address has street, city, and country.
If all constraints are not satisfied, the LDT design remains inconsistent, i.e., erroneous, unreliable, and sometimes invalid. Typically, practitioners consult with each other and try to resolve all constraints. It is also complicated and requires comprehensive analytical skills. Most of the time, it is impossible to resolve all constraints through manual deliberation. According to the U.S. Food & Drug Administration (FDA), this poses a severe threat to health care because patients’ health decisions are based on inconsistent LDT results.
4.2 Opt-LDT: An Optimized Laboratory-Developed Test Prototype
This subsection presents our proposed solution to the problems mentioned in Section 4.1. We label it opt-LDT (optimized-LDT), an independent module to configure LDTs. It can be easily integrated with existing LIMS because it does not require any dependent working modules. It is already discussed that the opt-LDT model of the LDT design process is a feature selection problem that has to comply with the given set of constraints. Violations of these constraints during the feature selection lead to an inconsistent LDT configuration. A natural solution to resolve inconsistencies is to automate the LDT configuration process. Moreover, generating an optimized configuration that models as a feature selection problem is not a new domain for the software industry. Many solutions are proposed to solve the problem, specifically Artificial Intelligence (AI) techniques such as optimization based on evolutionary computing, logic-based and ontological reasoning, swarm intelligence, and predictive analytics [23].
GA is a standard evolutionary computing algorithm with many successful applications [15], also widely employed to solve inconsistency issues in product configuration. Based on the existing literature, a GA-based opt-LDT solution is proposed to configure LDT feature selection and efficiently resolve inconsistencies to generate a consistent configuration that selects the optimal LDT feature subset.
To address our research inquiries, we employed the opt-LDT approach to enhance the previously unoptimized manual LDT configurations from a local hospital. In this paper, under non-disclosure agreement requirements, we will only show the more generic features of these configurations, such as PatientName and PatientAddress (a partial list is shown in Table 1), and avoid the more critical (disease-related) ones. In our hospital, a group of ten practitioners from three different clinical fields configured the LDT in two days. Initially, a discussion was conducted with practitioners to discuss and validate the LDT issues. As mentioned before, the clinicians concurred on the difficulty of selecting the feature subset; extended manual deliberation led them to make some compromises on feature constraints, and hence, they agreed that their final LDT product was inconsistent. The clinicians also did not give any guarantee regarding the health safety of their LDT for the patients.
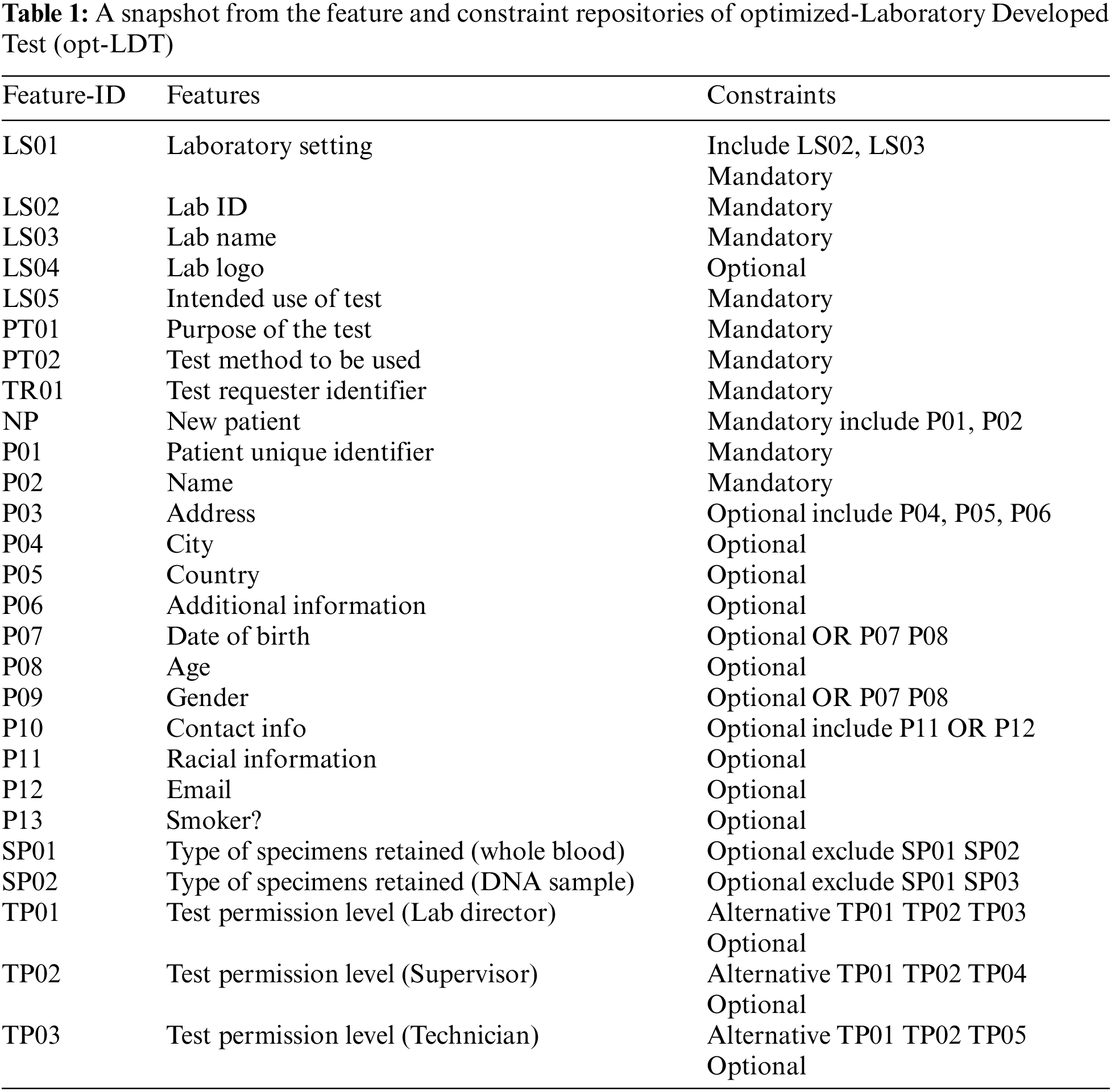
To solve this problem, we employed an opt-LDT for a hospital. The architecture of opt-LDT is shown in Fig. 1. opt-LDT primarily divides into two parts, i.e., repositories and GA-Engine. The Clinical Data Repository (CDR) stores all relevant medical data required to configure an LDT. It validates medical data from various standard medical sources. It also saves the potential features for LDT configurations. This feature set can be updated through reviews and feedback from clinicians and practitioners, research activities, or synchronization with other medical repositories. Two sub-repositories are populated from CDR, i.e., Constraint Repository (CR) and Features Repository (FR). For a particular LDT configuration, FR collects potential features such as Test Requester Identifier and Patient gender. At the same time, CR contains the applicable constraints on selected features, e.g., mandatory (Test Requester Identifier) and optional (Patient gender). FR stores 100 potential features for our experiments, and CR has 78 constraints. Both storages share an interface to help practitioners in an LDT design configuration stored in the LDT Design Configuration (LDC) repository.

Figure 1: Optimized-Laboratory Developed Test (opt-LDT)
Table 1 presents a subset of 27 potential features and constraints from FR and CR. Here, F-ID, Features, and Constraints represent the identifier, feature description, and applicable constraints. As already mentioned, the configuration of a complex LDT is a collaborative task involving multiple clinicians. Every clinician configures features of their domain which are stored in LDC. A clinician uses a unique credential (password and user name) to access the interface, which helps track the source dependency of the added test feature. After clinicians complete the LDT configuration, opt-LDT starts working by fetching the configuration from LDC. An objective function defined in Section 4.3 encodes the LDT design as an optimization problem.
Moreover, LDT configuration is mapped into a chromosomal representation (detailed in Section 4.3), generating the initial population with multiple candidates. Hyperparameters such as population size, cross-over function, cross-over fraction, and mutation rate are tuned. The chromosomal representation of the given LDT configuration with the optimal adjusted values of the hyperparameters is passed to the GA-Engine for final optimization. GA-Engine starts with generating the population of candidates with the tuned value of population size. The two fittest LDT configurations evaluated by the objective function are selected to reproduce further. Genetic operators, i.e., cross-over and mutation, are applied to these selected configurations to produce the next generation of offspring. The process repeats until one of the stopping criteria is met, such as an LDT configuration is acquired that satisfies the minimum function criteria, the maximum number of generations are completed, and the highest ranking LDT configuration fitness is met.
4.3 Formalizing GA Objective Function and Chromosome Encoding
GA objective function is centered on two activities: 1) maximizing the number of features selection and 2) prioritizing different types of inconsistencies. To understand the first activity, suppose an inconsistent LDT configuration is given as Conf = {LS01, …, TP01, TP02, …} (defined earlier). The simplest way to remove inconsistencies is to remove both inconsistent features, i.e., TP01 and TP02. However, such exclusion is illogical and could leave a small set of available features to complete the configuration. To prevent this in opt-LDT, we design the objective function to maximize feature selection and minimize inconsistencies (described next).
The second activity is to give weights to various inconsistencies and represent the real-world scenario in which clinicians give importance to those constraints’ violations that have more significance. We target four inconsistencies, i.e., exclude, include, alternative and mandatory. They are arranged based on the decreasing severity level, i.e., Mandatory, followed by Exclude, Alternative, and Include. Later, we normalize the weight values from 0–1. We randomly assign a weight of 0.4 to a mandatory and 0.3 to each of the alternatives, excluding or including violations. We did not assign weights to Optional and OR constraints because their selection does not introduce any inconsistencies.
The mathematical formalization of our objective function is as follows:
Suppose,
Þ = An inconsistent LDT design configuration
F = Potential feature set {F1, F2, …, Fn} configured in Þ
C = A set of constraints {C1, C2, …, Cn} applicable on Fi of Þ
W = Weights [0, 1] given to constraints Cn ∈ C and
I = {w1C1F1, w2C2F2, …, wjCjFj} represents the j inconsistencies, where ωk ∈ W and j are not always the same as n. Eq. (1) illustrates the maximization of selected features:
Also, the configured product should minimize constraints in C such that:
The given inconsistent LDT configuration is encoded as a chromosome. We assume this configuration has n features. We map these features to a chromosome with n genes. Bit strings (0 and 1) are used for encoding, where 1 shows the selection, and 0 shows the de-selection of a configured feature.
To select potentially useful candidates from a population, we use the standard stochastic uniform selection, which employs unbiased and proportionate selection with a minimal spread. It begins with the ordering of the population according to the fitness value. In each iteration, two individuals are selected, and crossover and mutation are applied to acquire the next set of chromosomes. We also use the typical GA replacement approach, which replaces a parent with its offspring in case the offspring has a higher fitness score than the parent. Otherwise, the candidate with the smallest fitness value in the population is replaced.
5 Dataset, Experiments, and Results
This section presents a detailed description of the dataset, experiments, and results. We performed these test experiments on a machine with Windows 7 OS, Intel Core i7 2.4 GHz processor, and 16 GB RAM.
5.1 Generating Inconsistent LDT Configurations Dataset
To test the proposed prototype, opt-LDT, we need an inconsistent LDT design configuration. We created ten randomized, inconsistent LDT configurations using the feature set outlined in Table 1. The configurations are required to run the GA experiments. We named them LDT-C01 to LDT-C10. We wrote Java code to analyze inconsistent configurations. We also counted the number of features and inconsistencies in the given configuration. Our main objective was to reduce inconsistencies, so we chose three configurations with the largest number of inconsistencies, namely LDT-C07 (74), LDT-C08 (70), and LDT-C06 (69). Additionally, to test the effectiveness of opt-LDT on a feature set with fewer inconsistencies, we included LDT-C02 (55). This also provided the opportunity to initialize the GA algorithm with input from clinicians. Table 2 shows details of these configurations, specifically the missing mandatory features, including the missing features and alternative/Exclude features that cause inconsistencies.
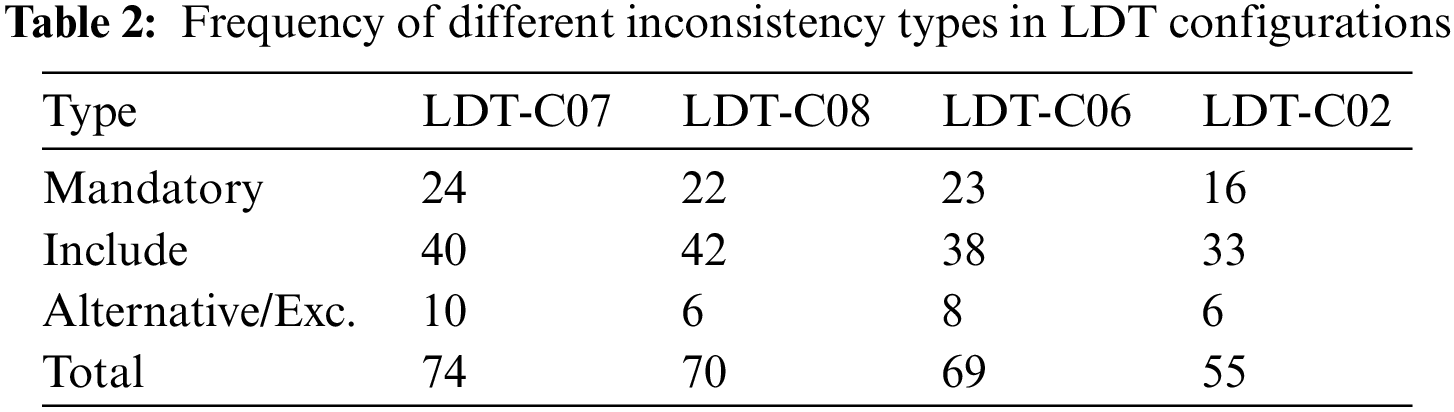
To give some detail about the inconsistencies, the relevant features related to these inconsistency types for LDT-C07 are shown next:
• Missing Mandatory Features: The mandatory features such as the intended use of the test, the purpose of the test, the test method to be used, the test requester identifier, the patient unique identifier, and sex are not selected in LDT-C07.
• Alternative/Exclude Features: The values ‘Whole blood’ and ‘DNA’ samples are both selected for the feature type of specimens retained, which introduces an exclude inconsistency because whole blood and DNA cannot co-exist in a consistent LDT configuration. Similarly, LDT-C07 contains both ‘lab director’ and ‘supervisor’ level permissions, which causes an alternative inconsistency.
• Missing Include Features: The feature Patient address is selected without ‘street,’ ‘city,’ and ‘country’ selection, which introduces an include inconsistency.
5.2.1 GA Parameters’ Tuning Results
We tune four GA parameters as shown in Table 3, on a selected inconsistent configuration. To determine the optimal values for the parameters, we employed a conventional method of parameter tuning by testing and evaluating various values prior to the actual run [24]. We arbitrarily chose LDT-C07 for this purpose. The experiments were repeated ten times to determine the best average value for each parameter setting [23], as shown in Table 3. We explored LDT-C07 using three standard mutation rates (0.01, 0.05, and 0.1) to identify the optimal mutation rate. These mutation rates were selected based on the rationale presented in [25]. With a mutation rate of 0.01, GA selects 55% of features and removes 90.4% of the inconsistencies, on average. With a 0.05 mutation rate, GA selected 61% of features and resolved 90.9% of the inconsistencies. However, a 0.1 value for mutation rate generates the optimal results, selects 87% of features, and resolves 91.9% of the inconsistencies.
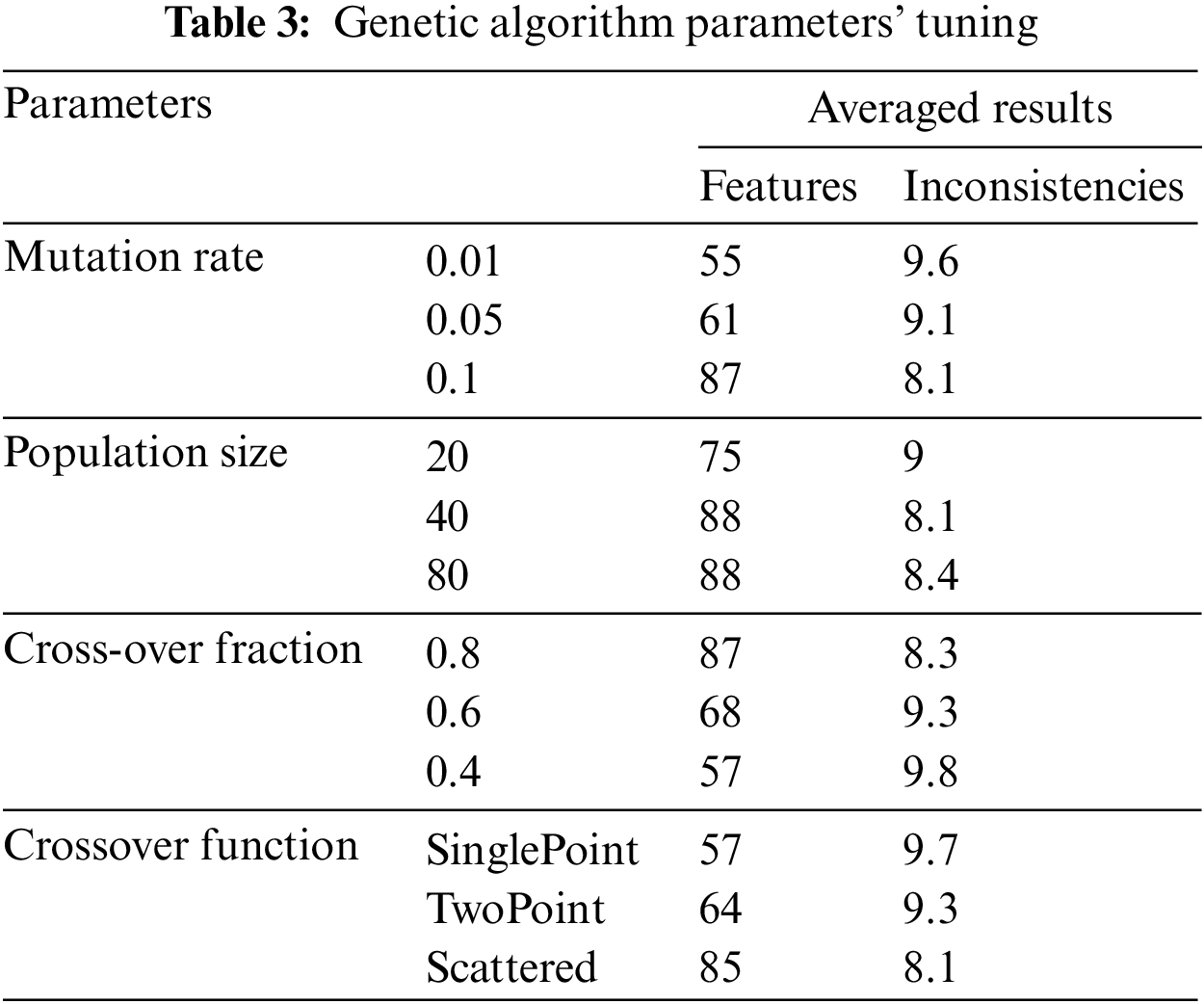
To find the optimal population size, we began with a low value and gradually increased it until the best value was found [26,27]. We experimented with LDT-C07 with small (20 features), medium (40 features), and large (80 features) population sizes. We determined these feature sets concerning the opt-LDT feature set. With a small population, GA selects 75% of features and resolves 91% of inconsistencies. On the other hand, for a large population, it selects 88% of features and resolves 91.6% of the inconsistencies. The optimal is a medium population that selects 88% of features and removes 91.9% of the inconsistencies.
Typical cross-over values lie between 0.5 and 1 [28]. However, 1 is exclusive [29]. In our experiments, we chose crossover fractions as 0.4, 0.6, and 0.8. GA selects 57% of features and removes 90.2% of inconsistencies on average, and with a 0.6 crossover fraction, GA selects 68% of features and resolves 90.7% of inconsistencies. The crossover fraction’s optimal value is 0.8, which selects 87% of features and resolves 91.7% of inconsistencies. Finally, we tested LDT-C07 with three standard functions for the crossover function, i.e., single-point, two-point, and scattered [30]. The single-point selects 57% of the features and removes 90.1% of inconsistencies, while the two-point selects 64% of the features and can remove 90.7% of inconsistencies. The scattered crossover function produces the optimal results, with 85% of the features and 91.9% consistent LDT configuration.
After running the GA parameter tuning experiments, we obtained the optimal values for mutation rate (0.1), crossover fraction (0.8), crossover function (scattered), and population size (medium-sized). We use these optimal values to run the final section of experiments.
5.2.2 Optimizing the Inconsistent LDT Configuration
After tuning the parameters, selected configurations are experimented with opt-LDT to obtain the optimization results and answer the research questions Q1 and Q2. Here, we optimize four inconsistent LDT configurations, i.e., LDT-C06, LDT-C07, LDT-C08, and LDT-C02. opt-LDT applies GA with the optimal parameter values, i.e., medium-sized (40) population, scattered crossover function, 0.1 mutation rate, and 0.8 crossover fraction. GA experiments are run ten times for each configuration to find an optimal, consistent configuration.
Initially, we answer the research question Q1, i.e., does GA have the potential to produce a consistent (conflicts-free) LDT configuration? For this, opt-LDT should maximize the number of selected features and minimize the number of inconsistencies in each configuration. Table 4 presents the results of the selection of potential features. Here, InitFtr and opt-LDTFtrSet represent the unoptimized and optimized (averaged over ten runs) feature set size, respectively, while %Incr represents the percentage increase. Our results show that opt-LDT significantly increases the number of features in each configuration by more than 100% in LDT-C07, LDT-C08, and LDT-C02, and approximately 50% in LDT-C06.
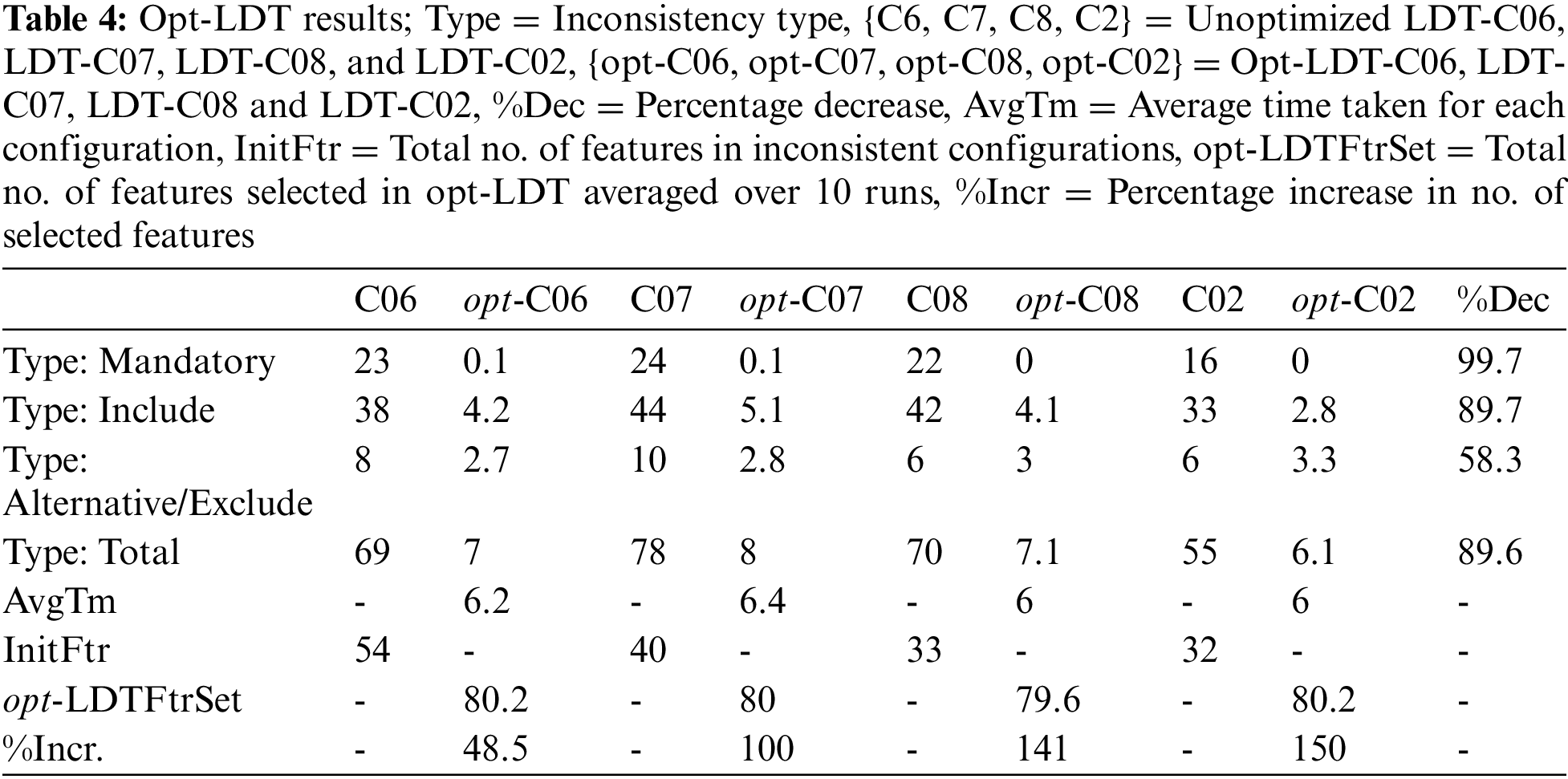
Table 4 also presents the results for minimizing inconsistencies. We observe considerable minimization in the number of inconsistencies for each inconsistency type in each configuration. The mandatory constraints are completely resolved, followed by the include constraints (90%) and alternative/exclude constraints (58.3%). Overall, opt-LDT can collectively resolve around 90% of constraints in all four configurations. Based on these results, we conclude that GA-based optimization can generate consistent LDT configurations that maximize the selected features by 100% and resolve almost 90% of constraints. These results are independent of the frequency of available features and inconsistencies in the initial unoptimized configurations.
A graphical representation of these results is shown in Fig. 2 for LDT-C06, LDT-C07, LDT-C08, and LDT-C02. In all graphs, R1–R10 show the runs of GA experiments. The upper solid black line illustrates the number of features chosen in the optimal configuration for each iteration, while the lower double line indicates the inconsistencies that were not resolved in the generated optimal configuration. The number of selected features in each iteration ranges from 72 to 86, surpassing the original feature set size of each configuration. Additionally, the number of unresolved inconsistencies ranges from 3 to 15, as compared to the 55–70 inconsistencies in the initial configurations. Finally, each run’s optimized values’ deviation is insignificant for both lines in each graph, indicating consistency in opt-LDT performance.

Figure 2: Opt-LDT results
We now answer research question Q2, i.e., is GA more efficient than a manual LDT configuration? For this, opt-LDT should minimize the time required to generate a consistent configuration. Table 4 (AvgTm) shows the time needed to produce an optimal, consistent configuration. One can observe that the average time to obtain an optimized configuration varies between 6 and 6.5 s. These times are negligible compared to a manual LDT configuration, which can easily extend over several days. Hence, GA technology is considerably efficient compared to the manual effort of LDT designers.
We statistically evaluate the optimization results to check the working of opt-LDT on all given LDT configurations. Table 5 presents the primary statistics; we are not expounding the table here because it is self-explanatory.
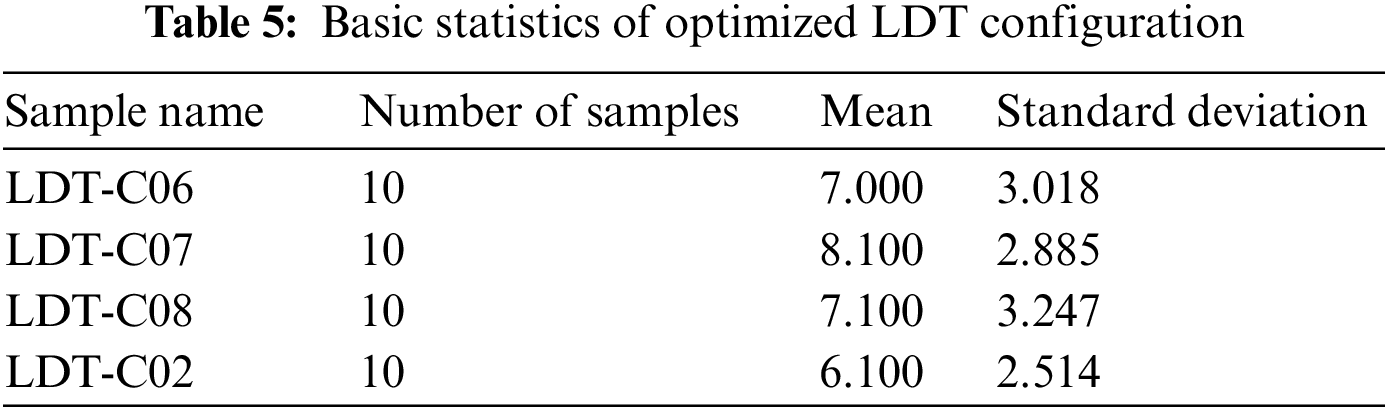
A t-test is used to determine if there is a statistically significant difference between the optimization results of the given configurations or opt-LDT optimized the given configurations with different/any number of inconsistencies. Our alternative hypothesis to test was “opt-LDT optimizes the all given configuration by resolving the existing inconsistencies.”
Table 6 presents the test summary based on a p-value threshold of 0.05. There is not a statistically significant difference between: {LDT-C06, LDT-C07, LDT-C08, and LDT-C02}, so we reject the null hypothesis and accept the alternative one that opt-LDT optimizes the given inconsistent configuration with any number of inconsistencies.
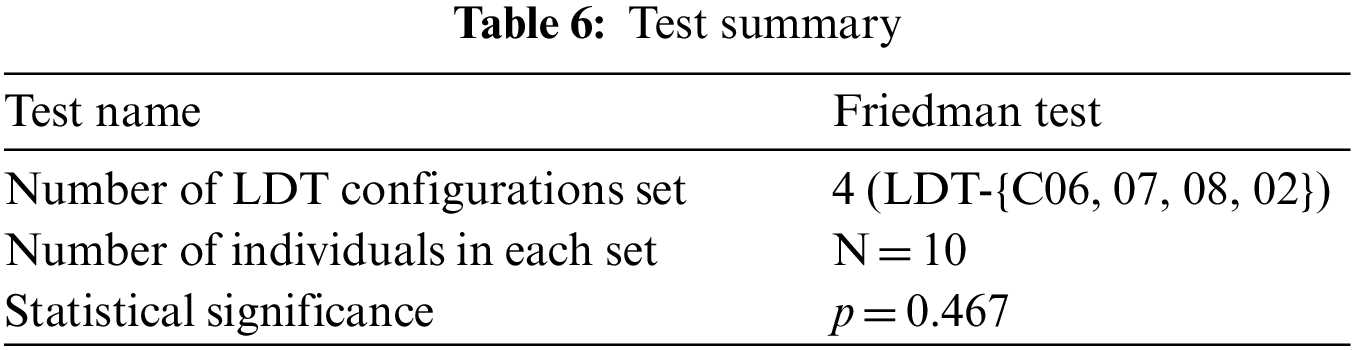
6 Evaluation, Practice Implication, and Limitations
Questionnaires collect information about participants’ knowledge and beliefs on a particular domain [31]. Similarly, we used it to analyze the different acceptability dynamics of opt-LDT by presenting the opt-LDT configurations for LDT-C06, LDT-C07, LDT-C08, and LDT-C02 to the practitioners for analysis and acquiring their feedback on these results through a subjective questionnaire, as shown in Table 7.
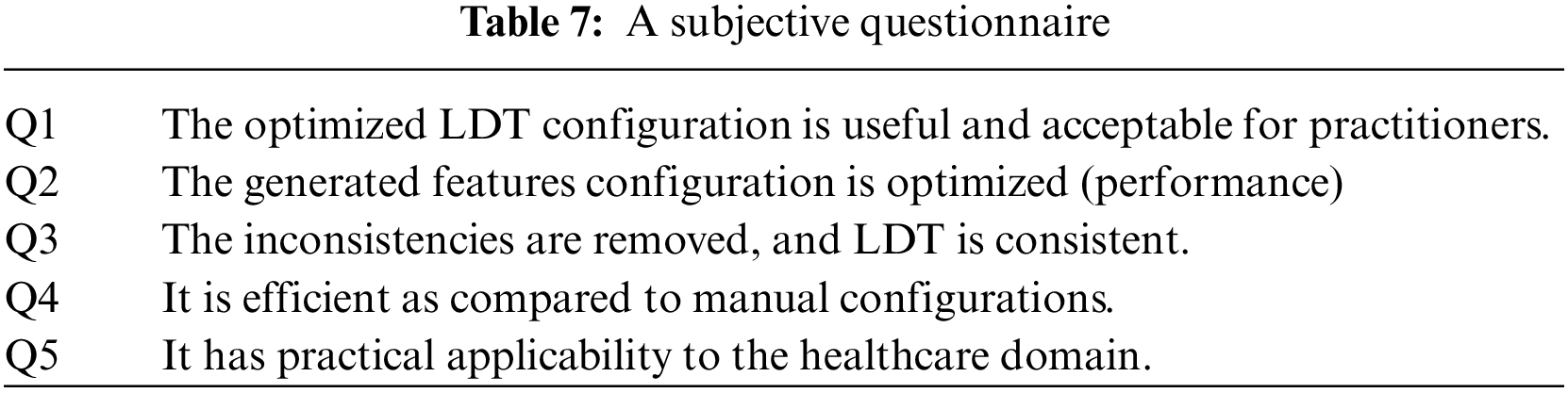
We designed this questionnaire according to the standard guidelines for questionnaire design [32]. This feedback was acquired anonymously from clinicians. The questionnaire is divided into four sections, each for feedback on LDT-C06, LDT-C07, LDT-C08, and LDT-C02. Each section has five questions about the results obtained through opt-LDT, marked on a scale of 1 (strongly disagree) to 5 (strongly agree). We circulated the questionnaire to our ten clinicians and calculated each question’s average response for each of our four configurations. As might have been expected, Q4 received an average rating of 5 for each configuration, i.e., all clinicians unanimously concurred (strongly agreed) on the efficiency of opt-LDT compared to their manual effort. The best subjective response is obtained for LDT-C02, with all questions getting an average score > 4.5 (average response is 4.8). The apparent reason for this is that, amongst all the configurations, the clinicians best understood the one they had designed, i.e., LDT-C02, compared to the other nine random configurations. Notwithstanding this, responses for Q1-Q5 for different configurations are all >3.6, with an average response of >4.1. This indicates the acceptability and concurrence of clinicians for the optimized opt-LDT results. Interestingly, after Q4, both Q1 and Q3 received the same average agreement rating of 4.3, indicating that clinicians unanimously concurred that opt-LDT resolves inconsistencies and produces optimized LDTs. The average rating for Q5 was also 4, indicating that clinicians are convinced of the practicality of opt-LDT. Furthermore, the work done in this paper will be submitted to DRAP for regulatory approval. At this point, we cannot predict the exact time frame for this activity. Considering these results, opt-LDT can significantly impact the LDT design and clinical validity process. Moreover, our work aptly addresses the LDT designers’ challenges and DRA’s concerns about LDTs.
Although opt-LDT is tested and validated in a real-time clinical environment, it still needs more testing to validate the working of opt-LDT with LDTs of different dynamics and complexities. Moreover, opt-LDT does not fully automate the encoding of the given LDT problem in a chromosome which is a tedious task, a partially automated prototype is used for problem encoding in this research work.
The LDTs are configured by selecting feature constraints from medical repositories available in existing LIMS. This configuration process is done manually due to the unavailability of automated support, which is time-consuming and prone to human errors. It leads to inconsistent design configurations in which standard constraints remain to be solved, and essential features could remain unselected. In this paper, we proposed opt-LDT, an optimization-based framework to automate and manage the LDT configuration, which can be easily integrated with existing LIMS and help the clinician generate a consistent LDT configuration. To validate the acceptability of opt-LDT, we implemented and tested it in a real-time healthcare setup. The results show that opt-LDT resolved 90% of the inconsistencies in 6.5 s and generated a consistent LDT configuration. The practicability and usefulness of the generated LDT configurations are also validated through a subjective questionnaire. Considering these results, opt-LDT can significantly impact the LDT design and clinical validity process. Moreover, these results will be submitted to DRAP for regulatory approval. This regularization will help LDT designers in solving the LDT configuration issues.
In the future, swarm intelligence algorithms could be implemented, including ant colony optimization and particle swarm optimization. Furthermore, the application of the proposed technique in futuristic healthcare applications based on 6G could also be studied [33]. The researchers can also study a link between LDT and Explainable AI [34].
Acknowledgement: The researchers would like to thank the Deanship of Scientific Research, Qassim University for funding the publication of this project.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. J. A. Lieberman, G. Pepper, S. N. Naccache, M. -L. Huang, K. R. Jerome et al., “Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories,” Journal of Clinical Microbiology, vol. 58, no. 8, pp. 550–576, 2020. [Google Scholar]
2. J. R. Genzen, J. S. Mohlman, J. L. Lynch, M. W. Squires and R. L. Weiss, “Laboratory-developed tests: A legislative and regulatory review,” Clinical Chemistry, vol. 63, no. 10, pp. 1575–1584, 2017. [Google Scholar] [PubMed]
3. A. E. Clark, J. Levy and F. M. Lee, “Laboratory-developed test regulation and the immunocompromised patient: Uncertainty ahead,” Current Opinion in Infectious Diseases, vol. 33, no. 4, pp. 304–311, 2020. [Google Scholar] [PubMed]
4. FDA, Laboratory Developed Tests, 2018. [Online]. Available: https://www.fda.gov/medical-devices/vitro-diagnostics/laboratory-developed-tests [Google Scholar]
5. V. Lapin, L. C. Mighion, C. P. da Silva, Y. Cuperus, L. J. H. Bean et al., “Regulating whole exome sequencing as a diagnostic test,” Human Genetics, vol. 135, no. 6, pp. 655–673, 2016. [Google Scholar] [PubMed]
6. A. K. Sarata and J. A. Johnson, “Regulation of clinical tests: In vitro diagnostic (IVD) devices, laboratory developed tests (LDTsand genetic tests,” Congressional Research Service–Technical Report, 2014. [Online]. Available: https://sgp.fas.org/crs/misc/R43438.pdf [Google Scholar]
7. LDTSolutions, Roche, 2022. [Online]. Available: http://ldtsolutions.roche.com/flipbook/index.html [Google Scholar]
8. P. J. Howanitz, “Errors in laboratory medicine: Practical lessons to improve patient safety,” Archives of Pathology and Laboratory Medicine, vol. 129, no. 10, pp. 1252–1261, 2005. [Google Scholar] [PubMed]
9. R. Fukuda and F. Takada, “Legal regulations on health-related direct-to-consumer genetic testing in 11 countries,” Kitasato Medical Journal, vol. 48, pp. 52–59, 2018. [Google Scholar]
10. DAWN, Unique Disease in Pakistan, 2016. [Online]. Available: http://www.dawn.com/news/1017290 [Google Scholar]
11. Thermo Scientific, “Confidence in results with data integrity,” [Online]. Available: https://assets.thermofisher.com/TFS-Assets/CMD/brochures/BR-64057-LC-MS-ClinQuan-MD-BR64057-EN.pdf [Google Scholar]
12. DRA, Drug regulatory authority of Pakistan, 2022. [Online]. Available: http://www.dra.gov.pk [Google Scholar]
13. C. Paszko and C. Turner, Laboratory Information Management Systems, Boca Raton, FL, USA: CRC Press, 2018. [Google Scholar]
14. D. O. Skobelev, T. M. Zaytseva, A. D. Kozlov, V. L. Perepelitsa and A. S. Makarova, “Laboratory information management systems in the work of the analytic laboratory,” Measurement Techniques, vol. 53, no. 10, pp. 1182–1189, 2011. [Google Scholar]
15. S. Katoch, S. S. Chauhan and V. Kumar, “A review on the genetic algorithm: Past, present, and future,” Multimedia Tools and Applications, vol. 80, pp. 8091–8126, 2021. [Google Scholar] [PubMed]
16. S. Mirjalili, “Genetic algorithm,” in Evolutionary Algorithms and Neural Networks, 1st ed., Cham, Switzerland: Springer, pp. 43–55, 2019. [Online]. Available: https://doi.org/10.1007/978-3-319-93025-1_4 [Google Scholar]
17. M. -C. Wu, C. -S. Lin, C. -H. Lin and C. -F. Chen, “Effects of different chromosome representations in developing genetic algorithms to solve DFJS scheduling problems,” Computers & Operations Research, vol. 80, pp. 101–112, 2017. [Google Scholar]
18. A. Hassanat, K. Almohammadi, E. Alkafaween, E. Abunawas, A. Hammouri et al., “Choosing mutation and crossover ratios for genetic algorithms—A review with a new dynamic approach,” Information, vol. 10, no. 12, pp. 1–36, 2019. [Google Scholar]
19. S. Mirjalili, J. S. Dong, A. S. Sadiq and H. Faris, “Genetic algorithm: Theory, literature review, and application in image reconstruction,” in Nature-Inspired Optimizers, Switzerland: Springer, pp. 69–85, 2020. [Online]. Available: https://doi.org/10.1007/978-3-030-12127-3_5 [Google Scholar]
20. U. S. Food & Drug Administration, “Overview of IVD regulation,” 2021. [Online]. Available: https://www.fda.gov/medical-devices/ivd-regulatory-assistance/overview-ivd-regulation [Google Scholar]
21. B. Chen, M. C. Gagnon, S. Shahangian, N. L. Anderson, D. A. Howerton et al., “Good laboratory practices for molecular genetic testing for heritable diseases and conditions,” MMWR Recommendations and Reports, vol. 58, no. RR-6, pp. 1–37, 2009. [Google Scholar] [PubMed]
22. LII, 42 CFR 493.1241–Standard: Test request, 2022. [Online]. Available: https://www.law.cornell.edu/cfr/text/42/493.1241 [Google Scholar]
23. U. Afzal, T. Mahmood, A. H. Khan, S. Jan, R. U. Rasool et al., “Feature selection optimization in software product lines,” IEEE Access, vol. 8, pp. 160231–160250, 2020. [Google Scholar]
24. H. Chung and K. -s. Shin, “Genetic algorithm-optimized long short-term memory network for stock market prediction,” Sustainability, vol. 10, no. 10, pp. 3765, 2018. [Google Scholar]
25. C. D. Lin, C. M. Anderson-Cook, M. S. Hamada, L. M. Moore and R. R. Sitter, “Using genetic algorithms to design experiments: A review,” Quality and Reliability Engineering International, vol. 31, no. 2, pp. 155–167, 2015. [Google Scholar]
26. Y. Deng, Y. Liu and D. Zhou, “An improved genetic algorithm with initial population strategy for symmetric TSP,” Mathematical Problems in Engineering, vol. 2015, Article ID 212794, 2015. [Google Scholar]
27. O. Roeva, S. Fidanova and M. Paprzycki, “Population size influence on the genetic and ant algorithms performance in case of cultivation process modeling,” in Recent Advances in Computational Optimization, Proc. 6th Workshop on Computational Optimization (WCO), Kraków, Poland, pp. 107–120, 2015. [Google Scholar]
28. A. Hassanat, K. Almohammadi, E. Alkafaween, E. Abunawas, A. Hammouri et al., “Choosing mutation and crossover ratios for genetic algorithms—A review with a new dynamic approach,” Information, vol. 10, no. 12, pp. 390, 2019. [Google Scholar]
29. Mathworks, “Vary mutation and crossover,” 2016. [Online]. Available: https://www.mathworks.com/help/gads/vary-mutation-and-crossover.html [Google Scholar]
30. S. Katoch, S. S. Chauhan and V. Kumar, “A review on genetic algorithm: Past, present, and future,” Multimedia Tools and Applications, vol. 80, pp. 8091–8126, 2021. [Google Scholar] [PubMed]
31. M. Schrepp and J. Thomaschewski. “Design and validation of a framework for the creation of user experience questionnaires,” International Journal of Interactive Multimedia and Artificial Intelligence, vol. 5, no. 7, pp. 88–95, 2019. [Google Scholar]
32. I. Brace, Questionnaire Design: How to Plan, Structure and Write Survey Material for Effective Market Research, 4th ed., London, UK: Kogan Page, 2018. [Google Scholar]
33. P. N. Srinivasu, M. F. Ijaz, J. Shafi, M. Woźniak and R. Sujatha, “6G driven fast computational networking framework for healthcare applications,” IEEE Access, vol. 10, pp. 94235–94248, 2022. [Google Scholar]
34. P. N. Srinivasu, N. Sandhya, R. H. Jhaveri and R. Raut, “From blackbox to explainable AI in healthcare: Existing tools and case studies,” Mobile Information Systems, vol. 2022, Article ID 8167821, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools