 Open Access
Open Access
ARTICLE
A Deep Learning Framework for Mass-Forming Chronic Pancreatitis and Pancreatic Ductal Adenocarcinoma Classification Based on Magnetic Resonance Imaging
1 Faculty of Information Engineering and Automation, Kunming University of Science and Technology, Kunming, 650504, China
2 Department of Radiology, Second Affiliated Hospital of Kunming Medical University, Kunming, 650101, China
3 School of Physics and Electronic Engineering, Yuxi Normal University, Yuxi, 653100, China
* Corresponding Authors: Jingang Hao. Email: ; Jianfeng He. Email:
(This article belongs to the Special Issue: Deep Learning in Computer-Aided Diagnosis Based on Medical Image)
Computers, Materials & Continua 2024, 79(1), 409-427. https://doi.org/10.32604/cmc.2024.048507
Received 10 December 2023; Accepted 11 February 2024; Issue published 25 April 2024
Abstract
Pancreatic diseases, including mass-forming chronic pancreatitis (MFCP) and pancreatic ductal adenocarcinoma (PDAC), present with similar imaging features, leading to diagnostic complexities. Deep Learning (DL) methods have been shown to perform well on diagnostic tasks. Existing DL pancreatic lesion diagnosis studies based on Magnetic Resonance Imaging (MRI) utilize the prior information to guide models to focus on the lesion region. However, over-reliance on prior information may ignore the background information that is helpful for diagnosis. This study verifies the diagnostic significance of the background information using a clinical dataset. Consequently, the Prior Difference Guidance Network (PDGNet) is proposed, merging decoupled lesion and background information via the Prior Normalization Fusion (PNF) strategy and the Feature Difference Guidance (FDG) module, to direct the model to concentrate on beneficial regions for diagnosis. Extensive experiments in the clinical dataset demonstrate that the proposed method achieves promising diagnosis performance: PDGNets based on conventional networks record an ACC (Accuracy) and AUC (Area Under the Curve) of 87.50% and 89.98%, marking improvements of 8.19% and 7.64% over the prior-free benchmark. Compared to lesion-focused benchmarks, the uplift is 6.14% and 6.02%. PDGNets based on advanced networks reach an ACC and AUC of 89.77% and 92.80%. The study underscores the potential of harnessing background information in medical image diagnosis, suggesting a more holistic view for future research.Keywords
Accurate differentiation between Mass-Forming Chronic Pancreatitis (MFCP) and Pancreatic Ductal Adenocarcinoma (PDAC) is crucial in clinical practice due to the substantial differences in treatment approaches and prognoses [1]. Both subtypes have similar features in various medical imaging modalities, presenting as localized pancreatic masses [2]. This similarity increases the risk of misdiagnosis [3]. For instance, some studies indicate that approximately 5% to 15% of pancreatitis is diagnosed as pancreatic cancer [4]. Accurate preoperative diagnosis is crucial for distinguishing MFCP from PDAC [5].
Radiologists accurately differentiate MFCP and PDAC without invasive procedures, basing their judgments on extensive experience and comprehensive references of multimodal data in the preoperative period. It is time-consuming and makes it impossible to ensure stable diagnosis in clinical practice. The application of deep learning in medical image analysis provides a solution to improve the accuracy and efficiency of diagnosis. There are three main research directions for deep learning-based image diagnosis of pancreatic lesions: 1) prior-free end-to-end diagnostic networks, 2) prior-injected cascade diagnostic networks, and 3) prior-injected parallel diagnostic networks.
Prior-free end-to-end diagnostic networks use original images as the training set for the diagnostic model, as shown in Fig. 1a. For example, Ziegelmayer et al. [6] used the VGG-19 [7] architecture, pre-trained on ImageNet [8], to accomplish the task of feature extraction and diagnostic differentiation between autoimmune pancreatitis (AIP) and PDAC. Such studies required large-scale datasets and more complex network structures to avoid the interference of redundant information. Notably, the relatively small percentage of the pancreatic lesion region in the image presents a challenge for prior-free networks, making capturing detailed information difficult.

Figure 1: Existing deep learning-based diagnostic frameworks for pancreatic lesions. (a) the prior-free diagnostic network, (b) the prior-injected diagnostic network, and (c) the prior difference guidance network (ours)
Prior-injected cascade diagnostic networks use a segmentation or detection model to identify the lesion region in original images, are used as the training set for the diagnostic model, as shown in Fig. 1b. For example, Si et al. [9] used a full end-to-end deep learning approach that consists of four stages: Image screening, pancreas localization, pancreas segmentation, and pancreas tumor diagnosis. Qu et al. [10] first reconstructed the pancreas region through anatomically-guided shape normalization, then used an instance-level contrast learning and balance adjustment strategy for the early diagnosis of pancreatic cancer. Li et al. [11] designed a multiple-instance-learning framework to extract fine-grained pancreatic tumor features, followed by an adaptive-metric graph neural network and causal contrastive mechanism for early diagnosis of pancreatic cancer. Chen et al. [12] designed a dual-transformation-guided comparative learning scheme based on intra-space-transformation consistency and inter-class specificity. This scheme aimed to mine additional supervisory information and extract more discriminative features to predict pancreatic cancer lymph node metastasis.
Prior-injected parallel diagnostic networks process the segmentation or detection task in a cascade network, running parallel to the diagnostic task. For example, Zhang et al. [13] first extracted the localization information of the tumor through the augmented feature pyramid network. They then enhanced this localization information with a self-adaptive feature fusion and dependencies computation module, enabling the simultaneous performance of pancreatic cancer detection and diagnosis tasks. Xia et al. [14] used a novel deep classification model with an anatomy-guided transformer to detect resectable pancreatic masses. They classified it as PDAC, other abnormalities (nonPDAC), and normal pancreas. Zhou et al. [15] proposed a meta-information-aware dual-path transformer consisting of a Convolutional Neural Network (CNN) based segmentation path and a transformer-based classification path. This design enabled the simultaneous handling of tasks related to detecting, segmenting, and diagnosing pancreatic lesion locations.
Prior-injected diagnostic networks align better with radiologists’ diagnostic mode. Focusing the analysis on the lesion region may avoid the interference of non-pathological changes in the image or irrelevant physiological information on the model training. However, these deep learning-based approaches have some limitations: 1) the diagnostic model’s performance strongly depends on the accuracy of segmentation or detection results, and biases in these results may mislead the diagnostic model, and 2) pancreatic lesions may cause nearby organs or tissues’ morphologic and physiologic alterations [16,17]. For example, PDAC, when infiltrating the duodenum, typically encircles the stomach and duodenal artery, resulting in bile duct dilation and pronounced jaundice. In contrast, the MFCP may not exhibit these effects [18]. The model, which relies primarily on the lesion region, may ignore contextually significant diagnostic information. Therefore, efficiently leveraging this prior information while preserving information integrity and minimizing redundancy presents a critical challenge.
For this purpose, the study involves the collection of an authentic dataset from MFCP and PDAC patients in a clinical environment. The dataset undergoes two initial exploratory experiments to assess the influence of prior information on diagnostic models’ performance. Such prior information, acquired before the deep learning model training, encompasses lesion regions in MFCP and PDAC, identified directly by radiologists through annotations based on their expertise, and background regions, which are calculated indirectly by masking these lesion areas. Preliminary experiments indicate that background regions, typically considered “noise” in deep learning, offer valuable clues essential for the diagnostic process.
Drawing on the insights, this study introduces the Prior Difference Guidance Network (PDGNet), as shown in Fig. 1c. Unlike existing models, the PDGNet utilizes decoupled lesion and background information to direct the model to concentrate on beneficial regions for diagnosis. The Prior Normalization Fusion (PNF) strategy, the component of this network, integrates the prior information of lesions and backgrounds with the original image before the data is fed into the model. The strategy enables the model to access richer contextual information than the original image. Additionally, the Feature Difference Guide (FDG) module, which employs comparative learning, is proposed. The module further utilizes the prior-augmented lesion and background information, capturing the difference between the lesion region’s and the background region’s augmented features. These differences guide the model to adjust the focus region adaptively according to the importance of the decisions, to achieve a more accurate identification and differentiation between MFCP and PDAC. The main contributions of this study are summarized as follows:
• The study introduces a novel diagnostic framework, the Prior Difference Guidance Network (PDGNet), which uniquely utilizes decoupled lesion and background information to improve the accuracy of differentiating between MFCP and PDAC.
• The study develops the Prior Normalization Fusion (PNF) strategy, an innovative approach within PDGNet that integrates the prior information of lesions and backgrounds with the original image before processing, to enrich the model’s input with a broader context.
• The study implements the Feature Difference Guide (FDG) module, introducing a comparative learning approach that exploits the differences between the augmented features of the lesion and background regions, to direct the model to concentrate on beneficial regions for diagnosis for decision-making adaptively.
2 Materials and Preliminary Analysis
The study investigates the impact of prior information on deep learning-assisted diagnosis for MFCP and PDAC tasks. Authentic datasets of MFCP and PDAC patients from clinical settings are collected. Based on these datasets, two validation experiments are designed: One to examine the influence of images without the lesion region on the diagnostic model, and the other to assess the effect of the background region on the diagnostic model.
A comprehensive dataset is collected from the Second Affiliated Hospital of Kunming Medical University, including arterial-phase abdominal Magnetic Resonance Imaging (MRI) sequences of 31 MFCP patients and 62 PDAC patients. The dataset includes 3,872 slices, with 375 slices annotated to indicate lesion regions. Fig. 2 illustrates the slice-image with the lesion region.

Figure 2: Illustration of MFCP and PDAC lesions. The top row shows the MFCP lesion slice-image, and the bottom row shows the PDAC lesion slice-image. (a) shows the original image, (b) Shows the lesion region with a masked background, (c) shows the lesion region after crop and resize, and (d) shows the background region with a masked lesion
Inclusion criteria: 1) Patients with MCFP and PDAC confirmed by surgery and/or biopsy histopathology, and 2) MRI scanning within 1 month before neoadjuvant chemotherapy or surgery.
Exclusion criteria: Lesions were poorly visualized or showed non-mass-like enhancement that was difficult to outline.
Scanning machine: Planar and dynamic enhancement scans of the upper abdomen were performed using a Siemens Sonata 1.5 Tesla (1.5 T) MR scanner.
Scanning sequence and parameters: Transverse, coronal, and sagittal scans were performed in VIBE sequence using gadopentetate dextran (0.2 ml/kg) during the arterial phase (25–30 s).
The lesion region annotation criteria: Initially, an experienced radiologist utilizes 3D Slicer software (https://www.slicer.org/) to label the entire tumor as comprehensively as possible, avoiding areas of necrosis, calcification, and gases that can obscure the lesion. To ensure accuracy, the labeled tumor area is subsequently reviewed by another radiologist.
This study involves randomly selecting 300 slice-images that contain lesion regions from the dataset. This selection establishes the base training for a preliminary diagnostic model for PDAC with MFCP. The percentage of slice-images without the lesion region in the training set is incrementally increased, to train multiple diagnostic models, as shown in Fig. 3a. These models undergo evaluation using the same test set, with specific experimental results presented in Table 1, and visualized as shown in Fig. 4a.

Figure 3: Schematic diagram of experimental designs: (a) Experiment 1 investigates the impact of non-lesion images on the diagnostic model. (b) Experiment 2 investigates the impact of the background region on the diagnostic model
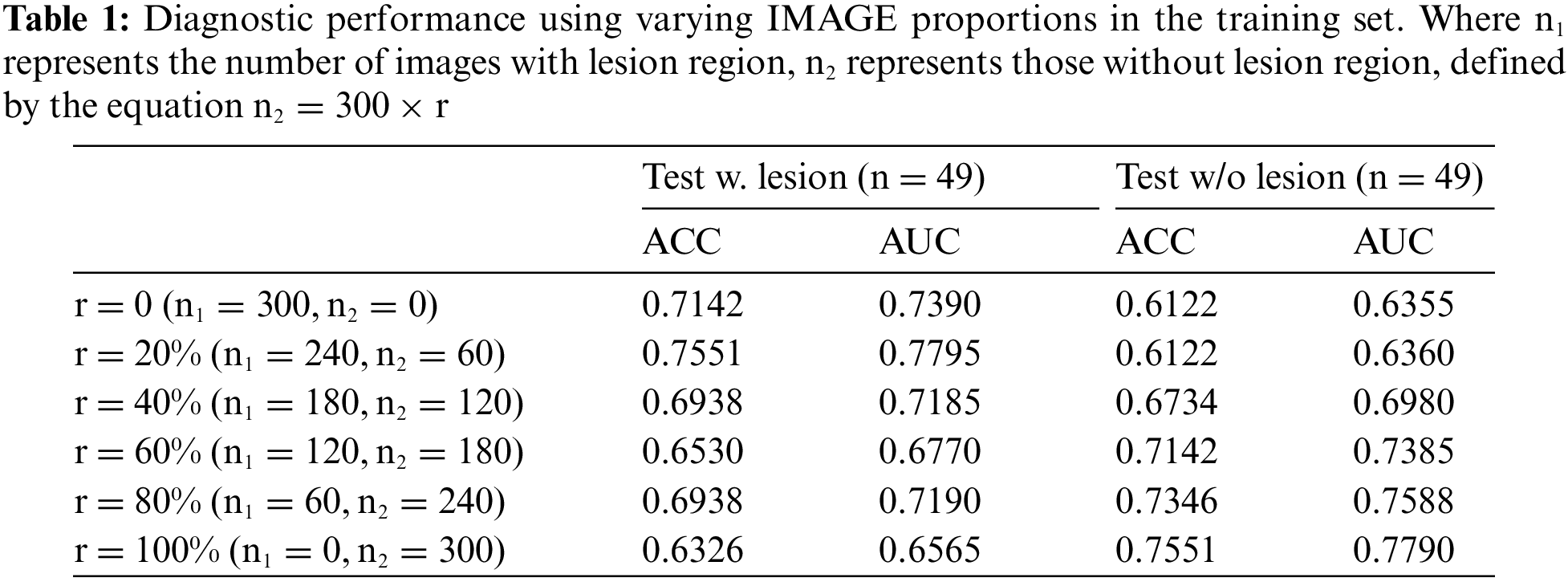

Figure 4: Visualization of experimental results: (a) ACC curve of the testing set as r varies with the proportion of non-lesion images in the training set, and (b) ACC curve of the testing set as r varies with the proportion of the background region in the training set
In another experiment, the lesion regions from 300 slice-images are extracted and utilized to create the new training set. The proportion of background region within this lesion region is progressively increased, to train several diagnostic models, as shown in Fig. 3b. These models are evaluated on the same test set, with specific experimental results presented in Table 2, and visualized as shown in Fig. 4b.
The VGG-11 architecture serves as the foundational diagnostic network for this study. All training sessions are conducted under uniform parameter settings. The slice-images designated for the training and test sets originate from distinct patients.
The experimental results lead to the following conclusions and insights: 1) the prior information plays an important role in deep learning-assisted diagnosis, according to the experimental results in Tables 1 and 2. The model’s performance fluctuates when the proportion of lesion and background region in the training data changes, 2) the model’s performance is not optimal when only lesion region images are used for training, according to the experimental results in Table 1. With the increase of non-lesion region images, the model’s performance is improved in some cases, indicating that it is beneficial to maintain a certain balance of diseased and non-diseased images for the diagnostic task of PDAC with MFCP, and 3) the model’s performance starts to decrease when the proportion of background region increases to a certain extent, which indicates that the background information holds significant value in the diagnostic task, according to the experimental results in Table 2. However, exceeding a specific percentage interferes with the model’s performance.
The insights from the comparative analysis of two sets of experiments inform subsequent model design enhancements. These improvements include: 1) a data augmentation strategy that maximizes the utilization of contextual information during training, to intensify the focus on identified lesion regions, thereby augmenting the recognition of these critical regions, and 2) an attention fusion module that enables it to dynamically adjust its focus on the lesion region and the relevant portions of the contextual regions, allowing for a more accurate diagnosis of PDAC and MFCP.
The analysis leads to the proposal of a Prior Difference Guidance Network (PDGNet), with its structure illustrated in Fig. 5. The Network consists of two main components: The Prior Normalization Fusion (PNF) strategy and the Feature Difference Guidance (FDG) module.

Figure 5: The structure of the prior difference guidance network (PDGNet), with two components: The prior normalization fusion (PNF) strategy and the feature difference guidance (FDG) module
3.1 Prior Normalization Fusion (PNF) Strategy
The Prior Normalization Fusion (PNF) strategy for data augmentation is proposed, as shown in Fig. 6. Before the data is input into the model, it tries to fuse the prior information of lesion and background with the original image, which enables the model to obtain richer contextual information than the original image when performing diagnosis.

Figure 6: The structure of the prior difference guidance network (PDGNet), with two components: The prior normalization fusion (PNF) strategy and the feature difference guidance (FDG) module
Specifically, the lesion region is initially selected based on the optimal background occupancy ratio (r = 60%), as determined in preliminary experiments. Subsequently, the background region is extracted by masking the lesion region in the original image. The original image is then overlaid with the prior images (both lesion and background). Normalization is conducted within the prior local region, considering only non-zero regions to prevent the diluted effect of a global homogeneous background on the normalized fused image.
Given an image
where
3.2 Feature Difference Guidance (FDG) Module
The PDGNet introduces a Feature Difference Guidance (FDG) module to utilize the prior-augmented lesion and background information further, as shown in Fig. 7. The module further utilizes the prior-augmented lesion and background information, capturing the difference between the lesion region’s and the background region’s augmented features. These differences guide the model to adjust the focus region adaptively according to the decisions’ importance.

Figure 7: The structure of the prior difference guidance network (PDGNet), with two components: The prior normalization fusion (PNF) strategy and the feature difference guidance (FDG) module
The module combines the original image with prior-augmented lesion and background information fusion images as inputs. This integration offers richer and multi-perspective contextual information for model training. Features
The channel-fused features
The overall framework is shown in Fig. 4, defines the backbone network comprising n blocks, with an FDG module added after each block. In the first n-1 blocks,
The training and test sets are divided by case with a 9:1 ratio to prevent mutual data leakage within the same case. The training set contains arterial-phase abdominal MRI sequences of 82 patients (27 MCFP, 55 PDAC), with a total of 3,432 slices (326 slices annotated with lesion areas), and the test set contains 11 cases (4 MCFP, 7 PDAC), with a total of 440 slices (49 slices annotated with lesion areas). A total of 440 slices (49 with lesion areas labeled).
For preprocessing, the image resolution is adjusted to 224 × 224, and image augmentation is applied using restricted contrast adaptive histogram equalization. In the experiments, the total number of epochs is set to 300. The learning rate is initialized at 1e-4 and dynamically adjusts using a cosine function, with a minimum value set at 1e-6 and a loop count of 50. An early-stopping mechanism is employed to prevent overfitting, terminating training if the loss value of the validation set does not decrease for 30 epochs. The batch size is 64, and the model is optimized using adaptive moment estimation (AdamW) [19] with a weight decay of 1e-3. Additionally, all experiments use the PyTorch framework on an NVIDIA GeForce RTX 4090 graphics processing unit.
This study employs a comprehensive evaluation of the diagnostic performance of the model using several metrics: Accuracy (ACC), area under the subject operating characteristic curve (AUC), sensitivity/recall (SEN/REC), specificity (SPE), precision (PREC) and F1 score (F1). These metrics are defined below:
Accuracy (ACC): Accuracy measures the proportion of all cases (both MFCP and PDAC) that are correctly identified by the model at a specific threshold, calculated as
Area Under the Curve (AUC): AUC refers to the area under the Receiver Operating Characteristic (ROC) curve, a graphical representation of a model’s diagnostic ability. It measures the model’s capability to discriminate between two classes (MFCP and PDAC) across all possible threshold values. A higher AUC value implies that the model performs better in distinguishing between negative (MFCP) and positive (PDAC) cases, regardless of any specific threshold set for classifying cases as positive or negative.
Sensitivity/Recall (SEN/REC): This metric quantifies the model’s ability to correctly identify positive cases (PDAC), calculated as
Specificity (SPE): Specificity measures the model’s ability to correctly identify negative cases (MFCP), calculated as
Precision (PREC): Precision reflects the proportion of cases identified as positive (PDAC) that are indeed PDAC, calculated as
F1 Score (F1): The F1 score is the harmonic mean of precision and recall, calculated as
TP, TN, FP, and FN represent the number of true-positive, true-negative, false-positive, and false-negative samples, respectively.
These metrics are intended to offer a holistic view of the model’s performance, covering various aspects of diagnostic accuracy. Each metric offers insights into different dimensions of the model’s effectiveness, ensuring a thorough evaluation of its capabilities in medical diagnosis.
4.3 Effectiveness of the Prior Normalization Fusion (PNF) Strategy
The study explores the effectiveness of the Prior Normalization Fusion (PNF) strategy by selecting ResNet-18 [20] as the baseline model, and comparing various data input types as the training set. These include the original image, the cropped and resized lesion region, the background region obtained by masking the lesion region, the lesion-augmented and background-augmented images obtained by the PNF strategy, and the lesion-augmented and background-augmented images obtained by the global normalization fusion strategy (GNF).
Furthermore, to examine the impact of attention mechanisms on the models, the study evaluates Squeeze-and-Excitation (SE) [21] without the prior information condition, Convolutional Block Attention Module (CBAM) [22], Channel Prior Convolutional Attention (CPCA) [23], and Vision Transformer (ViT) [24] based on the global spatial attention mechanism Self-Attention [25]. The corresponding results are shown in Table 3.
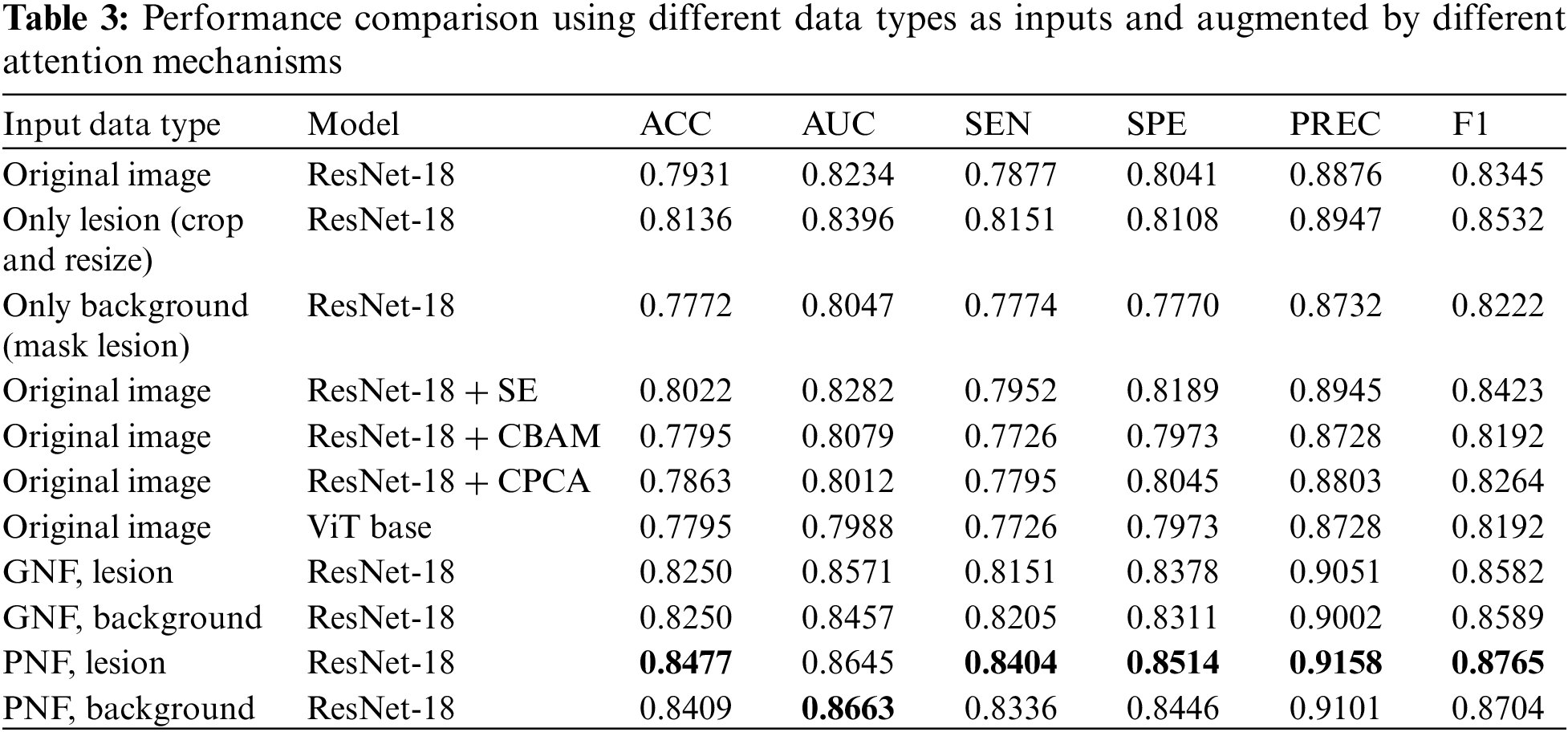
Table 3 indicates that ResNet-18, when trained with lesion-augmented and background-augmented images using the PNF strategy, surpasses the performance of models trained with images augmented by the GNF strategy or models trained with original images. In particular, the lesion-augmented image obtained by the PNF strategy achieves an ACC of 84.77%, a 5.46% improvement compared to the original image, and a 3.41% improvement compared to using only the lesion region. These results validate the superiority of the PNF strategy. Without utilizing the prior information, the SE attention mechanism improves the ACC of ResNet-18 to 80.22%, while the performance of CBAM, CPCA, and ViT is lower than that of the benchmark network model.
4.4 Effectiveness of the Feature Differential Guidance (FDG) Module
This study utilizes the lesion-augmented and background-augmented images obtained by the PNF strategy, and the original image as the training set, with ResNet-18 serving as the baseline model, to assess the role of the Feature Difference Guidance (FDG) module.
Additionally, the impact of various fusion strategies on diagnostic performance is examined. These strategies include: 1) Slicer-Dimension Concatenate: Connect the three images in the slicer dimension before modeling, 2) Channel-Dimension Concatenate: Connect the three images in the channel dimension, and 3) ResNet-18 + Feature Concatenate: Extract features using different encoders for each input image type, and then connect the features after each block of the model. The results are displayed in Table 4.
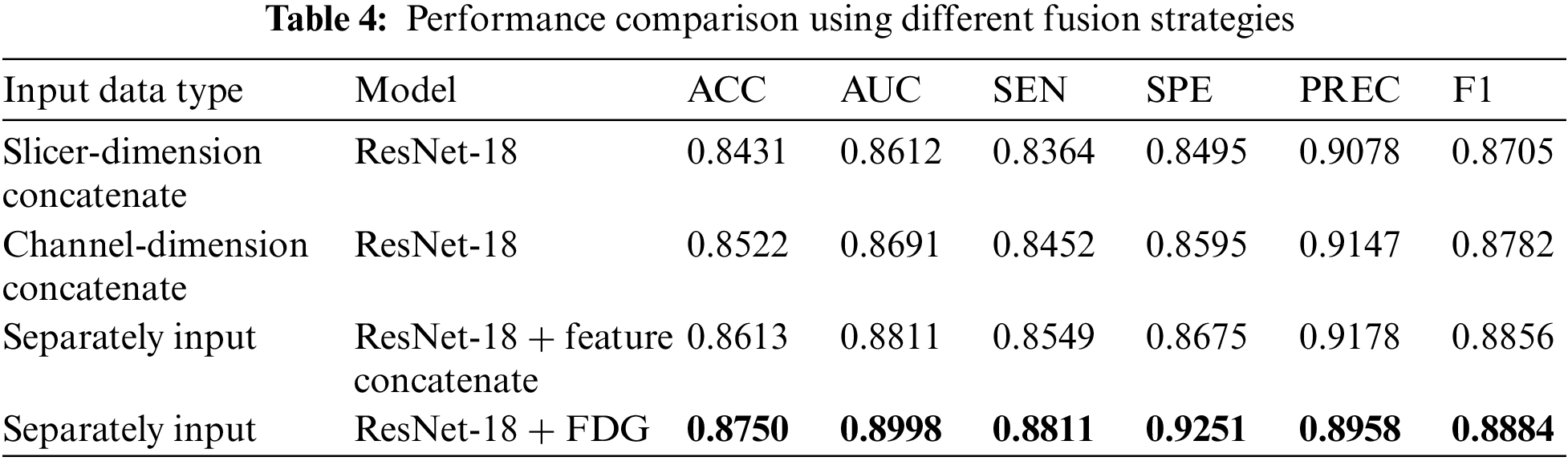
As described in Tables 3 and 4, ResNet-18, based on FDG modules, demonstrates the best performance with an ACC of 87.5%, higher than the other strategies. In addition, the other fusion strategies also brought performance improvements, reaching an ACC of 86.13% when feature linking was performed within the model.
The study conducts ablation experiments on four mainstream backbones, ResNet, ViT, Swin Transformer [26], and ConvNeXt [27], to further explore the benefits of the PNF strategy and the FDG module. The relevant results are listed in Table 5, and the ROC curves are shown in Fig. 8.
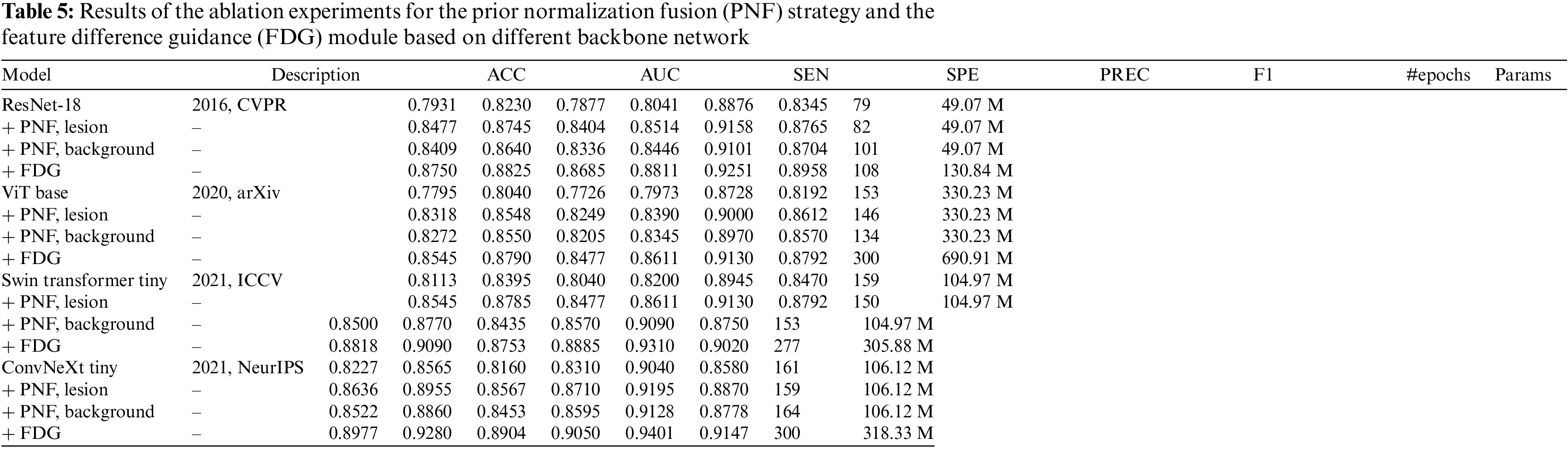

Figure 8: Average ROC curves and AUC values of the ablation experiments based on different backbone network
Table 5 and Fig. 8 illustrate that the implementation of the PNF strategy and the FDG module significantly improves the performance of models based on the CNN architecture, specifically ResNet-18 and ConvNeXt, as well as those based on the transformer architecture, such as ViT and Swin Transformer. This evidence underscores the effectiveness, superior generalization ability, and compatibility of these strategies across various network architectures.
4.6 Comparison with Other Methods
The study aims to differentiate between MFCP and PDAC using arterial phase MRI scans. The PDGNet is compared to other pancreatic lesion diagnostic models, including the prior-free diagnostic network by Ziegelmayer et al., which employed VGG-19 to distinguish between AIP and PDAC [6]. Si et al. [9] used a prior-injected diagnostic network, which was first trained using the U-Net32 [28] to train a pancreas segmentation model, and then input the segmentation results into ResNet34 to distinguish between five different pancreatic lesions. The study employs manual annotation instead of the segmentation results from U-Net.
As described in Table 6, the PDGNet based on ConvNeXt outperforms other models on all evaluation metrics for MFCP and PDAC classification tasks. This further demonstrates that the implemented strategies can effectively alleviate the problem of the difficulty of discriminative feature extraction.
The comparison may not be fair since the studies used different datasets, but it can still provide valuable references for future research.
The study investigates a concept frequently overlooked in existing research on deep learning for pancreatic lesion diagnosis: Background region considered as “noise” can actually provide valuable information for diagnostic models. As shown in Tables 1 and 2, the ACC and AUC of the diagnostic model reach 63.26% and 65.65%, even though the training set consists entirely of images without lesion regions. When masking the lesion region from the complete image containing the lesion region, the ACC and AUC are 67.34% and 71.67%, underlining the significance of the background region in the diagnostic modeling dataset.
Consequently, the Prior Normalization Fusion (PNF) strategy is proposed. The strategy, which fuses prior information before data input into the model, augments the feature recognizability of the prior (lesion and background) region while preserving the complete contextual details of the original image. As shown in Table 3, without utilizing the prior information, channel attention SE can only bring relatively limited performance improvement, with ACC and AUC increasing by 0.91% and 0.48%, respectively. In contrast, introducing spatial attention leads to a decline in model performance. This could be attributed to inherent noise in the image, causing a bias in the attention mechanism without prior information. However, the GNF and PNF strategies demonstrate significant performance gains, particularly the PNF strategy, which improves the ACC and AUC of the benchmark network model by 5.46% and 4.11%, respectively.
Otherwise, the study observes that both lesion-augmented and background-augmented images generated by the PNF strategy are able to improve the diagnostic model’s performance. To explore the potential of this prior-augmented information more deeply, a Feature Difference Guidance (FDG) module is introduced. The module combines the original image with the lesion-augmented and background-augmented images so that they jointly participate in the model training process. The superiority of this fusion strategy is further confirmed by the data in Table 5, where the FDG module demonstrates the best performance.
Ablation experiments on convolutional neural networks such as ResNet-18, ConvNeXt, and Transformer-based ViT, Swin Transformer, show that the proposed Prior Difference Guidance Network (PDGNet) with the PNF strategy and the FDG module achieve significant improvements on all four frameworks. Especially on ConvNeXt, the ACC and AUC of the model are improved to 89.77% and 92.80%, respectively.
In summary, the study confirms that the background region carries useful information for diagnosis, which the model should more fully utilize. The PDGNet, incorporating the PNF strategy and FDG module, significantly improves the diagnostic accuracy for MFCP and PDAC, uniquely leveraging prior information from lesion and background regions.
Although the model achieves excellent performance, it has some limitations. For example, the clinical datasets utilized might lack diversity and size. Nevertheless, the network demonstrates robustness and effectiveness in data diversity and size constraints by accurately extracting and analyzing key discriminative features. This offers promise for application in a wider range of clinical scenarios. Secondly, a notable shortcoming of the proposed approach is the extensive training time and large model parameters. Therefore, with continued optimization and algorithmic improvements, there is an expectation of significant reductions in training time and improvements in model efficiency. Thirdly, extensive testing in real-world clinical settings is yet to be conducted for the model. However, preliminary findings and the model’s theoretical design indicate that, with further refinement and validation, it will serve as an effective tool for assisted diagnosis in clinical environments. Future research will focus on collecting more clinical data to enhance the model’s generalization ability, exploring more efficient algorithms and network architectures to optimize the training process and minimize computational resource requirements, and conducting validations in actual clinical settings to confirm its effectiveness and feasibility. The ultimate goal is to improve the accuracy and reliability of automated diagnosis, aiming to implement these models in clinical practice and offering more effective diagnostic tools for physicians and patients.
This study proposes a novel approach for deep learning pancreatic lesion diagnostic research, focusing on the lesion region and fully utilizing the information in the background region. The study observes that even in background regions without obvious lesions, valuable information exists that helps with diagnosis. Drawing on this insight, the Prior Difference Guidance Network (PDGNet) significantly improves the performance of MFCP and PDAC diagnostic models through the Prior Normalization Fusion (PNF) strategy and the feature difference guidance (FDG) module.
The PNF strategy preserves the complete contextual information of the original image. It augments the feature recognizability of the prior region by fusing the prior information of the lesion and the background region. The FDG module, on the other hand, combines the original image with the augmented lesion and background fusion image so that both of them participate in the model’s training process, further improving the model’s accuracy. Ablation experiments conduct on various prominent deep learning networks, including ResNet-18, ConvNeXt, Vision Transformer (ViT), and Swin Transformer, substantiate the effectiveness of this approach.
In conclusion, the study emphasizes the importance of contextual information in deep learning pancreatic lesion diagnosis and proposes new methods to utilize this information more fully to improve model performance. The study provides a valuable reference for future medical image diagnosis. It suggests that scholars should not only focus on salient target regions but also pay full attention to the background information that is often overlooked.
Acknowledgement: The authors would like to express their gratitude to Prof. He and Prof. Hao for supervising of this study.
Funding Statement: This research is supported by the National Natural Science Foundation of China (No. 82160347); Yunnan Key Laboratory of Smart City in Cyberspace Security (No. 202105AG070010); Project of Medical Discipline Leader of Yunnan Province (D-2018012).
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, L.C. and K.B.; data curation, K.B. and Y.C.; investigation, L.C., K.B. and Y.C.; methodology, L.C.; formal analysis, L.C.; project administration, J.H. (Jianfeng He); supervision, J.H. (Jianfeng He) and J.H. (Jingang Hao); writing—original draft preparation, L.C.; writing—review and editing, L.C., J.H. (Jianfeng He) and J.H. (Jingang Hao). All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: The datasets generated during this study are not publicly available due to privacy and ethical considerations; however, anonymized data can be provided by the corresponding author upon reasonable request and with the approval of the Ethics Committee. Researchers interested in accessing the data should contact the corresponding author (Prof. Hao, kmhaohan@163.com) for further information.
Ethics Approval: The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Second Affiliated Hospital of Kunming Medical University (No. 2023-156).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Raimondi, A. B. Lowenfels, A. M. Morselli-Labate, P. Maisonneuve, and R. Pezzilli, “Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection,” Best Pract. Res. Clin. Gastroenterol., vol. 24, no. 3, pp. 349–358, 2010. doi: 10.1016/j.bpg.2010.02.007 [Google Scholar] [PubMed] [CrossRef]
2. F. J. Harmsen, D. Domagk, C. F. Dietrich, and M. Hocke, “Discriminating chronic pancreatitis from pancreatic cancer: Contrast-enhanced EUS and multidetector computed tomography in direct comparison,” Endosc. Ultrasound, vol. 7, no. 6, pp. 395, 2018. doi: 10.4103/eus.eus_24_18 [Google Scholar] [PubMed] [CrossRef]
3. Q. Yin et al., “Pancreatic ductal adenocarcinoma and chronic mass-forming pancreatitis: Differentiation with dual-energy MDCT in spectral imaging mode,” Eur. J. Radiol., vol. 84, no. 12, pp. 2470–2476, 2015. doi: 10.1016/j.ejrad.2015.09.023 [Google Scholar] [PubMed] [CrossRef]
4. T. Kennedy et al., “Incidence of benign inflammatory disease in patients undergoing Whipple procedure for clinically suspected carcinoma: A single-institution experience,” Am. J. Surg., vol. 191, no. 3, pp. 437–441, 2006. doi: 10.1016/j.amjsurg.2005.10.051 [Google Scholar] [PubMed] [CrossRef]
5. Y. Tajima, T. Kuroki, R. Tsutsumi, I. Isomoto, M. Uetani and T. Kanematsu, “Pancreatic carcinoma coexisting with chronic pancreatitis vs. tumor-forming pancreatitis: Diagnostic utility of the time-signal intensity curve from dynamic contrast-enhanced MR imaging,” World J. Gastroenterol, vol. 13, no. 6, pp. 858, 2007. doi: 10.3748/wjg.v13.i6.858 [Google Scholar] [PubMed] [CrossRef]
6. S. Ziegelmayer et al., “Deep convolutional neural network-assisted feature extraction for diagnostic discrimination and feature visualization in pancreatic ductal adenocarcinoma (PDAC) vs. autoimmune pancreatitis (AIP),” J. Clin. Med., vol. 9, no. 12, pp. 4013, 2020. doi: 10.3390/jcm9124013 [Google Scholar] [PubMed] [CrossRef]
7. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” presented at the 3rd Int. Conf. Learn. Rep., San Diego, CA, USA, May 7–9, 2015. [Google Scholar]
8. J. Deng, W. Dong, R. Socher, L. J. Li, K. Li, and L. Fei-Fei, “ImageNet: A large-scale hierarchical image database,” presented at the 2009 IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recog., Miami, Florida, USA, Jun. 20–25, 2009. doi: 10.1109/CVPR.2009.5206848. [Google Scholar] [CrossRef]
9. K. Si et al., “Fully end-to-end deep-learning-based diagnosis of pancreatic tumors,” Theranost., vol. 11, no. 4, pp. 1982–1990, 2021. doi: 10.7150/thno.52508 [Google Scholar] [PubMed] [CrossRef]
10. J. Qu, X. Wei, and X. Qian, “Generalized pancreatic cancer diagnosis via multiple instance learning and anatomically-guided shape normalization,” Med. Image Anal., vol. 86, pp. 102774, 2023. doi: 10.1016/j.media.2023.102774 [Google Scholar] [PubMed] [CrossRef]
11. X. Li, R. Guo, J. Lu, T. Chen, and X. Qian, “Causality-driven graph neural network for early diagnosis of pancreatic cancer in non-contrast computerized tomography,” IEEE Trans. Med. Imag., vol. 42, no. 6, pp. 1656–1667, 2023. doi: 10.1109/TMI.2023.3236162 [Google Scholar] [PubMed] [CrossRef]
12. X. Chen, W. Wang, Y. Jiang, and X. Qian, “A dual-transformation with contrastive learning framework for lymph node metastasis prediction in pancreatic cancer,” Med. Image Analy., vol. 85, pp. 102753, 2023. doi: 10.1016/j.media.2023.102753 [Google Scholar] [PubMed] [CrossRef]
13. Z. Zhang, S. Li, Z. Wang, and Y. Lu, “A novel and efficient tumor detection framework for pancreatic cancer via CT images,” presented at the 42nd Ann. Int. Conf. IEEE Eng. Med. Biol. Soc., Montreal, QC, Canada, Jul. 20–24, 2020. doi: 10.1109/embc44109.2020.9176172 [Google Scholar] [PubMed] [CrossRef]
14. Y. Xia et al., “Effective pancreatic cancer screening on non-contrast CT scans via anatomy-aware transformers,” presented at the 24th Int. Conf. Med. Image Comput. Comput. Assist. Intervent., Strasbourg, France, Sep. 27–Oct. 1, 2021. doi: 10.1109/embc44109.2020.9176172. [Google Scholar] [CrossRef]
15. B. Zhou et al., “Meta-information-aware dual-path transformer for differential diagnosis of multi-type pancreatic lesions in multi-phase CT,” presented at the 28th Int. Conf. Inf. Process. Med. Imag., San Carlos de Bariloche, Argentina, Jun. 18–23, 2023. doi: 10.1007/978-3-031-34048-2_10. [Google Scholar] [CrossRef]
16. W. Schima, G. Böhm, C. S. Rösch, A. Klaus, R. Függer and H. Kopf, “Mass-forming pancreatitis vs. pancreatic ductal adenocarcinoma: CT and MR imaging for differentiation,” Cancer Imag., vol. 20, no. 1, pp. 1–12, 2020. doi: 10.1186/s40644-020-00324-z [Google Scholar] [PubMed] [CrossRef]
17. S. B. Elsherif, M. Virarkar, S. Javadi, J. J. Ibarra-Rovira, E. P. Tamm and P. R. Bhosale, “Pancreatitis and PDAC: Association and differentiation,” Abdom. Radiol., vol. 45, no. 5, pp. 1324–1337, 2020. doi: 10.1007/s00261-019-02292-w [Google Scholar] [PubMed] [CrossRef]
18. H. A. Zeid et al., “Differentiating between mass-forming chronic pancreatitis and pancreatic ductal adenocarcinoma: A challenging clinical approach,” Int. J. Clin. Res., vol. 3, no. 1, pp. 276–284, 2022. doi: 10.38179/ijcr.v3i1.244. [Google Scholar] [CrossRef]
19. I. Loshchilov and F. Hutter, “Decoupled weight decay regularization,” presented at the 7th Int. Conf. Learn. Rep., New Orleans, LA, USA, May 6–9, 2019. [Google Scholar]
20. K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” presented at the 2016 IEEE Conf. Comput. Vis. Pattern Recogn., Las Vegas, NV, USA, Jun. 27–30, 2016. doi: 10.1109/CVPR.2016.90. [Google Scholar] [CrossRef]
21. J. Hu, L. Shen, and G. Sun, “Squeeze-and-excitation networks,” presented at the 2018 IEEE Conf. Comput. Vis. Pattern Recogn., Salt Lake City, UT, USA, Jun. 18–22, 2018. doi: 10.1109/CVPR.2018.00745. [Google Scholar] [CrossRef]
22. S. Woo, J. Park, J. Y. Lee, and I. S. Kweon, “CBAM: Convolutional block attention module,” presented at the 15th Eur. Conf. Comput. Vis., Munich, Germany, Sep. 8–14, 2018. doi: 10.1007/978-3-030-01234-2_1. [Google Scholar] [CrossRef]
23. H. Huang, Z. Chen, Y. Zou, M. Lu, and C. Chen, “Channel prior convolutional attention for medical image segmentation,” 2023. doi: 10.48550/arXiv.2306.05196. [Google Scholar] [CrossRef]
24. A. Dosovitskiy et al., “An image is worth 16 × 16 words: Transformers for image recognition at scale,” presented at the 9th Int. Conf. Learn. Rep., Austria, May 3–7, 2021. [Google Scholar]
25. A. Vaswani et al., “Attention is all you need,” presented at the Adv. Neural Inf. Process. Syst. 30: Ann. Conf. Neural Inf. Process. Syst., Long Beach, CA, USA, Dec. 4–9, 2017. doi: 10.5555/3295222.3295349. [Google Scholar] [CrossRef]
26. Z. Liu et al., “Swin transformer: Hierarchical vision transformer using shifted windows,” presented at the 2021 IEEE/CVF Int. Conf. Comput. Vis., Montreal, QC, Canada, Oct. 10–17, 2021. doi: 10.1109/ICCV48922.2021.00986. [Google Scholar] [CrossRef]
27. Z. Liu, H. Mao, C. Y. Wu, C. Feichtenhofer, T. Darrell and S. Xie, “A ConvNet for the, 2020s,” presented at the IEEE/CVF Conf. Comput. Vis. Pattern Recogn., New Orleans, LA, USA, Jun. 18–24, 2022. doi: 10.1109/CVPR52688.2022.01167. [Google Scholar] [CrossRef]
28. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” presented at the 18th Int. Conf. Med. Image Comput. Comput. Assist. Intervent., Munich, Germany, Oct. 5–9, 2015. doi: 10.1007/978-3-319-24574-4_28. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools