 Open Access
Open Access
ARTICLE
Secure Medical Image Retrieval Based on Multi-Attention Mechanism and Triplet Deep Hashing
School of Computer and Communication, Lanzhou University of Technology, Lanzhou, 730000, China
* Corresponding Author: Qiuyu Zhang. Email:
(This article belongs to the Special Issue: Emerging Trends and Applications of Deep Learning for Biomedical Signal and Image Processing)
Computers, Materials & Continua 2025, 82(2), 2137-2158. https://doi.org/10.32604/cmc.2024.057269
Received 13 August 2024; Accepted 01 November 2024; Issue published 17 February 2025
Abstract
Medical institutions frequently utilize cloud servers for storing digital medical imaging data, aiming to lower both storage expenses and computational expenses. Nevertheless, the reliability of cloud servers as third-party providers is not always guaranteed. To safeguard against the exposure and misuse of personal privacy information, and achieve secure and efficient retrieval, a secure medical image retrieval based on a multi-attention mechanism and triplet deep hashing is proposed in this paper (abbreviated as MATDH). Specifically, this method first utilizes the contrast-limited adaptive histogram equalization method applicable to color images to enhance chest X-ray images. Next, a designed multi-attention mechanism focuses on important local features during the feature extraction stage. Moreover, a triplet loss function is utilized to learn discriminative hash codes to construct a compact and efficient triplet deep hashing. Finally, upsampling is used to restore the original resolution of the images during retrieval, thereby enabling more accurate matching. To ensure the security of medical image data, a lightweight image encryption method based on frequency domain encryption is designed to encrypt the chest X-ray images. The findings of the experiment indicate that, in comparison to various advanced image retrieval techniques, the suggested approach improves the precision of feature extraction and retrieval using the COVIDx dataset. Additionally, it offers enhanced protection for the confidentiality of medical images stored in cloud settings and demonstrates strong practicality.Keywords
As medical imaging methods become increasingly widespread, the volume of medical imaging information has surged dramatically, creating critical challenges in storage, retrieval, information security, and optimal utilization for all healthcare institutions [1]. To explore more potential information from medical imaging data, similar medical image retrieval technologies have attracted widespread attention from relevant research experts and scholars [2]. For instance, content-based medical image retrieval (CBMIR) technology is capable of automatically extracting visual features from images [3–5]. Furthermore, deep hashing-based medical image retrieval technology provides a new approach to image feature extraction, maintaining the original semantic information of images by constructing high-quality compact binary hash codes [6]. Furthermore, to ensure the safety of medical imaging information stored in the cloud storage and prevent the leakage of personal, institutional, and national healthcare-related information, information security in medical imaging is also a critical issue that cannot be ignored.
Currently, deep hashing technology has become an essential approach for medical image retrieval. This method offers rapid retrieval capabilities and reduced storage expenses [7,8], enhancing the precision and effectiveness of similar medical image searches. For instance, TSDSH [9], MCRLDH [10], DUDH [11], SWTH [12], and DGSSH [13], etc.
In recent years, the advent of cloud storage solutions has significantly improved the ability of healthcare institutions to store medical images. Although the vast storage capacity and extensive computing power of cloud servers (CS) reduce the strain on local image storage and management, healthcare institutions consequently forfeit direct control over medical image data [14]. In the process of cloud storage for medical data, an increasingly serious issue is the illegal replication, modification, and forgery of medical data [15]. Therefore, it is essential to protect data integrity and prevent unauthorized access. Chang et al. [16] addressed cloud platform insecurities and adopted symmetric encryption to prevent unauthorized access or modification of patient data, ensuring medical data security during storage. Thus, safeguarding the privacy of medical images in the cloud necessitates encryption techniques, which are vital for secure medical image retrieval. Existing traditional encryption algorithms such as Data Encryption Standard (DES), Advanced Encryption Standard (AES), and Rivest-Shamir-Adleman (RSA) are not suitable for encrypting multimedia data. Homomorphic encryption (HE) is key to achieving data privacy computation, but currently, no feasible HE technology can encrypt large volumes of images, and HE is overly complex and time-consuming. Chaotic systems, known for their sensitivity to initial conditions, randomness, and ergodicity, are extensively used in multimedia data encryption.
To ensure the privacy and security of medical imaging data, this paper proposes a secure retrieval method for chest X-ray (CXR) images using deep hashing, a multi-attention mechanism, and lightweight encryption. The method, termed MATDH (Multi-Attention Triplet Deep Hashing), offers secure and efficient medical image retrieval. Key contributions are as follows:
1) CXR images are enhanced using contrast-limited adaptive histogram equalization, dynamically adjusting based on local contrast to better preserve details and improve image discriminability and expressiveness.
2) A triplet deep hashing model is designed, combining channel and enhanced spatial attention mechanisms to focus on local features, significantly improving the accuracy of important region extraction in CXR images.
3) Grounded in the principle of frequency domain encryption, a lightweight CXR image encryption method based on chaos theory is proposed. While securing medical images, the complexity of encryption is reduced, and the speed of both encryption and decryption processes is enhanced.
The paper is organized as follows: Section 2 reviews related research, Section 3 details the medical image secure retrieval method, Section 4 validates the scheme through experiments and compares its performance with existing methods, and Section 5 concludes the work.
2.1 Medical Image Retrieval Using Deep Hashing Techniques
With the excellent performance of artificial intelligence and deep hashing technology in various fields, deep hashing technology has also been extensively applied within the domain of medical image retrieval [17–20]. The medical image retrieval methods using deep hashing are generally divided into supervised deep hashing methods [8,21–23] and unsupervised deep hashing methods [18,24,25]. However, for high-precision medical image retrieval, unsupervised hashing methods face semantic gaps due to the lack of label information during network training, affecting the retrieval accuracy of medical images [26]. Therefore, for medical image datasets with complete label information, supervised deep hashing methods have become the optimal solution for constructing binary hash codes. For example, Wang et al. [8] proposed that the triplet constraint be directly integrated into medical image feature learning to capture the intricate relationships among medical images, while encoding-decoding networks are used to enhance the discriminative strength of the generated hash codes. However, this method cannot focus on the detail areas and important features of medical images because it does not enhance the images before retrieval. Fang et al. [22] proposed an attention-based triplet hashing network that effectively retains classification and limited sample information while learning binary hash codes. This method combines cross-entropy loss with triplet loss, simultaneously training similarity loss and classification loss to maintain classification information in hash codes, achieving maximum class discrimination and hash code distinguishability. However, the attention mechanism designed in this method still cannot effectively focus on the important areas of medical images.
2.2 Secure Medical Image Retrieval
Given that medical images contain patient information, which is considered highly sensitive data, hospitals or other healthcare institutions, as data owners, do not wish for every user to have access to and view these images. Instead, users are granted access only after authorization [27]. To ensure privacy and confidentiality during the medical image retrieval process, various security services are used, among which image encryption technology has become a crucial means of protecting medical image information from attacks [28–32]. Haddad et al. [28] utilized the cipher block chaining mode in AES and combined it with image watermarking technology to encrypt medical images, allowing tracking and controlling the reliability of medical images from encrypted or compressed domains. However, this method not only incurs high encryption costs and complexity but also introduces image quality loss. Guo et al. [29] developed a convolutional neural network (CNN) framework that protects privacy by allowing the use of homomorphic encryption technology for classifying and retrieving encrypted medical images, enhancing the security of the retrieval scheme. Nevertheless, the communication and computational costs significantly increase, making it unsuitable for use in resource-constrained scenarios. Kumar et al. [30] employed the idea of dual encryption to ensure the privacy of medical image retrieval processes. The first-level image encryption employs chaotic Arnold mapping to encrypt the images while preserving their statistical characteristics. The second level of encryption further boosts security and reduces the chances of cryptographic attacks. The use of a simple chaotic Arnold mapping in the first-level encryption cannot achieve perfect secrecy for query images, and encrypted visual content still provides relevant information about the true content of query images, posing a risk of attack. However, the methods used for classification and relevance scores may lead to privacy leaks and other issues.
A lightweight medical image encryption algorithm combining chaos-based frequency domain encryption and the PRESENT algorithm is proposed to ensure security and reduce costs in image retrieval. A triplet deep hashing method with multi-attention (MATDH), integrating channel and enhanced spatial attention mechanisms, improves the discriminability of CXR images. This secure medical image retrieval method enhances efficiency and accuracy while safeguarding data privacy.
Fig. 1 shows the system model for secure medical image retrieval, avoiding issues such as the leakage of medical images containing patient privacy by cloud servers.

Figure 1: System model
In Fig. 1, this model consists primarily of three entities: Cloud Server (CS), Data Owner (DO), and Data User (DU). The main responsibilities of the three entities in the system model are as follows:
1) Data Owner (DO): DO encrypts medical images using lightweight encryption and uploads them to the cloud. The images are enhanced via contrast-limited adaptive histogram equalization and processed to generate triplet deep hashing codes. DO then creates an image feature index set by linking image numbers with hashing codes, and uploading it to the cloud as a hash index table.
2) Cloud Server (CS): CS stores the encrypted images and the hash index table. Upon receiving a query, CS returns r semantically similar encrypted images to DU.
3) Data User (DU): DU generates a triplet deep hashing code for the query image, and submits it to CS, which performs similarity matching and returns r encrypted images. DU decrypts them and identifies the top-k similar images.
CXR images are crucial in medical diagnosis, and the image quality significantly affects accurate diagnosis. However, CXR images often suffer from insufficient contrast or uneven illumination, which affects the accuracy of feature extraction [33]. Therefore, to better display the details in CXR images and enhance the precision of feature extraction, this paper utilizes the contrast-limited adaptive histogram equalization technique applicable to color images [34] to enhance CXR images. This method dynamically adjusts the enhancement level based on the local contrast of CXR images, thereby better-preserving image details and avoiding issues such as excessive noise enhancement or detail loss that may occur with traditional grayscale histogram equalization methods [35].
The detailed process for enhancing CXR images includes the following steps:
Step 1: Convert the color image to an appropriate color space. Represented as Eq. (1):
Step 2: Separate the luminance channel. The luminance channel Y is extracted from the YUV color space, as shown in Eq. (2):
where W is the weight factor, where Wr, Wg, and Wb are the weights for the red, green, and blue channels.
Step 3: Enhance the image under the luminance channel using the contrast-limited adaptive histogram equalization technique applicable to color images.
Step 4: Reconstruct the luminance channel.
Step 5: Convert the enhanced color image back to the RGB color space, enabling it to serve as the correct input image for subsequent network models.
Fig. 2 shows the comparative images before and after the enhanced images of the proposed method.

Figure 2: Enhanced image vs. the original: (a) Original Image, (b) Enhanced Image
Fig. 2 clearly shows that the enhanced image exhibits clearer details compared to the pre-enhanced (unenhanced original CXR image) counterpart, without excessive distortion. This indicates that the method is effective and suitable for enhancing CXR and similar medical images.
3.3 Construction of Triplet Deep Hashing with Multi-Attention Mechanism
Within the feature extraction module shown in Fig. 1, based on the image characteristics of CXR images, this paper combines the channel attention mechanism [36] with the designed enhanced spatial attention mechanism. This combination enables the neural network to focus more on locally important features during feature extraction. Additionally, the triplet loss function is utilized to improve the discriminative ability of CXR image features and reduce the redundancy in the embedding space, thereby achieving triplet deep hashing. Fig. 3 shows the network architecture of MATDH.

Figure 3: The network structure of MATDH
During feature extraction, a UNet-based architecture is used [37], incorporating a dual attention mechanism in residual blocks to emphasize important local features of CXR images. The training combines triplet loss and reconstruction loss to enhance feature distinctiveness. As shown in Fig. 3, the feature encoder includes convolutional layers, maximum pooling layers, downsampling, residual blocks with channel and enhanced spatial attentions (RCESA), average pooling layers, full connectivity layers, and hash layers.
For the i-th CXR image, two large-kernel 2D convolutional layers are applied to capture local features, followed by a max pooling operation to downsize the image for better focus on local content. Four RCESA modules are then stacked to extract features, with downsampling after the first three modules to capture more abstract high-level features. The feature map from the fourth module is processed through an average pooling layer and a dense layer, producing a 1000-dimensional feature vector fi.
During feature decoding, alternating maximum pooling and upsampling operations reconstruct the feature maps from the encoding phase. A composite loss function integrates triplet loss, enhancing feature discriminability by ensuring similar samples are closer together than different ones. The multi-attention mechanism highlights important regions, allowing the network to focus on key features, thereby improving the distinction between similar and dissimilar samples. This approach retains essential information while minimizing redundancy. After minimizing the loss through the fully connected layer, a k-bit deep hash code representing CXR features is generated in the hashing layer, which includes a full connectivity layer and a hash function. The resulting hash codes are stored in a hash index table for future retrieval, the hash codes of the i-th CXR image can be represented as Eq. (3):
where bi is the k-bit hash code of the i-th CXR image Xi, bi ∈ {0, 1}, ω represents the trainable mapping in the fully connected layer, and h(x) denotes the hash function, defined as Eq. (4):
3.3.1 Multi-Attention Mechanism
To concurrently and adaptively determine the importance of key regions and various channels in CXR images, this paper combines the channel attention mechanism with the designed enhanced spatial attention mechanism in the residual block to form a multi-attention mechanism to achieve this goal. Fig. 4 shows the structure of the residual block (RCESA) with the multi-attention mechanism.

Figure 4: The structure of RCESA
In Fig. 4, incorporating residual blocks into the network allows for image feature extraction and helps tackle the issue of network degradation. Taking the intermediate feature vector Min extracted by the neural network as input, the weight of important areas is first enhanced through the enhanced spatial attention mechanism, and then its output serves as the basis for the channel attention mechanism operation to improve the accuracy of feature extraction. Specifically, within each residual block, different-sized convolutional layers are first used to extract multi-scale features to obtain the multi-scale feature vector MF. Then, the extracted features are subjected to max-pooling operation (used for further downsampling of the feature map after partial feature extraction to reduce network computational load and enhance the network model’s abstraction ability for important features of CXR images). Next, features are further extracted through a 3 × 3 convolutional layer, and bilinear interpolation is used for upsampling to match the feature map size with that of the input. The upsampled operation is then combined with the preceding feature map to generate the feature fusion vector MA. Feature fusion is performed through 1 × 1 convolutional operation, allowing the network to more effectively utilize features of varying scales. Ultimately, the feature vector after fusion is normalized to the [0, 1] range with the sigmoid activation function to obtain the weight matrix m, which is subsequently multiplied by the input feature Min to generate the weighted feature map as the output Mout of enhanced spatial attention operation, achieving enhanced spatial attention to enable the network model to utilize crucial regional information in input CXR images more effectively. This series of operations is represented by Eq. (5):
Subsequently, the feature vector Mout processed by the enhanced spatial attention mechanism is used as the input of the channel attention mechanism. Initially, the convolutional feature vector MG is obtained through two convolutional layers. This is followed by max-pooling and average-pooling to generate two vectorized representations. These vectors are then inputted into a shared multi-layer perceptron (MLP). The representations generated by the MLP are summed to form an attention vector, which is then multiplied with MG to produce the feature map MH with channel attention. This series of operations is represented by Eq. (6):
where σ is the sigmoid function, ϕ represents a trainable transformation within the MLP, ω1 and ω2 respectively represent max-pooling and average-pooling operations. After this step, the shortcut connection defined by Eq. (7) is applied to obtain the output of the RCESA.
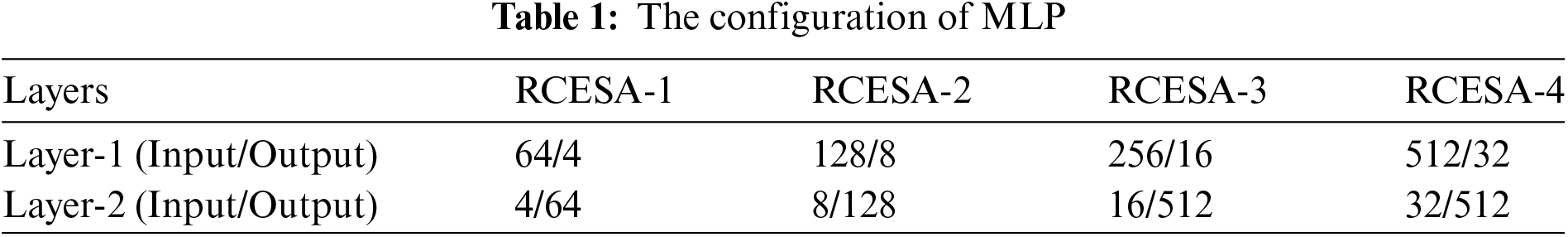
Each RCESA’s MLP comprises two convolutional layers with a kernel size of 1 × 1, referred to as Layer-1 and Layer-2. The detailed settings of the MLP are shown in Table 1.
To understand the complex relationships among the input samples, triplet constraints serve as the loss function guiding the training of deep networks, improving the distinguishing ability of the obtained deep feature hash codes. The post-training CXR images are represented as {X1, …, Xi, …, XM}, and the associated class labels are represented as {L1, …, Li, …, LM}, where Li∈{1, …, c}, and c represents the number of categories. Fig. 5 shows an example of triplet training.

Figure 5: An illustration of triplet learning
As shown in Fig. 5, the triplet loss is designed to increase similarity among samples from the same class while decreasing similarity between samples from different classes [38]. Mathematically, given a triplet unit {q, p, n}, where q represents the query sample, p represents the positive sample, and n represents the negative sample. The goal of triplet loss is to reduce the following items:
where d(q, p) is the distance between the query and the positive sample, d(q, n) is the distance between the query and the negative sample, and m represents a preset positive margin value.
In this study, given an image
where T indicates a total number of triplet units across the training images,
The specific learning algorithm for MATDH is shown in Algorithm 1.

3.4 Lightweight Image Encryption Algorithm
The construction of the encrypted image database utilizes a lightweight CXR image encryption method as shown in Fig. 6 to encrypt the original images.

Figure 6: Lightweight CXR image encryption processing
To overcome the shortcomings of simple encryption algorithms, this paper requires randomization of the original image before encrypting the coefficients of the Discrete Cosine Transform (DCT) in the image frequency domain. Firstly, in the spatial domain, use the two-dimensional Arnold transform to scramble the pixel blocks of the ordinary original image for encryption. Then, segment the image and perform DCT transformation on each image block to transition from the spatial representation to the frequency representation. Next, use the two-dimensional Logistic chaotic mapping to encrypt the DCT coefficients of each block. Finally, after the inverse, Discrete Cosine Transform (IDCT), secondary encryption uses the PRESENT algorithm with a 128-bit key to produce the final encrypted image. The specific steps for lightweight CXR image encryption are as follows:
Step 1: Apply a two-dimensional Arnold transformation to scramble and encrypt the pixels of the original image. With the definition of the two-dimensional Arnold transformation as shown in Eq. (10):
where b and c are arbitrary positive integers, and the matrix’s determinant needs to equal 1 to preserve the same region in the CXR image, the two-dimensional Arnold transformation iterates m times, generating a random image in each iteration. The values of parameters b, c, and m serve as the encryption key.
Step 2: Divide the scrambled image into 8 × 8 pixel blocks.
Step 3: Perform DCT processing on the segmented image blocks to transform them from the spatial domain to the frequency domain. The expression for the two-dimensional DCT of an M × N matrix is as shown in Eq. (11):
Step 4: Encrypt the DCT coefficients of each pixel block using two-dimensional Logistic chaotic mapping. The definition of two-dimensional Logistic chaotic mapping is as shown in Eq. (12):
where xn and yn are the two-state components at time n, r1, and r2 are control parameters, and δ is the coupling function.
Step 5: Perform IDCT on each pixel block to obtain the preliminarily encrypted image. The definition of the two-dimensional IDCT of an M × N matrix is as shown in Eq. (13):
Step 6: Utilize the PRESENT algorithm to perform secondary encryption on the preliminarily encrypted image, obtaining the final encrypted image.
4 Simulation Results and Analysis
The experimental hardware environment consists of CPU: Intel(R) Core(TM) i7-13700H CPU @2.20 GHz, GPU: NVIDIA GeForce RTX 4060 Laptop GPU, memory: 12 GB, software environment: Windows11, JetBrains PyCharm Community Edition 2023.2x64, Anaconda. The deep learning framework is PyTorch. This paper uses Adam to optimize the objective function, with a learning rate set to 0.001, batch size set to 32, 30 training iterations, and the margin threshold in triplet loss set to 0.2.
Dataset: This experiment evaluates the suggested approach using the public COVIDx dataset [39], which includes CXR images related to COVID-19. To enhance the model’s generalization ability during training, the 29,986 CXR images are divided into multiple subsets, which are alternately used as the test set to reduce the risk of overfitting. Additionally, the independent test set includes 400 medical images, including 200 negative images and 200 positive images for COVID-19.
Data preprocessing: CXR images are resized to 256 × 256 for uniformity, randomly cropped to 224 × 224, and horizontally flipped to enhance the model’s ability to handle spatial variations and orientation of chest structures.
4.2 Network Model and Performance Analysis
In this paper, experimental adjustments are made to the parameters of the network model, and accuracy and loss are evaluated. After multiple experiments, the deep network model based on the UNet architecture with optimal performance is obtained. Fig. 7 shows the training and testing curves for the network model.

Figure 7: The training and testing curves for the network model
As can be seen from Fig. 7, it can be observed that the training and testing accuracy curves initially converge but stabilize after a certain number of iterations, with no significant jumps or drastic fluctuations, indicating good generalization capability of the network model and no overfitting. After 30 iterations of training, the training accuracy is 94.56%, and the testing accuracy is 94.42%. Therefore, the network model adopted in this paper exhibits high performance and accuracy, with good retrieval performance.
4.3 Retrieval Performance Analysis
4.3.1 Retrieval Accuracy Analysis
Mean Average Precision (mAP) is commonly used to evaluate object detection models, considering the average precision (AP) across different categories; a higher mAP indicates more similar images at the top of the retrieved list. Additionally, P@H ≤ 2 is a key measure in image retrieval, emphasizing the use of shorter hash codes to capture similarity within the feature space. This paper calculates the average precision for instances where the Hamming distance between the query and database images is no more than 2. To mitigate bias from model initialization, five independent experiments were conducted.
To validate the CXR image retrieval performance of the proposed method, a comparison was conducted between our MATDH method and six other deep hashing methods, namely TCDH [8], SWTH [12], ATH [22], DPN [40], ASH [41], and DBDH [42]. For fairness, the comparison was conducted using the same data and parameter settings as MATDH. Table 2 shows mAP values for different methods across various hash code lengths.
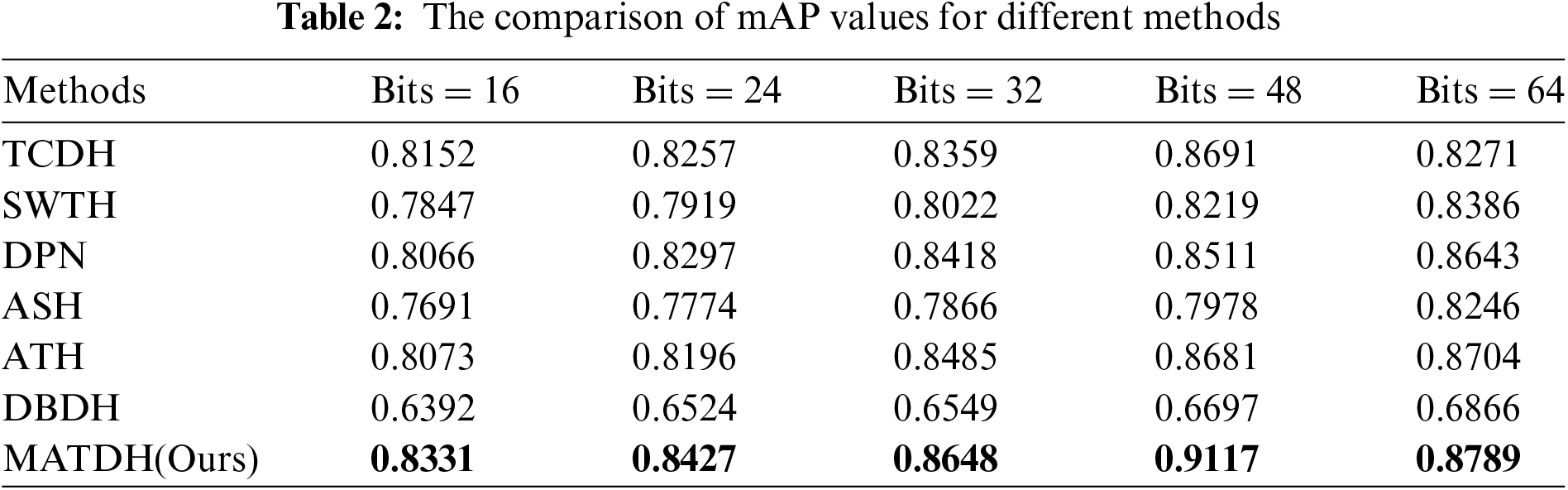
As can be seen from Table 2, it can be observed that MATDH outperforms the compared existing deep hashing methods. For example, with a 48-bit hash code length, MATDH (0.9117) enhances the mAP by approximately 4.2% over the top-performing deep learning technique, TCDH (0.8691). This is because the features extracted from CXR images after image enhancement under the dual attention mechanism are more accurate, and guided by the triplet loss function, the network can extract more discriminative features. Furthermore, after performing a t-test distribution calculation comparing the mAP of the MATDH method with that of six other methods, it was found that there is no significant difference between MATDH and ATH, while significant differences exist with the other five methods. This indicates that the MATDH method has statistical significance, providing important evidence for selecting and optimizing medical image retrieval methods. Fig. 8 shows the curve of P@H ≤ 2 for MATDH compared to six other methods.

Figure 8: Line graph comparing P@H ≤ 2 across various methods
As can be seen from Fig. 8, P@H ≤ 2 curves further illustrate the method’s effectiveness within a Hamming radius of 2. The accuracy curve in Fig. 8 shows that the proposed MATDH method consistently outperforms other advanced methods across hash code lengths from 16 to 64 bits. At a hash code length of 48 bits, the accuracy is approximately 20% higher than that of the DPN method. This suggests that the MATDH method is capable of retrieving a greater number of images from the same category when the Hamming radius is set to 2. Moreover, in comparison to other methods, the precision attained by MATDH consistently exceeds 0.85 as the number of hash bits increases. This highlights MATDH’s effectiveness in CXR image retrieval and its resilience to Hamming sorting.
Additionally, with a 48-bit hash code length, MATDH achieves the highest mAP values. This suggests that for CXR images, using 48-bit hash codes with MATDH can better preserve the main features of the images. Therefore, the MATDH method in this paper adopts a 48-bit deep hash code.
4.3.2 The Recall Rate, Precision Analysis
Besides relying on mAP, in medical image retrieval, accuracy for items positioned highest in the ranking is often a key priority for users. Therefore, this paper also uses the recall rate in the top-K retrieval results (Recall@K) and the precision of the top-N retrieval results (P@N) to evaluate the effectiveness of CXR image retrieval methods [8].
Fig. 9 shows the recall rate curves of top-k images across various techniques using a consistent 48-bit hash code length, and Table 3 presents the comparison results of P@1, P@5, and P@10 obtained through various methods using 48-bit deep hash codes in the COVIDx dataset. The XMIR method [43], which is not a deep hash method, is not concerned with the issue of hash code length.

Figure 9: Recall curve for different methods
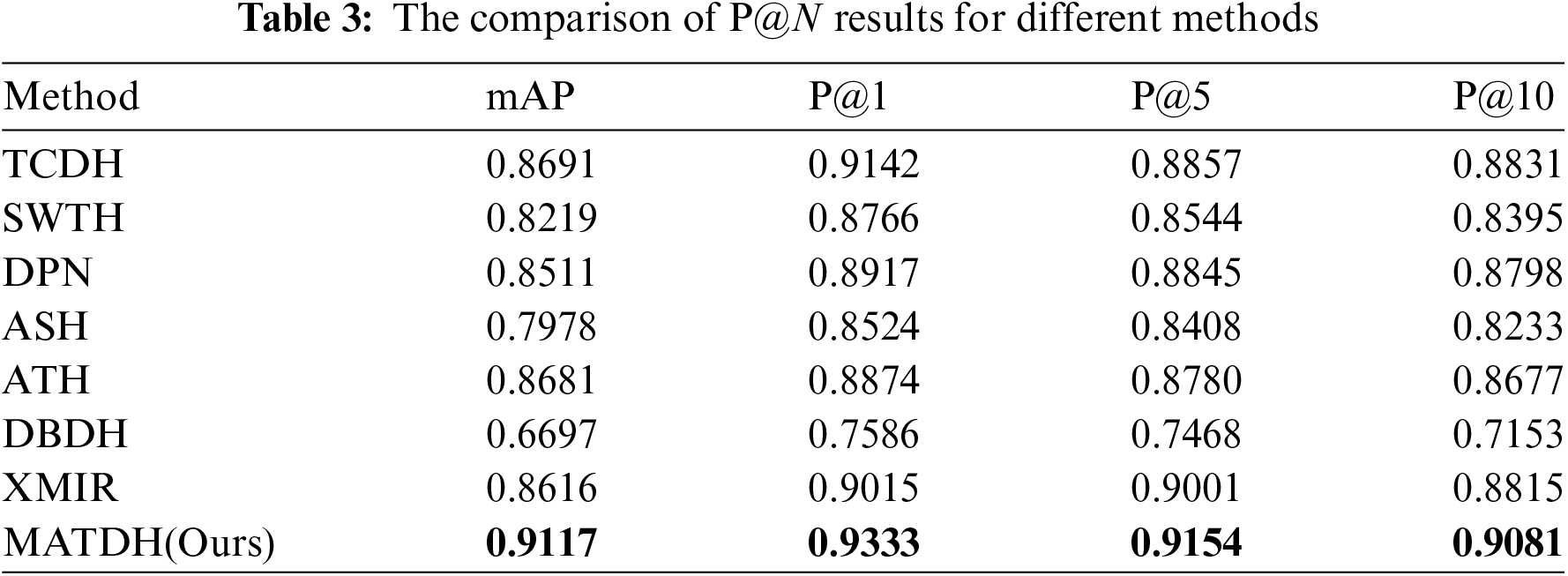
As shown in Fig. 9, the recall rate of MATDH remains consistently higher than that of other methods as the number of returned CXR images increases from 0 to 200. For example, when 180 images are returned, the recall rate is approximately 30% higher than that of the DBDH method. This indicates that MATDH retrieves a higher number of relevant images, consistent with the comparative analysis results.
As can be seen from Table 3, it is evident that when the limit on returned images is set, MATDH outperforms other methods. This is because, under the effect of the multi-attention mechanism, with the iterative training of deep neural networks, the constructed hash codes can more accurately represent images, thereby improving the accuracy of CXR image retrieval.
Although this study mainly uses the COVIDx dataset for experiments, the proposed MATDH method has the potential for clinical application. It can serve as an auxiliary diagnostic tool, helping doctors make faster and more accurate judgments on COVID-19 infections. The model can be integrated into hospital imaging systems for real-time radiological image analysis, improving early detection accuracy. Additionally, the proposed MATDH method can assist doctors in quickly and accurately retrieving medical imaging data, reducing the workload of data queries during treatment, especially when medical resources are limited.
This section uses the 48-bit deep hash codes to display the 10 most similar CXR images obtained through various methods. Fig. 10 shows the visualization results. The images within the red boxes represent those returned in the wrong categories compared to the query image.

Figure 10: The visualization results of the top 10 CXR images are recognized by different methods
As can be seen from Fig. 10, it can be observed that most of the images returned by MATDH compared to other methods are correct. This indicates that the retrieval results of MATDH are highly accurate, demonstrating that the multiple attention mechanism and optimized loss function used in MATDH outperform other advanced methods. Additionally, the visualization results of different methods further confirm the accuracy of the P@10 results.
4.5.1 Impact of Network Architecture and CXR Image Enhancement Methods
To objectively evaluate the network model and framework of this paper’s method, disintegration experiments are conducted through the removal or substitution of modules in the network architecture. For the network structure, the basic structure of ResNet18 in the UNet network used by the proposed method is replaced with AlexNet [44] and ResNet34 [45], and these two variants are respectively denoted as MATDH-AlexNet and MATDH-ResNet34. Furthermore, before extracting features from the deep network, the approach described in this paper does not enhance the original images, which are denoted as MATDH-NoEnhanced, and experiments are conducted using 48-bit deep hashing codes. The detailed data can be found in Table 4, where the top entries are emphasized in bold.
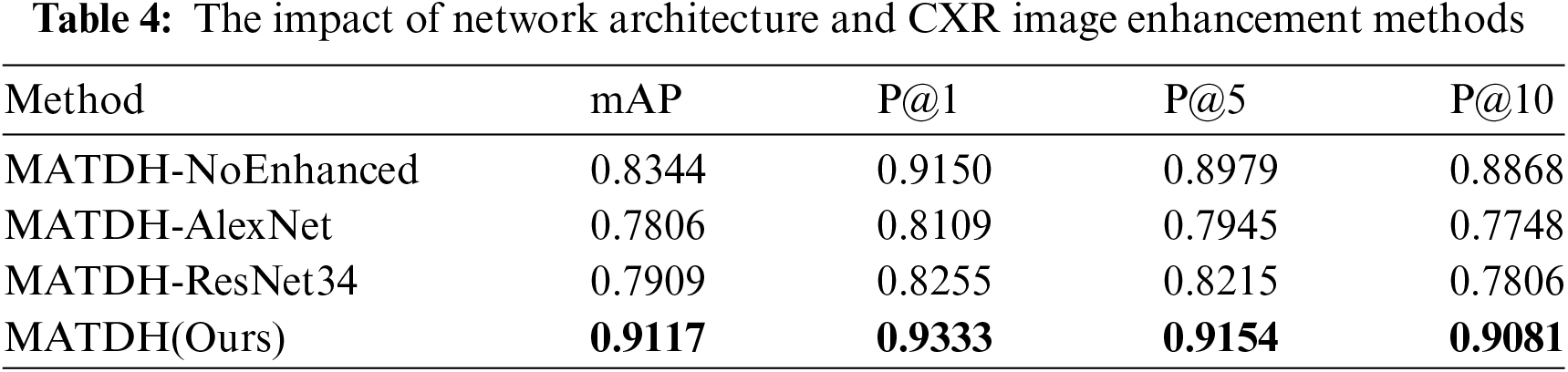
As can be seen from Table 4, MATDH enhances the retrieval performance of input CXR images by utilizing image enhancement, and it outperforms variant networks with different frameworks.
4.5.2 The Impact of Multi-Attention Mechanism
To examine how the dual attention mechanism within the RCESAs module influences the outcomes of CXR image retrieval experiments, this section modified the attention mechanisms used in the RCESAs module of the MATDH framework. One of the variants utilizes only the channel attention mechanism, referred to as MATDH-CA, while the other variant utilizes only the enhanced spatial attention mechanism, referred to as MATDH-ESA. Fig. 11 shows the comparison between MATDH and its variants that utilize various attention mechanisms for CXR image recovery from the COVIDx dataset, using a 48-bit hash code length, with the top outcomes emphasized in bold.
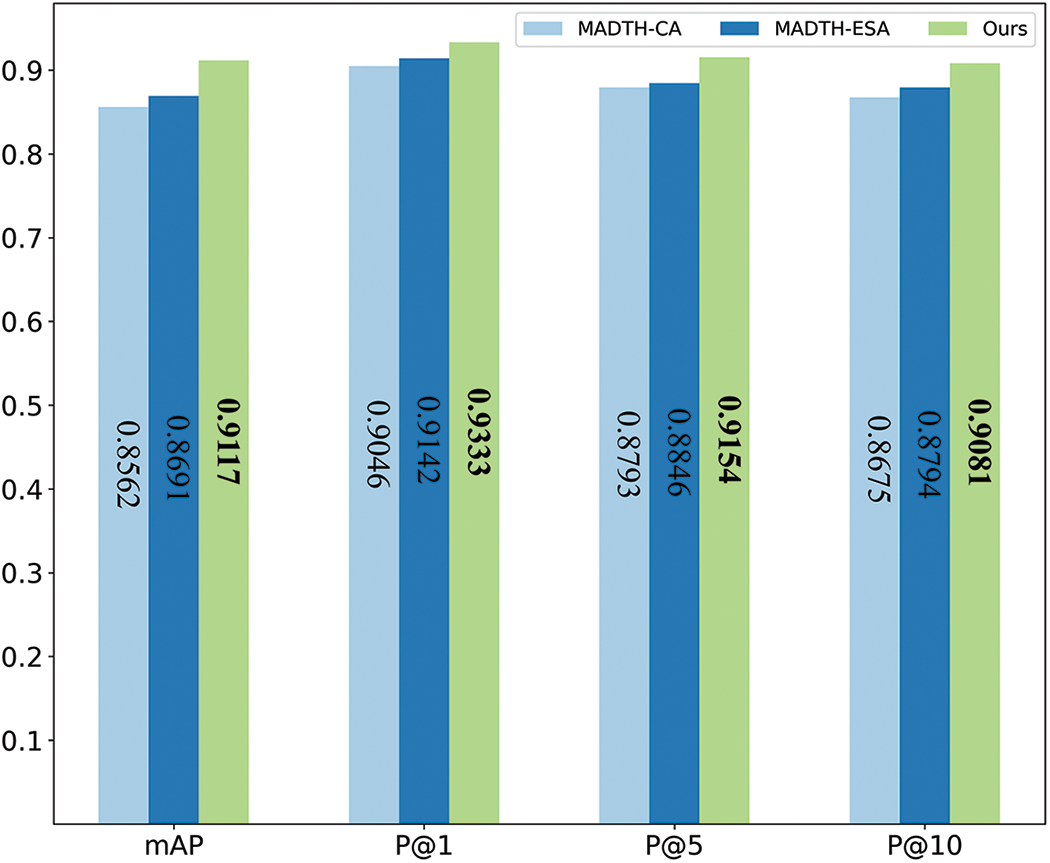
Figure 11: The comparison of MATDH and its variants
According to the results in Fig. 11, MATDH with a dual-attention mechanism outperforms the standalone CA and ESA mechanisms in retrieval performance. The specific contributions to this performance improvement can be summarized as follows:
1) Feature enhancement: The dual-attention mechanism effectively captures and enhances the correlation between useful feature channels, enabling the model to better identify features related to lesions in CXR images, thereby improving retrieval accuracy.
2) Redundant information suppression: This mechanism effectively reduces the influence of unnecessary data and noisy channels in CXR images, allowing the network to focus more on key information, thus enhancing the clarity of feature representation.
3) Dynamic attention adjustment: The dual-attention mechanism dynamically adjusts the focus of the deep neural network on different regions of CXR images, enabling the model to better capture important spatial features. This flexibility allows MATDH to maintain high retrieval performance across a diverse set of input images.
4) Combined effect: By integrating the above factors, the dual-attention mechanism significantly enhances the overall performance of MATDH, demonstrating its effectiveness and superiority in medical image retrieval tasks.
4.6 Encryption Performance Analysis
The cryptographic performance analysis in this section is demonstrated using three CXR images selected from the COVIDx dataset, as shown in Fig. 12, referred to as CXR-1, CXR-2, and CXR-3.

Figure 12: The three CXR images were used for encryption analysis: (a) CXR-1; (b) CXR-2; (c) CXR-3
4.6.1 Information Entropy Analysis
In cryptography, information entropy indicates the degree of unpredictability in image data. For a perfectly random image, it is essential that all grayscale values have an equal probability of occurrence. The formula for calculating information entropy is shown in Eq. (14):
where L represents the total number of grayscale levels in the image, while p(i) denotes the probability of occurrence for grayscale value i. For an image with p(i) = 256, when each pixel value in the image has an equal probability of occurrence, the entropy of the image can reach its maximum value of 8. A higher entropy indicates greater randomness in the image, leading to better encryption performance. Table 5 shows the compares the entropy between the original and encrypted images, both with dimensions of 512 × 512.

As can be seen from Table 5, the average entropy of the encrypted images generated by the encryption method presented in this paper is 7.9993, approaching the optimal value of 8. This suggests that the encryption technique presented here achieves strong performance, resulting in encrypted images that show significant randomness.
4.6.2 Comparative Performance Analysis with Existing Encryption Schemes
As the encryption method described in this study is a secure lightweight encryption algorithm, this section compares and analyzes it with existing schemes based on three evaluation indicators reflecting the lightweight and security of encryption algorithms: key space size, encryption speed, and information entropy. Table 6 shows the performance comparing results between the encryption method introduced here and various medical image encryption schemes [46–48].
From Table 6, it is evident that the encryption method in this paper shares the same key space as in Castro et al. [46], but is significantly smaller than that in Abdelfatah et al. [47] and Inam et al. [48]. This suggests that the proposed method is more resource-efficient, making it suitable for lightweight encryption of medical images. Although its key space is smaller, it still provides sufficient security (greater than 2100), enough to resist exhaustive attacks [48]. The encryption speed is higher than in Castro et al. [46] and Abdelfatah et al. [47] and slightly lower than in Inam et al. [48], due to frequency domain operations. These operations, while slowing down the process, offer better attack resistance. Additionally, the proposed method has higher entropy, indicating strong randomness and resistance to entropy attacks, making it a secure lightweight encryption algorithm for medical images compared to other schemes [46–48].
4.6.3 The Analysis Resists Different Types of Attacks
Building on the insights from the previous two sections, this part demonstrates the advantages of the encryption algorithm in resisting various types of attacks.
First, as discussed in Section 4.6.1, it is understood that the ciphertext generated by this algorithm exhibits high entropy, meaning that it is statistically close to a random distribution. This randomness complicates the process for attackers trying to obtain useful information through frequency analysis, as well as through statistical attacks such as known-plaintext or chosen-plaintext attacks. The existence of high-entropy ciphertext significantly enhances the algorithm’s defense against these forms of attacks.
Secondly, the analysis in Section 4.6.2 shows that the key space of this encryption algorithm is 2256, and such an extensive key range renders brute-force attacks virtually impossible. Even if attackers possess powerful computational resources, attempting to exhaust all possible keys within a reasonable time frame is still unlikely, providing additional security for the algorithm.
In summary, based on the aforementioned points, this encryption algorithm can be considered to possess strong security, effectively resisting brute-force attacks, statistical attacks, known-plaintext attacks, chosen-plaintext attacks, and chosen-ciphertext attacks, among other common attack methods. The high-entropy characteristic and the vast key space complement each other, ensuring the reliability and robustness of the encryption algorithm under various threats.
This paper proposes a secure technique for retrieving medical images utilizing a multi-attention mechanism and triplet deep hashing, addressing issues like poor feature extraction, low retrieval precision, and inadequate security in existing solutions. The approach enhances CXR images using contrast-limited adaptive histogram equalization, which reduces noise and highlights details for better feature extraction. The multi-attention mechanism dynamically allocates channel attention and focuses on important local features, improving retrieval accuracy through deep hash codes. A lightweight CXR image encryption method enhances system security while maintaining efficiency. A limitation is the small dataset of CXR disease types, with future work aimed at improving retrieval accuracy and security on more complex datasets.
Acknowledgement: The authors are grateful to all the editors and anonymous reviewers for their comments and suggestions and thank all the members who have contributed to this work with us.
Funding Statement: This work is supported by the National Natural Science Foundation of China (No. 61862041).
Author Contributions: The first author Shaozheng Zhang gives the main conception and writes of this paper. The second author Qiuyu Zhang mainly reviews and proposes revisions to the manuscript. The third author Jiahui Tang and the fourth author Ruihua Xu conducted the data collection. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data supporting the findings of this study can be obtained from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. M. K. Hasan, S. Islam, R. Sulaiman, S. Khan, A. A. Hashim and S. Habib, “Lightweight encryption technique to enhance medical image security on internet of medical things applications,” IEEE Access, vol. 9, pp. 47731–47742, 2021. doi: 10.1109/ACCESS.2021.3061710. [Google Scholar] [CrossRef]
2. P. Shamna, V. K. Govindan, and K. A. A. Nazeer, “Content-based medical image retrieval by spatial matching of visual words,” J. King Saud Univ.-Comput. Inf. Sci., vol. 34, no. 2, pp. 58–71, 2022. doi: 10.1016/j.jksuci.2018.10.002. [Google Scholar] [CrossRef]
3. Ş. Öztürk, E. Çelik, and T. Çukur, “Content-based medical image retrieval with opponent class adaptive margin loss,” Inf. Sci., vol. 637, no. 1, 2023, Art. no. 118938. doi: 10.1016/j.ins.2023.118938. [Google Scholar] [CrossRef]
4. V. H. Vu, “Content-based image retrieval with fuzzy clustering for feature vector normalization,” Multimed. Tools Appl., vol. 83, no. 2, pp. 4309–4329, 2024. doi: 10.1007/s11042-023-15215-1. [Google Scholar] [CrossRef]
5. X. Zhang, C. Bai, and K. Kpalma, “OMCBIR: Offline mobile content-based image retrieval with lightweight CNN optimization,” Displays, vol. 76, no. 5, 2023, Art. no. 102355. doi: 10.1016/j.displa.2022.102355. [Google Scholar] [CrossRef]
6. X. Chen et al., “Recent advances and clinical applications of deep learning in medical image analysis,” Med. Image Anal., vol. 79, 2022, Art. no. 102444. doi: 10.1016/j.media.2022.102444. [Google Scholar] [PubMed] [CrossRef]
7. S. M. Alizadeh, M. S. Helfroush, and H. Müller, “A novel Siamese deep hashing model for histopathology image retrieval,” Expert Syst. Appl, vol. 225, no. 4, 2023, Art. no. 120169. doi: 10.1016/j.eswa.2023.120169. [Google Scholar] [CrossRef]
8. L. Wang, Q. Wang, X. Wang, Y. Ma, L. Zhang and M. Liu, “Triplet-constrained deep hashing for chest X-ray image retrieval in COVID-19 assessment,” Neural Netw., vol. 173, no. 12, 2024, Art. no. 106182. doi: 10.1016/j.neunet.2024.106182. [Google Scholar] [PubMed] [CrossRef]
9. A. Hussain, H. Li, D. Ali, M. Ali, F. Abbas and M. Hussain, “An optimized deep supervised hashing model for fast image retrieval,” Image Vis. Comput., vol. 133, no. 10, 2023, Art. no. 104668. doi: 10.1016/j.imavis.2023.104668. [Google Scholar] [CrossRef]
10. C. Cui, H. Huo, and T. Fang, “Deep hashing with multi-central ranking loss for multi-label image retrieval,” IEEE Signal Process. Lett., vol. 30, pp. 135–139, 2023. doi: 10.1109/LSP.2023.3244516. [Google Scholar] [CrossRef]
11. D. Wu, Q. Dai, B. Li, and W. Wang, “Deep uncoupled discrete hashing via similarity matrix decomposition,” ACM Trans. Multimed. Comput., Commun., Appl., vol. 19, no. 1, 2023, Art. no. 22. doi: 10.1145/3524021. [Google Scholar] [CrossRef]
12. L. Peng, J. Qian, C. Wang, B. Liu, and Y. Dong, “Swin transformer-based supervised hashing,” Appl. Intell., vol. 53, no. 14, pp. 17548–17560, 2023. doi: 10.1007/s10489-022-04410-6. [Google Scholar] [CrossRef]
13. H. Zhou, Q. Qin, J. Hou, J. Dai, L. Huang and W. Zhang, “Deep global semantic structure-preserving hashing via corrective triplet loss for remote sensing image retrieval,” Expert. Syst. Appl., vol. 238, no. 2, 2024, Art. no. 122105. doi: 10.1016/j.eswa.2023.122105. [Google Scholar] [CrossRef]
14. D. Li, Q. Lü, X. Liao, T. Xiang, J. Wu and J. Le, “AVPMIR: Adaptive verifiable privacy-preserving medical image retrieval,” IEEE Trans. Depend. Secure Comput., vol. 21, no. 5, pp. 4637–4651, 2024. doi: 10.1109/TDSC.2024.3355223. [Google Scholar] [CrossRef]
15. K. N. Singh, O. P. Singh, A. K. Singh, and A. K. Agrawal, “WatMIF: Multimodal medical image fusion-based watermarking for telehealth applications,” Cogn. Comput., vol. 16, no. 4, pp. 1947–1963, 2024. doi: 10.1007/s12559-022-10040-4. [Google Scholar] [PubMed] [CrossRef]
16. J. Chang, Q. Ren, Y. Ji, M. Xu, and R. Xue, “Secure medical data management with privacy-preservation and authentication properties in smart healthcare system,” Comput. Netw., vol. 212, no. 1, 2022, Art. no. 109013. doi: 10.1016/j.comnet.2022.109013. [Google Scholar] [CrossRef]
17. E. Özbay and F. A. Özbay, “Interpretable pap-smear image retrieval for cervical cancer detection with rotation invariance mask generation deep hashing,” Comput. Biol. Med., vol. 154, no. 2, 2023, Art. no. 106574. doi: 10.1016/j.compbiomed.2023.106574. [Google Scholar] [PubMed] [CrossRef]
18. L. Xu, X. Zeng, B. Zheng, and W. Li, “Multi-manifold deep discriminative cross-modal hashing for medical image retrieval,” IEEE Trans. Image Process., vol. 31, no. 4, pp. 3371–3385, 2022. doi: 10.1109/TIP.2022.3171081. [Google Scholar] [PubMed] [CrossRef]
19. Y. Chen, Y. Tang, J. Huang, and S. Xiong, “Multi-scale triplet hashing for medical image retrieval,” Comput. Biol. Med., vol. 155, 2023, Art. no. 106633. doi: 10.1016/j.compbiomed.2023.106633. [Google Scholar] [PubMed] [CrossRef]
20. S. Suganyadevi, V. Seethalakshmi, and K. Balasamy, “A review on deep learning in medical image analysis,” Int. J. Multimed. Inf. Retr., vol. 11, no. 1, pp. 19–38, 2022. doi: 10.1007/s13735-021-00218-1. [Google Scholar] [PubMed] [CrossRef]
21. Y. Zhang, F. Xie, X. Song, Y. Zheng, J. Liu and J. Wang, “Dermoscopic image retrieval based on rotation-invariance deep hashing,” Med. Image Anal., vol. 77, no. 5, 2022, Art. no. 102301. doi: 10.1016/j.media.2021.102301. [Google Scholar] [PubMed] [CrossRef]
22. J. Fang, H. Fu, and J. Liu, “Deep triplet hashing network for case-based medical image retrieval,” Med. Image Anal., vol. 69, no. 4–5, 2021, Art. no. 101981. doi: 10.1016/j.media.2021.101981. [Google Scholar] [PubMed] [CrossRef]
23. Ş. Öztürk, “Class-driven content-based medical image retrieval using hash codes of deep features,” Biomed Signal Process. Control., vol. 68, no. 2, 2021, Art. no. 102601. doi: 10.1016/j.bspc.2021.102601. [Google Scholar] [CrossRef]
24. P. Huang, X. Zhou, Z. Wei, and G. Guo, “Energy-based supervised hashing for multimorbidity image retrieval,” in Med. Image Comput. Comput. Assist. Interv.—MICCAI 2021: 24th Int. Conf., Strasbourg, France, Springer International Publishing, 2021, pp. 205–214. doi: 10.1007/978-3-030-87240-3_20. [Google Scholar] [CrossRef]
25. Y. Qi, J. Gu, Y. Zhang, Y. Jia, and Y. Su, “Unsupervised deep hashing by joint optimization for pulmonary nodule image retrieval,” Measurement, vol. 159, no. 1, 2020, Art. no. 107785. doi: 10.1016/j.measurement.2020.107785. [Google Scholar] [CrossRef]
26. S. R. Dubey, “A decade survey of content based image retrieval using deep learning,” IEEE Trans. Circuits Syst. Video Technol., vol. 32, no. 5, pp. 2687–2704, 2021. doi: 10.1109/TCSVT.2021.3080920. [Google Scholar] [CrossRef]
27. A. Hafsa, M. Gafsi, J. Malek, and M. Machhout, “FPGA implementation of improved security approach for medical image encryption and decryption,” Sci. Program., vol. 2021, pp. 1–20, 2021. doi: 10.1155/2021/6610655. [Google Scholar] [CrossRef]
28. S. Haddad, G. Coatrieux, A. Moreau-Gaudry, and M. Cozic, “Joint watermarking-encryption-JPEG-LS for medical image reliability control in encrypted and compressed domains,” IEEE Trans. Inf. Foren. Secur., vol. 15, pp. 2556–2569, 2020. doi: 10.1109/TIFS.2020.2972159. [Google Scholar] [CrossRef]
29. C. Guo, J. Jia, K. K. R. Choo, and Y. Jie, “Privacy-preserving image search (PPISSecure classification and searching using convolutional neural network over large-scale encrypted medical images,” Comput. Secur., vol. 99, no. 11, 2020, Art. no. 102021. doi: 10.1016/j.cose.2020.102021. [Google Scholar] [CrossRef]
30. S. Kumar, S. K. Agarwal, and S. S. Ahmad, “A secure medical image retrieval technique using encrypted query image,” in 2022 2nd Int. Conf. Emerg. Front. Electr. Electron. Technol. (ICEFEET), Patna, India, IEEE, 2022, pp. 1–4. doi: 10.1109/ICEFEET51821.2022.9847826. [Google Scholar] [CrossRef]
31. D. Zhu, H. Zhu, X. Wang, R. Lu, and D. Feng, “An accurate and privacy-preserving retrieval scheme over outsourced medical images,” IEEE Trans. Serv. Comput., vol. 16, no. 2, pp. 913–926, 2023. doi: 10.1109/TSC.2022.3149847. [Google Scholar] [CrossRef]
32. G. Cai, X. Wei, and Y. Li, “Privacy-preserving CNN feature extraction and retrieval over medical images,” Int. J. Intell. Syst., vol. 37, no. 11, pp. 9267–9289, 2022. doi: 10.1002/int.22991. [Google Scholar] [CrossRef]
33. T. Rahman et al., “Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images,” Comput. Biol. Med., vol. 132, no. 2, 2021, Art. no. 104319. doi: 10.1016/j.compbiomed.2021.104319. [Google Scholar] [PubMed] [CrossRef]
34. G. Alwakid, W. Gouda, and M. Humayun, “Deep learning-based prediction of diabetic retinopathy using CLAHE and ESRGAN for enhancement,” Healthcare, vol. 11, no. 6, 2023, Art. no. 863. doi: 10.3390/healthcare11060863. [Google Scholar] [PubMed] [CrossRef]
35. J. Xiong et al., “Application of histogram equalization for image enhancement in corrosion areas,” Shock Vib., vol. 2021, no. 1, pp. 1–13, 2021. doi: 10.1155/2021/8883571. [Google Scholar] [CrossRef]
36. Z. Qin, P. Zhang, F. Wu, and X. Li, “FcaNet: Frequency channel attention networks,” in Proc. IEEE/CVF Int. Conf. Comput. Vis. (ICCV), 2021, pp. 783–792. [Google Scholar]
37. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. 18th Int. Conf. Med. Image Comput. Comput.-Assist. Interv, 2015, pp. 234–241. doi: 10.1007/978-3-319-24574-4_28. [Google Scholar] [CrossRef]
38. D. Yao, J. Sui, M. Wang, E. Yang, Y. Jiaerken and N. Luo, “A mutual multi-scale triplet graph convolutional network for classification of brain disorders using functional or structural connectivity,” IEEE Trans. Med. Imag., vol. 40, no. 4, pp. 1279–1289, 2021. doi: 10.1109/TMI.2021.3051604. [Google Scholar] [PubMed] [CrossRef]
39. L. Wang, Z. Q. Lin, and A. Wong, “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Sci. Rep., vol. 10, no. 1, 2020, Art. no. 19549. doi: 10.1038/s41598-020-76550-z. [Google Scholar] [PubMed] [CrossRef]
40. L. Fan, K. W. Ng, C. Ju, T. Zhang, and C. S. Chan, “Deep polarized network for supervised learning of accurate binary hashing codes,” in IJCAI’20: Proc. Twenty-Ninth Int. Conf. Int. Joint Conf. Artif. Intell., 2021, pp. 825–831. [Google Scholar]
41. J. Fang, Y. Xu, X. Zhang, Y. Hu, and J. Liu, “Attention-based saliency hashing for ophthalmic image retrieval,” in Proc. IEEE Int. Conf. Bioinf. Biomed., 2020, pp. 990–995. doi: 10.1109/BIBM49941.2020.9313536. [Google Scholar] [CrossRef]
42. X. Zheng, Y. Zhang, and X. Lu, “Deep balanced discrete hashing for image retrieval,” Neurocomputing, vol. 403, no. 3, pp. 224–236, 2020. doi: 10.1016/j.neucom.2020.04.037. [Google Scholar] [CrossRef]
43. B. Hu, B. Vasu, and A. Hoogs, “X-MIR: Explainable medical image retrieval,” in Proc. IEEE/CVF Winter Conf. Appl. Comput. Vis. (WACV), 2022, pp. 440–450. [Google Scholar]
44. E. Cortés and S. Sánchez, “Deep Learning Transfer with AlexNet for chest X-ray COVID-19 recognition,” IEEE Lat. Am. Trans., vol. 19, no. 6, pp. 944–951, 2021. doi: 10.1109/TLA.2021.9451239. [Google Scholar] [CrossRef]
45. Q. Zhuang, S. Gan, and L. Zhang, “Human-computer interaction based health diagnostics using ResNet34 for tongue image classification,” Comput. Methods Prog. Biomed, vol. 226, no. 3, 2022, Art. no. 107096. doi: 10.1016/j.cmpb.2022.107096. [Google Scholar] [PubMed] [CrossRef]
46. F. Castro, D. Impedovo, and G. Pirlo, “A medical image encryption scheme for secure fingerprint-based authenticated transmission,” Appl. Sci., vol. 13, no. 10, 2023, Art. no. 6099. doi: 10.3390/app13106099. [Google Scholar] [CrossRef]
47. R. I. Abdelfatah, H. M. Saqr, and M. E. Nasr, “An efficient medical image encryption scheme for (WBAN) based on adaptive DNA and modern multi chaotic map,” Multimed. Tools Appl., vol. 82, no. 14, pp. 22213–22227, 2023. doi: 10.1007/s11042-022-13343-8. [Google Scholar] [CrossRef]
48. S. Inam, S. Kanwal, R. Firdous, and F. Hajjej, “Blockchain based medical image encryption using Arnold’s cat map in a cloud environment,” Sci. Rep., vol. 14, no. 1, 2024, Art. no. 5678. doi: 10.1038/s41598-024-56364-z. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF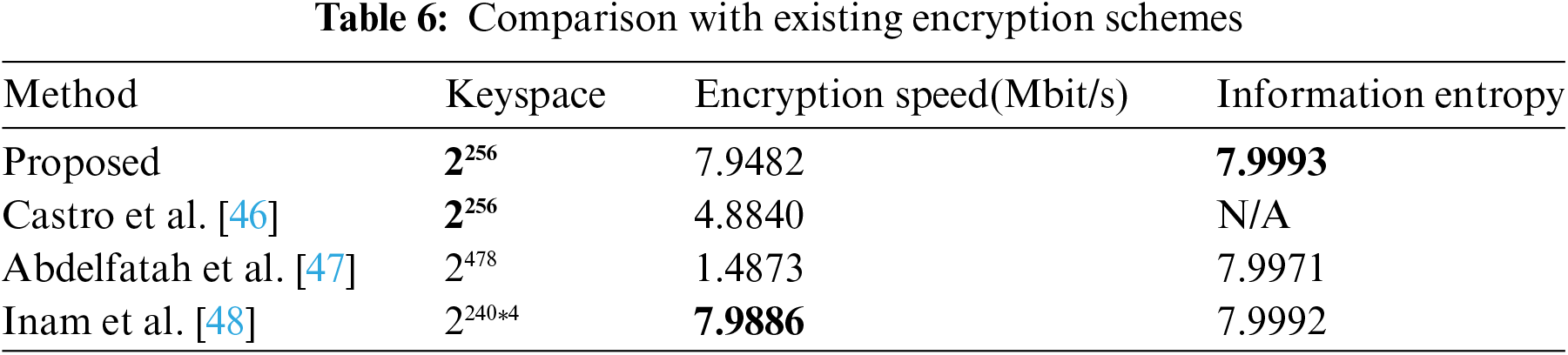
 Downloads
Downloads
 Citation Tools
Citation Tools