 Open Access
Open Access
ARTICLE
Harmonization of Heart Disease Dataset for Accurate Diagnosis: A Machine Learning Approach Enhanced by Feature Engineering
1 Department of Computer Science and Engineering, Metropolitan University, Sylhet, 3104, Bangladesh
2 Department of Statistics, Pabna University of Science and Technology, Pabna, 6600, Bangladesh
3 Department of Computer Science & Engineering, University of Aizu, Aizu-Wakamatsu, Fukushima, 956-8580, Japan
* Corresponding Author: Jungpil Shin. Email:
(This article belongs to the Special Issue: Advanced Medical Imaging Techniques Using Generative Artificial Intelligence)
Computers, Materials & Continua 2025, 82(3), 3907-3919. https://doi.org/10.32604/cmc.2025.061645
Received 29 November 2024; Accepted 24 January 2025; Issue published 06 March 2025
Abstract
Heart disease includes a multiplicity of medical conditions that affect the structure, blood vessels, and general operation of the heart. Numerous researchers have made progress in correcting and predicting early heart disease, but more remains to be accomplished. The diagnostic accuracy of many current studies is inadequate due to the attempt to predict patients with heart disease using traditional approaches. By using data fusion from several regions of the country, we intend to increase the accuracy of heart disease prediction. A statistical approach that promotes insights triggered by feature interactions to reveal the intricate pattern in the data, which cannot be adequately captured by a single feature. We processed the data using techniques including feature scaling, outlier detection and replacement, null and missing value imputation, and more to improve the data quality. Furthermore, the proposed feature engineering method uses the correlation test for numerical features and the chi-square test for categorical features to interact with the feature. To reduce the dimensionality, we subsequently used PCA with 95% variation. To identify patients with heart disease, hyperparameter-based machine learning algorithms like RF, XGBoost, Gradient Boosting, LightGBM, CatBoost, SVM, and MLP are utilized, along with ensemble models. The model’s overall prediction performance ranges from 88% to 92%. In order to attain cutting-edge results, we then used a 1D CNN model, which significantly enhanced the prediction with an accuracy score of 96.36%, precision of 96.45%, recall of 96.36%, specificity score of 99.51% and F1 score of 96.34%. The RF model produces the best results among all the classifiers in the evaluation matrix without feature interaction, with accuracy of 90.21%, precision of 90.40%, recall of 90.86%, specificity of 90.91%, and F1 score of 90.63%. Our proposed 1D CNN model is 7% superior to the one without feature engineering when compared to the suggested approach. This illustrates how interaction-focused feature analysis can produce precise and useful insights for heart disease diagnosis.Keywords
The heart is the most vital component of the human body out of every other part. The heart is essential to the function of several other vital organs. Other body parts including the brain, kidneys, etc., are affected if the heart is not functioning properly. Several factors increase the risk of heart disease [1]. Heart diseases include coronary artery disease, heart failure, endocarditis, myocarditis, and others. The major behavioral risk factors of heart disease include bad dietary habits, physical inactivity, consumption of tobacco products, and use of alcohol [2]. According to the World Health Organization (WHO), 17.9 million people die each year as a result of heart disease. The majority of cardiovascular disease (CVD) deaths occur in nations with low or middle incomes [3]. According to the 2021 report, 2552 CVD diseases are diagnosed every day, and someone dies every 34 s in the U.S.A. In the United States, heart disease and heart attacks cause around 1905 deaths every day. Seventy-five percent of mortality in developing nations like Bangladesh is caused by CVDs, which are a serious public health concern worldwide [4]. Blood, stress, and imaging tests are some of the methods used to identify cardiac disease. Proper diagnostic instruments, such as an echocardiogram, cardiac CT scan, cardiac MRI, or ECG, are crucial for detecting this condition in imaginary-based detection [5,6]. However, these tests are sometimes too costly and unfeasible for people in developing and developing countries. Physical and financial burdens can be decreased by detecting cardiac problems early. By 2030, there would be 23.6 million deaths from CVD overall, primarily from heart disease and stroke, according to another WHO study results [7]. To save lives and diminish the financial burden, advanced machine learning, and deep learning techniques should be used to identify this deadly disease. Data mining techniques are being used in healthcare to produce vast amounts of raw data, which exhibit various patterns and are undoubtedly vital for clinical diagnosis in the past few decades. Finding patterns in data is crucial for improving health decisions, early disease identification, preventing avoidable hospitalizations, and forbearing medical errors.
However, predicting heart disease requires meticulous assessment of several metrics, including diabetes, high blood pressure, excessive cholesterol, and an irregular pulse rate [8]. An automated prediction method may be more cost-effective and error-free for doctors based on its significant characteristics than a manual process to avoid such a complicated assessment to predict heart disease patients early and accurately [9,10]. Complex patterns in clinical data frequently emerge and influence patient prediction [11]. We process the data set as necessary to reduce it before running the ML model. To identify intricate patterns in data and predict the likelihood of diseases, numerous advanced statistical and ML techniques have recently been developed. This proposed data set contains both numerical and categorical features. These records are cleaned and filtered to eliminate and replace unnecessary data from the dataset before additional processing. To do this, we then highlight statistical feature interaction-based techniques that can detect complex patterns in the data. For feature engineering, we take into account numerical and categorical features separately. To capture the complex patterns of heart diseases, we employed correlation for numerical features and the chi-square test for categorical features. Since the feature interaction generates more features than the original data, we intend to reduce both the model’s dimension and complexity. To accomplish this, we used principal component analysis (PCA), a popular multivariate feature dimensioning technique that reduces dimension without disregarding data information. Then, we used numerous hyperparameter-based ML models, such as RF, Gradient Boosting, XGBoost, LightGBM, CatBoost, SVM, MLP, and their voting ensemble.
Finally, we proposed a customized one-dimensional convolution neural networks (1D CNN) model, which automatically learns the features and captures the hidden pattern, to reveal the data set’s intricate pattern. From a database of past heart disease cases, the improved system provides precise hidden information, trends, and connections related to heart disease. To help medical professionals make wise clinical judgments, it can also provide sophisticated answers to questions about heart disease diagnosis. The results demonstrated the surpassed power of the proposed strategy in achieving the specified prospecting goals. The diagnosis is made at every stage using the expertise and experience of the physician. This results in undesirable outcomes and exorbitant medical expenses for the therapies that patients receive. Thus, a system for automatic medical diagnosis would be quite helpful. Providing a thorough overview of the myriad data analysis techniques that can be used with these automated systems is the aim of our work. The suggested procedure for predicting heart disease is entirely depicted in Fig. 1.

Figure 1: Proposed heart disease prediction workflow
This paper is structured as follows: Section 1 provides a demonstration of the introduction; Section 2 demonstrates related works; Section 3 describes the proposed materials and methodology; Section 4 discusses the proposed feature engineering part; Section 5 discusses findings and comparative analysis; and Section 6 concludes and discusses future work.
Heart disease is the world’s leading cause of death in this decade, and it is caused by a complex interaction of hereditary risk factors, influences from the environment, and behavioral factors. As a result, many researchers work to find high-risk factors and make precise, early patient predictions. Saboor et al. [12] deployed a tuning-based classical machine learning algorithm based on GridSearch to study the prediction of heart disease. Six heart disease datasets were utilized, including the Cleveland, StatLog Heart, Z-Alizadeh Sani, Hungarian, Long Beach, VA, and Kaggle Framingham datasets. Their methodology involves comparing the performance matrix before and after the use of the data standardization technique. On the scaled dataset, the hyperparameter-tuned SVM achieved the highest accuracy score of 96.72% out of all the classifiers. Jindal et al. [13] used several ML models, including KNN, LR, and RF, on a dataset consisting of 304 patients and thirteen features obtained from the UCI repository. The goal of the study is to forecast the occurrence of heart disease. Out of all of them, KNN got the best accuracy rate of 88.52%. Mohan et al. [8] present a hybrid ML approach, the Hybrid Random Forest with Linear Model (HRFLM), to improve heart disease prediction accuracy. It combines the strengths of RF and a linear model. They only employed the Cleveland dataset out of several well-known integrated datasets, including Hungarian, Cleveland, VA Long Beach, and Switzerland. Because of its thorough records, this dataset is frequently used. Based on 14 features, the collection includes observations from 303 patients. The HRFLM model outperformed rival models like DT and SVM, with an accuracy of 88.7%, which is significant for medical prediction. It also enhanced performance like sensitivity and specificity. Although the HRFLM model displayed improved accuracy, it may be less flexible and unable to generalize to datasets with fewer attributes due to its dependence on all features. Also, the dataset was tiny (297 records after cleaning) and may not have been representative of the population at large when testing the model. Chitra et al. [14] introduce a system for predicting the occurrence of cardiovascular illness by categorizing patient data using support vector machine (SVM) and cascaded neural network (CNN) classifiers. In this work, the CNN classifier achieved an accuracy of 85%, outperforming the SVM, which had an accuracy of 82%. In terms of specificity and overall accuracy, CNN outperformed other techniques, suggesting that it may be more reliable for identifying disease-free patients. Mahmud et al. [15] present a framework for predicting cardiac failure through the integration of various ML (DT, KNN, RF, and GNB) algorithms. With an 87% accuracy rate, the metamodel surpassed competing models in recall, accuracy, and F1 score, suggesting it could improve the reliability of predictions. The goal of the study is to help doctors detect heart failure earlier and make more accurate predictions so that patients can get treatment when they need it. Hossain et al. [16] studied the use of several AI algorithms to predict cardiac disease early and correctly. In Bangladesh, they gathered primary data from various diagnostic, medical, and hospital. Following data collection, necessary pre-processing methods are used to ensure an optimal model fit, such as missing value treatment, deleting redundant and ambiguous data, and scaling. To determine the prominent features, they only employed the correlation-based feature selection with the best first search approach. The patients with heart disease are finally predicted using different AI algorithms (LR, NB, KNN, SVM, DT, RF, and MLP) and compared with all selected features. With the selected features, the RF classifiers obtained the highest accuracy score of 90% out of all the AI approaches. Chang et al.’s [17] research on heart disease prediction focuses on AI-based patient detection using an ML algorithm. To anticipate patients with heart disease, they created a healthcare application that uses machine learning algorithms, specifically the RF model, which has a greater accuracy of 83%. This paper made theoretical and practical contributions that improved the framework for patient diagnosis and helped the hospital and physicians alike. Jackins et al. [18] presented an AI-based intelligent clinical disease prediction system for diseases like breast cancer, diabetes, and coronary heart disease. They employed the Naïve Biased and Random Forest classification in contrast to K-mean clustering and DBSCAN to diagnose diseases. The accuracy of RF was the highest at 83.35% for the heart disease dataset, while the accuracy of the Bayesian model was 82.35%.
Feature engineering and model hypermeter-based patient identification are the main goals of our proposed heart disease prediction. To achieve the desired result, we first preprocess the data set, which includes replacing missing and null values using a robust method, detecting and replacing outliers, scaling features, and other things to make the model perform better than others. Following features engineering, we use PCA to reduce dimension complexity. Then, we use ML based on hyperparameter tuning and a 1D CNN model to achieve state-of-the-art performance. The subsequent part describes the specifics of the materials & methodology.
Five datasets on heart disease were used in this work, and they were gathered from Kaggle, a well-known machine learning data repository. This dataset’s integration, which captures an extensive range of patient attributes, makes it unique. There are 1190 instances of the common 11 traits in this collection. There is a nearly equal distribution in this dataset. Out of all the observations, there are 629 patients with the disease and 561 patients without. These datasets were collected and merged to help advance research on CAD-related disease using ML algorithms, and hopefully to ultimately advance clinical diagnosis and early treatment. The dataset includes various clinical and diagnostic features relevant to cardiovascular health. The patient’s age in years is represented by a numeric value that is recorded. With 1 denoting male and 0 denoting female, sex is a binary variable. Typical angina (1), atypical angina (2), non-anginal pain (3), and asymptomatic cases (4) are the numbers that indicate the nominal kind of chest pain. Serum cholesterol levels are measured in milligrams per deciliter (mg/dL), and resting blood pressure is measured in millimeters of mercury (mmHg). A binary variable that indicates whether or not fasting blood sugar surpasses 120 mg/dL is fasting blood sugar (1 = true, 0 = false). Estes’ criteria state that a resting ECG’s results are presented as nominal values: 0 for normal, 1 for aberrant ST-T waves, and 2 for likely or proven left ventricular hypertrophy. The highest heart rate that may be achieved is between 71 and 202. The binary variable of exercise-induced angina is 1 = yes, 0 = no. The details of the description of the heart disease data set are shown in Table 1.

In the preprocessing part, missing value handling is an important part of data analysis and ML. If it’s not done right, it can lead to biased models and wrong results. There are different ways to fill in missing values depending on the type of data and how it is distributed. When it comes to categorical and numerical traits, the method might be different. When it comes to categorical features, mode (the most usual value) is often used to fill in missing values. This makes sure that the numbers that are imputed are a good representation of how the categories are currently spread out in the feature.
4 Proposed Feature Engineering
By identifying the complex patterns in the dataset, we can forecast heart disease patients with ease and accuracy using the proposed feature engineering strategy. We used the correlation approach for numerical features and χ2 for categorical features so that the model could uncover the hidden pattern. The specifics are covered in the subsection that follows on feature selection based on correlation and χ2.
4.1 Correlation-Based Feature Selection
The statistical measurement of the connection between two variables is a correlation. The measure works well with variables that show a linear relationship with one another. The usual range for linear correlation scores is −1 to 1, where 0 denotes no link. The correlation coefficient is not affected by scaling or translation. As a result, this measure may have the same value for both features with distinct variances [19,20]. Let us consider

We hope to better understand the dynamics of heart disease by concentrating on these relationships to capture special synergistic effects that transcend the contributions of individual features. Combinations with lower correlation values showed weaker interactions, and thus, were less likely to provide additional predictive power.
4.2
Chi-square is a univariate feature selection method that can be used with categorical data as input and a categorical target variable as output. If there is a significant association between two category (nominal) variables, the

4.3 Feature Interaction Approach
Features interaction captures the intricate, non-linear pattern that a single feature can fail to reach improving model performance and lowering bias. Interactions learn automatically in models like DT, but they can be explicitly engineered or automatically identified in many models, such as linear or neural network models. It is possible to make models much more accurate by giving them a larger dataset through feature engineering using interaction terms. To mine more complex, nonlinear relationships, we need to add interaction terms [8]. If we denote two features
PCA is a multivariate analysis method that reduces dimensionality and complexity while identifying the key features that encapsulate the intricate pattern. PCA preserves the data’s fundamental structure by preserving its variance. It works by transforming the data into a new set of orthogonal components (principal components) that maximize variance [21,22]. If
4.5 Model’s Hyperparameter Tuning
By finding the best setup without overfitting, hyperparameter tuning makes models more accurate and useful in new situations. By trying out different setups, tuning helps find the model setup that works best for the provided information. Random Forest is a group of decision trees that work together as a whole. The gradient Boosting method builds sequential trees that fix mistakes made by earlier trees. Gradient boosting methods like XGBoost, LightGBM, and CatBoost are made to be fast and accurate. A classifier known as an SVM divides data into groups by identifying a hyperplane. MLP is a kind of neural network with programmable layers and activation functions. And a 1D CNN model that uses a refined technique to automatically learn from the feature. To find the model parameter, we applied gird-search and chose the best set of parameters to fit the model. Grid Search is an exhaustive parameters optimization method that evaluates every possible combination of hyperparameters within a predefined grid. It systematically tests all possible combinations in a specified hyperparameter space to find the best one [23]. Table 4 describes the specific final best hyperparameters.

5 Findings and Comparative Analysis
5.1 Evaluation Matrix and Computational Efficiency
The model’s performance is evaluated using a set of criteria that provide a thorough grasp of its diagnostic capability: accuracy, precision, recall, specificity, and F1 score. The proposed models were trained on a system specified as follows: Processor: Intel(R) Core (TM) i7-10510U CPU @ 1.80, 2.30 GHz; RAM: 16 GB; Operating System: Windows 11 Pro; Device Name: DESKTOP-SQ81EE1.
5.2 Model Performance Discussion
Different prediction metrics were shown in the study’s findings for both the suggested feature engineering and non-feature engineering approaches. We conducted our study using both the train test (70% and 30%) and the K-fold cross-validation approach. Model performance is displayed in Table 5 without feature engineering techniques, while performance matrices are displayed in Table 6 using feature engineering techniques. Before feature engineering, the RF model outperformed all other classification algorithms in terms of accuracy of 90.21%, precision of 90.40%, recall of 90.82%, specificity of 90.91%, and F1 score of 90.63% during the whole procedure. When assessing the effect of feature engineering on the model functionality, our proposed 1D CNN model achieved the greatest accuracy of 96.36%, precision of 96.45, recall of 96.36, specificity of 99.51%, and F1 score of 96.34%. In conclusion, all models’ performance was improved by feature engineering, but the 1D CNN stood out because of its remarkable accuracy, specificity, and F1 score gains, demonstrating its capacity for high-impact predictions. The ROC curve for the feature engineering method model is displayed in Fig. 2 of (a), while Fig. 2 of (b) displays the model without feature engineering.



Figure 2: (a) ROC curve of all models with feature engineering; (b) ROC curve of all models without feature engineering
A model is considered perfectly classified if its ROC value is 1.0, while it is considered to have no discrimination ability if its ROC value is 0.5. A ROC greater than 0.8 is generally regarded as favorable for binary classification. The performance metrics of several ML models, including the 1D CNN, for the prediction of heart disease using 10-fold cross-validation, are shown in Table 7. At 90.91%, RF has the greatest accuracy score. Therefore, in our 1D CNN model, the reliability of our findings is ensured by adding cross-validation for the RF model, which offers further validation of the outcomes. The model accuracy and loss curve for the proposed approach is displayed in Fig. 3 of (a) and (b), whereas the model accuracy and loss without feature engineering is displayed in Fig. 3 of (c) and (d). Table 8 compares the performance of the approach we propose with some recent state-of-the-art results.

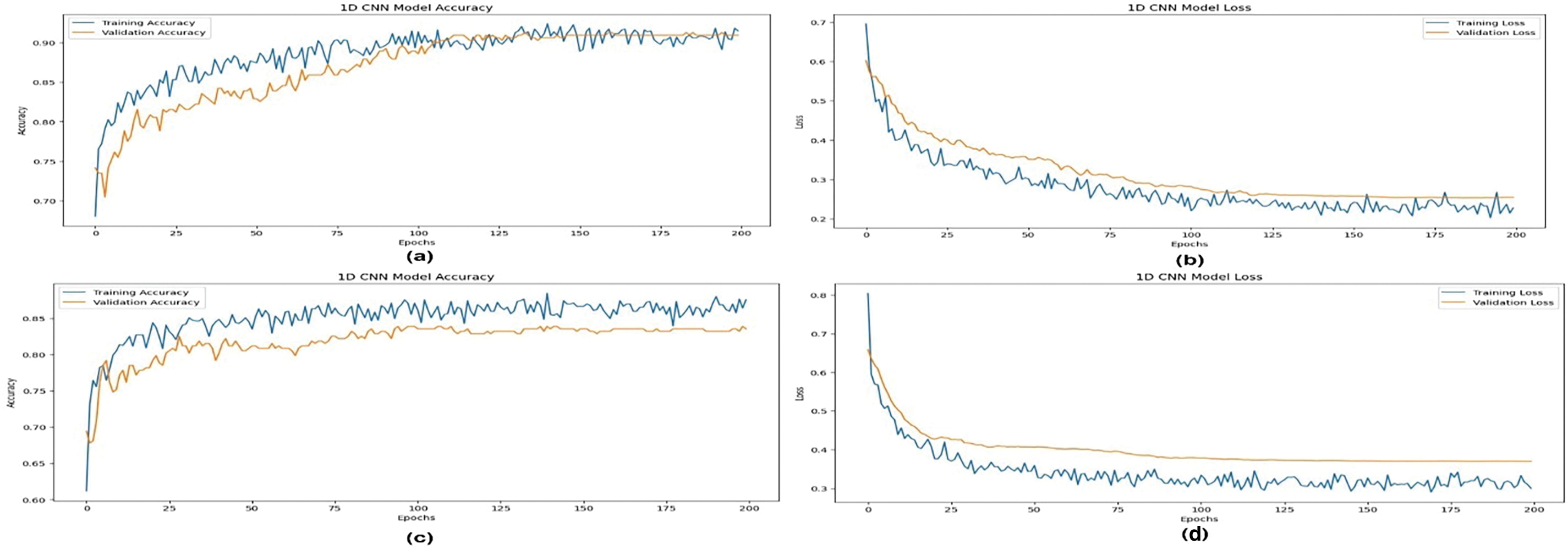
Figure 3: 1D CNN model accuracy (a) and loss (b) curve for with feature engineering and model accuracy (c) and loss (d) for without feature engineering
Finding the hidden pattern of heart disease in the health informatics data is our goal in this work. The dataset is initially preprocessed, including null and missing value replacement, robust outlier detection, etc., before delving deeply into the model application. Next, we attempt to determine how likely a feature is to cause the corresponding disease. Correlation and chi-squared feature interaction were then utilized to increase the model’s accuracy by mining the intricate hidden pattern, and the findings showed promise. To classify patients with and without the disease, we used several advanced machine learning models (RF, MLP, XGBoost, CatBoost, Light GBM, SVM, GB, Ensemble) that can capture the association with the target variable. Additionally, we used a 1D CNN model, which learns from features automatically without the need for manual feature engineering. We concluded that our proposed 1D CNN model has the greatest and most reliable outcomes, with accuracy, precision, recall, specificity, and F1 score of 96.36%, 96.45%, 99.51%, and 96.36%, respectively. This indicates state-of-the-art performance compared to recent work.
Acknowledgement: We would like to express our gratitude to the Division of Information Systems, School of Computer Science and Engineering, University of Aizu, Japan, for providing the funding and resources necessary to carry out the study.
Funding Statement: This study was supported by the Competitive Research Fund of the University of Aizu, Japan (Grant No. P-13).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Ruhul Amin, and Md. Jamil Khan, and Jungpil Shin; data collection: Ruhul Amin, and Md. Shamim Reza; analysis and interpretation of results: Ruhul Amin, and Md. Jamil Khan, and Tonway Deb Nath; draft manuscript preparation: Ruhul Amin, Md. Jamil Khan, and Tonway Deb Nath; review and editing: Jungpil Shin, and Md. Shamim Reza. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available at this link (https://www.kaggle.com/datasets/mexwell/heart-disease-dataset?resource=download) (accessed on 15 December 2024).
Ethics Approval: This study used secondary data that is publicly available and gathered from the Kaggle a well-known data repository. Therefore, formal ethics approval was not needed by established ethical principles for secondary data analysis.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Krishnaiah V, Narsimha G, Subhash N. Heart disease prediction system using data mining techniques and intelligent fuzzy approach: a review. Int J Comput Appl. 2016;136(2):43–51. doi:10.5120/ijca2016908409. [Google Scholar] [CrossRef]
2. El-Sofany H, Bouallegue B, El-Latif YMA. A proposed technique for predicting heart disease using machine learning algorithms and an explainable AI method. Sci Rep. 2024;14(1):23277. doi:10.1038/s41598-024-74656-2. [Google Scholar] [PubMed] [CrossRef]
3. Jyoti S, Ujma A, Dipesh S, Sunita S. Predictive data mining for medical diagnosis: an overview of heart disease prediction Sunita Soni. Int J Comput Appl. 2011;17(8):975–8887. doi:10.5120/2237-2860. [Google Scholar] [CrossRef]
4. Chowdhury MZI, Haque MA, Farhana Z, Anik AM, Chowdhury AH, Haque SM, et al. Prevalence of cardiovascular disease among bangladeshi adult population: a systematic review and meta-analysis of the studies. Vasc Health Risk Manag. 2018;14:165–81. doi:10.2147/VHRM.S166111. [Google Scholar] [PubMed] [CrossRef]
5. Di Carli MF, Dorbala S, Curillova Z, Kwong RJ, Goldhaber SZ, Rybicki FJ, et al. Relationship between CT coronary angiography and stress perfusion imaging in patients with suspected ischemic heart disease assessed by integrated PET-CT imaging. J Nucl Cardiol. 2007;14(6):799–809. doi:10.1016/j.nuclcard.2007.07.012. [Google Scholar] [PubMed] [CrossRef]
6. Klem I, Heitner JF, Shah DJ, Sketch MH, Behar V, Weinsaft J, et al. Improved detection of coronary artery disease by stress perfusion cardiovascular magnetic resonance with the use of delayed enhancement infarction imaging. J Am Coll Cardiol. 2006;47(8):1630–8. doi:10.1016/j.jacc.2005.10.074. [Google Scholar] [PubMed] [CrossRef]
7. Purushottam, Saxena K, Sharma R. Efficient heart disease prediction system. Procedia Comput Sci. 2016;85:962–9. doi:10.1016/j.procs.2016.05.288. [Google Scholar] [CrossRef]
8. Mohan S, Thirumalai C, Srivastava G. Effective heart disease prediction using hybrid machine learning techniques. IEEE Access. 2019;7:81542–54. doi:10.1109/ACCESS.2019.2923707. [Google Scholar] [CrossRef]
9. Chandrasekhar N, Peddakrishna S. Enhancing heart disease prediction accuracy through machine learning techniques and optimization. Processes. 2023;11(4):1210. doi:10.3390/pr11041210. [Google Scholar] [CrossRef]
10. Khan A, Qureshi M, Daniyal M, Tawiah K. A novel study on machine learning algorithm-based cardiovascular disease prediction. Health Soc Care Commun. 2023;2023:1–10. doi:10.1155/2023/1406060. [Google Scholar] [CrossRef]
11. Baghdadi NA, Farghaly Abdelaliem SM, Malki A, Gad I, Ewis A, Atlam E. Advanced machine learning techniques for cardiovascular disease early detection and diagnosis. J Big Data. 2023;10(1):1–29. doi:10.1186/s40537-023-00817-1. [Google Scholar] [CrossRef]
12. Saboor A, Usman M, Ali S, Samad A, Abrar MF, Ullah N. A method for improving prediction of human heart disease using machine learning algorithms. Mob Inf Syst. 2022;2022(15):1–9. doi:10.1155/2022/1410169. [Google Scholar] [CrossRef]
13. Jindal H, Agrawal S, Khera R, Jain R, Nagrath P. Heart disease prediction using machine learning algorithms. IOP Conf Series: Mater Sci Eng. 2021;1022(1):012072. doi:10.1088/1757-899X/1022/1/012072. [Google Scholar] [CrossRef]
14. Chitra R. Heart disease prediction system using supervised learning classifier. Bonfring Int J Softw Eng Soft Comput. 2013;3(1):1–7. doi:10.9756/BIJSESC.4336. [Google Scholar] [CrossRef]
15. Mahmud I, Kabir MM, Mridha MF, Alfarhood S, Safran M, Che D. Cardiac failure forecasting based on clinical data using a lightweight machine learning metamodel. Diagnostics. 2023;13(15):1–18. doi:10.3390/diagnostics13152540. [Google Scholar] [PubMed] [CrossRef]
16. Hossain MDI, Maruf MH, Khan MDAR, Prity FS, Fatema S, Ejaz MDS, et al. Heart disease prediction using distinct artificial intelligence techniques: performance analysis and comparison. Iran J Comput Sci. 2023;6(4):397–417. doi:10.1007/s42044-023-00148-7. [Google Scholar] [CrossRef]
17. Chang V, Bhavani VR, Xu AQ, Hossain MA. An artificial intelligence model for heart disease detection using machine learning algorithms. Healthcare Anal. 2022;2:100016. doi:10.1016/j.health.2022.100016. [Google Scholar] [CrossRef]
18. Jackins V, Vimal S, Kaliappan M, Lee MY. AI-based smart prediction of clinical disease using random forest classifier and Naive Bayes. J Supercomput. 2021;77(5):5198–219. doi:10.1007/s11227-020-03481-x. [Google Scholar] [CrossRef]
19. Nasir IM, Khan MA, Yasmin M, Shah JH, Gabryel M, Scherer R, et al. Pearson correlation-based feature selection for document classification using balanced training. Sensors. 2020;20(23):1–18. doi:10.3390/s20236793. [Google Scholar] [PubMed] [CrossRef]
20. Mohamad M, Selamat A, Krejcar O, Crespo RG, Herrera-Viedma E, Fujita H. Enhancing big data feature selection using a hybrid correlation-based feature selection. Electronics. 2021;10(23):1–24. doi:10.3390/electronics10232984. [Google Scholar] [CrossRef]
21. Reddy GT, Reddy MPK, Lakshmanna K, Kaluri R, Rajput DS, Srivastava G, et al. Analysis of dimensionality reduction techniques on big data. IEEE Access. 2020;8:54776–88. doi:10.1109/ACCESS.2020.2980942. [Google Scholar] [CrossRef]
22. Huang D, Jiang F, Li K, Tong G, Zhou G. Scaled PCA: a new approach to dimension reduction. Manage Sci. 2022;68(3):1678–95. doi:10.1287/mnsc.2021.4020. [Google Scholar] [CrossRef]
23. Fuadah YN, Pramudito MA, Lim KM. An optimal approach for heart sound classification using grid search in hyperparameter optimization of machine learning. Bioengineering. 2023;10(1):45. doi:10.3390/bioengineering10010045. [Google Scholar] [PubMed] [CrossRef]
24. Bharti R, Khamparia A, Shabaz M, Dhiman G, Pande S, Singh P. Prediction of heart disease using a combination of machine learning and deep learning. Comput Intell Neurosci. 2021;2021:1547. doi:10.1155/2021/8387680. [Google Scholar] [PubMed] [CrossRef]
25. Mauya J, Sahriar S, Akther S, Amin R, Ruhi S, Reza MS. Missing risk factor prediction in cardiovascular disease using a blended dataset and optimizing classification with a stacking algorithm. Eng Reports. 2024;7(1):e13034. doi:10.1002/eng2.13034. [Google Scholar] [CrossRef]
26. Anika S, Islam M, Palit A. Early prediction of coronary heart disease using hybrid machine learning models. Commun Comput Inform Sci. 2024;1995:63–75. doi:10.1007/978-3-031-51135-6. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF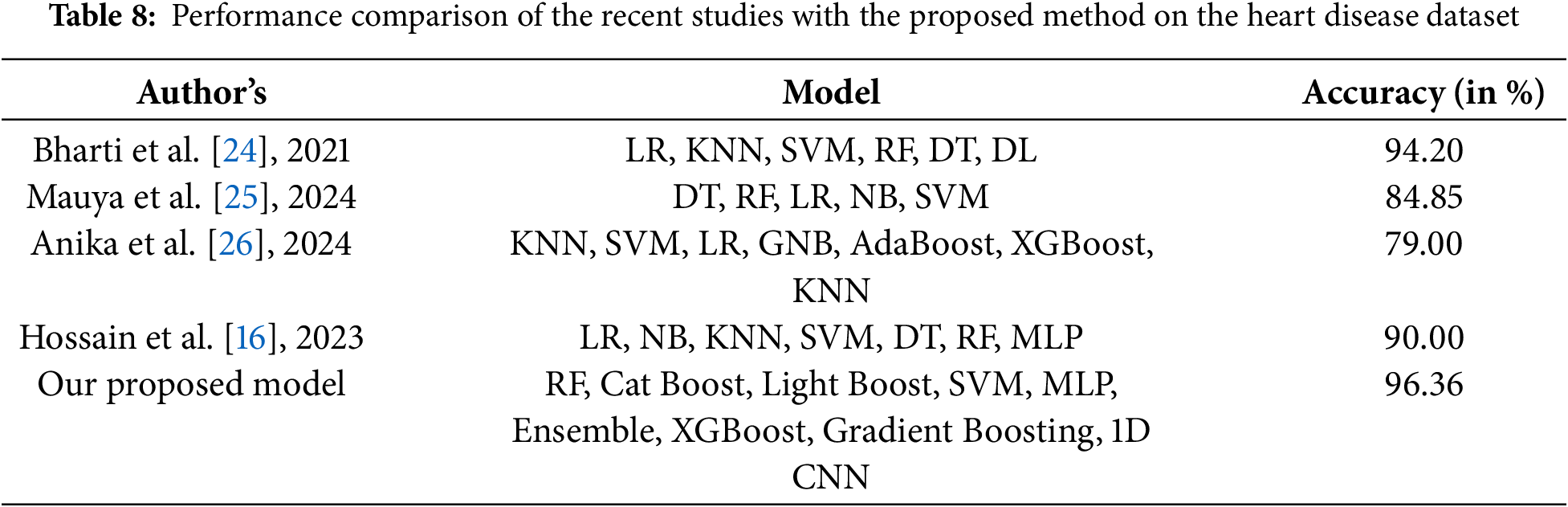
 Downloads
Downloads
 Citation Tools
Citation Tools