 Open Access
Open Access
ARTICLE
Leveraging Edge Optimize Vision Transformer for Monkeypox Lesion Diagnosis on Mobile Devices
1 Chitkara University Institute of Engineering and Technology, Chitkara University, Rajpura, 140401, Punjab, India
2 Centre for Research Impact and Outcome, Chitkara University, Rajpura, 140401, Punjab, India
3 Department of Computer Engineering & Applications, G.L.A. University, Mathura, 281406, India
4 Department of Data Science, University of Salford, Manchester, M54WT, UK
5 University Centre for Research and Development, Chandigarh University, Mohali, 140413, Punjab, India
6 Division of Research and Development, Lovely Professional University, Phagwara, 144411, India
7 Department of Management, College of Business Administration, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
* Corresponding Authors: Bhisham Sharma. Email: ; Surbhi Bhatia Khan. Email:
(This article belongs to the Special Issue: Medical Imaging Based Disease Diagnosis Using AI)
Computers, Materials & Continua 2025, 83(2), 3227-3245. https://doi.org/10.32604/cmc.2025.062376
Received 17 December 2024; Accepted 28 February 2025; Issue published 16 April 2025
Abstract
Rapid and precise diagnostic tools for Monkeypox (Mpox) lesions are crucial for effective treatment because their symptoms are similar to those of other pox-related illnesses, like smallpox and chickenpox. The morphological similarities between smallpox, chickenpox, and monkeypox, particularly in how they appear as rashes and skin lesions, which can sometimes make diagnosis challenging. Chickenpox lesions appear in many simultaneous phases and are more diffuse, often beginning on the trunk. In contrast, monkeypox lesions emerge progressively and are typically centralized on the face, palms, and soles. To provide accessible diagnostics, this study introduces a novel method for automated monkeypox lesion classification using the HMTNet (Hybrid Mobile Transformer Network). The convolutional layers and Vision Transformers (ViT) are combined to enhance the spatial features. In addition, we replace the classical MHSA (Multi-head self-attention) with the WMHSA (Window-based Multi-Head Self-Attention) to effectively capture long-range dependencies within image patches and depth-wise separable convolutions for local feature extraction. We trained and validated HMTNet on the two datasets for binary and multiclass classification. The model achieved 98.38% accuracy for multiclass classification using cross-validation and 99.25% accuracy for binary classification. These findings show that the model has the potential to be a useful diagnostic tool for monkeypox, especially in environments with limited resources.Keywords
Abbreviations
| HMTNet | Hybrid Mobile Transformer Network |
| ViT | Vision Transformers |
| MHSA | Multi-head self-attention |
| WMHSA | Window-based Multi-Head Self-Attention |
| DRC | Democratic Republic of the Congo |
| AI | Artificial intelligence |
| ML | Machine Learning |
| CNN | Convolutional neural network |
| BiLSTM | Bidirectional Long Short-Term Memory |
| MSLD | Monkeypox Skin Lesion Dataset |
| FVS | Fuzzified Voting Scheme |
| HPV | Human papilloma virus |
| MPV | Monkeypox virus |
The Mpox disease is triggered by the monkeypox virus, which is also responsible for cowpox, smallpox, and vaccinia viruses (used in smallpox vaccines). The DRC (Democratic Republic of the Congo) reported the first human case of monkeypox in 1970. Subsequently, the illness has been identified as endemic in multiple countries, including Central and West Africa [1]. Historically, Mpox has been an emerging public health problem in endemic countries, though recent outbreaks beyond the African region, including the large global outbreak in the year 2022. Approximately 107,725 cases of confirmed Mpox have been reported since the beginning of the outbreak. The Americas region is the largest, accounting for approximately 64% of cases, followed by Europe at 27% and the African Region at around 5.4%. In the year 2024, more than 20,000 Mpox cases have been reported across 13 AUMS (African Union Member States), with the DRC experiencing the highest burden, reporting over 16,000 cases and 501 deaths as of mid-2024. The signs of smallpox and other poxviruses are comparable to those of Mpox [2]. While most Mpox cases are cured within two to four weeks, serious complications can happen, especially in young children, mothers, and people with weak immune systems. Bacterial infections can produce diseases such as bronchopneumonia, sepsis, encephalitis, and corneal infections, which can result in blindness [3]. The West African clade has less efficient person-to-person transmission, and the Central African clade is more virulent, with mortality rates ranging from 3% to 10% [4]. It is mainly predominant in the DRC and the rest of the Congo Basin. The clade can transmit better from person to person and is often associated with more severe clinical outcomes and higher complication rates [5]. The clinical severity and transmission rates might be higher for one of the clades compared to the other.
Monkeypox and chickenpox share a similar rash, but the former’s lesions tend to develop more uniformly, while the latter’s lesions show up in multiple stages at once. Moreover, unlike chickenpox, lesions from monkeypox usually affect the face, palms, and soles. Because of these similarities, accurate diagnosis requires laboratory confirmation, especially when these diseases co-circulate. The clinical overlap of these conditions demands developing more sensitive diagnostic tools that can identify these conditions differently and make the process faster and more accurate [6]. Artificial intelligence (AI)/Machine Learning (ML) based technologies are increasingly being deployed within healthcare and other domains to enhance diagnostic accuracy, particularly in overlapping symptoms of these conditions [7]. AI methods, especially deep learning models, offer excellent robustness for binary multiclass classification. Recent work has combined CNNs with transfer learning to classify monkeypox lesions from digital images with an accuracy of over 90% [8]. There are morphological similarities between smallpox, chickenpox, and monkeypox, particularly in how they appear as rashes and skin lesions, which can sometimes make diagnosis challenging. Chickenpox lesions appear in many simultaneous phases and are more diffuse, often beginning on the trunk. In contrast, monkeypox lesions emerge progressively and are typically centralized on the face, palms, and soles. Monkeypox is similar in distribution but more clinically severe, with smallpox lesions being deeper and more uniform in development and primarily affecting the face and extremities.
In this study, we designed an HMTNet to achieve effective monkeypox image classification. The local spatial feature is extracted using MobileNet, and the transformer blocks provide global context. Furthermore, In the ViT module, we applied the WMHSA to assist spatial dependencies within Mpox image regions. To maintain low computational costs, it separates the image into non-overlapping windows, applies self-attention within each window, and records comprehensive region-based information. We evaluated the model on two datasets, and results are compared with several CNN and ViT-based method. At the same time lightweight nature of the models can be for edge devices and mobile applications.
The major contributions of this article include:
(1) Combining convolutional layers and vision transformers, this model provides a lightweight, accurate solution that is tailored for mobile and edge devices.
(2) We utilized WMHSA to handle both local and global dependencies in images, allowing the model to concentrate on both local image details and more general contextual information across image patches.
(3) We utilized a hierarchical feature aggregation, depth wise separable convolutions, and squeeze-and-excitation module. Which preserves computational efficiency while optimizing the model’s performance, which is crucial for real-time applications.
Remainder of the manuscript is as follows. Detail survey of the past methods is described in Section 2, meanwhile Section 3 elaborates the proposed method. In Section 4, quantitative results are presented and We concluded the proposed method in Section 5 with limitation and future scope.
In this section, we examine the latest developments in approaches for resource-constrained, real-time applications. In the research [9], they used Deoxyribonucleic acid (DNA) sequences to distinguish between Monkeypox virus (MPV) and human papillomavirus (HPV). Further, A BiLSTM (Bidirectional Long Short-Term Memory) was utilized for classification. The study [10] presents a combination of data mining and artificial intelligence techniques. They utilized 500 samples, with positive and negative cases infected by monkeypox. The experimental outcomes recall, accuracy, and precision are 91.1%, 88.91%, and 98.48%, respectively. The research investigation [11] identifies and classifies monkeypox using MonkeyNet inspired by DenseNet-201. A total of 770 photos were gathered from MSLD, which contains 107 chickenpox, 91 measles, 279 monkeypox, and 293 standard images. The testing for the original dataset had an accuracy of 91.91%. At the same time, on the augmented dataset model has a high accuracy of 98.91%. The strategy [12] utilized two datasets for robust training. Their method contains two steps: first, preprocessing, which selects the best features; then, there is the classification step, based on an ensemble of three classifiers. A Fuzzified Voting Scheme (FVS) combines the classifiers’ outputs to determine the final diagnosis. They obtained an accuracy of 97.2%, precision of 94.5%, recall of 93.1%, and F1-score of 94%. In another research, features are extracted from medical images and correctly identified using transfer learning and CNN. Their model attained the highest classification accuracy at 98.18% [13]. The research [14] utilized metaheuristic optimization to improve the model’s classification for the monkeypox lesions. They obtained performance measures through feature selection and a decision tree classifier: an F1-score of 0.92, sensitivity of 0.95, and specificity of 0.61. Four convolutional layers based on 2-D CNN were utilized to classify chickenpox and monkeypox. Their method achieved an accuracy of 99.60%. Furthermore, they confirm that the proposed CNN model performed is better than AlexNet, VGG16 and VGG19 [15].
With the help of the base models (InceptionV3, EfficientNet, and VGG16) and the SENet attention architecture, the method improved the classification accuracy of monkeypox. Hyperparameters like a learning rate of 0.0001, the Adam optimizer was used to fine-tune the deep learning models. With an accuracy of 98%, precision of 98.06%, recall of 97.97%, and an F1-score of 98.02%, the SENet+InceptionV3 model performed better than the other configurations [16]. In another study, they developed PoxNet22 using transfer learning to classify the monkeypox disease. They utilized the Monkeypox Skin Lesion Dataset (MSLD), which contains 3192 photos from the public, case studies, and internet portals. Results show that PoxNet22 performed very well with 100% recall, 100% accuracy, 100% precision, and 0% loss [17]. This research [18] developed a fusion system to identify monkeypox. They fused the LSTM with a CNN model to design a hybrid deep-learning model. This hybrid has a test accuracy and Cohen’s kappa score of 87% and 0.8222%, respectively. In the literature [19], Vision Transformers (ViT) and modified transfer learning models like M-VGG16, M-ResNet50, and M-ResNet101 were utilized for monkeypox lesion classification. They utilized a dataset of 2524 images from various studies and achieved a classification accuracy of 89% using M-ResNet50. The investigation [20] represents improved performance in the detection of monkeypox through an RN-50, principal component analysis (PCA), and MXGBoost. Their method achieved a precision of 97.41%. The study [21] used an attention-based MobileNetV2 model for monkeypox detection. The attention-based MobileNetV2 outperformed 92.28% accuracy in the extended MSID dataset, 98.19% in the original MSID dataset, and 93.33% in the Monkeypox Skin Lesion Dataset (MSLD) dataset. With the focus on identifying monkeypox, foot ulcers, and other mouth and oral disorders, new research was conducted in the literature [22]. They proposed a Choquet Fuzzy Integral-based Ensemble, CFI-Net. Results show that CFI-Net exhibited excellent performance by achieving an accuracy of 98.06%. The literature [23] proposed an FL-based framework based on deep learning to classify monkeypox lesions. Their framework consists of three main parts: (a) DL models, (b) An environment for security, and (c) a network to augment data samples for training. Publicly accessible datasets are used for the training of the ViT-B32 model and achieved an accuracy of 97.90%. The study [24] investigates the potential for using pre-trained deep-learning models to identify monkeypox. It provides an overview of the challenges in correctly diagnosing monkeypox. The performance of many pre-trained deep learning models is tested in the study, focusing on how well they can extract useful features from medical pictures for classification tasks. Compared to ResNet-50 and InceptionV3, EfficientNet-B7 showed significantly better monkeypox case detection, with a maximum accuracy of 94.6%. A paper presents MOX-NET [25], a multi-stage deep hybrid feature fusion and selection framework to classify monkeypox. The suggested model surpasses conventional methods with a high accuracy of 96.8%. The hybrid feature fusion and an efficient feature selection procedure highly enhance the model’s performance, making MOX-NET a reliable method for detecting monkeypox. In the research [26], the monkeypox identification application applies a MobileNetV2 deep learning model with a 93.5% classification accuracy in correctly identifying monkeypox from preprocessed medical images. The strong and light architecture of MobileNetV2, jointly applied with strong preprocessing methods, guarantees high performance, making it suitable for deployment in limited resource contexts. This level of precision marks a potential for incorporating the model into portable diagnostic devices to rapidly and accurately identify illnesses.
We developed a hybrid model by combining Vision Transformer (ViT) and MobileNetV3 to classify Mpox disease. The classical ViT calculates global attention using MHSA (multi-head self-attention), which is computationally expensive and may miss the skin’s edge and boundary region features. At the same time, in the standard convolution, a single filter is applied to the entire image channel. The proposed WMHSA calculates attention locally in the window of the patches to improve the feature map and reduce the computation burden. Moreover, the depthwise convolution applies a filter to each channel independently to improve the spatial feature of the skin lesion. First, images are resized and fed to a convolutional layer named Conv 3 × 3. It extracts basic features such as edges and textures. We obtain hierarchical features while reducing computational costs using MobileNetV3 blocks. The image’s resolution progressively drops as it moves through these blocks from 128 × 128 to 64 × 64, 32 × 32, and so on, making identifying patterns at different sizes. The processed features are then passed to Mobile ViT blocks, integrating transformer layers to enhance feature representation. The ViT encoder then refines feature extraction by dividing the processed image into smaller patches and adding positional embedding to retain spatial information. An essential component of this model is the WMHSA layer, which addresses the high computational cost of traditional transformers by processing the image in smaller “windows,” effectively capturing long-range dependencies while optimizing memory and processing efficiency. This approach makes the model concentrate on the important parts of the image without unnecessary computation, thus being more feasible for mobile devices. Finally, a global pooling layer aggregates the features, followed by a fully connected linear layer that outputs the classification result. The architecture of the proposed method is shown in Fig. 1. Using a local feature extraction module, our model efficiently extracts important patterns from the input image, including textures, edges, and fine-grained details. The MV3 block uses depth-wise separable convolutions, which divide the convolution into two stages: a pointwise (1 × 1) convolution to combine cross channel data and a depth-wise convolution to process each channel separately. For a tensor input X∈R^(H×W×C) the depth wise calculation is calculated as in Eq. (1).

Figure 1: The architecture of the proposed model
The convolution is applied to each channel independently, with n × n being the kernel size and pointwise (1 × 1) is calculated as in Eq. (2).
where
where
These components work together to guarantee that the MV3 block extracts rich, localized features in a lightweight way, which makes it appropriate for mobile-friendly applications such as image classification for monkeypox and the output is calculated as in Eq. (5).
The locally processed feature map
where P is number of pixels there are in each patch, the feature dimension for each patch is denoted by d. The total number of patches created is N = H × W/P. Through this transformation, the original large feature map is divided into smaller, easier-to-manage patches, each of which contains localized data. The WMHSA mechanism can more easily capture relationships within and between patches because these patches are processed independently during attention. The model can concentrate on both local details within patches and global dependencies between patches thanks to the unfolding step, which also makes global processing less complicated.
Using weighted aggregation, WMHSA assigns greater weight to specific layers while capturing global dependencies across various image patches. In order to increase accuracy, this attention mechanism ensures the model can comprehend both local and long-range relationships, and to determine the significance of the relationships between patches, each patch is processed through several self-attention layers that compute queries, keys, and values. Self-attention is used to record dependencies between patches for every layer l in the hierarchy and is calculated by in Eq. (7).
where the query, key, and value matrices for the l-th layer are
where L is the total number of attention layers and is the l-th layer’s learned weight, finally, WMHSA produces a globally attended feature map, H∈, that integrates the relationships between patches at different levels to contain rich global information. To restore the spatial structure, the patches are folded back into the original image representation after undergoing WMHSA processing. The information from each patch is accurately aligned within the image’s spatial grid thanks to this folding operation, which combines the processed patches. The folding operation mathematically converts the patch-based representation back into a feature map
This step is crucial for maintaining local and global patterns taken from the attention mechanism so that the model can use the improved features in later phases for classification. By ensuring that the global relationships recorded by WMHSA are incorporated into the image’s spatial context, folding helps the model produce more accurate predictions.
Another MV3 blocks are used to extract the local features, which include textures, edges, and minute details. By connecting remote areas of the image, the global feature which are modelled using WMHS that provide contextual understanding. Following the folding operation to reconstruct the global feature map
In order to minimize dimensional redundancy and prepare the fused feature map for later tasks like classification, the 1 × 1 convolution learns to efficiently weight and align the local and global features. The model performs better on challenging tasks like monkeypox detection because of to this fusion process, which enables it to comprehend both high-level dependencies and fine-grained details. It guarantees a comprehensive and computationally efficient final representation. When local and global features are fused, the resultant feature map,
where the learnable weights and biases of the FC layer are represented by
In this section, dataset description and quantitative results for binary and multi-class Monkeypox lesion diagnosis has been presented.
Through intensive manual searching, the article mainly gathered the monkeypox skin lesion dataset from publicly accessible case reports and websites. In order to preserve the aspect ratio, the photos were resized to 224 × 224 pixels and cropped to their area of interest. Of the 228 photos gathered, 102 are from the “Monkeypox” class, and the remaining 126 are from the “Others” class, which includes measles and chickenpox. We expanded the dataset size by using rotation, translation, reflection, shear, and scaling. Following augmentation, there were 5000 images in the Monkeypox and Others classes for binary classification.
The second dataset has four classes: Normal, Measles, Chickenpox (Cpox), and Monkeypox (Mpox). All of the image classes were gathered from online health websites. It consists of Chickenpox 107, monkeypox 279, measles 91, and normal 293 images. Data augmentation has been used to increase the size of images by 10-fold in each class. We performed each experiment on NVIDIA Quadro RTX-4000 GPU, which had a dual graphics card of 8 and 128 GB RAM. The script is written using Python 2.11 and executed on Windows 10 O.S. The Adam optimizer with an initial learning rate of 0.00001 is used to accelerate the training process, and the model is trained for 160 epochs in a mini-batch of 32.
4.2 Binary Class Confusion Matrix
The binary class classification confusion matrix is shown in Fig. 2. Our model correctly predicted 996 instances as Mpox and 4 as false negatives. The bottom-right quadrant indicates that the model correctly classified 989 as Normal, and the bottom-left, False Negatives, indicates that 11 Mpox cases were wrongly predicted as Normal. The proposed model can predict precisely 1985 out of 2000. We presented the performance measures in Table 1.

Figure 2: Result of binary class confusion matrix

Table 1 shows that each of the evaluation metrics Kappa, Recall, Precision, F1-score, and Accuracy, both classes have a Kappa statistic value of 98.5%, which indicates an excellent performance, being a measure of agreement between actual and expected classifications adjusted for chance. Similarly, the model’s high recall of 98.91% for the Mpox class and 99.60% for the Normal class indicates its ability to classify almost all positive examples correctly. Moreover, the precision scores of the model, 99.60% for Mpox and 98.90% for Normal indicate that it generates very accurate positive predictions with very few false positives. For both classes, the F1-score, representing the harmonic mean of precision and recall, is consistently 99.25%. Such balance suggests that the model is just as good in avoiding false positives as false negatives, making it reliable in practice.
4.3 Multiclass Confusion Matrix
The confusion matrix for multiclass is presented in Fig. 3. Table 3 shows that the Fold 1 model can predict 209 Cpox out of 214. For the Measles class, 176 out of 182 were correctly classified, except for a few misclassified as Cpox, Mpox, and Normal. The model performed notably well for Mpox, correctly classifying 551 out of 558 cases. In Fold 2, among the 215 cases correctly classified as Cpox, one was misclassified as Measles, Mpox, or Normal. In Measles, the model rightly predicted 177 out of 182 cases, with a small percentage misclassifying them into other categories.


Figure 3: Result of multiclass classification. (a) Fold 1; (b) Fold 2; (c) Fold 3; (d) Fold 4; (e) Fold 5
At the same time, in the Fold 3 out of 214 cases, the model correctly classified 212 cases as Cpox, misclassifying one case as Measles and one as Normal. In the case of measles, the model predicted 178 out of 182 cases. The model predicted 555 as Mpox out of 558, and misclassifications occurred in just 3 instances. In Fold 4, it correctly identified 213 out of 214 Cpox cases, misclassifying only 1 case as Normal. For Measles, it correctly identified 179 of the 182 cases and misclassified 3 others. Finally, in Fold 5 it misclassifies only one case as Normal out of 213. The model correctly classifies 182 out of 183 measles datasets, with just one mislabeled as mpox. Kappa, Recall, Precision, F1-score, and Accuracy are some of the performance metrics of the multiclass classification model, which evaluates five folds of cross-validation that are presented in Table 2. This thorough analysis highlights the model’s generalization and stability over subsets of the dataset.
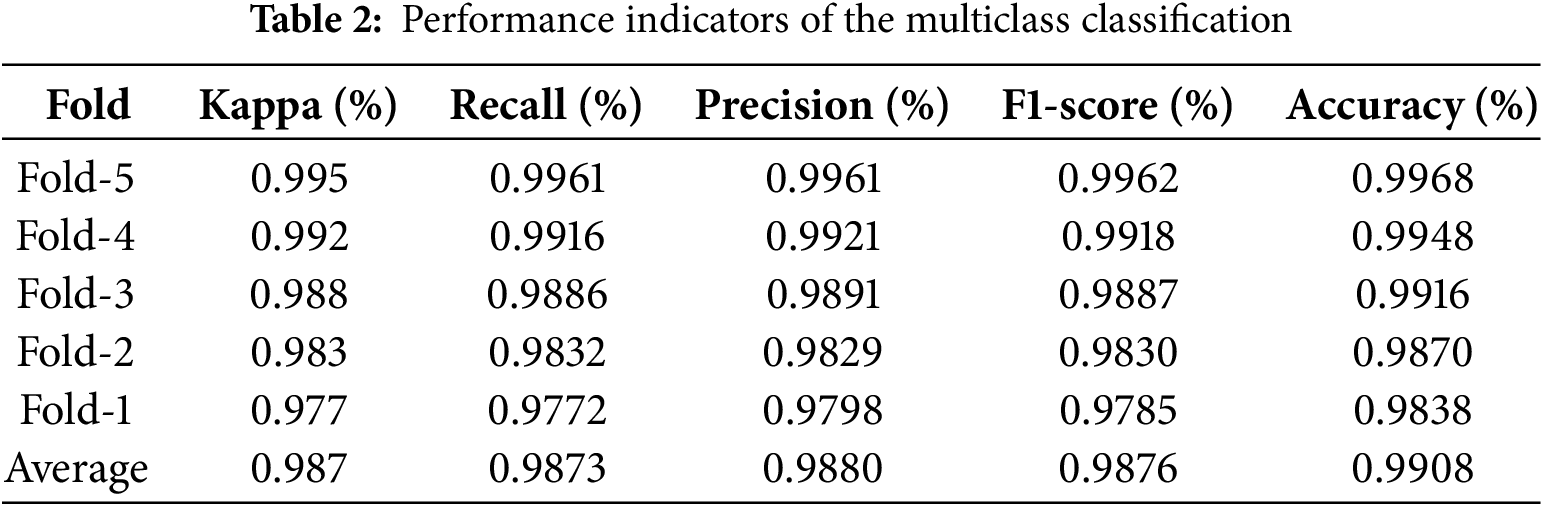
The fold-wise performance demonstrates the model’s robustness in individual runs. The model performs best for Fold-5 with a Kappa of 99.5%, Recall and Precision of 99.61%, F1-score of 99.62%, and Accuracy of 99.68%, showing nearly flawless categorization. Fold-4, though with slightly lower metrics, still shows excellent agreement, Kappa 99.2% and balanced classification metrics Recall 99.16%, Precision 99.21% and F1-score 99.18%. The values of metrics. Fold-1 holds the lowest scores Kappa 97.7%, Recall: 97.72%, Precision: 97.98%, F1-score: 97.85%, Accuracy: 98.38%. This decrease is quite normal since the categorization problem is inherently complex and because of probable variations in the data splits. The average performance across all folds shows the general consistency and reliability of the model. After controlling for chance, the average Kappa of 98.7% indicates very high agreement between the labels that were anticipated and the actual labels. The excellent ability of the model to correctly identify true positives and maintain the accuracy of the forecast is demonstrated by its average recall of 98.73% and precision of 98.80%, respectively.
The training and validation accuracy and loss curves are shown in Fig. 4. Fig. 4 shows that initial training and validation accuracy is less. After a few epochs, training accuracy increased by more than 99% on both datasets. Similarly, the initial training and validation loss for both datasets are high. It started decreasing and reached below 0.01.

Figure 4: Accuracy curve and loss curve. (a) Accuracy binary class; (b) Accuracy multiclass; (c) Loss binary class; (d) Loss multiclass, respectively
In this section, we present that quantitate results comparison with InceptionV3 [27], ResNet-50 [28], MobileNetV3 [29], SwinT [30] and SI-ViT [31]. We utilized same experimental condition to evaluate all the methods and binary class results are presented in Table 3.
Table 3 shows that with a Kappa of 98.50%, our model outperforms SwinT to 95.45% and SI-ViT to 96.23%. It has a better recall, 99.26%, and precision, 99.25%, than SI-ViT and SwinT, reflecting its ability to intensely recognize true positives and support high precision in positive predictions. Moreover, the proposed approach has an incredible F1-score of 99.25%, which also results in robust performance and a high accuracy level of 99.25%, which is greater than the rest and also supports it as excellent. Compared to conventional approaches such as ResNet-50, MobileNetV3, and InceptionV3, the suggested approach outperforms them highly. However, SI-ViT and SwinT perform closely, and finally, in total, the suggested approach represents a benchmark for accuracy, sensitivity, and reliability. Furthermore, we presented multi-class results in Table 4.

The results of Table 4 showed that the proposed model achieved the highest metrics scores, showing superiority in this task. With a Kappa value of 98.70%, the proposed method demonstrated exceptional agreement between its predictions and the ground truth while significantly outperforming results for SI-ViT at 95.27% and SwinT at 94.27%. It also achieved the highest precision of 98.80% and recall of 98.73%, which means it would identify all relevant instances without missing any while still making extremely reliable positive predictions. The F1-score of the proposed method is the highest among all the models at 98.76%, which means that its precision and recall performance are almost balanced. The proposed method achieves the highest accuracy of 99.08%, making it the most general efficient model. SI-ViT and SwinT are competitive compared to the other models, and on all metrics, SI-ViT outperforms SwinT by a small margin. For instance, SI-ViT obtains respectable scores of 96.88% Recall, 97.02% Precision, and 97.34% Accuracy but is still not comparable with the proposed method. With a recall of 95.14%, precision of 96.57%, and accuracy of 95.28%, SwinT is ranked second. The metrics above prove the efficiency of the proposed method while showing off the sophisticated ability of these models compared to the traditional architectures.
Fig. 5 shows examples of different dermatological conditions and grad-CAM visualizations of those conditions. Images in the first group are examples of chickenpox: fluid-filled blisters and characteristic rash caused by the Varicella zoster virus. The grad-CAM image of chickenpox may be of special interest when assessing the severity or spread of infection. This can illustrate the characteristic lesions, similar to chickenpox but typically more significant and have a central depression. In the image of monkeypox, a grad-CAM result is added to emphasize specific regions that show the presence of lesions in higher density. The third set of pictures is of the measles rash, which typically begins on the face and spreads outward to other body areas. The rash is usually flat, red spots that spread out and merge into a large, blotchy area.

Figure 5: The grad-CAM results of the proposed method
4.7 The ROC Based Comparison with SOTA
The ROC curve, as shown in Fig. 6a, compares the performances of several models for binary classification. A dashed, diagonal line performs an utterly random classifier AUC (Area Under the Curve). Each curve represents different models, with the performing model closer to the left upper-hand corner of the plot, which means low FPR and very high TPR. The AUC is used to evaluate each model’s discrimination ability. The AUC is scaled between 0.5, the worst-case scenario of random guessing, and 1.0, representing perfect classification. The highest AUC was that of HMTNet at 0.9931, followed by SI-ViT at 0.9817. MobileNetV3 has the lowest AUC but performed better than random guessing at 0.9163. This comparison shows that HMTNet is the best model for this classification task.

Figure 6: The ROC plot (a) Binary class and (b) Multiclass
Multiple Models performance is shown in Fig. 6b and evaluated one-vs.-all, based on ROC curve classification from more than two classes. Curves closer to the top-left corner mean better classification ability. AUC is also measured for each model, and higher values indicate a more discriminative power of a model. Our HMTNet scores with near-perfect results, at a level of 0.9986 AUC, topping other models. SI-ViT came next with an AUC value of 0.9876. Even with the lowest AUC of 0.9386, MobileNetV3 outperforms significantly. Overall, the results show that HMTNet is the best model for this multi-class classification task, though all other models performed well.
We performed the cross-sensor analysis of the proposed method to generalize the model. The model is trained on the Mpox Skin Lesion Dataset Version 2.0 (MSLD v2.0) [32]. This dataset contains six classes: mpox, chickenpox, measles, cowpox, hand-foot-mouth disease, and healthy. Furthermore, MSLD v2.0 has 755 original images and 10,570 augmented images. After training on the augmented dataset, we tested the binary and multiclass datasets used in the proposed study, and the results are presented in Table 5. Table 5 shows that the model achieved precision and accuracy of 97.83% and 97.81% on the binary class classification. At the same time, our model obtained precision and F1-score of 96.20% and 95.80%, respectively, for the multiclass classification.

4.8.1 Performance Evaluation on Noisy Images
The dataset used in the study is less noisy. To evaluate the model on the noisy data, we added Gaussian noise of different noise levels (NL) on the binary and multi-class datasets, and the results are presented in Table 6. We notice that the model achieved precision and Kappa values of 98.14% and 97.26% for binary classification on the NL = 10. At the same time, with the increase in NL, the HMTNet performance slightly decreases. Furthermore, on the multi-class dataset, HMTNet obtained 96.15% Kappa and 97.03% precision value. In addition, Kappa and precision scores of 94.25% and 95.10% were obtained for NL = 30. For NL = 50, the HMTNet obtained precision and Kappa scores of 91.62% and 92.68%, respectively.

4.8.2 Statistical Analysis of Performance
We performed a statistical p-value test on the multiclass dataset. We made following assumptions. Null Hypothesis (H0): The classification model does not perform better. Alternative Hypothesis (Ha): The classification model performs better. Confidence Value and p-value are calculated using Multiclass Classification results from Table 2 as given below:
Mean Accuracy (
where A is accuracy per each fold.
Standard Deviation (SD):
where Ai is the accuracy of each fold,
Standard Error (SE):
And 95% Confidence Interval: using Z = 1.96 for 95% confidence:
p-values are using the observed accuracy (
Z = 233.71
The Z-score is extremely large, making the p-value effectively 0, indicating the null hypothesis can be rejected. This confirmed model performed better, and it is statistically significant.
4.8.3 Effects of Different Components
We performed an ablation study using different components of the model on both datasets, and the results are presented in Table 7. Table 7 shows that MV3 precision on the first dataset (binary classification) has a precision value of 95.86%. At the same time, MV3+ViT (MHSA) improved the precision and kappa value by 1.26% and 2.15%, respectively. Moreover, the proposed MV3+ViT (WMSA) obtained 99.25% precision and 98.50% kappa score. Furthermore, on the second dataset, MV3 achieved 94.58% classification accuracy. At the same time, MV3+ViT (MHSA) improved the precision by 2.12%. Moreover, our MV3+ViT (WMSA) obtained 98.80 precision and 99.08% classification accuracy.

4.9 Performance Comparison with Other SOTA Methods
We compared the performance of the proposed method with the SOTA methods, as depicted in Table 8. Table shows that the performance of the classical CNN-based method is relatively less than that of ViT-based methods. In addition, federated learning achieved remarkable performance. Moreover, our CNN and ViT with WMSA obtained relatively better performance.
The research found that the HMTNet successfully addresses the requirement for accurate and easily accessible monkeypox diagnostic tools, particularly in settings with limited resources. Our model effectively captures local feature extraction and global context by combining transformer-based architecture with convolutional layers. This results in a high accuracy rate in differentiating monkeypox from other comparable dermatological conditions. The model’s performance demonstrates its resilience and dependability with a binary classification accuracy of 99.25% and a multiclass classification accuracy of 98.38%. Its lightweight design also makes it compatible with mobile and edge devices, providing a valuable way to help medical professionals quickly identify and treat monkeypox cases, essential for containing outbreaks. The suggested model will be improved in the future with an emphasis on increasing its effectiveness, usefulness, and practicality. By using dispersed datasets from many locations, federated learning will enhance model generalization while maintaining patient data security. The model’s lightweight design will make diagnosing monkeypox in real-time on mobile and edge devices easier for medical personnel, especially in environments with limited resources. Cross-validation across various areas will be expanded to improve dependability and determine region-specific modifications needed for broader adoption. The use of explainable AI methodologies to offer interpretable diagnostic insights will be one of the following usability improvements. This will help clinical decision-making and build confidence in AI-driven technologies. Additionally, efforts will be made to optimize the model through advanced quantization and pruning techniques to further reduce memory consumption and inference time. Ensuring seamless integration with existing healthcare systems and compliance with regulatory standards will also be key priorities.
Acknowledgement: The authors extend their heartfelt gratitude to the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R432), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: This research is supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R432), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: Conceptualization, Poonam Sharma, Dhirendra Prasad Yadav, Bhisham Sharma; Data curation, Dhirendra Prasad Yadav, Bhisham Sharma; Formal analysis, Surbhi Bhatia Khan, Ahlam Almusharraf; Investigation, Poonam Sharma, Bhisham Sharma; Methodology, Dhirendra Prasad Yadav, Poonam Sharma, Bhisham Sharma; Project administration, Surbhi Bhatia Khan, Ahlam Almusharraf; Resources, Surbhi Bhatia Khan, Ahlam Almusharraf; Software, Poonam Sharma, Bhisham Sharma; Visualization, Poonam Sharma, Bhisham Sharma; Writing—original draft, Poonam Sharma, Dhirendra Prasad Yadav; Writing—review & editing, Bhisham Sharma, Surbhi Bhatia Khan, Ahlam Almusharraf. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of multiclass classification in this study are openly available in (Mendeley data) at https://data.mendeley.com/datasets/r9bfpnvyxr/6 (accessed on 27 February 2025) and for binary class classification it is available in (Github repository) at https://github.com/mHealthBuet/Monkeypox-Skin-Lesion-Dataset?tab=readme-ov-file (accessed on 27 February 2025).
Ethics Approval: This study did not involve any human or animal subjects, and therefore, ethical approval was not required.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Bunge EM, Hoet B, Chen L, Lienert F, Weidenthaler H, Baer LR, et al. The changing epidemiology of human monkeypox—a potential threat? A systematic review. PLoS Negl Trop Dis. 2022;16(2):e0010141. doi:10.1371/journal.pntd.0010141. [Google Scholar] [PubMed] [CrossRef]
2. Sklenovská N, Van Ranst M. Emergence of monkeypox as the most important orthopoxvirus infection in humans. Front Public Health. 2018;6:383729. doi:10.3389/fpubh.2018.00241. [Google Scholar] [PubMed] [CrossRef]
3. Ganesan A, Arunagiri T, Mani S, Kumaran VR, Sk G, Elumalai S, et al. Mpox treatment evolution: past milestones, present advances, and future directions. Naunyn-Schmiedeberg’s Arch Pharmay. 2024 2024;398(2):1057–80. doi:10.1007/s00210-024-03385-0. [Google Scholar] [PubMed] [CrossRef]
4. Beer EM, Rao VB. A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Negl Trop Dis. 2019;13(10):e0007791. doi:10.1371/journal.pntd.0007791. [Google Scholar] [PubMed] [CrossRef]
5. Sorayaie Azar A, Naemi A, Babaei Rikan S, Bagherzadeh Mohasefi J, Pirnejad H, Wiil UK. Monkeypox detection using deep neural networks. BMC Infect Dis. 2023;23(1):438. doi:10.1186/s12879-023-08408-4. [Google Scholar] [PubMed] [CrossRef]
6. Sitaula C, Shahi TB. Monkeypox virus detection using pre-trained deep learning-based approaches. J Med Syst. 2022;46(11):78. doi:10.1007/s10916-022-01868-2. [Google Scholar] [PubMed] [CrossRef]
7. Asif S, Zhao M, Li Y, Tang F, Ur Rehman Khan S, Zhu Y. AI-based approaches for the diagnosis of Mpox: challenges and future prospects. Arch Comput Methods Eng. 2024;31(6):3585–617. doi:10.1007/s11831-024-10091-w. [Google Scholar] [CrossRef]
8. Jaradat AS, Al Mamlook RE, Almakayeel N, Alharbe N, Almuflih AS, Nasayreh A, et al. Automated monkeypox skin lesion detection using deep learning and transfer learning techniques. Int J Environ Res Public Health. 2023;20(5):4422. doi:10.3390/ijerph20054422. [Google Scholar] [PubMed] [CrossRef]
9. Alakus TB, Baykara M. Comparison of monkeypox and wart DNA sequences with deep learning model. Appl Sci. 2022;12(20):10216. doi:10.3390/app122010216. [Google Scholar] [CrossRef]
10. Saleh AI, Rabie AH. Human monkeypox diagnose (HMD) strategy based on data mining and artificial intelligence techniques. Comput Biol Med. 2023;152(2):106383. doi:10.1016/j.compbiomed.2022.106383. [Google Scholar] [PubMed] [CrossRef]
11. Bala D, Hossain MS, Hossain MA, Abdullah MI, Rahman MM, Manavalan B, et al. MonkeyNet: a robust deep convolutional neural network for monkeypox disease detection and classification. Neural Netw. 2023;161(2):757–75. doi:10.1016/j.neunet.2023.02.022. [Google Scholar] [PubMed] [CrossRef]
12. Rabie AH, Saleh AI. Monkeypox diagnosis using ensemble classification. Artif Intell Med. 2023;143(2):102618. doi:10.1016/j.artmed.2023.102618. [Google Scholar] [PubMed] [CrossRef]
13. Dahiya N, Sharma YK, Rani U, Hussain S, Nabilal KV, Mohan A, et al. Hyper-parameter tuned deep learning approach for effective human monkeypox disease detection. Sci Rep. 2023;13(1):15930. doi:10.1038/s41598-023-43236-1. [Google Scholar] [PubMed] [CrossRef]
14. Alharbi AH, Towfek SK, Abdelhamid AA, Ibrahim A, Eid MM, Khafaga DS, et al. Diagnosis of monkeypox disease using transfer learning and binary advanced dipper throated optimization algorithm. Biomimetics. 2023;8(3):313. doi:10.3390/biomimetics8030313. [Google Scholar] [PubMed] [CrossRef]
15. Uzun Ozsahin D, Mustapha MT, Uzun B, Duwa B, Ozsahin I. Computer-aided detection and classification of monkeypox and chickenpox lesion in human subjects using deep learning framework. Diagnostics. 2023;13(2):292. doi:10.3390/diagnostics13020292. [Google Scholar] [PubMed] [CrossRef]
16. Surati S, Trivedi H, Shrimali B, Bhatt C, Travieso-González CM. An enhanced diagnosis of monkeypox disease using deep learning and a novel attention model senet on diversified dataset. Multimodal Technol Interact. 2023;7(8):75. doi:10.3390/mti7080075. [Google Scholar] [CrossRef]
17. Yasmin F, Hassan MM, Hasan M, Zaman S, Kaushal C, El-Shafai W, et al. PoxNet22: a fine-tuned model for the classification of monkeypox disease using transfer learning. IEEE Access. 2023;11:24053–76. doi:10.1109/ACCESS.2023.3253868. [Google Scholar] [CrossRef]
18. Ahsan MM, Alam TE, Haque MA, Ali MS, Rifat RH, Nafi AAN, et al. Enhancing monkeypox diagnosis and explanation through modified transfer learning, vision transformers, and federated learning. Inform Med Unlocked. 2024;45(11):101449. doi:10.1016/j.imu.2024.101449. [Google Scholar] [CrossRef]
19. Uysal F. Detection of monkeypox disease from human skin images with a hybrid deep learning model. Diagnostics. 2023;13(10):1772. doi:10.3390/diagnostics13101772. [Google Scholar] [PubMed] [CrossRef]
20. Yadav S, Qidwai T. Machine learning-based monkeypox virus image prognosis with feature selection and advanced statistical loss function. Med Microecol. 2024;19:100098. doi:10.1016/j.medmic.2024.100098. [Google Scholar] [CrossRef]
21. Raha AD, Gain M, Debnath R, Adhikary A, Qiao Y, Hassan MM, et al. Attention to monkeypox: an interpretable monkeypox detection technique using attention mechanism. IEEE Access. 2024;12(1):51942–65. doi:10.1109/ACCESS.2024.3385099. [Google Scholar] [CrossRef]
22. Asif S, Zhao M, Li Y, Tang F, Zhu Y. CFI-Net: a choquet fuzzy integral based ensemble network with PSO-optimized fuzzy measures for diagnosing multiple skin diseases including mpox. IEEE J Biomed Health Inform. 2024;28(9):5573–86. doi:10.1109/JBHI.2024.3411658. [Google Scholar] [PubMed] [CrossRef]
23. Kundu D, Rahman MM, Rahman A, Das D, Siddiqi UR, Alam MGR, et al. Federated deep learning for monkeypox disease detection on GAN-augmented dataset. IEEE Access. 2024;12:32819–29. doi:10.1109/ACCESS.2024.3370838. [Google Scholar] [CrossRef]
24. Ren G. Monkeypox disease detection with pretrained deep learning models. Inf Technol Contr. 2023;52(2):288–96. doi:10.5755/j01.itc.52.2.32803. [Google Scholar] [CrossRef]
25. Maqsood S, Damaševičius R, Shahid S, Forkert ND. MOX-NET: multi-stage deep hybrid feature fusion and selection framework for monkeypox classification. Expert Syst Appl. 2024;255(8):124584. doi:10.1016/j.eswa.2024.124584. [Google Scholar] [CrossRef]
26. Mohan R, Damasevicius R, Taniar D, Raja NSM, Rajinikanth V. Automatic Monkeypox disease detection from preprocessed images using MobileNetV2. In: 2024 Tenth International Conference on Bio Signals, Images, and Instrumentation (ICBSII); 2024; Chennai, India. p. 1–4. [Google Scholar]
27. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016; Caesars Palace in Las Vegas, NV, USA. p. 2818–26. [Google Scholar]
28. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016; Caesars Palace in Las Vegas, NV, USA. p. 770–8. [Google Scholar]
29. Howard A, Sandler M, Chu G, Chen LC, Chen B, Tan M, et al. Searching for mobilenetv3. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2019; Seoul, Republic of Korea. p. 1314–24. [Google Scholar]
30. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021; Montreal, BC, Canada. p. 10012–22. [Google Scholar]
31. Zhang T, Feng Y, Feng Y, Zhao Y, Lei Y, Ying N, et al. Shuffle instances-based vision transformer for pancreatic cancer ROSE image classification. arXiv:2208.06833. 2022. [Google Scholar]
32. Ali SN, Ahmed MT, Jahan T, Paul J, Sakeef Sani SM, Noor N, et al. A web-based mpox skin lesion detection system using state-of-the-art deep learning models considering racial diversity. Biomed Signal Process Control. 2024;98(10369):106742. doi:10.1016/j.bspc.2024.106742. [Google Scholar] [CrossRef]
33. Alhasson HF, Almozainy E, Alharbi M, Almansour N, Alharbi SS, Khan RU. A deep learning-based mobile application for monkeypox detection. Appl Sci. 2023;13(23):12589. doi:10.3390/app132312589. [Google Scholar] [CrossRef]
34. Kundu D, Siddiqi UR, Rahman MM. Vision transformer based deep learning model for monkeypox detection. In: 2022 25th International Conference on Computer and Information Technology (ICCIT); 2022; Long Beach Hotel in Cox's Bazar, Bangladesh: IEEE. p. 1021–6. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools