 Open Access
Open Access
ARTICLE
LP-CRI: Label Propagation Immune Generation Algorithm Based on Clustering and Rebound Mechanism
1 College of Computer Science, Beijing Information Science and Technology University, Beijing, 100096, China
2 College of Communication Arts and Sciences, Beijing Information Science and Technology University, Beijing, 100096, China
* Corresponding Author: Kongyu Yang. Email:
(This article belongs to the Special Issue: Advanced Bio-Inspired Optimization Algorithms and Applications)
Computers, Materials & Continua 2025, 83(3), 5373-5391. https://doi.org/10.32604/cmc.2025.063311
Received 11 January 2025; Accepted 10 March 2025; Issue published 19 May 2025
Abstract
Many existing immune detection algorithms rely on a large volume of labeled self-training samples, which are often difficult to obtain in practical scenarios, thus limiting the training of detection models. Furthermore, noise inherent in the samples can substantially degrade the detection accuracy of these algorithms. To overcome these challenges, we propose an immune generation algorithm that leverages clustering and a rebound mechanism for label propagation (LP-CRI). The dataset is randomly partitioned into multiple subsets, each of which undergoes clustering followed by label propagation and evaluation. The rebound mechanism assesses the model’s performance after propagation and determines whether to revert to its previous state, initiating a subsequent round of propagation to ensure stable and effective training. Experimental results demonstrate that the proposed method is both computationally efficient and easy to train, significantly enhancing detector performance and outperforming traditional immune detection algorithms.Keywords
In the natural biological immune system, “auto” refers to normal cells or tissues, while “allo” refers to foreign entities such as viruses and necrotic cells [1]. Immune cells are capable of accurately differentiating between these two types, recognizing “foreign” entities and avoiding any interaction with “self” cells, thereby preserving the physiological equilibrium of the organism. The Artificial Immune System (AIS) [2] is a class of intelligent systems inspired by the principles and mechanisms of the biological immune system, leveraging advancements in various information processing and computational technologies. In AIS, the biological concepts of “auto” and “allo” are employed to define the classification of “self” and “non-self” [3], enabling the generation of artificial immune detectors to distinguish between these categories.
A mature immune detector [4] is one that has undergone thorough training and screening within the immune system to effectively recognize a specific antigen (i.e., an abnormal or invasive signal). It serves as the fundamental recognition component of the immune system, distinguishing between autologous and non-autologous samples, essentially functioning as a mature antibody. All mature detectors within the feature space collectively define a region of autologous distribution, which represents the characteristic profile of a normal sample [5]. Samples falling outside this region are considered abnormal (antigens), and their position within the space can be used to determine the type of abnormality, enabling accurate identification of abnormal samples dispersed across the space. The performance of the detector significantly influences the overall effectiveness of the immune system. Many immunization algorithms, such as Negative Selection Algorithms (NSA), which is the core of traditional immune-based methods, offer several advantages, including the absence of a need for a priori knowledge, robustness, and good parallelism [6]. However, these algorithms heavily rely on sufficient labeled samples. When labeled samples for abnormal instances are scarce and the labeled samples for normal instances are insufficient, the model faces the problem of data imbalance during training. Furthermore, in scenarios with insufficient labeled samples, NSA may struggle to generate enough defense mechanisms (antibodies) to fully cover the normal region of the feature space [7]. Although there may be an abundance of unlabeled samples, if their feature distribution significantly deviates from that of normal samples, the model may fail to effectively capture the characteristics of the normal samples. Additionally, these unlabeled samples may contain a considerable amount of noise or erroneous data, which could mislead the model during unsupervised learning or label propagation, ultimately affecting the performance of the detection system. In practice, the challenge of insufficient labeled samples is often encountered. For instance, in Intrusion Detection Systems (IDS) used in network security, anomalous traffic data typically only emerges during actual attacks, making it difficult to collect a sufficient amount of labeled anomaly data during normal conditions, as malicious events are relatively rare.
To address the aforementioned challenges, this paper proposes a label propagation-based immune generation algorithm, incorporating clustering and a rebound mechanism (LP-CRI), inspired by the concept of the affirmative selection algorithm. Initially, the samples are clustered, and label propagation is performed within each cluster, leveraging the similarity among samples within the same cluster. Since clustering may result in different categories of samples being grouped together, a confidence assessment is introduced to evaluate the unpropagated samples [8]. Samples with higher confidence are prioritized for inclusion in the propagation process. To mitigate the negative impact of noisy samples or incorrect labels during the propagation, a rebound mechanism is introduced: if the recognition performance deteriorates after a round of propagation, the mechanism is triggered, reverting to the previous state and initiating the next round of propagation to ensure optimal results. The rebound mechanism operates within each cluster independently, and in a given round of propagation, it is activated separately for each cluster.
In summary, the main contributions of this paper are as follows: (1) A new immune generation algorithm is designed to improve the final accuracy of the generated detector by expanding the labeling of a large number of samples using very few labeled samples based on clustering and rebound mechanism. (2) Experimental evaluations using publicly available datasets and comparisons with several well-performing models validate the effectiveness of the method in this paper.
Inspired by the principle of negative selection, Forrest et al. [9] first proposed the negative selection algorithm in 1994. The algorithm first represents the autosomal data to be protected and monitored as a multiset M, each member of which is a binary string of length L. The set of detectors S is a set of strings of length L. The set of detectors S is a set of strings of length L. The set of detectors S is a set of strings of length L. Then, a set of detectors D is generated, where the detectors S are strings that do not match the strings in M. Finally, the changes in M are monitored by comparing the detectors in D with M, and if a match is found, then an anomaly occurs in M. González et al. [10] proposed the Real Value Negative Selection Algorithm (RNSA). In this algorithm, antigens and detectors are defined as hyperspheres (immunorecognition spheres) in the feature space [2], and the similarity is measured by Manhattan distance. Ge [11,12] summarized the existing forms of artificial immunity algorithms and reviewed the forms, properties and applications of each type of algorithms. The difference between genetic algorithms and artificial immunization principles is compared, and the advantages and disadvantages of the two are contrasted. The focus of these algorithms is to utilize improved detector generation methods to improve training efficiency or detection accuracy. Wang [13] improved the negation selection algorithm by combining the genetic algorithm with the negation selection algorithm to improve the detection accuracy, computational efficiency, and maintain a high level of robustness and effectiveness. Praneet et al. [14] summarized various negative selection classification, representation, and matching techniques in anomaly detection, critically evaluated and classified existing literature to establish future research areas, and developed potential solutions. Siphesihle et al. [15] proposed a new method for the generation of Adversarial Artificial Immune Network (GAAINet) model for intrusion detection in the Internet of Things (IoT) systems. The model improves the quality of the Artificial Immune Network (AIN) detector by introducing a generator that generates fake intrusion samples from the latent space to spoof the classifier (or discriminator). Idris et al. [16] proposed an NSA algorithm based on differential evolution to optimize the distribution of detectors, which reduces the detection holes. Greensmith et al. [17,18] used a hazard model to better distinguish self/non-self cells, and discussed a new model for the detection of intrusions in the Internet of Things (IoT) systems. Non-self cells, and discussed that by simulating the function of dendritic cells, the concept of the hazard model is fully integrated into the actual immunodetector, which improves the efficiency of recognition between antibodies and antigens. Ostaszewski et al. [19] improved the traditional hyper-spherical detector, and introduced a variety of hyper-shape detectors, such as the hyper-ellipsoid and the hyper-rectangle, and these diversified detectors provide a more flexible detection range. These diverse detectors provide a more flexible detection range, effectively cover and monitor the target area, reduce the gaps and improve the detection accuracy. The methods mentioned above enhance the detection efficiency of immune systems by improving the detector generation algorithms, assuming the availability of sufficient labeled samples. However, in practice, it is often challenging to fully cover the feature space with training data due to the lack of adequately labeled samples. This limitation can result in undertraining, low detection rates, and difficulty in meeting performance expectations. In contrast, the LP-CRI approach only requires a small number of labeled samples at the outset. It then achieves label propagation through Density-Based Spatial Clustering of Applications with Noise (DBSCAN) clustering, mitigating the issue of insufficient labeled data while maintaining high detection performance.
In the field of machine learning, confidence learning theory can assess the consistency between predicted samples and known training samples and provide confidence information along with predicted labels. The confidence learning theory proposed by Curtis G. Northcutt et al. [20] can be used to perform identification of mislabels, characterize label noise, and be used for band-noise learning. The method is able to estimate the noise by characterizing and identifying labeling errors in the dataset, based on the principle of pruning noisy data, using probabilistic threshold counts, and training confidence on the examples by ranking them. Teng Shaohua et al. [21] proposed a transfer learning method for selecting confidence pseudo labels (TL-SCP). First, the prediction probability of the most likely category and the prediction probability of other categories are combined when evaluating the confidence of the pseudo-labels; second, the high-confidence labels are retained as much as possible in the label propagation process, and the updating of low-confidence labels is guided accordingly to reduce the propagation of mislabels. The experimental results show that the proposed model (TL-SCP) outperforms the existing models. Ying [22] utilized confidence theory to screen data and filter low-quality samples. First, a few classes in the original dataset are oversampled, and the samples after data balancing are screened based on confidence theory as a way to solve the error or noise samples that may be generated in the sampling process. It can be seen that the use of confidence theory, you can test, assess the credibility of the sample after the propagation of the label as well as filtering noise samples so that they do not propagate, the use of a small number of labeled samples to obtain a sizable number of training labeled samples, and ultimately improve the detection efficiency of the immunodetector purposes.
In the traditional Negative Selection Algorithm (NSA), Positive Selection Algorithm (PSA), the autosomal detector is generated using a limited number of autosomal samples [23]. Obviously, more labeled samples can improve the final result of the detector. However, it is difficult to meet this demand in practical applications. Therefore, LP-CRI improves the traditional PSA detector based on clustering and rebound mechanism (positive selection avoids the time-consuming process of autosomal comparison and saves a lot of time). The rapid expansion of a small number of existing labeled samples is achieved through label propagation, thus more fully training the immune detector and expanding the final coverage of the detector for the purpose of improving the efficiency of immune system detection [24]. The method identifies intrusions by analyzing network traffic data with the help of the idea of affirmative selection in the principle of immunity, and the system considers the normal behavior of the monitored network as self and the abnormal behavior as non-self [25]. After discovering a new type of intrusion and encoding it into the antibody library, the updating of the antibody library does not end; the antibody automatically evaluates and continuously optimizes the update, which ultimately achieves dynamic updating of the antibody library and the whole system.
As shown in the Fig. 1, the general framework of LP-CRI starts with data preprocessing. Then a cluster analysis is performed to divide the sample data into multiple subsets. Each subset undergoes label propagation, and the labels are subsequently updated. The process iterates until a predefined number of iterations is reached. The final step is to generate the final detector.
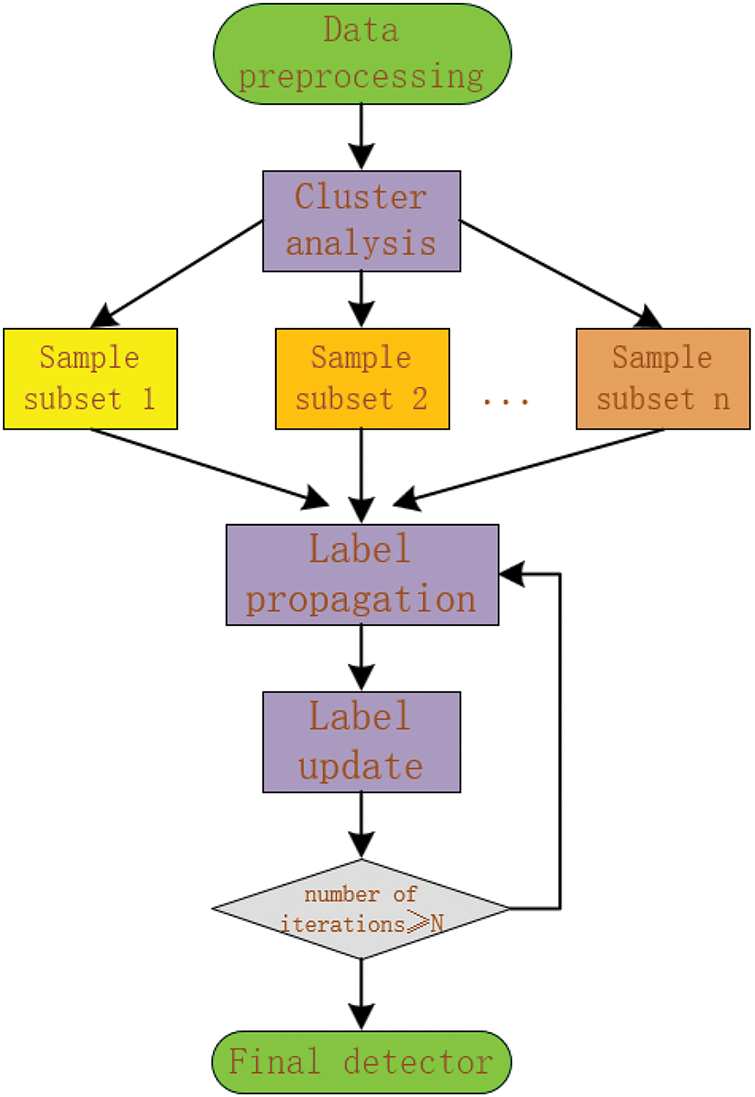
Figure 1: LP-CRI algorithm framework diagram
3.1 Clustering-Based Label Propagation
Different from the traditional NSA and PSA, LP-CRI requires only a small number of labeled samples in the initial stage. Subsequently, the clustered labeled influence propagation algorithm implements labeling expansion in each round of training, obtaining new labeled samples based on the expansion of the labeling propagation scheme, and realizing the expansion of the autosomal detector set.
In this method, there is a labeled set
In general, data points that are in close proximity to each other in the feature space are more likely to belong to the same class than randomly selected data samples [26]. Therefore, in this paper, clustering is performed on the set
In selecting a clustering algorithm for this study, DBSCAN was chosen due to its unique characteristics, particularly its ability to handle high levels of noise and data with irregular shapes. DBSCAN can effectively identify and manage noisy points by labeling them as “noise” rather than forcing them into clusters. Additionally, DBSCAN is based on density, allowing it to form clusters of arbitrary shapes, which is essential for real-world datasets, especially those containing highly nonlinear or complexly shaped samples. Unlike other clustering algorithms, such as k-means, DBSCAN does not require the number of clusters to be specified in advance. Instead, it adapts to the data distribution to determine the optimal number of clusters. In contrast, k-means struggles with noise and assumes uniformly distributed, circular clusters. For datasets with complex shapes, k-means may fail to capture the true cluster structure effectively. Furthermore, k-means requires the user to predefine the number of clusters [28], which can lead to suboptimal clustering results if prior knowledge is lacking. Therefore, DBSCAN is more suitable for the task at hand. DBSCAN has two key parameters: the domain radius Eps and the minimum number of points within the domain radius MinPts. DBSCAN parameter selection (Eps, MinPts): Wine (0.3,26), Stalog (0.3,28), Iris (0.5,8).
The DBSCAN clustering process is as follows. 1) Determine whether a point p is a core point.
After clustering, a number of clusters
where
The confidence of the labeling after sample propagation is further verified using a hypothesis test: the sample set
If
The label propagation process is shown in Fig. 2a–d:
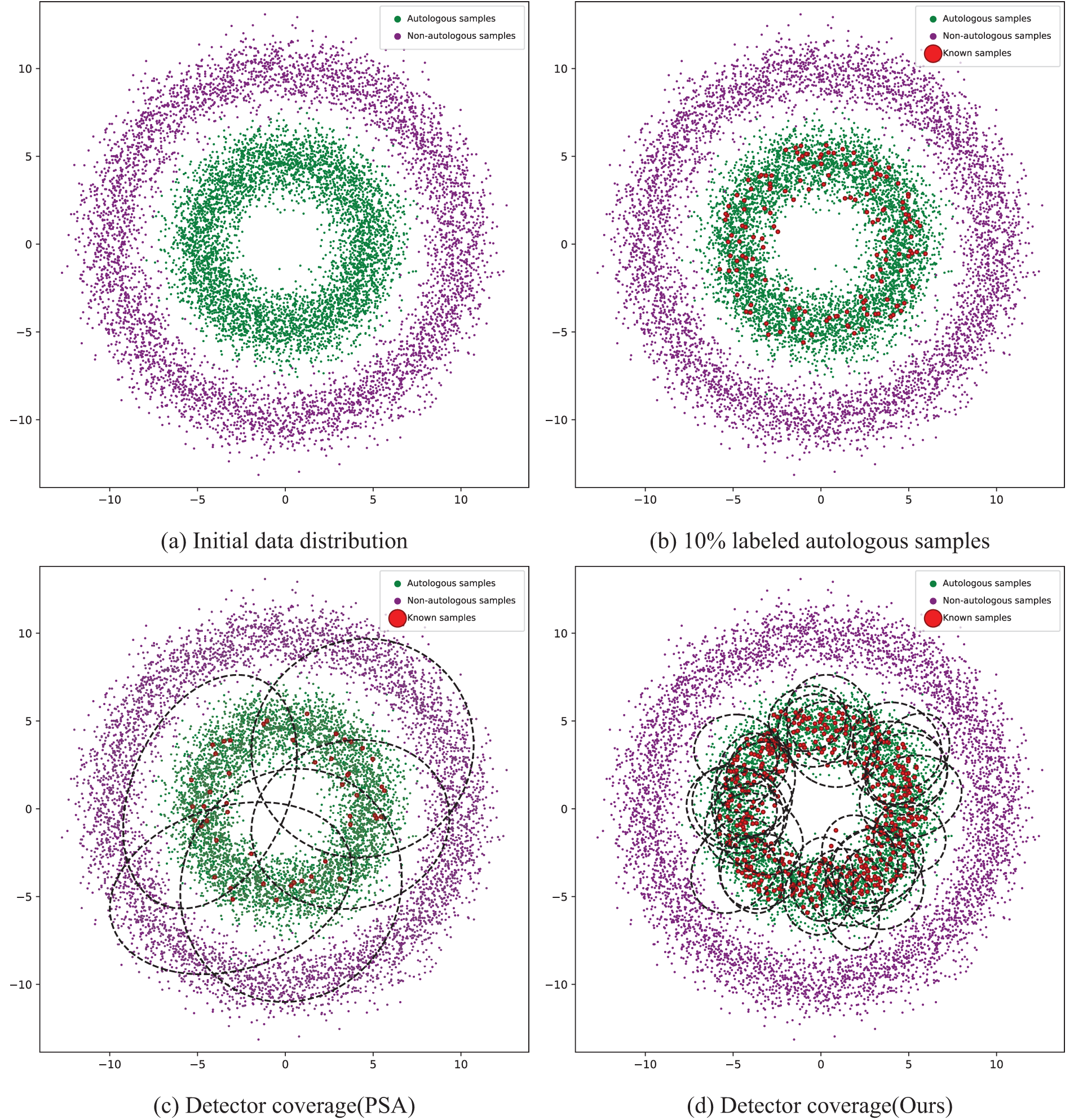
Figure 2: Schematic of label propagation
Fig. 2a represents the distribution of unlabeled samples in the original dataset, where the green dots in the inner circle indicate autosomal samples and the purple dots in the outer circle indicate non-autosomal samples. Fig. 2b represents replacing 10% of the unlabeled samples in the original sample set as labeled autosomal samples, i.e., the red dots indicate the portion. Fig. 2c represents the coverage of the detector trained with the traditional positive selection algorithm PSA based on 10% labeled autosomal samples before label propagation (the part covered by the black circle). Due to the lack of sufficient labeled samples, the detector covers both autosomal and non-autosomal samples, at which time the global accuracy of the detector is only about 70%. Fig. 2d indicates that after n rounds of label propagation, the detector’s coverage of autosomal samples (the part covered by black circles) reaches more than 99%. After label propagation, a large number of labeled samples suitable for immune detector training are generated, which effectively cover the region of autosomal samples, thus improving the final global detection rate.
After each round of training, we can adequately label the autosomal samples through label propagation. However, propagation errors inevitably occur during the training process; moreover, mislabeling events may also occur during the propagation process due to the errors of the original autosomal labeled samples themselves or the presence of noisy samples. With the propagation and the continuous addition of new samples, the detection error in the later stage will expand with the vicious propagation of the algorithm over and over again, resulting in the overall propagation efficiency of the algorithm becoming low. To solve the problem, this paper adopts the rollback detection mechanism to control this error.
Introducing the rebound mechanism: as shown in the Fig. 3, where n is the number of propagation rounds, K is the number of rebound rounds, m is the amount of error accumulation, and t is the error accumulation threshold. After the propagation starts, the number of propagation rounds (n = 0) and rebound rounds (K = 0) are initialized; the number of propagation rounds as well as rebound rounds are recorded before reaching the propagation limit. At the beginning of each propagation round, several newly labeled samples will be obtained. The number of samples generated in each iteration is not fixed, for the sample subset
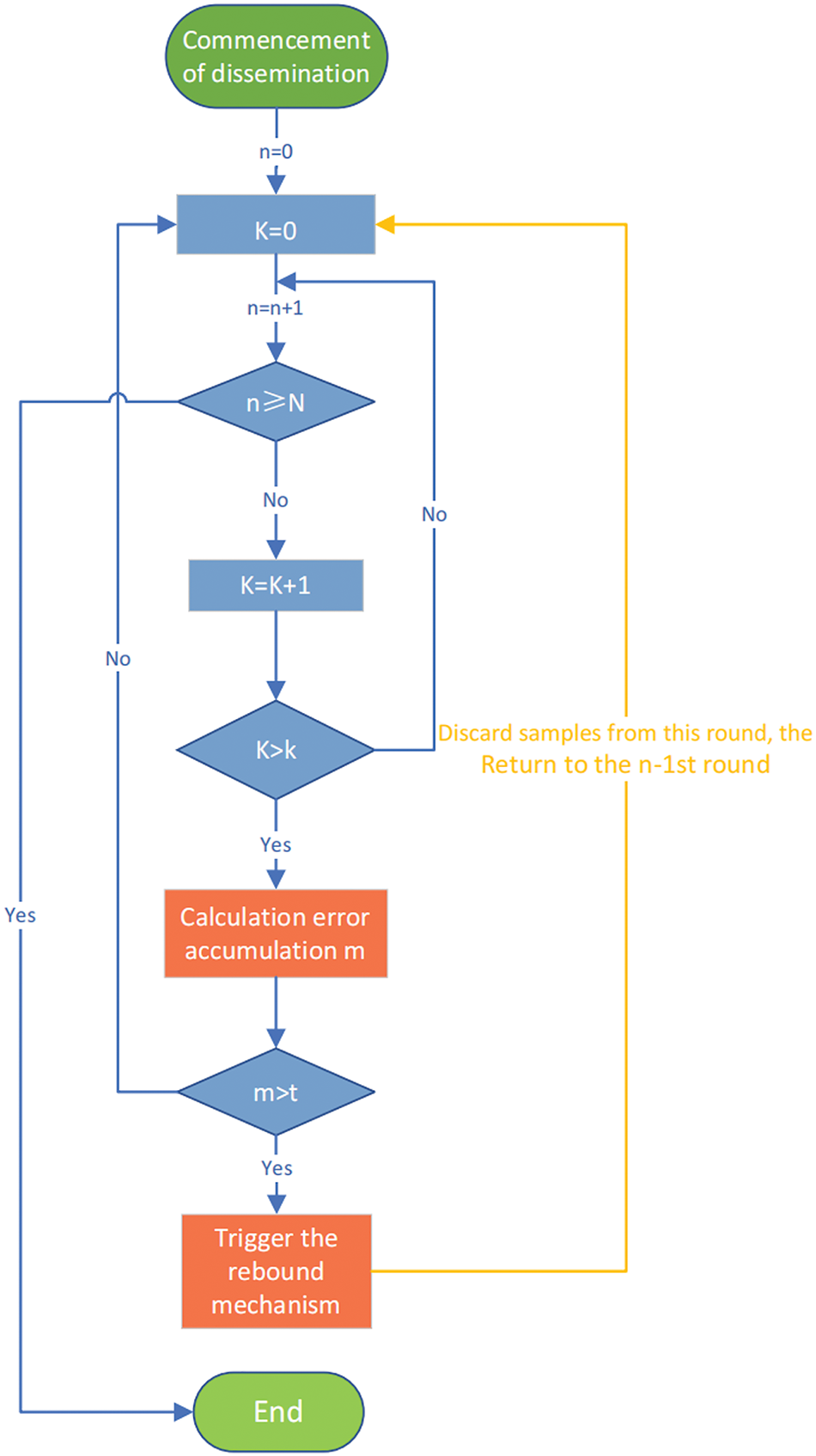
Figure 3: Rebound mechanism
The error record m and the error accumulation threshold t are defined as shown in Eqs. (7) and (8). In Eq. (7),
3.3 Updating of the Immuno-Antibody Pool
The LP-CRI algorithm relies on a small number of labeled samples to generate a large volume of labeled training samples through multiple rounds of label propagation, thus enriching the information in the antibody library. Each round of propagation not only introduces new samples but also evaluates and optimizes existing antibodies through a rebound mechanism, ensuring that the antibody library can adapt to emerging threats and changing environments in a timely manner. This dynamic updating process enables the antibody library to continuously capture new abnormal patterns and attacks, thereby enhancing the overall accuracy and efficiency of the detection system. Each mature detector is analogous to a mature antibody in the biological immune system. As the immune antibody library is updated and expanded, the overall performance and accuracy of the detection system improve. This process mirrors the antibody generation and optimization mechanism in the biological immune system, where continuous learning and adaptation create an effective defense system [29]. The update strategy for the immune-antibody library is as follows:
As shown in Fig. 4, the immune antibody library consists of two components: the total antibody library and the memory cell library. The total antibody library holds all antibodies, each carrying the antigen’s characteristics, such as type, self-adaptation value, and attributes. Each antibody has a flag bit, defaulting to 1. When an antibody is selected as a memory cell, its flag bit is set to 0. During intrusion detection, the total antibody library only uses the antibody with the flag bit 1 to recognize the antigen. The memory cell library is derived from the total antibody pool and contains antibodies that have matched at least N antigens, its self-antibody flag bit will be set to 0. When a memory cell is created, a timer starts, and once the survival time t is reached, the memory cell dies, and the flag bit of the corresponding antibody is reset to 1.
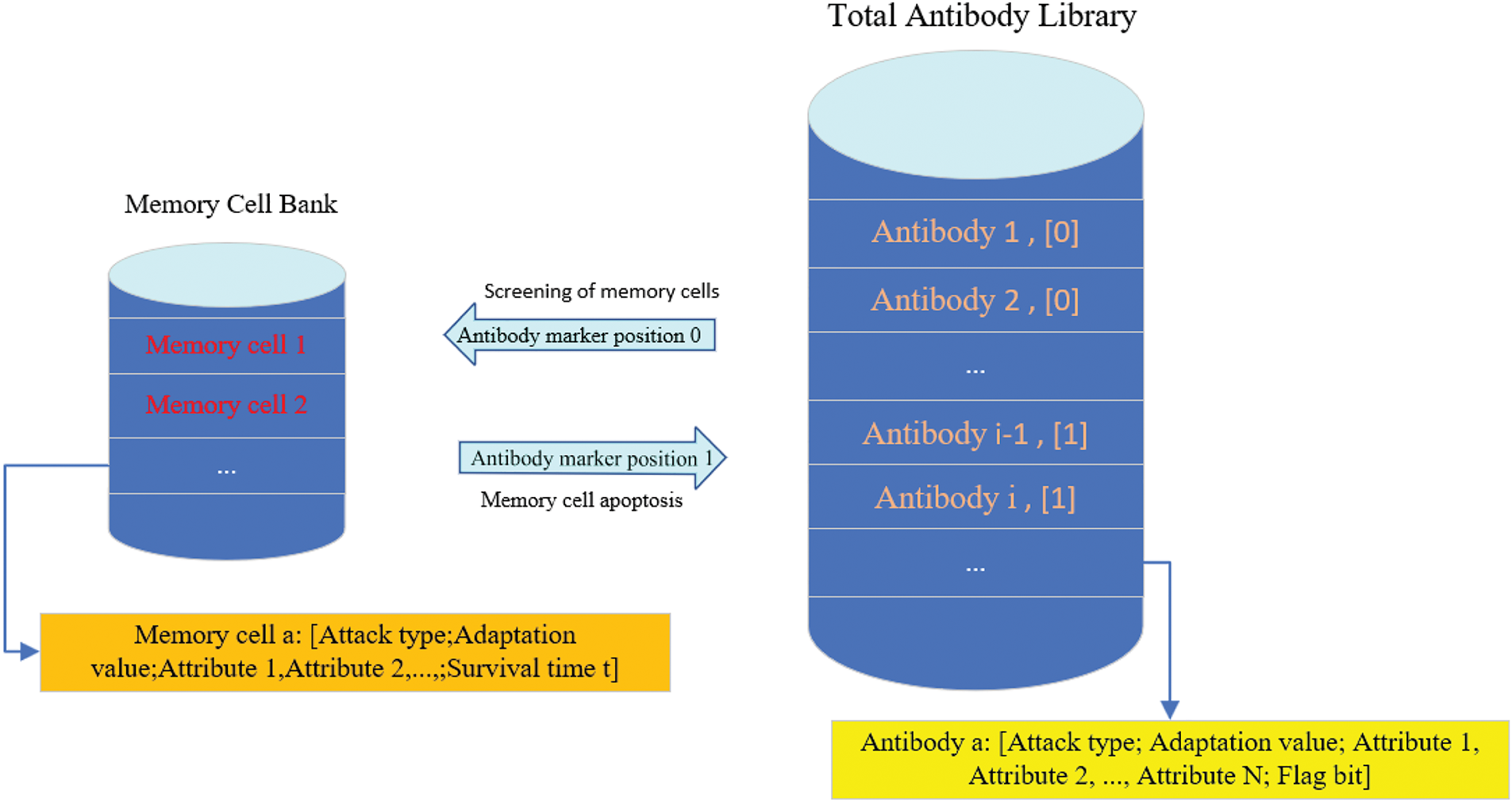
Figure 4: Antibody library composition
During intrusion detection, the system first checks the memory cell pool for antigen matches. If unsuccessful or the memory cell expires, it checks the total antibody pool. Successful matches result in antibodies being flagged and added to the memory cell pool. The total antibody and memory cell libraries are regularly evaluated and updated to keep the antibodies active and relevant, ensuring efficient and timely detection of intrusions with minimal resource usage. The steps are as follows: Step 1: Initialize both libraries, set survival time t, antigen count N, and flag bit. Step 2: Match antigens from the memory cell library first; if a match is found, mark the intrusion as detected. Step 3: If no match is found in the memory cell pool or the survival time t is reached, check the total antibody pool. Step 4: On a successful match, update the corresponding flag and add the antibody to the memory cell pool. Step 5: Periodically evaluate and update the libraries, eliminating unnecessary antibodies and adding new ones. Step 6: Repeat the process for continuous dynamic updating of the antibody library.
3.4 Label Propagation Immune Generation Algorithm
The time complexity of the Algorithm 1 is analyzed as follows: 1) The time complexity of the initialization is a constant

4 Experimentation and Analysis
The UCI (University of California, Irvine) Machine Learning Dataset Repository is a widely used dataset repository managed by the Center for Machine Learning and Intelligent Systems (ICS) at the University of California, Irvine, and is widely used for performance evaluation of immunization algorithms. In this paper, two higher detection accuracies, Stalog and Wine, and one lower detection accuracy, Iris, from this dataset repository are selected as the training and testing sets, as shown in the following Table 1.

The Stalog dataset has 14 features and is relatively balanced, with 383 self-samples and 307 non-self-samples. The Iris dataset, commonly used in classification tasks, contains 4 features, with 50 self-samples and 100 non-self-samples, making it imbalanced in terms of class distribution. Lastly, the Wine dataset consists of 13 features, with 59 self-samples and 119 non-self-samples, also showing some imbalance between classes.
In experimental research on intrusion detection, the Network Security Lab KDD(NSL-KDD) dataset is one of the most widely used and is therefore a suitable choice for experiments and comparisons. The NSL-KDD dataset was developed based on the KDDCup99 dataset, with several improvements made to address its shortcomings. First, the NSL-KDD dataset eliminates redundant data from KDDCup99 and avoids favoring duplicate records during training, leading to more accurate detection rates. Second, the number of records in both the training and test sets is more balanced, ensuring a more reasonable dataset composition. Each network connection record in the dataset is labeled as either normal or abnormal (attack). We took 5000, 10,000 and 100,000 data entries from the dataset, respectively, as shown in Table 2.

Metrics In the evaluation metrics section, we use accuracy (ACC), true positive Rate (TPR), false positive rate (FPR) and F1-score (F1-S):
In terms of interpretation, TP (True Positives): is the number of elements of autopositives that are correctly detected as autopositives, FP (False Positives): is the number of elements of non-autopositives that are incorrectly detected as autopositives, TN (True Negatives): the number of elements of non-autopositives that are correctly detected as non-autopositives, and FN (False Negatives):the number of elements that were incorrectly detected as non-autonomous (attacks) among the autonomous ones.
This chapter will compare with a set of detector generation algorithms, including the widely used variable-sized detectors (V-Detector), the newly proposed negative selection algorithm based on grid file of the feature space (GFNSA) and co-PSA in recent years, and the traditional positive selection algorithm (PSA).
• V-Detector [30]: A real-valued negation selection algorithm with a variable radius randomly generates a set of candidate detectors with a random detection radius, and calculates the distance between the detector and the autologous sample to obtain the detection radius of the detector [31].
• GFNSA [32]: On top of the NSA, the structure of the self-data set is preprocessed into a grid file format before the detectors are created. In addition, a unique grid ID is assigned to each detector, allowing it to be dynamically updated based on changes to the self-data in the grid structure.
• co-PSA: An algorithm that propagates labels to the entire data by expanding the size of the training set through label propagation algorithm (LPA) and selecting the samples with the highest similarity to the samples to be labeled from the labeling matrix for labeling.
• PSA: Immunology-inspired algorithms that identify data from candidate datasets that satisfy specific patterns or behaviors by mimicking the positive selection process of the biological immune system.
Table 3 shows the time complexity of several algorithms. Where

Compared with the traditional immunization algorithms, the clustering and bounce-based label propagation algorithm proposed in this paper achieves the training of a large number of labeled samples through a small number of existing labeled samples, which enhances the training and optimization ability of the immunodetector. We utilize the UCI dataset for experimental validation and compare it with four immunodetector generation algorithms, PSA, GFNSA, co-PSA and V-Detector, and the experimental results are shown in Tables 4–6. The results show that the LP-CRI in this paper has the highest global accuracy and high F1 score values in most scenarios. In addition, compared with the new algorithms GFNSA and co-PSA, this paper’s algorithm has better noise immunity. In the experimental data in this chapter, the salient parts of the model’s performance are boldly marked, as in the bolded portion of Tables 4–9.
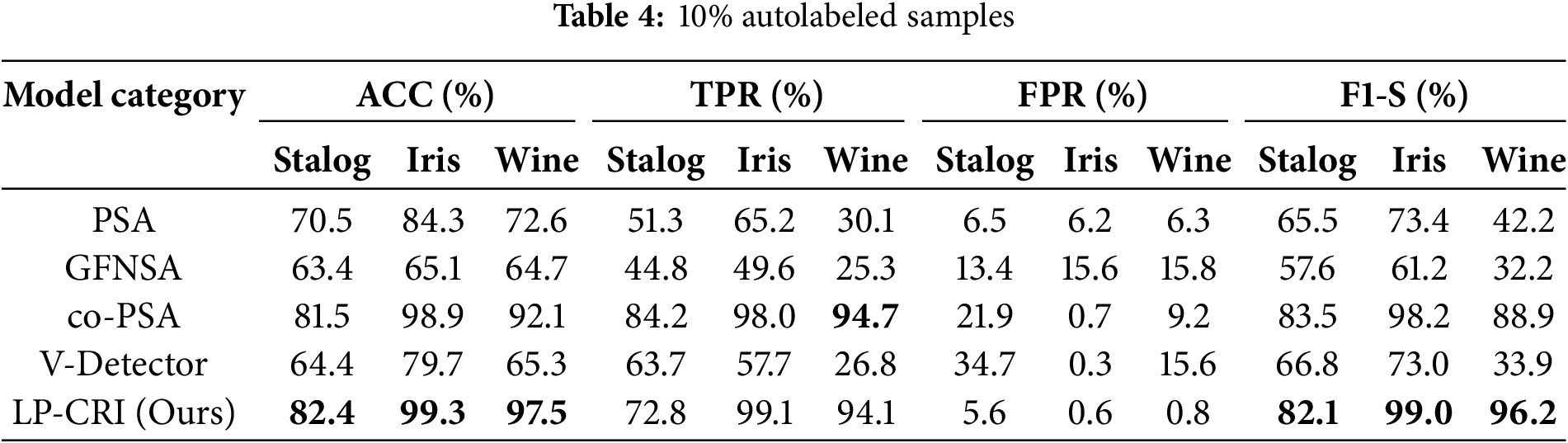
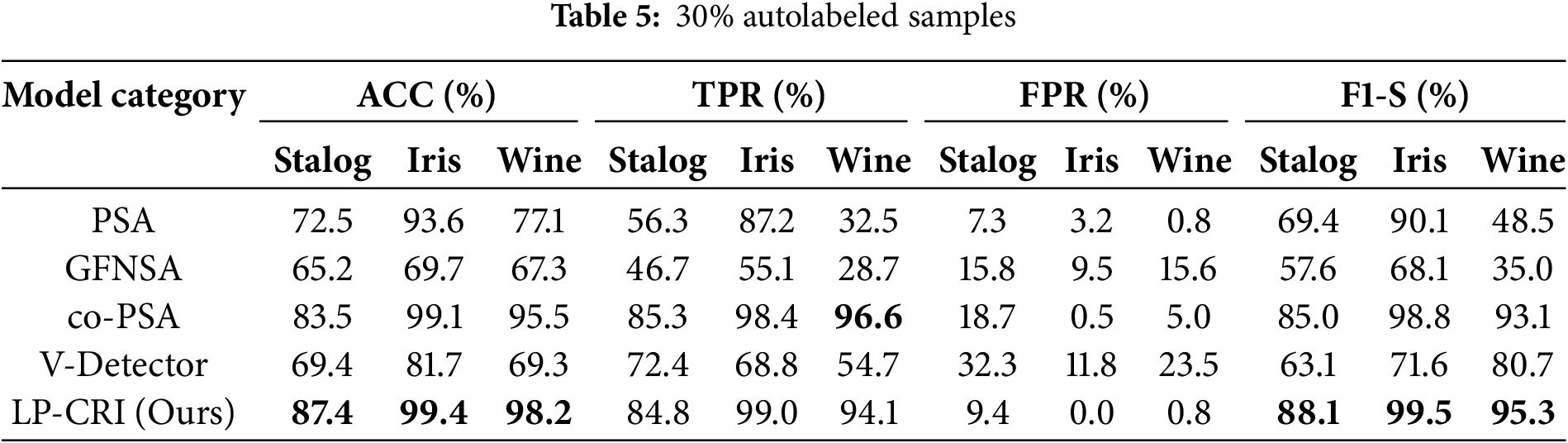
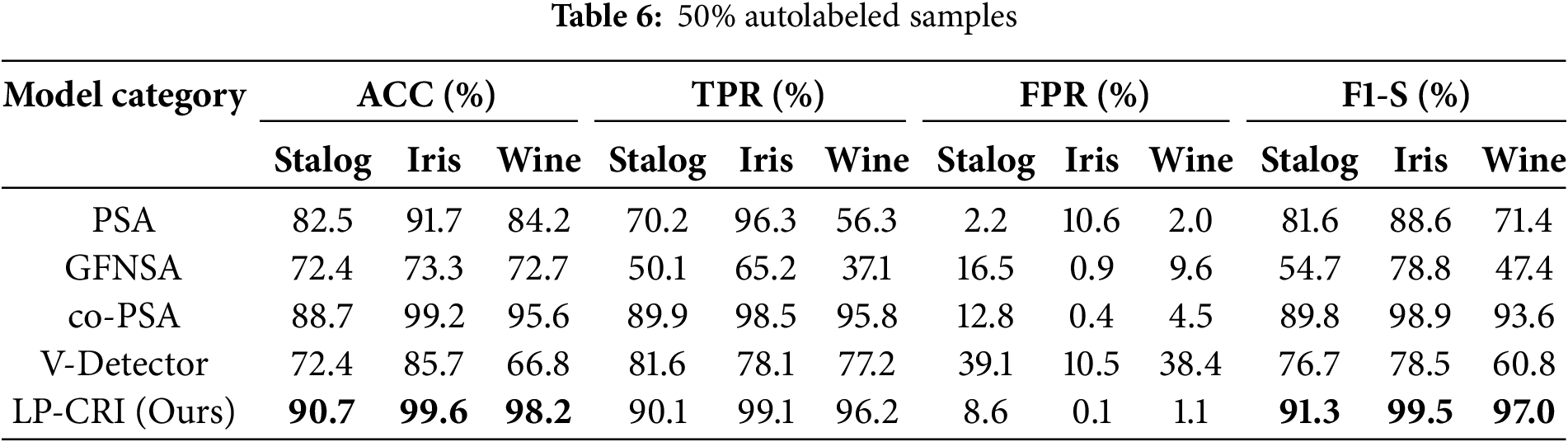
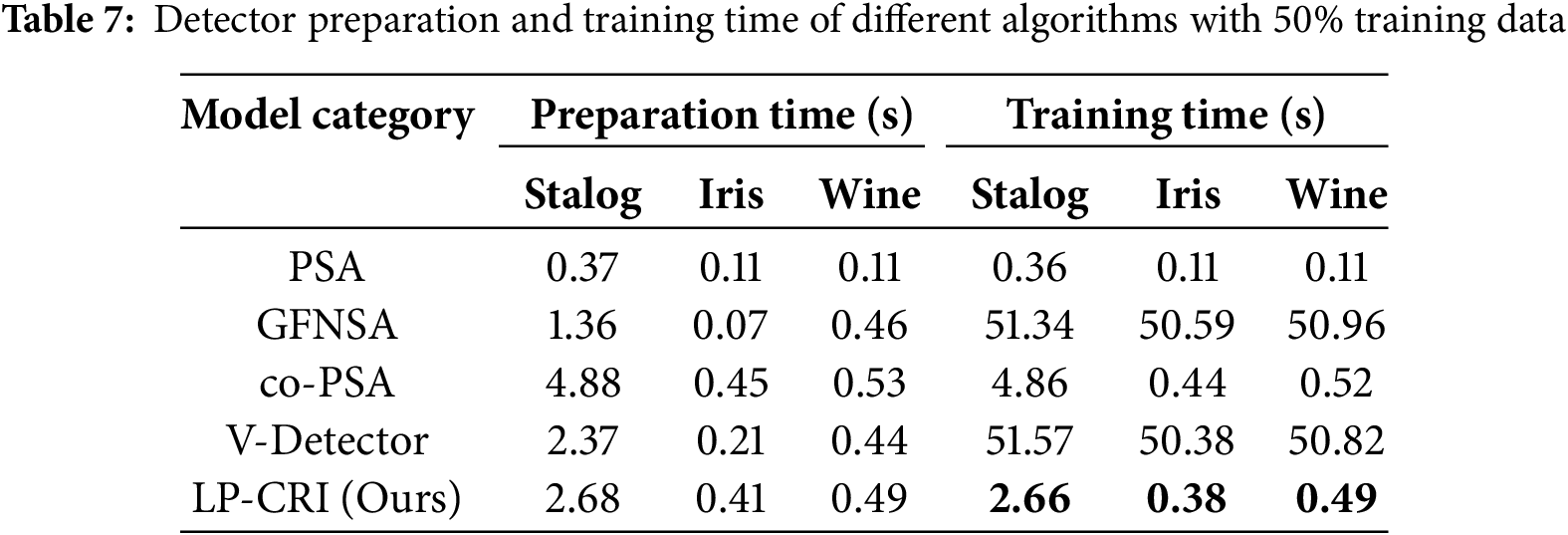
As can be seen from Tables 4–6, LP-CRI has the best Global Accuracy (ACC) and F1-s value in each dataset. Even with a small number of self-labeled samples (10% self-labeled samples), LP-CRI can still maintain a high detection rate. Compared to the traditional PSA, the global accuracy of LP-CRI is at least 10% higher; in the Wine dataset, the F1-S score of LP-CRI exceeds that of V-Detector and GFNSA algorithms by 60. This is due to the Wine dataset has a moderate sample size with a well-balanced distribution of labeled samples and shows high separability between categories. Additionally, the relatively low noise level in the Wine dataset reduces errors from incorrect labeling during propagation, allowing LP-CRI to maintain high-quality labels and improving the accuracy and stability of the detector. In addition, the algorithm in this paper is more balanced in terms of the True Positive Rate (TPR) of the self-sample and the False Positive Rate (FPR) of the non-self-sample. There is no situation where the detection rate of autologous samples is too low in PSA calculation, or the false detection rate of non-autologous samples is too high in the V-Detector algorithm: Compared with PSA, the true detection rate of autologous samples of LP-CRI is improved by more than 50% in some scenarios (e.g., the Wine dataset); compared with the V-Detector algorithm, the false detection rate of non-autologous samples is reduced by at least 20%. As can be seen in Table 7, LP-CRI has a much lower time cost than V-Detector and GFNSA while having a better detection rate. Although LP-CRI has a slightly higher time cost than PSA, it has a better detection rate and a balanced detection of autologous and non-autologous cells. LP-CRI trains an auto-detector by assigning labels to unknown samples through a label propagation mechanism. This approach can generate more mature auto-detectors to cover a wider range of auto-regions. Experimental results show that LP-CRI performs better in the task of immune recognition of normal/abnormal (auto/non-auto) differentiation, especially when the training data samples are very few.
Among several types of algorithms, co-PCA is similar in principle to the algorithm in this paper. Both use a small number of labeled samples from the same class to train a detector by label expansion. The difference is that co-PCA directly selects the sample with the greatest similarity to the sample to be labeled from the label matrix for propagation. Overfitting may occur due to the early completion of propagation caused by the high density of the distribution of a certain type of sample, resulting in too large a difference in the growth of the size between samples of different types. For key nodes on the category boundary, LPA may misclassify them and add them to the training set, resulting in all the samples originally belonging to the same category as the key node being misclassified. The LP-CRI algorithm proposed in this paper first clusters the sampled samples to find cluster members that are likely to belong to the same cluster. Label propagation is performed within each cluster obtained and the labeling results of each round of sampling are recorded. Finally, a hypothesis test and a back-propagation mechanism are used to retain labeled samples with high confidence. This avoids the problem in LPA, where the quality of newly expanded samples is unstable, which causes cumulative errors in subsequent label expansion, thereby reducing the quality of the expanded samples and affecting the final detector training results. As can be seen in Tables 4–6, compared to co-PSA, LP-CRI significantly improves both the ACC and F1-S score. In addition, LP-CRI has a lower FPR and is more stable and balanced in the detection of autologous and non-autologous samples. At the same time, it can be seen in Table 7 that LP-CRI requires less preprocessing and training time for each dataset. In terms of time cost, it is much lower than co-PSA. This is also due to the fact that the clustering algorithm reduces the time cost of processing data and avoids the impact of larger density categories on other categories.
4.4 Noise-Resistance Experiment
In addition to having a good detection rate, the also has good results in terms of noise immunity. In the Wine, Stalog and Iris datasets, 30% of the labeled autologs are used, and the labels of 5% to 50% of the autologs are randomly modified to simulate different noise scenarios. The experimental results are shown in Fig. 5. It can be seen that LP-CRI performs well in all cases, with only a significant decrease in amplitude when 45% of the sample data is noise. The overall detection rate decreased by 5%. Relatively stable performance on three datasets. Co-PSA, on the other hand, has a significant drop in detection rate when noise occurs, with an overall detection rate decrease of 17%. Experiments have proven that LP-CRI has better noise immunity than co-PSA. Based on hypothesis testing and noise learning, it can effectively reduce the error rate of label propagation.
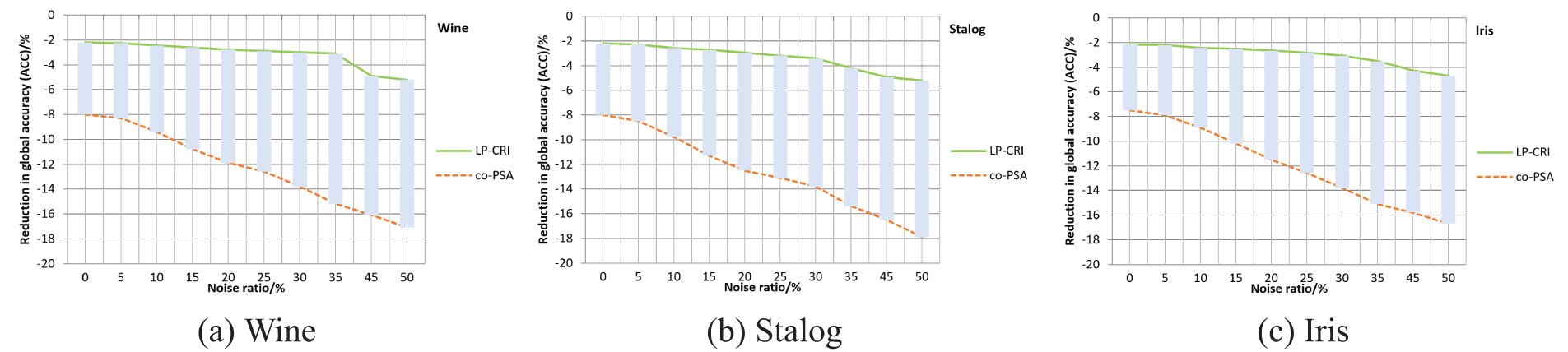
Figure 5: Comparison of noise reduction between LP-CRI and co-PSA
In summary, the proposed label propagation algorithm based on clustering and rebounding outperforms several immune algorithms, V-Detector, PSA, GFNSA and co-PSA, under different proportions of self-labelled samples and multi-class datasets. LP-CRI shows significant advantages in several metrics such as F1 score and ACC, while maintaining a good balance between the true detection rate of the self-sample and the false detection rate of the non-self-sample. In particular, it maintains stable detection performance in the case of sparse labeled samples or imbalanced data. In addition, LP-CRI has a relatively low time cost. Compared with the GFNSA and V-Detector algorithms, LP-CRI has both a better detection rate and a lower time cost. This shows that the algorithm in this paper can effectively use a small number of labeled autosomes to generate high-quality labeled samples through the label propagation mechanism, significantly improving the training effect and detection performance of the immune detector.
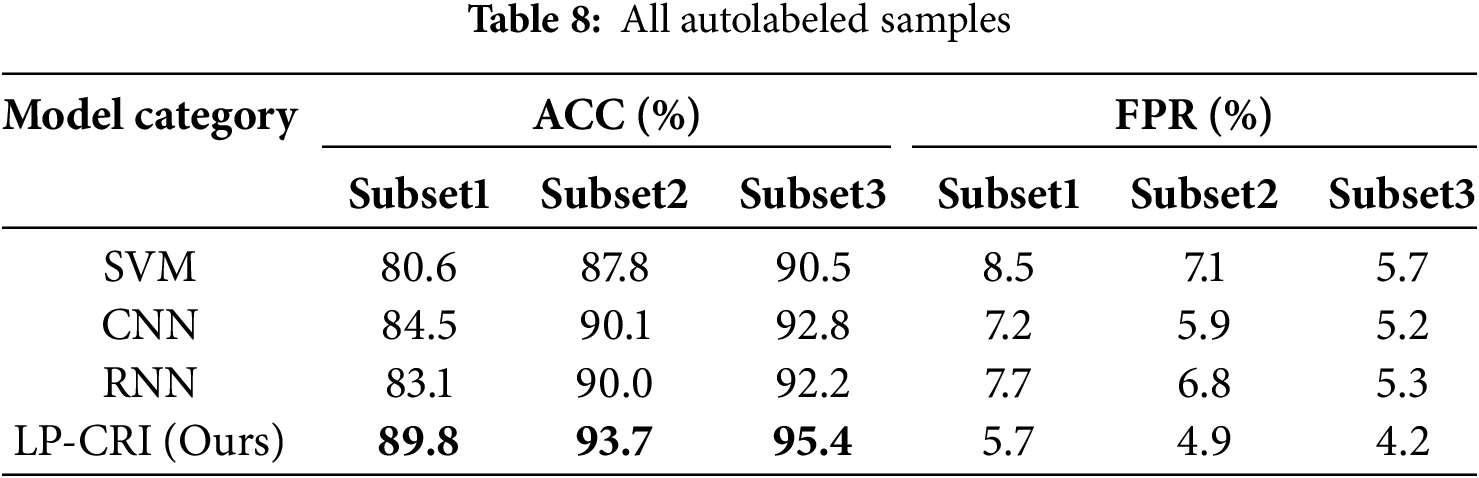
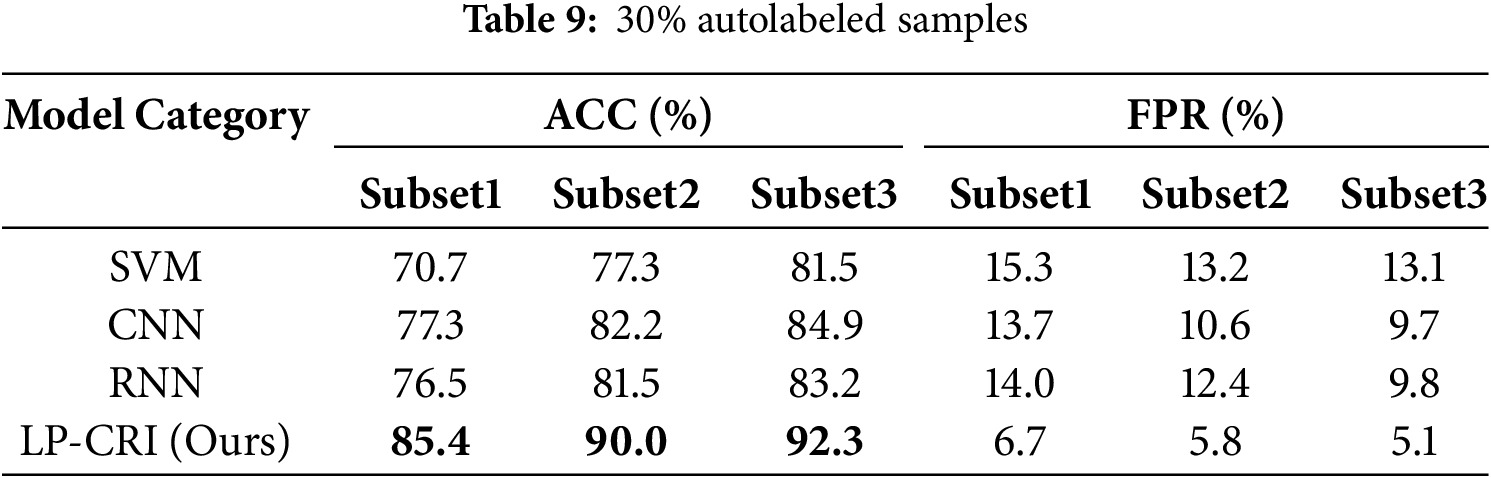
To verify the scalability and practical significance of the LP-CRI algorithm, we compared it with some common machines and deep learning algorithms. The models compared include Support Vector Machines (SVM), Convolutional Neural Networks (CNN), and Recurrent Neural Networks (RNN). The experimental results are shown in Tables 8 and 9. In Table 8, 30% of the labeled autologs are used and in Table 9, all the labeled autologs are used.
As can be seen from the Tables 8 and 9, LP-CRI still maintains a high global accuracy compared to other models. In addition, when the number of labeled drops to 30%, other models show a significant decline. In contrast, although the detection rate of LP-CRI has also decreased, the overall decrease is very small, and it maintains good detection stability. This is because the LP-CRI algorithm trains an auto-detector by assigning pseudo-labels to unknown samples through a label influence mechanism. It performs better in the task of distinguishing normal/abnormal (self-non/self) recognition, especially when the training data samples are very small or the dataset is unbalanced.
In this paper, we propose a label propagation immune generation algorithm based on clustering and rebound mechanism(LP-CRI). We improve the traditional algorithm from the perspective of clustering and label propagation. Unlike traditional methods such as PSA and NSA, in this paper, both labeled and unlabeled data are used to train the detector. We effectively expand the collection of labeled samples from both the self and non-self by label propagation, and introduce an antibody library optimization strategy to optimize the quality of antibodies and the use of computing resources. Experimental results show that LP-CRI performs well on multiple datasets and with different training sample ratios. In particular, its performance is significantly better than that of the traditional NSA and PSA algorithms when training samples are scarce or the dataset is unbalanced.
In future research, we will focus on two key areas. First, we aim to optimize the algorithm. To enhance its performance, we plan to further refine the clustering algorithm to reduce computational overhead and improve detection rates. Additionally, we will develop a more efficient backpropagation strategy to ensure faster and more effective model training. Second, we will explore the practical applications of the algorithm. Specifically, we intend to apply it to the field of network security and intrusion detection. Given the increasing frequency of network attacks, we will integrate the LP-CRI model with a generative adversarial network (GAN) to train a detection model capable of identifying adversarial network attacks. This will enable us to fully realize the potential of the algorithm.
Acknowledgement: We sincerely appreciate the participants who generously contributed their time and effort to this research. Their invaluable support played a crucial role in the success of our study. We are also deeply grateful to the funding agency for providing the necessary resources that made this work possible. Their assistance was fundamental to the completion of this project. Additionally, we extend our thanks to the anonymous reviewers for their thoughtful feedback and constructive suggestions, which greatly improved the quality of this manuscript.
Funding Statement: This work was granted by Key Project of Beijing Municipal Social Science Foundation (No. 15ZHA004) and Key Project of Beijing Municipal Social Science Foundation and Beijing Municipal Education Commission Social Science Program (No. SZ20231123202).
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Hao Huang and Kongyu Yang; methodology, Hao Huang and Kongyu Yang; software, Hao Huang; validation, Hao Huang; formal analysis, Kongyu Yang; investigation, Hao Huang; resources, Hao Huang and Kongyu Yang; data curation, Hao Huang; writing—original draft preparation, Hao Huang; writing—review and editing, Hao Huang and Kongyu Yang; visualization, Kongyu Yang; supervision, Kongyu Yang; project administration, Kongyu Yang; funding acquisition, Kongyu Yang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data openly available in a public repository. The data that support the findings of this study are openly available in UCI at https://archive.ics.uci.edu/datasetS (accessed on 7 March 2025) and NSL-KDD at https://www.unb.ca/cic/datasets/nsl.html (accessed on 7 March 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Zhou X, Tan W. An improved artificial immune negative selection algorithm. In: 2022 7th International Conference on Intelligent Computing and Signal Processing (ICSP); 2022; China. p. 237–41. [Google Scholar]
2. Lin Q, Zhu Q, Wang N, Huang P, Wang W, Chen J, et al. A multi-objective immune algorithm with dynamic population strategy. Swarm Evol Comput. 2019;50(4):100477. doi:10.1016/j.swevo.2018.12.003. [Google Scholar] [CrossRef]
3. Wang H, Gao X, Huang X, Song J. Anomaly detection based on improved negative selection algorithm. Comput Simulat. 2008;25(5):334–8. (In Chinese). doi:10.3969/j.issn.1006-9348.2008.05.085. [Google Scholar] [CrossRef]
4. Yang K. Theory and application of immune evolution. 2nd ed. Beijing, China: Beijing UNESCO Press; 2008. [Google Scholar]
5. Zheng D. Detection distribution method of v-detector based on grey wolf optimization. Intell Comput Applicat. 2022;12(11):34–40. (In Chinese). doi:10.3969/j.issn.2095-2163.2022.11.006. [Google Scholar] [CrossRef]
6. Jin ZZ, Liao MH, Xiao G. Review of negative selection algorithms. J Communicat. 2013;34(1):159–70. (In Chinese). [cited 2025 Mar 9] Available from: https://kns.cnki.net/kcms2/article/abstract?v=6h6U53PWxNQxhB1N8it-WHLIsjmf1s_YkMoq53bO73h7-bjnLeR5mpd7ZdhlcnLwjf6F33zBZ5YKyRfpf3WgIOn0k-095FvANo4vWdOetEINsaaRZYyHMBwxJCz3dYjabfXaLb0nrbhVUgzYHhn0HkhgF2DZR-MUzguw5i2ONyRjw-E8yXqg1VSgB0JyR5TG&uniplatform=NZKPT&language=CHS. [Google Scholar]
7. Hao X, Jiang W, Yuan Y. An improved v-detector algorithm for wireless sensor network intrusion detection technology based on immune system principle. In: Proceedings of 2016 3rd International Conference on Materials Engineering, Manufacturing Technology and Control (ICMEMTC 2016). China: School of Computer & Communication, Lanzhou University of Technology; 2016. p. 690–4. [Google Scholar]
8. Wu BW, Chen JS, Lu JG. Confidence learning based object detection label denoising. In: Proceedings of the 34th China Process Control Conference; State Key Laboratory of Industrial Control Technology, School of Control Science and Engineering. China: Zhejiang University; 2023. 419 p. [Google Scholar]
9. Forrest S, Perelson A, Allen L, Cherukuri R. Self-nonself discrimination in a computer. In: Proceedings of 1994 IEEE Computer Society Symposium on Research in Security and Privacy; 1994; China. p. 202–12. [Google Scholar]
10. González F, Dasgupta D, Niño LF. A randomized real-valued negative selection algorithm. Artif Immune Syst. 2003;2787:261–72. doi:10.1007/b12020. [Google Scholar] [CrossRef]
11. Ge H. Overview of immune algorithms. J South China Normal Univ (Nat Sci Med Edit). 2002;3(3):120–6. [Google Scholar]
12. Ge H. Comparison between immune algorithm and genetic algorithm. J Jinan Univ (Nat Sci Med Edit). 2003;24(1):22–5. [Google Scholar]
13. Wang J. Research and implementation of intrusion detection technology based on artificial immune algorithm and neural network [master’s thesis]. Lanzhou, China: Lanzhou Jiaotong University; 2022. [Google Scholar]
14. Praneet S, Bhupendra V. Negative selection in anomaly detection—a survey. Comput Sci Rev. 2023;48:100557. doi:10.1016/j.cosrev.2023.100557. [Google Scholar] [CrossRef]
15. Sithungu SP, Ehlers EM. Gaainet: a generative adversarial artificial immune network model for intrusion detection in industrial IoT systems. JAIT. 2022;13(5):456–61. doi:10.12720/jait.13.5.456-461. [Google Scholar] [CrossRef]
16. Idris I, Selamat A, Omatu S. Hybrid email spam detection model with negative selection algorithm and differential evolution. Eng Appl Artif Intell. 2014;28(5):97–110. doi:10.1016/j.engappai.2013.12.001. [Google Scholar] [CrossRef]
17. Kim J, Greensmith J, Twycross J, Aickelin U. Malicious code execution detection and response immune system inspired by the danger theory. 2010. doi:10.48550/arXiv.1003.4142. [Google Scholar] [CrossRef]
18. Greensmith J, Aickelin U, Cayzer S. Introducing dendritic cells as a novel immune-inspired algorithm for anomaly detection. Artif Immune Syst. 2005;3627:153–67. doi:10.1007/11536444. [Google Scholar] [CrossRef]
19. Ostaszewski M, Seredynski F, Bouvry P. Immune anomaly detection enhanced with evolutionary paradigms. In: Proceedings of the 8th Annual Conference on Genetic and Evolutionary Computation(GECCO ’06); 2006; New York, NY, USA: Association for Computing Machinery. p. 119–26. doi:10.1145/1143997.1144018. [Google Scholar] [CrossRef]
20. Northcutt CG, Jiang L, lsaac Chuang L. An introduction to confident learning: finding and learning with label errors in datasets. J Artif Intell Res (JAIR). 2021. doi:10.48550/arXiv.1911.00068. [Google Scholar] [CrossRef]
21. Teng SH, Zhou DG, Teng LY, Zhang W. Transfer learning for selecting confidence pseudo labels. J Jiangxi Normal Univ (Nat Sci Edit). 2024;48(1):31–44. [Google Scholar]
22. Ying D. Research on label propagation algorithm based on confidence evaluation and rollback mechanism and its application in network intrusion detection [master’s thesis]. China: Sichuan University; 2021. [Google Scholar]
23. Bereta M. Negative selection algorithm for unsupervised anomaly detection. Appl Sci. 2024;14(23):11040. doi:10.3390/app142311040. [Google Scholar] [CrossRef]
24. Sun P, Ban L, Jian H. Improved self-adaptive negative selection algorithm with double clustering for infrared target extraction. In: 2022 IEEE 5th International Conference on Automation, Electronics and Electrical Engineering (AUTEEE); 2022; China. p. 378–82. [Google Scholar]
25. Abid A, Khan MT, Haq IU, Anwar S, Iqbal J. An improved negative selection algorithm-based fault detection method. IETE J Res. 2022;68(5):3406–17. doi:10.1080/03772063.2020.1768158. [Google Scholar] [CrossRef]
26. Mo J, Yang H. Sampled value attack detection for busbar differential protection based on a negative selection immune system. J Mod Power Syst Clean Energy. 2023;11(2):421–33. doi:10.35833/MPCE.2021.000318. [Google Scholar] [CrossRef]
27. Bridges RA, Glass-Vanderlan TR, Iannacone MD, Vincent MS, Chen QG. A survey of intrusion detection systems leveraging host data. Associat Comput Mach. 2019 Nov;52(6):1–35. doi:10.1145/3344382. [Google Scholar] [CrossRef]
28. Wang Y, Jiang YM, Lan JL. Intrusion detection based on improved triple network and k-nearest neighbor algorithm. Comput Applicat. 2021;41(7):1996–2002. (In Chinese). doi:10.11772/j.issn.1001-9081.2020081217. [Google Scholar] [CrossRef]
29. Liang BL. A review of the application of artificial immunity in intrusion detection. Netw Secur Technol Applicat. 2023;(11):46–8. (In Chinese). doi:10.3969/j.issn.1009-6833.2023.11.019. [Google Scholar] [CrossRef]
30. Jingsha H, Song H, Nafei Z, Jiake G. Research and optimization of intrusion detection based on improved v-detector algorithm. Inform Netw Secur. 2020;20(12):19–27. (In Chinese). doi:10.3969/j.issn.1671-1122.2020.12.003. [Google Scholar] [CrossRef]
31. Ahlam K, Salim C, Reforgiato RD. Fuzzy optimized v-detector algorithm on apache spark for class imbalance issue of intrusion detection in big data. Neural Comput Appl. 2023;35(27):19821–45. doi:10.1007/s00521-023-08783-8. [Google Scholar] [CrossRef]
32. Yang T, Deng HL, Chen W, Wang Z. GF-NSA: a negative selection algorithm based on self grid file. In: Frontiers of manufacturing and design science, applied mechanics and materials. Vol. 44. China: Trans Tech Publications Ltd.; 2011. p. 3200–3. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools