 Open Access
Open Access
REVIEW
Generative Artificial Intelligence (GAI) in Breast Cancer Diagnosis and Treatment: A Systematic Review
1 Biomedical and Bioinformatics Engineering (BBE) Research Group, Centre for Multimodal Signal Processing (CMSP), Tunku Abdul Rahman University of Management and Technology (TAR UMT), Jalan Genting Kelang, Setapak, Kuala Lumpur, 53300, Malaysia
2 Department of Electrical and Electronics Engineering, Faculty of Engineering and Technology, Tunku Abdul Rahman University of Management and Technology (TAR UMT), Jalan Genting Kelang, Setapak, Kuala Lumpur, 53300, Malaysia
3 Sports Engineering Research Centre (SERC), Universiti Malaysia Perlis (UniMAP), Kampus Pauh Putra, Arau, 02600, Perlis, Malaysia
4 Faculty of Electronic Engineering and Technology, Universiti Malaysia Perlis (UniMAP), Kampus Pauh Putra, Arau, 02600, Perlis, Malaysia
5 Advanced Communication Engineering (ACE) Centre of Excellence, Universiti Malaysia Perlis (UniMAP), Kangar, 02600, Malaysia
6 Department of Pathology, Hospital Tuanku Fauziah, Jalan Tun Abdul Razak, Kangar, 01000, Malaysia
* Corresponding Author: Xiao Jian Tan. Email:
(This article belongs to the Special Issue: Advances in Artificial Intelligence and Generative AI: Impacts on Multidisciplinary Applications)
Computers, Materials & Continua 2025, 84(2), 2015-2060. https://doi.org/10.32604/cmc.2025.063407
Received 14 January 2025; Accepted 15 May 2025; Issue published 03 July 2025
Abstract
This study systematically reviews the applications of generative artificial intelligence (GAI) in breast cancer research, focusing on its role in diagnosis and therapeutic development. While GAI has gained significant attention across various domains, its utility in breast cancer research has yet to be comprehensively reviewed. This study aims to fill that gap by synthesizing existing research into a unified document. A comprehensive search was conducted following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, resulting in the retrieval of 3827 articles, of which 31 were deemed eligible for analysis. The included studies were categorized based on key criteria, such as application types, geographical distribution, contributing organizations, leading journals, publishers, and temporal trends. Keyword co-occurrence mapping and subject profiling further highlighted the major research themes in this field. The findings reveal that GAI models have been applied to improve breast cancer diagnosis, treatment planning, and outcome predictions. Geographical and network analyses showed that most contributions come from a few leading institutions, with limited global collaboration. The review also identifies key challenges in implementing GAI in clinical practice, such as data availability, ethical concerns, and model validation. Despite these challenges, the study highlights GAI’s potential to enhance breast cancer research, particularly in generating synthetic data, improving diagnostic accuracy, and personalizing treatment approaches. This review serves as a valuable resource for researchers and stakeholders, providing insights into current research trends, major contributors, and collaborative networks in GAI-based breast cancer studies. By offering a holistic overview, it aims to support future research directions and encourage broader adoption of GAI technologies in healthcare. Additionally, the study emphasizes the importance of overcoming implementation barriers to fully realize GAI’s potential in transforming breast cancer management.Keywords
GAI, a term that has gained attention recently across a wide spectrum of applications and domains, from experts to layman, from working adults to schooling children, regardless of formal working and/or leisure entertainment purposes, as well as daily errand planning. The emergence of consumer-facing products, for example, ChatGPT [1], StyleGAN [2], and/or MidJourney (AI image generator from text) [3] have revolutionizing the approaches of problem-solving amongst consumers in different facets of daily life. In clinical settings, GAI is not new however, some renowned architectures, for example, Generative Adversarial Networks (GAN), Recurrent Neural Networks (RNN), and Variational Autoencoders (VAE) have been used for decades in medical image analysis [4–6], diagnostic and treatment of cancers, as well as laboratory-related applications. These architectures are less scalable, compared to the state-of-the-art models, which have traditionally restricted their development to smaller scales in terms of parameters, data, and computational resources, formalizing the foundation models for modern architectures in GAI.
AI, the umbrella term that encompasses all computational algorithms with capabilities to perform task-specific activities that conventionally require human intelligence, for example, decision-making, pattern recognition, and learning from past experiences. The idea of mimicking human intelligence was first conceived by Alan Turing in 1950 [7] and later the phrase: “artificial intelligence”, was coined by John McCarthy in 1956, specifically to remark an important subject area in science and engineering, as a sub-element of machine intelligence [8]. Early AI systems, which are commonly regarded as expert systems, are mainly knowledge-driven where well-defined rules are used as the backbone of the system [9–11]. Thanks to the advancement in computational engineering, deep learning, a sub-element of AI, has made great strides in unsupervised learning of features from the sample/training datasets while performing task-specific activities (e.g., pattern recognition) automatically with promising performance that matches or even surpasses human performance [12]. Fig. A1 in Appendix A shows a brief timeline of AI advancement, while Fig. 1 shows the Venn diagram of AI and the respective sub-elements.
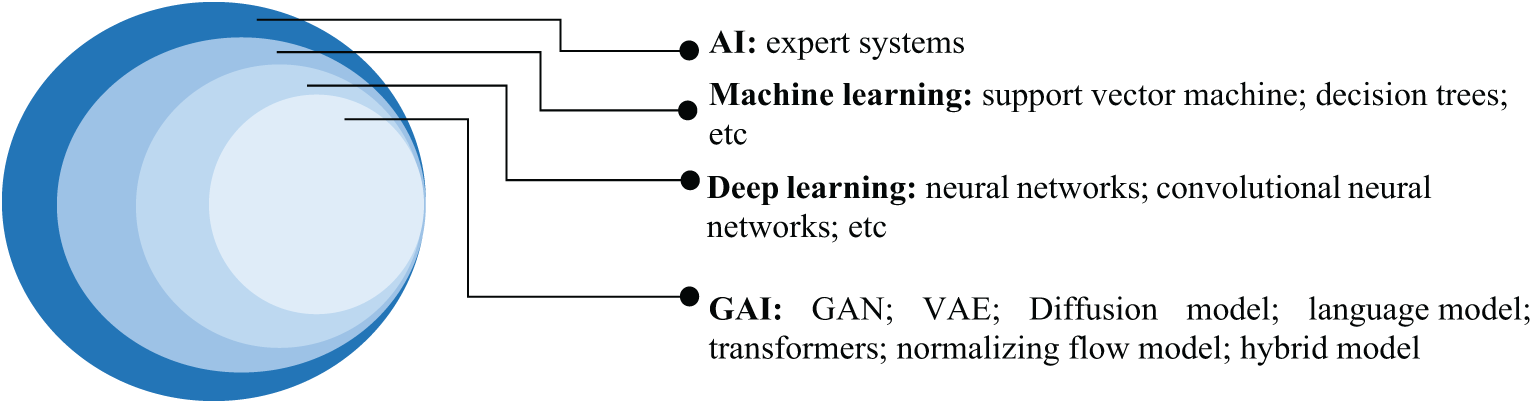
Figure 1: Venn diagram of AI and the respective sub-elements
In deep learning, the two primary models are discriminative and generative models (the main focus of this study). Understanding the differences between these two models is essential, as each serves distinct purposes and approaches within the field of AI, particularly in breast cancer research. A discriminative model is intended to model and formalize the relationship between the input features (i.e., learned features) and the output labels (i.e., detection results). A generative model however learns from the inherent dataset and focuses on the probabilistic generation of new outputs, instead of providing decisions on extant data, for example, classification and clustering of extant data. Unlike the discriminative model, the outputs from the GAI are often not replicable, such that the same prompt may result in different solutions. These solutions however remain valid, fulfilling the input prompt from the users. Therefore, the utilization of discriminative and generative models in clinical settings are different but complement one another. In breast cancer, discriminative models (e.g., convolutional neural networks) are widely used in detection and classification activities, for example, benign and malignant breast cancer classification [13–15]; prognosis and risk prediction typically on the likelihood of breast cancer recurrence or progression based on patient data [16,17]; outcome predictions that aid in predicting outcomes based on clinical, imaging, or genomic data [18], allowing clinicians to adjust treatment plans dynamically; and therapy optimization, for example, optimizing radiation or chemotherapy dosages by learning patterns in past treatment data [19,20]. Whereas GAI models (e.g., GAN) are often used in generative-oriented activities where creation or synthesis events are genuinely required, fulfilling the input prompt. GAI can improve breast lesion detection [21], facilitate image synthesis and data augmentation to generate augment datasets for training purposes [22,23], facilitate diagnosis and treatment procedures [24–26], enhance the quality of breast medical images [27], and explore the drug structures and results validation [28]. Fig. 2 shows the general concept of discriminative and generative models in deep learning.
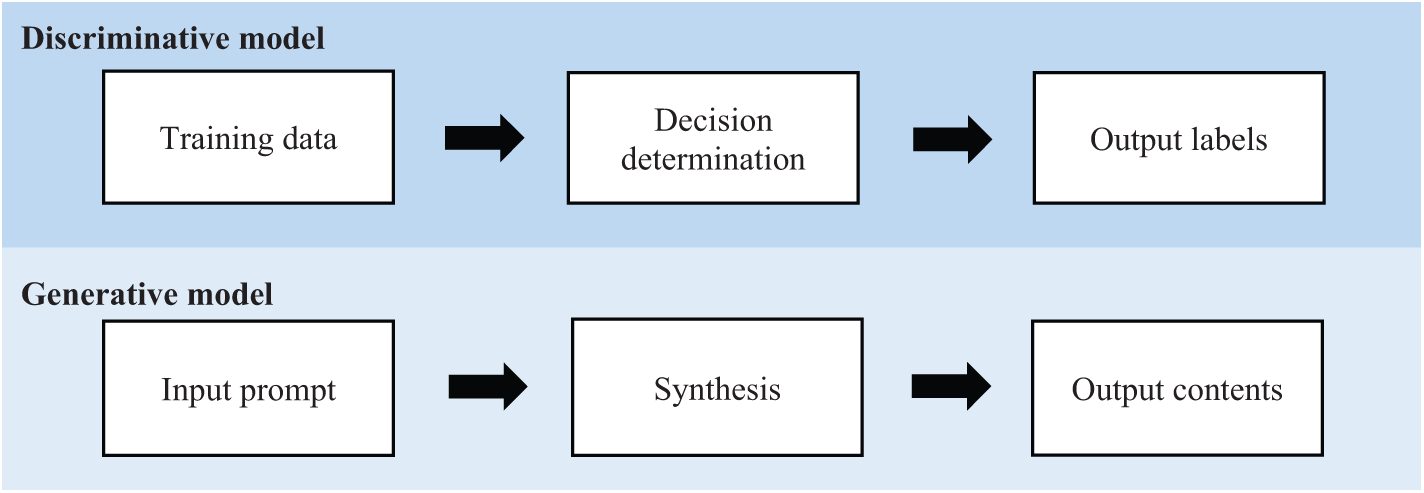
Figure 2: General concept of discriminative and generative models in deep learning (figure reproduced from [29] under Creative Commons CC BY license, Springer Nature)
In recent years, AI has become a ubiquitous term that is being used in a wide spectrum of domains with remarkable betterment. However, to adopt AI (or GAI) in the clinical setting, the system (so-called AI system) must be first defined and characterized, depending on the intended purposes associated with different risk categories. Defining AI systems is important, as this outlines clear boundaries and expectations for what the system is designed to do, how it operates, what the expected outcomes are, and the potential risks associated with it. Risk assessment is crucial, allowing users to understand, anticipate, and mitigate potential negative outcomes that could arise from AI usage, involving critical domains, for example, ethical consideration, safety and security, trust and public confidence, prevention of harm and legal risks, data privacy and security, as well as robustness and reliability. With the introduction of the AI Act, manufacturers are now required to identify which AI systems fall within the regulation scope and are consequently subject to act compliance and obligations. Table 1 shows the list of definitions of AI systems with respect to different sources.
Fig. 3 shows the classification of models in GAI, highlighting the respective architecture component and training method in each model. The architecture component defines how a model processes information and subsequently generates outputs, while the training method shapes the performance and effectiveness of a model.
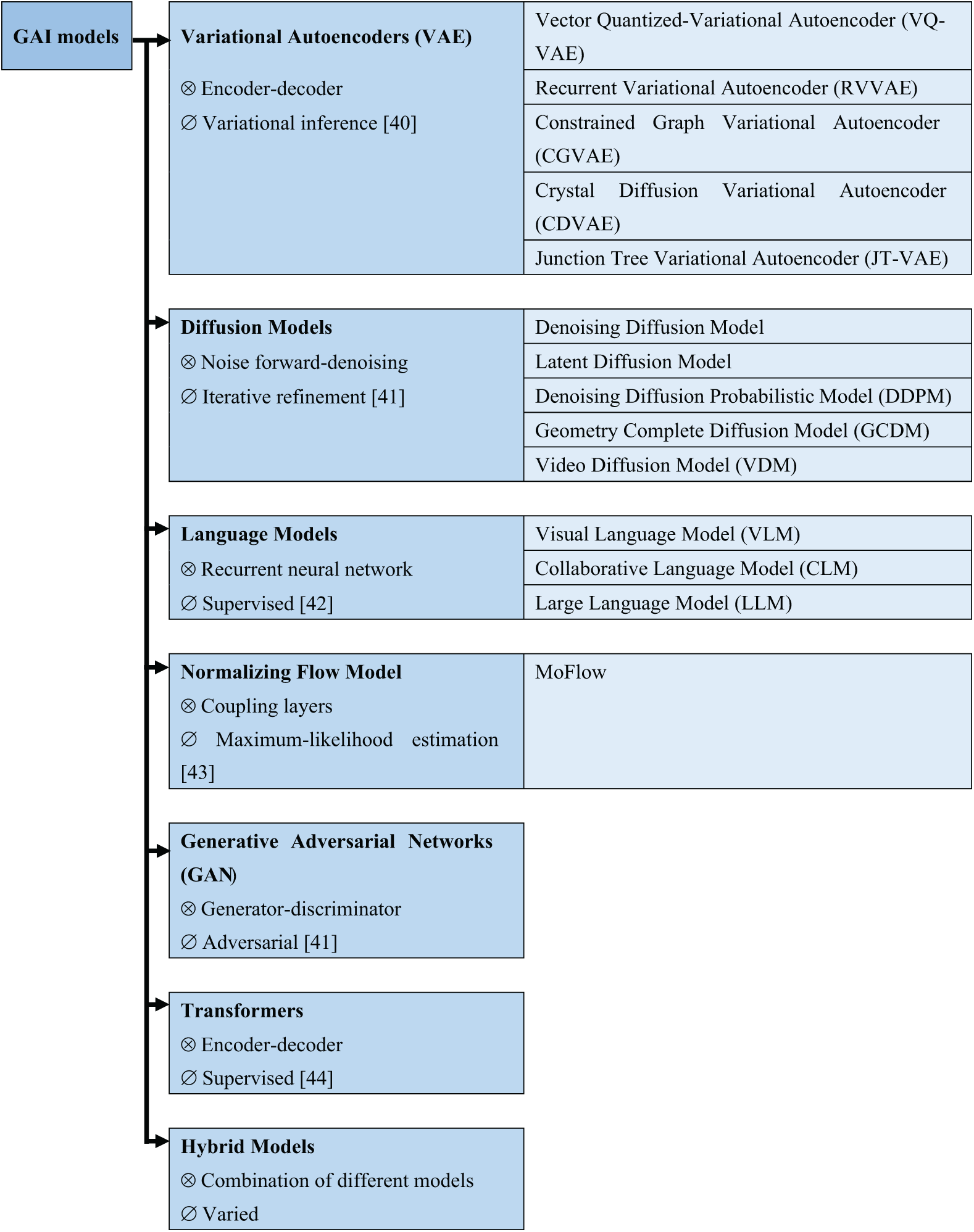
Figure 3: Classification of GAI models (figure adapted from [40–44] under Creative Commons CC BY license, MDPI), where “⊗” and “∅” denote architecture components and training methods, respectively
Briefly, the architecture of VAE is based on an encoder-decoder structure and uses variational inference in the training process [37,38]. VAEs are designed to learn compressed representations of input data by mapping it to a latent space. In the latent space, new samples can be generated by sampling and decoding from the learned distribution. A key highlight of VAEs is that the model incorporates a probabilistic framework, allowing them to generate diverse samples by drawing from a distribution rather than a fixed point, thus, making VAEs particularly suitable for tasks where a variety of output options is desirable, such as generating new images, reconstructing missing data, and/or performing anomaly detection.
GAN consists of two main components, namely, the generator and the discriminator. The primary role of the generator is to produce synthetic samples, for example, images, to deceive the discriminator, which evaluates whether a given sample is real or fake [39]. These two networks are trained adversarially, with the generator improving its output to fool the discriminator, and the discriminator improves capability in detecting fakes, formalizing a competition between the two leads (i.e., generator and discriminator), ultimately yielding highly realistic and diverse data. GANs have proven effective in areas, for example, image generation, video synthesis, and music composition. Despite their impressive results, GANs are known for being challenging to train, often requiring careful balancing between the generator and the discriminator to avoid issues like mode collapse.
Diffusion models involve a unique approach that starts with noisy data and progressively refines it through iterative denoising steps, which generate high-quality samples [41]. These models are trained by learning the dynamics of this noise-diffusion process, making them highly effective at tasks requiring precise, fine-grained detail, such as high-resolution image synthesis. The noising step inputs random noise into the data, and the denoising step gradually reconstructs the original signal through multiple iterations. Diffusion models have recently gained attention for their ability to outperform GANs in certain image generation tasks, specifically, in tasks in which the generation of detailed textures is required and successfully addresses some of the training instabilities seen in GANs.
Transformers employ an encoder-decoder architecture and make extensive use of self-attention mechanisms, which allow the model to capture long-range dependencies within data sequences [42]. Transformers are specifically effective in tasks that involve sequence data, for example, natural language processing, where the generation of coherent text or translation between languages is involved. The self-attention mechanism helps the model focus on relevant parts of the input sequence, making it highly scalable and efficient for tasks that involve handling large datasets with complex relationships. Notably, transformer models like Generative Pre-trained Transformers (GPTs) have demonstrated groundbreaking performance in text generation, translation, and summarization tasks.
Language models are typically built using RNNs or Long Short-Term Memory (LSTM) networks, which are designed to handle sequential data by predicting the next token in a sequence [43]. These models are trained through supervised learning and are specifically well-suited for generating natural language text, for example, completing sentences or writing paragraphs. The ability of language models to predict the next word in a sequence makes them crucial in applications like chatbots, translation systems, and automated content creation. With the rise of transformer-based models, language models have achieved even greater performance in generating coherent and contextually accurate text.
Normalizing Flow models use a sequence of invertible transformations, namely coupling layers, to transform data into a simpler distribution, for example, Gaussian distribution [44]. The key highlight of the normalizing flows is that the model preserves the exact probability density, allowing for accurate learning of complex distributions. By retaining density information in the data transformation process, this model is particularly suitable for tasks, for example, density estimation and probabilistic modeling. Normalizing Flow models are trained using maximum-likelihood estimation, making them useful in applications that require precise probability estimates, for example, anomaly detection or uncertainty quantification.
Hybrid models combine elements from multiple GAI models, allowing them to leverage the strengths of different models [40]. For example, a hybrid model can integrate the probabilistic sampling capabilities of VAEs with the adversarial training of GANs to achieve both diversity and realism in generated samples. By combining different architectures and training methods, hybrid models offer flexibility and can be tailored to meet specific generative goals, whether for image synthesis, text generation, or other creative tasks. These models are often designed to overcome the limitations of individual architectures, providing a more robust solution to GAI challenges.
1.3 Aims and Outline of the Study
Motivated by vigorous advancement in computation engineering and tremendous growth in world interest using GAI in breast cancer research, here, a systematic bibliographic survey focusing on GAI in breast cancer is presented. To ensure optimal search, no limiter is set on the publication year. The primary research questions of this study are: “What are the applications and impacts of generative AI in breast cancer diagnosis and treatment?” and “How has generative AI been utilized to enhance outcomes in breast cancer research, particularly in predictive modeling, diagnostics, and therapeutic development?”. The research question is framed as such to ensure the inclusion of all aspects of GAI models available within breast cancer. The present study is intended to focus on collating and synthesizing new insights and drawing constructive conclusions while highlighting the research gaps and challenges in the topic of interest. This study is organized as follows: Section 2 offers an overview of breast cancer diagnosis and treatment. Section 3 details the methodology employed in this systematic review. Section 4 presents synthesized findings from the included studies. Section 5 discusses self-assessment, limitations, challenges, and future directions. Finally, the conclusion is presented in Section 6.
2 Brief Descriptions of Diagnosis and Treatment of Breast Cancer
Here, the study primarily focuses on the diagnosis and treatment of breast cancer, formalizing the two main domains in cancer management. Diagnosis of breast cancer refers to the process of identifying the presence of cancer in the breast. This typically involves various tests and procedures, for example, clinical breast examination, breast imaging procedure, and biopsy grading [12,45]. In breast imaging procedures, imaging modalities such as non-ionizing radiation, gamma radiation (nuclear medicine), X-ray source, magnetic field, and ultrasound wave are commonly used [8,46]. Diagnosis of breast cancer is mainly to detect abnormal cells or tumors, determine their type, size, and stage, and assess whether the cancer has spread.
Treatment in breast cancer involves the medical management of the disease, including a combination of therapies, for example, surgery, chemotherapy, radiation therapy, hormone therapy, and targeted therapies. The choice of treatment is dependent on multi-facet factors, for example, the type and stage of the cancer, hormone receptor status, and overall health of the patient. The primary goal is to remove and/or destroy cancer cells and prevent recurrence. Briefly, some of the novel treatments, approved by the Food and Drug Administration (FDA) as targeted therapies for breast cancer are anti-estrogen, LH-RH analogs (goserelin and leuprolide), CDK4/6 inhibitor (Ribociclib, Palbociclib, and Abemaciclib), PI3Ki inhibitor (Pictilisib, Pilaralisib, and Voxtalisib), pan-PI3K inhibitor (Buparlisib), TKI (Neratinib and Lapatinib), and mAb [47].
The present study follows the PRISMA guidelines [48]. The review methodologies herein are adopted and adapted from the previously published protocol/works [49].
3.1 Information Sources and Search Strategy
The proposed search strategies are the output of collaborative discussion from a team comprised of engineering and medical experts. As the present study focuses on generative AI in breast cancer, the search strings included two elements: “generative AI” and “breast cancer”. These elements are intentionally formulated in a broad manner to ensure optimal inclusion of data from all aspects (including diagnosis and treatment) that primarily adopt/integrate generative AI in breast cancer. The search strategies included limiters, for example, English language and journal articles. No limiter is set on publication years to maximize the data retrieved. The ‘‘AND” and ‘‘OR” operators were used to formalize the search string in the present study (see Table 2). The systematic literature searches were last performed in December 2024 in four core databases: Scopus, Web of Science Core Collection, and PubMed, in lieu of search engines provided by specific publishers, such as SpringerLink, ScienceDirect, Multidisciplinary Digital Publishing Institute (MDPI), and Frontiers. Mendeley reference management software was used for reference management and related purposes.

Eligible studies were English-language original peer-reviewed journal research articles. Once a relevant journal research article was retrieved, the references of the articles were screened to explore potential articles that had not been identified in the initial search. This procedure iterates until no further articles are found. In Phase 1, only original peer-reviewed journal research articles were considered eligible. In Phase 2, screening was done on the abstract of the articles. The eligible materials were then subjected to full-text screening. In the full-text screening phase, a dual-independent approach was adopted, such that all articles were screened by two independent reviewers where the reviewers were blinded to the other’s decision. If necessary, a discussion is held between the independent reviewers for eligibility evaluation, especially when the articles partially fulfill the inclusion criteria. If required, a third reviewer was consulted to attain consensus. The main inclusion criteria are: (1) the journal articles were using generative AI and (2) the journal articles were focusing on breast cancer (or multiple cancers that must include breast cancer). The last search of this study was performed on 24 December 2024. Fig. 4 summarizes the flowchart of the systematic search process in accordance with the PRISMA guideline [48]. Based on the flowchart, using the search strings as in Table 2, a total of 3827 articles were first identified. Implementation of exclusion criteria of Phases 1 and 2, resulting in 31 articles eligible for the subsequent synthesis stage. In Phase 1, a total of 39 articles were excluded, whereas in Phase 2, only 4 articles were excluded. The complete PRISMA checklist is detailed in Tables A2 and A3 in Appendix C.
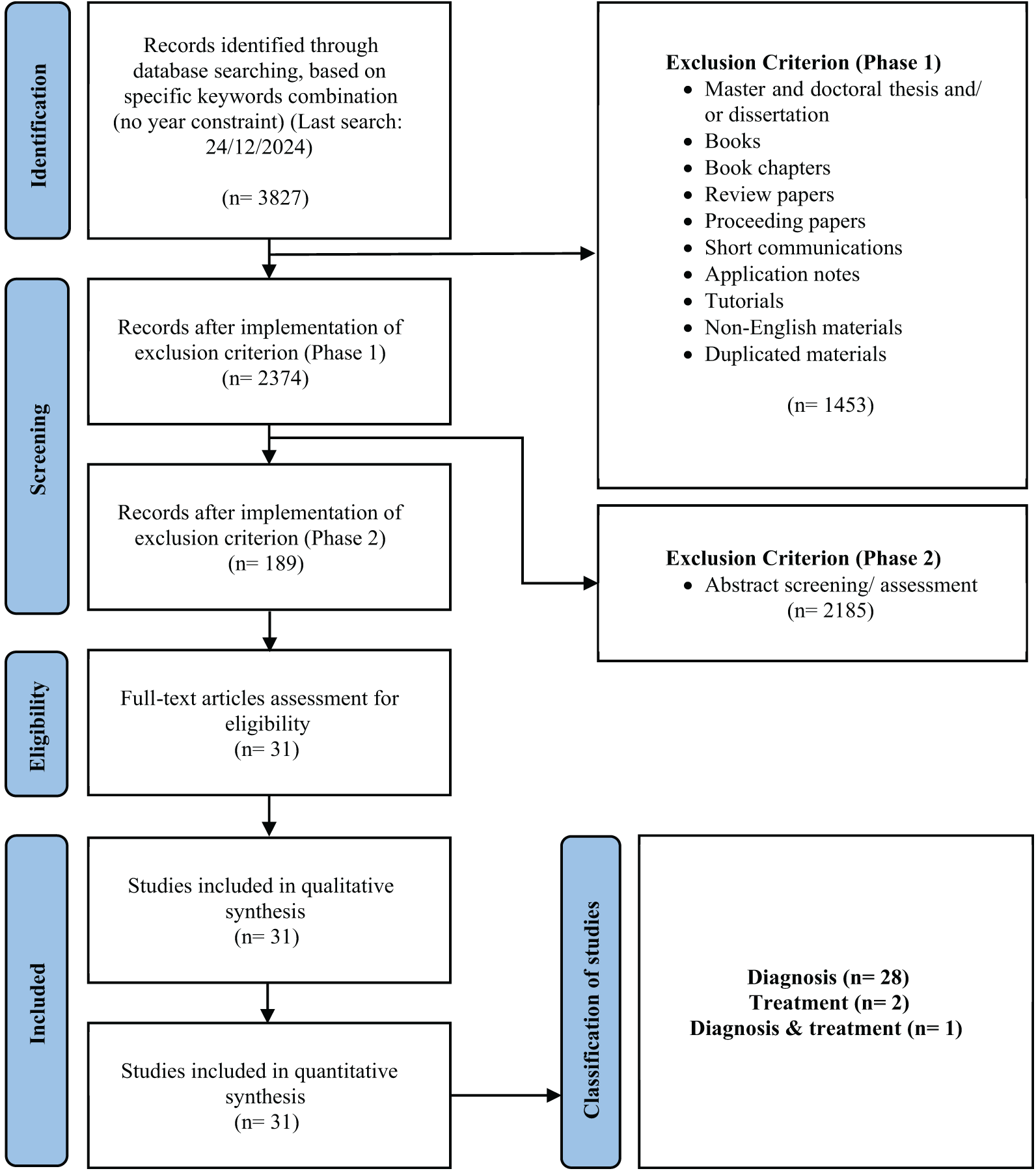
Figure 4: Flowchart of the systematic search in accordance with the PRISMA guideline [48]
Articles published in non-English languages were excluded. Materials, for example, master’s and doctoral theses or dissertations, books, book chapters, grey literature, conference papers, review articles, application notes, brief communications, tutorials, and duplicate works were excluded. Protocol articles were securitized separately for eligibility. Protocol articles that solely reported methodology and protocol in breast cancer, with no study outcomes, were excluded.
Qualitative and quantitative data from each included article were carefully distilled and systematized into a functional summary endpoint. Qualitative data, for example, type of modalities, dataset, methods, and findings were extracted, compiled, and tabulated using a table. Quantitative data, for example, statistical findings, co-occurrence analysis, and bibliometric information, were collated and systematized to offer a holistic view of the topic of interest. Charts, graphics, and tables were used as a medium to synthesize the mineable data. For all included articles, the affiliation and author name were manually disambiguated (for example, “Harvard Medical School” and “HMS” are both referring to the same affiliation) to avoid duplication in the analysis using software such as VOS Viewer [50].
Qualitative and quantitative findings from the included works were reported narratively via the descriptive approach, supported using charts, graphics, and tables. Statistical calculations and compilation of data were done using Microsoft Excel 2019. Co-occurrence analysis and bibliometric findings were generated using VOS Viewer 1.6.20 software for Windows [50]. Quantitative findings, for example, classification of included works was illustrated using a pie chart; geographical distribution was illustrated using a world map chart; distribution of most contributing journals and publishers using bar charts; temporal publication analysis using a line graph; subject profiling using a treemap; and keywords co-occurrence, co-authorship occurrence, and country-ship analysis using bibliometric networks.
3.5 Definition of Performance Metrics Used in the Literature
The confusion matrix is one of the most commonly used performance metrics from the body of the literature, which is specifically useful in summarizing the classification performance of a classifier with respect to the testing/validating data. Typically, the confusion matrix consists of a two-dimensional matrix, where one axis represents the true class of an object, and the other represents the class predicted by the classifier. Table 3 provides an example of a confusion matrix for a three-class classification task with classes A, B, and C. The first row of the matrix indicates that 15 objects belong to class A, with 12 correctly classified as A, one misclassified as B, and two as C.

In binary classification tasks, a simplified version of the confusion matrix is often used, where one class is designated as positive and the other as negative. The four cells of the matrix represent true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN), as shown in Table 4. Here, TP refers to the correctly predicted positive events, TN refers to the correctly predicted negative events, FP indicates the incorrectly predicted positive events, and FN represents the incorrectly predicted negative events.
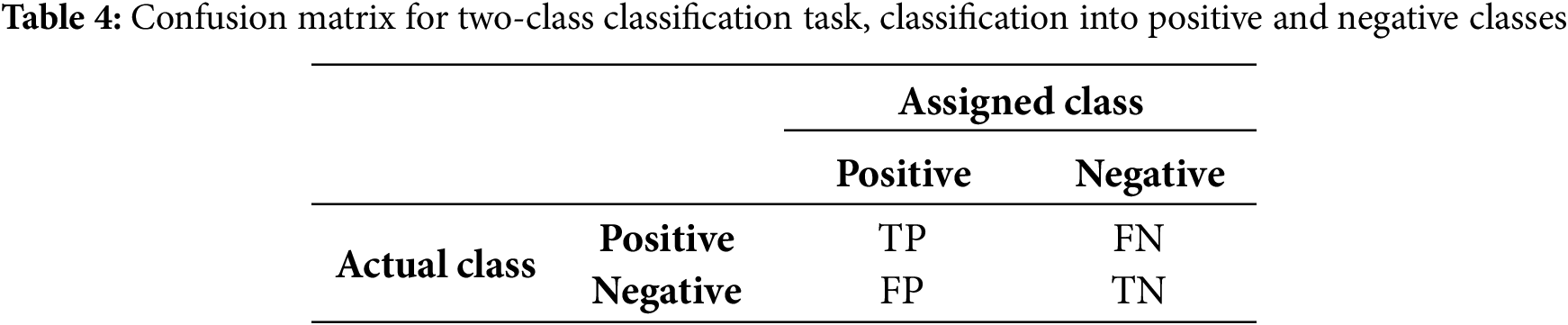
From the obtained TP, TN, FP, and FN, performance metrics, for example, Accuracy, Recall/Sensitivity, Precision, Specificity, F1-score, and Area under the curve (AUC) can be computed. High values denote a better performance and are preferable in the study. The equations for Accuracy, Recall/Sensitivity, Precision, Specificity, F1-score, and AUC are as follows (Eqs. (1)–(6)), where d in Eq. (6) denotes the differential element (the small change) in the false positive rate (i.e., 1-Specificity).
The p-value is a metric used in hypothesis testing to assess the statistical significance of an observed effect or outcome. It represents the probability of obtaining a test statistic as extreme as the one observed, assuming that the null hypothesis is true. The formula for calculating the p-value depends on the statistical test being used (e.g., z-test, t-test, chi-squared test, etc.). Eqs. (7)–(9) show the formula for calculation of the two-tailed test (commonly known as the z-test or t-test), one-tailed test, and chi-squared test, respectively.
where
where
where
Mean Squared Error (MSE) is a widely used metric for assessing the performance of regression models. It calculates the average of the squared differences between the actual values and the predicted values from the model. The MSE gauges how accurately the predicted values align with the true data points. It is always non-negative, with lower values indicating a better fit of the model to the data. Conversely, a higher MSE implies that the model’s predictions are further from the actual values. The formula for MSE is shown in Eq. (10).
where n denotes the number of data points (sample size);
Intersection over Union (IoU) is a metric frequently used to assess the performance of object detection, segmentation, and other tasks that require evaluating the spatial overlap between predicted and ground truth regions. It quantifies the overlap between two areas, such as the predicted bounding box (or segmentation mask) and the ground truth bounding box (or mask). Eq. (11) presents the IoU formula, where the Area of Overlap represents the region where the predicted and ground truth areas intersect (i.e., the shared portion of both regions), and the Area of Union refers to the total area covered by both the predicted and ground truth regions, excluding the overlap (i.e., the combined area of both regions).
The ideal IoU value is 1, indicating that the predicted region exactly matches the ground truth. To the opposite extreme (IoU of 0), the equation implies no overlap between the predicted and ground truth regions. Higher IoU is preferable in all cases, indicating better alignment between the predicted and actual regions, with values closer to 1 being ideal.
Mean IoU is an extension of IoU, mainly used for multi-class segmentation tasks. The Mean IoU computes the IoU for each class in a dataset and then takes the average over all classes. This gives a more comprehensive evaluation metric when dealing with multiple objects or regions across several classes. Eq. (12) shows the formula of Mean IoU, such that C denotes the number of classes; Area of Overlap for Class i and Area of Union for Class i denote the overlap and union areas for class i across all images or regions in the dataset.
The Mean IoU calculates the IoU for each class independently and then averages the results across all classes. The equation is particularly useful in semantic segmentation, where you need to evaluate how well the model predicts boundaries across multiple classes. Higher Mean IoU indicates better segmentation performance, with values closer to 1 indicating near-perfect segmentation.
The Intraclass Correlation Coefficient (ICC) is a statistic used to assess the reliability or agreement between measurements made by different observers, instruments, or measurement methods. ICC is especially useful when the measurements are made on the same subjects or items and can be used to evaluate consistency or conformity within groups of measurements. The ICC is commonly used in domains, for example, medicine, psychology, and medical image analysis, where multiple measurements or ratings are obtained for the same subjects under different conditions.
Eq. (13) shows the general form of the ICC formula, such that ICC (1,1) assesses absolute agreement between raters or measurements; ICC (2,1) assesses consistency across raters when raters are considered interchangeable; and ICC (3,1) assesses reliability when specific raters or instruments are used and generalization to other raters is not desired.
where “Between subject variance” denotes the variation between different subjects or items being measured, and “Within subject variance” denotes the variation between repeated measurements or raters for the same subject. ICC is mainly used in measuring the consistency or reliability of a measurement across different raters or instruments or to determine the level of agreement across two or more raters who provide ratings or scores for the same subjects.
The Peak Signal-to-Noise Ratio (PSNR) is a commonly used metric in image processing to assess the quality of a reconstructed or compressed image in relation to its original version. It measures the degree of distortion caused by lossy compression or other image processing methods by comparing the original and modified images. PSNR is particularly valuable for evaluating image compression algorithms, such as those used for image restoration through denoising or super-resolution techniques. The formula for PSNR is shown in Eq. (14).
where MAX denotes the maximum possible pixel value of the image. PSNR helps determine how much distortion or noise is introduced when compressing an image and is extremely useful in measuring the effectiveness of methods, for example, denoising, deblurring, and other image reconstruction techniques. A high PSNR value indicates that the processed image is closer to the original image, with less noise or distortion, whereas a low PSNR value implies that the processed image has higher distortion/noise, making it less similar to the original.
The Structural Similarity Index (SSIM) is a metric used to evaluate the similarity between two images. Unlike traditional methods, for example, MSE or PSNR, which focus on pixel-level differences, SSIM assesses image quality by considering structural information, resembling human visual perception. It compares the luminance, contrast, and structure of the images to determine their similarity. SSIM is especially beneficial in image processing tasks like compression, denoising, and enhancement, where preserving structural integrity is essential. The formula for SSIM is presented in Eq. (15).
where
The Kappa Value, also known as Cohen’s Kappa, is a statistical metric used to measure the degree of agreement between two raters or evaluators who categorize items into distinct groups. It is particularly valuable when assessing how well two different observers align in classifying the same items while accounting for the likelihood of agreement occurring by chance. Cohen’s Kappa quantifies the level of agreement between the two classifications, offering insight into whether the observed agreement surpasses what would be expected randomly. This metric is widely used in fields like psychology, medicine, and social sciences to assess the reliability of observers or raters. The formula for Cohen’s Kappa is shown in Eq. (16).
where
The Dice Coefficient, or Sorensen-Dice Index, is a statistical metric used to evaluate the similarity between two sets. It is widely used in image analysis and medical imaging to assess the accuracy of image segmentation and classification. The Dice Coefficient ranges from 0 to 1, where 0 signifies no overlap between the sets, and 1 denotes perfect overlap. The formula for the Dice Coefficient is shown in Eq. (17).
where A denotes the set of predicted positive samples; B denotes the set of ground truth positive samples;
Fréchet Inception Distance (FID) is a metric used to assess the quality of images produced by generative models, such as GANs. It calculates the distance between the distributions of real and generated images in a feature space, which is derived from a pre-trained Inception network, a specific type of convolutional neural network. The FID score evaluates both the quality and diversity of the generated images, making it a commonly used metric for evaluating generative models. The formula for FID is shown in Eq. (18).
where
4.1 Classification of the Articles in Terms of Generic Categories
Based on Fig. 4, the search string, as formalized in Table 2, resulted in 31 included articles. These articles were then characterized into two generic categories, namely diagnosis and treatment. Notice that one included article reported findings using GAI in both the categories. Henceforth, a new category, namely Diagnosis & Treatment (i.e., hybrid category) is created. Fig. 5 shows the classification of the 31 included articles. According to Fig. 5, the diagnosis category formalized the main body of the literature, accounting for 90.3% (28 articles) of the included articles. This is followed by articles in treatment and hybrid categories, respectively accounted for 6.5% (2 articles) and 3.2% (1 article). This could be attributed to the superiority of GAI in accomplishing task-specific events [8], for example, data generation and augmentation [22], abnormalities detection [51,52], image-to-image translation [53,54], which formalized the core foundation in computer-aided diagnostics.
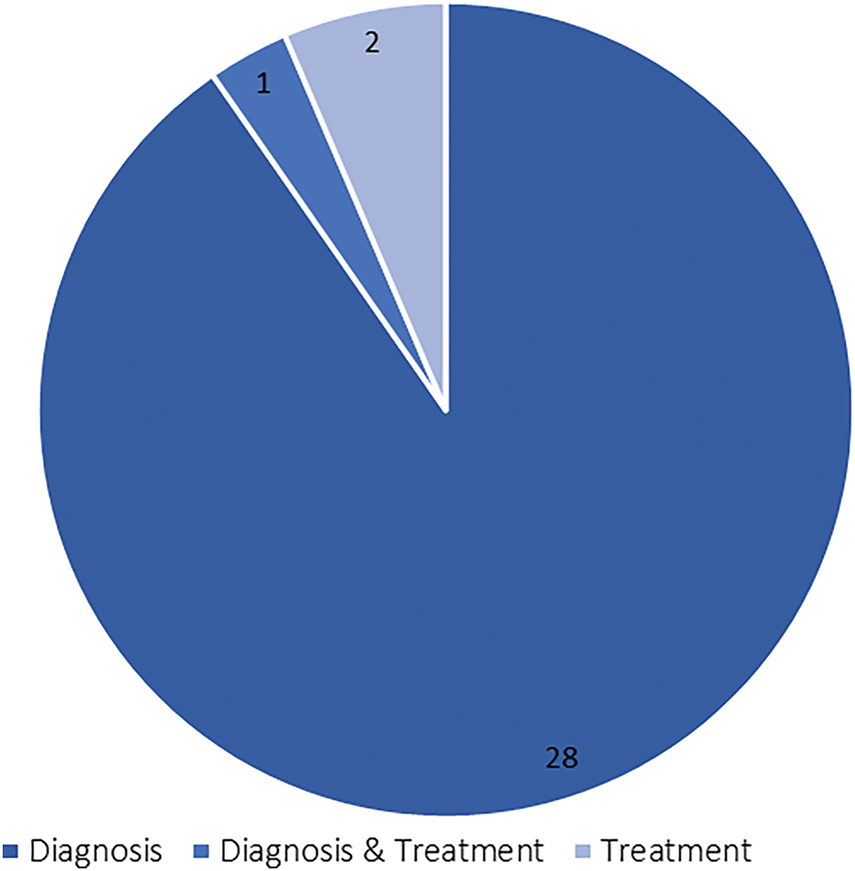
Figure 5: Analysis on the generic categories
In the treatment domain, personalized regimens may be required most of the time. The capability of GAI in the detection of non-clinical significance can result in overtreatment [12], subsequently leading to unnecessary clinical procedures and associated to morbidity [55], leading to lower interest in using GAI for breast cancer treatment purposes. Table A1 in Appendix B summarizes the details of the 31 included articles, comprised of useful information, for example, the generic category, modalities, dataset, methods, and core findings.
4.2 Geographical Scientometric Analysis
Fig. 6 shows the geographical scientometric analysis of the included articles. The geographical distribution of the contributing organizations in GAI in breast cancer research demonstrates a wide global engagement, with notable concentrations in certain regions. The USA leads with 8 contributions, highlighting its dominant role in AI-driven breast cancer research. China and India each follow with 4 contributions, indicating strong research activity in Asia. Other Asian countries such as Japan and Korea also show significant participation, with 3 and 2 contributions, respectively. European countries like Germany, Spain, Switzerland, and France collectively contribute 7 works, showcasing the region’s involvement in integrating AI with medical research. While Australia, Bangladesh, and the Netherlands each contribute 1 study, this distribution indicates active participation from both developed and developing nations. This wide geographical spread underscore the global importance and collaboration in leveraging AI for breast cancer diagnosis and treatment across diverse healthcare systems. To better illustrate the collaboration between different countries, a co-country-ship network analysis is generated, as in Fig. 7. Because of small number of included articles in this study, the co-country-ship network may look scatted. The network provided however remain valid in illustrating the collaborative relationship between the countries as aforementioned. The network reveals three distinct clusters, dominated by the USA, China, and Italy, with the USA at the core of the contributions. This is followed by China, establishing a close network with the United Kingdom. The third cluster features Italy, collaborating with Switzerland, forming the third strong partnership in the network.
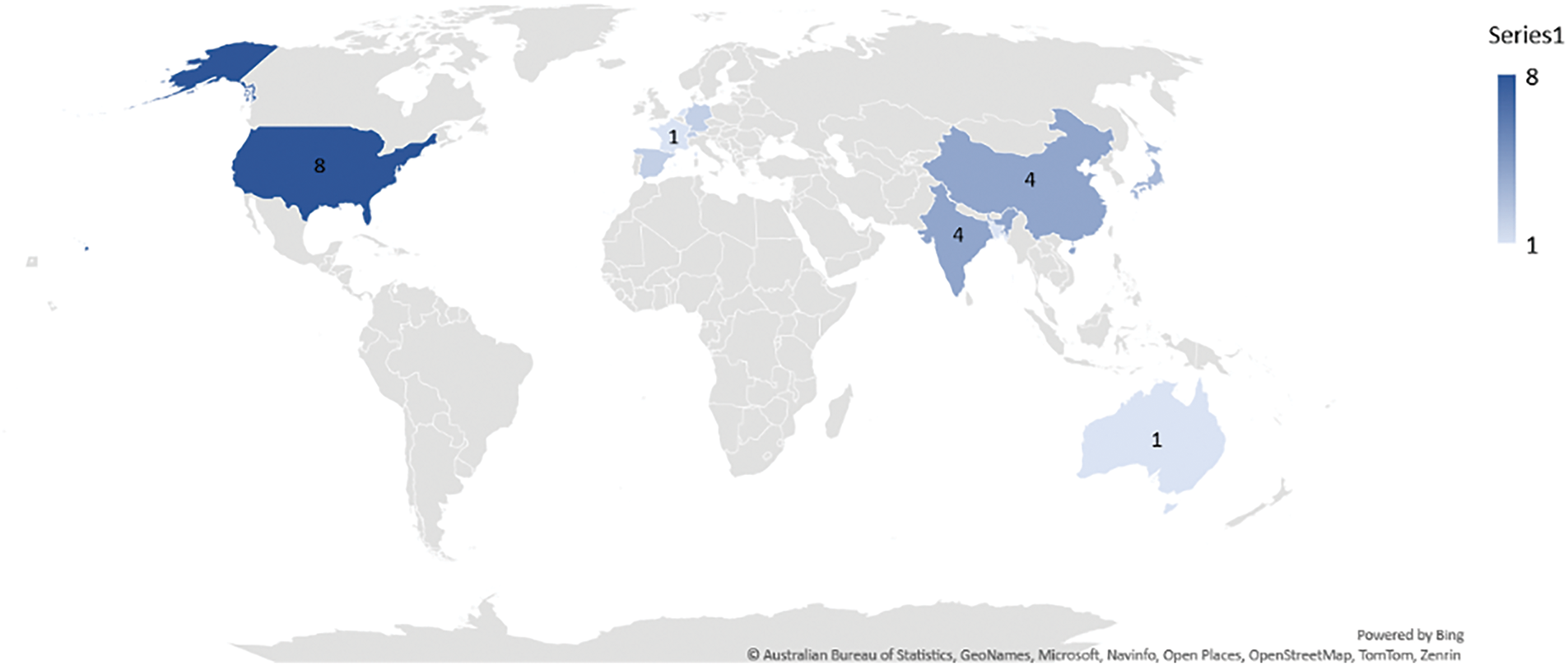
Figure 6: Geographical scientometric analysis of the included review works
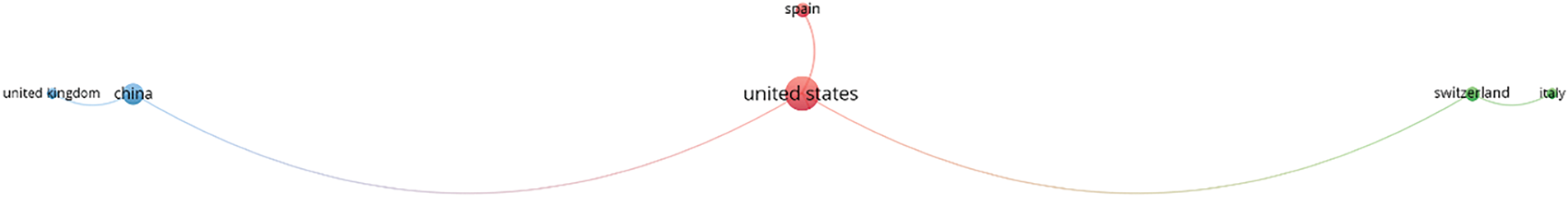
Figure 7: Co-country-ship network analysis
4.3 Distribution of the Most Contributing Journals
Fig. 8 shows the most contributing journals in reporting findings using GAI in breast cancer. Overall, a total of 29 journals were collated from the 31 included articles. Based on Fig. 8, insufficient evidences were found to substantiate or to support the argument on the most contributing journals herein. This is because the distribution of journals is found relatively scarce corresponding to the entire body of the literature. From the collected data, the journal, namely Journal of Medical Imaging and Dignostics, both ranked as the most contributing journals, but accounted only 6.5% (two articles), respectively, from the included articles. The scattered distribution may be due to the broad applicability of “diagnosis,” which aligns with the aims and scopes of various journals, for example, those shown in Fig. 8. It is important to note that diagnosis in breast cancer occurs at multiple stages, for example, during the examination phase using mammograms or during grading using histopathology images. These diverse diagnostic processes fit within the scope of numerous journals, leading to their widespread inclusion.

Figure 8: Analysis of most contributing journals
4.4 Distribution of the Most Contributing Publishers
As mentioned in Section 3.1, general databases, for example, Scopus, Web of Science Core Collection, and PubMed, were used for the literature search process. From all the included articles, 14 publishers were retrieved, such that the top five most contributing publishers are Springer, Elsevier, MDPI, SPIE, and Wolters Kluwer. Fig. 9 summarises the contributing publishers retrieved from the included articles. From the figure, Springer appeared to be the most contributing publisher, accounted 22.6% (7 articles) in total. This is followed by Elsevier (19.3%, 6 articles), MDPI (12.9%, 4 articles), SPIE (9.6%, 3 articles), and Wolters Kluwer (6.5%, 2 articles). In general, publishers maintain an oligopolistic hold on the publishing industry, a trend consistent with previous findings from an analysis spanning four decades (1973–2013). This analysis identified Elsevier, Taylor & Francis, and Springer as leading publishers across various topics. The same trend as well can be observed from some of the recent reviews in various fields, for example, agriculture, engineering, applied mathematics, and medicine. The oligopolistic structure of the publishing industry is likely to persist, as building a reputable publishing company requires significant time and resources. Furthermore, researchers may hesitate to submit their work to newer publishers to avoid the risks associated with potential predatory publishers.
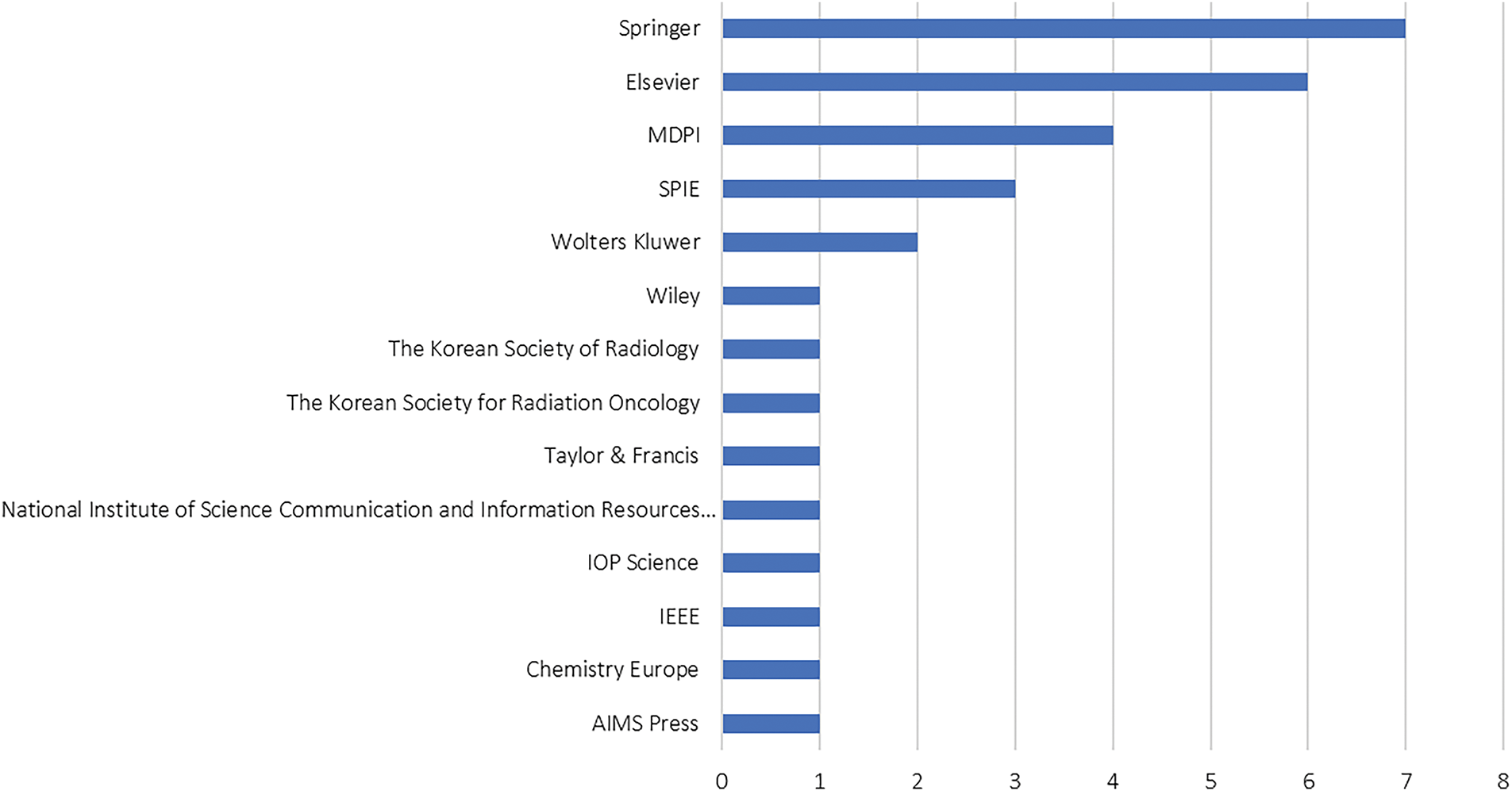
Figure 9: Analysis of most contributing publishers
4.5 Temporal Scientometric Analysis
Fig. 10 shows the temporal scientometric analysis for the included articles. In order to maximize the data retrieved pertaining to the proposed search string, no limiter is implemented on the publication year. Based on the figure, the temporal distribution demonstrates a continual increment in the topic of interest, with the first relevant publication in the year 2007. Relevant publication in GAI, specifically in breast cancer was not evident from the years 2008 to 2015, with limited growth observed. This slow start can be attributed to the early stage of AI development, where machine learning and AI applications in healthcare were still nascent, and there was limited access to large datasets, computing power, and expertise. However, from 2018 to 2022, there was a gradual increase in publications, reflecting the rise of deep learning techniques, particularly with the introduction of GAI in 2014. These advancements allowed for greater exploration in medical imaging, including tasks like cancer diagnosis, image segmentation, and data augmentation. The availability of better computational resources, such as GPUs, and the development of pre-trained models further fueled research efforts, leading to an increase in publications. The most significant surge occurred in 2023 and 2024, with the number of publications rising sharply to 11 and 9, respectively. This dramatic increase is likely due to the growing adoption of advanced generative AI models, such as YOLO and transformer-based architectures, which have proven highly effective in medical imaging applications. Researchers are increasingly recognizing the potential of AI in enhancing breast cancer diagnosis, classification, and even generating synthetic datasets for training purposes. This surge also reflects the broader trend of AI integration in healthcare, as interdisciplinary collaborations between AI researchers and medical professionals become more common, leading to more impactful studies and publications. Overall, the trend indicates that GAI’s role in breast cancer research is rapidly expanding, with further growth expected as AI technologies continue to evolve and find new applications in the medical field.
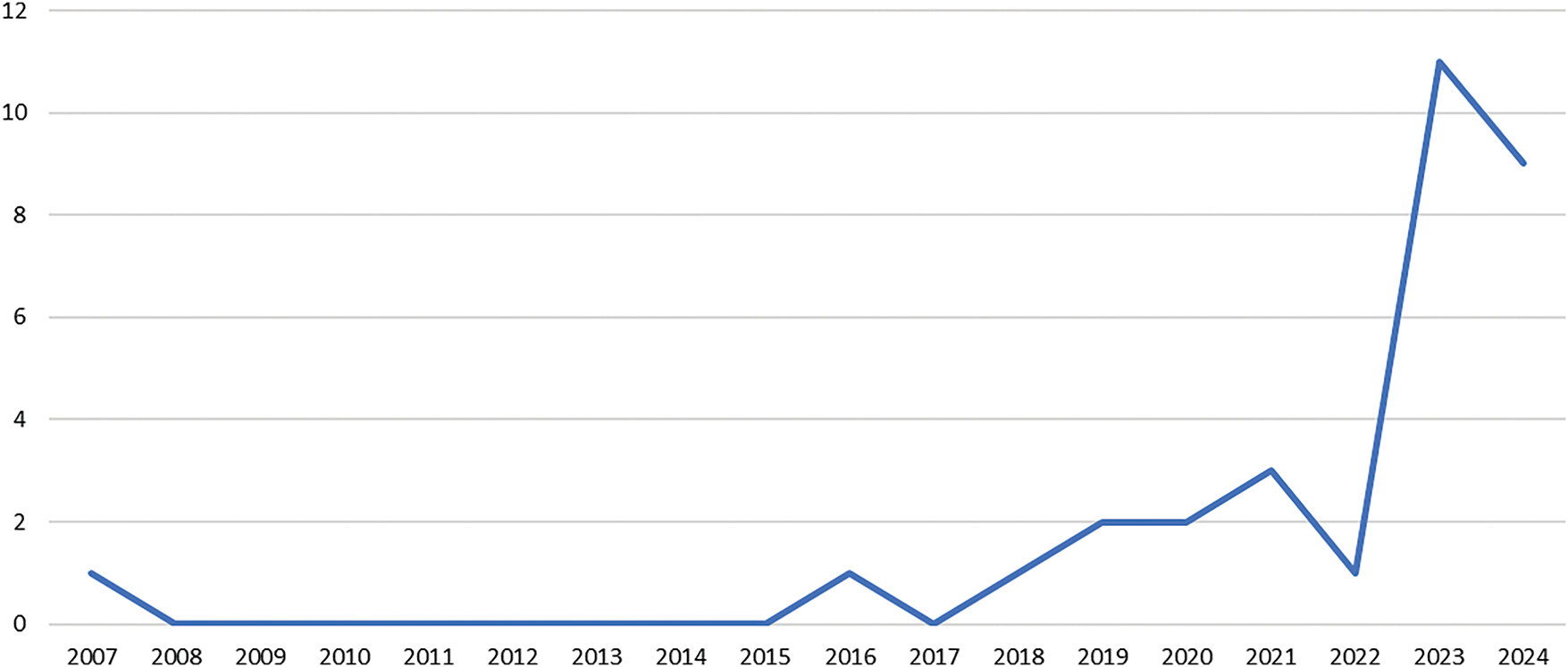
Figure 10: Temporal scientometric analysis of the included review works
Fig. 11 shows the subject areas profiling retrieved from the included articles in this study. Based on the figure, the subject area profiling for the 31 included works in GAI applied to breast cancer reveals a multidisciplinary research trend. Medicine is as the most dominant field, particularly in Radiology, Nuclear Medicine, and Imaging. This is then followed by Oncology, a sub-field of Medicine, reflecting the critical role of AI in medical imaging and cancer diagnosis. The presence of Computer Science areas, for example, AI Applications, Computer Vision, and Pattern Recognition highlights the use of computational methods in image analysis and diagnostics. Engineering fields, for example, Biomedical Engineering and Electrical Engineering underscore the integration of technology for enhanced healthcare solutions. Biochemistry, Genetics, and Molecular Biology, with emphasis on Cancer Research, and Materials Science also play crucial roles, pointing to advancements in the understanding and treatment of breast cancer through biomaterials and molecular studies. This multidisciplinary approach showcases the convergence of medicine, computer science, and engineering in leveraging AI to improve breast cancer diagnosis and treatment.
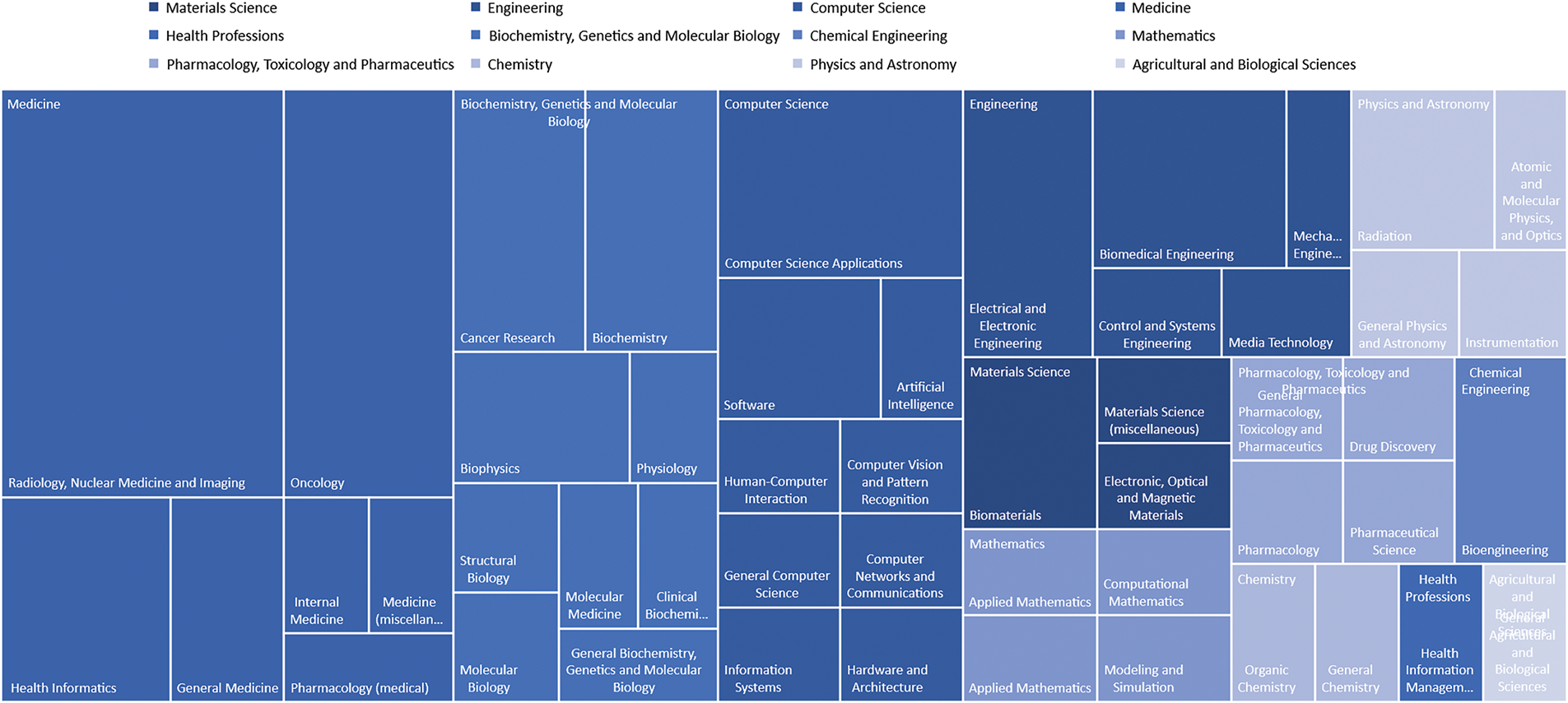
Figure 11: Subject area profiling
4.7 Keywords Co-Occurrence Mapping
Fig. 12 shows the thematic landscape of the included articles, focusing on GAI in breast cancer. The co-occurrence mapping of keywords reveals three distinct clusters, categorized by objects of interest, methodologies, and relevant sub-field approaches. The objects of interest cluster appear to be the most prominent cluster, indicated by the prevalence of keywords, for example, “breast,” “breast neoplasms,” and “breast tumour.” This is followed by the methodology cluster, characterized by keywords, for example, “artificial intelligence” and “controlled study.” The third cluster comprises keywords, for example, “image processing” and “machine learning”. Notably, the keyword “artificial intelligence” appears as the largest dot on the map, highlighting its widespread adoption across the included articles. This prominence is justifiable, given the literature search focuses on GAI in breast cancer. Additionally, the sub-field approaches, including “deep learning”, “generative adversarial networks”, and advancements in algorithms, signify significant innovations within the realm of artificial intelligence. From the perspective of objects of interest, the mapping indicates that topics such as “diagnostic imaging”, “mammography”, “breast neoplasms”, and “breast tumours” have garnered considerable attention from researchers over the past decades.
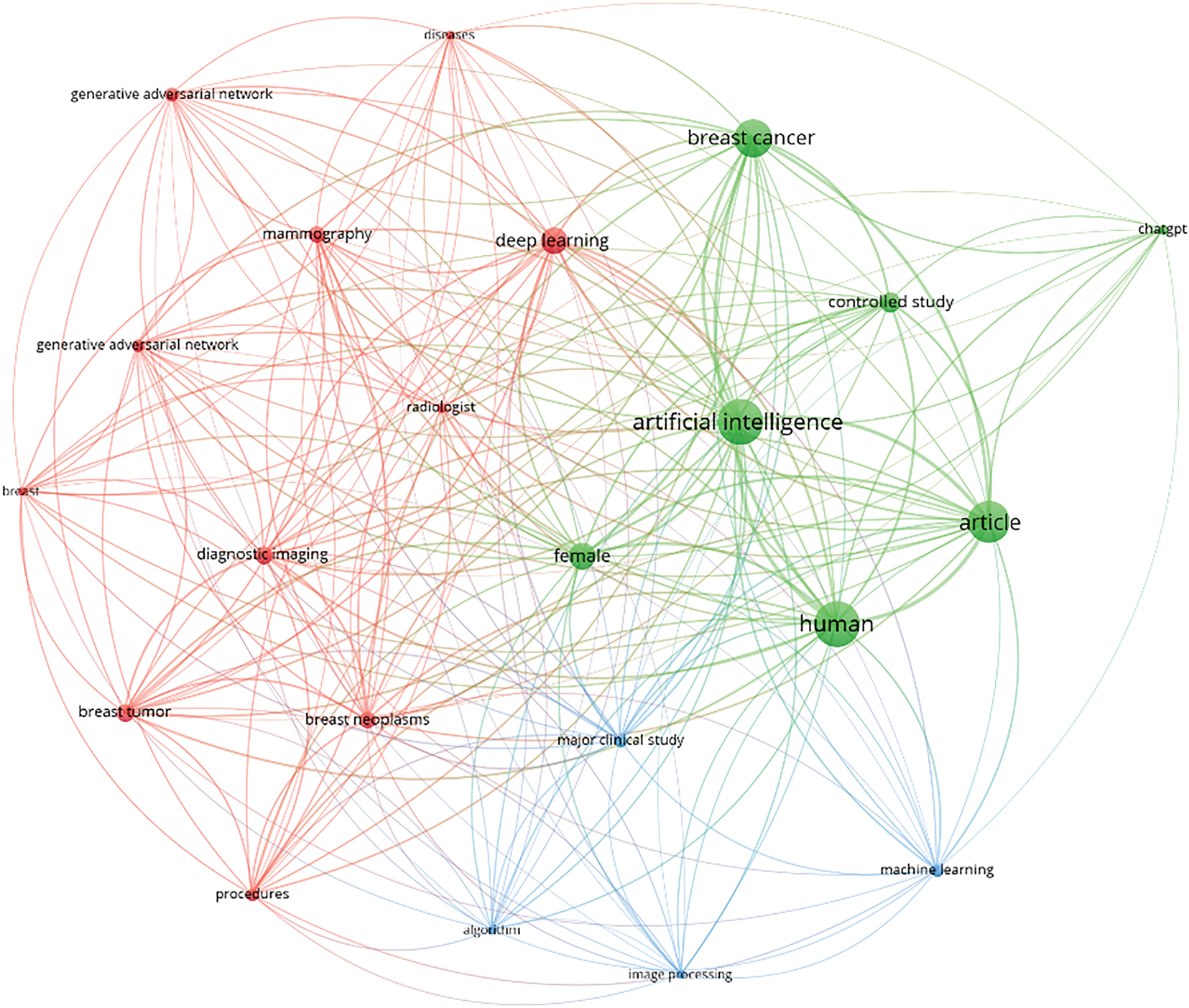
Figure 12: Keywords co-occurrence mapping
5 Self-Assessment, Limitations, Challenges, and Future Direction
The present study is a systematic review that focuses on the diagnosis and treatment of breast cancer. To maximize the data retrieved, no limiter is set on publication years. Here, a structured review methodology was used to collate, analyze, and synthesize the findings, highlighting patterns, trends, and the contents of included works in the topic of interest. A structured search strategy was proposed in compliance with the PRISMA guideline as detailed in Section 3.2. To affirm the quality of this study, an appraisal tool, namely A Measurement Tool to Assess Systematic Review (AMSTAR) is used to assess the quality of the present study in view of the content validity (see Table 5). AMSTAR is a self-appraisal tool that is commonly used to assess the quality of a systematic review, specifically to determine if the systematic reviews are comprehensive, have proper referencing, and are equipped with added value to the readers. The AMSTAR comprises 11 components for content validity. According to Table 5, the present study obtained one “no”, 10 “yes”, and one “NA”. Justifications were provided for each component to better support the evaluation outputs, as recommended by [8].

The findings of this study are subjected to several limitations. First, only full-text articles available in the English language are included in the synthesis process in accordance with the PRISMA guideline. Non-English material and/or articles without full text would be excluded in Phase 1 of the exclusion criteria. This may potentially introduce bias into the analysis, affecting the description of patterns and trends of the included articles in the topic of interest. Second, based on the proposed search string (Table 2), only articles utilizing GAI in breast cancer are retrieved. Therefore, emerging GAI models that are reported in other cancer types that potentially offer useful insight and utility in breast cancer may be excluded. Third, articles that were not indexed and populated by Scopus, Web of Science Core Collection, and PubMed databases were not included in the synthesis process. Lastly, grey literature is not included in the synthesis activities. This is mainly because the grey literature may lack rigorous peer review, subsequently resulting in inconsistent quality, associated with risk of bias, and challenging for reproducibility. However, considering the diversity and structured search methodology (in compliance with the PRISMA guideline) adopted here in retrieving articles within the topic of interest, the present study is confident with the findings synthesized from the included articles, such that the majority of the relevant literature has been included, and the findings accurately represent the current state of research.
5.3 Challenges in GAI Adoption in Clinical Settings
While recognizing GAI as the potential game-changer to meet the ever-increasing demand for quality healthcare in breast cancer, typically in the era of precision medicine, the technology is inevitable to inherit challenges in various spectrums for widespread clinical adoption. Awareness of the constraints of the current GAI-based systems and concerted efforts (from researchers and industry experts) are essential in pushing GAI technology to impact the future direction of breast cancer research.
Similar to all the discriminative models, the nature of data-driven in GAI-based systems is inevitable to bias [56,57], as this serves as the foundation for the generative models. Here, two biases are identified: data bias and algorithm bias. Data bias could potentially occur in the training phase when the training data comprises information from a specific cohort, while under-represented cohorts may be afflicted by the GAI outputs, assuming the output solutions fulfill the input prompts. For example, a GAI falsely identifies black patients as being healthier than white patients who suffered from the same illness when the cost of healthcare is used as the proxy. This is inaccurate as black patients receive less medical cost attention. Thus, injecting faulty perception into the training data, which subsequently reflects in the output solutions [58]. Likewise, countries with poor data registries, especially low-resource countries, are at risk of being under-represented in GAI-based systems, leading to skewed or inaccurate diagnostic and treatment recommendations [59,60]. If the generative model training data disproportionately reflects healthcare practices and/or patient demographics from high-resource countries, the GAI-based system may fail to adequately address the unique needs and/or health profiles of populations in low-resource regions. This can exacerbate existing healthcare disparities, as the outputs generated by such GAI models might misinterpret conditions prevalent in these under-represented cohorts. Consequently, bias in training data not only compromises the fairness and accuracy of GAI outputs but also raises ethical concerns about its application in global healthcare systems, particularly for vulnerable populations. Algorithm bias or mode collapse is defined as a situation where a GAI fails to capture the full diversity of the training data, resulting in repetitive or limited variations in the generated outputs [61]. Often, this bias originates from overfitting, where the generative models fail to accurately learn the underlying distribution of the training data, producing identical outputs with no variation [62].
Generalization and adaptation of GAI denote the model capability to produce a new and diverse output that is not directly replicated from the training data, implying the superiority of GAI in capturing the underpinning patterns and distributions of the training data and reflected in the output solution [63–66]. For example, a robust GAI shall demonstrate promising performance even when the input prompts are pertaining to breast histopathology images that are not presented in the training data. A well-generalized GAI can generate realistic and meaningful data across various scenarios, even when presented with inputs or conditions that have not been explicitly encountered during training. Generalization is crucial to avoid overfitting, ensuring that the model can generalize the patterns (from the training data) and adapt to unseen data while maintaining high performance in real-world applications. Considering the diversity of incoming patients (i.e., wide variety of races, ethnicity, nationality, and dietary backgrounds), GAIs that lack generalization are not reliable, increasing the risk associated, afflicting the diagnostic and treatment recommendations, ultimately, fragmenting the quality of healthcare service and exacerbate trust in GAI for clinical adoption. Adapting the GAI models to a new domain is now an ongoing challenge, where techniques, for example, fine-tuning, domain adaptation, and transfer learning are continued to be explored to address the present challenge.
Because of the generative nature of GAI, the model is susceptible to hallucination or confabulation, where the model itself expects a plausible output solution. In fact, the solution generated is unreasonable with respect to the training data [67,68]. The erroneous and/or misleading output solutions are not a result of bias, but the output solutions are factually incorrect, unrealistic, and/or completely unrelated to the training data. To date, the core reasons underpinning hallucination or confabulation are still mysterious, with early inference that training data embedded with fictional or contradictory content besides factual information may further promote the risk of hallucination [69]. This may suggest the importance of data cleaning (i.e., removal of outliers) in the training phase, to avoid skewed models due to disproportionate influence, distorted learning, and random noise resulting from the outliers. Considering the rich amount of training data in GAI, data cleaning is tedious and cumbersome however, especially for medical experts with mediocre knowledge of computational engineering and data analytics. Concerted efforts in data cleaning, data transparency, and algorithm error-checking mechanisms may be required to mitigate the aforementioned problem.
Transparency, explainability, and interpretability of an AI system in clinical settings are crucial, as medical decisions cannot be adopted solely from a “black box” model, which lacks sophisticated reasonings and justifications, with output solutions that are challenging to verify [70]. Unlike the discriminative model, the explainable model, for example, Explainable Artificial Intelligence (XAI) [71] in the generative model is still in its infancy with robust models yet to surface. Additionally, a recent study argues that the GAI (i.e., typically LLM) that gained interest across multiple spectrums due to its formal reasoning capabilities, particularly in mathematics may not be accurate. The findings argue the hypothesis with evidence proving the LLMs may not be capable of genuine local reasoning but, in fact, replicating the reasoning steps based on the observation in the training phase [72]. Transparency in GAI holds an important role, especially when multiple output solutions could be produced toward the same input prompts. Thus, sensemaking in fulfilling the input prompts is essential. To date, most of the GAIs are trained on a pre-trained model with respective fine-tuning performed in accordance with different circumstances or applications for local adaptation.
From a regulatory standpoint, the use of GAI-generated medical data introduces complex issues related to data privacy, intellectual property, and legal responsibilities, specifically, in the context of cross-border healthcare collaboration. Data privacy is a primary concern, as aforementioned, the data-driven GAI models often rely on large datasets, which may comprise sensitive patient information. Ensuring compliance with data protection regulations such as the General Data Protection Regulation (GDPR) in the European Union or the Health Insurance Portability and Accountability Act (HIPAA) in the United States becomes challenging in cross-border settings, where differing privacy standards may conflict. Additionally, the origins and handling of data used to train pre-trained models may lack transparency (i.e., black-box), raising ethical concerns about patient consent and data provenance. Intellectual property issues further complicate the landscape, as questions arise over the ownership of GAI-generated content, the rights to the original training data, and liability for errors or misuse of model outputs [73]. Events involving cross-border collaborations further exacerbate the situation, where intellectual property laws vary significantly between countries [74,75]. Legal responsibilities also present challenges, as determining accountability for adverse outcomes or misdiagnoses stemming from GAI-generated recommendations can be difficult, especially when models are developed in one jurisdiction and deployed in another. Differing healthcare regulations and liability frameworks across borders further complicate the establishment of clear accountability. Addressing these issues requires robust international frameworks to harmonize data privacy standards, clarify intellectual property rights, and establish legal accountability, ensuring the ethical and responsible use of GAI in global healthcare collaborations.
The adoption of GAI in clinical settings requires comprehensive technical support, planned preventive maintenance, and robust infrastructure readiness. This includes ensuring the availability of high-performance workstations and sufficient energy supply to handle the intensive computational demands of GAI models. Additionally, having skilled experts on hand is crucial to address daily technical difficulties and to provide manpower for routine maintenance and system checks. Cybersecurity is another critical aspect, as the integrity of GAI models must be safeguarded against potential hacking attempts that could compromise GAI accuracy and reliability [76]. Mitigation measures must also be in place to prevent malicious interference, for example, the introduction of noise or unauthorized alterations to the models, which could jeopardize healthcare services, afflicting the generative recommendations. Ensuring both technical and cybersecurity readiness is key to the safe and effective implementation of GAI in clinical environments. Considering all these requirements, ranging from infrastructure and energy to expert manpower and cybersecurity, the associated cost becomes a significant factor in the adoption process [77]. To keep healthcare affordable, GAI should not solely aim to eliminate occasional errors made by medical experts but should instead focus on fully automating specific procedures currently performed by experts. The primary goal of GAI in breast cancer research shall focus on enhancing clinical efficiency while avoiding unnecessary healthcare costs.
Misuse of GAI is not new, especially in domains involving entertainment and social media using techniques, for example, deepfakes. Considering the ease and robustness of the deepfake techniques in this era, the low cost of implementation, as well as sub-optimal legal boundaries, it is challenging to affirm a GAI is free from malicious actions if a comprehensive adversarial monitoring system is not in place [78]. Factors contributing to misuse of GAI in breast cancer, especially in diagnosis and treatment can be multi-factorial, especially involving enormous personal interests that may arise from conflicts, for example, malicious incidences involving insurance claims and tension between opting for optimal healthcare or medical profit. These underscore the vulnerability of GAI to malicious actions and exploitation, with potential threats that collectively hinder trust in GAI in clinical settings.
While GAI poses significant advancement and revolutionizes breast cancer research, typically in diagnosis and treatment stages, offering enhanced efficiency and productivity in clinical operations, awareness of the challenges of GAI adoption is important. The far-reaching risks of GAI, typically in bias, hallucination or confabulation, and misuse required concerted and sustained efforts from researchers and industry experts in developing comprehensive mitigation mechanisms and solutions to ensure responsible AI usage. The adoption of GAI can be performed in multiple stages, where continuous temporal assessment and quality improvement are implemented at different checkpoints [25], and allows interrogation and feedback from experts at different levels as well as consumers and the public, aiming for robust and reliable GAI adoption.
As GAI continues to evolve, its integration into clinical practice requires a multidisciplinary approach, addressing technical, ethical, legal, and practical considerations. Ensuring the successful deployment of GAI in breast cancer diagnosis and treatment will depend on collaborative efforts among researchers, policymakers, healthcare institutions, and clinical practitioners. This section outlines key focus areas and strategies for advancing GAI in healthcare.
From a researcher’s standpoint, the primary focus should be on developing GAI models that are robust against bias and capable of generalizing across diverse patient populations. This necessitates the creation of inclusive and representative training datasets, particularly from underrepresented cohorts and low-resource regions. Techniques such as domain adaptation, transfer learning, and federated learning should be explored to ensure GAI models perform reliably across varied demographics and healthcare settings. Additionally, XAI for generative models must be advanced to enhance clinicians’ trust in GAI-generated outputs. Developing interpretable models that provide clear justifications for their recommendations, along with visualization tools for decision-making processes, will be crucial for bridging the gap between AI and clinical practice. Furthermore, reducing hallucinations in GAI models is essential for improving reliability. This can be achieved through robust data-cleaning pipelines, outlier detection mechanisms, and error-checking algorithms that ensure the accuracy and consistency of generated solutions. Collaboration between AI developers and medical experts is critical to refining these processes and maintaining clinical relevance.
From a policymaker’s standpoint, establishing legal and ethical frameworks is essential for facilitating cross-border collaboration in GAI research and deployment. Regional and international alliances are necessary to create standardized regulations for data privacy, intellectual property, and legal accountability, ensuring the global adoption of GAI in healthcare. Open-access initiatives, such as DeepSeek, can serve as models for promoting transparency and equitable access to GAI technologies. Policymakers should also develop guidelines for data sharing, model ownership, and liability, ensuring that GAI systems are deployed responsibly and ethically. Effective collaboration among policymakers, researchers, and industry stakeholders is necessary to establish harmonized frameworks that balance technological innovation with patient rights and ethical considerations.
From a healthcare institution’s standpoint, a phased approach to GAI adoption should be considered for seamless integration into clinical workflows. Initially, GAI can be implemented in low-risk applications such as data augmentation, image synthesis, and preliminary diagnostic support, allowing healthcare providers to build confidence in the technology while minimizing risks to patient care. Establishing scalable and secure computational infrastructure is crucial for supporting the demands of GAI in clinical settings. This includes high-performance workstations, energy-efficient systems, and robust cybersecurity measures to protect sensitive patient data and maintain the integrity of GAI models. Additionally, comprehensive training programs and workshops should be provided to clinical practitioners to equip them with the necessary knowledge for effectively utilizing GAI technologies in medical practice.
From a clinical practitioner’s standpoint, collaboration with healthcare institutions is crucial in establishing strong data governance frameworks that comply with data privacy regulations such as GDPR and HIPAA. This includes verifying the provenance of training data used in GAI models and obtaining informed patient consent for data usage. Before fully integrating GAI into clinical workflows, clinicians should treat GAI-generated recommendations as supplementary tools, rather than definitive solutions. Rigorous validation of AI-generated outputs against clinical expertise and established guidelines is necessary to ensure patient safety and prevent over-reliance on AI systems. Clinical practitioners also play a vital role in advocating for the ethical use of AI by participating in policy discussions, addressing biases in AI models, and ensuring that GAI technologies prioritize patient welfare and equity. Continuous research and collaboration between clinicians and AI researchers are essential to refine GAI models and ensure their practical applicability in real-world healthcare settings.
Achieving full integration of GAI in clinical practice will require a multidisciplinary effort. Experts from various fields must exchange knowledge, break down research silos, and complement each other’s expertise to foster the development of innovative solutions. By addressing the technical, legal, and practical challenges, GAI has the potential to revolutionize breast cancer diagnosis and treatment, ultimately improving patient outcomes and advancing precision medicine.
In this study, a holistic birds-eye view of the research reviewing past literature with GAI in breast cancer is presented. Here, a thorough search string in compliance with the PRISMA guideline is proposed. The findings of the systematic review provide useful insight into the holistic view of how research communities contribute, the primary methods employed, the findings of included works, the publications trend, and the development of collaborative networks over time. Based on the analysis outcomes, this study highlighted: (1) the main research domain in GAI in breast cancer research fall within the diagnosis category, accounting for 90.3% (28 articles) of the included articles; (2) geographical scientometric analysis shows that USA leads the AI-driven breast cancer research with 8 contributions in the body of the literature; (3) the Journal of Medical Imaging and Dignostics are both ranked as the most contributing journals; (4) the publisher, namely Springer is found to be the most contributing publisher, accounted 22.6% (7 articles) in total; (5) based on the temporal scientometric analysis, from 2018 to 2022, there was a gradual increase in publications with the highest publications of 11 and 9 in 2023 and 2024, respectively; (6) in subject area profiling, the subject area of Medicine appears to be the most dominant field, specifically in Radiology, Nuclear Medicine, and Imaging; and (7) three broad thematic clusters found in keyword co-occurrence analysis, namely objects of interest, methodologies, and relevant sub-field approaches. The systematic review serves as a scientific communication, highlighting the research gap and challenges in the topic of interest. For newcomers to the field, this systematic review offers a comprehensive and timely overview, providing valuable insights into the intellectual landscape, understanding literature development, and outlining the potential challenges. For experienced researchers, it serves as a resource to stay updated, particularly in identifying relevant research areas that extend beyond their primary focus. For stakeholders, this systematic review can help prioritize research and funding to support impactful and urgent solutions through GAI integration in breast cancer research, typically in supporting diagnosis and treatment purposes.
Acknowledgement: The authors are grateful to all the editors and anonymous reviewers for their comments and suggestions.
Funding Statement: This study received financial support from the Fundamental Research Grant Scheme (FRGS) under grant number: FRGS/1/2024/ICT02/TARUMT/02/1, from the Ministry of Higher Education Malaysia. The Article Processing Charge (APC) was funded in part by the internal grant from the Tunku Abdul Rahman University of Management and Technology (TAR UMT) with grant number: UC/I/G2024-00129.
Author Contributions: Concept and design: Xiao Jian Tan, Wai Loon Cheor, and Khairul Shakir Ab Rahman. Acquisition of data: Xiao Jian Tan and Wai Loon Cheor. Analysis and interpretation of data: Xiao Jian Tan, Ee Meng Cheng, and Chee Chin Lim. Drafting the manuscript: Xiao Jian Tan. Statistical analysis: Xiao Jian Tan and Wai Loon Cheor. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.

Figure A1: Brief timeline of AI advancement. Note: SHRDLU is an early natural-language understanding computer program



References
1. Ray PP. ChatGPT: a comprehensive review on background, applications, key challenges, bias, ethics, limitations and future scope. Internet Things Cyber-Phys Syst. 2023;3(1):121–54. doi:10.1016/j.iotcps.2023.04.003. [Google Scholar] [CrossRef]
2. Karras T, Laine S, Aila T. A style-based generator architecture for generative adversarial networks. IEEE Trans Pattern Anal Mach Intell. 2021;43(12):4217–28. doi:10.1109/TPAMI.2020.2970919. [Google Scholar] [PubMed] [CrossRef]
3. Yin H, Zhang Z, Liu Y. The Exploration of integrating the midjourney artificial intelligence generated content tool into design systems to direct designers towards future-oriented innovation. Systems. 2023;11(12):566. doi:10.3390/systems11120566. [Google Scholar] [CrossRef]
4. Cetin I, Stephens M, Camara O, González BM. Attri-VAE: attribute-based interpretable representations of medical images with variational autoencoders. Comput Med Imaging Graph. 2023;104(6):102158. doi:10.1016/j.compmedimag.2022.102158. [Google Scholar] [PubMed] [CrossRef]
5. Liu X, Zhang L, Guo Z, Han T, Ju M, Xu B, et al. Medical image compression based on variational autoencoder. Math Probl Eng. 2022;2022(3):1–12. doi:10.1155/2022/7088137. [Google Scholar] [CrossRef]
6. Rakhmetulayeva S, Zhanabekov Z, Bolshibayeva A. Evaluation of modern generative networks for echocg image generation. Comput Mater Contin. 2024;81(3):4503–23. doi:10.32604/cmc.2024.057974. [Google Scholar] [CrossRef]
7. Turing AM. Computing machinery and intelligence. Mind. 1950;49:433–60. [Google Scholar]
8. Tan XJ, Cheor WL, Lim LL, Khairul Shakir AR, Ikamal Hisyam B. Artificial intelligence (AI) in breast imaging: a scientometric umbrella review. Diagnostics. 2022;12(12):1–35. doi:10.3390/diagnostics12123111. [Google Scholar] [PubMed] [CrossRef]
9. Collins C, Dennehy D, Conboy K, Mikalef P. Artificial intelligence in information systems research: a systematic literature review and research agenda. Int J Inf Manag. 2021;60(4):102383. doi:10.1016/j.ijinfomgt.2021.102383. [Google Scholar] [CrossRef]
10. Winston PH. Artificial intelligence. 3rd ed. Reading, MA, USA: Addison-Wesley; 1993 [Internet]. [cited 2025 May 14]. Available from: https://courses.csail.mit.edu/6.034f/ai3/rest.pdf. [Google Scholar]
11. Tan XJ, Cheor WL, Yeo KS, Leow WZ. Expert systems in oil palm precision agriculture: a decade systematic review. J King Saud Univ—Comput Inf Sci. 2022;34(4):1569–94. doi:10.1016/j.jksuci.2022.02.006. [Google Scholar] [CrossRef]
12. Bi WL, Ahmed H, Matthew BS, Maryellen LG, Nicolai JB, Alireza M, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–57. doi:10.3322/caac.21552. [Google Scholar] [PubMed] [CrossRef]
13. Srikantamurthy MM, Rallabandi VPS, Dudekula DB, Natarajan S, Park J. Classification of benign and malignant subtypes of breast cancer histopathology imaging using hybrid CNN-LSTM based transfer learning. BMC Med Imaging. 2023;23(1):1–15. doi:10.1186/s12880-023-00964-0. [Google Scholar] [PubMed] [CrossRef]
14. Chakravarthy S, Bharanidharan N, Khan SB, Kumar VV, Mahesh TR, Almusharraf A, et al. Multi-class breast cancer classification using CNN features hybridization. Int J Comput Intell Syst. 2024;17(1):191. doi:10.1007/s44196-024-00593-7. [Google Scholar] [CrossRef]
15. Jeslin T, Linsely JA. AGWO-CNN classification for computer-assisted diagnosis of brain tumors. Comput Mater Contin. 2022;71(1):171–82. doi:10.32604/cmc.2022.020255. [Google Scholar] [CrossRef]
16. Jiang B, Bao L, He S, Chen X, Jin Z, Ye Y. Deep learning applications in breast cancer histopathological imaging: diagnosis, treatment, and prognosis. Breast Cancer Res. 2024;26(1):137. doi:10.1186/s13058-024-01895-6. [Google Scholar] [PubMed] [CrossRef]
17. Arya N, Saha S. Deviation-support based fuzzy ensemble of multi-modal deep learning classifiers for breast cancer prognosis prediction. Sci Rep. 2023;13(1):1–10. doi:10.1038/s41598-023-47543-5. [Google Scholar] [PubMed] [CrossRef]
18. Qian L, Lu X, Haris P, Zhu J, Li S, Yang Y. Enhancing clinical trial outcome prediction with artificial intelligence: a systematic review. Drug Discov Today. 2025;30(4):104332. doi:10.1016/j.drudis.2025.104332. [Google Scholar] [PubMed] [CrossRef]
19. Salh CH, Ali AM. Automatic detection of breast cancer for mastectomy based on MRI images using Mask R-CNN and Detectron2 models. Neural Comput Appl. 2024;36(6):3017–35. doi:10.1007/s00521-023-09237-x. [Google Scholar] [CrossRef]
20. Azadinejad H, Farhadi RM, Shariftabrizi A, Rahmim A, Abdollahi H. Optimizing cancer treatment: exploring the role of AI in radioimmunotherapy. Diagnostics. 2025;15(3):397. doi:10.3390/diagnostics15030397. [Google Scholar] [PubMed] [CrossRef]
21. Lee J, Nishikawa RM. Improving lesion detection in mammograms by leveraging a Cycle-GAN-based lesion remover. Breast Cancer Res. 2024;26(1):1–16. doi:10.1186/s13058-024-01777-x. [Google Scholar] [PubMed] [CrossRef]
22. Kumar A, Sharma A, Singh AK, Singh SK, Saxena S. Data augmentation for medical image classification based on gaussian laplacian pyramid blending with a similarity measure. IEEE J Biomed Health Inform. 2023. doi:10.1109/JBHI.2023.3307216. [Google Scholar] [PubMed] [CrossRef]
23. Inan MSK, Hossain S, Uddin MN. Data augmentation guided breast cancer diagnosis and prognosis using an integrated deep-generative framework based on breast tumor’s morphological information. Inform Med Unlocked. 2022;37(1):101171. doi:10.1016/j.imu.2023.101171. [Google Scholar] [CrossRef]
24. Choi HS, Song JY, Shin KH, Chang JH, Jang BS. Developing prompts from large language model for extracting clinical information from pathology and ultrasound reports in breast cancer. Radiat Oncol J. 2023;41(3):209–16. doi:10.3857/roj.2023.00633. [Google Scholar] [PubMed] [CrossRef]
25. Floyd W, Kleber T, Carpenter DJ, Pasli M, Qazi J, Huang C, et al. Current strengths and weaknesses of ChatGPT as a resource for radiation oncology patients and providers. Int J Radiat Oncol Biol Phys. 2024;118(4):905–15. doi:10.1016/j.ijrobp.2023.10.020. [Google Scholar] [PubMed] [CrossRef]
26. James RJN, Andee K, David CQ, Neal SMC, Yuan L, Sagar AP. physician assessment of chatgpt and bing answers to american cancer society’s questions to ask about your cancer. Am J Clin Oncol. 2024;47(1):17–21. doi:10.1097/COC.0000000000001050. [Google Scholar] [PubMed] [CrossRef]
27. Fujioka T, Satoh Y, Imokawa T, Mori M, Yamaga E, Takashi K, et al. Proposal to improve the image quality of short-acquisition time-dedicated breast positron emission tomography using the pix2pix generative adversarial network. Diagnostics. 2022;12(12):3114. doi:10.3390/diagnostics12123114. [Google Scholar] [PubMed] [CrossRef]
28. Latha V, Gomathi V, Rajeshkanna A, Ram SH. Generating a potent inhibitor against MCF7 breast cancer cell through artificial intelligence based virtual screening and molecular docking studies. Indian J Biochem Biophys. 2023;60(11):844–56. doi:10.56042/ijbb.v60i11.6067. [Google Scholar] [CrossRef]
29. Banh L, Strobel G. Generative artificial intelligence. Electron Mark. 2023;33(1):1–17. doi:10.1007/s12525-023-00680-1. [Google Scholar] [CrossRef]
30. Fernández-Llorca D, Gómez E, Sánchez I, Mazzini G. An interdisciplinary account of the terminological choices by EU policymakers ahead of the final agreement on the AI Act: AI system, general purpose AI system, foundation model, and generative AI. Artif Intell Law. 2024:0123456789. doi:10.1007/s10506-024-09412-y. [Google Scholar] [CrossRef]
31. OECD. Recommendations of the council on artificial intelligence (adopted by the council at ministerial level). 2019 [Internet]. [cited 2025 May 14]. Available from: https://one.oecd.org/document/C/MIN(2019)3/FINAL/en/pdf. [Google Scholar]
32. European Commission. Impact assessment of the regulation on artificial intelligence. European Commission (SWD(2021) 84 final. 2021 [Internet]. [cited 2025 May 14]. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:52021SC0084. [Google Scholar]
33. Approach CG. The articles of the EU artificial intelligence act (25.11.2022) [Internet]. [cited 2025 May 14]. Available from: https://www.artificial-intelligence-act.com/Artificial_Intelligence_Act_Article_3_(Proposal_25.11.2022).html. [Google Scholar]
34. WilmerHale. The european parliament adopts the AI act [Internet]. [cited 2025 May 14]. Available from: https://www.wilmerhale.com/en/insights/blogs/wilmerhale-privacy-and-cybersecurity-law/20240314-the-european-parliament-adopts-the-ai-act#:~:text=“AIsystem”meansamachine,recommendations%2Cordecisionsthatcan. [Google Scholar]
35. OECD. Recommendations of the council on artificial intelligene (amended by the council on 8 november 2023). 2023 [Internet]. [cited 2025 May 14]. Available from: https://legalinstruments.oecd.org/en/instruments/%20OECD-LEGAL-0449. [Google Scholar]
36. OECD. Explanatory memorandum on the updated OECD definition of an AI system OpenAI (2022) Chatgpt [large language model]. 2024 [Internet]. [cited 2025 May 14]. Available from: https://www.oecd.org/en/publications/explanatory-memorandum-on-the-updated-oecd-definition-of-an-ai-system_623da898-en.html. [Google Scholar]
37. Kingma DP, Welling M. An introduction to variational autoencoders. Found Trends Mach Learn. 2019;12(4):307–92. doi:10.1561/2200000056. [Google Scholar] [CrossRef]
38. Girin L, Leglaive S, Bie X, Diard J, Hueber T, Alameda-Pineda X. Dynamical variational autoencoders: a comprehensive review. Found Trends Mach Learn. 2021;15(1–2):1–175. doi:10.1561/2200000089. [Google Scholar] [CrossRef]
39. Aggarwal A, Mittal M, Battineni G. Generative adversarial network: an overview of theory and applications. Int J Inf Manag Data Insights. 2021;1(1):100004. doi:10.1016/j.jjimei.2020.100004. [Google Scholar] [CrossRef]
40. Bandi A, Adapa PVSR, Kuchi YEVPK. The power of generative AI: a review of requirements, models, input-output formats, evaluation metrics, and challenges. Future Internet. 2023;15(8):260. doi:10.3390/fi15080260. [Google Scholar] [CrossRef]
41. Chen M, Mei S, Fan J, Wang M. An overview of diffusion models: applications, guided generation, statistical rates and optimization. arXiv:2404.07771. 2024. doi:10.48550/arXiv.2404.07771. [Google Scholar] [CrossRef]
42. Xu Y, Wei HP, Lin MX, Deng YY, Sheng KK, Zhang MD, et al. Transformers in computational visual media: a survey. Comput Vis Media. 2022;8:33–62. doi:10.1007/s41095-021-0247-3. [Google Scholar] [CrossRef]
43. Lin Z, Guan S, Zhang W, Zhang H, Li Y, Zhang H. Towards trustworthy LLMs: a review on debiasing and dehallucinating in large language models. Artif Intell Rev. 2024;57(9):243. doi:10.1007/s10462-024-10896-y. [Google Scholar] [CrossRef]
44. Vidal A, Wu FS, Tenorio L, Osher S, Nurbekyan L. Taming hyperparameter tuning in continuous normalizing flows using the JKO scheme. Sci Rep. 2023;13(1):1–11. doi:10.1038/s41598-023-31521-y. [Google Scholar] [PubMed] [CrossRef]
45. Tan XJ, Cheor WL, Cheng EM, Khairul Shakir AR, Wan Zuki Azman WM, Leow WZ. Breast cancer status, grading system, etiology, and challenges in Asia: an updated review. Oncologie. 2023;25(2):99–110. doi:10.1515/oncologie-2022-1011. [Google Scholar] [CrossRef]
46. Iranmakani S, Mortezazadeh T, Sajadian F, Ghaziani MF, Ghafari A, Khezerloo D, et al. A review of various modalities in breast imaging: technical aspects and clinical outcomes. Egyp J Radiol Nuclear Med. 2020;51(1):57. doi:10.1186/s43055-020-00175-5. [Google Scholar] [CrossRef]
47. Bhushan A, Gonsalves A, Menon JU. Current state of breast cancer diagnosis, treatment, and theranostics. Pharmaceutics. 2021;13(5):1–24. doi:10.3390/pharmaceutics13050723. [Google Scholar] [PubMed] [CrossRef]
48. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. 2021;88:105906. doi:10.1016/j.ijsu.2021.105906. [Google Scholar] [PubMed] [CrossRef]
49. Korn AR, Walsh-Bailey C, Pilar M, Sandler B, Bhattacharjee P, Moore WT, et al. Social determinants of health and cancer screening implementation and outcomes in the USA: a systematic review protocol. Syst Rev. 2022;11(1):1–8. doi:10.1186/s13643-022-01995-4. [Google Scholar] [PubMed] [CrossRef]
50. Perianes-Rodriguez A, Waltman L, Eck NJV. Constructing bibliometric networks: a comparison between full and fractional counting. J Informetr. 2016;10(4):1178–95. doi:10.1016/j.joi.2016.10.006. [Google Scholar] [CrossRef]
51. Kotei E, Thirunavukarasu R. Visual attention condenser model for multiple disease detection from heterogeneous medical image modalities. Multimed Tools Appl. 2024;83(10):30563–85. doi:10.1007/s11042-023-16625-x. [Google Scholar] [CrossRef]
52. Singh Y, Hathaway QA, Erickson BJ. Generative AI in oncological imaging: revolutionizing cancer detection and diagnosis. Oncotarget. 2024;15(1):607–8. doi:10.18632/oncotarget.28640. [Google Scholar] [PubMed] [CrossRef]
53. Zama S, Fujioka T, Yamaga E, Kubota K, Mori M, Katsuta L, et al. Clinical utility of breast ultrasound images synthesized by a generative adversarial network. Medicina. 2024;60(1):1–11. doi:10.3390/medicina60010014. [Google Scholar] [PubMed] [CrossRef]
54. Osuala R, Skorupko G, Lazrak N, Garrucho L, García E, Joshi S, et al. medigan: a Python library of pretrained generative models for medical image synthesis. J Med Imaging. 2023;10(6):061403. doi:10.1117/1.jmi.10.6.061403. [Google Scholar] [PubMed] [CrossRef]
55. Loeb S, Bjurlin M, Nicholson J, Tammela T, Penson D, Carter HB, et al. Overdiagnosis and overtreatment of prostate cancer. Eur Urol. 2014;65(6):1046–55. doi:10.1016/j.eururo.2013.12.062. [Google Scholar] [PubMed] [CrossRef]
56. Kim S, Lee CK, Kim SS. Large language models: a guide for radiologists. Korean J Radiol. 2024;25(2):126–33. doi:10.3348/kjr.2023.0997. [Google Scholar] [PubMed] [CrossRef]
57. Liu Y, Huang J, Li Y, Wang D, Xiao B. Generative AI model privacy: a survey. Artif Intell Rev. 2025;58(1):33. doi:10.1007/s10462-024-11024-6. [Google Scholar] [CrossRef]
58. Gordon R. Artificial intelligence predicts patients’ race from their medical images. MIT News. 2022 [Internet]. [cited 2025 Jun 17]. Available from: https://news.mit.edu/2022/artificial-intelligence-predicts-patients-race-from-medical-images-0520. [Google Scholar]
59. Zhao LH, Cao B, Borghi E, Chatterji S, Garcia-Saiso S, Rashidian A, et al. Data gaps towards health development goals, 47 low-and middle-income countries. Bull World Health Organ. 2022;100(1):40–9. doi:10.2471/BLT.21.286254. [Google Scholar] [PubMed] [CrossRef]
60. Nazer LH, Zatarah R, Waldrip S, Ke JXC, Moukheiber M, Khanna AK, et al. Bias in artificial intelligence algorithms and recommendations for mitigation. PLoS Digital Health. 2023;2(6):e0000278. doi:10.1371/journal.pdig.0000278. [Google Scholar] [PubMed] [CrossRef]
61. Rombach R, Blattmann A, Lorenz D, Esser P, Ommer B. High-resolution image synthesis with latent diffusion models. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2022; New Orleans, LA, USA. p. 10674–85. doi:10.1109/CVPR52688.2022.01042. [Google Scholar] [CrossRef]
62. González-Sendino R, Serrano E, Bajo J. Mitigating bias in artificial intelligence: fair data generation via causal models for transparent and explainable decision-making. Future Gener Comput Syst. 2024;155:384–401. doi:10.1016/j.future.2024.02.023. [Google Scholar] [CrossRef]
63. Bin Akhtar Z. Unveiling the evolution of generative AI (GAIa comprehensive and investigative analysis toward LLM models (2021–2024) and beyond. J Electr Sys Inform Technol. 2024;11(1):1–21. doi:10.1186/s43067-024-00145-1. [Google Scholar] [CrossRef]
64. Riveland R, Pouget A. Natural language instructions induce compositional generalization in networks of neurons. Nat Neurosci. 2024;27(5):988–99. doi:10.1038/s41593-024-01607-5. [Google Scholar] [PubMed] [CrossRef]
65. Peng W, Chen A, Huang W, Chen J, Xu H. Generalized aggregation index based collaborative fusion for medical diagnosis. Artif Intell Med. 2025;165(2):103128. doi:10.1016/j.artmed.2025.103128. [Google Scholar] [PubMed] [CrossRef]
66. Atto AM. Toward generalized artificial intelligence by assessment aggregation with applications to standard and extreme classifications. IEEE Trans Neural Netw Learn Syst. 2024;35(11):16659–70. doi:10.1109/TNNLS.2023.3297079. [Google Scholar] [PubMed] [CrossRef]
67. Ji Z, Lee N, Frieske R, Yu T, Su D, Xu Y, et al. Survey of hallucination in natural language generation. ACM Comput Surv. 2023;55(12):248. doi:10.1145/3571730. [Google Scholar] [CrossRef]
68. Susarla A, Gopal R, Thatcher JB, Sarker S. The janus effect of generative AI: charting the path for responsible conduct of scholarly activities in information systems. Inf Syst Res. 2023;34(2):399–408. doi:10.1287/isre.2023.ed.v34.n2. [Google Scholar] [CrossRef]
69. Dziri N, Milton S, Yu M, Zaiane O, Reddy S. On the origin of hallucinations in conversational models: is it the datasets or the models? In: NAACL 2022—2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Seattle, NY, USA: Association for Computational Linguistics; 2022. p. 5271–85. doi:10.18653/v1/2022.naacl-main.387. [Google Scholar] [CrossRef]
70. Marey A, Arjmand P, Alerab ADS, Eslami MJ, Saad AM, Sanchez N, et al. Explainability, transparency and black box challenges of AI in radiology: impact on patient care in cardiovascular radiology. Egyp J Radiol Nuclear Med. 2024;55(1):183. doi:10.1186/s43055-024-01356-2. [Google Scholar] [CrossRef]
71. Saranya A, Subhashini R. A systematic review of explainable artificial intelligence models and applications: recent developments and future trends. Decis Anal J. 2023;7(5):100230. doi:10.1016/j.dajour.2023.100230. [Google Scholar] [CrossRef]
72. Mirzadeh I, Alizadeh K, Shahrokhi H, Tuzel O, Bengio S, Farajtabar M. GSM-symbolic: understanding the limitations of mathematical reasoning in large language models. 2024. doi:10.48550/arXiv.2410.05229. [Google Scholar] [CrossRef]
73. Bhaskar P, Tiwari CK. Charming or chilling? A comprehensive review of ChatGPT’s in education sector. Int J Inf Learn Technol. 2025;32(10):1. doi:10.1108/IJILT-05-2024-0097. [Google Scholar] [CrossRef]
74. Wang M, Yan H, Ciabuschi F, Su C. Facilitator or inhibitor? The effect of host-country intellectual property rights protection on China’s technology-driven acquisitions. Int Bus Rev. 2023;32(6):102165. doi:10.1016/j.ibusrev.2023.102165. [Google Scholar] [CrossRef]
75. Al-Busaidi AS, Raman R, Hughes L, Albashrawi MA, Malik T, Dwivedi YK, et al. Redefining boundaries in innovation and knowledge domains: investigating the impact of generative artificial intelligence on copyright and intellectual property rights. J Innov Knowl. 2024;9(4):100630. doi:10.1016/j.jik.2024.100630. [Google Scholar] [CrossRef]
76. Humphreys D, Koay A, Desmond D, Mealy E. AI hype as a cyber security risk: the moral responsibility of implementing generative AI in business. AI Ethics. 2024;4(3):791–804. doi:10.1007/s43681-024-00443-4. [Google Scholar] [CrossRef]
77. Peretz-Andersson E, Tabares S, Mikalef P, Parida V. Artificial intelligence implementation in manufacturing SMEs: a resource orchestration approach. Int J Inf Manage. 2024;77(1):102781. doi:10.1016/j.ijinfomgt.2024.102781. [Google Scholar] [CrossRef]
78. Kaushik R, Veezhinathan SBK. IR-GAN: improved generative adversarial networks for infrared breast image segmentation. Quant Infrared Thermogr J. 2023;22(1):70–96. doi:10.1080/17686733.2023.2294598. [Google Scholar] [CrossRef]
79. Müller-Franzes G, Huck L, Bode M, Nebelung S, Kuhl C, Truhn D, et al. Diffusion probabilistic versus generative adversarial models to reduce contrast agent dose in breast MRI. Eur Radiol Exp. 2024;8(1):53. doi:10.1186/s41747-024-00451-3. [Google Scholar] [PubMed] [CrossRef]
80. Xu J, Luo Y, Wang C, Chen H, Tang Y, Xu Z, et al. A High-resolution prediction network for predicting intratumoral distribution of nanoprobes by tumor vascular and nuclear feature. Adv Intell Syst. 2024;6(3):2300592. doi:10.1002/aisy.202300592. [Google Scholar] [CrossRef]
81. Kuk LY, Kwong DLW, Chan WLW, Shea YF. Limitations of GPT-4 as a geriatrician in geri-oncology case conference: a case series. J Chin Med Assoc. 2024;87(2):148–50. doi:10.1097/JCMA.0000000000001032. [Google Scholar] [PubMed] [CrossRef]
82. Griewing S, Gremke N, Wagner U, Lingenfelder M, Kuhn S, Boekhoff J. Challenging ChatGPT 3.5 in senology—an assessment of concordance with breast cancer tumor board decision making. J Pers Med. 2023;13(10):1502. doi:10.3390/jpm13101502. [Google Scholar] [PubMed] [CrossRef]
83. Rao A, John KM, Meghana K, Michael P, Winston L, Keith JD, et al. Evaluating GPT as an adjunct for radiologic decision making: gPT-4 Versus GPT-3.5 in a breast imaging pilot. J Am Coll Radiol. 2023;20(10):990–7. doi:10.1016/j.jacr.2023.05.003. [Google Scholar] [PubMed] [CrossRef]
84. Lee J, Mustafaev T, Nishikawa RM. Impact of GAN artifacts for simulating mammograms on identifying mammographically occult cancer. J Med Imaging. 2023;10(5):1–11. doi:10.1117/1.jmi.10.5.054503. [Google Scholar] [PubMed] [CrossRef]
85. Colbert ZM, Ramachandran P. Auto-segmentation of thoracic organs in CT scans of breast cancer patients using a 3D U-net cascaded into 2D patchGANs. Biomed Phys Eng Express. 2023;9(5):055011. doi:10.1088/2057-1976/ace631. [Google Scholar] [PubMed] [CrossRef]
86. Jiao J, Xiao X, LiZ. dm–GAN: distributed multi-latent code inversion enhanced GAN for fast and accurate breast X-ray image automatic generation. Math Biosci Eng. 2023;20(11):19485–503. doi:10.3934/mbe.2023863. [Google Scholar] [PubMed] [CrossRef]
87. Zhou QW, Zuley M, Guo Y, Yang L, Nair B, Vargo A, et al. A machine and human reader study on AI diagnosis model safety under attacks of adversarial images. Nat Commun. 2021;12(1):7281. doi:10.1038/s41467-021-27577-x. [Google Scholar] [PubMed] [CrossRef]
88. Baccouche A, Garcia-Zapirain B, Castillo Olea C, Elmaghraby AS. Connected-UNets: a deep learning architecture for breast mass segmentation. npj Breast Cancer. 2021;7(1):1–12. doi:10.1038/s41523-021-00358-x. [Google Scholar] [PubMed] [CrossRef]
89. Tang YX, Zhang JL, He DD, Miao WF, Liu W, Li Y, et al. GANDA: a deep generative adversarial network conditionally generates intratumoral nanoparticles distribution pixels-to-pixels. J Control Release. 2021;336:336–43. doi:10.1016/j.jconrel.2021.06.039. [Google Scholar] [PubMed] [CrossRef]
90. Nguyen T, Bui V, Thai A, Lam V, Raub C, Chang LC, et al. Virtual organelle self-coding for fluorescence imaging via adversarial learning. J Biomed Opt. 2020;25(9):1–18. doi:10.1117/1.jbo.25.9.096009. [Google Scholar] [PubMed] [CrossRef]
91. Fujioka T, Mori M, Kubota K, Kikuchi Y, Katsuta L, Adachi M, et al. Breast ultrasound image synthesis using deep convolutional generative adversarial networks. Diagnostics. 2019;4(4):1–9. doi:10.3390/diagnostics9040176. [Google Scholar] [PubMed] [CrossRef]
92. Becker AS, Jendele L, Skopek O, Berger N, Ghafoor S, Marcon M, et al. Injecting and removing suspicious features in breast imaging with CycleGAN: a pilot study of automated adversarial attacks using neural networks on small images. Eur J Radiol. 2019;120:108649. doi:10.1016/j.ejrad.2019.108649. [Google Scholar] [PubMed] [CrossRef]
93. Grisoni F, Neuhaus CS, Gabernet G, Müller AT, Hiss JA, Schneider G. Designing anticancer peptides by constructive machine learning. ChemMedChem. 2018;13(13):1300–2. doi:10.1002/cmdc.201800204. [Google Scholar] [PubMed] [CrossRef]
94. Gimadiev TR, Madzhidov TI, Marcou G, Varnek A. Generative topographic mapping approach to modeling and chemical space visualization of human intestinal transporters. Bionanoscience. 2016;6(4):464–72. doi:10.1007/s12668-016-0246-5. [Google Scholar] [CrossRef]
95. Zhao X, Cheung LKW. Kernel-imbedded gaussian processes for disease classification using microarray gene expression data. BMC Bioinform. 2007;8(1):1–26. doi:10.1186/1471-2105-8-67. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools