 Open Access
Open Access
REVIEW
Transformers for Multi-Modal Image Analysis in Healthcare
1 Department of Biomedical Engineering, KMCT College of Engineering for Women, Kerala, 683104, India
2 MCA Department, Federal Institute of Science and Technology, Kerala, 683104, India
3 ECE Department, National Institute of Technology Calicut, Kerala, 683104, India
4 Electrical Engineering Department, College of Engineering, King Khalid University, Abha, 61421, Saudi Arabia
* Corresponding Author: Sameera V Mohd Sagheer. Email:
Computers, Materials & Continua 2025, 84(3), 4259-4297. https://doi.org/10.32604/cmc.2025.063726
Received 22 January 2025; Accepted 16 June 2025; Issue published 30 July 2025
Abstract
Integrating multiple medical imaging techniques, including Magnetic Resonance Imaging (MRI), Computed Tomography, Positron Emission Tomography (PET), and ultrasound, provides a comprehensive view of the patient health status. Each of these methods contributes unique diagnostic insights, enhancing the overall assessment of patient condition. Nevertheless, the amalgamation of data from multiple modalities presents difficulties due to disparities in resolution, data collection methods, and noise levels. While traditional models like Convolutional Neural Networks (CNNs) excel in single-modality tasks, they struggle to handle multi-modal complexities, lacking the capacity to model global relationships. This research presents a novel approach for examining multi-modal medical imagery using a transformer-based system. The framework employs self-attention and cross-attention mechanisms to synchronize and integrate features across various modalities. Additionally, it shows resilience to variations in noise and image quality, making it adaptable for real-time clinical use. To address the computational hurdles linked to transformer models, particularly in real-time clinical applications in resource-constrained environments, several optimization techniques have been integrated to boost scalability and efficiency. Initially, a streamlined transformer architecture was adopted to minimize the computational load while maintaining model effectiveness. Methods such as model pruning, quantization, and knowledge distillation have been applied to reduce the parameter count and enhance the inference speed. Furthermore, efficient attention mechanisms such as linear or sparse attention were employed to alleviate the substantial memory and processing requirements of traditional self-attention operations. For further deployment optimization, researchers have implemented hardware-aware acceleration strategies, including the use of TensorRT and ONNX-based model compression, to ensure efficient execution on edge devices. These optimizations allow the approach to function effectively in real-time clinical settings, ensuring viability even in environments with limited resources. Future research directions include integrating non-imaging data to facilitate personalized treatment and enhancing computational efficiency for implementation in resource-limited environments. This study highlights the transformative potential of transformer models in multi-modal medical imaging, offering improvements in diagnostic accuracy and patient care outcomes.Keywords
Contemporary medical practice relies extensively on diagnostic imaging techniques, which play a crucial role in providing vital insights into the anatomical structures of patients and their disease states. These imaging modalities encompass a range of technologies including Magnetic Resonance Imaging (MRI), Computed Tomography (CT), and Positron Emission Tomography (PET), each offering unique advantages in the diagnostic process. According to Zaidi et al. [1], MRI produces detailed images of soft tissues; CT is particularly effective for examining bone structures and identifying abnormalities; and PET captures metabolic activity for functional imaging. The integration of these complementary methods, referred to as multi-modal imaging, enables a comprehensive diagnostic assessment. This method improves the precision and effectiveness of medical decision making by offering a more comprehensive view of the health status of the patient. The analysis of multi-modal medical images poses numerous obstacles. Variations in resolution, contrast, and acquisition parameters among different modalities complicate direct comparisons. Furthermore, Wenderott et al. [2] highlighted that the immense quantity of information produced in medical environments surpasses the capabilities of the conventional analytical techniques and human interpreters. Artificial Intelligence (AI) has emerged as a promising approach for addressing these issues. By streamlining and enhancing image interpretation, AI can assist in overcoming the constraints of manual analysis, thus enhancing diagnostic accuracy and alleviating the workload of medical professionals [3–5].
In medical imaging, distinct sequences often capture crucial long-distance relationships and semantic contents, as shown in Fig. 1. These sequences are essential for accurately depicting the structural and functional aspects of the human organs. Consistency among organs ensures that medical images maintain an inherent structure, facilitating uniform visual interpretation. Disrupting or modifying these sequences can significantly impair the model performance by hindering the extraction of meaningful patterns. Consequently, maintaining the integrity of these sequences is crucial for achieving reliable and effective results in the analysis of medical images.
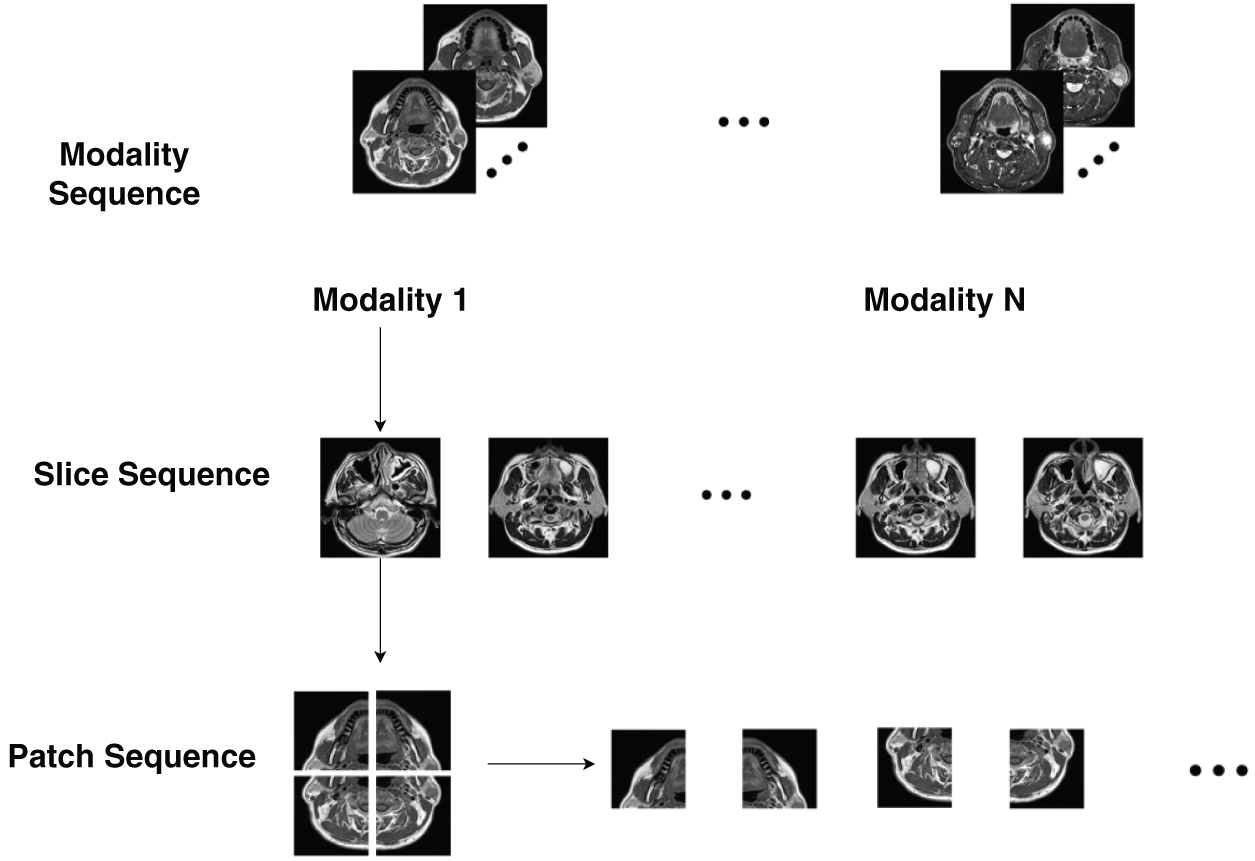
Figure 1: Comparison between natural and multi-modal medical images (adapted from Dai et al. [6])
CNNs have emerged as a crucial element of deep learning in the field of AI-driven medical imaging. These sophisticated networks demonstrate exceptional performance in various tasks, including the classification and segmentation of images, as well as the identification of anomalies. CNNs have become an essential tool in advancing AI applications within the medical imaging domain. Nevertheless, Li et al. [7] point out that CNNs have fundamental shortcomings in capturing long-distance relationships and comprehensive contextual information, which are essential for analyzing images across multiple modalities. The limited receptive field of CNNs, determined by their convolutional kernels, impedes their ability to detect relationships between spatially distant parts of an image. This limitation hampers capacity of CNNs to fully leverage the complementary aspects of multi-modal data. Advancements in interpretable AI, biomedical signal analysis, and biomechanics have contributed to improved medical imaging, gene selection, and interaction recognition [8–12]. The implementation of these denoising methods can enhance the robustness of multimodal medical-image analysis, thereby increasing the applicability of transformers in practical clinical environments [13–17]. Recent progress in areas such as skeleton-based human pose prediction, enhancing the resolution of retinal fundus images, improving the sensitivity of spin-exchange relaxation-free magnetometers, assessing image quality without reference using transformers, and parsing complex electronic medical records has made a substantial impact on the domains of computer vision, medical imaging, and biomedical signal processing [18–22]. Recent studies have explored deep learning-based approaches for biomedical signal processing, including Electrocardiogram (ECG) denoising, ultrasound imaging, dental plaque segmentation, and muscle fatigue detection [23–27].
Initially designed for Natural Language Processing (NLP) tasks, transformers have recently become increasingly popular in the field of computer vision owing to their ability to effectively model global relationships. Unlike CNNs, transformers employ a self-attention mechanism to process all input parts simultaneously, enabling them to capture the complex relationships between various image regions. This characteristic makes transformers particularly well-suited for multi-modal image analysis, where understanding spatial and semantic relationships across multiple imaging modalities is essential [28–31]. Recent progress in transformer-based models has demonstrated their capability to manage noisy data in medical imaging. For example, Naqvi et al. [32] investigated how transformers can improve image quality by minimizing noise, which aligns with our discussion on the resilience of the model to variations in image quality.
Transformers have shown significant potential in the field of medical imaging. Studies have revealed their effectiveness in various applications, including segmenting images, identifying diseases, and pinpointing anomalies. Through the use of self-attention mechanisms, transformers can more effectively learn and merge features from multiple modalities compared to conventional methods. For example, Lai et al. [33] explained tumor segmentation tasks: transformers can concurrently consider anatomical information from MRI and metabolic activity from PET, resulting in more precise tumor boundary delineation. This ability not only improves the precision of diagnoses but also offers crucial information about disease progression and response to treatments. Transformer-based models offer scalability and adaptability, which are vital in healthcare settings. These models can undergo initial training on large-scale datasets and subsequently be refined for particular applications using minimally labeled data, which is a typical situation in medical imaging because of the substantial costs and specialized knowledge required for data labeling. Additionally, Alsaad et al. [34] highlighted that the flexible architecture of transformers facilitates the seamless integration of various data types, including medical histories of the patients, laboratory test results, and genetic information. This adaptability opens up possibilities for developing comprehensive healthcare solutions that are centered around individual patients. The proposed framework for improving the clinical decision support through the integration of multimodal data is presented in Fig. 2. The process is initiated with the collection of information from diverse healthcare facilities, which is then consolidated using multimodal data-fusion techniques. Subsequently, AI modeling was applied to the aggregated data to derive crucial insights encompassing diagnosis, prognosis, risk evaluation, and treatment strategies. These valuable insights are then relayed to healthcare professionals, empowering them to make informed and effective decisions regarding patient care. Recent progress in compensating for magnetic fields in optically pumped magnetometers, guiding hematoma evacuation with imaging, employing multi-wavelength microscopy, classifying speech imagery using Electroencephalogram (EEG), and reconstructing visual stimuli from EEG signals has greatly enhanced biomedical imaging and neurological applications [35–38].
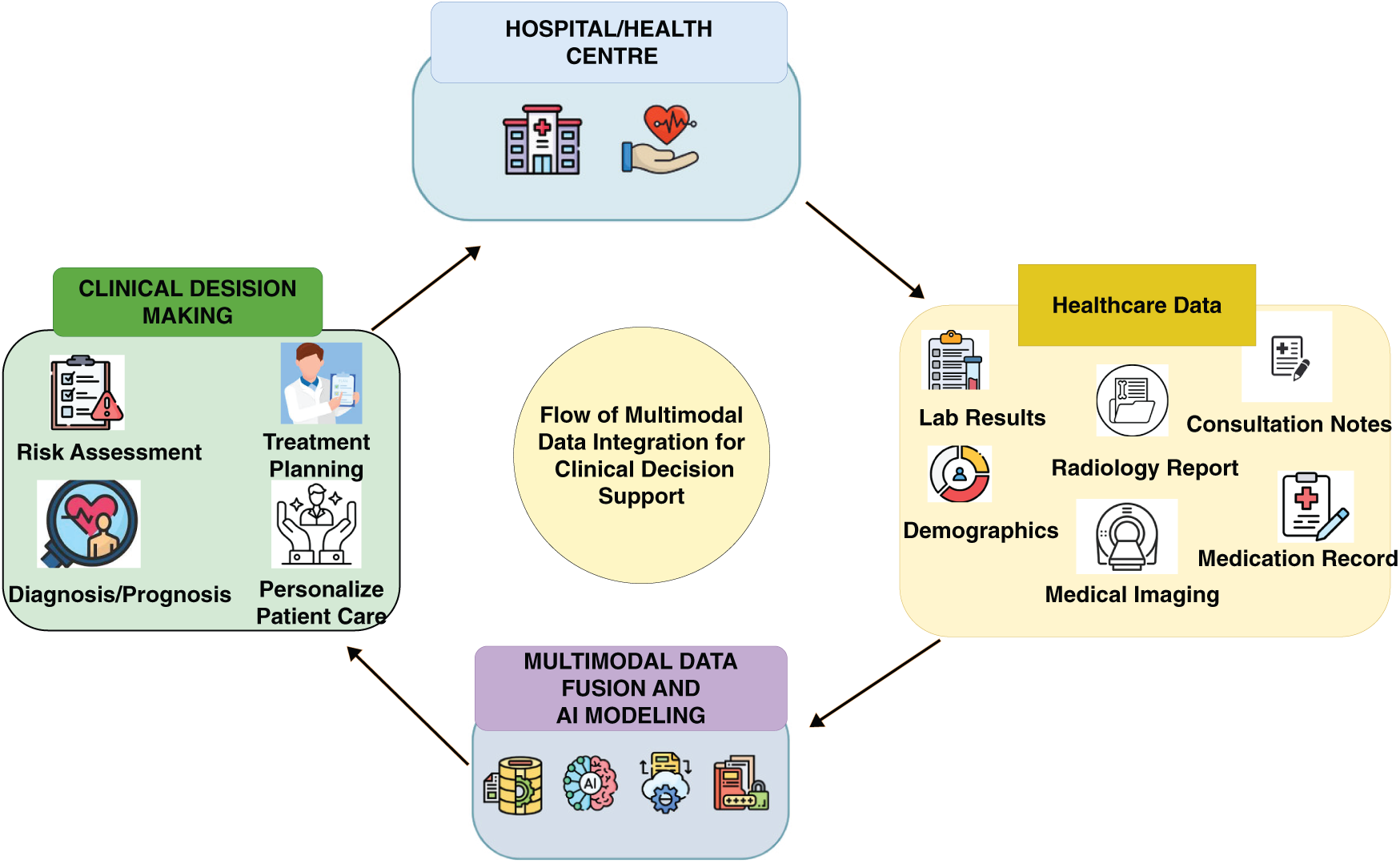
Figure 2: Flow of multimodal data integration for clinical decision support (adapted from Teoh et al. [39])
The interpretability of transformer models is another significant advantage. In clinical environments, it is crucial for AI systems to provide explainable results that clinicians can comprehend and trust. As referenced by Alshehri et al. [40] the attention maps generated by transformers highlight the input areas on which the model focuses, offering transparency in the decision-making processes. This interpretability not only aids in validating model predictions but also fosters confidence among healthcare professionals in adopting AI-assisted diagnostic tools.
Although transformer-based models offer numerous benefits, their implementation in clinical settings presents several hurdles. Transformers face significant constraints owing to their quadratic scaling with the input size, especially when processing high-resolution medical images, which results in considerable computational requirements. Researchers are currently developing more efficient transformer architectures such as Swin Transformers and Vision Transformers (ViTs) to reduce computational costs while maintaining performance levels, as noted by Xu et al. [41]. Furthermore, ensuring that these models can be applied across diverse patient groups and imaging protocols is essential for their widespread adoption in health care. Recent developments, such as the GLoG-CSUnet framework, enhance ViTs by incorporating flexible radiomic features, such as Gabor and Laplacian of Gaussian (LoG) filters. This method boosts the segmentation precision by capturing intricate anatomical details, highlighting the potential of feature-enhanced transformer models in medical image analysis [42–45].
This study investigated the potential of transformer-based models for analyzing multimodal images in healthcare. As mentioned by Dai et al. [6], we offer a thorough examination of their applications, emphasizing their capacity to integrate and examine data from various imaging modalities. It is important to note that this work does not present original experimental contributions but instead provides a comprehensive review and analysis of existing literature on transformer-based models in multimodal medical imaging.
The framework proposed in Fig. 3 utilizes transformer models featuring self-attention to analyze the inputs from various imaging modalities. These include MRI for soft tissue visualization, CT for bone structure examination, and PET for metabolic activity assessment. The ultimate goal of this multimodal approach is to achieve an accurate tumor segmentation.
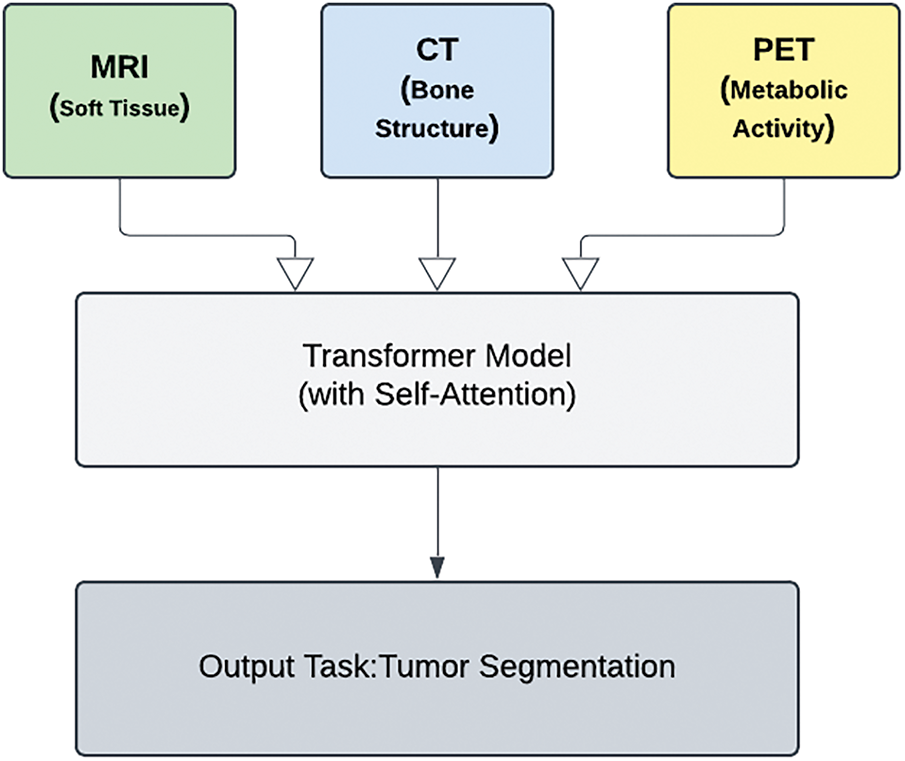
Figure 3: Transformer-based models in multi-modal medical imaging
Transformers represent a major advancement in the field of medical image analysis. By leveraging their distinctive attributes, healthcare systems can attain unparalleled levels of precision and effectiveness in diagnostics, ultimately resulting in improved patient outcomes, as discussed by Li et al. [46]. As this field continues to advance, future research should concentrate on optimizing transformer architectures for medical applications and integrating them with other AI-driven tools to create comprehensive healthcare ecosystems [47–49].
Table 1 presents an overview of various multi-modal medical image fusion techniques, highlighting their objectives and associated limitations.
Table 2 summarizes notable works on medical image segmentation, focusing on different deep learning models and their constraints.
Table 3 outlines key studies on cross-modality representation learning and image registration in medical imaging.
Table 4 provides insights into the use of transformers and attention mechanisms in medical imaging, emphasizing their challenges.
Healthcare has undergone significant changes owing to the incorporation of AI, which has revolutionized many diagnostic and therapeutic processes. Transformer-based models have emerged as particularly noteworthy among AI innovations, owing to their remarkable capacity to process and synthesize intricate, multidimensional data. This section delves into the essential concepts required to comprehend their functions in multimodal medical image analysis. It encompasses the examination of various medical imaging techniques, conventional image analysis methods, the emergence of transformers, and their implementation in multimodal medical imaging contexts [36].
2.1 Medical Imaging Modalities
In contemporary healthcare, medical imaging serves a crucial function by providing an in-depth understanding of the anatomical and functional conditions of the body. Various imaging techniques can be used to capture specific aspects of human physiology.
• MRI (Magnetic Resonance Imaging): MRI generates highly detailed, contrast-rich images of soft tissues that are crucial for detecting and evaluating various health issues, including neoplasms and disorders affecting the nervous system.
• CT (Computed Tomography): High-resolution cross-sectional imagery is particularly useful for identifying bone breaks, malignancies, and disorders affecting the blood vessels.
• PET (Positron Emission Tomography): Visualization of metabolic processes through functional imaging is commonly used in the fields of oncology and neurology.
Although individual modalities are effective on their own, integrating them improves the diagnostic precision by offering complementary data. Nevertheless, this multimodal strategy presents difficulties in merging and interpreting information and requires sophisticated computational techniques. Fig. 4 illustrates that, while individual modalities perform well on their own, combining them creates a multimodal approach that improves diagnostic precision by utilizing complementary information. Nevertheless, this method presents challenges in terms of data integration and interpretation, requiring sophisticated computational techniques.
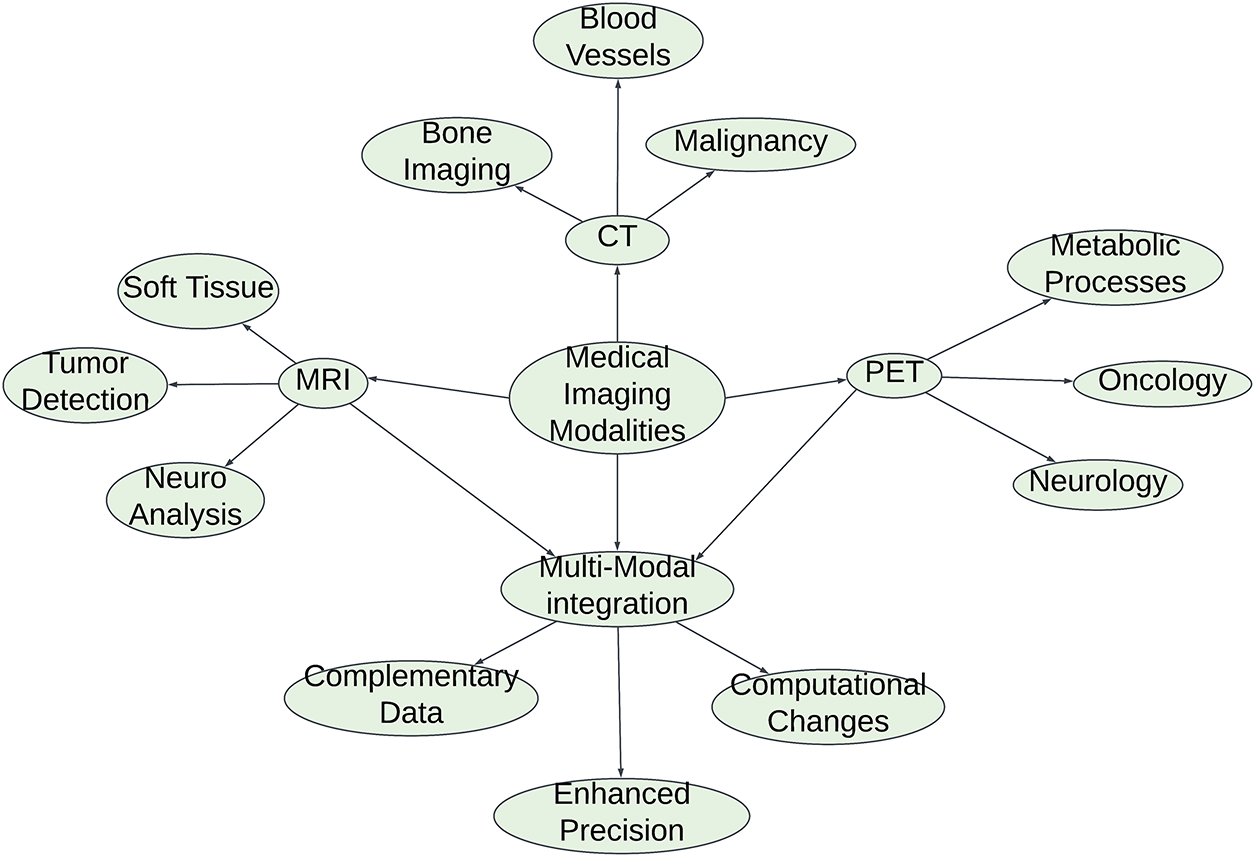
Figure 4: Medical imaging modalities and multi-modal integration
2.2 Traditional Approaches in Medical Image Analysis
Historically, medical image interpretation relied on hand-crafted features and traditional machine learning techniques, and the emergence of CNNs has marked a significant breakthrough, as these systems automated feature extraction processes and exhibited remarkable effectiveness in a range of applications, including:
• Image Classification: Assigning diagnostic labels based on visual patterns.
• Segmentation: Identifying and outlining specific anatomical components or diseased areas.
• Object Detection: Identifying and localizing specific abnormalities.
Despite the impressive achievements of CNNs, they have inherent limitations. The limited scope of their receptive fields hinders their capacity to detect long-distance relationships, which is essential for processing multimodal information. Furthermore, CNNs struggle to effectively combine diverse data types, as they handle each modality independently, thus constraining their effectiveness in tasks that require the comprehensive integration of multiple modalities [70–74].
2.3 Transformers: A Paradigm Shift
Originally designed for NLP, Transformers have brought about a significant shift in the field of AI by addressing the shortcomings of traditional models. Unlike CNNs, transformers utilize self-attention mechanisms to examine global connections throughout entire datasets. This ability allows them to recognize intricate relationships among diverse components of input data, making them exceptionally suitable for tasks demanding a thorough grasp of information. In the realm of computer vision, transformer models such as ViTs and Swin Transformers have demonstrated exceptional performance for
• Image Classification: Competing with or surpassing CNNs in accuracy.
• Object Detection and Segmentation: Providing enhanced precision by leveraging global context.
• Anomaly Detection: Identifying subtle, context-dependent irregularities.
These advancements have created new opportunities in the field of medical imaging, where it is essential to comprehend the spatial and semantic connections across various modalities [75–77].
2.4 Transformers in Multi-Modal Medical Imaging
The intricate nature of multimodal medical imaging data necessitates the development of models that can synthesize various types of information. In this field, transformers have shown exceptional performance by utilizing self-attention mechanisms to capture cross-modal relationships efficiently. This approach has resulted in notable progress in several areas including:
• Tumor Segmentation: Combining anatomical data from MRI scans with functional information from PET imaging to accurately determine tumor boundaries.
• Disease Detection: Improving diagnostic accuracy, especially for intricate disorders such as neurodegenerative conditions, by incorporating diverse types of input data.
• Anomaly Localization: Identifying abnormalities by integrating information across modalities.
Advanced models such as TransUNet and MedT, which integrate transformer-based architectures, have demonstrated remarkable performance in medical image segmentation. These advanced systems leverage self-attention mechanisms to extract multiscale contextual information, leading to more accurate and robust image analysis. Moreover, transformer models can be trained on large-scale datasets and then fine-tuned for specific applications using small amounts of labeled data, thereby addressing a common challenge in medical imaging. The modular design of these systems enables smooth integration of additional patient data, including electronic health records (EHRs), lab results, and genetic information. This integration establishes the foundation for personalized medical approaches [78].
2.5 Challenges and Future Directions
Despite their potential, transformer-based models face challenges in clinical deployment:
• High Computational Complexity: Processing high-resolution medical images using transformers requires a substantial amount of computational power and resources.
• Generalizability: To ensure broad adoption, it is essential to verify that the model functions effectively in diverse patient populations and across various imaging modalities.
Future studies will address these obstacles by streamlining transformer designs for greater efficiency and improving their ability to adapt to various clinical environments. The combination of transformers with other AI technologies is expected to result in holistic, patient-focused healthcare systems [79].
This literature review offers a comprehensive analysis of ongoing studies and methodologies pertaining to the application of transformer-based models in multimodal image analysis within healthcare. By exploring the current landscape, this section sheds light on the existing knowledge deficits, challenges, and prospective developments in the field. To methodically explore fundamental works, methodologies, comparative studies, and emerging trends, this review is structured into separate subsections [38].
This section discusses the approaches utilized to implement transformer-based models in multimodal medical image analysis. We aimed to provide a comprehensive analysis of the techniques, procedures, and key architectural decisions involved in developing and implementing transformer-based systems for medical image analysis. Our goal is to present a detailed exploration of the fundamental elements and factors to be considered when building these sophisticated analytical frameworks for healthcare imaging [80–83].
3.1.1 Transformer Architectures for Medical Image Analysis
In medical imaging, transformers demonstrate exceptional performance by capturing intricate patterns across different modalities and modeling global relationships. Their self-attention mechanisms allow for accurate segmentation and classification, thus outperforming conventional models. The following sections examine the influence of key architectures, including ViTs, swine transformers, and hybrid models, on the field of medical imaging.
ViTs (ViTs)
ViTs are one of the most significant architectures in image analysis, showcasing their capability in both classifying and segmenting medical images, which involves splitting images into patches and processing them using multihead self-attention mechanisms. This holistic method enables ViTs to recognize far-reaching connections, which is crucial for analyzing the complex patterns present in medical imagery. Li et al. [84] employed a dual-stream Vision Transformer to analyze gait using only a single, affordable RGB camera, highlighting the capability of transformers to derive medical insights from limited data. ViTs have demonstrated superior capabilities in certain image analysis tasks compared to CNNs, primarily due to their capacity to model relationships between distant pixels [66].
Swin Transformers (Swin-T)
Swin Transformers, developed as a model for extracting hierarchical features, excel in processing high-resolution imagery. In contrast to conventional ViTs, which use a fixed patch size for image analysis, Swin-T employs self-attention mechanisms based on windows to control the computational complexity. This design has shown remarkable effectiveness in medical imaging applications, such as segmenting tumors and classifying diseases, owing to its ability to process large-scale, high-resolution medical images efficiently [58]. Zhang et al. [85] introduced an innovative method to address the difficulties of segmenting brain tumors when MRI modalities are unavailable. Achieving precise segmentation of brain tumors using MRI is crucial for integrated analysis of multimodal images. Nonetheless, in clinical settings, obtaining a full set of MRIs is not always feasible, leading to significant performance drops in current multimodal segmentation techniques owing to missing modalities. In this study, the authors introduced the first approach to leverage the transformer for multimodal brain tumor segmentation, which remains effective regardless of the combination of available modalities. Specifically, the authors proposed a new multimodal Medical Transformer (mmFormer) designed for learning from incomplete multimodal data, featuring three key components: hybrid modality-specific encoders that connect a convolutional encoder with an intra-modal transformer to capture both local and global contexts within each modality; an intermodal transformer that establishes and aligns long-range correlations across modalities to derive modality-invariant features with global semantics related to the tumor region; and a decoder that progressively upsamples and merges these modality-invariant features to produce reliable segmentation. Additionally, auxiliary regularizers are incorporated into both the encoder and decoder to bolster the resilience of the model to missing modalities. The authors performed comprehensive experiments using the public BraTS 2018 dataset for brain tumor segmentation. The findings reveal that the proposed mmFormer surpasses the leading methods for incomplete multimodal brain tumor segmentation across nearly all subsets of missing modalities [85–87].
Hybrid Models Combining CNNs and Transformers
The integration of CNNs and transformers offers a synergistic approach: CNNs excel at efficient local feature extraction, whereas transformers excel at handling global dependence. This combined architecture has proven particularly valuable in multimodal medical image analysis, where integrating various data types, such as MRI, CT, and PET scans, is crucial. By merging these two architectural styles, significant enhancements were achieved in both segmentation and classification tasks [69].
3.1.2 Pre-Processing Techniques for Multi-Modal Medical Images
Multimodal medical imaging is based on a combination of diverse diagnostic imaging methods, including MRI, CT, and PET scans. Preparing the data through preprocessing is crucial for ensuring proper alignment, standardization, and overall quality improvement. This step addresses various issues, including misalignment, variations in intensity, and scarcity of labeled data, ultimately leading to enhanced model performance and improved diagnostic precision. Critical preprocessing techniques involve image registration, normalization, and data augmentation, which prepare the data for subsequent tasks, such as segmentation and feature extraction.
Image Registration
Image registration is a crucial preliminary step in multimodal medical image analysis. This method synchronizes different medical imaging modalities, including MRI, CT, and PET scans, with a common reference frame. By doing so, it ensures that anatomical structures identified in each modality correspond accurately, facilitating effective data integration. Various registration techniques, such as rigid and non-rigid methods, are employed based on the types of images and the level of precision required. The image registration process contributes to an improved model performance in multimodal fusion tasks by providing consistently aligned data for analysis. Accurate image alignment enables more precise interpretations from the combined data, which is vital for the efficacy of medical diagnostic tools. The alignment depicted in Fig. 5 guarantees that anatomical structures observed across different modalities correspond accurately, facilitating smooth integration of data [64].

Figure 5: Medical imaging modalities and multi-modal integration
Normalization and Standardization
Different modalities in multimodal medical imaging often have distinct intensity distributions, which create challenges when integrating data from various sources. To overcome this obstacle, normalization methods are frequently utilized to harmonize the pixel intensity across different modalities. For instance, MRI scans, which typically display lower intensity values than PET images, require normalization to ensure that the features from diverse imaging techniques are comparable. The standardization procedure is not just a technical necessity but also a vital element in ensuring that subsequent steps in feature extraction function efficiently and precisely across different modalities. It aids in creating consistent and comparable data for the model to learn from, thereby enabling the development of robust and high-performance models [63,88,89].
Data Augmentation for Medical Imaging
Data augmentation has become an essential strategy for overcoming challenges associated with the scarcity of labeled medical imagery. In healthcare, the shortage of labeled data remains a significant hurdle because acquiring a substantial amount of high-quality annotated images is often costly and time intensive. Various data augmentation techniques, including rotation, flipping, and scaling, have been employed to artificially increase the size of datasets. This expansion improves the model’s ability to effectively generalize. Through data augmentation, the models become more resilient to different image transformations and exhibit improved performance on unseen data. These augmentation techniques are specifically adapted for medical image datasets to ensure diverse representations of the anatomical structures and abnormalities. This modification is crucial for enhancing the resilience of the model and its ability to apply knowledge to diverse real-world situations [90]. The transformer-based framework for multimodal medical imaging was trained through a two-step process: pre-training followed by fine-tuning. Initially, during the pretraining stage, the model was exposed to a large-scale medical imaging dataset to acquire generalizable feature representations across various imaging modalities. This phase ensures that the transformer adeptly captures both the modality-specific and cross-modal relationships. In the fine-tuning stage, a diverse, task-specific dataset is employed to tailor the model for the intended medical imaging application. This dataset encompasses a broad spectrum of cases with different imaging conditions, anatomical structures, and pathological manifestations, thereby ensuring the robustness and generalizability of the model. Furthermore, domain-specific augmentation techniques and optimization strategies to boost model performance while mitigating overfitting. Additional details regarding the dataset size, diversity, and pre-training configurations can be found in the Methodology section.
3.1.3 Fusion Strategies for Multi-Modal Data
Data fusion techniques merge information from various sources to improve the model outcomes. One approach, known as early fusion, incorporates features from different data types during the initial processing stage, thus enabling the model to examine complementary information. Although this method is powerful for intricate tasks, it requires meticulous alignment and standardization. Another technique, late fusion, aggregates results from independent networks trained on single data types, providing ease of implementation and adaptability but potentially overlooking interactions between different modalities. To improve the interpretability of the transformer-based model for clinical use, attention-based visualization techniques to examine the decision-making process. specifically used attention heatmaps and Grad-CAM-like methods tailored for transformer architectures to pinpoint the most significant areas in the input data during predictions. These visual tools enable us to evaluate how the model focuses on various modalities and anatomical structures, thereby offering insights into its reasoning. By utilizing these techniques, it is ensured that the prediction of the model met clinical expectations, thereby enhancing transparency and trust among healthcare professionals. Furthermore, attention distributions across different layers to comprehend how interactions specific to each modality and cross-modal interactions contribute to the final decision. These visual explanations are essential to confirm the reliability of the model in practical medical applications. A detailed discussion of the interpretability and visualization outcomes is included in the Results section. Early fusion, a technique illustrated in Fig. 6, combines features from diverse data types during the initial processing phase. This approach enables the model to examine complementary data from the beginning.
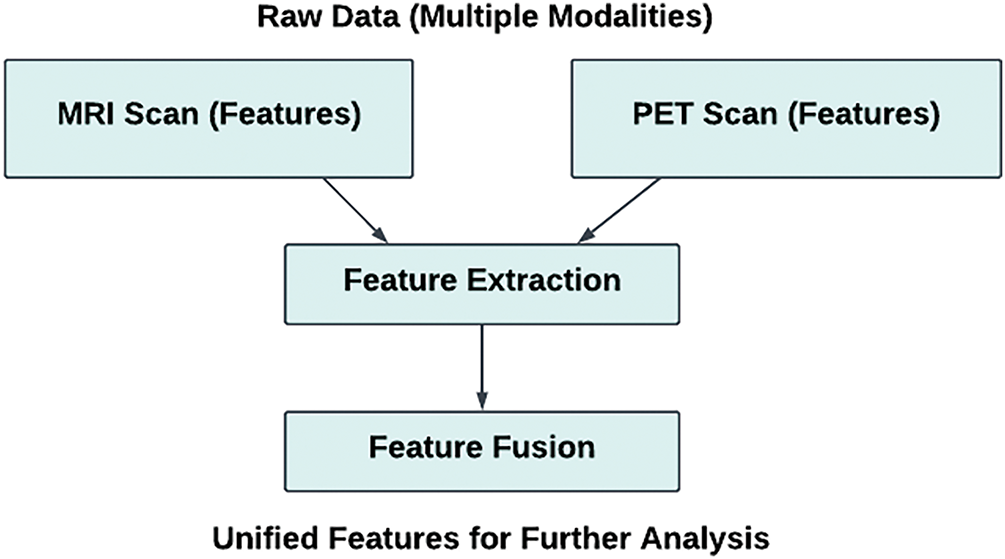
Figure 6: Feature-level fusion of MRI and PET scan data
Early Fusion (Feature-Level Fusion)
Early fusion, also known as feature-level fusion, combines features extracted from multiple modalities during the initial phases of processing before the network analyzes them. This method enables the model to concurrently evaluate the complementary attributes from multiple sources. Although this approach offers benefits, it also presents challenges. Sophisticated techniques for alignment and normalization are necessary to address issues stemming from modality-specific variations such as disparities in image quality, intensity, or spatial alignment. Nevertheless, early fusion remains a viable option when it’s crucial to incorporate diverse information from multiple modalities from the outset. For intricate challenges such as multi-modal segmentation or multi-class classification, this approach proves especially valuable. Integrating various data sources enables a more thorough understanding of the medical conditions being examined [51,73,91].
Late Fusion (Decision-Level Fusion)
In contrast to early fusion, late fusion combines the results or outputs from separate networks, each of which has been trained on different modalities that are processed through their own networks, with the results combined at the final stage using methods such as weighted averaging or majority voting. This approach, which is simpler and potentially advantageous when aligning modalities is difficult, has the drawback of not fully exploiting intermodal interactions during feature extraction. The simplicity of late fusion makes it a viable option when modal alignment is particularly challenging or when computational resources are constrained. However, learning complex relationships between modalities may not be optimal [52].
3.1.4 Self-Attention and Cross-Attention Mechanisms in Transformers
The self-attention mechanism in transformer models allows the system to focus on different parts of an image, effectively capturing long-range dependencies that are crucial for applications, such as tumor detection. Cross-attention, when analyzing data from multiple sources, enables the model to link relevant information across various modalities by integrating MRI and PET scan data. This combination improves the accuracy and effectiveness of the model in tasks such as defining tumor margins. Abidin et al. [92] underscored the significance of integrating various MRI modalities to enhance brain tumor segmentation. By combining different MRI sequences, a more comprehensive and precise depiction of tumors and adjacent brain structures can be achieved, which is vital for effective segmentation. Multi-modal MRI allows researchers to assess the efficacy of different segmentation algorithms and compare their outcomes. These comparative analyses have fostered the creation of new techniques, ultimately improving the precision of brain tumor segmentation. The Brain Tumor Segmentation (BraTS) Challenge dataset is a crucial benchmark for evaluating segmentation performance. This dataset comprises multiple MRI modalities, including T1, T2, T1ce, and FLAIR, along with meticulously annotated tumor-segmentation masks. It remains a vital resource for researchers and clinicians involved in glioma segmentation and brain tumor diagnosis.
Self-Attention in ViTs
Transformer models are characterized by their key component, namely self-attention. This capability enables the model to focus on different parts of an input image or sequence, regardless of their spatial connections, making it particularly useful for analyzing complex medical imagery. Self-attention facilitates the model’s ability to discern connections between distant image regions, thereby capturing essential long-range dependencies that are crucial for the precise segmentation or classification of complex structures in medical data. This capability is especially advantageous for tasks such as tumor identification, which requires comprehension of the interrelationships among diverse anatomical areas. The self-attention mechanism allows transformers to efficiently process these relationships, enabling a more precise and dependable analysis of medical imagery [68,75,93].
The attention mechanism between different input vectors is computed as follows [6]:
where
Cross-Attention for Multi-Modal Data
Cross-attention is a vital function of multimodal image analysis. In a multimodal framework, the model must grasp the interrelationships between different modalities, such as MRI and PET, which provide complementary information. Transformer models utilize cross-attention mechanisms to concentrate on the pertinent aspects of one modality while processing the other. This enables improved data integration because the model can correlate structural elements from one modality with functional or metabolic information from another. For example, in tumor segmentation, cross-attention enables the model to merge structural details from MRI with metabolic activity patterns from PET scans, resulting in more precise segmentation and diagnosis. A key advantage of transformers in multimodal medical image analysis is their capacity to capture and align relationships efficiently across different modalities [65].
3.1.5 Challenges and Future Directions in Transformer-Based Models for Healthcare
In the healthcare sector, transformer models encounter several obstacles, including intensive computational requirements, medical data inconsistencies, and limited applicability in various clinical environments. Ongoing studies aim to enhance these models by focusing on three key areas: boosting operational efficiency, strengthening resilience against data irregularities, and improving versatility. To overcome these obstacles and enhance the capabilities of transformer models in medical contexts, scientists are utilizing techniques like data augmentation and transfer learning.
Computational Efficiency and Scalability
The implementation of transformer models for high-resolution medical imaging presents significant computational hurdles. These models typically require extensive memory and processing capabilities, especially when processing large 3D medical datasets, which can be computationally demanding. Ongoing research is aimed at enhancing the computational efficiency of transformers. Strategies such as model pruning, distillation, and hybrid architectures are under investigation to decrease the parameter count while preserving high performance, while streamlining transformer architectures for deployment on hardware with limited resources, such as edge computing systems and mobile devices. This requires careful balancing of the model size with inference speed. These advancements are essential for enabling the implementation of transformer-based models in real-world healthcare settings, where computational resources may be constrained [38,67,79].
Data Quality and Noise Robustness
Noise and artifacts frequently degrade medical images, potentially hampering the effectiveness of the models trained on such data. To ensure practical applications in healthcare settings, transformer models must be able to withstand these imperfections. Current studies have explored strategies, such as adversarial training and noise reduction, to enhance model resilience when faced with noisy data. Researchers have strived to develop more dependable and stable solutions for real-world clinical applications by improving the model resistance to common imaging flaws. The widespread adoption of transformer models largely depends on their ability to handle noise because actual medical data often contain imperfections that must be considered during model development and implementation [94].
Generalizability Across Clinical Scenarios
A key issue is ensuring that transformer models can function effectively in various clinical settings. Bias in medical image datasets stemming from factors such as patient demographics and imaging techniques poses a significant challenge. Overcoming these obstacles is vital for models to perform consistently in different healthcare environments, and scientists are exploring methods such as data augmentation, domain adaptation, and transfer learning to mitigate biases in AI models. These efforts aimed to improve the flexibility of transformer models, allowing them to be used in a variety of clinical settings. This would enable the wider adoption and increase the impact of AI in the medical field. The success of AI systems in diverse real-world healthcare environments depends on their ability to effectively generalize [95–97].
The application of transformer-based models for multimodal medical image analysis has gained increasing attention in recent years. This section examines the primary insights from the existing research, emphasizing the efficacy, obstacles, and potential opportunities associated with incorporating transformers into healthcare applications [98].
3.2.1 Advancements in Medical Image Classification and Segmentation Using Transformers
ViTs have demonstrated exceptional performance in the realm of medical image analysis, particularly for tasks such as classification and segmentation. These capabilities are essential for interpreting various types of medical-data formats. Early studies have underscored the effectiveness of ViTs in detecting illnesses and outlining anatomical structures. Studies have shown that ViTs can effectively capture long-range pixel relationships by breaking down images into smaller segments and utilizing multihead self-attention mechanisms. The ability to recognize connections between distant elements is particularly advantageous in the field of medical imaging, where understanding the interrelations of spatially separated components is crucial [59].
Studies have shown that ViTs demonstrate superior performance compared with conventional CNNs in certain medical imaging tasks. This advantage is particularly evident in areas such as tumor segmentation and organ detection, especially when processing intricate images such as those obtained from MRI and CT scans. Unlike CNNs, which rely on localized receptive fields, the global attention mechanism employed by transformers enables them to detect intricate patterns across various image sections, which is a crucial feature in medical image analysis. Furthermore, numerous studies have shown that ViTs trained on large datasets demonstrate enhanced generalization capabilities when encountering unfamiliar images, highlighting their adaptability to different medical imaging modalities. In the field of medical image analysis, ViTs demonstrate a notable advantage owing to their ability to recognize intricate patterns across various parts of an image [66,79,99].
3.2.2 The Role of Swin Transformers in Handling High-Resolution Medical Images
Current studies have demonstrated the remarkable capabilities of swine transformers (Swin-T) in medical image analysis, especially for high-resolution applications. The hierarchical attention mechanism is a key strength of the Swin-T. Unlike ViTs, which use static patch sizes, Swin-T employs a window-based self-attention method that dynamically adjusts patch sizes, as shown in Fig. 7. This enables Swin-T to process high-resolution images, such as those from MRI and CT scans, more effectively. The hierarchical structure of Swin-T allows it to efficiently manage the computational requirements of high-resolution data while preserving essential spatial information [60].

Figure 7: Flowchart: Swin-T vs. ViTs in medical image analysis
Studies have shown that Swin-T outperforms traditional models across a range of medical imaging applications such as identifying lesions, outlining tumors, and categorizing diseases. The ability of the model to process high-resolution images efficiently makes it particularly well suited for analyzing medical images in real time. This capability is crucial in clinical settings where both speed and precision are vital factors to consider. Hussain et al. [100] advanced the segmentation of biomedical images by incorporating DenseNet-based attention mechanisms that focus on spatial and semantic channel guidance. This method enhances the extraction of features and the precision of segmentation, overcoming the shortcomings of conventional encoder-decoder models. Utilizing such sophisticated architectures can boost the model performance in medical imaging applications.
3.2.3 Hybrid Models Combining CNNs and Transformers for Enhanced Performance
Recent studies have highlighted a growing trend in merging CNNs and transformers. This combination leverages the advantages of both structural designs: CNNs are adept at identifying localized patterns, while transformer models are particularly effective in recognizing broader contextual relationships. Research has shown that these hybrid models can surpass the performance of individual CNNs or transformers when analyzing multimodal medical images.
Combining CNNs with transformers has shown remarkable success in applications that require the synthesis of information from multiple modalities, particularly when merging data from diverse medical imaging sources, such as MRI, CT, and PET scans. In this combined approach, CNNs efficiently extract localized features, whereas transformers process broader connections across different modalities. This strategy has led to notable advancements in applications such as multi-modal tumor segmentation and disease classification, where effectively combining complementary information from diverse modalities is crucial. The combined structural approach offers a promising avenue for improving both the accuracy and robustness of techniques used in medical image analysis [101–103].
3.2.4 Multi-Modal Data Fusion Strategies for Improved Diagnostic Accuracy
The successful integration of multimodal medical images plays a crucial role in improving diagnostic precision. Several strategies have been identified for incorporating multimodal data into transformer-based models, such as early, late, and intermediate fusion techniques. These methodologies are essential for effectively merging the information from diverse medical imaging modalities [55].
Early Fusion (Feature-Level Fusion)
Studies have demonstrated that early fusion–a technique for combining features from diverse modalities in the initial phase–enables models to develop integrated representations of complementary characteristics across various data types. This method is especially advantageous when working with heterogeneous data sources such as MRI and PET scans. Nevertheless, meticulous preprocessing is required to address the issues related to modality misalignment and intensity normalization. Although early fusion can be computationally intensive, it has been proven to improve model performance by facilitating acquisition of more comprehensive and complex representations of medical data [50].
Late Fusion (Decision-Level Fusion)
Late fusion is a widely used approach in multimodal medical image analysis, particularly when aligning the different modalities is difficult. This method involves processing each modality independently using separate neural networks before merging their final outputs. Current studies indicate that late fusion is particularly advantageous in situations with constrained computational resources, or when modality alignment is not feasible. However, this technique may not fully exploit the potential of the intermodal connections. Despite its simplicity, late fusion has shown efficacy in certain applications such as disease identification and classification, where the interplay between various modalities is less important [54].
Intermediate Fusion (Layer-Level Fusion)
Recently, a more sophisticated approach, called intermediate fusion, has gained prominence. This technique involves integrating features from diverse modalities within the middle layers of the neural networks. Models based on transformers employing self-attention mechanisms are particularly adept at implementing this fusion method. Studies have shown that intermediate fusion effectively captures the interrelationships between various modalities across different levels of abstraction, resulting in improved performance in classification and segmentation tasks. This method achieves a middle ground between the computational advantages of late fusion and the extensive feature representations of early fusion, offering a promising technique for examining multi-modal medical imagery [53,55,104].
3.2.5 Advancements in Self-Attention and Cross-Attention Mechanisms
Two key mechanisms are employed in multimodal image analysis using transformer models: self-attention and cross-attention. These methods allow the model to prioritize the key aspects of the input information, either within a single mode (self-attention) or between different modes (cross-attention). Using these techniques, the model can effectively prioritize and process important information from the input [105].
Self-Attention
Self-attention mechanisms in transformers enable the recognition and processing of extensive connections within medical images, which is crucial for the accurate segmentation and classification of anatomical components. Research has shown that this self-attention capability enhances transformers’ capacity to examine complex medical imagery, where relationships between distant regions are important. This characteristic makes transformers particularly adept at analyzing noncontiguous structures, such as detecting tumors or identifying anatomical features that span large portions of an image [106].
Cross-Attention
Cross-attention mechanisms have been demonstrated to be remarkably effective in multimodal frameworks. The transformer’s capabilities allow it to prioritize the crucial elements of one imaging modality while analyzing another, facilitating the integration of complementary data from various imaging techniques. For example, this approach can merge structural details from MRI with metabolic information from PET scans. The implementation of cross-attention has notably enhanced the precision of multimodal tasks such as tumor segmentation. In this scenario, various modalities provide essential but distinct insights into tumor size, location, and metabolic activity [107].
3.2.6 Challenges in Computational Efficiency and Scalability
Scientists have encountered notable obstacles when attempting to apply transformer-based models to high-resolution medical images, primarily because of the issues related to computational complexity and scalability. Despite their proven effectiveness, these models require substantial computing resources, particularly for processing three-dimensional medical imaging data. As illustrated in Fig. 8, current research efforts are focused on enhancing transformer efficiency using various methods, including model pruning, distillation, and the creation of hybrid architectures. The goal of these methods is to reduce the number of parameters and memory demands, thereby enhancing the efficiency and practical use of these models in medical imaging applications [7,108–110].

Figure 8: Challenges and strategies for improving Transformer models in medical imaging
Scientists are striving to improve transformer models for deployment in clinical settings, where real-time data analysis is essential. Studies have also explored the use of edge computing and mobile technologies to enable the deployment of transformer-based models in resource-constrained environments. Addressing these challenges is crucial to ensuring that transformer models are feasible and can be expanded for broad implementation across the healthcare industry.
3.2.7 Data Quality, Noise, and Robustness
Challenges related to data quality and noise continue to pose significant hurdles in medical image analysis. Current research has focused on enhancing the resilience of transformer models when confronted with defective or noisy datasets. Imperfections in medical images, such as noise generated by scanners and artifacts caused by motion, are common and can negatively impact the effectiveness of the algorithms used for image analysis. Research has demonstrated that transformer models are particularly vulnerable to these data imperfections, particularly when implemented in real-world clinical settings [111].
Researchers have explored multiple approaches to address these issues, such as adversarial training, noise reduction methods, and robust loss functions, with the goal of improving the ability of transformer models to handle noisy data. The objective is to improve the durability and dependability of these systems when implemented in healthcare settings where incomplete or distorted data are prevalent. Enhancing the robustness of transformers is a critical step toward their successful implementation in practical healthcare settings [75,112,113].
Evaluating the relative strengths of transformer-based models in multimodal medical image analysis has emerged as a crucial component in determining their efficacy and constraints compared with conventional deep learning techniques, particularly CNNs. This section provides a comparative evaluation of transformer architectures, CNNs-based methods, and hybrid solutions, focusing on their effectiveness in critical medical image analysis tasks including classification, segmentation, and multimodal data fusion. In addition, we investigate the trade-offs between these approaches in terms of computational performance, robustness, and versatility across various scenarios [114].
3.3.1 Transformer Models vs. CNNs in Medical Image Analysis
Transformers excel at managing tasks that require extensive context and long-range connections, whereas CNNs are more effective in handling localized features and processing smaller images. Although Transformers enhance segmentation capabilities, they require additional computational resources.
Performance Comparison in Image Classification
Transformers have demonstrated significant improvements over traditional CNNs in various image-classification tasks, particularly in situations requiring extensive context and long-range dependencies. CNNs have been widely adopted for medical image analysis, including applications in tumor detection, organ segmentation, and disease classification. However, these architectures have limitations. Although CNNs are adept at extracting localized features through convolution operations, they face challenges in capturing comprehensive relationships between distant image regions. This capability is particularly crucial in medical imaging, in which spatial correlations among structures can extend across substantial portions of an image. By contrast, transformers have shown promise in addressing these limitations and providing a more holistic approach to image analysis in medical applications [115]. Utilizing advanced frameworks such as Hussain & Shouno [116] can significantly improve the performance of medical image segmentation tasks. MAGRes-UNet introduces a multi-attention-gated residual U-net structure that incorporates multi-attention-gated residual blocks using activation functions such as Mish and ReLU, along with optimization techniques such as AdamW and Adam. This architecture overcomes the limitations of traditional encoder-decoder networks by effectively merging information from feature maps and capturing fine-scale contextual details, thus enhancing segmentation precision. The statistical significance analysis verifies that these improvements are not merely due to random fluctuations, but are a result of the superior capability of the model to capture global dependencies and multimodal relationships. A detailed breakdown of the performance metrics and significance tests is provided in the results section.
Conversely, ViTs operate by dividing images into patches and utilizing multihead self-attention mechanisms, allowing them to detect global connections across the entire image. Research comparing ViTs and CNNs has demonstrated that ViTs frequently surpass CNNs in tasks that require the recognition of patterns across distant areas, such as identifying large tumors or categorizing diseases with intricate patterns. Despite advancements in other techniques, CNNs remain more computationally efficient, particularly when processing images of smaller size or lower resolution, where the overall context is less important [57]. Transformer-based models have found successful applications beyond medical image analysis, particularly in various vision-based healthcare-monitoring systems. A significant example is their use in analyzing electrocardiograms (ECGs) to detect heart disease. In the research [117], scientists employed Vision Transformer architectures such as Google-ViT, Microsoft-Beit, and Swin-Tiny to classify ECG images. These models achieved impressive classification outcomes, underscoring the potential of transformers in interpreting ECG data to diagnose heart conditions [118,119].
Integrating Transformer models into vision-based healthcare monitoring systems provides notable benefits, including the ability to capture long-range dependencies and model global relationships within the visual data. These features can enhance the diagnostic precision and streamline monitoring processes across a range of healthcare applications.
Segmentation Tasks: Transformers vs. CNNs
In the field of image segmentation, where accurate identification of anatomical features or abnormalities is crucial, both transformers and CNNs exhibit distinct advantages. In the field of medical image segmentation, techniques based on CNNs, especially Fully Convolutional Networks (FCNs) and U-Net, have shown exceptional performance and effectiveness. These approaches excel in tasks such as delineating brain tumors, outlining organs, and lesion detection. CNNs excel in their capacity to effectively detect and process local patterns and formations, making them ideally suited for intricate segmentation tasks that demand precise boundary identification [56].
By contrast, transformers have become increasingly popular for segmentation tasks because of their capacity to model relationships across long distances. Vision Transformer and Swin Transformers have demonstrated enhanced segmentation accuracy by capturing contextual information from the remote areas of an image. This capability is particularly beneficial when segmenting intricate structures, such as the brain or organs, where transformers can identify spatial connections that are challenging for CNNs to detect, especially in cases involving large or irregularly shaped structures. Nevertheless, the computational demands of transformers, particularly ViTs, can be substantial when processing high-resolution images, potentially limiting their practical applications [62]. The Adversarial Vision Transformer framework boosts the segmentation of medical images by combining adversarial training with the transformers. This method enhances segmentation precision, particularly when annotations are scarce, by improving feature learning and strengthening the generalization [120–122].
3.3.2 Hybrid CNNs-Transformer Models
Architectures combining CNNs and transformers take advantage of the strengths of both components. These hybrid models utilize CNNs for their proficiency in detecting local patterns while simultaneously harnessing strength of the transformers in capturing broader contextual information. These hybrid architectures demonstrate exceptional performance in applications, such as multimodal tumor segmentation and disease classification. However, their high computational demands and complex training procedures necessitate careful optimization strategies.
Combining CNNs and Transformers for Enhanced Performance
Architectures that combine CNNs and transformers seek to leverage the strengths of both approaches. CNNs excel at identifying local patterns, whereas transformers are particularly skilled at recognizing long-distance connections within the data. Architectures such as the CNNs-Transformer and U-Net-Transformer strive to merge proficiency of the CNNs in local feature detection with the transformer’s ability to comprehend the global context. These hybrid approaches seek to combine the strengths of both neural network types to enhance overall performance [61].
Studies have demonstrated that combining CNNs and transformers into hybrid models can yield superior results compared with using either approach alone for certain medical imaging tasks. This involves examining multimodal medical imagery, which combines different imaging methods such as MRI, CT, and PET scans. In these scenarios, hybrid models excel by employing CNNs to efficiently extract localized features from each imaging modality, while simultaneously using transformers to capture global relationships across different modalities. This approach has proven particularly effective in applications such as multimodal tumor segmentation and disease classification, in which both localized details and broader contextual information play crucial roles [123].
Nevertheless, combining CNNs and transformers in hybrid models can be resource-intensive, necessitating careful network design to optimize the performance of both components. Moreover, integrating these distinct architectural approaches may present difficulties in model training and convergence, which presupposes that Fig. 9 demonstrates or exemplifies a hybrid CNNs-Transformer model or its implementation. If the image pertains to something else, the citation can be modified as needed.

Figure 9: Hybrid CNNs-Transformer model
3.3.3 Comparing Multi-Modal Data Fusion Techniques
Feature integration in multimodal networks can occur at various levels. One approach, known as early fusion, merges the features from various modalities at the input stage of the network. This method offers a comprehensive representation but requires advanced techniques for effectively aligning different modalities. Conversely, late fusion merges the outputs of separate networks, thereby offering computational simplicity at the cost of reduced precision. Striking a balance between these methods, intermediate fusion blends data at the middle layers, effectively capturing intermodal relationships while mitigating the drawbacks of both early and late fusion strategies.
Early Fusion vs. Late Fusion
The analysis of multimodal medical images often requires combining data from diverse imaging techniques, including MRI, CT, and PET scans. Researchers have suggested several integration approaches, with early fusion (at the feature level) and late fusion (at the decision level) being the most prevalent approaches. Early fusion combined features extracted from multiple modalities in the initial stages of the neural network, enabling the model to develop unified representations of complementary data. By contrast, late fusion merges the outputs of separate networks trained on individual modalities at a later point in the process [124].
Research comparing different approaches has demonstrated that early fusion can yield superior results in scenarios where fine-grained integration of complementary features from multiple modalities is necessary, such as multimodal tumor segmentation. Nevertheless, early fusion requires advanced techniques for alignment and normalization to address modality-specific variations, including differences in resolution, intensity, and spatial alignment.
In contrast, late fusion, which is computationally less complex and less affected by alignment issues, may not fully capitalize on the interactions between modalities. Research indicates that late fusion tends to be more effective when dealing with highly dissimilar modalities such as combining structural and functional imaging data. However, it may not achieve the same level of precision as early fusion when modalities are more complementary in nature [125].
Intermediate Fusion: A Balanced Approach
Intermediate fusion, which integrates features from various modalities in the mid-level layers of neural networks, is becoming increasingly popular as a balanced method. This approach offers the benefit of identifying cross-modal relationships without inundating the network with unprocessed data in its initial stages. Transformer-based architectures, which incorporate self-attention mechanisms, excel at intermediate fusion due to their capacity to identify complex relationships between modalities across various levels of abstraction [126].
Current studies have shown that intermediate fusion can achieve similar outcomes in multimodal applications, such as tumor segmentation and disease classification, effectively bridging the gap between early and late fusion approaches. This strategy is particularly effective when there is a need to capture both minute details and overarching dependencies in the merged data, as exemplified in multimodal imaging for tumor characterization [91,127,128].
3.3.4 Comparative Evaluation of Performance Metrics
Key metrics, such as accuracy, precision, recall, DSC, and IoU, were used to assess model performance. In tasks involving segmentation, CNNs show high precision and recall, whereas transformers perform well in intricate situations but struggle with smaller objects or structures. Hybrid approaches enhance both precision and recall and strike a balance between effectiveness and efficiency [129].
Accuracy, Precision, and Recall
To evaluate the efficacy of different models for analyzing medical images, crucial metrics include accuracy, precision, and recall. Although accuracy offers a broad overview of correct predictions, precision and recall are particularly crucial in healthcare settings because of the potentially serious implications of false positives and negatives [130].
For segmentation tasks, especially those involving the identification of specific anatomical structures, such as tumors or organs, CNNs typically demonstrate high precision and recall. By contrast, transformers may face challenges in achieving high recall, particularly when dealing with small or indistinct structures that are difficult to differentiate from the surrounding tissue [131].
Combining CNNs and Transformers in hybrid models has shown promising results in improving both accuracy and sensitivity. These combined methods employ CNNs to extract fine-grained local features while leveraging transformers to capture more comprehensive contextual information. Consequently, these hybrid models are more adept at delivering precise predictions while remaining responsive to smaller or less-obvious structures [132–134].
Dice Similarity Coefficient (DSC) and Intersection over Union (IoU)
In evaluating segmentation tasks, the Dice Similarity Coefficient (DSC) and Intersection over Union (IoU) are frequently employed as measures to evaluate the correspondence between predicted and ground truth masks. As illustrated in Fig. 10, transformer-based architectures, particularly ViTs and Swin Transformers, have demonstrated remarkable DSC and IoU results in specific segmentation contexts, especially when identifying large or complex structures [135].
Figure 10: Segmentation performance of Transformer models in DSC and IoU
Nevertheless, the DSC and IoU performances of transformer models can be significantly influenced by the input image resolution and the effectiveness of their attention mechanisms. In scenarios involving high-resolution images, the computational demands of transformers may restrict their capacity to handle large data volumes, potentially affecting their overall segmentation accuracy.
3.3.5 Computational Efficiency and Scalability
Owing to their quadratic complexity, the computational demands of transformers restrict their application in settings with limited resources. Although CNNs are more efficient, they struggle to capture the overall context. Hybrid models combine both architectures, achieving a compromise between the effectiveness and computational demands. This makes them well suited for analyzing medical images in real time [136].
Computational Complexity of Transformers
Despite their remarkable performance in numerous medical imaging tasks, transformer-based models continue to face significant challenges due to their high computational demands. Self-attention of the transformers component operates with quadratic complexity, resulting in significant resource consumption, particularly when processing extensive datasets or high-resolution three-dimensional images. For instance, ViTs and swine transformers demand considerable memory and processing capabilities, which may restrict their use in settings with limited resources, such as mobile devices or real-time clinical systems [137].
However, CNNs are more computationally efficient and can be utilized with less powerful hardware. The widespread use of CNNs in medical image analysis persists owing to their computational efficiency despite their limitations in capturing global relationships. To overcome these shortcomings, researchers developed hybrid models that combine CNNs with transformers. These integrated approaches aim to strike a balance between processing speed and effectiveness, thereby providing a viable option for analyzing medical images in real time [138–140].
3.4 Limitations of Transformer-Based Models in Medical Image Analysis
Although transformer-based models have shown remarkable progress and promise in medical image analysis, several obstacles hinder their widespread implementation and optimal use. These challenges encompass various aspects including computational efficiency, data needs, model resilience, and adaptability across different clinical contexts. This section delves into these limitations, offering a comprehensive examination of the hurdles that must be overcome to improve the effectiveness of transformers in medical imaging applications. This study investigates the intricacies of tackling these challenges to improve the real-world applicability of such models in medical environments [141].
3.4.1 Computational Complexity and Resource Demands
A major drawback of transformer architectures, especially ViTs and Swin Transformers, is their substantial computational demands; unlike CNNs, which employ localized receptive fields and are comparatively computationally efficient, transformer models rely on self-attention mechanisms that process every pair of positions within the input sequence. This results in a quadratic time complexity relative to the number of input elements, such as image patches. For substantial images, including 3D medical scans or high-resolution images, the computational load becomes increasingly challenging [142].
This computational expense also translates into substantial memory requirements. Transformers require considerable memory to store intermediate activations and attention maps during both training and inference phases. This can quickly become a constraining factor, particularly in scenarios involving large datasets or real-time processing. This issue is further amplified in medical imaging applications, where datasets may comprise 3D volumes containing hundreds or thousands of slices per image. Although numerous strategies have been employed to minimize complexity, including the use of efficient attention mechanisms and hierarchical methods, such as Swin Transformers, the substantial computational requirements continue to pose a major challenge to the widespread implementation of these technologies in clinical environments [143–146].
Solutions and Ongoing Research
Current research efforts are directed towards enhancing transformer models for use in medical imaging. Scientists are investigating various strategies to decrease the memory and processing demands of these models, including techniques such as model pruning, distillation, and low-rank approximation. Additionally, increasing attention is being paid to combined approaches that merge the benefits of CNNs and transformer architectures. These combined approaches enable efficient extraction of local features while leveraging the global contextual understanding provided by transformers [147].
3.4.2 Data Requirements and Labeling Challenges
A significant drawback of transformer models is their reliance on extensive high-quality datasets for training. CNNs can perform adequately with moderate amounts of labeled data through transfer learning. Transformers typically require vast quantities of labeled information to achieve comparable results, which presents a specific difficulty in the field of medical image analysis, where annotated datasets are often scarce and expensive to obtain, owing to the need for expert-provided labels [148].
Furthermore, medical image datasets frequently suffer from imbalances, with certain conditions or abnormalities being underrepresented. Similar to other deep learning architectures, transformers are prone to overfitting when faced with limited data, owing to their extensive number of trainable parameters. In the absence of sufficient data, transformers may struggle to generalize effectively, potentially resulting in suboptimal performance when applied in real-world clinical environments [149].
Data Augmentation and Synthetic Data Generation
Researchers are tackling the challenge of limited data by exploring techniques for data augmentation and creating synthetic data. To artificially expand the training datasets, methods such as rotation, scaling, and translation can be utilized, which helps improve the model’s capacity for generalization. Additionally, scientists are examining the potential of generative adversarial networks (GANs) and other approaches for creating synthetic data to produce lifelike medical images that can be used to train transformer models. Nevertheless, the production of high-quality synthetic data that accurately capture the diversity of real-world medical images remains a formidable challenge [20,150,151].
3.4.3 Overfitting and Generalization Issues
The versatility and strength of transformer models render them susceptible to overfitting, particularly when trained on small datasets. When a model learns to mimic training data instead of identifying general patterns, overfitting occurs, leading to suboptimal performance on unseen data. This problem is particularly significant in the field of medical image analysis, where there is a considerable diversity in patient characteristics, imaging protocols, and disease presentations [152].
Furthermore, transformer models often face challenges in adapting to new clinical environments or different medical facilities. As an example, a model developed using data from a single healthcare facility may not function effectively when applied to another facility with differing imaging techniques or patient demographics. The inability to generalize across various clinical settings poses a substantial obstacle to the practical implementation of transformer models in healthcare [36,153].
Transfer Learning and Domain Adaptation
Transfer learning is a commonly employed technique for addressing overfitting and improving the model generalization. This approach involves refining a model trained on one dataset using another. In the context of medical image analysis, this typically consists of initially training transformer models on large, publicly available datasets such as ImageNet or other medical image collections, and then fine-tuning them on smaller, more specialized datasets. Although transfer learning has shown promising results in some cases, its efficacy is largely dependent on the level of similarity between the source and target domains. To further enhance the adaptability of transformer models across various clinical contexts, researchers are investigating domain-adaptation techniques, such as adversarial training and domain-invariant feature learning [113].
3.4.4 Model Interpretability and Explainability
Transformer models face a major hurdle in terms of their lack of transparency and interpretability. Despite their effectiveness in identifying complex data patterns, the opaque nature of their decision-making process makes it challenging for healthcare professionals to rely on and comprehend model outputs. In medical imaging, this concern is especially crucial because comprehending the rationale behind a model’s decision is essential to safeguard patient health and facilitate informed clinical decision-making [154].
However, conventional approaches, such as CNNs, offer somewhat better interpretability, especially when used in conjunction with visualization methods, such as saliency maps or Grad-CAM. These methods emphasize the regions in the images that are most influential in determining the outputs of the model. Such interpretability approaches are essential for healthcare professionals who rely on transparent reasoning to make informed decisions based on a model’s findings [155].
Efforts in Enhancing Explainability
Researchers are actively working on developing techniques to understand how transformer models make decisions, with the aim of tackling the issue of interpretability. Researchers are investigating techniques such as attention heatmaps, which display the attention patterns of transformers, and feature attribution approaches, to shed light on the image areas that the model prioritizes. Furthermore, the incorporation of explainable AI (XAI) frameworks with transformer-based models can potentially enhance their interpretability in healthcare applications [75,156,157].
3.4.5 Handling Noisy and Incomplete Data
Medical imaging data frequently suffer from noise and artifacts that can adversely affect the effectiveness of the model. Noise can originate from various sources, including patient motion, differences in imaging equipment, and inherent flaws in the imaging technique. Although CNNs have demonstrated effectiveness in handling certain types of noise, transformer models generally exhibit greater sensitivity to noise because of their reliance on the global context. The capacity of a transformer to capture long-distance relationships makes it more susceptible to noise, particularly when large image areas are affected [158].
Furthermore, medical image datasets can be incomplete, with certain areas being missing or distorted. Transformer models, like other deep learning approaches, have difficulty handling incomplete data, and their performance may decline significantly if missing information is essential for the task at hand.
Techniques for Noise Reduction
Noisy, and incomplete data, respectively. One promising approach is adversarial training, which involves exposing the model to noisy data and employing regularization techniques to discourage overfitting to noise. Additionally, researchers are investigating techniques for image denoising and data imputation to tackle incomplete datasets and mitigate the detrimental effects of noise on model accuracy [159–161].
3.4.6 Clinical Adoption and Integration into Practice
Although transformer-based models show great potential in medical image analysis, their widespread adoption in clinical settings faces several obstacles. These models demand substantial computational power and infrastructure, which may be scarce in certain healthcare environments, particularly those with limited resources. Furthermore, implementing transformer models in real-world clinical scenarios requires thorough validation and regulatory approval given the paramount importance of patient safety [162].
Beyond the technical challenges, integrating transformer models into clinical workflows presents issues related to user confidence, acceptance by medical professionals, and the need for real-time processing. Many healthcare providers remain wary of AI-driven solutions because of concerns regarding reliability and interpretability. To gain widespread acceptance in clinical practice, transformers must demonstrate consistent and accurate results while seamlessly integrating into existing healthcare systems [163].
Future Directions for Clinical Integration
Future research in this area should focus on developing intuitive interfaces that allow medical professionals to engage with transformer-based models and provide them with accessible tools for model analysis and clinical guidance. As illustrated in Fig. 11, enhancing the computational performance of these models is essential to ensure their adaptability and practicality in healthcare settings. Successful integration into clinical practice necessitates joint efforts among AI experts, medical practitioners, and regulatory bodies [164].

Figure 11: Clinical adoption and integration of Transformer models
To summarize, transformer-based models exhibit substantial potential in medical image analysis; however, they face several obstacles. These include high computational demands, extensive data needs, tendency to overfit, and challenges in model interpretation and clinical integration. Nevertheless, ongoing research efforts are tackling these issues, with promising developments in hybrid models, transfer learning, and explainable AI, offering potential remedies. As these challenges are overcome, transformer models are expected to become increasingly important in the field of medical image analysis to improve diagnostic accuracy and patient care outcomes [67,79,80].
The results section examines the study outcomes, evaluates their significance, and highlights potential ramifications for subsequent investigations and medical applications. This thorough analysis is organized into several subsections to address various aspects of the study.
The findings of this study underscore the game-changing impact of transformers in analyzing multimodal medical images. There are diverse imaging modalities, such as MRI, CT, and PET scans, emission tomography (PET). This integration improves diagnostic accuracy by providing medical professionals with detailed insights into both anatomical and functional abnormalities and enables effective capturing of spatial and semantic connections, overcoming the constraints of conventional approaches. These innovations highlight the vital importance of transformers in modern healthcare systems [165].
Furthermore, the scalability of the transformer models offers a significant advantage. Transformers tackle issues related to scarce labeled medical datasets through initial training on extensive data collection and subsequent refinement for particular applications. This flexibility is especially beneficial in settings with limited resources, where acquiring labeled information can be both costly and time intensive. The demonstrated effectiveness of transformer-based frameworks in tumor segmentation, disease classification, and anomaly detection underscores their potential to revolutionize diagnostic processes [79,166,167].
4.2 Challenges and Limitations
Despite their promising applications, the clinical implementation of transformer-based models has several challenges. A primary concern is the computational complexity associated with processing high-resolution medical images. The computational demands of transformers in real-time applications and resource-limited settings are constrained by the quadratic growth of self-attention mechanisms relative to input length. This scaling characteristic requires significant processing power, which hinders the widespread implementation of these models.
Ensuring the applicability of transformer models across various patient groups and imaging protocols is another obstacle. Model performance can be affected by differences in imaging equipment, acquisition settings, and patient characteristics, which require extensive testing and refinement to guarantee reliability in different clinical contexts. Overcoming these hurdles requires cooperation among scientists, medical professionals, and industry partners to create transformer architectures that are efficient, resilient, and flexible [168].
4.3 Implications for Clinical Practice
Incorporating transformer-based models into medical settings can substantially enhance the quality of healthcare deliveries. By automating the analysis of multimodal medical images, these models reduce the workload of healthcare professionals, allowing them to focus on critical decision-making tasks. Improving the precision and speed of diagnostic processes can result in prompt medical intervention, ultimately enhancing patient care outcomes. Moreover, the interpretability of transformer models fosters trust and acceptance among medical professionals, generating attention maps that offer insights into their decision-making processes and promote transparency and accountability. This interpretability is crucial for integrating AI-driven tools into routine clinical practice, where explainable results are essential for informed decision making [169].
Future studies should address the shortcomings of transformer-based models to enhance their practical application in clinical settings. Initiatives to improve the computational efficiency, including the creation of streamlined transformer architectures and the use of specialized hardware, can enable real-time applications and implementation in settings with limited resources. Moreover, incorporating field-specific expertise and assumptions into the model design can boost the performance and decrease the need for extensive labeled datasets [170].
Another promising avenue involves integrating multimodal imaging data with non-imaging information, including, genetic data, and laboratory findings. This comprehensive strategy can provide more thorough insight into patient conditions and facilitate personalized treatment strategies and precision medicine. To maximize the potential of transformers in healthcare, it is essential to encourage collaboration among researchers, healthcare professionals, and policymakers [171].
4.5 Ethical and Regulatory Considerations
Implementing transformer-based models in healthcare requires a thorough consideration of ethical and regulatory issues. Safeguarding the confidentiality of patient information is essential, particularly when dealing with confidential health records. Adhering to regulations like HIPAA and GDPR is vital to safeguard patient rights and build confidence in AI-powered systems.
Moreover, it is essential to address potential biases in model predictions to ensure fair and equitable outcomes. Training datasets should encompass diverse populations to prevent systemic biases that could disproportionately affect certain demographic groups. To ensure that AI tools function as reliable and unbiased decision support systems, it is essential to consistently track and evaluate the model performance and identify and rectify any biases that may arise [172–174].
In conclusion, this discussion underscores the revolutionary impact of transformer-based models on multimodal medical image analysis, highlighting their potential to improve diagnostic accuracy, efficiency, and scalability. Although obstacles persist, advancements in model architecture and implementation have shown the potential for revolutionizing healthcare services. Continued research, teamwork, and inventiveness are essential for fully harnessing the capabilities of transformers, paving the way for more accurate, streamlined, and fair healthcare solutions [175].
The research highlights the potential of transformer-based models in multimodal medical imaging, but indicates that additional investigation is necessary to fully leverage their capabilities in clinical settings. Several crucial aspects require further examination to maximize the practical application of these models in healthcare.
1. Efficient Transformer Architectures: One major obstacle in implementing transformer models for medical image analysis is the substantial computational resources required, especially when dealing with high-resolution images. Ongoing research could explore the development of more efficient transformer designs, such as Swin Transformers or ViTs, aimed at decreasing computational requirements while preserving the necessary accuracy and performance for medical applications. Furthermore, using model compression strategies like quantization and pruning, along with optimizing inference through hardware acceleration (e.g., TPUs, GPUs), could improve scalability for real-time medical applications that involve large datasets.
2. Multi-Modal and Multi-Source Data Integration: Combining non-imaging data, including electronic health records EHRs, genetic information, and laboratory test results, with imaging data shows great potential. Researchers should explore how transformer models can efficiently merge multi-source data through cross-attention mechanisms or hybrid transformer architectures. Creating models that can dynamically emphasize pertinent features from various modalities might offer a more holistic understanding of the health of the patient. Furthermore, developing standardized multi-modal datasets for training and assessment would improve the generalizability of models in practical clinical settings.
3. Enhancing Generalization across Diverse Patient Populations: A significant hurdle exists in extending the applicability of transformer models across various patient cohorts and imaging techniques. Future studies should aim to enhance the resilience of these models by integrating domain-adaptation strategies, federated learning methods, and contrastive learning techniques. This approach will facilitate the widespread implementation of transformer models in various healthcare environments and enable them to effectively manage the inherent variability found in real-world medical datasets.
4. Explainability and Trust in Clinical Applications:The ability to understand and interpret AI models remains a crucial concern, particularly in medical settings. Research efforts should focus on enhancing interpretability using explainable AI (XAI) methods, including attention map visualization, saliency mapping, and uncertainty estimation techniques.This increased transparency will build confidence among medical professionals and contribute to the responsible and ethical deployment of AI systems in healthcare. Moreover, investigating transformer architectures with inherent interpretability features, such as prototype-based reasoning or decision-aware attention mechanisms, could bolster confidence in clinical applications.
Researchers proposed a multistream transformer framework designed to integrate non-imaging data by merging imaging modalities with clinical details, such as EHRs and genetic information. Non-imaging data are processed using specialized embeddings and cross-modal attention mechanisms are employed to enhance feature correlation, leading to better diagnosis and treatment suggestions. To ensure the generalizability of the model, the authors tested it on datasets from various healthcare institutions that used different imaging protocols and scanner types. Techniques such as transfer learning and fine-tuning are utilized for domain adaptation to enhance the robustness of the model across diverse clinical environments. Additional implementation and generalizability details are provided in the Experimental Evaluation section.
To summarize, the transformer-based method for multi-modal medical imaging presents significant benefits, but ongoing research must address several challenges. These include reducing computational costs, enhancing model adaptability, incorporating various data types, and developing more transparent models to facilitate wider acceptance in clinical settings [173,174].
Progress in transformer models for multimodal image analysis in healthcare represents a significant advancement in medical diagnostics and personalized treatment planning. By integrating information from various imaging modalities, transformers not only enhance diagnostic precision, but also allow clinicians to gain more comprehensive insights into patient conditions. This section summarizes the findings of the study and explores future directions, limitations, and broader implications for healthcare technology.
6.1 Conclusion and Vision for the Future
The incorporation of transformer models into multimodal image analysis has led to significant advancements in healthcare technology. By leveraging their unique capabilities and addressing their current limitations, these models have the capacity to revolutionize medical diagnostics and treatment strategies. To fully realize the benefits of these models, it is crucial for medical professionals, AI experts, and government officials to work together as research advances. This joint initiative strives to enhance the healthcare system by making transformer models more equitable, efficient, and patient-centered [176].
In conclusion, transformer models offer significant potential for bridging the gaps in multimodal medical imaging and driving revolutionary advancements in healthcare. By emphasizing ongoing innovation, ethical considerations, and practical applications, the medical field can leverage AI technology to enhance patient outcomes and set new standards in medical care. This study lays the groundwork for further investigation, encouraging future endeavors to fully realize the capabilities of transformers in healthcare settings.
Acknowledgement: The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Small Research Project under grant number RGP1/139/45.
Funding Statement: This research was supported by the Deanship of Research and Graduate Studies at King Khalid University under Small Research Project grant number RGP1/139/45.
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Sameera V Mohd Sagheer, Meghana K H; Literature search and data collection: Meghana K H, P M Ameer; Analysis and interpretation of findings: Sameera V Mohd Sagheer, P M Ameer, Muneer Parayangat; Draft manuscript preparation: Meghana K H, Mohamed Abbas; Critical revisions and scientific input: Muneer Parayangat, Mohamed Abbas; Final approval of the version to be published: All authors reviewed the results and approved the final version of the manuscript. All authors, including the corresponding author Sameera V Mohd Sagheer, reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets analyzed during this study are available from publicly accessible sources or upon reasonable request to the corresponding author.
Ethics Approval: Not Applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Zaidi H, Montandon ML, Alavi A. The clinical role of fusion imaging using PET, CT, and MR imaging. PET Clinics. 2008;3(3):275–91. doi:10.1016/j.cpet.2009.03.002. [Google Scholar] [PubMed] [CrossRef]
2. Wenderott K, Krups J, Zaruchas F, Weigl M. Effects of artificial intelligence implementation on efficiency in medical imaging-a systematic literature review and meta-analysis. npj Digit Med. 2024;7(1):265. doi:10.1038/s41746-024-01248-9. [Google Scholar] [PubMed] [CrossRef]
3. Jeong J, Kim S, Pan L, Hwang D, Kim D, Choi J, et al. Reducing the workload of medical diagnosis through artificial intelligence: a narrative review. Medicine. 2025;104(6):e41470. doi:10.1097/md.0000000000041470. [Google Scholar] [PubMed] [CrossRef]
4. Khalifa M, Albadawy M. AI in diagnostic imaging: Revolutionising accuracy and efficiency. Comput Methods Programs Biomed Update. 2024;5:100146. doi:10.1016/j.cmpbup.2024.100146. [Google Scholar] [CrossRef]
5. Oyeniyi J, Oluwaseyi P. Emerging trends in AI-powered medical imaging: enhancing diagnostic accuracy and treatment decisions. IJERSTE. 2024;13:2319–7463. [Google Scholar]
6. Dai Y, Gao Y, Liu F. Transmed: transformers advance multi-modal medical image classification. Diagnostics. 2021;11(8):1384. doi:10.3390/diagnostics11081384. [Google Scholar] [PubMed] [CrossRef]
7. Li M, Jiang Y, Zhang Y, Zhu H. Medical image analysis using deep learning algorithms. Front Public Health. 2023;11:1273253. doi:10.3389/fpubh.2023.1273253. [Google Scholar] [PubMed] [CrossRef]
8. Cai L, Fang H, Xu N, Ren B. Counterfactual Causal-effect intervention for interpretable medical visual question answering. IEEE T Med Imaging. 2024;43(12):4430–41. doi:10.1109/TMI.2024.3425533. [Google Scholar] [PubMed] [CrossRef]
9. Zhang C, Zhang S, Yin Y, Wang L, Li L, Lan C, et al. Clot removAl with or without decompRessive craniectomy under ICP monitoring for supratentorial IntraCerebral Hemorrhage (CARICHa randomized controlled trial. International Journal of Surgery. 2024;110(8):4804–9. doi:10.1097/JS9.0000000000001466. [Google Scholar] [PubMed] [CrossRef]
10. Huang H, Wu N, Liang Y, Peng X, Shu J. SLNL: a novel method for gene selection and phenotype classification. Int J Surg. 2022;37(9):6283–304. doi:10.1002/int.22844. [Google Scholar] [CrossRef]
11. Uesugi K, Mayama H, Morishima K. Analysis of rowing force of the water strider middle leg by direct measurement using a bio-appropriating probe and by indirect measurement using image analysis. Cyborg Bionic Syst. 2023;4:0061. doi:10.34133/cbsystems.0061. [Google Scholar] [PubMed] [CrossRef]
12. Wang H, Wang Y, Yan S, Du X, Gao Y, Liu H. Merge-and-split graph convolutional network for skeleton-based interaction recognition. Cyborg Bionic Syst. 2024;5(4):0102. doi:10.34133/cbsystems.0102. [Google Scholar] [PubMed] [CrossRef]
13. Long T, Song X, Han B, Suo Y, Jia L. In situ magnetic field compensation method for optically pumped magnetometers under three-axis nonorthogonality. IEEE T Instrum Meas. 2024;73:1–12. doi:10.1109/TIM.2023.3331425. [Google Scholar] [CrossRef]
14. Zhang C, Ge H, Zhang S, Liu D, Jiang Z, Lan C et al. Hematoma evacuation via image-guided para-corticospinal tract approach in patients with spontaneous intracerebral hemorrhage. Neurolo Therapy. 2021;10(2):1001–13. doi:10.1007/s40120-021-00279-8. [Google Scholar] [PubMed] [CrossRef]
15. Xu X, Luo Q, Wang J, Song Y, Ye H, Zhang X et al. Large-field objective lens for multi-wavelength microscopy at mesoscale and submicron resolution. Opto-Electron Adv. 2024;7(6):230212. doi:10.29026/oea.2024.230212. [Google Scholar] [CrossRef]
16. Pan H, Wang Y, Li Z, Chu X, Teng B, Gao H. A complete scheme for multi-character classification using EEG signals from speech imagery. IEEE T Bio-med Eng. 2024;71(8):2454–62. doi:10.1109/TBME.2024.3376603. [Google Scholar] [PubMed] [CrossRef]
17. Pan H, Li Z, Fu Y, Qin X, Hu J. Reconstructing visual stimulus representation from eeg signals based on deep visual representation model. IEEE T Hum-Mach Syst. 2024;54(6):711–22. doi:10.1109/THMS.2024.3407875. [Google Scholar] [CrossRef]
18. Cui Q, Ding Z, Chen F. Hybrid directed hypergraph learning and forecasting of skeleton-based human poses. Cyborg Bionic Syst. 2024;5(7):0093. doi:10.34133/cbsystems.0093. [Google Scholar] [PubMed] [CrossRef]
19. Jia Y, Chen G, Chi H. Retinal fundus image super-resolution based on generative adversarial network guided with vascular structure prior. Sci Report. 2024;14(1):22786. doi:10.1038/s41598-024-74186-x. [Google Scholar] [PubMed] [CrossRef]
20. Ma N, Fang X, Zhang Y, Xing B, Duan L, Lu J et al. Enhancing the sensitivity of spin-exchange relaxation-free magnetometers using phase-modulated pump light with external Gaussian noise. Optics Express. 2024;32(19):33378–90. doi:10.1364/OE.530764. [Google Scholar] [PubMed] [CrossRef]
21. Zhou M, Chen L, Wei X, Liao X, Mao Q, Wang H, et al. Perception-oriented U-shaped transformer network for 360-degree no-reference image quality assessment. IEEE T Broadcast. 2023;69(2):396–405. doi:10.1109/TBC.2022.3231101. [Google Scholar] [CrossRef]
22. Li Q, You T, Chen J, Zhang Y, Du C. LI-EMRSQL: linking information enhanced Text2SQL parsing on complex electronic medical records. IEEE T Reliab. 2024;73(2):1280–90. doi:10.1109/TR.2023.3336330. [Google Scholar] [CrossRef]
23. Bing P, Liu W, Zhai Z, Li J, Guo Z, Xiang Y et al. A novel approach for denoising electrocardiogram signals to detect cardiovascular diseases using an efficient hybrid scheme. Front Cardiovasc Med. 2024;11:1277123. doi:10.3389/fcvm.2024.1277123. [Google Scholar] [PubMed] [CrossRef]
24. Song W, Wang X, Guo Y, Li S, Xia B, Hao A. CenterFormer: a novel cluster center enhanced transformer for unconstrained dental plaque segmentation. IEEE T Multimed. 2024;26:10965–78. doi:10.1109/TMM.2024.3428349. [Google Scholar] [CrossRef]
25. Luan S, Yu X, Lei S, Ma C, Wang X, Xue X et al. Deep learning for fast super-resolution ultrasound microvessel imaging. Physic Med Biology. 2023;68(24):245023. doi:10.1088/1361-6560/ad0a5a. [Google Scholar] [PubMed] [CrossRef]
26. Yu X, Luan S, Lei S, Huang J, Liu Z, Xue X et al. Deep learning for fast denoising filtering in ultrasound localization microscopy. Physic Med Biology. 2023;68(20):205002. doi:10.1088/1361-6560/acf98f. [Google Scholar] [PubMed] [CrossRef]
27. Ou J, Li N, He H, He J, Zhang L, Jiang N. Detecting muscle fatigue among community-dwelling senior adults with shape features of the probability density function of sEMG. J NeuroEng Rehabil. 2024;21(1):196. doi:10.1186/s12984-024-01497-5. [Google Scholar] [PubMed] [CrossRef]
28. Lavanya P, Sasikala E. Deep learning techniques on text classification using Natural language processing (NLP) in social healthcare network: a comprehensive survey. In: 2021 3rd International Conference on Signal Processing and Communication (ICPSC); 2021 May 13–14; Coimbatore, India. p. 603–9. [Google Scholar]
29. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: transformers for image recognition at scale. arXiv:2010.11929. 2020. [Google Scholar]
30. Lahoud J, Cao J, Khan FS, Cholakkal H, Anwer RM, Khan S, et al. 3D vision with transformers: a survey. arXiv:2208.04309. 2022. [Google Scholar]
31. Pereira GA, Hussain M. A review of transformer-based models for computer vision tasks: capturing global context and spatial relationships. arXiv:2408.15178. 2024. [Google Scholar]
32. Naqvi RA, Haider A, Kim HS, Jeong D, Lee SW. Transformative noise reduction: leveraging a transformer-based deep network for medical image denoising. Mathematics. 2024;12(15):2313. doi:10.3390/math12152313. [Google Scholar] [CrossRef]
33. Lai AY, Perucho JA, Xu X, Hui ES, Lee EY. Concordance of FDG PET/CT metabolic tumour volume versus DW-MRI functional tumour volume with T2-weighted anatomical tumour volume in cervical cancer. BMC Cancer. 2017;17(1):1–8. doi:10.1186/s12885-017-3800-9. [Google Scholar] [PubMed] [CrossRef]
34. AlSaad R, Abd-Alrazaq A, Boughorbel S, Ahmed A, Renault MA, Damseh R, et al. Multimodal large language models in health care: applications, challenges, and future outlook. J Med Internet Res. 2024;26:e59505. [Google Scholar] [PubMed]
35. Zhao B, Li L, Zhang Y, Tang J, Liu Y, Zhai Y. Optically pumped magnetometers recent advances and applications in biomagnetism: a review. IEEE Sens J. 2023;23(17):18949–62. doi:10.1109/jsen.2023.3297109. [Google Scholar] [CrossRef]
36. Nerella S, Bandyopadhyay S, Zhang J, Contreras M, Siegel S, Bumin A, et al. Transformers in healthcare: a survey. arXiv:2307.00067. 2023. [Google Scholar]
37. Nerella S, Bandyopadhyay S, Zhang J, Contreras M, Siegel S, Bumin A, et al. Transformers and large language models in healthcare: a review. Artif Intell Med. 2024;154(6088):102900. doi:10.1016/j.artmed.2024.102900. [Google Scholar] [PubMed] [CrossRef]
38. Cho HN, Jun TJ, Kim YH, Kang H, Ahn I, Gwon H, et al. Task-specific transformer-based language models in health care: scoping review. JMIR Med Inform. 2024;12:e49724. [Google Scholar] [PubMed]
39. Teoh JR, Dong J, Zuo X, Lai KW, Hasikin K, Wu X. Advancing healthcare through multimodal data fusion: a comprehensive review of techniques and applications. PeerJ Comput Sci. 2024;10(9):e2298. doi:10.7717/peerj-cs.2298. [Google Scholar] [PubMed] [CrossRef]
40. Alshehri S, Alahmari KA, Alasiry A. A comprehensive evaluation of AI-assisted diagnostic tools in ENT medicine: insights and perspectives from healthcare professionals. J Pers Med. 2024;14(4):354. doi:10.3390/jpm14040354. [Google Scholar] [PubMed] [CrossRef]
41. Xu H, Xu Q, Cong F, Kang J, Han C, Liu Z, et al. Vision transformers for computational histopathology. IEEE Rev Biomed Eng. 2023;17:63–79. [Google Scholar]
42. Eghbali N, Bagher-Ebadian H, Alhanai T, Ghassemi MM. GLoG-CSUnet: enhancing vision transformers with adaptable radiomic features for medical image segmentation. arXiv:2501.02788. 2025. [Google Scholar]
43. Famiglini L. Enhancing the explainability and reliability of AI support for informed healthcare decisions [Ph.D. thesis]. Milano, Italy: Università degli Studi di Milano Bicocca; 2025. [Google Scholar]
44. Rashid M, Sharma M. AI-assisted diagnosis and treatment planning–a discussion of how AI can assist healthcare professionals in making more accurate diagnoses and treatment plans for diseases. In: AI in disease detection: advancements and applications. Hoboken, NJ, USA: Wiley-IEEE Press; 2025. p. 313–36. doi:10.1002/9781394278695.ch14. [Google Scholar] [CrossRef]
45. Maambo M. Assisted artificial intelligence medical diagnosis system for heart disease [master thesis]. Lusaka, Zambia: The University of Zambia; 2023. [Google Scholar]
46. Li J, Chen J, Tang Y, Wang C, Landman BA, Zhou SK. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med Image Anal. 2023;85(1):102762. doi:10.1016/j.media.2023.102762. [Google Scholar] [PubMed] [CrossRef]
47. Rashid Z, Ahmed H, Nadeem N, Zafar SB, Yousaf MZ. The paradigm of digital health: aI applications and transformative trends. Neural Comput Appl. 2025;37(17):11039–70. doi:10.1007/s00521-025-11081-0. [Google Scholar] [CrossRef]
48. Salehi F. The transformative role of artificial intelligence in the healthcare industry: a comprehensive analysis. Asian J Res Med Med Sci. 2024;6(1):62–70. [Google Scholar]
49. Balakrishna S, Solanki VK. A comprehensive review on ai-driven healthcare transformation. Ing Solidar. 2024;20(2):1–30. [Google Scholar]
50. Du J, Li W, Lu K, Xiao B. An overview of multi-modal medical image fusion. Neurocomputing. 2016;215:3–20. [Google Scholar]
51. Singh S, Gupta D. Detail enhanced feature-level medical image fusion in decorrelating decomposition domain. IEEE Trans Instrum Meas. 2020;70:1–9. doi:10.1109/tim.2020.3038603. [Google Scholar] [CrossRef]
52. Xiao G, Bavirisetti DP, Liu G, Zhang X, Xiao G, Bavirisetti DP, et al. Decision-level image fusion. In: Image fusion. Singapore: Springer; 2020. p. 149–70. [Google Scholar]
53. Hermessi H, Mourali O, Zagrouba E. Multimodal medical image fusion review: theoretical background and recent advances. Signal Process. 2021;183(4):108036. doi:10.1016/j.sigpro.2021.108036. [Google Scholar] [CrossRef]
54. Nair RR, Singh T, Basavapattana A, Pawar MM. Multi-layer, multi-modal medical image intelligent fusion. Multimedia Tools Appl. 2022;81(29):42821–47. doi:10.1007/s11042-022-13482-y. [Google Scholar] [CrossRef]
55. Khan SU, Khan MA, Azhar M, Khan F, Lee Y, Javed M. Multimodal medical image fusion towards future research: a review. J King Saud Univ—Comput Inf Sci. 2023;35(8):101733. doi:10.1016/j.jksuci.2023.101733. [Google Scholar] [CrossRef]
56. Parihar AS. A study on brain tumor segmentation using convolution neural network. In: 2017 International Conference on Inventive Computing and Informatics (ICICI); 2017 Nov 23–24; Coimbatore, India. p. 198–201. [Google Scholar]
57. Zhang Y, Liu H, Hu Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France. p. 14–24. [Google Scholar]
58. Xiao H, Li L, Liu Q, Zhu X, Zhang Q. Transformers in medical image segmentation: a review. Biomed Signal Process Control. 2023;84(12):104791. doi:10.1016/j.bspc.2023.104791. [Google Scholar] [CrossRef]
59. Khan A, Rauf Z, Khan AR, Rathore S, Khan SH, Shah NS, et al. A recent survey of vision transformers for medical image segmentation. arXiv:2312.00634. 2023. [Google Scholar]
60. Wei C, Ren S, Guo K, Hu H, Liang J. High-resolution Swin transformer for automatic medical image segmentation. Sensors. 2023;23(7):3420. doi:10.3390/s23073420. [Google Scholar] [PubMed] [CrossRef]
61. Liu X, Hu Y, Chen J. Hybrid CNN-Transformer model for medical image segmentation with pyramid convolution and multi-layer perceptron. Biomed Signal Process Control. 2023;86(5):105331. doi:10.1016/j.bspc.2023.105331. [Google Scholar] [CrossRef]
62. Ma J, Li F, Wang B. U-mamba: enhancing long-range dependency for biomedical image segmentation. arXiv:2401.04722. 2024. [Google Scholar]
63. van Tulder G, de Bruijne M. Learning cross-modality representations from multi-modal images. IEEE Trans Med Imaging. 2018;38(2):638–48. doi:10.1109/tmi.2018.2868977. [Google Scholar] [PubMed] [CrossRef]
64. Yu H, Zhou X, Jiang H, Kang H, Wang Z, Hara T, et al. Learning 3D non-rigid deformation based on an unsupervised deep learning for PET/CT image registration. In: Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging. Vol. 10953. Bellingham, WA, USA: Spie; 2019. p. 439–44. [Google Scholar]
65. Patel A, Tudosiu PD, Pinaya WHL, Cook G, Goh V, Ourselin S, et al. Cross attention transformers for multi-modal unsupervised whole-body pet anomaly detection. In: MICCAI Workshop on Deep Generative Models. Cham, Switzerland: Springer; 2022. p. 14–23. [Google Scholar]
66. He K, Gan C, Li Z, Rekik I, Yin Z, Ji W, et al. Transformers in medical image analysis. Intell Med. 2023;3(1):59–78. doi:10.1016/j.imed.2022.07.002. [Google Scholar] [CrossRef]
67. Xia K, Wang J. Recent advances of transformers in medical image analysis: a comprehensive review. MedComm-Future Med. 2023;2(1):e38. doi:10.1002/mef2.38. [Google Scholar] [CrossRef]
68. Papanastasiou G, Dikaios N, Huang J, Wang C, Yang G. Is attention all you need in medical image analysis? A review. IEEE J Biomed Health Inform. 2023;28(3):1398–411. doi:10.1109/jbhi.2023.3348436. [Google Scholar] [PubMed] [CrossRef]
69. Kim JW, Khan AU, Banerjee I. Systematic review of hybrid vision transformer architectures for radiological image analysis. medRxiv. 2024;13(6):3680. doi:10.1101/2024.06.21.24309265. [Google Scholar] [CrossRef]
70. Yadav AP, Patil S. Exploring hand gesture recognition techniques for enhanced control of bionic hands. In: 2024 International Conference on Emerging Smart Computing and Informatics (ESCI); 2024 Mar 5–7; Pune, India. p. 1–5. [Google Scholar]
71. Wang Y. Survey on deep multi-modal data analytics: collaboration, rivalry, and fusion. ACM Trans Multimed Comput Commun Appl (TOMM). 2021;17(1s):1–25. doi:10.1145/3408317. [Google Scholar] [CrossRef]
72. Gao J, Li P, Chen Z, Zhang J. A survey on deep learning for multimodal data fusion. Neural Comput. 2020;32(5):829–64. doi:10.1162/neco_a_01273. [Google Scholar] [PubMed] [CrossRef]
73. Duan J, Xiong J, Li Y, Ding W. Deep learning based multimodal biomedical data fusion: an overview and comparative review. Inf Fusion. 2024;112(9):102536. doi:10.1016/j.inffus.2024.102536. [Google Scholar] [CrossRef]
74. Boehm KM, Khosravi P, Vanguri R, Gao J, Shah SP. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22(2):114–26. doi:10.1038/s41568-021-00408-3. [Google Scholar] [PubMed] [CrossRef]
75. Lai T. Interpretable medical imagery diagnosis with self-attentive transformers: a review of explainable AI for health care. BioMedInformatics. 2024;4(1):113–26. doi:10.3390/biomedinformatics4010008. [Google Scholar] [CrossRef]
76. Kumar A, Kim J, Cai W, Fulham M, Feng D. Content-based medical image retrieval: a survey of applications to multidimensional and multimodality data. J Digit Imaging. 2013;26(6):1025–39. doi:10.1007/s10278-013-9619-2. [Google Scholar] [PubMed] [CrossRef]
77. Altaf F, Islam SM, Akhtar N, Janjua NK. Going deep in medical image analysis: concepts, methods, challenges, and future directions. IEEE Access. 2019;7:99540–72. doi:10.1109/access.2019.2929365. [Google Scholar] [CrossRef]
78. Mahdi MA, Ahamad S, Saad SA, Dafhalla A, Qureshi R, Alqushaibi A. Weighted fusion transformer for dual PET/CT head and neck tumor segmentation. IEEE Access. 2024;12:110905–19. doi:10.1109/access.2024.3439439. [Google Scholar] [CrossRef]
79. Rane N. Transformers for medical image analysis: applications, challenges, and future scope. 2023 Nov 2. doi:10.2139/ssrn.4622241. [Google Scholar] [CrossRef]
80. Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, et al. Transformers in medical imaging: a survey. Med Image Anal. 2023;88(1):102802. doi:10.1016/j.media.2023.102802. [Google Scholar] [PubMed] [CrossRef]
81. Palanisamy V, Thirunavukarasu R. Implications of big data analytics in developing healthcare frameworks—a review. J King Saud Univ-Comput Inform Sci. 2019;31(4):415–25. doi:10.1016/j.jksuci.2017.12.007. [Google Scholar] [CrossRef]
82. Sakr S, Elgammal A. Towards a comprehensive data analytics framework for smart healthcare services. Big Data Res. 2016;4(9):44–58. doi:10.1016/j.bdr.2016.05.002. [Google Scholar] [CrossRef]
83. Toennies KD. Guide to medical image analysis. Cham, Switzerland: Springer; 2017. [Google Scholar]
84. Li J, Wang Z, Wang C, Su W. GaitFormer: leveraging dual-stream spatial-temporal Vision Transformer via a single low-cost RGB camera for clinical gait analysis. Knowl Based Syst. 2024;295(9):111810. doi:10.1016/j.knosys.2024.111810. [Google Scholar] [CrossRef]
85. Zhang Y, He N, Yang J, Li Y, Wei D, Huang Y et al. mmformer: multimodal medical transformer for incomplete multimodal learning of brain tumor segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2022. p. 107–17. [Google Scholar]
86. Shi J, Yu L, Cheng Q, Yang X, Cheng KT, Yan Z. MFTrans: modality-masked fusion transformer for incomplete multi-modality brain tumor segmentation. IEEE J Biomed Health Inform. 2023;28(1):379–90. doi:10.1109/jbhi.2023.3326151. [Google Scholar] [PubMed] [CrossRef]
87. Karimijafarbigloo S, Azad R, Kazerouni A, Ebadollahi S, Merhof D. Mmcformer: missing modality compensation transformer for brain tumor segmentation. In: Medical Imaging with Deep Learning. London, UK: PMLR; 2024. p. 1144–62. [Google Scholar]
88. Didona D, Quaglia F, Romano P, Torre E. Enhancing performance prediction robustness by combining analytical modeling and machine learning. In: Proceedings of the 6th ACM/SPEC International Conference on Performance Engineering; 2015 Jan 28–Feb 4; Austin, TX, USA. p. 145–56. [Google Scholar]
89. Liang W, Tadesse GA, Ho D, Fei-Fei L, Zaharia M, Zhang C, et al. Advances, challenges and opportunities in creating data for trustworthy AI. Nat Mach Intell. 2022;4(8):669–77. doi:10.1038/s42256-022-00516-1. [Google Scholar] [CrossRef]
90. Abdollahi B, Tomita N, Hassanpour S. Data augmentation in training deep learning models for medical image analysis. Deep learners and deep learner descriptors for medical applications. Cham, Switzerland: Springer; 2020. p. 167–80. [Google Scholar]
91. Xu X, Li J, Zhu Z, Zhao L, Wang H, Song C, et al. A comprehensive review on synergy of multi-modal data and ai technologies in medical diagnosis. Bioengineering. 2024;11(3):219. doi:10.3390/bioengineering11030219. [Google Scholar] [PubMed] [CrossRef]
92. Abidin ZU, Naqvi RA, Haider A, Kim HS, Jeong D, Lee SW. Recent deep learning-based brain tumor segmentation models using multi-modality magnetic resonance imaging: a prospective survey. Front Bioeng Biotechnol. 2024;12:1392807. doi:10.3389/fbioe.2024.1392807. [Google Scholar] [PubMed] [CrossRef]
93. Li X, Li M, Yan P, Li G, Jiang Y, Luo H, et al. Deep learning attention mechanism in medical image analysis: basics and beyonds. Int J Netw Dyn Intell. 2023;2(1):93–116. doi:10.53941/ijndi0201006. [Google Scholar] [CrossRef]
94. Khanal B, Shrestha P, Amgain S, Khanal B, Bhattarai B, Linte CA. Investigating the robustness of vision transformers against label noise in medical image classification. arXiv:2402.16734. 2024. [Google Scholar]
95. Xu L, Sun H, Ni Z, Li H, Zhang S. MedViLaM: a multimodal large language model with advanced generalizability and explainability for medical data understanding and generation. arXiv:2409.19684. 2024. [Google Scholar]
96. Roy P. Enhancing real-world robustness in AI: challenges and solutions. J Recent Trends Comput Sci Eng (JRTCSE). 2024;12(1):34–49. doi:10.70589/jrtcse.2024.1.6. [Google Scholar] [CrossRef]
97. Singh RP, Hom GL, Abramoff MD, Campbell JP, Chiang MF, AAO Task Force on Artificial Intelligence. Current challenges and barriers to real-world artificial intelligence adoption for the healthcare system, provider, and the patient. Transl Vis Sci Technol. 2020;9(2):45–5. [Google Scholar] [PubMed]
98. Andrade-Miranda G, Jaouen V, Tankyevych O, Le Rest CC, Visvikis D, Conze PH. Multi-modal medical Transformers: a meta-analysis for medical image segmentation in oncology. Comput Med Imaging Graph. 2023;110(6):102308. doi:10.1016/j.compmedimag.2023.102308. [Google Scholar] [PubMed] [CrossRef]
99. Yuan F, Zhang Z, Fang Z. An effective CNN and Transformer complementary network for medical image segmentation. Pattern Recognit. 2023;136(11):109228. doi:10.1016/j.patcog.2022.109228. [Google Scholar] [CrossRef]
100. Hussain T, Shouno H, Mohammad MA, Marhoon HA, Alam T. DCSSGA-UNet: biomedical image segmentation with DenseNet channel spatial and semantic guidance attention. Knowl Based Syst. 2025;314(11):113233. doi:10.1016/j.knosys.2025.113233. [Google Scholar] [CrossRef]
101. Tang H, Chen Y, Wang T, Zhou Y, Zhao L, Gao Q, et al. HTC-Net: a hybrid CNN-transformer framework for medical image segmentation. Biomed Signal Process Control. 2024;88(9):105605. doi:10.1016/j.bspc.2023.105605. [Google Scholar] [CrossRef]
102. Xu Y, Quan R, Xu W, Huang Y, Chen X, Liu F. Advances in medical image segmentation: a comprehensive review of traditional, deep learning and hybrid approaches. Bioengineering. 2024;11(10):1034. doi:10.3390/bioengineering11101034. [Google Scholar] [PubMed] [CrossRef]
103. Rasool N, Bhat JI. Unveiling the complexity of medical imaging through deep learning approaches. Chaos Theory Applicati. 2023;5(4):267–80. [Google Scholar]
104. Azam MA, Khan KB, Salahuddin S, Rehman E, Khan SA, Khan MA, et al. A review on multimodal medical image fusion: compendious analysis of medical modalities, multimodal databases, fusion techniques and quality metrics. Comput Biol Med. 2022;144(3):105253. doi:10.1016/j.compbiomed.2022.105253. [Google Scholar] [PubMed] [CrossRef]
105. Gong H, Chen G, Liu S, Yu Y, Li G. Cross-modal self-attention with multi-task pre-training for medical visual question answering. In: Proceedings of the 2021 International Conference on Multimedia Retrieval; 2021 Nov 16–19; Taipei, Taiwan. p. 456–60. [Google Scholar]
106. Huang NY, Liu CX. Research on tumor detection and classification model based on self-attention mechanism. IEEE Access. 2024. [Google Scholar]
107. Wang J, Yu L, Tian S. Cross-attention interaction learning network for multi-model image fusion via transformer. Eng Appl Artif Intell. 2025;139(5):109583. doi:10.1016/j.engappai.2024.109583. [Google Scholar] [CrossRef]
108. Prabhod KJ, Gadhiraju A. Foundation models in medical imaging: revolutionizing diagnostic accuracy and efficiency. J Artif Intell Res Appl. 2024;4(1):471–511. [Google Scholar]
109. Ker J, Wang L, Rao J, Lim T. Deep learning applications in medical image analysis. IEEE Access. 2017;6:9375–89. doi:10.1109/access.2017.2788044. [Google Scholar] [CrossRef]
110. Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: overview, challenges and the future. In: Classification in BioApps: Automation of Decision Making. Cham, Switzerland: Springer; 2017. p. 323–50. [Google Scholar]
111. Chen B, Huang X, Liu Y, Zhang Z, Lu G, Zhou Z, et al. Attention-guided and noise-resistant learning for robust medical image segmentation. IEEE Trans Instrum Meas. 2024;73:1–13. doi:10.1109/tim.2024.3406804. [Google Scholar] [CrossRef]
112. Zhang A, Xing L, Zou J, Wu JC. Shifting machine learning for healthcare from development to deployment and from models to data. Nat Biomed Eng. 2022;6(12):1330–45. [Google Scholar] [PubMed]
113. Alijani S, Fayyad J, Najjaran H. Vision transformers in domain adaptation and domain generalization: a study of robustness. Neural Comput Appl. 2024;36(29):17979–8007. doi:10.1007/s00521-024-10353-5. [Google Scholar] [CrossRef]
114. Takahashi S, Sakaguchi Y, Kouno N, Takasawa K, Ishizu K, Akagi Y, et al. Comparison of vision transformers and convolutional neural networks in medical image analysis: a systematic review. J Med Syst. 2024;48(1):1–22. doi:10.1007/s10916-024-02105-8. [Google Scholar] [PubMed] [CrossRef]
115. Lu K, Xu Y, Yang Y. Comparison of the potential between transformer and CNN in image classification. In: ICMLCA 2021; 2nd International Conference on Machine Learning and Computer Application; 2021 Dec 17–19; Shenyang, China. p. 1–6. [Google Scholar]
116. Hussain T, Shouno H. MAGRes-UNet: improved medical image segmentation through a deep learning paradigm of multi-attention gated residual U-Net. IEEE Access. 2024;12:40290–310. doi:10.1109/access.2024.3374108. [Google Scholar] [CrossRef]
117. Kilimci ZH, Yalcin M, Kucukmanisa A, Mishra AK. Heart disease detection using vision-based transformer models from ECG images. arXiv:2310.12630. 2023. [Google Scholar]
118. Shah HA, Saeed F, Diyan M, Almujally NA, Kang JM. ECG-TransCovNet: a hybrid transformer model for accurate arrhythmia detection using electrocardiogram signals. CAAI Trans Intell Technol. 2024. [Google Scholar]
119. El-Ghaish H, Eldele E. ECGTransForm: empowering adaptive ECG arrhythmia classification framework with bidirectional transformer. Biomed Signal Process Control. 2024;89(1):105714. doi:10.1016/j.bspc.2023.105714. [Google Scholar] [CrossRef]
120. Wang Z, Zhao W, Ni Z, Zheng Y. Adversarial vision transformer for medical image semantic segmentation with limited annotations. In: 33rd British Machine Vision Conference; 2022 Nov 21–24; London, UK. [Google Scholar]
121. Zhao B, Feng J, Wu X, Yan S. A survey on deep learning-based fine-grained object classification and semantic segmentation. Int J Automa Comput. 2017;14(2):119–35. doi:10.1007/s11633-017-1053-3. [Google Scholar] [CrossRef]
122. Ouyang C, Biffi C, Chen C, Kart T, Qiu H, Rueckert D. Self-supervision with superpixels: training few-shot medical image segmentation without annotation. In: Computer Vision–ECCV 2020: 16th European Conference; 2020 Aug 23–28; Glasgow, UK. p. 762–80. [Google Scholar]
123. Zhou Q, Ye S, Wen M, Huang Z, Ding M, Zhang X. Multi-modal medical image fusion based on densely-connected high-resolution CNN and hybrid transformer. Neural Comput Appl. 2022;34(24):21741–61. doi:10.1007/s00521-022-07635-1. [Google Scholar] [CrossRef]
124. Yang Q, Zhao Y, Cheng H. MMLF: multi-modal multi-class late fusion for object detection with uncertainty estimation. arXiv:2410.08739. 2024. [Google Scholar]
125. Zhang Y, Yang J, Tian J, Shi Z, Zhong C, Zhang Y et al. Modality-aware mutual learning for multi-modal medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference; 2021 Sep 27–Oct 1; Strasbourg, France. p. 589–99. [Google Scholar]
126. Harouni A, Karargyris A, Negahdar M, Beymer D, Syeda-Mahmood T. Universal multi-modal deep network for classification and segmentation of medical images. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018 Apr 4–7; Washington, DC, USA. p. 872–6. [Google Scholar]
127. Pan C, Zhou P, Tan J, Sun B, Guan R, Wang Z, et al. Liver tumor detection via a multi-scale intermediate multi-modal fusion network on MRI images. In: 2021 IEEE International Conference on Image Processing (ICIP); 2021 Sep 19–22; Anchorage, AK, USA. p. 299–303. [Google Scholar]
128. Kumar RR, Priyadarshi R. Denoising and segmentation in medical image analysis: a comprehensive review on machine learning and deep learning approaches. Multimed Tools Appl. 2025;84(12):10817–75. doi:10.1007/s11042-024-19313-6. [Google Scholar] [CrossRef]
129. Kumar B, Singh SP, Mohan A, Anand A. Performance of quality metrics for compressed medical images through mean opinion score prediction. J Med Imaging Health Infor. 2012;2(2):188–94. [Google Scholar]
130. Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep learning in medical image analysis: challenges and applications. 1st ed. Cham, Switzerland: Springer; 2020. p. 3–21. [Google Scholar]
131. Moeskops P, Wolterink JM, Van Der Velden BH, Gilhuijs KG, Leiner T, Viergever MA, et al. Deep learning for multi-task medical image segmentation in multiple modalities. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2016: 19th International Conference; 2016 Oct 17–21; Athens, Greece. p. 478–86. [Google Scholar]
132. Almotairi S, Badr E, Abdelbaky I, Elhakeem M, Abdul Salam M. Hybrid transformer-CNN model for accurate prediction of peptide hemolytic potential. Sci Rep. 2024;14(1):14263. doi:10.1038/s41598-024-63446-5. [Google Scholar] [PubMed] [CrossRef]
133. Jablonka KM, Ongari D, Moosavi SM, Smit B. Big-data science in porous materials: materials genomics and machine learning. Chem Rev. 2020;120(16):8066–129. doi:10.1021/acs.chemrev.0c00004. [Google Scholar] [PubMed] [CrossRef]
134. Sadybekov AV, Katritch V. Computational approaches streamlining drug discovery. Nature. 2023;616(7958):673–85. doi:10.1038/s41586-023-05905-z. [Google Scholar] [PubMed] [CrossRef]
135. Yuan Y, Du Y, Ma Y, Lv H. DSC-Net: enhancing blind road semantic segmentation with visual sensor using a dual-branch swin-CNN architecture. Sensors. 2024;24(18):6075. doi:10.3390/s24186075. [Google Scholar] [PubMed] [CrossRef]
136. Katharopoulos A, Vyas A, Pappas N, Fleuret F. Transformers are RNNs: fast autoregressive transformers with linear attention. In: International Conference on Machine Learning. London, UK: PMLR; 2020. p. 5156–65. [Google Scholar]
137. Selvan R, Schön J, Dam EB. Operating critical machine learning models in resource constrained regimes. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland: Springer; 2023. p. 325–35. [Google Scholar]
138. Wang J, Wang S, Zhang Y. Deep learning on medical image analysis. CAAI Trans Intell Technol. 2025;10(1):1–35. [Google Scholar]
139. Wang Y, Kung L, Byrd TA. Big data analytics: understanding its capabilities and potential benefits for healthcare organizations. Technol Forecast Soc Change. 2018;126(4):3–13. doi:10.1016/j.techfore.2015.12.019. [Google Scholar] [CrossRef]
140. Bekbolatova M, Mayer J, Ong CW, Toma M. Transformative potential of AI in healthcare: definitions, applications, and navigating the ethical landscape and public perspectives. Healthcare. 2024;12(2):125. doi:10.3390/healthcare12020125. [Google Scholar] [PubMed] [CrossRef]
141. Henry EU, Emebob O, Omonhinmin CA. Vision transformers in medical imaging: a review. arXiv:2211.10043. 2022. [Google Scholar]
142. Perera S, Navard P, Yilmaz A. SegFormer3D: an efficient Transformer for 3D medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2024 Jun 16–22; Seattle, WA, USA. p. 4981–8. [Google Scholar]
143. Bulatov A, Kuratov Y, Burtsev M. Recurrent memory transformer. Adv Neural Inf Process Syst. 2022;35:11079–91. [Google Scholar]
144. Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M. Transformers in vision: a survey. ACM Comput Surv (CSUR). 2022;54(10s):1–41. doi:10.1145/3505244. [Google Scholar] [CrossRef]
145. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2021 Oct 11–17; Montreal, QC, Canada. p. 10012–22. [Google Scholar]
146. Heidari M, Azad R, Kolahi SG, Arimond R, Niggemeier L, Sulaiman A, et al. Enhancing efficiency in vision transformer networks: design techniques and insights. arXiv:2403.19882. 2024. [Google Scholar]
147. Hamlomo S, Atemkeng M, Brima Y, Nunhokee C, Baxter J. Advancing low-rank and local low-rank matrix approximation in medical imaging: a systematic literature review and future directions. arXiv:2402.14045. 2024. [Google Scholar]
148. Kici D, Bozanta A, Cevik M, Parikh D, Başar A. Text classification on software requirements specifications using transformer models. In: Proceedings of the 31st Annual International Conference on Computer Science and Software Engineering; 2021 Nov 22–25; Toronto, ON, Canada. p. 163–72. [Google Scholar]
149. Xu Z, Liu R, Yang S, Chai Z, Yuan C. Learning imbalanced data with vision transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2023 Jun 17–24; Vancouver, QC, Canada. p. 15793–803. [Google Scholar]
150. Koetzier LR, Wu J, Mastrodicasa D, Lutz A, Chung M, Koszek WA, et al. Generating synthetic data for medical imaging. Radiology. 2024;312(3):e232471. doi:10.1148/radiol.232471. [Google Scholar] [PubMed] [CrossRef]
151. Shah D, Khan MAU, Abrar M, Amin F, Alkhamees BF, AlSalman H. Enhancing the quality and authenticity of synthetic mammogram images for improved breast cancer detection. IEEE Access. 2024;12:12189–98. doi:10.1109/access.2024.3354826. [Google Scholar] [CrossRef]
152. Shao R, Bi XJ. Transformers meet small datasets. IEEE Access. 2022;10:118454–64. doi:10.1109/access.2022.3221138. [Google Scholar] [CrossRef]
153. Denecke K, May R, Rivera-Romero O. Transformer models in healthcare: a survey and thematic analysis of potentials, shortcomings and risks. J Med Syst. 2024;48(1):23. doi:10.1007/s10916-024-02043-5. [Google Scholar] [PubMed] [CrossRef]
154. Maruthi S, Dodda SB, Yellu RR, Thuniki P, Reddy SRB. Language Model Interpretability-explainable AI methods: exploring explainable AI methods for interpreting and explaining the decisions made by language models to enhance transparency and trustworthiness. Australian J Mach Learn Res Appl. 2022;2(2):1–9. doi:10.23880/art-16000110. [Google Scholar] [CrossRef]
155. Pillai V. Enhancing transparency and understanding in AI decision-making processes. Iconic Res Eng J. 2024;8(1):168–72. [Google Scholar]
156. Bhati D, Neha F, Amiruzzaman M. A survey on explainable artificial intelligence (xai) techniques for visualizing deep learning models in medical imaging. J Imaging. 2024;10(10):239. doi:10.3390/jimaging10100239. [Google Scholar] [PubMed] [CrossRef]
157. Mir AN, Rizvi DR, Ahmad MR. Enhancing histopathological image analysis: an explainable vision transformer approach with comprehensive interpretation methods and evaluation of explanation quality. Eng Appl Artif Intell. 2025;149:110519. doi:10.1016/j.engappai.2025.110519. [Google Scholar] [CrossRef]
158. Li S, Sui X, Luo X, Xu X, Liu Y, Goh R. Medical image segmentation using squeeze-and-expansion transformers. arXiv:2105.09511. 2021. [Google Scholar]
159. Abiri N, Linse B, Edén P, Ohlsson M. Establishing strong imputation performance of a denoising autoencoder in a wide range of missing data problems. Neurocomputing. 2019;365(6):137–46. doi:10.1016/j.neucom.2019.07.065. [Google Scholar] [CrossRef]
160. Thakur RS, Chatterjee S, Yadav RN, Gupta L. Image de-noising with machine learning: a review. IEEE Access. 2021;9:93338–63. doi:10.1109/access.2021.3092425. [Google Scholar] [CrossRef]
161. Ma Q, Lee WC, Fu TY, Gu Y, Yu G. MIDIA: exploring denoising autoencoders for missing data imputation. Data Min Knowl Discov. 2020;34(6):1859–97. doi:10.1007/s10618-020-00706-8. [Google Scholar] [CrossRef]
162. Chow JC, Wong V, Li K. Generative pre-trained transformer-empowered healthcare conversations: current trends, challenges, and future directions in large language model-enabled medical chatbots. BioMedInformatics. 2024;4(1):837–52. doi:10.3390/biomedinformatics4010047. [Google Scholar] [CrossRef]
163. Blezek DJ, Olson-Williams L, Missert A, Korfiatis P. AI integration in the clinical workflow. J Digit Imaging. 2021;34(6):1435–46. doi:10.1007/s10278-021-00525-3. [Google Scholar] [PubMed] [CrossRef]
164. Rahman AA, Agarwal P, Noumeir R, Jouvet P, Michalski V, Kahou SE. Empowering clinicians with medical decision Transformers: a framework for sepsis treatment. arXiv:2407.19380. 2024. [Google Scholar]
165. Shokrollahi Y, Yarmohammadtoosky S, Nikahd MM, Dong P, Li X, Gu L. A comprehensive review of generative AI in healthcare. arXiv:2310.00795. 2023. [Google Scholar]
166. Gao Y, Zhou M, Liu D, Yan Z, Zhang S, Metaxas DN. A data-scalable transformer for medical image segmentation: architecture, model efficiency, and benchmark. arXiv:2203.00131. 2022. [Google Scholar]
167. Kocharekar AM, Datta S, Padmanaban, Rajakumar R. Comparative analysis of vision Transformers and CNN-based models for enhanced brain tumor diagnosis. In: 2024 3rd International Conference on Automation, Computing and Renewable Systems (ICACRS); 2024 Dec 4–6; Pudukkottai, India. p. 1217–23. [Google Scholar]
168. Sai S, Gaur A, Sai R, Chamola V, Guizani M, Rodrigues JJ. Generative AI for transformative healthcare: a comprehensive study of emerging models, applications, case studies and limitations. IEEE Access. 2024;12:31078–106. doi:10.1109/access.2024.3367715. [Google Scholar] [CrossRef]
169. Gonçalves T, Rio-Torto I, Teixeira LF, Cardoso JS. A survey on attention mechanisms for medical applications: are we moving toward better Algorithms? IEEE Access. 2022;10:98909–35. doi:10.21203/rs.3.rs-1594205/v1. [Google Scholar] [CrossRef]
170. Yang X, Bian J, Hogan WR, Wu Y. Clinical concept extraction using transformers. J Am Med Inform Assoc. 2020;27(12):1935–42. [Google Scholar] [PubMed]
171. Yu H, Fan L, Li L, Zhou J, Ma Z, Xian L, et al. Large language models in biomedical and health informatics: a review with bibliometric analysis. J Healthcare Inform Res. 2024;8(4):658–711. doi:10.1007/s41666-024-00171-8. [Google Scholar] [PubMed] [CrossRef]
172. Agarwal S, Peta SB. Balancing technology and privacy: securing patient data in healthcare under HIPAA regulations. TechRxiv. 2024. [Google Scholar]
173. Williamson SM, Prybutok V. Balancing privacy and progress: a review of privacy challenges, systemic oversight, and patient perceptions in AI-driven healthcare. Appl Sci. 2024;14(2):675. doi:10.3390/app14020675. [Google Scholar] [CrossRef]
174. Almeida D, Barr N. Innovations in health data protection ethical, legal, and technological perspectives in a global context: ai-powered diagnosis systems and health data innovation. In: Navigating Privacy, Innovation, and Patient Empowerment Through Ethical Healthcare Technology. Hershey, PA, USA: IGI Global Scientific Publishing; 2025. p. 171–96 doi:10.4018/979-8-3693-7630-0.ch007. [Google Scholar] [CrossRef]
175. Tang W, He F, Liu Y, Duan Y. MATR: multimodal medical image fusion via multiscale adaptive transformer. IEEE Trans Image Process. 2022;31(12):5134–49. doi:10.1109/tip.2022.3193288. [Google Scholar] [PubMed] [CrossRef]
176. Roy SK, Deria A, Hong D, Rasti B, Plaza A, Chanussot J. Multimodal fusion transformer for remote sensing image classification. IEEE Trans Geosci Remote Sens. 2023;61:1–20. doi:10.1109/tgrs.2023.3286826. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF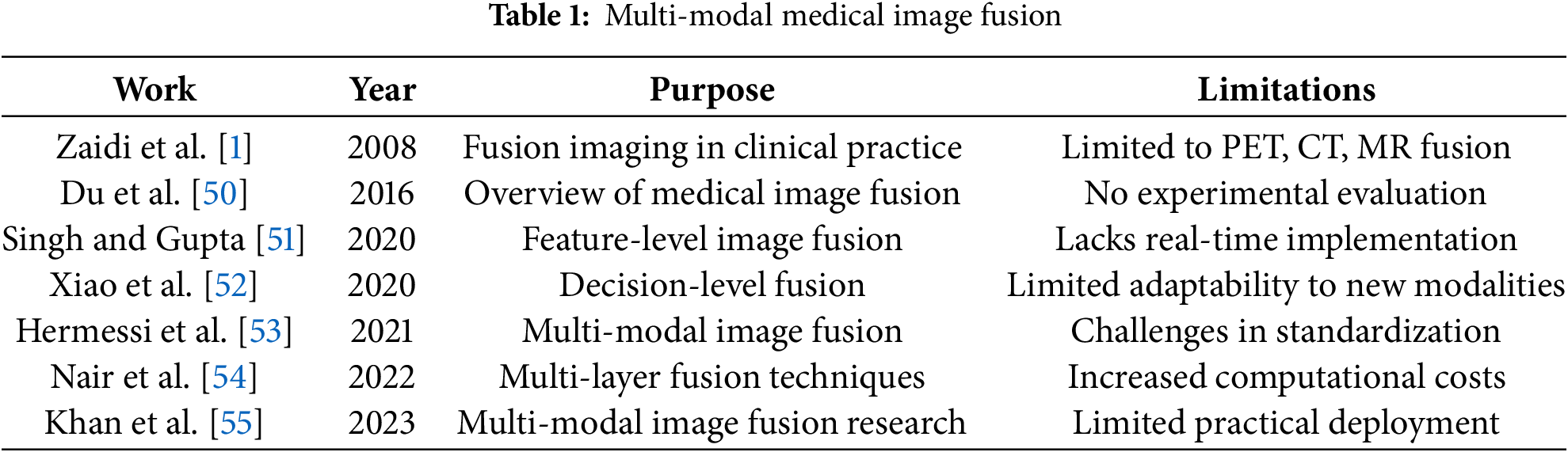
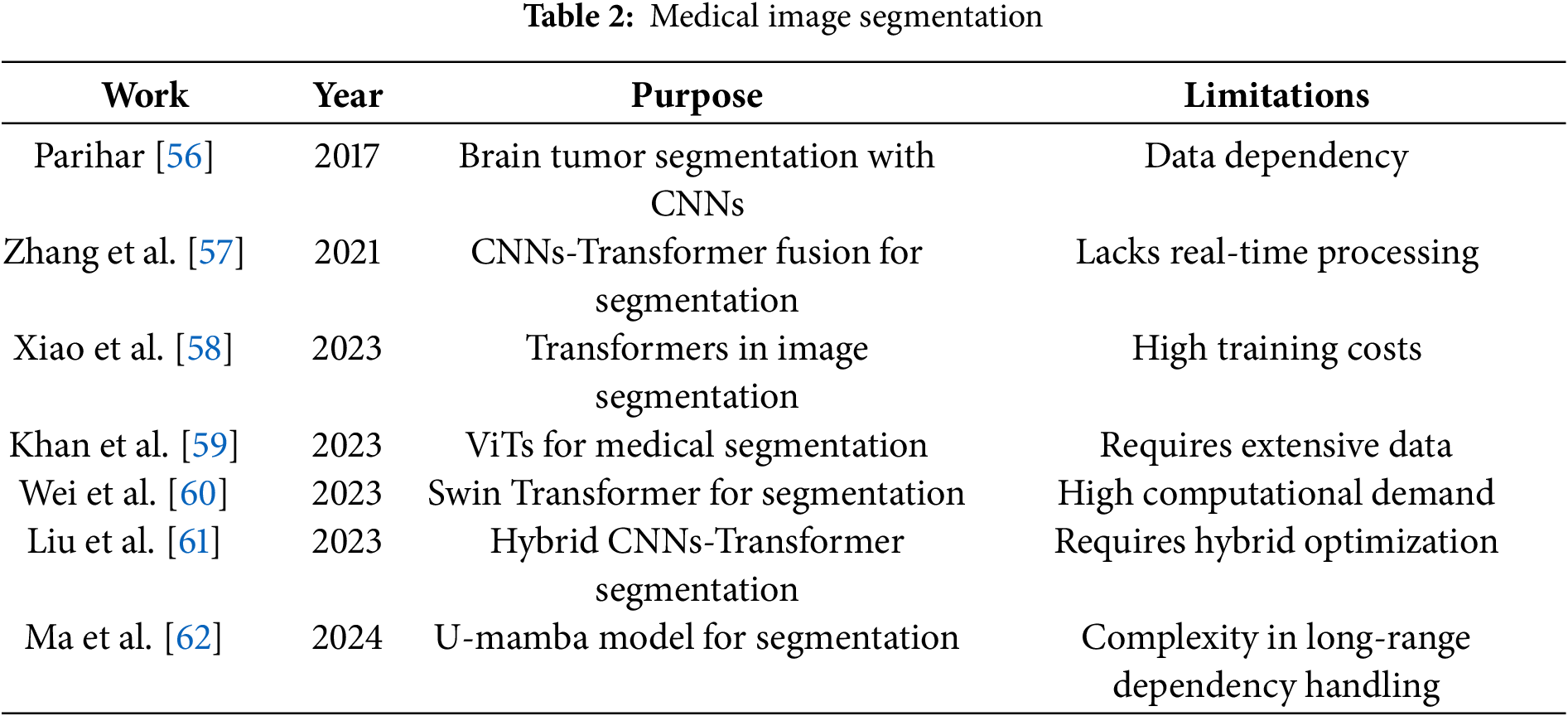
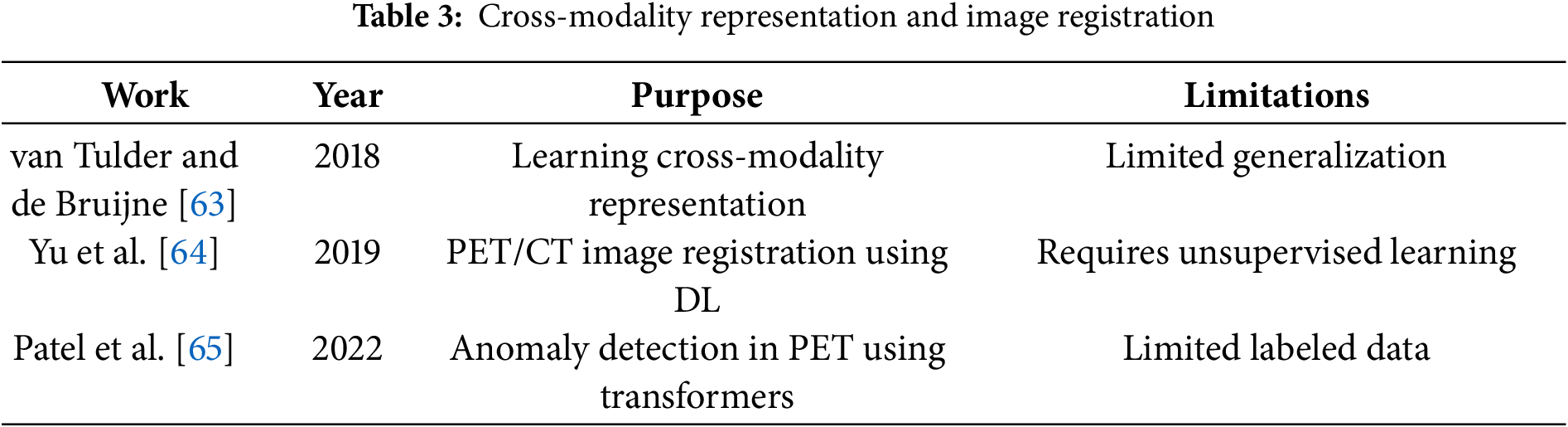
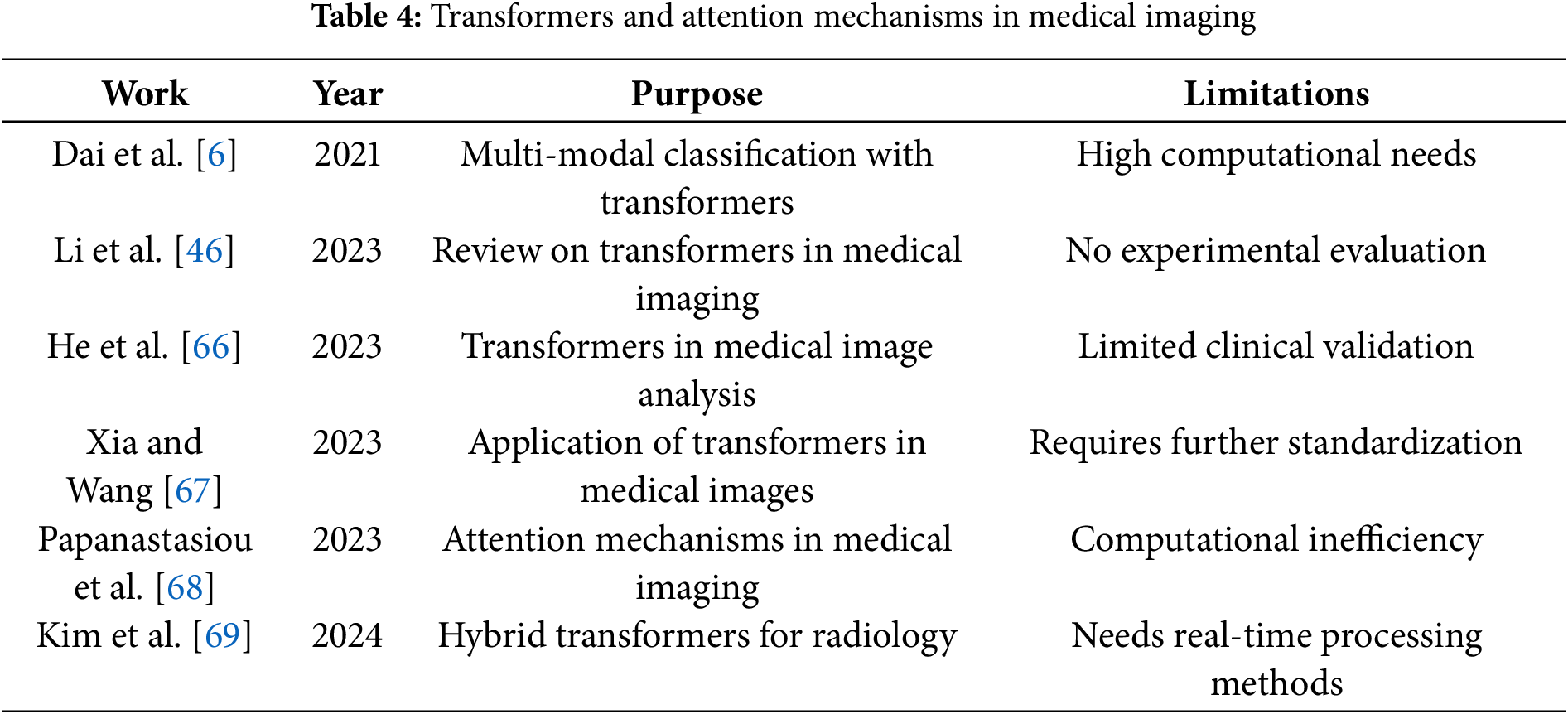
 Downloads
Downloads
 Citation Tools
Citation Tools