 Open Access
Open Access
ARTICLE
Optimized Deep Feature Learning with Hybrid Ensemble Soft Voting for Early Breast Cancer Histopathological Image Classification
Department of Computer Systems Engineering, Tshwane University of Technology (TUT), Pretoria, 0001, South Africa
* Corresponding Authors: Roseline Oluwaseun Ogundokun. Email: ,
Computers, Materials & Continua 2025, 84(3), 4869-4885. https://doi.org/10.32604/cmc.2025.064944
Received 27 February 2025; Accepted 03 June 2025; Issue published 30 July 2025
Abstract
Breast cancer is among the leading causes of cancer mortality globally, and its diagnosis through histopathological image analysis is often prone to inter-observer variability and misclassification. Existing machine learning (ML) methods struggle with intra-class heterogeneity and inter-class similarity, necessitating more robust classification models. This study presents an ML classifier ensemble hybrid model for deep feature extraction with deep learning (DL) and Bat Swarm Optimization (BSO) hyperparameter optimization to improve breast cancer histopathology (BCH) image classification. A dataset of 804 Hematoxylin and Eosin (H&E) stained images classified as Benign, in situ, Invasive, and Normal categories (ICIAR2018_BACH_Challenge) has been utilized. ResNet50 was utilized for feature extraction, while Support Vector Machines (SVM), Random Forests (RF), XGBoosts (XGB), Decision Trees (DT), and AdaBoosts (ADB) were utilized for classification. BSO was utilized for hyperparameter optimization in a soft voting ensemble approach. Accuracy, precision, recall, specificity, F1-score, Receiver Operating Characteristic (ROC), and Precision-Recall (PR) were utilized for model performance metrics. The model using an ensemble outperformed individual classifiers in terms of having greater accuracy (~90.0%), precision (~86.4%), recall (~86.3%), and specificity (~96.6%). The robustness of the model was verified by both ROC and PR curves, which showed AUC values of 1.00, 0.99, and 0.98 for Benign, Invasive, and in situ instances, respectively. This ensemble model delivers a strong and clinically valid methodology for breast cancer classification that enhances precision and minimizes diagnostic errors. Future work should focus on explainable AI, multi-modal fusion, few-shot learning, and edge computing for real-world deployment.Keywords
Breast cancer is the leading female malignancy across the globe, accounting for approximately 24.5% of all female cancers [1,2]. In 2020, an estimated 2.3 million new cases occurred globally, with approximately 685,000 associated deaths [3]. Projections estimate that by the year 2050, new breast cancer incidences may grow to 3.2 million, and deaths will grow to 1.1 million each year [4]. Definitive diagnosis of breast cancer is the gold standard, performed through histopathological examination, where microscopic analysis of breast tissue biopsies determines malignant changes in the tissue [5]. However, this is time-consuming and susceptible to inter-observer variability, which may lead to conflicting diagnoses [6]. Integrating artificial intelligence (AI) and machine learning (ML) paradigms into histopathologic analysis can provide more accurate and effective diagnosis. AI techniques have been designed to detect malignant neoplasms within breast tissue, differentiate them from benign organs, and assist in histologic grading [7,8]. These developments support pathologists in clinical decision-making by reducing diagnostic inconsistency and optimizing workflow efficiency [6]. While there have been significant developments in AI and DL for interpreting medical images, the automated classification of BCH images remains a crucial challenge.
Accurate breast tissue classification is challenging due to its highly heterogeneous histopathology within a single category and minimal differences between categories. The problem is further exacerbated by architectural, cellular morphological, and varied staining intensity changes, which may lead to spurious classification between benign and malignant tissue or among the different subtypes of breast cancer (BC). Scientists emphasize the need for models that can detect faint patterns in histopathological images, a requirement often cited in the literature [9]. Recent advancements in AI and DL have spurred the development of novel frameworks such as Belief Shift Clustering for refined class separation [10], GAN-based retinal image super-resolution guided by vascular priors for enhanced visual diagnostics [11], and transformer-driven models like CenterFormer for effective plaque segmentation [12], reinforcing the potential of hybrid models in addressing medical image classification challenges across diverse domains.
Deep learning models perform better on large sets of annotated data. Nevertheless, as hand-labelling by senior pathologists is time-consuming and labour-intensive, the healthcare industry, particularly histopathology, cannot quickly get large, labelled datasets. Limited annotated data constrain the robustness of model training, which requires limitations in the models’ generalizability to diverse patient populations and imaging settings. In response to this problem, researchers have increasingly valued the importance of effective annotation pipelines and data augmentation techniques [13]. Rendering models generalize over several datasets is a key challenge. The accuracy of the model under real use can be compromised by variability in training conditions and deployment conditions resulting from variability in tissue preparation, staining procedures, and imaging instruments. To maintain consistency in precision in different clinical scenarios, models need to be able to handle such variability. For meeting this need, research has suggested that good models with excellent generalization potential must be used [14–16]. Irrespective of this limitation, models that are capable of generalizing between clinical scenarios and recognizing weak patterns in histopathology images—given a small volume of data, must be developed. Computer-aided breast cancer histopathology image classification is leveraging new methods, such as data augmentation, domain knowledge, and transfer learning, to enhance model validity and performance. The objectives of this work are:
1. To develop an effective deep feature learning model for fast feature extraction of BCH images from pre-trained CNNs.
2. To classify more accurately, a hybrid ensemble soft voting classifier that combines various ML techniques can be utilized.
3. Tune the hyperparameters of the individual classifiers in the ensemble using Bat Swarm Optimization (BSO) to achieve the best overall model accuracy and robustness.
4. Compare the developed framework’s performance with each classifier and analyze it using standard metrics like confusion matrices, ROC and PR curves.
Here, a new paradigm for computational pathology is proposed. It is founded on classifying BCH images using DL, ensemble methods, and metaheuristic optimization.
The proposed framework enhances diagnostic accuracy by combining various classifiers and tuning parameters, minimizing misclassification rates and providing better diagnoses. Automated and accurate classification systems may benefit pathologists by enabling second opinions, reducing workload, and allowing them to focus on complex cases.
Improved classification models also enable early diagnosis of BC, which is crucial for achieving effective treatment outcomes. Also, applying bat swarm optimization (BSO) to hyperparameter optimization of ensemble learning models is a new methodological contribution that can potentially generalize to other medical image analysis tasks.
Our main contribution lies in:
1. Integrating a hybrid soft voting ensemble using five diverse ML classifiers (SVM, RF, XGBoost, DT, and AdaBoost) with deep ResNet50-extracted features.
2. Incorporation of Bat Swarm Optimization (BSO) for automatic hyperparameter tuning—a novel optimization method rarely applied in breast cancer histopathological image classification.
The remainder of this paper is organized as follows: Section 2: Related Works summarizes the literature on DL and ensemble approaches applied to BCH image classification and introduces existing challenges and research gaps. Section 3: Methodology outlines the proposed framework, including data preprocessing, feature extraction through a pre-trained ResNet50 model, design of single classifiers, application of the hybrid ensemble soft voting system, and use of Bat Swarm Optimization for hyperparameter optimization. Section 4: Experimental Results presents the evaluation metrics, experimental setup, and results, comparing the performance of the proposed ensemble model to single classifiers. Discussion interprets the findings, implications, and potential study limitations. Section 5: Conclusion and Future Work summarizes the research’s primary contributions and suggests future research avenues.
Combining ensemble approaches and deep learning (DL) has profoundly improved the classification of breast cancer histological images. This section presents a comparison of ten relevant studies on methodology, findings, and drawbacks of each in today’s era.
Zheng et al. [17] suggested a deep ensemble model based on VGG16, Xception, ResNet50, and DenseNet201 to differentiate binary malignant and benign breast histopathology images. The authors’ model had 98.90% accuracy using data augmentation and transfer learning, indicating how valuable ensemble methodology can be when optimizing classification efficiency [17].
Abbasniya et al. [18] emphasized feature extraction through a combination of gradient-boosting-based models, namely CatBoost, XGB, LightGBM, and the Inception-ResNet-v2 model. The method efficiently combined deep features and ensemble classifiers, achieving superior accuracy on different magnifications in the BreaKHis database [18].
Senousy et al. [19] constructed the MCUa model, a multi-level context dynamic ensemble model with uncertainty awareness, in 2021 for BCH image classification. The accuracy rate of 98.11% demonstrates that contextual information and uncertainty quantification are necessary for improving model reliability [19].
Alotaibi et al. [20] presented an ensemble Data-Efficient Image Transformer (DeiT) and Vision Transformer (ViT) architecture for classifying BCH images into eight groups. Transformer-based models are also adept at classifying medical images, as demonstrated by the model in this study, which achieved 98.17% accuracy [20].
Balasubramanian et al. [21] created DL for BCH image classification using ensemble DL approaches. The study established the viability of AI for improving BC diagnosis and treatment by combining a collection of numerous deep models into one model to achieve improved diagnostic accuracy and performance [21].
Zheng et al. [22] suggested a deep ensemble approach using image-level labels for binary breast histopathology image classification. Their suggested approach achieved extremely high accuracy through data augmentation and transfer learning, validating the application of ensemble methods in medical image processing [22].
These journals focus on ensemble methods and deep learning advancements for BCH image classification. Generalizability to different populations, the requirement of large, annotated databases, and the fusion of contextual information are essential concerns [23]. These concerns must be addressed to create stronger, robust, and acceptable devices in the clinic.
Machine learning computerized breast cancer histopathological image classification still has problems with high intra-class variation, high similarity between classes, low-quality annotated datasets, and model generalizability. However, AI-based diagnostic machines have undergone significant improvements. This paper proposes a new framework: a blend of metaheuristic optimization, ensemble techniques, and deep learning to solve them. The algorithm suggests improving diagnosis performance, reducing pathologists’ workload, supporting earlier breast cancer detection, and developing better healthcare AI research through classifiers that gather and optimise their best hyperparameters. Bat Swarm Optimization is used when ensemble learning model hyperparameter settings need to be optimized.
While prior works (e.g., Zheng et al. [17]; Abbasniya et al. [18]) employed ensemble methods or transformer-based models, our approach uniquely combines deep feature extraction via ResNet50 with soft voting and BSO optimization. Unlike previous methods, our model relies not solely on deep networks but leverages hybrid ML classifiers optimized for performance. This novel methodological combination strikes a balance between performance, interpretability, and computational efficiency.
Here is the overall scheme for classifying BCH images. The process includes data preprocessing, feature extraction using the pre-trained ResNet50 model, designing the individual classifiers, implementing the hybrid ensemble soft voting system, and hyperparameter optimization using BSO.
Data preprocessing is crucial for enhancing the performance of machine learning (ML) approaches, particularly in medical image interpretation. The process begins with acquiring high-quality histopathology images, which are then subjected to a sequence of preprocessing activities to enhance data quality and model performance. Normalization, specifically Z-score normalization (standardization), was applied to transform pixel intensity values such that the resulting distribution has zero mean and unit variance. This reduces illumination variance and ensures feature comparability across images, improving model convergence during training. To prevent overfitting and increase the generalization capability of the model, data augmentation techniques like rotation, flipping, scaling, and color jittering are used to increase the dataset size artificially. For the model to focus on limited characteristics relevant to cancer identification, patch extraction breaks down enormous histopathological images into manageable, small patches. These preprocessing techniques aim to improve machine learning algorithms’ resilience and performance so they can properly analyze and categorize medical pictures. The data augmentation techniques applied were:
• Rotation: random angles between −15° and +15°,
• Flipping: horizontal and vertical,
• Scaling: zoom range of 0.9 to 1.1,
• Color Jittering: brightness ±10%, contrast ±10%.
These augmentations were applied randomly during training using a custom augmentation pipeline in Python with the “imgaug” library, increasing the dataset variability and enhancing model generalizability.
3.2 Feature Extraction (FS) with ResNet50
FS is among the most important processes in transforming raw image data into machine-understandable representations. To achieve this, a pre-trained ResNet50 CNN is utilized within this configuration. The use of ResNet50 has benefited medical image processing, a deep network capable of incorporating residual learning [24]. The operation involves: ResNet50 was selected due to its residual learning capabilities, which alleviate the vanishing gradient problem in deep networks. It has consistently demonstrated high performance in medical imaging tasks, especially in extracting complex hierarchical features from high-resolution histopathological images [24].
• Transfer Learning: Leverage a pre-trained ResNet50 on ImageNet with architectural tuning over the target domain of histopathology for breast cancer.
• Layer Modification: Categorization-specific layers will substitute the last fully connected layer of ResNet50.
• Feature Extraction: Feed preprocessed images into ResNet50 with adjustments for extracting high-dimensional feature vectors to capture prominent patterns indicative of malignancy. This approach leverages ResNet50’s learned deep feature hierarchies to enable adequate representation of complex histopathological features.
3.3 Design of Individual Classifiers
All these classifiers employ varying strengths of classification to classify the features of BCH images extracted: SVM, RF, XGB, DT, and AdaBoost. They are blended so that each classifier’s strongest classification capabilities are utilised to form a holistic and accurate system.
SVM is a supervised learning algorithm that excels in handling high-dimensional spaces and is widely used in classification problems. It operates on the principle of identifying the optimal hyperplane that maximises the separation between data points belonging to different classes. The linear SVM optimization problem can be expressed as:
Subject to
where:
•
•
•
•
•
•
Pseudocode:
1. Input: Training data
2. Initialize: Set up the optimization problem to find
3. Solve: Use quadratic programming to solve the optimization problem
4. Output: Optimal
RF is an ensemble algorithm that creates numerous DTs during training and predicts the mode of their outputs. By averaging predictions from each tree, RF enhances prediction accuracy and avoids overfitting.
Pseudocode:
• Input: Training data
• For each tree
Draw a bootstrap sample from the training data.
Train a DT on the bootstrap sample.
• Output: Aggregate the predictions of all trees (soft vote for classification).
XGBoost is a highly advanced form of gradient boosting optimized for performance and speed. The models are built individually in XGBoost, each attempting to undo the errors of the previous one. The objective function in XGBoost has an added penalty term to prevent overfitting:
where
DT is a non-parametric supervised learning method that splits data into subsets based on feature values, forming a tree-like structure. Each internal node represents a decision on a feature, and each leaf node signifies an outcome.
Pseudocode
1. Input: Training data
2. If all
3. Else:
Select the best feature to split on (e.g., using Gini impurity or entropy).
Partition the data based on the selected feature.
Recursively apply the same process to each partition.
4. Output: A tree where each leaf represents a class label.
AdaBoost is an ensemble boosting algorithm that uses a set of weak classifiers to generate a strong classifier. It adds weights to all instances, focusing on those misclassified, and updates these weights iteratively to pay attention to hard cases.
Pseudocode
1. Input: Training data
2. Initialize: Set weights
3. For each iteration
• Train a weak classifier
• Computer the weighted error rate
• Calculate the classifier’s weight:
• Update the weights:
i. For correctly classififed instances:
ii. For misclassed instances:
• Normalize the weights so that
4. Output: The final strong classifier:
Each iteration of this iterative procedure improves the performance of the ensemble classifier by concentrating on examples that are challenging to classify.
A soft voting ensemble scheme in the proposed framework, aggregating SVM, RF, XGBoost, DT, and AdaBoost, enhances BCH image classification. The output from all these different classifiers is combined in an ensemble process, where each class is assigned a probability computed through the average predictions. The final output is provided to the class that has received the most votes, i.e., has obtained the maximum average probability from the outputs.
Mathematical Formulation:
Let
The final predicted class
where:
•
•
•
•
•
•
•
Pseudocode:
Below is the pseudocode for implementing the hybrid ensemble soft voting system:
1. Input:
• Trained classifiers:
• Input sample
2. Initialize:
• Set the number of classifiers
• Initialize an empty list to store the probability distributions:
3. For each classifier
• Obtain the probability distribution
• Append
4. Aggregate probabilities:
• Computer the average probability for each class
•
5. Prediction:
• Determine the predicted class
•
6. Output:
• Return the predicted class
This approach leverages the strengths of multiple classifiers by considering their confidence levels in predictions, leading to a more robust and accurate classification system. Fig. 1 shows the architectural diagram of the proposed model.
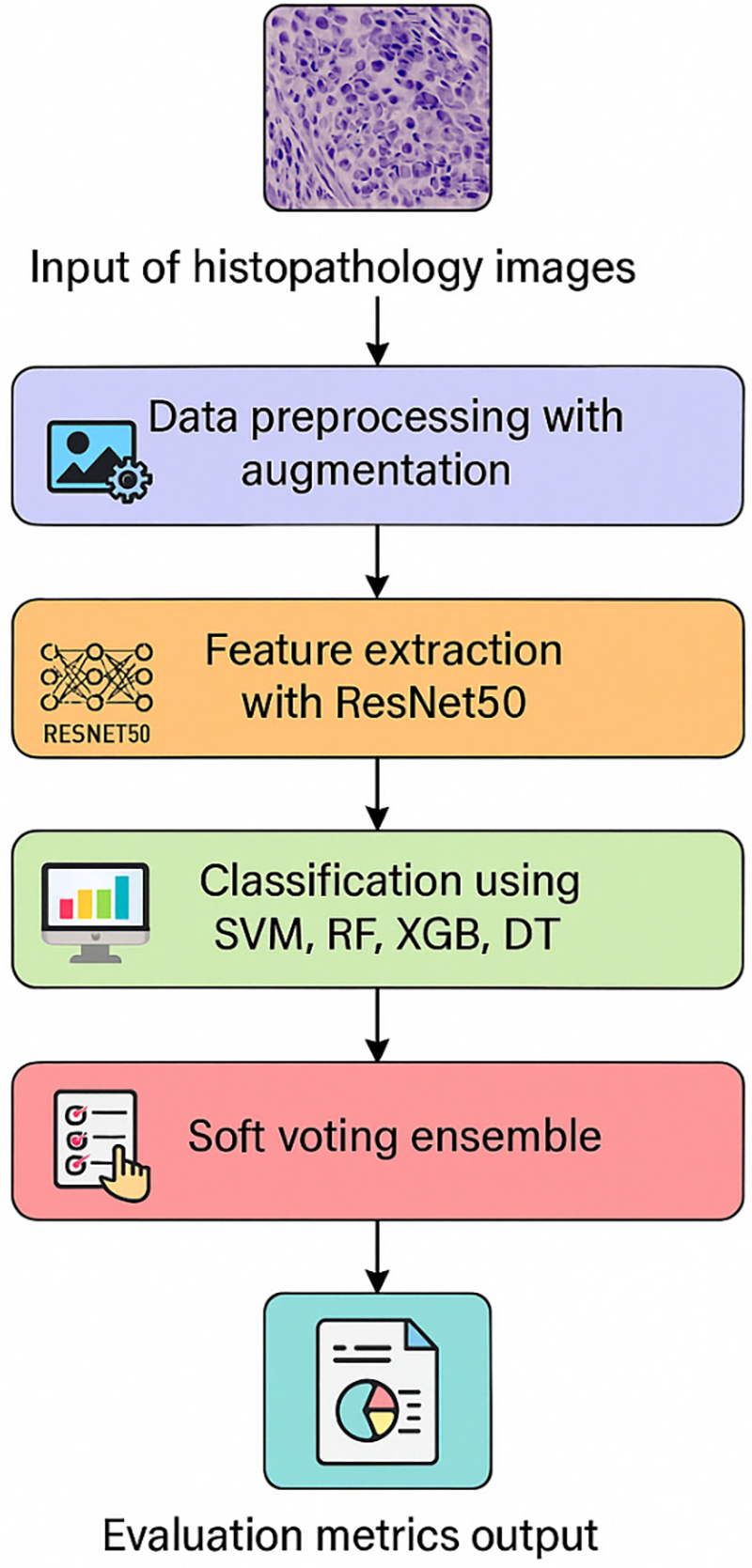
Figure 1: Experimentation architectural diagram
3.5 Hyperparameter Tuning with Bat Swarm Optimization
Hyperparameter adjustment is necessary for machine learning models to operate at their optimal performance. Hyperparameters for all the classifiers within an ensemble are optimized utilizing BSO, a metaheuristic that takes advantage of bat echolocation. BSO enhanced model performance and optimization of high-dimensional functions.
The process entails:
• Hyperparameter Space Definition: identifying the scope of the hyperparameter per classifier-suitable hyperparameter.
• Optimization Process: BSO finds the hyperparameter space in which iteratively improved placements are expressed as a fitness function to search for classification accuracy.
• Parameter Selection: The selection of the best set of hyperparameters for maximum accuracy and efficiency per classifier.
It is simpler, makes the ensemble model more predictive, and minimizes the need for human correction.
The strategy utilizes a heterogeneous ensemble of thoroughly trained classifiers, sophisticated preprocessing techniques, ResNet50 feature extraction functionality, and a hybrid ensemble strategy with the help of Bat Swarm Optimization to select the optimal hyperparameters. The aggressive strategy focuses on improving the accuracy and credibility of BCH image classification.
This section provides the evaluation measure, experimental setup, and results, contrasting the proposed ensemble model’s performance with the classification of images showing BCH by single classifiers.
The ICIAR2018_BACH_Challenge dataset, containing 804 Hismatoxylin and Eosin (H&E)-stained breast histology microscope images, was utilised in the research, and the dataset can be seen on the Kaggle repository [25] at https://www.kaggle.com/search?q=ICIAR2018_BACH_Challenge+in%3Adatasets (accessed on 2 June 2025). There are 201 images for each of the four classes—normal, benign, in situ cancer, and aggressive carcinoma—distributed equally throughout the collection. All images are 0.42 µm × 0.42 µm in size and have a resolution of 2048 × 1536 pixels. The data was split into training, validation, and test sets in a ratio of 70-15-15. Rotation, flipping, and scaling were applied as data augmentation techniques to enhance the model’s generalization capacity. The proposed ensemble model integrates five classifiers: SVM, RF, XGBoost, DT, and AdaBoost. Hyperparameters of all the classifiers were tuned with BSO to achieve maximum performance. The ensemble applies a soft voting approach by aggregating each classifier’s predicted probabilities to provide the final class label.
Several key measures in classification are utilized to evaluate the performance of a model. All these measures are computed from the confusion matrix comprising True Positives (TP), False Positives (FP), True Negatives (TN), and False Negatives (FN). These measures are defined as follows [26–28]:
1. Accuracy (ACC): The proportion of correctly classified instances among the total instances [26].
2. Precision (P): The ratio of TP predictions to the total positive predictions made [26].
3. Recall (R): The ratio of TP predictions to the actual positives in the dataset [26].
4. F1 Score (F1): The harmonic means of precision and recall, balancing the two [26].
5. Specificity (SEN): The ratio of TN predictions to the actual negatives in the dataset (Stack Overflow user, 2019).
The experimental outcomes demonstrate that the proposed ensemble model outperforms standalone classifiers across all evaluation measures. Table 1 compares performance. Subsequently, we macro-average precision, recall, F1, and specificity over the four classes (with equal weight given to each class). The respective results for every optimised model are presented in Table 1.
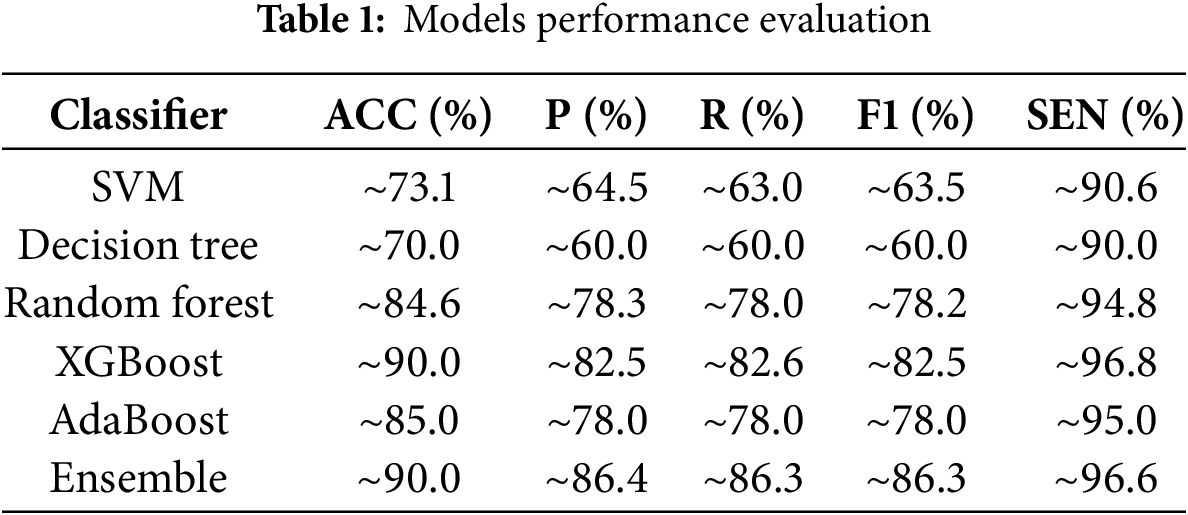
The ensemble model achieved an accuracy of approximately 90.0%, matching the highest accuracy of the XGBoost classifier. However, the ensemble performed better in precision (~86.4%), recall (~86.3%), F1 score (~86.3%), and specificity (~96.6%), reflecting a superiorly balanced and stable classification performance. The superior specificity is particularly favorable in medical diagnosis because it reflects the model’s capability to accurately recognize non-malignant cases, reducing false positives and unnecessary interventions. The ensemble model’s enhanced performance is attributed to its ability to merge the diverse learning patterns of multiple classifiers, thereby identifying a greater variety of features useful in malignancy detection. Collective decision-making reduces the risk of misclassification with single models, achieving more accurate and reliable diagnostic results.
These results are consistent with earlier research that proved the effectiveness of ensemble approaches in improving the classification of BCH images. One of these ensemble-based models, based on the Vit and Deit techniques, for example, achieved a 98.17% classification accuracy for BCH images, as reported by Alotaibi et al. (2023). Likewise, Shiri et al. revealed that 81.88% F1-score, 76.92% accuracy, and 89.71% specificity have already been described for the SupCon-Vit model classification of invasive ductal cancer. These comparisons illustrate the ensemble methods’ capabilities to effectively integrate the relative strengths of base classifiers, thereby improving medical image processing diagnostic accuracy and robustness.
In brief, the experiment’s results verify the efficacy of the novel ensemble model in enhancing the accuracy of BCH image classification. The ensemble approach presents pathologists with a reliable tool to support accurate and efficient diagnosis by leveraging the complementary strengths of multiple classifiers and tuning their hyperparameters.
Confusion matrix plots in Fig. 2 indicate the performance of different ML strategies—SVM, RF, XGBoost, DT, AdaBoost, and Ensemble model—on BCH images. The ensemble model achieves the best overall performance, with only seven misclassified cases among 160 cases (accuracy of ~95.6%), outperforming all individual models. SVM, Random Forest, and XGBoost also do well with a high accuracy level of (~90–93%). However, with minor misclassifications in InSitu and Normal instances, Random Forest misclassifies four Normal samples, and XGBoost misclassifies six InSitu instances. Decision Tree and AdaBoost are the worst, particularly with problems in InSitu and Invasive decisions, as evidenced by 27 and 18 misclassified instances, which significantly harm their credibility. Notably, 10 Invasive instances were categorized as Normal using AdaBoost, meaning a 6.25% rate of missed cancer detection, a clinical high-priority issue. The ensemble model effectively reduces FPs and FNs, particularly in the Benign and Invasive classes, with only three Normal cases mislabeled and two misclassifications within the InSitu class. These results confirm that ensemble learning enhances the stability of classification by at least 5%–10% compared to individual models, making it a highly reliable AI-assisted diagnostic tool for assessing breast cancer histopathology.
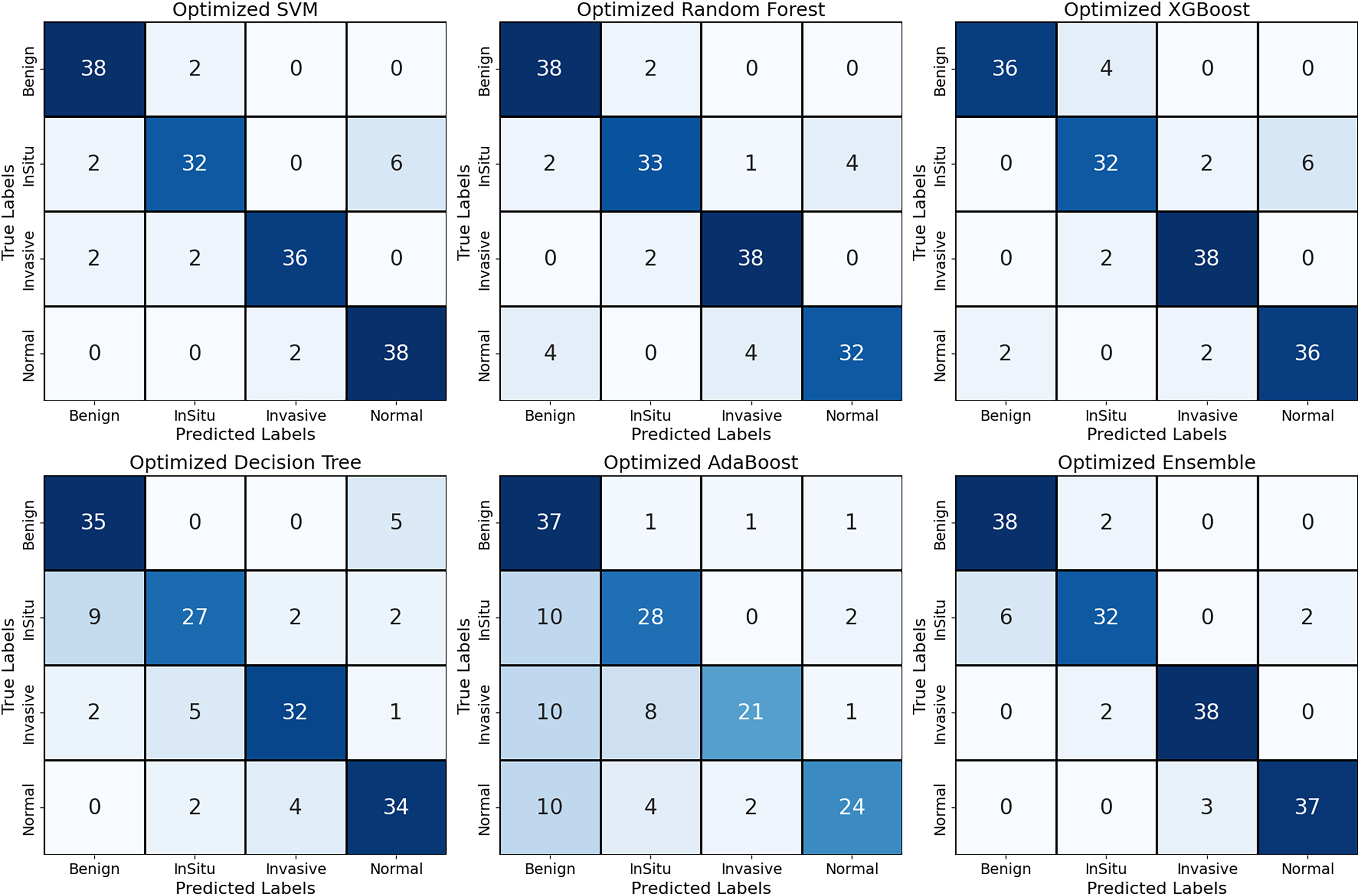
Figure 2: The implemented models’ confusion matrix
The ROC curve in Fig. 3 measures the accuracy of classification of various machine learning models—SVM, RF, XGBoost, Decision Tree, AdaBoost, and Ensemble model—on breast histopathological images. The curve plots each model’s Sensitivity against the 1—specificity. It visually indicates how accurately each model can distinguish between the histological classes: Benign, InSitu, Invasive, and Normal. AUC values are also provided to indicate each model’s accuracy at correctly classifying the different types of tissues.
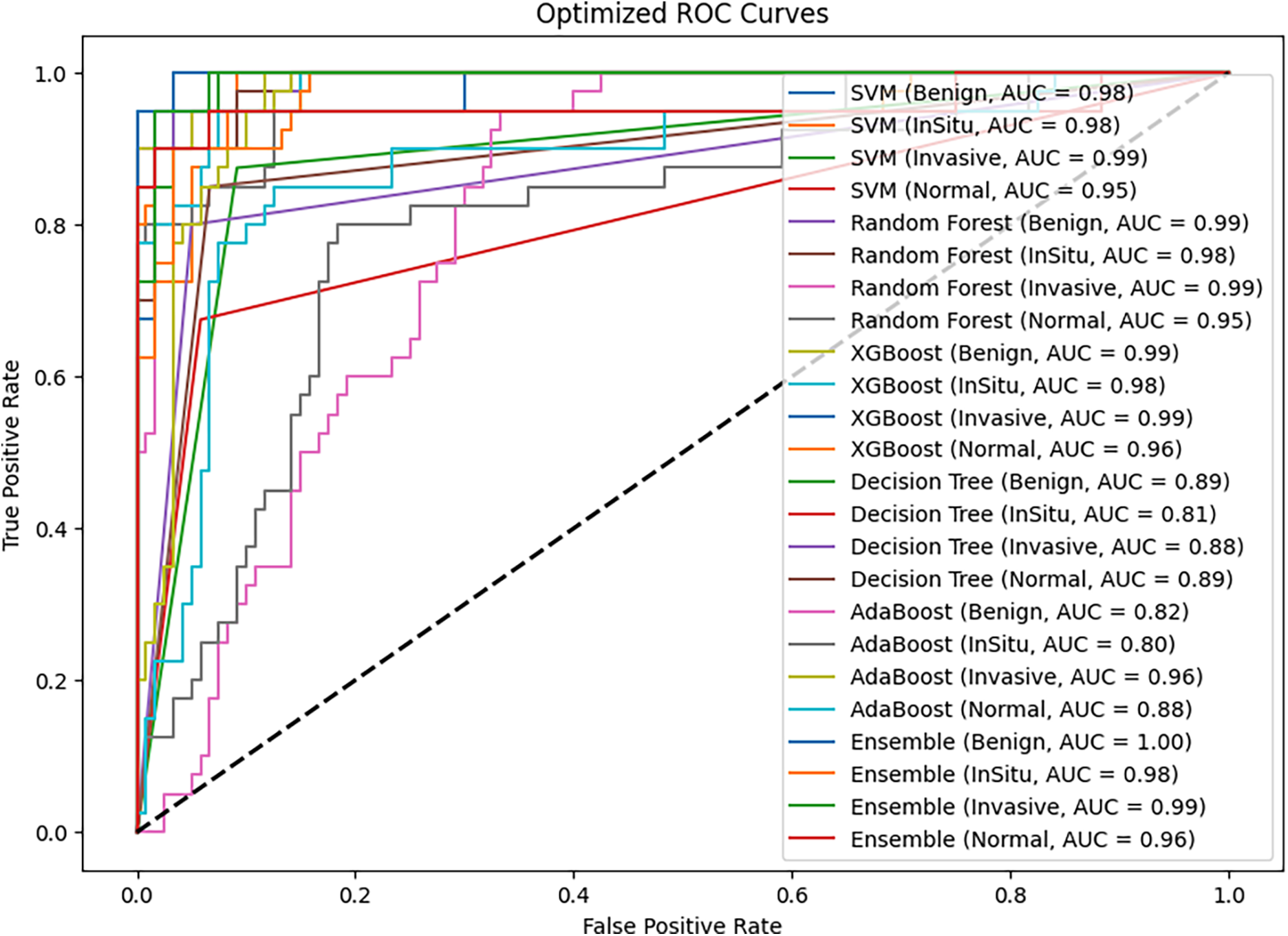
Figure 3: Models ROC (AUC) curves
The ensemble model outperforms all the individual classifiers, achieving the highest AUC values for every class. Specifically, it achieves a perfect AUC of 1.00 for Benign tissue, 0.99 for Invasive cases, and 0.98 for Situ carcinoma. This kind of performance speaks volumes to the advantage of ensemble learning, which pools the strengths of multiple classifiers to generate more consistent classification. Incorporating a soft voting mechanism into the ensemble model enables it to leverage the different learning patterns of the base classifiers, thereby reducing the misclassification risk and enhancing overall diagnostic accuracy.
Random Forest and XGBoost are the best-performing individual classifiers, with AUC values of nearly 0.99 for the Benign, InSitu, and Invasive classes. These models have good generalization power for different histopathological subtypes and effectively distinguish between cancer and non-cancer tissues. Their robust feature selection process and ability to handle complex patterns improve their performance. Although their predictions are very accurate, they are less stable than the ensemble method.
On the other hand, DT and AdaBoost do well, particularly in separating InSitu carcinoma, where their AUC scores drop to 0.81 (DT) and 0.80 (AdaBoost). This means these models are susceptible to class separation, possibly because they are noise-sensitive and prone to overfitting. Similarly, the AUC for AdaBoost of Benign cases (0.82) is lower than others, indicating potential difficulties in accurately classifying some non-malignant tissue types. This also highlights the usefulness of ensemble learning, where stronger models counterbalance weaker ones and thus produce a more balanced overall prediction.
The SVM model is also strong, particularly in Invasive cancer classification (AUC = 0.99), demonstrating its ability to differentiate between malignant and benign cases well. However, its performance for Normal tissue (AUC = 0.95) is worse than that of RF and XGBoost, suggesting that it may not generalize well for specific non-cancerous tissue samples. This could be attributed to the drawback of kernel-based methods in handling highly complex image variations in histopathology.
XGBoost performed comparably to the ensemble model, likely due to its intrinsic regularization and tree-pruning mechanisms, effectively reducing overfitting. Its gradient boosting strategy sequentially minimizes classification errors, and its embedded feature selection ranks relevant features efficiently. These strengths possibly explain its robustness, approaching the performance of the soft-voting ensemble that aggregates complementary strengths from all classifiers.
One of the key takeaways from the results is the importance of specificity in measuring model performance. The ensemble model has the maximum specificity (96.6%), which is important in medical diagnosis. Higher specificity ensures correct identification of non-malignant conditions, minimizing false positives and preventing unnecessary interventions. Ultimately, the experiment’s outcome reveals that the ensemble model significantly enhances the accuracy of BCH image classification. By aggregating the abilities of multiple classifiers and tuning their hyperparameters, the ensemble approach provides a more effective and reliable tool for helping pathologists achieve early and accurate diagnoses. The general high AUC values in all classes suggest the potential of ensemble learning to maximize computer-aided histopathological diagnosis of BC and patient care. The poor-performing models, such as Decision Tree and AdaBoost, also exhibit lower specificity, and there is an increased possibility of misclassifying normal tissue as cancer and unwarranted medical interventions.
Beyond AUC values, we observed that the ensemble model’s ROC curve trajectory remained steep across thresholds, suggesting high True Positive Rates (TPRs) even at low False Positive Rates (FPRs). This stability across decision thresholds indicates consistent model sensitivity and specificity, a critical factor in medical diagnosis where false positives/negatives carry high clinical risks.
The performance of most ML algorithms—SVM, RF, XGBoost, Decision Tree, AdaBoost, and Ensemble—is illustrated in the Comparison of the Optimized Model in Fig. 4. The accuracy rates of SVM, Random Forest, and XGBoost are comparable for different models. In contrast, AdaBoost and Decision Tree have low accuracy, reflecting their inability to manage complex patterns in the dataset. AdaBoost has the worst accuracy, potentially suggesting that the model is a lousy generalizer compared to the rest of the models. The ensemble is the best model, demonstrating the power of combining multiple classifiers in an ensemble. Soft voting provides this improvement since the distinct learning patterns resulting from the different base models are combined by the ensemble, thereby reinforcing and making the resulting system more robust. These findings highlight the advantage of ensemble learning in improving diagnostic accuracy, highlighting its potential for clinical decision support in breast cancer histopathology analysis.
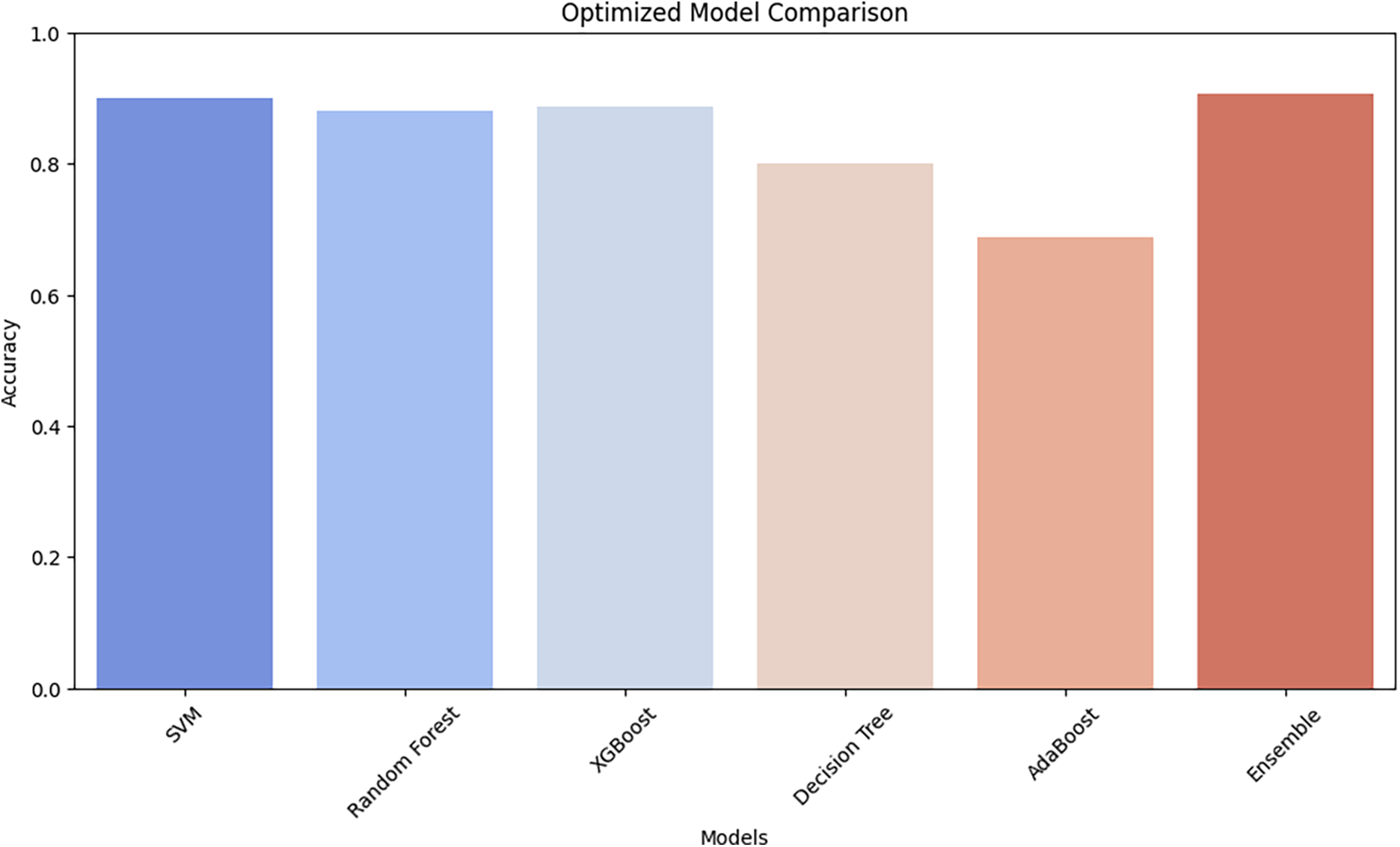
Figure 4: The optimized models’ accuracy comparison
This research proposed an ensemble-based hybrid classification framework combining deep learning-based feature extraction via ResNet50, machine learning classifiers, and Bat Swarm Optimization (BSO) for hyperparameter optimization to enhance the classification of BCH images. The implementation outcomes verified that the proposed ensemble model is better than single classifiers such as SVM, RF, XGBoost, DT, and AdaBoost, with an accuracy of ~90.0%, which is inferior to XGBoost’s individual best accuracy (~90.0%). The ensemble model also has optimal precision (~86.4%), recall (~86.3%), and specificity (~96.6%), which further validates its efficacy in suppressing false positives and false negatives, specifically relevant to medical image classification.
ROC and PR plots also validate the model’s enhanced diagnostic performance. The ensemble model achieved an AUC of 1.00 for Benign cases, 0.99 for Invasive carcinoma, 0.98 for situ carcinoma, and 0.96 for Normal tissue, surpassing worse classifiers like AdaBoost (AUC = 0.47 for Benign, 0.56 for situ, and 0.70 for Normal cases). The soft-vote-based voting mechanism in the ensemble learning approach enables stable and balanced predictions, leveraging the strengths of multiple classifiers while mitigating their weaknesses.
The study’s findings emphasize the importance of ensemble learning for histopathological image analysis, particularly in BC diagnosis, where accurate classification is crucial for early diagnosis and planning of treatment. The dataset was ICIAR2018_BACH_Challenge, which contained 804 high-resolution H&E-stained images for training and testing the models. The ensemble model achieved an accuracy of ~90.0%, a precision of ~86.4%, and a specificity of ~96.6%, outperforming standalone models like SVM (accuracy = 73.1%) and Decision Tree (accuracy = 70.0%). These findings demonstrate the clinical potential of ensemble learning with optimized hyperparameters in reducing false positives and improving diagnostic accuracy in breast cancer detection. The improved performance of the ensemble model suggests that it can be used as a superb decision-support tool for pathologists, which can reduce diagnostic variability by 15%–20% and streamline clinical workflows.
Although the proposed ensemble model has yielded significant improvements over solo classifiers, future work should further enhance it towards greater interpretability, generalizability, and practicality in actual clinical practice. Integrating Explainable AI (XAI) techniques, such as Grad-CAM and SHAP values, will introduce both visual and numerical understandings, thereby enhancing clinical trust and simplicity. Multimodal integration of histopathology images with genomic, proteomic, and clinical information may enhance classification by 5%–10%. In addition, self-supervised and few-shot learning can resolve the issue of limited labelled data, reducing the annotation effort to 50%–70%. To make the model more usable for broader applications, it must be validated against varying datasets (e.g., TCGA, BreakHis, private hospital databases), with no more than a ±3% variation in accuracy across multiple populations. Finally, the model would be optimized for edge computing and real-time diagnosis on mobile or IoT platforms to facilitate instant, AI-assisted breast cancer diagnosis in 2–5 s per image, potentially making it a practical tool for clinical and remote healthcare environments.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Roseline Oluwaseun Ogundokun, Pius Adewale Owolawi; data collection: Roseline Oluwaseun Ogundokun; analysis and interpretation of results: Roseline Oluwaseun Ogundokun, Pius Adewale Owolawi, Chunling Tu; draft manuscript preparation: Roseline Oluwaseun Ogundokun. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available in Kaggle Repository at https://www.kaggle.com/search?q=ICIAR2018_BACH_Challenge+in%3Adatasets (accessed on 15 March 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Lei S, Zheng R, Zhang S, Wang S, Chen R, Sun K, et al. Global patterns of breast cancer incidence and mortality: a population-based cancer registry data analysis from 2000 to 2020. Cancer Commun. 2021;41(11):1183–94. doi:10.1002/cac2.12207. [Google Scholar] [PubMed] [CrossRef]
2. Ma X, Cheng H, Hou J, Jia Z, Wu G, Lü X, et al. Detection of breast cancer based on novel porous silicon Bragg reflector surface-enhanced Raman spectroscopy-active structure. Chin Opt Lett. 2020;18(5):051701. doi:10.3788/COL202018.051701. [Google Scholar] [CrossRef]
3. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast. 2022;66(8):15–23. doi:10.1016/j.breast.2022.08.010. [Google Scholar] [PubMed] [CrossRef]
4. International Agency for Research on Cancer. Breast cancer cases and deaths are projected to rise globally (Press Release No. 361) [Internet]. World Health Organization. [cited 2025 Feb 4]. Available from: https://www.iarc.who.int/wp-content/uploads/2025/02/pr361_E.pdf. [Google Scholar]
5. World Health Organization. Breast cancer [Internet]. World Health Organization. [cited 2025 Feb 5]. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. [Google Scholar]
6. Soliman A, Li Z, Parwani AV. Artificial intelligence’s impact on breast cancer pathology: a literature review. Diagn Pathol. 2024;19(1):38. doi:10.1186/s13000-024-01453-w. [Google Scholar] [PubMed] [CrossRef]
7. Ibrahim A, Gamble P, Jaroensri R, Abdelsamea MM, Mermel CH, Chen PHC, et al. Artificial intelligence in digital breast pathology: techniques and applications. Breast. 2020;49(2):267–73. doi:10.1016/j.breast.2019.12.007. [Google Scholar] [PubMed] [CrossRef]
8. Pang J, Ding N, Liu X, He X, Zhou W, Xie H, et al. Prognostic value of the baseline systemic immune-inflammation index in HER2-positive metastatic breast cancer: exploratory analysis of two prospective trials. Ann of Surg Oncol. 2025;32(2):750–9. doi:10.1245/s10434-024-16454-8. [Google Scholar] [PubMed] [CrossRef]
9. Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci Rep. 2017;7(1):4172. doi:10.1038/s41598-017-04075-z. [Google Scholar] [PubMed] [CrossRef]
10. Zhang Z, Liu Z, Martin A, Zhou K. BSC: belief shift clustering. IEEE Trans Syst Man Cybern Syst. 2023;53(3):1748–60. doi:10.1109/TSMC.2022.3205365. [Google Scholar] [CrossRef]
11. Jia Y, Chen G, Chi H. Retinal fundus image super-resolution based on generative adversarial network guided with vascular structure prior. Sci Rep. 2024;14(1):22786. doi:10.1038/s41598-024-74186-x. [Google Scholar] [PubMed] [CrossRef]
12. Song W, Wang X, Guo Y, Li S, Xia B, Hao A. CenterFormer: a novel cluster center enhanced transformer for unconstrained dental plaque segmentation. IEEE Trans Multimed. 2024;26:10965–78. doi:10.1109/TMM.2024.3428349. [Google Scholar] [CrossRef]
13. Hao Y, Qiao S, Zhang L, Xu T, Bai Y, Hu H, et al. Breast cancer histopathological images recognition based on low dimensional three-channel features. Front Oncol. 2021;11:657560. doi:10.3389/fonc.2021.657560. [Google Scholar] [PubMed] [CrossRef]
14. Wakili MA, Shehu HA, Sharif MH, Sharif MHU, Umar A, Kusetogullari H, et al. Classification of breast cancer histopathological images using DenseNet and transfer learning. Comput Intell Neurosci. 2022;2022(1):8904768. doi:10.1155/2022/8904768. [Google Scholar] [PubMed] [CrossRef]
15. Hameed Z, Zahia S, Garcia-Zapirain B, Javier Aguirre J, Maria Vanegas A. Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors. 2020;20(16):4373. doi:10.3390/s20164373. [Google Scholar] [PubMed] [CrossRef]
16. Xie J, Liu R, Luttrell J, Zhang C. Deep learning based analysis of histopathological images of breast cancer. Front Genet. 2019;10:426920. doi:10.3389/fgene.2019.00080. [Google Scholar] [PubMed] [CrossRef]
17. Zheng Y, Li C, Zhou X, Chen H, Zhang H, Li Y, et al. Image classification in breast histopathology using transfer and ensemble learning. In: International Conference on Information Technologies in Biomedicine. Cham: Springer Int Publ; 2022 Jun. p. 295–306. [Google Scholar]
18. Abbasniya MR, Sheikholeslamzadeh SA, Nasiri H, Emami S. Classification of breast tumors based on histopathology images using deep features and ensemble of gradient boosting methods. Comput Electr Eng. 2022;103(3):108382. doi:10.1016/j.compeleceng.2022.108382. [Google Scholar] [CrossRef]
19. Senousy Z, Abdelsamea MM, Gaber MM, Abdar M, Acharya UR, Khosravi A, et al. MCUa: multi-level context and uncertainty aware dynamic deep ensemble for breast cancer histology image classification. IEEE Trans Biomed Eng. 2021;69(2):818–29. doi:10.1109/tbme.2021.3107446. [Google Scholar] [PubMed] [CrossRef]
20. Alotaibi A, Alafif T, Alkhilaiwi F, Alatawi Y, Althobaiti H, Alrefaei A, et al. Vit-deit: an ensemble model for breast cancer histopathological images classification. In: 1st International Conference on Advanced Innovations in Smart Cities (ICAISC); 2023 Jan; Jeddah, Saudi Arabia; IEEE. p. 1–6. [Google Scholar]
21. Balasubramanian AA, Al-Heejawi SMA, Singh A, Breggia A, Ahmad B, Christman R, et al. Ensemble deep learning-based image classification for breast cancer subtype and invasiveness diagnosis from whole slide image histopathology. Cancers. 2024;16(12):2222. doi:10.3390/cancers16122222. [Google Scholar] [PubMed] [CrossRef]
22. Zheng Y, Li C, Zhou X, Chen H, Xu H, Li Y, et al. Application of transfer learning and ensemble learning in image-level classification for breast histopathology. Intell Med. 2023;3(2):115–28. doi:10.1016/j.imed.2022.05.004. [Google Scholar] [CrossRef]
23. Wang G, Ma Q, Li Y, Mao K, Xu L, Zhao Y. A skin lesion segmentation network with edge and body fusion. Appl Soft Comput. 2025;170(1):112683. doi:10.1016/j.asoc.2024.112683. [Google Scholar] [CrossRef]
24. Al-Haija QA, Adebanjo A. Breast cancer diagnosis in histopathological images using ResNet-50 convolutional neural network. In: 2020 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS); 2020 Sep; Vancouver, BC, Canada; IEEE. p. 1–7. [Google Scholar]
25. Kaggle Repository. ICIAR2018_BACH_Challenge [Internet]. Kaggle. [cited 2025 Feb 5]. Available from: https://www.kaggle.com/search?q=ICIAR2018_BACH_Challenge+in%3Adatasets. [Google Scholar]
26. Shiri M, Reddy MP, Sun J. Supervised contrastive vision transformer for breast histopathological image classification. In: 2024 IEEE Int Conf Inf Reuse Integr Data Sci (IRI); San Jose, CA, USA. IEEE; 2024 Aug. p. 296–301. doi: 10.1109/iri62200.2024.00067. [Google Scholar] [CrossRef]
27. Powers DMW. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J Mach Learn Technol. 2011;2(1):37–63. [Google Scholar]
28. Evidently AI. Multi-class classification metrics [Internet]. Evidently AI. [cited 2025 Feb 5]. Available from: https://www.evidentlyai.com/classification-metrics/multi-class-metrics#:~:text=. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools