 Open Access
Open Access
ARTICLE
A Quality of Service Analysis of FPGA-Accelerated Conv2D Architectures for Brain Tumor Multi-Classification
1 Laboratory of Electronics and Microelectronics (EµE), Faculty of Sciences of Monastir, University of Monastir, Monastir, 5000, Tunisia
2 Laboratory of Automation and Electrical Systems and Environment, Monastir National School of Engineers (ENIM), University of Monastir, Monastir, 5035, Tunisia
3 Department of Computer Science, Faculty of Science, Northern Border University, Arar, 73222, Saudi Arabia
4 Department of Science and Technology College of Ranyah, Taif University, P.O. Box 11099, Taif, 21944, Saudi Arabia
* Corresponding Author: Ayoub Mhaouch. Email:
Computers, Materials & Continua 2025, 84(3), 5637-5663. https://doi.org/10.32604/cmc.2025.065525
Received 15 March 2025; Accepted 16 June 2025; Issue published 30 July 2025
Abstract
In medical imaging, accurate brain tumor classification in medical imaging requires real-time processing and efficient computation, making hardware acceleration essential. Field Programmable Gate Arrays (FPGAs) offer parallelism and reconfigurability, making them well-suited for such tasks. In this study, we propose a hardware-accelerated Convolutional Neural Network (CNN) for brain cancer classification, implemented on the PYNQ-Z2 FPGA. Our approach optimizes the first Conv2D layer using different numerical representations: 8-bit fixed-point (INT8), 16-bit fixed-point (FP16), and 32-bit fixed-point (FP32), while the remaining layers run on an ARM Cortex-A9 processor. Experimental results demonstrate that FPGA acceleration significantly outperforms the CPU (Central Processing Unit) based approach. The obtained results emphasize the critical importance of selecting the appropriate numerical representation for hardware acceleration in medical imaging. On the PYNQ-Z2 FPGA, the INT8 achieves a 16.8% reduction in latency and 22.2% power savings compared to FP32, making it ideal for real-time and energy-constrained applications. FP16 offers a strong balance, delivering only a 0.1% drop in accuracy compared to FP32 (94.1% vs. 94.2%) while improving latency by 5% and reducing power consumption by 11.1%. Compared to prior works, the proposed FPGA-based CNN model achieves the highest classification accuracy (94.2%) with a throughput of up to 1.562 FPS, outperforming GPU-based and traditional CPU methods in both accuracy and hardware efficiency. These findings demonstrate the effectiveness of FPGA-based AI acceleration for real-time, power-efficient, and high-performance brain tumor classification, showcasing its practical potential in next-generation medical imaging systems.Keywords
The rapid advancements in medical image analysis, particularly in cancer detection, have led to an increasing reliance on Convolutional Neural Networks (CNNs) for accurate and efficient classification. In particular, brain cancer classification from 2D MRI (Magnetic Resonance Imaging) slices has become a crucial application of CNNs, where precision and speed are critical for timely diagnosis [1,2]. Real-time classification of these 2D MRI slices presents distinct computational and clinical challenges. In diagnostic workflows, rapid processing of sequential 2D MRI slices is essential for efficient tumor localization and multi-classification, particularly in time-sensitive scenarios like emergency stroke assessment. However, this task is complicated by MRI specific artifacts including slice-to-slice intensity variations, partial volume effects at tumor boundaries, and in-plane susceptibility distortions that degrade classification accuracy [3]. While deep learning models achieve high accuracy in offline analysis, their computational demands especially when using convolutional operations (Conv2D) present significant challenges in terms of processing time, resource utilization, and energy consumption [4,5]. This challenge becomes more pronounced when dealing with resource-constrained environments such as embedded systems or real-time medical applications [6], where the models’ computational requirements often prevent real-time performance when processing streaming 2D MRI data.
The growing complexity of CNNs has highlighted the need for hardware acceleration to achieve faster processing and improved energy efficiency. Hardware implementations of CNN operations, such as Conv2D, are particularly beneficial in alleviating the strain on traditional software-based methods [7–9]. Despite their potential, the choice of numerical precision (e.g., 8-bit fixed-point vs. 16-bit or 32-bit fixed-point) introduces trade-offs that affect both computational efficiency and classification accuracy. The motivation for this study arises from the need to optimize hardware-accelerated Conv2D implementations, balancing precision, performance, and resource utilization to meet the stringent demands of real-time brain cancer classification.
The need for hardware-accelerated Conv2D operations arises from the requirement for real-time or near-real-time classification of MRI images in edge computing environments. MRI image acquisition rates vary depending on the scanning protocol and resolution, with standard sequences generating new images every few seconds [10]. For practical deployment on an edge device, the classification process must keep pace with image acquisition to enable real-time decision support without introducing significant delays. Traditional software-based CNN inference on general-purpose processors may fail to meet these timing constraints due to computational complexity, particularly in resource-constrained edge devices [11]. Hardware acceleration on FPGA offers a viable solution by optimizing the balance between precision, performance, and resource utilization.
Fixed-point precision (e.g., 8-bit) offers significant advantages in terms of reducing hardware demands and improving inference speed, making it a promising choice for resource-constrained environments such as edge devices. However, this precision comes at the cost of potentially reduced classification accuracy, which is especially critical in high-stakes applications like cancer detection [12,13]. On the other hand, floating-point precision (16-bit or 32-bit) provides higher accuracy, essential for complex classification tasks, but it comes with increased resource consumption and latency, which can hinder real-time performance on limited hardware [14]. Balancing these trade-offs is crucial for the successful implementation of hardware-accelerated convolutional neural networks (CNNs) in medical diagnostics, where both speed and accuracy are paramount. Recent studies have delved into optimizing hardware architectures for power efficiency and resource awareness, offering solutions to these challenges while maintaining the necessary performance levels for real-time, accurate classifications [15].
This study aims to explore the impact of different numerical precision formats on hardware-accelerated Conv2D operations for brain cancer classification using CNNs. Specifically, we analyze the trade-offs between 8-bit fixed-point and 16-bit/32-bit fixed-point implementations, focusing on classification accuracy, processing time, and resource utilization. The objective is to identify an optimized hardware design that provides the best balance of performance and resource efficiency, while maintaining the accuracy necessary for reliable brain cancer detection. The contributions of this work are:
• An convolutional neural network (CNN) is proposed for the Multi-classification of brain cancer in MRI images.
• A novel hybrid approach where the first Conv2D layer is implemented in FPGA hardware, leveraging different numerical precision strategies (INT8, FP16, and FP32), while the rest of the network runs on the ARM Cortex-A9 CPU.
• A detailed Quality of Service (QoS) analysis of hardware-accelerated Conv2D operations for brain cancer Multi-classification, comparing the impact of various numerical precisions on the classification performance.
The remainder of this paper is organized as follows: Section 2 reviews related works in hardware-accelerated CNNs, focusing on the use of Conv2D operations in medical image classification. Section 3 outlines the methodology and experimental setup, including details on the hardware implementation and precision configurations. Section 4 presents the results and discusses the impact of different numerical precisions on classification accuracy, processing time, and resource usage. Finally, Section 5 concludes the paper, summarizing the findings and suggesting directions for future research.
The integration of hardware acceleration in convolutional neural networks (CNNs) has emerged as a pivotal advancement in the field of medical imaging, particularly in the diagnosis and treatment of brain cancer. The ability to process vast amounts of data rapidly and efficiently has opened new avenues for research and clinical applications. This literature review aims to synthesize the existing body of knowledge regarding hardware-accelerated CNNs in the context of brain cancer, highlighting significant contributions, methodologies, and the implications of these technologies. Several studies have conducted comparative analyses of different hardware implementations of CNNs, focusing on precision and execution time [14,15]. These studies provide valuable insights into the effectiveness of various approaches in the context of brain tumor detection.
FPGA implementations of CNNs have gained prominence due to their flexibility and reconfigurability. Researchers have explored various architectures for implementing Conv2D layers on FPGAs, focusing on optimizing resource utilization and execution time. Pacini et al. presented an FPGA-based CNN accelerator that achieved a substantial reduction in execution time while maintaining classification accuracy for brain tumor detection [16]. Their work highlighted the trade-offs between precision and speed, revealing that quantizing weights and activations to lower bit-widths (e.g., 8-bit) could significantly enhance performance without compromising diagnostic efficacy.
Quantizing CNNs to 8 bits has been shown to maintain acceptable levels of accuracy while significantly improving execution speed. A comparative analysis by Neiso et al. demonstrated that 8-bit quantization could yield performance improvements of up to 4× in execution time without a substantial drop in classification accuracy for brain tumor detection tasks [17]. The authors noted that while some information loss occurs due to quantization, the overall impact on model performance can be mitigated through careful training strategies, such as using mixed-precision training.
Rayapati et al. [18] proposed an FPGA-based hardware-software co-design to accelerate brain tumor segmentation using a combination of Otsu’s binarization and the Watershed algorithm. Their approach leverages the strengths of a System-on-Chip (SoC) architecture by offloading the highly parallelizable Otsu algorithm to the programmable logic (PL), while executing the more complex, sequential Watershed method on the processor system (PS). By optimizing the hardware implementation with pipelining, loop unrolling, and the use of Digital Signal Processing/Mathematical (DSP/MATH) blocks, they achieved a 1.97× speed-up over a CPU-only solution, driven primarily by a 1973× reduction in latency when moving the Otsu module to the FPGA. Their work demonstrates the effectiveness of hybrid FPGA architectures for segmentation tasks. In contrast, our work focuses on CNN-based brain tumor multi-classification, where we accelerate the Conv2D layers using a quantization-aware, hybrid hardware strategy to improve inference speed and efficiency in classification tasks.
Recent advances in FPGA-based hardware acceleration for convolutional neural networks (CNNs) have demonstrated significant potential in enhancing the performance of medical imaging applications, particularly for brain tumor detection. Our work builds upon this foundation by focusing specifically on Conv2D optimization a computational bottleneck in CNNs through fixed-point implementations at 8-bit, 16-bit, and 32-bit precision levels. While 8-bit quantization is a common technique to reduce computational complexity, our contribution goes further by offering a precision-aware design space exploration supported by a comprehensive Quality-of-Service (QoS) analysis. This includes trade-offs between execution time, resource utilization, and classification accuracy, with consideration for real-world clinical applicability.
Furthermore, unlike prior works such as those by Pacini et al. [16] and Neiso et al. [17], which implement full hardware accelerators by mapping the entire CNN onto the FPGA fabric, our approach adopts a selective hybrid Conv2D acceleration strategy. Specifically, we offload only the most computationally intensive Conv2D layer to the FPGA, while executing the remaining layers such as pooling and dense in software on the ARM processor. This enables a flexible and scalable architecture that significantly reduces hardware resource usage and power consumption without sacrificing performance. The fully-mapped implementations in [16,17] often suffer from limited adaptability to different CNN architectures and may face scalability challenges on resource-constrained FPGAs. In contrast, our method achieves real-time inference with a lighter hardware footprint, making it more practical for embedded medical applications where both computational efficiency and architectural flexibility are critical. These design decisions clearly distinguish our contribution from existing literature and emphasize its novelty, efficiency, and relevance for next-generation medical imaging systems.
In the next section, we present the methodology and experimental setup used to implement and evaluate our proposed brain cancer multi-classification system. This includes the CNN model design, the hybrid hardware acceleration strategy for Conv2D layers, and the experimental framework used to assess performance across different quantization levels.
3 Methodology and Experimental Setup
In this section, an overview of the proposed AI-based brain cancer classification is provided, including details of the dataset and training parameters. Additionally, an overview of the proposed hardware acceleration of Conv2D is presented to enhance the performance of the brain cancer classification model.
3.1 Proposed Brain Cancer Multi-Classification
Methodology
The proposed method for brain cancer classification is structured around a convolutional neural network (CNN) architecture, designed to classify brain MRI images into four distinct categories: glioma, meningioma, no tumor, and pituitary tumors. The overall workflow consists of three main stages: data preprocessing, model architecture design, and training and evaluation of the model. Fig. 1 presents the workflow of the proposed AI-Model for brain cancer classification.
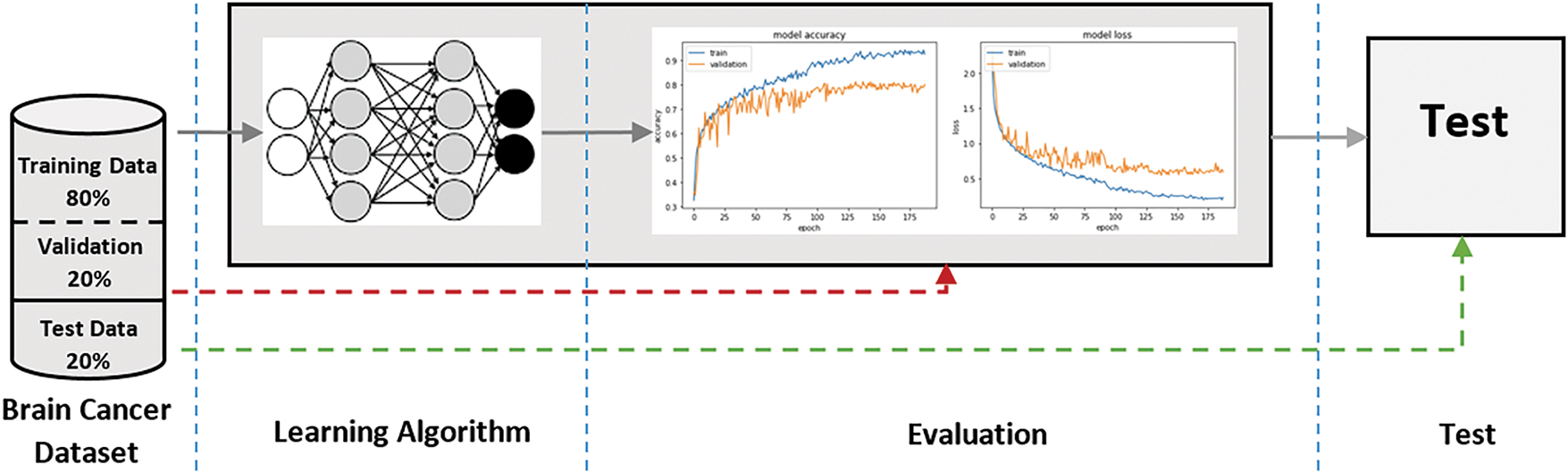
Figure 1: Workfow of the proposed AI-Model for brain cancer classification
The learning algorithm employed in this study is based on a supervised learning framework, which is particularly suitable for classification tasks. The CNN architecture is designed to automatically learn features from the input images, thereby eliminating the need for manual feature extraction. The training process involves feeding the model with labeled MRI images, allowing it to learn the distinguishing characteristics of each tumor type through backpropagation and optimization.
The training of the CNN is performed using the Adam optimizer, which is known for its efficiency in handling sparse gradients and its adaptive learning rate capabilities. The loss function utilized is categorical cross-entropy, which is appropriate for multi-classification problems. The model is trained over a series of epochs, and the performance is evaluated using a validation set to prevent overfitting.
To ensure the robustness of the model, a stratified k-fold cross-validation approach is adopted. This method allows for the effective utilization of the dataset by dividing it into k subsets, or folds. In each iteration, one fold is used as the validation set while the remaining k-1 folds serve as the training set. This process is repeated k times, providing a comprehensive evaluation of the model’s performance across different subsets of the data.
The performance metrics employed to evaluate the model include accuracy, precision, recall, and F1-score. These metrics provide a holistic view of the model’s classification capabilities, allowing for a deeper understanding of its strengths and weaknesses. Additionally, confusion matrices are generated to visualize the classification performance across the different tumor categories, facilitating the identification of any potential misclassifications.
Dataset and Preprocessing
The dataset employed in this study comprises a total of 7023 MRI images, categorized into four classes: glioma (1621 images), meningioma (1645 images), no tumor (2000 images), and pituitary tumors (1757 images). These images were sourced from a publicly accessible MRI dataset, Msoud [19], obtained from the Kaggle repository. The Msoud dataset integrates three publicly available datasets: Figshare [20], SARTAJ [21], and BR35H [22], ensuring a diverse representation of each tumor type (Fig. 2).
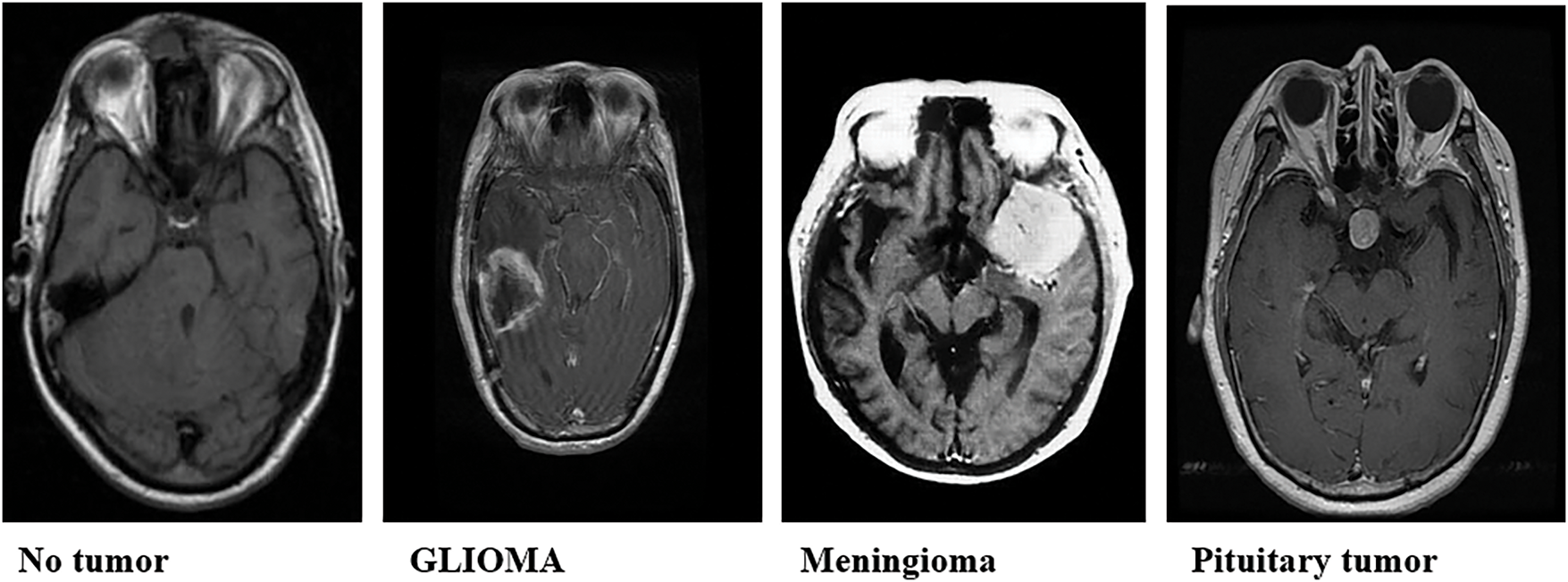
Figure 2: Sample brain tumor classification labels: glioma, meningioma, and pituitary tumor
To facilitate robust model evaluation, the dataset was split into training and testing sets using an 80:20 ratio, a standard practice in machine learning. Before partitioning, a duplication analysis was performed, and 297 duplicate images identified across the combined sources were removed to ensure a strict separation between training and testing sets. This step was essential to prevent data leakage and ensure the integrity of the evaluation process. The diversity and balance of the dataset are critical for training a reliable model capable of generalizing effectively to unseen data.
Prior to training the model, a series of preprocessing steps were undertaken to enhance image quality and facilitate effective learning. The preprocessing pipeline includes image resizing and data augmentation. All MRI images were resized to a uniform dimension of 256 × 256 pixels, ensuring consistent input dimensions for the CNN and enabling efficient processing across the dataset.
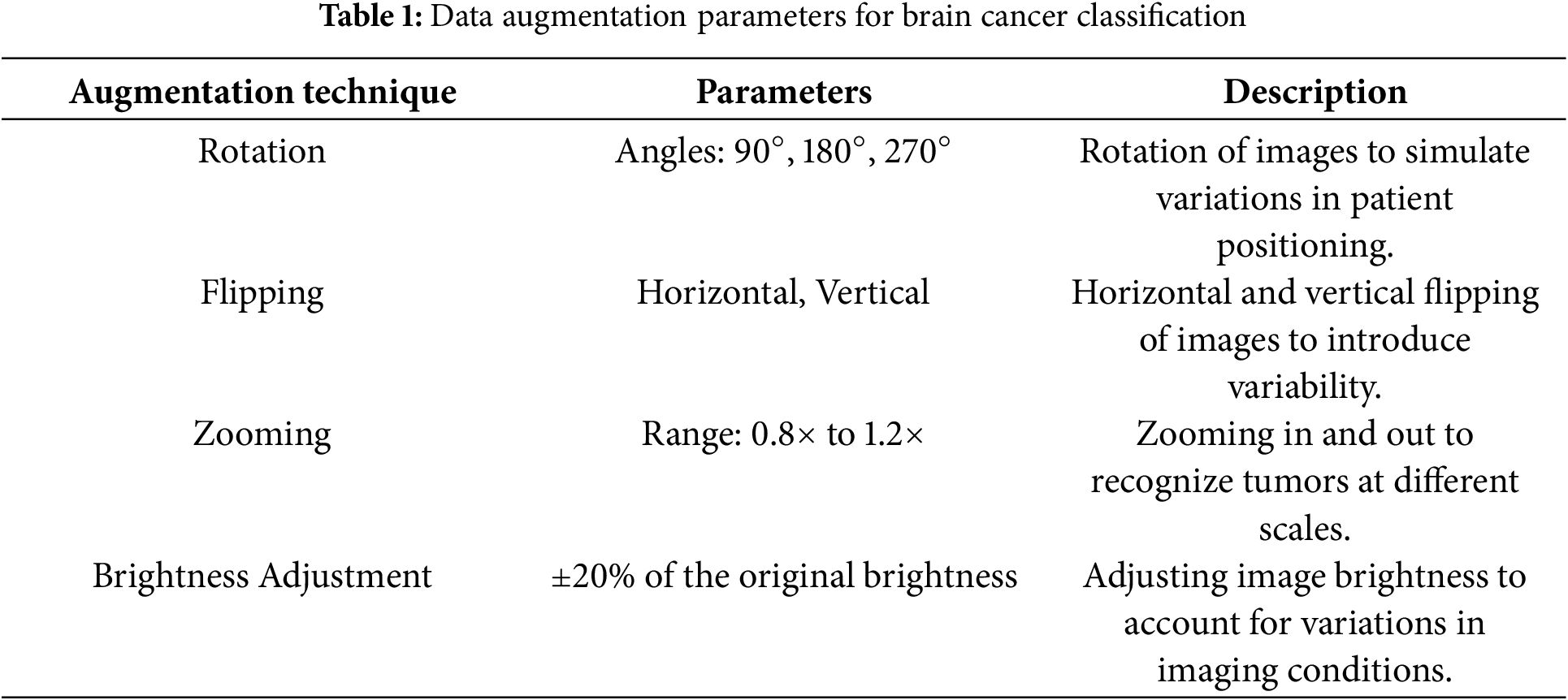
To enhance the model’s robustness and prevent overfitting, data augmentation techniques are employed. The augmentation strategies include: Rotation, Flipping, Zooming, and Brightness Adjustment. Rotation involves rotating images by specific angles, such as 90°, 180°, and 270°, to simulate variations in patient positioning. Flipping includes both horizontal and vertical flips, introducing additional variability to help the model handle different image orientations. Zooming, applied within a range such as 0.8× to 1.2× of the original size, ensures the model can recognize tumors at different scales. Lastly, Brightness Adjustment, typically altering the brightness by ±20%, accounts for variations in imaging conditions, enabling the model to generalize better across diverse datasets. These augmentation techniques collectively improve the model’s ability to adapt to real-world scenarios and enhance its predictive accuracy.
These augmentation techniques are applied in real-time during the training process, effectively increasing the diversity of the training dataset without the need for additional data collection. Table 1 summarizes the data augmentation strategies used to enhance the model’s ability to generalize and improve its robustness.
3.2 Proposed CNN Architecture for Brain Cancer Multi-Classification
The proposed CNN architecture is designed to effectively capture the spatial hierarchies in the MRI images, enabling accurate multi-classification. The architecture comprises four convolutional layers, each followed by a max-pooling layer, culminating in a fully connected output layer. Fig. 3 illustrate the proposed CNN architecture for Brain Cancer Classification (Table 2).
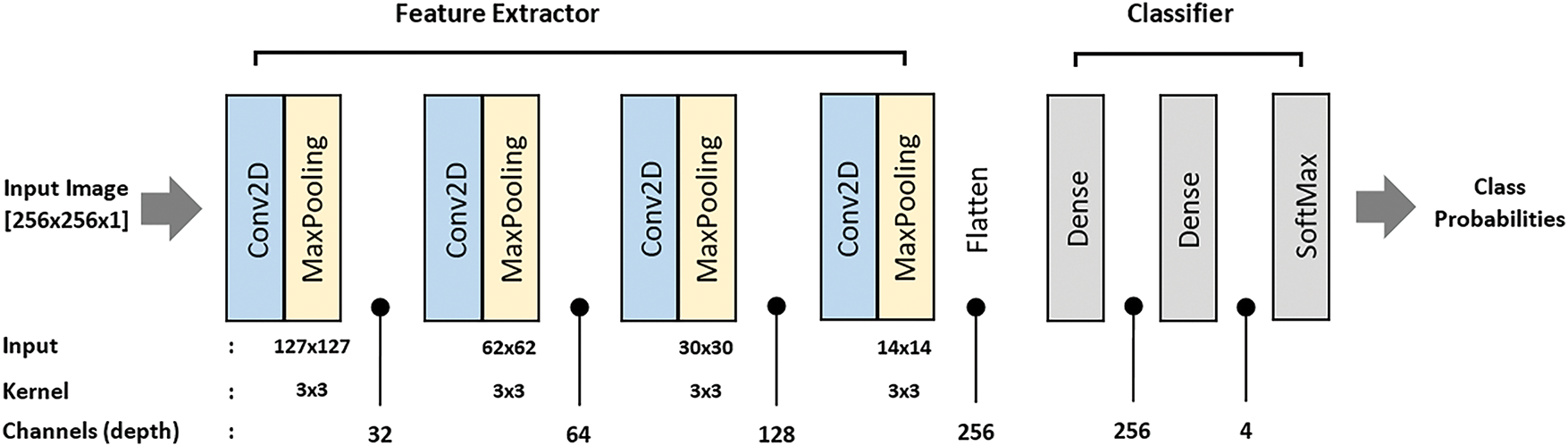
Figure 3: Proposed CNN architecture for brain cancer classification
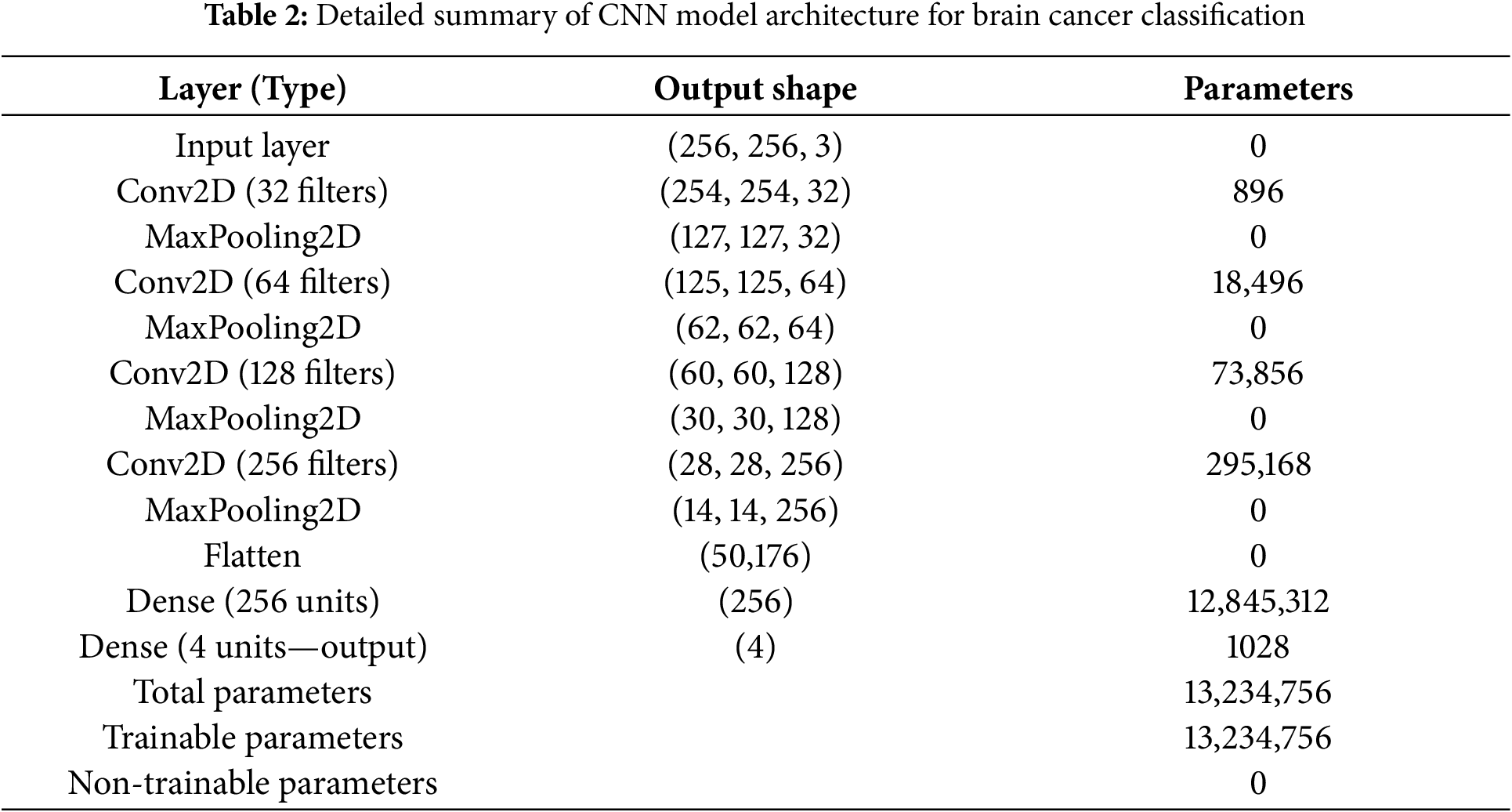
The architecture begins with an input shape of (256, 256, 1), representing grayscale images. It comprises three convolutional layers, each paired with a max-pooling layer, followed by fully connected layers for classification. The first convolutional layer uses 32 filters with a kernel size of (3, 3) and ReLU activation to extract low-level features such as edges and textures. This is followed by a max-pooling layer with a pool size of (2, 2), which reduces the spatial dimensions of the feature maps while retaining key features and lowering computational complexity.
The second convolutional layer employs 64 filters with a (3, 3) kernel and ReLU activation to extract more complex features by combining those from the first layer. A second max-pooling layer, also with a pool size of (2, 2), further reduces the feature map dimensions. The third convolutional layer uses 128 filters, a (3, 3) kernel, and ReLU activation to capture high-level representations of the input images, which is followed by a third max-pooling layer to prepare the feature maps for the fully connected layers. The fourth convolution layer with 256 filters further increases the number of extracted features, resulting in an output shape of (28, 28, 256), followed by a MaxPooling2D layer that reduces it to (14, 14, 256). All convolutional layers use a padding of 2 pixel on all sides (top, bottom, left, right) to preserve the spatial dimensions of the feature maps throughout the network. This setting helps maintain spatial consistency between layers and supports effective feature extraction.
A flatten layer then converts the high-dimensional feature maps into a one-dimensional vector, transitioning the data to the dense layers. The first dense layer, with 256 units and ReLU activation, learns complex patterns from the flattened feature vector. Finally, the output layer is a dense layer with 4 units and a softmax activation function, which outputs probabilities for each of the four classes, enabling multi-classification. This hierarchical structure ensures the model captures increasingly complex features at each stage, optimizing its ability to classify MRI images accurately.
The training procedure involves several key steps to ensure effective learning and robust performance. First, the model is compiled using the Adam optimizer and categorical cross-entropy loss function, with a learning rate set to 0.001, a commonly used value for training CNNs. Next, batch size and epochs are configured, with a batch size of 32 chosen to balance training speed and model performance, and the number of epochs set to 50. Early stopping is implemented to halt training if the validation loss fails to improve for a specified number of epochs. During model training, the augmented training dataset is used, and validation is performed on a hold-out validation set. The model’s weights are updated using gradients computed through backpropagation, enabling it to learn optimal parameters for classification. Throughout the process, performance monitoring is conducted by tracking training and validation loss and accuracy, which helps assess learning progress and make necessary adjustments. Finally, the model is tested on a separate test set that was not used during training or validation. This evaluation step is essential for determining the model’s generalization capabilities, with performance metrics on the test set providing critical insights into its effectiveness in classifying unseen MRI images.
3.3 Proposed Hardware-Acceleration
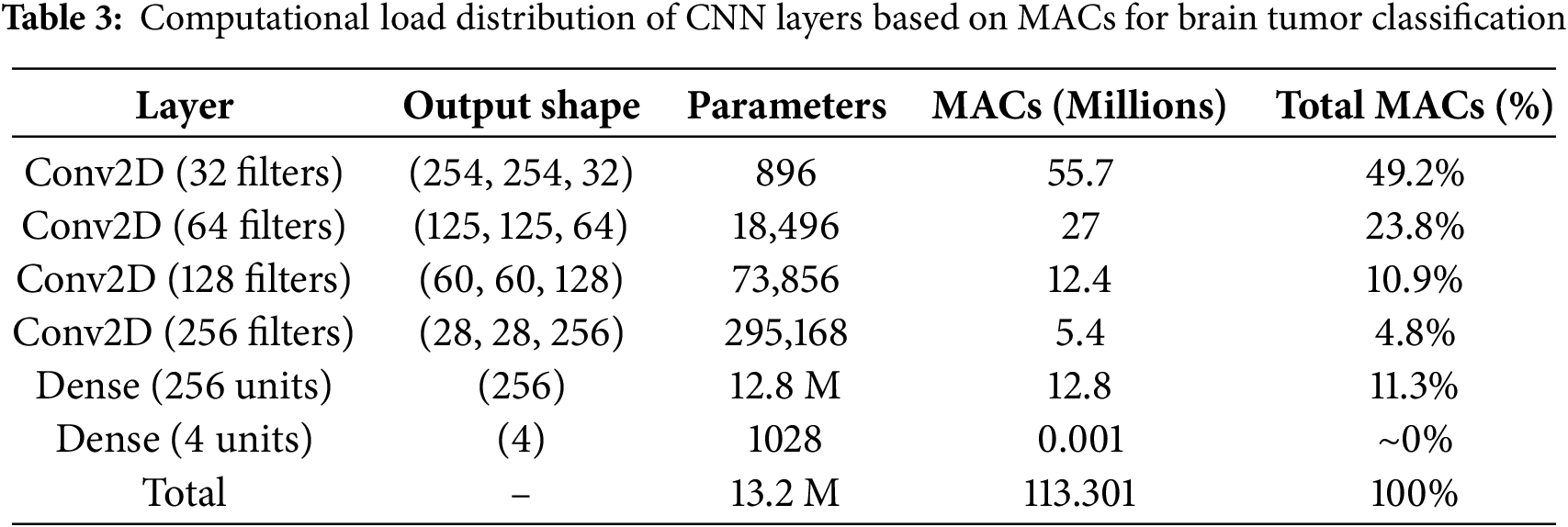
To determine which parts of the model would benefit most from hardware acceleration, we analyzed the computational load distribution across individual layers. This analysis helps identify performance bottlenecks and guides our strategy for partial hardware acceleration, where only the most computationally intensive layers are offloaded to hardware accelerators (e.g., FPGAs or ASICs), while the remaining layers continue to run on general-purpose processors. Table 3 shows a breakdown of the number of operations (measured in Multiply-Accumulates (MACs)) for each major layer in the model when run on a CPU. The percentage contribution of each layer to total computation is also provided.
In the proposed CNN architecture for brain tumor classification, a detailed analysis of computational load reveals that the first Conv2D layer is the most computationally intensive. It alone accounts for approximately 49.2% of the total MAC operations, as shown in Table 3. This is primarily due to its large input size (256 × 256 × 3) and the relatively high number of filters (32), resulting in 55.7 million MACs. The number of Multiply-Accumulate operations (MACs) in a convolutional layer is computed as [23]:
where:
•
•
•
•
The total MACs for the entire network are approximately 113.3 million, distributed across the convolutional and dense layers. The subsequent Conv2D layers consume significantly fewer MACs due to reduced spatial dimensions after pooling, with the second, third, and fourth Conv2D layers contributing only 23.8%, 10.9%, and 4.8% of total MACs, respectively. Although the Dense layer with 256 units has a large number of parameters, its contribution to total MACs is only 11.3%. Given the limited resources of the target FPGA (PYNQ-Z2) and the goal of minimizing latency and power consumption, accelerating only the first Conv2D layer provides the most impactful performance gain while keeping hardware utilization efficient. This selective acceleration strategy optimally balances performance, energy efficiency, and hardware constraints, making it well-suited for real-time medical imaging applications.
The first Conv2D layer is identified as the most computationally demanding component of the CNN. It processes high-resolution MRI inputs and extracts low-level features such as edges and textures, requiring a large number of Multiply-Accumulate (MAC) operations. Given the large input dimensions and filter depth, this layer alone accounts for nearly 50% of the total computational load, as detailed in Section 3.2. Accelerating this layer significantly reduces overall inference time and power consumption, making it particularly advantageous in resource-constrained environments such as edge devices.
To address these challenges, we propose a hybrid approach for brain cancer multi-classification, where the first Conv2D layer is implemented in hardware, leveraging different numerical precision strategies: 8-bit Integer, 16-bit Fixed-Point, and 32-bit Fixed-Point, with the 32-bit representation being the maximum data width for CPU processing. This hardware-based solution is designed to optimize the computational load of the first layer. The remaining layers, including MaxPooling, additional Conv2D layers, Flatten, and Dense layers, are executed in software. By using this hybrid architecture, we leverage hardware acceleration to optimize the most computation-heavy layer, while maintaining flexibility and scalability by executing the remaining layers in software. This approach ensures both performance and accuracy, providing a balanced solution for real-time brain cancer multi-classification on edge devices. Fig. 4 presents the proposed hybrid approach for brain cancer multi-classification.
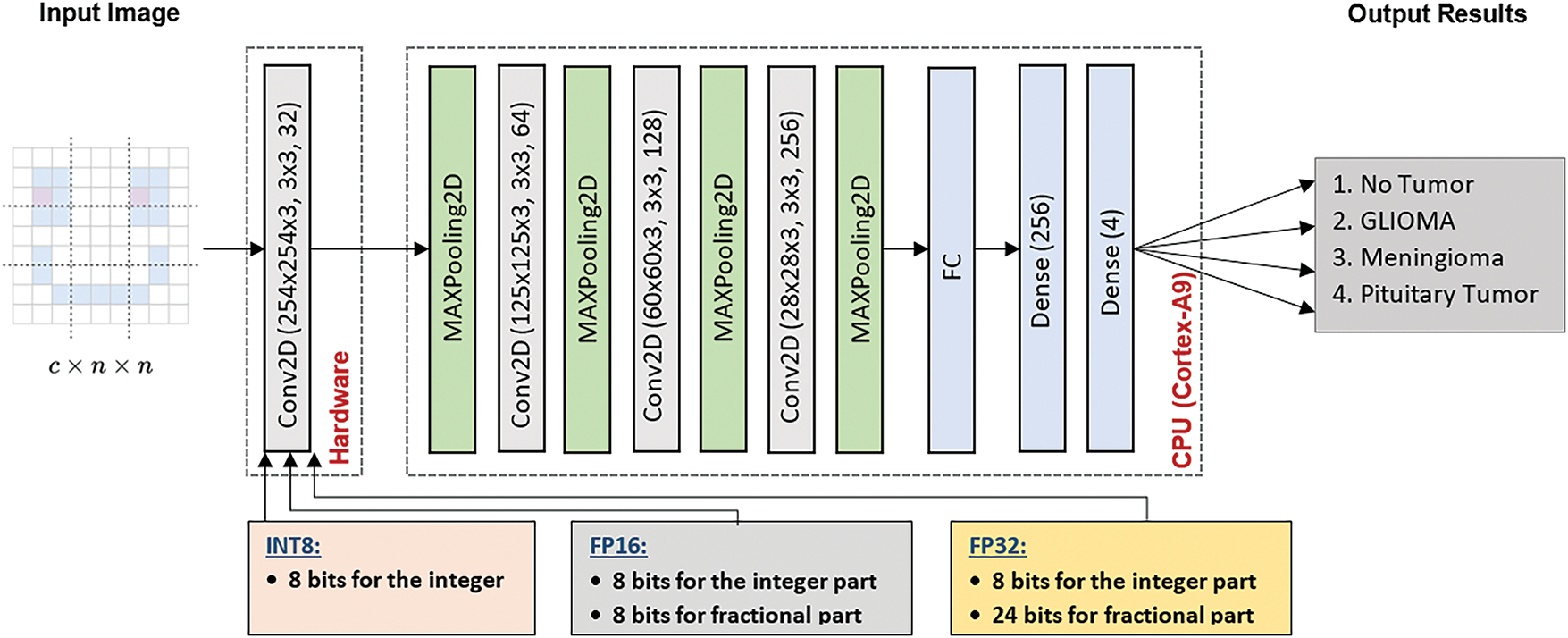
Figure 4: Proposed hybrid inference framework for brain cancer multi-classification
In this study, we implement a CNN architecture tailored for brain cancer classification using a hybrid hardware-software approach across three different hardware configurations. Each configuration focuses on accelerating only the first Conv2D layer on FPGA hardware while varying the numerical precision: 8-bit Integer (INT8), 16-bit Fixed-Point (FP16), and 32-bit Fixed-Point (FP32). The primary objective is to enhance computational throughput, minimize power consumption, and preserve high classification accuracy key requirements for real-time, resource-constrained medical applications. By systematically analyzing the trade-offs among numerical formats, we demonstrate how precision scaling affects performance, efficiency, and resource utilization in FPGA-based acceleration for medical imaging. Fig. 4 illustrates the proposed hybrid deployment strategy that integrates both hardware and software components. The first Conv2D layer is offloaded to FPGA hardware for acceleration using three numerical precision formats: 8-bit Integer (INT8), 16-bit Fixed-Point (FP16), and 32-bit Fixed-Point (FP32). Meanwhile, the subsequent layers including pooling, additional Conv2D operations, flattening, and fully connected layers are executed on the ARM Cortex-A9 processor. This hybrid architecture is designed to optimize inference speed, energy efficiency, and scalability, making it highly suitable for embedded medical imaging systems.
Fig. 5 illustrates the proposed hardware architecture implemented on the PYNQ-Z2 platform for accelerating the first Conv2D layer in the CNN model. The design includes modules for input feature map buffering, weight memory storage, bias integration, fixed-point arithmetic units, padding logic, and output formatting. The architecture is parameterizable to support multiple numerical formats 8-bit Integer (INT8), 16-bit Fixed-Point (FP16), and 32-bit Fixed-Point (FP32) providing scalable deployment depending on application requirements. This implementation targets reduced latency and power consumption for real-time medical image analysis in embedded systems.
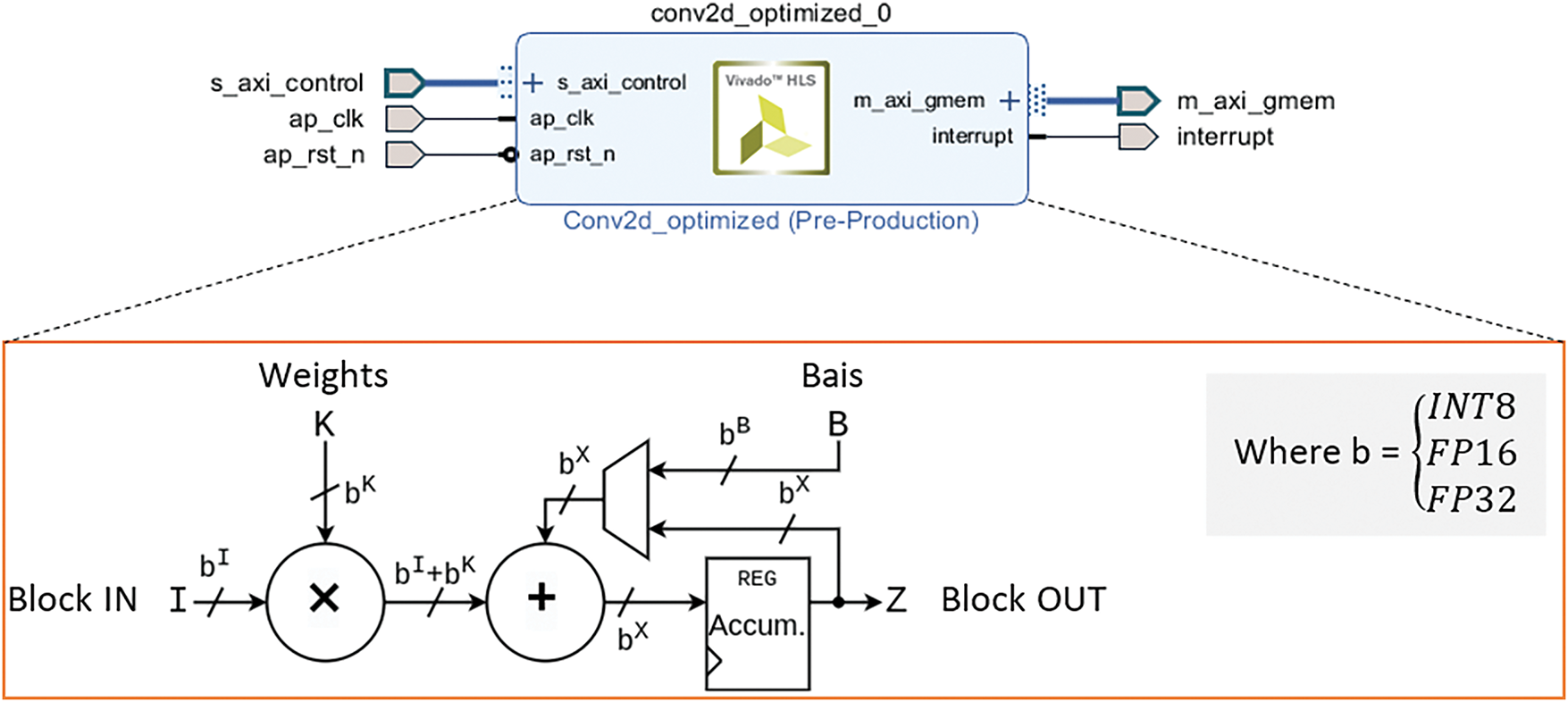
Figure 5: FPGA-based hardware architecture for accelerating Conv2D operation
For the proposed hardware acceleration, Fig. 5 details the internal architecture of the Conv2D operation mapped onto the FPGA. The process begins with the element-wise multiplication of input feature blocks (Block_in) and corresponding filter weights. This core convolution step generates partial products, which are computed using configurable arithmetic units that support INT8, FP16, and FP32 data formats. This flexibility allows for balancing accuracy, computation speed, and FPGA resource usage. After the multiplication step, the partial products are summed and then a bias is added to the accumulated result. The bias term, which is a constant value associated with each filter, helps to adjust the output of the convolution and is crucial for improving the model’s accuracy during training. The sum of the products and bias is then passed to an accumulator register. This register plays a key role in storing and summing the intermediate results of the convolution, which is especially important for managing the multiple filter operations that occur in parallel across the input block. The accumulated result is stored in Block_out, which represents the final feature map produced after applying the convolution operation. The output precision of Block_out is determined by the selected data representation, such as int8, FP16, or FP32. Each of these data types has different trade-offs in terms of speed and resource usage, with int8 offering the fastest computation but lower accuracy, and FP16/FP32 providing higher precision at the cost of increased resource usage.
This hardware architecture is designed for high efficiency, leveraging parallel processing and optimized data flow to accelerate the Conv2D operation. By offloading the most demanding computations to hardware, it significantly reduces the overall execution time, making it suitable for real-time applications, especially on resource-constrained edge devices. The proposed approach ensures that the trade-off between speed, precision, and resource utilization is optimized, allowing the architecture to handle large-scale medical image classification tasks efficiently while maintaining high accuracy. The proposed Conv2D for brain cancer multi-classification is presented in Algorithm 1.
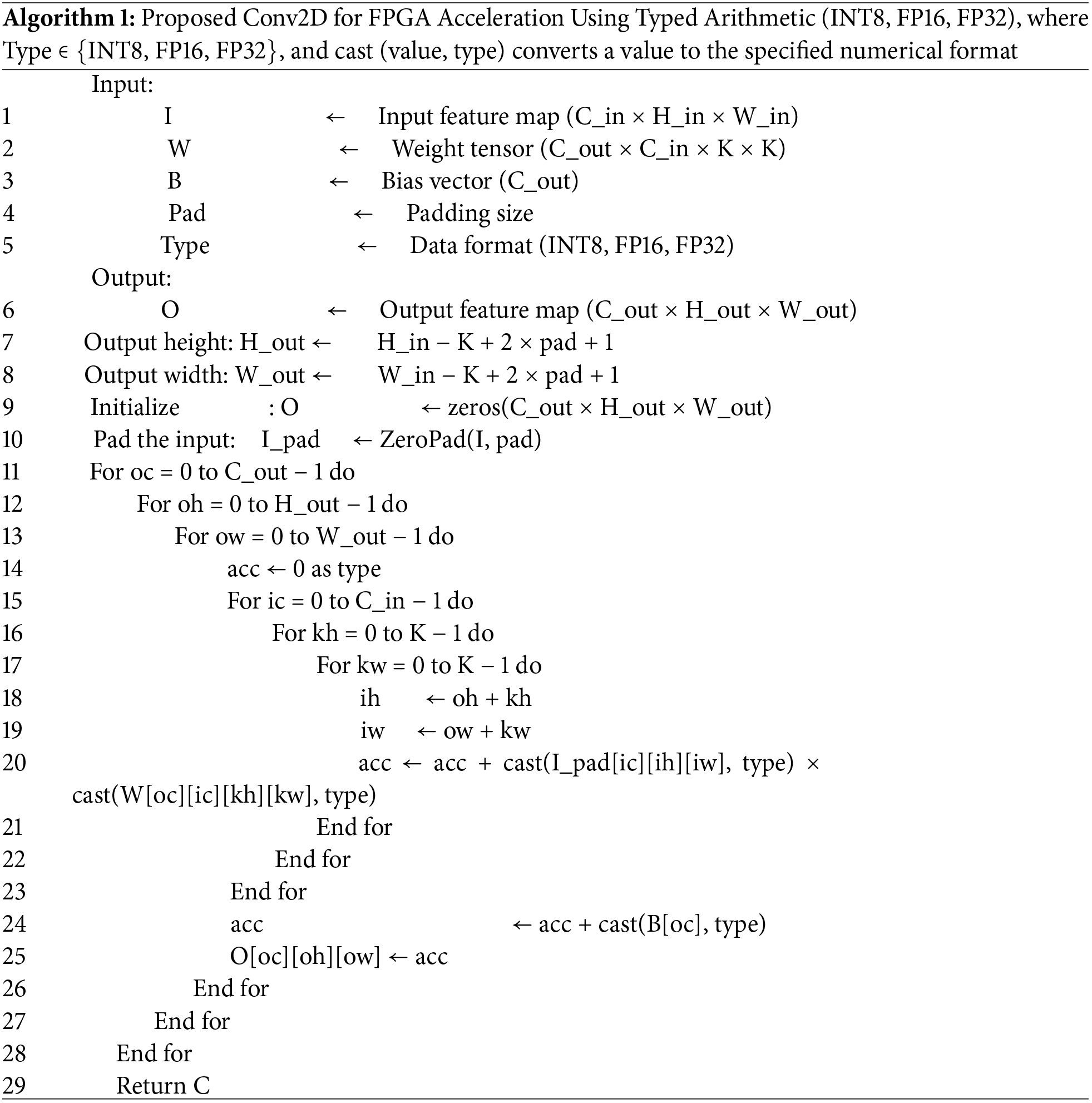
Fig. 6 illustrates the proposed hybrid implementation for brain cancer multi-classification, where the first Conv2D layer is hardware-accelerated with varying data representations: 8-bit Integer (int8), 16-bit Fixed-Point (FP16), and 32-bit Fixed-Point (FP32). This hardware-based Conv2D operation optimizes the computational load by utilizing different precision strategies, offering a trade-off between speed, accuracy, and resource utilization. The hardware implementation is designed to perform the computationally intensive convolution efficiently.
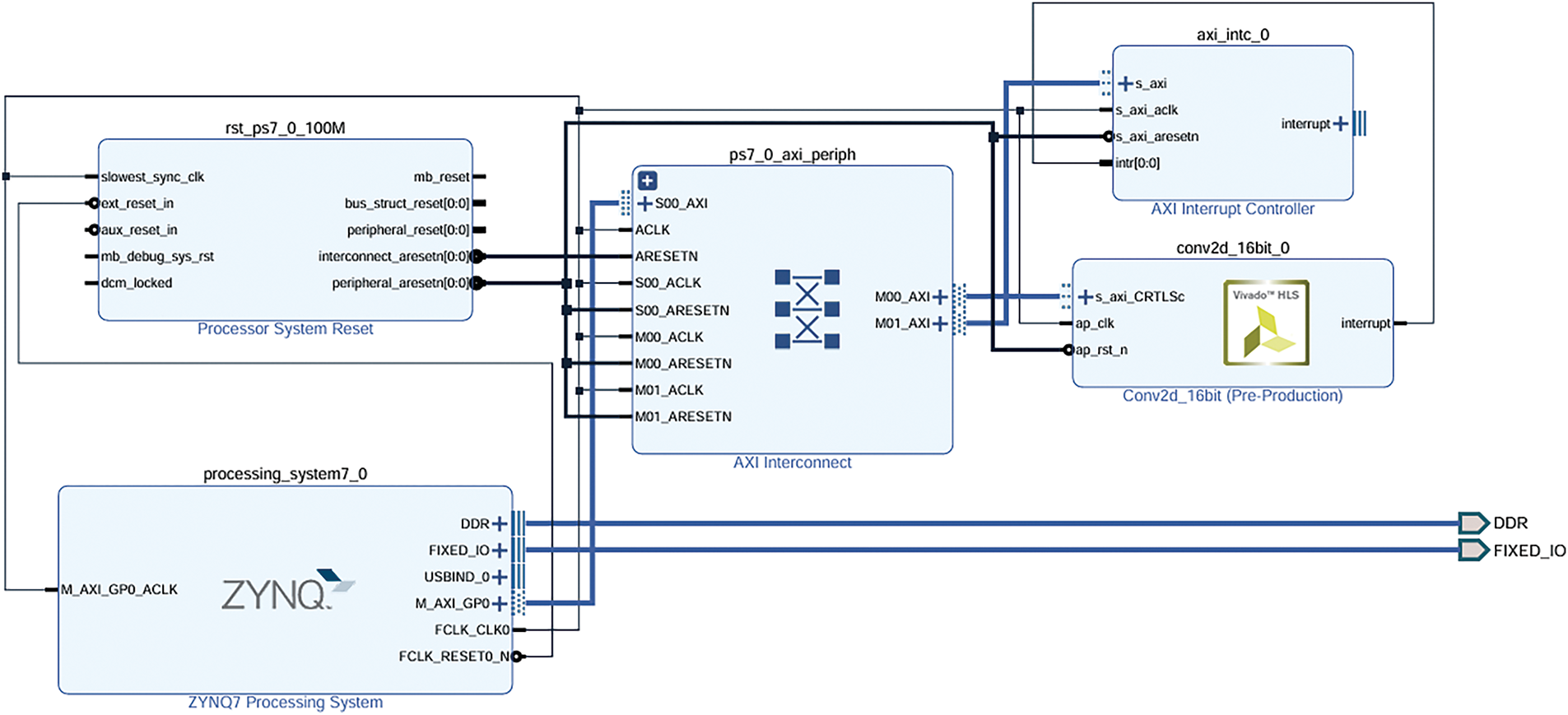
Figure 6: Proposed design of a hybrid implementation for the brain cancer multi-classification
The remaining layers of the CNN, including MaxPooling, additional Conv2D layers, Flatten, and Dense layers, are executed in software using the Zynq processing unit (CPU). This hybrid architecture leverages the flexibility and scalability of the Zynq processing system to handle the less computationally demanding layers, while hardware acceleration is applied to the most intensive part, ensuring high performance and energy efficiency. This combination of hardware and software execution enables the system to meet the real-time requirements of brain cancer multi-classification on resource-constrained edge devices.
In the next section, we present the results and performance analysis of the proposed system. This includes a detailed evaluation of the classification model’s effectiveness, followed by an assessment of the hybrid CPU-FPGA implementation. Metrics such as execution time, resource utilization, power consumption, and classification accuracy are analysed to demonstrate the trade-offs and benefits of the hardware-software co-design.
4 Results and Performance Analysis
In this section, we provide an evaluation of the proposed model for brain cancer multi-classification. The performance of the model is tested on two different embedded system configurations. The first test evaluates the model’s performance on a CPU-based system using the Cortex-A9 processor. This test serves as a baseline to assess the efficiency of the software implementation on a general-purpose processor. The second test involves a hybrid implementation, combining the processing power of the Cortex-A9 CPU with the XC7Z020 FPGA for hardware acceleration. In this configuration, the most computationally intensive Conv2D layer is offloaded to the FPGA, while the remaining layers are executed on the CPU. This hybrid setup aims to exploit the strengths of both hardware and software to achieve optimal performance in terms of speed, power efficiency, and classification accuracy.
4.1 Evaluation of the Proposed Model
The evaluation of the proposed model was conducted using a k-fold cross-validation approach, where the dataset was divided into five folds to ensure a comprehensive assessment of the model’s generalization capabilities. During each fold, the model was trained on k−1 folds and validated on the remaining fold, with performance metrics averaged across all folds. Table 4 shows the performance evaluation of the proposed brain cancer classification by CNN model.
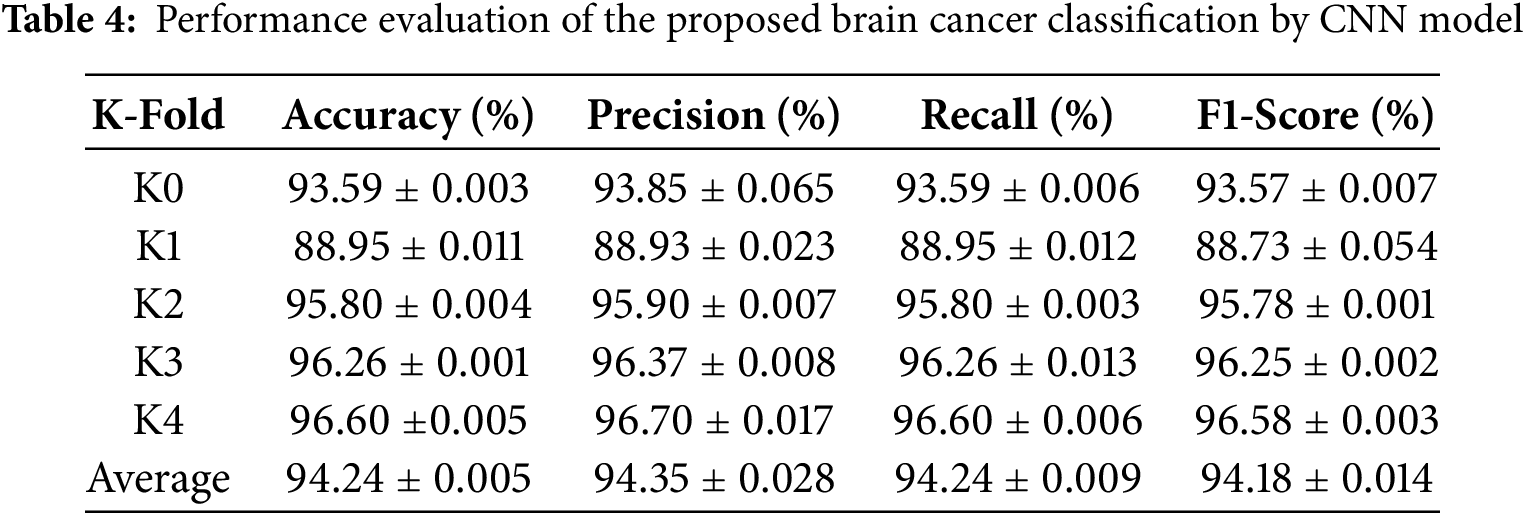
Key evaluation metrics, including accuracy, precision, recall, and F1-score, were calculated to provide a holistic understanding of the model’s performance. The model’s predictions on the test set were compared with ground truth labels, and a confusion matrix was generated to visualize the classification results for each class. The confusion matrix helped identify any misclassifications, highlighting areas for improvement. Furthermore, training and validation accuracy and loss were plotted for each fold to monitor the model’s learning behavior and detect potential overfitting or underfitting. The visualizations demonstrated the model’s ability to converge effectively, with validation metrics closely following the training metrics, indicating strong generalization capabilities (Fig. 7).
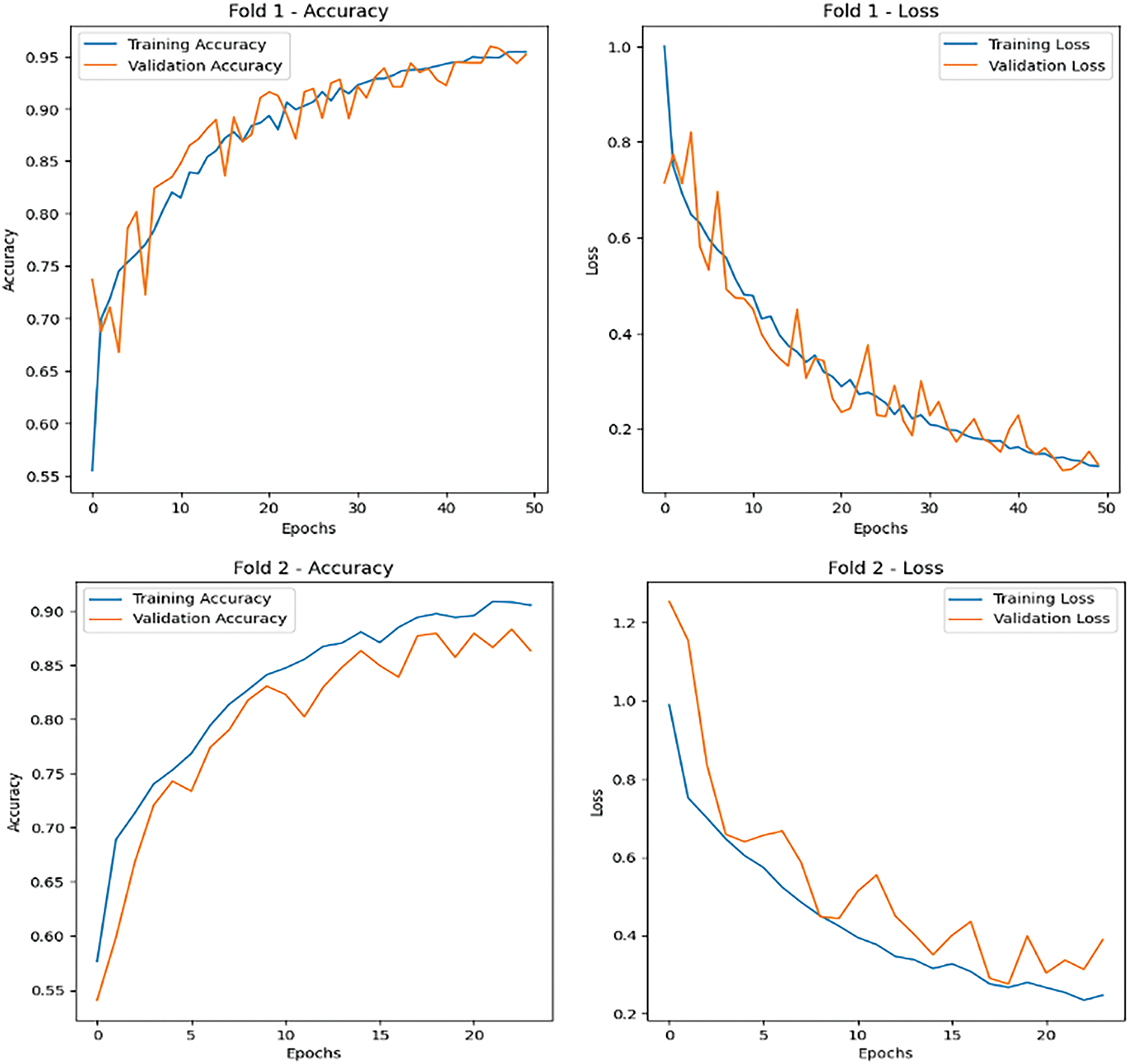
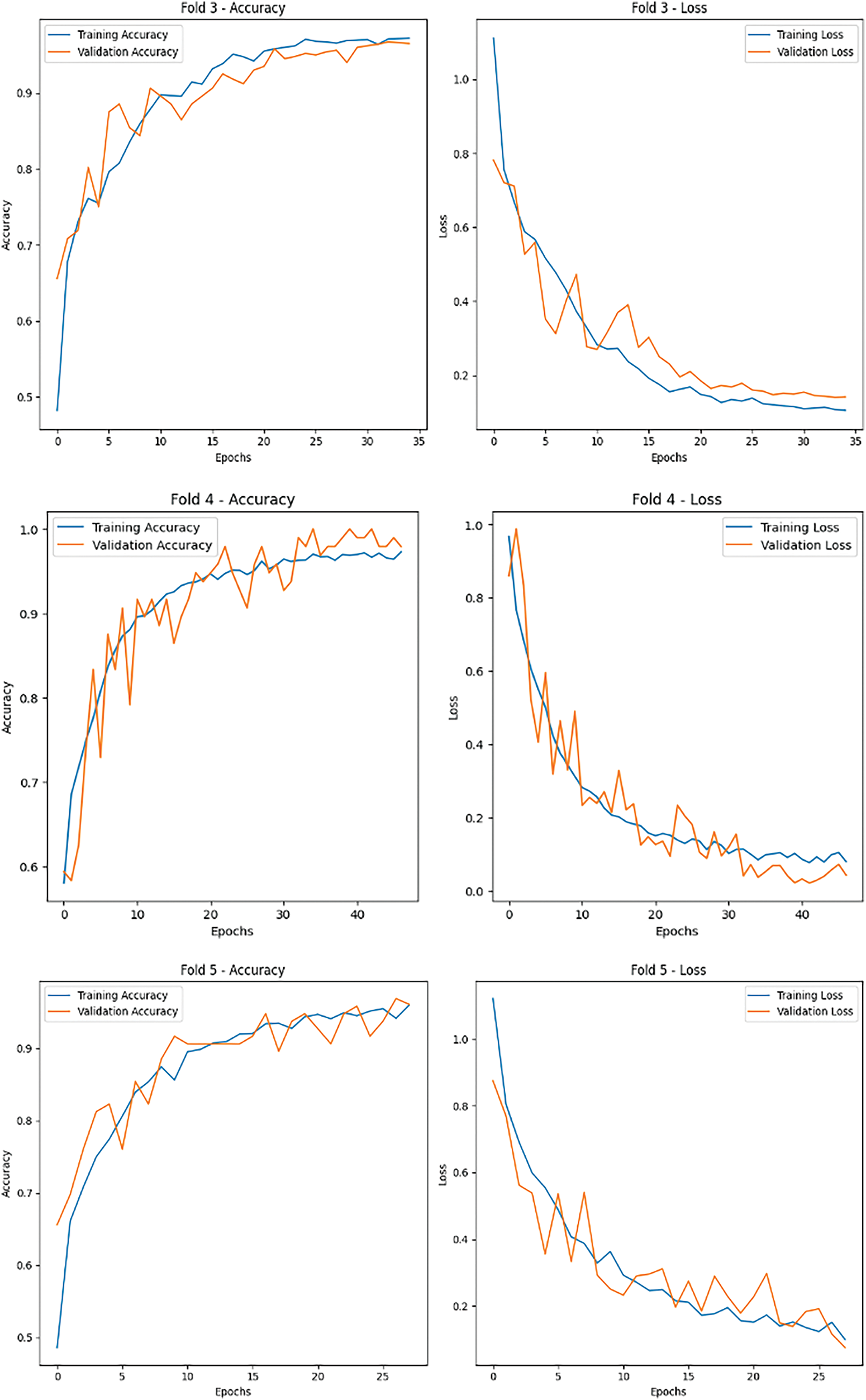
Figure 7: Accuracy and loss metrics for the proposed brain cancer classification
The results of the evaluation process provided critical insights into the model’s effectiveness in classifying unseen MRI images, showcasing its robustness and suitability for real-world applications (Fig. 8). The use of cross-validation and performance monitoring ensured that the model was thoroughly tested and optimized for reliable tumor classification.
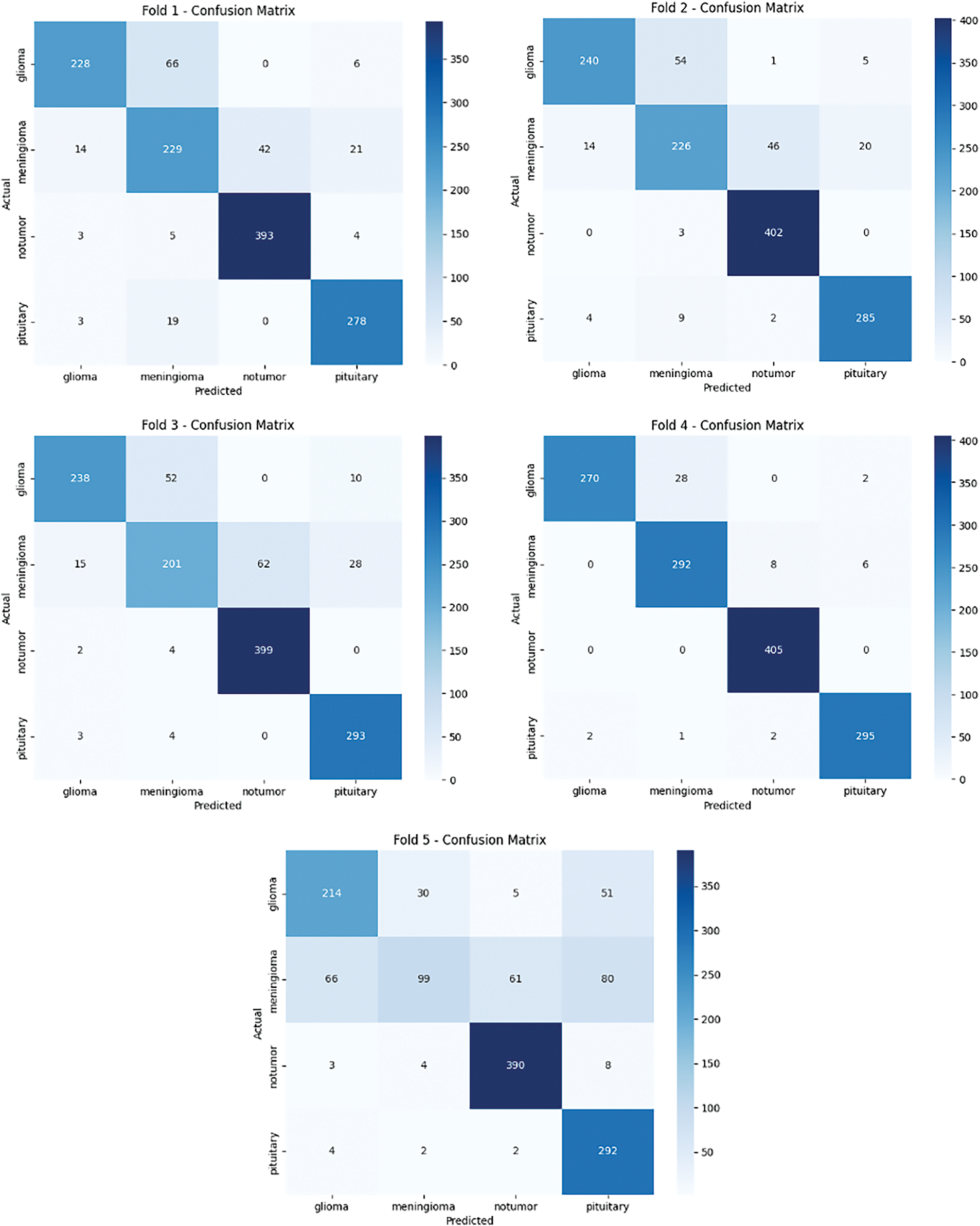
Figure 8: Confusion matrix for the proposed brain cancer classification
The ROC curves demonstrate strong and consistent classification performance across all five folds, with all curves positioned close to the top-left corner of the plot, reflecting high diagnostic accuracy (Fig. 9). The model achieved an average AUC of 0.9664, with individual fold performances ranging from 0.8895 (K1) to 0.9670 (K4), indicating excellent discrimination capability. The tight clustering of the curves suggests reliable generalization across different data partitions, with minimal variability between folds. Precision and recall values remained consistently high (all > 0.88), confirming the model’s ability to accurately identify positive cases while minimizing false positives. These results underscore the CNN model’s robustness and effectiveness for brain cancer classification, supported by high reproducibility across cross-validation folds.
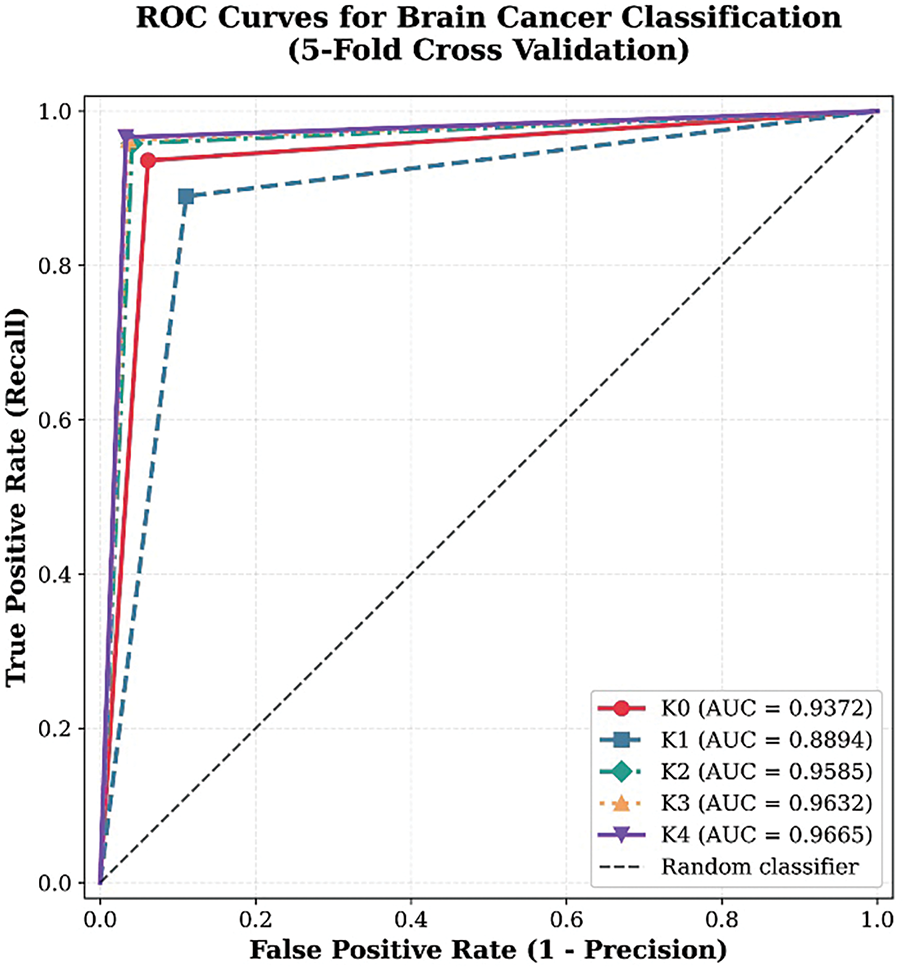
Figure 9: ROC Curves for proposed brain cancer classification
Comparative Analysis
The performance of the proposed CNN model for brain tumor classification is compared with several state-of-the-art methods using the same dataset (Table 5). The evaluation metrics include Accuracy, Precision, Recall, and F1-Score, which are widely used to assess the effectiveness of classification models in medical imaging. The proposed model employs K-Fold cross-validation (K = 5) to ensure robustness and generalizability. The comparison includes methods such as transfer learning with ResNet, hybrid CNN-SVM approaches, traditional machine learning techniques (e.g., SVM and k-NN), and CNN-based methods. The results demonstrate that the proposed CNN model achieves competitive performance, outperforming traditional machine learning methods and closely matching or exceeding the performance of advanced deep learning approaches. This highlights the effectiveness of the proposed method for accurate and reliable brain tumor classification.
4.2 Evaluation on Cortex-A9 CPU
In this evaluation, the proposed CNN model for brain cancer multi-classification is executed entirely on the Cortex-A9 CPU to establish a baseline performance analysis. The Cortex-A9, part of the Xilinx Zynq-7000 SoC, is selected due to its widespread use in embedded systems, offering a balance between computational capability and power efficiency. This processor supports the ARMv7-A architecture, which is optimized for embedded applications, making it suitable for testing software-based deep learning models before implementing hardware acceleration.
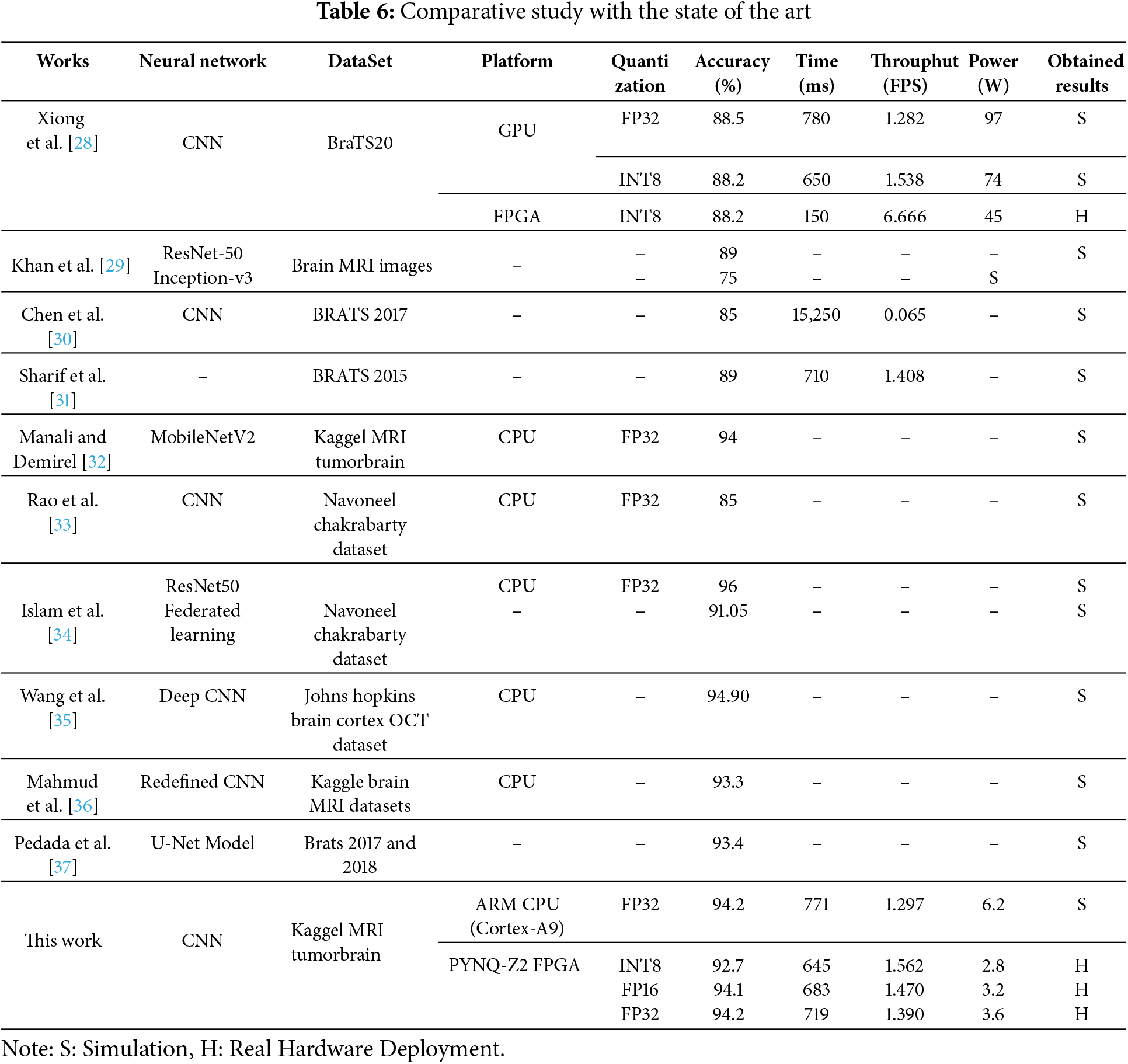
The evaluation focuses on measuring inference time, power consumption, and classification accuracy when executing the entire CNN model purely in software. All operations, including convolutional layers, pooling layers, flattening, and fully connected layers, are processed sequentially on the CPU. Given the computational complexity of deep learning models, running the CNN solely on the Cortex-A9 presents several challenges. The limited processing power results in increased latency, making it difficult to meet real-time classification requirements. Additionally, the high computational load leads to greater power consumption, which can be a constraint for resource-constrained edge devices. Table 6 illustrate the performace evaluation of proposed CNN model for brain tumor Multi-Classification.
The performance of the proposed CNN model for brain tumor multi-classification is evaluated on the Cortex-A9 CPU using 32-bit fixed-point (FP32) precision. The key performance metrics analyzed include classification accuracy, inference time, and power consumption. The results indicate that the model achieves an accuracy of 94.2%, demonstrating its effectiveness in distinguishing between different brain tumor classes. However, the execution time on the Cortex-A9 CPU is 771 ms per inference, which is relatively high for real-time applications. Additionally, the power consumption is measured at 6.2 W, highlighting the need for optimization in resource-constrained environments.
These findings emphasize the limitations of running a computationally intensive CNN model entirely in software on an embedded CPU. The high latency and power consumption make it challenging to deploy the model in real-time edge computing applications, particularly in medical imaging where fast and efficient processing is crucial. To address these limitations, a hybrid hardware-software approach is explored, leveraging FPGA acceleration for the most computationally demanding operations while maintaining classification accuracy. The next section analyzes the performance improvements achieved through this hybrid implementation.
4.3 Evaluation on Hybrid CPU-FPGA Implementation
To implement and evaluate the hardware-accelerated Conv2D layers in the proposed AI-based brain cancer classification model, a structured hardware design and verification process was followed. The implementation workflow was carried out using Vivado High-Level Synthesis (HLS), Vivado HLx, and PYNQ-Z2 for accuracy and real-time testing.
The first step involved designing and optimizing the hardware-accelerated Conv2D operations using Vivado High-Level Synthesis (HLS). The HLS environment allowed for efficient design space exploration, enabling modifications and optimizations of the bit-width representation for the Conv2D computations (Fig. 10). The proposed architecture was implemented with three different numerical precisions:
• 8-bit integer representation for fast and low-power computations.
• 16-bit hybrid representation (8-bit integer, 8-bit fractional) for a balance between speed and precision.
• 32-bit fixed-point representation (8-bit integer, 24-bit fractional) for maximum accuracy.
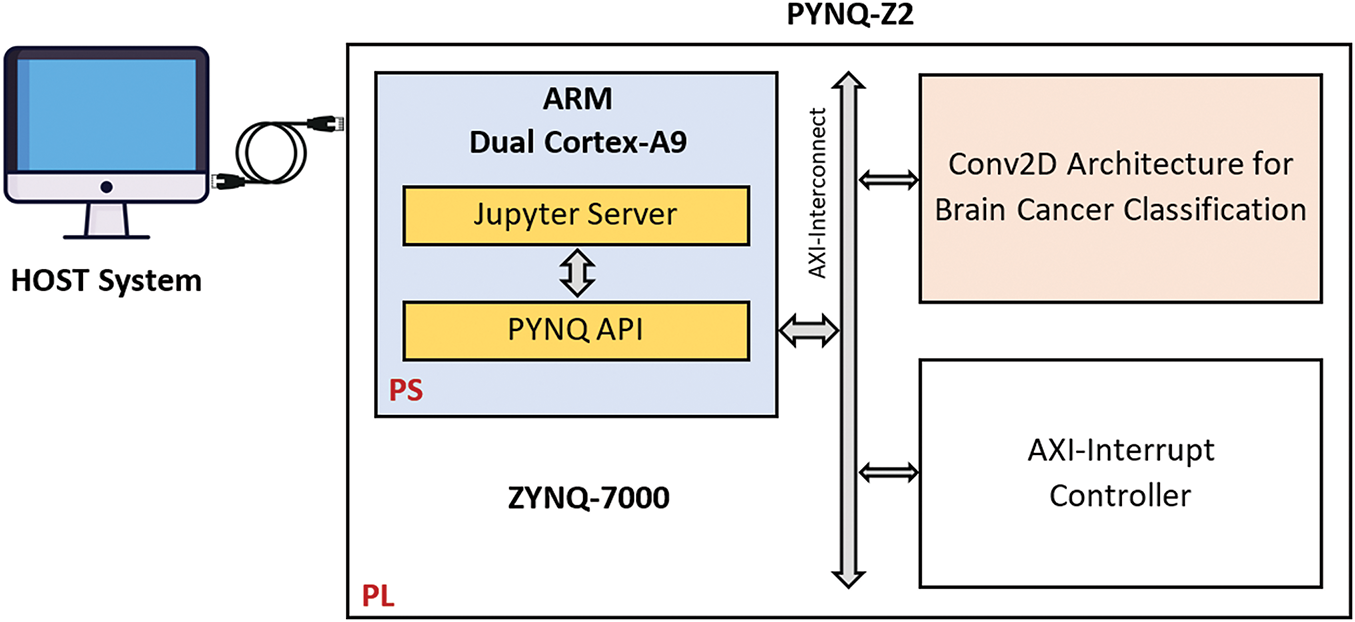
Figure 10: Synoptic flow of Hardware/software implementation on PYNQ-Z2
Various optimizations, such as loop unrolling, pipelining, and dataflow optimizations, were applied to improve the latency and throughput of the Conv2D hardware module. Once the optimized hardware design was verified at the C simulation and C synthesis levels, the RTL (Register Transfer Level) implementation was exported for further integration.
The next phase involved integrating the optimized Conv2D hardware module into a complete hardware processing system. This was accomplished using Vivado HLx, where the proposed Conv2D IP core was interfaced with the Zynq Processing System (PS). The AXI (Advanced eXtensible Interface) interconnect was used to enable efficient communication between the programmable logic (PL) and processing system (PS) of the Xilinx Zynq SoC. Once the system design was finalized, a bitstream file was generated and deployed to the FPGA hardware. Table 7 illustrate FPGA Resource Utilization for the Proposed Conv2D on PYNQ-Z2.
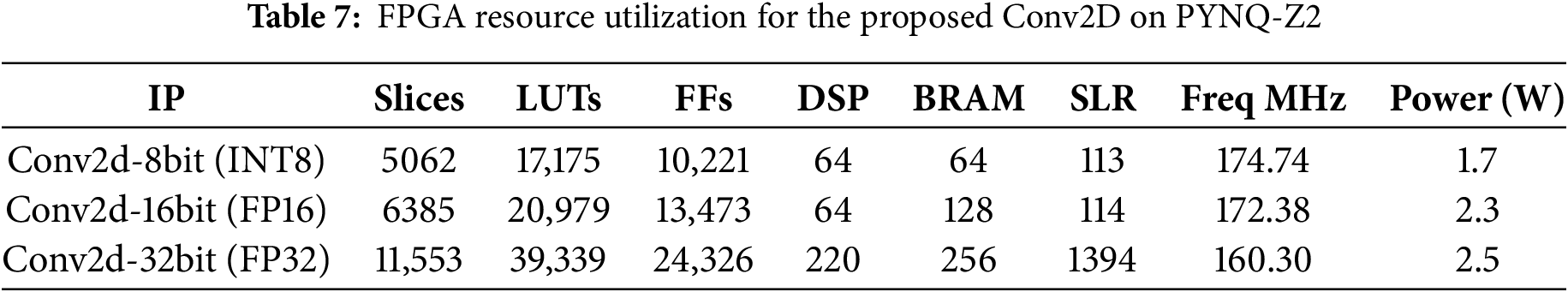
A detailed comparison of the three Conv2D configurations (INT8, FP16, and FP32) on the PYNQ-Z2 platform highlights key trade-offs in terms of latency, cost, power consumption, and resource utilization. The Conv2D-8bit (INT8) implementation stands out with the lowest latency, highest clock frequency (174.74 MHz), and minimal power consumption (1.7 W). Due to its efficient use of FPGA resources (slices, Look-Up Tables “LUTs”, Digital Signal Processing blocks “DSPs”, Super Logic Region “SLR”, and Block RAM “BRAM”), it enables fast and energy-efficient execution, making it particularly suitable for real-time, resource-constrained applications such as medical diagnostics (Table 7).
In contrast, the FP16 (16-bit floating-point) version provides improved numerical precision at the cost of moderately higher resource usage and power consumption (2.3 W). It offers a balanced compromise between performance and precision, maintaining reasonable latency and complexity. The FP32 (32-bit floating-point) variant, while delivering the highest precision, comes with significant trade-offs. It requires substantially more FPGA resources (more than double the LUTs and BRAM compared to INT8), operates at a lower frequency (160.30 MHz), and consumes more power (2.5 W). These factors result in increased latency and hardware overhead, limiting its suitability for deployment on lightweight FPGA platforms. Overall, the INT8 implementation offers the best balance between speed, energy efficiency, and cost, making it ideal for real-time systems where rapid response and hardware efficiency are critical. Meanwhile, FP16 and FP32 may be reserved for specific use cases that demand higher numerical accuracy and where sufficient FPGA resources are available.
The decision to accelerate only the first Conv2D layer on the FPGA was driven by the high computational load associated with processing full-resolution input images in medical imaging tasks. As this initial layer operates on the largest feature maps, it benefits most from hardware acceleration. Subsequent layers, which handle progressively smaller feature maps, were executed on the CPU to balance execution time and FPGA resource constraints (slices, LUTs, DSPs, BRAM). This hybrid strategy ensures low latency and efficient resource utilization while maintaining high classification accuracy, making it particularly suitable for real-time medical diagnostics where both performance and energy efficiency are critical (Table 6).
To improve the efficiency of the proposed CNN model for brain cancer multi-classification, a hybrid implementation is deployed on the PYNQ-Z2 FPGA. In this approach, the first Conv2D layer, which is the most computationally intensive, is offloaded to FPGA hardware, while the remaining layers are executed on the ARM Cortex-A9 processor. The model is evaluated using three different numerical precision strategies: INT8, FP16, and FP32, allowing a trade-off between accuracy, inference time, and power consumption.
The experimental results, as presented in Table 6, demonstrate that hardware acceleration significantly enhances the inference speed and reduces power consumption compared to the purely CPU-based implementation. Specifically, the INT8 quantized model achieves the lowest inference time of 645 ms with a power consumption of 2.8 W, at the cost of a slight drop in accuracy to 92.7%. The FP16 implementation balances accuracy and efficiency, achieving 94.1% accuracy with an inference time of 683 ms and a power consumption of 3.2 W. Finally, the FP32 precision model achieves the highest accuracy of 94.2%, matching the CPU-based results, while reducing inference time to 719 ms and power consumption to 3.6 W. Fig. 11 illustrates the trade-offs between accuracy, processing speed, and hardware resource utilization in our proposed hardware-accelerated Conv2D implementations using different numerical data representations: INT8, Fixed-Point 16 (FP16), and Fixed-Point 32 (FP32). These trade-offs are crucial in determining the optimal balance between computational efficiency and classification performance for real-time brain cancer multi-classification on embedded systems.
Figure 11: Performance Trade-offs of Conv2D acceleration across quantization levels on FPGA
The results of our proposed hybrid implementation demonstrate a well-balanced trade-off among precision, speed, and resource utilization, as illustrated in Fig. 11. This figure summarizes how different quantization strategies, image sizes, and architectural choices directly affect the performance metrics of Conv2D operations on hardware. Lower-bit quantization like INT8 reduces computational complexity, resulting in faster processing and lower power consumption, but may introduce a slight degradation in model accuracy. Conversely, higher precision formats such as FP32 offer improved numerical accuracy but significantly increase latency and power usage due to higher resource demands.
Our experiments validate this trade-off clearly. Implemented on a PYNQ-Z2 FPGA, INT8 quantization achieves a strong throughput of 1.562 FPS with the lowest power consumption (2.8 W) and shortest inference time (645 ms), confirming its suitability for power-constrained, real-time medical diagnostic applications. Although INT8 resulted in a slight drop in accuracy (92.7%), the gain in efficiency justifies its use for embedded systems. Meanwhile, FP16 serves as a middle ground, offering 94.1% accuracy, moderate latency (683 ms), and power usage (3.2 W). This format delivers a favorable compromise between computational load and inference quality. Finally, FP32, despite its higher demands, achieved the highest accuracy (94.2%), closely matching the results obtained on the ARM Cortex-A9 CPU. However, this came at the cost of increased power (3.6 W) and latency (719 ms), making FP32 more suitable for precision-critical scenarios rather than real-time execution.
When benchmarked against state-of-the-art methods in Table 6, our approach stands out for achieving high accuracy comparable to advanced deep learning models (e.g., ResNet50, MobileNetV2) while significantly outperforming them in inference speed and power efficiency, particularly on FPGA. For example, while Xiong et al. [28] achieved similar accuracy on GPU and FPGA, our implementation delivers faster inference and lower power use. Additionally, many CPU-based implementations listed in the literature lack real-time capability or detailed power metrics, making our results more applicable to deployable embedded medical systems.
These findings validate the effectiveness and flexibility of our hybrid hardware-software design, which allows tuning the quantization level based on the constraints of the target application. Whether the goal is to maximize accuracy (FP32), balance performance and energy (FP16), or prioritize efficiency for real-time deployment (INT8), our architecture supports scalable optimization, making it a strong candidate for practical use in portable brain tumor detection systems.
To assess the effectiveness of our proposed hybrid implementation, we compare its performance with state-of-the-art approaches, as summarized in Table 6. The comparison considers key evaluation metrics, including accuracy, inference time, and power consumption, across different platforms such as GPUs, CPUs, and FPGAs.
Our proposed method, implemented on the PYNQ-Z2 FPGA, demonstrates significant advantages over recent state-of-the-art works in terms of accuracy (Fig. 12), energy efficiency, and real-world performance (Fig. 13). With FP32 quantization, our approach achieves 94.2% accuracy, which notably surpasses the 88.5% reported by Xiong et al. [28] on GPU, 89% by Khan et al. [29], and 85% by Chen et al. [30]. Even when quantized to INT8, our system maintains a high accuracy of 92.7% while consuming only 2.8 W of power, compared to Xiong et al.’s FPGA implementation which consumes 45 W but yields lower accuracy (88.2%). Additionally, our method attains a throughput of 1.562 frames per second (FPS) with INT8 quantization, outperforming Xiong et al. on FPGA (1.282 FPS with FP32) and Sharif et al. [31] (1.408 FPS), the latter lacking detailed hardware specifications. Unlike Khan et al. and Sharif et al., whose results are based on simulations without real hardware deployment, our approach has been validated on actual hardware (PYNQ-Z2 FPGA combined with an ARM Cortex-A9 CPU), making it a more reliable solution for resource-constrained embedded applications. By accelerating the most computationally intensive Conv2D layer in hardware while executing the rest of the CNN in software, our hybrid implementation strikes an optimal balance between accuracy, inference latency, and power consumption, enabling real-time performance suitable for edge-based medical diagnostics. Furthermore, although recent studies by Rao et al. [33], Islam et al. [34], Wang et al. [35], Mahmud et al. [36], and Pedada et al. [37] report high classification accuracies between 91% and 96%, these works primarily run on general-purpose CPUs and lack critical deployment metrics such as inference latency, power consumption, and throughput. This absence of hardware-oriented evaluation limits their applicability in practical, portable medical systems where both accuracy and system-level efficiency are essential. Overall, our approach offers a comprehensive and hardware-validated solution with a superior trade-off between performance and energy efficiency, making it particularly well-suited for embedded medical applications requiring real-time processing.
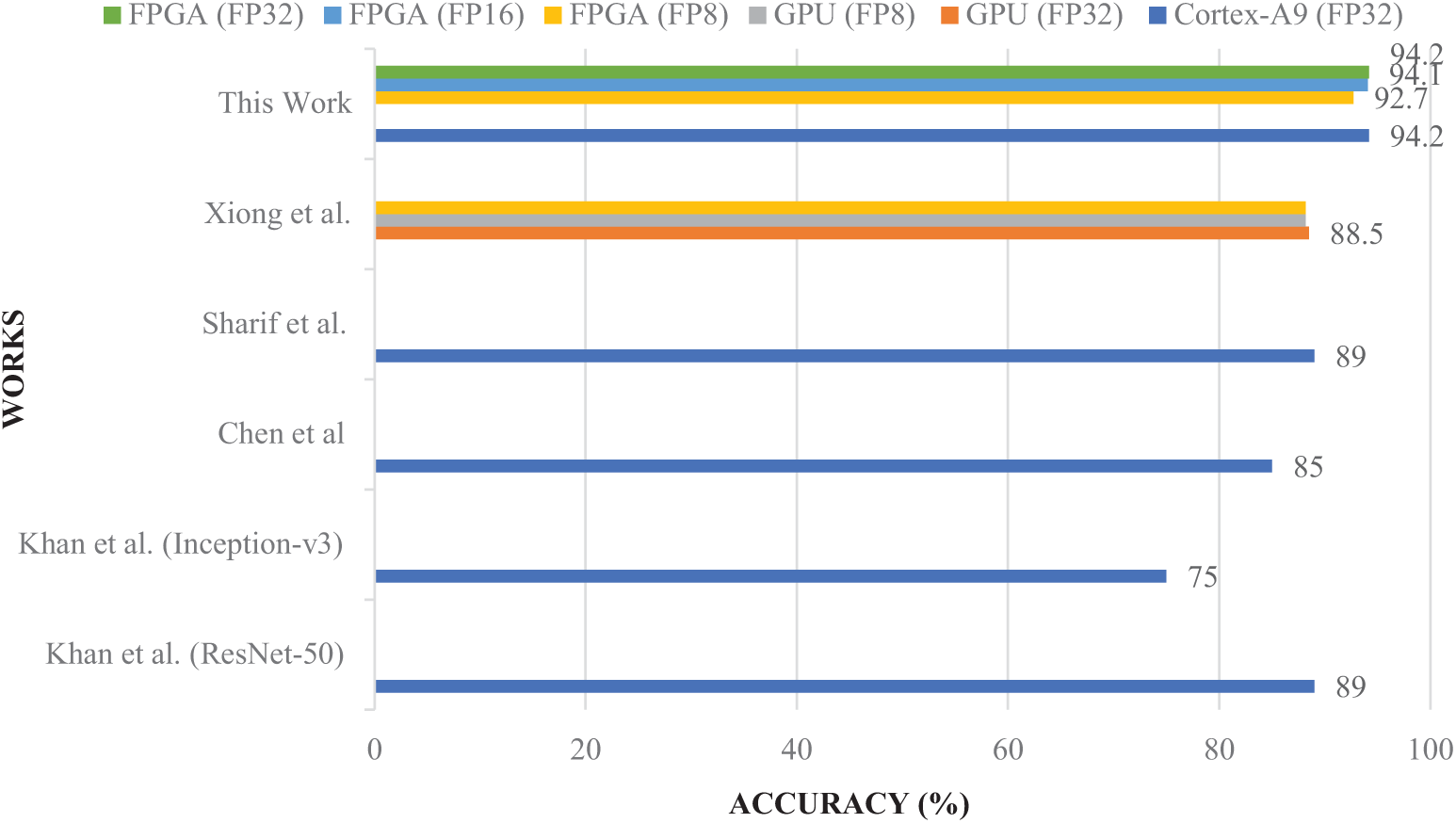
Figure 12: Accuracy comparison of the proposed model with state-of-the-art methods [27–30]
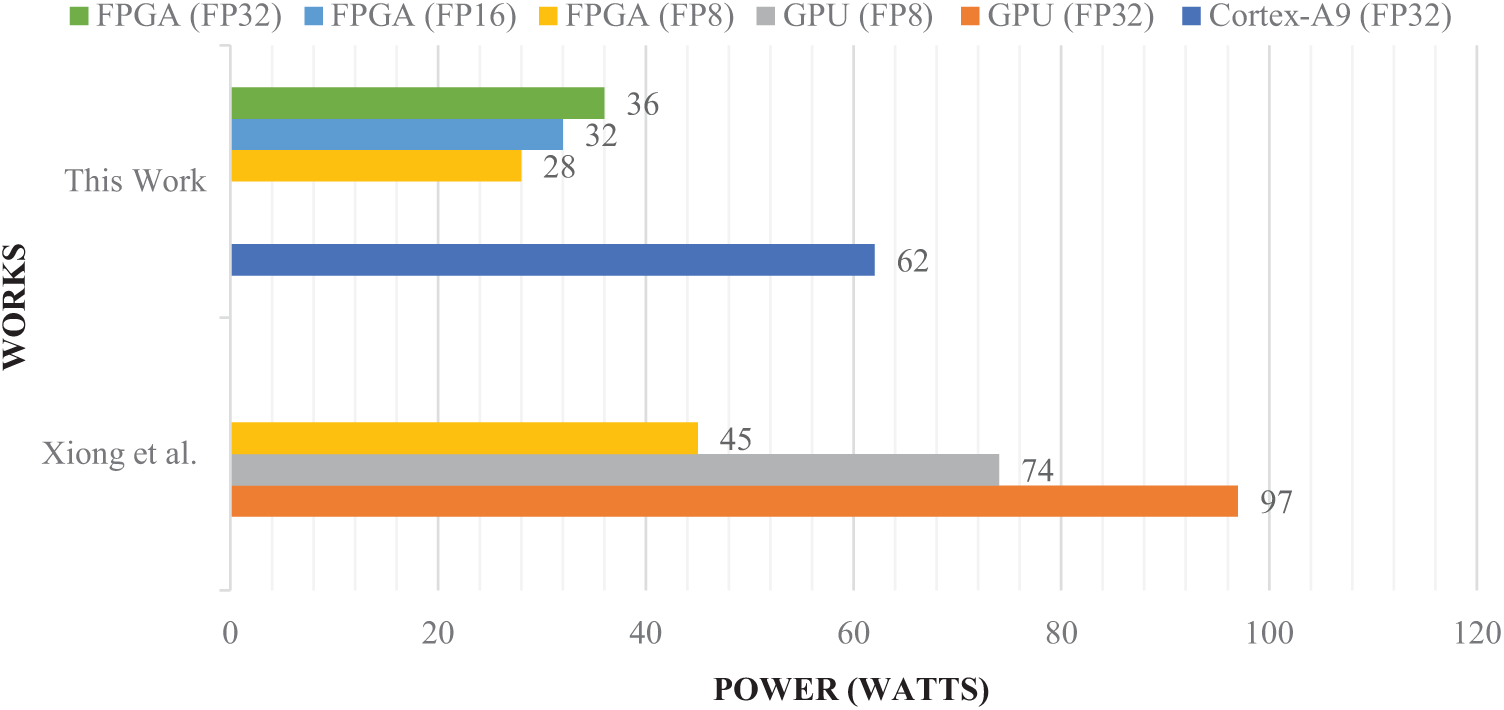
Figure 13: Power consumption comparison of the proposed model with state-of-the-art methods [27]
Deploying the hybrid model for brain cancer classification, implemented on the ARM Cortex-A9 and XC7Z020 FPGA, in mobile devices and other highly constrained devices is feasible for our work. The combination of the Cortex-A9 processor and FPGA-based acceleration allows for a highly efficient system that balances computational power with low power consumption key requirements for mobile and highly constrained devices. Our model’s low power consumption, particularly when using INT8 quantization, ensures that it can operate effectively within the strict power limits of these devices, such as smartphones or portable diagnostic tools. The high accuracy of 94.2% with FP32 and 92.7% with INT8 quantization ensures that the system can deliver reliable brain cancer classification results in real-time, an essential feature for medical applications. Moreover, the real-time processing capability of 1.562 FPS with INT8 quantization ensures the model can process data quickly enough for mobile use cases that require immediate feedback. The compact nature of the ARM Cortex-A9 and XC7Z020 FPGA platforms also makes them suitable for integration into small, resource-constrained devices. The scalability of the model, with its varying quantization options, further allows for fine-tuning to achieve an optimal balance between accuracy, power consumption, and processing speed based on the device’s constraints. While challenges like optimizing the system for even lower power consumption in ultra-compact devices remain, the hybrid model represents a promising solution for brain cancer classification in mobile and highly constrained medical devices, offering both efficiency and performance within the limitations of these environments.
The proposed hybrid hardware-software CNN acceleration strategy holds strong potential for clinical integration, particularly in point-of-care and portable diagnostic devices. Its low-latency and energy-efficient design makes it suitable for real-time inference in resource-constrained environments such as rural clinics, emergency units, and mobile screening platforms. Deployment can be streamlined through integration with existing imaging modalities like MRI scanners, where the FPGA-based module can be embedded directly within the acquisition hardware or connected via edge-computing interfaces. Future work will involve developing a user-friendly interface, validating performance with larger clinical datasets, and complying with medical device regulations (e.g., FDA, CE marking). Furthermore, with support for multiple numerical formats (INT8, FP16, FP32), the design allows scalability across a spectrum of clinical workloads, from preliminary triage to detailed diagnostic support, facilitating broader adoption in next-generation AI-assisted medical imaging systems.
In this work, we proposed a hybrid hardware-software CNN architecture for brain cancer multi-classification, leveraging FPGA acceleration for the first Conv2D layer while executing the remaining layers on an embedded Cortex-A9 CPU. Our approach effectively balances accuracy, computational efficiency, and power consumption, making it suitable for real-time medical diagnostics on edge devices. Through extensive evaluations, we demonstrated that hardware acceleration significantly improves inference speed and reduces power consumption compared to a purely software-based implementation. The PYNQ-Z2 FPGA implementation using INT8, FP16, and FP32 quantization strategies showcased a trade-off between accuracy and efficiency, with FP16 offering the best balance (94.1% accuracy, 683 ms inference time, and 32 W power consumption). Compared to state-of-the-art methods, our model achieves higher classification accuracy (94.2%) while consuming significantly less power than GPU-based solutions. Our work highlights the potential of hybrid CNN implementations for edge-based medical AI, paving the way for more efficient and accurate real-time tumor classification solutions.
To further enhance the performance and efficiency of our model, future work will focus on extending the current FPGA-based acceleration to a full Convolutional Neural Network (CNN) implementation. This will involve optimizing all layers of the CNN, from convolutions to fully connected layers, on FPGA hardware to further enhance performance and resource efficiency. Additionally, real-time testing in clinical environments will be conducted to validate the system’s practicality and reliability in actual medical imaging workflows. This will include evaluating the model’s robustness across different brain tumor datasets, testing scalability for larger images, and assessing the real-time inference capabilities within operational healthcare settings.
Acknowledgement: The authors extend their appreciation to the Deanship of Scientific Research at Northern Border University, Arar, Saudi Arabia for funding this research work through the project number NBU-FFR-2025-432-03.
Funding Statement: This research is supported by Northern Border University Researchers Supporting Project number (NBU-FFR-2025-432-03), Northern Border University, Arar, Saudi Arabia.
Author Contributions: Ayoub Mhaouch: Software & hardware development, validation, data analysis, writing—review & editing. Wafa Gtifa: Conceptualization, methodology, investigation, writing—original draft. Turke Althobaiti: Funding acquisition, visualization, review of revisions. Hamzah Faraj: Funding acquisition, visualization, reviewer comment integration. Mohsen Machhout: Supervision, project administration, technical review. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset used in this study is publicly available on Kaggle at: https://www.kaggle.com/datasets/ (accessed on 15 June 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Tandel GS, Biswas M, Kakde OG, Tiwari A, Suri HS, Turk M, et al. A review on a deep learning perspective in brain cancer classification. Cancers. 2019;11(1):111. doi:10.3390/cancers11010111. [Google Scholar] [PubMed] [CrossRef]
2. Nazir M, Shakil S, Khurshid K. Role of deep learning in brain tumor detection and classification (2015 to 2020a review. Comput Med Imag Graph. 2021;91:101940. doi:10.1016/j.compmedimag.2021.101940. [Google Scholar] [PubMed] [CrossRef]
3. Deng L, Li G, Han S, Shi L, Xie Y. Model compression and hardware acceleration for neural networks: a comprehensive survey. Proc IEEE. 2020;108(4):485–532. doi:10.1109/JPROC.2020.2976475. [Google Scholar] [CrossRef]
4. Wang Y, Han Y, Wang C, Song S, Tian Q, Huang G. Computation-efficient deep learning for computer vision: a survey. Cybern Intell, 2024:1–24. doi:10.26599/CAI.2024.9390002. [Google Scholar] [CrossRef]
5. Canziani A, Paszke A, Culurciello E. An analysis of deep neural network models for practical applications. arXiv:1605.07678. 2016. [Google Scholar]
6. Vardhana M, Arunkumar N, Lasrado S, Abdulhay E, Ramirez-Gonzalez G. Convolutional neural network for bio-medical image segmentation with hardware acceleration. Cogn Syst Res. 2018;50:10–4. doi:10.1016/j.cogsys.2018.03.005. [Google Scholar] [CrossRef]
7. Azghadi MR, Lammie C, Eshraghian JK, Payvand M, Donati E, Linares-Barranco B, et al. Hardware implementation of deep network accelerators towards healthcare and biomedical applications. IEEE Trans Biomed Circuits Syst. 2020;14(6):1138–59. doi:10.1109/TBCAS.2020.3036081. [Google Scholar] [PubMed] [CrossRef]
8. Alcaín E, Fernández PR, Nieto R, Montemayor AS, Vilas J, Galiana-Bordera A, et al. Hardware architectures for real-time medical imaging. Electronics. 2021;10(24):3118. doi:10.3390/electronics10243118. [Google Scholar] [CrossRef]
9. Garifulla M, Shin J, Kim C, Kim WH, Kim HJ, Kim J, et al. A case study of quantizing convolutional neural networks for fast disease diagnosis on portable medical devices. Sensors. 2021;22(1):219. doi:10.3390/s22010219. [Google Scholar] [PubMed] [CrossRef]
10. Xu Y, Luo J, Sun W. Flare: an FPGA-based full precision low power CNN accelerator with reconfigurable structure. Sensors. 2024;24(7):2239. doi:10.3390/s24072239. [Google Scholar] [PubMed] [CrossRef]
11. Sze V, Chen YH, Yang TJ, Emer JS. Efficient processing of deep neural networks: a tutorial and survey. Proc IEEE. 2017;105(12):2295–329. doi:10.1109/JPROC.2017.2761740. [Google Scholar] [CrossRef]
12. Oh S, Shi Y, Del Valle J, Salev P, Lu Y, Huang Z, et al. Energy-efficient Mott activation neuron for full-hardware implementation of neural networks. Nat Nanotechnol. 2021;16(6):680–7. doi:10.1038/s41565-021-00874-8. [Google Scholar] [PubMed] [CrossRef]
13. Mohaidat T, Khalil K. A survey on neural network hardware accelerators. IEEE Trans Artif Intell. 2024;5(8):3801–22. doi:10.1109/TAI.2024.3377147. [Google Scholar] [CrossRef]
14. Tran VD, Xuan Hieu Le T, Tran TD, Luan Pham H, Duong Le VT, Hai Vu T, et al. Exploring the limitations of Kolmogorov-Arnold networks in classification: insights to software training and hardware implementation. In: Proceedings of the 2024 Twelfth International Symposium on Computing and Networking Workshops (CANDARW); 2024 Nov 26–29; Naha, Japan. Piscataway, NJ, USA: IEEE; 2024. p. 110–6. doi:10.1109/CANDARW64572.2024.00026. [Google Scholar] [CrossRef]
15. Alimisis V, Anastasios Serlis E, Papathanasiou A, Eleftheriou NP, Sotiriadis PP. Power-efficient analog hardware architecture of the learning vector quantization algorithm for brain tumor classification. IEEE Trans Very Large Scale Integr VLSI Syst. 2024;32(11):1969–82. doi:10.1109/TVLSI.2024.3447903. [Google Scholar] [CrossRef]
16. Pacini F, Pacini T, Lai G, Zocco AM, Fanucci L. Design and evaluation of CPU-, GPU-, and FPGA-based deployment of a CNN for motor imagery classification in brain-computer interfaces. Electronics. 2024;13(9):1646. doi:10.3390/electronics13091646. [Google Scholar] [CrossRef]
17. Neiso MK, Muchuka NM, Mambo SM. FPGA-based implementation of a resource-efficient UNET model for brain tumour segmentation. Int J Adv Comput Sci Appl. 2024;15(1):622–29. doi:10.14569/ijacsa.2024.0150161. [Google Scholar] [CrossRef]
18. Rayapati V, Gogireddy RKR, Gandi AK, Gajawada S, Sanampudi GKR, Rao N. FPGA-based hardware software co-design to accelerate brain tumour segmentation. In: Proceedings of the 2024 IEEE International Symposium on Circuits and Systems (ISCAS); 2024 May 19–22; Singapore. Piscataway, NJ, USA: IEEE; 2024. p. 1–5. doi:10.1109/ISCAS58744.2024.10558230. [Google Scholar] [CrossRef]
19. Nickparvar M. Brain Tumor MRI Dataset [Internet]. [cited 2025 Jun 15]. Available from: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset. [Google Scholar]
20. Cheng J. Brain tumor dataset [Internet]. [cited 2025 Jun 15]. Available from: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427. [Google Scholar]
21. Kaggle. Brain tumor classification (MRI) [Internet]. [cited 2025 Jun 15]. Available from: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri. [Google Scholar]
22. Hamada A. Br35H: brain tumor detection [Internet]. [Internet]. [cited 2025 Jun 15]. Available from: https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection. [Google Scholar]
23. Verma R, Vishwakarma SK, Bodade RM, Kumar P. MAC-DNN: an optimized hardware implementation of MAC unit in neuron engine for deep neural network applications. IETE J Res. 2025;71(3):919–26. doi:10.1080/03772063.2024.2448584. [Google Scholar] [CrossRef]
24. Afshar P, Mohammadi A, Plataniotis KN. Brain tumor type classification via capsule networks. In: Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP); 2018 Oct 7–10; Athens, Greece. Piscataway, NJ, USA: IEEE; 2018. p. 3129–33. doi:10.1109/ICIP.2018.8451379. [Google Scholar] [CrossRef]
25. Deepak S, Ameer PM. Automated categorization of brain tumor from MRI using CNN features and SVM. J Ambient Intell Humaniz Comput. 2021;12(8):8357–69. doi:10.1007/s12652-020-02568-w. [Google Scholar] [CrossRef]
26. Amin J, Sharif M, Haldorai A, Yasmin M, Nayak RS. Brain tumor detection and classification using machine learning: a comprehensive survey. Complex Intell Syst. 2022;8(4):3161–83. doi:10.1007/s40747-021-00563-y. [Google Scholar] [CrossRef]
27. Abiwinanda N, Hanif M, Hesaputra ST, Handayani A, Mengko TR. Brain tumor classification using convolutional neural network. In: World Congress on Medical Physics and Biomedical Engineering 2018; Singapore: Springer Nature; 2018. p. 183–9. doi:10.1007/978-981-10-9035-6_33. [Google Scholar] [CrossRef]
28. Xiong S, Wu G, Fan X, Feng X, Huang Z, Cao W, et al. MRI-based brain tumor segmentation using FPGA-accelerated neural network. BMC Bioinform. 2021;22(1):421. doi:10.1186/s12859-021-04347-6. [Google Scholar] [PubMed] [CrossRef]
29. Khan HA, Jue W, Mushtaq M, Mushtaq MU. Brain tumor classification in MRI image using convolutional neural network. Math Biosci Eng. 2020;17(5):6203–16. doi:10.3934/mbe.2020328. [Google Scholar] [PubMed] [CrossRef]
30. Chen S, Ding C, Liu M. Dual-force convolutional neural networks for accurate brain tumor segmentation. Pattern Recognit. 2019;88(suppl_5):90–100. doi:10.1016/j.patcog.2018.11.009. [Google Scholar] [CrossRef]
31. Sharif M, Amin J, Raza M, Anjum MA, Afzal H, Ali Shad S. Brain tumor detection based on extreme learning. Neural Comput Appl. 2020;32(20):15975–87. doi:10.1007/s00521-019-04679-8. [Google Scholar] [CrossRef]
32. Manalı D, Demirel H. Comparison of vision transformer with convolutional neural networks for brain cancer classification. In: Proceedings of the 2025 IEEE 17th International Conference on Computer Research and Development (ICCRD); 2025 Jan 17–19; Shangrao, China. Piscataway, NJ, USA: IEEE; 2025. p. 78–84. doi:10.1109/ICCRD64588.2025.10963037. [Google Scholar] [CrossRef]
33. Rao KN, Khalaf OI, Krishnasree V, Kumar AS, Alsekait DM, Priyanka SS, et al. An efficient brain tumor detection and classification using pre-trained convolutional neural network models. Heliyon. 2024;10(17):e36773. doi:10.1016/j.heliyon.2024.e36773. [Google Scholar] [PubMed] [CrossRef]
34. Islam M, Reza MT, Kaosar M, Parvez MZ. Effectiveness of federated learning and CNN ensemble architectures for identifying brain tumors using MRI images. Neural Process Lett. 2023;2022(4):1–31. doi:10.1007/s11063-022-11014-1. [Google Scholar] [PubMed] [CrossRef]
35. Wang N, Lee CY, Park HC, Nauen DW, Chaichana KL, Quinones-Hinojosa A, et al. Deep learning-based optical coherence tomography image analysis of human brain cancer. Biomed Opt Express. 2022;14(1):81–8. doi:10.1364/BOE.477311. [Google Scholar] [PubMed] [CrossRef]
36. Mahmud MI, Mamun M, Abdelgawad A. A deep analysis of brain tumor detection from MR images using deep learning networks. Algorithms. 2023;16(4):176. doi:10.3390/a16040176. [Google Scholar] [CrossRef]
37. Pedada KR, Bhujanga Rao A, Patro KK, Allam JP, Jamjoom MM, Abdel Samee N. A novel approach for brain tumour detection using deep learning based technique. Biomed Signal Process Control. 2023;82(2):104549. doi:10.1016/j.bspc.2022.104549. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF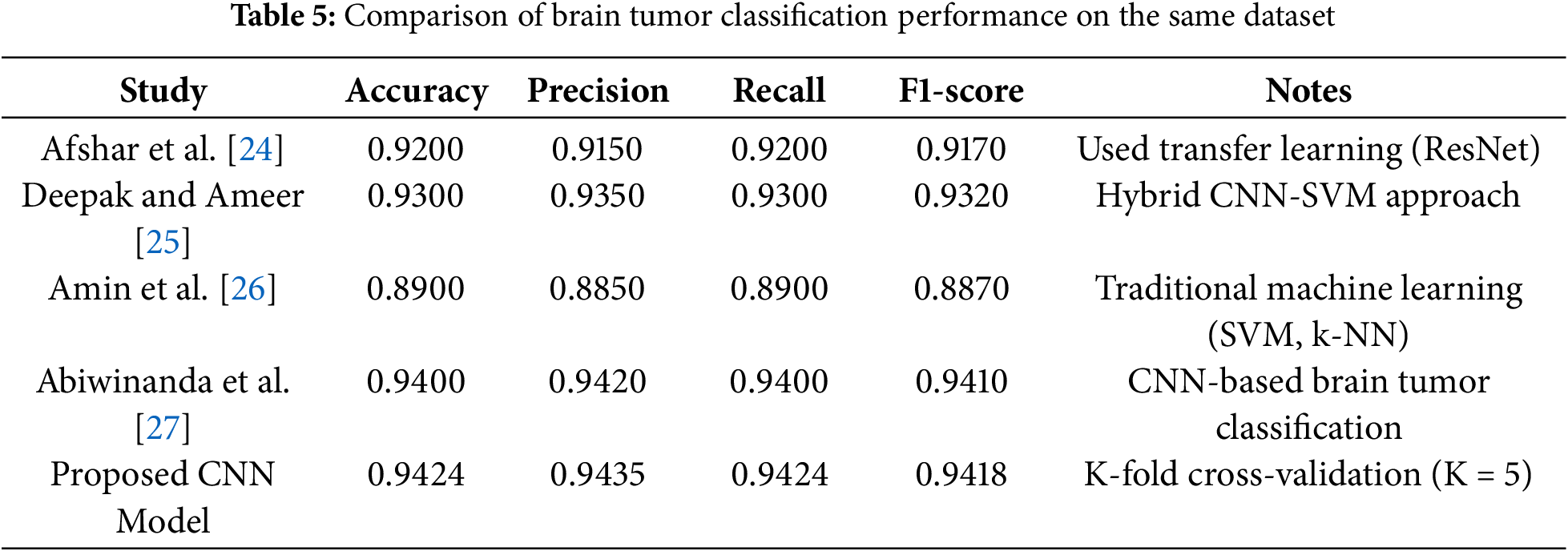
 Downloads
Downloads
 Citation Tools
Citation Tools