 Open Access
Open Access
ARTICLE
3D Echocardiogram Reconstruction Employing a Flip Directional Texture Pyramid
PG & Research Department of Computer Science, SadakathullahAppa College, Affiliation of Manonmaniam Sundaranar University, Tirunelveli, 627012, Tamil Nadu, India
* Corresponding Author: C. Preethi. Email:
Computer Systems Science and Engineering 2023, 45(3), 2971-2988. https://doi.org/10.32604/csse.2023.033423
Received 16 June 2022; Accepted 18 August 2022; Issue published 21 December 2022
Abstract
Three dimensional (3D) echocardiogram enables cardiologists to visualize suspicious cardiac structures in detail. In recent years, this three-dimensional echocardiogram carries important clinical value in virtual surgical simulation. However, this 3D echocardiogram involves a trade-off difficulty between accuracy and efficient computation in clinical diagnosis. This paper presents a novel Flip Directional 3D Volume Reconstruction (FD-3DVR) method for the reconstruction of echocardiogram images. The proposed method consists of two main steps: multiplanar volumetric imaging and 3D volume reconstruction. In the creation of multiplanar volumetric imaging, two-dimensional (2D) image pixels are mapped into voxels of the volumetric grid. As the obtained slices are discontinuous, there are some missing voxels in the volume data. To restore the structural and textural information of 3D ultrasound volume, the proposed method creates a volume pyramid in parallel with the flip directional texture pyramid. Initially, the nearest neighbors of missing voxels in the multiplanar volumetric imaging are identified by 3D ANN (Approximate Nearest Neighbor) patch matching method. Furthermore, a flip directional texture pyramid is proposed and aggregated with distance in patch matching to find out the most similar neighbors. In the reconstruction step, structural and textural information obtained from different flip angle directions can reconstruct 3D volume well with the desired accuracy. Compared with existing 3D reconstruction methods, the proposed Flip Directional 3D Volume Reconstruction (FD-3DVR) method provides superior performance for the mean peak signal-to-noise ratio (40.538 for the proposed method I and 39.626 for the proposed method II). Experimental results performed on the cardiac datasets demonstrate the efficiency of the proposed method for the reconstruction of echocardiogram images.Keywords
Medical imaging plays an essential role in the healthcare continuum from early diagnosis and selection of treatment to image-guided interventions [1]. Over the years, 2D Ultrasound has been explored as a mainstream imaging modality among CT (Computed Tomography), MRI (Magnetic Resonance Imaging), and PET (Positron Emission Tomography) due to its indispensable features, including cost-effective, faster, non-ionizing radiation and flexibility [2]. 2D Cardiac ultrasound or 2D echocardiogram becomes an invaluable tool for the assessment of abnormalities in a mitral valve cardiac pathology.
Although conventional 2D echocardiogram is diffused into the clinical practice, three-dimensional echocardiogram [3] allows the medical professionals to examine the anatomy of patients and permits virtual run-through of the complicated surgical procedures [4,5]. In the case of the mitral valve, 3D echocardiogram reconstruction provides the ability to visualize the complete dimensionality of the prolapsed mitral valve from multiple views (Parasternal Short Axis (PSAX), Apical 4-Chamber (A4C), and Apical 2-Chamber (A2C) [6]. However, there are some common issues concerning 3D reconstruction including missing gaps and interpolation errors. To overcome these drawbacks, a new method is proposed to reconstruct a 3D volume, which provides the best visualization for the simulation of virtual surgery [7].
The main objective of the proposed approach is to generate the three-dimensional volume and reconstruct the missing areas using the structural and texture information of the volume. The contributions of the proposed work are summarized as follows:
a) The first stage constitutes the creation of multiplanar volumetric imaging using three cardiac views (Parasternal Short Axis (PSAX), apical 4-Chamber (A4C), and apical 2-Chamber (A2C) respectively.
b) In the second stage, the 3D approximate nearest neighbor would be applied to shorten the search for the nearest neighborhood voxels of missing regions. To obtain the most similar voxels, a novel Flip Oriented 3D Volume Reconstruction (FD-3DVR) method is proposed which extracts the textural information along the top, bottom, left, and right positions around the missing voxel.
c) In the third stage, volume and texture directional pyramids are generated to reconstruct the missing regions of the echocardiogram iteratively.
d) Finally, the proposed FD-3DVR method is analyzed using four different quality metrics namely, peak signal-to-noise ratio (PSNR), feature similarity index (FSIM), structural content (SC), and average difference (AD).
The rest of the sections are organized as follows: Section 2 describes the related works about 3D reconstruction techniques. Section 3 represents the methodology of 3D Volume Reconstruction followed by experimental results and analysis in Section 4. Finally, Section 5 highlights the work summary and discussions.
In terms of state-of-the-art approaches, plenty of techniques have been proposed for performing 3D ultrasound reconstruction. Generally, volume reconstruction methods fall into three categories based on implementation [8]: Pixel-Based Methods (PBM), Voxel-Based Methods (VBM), and Function-Based Methods (FBM). In VBM, every voxel in the 3D volume grid is assigned with the value of the nearest pixel in 2D ultrasound input slices. Voxel Nearest Neighbor (VNN) and Distance Weighted (DW) are the most common implementation method in VBM. VNN traverses each voxel in the structured 3D volume grid at a time and assigns a value with the nearest pixel from the acquired 2D slices. Although its downside is that, when the distance between voxel to pixel is too large, reconstruction artifacts including the misalignment of gaps can be observed. Distance Weighted method proceeds across each voxel and replaces the average value of the voxel based on the weighted distance between the pixel and voxel. However, the selected voxels smooth the 3D volume causing information loss in the reconstructed volume.
Pixel-Based Methods (PBM) are used to reconstruct the missing regions in 3D volume by traversing across all the pixels in the 2D ultrasound slices. Pixel-Nearest Neighbor (PNN) and Direct Frame Interpolation (DFI) [9] are the most common forms of pixel based methods. The neighboring voxels are selected based on the parameter valve that defines the distance from the missing voxel to be filled. Besides that, some artifacts within the reconstructed volume have been observed on the edges with this method. Function-Based Methods (FBM) always reconstruct the data in 3D volume depending on functional relationships between voxels. The most widely used FBM methods are Rayleigh interpolation [10] and Radial Basis Function (RBF) [11]. These methods have been reported with moderate reconstruction precision and over-fitting problem are also unavoidable.
Wen et al. [12] proposed fast marching method (FMM) for reconstructing 3D ultrasound volume, which initially determines the volume grid and then fills the empty voxels in the boundary using the forward approach. Conversely, the reverse approach iteratively fills the empty voxels in an inner boundary. Although the problem of obtaining accurate reconstruction results in less computational time remains unresolved. Wen et al. [13] have proposed an adaptive kernel regression (KR) for the reconstruction of freehand ultrasound images employing a smoothing parameter of the kernel. The difficulties stemming from over smoothening the 3D volume, causing some information loss on the original 2D slices. Cong et al. [14] have proposed a Global Patch Matching (GPM) Method using two strategies bin-filling and hole-filling. Upon comparison with other PNN, VNN, and KR methods they have reported that the GPM reconstruction method preserves structural distribution effectively. Most importantly, these KR and GPM methods could not address texture preservation, which is necessary for volume reconstruction.
The problems of existing methods such as inaccurate voxel reconstruction, over smoothing, lack of texture preservation, and time complexity can be resolved by the proposed flip directional 3D volume reconstruction method. Here 3D approximate nearest neighbor method is used to decrease the time complexity for nearest neighborhood voxels. Volume and flip texture pyramids are used for the reconstruction of missing regions in the volume to preserve the structural and texture directional information of the original slices. The main purpose of this work is to create three-dimensional echocardiogram images in parallel with the reconstruction of missing regions using the proposed method to obtain a desired level of quality in 3D visualization.
3 Methodology of 3D Volume Reconstruction
3.1 Multiplanar Volumetric Imaging
Multiplanar volumetric imaging is standard clinical practice for many modalities namely CT, MRI, and ultrasound datasets. This display format permits examination of the 2D ultrasound slices in three orthogonal planes representing sagittal, axial, and coronal planes. Such an interactive volumetric display involves mapping the 2D cardiac ultrasound pixels into a regularly arranged volume grid. Initially, an empty volume grid is generated and a stack of three cardiac views (Parasternal Short Axis (PSAX), Apical 4-Chamber (A4C), and Apical 2-Chamber (A2C) are mapped based on position to display 3D volume. The mapping relationship [15] between the ultrasound scan coordinate system and 3D volume coordinates can be defined as
where
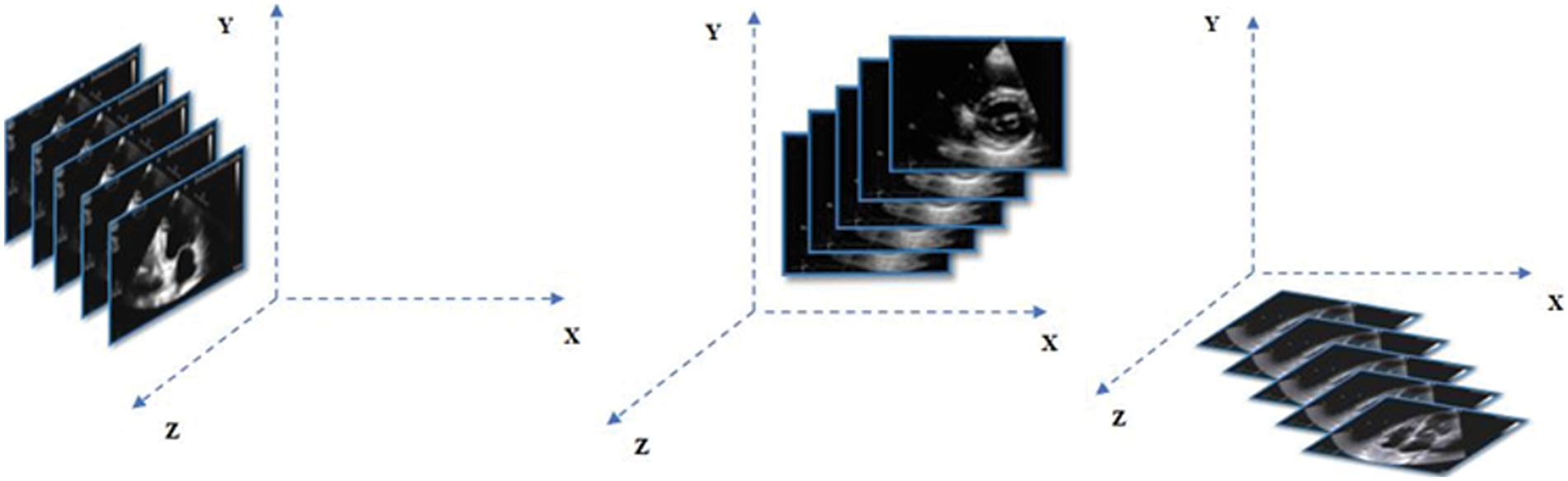
Figure 1: Schematic multiplanar volumetric arrangement of cardiac ultrasound along the X, Y, and Z axis
As the typical acquisition of ultrasound scan images is distorted with misalignment of pixels, there is missing data in the volume. Hence these missing values need to be reconstructed according to the nearest neighboring voxels in volume data.
3.2 Overview of Proposed Flip Oriented 3D Volume Reconstruction (FO-3DVR) Method
An efficient 3D reconstruction method is a significant requirement for volumetric cardiac imaging. Most of the existing approaches rely on single-dimensional textural features to build the 3D volume. Assume there are L levels in volume, and missing voxels at
Fig. 2 depicts the complete map of the proposed 3D reconstruction method. The proposed methodology is performed in two steps. The first step is, searching for the nearest neighbor of the missing voxels. Similar neighboring voxels are obtained through the 3D Approximate Nearest Neighbor (ANN) matching method [16]. The most similar voxels are obtained through the texture features in all flip directions and information is aggregated to the voxel distance calculated by the 3D ANN method.
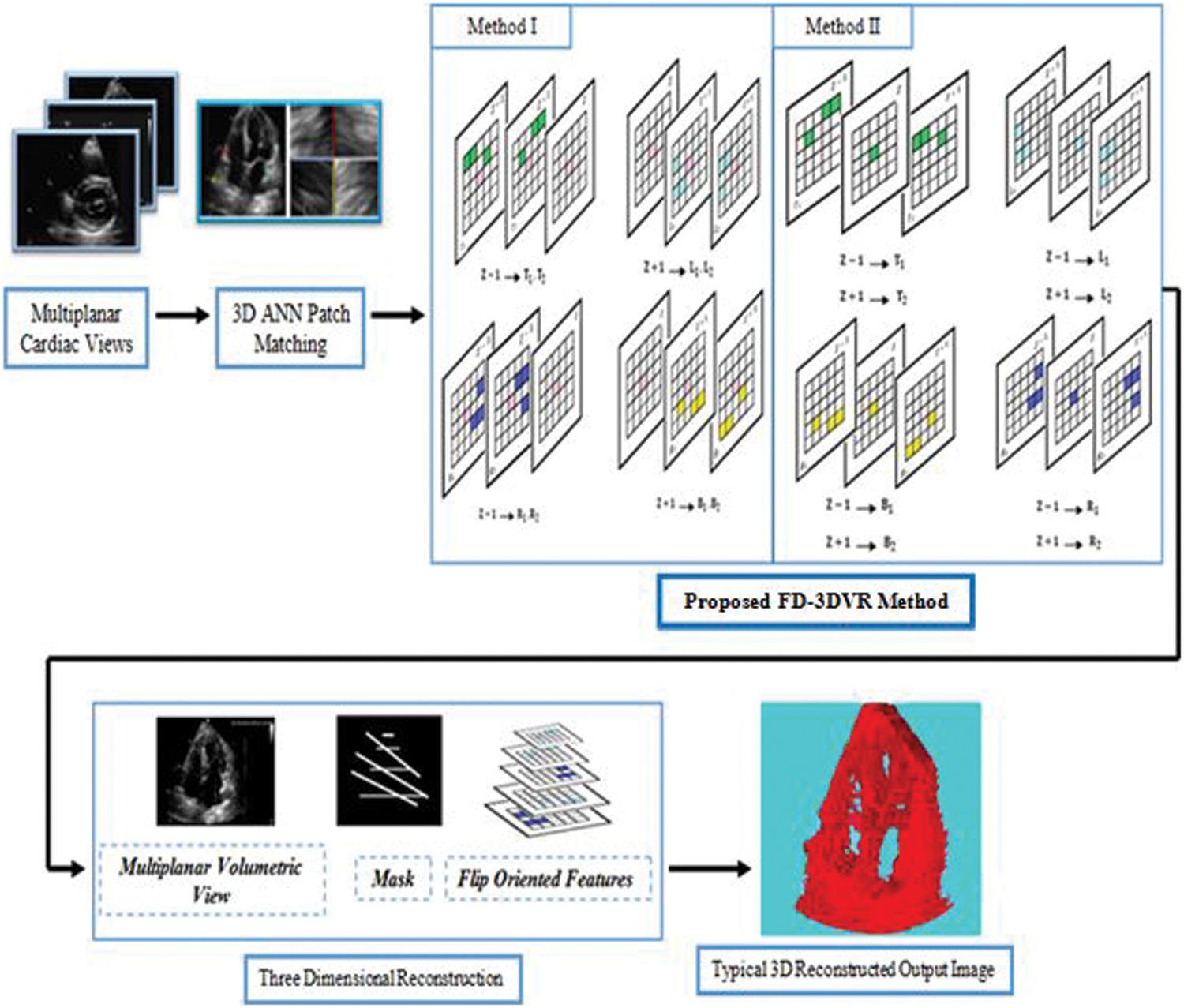
Figure 2: A complete map of the proposed flip directional 3d volume reconstruction (fd-3dvr) method
Next, in the reconstruction step, missing voxels in 3D ultrasound volume are reconstructed by propagating the pyramid scheme at intensity level and texture level. The basic idea of the proposed approach is to reconstruct the missing voxels of 3D ultrasound slices in terms of volume, corresponding mask, and flip directional textures through iterative down-sampling.
The non-local 3D patch matching method finds similar patches of the missing voxels in 3D ultrasound volume by randomly searching the entire volume into consideration. Let
where
3.2.2 Flip Directional Texture Pyramid (FDTP)
The volume texture pyramid aims to reconstruct missing areas by subsampling the texture features for the x, y, and z directions. This technique lacks reconstruction quality by the missing pixels. Hence in this study, we propose a pyramid that captures all the texture attributes in all the eight directions which we refer to as Flip Directional Texture Pyramid (FDTP). The angle range is subdivided into eight sub-ranges between
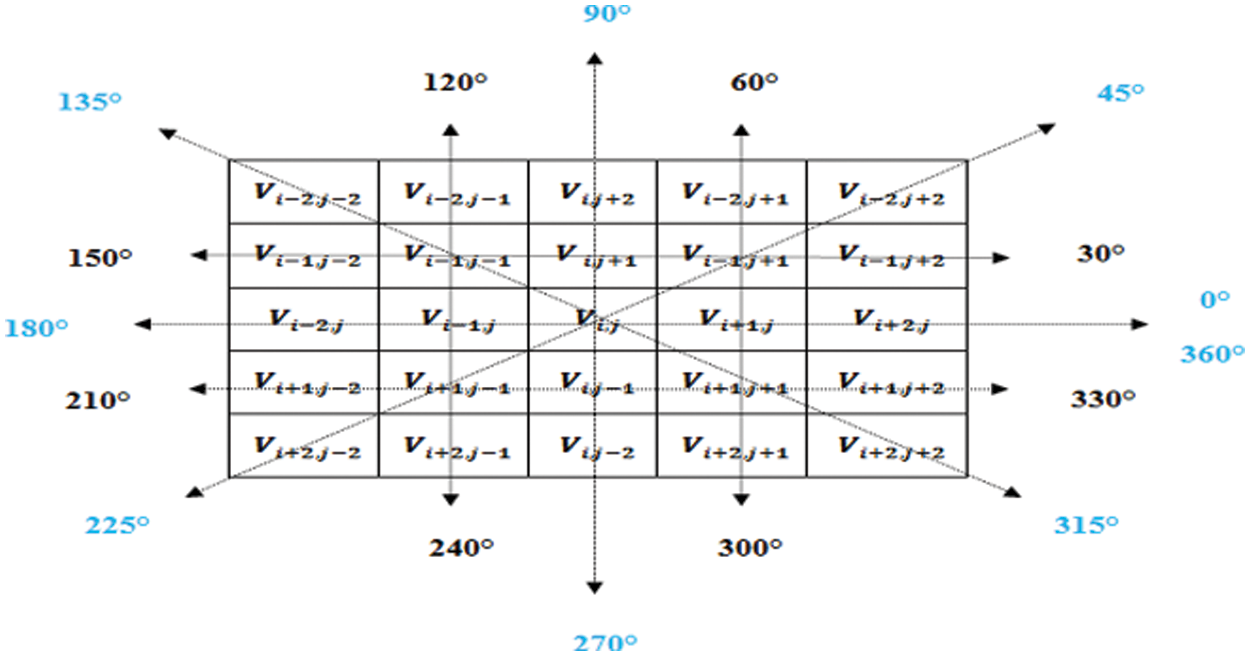
Figure 3: Directions regarded for the proposed flip directional texture pyramid
As noted, texture features are identified by averaging the absolute value of all gradient derivatives over a correspondent spatial neighborhood
where
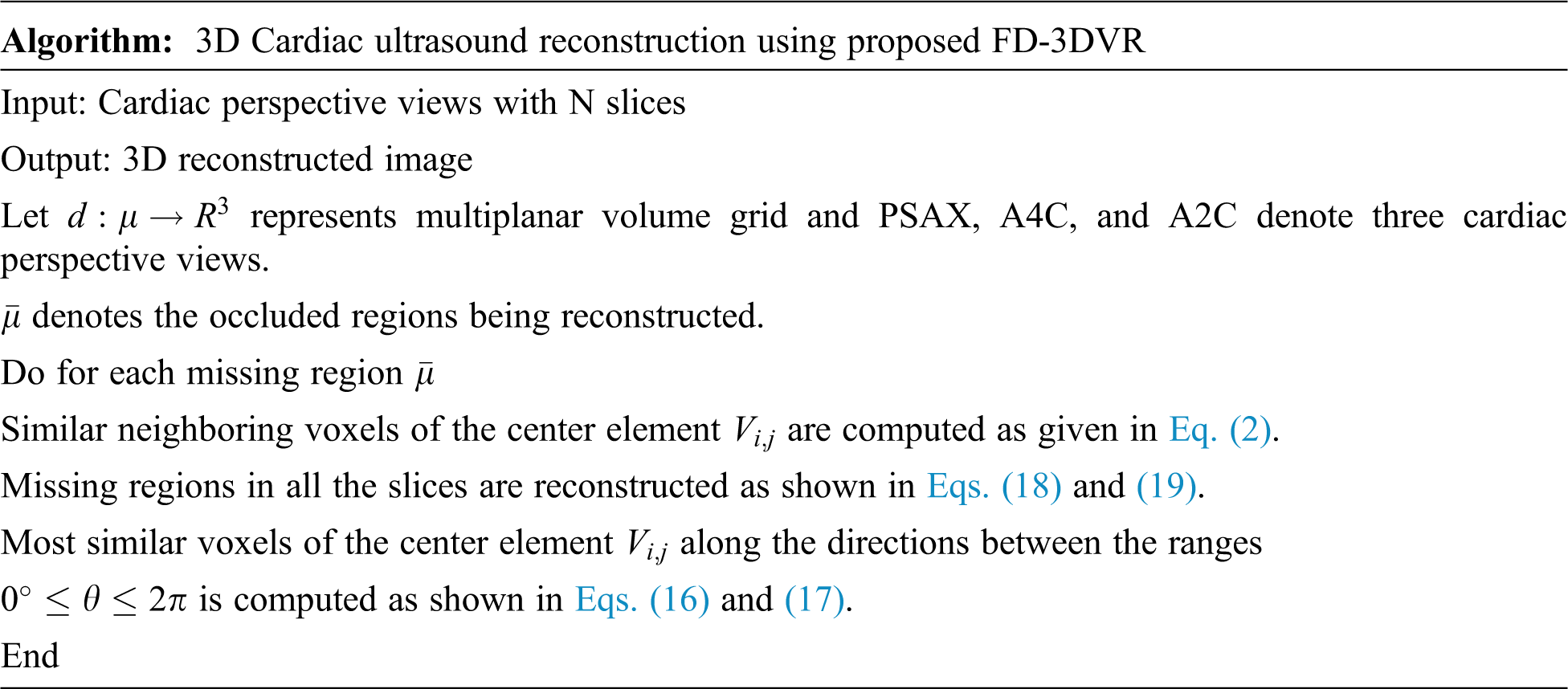
(i) Inter Voxel Based Flip Directional Texture Pyramid (Method I)
This proposed method results in the extraction of textural attributes of most similar neighboring voxels by taking up both the top and right level intensities from
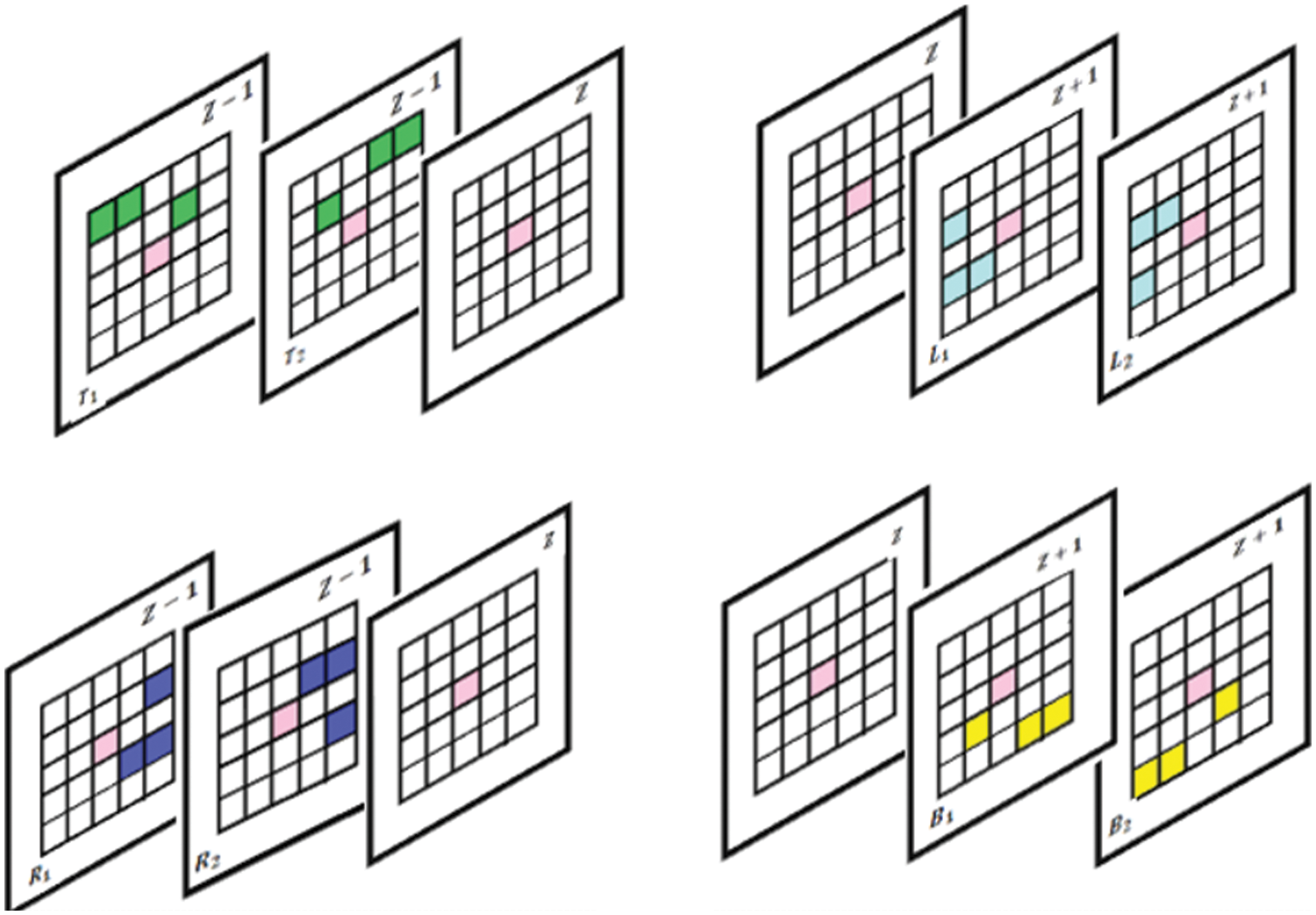
Figure 4: Proposed slice alignment for inter voxel based flip directional texture pyramid
The first row represents the top and left level and the second row indicates right and bottom level intensities.
In a single slice
where
where
The next bottom set of intensities is constructed below the center element
where
where
Similarly, the set of intensities is also extended along the right and left directions in slice
where
where
where
where
Thus, the flip directional feature for the patch P is estimated by taking the maximum value among each pair of directions and it is represented in Eqs. (12)–(15) respectively.
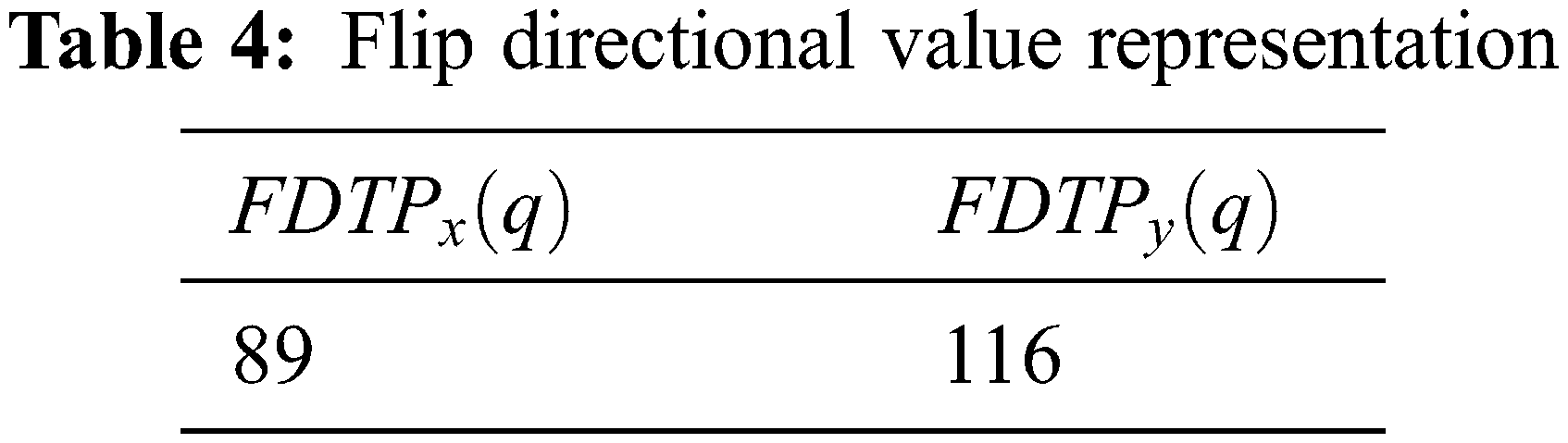
The resultant flip directional texture pyramid for three-dimensional reconstructions is computed using the Eqs. (16) and (17).
Let us consider the following representation for estimating
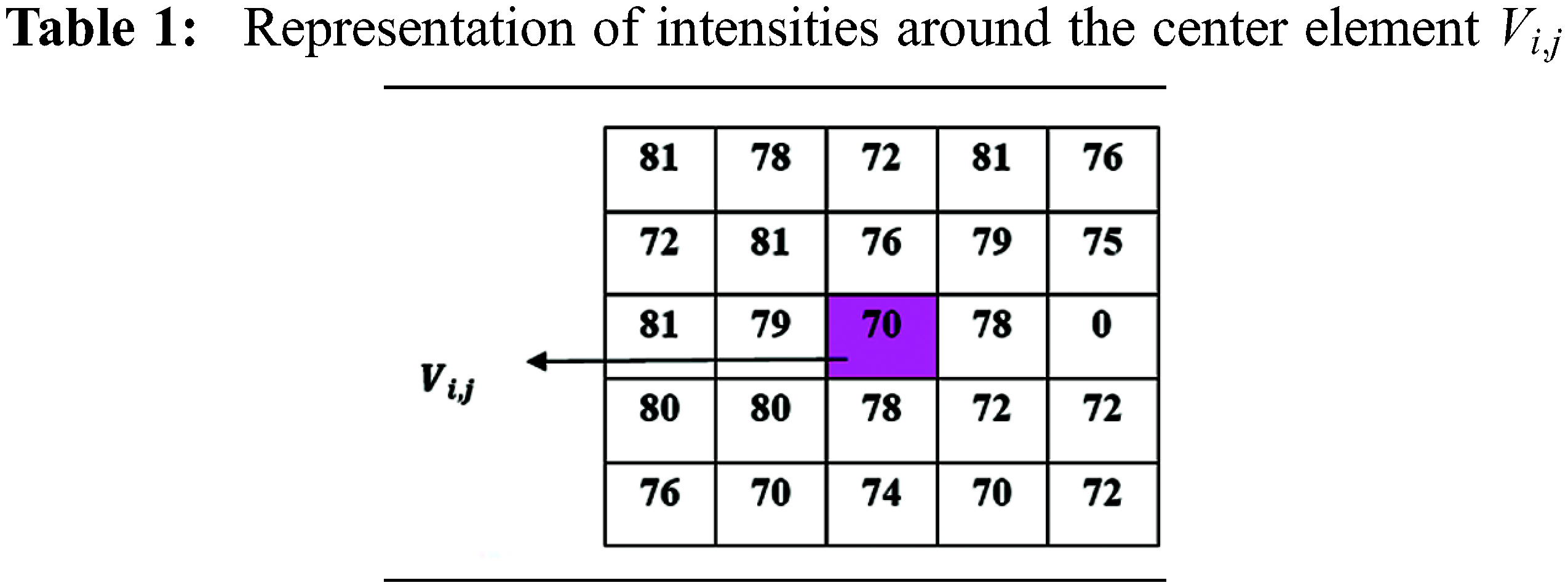
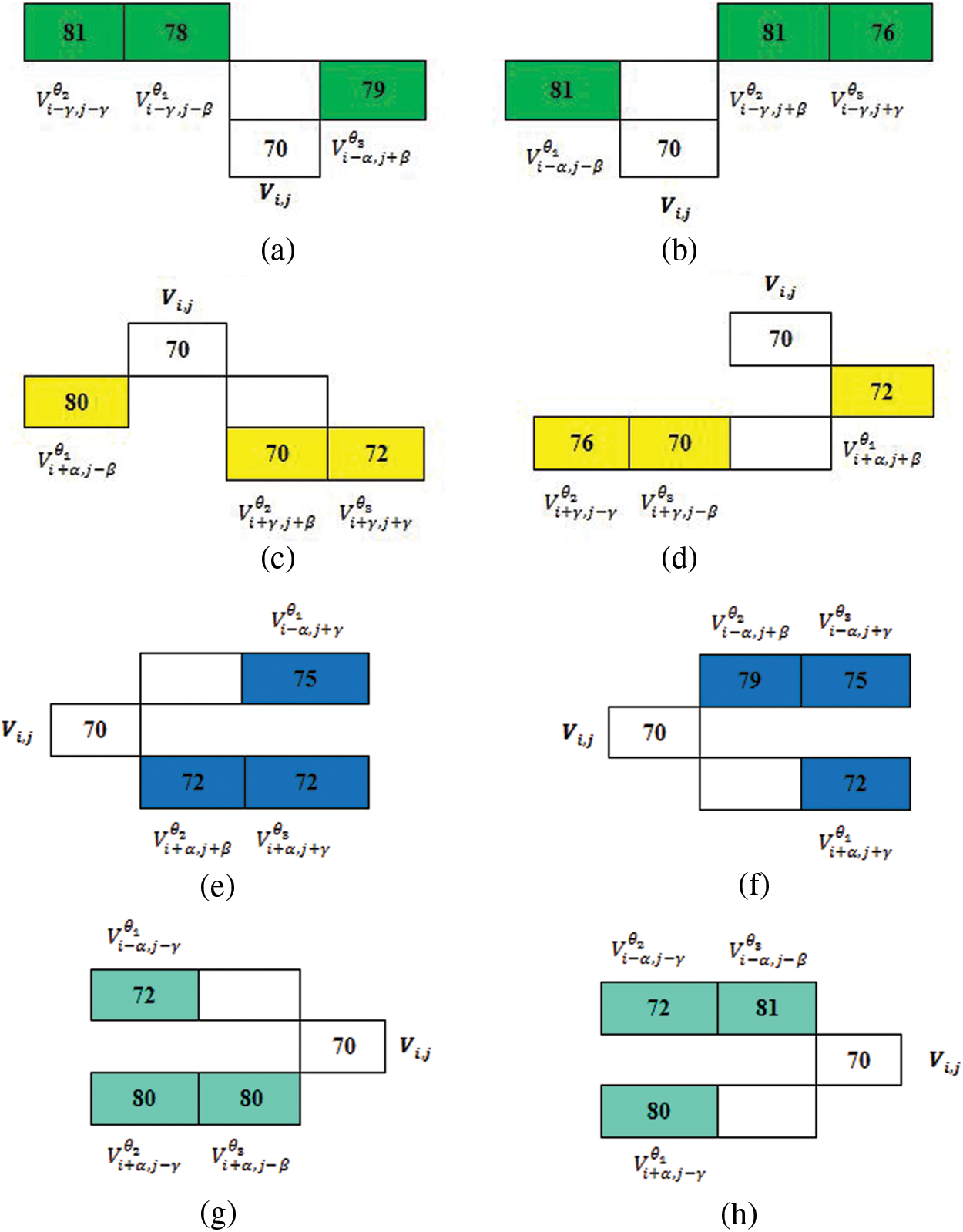
Figure 5: (a) and (b) Top
Using the above representation, flip directional values along the directions between the ranges
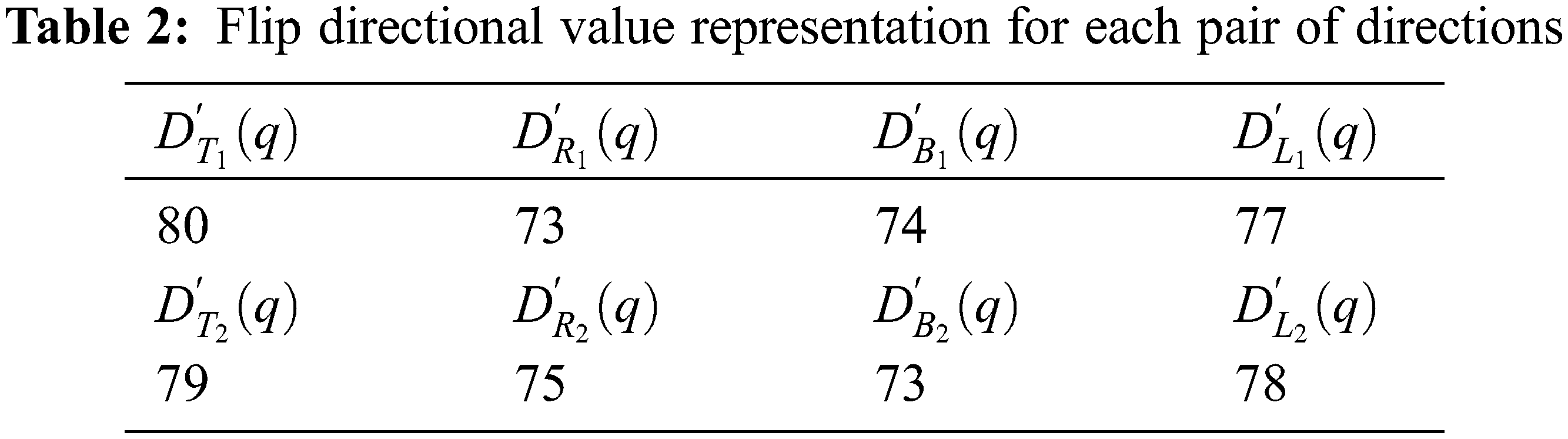
Now, the maximum value representation for the above pair of directions is shown in the Tab. 3.

The flip directional texture pyramid
(ii) Intra Voxel Based Flip Directional Texture Pyramid (Method II)
This method almost follows the same strategy as the aforementioned method, but the difference lies in the fact that here, textural features are extracted by constructing the initial top level intensities from the previous slice and next top level intensities from the successor slice in the range between
3.3 Reconstruction of Volumetric Imaging
Once the volume pyramid and proposed flip directional texture pyramid are built, three-dimensional volume reconstruction of the cardiac ultrasound procedure follows. Assuming there are N levels in the pyramid, in particular, every missing region is reconstructed in terms of structure and flip directional features as per Eqs. (18) and (19).
The same reconstruction procedure is repeated iteratively until the convergence of the finest resolution level. Therefore, the proposed reconstruction methods create a 3D volume and interpolate the missing regions in terms of structure and flip directional textures. The overall algorithm for the proposed method is presented here.
4 Experimental Results and Analysis
In this section, experiments were conducted with the proposed Flip Oriented 3D Volume Reconstruction (FO-3DVR) Method for the echocardiogram images. The images are obtained from a publicly available echocardia database [18]. The database contains more than 10,000 echocardiogram images and videos presenting various pathologies of the heart such as mitral valve disease, aortic valve prolapse, anterior myocardial infarction, etc., and the database also presents the visualization of cardiac views from different angles and orientations. A total of 200 prolapsed mitral valve echo images in three perspective views, Parasternal Short Axis (PSAX), Apical 4-Chamber (A4C), and Apical 2-Chamber (A2C) are chosen for our implementation. The current implementation is carried out using MATLAB 2019a.
The experiment is processed on five different ultrasound volumes with the four different predefined masks with a dimension of 240
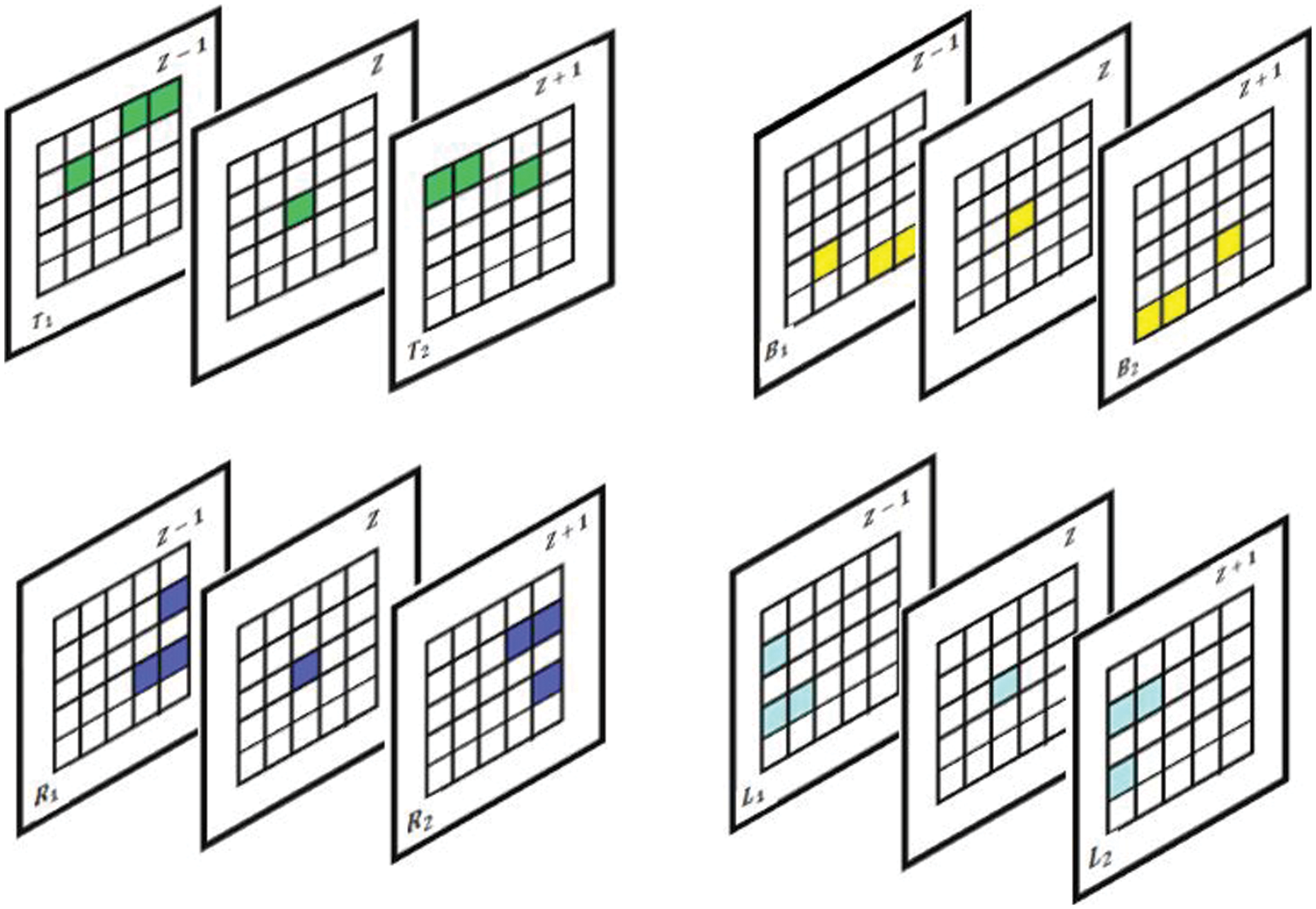
Figure 6: Proposed slice alignment for intra voxel based flip directional texture pyramid
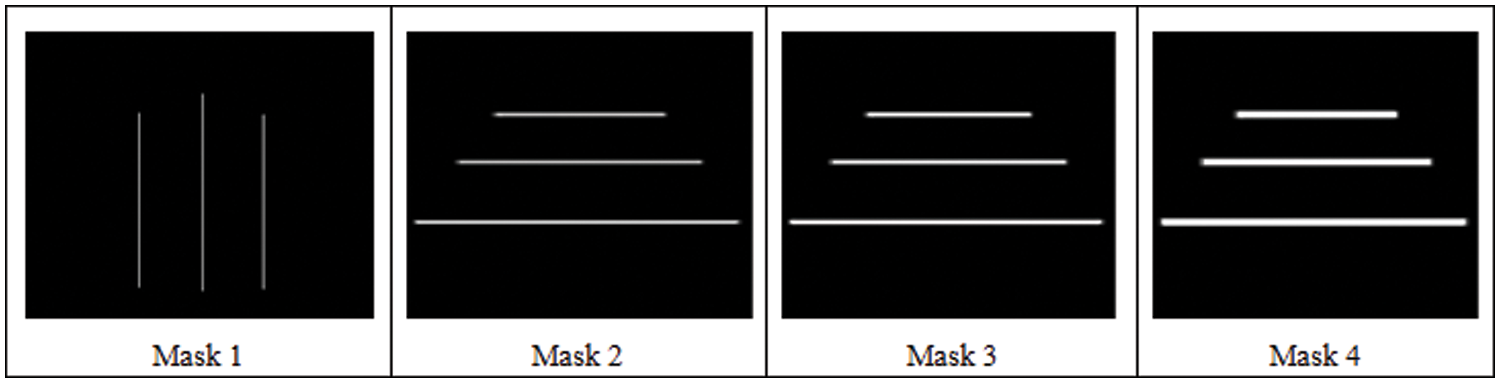
Figure 7: Four predefined masks used in the analysis
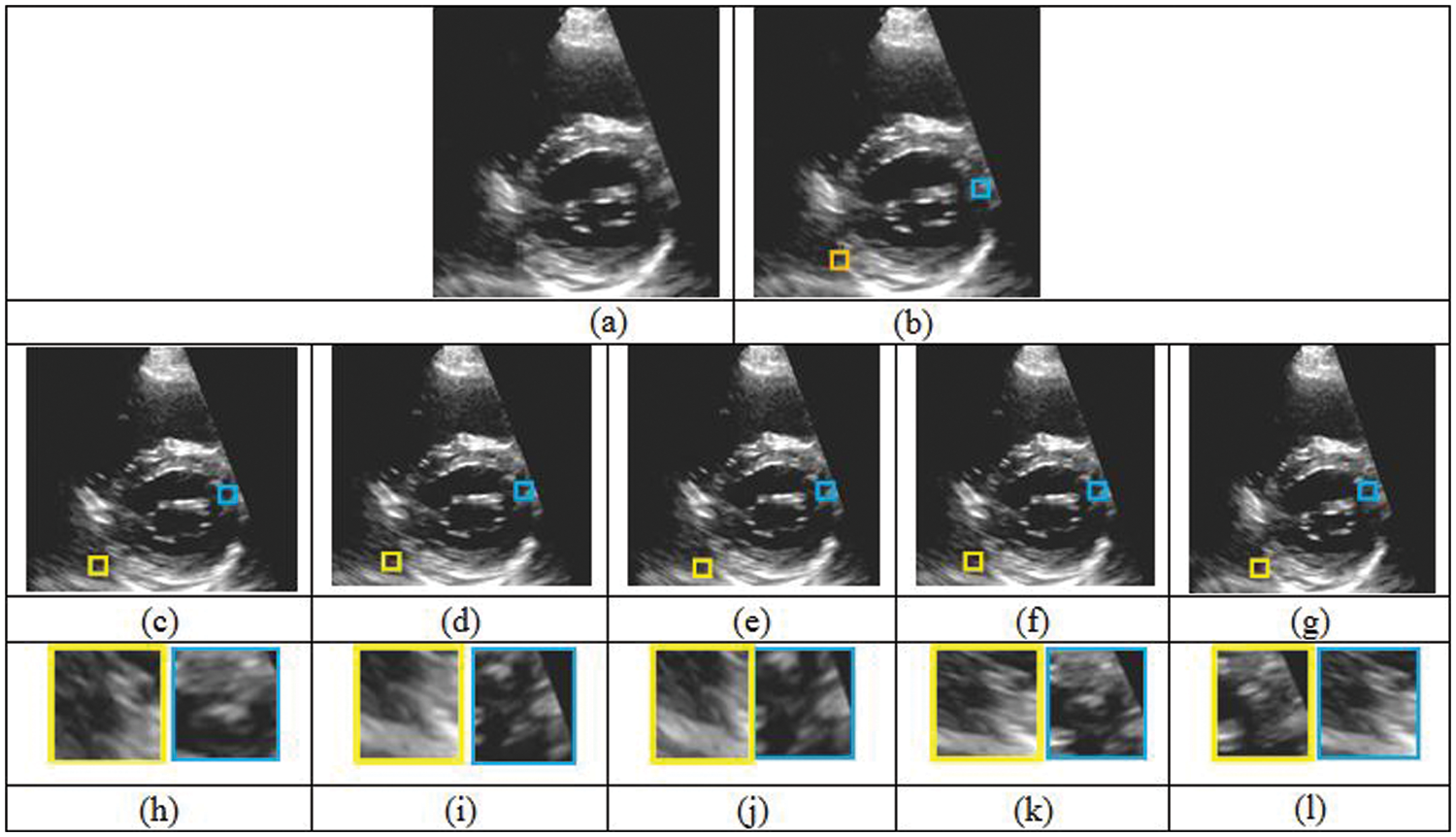
Figure 8: Comparison of five different reconstruction results. (a) Original cardiac ultrasound in PSAX view (b) ultrasound volume with missing regions. (c)–(g) Reconstruction results of FMM, MVR, NEWSON, Proposed Method I and II. (h)–(l) Magnified regions corresponding to the second row results
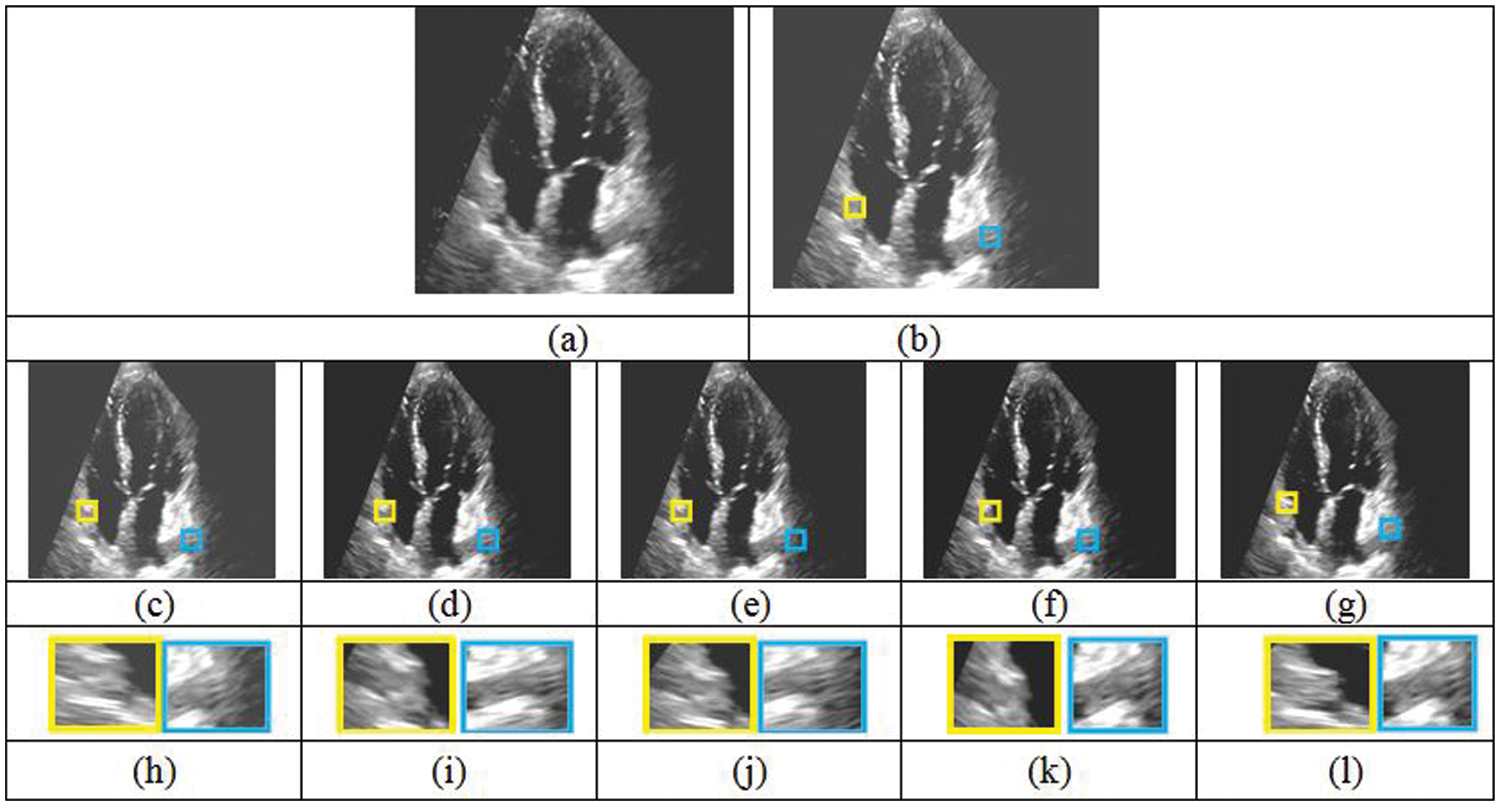
Figure 9: Comparison of five different reconstruction results. (a) Original cardiac ultrasound in A2C view (b) ultrasound volume with missing regions. (c)–(g) Reconstruction results of FMM, MVR, NEWSON, Proposed Method I and II. (h)–(l) Magnified regions corresponding to the second row results
The 3D reconstructed cardiac ultrasound image is shown in Fig. 10.
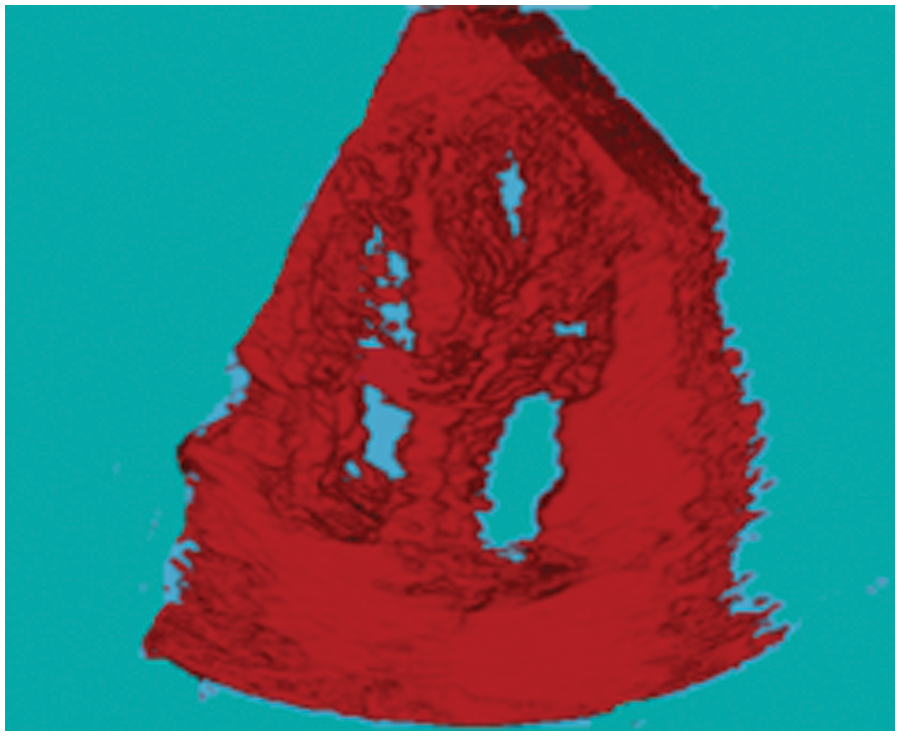
Figure 10: 3D Reconstructed cardiac ultrasound image
Tabs. 5–8 depicts the PSNR (Peak Signal-to-Noise Ratio) and FSIM (Feature Similarity Index) values of the proposed 3D reconstruction method compared against three different algorithms. Here, the two methods of the proposed approach are tested with four masks in terms of volume. From the observing performance analysis, it is evident that the proposed FD-3DVR method outperforms the other methods concerning higher PSNR and FSIM values. The graphical analysis for five volumes with mask 3 is shown in Fig. 11.
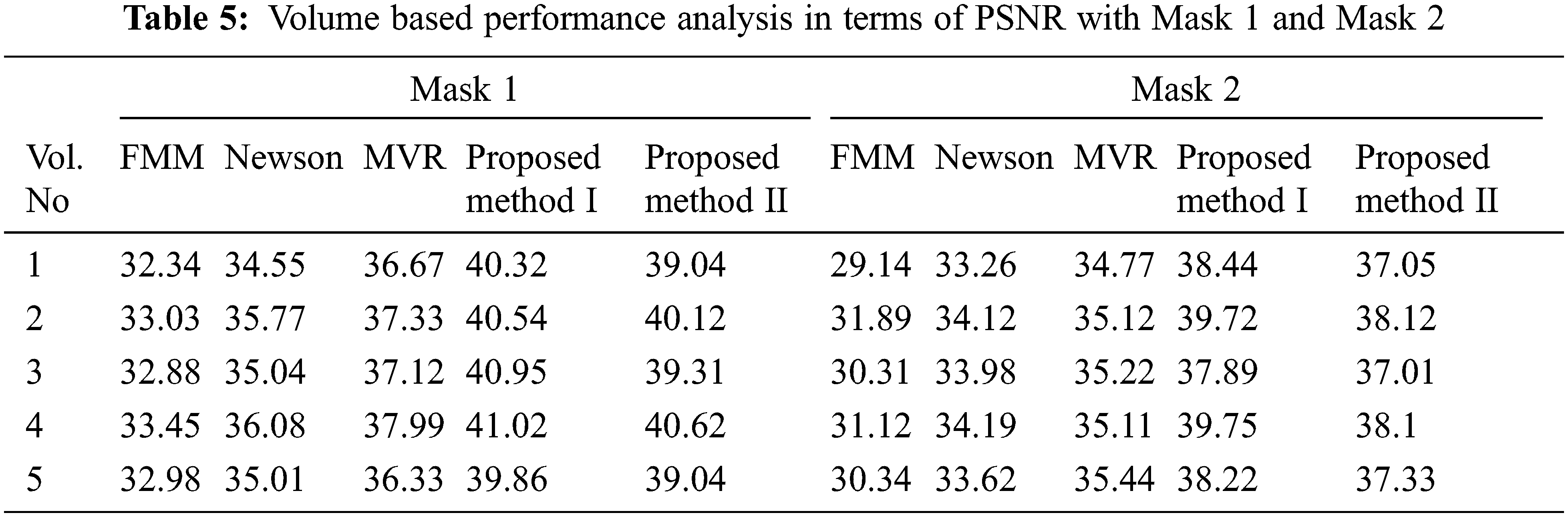
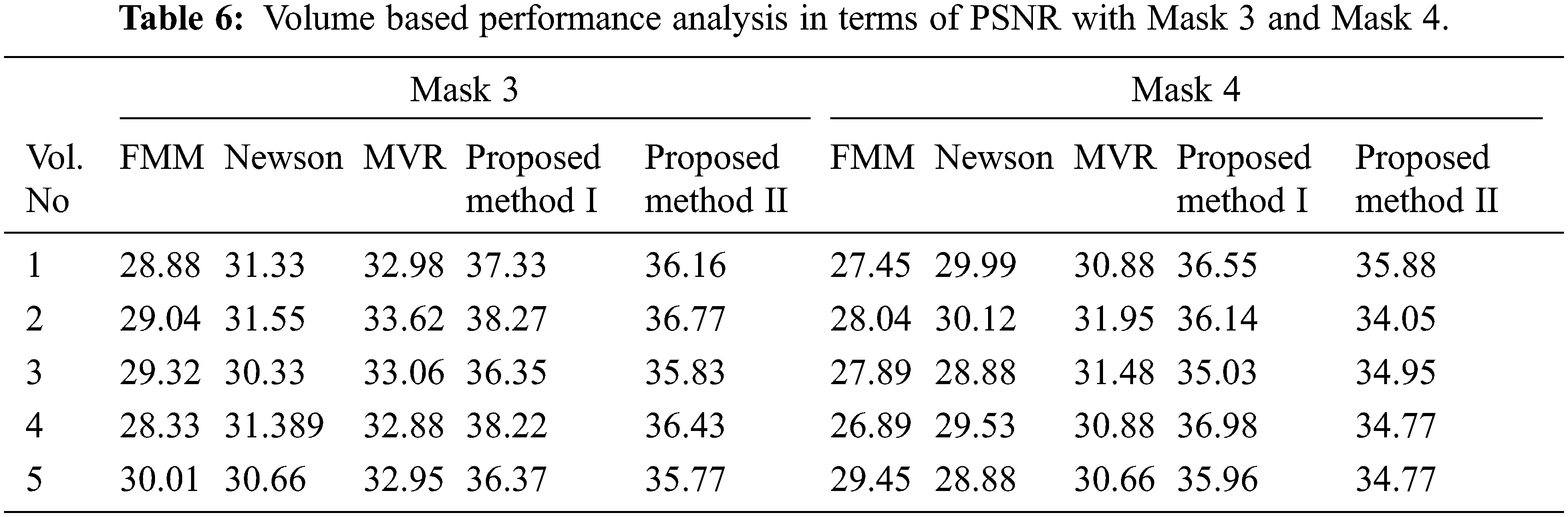
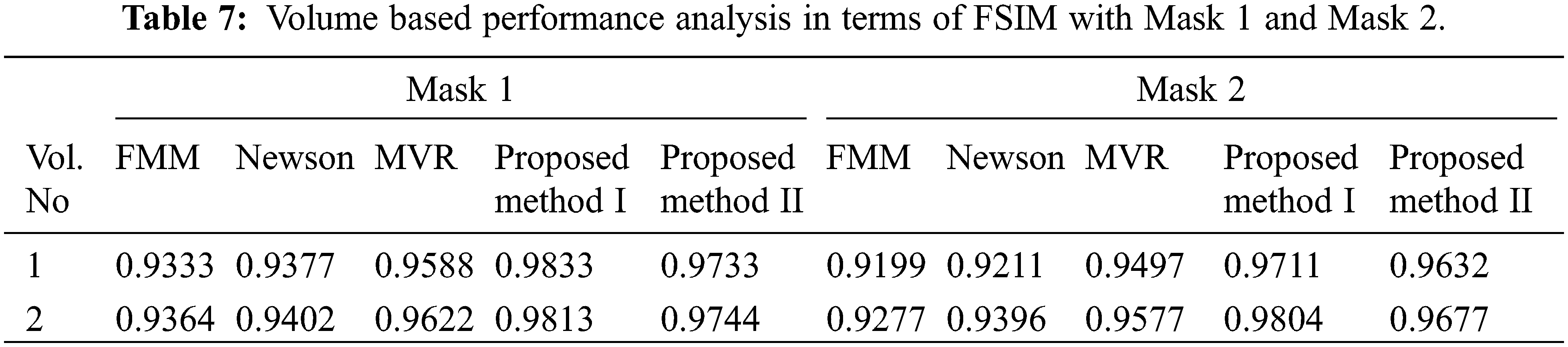
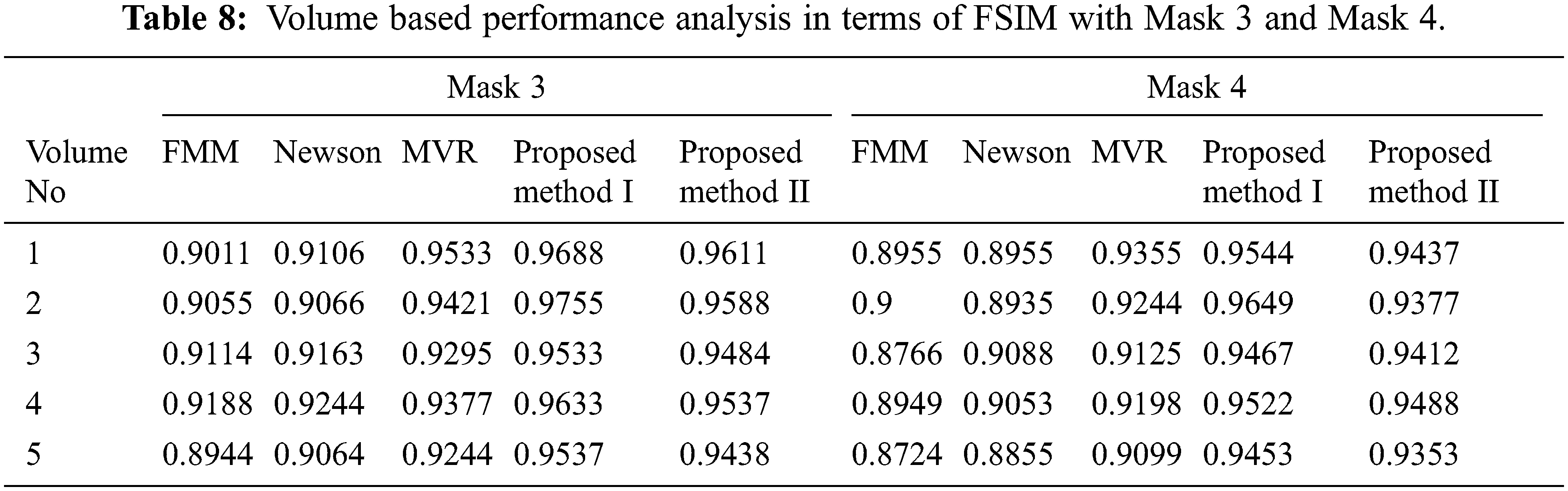
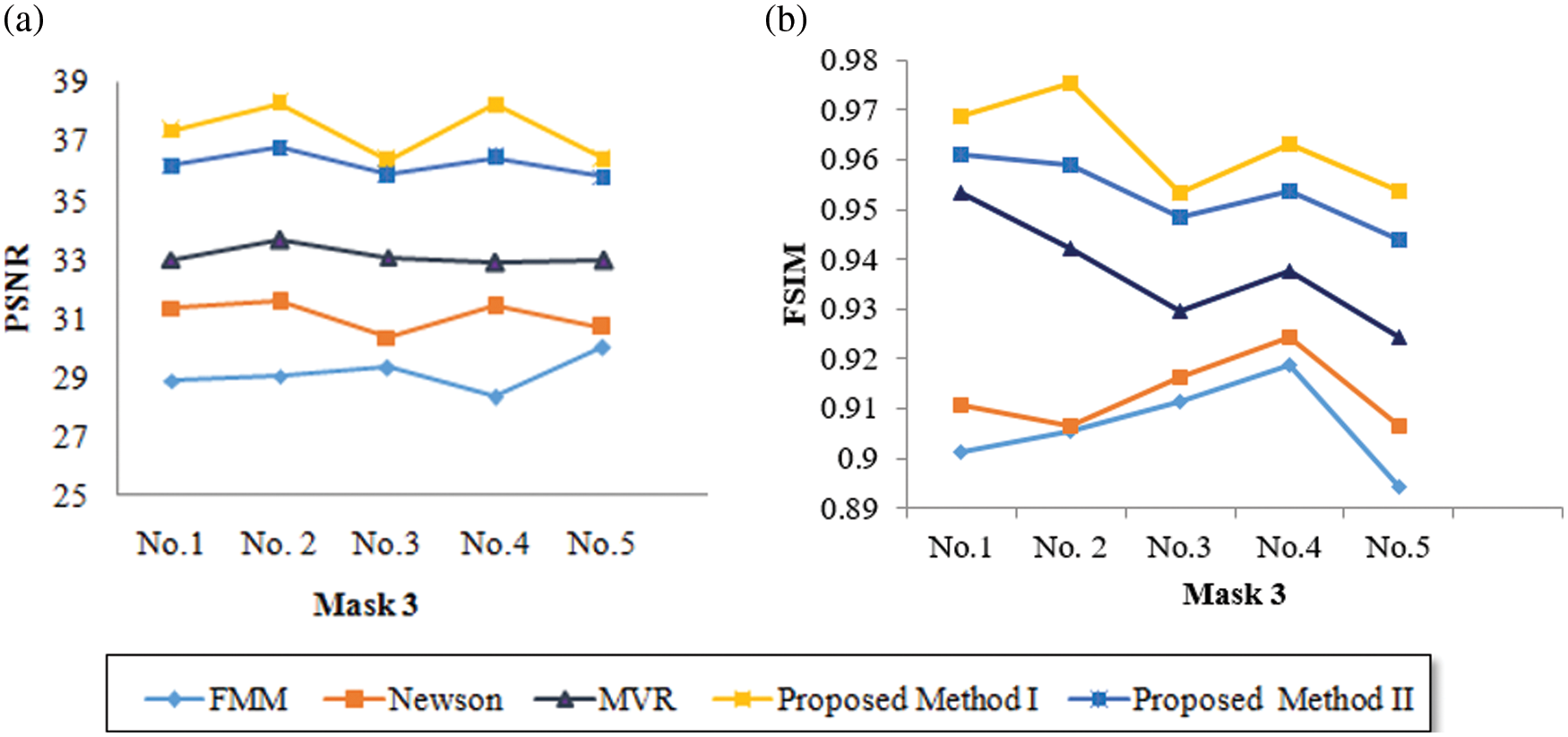
Figure 11: Graphical plot analysis for five volumes based on (a) PSNR and (b) FSIM with mask 3
Tab. 9 presents an overall performance analysis of 3D reconstruction results for all the five reconstruction methodologies. Compared to the other techniques, the observed results of the proposed method I and II provides an improvement in mPSNR (mean PSNR, average PSNR values of all the five volumes for the corresponding mask) and mFSIM (mean FSIM, average FSIM values of all the five volumes for the corresponding mask). Moreover, numerical analysis concerning average difference (AD) and structural content (SC) are tabulated in Tab. 10. From the table, it is clear that the average difference of proposed methods I and II outperforms the other methods, particularly with a low difference of 0.25124 and 0.28136 for mask 3. Also, the proposed method achieved maximum structural content similarities for all the masks, particularly with higher values of 0.9975 and 0.9969 for mask 3. The graphical plot analysis of the proposed method in comparison with existing methods is shown in Fig. 12.
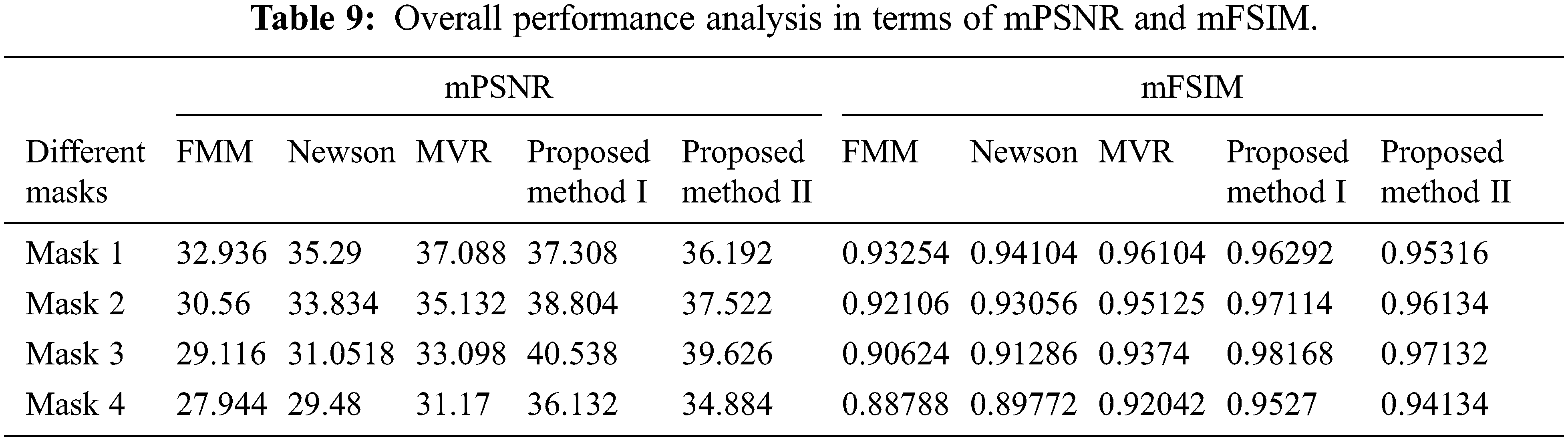
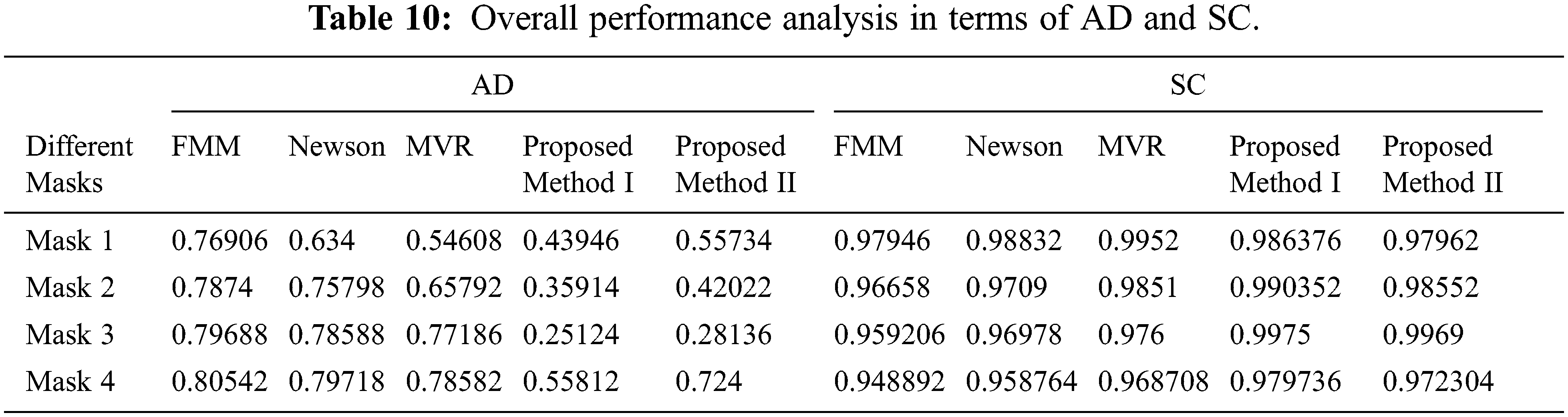
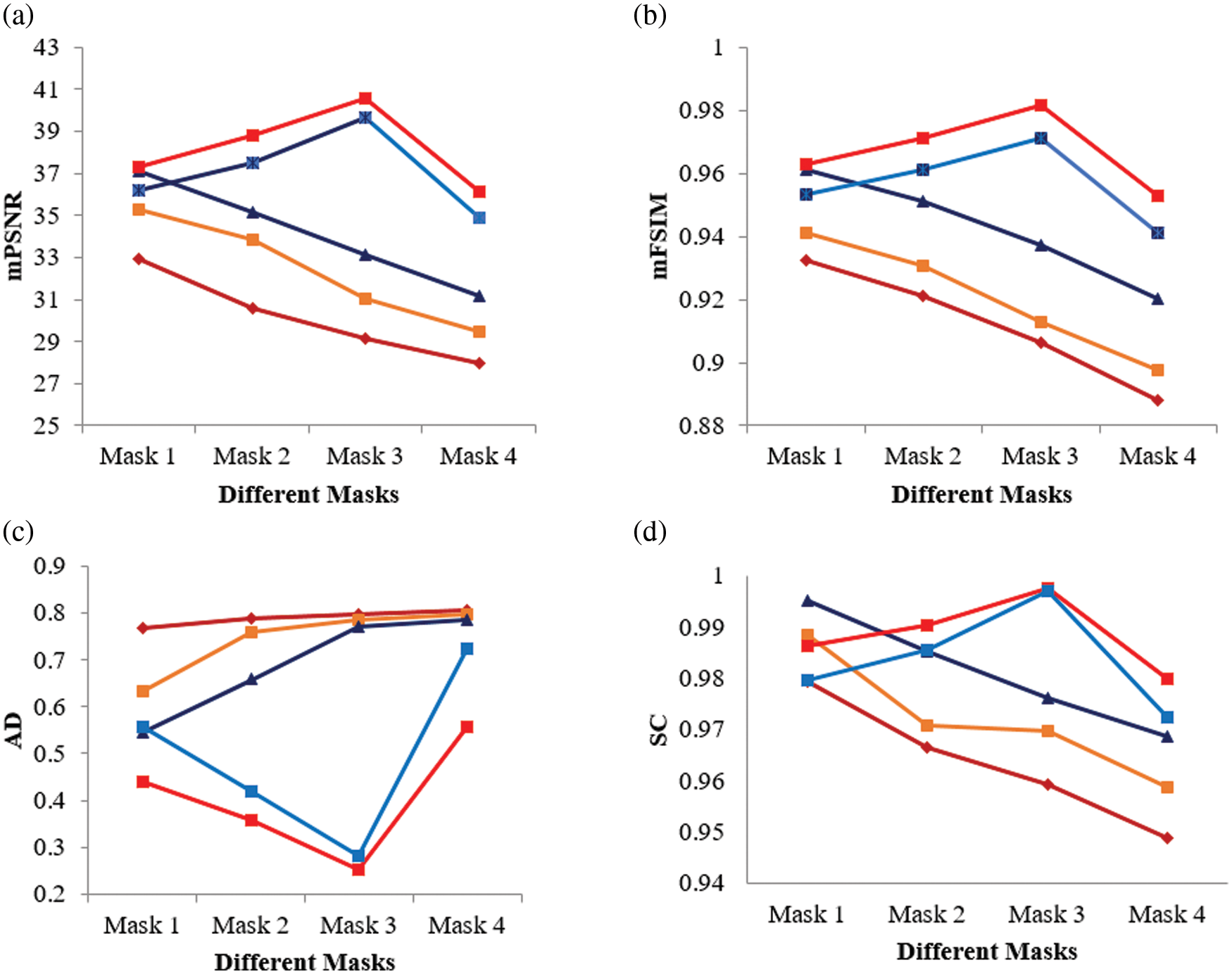
Figure 12: Graphical plot of performance metrics for proposed and existing methods (a) mPSNR (b) mFSIM (c) AD (d) SC
In this paper, a novel Flip Oriented 3D Volume Reconstruction (FD-3DVR) Method has been proposed for reconstructing the cardiac ultrasound (echocardiogram) images. The processing pipeline starts with the generation of a 3D volume grid using three cardiac views namely parasternal short axis, apical 2 chamber, and apical 4 chamber. The pixels of these 2D cardiac views are mapped into voxels of the volume grid according to the position. During this mapping, missing data is usually found in the reconstructed volume as the ultrasound scan images are discontinuous with the unequal separation in slices. This condition will affect the diagnosis of the exact location of the damaged mitral valve. Hence the proposed method effectively reconstructs the missing regions using neighborhood voxels. These neighborhood voxels are obtained by finding similar patches through the 3D ANN matching method. For finding the most similar neighborhood voxels, flip directional texture information is calculated and also incorporated into the voxel distance calculation. In this way, the presented work reconstructs the missing regions and permits the reconstructed regions to blend naturally with the neighboring voxels. According to the experimental results, the proposed FD-3DVR method can reconstruct the 3D cardiac image with fewer artifacts and provides better performance than the traditional FMM, MVR, and NEWSON methods in terms of higher PSNR and feature similarity values. The computational complexity of the proposed method, however, is primarily due to the searching procedure for similar matching voxels across the entire volumetric grid. With the development of GPU-based acceleration techniques, computational complexity will be reduced in the future. Along with flip directional features, other features will be considered to improve the efficiency in echocardiogram reconstruction.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Mustafa, M. Alshare, D. Bhargava, R. Neware, B. Singh et al., “Perceived security risk based on moderating factors for blockchain technology applications in cloud storage to achieve secure healthcare systems,” Computational and Mathematical Methods in Medicine, vol. 2022, pp. 1–10, 2022. [Google Scholar]
2. Q. Huang and Z. Zeng, “A review on real-time 3D ultrasound imaging technology,” BioMed Research International, vol. 2017, no. 1, pp. 1–20, 2017. [Google Scholar]
3. T. Shiota, “3D echocardiography: The present and the future,” Journal of Cardiology, vol. 52, no. 3, pp. 169–185, 2008. [Google Scholar]
4. M. Mustafa, S. Alzubi and M. Alshare, “The moderating effect of demographic factors acceptance virtual reality learning in developing countries in the Middle East,” in Int. Conf. on Advances in Computing and Data Sciences, Communications in Computer and Information Science book series, Singapore, Springer, vol. 1244, pp. 12–23, 2020. [Google Scholar]
5. H. Arshad, M. Mustafa and H. B. Zaman, “Design of vibratory haptic interface model (VHIM) for autistic children’s social interaction,” Asian Journal of Information Technology, vol. 14, no. 3, pp. 111–116, 2015. [Google Scholar]
6. R. Wang, Z. Fang, J. Gu, Y. Guo, S. Zhou et al., “High-resolution image reconstruction for portable ultrasound imaging devices,” EURASIP Journal on Advances in Signal Processing, vol. 2019, no. 1, pp. 56, 2019. [Google Scholar]
7. M. Mustafa, H. Arshad and H. B. Zaman, “Framework methodology of the autism children—vibratory haptic interface (AC-VHI),” in 2013 Int. Conf. on Advanced Computer Science Applications and Technologies, Kuching, Malaysia, pp. 201–206, 2013. [Google Scholar]
8. F. Mohamed and C. V. Siang, “A survey on 3D ultrasound reconstruction techniques,” in Artificial Intelligence – Applications in Medicine and Biology, M. Antonio Aceves-Fernandez (ed.London: IntechOpen, 2019. [Google Scholar]
9. D. Miller, C. Lippert, F. Vollmer, O. Bozinov, L. Benes et al., “Comparison of different reconstruction algorithms for three-dimensional ultrasound imaging in a neurosurgical setting: Comparison 3D US reconstruction algorithms,” International Journal of Medical Robotics and Computer Assisted Surgery, vol. 8, no. 3, pp. 348–359, 2012. [Google Scholar]
10. H. B. Liu, X. Wang, X. J. Wu and W. Y. Qiang, “Surface reconstruction based on radial basis functions network,” in Int. Symposium on Neural Networks, Advances in Neural Networks, Lecture Notes in Computer Science, Berlin, Heidelberg, Springer, vol. 3973, pp. 1242–1247, 2006. [Google Scholar]
11. J. M. Sanches and J. S. Marques, “A Rayleigh reconstruction/interpolation algorithm for 3D ultrasound,” Pattern Recognition Letters, vol. 21, no. 10, pp. 917–926, 2000. [Google Scholar]
12. T. Wen, Q. Zhu, W. Qin, L. Li, F. Yang et al., “An accurate and effective FMM-based approach for freehand 3D ultrasound reconstruction,” Biomedical Signal Processing and Control, vol. 8, no. 6, pp. 645–656, 2013. [Google Scholar]
13. T. Wen, F. Yang, J. Gu, S. Chen, L. Wang et al., “An adaptive kernel regression method for 3D ultrasound reconstruction using speckle prior and parallel GPU implementation,” Neurocomputing, vol. 275, pp. 208–223, 2018. [Google Scholar]
14. W. Cong, D. Ai, H. Song, G. Chen, X. Liang et al., “Global patch matching (GPM) for freehand 3D ultrasound reconstruction,” BioMedical Engineering Online, vol. 16, no. 1, pp. 1–26, 2017. [Google Scholar]
15. A. Uus, T. Zhang, L. H. Jackson, T. A. Roberts, M. A. Rutherford et al., “Deformable slice-to-volume registration for motion correction of fetal body and placenta MRI,” IEEE Transactions on Medical Imaging, vol. 39, no. 9, pp. 2750–2759, 2020. [Google Scholar]
16. Z. Cai, C. Wang, C. Wen and J. Li, “3D-PatchMatch: An optimization algorithm for point cloud completion,” in 2015 2nd IEEE Int. Conf. on Spatial Data Mining and Geographical Knowledge Services (ICSDM), Fuzhou, China, pp. 157–161, 2015. [Google Scholar]
17. C. Barnes, E. Shechtman and A. Finkelstein, “PatchMatch: A randomized correspondence algorithm for structural image editing,” ACM Transactions on Graphics, vol. 28, no. 3, pp. 24:1–24:11, 2009. [Google Scholar]
18. Echocardia, The Echocardiogram Resource. [Online]. Available: https://echocardia.com/. [Google Scholar]
19. M. H. R. Cardinal, G. Soulez, J. C. Tardif, J. Meunier and G. Cloutier, “Fast-marching segmentation of three-dimensional intravascular ultrasound images: A pre- and post-intervention study,” Medical Physics, vol. 37, no. 7, pp. 3633–3647, 2010. [Google Scholar]
20. A. Newson, A. Almansa, M. Fradet, Y. Gousseau and P. Pérez, “Video inpainting of complex scenes,” SIAM Journal on Imaging Sciences, vol. 7, no. 4, pp. 1993–2019, 2014. [Google Scholar]
21. J. Dong, W. Cong, D. Ai, Y. Chu, Y. Huang et al., “Multiresolution cube propagation for 3-D ultrasound image reconstruction,” IEEE Transactions on Computational Imaging, vol. 5, no. 2, pp. 251–261, 2019. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools