 Open Access
Open Access
ARTICLE
A Unified U-Net-Vision Mamba Model with Hierarchical Bottleneck Attention for Detection of Tomato Leaf Diseases
School of Computing and Information Technology, Murang’a University of Technology, Murang’a, 75-10200, Kenya
* Corresponding Author: Geoffry Mutiso. Email:
(This article belongs to the Special Issue: Advances in Artificial Intelligence for Engineering and Sciences)
Journal on Artificial Intelligence 2025, 7, 275-288. https://doi.org/10.32604/jai.2025.069768
Received 30 June 2025; Accepted 11 August 2025; Issue published 05 September 2025
Abstract
Tomato leaf diseases significantly reduce crop yield; therefore, early and accurate disease detection is required. Traditional detection methods are laborious and error-prone, particularly in large-scale farms, whereas existing hybrid deep learning models often face computational inefficiencies and poor generalization over diverse environmental and disease conditions. This study presents a unified U-Net-Vision Mamba Model with Hierarchical Bottleneck Attention Mechanism (U-net-Vim-HBAM), which integrates U-Net’s high-resolution segmentation, Vision Mamba’s efficient contextual processing, and a Hierarchical Bottleneck Attention Mechanism to address the challenges of disease detection accuracy, computational complexity, and efficiency in existing models. The model was trained on the Tomato Leaves and PlantVillage combined datasets from Kaggle and achieved 98.63% accuracy, 98.24% precision, 96.41% recall, and 97.31% F1 score, outperforming baseline models. Simulation tests demonstrated the model’s compatibility across devices with computational efficacy, ensuring its potential for integration into real-time mobile agricultural applications. The model’s adaptability to diverse datasets and conditions suggests that it is a versatile and high-precision instrument for disease management in agriculture, supporting sustainable agricultural practices. This offers a promising solution for crop health management and contributes to food security.Keywords
Crop diseases pose a significant threat to global food security by affecting crop yield, quality, and profitability. Tomatoes are among the most widely grown crops worldwide and are severely affected by various diseases. Tomato leaf diseases, such as bacterial spots, early blight, and leaf mold, directly affect tomato production, prompting farmers to rely on pesticides and other protective measures that increase production costs. The excessive use of pesticides poses a significant risk to the environment and human health. With the rising demand for food and crops susceptibility to climate change, the need to develop effective crop disease management strategies for sustainable agriculture has increased in most nations.
Traditional disease detection methods, such as visual inspections by farmers or agricultural experts, are laborious, error-prone, and hardly scalable. Most of the time, these traditional methods of detecting crop diseases fail to identify diseases at their early stages, leading to widespread infections and devastating losses. Hence, innovations in automated and scalable approaches for disease detection and management are urgently required.
Recent advances in Artificial Intelligence (AI) and Machine Learning (ML) have presented deep learning models as potential tools for image-based disease diagnosis and classification [1–5]. Convolutional Neural Networks (CNNs) have shown promise in automating feature extraction for plant disease recognition [6–9]. For instance, Ferentinos [9] successfully applied CNNs to tomato leaf images from the PlantVillage dataset, achieving a classification accuracy of above 99.53%. However, they are usually hampered by environmental variability, such as lighting conditions and subtle differences in symptoms, which undermine their accuracy in real-world applications. Recent attention-based models, such as Vision Transformers (ViTs) have improved contextual awareness but require high computational resources, which limits their applicability in resource-constrained agricultural settings [10–12]. A study conducted by Boukabouya et al. [12] demonstrated the benefits of ViT-based models in enhancing disease detection accuracy on tomato crops under variable conditions. However, ViT based models are noted to have high computational demands making them impractical for edge deployment in low-resource farms. Vision Mamba (ViM) has partially solved some of these challenges by introducing a bidirectional state-space model (SSM) for improved computational efficiency and accuracy as noted by Gu et al. [13]. However, current solutions, including ViM and hybrid CNN-Transformer models [14–16], are often constrained by their adaptation capabilities, failing to achieve the trade-off between spatial resolution and contextual understanding essential for robust disease detection.
The primary objective of this study was to develop and evaluate a new model that overcomes the challenges of manual inspection inefficiencies, scalability limitations, environmental variability, and high computational complexity in tomato leaf disease detection by combining high-resolution spatial feature extraction of U-Net with efficient contextual processing of ViM and a Hierarchical Bottleneck Attention Mechanism (HBAM). This study proposed a new deep learning model called U-Net-ViM-HBAM to improve the identification and classification of diseases in tomato leaves accurately and efficiently, and hence provide a scalable solution for precision agriculture.
This study contributes to the field of computer vision and deep learning by presenting a unified model that combines U-Net, ViM and HBAM for disease detection. The proposed model is lightweight and modular, making it practical for deployment in real-time and edge computing environments. Additionally, with minimal adjustment and retraining of the model, it can also be applied to domains including medical imaging, industrial inspection, and environmental monitoring.
This paper is organized as follows: Section 2 reviews related works, emphasizing the existing gaps in crop disease detection; Section 3 explains the methodology in detail, including the architecture of the U-Net-ViM-HBAM model, datasets, and evaluation metrics; Section 4 presents the results and discusses the performance of the models, including a comparison with baseline models; and finally, Section 5 concludes this paper by summarizing its contributions and future research directions.
Recent advances in deep learning have significantly impacted crop disease detection, demonstrating accuracy and efficiency in detecting diseases in crops such as tomatoes. Although initially developed for biomedical applications, U-Net is remarkably effective in pixel-level image segmentation. In these studies [7,17–19], the authors utilized U-Net to develop disease detection models that demonstrated higher accuracy than traditional Convolutional Neural Networks (CNNs). The encoder-decoder structure of U-Net, with skip connections, saves important spatial information required when identifying diseased regions on tomato leaves. This capacity for high-resolution segmentation has made U-Net a popular choice in scenarios that require detailed feature extraction. However, its lack of contextual comprehension limits its effectiveness in complex agricultural settings, where symptoms vary in terms of scale and conditions.
Transformer-based architectures, such as ViT and ViM, have been used as strong tools to overcome these limitations. A study by Barman et al. proposed ViT-SmartAgri. In their study, the proposed vision transformer-based model for tomato leaf disease classification demonstrated high generalization performance under uncontrolled field conditions [11]. Sun et al. [10] also proposed SE-ViT model for diagnosing sugarcane leaf diseases. Their model achieved an accuracy of 97.26% on the PlantVillage dataset. Despite the observed high performance of these transformers-based models, they usually require significant computational resources and may not be suitable for deployment in low-power environments. Unlike traditional Vision Transformers, ViM considers a bidirectional state-space model (SSM) to compute attention mechanisms effectively. In a previous study [18], Shi et al. demonstrated the significance of ViM in tomato blight disease spot detection. Their study combined U-Net with Vision Mamba and ConvNeXt (VMC-Unet) to address the challenges of oversegmentation and undersegmentation. Their proposed model achieved 97.82%, 87.94%, and 86.75% accuracy, F1 score, and Mean Intersection over Union (mIoU), respectively. These results outperformed classical segmentation [18]. ViM enables the model to recognize patterns specific to diseases while maintaining computational efficiency for real-time applications in resource-constrained environments. Although ViM is strong in contextual analysis, it does not perform well in capturing fine spatial details in settings with subtle or poorly defined disease symptoms [20]. Furthermore, its dependence on high-quality and consistent input data makes its application in field conditions difficult owing to variations in lighting, image resolution and environmental factors.
The introduction of selective attention mechanisms partially addressed these challenges. In this study [2], Alirezazadeh et al. improved the accuracy of plant disease detection using a Bottleneck Attention Mechanism, specifically the Convolutional Bottleneck Attention Module (CBAM), which enhances the representation power of CNN networks for plant disease classification. The models they developed, that is MobileNetV2 + CBAM and EfficientNetB0 + CBAM, achieved high accuracies of 83.99% and 86.89%, respectively, compared to MobileNetV2 and EfficientNetB0, which had lower accuracies by 1.93% and 1.07%, respectively. The Hierarchy of Bottleneck Attention Mechanism improves the model’s ability to isolate and highlight disease-specific features by filtering irrelevant information. Its hierarchical structure allows for refined feature extraction in a multiscale manner, from large lesions to minute discolorations, thereby enhancing both the accuracy and computational efficiency [21]. HBAM has significant potential to overcome the noise and inconsistencies typical of agricultural datasets, thereby improving the accuracy of crop disease detection. However, its integration into hybrid architectures that are practically deployable remains underexplored, limiting its scalability and effectiveness in diverse, real-world scenarios.
Despite the significant development of deep learning models for crop disease detection, several gaps still exist. U-Net has been excellent in high-resolution segmentation but suffers from contextual comprehension and is thus less effective under diverse field conditions. Vision Mamba has been introduced for contextual analysis but suffers from a lack of spatial precision necessary to detect the subtle symptoms of diseases and is sensitive to inconsistent input quality in terms of light and image resolution. Although the Hierarchical Bottleneck Attention Mechanism improves feature refinement and computational efficiency, the practical deployment of HBAM in hybrid architectures remains limited because of its scalability and adaptability to real-world agricultural variability. Traditional hybrid models that combine segmentation, contextual analysis, and attention mechanisms frequently encounter high computational complexity and poor generalization over varied environmental and disease conditions. Therefore, a strong and adaptive hybrid model is required.
This section presents the methodology used to develop, train, and evaluate the U-Net-ViM-HBAM hybrid model for detecting and classifying eleven (11) distinct classes in the combined dataset. The methodology includes dataset collection, data preprocessing techniques, experimental materials, model development, mathematical formulation, and training and evaluation processes.
3.1 Data Collection and Preprocessing
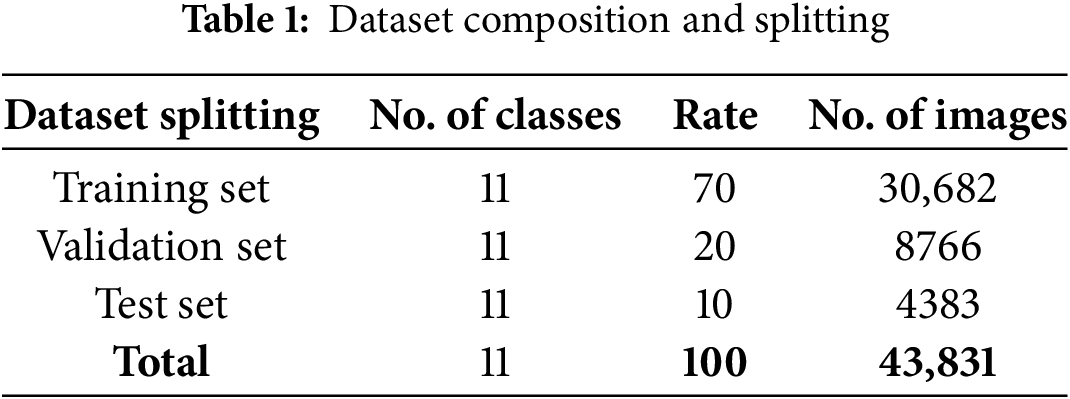
The combined dataset used in this study included the Tomato Leaves Dataset with 25,671 images and the PlantVillage Dataset with 18,160 tomato leaf images giving a total of 43,831 images. The datasets were sourced from Kaggle and included 11 distinct classes. The diseases included bacterial spots, early blight, late blight, leaf mold, Septoria leaf spot, spider mite spot, target spot, yellow leaf curl virus, powdery mildew, and mosaic virus. Table 1 shows the composition of the final curated dataset after combining images from the two datasets. The data were split in a ratio of 70:20:10 (30,682, 8766, 4383) for the training, validation, and testing sets, respectively. The use of two datasets that vary in the presentation of diseases helped improve the generalizability of the model with respect to different environmental conditions and symptom manifestations.
Data preprocessing is imperative for the consistency and quality of the training and validation steps. This involved resizing all images to a standard resolution of 256 × 256 pixels, normalizing the pixel values for better computational efficiency, and applying noise-reduction filters to clarify the features. The model was made resilient through data augmentation by rotation, flipping, and zooming. These techniques artificially increased the dataset size so that the model could handle scale, orientation, and lighting variations that are typical of real-world agricultural imagery.
The development and evaluation of the U-Net-ViM-HBAM model were performed using a robust base of software tools and computational resources. The tools used included Google Colab, OpenCV, TensorFlow, Keras, and Scikit-learn. The computational resources utilized were Google Colab’s cloud infrastructure with an NVIDIA Tesla K80 GPU and a local machine equipped with an Intel Core i7 processor, 16 GB RAM, and an NVIDIA GeForce GTX 1650 GPU. Python was chosen as the programming language because of its rich ecosystem, including TensorFlow and Keras for building the deep learning models, OpenCV for image preprocessing, and Scikit-learn for evaluating the performance of the developed models.
The experiments were conducted using Google Colab, which provided GPU acceleration for training and testing. This cloud-based platform ensures easy access to state-of-the-art computing resources, including the NVIDIA Tesla K80 GPU. This hardware-software synergy between powerful laptops, complemented by efficient software tools, allows for effortless experimentation and fine-tuning of the model.
The U-Net-ViM-HBAM unified model combines three key components: U-Net for spatial segmentation, Vision Mamba for contextual analysis, and a Hierarchical Bottleneck Attention Mechanism that guides feature extraction selectively. The components were selected based on their complementary architectural strengths. Each component contributes uniquely to the model’s overall performance. The novelty of the proposed model lies in its ability to combine the three powerful techniques namely U-Net, ViM, and HBAM into a unified architecture. This combination had not yet been explored for tomato leaf disease detection. U-Net is effective for localized lesion detection due to its ability to preserve fine details through skip connections. Vision Mamba improves the model’s ability to understand the context of an image, which U-Net cannot do alone. The attention mechanism enhances the most important features and reduces background noise. We assumed that tomato leaf disease symptoms can have both local patterns, such as spots, and broader patterns, such as color changes, and that attention mechanisms can help to highlight these patterns. Based on this, we designed a model that combines spatial details, context awareness, and attention refinement to improve the accuracy and generalization.
The U-Net encoder captures the spatial features of the input images using convolutional layers and max pooling. Maintaining skip connections helped preserve the high-resolution details that are crucial for accurately segmenting disease-affected regions. The encoder comprises of five convolutional blocks with progressively increasing filter sizes [64, 128, 256, 512, 1024]. Batch normalization and ReLu activation were performed after each block. The incremental architecture adopted from the standard U-Net design enables the model to progressively learn abstract features in deeper layers while simultaneously reducing the spatial resolution. Vision Mamba further processes the encoded features using a bidirectional state-space model to capture contextual patterns at both local and global levels. This contextual analysis is valuable for distinguishing between diseases that are visually similar.
The encoded features were further refined using a Hierarchical Bottleneck Attention Mechanism that applies sequential channel and spatial attention modules arranged hierarchically across three spatial scales. Fig. 1 illustrates the internal HBAM architecture, which consists of a channel attention module that aggregates global spatial information using global average pooling, followed by a spatial attention module that uses 7 × 7 convolution to capture spatial dependencies. The Channel Attention Module determines the important features by computing inter-channel relationships through global average pooling, thereby emphasizing channels associated with disease-relevant features. The Spatial Attention Module focuses on the location of these features by applying a spatial filter that highlights the key regions within the image. This attention is repeated across three special scales, as shown in Fig. 1. The HBAM selectively enhances disease-specific features while suppressing irrelevant information, thereby reducing computational overhead. It progressively narrows the focus, allowing the model to prioritize subtle patterns that are indicative of specific diseases.
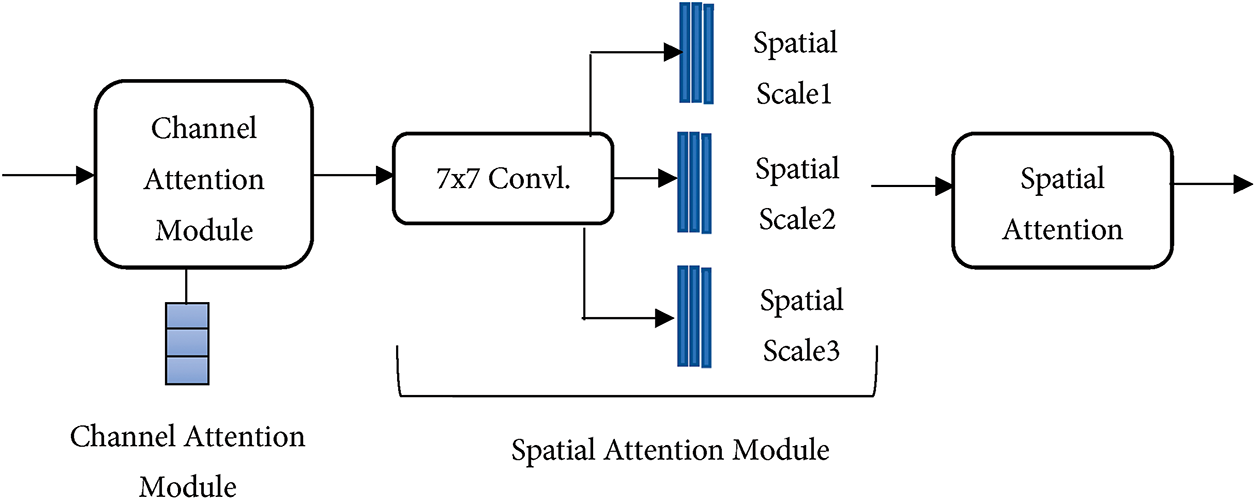
Figure 1: HBAM architecture
The U-Net decoder reconstructs spatial information by upsampling the refined features and integrating skip connections from the encoder at each stage to restore the spatial information that may be lost during downsampling. This ensures accurate localization of disease symptoms. Finally, the classification layer assigns disease labels by flattening the output feature map and passing it through fully connected layers. Fig. 2 illustrates the architecture of the U-Net-ViM-HBAM model. It shows the sequential integration of U-Net, ViM, and HBAM. The U-Net encoder extracts spatial details, whereas the ViM encoder captures the contextual information. In the center, the Hierarchical Bottleneck Attention Mechanism refines these features by emphasizing disease-specific patterns and reducing noise. Reconstruction begins with the ViM decoder, whose output is processed by the U-Net decoder to restore spatial information, culminating in a classification layer that assigns disease labels.
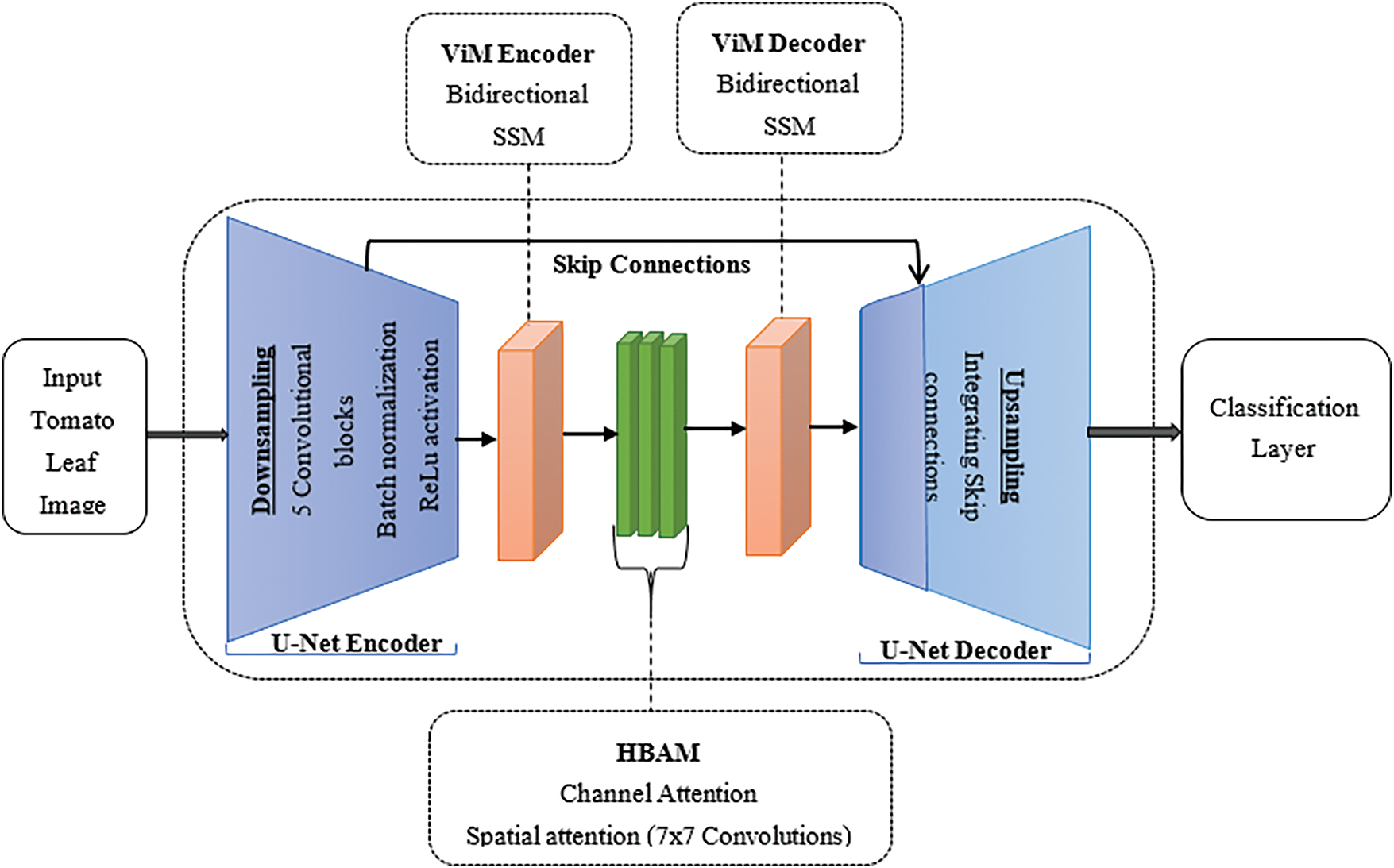
Figure 2: U-Net-ViM-HBAM model architecture
3.4 Mathematical Formulation of the Unified Model
The U-Net-ViM-HBAM model integrates U-Net for spatial segmentation, ViM for contextual processing, and HBAM for the enhancement of selective features. The unified model follows a hierarchical transformation process and is mathematically represented by Eq. (1), which models the flow from the input image to disease classification by integrating spatial and contextual features. Let X be the input image and X′ the final output prediction.
where:
• X = input image (tomato leaf image).
• Encoder(X) = extracts spatial features using the U-Net encoder from the input image.
• ViM() = processes the encoded features for contextual understanding using Vision Mamba.
• HBAM() = refines the features using the Hierarchical Bottleneck Attention Mechanism.
• Decoder() = reconstructs the spatial information using transposed convolutions of the U-Net’s decoder.
• ClassificationLayer() = outputs the final disease classification.
This formulation allows the model to simultaneously learn both low and high-level features, enhanced by attention-driven refinements, ultimately resulting in more robust and accurate disease detection.
The pretrained U-Net encoder was loaded directly, and ViM was added after the U-Net encoder outputs. HBAM was used to refine the ViM features. Finally, the U-Net decoder from the pretrained U-Net model was loaded to reconstruct spatial information and disease-affected regions. The model was trained using the Adam optimizer with a learning rate of 0.001 and batch size of 32. In addition, dropout layers and L2 regularization were used to prevent the overfitting of the model. The performance metrics used to evaluate the model were accuracy, precision, recall, and F1 score. These evaluation metrics provided a holistic view of the ability of the model to detect and classify tomato leaf diseases into their respective classes. Confusion matrices were also created to examine the classification accuracy of each disease category, which helped identify areas that required improvement. Simulations were also performed on devices with varying computational capacities to test the adaptability and efficiency of the model. Simulations were conducted to ensure adequate deployment of the model in resource-constrained environments, such as small-scale farms.
This section presents the results of the unified U-Net-ViM-HBAM model for detecting and classifying 11 classes of tomato leaf diseases. It includes performance metrics, comparative analyses, and insights into the adaptability of the model in simulated and practical agricultural scenarios.
The development of the unified U-Net-ViM-HBAM model involved iterative optimization of its components. The U-Net layers were precisely tuned to learn the spatial complexities that could help differentiate between healthy and infected leaf areas. Vision Mamba provided an analysis using its bidirectional state space model (SSM) for contextual assessments of images. HBAM was introduced to selectively amplify disease-related features and filter out useless background information, thereby minimizing noise and computational demands. The results demonstrated that integrating these components significantly improved the model’s ability to handle complex datasets, establishing a unified architecture as an effective approach for crop disease detection.
4.2 Testing and Validation Results
The performance of the U-Net-ViM-HBAM model was assessed using a curated dataset derived from the Tomato Leaves and PlantVillage datasets. Fig. 3 summarizes the performance metrics, including accuracy, precision, recall, and F1 score. The model achieved an overall accuracy of 98.6% across the datasets, with 98.24% precision, 96.41% recall, and 97.31% F1 score for most of the disease classes. As shown in Fig. 4, diseases with more subtle visual symptoms, such as “Leaf Mold” and “Powdery Mildew”, had slightly lower metrics, which rationalized the areas for further refinement of the model.
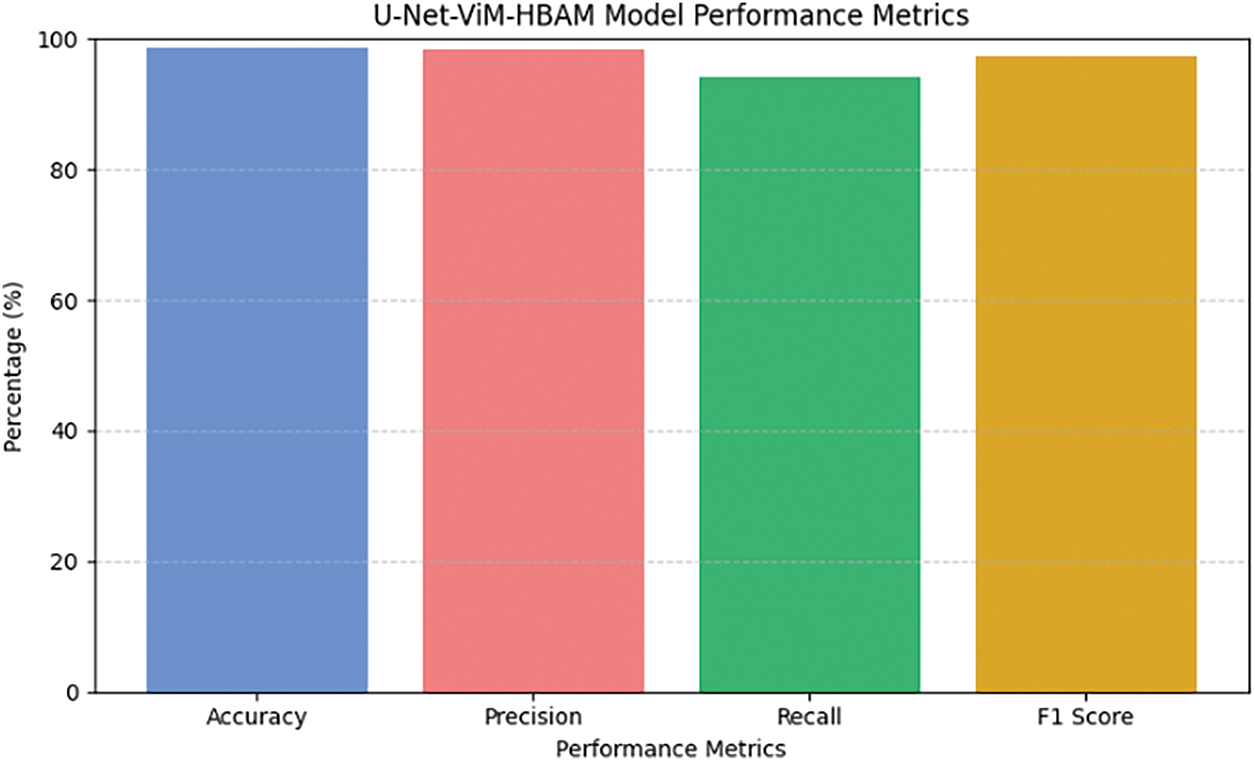
Figure 3: U-Net-ViM-HBAM model performance metrics
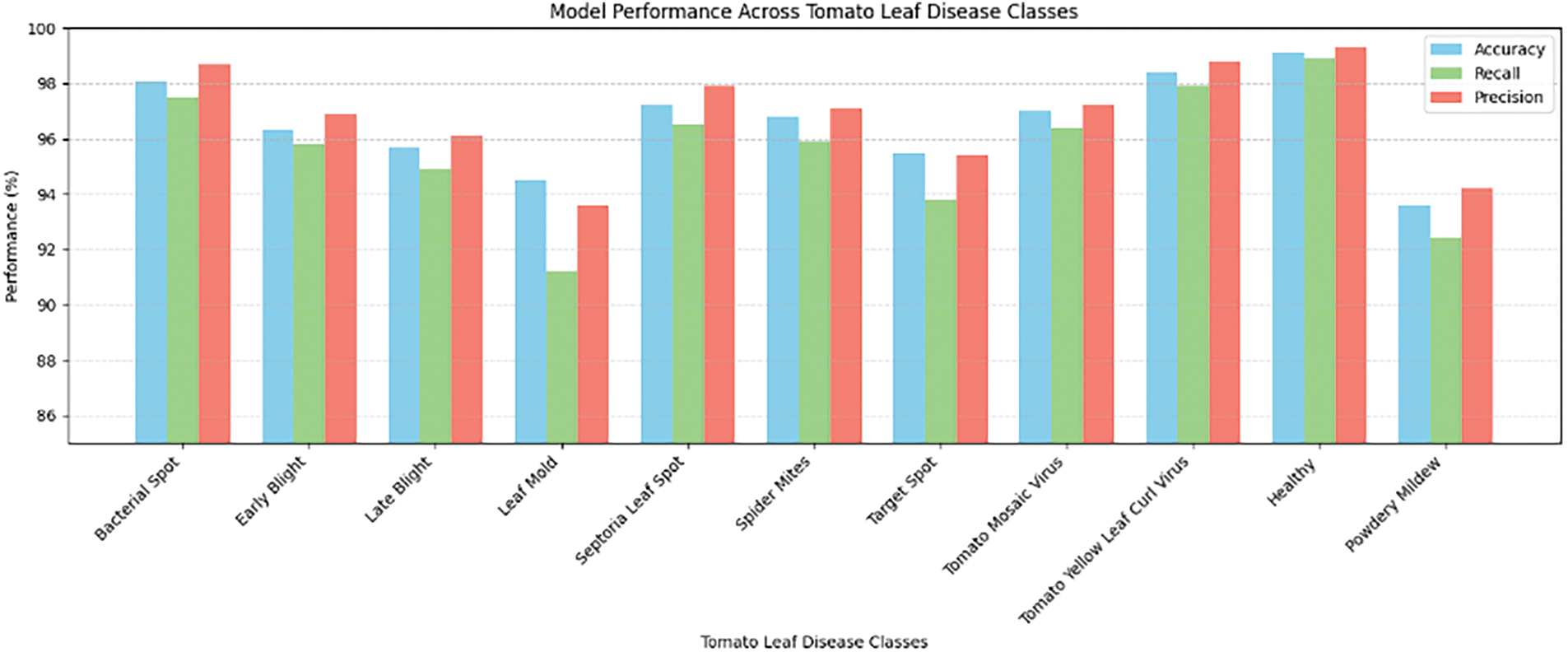
Figure 4: Model performance across tomato leaf disease classes
4.3 Comparative Analysis with Existing Models
To validate the performance of the U-Net-ViM-HBAM model, it was compared with some baseline models, including ViT Mamba and ResNet. Table 2 lists the comparative metrics. The U-Net-ViM-HBAM model outperformed all baseline models in terms of F1 score, precision, and accuracy. The metrics showed that the model might miss some disease cases, such as those with very close visual similarities, as it achieved a recall of 96.41%. However, the precision was high (98.24%), indicating that the model is precise and that when a disease is classified, there is a high confidence that the prediction is accurate. This demonstrates the benefits of combining spatial segmentation, contextual processing, and selective attention mechanisms into a single framework for agricultural disease detection.
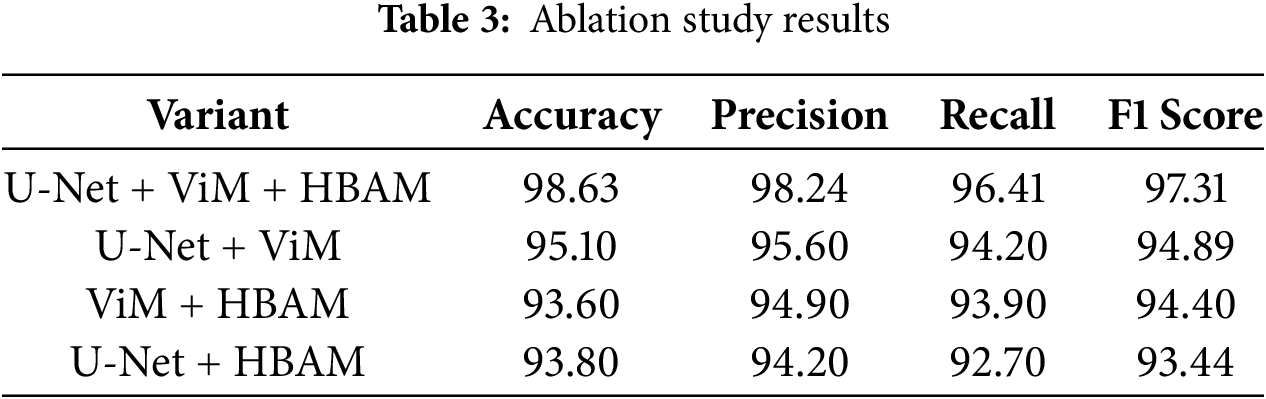
To further evaluate the contribution of each component, we performed an ablation study by selectively removing one component at a time and observing the performance. As shown in Table 3, the complete model achieved the highest overall performance. When HBAM was excluded, we noted that the precision dropped by −2.64%, indicating that HBAM is significant in enhancing the saliency of disease-specific features. However, when Vision Mamba was removed, a decline in recall was noted, confirming the strength of ViMs in modeling long-range dependencies and contextual variations. Finally, we removed the U-Net component, and its absence caused the most significant decrease in accuracy. This confirms that all three components are significant.
To provide a clearer view of the model’s performance in classification, a confusion matrix was generated Fig. 5 shows tiny misclassifications across classes, which shows tiny misclassifications across classes, indicating the model’s specificity and sensitivity.
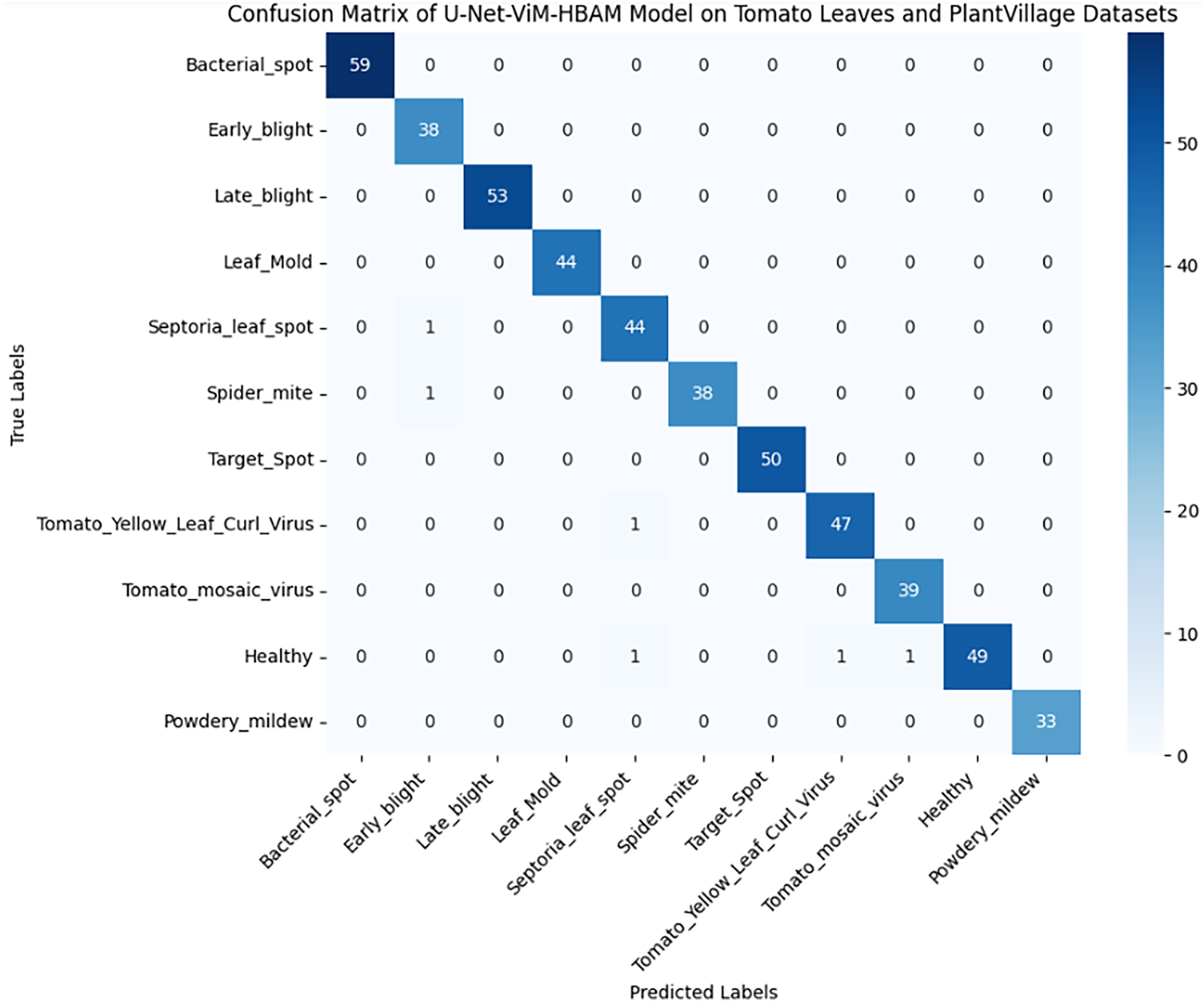
Figure 5: Confusion matrix of U-Net-ViM-HBAM model
4.5 Impact of Loss Functions on Pixel Accuracy
In this study, the effects of different loss functions, such as Cross-Entropy, Dice Loss, Focal Loss, and Label Smoothing, were compared on pixel accuracy over 100 epochs. Fig. 6 illustrates the convergence trends, where the Dice and Focal losses behave better in terms of maintaining high pixel accuracy during training.
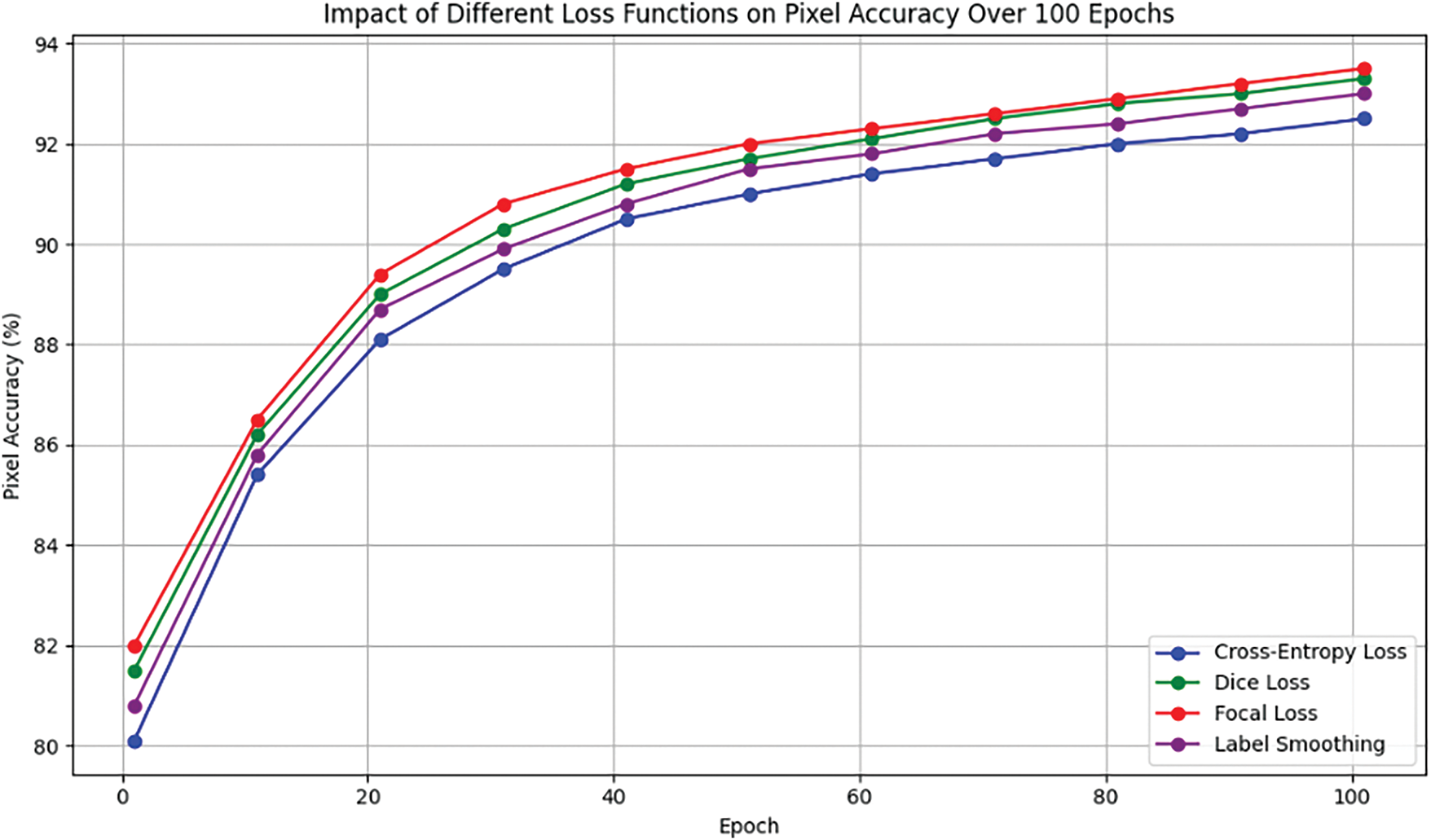
Figure 6: Impact of loss functions on pixel accuracy
The U-Net-ViM-HBAM model was shown to be robust and user-friendly when tested under simulated and real-world conditions. The model maintained high accuracy with minimal variations in processing time across devices, proving that it is suitable for mobile deployment with minimal computational resources. Fig. 7 illustrates the performance of the U-Net-ViM-HBAM model compared to the baseline models over different types of devices.
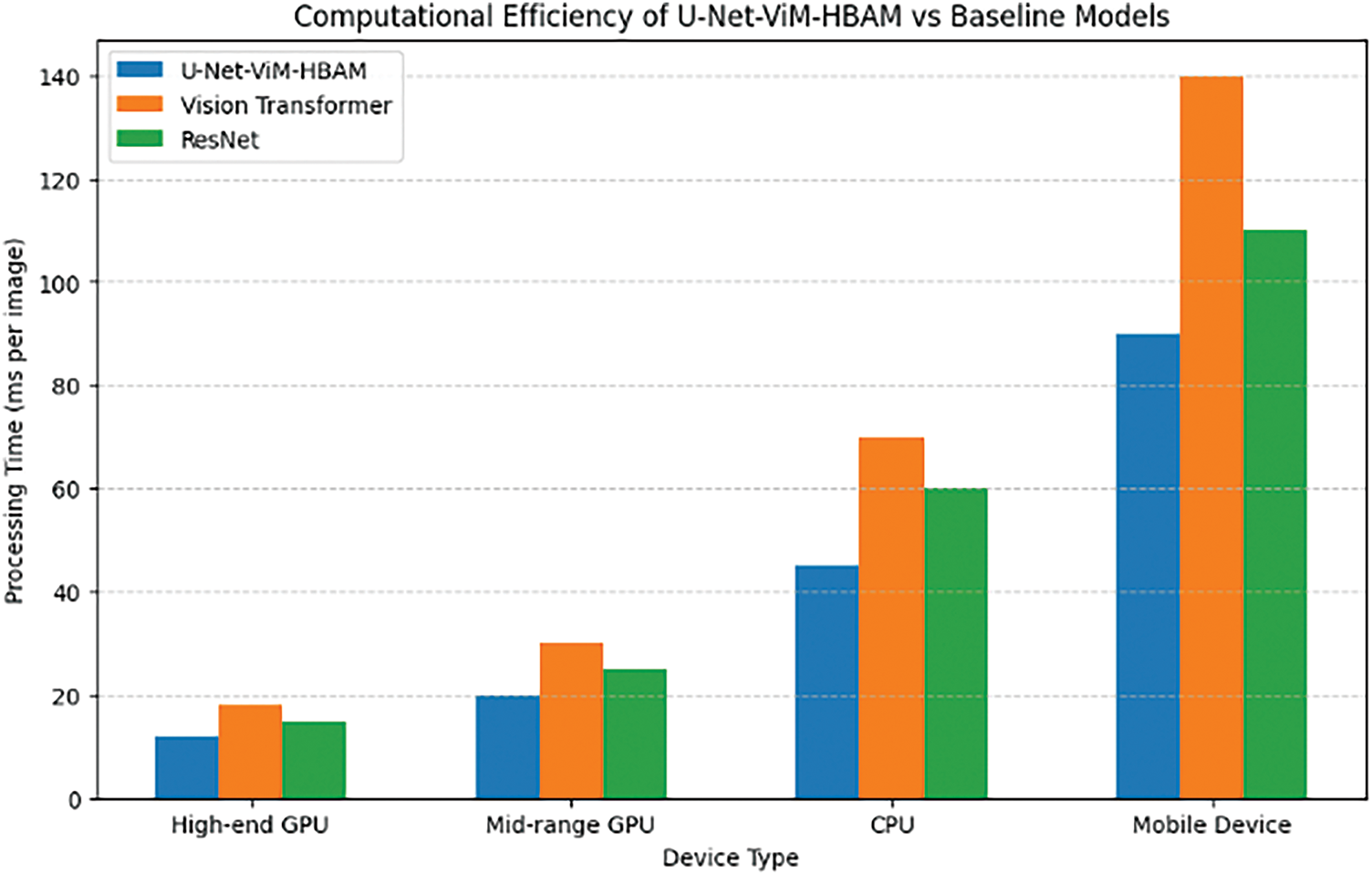
Figure 7: Computational efficiency of U-Net-ViM-HBAM model vs. baseline models
The U-Net-ViM-HBAM model showed tremendous improvement in the task of crop disease detection, particularly for detecting tomato leaf diseases. However, although the model was trained for the identification and classification of tomato leaf diseases, the architecture of the model allows adaptation to other crops with proper retraining of the model. Its hybrid architecture, which combines segmentation features from U-Net, contextual understanding from Vision Transformer, and selective attention mechanisms from HBAM, presents an effective way to address the critical challenges in identifying diseases under diverse and complex agricultural conditions. Superior performance metrics, such as a high accuracy of 98.63%, showed the model’s overall correctness in the identification and classification of tomato leaf diseases, the model’s precision of 98.24% reflected consistency in ensuring true positives and avoiding false positives, and a recall of 96.41% showed the model’s ability to correctly identify and classify most tomato leaf diseases. Although the recall slightly lower, the consistency of the high performance across other metrics validated the trained model.
The integration of U-Net, ViM, and HBAM significantly improved disease detection and classification accuracy and reduced the computational complexity under complex imaging conditions. The consistent performance of the model on diverse datasets and devices attests to its adaptability and potential for broader applications in precision agriculture, including its scalability to other crops and regions.
The model was also tested on mobile-compatible simulations, demonstrating its adaptability to mobile platforms, which highlights the model’s potential for deployment in resource-constrained environments, empowering smallholder farmers with accessible tools for early stage disease detection. These findings indicate that this model is in line with the principles of precision agriculture and provides data-driven, targeted interventions that reduce resource wastage and improve crop health management. Although the combined dataset used in this study is both public and anonymized, future applications involving real-world implementation may require strict data privacy protocols and informed consent procedures. The socio-economic implications of automating disease detection should be considered, particularly to ensure equitable access to such technologies for smallholder farmers. The proposed model is promising; however, it may underperform when subjected to uneven lighting, blurred images, and scenarios involving multiple diseases on a single leaf.
The proposed U-Net-ViM-HBAM model is a robust hybrid architecture that was successfully developed and validated for the detection and classification of diseases in tomato leaves with improved accuracy and efficiency compare to existing models. Thus, this model was systematically tested through validation and exhibited better performance in terms of accuracy, precision, and adaptability for diverse crop diseases, showing great promise as an instrument for precision agriculture. It addressed the computational inefficiencies and poor generalization over diverse environmental and disease detection shortcomings of individual deep learning models and charted a course for incorporating artificial intelligence into sustainable agricultural practices. From technologically advanced to resource-constrained environments, the model’s scalability and portability make it relevant to diverse agricultural settings in developing countries.
The unified U-Net-ViM-HBAM model is a state-of-the-art technology for detecting and classifying crop diseases. This helps to cover the growth and diversity of food demand. This study found its background in artificial intelligence and agriculture, but at the same time, it opened possibilities for new inventions in crop health management and productivity assurance for sustainability. This opens avenues for further innovation in the integration of AI and IoT technologies for real-time crop health management. These contributions represent a pivotal step toward ensuring global food security and sustainable agricultural productivity in the future.
In the future, we will prioritize enhancing the model performance, especially recall, for diseases characterized by subtle visual symptoms, including Leaf Mold and Powdery Mildew. This will involve experimenting with weighted loss functions to penalize false negatives more heavily, utilizing advanced augmentation techniques to increase symptom diversity and poor image conditions, and exploring contrastive learning to improve feature discrimination. These diseases exhibited marginally lower detection accuracies. We will also focus on improving recall for visually similar diseases. Future research should consider aspects such as uneven lighting, blurred images, and scenarios involving multiple diseases on a single leaf, and modify the model to maintain its performance across diverse agricultural settings. Further research is required to study how to adapt HBAM-based architectures for real-time field deployment, including mobile platforms and edge AI devices, to ensure performance and scalability across diverse agricultural environments.
Acknowledgement: We extend our sincere gratitude to everyone who supported this work.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: In this study, all the research work was conducted by Geoffry Mutiso and John Ndia. The first author contributed in the following areas; introduction, related works, methodology, results, discussion, and conclusion. The second author contributed to introduction, discussion and conclusion. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset modified and used in this study will be available upon request from the authors.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wuzhao R, Gong G, Cao S, Chen C, Wang W. Research on image classification of pathology based on deep learning. J Intell Knowl Eng. 2024;2(1):76. doi:10.62517/jike.202404111. [Google Scholar] [CrossRef]
2. Alirezazadeh P, Schirrmann M, Stolzenburg F. Improving deep learning-based plant disease classification with attention mechanism. Gesunde Pflanz. 2023;75(1):49–59. doi:10.1007/s10343-022-00796-y. [Google Scholar] [CrossRef]
3. Nakhale SR, Asutkar DS. Deep learning-based leaf disease detection in crop using images for agricultural application. Int J Innov Res Eng. 2024;146–50. [Google Scholar]
4. Hussain AA, Nair PS. Deep learning approach: precision agriculture advancements through accurate segmentation of crop and weed density. In: Proceedings of the 2024 2nd International Conference on Advancement in Computation & Computer Technologies (InCACCT); 2024 May 9–10; Gharuan, India. Piscataway, NJ, USA: IEEE; 2024. p. 154–60. [Google Scholar]
5. Dey P, Mahmud T, Nahar SR, Hossain MS, Andersson K. Plant disease detection in precision agriculture: deep learning approaches. In: Proceedings of the 2024 2nd International Conference on Intelligent Data Communication Technologies and Internet of Things (IDCIoT); 2024 Jan 4–6; Bengaluru, India. Piscataway, NJ, USA: IEEE; 2024. p. 661–7. [Google Scholar]
6. Upadhya NM. Classification and detection of plant disease using CNN and machine learning [Internet]. [cited 2024 Aug 10]. Available from: https://ijsrem.com/download/classification-and-detection-of-plant-disease-using-cnn-and-machine-learning/. [Google Scholar]
7. Li X, Ding J, Tang J, Guo F. Res2Unet: a multi-scale channel attention network for retinal vessel segmentation. Neural Comput Appl. 2022;34(14):12001–15. doi:10.1007/s00521-022-07086-8. [Google Scholar] [CrossRef]
8. Zhang T, Zhu J, Zhang F, Zhao S, Liu W, He R, et al. Residual swin transformer for classifying the types of cotton pests in complex background. Front Plant Sci. 2024;15:1445418. doi:10.3389/fpls.2024.1445418. [Google Scholar] [PubMed] [CrossRef]
9. Ferentinos KP. Deep learning models for plant disease detection and diagnosis. Comput Electron Agric. 2018;145:311–8. [Google Scholar]
10. Sun C, Zhou X, Zhang M, Qin A. SE-VisionTransformer: hybrid network for diagnosing sugarcane leaf diseases based on attention mechanism. Sensors. 2023;23(20):8529. doi:10.3390/s23208529. [Google Scholar] [PubMed] [CrossRef]
11. Barman U, Sarma P, Rahman M, Deka V, Lahkar S, Sharma V, et al. ViT-SmartAgri: vision transformer and smartphone-based plant disease detection for smart agriculture. Agronomy. 2024;14(2):327. doi:10.3390/agronomy14020327. [Google Scholar] [CrossRef]
12. Boukabouya RA, Moussaoui A, Berrimi M. Vision transformer based models for plant disease detection and diagnosis. In: Proceedings of the 2022 5th International Symposium on Informatics and its Applications (ISIA); 2022 Nov 29–30; M’sila, Algeria. Piscataway, NJ, USA: IEEE; 2022. p. 1–6. [Google Scholar]
13. Gu A, Dao T. Mamba: linear-time sequence modeling with selective state spaces. arXiv:2312.00752. 2023. [Google Scholar]
14. Christakakis P, Giakoumoglou N, Kapetas D, Tzovaras D, Pechlivani EM. Vision transformers in optimization of AI-based early detection of botrytis cinerea. AI. 2024;5(3):1301–23. doi:10.3390/ai5030063. [Google Scholar] [CrossRef]
15. Kumaar DM, Palani S. ResNet50 Integrated vision transformer for enhanced plant disease classification. In: 2024 3rd International Conference on Artificial Intelligence for Internet of Things (AIIoT); 2024 May 17–19; Vellore, India. Piscataway, NJ, USA: IEEE; 2024. p. 1–6. [Google Scholar]
16. Yuan Q, Zou S, Wang H, Luo W, Zheng X, Liu L, et al. A lightweight pine wilt disease detection method based on vision transformer-enhanced YOLO. Forests. 2024;15(6):1050. doi:10.3390/f15061050. [Google Scholar] [CrossRef]
17. Guo R, Zhang R, Zhou H, Xie T, Peng Y, Chen X, et al. CTDUNet: a multimodal CNN-transformer dual u-shaped network with coordinate space attention for Camellia oleifera pests and diseases segmentation in complex environments. Plants. 2024;13(16):2274. doi:10.3390/plants13162274. [Google Scholar] [PubMed] [CrossRef]
18. Shi D, Li C, Shi H, Liang L, Liu H, Diao M. A hierarchical feature-aware model for accurate tomato blight disease spot detection: Unet with Vision Mamba and ConvNeXt perspective. Agronomy. 2024;14(10):2227. doi:10.3390/agronomy14102227. [Google Scholar] [CrossRef]
19. Zhang S, Zhang C. Modified U-net for plant diseased leaf image segmentation. Comput Electron Agric. 2023;204(14):107511. doi:10.1016/j.compag.2022.107511. [Google Scholar] [CrossRef]
20. Rahman MM, Tutul AA, Nath A, Laishram L, Jung SK, Hammond T. Mamba in vision: a comprehensive survey of techniques and applications. arXiv:2410.03105. 2024. [Google Scholar]
21. Tang L, Yi J, Li X. Improved multi-scale inverse bottleneck residual network based on triplet parallel attention for apple leaf disease identification. J Integr Agric. 2024;23(3):901–22. doi:10.1016/j.jia.2023.06.023. [Google Scholar] [CrossRef]
22. Bai M, Di X, Yu L, Ding J, Lin H. A pine wilt disease detection model integrated with mamba model and attention mechanisms using UAV imagery. Remote Sens. 2025;17(2):255. doi:10.3390/rs17020255. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF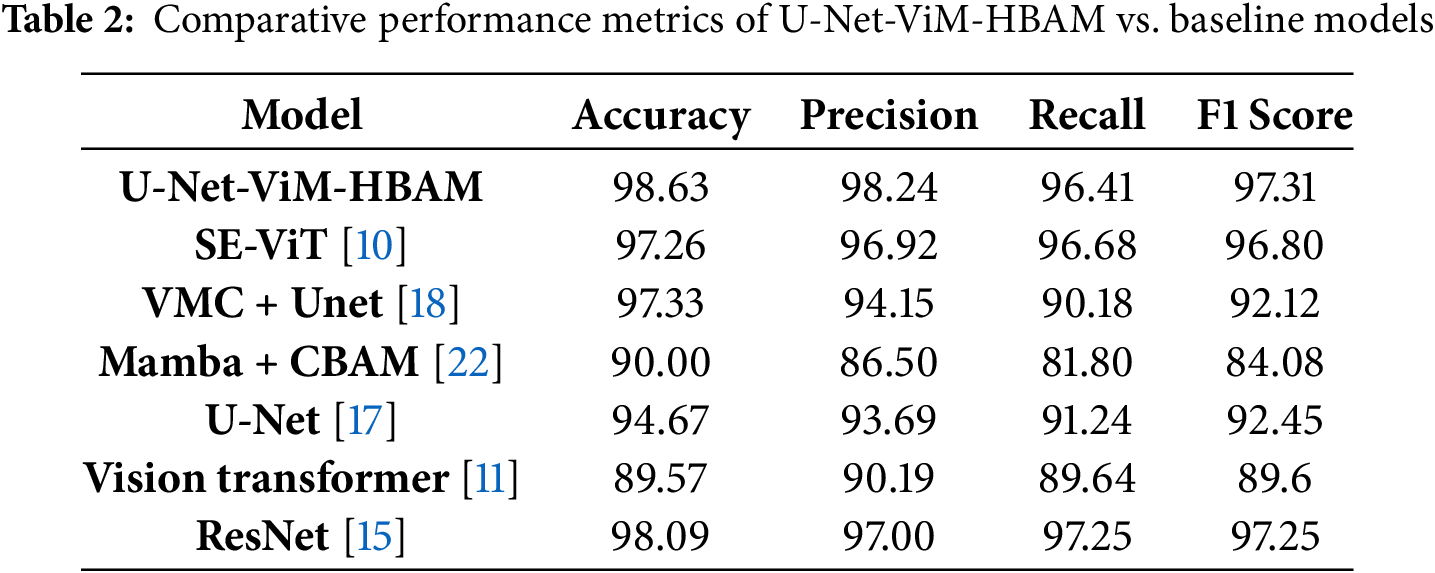
 Downloads
Downloads
 Citation Tools
Citation Tools