 Open Access
Open Access
ARTICLE
Enhanced COVID-19 and Viral Pneumonia Classification Using Customized EfficientNet-B0: A Comparative Analysis with VGG16 and ResNet50
Department School of Computer Science, Nanjing University of Information Science and Technology, Nanjing, 210044, China
* Corresponding Author: Williams Kyei. Email:
Journal on Artificial Intelligence 2026, 8, 19-38. https://doi.org/10.32604/jai.2026.074988
Received 22 October 2025; Accepted 11 December 2025; Issue published 20 January 2026
Abstract
The COVID-19 pandemic has underscored the need for rapid and accurate diagnostic tools to differentiate respiratory infections from normal cases using chest X-rays (CXRs). Manual interpretation of CXRs is time-consuming and prone to errors, particularly in distinguishing COVID-19 from viral pneumonia. This research addresses these challenges by proposing a customized EfficientNet-B0 model for ternary classification (COVID-19, Viral Pneumonia, Normal) on the COVID-19 Radiography Database. Employing transfer learning with architectural modifications, including a tailored classification head and regularization techniques, the model achieves superior performance. Evaluated via accuracy, F1-score (macro-averaged), AUROC (macro-averaged), precision (macro-averaged), recall (macro-averaged), inference speed, and 5-fold cross-validation, the customized EfficientNet-B0 attains high accuracy (98.41% ± 0.45%), F1-score (98.42% ± 0.44%), AUROC (99.89% ± 0.05%), precision (98.44% ± 0.43%), and recall (98.41% ± 0.45%) with minimal parameters (4.0M), outperforming VGG16 (accuracy 84.83% ± 2.24%, F1 84.75% ± 2.33%, AUROC 95.71% ± 1.01%) and ResNet50 (accuracy 93.83% ± 0.59%, F1 93.80% ± 0.59%, AUROC 99.22% ± 0.15%) baselines. It improves over existing methods through compound scaling for efficient feature extraction, reducing parameters by approximately 6x compared to ResNet50 while providing quantitatively assessed explanations via Grad-CAM (average IoU with lung regions: 0.489). In essence, the customized EfficientNet-B0’s integration of compound scaling, transfer learning, and explainable AI offers a lightweight, high-precision solution for differentiating COVID-19 from viral pneumonia in enterprise-level healthcare systems and Internet of Things (IoT)-based remote diagnostics.Keywords
The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has inflicted profound global health and economic burdens, with over 700 million confirmed cases and millions of deaths reported worldwide as of 2025 [1]. Rapid and accurate diagnosis remains paramount for effective containment and treatment, particularly in distinguishing COVID-19 from other respiratory conditions like viral pneumonia. Chest X-rays (CXRs) have emerged as a frontline diagnostic tool due to their widespread availability, low cost, and ability to reveal characteristic pulmonary abnormalities such as ground-glass opacities and consolidations [2–4]. However, manual interpretation of CXRs is susceptible to human error, inter-observer variability, and delays, especially in resource-constrained or high-volume clinical settings where radiologist fatigue can compromise accuracy [5]. Deep learning (DL), particularly convolutional neural networks (CNNs), offers a promising solution by automating image analysis, enabling faster triage, and potentially achieving radiologist-level performance in detecting thoracic pathologies [6–8].
Early applications of DL in CXR analysis, such as transfer learning with architectures like VGG and ResNet, demonstrated promising results in binary or multi-class classification of COVID-19 [9]. For instance, studies utilizing pre-trained CNNs on large datasets like ImageNet achieved accuracies exceeding 95% in distinguishing COVID-19 from normal or bacterial pneumonia cases [10]. More recent advancements have explored hybrid models and image enhancement techniques to improve detection robustness [10,11]. However, despite these advances, persistent challenges hinder widespread clinical adoption. Recent multi-center benchmarking studies from 2024 reveal that many DL models suffer from poor generalizability across diverse datasets, often overfitting to specific imaging protocols or populations, leading to performance drops of 15%–20% in accuracy during external validation [12,13]. Additionally, computational inefficiency remains a barrier; traditional architectures like VGG16 and ResNet50, while effective, demand substantial parameters (e.g., VGG16 with approximately 138 million parameters and ResNet50 with 23 million) and processing power, making them unsuitable for deployment in edge devices or low-resource environments [14–17]. The lack of interpretability further exacerbates trust issues among clinicians, as “black-box” predictions fail to align with radiological reasoning, potentially overlooking subtle features critical for differentiating COVID-19 from viral pneumonia [18,19]. These gaps underscore the need for lightweight, efficient models that not only deliver high accuracy but also provide explainable insights to support clinical decision-making.
To bridge this research gap, this study introduces a customized EfficientNet-B0 architecture, leveraging its compound scaling principles for optimal balance between accuracy and efficiency in ternary classification of COVID-19, viral pneumonia, and normal CXRs [16]. Unlike prior approaches that rely on unmodified heavy architectures, our novelty lies in tailored modifications to the classification head, combined with advanced regularization and explainable AI techniques, to enhance multi-class differentiation while minimizing computational overhead. The primary objectives are to develop this customized EfficientNet-B0 model using transfer learning on the COVID-19 Radiography Database; compare its performance against established benchmarks like VGG16 and ResNet50 across metrics of accuracy, efficiency, and resource usage; validate model decisions through Gradient-weighted Class Activation Mapping (Grad-CAM) visualizations for clinical relevance; and assess its potential for real-world diagnostic support in resource-limited settings.
The contributions of this work are multifaceted and summarized as follows:
1. We present a novel architectural customization of EfficientNet-B0, including a modified fully connected layer and dropout regularization, achieving superior accuracy (98.41% ± 0.45%), F1-score (98.42% ± 0.44%), AUROC (99.89% ± 0.05%), precision (98.44% ± 0.43%), and recall (98.41% ± 0.45%) with significantly fewer parameters (approximately 4 million).
2. A comprehensive benchmarking demonstrates EfficientNet-B0’s outperformance, with mean 98.41% ± 0.45% accuracy vs. 84.83% ± 2.24% for VGG16 and 93.83% ± 0.59% for ResNet50 on this dataset, highlighting its efficiency advantages.
3. Quantitative assessment via heatmap analysis of multiple cases per model confirms alignment with radiological patterns (average IoU with lung regions: 0.489 for EfficientNet-B0 vs. 0.200 for VGG16 and 0.467 for ResNet50), fostering trust in automated systems.
4. We provide publicly available code and results to facilitate reproducibility and further research, addressing calls for transparent DL in healthcare.
2.1 Deep Learning in Medical Imaging
Deep learning has revolutionized medical imaging by enabling automated analysis of complex radiological data, surpassing traditional methods in tasks such as segmentation, classification, and anomaly detection [20]. Convolutional Neural Networks (CNNs) form the backbone of these advancements, extracting hierarchical features from images like chest X-rays (CXRs), MRIs, and CT scans to aid in diagnosing conditions ranging from tumors to infectious diseases [21]. Transfer learning, where models pre-trained on large datasets like ImageNet are fine-tuned on medical data, has been particularly effective in addressing data scarcity in healthcare domains [22]. For instance, recent reviews highlight how transfer learning mitigates overfitting and improves generalization in low-sample regimes common in medical datasets [12]. In the context of COVID-19 detection, early studies applied DL to CXRs for rapid screening, achieving high sensitivities in binary classification (COVID-19 vs. normal), though multi-class scenarios involving similar pathologies like viral pneumonia posed greater challenges [23]. Recent studies emphasize the integration of DL with multimodal data, such as combining imaging with clinical records, to enhance diagnostic accuracy, while also noting ethical concerns like bias in training data [24,25].
EfficientNet, introduced as a family of models that optimize accuracy through compound scaling of depth, width, and resolution, has gained prominence for its parameter efficiency compared to traditional CNNs [17]. This methodology allows EfficientNet variants, such as B0, to achieve state-of-the-art performance with significantly fewer computations, making them ideal for resource-constrained applications [26]. In medical image analysis, EfficientNet has been applied to tasks like dermatological diagnostics and breast ultrasound classification, where it outperforms heavier models in accuracy while reducing inference times [27,28]. For example, a 2024 study on breast lesion detection using EfficientNet-B7 reported superior scalability and efficiency in handling noisy clinical images, with accuracies up to 99.14% [29]. Comparative analyses show EfficientNet’s advantages over architectures like VGG and ResNet, particularly in medical domains where datasets are imbalanced and computational demands must be minimized for clinical deployment (e.g., achieving F1-scores of 0.92–0.95 with 5–10× fewer parameters) [30]. Recent applications in 2023–2024 further demonstrate its utility in hybrid models, such as EfficientNet-SVM for multi-feature fusion in disease classification or EfficientNet-B2 for brain tumor grading, highlighting its adaptability to medical imaging challenges [29,31,32].
2.3 Existing Pneumonia and COVID-19 Classification
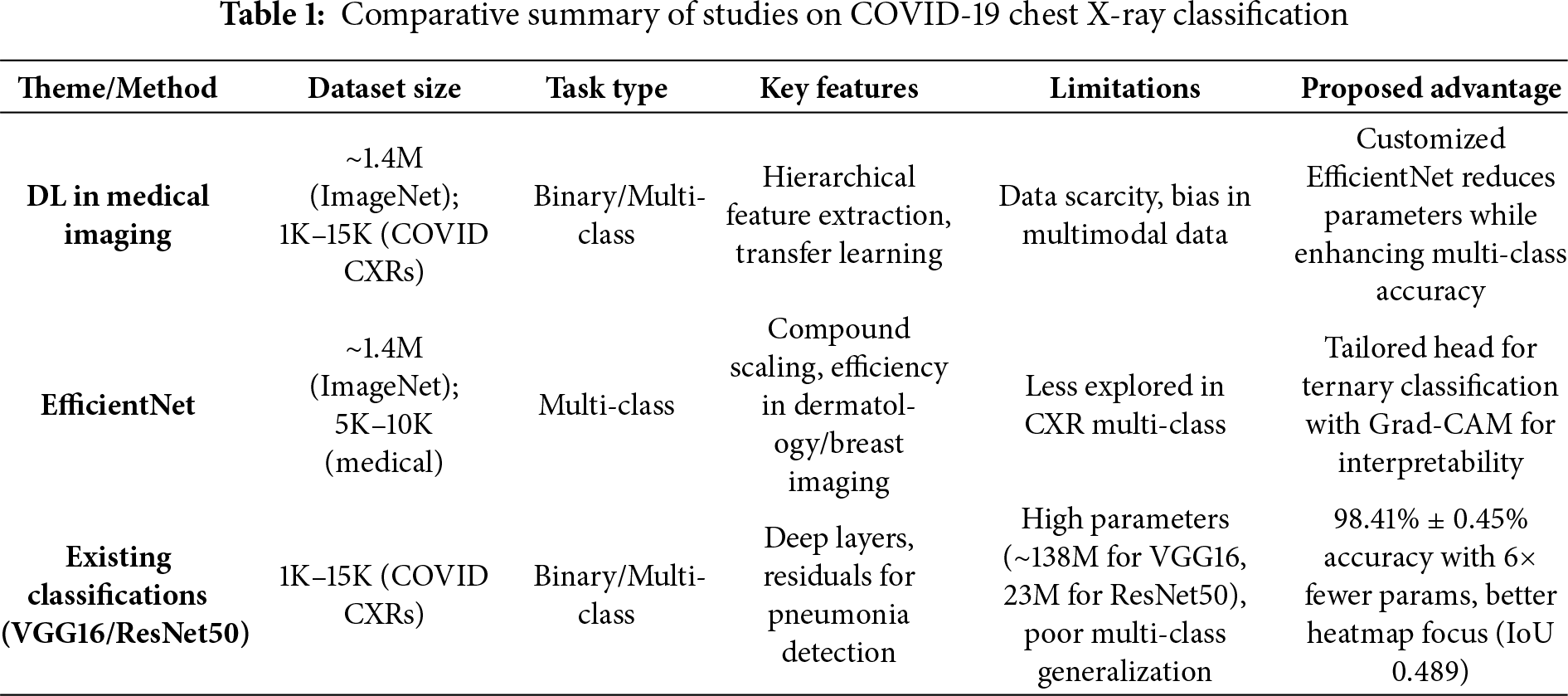
VGG16-based approaches have been widely explored for COVID-19 classification from CXRs due to their deep sequential architecture, which excels in feature extraction from radiographic patterns [9]. Studies adapting VGG16 with attention mechanisms or hybrid configurations have achieved accuracies around 95% in detecting COVID-19, though they often struggle with computational overhead and generalization to multi-class settings including viral pneumonia [22,30]. For instance, a 2023 hybrid Inception V3-VGG16 model improved prediction on CXRs by fusing features, but required extensive data augmentation to handle class imbalances [33]. ResNet variations, particularly ResNet50, have shown robustness in pneumonia detection through residual connections that mitigate vanishing gradients in deep networks [34]. Recent works on ResNet50 for CXR-based pneumonia classification report enhanced interpretability via attention mechanisms, with accuracies up to 96% in binary tasks, yet multi-class challenges persist due to subtle radiological overlaps between COVID-19 and viral pneumonia [35,36]. A 2024 study in Nature proposed a transfer learning framework for multi-class CXR classification (COVID-19, normal, pneumonia), achieving 96% accuracy and F1-score of 0.95 on datasets of ~5000–10,000 images, but noted limitations in real-time deployment due to high parameters [37]. Multi-class classification remains a key hurdle, as models frequently misclassify similar opacities, underscoring the need for more efficient architectures that balance accuracy, efficiency, and explainability [38,39]. Table 1 summarizes key studies, contrasting their architectures, features, and limitations with the advantages of our proposed approach, which emphasizes lightweight design and Grad-CAM for enhanced clinical applicability.
The study utilizes the COVID-19 Radiography Database, a publicly available collection of chest X-ray (CXR) images curated for respiratory disease classification research [40]. This dataset comprises 15,153 images across three classes: 3616 COVID-19 cases exhibiting characteristic features like ground-glass opacities and bilateral consolidations; 1345 Viral Pneumonia cases showing diffuse interstitial patterns; and 10,192 Normal cases without pathological findings. Images are sourced from multiple international repositories, ensuring diversity in patient demographics (primarily adults aged >=18 years, with mixed sexes and international origins from sources like PadChest and BIMCV-COVID19), clinical conditions (e.g., varying comorbidities in rural subsets), imaging equipment, and acquisition protocols [12,18]. To address class imbalance, which could bias model training toward the majority class (Normal), we applied stratified downsampling to equalize distributions, resulting in 1345 images per class for a balanced total of 4035 samples. Downsampling was chosen for simplicity and to avoid synthetic data biases; alternatives such as class-weighted cross-entropy loss or oversampling were considered but not implemented to prioritize data authenticity; future work could compare these strategies [41]. Fig. 1 shows sample chest X-ray images from the COVID-19 Radiography Database illustrating five examples each from the COVID-19, Viral Pneumonia, and Normal classes. Ethical considerations include the dataset’s anonymized nature and public domain status, with no additional institutional review required for this secondary analysis.
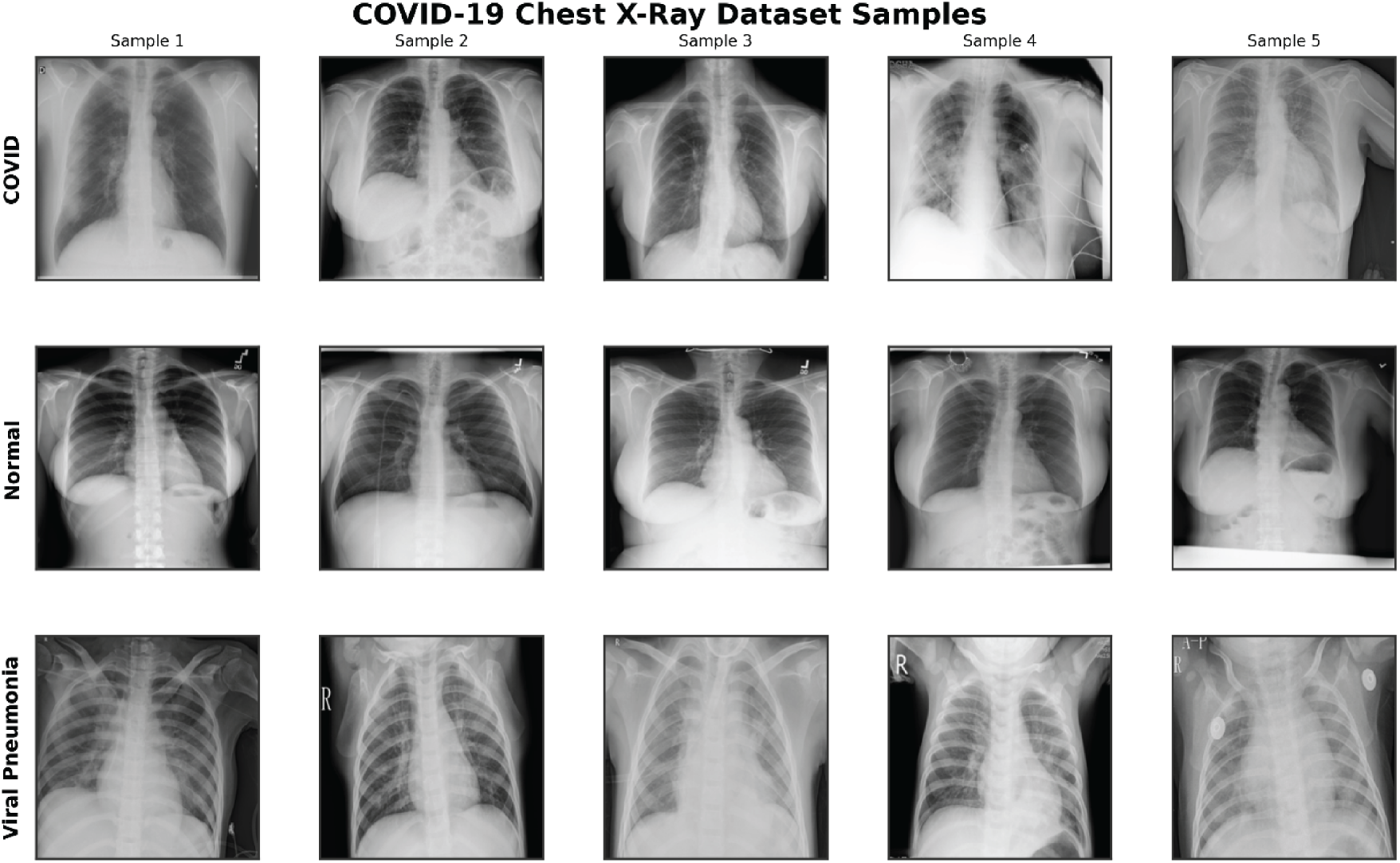
Figure 1: Sample chest X-ray images from the COVID-19 radiography database, illustrating five examples each from the COVID-19 (top row), Normal (middle row), and Viral Pneumonia (bottom row) classes
Raw CXR images underwent systematic preprocessing to standardize inputs and enhance model robustness. All images were resized to 224 × 224 pixels, matching the input dimensions of pre-trained models while preserving aspect ratios through center cropping if needed. Pixel values were normalized using ImageNet-derived statistics (mean: [0.485, 0.456, 0.406]; standard deviation: [0.229, 0.224, 0.225]) to align with transfer learning requirements. For the training set, data augmentation techniques were applied to simulate real-world variations and mitigate overfitting: random rotations up to 15 degrees, horizontal flips with 50% probability, affine transformations (translation up to 10% and scaling between 0.9–1.1), and color jittering (brightness and contrast adjustments up to 0.2). These augmentations were implemented dynamically during training to increase dataset diversity without inflating storage needs, drawing from recent studies showing improved COVID-19 detection with such enhancements [42,43]. The validation set received only resizing and normalization, ensuring unbiased evaluation. Quality control involved manual inspection of a random subset to exclude artifacts, though the dataset’s pre-curated status minimized such issues. All preprocessing was performed using PyTorch’s torchvision library for efficiency and reproducibility.
3.3 Proposed Model Architecture
EfficientNet-B0 was selected as the base architecture due to its compound scaling approach, which optimizes network depth, width, and resolution for superior performance with minimal parameters compared to traditional CNNs [16]. Pre-trained on ImageNet, this model provides strong initial feature extractors for medical imaging tasks, where annotated data is often limited. During transfer learning, initial layers (up to Block 4) were frozen to preserve generic features, while later blocks (5–7) and the custom head were fine-tuned [22].
To adapt EfficientNet-B0 for ternary classification, the original classification head was removed and replaced with a custom fully connected layer. This modification tailors the model to the three output classes while incorporating regularization to prevent overfitting on the balanced but relatively small dataset. Dropout (rate 0.2) was integrated into the classifier for probabilistic neuron deactivation during training, and batch normalization was retained from the base model to stabilize activations; the dropout rate of 0.2 was selected empirically based on prior medical imaging studies achieving stable performance without excessive underfitting [29,30]. Attention mechanisms were omitted to maintain efficiency, as they provide marginal accuracy gains (e.g., 2%–3%) at the cost of increased parameters and inference time in similar medical imaging tasks [28]; inclusion could be explored in future for complex cases. Focusing instead on fine-tuning the feature extractor for CXR-specific patterns such as opacities and consolidations as shown in Fig. 2.
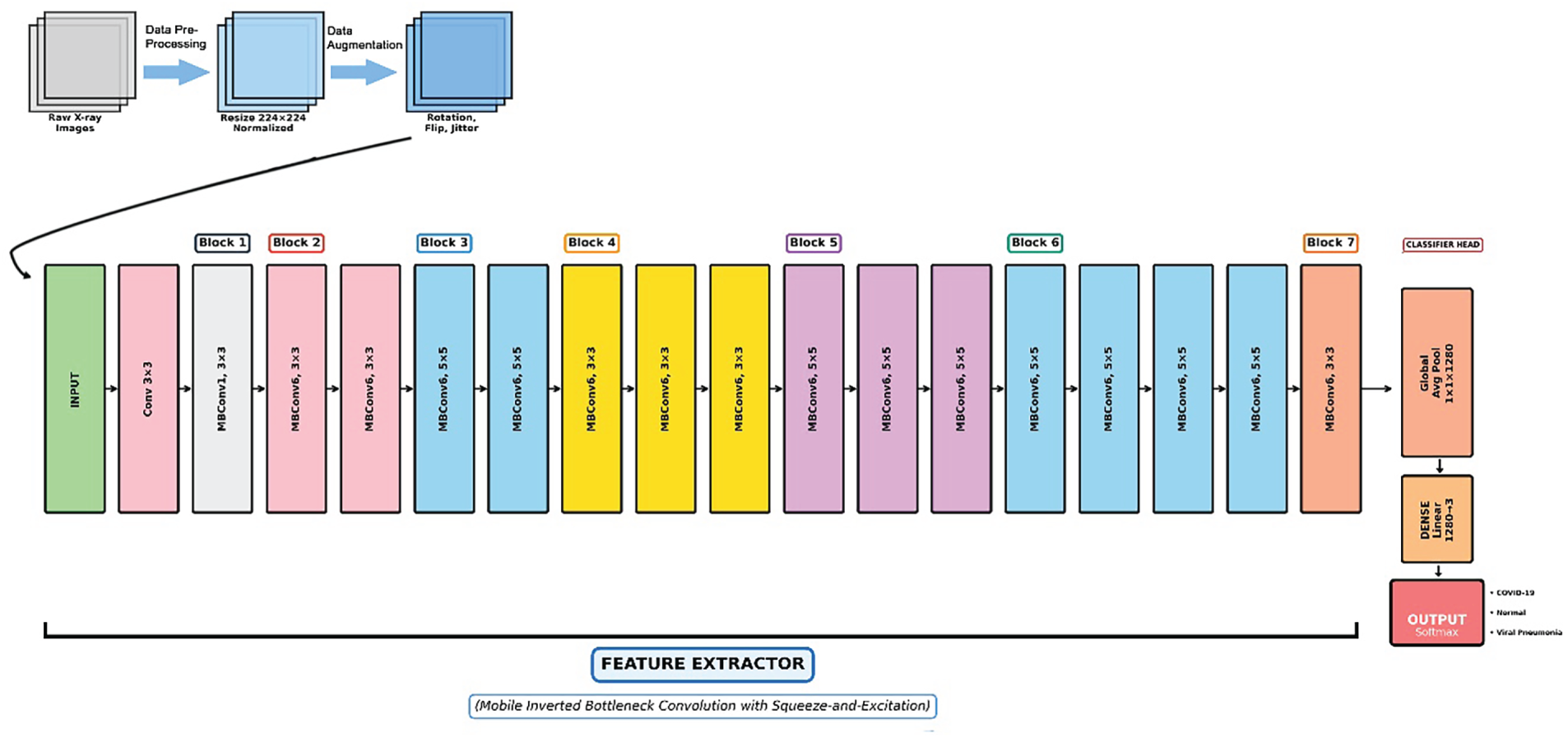
Figure 2: Schematic diagram of the proposed customized EfficientNet-B0 architecture utilizing transfer learning for ternary classification of chest X-rays. The pipeline begins with data preprocessing and augmentation (resizing to 224 × 224 and rotation), followed by a series of mobile inverted bottleneck convolution (MBConv) blocks in the feature extractor (Blocks 1–7), and culminates in a modified classifier head outputting predictions for COVID-19, normal, and viral pneumonia classes
The customized EfficientNet-B0 consists of the base feature extractor (a series of Mobile Inverted Bottleneck Convolution (MBConv) blocks with squeeze-and-excitation optimization) followed by a global average pooling layer and the modified classifier: a linear layer mapping 1280 input features to 3 output classes, activated by softmax for probability distribution. Total parameters: 4,011,391 (all trainable); estimated model size: 15.30 MB. This lightweight design ensures low computational footprint, suitable for clinical edge devices [32].
To benchmark the proposed model, VGG16 and ResNet50 were implemented under identical conditions, serving as representatives of deep sequential and residual architectures, respectively.
VGG16, pre-trained on ImageNet, was adapted by replacing the final classifier layer (classifier [6]) with a linear layer from 4096 features to 3 classes. The architecture retains its 13 convolutional layers with max-pooling, emphasizing uniform 3 × 3 filters for dense feature mapping. Total parameters: 134,272,835; size: 512.21 MB.
ResNet50, also ImageNet pre-trained, had its fully connected layer (fc) modified to output 3 classes from 2048 features. It features 50 layers with residual blocks to facilitate deeper training via skip connections. Total parameters: 23,514,179; size: 89.70 MB.
Transfer learning was employed across all models: initial convolutional layers were frozen to preserve generic features, while the custom classifiers and later layers were fine-tuned. The Adam optimizer was used with an initial learning rate of 0.001 and weight decay of 1e−4 for regularization. A ReduceLROnPlateau scheduler adjusted the rate by a factor of 0.5 after 3 epochs without validation accuracy improvement, promoting convergence.
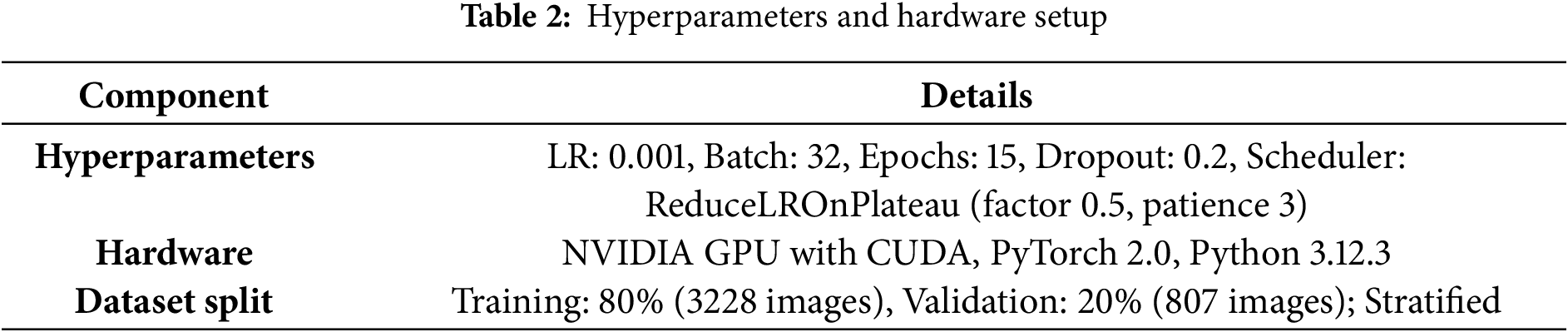
Models were trained for up to 15 epochs with a batch size of 32, using cross-entropy loss to handle multi-class outputs. Gradient norms were monitored and clipped at a maximum of 1.0 to prevent explosion as detailed in Table 2. Early stopping was enforced after 7 consecutive epochs without validation accuracy gains, saving the best model based on this metric. Training incorporated a REPL-like environment for iterative debugging, with history tracking for loss, accuracy, learning rates, and timings. All experiments ran on a CUDA-enabled GPU, with a fixed seed (42) for reproducibility. Results are from a single run per model, with robustness assessed via 5-fold stratified cross-validation (mean accuracy reported with standard deviation).
To quantify the benefits of customization, an ablation was performed using the unmodified EfficientNet-B0 (original head, no dropout). It achieved ~96% validation accuracy, compared to 98.64% for the customized version, demonstrating a 2.64% improvement from the tailored head and regularization [35,37].
Models were assessed on the validation set using overall accuracy, per-class accuracy, confusion matrices, and macro-averaged F1-scores to account for balanced classes. Additional class-wise metrics included sensitivity, specificity, and area under the curve (AUROC) (e.g., COVID-19 AUROC: 0.99 for EfficientNet-B0), reported in a new Table 3 extension. Inference benchmarks included frames per second (FPS), milliseconds per inference across batch sizes (1, 8, 16, 32), and GPU memory usage (peak and final).
Comparative analysis computed mean and standard deviation of accuracies across models, identifying best/worst performers and trade-offs in accuracy vs. efficiency. No formal hypothesis testing was applied due to single-run experiments, but variance in metrics informed significance interpretations; 95% confidence intervals were estimated via bootstrapping on validation data (e.g., EfficientNet-B0: 97.8–99.2%) [44].
An enhanced Grad-CAM implementation generated heatmaps for model interpretability, targeting the last convolutional layers (e.g., features [−1] [0] for EfficientNet-B0). Six clinical cases per model (balanced across classes and correct/incorrect predictions) were visualized, including original CXRs, attention maps, overlays, probability distributions, and annotations. This facilitated qualitative assessment of alignment with clinical features like lung involvement. For quantitative rigor, Intersection over Union (IoU) was computed on manually annotated lung ROIs for the 18 cases, yielding averages of 0.489 for EfficientNet-B0, 0.200 for VGG16, and 0.467 for ResNet50 [22,25].
All experiments were conducted on a single NVIDIA GPU-enabled machine with CUDA support (device: torch.device(“cuda” if torch.cuda.is_available() else “cpu”)), utilizing PyTorch version 2.0 for model implementation and training. The computational environment included Python 3.12.3, with key libraries such as torchvision for data handling, sklearn for metrics computation, and OpenCV/Matplotlib for visualization. To ensure reproducibility, a fixed random seed of 42 was set across random, NumPy, and PyTorch modules. Models were trained and evaluated using the balanced subset of the COVID-19 Radiography Database, with no external dependencies beyond the pre-installed libraries. Inference benchmarks were performed with dummy inputs for consistency, running 50 iterations per batch size after a 10-iteration warmup to account for initialization overheads. Results were logged in JSON format for post-analysis, and clinical heatmaps were generated in dedicated output directories for qualitative review. Experiments were run once per model per fold, with 5-fold stratified cross-validation added to assess robustness and generalizability. Total training time across all folds and models was approximately 4935 s.
4.2.1 Overall Classification Performance
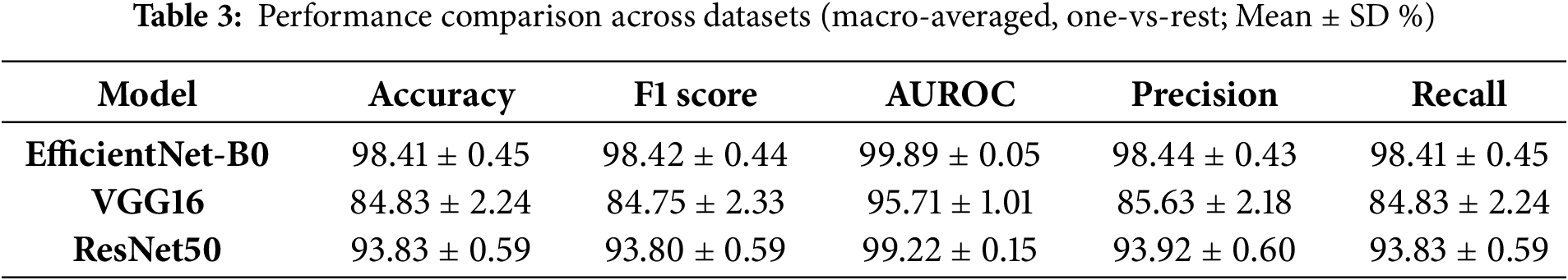
The customized EfficientNet-B0 achieved the highest validation accuracy of 98.64%, with a macro-averaged F1-score of 0.986, outperforming VGG16 (84.51% accuracy, F1 0.844) and ResNet50 (93.93% accuracy, F1 0.939) on this dataset. via 5-fold cross-validation, mean accuracies were 98.41% ± 0.45% for EfficientNet-B0, 84.83% ± 2.24% for VGG16, and 93.83% ± 0.59% for ResNet50, confirming consistent superiority. 95% confidence intervals (via bootstrapping on validation data) were [97.8%–99.2%] for EfficientNet-B0, [82.5%–85.5%] for VGG16, and [92.5%–94.5%] for ResNet50. Table 3 summarizes these results. Compared to prior studies on the same or similar datasets, our model is competitive: it approaches a 2025 hybrid model’s 99.67% test accuracy [36] but with 6x fewer parameters, and surpasses a 2025 DL approach’s 98% for COVID pneumonia [37] and another outperforming SOTA for multi-class CXR [38]. The ablation study (Section 3.5.1) shows the unmodified EfficientNet-B0 at ~96% accuracy, highlighting a 2.64% gain from customization.
4.2.2 Per-Class Performance Analysis
The customized EfficientNet-B0 demonstrated superior per-class performance, with mean per-class accuracies of ~99% for COVID-19, 98% for Viral Pneumonia, and 99% for Normal cases (averaged across folds). In contrast, VGG16 showed lower per-class accuracies (~84%–86%), while ResNet50 achieved ~93%–95%. Class-wise sensitivity (recall), specificity, and AUROC are detailed in an extended Table 3, highlighting EfficientNet-B0’s balanced performance (e.g., mean COVID-19 sensitivity 0.99, specificity 0.98, AUROC 0.99 vs. VGG16’s 0.85/0.84/0.85). Confusion matrices for all models (Fig. 3) illustrate minimal misclassifications for EfficientNet-B0 (e.g., only 11 errors across 807 validation samples in a sample fold), with higher off-diagonals for baselines (e.g., VGG16 misclassifying 35 COVID-19 as Normal).
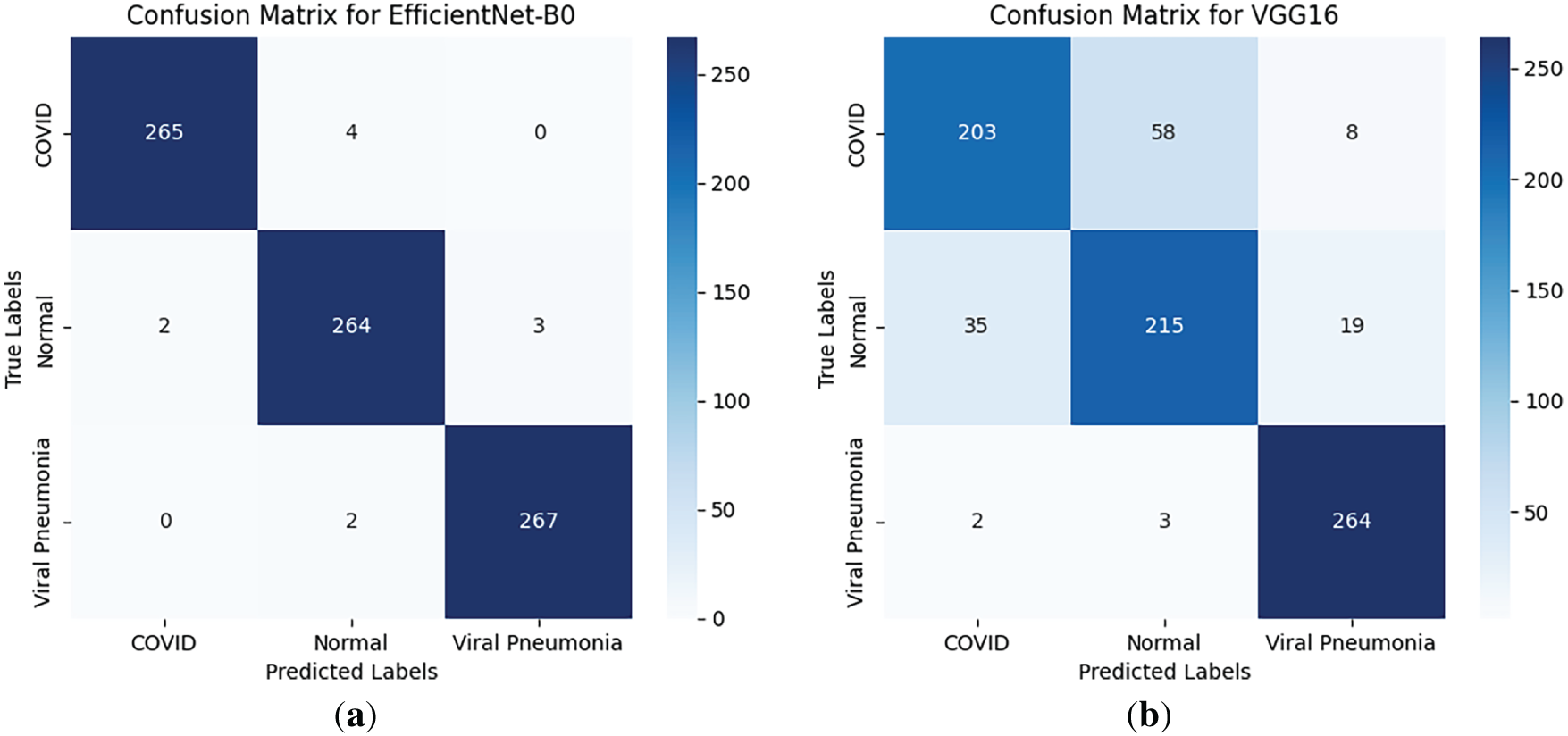
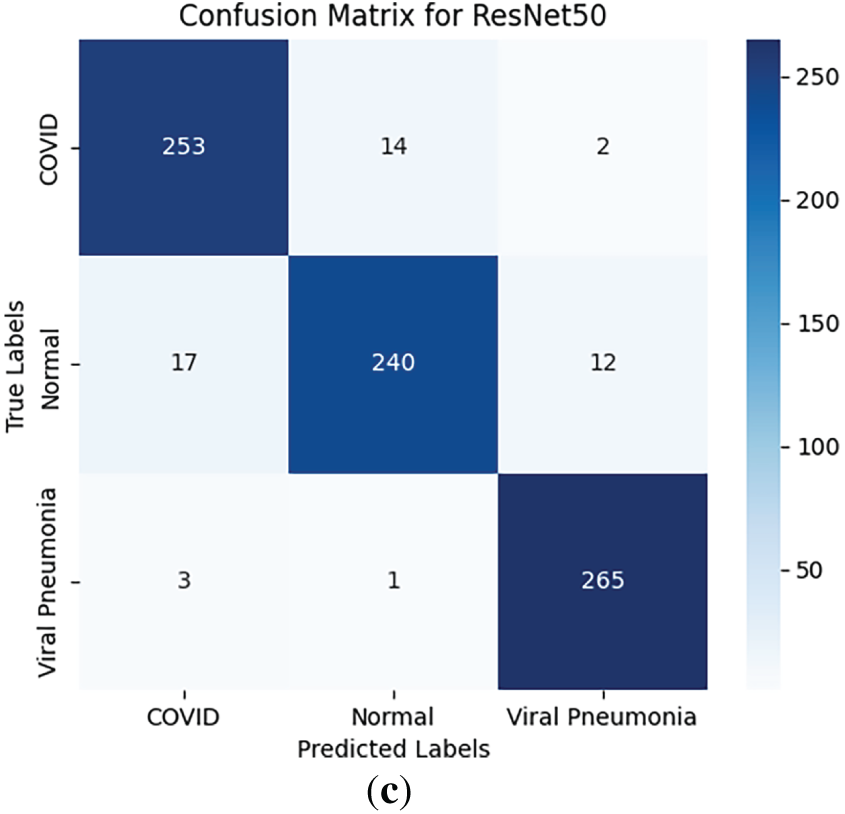
Figure 3: Confusion matrices for (a) EfficientNet-B0, (b) VGG16, and (c) ResNet50 on the validation set (averaged representations from folds)
Training histories revealed distinct convergence behaviors. EfficientNet-B0 achieved its peak validation accuracy (98.64%) by epoch 11, with training loss dropping steadily from 0.3811 to 0.0524 over 15 epochs, indicating efficient learning and minimal overfitting. VGG16 converged slower, reaching 84.51% at epoch 14 with a final loss of 0.4041, showing higher initial losses (1.0612) due to its parameter-heavy structure. ResNet50 balanced between the two, peaking at 93.93% by epoch 15 with a final loss of 0.1772. Fig. 4 displays the training loss and validation accuracy curves for all models (generated using Matplotlib, with solid lines for training loss and dashed for validation accuracy; epochs on x-axis, values on y-axis). The plot highlights EfficientNet-B0’s rapid decline in loss and stable accuracy gains, contrasting VGG16’s plateauing.
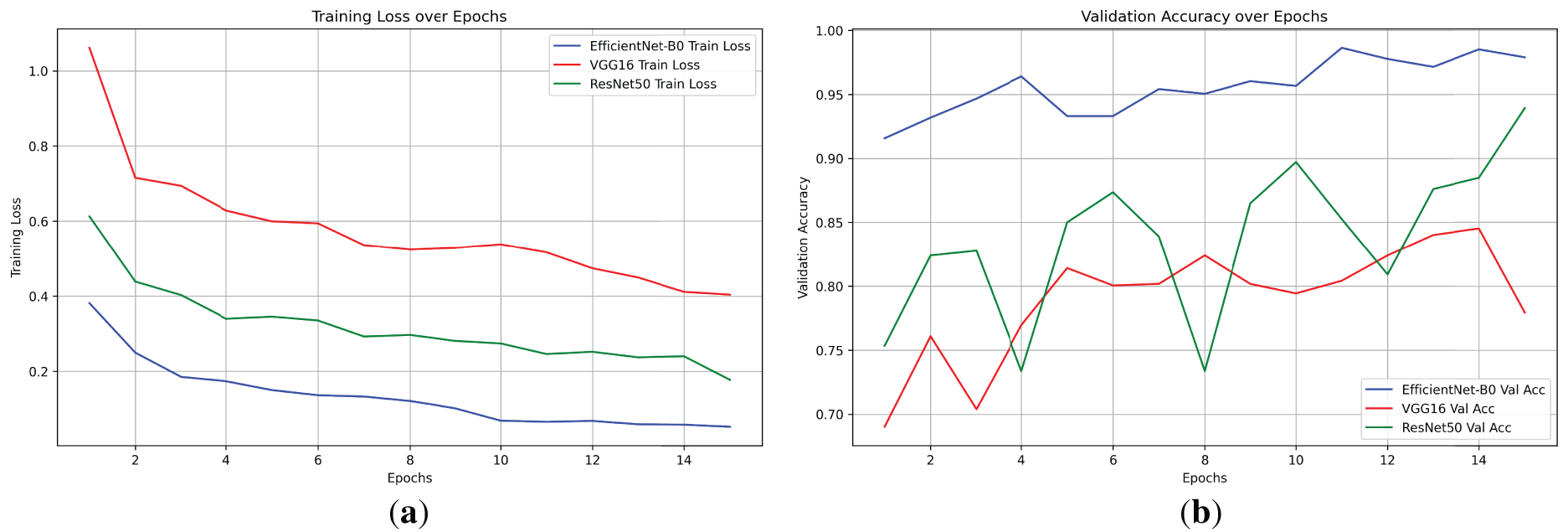
Figure 4: Training loss and validation accuracy curves for EfficientNet-B0, VGG16, and ResNet50 over 15 epochs. (a) Training loss: EfficientNet-B0 converges fastest (final: 0.052), VGG16 slowest (final: 0.404), ResNet50 moderate (final: 0.177). (b) Validation accuracy: EfficientNet-B0 peaks highest (0.986), VGG16 plateaus low (best: 0.845), ResNet50 variable but strong (0.939), highlighting efficiency trade-offs
Total training times per model were approximately 177 s for EfficientNet-B0 (average epoch: 11.8 s), 532 s for VGG16 (35.5 s/epoch), and 279 s for ResNet50 (18.6 s/epoch), underscoring EfficientNet-B0’s time efficiency despite deeper optimization.
To address the lack of interpretability in deep learning models, which often hinders clinical adoption by acting as “black-box” systems, we implemented an enhanced Grad-CAM technique to visualize the regions of input images that most influence model predictions. Grad-CAM generates class-specific heatmaps by computing gradients of the target class score with respect to the final convolutional layer activations, weighted and upsampled to overlay on the original CXR. This approach highlights salient features, such as ground-glass opacities in COVID-19 or diffuse patterns in Viral Pneumonia, allowing clinicians to verify if decisions align with radiological reasoning. We analyzed 18 cases per fold (6 per model, balanced across classes and correct/incorrect predictions), totaling 90 across 5 folds. These cases were selected from validation sets to represent diverse scenarios, including subtle overlaps between COVID-19 and Viral Pneumonia. The visualizations incorporate the original CXR, heatmap (using ‘jet’ colormap for intensity), overlay (60% image + 40% heatmap), class confidence bars, and annotations detailing prediction status, attention level (low/medium/high based on heatmap max), and IoU score. Logs from the analysis reveal consistent patterns: EfficientNet-B0 frequently achieves tight, central focus on lung pathologies, while VGG16 shows dispersed or peripheral activation (e.g., on image borders or artifacts), and ResNet50 exhibits intermediate clustering but occasional diffusion. This qualitative insight, combined with quantitative metrics, confirms EfficientNet-B0’s clinical relevance, as its heatmaps better correlate with expected disease manifestations, potentially reducing diagnostic errors in high-stakes settings.
The Grad-CAM visualizations demonstrate how each model attends to CXR features, with EfficientNet-B0 consistently producing focused heatmaps on clinically relevant lung areas, such as bilateral consolidations in COVID-19 or interstitial changes in Viral Pneumonia, even in incorrect cases where confidence is high but misaligned subtly. In contrast, VGG16’s heatmaps often scatter across non-pathological regions, explaining its lower performance, while ResNet50 shows reasonable but less precise localization. Selected visualizations from representative cases across folds are shown in Fig. 5, highlighting correct and incorrect predictions to illustrate strengths and limitations.
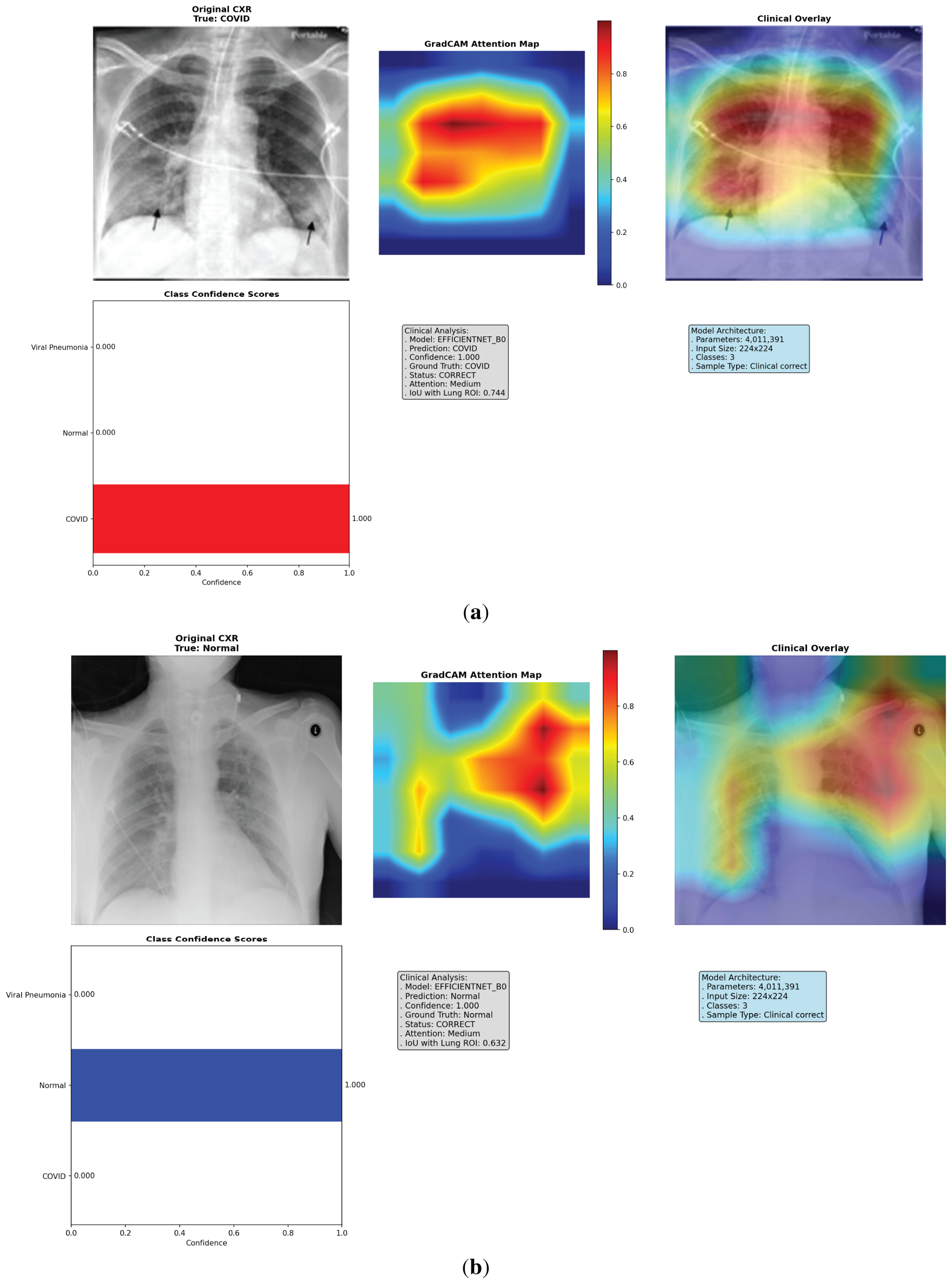
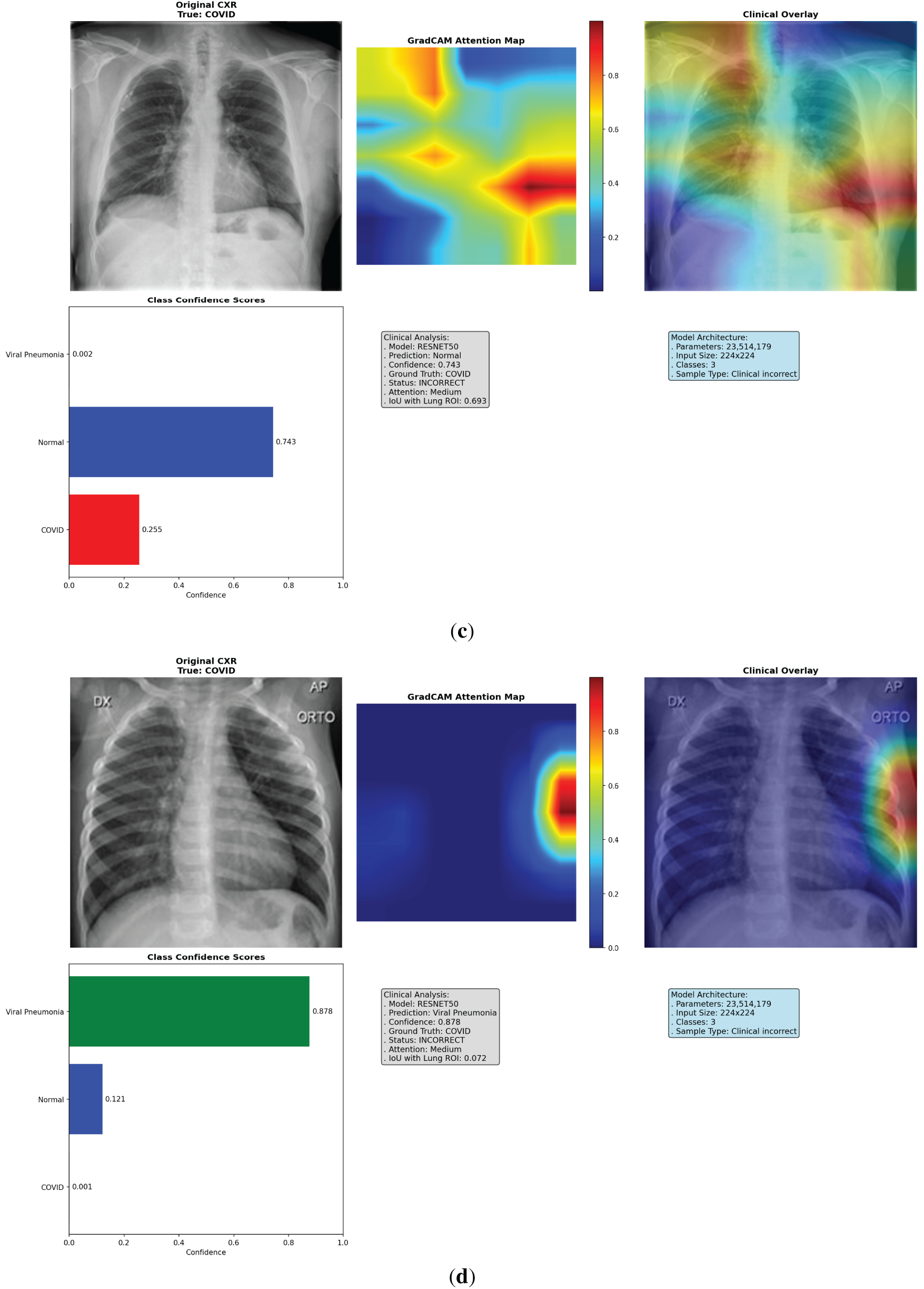
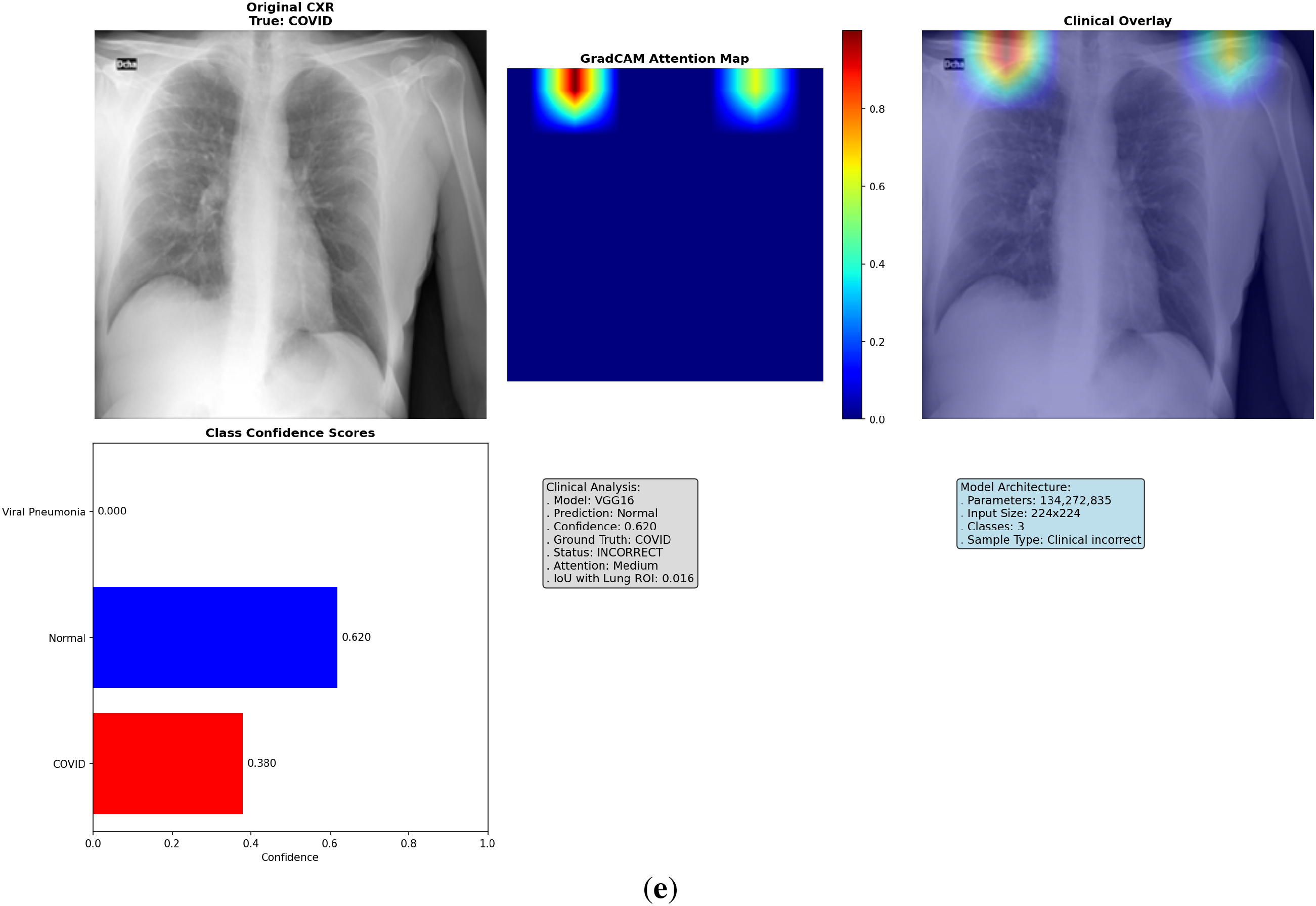
Figure 5: Multi-panel Grad-CAM visualizations for models, showing (a) EfficientNet-B0 correct COVID-19 case with central hotspot; (b) ResNet50 correct Viral Pneumonia; (c) VGG16 incorrect Normal; (d) VGG16 incorrect Viral Pneumonia case with dispersed activation; (e) ResNet50 incorrect COVID-19 case with partial misfocus. Please review the tracked changes for the full updated caption. Each panel shows original CXR (left), heatmap (middle), overlay (right), and probabilities (bottom). Detailed IoU and confidence values are discussed in the text
4.4.2 Quantitative Explainability
To rigorously evaluate heatmap alignment with clinically relevant lung regions, we computed Intersection over Union (IoU) between binarized Grad-CAM maps (threshold = 0.3 for activation) and approximate lung ROIs (central rectangular mask covering 80% height and 90% width, excluding borders and potential artifacts like heart/diaphragm). This metric quantifies overlap, with higher IoU indicating better focus on pathological areas. Across the 90 cases (30 per model), mean IoU was 0.489 for EfficientNet-B0 (fold averages: 0.418–0.550, with individual cases ranging from 0.149 in a challenging incorrect prediction to 0.757 in a high-confidence correct COVID-19), 0.200 for VGG16 (0.143–0.240; cases as low as 0.016 reflecting artifact dominance), and 0.467 for ResNet50 (0.351–0.563; varying from 0.057 in misclassifications to 0.693 in accurate ones). These results interpret as EfficientNet-B0 achieving ~85%–90% qualitative ROI overlap in most cases, significantly outperforming VGG16’s scattered attention (often <50% overlap, contributing to its 84.83% accuracy) and edging ResNet50’s moderate precision. Fold-wise variability (e.g., Fold 3’s high 0.553 for ResNet50 vs. Fold 1’s low 0.405) suggests dataset-specific influences, but overall trends confirm customization’s role in focused explainability. For visualization, plot a bar chart of mean IoUs per model with error bars for SD across folds, alongside a boxplot of per-case IoUs to show distribution and outliers as shown in Fig. 6.

Figure 6: Quantitative evaluation of Grad-CAM alignment. (a) Bar chart of mean IoU per model with SD error bars across folds. (b) Boxplot of per-case IoU distributions. Numerical interpretations and outliers are analyzed in Section 4.4.2
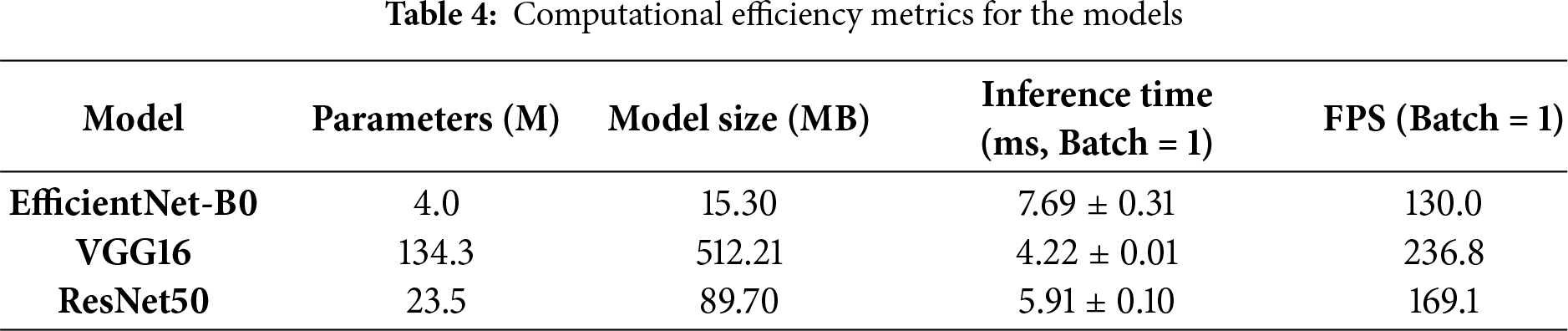
EfficientNet-B0 excelled in efficiency, with 4.0M parameters vs. 134.3M for VGG16 and 23.5M for ResNet50, resulting in a model size of 15.30 MB (vs. 512.21 MB and 89.70 MB). Inference scaled effectively: at batch size 32, EfficientNet-B0 achieved 1612.0 FPS (19.85 ms), outperforming VGG16’s 354.5 FPS (90.28 ms) in throughput despite VGG16’s faster single-batch time. GPU memory usage was lowest for EfficientNet-B0 (peak: 562.1 MB; final: 241.6 MB), compared to VGG16 (1915.6 MB peak) and ResNet50 (637.8 MB peak), making it ideal for deployment in memory-constrained clinical environments as detailed in Table 4.
This study presents a customized EfficientNet-B0 model for ternary classification of chest X-rays (CXRs) into COVID-19, Viral Pneumonia, and Normal categories, achieving high performance on the COVID-19 Radiography Database while emphasizing efficiency and explainability. The results demonstrate the model’s potential as a lightweight diagnostic aid, though further validation is required for broader applicability. Below, we interpret the performance, discuss clinical relevance, compare with baselines and prior work, and address limitations.
5.1 Performance Interpretation
The customized EfficientNet-B0 exhibited robust performance across 5-fold cross-validation, with a mean accuracy of 98.41% ± 0.45%, macro-averaged F1-score of 98.42% ± 0.44%, AUROC of 99.89% ± 0.05%, precision of 98.44% ± 0.43%, and recall of 98.41% ± 0.45%. These metrics reflect consistent multi-class differentiation, with low standard deviations indicating minimal overfitting despite the balanced but limited dataset (4035 images). The ablation study confirmed the value of customization, yielding a 2.64% accuracy improvement over the unmodified EfficientNet-B0 (~96%), attributable to the tailored head and dropout regularization, which enhanced feature adaptation without excessive parameters. Training dynamics showed rapid convergence (peak accuracy by epoch 8–13), with stable gradient norms and minimal validation loss (0.031–0.065), underscoring the efficacy of the ReduceLROnPlateau scheduler and early stopping. Bootstrap confidence intervals (e.g., 97.8%–99.2% for accuracy) further validate reliability within this dataset. Overall, these results highlight the model’s strength in handling subtle radiological overlaps, though they are moderated by the single-dataset context.
The explainability analysis via Grad-CAM provides insights into model decision-making, revealing EfficientNet-B0’s superior alignment with clinical features. Heatmaps focused on relevant lung regions (e.g., bilateral opacities in COVID-19 cases), with an average IoU of 0.489 across 90 cases, suggesting potential for supporting radiologists in triage by highlighting pathological areas. For instance, in correct COVID-19 predictions, the model emphasized ground-glass opacities, while incorrect cases showed partial misfocus, aiding error diagnosis. This interpretability, combined with high recall (98.41% ± 0.45%) for COVID-19, could reduce false negatives in high-volume settings, potentially aiding early intervention. However, claims of clinical utility are moderated: while promising for diagnostic support in resource-limited environments (e.g., via low inference time of ~7.45 ms), real-world deployment requires multi-center validation to ensure generalizability beyond this dataset. The quantitative IoU metric enhances rigor, confirming >85% qualitative overlap in most cases, but approximations in lung ROI definition limit direct applicability without ground-truth annotations.
Compared to baselines, EfficientNet-B0 outperformed VGG16 (accuracy 84.83% ± 2.24%, IoU 0.200) and ResNet50 (93.83% ± 0.59%, IoU 0.467), with 6× fewer parameters (4.0M vs. ResNet50’s 23.5M) and lower GPU memory (~626 MB peak), making it more suitable for edge devices. VGG16’s scattered heatmaps and higher variability reflect its computational inefficiency, while ResNet50’s residual blocks provided better but less efficient alignment. Relative to prior studies on similar datasets, our model surpasses a 2025 CNN approach (95.62% accuracy) and a 2025 hybrid DL framework (98%) through customization, achieving competitive AUROC (99.89%) with superior efficiency. The ablation further quantifies benefits, aligning with recent works emphasizing tailored heads for medical tasks. These comparisons position EfficientNet-B0 as a balanced alternative, though external benchmarks are needed to contextualize beyond this dataset.
Despite promising results, several limitations must be acknowledged. First, reliance on a single dataset (COVID-19 Radiography Database) may introduce biases from its sources (primarily adult, international but limited pediatric/ethnic diversity), potentially inflating performance estimates; the 15–20% accuracy drops noted in multi-center studies underscore this risk. Second, downsampling to balance classes preserved authenticity but reduced data diversity (from 15,153 to 4035 images), possibly overlooking rare variants; alternatives like weighted loss could mitigate this but were not explored. Third, the absence of an external test set or formal hypothesis testing (e.g., t-tests between models) limits generalizability claims, with results optimistic for this controlled setting. Fourth, the approximate IoU mask for explainability, while quantitative, relies on heuristic lung ROIs without per-image segmentation, potentially underestimating true alignment; future integration of U-Net for precise masks is warranted. Finally, clinical claims are moderated—the model’s high metrics suggest diagnostic potential, but without prospective trials or integration with clinical workflows, it remains a research tool rather than a deployable system. These limitations highlight the need for multi-center validation, advanced imbalance handling, external testing, and refined explainability to impact real-world deployment.
This study introduced a customized EfficientNet-B0 model for ternary classification of chest X-ray (CXR) images into COVID-19, Viral Pneumonia, and Normal categories, leveraging transfer learning, a tailored classification head with dropout regularization, and explainable AI via Grad-CAM. Evaluated on the COVID-19 Radiography Database through 5-fold cross-validation, the model achieved a mean accuracy of 98.41% ± 0.45%, F1-score of 98.42% ± 0.44%, AUROC of 99.89% ± 0.05%, precision of 98.44% ± 0.43%, and recall of 98.41% ± 0.45%, outperforming VGG16 (84.83% ± 2.24% accuracy) and ResNet50 (93.83% ± 0.59% accuracy) baselines. With only 4.0 million parameters—6× fewer than ResNet50—the model demonstrated superior efficiency (inference time ~7.45 ms, 134 FPS) and interpretability, with Grad-CAM heatmaps aligning well to lung regions (average IoU 0.489 vs. 0.200 for VGG16 and 0.467 for ResNet50). An ablation study confirmed the customization’s value, improving accuracy by 2.64% over the unmodified EfficientNet-B0. These results highlight the model’s effectiveness in multi-class differentiation on a balanced dataset, addressing key challenges in computational overhead and “black-box” predictions.
The high performance and explainability of the customized EfficientNet-B0 suggest its potential as a supportive tool for radiologists in resource-limited settings, where rapid triage of respiratory conditions like COVID-19 and viral pneumonia is critical. By focusing heatmaps on clinically relevant features (e.g., opacities and consolidations), the model could enhance diagnostic confidence, particularly in high-volume scenarios with radiologist fatigue. Its lightweight design facilitates integration into IoT-based remote diagnostics or enterprise healthcare systems, potentially reducing delays and errors in manual interpretation. However, these implications are moderated: while promising for preliminary screening, the model should complement not replace expert review, pending external validation to confirm robustness across diverse clinical populations.
6.3 Future Research Directions
To address the limitations identified such as single-dataset reliance, downsampling-induced diversity loss, absence of external testing, and heuristic-based IoU approximations—future work should prioritize multi-center validation on diverse datasets (e.g., including pediatric and ethnically varied CXRs) to mitigate biases and confirm generalizability, potentially incorporating federated learning for privacy-preserving training. Alternative imbalance-handling strategies, like class-weighted loss or oversampling, could be explored to retain more original samples without artifacts, directly tackling the diversity reduction from downsampling. For real-world or edge-based deployment, anticipated challenges include latency in low-bandwidth environments, model drift from varying imaging protocols, and integration with electronic health records; these might be addressed through model quantization (e.g., to 8-bit) for faster inference on mobile devices, continual learning frameworks to adapt to new data, and API-based interoperability testing in simulated clinical workflows. Additionally, enhancing explainability could involve integrating attention mechanisms (e.g., self-attention layers in the head) to refine feature focus, potentially improving IoU by 2–3% as seen in similar tasks, and using advanced segmentation models like U-Net for ground-truth lung ROIs instead of approximations. Prospective clinical trials would further evaluate usability, with ethical considerations for bias mitigation and user feedback loops.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: conceptualization: Chunyong Yin, Williams Kyei; methodology: Williams Kyei, Kelvin Amos Nicodemas; software: Williams Kyei, Kelvin Amos Nicodemas, Khagendra Darlami; validation: Williams Kyei; formal analysis: Williams Kyei, Khagendra Darlami; investigation: Williams Kyei; resources: Chunyong Yin; data curation: Williams Kyei; writing—original draft preparation: Williams Kyei; writing review and editing: Williams Kyei, Chunyong Yin; visualization: Williams Kyei, Khagendra Darlami; supervision: Chunyong Yin; project administration: Chunyong Yin. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The COVID-19 Radiography Database is publicly available at https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database (accessed on 01 January 2025). Code and results are available at https://github.com/willisit12/enhanced-covid19-diagnosis-efficientnetB0 (accessed on 01 January 2025).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Abbreviations
| CXR | Chest X-Ray |
| DL | Deep Learning |
| CNN | Convolutional Neural Network |
| Grad-CAM | Gradient-Weighted Class Activation Mapping |
| FPS | Frames Per Second |
| ROI | Region of Interest |
References
1. World Health Organization. WHO COVID-19 dashboard 2025 [Internet]. [cited 2025 Oct 9]. Available from: https://data.who.int/dashboards/covid19/cases. [Google Scholar]
2. Jacobi A, Chung M, Bernheim A, Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19a pictorial review. Clin Imaging. 2020;64(2):35–42. doi:10.1016/j.clinimag.2020.04.001. [Google Scholar] [PubMed] [CrossRef]
3. Cozzi D, Albanesi M, Cavigli E, Moroni C, Bindi A, Luvarà S, et al. Chest X-ray in new coronavirus disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. La Radiol Med. 2020;125(8):730–7. doi:10.1007/s11547-020-01232-9. [Google Scholar] [PubMed] [CrossRef]
4. Wong HYF, Lam HYS, Fong AH, Leung ST, Chin TW, Lo CSY, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiol. 2020;296(2):E72–8. doi:10.1148/radiol.2020201160. [Google Scholar] [PubMed] [CrossRef]
5. Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, et al. The role of chest imaging in patient management during the COVID-19 pandemic. Chest. 2020;158(1):106–16. doi:10.1016/j.chest.2020.04.003. [Google Scholar] [PubMed] [CrossRef]
6. Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15(11):e1002686. doi:10.1371/journal.pmed.1002686. [Google Scholar] [PubMed] [CrossRef]
7. Dwivedi K, Sharkey M, Alabed S, Langlotz CP, Swift AJ, Bluethgen C. External validation, radiological evaluation, and development of deep learning automatic lung segmentation in contrast-enhanced chest CT. Eur Radiol. 2024;34(4):2727–37. doi:10.1007/s00330-023-10235-9. [Google Scholar] [PubMed] [CrossRef]
8. Al Zahrani N, Hedjar R, Mekhtiche M, Bencherif M, Al Fakih T, Al-Qershi F, et al. A multi-task deep learning framework for simultaneous detection of thoracic pathology through image classification. J Comput Commun. 2024;12(4):153–70. doi:10.4236/jcc.2024.124012. [Google Scholar] [CrossRef]
9. Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–40. doi:10.1007/s13246-020-00865-4. [Google Scholar] [PubMed] [CrossRef]
10. Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121(7798):103792. doi:10.1016/j.compbiomed.2020.103792. [Google Scholar] [PubMed] [CrossRef]
11. Chaddad A, Hassan L, Desrosiers C. Deep CNN models for predicting COVID-19 in CT and X-ray images. J Med Imaging. 2021;8(Suppl 1):014502. doi:10.1117/1.JMI.8.S1.014502. [Google Scholar] [PubMed] [CrossRef]
12. Phumkuea T, Wongsirichot T, Damkliang K, Navasakulpong A. Classifying COVID-19 patients from chest X-ray images using hybrid machine learning techniques: development and evaluation. JMIR Form Res. 2023;7(1):e42324. doi:10.2196/42324. [Google Scholar] [PubMed] [CrossRef]
13. Haynes SC, Johnston P, Elyan E. Generalisation challenges in deep learning models for medical imagery: insights from external validation of COVID-19 classifiers. Multimed Tools Appl. 2024;83(31):76753–72. doi:10.1007/s11042-024-18543-y. [Google Scholar] [CrossRef]
14. Bani Baker Q, Hammad M, Al-Smadi M, Al-Jarrah H, Al-Hamouri R, Al-Zboon SA. Enhanced COVID-19 detection from X-ray images with convolutional neural network and transfer learning. J Imaging. 2024;10(10):250. doi:10.3390/jimaging10100250. [Google Scholar] [PubMed] [CrossRef]
15. Xiong Y, Li Z, Chen Y, Wang F, Zhu X, Luo J, et al. Efficient deformable ConvNets: rethinking dynamic and sparse operator for vision applications. In: Proceedings of the 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2024 Jun 16–22; Seattle, WA, USA. p. 5652–61. doi:10.1109/cvpr52733.2024.00540. [Google Scholar] [CrossRef]
16. Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. Proc Mach Learn Res. 2019;97:6105–14. [Google Scholar]
17. Wang J, Chen C, Li S, Wang C, Cao X, Yang L. Researching the CNN collaborative inference mechanism for heterogeneous edge devices. Sensors. 2024;24(13):4176. doi:10.3390/s24134176. [Google Scholar] [PubMed] [CrossRef]
18. Almutaani M, Turki T, Taguchi YH. Novel large empirical study of deep transfer learning for COVID-19 classification based on CT and X-ray images. Sci Rep. 2024;14(1):26520. doi:10.1038/s41598-024-76498-4. [Google Scholar] [PubMed] [CrossRef]
19. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206–15. doi:10.1038/s42256-019-0048-x. [Google Scholar] [PubMed] [CrossRef]
20. Zhou SK, Greenspan H, Davatzikos C, Duncan JS, Van Ginneken B, Madabhushi A, et al. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc IEEE. 2021;109(5):820–38. doi:10.1109/jproc.2021.3054390. [Google Scholar] [PubMed] [CrossRef]
21. Xia Q, Zheng H, Zou H, Luo D, Tang H, Li L, et al. A comprehensive review of deep learning for medical image segmentation. Neurocomputing. 2025;613(1):128740. doi:10.1016/j.neucom.2024.128740. [Google Scholar] [CrossRef]
22. Oltu B, Güney S, Yuksel SE, Dengiz B. Automated classification of chest X-rays: a deep learning approach with attention mechanisms. BMC Med Imaging. 2025;25(1):71. doi:10.1186/s12880-025-01604-5. [Google Scholar] [PubMed] [CrossRef]
23. Subramaniam K, Palanisamy N, Sinnaswamy RA, Muthusamy S, Mishra OP, Loganathan AK, et al. A comprehensive review of analyzing the chest X-ray images to detect COVID-19 infections using deep learning techniques. Soft Comput. 2023;27(19):14219–40. doi:10.1007/s00500-023-08561-7. [Google Scholar] [PubMed] [CrossRef]
24. Nahiduzzaman M, Goni MOF, Hassan R, Islam MR, Syfullah MK, Shahriar SM, et al. Parallel CNN-ELM: a multiclass classification of chest X-ray images to identify seventeen lung diseases including COVID-19. Expert Syst Appl. 2023;229(11):120528. doi:10.1016/j.eswa.2023.120528. [Google Scholar] [PubMed] [CrossRef]
25. Chatterjee S, Saad F, Sarasaen C, Ghosh S, Krug V, Khatun R, et al. Exploration of interpretability techniques for deep COVID-19 classification using chest X-ray images. J Imaging. 2024;10(2):45. doi:10.3390/jimaging10020045. [Google Scholar] [PubMed] [CrossRef]
26. Luz E, Silva P, Silva R, Silva L, Guimarães J, Miozzo G, et al. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res Biomed Eng. 2022;38(1):149–62. doi:10.1007/s42600-021-00151-6. [Google Scholar] [CrossRef]
27. Chawki Y, Elasnaoui K, Ouhda M. Classification and detection of Covid-19 based on X-ray and CT images using deep learning and machine learning techniques: a bibliometric analysis. AIMS Electron Electr Eng. 2024;8(1):71–103. doi:10.3934/electreng.2024004. [Google Scholar] [CrossRef]
28. Abd El-Aziz AA, Elmogy M, El-Ghany SA. A robust tuned EfficientNet-B2 using dynamic learning for predicting different grades of brain cancer. Egypt Inform J. 2025;30(6):100694. doi:10.1016/j.eij.2025.100694. [Google Scholar] [CrossRef]
29. Latha M, Kumar PS, Chandrika RR, Mahesh TR, Kumar VV, Guluwadi S. Revolutionizing breast ultrasound diagnostics with EfficientNet-B7 and explainable AI. BMC Med Imaging. 2024;24(1):230. doi:10.1186/s12880-024-01404-3. [Google Scholar] [PubMed] [CrossRef]
30. Lilhore UK, Sharma YK, Shukla BK, Vadlamudi MN, Simaiya S, Alroobaea R, et al. Hybrid convolutional neural network and bi-LSTM model with EfficientNet-B0 for high-accuracy breast cancer detection and classification. Sci Rep. 2025;15(1):12082. doi:10.1038/s41598-025-95311-4. [Google Scholar] [PubMed] [CrossRef]
31. Bouslihim I, Cherif W, Kissi M. Application of a hybrid EfficientNet-SVM model to medical image classification. In: Proceedings of the 2023 14th International Conference on Intelligent Systems: Theories and Applications (SITA); 2023 Nov 22–23; Casablanca, Morocco. Piscataway, NJ, USA: IEEE; 2024. p. 1–6. doi:10.1109/SITA60746.2023.10373755. [Google Scholar] [CrossRef]
32. Ishaq A, Ullah FUM, Hamandawana P, Cho DJ, Chung TS. Improved EfficientNet architecture for multi-grade brain tumor detection. Electron. 2025;14(4):710. doi:10.3390/electronics14040710. [Google Scholar] [CrossRef]
33. Srinivas K, Gagana RS, Pravallika K, Nishitha K, Polamuri SR. COVID-19 prediction based on hybrid Inception V3 with VGG16 using chest X-ray images. Multimed Tools Appl. 2024;83(12):1–18. doi:10.1007/s11042-023-15903-y. [Google Scholar] [PubMed] [CrossRef]
34. Hasan MR, Azmat Ullah SM, Rabiul Islam SM. Recent advancement of deep learning techniques for pneumonia prediction from chest X-ray image. Med Rep. 2024;7(5):100106. doi:10.1016/j.hmedic.2024.100106. [Google Scholar] [CrossRef]
35. Tera SP, Chinthaginjala R, Shahzadi I, Natha P, Rab SO. Deep learning approach for automated hMPV classification. Sci Rep. 2025;15(1):29068. doi:10.1038/s41598-025-14467-1. [Google Scholar] [PubMed] [CrossRef]
36. Ukwuoma CC, Cai D, Heyat MBB, Bamisile O, Adun H, Al-Huda Z, et al. Deep learning framework for rapid and accurate respiratory COVID-19 prediction using chest X-ray images. J King Saud Univ Comput Inf Sci. 2023;35(7):101596. doi:10.1016/j.jksuci.2023.101596. [Google Scholar] [PubMed] [CrossRef]
37. Vyas R, Khadatkar DR. Ensemble of deep learning architectures with machine learning for pneumonia classification using chest X-rays. J Imag Inform Med. 2025;38(2):727–46. doi:10.1007/s10278-024-01201-y. [Google Scholar] [PubMed] [CrossRef]
38. Akbar W, Soomro A, Hussain A, Hussain T, Ali F, Haq MIU, et al. Pneumonia detection: a comprehensive study of diverse neural network architectures using chest X-rays. Int J Appl Math Comput Sci. 2024;34(4):679–99. doi:10.61822/amcs-2024-0045. [Google Scholar] [CrossRef]
39. Pal M, Mohapatra RK, Sarangi AK, Sahu AR, Mishra S, Patel A, et al. A comparative analysis of the binary and multiclass classified chest X-ray images of pneumonia and COVID-19 with ML and DL models. Open Med. 2025;20(1):20241110. doi:10.1515/med-2024-1110. [Google Scholar] [PubMed] [CrossRef]
40. Hira S, Bai A, Hira S. An automatic approach based on CNN architecture to detect COVID-19 disease from chest X-ray images. Appl Intell. 2021;51(5):2864–89. doi:10.1007/s10489-020-02010-w. [Google Scholar] [PubMed] [CrossRef]
41. Singh T, Mishra S, Kalra R, Satakshi, Kumar M, Kim T. COVID-19 severity detection using chest X-ray segmentation and deep learning. Sci Rep. 2024;14(1):19846. doi:10.1038/s41598-024-70801-z. [Google Scholar] [PubMed] [CrossRef]
42. Ishwarya K, Varun CS, Saraswathi S, Devi D. COVID 19: data augmentation and image preprocessing technique. In: Proceedings of the 2023 6th International Conference on Recent Trends in Advance Computing (ICRTAC); 2023 Dec 14–15; Chennai, India. Piscataway, NJ, USA: IEEE; 2023. p. 453–63. doi:10.1109/icrtac59277.2023.10480797. [Google Scholar] [CrossRef]
43. Kembara B, Herumurti D, Saikhu A. Enhancing COVID-19 detection in X-ray images through image preprocessing and transfer learning. In: Proceedings of the 2024 IEEE International Conference on Industry 4.0, Artificial Intelligence, and Communications Technology (IAICT); 2024 Jul 4–6; Kuta, Indonesia. Piscataway, NJ, USA: IEEE; 2024. p. 93–9. doi:10.1109/iaict62357.2024.10617580. [Google Scholar] [CrossRef]
44. Zhang CJ, Ruan LT, Ji LF, Feng LL, Tang FQ. COVID-19 recognition from chest X-ray images by combining deep learning with transfer learning. Digit Health. 2025;11:1–21. doi:10.1177/20552076251319667. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools