 Open Access
Open Access
ARTICLE
Identification of Circular RNA hsa-circ-0006969 as a Novel Biomarker for Breast Cancer
1 Department of Beijing National Biochip Research Center Sub-Center in Ningxia, General Hospital of Ningxia Medical University, Yinchuan, 750004, China
2 College of Clinical Medicine, Ningxia Medical University, Yinchuan, 750004, China
3 Institute of Medical Sciences, General Hospital of Ningxia Medical University, Yinchuan, 750004, China
4 Department of Oncological Surgery, General Hospital of Ningxia Medical University, Yinchuan, 750004, China
* Corresponding Authors: Jinping Li. Email: ; Xu Zhang. Email:
# These authors contribute equally to this work
(This article belongs to the Special Issue: Biomarkers for Breast Cancer Diagnosis and Treatment Selection: from Basic Research to Practice)
Oncologie 2022, 24(4), 789-801. https://doi.org/10.32604/oncologie.2022.026589
Received 14 September 2022; Accepted 08 December 2022; Issue published 31 December 2022
Abstract
Background: To investigate the characteristics of circular RNA hsa-circ-0006969 in breast cancer and identify it as a novel biomarker for breast cancer. Methods: Three breast cancer (BC) patient tissues were selected to perform human circRNA microarray analysis. GeneSpring 13.0 (Agilent) software was applied for analyzing the data. Another 116 BC patients were recruited for verification. Hsa-circ-0006969 was found as a potential circRNA for BC diagnostic biomarker. The structure of hsa-circ-0006969 was predicted by circPrimer1.2 software. MiRanda v3.3, RNA hybrid 2.1, and Cytoscape 3.6.0 were used for predicting the networks of circRNA-miRNA. T-test, Curve regression, and ROC analysis were applied to certify the diagnostic values of hsa-circ-0006969. Using SPSS25.0 software to perform the Statistical analysis. Results: 546 higher expression and 1475 lower expression circRNAs were identified. Four circRNAs were filtrated for further verification. Hsa-circ-0006969 was significantly low expressed in BC tissues and peripheral blood. Hsa-circ-0006969 was confirmed as higher diagnostic correlation with BC tissues (AUC = 0.965) and peripheral blood (AUC = 0.842), with Grade 1 (AUC = 0.639), ER-positive (AUC = 0.612), TNM I (AUC = 0.693), TNM II (AUC = 0.712), TNM III (AUC = 0.757), TNM early stage(I, II) (AUC = 0.709). Hsa-circ-0006969 presented more effective diagnostic values for tumor metastasis (AUC = 0.784) compared with CA153 (AUC = 0.752). Conclusion: Hsa-circ-0006969 could be a novel biomarker for the diagnosis and treatment of BC.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| TNM | Tumor Node Metastasis |
| HER-2 | Human epidermal growth factor receptor-2 |
| CEA | Carcino-embryonic antigen |
| CA153 | Carbohydrate antigen 15-3 |
| AUC | Area under the curve |
| CI | Confidence interval |
| NA | NOT was applicable |
Breast cancer (BC) is the most common cancer which is a leading cause of mortality in women [1]. In 2020, about 2.26 million new cases of female BC were diagnosed and accounted for 11.7% of new cases of cancer [2,3]. Although treatment of surgery, radiotherapy, and chemotherapy was carried out, the burden of breast cancer is still increasing [4]. Therefore, discovering new and effective biomarkers and developing target therapeutic strategies for the early diagnosis and treatment of BC became urgent.
Circular RNA(circRNA) is one type of covalently closed noncoding RNA which have a closed loop structure [5]. A number of miRNA binding sites were found on the circRNAs sequences which often regulate gene expression in eukaryotes. Recent research indicates that many circRNAs are cell-type-specific expressions and are associated with physiological development and various diseases [6]. Recent reports have shown that circRNA is involved in cancer cells’ metastasis in a variety of ways. For example, hsa_circ_001783 promoted the progression of BC cells through sponging miR-200c-3p [7]. Knockdown of hsa_circ_0110389 could restrain GC growth in vivo via miR-127-5p- miR-136-5p-SORT1 pathway [8]. Unfortunately, the functions of the vast majority of circRNAs are still not clear.
In this study, the circRNAs was profiled in breast cancer and para-carcinoma tissue using microarray analysis. Then verified the differential expressions of circRNAs in other cohorts of BC tissues and peripheral blood specimens. On account of the association studies of the target circRNA and the BC patient’s clinical characteristics, we confirmed hsa-circ-0006969 as a potential biomarker for breast cancer. Further analysis is being performed to evaluate the diagnostic value of hsa-circ-0006969 in breast cancer.
2.1 Participants and Specimens
Both cancer tissues and para-carcinoma samples were obtained from the oncology surgery department of the General Hospital of Ningxia Medical University. The study was approved by the General Hospital of Ningxia Medical University Ethics Committee (No. 2018-118). Every patient has signed the informed consent to authorize the use of their samples.19 pair of cancer tissues and para-carcinoma samples and 100 BC peripheral blood was collected from March 2020 to August 2021. The age of patients ranged between 25–75 years old (average 48.26 ± 8.32 years). Every patient has not received radiotherapy and chemotherapy treatment before sample collection.
Peripheral blood samples were obtained from the vein with an EDTA plain tube (2 mL) after overnight fasting. Cancer tissues and para-carcinoma samples were obtained from patients who underwent surgical breast resection. The para-carcinoma samples were located >5 cm from the tumor part. Healthy persons were recruited from the physical examination department in the General Hospital of Ningxia Medical University as control.
2.2 RNA Isolation and RNase R Treatment
Tissue RNA was extracted from cancer and para-carcinoma tissues with trizol reagent (Invitrogen, Carlsbad, USA). Peripheral blood RNA was extracted using a total RNA rapid extraction kit (Bioteke, Beijing, China). The RNA was measured using nanodrop 2000 (Thermo Scientific, Waltham, USA). RNA integrity test was performed by 1.3% agarose gel electrophoresis (120 V, 15 min, 1 × buffer). Total RNA was incubated with 10 U of RNase R (Geneseed, Guangzhou, China) at 37°C for 30 min. Finally, the expression of β-actin, ARHGEF28, and hsa-circ-0006969 was detected by RT-qPCR.
Three pair tumor and para-carcinoma tissues in BC patients were obtained for circRNA expression analysis using Human CircRNA Array v2 microarray (Beijing Capital Bio Biotechnology Corporation, Beijing, China). The circRNA microarray data were analyzed by GeneSpring 13.0 software (Agilent). To improve the screening efficiency, candidate circRNAs were screened with the filter criteria Fold change(FC) ≥4, p-value < 0.05, and original fluorescence value ≥100.
Total RNA was synthesized to cDNA with the revert aid first-strand cDNA synthesis kit (Thermo Scientific, Waltham, USA). Primers were synthesized by Sangon Biotech (Shanghai) Co, Ltd., primers are listed in Table S1. Light Cycler480 II quantitative system (Roche, Rotkreuz, Switzerland) was used for RT-qPCR. The reaction volume includes 2 µl cDNA, 0.8 µl sense primer, 0.8 µl reverse primer, 10 µl TB-Green Premix Ex Taq (TAKARA, Kyoto, Japanese), replenish ddH2O to 20 µl. The program is 94˚C for 10 s, 40 cycles of 94˚C for 15 s, 55˚C for 15 s, and an annealing temperature for 30 s. β-actin was used as an internal control. Each reaction was repeated 3 times and the results of relative expression were calculated using 2−ΔΔCq method [9].
CircRNA microarray data was obtained using Feature Extraction software (CapitalBio, Beijing, China). Normalization, fold change and P value were performed by GeneSpring software V13.0 (Agilent). Heat map, ScatterPlot, and VolcanoPlot analysis were performed through the Omic Studio tools (https://www.omicstudio.cn/tool). CircRNA structures were predicted by circPrimer2.0 software [10]. Miranda v3.3a (http://miranda.org.uk/) and Targetscan (http://www.targetscan.org) were utilized to predict the target miRNAs and binding sites of hsa-circ-0006969 [11]. The circRNA-miRNA interaction networks were performed using Cytoscape 3.6.0 (https://cytoscape.org/). The biological processes, cellular components, and molecular functions of circRNAs were annotated through GO analysis and the genecards database(https://www.genecards.org/). KEGG analysis was performed to determine the biological pathways’ target genes. Workflow and analysis tools were shown in Figure S1.
SPSS25.0 software (IBM, Almunk, USA) and GraphPad Prism version 9.0 (GraphPad Software, San Diego, USA) were applied for statistical analysis. The continuous data were presented as mean ± SD and tested using a t-test. Receiver operating characteristic (ROC) analysis was used to evaluate the diagnostic value of circRNA. Curve regression analysis was used to assess the correlation of circRNA and the BC pathological characteristics. Differences were considered significant if p < 0.05 (*p < 0.05; **p < 0.01).
3.1 Profiling of CircRNAs Expressions of BC Patients
With circRNA microarray, the differentially expressed circRNAs in 3 paired BC tissue specimens (BC1–BC3) and para-carcinoma tissue specimens (PC1–PC3) were analyzed. A total of 2021 differentially expressed circRNAs were identified in BC tissue compared with para-carcinoma tissue. With the screening criteria as FC ≥2 and p-value < 0.05, 546 circRNAs were up-regulated and 1475 circRNAs were down-regulated (Figs. 1A–1C). With the screening criteria set as FC ≥4, p-value < 0.05, and original fluorescence value ≥500, finally 21 higher expressed and 14 lower expressed circRNAs were obtained (Fig. 1D).
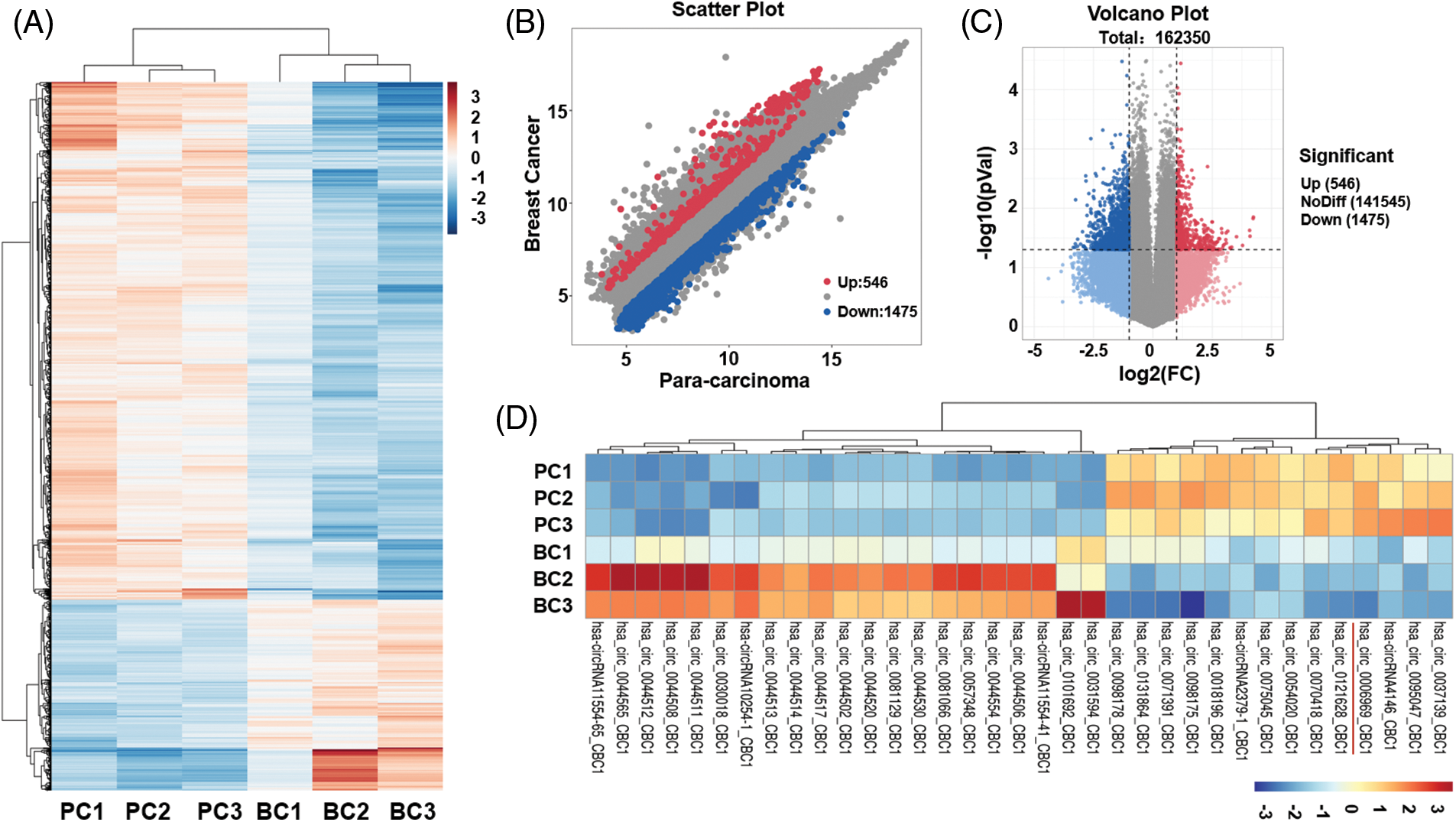
Figure 1: Expressions of circRNAs of BC tissues
(A) circRNA microarray assay analysis of the specific expressions of circRNAs in 3 paired breast cancer (BC) and para-carcinoma tissues (PC). Upregulated circRNA was indicated by “red”, downregulated circRNA was indicated by “blue”, and no significant difference was shown by “white”. (B) Scatter plot revealed the results of microarray assay; (C) Volcano plot showed the results of microarray assay; (D) Cluster diagram of differentially expressed circRNAs with the filter criteria as FC ≥4, p-value < 0.01, and original fluorescence value ≥500.
3.2 Verification of the Candidate CircRNAs
Based on the gene function annotation and prediction, four circRNA candidates were selected for further identification. The 4 circRNAs include two higher expressed circRNAs: hsa-circ-0044513, hsa-circ-0101692, and two lower expressed circRNAs: hsa-circ-0006969, hsa-circ-0054020. RT-qPCR was then performed for validation of the circRNAs in an independent cohort of 16 cases of BC samples. The results confirmed that hsa-circ-0006969 has a good consistency with BC samples. ROC curve analysis results illustrated that hsa-circ-0006969 has a high diagnostic value for BC tissues (AUC = 0.965, p = 0.0001), indicating that hsa-circ-0006969 has a good association with BC (Fig. 2).
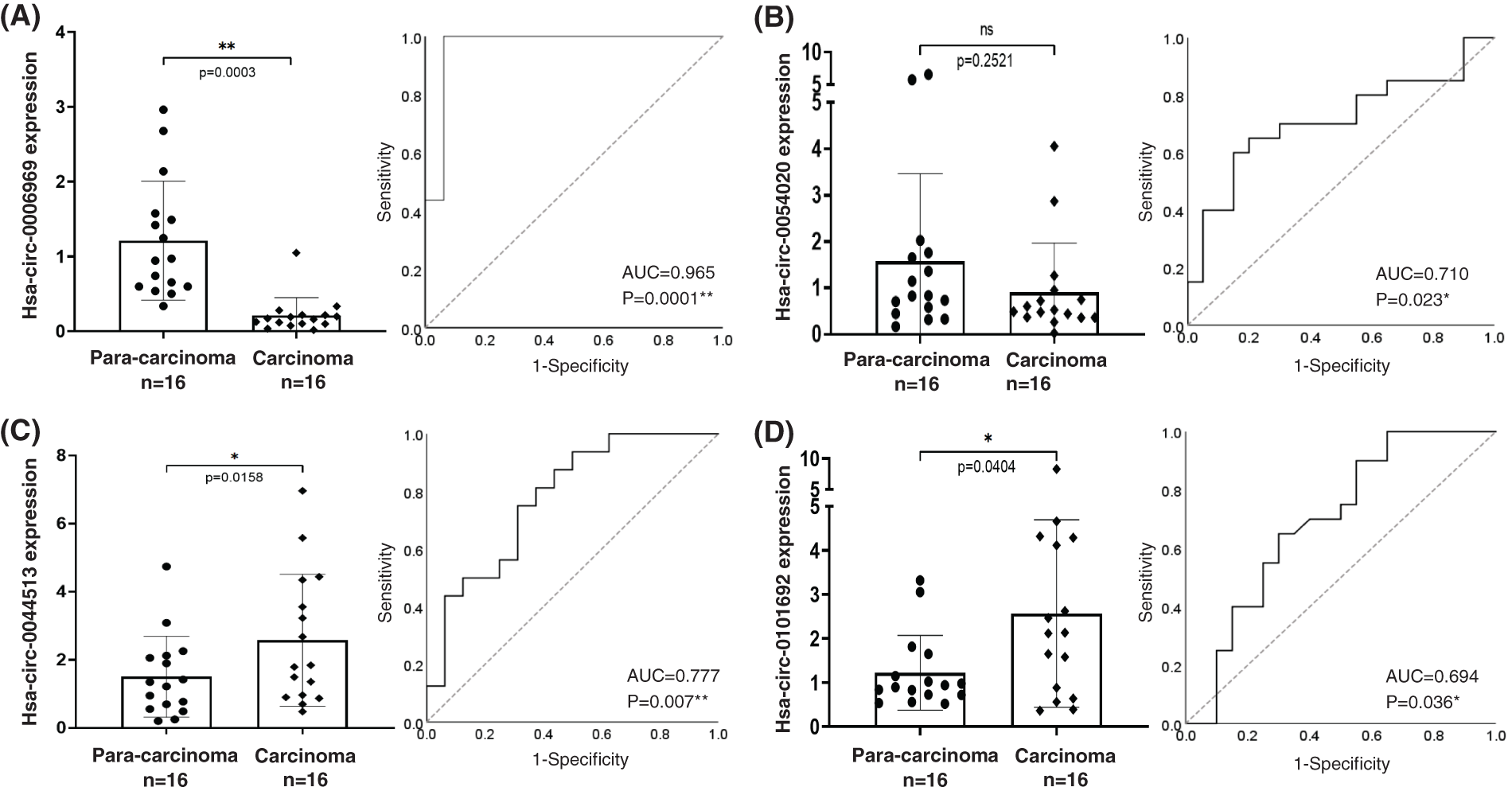
Figure 2: The diagnostic capability of 4 candidate circRNAs
After bio-information analysis, 4 circRNA candidates were selected for further identification, including 2 upregulated and 2 downregulated circRNAs. RT-qPCR validated the four circRNAs in an independent cohort including 16 cases of para-carcinoma and carcinoma tissues; (A) RT-qPCR and ROC of hsa-circ-0006969; (B) RT-qPCR and ROC of hsa-circ-0054520; (C) RT-qPCR and ROC of hsa-circ-0044513; (D) RT-qPCR and ROC of hsa-circ-0101692; ns, no significant, *p < 0.05, **p < 0.01.
3.3 The Diagnostic Values of hsa-circ-0006969 in BC
Specific primers were designed according to the reverse cleaved sites and linear sites of hsa-circ-0006969 (Fig. 3A). Combined with RNase R digestion, agarose gel electrophoresis, and RT-qPCR, hsa-circ-0006969 was proved to be a circRNA (Figs. 3B, 3C). Meanwhile, 100 BC patients and 50 healthy donator’s peripheral blood samples were collected. RT-qPCR results indicated that the expression of hsa-circ-0006969 was significantly decreased in the peripheral blood sample (Fig. 3D). The AUC value of hsa-circ-0006969 was 0.842 with 79.0% sensitivity and 80.4% specificity in BC patients’ peripheral blood (Fig. 3E). These results demonstrated that hsa-circ-0006969 has a high diagnostic value in BC.
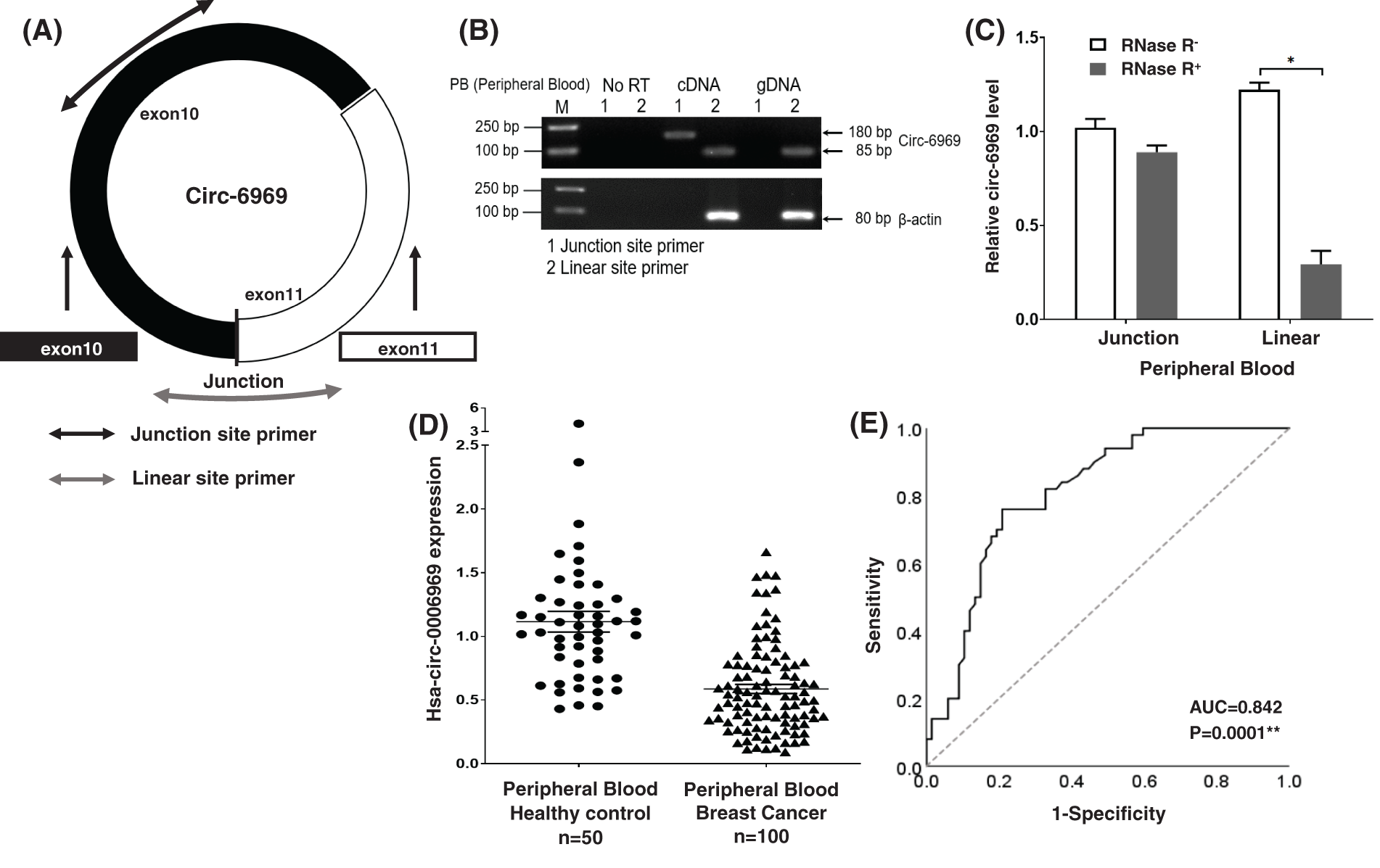
Figure 3: Validation and characteristics of hsa-circ-0006969 in peripheral blood
(A) Schematic illustration showing junction site and linear site primers in hsa-circ-0006969. The presence of hsa-circ-0006969 was validated by RT-qPCR using junction site and linear site primers. Junction was head-to-tail hsa-circ-0006969 splicing sites; (B) Verification of the hsa-circ-0006969 expression in peripheral blood by RT-qPCR in gDNA, RNA, and cDNA. Junction site, and linear site primers were used; (C) RT-qPCR analysis of hsa-circ-0006969 expression in peripheral blood after treated or not with RNase R digestion, *p < 0.05 (RNase R- vs. RNase R+); (D) Expression of hsa-circ-0006969 quantified by qPCR in peripheral blood (BC peripheral blood, n = 100; healthy person peripheral blood, n = 50); (E) The ROC analysis of hsa-circ-0006969; *p < 0.05, **p < 0.01.
3.4 Correlation Analysis of hsa-circ-0006969 in BC
The association between hsa-circ-0006969 and BC clinical characteristics was verified in 100 patients. The results show that the expression of hsa-circ-0006969 was significantly related to triple-negative breast cancer (p < 0.05), ER-positive (p < 0.05), tumor grading (p < 0.05), TNM stage (p < 0.01), and tumor metastasis (p < 0.01) (Table 1). Curve regression analysis revealed that hsa-circ-0006969 was also had a significant correlation with CA153 in peripheral blood (p < 0.05) (Fig. 4).
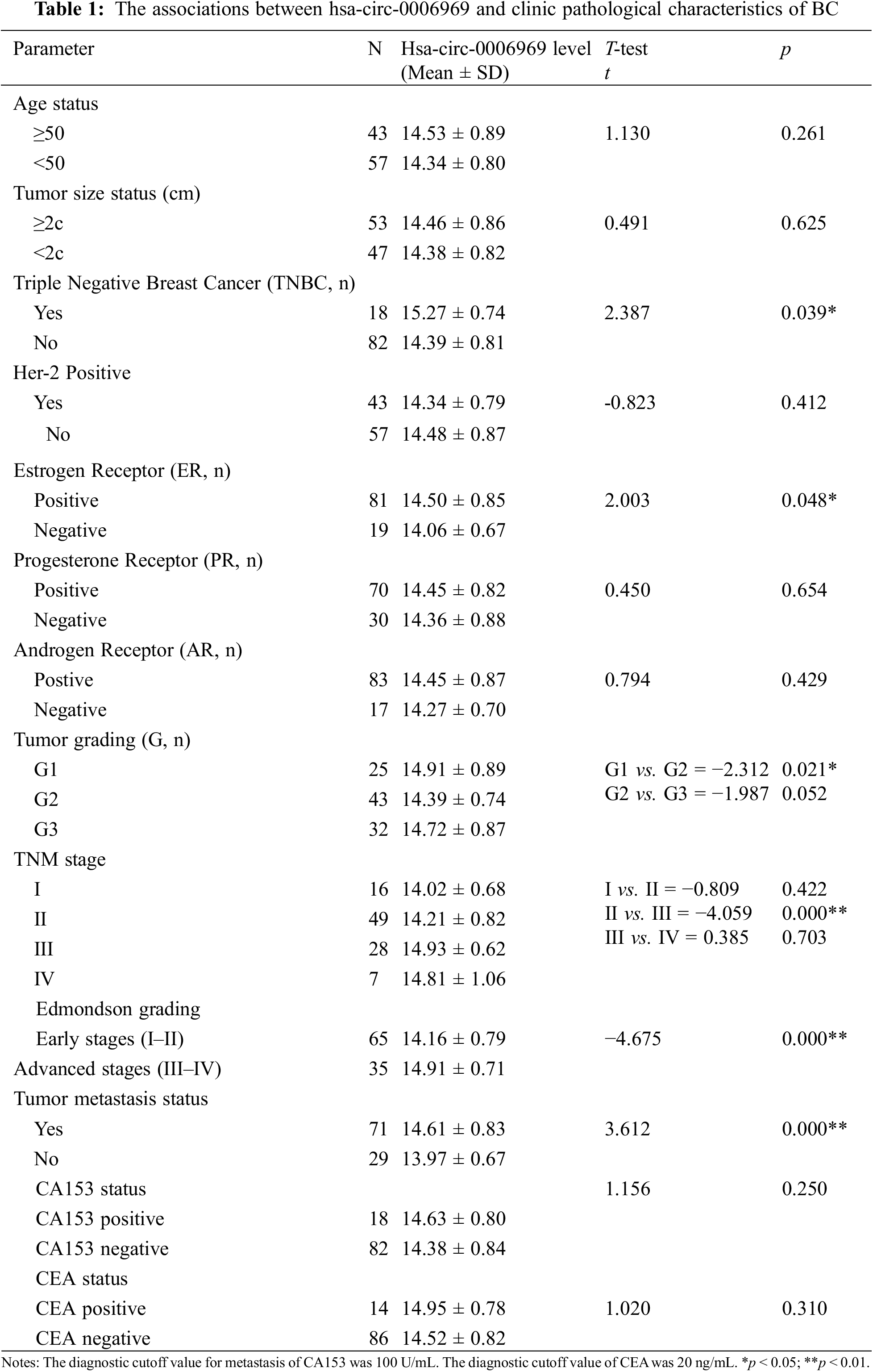
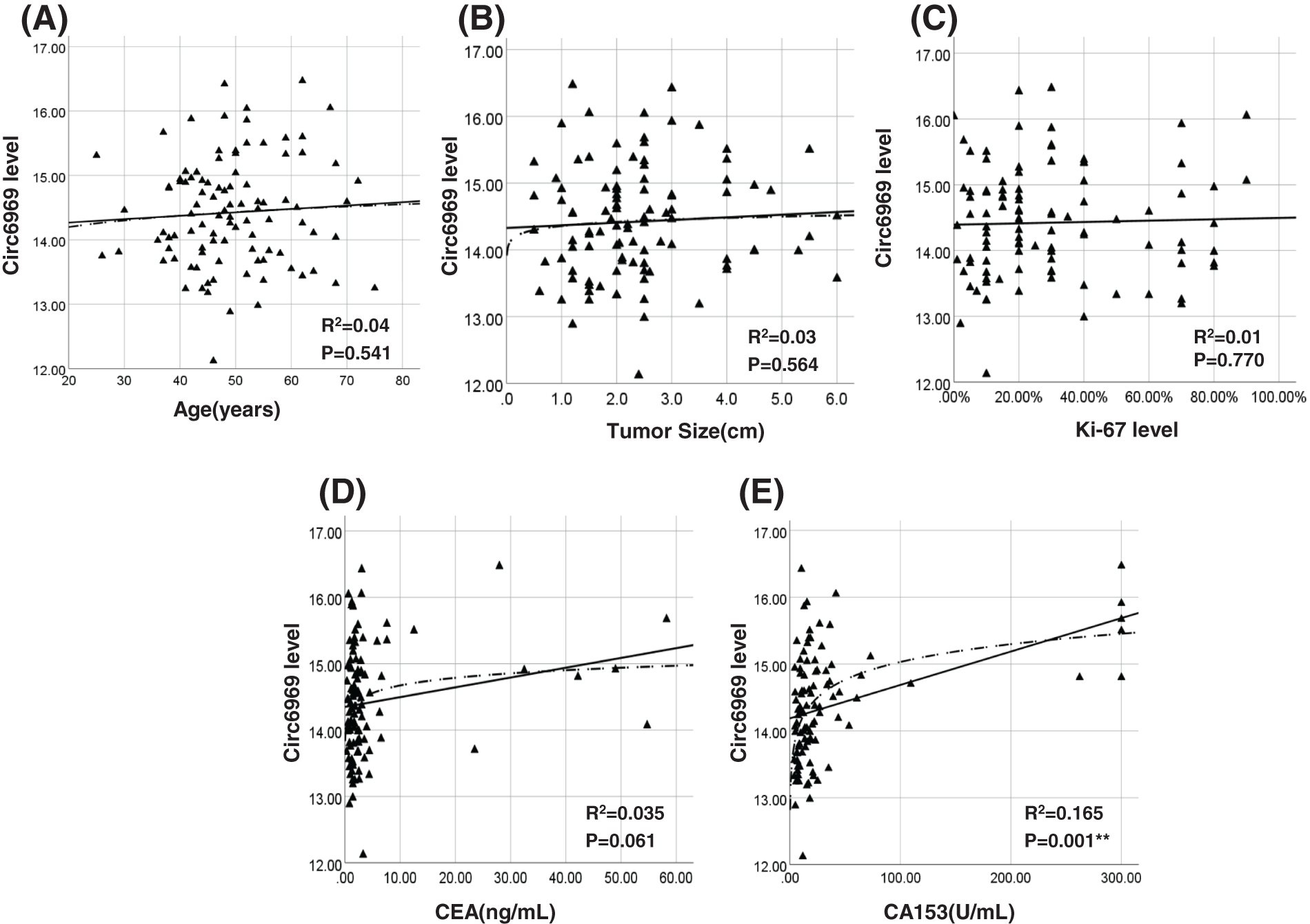
Figure 4: Association analysis of hsa-circ-0006969 in BC peripheral blood
(A–E) The association analysis of hsa-circ-0006969 with clinical pathological characteristics of 100 BC patients. (A) hsa-circ-0006969 vs. age, (B) hsa-circ-0006969 vs. tumor size, (C) hsa-circ-0006969 vs. Ki-67 level, (D) hsa-circ-0006969 vs. CEA level, (E) hsa-circ-0006969 vs. CA153 level; *p < 0.05, **p < 0.01.
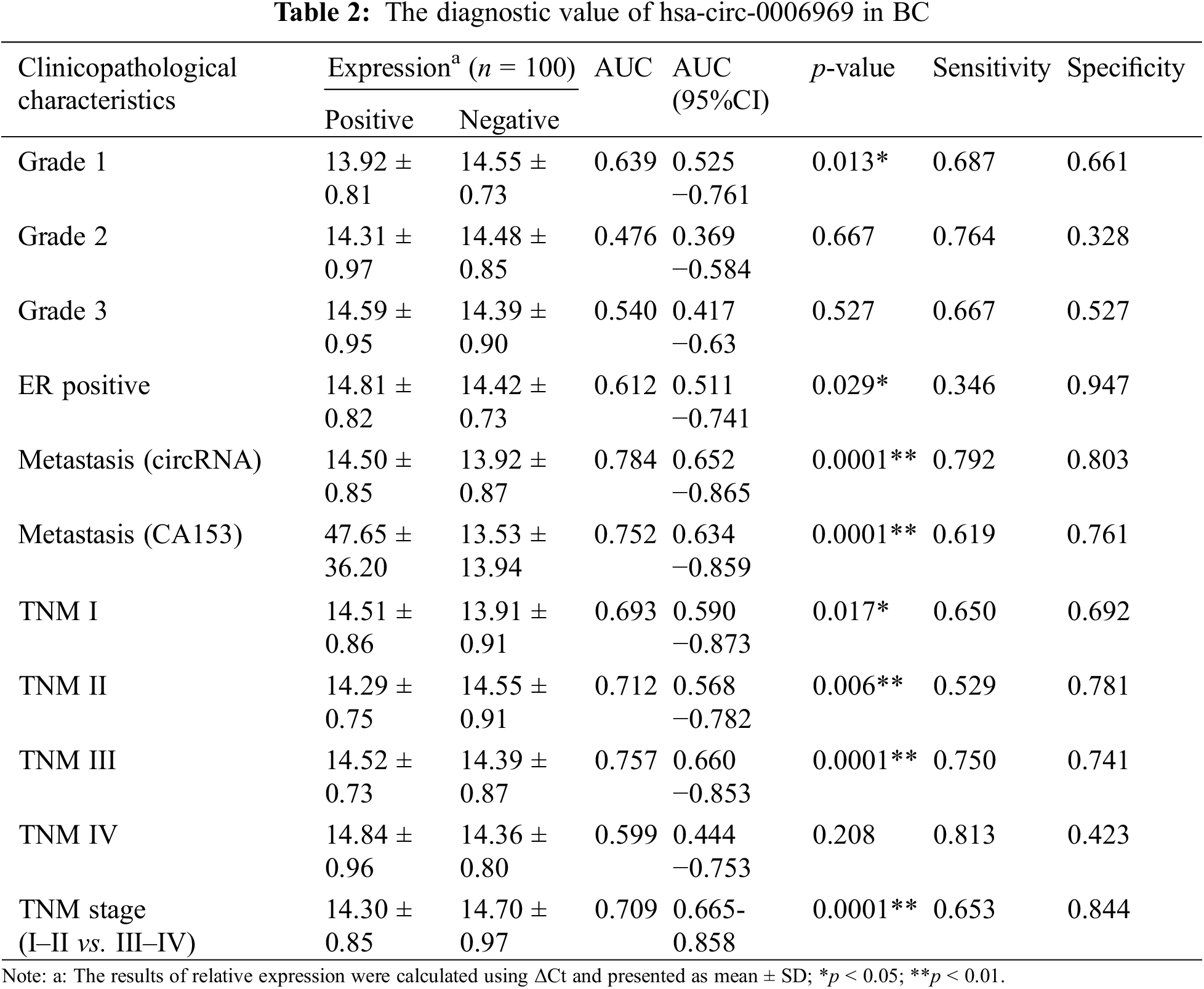
To determine the diagnostic values of hsa-circ-0006969 in BC patient peripheral blood, ROC curve analysis was applied. The results proved that the AUC of hsa-circ-0006969 for diagnosis of High differentiated tumor (Grade 1) was 0.639 (p = 0.013), for moderately differentiated tumor (Grade 2) was 0.476(p = 0.667) and for low differentiated tumor (Grade 3) was 0.540 (p = 0.527). The AUC of hsa-circ-0006969 for the diagnosis of ER-positive patients was 0.612 (p = 0.029). Compared with CA153, the AUC of hsa-circ-0006969 for tumor metastasis was 0.784 (p = 0.0001). Meanwhile, The AUC of hsa-circ-0006969 for diagnosis of TNM I, TNM II, TNM III, TNM IV and TNM early stage (I, II) were 0.693 (p = 0.017), 0.712 (p = 0.006), 0.757 (p = 0.0001), 0.599 (p = 0.208) and 0.709 (p = 0.0001). (Table 2). The results suggested that hsa-circ-0006969 has high diagnostic values for BC.
3.5 Bioinformatics Analysis of hsa-circ-0006969
The binding miRNA of hsa-circ-0006969 was predicted by miRanda and RNAhybrid software. The results proved that 56 microRNAs have potential binding sites with hsa-circ-0006969 (Fig. 5A). After overlapping the two databases, 6 miRNAs were identified with a significant association with hsa-circ-0006969 (Fig. 5B and Table S2). The biological processes of these miRNAs were mainly related to the cell cycle, cell proliferation, cell death, mRNA transport, and apoptotic (Figs. 5C, 5D). The results indicated that hsa-circ-0006969 may intervene in BC cell differentiation and proliferation through binding target miRNAs.
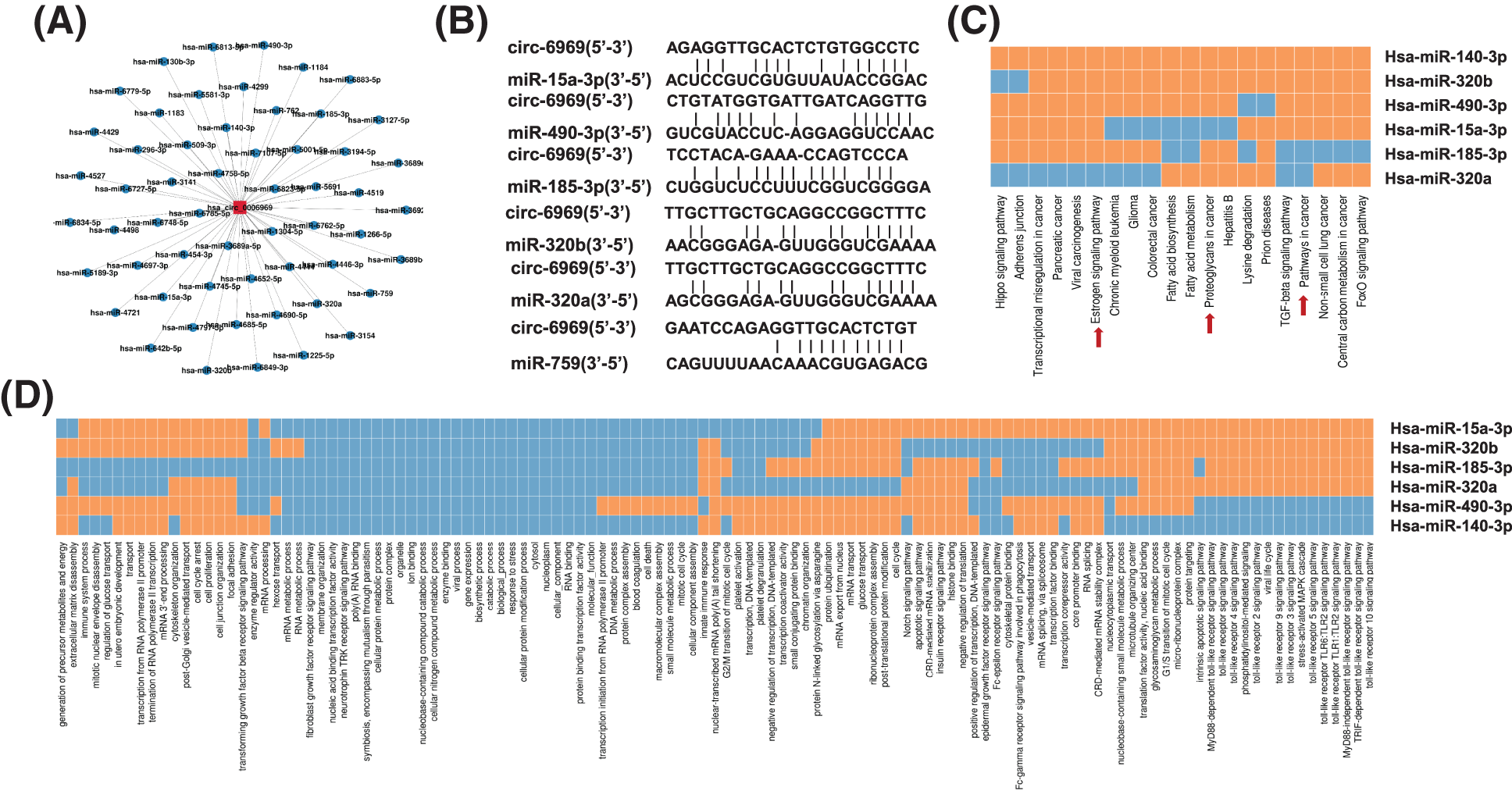
Figure 5: The biological structure and potential function of hsa-circ-0006969
(A) All candidate binding miRNAs of hsa-circ-0006969; (B) The top 6 candidate binding miRNAs and binding sites of hsa-circ-0006969 associated with cancer cell cycle, proliferation and apoptosis; (C–D) GO and KEGG database predicts the main signal biological process and pathway of top 6 candidate target miRNAs; miRNA target mRNA positive correlation was indicated by “orange”, miRNA target miRNA negative correlation was indicated by “blue”.
CircRNA is a non-coding RNA with a closed loop and the structure lack of 3’ and 5’ ends. The expression of circRNA has tissue-specific stability in most cancer tissue and blood [12]. CircRNA often regulates mRNA expression as a ceRNA or miRNA “sponge”. Previous research reported that circRNA is involved in many biological processes through multiple mechanisms [13]. CircRNA could influence cancer cell differentiation, proliferation, migration, protein cracking, and apoptosis based on the function of ceRNA or miRNA “sponge” [14–16]. These properties of circRNA allow it to become an ideal tumor biomarker for the diagnosis and therapy of BC cancer.
The higher morbidity and recurrence of BC are a serious threat to human health. Early diagnoses are helpful in the treatment of BC patients. Due to the unique molecular structure and expression specificity, circRNAs have the potential to be diagnostic markers or therapeutic targets for BC. Jahani et al found that circRNAs were expressed in BC cells in a cell-type and stage-specific manner [17]. However, the diagnostic value of circRNAs in BC requires further investigation. The clear and definite functions of circRNAs help us understand the occurrence mechanism of BC profoundly [18,19]. Precious research reported that circSEPT9-miR-637-LIF axis could facilitate the carcinogenesis and development of triple-negative breast cancer [20]. CircBCBM1 over-expression in primary cancerous tissues was associated with shorter brain metastasis-free survival (BMFS) of BC patients [21]. Screening the specificity circRNAs and considering their clinical correlation analysis will help to identify their role in the occurrence of cancer [22].
In this study, the circRNA expression profiling in Breast cancer was conducted by circRNA microarray. After the cluster analysis and GO analysis, 546 higher expressed and 1475 lower expressed circRNAs were identified in BC tumor tissues. With the filter criteria, 4 candidate circRNAs were screened for further study. RT-qPCR validation results suggest hsa-circ-0006969 has good consistency in BC tissue and peripheral blood. ROC curve analysis identified the diagnostic value of hsa-circ-0006969 in BC. The bioinformatics analysis revealed that hsa-circ-0006969 has 6 target miRNAs and the molecular biological function of these 6 miRNAs was associated with cell cycle and cell proliferation. The result indicated that hsa-circ-0006969 intervenes in BC cell differentiation and proliferation by binding the 6 target miRNAs as a “sponge”.
Furthermore, the structural and functional annotation analysis revealed that hsa-circ-0006969 was transcribed from the exon 10 and 11 regions of the human ARHGEF28 gene. ARHGEF28 was a RhoA-specific guanine nucleotide exchange factor that was involved in cell aggregation, apoptosis, and motility by influencing growth factor receptors [23]. A previous study reported that hsa-circ-0006969 was found in the cerebellum, Hela cell, and HepG2 cell [24]. This study positively demonstrated the relationship between hsa-circ-0006969 and BC first. After analyzing the association between hsa-circ-0006969 and 119 BC patients’ clinical characteristics, the diagnostic accuracy of hsa-circ-0006969 was identified in BC. Followed by ROC curve analysis further confirmed that hsa-circ-0006969 might be applied as a potential biomarker for BC diagnosis and treatment.
In conclusion, this study provided evidence that hsa-circ-0006969 is a lower expression both in BC tissues and peripheral blood samples. hsa-circ-0006969 may play a role as a promising potential biomarker for BC diagnosis and treatment. However, further study is needed to elucidate the underlying mechanisms.
Authorship: Wang LB planned the project, revised and polished the article, and adjusted all aspects of the work. Zhang X planned the project, executedall experiments, and wrote and revised the article. Li JP planned the project, revised the article, participated in circRNA Microarray analysis and screening candidate circRNAs. Li XH circRNA expression in specimens, and did ROC curve analysis. Tian JH did the circRNA Microarray data analysis and screening specific circRNAs. Yu JJ did the circRNA genechip analysis and statistical analysis. Huang Q prepared a table and supplementary file. Ma R collected tissue and peripheral blood samples and total RNA and cDNA extraction. Wang J prepared reagent and instrument, designed and screened primers. Cao J collected clinicopathological characteristics of patients with breast cancer.
Ethics Approval and Consent to Participate: The study protocol was reviewed and approved by the Ethics Committees of the General Hospital of Ningxia Medical University Ethics Committee (No: 2018-118). Informed consent was obtained from all individual participants included in the study.
Availability of Data and Materials: We guarantee the authenticity and validity of all data and results. Open up some of the raw data uploads as supplementary files.
Consent for Publication: I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and is not under consideration for publication elsewhere, in whole or in part. The manuscript is approved by all authors for publication.
Funding Statement: This study was supported by the National Natural Science Foundation of China (No. 81860470), the Science Research Project of Ningxia Higher Education (No. NGY2018-91), the Foreign Science and Technology Cooperation Projects of Ningxia Autonomous Region Key R&D Programs (No. 2019BFH02012), the First-Class Discipline Construction Project of Ningxia Medical University Clinical Medicine in 2022, and the Fifth Group of Ningxia Young Scientific and Technological Talents Lifting Project (NXKJTJGC2020080).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Trayes, K. P., Cokenakes, S. E. H. (2021). Breast cancer treatment. American Family Physician, 104(2), 171–178. [Google Scholar]
2. Akram, M., Iqbal, M., Daniyal, M., Khan, A. U. (2017). Awareness and current knowledge of breast cancer. Biological Research, 50(1), 33. DOI 10.1186/s40659-017-0140-9. [Google Scholar] [CrossRef]
3. Lambertini, M., Santoro, L., del Mastro, L., Nguyen, B., Livraghi, L. et al. (2016). Reproductive behaviors and risk of developing breast cancer according to tumor subtype: A systematic review and meta-analysis of epidemiological studies. Cancer Treatment Reviews, 49, 65–76. DOI 10.1016/j.ctrv.2016.07.006. [Google Scholar] [CrossRef]
4. Rossi, L., Mazzara, C., Pagani, O. (2019). Diagnosis and treatment of breast cancer in young women. Current Treatment Options in Oncology, 20(12), 86. DOI 10.1007/s11864-019-0685-7. [Google Scholar] [CrossRef]
5. Li, J., Sun, D., Pu, W., Wang, J., Peng, Y. (2020). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends in Cancer, 6(4), 319–336. DOI 10.1016/j.trecan.2020.01.012. [Google Scholar] [CrossRef]
6. Zhang, M., Bai, X., Zeng, X., Liu, J., Liu, F. et al. (2021). CircRNA-miRNA-mRNA in breast cancer. Clinica Chimica Acta, 523(Suppl 1), 120–130. DOI 10.1016/j.cca.2021.09.013. [Google Scholar] [CrossRef]
7. Liu, Z., Zhou, Y., Liang, G., Ling, Y., Tan, W. et al. (2019). Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death & Disease, 10(2), 55. DOI 10.1038/s41419-018-1287-1. [Google Scholar] [CrossRef]
8. Liang, M., Yao, W., Shi, B., Zhu, X., Cai, R. et al. (2021). Circular RNA hsa_circ_0110389 promotes gastric cancer progression through upregulating SORT1 via sponging miR-127-5p and miR-136-5p. Cell Death & Disease, 12(7), 639. DOI 10.1038/s41419-021-03903-5. [Google Scholar] [CrossRef]
9. Xie, J., Chen, L., Sun, Q., Li, H., Wei, W. et al. (2022). An immune subtype-related prognostic signature of hepatocellular carcinoma based on single-cell sequencing analysis. Sedentary Life and Nutrition, 14(7), 3276–3292. DOI 10.18632/aging.204012. [Google Scholar] [CrossRef]
10. Zhong, S., Feng, J. (2022). CircPrimer 2.0: A software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinformatics, 23(1), 215. DOI 10.1186/s12859-022-04705-y. [Google Scholar] [CrossRef]
11. Agarwal, V., Bell, G. W., Nam, J. W., Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4, e05005. DOI 10.7554/eLife.05005. [Google Scholar] [CrossRef]
12. Wang, S., Zhang, K., Tan, S., Xin, J., Yuan, Q. et al. (2021). Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Molecular Cancer, 20(1), 13. DOI 10.1186/s12943-020-01298-z. [Google Scholar] [CrossRef]
13. Chen, X., Lu, Y. (2021). Circular RNA: Biosynthesis in vitro. Frontiers in Bioengineering and Biotechnology, 9, 787881. DOI 10.3389/fbioe.2021.787881. [Google Scholar] [CrossRef]
14. Huang, X. Y., Huang, Z. L., Huang, J., Xu, B., Huang, X. Y. et al. (2020). Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. Journal of Experimental & Clinical Cancer Research, 39(1), 20. DOI 10.1186/s13046-020-1529-9. [Google Scholar] [CrossRef]
15. Huang, Z. L., Huang, X. Y., Huang, J., Huang, X. Y., Xu, Y. H. et al. (2021). Multiple omics integration reveals key circular RNAs in hepatocellular carcinoma. Frontiers in Oncology, 11, 621353. DOI 10.3389/fonc.2021.621353. [Google Scholar] [CrossRef]
16. Zhong, J. X., Kong, Y. Y., Luo, R. G., Xia, G. J., He, W. X. et al. (2021). Circular RNA circ-ERBB2 promotes HER2-positive breast cancer progression and metastasis via sponging miR-136-5p and miR-198. Journal of Translational Medicine, 19(1), 455. DOI 10.1186/s12967-021-03114-8. [Google Scholar] [CrossRef]
17. Jahani, S., Nazeri, E., Majidzadeh-A, K., Jahani, M., Esmaeili, R. (2020). Circular RNA; a new biomarker for breast cancer: A systematic review. Journal of Cellular Physiology, 235(7–8), 5501–5510. DOI 10.1002/jcp.29558. [Google Scholar] [CrossRef]
18. Li, Z., Chen, Z., Hu, G., Jiang, Y. (2019). Roles of circular RNA in breast cancer: Present and future. American Journal of Translational Research, 11(7), 3945–3954. [Google Scholar]
19. Rao, A. K. D. M., Arvinden, V. R., Ramasamy, D., Patel, K., Meenakumari, B. et al. (2021). Identification of novel dysregulated circular RNAs in early-stage breast cancer. Journal of Cellular and Molecular Medicine, 25(8), 3912–3921. DOI 10.1111/jcmm.16324. [Google Scholar] [CrossRef]
20. Zheng, X., Huang, M., Xing, L., Yang, R., Wang, X. et al. (2020). The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Molecular Cancer, 19(1), 73. DOI 10.1186/s12943-020-01183-9. [Google Scholar] [CrossRef]
21. Fu, B., Liu, W., Zhu, C., Li, P., Wang, L. et al. (2021). Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. International Journal of Biological Sciences, 17(12), 3104–3117. DOI 10.7150/ijbs.58916. [Google Scholar] [CrossRef]
22. Xie, J., Li, H., Chen, L., Cao, Y., Hu, Y. et al. (2021). A novel pyroptosis-related lncRNA signature for predicting the prognosis of skin cutaneous melanoma. International Journal of General Medicine, 14, 6517–6527. DOI 10.2147/IJGM.S335396. [Google Scholar] [CrossRef]
23. Jaafar, R., Mnich, K., Dolan, S., Hillis, J., Almanza, A. et al. (2018). RIP2 enhances cell survival by activation of NF-ĸB in triple negative breast cancer cells. Biochemical and Biophysical Research Communications, 497(1), 115–121. DOI 10.1016/j.bbrc.2018.02.034. [Google Scholar] [CrossRef]
24. Rybak-Wolf, A., Stottmeister, C., Glažar, P., Jens, M., Pino, N. et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell, 58(5), 870–885. DOI 10.1016/j.molcel.2015.03.027. [Google Scholar] [CrossRef]
Appendix
Table S1:Primer sequences of quantitative real-time PCR
Table S2:Predicted miRNA response elements regarding the top 6 of hsa-circ-0006969
Figure S1: Workflow and analysis tools
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools