 Open Access
Open Access
ARTICLE
Identification of microbial metabolites that accelerate the ubiquitin-dependent degradation of c-Myc
1 Bioprobe Application Research Unit, RIKEN CSRS, Saitama, 351-0198, Japan
2 Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, 113-8510, Japan
3 Chemical Biology Research Group, RIKEN CSRS, Saitama, 351-0198, Japan
4 Chemical Resource Development Research Unit, RIKEN CSRS, Saitama, 351-0198, Japan
5 Molecular Structure Characterization Unit, RIKEN CSRS, Saitama, 351-0198, Japan
6 Japan Biological Informatics Consortium (JBIC), Tokyo, 135-8073, Japan
7 Department of Life Science and Medical Bioscience, School of Advanced Science and Engineering, Waseda University, Tokyo, 162-8480, Japan
8 Medical-Industrial Translational Research Center, Fukushima Medical University, Fukushima, 960-1295, Japan
9 Guangdong Engineering Research Center of Oral Restoration and Reconstruction, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou, China
10 KingMed School of Laboratory Medicine, Guangzhou Medical University, Guangzhou, China
11 Department of Pharmaceutical Sciences, University of Shizuoka, Shizuoka, 422-8526, Japan
* Corresponding Authors: HIROYUKI OSADA. Email: ; NOBUMOTO WATANABE. Email:
(This article belongs to the Special Issue: Approach from Chemical Biology for Cancer Research)
Oncology Research 2023, 31(5), 655-666. https://doi.org/10.32604/or.2023.030248
Received 28 March 2023; Accepted 15 June 2023; Issue published 21 July 2023
Abstract
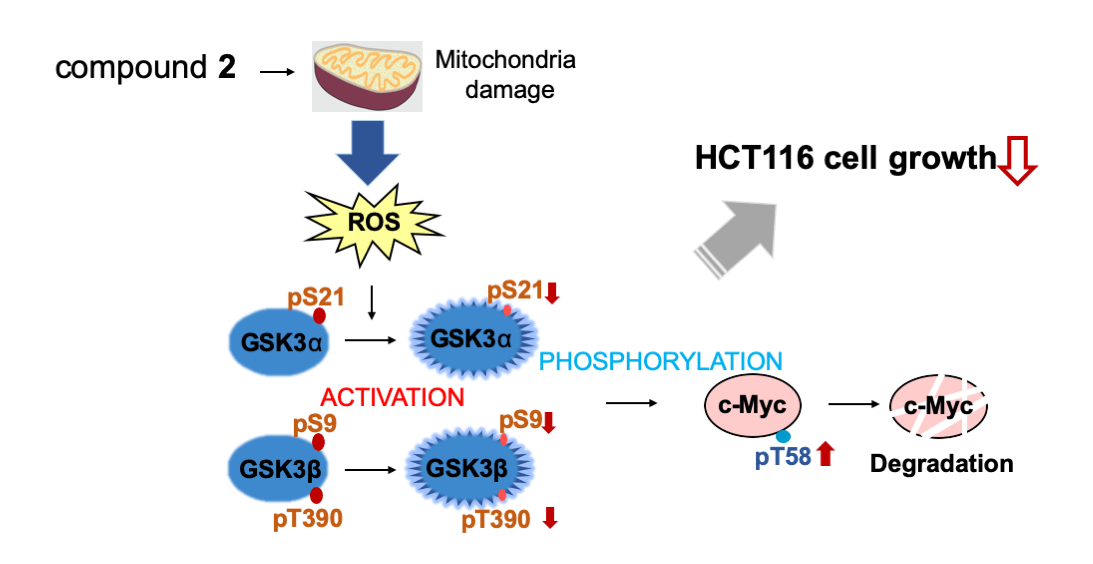
Myc belongs to a family of proto-oncogenes that encode transcription factors. The overexpression of c-Myc causes many types of cancers. Recently, we established a system for screening c-Myc inhibitors and identified antimycin A by screening the RIKEN NPDepo chemical library. The specific mechanism of promoting tumor cell metastasis by high c-Myc expression remains to be explained. In this study, we screened approximately 5,600 microbial extracts using this system and identified a broth prepared from Streptomyces sp. RK19-A0402 strongly inhibits c-Myc transcriptional activity. After purification of the hit broth, we identified compounds closely related to the aglycone of cytovaricin and had a structure similar to that of oligomycin A. Similar to oligomycin A, the hit compounds inhibited mitochondrial complex V. The mitochondria dysfunction caused by the compounds induced the production of reactive oxygen species (ROS), and the ROS activated GSK3α/β that phosphorylated c-Myc for ubiquitination. This study provides a successful screening strategy for identifying natural products as potential c-Myc inhibitors as potential anticancer agents.
Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileCite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools