 Open Access
Open Access
REVIEW
Igniting Cold Tumors: Multi-Omics-Driven Strategies to Overcome Immune Evasion and Restore Immune Surveillance
1 The First Clinical Medical College, Nanjing University of Chinese Medicine, Nanjing, 210023, China
2 School of Chinese Medicine, Nanjing University of Chinese Medicine, Nanjing, 210023, China
3 Department of Gastroenterology and Hepatology, Jinling Hospital, Affiliated Hospital of Medical School, Nanjing University, Nanjing, 210016, China
4 School of Acupuncture and Tuina, School of Regimen and Rehabilitation, Nanjing University of Chinese Medicine, Nanjing, 210023, China
* Corresponding Authors: Ziyun Li. Email: ; Fangyu Wang. Email:
# These co-first authors contribute equally to this manuscript
(This article belongs to the Special Issue: Multi-Omics Approaches for Precision Medicine)
Oncology Research 2025, 33(10), 2857-2902. https://doi.org/10.32604/or.2025.066805
Received 17 April 2025; Accepted 08 July 2025; Issue published 26 September 2025
Abstract
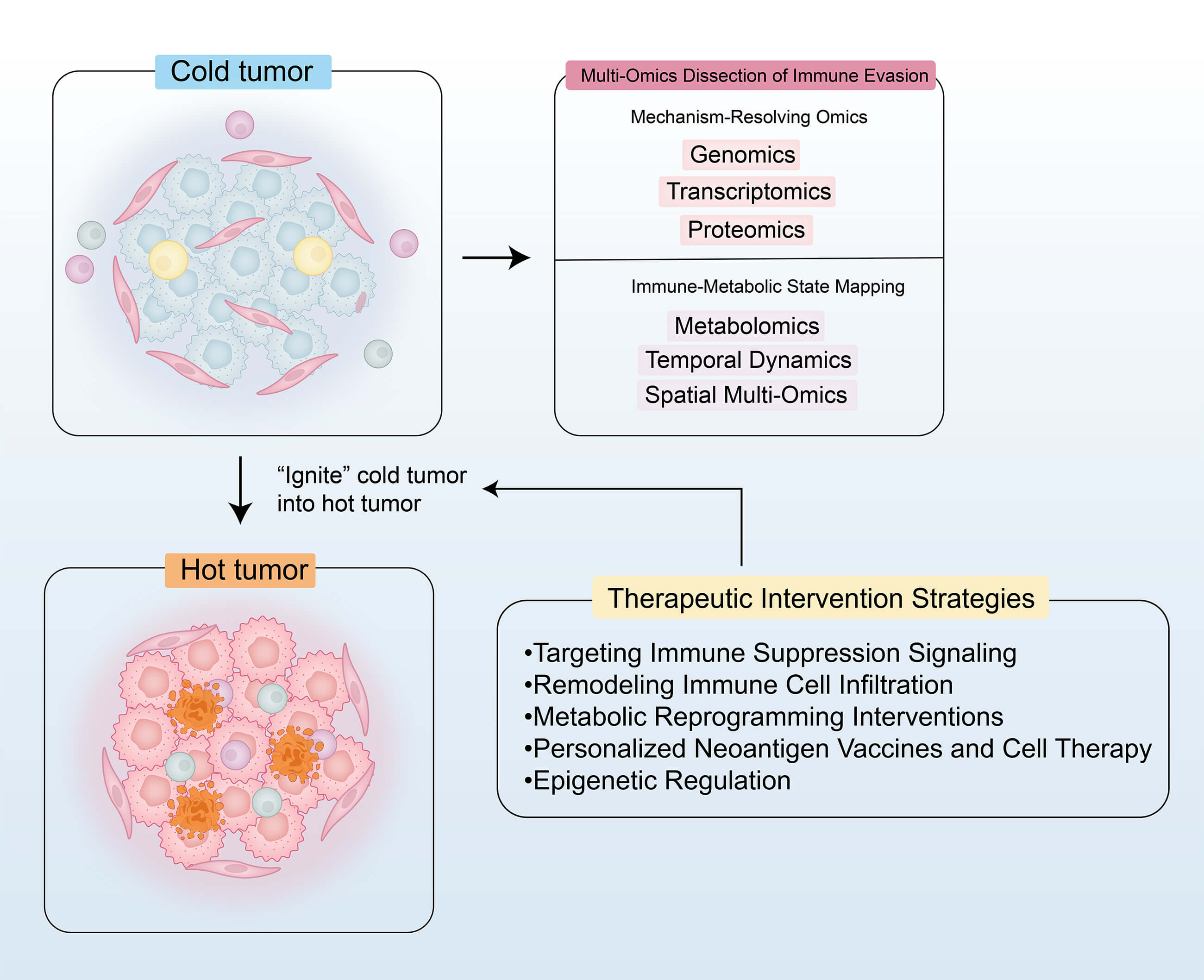
Cold tumors, defined by insufficient immune cell infiltration and a highly immunosuppressive tumor microenvironment (TME), exhibit limited responsiveness to conventional immunotherapies. This review systematically summarizes the mechanisms of immune evasion and the therapeutic strategies for cold tumors as revealed by multi-omics technologies. By integrating genomic, transcriptomic, proteomic, metabolomic, and spatial multi-omics data, the review elucidates key immune evasion mechanisms, including activation of the WNT/β-catenin pathway, transforming growth factor-β (TGF-β)–mediated immunosuppression, metabolic reprogramming (e.g., lactate accumulation), and aberrant expression of immune checkpoint molecules. Furthermore, this review proposes multi-dimensional therapeutic strategies, such as targeting immunosuppressive pathways (e.g., programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1) inhibitors combined with TGF-β blockade), reshaping the TME through chemokine-based therapies, oncolytic viruses, and vascular normalization, and metabolic interventions (e.g., inhibition of lactate dehydrogenase A (LDHA) or glutaminase (GLS)). In addition, personalized neoantigen vaccines and engineered cell therapies (e.g., T cell receptor-engineered T (TCR-T) and natural killer (NK) cells) show promising potential. Emerging evidence also highlights the role of epigenetic regulation (e.g., histone deacetylase (HDAC) inhibitors) and N6-Methyladenosine (m6A) RNA modifications in reversing immune evasion. Despite the promising insights offered by multi-omics integration in guiding precision immunotherapy, challenges remain in clinical translation, including data heterogeneity, target-specific toxicity, and limitations in preclinical models. Future efforts should focus on coupling dynamic multi-omics technologies with intelligent therapeutic design to convert cold tumors into immunologically active (“hot”) microenvironments, ultimately facilitating breakthroughs in personalized immunotherapy.Graphic Abstract
Keywords
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools