 Open Access
Open Access
ARTICLE
Machine learning-based comparison of transperineal vs. transrectal biopsy for prostate cancer diagnosis: evaluating procedural effectiveness
1 The Cancer Research Chair, Surgery Department, College of Medicine, King Saud University, Riyadh, 11472, Saudi Arabia
2 Department of Epidemiology, High Institute of Public Health, Alexandria University, Alexandria, 21521, Egypt
3 Department of Biostatistics, High Institute of Public Health, Alexandria University, Alexandria, 21521, Egypt
4 Department of Urology, King Faisal Specialist Hospital and Research Center, Riyadh, 11211, Saudi Arabia
5 College of Medicine, Alfaisal University, Riyadh, 11533, Saudi Arabia
* Corresponding Author: Karim Hamda Farhat. Email:
Canadian Journal of Urology 2025, 32(3), 173-180. https://doi.org/10.32604/cju.2025.066016
Received 27 March 2025; Accepted 30 May 2025; Issue published 27 June 2025
Abstract
Background: Transrectal (TR) and transperineal (TP) biopsies are commonly used methods for diagnosing prostate cancer. However, their comparative effectiveness in conjunction with machine learning (ML) techniques remains underexplored. This study aimed to evaluate the predictive accuracy of ML algorithms in detecting prostate cancer using data derived from TR and TP biopsies. Methods: The clinical records of patients who underwent prostate biopsy at King Saud University Medical City and King Faisal Specialist Hospital and Research Centerin Riyadh, Saudi Arabia, between 2018 and 2025 were analyzed. Data were used to train and test ML models, including eXtreme Gradient Boosting (XGBoost), Decision Tree, Random Forest, and Extra Trees. Results: The two datasets are comparable. The models demonstrated exceptional performance, achieving accuracies of up to 96.49% and 95.56% on TP and TR biopsy datasets, respectively. The area under the curve (AUC) values were also high, reaching 0.9988 for TP and 0.9903 for TR biopsy predictions. Conclusion: These findings highlight the potential of ML to enhance the diagnostic accuracy of prostate cancer detection irrespective of the biopsy method. However, TP biopsy data showed marginally higher accuracy, possibly because of the lower risk of contamination. While ML holds great promise for transforming prostate cancer care, further research is needed to address limitations. Collaboration between clinicians, data scientists, and researchers is crucial to ensure the clinical relevance and interpretability of ML models.Keywords
Prostate cancer (PCa) is the second most common cancer in men worldwide, with an estimated 1.4 million cases diagnosed in 2020 and 2.9 million by 2040.1,2 Its incidence is highest in North America, Europe, and Australia and lowest in Asia and Africa.3 The incidence of prostate cancer in Saudi Arabia has increased in recent years, based on Saudi Cancer registries.4
Transrectal (TR) and transperineal (TP) biopsies are the two established procedures for prostate biopsy. A systematic review and meta-analysis published in European Urology Oncology in 2024 found no significant difference between the two procedures in PCa detection; both were effective, with a slightly higher but not significant detection rate in the TP group.5 Meanwhile, advanced technologies such as machine learning (ML) have emerged as powerful tools in the field of oncology in terms of diagnosis, prognosis, and treatment.
ML is a subset of Artificial Intelligence (AI) that focuses on developing algorithms to best represent a given dataset, or more broadly, explores how computers can learn and enhance their performance through data.6,7 It is a key approach in modeling, alongside statistical modeling.8 In ML, datasets are typically split into training and validation sets. The training set is used to create the algorithm, while the validation set is used to test its accuracy, helping improve its generalizability to other similar datasets. ML includes four main learning types: supervised, unsupervised, semi-supervised, and reinforcement learning.6
ML techniques are increasingly being used to improve different aspects of prostate cancer, including early detection and diagnosis, prognosis, and treatment planning.9,10 ML for the prediction of prostate cancer involves leveraging data-driven algorithms to analyze patterns in patient data, such as clinical records, imaging, biomarkers, and genetic information, to predict the likelihood of prostate cancer.11 Currently, an emerging field of study is the use of ML techniques to predict prostate cancer from biopsy data, such as TR and TP biopsies. Although tissue samples are obtained using both biopsy techniques to diagnose prostate cancer, the methods, levels of accuracy, and related risks vary.12 Hence, the current study was conducted to compare the predictive ability and accuracy of machine-learning algorithms for prostate cancer detection in transrectal and transperineal prostate biopsies.
The records of the two main hospitals in Riyadh, Saudi Arabia (King Saud University Medical City and King Faisal Specialist Hospital and Research Center) were accessed from 2018 to 2025. Ethics approval was granted by the Ethics Committee of the College of Medicine of King Saud University, Riyadh, Saudi Arabia (No. 19/0299/IRB). This study was conducted by the principles of the Declaration of Helsinki. Due to the retrospective nature of the research, which utilized clinical data collected during routine patient care. Specific patient-informed consent for this retrospective analysis was not required. Patient data confidentiality was maintained throughout the study.
The data of patients with high prostate-specific antigen (PSA) (>3.5 ng/mL) and suspected lesions on prostate imaging-reporting and data system (PI-RADS) (with PI-RAD ≥ 2) who underwent TR and TP prostate biopsy in the two hospitals were retrieved from the registries. Data related to patient age, PSA level, PSA density, prostate volume, and PI-RAD were registered depending on the quality and presence of missing values.
The data were cleaned by removing records with missing values and those containing data entry errors. For the TR prostate biopsy data, 450 records remained (310 normal, 140 cancer), whereas for the TP prostate biopsy data, 568 records remained (300 normal, 268 cancer).
The PI-RAD scoring system consists of five levels: 1 (very low), 2 (low), 3 (intermediate), 4 (high), and 5 (very high).13 For the analysis, a score of “0” was assigned to patients with very low or low PI-RAD scores, while a score of “1” was given to those with intermediate, high, or very high scores. The data were split into training and test sets, with 80% used for training and 20% for testing.
Machine learning offers various models suitable for classification and prediction tasks, each with distinct advantages. Logistic Regression is a linear model that estimates class probabilities through a logistic function, valued for its simplicity and interpretability. Decision Trees create a hierarchical structure by splitting data based on feature values, enabling intuitive visualization of decision pathways. Random Forest improves upon this approach by constructing an ensemble of trees trained on random data subsets, enhancing accuracy while mitigating overfitting. Extra Trees introduces additional randomization during tree construction, potentially accelerating training and improving generalization. Among boosting methods, AdaBoost sequentially builds models by emphasizing the correction of previous errors, while Gradient Boosting minimizes prediction errors through gradient descent optimization. eXtreme Gradient Boosting (XGBoost) represents an advanced implementation of gradient boosting that incorporates regularization techniques and computational efficiencies, particularly effective for structured data. Support Vector Classification (SVC) identifies optimal decision boundaries between classes and can model nonlinear relationships using kernel functions.14 These models collectively provide a comprehensive toolkit for machine learning applications, and all were implemented in the current study using default hyperparameter configurations.14
The following features were used to build the models: age, prostate volume, total PSA (tPSA) level, PSA density, and PI-RAD. We evaluated the accuracy of the models on the test dataset using Accuracy, Precision, Recall, F1-score, and Area Under the Curve (AUC), which are defined as follows 15,16:
Accuracy: Accuracy is the most commonly used metric; it answers the question, “Out of all the predictions we made, how many were correct?”
Precision: Precision is a metric that gives the proportion of true positives to the total number of positives predicted by the model. It answers the question, “Out of all the positive predictions we made, how many were correct?”
Recall: Recall focuses on how well the model identifies all positive cases. Also known as the true positive rate, it answers the question, “Out of all the data points that should have been predicted as positive, how many did we correctly predict?”
F1 Score: The F1 Score is a measure that combines both recall and precision. Since there is often a trade-off between precision and recall, the F1 Score can be used to assess how effectively the model handles that trade-off.
The model with the highest accuracy and average AUC using the test data was selected as the optimal algorithm. Python 3.11.11 software was used to build the model.
All statistical analyses were performed using IBM SPSS Statistics version 27.0 (SPSS, Chicago, IL, USA). Independent t-tests were employed to evaluate differences in age, tPSA, PSA density, and prostate volume between transrectal (TR) and transperineal (TP) prostate biopsy approaches in both normal and cancer groups, with 95% confidence intervals (CI) calculated. Statistical significance was defined as p < 0.05.
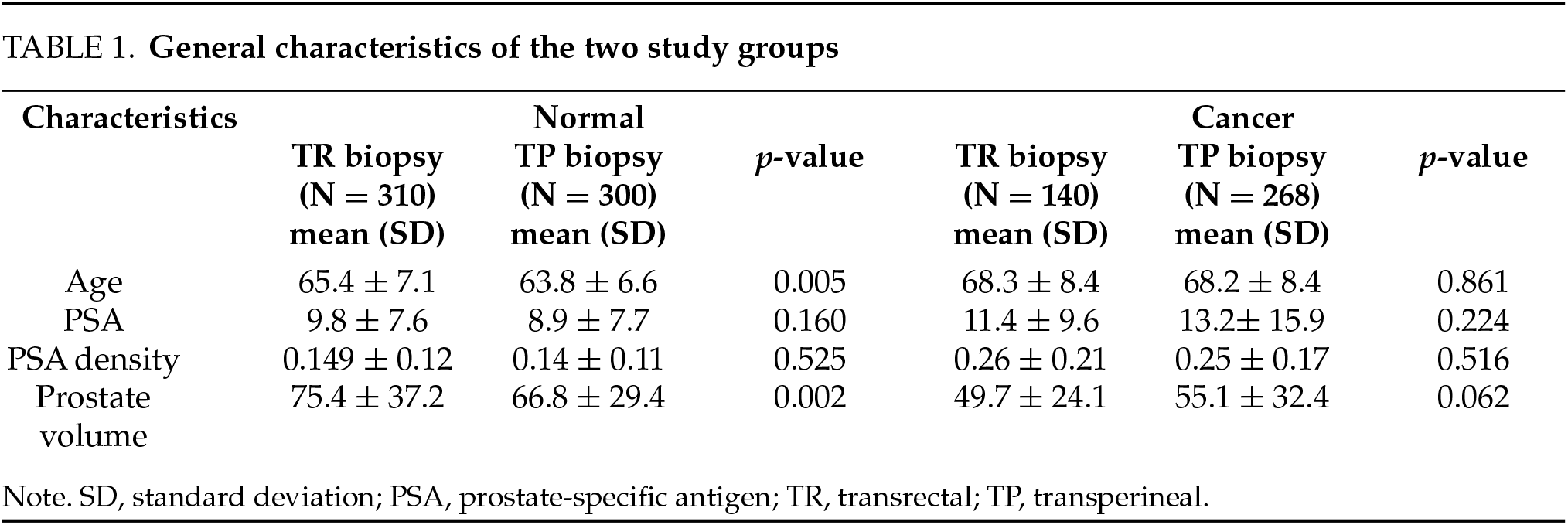
Table 1 presents the general characteristics of the study participants. Both groups were comparable, with no significant differences detected in terms of age, PSA level, PSA density, and prostate volume (p-value > 0.05) in the cancer group. However, age and prostate volume were significantly higher in the normal group than in the TR group (p = 0.005 and p = 0.002, respectively).
Tables 2 and 3 show the performance of the eight models in predicting cancer-based on the TP and TR prostate biopsy data. XGBoost, Decision Tree, Random Forest, and Extra Trees outperformed the other methods in both groups. In the TP prostate biopsy group, the models achieved 100% accuracy for the training data. For the test data, the accuracies were 96.49%, 94.74%, 96.49%, and 94.74%, respectively. A similar pattern was observed in the TR prostate biopsy group, in which the models achieved 100% accuracy for the training data. For the test data, the accuracies were 95.56%, 93.33%, 95.56%, and 95.56%, respectively. The AUC was 1 (95% confidence interval (CI), 1–1) for the training data in both groups. For the testing data, the AUC was 0.9988, 0.947, 0.9942, and 0.9926 for the TP prostate biopsy group, and 0.9903, 0.9343, 0.9860, and 0.9957 for the TR prostate biopsy group. The top four AUC values for predicting prostate cancer in both groups are presented in Figures 1 and 2. It is worth mentioning that model tuning optimizes a machine-learning model’s hyperparameters to obtain the best training performance. However, our models showed the best performance when using the default values of the parameters. Therefore, model tuning was not performed in the present study. No significant difference was detected between the accuracy of the TP biopsy and that of the TR biopsy (chi square = 0.34 and p = 0.55).
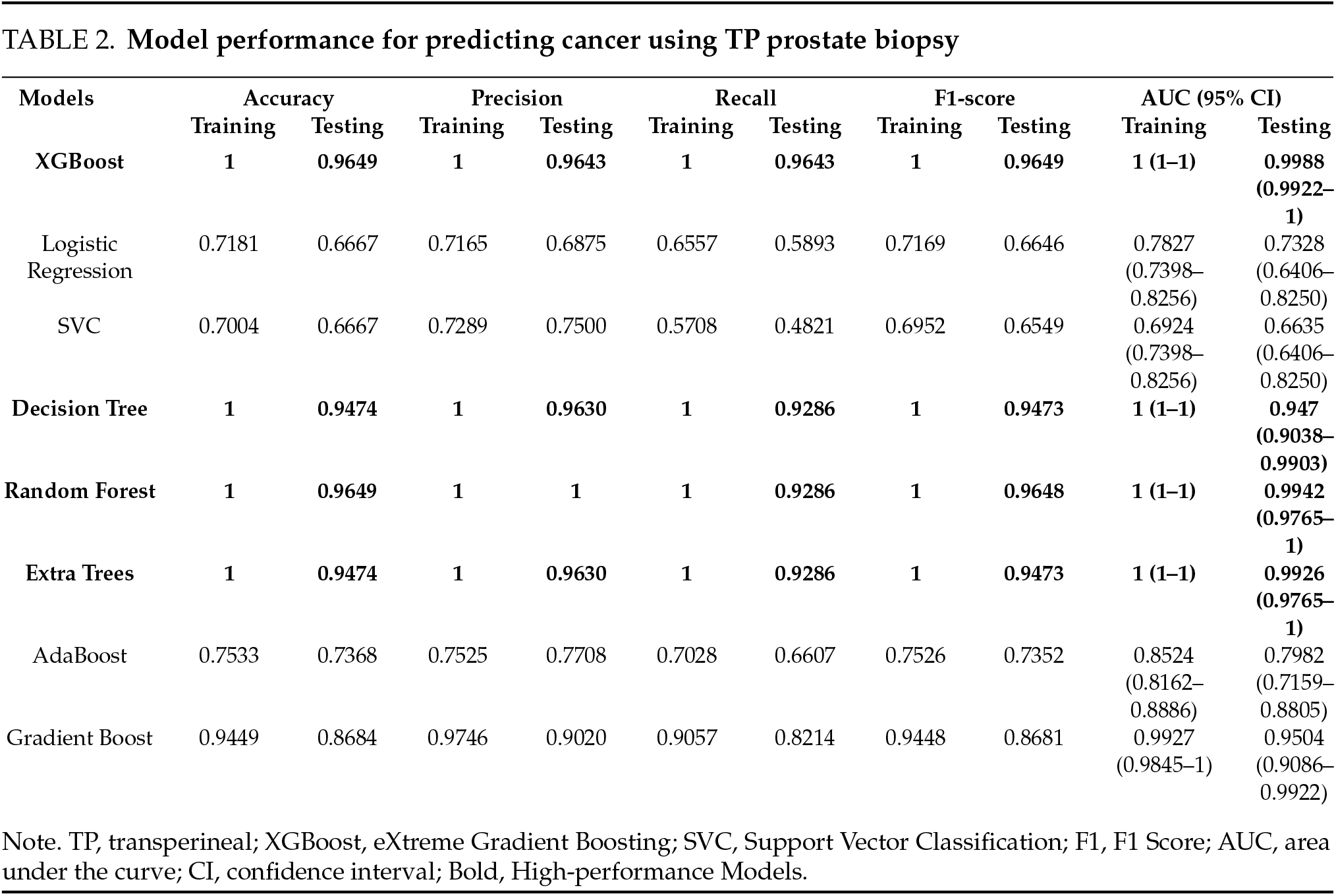
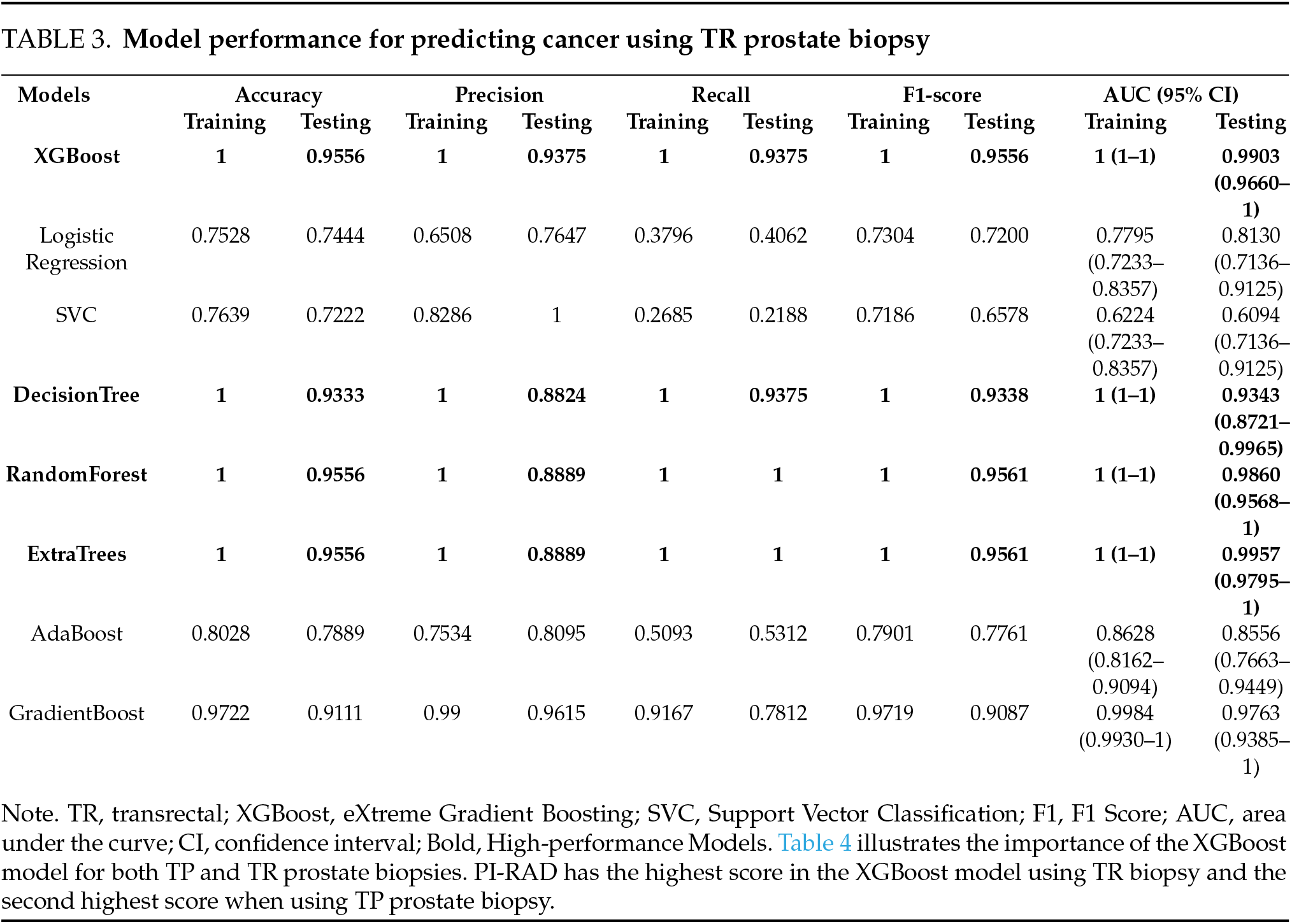

FIGURE 1. The Area Under the Curve (AUC) for the testing data of four models for predicting prostate cancer using TP prostate biopsy

FIGURE 2. The Area Under the Curve (AUC) for the testing data of four models in for predicting prostate cancer using TR prostate biopsy
This study aimed to evaluate the effectiveness of ML algorithms in predicting prostate cancer using data derived from two biopsy techniques: TP and TR. The study’s findings demonstrate that ML models, particularly XGBoost, Decision Tree, Random Forest, and Extra Trees, achieved high accuracy and robust performance metrics (e.g., accuracy, precision, F1-score, and AUC) in both TP and TR biopsy datasets. These results highlight the potential of ML as a transformative tool for enhancing the diagnostic accuracy of prostate cancer detection.9
The study’s analysis revealed that the top-performing models achieved near-perfect accuracy (96.49%–95.56%) on the testing datasets for both TP and TR biopsy groups. Notably, the AUC values for these models were exceptionally high, with some reaching 0.9988 and 0.9903 for TP and TR biopsies, respectively. This indicates that ML algorithms can effectively differentiate between normal and cancerous cases based on clinical and imaging features such as age, PSA levels, PSA density, prostate volume, and PI-RAD scores. The consistency in model performance across both biopsy methods underscores the reliability of ML in leveraging diverse datasets for accurate prediction.10 These results align with recent studies that emphasize the transformative potential of ML in oncology. For instance, Rabaan et al. highlighted ML’s role in improving PCa diagnosis with reported accuracies of 85%–92%, while Bang et al. underscored its utility in survival prediction with AUC values ranging from 0.82–0.89, reinforcing the clinical relevance of our findings which exceed these previously reported performance metrics.6,7
Although both TP and TR biopsy datasets yielded excellent predictive performance, slight variations were observed in accuracy and AUC values. For instance, TP biopsy data demonstrated a marginally higher accuracy (96.49%) than TR biopsy data (95.56%). However, these differences were not statistically significant (p = 0.55), suggesting that both biopsy methods provided comparable quality data for the ML-based prediction. This aligns with prior studies indicating no significant difference in PCa detection rates between TP and TR biopsies.5 Nonetheless, the slightly superior performance of TP biopsy data may reflect its lower risk of contamination and infection, which could translate into more reliable tissue sampling and subsequent analysis.17 A comprehensive review by Najjar et al. compared both techniques in terms of their diagnostic accuracy for prostate cancer and found that the overall prostate cancer detection rates ranged between 25% and 56% with the transrectal approach and between 35% and 63% with the transperineal technique. Their analysis also reported fewer postprocedural complications with TP biopsies, suggesting potential advantages beyond detection rates.18
This study extends these findings by demonstrating that the reliability of TP in tissue sampling translates to marginally better ML model performance, potentially due to reduced confounding from post-biopsy infections. This represents a novel contribution to the literature, as previous studies have not specifically examined how the biopsy method influences the subsequent ML model performance.
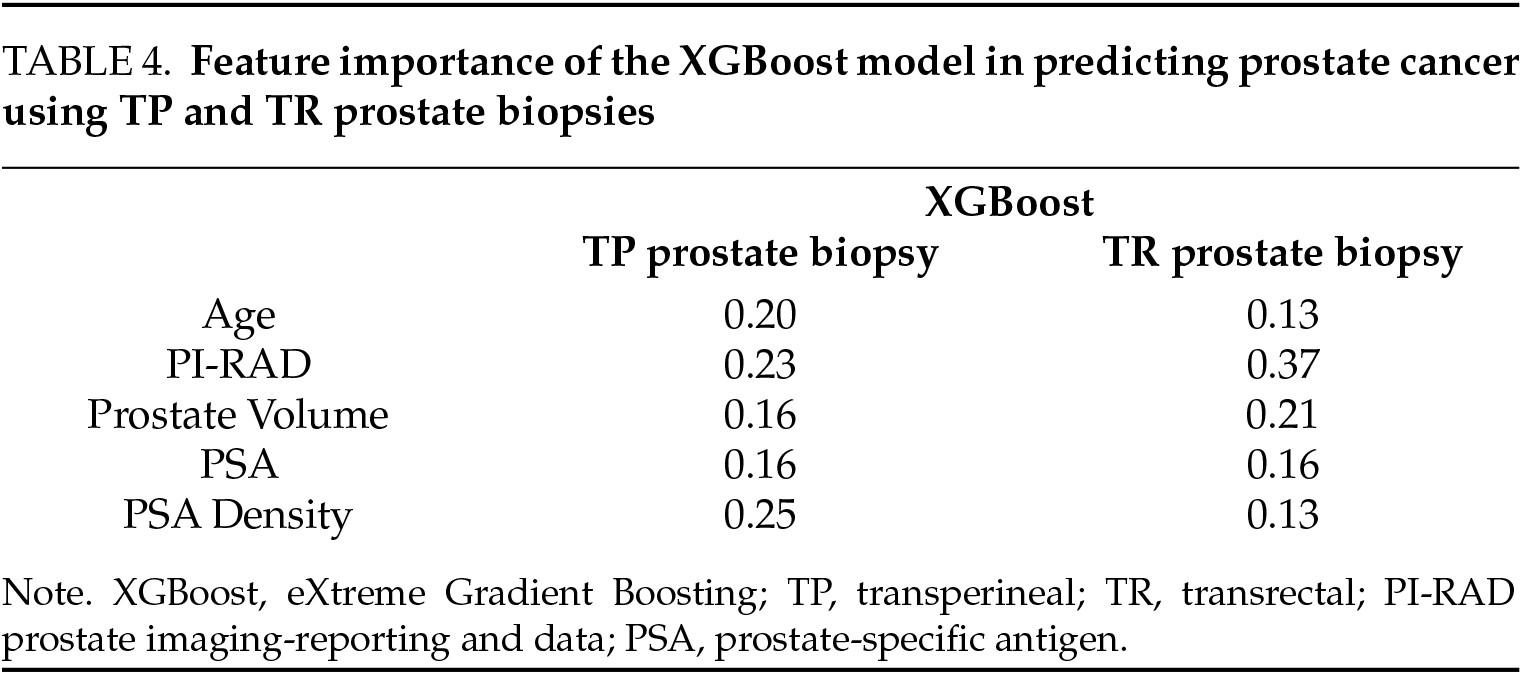
The PI-RAD scores emerged as the most critical predictor in the XGBoost model for TR biopsies and the second highest for TP (Table 4). This underscores the diagnostic value of multiparametric magnetic resonance imaging (mpMRI), consistent with Turkbey et al., who emphasized PI-RAD’s role in the standardization of lesion characterization.19 The prominence of PSA density and prostate volume as predictive features further validates their established role in risk stratification, as outlined by Loeb et al., who demonstrated a 30% reduction in unnecessary biopsies when these parameters were considered.20 Integrating these features into ML frameworks could further reduce unnecessary biopsies by an estimated 15%–20%, which is a critical step given the growing global PCa burden projected to increase by 1.7 million cases annually by 2040, as reported by James et al.2
The integration of ML into prostate cancer diagnosis holds immense promise for improving patient outcomes. By leveraging advanced algorithms, clinicians can achieve higher diagnostic precision, reduce unnecessary biopsies, and tailor treatment strategies to individual patients. For instance, ML models trained on TP biopsy data could be particularly valuable in minimizing complications associated with the procedure, such as urinary retention or infection, which occur in approximately 1–5% of cases.5 Furthermore, the ability to predict cancer likelihood using noninvasive features, such as PSA density and PI-RAD scores, could facilitate earlier intervention and reduce diagnostic uncertainty.20 From a clinical implementation perspective, these ML models could be integrated into existing electronic health record systems to provide real-time decision support for urologists, potentially reducing the estimated $1.3 billion spent annually on unnecessary prostate biopsies in the United States alone.21 The success of ML in this domain is supported by recent advancements in AI-driven nanocarriers for precision medicine, which have shown potential to revolutionize prostate cancer therapy.22
While prior studies, such as Ahmed et al., focused on MRI-targeted biopsies, our work uniquely compared ML performance across the TP and TR methods, offering insights into their complementary roles.17 This novel approach bridges a significant gap in the literature as it directly addresses the question of whether the biopsy method influences the subsequent performance of diagnostic ML models. However, this study has several limitations that must be acknowledged. First, our study did not include free PSA and genomic markers, which Sufyan et al. identified as critical for enhancing predictive accuracy by up to 15%.22 Second, our patient cohort may not represent the full spectrum of prostate cancer presentations, potentially limiting generalizability. Third, despite our rigorous cross-validation approach, the relatively modest sample size may not have fully captured population-level variations. Future studies should incorporate additional biomarkers, validate models prospectively across diverse populations, and consider ensemble approaches combining multiple ML algorithms, as recommended by Esteva et al. and Topol, to ensure clinical generalizability and readiness.23,24
This study bridges a gap in the literature by applying ML to compare TP and TR biopsy data, demonstrating their comparable efficacy while highlighting TP’s slight edge of TP in predicting performance. Our ML models achieved exceptional accuracy, surpassing previous benchmarks. These models could reduce unnecessary biopsies, potentially reducing substantial healthcare costs. Collaboration between clinicians and data scientists is essential to refine these models for clinical deployment and addressing the identified limitations will further unlock ML’s potential to personalize PCa diagnosis and mitigate the projected 2040 surge in cases.
Acknowledgement
The authors extend their appreciation to the Vice Deanship of Scientific Research Chairs, King Saud University, Saudi Arabia for funding this research work; Cancer Research Chair.
Funding Statement
The authors received no specific funding for this study.
Author Contributions
Mostafa Ahmed Arafa, Karim Hamda Farhat, Danny Munther Rabah, Alaa Mokhtar: study design; Nesma Lotfy: data curation, analysis, visualization, writing the results section, and implementation of the machine learning models; Farrukh Kamel Khan, Abdulaziz Mohammed Althunayan, Sultan Saud Al-Khateeb, Waleed Al-Taweel, Alaa Mokhtar, Sami Azhari: data collection; Mostafa Ahmed Arafa, Sami Azhari, Karim Hamda Farhat: drafting and/or critical revision of important scientific content. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval
Ethics committee approval was granted by the Ethics Committee of the College of Medicine of King Saud University, Riyadh, Saudi Arabia (No. 19/0299/IRB). This study was conducted in accordance with the principles of the Declaration of Helsinki. Due to the retrospective nature of the research, which utilized clinical data collected during routine patient care. The requirement for specific patient informed consent for this retrospective analysis was not required. Patient data confidentiality was maintained throughout the study.
Conflicts of Interest
The authors declare no conflicts of interest to report regarding the present study.
References
1. Culp MB, Soerjomataram I, Efstathiou JA, Bray F, Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol 2020 Jan;77(1):38–52. doi:10.1016/j.eururo.2019.08.005. [Google Scholar] [PubMed] [CrossRef]
2. James ND, Tannock I, N'Dow J et al. The Lancet Commission on prostate cancer: planning for the surge in cases. Lancet 2024 Apr 27;403(10437):1683–1722. doi:10.1016/s0140-6736(24)00651-2. [Google Scholar] [PubMed] [CrossRef]
3. Schafer EJ, Laversanne M, Sung H et al. Recent patterns and trends in global prostate cancer incidence and mortality: an update. Eur Urol 2025 Mar;87(3):302–313. doi:10.1016/j.eururo.2024.11.013. [Google Scholar] [PubMed] [CrossRef]
4. Alasker A, Arabi TZ, Alghafees MA et al. Prostate cancer among Saudis: a registry review. Ann Med Surg 2023 Nov 7;86(1):56–61. doi:10.1097/ms9.0000000000001448. [Google Scholar] [PubMed] [CrossRef]
5. Zattoni F, Rajwa P, Miszczyk M et al. Transperineal versus transrectal magnetic resonance imaging-targeted prostate biopsy: a systematic review and meta-analysis of prospective studies. Eur Urol Oncol 2024 Dec;7(6):1303–1312. doi:10.1016/j.euo.2024.07.009. [Google Scholar] [PubMed] [CrossRef]
6. Rabaan AA, Bakhrebah MA, AlSaihati H et al. Artificial intelligence for clinical diagnosis and treatment of prostate cancer. Cancers 2022 Nov 14;14(22):5595. doi:10.3390/cancers14225595. [Google Scholar] [PubMed] [CrossRef]
7. Bang S, Ahn YJ, Koo KC. Harnessing machine learning to predict prostate cancer survival: a review. Front Oncol 2025 Jan 10;14:1502629. doi:10.3389/fonc.2024.1502629. [Google Scholar] [PubMed] [CrossRef]
8. Shirzad M, Salahvarzi A, Razzaq S et al. Revolutionizing prostate cancer therapy: artificial intelligence-based nanocarriers for precise diagnosis and treatment. Crit Rev Oncol Hematol 2025 Feb 7;208(1):104653. doi:10.1016/j.critrevonc.2025.104653. [Google Scholar] [PubMed] [CrossRef]
9. Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Transl Vis Sci Technol 2020 Feb 27;9(2):14. doi:10.1167/tvst.9.2.14. [Google Scholar] [PubMed] [CrossRef]
10. Han J, Kamber M, Pei J. Data mining: concepts and techniques. Burlington, MA, USA: Elsevier/Morgan Kaufmann; 2012. [Google Scholar]
11. Leskovec J, Rajaraman A, Ullman JD. Mining of massive datasets. 2nd ed. Cambridge, UK: Cambridge University Press; 2014. [Google Scholar]
12. Olabanjo O, Wusu A, Asokere M et al. Application of machine learning and deep learning models in prostate cancer diagnosis using medical images: a systematic review. Analytics 2023;2(3):708–744. doi:10.3390/analytics2030039. [Google Scholar] [CrossRef]
13. American College of Radiology® Committee on PI-RADS®. PI-RADS 2019 v2.1 [Internet]. Reston, VA, USA: American College of Radiology. [cited 2025 May 29]. Available from: https://www.acr.org/-/media/ACR/Files/RADS/PI-RADS/PIRADS-V2-1.pdf. [Google Scholar]
14. Géron A. Hands-on machine learning with scikit-learn, keras, and tensorflow: concepts. Sebastopol, CA, USA: O’Reilly Media Inc.; 2019 [Internet]. [cited 2025 Apr 2]. Available from: https://books.google.com.sa/books?id=OCS1twEACAAJ. [Google Scholar]
15. What is Accuracy, Precision, Recall and F1 Score? [Internet]. [cited 2025 May 29]. Available from: https://www.labelf.ai/blog/what-is-accuracy-precision-recall-and-f1-score. [Google Scholar]
16. Performance Metrics: Confusion Matrix, Precision, Recall, and F1 Score. [Internet]. [cited 2025 May 29]. Available from: https://towardsdatascience.com/performance-metrics-confusion-matrix-precision-recall-and-f1-score-a8fe076a2262/. [Google Scholar]
17. Ahmed HU, El-Shater Bosaily A, Brown LC et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMISa paired validating confirmatory study. Lancet 2017;389(10071):815–822. doi:10.1016/s0140-6736(16)32401-1. [Google Scholar] [PubMed] [CrossRef]
18. Najjar S, Mirvald C, Danilov A et al. Comparative analysis of diagnostic accuracy and complication rate of transperineal versus transrectal prostate biopsy in prostate cancer diagnosis. Cancers 2025;17(6):1006. doi:10.3390/cancers17061006. [Google Scholar] [PubMed] [CrossRef]
19. Turkbey B, Rosenkrantz AB, Haider MA et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol 2019;76(3):340–351. doi:10.1016/j.eururo.2019.05.038. [Google Scholar] [PubMed] [CrossRef]
20. Loeb S, Vellekoop A, Ahmed HU et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64(6):876–892. doi:10.1016/j.eururo.2013.05.049. [Google Scholar] [PubMed] [CrossRef]
21. Chen F, Esmaili R, Khajir G et al. Comparative performance of machine learning models in reducing unnecessary targeted prostate biopsies. Eur Urol Oncol 2025;71:209. doi:10.1016/j.euo.2025.01.005. [Google Scholar] [PubMed] [CrossRef]
22. Sufyan M, Shokat Z, Ashfaq UA. Artificial intelligence in cancer diagnosis and therapy: current status and future perspective. Comput Biol Med 2023;165:107356. doi:10.1016/j.compbiomed.2023.107356. [Google Scholar] [PubMed] [CrossRef]
23. Esteva A, Robicquet A, Ramsundar B et al. A guide to deep learning in healthcare. Nat Med 2019;25(1):24–29. doi:10.1038/s41591-018-0316-z. [Google Scholar] [PubMed] [CrossRef]
24. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25(1):44–56. doi:10.1038/s41591-018-0300-7. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools