 Open Access
Open Access
ARTICLE
An Effective Lung Cancer Diagnosis Model Using Pre-Trained CNNs
1 Department of Business Information Technology, Princess Sumaya University for Technology, Amman, 11941, Jordan
2 Department of Computer Engineering, Al Yamamah University, Riyadh, 11512, Saudi Arabia
3 Department of Computer Science, John Jay College and the Graduate Center, The City University of New York, New York, NY 10019, USA
4 Department of Computer Science, John Jay College at the City University of New York, New York, NY 10019, USA
5 LIMOSE Laboratory, Department of Computer Science, M’hamed Bougara University, Boumerdes, 35000, Algeria
6 Department of Professional Security Studies, New Jersey City University, Jersey City, NJ 07305, USA
* Corresponding Author: Majdi Rawashdeh. Email:
(This article belongs to the Special Issue: Exploring the Impact of Artificial Intelligence on Healthcare: Insights into Data Management, Integration, and Ethical Considerations)
Computer Modeling in Engineering & Sciences 2025, 143(1), 1129-1155. https://doi.org/10.32604/cmes.2025.063765
Received 23 January 2025; Accepted 13 March 2025; Issue published 11 April 2025
Abstract
Cancer is a formidable and multifaceted disease driven by genetic aberrations and metabolic disruptions. Around 19% of cancer-related deaths worldwide are attributable to lung and colon cancer, which is also the top cause of death worldwide. The malignancy has a terrible 5-year survival rate of 19%. Early diagnosis is critical for improving treatment outcomes and survival rates. The study aims to create a computer-aided diagnosis (CAD) that accurately diagnoses lung disease by classifying histopathological images. It uses a publicly accessible dataset that includes 15,000 images of benign, malignant, and squamous cell carcinomas in the lung. In addition, this research employs multiscale processing to extract relevant image features and conducts a comprehensive comparative analysis using four Convolutional Neural Network (CNN) based on pre-trained models such as AlexNet, VGG (Visual Geometry Group)16, ResNet-50, and VGG19, after hyper-tuning these models by optimizing factors such as batch size, learning rate, and epochs. The proposed (CNN + VGG19) model achieves the highest accuracy of 99.04%. This outstanding performance demonstrates the potential of the CAD system in accurately classifying lung cancer histopathological images. This study contributes significantly to the creation of a more precise CNN-based model for lung cancer identification, giving researchers and medical professionals in this vital sector a useful tool using advanced deep learning techniques and publicly available datasets.Keywords
Cell growth, which is not normal in any area of the body, causes the occupation of space and leads to the development of a condition known as cancer. If these unusual cells develop in the lung area, it can result in lung cancer. The World Health Organization (WHO) states that lung cancer is classified as a non-communicable disease, and it is the fifth highest cause of death globally when considering all possible causes, including trachea and bronchus cancer [1]. Over the past few decades, lung cancer has emerged as the most prevalent cancer globally, affecting a significant number of people and resulting in a death rate close to 1.59 million annually. Based on [2], lung cancer is predicted to have the highest incidence of new cases (222,500 cases) among all cancers in the United States in 2017 when taking men and women together, as well as the highest mortality rate (155,870 deaths) for both sexes. It will rank second in terms of cause of death for both men and women, coming in second only to breast cancer for women and prostate cancer for men [3]. The CNN (Convolutional Neural Network)-based methods used for classifying lung nodules are shown in Fig. 1. Many lives can be saved when cancer cells are diagnosed early. Due to the differences in the morphological patterns of lung tissues, it is challenging to precisely identify every nodule using these methods. Therefore, determining whether a lung nodule is benign or malignant depends on its discovery. The accidental failure in the CT screening process is precisely located using the CAD approach. Even while it can be difficult to accurately predict diseases using human interpretations, it is not always desirable to obtain precise findings since it can occasionally lead to missprophecies. It will be simpler to anticipate the impacted location for system-aided exams. In addition, imaging methods, including magnetic resonance imaging (MRI), CT scans, and X-rays, can be utilized to visualize the affected area. Every method of detection is exposed to radiation, and each has pros and cons of its own. Increased radiation doses have an impact on the body, and decreasing radiation doses result in hazy images. As a result, a thorough method for applying computer diagnostics to human healthcare has emerged. Two primary classification methods used for training and evaluating datasets to meet system requirements are ANN and SVM, which are utilized to improve the accuracy of CAD systems. Based on the findings, SVM classifier efficiency has significantly reduced erroneous rate densities and achieved 97% sensitivity and accuracy when compared to ANN classifiers [4].
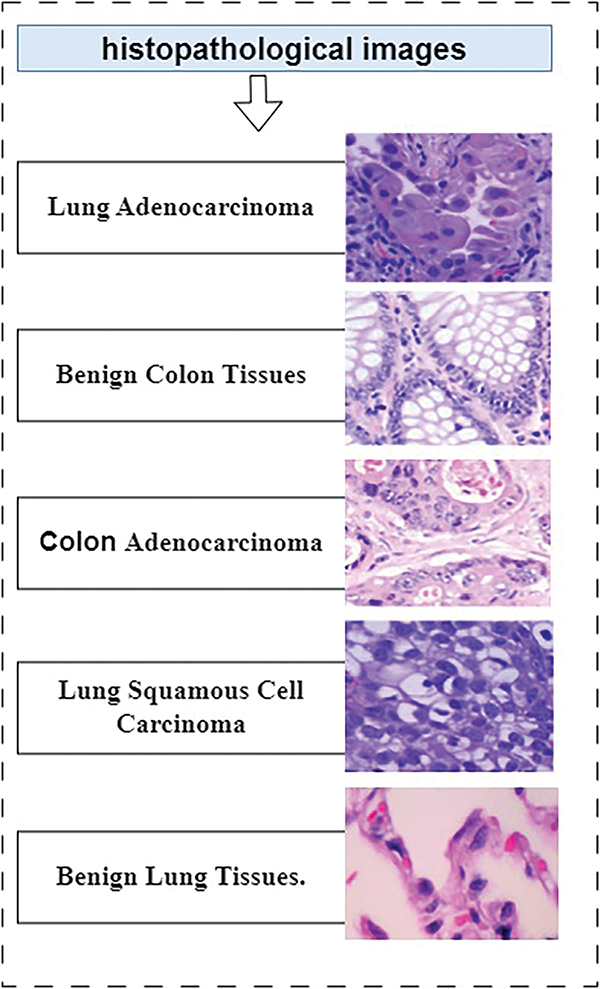
Figure 1: Types of cancer in histopathological images
The need for this work is highlighted by the severity and complexity of cancer as a disease, particularly lung cancer, which is among the leading causes of cancer-related deaths worldwide. The 5-year survival rate for lung cancer is alarmingly low at 19%, highlighting the critical importance of early detection in improving treatment outcomes and overall survival. The costs associated with lung nodule screening are high, and because nodules vary widely in size and shape, it can be challenging to identify abnormalities. However, Physicians now depend heavily on computer-aided diagnostic (CAD) technology to help them with this difficult task.
If experiments on machine learning-based digital pathology photo categorization prove fruitful, these technologies can see increased use in pathology clinics. Future predictions indicate a rapid rise in the application of machine learning-based solutions and AI, especially in the field of pathology. Lung cancer is the deadliest kind of disease; however, patients’ chances are greatly improved by early detection. Determining whether lung nodules need a biopsy to check for benign or malignant nature is now best achieved through low-dose computed tomography. This method has a somewhat significant risk of false positives in clinical settings. It usually requires many possibly malignant nodules to be identified to perform a biopsy, which leads to a significant number of unnecessary biopsies being performed on individuals who aren’t cancer patients.
All acronyms listed in Table 1 reflect the interdisciplinary nature of research in this domain, where lung and colon cancer-based medical imaging techniques converge.

This study achieves several significant milestones:
• It combines (CNN + VGG19) model for detecting lung and colon cancer, integrating deep feature extraction, high-performance filtering, and ensemble learning strategy, providing an efficient and effective solution.
• It compares and analyzes the performance of pre-trained models (CNN, VGG16, VGG19, ResNet-50, and CNN+VGG19).
• It develops a unique feature extraction technique, using various transfer learning models to extract robust features, enhancing the accuracy of cancer diagnosis.
• It demonstrates the effectiveness of the proposed model lung and colon diagnosis by conducting an extensive evaluation of the LC25000 dataset utilizing an extensive range of metrics, contributing to the advancement of AI-based healthcare applications.
Lastly, the manuscript is organized as follows: Section 2 provides a comprehensive review of existing methods for classifying lung and colon cancer images, highlighting their strengths and weaknesses. Section 3 presents a detailed description of the proposed methodology, including tools and techniques for analyzing cancer images. Section 4 covers results and discussion and provides experimental details, such as parameters and results of the proposed model with metrics and a comparison of existing methods. Finally, Section 5 concludes with a summary of findings and contributions, limitations, potential areas of improvement, and research directions and applications.
CNNs have been used since the late 1980s to address a variety of visual issues. In 1989, LeCun et al. recognized handwritten zip codes using ConvNet, an initial replication approach in multilayered CNN [5,6]. Then, Khehrah et al. [7] reported a technique for finding lung lesions on CT scans that uses shape-based and statistical characteristics. In addition, Lung nodule identification utilizing texture and form characteristics in an artificial neural network was introduced in a different study [8]. The model attained an accuracy of 89.62%. Analogously, a 96.67% accurate model for lung cancer diagnosis based on ANN was proposed by Nasser et al. [9]. A neural network-based lung cancer detection method with 96.67% accuracy using CT images was presented by Mohandass et al. [10]. In addition, LeNet-5 was proposed in 1998 by LeCun et al. [11], an enhanced version of ConvNet, for character classification in documents. In a study done by researchers in reference [12], they used thoracic CT scans to distinguish between malignant pulmonary and benign nodules using the LeNet-5 model. The experimental results were obtained using the datasets from the Image Database Resource Initiative (IDRI) and the Lung Image Database Consortium(LIDC). The model classification was evaluated using a 10-folder cross-validation. The accuracy of the LeNet-5 in differentiating between benign and malignant nodules was 97.041%, and it achieved an accuracy of 96.685% in distinguishing between serious malignancies and mild malignancies. In addition, another study [13] used a hybrid convolutional neural network based on a mix of LeNet and AlexNet to classify lung nodules. The hybrid design made use of both the LeNet layers and the AlexNet parameters. The LIDC-IDRI dataset provided 1018 CT images in total, which were used for the training and evaluation of the agile convolutional neural network. Achieving great accuracy requires careful consideration of several factors, including learning rate, batch size, weight initialization, and kernel size. The framework obtained 0.822 accuracy and 0.877 for the area under the curve (AUC) using 7 × 7 kernel size, 32 batch size, and 0.005 learning rate. In this work, Dropout and Gaussian were also used. In 2012, Sharma et al. released AlexNet [14]. AlexNet won the 2012 ILSVRC, surpassing competitors with a top 5 error rate of 15.3%, demonstrating a widespread advancement in CNN performance. The researchers in [15] proposed two designs for the categorization of pulmonary nodules: hybrid 3D-CNN and straight 3D-CNN. Although the classifiers in the two models were different, the feature extraction technique remained the same. The pulmonary CT images were classified Using the 3D-CNN model’s SoftMax classifier, while the hybrid 3D-CNN employed an SVM classifier based on the radial basis function (RBF). The results of the experiment showed that the accuracy of straight 3D-CNN models and the hybrid 3D-CNN was higher. However, when compared to a SoftMax classifier for the 3D-CNN model, the strategy achieved 91.9% precision, 88.53% sensitivity, 94.23% specificity, and 91.8% accuracy.
Ali et al. [16] made use of convolutional neural networks to classify lung tumors. A final fully linked layer, a dropout layer, a pooling layer, and two convolution layers comprise the proposed CanNet architecture. The accuracy achieved by artificial neural networks was 72.50%, while LeNet achieved an accuracy of 56.0%, and CanNet achieved an accuracy of 76.00%. Compared to the LeNet and ANN networks, the CanNet model performed better. Lin and colleagues [17] proposed a model for classifying lung nodules that use a convolutional neural network with Taguchi architecture. The most important benefit of the Taguchi approach is the valuable information that can be discovered with fewer tests. Compared to other methods, the experimental findings showed that the Taguchi-based model employed by AlexNet reduced the training time. Al-Yasriy et al. [18] presented a computer-aided method for lung cancer diagnosis and classification that uses an AlexNet-based convolutional neural network architecture. The CT scan dataset for lung cancer was gathered from Iraqi hospitals and utilized for system testing and training. The system’s results include a 96.403% F1 score, 95% specificity, 95.714% sensitivity, 97.102% precision, and 93.548% accuracy. The impact of Groups of Visual Geometry 19 (VGG19) on the 2014 ImageNet Challenge and 16 (VGG16) was examined by Patil et al. [19]. There were 16 weight layers in VGG16 and 19 weight layers in VGG19. The convolutional filter size of 3 × 3 was employed in both networks, resulting in enhancements in the image recognition process, whereas 224 × 224 RGB images made up the input size. The experimental findings demonstrated how depth representation improved state-of-the-art accuracy and helped categorize difficulties. Compared to alternative methods, both networks demonstrated great accuracy when evaluated on additional datasets. In a different study [20], a deep learning genetic algorithm was utilized to provide a method for early lung cancer detection. Three strategies were utilized in the preprocessing method: the picture was cropped into 227 × 227 for VGG19 and 224 × 224 for VGG16 for the AlexNet architecture; The contrast of the original image was enhanced by applying the histogram stretching technique; and the Noise was removed using a Wiener filter. Three CNN designs were utilized to handle images obtained from low-dose computed tomography (LDCT): AlexNet, VGG19, and VGG16. The most pertinent traits were chosen using a genetic algorithm. In the end, the classification of pulmonary lung nodules was studied using SVM classifiers, decision trees, and K-nearest neighbor. A dilated convolution network-based accurate lung segmentation technique and Geng et al. [21] reported VGG-16. The size of the corresponding field was shown to expand using dilated convolution, which employed a dilation rate parameter. Multi-scale convolution features were fused using the hyper-column features methodology to improve the robustness of the lung segmentation method. The ReLU (rectified linear unit) activation function and multilayer perceptron (MLP) were employed after the updated VGG16. A dice similarity coefficient of 0.9867 was attained by the procedure. Table 2 presents a comprehensive collection of lung cancer imaging datasets comprising various image types, including CT scans, X-rays, and histopathology images. For the purpose of creating and validating computer-aided diagnosis methods for lung cancer, these databases provide researchers and physicians with useful resources. A boosting strategy for VGG16 was presented in a different study [22] based on VGG16 to determine the pathogenic varieties of lung cancer. VGG16-T was made up of a 3 × 3 kernel size of five layers. The boosting technique was discovered to improve efficiency and build a powerful classifier using just three weak classifiers. In the end, CT scans were utilized to identify the pathogenic type of lung cancer and the SoftMax function. Compared to the other methods, DenseNet, AlexNet, ResNet-34, and the VGG16-T testing results with boosting yielded an accuracy of 85%. Table 3 includes an overview of earlier relevant work. It is claimed that deep knowledge [7–10] is needed to get handmade characteristics, which is one of the limits of earlier studies.
The research [23–26] used unbalanced datasets and smaller picture sizes. Research works [12,13,15,27] concentrated on hybrid strategies that increased model complexity, whereas research works [28–30] employed various architectures to increase accuracy. A novel deep learning technique was proposed in [31] by altering the DenseNet201 model and incorporating additional layers into the original DenseNet framework to detect lung cancer. Maurya et al. [32] compared twelve machine learning algorithms using clinical data and found K-Nearest Neighbor and Bernoulli Naive Bayes as the most effective for early prediction. The results are based on classification and heat map correlation analysis. Hossain et al. [33] compared Machine Learning (ML) models for lung cancer prediction, finding tree-based models balance accuracy and interpretability best, while neural networks excel but lack transparency. Key predictors include age, smoking history, and symptoms. The study called for larger datasets and improved interpretability for clinical use. Javed et al. [34] reviewed deep learning techniques, especially CNNs, for lung cancer detection and classification using medical imaging. They analyzed research from 2015–2024. The findings emphasized that deep convolutional neural networks play a vital role in improving diagnosis and reducing the burden on medical practitioners. Deep learning techniques, mainly CNNs, for lung cancer detection using medical imaging were discussed in [35]. The authors analyzed key datasets, compared studies, and explored explainable AI for real-world use.
This study presents a novel Computer Aided Diagnostic (CAD) System aimed at accurately classifying histopathological images for lung tissues to assist in the early diagnosis of lung cancer. Therefore, a robust detection system is necessary to identify lung cancer in its early stages. Hence, this study proposes a robust detection system using advanced deep learning algorithms and optimizers. The methodology encompasses dataset preparation, multi-scale feature processing, feature fusion, and hyperparameter tuning. Specifically, this study employs state-of-the-art algorithms, including AlexNet, VGG16, ResNet-50, VGG19, and the proposed model (CNN + VGG19). These models are used in conjunction with RMSprop, SGD, and Adam optimizers to enhance accuracy. Fig. 2 illustrates the process of using these algorithms and optimizers for lung cancer classification.
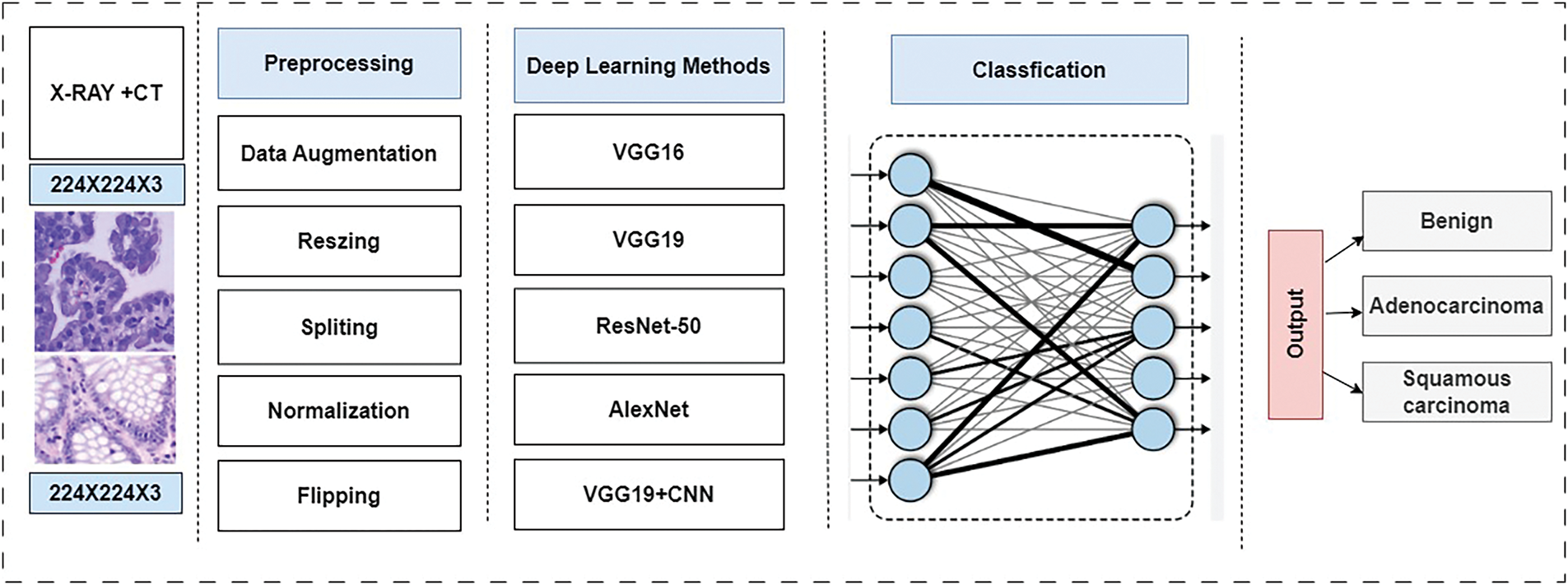
Figure 2: Block diagram for multiclassification based on pre-trained models
AlexNet to classify lung nodules. The hybrid design made use of both the LeNet layers and the AlexNet parameters. The LIDC-IDRI dataset provided 1018 CT images in total, which were used for the training and evaluation of the agile convolutional neural network. Achieving great accuracy required careful consideration of several factors, including learning rate, batch size, weight initialization, and kernel size. The framework obtained 0.822 accuracy and 0.877 AUC using 7 × 7 kernel size, 32 batch size, and 0.005 learning rate. In this work, Dropout and Gaussian were also used. In 2012, Sharma et al. released AlexNet [14]. AlexNet won the 2012 ILSVRC, surpassing competitors with a top 5 error rate of 15.3%, demonstrating a widespread advancement in CNN performance. The researchers in [15] proposed two designs for the categorization of pulmonary nodules: hybrid 3D-CNN and straight 3D-CNN. Although the classifiers in the two models were different, the feature extraction technique remained the same. The pulmonary CT images were classified Using the 3D-CNN model’s SoftMax classifier, while the hybrid 3D-CNN employed an SVM classifier based on the radial basis function (RBF). The results of the experiment showed that the accuracy of straight 3D-CNN models and the hybrid 3D-CNN was higher. However, in comparison to a SoftMax classifier for the 3D-CNN model, the strategy achieved 91.9% precision, 88.53% sensitivity, 94.23% specificity, and 91.8% accuracy.
Ali et al. [16] made use of convolutional neural networks to classify lung tumors. A final fully linked layer, a dropout layer, and a pooling layer, and two convolution layers made up the proposed CanNet architecture. The accuracy achieved by artificial neural networks was 72.50%, while LeNet achieved an accuracy of 56.0%, and CanNet achieved an accuracy of 76.00%. In comparison to the LeNet and ANN networks, the CanNet model performed better. Lin and colleagues [17] proposed a model for classifying lung nodules that use a convolutional neural network with Taguchi architecture. The most important benefit of the Taguchi approach is the valuable information that can be discovered with fewer tests. Compared to other methods, the experimental findings showed that the Taguchi-based model employed by AlexNet reduced the training time. Al-Yasriy et al. [18] presented a computer-aided method for lung cancer diagnosis and classification that uses an AlexNet-based convolutional neural network architecture. The CT scan dataset for lung cancer was gathered from Iraqi hospitals and utilized for system testing and training. The system’s results include a 96.403% F1 score, 95% specificity, 95.714% sensitivity, 97.102% precision, and 93.548% accuracy. The impact of Groups of Visual Geometry 19 (VGG19) on the 2014 ImageNet Challenge and 16 (VGG16) was examined by Patil et al. [19]. There were 16 weight layers in VGG16 and 19 weight layers in VGG19. The convolutional filter size of 3 × 3 was employed in both networks, resulting in enhancements in the image recognition process, whereas 224 × 224 RGB images made up the input size. The experimental findings demonstrated how depth representation improved state-of-the-art accuracy and helped categorize difficulties. Compared to alternative methods, both networks demonstrated great accuracy when evaluated on additional datasets. Elnakib et al. [20] employed a deep learning genetic algorithm to provide a method for early lung cancer detection. Three strategies were utilized in the preprocessing method: the picture was cropped into 227 × 227 for VGG19 and 224 × 224 for VGG16 for the AlexNet architecture; The contrast of the original image was enhanced by applying the histogram stretching technique; and the Noise was removed using a Wiener filter. Three CNN designs were utilized to handle images obtained from low-dose computed tomography (LDCT): AlexNet, VGG19, and VGG16. The most pertinent traits were chosen using a genetic algorithm. In the end, the classification of pulmonary lung nodules was studied using K-nearest neighbor SVM classifiers, decision trees, and (KNN). A dilated convolution network-based accurate lung segmentation technique and VGG-16 were reported by Geng et al. [21]. The size of the corresponding field was shown to expand using dilated convolution, which employed a dilation rate parameter. Multi-scale convolution features were fused using the hyper-column features methodology to improve the robustness of the lung segmentation method. The ReLU activation function and multilayer perceptron (MLP) were employed after the updated VGG16. A dice similarity coefficient of 0.9867 was attained by the procedure. Table 3 presents a comprehensive collection of lung cancer imaging datasets comprising various image types, including CT scans, X-rays, and histopathology images. For the purpose of creating and validating computer-aided diagnosis methods for lung cancer, these databases provide researchers and physicians with useful resources. A boosting strategy for VGG16 was presented in a different study [22] based on VGG16 to determine the pathogenic varieties of lung cancer. VGG16-T was made up of a 3 × 3 kernel size of five layers. The boosting technique was discovered to improve efficiency and build a powerful classifier using just three weak classifiers. In the end, CT scans were utilized to identify the pathogenic type of lung cancer and the SoftMax function. Compared to the other methods, DenseNet, AlexNet, ResNet-34, and the VGG16-T testing results with boosting yielded an accuracy of 85%. Table 3 includes an overview of earlier relevant work. It is claimed that deep knowledge [7–10] is needed to get handmade characteristics, which is one of the limits of earlier studies.
The research [23–25] used unbalanced datasets and smaller picture sizes. Research works [12,13,15] concentrated on hybrid strategies that increased model complexity, whereas research works [28–30] employed various architectures to increase accuracy. Classifying histopathological images for lung tissues to assist the early diagnosis of lung cancer. Therefore, a robust detection system is necessary to identify lung cancer in its early stages. This study proposes a robust detection system using advanced deep learning algorithms and optimizers. The methodology encompasses dataset preparation, multi-scale feature processing, feature fusion, and hyperparameter tuning. Specifically, this study employs state-of-the-art algorithms, including AlexNet, VGG16, ResNet-50, VGG19, and the proposed model (CNN + VGG19). These models are used in conjunction with RMSprop, SGD, and Adam optimizers to enhance accuracy. Fig. 2 illustrates the process of using these algorithms and optimizers for lung cancer classification.
This study proposes a novel Computer Aided Diagnostic (CAD) System to accurately classify histopathological images for lung tissues to assist in early lung cancer detection. Therefore, a robust detection system is necessary to diagnose lung cancer in its early stages. This approach efficiently compares various models’ performance in lung and colon cancer image classification tasks. Fig. 3 illustrates the methodology employed in the research, which combines a conventional neural network (CNN) with a pre-trained VGG19 model. In addition, for the training and testing model, this study uses the LC25000 dataset, allocating 75% for training, 12.5% for testing, and 12.5% for validation. It also implements 5-fold cross-validation, where the dataset is divided into five-fold, and one-fold is used for testing to ensure robustness and generalizability. In contrast, the remaining folds are used for training and validation. In addition, the proposed method allows to evaluate the performance of various models, including AlexNet, VGG16, and ResNet-50.
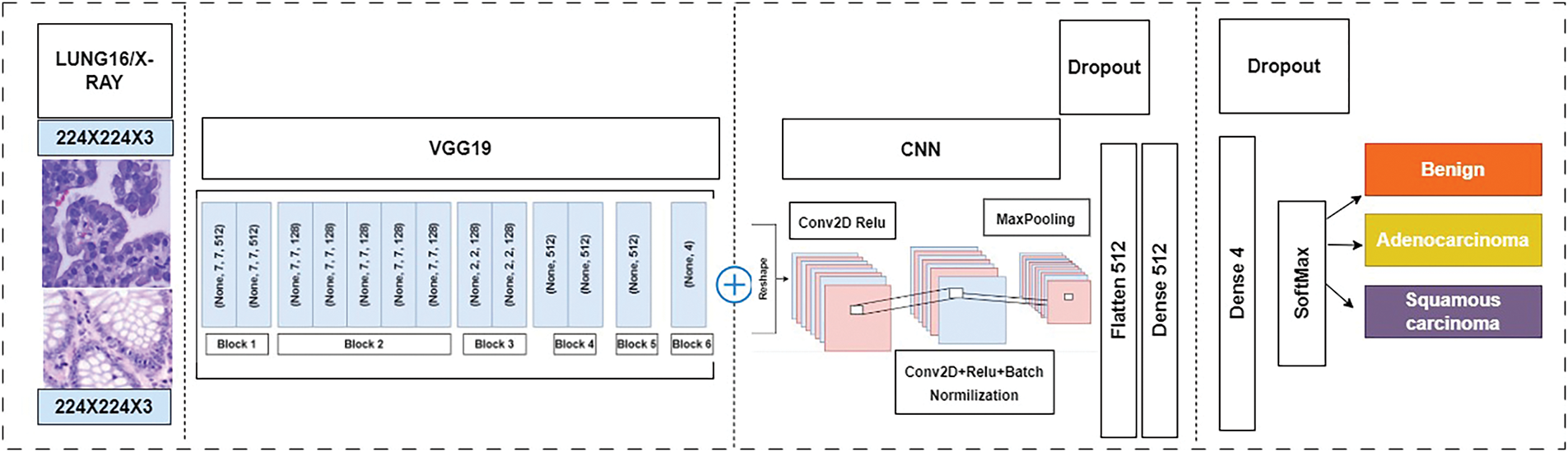
Figure 3: Proposed model combined CNN + VGG19 for lung cancer detection
Fig. 2 illustrates the process of using these algorithms and optimizers for lung cancer classification [54].
This approach efficiently compares various models’ performance in lung and colon cancer image classification tasks. Fig. 3 [54] illustrates the methodology employed in the research, which combines a conventional neural network (CNN) with a pre-trained VGG19 model that was initially proposed in [54]. In addition, for the training and testing model, this study utilizes the LC25000 dataset, allocating 75% for training, 12.5% for testing, and 12.5% for validation. It also implements five-fold cross-validation, where the dataset is divided into five-fold, and one-fold is utilized for testing to ensure robustness and generalizability. In contrast, the remaining folds were used for training and validation. In addition, the proposed method allows to evaluate the performance of various models, including AlexNet, VGG16, and ResNet-50.
A. Dataset Preparation
The datasets that are used are first augmented, and the dataset is a comprehensive collection of 25,000 images with five distinct classes. Each class contains 5000 images to ensure that there is a balance between the classes and that more photographs are utilized than necessary. The images are high quality with a pixel size of 768 × 768 and are stored in jpeg format for efficient compression and transfer. The dataset was converted into a single zip file with a total size of 1.85 GB to facilitate easy access and download. Once downloaded and extracted, the dataset is organized into a clear and intuitive folder structure. The main folder lung_colon_image_set contains two primary subfolders: colon_image_sets and lung_image_sets. These subfolders are further divided into secondary subfolders, each containing specific types of images. Within the colon_image_sets folder, there are two subfolders: ‘colon_aca’, housing 5000 images of colon adenocarcinomas, and colon_n, containing 5000 images of benign colonic tissues. In the lung_image_sets there are three folders: (lung_aca, 5000 images of adenocarcinomas), ‘lung_aca’, featuring 5000 images of lung squamous cell carcinomas, and ‘lung_n’, with 5000 images of benign lung tissues [48]. Fig. 1 shows the category of lung and colon histopathology cancer diseases, and Fig. 4 depicts the random samples from the LC25000 dataset.

Figure 4: Number of layers and blocks in combined approach (CNN + VGG19)
B. Feature Fusion
All the Preprocessing Techniques are used with the help of an image data generator. The processing techniques used in the Image Data Generator are applied only to the training dataset.
i. Pixel Normalization
The normalization process involves a simple element-wise division of the pixel values by 255, the maximum possible value, applied uniformly across all color channels, regardless of the image’s original dynamic range [9].
ii. Pixel Centering
The image data processing pipeline involves a sequence application of normalization and centering transformations. Normalization rescales the pixel values to a standardized range, typically between 0 and 1, to mitigate feature dominance and facilitate model convergence. Subsequent centering involves subtracting the mean value from the normalized pixel values that remain within the desired range, maintaining the image’s visual integrity and preventing erroneous reading. Centering requires calculating the mean pixel value before subtracting it from the pixel values. The mean can be computed using various methods, including:
• Per Image Mean
• Per Mini Batch Mean (under SGD)
• Per Training Dataset Mean
In addition, the mean pixel value per image is calculated. For color images, centering can be applied either globally or locally:
Global centering: subtracting the mean pixel value computed across all color channels.
Local centering: subtracting the mean pixel value computed separately for each color channel
iii. Pixel Standardization
The distribution of pixel values often exhibits a normal or Gaussian profile characterized by a bell-shaped curve. The distribution can manifest at various levels, contains per image, per min batch, across the entire training, globally color channels, and locally per color channel. Transferring the pixel value distribution into a standard Gaussian distribution can be beneficial, involving both centering and scaling operations. This process is known as standardization. Standardization is often preferred over normalization and centering alone, as it yields zero-centered values with small input values, typically ranging from −3 to 3, depending on the dataset specifics [9].
C. Combined CNN + VGG19
This study combines CNN + VGG19 model lung and colon cancer diagnosis from histological images utilizing a VGG19 pre-trained model augmented with additional layers for enhanced feature extraction and classification. The architecture of the proposed model is illustrated in Fig. 5, with details of the layers and blocks used. The proposed model takes input image size 768 × 768 pixels, derived from lung and colon histological datasets. Initially, the VGG19 model performs preliminary feature extraction. This is followed by two custom CNN blocks designed for further feature refinement. Each CNN block contains Convolutional layers with ReLU activation, Batch normalization, Max pooling, and dropout layers to prevent overfitting. Following these CNN blocks, a flattened layer transforms the multi-dimensional output into a one-dimensional vector, which is then fed into a dense layer containing 512 neurons. The dense layer, equipped with a dropout mechanism, serves as the primary classifier, and the final dense layer, with neurons corresponding to the number of classes, uses a SoftMax activation function to classify the images for three classes.

Figure 5: Distribution of dataset
D. Hyperparameter Tuning
CNN comprises numerous hyperparameters and parameters such as weights, neurons, layers, stride, filter size and biases, learning rate, and optimization functions. Convolution layers are important for extracting deep features from the images [46]. Two types of filters: small filters used for filters focused on fine-grained details and large filters used for coarse-grained features.
The pioneering LeNet model, introduced by LeCun et al. in 1988, employed a 5 × 5 convolutional filter with a stride of 2 for zip code recognition, followed by a 2 × 2 pooling layer with a stride of 2; this groundbreaking work laid the foundation for future advancements. A seminal moment in deep learning occurred in 2012 when AlexNet, a convolutional neural network (CNN) architecture, triumphed in the ImageNet Large-scale visual recognition challenge (ILSVRC). This achievement marked a paradigm shift in computer vision, as AlexNet’s innovative design comprised five convolution layers. Three pooling layers and fully connected (FC) layers introduce the rectified linear unit (ReLU) activation function to CNNs. The visual Geometry Group (VGG) network, another influential architecture, is available in two variants: VGG18, consisting of 16 layers, and VGG19, comprising 19 layers. As the number of layers increases, the filter size decreases, demonstrating a tradeoff between depth and granularity. This research uses a suite of esteemed CNN architectures, including AlexNet, VGG16, ResNet-50, and VGG19, to classify lung cancer using the LC25000 dataset.
This research uses a suite of esteemed CNN architectures, including AlexNet, VGG16, ResNet-50, and VGG19, to classify lung cancer using the LC25000 dataset. The basic configuration of these pre-trained models is summarized in Table 4. The pros and cons of CNN-based pre-trained methods are discussed in Table 5, highlighting their advantages in training, improved performance, and high accuracy while also noting their limitations, including high memory requirements, slow training, and impractical training times for real-world applications.


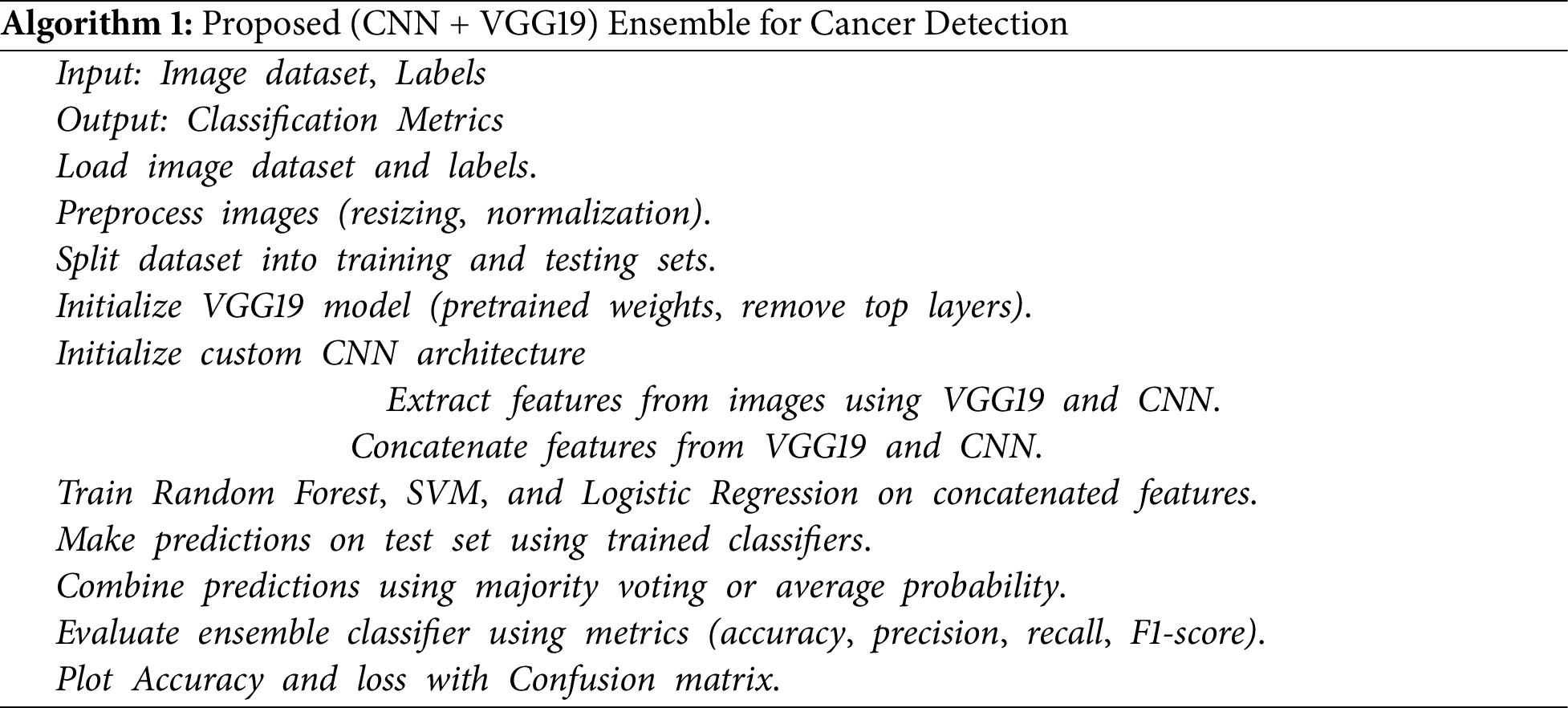
This study uses the LC25000 dataset to comprehensively evaluate the performance of the proposed model (CNN + VGG19) as shown in Algorithm 1, against the other fine-tuned or pre-trained models for lung cancer detection. Specifically, it trains the model on a three-class classification problem encompassing Adenocarcinoma, Squamous Cell Carcinoma, and Benign. The experimental design allocates 75% of the dataset for training and rigorously evaluates the model performance on the test set 12.5% using the metrics listed in Table 6. In addition, this research investigates implemented 5-fold cross-validation to ensure robustness and generalizability. Table 6 indicates all pre-trained models performed exceptionally well on the LC25000 dataset, achieving accuracy rates exceeding 90%. The proposed model achieved the highest accuracy, 99.04%. Surpassing the performance of individual pre-trained models. These findings demonstrate the effectiveness of using pre-trained models and combining CNN architectures for lung cancer detection.

This study comprehensively compares state-of-the-art CNN architectures—AlexNet, VGG16, VGG19, and ResNet-50 for lung cancer detection, utilizing various optimizers (SGD, Adam, and RMSprop). The performance of each architecture is thoroughly evaluated and validated in terms of accuracy, employing binary cross-entropy loss. The respective detection algorithms achieve the results presented. The performance is examined with Python 3.8, the Keras tool, a batch size of 20, 50 epochs, and a learning rate of 0.001. Several optimizers are utilized to analyze each design’s performance, and statistical metrics such as accuracy, specificity, sensitivity, and F1 score, as well as negative predictive value, false omission rate, and predictive value, are examined to determine how well each architecture detects lung cancer. This thorough examination facilitates the section on the best strategy for lung cancer diagnosis by illuminating the advantages and disadvantages of each design and optimizer combination.
This study assesses how well the indicated method classified nodules based on several well-recognized standards [15,27]. These metrics evaluate how well the model differentiates between benign lung tissue, lung squamous cell cancer, colon adenocarcinoma, lung adenocarcinoma, and benign lung tissue.
The capacity of the model to reliably detect malignant situations is observed in the image.
A key aspect of accurate sickness detection is the illustration’s ability to show how effectively the model can consistently identify instances of illness. A crucial measure, accuracy, shows the total percentage of correctly classified data. This study evaluates its specificity, AUC, and F1 score to assess the proposed model classification performance on lung and colon; specificity measures the accurate identification of lung and colon cancer, while AUC under the receiver operating characteristic (ROC) evaluates the model’s ability to distinguish between lung and colon. The following assessment metrics can be calculated in Table 6.
Table 6 presents various metrics utilized to evaluate the performance of lung cancer detection models. These metrics provide insights into different aspects of the model’s performance, enabling a comprehensive understanding of its strengths and weaknesses. The terms TP, TN, FN, and FP are employed in the subsequent calculations. Nodules that are correctly identified as benign and malignant are considered true positives. True negativity, or TN, is a metric that measures the number of benign nodules that are accurately recognized. The number of malignant nodules that were mistakenly diagnosed as benign is known as false negative, or FN. The term “false positive,” or “FP,” describes the quantity of benign nodules that were incorrectly classified as cancerous.
The novelty of this study lies in the integration of CNN and VGG19 for lung and colon cancer detection, combining deep feature extraction, high-performance filtering, and ensemble learning to enhance diagnostic accuracy. Unlike existing studies, it introduces a unique feature extraction technique using multiple transfer learning models for robust feature representation. In addition, a comprehensive evaluation of the LC25000 dataset across various metrics validates the effectiveness of the proposed approach.
Figs. 6 and 7 depict a comprehensive evaluation of the proposed CNN + VGG19 model and its comparison to other pre-trained CNN-based models, namely AlexNet, VGG16, VGG19, and ResNet50, for the classification of lung and colon cancer. The performance metrics demonstrated in Fig. 7 reveal that the proposed CNN + VGG19 approach surpassed other models, achieving an exceptional accuracy of 99.04%. VGG19 secured the second-highest accuracy of 99.02%, closely trailing the proposed model. This remarkable performance highlights the effectiveness of the CNN + VGG19 model in using the strengths of both architectures to improve cancer classification accuracy. The findings indicate that the proposed model’s ability to learn and adapt to complex patterns in medical images can lead to enhanced diagnostic precision, making it a valuable tool for clinical applications. Future research directions can include exploring the model’s generalizability across diverse cancer types and investigating the impact of transfer learning on its performance.
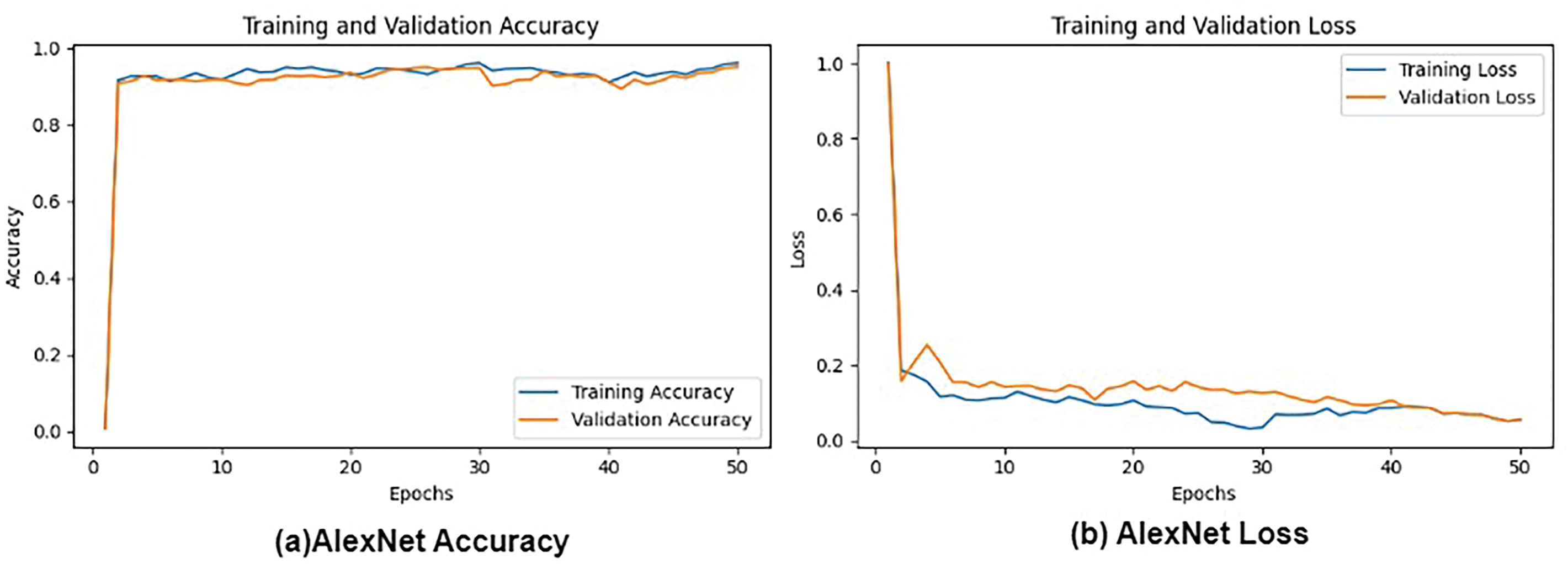
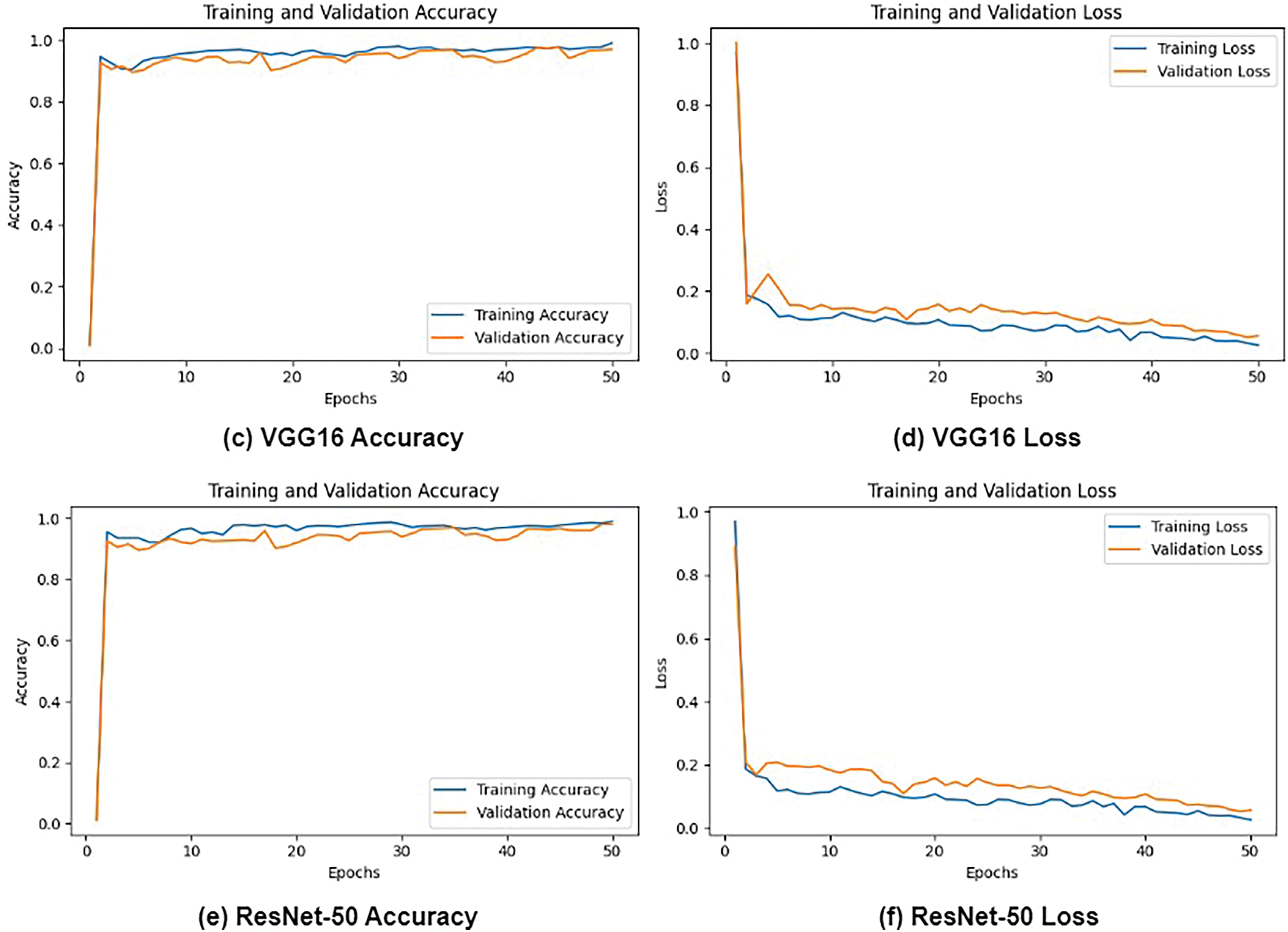
Figure 6: Loss and Accuracy: (a) AlexNet Accuracy, (b) AlexNet Loss and, (c) VGG16 Accuracy, (d) VGG16 Loss, (e) ResNet-50 Accuracy, and (f) ResNet-50 loss
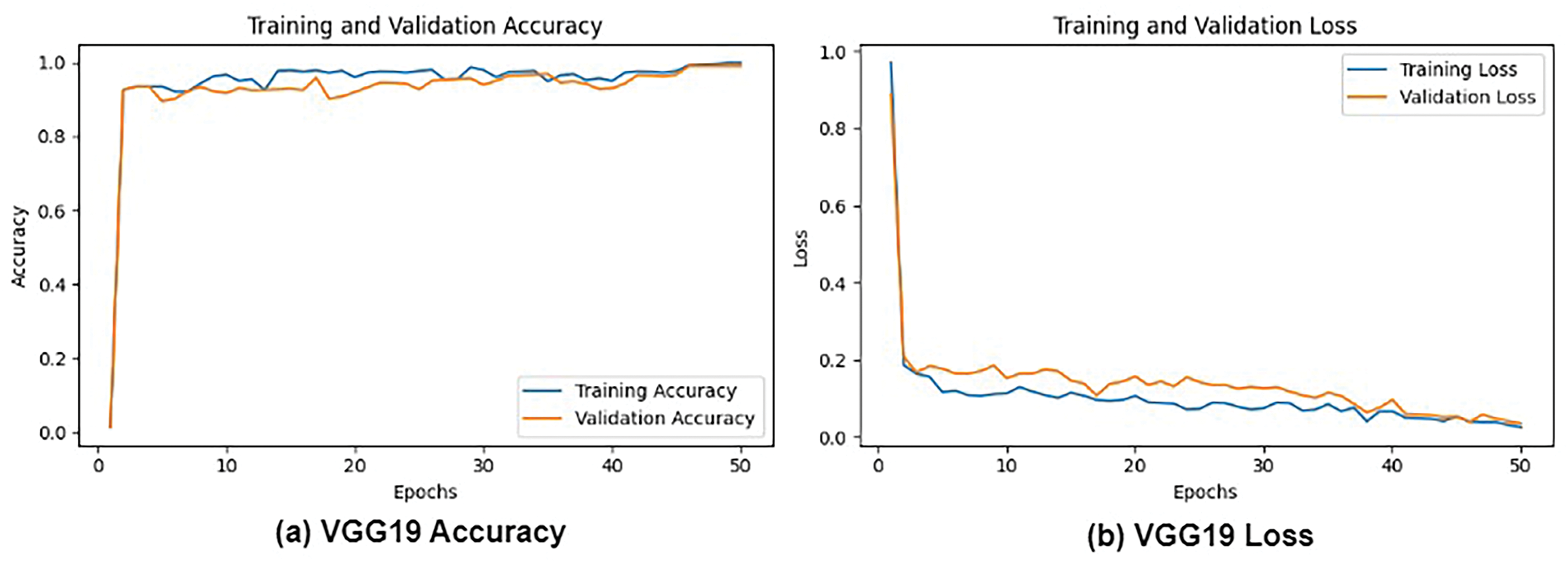

Figure 7: Loss and accuracy: (a) VGG19 Accuracy, (b) VGG19 Loss and, (c) CNN + VGG19 Accuracy, and (d) CNN+VGG19 Loss
Fig. 8 presents the confusion matrices for all evaluated models, providing a detailed comparison of the performance of AlexNet, VGG16, VGG19, ResNet50, and the proposed CNN+ VGG19 model in classifying three types of cancer diseases: Lung_n (normal lung tissue), Lung_aca (lung adenocarcinoma), and Lung_scc (lung squamous cell carcinoma). The confusion matrices reveal that the proposed CNN + VGG19 model achieves outstanding results, demonstrating superior performance across all three cancer types. The proposed model exhibits exceptional accuracy in classifying Lung_scc, with significantly higher true positive and lower false negative rates than the other models. The results also indicate that the proposed model distinguishes between normal lung tissue (Lung_n) and cancerous images, achieving a high true negative rate and low false positive rate. This is critical for reducing the likelihood of misdiagnosis and ensuring accurate cancer detection. The results demonstrate exceptional diagnostic performance of the proposed model, with AUC values exceeding 0.99 for all three classes.
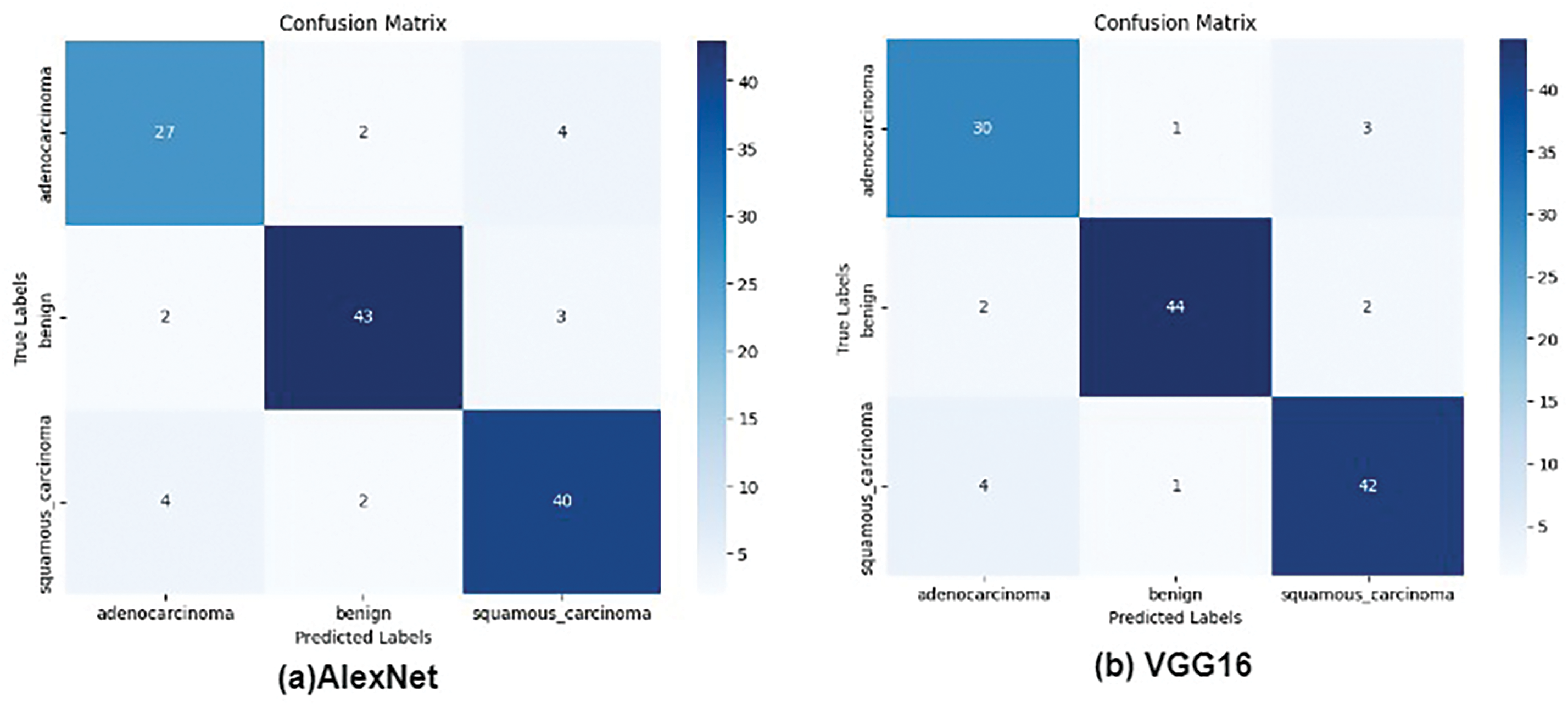
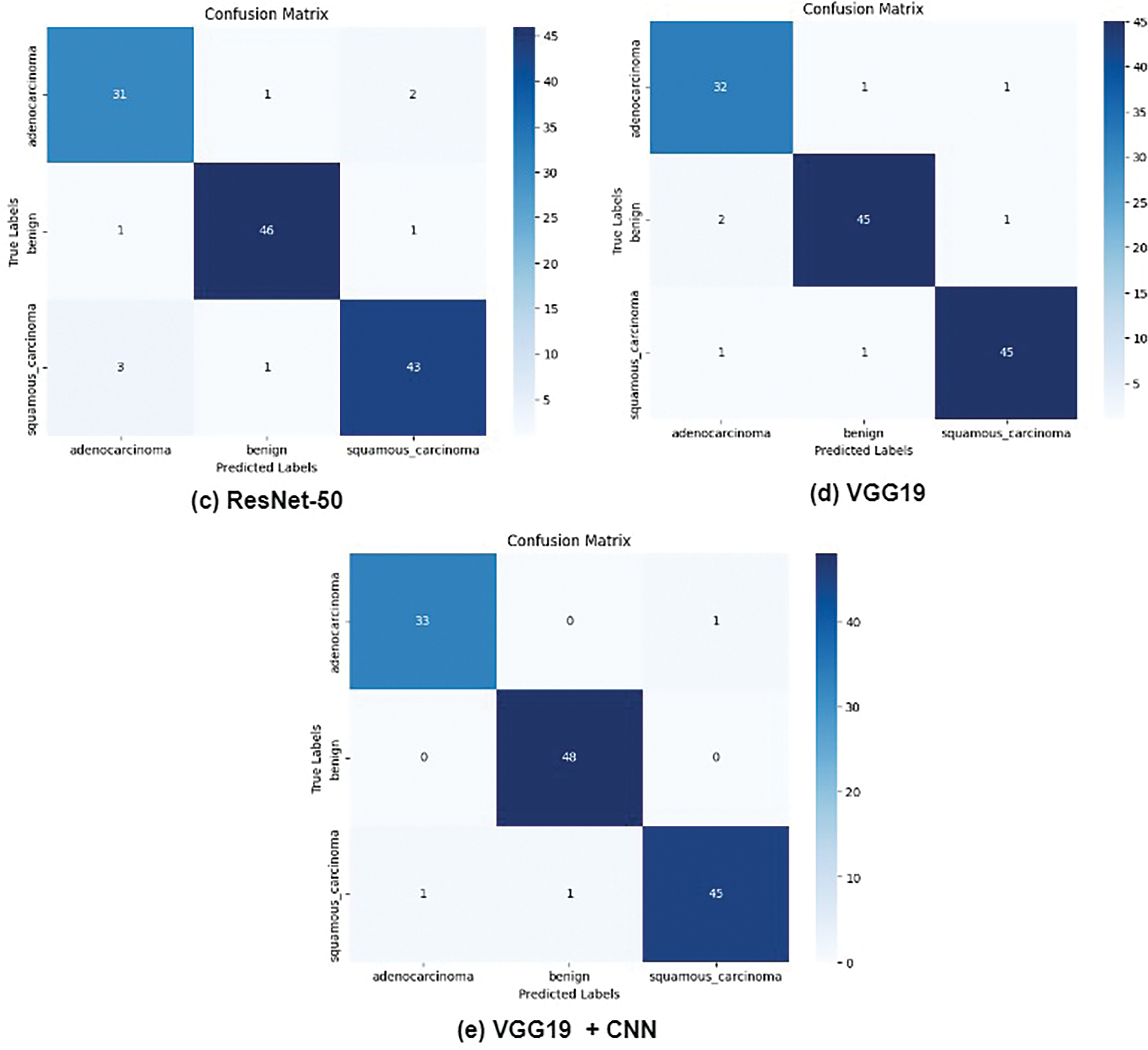
Figure 8: True and false prediction of pre-trained models for cancer classes
Fig. 9 presents the validation performance of various hyperparameters on pre-trained models (AlexNet, VGG16, VGG19, and ResNet-50) for lung cancer detection using different optimizers (SGD, Adam, and RMSprop). The proposed approach achieved outstanding results with Adam (99.04% accuracy) and RMSprop (99.03% accuracy), but SGD performance is slightly low.

Figure 9: Validation performance with optimizers (Adam, RMSprop, and SGD)
Fig. 10 compares the actual cancer images and the predicted images generated by the proposed CNN+ VGG19 model for lung cancer detection. The figure demonstrates the exceptional performance of the proposed model in accurately detecting and predicting lung cancer, particularly for lung adenocarcinoma (lung_aca) and lung squamous cell carcinoma (lung_scc).
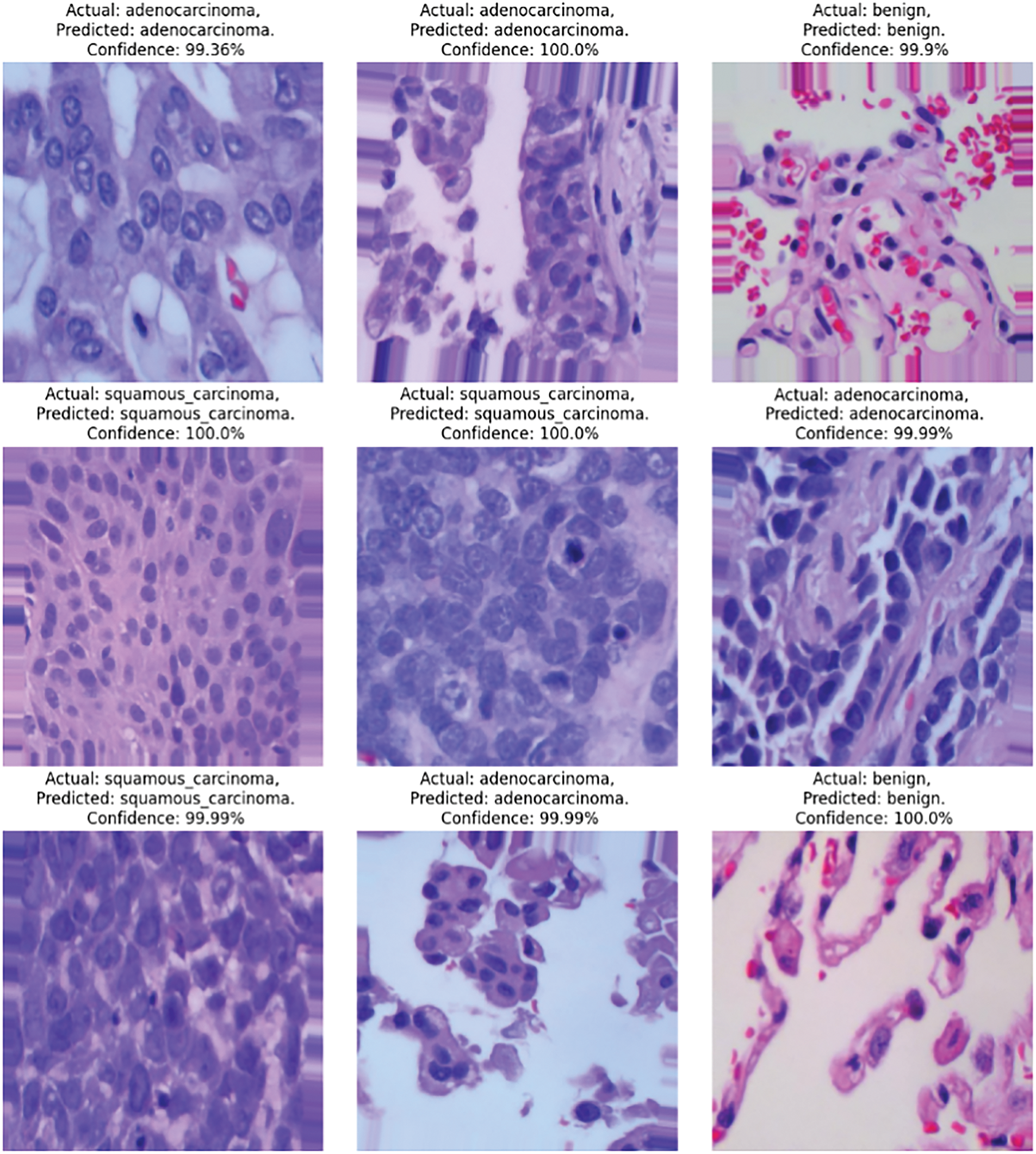
Figure 10: Proposed model prediction on lung cancer detection
Fig. 11 illustrates the training and testing accuracy of various deep learning models, where the graph indicates a visual comparison of the performance of these models, highlighting the superiority of the proposed (VGG19 + CNN) model. The training accuracy curves demonstrate that the proposed model achieves rapid convergence and consistently high accuracy throughout the training process, indicating effective learning and adaptation to the dataset. In contrast, the other models exhibit slower convergence and lower training accuracy, indicating limitations in their ability to capture complex patterns in the data.

Figure 11: Training and testing of CNNs-based pretrained models
The study evaluates the proposed CNN + VGG19 model against pre-trained models (AlexNet, VGG16, VGG19, and ResNet50) for lung and colon cancer classification. Results show that CNN + VGG19 outperforms all models, achieving 99.04% accuracy, with VGG19 closely following at 99.02%. Confusion matrices highlight its superior classification of cancer types, reducing false positives and misdiagnoses. The model performs exceptionally well with Adam and RMSprop optimizers, as shown in the validation tests. ROC curves confirm strong class separation, while training accuracy graphs demonstrate rapid convergence and high performance, highlighting the model’s effectiveness in medical diagnostics.
Table 7 shows that the proposed (CNN + VGG19) model achieves the highest performance across all classes, with F1-scores of 99% for Lung_n and Lung_scc, and 98% for Lung_aca. ResNet50 and VGG19 also perform well, achieving F1 scores above 97% for all classes. In contrast, AlexNet and VGG16 have relatively lower performance, with F1 scores ranging from 94% to 96% across classes.

Table 8 lists an analysis of the proposed study compared to previous techniques. The table discusses how researchers have proposed various deep learning-based methods for cancer diagnosis using histopathological images, achieving high accuracy rates. For instance, Masud et al. and Hatuwal et al. reported accuracy rates of 96.38% and 97.33%, respectively, using CNN-based approaches for lung cancer diagnosis, whereas the proposed method, combining CNN and VGG19, outperforms existing methods with an accuracy rate of 99.04% for lung and colon cancer diagnosis.
5 Conclusion and Future Research Directions
This study aims to develop an automated detection and classification system for lung and colon cancer images to localize the cells accurately. It employs transfer learning-based feature extraction. The proposed method combines CNN with VGG19 to enhance performance. TensorFlow with Keras API is applied on Google Laboratory to evaluate performance metrics such as precision, recall specificity, F1 score, and accuracy. The proposed system, using the fusion features of the VGG-19 model and handcrafted features, achieved a precision of 98%, recall of 98.64%, and accuracy of 99.04% for early diagnosis of the LC25000 dataset images.
Although this research provides valuable insights into the classification of lung and colon cancer, it has several limitations and warrants consideration; firstly, the dataset’s properties, including size, quality, and diversity, can significantly impact the efficiency of the models. This study recommends using larger and more diverse datasets, potentially integrating data from multiple sources to enhance model generalization and robustness to alleviate these constraints. In addition, it focuses only on image-based classification, ignoring potential interaction with other modalities such as genomic, clinical, or radiological data. Future research should explore multimodal approaches to improve classification accuracy, adaptability, and patient outcomes. Integrating diverse data sources can uncover complex patterns and relationships, leading to more accurate diagnoses and personalized treatment plans. In addition, this research employs explainable (XAI) techniques to enhance model interpretability, which remains challenging, particularly in complex medical domains. Further research is essential to develop more interpretable models and refine existing XAI methods, providing deep insights into model decision-making processes and facilitating trust in AI-driven diagnostics. In addition, the study concentrates on two types of cancer: lung and colon. Expanding the proposed approach to encompass additional forms of cancer will significantly enhance its relevance and impact in oncology; investigating the model transferability across diverse healthcare settings, patient demographics, and cancer types is critical for real-world deployment and broad applicability. It can further advance the accuracy, interpretability, adaptability, and clinical utility of the models, ultimately improving patient outcomes and transforming cancer diagnosis and treatment by addressing these limitations and exploring new avenues of research.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: Conceptualization & Design: Majdi Rawashdeh, Meryem Abouali, and Muath A. Obaidat. Data Collection: Majdi Rawashdeh and Dhia Eddine Salhi. Methodology: Majdi Rawashdeh, Meryem Abouali, and Muath A. Obaidat. Formal Analysis: Majdi Rawashdeh and Kutub Thakur. Supervision: Majdi Rawashdeh. Writing: Majdi Rawashdeh, Muath A. Obaidat, Meryem Abouali, Dhia Eddine Salhi, and Kutub Thakur. Review and Editing: Majdi Rawashdeh and Muath A. Obaidat. Discussion: Majdi Rawashdeh, Muath A. Obaidat, Meryem Abouali, Dhia Eddine Salhi, and Kutub Thakur. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data is available upon request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA A Cancer J Clinicians. 2021;71(1):7–33. doi:10.3322/caac.21654. [Google Scholar] [PubMed] [CrossRef]
2. Ghasemi S, Akbarpour S, Farzan A, Ali Jabraeil Jamali M. Automatic pulmonary nodule detection on computed tomography images using novel deep learning. Multimed Tools Appl. 2024;83(18):55147–73. doi:10.1007/s11042-023-17502-3. [Google Scholar] [CrossRef]
3. Trung NT, Trinh DH, Trung NL, Luong M. Low-dose CT image denoising using deep convolutional neural networks with extended receptive fields. Signal Image Video Processing. 2022;16(7):1963–71. doi:10.1007/s11760-022-02157-8. [Google Scholar] [CrossRef]
4. Wang Q, Zuo M. A novel variational optimization model for medical CT and MR image fusion. Signal Image Video Process. 2023;17(1):183–90. doi:10.1007/s11760-022-02220-4. [Google Scholar] [CrossRef]
5. Jiang H, Ma H, Qian W, Gao M, Li Y. An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J Biomed Health Inform. 2017;22(4):1227–37. doi:10.1109/JBHI.2017.2725903. [Google Scholar] [PubMed] [CrossRef]
6. Gupta P, Shukla AP. Improving accuracy of lung nodule classification using AlexNet model. In: 2021 International Conference on Innovative Computing, Intelligent Communication and Smart Electrical Systems (ICSES); 2021 Sep 24–25; Chennai, India: IEEE; 2021. p. 1–6. doi:10.1109/ICSES52305.2021.9633903. [Google Scholar] [CrossRef]
7. Khehrah N, Farid MS, Bilal S, Khan MH. Lung nodule detection in CT images using statistical and shape-based features. J Imaging. 2020;6(2):6. doi:10.3390/jimaging6020006. [Google Scholar] [PubMed] [CrossRef]
8. Xie Y, Zhang J, Xia Y, Fulham M, Zhang Y. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inf Fusion. 2018;42(2):102–10. doi:10.1016/j.inffus.2017.10.005. [Google Scholar] [CrossRef]
9. Nasser IM, Abu-Naser SS. Lung cancer detection using artificial neural network. Int J Eng Inf Syst. 2019;3(3):17–23. [Google Scholar]
10. Mohandass G, Hari Krishnan G, Selvaraj D, Sridhathan C. Lung cancer classification using optimized attention-based convolutional neural network with DenseNet-201 transfer learning model on CT image. Biomed Signal Process Contr. 2024;95(3):106330. doi:10.1016/j.bspc.2024.106330. [Google Scholar] [CrossRef]
11. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–324. doi:10.1109/5.726791. [Google Scholar] [CrossRef]
12. Zhang S, Sun F, Wang N, Zhang C, Yu Q, Zhang M, et al. Computer-aided diagnosis (CAD) of pulmonary nodule of thoracic CT image using transfer learning. J Digit Imaging. 2019;32(6):995–1007. doi:10.1007/s10278-019-00204-4. [Google Scholar] [PubMed] [CrossRef]
13. Zhao X, Liu L, Qi S, Teng Y, Li J, Qian W. Agile convolutional neural network for pulmonary nodule classification using CT images. Int J Comput Assist Radiol Surg. 2018;13(4):585–95. doi:10.1007/s11548-017-1696-0. [Google Scholar] [PubMed] [CrossRef]
14. Sharma G, Jadon VK. Classification of image with convolutional neural network and TensorFlow on CIFAR-10 dataset. In: International Conference on Women Researchers in Electronics and Computing; 2023; Singapore: Springer Nature. [Google Scholar]
15. Polat H, Danaei Mehr H. Classification of pulmonary CT images by using hybrid 3D-deep convolutional neural network architecture. Appl Sci. 2019;9(5):940. doi:10.3390/app9050940. [Google Scholar] [CrossRef]
16. Ali AS, Iqbal MM, Khan AH, Hameed N, Bibi S. Lung cancer detection using convolutional neural networks from computed tomography images. J Comput Biomed Inform. 2023;6(1):133–43. [Google Scholar]
17. Lin CJ, Li YC. Lung nodule classification using taguchi-based convolutional neural networks for computer tomography images. Electronics. 2020;9(7):1066. doi:10.3390/electronics9071066. [Google Scholar] [CrossRef]
18. Al-Yasriy HF, AL-Husieny MS, Mohsen FY, Khalil EA, Hassan ZS. Diagnosis of lung cancer based on CT scans using CNN. IOP Conf Ser: Mater Sci Eng. 2020;928(2):022035. doi:10.1088/1757-899X/928/2/022035. [Google Scholar] [CrossRef]
19. Patil NC. Lung cancer detection using CNN VGG19+ model. Jes. 2024;20(3):541–50. doi:10.52783/jes.2981. [Google Scholar] [CrossRef]
20. Elnakib A, Amer HM, Abou-Chadi FEZ. Early lung cancer detection using deep learning optimization. Int J Onl Eng. 2020;16(6):82–94. doi:10.3991/ijoe.v16i06.13657. [Google Scholar] [CrossRef]
21. Geng L, Zhang S, Tong J, Xiao Z. Lung segmentation method with dilated convolution based on VGG-16 network. Comput Assist Surg. 2019;24(sup2):27–33. doi:10.1080/24699322.2019.1649071. [Google Scholar] [PubMed] [CrossRef]
22. Pang S, Meng F, Wang X, Wang J, Song T, Wang X, et al. VGG16-T: a novel deep convolutional neural network with boosting to identify pathological type of lung cancer in early stage by CT images. Int J Comput Intell Syst. 2020;13(1):771–80. doi:10.2991/ijcis.d.200608.001. [Google Scholar] [CrossRef]
23. Agarwal A, Patni K, Rajeswari D. Lung cancer detection and classification based on alexnet CNN. In: Proceedings of the 2021 6th International Conference on Communication and Electronics Systems (ICCES); 2021 Jul 8–10; Coimbatre, India. p. 1390–7. [Google Scholar]
24. Saha A, Ganie SM, Pramanik PKD, Yadav RK, Mallik S, Zhao Z. VER-Net: a hybrid transfer learning model for lung cancer detection using CT scan images. BMC Med Imaging. 2024;24(1):120. doi:10.1186/s12880-024-01238-z. [Google Scholar] [PubMed] [CrossRef]
25. Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: cnn architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–98. doi:10.1109/TMI.2016.2528162. [Google Scholar] [PubMed] [CrossRef]
26. Zheng G, Han G, Soomro NQ. An inception module CNN classifiers fusion method on pulmonary nodule diagnosis by signs. Tsinghua Sci Technol. 2020;25(3):368–83. doi:10.26599/TST.2019.9010010. [Google Scholar] [CrossRef]
27. Lin H, Tao S, Song S, Liu H. An improved yolov3 algorithm for pulmonary nodule detection. In: 2021 IEEE 4th Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC); 2021 Jun 18–20; Chongqing, China: IEEE; 2021. p. 1068–72. doi:10.1109/imcec51613.2021.9482291. [Google Scholar] [CrossRef]
28. Bu Z, Zhang X, Lu J, Lao H, Liang C, Xu X, et al. Lung nodule detection based on YOLOv3 deep learning with limited datasets. Mol Cell Biomech. 2022;19(1):17–28. doi:10.32604/mcb.2022.018318. [Google Scholar] [CrossRef]
29. Zhang X, Lee VCS, Rong J, Liu F, Kong H. Multi-channel convolutional neural network architectures for thyroid cancer detection. PLoS One. 2022;17(1):e0262128. doi:10.1371/journal.pone.0262128. [Google Scholar] [PubMed] [CrossRef]
30. Leo M, Cacagnì P, Signore L, Benincasa G, Laukkanen MO, Distante C. Improving colon carcinoma grading by advanced CNN models. In: International Conference on Image Analysis and Processing; 2022; Cham, Switzerland: Springer. p. 233–44. [Google Scholar]
31. Lanjewar M, Panchbhai KG, Panem C. Lung cancer detection from CT scans using modified DenseNet with feature selection methods and ML classifiers. Expert Syst Appl. 2023;224(8):119961. doi:10.1016/j.eswa.2023.119961. [Google Scholar] [CrossRef]
32. Maurya SP, Sisodia PS, Mishra R, Singh DP. Performance of machine learning algorithms for lung cancer prediction: a comparative approach. Sci Rep. 2024;14(1):18562. doi:10.1038/s41598-024-58345-8. [Google Scholar] [PubMed] [CrossRef]
33. Hossain N, Anjum N, Alam M, Rahman MH, Taluckder MS, et al. Performance of machine learning algorithms for lung cancer prediction: a comparative study. Int J Med Sci Public Health Res. 2024;5(11):41–55. doi:10.37547/ijmsphr/Volume05Issue11-05. [Google Scholar] [CrossRef]
34. Javed R, Abbas T, Khan AH, Daud A, Bukhari A, Alharbey R. Deep learning for lungs cancer detection: a review. Artif Intell Rev. 2024;57(8):197. doi:10.1007/s10462-024-10807-1. [Google Scholar] [CrossRef]
35. Liz-López H, de Sojo-Hernández ÁA, D’Antonio-Maceiras S, Díaz-Martínez MA, Camacho D. Deep learning innovations in the detection of lung cancer: advances, trends, and open challenges. Cogn Comput. 2025;17(2):67. doi:10.1007/s12559-025-10408-2. [Google Scholar] [CrossRef]
36. Perumal V, Narayanan V, Rajasekar SJS. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl Intell. 2021;51(1):341–58. doi:10.1007/s10489-020-01831-z. [Google Scholar] [PubMed] [CrossRef]
37. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296(2):65–71. [Google Scholar]
38. Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19). Eur Radiol. 2021;31(8):6096–104. doi:10.1007/s00330-021-07715-1. [Google Scholar] [PubMed] [CrossRef]
39. Xu X, Jiang X, Ma C, Du P, Li X, Lv S, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–9. doi:10.1016/j.eng.2020.04.010. [Google Scholar] [PubMed] [CrossRef]
40. Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans Comput Biol Bioinform. 2021 Nov 1–Dec;18(6):2775–80. doi:10.1109/TCBB.2021.3065361.. [Google Scholar] [PubMed] [CrossRef]
41. Wang B, Jin S, Yan Q, Xu H, Luo C, Wei L, et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system. Appl Soft Comput. 2021;98(10223):106897. doi:10.1016/j.asoc.2020.106897. [Google Scholar] [PubMed] [CrossRef]
42. Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK. Diagnosing COVID19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv:2004.00038. 2020. [Google Scholar]
43. Zhang J, Xie Y, Li Y, Shen C, Xia Y. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv:2003.12338. 2020. [Google Scholar]
44. Khalifa NEM, Taha MHN, Hassanien AE, Elghamrawy S. Detection of coronavirus (COVID-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest X-ray dataset. arXiv:2004.01184. 2020. [Google Scholar]
45. Hammoudi K, Benhabiles H, Melkemi M, Dornaika F, Arganda-Carreras I, Collard D, et al. Deep learning on chest X-ray images to detect and evaluate pneumonia cases at the era of COVID-19. arXiv:2004.03399. 2020. [Google Scholar]
46. Farooq M, Hafeez A. COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs. arXiv:2003.14395. 2020. [Google Scholar]
47. Sultana Z, Foysal M, Islam S, Al Foysal A. Lung cancer detection and classification from chest CT images using an ensemble deep learning approach. In: 2024 6th International Conference on Electrical Engineering and Information & Communication Technology (ICEEICT); 2024 May 2–4; Dhaka, Bangladesh: IEEE; 2024. p. 364–9. doi:10.1109/ICEEICT62016.2024.10534468. [Google Scholar] [CrossRef]
48. Borkowski A, Bui M, Thomas L, Wilson C, DeLand L, Mastorides S. Lung and colon cancer histopathological image dataset (lc25000). arXiv:1912.12142. 2019. [Google Scholar]
49. Lu Y, Liang H, Shi S, Fu X. Lung cancer detection using a dilated CNN with VGG16. In: 2021 4th International Conference on Signal Processing and Machine Learning; 2021; Beijing, China: ACM. p. 45–51. doi:10.1145/3483207.3483215. [Google Scholar] [CrossRef]
50. Bhattacharyya N, Satish AS. A study of accuracy in detection of lung cancer through CNN models. Des Eng. 2021;9:1250–70. [Google Scholar]
51. Ramana K, Kumar MR, Sreenivasulu K, Gadekallu TR, Bhatia S, Agarwal P, et al. Early prediction of lung cancers using deep saliency capsule and pre-trained deep learning frameworks. Front Oncol. 2022;12:886739. doi:10.3389/fonc.2022.886739. [Google Scholar] [PubMed] [CrossRef]
52. Shandilya S, Nayak SR. Analysis of lung cancer by using deep neural network. In: Innovation in Electrical Power Engineering, Communication, and Computing Technology: Proceedings of Second IEPCCT 2021; 2022; Singapore: Springer. [Google Scholar]
53. Abd Al-Ameer AA, Abdulameer Hussien G, Al Ameri HA. Lung cancer detection using image processing and deep learning. Indones J Electr Eng Comput Sci. 2022;28(2):987. doi:10.11591/ijeecs.v28.i2.pp987-993. [Google Scholar] [CrossRef]
54. Rawashdeh M, Obaidat MA, Abouali M, Thakur K. A deep learning-driven approach for detecting lung and colon cancer using pre-trained neural networks. In: 2024 IEEE 21st International Conference on Smart Communities: Improving Quality of Life using AI, Robotics and IoT (HONET); 2024 Dec 3–5; Doha, Qatar: IEEE; 2024. p. 183–8. doi:10.1109/HONET63146.2024.10822988. [Google Scholar] [CrossRef]
55. Masud M, Sikder N, Nahid AA, Bairagi AK, AlZain MA. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors. 2021;21(3):748. doi:10.3390/s21030748. [Google Scholar] [PubMed] [CrossRef]
56. Kumar N, Sharma M, Singh VP, Madan C, Mehandia S. An empirical study of handcrafted and dense feature extraction techniques for lung and colon cancer classification from histopathological images. Biomed Signal Process Contr. 2022;75(1):103596. doi:10.1016/j.bspc.2022.103596. [Google Scholar] [CrossRef]
57. Mangal S, Chaurasia A, Khajanchi A. Convolution neural networks for diagnosing colon and lung cancer histopathological images. arXiv:2009.03878. 2020. [Google Scholar]
58. Liang M, Ren Z, Yang J, Feng W, Li B. Identification of colon cancer using multi-scale feature fusion convolutional neural network based on shearlet transform. IEEE Access. 2020;8:208969–77. doi:10.1109/ACCESS.2020.3038764. [Google Scholar] [CrossRef]
59. Bukhari SUK, Syed A, Bokhari SKA, Hussain SS, Armaghan SU, Shah SSH. The histological diagnosis of colonic adenocarcinoma by applying partial self-supervised learning. MedRxiv. 2020. doi:10.1101/2020.08.15.20175760. [Google Scholar] [CrossRef]
60. Sarwinda D, Bustamam A, Paradisa RH, Argyadiva T, Mangunwardoyo W. Analysis of deep feature extraction for colorectal cancer detection. In: 2020 4th International Conference on Informatics and Computational Sciences (ICICoS); 2020 Nov 10–11; Semarang, Indonesia: IEEE; 2020. p. 1–5. doi:10.1109/icicos51170.2020.9298990. [Google Scholar] [CrossRef]
61. Hatuwal BK, Thapa HC. Lung cancer detection using convolutional neural network on histopathological images. Int J Comput Trends Technol. 2020;68(10):21–4. doi:10.14445/22312803/IJCTT-V68I10P104. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools