 Open Access
Open Access
ARTICLE
A Lightweight Explainable Deep Learning for Blood Cell Classification
1 Faculty of Information Technology, Vinh Long University of Technology Education, Vinh Long, 85000, Vietnam
2 Center for Technology Application and Transfer, Vinh Long University of Technology Education, Vinh Long, 85000, Vietnam
3 Faculty of Information Technology, Vinh Long University of Technology Education, Vinh Long, 85000, Vietnam
* Corresponding Author: Anh-Cang Phan. Email:
(This article belongs to the Special Issue: Intelligent Medical Decision Support Systems: Methods and Applications)
Computer Modeling in Engineering & Sciences 2025, 145(2), 2435-2456. https://doi.org/10.32604/cmes.2025.070419
Received 15 July 2025; Accepted 09 October 2025; Issue published 26 November 2025
Abstract
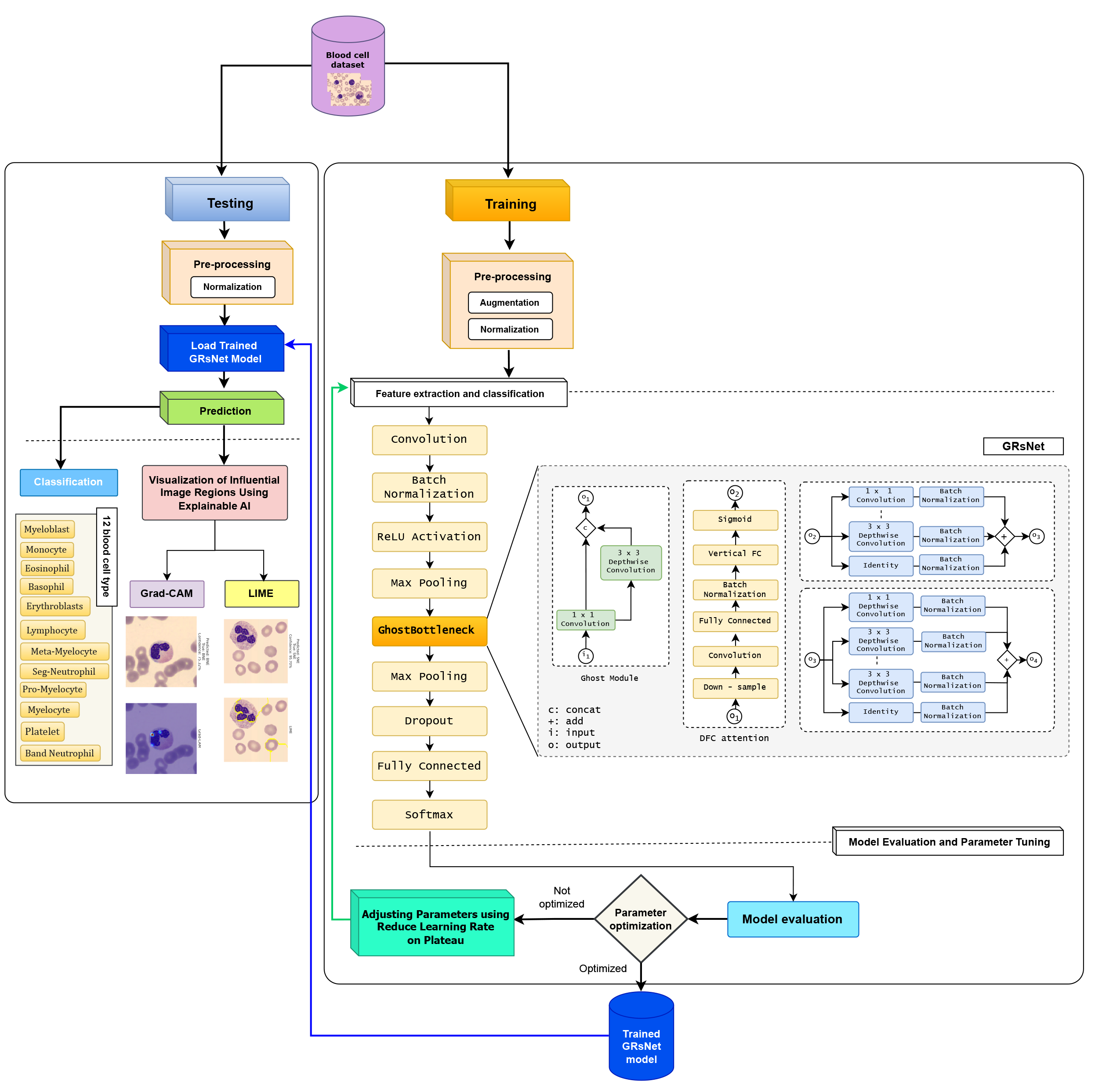
Blood cell disorders are among the leading causes of serious diseases such as leukemia, anemia, blood clotting disorders, and immune-related conditions. The global incidence of hematological diseases is increasing, affecting both children and adults. In clinical practice, blood smear analysis is still largely performed manually, relying heavily on the experience and expertise of laboratory technicians or hematologists. This manual process introduces risks of diagnostic errors, especially in cases with rare or morphologically ambiguous cells. The situation is more critical in developing countries, where there is a shortage of specialized medical personnel and limited access to modern diagnostic tools. High testing costs and delays in diagnosis hinder access to quality healthcare services. In this context, the integration of Artificial Intelligence (AI), particularly Explainable AI (XAI) based on deep learning, offers a promising solution for improving the accuracy, efficiency, and transparency of hematological diagnostics. In this study, we propose a Ghost Residual Network (GRsNet) integrated with XAI techniques such as Gradient-weighted Class Activation Mapping (Grad-CAM), Local Interpretable Model-Agnostic Explanations (LIME), and SHapley Additive exPlanations (SHAP) for automatic blood cell classification. These techniques provide visual explanations by highlighting important regions in the input images, thereby supporting clinical decision-making. The proposed model is evaluated on two public datasets: Naturalize 2K-PBC and Microscopic Blood Cell, achieving a classification accuracy of up to 95%. The results demonstrate the model’s strong potential for automated hematological diagnosis, particularly in resource-constrained settings. It not only enhances diagnostic reliability but also contributes to advancing digital transformation and equitable access to AI-driven healthcare in developing regions.Graphic Abstract
Keywords
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools