 Open Access
Open Access
REVIEW
Aptamer based detection and separation platforms for ochratoxin A: A systematic review
1 Laboratory of Bioanalysis, Institute of Biochemistry & Biophysics, University of Tehran, Tehran, Iran
2 Baqiyatallah Research Center for Gastroenterology and Liver Diseases (BRCGL), Baqiyatallah University of Medical Sciences, Tehran, Iran
3 Applied Virology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran
4 Molecular Biology Research Center, Systems Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran
* Corresponding Authors: REZA RANJBAR. Email: ; MOHAMMAD HEIAT. Email:
BIOCELL 2022, 46(12), 2537-2557. https://doi.org/10.32604/biocell.2022.021373
Received 11 January 2022; Accepted 11 April 2022; Issue published 10 August 2022
Abstract
Ochratoxin A (OTA), one of the most dangerous mycotoxins for human health, has been subjected to numerous studies for separation and detection in minimal amounts. Aptamers as novel recognition elements have been employed to fabricate ultrasensitive biosensors for the detection of OTA and designing delicate analytical tools. This review attempted to comprehensively examine all reported aptamer-based detection and separation platforms for ochratoxin. The most relevant databases were considered to discover all specific aptamers for dealing with OTA. Aptamer-based detection and separation devices specified for OTA were searched for, analyzed, discussed, and classified based on their specifications. The optical aptasensors have gathered a higher interest than electrochemical aptasensors, which can achieve a lower limit of detections. Moreover, some extraction platforms based on these aptamers were also found. However, aptamer-based devices seem to have some challenges in their application.Keywords
Ochratoxins belong to a family of toxic secondary metabolites produced by several species of fungi such as Aspergillus and Penicillium spp. Ochratoxins A, B, and C are members of this family with abundance in specimens. Ochratoxins are ubiquitous fungal toxins in a wide variety of poorly stored agricultural supplies, ranging from cereal grains to dried fruits to wine and coffee (Bui-Klimke and Wu, 2015). They threaten human and animal health by impacting food and nutrition safety and can affect food and agricultural economics (Heussner and Bingle, 2015).
The structure of ochratoxin A (OTA), a chlorophenolic mycotoxin, is chemically stable (Fig. 1) and comprises a phenylalanine moiety and dihydro-isocoumarin ring.

Figure 1: Chemical structure of ochratoxin A (OTA).
OTA exerts several toxic effects such as immunotoxicity, nephrotoxicity, hepatotoxicity, neurotoxicity, teratogenicity, and carcinogenicity (el Khoury and Atoui, 2010). It enters the human food chain through livestock products as a result of animal feeding on contaminated nutrition.
Children who consume large amounts of milk daily may have higher daily intakes of OTA (Muñoz et al., 2014).
Avoiding the risk of OTA exposure and the detection and quantitation of OTA levels are issues of great significance.
Currently, there are various analytical methods to detect OTA in as low as microgram levels. The most typical methods to detect and analyze OTA include immunological, such as enzyme-linked immunosorbent assay (ELISA) (Sun et al., 2019), Radio-immunoassay (RIA) (Rousseau et al., 1985), and chromatographic methods such as thin-layer chromatography (Pittet and Royer, 2002), liquid chromatography (LC), liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) (Chung and Kwong, 2019), gas chromatography (Olsson et al., 2002), and high-performance liquid chromatography (HPLC) (Sibanda et al., 2001).
In addition to their high cost, these methods take a long time and require a trained person and specific instruments to perform.
With the increasing need to develop a simple method to detect and separate OTA, various biosensors have been developed to detect this toxin in cereal products, food materials, and beverages. The development of novel biosensors has created an opportunity in the agriculture and food industries to improve food quality and safety assurance.
Hitherto numerous antibody-based biosensors have been reported for the detection of OTA (Huang et al., 2017; Ren et al., 2018; Alhamoud et al., 2019). In most biosensors, antibodies are used to identify targets. However, several new recognition elements have been introduced as alternatives for antibodies.
Aptamers, oligonucleotides, or peptide molecules that bind to a specific target can be isolated against amino acids, drugs, proteins, and other molecules. These are potent in their use in a variety of tools, including biosensors and analytical techniques (Zhao et al., 2008). Aptamers, known as chemical antibodies (Zhou et al., 2016a), are worthwhile alternatives for antibodies with greater benefits. They can bind specifically with a high affinity to their targets, ranging from ions to a complex targets such as whole cells. The specificity of aptamers is such that they can even distinguish between chiral molecules. These characteristics have enabled aptamers into a promising tool to construct diagnostic and analytical platforms with low limits of detection ranging from picomoles to nanomoles of targets (Song et al., 2008).
An interesting issue to address OTA contaminations is the development of aptasensors and biosensors. Some studies applied OTA aptamer for separation approaches such as aptamer-assisted real-time polymerase chain reaction (PCR), HPLC, suspension arrays, microfluidic devices, and affinity columns.
In © present review, we have tried to systematically study and classify OTA-specific isolated aptamers based on their applications and specifications.
Search for available OTA-specific aptamers and their separation and detection platforms
Data mining and searches were carried out in PubMed and Scopus for articles from 2008 to 2020. The main search keywords included OTA-specific aptamers, aptamer-based biosensors, aptamer-based detection, OTA aptasensors, ochratoxin A separation, ochratoxin A analysis, and other similar compositions. Keywords were selected using the medical subject headings (MeSH) terms and composed using proper Boolean operators (AND/OR/NOT).
The data mining was performed independently by two researchers, and the results were checked by the other researchers.
The following inclusion criteria were considered: only articles published in the English language, articles with methods for separation or detection of OTA, and use of aptamer-based sensors.
For data collection and extraction, selected publications that met inclusion criteria were reviewed. The methods were categorized and reviewed based on the separation approach, sensor platform, label, detection limit, detection range, and sequences of aptamers. The analytical methods used to measure OTA were also evaluated.
A total of 269 records (up to October 2020) were recovered; among these, 15 studies were removed after the first screening. The remaining 254 studies were reviewed in detail. Studies addressing other aspects of the title and deviation from inclusion criteria were excluded. The remaining studies that met the inclusion criteria and were classified for inclusion in the systematic review (Fig. 2).

Figure 2: PRISMA flow-diagram.
OTA structure was prepared from (PDB: 6J2W), and the tertiary structures of OTA-specific aptamers were designed according to the findings in our recent article. In summary, Secondary structures of four OTA-specific aptamers were predicted using the Mfold web server (Zuker, 2003) (version 3.0, http://www.unafold.org/mfold/applications/dna-folding-form.php). Then, based on the resulting secondary structure of the Vienna file format, their PDB structure was predicted using RNA composer (Antczak et al., 2016) and refined by UCSF Chimera (Pettersen et al., 2004) and discovery studio visualizer. In summary, the uracil was replaced by thymine (T), and the ribose sugars were substituted with deoxyribose sugars in the primary chain. The OTA was docked on each aptamer by HDOCK webserver (Yan et al., 2020) to predict complex models. Then, discovery studio visualizer software (2016) was employed to find the most appropriate and final structure of OTA and each aptamer complex and to monitor possible intermolecular interactions.
Specific Aptamers against OTA
Some aptamers have been reported for specific detection of OTA (Table 1).
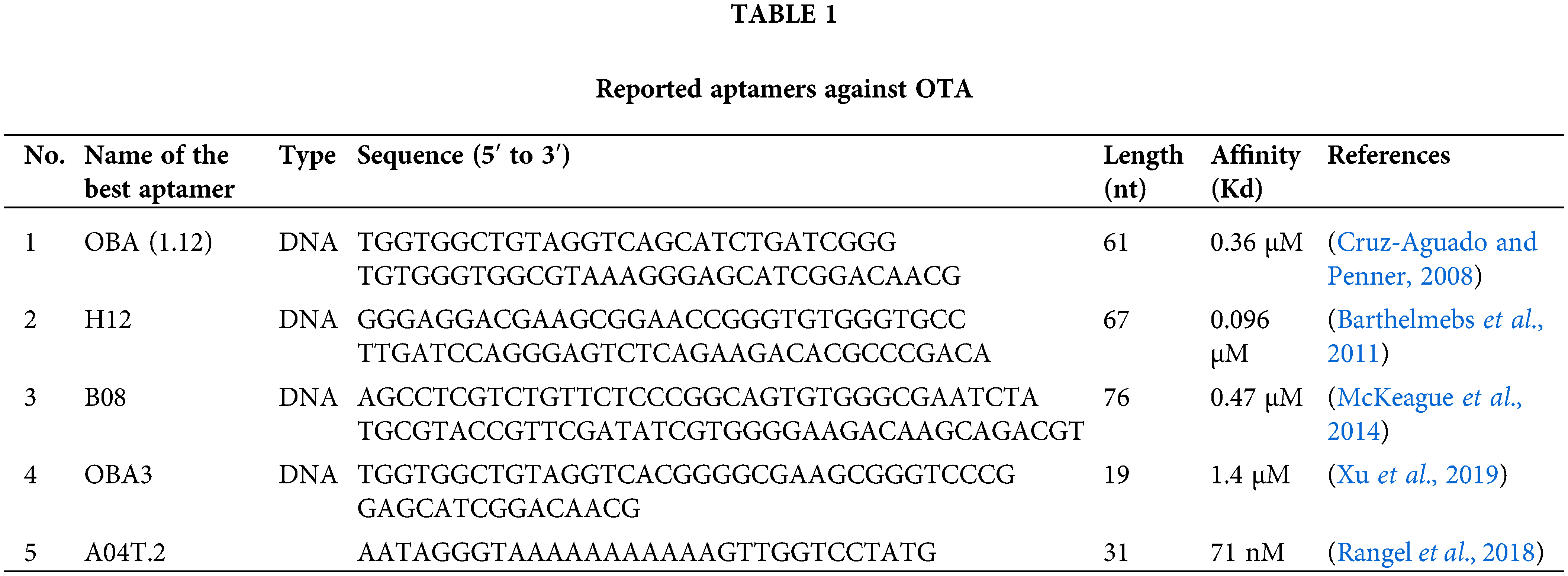
Cruz-Aguado and Penner isolated 13 DNA aptamers against OTA by SELEX after twelve reiterative rounds, between which an aptamer named OTA binding aptamer (OBA; 1.12) showed the highest affinity (Kd = 0.36 μM). They also developed buffer conditions by adding divalent cations to reduce the Kd value of the aptamer to 50 nM. Their results found that the Kd value could be reduced by substituting calcium for magnesium in the buffer.
Presumably, OTA forms a coordination complex with magnesium or calcium with the aid of its carboxyl and 8-hydroxyl groups, and this complex increases the binding to the aptamer. This buffer was subsequently used for the separation of the OTA from wheat samples using an aptamer affinity column. The column removed more than 97% of OTA from 1 mL of a 100 nM OTA solution (Cruz-Aguado and Penner, 2008). Other scientists applied a structure-guided post-SELEX approach to improve the affinity of this reported aptamer. They achieved this goal by forming a novel hairpin structure containing an intramolecular triple helix in the aptamer structure by mutating T and adenine (A) in critical points in aptamer sequence to cytosine (C) and guanine (G), respectively, and adding a C in the 5’–terminus of the aptamer. Using such an approach increased the affinity aptamers’ by up to 50-fold (Xu et al., 2019).
In 2011, another research group (Barthelmebs et al., 2011) isolated five aptamers against OTA by employing in vitro SELEX method. They selected an aptamer with the lowest IC50 (0.051 µg/mL), called H12, with Kd of 96 nM as the best one (Barthelmebs et al., 2011). They found two important conserved sequences (GGGTGTGGG) and (AGGGAGT) in the stem region and the single-strand terminal loop, respectively, in these aptamers for binding to OTA in the stem and loop regions similar to aptamer OBA (Cruz-Aguado and Penner, 2008).
A08 and B08, were other novel aptamers reported for the detection of OTA with relatively high affinity (Kd = 290 ± 150 and 110 ± 50 nM respectively), after fifteen reiterative rounds of SELEX (McKeague et al., 2014) without any similarity with previously reported aptamers (Cruz-Aguado and Penner, 2008; Barthelmebs et al., 2011).
Recently, a study used the in vitro evolution technique and introduced a threose nucleic acid aptamer for OTA with high stability and low Kd (71 nM) (Rangel et al., 2018).
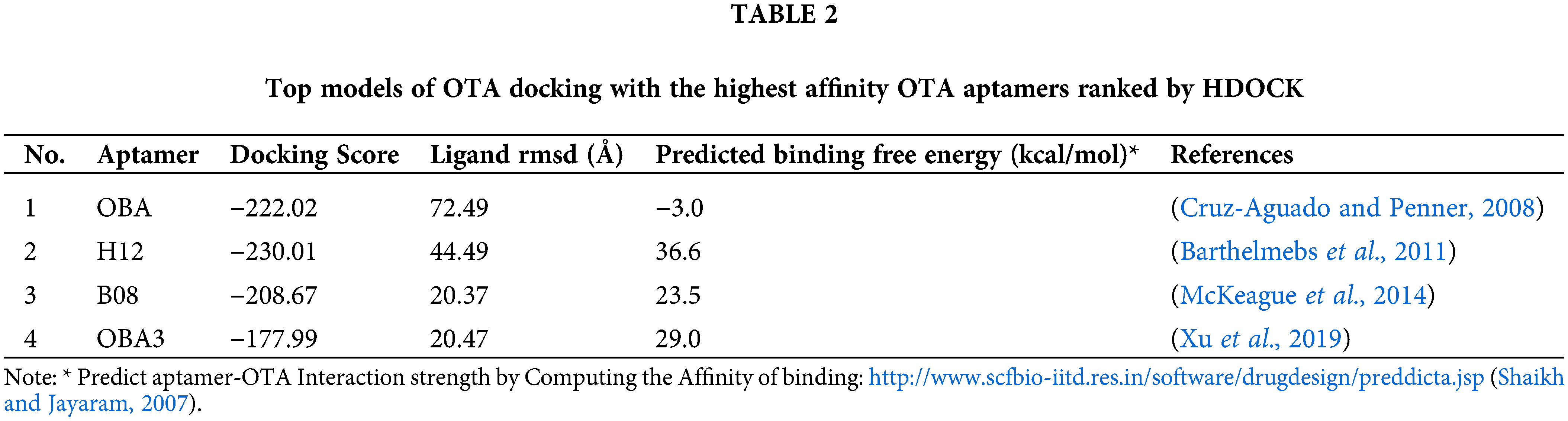
In silico investigation on binding structures confers valuable data about intermolecular interactions. Knowledge of the interaction mechanism can help us design purposeful diagnostic and analytical systems. Each binding—partial or complete binding or pocket formation—of the aptamer has features that help researchers to design their devices intelligently and accurately.
To know more about OTA-aptamer interactions, the OTA molecule was docked on reported aptamers (aptamers with the highest affinity in each study). Docking of OTA on its specific aptamers by the HDOCK web server (http://hdock.phys.hust.edu.cn/) (Yan et al., 2020) resulted in over 100 probable models. From these structures, the top models ranked by HDOCK were summarized in Table 2. We also calculated the strength of interaction of each top model of docking of OTA with the highest affinity OTA aptamers ranked by HDOCK using the PreDDICTA tool (https://www.scfbio-iitd.res.in/software/drugdesign/preddictanew.jsp) http://www.scfbio-iitd.res.in/software/drugdesign/preddictanew.jsp and compared them (Shaikh and Jayaram, 2007). The OBA (1.12) (Cruz-Aguado and Penner, 2008) showed the lowest predicted binding free energy (−3.0 kcal/mol).
After minimizing the energy of these models for each aptamer, final models were achieved and considered for further analysis (Fig. 3). Hydrogen bonds in the final complex structures was monitored by Discovery studio visualizer software (2016) (Wallace et al., 1995). The illustration revealed the possible hydrogen bonds between OTA and its aptamers (Fig. 3 and Table 3).

Figure 3: Illustration of docking Ochratoxin A (OTA) (purple) on its specific aptamers (white tubes): a) OTA binding aptamer (OBA; Cruz-Aguado and Penner, 2008), b) H12 (Barthelmebs et al., 2011), c) B08 (McKeague et al., 2014) and d) OBA3 (Xu et al., 2019). The OTA bonded nucleotides of each aptamer are displayed in the magnified form on the right side of each figure. Green dashed lines: Conventional hydrogen, Blue dashed lines: Carbon-hydrogen bonds.
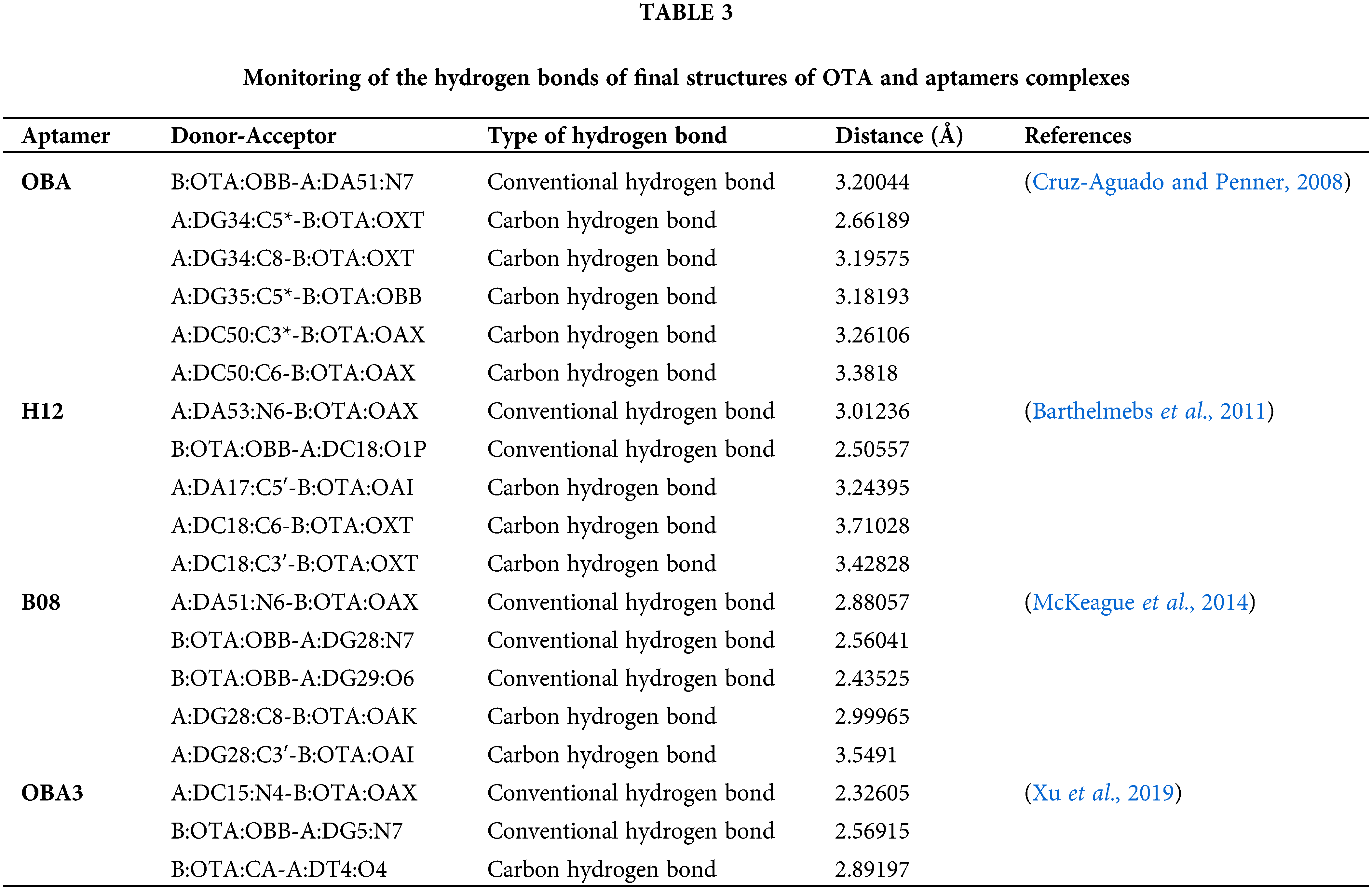
Docking results revealed that these aptamers could surround and bind OTA through some hydrogen bonds by creating binding pockets. The carbonyl oxygen and hydrogens from the amide and chlorine of OTA showed high potential for forming intermolecular hydrogen bonds between the OTA and the aptamers.
Different binding interactions could explain their distinct predicted affinity for binding OTA (Table 2). Some studies showed comparable binding strength of carbon hydrogen bonds to that of the conventional hydrogen bonds. The attractive dispersion interaction could explain the unusual stabilities of these hydrogen bonds (Ghosh et al., 2020). However, aptamers are more complex to be interpreted with preliminary molecular studies, and such a basic analysis cannot totally disclose the mechanism of aptamer-target binding.
Various aptamer-based sensors have been developed for the quantitative measurement of OTA. In this regard, electrochemical, optical, and mass-sensitive transducers have been used to fabricate aptasensors. In addition to the biosensors, some other approaches such as aptamer assisted real-time PCR, HPLC, suspension array, microfluidic devices, and extraction columns have been developed.
All studies can be divided into two main categories of detection (aptasensor or non-biosensor detection methods) and separation.
Most of the fabricated aptasensors for OTA have been developed based on the first reported aptamer (OBA, Kd = 0.36 µM) (Cruz-Aguado and Penner, 2008). Of reported aptasensors based on OBA aptamer, 38 were electrochemical (Table 4), and 70 were optical aptasensor (Table 5). An optical aptasenor was also fabricated based on B08 (McKeague et al., 2014).

Other studies (17 original studies) have used either the first reported aptamer (OBA aptamer, 15 studies) (Cruz-Aguado and Penner, 2008) or H12 aptamer (two bioassays including direct and indirect Enzyme-Linked Aptamer Assay) (Barthelmebs et al., 2011) in non-biosensor detection approaches, isolation, and extraction.
In the following, the most prominent studies in two categories of aptasensors and non-biosensor aptamer-based detection and separation methods were discussed. We have also mentioned a few reported lateral flow and Lab-on-Chip-based assays. However, all the relevant studies are listed in the Tables.
a) Aptasensors
i) Electrochemical Aptasensors
In electrochemical biosensors, the biologically active materials are combined with an electrochemical sensing element transducer. The transducer transduces a chemical signal into an electrical signal by amperometric, voltammetric, potentiometric, or conductometric methods. There are thirty-eight reports on electrochemical aptrasensors for the detection of OTA (Table 4).
In these studies, gold electrodes were employed as the working electrode in twenty electrochemical aptasensors (n = 20) that exhibited favorable outcomes using this type of electrode. The advantages of this electrode include high surface area, ease of synthesis, preparation, and modification, tunable pore size, conductivity, and a bicontinuous open pore network (Angnes et al., 2000).
The other popular electrode for this type of OTA specific electrochemical aptasensor (n = 10) is a screen-printed carbon electrode (SPCE) with a wide potential window, low background current, and low-cost properties. SPCE consists of a reference electrode, a counter electrode, and a carbon working electrode. The area, thickness, and composition of these electrodes can be readily controlled. Catalysts can be simply incorporated by pasting them to the screen printing ink (Cumba et al., 2020).
Glassy carbon electrodes (GCEs), which were employed in six of the OTA electrochemical biosensors are less sensitive than GE because of having a wider window of electrochemical activity. Since these electrodes are highly resistant to heat and corrosion and can be easily cleaned and polished due to their tightly knit atomic structure and glassy exterior, they are widely applied in electrochemical biosensors.
The indium tin oxide electrode exhibited a wide range of current density implemented in two of the reported devices (n = 2) (Bouden et al., 2016).
The electrochemical properties of electrodes could be improved by various techniques. Nanomaterials were used to develop electrochemical sensors by signal amplification to improve both sensitivity and selectivity. These materials produce a synergistic effect among catalytic activity, conductivity, and biocompatibility to accelerate the signal transduction. The surface of nanomaterials could be efficiently activated and easily functionalized, making them an ideal surface for immobilization of biomolecules such as enzymes and bioreceptors, including aptamers, antibodies, and also electroactive labels.
For example, Chrouda et al. (2015) used an electrochemically oxidated carboxyl end of the long spacer chain of polyethylene glycol (NH2-PEG-COOH) on a boron-doped diamond (BDD) microcell through which sensitivity of the system was increased to the range of pg/mL. For immobilization of biomolecules on the BDD surface of microcell, that is inactive, the surface should be activated by reactive groups. This procedure helps to control the surface easily. In their aptasensor, immobilized amino-aptamer on PEG was utilized. In the presence of OTA, the conformation of the aptamer changed to G-quadruplex structures, and the electron transfer rate of the redox probe was decreased. Their results showed a wide linear range (0.00001–0.0132 ng/mL) with a LOD of 0.00001 ng/mL. This sensor needed a shorter time (1 h) to detect OTA in real samples of rice in comparison with the other designed methods using the same aptamer (6–16 h).
Kaur et al. (2019) fabricated a simple and efficient functionalized graphene (f-graphene) doped chitosan (CS) nanocomposites based electrochemical aptasensor for the detection of OTA. The use of f-graphene increased the electroactive surface area of the electrode, and CS prevented the leaching of the aptamer molecules. CS is a suitable matrix due to its biocompatibility, hydrophilicity, non-toxicity, excellent mechanical stability, cost-effectiveness, and availability of reactive functional groups for chemical modifications. Besides, f-graphene increased the electroactive surface area of the electrode. The dual use of CS and f-graphene overall improved the sensor performance. This aptasensor exhibited LOD of 1 fg/mL for standard and 0.01 ng/mL for real samples within a response time of 8 min.
Yang et al. (2019) developed a label-free ultrasensitive electrochemical aptasensor based on NH2/Janus particles for the detection of OTA. Janus particles are special types of nanoparticles (NPs) with two or more surfaces having distinct physical or chemical properties. This allows two different types of chemicals to occur on the same particle. They prepared Janus particles by coating a layer of Au onto the hemisphere of amino polystyrene microspheres and immobilized the thiolated OTA aptamer on the Au layer. This aptasensor showed a very low LOD (0.0000001 ng/mL: 3.3 × 10−3 pM) and a wide dynamic linear range of OTA concentration (0.000004–4.0381 ng/mL: 1 × 10−5 to 10 nM) with high selectivity due to highly specific molecular recognition between OTA and aptamer and also good stability and reproducibility. They also applied this electrochemical sensor for the detection of OTA in red wine.
The common issue in these devices is employing nanomaterials. Nanoparticles provide a more accessible surface for immobilization of biorecognition elements which increase the sensitivity of biosensor to as low as pico/femtomolar levels.
Interestingly, there were some devices with an extremely low limit of detection (LOD) down to pico/femtogram/mL of OTA. The key point of some of these devices is the use of amplification techniques that enhance the signal to fabricating ultrasensitive biosensors such as PCR, real-time PCR, and recently, isothermal amplification-detection strategies such as loop-mediated isothermal amplification (LAMP).
For the first time, Yuan et al. (2014) integrated the LAMP technique with an electro-chemiluminescent (ECL) system to fabricate an ultrasensitive aptasensor. They immobilized a dsDNA composed of an OTA aptamer and its capture DNA on the electrode. With the presence of OTA, some of the aptamers were separated, and the remaining aptamers on the electrode served as an inner primer to initiate the LAMP reaction. The amplification procedure was tracked by monitoring the intercalation of DNA-binding Ru(phen)32+ ECL indicators into newly formed amplicons. Therefore, the presence of more OTA was equal to the release of more aptamer, less remaining aptamer on the electrode for producing LAMP amplicons, less Ru(phen)32+ intercalating into the formed amplicons, and thus increasing the ECL signal. They achieved a detection limit as low as 10 fM of OTA with good reproducibility and stability.
The integration of nanomaterials with these nucleic acid amplification strategies also could have a positive impact on the sensitivity of aptasensors.
Wang et al. (2017b) applied NH2@Cobalt-Metal–organic frameworks (MOFs) with their μ3-O linked trigonal prism structures to construct a sensitive biosensor. These frameworks efficiently possess intrinsic electrocatalytic activities for redox molecules, like thionine, which could intercalate into grooves of dsDNA via electrostatic adsorption and generate ultrasensitive square wave voltammetry signal. For amplification of the signal, they also employed a versatile exonuclease I (Exo I)-assisted target recycling for the production of sufficient numbers of cDNA sequences. Exo I catalyzes the hydrolysis of ssDNA instead of dsDNA from its 3′ end and digests the aptamer in the OTA-aptamer complex to release OTA again for further reaction. In brief, the gold nanoparticles (AuNPs), which effectively promote the electron transfer, were linked to the surface of NH2-Co-MOFs on the Au electrode. Biotin-modified OTA aptamer and cDNA were immobilized onto streptavidin magnetic beads. A series of OTA concentrations were incubated, and then Exo I-assisted target recycling reaction was performed. Eventually, the aptamer was dissociated from the matched double-stranded DNA in the presence of OTA and digested by Exo I, which led to the release of OTA. Then, the released OTA participated in another specific recognition reaction with the remaining aptamers, achieving the target recycling and signal amplification. The dissociated cDNA in the supernatant solution was collected. The mixture of the collected complementary DNA, and SH-Capture probe was incubated on the Au electrode to form the ternary DNA Y-junction structure on the NH2-Co-MOFs sensing surface. At the signal generation step, large amounts of thionine could intercalate into the three complementary DNA sequences. Further hybridization of the SH-Capture probe and cDNA to form a “Y” junction structure on the electrode surface resulted in significant signal recovery. They successfully applied this sensor to determine OTA in red wine with a linear range of 0.000001 to 1 ng/mL and LOD of 0.00000033 ng/mL (0.33 fg/mL). It also showed 90.0–105.0% recoveries for OTA in the red wine.
ii) Optical Aptasensors
Based on our search, 70 papers have reported optical aptrasensors for the detection of OTA (Table 5). Here we have attempted to discuss the most prominent construction.

A sensitive luminescent aptasensor was designed by Wu et al. (2011a). They utilized aptamer-conjugated magnetic nanoparticles (MNPs) and sodium yttrium fluoride (NaYF4): Yb (Ytterbium), Erbium (Er) labeled upconversion nanoparticles (UCNPs) as the recognition element and highly sensitive label, respectively. They immobilized OBA aptamer (Cruz-Aguado and Penner, 2008) on the surface of Fe3O4 MNPs hybridized with UCNPs coated with complementary DNA. Once OTA was bound to the aptamer, the complementary ssDNA was released, and subsequently, the luminescent signal was reduced. By fabricating this simple method, they could achieve very low LOD (0.0001 ng/mL) and a wide linearity range (0.0001–0.1 ng/mL) (Fig. 4a).

Figure 4: Schematic illustration of the most sensitive reported optical aptasensors. a) The immobilized aptamer on the surface of magnetic nanoparticles (MNPs) hybridized with cDNA immobilized on sodium yttrium fluoride (NaYF4): Yb (Ytterbium), Erbium (Er) labeled upconversion nanoparticles. Once OTA was bound to the aptamer, the cDNA was released, and subsequently, the luminescent signal was reduced (Wu et al., 2011a). b) Simultaneous detection of ochratoxin A (OTA) and fumonisin B1 (FB1) based on aptamer−SPCMs suspension arrays (Yue et al., 2014).
Song et al. (2018) introduced a Surface-Enhanced Raman Spectroscopy (SERS)-based aptasensor for the detection of OTA by hybridizing cDNA coated Fe3O4@Au magnetic nanoparticles (MGNPs), and the detection of OBA aptamer (Cruz-Aguado and Penner, 2008) modified Au@Ag nanoprobes labeled with the Raman reporter 5,5′-Dithiobis(2-nitrobenzoic acid) (DNTB) (Au-DTNB@Ag NPs). In the absence of OTA, the peak of SERS was high, but with the presence of OTA, due to the binding of OTA to the aptamer and the release of cDNA, the signal decreased proportionally to the concentration of OTA. This platform showed picogram levels (0.48 pg/mL) of LOD with good recovery and accuracy in evaluating real samples of wine and coffee.
A label-free fluorescent aptasensor which was developed by Lv et al. (2014), gives one of the widest reported dynamic ranges of detection. In this highly-sensitive (0.025 ng/mL) and selective fluorescent sensor, they applied PicoGreen (PG), an asymmetric cyanine dye, which exhibits its fluorescence property only after ultra-selective binding to the minor groove of dsDNA. In this platform, if there is no OTA, the ssDNA aptamers (Cruz-Aguado and Penner, 2008) hybridize to cDNAs. The PG dyes can bind the formed dsDNA and exhibit fluorescence. In the presence of OTA, the aptamer binds to the target and generates a G-quadruplex structure, and the signal intensity decreases. In this approach, a wide dynamic range (1 to 100000 ng/mL) was achieved to determine OTA concentrations in beer samples.
Yue et al. (2014) developed a simple and novel aptamer-based photonic crystal encoded suspension which could simultaneously recognize and quantify OTA and fumonisin B1 (FB1) in cereal samples by immobilizing the OBA aptamers (Cruz-Aguado and Penner, 2008) on the surfaces of different kinds of silica photonic crystal microsphere (SPCM) by a covalent bond. The different OTA and FB1 aptamer probes hybridized with their fluorescence-labeled complementary DNA and those immobilized on the surfaces of SPCMs could bind to their specific targets (OTA and FB1). Binding each aptamer to its target induced a structural switch of the aptamer, causing the release of the fluorescence-labeled aptamer complementary DNA and a marked decrease in fluorescent intensity of each kind of SPCM (Fig. 4b).
One interesting method in the line of optical aptasensors was developed by Song et al. (2018). They utilized a dual-color fluorescence resonance energy transfer (FRET) based aptasensor for simultaneous detection of the mycotoxins aflatoxin M1 (AFM1) and OTA. They labeled AFM1 and OTA aptamers with two different fluorophores as the signaling probes. In the blank samples, the aptamers were hybridized to their specific quencher labeled cDNA, resulting in weak fluorescent signals. Simultaneously, the fluorescent labels of the aptamers bound to their targets in milk samples could produce strong signals under optimized conditions, as this aptasensor selectively detected AFM1 and OTA with a LOD value of 0.021 ng/mL over a wide linear range (0.001–1000 ng/mL).
In another study, an optical aptasensor was constructed based on a B08 aptamer by McKeague et al. (2014). In this diagnostic system, the SYBR Green I dye, which can intercalate to the minor groove of dsDNA, was used to enhance fluorescent emission. In their designed sensor, OTA competed with SYBR Green I for binding to aptamer, and consequently led to a decline in SYBR Green I fluorescence emission. SYBR Green intercalates with the free DNA aptamer and fluoresces at 520 nM. However, increasing concentrations of the OTA and binding to aptamers displaces SYBR Green I and a concentration-dependent loss of emitted fluorescence. This label-free SYBR Green I-based aptasensor showed Kd (9 nM) and linear range in the nanomolar scales (9–100 nM).
There are some reports that introduced lateral flow assay (LFAs)-based optical biosensors (Anfossi et al., 2012; Anfossi et al., 2013; Moon et al., 2013; Zhou et al., 2016b; Jiang et al., 2017; Ren et al., 2018; Velu and DeRosa, 2018; Oh et al., 2019; Hao et al., 2021; Zhao et al., 2021).
LFAs are popular among commercial paper-based assay products for quality control, food safety assessment, and in medical and clinical centers and laboratories (Majdinasab et al., 2022). However, this method has limited sensitivity in the detection of low concentrations of analytes. The samples also require pretreatment, including sample extraction, filtration, and dilution in some analytes, such as complex matrix or solid samples. Thus, LFA strips are not ideal for these samples. Positive results of LFA tests also need confirmatory analysis (Majdinasab et al., 2022).
b) Non-biosensor Aptamer based separation and detection methods
Due to the specificity and selectivity of the OTA aptamer isolated by Cruz-Aguado and Penner (OBA aptamer) (Cruz-Aguado and Penner, 2008), several studies have applied it as a detection element in a column to isolate, extract and clean up this toxin from samples, including wine, beer, or food (Table 6).

In some studies, various oligosorbents on solid-phase columns were constructed, such as magnetic nanospheres (MNS) (Wu et al. 2011b), coupling gel for the preparation of the aptamer-SPE (solid-phase extraction) columns (de Girolamo et al., 2011), cyanogen bromide-activated sepharose (Hadj Ali and Pichon, 2014), chitosan magnetic nanoparticles (Wang et al., 2020), or monolithic columns with high coverage density of aptamer (Chen et al., 2018; Yu et al., 2018; Lyu et al., 2020).
i) Liquid chromatography
In one of the considerable methods, an ultra-fast liquid chromatography with tandem mass spectrometry method was reported based on aptamer-affinity column and vortex-assisted solid-liquid microextraction, which can promote the diffusive motion of analyte from the sample to the extraction solvent (Yang et al., 2014). In another method, polyhedral oligomeric silsesquioxane (POSS) was applied to construct aptamer monoliths. POSS is a special organic-inorganic material with highly stereoscopic nano-cage and massive function sites that provide high coverage density of aptamer. Polyethyleneimine (PEI) was also used for the immobilization of AuNPs aptamer due to abundant amino groups. This fabrication resulted in a highly efficient, sensitive, and selective recognition system (LOD = 0.06 ng/mL) (Yu et al., 2018).
In a similar study, a POSS-PEI monolith was prepared with 2,4,6-trichloro-1,3,5-triazine (TCT) as a linker for binding high levels of the aptamer that enable them to achieve a LOD as low as 0.01 ng/mL in beer samples (Yu et al., 2019).
ii) Polymerase chain reaction-based assays
Real-time PCR is an amplification method and a high throughput screening method that could shorten operation time, decrease detection limit for low concentration analyte samples and have a favorable reproducibility.
The most sensitive device was reported with a LOD of 0.000001 ng/mL (1 fg/mL) for the detection of OTA by employing real-time PCR (Ma et al., 2013). It has shown a satisfactory recovery rate (99–112%) in red wine samples. The recruited strategy was based on the conformational change of the OBA aptamer (Cruz-Aguado and Penner, 2008). At first, the biotinylated aptamers were immobilized on the surface of the streptavidin-coated PCR tubes. Then, in the absence of the OTA, ssDNA aptamer was hybridized with complementary DNA strands and subjected to the same treatment as the PCR template. The forward and reverse primers and other PCR components were added and the PCR procedure was performed. Then, the emitted fluorescence was measured after each annealing step. In the presence of OTA, a structural switch of the aptamer was induced by the target binding, leading to the formation of an antiparallel G-quadruplex, which resulted in complementary ssDNA release. This could reduce the amount of the template for amplification and increase the cycle threshold (Ct). Thus, the concentration of the OTA was measured by the change in the PCR amplification (Ma et al., 2013).
iii) Enzyme-Linked Aptamer Assay (ELAA)
ELAA is employed for biorecognition of analytes by replacing antibody with aptamer, with its benefits over antibody through (Vargas-Montes et al., 2019).
Among other reported aptamers, H12 aptamer (Barthelmebs et al., 2011) was also applied for the detection of OTA. The H12 aptamer was used for both direct and indirect ELAA for detection of OTA in spiked red wine samples by Barthelmebs et al. (2011). In this approach, the fluorescein-labeled aptamers are specifically bound to biotinylated OTA. This complex could attach to the OTA-HRP conjugate, and the emitted fluorescence could be simply detected (Barthelmebs et al., 2011).
Miniaturization is one of the important issues in the fabrication of biosensors to produce portable and user-friendly devices. Lab-on-a-chip (LoC) is also recently considered in this field. Some Efforts have also been made to design LoC-aptasensors for the detection of OTA.
Recently, a novel ACSB was described for the detection of OTA via FRET with an LOD of 0.005 ng/m in a linearity range of 0.01–10 ng/mL. They immobilized a Cy3-labeled OTA-specific biotinylated aptamer on an epoxy-coated chip via streptavidin-biotin binding. A black hole quencher 2 (BHQ2) labeled complementary DNA strand to OTA aptamer. In the presence of OTA, the Cy3-labeled OTA aptamer bound specifically to OTA and led to the physical separation of Cy3 and BHQ2, which resulted in an increase in fluorescence signal. This aptasensor was tested in rice samples spiked with OTA with a mean recovery rate of 91% (Li et al., 2021).
A fluorescent label-free LoC aptamer portable assay, integrated into the microfluidic network, was constructed on a single glass substrate, comprising an array of amorphous silicon photosensors and a long pass interferential filter. The employed fluorescent molecule was a “light switch” complex [Ru(phen)2(dppz)](2+) which intercalated between the base pairs of the aptamer. The aptamer was directly anchored into a layer of poly(2-hydroxyethyl methacrylate) polymer brushes grown inside the channels. The presence of OTA changed the aptamer conformation and released the fluorophore, causing a decline in fluorescence. This device performed detection in 5 min with an LOD of 1.3 ng/mL and 5–200 ng/mL linear detection in real samples (beer and wheat samples) (Costantini et al., 2019).
Nekrasov et al. (2022) reported an advanced aptasensor based on an array of graphene field-effect transistors integrated on a single silicon chip. Graphene with electrochemical, thermal, optical, electronic, and mechanical properties holds enormous potential for LOC devices. The G-rich aptamer was covalently attached to the graphene surface via pyrenebutanoic acid, succinimidyl ester (PBASE) chemistry. PBASE created efficient π-π stacking to graphene via an electric field stimulation. In the absence of OTA, the aptamer strands created π-π stacking on graphene, and after the addition of OTA molecules, aptamers reconfigured in G-quadruplex to bind OTA molecule. This fast assay (10 s) graphene-aptasensor showed an LOD of 1.4 pM for OTA with a demonstrated performance of wine in real-time.
An ultrasensitive label-free liquid crystal (LCs) OTA aptasensor was also designed with the lowest reported LOD (0.63 Am) based on the conformational switch of the immobilized π-shaped aptamer on the glass substrate in the presence of the OTA. A shift in the orientation of LCs from random to a homeotropic state altered the optical appearance of the aptasensor platform, which could be examined by polarized light microscopy for the detection of OTA in grape juice, coffee, and human serum samples (Khoshbin et al., 2021).
OTA-specific recognition aptamers show good affinity, making them applicable recognition elements in different devices. Docking results reveal that these aptamers could surround the OTA through some hydrogen bonds by creating binding pockets. However, a logical and straight relationship between docking scores and monitored hydrogen bonds has not yet been found, possibly because aptamer-target interaction has more intricacy and requires more in silico analyses to discover critical features in this interaction.
Aptamers as alternative recognition elements to antibodies exhibit several advantages, including the ease of in vitro isolation, high specificity and selectivity, longer duration of stability, ability to isolate a variety of small, toxic, and non-immunogenic molecules, amenable to modification, rare immunogenicity, and limited batch-to-batch variation. Aptamer-based systems also have several advantages over conventional methods due to their high selectivity and sensitivity, low cost, and stability.
Overview of this database could show that, by employing signal amplification strategies such as nanomaterials for increasing surface accessible area or catalytic effects on reaction, could be ultrasensitive devices with LODs as low as pico/femto level. Most of the aptasensors show reproduced responses with a good confidence. They are also applied frequently for the detection of real samples such as beverages, particularly wine, beer, and also cereals such as wheat. This could confirm the efficiency of these devices for consumers.
Nucleic acid amplification strategies are the other techniques for enhancing signals and, consequently, increasing sensitivity. PCR, real-time PCR and recently, LAMP as isothermal amplification-detection strategies were employed in OTA-specific aptasensors. Utilizing other nucleic acid amplification techniques such as helicase-dependent amplification, strand displacement amplification, rolling circle amplification, and recombinase polymerase amplification could be considered for further aptasensor fabrication.
Gold electrodes act as favorable working electrodes among electrochemical OTA aptasensors due to their unique properties such as good conductivity and chemical inertness.
Aptamers as alternative recognition elements to antibodies exhibit several advantages, including the ease of in vitro isolation, high specificity, and selectivity, long duration of stability, ability to isolate a variety of small, toxic, and non-immunogenic molecules, the capability of modification, rare immunogenicity, and limited batch-to-batch variation. Aptamer-based systems also have several advantages over conventional methods due to their high selectivity and sensitivity, low cost, and stability. However, the application of aptamer as a recognition element has some challenges. The most important issue is maintaining the desired affinity of the aptamer. Changing conditions in different detection methods could impact the affinity of the aptamer. For instance, immobilization techniques could result in microstructure changes which may alter the binding affinity. Other circumstances such as ionic strength and pH could influence aptamer configuration which may reduce its affinity properties. On the other hand, complex samples such as foods include various components such as nucleases which may have an adverse effect on the structure of aptamers and affect their activity.
Despite hundreds of reported separation methods and aptasensors with very low LODs for OTA, none have received approval for commercial applications, posing major challenges for improving research prototypes to reliable instruments.
Achieving the simple, rapid, sensitive, specific, and cost-effective methods for measuring OTA are the primary requisites for industrial, analytical, and medical assessments. Numerous aptamers are just capable of detecting samples in the aqueous solutions but not in solid analytes. It is necessary to improve some aspects, including ease of application, sample preparation, and cost of the production to reduce the gap between research and large-scale industrial applications (Schmitz et al., 2020).
Almost all of the sensors and separation approaches have used ssDNA aptamer reported by Cruz-Aguado and Penner (2008). They demonstrated a well-designed isolated aptamer which can meet merits for application.
Electrochemical and optical biosensors are two types of aptasensors developed for detection of OTA. Although the electrochemical methods have shown the highest sensitivity and selectivity, the optical aptasensor exhibited the widest linear ranges. However, several ultra-sensitive aptasensors have used nanomaterials to increase accessible surface area to enhance the OTA-aptamer interaction for signal amplification.
The current systematic review demonstrated the possibility, simplicity, and high selectivity of using aptamers to detect and analyze OTA in different real samples that are highly needed in agriculture, food industry, and water management.
Acknowledgement: The authors would like to thank the Clinical Research Development Unit of Baqiyatallah Hospital, Tehran, Iran, for guidance and advice.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: M. Heiat and R. Ranjbar; data collection, analysis and interpretation of results, draft manuscript preparation: R. Torabi, A. A. Rezvanipour, H. Esmaeili Gouvarchinghaleh. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest:The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abnous K, Danesh NM, Alibolandi M, Ramezani M, Taghdisi SM (2017). Amperometric aptasensor for ochratoxin A based on the use of a gold electrode modified with aptamer, complementary DNA, SWCNTs and the redox marker Methylene Blue. Microchimica Acta 184: 1151–1159. DOI 10.1007/s00604-017-2113-7. [Google Scholar] [CrossRef]
Alhamoud Y, Yang D, Kenston SSF, Liu G, Liu L et al. (2019). Advances in biosensors for the detection of ochratoxin A: Bio-receptors, nanomaterials, and their applications. Biosensors and Bioelectronics 141: 111418. DOI 10.1016/j.bios.2019.111418. [Google Scholar] [CrossRef]
Anfossi L, di Nardo F, Giovannoli C, Passini C, Baggiani C (2013). Increased sensitivity of lateral flow immunoassay for ochratoxin A through silver enhancement. Analytical and Bioanalytical Chemistry 405: 9859–9867. DOI 10.1007/s00216-013-7428-6. [Google Scholar] [CrossRef]
Anfossi L, Giovannoli C, Giraudi G, Biagioli F, Passini C, Baggiani C (2012). A lateral flow immunoassay for the rapid detection of ochratoxin A in wine and grape must. Journal of Agricultural and Food Chemistry 60: 11491–11497. DOI 10.1021/jf3031666. [Google Scholar] [CrossRef]
Angnes L, Richter EM, Augelli MA, Kume GH (2000). Gold electrodes from recordable CDs. Analytical Chemistry 72: 5503–5506. DOI 10.1021/ac000437p. [Google Scholar] [CrossRef]
Antczak M, Popenda M, Zok T, Sarzynska J, Ratajczak T, Tomczyk K, Adamiak RW, Szachniuk M (2016). New functionality of RNAComposer: An application to shape the axis of miR160 precursor structure. Acta Biochimica Polonica 63: 737–744. DOI 10.18388/abp.2016_1329. [Google Scholar] [CrossRef]
Armstrong-Price DE, Deore PS, Manderville RA (2020). Intrinsic “Turn-On” aptasensor detection of ochratoxin A using energy-transfer fluorescence. Journal of Agricultural and Food Chemistry 68: 2249–2255. DOI 10.1021/acs.jafc.9b07391. [Google Scholar] [CrossRef]
Barthelmebs L, Jonca J, Hayat A, Prieto-Simon B, Marty JL (2011). Enzyme-linked aptamer assays (ELAAsbased on a competition format for a rapid and sensitive detection of Ochratoxin A in wine. Food Control 22: 737–743. DOI 10.1016/j.foodcont.2010.11.005. [Google Scholar] [CrossRef]
Bi X, Luo L, Li L, Liu X, Chen B, You T (2020). A FRET-based aptasensor for ochratoxin A detection using graphitic carbon nitride quantum dots and CoOOH nanosheets as donor-acceptor pair. Talanta 218: 121159. DOI 10.1016/j.talanta.2020.121159. [Google Scholar] [CrossRef]
Bonel L, Vidal JC, Duato P, Castillo JR (2011). An electrochemical competitive biosensor for ochratoxin A based on a DNA biotinylated aptamer. Biosensors and Bioelectronics 26: 3254–3259. DOI 10.1016/j.bios.2010.12.036. [Google Scholar] [CrossRef]
Bouden S, Dahi A, Hauquier F, Randriamahazaka H, Ghilane J (2016). Multifunctional indium tin oxide electrode generated by unusual surface modification. Scientific Reports 6: 36708. DOI 10.1038/srep36708. [Google Scholar] [CrossRef]
Bui-Klimke TR, Wu F (2015). Ochratoxin A and human health risk: A review of the evidence. Critical Reviews in Food Science and Nutrition 55: 1860–1869. DOI 10.1080/10408398.2012.724480. [Google Scholar] [CrossRef]
Chen J, Fang Z, Liu J, Zeng L (2012). A simple and rapid biosensor for ochratoxin A based on a structure-switching signaling aptamer. Food Control 25: 555–560. DOI 10.1016/j.foodcont.2011.11.039. [Google Scholar] [CrossRef]
Chen J, Zhang X, Cai S, Wu D, Chen M, Wang S, Zhang J (2014a). A fluorescent aptasensor based on DNA-scaffolded silver-nanocluster for ochratoxin A detection. Biosensors and Bioelectronics 57: 226–231. DOI 10.1016/j.bios.2014.02.001. [Google Scholar] [CrossRef]
Chen Y, Chen M, Chi J, Yu X, Chen Y, Lin X, Xie Z (2018). Aptamer-based polyhedral oligomeric silsesquioxane (POSS)-containing hybrid affinity monolith prepared via a “one-pot” process for selective extraction of ochratoxin A. Journal of Chromatography A 1563: 37–46. DOI 10.1016/j.chroma.2018.05.044. [Google Scholar] [CrossRef]
Chen Y, Ding X, Zhu D, Lin X, Xie Z (2019). Preparation and evaluation of highly hydrophilic aptamer-based hybrid affinity monolith for on-column specific discrimination of ochratoxin A. Talanta 200: 193–202. DOI 10.1016/j.talanta.2019.03.053. [Google Scholar] [CrossRef]
Chen Y, Yang M, Xiang Y, Yuan R, Chai Y (2014b). Binding-induced autonomous disassembly of aptamer-DNAzyme supersandwich nanostructures for sensitive electrochemiluminescence turn-on detection of ochratoxin A. Nanoscale 6: 1099–1104. DOI 10.1039/C3NR05499C. [Google Scholar] [CrossRef]
Chrouda A, Sbartai A, Baraket A, Renaud L, Maaref A, Jaffrezic-Renault N (2015). An aptasensor for ochratoxin A based on grafting of polyethylene glycol on a boron-doped diamond microcell. Analytical Biochemistry 488: 36–44. DOI 10.1016/j.ab.2015.07.012. [Google Scholar] [CrossRef]
Chu X, Dou X, Liang R, Li M, Kong W, Yang X, Luo J, Yang M, Zhao M (2016). A self-assembly aptasensor based on thick-shell quantum dots for sensing of ochratoxin A. Nanoscale 8: 4127–4133. DOI 10.1039/C5NR08284F. [Google Scholar] [CrossRef]
Chung SWC, Kwong KP (2019). Determination of ochratoxin A at parts-per-trillion levels in cereal products by immunoaffinity column cleanup and high-performance liquid chromatography/mass spectrometry. Journal of AOAC International 90: 773–777. DOI 10.1093/jaoac/90.3.773. [Google Scholar] [CrossRef]
Costantini F, Lovecchio N, Ruggi A, Manetti C, Nascetti A, Reverberi M, de Cesare G, Caputo D (2019). Fluorescent label-free aptasensor integrated in a lab-on-chip system for the detection of ochratoxin A in beer and wheat. ACS Applied Bio Materials 2: 5880–5887. DOI 10.1021/acsabm.9b00831. [Google Scholar] [CrossRef]
Cruz-Aguado JA, Penner G (2008). Determination of ochratoxin a with a DNA aptamer. Journal of Agricultural and Food Chemistry 56: 10456–10461. DOI 10.1021/jf801957h. [Google Scholar] [CrossRef]
Cumba LR, Camisasca A, Giordani S, Forster RJ (2020). Electrochemical properties of screen-printed carbon nano-onion electrodes. Molecules 25: 3884. DOI 10.3390/molecules25173884. [Google Scholar] [CrossRef]
Dai S, Wu S, Duan N, Chen J, Zheng Z et al. (2017). An ultrasensitive aptasensor for Ochratoxin A using hexagonal core/shell upconversion nanoparticles as luminophores. Biosensors and Bioelectronics 91: 538–544. DOI 10.1016/j.bios.2017.01.009. [Google Scholar] [CrossRef]
de Girolamo A, McKeague M, Miller JD, DeRosa MC, Visconti A (2011). Determination of ochratoxin A in wheat after clean-up through a DNA aptamer-based solid phase extraction column. Food Chemistry 127: 1378–1384. DOI 10.1016/j.foodchem.2011.01.107. [Google Scholar] [CrossRef]
El-Moghazy AY, Amaly N, Istamboulie G, Nitin N, Sun G (2020). A signal-on electrochemical aptasensor based on silanized cellulose nanofibers for rapid point-of-use detection of ochratoxin A. Mikrochimica Acta 187: 535. DOI 10.1007/s00604-020-04509-y. [Google Scholar] [CrossRef]
el Khoury A, Atoui A (2010). Ochratoxin a: General overview and actual molecular status. Toxins 2: 461–493. DOI 10.3390/toxins2040461. [Google Scholar] [CrossRef]
Evtugyn G, Porfireva A, Stepanova V, Kutyreva M, Gataulina A, Ulakhovich N, Evtugyn V, Hianik T (2013). Impedimetric aptasensor for ochratoxin A determination based on Au nanoparticles stabilized with hyper-branched polymer. Sensors 13: 16129–16145. DOI 10.3390/s131216129. [Google Scholar] [CrossRef]
Galarreta BC, Tabatabaei M, Guieu V, Peyrin E, Lagugne-Labarthet F (2013). Microfluidic channel with embedded SERS 2D platform for the aptamer detection of ochratoxin A. Analytical and Bioanalytical Chemistry 405: 1613–1621. DOI 10.1007/s00216-012-6557-7. [Google Scholar] [CrossRef]
Gao J, Chen Z, Mao L, Zhang W, Wen W, Zhang X, Wang S (2019). Electrochemiluminescent aptasensor based on resonance energy transfer system between CdTe quantum dots and cyanine dyes for the sensitive detection of Ochratoxin A. Talanta 199: 178–183. DOI 10.1016/j.talanta.2019.02.044. [Google Scholar] [CrossRef]
Ghosh S, Chopra P, Wategaonkar S (2020). C-H.S interaction exhibits all the characteristics of conventional hydrogen bonds. Physical Chemistry Chemical Physics 22: 17482–17493. DOI 10.1039/D0CP01508C. [Google Scholar] [CrossRef]
Guo Z, Ren J, Wang J, Wang E (2011). Single-walled carbon nanotubes based quenching of free FAM-aptamer for selective determination of ochratoxin A. Talanta 85: 2517–2521. DOI 10.1016/j.talanta.2011.08.015. [Google Scholar] [CrossRef]
Hadj Ali W, Pichon V (2014). Characterization of oligosorbents and application to the purification of ochratoxin A from wheat extracts. Analytical and Bioanalytical Chemistry 406: 1233–1240. DOI 10.1007/s00216-013-7509-6. [Google Scholar] [CrossRef]
Hao L, Chen J, Chen X, Ma T, Cai X, Duan H, Leng Y, Huang X, Xiong Y (2021). A novel magneto-gold nanohybrid-enhanced lateral flow immunoassay for ultrasensitive and rapid detection of ochratoxin A in grape juice. Food Chemistry 336: 127710. DOI 10.1016/j.foodchem.2020.127710. [Google Scholar] [CrossRef]
Hao L, Wang W, Shen X, Wang S, Li Q, An F, Wu S (2020). A fluorescent DNA hydrogel aptasensor based on the self-assembly of rolling circle amplification products for sensitive detection of ochratoxin A. Journal of Agricultural and Food Chemistry 68: 369–375. DOI 10.1021/acs.jafc.9b06021. [Google Scholar] [CrossRef]
Hayat A, Haider W, Rolland M, Marty JL (2013a). Electrochemical grafting of long spacer arms of hexamethyldiamine on a screen printed carbon electrode surface: Application in target induced ochratoxin A electrochemical aptasensor. Analyst 138: 2951–2957. DOI 10.1039/c3an00158j. [Google Scholar] [CrossRef]
Hayat A, Mishra RK, Catanante G, Marty JL (2015). Development of an aptasensor based on a fluorescent particles-modified aptamer for ochratoxin A detection. Analytical and Bioanalytical Chemistry 407: 7815–7822. DOI 10.1007/s00216-015-8952-3. [Google Scholar] [CrossRef]
Hayat A, Sassolas A, Marty JL, Radi AE (2013b). Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 103: 14–19. DOI 10.1016/j.talanta.2012.09.048. [Google Scholar] [CrossRef]
He Y, Tian F, Zhou J, Jiao B (2019). A fluorescent aptasensor for ochratoxin A detection based on enzymatically generated copper nanoparticles with a polythymine scaffold. Mikrochimica Acta 186: 199. DOI 10.1007/s00604-019-3314-z. [Google Scholar] [CrossRef]
Hernández Y, Lagos LK, Galarreta BC (2020). Development of a label-free-SERS gold nanoaptasensor for the accessible determination of ochratoxin A. Sensing and Bio-Sensing Research 28: 100331. DOI 10.1016/j.sbsr.2020.100331. [Google Scholar] [CrossRef]
Heussner AH, Bingle LEH (2015). Comparative ochratoxin toxicity: A review of the available data. Toxins 7: 4253–4282. DOI 10.3390/toxins7104253. [Google Scholar] [CrossRef]
Huang B, Xiao L, Dong H, Zhang X, Gan W, Mahboob S, Al-Ghanim KA, Yuan Q, Li Y (2017). Electrochemical sensing platform based on molecularly imprinted polymer decorated N,S co-doped activated graphene for ultrasensitive and selective determination of cyclophosphamide. Talanta 164: 601–607. DOI 10.1016/j.talanta.2016.11.009. [Google Scholar] [CrossRef]
Huang H, Wang D, Zhou Y, Wu D, Liao X, Xiong W, Du J, Hong Y (2021). Multiwalled carbon nanotubes modified two dimensional MXene with high antifouling property for sensitive detection of ochratoxin A. Nanotechnology 32: 455501. DOI 10.1088/1361-6528/ac1a42. [Google Scholar] [CrossRef]
Huang L, Wu J, Zheng L, Qian H, Xue F, Wu Y, Pan D, Adeloju SB, Chen W (2013). Rolling chain amplification based signal-enhanced electrochemical aptasensor for ultrasensitive detection of ochratoxin A. Analytical Chemistry 85: 10842–10849. DOI 10.1021/ac402228n. [Google Scholar] [CrossRef]
Jiang D, Huang C, Shao L, Wang X, Jiao Y, Li W, Chen J, Xu X (2020). Magneto-controlled aptasensor for simultaneous detection of ochratoxin A and fumonisin B1 using inductively coupled plasma mass spectrometry with multiple metal nanoparticles as element labels. Analytica Chimica Acta 1127: 182–189. DOI 10.1016/j.aca.2020.06.057. [Google Scholar] [CrossRef]
Jiang H, Li X, Xiong Y, Pei K, Nie L, Xiong Y (2017). Silver nanoparticle-based fluorescence-quenching lateral flow immunoassay for sensitive detection of ochratoxin A in grape juice and wine. Toxins 9: 83. DOI 10.3390/toxins9030083. [Google Scholar] [CrossRef]
Jiang L, Qian J, Yang X, Yan Y, Liu Q, Wang K, Wang K (2014). Amplified impedimetric aptasensor based on gold nanoparticles covalently bound graphene sheet for the picomolar detection of ochratoxin A. Analytica Chimica Acta 806: 128–135. DOI 10.1016/j.aca.2013.11.003. [Google Scholar] [CrossRef]
Kaur N, Bharti A, Batra S, Rana S, Rana S et al. (2019). An electrochemical aptasensor based on graphene doped chitosan nanocomposites for determination of Ochratoxin A. Microchemical Journal 144: 102–109. DOI 10.1016/j.microc.2018.08.064. [Google Scholar] [CrossRef]
Khoshbin Z, Abnous K, Taghdisi SM, Verdian A (2021). A novel liquid crystal-based aptasensor for ultra-low detection of ochratoxin a using a π-shaped DNA structure: Promising for future on-site detection test strips. Biosensors and Bioelectronics 191: 113457. DOI 10.1016/j.bios.2021.113457. [Google Scholar] [CrossRef]
Kim K, Jo EJ, Lee KJ, Park J, Jung GY, Shin YB, Lee LP, Kim MG (2020). Gold nanocap-supported upconversion nanoparticles for fabrication of a solid-phase aptasensor to detect ochratoxin A. Biosensors & Bioelectronics 150: 111885. DOI 10.1016/j.bios.2019.111885. [Google Scholar] [CrossRef]
Kuang H, Chen W, Xu D, Xu L, Zhu Y, Liu L, Chu H, Peng C, Xu C, Zhu S (2010). Fabricated aptamer-based electrochemical “signal-off” sensor of ochratoxin A. Biosensors and Bioelectronics 26: 710–716. DOI 10.1016/j.bios.2010.06.058. [Google Scholar] [CrossRef]
Lee B, Park JH, Byun JY, Kim JH, Kim MG (2018). An optical fiber-based LSPR aptasensor for simple and rapid in-situ detection of ochratoxin A. Biosensors and Bioelectronics 102: 504–509. DOI 10.1016/j.bios.2017.11.062. [Google Scholar] [CrossRef]
Lee J, Jeon CH, Ahn SJ, Ha TH (2014). Highly stable colorimetric aptamer sensors for detection of ochratoxin A through optimizing the sequence with the covalent conjugation of hemin. Analyst 139: 1622–1627. DOI 10.1039/C3AN01639K. [Google Scholar] [CrossRef]
Li Y, Peng Z, Li Y, Xiao M, Tan G et al. (2021). An aptamer-array-based sample-to-answer biosensor for ochratoxin A detection via fluorescence resonance energy transfer. Chemosensors 9: 309. DOI 10.3390/chemosensors9110309. [Google Scholar] [CrossRef]
Li Y, Zhang N, Wang H, Zhao Q (2020). Fluorescence anisotropy-based signal-off and signal-on aptamer assays using lissamine rhodamine B as a label for ochratoxin A. Journal of Agricutural and Food Chemistry 68: 4277–4283. DOI 10.1021/acs.jafc.0c00549. [Google Scholar] [CrossRef]
Lin C, Zheng H, Sun M, Guo Y, Luo F, Guo L, Qiu B, Lin Z, Chen G (2018). Highly sensitive colorimetric aptasensor for ochratoxin A detection based on enzyme-encapsulated liposome. Analytica Chimica Acta 1002: 90–96. DOI 10.1016/j.aca.2017.11.061. [Google Scholar] [CrossRef]
Liu B, Huang R, Yu Y, Su R, Qi W, He Z (2018a). Gold nanoparticle-aptamer-based lspr sensing of ochratoxin A at a widened detection range by double calibration curve method. Frontiers in Chemistry 6: 94. DOI 10.3389/fchem.2018.00094. [Google Scholar] [CrossRef]
Liu F, Ding A, Zheng J, Chen J, Wang B (2018b). A label-free aptasensor for ochratoxin A detection based on the structure switch of aptamer. Sensors 18: 1769. DOI 10.3390/s18061769. [Google Scholar] [CrossRef]
Liu L, Tanveer ZI, Jiang K, Huang Q, Zhang J et al. (2019). Label-free fluorescent aptasensor for ochratoxin-a detection based on CdTe quantum dots and (N-Methyl-4-pyridyl) porphyrin. Toxins 11: 447. DOI 10.3390/toxins11080447. [Google Scholar] [CrossRef]
Liu R, Huang Y, Ma Y, Jia S, Gao M et al. (2015). Design and synthesis of target-responsive aptamer-cross-linked hydrogel for visual quantitative detection of ochratoxin A. ACS Applied Materials & Interfaces 7: 6982–6990. DOI 10.1021/acsami.5b01120. [Google Scholar] [CrossRef]
Liu R, Wu H, Lv L, Kang X, Cui C, Feng J, Guo Z (2018c). Fluorometric aptamer based assay for ochratoxin A based on the use of exonuclease III. Mikrochimica Acta 185: 254. DOI 10.1007/s00604-018-2786-6. [Google Scholar] [CrossRef]
Luan Y, Chen J, Li C, Xie G, Fu H, Ma Z, Lu A (2015). Highly sensitive colorimetric detection of ochratoxin A by a label-free aptamer and gold nanoparticles. Toxins 7: 5377–5385. DOI 10.3390/toxins7124883. [Google Scholar] [CrossRef]
Lv L, Cui C, Xie W, Sun S, Ji S, Tian J, Guo Z (2019). A label-free aptasensor for turn-on fluorescent detection of ochratoxin A based on aggregation-induced emission probe. Methods and Applications in Fluorescence 8: 15003. DOI 10.1088/2050-6120/ab4edf. [Google Scholar] [CrossRef]
Lv Z, Chenn A, Liu J, Guan Z, Zhou Y et al. (2014). A simple and sensitive approach for ochratoxin A detection using a label-free fluorescent aptasensor. PLoS One 9: e85968. DOI 10.1371/journal.pone.0085968. [Google Scholar] [CrossRef]
Lyu H, Sun H, Zhu Y, Wang J, Xie Z, Li J (2020). A double-recognized aptamer-molecularly imprinted monolithic column for high-specificity recognition of ochratoxin A. Analytica Chimica Acta 1103: 97–105. DOI 10.1016/j.aca.2019.12.052. [Google Scholar] [CrossRef]
Ma C, Wu K, Zhao H, Liu H, Wang K, Xia K (2018). Fluorometric aptamer-based determination of ochratoxin A based on the use of graphene oxide and RNase H-aided amplification. Mikrochimica Acta 185: 347. DOI 10.1007/s00604-018-2885-4. [Google Scholar] [CrossRef]
Ma W, Yin H, Xu L, Xu Z, Kuang H, Wang L, Xu C (2013). Femtogram ultrasensitive aptasensor for the detection of Ochratoxin A. Biosensors and Bioelectronics 42: 545–549. DOI 10.1016/j.bios.2012.11.024. [Google Scholar] [CrossRef]
Majdinasab M, Badea M, Marty JL (2022). Aptamer-based lateral flow assays: Current trends in clinical diagnostic rapid tests. Pharmaceuticals 15: 90. DOI 10.3390/ph15010090. [Google Scholar] [CrossRef]
Mazaafrianto DN, Ishida A, Maeki M, Tani H, Tokeshi M (2019). An electrochemical sensor based on structure switching of dithiol-modified aptamer for simple detection of ochratoxin A. Analytical Sciences 35: 1221–1226. DOI 10.2116/analsci.19P240. [Google Scholar] [CrossRef]
McKeague M, Velu R, Hill K, Bardoczy V, Meszaros T, DeRosa MC (2014). Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 6: 2435–2452. DOI 10.3390/toxins6082435. [Google Scholar] [CrossRef]
Mishra RK, Hayat A, Catanante G, Istamboulie G, Marty JL (2016). Sensitive quantitation of Ochratoxin A in cocoa beans using differential pulse voltammetry based aptasensor. Food Chemistry 192: 799–804. DOI 10.1016/j.foodchem.2015.07.080. [Google Scholar] [CrossRef]
Mishra RK, Hayat A, Catanante G, Ocaña C, Marty JL (2015). A label free aptasensor for Ochratoxin A detection in cocoa beans: An application to chocolate industries. Analytica Chimica Acta 889: 106–112. DOI 10.1016/j.aca.2015.06.052. [Google Scholar] [CrossRef]
Modh H, Scheper T, Walter JG (2017). Detection of ochratoxin A by aptamer-assisted real-time PCR-based assay (Apta-qPCR). Engineering in Life Sciences 17: 923–930. DOI 10.1002/elsc.201700048. [Google Scholar] [CrossRef]
Moon J, Kim G, Lee S (2013). Development of nanogold-based lateral flow immunoassay for the detection of ochratoxin A in buffer systems. Journal of Nanoscience and Nanotechnology 13: 7245–7249. DOI 10.1166/jnn.2013.8099. [Google Scholar] [CrossRef]
Muñoz K, Blaszkewicz M, Campos V, Vega M, Degen GH (2014). Exposure of infants to ochratoxin A with breast milk. Archives of Toxicology 88: 837–846. DOI 10.1007/s00204-013-1168-4. [Google Scholar] [CrossRef]
Nameghi MA, Danesh NM, Ramezani M, Hassani FV, Abnous K, Taghdisi SM (2016). A fluorescent aptasensor based on a DNA pyramid nanostructure for ultrasensitive detection of ochratoxin A. Analytical and Bioanalytical Chemistry 408: 5811–5818. DOI 10.1007/s00216-016-9693-7. [Google Scholar] [CrossRef]
Nan M, Bi Y, Xue H, Xue S, Long H et al. (2019). Rapid determination of Ochratoxin A in grape and its commodities based on a label-free impedimetric aptasensor constructed by layer-by-layer self-assembly. Toxins 11: 71. DOI 10.3390/toxins11020071. [Google Scholar] [CrossRef]
Nekrasov N, Jaric S, Kireev D, Emelianov AV, Orlov AV, Gadjanski I, Nikitin PI, Akinwande D, Bobrinetskiy I (2022). Real-time detection of ochratoxin A in wine through insight of aptamer conformation in conjunction with graphene field-effect transistor. Biosensors and Bioelectronics 200: 113890. DOI 10.1016/j.bios.2021.113890. [Google Scholar] [CrossRef]
Oh HK, Joung HA, Jung M, Lee H, Kim MG (2019). Rapid and simple detection of ochratoxin A using fluorescence resonance energy transfer on lateral flow immunoassay (FRET-LFI). Toxins 11: 292. DOI 10.3390/toxins11050292. [Google Scholar] [CrossRef]
Olsson J, Börjesson T, Lundstedt T, Schnurer J (2002). Detection and quantification of ochratoxin A and deoxynivalenol in barley grains by GC-MS and electronic nose. International Journal of Food Microbiology 72: 203–214. DOI 10.1016/S0168-1605(01)00685-7. [Google Scholar] [CrossRef]
Park JH, Byun JY, Mun H, Shim WB, Shin YB, Li T, Kim MG (2014). A regeneratable, label-free, localized surface plasmon resonance (LSPR) aptasensor for the detection of ochratoxin A. Biosensors and Bioelectronics 59: 321–327. DOI 10.1016/j.bios.2014.03.059. [Google Scholar] [CrossRef]
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). UCSF Chimera–A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25: 1605–1612. DOI 10.1002/(ISSN)1096-987X. [Google Scholar] [CrossRef]
Pittet A, Royer D (2002). Rapid, low cost thin-layer chromatographic screening method for the detection of ochratoxin A in green coffee at a control level of 10 μg/kg. Journal of Agricultural and Food Chemistry 50: 243–247. DOI 10.1021/jf010867w. [Google Scholar] [CrossRef]
Prabhakar N, Matharu Z, Malhotra BD (2011). Polyaniline Langmuir-Blodgett film based aptasensor for ochratoxin A detection. Biosensors and Bioelectronics 26: 4006–4011. DOI 10.1016/j.bios.2011.03.014. [Google Scholar] [CrossRef]
Qian J, Ren C, Wang C, Chen W, Lu X, Li H, Liu Q, Hao N, Li H, Wang K (2018). Magnetically controlled fluorescence aptasensor for simultaneous determination of ochratoxin A and aflatoxin B1. Analytica Chimica Acta 1019: 119–127. DOI 10.1016/j.aca.2018.02.063. [Google Scholar] [CrossRef]
Qian J, Wang K, Wang C, Hua M, Yang Z, Liu Q, Mao H, Wang K (2015). A FRET-based ratiometric fluorescent aptasensor for rapid and onsite visual detection of ochratoxin A. Analyst 140: 7434–7442. DOI 10.1039/C5AN01403D. [Google Scholar] [CrossRef]
Qian M, Hu W, Wang L, Wang L, Dong Y (2020). A non-enzyme and non-label sensitive fluorescent aptasensor based on simulation-assisted and target-triggered hairpin probe self-assembly for ochratoxin A detection. Toxins 12: 376. DOI 10.3390/toxins12060376. [Google Scholar] [CrossRef]
Rangel AE, Chen Z, Ayele TM, Heemstra JM (2018). In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Research 46: 8057–8068. DOI 10.1093/nar/gky667. [Google Scholar] [CrossRef]
Ren X, Lu P, Feng R, Zhang T, Zhang Y et al. (2018). An ITO-based point-of-care colorimetric immunosensor for ochratoxin A detection. Talanta 188: 593–599. DOI 10.1016/j.talanta.2018.06.004. [Google Scholar] [CrossRef]
Rivas L, Mayorga-Martinez CC, Quesada-Gonzalez D, Zamora-Galvez A, de la Escosura-Muniz A, Merkoci A (2015). Label-free impedimetric aptasensor for ochratoxin-A detection using iridium oxide nanoparticles. Analytical Chemistry 87: 5167–5172. DOI 10.1021/acs.analchem.5b00890. [Google Scholar] [CrossRef]
Rousseau DM, Slegers GA, van Peteghem CH (1985). Radioimmunoassay of ochratoxin A in barley. Applied and Environmental Microbiology 50: 529–531. DOI 10.1128/aem.50.2.529-531.1985. [Google Scholar] [CrossRef]
Samokhvalov AV, Safenkova IV, Eremin SA, Zherdev AV, Dzantiev BB (2017). Use of anchor protein modules in fluorescence polarisation aptamer assay for ochratoxin A determination. Analytica Chimica Acta 962: 80–87. DOI 10.1016/j.aca.2017.01.024. [Google Scholar] [CrossRef]
Schmitz FRW, Valério A, de Oliveira D, Hotza D (2020). An overview and future prospects on aptamers for food safety. Applied Microbiology and Biotechnology 104: 6929–6939. DOI 10.1007/s00253-020-10747-0. [Google Scholar] [CrossRef]
Shaikh SA, Jayaram B (2007). A swift all-atom energy-based computational protocol to predict DNA−Ligand binding affinity and ΔTm. Journal of Medicinal Chemistry 50: 2240–2244. DOI 10.1021/jm060542c. [Google Scholar] [CrossRef]
Shen P, Li W, Ding Z, Deng Y, Liu Y, Zhu X, Cai T, Li T, Zheng T (2018). A competitive aptamer chemiluminescence assay for ochratoxin A using a single silica photonic crystal microsphere. Analytical Biochemistry 554: 28–33. DOI 10.1016/j.ab.2018.05.025. [Google Scholar] [CrossRef]
Shen P, Li W, Liu Y, Ding Z, Deng Y, Zhu X, Jin Y, Li Y, Li J, Zheng T (2017). High-throughput low-background G-quadruplex aptamer chemiluminescence assay for Ochratoxin A using a single photonic crystal microsphere. Analytical Chemistry 89: 11862–11868. DOI 10.1021/acs.analchem.7b03592. [Google Scholar] [CrossRef]
Sheng L, Ren J, Miao Y, Wang J, Wang E (2011). PVP-coated graphene oxide for selective determination of ochratoxin A via quenching fluorescence of free aptamer. Biosensors and Bioelectronics 26: 3494–3499. DOI 10.1016/j.bios.2011.01.032. [Google Scholar] [CrossRef]
Sibanda L, de Saeger S, Bauters TG, Nelis HJ, van Peteghem C (2001). Development of a flow-through enzyme immunoassay and application in screening green coffee samples for ochratoxin A with confirmation by high-performance liquid chromatography. Journal of Food Protection 64: 1597–1602. DOI 10.4315/0362-028X-64.10.1597. [Google Scholar] [CrossRef]
Somerson J, Plaxco KW (2018). Electrochemical Aptamer-based sensors for rapid point-of-use monitoring of the mycotoxin ochratoxin A directly in a food stream. Molecules 23: 912. DOI 10.3390/molecules23040912. [Google Scholar] [CrossRef]
Song D, Yang R, Fang S, Liu Y, Long F et al. (2018). SERS based aptasensor for ochratoxin A by combining Fe3O4@Au magnetic nanoparticles and Au-DTNB@Ag nanoprobes with multiple signal enhancement. Mikrochimica Acta 185: 491. DOI 10.1007/s00604-018-3020-2. [Google Scholar] [CrossRef]
Song S, Wang L, Li J, Fan C, Zhao J (2008). Aptamer-based biosensors. Trends in Analytical Chemistry 27: 108–117. DOI 10.1016/j.trac.2007.12.004. [Google Scholar] [CrossRef]
Suea-Ngam A, Howes PD, Stanley CE, deMello AJ (2019). An ochratoxin A detection. ACS Sensors 4: 1560–1568. DOI 10.1021/acssensors.9b00237. [Google Scholar] [CrossRef]
Sun AL, Zhang YF, Sun GP, Wang XN, Tang D (2017). Homogeneous electrochemical detection of ochratoxin A in foodstuff using aptamer-graphene oxide nanosheets and DNase I-based target recycling reaction. Biosensors and Bioelectronics 89: 659–665. DOI 10.1016/j.bios.2015.12.032. [Google Scholar] [CrossRef]
Sun Z, Wang X, Tang Z, Chen Q, Liu X (2019). Development of a biotin-streptavidin-amplified nanobody-based ELISA for ochratoxin A in cereal. Ecotoxicology and Environmental Safety 171: 382–388. DOI 10.1016/j.ecoenv.2018.12.103. [Google Scholar] [CrossRef]
Taghdisi SM, Danesh NM, Beheshti HR, Ramezani M, Abnous K (2016). A novel fluorescent aptasensor based on gold and silica nanoparticles for the ultrasensitive detection of ochratoxin A. Nanoscale 8: 3439–3446. DOI 10.1039/C5NR08234J. [Google Scholar] [CrossRef]
Tang J, Huang Y, Cheng Y, Huang L, Zhuang J, Tang D (2018). Two-dimensional MoS2 as a nano-binder for ssDNA: Ultrasensitive aptamer based amperometric detection of Ochratoxin A. Mikrochimica Acta 185: 162. DOI 10.1007/s00604-018-2706-9. [Google Scholar] [CrossRef]
Tian F, Zhou J, Fu R, Cui Y, Zhao Q, Jiao B, He Y (2020). Multicolor colorimetric detection of ochratoxin A via structure-switching aptamer and enzyme-induced metallization of gold nanorods. Food Chemistry 320: 126607. DOI 10.1016/j.foodchem.2020.126607. [Google Scholar] [CrossRef]
Tian J, Wei W, Wang J, Ji S, Chen G, Lu J (2018). Fluorescence resonance energy transfer aptasensor between nanoceria and graphene quantum dots for the determination of ochratoxin A. Analytica Chimica Acta 1000: 265–272. DOI 10.1016/j.aca.2017.08.018. [Google Scholar] [CrossRef]
Tong P, Zhang L, Xu JJ, Chen HY (2011). Simply amplified electrochemical aptasensor of ochratoxin A based on exonuclease-catalyzed target recycling. Biosensors and Bioelectronics 29: 97–101. DOI 10.1016/j.bios.2011.07.075. [Google Scholar] [CrossRef]
Vargas-Montes M, Cardona N, Moncada DM, olina DA, Zhang Y et al. (2019). Enzyme-linked aptamer assay (ELAA) for detection of toxoplasma ROP18 protein in human serum. Frontiers in Cellular and Infection Microbiology 9: 1606. DOI 10.3389/fcimb.2019.00386. [Google Scholar] [CrossRef]
Velu R, DeRosa MC (2018). Lateral flow assays for Ochratoxin A using metal nanoparticles: comparison of “adsorption-desorption” approach to linkage inversion assembled nano-aptasensors (LIANA). Analyst 143: 4566–4574. DOI 10.1039/C8AN00963E. [Google Scholar] [CrossRef]
Wallace AC, Laskowski RA, Thornton JM (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, Design and Selection 8: 127–134. DOI 10.1093/protein/8.2.127. [Google Scholar] [CrossRef]
Wang C, Dong X, Liu Q, Wang K (2015a). Label-free colorimetric aptasensor for sensitive detection of ochratoxin A utilizing hybridization chain reaction. Analytica Chimica Acta 860: 83–88. DOI 10.1016/j.aca.2014.12.031. [Google Scholar] [CrossRef]
Wang C, Qian J, Wang K, Yang X, Liu Q, Hao N, Wang C, Dong X, Huang X (2016a). Colorimetric aptasensing of ochratoxin A using Au@Fe3O4 nanoparticles as signal indicator and magnetic separator. Biosensors and Bioelectronics 77: 1183–1191. DOI 10.1016/j.bios.2015.11.004. [Google Scholar] [CrossRef]
Wang Q, Chen M, Zhang H, Wen W, Zhang X et al. (2016b). Enhanced electrochemiluminescence of RuSi nanoparticles for ultrasensitive detection of ochratoxin A by energy transfer with CdTe quantum dots. Biosensors and Bioelectronics 79: 561–567. DOI 10.1016/j.bios.2015.12.098. [Google Scholar] [CrossRef]
Wang R, Xiang Y, Zhou X, Liu LH, Shi H (2015b). A reusable aptamer-based evanescent wave all-fiber biosensor for highly sensitive detection of Ochratoxin A. Biosensors and Bioelectronics 66: 11–18. DOI 10.1016/j.bios.2014.10.079. [Google Scholar] [CrossRef]
Wang S, Niu R, Yang Y, Zhou X, Luo S, Zhang C, Wang Y (2020). Aptamer-functionalized chitosan magnetic nanoparticles as a novel adsorbent for selective extraction of ochratoxin A. International Journal of Biological Macromolecules 153: 583–590. DOI 10.1016/j.ijbiomac.2020.03.035. [Google Scholar] [CrossRef]
Wang S, Zhang Y, Pang G, Zhang Y, Guo S (2017a). Tuning the aggregation/disaggregation behavior of graphene quantum dots by structure-switching aptamer for high-sensitivity fluorescent ochratoxin A sensor. Analytical Chemistry 89: 1704–1709. DOI 10.1021/acs.analchem.6b03913. [Google Scholar] [CrossRef]
Wang Z, Duan N, Hun X, Wu S (2010). Electrochemiluminescent aptamer biosensor for the determination of ochratoxin A at a gold-nanoparticles-modified gold electrode using N-(aminobutyl)-N-ethylisoluminol as a luminescent label. Analytical and Bioanalytical Chemistry 398: 2125–2132. DOI 10.1007/s00216-010-4146-1. [Google Scholar] [CrossRef]
Wang Z, Yu H, Han J, Xie G, Chen S (2017b). Rare Co/Fe-MOFs exhibiting high catalytic activity in electrochemical aptasensors for ultrasensitive detection of ochratoxin A. Chemical Communications 53: 9926–9929. DOI 10.1039/C7CC05327D. [Google Scholar] [CrossRef]
Wei M, Wang C, Xu E, Chen J, Xu X, Wei W, Liu S (2019). A simple and sensitive electrochemiluminescence aptasensor for determination of ochratoxin A based on a nicking endonuclease-powered DNA walking machine. Food Chemistry 282: 141–146. DOI 10.1016/j.foodchem.2019.01.011. [Google Scholar] [CrossRef]
Wei M, Xin L, Feng S, Liu Y (2020). Simultaneous electrochemical determination of ochratoxin A and fumonisin B1 with an aptasensor based on the use of a Y-shaped DNA structure on gold nanorods. Mikrochimica Acta 187: 102. DOI 10.1007/s00604-019-4089-y. [Google Scholar] [CrossRef]
Wei M, Zhang W (2018). The determination of Ochratoxin A based on the electrochemical aptasensor by carbon aerogels and methylene blue assisted signal amplification. Chemistry Central Journal 12: 45. DOI 10.1186/s13065-018-0415-4. [Google Scholar] [CrossRef]
Wei Y, Zhang J, Wang X, Duan Y (2015). Amplified fluorescent aptasensor through catalytic recycling for highly sensitive detection of ochratoxin A. Biosensors and Bioelectronics 65: 16–22. DOI 10.1016/j.bios.2014.09.100. [Google Scholar] [CrossRef]
Wu H, Liu R, Kang X, Liang C, Lv L, Guo Z (2017). Fluorometric aptamer assay for ochratoxin A based on the use of single walled carbon nanohorns and exonuclease III-aided amplification. Mikrochimica Acta 185: 27. DOI 10.1007/s00604-017-2592-6. [Google Scholar] [CrossRef]
Wu J, Chu H, Mei Z, Deng Y, Xue F, Zheng L, Chen W (2012). Ultrasensitive one-step rapid detection of ochratoxin A by the folding-based electrochemical aptasensor. Analytica Chimica Acta 753: 27–31. DOI 10.1016/j.aca.2012.09.036. [Google Scholar] [CrossRef]
Wu K, Ma C, Zhao H, Chen M, Deng Z (2019). Sensitive aptamer-based fluorescene assay for ochratoxin A based on RNase H signal amplification. Food Chemistry 277: 273–278. DOI 10.1016/j.foodchem.2018.10.130. [Google Scholar] [CrossRef]
Wu K, Ma C, Zhao H, He H, Chen H (2018a). Label-free G-quadruplex aptamer fluorescence assay for ochratoxin A using a thioflavin T probe. Toxins 10: 198. DOI 10.3390/toxins10050198. [Google Scholar] [CrossRef]
Wu S, Duan N, Wang Z, Wang H (2011a). Aptamer-functionalized magnetic nanoparticle-based bioassay for the detection of ochratoxin A using upconversion nanoparticles as labels. Analyst 136: 2306–2314. DOI 10.1039/c0an00735h. [Google Scholar] [CrossRef]
Wu S, Liu L, Duan N, Wang W, Yu Q, Wang Z (2018b). A test strip for ochratoxin A based on the use of aptamer-modified fluorescence upconversion nanoparticles. Mikrochimica Acta 185: 497. DOI 10.1007/s00604-018-3022-0. [Google Scholar] [CrossRef]
Wu X, Hu J, Zhu B, Lu L, Huang X, Pang D (2011b). Aptamer-targeted magnetic nanospheres as a solid-phase extraction sorbent for determination of ochratoxin A in food samples. Journal of Chromatography A 1218: 7341–7346. DOI 10.1016/j.chroma.2011.08.045. [Google Scholar] [CrossRef]
Xiao MW, Bai XL, Liu YM, Yang L, Liao X (2018). Simultaneous determination of trace Aflatoxin B1 and Ochratoxin A by aptamer-based microchip capillary electrophoresis in food samples. Journal of Chromatography A 1569: 222–228. DOI 10.1016/j.chroma.2018.07.051. [Google Scholar] [CrossRef]
Xie S, Chai Y, Yuan Y, Bai L, Yuan R (2014). Development of an electrochemical method for ochratoxin A detection based on aptamer and loop-mediated isothermal amplification. Biosensors and Bioelectronics 55: 324–329. DOI 10.1016/j.bios.2013.11.009. [Google Scholar] [CrossRef]
Xu G, Zhao J, Liu N, Yang M, Zhao Q, Li C, Liu M (2019). Structure-guided post-SELEX optimization of an ochratoxin A aptamer. Nucleic Acids Research 47: 5963–5972. DOI 10.1093/nar/gkz336. [Google Scholar] [CrossRef]
Yan Y, Tao H, He J, Huang S-Y (2020). The HDOCK server for integrated protein-protein docking. Nature Protocols 15: 1829–1852. DOI 10.1038/s41596-020-0312-x. [Google Scholar] [CrossRef]
Yang C, Lates V, Prieto-Simon B, Marty JL, Yang X (2012). Aptamer-DNAzyme hairpins for biosensing of Ochratoxin A. Biosensors and Bioelectronics 32: 208–212. DOI 10.1016/j.bios.2011.12.011. [Google Scholar] [CrossRef]
Yang C, Lates V, Prieto-Simon B, Marty JL, Yang X (2013). Rapid high-throughput analysis of ochratoxin A by the self-assembly of DNAzyme-aptamer conjugates in wine. Talanta 116: 520–526. DOI 10.1016/j.talanta.2013.07.011. [Google Scholar] [CrossRef]
Yang C, Wang Y, Marty JL, Yang X (2011). Aptamer-based colorimetric biosensing of Ochratoxin A using unmodified gold nanoparticles indicator. Biosensors and Bioelectronics 26: 2724–2727. DOI 10.1016/j.bios.2010.09.032. [Google Scholar] [CrossRef]
Yang L, Zhang Li R, Lin C, Guo L, Qiu B, Lin Z, Chen G (2015). Electrochemiluminescence biosensor for ultrasensitive determination of ochratoxin A in corn samples based on aptamer and hyperbranched rolling circle amplification. Biosensors and Bioelectronics 70: 268–274. DOI 10.1016/j.bios.2015.03.067. [Google Scholar] [CrossRef]
Yang X, Hu Y, Kong W, Chu X, Yang M, Zhao M, Ouyang Z (2014). Ultra-fast liquid chromatography with tandem mass spectrometry determination of ochratoxin A in traditional Chinese medicines based on vortex-assisted solid-liquid microextraction and aptamer-affinity column clean-up. Journal of Separation Science 37: 3052–3059. DOI 10.1002/jssc.201400635. [Google Scholar] [CrossRef]
Yang YJ, Zhou Y, Xing Y, Zhang GM, Zhang Y et al. (2019). A Label-free aptasensor based on Aptamer/NH2 Janus particles for ultrasensitive electrochemical detection of Ochratoxin A. Talanta 199: 310–316. DOI 10.1016/j.talanta.2019.02.015. [Google Scholar] [CrossRef]
Yin X, Wang S, Liu X, He C, Tang Y, Li Q, Liu J, Su H, Tan T, Dong Y (2017). Aptamer-based colorimetric biosensing of ochratoxin A in fortified white grape wine sample using unmodified gold nanoparticles. Analytical Science 33: 659–664. DOI 10.2116/analsci.33.659. [Google Scholar] [CrossRef]
Yu X, Lai S, Wang L, Chen Y, Lin X, Xie Z (2019). Preparation of aptamer-bound polyamine affinity monolithic column via a facile triazine-bridged strategy and application to on-column specific discrimination of ochratoxin A. Journal of Separation Science 42: 2272–2279. DOI 10.1002/jssc.201900175. [Google Scholar] [CrossRef]
Yu X, Song H, Huang J, Chen Y, Dai M, Lin X, Xie Z (2018). An aptamer@AuNP-modified POSS-polyethylenimine hybrid affinity monolith with a high aptamer coverage density for sensitive and selective recognition of ochratoxin A. Journal of Materials Chemistry B 6: 1965–1972. DOI 10.1039/C7TB03319B. [Google Scholar] [CrossRef]
Yuan Y, Wei S, Liu G, Xie S, Chai Y, Yuan R (2014). Ultrasensitive electrochemiluminescent aptasensor for ochratoxin A detection with the loop-mediated isothermal amplification. Analytica Chimica Acta 811: 70–75. DOI 10.1016/j.aca.2013.11.022. [Google Scholar] [CrossRef]
Yue S, Jie X, Wei L, Bin C, Dou DW, Yi Y, QingXia L, JianLin J, TieSong Z (2014). Simultaneous detection of ochratoxin A and fumonisin B1 in cereal samples using an aptamer-photonic crystal encoded suspension array. Analytical Chemistry 86: 11797–11802. DOI 10.1021/ac503355n. [Google Scholar] [CrossRef]
Zejli H, Goud KY, Marty JL (2018). Label free aptasensor for ochratoxin A detection using polythiophene-3-carboxylic acid. Talanta 185: 513–519. DOI 10.1016/j.talanta.2018.03.089. [Google Scholar] [CrossRef]
Zhang J, Zhang X, Yang G, Chen J, Wang S (2013). A signal-on fluorescent aptasensor based on Tb3+ and structure-switching aptamer for label-free detection of Ochratoxin A in wheat. Biosensors and Bioelectronics 41: 704–709. DOI 10.1016/j.bios.2012.09.053. [Google Scholar] [CrossRef]
Zhao Q, Geng X, Wang H (2013). Fluorescent sensing ochratoxin A with single fluorophore-labeled aptamer. Analytical and Bioanalytical Chemistry 405: 6281–6286. DOI 10.1007/s00216-013-7047-2. [Google Scholar] [CrossRef]
Zhao Q, Li X-F, Shao Y, Le XC (2008). Aptamer-based affinity chromatographic assays for thrombin. Analytical Chemistry 80: 7586–7593. DOI 10.1021/ac801206s. [Google Scholar] [CrossRef]
Zhao Q, Lv Q, Wang H (2014). Identification of allosteric nucleotide sites of tetramethylrhodamine-labeled aptamer for noncompetitive aptamer-based fluorescence anisotropy detection of a small molecule, ochratoxin A. Anaytical Chemistry 86: 1238–1245. DOI 10.1021/ac4035532. [Google Scholar] [CrossRef]
Zhao X, Jin X, Lin Z, Guo Q, Liu B et al. (2021). Simultaneous rapid detection of aflatoxin B1 and ochratoxin A in spices using lateral flow immuno-chromatographic assay. Foods 10: 2738. DOI 10.3390/foods10112738. [Google Scholar] [CrossRef]
Zheng F, Ke W, Shi L, Liu H, Zhao Y (2019). Plasmonic Au-Ag Janus nanoparticle engineered ratiometric surface-enhanced Raman scattering aptasensor for ochratoxin A detection. Analytical Chemistry 91: 11812–11820. DOI 10.1021/acs.analchem.9b02469. [Google Scholar] [CrossRef]
Zhou G, Wilson G, Hebbard H, Duan W, Liddle C, George J, Qiao L (2016a). Aptamers: A promising chemical antibody for cancer therapy. Oncotarget 7: 13446–13463. DOI 10.18632/oncotarget.7178. [Google Scholar] [CrossRef]
Zhou W, Kong W, Dou X, Zhao M, Ouyang Z, Yang M (2016b). An aptamer based lateral flow strip for on-site rapid detection of ochratoxin A in Astragalus membranaceus. Journal of Chromatography B 1022: 102–108. DOI 10.1016/j.jchromb.2016.04.016. [Google Scholar] [CrossRef]
Zhu C, Liu D, Li Y, Shen X, Ma S, Liu Y, You T (2020). Ratiometric electrochemical aptasensor for ultrasensitive detection of Ochratoxin A based on a dual signal amplification strategy: Engineering the binding of methylene blue to DNA. Biosensors and Bioelectronics 150: 111814. DOI 10.1016/j.bios.2019.111814. [Google Scholar] [CrossRef]
Zuker M (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. DOI 10.1093/nar/gkg595. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools