 Open Access
Open Access
ARTICLE
Comprehensive bioinformatics analysis of CYB561 expression in breast cancer: Link between prognosis and immune infiltration
1 Department of Radiation Oncology, Affiliated Hospital of Guilin Medical University, Guilin, 541001, China
2 Guangxi Key Laboratory of Molecular Medicine in Liver Injury and Repair, Affiliated Hospital of Guilin Medical University, Guilin, 541001, China
3 Guangxi Health Commission Key Laboratory of Basic Research in Sphingolipid Metabolism Related Diseases, Affiliated Hospital of Guilin Medical University, Guilin, 541001, China
4 Department of Information, Affiliated Hospital of Guilin Medical University, Guilin, 541001, China
5 Department of Radiation Oncology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 53002, China
6 Department of Nuclear Medicine, Affiliated Hospital of Guilin Medical University, Guilin, 541001, China
* Corresponding Author: MEI DENG. Email:
# These authors have contributed equally to this work and share first authorship
BIOCELL 2023, 47(5), 1021-1037. https://doi.org/10.32604/biocell.2023.027103
Received 14 October 2022; Accepted 16 January 2023; Issue published 10 April 2023
Abstract
Background: Cytochrome b561 (CYB561) plays a critical role in neuroendocrine function, cardiovascular regulation, and tumor growth; however, the prognostic value of CYB561 in patients with breast cancer and the relationship between CYB561 expression and immune infiltration in breast cancer remain unclear. Methods: The mRNA expression and clinical data of patients with breast cancer were obtained from The Cancer Genome Atlas database. Functional enrichment analysis was used to explore underlying biological functions associated with CYB561. The methylation status of CYB561 was analyzed using the MethSurv database. The enrichment score of immune cell infiltration for CYB561 in breast cancer was calculated using single-sample gene set enrichment analysis. The prognostic value of CYB561 was evaluated using Kaplan-Meier method and Cox regression analysis. Based on the results of the multivariate Cox analysis, a nomogram was constructed to predict the effect of CYB561 expression on overall survival (OS). Results: The results showed that CYB561 was highly expressed in breast cancer tissues. Hypomethylation of CYB561 is associated with an unfavorable prognosis. In multivariate Cox regression analysis, CYB561 was an independent prognostic factor for OS. Functional enrichment analysis indicated that estrogen signaling pathway, inflammatory response, KRAS signaling pathway, epithelial-mesenchymal transition, leukocyte migration, and regulation of lymphocyte activation were strongly enriched in the low CYB561 expression group. Additionally, CYB561 expression was negatively correlated with immune infiltration of B cells, plasmacytoid dendritic cells, dendritic cells, and neutrophils. Conclusion: CYB561 may serve as a potential biomarker for breast cancer diagnosis and prognosis.Keywords
Supplementary Material
Supplementary Material FileAccording to the Global Burden of Cancer Report, breast cancer (BRCA) surpassed lung cancer as the most common cancer worldwide by 2020, with an estimated 2.3 million new cases and 685,000 deaths (Sung et al., 2021). Although treatment strategies for BRCA are well-developed, the tumor-related mortality rate remains relatively high. Owing to the limitations of individualized tumor therapy, the development of new methods to screen high-risk groups and provide accurate prognoses has become an urgent need for BRCA patients.
Cytochrome b561 (CYB561) is a protein family composed of six transmembrane helix proteins, and their structural characteristics and sequence are conserved at the protein level (Bashtovyy et al., 2003). CYB561 is a prototype of the diheme cytochrome family, which is widely distributed in plant and animal species (Tsubaki et al., 2005). Ascorbate (ASC) is the main electron donor for enzymatic reactions in the redox processes of plants and animals (Foyer and Noctor, 2011). For the maintenance of normal neuroendocrine function, CYB561 supports the regeneration of ASC, which is an indispensable process for dopamine β-hydroxylation (Berczi et al., 2005; van den Berg et al., 2018). CYB561 is also involved in tumor suppression and cooperates with ASC to inhibit lung cancer cell growth (Ohtani et al., 2007). In a meta-analysis of single-gene prognostic biomarkers for ovarian cancer, low CYB561 expression levels indicated a poor prognosis (Willis et al., 2016). Additionally, an alternate promoter of CYB561 was associated with clinical outcomes in patients with endometrial cancer, and decreased CYB561 expression was predictive of inferior overall survival (OS) (Sun et al., 2022). In contrast, CYB561 was observed to be highly expressed in metastatic prostate cancer cells and to promote the proliferation and metastatic potential of prostate cancer cells (Olarte and Bagamasbad, 2020). However, the prognostic value of CYB561 expression and its correlation with immune infiltration in BRCA remain unclear.
In the present study, our aim was to investigate the relationship linking CYB561 expression and patient outcomes, pathological characteristics, and infiltration abundances of immune cells using bioinformatics, which may aid in designing individualized therapeutic strategies.
The RNAseq data with clinical information of BRCA patients were collected from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/), and the Genotype Tissue Expression Project (GTEx) databases (https://commonfund.nih.gov/gtex). Subsequently, the fragments per kilobase of transcript per million fragments mapped (FPKM) data (level 3) were converted to transcripts per million (TPM) reads. To verify the prognostic value of CYB561 expression in BRCA patients, the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset was also obtained from the cBioPortal database (http://www.cbioportal.org/). The mRNA expression data of various types of cancer were collected from the UCSC Xena (https://xenabrowser.net/datapages/) and GTEx databases.
Differentially expressed gene (DEG) analysis
All BRCA patients in the TCGA cohort were divided into two groups (high vs. low expression) based on the median CYB561 expression level. The DEGs between the two groups were identified using the R package DESeq2 with an adjusted p-value < 0.05 and |log2-fold-change (FC)| > 1.5 as the threshold (Love et al., 2014). Spearman’s correlation analysis was applied to assess the correlations between the CYB561 expression and the top 10 DEGs in the two groups.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were applied to calculate the enrichment terms of the DEGs by using the R package GO plot (version 1.0.2), based on p < 0.05, |log2FC| > 1. We used the R package ClusterProfiler for Gene set enrichment analysis (GSEA) to identify potential CYB561-related functions and pathways (Yu et al., 2012). Only enrichment in function or pathway with an adjusted p-value < 0.05 and a false discovery rate (FDR) < 0.25 were considered statistically significant.
The methylation status of the CYB561 promoter in BRCA was identified using the UALCAN database (http://ualcan.path.uab.edu) (Chandrashekar et al., 2017). Correlations between the expression of CYB561 and DNA methyltransferases were calculated using Spearman’s correlation analysis. The heatmap of DNA methylation sites and the prognostic value of CYB561-methylation levels were obtained from a web-based MethSurv methylation analysis tool (https://biit.cs.ut.ee/methsurv/) (Modhukur et al., 2017).
Immune cell infiltration analysis
The relative infiltration of immune cells in the BRCA microenvironment was analyzed using single-sample gene set enrichment analysis (ssGSEA), which was implemented using the GSVA package. Spearman’s correlation analysis was utilized to explore the correlation between various immune cell subsets and CYB561 expression. Next, the Wilcoxon rank sum test was used to compare immune cell infiltration between CYB561 high and low groups, and a p-value < 0.05 was set as the threshold. Additionally, the correlations between CYB561 expression and tumor-related immune checkpoint genes were evaluated using the R package IOBR (Zeng et al., 2021).
The BRCA patients were divided into high and low CYB561 expression groups based on the median value. Overall survival (OS) and disease-specific survival (DSS) were calculated using the Kaplan–Meier method with the log-rank test. Subsequently, we used univariate and multivariate Cox regression analysis to evaluate the effect of CYB561 expression on OS and DSS in the context of other clinicopathological variables.
Nomogram construction and validation
The results of the multivariate Cox analysis were used to construct a nomogram that predicts the OS probability. The bootstrap validation method was used to calculate the consistency index (C-index) to estimate the bias-corrected or overfitting-corrected prediction accuracy of the model. The nomogram and calibration plots were generated with the package RMS of R software. The prediction performance of the model was also evaluated by using R package timeROC through time-dependent receiver operating characteristic (ROC) curves.
Statistical analysis was performed using code written in R programming language (version 3.6.3) (https://www.r-project.org/). The Wilcoxon rank sum test and paired sample t-test were used to evaluate CYB561 expression in unpaired and paired samples, respectively. Wilcoxon rank-sum test and logistic regression were applied to calculate the associations between CYB561 expression and clinical variables. In all tests, p-value < 0.05 was considered statistically significant.
In our study, a total of 1065 BRCA cases with RNA-sequencing data and matched clinical information were included. Among them, 110 cases had paired tumor and adjacent normal tissue samples. The clinicopathological characteristics of the cases are shown in Suppl. Table S1.
Overexpression of CYB561 in BRCA
We first compared the expression levels of CYB561 in 33 tumor tissues and corresponding normal tissues. As shown in Fig. 1A, the expression of CYB561 was significantly high in most types of cancers, such as bladder urothelial carcinoma, cholangiocarcinoma, and colon adenocarcinoma (Fig. 1A). Next, we evaluated the expression of CYB561 in BRCA samples. The results showed that CYB561 was highly expressed in BRCA compared to normal breast tissues in both unpaired and paired tests (p < 0.001; Figs. 1B and 1C). Additionally, the ROC curve showed that CYB561 expression had great diagnostic accuracy to distinguish BRCA tissues from normal tissues, with an area under the curve (AUC) of 0.919 (95% confidence interval [CI] = 0.902–0.935) (Fig. 1D).
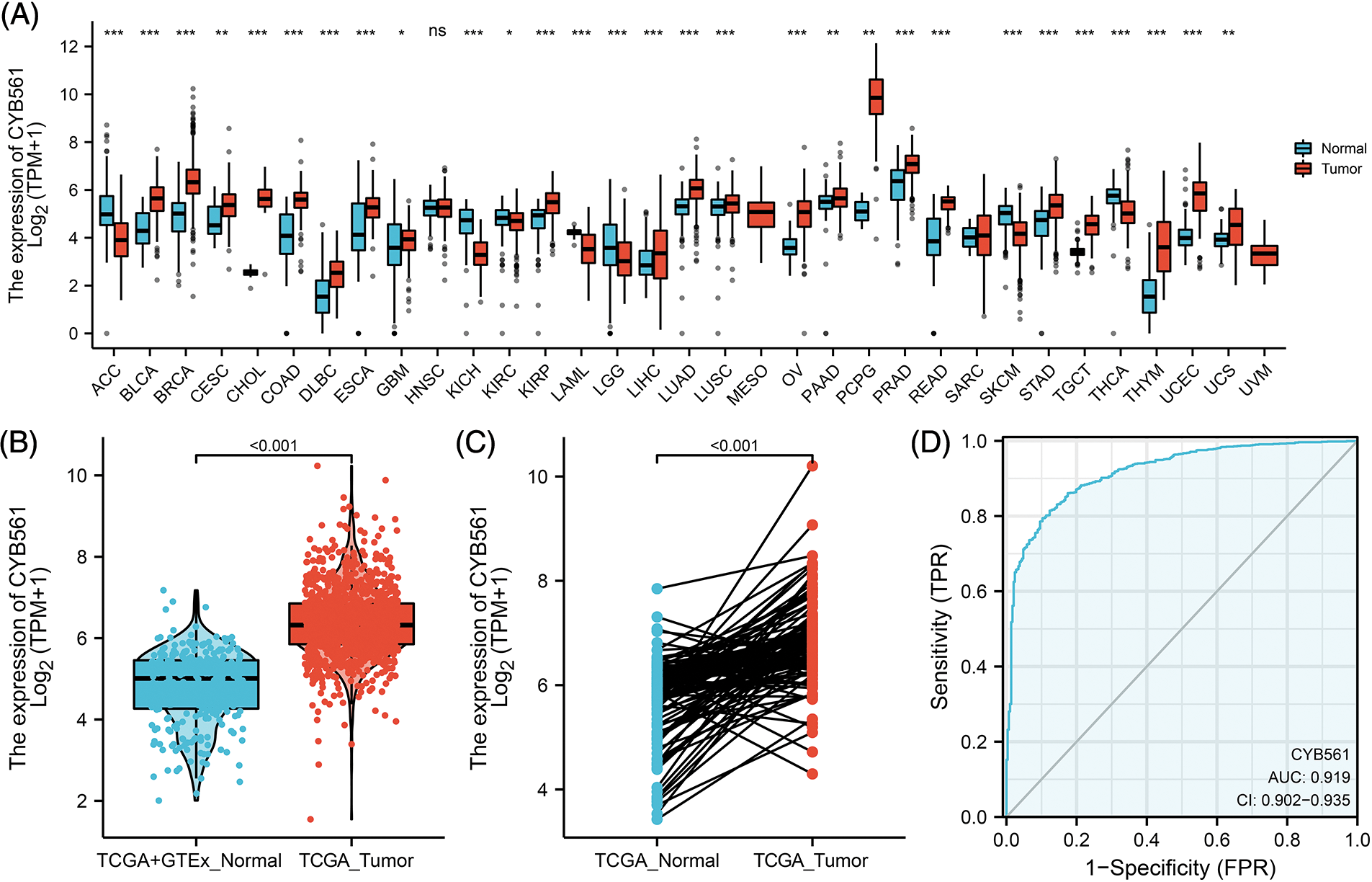
Figure 1: Expression of CYB561 in BRCA and normal tissues. (A) Expression of CYB561 in 33 types of cancer and normal tissues. (B) Expression of CYB561 in BRCA and normal tissues. (C) Expression of CYB561 in BRCA and matched normal tissues. (D) ROC curve shows that CYB561 has good discrimination ability between cancer and normal tissues. BRCA, breast cancer; TCGA, The Cancer Genome Atlas; GTEx, Genotype Tissue Expression Project; ROC, receiver operating characteristic. *p < 0.05, ** p < 0.01, and ***p < 0.001.
Association between CYB561 expression and clinicopathologic features
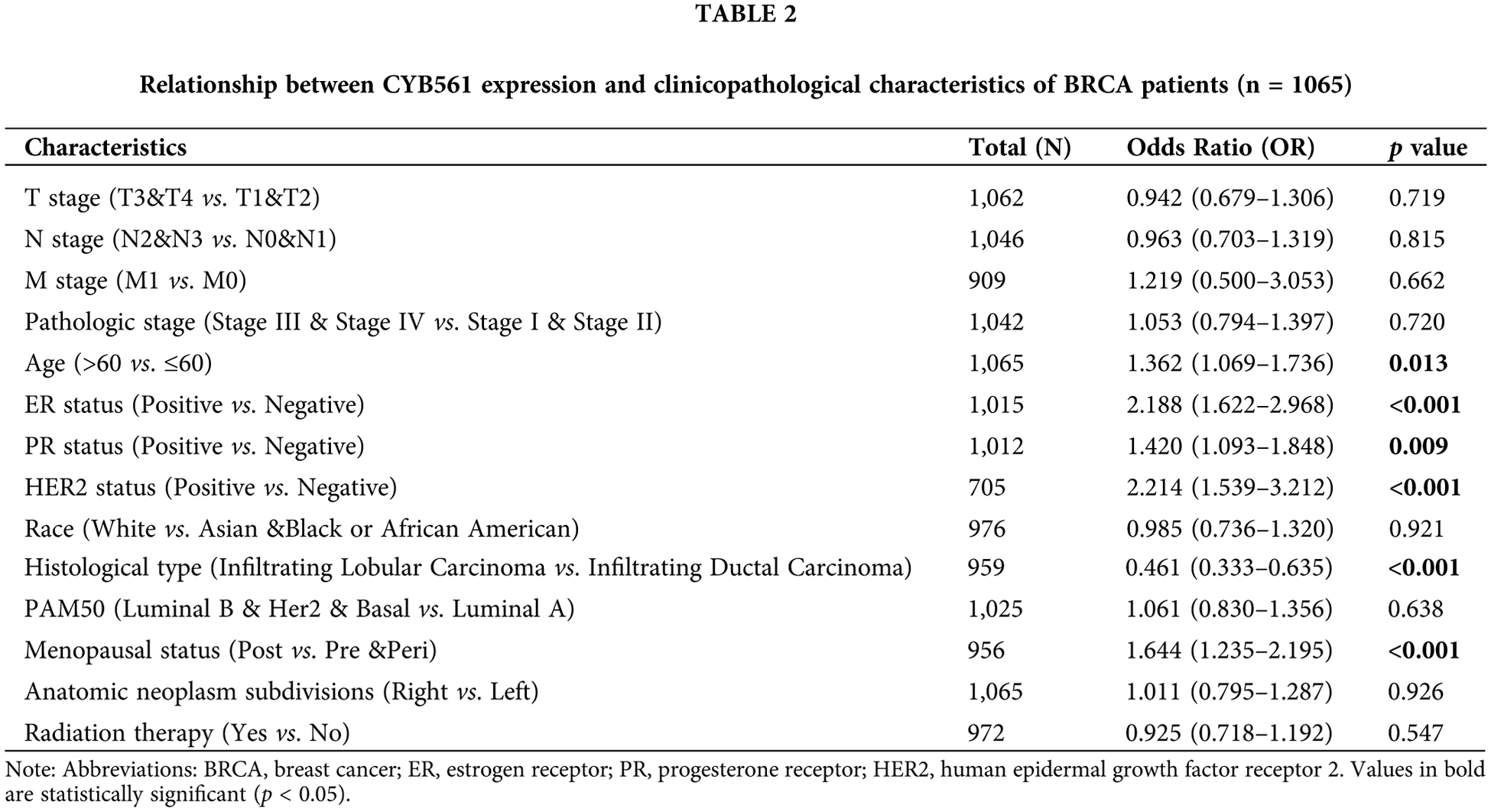
To clarify the significance of CYB561 expression, we then evaluated the relationship between CYB561 expression and clinicopathological features of the patients. As shown in Table 1 and Figs. 2A–2I, elevated expression of CYB561 was significantly correlated with histological type (p < 0.001), estrogen receptor (ER) status (p < 0.001), progesterone receptor (PR) status (p = 0.012), human epidermal growth factor receptor-2 (HER-2) status (p < 0.001), and PAM50 (p < 0.001). Moreover, the results of a univariate logistic regression analysis revealed that overexpression of CYB561 was remarkably associated with age (odds ratio [OR] = 1.362, 95% CI = 1.069–1.736, p = 0.013), ER status (OR = 2.188, 95% CI = 1.622–2.968, p < 0.001), PR status (OR = 1.420, 95% CI = 1.093–1.848, p = 0.009), histological type (OR = 0.461, 95% CI = 0.333–0.635, p < 0.001), and menopausal status (OR = 1.644, 95% CI = 1.235–2.195, p < 0.001) (Table 2).
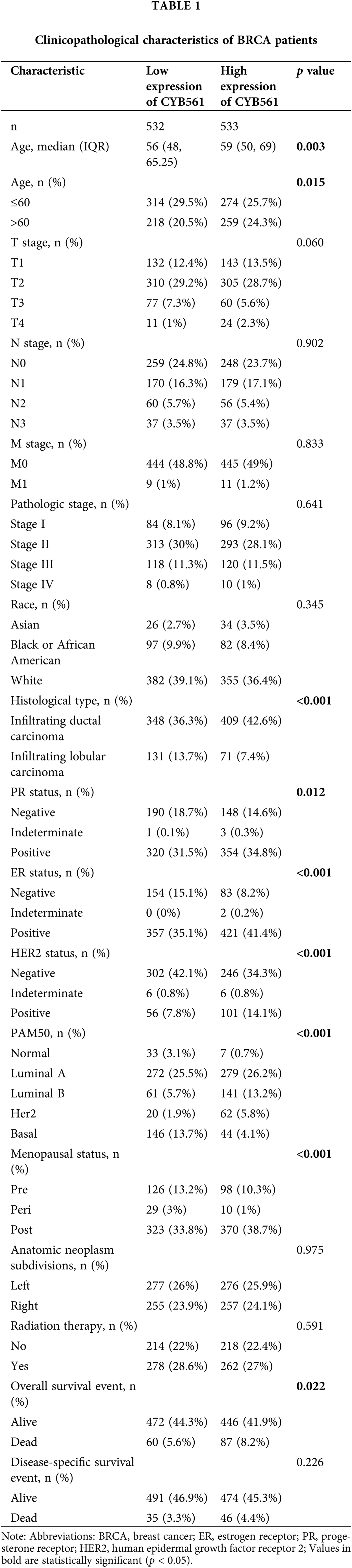
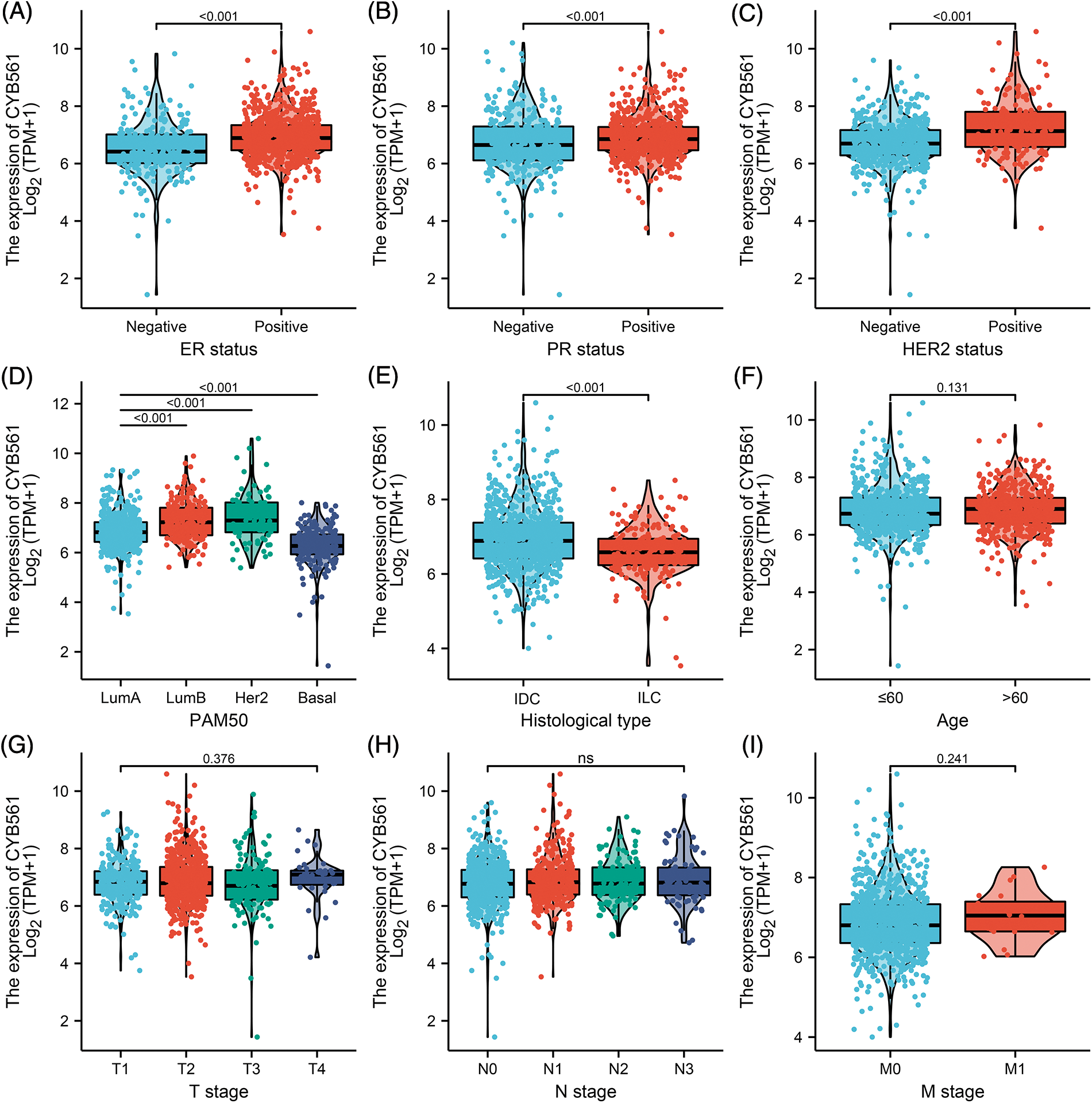
Figure 2: Association of CYB561 expression with clinicopathologic characteristics. Data are shown for (A) ER status; (B) PR status; (C) HER2 status; (D) PAM50; (E) histological type; (F) age; (G) T stage; (H) N stage; (I) M stage. LumA, Luminal A; LumB, Luminal B; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Identification of DEGs in BRCA
We used the DEseq2 package to identify DEGs between CYB561 high- and low-expression groups and identified a total of 267 DEGs, of which 69 were upregulated and 198 were downregulated (adjusted p < 0.05, |log2FC | > 1.5; Fig. 3A). Fig. 3B showed correlations between CYB561 expression and the top 10 DEGs, which included CSN2, LALBA, CPLX2, FTHL17, CHGA, MUC2, CARTPT, XAGE2, SYT4, and NCAN.
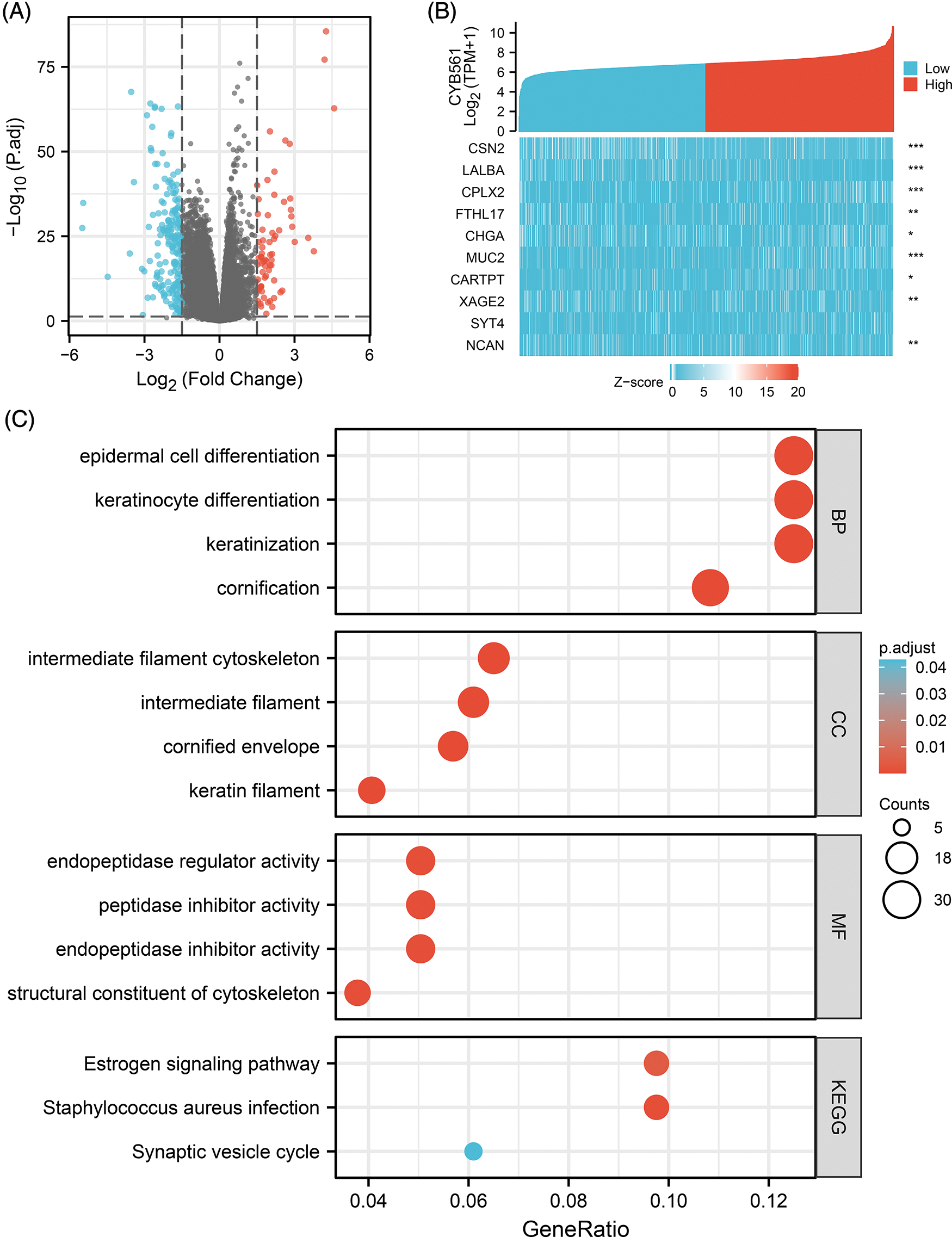
Figure 3: CYB561-related DEGs and functional enrichment analysis of these DEGs. (A) Volcano plot of DEGs between the high and low CYB561 expression groups. Blue and red dots represent downregulated and upregulated DEGs, respectively. (B) Heatmap of the top 10 DEGs. (C) Representative results of GO and KEGG analysis. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. *p < 0.05, **p < 0.01, and ***p < 0.001.
Functional enrichment analysis
To explore the role of CYB561 dysregulation in the progenesis of BRCA, functional enrichment analysis including GO, KEGG and GSEA were performed. The results of GO term enrichment analysis of DEGs indicated that significantly enriched GO terms included epidermal cell differentiation, keratinocyte differentiation, intermediate filament cytoskeleton, intermediate filament, endopeptidase regulator activity, peptidase inhibitor activity, endopeptidase inhibitor activity, and structural constituents of the cytoskeleton. KEGG pathway analysis suggested that DEGs may take part in the estrogen signaling pathway, Staphylococcus aureus infection, and synaptic vesicle cycle (Fig. 3C, Suppl. Tables S2 and S3).
Furthermore, the results of GSEA indicated that the inflammatory response, KRAS signaling pathway, epithelial-mesenchymal transition (EMT), and interferon gamma response were strongly enriched in the low CYB561 expression group (Figs. 4A–4D). Besides, in the low CYB561 expression group, the biological process terms associated immunity, such as leukocyte migration and regulation of lymphocyte activation, were notably enriched.
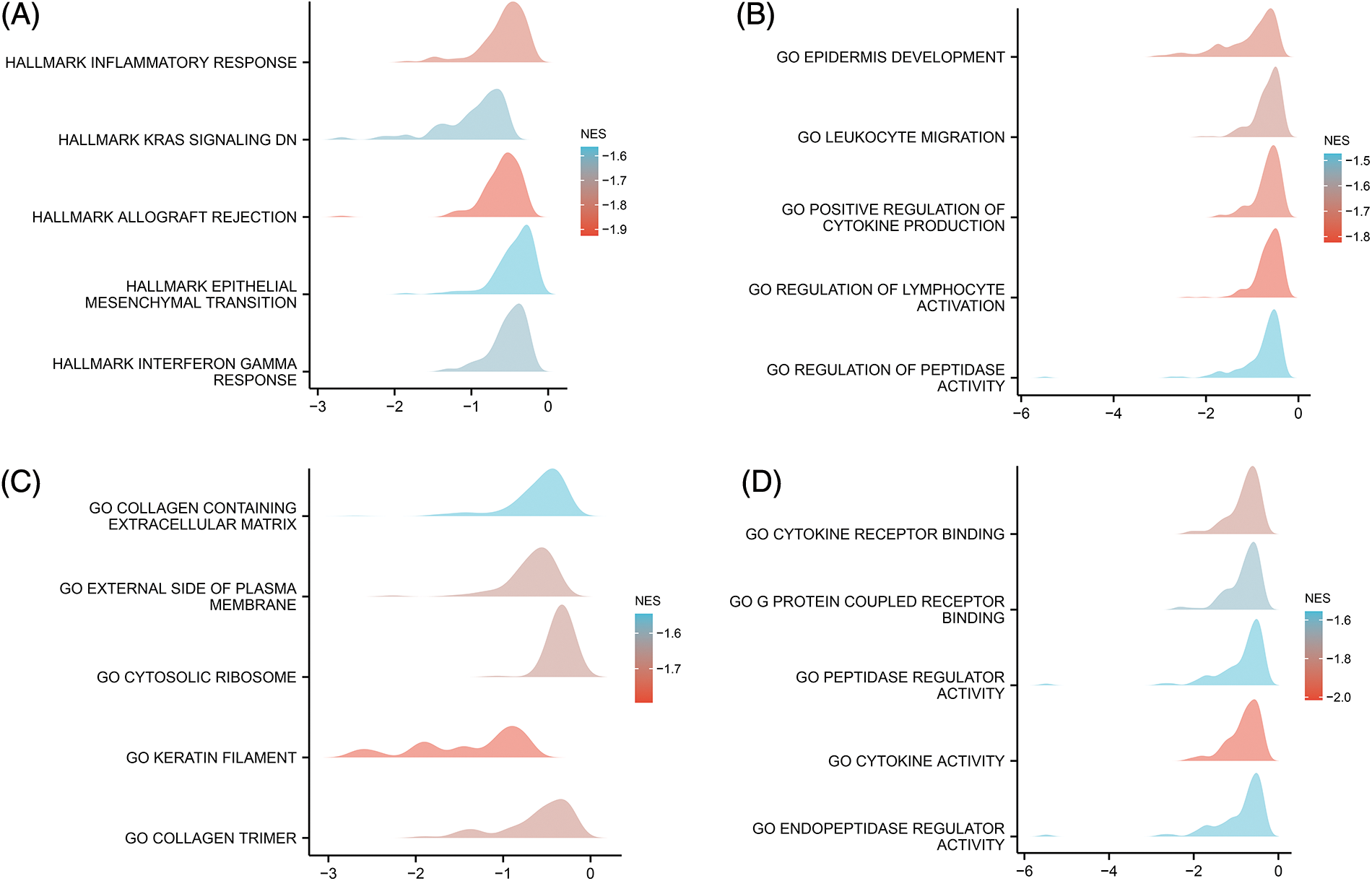
Figure 4: Pathway and functional enrichment were assessed by GSEA. (A) GSEA was performed using the MSigDB Hallmark gene sets. (B) GSEA was performed using the MSigDB bp of GO gene sets. (C) GSEA was performed using the MSigDB cc of GO gene sets. (D) GSEA was performed using the MSigDB mf of GO gene sets. GSEA, Gene Set enrichment analysis; MSigDB, Molecular Signatures Database; NES, normalized enrichment score; GO, Gene Ontology; bp, biological process; cc, cellular component; mf, molecular function.
DNA methylation status of CYB561
To determine the mechanism of CYB561 dysregulation, we attempted to evaluate the methylation status of the CYB561 promoter. The expression level of CYB561 showed a significant positive correlation with the expression levels of DNMT1 and DNMT3A (all p < 0.05) (Figs. 5A–5C). Heatmap showed methylation levels of individual methylation sites on CYB561 DNA sequence in breast cancer tissues (Fig. 5D). Furthermore, Kaplan–Meier survival curves revealed unfavorable survival of patients with lower methylation levels at cg14204735 in TSS1500 region, cg04987499 in TSS1500 region, cg22207733 in TSS200 and TSS1500 region, cg27279583 in the body, cg26300322 in the body, and cg18499750 in the 3′UTR of CYB561 (Figs. 6A–6F).
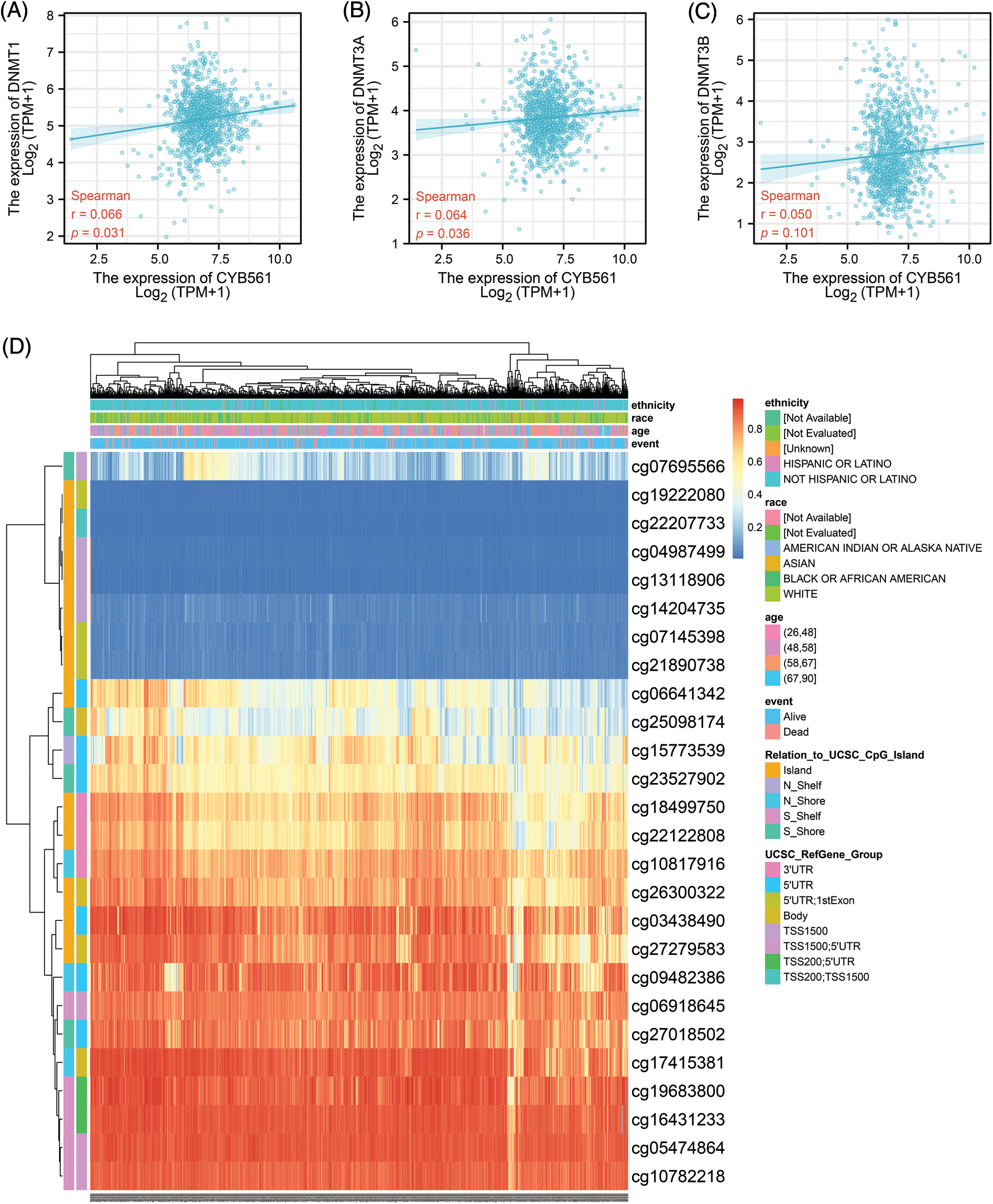
Figure 5: Methylation status of CYB561 in BRCA patients. (A) Analysis of correlation between CYB561 and DNTM1expression levels. (B) Analysis of correlation between CYB561 and DNTM3A expression levels. (C) Analysis of correlation between CYB561 and DNTM3B expression levels. (D) Heatmap of methylation status of serval methylation sites of CYB561. BRCA, breast cancer.
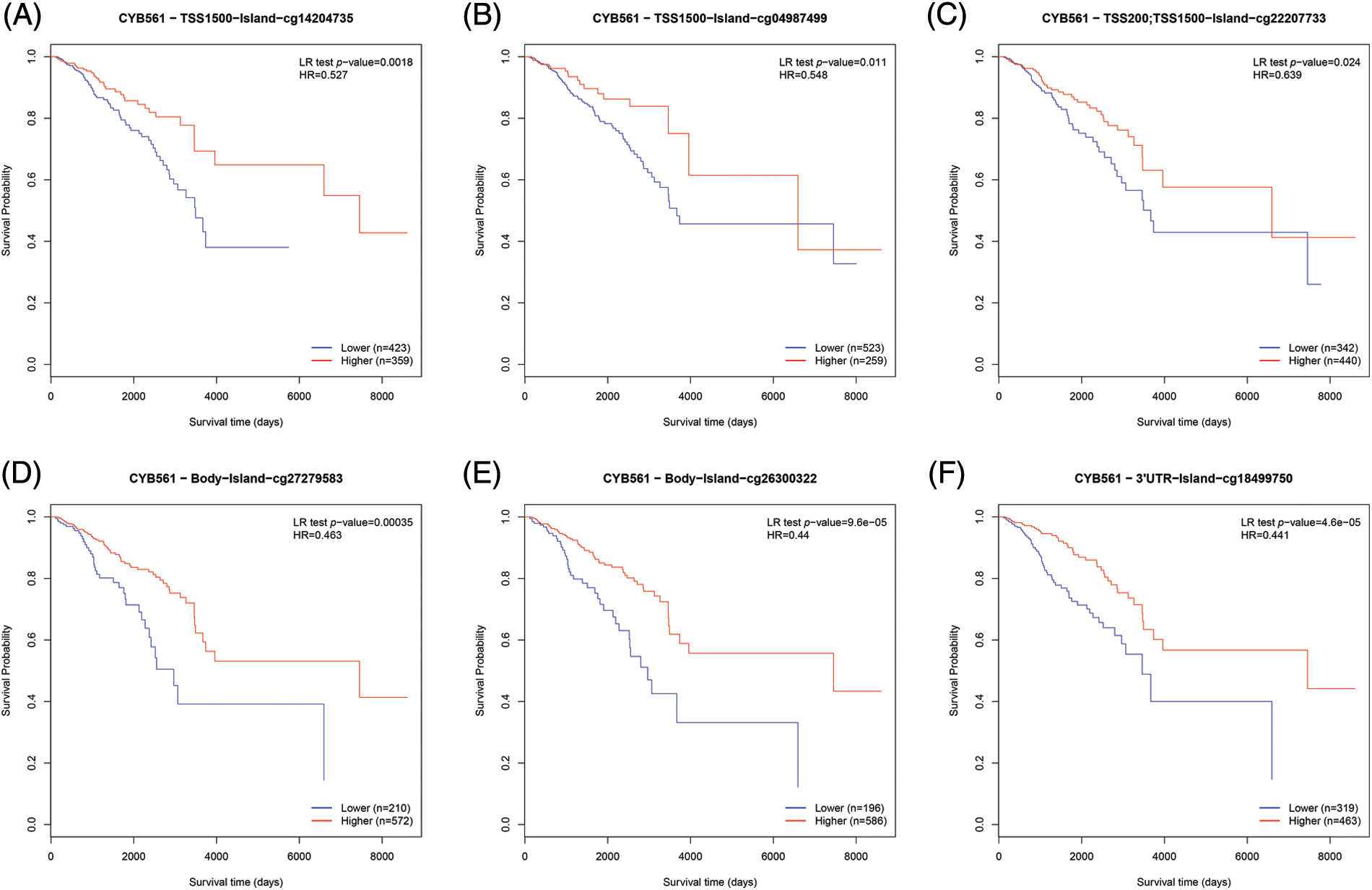
Figure 6: Effect of DNA methylation status of CYB561 on prognosis of BRCA patients. (A–F) Kaplan–Meier survival curves of CYB561 methylation sites. BRCA, breast cancer.
Correlation of CYB561 expression with immune infiltration level in BRCA
Given the interesting results we found, we further investigated the correlation of CYB561 expression with infiltrating immune cells in BRCA. CYB561 expression was notably correlated with 20 out of 24 types of immune infiltrating cells (Fig. 7A). B cells, plasmacytoid dendritic cells (pDCs), dendritic cells (DCs), and neutrophils were more infiltrated in the CYB561 low expression group (all p < 0.001) (Figs. 7B–7E). Moreover, CYB561 expression had a significant negative correlation with the immune infiltration of B cells, pDCs, DCs, and neutrophils (all p < 0.001) (Figs. 7F–7I). Furthermore, multiple immune checkpoint-related molecules, such as CTLA4, IDO1, LAG3, PDCD1, PDCD1LG2 (also known as PD-L2), TIGIT, and VTCN1 were observed to be more highly expressed in the CYB561 high expression group (Fig. 7J).
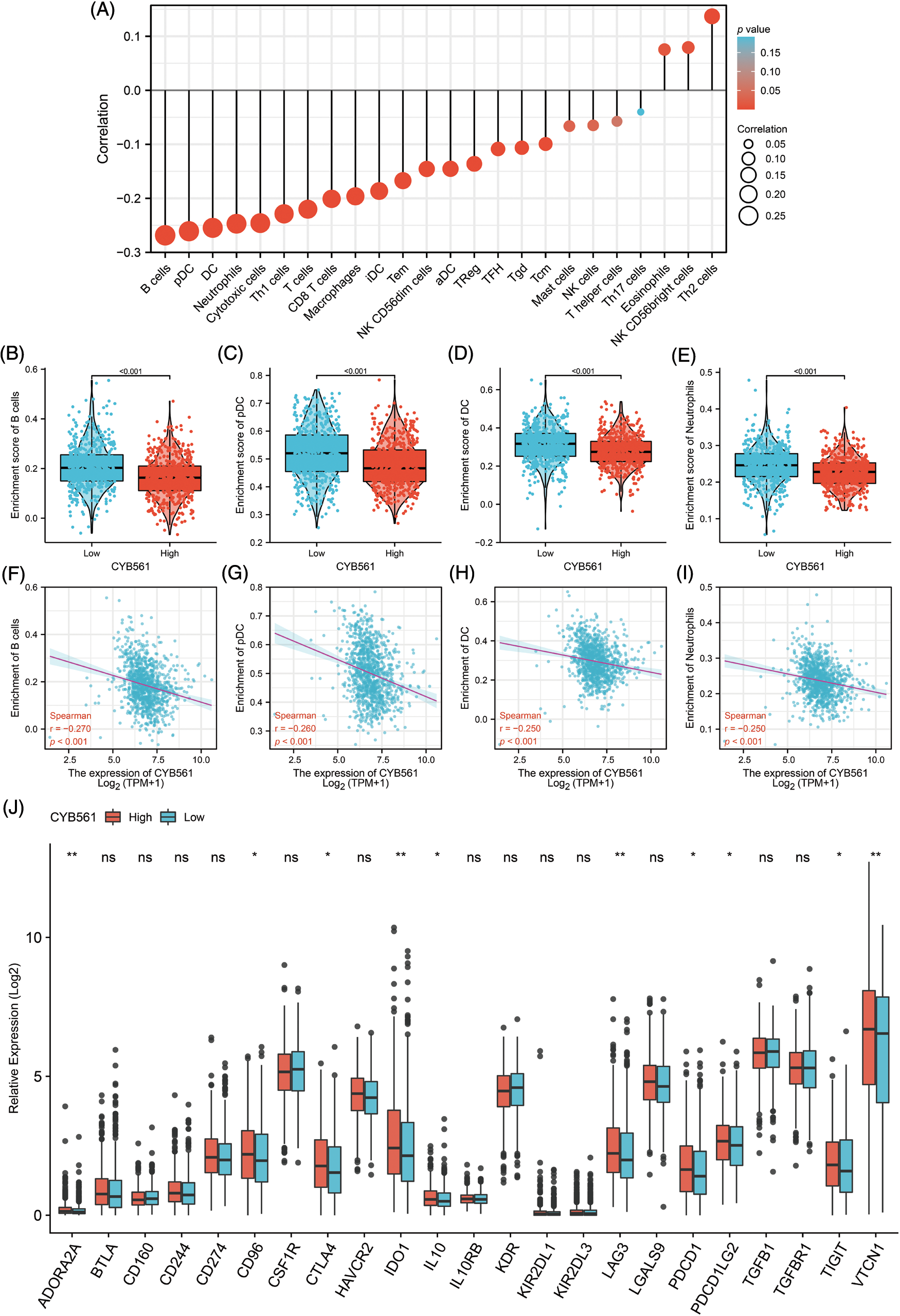
Figure 7: Correlation between CYB561 expression and immune cell infiltration in BRCA. (A) Correlation between CYB561 expression and immune cell infiltration. The size of the black circles corresponds to the absolute values of the Spearman’s correlation coefficient. (B–E) Comparison of the relative enrichment scores of immune cells (including B cells, pDCs, DCs, and Neutrophils) between the high- and low-CYB561 expression groups. (F–I) Analysis of the correlation between the relative enrichment scores of immune cells (including B cells, pDCs, DCs, and Neutrophils) and CYB561 expression levels. (J) Comparison of the expression of immune checkpoint-related genes between the high- and low-CYB561 expression groups. BRCA, breast cancer; pDCs, plasmacytoid dendritic cells; DCs, dendritic cells. *p < 0.05, and **p < 0.01.
Prognostic value of CYB561 in BRCA patients
Subsequently, we examined the prognostic significance of CYB561 expression in BRCA. In the TCGA cohort, BRCA patients with high CYB561 expression had significantly worse OS and DSS compared to those with low CYB561 expression [OS: hazard ratio (HR) = 1.79, 95% CI = 1.29–2.48, p < 0.001; DSS: HR = 1.65, 95% CI = 1.06–2.58, p = 0.026] (Figs. 8A and 8B). Similarly, in the METABRIC cohort, patients with increased CYB561 expression were remarkably associated with the decline in OS (HR = 1.32, 95% CI = 1.17–1.48, p < 0.001) (Fig. 8C). Additionally, regardless of the OS or DSS, BRCA patients with increased CYB561 expression had notably unfavorable survival probabilities in several subgroups, including ER positive, PR negative, T stage 1–3, N stage 0–2, M stage 0, and pathologic stages II–III Subgroups (all p < 0.05) (Figs. 8D–8O).
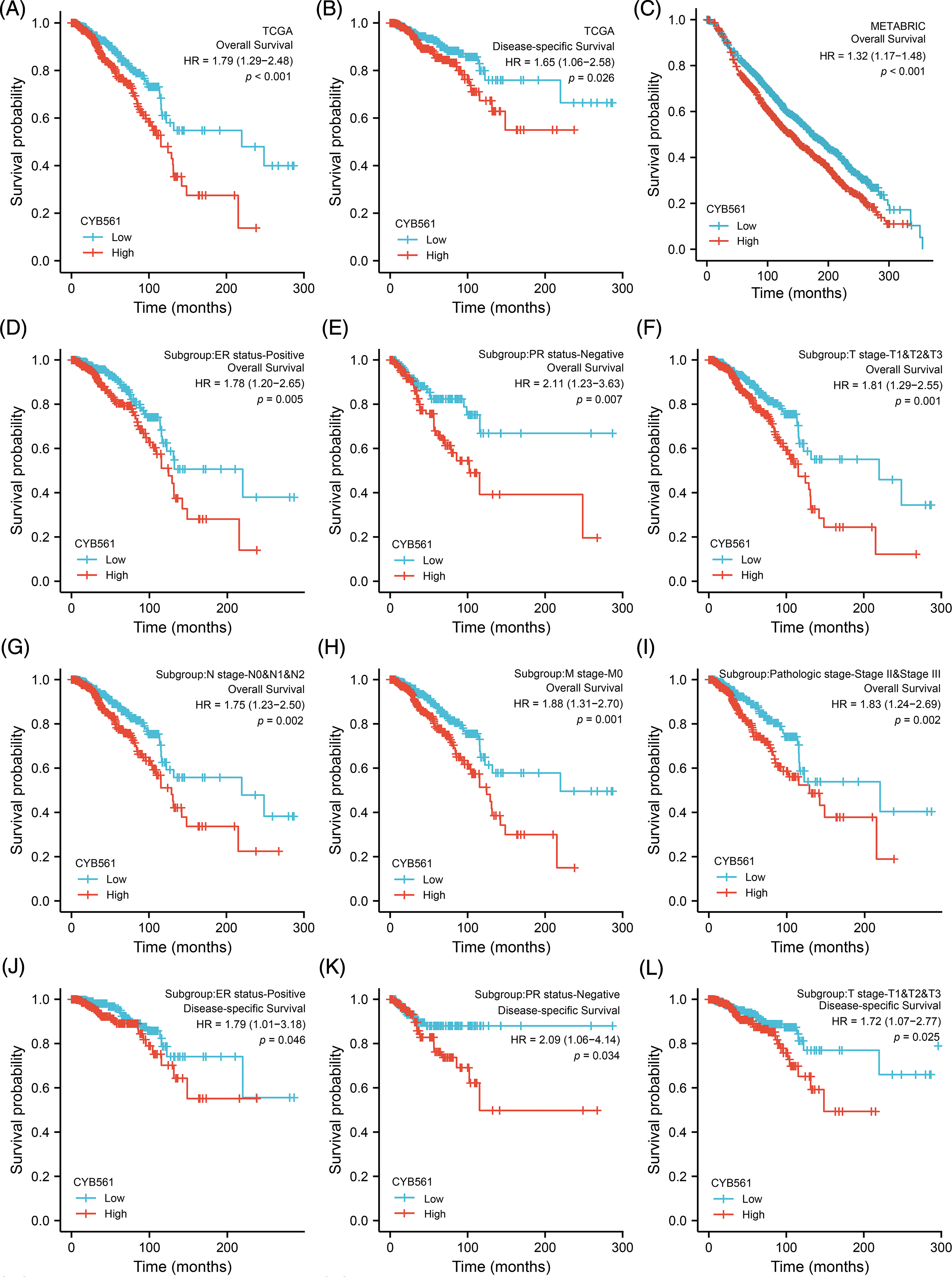
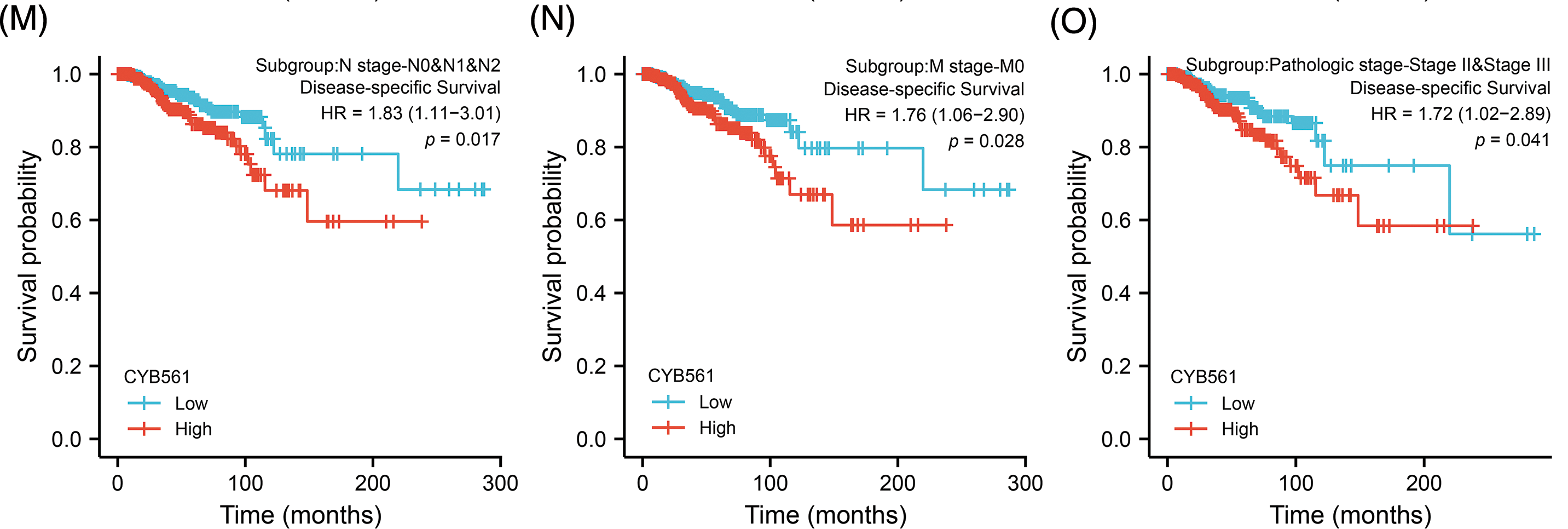
Figure 8: Prognostic values of CYB561 expression in BRCA patients. Effect of CYB561 expression on overall survival (A) and disease-specific survival (B) of BRCA patients in the TCGA database. Effect of CYB561 expression on overall survival (C) of BRCA patients in the METABRICdatabase. (D–I) Overall survival curves of ER positive; PR positive; T1, T2 and T3; N0, N1, and N2; M0; stages II and III subgroups of BRCApatients with high and low CYB561 expression. (J–O) Disease-specific survival curves of ER positive; PR positive; T1, T2 and T3; N0, N1, andN2; M0; stages II and III subgroups of BRCA patients with high and low CYB561 expression. BRCA, breast cancer; TCGA, The CancerGenome Atlas; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; ER, estrogen receptor; PR, progesteronereceptor.
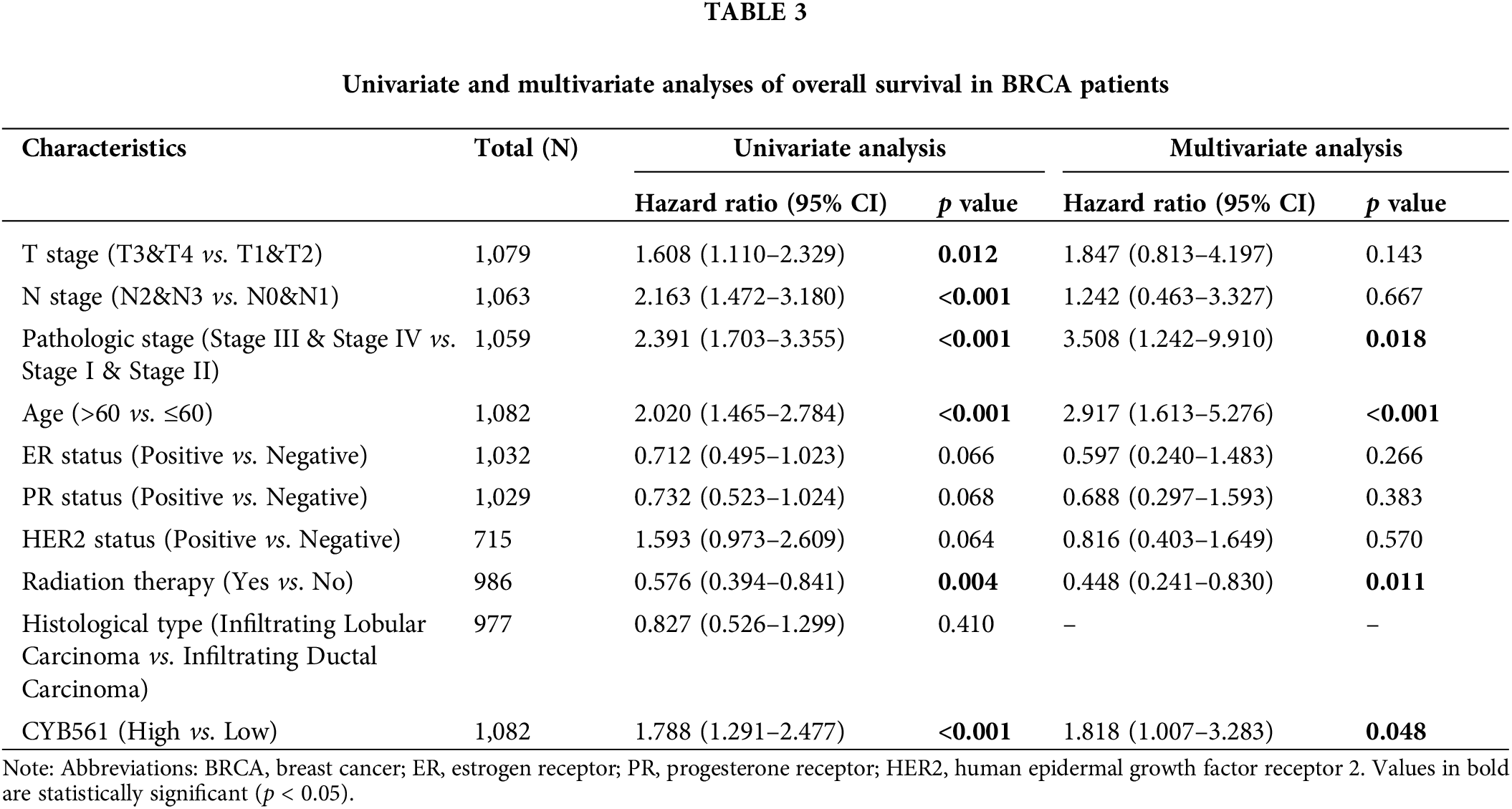
Moreover, multivariate Cox regression analysis indicated that CYB561 expression (adjusted HR = 1.818, 95% CI = 1.007–3.283, p = 0.48), pathological stage (adjusted HR = 3.508, 95% CI = 1.242–9.910, p = 0.018), age (adjusted HR = 2.917, 95% CI = 1.613–5.272, p < 0.001), and radiation therapy (adjusted HR = 0.488, 95% CI = 0.241–0.830, p = 0.011) were independent prognostic markers for OS in BRCA patients (Table 3). Likewise, for DSS, age (adjusted HR = 1.731, 95% CI = 1.033–2.901, p = 0.037), and histological type (adjusted HR = 0.381, 95% CI = 0.156–0.929, p = 0.034) were identified as independent prognostic indicators (Suppl. Table S4).
Construction and evaluation of a nomogram
To further predict prognostic risk in BRCA patients, a nomogram was developed using the results of multivariate COX analysis (Fig. 9A). In the multivariate analysis, pathologic stage, age, radiation therapy, and CYB561 expression were independent predictors of OS, and these variables were included in the nomogram. According to the nomogram, these factors were used to calculate a weighted total score for each patient and to estimate the patient’s 1-, 3-, and 5-year OS. The obtained model was verified internally using the bootstrap resampling method (1000 repetitions), with a C-index of 0.771 (95% CI: 0.745–0.797). The bias-corrected lines shown in the calibration plots were very close to the ideal curve, indicating that the predicted values agreed well with the observed values (Figs. 9B–9D). Additionally, the time-dependent ROC curve was generated, and the AUC of the nomogram model for predicting OS probability at 1 year, 3 years and 5 years was 0.861, 0.748, and 0.757, respectively (Fig. 9E).
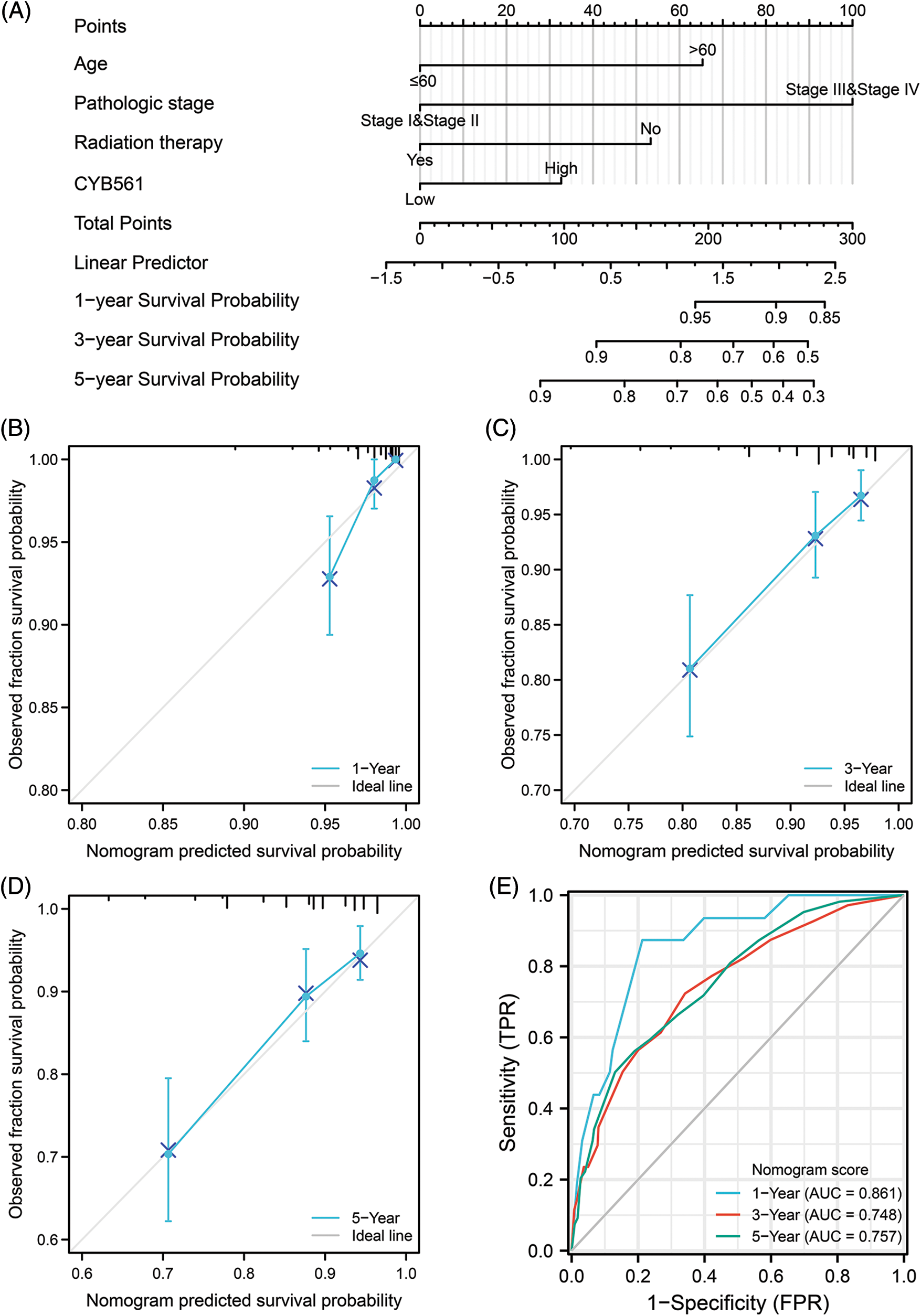
Figure 9: A nomogram for predicting overall survival of BRCA patients and its predictive performance. (A) A nomogram for predicting overall survival of BRCA patients. (B–D) Calibration plots of nomogram for 1-, 3-, and 5-year overall survival, respectively. (E) Time-dependent ROC curve of nomogram. BRCA, breast cancer.
As a transmembrane electron transfer protein, human CYB561 has a physiological function that is closely related to ASC. The ASC-dependent β-hydroxylation of dopamine is an important process in the synthesis of norepinephrine (Zhang et al., 2014; van den Berg et al., 2018). CYB561 exhibits ASC-mediated Fe3+ reductase activity, which provides reduced iron for transmembrane transport (Su and Asard, 2006; Berczi et al., 2007). Currently, few studies have described the relationship between CYB561 and malignant tumors, especially BRCA, and CYB561 expression status, potential function, and ability of predicting prognosis in BRCA remain unclear.
In our study, bioinformatics analysis was performed using high-throughput RNA-seq data obtained from TCGA database to evaluate the methylation status, immune infiltration score, and prognostic value of CYB561 in BRCA patients. Our results showed that CYB561 is highly expressed in BRCA and may be a potential negative prognostic biomarker, which is consistent with previous studies on CYB561 expression in BRCA (Yang et al., 2021; Zhou et al., 2022). In contrast, overexpression of CYB561 has been reported to be a marker for good prognosis in ovarian serous carcinoma (Willis et al., 2016).
Since the biological function of CYB561 in BRCA is still unclear, functional enrichment analysis was conducted. Our findings revealed that CYB561 may act on BRCA through EMT, KRAS signaling pathway, and estrogen signaling pathway. EMT plays an important role in the development and progression of cancer, including carcinogenesis, metastasis, and drug resistance (Nieto et al., 2016; Zhang and Weinberg, 2018). As a proto-oncogene, previous studies demonstrated that elevated KRAS expression was associated with BRCA progression and can be used as a prognostic factor of BRCA (Banys-Paluchowski et al., 2020; Liang et al., 2021). Moreover, KRAS can promote the proliferation, apoptosis, and invasion of BRCA via EMT (Jung et al., 2019; Zhang et al., 2020). Additionally, it has been reported that estrogen contributes to regulate the proliferation and differentiation of normal mammary epithelial cells (Saha et al., 2019). Persistent estrogen exposure, or dysregulated estrogen signaling, has been implicated in many types of cancers, including breast, endometrial, ovarian, and prostate cancers (Liang and Shang, 2013). As a mammary gland carcinogen, estrogen participates in the initiation, promotion, and progression of breast cancer (Yager and Davidson, 2006). Furthermore, our results also suggested that CYB561 may be involved in inflammation-related pathways, such as inflammatory response, interferon gamma response, Staphylococcus aureus infection, and leukocyte migration. Numerous studies have confirmed that chronic inflammation plays a crucial role in different stages of tumorigenesis, including initiation, proliferation, angiogenesis, and metastasis (Diakos et al., 2014; Hanahan, 2022). Inflammation is a threat to the long-term survival of BRCA patients (Mahmoud et al., 2012). A recent clinical study showed that the use of nonsteroidal anti-inflammatory drugs can prevent the progression of biopsy-proven benign breast tumor to breast cancer (Sherman et al., 2020). Interestingly, it has been shown that there is an association between Staphylococcus aureus infection at the surgical site and breast cancer recurrence (O’Connor et al., 2020). Leukocyte migration is critical for the maintenance and regulation of immune responses and is involved in inflammatory responses and the composition of the tumor microenvironment (Kameritsch and Renkawitz, 2020).
As one of the most common epigenetic mechanisms, DNA methylation regulates gene expression (Moore et al., 2013). To our knowledge, there are no studies available to date on the methylation status of CYB561 in tumors. Thus, we attempted to study the underlying mechanism of overexpression of CYB561 in BRCA. Our findings showed that several methylated sites on the DNA sequence of the CYB561 gene are hypomethylated, which were strongly associated with poor prognosis in BRCA patients. This suggests that the methylation of CYB561 may be introduced by DNTM1 and DNMT3A.
The tumor microenvironment (TME) is thought to play an important regulatory role in tumorigenesis and progression (Quail and Joyce, 2013). Tumor-infiltrating immune cells are an integral part of the TME and can recognize, inhibit, or promote tumor progression (Ostrand-Rosenberg, 2008). The number of tumor-infiltrating lymphocytes (TILs) often indicates the prognosis of BRCA patients. Increased TILs predict a response to neoadjuvant chemotherapy in most molecular subtypes of BRCA and are particularly associated with survival benefits in HER2-positive and triple-negative breast cancer (TNBC) (Denkert et al., 2018; Loi et al., 2019; Park et al., 2019). TILs comprise a group of heterogeneous immune cells, usually dominated by T cells, but also include B cells, NK cells, and myeloid-lineage cells such as macrophages, neutrophils, and mast cells. In our study, we found that B cells infiltrated more in the CYB561 low expression group. A previous study showed that the density of tumor-infiltrating B cells was increased in untreated BRCA compared to normal breast tissues and was strongly related to higher tumor grade, higher proliferation, and hormone receptor negativity (Garaud et al., 2019). Besides, B cells have been reported to be associated with poor prognosis in BRCA (Shariati et al., 2020). In contrast, a higher total number of infiltrating B cells in BRCA have previously been related to improved clinical prognosis (Mahmoud et al., 2012). One possible explanation for these divergent results may be the differences in the samples and assays used. pDCs and DCs, as important components of innate immunity system, have been observed to have inhibitory effects on BRCA cell proliferation (Wu et al., 2016). Neutrophils are the most abundant circulating leukocytes in the human body, which can regulate not only the adaptive immune system, but also the innate immune system (Rosales, 2020). Growing evidence suggests that neutrophils may play an important role in the progression and metastasis of BRCA (Coffelt et al., 2015; Park et al., 2016). In a previous meta-analysis, elevated neutrophil-to-lymphocyte ratio in the peripheral blood of BRCA patients was associated with an adverse OS and DFS (Ethier et al., 2017).
Immune evasion is a key factor in tumor progression. Recently, immunotherapy, especially immune checkpoint inhibitors (ICIs), has emerged as an important treatment for many malignant tumors (Moujaess et al., 2019). The programmed cell death receptor-1/ligand 1 (PD-1/L1) and cytotoxic T-lymphocyte associated protein-4 (CTLA-4) are the most important targets of ICIs. PD-L1 is highly expressed in a variety of tumors and can predict the clinical response to PD-1-targeted immunotherapy (Zou and Chen, 2008). Since BRCA has not traditionally been considered a highly immunogenic disease, the efficacy of ICIs is largely limited by low response rates (Tokumaru et al., 2020). TNBC is by far the most promising molecular subtype of BRCA for immune checkpoint blockade. A previous study demonstrated that patients with TNBC who were PD-L1-positive had longer median OS than those who were PD-L1-negative (AiErken et al., 2017). Although TNBC has a higher response rate to ICIs than other subtypes of BRCA, the efficacy of ICIs remains low, with response rates ranging from approximately 5% to 23% (Keenan and Tolaney, 2020). Therefore, we must explore novel and effective targets for immune checkpoint blockade in BRCA. In our study, we found that elevated expression of CYB561 was closely related to the increased expression of CTLA4, IDO1, LAG3, PDCD1, PDCD1LG2, and TIGIT, which exhibited different immunomodulatory effects in different tissue sites. These results suggest that patients with high expression of CYB561 may be more likely to benefit from immunotherapy.
Although our study provides novel insights into the relationship between CYB561 expression and BRCA from multiple perspectives and levels by mining public databases, there are still several limitations that warrant consideration. First, given that our study was mainly based on public databases, selection bias may affect our findings. Second, some important clinical features, such as surgical approach and adjuvant chemotherapy regimens, could not be collected due to limitations in the public databases; hence, we need to validate the results using a patient cohort in our center. Third, considering that all the results were obtained using bioinformatics, further systematic experimental studies are required to unravel the underlying biological mechanisms of CYB561 in BRCA.
In summary, all the data in our study demonstrate that elevated CYB561 is strongly linked with aggressive clinicopathologic characteristics, unfavorable immune infiltration, and poor patient outcomes, which may be a novel prognostic biomarker for BRCA patients. However, more studies are needed to explore the function of CYB561 in BRCA and more importantly, how other molecules contribute to this function in tumorigenesis and progression.
Funding Statement: This study was supported by grants from the National Natural Science Foundation of China (Grant No. 82060483), Guangxi Research Foundation for Science & Technology Base and Talent Special (Grant No. AD19110079), and Natural Science Foundation of Guangxi Province (Grant No. 2020GXNSFBA238002).
Author Contributions: The authors confirm contribution to the paper as follows: wrote the manuscript: XY; data analysis: H-XW and CX; data collection: BZ and M-LL; the technical support: BZ and JQ; conceived the idea, designed the study, and revised the manuscript: MD. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets analyzed during the current study are available in the following repositories: The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), Genotype Tissue Expression Project (GTEx) database (https://commonfund.nih.gov/gtex), cBioPortal database (http://www.Cbioportal.org/), UCSC Xena database (https://xenabrowser.net/datapages/), MethSurv methylation analysis tool (https://biit.cs.ut.ee/methsurv/).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at DOI: 10.32604/biocell.2023.027103.
References
AiErken N, Shi HJ, Zhou Y, Shao N, Zhang J, Shi Y, Yuan ZY, Lin Y (2017). High PD-L1 expression is closely associated with tumor-infiltrating lymphocytes and leads to good clinical outcomes in Chinese triple negative breast cancer patients. International Journal of Biological Sciences 13: 1172–1179. https://doi.org/10.7150/ijbs.20868 [Google Scholar] [PubMed] [CrossRef]
Banys-Paluchowski M, Milde-Langosch K, Fehm T, Witzel I, Oliveira-Ferrer L, Schmalfeldt B, Muller V (2020). Clinical relevance of H-RAS, K-RAS, and N-RAS mRNA expression in primary breast cancer patients. Breast Cancer Research and Treatment 179: 403–414. https://doi.org/10.1007/s10549-019-05474-8 [Google Scholar] [PubMed] [CrossRef]
Bashtovyy D, Berczi A, Asard H, Pali T (2003). Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma 221: 31–40. https://doi.org/10.1007/s00709-002-0065-0 [Google Scholar] [PubMed] [CrossRef]
Berczi A, Su D, Asard H (2007). An Arabidopsis cytochrome b561 with trans-membrane ferrireductase capability. FEBS Letters 581: 1505–1508. https://doi.org/10.1016/j.febslet.2007.03.006 [Google Scholar] [PubMed] [CrossRef]
Berczi A, Su D, Lakshminarasimhan M, Vargas A, Asard H (2005). Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Archives of Biochemistry and Biophysics 443: 82–92. https://doi.org/10.1016/j.abb.2005.09.006 [Google Scholar] [PubMed] [CrossRef]
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S (2017). UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19: 649–658. https://doi.org/10.1016/j.neo.2017.05.002 [Google Scholar] [PubMed] [CrossRef]
Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K et al. (2015). IL-17-producing γδT cells and neutrophils conspire to promote breast cancer metastasis. Nature 522: 345–348. https://doi.org/10.1038/nature14282 [Google Scholar] [PubMed] [CrossRef]
Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI et al. (2018). Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. The Lancet Oncology 19: 40–50. https://doi.org/10.1016/S1470-2045(17)30904-X [Google Scholar] [PubMed] [CrossRef]
Diakos CI, Charles KA, McMillan DC, Clarke SJ (2014). Cancer-related inflammation and treatment effectiveness. The Lancet Oncology 15: e493–e503. https://doi.org/10.1016/S1470-2045(14)70263-3 [Google Scholar] [PubMed] [CrossRef]
Ethier JL, Desautels D, Templeton A, Shah PS, Amir E (2017). Prognostic role of neutrophil-to-lymphocyte ratio in breast cancer: A systematic review and meta-analysis. Breast Cancer Research 19: 2. https://doi.org/10.1186/s13058-016-0794-1 [Google Scholar] [PubMed] [CrossRef]
Foyer CH, Noctor G (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiology 155: 2–18. https://doi.org/10.1104/pp.110.167569 [Google Scholar] [PubMed] [CrossRef]
Garaud S, Buisseret L, Solinas C, Gu-Trantien C, de Wind A et al. (2019). Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 5: e129641. https://doi.org/10.1172/jci.insight.129641 [Google Scholar] [PubMed] [CrossRef]
Hanahan D (2022). Hallmarks of cancer: New dimensions. Cancer Discovery 12: 31–46. https://doi.org/10.1158/2159-8290.CD-21-1059 [Google Scholar] [PubMed] [CrossRef]
Jung SY, Malhotra P, Nguyen KC, Salzman D, Qi Y, Pak EH, King J, Vlashi E, Ann D, Weidhaas JB (2019). The KRAS-variant and its impact on normal breast epithelial cell biology. Cell Death & Differentiation 26: 2568–2576. https://doi.org/10.1038/s41418-019-0320-y [Google Scholar] [PubMed] [CrossRef]
Kameritsch P, Renkawitz J (2020). Principles of leukocyte migration strategies. Trends in Cell Biology 30: 818–832. https://doi.org/10.1016/j.tcb.2020.06.007 [Google Scholar] [PubMed] [CrossRef]
Keenan TE, Tolaney SM (2020). Role of immunotherapy in triple-negative breast cancer. Journal of the National Comprehensive Cancer Network 18: 479–489. https://doi.org/10.6004/jnccn.2020.7554 [Google Scholar] [PubMed] [CrossRef]
Liang J, Shang Y (2013). Estrogen and cancer. Annual Review of Physiology 75: 225–240. https://doi.org/10.1146/annurev-physiol-030212-183708 [Google Scholar] [PubMed] [CrossRef]
Liang H, Zhou G, Lv L, Lu J, Peng J (2021). KRAS expression is a prognostic indicator and associated with immune infiltration in breast cancer. Breast Cancer 28: 379–386. https://doi.org/10.1007/s12282-020-01170-4 [Google Scholar] [PubMed] [CrossRef]
Loi S, Drubay D, Adams S, Pruneri G, Francis PA et al. (2019). Tumor-infiltrating lymphocytes and prognosis: A pooled individual patient analysis of early-stage triple-negative breast cancers. Journal of Clinical Oncology 37: 559–569. https://doi.org/10.1200/JCO.18.01010 [Google Scholar] [PubMed] [CrossRef]
Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 550. https://doi.org/10.1186/s13059-014-0550-8 [Google Scholar] [PubMed] [CrossRef]
Mahmoud SM, Lee AH, Paish EC, Macmillan RD, Ellis IO, Green AR (2012). The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Research and Treatment 132: 545–553. https://doi.org/10.1007/s10549-011-1620-1 [Google Scholar] [PubMed] [CrossRef]
Modhukur V, Iljasenko T, Metsalu T, Lokk K, Podar TL, Vilo (2017). MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 110: 277–288. https://doi.org/10.2217/epi-2017-0118 [Google Scholar] [PubMed] [CrossRef]
Moore LD, Le T, Fan G (2013). DNA methylation and its basic function. Neuropsychopharmacology 38: 23–38. https://doi.org/10.1038/npp.2012.112 [Google Scholar] [PubMed] [CrossRef]
Moujaess E, Haddad FH, Eid R, Kourie HP (2019). The emerging use of immune checkpoint blockade in the adjuvant setting for solid tumors a review. Immunotherapy 11: 1409–1422. https://doi.org/10.2217/imt-2019-0087 [Google Scholar] [PubMed] [CrossRef]
Nieto MA, Huang RY, Jackson RA, Thiery JP (2016). EMT: 2016. Cell 166: 21–45. https://doi.org/10.1016/j.cell.2016.06.028 [Google Scholar] [PubMed] [CrossRef]
Ohtani S, Iwamaru A, Deng W, Ueda K, Wu G et al. (2007). Tumor suppressor 101F6 and ascorbate synergistically and selectively inhibit non-small cell lung cancer growth by caspase-independent apoptosis and autophagy. Cancer Research 67: 6293–6303. https://doi.org/10.1158/0008-5472.CAN-06-3884 [Google Scholar] [PubMed] [CrossRef]
Olarte KCV, Bagamasbad PD (2020). SAT-132 the secretory vesicle membrane protein, CYB561, promotes the growth and metastatic potential of castration-resistant neuroendocrine prostate cancer. Journal of the Endocrine Society 4: SAT-132. https://doi.org/10.1210/jendso/bvaa046.1194 [Google Scholar] [CrossRef]
Ostrand-Rosenberg S (2008). Immune surveillance: A balance between protumor and antitumor immunity. Current Opinion in Genetics & Development 18: 11–18. https://doi.org/10.1016/j.gde.2007.12.007 [Google Scholar] [PubMed] [CrossRef]
O’Connor R, Kiely PA, Dunne CP (2020). The relationship between post-surgery infection and breast cancer recurrence. Journal of Hospital Infection 106: 522–535. https://doi.org/10.1016/j.jhin.2020.08.004 [Google Scholar] [PubMed] [CrossRef]
Park JH, Jonas SF, Bataillon G, Criscitiello C, Salgado R et al. (2019). Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Annals of Oncology 30: 1941–1949. https://doi.org/10.1093/annonc/mdz395 [Google Scholar] [PubMed] [CrossRef]
Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR et al. (2016). Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Science Translational Medicine 8: 361ra138. https://doi.org/10.1126/scitranslmed.aag1711 [Google Scholar] [PubMed] [CrossRef]
Quail DF, Joyce JA (2013). Microenvironmental regulation of tumor progression and metastasis. Nature Medicine 19: 1423–1437. https://doi.org/10.1038/nm.3394 [Google Scholar] [PubMed] [CrossRef]
Rosales C (2020). Neutrophils at the crossroads of innate and adaptive immunity. The Journal of Leukocyte Biology 108: 377–396. https://doi.org/10.1002/JLB.4MIR0220-574RR [Google Scholar] [PubMed] [CrossRef]
Saha T, Makar S, Swetha R, Gutti G, Singh SK (2019). Estrogen signaling: An emanating therapeutic target for breast cancer treatment. European Journal of Medicinal Chemistry 177: 116–143. https://doi.org/10.1016/j.ejmech.2019.05.023 [Google Scholar] [PubMed] [CrossRef]
Shariati S, Mehdipour F, Samadi M, Rasolmali R, Talei A-R, Ghaderi A (2020). The balance of regulatory and stimulatory B cell subsets in breast cancer draining lymph nodes correlates with tumor prognostic factors. Life Sciences 257: 118117. https://doi.org/10.1016/j.lfs.2020.118117 [Google Scholar] [PubMed] [CrossRef]
Sherman ME, Vierkant RA, Kaggal S, Hoskin TL, Frost MH et al. (2020). Breast cancer risk and use of nonsteroidal anti-inflammatory agents after a benign breast biopsy. Cancer Prevention Research 13: 967–976. https://doi.org/10.1158/1940-6207.CAPR-20-0178 [Google Scholar] [PubMed] [CrossRef]
Su D, Asard H (2006). Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS Journal 273: 3722–3734. https://doi.org/10.1111/j.1742-4658.2006.05381.x [Google Scholar] [PubMed] [CrossRef]
Sun D, Zhang A, Gao B, Zou L, Huang H, Zhao X, Xu D (2022). Identification of alternative splicing-related genes CYB561 and FOLH1 in the tumor-immune microenvironment for endometrial cancer based on TCGA data analysis. Frontiers in Genetics 13: 770569. https://doi.org/10.3389/fgene.2022.770569 [Google Scholar] [PubMed] [CrossRef]
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 71: 209–249. https://doi.org/10.3322/caac.21660 [Google Scholar] [PubMed] [CrossRef]
Tokumaru Y, Joyce D, Takabe K (2020). Current status and limitations of immunotherapy for breast cancer. Surgery 167: 628–630. https://doi.org/10.1016/j.surg.2019.09.018 [Google Scholar] [PubMed] [CrossRef]
Tsubaki M, Takeuchi F, Nakanishi N (2005). Cytochrome b561 protein family: Expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochimica et Biophysica Acta (BBA)—Proteins and Proteomics 1753: 174–190. https://doi.org/10.1016/j.bbapap.2005.08.015 [Google Scholar] [PubMed] [CrossRef]
van den Berg MP, Almomani R, Biaggioni I, van Faassen M, van der Harst P et al. (2018). Mutations in CYB561 causing a novel orthostatic hypotension syndrome. Circulation Research 122: 846–854. https://doi.org/10.1161/CIRCRESAHA.117.311949 [Google Scholar] [PubMed] [CrossRef]
Willis S, Villalobos VM, Gevaert O, Abramovitz M, Williams C, Sikic BI, Leyland-Jones B (2016). Single gene prognostic biomarkers in ovarian cancer: A meta-analysis. PLoS One 11: e0149183. https://doi.org/10.1371/journal.pone.0149183 [Google Scholar] [PubMed] [CrossRef]
Wu J, Li S, Yang Y, Zhu S, Zhang M, Qiao Y, Liu YJ, Chen J (2016). TLR-activated plasmacytoid dendritic cells inhibit breast cancer cell growth in vitro and in vivo. Oncotarget 8: 11708–11718. https://doi.org/10.18632/oncotarget.14315 [Google Scholar] [PubMed] [CrossRef]
Yager JD, Davidson NE (2006). Estrogen carcinogenesis in breast cancer. The New England Journal of Medicine 354: 270–282. https://doi.org/10.1056/NEJMra050776 [Google Scholar] [PubMed] [CrossRef]
Yang X, Zhao Y, Shao Q, Jiang G (2021). Cytochrome b561 serves as a potential prognostic biomarker and target for breast cancer. International Journal of General Medicine 14: 10447–10464. https://doi.org/10.2147/IJGM.S338878 [Google Scholar] [PubMed] [CrossRef]
Yu G, Wang LG, Han Y, He QY (2012). Cluster profiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 16: 284–287. https://doi.org/10.1089/omi.2011.0118 [Google Scholar] [PubMed] [CrossRef]
Zeng D, Ye Z, Shen R, Yu G, Wu J et al. (2021). IOBR: Multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Frontiers in Immunology 12: 687975. https://doi.org/10.3389/fimmu.2021.687975 [Google Scholar] [PubMed] [CrossRef]
Zhang K, Deacon DC, Rao F, Schork AJ, Fung MM, Waalen J, Schork NJ, Nievergelt CM, Chi NC, O'Connor DT (2014). Human heart rate: Heritability of resting and stress values in twin pairs, and influence of genetic variation in the adrenergic pathway at a microribonucleic acid (microRNA) motif in the 3′-UTR of cytochrome b561. Journal of the American College of Cardiology 63: 358–368. https://doi.org/10.1016/j.jacc.2013.09.025 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Liu JL, Wang J (2020). KRAS gene silencing inhibits the activation of PI3K-Akt-mTOR signaling pathway to regulate breast cancer cell epithelial-mesenchymal transition, proliferation and apoptosis. European Review for Medical and Pharmacological Sciences 24: 3085–3096. https://doi.org/10.26355/eurrev_202003_20673 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Weinberg RA (2018). Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Frontiers of Medicine 12: 361–373. https://doi.org/10.1007/s11684-018-0656-6 [Google Scholar] [PubMed] [CrossRef]
Zhou X, Shen G, Ren D, Guo X, Han J et al. (2022). Expression and clinical prognostic value of CYB561 in breast cancer. Journal of Cancer Research and Clinical Oncology 148: 1879–1892. https://doi.org/10.1007/s00432-022-03928-z [Google Scholar] [PubMed] [CrossRef]
Zou W, Chen L (2008). Inhibitory B7-family molecules in the tumour microenvironment. Nature Reviews Immunology 8: 467–477. https://doi.org/10.1038/nri2326 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Table S1. Baseline characteristics of patients (n = 1065)
Table S2. DEGs enriched Gene Ontology (GO) terms
Table S3. DEGs enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) terms
Table S4. Univariate and multivariate analysis of disease-specific survival in patients with breast cancer
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools