 Open Access
Open Access
SHORT COMMUNICATION
RNF213 Formed and Decorated Membrane-Based Structures in U-2 OS Cells
Department of Food Science and Nutrition, School of Food Science and Nutrition, Mukogawa Women’s University, Nishinomiya, Hyogo, 663-8558, Japan
* Corresponding Author: TOSHIYUKI HABU. Email:
(This article belongs to the Special Issue: Advanced Cell Signaling Pathways in Health and Disease)
BIOCELL 2026, 50(1), 1 https://doi.org/10.32604/biocell.2025.071798
Received 30 June 2025; Accepted 22 September 2025; Issue published 23 January 2026
Abstract
RING protein 213 (RNF213), the susceptibility gene for Moyamoya disease (MMD), possesses two active AAA+ ATPase (ATPases Associated with diverse cellular Activities) modules, a RING, and RNF213-ZNFX1 finger (RZ finger) domains. Several RNF213 variants have been reported in MMD patients, including the p.R4810K variant (rs112735431), which is a founder polymorphism associated with MMD in East Asia. To elucidate the function of RNF213 and its variant, we investigated the localization of RNF213 and the R4810K variant in this study. RNF213 induced circular hole structures near the nucleus, similar to lipid droplets (LDs), in U-2 OS cells. The holes decorated with tagged RNF213 protein were colocalized with mCherry-RAB5A and mCherry-peroxisome, but not mCherry-RAB9A and other organelles. RNF213 decorated the holes, and the structures were growing and changing to cylindrical objects, but not the R4810K variant efficiently. Furthermore, both AAA+ ATPase modules and the ubiquitin ligase domain of RNF213 were required for the formation of the hole structures and cylindrical objects. These findings suggest that RNF213 activity may be involved in the formation of organelle-like structures, and this structure formation by RNF213 may be responsible for vascular functions.Keywords
Moyamoya disease (MMD) is a disorder characterized by internal dysfunctions in the internal carotid artery and its branches within the circle of Willis [1]. RING protein 213 (RNF213), the susceptibility gene for MMD, shows ubiquitin E3 ligase activity. Several rare RNF213 variants have been reported in MMD patients, including the p.R4810K variant (rs112735431) [2,3]. The Rnf213 knockout mice exhibited significant reductions in pericytes and impairment of blood-brain barrier function in the cortex, and promoted pathological angiogenesis [4–6]. RNF213 knockdown HUVEC cells showed no distinct changes regarding angiogenic activity, although HUVEC cells expressing RNF213 R4810K mutant influenced the angiogenic activity in endothelial cells. These phenomena indicated that RNF213 might play a role in the angiogenesis pathway in MMD endothelial cells.
RNF213 possessed two active AAA+ ATPase modules, a CBM20 carbohydrate-binding domain, a RING, and RZ finger domains [7,8]. RNF213 interacted and showed ubiquitylation activity with several E2 enzymes. The nucleotide variations of RNF213 were observed within the RING and RZ finger domains [3]. These enzymatic activities were required for functions and localization on lipid droplets, as well as for lipotoxicity, inflammation, and pathogen recognition [8–11].
Herein, we have found that RNF213 ectopic expression leads to the formation of a circular structure and increases the number of LD-like structures in U-2 OS cells. The formation of a circular structure was immature and inefficient in U-2 OS cells expressing the R4810 K variant. We propose that the immature circular structure of the R4810K variant of RNF213 might cause impairment of endothelial cells in MMD.
2.1 Cell Culture and Plasmid Transfection
All mycoplasma-free U-2 OS cells have been purchased from the JCRB Cell Bank (Japan), and the cells have been maintained in suitable media containing the antimycotic reagent Amphotericin B to protect against mycoplasma contamination. Tagged RNF213 stably expressing U-2 OS cells were prepared as described previously [12]. Briefly, the U-2 OS cells were grown under standard conditions in RPMI media (Fujifilm Wako Chemical, Osaka, Japan, catalog number 189-02025) supplemented with 10% FBS (Biosera) and penicillin, streptomycin, and Amphotericin B mixture (Fujifilm Wako Chemical, catalog number 161-23181). U-2 OS AAVS1-tetR cells were generated in the AAVS1 locus with the CRISPR-Cas9 system using the pZDonor-AAVS1 Puromycin plasmid (Merck, Darmstadt, Germany, catalog number D4696), in which a tet repressor cassette is subcloned between AAVS1 exons [13]. Stable cells expressing RNF213 protein were selected using Puromycin (Merck, Darmstadt, Germany, catalog number D7255), and protein expression was confirmed by western blotting [12]. Mycoplasma-free HUVEC cells were purchased from Thermo Fisher Scientific (catalog number C01510C). The medium 200 supplemented with low-serum growth supplement (Thermo Fisher Scientific Inc., Waltham, MA, USA, catalog numbers M200500 and S00310) was purchased from Thermo Fisher Scientific. Lipofectamine LTX (Thermo Fisher Scientific Inc., catalog number A12621) was used for plasmid transfection. The 4.0 × 105 cells were seeded into the well of a 6-well plate, and transfection was performed after 6 or 12 h. After 24 h, 1.5 × 105 cells were replated into wells of a 6-well plate and analyzed 30–60 h after transfection.
Recombinant adenoviruses were produced using the ViraPower Adenovirus kit (Thermo Fisher Scientific Inc., catalog number V49320) according to the manufacturer’s protocol. The amplified recombinant adenovirus was titrated, and the expression of the mCherry-tagged gene (LifeAct, EMBP, LAMP1, RAB 5A, 9A coding sequences, peroxisomes targeting signal, ER, Golgi targeting signal sequences) [14] was monitored.
Cells expressing TurboGFP and mCherry-tagged proteins were grown on poly-L-lysine (Merck, catalog number P8920)-coated coverslips. The cells were fixed with 4% formaldehyde in PHEM for 20 min on ice. DNA was visualized with 50 ng/mL Hoechst 33324 (Nacalai Tesque, Kyoto, Japan, catalog number 19173-41) for 5 min.
Cells were grown in the media with lipid-depleted FBS (BioWes, Nuaillé, France, catalog number S181L). The cells are washed twice with PBS and then fixed with 4% paraformaldehyde (Fujifilm Wako Chemical, catalog number 161-20141) in phosphate buffer (pH 7.0) for 10 min at room temperature. The fixed cells are rinsed with deionized water (ddH2O) and then incubated in 60% isopropanol for 1 min at room temperature. The cells are stained with 60% Oil Red O (Fujifilm Wako Chemical, catalog number 154-02072) solution for 15 min. The stained cells are washed three times with tap water, then dried at 32°C. The staining images are captured under BioZero microscopy (Keyence, Osaka, Japan). The incorporated dye is extracted with isopropanol, and then the solvent is directly measured for absorbance at 510 nm with an Infinite 200 PRO plate reader (TECAN, Groedig, Austria).
Anti-PDI and anti-mCherry antibodies were purchased from Abcam (Cambridge, UK, catalog number ab4644) and Takara Bio Inc. (Otsu City, Japan, catalog number 632543), respectively. Anti-RNF213 were previously reported [11].
Localization analysis of the RNF213 protein in HeLa cells was performed using an anti-RNF213 antibody as previously described [12,15]. The results of indirect immunostaining showed that the RNF213 staining pattern spread and was punctate throughout the cytoplasm in HeLa cells. Sugihara et al. reported that RNF213 stabilized cytoplasmic lipid droplets and localized on them [9]. To reveal RNF213 functions and localizations, expression of mCherry-tagged RNF213 proteins (WT and R4810K) was stably regulated under a Doxycycline-inducible promoter in U-2 OS cells, and the induced protein after adding Doxycycline was observed (Fig. 1a,b). The mCherry-tagged RNF213 protein was localized around the circular hole structures on the periphery of the nucleus (Fig. 1a). The mCherry-tagged RNF213 protein was fully decollated, and some fractions of the RNF213 protein were punctate close to the RNF213 decollated circular structures. These circular structures were continuously observed, and the number gradually increased. These observations were also confirmed in the localization study using TurboGFP-tagged RNF213 protein (Fig. A1a), indicating that RNF213 specifically localized to the surrounding circular structures.
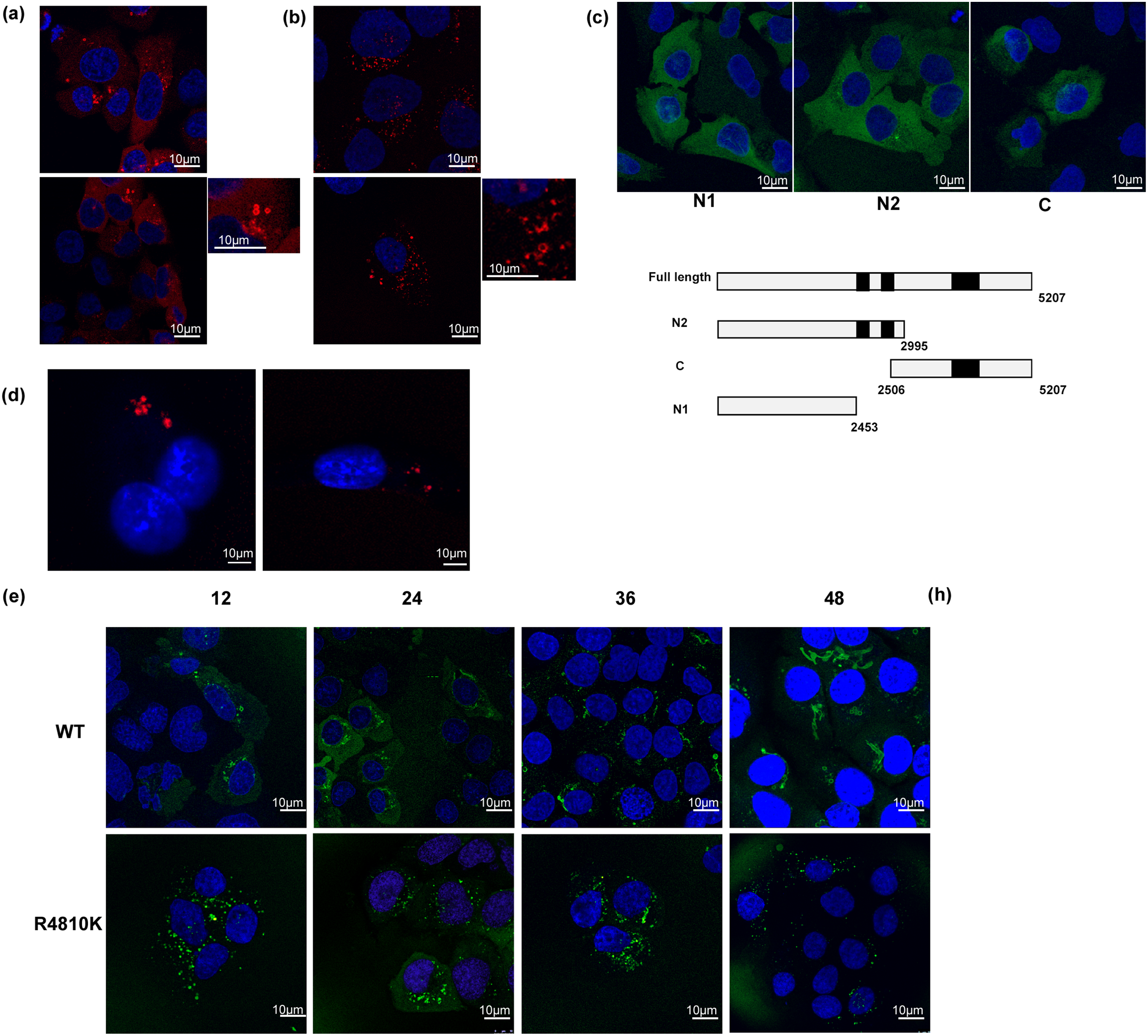
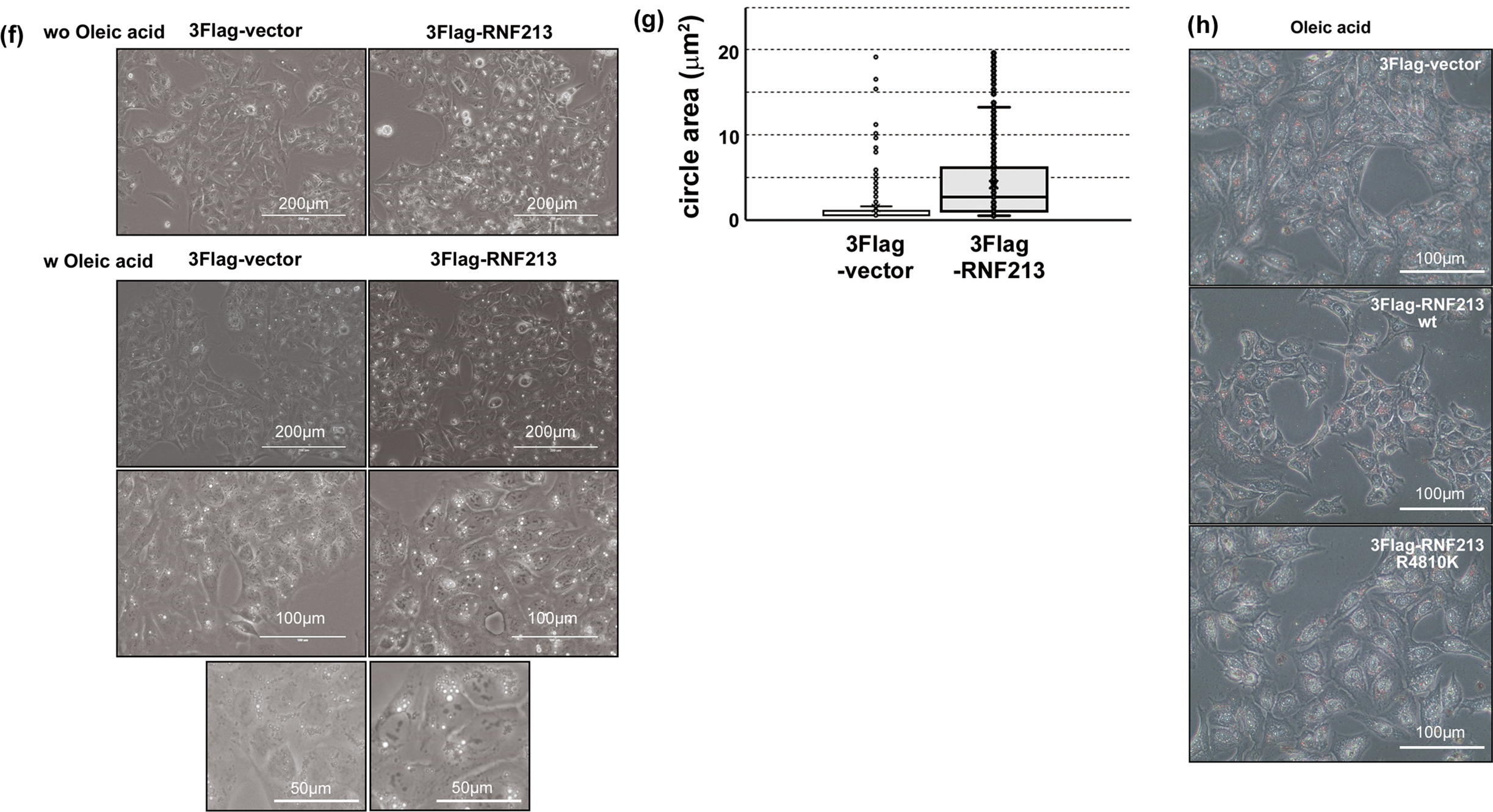
Figure 1: Localization of RNF213. (a) Localization of mCherry-tagged wild-type RNF213. The scale bars indicate 10 µm. Red: mCherry-tagged RNF213, Blue: DNA. The magnified image was shown in the right panel. (b) mCherry-tagged RNF213 R4810K variant. The scale bars indicate 10 µm. Red: mCherry-tagged RNF213, Blue: DNA. The magnified image was shown in the right panel. (c) Localization of TurboGFP-tagged deletion mutants of RNF213. Schematic illustration of deletion mutants. The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Blue: DNA (d) Expression of mCherry-tagged RNF213 proteins in HUVEC cells, wild-type RNF213 protein (left), R4810K variant (right). The scale bars indicate 10 µm. Red: mCherry-tagged RNF213, Blue: DNA (e) Time-dependent cylinder structure formation of tagged wild-type RNF213 protein, wild-type RNF213 protein (upper), R4810K variant (lower). The digital images were captured at the indicated times. The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Blue: DNA (f) RNF213-induced LD-like structures without fatty acid (upper panels), and with oleic acid (lower, middle, and lower panels) under bright field microscopy. The scale bars indicate 200 µm in photos without fatty acid (upper) and with oleic acid (upper). The scale bars indicate 100 µm in photos with oleic acid (middle). The scale bars indicate 50 µm in photos with oleic acid (lower). (g) The circle area size of the LD-like structure in the absence of fatty acid. The area sizes were measured using ImageJ software. 100 circles in independent cells are measured in independent experiments. (h) Oil Red O staining of LD structures with oleic acid under bright field microscopy. The scale bars indicate 100 µm
To investigate the relationship between RNF213 function and circular structures, a series of truncated RNF213s was expressed in U-2 OS cells. A fragment N1without AAA+ modules and RING domain, and a fragment N2 containing two AAA+ modules fused with TurboGFP proteins, did not decollate the circular structures (Fig. 1c). A fragment C1 with the RING domain also did not decollate the circular structures (Fig. 1c). These results indicated that both ATPase and ubiquitin ligase activities might be required for RNF213-decollating circular structures.
Expression of the mCherry-tagged RNF213 protein R4810K variant was stably regulated under a Doxycycline-inducible promoter in U-2 OS cells (Fig. 1b). The induced protein after adding Doxycycline was observed to be at a similar level to the wild-type protein (Fig. A1b). Although the mCherry-tagged R4810K variant showed punctate close to the nucleus as wild type, the R4810K variant could not fully decollate the circular structure (Fig. 1b). These differences were also observed in HUVEC cells overexpressing mCherry-tagged RNF213 proteins (Fig. 1d). In these cells, wild-type RNF213 protein decollated filled-circular structures but not hollow ones. On the other hand, the R4810K variant formed smaller dots (Fig. 1d). These dots and the immature circular structures were observed thoroughly during the observation in R4810K variant expressing cells (Fig. A1c). These results suggest that the R4810K variant may be unable to decollate and assemble on the circular structures compared to the wild-type RNF213 protein in U-2 OS and HUVEC cells.
Post-induction 36 h, circular hole structures by TurboGFP-tagged RNF213 protein appeared to elongate and change the structure into cylinder-like and sheet-like structures (Fig. 1e, upper and Fig. A1c). At 48 h, the cylinder-like structures elongated further and surrounded half of the nucleus (Fig. 1e, upper). These elongated cylinder structures were not observed in the study using the R4810K variant (Fig. 1e, lower and Fig. A1c). Interestingly, cells with more than 2 circular hole structures were rarely observed in R4810K variant expressing cells. However, cells with more than 2 circular hole structures were observed to become increasingly dependent on time (Fig. A1c). The observations suggested that RNF213 might form cylinder-like structures from circular ones. Additionally, these results indicated that the R4810K variant may be unable to form cylinder- and sheet-like structures compared to the RNF213 wild-type protein in U-2 OS cells because of the immature and lower number of circular hole structures in R4810K variant expressing cells.
To address the circular structure decorated by the RNF213 protein, organelle-specific targeting mCherry proteins (targeting to actin, tubulin, endoplasmic reticulum (ER), Golgi apparatus, peroxisomes, and endosomes) were coexpressed in TurboGFP-tagged RNF213 protein overexpressing U-2 OS cells. In LifeAct-mCherry protein or EMTB-mCherry protein-expressing cells, TurboGFP-tagged RNF213 protein was observed independently on these filamentous structures (Fig. A1d). In ER-targeting mCherry protein-expressing cells, signals of TurboGFP-tagged RNF213 proteins were not observed on ER structures. However, RNF213 signals were observed near ER signals (Fig. A1d). TurboGFP-tagged RNF213 proteins seemed to be close to the Golgi apparatus (Fig. A1e) in every cell. These observations suggest that the circular structure of RNF213 may be dependent on the functions of Golgi structures. Interestingly, the mCherry-fused peroxisome targeting signal sequence [16], which is imported into the peroxisome matrix by being recognized by the PEX5 protein, is distributed patchily on TurboGFP-RNF213, decollating circular structures (Fig. A1e, middle and magnified images). Some of the endosome protein RAB5A signal overlapped with TurboGFP RNF213 signals, but RAB9A signals did not (Fig. A1f). These observations suggested that RNF213 protein-decollating structures may be monolayer lipid structures, similar to those found in peroxisomes. These observations indicated that RNF213 may be associated with early and endosomal-mediated membrane-based structures.
Localization and organelle-staining studies suggested that RNF213 may play a role in lipid-based membrane maintenance. To clarify functional interactions between RNF213 function and the structures, RNF213-overexpressing cells were observed under bright-field microscopy. In Flag-tagged RNF213-overexpressing U-2 OS cells, lipid droplet (LD) -like structures were observed to be larger than in control cells (Fig. 1f,g). Furthermore, in the presence of fatty acid, the size of these LD-like structures increased (Fig. 1f, lower). However, the number of LD-like structures did not increase (Fig. 1f).
Furthermore, uptake of fatty acids was traced by Oil Red O staining. RNF213 overexpressing cells were stained with a dye similar to that of control cells, although the size of LD-like structures increased (Fig. 1h). These results were also observed in measuring the extracted dye from the uptake cells. (Fig. A1g) In addition, the R4810K variant of RNF213-overexpressing cells also stained with Oil Red O dye, similar to control and wild-type RNF213 overexpressing cells. The dye-negative LD-like structures were observed in RNF213-overexpressing cells (Fig. 1h). These observations suggested that RNF213 might regulate and maintain lipid-based membrane structures, but not lipid accumulation and metabolism. The LD-like structures were observed in more than 10 holes in one cell, but the RNF213 decorating structures were observed in fewer than 3 holes. In the absence of fatty acids, the signals of TurboGFP-RNF213 WT overlapped with LD were not observed in our experiments (data not shown). These observations suggested that RNF213 may induce lipid-based membrane structures, but not LD. The LD-like structure might be different from the RNF213 decollated circular structure.
In this study, RNF213 induced circular hole structures near the nucleus, similar to lipid droplets (LDs), in U-2 OS cells. RNF213 consists of two active AAA+ ATPase modules and a RING and RZ finger domains. RNF213 interacted and showed ubiquitylation activity with several E2 enzymes. In this study, ectopic expression of RNF213 could decollate circular hole structures, and these structures elongated and formed sheets and cylinder structures. On the other hand, the R4810K variant of RNF213 could not decollate like the wild-type NF213 protein. Furthermore, truncated deletion mutants of RNF213 could not decollate one. The holes decorated with tagged RNF213 protein were colocalized with mCherry-RAB5A and mCherry-peroxisome, but not mCherry-RAB9A and other organelles. RNF213 decorated the holes, and the structures were growing and changing to cylindrical objects, but not the R4810K variant efficiently. Both AAA+ ATPase and ubiquitin ligase activities of RNF213 were required for the formation of the hole structures and cylindrical objects. Our previous studies have shown that RNF213 forms a dimer through the RING domain. The amino-terminal fragment of the RNF213 protein, lacking AAA+ ATPase domains, forms multimers using recombinant proteins (data not shown). The purified RNF213 protein from HEK293 cells forms a ~20 nm diameter ring-shaped oligomer [17]. The observed circular structures in U-2 OS cells were ~5 µm in diameter and formed large, cylindrical, and sheet-like structures. The R4810K variant exhibited a lower level of ATPase activity using immunoprecipitated protein from HEK293 cells, and the Lysine residue at the 4810 amino acid position might affect AAA+ ATPase modules [3]. The ATPase activities of 2 ATPase modules are required for the assembly and disassembly of the ~20 nm diameter ring-shaped oligomer [17]. These findings suggest that the RNF213 protein may serve as a hub for the formation of large lipid-based structures through the ATPase activity. These findings suggest that RNF213 activity may be involved in the formation of membrane-based structures.
Overall, ectopic expression of wild-type RNF213 protein may induce the formation of circular structures in U-2 OS cells. The RNF213 inducible structures were growing and forming sheet and cylinder structures. The signals surrounding circular structures overlapped with components of peroxisomes and RAB3A, 5A endosomes, which have a monolayer lipid structure. Because the structures were not identified under bright field in RNF213-overexpressing cells, further investigation may be required. The bright field observations revealed an increased size of LD-like structures in the absence and presence of fatty acids, but not the number of LD-like structures. These structures did not induce the accumulation of triacylglycerol because they were not stained by Oil Red O dye. These results indicated that the R4810K variant might be functionally defective. These enzymatic activities may organize cellular functions on the membrane of lipid droplets, thereby increasing sensitivity against lipotoxicity.
Acknowledgement: We thank Akio Koizumi, Harada Kouji H, and Hatasu Kobayashi for their kind help.
Funding Statement: This work was supported by grants (#15611607) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.H.
Availability of Data and Materials: The data and materials that support the findings of this study are available from the Corresponding Author, Toshiyuki Habu, upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The author declares no conflicts of interest to report regarding the present study.
A list of nonstandard abbreviations
| MMD | Moyamoya disease |
| RNF213 | RING protein 213 |
| RING | Really Interesting New Gene |
| RZ finger | RNF213-ZNFX1 finger |
| HUVEC | Human Umbilical Vein Endothelial Cells |
| RPMI | Roswell Park Memorial Institute medium |
| CRISPR-Cas9 | Clustered regularly interspaced short palindromic repeat- CRISPR-associated proteins 9 |
| AAVS1 | Adeno-associated virus integration site 1 |
| AAA | ATPases Associated with Diverse Cellular Activities |
Appendix A
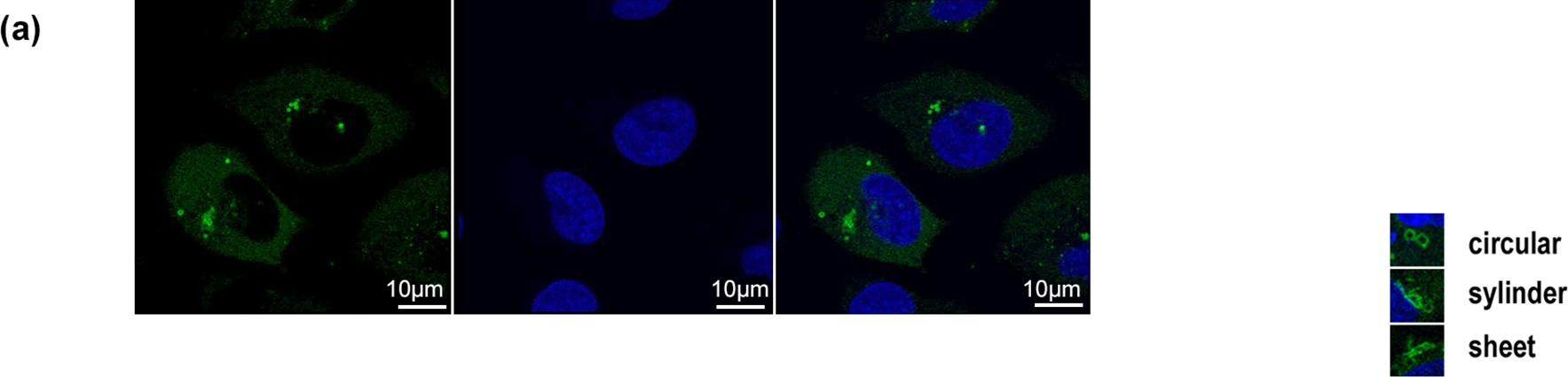
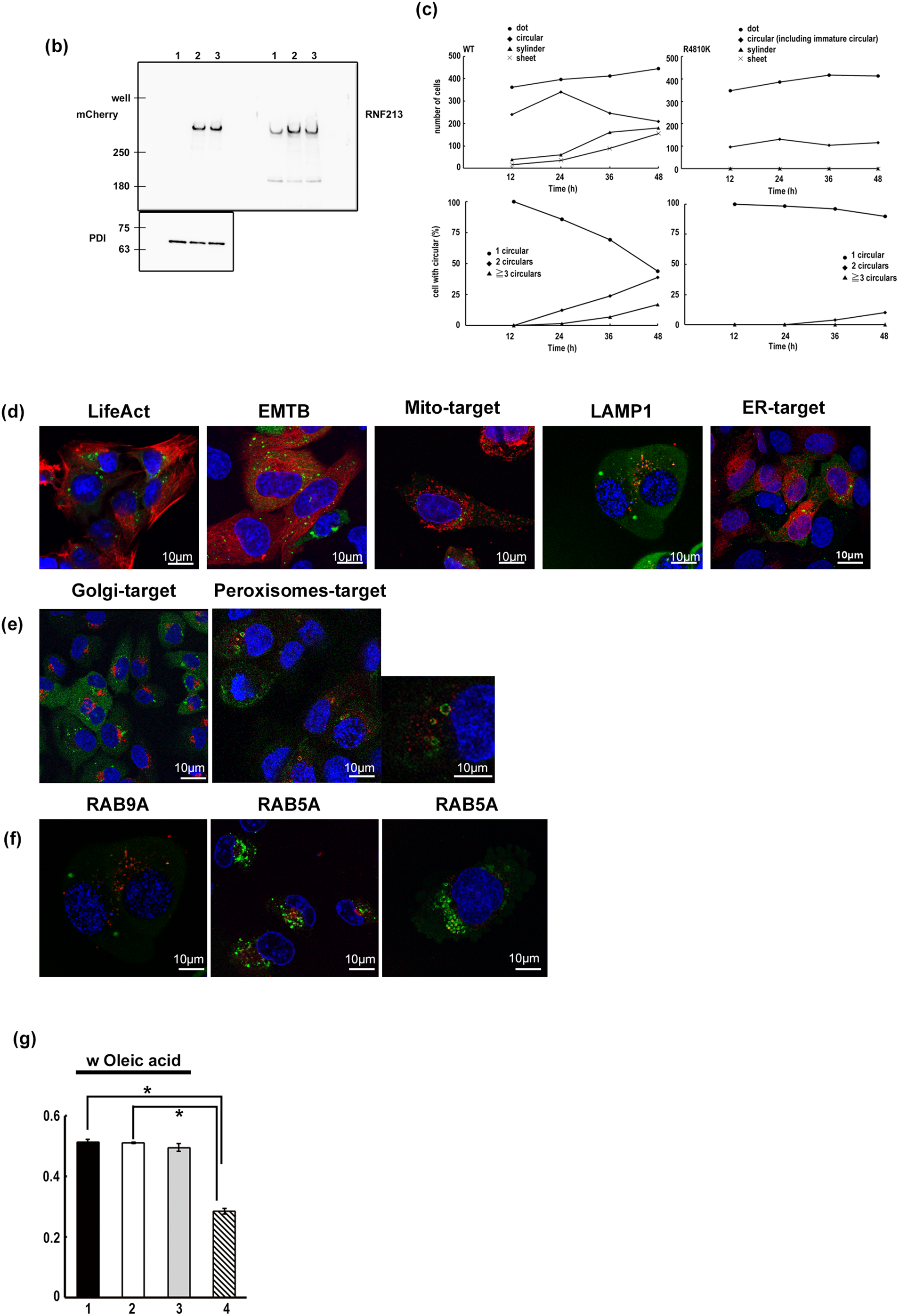
Figure A1: Localization of RNF213. (a) Localization of TurboGFP-tagged wild-type RNF213 (left), DNA (middle), and merge (right). The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Blue: DNA. (b) Western blotting analysis of tagged RNF213 proteins with anti-mCherry, anti-RNF213, and anti-PDI (internal reference) antibodies. (c) Time-dependent formation of circular, cylindrical, and sheet structures (upper graphs, left: wild type RNF213, right: R4810K RNF213). At the indicated time, TurboGFP signals were monitored and counted. Over five hundred cells were counted. The cells with circular structures were monitored in the upper graph. The cells were classified by the number of circular structures (lower graphs, left: wild type RNF213, right: R4810K RNF213). (d) Localization of TuroboGFP-tagged wild-type RNF213 protein with organelles targeting mCherry proteins: LifeAct for Actin, EMBP for microtubules, Mito-target for mitochondria, LAMP1 for lysosomes, ER target sequence for endoplasmic reticulum. The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Red: mCherry-tagged the indicated proteins, Blue: DNA. (e) Localization of TurboGFP-tagged wild-type RNF213 protein with organelles targeting mCherry protein: Golgi-target for Golgi body, Peroxisomes-target for peroxisomes. The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Red: mCherry-tagged the indicated proteins, Blue: DNA. The magnified image was shown in the right panel. (f) Localization of TurboGFP-tagged wild-type RNF213 protein with organelles targeting mCherry protein: RAB9A for late endosomes, RAB5A for early endosomes. The scale bars indicate 10 µm. Green: TurboGFP-tagged RNF213, Red: mCherry-tagged RAB proteins, Blue: DNA. (g) Measurement of Oil Red O staining. Extracted Oil Red O dye was analyzed by Absorbance 540 nm, and three independent results were subjected (n = 3; *p < 0.03, by Student’s t-test). 1: Flag only, 2: wild-type Flag-tagged RNF213, 3: R4810K Flag-tagged RNF213, 4: blank
References
1. Ihara M, Yamamoto Y, Hattori Y, Liu W, Kobayashi H, Ishiyama H, et al. Moyamoya disease: diagnosis and interventions. Lancet Neurol. 2022;21(8):747–58. doi:10.1016/s1474-4422(22)00165-x. [Google Scholar] [PubMed] [CrossRef]
2. Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One. 2010;6(7):e22542. doi:10.1371/journal.pone.0022542. [Google Scholar] [PubMed] [CrossRef]
3. Kobayashi H, Matsuda Y, Hitomi T, Okuda H, Shioi H, Matsuda T et al. Biochemical and functional characterization of RNF213 (Mysterin) R4810K, a susceptibility mutation of moyamoya disease, in angiogenesis in vitro and in vivo. J Am Heart Assoc. 2015;4(7):e002146. doi:10.1161/jaha.115.002146. [Google Scholar] [PubMed] [CrossRef]
4. Li W, Niu X, Dai Y, Wu X, Li J, Sheng W. Rnf-213 Knockout induces pericyte reduction and blood-brain barrier impairment in mouse. Mol Neurobiol. 2023;7:1–13. doi:10.21203/rs.3.rs-2526175/v1. [Google Scholar] [CrossRef]
5. Roy V, Ross JP, Pépin R, Ghio SC, Brodeur A, Deschênes LT, et al. Moyamoya disease susceptibility gene RNF213 regulates endothelial barrier function. Stroke. 2022;53(4):1263–75. doi:10.1161/strokeaha.120.032691. [Google Scholar] [PubMed] [CrossRef]
6. Ye F, Niu X, Liang F, Dai Y, Liang J, Li J, et al. RNF213 loss-of-function promotes pathological angiogenesis in moyamoya disease via the Hippo pathway. Brain. 2023;146(11):4674–89. doi:10.1093/brain/awad225. [Google Scholar] [PubMed] [CrossRef]
7. Otten EG, Werner E, Crespillo-Casado A, Boyle KB, Dharamdasani V, Pathe C, et al. Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature. 2021;594(7861):111–6. doi:10.1038/s41586-021-03566-4. [Google Scholar] [PubMed] [CrossRef]
8. Crespillo-Casado A, Pothukuchi P, Naydenova K, Yip MCJ, Young JM, Boulanger J, et al. Recognition of phylogenetically diverse pathogens through enzymatically amplified recruitment of RNF213. EMBO Rep. 2024;25(11):4979–5005. doi:10.1101/2024.09.24.614677. [Google Scholar] [CrossRef]
9. Sugihara M, Morito D, Ainuki S, Hirano Y, Ogino K, Kitamura A, et al. The AAA+ ATPase/ubiquitin ligase mysterin stabilizes cytoplasmic lipid droplets. J Cell Biol. 2019;218(3):949–60. doi:10.1083/jcb.201712120. [Google Scholar] [PubMed] [CrossRef]
10. Piccolis M, Bond LM, Kampmann M, Pulimeno P, Chitraju C, Jayson CBK, et al. Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Mol Cell. 2019;74(1):32–44.e8. doi:10.1016/j.molcel.2019.01.036. [Google Scholar] [PubMed] [CrossRef]
11. Zhou X, Zhang H, Wang Y, Wang D, Lin Z, Zhang Y, et al. Shigella effector IpaH1.4 subverts host E3 ligase RNF213 to evade antibacterial immunity. Nat Commun. 2025;16(1):3099. doi:10.1038/s41467-025-58432-y. [Google Scholar] [PubMed] [CrossRef]
12. Habu T, Harada KH. UBC13 is an RNF213-associated E2 ubiquitin-conjugating enzyme, and Lysine 63-linked ubiquitination by the RNF213-UBC13 axis is responsible for angiogenic activity. Faseb Bioadvances. 2021;3(4):243–58. doi:10.1096/fba.2019-00092. [Google Scholar] [PubMed] [CrossRef]
13. Habu T, Ishikawa H, Kim J. Gulo gene locus, a new gene editing locus for mammalian cells. Biotechnol J. 2022;17:2100493. doi:10.1002/biot.202100493. [Google Scholar] [PubMed] [CrossRef]
14. Goedhart J, Von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun. 2012;3:751. doi:10.1038/ncomms1738. [Google Scholar] [PubMed] [CrossRef]
15. Hitomi T, Habu T, Kobayashi H, Okuda H, Harada KH, Osafune K, et al. Downregulation of Securin by the variant RNF213 R4810K (rs112735431, G>A) reduces angiogenic activity of induced pluripotent stem cell-derived vascular endothelial cells from moyamoya patients. Biochem Bioph Res Co. 2013;438(1):13–9. doi:10.1016/j.bbrc.2013.07.004. [Google Scholar] [PubMed] [CrossRef]
16. Brocard C, Hartig A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochimica et Biophysica Acta (BBA). Mol Cell Res. 2006;1763(12):1565–73. doi:10.1016/j.bbamcr.2006.08.022. [Google Scholar] [PubMed] [CrossRef]
17. Morito D, Nishikawa K, Hoseki J, Kitamura A, Kotani Y, Kiso K, et al. Moyamoya disease-associated protein mysterin/RNF213 is a novel AAA+ ATPase, which dynamically changes its oligomeric state. Sci Rep. 2014;4(1):4442. doi:10.1038/srep04442. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools