 Open Access
Open Access
ARTICLE
Lightweight Multi-scale Convolutional Neural Network for Rice Leaf Disease Recognition
1 College of Information Technology, Jilin Agricultural University, Changchun, 130118, China
2 Jilin Province Agricultural Internet of Things Technology Collaborative Innovation Center, Changchun, 130118, China
3 Jilin Province Intelligent Environmental Engineering Research Center, Changchun, 130118, China
4 Jilin Province Information Technology and Intelligent Agriculture Engineering Research Center, Changchun, 130118, China
5 Department of Agricultural Economics and Animal Production, University of Limpopo, Private Bag X 1106, Sovenga, 0727, Polokwane, South Africa
* Corresponding Author: Yu Sun. Email:
Computers, Materials & Continua 2023, 74(1), 983-994. https://doi.org/10.32604/cmc.2023.027269
Received 13 January 2022; Accepted 23 March 2022; Issue published 22 September 2022
Abstract
In the field of agricultural information, the identification and prediction of rice leaf disease have always been the focus of research, and deep learning (DL) technology is currently a hot research topic in the field of pattern recognition. The research and development of high-efficiency, high-quality and low-cost automatic identification methods for rice diseases that can replace humans is an important means of dealing with the current situation from a technical perspective. This paper mainly focuses on the problem of huge parameters of the Convolutional Neural Network (CNN) model and proposes a recognition model that combines a multi-scale convolution module with a neural network model based on Visual Geometry Group (VGG). The accuracy and loss of the training set and the test set are used to evaluate the performance of the model. The test accuracy of this model is 97.1% that has increased 5.87% over VGG. Furthermore, the memory requirement is 26.1 M, only 1.6% of the VGG. Experiment results show that this model performs better in terms of accuracy, recognition speed and memory size.Keywords
In recent years, with the continuous development of computer technology, deep learning with powerful learning capabilities has been widely used in the field of computer vision, leading to progress in the research on intelligent recognition of leaf diseases. Image-based disease recognition is essentially an image classification problem, and the application of deep convolutional neural networks in the field of image recognition is a new research hotspot. In the work of DL, Barman et al. [1] proposed a low-cost smartphone-based image acquisition technology and constructed a self-structured classifier, which has higher verification accuracy and a shorter average time than MobileNet CNN calculation, but the model has obvious overfitting problems. Zhang et al. [2] used a dual-path convolutional neural network to screen corn ears. Tang et al. [3] proposed a lightweight CNN model to diagnose grape diseases. This method successfully reduced the model size from 227.5 to 4.2 MB. Wang et al. [4] established an effective neural network model, to identify common pests in agriculture and forestry. This model effectively reduced the training time and achieved recognition accuracy of 92.63%. Yao et al. [5] developed a machine vision-based automated monitoring system for pest trapping that can automatically identify and count five target pests in the image. Wu et al. [6] optimised the detection and enumeration of wheat grains using the migration method. With an average accuracy of 91%, the model can effectively count the number of grains. Qiao et al. [7] used unmanned aerial vehicles (UAV) to collect images of the field environment and proposed a novel network structure called MmNet (Mikania micrantha network). This model is mainly used to identify Mikania micrantha Kunth, and it has a simple structure with high accuracy. Waheed et al. [8] optimised the DenseNet model for disease identification and classification in corn leaves and its performance is close to the established CNN architecture. Kozłowski et al. [9] employed computer vision methods and deep neural network CNN for barley quality evaluation in the barley industry, but it is suitable only for quality evaluation of a small number of barley kernel samples. Xiao et al. [10] proposed a method for identifying rice blast. By extracting features, a three-layer back propagation (BP) neural network model was constructed to solve the problem of low accuracy of artificial identification of rice blast. Lu et al. [11] suggested a technique to enhance CNN’s DL capabilities and classify 10 common rice diseases. Dyrmann et al. [12] used CNNs to distinguish the early growth stages of plants. The model was tested on 22 different plant images. The model’s accuracy in classifying individual species was very low, resulting in an average accuracy rate of only 86.2%. Alenezi [13] used a combination of CNN with parallax attention mechanism (PAM) via graph-cut algorithms to solve the dehazing problem. Zhang et al. [14] focused on designing a new CNN network structure to improve the detection accuracy of spatial-domain steganography. Rao et al. [15] proposed a bi-linear convolution neural network (Bi-CNNs) for identifying and classifying plant leaf diseases. Then, VGG and pruned Residual neural network (ResNet) were fine-tuned and utilised as feature extractors and connected with fully linked dense networks. Chen et al. [16] proposed a region of interest (RoI)-based deep convolutional representation, for instance retrieval. Fang et al. [17] generated samples and training in an image recognition model based on CNN using deep convolution generative adversarial networks (DCGAN).
Rice diseases seriously impact the yield and quality and many diseases start from leaves. Therefore, timely and accurately identifying the types of rice leaf diseases is the key to comprehensive disease prevention. Although the use of chemical pesticides can control plant diseases, due to a wide variety of diseases, it is easy to misjudge without experience. The following information can help to identify rice diseases. Jiang et al. [18] proposed a CNN-SVM (support vector machine) hybrid algorithm. This approach can detect four major rice illnesses, with an average accuracy rate of 96.8%. Rahman et al. [19] used a basic CNN model to identify 1426 pictures of rice pests and illnesses, including eight different rice pests and diseases. The model’s accuracy is 93.3%, while the size is only 1% of VGG. Shah et al. [20] introduced various image processing and machine learning algorithms for recognising rice plant illnesses. There are also a number of recognition approaches, including feature recognition based on texture morphology and others that employ CNN-related technologies [21–28]. The accuracy of rice leaf disease identification and classification by lightweight models can still be improved. Some of the studies mentioned above mainly focused on accurate plant disease recognition and classification. For this purpose, they implemented various types of CNN architectures. In some studies, ensemble of multiple neural network architectures have been used. These studies played an important role for automatic and accurate recognition and classification of plant diseases. However, they did not consider the impact of the large number of parameters of these high performing CNN models in real life mobile application deployment. Moreover, in some studies, reducing the number of parameters in the model makes the model lightweight, while the accuracy of recognition and classification is not satisfactory. Since the reduction of the number of parameters in a CNN model reduces its learning capability, one needs to make a trade-off between memory requirement and classification accuracy to build such a model.
To address the above issue, in this research, we propose a new model structure. Specifically, we choose VGG as the backbone. In addition, to reduce the weight of the convolutional neural network recognition model, we combine the backbone with a multi-scale convolution module. Through comparative experiments, the classification of diseased rice leaves shows that the model we designed is more effective than its corresponding structure. Furthermore, the results show that the model can improve accuracy while reducing memory requirements to meet application requirements.
Using the rice disease public image data provided in the Kaggle database [29], a variety of disease images are selected and organised into a new data set, including healthy leaves and three types of rice leaf disease images. Rice diseases occurrence depends on many factors such as temperature, humidity, rainfall, variety of rice plants, season, nutrition, etc. Brown spot is characterized by brown lesions on the leaves that develop brown dots, irregular dark brown lesions appear on the leaves of leaf blast, and the lesion part of leaf burn is bright yellow. When the following four types of image data are examined, the data set is less diseased and difficult to differentiate. The more similar the photographs are, the harder it is to classify them. Concurrently, we discovered that several of the perplexing data categories had been improperly assigned throughout the sorting process. After processing this part of the data, we will choose these four common leaf images to train and evaluate our model. Next, the sorted original image data is divided into three sets: a train set, a validation set and a test set in 8:1:1 ratio. Due to the small amount of image data, the number of various types of samples varies from 300 to 700. The imbalance of the sample distribution will result in deviations in the model’s recognition of each category after training, and the small number of sample data will be enhanced. The original data image is shown in Fig. 1.

Figure 1: Common rice leaf diseases
To improve the classification accuracy of the network, this paper uses data enhancement to expand the data volume. Data enhancement algorithms, including flipping, rotating, scaling, contrast enhancement and colour enhancement, were used in this paper. After enhancement, each category has around 3,000 images. The final expanded image data set has 12,229 samples. Tab. 1 shows the name and the number of images of each kind of grape leaf disease.
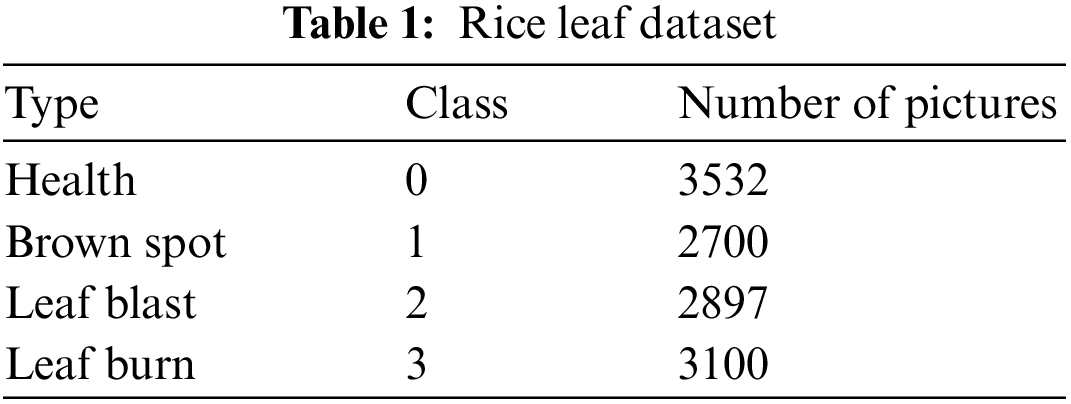
In the 2014 ILSVRC (ImageNet Large Scale Visual Recognition Challenge) competition, VGG won the second place in the classification project and the first place in the positioning project [30]. Its network structure is very simple; the entire network uses the same size convolution core size (3 × 3) and maximum pooling size (2 × 2). Using several consecutive 3 × 3 convolution kernels, a stacked small convolution kernel outperforms a large convolution kernel for a given receptive field because multiple non-linear layers can increase the depth of the network to ensure that more complex patterns are learned with fewer parameters. However, VGG consumes more computing resources and parameters, resulting in more memory usage. The VGG network structure is shown in Fig. 2.
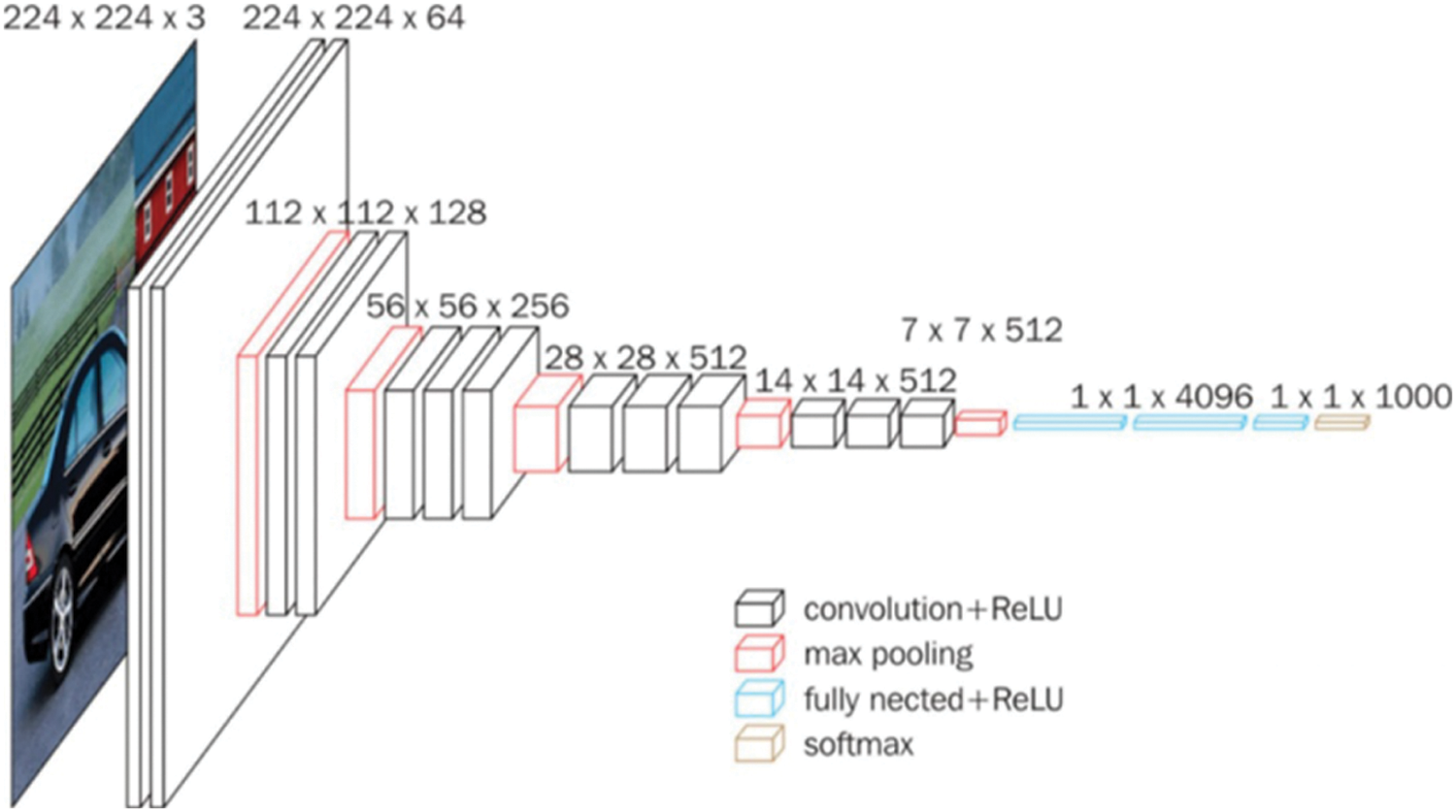
Figure 2: VGG convolutional neural network
The model parameters of the VGG network structure are large, and the training time is long. Therefore, we used some methods to improve the model, and new architecture was built.
We discuss below the building blocks of the network.
Inception [31] is a structure proposed by GoogLeNet to expand the depth and width of the network and improve the performance of deep neural networks. First, convolve the input features of different sizes, such as 3 × 3, 5 × 5, 1 × 1 and max pooling. For four branches, use filters of different sizes for convolution or pooling and finally stitch together in the feature dimension. Convolution on multiple scales simultaneously can extract the features of different scales and richer kinds, resulting in a more accurate final classification judgement. We will learn from the inception module and two neural network structures of VGG, combining their advantages and propose a neural network structure that reduces the size of the model as much as possible while ensuring accuracy.
Compared with the fully connected layer, global average pooling technology is a simpler choice of convolutional structure for establishing the relationship between feature maps and categories. It accepts images of any size and better matches the category with the feature map of the last convolutional layer, reducing the number of parameters. We will use the global average pooling operation to replace the fully connected layer in the neural network.
Batch normalization (BN) was proposed to solve Internal Covariate Shift (ICF), that is, in the process of deep network training, the process of internal node data distribution changes due to changes in the parameters of the network [32]. In DL, due to the large memory requirements of the full batch training method, each round of training time is considerably long. Therefore, we add the BN layer to the back of the partial convolutional layer. BN normalises the input of each layer of the network to ensure that the mean and variance of the input distribution are fixed within a certain range, thus reducing the ICF problem in the network, alleviating the disappearance of the gradient to a certain extent and accelerating the model convergence. In addition, BN makes the network more robust to parameters and activation functions and reduces the complexity of the neural network model training and parameter adjustment. Finally, in the BN training process, the mini-batch mean/variance is used as the overall sample statistic estimation and the introduction of random noise, which has a regularising effect on the model to a certain extent.
The low-level convolution extracts simple information, such as edges, colours and textures in the deep convolutional neural network, while the high-level convolution completes the abstraction of features. The following issues need to be considered when identifying different diseases on the leaf part: At the early stage of disease onset, the disease spots are tiny, making it difficult to capture detailed textures, causing overfitting of the model and increasing the difficulty of model training. Tiny colour differences are the key to distinguish different diseases. Different diseases exhibit similar colour, texture, and contour features at a certain stage of disease onset. The same disease varies significantly at different stages of disease onset. The integrated extraction of multiple features is the key to characterize the dynamic changes of diseases. Therefore, convolutional kernels of different sizes are set in the model for improving the response of the network to different features.
The improvement in the inception module uses the 7 × 7 convolution kernel on the original inception structure instead of max pooling and adds 1 × 1 convolution before 3 × 3, 5 × 5 and 7 × 7 convolution for dimensionality reduction. By using 7 × 7 convolution kernels instead of max pooling to extract features, the number of parameters can be reduced and the adaptability of the network to the scale can be increased. At the same time, four scales of convolution kernels are used to extract information on different scales, and the sample image features are extracted in parallel before being merged into the same tensor to continue downstream. Without changing the size of the receptive field of the feature layer, use 1 × 1 convolution to reduce the depth of the feature layer to minimise calculations. Connecting the rectified linear unit (ReLU) activation function after 1 × 1 convolution can also increase the nonlinearity of the network to a certain extent. The improved inception module is shown in Fig. 3.
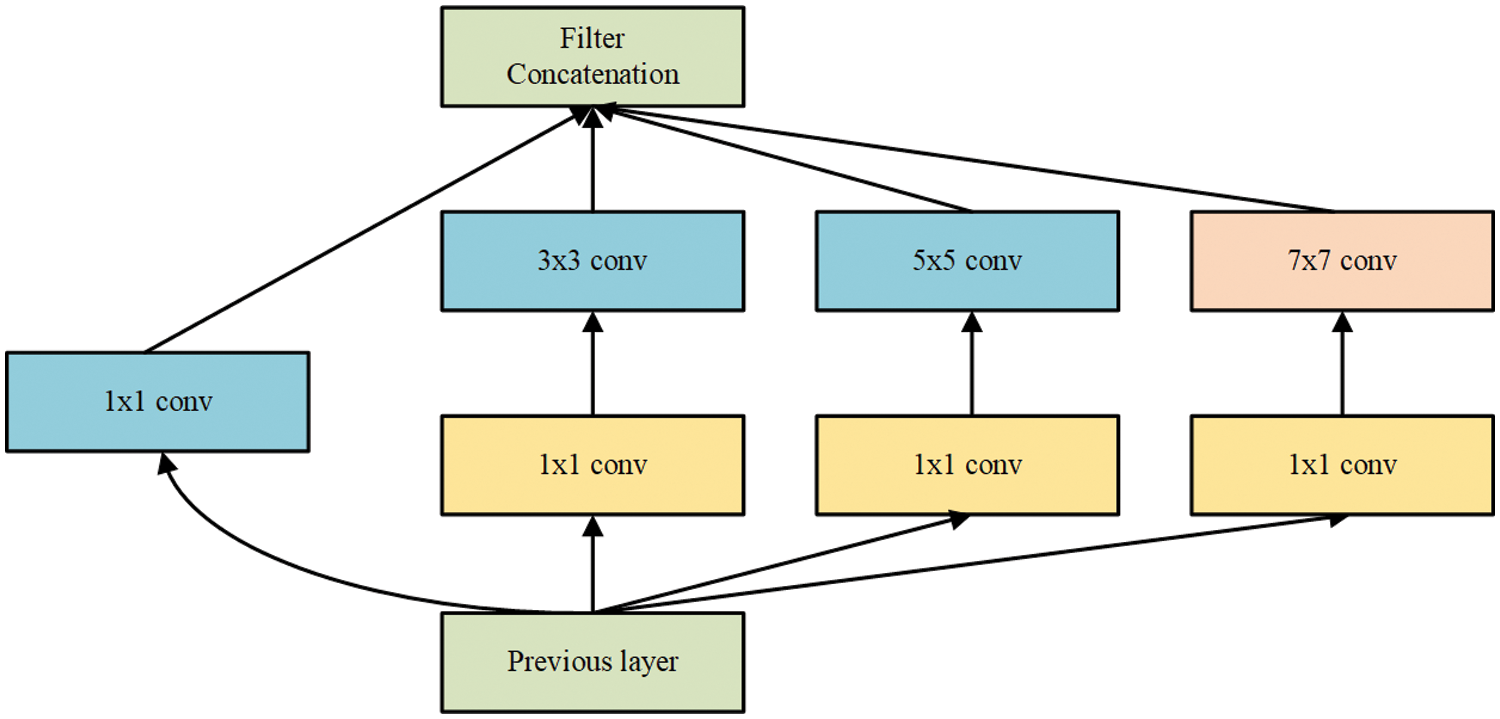
Figure 3: Improved inception module
The input image size is 224 × 224 × 3, and the convolution kernel size of the first convolutional layer is 7 × 7. As inspired by the inception module, a 1 × 1 convolutional layer is added after the first convolutional layer to reduce the dimensionality and calculation of the input. After adding the improved inception module, the max pooling layer has an uneven advantage in features. Compared with VGG, only one convolutional kernel size is applied, which indicates more information. In addition, using the global average pooling operation rather than a fully connected layer can significantly reduce the parameter size, save hardware resources and improve training speed. Finally, add two BN layers to the convolutional layer to accelerate neural network training and stabilise model training. The neural network structure is shown in Figs. 4 and 5.
Figure 4: The process of model operation
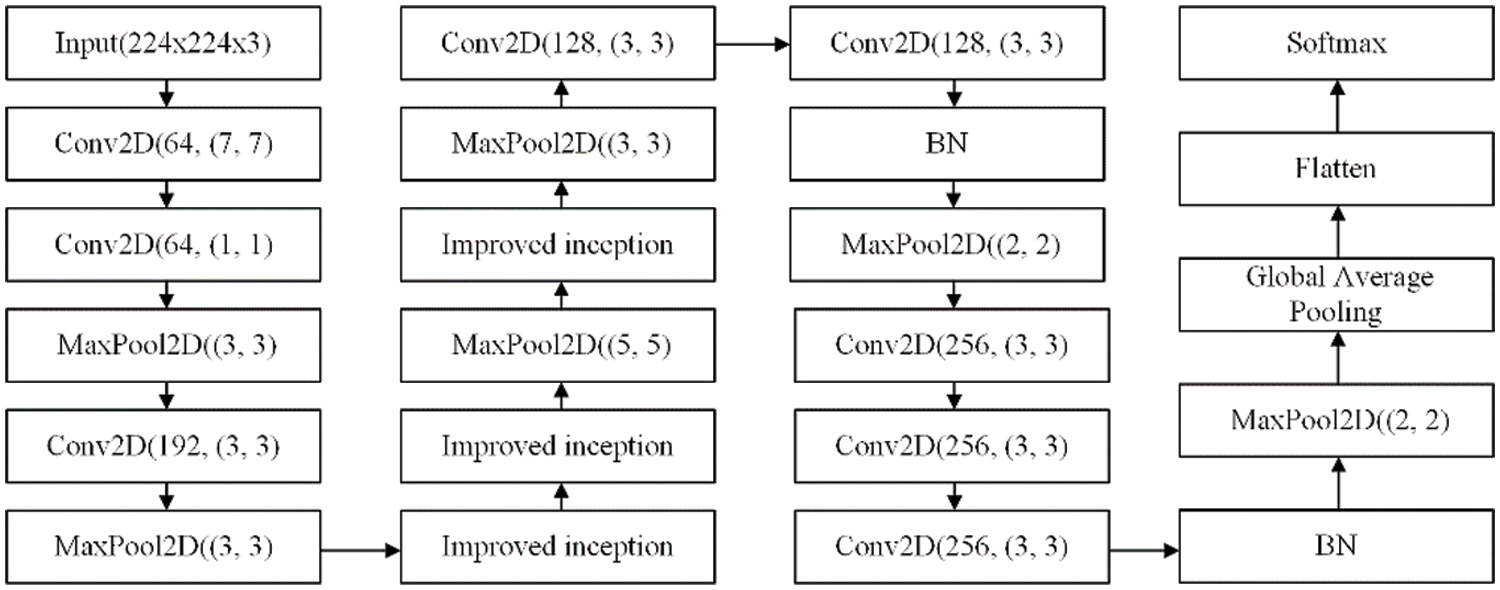
Figure 5: The neural network architecture
The test platform is desktop computers and the software testing environment is windows 10, a 64-bit system. It is equipped with a processor Intel Core i5-4590 clocked at 3.3 GHz, 8 GB of RAM, graphics NVIDIA Ge Force GT 705, all running under the python programming language and using DL framework TensorFlow-gpu2.2 and keras2.4.3.
During training or model evaluation, normalisation is achieved by dividing each pixel by 255.0, resulting in the range of 0 to 1. Then, such multi-channel images are used as input to the CNN model. The initialisation of the weights will have an impact on the network performance. In this paper, the Xavier method is chosen to initial convolutional layer Conv1. The selected optimiser was stochastic gradient descent (SGD) with a momentum of 0.9, a learning rate of 0.0001, a training batch of 32, an input image size of 224 × 224 × 3 and each model update iteration of 50 times. Each input iteration is randomly shuffled during the training process to ensure that the model completes the training quickly. During the training process, setting callback function to reduce the learning rate and early stopping to end the training early.
The lightweight model requires the recognition model to respond rapidly while maintaining accuracy and compressing the model’s memory requirements as much as possible. As a result, the average recognition accuracy, memory needs and average forward processing time are utilised to evaluate the performance of the recognition system.
The accuracy of the results is ensured by training of the model repeatedly. After the model is trained, its performance is evaluated and compared based on the accuracy and loss curve. The average recognition accuracy is the most important indicator to test the model’s performance, and the loss function uses cross-entropy.
where
Fig. 6 depicts the accuracy and loss value of each model iteration in this paper. The model reduces three times learning rate during the training process and finally triggers the callback function to complete the training at the fifteenth iteration.
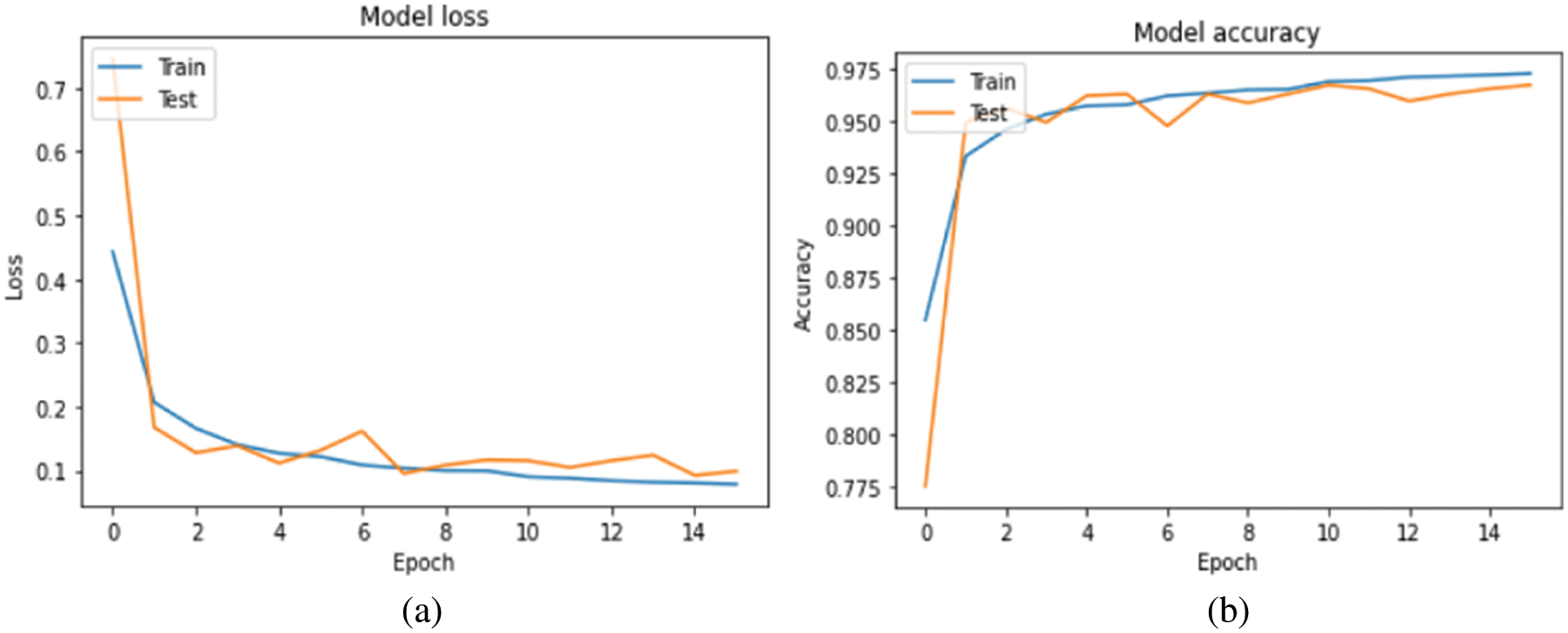
Figure 6: (a) Loss plot and (b) Accuracy plot
In addition, the model in this paper was evaluated with the help of a confusion matrix. Fig. 7 shows the classification results for each category in the normalised confusion matrix of the model. The figure shows that a small number of brown-spot infected leaves mingled with healthy leaves. The main reason for this situation is that some brown-spot leaves are in the early stages of onset and the characteristics are not obvious. Some healthy leaves also have small spots that are not obvious. There is only one image classification error for burnt leaves and blast leaves. This result explains the recognition ability of the model in this article.
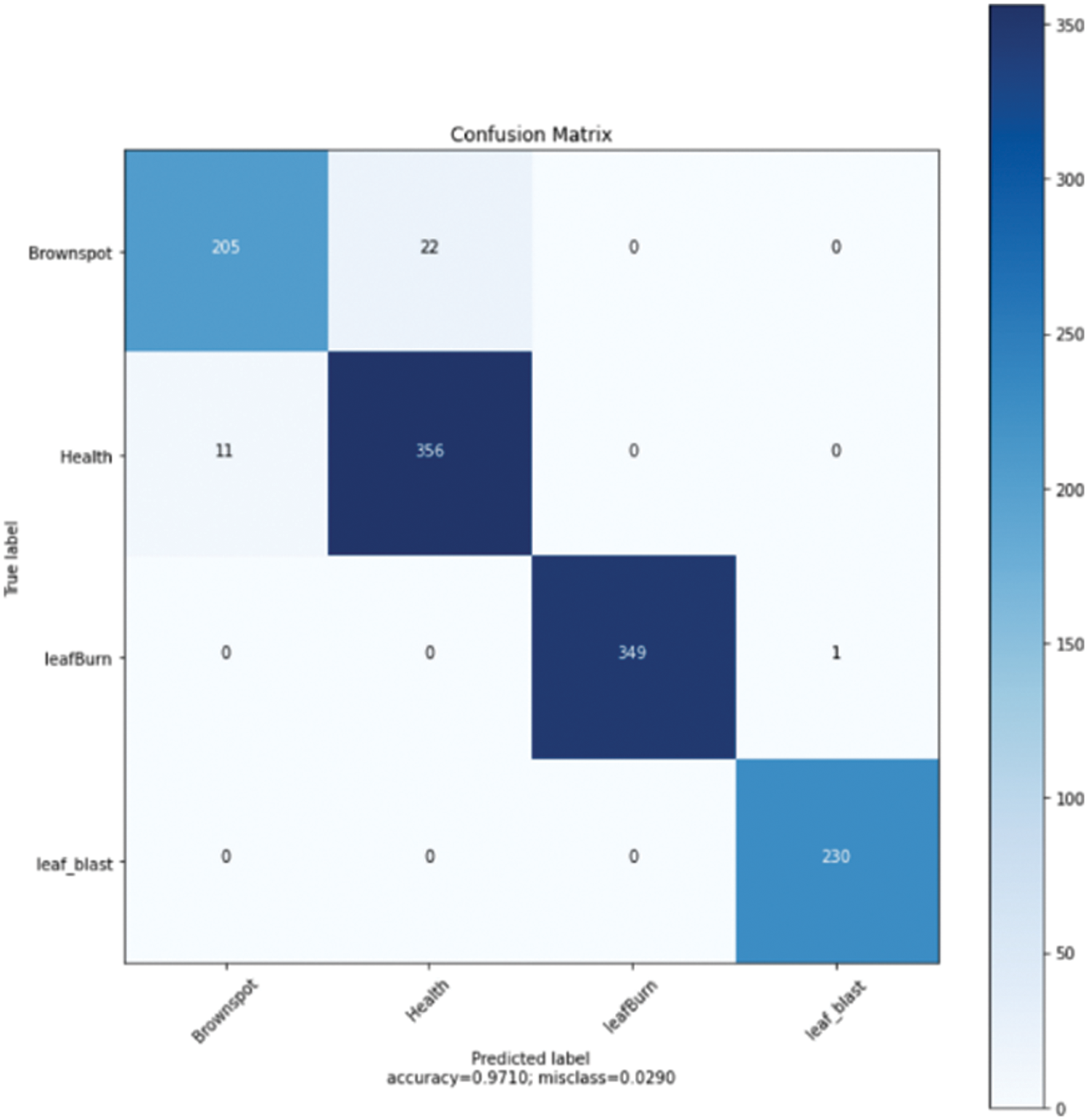
Figure 7: Confusion matrix
In addition to classification accuracy, speed is another important performance indicator. In some application scenarios, speed is a crucial evaluation indicator. The average forward processing time (AFT) represents the time required to predict a certain image on the same hardware [3].
The number of test rounds is n, and the time required for each forward propagation is t in milliseconds.
The comparison experiment uses transfer learning, mainly focuses on four neural networks: VGG16, GoogLeNet (inception v3), MobileNet v1 and improved-ShuffleNet V1 [3]. Fine tuning, transfer learning and training from scratch have been implemented to assess their performance. In these four architectures, fine tuning the model while training has shown the best performance. The experimental results are shown in Tab. 2.
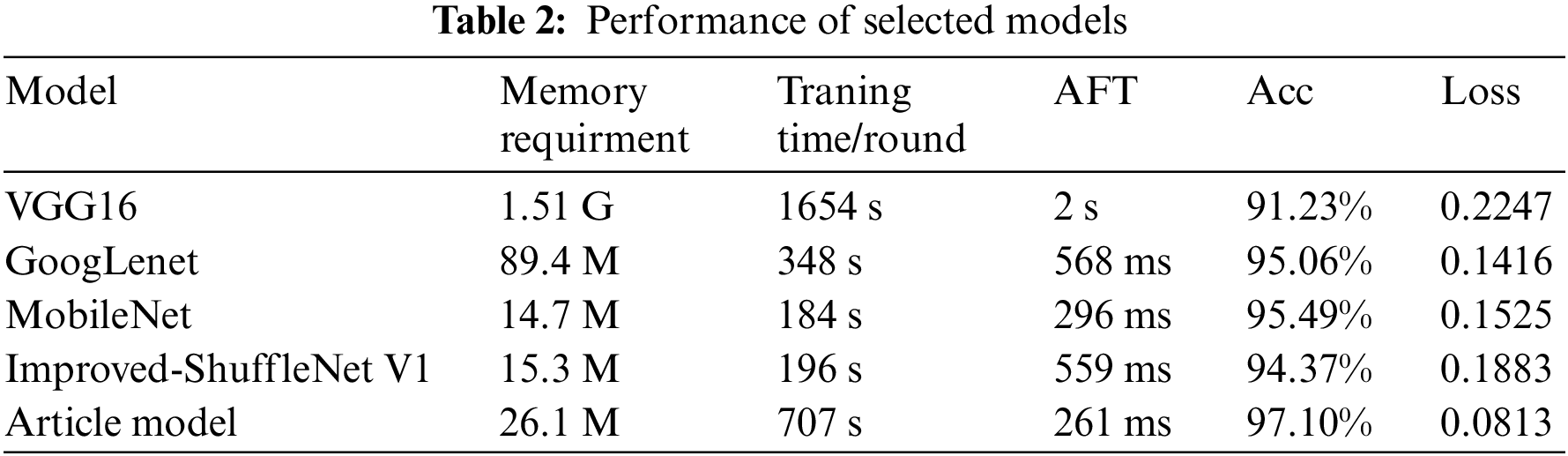
The VGGNet-based model involves a large number of parameters, so the training time of the model is long, and the memory requirement of the model is also large. However, the memory requirement of the model in this paper is only 1.6% of the VGGNet model, which can completely fulfil the need to identify disease images under the mobile platform in production practice. The accuracy of the model in this paper indicates that the model is more accurate in identifying different diseases, however, multiple convolution operations increase the training time of the model due to the use of convolution kernels of multiple sizes. The improved-ShuffleNet V1 model had high classification accuracy in the PlantVillage dataset, but only 94.37% accuracy in the rice disease classification task. Both the GoogLeNet and MobileNet models have significantly reduced the number of parameters and downsized the memory requirement to less than 100 M while slightly improving the accuracy of the model, but the accuracy of the model in this paper is 2.04% and 1.61% higher than the above two models, respectively. Due to the depth separable convolution unit, MobileNet employs a large number of small-size convolution operations and frequent feature stitching involved in the network. As a result, the recognition time is slightly longer. Compared with network structures, such as MobileNet, the model in this paper has the same image input size, small memory requirements, shorter recognition time and the highest accuracy.
This paper proposes a rice leaf disease classification model combining VGG and inception v1 variant structures. This network combines the advantages of VGG and the inception module to obtain a practical model for classifying leaf diseases. The author optimised the structure of VGG16, which significantly improved the recognition speed and reduced memory requirements. The optimal batch size established through experiments and the initial learning rate makes the model more stable and accurate. To check the superiority of this model, we compared it with other CNNs. Experiments show that our network architecture has the most advanced performance. The classification accuracy of this model reaches 97.10%. To further improve the accuracy of rice disease recognition, we still need more high-quality rice disease image samples. The model can be further expanded to classify more than four types of rice leaf diseases and applied to mobile platforms.
Funding Statement: This research was supported by National key research and development program sub-topics [ 2018YFF0213606-03 (Mu Y., Hu T. L., Gong H., Li S. J. and Sun Y. H.) http://www.most.gov.cn], Jilin Province Science and Technology Development Plan focuses on research and development projects [20200402006NC (Mu Y., Hu T. L., Gong H. and Li S.J.) http://kjt.jl.gov.cn], Science and technology support project for key industries in southern Xinjiang [2018DB001 (Gong H., and Li S.J.) http://kjj.xjbt.gov.cn], Key technology R & D project of Changchun Science and Technology Bureau of Jilin Province [21ZGN29 (Mu Y., Bao H. P., Wang X. B.) http://kjj.changchun.gov.cn].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. U. Barman, R. D. Choudhury, D. Sahu and G. G. Barman, “Comparison of convolution neural networks for smartphone image based real time classification of citrus leaf disease,” Computers and Electronics in Agriculture, vol. 177, no. 4, pp. 105661, 2020. [Google Scholar]
2. J. Zhang, Q. Ma, X. Cui, H. Guo, K. Wang et al., “High-throughput corn ear screening method based on two-pathway convolutional neural network,” Computers and Electronics in Agriculture, vol. 175, no. 9, pp. 105525, 2020. [Google Scholar]
3. Z. Tang, J. Yang, Z. Li and F. Qi, “Grape disease image classification based on lightweight convolution neural networks and channelwise attention,” Computers and Electronics in Agriculture, vol. 178, no. 2, pp. 105735, 2020. [Google Scholar]
4. J. Wang, Y. Li, H. Feng, L. Ren, X. Du et al., “Common pests image recognition based on deep convolutional neural network,” Computers and Electronics in Agriculture, vol. 179, no. 1, pp. 105834, 2020. [Google Scholar]
5. Q. Yao, J. Feng, J. Tang, W. G. Xu, X. H. Zhu et al., “Development of an automatic monitoring system for rice light-trap pests based on machine vision,” Journal of Integrative Agriculture, vol. 19, no. 10, pp. 2500–2513, 2020. [Google Scholar]
6. W. Wu, T. -L. Yang, R. Li, C. Chen, T. Liu et al., “Detection and enumeration of wheat grains based on a deep learning method under various scenarios and scales,” Journal of Integrative Agriculture, vol. 19, no. 8, pp. 1998–2008, 2020. [Google Scholar]
7. X. Qiao, Y. Z. Li, G. Y. Su, H. K. Tian, S. Zhang et al., “MmNet: Identifying mikania micrantha kunth in the wild via a deep convolutional neural network,” Journal of Integrative Agriculture, vol. 19, no. 5, pp. 1292–1300, 2020. [Google Scholar]
8. A. Waheed, M. Goyal, D. Gupta, A. Khanna, A. E. Hassanien et al., “An optimized dense convolutional neural network model for disease recognition and classification in corn leaf,” Computers and Electronics in Agriculture, vol. 175, no. 12, pp. 105456, 2020. [Google Scholar]
9. M. Kozłowski, P. Górecki and P. M. Szczypiński, “Varietal classification of barley by convolutional neural networks,” Biosystems Engineering, vol. 184, no. 1, pp. 155–165, 2019. [Google Scholar]
10. M. Xiao, Y. Ma, Z. Feng, Z. Deng, S. Hou et al., “Rice blast recognition based on principal component analysis and neural network,” Computers and Electronics in Agriculture, vol. 154, no. 4, pp. 482–490, 2018. [Google Scholar]
11. Y. Lu, S. Yi, N. Zeng, Y. Liu and Y. Zhang, “Identification of rice diseases using deep convolutional neural networks,” Neurocomputing, vol. 267, no. 1, pp. 378–384, 2017. [Google Scholar]
12. M. Dyrmann, H. Karstoft and H. S. Midtiby, “Plant species classification using deep convolutional neural network,” Biosystems Engineering, vol. 151, no. 1, pp. 72–80, 2016. [Google Scholar]
13. F. Alenezi, “Image dehazing based on pixel guided CNN with pam via graph cut,” Computers, Materials & Continua, vol. 71, no. 2, pp. 3425–3443, 2022. [Google Scholar]
14. R. Zhang, F. Zhu, J. Liu and G. Liu, “Depth-wise separable convolutions and multi-level pooling for an efficient spatial CNN-based steganalysis,” IEEE Transactions on Information Forensics and Security, vol. 15, no. 1, pp. 1138–1150, 2020. [Google Scholar]
15. D. S. Rao, R. B. Ch, V. S. Kiran, N. Rajasekhar, K. Srinivas et al., “Plant disease classification using deep bilinear CNN,” Intelligent Automation & Soft Computing, vol. 31, no. 1, pp. 161–176, 2022. [Google Scholar]
16. J. Chen, Z. Zhou, Z. Pan and C. Yang, “Instance retrieval using region of interest based CNN features,” Journal of New Media, vol. 1, no. 2, pp. 87–99, 2019. [Google Scholar]
17. W. Fang, F. H. Zhang, V. S. Sheng and Y. W. Ding, “A method for improving CNN-based image recognition using DCGAN,” Computers, Materials & Continua, vol. 57, no. 1, pp. 167–178, 2018. [Google Scholar]
18. F. Jiang, Y. Lu, Y. Chen, D. Cai and G. Li, “Image recognition of four rice leaf diseases based on deep learning and support vector machine,” Computers and Electronics in Agriculture, vol. 179, no. 2, pp. 105824, 2020. [Google Scholar]
19. C. R. Rahman, P. S. Arko, M. E. Ali, M. A. I. Khan, S. H. Apon et al., “Identification and recognition of rice diseases and pests using convolutional neural networks,” Biosystems Engineering, vol. 194, no. 1, pp. 112–120, 2020. [Google Scholar]
20. J. P. Shah, H. B. Prajapati and V. K. Dabhi, “A survey on detection and classification of rice plant diseases,” in 2016 IEEE Int. Conf. on Current Trends in Advanced Computing (ICCTAC), Bangalore, India, pp. 1–8, 2016. [Google Scholar]
21. S. Phadikar and J. Sil, “Rice disease identification using pattern recognition techniques,” in 2008 11th Int. Conf. on Computer and Information Technology, Khulna, Bangladesh, pp. 420–423, 2016. [Google Scholar]
22. V. K. Shrivastava, M. K. Pradhan, S. Minz and M. P. Thakur, “Rice plant disease classification using transfer learning of deep convolution neural network,” International Archives of the Photogrammetry, Remote Sensing & Spatial Information Sciences, vol. 3, no. 6, pp. 631–635, 2019. [Google Scholar]
23. S. Phadikar, J. Sil and A. K. Das, “Classification of rice leaf diseases based on morphological changes,” International Journal of Information and Electronics Engineering, vol. 2, no. 3, pp. 460–463, 2012. [Google Scholar]
24. S. Phadikar, J. Sil and A. K. Das, “Rice diseases classification using feature selection and rule generation techniques,” Computers and Electronics in Agriculture, vol. 90, no. 2, pp. 76–85, 2013. [Google Scholar]
25. K. Bashir, M. Rehman and M. Bari, “Detection and classification of rice diseases: An automated approach using textural features,” Mehran University Research Journal of Engineering & Technology, vol. 38, no. 1, pp. 239–250, 2019. [Google Scholar]
26. M. J. Hasan, S. Mahbub, M. S. Alom and M. A. Nasim, “Rice disease identification and classification by integrating support vector machine with deep convolutional neural network,” in 2019 1st Int. Conf. on Advances in Science, Engineering and Robotics Technology (ICASERT), Dhaka, Bangladesh, pp. 1–6, 2019. [Google Scholar]
27. Y. Lu, S. Yi, N. Zeng, Y. Liu and Y. Zhang, “Identification of rice diseases using deep convolutional neural networks,” Neurocomputing, vol. 267, no. 1, pp. 378–384, 2017. [Google Scholar]
28. K. Kiratiratanapruk, P. Temniranrat, A. Kitvimonrat, W. Sinthupinyo and S. Patarapuwadol, “Using deep learning techniques to detect rice diseases from images of rice fields,” in Int. Conf. on Industrial, Engineering and Other Applications of Applied Intelligent Systems, Kitakyushu, Japan, pp. 225–237, 2020. [Google Scholar]
29. https://www.kaggle.com/minhhuy2810/rice-diseases-image-dataset. [Google Scholar]
30. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in Int. Conf. on Learning Representations, San Diego, pp. 1–14, 2015. [Google Scholar]
31. C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed et al., “Going deeper with convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, New York, NY, USA, pp. 1–9, 2015. [Google Scholar]
32. S. Ioffe and C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” in Int. Conf. on Machine Learning, Lille, France, pp. 448–456, 2015. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools