 Open Access
Open Access
ARTICLE
A Hybrid Deep Learning Approach to Classify the Plant Leaf Species
1 Department of CS&SE, Islamic International University, Islamabad, 44000, Pakistan
2 Department of IT Services, University of Okara, Okara, 56310, Pakistan
3 Faculty of Science and Arts Belqarn, University of Bisha, Sabtul Alaya, 61985, Saudi Arabia
4 Department of CS, University of Okara, Okara, 56310, Pakistan
5 Department of Computer Science & Engineering, University of Engineering & Technology Lahore, Narowal, Campus, Narowal, 51601, Pakistan
6 Electrical Engineering Department, College of Engineering, King Khalid University, Abha, 61421, Saudi Arabia
7 Research Center for Advanced Materials Sciences (RCAMS), King Khalid University, Abha, Saudi Arabia
* Corresponding Author: Mubbashar Saddique. Email:
Computers, Materials & Continua 2023, 76(3), 3897-3920. https://doi.org/10.32604/cmc.2023.040356
Received 15 March 2023; Accepted 30 May 2023; Issue published 08 October 2023
Abstract
Many plant species have a startling degree of morphological similarity, making it difficult to split and categorize them reliably. Unknown plant species can be challenging to classify and segment using deep learning. While using deep learning architectures has helped improve classification accuracy, the resulting models often need to be more flexible and require a large dataset to train. For the sake of taxonomy, this research proposes a hybrid method for categorizing guava, potato, and java plum leaves. Two new approaches are used to form the hybrid model suggested here. The guava, potato, and java plum plant species have been successfully segmented using the first model built on the MobileNetV2-UNET architecture. As a second model, we use a Plant Species Detection Stacking Ensemble Deep Learning Model (PSD-SE-DLM) to identify potatoes, java plums, and guava. The proposed models were trained using data collected in Punjab, Pakistan, consisting of images of healthy and sick leaves from guava, java plum, and potatoes. These datasets are known as PLSD and PLSSD. Accuracy levels of 99.84% and 96.38% were achieved for the suggested PSD-SE-DLM and MobileNetV2-UNET models, respectively.Keywords
Plants are essential for the survival of all life on Earth. Among other things, they provide us with oxygen to breathe, food, and medicine. Every living thing depends on them [1]. Plants are a vital part of our planet’s ecosystem, and there are around 391,000 vascular plant species worldwide [2]. Detecting and eliminating weeds, for example, necessitates accurate identification of plant species utilizing automated methods that rely on human expertise [3].
It is impossible to examine the planet’s plant species and discover certain features that allow botanists to distinguish between them [4]. Hand-species identification can be time intensive and error-prone even when performed by experts in a given plant taxon, making it difficult to scale up to high-throughput needs.
A biologist requires assistance classifying plant species that look similar but differ from one another. When botanists use manual identification methods, they look to the plants’ well-described characteristics as the basis for making an identification. It will be asked to provide information about the plant at each identification stage. A polytomous or dichotomous key is used in the next stage of the identification process [5]. Let us pretend the basic design could use some work. When the botanical sample lacks the necessary characters or technical information is not readily available, choosing the correct answer can be challenging. They devote much effort to learning about and identifying the features of various plant species [6]. Characteristics such as the venation pattern, leaf shape, and texture can help determine the identity of a mysterious plant. When these characteristics are investigated in depth, they point to the desired species. The taxonomic expertise needed to identify a plant species in its natural habitat is outside the scope of most people’s experience or education. Because of this, both amateur and expert taxonomists find it challenging to use conventional methods for identifying plant species. Knowing the species can be time-consuming, even for the most seasoned botanists. Botanical identification and categorization should be computerized or otherwise mechanized.
Leaf shape is frequently used in automated plant classification systems. Improves in computer vision have made it practical to employ AI for id purposes [7]. Artificial intelligence (AI) techniques of the present day, such as deep learning [5], lay a firm groundwork for supervised learning. In recent years, Deep Learning (DL) has been widely applied in agriculture [8–10] to improve crop yields through adaptation, control, and maintenance. Specific complex problems, including pattern recognition, identifying plant leaves, recognizing emotions in a person’s face, medical diagnosis, and smart cities can be made more apparent with deep learning [11–15]. To identify illnesses in grape plants, Ji et al. [16] suggested a Convolutional Neural Network (CNN) based disease detection method employing the PlantVillage Dataset. This approach’s validation and testing accuracies were 99.17% and 98.57%, respectively. Similar to how Jiang et al. [17] proposed a real-time deep learning-based technique using CNNs to classify apple leaf diseases on the Apple Leaf Disease Dataset (ALDD). Lasseck [18] researched classifying plant species using an ensemble deep-learning network. They used the LifCLEF2017 dataset, which contains over 10,000 photos from various plant species, and reached an accuracy of 92.6%. Using deep learning, Barré et al. [5] proposed a model called LeafNet to identify plant species. Scientists utilized LeafSnap to take pictures of 184 different plant species, Foliage to take photos of 60 other plant species, and the Flavia dataset to take photographs of 32 different plant species. On the LeafSnap dataset, the proposed technique scored 86.3% top-1 and 97.8% top-5 accuracy; on the Foliage species dataset, it achieved 95.8% top-1 and 99.6% top-5 accuracy; and on the Flavia dataset, it reached 97.9% top-1 and 99.9% accuracy. To identify the diverse plant species, the group of researchers led by Wei Tan et al. [19] created a D-Leaf deep learning model. They developed their dataset of 30 different plant species, and they looked into both the pre-trained and fine-tuned AlexNet models. The proposed D-Leaf model had an accuracy of 94.88%, whereas the pre-trained AlexNet model had an accuracy of 93.26% and the fine-tuned AlexNet model had an accuracy of 95.54%. Kaur et al. [20] used Support Vector Machine (SVM) to develop a model for identifying plant species. Tiwari [21] analyzed the efficacy of Deep Neural Networks (DNNs) and CNN for plantspecies classification. They compiled data on forty different types of plants. With 15 plant species from Sweden as their basis, the researchers performed a 93.26% accuracy rate. DNN was found to have achieved 91.17% accuracy, whereas CNN reached 95.58%.
Many problems exist in the literature concerning plant leaf species identification and segmentation. The first problem in the literature was that the existing studies used only the healthy plant leaf species for the classification and segmentation; they did not classify and segment the diseased plant leaf species. Therefore, there was a need to develop a system that could identify or segment the healthy plant leaf species and the diseased leaves. The second problem was that no single study in the literature could identify or segment the guava, java plum, and potato leaf species. In literature, most work is done on indoor plant leaf species or small plant segmentation. Another significant gap in the literature was the non-availability of guava, java plum, and potato leaf species datasets for classification and segmentation purposes. There was a dire need to develop the datasets of classification and segmentation of potato, guava, and java plum leaf species. These datasets can be helpful for future research on the plant leaf species classification and segmentation of healthy and diseased leaves. The last and important problem in the literature was that the existing methods’ efficiency needed to improve accuracy. This study was conducted to address the problems mentioned above.
For this purpose, the Plant Species Dataset (PLSD) and Plant Species Segmentation Dataset (PLSSD) were developed. The Plant Species Dataset (PLSD), which included healthy and diseased leaves of guava, potato, and java plum for classification, and PLSSD generated for semantic segmentation of the above classes, were used to tackle the above problems. A hybrid deep-learning model was developed for the guava, java plum, and potato plant leaf species. A unique segmentation model based on MobileNetV2-UNET was developed in the first stage to segment the plant leaf species. We created the Plant Species Detection Stacking Ensemble Deep Learning Model (PSD-SE-DLM) to categorize plant leaf species further to analyze data from guava, potato, and java plum plants. These findings are the main contributions of the current study:
1. A Plant Leaf Segmentation based on the MobileNetV2-UNET-based technique is developed to segment the guava, java plum, and potato healthy and diseased leaves. The MobileNetV2 is used as an encoder and UNET as a decoder.
2. A Plant Species detection using Stacking Ensemble Deep Learning Model (PSD-SE-DLM) is developed based on MobileNetV2, InceptionV3, and ResNet50 models to classify the guava, java plum, and potato leaves. The proposed model is trained and tested on the healthy and infected plant leaf species to ensure the diversity of the model.
3. Two first-ever plant species datasets are developed to segment and classify the guava, java plum, and potato plant leaf species from the central Punjab region of Pakistan.
Regarding computer vision, investigations of plant categorization using image processing have recently emerged as a hot area. The literature contains many datasets to test various plant categorization methods. These datasets include ICL [22], MalayaKew [23], PlantCLEF [24], LeafSnap [25], Swedish [26], Foliage [27], and Flavia [28]. A wide range of difficulties, including fine-grained complexity, imbalanced distribution, substantial intraclass variability, modest interclass variability, and noisy pictures, are well-illustrated by these datasets, which reflect the issue area.
2.1 Flowers Species Identification
The published literature on plant species recognition from leaf photos included several works utilizing deep learning techniques. Nguyen et al. [29] investigated GoogLeNet [30], CaffeNet [31], and AlexNet [32] deep learning models to detect different flower species. The researchers used the PlantCLEF2015 dataset of 967 flower species. The proposed method obtained 66.6% accuracy on GoogLeNet, 54.84% on CaffeNet, and 50.60% on AlexNet. Gogul et al. [33] also investigated using InceptionV3, OverFeat, and Xception deep learning methods to distinguish the multiple flower species. The researchers used the Flowers28 dataset of 28 different species of flowers. The OverFeat model achieved 85.71% accuracy, InceptionV3 got 92.41%, and Xception obtained 90.18% accuracy. Xiao et al. [34] investigated InceptionV3 and ResNet50 deep learning models to detect the 1000 plant species. This study utilized both the PlantCLEF and Oxford Flower datasets. For comparison, the InceptionV3 model was 69.5% accurate on the PlantCLEF dataset and 92.8% correct on the Oxford Flower dataset. PlantCLEF achieved 68%, and the Oxford Flower dataset obtained 92.4% accuracy with the ResNet50 deep learning model.
2.2 Grape Leaf Species Classification
Research conducted by Pereira et al. [35] investigated the AlexNet model based on a convolutional neural network to classify the different varieties of grapes. The researchers used two datasets for the research. The first dataset was developed by merging the DRVG and DRGV2018 datasets and Flavia Leaf as the second dataset. The merger dataset DRVG obtained 77.30% accuracy and 89.75% on the Flavia Leaf dataset.
2.3 Different Plant Leaf Species Classification
Mahmudul Hassan et al. [36] proposed a CNN to identify the different plant species. The researchers used Flavia, MK, and LeafSnap datasets. The MK consisted of MK-D1 and MK-D2. The proposed method achieved 99.15% accuracy on MK-D1 and 99.43% on the MK-D2 dataset. The Flavia dataset gained 99.75% accuracy, while the LeafSnap dataset obtained 89.17% accuracy. Erkan et al. [3] proposed an ODC model based on the artificial bee colony method to detect the different plant species and handwritten digit classification. The researchers used the Folio Leaf image dataset for plant species identification and the MNIST digit image dataset for handwritten digit classification. The bilateral CNN was studied by Pearline et al. [37] using machine learning classifiers. DenseNet-121, MobileNet, and Xception deep learning models extracted the features. Regarding accuracy, the suggested technique attained 98.2% on the Folio Leaf Image dataset and 99.21% on the MNIST dataset. Then different machine learning classifiers such as Support Vector Machine (SVM), Multi-Layer Perceptron (MLP), Behavioral Cloning (BC), Random Forest (RF) classifier, Classification and Regression Tree (CART), K-Nearest Neighbors (KNN), Naïve Bayes (NB), Multiple Linear Regression (MLR), and Latent Dirichlet Allocation (LDA) [38–45] were used. It was observed that the MLR classifier performed better compared to other classifiers. MLR attained 98.71% accuracy on Flavia, 96.38% on Folio, 99.14% on Swedish Leaf, and obtained 99.39% accuracy on a self-created dataset.
2.4 Multiple Plant Leaf Species Segmentation
Leaf segmentation with a complex background was also a challenging and complicated task. Few researchers worked on different plant leaves segmentation, such as Wang et al. [46], which combines pre-segmentation and morphological operations. After leafstalk removal, seven Hu geometric moments and sixteen Zernike moments are retrieved as shape characteristics from segmented binary pictures. A self-collected dataset of 1200 images were used, with 20 classes including 60 leaf samples for each class with complex background. They segmented the willow, plum, London plane tree, China redbud, chestnut, laurel, rose bush, hazel, panicled goldrain tree, sweetgum, honeysuckle, donglas fir, maple, arrowwood, tulip tree, ginkgo, photinia, camphor tree, seating, and Chinese allspice and got 92.6% accuracy. Research conducted by Itakura et al. [47] in which automatic leaf segmentation and parameters of plant structure were retrieved using point cloud 3D images. The leaves were segmented automatically with 3D models combined with 2D and 3D point-cloud processing techniques. The researchers used small plants such as Japanese sacandra, umbellate, kangaroo vine, council tree, dwarf schefflera, hydrangea, and pothos. The proposed 3D model achieved 86.9% accuracy. Research conducted by Yang et al. [48] in which 15 species segmentation was performed using Mask RCNN deep learning model used VGG16 deep learning model. They used a dataset of fifteen species and two thousand and five hundred images. The species included gardenia jasminoides, callisia fragrans, psidium littorale, Osmanthus fragrans, bixa Orellana, ficus macrocarpa, calathea makoyana, rauvolfia verticillate, Ardisia quinquegona, baccaurea ramiflora, synesepalum dulcificum, Hydnocarpus anthelminthic, daphne odora, dracaena surculose, and mussaenda pubesens. The proposed method achieved 91.5% accuracy. Nikbakhsh et al. [49] developed a new unsupervised approach based on G-mutual information for a fusion of plant leaf image segmentation. They evaluated the proposed method by generating a new leaf image dataset with natural scenes having different orientations and positions. The researchers used k-means, Self-Organizing Map (SOM), c-means, and fuzzy leaf segmentation methods to fust the results.
The literature on plant leaf species identification and segmentation needs to be improved with difficulties. The first issue with the available research is that only healthy plant leaf species were utilized for categorization and segmentation. Diseased plant leaf species were not included in these investigations. As a result, it became necessary to design a method for distinguishing between healthy and diseased leaves of the same plant species. The second issue was that the guava, java plum, and potato leaf species could not be uniquely identified or segmented by any one study in the literature. Most studies focus on the leaves of houseplants or the division of very small plants. The lack of information for guava, java plum, and potato leaf species meant that researchers had to develop their methods for classifying and segmenting these plants. Datasets were urgently required to classify and segment potato, guava, and java plum leaves. These datasets will be the potential for further studies, including the identification of plant species and the segmentation of both healthy and damaged leaves. The last major issue in the literature was that the efficiency of existing approaches needed to be enhanced to achieve desired results. This research aimed to provide solutions to the issues mentioned above.
This study suggests a hybrid deep-learning method for plant leaf species identification. A segmentation model based on MobileNetV2 and UNET is first used to segment the leaves of guava, java plum, and potato. Then the plant species detection stacking ensemble deep learning model (PSD-SE-DLM) is used to classify the species of those leaves. In total, there are two stages to the proposed method. Fig. 1 is a flowchart depicting the suggested procedure.
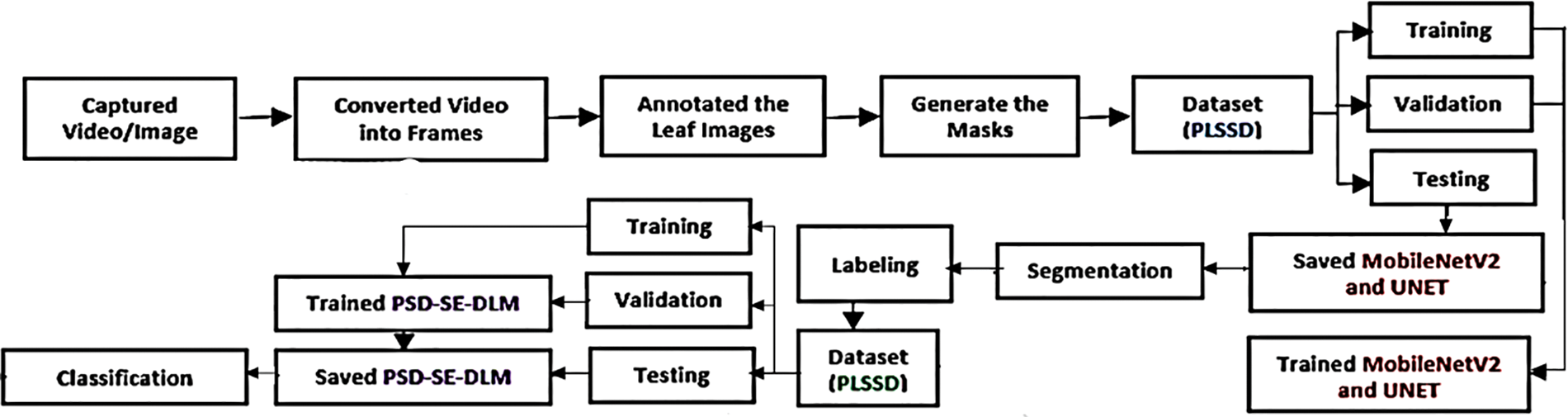
Figure 1: Flowchart of the proposed hybrid deep learning model
We created two distinct datasets using PLSSD for MobileNetV2-UNET training and Plant Species Dataset (PLSD) for PSD-SE-DLM training. A hybrid approach was trained and tested on two datasets during the present investigation. The datasets are described in depth in the following section:
3.1.1 The Plant Species Dataset (PLSD)
Within the province of Punjab in central Pakistan, a new Plant Leaf Species Dataset (PLSD) has been compiled. As seen in Fig. 2, the PLSD data set contained uninfected and infected leaf samples of guava, potato, and java plum. Our real-time dataset was compiled using video and still photos. Variations in the real-time dataset were made using various capture devices, including smartphones, digital cameras, and drones. Cell phones and digital cameras had a capture distance of 1 to 2 feet, but the drone was 5 to 10 feet. The fluttering of plant leaves distorted videos and stills captured by a fanning drone; to counteract this, we kept the plants safe from the drone. Okara is a district in Central Punjab, Pakistan, chosen for its abundance of potato, guava, and java plum farms. We zero down on the Okara district’s Coroda, Mozika, and Sante potato kinds. From November 2020 to January 2021, various adapted potato cultivars were planted on the farmland and given ample exposure to the sun. Leaves from Choti Surahi, Bari Surahi, Gola, Golden, and Sadabahar guava and java plum plants were selected. Images and videos were taken at different times of day, in varied lighting settings, during different seasons (summer, winter, spring, and fall for guava and java plum), and under diverse weather circumstances to track how these factors affected disease prevalence and severity. The dataset was diverse, using different image-capturing devices at various resolutions. Plant pathologists classified the pictures into guava, potato, and guava leaf categories. The PLSD dataset includes 5,680 photos of healthy and diseased leaves from guava, potato, and java plum. Table 1 shows that the plant leaf species dataset included 1900 photos of guava leaves, 1900 images of potato leaves, and 1880 images of java plum leaves. The PLSD dataset can be accessed from https://drive.google.com/drive/folders/1_Q5yuT0vwWR-301-SGgChiwNNrO4O_nI?usp=sharing, accessed on 31 January 2023.
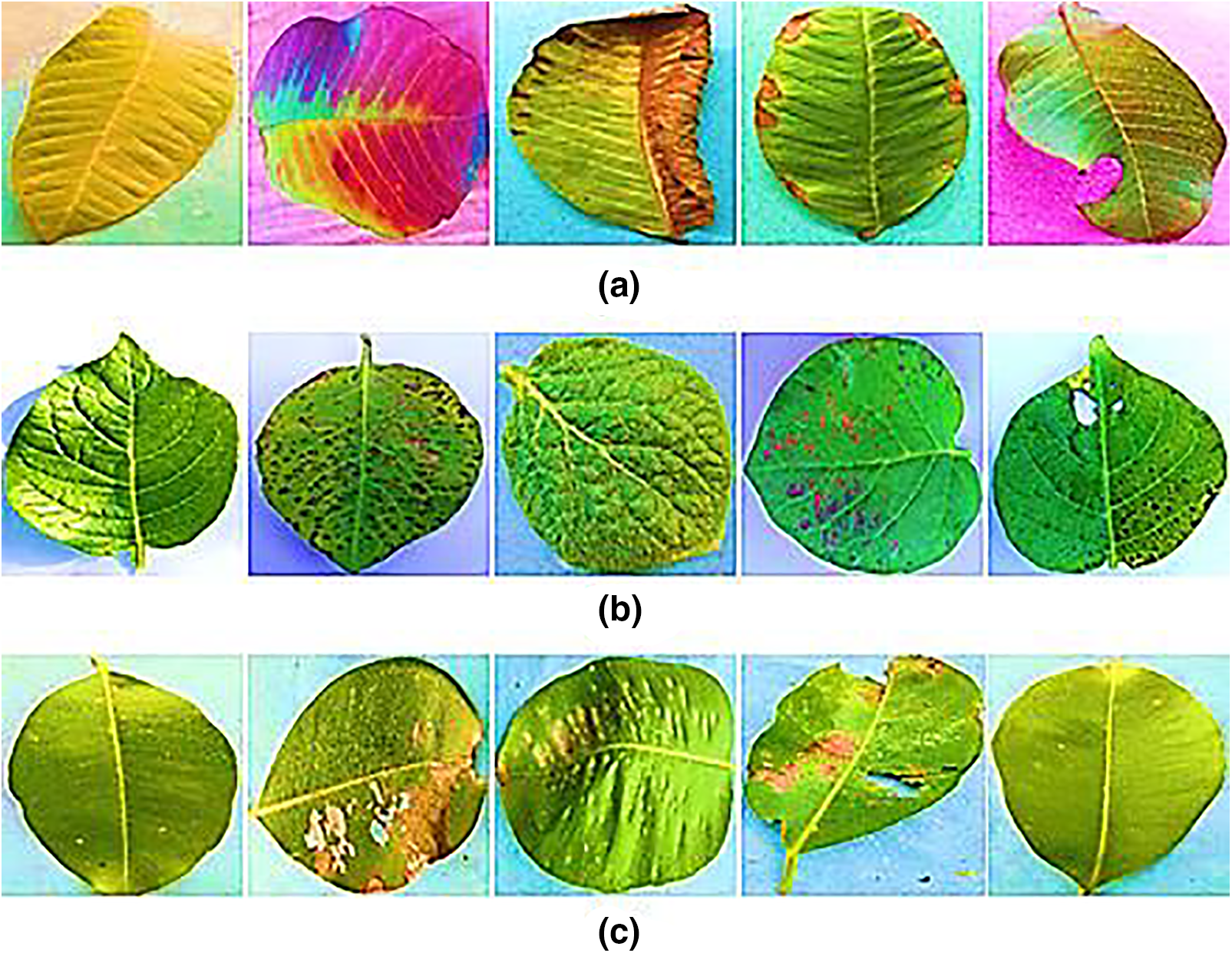
Figure 2: (a) Guava leaf (b) potato leaf (c) java plum leaf species
3.1.2 The Plant Leaf Species Segmentation Dataset (PLSSD)
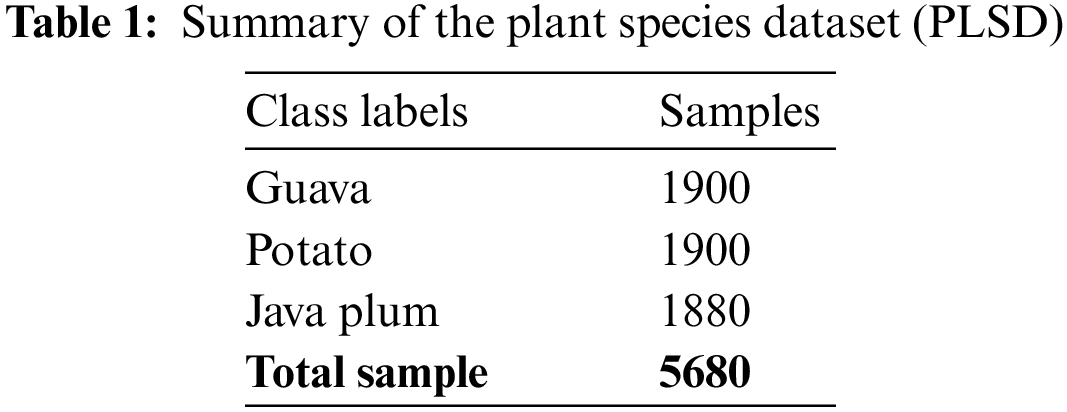
As shown in Fig. 2, we used the Plant Leaf Species Dataset (PLSD) to select healthy and diseased guava, java plum, and potato leaves. Next, label the images with the help of the LabelMe semantic app. The dataset comprised a wide variety of guava, java plum, and potato leaf varieties. The masks of multi-class images were built using annotations produced with Python code. The raw photos and masks were utilized to generate the Plant Leaf Species Segmentation Dataset (PLSSD). Thirteen hundred and twenty-nine photos and an equal number of masks made up each dataset category. As demonstrated in Table 2 and illustrated in Fig. 3, the PLSSD dataset included 3987 images and masks corresponding to those images. The PLSSD dataset can be accessed from https://drive.google.com/drive/folders/17HyqqzJUVdZhfw8MgesD2mJO-b31pYb2?usp=share_link, accessed on 31 January 2023.
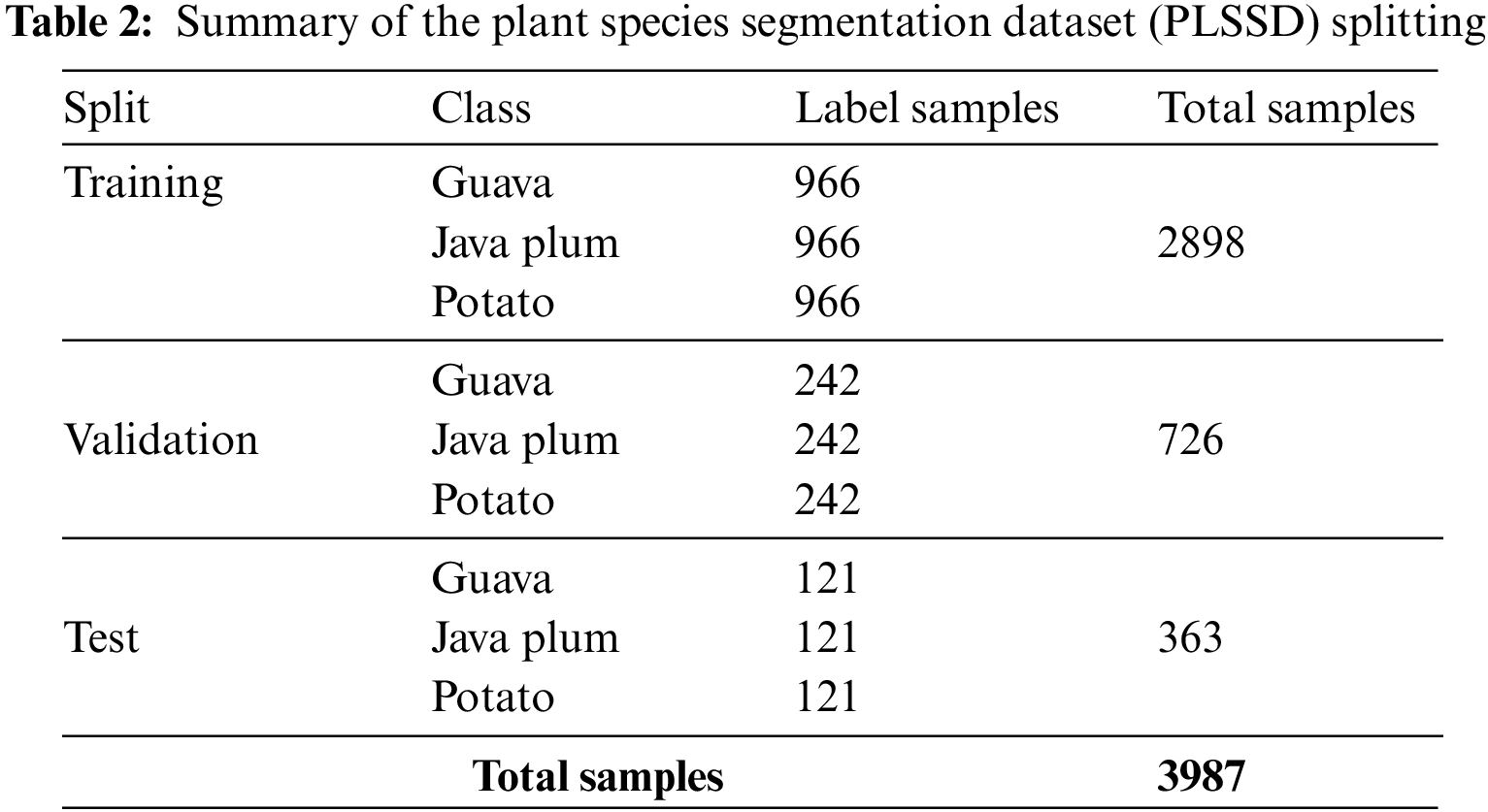
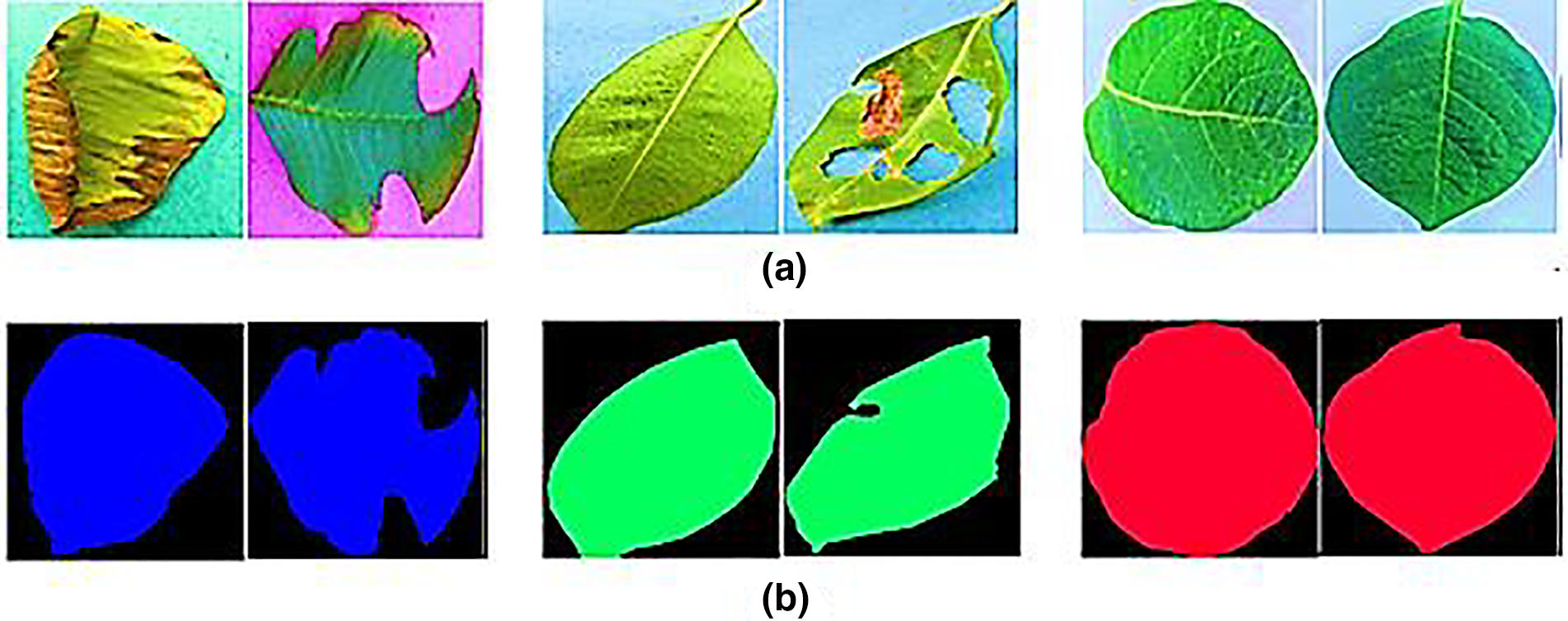
Figure 3: (a) Guava, java plum, and potato leaf images (b) guava, java plum, and potato leaf masks
Images from the PLSD and PLSSD were pre-processed to ensure more consistent classification results and improved feature extraction. Images were extracted using the python code from the plant video footage. The photos were then resized to 224 × 224 pixels using python scripts that clipped out the excess background.
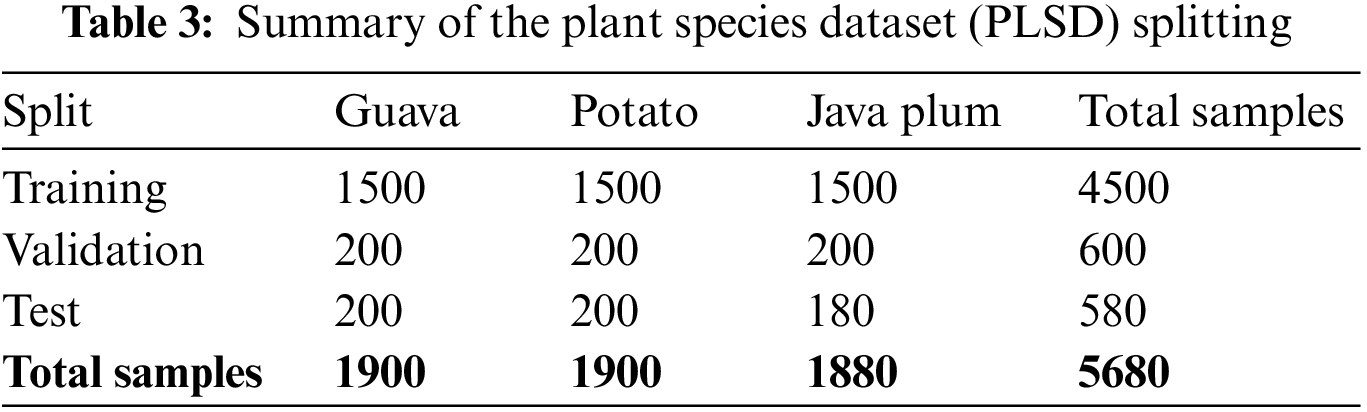
Training, validation, and testing sets were each created from the full PLSD dataset. We trained a PDDCNN model with the training dataset and then used validation and test datasets to assess the model’s efficacy. The datasets were divided into a training set of 79.23%, a validation set of 10.56%, and a testing set of 10.21%. As can be seen in Table 3, the PLSD dataset contains 4500 images, 600 of which were utilized for training and validation and 580 for testing. The current research used training photos labeled guava, potato, and java plum while keeping the same 79.23% image ratios. Regarding PLSD, the remaining 20.77% of fresh photos were divided into a validation and testing group with ratios of 10.55% and 10.21%. The proposed Plant Species Detection using Stacking Ensemble Deep Learning Model (PSD-SE-DLM) was trained on a training set to classify and predict the class label of each training image.
We split the Plant Leaf Species Segmentation Dataset (PLSSD) into three parts: training, validation, and testing. The total number of images utilized in the PLSSD training set was 2,898; 966 were assigned to each of the three classes (guava, java plum, and potato). In addition, there were 726 images in the validation set, with 242 representing each class. However, there were 363 inference samples in the testing set, with 121 examples in each category. As can be seen in Table 2, the PLSSD contained 3987 samples across all classes.
3.4.1 The MobileNetV2-UNET Segmentation Model Architecture for Plant Leaf Species
The network classifies images by assigning them a category (or label). It’s possible to know the shape of an object, which pixel corresponds to which object, and so on, but it’s not always possible. Each pixel in the image should be assigned a category in this situation. Segmentation is breaking down a problem into smaller, more manageable pieces. A segmentation model provides more information about the image than a generalized one.
The VGG-Net [50] network is the core feature extraction network in the traditional encoding-decoding UNET approach for semantic segmentation networks. However, the model is computationally intensive for embedded devices due to its complicated network topology and excessive weight parameters. Sandler et al. [51] created a convolutional neural network called MobileNetV2 specifically for mobile devices. MobileNet is the foundation upon which MobileNetV2 is built. Inverse residual structure, combined with depthwise separable convolution, dramatically reduces the loss of low-dimensional spatial information while simultaneously reducing the network parameters, allowing the overall network to run faster and meet the real-time requirements of the embedded platform. By fusing the MobileNetV2 network with the UNET semantic segmentation model, we get the MobileNetV2-UNET model. The basic outline of the model is shown in Fig. 4. In the encoder portion of the model, we see that the conventional VGG convolutional network has been replaced by MobileNetV2’s 17-layer inverted residual block, which allows us to acquire shallower features from the UNET. The decoding portion (right side of Fig. 4) is a feature recovery process in which the feature recovery layers are combined with the extracted features from the coding portion through multiple concatenate operations, as shown. It is done to reduce model parameter calculation, improve model feature extraction efficiency, and decrease information loss caused by image compression. A technique called “feature recovery” is implemented. We have also reduced the filter sizes of the proposed model, such as 16, 32, 32, 64 and 320; this will help to reduce the computational cost and inference speed, as shown in Fig. 4.
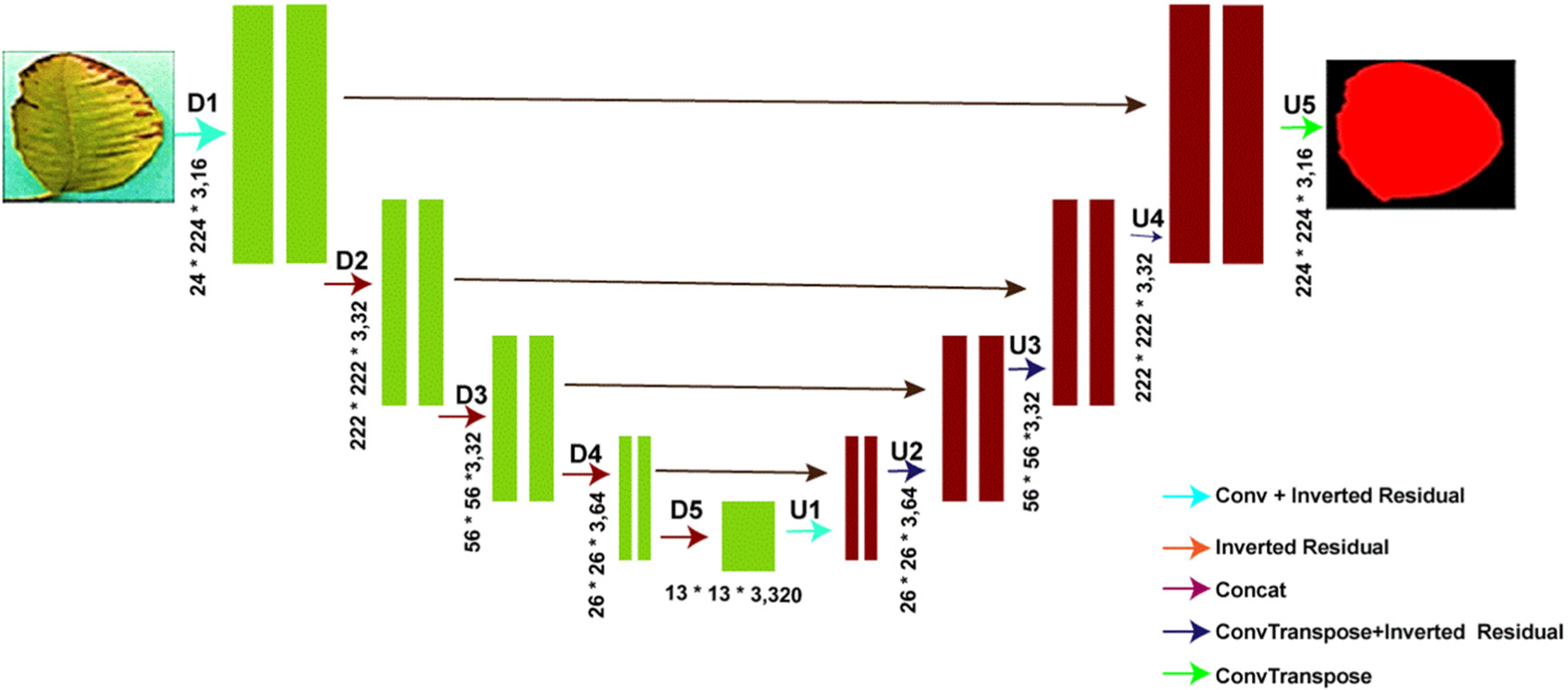
Figure 4: MobileNetV2-UNET architecture
The UNET architecture’s MobileNetV2 encoder is advantageous for semantic segmentation. MobileNetV2 is optimized for low-resource devices. Optimizations and depthwise separable convolutions minimize computational complexity and model size. Its efficient and lightweight architecture makes it suitable for real-time applications and deployment on devices with limited processing resources. MobileNetV2’s lightweight nature speeds up inference compared to heavier models. Autonomous vehicles, robots, and interactive systems benefit from real-time performance. MobileNetV2’s compact architecture uses less memory for model parameters and activations. This benefit is substantial on embedded devices and edge computing settings with limited memory. MobileNetV2, pre-trained on large-scale picture classification datasets, learns powerful and discriminative visual features. MobileNetV2, as the encoder in the U-Net architecture, allows the model to use its high-level feature representations for semantic segmentation. It improves segmentation by capturing contextual information. MobileNetV2 can transfer learning from picture categorization pretraining. U-Net models trained on smaller segmentation datasets can benefit from MobileNetV2’s generalization and feature extraction by employing it as the encoder. It can boost performance and training convergence. MobileNetV2’s design makes it versatile for diverse applications and datasets. It handles photos of different scales accurately and efficiently. MobileNetV2 does well in semantic segmentation despite being lightweight. It captures precise features and boundaries in segmented regions with reasonable accuracy. Resource efficiency, real-time processing, and accurate segmentation make it an attractive application option.
3.4.2 The Proposed Plant Species Detection Using Stacking Ensemble Deep Learning Model (PSD-SE-DLM)
The goal of ensemble learning [52] is to improve the performance of a model by consolidating the predictions generated by multiple models or to lower the likelihood of making an incorrect selection. For instance, in the gradient-boosting ensemble technique [53], model development occurs through a continuous process of reflecting on and gaining wisdom from previous errors. Suppose one of the models produces inaccurate predictions. In that case, the subsequent models will attempt to make up for it by doing relatively well on the dataset, thereby boosting the overall performance of the ensemble. The two essential characteristics that are anticipated to be possessed by a method are bias and variance [54]. The ensemble method tends to reduce both characteristics by combining individual models, resulting in a robust learner that is much more responsive and fewer data sensitive.
Bagging, boosting, and stacking are alternative strategies for merging different types of learners, as shown in Fig. 5 [55]. Instead of bagging and boosting, stacking trains the tier-2 (meta classifier) learner by simultaneously integrating the predictions from many independent models trained as base/tier-1 learners. Stacking can accomplish [55] independence amongst varied learners by simultaneously merging base models and achieving [55] dependence between learners by progressively introducing the meta-learner. As a consequence, it results in improved precision of the forecast and a reduced likelihood of overfitting. As for model combination, a meta-classifier that combines the predictions of the base learners is known as the stacking approach. Regarding image classification problems, deep stack ensemble models excel in accuracy, generalization, robustness, uncertainty estimates, flexibility in ensemble variants, scalability, and interpretability. For these reasons, deep stack ensembles are an effective method for improving the state-of-the-art image classification results. To build a good model using the stacking technique, you must select the base learner to use carefully. The foundational learner is chosen to utilize several deep learning methods, including MobileNetV2 [51], InceptionV3 [56], and ResNet50 [57]. These algorithms ensure that the classification accuracy is maximized at the same time. Our model achieves the highest accuracy possible by combining MobileNetV2, InceptionV3, and ResNet50 into a single algorithm. When it comes to combining models, the stacking approach refers to a metaclassifier that integrates the predictions made by several basic learners, as shown in Fig. 6.
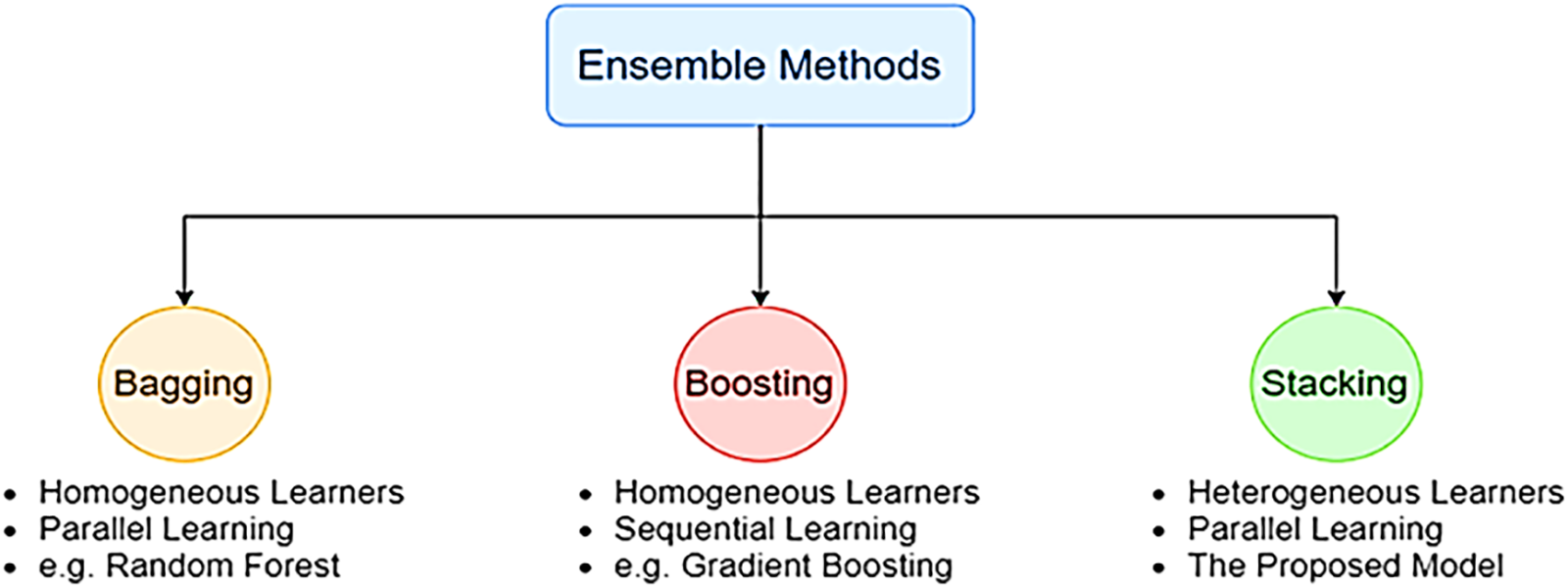
Figure 5: Ensemble learning techniques
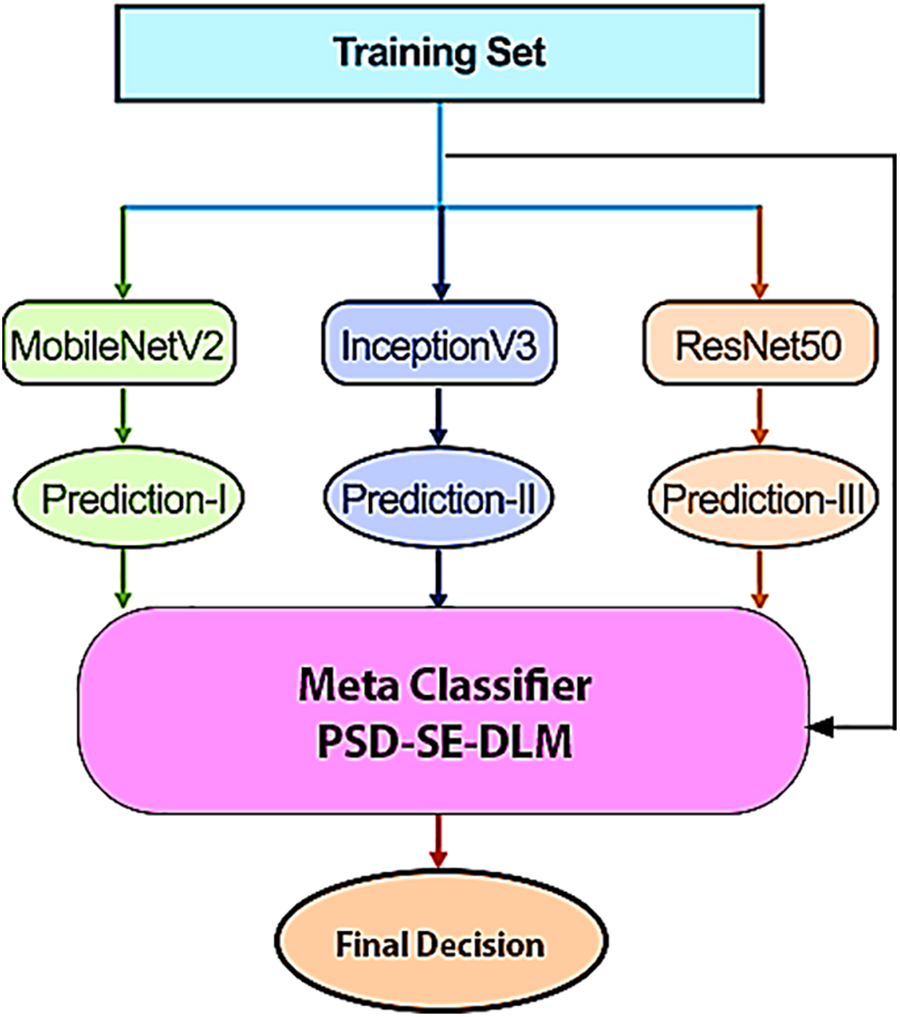
Figure 6: The proposed PSD-SE-DLM technique
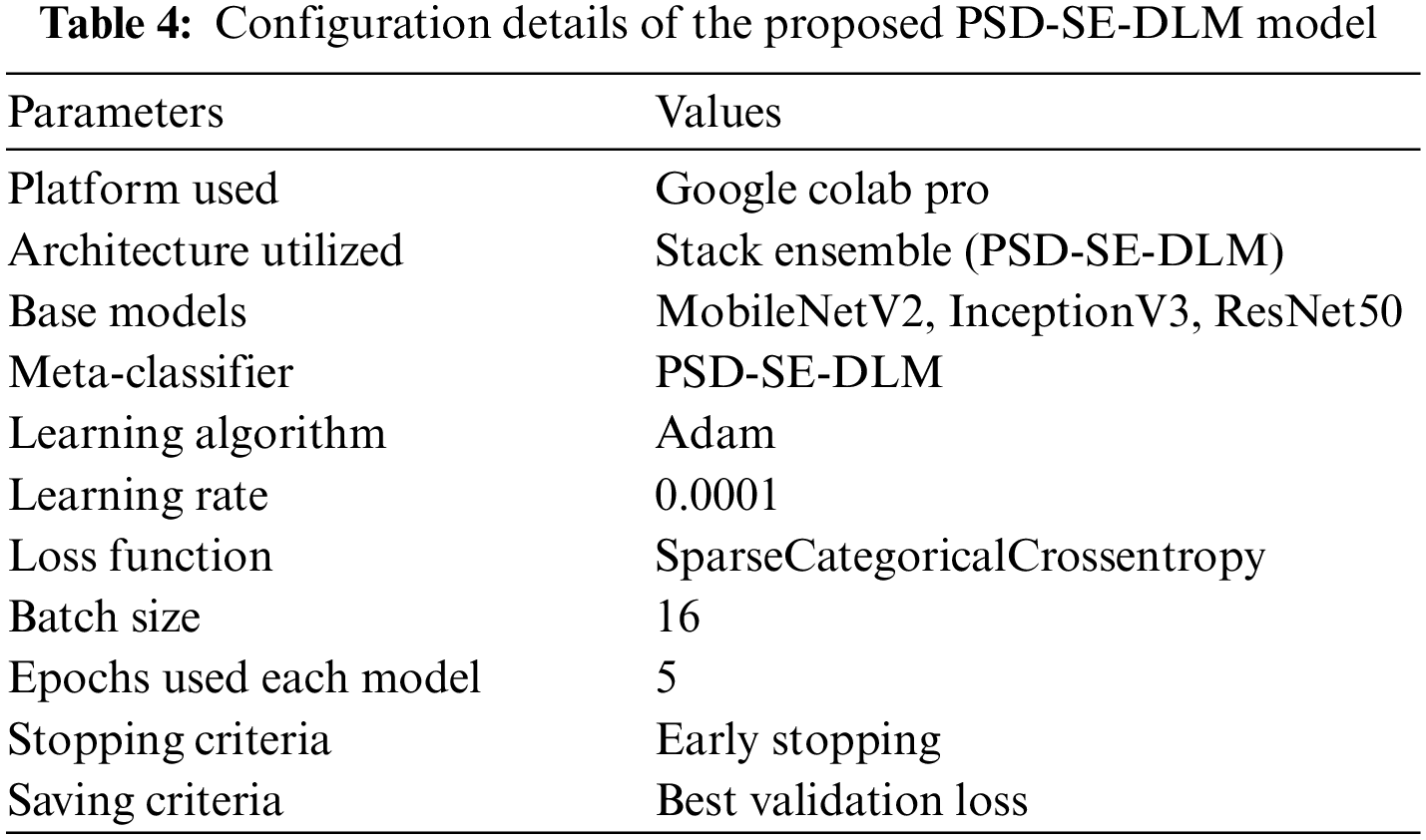
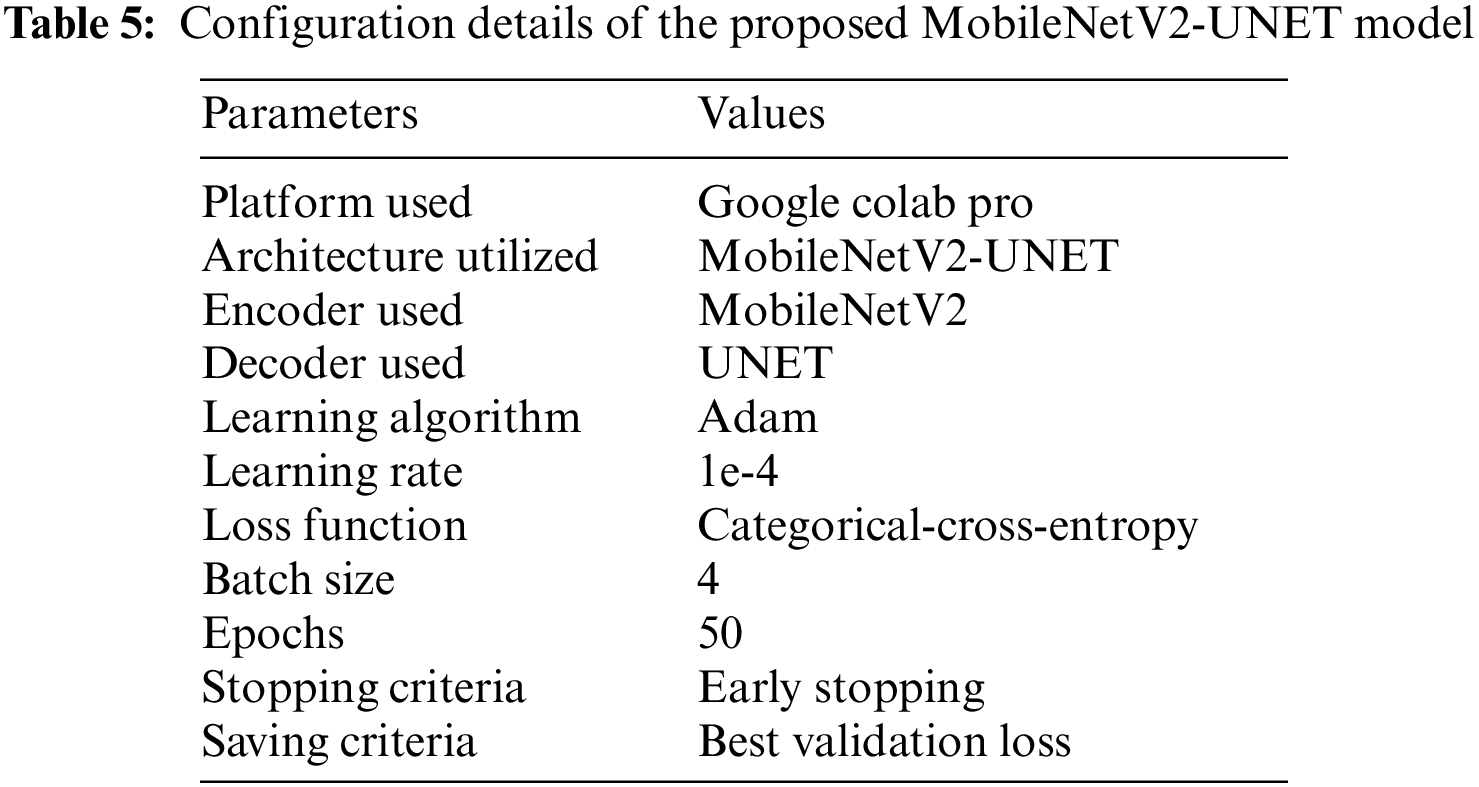
The proposed models were trained and tested using a Google Colab Pro account with powerful Graphical Processing Units (GPUs) without configuration requirements. The transfer learning deep learning models were used for training. We conducted four experiments in which three base models were used then a stacking ensemble model was developed using the output of the base models. All experiments of proposed PSD-SE-DLM used Adam optimizer with a learning rate of 0.0001, and SparseCategoricalCrossentropy loss functions were used for compiling all the models. We used 16 batch size, early stopping, saved the best val_loss model, and 5 epochs to train all the models, as shown in Table 4. The proposed MobileNetV2-UNET model used 4 batch size, 50 epochs, early stopping, and saved the best val_loss model, as depicted in Table 5.
A classifier’s accuracy is determined by dividing the total number of correct classifications by the number of proper categories overall.
When evaluating a model’s performance, it is important to remember that classification accuracy is not always an accurate indicator. Because the model is not learning anything and all samples are expected to be of the highest quality, expecting a high accuracy rate makes no sense. When the distribution of classes is unbalanced, this is one possibility. Following on the heels of this definition, precision refers to the inconsistencies found when you repeatedly measure the same object with the same equipment. One of these measurements is precision, which is defined as:
Another critical parameter is recall, the proportion of input samples from a class that the model correctly predicts. Calculating the recall is as simple as:
Precision and recall can be measured by using a statistic known as the F1 score:
Classifiers are plotted on a receiver operating characteristic curve (ROC) based on the work of their cutoff limit. An outstanding model’s cutoff threshold can be determined using the well-known ROC curve. It appears that the TPR against the FPR for various limit values.
The experimental results focused on the following:
1. The performance of the Proposed MobileNetV2-UNET was evaluated using Plant Species Segmentation Dataset (PLSSD).
2. In addition, PLSD was used to evaluate the effectiveness of a new model for plant species detection that uses a stacking ensemble of deep learning models.
3. The proposed PSD-SE-DLM model is compared with the state-of-the-art models.
4.1 The Performance Analysis of the MobileNetV2-UNET Plant Leaf Species Segmentation
Fig. 7a depicts the proposed model’s training and validation set loss and accuracy performance, as shown in the performance graph. A batch size of 04 was used, with 50 training epochs and employed an Adam optimizer. After 28 epochs, the training loss dropped from 10.30 percent to 5.91 percent. Validation losses begin at 6.45 percent and gradually drop to 5.95 percent at the end of the era. Fig. 7b shows the proposed MobileNetV2-UNET training and validation accuracy. During the last period, the training accuracy was 89.47 percent, while the validation accuracy was 96.53 percent and gradually climbed until it reached 96.74 percent at the end. Fig. 8 shows the results of the MobileNetV2-UNET segmentation model prediction on the test set. The proposed method obtained 96.38% accuracy on the test set, as shown in Table 6. The proposed segmentation method performed excellent segmentation of the complex and diseased leaves of guava, java plum, and potato. All metrics indicate that the MobileNetV2-UNET plant leaf segmentation technology performs exceptionally well.
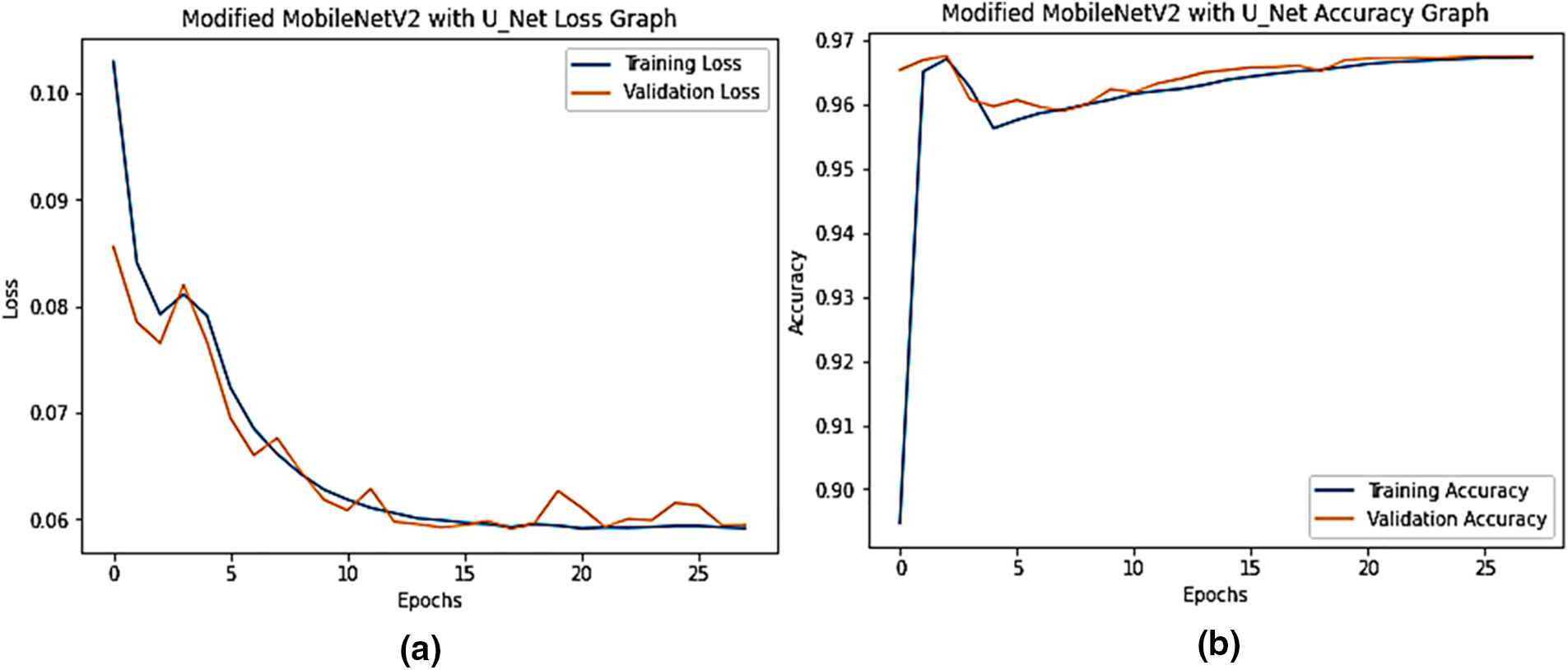
Figure 7: The proposed MobileNetV2-UNET accuracy and loss graph
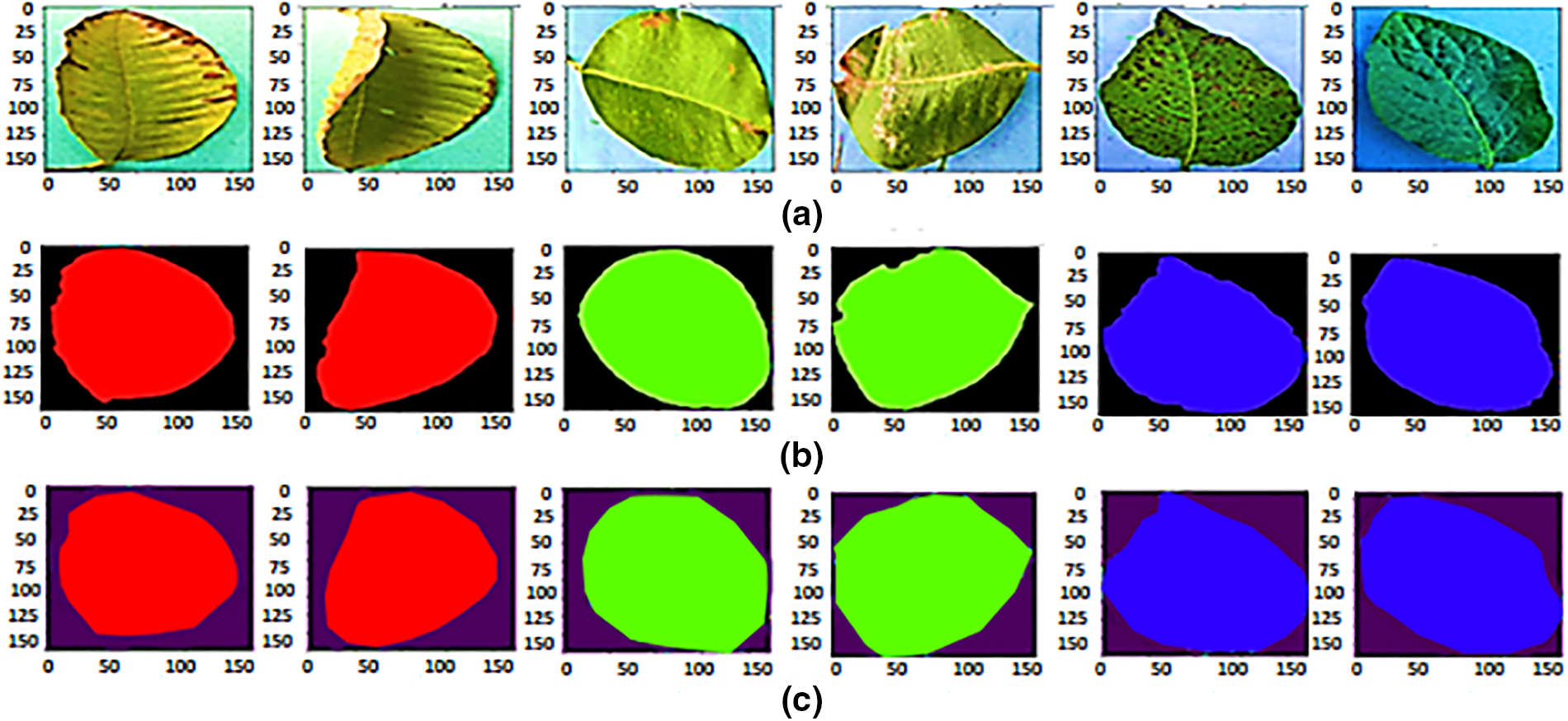
Figure 8: (a) Actual images (b) ground truth, and (c) predications of guava, java plum, and potato

The suggested MobileNetV2-UNET model accurately segmented healthy and diseased guava, potato, and java plum leaves (96.36%). As shown in Fig. 9, the suggested method successfully segmented damaged leaves from their outside boundaries but failed to appropriately segment the hole, fold over, and the damaged leaf from its interior. The suggested model performed exceptionally well when segmenting healthy and diseased leaves from potatoes, java plums, and potatoes.
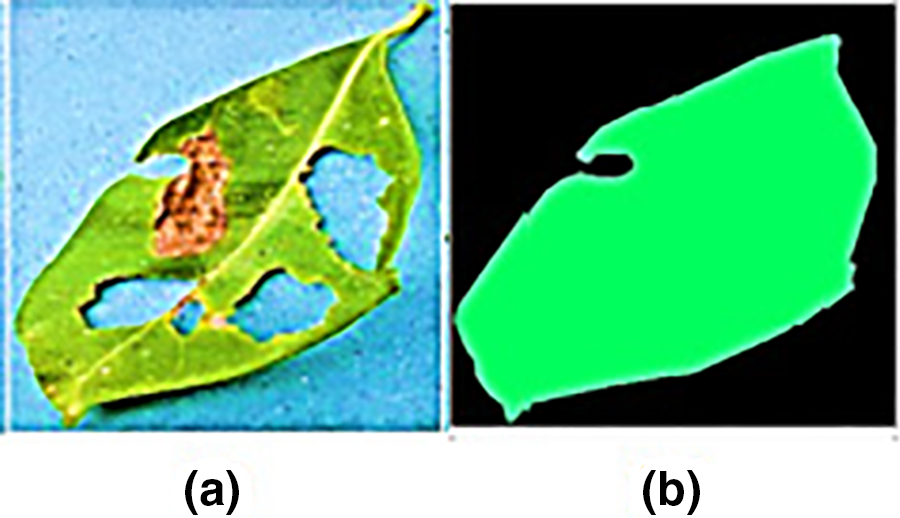
Figure 9: Proposed model prediction holes in leaf center
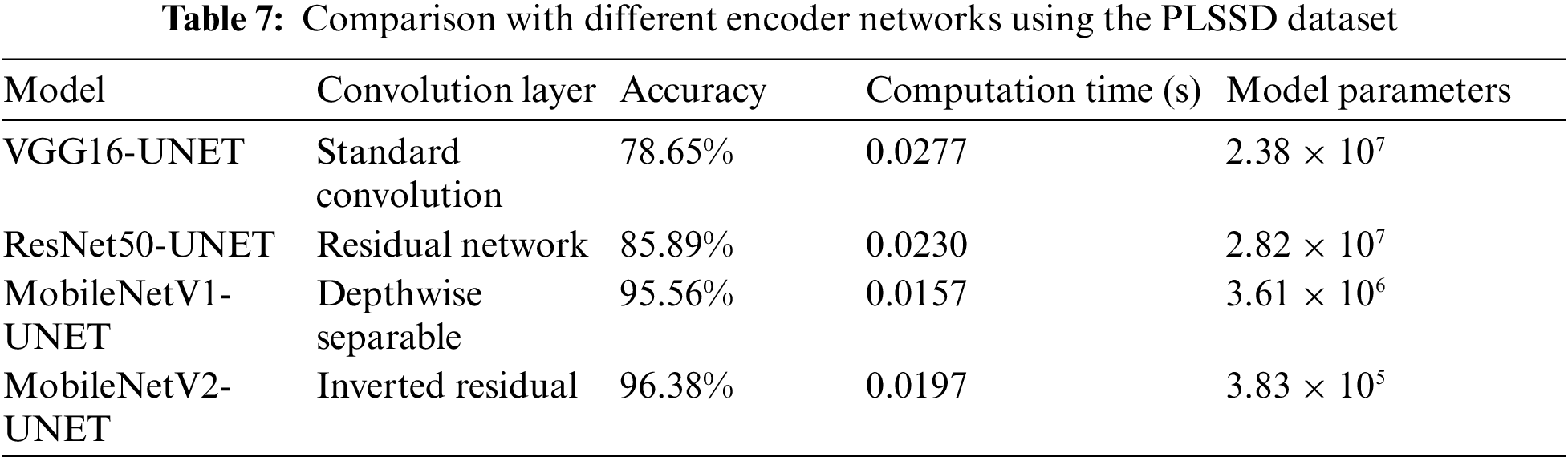
4.1.1 Comparison with Different Encoder Networks Using the PLSSD Test Set
The U-Net design evaluated various encoders, including VGG16, ResNet50, MobileNetV1, and MobileNetV2. The accuracy, calculation time, and model parameters of various encoder networks are compared in Table 7 using the PLSSD test set. The VGG16-UNET model, which comes first, uses the more common convolution layers. It takes 0.0277 s to calculate and has an accuracy of 78.65% on the PLSSD test set. The model’s relatively high complexity is reflected in its large number of parameters (2.38 × 107). ResNet50-UNET, the second model, is built on residual networks. Compared to VGG16-UNET, its accuracy is 85.89%, which is greater. The model parameters are bigger at 2.82 × 107, but the computation time is reduced at 0.0230 s. MobileNetV1-UNET, the third model, uses convolution layers that are depth-separable. On the PLSSD test set, it achieves a much greater accuracy of 95.56%. The model contains 3.61 × 106 parameters and a shorter computation time of 0.0157 s, indicating a more compact design. The convolution layers of the fourth model, MobileNetV2-UNET, are inverted residual blocks. It outperforms every other model tested by a wide margin, with an accuracy of 96.38%. While the 0.0197-s computing time is slightly longer than before, the 3.83 × 105-parameter model is much more manageable. Comparing the encoder networks shows that MobileNetV1-UNET and MobileNetV2-UNET obtain greater accuracy rates on the PLSSD test set, while VGG16-UNET and ResNet50-UNET have comparatively lower accuracies. While MobileNetV2-UNET stands out for its drastically decreased model parameters, MobileNetV1-UNET exhibits a more efficient computation time. Researchers and practitioners can use these results to select an encoder network that balances accuracy, computational economy, and model complexity for their unique jobs.
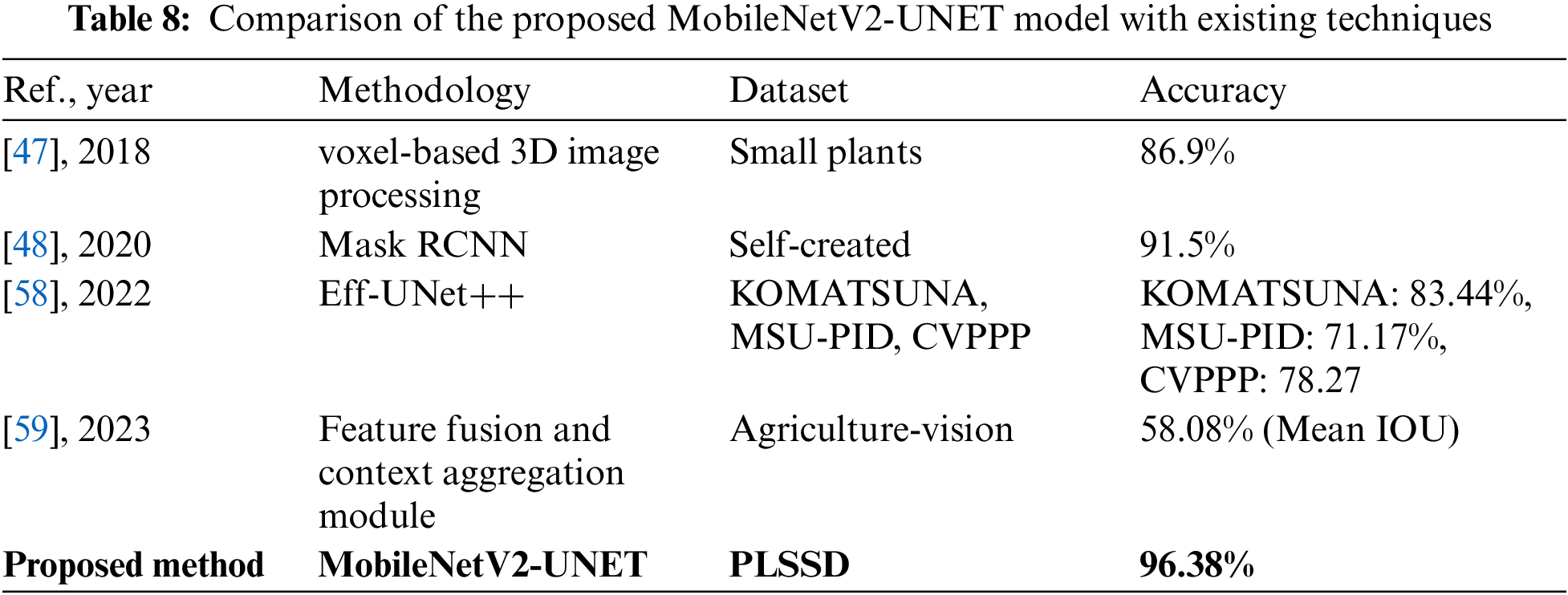
4.1.2 Comparison Analysis of the Proposed MobileNetV2-UNET Method with the Existing Techniques
Table 8 contrasts the accuracy, dataset size, and approach of existing strategies with those of the proposed MobileNetV2-UNET leaf species segmentation model. Using voxel-based 3D image processing, researchers in [47] improved accuracy to 86.9% on the Small Plants dataset. When compared to MobileNetV2-UNET, this technique took a new tack. In [48], researchers used the Mask RCNN method on a dataset they constructed and got an accuracy of 91.5%. It was successful but differed from MobileNetV2-UNET in terms of approach and dataset. The Eff-UNet++ model was first presented in [58] 2022 and tested using the KOMATSUNA, MSU-PID, and CVPPP datasets. The results showed an accuracy of 83.44% for KOMATSUNA, 71.17% for MSU-PID, and 78.27% for CVPPP. While comparable to MobileNetV2-UNET in terms of results, this method and dataset differ. The Agriculture-Vision dataset assessed the efficacy of a strategy presented in a 2023 study [59] that relied on a feature fusion and context aggregation module. Mean IOU accuracy was recorded at 58.08% using this method. Despite using novel approaches, this method needed to be more precise than MobileNetV2-UNET. Table 8 shows that the suggested MobileNetV2-UNET model achieved an amazing accuracy of 96.38% when tested on the PLSSD dataset. It reveals that MobileNetV2-UNET performs better than the existing leaf species segmentation methods.
4.2 The Performance Analysis of the Proposed Plant Species Detection Using Stacking Ensemble Deep Learning Model (PSD-SE-DLM)
We also examined the proposed Plant Species Detection performance utilizing the Stacking Ensemble Deep Learning Model (PSD-SE-DLM). As shown in Fig. 10, the training accuracy began at 94.97% after the first epoch and achieved its maximum (100%) following the final epoch of the algorithm. During the start to final epoch, PSD-SE-DLM training loss dropped from 39.18 to 0.52, and PSD-SE-DLM validation loss dropped from 14.0 to 2.40. At the end of the last epoch, the validation accuracy dropped from 99.32% to 99.16%.
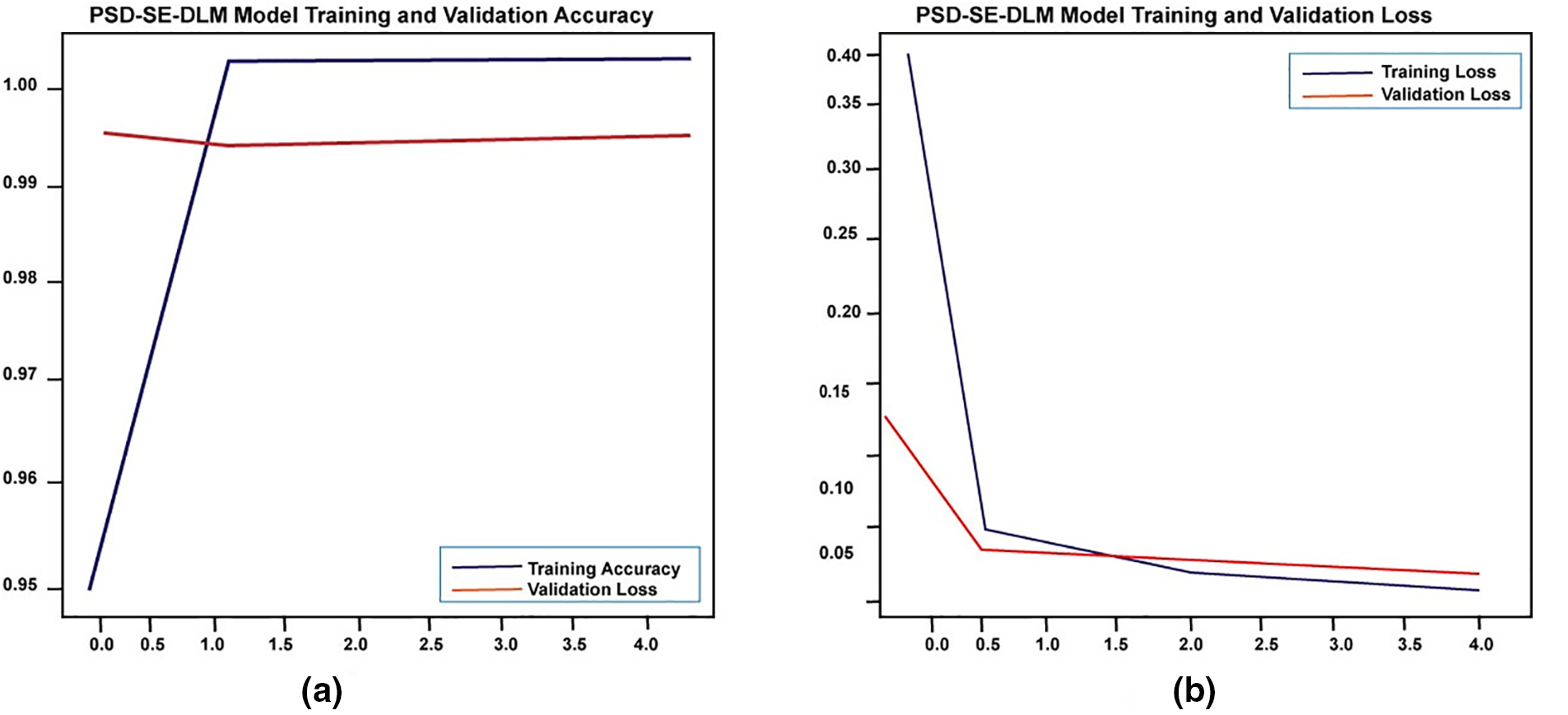
Figure 10: The proposed PSD-SE-DLM model accuracy and loss graph
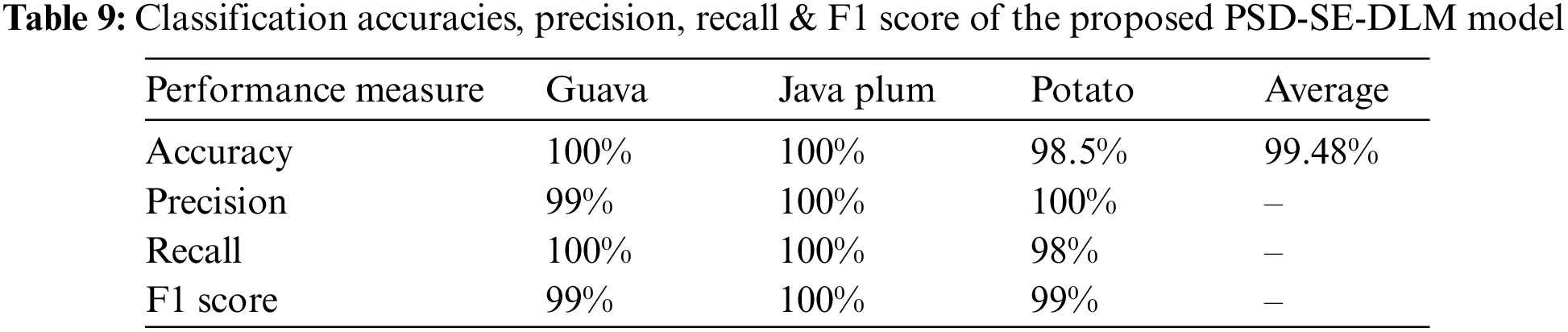
Table 9 shows the PSD-SE-DLM model’s precision, recall, F1 score, and accuracy based on the test set’s unseen data. Table 9 shows that the PSD-SE-DLM model performed exceptionally well on the test set. The guava plant leaf species got 100% recall and 99% precision and recall. The java plum classes had 100% precision, recall, and f1 scores, while the potato class had 100% precision, 98% recall, and 99% f1 scores. The proposed model received 100% accuracy in guava and java plum classes, 98.5% on potato leaf species, and 99.48% overall accuracy in all categories. We found that the proposed PSD-SE-DLM model outperformed state-of-the-art methods in every category we tested.
As shown in Fig. 11, the performance of the PSD-SE-DLM model was also validated using the confusion matrix on the test set. All test photos of guavas and java plums were successfully identified, demonstrating the efficacy of the stacking ensemble model. In comparison, 197 out of 200 photos for potato leaves were appropriately identified, while three were incorrectly labeled as guava. The confusion matrix depicted that the PSD-SE-DLM model achieved 99.48% accuracy and 0.52% misclassification accuracy. The confusion matrix of the proposed model reflected the excellent classification accuracy of all the plant species classes.
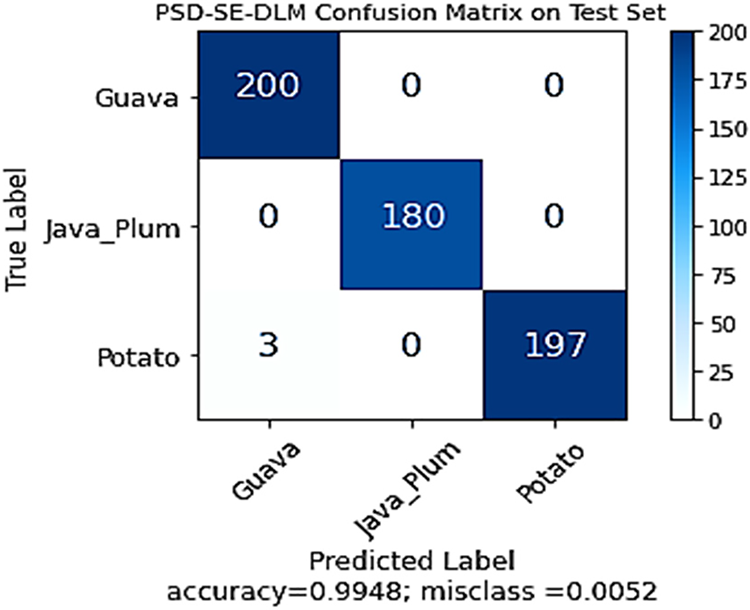
Figure 11: The proposed PSD-SE-DLM model confusion matrix on test set
As demonstrated in Fig. 12, the performance of the PSD-SE-DLM base model may also be evaluated using the ROC curve. By contrasting the true positive rate (TPR) with the false positive rate (FPR) at specific cutoffs, we may see the receiver operating characteristic (ROC). According to [60], if your classification problem has an area under the curve (AUC) of 80–90, you are doing well; if it has an AUC of 90–100, you are doing well. Classes like guava and java plum had about 100%, while the potato class got 99% area under the curve, demonstrating the superior performance of the PSD-SE-DLM model.
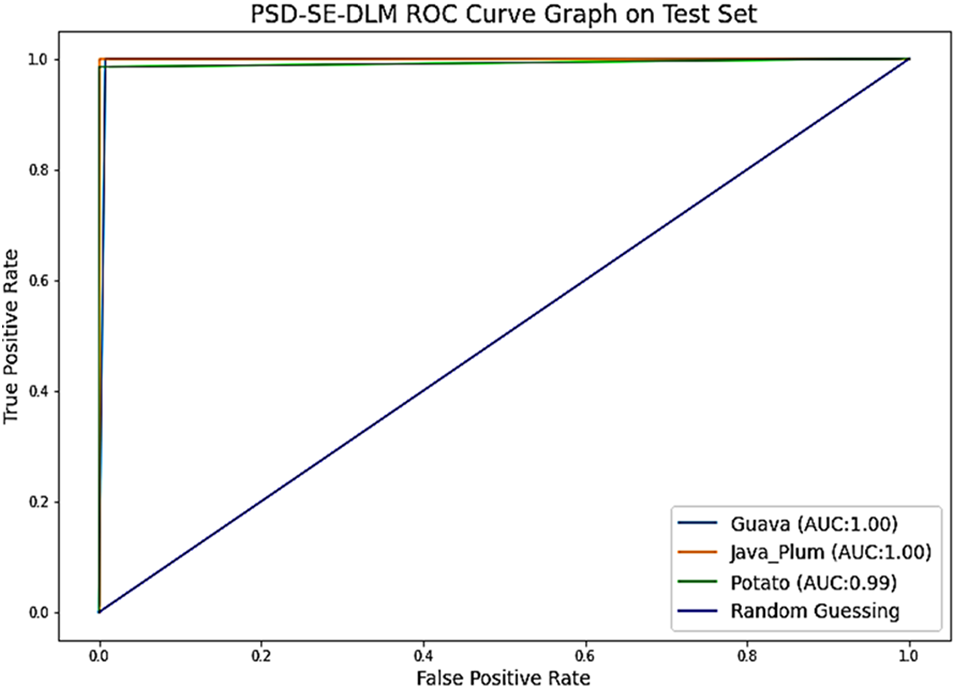
Figure 12: The proposed PSD-SE-DLM model ROC graph on test set
All the evaluation measures, such as accuracy, precision, recall, F1 score, and roc curve, depicted that the proposed PSD-SE-DLM model had performed exceptionally well in classifying test data.
4.3 Comparison Analysis of the Proposed PSD-SE-DLM Method with the Existing Techniques
No study has been found in the literature that worked on guava or potato or java plum, or a combination of three plant species. Therefore, a comparison of this research is not possible. We compare our model with the other plant leaf species. The study presented in [17] proposed an ensemble model which can detect multiple plant leaf species and got 92.60% accuracy, as illustrated in Table 10. Szededy et al. [30] got 92.8% accuracy on InceptionV3, and the ResNet50 model obtained 92.4% accuracy. Gogul and Kumar [33] studied CART, KNN, NB, MLR, and LDA classifiers and obtained 98.71% accuracy using the MLR classifier. The suggested ensemble model achieved an accuracy of 99.48% for identifying the leaves of guava, potato, and java plum plants using the stack ensemble technique. The proposed ensemble model used the stack ensemble technique for the guava, potato, and java plum leaf species and achieved 99.48% accuracy, as shown in Table 10. The results showed that the proposed PSD-SE-DLM model achieved higher accuracy than the existing studies.
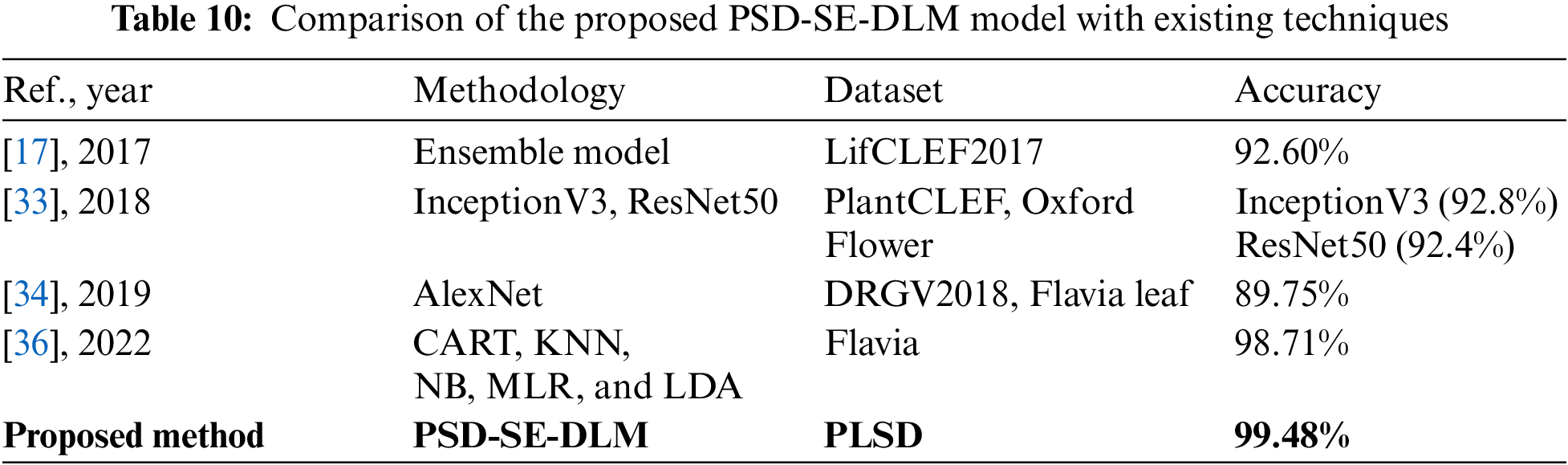
4.4 Comparison Analysis of the Proposed PSD-SE-DLM Method with the State-of-the-Art Techniques
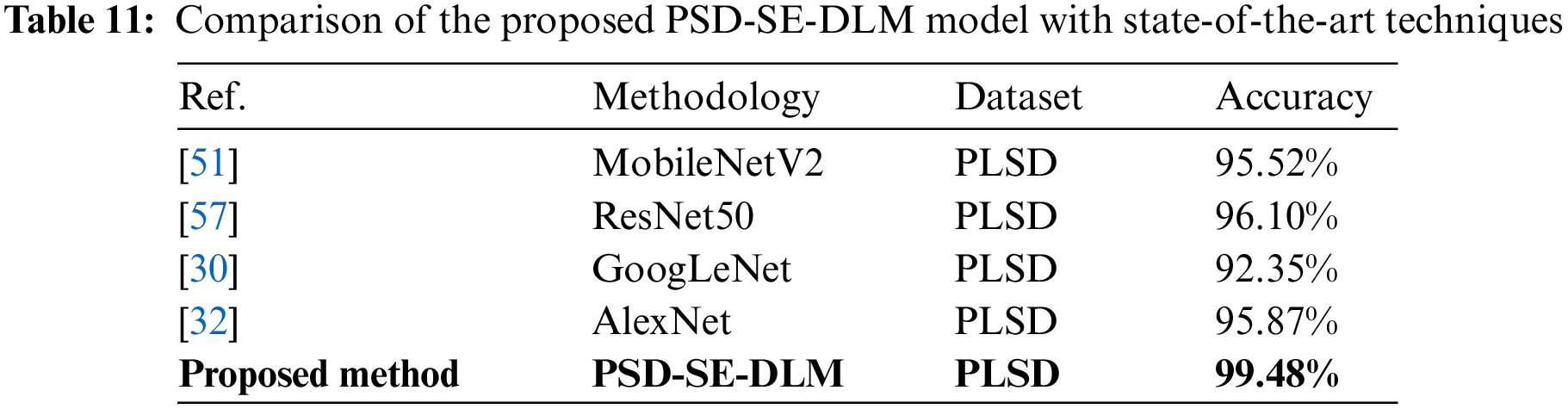
The proposed PSD-SE-DLM model was put to the test on the PLSD dataset by combining it with the MobileNetV2 [51], ResNet50 [57], GoogLeNet [30], and AlexNet [32] models using transfer learning. Thus, there was necessary to be consistent between studies concerning the environment and the methods used to enhance the data. The accuracy of modern deep learning methods is displayed in Table 11. As shown in Table 11, the accuracy of the MobileNetV2 model was 95.52%, ResNet50 was 96.10%, GoogLeNet was 92.35%, and AlexNet was 95.87%, and that of the suggested PSD-SE-DLM model was 99.48%. As shown in Table 11, the findings demonstrated that the proposed PDDCNN model achieved the best accuracy (99.75%) compared to other state-of-the-art models.
Plant leaf segmentation is a difficult task. Despite progress, this area has several limits. This study limited to images of plant leaves of guava, java plum, and potatoes. Plant canopy leaves overlap, making them hard to distinguish. Occlusion can misclassify or partially segment leaves, causing segmentation errors. Leaf forms, textures, and curves vary. The proposed method struggles with leaves with irregular edges, complicated patterns, or fine structures. Complex leaf structures may challenge the proposed. Segmentation depends on the lighting. Uneven lighting, shadows, and reflections can make leaves hard to identify from the background. The proposed method may struggle with illumination differences between photos or locales. Some leaves have colors or textures that match their surroundings. Due to this resemblance, the proposed MobileNetV2-UNET method may misclassify or merge leaves with the background, resulting in mistakes. Sensor noise, picture acquisition circumstances, and image compression can cause distortions and flaws in plant leaf photographs. Noise sources might skew the proposed segmentation findings. Accurate segmentation models require lots of labeled training data. However, collecting high-quality annotated data for plant leaf segmentation takes much work. More diversified and annotated datasets may hamper segmentation algorithm development and generalization. Leaf forms, sizes, hues, and textures vary per species. Due to leaf features, building a universal segmentation model that works across plant species takes much work. High-resolution photos and big datasets demand computationally intensive segmentation techniques. It can hinder these algorithms’ real-time or efficient deployment, especially in resource-constrained applications.
Though promising, the PSD-SE-DLM approach has limitations. The proposed leaf species identification system may need more training data. Same-species leaves vary in form, size, color, texture, and other characteristics. PSD-SE-DLM may misclassify or under-classify intra-class variability if it cannot capture it. This variability demands a robust model architecture and a diversified training dataset with many leaf variants within each species. However, various plants may have similar leaves. The approach may need help differentiating closely related species with comparable leaf features. Higher-level contextual information or multi-modal data sources may be needed to distinguish comparable species.
Identification and segmenting plant species based upon observation of potato, guava, and java plum leaves, was complex and time-consuming. One model was employed to segment the guava, potato, and java plum leaf species, while the other was utilized to identify the guava, java plum, and potato leaves. The initial model segments the diseased and healthy leaves of guava, potato, and java plum using MobileNetV2-UNET. To categorize java plum, potato, and guava plant leaf species, the second PSD-SE-DLM model was used. MobileNetV2, InceptionV3, and ResNet50 deep learning models were employed in the stacking ensemble PSDSE-DLM model. We observed the efficiency of all deep learning models on the Plant Leaf Species Segmentation Dataset (PLSSD) and the Plant Leaf Species Dataset (PLSD) datasets. Both datasets relied on video and stills captured in the guava, potato, and java plum farms. The proposed model was automated entirely compared to the current state-of-the-art leaf species detection methods. For training purposes, plant leaves were photographed and labeled manually. As an additional benefit, the proposed model possesses significance in real-time to identifying different types of plant leaves. However, a lot of work requires observing environmental conditions and growing various species across other regions. As a result of environmental changes, new species and disease appearances in diverse plant leaf species will be the central core of future research.
Acknowledgement: The authors are thankful to the Deanship of Scientific Research at University of Bisha for supporting this work through the Fast-Track Research Support Program.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University (KKU) for funding this work through the Research Group Program under the Grant Number: (R.G.P.2/382/44).
Author Contributions: All the authors contributed to the design and methodology of this study, the assessment of outcomes and the writing of the manuscript.
Availability of Data and Materials: All the relevant data are within the paper and its supporting information files.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. A. Kaya, A. S. Keceli, C. Catal, H. Y. Yalic, H. Temucin et al., “Analysis of transfer learning for deep neural network based plant classification models,” Computers and Electronics in Agriculture, vol. 158, no. 3, pp. 20–29, 2019. [Google Scholar]
2. K. Willis, “State of the World’s Plants 2017 Report,” London: Royal Botanic Gardens Kew, 2017 [Google Scholar]
3. U. Erkan, A. Toktas and D. Ustun, “Hyperparameter optimization of deep CNN classifier for plant species identification using artificial bee colony algorithm,” Journal of Ambient Intelligence and Humanized Computing, vol. 148, no. 10, pp. 1–12, 2022. [Google Scholar]
4. D. Bisen, “Deep convolutional neural network based plant species recognition through features of leaf,” Multimedia Tools and Applications, vol. 80, no. 4, pp. 6443–6456, 2021. [Google Scholar]
5. P. Barré, B. C. Stöver, K. F. Müller and V. Steinhage, “Leafnet: A computer vision system for automatic plant species identification,” Ecological Informatics, vol. 40, pp. 50–56, 2017. [Google Scholar]
6. S. Y. Arafat, M. I. Saghir, M. Ishtiaq and U. Bashir, “Comparison of techniques for leaf classification,” in 2016 Sixth Int. Conf. on Digital Information and Communication Technology and its Applications (DICTAP), Konya, Turkey, IEEE, pp. 136–141, 2016. [Google Scholar]
7. M. Hassaballah and A. I. Awad, Deep Learning in Computer Vision: Principles and Applications. Boca Raton, Florida: CRC Press, 2020. [Google Scholar]
8. S. W. Chen, S. S. Shivakumar, S. Dcunha, J. Das, E. Okon et al., “Counting apples and oranges with deep learning: A data-driven approach,” IEEE Robotics and Automation Letters, vol. 2, no. 2, pp. 781–788, 2017. [Google Scholar]
9. P. A. Dias, A. Tabb and H. Medeiros, “Multispecies fruit flower detection using a refined semantic segmentation network,” IEEE Robotics and Automation Letters, vol. 3, no. 4, pp. 3003–3010, 2018. [Google Scholar]
10. J. Rashid, I. Khan, G. Ali, S. H. Almotiri, M. A. AlGhamdi et al., “Multi-level deep learning model for potato leaf disease recognition,” Electronics, vol. 10, no. 17, pp. 2064, 2021. [Google Scholar]
11. A. Krizhevsky, I. Sutskever and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, no. 6, pp. 84–90, 2017. [Google Scholar]
12. J. Rashid, M. Ishfaq, G. Ali, M. R. Saeed, M. Hussain et al., “Skin cancer disease detection using transfer learning technique,” Applied Sciences, vol. 12, no. 11, 5714, 2022. [Google Scholar]
13. M. Sohail, G. Ali, J. Rashid, I. Ahmad, S. H. Almotiri et al., “Racial identity-aware facial expression recognition using deep convolutional neural networks,” Applied Sciences, vol. 12, no. 1, 88, 2022. [Google Scholar]
14. J. Rashid, I. Khan, G. Ali, S. U. Rehman, F. Alturise et al., “Real-time multiple guava leaf disease detection from a single leaf using hybrid deep learning technique,” Computers, Materials & Continua, vol. 74, no. 1, pp. 1235–1257, 2023. [Google Scholar]
15. H. Chu, M. R. Saeed, J. Rashid, M. T. Mehmood, I. Ahmad et al., “Deep learning method to detect the road cracks and potholes for smart cities,” Computers, Materials & Continua, vol. 75, no. 1, pp. 1863–1881, 2023. [Google Scholar]
16. M. Ji, L. Zhang and Q. Wu, “Automatic grape leaf diseases identification via unitedmodel based on multiple convolutional neural networks,” Information Processing in Agriculture, vol. 7, no. 3, pp. 418–426, 2020. [Google Scholar]
17. P. Jiang, Y. Chen, B. Liu, D. He and C. Liang, “Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks,” IEEE Access, vol. 7, pp. 59069–59080, 2019. [Google Scholar]
18. M. Lasseck, “Image-based plant species identification with deep convolutional neural networks,” CLEF (Working Notes), pp. 1–11, 2017. [Google Scholar]
19. J. W. Tan, S. W. Chang, S. Binti Abdul Kareem, H. J. Yap and K. T. Yong, “Deep learning for plant species classification using leaf vein morphometric,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 17, no. 1, pp. 82–90, 2018. [Google Scholar] [PubMed]
20. S. Kaur and P. Kaur, “Plant species identification based on plant leaf using computer vision and machine learning techniques,” Journal of Multimedia Information System, vol. 6, no. 2, pp. 49–60, 2019. [Google Scholar]
21. S. Tiwari, “A comparative study of deep learning models with handcraft features and non-handcraft features for automatic plant species identification,” International Journal of Agricultural and Environmental Information Systems (IJAEIS), vol. 11, no. 2, pp. 44–57, 2020. [Google Scholar]
22. M. Šulc and J. Matas, “Fine-grained recognition of plants from images,” Plant Methods, vol. 13, no. 1, pp. 1–14, 2017. [Google Scholar]
23. S. H. Lee, C. S. Chan, P. Wilkin and P. Remagnino, “Deep-plant: Plant identification with convolutional neural networks,” in 2015 IEEE Int. Conf. on Image Processing (ICIP), QC, Canada, IEEE, pp. 452–456, 2015. [Google Scholar]
24. A. Joly, H. Goëau, H. Glotin, C. Spampinato, P. Bonnet et al., “LifeCLEF 2015: Multimedia life species identification challenges,” in Int. Conf. of the Cross-Language Evaluation Forum for European Languages, Bologna, Italy, Springer, pp. 462–483, 2015. [Google Scholar]
25. N. Kumar, P. N. Belhumeur, A. Biswas, D. W. Jacobs, W. J. Kress et al., “Leafsnap: A computer vision system for automatic plant species identification,” in European Conf. on Computer Vision, Florence, Italy, Springer, pp. 502–516, 2012. [Google Scholar]
26. O. Söderkvist, “Computer vision classification of leaves from swedish trees,” Ph.D. dissertation, Linkoping University, Sweden, 2001. [Google Scholar]
27. A. Kadir, L. E. Nugroho, A. Susanto and P. I. Santosa, “Performance improvement of leaf identification system using principal component analysis,” International Journal of Advanced Science and Technology, vol. 44, no. 11, pp. 113–124, 2012. [Google Scholar]
28. S. G. Wu, F. S. Bao, E. Y. Xu, Y. X. Wang, Y. F. Chang et al., “A leaf recognition algorithm for plant classification using probabilistic neural network,” in 2007 IEEE Int. Symp. on Signal Processing and Information Technology, Giza, Egypt, IEEE, pp. 11–16, 2007. [Google Scholar]
29. T. T. N. Nguyen, V. Le, T. Le, V. Hai, N. Pantuwong et al., “Flower species identification using deep convolutional neural networks,” in AUN/SEED-Net Regional Conf. for Computer and Information Engineering, Bangkok, Thailand, pp. 1–6, 2016. [Google Scholar]
30. C. Szededy, W. Liu, Y. Jia, P. Sermanet, S. Reed et al., “Going deeper with convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, pp. 1–9, 2015. [Google Scholar]
31. Y. Jia, E. Shelhamer, J. Donahue, S. Karayev, J. Long et al., “Caffe: Convolutional architecture for fast feature embedding,” in Proc. of 22nd ACM Int. Conf. on Multimedia, New York, USA, pp. 675–678, 2014. [Google Scholar]
32. A. Krizhevsky, I. Sutskever and G. Hinton, “Imagenet classification with deep convolutional networks,” Communications of the ACM, vol. 60, no. 6, pp. 84–90, 2017. [Google Scholar]
33. I. Gogul and V. S. Kumar, “Flower species recognition system using convolution neural networks and transfer learning,” in 2017 Fourth Int. Conf. on Signal Processing, Communication and Networking (ICSCN), Chennai, India, IEEE, pp. 1–6, 2017. [Google Scholar]
34. Q. Xiao, G. Li, L. Xie and Q. Chen, “Real-world plant species identification based on deep convolutional neural networks and visual attention,” Ecological Informatics, vol. 48, no. 3, pp. 117–124, 2018. [Google Scholar]
35. C. S. Pereira, R. Morais and M. J. Reis, “Deep learning techniques for grape plant species identification in natural images,” Sensors, vol. 19, no. 22, 4850, 2019. [Google Scholar] [PubMed]
36. S. Mahmudul Hassan and A. Kumar Maji, “Identification of plant species using deep learning,” in Proc. of Int. Conf. on Frontiers in Computing and Systems, West Bengal, India, Springer, pp. 115–125, 2021. [Google Scholar]
37. S. A. Pearline and V. S. Kumar, “Performance analysis of real-time plant species recognition using bilateral network combined with machine learning classifier,” Ecological Informatics, vol. 67, 101492, 2022. [Google Scholar]
38. H. Taud and J. Mas, “Multilayer perceptron (MLP),” Geomatic Approaches for Modeling Land Change Scenarios, pp. 451–455, 2018. [Google Scholar]
39. F. Torabi, G. Warnell and P. Stone, “Behavioral cloning from observation,” arXiv:1805.01954, 2018. [Google Scholar]
40. J. Ali, R. Khan, N. Ahmad and I. Maqsood, “Random forests and decision trees,” International Journal of Computer Science Issues (IJCSI), vol. 9, no. 5, 272, 2012. [Google Scholar]
41. Y. Shao and R. S. Lunetta, “Comparison of support vector machine, neural network, and CART algorithms for the land-cover classification using limited training data points,” ISPRS Journal of Photogrammetry and Remote Sensing, vol. 70, no. 22, pp. 78–87, 2012. [Google Scholar]
42. Q. Kuang and L. Zhao, “A practical GPU based KNN algorithm,” in Proc. of 2009 Int. Symp. on Computer Science and Computational Technology (ISCSCI 2009), Kuala Lumpur, Malaysia, IEEE, pp. 151, 2009. [Google Scholar]
43. M. M. Saritas and A. Yasir, “Performance analysis of ANN and naive bayes classification algorithm for data classification,” International Journal of Intelligent Systems and Applications in Engineering, vol. 7, no. 2, pp. 88–91, 2019. [Google Scholar]
44. M. Ali, R. Prasad, Y. Xiang and R. Deo, “Near real-time significant wave height forecasting with hybridized multiple linear regression algorithms,” Renewable and Sustainable Energy Reviews, vol. 132, 110003, 2020. [Google Scholar]
45. J. Yang, H. Yu and W. Kunz, “An efficient LDA algorithm for face recognition,” in Proc. of the Int. Conf. on Automation, Robotics, and Computer Vision (ICARCV 2000), Singapore, IEEE, pp. 34–47, 2000. [Google Scholar]
46. X. F. Wang, D. S. Huang, J. X. Du, H. Xu and L. Heutte, “Classification of plant leaf images with complicated background,” Applied Mathematics and Computation, vol. 205, no. 2, pp. 916–926, 2008. [Google Scholar]
47. K. Itakura and F. Hosoi, “Automatic leaf segmentation for estimating leaf area and leaf inclination angle in 3D plant images,” Sensors, vol. 18, no. 10, 3576, 2018. [Google Scholar] [PubMed]
48. K. Yang, W. Zhong and F. Li, “Leaf segmentation and classification with a complicated background using deep learning,” Agronomy, vol. 10, no. 11, pp. 1721, 2020. [Google Scholar]
49. N. Nikbakhsh, Y. Baleghi and H. Agahi, “A novel approach for unsupervised image segmentation fusion of plant leaves based on g-mutual information,” Machine Vision and Applications, vol. 32, no. 1, pp. 1–12, 2021. [Google Scholar]
50. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv:1409.1556, 2014. [Google Scholar]
51. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. C. Chen, “MobileNetV2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 4510–4520, 2018. [Google Scholar]
52. H. Yang and P. A. Bath, “The use of data mining methods for the prediction of dementia: Evidence from the english longitudinal study of aging,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 2, pp. 345–353, 2019. [Google Scholar] [PubMed]
53. M. Wang, H. Wang, J. Wang, H. Liu, R. Lu et al., “A novel model for malaria prediction based on ensemble algorithms,” PLoS One, vol. 14, no. 12, e0226910, 2019. [Google Scholar] [PubMed]
54. E. L. Ray and N. G. Reich, “Prediction of infectious disease epidemics via weighted density ensembles,” PLoS Computational Biology, vol. 14, no. 2, e1005910, 2018. [Google Scholar] [PubMed]
55. T. K. Yamana, S. Kandula and J. Shaman, “Superensemble forecasts of dengue outbreaks,” Journal of the Royal Society Interface, vol. 13, no. 123, 20160410, 2016. [Google Scholar] [PubMed]
56. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna, “Rethinking the inception architecture for computer vision,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NV, USA, pp. 2818–2826, 2016. [Google Scholar]
57. K. He, X. Zhang, S. Ren and J. Sun, “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NV, USA, pp. 770–778, 2016. [Google Scholar]
58. S. Baghat, M. Kokare, V. Haswani, P. Hambarde and R. Kamble, “Eff-UNet++: A novel architecture for plant leaf segmentation and counting,” Ecological Informatics, vol. 68, 101583, 2022. [Google Scholar]
59. S. D. Khan, L. Alarabi and S. Basalamah, “Segmentation of farmlands in aerial images by deep learning framework with feature fusion and context aggregation modules,” Multimedia Tools and Applications, vol. 2, no. 5, pp. 1–20, 2023. [Google Scholar]
60. S. Safari, A. Baratloo, M. Elfil and A. Negida, “Evidence based emergency medicine; Part 5 Receiver operating curve and area under the curve,” Emergency, vol. 4, no. 2, pp. 111–113, 2016. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools