 Open Access
Open Access
ARTICLE
Multi-Scale Vision Transformer with Dynamic Multi-Loss Function for Medical Image Retrieval and Classification
Department of Computer Science, College of Computer Science, King Khalid University, Abha, 61421, Saudi Arabia
* Corresponding Author: Mohamed Ghouse. Email:
(This article belongs to the Special Issue: Emerging Trends and Applications of Deep Learning for Biomedical Signal and Image Processing)
Computers, Materials & Continua 2025, 83(2), 2221-2244. https://doi.org/10.32604/cmc.2025.061977
Received 07 December 2024; Accepted 04 March 2025; Issue published 16 April 2025
Abstract
This paper introduces a novel method for medical image retrieval and classification by integrating a multi-scale encoding mechanism with Vision Transformer (ViT) architectures and a dynamic multi-loss function. The multi-scale encoding significantly enhances the model’s ability to capture both fine-grained and global features, while the dynamic loss function adapts during training to optimize classification accuracy and retrieval performance. Our approach was evaluated on the ISIC-2018 and ChestX-ray14 datasets, yielding notable improvements. Specifically, on the ISIC-2018 dataset, our method achieves an F1-Score improvement of +4.84% compared to the standard ViT, with a precision increase of +5.46% for melanoma (MEL). On the ChestX-ray14 dataset, the method delivers an F1-Score improvement of 5.3% over the conventional ViT, with precision gains of +5.0% for pneumonia (PNEU) and +5.4% for fibrosis (FIB). Experimental results demonstrate that our approach outperforms traditional CNN-based models and existing ViT variants, particularly in retrieving relevant medical cases and enhancing diagnostic accuracy. These findings highlight the potential of the proposed method for large-scale medical image analysis, offering improved tools for clinical decision-making through superior classification and case comparison.Keywords
Medical image retrieval has become a vital tool in clinical diagnostics, enabling physicians to compare patient data with historical cases to improve diagnostic accuracy [1,2]. Traditional image retrieval techniques, particularly those based on Convolutional Neural Networks (CNNs), have demonstrated effectiveness by capturing local features and hierarchical spatial patterns [3–5]. However, these methods often struggle to capture the global context of images, which is crucial for medical imaging tasks that require a comprehensive analysis of both local and global structures [6,7].
Vision Transformers (ViTs) [6] have gained significant attention due to their self-attention mechanisms, which enhance context modeling and improve performance in tasks requiring a holistic understanding of data [7,8]. Recent studies have demonstrated the effectiveness of ViTs in medical imaging, particularly in handling complex and heterogeneous datasets [7,9]. Despite these advancements, challenges persist in efficiently integrating multi-scale information and optimizing models for small-object representation [10].
A fundamental challenge in medical image retrieval lies in managing the complexity and diversity of medical datasets, which often involve multi-label classifications [11]. Datasets such as ISIC-2018 [12,13] and ChestX-ray14 [1] pose unique difficulties due to the wide range of disease categories and the need for models to generalize across varying conditions. Fig. 1 presents sample images from these datasets, highlighting their complexity and diversity. Traditional CNN-based approaches often struggle to effectively capture both global and local features, particularly in multi-label classification settings [11,14].
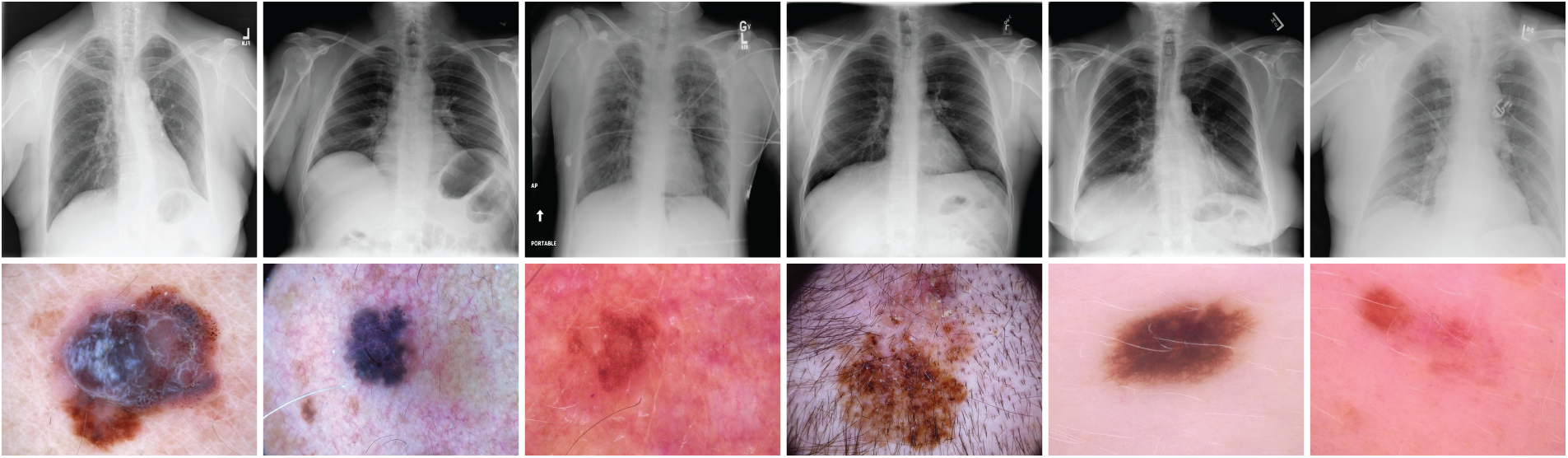
Figure 1: Sample images from the ISIC-2018 [12] and ChestX-ray14 [1] datasets. These examples illustrate the diversity of medical conditions, including skin lesions (melanoma, basal cell carcinoma) and thoracic pathologies (pneumonia, cardiomegaly), emphasizing the need for robust models capable of accurate image classification and retrieval
To address these limitations, recent research has focused on enhancing network architectures and loss functions. Multi-scale context-aware attention models [15] and improved U-Net variants [16,17] have been proposed to enhance feature extraction in medical images. Furthermore, efficient segmentation techniques integrating knowledge distillation [18] and lightweight models [19] have demonstrated promising results in skin lesion analysis.
In this paper, we present a novel approach for medical image classification and retrieval that integrates a multi-scale encoding mechanism [20] with a Vision Transformer (ViT) architecture [6]. Our method is evaluated on the ISIC-2018 [12,13] and ChestX-ray14 [1] datasets. The multi-scale encoding enhances the model’s ability to capture both fine-grained and global features, leading to improved classification and retrieval performance. Additionally, we introduce a dynamic multi-loss function that adapts to different training stages, optimizing classification accuracy, feature space structuring for retrieval, and overall model robustness. By adjusting the contributions of various loss components, including cross-entropy loss [21], triplet loss [22], contrastive loss [23], and distillation loss [24], our approach achieves a balanced learning process, resulting in enhanced performance across both datasets.
Our method is tested on two widely used medical image datasets: ISIC-2018, which focuses on skin lesion classification [12,13], and ChestX-ray14, which involves the identification of various thoracic diseases [1]. The ISIC-2018 dataset presents challenging lesion categories such as melanoma, basal cell carcinoma, and benign keratosis [12], while the ChestX-ray14 dataset contains diverse pathologies, including atelectasis, edema, and pneumonia [1]. Our approach achieves higher precision, recall, and F1-scores across both datasets, significantly outperforming existing methods [7,8].
Experimental results demonstrate that the proposed approach not only achieves high classification accuracy but also excels in medical image retrieval tasks, which are critical for case-based reasoning in clinical applications. The integration of multi-scale encoding with the ViT framework [6], combined with the dynamic multi-loss function, results in a well-structured feature space that enhances both diagnostic accuracy and retrieval performance. As shown in ‘Results and Analysis’ Section 5, our approach surpasses traditional CNN-based models and existing ViT variants [7,8], demonstrating its potential for large-scale medical image analysis.
Convolutional Neural Networks (CNNs) have long served as the backbone for image recognition and retrieval tasks, largely due to their hierarchical architectures that capture both low-level and high-level features effectively [3–5]. Pioneering architectures such as VGGNet [3], GoogLeNet [4], and ResNet [5] have substantially advanced the field by introducing innovative techniques to enhance performance and training efficiency. VGGNet underscored the significance of network depth in feature extraction, GoogLeNet optimized computational efficiency via Inception modules, and ResNet mitigated the degradation problem in deep networks through residual learning, enabling the effective training of networks with over 100 layers. Despite these advancements, CNNs inherently struggle to capture long-range dependencies and global contextual information because of their localized receptive fields.
Lightweight CNN models such as DenseNet [25], MobileNet [26], SqueezeNet [27], and Fast&Focused-Net [10] have made notable contributions to image retrieval and classification, particularly in resource-constrained settings. DenseNet improves feature reuse through dense connectivity, MobileNet lowers computational cost with depthwise separable convolutions, SqueezeNet achieves high accuracy with fewer parameters via Fire modules, and Fast&Focused-Net enhances small object encoding through a Volume-wise Dot Product (VDP) layer. Nonetheless, these traditional CNN approaches often fall short in generalizing to complex datasets characterized by high intra-class variance and inter-class similarities.
More recently, Vision Transformers (ViTs) have emerged as a powerful alternative by employing self-attention mechanisms to capture long-range dependencies and global features more effectively [6]. Unlike CNNs, ViTs treat images as sequences of patches and use attention to focus on the most relevant regions, a strategy that is particularly beneficial in medical image analysis where both local details and global context are critical [7]. However, standard ViTs typically require large-scale training datasets due to their limited inductive biases, which can lead to overfitting when applied to smaller medical datasets.
In medical imaging, ViTs have been increasingly adopted across various applications. For instance, they have been applied to tumor classification in MRI scans, where they effectively differentiate between tumor types. In segmentation tasks, models such as TransUNet have improved the delineation of anatomical structures [28,29]. Additionally, ViTs have demonstrated utility in MRI reconstruction and automated report generation in telehealth systems [28]. Nonetheless, a key limitation of ViTs in this domain is their reduced capacity to integrate multi-scale feature representations, which are crucial for segmenting small lesions and detecting subtle abnormalities.
For multi-label datasets such as ISIC-2018 [12,13] and ChestX-ray14 [1], current ViT-based approaches show promising results yet continue to face challenges including difficulties in capturing fine-grained details, high computational demands, and reliance on large annotated datasets. Some studies have attempted to overcome these issues by integrating CNN-like hierarchical feature extraction into ViTs [7] or by incorporating attention mechanisms into CNNs [14], but a comprehensive multi-scale approach remains underexplored.
Loss functions play a critical role in optimizing deep learning models for classification and retrieval tasks. Loss functions from deep metric learning, such as contrastive loss [23], triplet loss [22], and cross-entropy loss [21], enhance retrieval performance by structuring the feature space. Additionally, knowledge distillation [24] has been used to transfer insights from larger models to smaller ones, thereby improving efficiency [18]. Recently, dynamic multi-loss functions that adaptively adjust weight contributions during training have shown promise in balancing classification accuracy with retrieval performance. However, existing multi-loss strategies have yet to fully exploit training stage-dependent weight adjustments, which could further enhance model performance [7].
In medical image segmentation, architectures such as U-Net [30] and its variants, including H-DenseUNet [31], V-Net [32], and U-Net++ [30], have been widely employed for lesion segmentation. Efforts to improve these models have focused on integrating attention mechanisms [33] and using hybrid preprocessing techniques [34]. Despite these enhancements, challenges persist in efficiently capturing multi-scale contextual information. Similarly, disease classification models like CheXNet [35] have achieved performance comparable to that of radiologists in pneumonia detection, yet their dependence on fixed-scale feature extraction limits their adaptability to diverse datasets.
Our proposed approach addresses these limitations by integrating multi-scale encoding within a ViT framework and employing a dynamic multi-loss function. Unlike traditional CNNs, which struggle with long-range dependencies, or standard ViTs, which lack hierarchical feature extraction, our model explicitly incorporates multi-scale representations. Furthermore, by dynamically adjusting loss function weights during different training stages, our method optimizes both classification and retrieval performance, making it particularly effective for complex medical image datasets.
In this work, we propose a novel approach that integrates a multi-scale Vision Transformer (ViT) architecture [6] with a dynamic multi-loss function to address the challenges inherent in medical image retrieval and classification tasks. The method leverages multi-scale image encoding to effectively capture both fine-grained and global features, while the dynamic loss scheme balances multiple learning objectives throughout the training process, enhancing both classification accuracy and retrieval performance.
Our approach begins by generating scaled versions of each input image by progressively downscaling it until the smallest side is greater than 16 pixels, similar to multi-scale techniques used in medical image analysis [15]. At each scale, the image is partitioned into patches of size
This sequence is fed into a stack of multi-head self-attention encoder layers within the ViT architecture [6]. Each encoder layer consists of multi-head attention mechanisms and feed-forward networks that enable the model to learn long-range dependencies and global context. The output corresponding to the [CLS] token is extracted after the encoding layers and processed through a feed-forward layer to produce an embedding vector. This embedding is utilized for image retrieval tasks and serves as input for computing loss components like triplet loss [22] and contrastive loss [23].
An overview of the proposed architecture is depicted in Fig. 2.
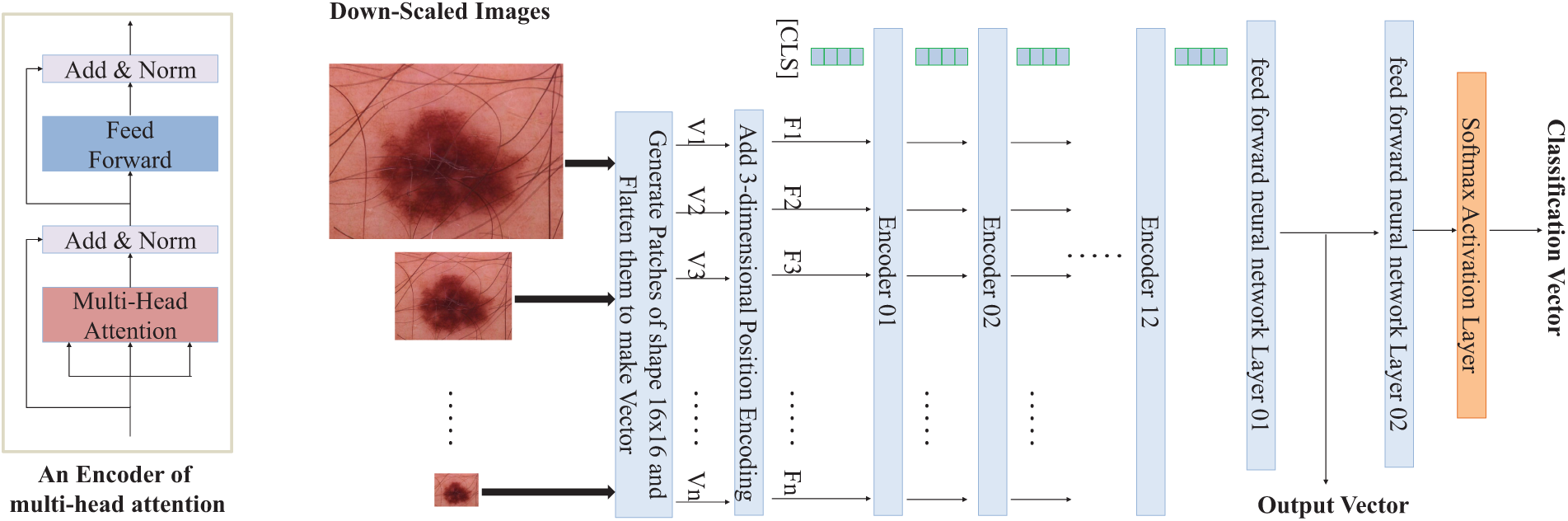
Figure 2: Overview of the proposed multi-scale Vision Transformer architecture for medical image classification and retrieval tasks. The input image is downscaled multiple times, divided into patches, and processed through a series of multi-head attention layers with positional encoding added. The final output vector is used for image retrieval and classification tasks, leveraging a dynamic multi-loss function
For classification tasks, the embedding vector is passed through an additional feed-forward layer followed by a softmax activation function to generate class probabilities. The cross-entropy loss [21] is computed using these probabilities and the ground truth labels.
3.2 Dynamic Multi-Loss Function
To balance the model’s performance on both classification and retrieval tasks, we employ a combined loss function that integrates cross-entropy loss (
Here,
The training process is structured into multiple phases, each focusing on different aspects of the model’s capabilities. In the early phases, the emphasis is on feature embedding and retrieval performance, facilitated by higher weights on metric learning losses and a relatively higher learning rate to encourage exploration of the feature space [7]. In subsequent phases, the focus shifts toward enhancing classification accuracy by increasing the weight of the cross-entropy loss and gradually reducing the learning rate to refine the model’s predictions.
An exponential decay schedule is applied to the learning rate across the training phases to ensure smooth convergence and adapt the model to the data’s complexities [22]. This dynamic adjustment of both loss weights and learning rates allows the model to balance and optimize multiple objectives effectively.
By integrating multi-scale encoding [15], a dynamic multi-loss function [7], and a phased training strategy, our proposed method effectively captures both global and local features. This leads to enhanced performance in medical image classification and retrieval tasks, particularly when dealing with complex and diverse datasets like ISIC-2018 [12] and ChestX-ray14 [1].
In this section, we provide a detailed description of the datasets used in our experiments, the evaluation metrics employed to assess the performance of our proposed method, and the hyperparameter settings configured during training.
We conducted experiments using two widely recognized medical imaging datasets: ISIC-2018 [12,13] and ChestX-ray14 [1]. Fig. 1 presents sample images from both datasets, illustrating the diversity and complexity of skin lesions and chest pathologies.
The International Skin Imaging Collaboration (ISIC) 2018 dataset [12] is a comprehensive collection of 10,015 high-resolution dermoscopic images designed to support automated skin cancer diagnosis. It includes seven lesion classes: Melanoma (MEL), Melanocytic Nevus (NV), Basal Cell Carcinoma (BCC), Actinic Keratoses and Intraepithelial Carcinoma (AKIEC), Benign Keratosis (BKL), Dermatofibroma (DF), and Vascular Lesions (VASC). Released as part of the ISIC 2018 challenge [13], this dataset is widely used for benchmarking lesion segmentation, feature extraction, and disease classification algorithms. Key challenges include class imbalance, variations in image quality, and significant visual similarities between lesion types [13]. To maintain consistency with prior studies, we utilized the official training, validation, and test splits provided by the ISIC challenge [7,13].
The ChestX-ray14 dataset [1], compiled by Wang et al. [1] from the National Institutes of Health Clinical Center, is one of the largest publicly available chest X-ray databases. It consists of 112,120 frontal-view X-ray images from 30,805 unique patients, annotated with 14 common thoracic conditions: Atelectasis (ATL), Cardiomegaly (CARD), Effusion (EFF), Infiltration (INF), Mass (MAS), Nodule (NOD), Pneumonia (PNEU), Pneumothorax (PNE), Consolidation (CONS), Edema (EDE), Emphysema (EMP), Fibrosis (FIB), Pleural Thickening (PLT), and Hernia (HER). This dataset has been extensively used for training and evaluating deep learning models in chest pathology detection and classification [1,11,14,35]. Its primary challenges include multi-label annotations, high inter-class similarity, and variations in image quality [1]. To ensure fair comparison with existing methods, we followed the official training and testing splits provided by Wang et al. [1,14,35].
To comprehensively evaluate the performance of our proposed method on both classification and retrieval tasks, we employed a set of widely used metrics, consistent with prior studies [7,11].
For classification tasks, we used:
• Precision: The ratio of true positive predictions to the total number of positive predictions, measuring the model’s accuracy in identifying relevant instances [11].
• Recall: The ratio of true positive predictions to the total number of actual positive instances, indicating the model’s ability to capture all relevant cases [11].
• F1-Score: The harmonic mean of precision and recall, providing a single measure that balances both metrics [11].
• Area Under the ROC Curve (AUC): Measures the ability of the model to distinguish between classes across all classification thresholds, with higher values indicating better performance [11].
For retrieval tasks, we utilized:
• Precision at K (P@K): The proportion of relevant images among the top K retrieved images, assessing the quality of the retrieval results [7].
• Recall at K (R@K): The proportion of relevant images retrieved among the top K, relative to the total number of relevant images, indicating the retrieval system’s completeness [7].
• Mean Average Precision (mAP): The average of precision values computed at the point of each relevant image retrieved, providing a summary of the precision-recall curve [7].
These metrics provide a comprehensive assessment of the model’s performance in both accurately classifying medical images and effectively retrieving similar images for clinical reference. The use of multiple evaluation measures ensures a thorough analysis of the model’s strengths and weaknesses in different aspects of medical image analysis [7].
Our model was implemented using the PyTorch framework and trained on NVIDIA Tesla V100 GPUs. The training process was divided into three phases, each consisting of 70 epochs, totaling 210 epochs, similar to the training strategies employed in previous studies [7].
The key hyperparameters used in the experiments are as follows:
• Optimizer: We used the Adam optimizer for training, with parameters
• Initial Learning Rate (
• Learning Rate Decay: The learning rate decayed exponentially in each phase as defined by:
This dynamic learning rate schedule was designed to facilitate the model’s convergence and performance across different training phases [7].
• Batch Size: Set to 32 for both datasets.
• Loss Weights: The dynamic multi-loss function employed weights for cross-entropy loss (
• Image Preprocessing: Images were resized to a fixed size while maintaining aspect ratio and normalized using the mean and standard deviation of the ImageNet dataset [36].
• Data Augmentation: To enhance model generalization, random horizontal and vertical flips and rotations up to 15 degrees were applied during training, as commonly used in prior works [6,14].
• Patch Size: Each image was divided into patches of size
• Positional Encoding: A 3-dimensional positional encoding vector representing scale, X-coordinate, and Y-coordinate was added to each patch embedding to capture spatial and scale information, following the methodology in [6].
• Regularization: Dropout with a rate of 0.1 was applied to prevent overfitting.
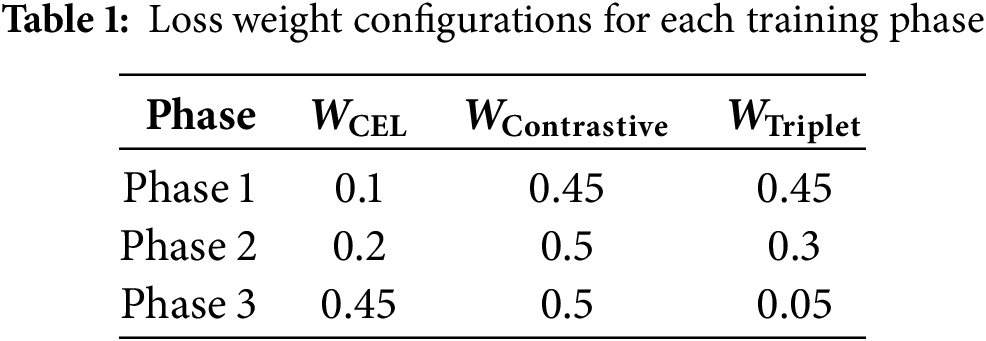
The hyperparameters were selected based on preliminary experiments and tuned to achieve optimal performance on both datasets. The dynamic adjustment of loss weights and learning rates was critical in balancing the multiple objectives of classification accuracy and retrieval effectiveness [7].
In this section, we present the experimental results of our proposed multi-scale Vision Transformer (ViT) architecture with a dynamic multi-loss function on the ISIC-2018 and ChestX-ray14 datasets. We compare our method with several state-of-the-art models, including traditional Convolutional Neural Networks (CNNs) and other transformer-based architectures, to demonstrate the effectiveness of our approach in both classification and retrieval tasks. We also perform ablation studies to assess the contribution of each component of our method.
5.1 Classification Performance
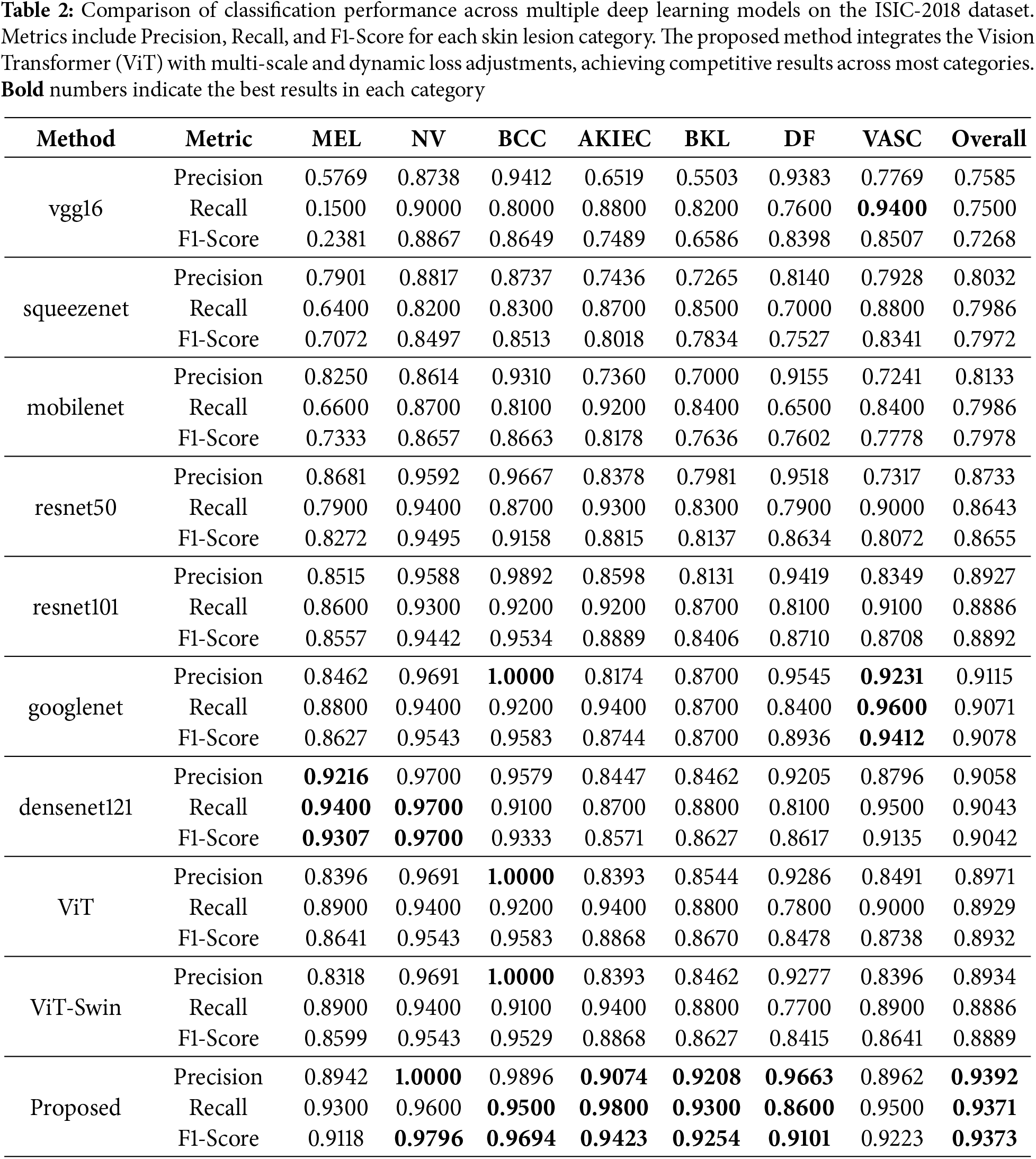
Table 2 summarizes the classification performance of different models on the ISIC-2018 dataset. The proposed method achieves the highest overall precision, recall, and F1-score, significantly outperforming traditional CNNs like VGG16 and ResNet50, as well as the standard ViT and its variants.
Figs. 3a, 4a, and 5a illustrate the precision, recall, and F1-score comparisons for each skin lesion category. The proposed method consistently outperforms other models across all categories, with particularly notable improvements in detecting melanoma (MEL) and basal cell carcinoma (BCC), which are critical for early diagnosis and treatment.
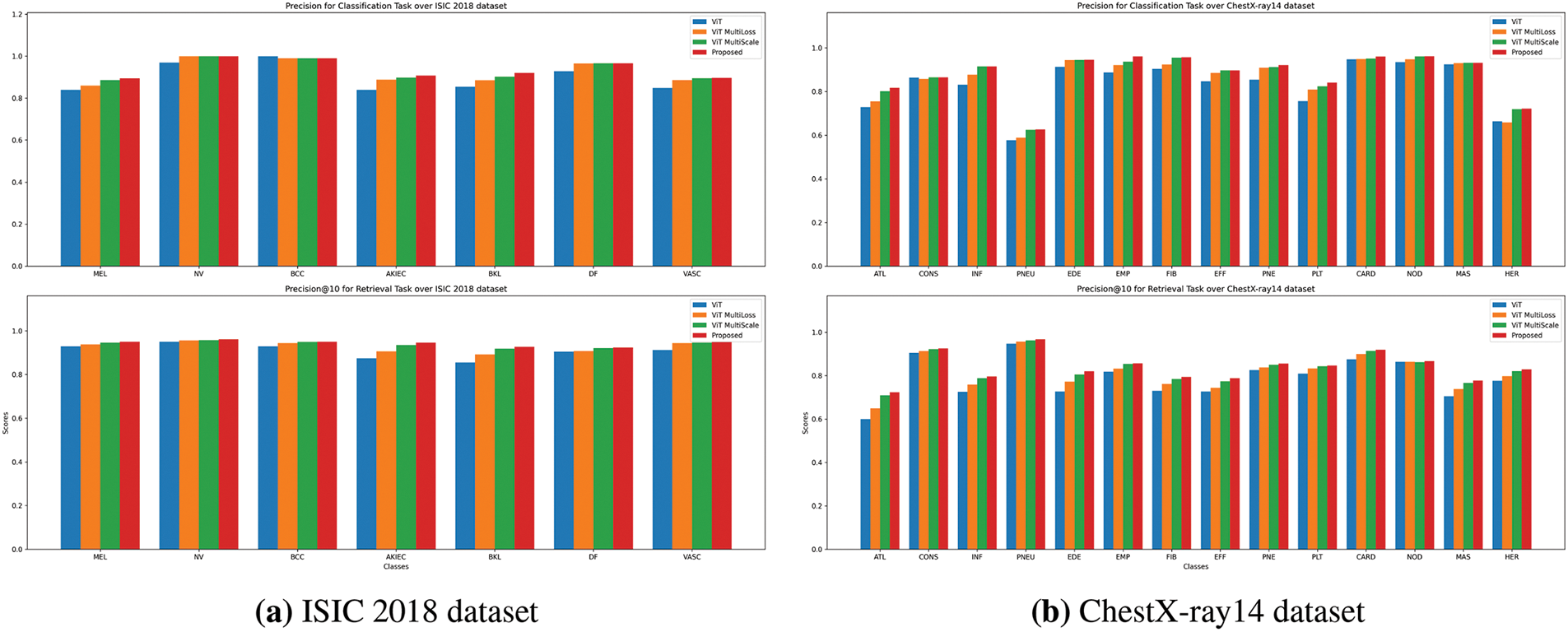
Figure 3: Precision comparison for Classification and Retrieval tasks. The precision scores are presented for both tasks (classification on top, retrieval on bottom) using ViT, ViT MultiScale, ViT MultiLoss, and the proposed method. The results highlight the superior performance of the proposed method across most lesion categories
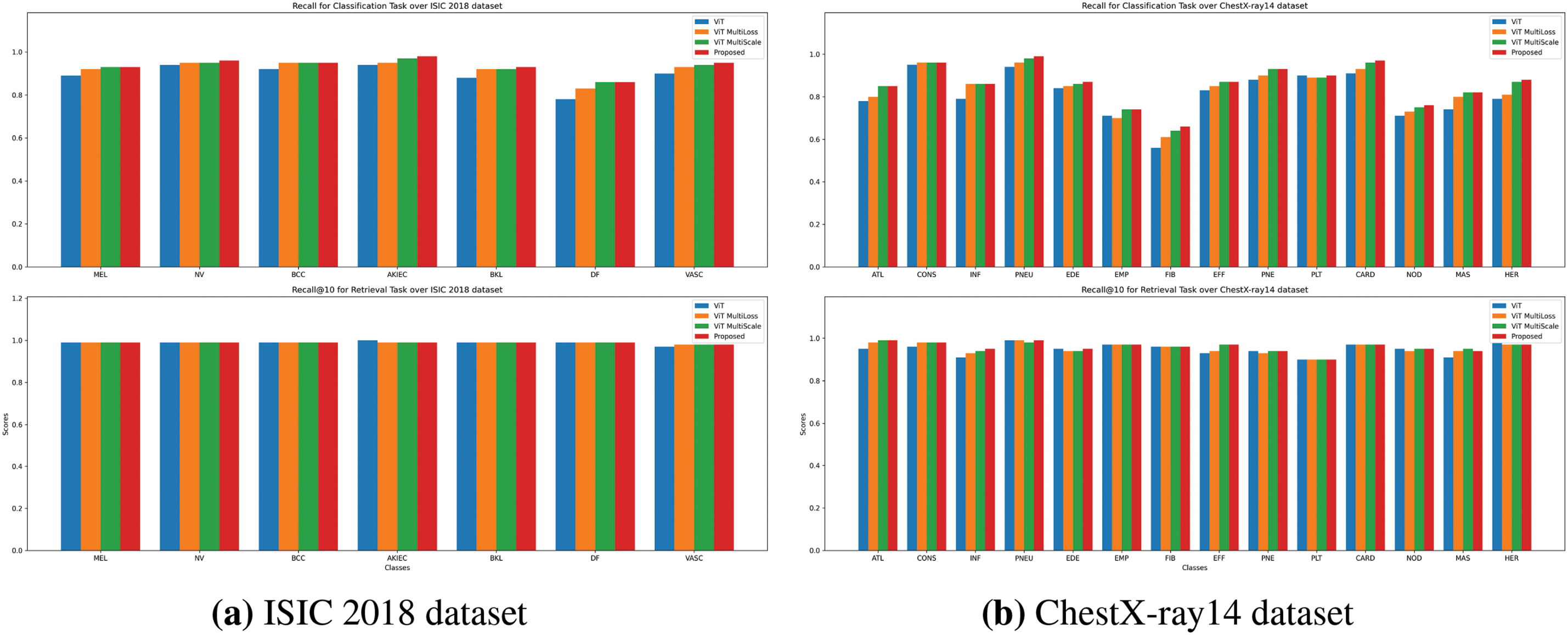
Figure 4: Recall comparison for Classification and Retrieval tasks. Recall scores for both tasks (classification on top, retrieval on bottom) are compared across the ViT, ViT MultiScale, ViT MultiLoss, and proposed method. The proposed method consistently shows improved recall, especially for challenging lesion categories
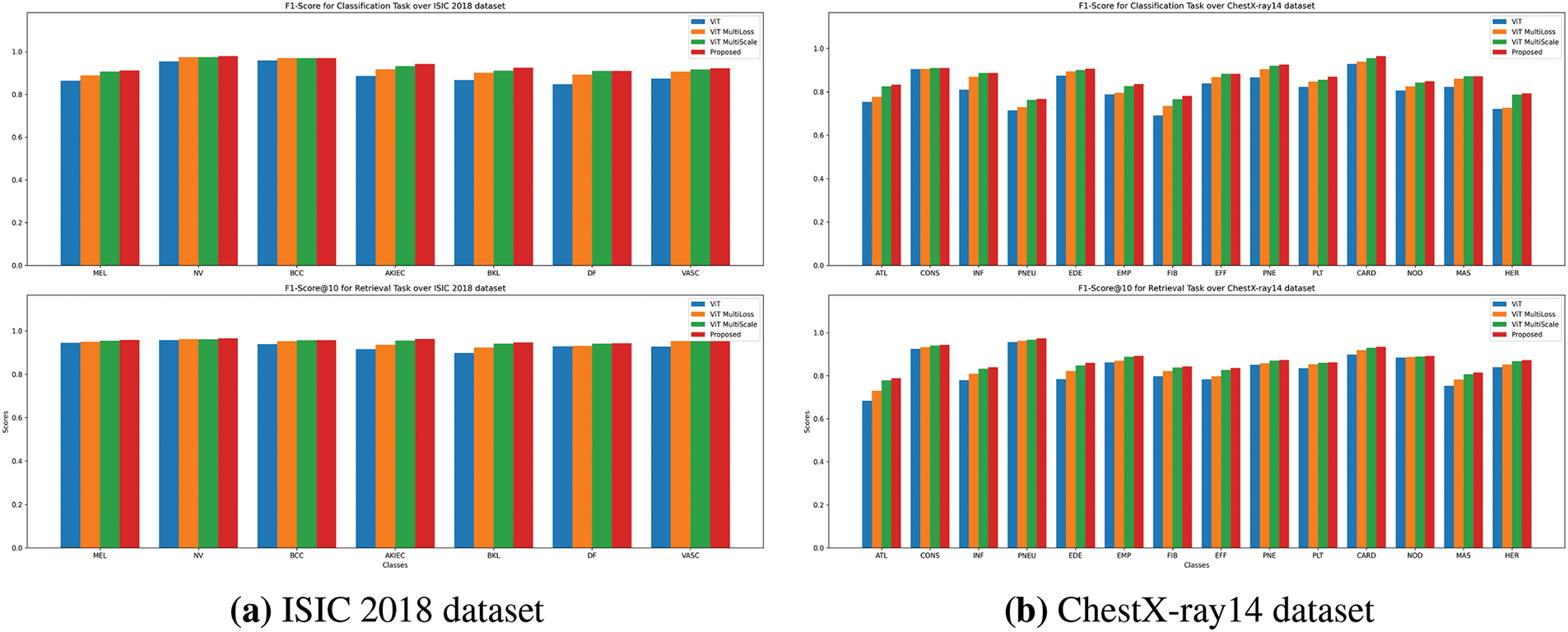
Figure 5: F1-Score comparison for Classification and Retrieval tasks. The bar charts illustrate F1-scores for both classification (top) and retrieval (bottom) tasks, comparing performance across ViT, ViT MultiScale, ViT MultiLoss, and proposed method. The proposed method achieves higher F1-Scores, indicating balanced precision and recall across categories
The normalized confusion matrix in Fig. 6a provides a visual representation of the classification accuracy for each class. The high values along the diagonal indicate that the model accurately classifies most samples, while the low off-diagonal values suggest minimal misclassifications. The Receiver Operating Characteristic (ROC) curves in Fig. 7a further demonstrate the model’s strong ability to distinguish between different lesion types, with Area Under the Curve (AUC) values exceeding 0.92 for all classes.
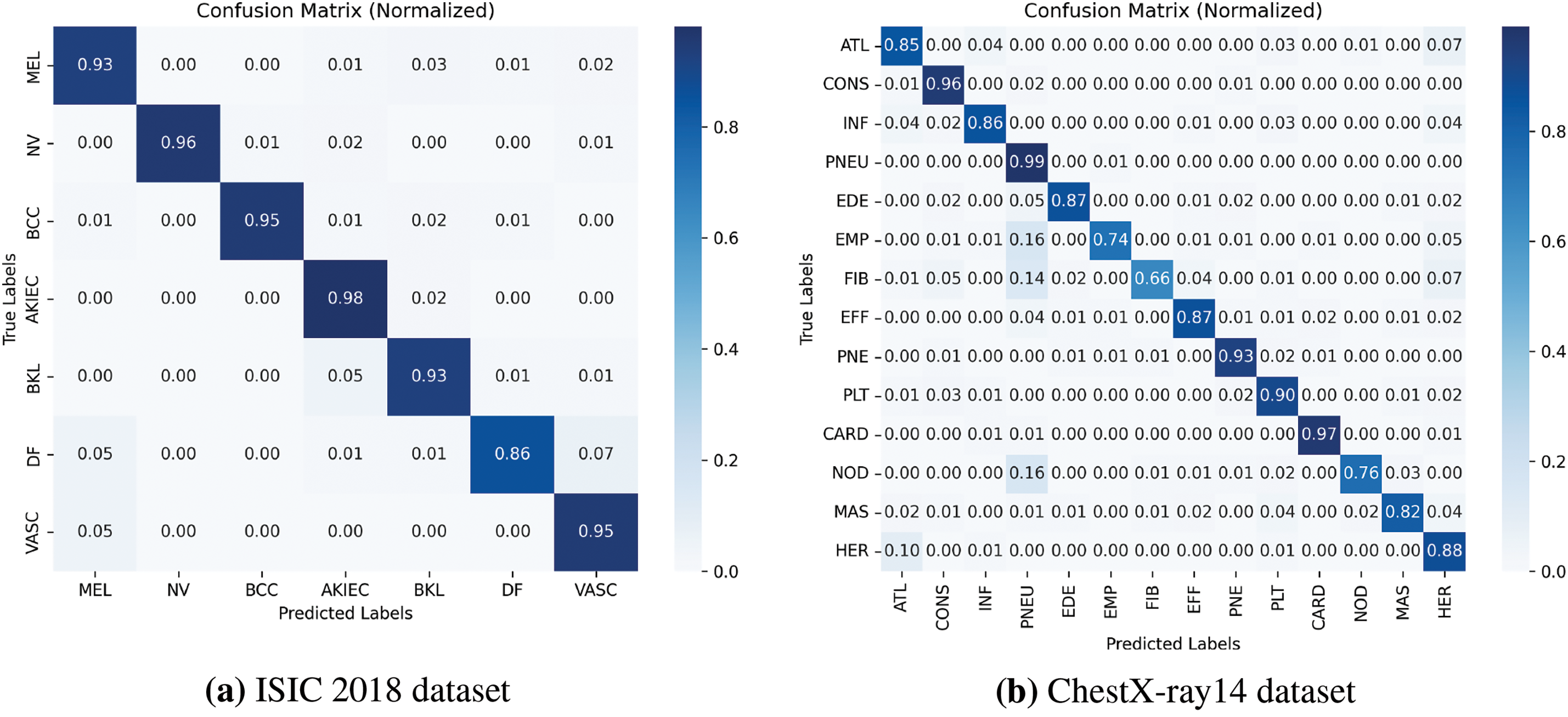
Figure 6: Normalized Confusion Matrix for classification task using the proposed method. This figure shows the classification accuracy for each skin lesion category, normalized to highlight the distribution of correct and incorrect predictions
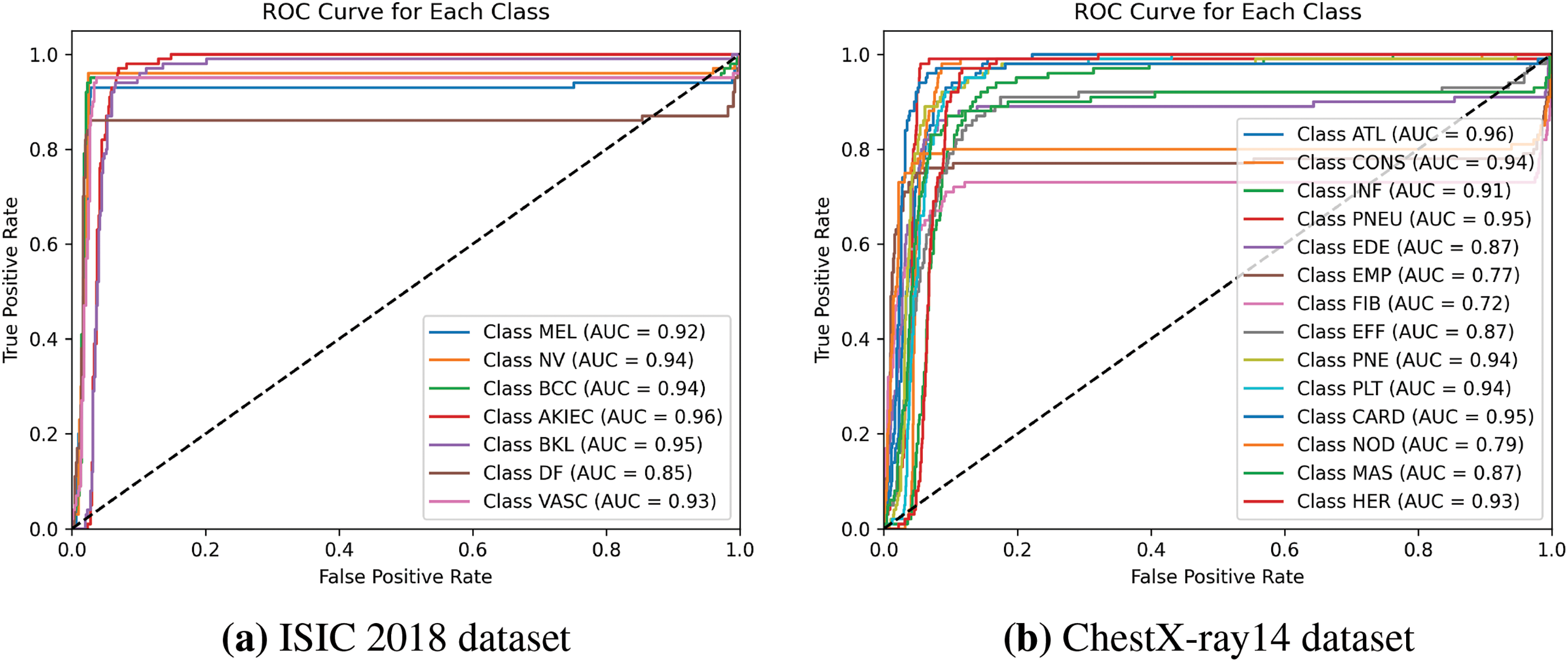
Figure 7: ROC Curve for each skin lesion class for classification task using the proposed method. The Receiver Operating Characteristic (ROC) curve and corresponding Area Under the Curve (AUC) values are plotted for each lesion category, showcasing the model’s ability to distinguish between classes
Table 3 presents the classification results on the ChestX-ray14 dataset. Our proposed method achieves superior performance with higher precision, recall, and F1-score compared to other models. The improvement is particularly significant for pathologies such as cardiomegaly (CARD) and pneumonia (PNEU).
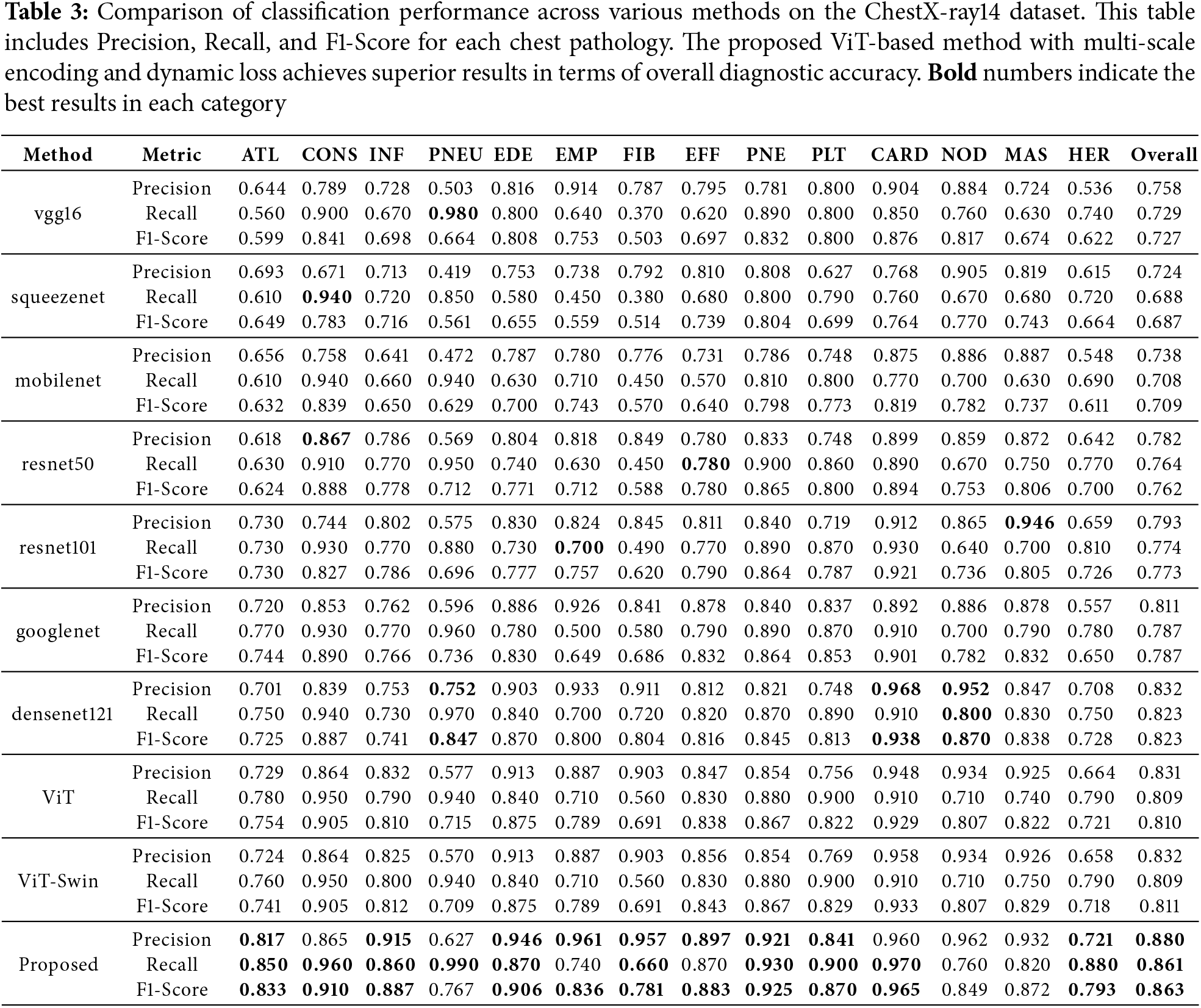
Figs. 3b, 4b, and 5b display the performance metrics for each chest pathology. The proposed method shows marked improvements in detecting diseases that are challenging due to visual similarity or class imbalance, such as infiltration (INF) and edema (EDE).
The confusion matrix in Fig. 6b illustrates the classification performance across all pathologies. The ROC curves in Fig. 7b highlight the model’s discriminative power, with high AUC values indicating strong classification capabilities.
Table 4 presents the retrieval performance on the ISIC-2018 dataset at different top-K levels (K = 2, 5, 10). The proposed method achieves the highest precision and recall at all levels, demonstrating its effectiveness in retrieving relevant images for clinical reference.
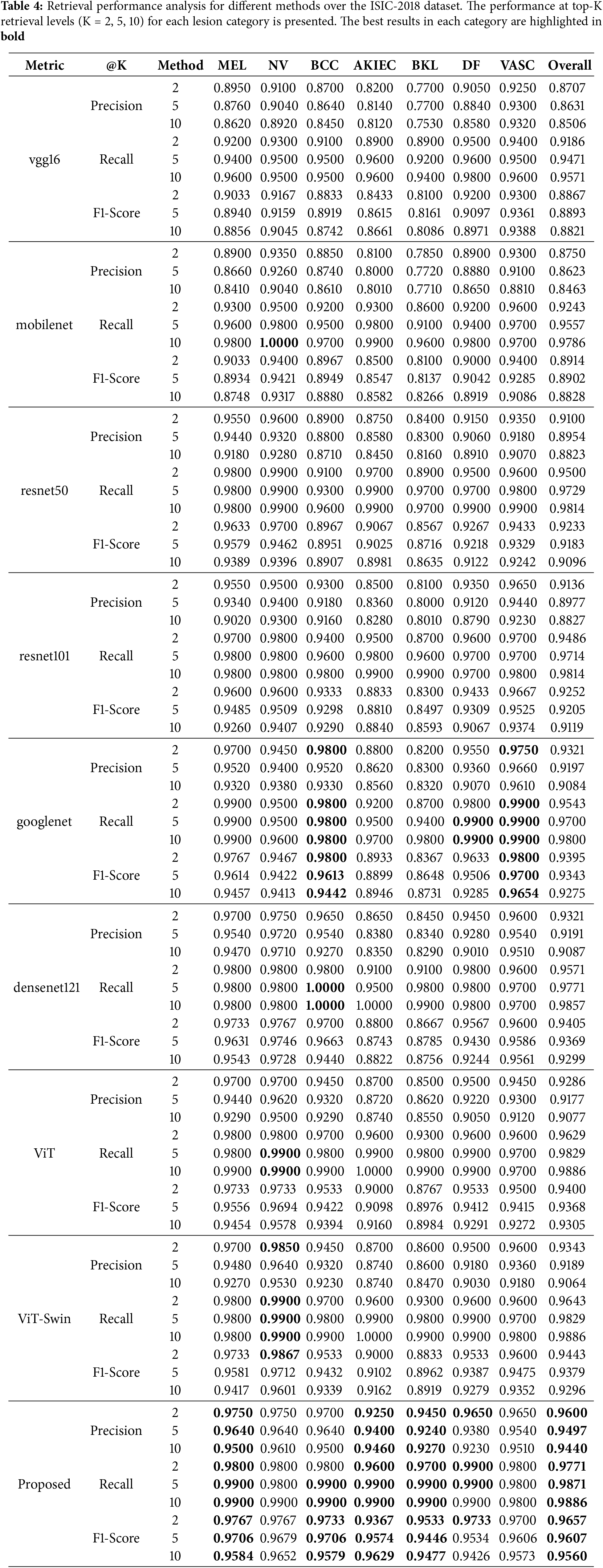
Figs. 3a and 4a illustrate the precision and recall for retrieval tasks across different models. The proposed method consistently outperforms others, particularly for melanoma (MEL) and actinic keratosis (AKIEC), which are crucial for accurate diagnosis.
The retrieval results on the ChestX-ray14 dataset are shown in Table 5. Our method achieves superior retrieval precision and recall at various top-K levels, highlighting its ability to retrieve clinically relevant images across multiple pathologies.
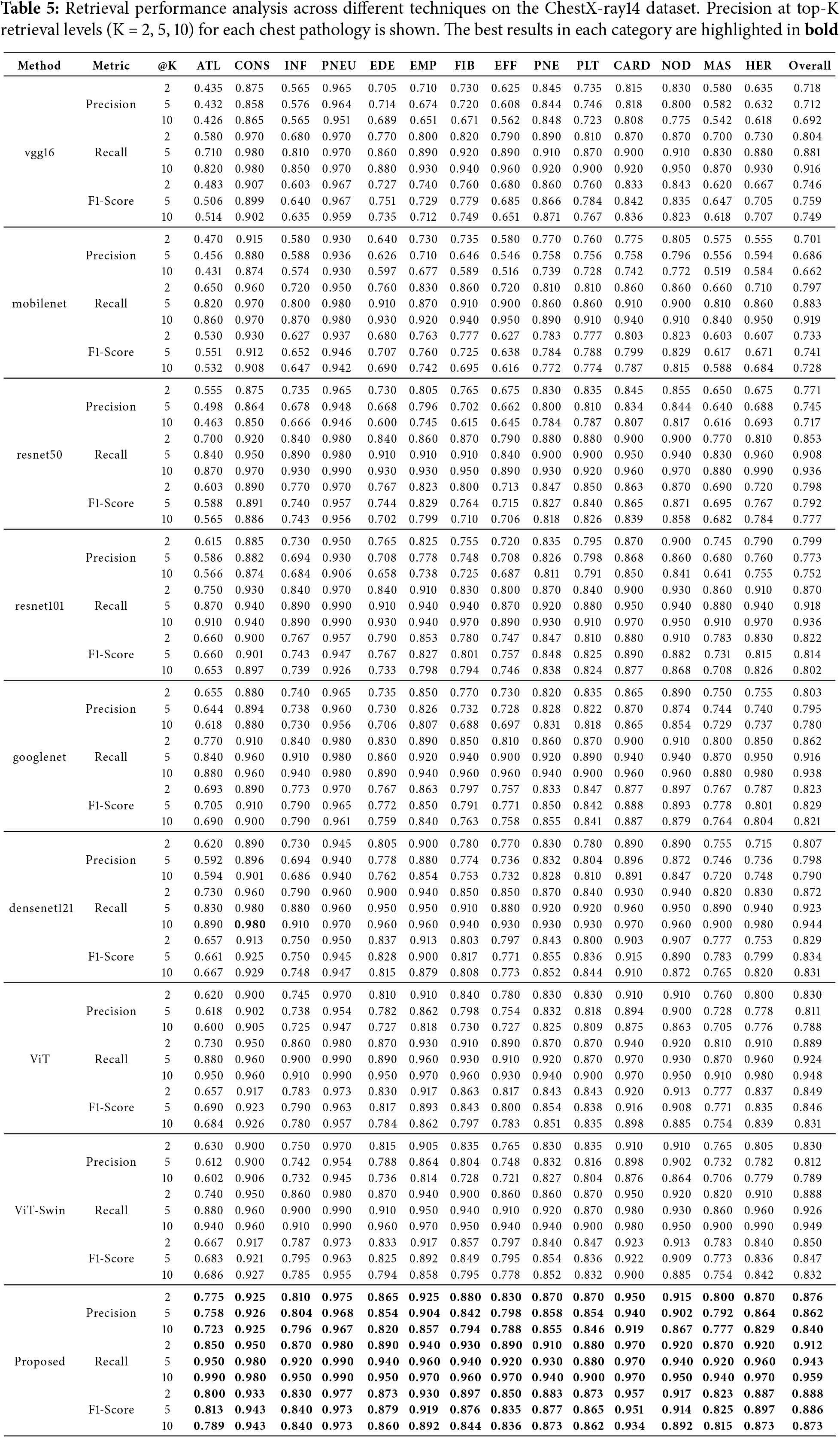
As depicted in Figs. 3b and 4b, the proposed method shows enhanced retrieval performance for diseases such as pneumothorax (PNEU) and cardiomegaly (CARD), which are critical for patient care.
To evaluate the contribution of each component of our proposed method, we conducted ablation studies by comparing the performance of the baseline ViT model, ViT with multi-scale encoding (ViT MultiScale), ViT with the dynamic multi-loss function (ViT MultiLoss), and the full proposed method.
5.3.1 Impact of Multi-Scale Encoding
Tables 6 and 7 show that incorporating multi-scale encoding into the ViT architecture (ViT MultiScale) leads to improved classification and retrieval performance compared to the baseline ViT model. This enhancement is attributed to the model’s ability to capture fine-grained details and global context by processing images at multiple scales.
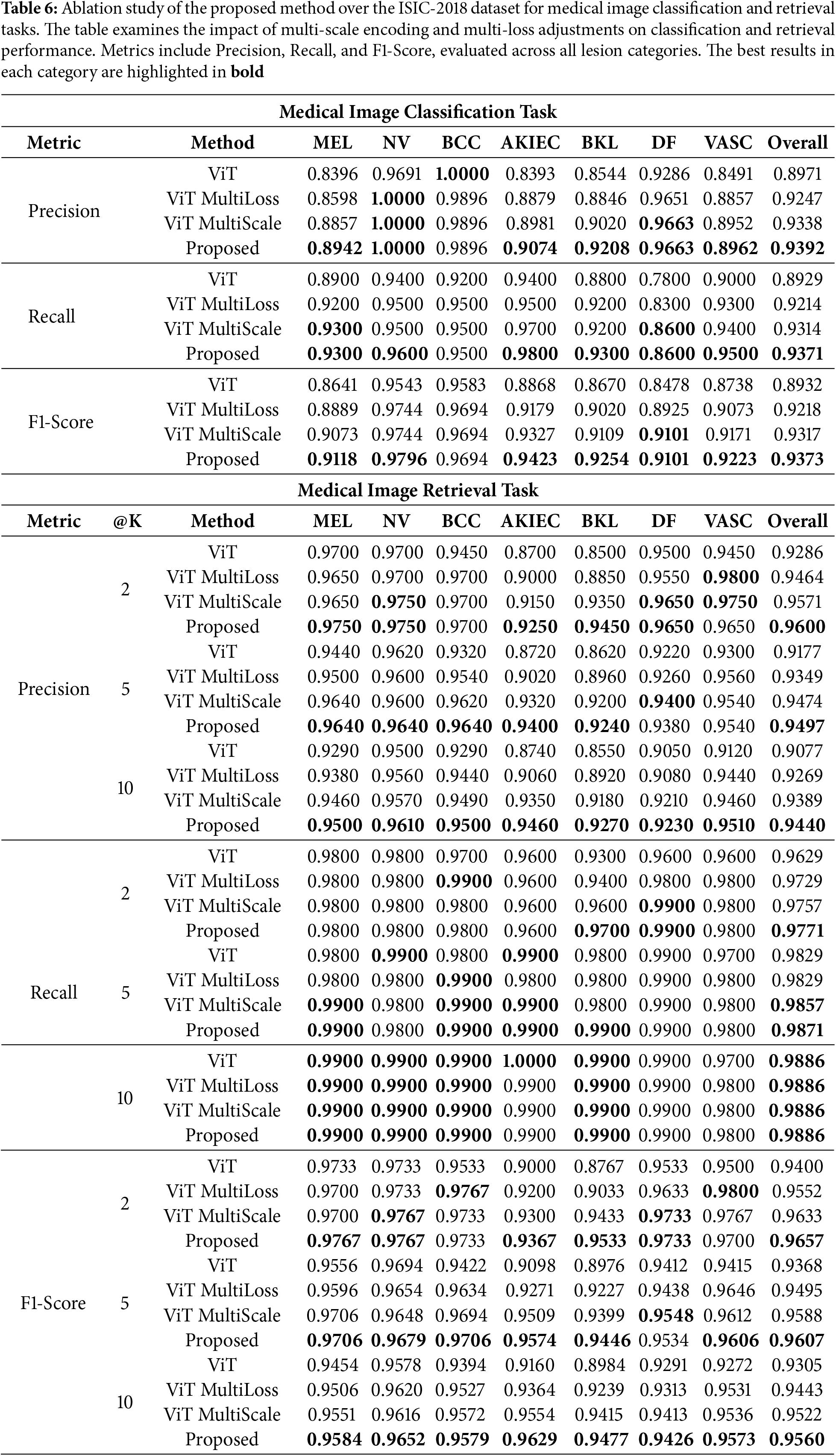
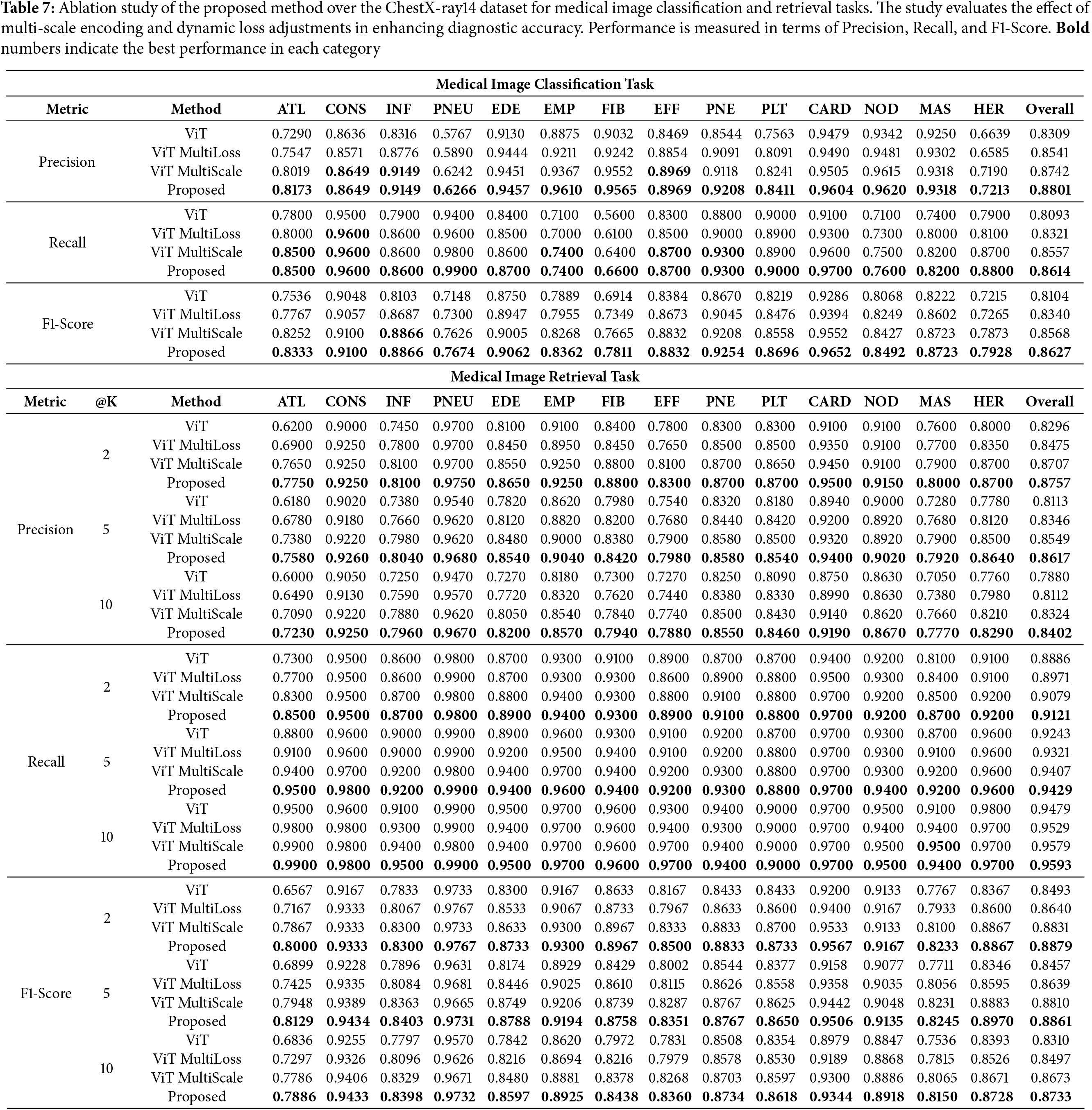
5.3.2 Impact of Dynamic Multi-Loss Function
The results also indicate that applying the dynamic multi-loss function (ViT MultiLoss) improves performance over the baseline by better balancing the learning objectives of classification and retrieval tasks. The adaptive weighting of loss components allows the model to focus on different aspects during training, enhancing overall robustness.
The full proposed method, which integrates both multi-scale encoding and the dynamic multi-loss function, achieves the highest performance in both datasets. This demonstrates the synergistic effect of combining these two components, leading to significant improvements over individual enhancements.
The experimental results validate the effectiveness of our proposed method in medical image classification and retrieval tasks. The integration of multi-scale encoding enables the model to capture important features at different resolutions, which is particularly beneficial for medical images where lesions and pathologies may vary greatly in size and appearance.
The dynamic multi-loss function allows the model to balance multiple learning objectives, optimizing for both classification accuracy and retrieval effectiveness. By adjusting the loss weights during training, the model can adapt to the complexities of the data, improving generalization and robustness.
The superior performance of our method over traditional CNNs and existing transformer-based models underscores the potential of combining multi-scale processing with advanced training strategies. This approach addresses the challenges posed by complex medical datasets, such as class imbalance and high inter-class similarity.
Overall, the proposed method demonstrates significant advancements in medical image analysis, offering improved tools for clinicians in diagnosis and decision-making processes.
This paper presents a novel multi-scale Vision Transformer (ViT) architecture with a dynamic multi-loss function for medical image classification and retrieval. By integrating multi-scale encoding, our approach effectively captures both fine-grained and global features, while the dynamic loss function adaptively balances multiple learning objectives.
Extensive experiments on the ISIC-2018 and ChestX-ray14 datasets demonstrate that our method consistently outperforms existing CNN-based and transformer-based models. The results confirm improved classification accuracy, precision, and retrieval performance, making our approach a valuable tool for medical image analysis.
Despite its effectiveness, the model’s performance is influenced by dataset diversity and computational complexity. Future research could focus on extending the approach to other imaging modalities, improving optimization strategies, and enhancing model interpretability for real-world clinical adoption.
Our work contributes to advancing deep learning techniques in medical imaging, with the potential to assist healthcare professionals in more accurate diagnoses and improved patient care.
Acknowledgement: The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through small group research under grant number RGP1/278/45.
Funding Statement: This research was funded by the Deanship of Research and Graduate Studies at King Khalid University through small group research under grant number RGP1/278/45.
Author Contributions: The authors confirm their contribution to the paper as follows: Study conception and design: Omar Alqhatani, Mohamed Ghouse. Data collection: Asfia Sabahath, Omer Bin Hussain. Analysis and interpretation of results: Omar Alqhatani, Mohamed Ghouse, Asfia Sabahath. Draft manuscript preparation: Mohamed Ghouse, Arshiya Begum. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All datasets used in this study are publicly available and properly cited in the manuscript. Further details and links can be provided upon request.
Ethics Approval: This study did not involve any human or animal subjects. Hence, ethical approval is not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. arXiv:1705.02315. 2017. [Google Scholar]
2. Harley AW, Ufkes A, Derpanis KG. Evaluation of deep convolutional nets for document image classification and retrieval. arXiv:1502.07058. 2015. [Google Scholar]
3. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556. 2015. [Google Scholar]
4. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. arXiv:1409.4842. 2014. [Google Scholar]
5. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016; Las Vegas, NV, USA: IEEE. p. 770–8. [Google Scholar]
6. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: transformers for image recognition at scale. In: International Conference on Learning Representations; 2021; Vienna, Austria. [cited 2025 Jan 20]. Available from: https://arXiv.org/abs/2010.11929. [Google Scholar]
7. Pundhir A, Sagar S, Singh P, Raman B. Echoes of images: multi-loss network for image retrieval in vision transformers. Med Biolo Eng Comput. 2024;62(7):2037–58. doi:10.1007/s11517-024-03055-6. [Google Scholar] [CrossRef]
8. Ashraf SMN, Mamun MA, Abdullah HM, Alam MGR. SynthEnsemble: a fusion of CNN, vision transformer, and hybrid models for multi-label chest X-ray classification. In: 2023 26th International Conference on Computer and Information Technology (ICCIT); 2023. p. 1–6. [Google Scholar]
9. Shen T, Li X. Automatic polyp image segmentation and cancer prediction based on deep learning. Front Oncol. 2023;12:1087438. doi:10.3389/fonc.2022.1087438. [Google Scholar] [CrossRef]
10. Ali T, Roy PP, Saini R. Fast&Focused-Net: enhancing small object encoding with VDP layer in deep neural networks. IEEE Access. 2024;12:130603–16. doi:10.1109/ACCESS.2024.3447888. [Google Scholar] [CrossRef]
11. Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep. 2019;9(1):6381. doi:10.1038/s41598-019-42294-8. [Google Scholar] [CrossRef]
12. Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5(1):180161. doi:10.1038/sdata.2018.161. [Google Scholar] [CrossRef]
13. Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, et al. Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the international skin imaging collaboration (ISIC). arXiv:1902.03368. 2019. [Google Scholar]
14. Innat M, Hossain MF, Mader K, Kouzani AZ. A convolutional attention mapping deep neural network for classification and localization of cardiomegaly on chest X-rays. Sci Rep. 2023;13(1):6247. doi:10.1038/s41598-023-32611-7. [Google Scholar] [CrossRef]
15. Alam MS, Wang D, Liao Q, Sowmya A. A multi-scale context aware attention model for medical image segmentation. IEEE J Biomed Health Inform. 2023;27(8):3731–9. doi:10.1109/JBHI.2022.3227540. [Google Scholar] [CrossRef]
16. Nampalle KB, Pundhir A, Jupudi PR, Raman B. Towards improved U-Net for efficient skin lesion segmentation. Multimed Tools Appl. 2024;83(28):71665–82. doi:10.1007/s11042-024-18334-5. [Google Scholar] [CrossRef]
17. Liang S, Tian S, Yu L, Kang X. Improved U-Net based on contour attention for efficient segmentation of skin lesion. Multimed Tools Appl. 2023;83(11):33371–91. doi:10.1007/s11042-023-16759-y. [Google Scholar] [CrossRef]
18. Zhang Z, Lu B. Efficient skin lesion segmentation with boundary distillation. Med Biol Eng Comput. 2024;62(9):2703–16. doi:10.1007/s11517-024-03095-y. [Google Scholar] [CrossRef]
19. Hao S, Wu H, Jiang Y, Ji Z, Zhao L, Liu L, et al. GSCEU-Net: an end-to-end lightweight skin lesion segmentation model with feature fusion based on U-Net enhancements. Information. 2023;14(9):486. doi:10.3390/info14090486. [Google Scholar] [CrossRef]
20. Ali T, Siddiqui MFH, Shahab S, Roy PP. GMIF: a gated multiscale input feature fusion scheme for scene text detection. IEEE Access. 2022;10:93992–4006. [Google Scholar]
21. Rubinstein RY. The cross-entropy method for combinatorial and continuous optimization. Methodol Comput Appl Probab. 1999;1(2):127–90. [Google Scholar]
22. Schroff F, Kalenichenko D, Philbin J. FaceNet: a unified embedding for face recognition and clustering. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015; Boston, MA, USA. p. 815–23. [Google Scholar]
23. Hadsell R, Chopra S, LeCun Y. Dimensionality reduction by learning an invariant mapping. In: 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06); 2006; New York City, NY, USA. vol. 2, p. 1735–42. [Google Scholar]
24. Hinton G, Vinyals O, Dean J. Distilling the knowledge in a neural network. arXiv:1503.02531. 2015. [Google Scholar]
25. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017; Honolulu, HI, USA: IEEE. p. 2261–9. [Google Scholar]
26. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv:1704.04861. 2017. [Google Scholar]
27. Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. arXiv:1602.07360. 2016. [Google Scholar]
28. Al-hammuri K, Gebali F, Kanan A, Thirumarai Chelvan I. Vision transformer architecture and applications in digital health: a tutorial and survey. Visual Comput Indust, Biomed Art. 2023;6(1):1–14. doi:10.1186/s42492-023-00140-9. [Google Scholar] [CrossRef]
29. Halder A, Gharami S, Sadhu P, Singh PK, Wozniak M, Ijaz MF. Implementing vision transformer for classifying 2D biomedical images. Sci Rep. 2024;14(1):12567. doi:10.1038/s41598-024-63094-9. [Google Scholar] [CrossRef]
30. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. (DLMIA 2018, ML-CDS 2018); 2018; Granada, Spain. p. 3–11. [Google Scholar]
31. Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imag. 2018;37(12):2663–74. doi:10.1109/TMI.2018.2845918. [Google Scholar] [CrossRef]
32. Milletari F, Navab N, Ahmadi SA. V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV); 2016; Stanford, CA, USA: IEEE. p. 565–71. [Google Scholar]
33. Ramadan R, Aly S. DGCU–Net: a new dual gradient-color deep convolutional neural network for efficient skin lesion segmentation. Biomed Signal Process Control. 2022;77(1):103829. doi:10.1016/j.bspc.2022.103829. [Google Scholar] [CrossRef]
34. Malik S, Akram T, Ashraf I, Rafiullah M, Ullah M, Tanveer J. A hybrid preprocessor DE-ABC for efficient skin-lesion segmentation with improved contrast. Diagnostics. 2022;12(11):2625. doi:10.3390/diagnostics12112625. [Google Scholar] [CrossRef]
35. Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv:1711.05225. 2017. [Google Scholar]
36. Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009; Miami, FL, USA: IEEE. p. 248–55. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools