 Open Access
Open Access
REVIEW
A Comprehensive Review of Multimodal Deep Learning for Enhanced Medical Diagnostics
1 Computer Science Department, Faculty of Computers and Artificial Intelligence, Damietta University, New Damietta, 34517, Egypt
2 Faculty of Computer Science and Engineering, New Mansoura University, Dakhlia, 35516, Egypt
3 Computer Science Department, Faculty of Graduate Studies for Statistical Research, Cairo University, Giza, 12613, Egypt
* Corresponding Author: Ahmed Ismail Ebada. Email:
(This article belongs to the Special Issue: Multi-Modal Deep Learning for Advanced Medical Diagnostics)
Computers, Materials & Continua 2025, 84(3), 4155-4193. https://doi.org/10.32604/cmc.2025.065571
Received 17 March 2025; Accepted 18 June 2025; Issue published 30 July 2025
Abstract
Multimodal deep learning has emerged as a key paradigm in contemporary medical diagnostics, advancing precision medicine by enabling integration and learning from diverse data sources. The exponential growth of high-dimensional healthcare data, encompassing genomic, transcriptomic, and other omics profiles, as well as radiological imaging and histopathological slides, makes this approach increasingly important because, when examined separately, these data sources only offer a fragmented picture of intricate disease processes. Multimodal deep learning leverages the complementary properties of multiple data modalities to enable more accurate prognostic modeling, more robust disease characterization, and improved treatment decision-making. This review provides a comprehensive overview of the current state of multimodal deep learning approaches in medical diagnosis. We classify and examine important application domains, such as (1) radiology, where automated report generation and lesion detection are facilitated by image-text integration; (2) histopathology, where fusion models improve tumor classification and grading; and (3) multi-omics, where molecular subtypes and latent biomarkers are revealed through cross-modal learning. We provide an overview of representative research, methodological advancements, and clinical consequences for each domain. Additionally, we critically analyzed the fundamental issues preventing wider adoption, including computational complexity (particularly in training scalable, multi-branch networks), data heterogeneity (resulting from modality-specific noise, resolution variations, and inconsistent annotations), and the challenge of maintaining significant cross-modal correlations during fusion. These problems impede interpretability, which is crucial for clinical trust and use, in addition to performance and generalizability. Lastly, we outline important areas for future research, including the development of standardized protocols for harmonizing data, the creation of lightweight and interpretable fusion architectures, the integration of real-time clinical decision support systems, and the promotion of cooperation for federated multimodal learning. Our goal is to provide researchers and clinicians with a concise overview of the field’s present state, enduring constraints, and exciting directions for further research through this review.Keywords
The integration of deep learning and big data has significantly transformed numerous fields, including healthcare. Recent advances in machine learning, particularly deep neural networks, have enabled the extraction of high-level features from complex datasets, such as images, text, and omics data. Healthcare domains, especially medical diagnostics, have benefited from these advances in areas such as computer-aided diagnosis, biomarker discovery, and personalized treatment planning.
Unimodal systems, which rely on a single type of data (e.g., clinical text or medical imaging), often exhibit limited effectiveness and restricted applicability. They cannot fully understand the complexity of a patient’s condition and may overlook important indicators that are apparent in other modalities. For example, a chest X-ray may show anomalies, but the interpretation could be inaccurate or misleading if it is not accompanied by contextual information, such as test findings, patient history, or symptoms. Furthermore, unimodal models are typically less reliable and more vulnerable to errors or missing data in their modality. These drawbacks highlight the necessity of models that combine data from multiple sources [1]. Multimodal deep learning, on the other hand, provides a revolutionary method by combining various disparate data sources into a unified analytical framework, which results in a more thorough and sophisticated comprehension of a patient’s health. This methodology improves biomarker identification, personalized treatment planning, and diagnostic accuracy by capturing complementary information from many modalities. A multifaceted view that goes beyond the potential of individual modalities is made possible by the integration of data from clinical notes, medical imaging, genetic profiles, and sensor outputs. Multimodal learning has shown notable advancements in disease detection, prognosis, and patient outcome prediction, especially when applied to complex and multivariate situations like cardiovascular diseases and cancer. This integrative approach advances clinical decision-making and precision medicine by helping researchers and clinicians identify links and patterns that might otherwise go unnoticed [2].
Despite this potential, significant clinical obstacles highlight the need to adopt such integrative technologies. No statistics have adequately captured the complexity of human health. Genetic, environmental, and behavioral factors all play a role in diseases, including cancer, diabetes, and neurodegenerative disorders. In addition, the amount of medical data is predicted to double every 73 days, placing clinicians at risk of cognitive overload in the absence of suitable analytical assistance. Additionally, the need for customized care that accounts for each patient’s particular biological characteristics and lifestyle choices is increasing, forcing healthcare organizations to abandon the use of one-size-fits-all approaches. By making scalable, context-aware, and patient-specific modeling possible, multimodal deep learning can address these issues.
We use a structured methodology that includes a systematic literature review of peer-reviewed articles published between 2022 and 2025, focusing on works that use state-of-the-art architecture like graph neural networks and attention-based models for data fusion. In this review, we critically assessed algorithmic innovations and their clinical impact, focusing on studies that demonstrate clear performance improvements across multiple medical conditions.
The scope of this review is to provide a thorough and critical analysis of recent advancements in multimodal deep learning for medical diagnostics, as well as its methodological evolution, real-world clinical applications, and related challenges.
Deep learning methods like CNNs (convolutional neural networks) [3] and RNNs (recurrent neural networks) [4] Can efficiently combine clinical text, physiological signals, genomic information, and medical images. This enables the identification of latent associations that aid in diagnosing and treating individualized diseases. Modern advances in deep learning, such as graph neural networks [5] and attention processes [6], have created new opportunities for combining and analyzing multimodal medical data. These innovative techniques have shown promise in better capturing contextual data and complex relationships to improve healthcare and enable more personalized and efficient care. Multimodal healthcare, which is becoming a major influence in the medical field, aims to use information technology to transform clinical procedures. It has drawn a lot of interest from academics and professionals as a possible approach to tackling important disease diagnosis issues in areas with unequal access to medical resources. The emergence of multimodal healthcare addresses pressing medical challenges. First, human health is too complex to be reduced to a single test or metric. No single diagnostic method can adequately account for the complex network of genetic, environmental, and behavioral factors that contribute to cancer [7], Diabetes [8], and neurodegenerative disorders [9]. Second, there are opportunities and challenges associated with the explosion of healthcare data, which is predicted to double every few years. Clinicians are at risk of becoming overwhelmed by a sea of disparate facts if no mechanisms are in place to synthesize these data. Furthermore, in a time when people vary greatly in terms of genetic composition, lifestyle, and reactions to treatment, patients now demand more proactive and individualized care. Multimodal healthcare satisfies this need by customizing interventions to each patient’s specific profile, improving results while reducing trial and error.
Several recent reviews have discussed various facets of multimodal medical AI (Artificial Intelligence). However, their coverage is frequently limited or devoid of critical analysis. For example, Muhammad et al. [10] introduced several topics related to multimodal signal fusion for intelligent medical devices. The survey article was mainly focused on IoMT. It included several important topics about IoMT (Internet of Medical Things) applications and smart healthcare difficulties. The review highlights four main limitations: its narrow focus may exclude some fusion techniques in smart healthcare, selection bias in study criteria may distort results, findings may become outdated due to rapid technological advancements, and sensor data variability could impact the validity of synthesized insights. Amal et al. [11] presented the applications and scope of machine learning and multimodal data concerning cardiovascular healthcare. The challenges of multimodal data fusion were also briefly covered by the writers. The study identified four main limitations: (1) a stated lack of conflicts of interest, although there may still be concerns about the independence of the results, (2) limited generalizability because of the study’s narrow demographic focus, (3) potential biases from the use of genetic and electronic health data, and (4) difficulties reproducing the intricate machine learning framework, which prevents practical application. In the opinion of Stahlschmidt et al. [12] Biomedical data is becoming more and more multimodal, offering a useful source of hidden information that is inaccessible using single-modality methodologies. The complex link between the modalities can be captured by deep learning approaches, which can combine multimodal data. Transfer learning is a good approach for multimodal medical big data, according to the authors. Although it provides a comprehensive review of multimodal deep learning techniques for biological data fusion, the research has some significant drawbacks. It is limited in its applicability to performance evaluation because it excludes quantitative comparisons between approaches and rigorous benchmarking. There is little technical depth and no emphasis on algorithmic implementation or reproducibility, as well as little help in choosing suitable fusion algorithms for biomedical applications. There is only a cursory recognition of real-world deployment issues, including infrastructure, privacy, and missing data. Moreover, the evaluation might already be out of date because of how quickly the subject is evolving, particularly with the introduction of more recent architectures like transformers. Pei et al. [13] discussed the main features of medical multimodal fusion techniques, including supported medical data, diseases, target samples, and implementation performance, and examined the effectiveness of current multimodal fusion pre-training algorithms. Furthermore, this paper outlines the primary obstacles and objectives of the most recent developments in multimodal medical convergence. It has various significant drawbacks. In contrast to providing in-depth technical insights or critical assessments of the approaches offered, it emphasizes publication trends. The lack of a defined research selection technique in the report compromises reproducibility and transparency. In addition, it offers little clinical or practical viewpoints and scant attention to how models function in actual environments. Evaluation metrics are also barely mentioned, and the section on future initiatives is vague and short. Lastly, the review feels old in some ways because it ignores some of the most recent developments in multimodal learning. Recently, Shaik et al. [14] concentrated on algorithmic techniques for managing multimodal data, including rule-based systems, feature selection, natural language processing, and data fusion using machine learning and deep learning techniques. Additionally, several smart healthcare concerns were discussed. After that, they suggest a general framework for combining multimodal medical data that is consistent with the DIKW (data to information to knowledge to wisdom) paradigm. This study overemphasizes the DIKW framework, offers little technical depth, and lacks a defined review approach. It lacks a comparative study of approaches, under-represents newer developments, such as transformer models, and offers little information about clinical integration. The future directions are imprecise and high-level, limiting the paper’s practical value.
This review makes three main contributions. First, we provide a taxonomy of multimodal learning strategies based on recent architectural developments that are specific to the medical field. Second, we evaluate the application of these techniques in real-world clinical settings, ranging from genetics to radiology, emphasizing examples where they have produced quantifiable gains in diagnostic performance. Third, we point out remaining challenges and suggest future lines of studies, most with a focus on interpretability, standardization, and ethical issues, to steer the creation of reliable, implementable solutions. We hope to accomplish this by providing a unique and practical viewpoint that transcends traditional literature reviews, thereby bridging the gap between clinical utility and machine learning innovation.
The rest of this review is organized to make it easier to navigate. Section 2 provides basic information and fundamental ideas, including important architectural frameworks and unimodal and multimodal techniques in medical diagnostics. Section 3 explains the different kinds of medical data used in multimodal learning. Section 4 examines the latest deep-learning methods used in medical diagnosis. Section 5 discusses the methods used to integrate multimodal data. Section 6 demonstrates how these techniques are used in actual healthcare settings. Section 7 discusses the main issues facing multimodal deep learning is facing, and Section 8 proposes possible fixes. Lastly, Section 9 describes upcoming research avenues and new developments in this rapidly changing field.
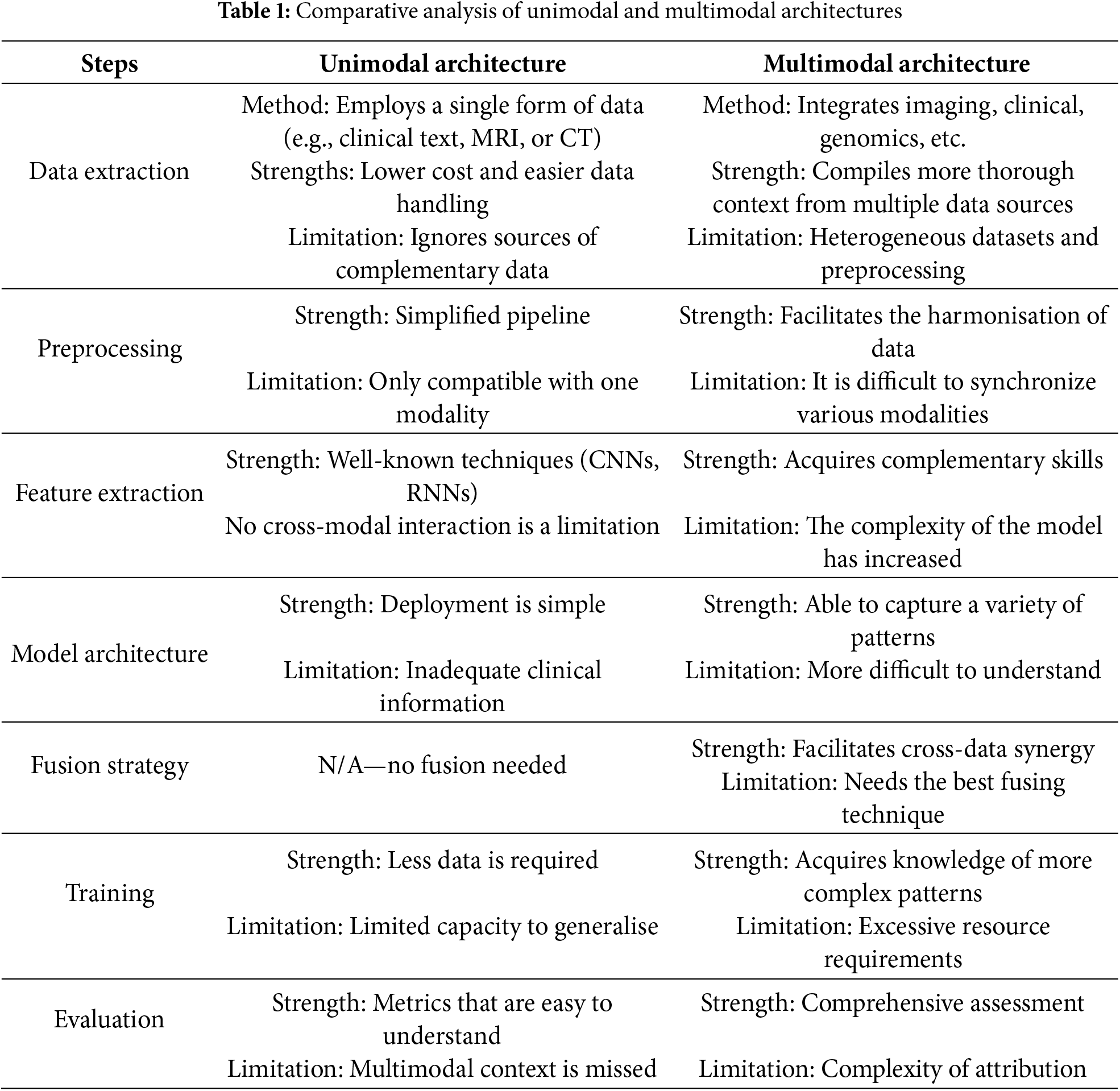
2.1 Unimodal in Medical Diagnostics
Unimodal medical diagnostics refers to the usage of a single diagnostic or imaging technique for the evaluation and diagnosis of medical disorders. This method has been a mainstay of healthcare for many years, and various approaches offer insightful information about patient health. Many unimodal diagnostic techniques are frequently employed in clinical settings, including computed tomography (CT), magnetic resonance imaging (MRI), ultrasound, and X-rays [15]. Physicians can see inside structures and spot problems using these imaging methods without undergoing invasive treatments. These unimodal diagnostic methods’ capabilities have been greatly increased in recent years using artificial intelligence (AI) and machine learning [16]. For example, deep neural networks (DNNs) have demonstrated state-of-the-art performance in image classification tasks, offering doctors diagnostic assistance when examining medical images [17].
2.1.1 Performance and Applications
In medical diagnostics, unimodal models are frequently employed, especially for imaging-based tasks like pulmonary abnormality prediction, breast mass categorisation, and knee osteoarthritis detection. For instance, knee osteoarthritis has been diagnosed using only imaging data using deep learning models such as InceptionV3 and EfficientNetv2, which have demonstrated remarkable accuracy (up to 0.75 for 3-class severity classification) and, in certain situations, outperform more intricate multimodal models when only imaging features are considered [18]. In a similar vein, unimodal machine learning systems that use ultrasonic features have demonstrated performance in breast mass categorisation that is on par with human experts (AUC 0.82–0.84) [19].
Unimodal diagnostic models, which rely on a single data source, offer certain methodological and practical advantages, such as simplicity, lower computational cost, and easier implementation. The main advantages of these systems are their ease of use and computational effectiveness. Because these models do not require the synchronization or integration of numerous data sources, they are typically simpler to build and implement than multimodal systems [19]. Unimodal techniques are therefore frequently less computationally demanding, making them appropriate for environments with constrained computational resources. Their great performance at baseline is another asset. Unimodal models can compete with or even surpass multimodal systems in situations where the selected modality is informative, such as high-resolution imaging in radiology or unique molecular fingerprints in genomics [20]. This is particularly true for well-characterized disorders where most of the diagnostic information required is extracted by a single data type. Additionally, unimodal diagnostics are more appealing due to their practicality and clinical relevance. Most clinical workflows in use today are built on single-modality data, including histopathology slides, blood tests, or radiography pictures [21]. Therefore, unimodal models can be easily incorporated into current diagnostic workflows without requiring significant infrastructure modifications.
Although unimodal diagnostics offer many benefits, they also have certain drawbacks [22,23]. Their limited scope of knowledge is a major concern. These models might overlook supplementary information that could improve diagnostic accuracy if they only used one data modality, particularly in complicated or heterogeneous circumstances. For instance, a more thorough understanding of disease pathophysiology can be obtained by integrating imaging data with genetic or clinical information. However, limited generalizability presents another difficulty. When the modality is absent or distorted, unimodal models may not work as intended or are frequently sensitive to data quality changes. This dependence on a single source may reduce robustness, particularly in clinical settings where data are noisy or lacking. Finally, accuracy and false-positive rates are important issues. Compared to multimodal systems, which are better able to cross-validate signals across data types, unimodal biomarker-based diagnostics have been linked to increased false-positive rates and decreased precision in fields such as oncology. This restriction may cause patients to feel anxious and require needless procedures.
However, unimodal diagnostics have certain drawbacks. A more thorough diagnostic approach is frequently necessary because of the complexity of many medical diseases. As a result, multimodality imaging approaches have been developed, combining data from many imaging modalities to provide a more comprehensive view of a patient’s state [24]. Although multimodal approaches are becoming increasingly popular, continuous research is still improving unimodal diagnostic methods. For instance, single-modality diagnostic tools are becoming more sensitive and specific due to developments in quantum biosensors [25].
2.2 Multimodal in Medical Diagnostics
Multimodal medical diagnostics is a new field that integrates data from several data sources and imaging modalities to provide a more thorough understanding of patient situations and increase diagnosis accuracy. This method offers a synergistic effect in clinical diagnosis and medical research by utilizing the complementary nature of several imaging modalities and data kinds [26]. Combining many modalities, including pathological slides, radiological scans, and genomic data, enables a more comprehensive understanding of the patient’s situation. Recent developments in artificial intelligence, including deep learning-based methods for multimodal fusion, have greatly improved multimodal medical diagnostics [27]. Research has demonstrated, for example, that AI models such as GPT-4V can attain greater diagnostic accuracy when given multimodal inputs as opposed to single-modality inputs [28]. Curiously, multimodal medical diagnostics encompasses more than just conventional imaging modalities. New technologies like upconversion nanoparticles (UCNPs) are being investigated for their potential use in targeted therapies and multimodal cancer imaging [26]. Furthermore, chances to further advance precision oncology beyond genomics and conventional molecular approaches are presented by the combination of improved molecular diagnostics, radiographic and histological imaging, and coded clinical data [29]. Because of their increased precision and dependability, multimodal medical diagnostics can be considered a potential new area in healthcare. Still, there are obstacles to overcome, such as the requirement for strong image fusion algorithms, the management of partial multimodal data, and the resolution of privacy and ethical issues [30].
Multimodal architecture typically follows a structured pipeline, starting with feature extraction from several data modalities, including text, audio, pictures, and sensor inputs. This includes unstructured text (such as clinical notes and chief complaints), structured clinical information (such as laboratory findings and patient demographics), and imaging data (such as MRI, CT, PET (positron emission tomography), and ultrasound) in medical applications [31]. Word embedding models use textual analysis; however, convolutional neural networks (CNNs) are frequently used for visual and audio data [32]. For instance, combining genetic, pathological, radiological, and clinical data in the diagnosis of cancer offers a thorough description of disease phenotypes [33].
Following initial data extraction, each modality is subjected to specific preprocessing: structured and unstructured textual data are tokenized or converted into embeddings, whereas imaging data are usually normalized and segmented. The use of embedding layers in advanced deep learning architectures allows for the cooperative processing of visual and textual tokens in later stages by transforming inputs, including text and images, into a single representation space.
Data fusion is a crucial stage in multimodal pipelines that combine diverse information sources to create richer representations. There are three different levels of fusion: data-level fusion, which combines low-level features or raw data early on; feature level fusion, which incorporates modality-specific features into neural network architectures, frequently using transformer or convolutional layers; and decision-level fusion, which combines outputs from independently trained models, usually using ensemble techniques or voting strategies. Research has demonstrated that feature-level fusion often outperforms late fusion methods, especially in deep-learning models. Furthermore, other frameworks use methods like deep unfolding operators, which incorporate sparse priors and structured learning principles into the network design to include domain knowledge [34]. Feature extraction, fusion, and classification are becoming less distinct in modern multimodal systems, leading to unified designs that capture inter- and intra-modal interactions. Transformer-based models and convolutional neural networks (CNNs) are both commonly used; the latter is particularly good at joint multimodal representation learning using attention mechanisms. Complex anatomical structures and specialized designs, such as Multi ResU Net, have shown improved biomedical image segmentation performance. Furthermore, by including medical expertise at different stages of the model, knowledge-augmented networks can significantly increase diagnostic accuracy [35]. These models must be trained on extensive datasets with annotations and inputs that are synchronized across modalities. Standard criteria, including accuracy, sensitivity, and the area under the receiver operating characteristic curve (AUC), were used to assess performance, and multimodal models routinely outperformed baseline models with only one modality. The recent change toward unified multitask architectures that can manage issues like noisy data, missing modalities, and privacy concerns recent trend. Methods like hybrid secure models and deep hashing are being investigated to improve multimodal systems’ security, generalisability, and resilience [36,37].
To ensure stable cross-modal associations, recent research emphasizes cross-modality techniques that align and synthesize data across modalities. Cross-modality learning, such as Cross-Modality Optimal Transport (CMOT), aligns disparate modalities into a shared latent space, enabling more reliable classification and missing modality inference in complex settings like cancer and cell-type identification [38]. Synthesis methods address data incompleteness by generating missing modalities from available ones [39]. Frameworks like MultiFuseNet, which integrate multiple screening test modalities, have proven effective in cervical dysplasia diagnosis [40], while MADDi employs cross-modal attention mechanisms for high-accuracy differentiation in Alzheimer’s disease stages [41]. Stability is further reinforced through methods like quaternion-based spatial learning and deep co-training, which improve segmentation performance across poorly annotated modalities [42,43]. Additionally, empirical approaches such as multimodally-additive function projection (EMAP) are used to determine whether improvements stem from true cross-modal interactions or dominant unimodal contributions [44], reinforcing the importance of evaluating and sustaining robust cross-modal coherence across all stages of multimodal processing. Table 1 presents a comparative analysis of unimodal and multimodal architectures, highlighting their respective strengths, limitations.
3 Overview of Medical Data Types
Multimodal deep learning extends these advantages by integrating data from multiple sources, including EHRs, radiological imaging, histological/pathological imaging, and omics data, as illustrated in Fig. 1. These modalities include raw, unstructured data that are unique to their formats. The data is processed using feature extraction techniques applied to medical imaging, wearable device data, and structured EHRs to generate valuable clinical insights. In the context of medical diagnostics, several major data modalities contribute to this process.
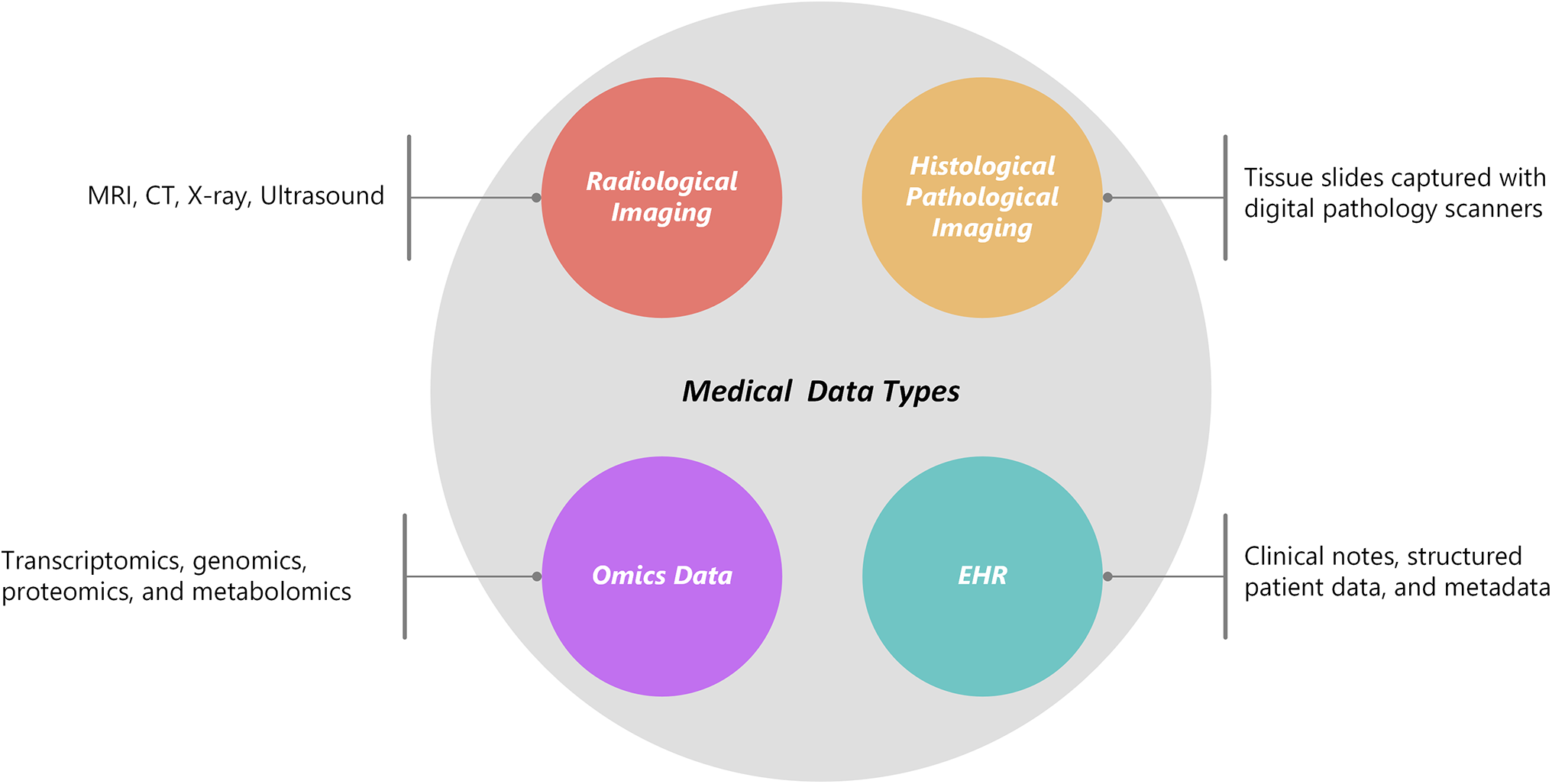
Figure 1: Medical data types
Medical 3D volumetric images are usually created from a stack of 2D slices with a specified thickness, representing a specific region of interest within the body. These individual slices can be processed and analyzed separately (as 2D images) or collectively (as 3D volumes) to extract vital information. Most medical imaging data are stored as 2D image slices in the Digital Imaging and Communications in Medicine (DICOM) format after acquisition [45]. This includes patient metadata, imaging procedure information, devices used for image acquisition, and imaging protocol settings. Radiological imaging could include MRI, CT, X-ray, or ultrasound.
X-rays are 2D grayscale radiograph pictures by nature. Five levels of attenuation can be distinguished using conventional radiography: air, fat, soft tissue, bone, and metal. The air looks dark on radiographs because most X-rays may flow through it because of its density, while much denser metal appears dazzling white because it absorbs most of the energy from the X-ray beam. The idea behind X-ray imaging is that different types of bodily tissues attenuate X-rays differently [46]. Different colors of gray are displayed for fat, soft tissue, and bone; fat is darker than soft tissue, and bone is lighter. However, X-rays are often used as a screening technique because they do not provide sufficient spatial depth information for definitive diagnosis. X-rays have been used in numerous recent studies for cardiology prediction tasks, including auxiliary conduction circuit analysis, pulmonary edema assessment [47], and cardiomegaly identification [48].
CT stands for Computed Tomography. CT scans are a common option for medical diagnostics because they provide high-resolution imaging, accessibility, affordability, and speed. The ability of ionizing radiation to differentiate soft tissues and expose patients to ionizing radiation [49]. The human body’s detailed cross-sectional images can be obtained from Computed Tomography (CT) scans [50]. These scans use radiographic projections obtained from various angles to reassemble several successive 2D slices, creating 3D picture volumes. CT is a flexible imaging method that is mostly used to find structural anomalies, to find tumors, to diagnose cardiac issues, and to image the brain for different neurological disorders. It is frequently used in cancer diagnosis [51], therapy planning [52], respiratory therapy [53], and cardiovascular research [54].
MRI facilitates tissue-specific reconstruction by measuring magnetization in both the longitudinal and transverse directions [55]. Without ionizing radiation, this method produces fine-grained images of internal structures, soft tissues, and organs. Numerous brain illnesses, such as Parkinson’s disease [56], multiple sclerosis [57], and Alzheimer’s disease [58], can be studied using this method.
Doppler techniques, which provide useful color overlays on grayscale images, are frequently used to observe blood flow and evaluate velocity [59]. Ultrasound is a preferred option for obstetrics and gynecology because of its noninvasiveness and absence of ionizing radiation [60]. And check numerous organs (liver, kidneys, etc.) [61] for possible problems. In addition, ultrasonography is crucial for monitoring the course of diseases and directing exacting surgical operations [62].
3.2 Histological/Pathological Imaging
Tissue slides were captured with digital pathology scanners. Histological and pathological imaging involves the examination of tissue samples under a microscope. These tissue slides are now digitally transformed into high-resolution images because of the development of digital pathology scanners, which provide sophisticated analysis, sharing, and storage.
3.3 Omics Data: Transcriptomics, Genomics, Proteomics, and Metabolomics
Omics technologies are a collection of high-throughput techniques used to investigate biological molecules at the system level. Proteomics analyses protein profiles, metabolomics maps small-molecule compounds, transcriptomics analyses RNA expression, and genomics decodes DNA sequences [63]. Therapeutic medications and possible risk protein biomarkers for the prevention and treatment of cancer. The proposed study [64] combined extensive genome, transcriptomics, proteomics, and metabolomics data. Thirty-six possible druggable proteins for cancer prevention were identified in subsequent investigations. Furthermore, a review of more than 3.5 million electronic health records revealed medications associated with either a higher or lower risk of cancer, providing new information for treatment approaches.
3.4 Electronic Health Records (EHRs)
EHRs are digital repositories that contain patient health data, such as imaging reports, lab findings, clinical notes, and demographic metadata. EHRs have emerged as a key component of contemporary healthcare, facilitating research, population health management, and data-driven decision-making. EHRs contain a multitude of information, both structured (such as vital signs and diagnoses) and unstructured (such as free-text notes) [65].
By aligning these heterogeneous data types, multimodal deep learning models aim to improve diagnostic accuracy and uncover complex disease mechanisms. Techniques vary widely, from early fusion (e.g., concatenating feature vectors) to late fusion (e.g., combining model output).
These various data types require efficient integration techniques that are becoming increasingly prevalent in contemporary multimodal datasets. Representative examples are presented in Table 2.
4 State of the Art in Deep Learning Techniques for Medical Diagnostics
Deep learning’s amazing capacity to extract hierarchical representations directly from raw and high-dimensional data has greatly benefited healthcare research. This paradigm change has made it easier to create systems that can assess complicated medical data in a highly accurate, automated, and scalable manner. In addition to improving diagnostic and prognostic activities, deep learning’s incorporation into the healthcare industry has set the stage for intelligent decision support systems. A comparative review of previous research using deep learning in medical diagnostics is presented in Table 3, which also highlights the methodological frameworks, goals, and inherent constraints identifying the model type of each study.
Three key advantages of deep learning, automated feature extraction, scalability, and transfer learning, are primarily responsible for their efficacy in medical applications. These benefits have made deep learning architectures indispensable for handling the growing complexity of contemporary healthcare data. Automated feature extraction eliminates the need for handcrafted features and enables deep neural networks to extract important patterns from raw data input. The hierarchical structure of these models makes it easier to learn progressively more abstract representations, leading to a deeper comprehension of the underlying facts [73]. Scalability is one of deep learning’s other main advantages in the medical field. Large-scale, heterogeneous medical datasets can be used to train deep models due to the availability of high-performance computing infrastructure. This potential has sparked the creation of hybrid deep learning frameworks that can effectively handle high-dimensional data while simultaneously tackling issues like computational security, data privacy, and interoperability in medical settings [74]. Transfer learning has increased deep learning’s applicability even more, especially in scenarios with limited labeled medical data. Researchers can optimize these architectures for particular healthcare problems using models that have already been trained on sizable general-purpose datasets. This improves performance and speeds up convergence [75]. When combined, these key components, scalable model development, automated feature extraction, and the thoughtful application of transfer learning, have established deep learning as an essential instrument in medical research. The creation of next-generation medical technologies that are not only more precise and effective but also better able to adjust to the intricacies of actual clinical settings depends on these developments. Building on this fundamental summary of deep learning’s influence in healthcare, a more thorough analysis of the core architectures, such as Transformers, Recurrent Neural Networks, and Convolutional Neural Networks, is necessary to comprehend their unique contributions, advantages, and uses in the medical field.
4.1 Convolutional Neural Networks (CNN)
Deep learning has greatly improved medical diagnoses by increasing the precision and effectiveness of image processing, especially when CNNs are used. CNNs have transformed medical image analysis and diagnostics. CNNs have outperformed traditional computer-aided detection (CAD) systems in various tasks, including segmentation, object detection, and image classification [76]. CNNs can automatically learn complex image features, removing the need for manually engineered feature extraction, a major advantage over traditional machine learning techniques [77]. CNNs have been used in a variety of imaging modalities in medical diagnostics, such as MRI, CT, X-ray, and histopathology. The excellent accuracy of CNNs in analyzing medical pictures may help radiologists and physicians make more accurate and timely diagnoses [78]. CNN’s performance in image recognition tasks, as exemplified by models like AlexNet and GoogleNet, which have been successfully applied to medical images, has fueled their popularity in medical diagnostics [79]. These networks have played a key role in helping doctors make more accurate diagnoses by automating the examination of complicated medical images [80]. CT scans are used to detect and segment pelvic and omental lesions in patients with ovarian cancer [81].
More recent works have combined radiology images with text data (radiology reports) to augment understanding for Multimodality. The researchers proposed a framework [82] combines survival prediction, clinical variable selection, and 3D CNN-based feature extraction for the prognosis of renal cell cancer. It uses a deep learning model with Logistic Hazard-based loss for survival prediction, chooses clinical variables using Spearman and random forest scores, and predicts tumor ISUP grades from CT images. For best results, variable selection is fine-tuned through nine experiments. Other researchers [83] produced an improved CNN model is presented that overcomes data fusion and feature extraction restrictions to better multimodal medical image segmentation. Other studies [84] have combined MRI and CT imaging to create deep learning-based diagnostic models for osteoporosis prediction. Utilizing both unimodal and multimodal strategies. To construct the findings part of radiology reports, radiological images [4] and patient indication text in a multimodal strategy for automatic report generation were integrated using chest X-ray (CXR) imaging.
Digital pathology involves extremely high-resolution whole-slide images (WSIs). CNNs and attention-based methods have been used to localize regions of interest, with further integration of patient metadata for contextual interpretation. Researchers [85] examined the connection between Tumor mutational burden (TMB) clinical variables, gene expression, and image features by analyzing histopathological pictures, clinical data, and molecular data from The Cancer Genome Atlas (TCGA). To go beyond conventional unimodal methods for multimodal breast cancer diagnosis [86]. The researchers investigate the integration of histopathological pictures with non-image data. Enhancing diagnostic accuracy, clinician confidence, and patient involvement, the study highlights the significance of transparent AI decision-making by utilizing Explainable AI (XAI). Researchers combined genomic data [87] with histopathological images, a multimodal CNN-ensemble method for early and precise pancreatic cancer identification. The model uses feature fusion techniques, deep learning survival models, and ensemble CNNs to improve tumour segmentation, classification, and survival prediction.
Joint analyses of radiological and histological data have shown improved classification and staging results in cancer diagnostics. For instance, automated tumor detection can benefit from both imaging modalities radiology provides macroscale structural context, while histology validates microscale cellular anomalies.
It’s crucial to remember that, despite their enormous potential, CNNs have drawbacks. Large volumes of well-annotated training data are required, which can be costly and challenging to acquire in medical contexts. This is one major problem. To overcome this, transfer learning approaches have been investigated, in which CNNs that have already been trained on non-medical pictures are adjusted for particular medical tasks [88]. Researchers are also looking at self-supervised learning and transformer networks as ways to further enhance performance and lower data needs [89].
4.2 Recurrent Neural Networks (RNNs)
The use of deep learning methods, especially Recurrent Neural Networks (RNNs), has become essential for improving medical diagnosis. RNNs are skilled at handling multivariate time-series data, which is common in clinical environments like intensive care units (ICUs). This is especially true of those that use Long Short-Term Memory (LSTM) units. An innovative work [90] empirically assessed how well LSTMs can identify patterns in clinical parameters and demonstrated that they can categorize several diseases using only raw time-series data. According to their findings, LSTMs outperformed conventional machine learning models, providing a solid basis for the application of deep learning in medical diagnostics. The potential of deep learning approaches to forecasting violent episodes during patient admissions has been investigated in the field of psychiatric care. Their research demonstrated the superiority of deep learning over traditional techniques by using clinical text data stored in Electronic Health Records (EHRs) to achieve state-of-the-art predicted accuracy performance. The RNN-SURV model outperformed state-of-the-art methods in terms of the concordance index (C-index) in survival analysis, demonstrating superior performance in calculating risk scores and survival functions for individual patients [91]. RNNs have also shown promise in solving problems outside the mainstream of medical diagnosis. For example, a Modified Long Short-Term Memory (MLSTM) model has been constructed to predict new cases, fatalities, and recoveries in the COVID-19 pandemic setting, outperforming traditional LSTM and Logistic Regression models [92].
Increasingly, multimodal deep learning, which combines information from various medical sources, is being used to improve the precision and effectiveness of diagnosis. When used in combination with other deep learning models, recurrent neural networks (RNNs) are crucial for analyzing temporal and sequential medical data, thereby enhancing disease categorization and identification. RNNs are particularly good at interpreting sequential or temporal data, such as time-series signals (e.g., ECG) or dynamic imaging (e.g., ultrasound movies). When combined with other models, such as CNNs and autoencoders, RNNs improve temporal pattern identification and feature extraction, which is important for applications like video-based diagnostics, disease progression prediction, and cardiac MRI segmentation [93,94]. Reported prediction accuracies of up to 98% [95] have been achieved in image recognition and sequential data processing using hybrid frameworks that combine RNNs with CNNs and autoencoders.
Recurrent neural networks (RNNs) are a type of deep learning model that has shown remarkable efficacy in a variety of medical applications, most notably in tumor categorization and detection. The applications of these models to multimodal imaging modalities, like PET-MRI and PET-CT, have demonstrated their capacity to enhance diagnostic robustness and precision, enabling more precise and trustworthy tumor evaluations [96,97]. In addition, RNN-integrated multimodal fusion models have substantially enhanced the sensitivity and accuracy of disease recognition tasks, outperforming conventional single-modality methods. This enhancement reaches important domains, such as Alzheimer’s disease and heart disorders, where RNNs obtain high classification accuracy when combined with multimodal neuroimaging and clinical data [33]. These developments facilitate early prognosis and diagnosis, laying the groundwork for prompt clinical interventions. Multimodal deep learning frameworks have several advantages [98], such as improved accuracy, reduced diagnostic time and expense, and increased resilience to noise and adversarial attacks—all of which are critical in clinical contexts. However, there are still issues regarding enhancing model interpretability, refining data fusion techniques, and incorporating expert medical knowledge to increase diagnostic accuracy. To properly use deep learning models in medical diagnostics, these constraints must be overcome [99].
Graph Neural Networks (GNNs) have demonstrated a great deal of promise in improving medical diagnosis. GNNs are especially well-suited for integrating various medical data types because they effectively blend graph structure representations with deep learning’s outstanding prediction accuracy [100]. In cancer research, where data range across several dimensions, modalities, and resolutions—from digital histopathology slides and genetic data to screening and diagnostic imaging, this method is particularly helpful [101]. Graph Neural Networks (GNNs) are becoming a crucial tool for combining and evaluating these multimodal datasets, providing deeper insights and increased accuracy, particularly in intricate domains like neurodegenerative illnesses and oncology. Deep reinforcement learning (DRL) combined with GNNs has further increased the potential of models for use in medical diagnostics. This combination can lead to more reliable and accurate diagnostic tools by strengthening the application of GNNs and improving the formulation of DRL [102] For example, an AI-powered model that uses neural network optimization, multilevel thresholding, and image preprocessing has 92% accuracy in classifying various forms of brain tumors [103]. Incorporating protein-protein interaction networks to combine omics data with imaging features. The researchers introduced [104] the integration of multi-omics data in biomedical research using graph-based machine learning techniques, namely graph neural networks (GNNs). Multi-omics techniques, whether used at bulk or single-cell resolution, aid in finding biomarkers, predicting treatment response, and gaining a mechanistic understanding of cellular and microenvironmental processes. Multi-omics information [105], such as transcriptomics, proteomics, epigenomics, and genomics, provide a thorough understanding of cellular signaling pathways. Because they can naturally integrate and represent multi-omics data as a biologically meaningful multi-level signaling graph and interpret multi-omics data using graph node and edge ranking analysis, graph AI models, which have been widely used to analyze graph-structure datasets, are perfect for integrative multi-omics data analysis.
In summary, multimodal deep learning using GNNs is an effective approach for medical diagnostics due to its efficient integration and analysis of many kinds. This technology could improve patient outcomes by streamlining workflows and reducing interpretation time [106]. However, for a smooth transition into clinical practice, issues including data heterogeneity, model interpretability, and regulatory compliance must be resolved [107].
4.4 Generative Adversarial Networks (GANs)
Adversarial Generative Networks (GANs) have greatly improved medical imaging by producing realistic synthetic images for data augmentation, which has improved segmentation and classification, particularly for rare disorders [108,109]. They provide thorough diagnosis and personalized care by promoting multimodal analysis through image-to-image translation and cross-modality synthesis [110,111]. In clinical settings, GANs improve tasks like segmentation, reconstruction, and denoising in the diagnosis of diseases like Alzheimer’s and myocarditis [112,113]. One of the primary benefits of GANs is their capacity to produce realistic synthetic data in medical diagnostics, which can be applied to data augmentation and to solve the problem of medical imaging’s sparse datasets [114]. This is especially helpful when training AI-based computer-aided diagnostic systems because performance improvement requires multiple different data types [115]. In addition, in tasks involving image augmentation, denoising, and super-resolution, GANs have demonstrated promise. These tasks can increase picture quality and lower radiation exposure in specific imaging modalities [116]. Although GANs have made impressive strides in medical imaging applications, obstacles remain. More dependable and consistent outcomes require addressing problems such as mode collapse, non-convergence, and instability during training [117]. Additionally, it is crucial to guarantee that GANs learn the statistics essential to objective picture quality assessment and medical imaging applications [118]. As this area of study develops, GANs could transform medical diagnostics by facilitating more precise and effective picture synthesis, analysis, and interpretation in a variety of modalities [119].
Transformer-based models use embedding layers and attention processes to transform different inputs (such as text, images, and structured data) into cohesive representations for multimodal data analysis. These models provide a comprehensive comprehension of patient data by learning both intra- and intermodal interactions. For instance, models that combine laboratory results, clinical histories, and radiographs into a single diagnostic framework using visual and text tokens and bidirectional attention blocks outperform models that use just one input type or analyze modalities independently [120,121]. Interestingly, some studies have explored hybrid techniques that combine the strengths of Transformers and Convolutional Neural Networks (CNNs). For example, the HybridCTrm network outperformed fully CNN-based approaches in multimodal medical picture segmentation tasks [122]. This demonstrates that using both local and global feature representations enhances performance. Among Transformer-based designs, Vision Transformers (ViT) and other devices have demonstrated exceptional performance in jobs involving medical image interpretation tasks. For example, to overcome restrictions such as the absence of cross-modal feature interaction and local feature extraction, a unique lightweight cross-Transformer based on a cross-multiaxis mechanism has been developed for multimodal medical picture fusion [123]. An ensemble method that included the ViT and EfficientNet-V2 models outperformed the standalone models in brain tumor classification, achieving an impressive 95% accuracy [124]. Leveraging self-attention to correlate genomic markers with imaging signatures. A new deep learning model called DeepFusionCDR [125] combines drug chemical structures with multi-omics data from cell lines to forecast cancer drug responses (CDRs). For analyzing the chemical structures of drugs using transformers that are specialized in SMILES. A transformer-based deep learning model called DeePathNet [126] was developed for the processing of multi-omics data in cancer research. It combines information about cancer-specific pathways to enhance subtype identification, cancer classification, and treatment response prediction.
Thus, transformer-based multimodal deep learning techniques have demonstrated notable progress in medical diagnosis. These techniques have demonstrated promise in several applications, such as the diagnosis of Parkinson’s disease, classification of brain tumors, and detection of cancer [127]. By combining various data types and using transformer structures, these methods provide better interpretability and diagnostic accuracy, as well as the possibility of optimizing clinical operations. To fully exploit these technologies in healthcare, further study and cooperation between medical professionals and AI specialists are essential as the field develops. Enhancing early Parkinson’s disease detection through multimodal deep learning and explainable AI: insights from the PPMI database.
4.6 Autoencoders and Variational Autoencoders (VAEs)
VAEs have been effectively used in biomedical informatics applications, such as large-scale biological sequence analysis, integrated multi-omics data analytics, and medical image classification and segmentation [128]. Variational Autoencoders (VAEs) have shown great promise in medical diagnostics through improved interpretability, representation learning, and multimodal data fusion. They have made it possible to grade gliomas accurately using interpretable MRI-based characteristics, and expedited the screening process for cognitive impairment using a variety of data sources [129] and enhanced the detection of early cardiac disease by combining imaging and clinical data [130]. Additionally, VAEs addressed data scarcity by producing synthetic eye-tracking data [131] and performed better than conventional approaches in deriving strong representations from metabolomics and protein data [132,133]. Additional model improvements, like adversarial training and attention processes, improved performance in tasks involving face analysis and cancer detection [134].
4.7 Explainable AI (XAI) in Deep Learning
XAI is essential for increasing the transparency and reliability of deep learning models, particularly in high-stakes medical applications. The goal of this study was to clarify the data underlying the deep learning black-box model, thereby exposing the decision-making process [135,136]. In the healthcare industry, where every choice or judgment has associated dangers, this is especially crucial. By assisting doctors in comprehending and interpreting AI-generated data, XAI procedures can increase their trust in the technology’s dependability [137]. The development of innovative techniques for COVID-19 classification models, which offer both quantitative and qualitative visualizations to improve doctors’ comprehension and decision-making, is an example of recent developments in XAI for medical applications [138]. Researchers have also investigated XAI approaches for regression models (XAIR), which tackle the particular difficulties in comprehending predictions for continuous output [139]. Researchers have proposed frameworks for identifying XAI strategies in deep learning-based medical image analysis to advance the field. These frameworks classify methods according to certain XAI criteria and anatomical location [140]. In the end, such efforts aim to create more dependable and understandable AI-driven diagnostic tools by standardizing and enhancing the use of XAI in healthcare.
In medical diagnostics, multimodal integration combines various data types discussed in Section 3, including clinical text, imaging, molecular profiles, and structured electronic health records, to improve the accuracy of diagnosis and offer a thorough grasp of patient health [152]. These modalities [153] include radiological and histological images, omics data (e.g., proteomics, genomes), structured variables from electronic health records, unstructured clinical narratives, and even audio or video recordings of clinical conversations. Multimodal integration can be further extended in smart healthcare systems to incorporate contextual and behavioral data that reflect environmental and lifestyle factors. Combining such diverse data sources allows for uncovering hidden patterns, correlations, and interdependencies essential for identifying risk factors, predicting disease progression, enhancing treatment regimens, and implementing preventative measures. The deep learning methods described in Section 4 enable the integration of these various modalities by providing scalable frameworks for learning joint representations and facilitating end-to-end predictive modeling. Typically, integration involves modality-specific feature extraction (e.g., Random Forests for structured data, CNNs or vision transformers for images, and transformer-based encoders for clinical text), followed by fusion algorithms that align and merge the results. Table 4 classifies and summarizes contemporary fusion approaches based on their underlying processes and medical applications, while Fig. 2 illustrates the advantages and disadvantages of each fusion type (data-level, feature-level, and decision-level). In certain applications, the uncertainty present in real-world clinical data is managed through probabilistic reasoning (e.g., Bayesian models) [154]. Despite the revolutionary promise of multimodal systems, there are certain challenges, such as temporal misalignment, data heterogeneity, and the lack of standardized validation frameworks [155]. Additionally, recent research has indicated that performance improvement does not necessarily involve simply adding more modalities. For instance, ChatGPT-4V performed worse on diagnostic tasks than its text-only counterpart, despite having access to both visual and textual input [156]. Unlocking the full potential of multimodal diagnostics in clinical settings requires addressing these limitations [157]. This section describes the primary types of multimodal fusion: data-level, feature-level, hybrid fusion, decision-level, and model-based fusion, and provides a thorough analysis of each implementation strategy.
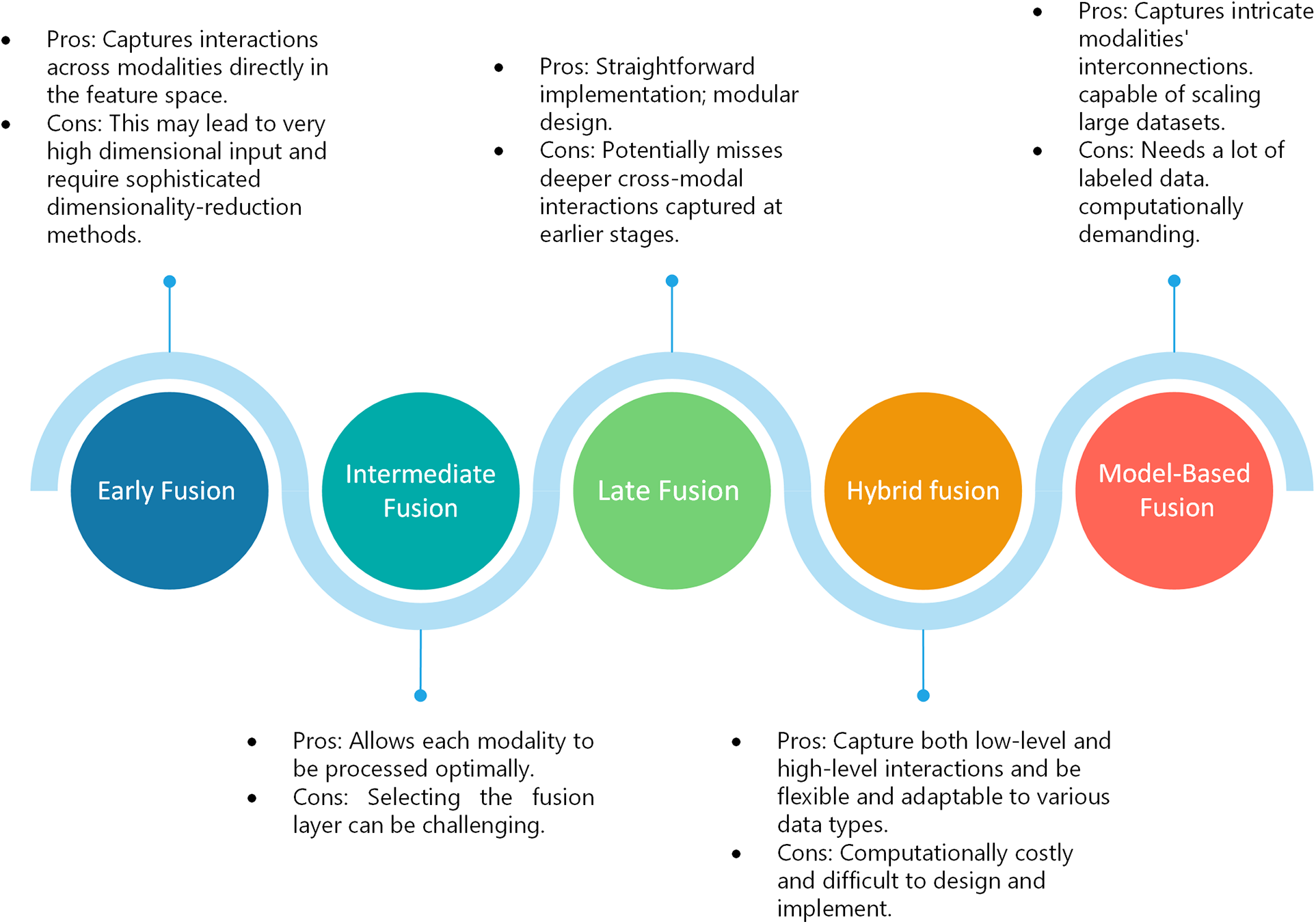
Figure 2: Multimodal fusion types
5.1 Early Fusion (Feature-Level Fusion)
Raw data or extracted features from multiple modalities are combined early.
Modality-specific networks process each input type separately, producing latent representations. These representations are merged at an intermediate layer. The study [158] proposed a thorough analysis of state-of-the-art methods and a complex classification scheme that allows for a better-informed choice of fusion strategies for biological applications, as well as an investigation of novel approaches. Kumar et al. [159] proposed a fusion technique that combines information or characteristics from many modalities to produce improved images. This is consistent with intermediate fusion, which combines the properties of modalities after they are initially processed independently. Manifold learning-based dimensionality reduction [160] was introduced in an intermediate multimodal fusion network. Using 1D-CNN and 2D-CNN, the multimodal network creates independent representations from biometric inputs and facial landmarks. A multi-stage intermediate fusion method for classifying NSCLC [161] subtypes from CT and PET images are presented. The proposed method employs voxel-wise fusion to use complementary information across different abstraction levels while maintaining spatial correlations, integrating the two modalities at different phases of feature extraction.
5.3 Late Fusion (Decision-Level Fusion)
Individual models produce modality-specific predictions or embeddings, which are combined to reach a final decision. Using the late fusion technique [162] clinical information and CT images are combined to diagnose chronic kidney disease (CKD). The model achieves accuracy comparable to that of a human expert and shows promise as a trustworthy diagnostic tool for medical practitioners by independently analyzing modalities and combining them at the decision level. To enhance the detection and diagnosis of heart disorders, Ref. [163] introduces a model that integrates 12-lead ECG imaging data and EHR data. By addressing the drawbacks of using electrocardiogram (ECG) data alone, which might not be definitive in predicting cardiac normality and abnormality, the proposed late fusion strategy aims to attain improved accuracy in the classification of cardiac diseases compared to unimodal approaches. FH-MMA [164] combines relational, sequential, and image information at the decision level using late fusion, a privacy-preserving, multimodal analytics framework. The diagnostic accuracy, computational efficiency, and scalability of FH-MMA can be significantly increased using FLE and attention methods to investigate and contrast multimodal fusion approaches, with an emphasis on late fusion [165] in the context of cancer research. The late fusion technique is used. It continuously beats unimodal models by combining data from several modalities, demonstrating the potential of multimodal fusion to enhance patient outcome forecasts. A thorough comparison of data fusion techniques in smart healthcare [166] highlights the importance of seamless integration and analysis of diverse healthcare data. They used three types of fusion: early, intermediate, and late.
Integrate early, intermediate, and late fusion techniques to exploit their advantages. The VAEs [167] provide a common latent space in which both structured and image data are represented. The squeeze-and-Excitation block (SE-Block) and Convolutional Block Attention Module (CBAM) attention processes ensure that, during fusion, the most essential aspects of both modalities are highlighted. Transformer encoders enhance the structured data representation, making it easier to integrate with picture data. The MDL-Net [168] integrates the disease-induced region-aware learning (DRL) and multi-fusion joint learning (MJL) modules to improve the early determination of brain areas linked to Alzheimer’s disease (AD) and provide an accurate and comprehensible diagnosis. The MDL-Net was created to address interpretability problems in multimodal fusion and inherent diversity among multimodal neuroimages. Improve feature representation using the latent space and local and global learning. Golcha et al. [169] introduced an enhanced health monitoring system that combines feature-level fusion and decision-level fusion to overcome the drawbacks of single-modal systems that improves patient quality of life, reduces healthcare expenses, and transforms the management of chronic diseases. A novel hybrid pre-processing method [170] called Laplacian Filter + Discrete Fourier Transform (LF + DFT) was proposed to improve medical images before fusion. This method efficiently detects significant discontinuities and adjusts image frequencies from low to high by emphasizing important details, capturing minute details, and sharpening edge details. To integrate multimodal EHR data, a hybrid fusion [171] is used, which combines early fusion, joint fusion (intermediate fusion), and late fusion. This method handles the heterogeneous nature of EHR data and enhances clinical risk prediction by utilizing the advantages of various fusion procedures.
It takes advantage of sophisticated models to implicitly merge modalities, such as transformers and graph neural networks. To overcome the difficulties associated with multimodal fusion in healthcare, the proposed model-based fusion architecture [172] uses multiplexed graphs and graph neural networks (GNNs). The proposed system provides state-of-the-art performance on benchmark and clinical datasets by adaptively modeling complicated interactions between modalities via embedding the fusion process within the GNN architecture. A Neural Architecture Search (NAS) [173] the method is presented in the AutoFM framework to automatically create the best model architectures for multimodal EHR data. This method demonstrates how model-based fusion can improve healthcare services through deep learning while reducing the dependency on manually created models.
Selecting an appropriate fusion strategy often depends on the complexity, dimensionality, and correlation structure of the modalities involved. In clinical practice, late fusion is common due to simpler model interpretability and the feasibility of using existing modality-specific analysis pipelines.
5.6 A Comparison of Recent Modern Multimodal Models (2023–2025)
Several multimodal designs are state-of-the-art (SOTA) in various medical diagnostic sectors. Table 5 presents a carefully selected collection of clinical use cases from recent studies that illustrate the real-world significance of multimodal deep learning in healthcare. These examples include various activities across diseases, such as breast cancer, lung cancer, and interstitial lung disease, including early detection, categorization, and survival prediction. Each case illustrates the increasing influence of multimodal AI in real-world medical conditions by providing the fusion method employed, performance data, and reported results. Cahan et al. [174] developed an intermediate fusion model for pulmonary embolism prognosis using TabNet and bilinear attention. It achieved an AUC of 0.96 with 90% sensitivity and 94% specificity, demonstrating performance improvement by integrating structured and picture data. For breast cancer classification, Hussain et al.’s late fusion SE-ResNet50 + ANN framework achieved an AUC of 0.965, which was significantly higher than that of unimodal baselines [175]. Similarly, Huang et al. outperformed professional radiologists with an accuracy of 88.5% and an AUC of 0.957 in their demonstration of a residual learning and Multilayer Perceptron (MLP) attention model for lung cancer invasiveness prediction [176]. The ILDIM-MFAM model for interstitial lung disease diagnosis was created by Zhong et al. [177] by combining CNNs, Bi-LSTMs, and Transformer blocks to improve the F1-score and AUC while preserving a low computational cost appropriate for clinical use. To predict pan-cancer survival, Gao et al. [178] presented an interpretable bridging fusion model that was successful in missing modality settings and validated across 12 cancer types. To further explore the adaptability of fusion, Kumar and Sharma [179] presented a late fusion CNN framework tailored for the study of liver, lung, and breast cancer, with respective AUCs of 0.92, 89% accuracy, and F1-scores of 0.87. Noaman et al. [180] applied early hybrid CNN fusion to histological pictures and clinical metadata for early lung cancer detection and obtained a sensitivity of 94%, specificity of 91%, and AUC of 0.95. In a related field, Yao et al. [181] used a late fusion model that included Vision Transformers and Natural Language Processing (NLP) modules to merge radiological imagery with EHR text. They achieved an AUC of 0.90 for breast cancer and an accuracy of 91% for lung cancer. Atrey et al. [182] achieved 93% accuracy, 90% sensitivity, and 92% specificity for breast cancer using residual neural networks and conventional machine learning classifiers in conjunction with early fusion between ultrasound and mammography data. To classify lung and colon cancers, Uddin et al. [183] developed an intermediate fusion technique based on EfficientNet and ResNet, which produced AUC scores of 0.94 and 0.96, respectively. Furthermore, Sharma et al. [184] combined ResNet and DenseNet to create a knowledge transfer-driven ensemble framework that achieved 96% accuracy and an AUC of 0.97 for the delineation of lung cancer. Lastly, Zhang et al. [185] achieved 92% sensitivity and an AUC of 0.94 for the identification of early-stage lung cancer by combining CNN-based CT imaging with liquid biopsy data in a late fusion scheme.

Collectively, these models demonstrate the rapid progress and efficacy of multimodal fusion in improving diagnostic accuracy. They demonstrated that the fusion approach (early, intermediate, or late), integration depth, and application of sophisticated architectures, such as transformers and attention mechanisms, significantly impact performance benefits. Thus, these studies indicate a substantial advancement toward high-performance, interpretable, and clinically feasible AI solutions for medical diagnostics.
Healthcare is transforming due to deep learning (DL), which enables more precise diagnosis, focused therapies, and improved patient outcomes. DL’s use extends beyond algorithmic implementation, requiring careful consideration of its potential therapeutic applications. Fig. 3 shows a variety of healthcare use cases influenced by DL techniques.
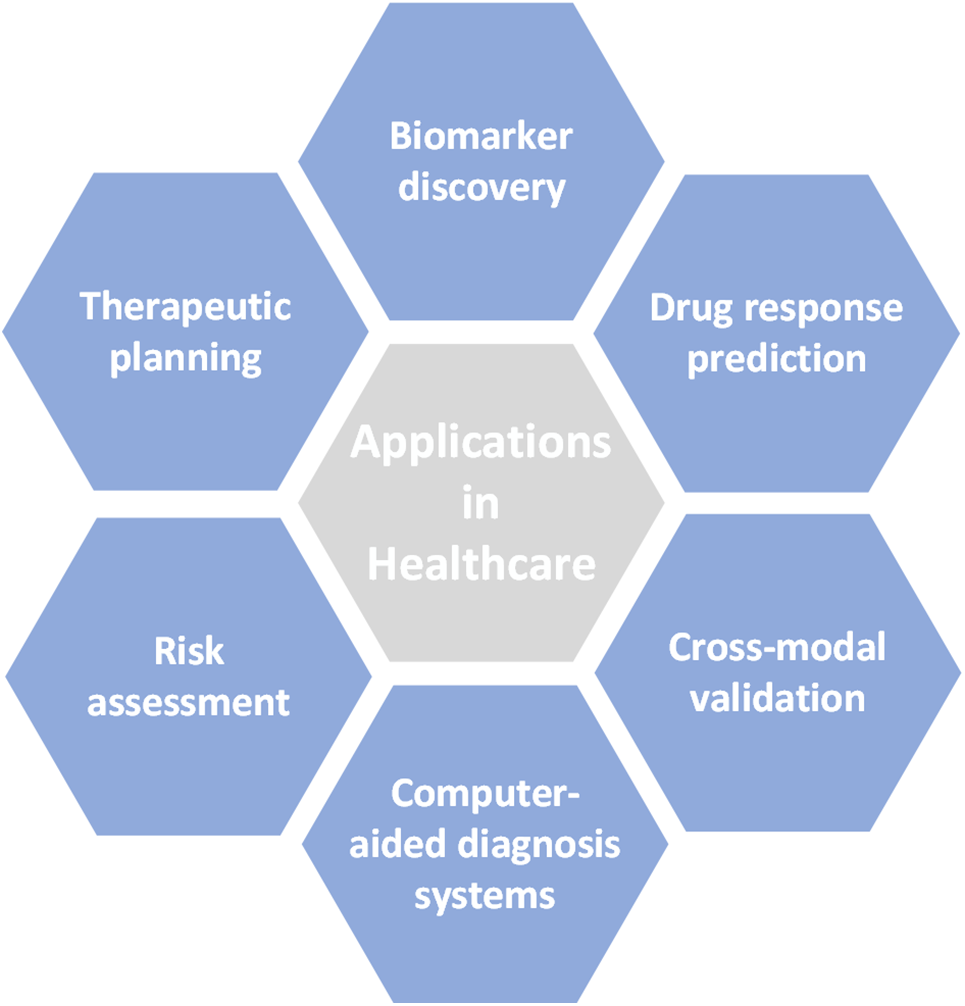
Figure 3: Healthcare application
6.1 Precision Medicine and Patient Stratification
The ability to stratify patients into clinically significant subgroups using DL frameworks to integrate multi-omic data has sped up advancements in precision medicine. For instance, tumors can be easily classified into molecularly different groups when radiological imaging and transcriptome profiles are combined. Prognosis, treatment choice, and therapeutic results are all significantly impacted by this classification.
DL-based integrative analysis of gene expression data and imaging-derived characteristics identified new biomarkers for early disease detection. In addition to being helpful for diagnosis, these biomarkers provide insight into the pathophysiology of diseases and may help direct the creation of focused treatments. However, the variety of patient data and the requirement for sizable, annotated datasets make it difficult to use these findings in clinical practice.
6.1.2 Drug Response Prediction
Multimodal DL models use data from gene expression, tumor imaging, and treatment histories to infer drug responses specific to individual patients. This method lessens the need for trial and testing treatment strategies, thereby improving the personalization of therapy. However, it is unclear whether these models can be applied to various populations and healthcare systems, emphasizing the necessity of cross-cohort validation.
6.2 Enhanced Disease Diagnosis and Prognosis
The integration and validation of data from several modalities have greatly enhanced the identification of diseases through DL, resulting in more trustworthy clinical judgments.
DL models can improve diagnostic specificity by integrating radiological imaging and histological data, especially in neurology, cardiovascular disease, and oncology. A comprehensive approach to patient health is crucial, as demonstrated by the superior performance of these cross-modal systems compared with conventional single-modality models. However, issues such as data standardization and alignment continue to exist.
6.2.2 Computer-Aided Diagnosis Systems
Multimodal computer-aided diagnosis (CAD) systems, which integrate imaging, omics data, and patient history, have demonstrated efficacy in complicated diagnostic tasks, including chronic condition management and Alzheimer’s disease progression tracking. Although these technologies are useful resources for clinical workflows, their incorporation into practical environments requires thorough validation and clinician assistance.
6.3 Personalized Healthcare and Clinical Decision Support
Clinical workflows are changing because of DL-powered systems’ customized decision support tools, which closely match unique patient profiles.
Advanced DL models can use genetic data, medical imaging, EHR metadata, and blood test results to evaluate the risk of critical health events (such as myocardial infarction or stroke). By proactively directing healthcare interventions, these risk classification technologies can lower morbidity and mortality. Nonetheless, the interpretability and openness of these models remain significant barriers to clinical implementation.
Analyzing treatment outcomes from patients with comparable multimodal profiles allows DL to be used for personalized therapeutic planning, which recommends the optimal plan of action. Although this approach improves treatment precision, it requires thorough model training on various representative datasets to prevent biases and guarantee impartiality in decision-making.
7 Challenges in Multimodal Deep Learning
Multimodal deep learning has transformative potential in industries such as healthcare, but its broad and successful implementation is hampered by several intricate issues. Data heterogeneity and quality, computational complexity, cross-modal alignment, interpretability and explainability, and privacy and ethical issues are some of the interconnected domains that these difficulties encompass, as shown in Fig. 4. Each of these problems affects the clinical applicability, generalisability, and dependability of the produced systems, in addition to making model development more difficult.
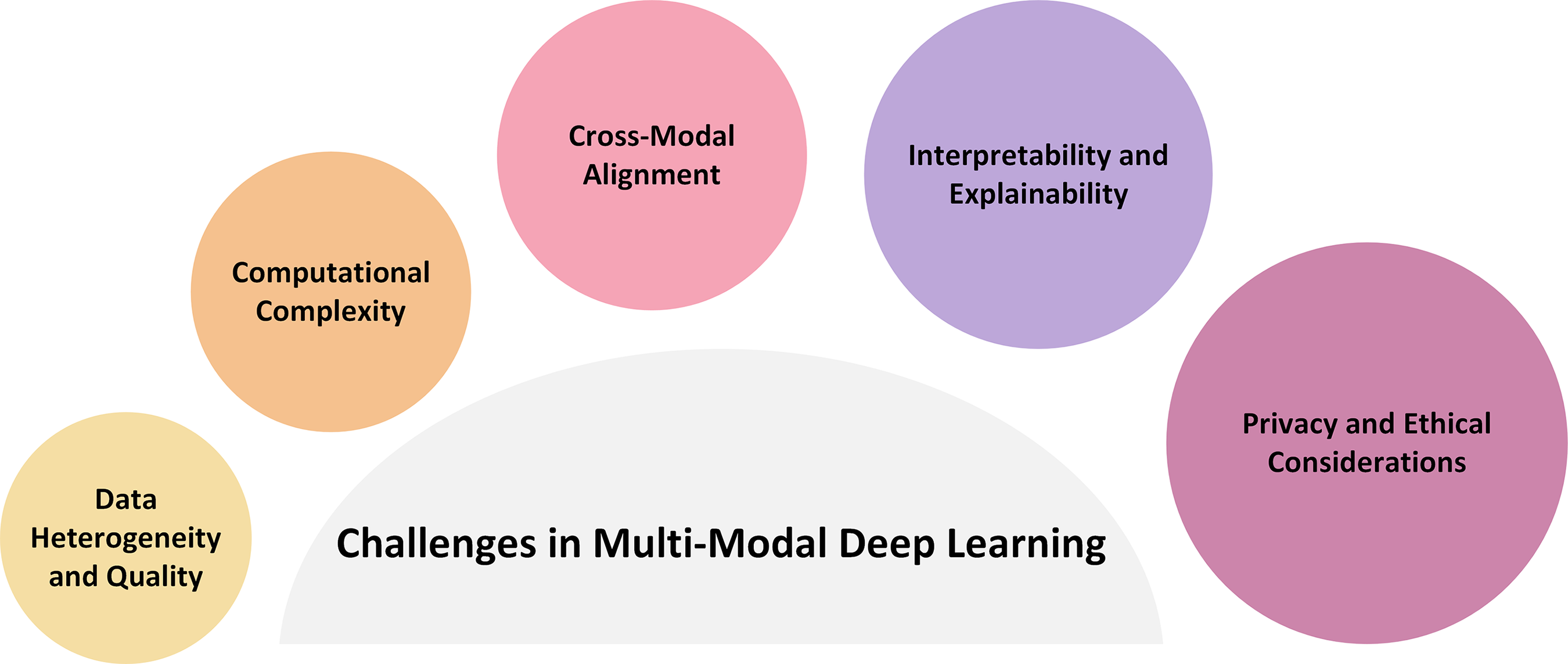
Figure 4: Multimodal challenges
7.1 Data Heterogeneity and Quality
Imaging modalities, electronic health records, and genomic sequences are only a few of the multiple sources of medical data, each with its own format, resolution, and completeness levels. For instance, confounding variability may be introduced by varied resolution in radiological imaging or batch effects in high-throughput sequencing. These discrepancies compromise the repeatability of multimodal models and increase the difficulty of data integration. This problem becomes more difficult in the absence of standard preprocessing techniques throughout organizations. In addition to technical fixes such as domain adaptation and data harmonization, overcoming these obstacles calls for cooperative efforts to create cross-institutional data standards.
An extensive amount of computational resources is available to integrate and analyze multimodal information, which frequently includes high-dimensional, large-scale data such as gigapixel histopathology images or whole-genome sequencing. When models must be trained jointly across modalities, complexity increases, which imposes a burden on processing and memory capacities. Although distributed learning frameworks and technology advancements like GPU/TPU clusters provide some respite, the discipline still lacks commonly used techniques for scalable, resource-efficient multimodal learning. The gap between research and clinical translation may grow as a result of this obstacle for organizations with inadequate computational infrastructure.
The accurate alignment of many kinds of data is one of the most technically challenging parts of multimodal learning. For example, sophisticated modeling techniques are required to align temporal EHR data with static genetic markers or transfer pixel-level information from histopathology slides to corresponding radiographic pictures. Noise from misalignment can reduce feature fusion’s efficacy and produce less-than-ideal predictions. Although robust, generalizable ways are still being investigated, recent research has explored solutions, including contrastive learning and attention mechanisms, to improve alignment.
7.4 Interpretability and Explainability
Deep learning models are frequently criticized for their lack of transparency despite their predictive capacity. This is a critical issue in the healthcare industry because clinical decision-making necessitates accountability. Clinicians’ trust and uptake of AI solutions are hampered by black-box models. The development of explainable AI (XAI) techniques, such as saliency maps, attention visualizations, and counterfactual reasoning, is therefore clinically necessary rather than just technically necessary. More reliable and context-aware XAI approaches are required because many current interpretability techniques provide little insight into multimodal interactions and frequently falter under rigorous validation.
7.5 Privacy and Ethical Considerations
The presence of sensitive personal information in multimodal healthcare data raises serious ethical and legal issues. Although adherence to regulations like the GDPR (EU) and HIPAA (US) is crucial, it complicates data sharing and model training. There are encouraging opportunities to develop ethical models using emerging privacy-preserving methods, such as safe multi-party computation, federated learning, and differential privacy. Nevertheless, these approaches provide new difficulties, such as decreased performance, communication overhead, and problems with model convergence. Utility and privacy balance remains a hot topic of ethical and technical discussion.
8 Strategies to Overcome Challenges
To successfully traverse the complexity of multimodal biological data integration, interdisciplinary cooperation, regulatory adaptability, and ongoing technical development are essential. The following tactics offer a way to overcome important constraints, with a focus on not only execution but also the justification and anticipated results of each strategy.
Effective integration of different datasets necessitates stringent harmonization methods. Different platforms or institutions’ approaches to data collection can seriously impede downstream analysis.
• Protocol standardization is essential to guaranteeing dataset comparability. Preprocessing pipelines can reduce sources of bias or technological artifacts by incorporating domain expertise.
• Advanced normalization methods, like ComBat, are very useful in omics research to address batch effects, which are systematic non-biological fluctuations that can mask real biological signals if left unchecked. These techniques improve the generalisability of the model and the reliability of the data.
8.2 Efficient Model Architectures
Model scalability becomes a critical issue when biomedical datasets increase in size and complexity.
• Techniques for compressing models, pruning, quantization, and knowledge distillation allow deep learning models to be implemented in contexts with limited resources without suffering appreciable performance degradation. These techniques also improve energy efficiency and model interpretability.
• Distributed and parallel training Architecture enables effective management of huge datasets. These systems provide iterative experimentation and hyperparameter adjustment that are frequently not feasible in single-machine situations, going beyond simple computing acceleration.
8.3 Robust Feature Alignment and Fusion
Accurate spatial and semantic alignment is essential for the successful integration of multimodal data.
• Image registration algorithms, such as ANTs and elastix, are essential for lining up anatomical features in various imaging modalities. For tasks such as morphological comparison and lesion identification, high-fidelity alignment maintains the essential spatial relations.
• Attention-based fusion methods dynamically determine the relative significance of each modality, allowing for more task-specific and sophisticated integration. In clinical settings, when not all data modalities have the same diagnostic weight, this is advantageous.
8.4 Explainable AI (XAI) Methods
The interpretability of models is essential for the transparency and trustworthiness of AI-driven healthcare decision-making.
• Tools for post hoc explanations, such as saliency maps, CAMs, and LIME, help reveal which features influence model predictions. Allows clinicians to gain insights into the reasoning process.
• Architectures with inherent interpretability, Attention-based models, for example, include transparency into the actual learning process, thereby promoting greater confidence and making regulatory adoption easier.
8.5 Privacy-Preserving Techniques
Powerful privacy-preserving procedures are required when handling sensitive medical data to adhere to legal and ethical requirements.
• Federated learning provides a paradigm change by allowing local data retention and collaborative model training across decentralized data sources. This method is especially attractive for hospital networks that are ordinarily prohibited from exchanging patient information due to privacy issues.
• Differential privacy presents statistically valid approaches for data anonymization that guarantee that individual-level information cannot be deduced from aggregate statistics or model outputs. This improves public trust and moral integrity.
9.1 Unified Benchmarking and Standardization
A major obstacle to comparing multimodal models across various activities and domains is the lack of standardized benchmarks and evaluation methodologies. To address this need, it is crucial to create extensive, superior, and publicly available multimodal datasets. These benchmarks must include a broad spectrum of clinical situations and techniques to provide a reliable evaluation of the performance and generalisability of the model. Additionally, standardization can guarantee that performance gains are the result of model innovation rather than dataset-specific artifacts and reduce biases brought about by dataset variability.
9.2 Real-Time Analysis and Clinical Deployment
Even with recent advancements, real-time multimodal analysis in clinical settings that require quick decisions, such as stroke diagnosis or trauma reaction, is still difficult to accomplish. Real-time inference necessitates smooth interaction with clinical hardware and software systems in addition to algorithmic efficiency. Future studies should focus on increasing data flow and decreasing computing latency without sacrificing diagnostic precision. This is important because it can revolutionize emergency treatment by facilitating prompt, well-informed decision-making.
9.3 Integration of Additional Modalities
The majority of multimodal frameworks in use today are mainly concerned with imaging, histology, and omics data. Richer contextual information is included by broadening the input spectrum to include physiological signals (EEG, ECG), sensor data (e.g., wearables, IoT-enabled devices), and longitudinal patient records. This increase is not only additive; it offers a more comprehensive and temporally-aware view of disease processes. Early diagnosis, better risk assessment, and the modeling of intricate temporal patterns in chronic illnesses can be made possible by integrating different modalities.
9.4 Personalization and Adaptive Learning
Multimodal AI’s ability to continuously learn and adjust to the unique paths of each patient will determine its future. Diagnostic and prognostic outputs could be improved over time by adaptive models that change in reaction to incoming data streams (such as those from home monitoring devices or routine exams). Although this transition from static to dynamic modeling has potential applications in precision medicine, it also brings up important issues regarding model stability, validation in dynamic situations, and the dangers of overfitting to noise rather than signal.
9.5 Regulatory Frameworks and Ethical AI
As multimodal AI systems start to influence critical clinical judgements, strict regulatory control is becoming increasingly necessary. Demonstrating technical proficiency is insufficient; models also need to adhere to criteria for clinical safety, explainability, and openness. This calls for interdisciplinary cooperation between regulatory agencies, medical practitioners, and AI researchers. Furthermore, to build public confidence and guarantee equitable deployment, ethical issues, including algorithmic bias, data privacy, and informed consent, need to be addressed early.
Multimodal deep learning is at the forefront of changing healthcare through the integration of multi-omic transcriptomic datasets, histology data, and radiological pictures. This convergence makes it possible to comprehend patient health in a more thorough and nuanced way, which could greatly improve diagnostic precision, simplify complicated data processing, and allow for completely personalized medication.
This review offers a focused and organized evaluation of current developments, highlighting the latest models, data fusion techniques, and recent developments that have influenced this rapidly developing field. In contrast to earlier studies, this paper provides an in-depth discussion of fusion approaches and their relative benefits and drawbacks, a comparative synthesis across both unimodal and multimodal architectures, and a comprehensive evaluation of contemporary multimodal datasets.
This review is essential because it addresses the fragmentation of the literature across biological applications and AI subfields and provides an overview for academics and clinicians. Although there have been recent beneficial developments, limited model interpretability, scalability, and data heterogeneity still exist. These challenges underline how urgent it is to conduct more research on explainable AI, privacy-preserving techniques, and powerful data harmonization. In addition, this work highlights the significance of interdisciplinary collaboration among computer scientists, healthcare professionals, and decision-makers by outlining future research areas that connect technical improvement with clinical practicality. This review provides a fundamental resource for developing multimodal AI in healthcare by addressing important outstanding topics and integrating various lines of research. As this field grows, we expect to see AI-driven solutions that are not only precise and scalable but also ethical and widely applicable, thereby bringing in a new era of data-driven, personalized healthcare.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Aya M. Al-Zoghby: Conceptualization of the review topic, overall supervision, and final editing of the manuscript. Ahmed Ismail Ebada: Co-conceptualized the review topic and authored the sections on Introduction, Background and Foundation, and Overview of Medical Data Types. Aya S. Saleh: Conducted a comprehensive literature review, synthesized key findings, and drafted the sections on State of the Art in Deep Learning Techniques, Data Integration Strategies, Applications for Healthcare, and Challenges in Multimodal Deep Learning. Mohammed Abdelhay: Designed the comparative analysis framework, created all visual figures and tables, and contributed to the section on Strategies to Overcome Challenges. Wael A. Awad: Critically reviewed and revised the manuscript for academic rigor, managed references, and contributed to the Conclusion and Future Directions sections. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Luo N, Zhong X, Su L, Cheng Z, Ma W, Hao P. Artificial intelligence-assisted dermatology diagnosis: from unimodal to multimodal. Comput Biol Med. 2023;165(9):107413. doi:10.1016/j.compbiomed.2023.107413. [Google Scholar] [PubMed] [CrossRef]
2. Baltrušaitis T, Ahuja C, Morency LP. Multimodal machine learning: a survey and taxonomy. IEEE Trans Pattern Anal Mach Intell. 2019;41(2):423–43. doi:10.1109/TPAMI.2018.2798607. [Google Scholar] [PubMed] [CrossRef]
3. Yildirim-Yayilgan S, Arifaj B, Rahimpour M, Hardeberg JY, Ahmedi L. Pre-trained CNN based deep features with hand-crafted features and patient data for skin lesion classification. In: Intelligent technologies and applications. Berlin/Heidelberg, Germany: Springer; 2021. p. 151–62. doi:10.1007/978-3-030-71711-7_13. [Google Scholar] [CrossRef]
4. Shen H. Enhancing diagnosis prediction in healthcare with knowledge-based recurrent neural networks. IEEE Access. 2023;11:106433–42. doi:10.1109/access.2023.3319502. [Google Scholar] [CrossRef]
5. Zhang L, Zhao Y, Che T, Li S, Wang X. Graph neural networks for image-guided disease diagnosis: a review. iRADIOLOGY. 2023;1(2):151–66. doi:10.1002/ird3.20. [Google Scholar] [CrossRef]
6. Kandhro IA, Manickam S, Fatima K, Uddin M, Malik U, Naz A, et al. Performance evaluation of E-VGG19 model: enhancing real-time skin cancer detection and classification. Heliyon. 2024;10(10):e31488. doi:10.1016/j.heliyon.2024.e31488. [Google Scholar] [PubMed] [CrossRef]
7. Alshuhri MS, Al-Musawi SG, Al-Alwany AA, Uinarni H, Rasulova I, Rodrigues P, et al. Artificial intelligence in cancer diagnosis: opportunities and challenges. Pathol Res Pract. 2024;253:154996. doi:10.1016/j.prp.2023.154996. [Google Scholar] [PubMed] [CrossRef]
8. MacKenzie SC, Sainsbury CAR, Wake DJ. Diabetes and artificial intelligence beyond the closed loop: a review of the landscape, promise and challenges. Diabetologia. 2024;67(2):223–35. doi:10.1007/s00125-023-06038-8. [Google Scholar] [PubMed] [CrossRef]
9. Gadhave DG, Sugandhi VV, Jha SK, Nangare SN, Gupta G, Singh SK, et al. Neurodegenerative disorders: mechanisms of degeneration and therapeutic approaches with their clinical relevance. Ageing Res Rev. 2024;99:102357. doi:10.1016/j.arr.2024.102357. [Google Scholar] [PubMed] [CrossRef]
10. Muhammad G, Alshehri F, Karray F, El Saddik A, Alsulaiman M, Falk TH. A comprehensive survey on multimodal medical signals fusion for smart healthcare systems. Inf Fusion. 2021;76(8):355–75. doi:10.1016/j.inffus.2021.06.007. [Google Scholar] [CrossRef]
11. Amal S, Safarnejad L, Omiye JA, Ghanzouri I, Cabot JH, Ross EG. Use of multi-modal data and machine learning to improve cardiovascular disease care. Front Cardiovasc Med. 2022;9:840262. doi:10.3389/fcvm.2022.840262. [Google Scholar] [PubMed] [CrossRef]
12. Stahlschmidt SR, Ulfenborg B, Synnergren J. Multimodal deep learning for biomedical data fusion: a review. Brief Bioinform. 2022;23(2):bbab569. doi:10.1093/bib/bbab569. [Google Scholar] [PubMed] [CrossRef]
13. Pei X, Zuo K, Li Y, Pang Z. A review of the application of multi-modal deep learning in medicine: bibliometrics and future directions. Int J Comput Intell Syst. 2023;16(1):44. doi:10.1007/s44196-023-00225-6. [Google Scholar] [CrossRef]
14. Shaik T, Tao X, Li L, Xie H, Velásquez JD. A survey of multimodal information fusion for smart healthcare: mapping the journey from data to wisdom. Inf Fusion. 2024;102(1):102040. doi:10.1016/j.inffus.2023.102040. [Google Scholar] [CrossRef]
15. Khan U, Yasin A, Abid M, Shafi I, Khan SA. A methodological review of 3D reconstruction techniques in tomographic imaging. J Med Syst. 2018;42(10):190. doi:10.1007/s10916-018-1042-2. [Google Scholar] [PubMed] [CrossRef]
16. Shivahare BD, Singh J, Ravi V, Chandan RR, Alahmadi TJ, Singh P, et al. Delving into machine learning’s influence on disease diagnosis and prediction. Open Public Health J. 2024;17(1):e18749445297804. doi:10.2174/0118749445297804240401061128. [Google Scholar] [CrossRef]
17. Fernandes FE, Yen GG. Automatic searching and pruning of deep neural networks for medical imaging diagnostic. IEEE Trans Neural Netw Learn Syst. 2021;32(12):5664–74. doi:10.1109/TNNLS.2020.3027308. [Google Scholar] [PubMed] [CrossRef]
18. Yu Teh X, Shan Qing Yeoh P, Wang T, Wu X, Hasikin K, Wee Lai K. Knee osteoarthritis diagnosis with unimodal and multi-modal neural networks: data from the osteoarthritis initiative. IEEE Access. 2024;12:146698–717. [Google Scholar]
19. Pfob A, Sidey-Gibbons C, Barr RG, Duda V, Alwafai Z, Balleyguier C, et al. The importance of multi-modal imaging and clinical information for humans and AI-based algorithms to classify breast masses (INSPiRED 003an international, multicenter analysis. Eur Radiol. 2022;32(6):4101–15. doi:10.1007/s00330-021-08519-z. [Google Scholar] [PubMed] [CrossRef]
20. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. doi:10.1038/nature21056. [Google Scholar] [PubMed] [CrossRef]
21. Topol E. Deep medicine: how artificial intelligence can make healthcare human again. Paris, France: Hachette UK; 2019. [Google Scholar]
22. Shetty S, Ananthanarayana VS, Mahale A. Multimodal medical tensor fusion network-based DL framework for abnormality prediction from the radiology CXRs and clinical text reports. Multimed Tools Appl. 2023;82(48):1–48. doi:10.1007/s11042-023-14940-x. [Google Scholar] [PubMed] [CrossRef]
23. Wu N, Wong KY, Yu X, Zhao JW, Zhang XY, Wang JH, et al. Multispectral 3D DNA machine combined with multimodal machine learning for noninvasive precise diagnosis of bladder cancer. Anal Chem. 2024;96(24):10046–55. doi:10.1021/acs.analchem.4c01749. [Google Scholar] [PubMed] [CrossRef]
24. Martí-Bonmatí L, Sopena R, Bartumeus P, Sopena P. Multimodality imaging techniques. Contrast Media Mol. 2010;5(4):180–9. doi:10.1002/cmmi.393. [Google Scholar] [PubMed] [CrossRef]
25. Das S, Mazumdar H, Khondakar KR, Mishra YK, Kaushik A. Review—quantum biosensors: principles and applications in medical diagnostics. ECS Sens Plus. 2024;3(2):025001. doi:10.1149/2754-2726/ad47e2. [Google Scholar] [CrossRef]
26. Yang D, Li C, Lin J. Multimodal cancer imaging using lanthanide-based upconversion nanoparticles. Nanomed. 2015;10(16):2573–91. doi:10.2217/nnm.15.92. [Google Scholar] [PubMed] [CrossRef]
27. Feng X, Shu W, Li M, Li J, Xu J, He M. Pathogenomics for accurate diagnosis, treatment, prognosis of oncology: a cutting edge overview. J Transl Med. 2024;22(1):131. doi:10.1186/s12967-024-04915-3. [Google Scholar] [PubMed] [CrossRef]
28. Schubert MC, Lasotta M, Sahm F, Wick W, Venkataramani V. Evaluating the multimodal capabilities of generative ai in complex clinical diagnostics. medRxiv. 2023;11:1–6. [Google Scholar]
29. Boehm KM, Khosravi P, Vanguri R, Gao J, Shah SP. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22(2):114–26. doi:10.1038/s41568-021-00408-3. [Google Scholar] [PubMed] [CrossRef]
30. Azam MA, Khan KB, Salahuddin S, Rehman E, Khan SA, Khan MA, et al. A review on multimodal medical image fusion: compendious analysis of medical modalities, multimodal databases, fusion techniques and quality metrics. Comput Biol Med. 2022;144(3):105253. doi:10.1016/j.compbiomed.2022.105253. [Google Scholar] [PubMed] [CrossRef]
31. Li Y, Pan L, Peng Y, Li X, Wang X, Qu L, et al. Application of deep learning-based multimodal fusion technology in cancer diagnosis: a survey. Eng Appl Artif Intell. 2025;143(7793):109972. doi:10.1016/j.engappai.2024.109972. [Google Scholar] [CrossRef]
32. Tian H, Tao Y, Pouyanfar S, Chen SC, Shyu ML. Multimodal deep representation learning for video classification. World Wide Web. 2019;22(3):1325–41. doi:10.1007/s11280-018-0548-3. [Google Scholar] [CrossRef]
33. Guo D, Lu C, Chen D, Yuan J, Duan Q, Xue Z, et al. A multimodal breast cancer diagnosis method based on knowledge-augmented deep learning. Biomed Signal Process Control. 2024;90(3):105843. doi:10.1016/j.bspc.2023.105843. [Google Scholar] [CrossRef]
34. Marivani I, Tsiligianni E, Cornelis B, Deligiannis N. Multimodal deep unfolding for guided image super-resolution. IEEE Trans Image Process. 2020;29:8443–56. doi:10.1109/TIP.2020.3014729. [Google Scholar] [PubMed] [CrossRef]
35. Talreja V, Valenti MC, Nasrabadi NM. Deep hashing for secure multimodal biometrics. IEEE Trans Inf Forensics Secur. 2020;16:1306–21. doi:10.1109/TIFS.2020.3033189. [Google Scholar] [CrossRef]
36. Seng JKP, Ang KL. Multimodal emotion and sentiment modeling from unstructured big data: challenges, architecture & techniques. IEEE Access. 2019;7:90982–98. [Google Scholar]
37. Chai W, Wang G. Deep vision multimodal learning: methodology, benchmark, and trend. Appl Sci. 2022;12(13):6588. doi:10.3390/app12136588. [Google Scholar] [CrossRef]
38. Alatkar SA, Wang D. CMOT: cross-modality optimal transport for multimodal inference. Genome Biol. 2023;24(1):163. doi:10.1186/s13059-023-02989-8. [Google Scholar] [PubMed] [CrossRef]
39. Huang Y, Shao L, Frangi AF. Cross-modality image synthesis via weakly coupled and geometry co-regularized joint dictionary learning. IEEE Trans Med Imaging. 2018;37(3):815–27. doi:10.1109/TMI.2017.2781192. [Google Scholar] [PubMed] [CrossRef]
40. Chen T, Ma X, Ying X, Wang W, Yuan C, Lu W, et al. Multi-modal fusion learning for cervical dysplasia diagnosis. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019 Apr 8–11; Venice, Italy. p. 1505–9. doi:10.1109/isbi.2019.8759303. [Google Scholar] [CrossRef]
41. Golovanevsky M, Eickhoff C, Singh R. Multimodal attention-based deep learning for Alzheimer’s disease diagnosis. J Am Med Inform Assoc. 2022;29(12):2014–22. doi:10.1093/jamia/ocac168. [Google Scholar] [PubMed] [CrossRef]
42. Zhu L, Chan LL, Ng TK, Zhang M, Ooi BC. Deep co-training for cross-modality medical image segmentation. In: Proceedings of the 26th European Conference on Artificial Intelligence(ECAI 2023); 2023 Sep 30–Oct 4; Kraków, Poland. Frontiers in artificial intelligence and applications. Vol. 372. Amsterdam, Netherlands: IOS Press; 2023. doi:10.3233/FAIA230633. [Google Scholar] [CrossRef]
43. Chen J, Huang G, Yuan X, Zhong G, Zheng Z, Pun CM, et al. Quaternion cross-modality spatial learning for multi-modal medical image segmentation. IEEE J Biomed Health Inform. 2024;28(3):1412–23. doi:10.1109/jbhi.2023.3346529. [Google Scholar] [PubMed] [CrossRef]
44. Hessel J, Lee L. Does my multimodal model learn cross-modal interactions? It’s harder to tell than you might think! In: Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2020 Nov 16–20; Online. p. 861–77. doi:10.18653/v1/2020.emnlp-main.62. [Google Scholar] [CrossRef]
45. Shakya AK, Vidyarthi A. Comprehensive study of compression and texture integration for digital imaging and communications in medicine data analysis. Technologies. 2024;12(2):17. doi:10.3390/technologies12020017. [Google Scholar] [CrossRef]
46. Klein JS, Brant WE, Helms CA, Vinson EN. Brant and helms’ fundamentals of diagnostic radiology. Philadelphia, PA, USA: Lippincott Williams and Wilkins; 2019. [Google Scholar]
47. Miger K, Overgaard Olesen AS, Grand J, Fabricius-Bjerre A, Sajadieh A, Høst N, et al. Computed tomography or chest X-ray to assess pulmonary congestion in dyspnoeic patients with acute heart failure. ESC Heart Fail. 2024;11(2):1163–73. doi:10.1002/ehf2.14688. [Google Scholar] [PubMed] [CrossRef]
48. Ayalew AM, Enyew B, Bezabh YA, Abuhayi BM, Negashe GS. Early-stage cardiomegaly detection and classification from X-ray images using convolutional neural networks and transfer learning. Intell Syst Appl. 2024;24(1):200453. doi:10.1016/j.iswa.2024.200453. [Google Scholar] [CrossRef]
49. Goldman LW. Principles of CT and CT technology. J Nucl Med Technol. 2007;35(3):115–28. doi:10.2967/jnmt.107.042978. [Google Scholar] [PubMed] [CrossRef]
50. MacDonald D. Computed tomography. In: Oral and maxillofacial radiology: a diagnostic approach. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2019. doi:10.1002/9781119218739.ch4. [Google Scholar] [CrossRef]
51. Khan M, Shiwlani A, Qayyum MU, Sherani AMK, Hussain HK. Revolutionizing healthcare with AI: innovative strategies in cancer medicine. Int J Multidiscip Sci Arts. 2024;3(2):316–24. doi:10.47709/ijmdsa.v3i1.3922. [Google Scholar] [CrossRef]
52. Kalsi S, French H, Chhaya S, Madani H, Mir R, Anosova A, et al. The evolving role of artificial intelligence in radiotherapy treatment planning—a literature review. Clin Oncol. 2024;36(10):596–605. doi:10.1016/j.clon.2024.06.005. [Google Scholar] [PubMed] [CrossRef]
53. Al-Anazi S, Al-Omari A, Alanazi S, Marar A, Asad M, Alawaji F, et al. Artificial intelligence in respiratory care: current scenario and future perspective. Ann Thorac Med. 2024;19(2):117–30. doi:10.4103/atm.atm_192_23. [Google Scholar] [PubMed] [CrossRef]
54. Milosevic M, Jin Q, Singh A, Amal S. Applications of AI in multi-modal imaging for cardiovascular disease. Front Radiol. 2024;3:1294068. doi:10.3389/fradi.2023.1294068. [Google Scholar] [PubMed] [CrossRef]
55. Grover VPB, Tognarelli JM, Crossey MME, Cox IJ, Taylor-Robinson SD, McPhail MJW. Magnetic resonance imaging: principles and techniques: lessons for clinicians. J Clin Exp Hepatol. 2015;5(3):246–55. doi:10.1016/j.jceh.2015.08.001. [Google Scholar] [PubMed] [CrossRef]
56. Hussain MM, Murugaananda S, Purnachand K. A study on deep learning techniques for Parkinson’s disease detection. In: 2024 International Conference on Advancements in Smart, Secure and Intelligent Computing (ASSIC); 2024 Jan 27–29; Bhubaneswar, India. doi:10.1109/ASSIC60049.2024.10507957. [Google Scholar] [CrossRef]
57. Yousef H, Malagurski Tortei B, Castiglione F. Predicting multiple sclerosis disease progression and outcomes with machine learning and MRI-based biomarkers: a review. J Neurol. 2024;271(10):6543–72. doi:10.1007/s00415-024-12651-3. [Google Scholar] [PubMed] [CrossRef]
58. Pradhan N, Sagar S, Singh AS. Analysis of MRI image data for Alzheimer disease detection using deep learning techniques. Multimed Tools Appl. 2024;83(6):17729–52. doi:10.1007/s11042-023-16256-2. [Google Scholar] [CrossRef]
59. Carovac A, Smajlovic F, Junuzovic D. Application of ultrasound in medicine. Acta Inform Med. 2011;19(3):168–71. doi:10.5455/aim.2011.19.168-171. [Google Scholar] [PubMed] [CrossRef]
60. Merz E, Abramowicz JS. 3D/4D ultrasound in prenatal diagnosis: is it time for routine use? Clin Obstet Gynecol. 2012;55(1):336–51. doi:10.1097/grf.0b013e3182446ef7. [Google Scholar] [PubMed] [CrossRef]
61. Gao J, Flick A, Allen A, Krasnoff M, Kinder D, Nguyen T. Variability in liver size measurements using different view angles in ultrasound imaging. J Ultrasound Med. 2024;43(12):2345–55. doi:10.1002/jum.16570. [Google Scholar] [PubMed] [CrossRef]
62. Farhoudi N, Laurentius LB, Magda J, Reiche CF, Solzbacher F. Micromechanical resonators for ultrasound-based sensors. Meet Abstr. 2021;MA2021-01(59):1593. doi:10.1149/ma2021-01591593mtgabs. [Google Scholar] [CrossRef]
63. Cen X, Dong W, Lv W, Zhao Y, Dubee F, Mentis AA, et al. Towards interpretable imaging genomics analysis: methodological developments and applications. Inf Fusion. 2024;102(2):102032. doi:10.1016/j.inffus.2023.102032. [Google Scholar] [CrossRef]
64. Li Q, Song Q, Chen Z, Choi J, Moreno V, Ping J, et al. Large-scale integration of omics and electronic health records to identify potential risk protein biomarkers and therapeutic drugs for cancer prevention and intervention. medRxiv. Forthcoming 2024;9:4285. doi:10.1101/2024.05.29.24308170. [Google Scholar] [PubMed] [CrossRef]
65. Nowrozy R, Ahmed K, Kayes ASM, Wang H, McIntosh TR. Privacy preservation of electronic health records in the modern era: a systematic survey. ACM Comput Surv. 2024;56(8):1–37. doi:10.1145/3653297. [Google Scholar] [CrossRef]
66. Zhou Y, Huang S, Fries JA, Youssef A, Amrhein TJ, Chang M, et al. RadFusion: benchmarking performance and fairness for multimodal pulmonary embolism detection from CT and HER. arXiv:2111.11665. 2021. [Google Scholar]
67. Bae S, Kyung D, Ryu J, Cho E, Lee G, Kweon S, et al. EHRXQA: a multi-modal question answering dataset for electronic health records with chest X-ray images. In: Proceedings of the 37th International Conference on Neural Information Processing Systems, NIPS’23; 2023 Dec 10–16; New Orleans, LA, USA. [Google Scholar]
68. Huang SC, Huo Z, Steinberg E, Chiang CC, Lungren MP, Langlotz CP, et al. INSPECT: a multimodal dataset for pulmonary embolism diagnosis and prognosis. In: Proceedings of the 37th International Conference on Neural Information Processing Systems, NIPS’23; 2023 Dec 10–16; New Orleans, LA, USA. [Google Scholar]
69. Zhang S, Xu Y, Usuyama N, Xu H, Bagga J, Tinn R, et al. A multimodal biomedical foundation model trained from fifteen million image-text pairs. arXiv:2303.00915. 2023. [Google Scholar]
70. Mota T, Verdelho MR, Araújo DJ, Bissoto A, Santiago C, Barata C. MMIST-ccRCC: a real world medical dataset for the development of multi-modal systems. In: 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2024 Jun 17–18; Seattle, WA, USA. p. 2395–403. doi:10.1109/CVPRW63382.2024.00246. [Google Scholar] [CrossRef]
71. Tripathi A, Waqas A, Yilmaz Y, Rasool G. HoneyBee: a scalable modular framework for creating multimodal oncology datasets with foundational embedding models. arXiv:2405.07460. 2024. [Google Scholar]
72. Xie Y, Zhou C, Gao L, Wu J, Li X, Zhou HY, et al. MedTrinity-25M: a large-scale multimodal dataset with multigranular annotations for medicine. [cited 2025 Jun 1]. Available from: https://yunfeixie233.github.io/MedTrinity-25M/. [Google Scholar]
73. Yuan X. Research on intelligent analysis and recognition system of medical data based on deep learning. Med Insights. 2025;2(1):1–10. doi:10.70088/hqjawt58. [Google Scholar] [CrossRef]
74. Ali A, Ali H, Saeed A, Ahmed Khan A, Tin TT, Assam M, et al. Blockchain-powered healthcare systems: enhancing scalability and security with hybrid deep learning. Sensors. 2023;23(18):7740. doi:10.3390/s23187740. [Google Scholar] [PubMed] [CrossRef]
75. Yenumala A, Zhang X, Lo D. Towards more robust and scalable deep learning systems for medical image analysis. In: 2024 IEEE International Conference on Big Data (BigData); 2024 Dec 15–18; Washington, DC, USA. p. 7577–85. doi:10.1109/BigData62323.2024.10825626. [Google Scholar] [CrossRef]
76. Gao J, Jiang Q, Zhou B, Chen D. Convolutional neural networks for computer-aided detection or diagnosis in medical image analysis: an overview. Math Biosci Eng. 2019;16(6):6536–61. doi:10.3934/mbe.2019326. [Google Scholar] [PubMed] [CrossRef]
77. Cheng PM, Montagnon E, Yamashita R, Pan I, Cadrin-Chênevert A, Perdigón Romero F, et al. Deep learning: an update for radiologists. RadioGraphics. 2021;41(5):1427–45. doi:10.1148/rg.2021200210. [Google Scholar] [PubMed] [CrossRef]
78. Abdelhafiz D, Yang C, Ammar R, Nabavi S. Deep convolutional neural networks for mammography: advances, challenges and applications. BMC Bioinformatics. 2019;20(Suppl 11):281. doi:10.1186/s12859-019-2823-4. [Google Scholar] [PubMed] [CrossRef]
79. Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B. 3D deep learning on medical images: a review. Sensors. 2020;20(18):5097. doi:10.3390/s20185097. [Google Scholar] [PubMed] [CrossRef]
80. Farhad M, Masud MM, Beg A, Ahmad A, Ahmed L. A review of medical diagnostic video analysis using deep learning techniques. Appl Sci. 2023;13(11):6582. doi:10.3390/app13116582. [Google Scholar] [CrossRef]
81. Buddenkotte T, Rundo L, Woitek R, Escudero Sanchez L, Beer L, Crispin-Ortuzar M, et al. Deep learning-based segmentation of multisite disease in ovarian cancer. Eur Radiol Exp. 2023;7(1):77. doi:10.1186/s41747-023-00388-z. [Google Scholar] [PubMed] [CrossRef]
82. Mahootiha M, Qadir HA, Bergsland J, Balasingham I. Multimodal deep learning for personalized renal cell carcinoma prognosis: integrating CT imaging and clinical data. Comput Methods Programs Biomed. 2024;244:107978. doi:10.1016/j.cmpb.2023.107978. [Google Scholar] [PubMed] [CrossRef]
83. Han S, Wang Y, Wang Q. Multimodal medical image segmentation algorithm based on convolutional neural networks. In: 2024 Second International Conference on Networks, Multimedia and Information Technology (NMITCON); 2024 Aug 9–10; Bengaluru, India. doi:10.1109/NMITCON62075.2024.10698930. [Google Scholar] [CrossRef]
84. Küçükçiloğlu Y, Şekeroğlu B, Adalı T, Şentürk N. Prediction of osteoporosis using MRI and CT scans with unimodal and multimodal deep-learning models. Diagn Interv Radiol. 2024;30(1):9–20. doi:10.4274/dir.2023.232116. [Google Scholar] [PubMed] [CrossRef]
85. Li J, Liu H, Liu W, Zong P, Huang K, Li Z, et al. Predicting gastric cancer tumor mutational burden from histopathological images using multimodal deep learning. Brief Funct Genomics. 2024;23(3):228–38. doi:10.1093/bfgp/elad032. [Google Scholar] [PubMed] [CrossRef]
86. Abdullakutty F, Akbari Y, Al-Maadeed S, Bouridane A, Hamoudi R. Advancing histopathology-based breast cancer diagnosis: insights into multi-modality and explainability. arXiv:2406.12897. 2024. [Google Scholar]
87. Kavitha A, Dhanush Sriram R, ArunKumar R. Multi-modal CNN-ensemble learning with pansegnet for early and accurate pancreatic cancer analysis. In: 2024 3rd International Conference on Automation, Computing and Renewable Systems (ICACRS); 2024 Dec 4–6; Pudukkottai, India. p. 1420–7. doi:10.1109/ICACRS62842.2024.10841746. [Google Scholar] [CrossRef]
88. Ponzio F, Urgese G, Ficarra E, Di Cataldo S. Dealing with lack of training data for convolutional neural networks: the case of digital pathology. Electronics. 2019;8(3):256. doi:10.3390/electronics8030256. [Google Scholar] [CrossRef]
89. Willemink MJ, Roth HR, Sandfort V. Toward foundational deep learning models for medical imaging in the New Era of transformer networks. Radiol Artif Intell. 2022;4(6):e210284. doi:10.1148/ryai.210284. [Google Scholar] [PubMed] [CrossRef]
90. Lipton ZC, Kale DC, Elkan C, Wetzel R. Learning to diagnose with LSTM recurrent neural networks. In: 4th International Conference on Learning Representations, ICLR 2016—Conference Track Proceedings; 2016 May 2–4; San Juan, Puerto Rico. [Google Scholar]
91. Giunchiglia E, Nemchenko A, van der Schaar M. RNN-SURV: a deep recurrent model for survival analysis. In: Proceedings of 27th International Conference on Artificial Neural Networks; 2018 Oct 4–7; Rhodes, Greece. doi:10.1007/978-3-030-01424-7_3. [Google Scholar] [CrossRef]
92. Kumar RL, Khan F, Din S, Band SS, Mosavi A, Ibeke E. Recurrent neural network and reinforcement learning model for COVID-19 prediction. Front Public Health. 2021;9:744100. doi:10.3389/fpubh.2021.744100. [Google Scholar] [PubMed] [CrossRef]
93. Jo T, Nho K, Saykin AJ. Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci. 2019;11:220. doi:10.3389/fnagi.2019.00220. [Google Scholar] [PubMed] [CrossRef]
94. Liu X, Qiu H, Li M, Yu Z, Yang Y, Yan Y. Application of multimodal fusion deep learning model in disease recognition. In: 2024 IEEE 2nd International Conference on Sensors, Electronics and Computer Engineering (ICSECE); 2024 Aug 29–31; Jinzhou, China. p. 1246–50. doi:10.1109/ICSECE61636.2024.10729504. [Google Scholar] [CrossRef]
95. Sufian MA, Niu M. Hybrid deep learning for computational precision in cardiac MRI segmentation: integrating autoencoders, CNNs, and RNNs for enhanced structural analysis. Comput Biol Med. 2025;186(2):109597. doi:10.1016/j.compbiomed.2024.109597. [Google Scholar] [PubMed] [CrossRef]
96. Guo Z, Li X, Huang H, Guo N, Li Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci. 2019;3(2):162–9. doi:10.1109/trpms.2018.2890359. [Google Scholar] [PubMed] [CrossRef]
97. Hussain D, Al-masni MA, Aslam M, Sadeghi-Niaraki A, Hussain J, Gu YH, et al. Revolutionizing tumor detection and classification in multimodality imaging based on deep learning approaches: methods, applications and limitations. J X Ray Sci Technol Clin Appl Diagn Ther. 2024;32(4):857–911. doi:10.3233/xst-230429. [Google Scholar] [PubMed] [CrossRef]
98. Pandey RP, Rengarajan A, Awasthi A. Automated multimodal medical diagnostics using deep learning frameworks. In: 2024 International Conference on Optimization Computing and Wireless Communication (ICOCWC); 2024 Jan 29–30; Debre Tabor, Ethiopia. doi:10.1109/ICOCWC60930.2024.10470876. [Google Scholar] [CrossRef]
99. Yan K, Li T, Lobo Marques JA, Gao J, Fong SJ. A review on multimodal machine learning in medical diagnostics. Math Biosci Eng. 2023;20(5):8708–26. doi:10.3934/mbe.2023382. [Google Scholar] [PubMed] [CrossRef]
100. Gogoshin G, Rodin AS. Graph neural networks in cancer and oncology research: emerging and future trends. Cancers. 2023;15(24):5858. doi:10.3390/cancers15245858. [Google Scholar] [PubMed] [CrossRef]
101. Waqas A, Tripathi A, Ramachandran RP, Stewart PA, Rasool G. Multimodal data integration for oncology in the era of deep neural networks: a review. Front Artif Intell. 2024;7:1408843. doi:10.3389/frai.2024.1408843. [Google Scholar] [PubMed] [CrossRef]
102. Munikoti S, Agarwal D, Das L, Halappanavar M, Natarajan B. Challenges and opportunities in deep reinforcement learning with graph neural networks: a comprehensive review of algorithms and applications. IEEE Trans Neural Netw Learn Syst. 2024;35(11):15051–71. doi:10.1109/TNNLS.2023.3283523. [Google Scholar] [PubMed] [CrossRef]
103. Sinha A, Kumar T. Enhancing medical diagnostics: integrating AI for precise brain tumour detection. Procedia Comput Sci. 2024;235(16):456–67. doi:10.1016/j.procs.2024.04.045. [Google Scholar] [CrossRef]
104. Valous NA, Popp F, Zörnig I, Jäger D, Charoentong P. Graph machine learning for integrated multi-omics analysis. Br J Cancer. 2024;131(2):205–11. doi:10.1038/s41416-024-02706-7. [Google Scholar] [PubMed] [CrossRef]
105. Zhang H, Cao D, Chen Z, Zhang X, Chen Y, Sessions C, et al. mosGraphGen: a novel tool to generate multi-omics signaling graphs to facilitate integrative and interpretable graph AI model development. Bioinform Adv. 2024;4(1):vbae151. doi:10.1093/bioadv/vbae151. [Google Scholar] [PubMed] [CrossRef]
106. Thakur GK, Thakur A, Kulkarni S, Khan N, Khan S. Deep learning approaches for medical image analysis and diagnosis. Cureus. 2024;16(5):e59507. doi:10.7759/cureus.59507. [Google Scholar] [PubMed] [CrossRef]
107. Li Y, El Habib Daho M, Conze PH, Zeghlache R, Le Boité H, Tadayoni R, et al. A review of deep learning-based information fusion techniques for multimodal medical image classification. Comput Biol Med. 2024;177(6):108635. doi:10.1016/j.compbiomed.2024.108635. [Google Scholar] [PubMed] [CrossRef]
108. Ahmadi Golilarz H, Azadbar A, Alizadehsani R, Gorriz JM. GAN-MD: a myocarditis detection using multi-channel convolutional neural networks and generative adversarial network-based data augmentation. CAAI Trans Intel Tech. 2024;9(4):866–78. doi:10.1049/cit2.12307. [Google Scholar] [CrossRef]
109. Sorin V, Barash Y, Konen E, Klang E. Creating artificial images for radiology applications using generative adversarial networks (GANs)—a systematic review. Acad Radiol. 2020;27(8):1175–85. doi:10.1016/j.acra.2019.12.024. [Google Scholar] [PubMed] [CrossRef]
110. Ali M, Ali M, Hussain M, Koundal D. Generative adversarial networks (GANs) for medical image processing: recent advancements. Arch Comput Meth Eng. 2025;32(2):1185–98. doi:10.1007/s11831-024-10174-8. [Google Scholar] [CrossRef]
111. Makhlouf A, Maayah M, Abughanam N, Catal C. The use of generative adversarial networks in medical image augmentation. Neural Comput Appl. 2023;35(34):24055–68. doi:10.1007/s00521-023-09100-z. [Google Scholar] [CrossRef]
112. Wang R, Bashyam V, Yang Z, Yu F, Tassopoulou V, Chintapalli SS, et al. Applications of generative adversarial networks in neuroimaging and clinical neuroscience. NeuroImage. 2023;269(1):119898. doi:10.1016/j.neuroimage.2023.119898. [Google Scholar] [PubMed] [CrossRef]
113. Jeong JJ, Tariq A, Adejumo T, Trivedi H, Gichoya JW, Banerjee I. Systematic review of generative adversarial networks (GANs) for medical image classification and segmentation. J Digit Imag. 2022;35(2):137–52. doi:10.1007/s10278-021-00556-w. [Google Scholar] [PubMed] [CrossRef]
114. Sabnam S, Rajagopal S. Application of generative adversarial networks in image, face reconstruction and medical imaging: challenges and the current progress. Comput Meth Biomech Biomed Eng Imag Vis. 2024;12(1):2330524. doi:10.1080/21681163.2024.2330524. [Google Scholar] [CrossRef]
115. Koshino K, Werner RA, Pomper MG, Bundschuh RA, Toriumi F, Higuchi T, et al. Narrative review of generative adversarial networks in medical and molecular imaging. Ann Transl Med. 2021;9(9):821. doi:10.21037/atm-20-6325. [Google Scholar] [PubMed] [CrossRef]
116. Shin Y, Yang J, Lee YH. Deep generative adversarial networks: applications in musculoskeletal imaging. Radiol Artif Intell. 2021;3(3):e200157. doi:10.1148/ryai.2021200157. [Google Scholar] [PubMed] [CrossRef]
117. Chen H. Challenges and corresponding solutions of generative adversarial networks (GANsa survey study. J Phys Conf Ser. 2021;1827(1):012066. doi:10.1088/1742-6596/1827/1/012066. [Google Scholar] [CrossRef]
118. Kelkar VA, Gotsis DS, Brooks FJ, Kc P, Myers KJ, Zeng R, et al. Assessing the ability of generative adversarial networks to learn canonical medical image statistics. IEEE Trans Med Imag. 2023;42(6):1799–808. [Google Scholar]
119. Mao YJ, Zha LW, Tam AY, Lim HJ, Cheung AK, Zhang YQ, et al. Endocrine tumor classification via machine-learning-based elastography: a systematic scoping review. Cancers. 2023;15(3):837. doi:10.3390/cancers15030837. [Google Scholar] [PubMed] [CrossRef]
120. Zhou HY, Yu Y, Wang C, Zhang S, Gao Y, Pan J, et al. A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat Biomed Eng. 2023;7(6):743–55. doi:10.1038/s41551-023-01045-x. [Google Scholar] [PubMed] [CrossRef]
121. Khader F, Müller-Franzes G, Wang T, Han T, Tayebi Arasteh S, Haarburger C, et al. Multimodal deep learning for integrating chest radiographs and clinical parameters: a case for transformers. Radiology. 2023;309(1):e230806. doi:10.1148/radiol.230806. [Google Scholar] [PubMed] [CrossRef]
122. Sun Q, Fang N, Liu Z, Zhao L, Wen Y, Lin H. HybridCTrm: bridging CNN and transformer for multimodal brain image segmentation. J Healthc Eng. 2021;2021:7467261. doi:10.1155/2021/7467261. [Google Scholar] [PubMed] [CrossRef]
123. Xie X, Zhang X, Tang X, Zhao J, Xiong D, Ouyang L, et al. MACTFusion: lightweight cross transformer for adaptive multimodal medical image fusion. IEEE J Biomed Health Inform. 2025;29(5):3317–28. doi:10.1109/jbhi.2024.3391620. [Google Scholar] [PubMed] [CrossRef]
124. Gasmi K, Ben Aoun N, Alsalem K, Ltaifa IB, Alrashdi I, Ammar LB, et al. Enhanced brain tumor diagnosis using combined deep learning models and weight selection technique. Front Neuroinform. 2024;18:1444650. doi:10.3389/fninf.2024.1444650. [Google Scholar] [PubMed] [CrossRef]
125. Hu X, Zhang P, Zhang J, Deng L. DeepFusionCDR: employing multi-omics integration and molecule-specific transformers for enhanced prediction of cancer drug responses. IEEE J Biomed Health Inform. 2024;28(10):6248–58. doi:10.1109/JBHI.2024.3417014. [Google Scholar] [PubMed] [CrossRef]
126. Cai Z, Poulos RC, Aref A, Robinson PJ, Reddel RR, Zhong Q. DeePathNet: a transformer-based deep learning model integrating multiomic data with cancer pathways. Cancer Res Commun. 2024;4(12):3151–64. doi:10.1158/2767-9764.crc-24-0285. [Google Scholar] [PubMed] [CrossRef]
127. Dentamaro V, Impedovo D, Musti L, Pirlo G, Taurisano P. Enhancing early Parkinson’s disease detection through multimodal deep learning and explainable AI: insights from the PPMI database. Sci Rep. 2024;14(1):20941. doi:10.1038/s41598-024-70165-4. [Google Scholar] [PubMed] [CrossRef]
128. Wei R, Mahmood A. Recent advances in variational autoencoders with representation learning for biomedical informatics: a survey. IEEE Access. 2020;9:4939–56. [Google Scholar]
129. Cheng J, Gao M, Liu J, Yue H, Kuang H, Liu J, et al. Multimodal disentangled variational autoencoder with game theoretic interpretability for glioma grading. IEEE J Biomed Health Inform. 2022;26(2):673–84. doi:10.1109/JBHI.2021.3095476. [Google Scholar] [PubMed] [CrossRef]
130. Tang Z, Tang S, Wang H, Li R, Zhang X, Zhang W, et al. S2VQ-VAE: semi-supervised vector quantised-variational AutoEncoder for automatic evaluation of trail making test. IEEE J Biomed Health Inform. 2024;28(8):4456–70. doi:10.1109/JBHI.2024.3407881. [Google Scholar] [PubMed] [CrossRef]
131. Elbattah M, Loughnane C, Guérin JL, Carette R, Cilia F, Dequen G. Variational autoencoder for image-based augmentation of eye-tracking data. J Imaging. 2021;7(5):83. doi:10.3390/jimaging7050083. [Google Scholar] [PubMed] [CrossRef]
132. Gomari DP, Schweickart A, Cerchietti L, Paietta E, Fernandez H, Al-Amin H, et al. Variational autoencoders learn transferrable representations of metabolomics data. Commun Biol. 2022;5(1):645. doi:10.1038/s42003-022-03579-3. [Google Scholar] [PubMed] [CrossRef]
133. Tian H, Jiang X, Trozzi F, Xiao S, Larson EC, Tao P. Explore protein conformational space with variational autoencoder. Front Mol Biosci. 2021;8:781635. doi:10.3389/fmolb.2021.781635. [Google Scholar] [PubMed] [CrossRef]
134. Rocha MB, Krohling RA. VAE-GNA: a variational autoencoder with Gaussian neurons in the latent space and attention mechanisms. Knowl Inf Syst. 2024;66(10):6415–37. doi:10.1007/s10115-024-02169-5. [Google Scholar] [CrossRef]
135. Chaddad A, Peng J, Xu J, Bouridane A. Survey of explainable AI techniques in healthcare. Sensors. 2023;23(2):634. doi:10.3390/s23020634. [Google Scholar] [PubMed] [CrossRef]
136. Sindiramutty SR, Tee WJ, Balakrishnan S, Kaur S, Thangaveloo R, Jazri H, et al. Explainable AI in healthcare application. In: Advances in explainable AI applications for smart cities. Hershey, PA, USA: IGI Global Scientific Publishing; 2024. p. 123–76. doi:10.4018/978-1-6684-6361-1.ch005. [Google Scholar] [CrossRef]
137. Hulsen T. Explainable artificial intelligence (XAIconcepts and challenges in healthcare. AI. 2023;4(3):652–66. doi:10.3390/ai4030034. [Google Scholar] [CrossRef]
138. Ye Q, Xia J, Yang G. Explainable AI for COVID-19 CT classifiers: an initial comparison study. In: 2021 EEE 34th International Symposium on Computer-Based Medical Systems (CBMS); 2021 Jun 7–9; Aveiro, Portugal. p. 521–6. doi:10.1109/cbms52027.2021.00103. [Google Scholar] [CrossRef]
139. Letzgus S, Wagner P, Lederer J, Samek W, Müller KR, Montavon G. Toward explainable artificial intelligence for regression models: a methodological perspective. IEEE Signal Process Mag. 2022;39(4):40–58. doi:10.1109/MSP.2022.3153277. [Google Scholar] [CrossRef]
140. van der Velden BHM, Kuijf HJ, Gilhuijs KGA, Viergever MA. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Image Anal. 2022;79:102470. doi:10.1016/j.media.2022.102470. [Google Scholar] [PubMed] [CrossRef]
141. Vishwa Priya V, Chattu P, Sivasankari K, Pisal DT, Renuka Sai B, Suganthi D. Exploring convolution neural networks for image classification in medical imaging. In: 2024 International Conference on Intelligent and Innovative Technologies in Computing, Electrical and Electronics (IITCEE); 2024 Jan 24–25; Bangalore, India. doi:10.1109/IITCEE59897.2024.10467794. [Google Scholar] [CrossRef]
142. Kourounis G, Elmahmudi AA, Thomson B, Hunter J, Ugail H, Wilson C. Computer image analysis with artificial intelligence: a practical introduction to convolutional neural networks for medical professionals. Postgrad Med J. 2023;99(1178):1287–94. doi:10.1093/postmj/qgad095. [Google Scholar] [PubMed] [CrossRef]
143. Said MMR, Islam MSB, Sumon MSI, Vranic S, Al Saady RM, Alqahtani A, et al. Innovative deep learning architecture for the classification of lung and colon cancer from histopathology images. Appl Comput Intell Soft Comput. 2024;2024(1):5562890. doi:10.1155/2024/5562890. [Google Scholar] [CrossRef]
144. Dasgupta S, Saha B. Big data analysis on medical field for drug recommendation using apriori algorithm and deep learning. Multimed Tools Appl. 2024;83(35):83029–51. doi:10.1007/s11042-024-18832-6. [Google Scholar] [CrossRef]
145. Yang M, Matan-Lithwick S, Wang Y, De Jager PL, Bennett DA, Felsky D. Multi-omic integration via similarity network fusion to detect molecular subtypes of ageing. Brain Commun. 2023;5(2):fcad110. doi:10.1093/braincomms/fcad110. [Google Scholar] [PubMed] [CrossRef]
146. Gong P, Cheng L, Zhang Z, Meng A, Li E, Chen J, et al. Multi-omics integration method based on attention deep learning network for biomedical data classification. Comput Methods Programs Biomed. 2023;231(S3):107377. doi:10.1016/j.cmpb.2023.107377. [Google Scholar] [PubMed] [CrossRef]
147. Wang Z, Lin R, Li Y, Zeng J, Chen Y, Ouyang W, et al. Deep learning-based multi-modal data integration enhancing breast cancer disease-free survival prediction. Precis Clin Med. 2024;7(2):pbae012. doi:10.1093/pcmedi/pbae012. [Google Scholar] [PubMed] [CrossRef]
148. Zhang H, Huang D, Chen Y, Li F. GraphSeqLM: a unified graph language framework for omic graph learning. arXiv:2412.15790. 2024. [Google Scholar]
149. Cai L, Ma X, Ma J. Integrating scRNA-seq and scATAC-seq with inter-type attention heterogeneous graph neural networks. Brief Bioinform. 2024;26(1):bbae711. doi:10.1093/bib/bbae711. [Google Scholar] [PubMed] [CrossRef]
150. Mohammed S, Fiaidhi J, Martinez AS. Using meta-transformers for multimodal clinical decision support and evidence-based medicine. medRxiv. 2024;2024(3):1–8. doi:10.1101/2024.08.14.24312001. [Google Scholar] [CrossRef]
151. Wu Y, Xie L. AI-driven multi-omics integration for multi-scale predictive modeling of genotype-environment-phenotype relationships. Comput Struct Biotechnol J. 2025;27(3):265–77. doi:10.1016/j.csbj.2024.12.030. [Google Scholar] [PubMed] [CrossRef]
152. AlSaad R, Abd-Alrazaq A, Boughorbel S, Ahmed A, Renault MA, Damseh R, et al. Multimodal large language models in health care: applications, challenges, and future outlook. J Med Internet Res. 2024;26:e59505. doi:10.2196/59505. [Google Scholar] [PubMed] [CrossRef]
153. Ramzan M, Saeed MU, Ali G. Enhancing anemia detection through multimodal data fusion: a non-invasive approach using EHRs and conjunctiva images. Discov Artif Intell. 2024;4(1):100. doi:10.1007/s44163-024-00196-3. [Google Scholar] [CrossRef]
154. Fischer R, Dinklage A, Pasch E. Bayesian modelling of fusion diagnostics. Plasma Phys Control Fusion. 2003;45(7):1095–111. doi:10.1088/0741-3335/45/7/304. [Google Scholar] [CrossRef]
155. Steyaert S, Pizurica M, Nagaraj D, Khandelwal P, Hernandez-Boussard T, Gentles AJ, et al. Multimodal data fusion for cancer biomarker discovery with deep learning. Nat Mach Intell. 2023;5(4):351–62. doi:10.1038/s42256-023-00633-5. [Google Scholar] [PubMed] [CrossRef]
156. Hirosawa T, Harada Y, Tokumasu K, Ito T, Suzuki T, Shimizu T. Evaluating ChatGPT-4’s diagnostic accuracy: impact of visual data integration. JMIR Med Inform. 2024;12:e55627. doi:10.2196/55627. [Google Scholar] [PubMed] [CrossRef]
157. Chang JS, Kim H, Baek ES, Choi JE, Lim JS, Kim JS, et al. Continuous multimodal data supply chain and expandable clinical decision support for oncology. npj Digit Med. 2025;8(1):128. doi:10.1038/s41746-025-01508-2. [Google Scholar] [PubMed] [CrossRef]
158. Lilhore UK, Simaiya S. Integrating multimodal data fusion for advanced biomedical analysis. In: Multimodal data fusion for bioinformatics artificial intelligence. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2025. p. 127–45. doi: 10.1002/9781394269969.ch7. [Google Scholar] [CrossRef]
159. Kumar V, Joshi K, Kumar R, Anandaram H, Bhagat VK, Baloni D, et al. Multi modalities medical image fusion using deep learning and metaverse technology: healthcare 4.0 a futuristic approach. Biomed Pharmacol J. 2023;16(4):1949–59. doi:10.13005/bpj/2772. [Google Scholar] [CrossRef]
160. Bodaghi M, Hosseini M, Gottumukkala R. A multimodal intermediate fusion network with manifold learning for stress detection. In: 2024 IEEE 3rd International Conference on Computing and Machine Intelligence (ICMI); 2024 Apr 13–14; Mt Pleasant, MI, USA. doi:10.1109/ICMI60790.2024.10586177. [Google Scholar] [CrossRef]
161. Aksu F, Gelardi F, Chiti A, Soda P. Multi-stage intermediate fusion for multimodal learning to classify non-small cell lung cancer subtypes from CT and PET. Pattern Recognit Lett. 2025;193(9):86–93. doi:10.1016/j.patrec.2025.04.001. [Google Scholar] [CrossRef]
162. Tatiparti S, Reddy TBP. Deep multi-modal fusion of clinical and non-clinical data using early submission for enhanced kidney disease prediction. In: Data science and intelligent computing techniques. Delhi, India: Soft Computing Research Society Publications; 2023. p. 711–21. doi:10.56155/978-81-955020-2-8-63. [Google Scholar] [CrossRef]
163. Patel KK, Kanodia A, Kumar B, Gupta R. Multi-modal data fusion based cardiac disease prediction using late fusion and 2D CNN architectures. In: 2024 11th International Conference on Signal Processing and Integrated Networks (SPIN); 2024 Mar 21–22; Noida, India. p. 279–84. doi:10.1109/SPIN60856.2024.10512195. [Google Scholar] [CrossRef]
164. Begum US. Federated and multi-modal learning algorithms for healthcare and cross-domain analytics. Patterniq Min. 2024;1(4):38–51. doi:10.70023/sahd/241104. [Google Scholar] [CrossRef]
165. Nikolaou N, Salazar D, RaviPrakash H, Gonçalves M, Mulla R, Burlutskiy N, et al. Quantifying the advantage of multimodal data fusion for survival prediction in cancer patients. bioRxiv. 2024;19(6):3735. doi:10.1101/2024.01.08.574756. [Google Scholar] [CrossRef]
166. Karbout K, El Ghazouani M, Lachgar M, Hrimech H. Multimodal data fusion techniques in smart healthcare. In: 2024 International Conference on Global Aeronautical Engineering and Satellite Technology (GAST); 2024 Apr 24–26; Marrakesh, Morocco. doi:10.1109/GAST60528.2024.10520803. [Google Scholar] [CrossRef]
167. Wang J, Li J, Wang R, Zhou X. VAE-driven multimodal fusion for early cardiac disease detection. IEEE Access. 2024;12:90535–51. [Google Scholar]
168. Qiu Z, Yang P, Xiao C, Wang S, Xiao X, Qin J, et al. 3D multimodal fusion network with disease-induced joint learning for early Alzheimer’s disease diagnosis. IEEE Trans Med Imaging. 2024;43(9):3161–75. doi:10.1109/TMI.2024.3386937. [Google Scholar] [PubMed] [CrossRef]
169. Golcha R, Khobragade P, Talekar A. Multimodal deep learning for advanced health monitoring a comprehensive approach for enhanced precision and early disease detection. In: 2024 5th International Conference on Innovative Trends in Information Technology (ICITIIT); 2024 Mar 15–16; Kottayam, India. doi:10.1109/ICITIIT61487.2024.10580622. [Google Scholar] [CrossRef]
170. Muhammad Danyal M, Shah Khan S, Shah Khan R, Jan S, Rahman NU. Enhancing multi-modality medical imaging: a novel approach with Laplacian filter + discrete Fourier transform pre-processing and stationary wavelet transform fusion. J Intell Med Healthcare. 2024;2:35–53. doi:10.32604/jimh.2024.051340. [Google Scholar] [CrossRef]
171. Wang Y, Yin C, Zhang P. Multimodal risk prediction with physiological signals, medical images and clinical notes. Heliyon. 2024;10(5):e26772. doi:10.1016/j.heliyon.2024.e26772. [Google Scholar] [PubMed] [CrossRef]
172. D’Souza NS, Wang H, Giovannini A, Foncubierta-Rodriguez A, Beck KL, Boyko O, et al. Fusing modalities by multiplexed graph neural networks for outcome prediction from medical data and beyond. Med Image Anal. 2024;93(2):103064. doi:10.1016/j.media.2023.103064. [Google Scholar] [PubMed] [CrossRef]
173. Cui S, Wang J, Zhong Y, Liu H, Wang T, Ma F. Automated fusion of multimodal electronic health records for better medical predictions. In: Proceedings of the 2024 SIAM International Conference on Data Mining (SDM). Philadelphia, PA, USA: Society for Industrial and Applied Mathematics; 2024. p. 361–9. doi:10.1137/1.9781611978032.41. [Google Scholar] [PubMed] [CrossRef]
174. Cahan N, Klang E, Marom EM, Soffer S, Barash Y, Burshtein E, et al. Multimodal fusion models for pulmonary embolism mortality prediction. Sci Rep. 2023;13(1):7544. doi:10.1038/s41598-023-34303-8. [Google Scholar] [PubMed] [CrossRef]
175. Hussain S, Ali Teevno M, Naseem U, Betzabeth Avendaño Avalos D, Cardona-Huerta S, Gerardo Tamez-Peña J. Multiview multimodal feature fusion for breast cancer classification using deep learning. IEEE Access. 2024;13:9265–75. [Google Scholar]
176. Huang H, Zheng D, Chen H, Wang Y, Chen C, Xu L, et al. Fusion of CT images and clinical variables based on deep learning for predicting invasiveness risk of stage I lung adenocarcinoma. Med Phys. 2022;49(10):6384–94. doi:10.1002/mp.15903. [Google Scholar] [PubMed] [CrossRef]
177. Zhong B, Zhang R, Luo S, Zheng J. ILDIM-MFAM: interstitial lung disease identification model with multi-modal fusion attention mechanism. Front Med. 2024;11:1446936. doi:10.3389/fmed.2024.1446936. [Google Scholar] [PubMed] [CrossRef]
178. Gao F, Ding J, Gai B, Cai D, Hu C, Wang FA, et al. Interpretable multimodal fusion model for bridged histology and genomics survival prediction in pan-cancer. Adv Sci. 2025;12(17):e2407060. doi:10.1002/advs.202407060. [Google Scholar] [PubMed] [CrossRef]
179. Kumar S, Sharma S. An improved deep learning framework for multimodal medical data analysis. Big Data Cogn Comput. 2024;8(10):125. doi:10.3390/bdcc8100125. [Google Scholar] [CrossRef]
180. Noaman NF, Kanber BM, Al Smadi A, Jiao L, Alsmadi MK. Advancing oncology diagnostics: ai-enabled early detection of lung cancer through hybrid histological image analysis. IEEE Access. 2024;12:64396–415. [Google Scholar]
181. Yao Z, Lin F, Chai S, He W, Dai L, Fei X. Integrating medical imaging and clinical reports using multimodal deep learning for advanced disease analysis. In: 2024 IEEE 2nd International Conference on Sensors, Electronics and Computer Engineering (ICSECE); 2024 Aug 29–31; Jinzhou, China. p. 1217–23. doi:10.1109/ICSECE61636.2024.10729527. [Google Scholar] [CrossRef]
182. Atrey K, Singh BK, Bodhey NK. Integration of ultrasound and mammogram for multimodal classification of breast cancer using hybrid residual neural network and machine learning. Image Vis Comput. 2024;145(1):104987. doi:10.1016/j.imavis.2024.104987. [Google Scholar] [CrossRef]
183. Uddin AH, Chen YL, Akter MR, Ku CS, Yang J, Por LY. Colon and lung cancer classification from multi-modal images using resilient and efficient neural network architectures. Heliyon. 2024;10(9):e30625. doi:10.1016/j.heliyon.2024.e30625. [Google Scholar] [PubMed] [CrossRef]
184. Sharma A, Singh SK, Kumar S, Preet M, Gupta BB, Arya V, et al. Revolutionizing healthcare systems: synergistic multimodal ensemble learning & knowledge transfer for lung cancer delineation & taxonomy. In: 2024 IEEE International Conference on Consumer Electronics (ICCE); 2024 Jan 6–8; Las Vegas, NV, USA. doi:10.1109/ICCE59016.2024.10444476. [Google Scholar] [CrossRef]
185. Zhang Y, Sun B, Yu Y, Lu J, Lou Y, Qian F, et al. Multimodal fusion of liquid biopsy and CT enhances differential diagnosis of early-stage lung adenocarcinoma. npj Precis Oncol. 2024;8(1):50. doi:10.1038/s41698-024-00551-8. [Google Scholar] [PubMed] [CrossRef]
186. Dai J, Li X, Xing R, Zheng L. Graph-driven multimodal information model for robust feature fusion. In: 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2024 Dec 3–6; Lisbon, Portugal. p. 5735–42. doi:10.1109/BIBM62325.2024.10822672. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF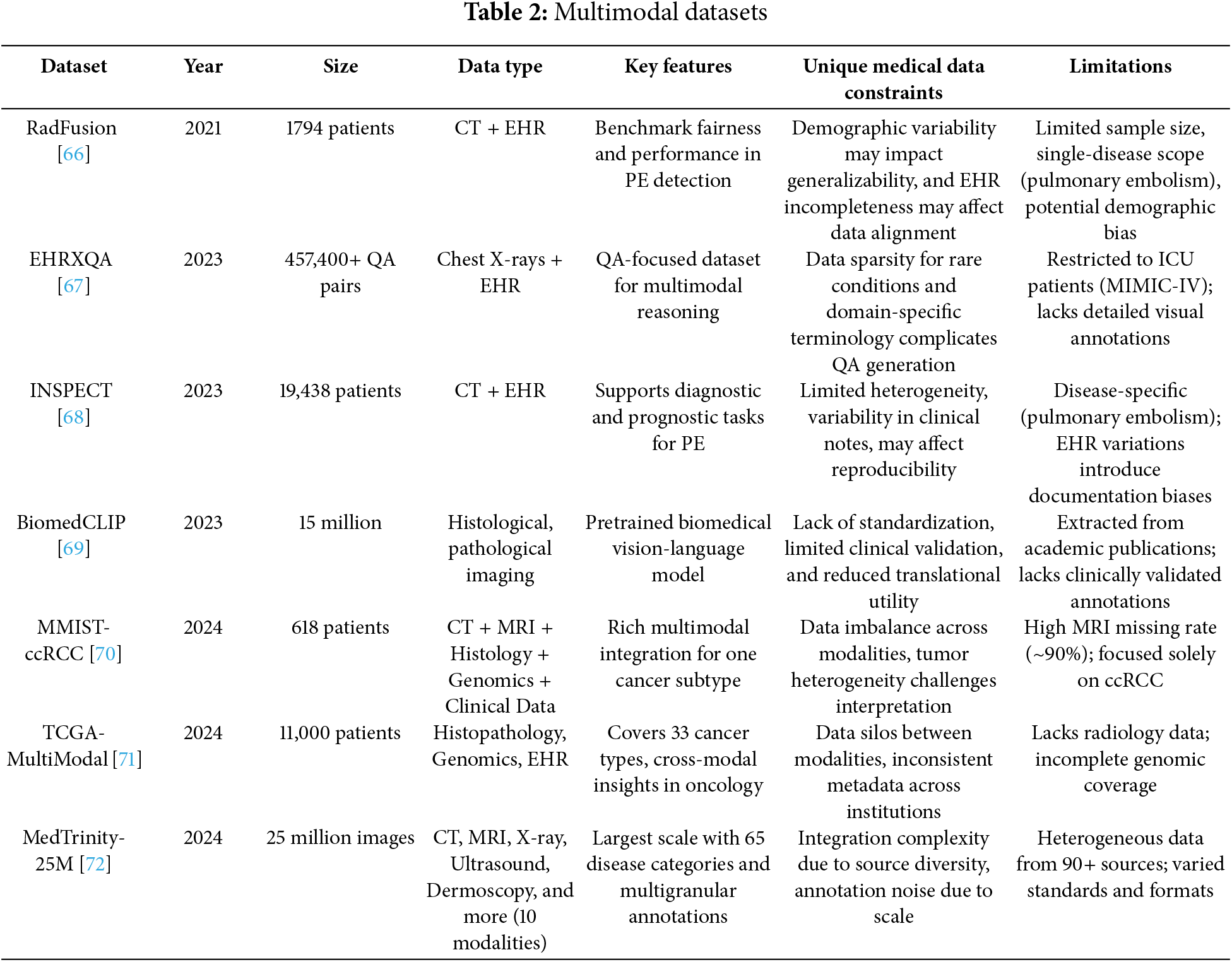
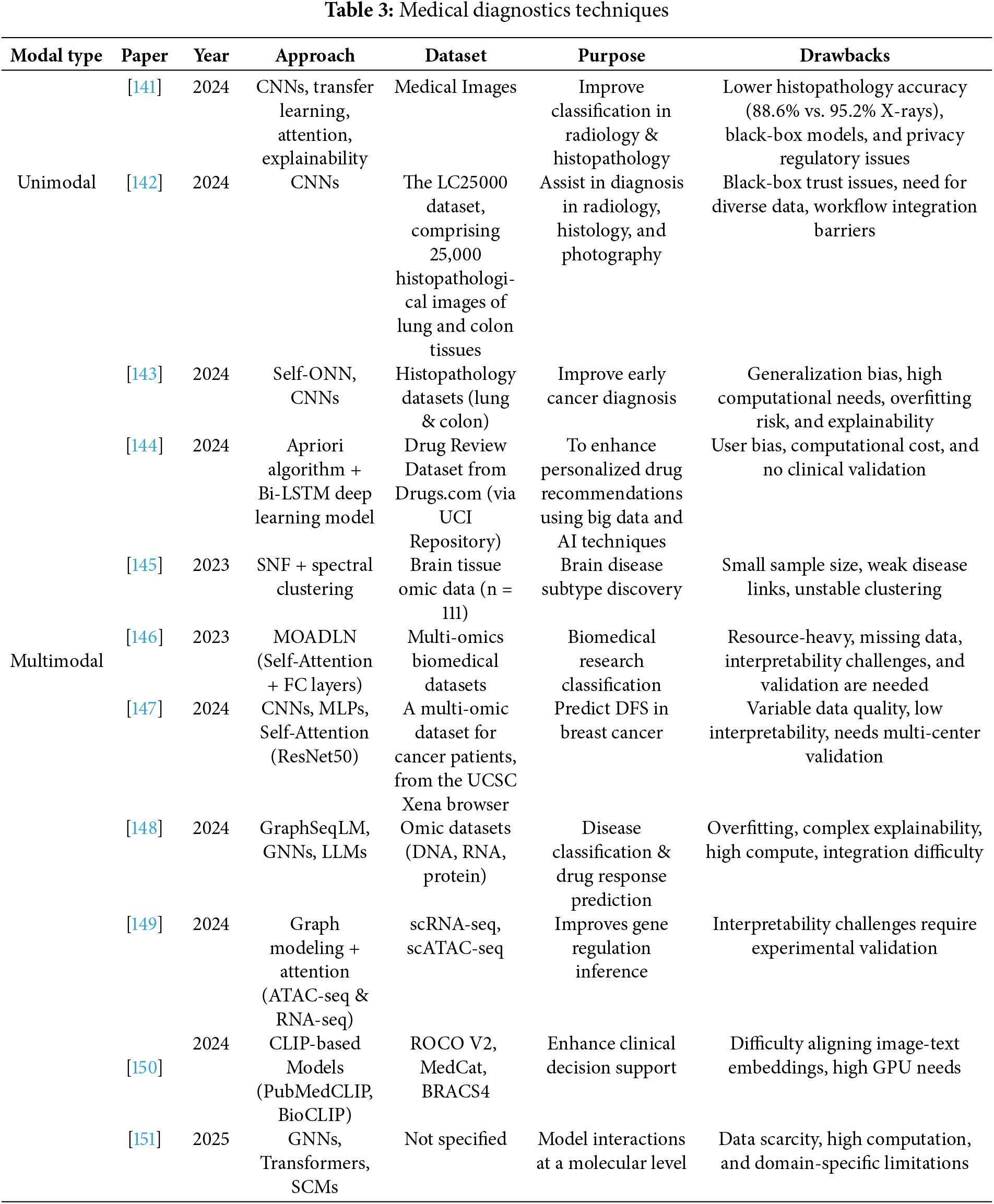
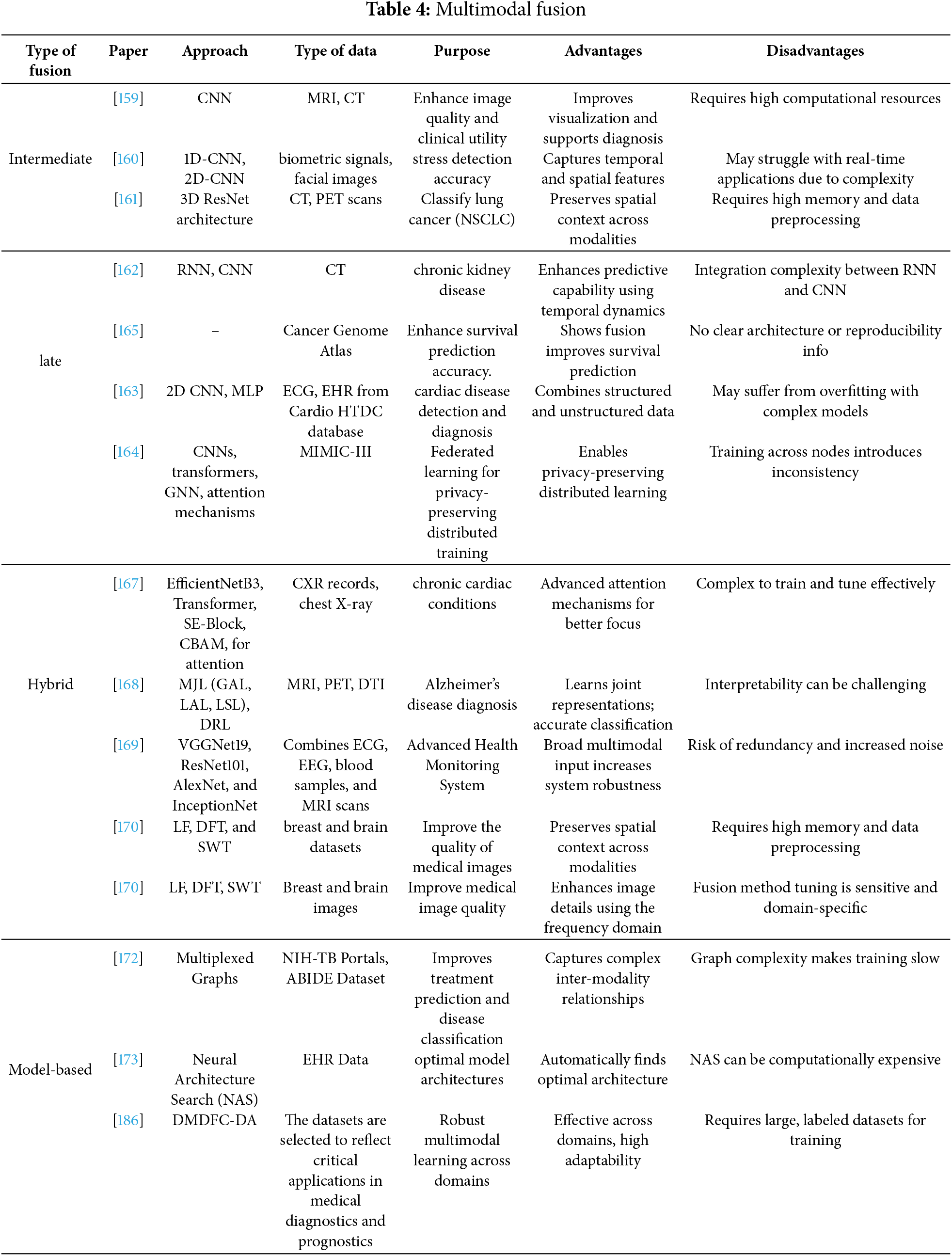
 Downloads
Downloads
 Citation Tools
Citation Tools