 Open Access
Open Access
ARTICLE
RE-UKAN: A Medical Image Segmentation Network Based on Residual Network and Efficient Local Attention
School of Electronic Information Engineering, Shanghai Dianji University, Shanghai, 201306, China
* Corresponding Author: Jie Jia. Email:
(This article belongs to the Special Issue: Deep Learning: Emerging Trends, Applications and Research Challenges for Image Recognition)
Computers, Materials & Continua 2026, 86(3), 94 https://doi.org/10.32604/cmc.2025.071186
Received 01 August 2025; Accepted 19 November 2025; Issue published 12 January 2026
Abstract
Medical image segmentation is of critical importance in the domain of contemporary medical imaging. However, U-Net and its variants exhibit limitations in capturing complex nonlinear patterns and global contextual information. Although the subsequent U-KAN model enhances nonlinear representation capabilities, it still faces challenges such as gradient vanishing during deep network training and spatial detail loss during feature downsampling, resulting in insufficient segmentation accuracy for edge structures and minute lesions. To address these challenges, this paper proposes the RE-UKAN model, which innovatively improves upon U-KAN. Firstly, a residual network is introduced into the encoder to effectively mitigate gradient vanishing through cross-layer identity mappings, thus enhancing modelling capabilities for complex pathological structures. Secondly, Efficient Local Attention (ELA) is integrated to suppress spatial detail loss during downsampling, thereby improving the perception of edge structures and minute lesions. Experimental results on four public datasets demonstrate that RE-UKAN outperforms existing medical image segmentation methods across multiple evaluation metrics, with particularly outstanding performance on the TN-SCUI 2020 dataset, achieving IoU of 88.18% and Dice of 93.57%. Compared to the baseline model, it achieves improvements of 3.05% and 1.72%, respectively. These results fully demonstrate RE-UKAN’s superior detail retention capability and boundary recognition accuracy in complex medical image segmentation tasks, providing a reliable solution for clinical precision segmentation.Keywords
Medical image segmentation plays a critical role in modern medical imaging [1]. In this way, it is possible for professionals to locate the areas of lesions more accurately and to supply a scientific basis for treatment planning and the progress of the disease [2]. Traditional medical image segmentation methods have attained considerable success, relying on edge and texture features and requiring extensive human intervention to limit their applicability and accuracy [3]. As deep learning technology has advanced rapidly, neural network-based segmentation methods are now taking over traditional approaches and have become the main technology used in medical image segmentation [4].
U-Net [5] brought notable progress to medical image segmentation in 2015. Its distinctive U-shaped symmetric structure and skip-connection configurations laid the foundation for this advancement, as they well balance retention of fine-grained features and extraction of contextual information. Subsequent research has produced numerous improved models based on the U-shaped architecture. Attention U-Net [6] introduces attention mechanisms to better focus on target areas. U-Net++ [7] utilizes nested dense connections, and U-NeXt [8] and Rolling U-Net [9] integrate convolutions with multilayer perceptrons (MLPs). The benefits of these models are mainly for the enhancement of segmentation performance by increasing what is known as target region awareness, improving multi-scale feature fusion as well as improving the balance between local and global modeling capabilities. Nevertheless, these methods have limitations in certain specific tasks, especially in the case of complex situations such as contrast deficiency of tissues and blurring of edges where improvements in nonlinear capabilities and detail conservation are required. The recently proposed U-KAN [10], building upon the U-Net architecture, enhances the model’s nonlinear representation capability and interpretability by incorporating Kolmogorov-Arnold Networks (KANs) [11], thereby offering a new perspective for medical image segmentation. However, U-KAN still exhibits notable shortcomings when addressing challenges unique to medical images. As network depth increases, the model suffers from gradient vanishing and performance degradation, which limits further enhancement of its feature extraction capabilities. Furthermore, the efficacy of feature downsampling in preserving spatial detail information is constrained, leading to suboptimal segmentation accuracy for edge structures and minute lesions.
The proposed RE-UKAN model systematically addresses key challenges in medical image segmentation. To address the issue of instability in deep network training, this model uses U-KAN as the base framework and incorporates residual networks in the encoder. The design establishes a shortcut for gradient propagation through cross-layer identity mappings, thereby enabling the network to learn residual functions rather than directly fitting target mappings. This effectively mitigates gradient vanishing, ensuring training stability for deep networks. The addition of residual modules significantly extends network depth, avoids degradation issues, enhances the extraction of multi-level semantic information, and improves the modeling of complex pathological structures. To address the issue of feature detail loss, Efficient Local Attention (ELA) [12] has been integrated. This module has been shown to enhance focus on critical regions while maintaining computational efficiency. It adaptively amplifies feature in edge areas and minute structures, effectively suppressing detail loss during the process of downsampling. This enhancement leads to an improvement in the accuracy of tissue segmentation, particularly in cases where the tissues exhibit low contrast and blurred boundaries. The RE-UKAN model demonstrates a dual enhancement in training stability and detail preservation through the synergistic interaction of residual networks and ELA. This enhancement is achieved while retaining the nonlinear modeling advantages characteristic of the U-KAN model. This approach offers a more reliable solution for complex medical image segmentation tasks.
The remaining part of this article is organized as follows: In Section 2, this article will conduct a review of the existing literature on medical image segmentation methods and also analyze the current technical status in this field. In Section 3, the RE-UKAN model proposed will elaborate on its overall architecture and various areas for improvement. Section 4 will discuss and analyze the experimental results, which will include the data used in the experiments and the evaluation methods. Finally, Section 5 will integrate the existing research results and describe what the prospects of future research will be like.
With the rapid advancement of deep learning, in the field of medical image segmentation, U-Net based on the encoder-decoder architecture and its variants have shown significant advantages. The H-DenseUNet [13] model uses Dense modules as a fundamental unit of computation, while DUNet [14] introduces deformable convolutions, thereby enabling the network to adaptively adjust the shapes of its receptive fields. The nnU-Net [15] employs an adaptive configuration framework, which automatically optimizes the network architecture and the adopted training strategy based on the characteristics of the dataset. Nevertheless, these methods mainly rely on convolutional neural networks (CNNs) as their fundamental operational units. The local receptive fields inherent in CNNs constrain the model’s capacity to discern global contextual information.
To overcome the limitations of convolution operations, researchers shifted their research focus to architectures with global perception capabilities. They successfully introduced the Vision transformer into the field of medical image segmentation and proposed models like TransUNet [16] and UNETR [17]. The ability of global dependency modeling has been greatly enhanced by leveraging the self-attention mechanisms. However, this global modeling capability also poses some challenges, such as high computational complexity and a relatively strong reliance on large amounts of data. To balance modeling capability and computational efficiency, state space models represented by Mamba attracted attention. Subsequent models such as U-Mamba [18] and VM-UNet [19] emerged by taking advantage of its linear computational complexity and powerful sequence modeling capabilities, it has opened up a new direction for the development of U-shaped architecture.
The model mentioned above, by improving the network architecture and adjusting the optimization strategy, ultimately achieved a relatively satisfactory segmentation performance. However, their core components (e.g., Convolution, Transformer, and MLPs) can only model relationships among different channels within a linear latent space, struggling to capture complex nonlinear patterns effectively. The introduction of KANs offers a novel approach to addressing this challenge. As the first model to integrate KANs into the U-Net framework, U-KAN replaces traditional activation functions with spline-based functions, which improves the model’s ability to model complex spatial dependencies, thus laying a crucial foundation for this study.
2.2 Attention Mechanism in Deep Learning
In fields such as image processing and natural language processing, the integration of attention mechanisms can significantly enhance the effectiveness of deep learning models. In recent years, SENet [20] has received much attention for its introduction of channel attention mechanisms in convolutional blocks, demonstrating considerable potential for performance enhancement. Subsequently, CBAM [21] integrates the attention of the channel dimension and the spatial dimension. It adopts parallel pooling and convolution operations to enhance the ability of feature selection. However, when implementing the dual-branch architecture, computational overhead is introduced. To achieve a balance between performance and efficiency, ECA-Net [22] introduces a local cross-channel interaction mechanism that uses 1D convolutions to generate attention weights. This approach significantly improves performance and minimizes computational requirements. CA [23] embeds the position information into the channel Attention, so that the module can capture broader spatial context information. Triplet Attention [24] establishes cross-dimensional interaction through rotation operations, thus achieving effective feature calibration without reducing the dimension.
Although the attention mechanisms mentioned earlier have improved the model’s performance in various dimensions, they still have certain limitations when dealing with remote spatial dependencies. To address this issue, Xu et al. proposed ELA, which can effectively capture remote spatial dependencies within images [12]. And it can also avoid the performance degradation caused by the reduction of channels. Its most significant advantage is that it can guide the network to focus more precisely on the key image details, greatly improving the segmentation accuracy. ELA does not require channel dimensionality reduction, thus maintaining the simplicity of the model and enhancing the computational efficiency.
2.3 Residual Network Based Applications
In the field of image segmentation, it has been found that increasing the depth of the network is an effective way to improve model performance. However, overly deep networks are prone to issues such as gradient vanishing and network degradation, which eventually limit the improvement of model performance. The introduction of residual networks effectively addressed this challenge by combining identity mapping, which simplifies the training process for deep networks and enables the construction and training of particularly deep architectures.
The combination of residual networks and classical segmentation structures has led to the creation of many successful models. ResUnet [25] added residual blocks to U-Net, which not only retained the original feature extraction ability of the network but also effectively alleviated the problem of vanishing gradients. R2U-Net [26] combined convolutional layers with residual connections, enhancing feature reuse and boundary segmentation accuracy through recurrent residual units. As development progressed, the application of residual mechanisms became increasingly complex. The DeepLabv3+ [27] model achieves the fusion of multi-scale features by integrating residual networks and spatial pyramid pooling. High-Resolution Network (HRNet) [28] has demonstrated a relatively high level of proficiency in pose estimation and semantic segmentation. It achieves such an effect by retaining high resolution while integrating residual connections. These technological advancements have had a significant impact on the performance of deep networks in image segmentation tasks.
The overall structure of the RE-UKAN model is shown in Fig. 1. This model has been optimized based on the U-KAN network architecture and adopts a two-stage encoder-decoder structure. In the encoder part, the Residual-ELA stage is the core stage of feature extraction, which integrates the residual module and the ELA Block. These modules enhance the gradient flow and also enable local feature fusion. The Tokenized KAN stage of the encoder introduces the ELA Block to boost feature processing performance. In the decoder part, the Tokenized KAN stage is the same as the encoder stage, and the ELA Block is introduced for fine-grained feature reconstruction. The convolution stage of the decoder adopts standard convolution operations for feature upsampling and restores the spatial information. The structural design of the RE-UKAN model can well alleviate the problem of vanishing gradients, reduce the loss of features, and improve the ability of the model to capture features.
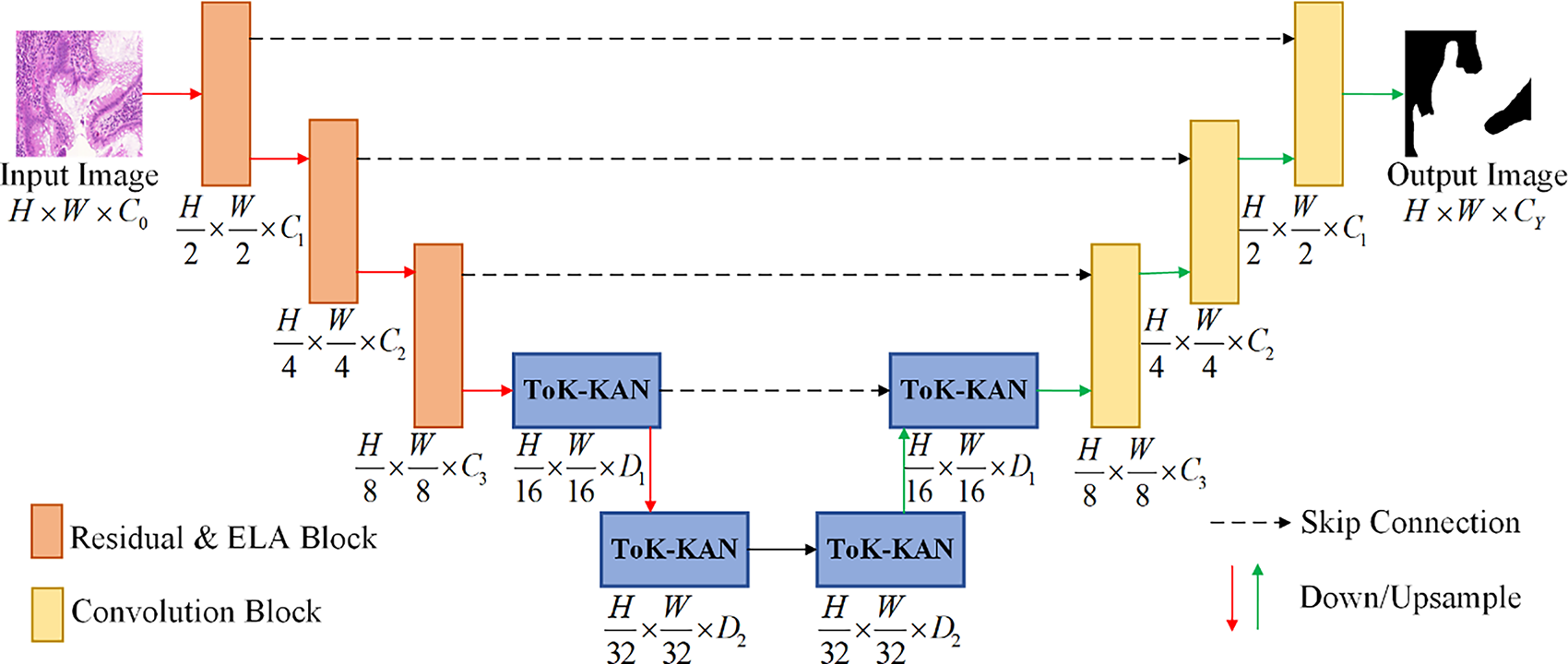
Figure 1: The overall structure diagram of RE-UKAN
In order to address the loss of detail in the U-KAN model caused by limited receptive fields during feature downsampling, and to enhance the model’s ability to represent complex structures and boundaries in medical images, the ELA Block is introduced. This module employs a lightweight design featuring 1D convolutions and group normalization, achieving significant performance improvements through its streamlined architecture.
The structure of ELA is shown in Fig. 2. Firstly, ELA employs strip pooling as opposed to traditional spatial global pooling in order to capture spatially long-range dependencies with precise positional information along both height and width directions. For the inputs
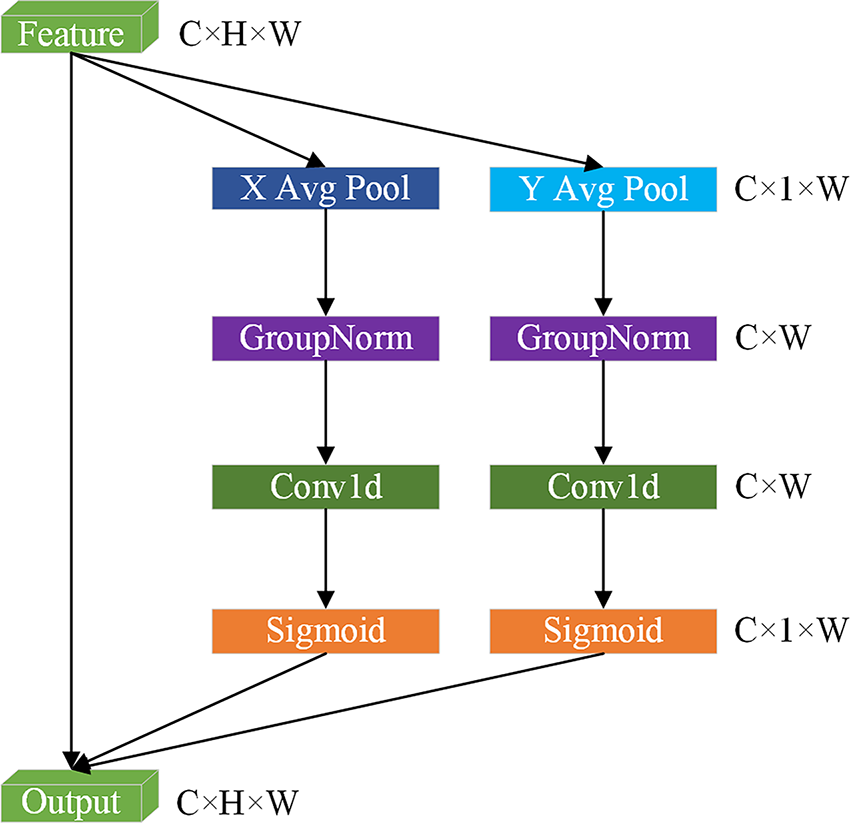
Figure 2: The schematic of Efficient Local Attention (ELA)
In the same way, the output of the
Subsequently, 1D convolutions independently process features in two directions, thereby reducing computational complexity while preserving feature discriminative power. Group normalization (denoted as
As described above,
However, the integration of the attention mechanism may result in an escalation of model intricacy, which could potentially lead to overfitting phenomena. Consequently, a dropout mechanism has been incorporated into the ELA Block to balance the model complexity and reduce the dependence on a single feature. This strategy preserves the performance gains from the attention mechanism while ensuring stable segmentation performance on new data.
3.3 Residual Module in the Encoder Layer
In order to overcome the issues of gradient vanishing and model degradation encountered by the U-KAN network when processing deep architectures, we introduced a residual module into the encoder and incorporated an ELA Block into the residual module. As illustrated in Fig. 3a, the standard convolutional module in the convolutional stage of the decoder consists of two 3 × 3 convolutional blocks with ReLU activation functions and a batch normalization layer.
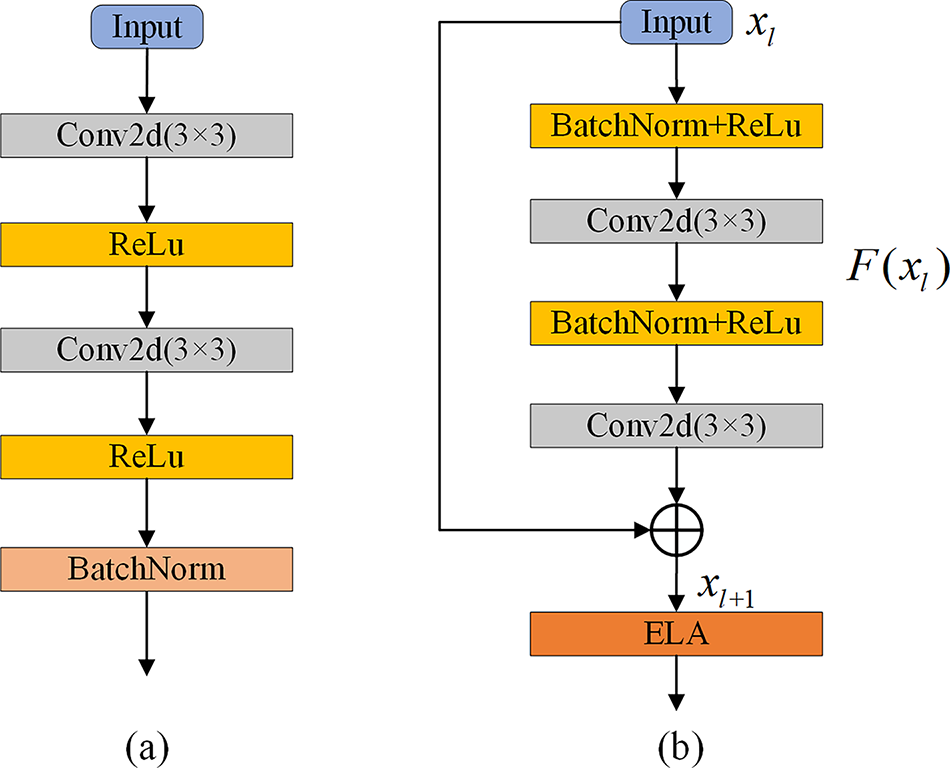
Figure 3: The components of the Residual module in RE-UKAN. (a) Convolution module for the decoder convolution stage, (b) Residual module for the encoder Residual-ELA stage
The residual module for the Residual-ELA stage of the encoder is illustrated in Fig. 3b. In the Residual-ELA stage of the coding path, each layer contains a residual unit, which aims to effectively improve the depth and expressiveness of the network. Each residual unit consists of two 3 × 3 convolutional blocks, two batch normalization layers, and the ReLU activation function, guaranteeing the stability of the training process. In the first residual unit, we halve the spatial resolution of the feature map through stride convolution. This approach reduces computational effort while enabling the encoder to extract more accurate image features during the downsampling process. The formulas for the residual block are represented by Eqs. (6) and (7).
where
The ELA Block is introduced into the network subsequent to the residual output for the purpose of further processing of features. The expansion of the receptive field enables the network to capture the global background more effectively and retain local details. The stacking of residual modules expands the receptive field of the network, allowing it to process more information and retain richer spatial details. This process provides the decoder with more abundant features, enabling the network to effectively capture information about the global structure while retaining local details. This leads to significantly improved segmentation performance.
In order to enhance the dynamic modeling ability of the model for high-level semantic features at the bottleneck layer, improvements were made to the Tokenized KAN Block based on U-KAN, as illustrated in Fig. 4. This module tokenizes the output features of the encoder’s Residual-ELA stage through overlapping patch embedding, using a 3 × 3 convolutional kernel with a stride of 2 to convert the spatial feature map into serialized tokens. The tokenized features are processed through KAN layers to establish complex nonlinear mapping relationships, effectively capturing the intricate dependencies between tokens.
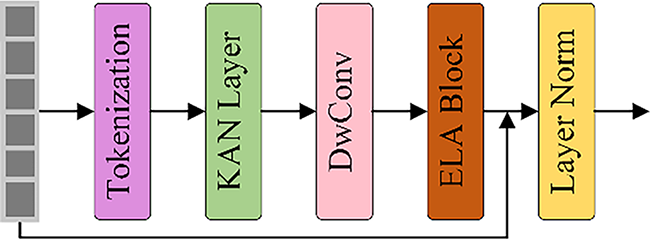
Figure 4: The diagram of Tokenized KAN Block in RE-UKAN
The improvement of lightweight features is achieved through the utilization of separable convolution, which combines the 3 × 3 and the block convolution mechanism. This method significantly reduces the complexity of model parameters and calculations when extracting spatial features. The ELA block introduces a window attention mechanism. It can establish pixel-level dependencies within local areas and also combines residual connections for the superposition of linear features. Layer normalization performs standardized operations on the output features to ensure the stability of training. Then, the regularized features are passed to the next module. The decoder generates the final high-resolution segmentation map by means of progressive upsampling.
In summary, the RE-UKAN model has two key differences in architecture from the U-KAN baseline model to address the inherent limitations of U-KAN. Although U-KAN enhances nonlinear approximation capabilities, its encoder is still prone to vanishing gradients and model degradation when scaled to greater depths. The residual module introduced in RE-UKAN effectively alleviates this problem through identity mapping, enabling the construction of deeper networks and the extraction of abundant hierarchical features. Then, U-KAN lacks an effective mechanism for preserving spatial details during the downsampling process, leading to the loss of information on marginal and minor lesions. The integrated ELA Block in RE-UKAN is specifically designed to enhance the capture of long-range spatial dependencies and detailed features. Therefore, RE-UKAN is not merely an incremental improvement but systematically enhances the training stability of the model and spatial detail perception while retaining the nonlinear advantages of KAN.
To evaluate RE-UKAN’s generalization ability systematically in multi-organ and multi-modal scenarios, comparative experiments were conducted on four public datasets. These datasets contain diverse anatomical structures, including cell nuclei, glands, polyps and thyroid nodules. Furthermore, the experiments involved multiple imaging modalities, including microscopy, endoscopy and ultrasound. Representative images from the four datasets are displayed in Fig. 5.
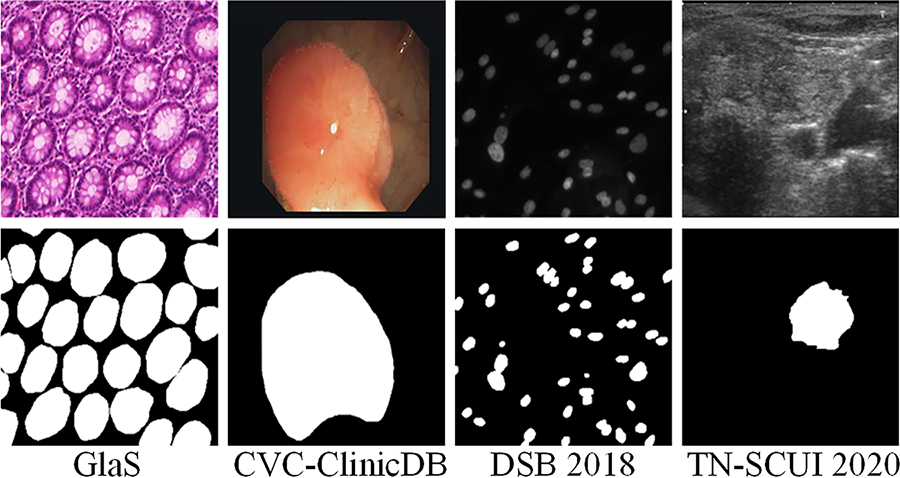
Figure 5: The example images of four datasets. The original images are shown in the upper part and the markers corresponding to the image are shown below
The GlaS dataset is a collection for colorectal gland segmentation, comprising 165 H&E-stained slides. The images originally had a resolution of 775 × 522 pixels; however, all were resized to 512 × 512 pixels to ensure experimental consistency.
The CVC-ClinicDB dataset is composed of 612 RGB frames, which were collected from 29 colonoscopy sequences for the purpose of early polyp detection. The original resolution of the image is 384 × 288, and it has been preprocessed and resized to 256 × 256. Pixel-level polyp masks are also provided.
The DSB 2018 dataset is composed of 670 microscopic RGB images, each of which is annotated with instance-level cell nucleus labels. The images were originally subject to variation in resolution, but were uniformly resized to 512 × 512 for experimental purposes, with the objective of evaluating the performance of the model in scenarios involving the segmentation of cell nuclei.
The TN-SCUI 2020 dataset is currently the largest publicly available thyroid ultrasound nodule segmentation dataset, comprising 4554 2D ultrasound images. Prior to the initiation of the preprocessing stage, a uniform resizing of all images was conducted to a dimension of 256 × 256.
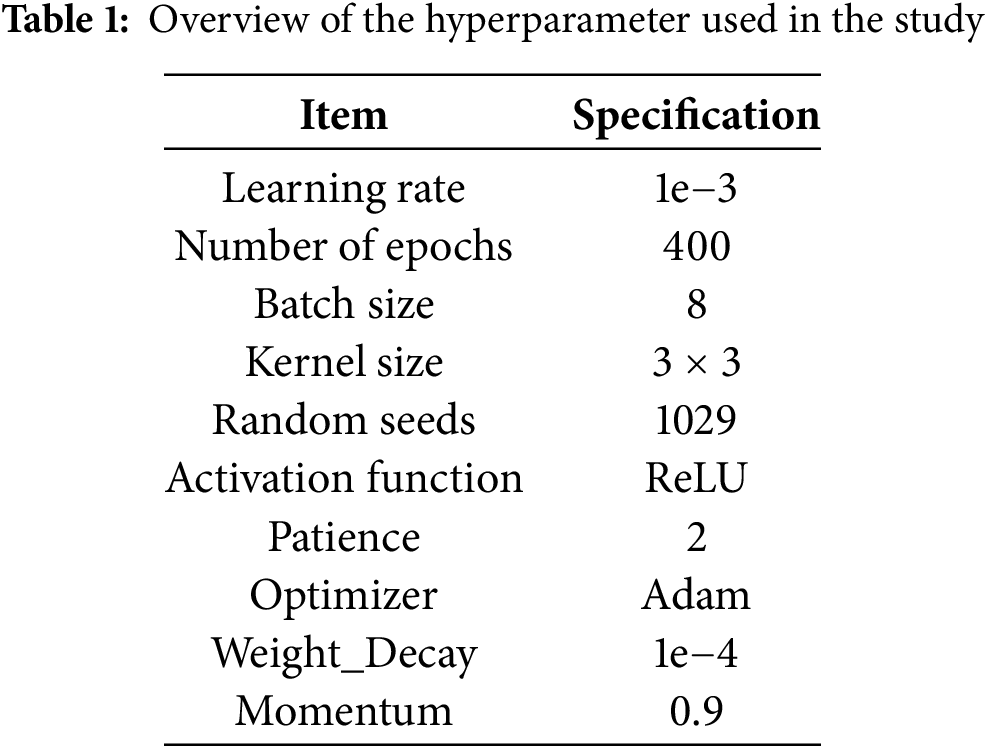
In this study, the implementation of the RE-UKAN model is based on Python 3.10 and the PyTorch 2.0 framework. All experiments were conducted on a device equipped with an NVIDIA Tesla P40 GPU featuring 24 GB of VRAM, running CUDA driver version 12.1 on Windows 11. During the training process, the input images were consistently resized to 256 × 256 pixels, and the segmentation map generated by the model corresponded to this input dimension. The dataset was randomly partitioned into three parts: 80% as the training set, 10% as the validation set, and 10% as the test set. In the model training, the Adam optimizer was adopted. The initial learning rate of this optimizer was set at 1e−3. Meanwhile, the cosine annealing learning rate scheduler was also used to dynamically adjust the learning rate. The minimum learning rate of this scheduler is 1e−5. The loss function integrates the concepts of Binary Cross-Entropy (BCE) and dice loss, enhancing the model’s balanced performance in the domains of foreground and background recognition. To ensure adequate model fitting, all experiments were trained for 400 epochs to guarantee sufficient convergence. Table 1 provides detailed settings of hyperparameters.
During the evaluation phase, three metrics were employed to assess the model’s performance across the four datasets: average Intersection over Union (IoU), average Dice coefficient, and 95% Hausdorff distance (HD95). The Dice coefficient is calculated by Eq. (8) to represent it, where
IoU, which indicates the ratio of predicted area to actual area, is calculated as shown in Eq. (9).
In medical image segmentation, HD95 is a commonly used evaluation metric that is particularly applicable to the evaluation of boundary accuracy. Unlike the Dice coefficient, which is more sensitive to region overlap inside the mask, HD95 focuses on the accuracy of segmented object boundaries. It measures segmentation quality by calculating the maximum distance between the predicted segmentation and the true segmentation boundary, and is able to reduce the effect of outliers or small errors. Therefore, HD95 is more suitable for evaluating segmentation tasks such as organ or tumor boundaries, where accurate boundary representations are important for clinical decision-making. HD95 is derived by Eqs. (10)–(12).
As demonstrated in Fig. 6, which presents the training outcomes across different experimental rounds, it can be observed that the number of training epochs to 400 is more appropriate for ensuring the model’s convergence and stable segmentation performance.
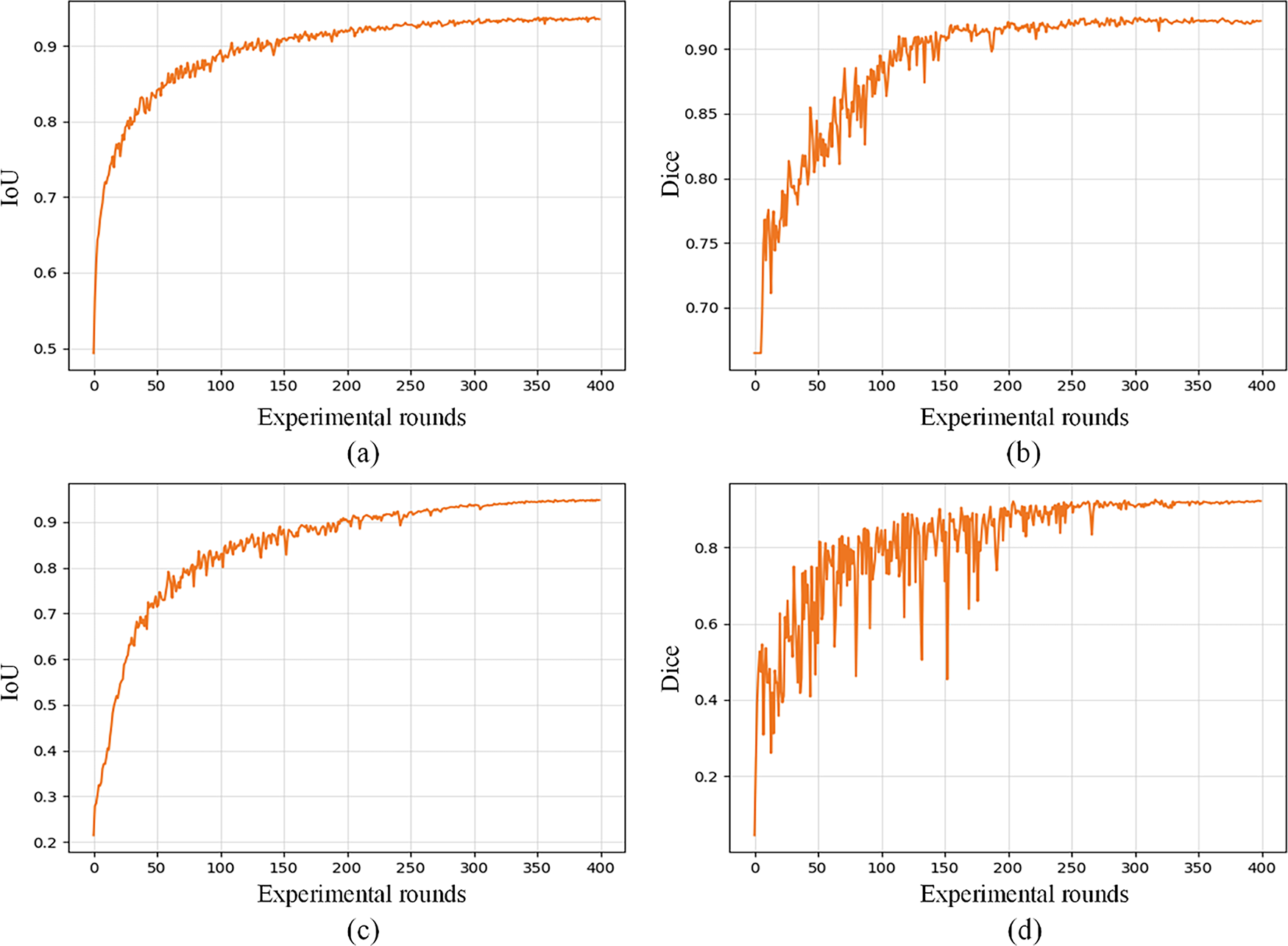
Figure 6: The curves obtained by RE-UKAN on the GlaS dataset (a,b) and the CVC-ClinicDB dataset (c,d). (a) variation curve of Dice on GlaS dataset, (b) variation curve of IoU on GlaS dataset, (c) variation curve of Dice on CVC-ClinicDB dataset, and (d) variation curve of IoU on CVC-ClinicDB dataset. The abscissa indicates the experimental rounds, and the ordinate indicates the numerical value of each index
4.4 Comparison with Other Image Segmentation Methods
In order to systematically evaluate the segmentation performance of RE-UKAN, seven of the most representative U-shaped networks were selected for comparison across four experimental datasets.
As illustrated in Fig. 7, the proposed RE-UKAN method is shown to be effective in achieving accurate segmentation results, with comparable performance to other approaches when applied to each dataset. The figure demonstrates that RE-UKAN consistently produces segmentation results closest to the ground truth across all datasets. In the GlaS gland segmentation task, compared to discontinuities or excessive smoothing observed at blurred boundaries in other methods, RE-UKAN generates continuous and precise gland contours, effectively preserving complex morphological structures. In the CVC-ClinicDB polyp segmentation task, both the baseline model U-KAN and RE-UKAN achieved segmentation results similar to those of the ground truth mask. However, RE-UKAN performed better in handling the fine details of polyp edges. In the DSB2018 cell nucleus segmentation and TN-SCUI 2020 thyroid nodule segmentation tasks, RE-UKAN was particularly proficient in capturing tiny targets and irregular-shaped structures. This results in a significant reduction in segmentation omissions and boundary localization errors, while also exhibiting exceptional detail preservation. It is clearly evident from these visualized results that our method, through the joint design of the residual module and ELA, has significant advantages in enhancing the model’s perception of complex boundaries and its ability to preserve spatial details.
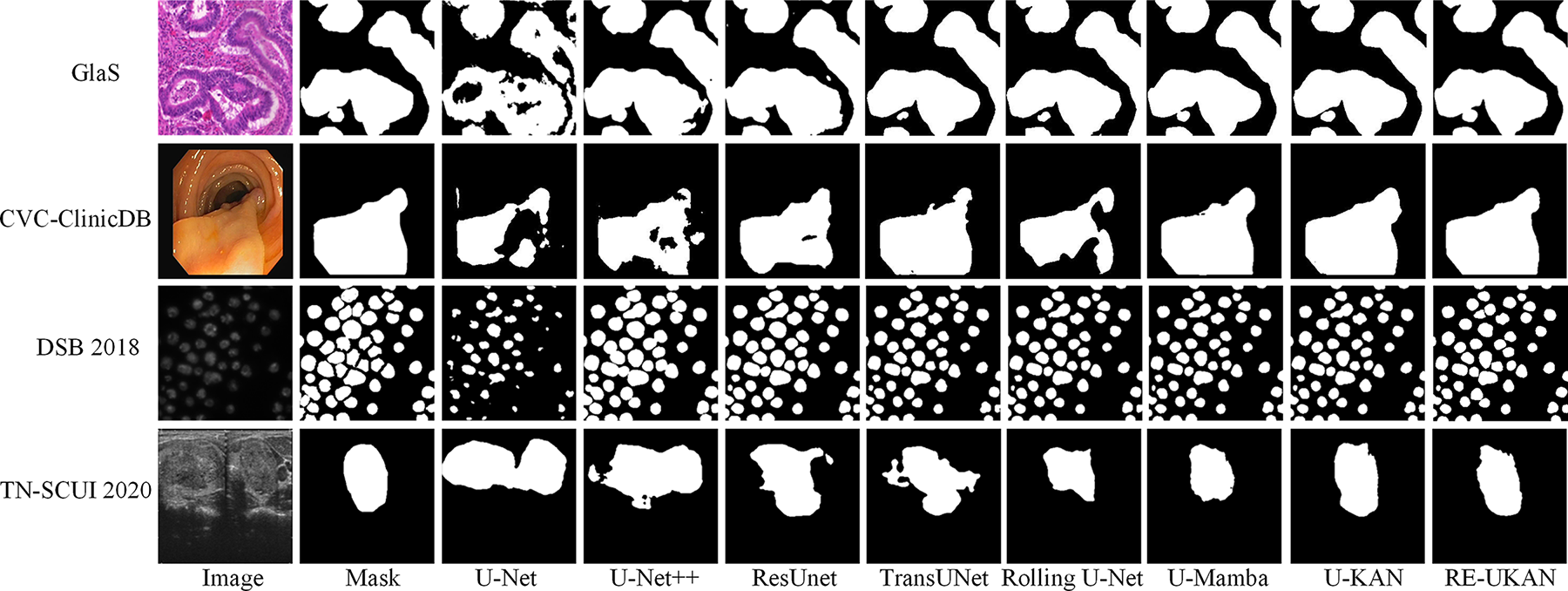
Figure 7: The output results of different methods on the four datasets
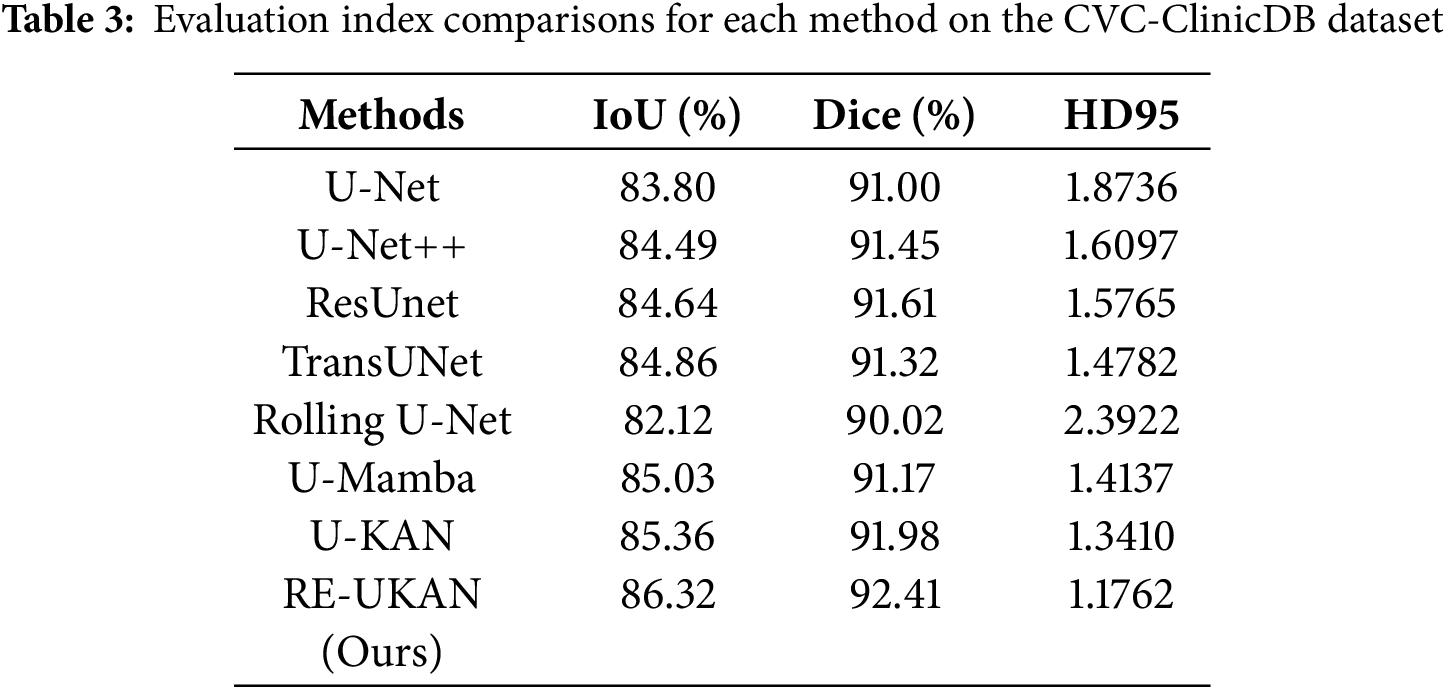
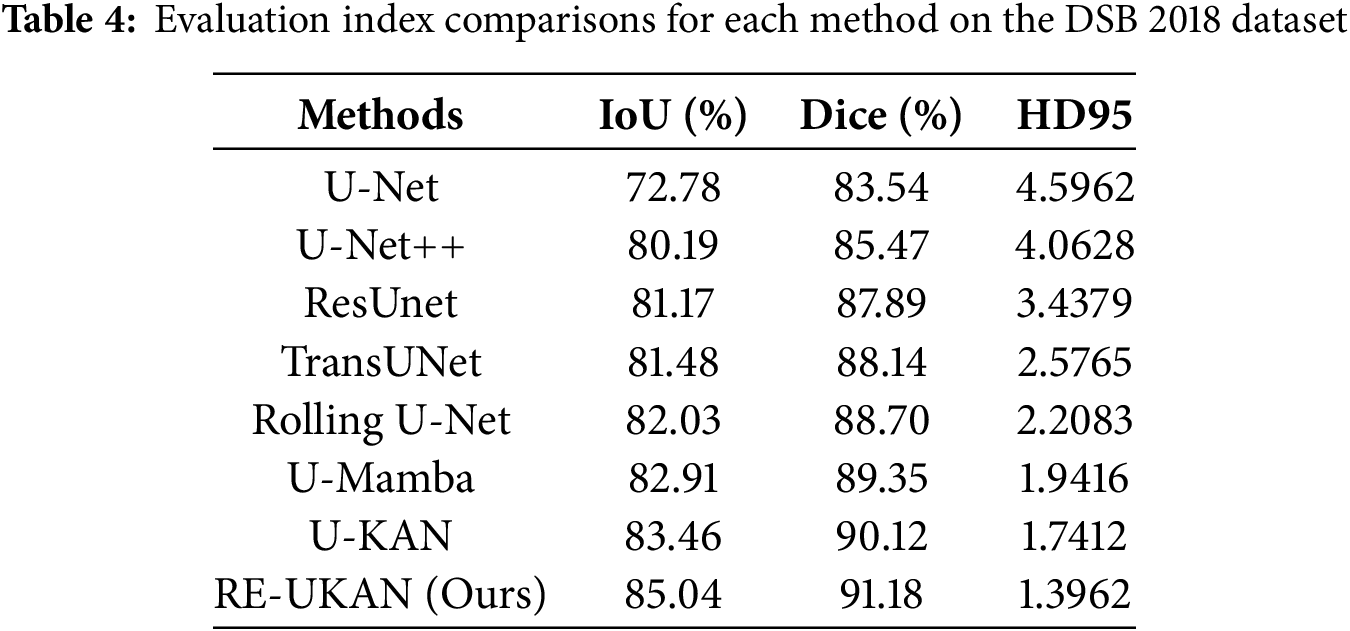
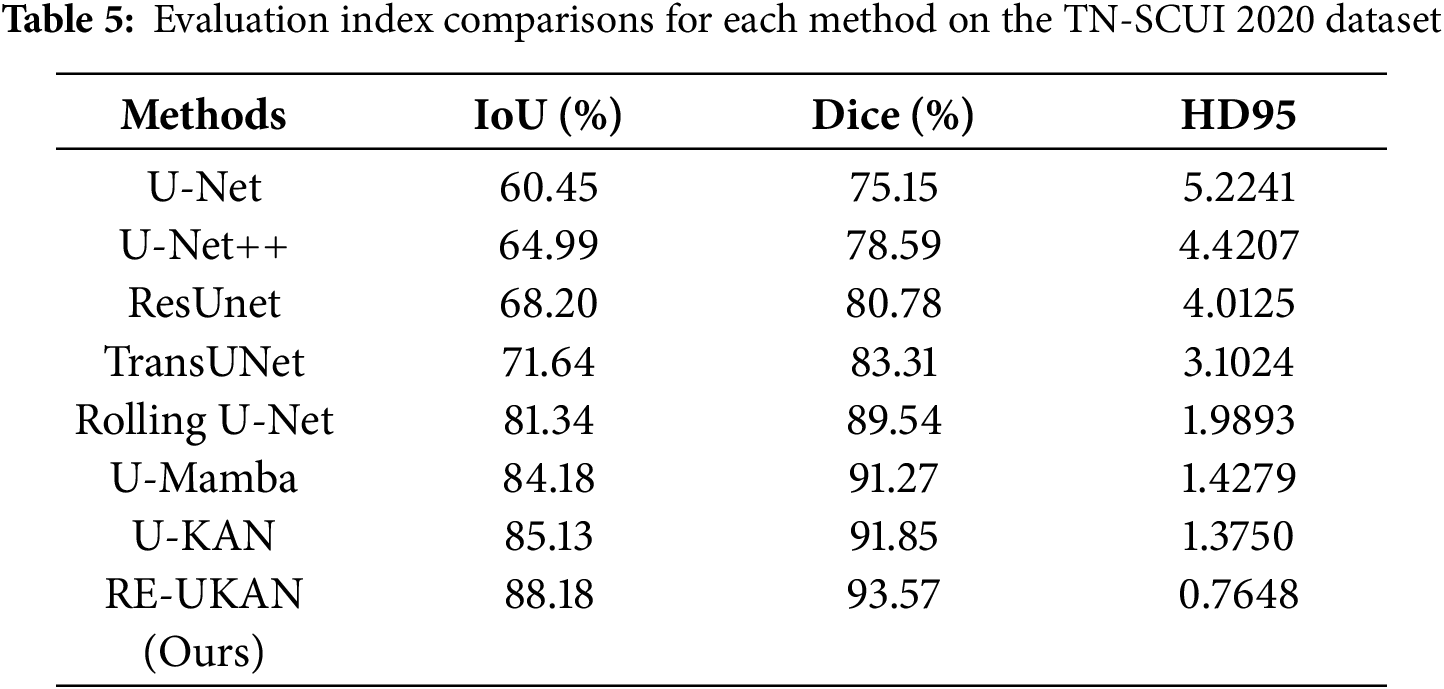
To explain the performance of each method in different tasks in detail, Tables 2–5 summarize the metric comparisons of all benchmark models across four datasets.
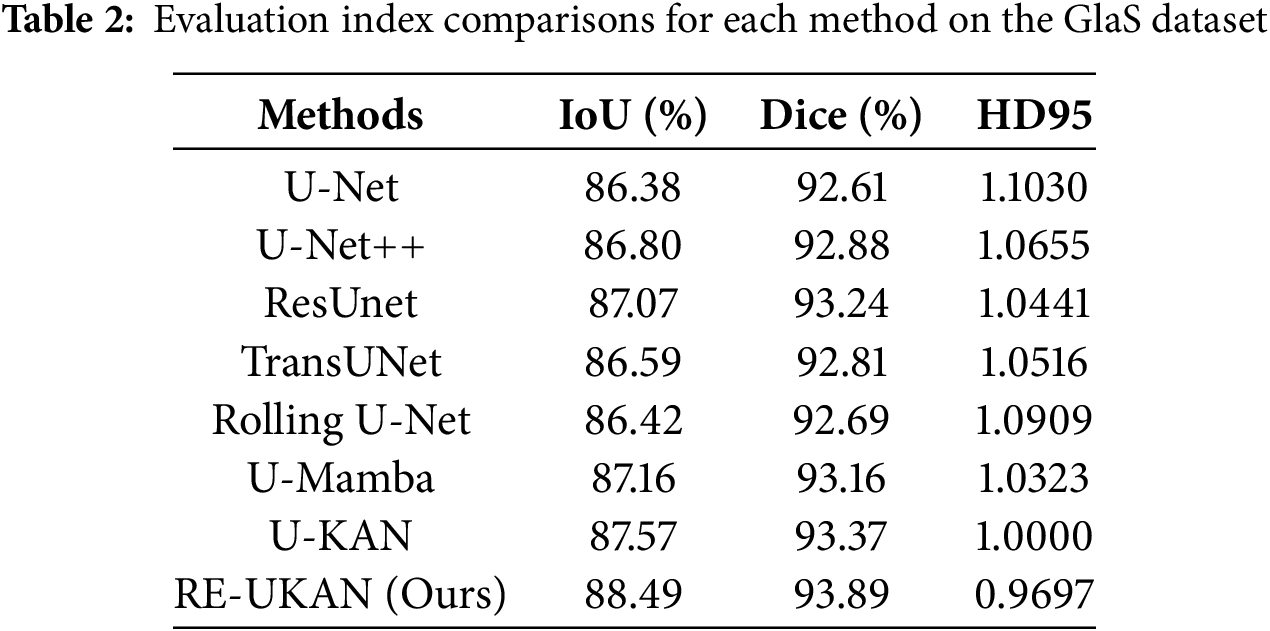
Overall, RE-UKAN shows the optimal performance among all evaluation metrics. As shown in Table 2, RE-UKAN achieved an Aver_Dice of 93.89% on the GlaS dataset, which is 0.52% higher than that of the baseline U-KAN model. This outcome indicated the enhanced efficacy of the model in segmenting intricate glandular structures. Particularly on the boundary precision metric HD95, RE-UKAN achieved 0.9697, significantly outperforming other comparison methods, highlighting the advantage of the ELA Block in preserving edge details.
As shown in Tables 3 and 4, RE-UKAN exhibits substantial performance enhancements on both the CVC-ClinicDB and DSB 2018 datasets. In the polyp segmentation task on CVC-ClinicDB, the model attains Aver_Dice of 92.41% and Aver_IoU of 86.32%. Compared with the baseline model U-KAN, Aver_Dice and Aver_IoU increased by 0.43% and 0.96%. This performance improvement was obvious in the nuclear segmentation task of the DSB 2018 dataset, with Aver_Dice and Aver_IoU increased by 1.06% and 1.58%. The boundary segmentation metric HD95 has also decreased significantly, from 1.7412 in the U-KAN model to 1.3962, a reduction of approximately 20%. These results fully demonstrate that by introducing residual module, RE-UKAN effectively stabilizes deep network training. By working in synergy with the ELA Block, the model was making significant progress in overall segmentation accuracy and achieving breakthrough results in preserving boundary details. Thus, it effectively resolves the limitations of the baseline model in feature extraction and spatial detail retention.
As shown in Table 5, RE-UKAN exhibits remarkable performance on the TN-SCUI 2020 dataset, attaining Aver_Dice of 93.57% and Aver_IoU of 88.18%. This result substantially outperforms traditional methods like U-Net and U-Net++. It also shows notable improvements over the baseline U-KAN model, with gains of 1.72% in Aver_Dice and 3.05% in Aver_IoU. More importantly, in terms of boundary segmentation accuracy, RE-UKAN achieves an HD95 metric of 0.7648, representing a substantial 44.4% reduction compared to U-KAN’s 1.3750. This result clearly shows that through architectural optimization, RE-UKAN achieves breakthrough progress in boundary detail handling while maintaining overall segmentation performance enhancement. This model provides an effective technical solution to solve the problems of blurred boundaries and segmentation obstacles in medical images. It has crucial clinical value for lesion measurement and surgical planning.
In order to comprehensively analyze the contribution of each component module in RE-UKAN to segmentation performance, a series of systematic ablation experiments were designed using two representative datasets: GlaS and CVC-ClinicDB. For the sake of clarity, the residual module is denoted as Res, the ELA Block as ELA, and RE module refers to the combined improvement introduced in this paper. Here it should be noted that all ablation results use Aver_Dice and Aver_IoU as evaluation metrics. The optimal combination is highlighted in color, so that the performance gains brought by different modules can be compared intuitively.
4.5.1 Ablation Studies on Proposed Methods
The experimental results indicated that each enhanced module exhibited a positive impact on the improvement of model performance. As shown in Fig. 8, on the CVC-ClinicDB dataset, compared with the baseline model U-KAN, U-KAN+Res has increased by 0.25% and 0.61% in Aver_Dice and Aver_IoU. This finding serves to verify the efficacy of the residual module in enhancing gradient propagation and maintaining the stability of deep network training. The performance improvement of U-KAN+ELA was more significant, with its Aver_Dice increasing by 0.33% and Aver_IoU increasing by 0.7%. This discovery indicates that the ELA Block improved the representation accuracy of the model for edge details and fine structures by enhancing the ability to capture local features. The RE-UKAN model, which combines residual modules with the ELA Block, was ultimately found to achieve optimal performance. On the GlaS dataset, Aver_Dice and Aver_IoU were improved by 0.52% and 0.92%. On the CVC-ClinicDB dataset, the Aver_Dice and Aver_IoU increased by 0.43% and 0.96%, respectively. In summary, the proposed improvements not only significantly enhance segmentation accuracy but also maintain balanced development in all performance metrics.
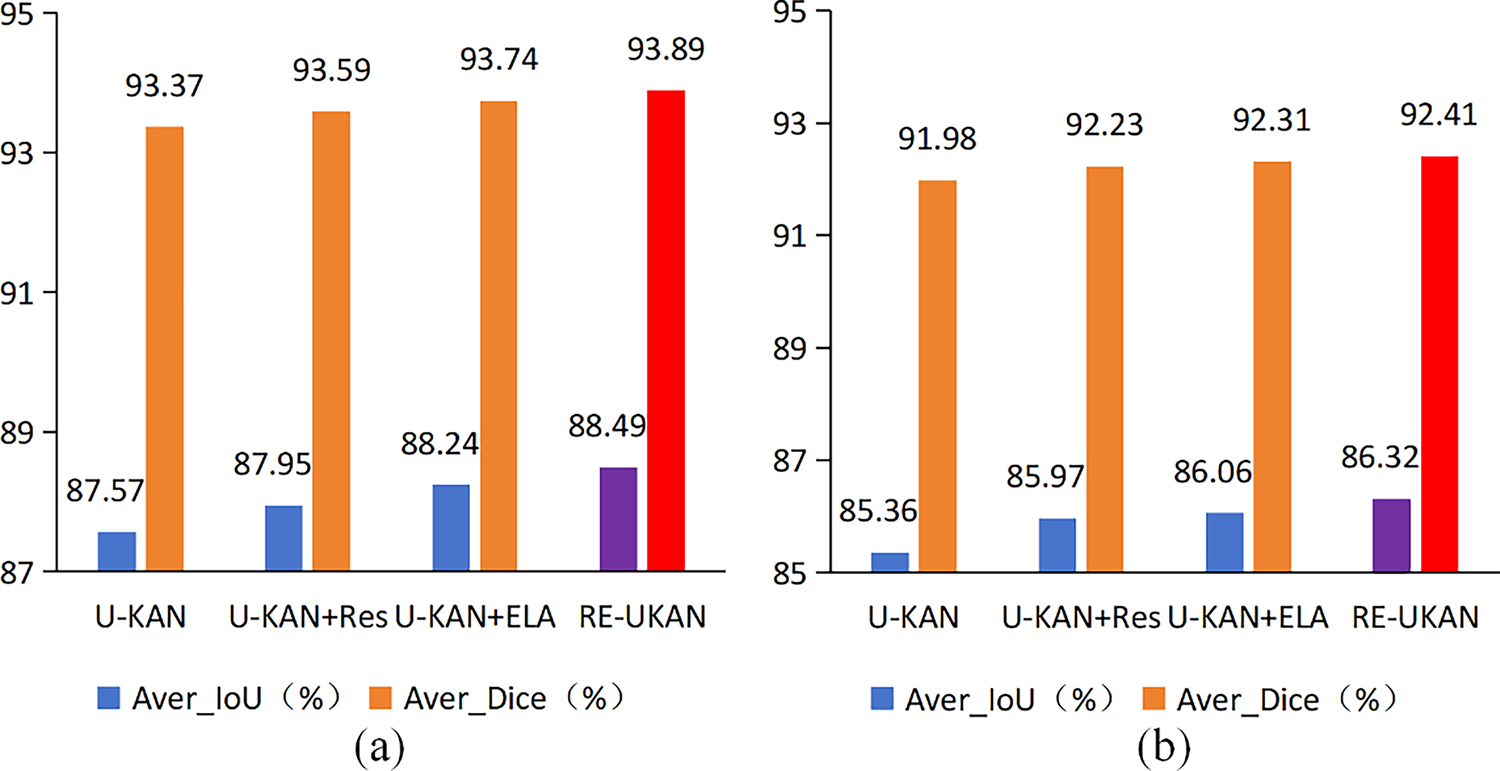
Figure 8: Ablation experiments of the proposed method on the GlaS dataset (a) and the CVC-ClinicDB dataset (b)
4.5.2 Ablation Studies on the Number of RE Module in Encoder Layer
The previous comparison experiments have shown that RE module can efficiently extract additional low-scale features and capture the complex details of the feature map, facilitating enhanced segmentation. We conducted additional ablation experiments on the number of RE module in the encoder layer. In the ablation experiments, the number of residual modules was set to 0, 1, 2, and 3, with each RE module consisting of one residual module and the ELA Block. The results of the ablation experiment are presented in Fig. 9. As illustrated in Fig. 9a, the highest values for both Aver_Dice and Aver_IoU are observed when the number of RE module is set to 3. When the number of RE module is 1, the segmentation results exhibit some fluctuations. As illustrated in Fig. 9b, the values of Aver_Dice and Aver_IoU demonstrate a gradual increase with the addition of RE module. The highest values of Aver_Dice and Aver_IoU are attained when the number of RE modules reaches 3. It can thus be concluded that the optimal number of RE modules for RE-UKAN is 3.
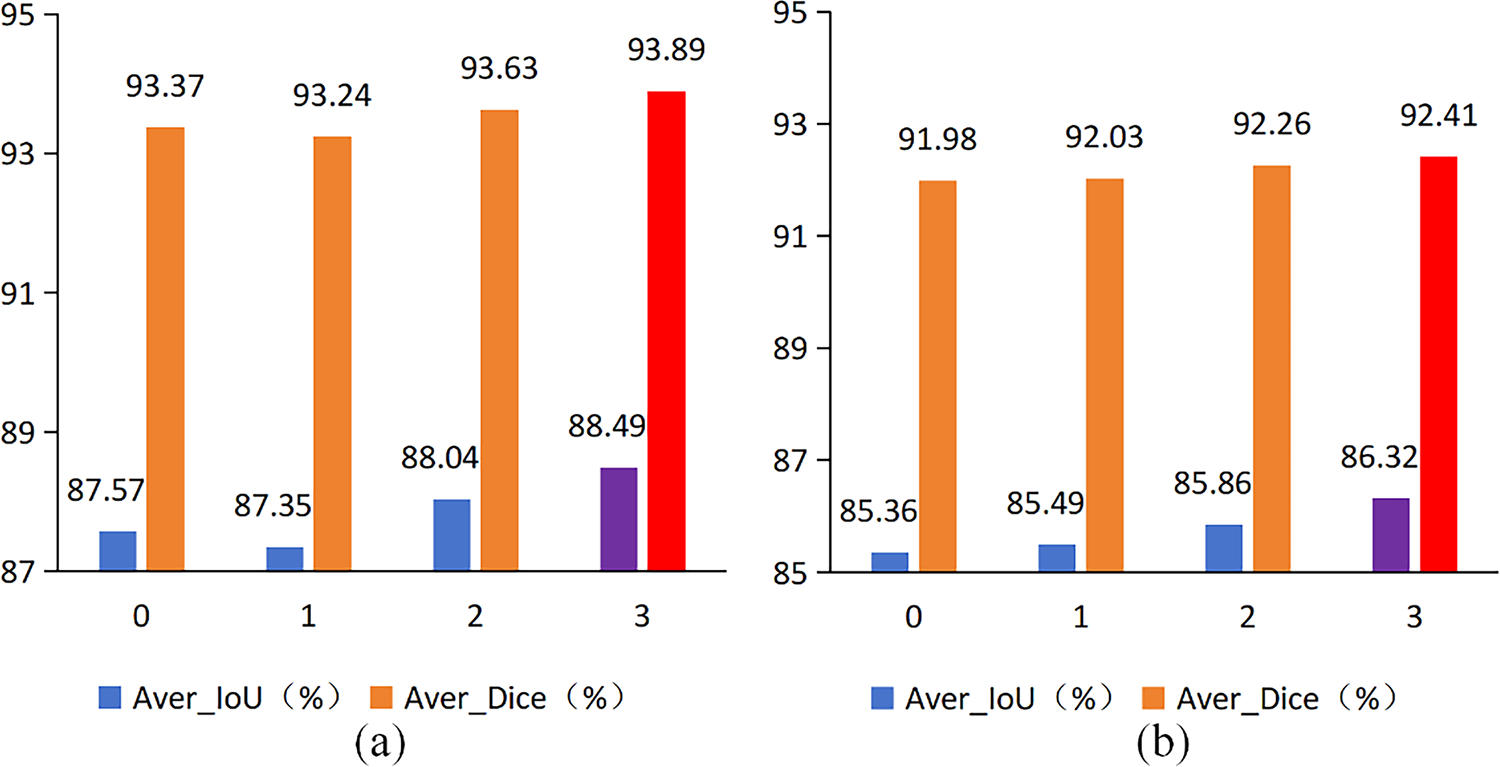
Figure 9: Ablation experiments of the number of RE module on the GlaS dataset (a) and the CVC-ClinicDB dataset (b)
This study presents RE-UKAN, a refined model leveraging the residual network and the ELA Block. This model takes U-KAN as the basic framework and integrates the residual modules and ELA Block in the encoder stage. The objective of this integration is to mitigate the loss of feature information during downsampling, thereby improving the precision of image segmentation. At the same time, this design effectively alleviates the vanishing gradient problem caused by the increased network depth. Experimental results from multiple medical image segmentation datasets show that RE-UKAN achieves optimal segmentation performance, with an Aver_IoU of 87.01%. This represents a significant 1.63% improvement over the baseline U-KAN model, which validates its effectiveness and superiority in complex medical image segmentation tasks. Ablation studies further confirm the synergistic effect of the residual module and the ELA Block. The model’s organic integration facilitates the enhancement of training stability and detail preservation capabilities, while retaining the nonlinear modelling advantages of U-KAN. This renders the model a more reliable solution for complex medical image segmentation tasks. Currently, RE-UKAN has only been trained and validated on 2D images. In the future, the network will be expanded to 3D medical images. This can enhance the model’s generalization capabilities across different imaging protocols and anatomical regions throughout the body. Moreover, the prevailing framework is characterised by protracted training times and sub-optimal inference speeds. Subsequent work will systematically evaluate temporal performance metrics and further improve the model. This will lay a foundation for its deployment in real-time clinical scenarios.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Jie Jia and Bo Li; methodology, Jie Jia; software, Jie Jia; validation, Jie Jia, Peiwen Tan and Xinyan Chen; formal analysis, Peiwen Tan and Dongjin Li; investigation, Jie Jia; resources, Bo Li; data curation, Xinyan Chen and Dongjin Li; writing—original draft preparation, Jie Jia; writing—review and editing, Jie Jia and Bo Li; visualization, Jie Jia; supervision, Bo Li; project administration, Bo Li. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available in the following datasets: The GlaS Dataset, which provides gland segmentation data, is available at https://warwick.ac.uk/services/gov/calendar/section2/regulations/computing/ (accessed on 01 August 2025); the CVC-ClinicDB Dataset, which provides polyp segmentation data, is available at https://polyp.grand-challenge.org/CVCClinicDB/ (accessed on 01 August 2025); the DSB 2018 Dataset, which provides cell nucleus segmentation data, is available at https://www.kaggle.com/c/data-science-bowl-2018 (accessed on 01 August 2025); and the TN-SCUI 2020 Dataset, which provides thyroid nodule segmentation data, is available at https://tn-scui2020.grand-challenge.org/ (accessed on 01 August 2025).
Ethics Approval: All the medical image data used in this study are from publicly available datasets and do not involve any ethical issues. All the data in the study are authentic and reliable, without any tampering or falsification. This paper has not been published elsewhere and is not simultaneously submitted to any other journals or conferences.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Liu X, Song L, Liu S, Zhang Y. A review of deep-learning-based medical image segmentation methods. Sustainability. 2021;13(3):1224. doi:10.3390/su13031224. [Google Scholar] [CrossRef]
2. Cheng J, Li H, Li D, Hua S, Sheng VS. A survey on image semantic segmentation using deep learning techniques. Comput Mater Contin. 2023;74(1):1941–57. doi:10.32604/cmc.2023.032757. [Google Scholar] [CrossRef]
3. Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi AK. Medical image segmentation using deep learning: a survey. IET Image Process. 2022;16(5):1243–67. doi:10.1049/ipr2.12419. [Google Scholar] [CrossRef]
4. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi:10.1016/j.media.2017.07.005. [Google Scholar] [PubMed] [CrossRef]
5. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; 2015 Oct 5–9; Munich, Germany. p. 234–41. doi:10.1007/978-3-319-24574-4_28. [Google Scholar] [CrossRef]
6. Oktay O, Schlemper J, Le Folgoc L, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas. arXiv:1804.03999. 2018. [Google Scholar]
7. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. UNet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham, Switzerland: Springer International Publishing; 2018. p. 3–11. doi:10.1007/978-3-030-00889-5_1. [Google Scholar] [CrossRef]
8. Valanarasu JMJ, Patel VM. UNeXt: MLP-based rapid medical image segmentation network. In: Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; 2022 Sep 18–22; Singapore. p. 23–33. doi:10.1007/978-3-031-16443-9_3. [Google Scholar] [CrossRef]
9. Liu Y, Zhu H, Liu M, Yu H, Chen Z, Gao J. Rolling-unet: revitalizing MLP’s ability to efficiently extract long-distance dependencies for medical image segmentation. Proc AAAI Conf Artif Intell. 2024;38(4):3819–27. doi:10.1609/aaai.v38i4.28173. [Google Scholar] [CrossRef]
10. Li C, Liu X, Li W, Wang C, Liu H, Liu Y, et al. U-KAN makes strong backbone for medical image segmentation and generation. arXiv:2406.02918. 2024. [Google Scholar]
11. Liu Z, Wang Y, Vaidya S, Ruehle F, Halverson J, Soljačić M, et al. Kan: kolmogorov-Arnold networks. arXiv: 2404.19756. 2024. [Google Scholar]
12. Xu W, Wan Y. ELA: efficient local attention for deep convolutional neural networks. arXiv:2403.01123. 2024. [Google Scholar]
13. Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging. 2018;37(12):2663–74. doi:10.1109/TMI.2018.2845918. [Google Scholar] [PubMed] [CrossRef]
14. Jin Q, Meng Z, Pham TD, Chen Q, Wei L, Su R. DUNet: a deformable network for retinal vessel segmentation. Knowl Based Syst. 2019;178:149–62. doi:10.1016/j.knosys.2019.04.025. [Google Scholar] [CrossRef]
15. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203–11. doi:10.1038/s41592-020-01008-z. [Google Scholar] [PubMed] [CrossRef]
16. Chen J, Mei J, Li X, Lu Y, Yu Q, Wei Q, et al. TransUNet: rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med Image Anal. 2024;97:103280. doi:10.1016/j.media.2024.103280. [Google Scholar] [PubMed] [CrossRef]
17. Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, et al. UNETR: transformers for 3D medical image segmentation. In: Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV); 2022 Jan 3–8; Waikoloa, HI, USA. p. 1748–58. doi:10.1109/WACV51458.2022.00181. [Google Scholar] [CrossRef]
18. Ma J, Li F, Wang B. U-mamba: enhancing long-range dependency for biomedical image segmentation. arXiv: 2401.04722. 2024. [Google Scholar]
19. Ruan J, Li J, Xiang S. VM-UNet: vision mamba UNet for medical image segmentation. ACM Trans Multimedia Comput Commun Appl. 2025;10:1–17. doi:10.1145/3767748. [Google Scholar] [CrossRef]
20. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18–23; Salt Lake City, UT, USA. p. 7132–41. doi:10.1109/CVPR.2018.00745. [Google Scholar] [CrossRef]
21. Woo S, Park J, Lee JY, Kweon IS. CBAM: convolutional block attention module. In: Proceedings of the Computer Vision—ECCV 2018; 2018 Sep 8–14; Munich, Germany. p. 3–19. doi:10.1007/978-3-030-01234-2_1. [Google Scholar] [CrossRef]
22. Wang Q, Wu B, Zhu P, Li P, Zuo W, Hu Q. ECA-net: efficient channel attention for deep convolutional neural networks. In: Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13–19; Seattle, WA, USA. p. 11531–9. doi:10.1109/cvpr42600.2020.01155. [Google Scholar] [CrossRef]
23. Hou Q, Zhou D, Feng J. Coordinate attention for efficient mobile network design. In: Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20–25; Nashville, TN, USA. p. 13708–17. doi:10.1109/cvpr46437.2021.01350. [Google Scholar] [CrossRef]
24. Misra D, Nalamada T, Arasanipalai AU, Hou Q. Rotate to attend: convolutional triplet attention module. In: Proceedings of the 2021 IEEE Winter Conference on Applications of Computer Vision (WACV); 2021 Jan 3–8; Waikoloa, HI, USA. p. 3138–47. doi:10.1109/WACV48630.2021.00318. [Google Scholar] [CrossRef]
25. Xiao X, Lian S, Luo Z, Li S. Weighted res-UNet for high-quality retina vessel segmentation. In: Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME); 2018 Oct 19–21; Hangzhou, China. p. 327–31. doi:10.1109/ITME.2018.00080. [Google Scholar] [CrossRef]
26. Alom MZ, Yakopcic C, Taha TM, Asari VK. Nuclei segmentation with recurrent residual convolutional neural networks based U-Net (R2U-Net). In: Proceedings of the NAECON 2018—IEEE National Aerospace and Electronics Conference; 2018 Jul 23–26; Dayton, OH, USA. p. 228–33. doi:10.1109/NAECON.2018.8556686. [Google Scholar] [CrossRef]
27. Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the Computer Vision—ECCV 2018; 2018 Oct 8–14; Munich, Germany. p. 833–51. doi:10.1007/978-3-030-01234-2_49. [Google Scholar] [CrossRef]
28. Wang J, Sun K, Cheng T, Jiang B, Deng C, Zhao Y, et al. Deep high-resolution representation learning for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2021;43(10):3349–64. doi:10.1109/tpami.2020.2983686. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools